Abstract
Influenza viruses circulated at very low levels during the beginning of the COVID-19 pandemic, and population immunity against these viruses is low. An H3N2 strain (3C.2a1b.2a2) with a hemagglutinin (HA) that has several substitutions relative to the 2021–22 H3N2 vaccine strain is dominating the 2021–22 Northern Hemisphere influenza season. Here, we show that one of these substitutions eliminates a key glycosylation site on HA and alters sialic acid binding. Using glycan array profiling, we show that the 3C.2a1b.2a2 H3 maintains binding to an extended biantennary sialoside and replicates to high titers in human airway cells. We find that antibodies elicited by the 2021–22 Northern Hemisphere influenza vaccine poorly neutralize the 3C.2a1b.2a2 H3N2 strain. Together, these data indicate that 3C.2a1b.2a2 H3N2 viruses efficiently replicate in human cells and escape vaccine-elicited antibodies.
Keywords: influenza virus, antigenic mismatch, influenza vaccine, antibody
Graphical abstract
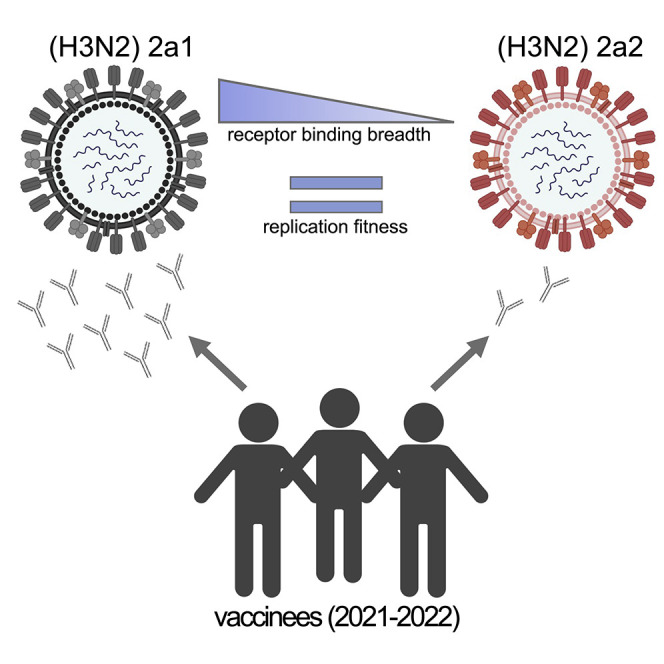
Bolton et al. show that recent human H3N2 influenza viruses have hemagglutinin substitutions that alter sialic acid binding and reduce antibody binding. These viruses are currently spreading widely around the world.
Introduction
Population immunity against influenza viruses is likely low since these viruses have not circulated widely during the COVID-19 pandemic (Laurie and Rockman, 2021). Social distancing, mask wearing, and decreases in international travel have likely contributed to reduced global circulation of influenza viruses (Koutsakos et al., 2021). As COVID-19-related restrictions are eased or lifted, it is likely that influenza viruses will circulate widely due to lack of infection-induced population immunity over the past 2 years. In the United States and other parts of the world, a unique H3N2 clade, 3C.2a1b.2a2 (herein 2a2), began circulating at high levels toward the end of 2021 (Figure 1A). This clade emerged early in the COVID-19 pandemic and almost completely displaced other H3N2 clades in Europe, Oceania, South Asia, West Asia, and North America in 2021. Influenza virus activity decreased in the United States during the Omicron SARS-CoV-2 wave in early 2022 but has increased substantially during the spring of 2022 as Omicron cases have decreased. Viruses within the 2a2 H3N2 clade have several substitutions in key antigenic sites on hemagglutinin (HA) relative to the 2021–22 Northern Hemisphere H3N2 vaccine strain, a 3C.2a1b.2a1 (herein 2a1) virus (Table 1 ). Vaccine effectiveness against 2a2 was found to be low during an outbreak on a college campus in the United States during November 2021 (Delahoy et al., 2021). Interim estimates suggest that the 2021–22 influenza vaccine did not reduce the risk for outpatient respiratory illness caused by H3N2 viruses in the United States (Chung et al., 2022). It is therefore important to understand the virological and antigenic properties of 2a2 H3N2 viruses.
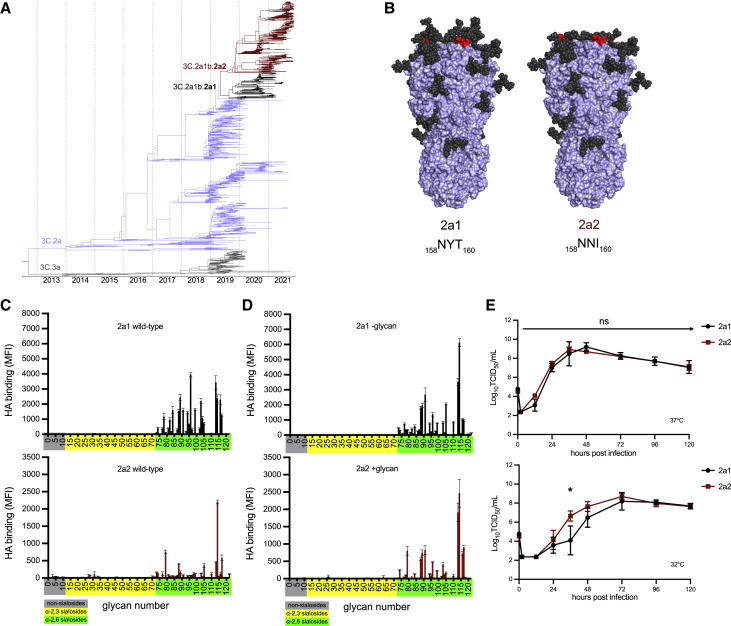
Figure 1.
H3N2 variant lineage 2a2 replicates efficiently in human cells despite changes in HA receptor specificity
(A) Nextstrain phylogenetic analysis of the HA gene of contemporary (2019–21) H3N2 viruses.
(B) Amino acid differences at residues 158–160 are denoted (red), and glycan loss is modeled on the crystal structure of an H3 HA trimer (PDB: 4O5I) for 2a1 and 2a2 HAs.
(C and D) Binding of wild-type (C) and mutant (D) recombinant HAs to a sialoside microarray. Printed glycans include those with α2,3 sialic acid linkages (yellow), α2,6 sialic acid linkages (green), or no sialic acid linkage (gray). Bars represent the mean fluorescent intensity with standard error bars. Mutant 2a1 HA proteins (D) had N159 and I160 substitutions to remove glycosylation motif, and mutant 2a2 HA proteins (D) had Y159 and T160 substitutions to add glycosylation motif. Glycan numbers are shown on the x axes.
(E) Infectious virus production after an MOI = 0.01 infection of primary differentiated human nasal epithelial cell (hNEC) cultures incubated at 37°C (top) or 32°C (bottom). Virus titers were measured by TCID50 assay using MDCK cells. Data at each time point are the mean/SEM of three separate wells. Curves are representative of two independent experiments. A repeated measures MANOVA followed by a Bonferonni post-test was completed (∗p ≤ 0.05).
Table 1.
Amino acid residue differences between 2a1 and 2a2 HAs
| H3N2 virus clade | ||
|---|---|---|
| Residue number | 2a1 | 2a2 |
| 9 (signal peptide) | Y | N |
| 156 | H | H/S |
| 159 | Y | N |
| 160 | T | I |
| 164 | L | Q |
| 171 | N | K |
| 186 | S | D |
| 190 | D | N |
Results and discussion
Viruses within the 2a2 H3N2 clade possess a T160I HA substitution that is expected to eliminate a glycosylation site at amino acids 158–160 in antigenic site B (Figure 1B). HAs from a 2a2 virus (A/Bangladesh/1450/2020) migrated with a lower molecular weight compared to HAs from a 2a1 virus (A/Cambodia/e0826360/2020) (Figure S1A). Recombinant HA proteins engineered to possess I160 (2a2 and 2a1 –glycan) also migrated with a lower molecular weight compared to HAs with T160 (2a1 and 2a2 +glycan) (Figure S1B), consistent with a loss of a glycosylation site with the T160I substitution. Influenza virus entry into human cells is mediated by HA binding to α2,6-linked sialoglycans (Thompson and Paulson, 2021), and previous studies have demonstrated that the 158–160 HA glycosylation site impacts HA sialic acid binding specificity (Zeng et al., 2020). To determine if the 2a2 H3N2 HA has altered receptor specificity, we measured HA binding to a panel of α2,3- and α2,6-linked sialoside glycans (Figures 1C and Table S1). As expected, HAs from the predecessor 2a1 H3N2 virus and the 2a2 H3N2 virus did not bind strongly to α2,3-linked sialosides or those lacking sialic acid altogether (non-sialosides). Consistent with previous studies (Peng et al., 2017), the predecessor 2a1 H3N2 HA bound to many α2,6-linked sialoside glycans, with preference to those with extended poly-LacNAc chains and/or branched structures. Strikingly, the 2a2 H3N2 HA bound poorly to most α2,6-linked sialoside glycans, but it maintained strong binding to an extended biantennary sialoside. We also measured the binding of 2a1 and 2a2 mutant HAs that we generated via site-directed mutagenesis to either remove (in the case of 2a1) or add (in the case of 2a2) a glycosylation motif at positions 158–160 (Figure 1D). Interestingly, mutant 2a1 HA with N159 and I160 (2a1 –glycan) had a similar binding profile to the wild-type 2a2 HA, with a strong preference for an extended biantennary sialoside (#114, Table S1). Conversely, the 2a2 HA engineered with Y159 and T160 (2a2 +glycan) maintained this preference for the extended biantennary sialoside, but it also bound with broader specificity than the wild-type 2a2 HA to multiple α2,6-linked sialoside glycans (Figure 1D). These data suggest a role for the 158–160 glycan in shaping the binding specificity of the 2a2 H3N2 HAs for sialoside receptors, although we did not evaluate other encoded HA substitutions that likely impact binding specificity as well.
The types and prevalence of different sialo-linked receptors in the human respiratory tract are incompletely understood, and therefore it is difficult to interpret how changes in sialoside glycan specificity may affect viral fitness. We measured viral growth kinetics over 120 h in primary human nasal epithelial cells (hNECs) (Figure 1E) and found that both 2a1 and 2a2 H3N2 viruses replicated well in hNEC cultures at 37°C, with no significant differences in infectious virus peak titers or kinetics of virus production between the two viruses (Figure 1E, top). When we measured growth kinetics of the two viruses in hNEC cultures incubated at 32°C, which is a temperature more consistent with that of the upper respiratory tract, we again found that both viruses replicated well, but at this lower temperature the 2a2 H3N2 virus replicated with slightly faster kinetics than 2a1 H3N2, though both viruses reached the same peak titer at the same time (Figure 1E, bottom). Additionally, both viruses replicated to similar levels in MDCK-S and hCK transformed cell lines engineered to support human-adapted influenza virus replication (Figures S1B and S1C). These data indicate that 2a2 H3N2 viruses, despite a decrease in receptor specificity breadth, replicate at least as efficiently as 2a1 H3N2 viruses in both primary and transformed cell culture systems.
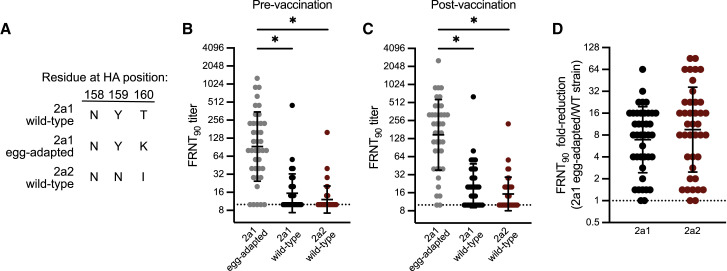
We previously demonstrated that the addition of a 158–160 H3 glycan resulted in a major antigenic change during the 2014–15 season (Chambers et al., 2015; Zost et al., 2017), which likely contributed to low vaccine effectiveness that season (Zimmerman et al., 2016). Most influenza vaccines are based on egg-adapted influenza virus antigens, and we previously reported that mutations at HA residues 158–160 can occur as a result of egg adaptation (Zost et al., 2017). To determine if the 2a2 H3N2 virus is antigenically distinct from the 2021–22 Northern Hemisphere H3N2 component, we completed neutralization assays using serum collected from 40 individuals before and 27–28 days after receiving the egg-adapted Flulaval Quadrivalent vaccine. We tested antibody neutralization of wild-type and egg-adapted 2a1 H3N2 strains, as well as the 2a2 H3N2 strain. Importantly, the wild-type 2a1 HA encodes amino acids Asn, Tyr, and Thr at positions 158–160, which leads to a glycosylation site, whereas the egg-adapted 2a1 HA encodes amino acids Asn, Tyr, and Lys at positions 158–160, which abrogates this glycosylation site (Figure 2A). Egg-adapted 2a1 viruses possess one additional substitution (S186R) in HA relative to wild-type 2a1 viruses. We found that most individuals possessed antibodies that neutralized the egg-adapted 2a1 strain but not the wild-type version of this strain or the 2a2 strain (Figures 2B and 2C). Neutralizing antibodies against the egg-adapted vaccine strain were higher relative to the other strains tested before vaccination (Figure 2B) and were boosted ∼2-fold after vaccination (Figure 2C). Neutralizing antibodies against wild-type 2a1 and the 2a2 strain were very low before vaccination and remained undetectable in many individuals (35% to wild-type 2a1; 55% to 2a2) following vaccination (Figure 2C). On average, neutralizing antibody titers were reduced ∼10-fold comparing the 2a2 H3N2 strain relative to the egg-adapted 2a1 vaccine strain (Figure 2D), which is likely an underestimation since samples with an undetectable neutralization titer were arbitrarily assigned a neutralization titer of 10 (the limit of detection in our assays). We stratified individuals based on prior year (2020–21) vaccination status, and we found that neutralization titers to these viruses were comparable between individuals regardless of vaccination the previous year (Figure S2). It is likely that the loss of the glycan at HA residues 158–160 primarily contributed to the antigenic mismatch observed in our experiments, although other HA substitutions that differ between 2a1 and 2a2 H3N2 viruses (Table 1) may be involved.
Figure 2.
Potential H3N2 antigenic mismatch for the 2021–22 Northern Hemisphere influenza season
(A) Amino acid residues encoded by egg-adapted and wild-type circulating strains at HA positions 158, 159, and 160, an N-linked glycosylation site.
(B and C) Virus neutralization titers to the vaccine virus and circulating viruses prior to vaccination (B) and 27–28 days following vaccination (C) in a group of vaccinees in Oct–Nov 2021 (n = 40). Neutralization titers are plotted as the geometric mean titer (GMT) of two independent experiments. Lines represent the GMT and standard deviation among vaccinees. A Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparisons test was completed on log2 transformed titers (∗p ≤ 0.05).
(D) Fold-reduction in neutralizing antibodies to circulating strains when compared to the vaccine virus in a group of vaccinees (n = 40).
Our studies suggest that the 2a2 H3N2 replicates efficiently in human airway cells and can partially circumvent antibodies elicited by egg-adapted 2021–22 Northern Hemisphere influenza vaccines. We were hopeful that the egg-adapted H3N2 vaccine strain would be closely matched to the 2a2 H3N2 strain since both of these strains lack a glycan at residues 158–160 of HA. However, our data indicate that these H3N2 strains are antigenically mismatched, likely due to amino acid differences at HA residues 159 and 160. The 2a2 H3N2 strain lost the 158–160 antigenic site B glycan through a T160I substitution, while the egg-adapted 2021–22 H3N2 strain lost it through a T160K substitution. Previous studies have demonstrated that amino acid differences at HA residues 158–160 can impact antigenicity (Koel et al., 2013), and it is likely that the amino acid differences at residue 160 contribute to this vaccine mismatch. While cases of 2a2 H3N2 infections are again quickly rising in the United States and other parts of the world, it is possible that other clades of H3N2 will become predominant in the future. It is also possible that H1N1 or influenza B viruses might dominate later in the 2021–22 season. Studies have clearly shown that seasonal influenza vaccines consistently prevent hospitalizations and deaths even in years where there are large antigenic mismatches (Ferdinands et al., 2021). Influenza vaccinations will be crucial for reducing hospitalizations as SARS-CoV-2 and 2a2 H3N2 viruses co-circulate in the coming months.
Limitations of the study
Further studies will be needed to evaluate antibody responses elicited by other vaccine types that employ wild-type 2a1 H3N2 strains, such as cell-based or recombinant protein vaccines (Gouma et al., 2020a). While there is a potential antigenic mismatch between wild-type 2a1 HA and the 2a2 HA, it is possible that glycan shielding from the 158–160 glycan on wild-type vaccine antigens may promote antibody responses directed against epitopes other than antigenic site B of HAs. It is worth noting that most vaccinees in our study were relatively young and healthy, and additional studies will need to be completed with larger cohorts to better evaluate the 2021–22 Northern Hemisphere vaccine in individuals with different birth years. Further studies need to be completed to evaluate the antigenic effect of individual HA substitutions that differ between 2a1 and 2a2 H3N2 viruses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Peroxidase-conjugated Goat Affinity Purified Antibody to Mouse IgG (whole molecule) | MP Biomedical | Cat. 0855563; RRID: AB_2334548 |
| Mouse anti-nucleoprotein antibody [IC5-1B7] | BEI Resources | Cat. NR-43899 |
| IRDye 800CW Donkey anti-Mouse IgG Secondary Antibody | Licor | Cat. 926-32212; RRID: AB_621847 |
| Monoclonal Anti-HA antibody produced in mouse [clone HA-7] | Sigma | Cat. 59658 |
| Bacterial and virus strains | ||
| A/Cambodia/e0826360/2020 (wild-type) | Produced for this paper | N/A |
| A/Cambodia/e0826360/2020 S186R + T160K (egg-adapted) | Produced for this paper | N/A |
| A/Bangladesh/1450/2020 | Produced for this paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TPCK-treated trypsin | Sigma | Cat. 4370285 |
| TrueBlue TMB Peroxidase Substrate | KPL-Seracare | Cat. 5510-0030 |
| Modified Eagle Medium | Corning | Cat. MT10-010-CM |
| Dulbecco’s Modified Eagle Medium | Corning | Cat. MT10-013-CM |
| FreeStyle 293 Expression Medium | Thermo Fisher | Cat. 12338026 |
| Opti-MEM | Thermo Fisher | Cat. 31985062 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat. F0926-100ML |
| Newborn Calf Serum | Gibco | Cat. 16010167 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher | Cat. 11668019 |
| Paraformaldehyde 32% solution | Electron Microscopy Sciences | Cat. 15714-S |
| Triton X-100 | Sigma-Aldrich | Cat. 10789704001 |
| Fat-free milk, dry powder | Dot Scientific | Cat. DSM17200-1000 |
| 293fectin transfection reagent | Thermo Fisher | Cat. 12347019 |
| Ni-NTA agarose | QIAGEN | Cat. 30210 |
| RDE (II) | Denka Seiken Co. Ltd. | Cat. 370013 |
| HEPES buffer (1M) | Gibco | Cat. 15630080 |
| Gentamicin | Gibco | Cat. 15710064 |
| Avicel RC-591 | DuPont | Gift from DuPont |
| 10% Bis-Tris mini protein gel | Invitrogen | Cat. NP0301BOX |
| 4-12% Bis-Tris mini protein gel | Invitrogen | Cat. NP0321BOX |
| SimplyBlue SafeStain | Invitrogen | Cat. LC6065 |
| A/Cambodia/e0826360/2020 WT recombinant HA | Produced for this paper | N/A |
| A/Cambodia/e0826360/2020 Y159N + T160I recombinant HA | Produced for this paper | N/A |
| A/Bangladesh/1450/2020 WT recombinant HA | Produced for this paper | N/A |
| A/Bangladesh/1450/2020 N159Y + I160T recombinant HA | Produced for this paper | N/A |
| Sialoside glycan array | Laboratory of James Paulson, Scripps Research, CA, USA | Peng et al., 2017 |
| Critical commercial assays | ||
| QuikChange II-XL Site-Directed Mutagenesis Kit | Agilent | Cat. 200521 |
| Experimental models: Cell lines | ||
| 293T | ATCC | Cat. CRL-3216, RRID:CVCL_0063 |
| 293F | Thermo Fisher | Cat. R79007 |
| Madin-Darby canine kidney (MDCK) | NIH | N/A |
| MDCK-Siat1 | NIH | N/A |
| HCK | Laboratory of Yoshihiro Kawaoka, University of Wisconsin-Madison, WI, USA | Takada et al., 2019 |
| Recombinant DNA | ||
| pHW2000_A/Cambodia/e0826360/2020 HA (for virus production) | Produced for this paper | EPI_ISL_806547 |
| pHW2000_A/Cambodia/e0826360/2020 HA (EPI_ISL_806547) with egg-adapted substitutions: T160K & S186R (for virus production) | Produced for this paper | N/A |
| pHW2000_A/Bangladesh/1450/2020 HA (for virus production) | Produced for this paper | N/A |
| pHW2000_A/Cambodia/e0826360/2020 NA (for virus production) | Produced for this paper | N/A |
| pCMV-Sport6_A/Cambodia/e0826360 /2020 WT recombinant HA | Produced for this paper | N/A |
| pCMV-Sport6_A/Cambodia/e0826360 /2020_Y159N + T160I recombinant HA | Produced for this paper | N/A |
| pCMV-Sport6_A/Bangladesh/1450/2020 WT recombinant HA | Produced for this paper | N/A |
| pCMV-Sport6_A/Bangladesh/1450/2020 N159Y + I160T recombinant HA | Produced for this paper | N/A |
| Software and algorithms | ||
| Prism (v.9.2) | GraphPad Software | www.graphpad.com/scientific-software/prism/ |
| PyMOL (v.2.2.3) | Schrödinger | https://pymol.org |
| Other | ||
| Amicon Ultra centrifugal filters. 30K MWCO | Millipore | Cat. UFC9030 |
| Immunospot S6 Universal Analyzer | CTL | https://immunospot.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Scott E. Hensley (hensley@pennmedicine.upenn.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subjects details
Human serum samples
Serum samples from 40 adults (ages 21-71) were collected at the time of and 27-28 days after vaccination with Flulaval Quadrivalent influenza virus vaccine (GlaxoSmithKline) between October-December 2021. This study was approved by the Institutional Review Board of the University of Pennsylvania.
Cell lines and primary cells
MDCK-SIAT1 (MDCK-S) cells were cultured in Minimal Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS), and 293T cells were cultured in Dulbecco’s Minimal Essential Medium (DMEM) with 10% FBS. hCK cells (Takada et al., 2019) were cultured in MEM supplemented with 10% newborn calf serum. All transformed cell lines were cultured at 37C in humidified incubators supplemented with 5% CO2. Primary Human Nasal Epithelial Cells (hNECs, Promocell, Heidelberg, Germany) were plated and cultured in serum-free Airway Epithelial Growth Medium (Promocell) with SupplementPack Airway Epithelial Cell Growth Medium without antibiotics. The 24 well plate Transwell inserts were coated with 0.03 mg/mL Collagen I, Rat Tail (Gibco). Cells were plated and cultured with Airway media on the apical and basolateral surface with media changes at 24 h after cell plating and every 48 h afterwards. After 7-10 days, confluence was assessed by light microscopy and by determining transepithelial electrical resistance (TEER). When fully confluent (TEER> 400), both apical and basolateral media were removed and ALI Differentiation media (Stem Cell Technologies, Pneumacult ALI Basal Medium supplemented with 1X ALI Maintenance Supplement (StemCell Technologies), 048 ug/mL Hydrocortisone solution (StemCell Technologies), and 4 μg/mL Heparin sodium salt in PBS (StemCell Technologies) was replaced in the basolateral chamber only. Media was changed every 48 h and the apical surface of the cultures was intermittently washed with PBS to remove excess mucus. Full differentiation occurred in approximately 4 weeks and cells were considered fully differentiated when there was presence of mobile cilia on the cell surface visible with light microscopy.
Method details
Viruses
Influenza viruses were generated by an 8 plasmid reverse genetics system as previously described (Hoffmann et al., 2000) with internal segments from A/Puerto Rico/8/1934. The wild-type HA and NA sequences for A/Cambodia/e0826360/2020 (3C.2a1b.2a1) and the HA sequence for A/Bangladesh/1450/2020 (3C.2a1b.2a2) were synthesized as gene fragments (IDT) and cloned into the pHW2000 reverse genetics plasmid by restriction digest cloning. Viruses were launched by transient transfection of 8 plasmids encoding each of the influenza virus genome segments in co-cultures of 293T and MDCK-S cells. Virus stocks were passaged one time in MDCK-S cells and aliquots were stored at -80°C. All virus stocks were sequence confirmed by Sanger sequencing.
Recombinant proteins
Soluble recombinant HA trimers were produced as previously described (Whittle et al., 2014). HA mammalian expression plasmids encoded for a codon-optimized HA sequence followed by a T4 fibritin FoldOn trimerization domain, an AviTag biotinylation sequence, and a 6xHistidine tag at the C-terminus in place of the transmembrane domain and cytoplasmic tail of HA. Proteins were produced by transient transfection in 293F cells (Thermo Fisher), and purified using Ni-NTA agarose beads (Qiagen) using gravity flow chromatography columns (Bio-Rad). Site-directed mutants were generated in HA expression plasmids using the QuikChange II-XL kit according to manufacturer’s instructions (Aglient). All plasmids were sequence confirmed prior to transfection, and all proteins were checked for integrity and the absence of contaminants by SDS-PAGE.
Glycan arrays
Glycans were synthesized and printed on microarray chips as previously described (Peng et al., 2017). Recombinant HA protein (50 μg/mL), a mouse anti-histidine tag antibody, and an Alexa647-conjugated anti-mouse antibody were pre-complexed in a 4:2:1 ratio on ice for 15 min in 100 μL PBS-T. Pre-complexed HAs were incubated on the microarray surface for 90 min in a humidified chamber.
Virus growth curves
The hNECs were acclimated to 32°C or 37°C for 48hrs before infection. The apical surface was washed three times with PBS and the basolateral media was changed at the time of infection. The hNEC cultures were inoculated at an ∼ MOI of 0.01 in the apical chamber and incubated at 32°C incubator for 2 h. The apical surface of the hNEC culture was washed three times with PBS+. At the indicated times, 100 μL of IM without N-acetyl trypsin was added to the apical surface of the hNECs for 5 min at 32°C, the IM was harvested and stored at -80°C. Basolateral media was changed every 24hrs post infection for the duration of the experiment. Infectious virus titers in the apical supernatants were measured with TCID50 assay on MDCK cells (Powell and Pekosz, 2020).
For growth assays in MDCK-S and hCK cells, triplicate wells of a 6-well plate were seeded with 106 cells/well. 12 h later, cells were washed 2 times with serum free MEM (SF-MEM) and then infected with virus at a multiplicity of infection of 0.0001 and incubated at 37°C for 1 h. Following virus adsorption, inoculum was removed and cells were washed twice with SF-MEM. MEM supplemented with 5 mM HEPES buffer, 50 μg/mL gentamycin-sulfate, and 1 μg/mL TPCK-treated trypsin (growth media) was then added to wells, and plates were incubated at 37C + 5% CO2. At designated times post-infection, a 400 μL aliquot was removed from each well, clarified by centrifugation, and frozen at -80°C. Media depleted from wells was replaced with growth media.
Focus reduction neutralization test (FRNT)
FRNT assays were completed as previously described (Gouma et al., 2020b). Prior to testing, serum samples were treated with receptor destroying enzyme (RDE) (Denka-Seiken) for two hours at 37°C and then inactivated for 30 min at 56°C. On the day of the assay, serum samples were serially diluted in serum-free MEM and mixed with a virus dilution that resulted in 300-500 focus forming units (FFU) per well in virus only control wells. 96-well tissue culture plates containing MDCK-S cells plated the previous day at 25,000 cells/well were washed three times with serum-free MEM. Virus-antibody mixtures were added to washed cells and incubated for 1 h at 37C. Virus-antibody mixtures were removed from cells, cells were washed with serum-free MEM and media was replaced with MEM supplemented with 5 mM HEPES buffer, 50 μg/mL gentamycin-sulfate, and 1.25% Avicel. Plates were incubated for 18 h in a 37°C incubator supplied with 5% CO2. For visualizing infected cells, media was removed, and cells were fixed with 4% paraformaldehyde for 1 h at 4°C. Fixed cells were permeabilized with 0.5% Triton X-100 for 7 min, and then blocked for 1 h with 5% milk in PBS. A mouse anti-nucleoprotein antibody (clone IC5-1B7), and then a peroxidase-conjugated goat anti-mouse antibody were each diluted in blocking buffer and incubated on plates for 1 h after each other. Plates were washed 5 times with distilled water after blocking, primary, and secondary steps. TrueBlue TMB substrate (KPL) was added to plates and incubated in the dark for 1 h for foci development. Substrate was flicked out of wells, and plates were thoroughly dried before visualization on an ImmunoSpot S6 plate reader. FRNT90 titers were defined as the highest reciprocal serum dilution that inhibited ≥90% of virus, according to virus only control wells on each plate.
Western blot and SDS-PAGE analysis of HA
For Western blots, viruses were expanded on MDCK-Siat1 cells and concentrated using Amicon Ultra-0.5 30 kDa MWCO centrifugal filters (Millipore). Virus samples were run on a 10% Bis-Tris gel (Invitrogen) under reducing conditions and transferred to a nitrocellulose membrane (Invitrogen), then probed with an anti-HA tag primary (clone HA-7, cat. 59658, Sigma-Aldrich) and an IRDye-conjugated anti-mouse secondary antibody (cat. 926-32212, Licor).
For SDS-PAGE of recombinant HA proteins, 1 microgram of each HA was run on a 4-12% Bis-Tris gel (Invitrogen) under reducing conditions. Gel was stained with Coomassie SimplyBlue SafeStain (Invitrogen) to visualize HA proteins.
Quantification and statistical analysis
For neutralization titer determination, the geometric mean titer determined from two independent experiments was plotted for each sample. Bars were plotted to show the geometric mean and standard deviation among samples at a given timepoint for each virus. Differences in neutralization titers between viruses were compared using a Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparisons post-test on log10-transformed titers. Infectious titers (either TCID50 or FFU) were plotted for virus growth curves, with error bars representing the standard deviation among triplicate wells at each timepoint. Differences in virus growth curves were analyzed by either a repeated measures MANOVA followed by a Bonferroni post-test or by a Welch’s t-test at each timepoint on log10-transformed titers, as indicated in figure legends. All statistical analyses were performed in GraphPad Prism softSware.
Acknowledgments
We thank Yoshihiro Kawaoka for providing hCK cells and Sarah Cobey for providing comments on the manuscript. We thank Penn’s Immune Health for processing human samples for this study. This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. 75N93021C00015, Contract No. 7N593021C00045, and Grant Nos. 1R01AI108686 and 1R01AI114730. S.E.H. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Author contributions
Conceptualization, M.J.B., E.M.D., and S.E.H.; investigation, M.J.B., J.T.O., R.M., N.J.S., J.W., M.A., and C.F.; coordination and processing of clinical samples, A.R.G. and E.M.D.; writing – original draft, M.J.B. and S.E.H.; writing – review & editing, M.J.B., S.E.H., J.C.P., and A.P.; supervision, S.E.H., J.C.P., and A.P.; funding acquisition, S.E.H., J.C.P., and A.P.
Declaration of interests
S.E.H. reports receiving consulting fees from Sanofi Pasteur, Lumen, Novavax, and Merck.
Published: May 31, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110897.
Supplemental information
Data and code availability
-
-
The published paper includes all data generated or analyzed during the study.
-
-
This paper does not report original code.
-
-
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Chambers B.S., Parkhouse K., Ross T.M., Alby K., Hensley S.E. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014-2015 influenza season. Cell Rep. 2015;12:1–6. doi: 10.1016/j.celrep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.R., Kim S.S., Kondor R.J., Smith C., Budd A.P., Tartof S.Y., Florea A., Talbot H.K., Grijalva C.G., Wernli K.J., et al. Interim estimates of 2021-22 seasonal influenza vaccine effectiveness - United States, February 2022. MMWR Morb Mortal Wkly Rep. 2022;71:365–370. doi: 10.15585/mmwr.mm7110a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahoy M.J., Mortenson L., Bauman L., Marquez J., Bagdasarian N., Coyle J., Sumner K., Lewis N.M., Lauring A.S., Flannery B., et al. Influenza A(H3N2) outbreak on a university campus - Michigan, October-November 2021. MMWR Morb. Mortal Wkly Rep. 2021;70:1712–1714. doi: 10.15585/mmwr.mm7049e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinands J.M., Thompson M.G., Blanton L., Spencer S., Grant L., Fry A.M. Does influenza vaccination attenuate the severity of breakthrough infections? A narrative review and recommendations for further research. Vaccine. 2021;39:3678–3695. doi: 10.1016/j.vaccine.2021.05.011. [DOI] [PubMed] [Google Scholar]
- Gouma S., Anderson E.M., Hensley S.E. Challenges of making effective influenza vaccines. Annu. Rev. Virol. 2020;7:495–512. doi: 10.1146/annurev-virology-010320-044746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouma S., Weirick M., Hensley S.E. Antigenic assessment of the H3N2 component of the 2019-2020 Northern Hemisphere influenza vaccine. Nat. Commun. 2020;11:2445. doi: 10.1038/s41467-020-16183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Neumann G., Kawaoka Y., Hobom G., Webster R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U S A. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel B.F., Burke D.F., Bestebroer T.M., van der Vliet S., Zondag G.C.M., Vervaet G., Skepner E., Lewis N.S., Spronken M.I.J., Russell C.A., et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Koutsakos M., Wheatley A.K., Laurie K., Kent S.J., Rockman S. Influenza lineage extinction during the COVID-19 pandemic? Nat. Rev. Microbiol. 2021;19:741–742. doi: 10.1038/s41579-021-00642-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie K.L., Rockman S. Which influenza viruses will emerge following the SARS-CoV-2 pandemic? Influenza Other Respir. Viruses. 2021;15:573–576. doi: 10.1111/irv.12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W., de Vries R.P., Grant O.C., Thompson A.J., McBride R., Tsogtbaatar B., Lee P.S., Razi N., Wilson I.A., Woods R.J., Paulson J.C. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe. 2017;21:23–34. doi: 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell H., Pekosz A. Neuraminidase antigenic drift of H3N2 clade 3c.2a viruses alters virus replication, enzymatic activity and inhibitory antibody binding. PLoS Pathog. 2020;16:e1008411. doi: 10.1371/journal.ppat.1008411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K., Kawakami C., Fan S., Chiba S., Zhong G., Gu C., Shimizu K., Takasaki S., Sakai-Tagawa Y., Lopes T.J.S., et al. A humanized MDCK cell line for the efficient isolation and propagation of human influenza viruses. Nat. Microbiol. 2019;4:1268–1273. doi: 10.1038/s41564-019-0433-6. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Paulson J.C. Adaptation of influenza viruses to human airway receptors. J. Biol. Chem. 2021;296:100017. doi: 10.1074/jbc.rev120.013309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J.R.R., Wheatley A.K., Wu L., Lingwood D., Kanekiyo M., Ma S.S., Narpala S.R., Yassine H.M., Frank G.M., Yewdell J.W., et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 2014;88:4047–4057. doi: 10.1128/jvi.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Yau L.F., Lin Z., Xia X., Yang Z., Wang J.R., Song W., Wang X. Characterization and evolutionary analysis of a Novel H3N2 influenza A virus glycosylation motif in Southern China. Front. Microbiol. 2020;11:1318. doi: 10.3389/fmicb.2020.01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman R.K., Nowalk M.P., Chung J., Jackson M.L., Jackson L.A., Petrie J.G., Monto A.S., McLean H.Q., Belongia E.A., Gaglani M., et al. 2014-2015 influenza vaccine effectiveness in the United States by vaccine type. Clin. Infect. Dis. 2016;63:1564–1573. doi: 10.1093/cid/ciw635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Parkhouse K., Gumina M.E., Kim K., Diaz Perez S., Wilson P.C., Treanor J.J., Sant A.J., Cobey S., Hensley S.E. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. U S A. 2017;114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
-
The published paper includes all data generated or analyzed during the study.
-
-
This paper does not report original code.
-
-
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.