Abstract
The microbiome, which refers to the microbiota within a host and their collective genomes, has recently been demonstrated to play a critical role in cancer progression, metastasis, and therapeutic response. The microbiome is known to affect host immunity, but its influence on human papilloma virus (HPV) gynecologic malignancies remains limited and poorly understood. To date, studies have largely focused on the cervicovaginal microbiome; however, there is growing evidence that the gut microbiome may interact and substantially affect therapeutic response in gynecologic cancers. Importantly, new developments in microbiome sequencing and advanced bioinformatics technologies have enabled rapid advances in our understanding of the gut and local tumor microbiota. In this review, we examine the evidence supporting the role of the microbiome in HPV-associated cervical intraepithelial neoplasia (CIN) and cervical cancer, explore characteristics that influence and shape the host microbiota that impact HPV-driven carcinogenesis, and highlight potential approaches and considerations for future and ongoing research of the microbiome's effect on HPV-associated cancer.
Keywords: cervical dysplasia, cervical cancer, gynecologic cancer, cervical microbiota, cervicovaginal microbiota, microbiome, gut microbiome, human papillomavirus (HPV)
INTRODUCTION
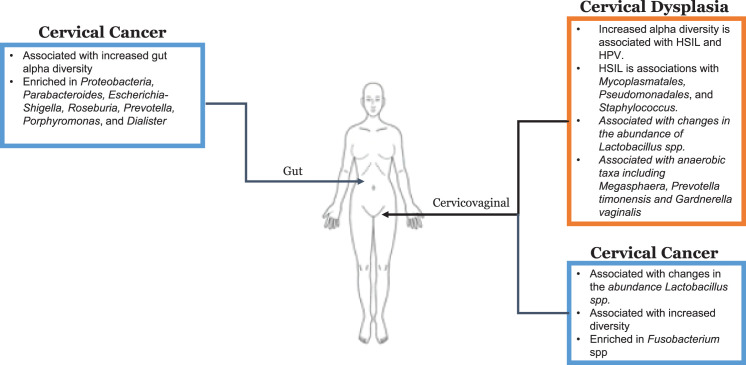
Persistent exposure to human papilloma virus (HPV) is the well-established antecedent to many gynecologic cancers.[1,2] HPV is considered to be responsible for 99.7% of cervical cancers, up to 70% of squamous cell carcinomas of the vulva, and 60% of squamous cell carcinomas of the vagina.[3] Additionally, there appears to be a high prevalence of HPV DNA in ovarian cancer cases, although the precise role of HPV in ovarian cancer remains unknown.[3–5] Because most microbiome studies exploring HPV-related gynecologic cancers have reported on cervical dysplasia and cervical cancer, these will be the main focus of this review (Fig. 1).
Figure 1.
The gut and cervicovaginal microbiome of cervical cancer and cervical dysplasia. CIN: cervical intraepithelial neoplasia; LSIL: low grade squamous intraepithelial lesions; HSIL: high grade squamous intraepith elial lesions; spp: species; HPV: human papilloma virus.
HPV 16 and 18 are responsible for most HPV-related gynecologic cancers.[3] It is thought that host-dependent immunologic status and HPV-induced immune evasion are responsible for a persistent HPV infection and thus a predisposition to cancer.[5] However, a persistent HPV infection alone is inadequate for cancer formation. Factors unique to the individual mucosal sites such as epithelial surface integrity, immune regulation, and the local microbiota play an important role in HPV carcinogenesis.[6–8] Additionally, HPV carcinogenesis is contingent on the patient's immune system's ability to recognize tumor antigens and clear an oncogenic HPV infection.[9] High-risk HPV downregulates interferon signaling, which favors HPV persistence and the development of precursor lesions.[9,10] HPV integration into the host cellular genome permits permanent expression of viral oncoproteins, stimulating cell transformation into cancerous cells. HPV infects mature mucocutaneous epithelial cells, producing viral particles that initiate the production of oncoproteins E6 and E7. These oncoproteins cause a disruption in normal cell-cycle control by interfering with cell death and promoting continued cell proliferation and uncontrolled cell division—ultimately leading to genetic damage.[3,11] Given that host-dependent immunologic status and HPV-induced immune evasion are responsible for a persistent HPV infection, the gut and local tumor microbiome has drawn attention. The microbiome has been purported to modify host immunity by regulating several immunologic pathways, thereby affecting patients' risk of various cancers and their treatment outcomes.[12,13] A more diverse, abundant gut microbiota, with a distinct composition, is believed to enhance the immune response by priming the activation of antitumor T cells.[14] In this review, we assess the evidence for the role of the gut and tumor microbiota in HPV gynecologic cancers and highlight potential approaches and considerations for future and ongoing research into the relationship between the microbiome and HPV-related cancer.
METHODS
A systematic literature search was conducted in PubMed to find relevant studies published up to December 1, 2019. The following combination of search keywords was used: [vaginal (OR) cervical (OR) cervicovaginal (OR) gut (OR) gastrointestinal] (AND) [dysplasia (OR) cancer (OR) neoplasia (OR) carcinoma (OR) malignancy (OR) CIN] (AND) [microbiome (OR) microbiota]. Inclusion criteria were used to eliminate inappropriate publications and to identify studies to be reviewed. The inclusion criteria for accepting articles included the use of those that used gram staining, 16S ribosomal RNA (rRNA) amplicon sequencing, whole-genome shotgun sequencing, or metagenomic sequencing. With application of inclusion criteria, and after examining titles and abstracts of all identified articles, our search yielded nine studies with potential relevance for the study topic and for inclusion in this review.
Cervicovaginal Microbiome Factors in Cervical Intraepithelial Neoplasia (CIN)
A state of cervicovaginal polybacterial dysbiosis and chronic local inflammation encourages the perseverance of HPV,[15] which ultimately promotes the development of cervical dysplasia and carcinogenesis.[16–21] As shown in the Table 1, the role of the cervicovaginal microbiome in HPV-driven diseases has been broadly studied in CIN and cervical cancer. Persistent HPV infections are thought to trigger an innate immune response, resulting in the suppression of infected cervicovaginal mucosal cells.[22–24] Cervicovaginal dysbiotic states lead to an altered metabolic profile and reduced cervicovaginal barrier function. This dysbiotic state is associated with an increased acquisition of high-risk HPV, cervical dysplasia, and ultimately, cervical cancer.[23,25] One of the first studies to report this phenomena characterized the CIN microbiota using laboratory culture. This study reported on the presence of “abnormal vaginal flora,” which future studies would later corroborate with 16S rRNA analysis.[26] This focus on exploring the relative abundance of bacteria in the cervicovaginal epithelium eventually led to the concept and assignment of community-state types (CSTs) based on the richness of Lactobacilli species.[13–15] The depletion of specific Lactobacilli species—for example Lactobacillus crispatus, Lactobacillus gasseri, or Lactobacillus jensenii—has been associated with a predisposition toward bacterial vaginosis and other proinflammatory states, consequently leading to DNA cell damage and potentially carcinogenic changes.[22,27–32] The most unique CSTs found in women with CIN were CSTs characterized by Lactobacillus depletion, anaerobic bacteria predominance, and Lactobacillus iners dominance.[29] An increase in vaginal microbiome diversity and richness is associated with increased acquisition of HPV infection and persistence as well as higher CIN severity.[33–35] Additionally, multiple studies reported the bacteria Sneathia to be significantly enriched in CIN samples, although our understanding of its pathogenic or protective role remains somewhat lacking.[36,37]
Table 1.
Summary of articles discussing the microbiome and HPV gynecologic cancer
|
Author
|
Participants
|
Microbial Analysis
|
Key Bacterial Organisms
|
Comments
|
| Studies on the Cervicovaginal Microbiome and CIN | ||||
| Guijon et al26 | CIN positive (n = 106) Control (n = 79) | Culture, gram staining | Mycoplasma hominis | HPV, abnormal vaginal flora, and M. hominis, significantly associated with CIN. |
| Piyathilake et al34 | CIN3 (n = 132) CIN2 (n = 208) CIN1 (n = 90) | V4 16S rRNA | Lactobacillus iners | α-and β-diversity was not significantly associated with disease status. L. iners abundance was associated with increased disease severity. |
| Klein et al35 | LSIL (n = 72) HSIL (n = 50) Control (n = 23) | V4 16S rRNA | Mycoplasmatales, Pseudomonadales, Fusobacteria, Staphylococcus | Increased α-diversity was associated with HSIL and HPV. Brush samples from HSIL patients revealed unique associations with Mycoplasmatales, Pseudomonadales, and Staphylococcus. |
| Studies on the Cervicovaginal Microbiome, CIN, and Cervical Cancer | ||||
| Mitra et al33 | LSIL (n = 52) HSIL (n = 92) CC (n = 5) Control (n = 20) | V1-V2 16S rRNA | Lactobacillus spp., Lactobacillus crispatus, Sneathia sanguinegens | Lactobacillus depletion, high diversity and species richness was associated with increasing disease severity and high-risk HPV positivity. |
| Mitra et al32 | CIN2 (n = 87) | V1-V2 16S rRNA | Lactobacillus spp., Megasphaera, Prevotella timonensis, Gardnerella vaginalis | Lactobacillus-dominant microbiome at baseline is more likely to have regression of CIN2 at 12 months. Lactobacillus spp. depletion and presence of specific anaerobic taxa including Megasphaera, Prevotella timonensis, and Gardnerella vaginalis are associated with CIN2 persistence and slower regression. |
| Audirac-Chalifour et al37 | HPV− control (n = 10) HPV+ control (n = 10) SIL HPV+ (n = 4) CC HPV+ (n = 8) | V3-V4 16S rRNA | Lactobacillus iners, Sneathia spp., Fusobacterium spp. | Increased α-diversity in CC and SIL with unique β-diversities at every stage of CC. All four study groups were dominated by a single distinct population of bacteria: L. crispatus, L. iners, Sneathia spp., and Fusobacterium spp. were dominant in HPV-negative samples, HPV-positive samples, SIL samples, and CC samples, respectively. |
| Łaniewski et al36 | Control (n = 51) LSIL (n = 12) HSIL (n = 27) CC (n = 10) | V4 16S rRNA | Lactobacillus spp., Sneathia spp. | Decreased abundance of Lactobacillus spp. and increased microbiome diversity was associated with increasing severity of cervical neoplasm and CC. |
| Kwon et al45 | CIN (n = 17) CC (n = 12) Control (n = 18) | Whole-genome sequencing | Alkaliphilus, Pseudothermotoga, Wolbachia, Lactobacillus, Staphylococcus, Candidatus Endolissoclinum | Diversity was not significantly associated with disease status. CC and CIN were each significantly enriched with bacteria unique to the other disease status. |
| Studies on the Gut (Fecal) Microbiome and Cervical Cancer | ||||
| Wang et al50 | ICC (n = 8) Control (n = 5) | V4 16S rRNA | Proteobacteria, Parabacteroides, Escherichia-Shigella, Roseburia | Increased α-diversity (NS) and differing β-diversity of gut microbiome in CC versus control. Seven genera differentiated significantly in relative abundance between CC and controls. |
| Sims et al51 | ICC (n = 42) Control (n = 46) | V4 16S rRNA | Prevotella, Porphyromonas, Dialister | Increased α-diversity and differing β-diversity in CC versus control. CC patients exhibited significantly enriched Prevotella, Porphyromonas, and Dialister when compared to age, race, and BMI-matched controls. |
CC: cervical cancer; HSIL: high-grade squamous intraepithelial lesion; ICC: invasive cervical cancer; LSIL: low-grade squamous intraepithelial lesion; SIL: squamous intraepithelial lesion.
The most common type of cervicovaginal dysbiosis (defined as a cervicovaginal microbiome not dominated by Lactobacilli) is bacterial vaginosis.[38] Bacterial vaginosis is characterized by a persistent decrease in Lactobacilli and an increase in fastidious anaerobes.[23] Lactobacillus species seem to serve largely as protective organisms, with some exceptions. L. iners is the most commonly reported Lactobacillus-dominated CST detected in women diagnosed with CIN.[32] The role of this bacteria in cervicovaginal health is uncertain, since it can be identified in normal conditions as well as in states of vaginal dysbiosis, such as bacterial vaginosis.[39] L. iners is believed to have more complex nutritional requirements and variable morphology than do other Lactobacillus species. Additionally, in contrast to other Lactobacilli, L. iners has an unusually small genome, suggestive of a symbiotic or parasitic lifestyle.[39,40] These characteristics have led some to suggest that L. iners may have clonal variants that in some cases promote health and in other cases are associated with dysbiosis and a predisposition to disease.[39]
Conversely, L. crispatus dominance has been strongly associated with a healthy cervicovaginal microbiome. In African women, Borgdorff et al.[41] found an L. crispatus–dominated vaginal microbiota to be associated with a lower prevalence of HPV, HIV, HSV-2, and bacterial sexually transmitted infections (Neisseria gonorrhoeae, Chlamydia trachomatis, and Trichomonas vaginalis). Interestingly, Gajer et al.[42] found that vaginal communities dominated by L. crispatus can convert to a community state dominated by L. iners or to a community state characterized by a small abundance of either L. crispatus or L. iners.[39] This finding suggests that Lactobacilli CSTs are likely to transition to CSTs characteristic of CIN patients. This likely depends largely on a particular Lactobacilli species' ability to adapt to a variety of pH environments. This association between bacterial vaginosis and CIN has been extensively studied.[43] Studies suggest that a decreased abundance of lactic acid–producing Lactobacilli resulting in abnormally high vaginal pH (> 4.5) can cause bacterial overgrowth and a decrease in protective flora, thereby resulting in weakened defense mechanisms to fend off viral infections such as HPV.[36] Additionally, bacterial vaginosis is associated with increased levels of proinflammatory cytokines such as IL-1β and decreased levels of the anti-inflammatory molecule SLPI (i.e., secretory leukocyte protease inhibitor), supporting the theory that immune changes associated with bacterial vaginosis can result in an increased susceptibility to HPV and the development of high-grade cervical dysplasia.[44]
Cervicovaginal Microbiome Factors in Cervical Cancer
Limited studies of women with cervical cancer have found the cervicovaginal microbiota to have increased overall bacterial diversity, increased predominance of Fusobacterium species, and decreased abundance of Lactobacillus species. As in women with CIN, the cervicovaginal microbiome in cervical cancer patients resembles that of women with bacterial vaginosis.[37] Although found in both CIN and cervical cancer, Fusobacterium predominance was more commonly observed in cervical cancer patients and was shown to be correlated with a cytokine pattern of increased levels of IL-4 and TGF-β1 mRNA, suggesting a local immunosuppression state and supporting the concept of microbiota immune system modification.[37] Another study using shotgun metagenomic sequencing found that the cervical microbiome community compositions and their metagenomics profiles differed between patients with cervical cancer and individuals without cancer, but the researchers cautioned that larger additional whole-genome shotgun studies are required to confirm these associations.[45]
Gut Microbiome Factors in Invasive Cervical Cancer
A review by Chase et al.[46] pointed out a lack of research involving the association between the gut microbiome and gynecologic cancers. Table 1 provides a summary of cervical cancer-related gut microbiome studies. The gut microbiome is proposed to affect host immunity by modifying various immunologic pathways, thus impacting carcinogenesis and treatment outcomes in various malignancies.[47–49]
Two recently published studies have explored the relationship between the gut microbiome and cervical cancer. Wang et al.[50] compared the gut microbiome between eight women with cervical cancer and five healthy women. Women with cervical cancer had a higher gut microbiota α-diversity (the number of distinct species present and whether distinct species are evenly represented), although this difference was not statistically significant. The researchers reported a difference in β-diversity (differences in taxonomic abundance profiles between different samples) between cervical cancer patients and the control group. The researchers also reported significant differences in several taxa between cervical cancer patients and controls, chiefly that members of the phylum Proteobacteria were higher in cervical cancer patients.[50] In a larger analysis of the gut microbiome in cervical cancer patients versus women without cervical cancer, our group observed a statistically significant higher α-diversity in cervical cancer patients than in healthy controls.[51] We also observed a statistically significant difference in β-diversity between women with cervical cancer and women without cervical cancer, confirming compositional differences in the gut microbiota between these two groups. Our ongoing studies suggest that both the cervical and gut microbiomes are associated with treatment response and outcomes in cervical cancers.[52,53]
CONCLUSION AND FUTURE DIRECTIONS
There is growing evidence of the role of the cervicovaginal and gut microbiota in HPV gynecologic malignancies. Future studies are needed to investigate how the microbiome affects the immune response and response to therapy in HPV-related gynecologic cancers and to examine whether manipulation of microbiota can improve response to therapy in preclinical models. Such research will require distinct considerations that vary among different diseases and patient populations. Further elucidation of the underlying degree of cervicovaginal and gut microbiome dysbiosis with current advances in microbiome sequencing technologies might influence how these sites may be amenable to modulation.
Advances in microbiome sequencing technologies and bioinformatics have fueled advances in our understanding of the human microbiome[54,55]; 16S ribosomal RNA (rRNA) amplicon sequencing is the most common method used. This method sequences a specific 30S ribosome subunit of the human microbiome that is unique to prokaryotes and has regions that vary greatly between different species of bacteria and clusters the identified bacteria into operational taxonomic units.[12,56] This technique can be used to quantify diversity metrics such as α-diversity and β-diversity. Additionally, this method can give the relative abundance of specific bacterial taxa and provide genus-level identification.[56] In contrast, whole-genome shotgun sequencing, or metagenomic sequencing, involves deciphering and sequencing broad regions of the entire genomes of all microbes in a given sample.[12,57] Whereas 16S rRNA sequencing has inherent disadvantages, including the inability to provide species-level identification, metagenomic sequencing has the enhanced ability to provide deeper resolution and allows for superior species-level identification, understanding into genome functionality and structure, and the ability to identify all microbes (including viruses, fungi, protozoa, and archaea) in a given sample. Given the substantially higher cost in terms of both time and money associated with metagenomic sequencing, however, future investigations must use whole-genome shotgun sequencing in conjunction with the more cost-effective 16S rRNA method as a complementary approach in order to fully to investigate the role of the human microbiome in HPV carcinogenesis.57 Ultimately, through these new approaches and with validation in preclinical models, the integration of these data will produce actionable strategies geared toward targeting and manipulating the microbiome in order to improve HPV gynecologic cancer therapy.
We are only beginning to understand the microbiome and its role in gynecologic health and disease. Questions still remain regarding how microbial dysbiosis fits in relation to the many known and established risk factors associated with HPV-driven gynecologic malignancies. Current evidence indicates that an impaired microbiota can directly affect immunity by affecting a variety of immunologic pathways. Owing to the vastness of the metagenome, future investigation will focus on mechanistic studies that describe the relationship between the microbiota and the immune response to HPV infection, clearance, and carcinogenesis. We are a long way from completely understanding associations between the microbiota, HVP, and gynecologic cancer, but the human microbiota offers new opportunities to implement strategies that could forever change response to cancer therapy through various mechanisms. The microbiome therefore continues to merit a special focus in order to improve our understanding of its role in HPV-related gynecologic cancer.
Acknowledgments
The funding sources were not involved in the research hypothesis development, study design, data analysis, or manuscript writing. Data access was limited to the authors of this manuscript. The human subjects who participated in any of the referenced studies are gratefully acknowledged.
Funding Statement
Source of Support: This research was supported in part by the National Institutes of Health (NIH) through MD Anderson's Cancer Center Support Grant P30 CA016672 and the NIH T32 grant 5T32 CA101642-14 (Travis T. Sims). This research was partially funded by the MD Anderson HPV-Related Cancers Moonshot (Ann Klopp). Editorial support was provided by Bryan Tutt in Scientific Publications Services, Research Medical Library.
Footnotes
Conflicts of Interest: None.
References
- 1.Hammer A, Rositch A, Qeadan F, Gravitt PE, Blaakaer J. Age-specific prevalence of HPV16/18 genotypes in cervical cancer: A systematic review and meta-analysis. Int J Cancer . 2016;138:2795–2803. doi: 10.1002/ijc.29959. [DOI] [PubMed] [Google Scholar]
- 2.Grover S, Bvochora-Nsingo M, Yeager A, et al. Impact of human immunodeficiency virus infection on survival and acute toxicities from chemoradiation therapy for cervical cancer patients in a limited-resource setting. Int J Radiat Oncol Biol Phys . 2018;101:201–210. doi: 10.1016/j.ijrobp.2018.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chi DS, Berchuck A, Dizon DS, Yashar CM. Principles and Practice of Gynecologic Oncology 7th ed. Philadelphia, PA: Wolters Kluwer; 2017. [Google Scholar]
- 4.Rosa MI, Silva GD, de Azedo Simões PWT, et al. The prevalence of human papillomavirus in ovarian cancer: a systematic review. Int J Gynecol Cancer . 2013;23:437–441. doi: 10.1097/IGC.0b013e318280f3e0. [DOI] [PubMed] [Google Scholar]
- 5.Conesa-Zamora P. Immune responses against virus and tumor in cervical carcinogenesis: Treatment strategies for avoiding the HPV-induced immune escape. Gynecol Oncol . 2013;131:480–488. doi: 10.1016/j.ygyno.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 6.Schiffman M, Doorbar J, Wentzensen N, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers . 2016;2:16086. doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 7.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog . 2009;5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes JV, de Medeiros Fernandes TAA, de Azevedo JCV, et al. Link between chronic inflammation and human papillomavirus-induced carcinogenesis (review) Oncol Lett . 2015;9:1015–1026. doi: 10.3892/ol.2015.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-B-responsive genes in cervical keratinocytes. J Virol . 2001;75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heller C, Weisser T, Mueller-Schickert A, et al. Identification of key amino acid residues that determine the ability of high risk hpv16-e7 to dysregulate major histocompatibility complex class I expression. J Biol Chem . 2011;286:10983–10997. doi: 10.1074/jbc.M110.199190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okunade KS. Human papillomavirus and cervical cancer. J Obstet Gynaecol . 2019]. [published online ahead of print December 11. [DOI] [PubMed]
- 12.McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol . 2019;20:e77–e91. doi: 10.1016/S1470-2045(18)30952-5. [DOI] [PubMed] [Google Scholar]
- 13.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell . 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science . 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao W, Weng J, Gao Y, Chen X. Comparison of the vaginal microbiota diversity of women with and without human papilloma virus infection–a cross-sectional study. BMC Infect Dis . 2013;13:271. doi: 10.1186/1471-2334-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brotman RM, Shardell MD, Gajer P, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis . 2014;210:1723–1733. doi: 10.1093/infdis/jiu330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King CC, Jamieson DJ, Wiener J, et al. Bacterial vaginosis and the natural history of human papillomavirus. Infect Dis Obstet Gynecol . 2011. 2011. [DOI] [PMC free article] [PubMed]
- 18.Marullo R, Werner E, Zhang H, Chen GZ, Shin DM, Doetsch PW. HPV16 E6 and E7 proteins induce a chronic oxidative stress response via NOX2 that causes genomic instability and increased susceptibility to DNA damage in head and neck cancer cells. Carcinogenesis . 2015;36:1397–1406. doi: 10.1093/carcin/bgv126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra A, MacIntyre DA, Lee YS, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep . 2015;5:1–12. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balkwill F, Mantovani A. Inflammation and cancer: Back to Virchow? Lancet . 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 21.Balkwill FR, Mantovani A. Cancer-related inflammation: Common themes and therapeutic opportunities. Semin Cancer Biol . 2012;22:33–40. doi: 10.1016/j.semcancer.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 22.van de Wijgert JHHM, Borgdorff H, Verhelst R, et al. The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS One . 2014;9:e105998. doi: 10.1371/journal.pone.0105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Wijgert JHHM, Gill AC, Chikandiwa A, et al. Human papillomavirus infection and cervical dysplasia in HIV-positive women: potential role of the vaginal microbiota. AIDS . 2020;34:115–125. doi: 10.1097/QAD.0000000000002381. [DOI] [PubMed] [Google Scholar]
- 24.Karim R, Tummers B, Meyers C, et al. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response. PLoS Pathog . 2013;9:e1003384. doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borgdorff H, Gautam R, Armstrong SD, et al. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal Immunol . 2016;9:621–633. doi: 10.1038/mi.2015.86. [DOI] [PubMed] [Google Scholar]
- 26.Guijon F, Paraskevas M, Rand F, Heywood E, Brunham R, McNicol P. Vaginal microbial flora as a cofactor in the pathogenesis of uterine cervical intraepithelial neoplasia. Int J Gynaecol Obstet . 1992;37:185–191. doi: 10.1016/0020-7292(92)90379-w. [DOI] [PubMed] [Google Scholar]
- 27.Kyrgiou M, Mitra A, Moscicki A-B. Does the vaginal microbiota play a role in the development of cervical cancer? Transl Res . 2017;179:168–182. doi: 10.1016/j.trsl.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A . 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brotman RM, Shardell MD, Gajer P, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis . 2014;210:1723–1733. doi: 10.1093/infdis/jiu330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balkwill FR, Capasso M, Hagemann T. The tumor microenvironment at a glance. J Cell Sci . 2012;125:5591–5596. doi: 10.1242/jcs.116392. [DOI] [PubMed] [Google Scholar]
- 31.King CC, Jamieson DJ, Wiener J, et al. Bacterial vaginosis and the natural history of human papillomavirus. Infect Dis Obstet Gynecol . 2011;2011:319460. doi: 10.1155/2011/319460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitra A, MacIntyre DA, Marchesi JR, Lee YS, Bennett PR, Kyrgiou M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: what do we know and where are we going next? Microbiome . 2016;4:58. doi: 10.1186/s40168-016-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitra A, MacIntyre DA, Lee YS, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep . 2015;5:16865. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piyathilake CJ, Ollberding NJ, Kumar R, Macaluso M, Alvarez RD, Morrow CD. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high-risk human papillomaviruses. Cancer Prev Res (Phila) . 2016;9:357–366. doi: 10.1158/1940-6207.CAPR-15-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein C, Gonzalez D, Samwel K, et al. Relationship between the cervical microbiome, HIV status, and precancerous lesions. mBio . 2019;10:e02785–18. doi: 10.1128/mBio.02785-18. Published 2019 Feb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Łaniewski P, Barnes D, Goulder A, et al. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep . 2018;8:7593. doi: 10.1038/s41598-018-25879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PLoS One . 2016;11:e0153274. doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Wijgert JHHM, Jespers V. The global health impact of vaginal dysbiosis. Res Microbiol . 2017;168:859–864. doi: 10.1016/j.resmic.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Petrova MI, Reid G, Vaneechoutte M, Lebeer S. Lactobacillus iners: Friend or foe? Trends Microbiol . 2017;25:182–191. doi: 10.1016/j.tim.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Macklaim JM, Gloor GB, Anukam KC, Cribby S, Reid G. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc Natl Acad Sci U S A . 2011;108:4688–4695. doi: 10.1073/pnas.1000086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borgdorff H, Tsivtsivadze E, Verhelst R, et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J . 2014;8:1781–1793. doi: 10.1038/ismej.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med . 2012;4 doi: 10.1126/scitranslmed.3003605. 132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillet E, Meys JFA, Verstraelen H, et al. Association between bacterial vaginosis and cervical intraepithelial neoplasia: systematic review and meta-analysis. PLoS One . 2012;7:e45201. doi: 10.1371/journal.pone.0045201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell C, Marrazzo J. Bacterial vaginosis and the cervicovaginal immune response. Am J Reprod Immunol . 2014;71:555–563. doi: 10.1111/aji.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon M, Seo S-S, Kim MK, Lee DO, Lim MC. Compositional and functional differences between microbiota and cervical carcinogenesis as identified by shotgun metagenomic sequencing. Cancers . 2019;11:309. doi: 10.3390/cancers11030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chase D, Goulder A, Zenhausern F, Monk B, Herbst-Kralovetz M. The vaginal and gastrointestinal microbiomes in gynecologic cancers: A review of applications in etiology, symptoms and treatment. Gynecol Oncol . 2015;138:190–200. doi: 10.1016/j.ygyno.2015.04.036. [DOI] [PubMed] [Google Scholar]
- 47.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell . 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol . 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol . 2019;20:e77–e91. doi: 10.1016/S1470-2045(18)30952-5. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Wang Q, Zhao J, et al. Altered diversity and composition of the gut microbiome in patients with cervical cancer. AMB Express . 2019;9:40. doi: 10.1186/s13568-019-0763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims TT, Colbert LE, Zheng J, et al. Gut microbial diversity and genus-level differences identified in cervical cancer patients versus healthy controls. Gynecol Oncol . 2019;155:237–244. doi: 10.1016/j.ygyno.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colbert LE, Medrano AYD, Mikkelson MD, et al. Clonal expansion of antigen specific T-cells during radiation therapy for HPV associated cervical cancers is regulated by the vaginal microbiome. Int J Radiat Oncol Biol Phys . 2018;102:S24. [Google Scholar]
- 53.Sims TT, Colbert LE, Karpinets T, et al. Compositional and temporal changes of the gut microbiome in women with cervical cancer undergoing chemoradiation: Does it predict response? Society of Gynecologic Oncology (SGO) Annual Meeting March 2831 2020. https://sgo.confex.com/sgo/2020/meetingapp.cgi/Paper/15081 Accessed May 7, 2020.
- 54.Navas-Molina JA, Peralta-Sánchez JM, González A, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol . 2013;531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature . 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D'Amore R, Ijaz UZ, Schirmer M, et al. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genomics . 2016;17:55. doi: 10.1186/s12864-015-2194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun . 2016;469:967–977. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]