Abstract
Cancer is a complicated disease attributed to multifactorial changes, which causes difficulties with treatment strategies. Various factors have been regarded as the main contributors, and infectious etiological factors have recently attracted interest. Several microbiomes contribute to carcinogenesis, cancer progression, and modulating cancer treatment by inducing cancerous epithelial cells and chronic inflammation. Most of our knowledge on the role of microbiota in tumor oncogenesis and clinical efficiency is associated with the intestinal microbiome. However, compelling evidence has also confirmed the contribution of the intratumor microbiome in cancer. Indeed, the findings of clinical tumor samples, animal models, and studies in vitro have revealed that many intratumor microbiomes promote tumorigenesis and immune evasion. In addition, the intratumor microbiome participates in regulating the immune response and even affects the outcomes of cancer treatment. This review summarizes the interplay between the intratumor microbiota and cancer, focusing on the contribution and mechanism of intratumor microbiota in cancer initiation, progression, and potential applications to cancer therapy.
Keywords: Intratumor microbiome, Immune system, Anticancer treatment
Introduction
The human body comprises a mixture of mammalian and microbial cells, with the latter exceeding the former by nearly tenfold. The microbial genetic repertoire is approximately 100-fold more abundant than that of the human host [1]. Beyond bacteria, the human commensal microbiome consists of viruses, archaea, fungi, and other eukaryotic species [2]. Commensal microbes inhabit at all mucosal barrier surfaces, with the distal gastrointestinal (GI) tract residing the most abundant population [3]. The commensal microbiome is physiologically beneficial to the human host, but perturbed microbiota components or a disrupted mucosal environment could drive immune pathology and systemic inflammation [4] that affects human health. Microbiome dysbiosis contributes to the development of enteritis, pneumonia, and cancer [5, 6].
Cancer is a threat to human health worldwide owing to the high morbidity and mortality rates. All cancer cells are characterized by common hallmarks, including transformation, unrestricted growth, and progression [7–10]. Various factors have been identified that contribute to cancer initiation and progression, including gene mutations, suppressed immune responses, and a complex tumor microenvironment (TME) [11–14]. The tumorigenic and immunomodulatory roles of abnormal microbiomes are now recognized. The existence of the microbiome in tumor sites has been widely validated and accepted [15], and their effects on oncogenesis and progression have been extensively studied [1, 2, 16]. The interplay between the commensal microbiome and clinical treatment efficacy has also been proposed [2, 17]. The intimate interconnection between cancer and microbiota was documented as early as 1550 BCE when tumors were treated by incisions and poultices [18]. However, early attempts to apply microbiota to cancer treatment failed [18–20]. A limited mechanistic foundation might explain this, as technology that could detect low microbiome biomass was restricted. Current research into microbiota and cancer is supported by methods and technologies such as immunohistochemistry, quantitative PCR, immunofluorescence, fluorescence in situ hybridization, electron microscopy, and 16S rRNA sequencing [15].
The contribution of gut microbiota in cancer initiation, progression, and drug resistance has been thoroughly investigated. The gut microbiota can affect responses to chemo- and immunotherapeutic agents by modulating their efficacy or toxicity [21–25]. Therapeutic interventions to modulate microbiota composition to improve immunotherapy efficacy in mouse models have been promising [17, 26–28]. Subsequent endeavors have also translated preclinical findings into early-stage clinical tests with encouraging outcomes [29–31]. Apart from the gut microbiota, the existence and functional importance of intratumor microbiota in cancer remain contentious [15]. This review summarizes the roles of the intratumor microbiota in the tumor microenvironment, responses to therapies, and potential strategies that might facilitate better outcomes of cancer treatment.
Intratumor microbiome
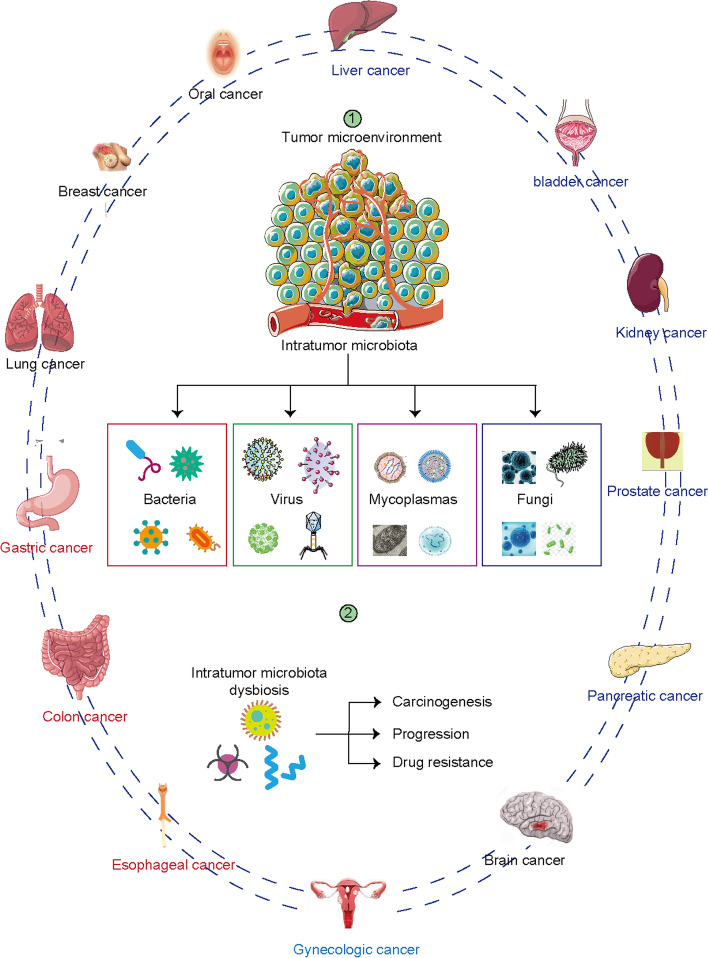
The intratumor microbiota has received less attention than the gut microbiome. In contrast to intestinal cancer, little is known about correlations between intratumor microbiota and other cancers. However, despite the paucity of studies, the composition of the intratumor microbiota is associated with many types of cancer. Organs and tissues, including the esophagus, lung, breast, prostate, bladder, stomach, kidney, liver, and pancreas, were previously considered sterile. However, next-generation sequencing (NGS) revealed that these organs harbor low-biomass microbial populations [15, 32, 33]. The intratumor microbiome is a major constituent of the tumor microenvironment that affects tumorigenesis, disease progression, drug resistance, and prognosis [34] (Fig. 1).
Fig. 1.
Intratumor microbiota niches across cancer types. Microbiota are detected in multiple solid tumors, including liver, bladder, kidney, prostate, pancreatic, brain, esophageal, colon, gastric, lung, breast, oral and gynecologic cancers. The intratumor microbiome has been convinced to contribute to the carcinogenesis, cancer progression and drug resistance
Intratumor tumorigenic bacteria
The human microbiome comprises bacteria, fungi, viruses, and mycoplasmas [35]. Epidemiological, basic, and clinical findings have established a link between intratumor bacteria and increased risk of cancers, suggesting that intratumor bacteria are high-risk factors for many cancers, including oral, lung, pancreatic, prostate, esophageal, bladder, colon, and gastric cancers [15, 36] (Table 1).
Table 1.
Summary of intratumor bacteria in cancerous tissues and their roles in oncogenesis, progression, and prognosis in cancers
| Genus | Status | Cancer | Samples | Role | Mechanism | Refs |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| Thermus and Ralstonia | Dysbiosis | Lung | Adenocarcinoma and squamous cell carcinoma | Cancer histology | NT | |
| Legionella | Enriched | Primary and metastatic lung tumor tissues | Oncogenesis metastasis | NT | [37] | |
| Acidovorax | Enriched | Lung cancer tissues with or without TP53 mutation | Linked with TP53 mutation | [38] | ||
| Staphylococcus | Decreased | Tissues from lung cancer patients with unilateral lobar masses and healthy controls | [39] | |||
| Anaerococcus, Caulobacter, Streptococcus, and Propionibacterium | Decreased | Breast | Tissues from Breast cancer patients, predisposed to breast cancer, and healthy controls | Negatively correlated with oncogenic immune features; positively associated with T-cell activation-related genes | [40] | |
| Bacteroidetes and Firmicutes | Lower ratio | Benign and breast cancer | [41] | |||
| Fusobacterium nucleatum | Enriched | Benign and breast cancer | Poor prognosis | Oncogenesis and suppressed immune response | [42] | |
| Bacteroides fragilis | Enriched | Cancer progression | BFT drives epithelial hyperplasia in the mammary gland | [43] | ||
| Lactobacillus fermentum | Enriched | Esophageal | Tissues from esophageal cancer patients and healthy controls | Cancer screen | NT | [44] |
| Campylobacter species | Enriched | Esophageal adenocarcinoma and control tissues | Prognosis | NT | [45] | |
| F. nucleatum | Enriched | Esophageal cancer and normal tissues | Prognosis | NT | [46] | |
| Bacteroidetes/Fusobacteria/Spirochaetes | Decreased | Esophageal cancer and normal tissues | NT | [47] | ||
| Actinobacteria | Enriched | Esophageal cancer and normal tissues | NT | [48] | ||
| P. gingivalis | Enriched | Esophageal tissues from ESCC patients and normal controls | Progression and prognosis | NT | [49] | |
| Fusobacterium nucleatum | Enriched | Resected ESCC samples | Chemoresistance | NT | [50] | |
| Fusobacterium and less Streptococcus | Dysbiosis | ESCC tumor tissues and normal tissues | Oncogenesis | NT | [51] | |
| H. pylori | Enriched | Gastric | Gastric cancer and normal tissues | Oncogenesis | Promote p53 degradation and immune evasion | [52–55] |
| E.coli, butyrate-producing bacterium SM4/1, Oscillatoria | Enriched | Bladder | Tumor samples with muscle invasive bladder carcinoma (n = 400) | Poor prognosis | Positively correlates with EMT-associated genes | [56] |
| Staphylococcaceae | Enriched | Prostate | prostatic tumor, peritumor and nontumor tissues | Oncogenesis | [57] | |
| Pseudomonas, Escherichia, Acinetobacter, Propionibacterium spp. | Enriched | frozen radical prostate samples from tumor and adjacent benign tissue | Oncogenesis | [58] | ||
| Enriched | Propionibacterium acnes spp | Prostate tissue inflammation | [59] | |||
| Proteobacteria | Enriched | Prostatic tumor tissues | Oncogenesis | [60] | ||
| Enterobacteriaceae Pseudomonadaceae | Enriched | Pancreatic | Pancreatic ductal adenocarcinoma tissues and normal human pancreas | Chemotherapy resistance | Metabolize chemotherapy drugs | [32] |
| Proteobacteria Bacteroidetes Firmicutes | Enriched | Pancreatic cancer and normal tissues | Tumor progression | Modulating M1 macrophage/Th1 differentiation, that affect CD8 + T cell function | [61] | |
| Pseudoxanthomonas Streptomyces Saccharopolyspora Bacillus clausii | Enriched | pancreatic adenocarcinoma (PDAC) patients with short-term survival (STS, < 5 years) and long-term survival (LTS, > 5 years) | Prognosis | Elevated infiltration and activation of CD8 T cells | [62] | |
| F. nucleatum | Enriched | Mouth | Oral squamous cell carcinoma and normal oral tissues | Predictor | Promotes EMT transition | [63] |
| Abundance of Firmicutes (especially Streptococcus) and Actinobacteria (especially Rothia) | Bacterial dysbiosis | Oral cancers and anatomically matched contralateral normal tissue | Promote oncogenesis and progression | [64] | ||
| Fusobacterium/Prevotella | Enriched | Oral squamous cell carcinoma tissues and adjacent non-tumor mucosa 5 cm distant | Oncogenesis | NT | [65] | |
| Peptostreptococcus | Enriched | Tumor samples from patients with OSCC | Better prognosis | NT | [66] | |
| F. nucleatum | Enriched | Colon | Colorectal cancer and paired normal tissues | Oncogenesis and progression |
Activates β-catenin signaling; Lower density of CD3+ T cells; Recruits immuno- suppressive cells; Inactivation of NK and T cells |
[67–73] |
| Escherichia coli and Bacteroides fragilis | Enriched | Familial adenomatous polyposis samples and healthy controls | Promotes cancer initiation | that secrete oncotoxins | [34] | |
| Fusobacterium | Enriched | Paired primary colorectal and metastatic tumors | Cancer metastasis | NT | [70] | |
| Helicobacter spp | Enriched | Bile duct | Bile duct cancer tissues | Oncogenesis | NT | [74] |
| Fusobacterium nucleatum, Escherichia coli, and Enterobacter sp. | Enriched | Gallbladder | Bile samples from patients with gallbladder cancer and cholelithiasis | Oncogenesis | NT | [75] |
| Decreased Nesterenkonia, and increased Methylophilaceae, Fusobacterium, Prevotella, Actinomyces, Novosphingobium, and H. pylori | Bacteria dysbiosis | Extrahepatic cholangiocarcinoma | Tissues from extrahepatic cholangiocarcinoma (ECCA) and benign biliary pathology (BBP) cohorts | Oncogenesis | NT | [76] |
| Helicobacter bilis | Enriched | Extrahepatic cholangiocarcinoma | Tissues from extrahepatic cholangiocarcinoma (ECCA) and benign biliary pathology (BBP) cohorts | Oncogenesis | NT | [77] |
| Bifidobacteriaceae Enterobacteriaceae Enterococcaceae | Enriched | Cholangiocarcinoma | primary CCA tumors and matched normal tissues | Oncogenesis | NT | [78] |
| Helicobacter species | Enriched | Hepatocellular carcinoma | Liver samples from patients with hepatocellular carcinoma, non-cirrhotic chronic hepatitis C, and healthy controls | Oncogenesis | NT | [79] |
| Sneathia and Lactobacillus | Dysbiosis | Cervix | [80] | |||
| Fusobacterium spp | Enriched | Samples from patients with squamous intraepithelial lesions (SIL) and cervical cancer | Oncogenesis | NT | [81] | |
| L. gasseri | Enriched | Oncogenesis | NT | [82] | ||
| Atopobium, Porphyromonas, Dialister, Peptoniphilus, Ruminococcus, Anaerotruncus, Anaerostipes, Treponema, Bacteroides and Arthrospira | Enriched | Endometrium | Uterine samples from cancer and benign disease | Oncogenesis | Modulating the vaginal pH | [83] |
| Brucella, Mycoplasma, Chlamydia spp. | Enriched | Ovary | Ovarian tumor | Oncogenesis | [84] | |
| Proteobacteria | Enriched | Ovarian cancer tissues and normal distal fallopian tube tissues | Cancer initiation and progression | Modulating immune response | [85] | |
| Actinomyces and Parvimonas | Dysbiosis | Head and neck squamous cell carcinomas (OSCC) | Paired normal and tumor resected OSCC specimens | Tumor stage | NT | [86] |
Gastrointestinal cancer
Among all tumors, gastrointestinal malignancies have received the most attention because of the abundance of bacterial residues in the gut [3]. Gut bacteria dysbiosis occurs in patients with adenoma or colorectal cancer (CRC), as populations of F. nucleatum [67–69], Escherichia coli, B. fragilis [34] and Fusobacterium [70] are increased, whereas those of Ruminococcus, Bifidobacterium, and Streptococcus species are decreased [87]. Intratumor bacterial dysbiosis is causally correlated with the oncogenesis of CRC [34, 63, 79, 88], metastasis [70], immune evasion [71–73], and drug resistance [89, 90]. The interplay between intratumor bacteria and liver/biliary tract cancers has been extensively investigated because the liver and biliary tract are exposed to the gastrointestinal microbiome through the gut–liver axis. Bifidobacteriaceae, Enterobacteriaceae, and Enterococcaceae are enriched in tumor samples from patients with cholangiocarcinoma [78]. The population of Nesterenkonia is decreased, whereas those of Helicobacter bilis, Fusobacterium, Methylophilaceae, Prevotella, Novosphingobium, Actinomyces, and H. pylori are increased in patients with extrahepatic cholangiocarcinoma (ECCA), compared to those with benign biliary pathology (BBP) [76, 77]. The abundance of Helicobacter species is high in hepatocellular carcinoma tissues [79].
Abnormal bacterial abundance such as increased F. nucleatum [63] and Fusobacterium/Prevotella [65] and decreased Streptococcus/Rothia [64] in the oral cavity might be a risk factor for oral cancer. Bacterial dysbiosis also correlates with the prognosis of patients [66]. Intratumor bacteria have also been detected in other gastrointestinal cancers, and dysbiosis causally correlates with carcinogenesis, progression, suppressed immune response, and drug resistance. Helicobacter pylori was one of the first bacterial species to be directly associated with the oncogenesis of gastric cancer [52–55]. Bacterial dysbiosis [47–49, 51] promotes chemoresistance in esophageal cancer [50]. The bacterial biomass in esophageal tissues can distinguish cancer cohorts [45], cancer types [44], tumor stage, and prognosis [46]. In addition, Proteobacteria, Bacteroidetes, and Firmicutes are abundant in tissues from mouse models and humans with pancreatic cancer [61]. The presence and dysbiosis of bacteria in pancreatic cancer contribute to oncogenesis, immune evasion, resistance to chemotherapy [32, 61] and even affects patient prognosis [62]. Dysbiosis also occurs in cancers of other parts, such as the bile duct [74] and gallbladder [75].
Genitourinary cancers
Intratumor bacteria participate in the development of cancer in genitourinary organs that were previously considered sterile [15]. Bacterial biomass has been recognized in tissues [59] as well as in frozen samples of prostate tumors and adjacent benign tissues after radical prostatectomy. Over 40 unique bacterial genera have been identified [58] in freshly resected prostate tissues. The abundance of Staphylococcaceae [57, 60] and Propionibacterium acnes spp. [59] is increased, and the biomass of Actinobacteria, Firmicutes, Proteobacteria, Lactobacillales, and Streptococcaceae is decreased [57, 60]. Intratumor bacteria with increased E. coli, the butyrate-producing bacterium SM4/1, and Oscillatoria indicate a poor prognosis of bladder cancer [56].
Gynecological cancers
Microbiota in the lower female reproductive tract protects the endometrium, ovary, and fallopian tubes from pathogen attack and sustains homeostasis [91]. However, bacterial dysbiosis contributes to gynecological malignancies. An increased abundance of Atopobium, Dialister, Porphyromonas, Peptoniphilus, Anaerotruncus, Ruminococcus, Anaerostipes, Treponema, Bacteroides, and Arthrospira promotes the carcinogenesis of endometrial cancer [83]. Cervicovaginal microbiome dysbiosis correlates with a high risk of ovarian cancer [92]. The prevalence of Brucella, Mycoplasma, and Chlamydia spp. has been confirmed in samples from patients with ovarian cancer [84]. Both diversity and richness are lower in ovarian cancer tissues than in normal distal fallopian tube samples, with a decrease in Proteobacteria abundance [85]. Bacterial dysbiosis with increased abundance of Sneathia, Lactobacillus gasseri [80, 82] and Fusobacterium spp. [81], and decreased Lactobacillus biomass [80] promotes the oncogenesis of cervical cancer, which is the most prevalent malignancy associated with human papillomavirus (HPV).
Other cancers
In addition to being identified in tumors that arise from mucosal organs, intratumor bacteria have also have been identified in lung, breast, bone, melanoma cancers, glioblastoma multiforme (GBM), and head and neck squamous cell carcinomas [15, 37–41, 86]. Other bacteria, such as B. fragilis [43] and F. nucleatum [42], also contribute to breast cancer progression. Intratumor bacterial dysbiosis correlates with a high risk of lung cancer in clinical samples [37–39]. Moreover, bacterial dysbiosis correlates with TP53 mutations [38], cancer metastasis [37], and cancer histology [37]. In addition to lung cancer tissues, bacterial profiles differ in saliva, sputum, bronchoscopic samples, and bronchoalveolar lavage fluid (BALF) between patients with lung cancer and healthy controls [37, 39, 93] (Table 1). However, associations between intratumor bacteria and other types of cancers have not been extensively investigated. The profiles of intratumor bacteria in different types of cancer are distinct, with abundance and diversity being the highest in breast tumors [15].
Intratumor non-bacterial microbiome
Mycoplasma, fungi, archaea, protists, and viruses are also microbiome components [94]. Investigations into the roles of microbes in cancers have mainly focused on bacteria [95]. However, other types of microbes, such as mycoplasmas, fungi, and viruses, also play roles in cancer [96, 97].
Intratumor mycoplasmas
The interplay between mycoplasmas and malignancy was discovered during the 1960s [97]. Mycoplasma infection is prevalent in colon carcinoma and gastric, esophageal, lung, breast, prostate, ovarian, cervical, kidney, pancreatic cancers, and glioma [84, 98–102]. A direct comparison of samples from patients with small-cell lung carcinoma and healthy controls found significant mycoplasma accumulation in cancer tissues [103]. Furthermore, mycoplasma infection induces transformation and tumorigenicity in the normal human lung cell line, BEAS-2B, and promotes lung cancer angiogenesis by elevating bone morphogenetic protein 2 (BMP2) levels [16]. Mycoplasma infection induces the malignant transformation of other human cell lines, such as A549 (lung) [16], benign prostate hyperplasia (BPH)-1 [36], blood cells [104], SK-UT-1B (uterus) [105], and BE-M17 (neurons) [106]. Although mycoplasmas have malignant potential and are prevalent in various types of cancer, their pathological role in tumorigenesis remains controversial. Besides oncogenesis, mycoplasma infection contributes to drug resistance [32].
Intratumor fungi
Fungi correlate with cancer risk [96]. Fungi are approximately 3,000-fold more abundant in pancreatic ductal adenocarcinoma (PDA) than in normal pancreatic tissues from mice model and human samples, and most of them comprise Malassezia spp. This fungus accelerates oncogenesis in mouse models of PDA, and ablating it represses tumor growth and progression [96]. Mechanistically, Malassezia binds through its surface glycans to mannose-binding lectin (MBL) to activate the complement cascade, resulting in oncogenic progression [96]. A fluorescence-tagged fungal strain introduced into the gut of a mouse model that was detectable in the pancreas after 30 min, suggesting that intratumor fungi are translocated from the intestine [107]. Candida infection is causally linked to cancer risk. Several putative mechanisms might explain their contribution to oncogenesis. Candida produces nitrosamines that alter cell proliferation [108] and secretes cytokines such as tumor necrosis factor-alpha (TNF-α) and interleukin (IL) 18 (IL-18) that modulate the immune response and promote tumor cell adhesion to epithelial cells [109]. Fungi have also been detected in prostate, esophageal, gastric, skin, oral, lung, and colon cancers [60, 107, 108]. However, the underlying mechanisms remain unclear and await further investigation.
Intratumor viruses
As in the case of other microbiomes, virome infection closely correlates with cancer in solid tumors of colon, hepatocellular, oral, breast, cervical, esophageal, gastric, and lung cancers [110–116]. To date, the following viruses have been identified as being cancer-related: Epstein-Barr virus, Kaposi sarcoma herpes virus, HPV, human T-cell lymphotropic virus, hepatitis B virus, hepatitis C virus, and Merkel cell polyomavirus [25]. A causal effect of HPV on cervical cancer oncogenesis has been confirmed [80], and HPV infection also correlates with the progression of head and neck cancers, esophageal squamous cell carcinoma (ESCC), and bladder cancers [114, 116]. Virome infection also directly causes esophageal squamous cell carcinoma, including HPV, Epstein-Barr virus, and polyoma viruses [114]. Hepatitis B and C viral infections lead to liver cancer [79] and cholangiocarcinoma [112]. Bacteriophages might also be involved in cancer development. Multiple Streptococcus-specific bacteriophages and a Vibrio-inhabiting bacteriophage have been detected in the gut of patients with CRC compared with controls [117].
Mechanisms of intratumor microbiome impacts on tumorigenesis
Intratumor microbiome dysbiosis and its clinical significance have been confirmed in clinical samples, but the underlying mechanism remains obscure. The interplay is complex between cancer and the intratumor microbiome, which affects cancer growth and spread by promoting cancer development mainly by increasing mutagenesis, modulating oncogenes or oncogenic pathways, and affecting the immune response.
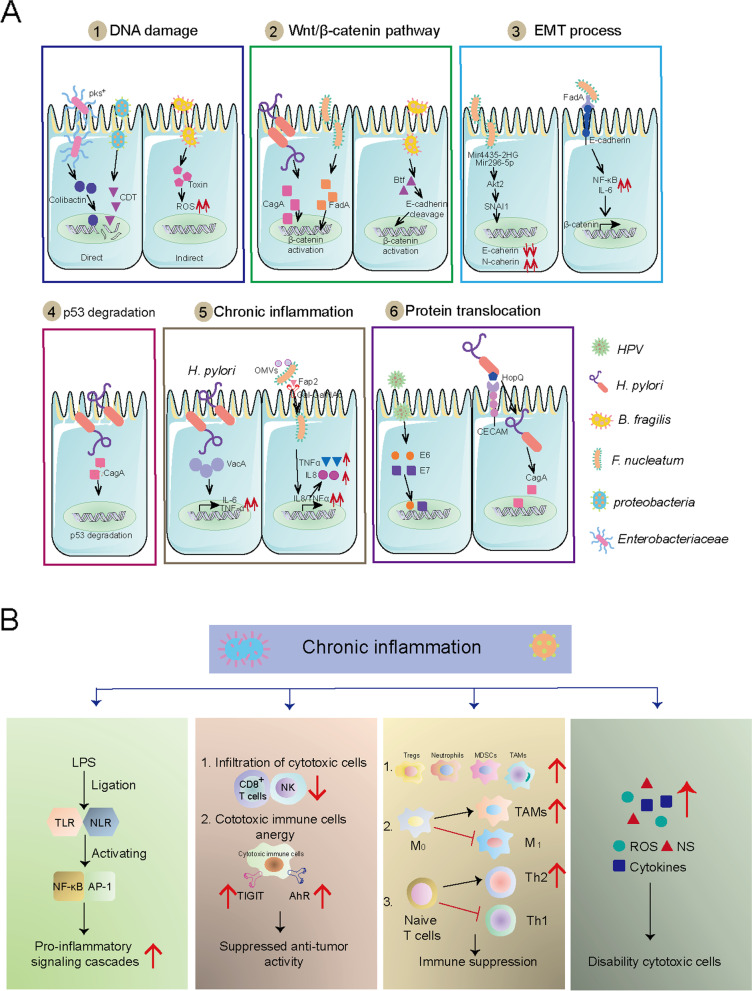
Many bacteria have evolved to acquire the ability to damage DNA, which could lead to mutational events and eventually contribute to carcinogenesis. Enterobacteriaceae, such as B2 group E. coli [118], secrete colibactin and directly induce DNA damage, resulting in colon cancer tumorigenesis [119]. Bacteria with similar functions include B. fragilis [34], H. pylori [55] and ε- and γ-proteobacteria [119]. Mechanistically, colibactin and cytolethal-distending toxin (CDT) can directly induce DNA damage [119], whereas Bft functions indirectly by producing high levels of reactive oxygen species (ROS) [120]. Chronically high ROS levels can outpace the host DNA repair, and finally results in DNA damage and mutations [38, 88].
Intratumor bacteria are involved in carcinogenesis by producing proteins that participate in host pathways. Among these, the Wnt/β-catenin pathway, an oncogenic signaling pathway in cancer, is altered in many malignancies and is involved in cancer stemness, polarity, and growth [121–123]. This might be because β-catenin activation induces the upregulation of genes involved in cellular proliferation, survival, apoptosis, and migration [121–123]. Several cancer-associated bacteria contribute to activating Wnt/β-catenin signaling. Examples include H. pylori, which produces cytotoxin-associated gene A (CagA) protein [52, 124], F. nucleatum, which expresses Fn secretes an adhesin A (FadA) [125]and enterotoxigenic B. fragilis, which secretes Bft [43]. Mechanistically, CagA can pass into the cytoplasm of host cells and induce gastric cancer by affecting the β-catenin pathway [52, 124], and FadA induces carcinogenesis by activating the β-catenin pathway [125]. Similarly, enterotoxigenic B. fragilis produces Bft that stimulates E-cadherin cleavage and subsequently induces β-catenin activation (Fig. 2A) [43].
Fig. 2.
Potential molecular mechanisms by which intratumor microbiota promote carcinogenesis. A. Microbiome contributed to the tumorigenesis through inducing DNA damage, Wnt/β-catenin pathway, EMT process, p53 degradation, chronic inflammation and protein translocation. B. The chronic inflammation that induced by intratumor microbiota include cancer-associated inflammation, cancer-associated cytokines and ROS/NS production, inhibited cytotoxic immune cells infiltration and function and enhanced immunosuppressive cells infiltration and polarization
Intratumor bacteria, such as F. nucleatum [63] and H. pylori [52] can drive carcinogenesis by inducing degradation of the tumor suppressor gene p53. Other mechanisms explain the tumorigenicity of H. pylori, including chronic inflammatory responses and epithelial-mesenchymal transition (EMT) modulation. Helicobacter pylori can drive cytotoxicity and chronic inflammation via IL1β, TNFα, and the interferon-gamma (IFN) γ-stimulated Th1-type response [126] or by stimulating TNF-α and IL-6 by secreting vacuolating toxin A (VacA) [55]. Helicobacter pylori can modulate host cells through the bacterial protein CagA, which can directly translocate into gastric epithelial cells through the type 4 secretion system (T4SS) [115]. Fusobacterium nucleatum is an oncogenic factor in solid tumors, including breast and colon cancer [42, 63, 68]. Mechanistically, F. nucleatum stimulates the activation of pro-inflammatory cascades mediated by nuclear factor kappa B (NF-κB) and IL-6, which might facilitate oral squamous cell cancer (OSCC) cell invasion [127]. Outer membrane vesicles (OMVs) produced by F. nucleatum can drive chronic inflammation by stimulating colonic epithelial cells to secrete IL-8 and TNF-α [128]. In addition, F. nucleatum can induce EMT to drive carcinogenesis [42, 63] (Fig. 2A).
Chronic high-grade inflammation is another mechanism that explains intratumor microbiome-induced oncogenesis [25–27]. Numerous cancer-associated microbiomes induce pro-tumor immune responses. According to data from clinical tissues and animal models, intratumor microbes inhibit innate and adaptive immune systems [15, 129]. The commensal microbiome can stimulate Toll-like receptors (TLRs) via lipopolysaccharide (LPS) that promote proinflammatory signaling cascades to enable a cancer-associated microenvironment. Such bacteria include commensal gram-negative gut bacteria in cholangiocarcinoma [130] and F. nucleatum [128]. Activator protein 1 (AP-1) and NF-kB are located downstream of the LPS–TLR axis [131]. Commensal microbes stimulate the production of cancer-associated-cytokines that often have deleterious consequences for tumor progression via activation of the interleukin-23 (IL-23)-IL-17 axis [129], the TNF-α/TNF receptor axis [55, 130], IL-6 family signaling [55, 66, 130], IL-10, IL-8, IL-18, monocyte chemoattractant protein-1 (MCP-1) [125, 132], signal transducer and activator of transcription 3 (STAT3) [55, 133], and the production of ROS [88] and nitrogen species (NS) [38, 120]. Intratumor microbes can directly inhibit anti-tumor immunity by inhibiting cytotoxic immune cell infiltration [42, 61, 71, 131] and blocking their ability to kill tumor cells [124]. Examples include T cell immunoreceptor with Ig and ITIM domains (TIGIT) that is expressed on some T cells and natural killer cells [73] and aryl hydrocarbon receptors (AhRs) expressed on T cells [134]. Commensal microbes recruit abundant inflammatory cells, including tumor-associated macrophages (TAMs), regulatory T cells (Tregs), granulocytes, Vγ6 + Vδ1 + γδ T cells, and myeloid-derived suppressor cells (MDSCs), which results in a pro-inflammatory environment [61, 72, 129, 131, 135, 136] (Fig. 2B).
The mechanisms of nonbacterial cancer-associated microbial action have not been investigated in detail. Only a few mechanisms might explain non-bacterial tumorigenesis. For example, HPV expresses oncoproteins E6 and E7, which can integrate into the host genome [80, 116], where they trigger the amplification of specific oncogenic genes that induce tumorigenesis in cervical cancer. The putative mechanism of bacteriophage alterations might be through initiating the genetic exchange, which enables ecological adaptations and community networking within hosts, thereby affecting cancer [137]. However, a direct effect of phages on carcinogenesis has not yet been identified. Mycoplasma can induce transformation and tumorigenicity by elevating levels of BMP2, which then increases cell proliferation and migration, and represses apoptosis (Fig. 2A, 3 and 4) [16].
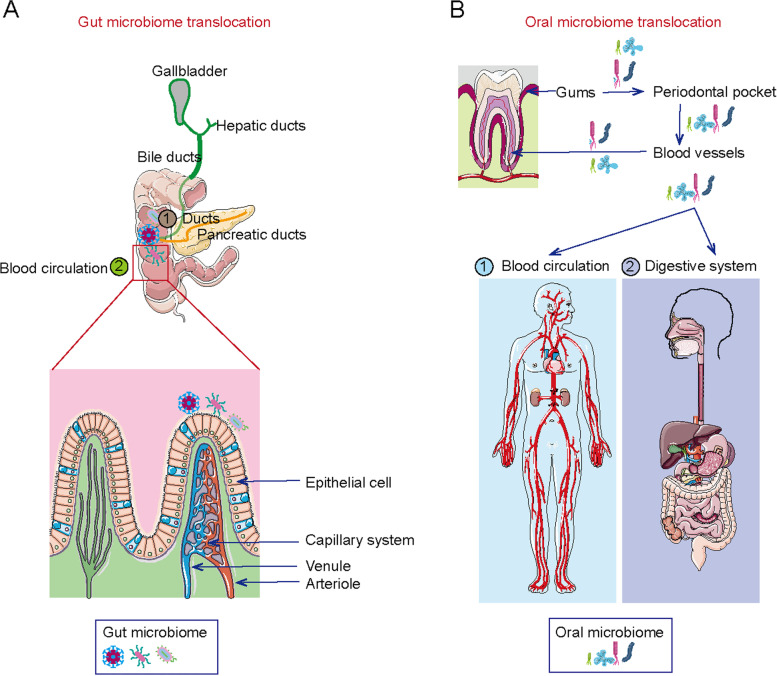
Fig. 3.
The potential source of intratumor microbiota. A. Microbiome may translocate from intestine to the tumor sites, which depends on blood circulation and ducts translocation. B. Oral microbiome may be another origin of intratumor microbiota. And blood circulation and digestive system are the main pathways
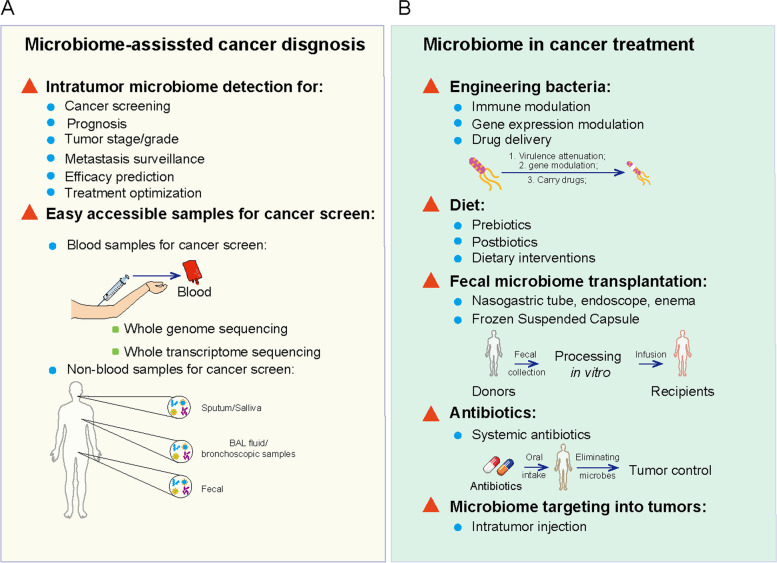
Fig. 4.
The utilization of intratumor microbiota data in cancer screen and treatment. A. Data from clinical samples may facilitate the development of new cancer screen and prognosis, including microbiota patterns from tumor sites and easily accessible samples. B. Intratumor microbiota may be applied for cancer treatment, including engineering bacteria, diet modulation, fecal microbiome transplantation, antibiotics and intratumor microbiome injection
Microbiota translocation
The commensal microbiome inhabits the mouth, skin, reproductive organs, and gastrointestinal tract in humans [21, 138]. One explanation for the inhabitation of microbiota in tumor sites and the source of this intratumor microbiota is their translocation from the intestine [139].
Gut translocation
The gut is the main organ colonized by commensal microorganisms, comprising 3.93 × 1013 bacteria [22]. Physically, human and gut microbes inherently coexist symbiotically, and this is partly maintained by spatial separation, explaining the presumed sterility of organs, including the pancreas, breast, kidney, and lung [140]. However, bacterial biomass has been revealed based on the 16S rRNA sequences of bacteria in these organs [15, 61, 141]. Clinical data have revealed that microbiome expression overlaps between tumor tissues and fecal samples, indicating that the intestine is the source of intratumor microbiomes [87, 142]. Despite scant evidence, the microbiome is translocated from the gut into sites in several solid tumors. For example, bacterial profiles are similar in the duodenum and gut, indicating that bacteria can translocate from the gastrointestinal tract into the pancreas [32, 61]. This has been validated in mouse models given fluorescently labeled oral Enterococcus faecalis, which confirmed the translocation of bacteria from the gut into the pancreas [61].
The translocation of gram-negative bacteria from the gut, resulting from primary sclerosing cholangitis (PSC) or colitis-induced intestinal barrier dysfunction, finally drives the development of cholangiocarcinoma (CCA) [130]. Translocation of the gut microbiome to the gastrointestinal organ is further validated by the fact that Fusobacteria are maintained in distal liver metastases [70]. Fusobacterium and its associated microbiome comprising Bacteroides, Selenomonas, and Prevotella species overlap in microbiomes in primary colon cancer tissues and paired metastatic liver tumor sites [70]. The microbiome might be translocated from the intestinal tract into other organs via the blood circulation and/or bile, hepatic, and pancreatic ducts [61, 143].
Oral translocation
The oral cavity has been regarded as the origin of the intratumoral microbiome [127, 144]. As the entry portal for the gastrointestinal tract, the oral cavity is connected to the respiratory tract, and microbiota residing in the oral cavity can disseminate into the respiratory and gastrointestinal tracts [144]. Therefore, dysbiosis of the oral microbiota, such as periodontal disease, can contribute to the respiratory tract and gastrointestinal cancers, including esophageal cancer [33, 49], head and neck squamous cell carcinoma [145], lung [146], gastric [144], and colorectal [147] types. For example, the esophageal microbiota is similar to the oral microbiota, as it includes Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria, and TM7, indicating the translocation of microbiota from the oral cavity to the esophagus [44]. Furthermore, the results of organisms cultured from aspirates obtained from patients with esophageal cancer and healthy persons during esophagoscopy revealed that Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus spp in the esophagus overlap with oral microbiota [33, 114, 148–150]. Additionally, oral Prevotella intermedia, Tannerella forsythia, and Treponema denticola possess peptidyl arginine deaminase (PAD) that induces P53 mutations in pancreatic cancer [151].
Periodontal disease, characterized by dysbiosis in oral microbiota, is associated with the risk of genitourinary cancers, including bladder and prostate cancers [49, 141, 144–147, 152, 153]. Likewise, a comparison of microbiota in chronic prostatitis and BPH revealed overlapping P. gingivalis, Treponema denticola, and E. coli in the oral cavity and prostatic tumor sites [118]. The detection of oral microbiota in genitourinary organs suggests a potential role of oral microbiota in the initiation and progression of genitourinary cancers through the oral-genitourinary axis. Although epidemiological studies have identified interplay between periodontitis and various types of cancers and a close correlation between them, the causal impact of oral microbiota on genitourinary cancers remains mechanistically speculative. This is because the periodontium where the oral microbiota originally resides is located far from the genitourinary organs. The migration of oral microbiota from the oral cavity to distant organs has been investigated. The translocation of oral bacteria to the respiratory tract and gastrointestinal organs might be via the bloodstream and the digestive tract [154]. The proposed concept of oral bacterial translocation via the bloodstream is based on the anatomical structure and the proximity of the periodontal pockets to the bloodstream.
Potential clinical application of intratumor microbiota
Cancer screening and diagnostics
The microbiota species significantly differ between tumor and healthy tissues, and some bacteria are causally associated with cancer development [155]. This indicates that the intratumor microbiota could function as a biomarker for cancer screening [139] [40]. Examples include intratumor microbiome-derived personalized data that can distinguish patients with esophageal [47], pancreatic [35], lung [93] and oral [64] cancers from healthy persons. In addition to cancer diagnosis, microbiome signatures also differ among tumor stages, tumor grades, cancer scores [60, 81], gene mutations (such as estrogen, progesterone receptors, and Her2) [156] or regions [157]. Furthermore, the unique intratumor microbiome signature correlates with distant tumor metastasis [42, 63] and responses to chemotherapy [50, 89]. Profiling the intratumor microbiome might offer potential for tumor prognostic evaluation. Indeed, intratumor microbiome diversity is higher and Pseudoxanthomonas, Streptomyces, Saccharopolyspora, and B. clausii are upregulated in patients who have survived PDA over the long-term [62]. Similarly, F. nucleatum signatures have been identified in esophageal cancer and the correlation with prognosis [46].
As understanding of the intratumor microbiome effects on cancer pathogenesis deepens, the application of these profiles to precision oncology is highly attractive. However, obtaining organ biopsies from healthy humans is unethical, and obtaining samples for cancer screening is difficult. Alternatives to inaccessible and unethical biopsies in healthy humans include saliva, sputum, bronchoscopy samples, and BALF [37, 39, 93] to screen for lung cancer, the tongue microbiome to screen for pancreatic cancer [158], and the oral microbiome to screen for esophageal cancer [159]. The existence of microbiome in blood is verified in colorectal and breast cancer patients according to the data from The Cancer Genome Atlas (TCGA) [160]. The blood microbiome is unique and can distinguish cancer based on whole-genome and -transcriptome sequencing data for 33 types of cancer from treatment-naive patients (18,116 samples) in TCGA [139].
Systematic characterization of the microbiota provides an opportunity to explore new methods for non-human microorganism-derived molecules in cancer diagnosis. Moreover, new strategies involving easily accessible samples are needed to screen for cancer without invasive testing. However, many challenges must be tackled for intratumor microbiome-based clinical diagnoses, such as inaccessible tumor tissues, relatively low microbiome biomass, and easy contamination. In addition, the specificity, prevalence, and stability of the intratumor microbiome during cancer treatment must be addressed before clinical deployment.
Intratumor microbiome modulation and clinical efficacy
Cancers have a dismal prognosis, and outcomes have improved little with modern [161–164] and traditional [59, 165, 166] therapeutic strategies. Apart from tumor cell-intrinsic mechanisms, discovering the intratumor microbiome and elucidating host-microbe interactions present opportunities for intervention [1, 2, 167]. Increasing evidence suggest that microbiota modulation is currently recognized as a novel and important adjunct to enhance anticancer therapies [27, 168, 169]. Efforts to use microbiota for cancer treatment during the twentieth century [20] were unsuccessful. The intratumor microbiome has recently been extensively investigated [15] and tumor-targeting bacteria such as Salmonella typhimurium strain VNP20009 [170], Listeria monocytogenes [171] and Listeria spp. [172] can selectively eliminate tumor cells. Owing to their ability to selectively colonize in tumors or tumor-driven lymph nodes to inhibit tumor growth, preclinical studies have evaluated treatment efficiency in mouse models [170, 172]. The promising anti-tumor responses in preclinical studies have led to the selection of several bacterial strains for evaluation in patients with tumors [173, 174]. However, the clinical outcomes are unsatisfactory; only objective responses or minor and transient side effects have been identified in clinical trials [173, 175]. The discrepancies between outcomes in preclinical researches and clinical trials might be explained by differences in tumor structures and growth rates, which could change bacterial penetration, proliferation, and clearance within tumors, as well as in peripheral circulation.
Also, live bacteria could be attenuated and further reprogrammed to produce and deliver anticancer agents based on clinical requirements [176, 177]. Tumor-targeting bacteria offer several advantages as delivery vectors, including improved penetrability of tumor sites, maximized activities of chemotherapeutic agents, and reduced systemic toxicity. Regulating bacterial gene expression might further modulate the accumulation of anti-tumor payloads at tumor sites and control continuous drug delivery [132]. Several strategies have been developed to selectively deliver tumor-targeting bacteria, including cytokines, chemotherapeutic agents, prodrug-converting enzymes, small interfering RNAs (siRNAs), and immunomodulators [95]. These have enhanced anti-tumor responses and reduced nonspecific side effects in tumor models [178–180]. Considering the role of the intratumor microbiota in modulating host immunity [102, 181], it could probably influence responses to and toxicity of various types of cancer therapy. Nevertheless, direct control of intratumor microbiome modulation is still a long way off. Obstacles need to be overcome, such as controlling microbiome toxicity, inaccessible microbiomes in tumor sites, potential side effects and the accuracy of microbiome biomass delivery into tumor sites.
Since the oral cavity and intestine are recognized as the primary sources of the intratumor microbiome, modulation of the microbiome in the gut might reshape that in tumors and affect cancer therapies. Cross-talk between the gut and intratumor microbiomes has been identified in pancreatic cancer [61, 62, 96]. Thus, gut microbial modulation via antibiotics, diet, and fecal microbiota transplantation (FMT) might have the potential as a powerful immunotherapeutic modality. Antimicrobial therapy in cancer is limited to address or prevent known cancer-associated microbiomes, such as HPV, H. pylori, HBV/HCV, Epstein-Barr virus, and polyoma virus-induced cancers [79, 80, 112, 114, 116]. Nevertheless, systemic antibiotics are not always beneficial, as they can weaken the immune checkpoint blockade (ICB) and result in a poor prognosis [182, 183]. Prebiotics, postbiotics, and dietary interventions are also regarded as promising strategies to improve anti-tumor immunity and therapy responses to cancers in both mouse models [17, 184] and clinical trials (NCT03870607, NCT03950635) [136, 184]. However, collecting dietary data has restrained elucidation of the causal mechanisms underlying this strategy. Instead, metabolomic data that can reveal dietary intake and concomitant small-molecule effectors might serve as a substitute for mechanistic exploration. In addition, the gut microbiota has been modulated using FMT to enhance ICB efficacy. The results of mouse models [21, 142] and a clinical trial (NCT03353402) have been promising [23, 30, 31, 136]. Many factors complicate the use of FMT in clinical cancer treatment, such as the complexity of monoclonal bacterial strains, multiplexed consortia, antibiotic preconditioning, administration routes, the modulation frequency, and dietary recommendations. However, the long-term efficacy and stability of FMT in cancer treatment have not been evaluated.
Concluding remarks
The expression of the intratumor microbiome in patients with cancer has gradually been revealed due to technological developments [15]. Although many intratumor microbiome dysbiosis in solid cancers contribute to oncogenesis, progression, and drug resistance, the direct causal roles and underlying mechanisms of the intratumor microbiome remain ambiguous. Gaining sufficient insight into modes of action through which the microbiome might function as a biotherapeutic agent is important for patient prediction and the successful, rational development of microbiome-modulating therapies to enhance clinical treatment effects. Efforts have been targeted towards the application of gut microbiota to modulation-based cancer therapies because of cross-talk between intratumor and gut microbiota. Modulating the gut microbiome to treat cancer has been attempted, but causal mechanisms of adjuvants are difficult to reveal in complex environments. Identifying monoclonal bacterial strains that are beneficial to the anti-tumor response is imperative. Tumor-specific microbiomes have been confirmed [15]. Therefore, precisely characterizing the components of the tumor microbiome would provide valuable insights that might facilitate the development of tumor-specific treatments without severe side effects.
Acknowledgements
Not applicable
Abbreviations
- TME
Tumor microenvironment
- NGS
Next-generation sequencing
- CRC
Colorectal cancer
- ECCA
Extrahepatic cholangiocarcinoma
- HPV
Human papillomavirus
- GBM
Glioblastoma multiforme
- BALF
Bronchoalveolar lavage fluid
- BMP2
Bone morphogenetic protein 2
- BPH-1
Benign prostate hyperplasia
- PDA
Pancreatic ductal adenocarcinoma
- MBL
Mannose-binding lectin
- TNF-α
Tumor necrosis factor-alpha
- IL-18
Interleukin (IL) 18
- ESCC
Esophageal squamous cell carcinoma
- CDT
Cytolethal-distending toxin
- ROS
Reactive oxygen species
- CagA
Cytotoxin-associated gene A
- FadA
Fn secretes an adhesin A
- IFN γ
Interferon-gamma (IFN) γ
- VacA
Vacuolating toxin A
- T4SS
The type 4 secretion system
- NF-κB
Nuclear factor kappa B
- OMVs
Outer membrane vesicles
- TLRs
Toll-like receptors
- LPS
Lipopolysaccharide
- AP-1
Activator protein 1
- MCP-1
Monocyte chemoattractant protein-1
- NS
Nitrogen species
- AhRs
Aryl hydrocarbon receptors
- TAMs
Tumor-associated macrophages
- MDSCs
Myeloid-derived suppressor cells
- PSC
Primary sclerosing cholangitis
- CCA
Cholangiocarcinoma
- PAD
Peptidyl arginine deaminase
- siRNAs
Interfering RNAs
- FMT
Fecal microbiota transplantation
- ICB
Immune checkpoint blockade
- EMT
Epithelial-mesenchymal transition
- OSCC
Oral squamous cell cancer
Author contributions
Jinyan Liu drafted the article and prepared the figures. Yi Zhang revised the manuscript.
Funding
This work was supported by grants from the Program of the Major Research Plan of the Nationoal Natural Science Foundation of China (Grant No. 91942314), the National Natural Science Foundation of China (Grant Nos. 82001659), the followship of China postdoctoral Science Foundation (Grant No. 2021M692914) and the Major Public Welfare Projects in Henan Province (Grant No. 201300310400).
Availability of data and materials
Data sharing not applicable to this article as no data-sets were generated or analyzed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Helmink BA, et al. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 2.Buchta Rosean C, et al. Impact of the microbiome on cancer progression and response to anti-cancer therapies. Adv Cancer Res. 2019;143:255–294. doi: 10.1016/bs.acr.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone CA, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva MJ, et al. The multifaceted role of commensal microbiota in homeostasis and gastrointestinal diseases. J Immunol Res. 2015;2015:321241. doi: 10.1155/2015/321241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.López de Andrés J. Cancer stem cell secretome in the tumor microenvironment: a key point for an effective personalized cancer treatment. J Hematol Oncol. 2020;13(1):136. doi: 10.1186/s13045-020-00966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balian A, Hernandez FJ. Nucleases as molecular targets for cancer diagnosis. Biomark Res. 2021;9(1):86. doi: 10.1186/s40364-021-00342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldea M, et al. Overcoming Resistance to Tumor-Targeted and Immune-Targeted Therapies. Cancer Discov. 2021;11(4):874–899. doi: 10.1158/2159-8290.CD-20-1638. [DOI] [PubMed] [Google Scholar]
- 12.Xu T, et al. Dysregulated tumor-associated macrophages in carcinogenesis, progression and targeted therapy of gynecological and breast cancers. J Hematol Oncol. 2021;14(1):181. doi: 10.1186/s13045-021-01198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y, et al. The key to immunotherapy: how to choose better therapeutic biomarkers for patients with non-small cell lung cancer. Biomark Res. 2022;10(1):9. doi: 10.1186/s40364-022-00355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong S, Dong L, Cheng L. Neutrophils in cancer carcinogenesis and metastasis. J Hematol Oncol. 2021;14(1):173. doi: 10.1186/s13045-021-01187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nejman D, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang S, et al. Mycoplasma infection transforms normal lung cells and induces bone morphogenetic protein 2 expression by post-transcriptional mechanisms. J Cell Biochem. 2008;104(2):580–594. doi: 10.1002/jcb.21647. [DOI] [PubMed] [Google Scholar]
- 17.Zhang SL, et al. Pectin supplement significantly enhanced the anti-PD-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics. 2021;11(9):4155–4170. doi: 10.7150/thno.54476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoption Cann SA, van Netten JP, van Netten C. Dr William Coley and tumour regression: a place in history or in the future. Postgrad Med J. 2003;79(938):672–80. [PMC free article] [PubMed] [Google Scholar]
- 19.Starnes CO. Coley’s toxins in perspective. Nature. 1992;357(6373):11–2. [DOI] [PubMed]
- 20.Livingston-Wheeler therapy. CA Cancer J Clin, 1990. 40(2): p. 103–8. [DOI] [PubMed]
- 21.Matson V, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gut Bacteria Shape Therapeutic Response Cancer Discov. 2018;8(2):134. doi: 10.1158/2159-8290.CD-ND2018-001. [DOI] [PubMed] [Google Scholar]
- 23.Jin Y, et al. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J Thorac Oncol. 2019;14(8):1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, et al. Hepatic NOD2 promotes hepatocarcinogenesis via a RIP2-mediated proinflammatory response and a novel nuclear autophagy-mediated DNA damage mechanism. J Hematol Oncol. 2021;14(1):9. doi: 10.1186/s13045-020-01028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai R, et al. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark Res. 2020;8:34. doi: 10.1186/s40364-020-00209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong M, et al. Diosgenin promotes antitumor immunity and PD-1 antibody efficacy against melanoma by regulating intestinal microbiota. Cell Death Dis. 2018;9(10):1039. doi: 10.1038/s41419-018-1099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopalakrishnan V, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutta D, Lim SH. Bidirectional interaction between intestinal microbiome and cancer: opportunities for therapeutic interventions. Biomark Res. 2020;8:31. doi: 10.1186/s40364-020-00211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gut Microbiome Manipulation May Facilitate Immunotherapy Response Cancer Discov. 2021;11(2):221. [Google Scholar]
- 30.Davar D, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371(6529):595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baruch EN, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371(6529):602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 32.Geller LT, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau WF, et al. Oesophageal microbial flora in carcinoma of the oesophagus. Aust N Z J Surg. 1981;51(1):52–55. doi: 10.1111/j.1445-2197.1981.tb05905.x. [DOI] [PubMed] [Google Scholar]
- 34.Dejea CM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickson I. Microbiome promotes pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2018;15(6):328. doi: 10.1038/s41575-018-0013-x. [DOI] [PubMed] [Google Scholar]
- 36.Namiki K, et al. Persistent exposure to Mycoplasma induces malignant transformation of human prostate cells. PLoS ONE. 2009;4(9):e6872. doi: 10.1371/journal.pone.0006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17(1):163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greathouse KL, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19(1):123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu HX, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer. 2018;142(4):769–778. doi: 10.1002/ijc.31098. [DOI] [PubMed] [Google Scholar]
- 40.Tzeng A, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 2021;13(1):60. doi: 10.1186/s13073-021-00874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hieken TJ, et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci Rep. 2016;6:30751. doi: 10.1038/srep30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parhi L, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. 2020;11(1):3259. doi: 10.1038/s41467-020-16967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parida S, et al. A Procarcinogenic Colon Microbe Promotes Breast Tumorigenesis and Metastatic Progression and Concomitantly Activates Notch and β-Catenin Axes. Cancer Discov. 2021;11(5):1138–1157. doi: 10.1158/2159-8290.CD-20-0537. [DOI] [PubMed] [Google Scholar]
- 44.Lv J, et al. Alteration of the esophageal microbiota in Barrett’s esophagus and esophageal adenocarcinoma. World J Gastroenterol. 2019;25(18):2149–61. [DOI] [PMC free article] [PubMed]
- 45.Kaakoush NO, et al. Is Campylobacter to esophageal adenocarcinoma as Helicobacter is to gastric adenocarcinoma? Trends Microbiol. 2015;23(8):455–462. doi: 10.1016/j.tim.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Yamamura K, et al. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res. 2016;22(22):5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 47.Yang W, et al. Tumor-Associated Microbiota in Esophageal Squamous Cell Carcinoma. Front Cell Dev Biol. 2021;9:641270. doi: 10.3389/fcell.2021.641270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D, et al. Characterization of the Esophageal Microbiota and Prediction of the Metabolic Pathways Involved in Esophageal Cancer. Front Cell Infect Microbiol. 2020;10:268. doi: 10.3389/fcimb.2020.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao S, et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer. 2016;11:3. doi: 10.1186/s13027-016-0049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, et al. Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br J Cancer. 2021;124(5):963–974. doi: 10.1038/s41416-020-01198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao D, et al. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-risk region of China. Cancer. 2019;125(22):3993–4002. doi: 10.1002/cncr.32403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buti L, et al. Helicobacter pylori cytotoxin-associated gene A (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc Natl Acad Sci U S A. 2011;108(22):9238–9243. doi: 10.1073/pnas.1106200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatakeyama M. Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(4):196–219. doi: 10.2183/pjab.93.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1(8336):1273–1275. [PubMed] [Google Scholar]
- 55.Baik SC, et al. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996;56(6):1279–1282. [PubMed] [Google Scholar]
- 56.Li WT, et al. The Bladder Microbiome Is Associated with Epithelial-Mesenchymal Transition in Muscle Invasive Urothelial Bladder Carcinoma. Cancers (Basel) 2021;13(15):3649. doi: 10.3390/cancers13153649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cavarretta I, et al. The Microbiome of the Prostate Tumor Microenvironment. Eur Urol. 2017;72(4):625–631. doi: 10.1016/j.eururo.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 58.Feng Y, et al. Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. BMC Genomics. 2019;20(1):146. doi: 10.1186/s12864-019-5457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen RJ, et al. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? J Urol. 2005;173(6):1969–1974. doi: 10.1097/01.ju.0000158161.15277.78. [DOI] [PubMed] [Google Scholar]
- 60.Banerjee S, et al. Microbiome signatures in prostate cancer. Carcinogenesis. 2019;40(6):749–764. doi: 10.1093/carcin/bgz008. [DOI] [PubMed] [Google Scholar]
- 61.Pushalkar S, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018;8(4):403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riquelme E, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178(4):795–806.e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang S, et al. Fusobacterium nucleatum promotes epithelial-mesenchymal transiton through regulation of the lncRNA MIR4435-2HG/miR-296-5p/Akt2/SNAI1 signaling pathway. Febs j. 2020;287(18):4032–4047. doi: 10.1111/febs.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt BL, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE. 2014;9(6):e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pushalkar S, et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12:144. doi: 10.1186/1471-2180-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng DW, et al. Biomaterial-mediated modulation of oral microbiota synergizes with PD-1 blockade in mice with oral squamous cell carcinoma. Nat Biomed Eng. 2021;6(1):32–43. [DOI] [PubMed]
- 67.Bundgaard-Nielsen C, et al. The presence of bacteria varies between colorectal adenocarcinomas, precursor lesions and non-malignant tissue. BMC Cancer. 2019;19(1):399. doi: 10.1186/s12885-019-5571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castellarin M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kostic AD, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bullman S, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mima K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1(5):653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gur C, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou D, et al. Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: a meta-analysis. Eur J Gastroenterol Hepatol. 2013;25(4):447–54. doi: 10.1097/MEG.0b013e32835c0362. [DOI] [PubMed] [Google Scholar]
- 75.Tsuchiya Y, et al. Metagenomics of Microbial Communities in Gallbladder Bile from Patients with Gallbladder Cancer or Cholelithiasis. Asian Pac J Cancer Prev. 2018;19(4):961–967. doi: 10.22034/APJCP.2018.19.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Avilés-Jiménez F, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infect. 2016;22(2):178.e11–178.e22. doi: 10.1016/j.cmi.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 77.Segura-López FK, et al. Infection with Helicobacter bilis but not Helicobacter hepaticus was Associated with Extrahepatic Cholangiocarcinoma. Helicobacter. 2015;20(3):223–230. doi: 10.1111/hel.12195. [DOI] [PubMed] [Google Scholar]
- 78.Chng KR, et al. Tissue Microbiome Profiling Identifies an Enrichment of Specific Enteric Bacteria in Opisthorchis viverrini Associated Cholangiocarcinoma. EBioMedicine. 2016;8:195–202. doi: 10.1016/j.ebiom.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rocha M, et al. Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut. 2005;54(3):396–401. doi: 10.1136/gut.2004.042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Łaniewski P, et al. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep. 2018;8(1):7593. doi: 10.1038/s41598-018-25879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Audirac-Chalifour A, et al. Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PLoS ONE. 2016;11(4):e0153274. doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Norenhag J, et al. The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG. 2020;127(2):171–180. doi: 10.1111/1471-0528.15854. [DOI] [PubMed] [Google Scholar]
- 83.Walther-António MR, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016;8(1):122. doi: 10.1186/s13073-016-0368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan PJ, et al. Prevalence of mycoplasma conserved DNA in malignant ovarian cancer detected using sensitive PCR-ELISA. Gynecol Oncol. 1996;63(2):258–260. doi: 10.1006/gyno.1996.0316. [DOI] [PubMed] [Google Scholar]
- 85.Zhou B, et al. The biodiversity Composition of Microbiome in Ovarian Carcinoma Patients. Sci Rep. 2019;9(1):1691. doi: 10.1038/s41598-018-38031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. 2017;9(1):14. doi: 10.1186/s13073-017-0405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garrett WS. The gut microbiota and colon cancer. Science. 2019;364(6446):1133–1135. doi: 10.1126/science.aaw2367. [DOI] [PubMed] [Google Scholar]
- 88.Goodwin AC, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(37):15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu T, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170(3):548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang S, et al. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):14. doi: 10.1186/s13046-018-0985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Łaniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. 2020;17(4):232–250. doi: 10.1038/s41585-020-0286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nené NR, et al. Association between the cervicovaginal microbiome, BRCA1 mutation status, and risk of ovarian cancer: a case-control study. Lancet Oncol. 2019;20(8):1171–1182. doi: 10.1016/S1470-2045(19)30340-7. [DOI] [PubMed] [Google Scholar]
- 93.Yan X, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5(10):3111–3122. [PMC free article] [PubMed] [Google Scholar]
- 94.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Howell LM, Forbes NS. Bacteria-based immune therapies for cancer treatment. Semin Cancer Biol. 2021;S1044-579X(21)00231–5. [DOI] [PubMed]
- 96.Aykut B, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cimolai N. Do mycoplasmas cause human cancer? Can J Microbiol. 2001;47(8):691–697. [PubMed] [Google Scholar]
- 98.Huang S, et al. Mycoplasma infections and different human carcinomas. World J Gastroenterol. 2001;7(2):266–269. doi: 10.3748/wjg.v7.i2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yacoub E, et al. The Relationship between Mycoplasmas and Cancer: Is It Fact or Fiction ? Narrative Review and Update on the Situation. J Oncol. 2021;2021:9986550. [DOI] [PMC free article] [PubMed]
- 100.Klein C, et al. Mycoplasma Co-Infection Is Associated with Cervical Cancer Risk. Cancers (Basel) 2020;12(5):1093. doi: 10.3390/cancers12051093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pehlivan M, et al. Can mycoplasma-mediated oncogenesis be responsible for formation of conventional renal cell carcinoma? Urology. 2005;65(2):411–414. doi: 10.1016/j.urology.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 102.Baghdadi J, et al. Microbiome, innate immunity, and esophageal adenocarcinoma. Clin Lab Med. 2014;34(4):721–732. doi: 10.1016/j.cll.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pehlivan M, et al. Does Mycoplasma sp. play role in small cell lung cancer? Lung Cancer. 2004;45(1):129–30. doi: 10.1016/j.lungcan.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 104.Zhang S, et al. Mycoplasma fermentans infection promotes immortalization of human peripheral blood mononuclear cells in culture. Blood. 2004;104(13):4252–4259. doi: 10.1182/blood-2004-04-1245. [DOI] [PubMed] [Google Scholar]
- 105.Polianskaia GG, Efremova TN, Ender NA. Effect of mycoplasma contamination of the human uterine leiomyosarcoma cell line SK-UT-1B on karyotype structure. Tsitologiia. 1998;40(1):23–30. [PubMed] [Google Scholar]
- 106.Ji Y, Karbaschi M, Cooke MS. Mycoplasma infection of cultured cells induces oxidative stress and attenuates cellular base excision repair activity. Mutat Res Genet Toxicol Environ Mutagen. 2019;845:403054. doi: 10.1016/j.mrgentox.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dambuza IM, Brown GD. Fungi accelerate pancreatic cancer. Nature. 2019;574(7777):184–185. doi: 10.1038/d41586-019-02892-y. [DOI] [PubMed] [Google Scholar]
- 108.Sanjaya PR, et al. Candida in oral pre-cancer and oral cancer. Med Hypotheses. 2011;77(6):1125–1128. doi: 10.1016/j.mehy.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 109.Ramirez-Garcia A, et al. Candida albicans increases tumor cell adhesion to endothelial cells in vitro: intraspecific differences and importance of the mannose receptor. PLoS ONE. 2013;8(1):e53584. doi: 10.1371/journal.pone.0053584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cai HZ, et al. Preliminary assessment of viral metagenome from cancer tissue and blood from patients with lung adenocarcinoma. J Med Virol. 2021;93(8):5126–5133. doi: 10.1002/jmv.26887. [DOI] [PubMed] [Google Scholar]
- 111.Mollerup S, et al. High-Throughput Sequencing-Based Investigation of Viruses in Human Cancers by Multienrichment Approach. J Infect Dis. 2019;220(8):1312–1324. doi: 10.1093/infdis/jiz318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.An J, et al. Comprehensive characterization of viral integrations and genomic aberrations in HBV-infected intrahepatic cholangiocarcinomas. Hepatology. 2021;75(4):997–1011. [DOI] [PubMed]
- 113.Enokida T, Moreira A, Bhardwaj N. Vaccines for immunoprevention of cancer. J Clin Invest. 2021;131(9):e146956. doi: 10.1172/JCI146956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.El-Zimaity H, et al. Risk factors for esophageal cancer: emphasis on infectious agents. Ann N Y Acad Sci. 2018;1434(1):319–332. doi: 10.1111/nyas.13858. [DOI] [PubMed] [Google Scholar]
- 115.Odenbreit S, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287(5457):1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 116.Cantalupo PG, Katz JP, Pipas JM. Viral sequences in human cancer. Virology. 2018;513:208–216. doi: 10.1016/j.virol.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nakatsu G, et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology. 2018;155(2):529–541.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 118.Estemalik J, et al. Simultaneous Detection of Oral Pathogens in Subgingival Plaque and Prostatic Fluid of Men With Periodontal and Prostatic Diseases. J Periodontol. 2017;88(9):823–829. doi: 10.1902/jop.2017.160477. [DOI] [PubMed] [Google Scholar]
- 119.Arthur JC, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5:4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kipanyula MJ, et al. Signaling pathways bridging microbial-triggered inflammation and cancer. Cell Signal. 2013;25(2):403–416. doi: 10.1016/j.cellsig.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 121.Bugter JM, Fenderico N, Maurice MM. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat Rev Cancer. 2021;21(1):5–21. doi: 10.1038/s41568-020-00307-z. [DOI] [PubMed] [Google Scholar]
- 122.Daulat AM, Borg JP. Wnt/Planar Cell Polarity Signaling: New Opportunities for Cancer Treatment. Trends Cancer. 2017;3(2):113–125. doi: 10.1016/j.trecan.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 123.He J, et al. Inhibition of SALL4 reduces tumorigenicity involving epithelial-mesenchymal transition via Wnt/β-catenin pathway in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2016;35(1):98. doi: 10.1186/s13046-016-0378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abreu MT, Peek RM., Jr Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146(6):1534–1546.e3. doi: 10.1053/j.gastro.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bagheri N, Salimzadeh L, Shirzad H. The role of T helper 1-cell response in Helicobacter pylori-infection. Microb Pathog. 2018;123:1–8. doi: 10.1016/j.micpath.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 127.Han YW, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68(6):3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Engevik MA, et al. Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. mBio. 2021;12(2):e02706. doi: 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jin C, et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell. 2019;176(5):998–1013.e16. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang Q, et al. Gut Microbiome Directs Hepatocytes to Recruit MDSCs and Promote Cholangiocarcinoma. Cancer Discov. 2021;11(5):1248–1267. doi: 10.1158/2159-8290.CD-20-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang Y, et al. Role of the microbiome in occurrence, development and treatment of pancreatic cancer. Mol Cancer. 2019;18(1):173. doi: 10.1186/s12943-019-1103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10(11):785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Safarzadeh E, et al. STAT3 Silencing and TLR7/8 Pathway Activation Repolarize and Suppress Myeloid-Derived Suppressor Cells From Breast Cancer Patients. Front Immunol. 2020;11:613215. doi: 10.3389/fimmu.2020.613215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Y, et al. IL-2 regulates tumor-reactive CD8(+) T cell exhaustion by activating the aryl hydrocarbon receptor. Nat Immunol. 2021;22(3):358–369. doi: 10.1038/s41590-020-00850-9. [DOI] [PubMed] [Google Scholar]
- 135.Campbell C, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581(7809):475–479. doi: 10.1038/s41586-020-2193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lam KC, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184(21):5338–5356.e21. doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Putra RD, Lyrawati D. Interactions between Bacteriophages and Eukaryotic Cells. Scientifica (Cairo) 2020;2020:3589316. doi: 10.1155/2020/3589316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tanoue T, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565(7741):600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 139.Poore GD, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Charlson ES, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186(6):536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Markowski MC, et al. The Microbiome and Genitourinary Cancer: A Collaborative Review. Eur Urol. 2019;75(4):637–646. doi: 10.1016/j.eururo.2018.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gopalakrishnan V, et al. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fritz S, et al. Bacterial translocation and infected pancreatic necrosis in acute necrotizing pancreatitis derives from small bowel rather than from colon. Am J Surg. 2010;200(1):111–117. doi: 10.1016/j.amjsurg.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 144.Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control. 2012;23(3):399–404. doi: 10.1007/s10552-011-9892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Eliot MN, et al. Periodontal disease and mouthwash use are risk factors for head and neck squamous cell carcinoma. Cancer Causes Control. 2013;24(7):1315–1322. doi: 10.1007/s10552-013-0209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zeng XT, et al. Periodontal Disease and Incident Lung Cancer Risk: A Meta-Analysis of Cohort Studies. J Periodontol. 2016;87(10):1158–1164. doi: 10.1902/jop.2016.150597. [DOI] [PubMed] [Google Scholar]
- 147.Koliarakis I, et al. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int J Mol Sci. 2019;20(17):4146. doi: 10.3390/ijms20174146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Finlay IG, et al. Microbial flora in carcinoma of oesophagus. Thorax. 1982;37(3):181–184. doi: 10.1136/thx.37.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Norder Grusell E, et al. The cultivable bacterial flora of the esophagus in subjects with esophagitis. Scand J Gastroenterol. 2018;53(6):650–656. doi: 10.1080/00365521.2018.1457712. [DOI] [PubMed] [Google Scholar]
- 150.Norder Grusell E, et al. Bacterial flora of the human oral cavity, and the upper and lower esophagus. Dis Esophagus. 2013;26(1):84–90. doi: 10.1111/j.1442-2050.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 151.Öğrendik M. Oral bacteria in pancreatic cancer: mutagenesis of the p53 tumour suppressor gene. Int J Clin Exp Pathol. 2015;8(9):11835–11836. [PMC free article] [PubMed] [Google Scholar]
- 152.Michaud DS, Izard J. Microbiota, oral microbiome, and pancreatic cancer. Cancer J. 2014;20(3):203–206. doi: 10.1097/PPO.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dizdar O, et al. Increased cancer risk in patients with periodontitis. Curr Med Res Opin. 2017;33(12):2195–2200. doi: 10.1080/03007995.2017.1354829. [DOI] [PubMed] [Google Scholar]
- 154.Bourgeois D, et al. Periodontal Pathogens as Risk Factors of Cardiovascular Diseases, Diabetes, Rheumatoid Arthritis, Cancer, and Chronic Obstructive Pulmonary Disease-Is There Cause for Consideration? Microorg. 2019;7(10):424. doi: 10.3390/microorganisms7100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang Y, et al. Crosstalk between autophagy and microbiota in cancer progression. Mol Cancer. 2021;20(1):163. doi: 10.1186/s12943-021-01461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fernández MF, et al. Breast Cancer and Its Relationship with the Microbiota. Int J Environ Res Public Health. 2018;15(8):1747. doi: 10.3390/ijerph15081747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thyagarajan S, et al. Comparative analysis of racial differences in breast tumor microbiome. Sci Rep. 2020;10(1):14116. doi: 10.1038/s41598-020-71102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lu H, et al. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J Oral Microbiol. 2019;11(1):1563409. doi: 10.1080/20002297.2018.1563409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Peters BA, et al. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res. 2017;77(23):6777–6787. doi: 10.1158/0008-5472.CAN-17-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dohlman AB, et al. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe. 2021;29(2):281–298.e5. doi: 10.1016/j.chom.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bagchi S, Yuan R, Engleman EG. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu Rev Pathol. 2021;16:223–249. doi: 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 162.Cheng SS, et al. The design and development of covalent protein-protein interaction inhibitors for cancer treatment. J Hematol Oncol. 2020;13(1):26. doi: 10.1186/s13045-020-00850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen R, et al. Emerging therapeutic agents for advanced non-small cell lung cancer. J Hematol Oncol. 2020;13(1):58. doi: 10.1186/s13045-020-00881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou S, et al. The landscape of bispecific T cell engager in cancer treatment. Biomark Res. 2021;9(1):38. doi: 10.1186/s40364-021-00294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu M, et al. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J Hematol Oncol. 2022;15(1):24. doi: 10.1186/s13045-022-01242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krasniqi E, et al. MicroRNA-based signatures impacting clinical course and biology of ovarian cancer: a miRNOmics study. Biomark Res. 2021;9(1):57. doi: 10.1186/s40364-021-00289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.He Y, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. 2021;33(5):988–1000.e7. doi: 10.1016/j.cmet.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 168.Yi M, et al. Gut microbiome modulates efficacy of immune checkpoint inhibitors. J Hematol Oncol. 2018;11(1):47. doi: 10.1186/s13045-018-0592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang R, et al. Gut microbiome, liver immunology, and liver diseases. Cell Mol Immunol. 2021;18(1):4–17. doi: 10.1038/s41423-020-00592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Luo X, et al. Antitumor effect of VNP20009, an attenuated Salmonella, in murine tumor models. Oncol Res. 2001;12(11–12):501–508. doi: 10.3727/096504001108747512. [DOI] [PubMed] [Google Scholar]
- 171.Le DT, et al. Safety and survival with GVAX pancreas prime and Listeria Monocytogenes-expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J Clin Oncol. 2015;33(12):1325–1333. doi: 10.1200/JCO.2014.57.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Quispe-Tintaya W, et al. Nontoxic radioactive Listeria(at) is a highly effective therapy against metastatic pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110(21):8668–8673. doi: 10.1073/pnas.1211287110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Heimann DM, Rosenberg SA. Continuous intravenous administration of live genetically modified salmonella typhimurium in patients with metastatic melanoma. J Immunother. 2003;26(2):179–180. doi: 10.1097/00002371-200303000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nemunaitis J, et al. Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer Gene Ther. 2003;10(10):737–44. [DOI] [PubMed]
- 175.Toso JF, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20(1):142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ozdemir T, et al. Synthetic Biology and Engineered Live Biotherapeutics: Toward Increasing System Complexity. Cell Syst. 2018;7(1):5–16. doi: 10.1016/j.cels.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 177.Thamm DH, et al. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clin Cancer Res. 2005;11(13):4827–4834. doi: 10.1158/1078-0432.CCR-04-2510. [DOI] [PubMed] [Google Scholar]
- 178.Dai Y, et al. Construction of an inducible cell-communication system that amplifies Salmonella gene expression in tumor tissue. Biotechnol Bioeng. 2013;110(6):1769–1781. doi: 10.1002/bit.24816. [DOI] [PubMed] [Google Scholar]
- 179.Felgner S, et al. aroA-Deficient Salmonella enterica Serovar Typhimurium Is More Than a Metabolically Attenuated Mutant. mBio. 2016;7(5):e01220. doi: 10.1128/mBio.01220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Manuel ER, et al. Enhancement of cancer vaccine therapy by systemic delivery of a tumor-targeting Salmonella-based STAT3 shRNA suppresses the growth of established melanoma tumors. Cancer Res. 2011;71(12):4183–4191. doi: 10.1158/0008-5472.CAN-10-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Montalban-Arques A, et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe. 2021;29(10):1573–1588.e7. doi: 10.1016/j.chom.2021.08.001. [DOI] [PubMed] [Google Scholar]
- 182.Pinato DJ, et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients With Cancer. JAMA Oncol. 2019;5(12):1774–1778. doi: 10.1001/jamaoncol.2019.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Derosa L, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29(6):1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Y, et al. Prebiotic-Induced Anti-tumor Immunity Attenuates Tumor Growth. Cell Rep. 2020;30(6):1753–1766.e6. doi: 10.1016/j.celrep.2020.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no data-sets were generated or analyzed during the current study.