Abstract
Breast tumors have their own specific microbiota, distinct from normal mammary gland tissue. Breast cancer patients that present with locally advanced disease often undergo neoadjuvant chemotherapy to reduce tumor size prior to surgery to allow breast conservation or limit axillary lymph node dissection. The purpose of our study was to evaluate whether neoadjuvant chemotherapy modulates the tumor microbiome and the potential impact of microbes on breast cancer signaling. Using snap-frozen aseptically collected breast tumor tissue from women that underwent neoadjuvant chemotherapy (n=15) or women with no prior therapy at time of surgery (n=18), we performed 16S rRNA sequencing to identify tumoral bacterial populations. We also stained breast tumor microarrays to confirm presence of identified microbiota. Using bacteria-conditioned media, we determined the effect of bacterial metabolites on breast cancer cell proliferation and doxorubicin therapy responsiveness. We show chemotherapy administration significantly increased breast tumor Pseudomonas. Primary breast tumors from patients that developed distant metastases displayed increased tumoral abundance of Brevundimonas and Staphylococcus. We confirmed presence of Pseudomonas in breast tumor tissue by immunohistochemical staining. Treatment of breast cancer cells with Pseudomonas aeruginosa conditioned media differentially effected proliferation in a dose-dependent manner and modulated doxorubicin-mediated cell death. Our results indicate chemotherapy shifts breast tumor microbiome and specific microbes correlate with tumor recurrence. Further studies with a larger patient cohort may provide greater insights into the role of microbiota in therapeutic outcome and develop novel bacterial biomarkers that could predict distant metastases.
Implications:
Breast tumor microbiota are modified by therapy and affects molecular signaling.
Introduction
Previous studies have identified the presence of breast-specific microbiome (1–5). Mammary gland samples obtained from Canadian or Irish women undergoing lumpectomies, mastectomies, or breast reduction surgeries displayed bacterial taxa differences. Canadian women’s breast tissue had a high proportional abundance of Bacillus, Acinetobacter, Enterobacteriaceae, Pseudomonas, Staphylococcus, Propionibacterium, and Prevotella. When compared with Canadian breast samples the mammary tissue from Irish women displayed high Listeria welshimeri, over a 3-fold higher abundance of Enterobacteriaceae, 2-fold higher Staphylococcus, and a 2-fold higher abundance of Propionibacterium. Mammary gland samples from Irish and Canadian women displayed similar (5–6%) abundance of Pseudomonas genus level taxa (1).
Another study investigating the microbiome of normal mammary gland tissue compared with breast tumor adjacent mammary gland tissue, indicated increased relative abundance of Staphylococcus in mammary gland tissue of cancer patients. This study did not control for patient body weight (2). Another study which reported patient BMI, breast tissue was obtained adjacent to either benign or malignant breast tumors and mammary gland tissue was sequenced to identify changes in microbiota. Mammary gland tissue from patients with malignant disease had decreased levels of Lactobacillus, suggesting breast tumorigenesis may modify mammary gland microbiota (6). Recently our group demonstrated the plasticity of the mammary gland microbiome using a dietary approach in a non-human primate model. We showed that monkeys consuming a Mediterranean diet had distinctively different breast microbiota populations than monkeys consuming a Western diet that was modulated independently of the gut microbiome and systemic effects (5).
Breast tumors also display a distinct microbiota population when compared with mammary tissue. Non-cancerous tumor adjacent tissue (n=72) and breast tumor (n=668) TCGA data were mined and non-overlapping sequences were mapped to bacterial genomes (7). Bacterial 16S ribosomal genes were identified from these non-overlapping sequences and prevalent bacterial populations quantified (exclusion criteria included: male breast cancers, neoadjuvant chemotherapy treated, metastatic samples, and history of prior breast cancer). Tumor tissue was enriched with bacteria from Proteobacteria phylum when compared with non-cancerous adjacent tissue. This study also did not investigate the impact of patient body weight on tumor microbial populations. Another study that isolated DNA and performed 16S sequencing from formalin-fixed paraffin-embedded breast tumors (ER+ n=50, HER2+ n=34, and triple negative n=40) and normal tissue from breast reduction surgeries (n=20) indicate elevated Proteobacteria in tumor samples and specific changes in bacteria populations by subtype (8). This study did not have access to de-identified data and could not correlate breast tumor bacterial populations with BMI or relapse-free survival.
Neoadjuvant chemotherapy, usually a combination of anthracycline (Adriamycin or doxorubicin), alkylating agents (cyclophosphamide), and taxanes (Taxotere), are often used in locally advanced breast cancer patients to reduce tumor size and limit lymph node involvement before surgery to enable breast conservative surgery and minimize surgery in the axilla (9, 10). Whether chemotherapy can modulate breast tumor microbiota is unknown. Moreover, whether the tumor bacterial populations impact therapeutic responsiveness or the development of distant metastases is undetermined.
To determine whether chemotherapy modulates breast tumor microbiota, we obtained snap-frozen aseptically collected tumor tissue from breast cancer patients that underwent neoadjuvant chemotherapy or tumors from women with no prior therapy at time of surgery. Our sample set is clinically annotated enabling correlation of primary tumor microbiota populations with tumor recurrence and obesity. Our data indicates that neoadjuvant chemotherapy increased tumoral Pseudomonas abundance and decreased Streptococcus populations. The development of distant metastases correlated with increased primary tumor abundance of Brevundimonas and Staphylococcus. Moreover, using a secondary breast tumor microarray sample set, we validated Pseudomonas presence by immunohistochemistry. We also used P. aeruginosa conditioned media to demonstrate differential molecular signaling activation in breast cancer cells depending on the concentration of bacterial-secreted metabolites to impact cell growth, apoptosis, and chemotherapy responsiveness. Taken together these data suggest that distinct intra-tumoral microbiota populations are present, and that the composition of bacterial populations can be modified by therapy to impact breast cancer cell signaling and drug responsiveness.
Material and Methods
Study approval and breast tumor tissue procurement.
This study was approved by our institutional review board (IRB00045734) in accordance with HHS regulations for the protection of human research subjects. Subjects were retrospectively identified as those in the Sentinel Lymph Node Mapping and Surgical Outcomes (IRB00008131) database who were female and diagnosed with invasive ductal carcinoma. In order for subjects to be included, they must have provided written consent for the Advanced Tumor/Tissue Bank (BG04–104) and have tumor tissue for research in the Tumor Bank. Patient demographics, preoperative variables, surgical details and clinical outcomes were also collected. Patient characteristics are described in Table 1. Breast tumor specimens provided to us by the pathologist in the OR Path Lab within 45–60 minutes from surgery are snap-frozen. A designation of tumor or normal is made by visual gross inspection by the pathologist before tumor samples are submitted to the Tumor Bank.
Table 1.
Breast Cancer Patient Characteristics. Numbers for patient age and BMI represent mean values ± standard deviation. There are no significant differences between treatment groups.
| Pretreatment Group n=18 | Neoadjuvant Chemotherapy Group n=15 | Tumor Recurrence Group n=9 | |
|---|---|---|---|
|
| |||
| Age (years) | 65.3±8.9 | 58.9±10.1 | 64.3±7.9 |
|
| |||
| BMI | 32.7±9.3 | 30.4±7.5 | 33.7±7.7 |
|
| |||
| Ethnicity | |||
| Caucasian | 83% | 80% | 100% |
| African American | 17% | 20% | 0% |
|
| |||
| Tumor Classification | |||
| Ductal | 89% | 80% | 88% |
| Lobular | 11% | 7% | 12% |
| Other | 0% | 13% | 0% |
|
| |||
| Estrogen Receptor-α positive (ER+) | 39% | 21.5% | 38% |
|
| |||
| ER-, Progesterone Receptor + (PR+) | 0% | 7% | 0% |
|
| |||
| Triple Negative Breast Cancer (ER-, PR-, HER2-) | 39% | 43% | 25% |
|
| |||
| ER+/HER2+ | 11% | 7% | 25% |
|
| |||
| HER2+ | 11% | 21.5% | 12% |
DNA extraction, PCR, 16s rRNA sequencing, and data processing.
Breast tumor specimens (n=33) were placed into a MoBio PowerMag Soil DNA Isolation Bead Plate. DNA was extracted following the manufacturer’s protocol. Bacterial 16S sequencing, data analysis, and interpretation were performed by Microbiome Insights, Vancouver, Canada. Bacterial 16S rRNA genes were PCR-amplified with dual-barcoded primers targeting the V4 region, following the methods of (11). Amplicons were sequenced with an Illumina MiSeq using the 250-bp paired-end kit (v.2). Sequences were taxonomically classified using Greengenes (v. 13_8) reference database, and clustered into 97%-similarity operational taxonomic units (OTUs) with the mothur (v. 1.39.5) software package (12), following the recommended procedure (https://www.mothur.org/wiki/MiSeq_SOP; accessed Nov 2017). The potential for contamination was addressed by co-sequencing DNA amplified from specimens and from four each of template-free controls and extraction kit reagents processed the same way as the specimens. Two positive controls, consisting of cloned SUP05 DNA, were also included (number of copies = 2 × 106). Samples with less than 100 OTU counts were excluded from analysis.
To estimate beta diversity across samples, we excluded OTUs occurring in fewer than 10% of the samples with a count of less than three and computed Bray-Curtis indices. We visualized beta diversity, emphasizing differences across samples, using non-metric multidimensional (NMDS) ordination. Variation in community structure was assessed with permutational multivariate analyses of variance with treatment group as the main fixed factor and using 4,999 permutations for significance testing. All analyses were conducted using R software. Proportional abundance of selected microbiota were graphed and multiple unpaired t-tests performed (correcting for multiple comparisons using Holm-Sidak method) was used and a value of p ≤ 0.05 was considered statistically significant.
Immunohistochemistry.
A tissue microarray (TMA) consisting of normally mammary gland tissue (n=10), primary breast tumor tissue (n=50), and lymph node metastases (n=40) was purchased from US Biomax, Inc. (Catalog BR1008a). TMA was stained against P. aeruginosa antibody (Abcam; Catalog #ab68538) in a 1:100 dilution using the Dako Envision Plus IHC staining kit.
Pseudomonas aeruginosa conditioned media (P-CM).
P. aeruginosa (ATCC 27853) was cultured overnight in tryptic soy broth at 37°C in an orbital shaker. Media was spun down at 1000rpm for 10 minutes to pellet the P. aeruginosa, which was discarded. The supernatant was filtered using a Corning disposable vacuum filtration system with a 0.22 micron pore size any bacterial particulates. Media was stored at 4°C degrees and used within 24–48 hours of harvest.
Cell line acquisition and growth conditions.
MDA-MB-231, 4T1 and the ZR-75–1 breast cancer cell lines were previously purchased from American Tissue Culture Collection (Manassas, VA). The 67NR cell line were generously provided by Dr. Andrea Mastro at Pennsylvania State University. MCF7 clone 2 cells were obtained from Dr. Robert Clarke at Georgetown University. MDA-MB-231 human triple negative breast cancer cells, 4T1 murine triple negative breast cancer cells, ZR-75–1 human ER+ breast cancer cells, and 67NR murine ER+ breast cancer cells were grown in RMPI media with 10% FBS at 37°C (basal growth conditions). MCF-7 human ER+ breast cancer cells were grown in DMEM media with 10% FBS at 37°C (basal growth conditions). Cells were passaged for approximately 3 months before new cell stock was obtained from liquid nitrogen storage. 4T1 and the MDA-MB-231 cell lines were last authenticated by IDEXX BioAnalytics using short tandem repeat (STR) analysis, in November 2018 (Columbia, MO). The 4T1 was confirmed to be of mouse origin and no mammalian interspecies contamination was detected. The MDA-MB-231 was confirmed to be of human origin and no interspecies contamination was detected. The MDA-MB-231 cells 100% identity match consistent with the cell line of origin. Mycoplasma contamination are measured in cell culture media supernatant by PCR. Primer sequences are as follows: CGCCTGAGTAGTACGTTCGC;CGCCTGAGTAGTACGTACGC;TGCCTGAGTAGTACATTCGC;TGCCTGGGTAGTACATTCGC;CGCCTGGGTAGTACATTCGC;CGCCTGAGTAGTATGCTCGC;GCGGTGTGTACAAGACCCGA;GCGGTGTGTACAAAACCCGA;GCGGTGTGTACAAACCCCGA.
In vitro cell index measurement.
Breast cancer cells (5 × 105) were plated in an ACEA xCELLigence system for 24 hours before treated with 5%, 10%, or 20% P-CM for an additional 48 hours. In another set of experiments breast cancer cells were plated in an ACEA xCELLigence system for 24 hours before treated with 10% or 20% P-CM +/− 1 μg/mL doxorubicin for an additional 48 hours. Cell index was measured by electrical impedance every 6 hours.
In vitro P. aeruginosa metabolite replacement cell index measurement.
MDA-MB-231 cells (5 × 105) in basal growth conditions cells were plated in an ACEA xCELLigence system for 24 hours before treated with 10 μg/mL pyocyanin (Catalog #R9532, Sigma-Aldrich), 10 μg/mL lectin (Catalog #L9895, Sigma-Aldrich), 10 ng/mL exotoxin A (Catalog #P0184, Sigma-Aldrich), 10 μg/mL phospholipase D (Catalog #P0065, Sigma-Aldrich), or 1 μg/mL LPS (Catalog #L9143, Sigma-Aldrich) ± 1 μg/mL doxorubicin for an additional 24 hours. Cell index was measured by electrical impedance every 6 hours.
Western blot hybridization.
MDA-MB-231, 4T1, MCF7, and ZR-75-1 cells were plated for 24 hours before treated with 5%, 10%, or 20% P-CM ± 1 μg/mL doxorubicin for an additional 24 hours. Treated cells were harvested in RIPA buffer, protein was size fractionated by gel electrophoresis, and transferred to a nitrocellulose membrane. Membranes were blocked in blotto for 30 minutes then incubated overnight at 4°C with primary antibodies (pAkt Ser473, Akt (total), cleaved caspase-7, and β-actin from Cell Signaling Technologies, dilution 1:1000). The next day, membranes were washed and incubation with polyclonal HRP-conjugated secondary antibodies. Immunoreactive products were visualized by chemoluminescence (SuperSignal Femto West, Pierce Biotechnology) and quantified by densitometry using the Bio-Rad digital densitometry software. Western blots are shown in figures as cropped images.
Results
Neo-adjuvant chemotherapy shifts tumor microbiota.
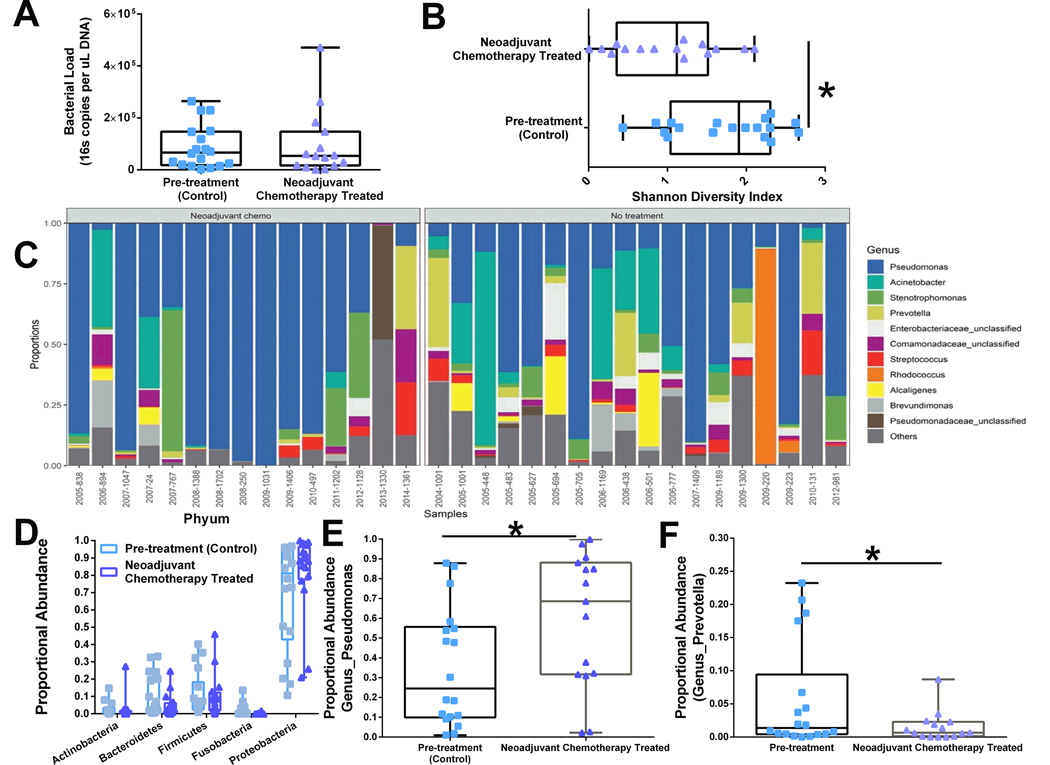
Bacterial load measured by qPCR targeting the V4 region of 16S gene indicate similar levels of total bacteria in untreated and neoadjuvant chemotherapy treated patients (Figure 1A), demonstrating that chemotherapy administration does not reduce total bacterial presence in the tumor tissue. However, analysis of microbiota diversity (Shannon index) indicates that neoadjuvant chemotherapy administration significantly reduces bacterial diversity within the tumor (Figure 1B). OTU abundances were aggregated based upon genus-level organization and grouped by treatment (untreated (n= 18) vs neoadjuvant chemotherapy (n=15)). Relative abundance of bacterial genera in different tumor samples is visualized by bar plots. Each bar represents a single tumor and each colored box a bacterial taxon. The height of a color box represents the relative abundance of that organism within the sample (Figure 1C). Analysis of phylum proportional abundance indicates that tumors from neoadjuvant chemotherapy treated patients do not display any significant regulation of bacteria at phylum level classification (Figure 1D). However, analysis of genus-level differences indicate significant increase in tumor Pseudomonas abundance (Figure 1E) and decreased tumoral Prevotella abundance (Figure 1F) from breast cancer patients treated with neoadjuvant chemotherapy.
Figure 1.
Neoadjuvant chemotherapy modulates breast tumor microbiota. Tumor 16S sequencing results were grouped by whether patient was treated with neoadjuvant chemotherapy before definitive surgery (n=15) or whether patient underwent surgery first (n= 18). A. Total bacterial load was quantified by 16S RT-PCR and graphed as 16S copies per uL of DNA. Neoadjuvant chemotherapy did not change total levels of bacteria within a tumor. B. Shannon diversity. Neoadjuvant chemotherapy significantly reduced the bacterial diversity within the tumor. C. Relative abundance of bacterial genera in different tumor samples is visualized by bar plots. Each bar represents a single tumor and each colored box a bacterial taxon. The height of a color box represents the relative abundance of that organism within the sample. “Other” represents lower abundance taxa. Approximately 25% of total tumor microbiota comprises of 105 genus level taxa with less than 1% proportional abundance. D. Primary breast tumors from patients that received neoadjuvant chemotherapy display no broad spectrum differences in bacterial phylum level proportional abundances. At the genus level, neoadjuvant chemotherapy treated tumors have elevated proportional abundance of Pseudomonas (E) and decreased levels of Prevotella (F) taxa when compared with untreated breast tumors. *p<0.05 and error bars show the min to max distribution.
Specific primary tumor microbiota correlate with distant metastases.
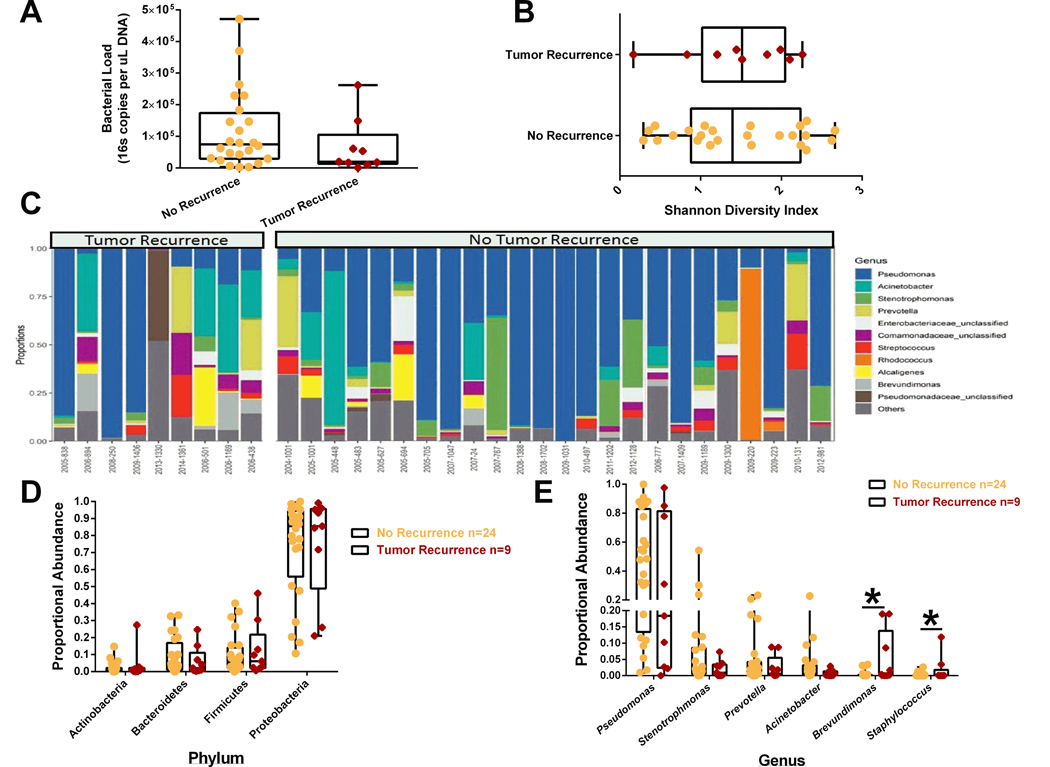
No significant differences in bacterial load (Figure 2A), Shannon diversity (Figure 2B), or phylum level proportional abundance (Figure 2D) were observed in patients that developed metastases. We aggregated OTU abundances into genus level grouping and segregated tumor samples based upon whether patients developed metastatic disease (Figure 2C). At the genus level (Figure 2E), primary breast tumor proportional abundance Brevunimonas and Staphylococcus were increased in patients that developed metastatic disease. There was no observed changes in the proportional abundances of Pseudomonas or Prevotella genus taxa (microbes regulated by neoadjuvant chemotherapy) by tumor recurrence.
Figure 2.
Primary breast tumor microbiota may predict tumor recurrence. Tumor 16S sequencing results were grouped by whether patient developed metastases within 5 years of primary tumor resection surgery (n=9) or whether patient did not develop metastases (n= 24). A. Total bacterial load was not different between primary tumors that displayed recurrence. B. Shannon diversity. Later on tumor recurrence did not impact bacterial diversity within the primary tumor. C. Relative abundance of bacterial genera in different tumor samples is visualized by bar plots. Each bar represents a single tumor and each colored box a bacterial taxon. The height of a color box represents the relative abundance of that organism within the sample. “Other” represents lower abundance taxa. D. Primary breast tumors from patients that later on developed metastases did not display differences in bacterial phylum level proportional abundances. E. At the genes level, primary breast tumors from patients that later on develop metastases have elevated proportional abundance of Brevundimonas and Staphylococcus taxa. *p<0.05 and error bars show the min to max distribution.
Patient obesity modifies tumoral microbiome populations.
We aggregated OTU abundances into genus level grouping and segregated tumor samples based upon patient BMI (Supplemental Figure S1A). Looking at broad phylum level differences, obesity significantly decreased bacteriodetes phylum level abundance in breast tumor tissue (Supplemental Figure S1B). There was also a trend for increased tumoral firmicutes abundance (p=0.1), partially correlating with observed obesity effects in the gut microbiome. At the family level, obesity significantly decreased tumor Comamonadaceae (Supplemental Figure S1C) and Ruminococcacea (Supplemental Figure S1D) proportional abundance. At the genus level, obesity increased tumoral Enterobacteriaceae_unclassified abundance (Supplemental Figure S1E).
P. aeruginosa laden breast tumor epithelial cells are observed in primary breast tumors and lymph node metastases.
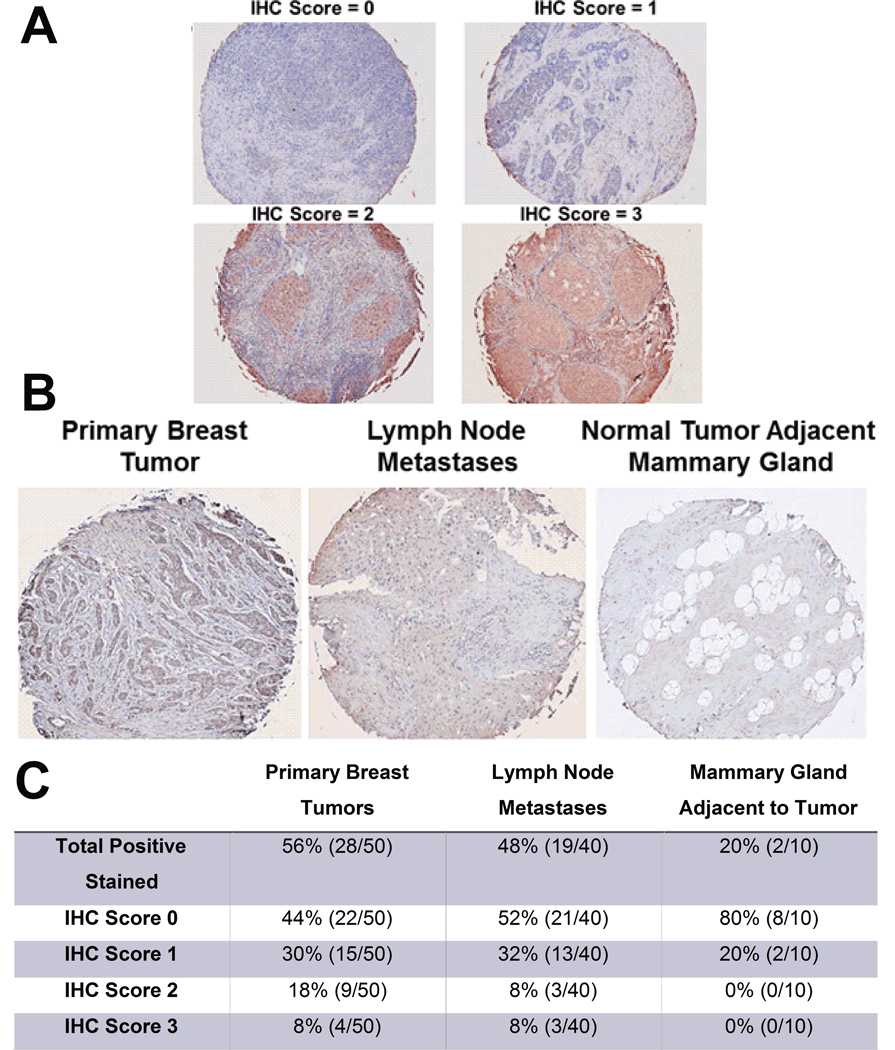
Paraffin-embedded primary breast tumors, breast tumor lymph node metastases, and tumor adjacent breast tissue was stained against a P. aeruginosa specific antibody. See Figure 3A for primary breast tumors representative images demonstrating gradient of P. aeruginosa staining positivity associated with each IHC score. Representative primary breast tumor, lymph node metastases, and normal adjacent mammary gland tissue stained for P. aeruginosa is shown in Figure 3B. IHC quantification is shown in Figure 3C. Approximately, 56% of the primary tumors stained positive for P. aeruginosa compared with 20% of the normal surrounding mammary gland tissue. Lymph node metastases also had higher levels of P. aeruginosa with 48% of the cases staining positive. There was also differential staining intensity within positively stained tumor sections. Approximately 26% of tissue sections from primary breast tumors had a 2 or higher IHC score, suggesting a gradient of tumoral bacterial infection that may result in varied molecular signaling impacting cancer cell survival or apoptosis.
Figure 3.
Elevated P. aeruginosa abundance in primary breast tumors and lymph node metastases when compared with normal surrounding mammary gland tissue. A. Representative images of primary breast tumors with differential P. aeruginosa IHC scoring B. Representative images of primary breast tumors and lymph node metastases, and normal tumor adjacent mammary gland tissue stained with an anti-Pseudomonas aeruginosa antibody. C. Table of the quantified P. aeruginosa tissue staining. Percentages and total number of cases shown. n=10–50.
Bacteria-produced bioactive compounds may modify breast cancer proliferation and chemotherapy responsiveness.
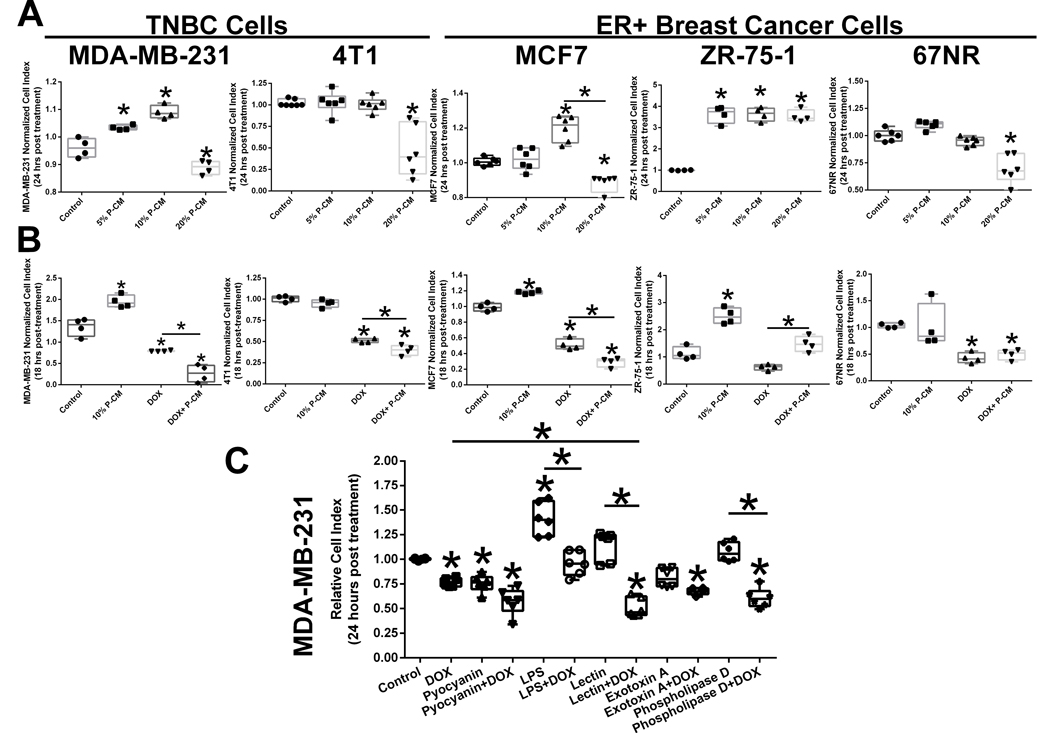
To determine whether P. aeruginosa secreted metabolites modulate breast cancer cell signaling, we used varying doses of P-CM to treat MDA-MB-231, 4T1, MCF7, ZR-75–1, and 67NR cells. We demonstrated that low doses of P-CM (5–10%) stimulate breast cancer cell growth within 24 hours of administration in MDA-MB-231, MCF7, and ZR-75–1 cell lines, while higher doses of P-CM (20%) reduce cell viability in the MDA-MB-231, 4T1, MCF7, and 67NR cell lines (Figure 4A). High dose P-CM (20%) stimulated cell growth in the ZR-75–1 cell line only. We also combined chemotherapy with P-CM to determine whether bacterial metabolites modify drug responsiveness. P-CM enhanced doxorubicin (DOX)-mediated cell death in the MDA-MB-231, 4T1, and the MCF7 cell lines. Combining DOX with 10% P-CM had no negative effect in the 67NR cell line. However, P-CM reduced DOX-killing efficacy in the ZR-75–1 cell line, possibly suggesting a cell line specific phenotype. However for the majority of the cell line tested, the data indicates that while low dose P-CM may stimulate growth other secreted factors potentiate chemotherapy cancer cell killing (Figure 4B). To further identify which of the potential P. aeruginosa metabolites may be having these effects, we treated MDA-MB-231 cells with LPS, pyocyanin, lectin, phospholipase D, and exotoxin A in the presence or absence of DOX (Figure 4C). LPS stimulated MDA-MB-231 cell proliferation while pyocyanin P. aeruginosa-derived metabolites potentiated cancer cell death. Lectin while having no significant overall effect alone on MDA-MB-231 proliferation, when combined with doxorubicin potentiated chemotherapy-mediated breast cancer cell killing.
Figure 4.
Secreted P. aeruginosa metabolites effects breast cancer cell survival and doxorubicin responsiveness. A. Differential effects of P. aeruginosa conditioned media (P-CM) on MDA-MB-231 human triple negative breast cancer cell, 4T1 murine triple negative breast cancer cell, MCF7 human ER+ breast cancer cell, ZR-75–1 human ER+ breast cancer cell, and 67NR murine ER+ breast cancer cell viability. n=4–6; *p<0.05. B. Combining P-CM and doxorubicin (DOX) enhances anti-cancer chemotherapy effects on MDA-MB-231 human triple negative breast cancer cell, 4T1 murine triple negative breast cancer cell, MCF7 human ER+ breast cancer cell, ZR-75–1 human ER+ breast cancer cell, and 67NR murine ER+ breast cancer cells. n=4; C. Administration of individual P. aeruginosa secreted metabolites pyocyanin, lectin (PA-I), lipopolysaccharide (LPS), exotoxin A, and phospholipase D on MDA-MB-231 cells viability and doxorubicin responsiveness. n=6; *p<0.05.
Potential molecular mechanisms driving the dose-dependent duality of P-CM.
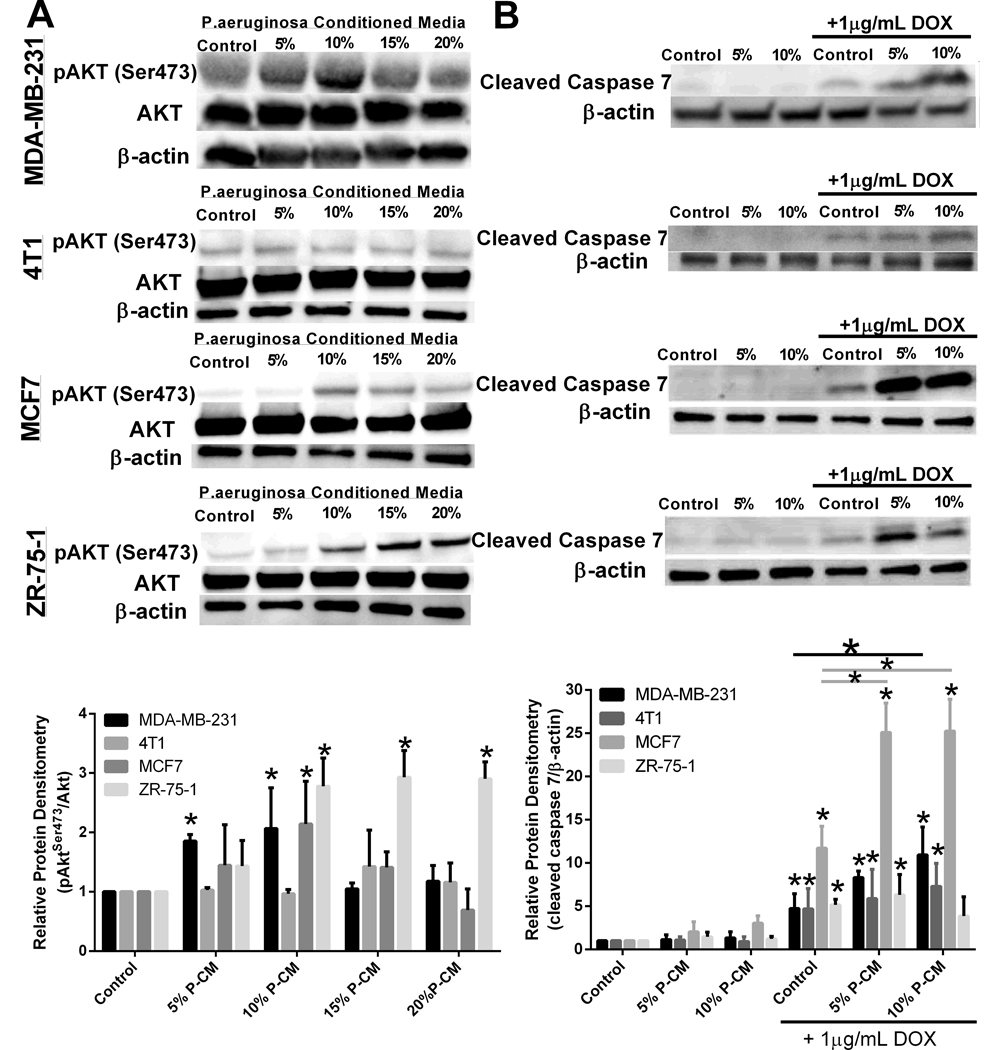
Treatment of MDA-MB-231 and MCF7 breast cancer cells with P-CM resulted in a dose-dependent stimulation phosphorylated Akt (Ser473), whereas all doses of P-CM stimulated pAkt in the ZR-75–1 breast cancer cells (Figure 5A). We did not observe pAkt activation in 4T1 or 67NR cell lines, both of which were not growth stimulated by low dose P-CM (Figure 4A). When combining P-CM with doxorubicin, protein analysis indicates that P-CM enhances chemotherapy mediated killing through modulation of apoptosis. Treatment with 10% P-CM doxorubicin significantly increased doxorubicin-induced cleaved caspase-7 protein levels in the MDA-MB-231 and MCF7 breast cancer cell lines when compared with doxorubicin treatment alone (Figure 5B). P-CM treatment did not negatively affect doxorubicin apoptosis induction in the 4T1 cell line. However, treatment with 10% P-CM reduced doxorubicin-mediated cleaved caspase 7 in the ZR-75–1 breast cancer cell line.
Figure 5.
Apoptotic and proliferative molecular signaling pathways modulated by secreted P. aeruginosa metabolites on breast cancer cells. A. Treatment with low dose P-CM stimulates pro-survival Akt signaling in MDA-MB-231 cells. n=3 *p<0.05. B. Administration of P-CM with DOX increases cleaved caspase 7 or cleaved caspase 3 protein expression in breast cancer cells, indicating P-CM enhances apoptosis when combined with chemotherapy n=3; *p<0.05.
Discussion
The human body contains slightly more bacterial cells than human cells (1.3 bacterial cells to each human cell in the body), according to new estimates refuting the previously estimated 10:1 ratio established in the 1970’s (13). While the majority of the bacteria biomass is contained within the intestinal track, microbes in lower abundance have been identified in other organs located outside the gastrointestinal tract, including the mammary gland (1). Our group recently identified that diet can modulate the mammary gland microbiota population, suggesting the plasticity of the breast microbiome (5). Moreover, recent studies demonstrate that breast tumors display distinct bacterial populations when compared with the surrounding mammary gland tissue microbiota (2–4, 7). While other reports have highlighted the presence of bacteria in breast tumor tissues, these studies have not shown the impact of therapy on the tumor microbiome. Moreover, these initial breast tumor microbiome studies, for the most part, fail to indicate the possible functional relevance of these microbes in the tumor microenvironment. In the study by Urbaniak et al. (2), authors demonstrated that the E.coli found in the surrounding mammary gland tissue can stimulate DNA breakage which could induce breast tumorigenesis indicating a possible role of the microbiota in breast tumor formation. Our study aimed to determine impact of breast tumor microbiota on therapeutic responsiveness and breast cancer outcomes. Our 16S sequencing data of breast tumors from untreated or neoadjuvant chemotherapy treated patients indicates that chemotherapy increases the tumor proportional abundance of Pseudomonas. Normal mammary gland tissue display a low proportional abundance (approximately 5%; for reference see Urbaniak et al. (1)) of Pseudomonas, while breast tumor tissue contains elevated (approximately 20%) Pseudomonas. Neoadjuvant chemotherapy further increases the proportional abundance of Pseudomonas to 85%, suggesting chemotherapy induces preferential growth or survival of these bacteria.
While Pseudomonas genus contains over 140 species, most of these are found in water or soil (14). Pseudomonas genus contains only a few animal pathogenic strains: P. aeruginosa (wound infections, cystic fibrosis, and hospital-based infections in humans), P. oryzihabitans (sepsis in humans), and P. plecoglossicida (hemorrhagic ascites in fish). While we currently do not know the strain of Pseudomonas found in the 16S sequencing results from the neoadjuvant chemotherapy treated breast tumors, we anticipate that it is most likely P. aeruginosa. The TMA staining data using a P. aeruginosa specific antibody indicates that this species is found in breast tumor samples. Future studies will focus on the isolation and characterization of breast tumor specific Pseudomonas populations. P. aeruginosa is implicated in hospital infections (15). While P. aeruginosa infection is well studied in ventilator-based infections, chronic wounds, and cystic fibrosis, the impact in tumorigenesis is unknown. Molecular evidence supplied by the P. aeruginosa wound healing field suggests that P. aeruginosa biofilm and secreted factors may play important roles in various signaling pathways including cell death, proliferation, chemotherapy responsiveness, and/or development of metastases. The signaling mechanism stimulated by P. aeruginosa secreted factor may be dependent on concentration driven by tumoral P. aeruginosa abundance. P. aeruginosa secretes numerous proteases, lipases, lipopolysaccharides, quinolones, pyocyanin, Cif, or hemolysin-coregulated proteins (16–20). It also forms multicellular biofilms which are resistant to biotic and abiotic stressors. Literature showed that P. aeruginosa can grow in the presence of chemotherapy and that doxorubicin stimulated P. aeruginosa biofilm production (21). Therefore, chemotherapy administration may offer selective pressure killing off other bacterial populations while concurrently enabling Pseudomonas outgrowth in the tumor microenvironment. This is supported by the similar total bacterial counts in each treatment group (untreated versus neoadjuvant chemotherapy) but lower alpha diversity scores in the chemotherapy treated tumors, e.g. same amount of bacteria but decreased variety of strains expressed.
P. aeruginosa secreted factors may also directly impact breast cancer cell proliferative signaling. P. aeruginosa type VI secretion phospholipase D effector (PldA) was shown to effect host lung epithelial cell signaling through PI3K/Akt activation (18). P. aeruginosa is also a potent producer of lipopolysaccharide (LPS) (22). LPS-mediated TLR4 stimulation was previously shown to promote breast cancer metastases through Akt activation (23). We show that P. aeruginosa conditioned media at low concentrations (5–10%) stimulate MDA-MB-231, ZR-75–1, and MCF7 breast cancer cell growth and activate pro-tumorigenic Akt signaling, suggesting that the mid-level proportional abundance (20%) of Pseudomonas found in untreated breast tumors may promote tumor survival and growth. Treatment of MDA-MB-231 cells with LPS promoted proliferation while administration of cancer cells with phospholipase D (albeit derived from Streptomyces) did not affect MDA-MB-231 viability, suggesting the pro-growth mediated by P-CM may be due to LPS secretion.
Literature indicates that hypoxia (low oxygen concentrations often found in solid tumors) decreases the virulence capacity of P. aeruginosa, as indicated by decreased pyocyanin secretion (24). Pyocyanin is a potent reactive oxygen species (ROS) inducer. In the cancer field ROS is often considered diphasic; Low levels of ROS are pro-tumorigenic while higher ROS concentrations promote cell death. Therefore, intra-tumoral P. aeruginosa subjected to hypoxia would decrease pyocyanin production, promoting lower levels of ROS and pro-survival signaling. The decreased virulence observed by P. aeruginosa subjected to hypoxia may also give evidence to why elevated tumoral levels of bacteria do not result in systemic infection. We demonstrated that pyocyanin administration reduced MDA-MB-231 breast cancer cell growth with an observed trend to enhance doxorubicin-killing, suggesting a possible ROS-dependent mechanism of P-CM potentiation of chemotherapy observed in the MDA-MB-231 cells.
In the tumor microenvironment, P. aeruginosa may have potent immunomodulatory effects. P. aeruginosa secretes numerous products including zinc metalloproteases (AprA and LasB to combat host immune response (16)), Cif (inhibits CFTR and TAP1, preventing antigen presentation (20)), pyocyanin (inhibits lymphocyte activity (17)), among others that can affect immune cell recruitment and function. This suggests that tumor specific P. aeruginosa produced bioactive compounds may enhance cancer immunoavoidance (25).
Using P-CM, we show that P. aeruginosa secreted factors enhance doxorubicin killing capacity. Transcriptomic analysis of doxorubicin effects on P. aeruginosa indicate doxorubicin increased PqsH gene expression (21). PqsH is a FAD-dependent monooxygenase that is required for the production of the Pseudomonas quinolone signal (PQS) (26). PQS, while predominately implicated in quorum sensing, has been linked with p38 MAPK signaling (pro-apoptotic) and NFκB inhibition (27). Doxorubicin treatment can subsequently activate NFκB signaling initiating an inherent pathway promoting therapeutic resistance (28). Therefore, PQS-mediated inhibition of NFκB may enhance doxorubicin-mediating killing. PQS also serves as a ferric iron chelator, where exogenous PQS treatment imitates iron starvation (29). Cancer cells exhibit “iron addiction” and therapies to reduce iron levels were previously shown to suppress breast tumor growth (30, 31), suggesting another possible molecular mechanism by which Pseudomonas secreted compounds may potentiate chemotherapy responsiveness.
Due to the annotation of breast tumor samples, we were able to compare primary breast tumor bacteria populations between patients that displayed tumor recurrence (within 5 years of surgery) and those patients that did not have recurrence. We demonstrate elevated Brevundimonas and Staphylococcus in primary tumors from breast cancer patients that later developed metastatic disease. Previous human breast tumor microbiome studies have identified elevated Brevundimonas prevalence in all subtypes (ER+, HER2+, and TNBC) of breast tumors when compared with non-matched control breast tissue (8). However, to our knowledge, this is the first report suggesting elevated Brevundimonas correlates with the development of metastases. Further studies are needed to explore the potential impact of breast tumor Brevundimonas on cancer cell migration and metastatic signaling cascades.
Many criticisms for tumor microbiome studies include the possibility of sample contamination skewing outcomes (32). Low microbial abundance in tissue, such as mammary gland or breast tumors, would be highly sensitive for sample contamination being detected as real. These outside bacterial contaminations may occur in the surgical suite, at the pathologist, in the tumor bank storage, during DNA isolation, or during the paraffin embedding process just to name a few possibilities. Our study takes into consideration all of these variables. First, our breast tumor sample set was collected over a 9 year period (2004–2013) limiting any possible contaminations that could have occurred either in the surgical suite or at the pathologist. Having the same bacteria populations at all of these locations over a 9 year period would be rare. Secondly, to control for any possible tumor bank storage or DNA isolation contamination we used a tumor microarray purchased from an outside company to serve as a validation cohort for our significantly regulated microbes that were detected by 16S sequencing. Furthermore, any environmental contamination that could occur during DNA isolation of the samples would be across all the groups since these analysis were performed as batched samples and therefore would not be differentially expressed depending on treatment group. Our tumor bank samples also serve as a control for any possible contamination in the paraffin embedding process in the tumor microarray cohort. These contingencies for outside contamination overall strengthen our study, thereby demonstrating that breast tumors have their own microbiota population and that these bacterial populations are modified by neoadjuvant chemotherapy which modulates breast cancer cell signaling to impact outcome. Furthermore, we also show functional relevance of breast tumor specific P. aeruginosa modulating breast cancer cell proliferation and doxorubicin sensitivity.
We demonstrate that therapeutic modalities shift the tumor microbiota suggesting a role of certain bacteria in potentially increasing the response rate to neoadjuvant chemotherapy and improved cancer outcomes. These data may also be extrapolated to suggest tumor microbiota may impact outcomes such as improved disease-free survival and/or overall survival. Our observation that certain primary breast tumor microbiota populations have increased likelihood of breast cancer recurrence suggests that modulating tumor microbiome could potentially decrease the rate of tumor recurrence. However, further studies with increased samples are needed to more clearly delineate the role of tumor microbiota in metastases. In this retrospective analysis of operable breast cancer, breast tumor microbiota may improve responsiveness to neoadjuvant chemotherapy by increasing therapeutic efficacy. Further research is warranted to evaluate the possible role of microbiome in improving breast cancer outcomes.
Supplementary Material
Acknowledgements:
DSP is supported by K22CA181274. KLC is supported by the Chronic Disease Research Fund, an American Cancer Society Research Scholar grant (RSG-16–204-01-NEC), and a Career Catalyst grant from the Susan G. Komen Foundation (CCR18547795). Shared Resource services were provided by the Wake Forest Baptist Comprehensive Cancer Center’s NCI Cancer Center Support Grant P30CA012197.
Footnotes
Conflict of interest statement: Authors declare that no conflict of interest exists.
References:
- 1.Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, et al. Microbiota of human breast tissue. Appl Environ Microbiol. 2014;80(10):3007–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl Environ Microbiol. 2016;82(16):5039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Altemus J, Niazi F, Green H, Calhoun BC, Sturgis C, et al. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget. 2017;8(50):88122–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci Rep. 2016;6:30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shively CA, Register TC, Appt SE, Clarkson TB, Uberseder B, Clear KYJ, et al. Consumption of Mediterranean versus Western diet leads to distinct mammary gland microbiome populations: Implications for breast cancer. Cell Reports. 2018;25(1):47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–405. [PubMed] [Google Scholar]
- 7.Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS One. 2017;12(11):e0188873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct Microbial Signatures Associated With Different Breast Cancer Types. Front Microbiol. 2018;9:951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubovszky G, Horvath Z. Recent Advances in the Neoadjuvant Treatment of Breast Cancer. J Breast Cancer. 2017;20(2):119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones SE, Savin MA, Holmes FA, O’Shaughnessy JA, Blum JL, Vukelja S, et al. Phase III trial comparing doxorubicin plus cyclophosphamide with docetaxel plus cyclophosphamide as adjuvant therapy for operable breast cancer. J Clin Oncol. 2006;24(34):5381–7. [DOI] [PubMed] [Google Scholar]
- 11.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8):e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iglewski BH. Pseudomonas. In: th, Baron S, editors. Medical Microbiology. Galveston (TX)1996. [Google Scholar]
- 15.Fazeli H, Akbari R, Moghim S, Narimani T, Arabestani MR, Ghoddousi AR. Pseudomonas aeruginosa infections in patients, hospital means, and personnel’s specimens. J Res Med Sci. 2012;17(4):332–7. [PMC free article] [PubMed] [Google Scholar]
- 16.Casilag F, Lorenz A, Krueger J, Klawonn F, Weiss S, Haussler S. The LasB Elastase of Pseudomonas aeruginosa Acts in Concert with Alkaline Protease AprA To Prevent Flagellin-Mediated Immune Recognition. Infect Immun. 2016;84(1):162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall S, McDermott C, Anoopkumar-Dukie S, McFarland AJ, Forbes A, Perkins AV, et al. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins (Basel). 2016;8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe. 2014;15(5):600–10. [DOI] [PubMed] [Google Scholar]
- 19.Nakagami G, Minematsu T, Morohoshi T, Yamane T, Kanazawa T, Huang L, et al. Pseudomonas aeruginosa quorum-sensing signaling molecule N-3-oxododecanoyl homoserine lactone induces matrix metalloproteinase 9 expression via the AP1 pathway in rat fibroblasts. Biosci Biotechnol Biochem. 2015;79(10):1719–24. [DOI] [PubMed] [Google Scholar]
- 20.Stanton BA. Effects of Pseudomonas aeruginosa on CFTR chloride secretion and the host immune response. Am J Physiol Cell Physiol. 2017;312(4):C357–C66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groizeleau J, Rybtke M, Andersen JB, Berthelsen J, Liu Y, Yang L, et al. The anti-cancerous drug doxorubicin decreases the c-di-GMP content in Pseudomonas aeruginosa but promotes biofilm formation. Microbiology. 2016;162(10):1797–807. [DOI] [PubMed] [Google Scholar]
- 22.Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol. 2007;297(5):277–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Yin J, Shen W, Gao R, Liu Y, Chen Y, et al. TLR4 Promotes Breast Cancer Metastasis via Akt/GSK3beta/beta-Catenin Pathway upon LPS Stimulation. Anat Rec (Hoboken). 2017;300(7):1219–29. [DOI] [PubMed] [Google Scholar]
- 24.Schaible B, Rodriguez J, Garcia A, von Kriegsheim A, McClean S, Hickey C, et al. Hypoxia Reduces the Pathogenicity of Pseudomonas aeruginosa by Decreasing the Expression of Multiple Virulence Factors. J Infect Dis. 2017;215(9):1459–67. [DOI] [PubMed] [Google Scholar]
- 25.Sadikot RT, Blackwell TS, Christman JW, Prince AS. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171(11):1209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184(23):6472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J, Cheng J, Wang Y, Shen X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front Cell Infect Microbiol. 2018;8:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JY, Jung HH, Ahn S, Bae S, Lee SK, Kim SW, et al. The relationship between nuclear factor (NF)-kappaB family gene expression and prognosis in triple-negative breast cancer (TNBC) patients receiving adjuvant doxorubicin treatment. Sci Rep. 2016;6:31804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14(1):87–96. [DOI] [PubMed] [Google Scholar]
- 30.Manz DH, Blanchette NL, Paul BT, Torti FM, Torti SV. Iron and cancer: recent insights. Ann N Y Acad Sci. 2016;1368(1):149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoke EM, Maylock CA, Shacter E. Desferal inhibits breast tumor growth and does not interfere with the tumoricidal activity of doxorubicin. Free Radic Biol Med. 2005;39(3):403–11. [DOI] [PubMed] [Google Scholar]
- 32.Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS. Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends Microbiol. 2019;27(2):105–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.