Abstract
One goal of precision medicine is to identify mutations within individual tumors to design targeted treatment approaches. This report details the use of genomic testing to select a targeted therapy regimen of erlotinib and rapamycin for a pediatric anaplastic oligodendroglioma refractory to standard treatment, achieving a 33-month sustained response. Immunohistochemical analysis of total and phosphorylated protein isoforms showed abnormal signaling consistent with detected mutations, while revealing heterogeneity in per-cell activation of signaling pathways in multiple subpopulations of tumor cells throughout the course of disease. This case highlights molecular features that may be relevant to designing future targeted treatments.
Keywords: brain tumor, EGFR, mTOR, neuro-oncology, pediatric oncology, targeted therapy
1 ∣. INTRODUCTION
Genomic profiling is a valuable tool in designing precision medicine approaches. Responses to therapy and the recurrence of targeted tumors can be strongly affected by intratumoral heterogeneity at the DNA, RNA, or protein levels.1 Here, we detail an unusual clinical response following such an approach in a pediatric anaplastic oligodendroglioma refractory to standard treatment. A combination erlotinib/rapamycin course of treatment selected based on sequencing analyses resulted in substantial tumor regression sustained for 33 months followed by recurrence. To better understand whether the drugs targeted a unified cellular phenotype or multiple subpopulations, additional cyclic immunostaining analyses of resected tissue were performed, revealing variable levels of signaling activity among tumor cells.
2 ∣. RESULTS
The patient, a 10-year-old female, first presented with left-sided weakness and numbness and was found to have a large infiltrating mass involving the right perisylvian frontal and temporal cortex causing mass effect. A biopsy of the lesions revealed an anaplastic oligodendroglioma (WHO Grade III), IDH wild type and negative for 1p/19q codeletion. Histologic sections showed an infiltrating neoplasm composed of cells with round-to-ovoid nuclei and perinuclear halos, with secondary structure formation around blood vessels and perineuronal satellitosis. Frequent mitoses were seen (10/10 high-power field [HPF]). Neither microvascular proliferation nor necrosis were found. The tumor cells were immunoreactive for glial fibrillary acidic protein (GFAP) and S100B. FoundationOne testing revealed mutations in epidermal growth factor receptor (EGFR [R222C]), F-box/WD repeat-containing protein 7 (FBXW7), a member of the SCF complex predicted to sensitize cells to rapamycin, as well as mutations in KRAS, BCOR, and TERT. The mutation profile, suggestive of an astrocytic lineage, was similar to a previously reported case of an anaplastic oligodendroglioma in a pediatric patient.2 The specific EGFR mutation found here was rarely seen in study of low-grade astrocytomas (one tumor, n = 46),2 but has been frequently reported in both primary and recurrent high-grade gliomas (n = 16).3 Intratumoral heterogeneity has been well described in recurrent oligodendrogliomas, specifically including mutations in the TERT promoter (n = 12).4 Though mutations in FBXW7 have been reported in high-grade gliomas (n = 1000),5 published reports of concurrent EGFR and FBXW7 mutations in pediatric brain tumors were not found. It remains unclear how common the specific combination of mutations and tumor type reported here is, as genetic profiling has been completed on comparatively smaller numbers of low-grade pediatric tumors.
Changes in radiographic appearance over the course of treatment are shown in Figure S1. The patient was initially treated with standard of care 50 Gy cranial radiation to temporal lobe with boost to 60 Gy with concurrent temozolomide during radiation. Magnetic resonance imaging (MRI) following radiation showed progression and the patient was started on maintenance temozolomide, bevacizumab, and irinotecan. Seven months later, the tumor further progressed and the patient was switched to procarbazine, lomustine, and vincristine. Four months later, the patient was wheelchair bound and made “do not resuscitate,” as scans showed further progression. Based on FoundationOne sequencing and safety results from a study in 19 patients with low-grade glioma,6 the patient was started on erlotinib, an EGFR inhibitor, at 65 mg/m2/day PO daily and rapamycin, an mechanistic target of rapamycin (mTOR) inhibitor, 0.8 mg/m2/dose twice daily. Troughs maintained level between 10 and 15 ng/mL. The patient showed immediate response and tumor regression. She continued showing response for 33 months (through 48 months post diagnosis) with excellent blood counts without neutropenia or need for transfusions. She had an occasional mild intermittent rash, but rapidly became steroid independent and, with frequent therapy, physical functioning improved. At the 51-month scan, despite ongoing therapy, the tumor showed growth. A repeat biopsy and molecular tests did not detect the EGFR [R222C] mutation, but redetected the KRAS and FBXW7 mutations. Based on these mutations, treatment was switched from erlotinib to trametinib, a mitogen-activated protein kinase kinase (MEK) inhibitor at 0.0125 mg/kg/dose daily, with continued rapamycin. Due to delays in obtaining insurance approval, the patient was on rapamycin alone for 8 weeks before starting the combination therapy.
The patient quickly began to show increased toxicity with severe rash, fatigue, and severe diarrhea and dehydration leading to four hospital admissions. Radiographic progression was seen 2 months post starting the new regimen, but stabilized for 4 months after the trametinib dose was increased to 0.025 mg/kg/dose. However, neurologic status started to decline and she began to have seizures. The patient then developed a bowel perforation that required discontinuation of the medication. Her tumor progressed while chemotherapy was held, and the patient passed away 62 months after initial tumor presentation.
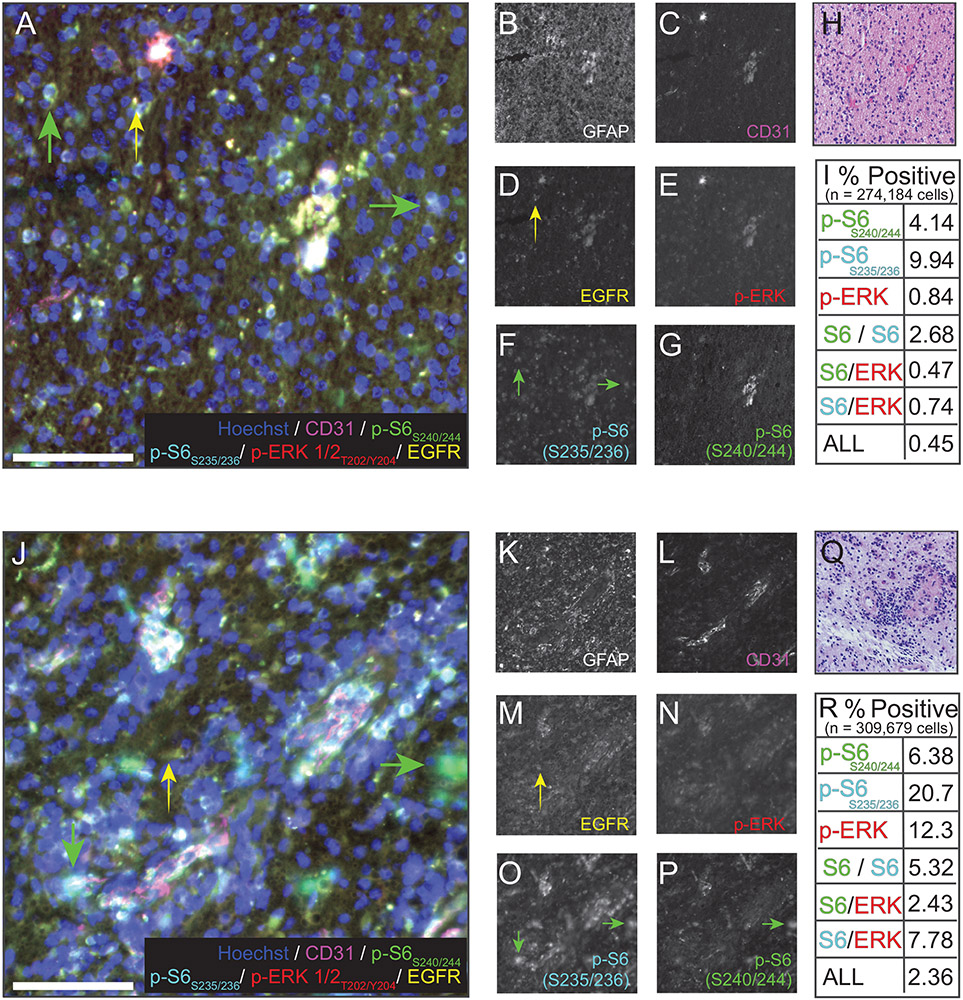
To ascertain whether the mutations detected by sequencing corresponded to detectable activity at the protein level, and whether pathways were coactivated in individual cells, cyclic multiplexed immunostaining was conducted on resected tissue from both of the timepoints at which FoundationOne sequencing was performed (Figure 1; extended methods in Supporting Materials).7,8 The epitopes detected by antibody staining were GFAP, CD31, total EGFR, and phosphorylated forms of ribosomal protein S6 and ERK. Activity of the RAS/RAF/MEK pathway was assayed via staining for p-ERK1/2 T202/Y204 (Figure 1A,E,J,N).9 Rapamycin-sensitive mTOR pathway activity was probed by staining for both p-S6 S240/244 (Figure 1A,G,J,P), expected to be mTORC1-dependent and thus rapamycin-sensitive, and p-S6 S235/236, indicative of signaling through either the RAS/RAF/MEK/ERK pathway or mTOR complexes (Figure 1A,F,J,O).10 Immunostaining of fields containing histologically abnormal cells was positive for each of the parameters measured, indicating that both RAS and mTOR signaling were active within the tumor mass. Consistent with the differences in sequencing data between first and second resections, an increase in cells with intense p-ERK or p-S6 staining was observed in the second resection relative to the first. While some cells were positive for both EGFR and a select phosphoprotein, multiple cells were also detectable that were highly positive for only EGFR or specific phosphoproteins (arrows in Figure 1A,J), with all possible combinations of dual or triple phosphoprotein labeling observed at variable abundance (quantified in Figure 1I,R). These data suggest that the tumor cells were heterogeneous in their activation of signaling events targeted by the therapies described.
FIGURE 1.
Cyclic immunofluorescence imaging and quantification correlate with sequencing data. Representative 40× immunofluorescence images (stitched fields) of seven-parameter cyclic immunofluorescence on first (A-I) and second (J-R) tumor resections. A and I show six-parameter images of nuclei (Hoechst, blue), CD31 (magenta), p-S6S240/244 (green), pS6S235/236 (cyan) p-ERK1/2T202/Y204, and EGFR (yellow). B-G and K-P show individual channels as grayscale images. Yellow arrowheads point to cells positive for EGFR, but not a phosphorylated protein. Green arrows point to cells positive for a phosphorylated protein, but not EGFR. Scale bars = 100μm. The histological staining used to identify region of interest is shown in H and Q, respectively. Quantification for phospho-signalingin all the cells in all the fields measured is summarized in panels I and R. Cells identified as singly positive for an individual phosphoprotein, combinations of two phosphoproteins, or positive for all three were found in both resections, but were more abundant in the second (recurrence) resection, consistent with genomic profiling
3 ∣. DISCUSSION
The striking response of this patient to targeted therapies selected based on genomic data suggests possible therapeutic strategies for future cases with similar profiles. As EGFR, KRAS, and mTOR may interact with similar and overlapping downstream signaling molecules, one key question was whether the drugs used in combination targeted the same population of cells or exerted overlapping effects on cell subpopulations with differing downstream signaling activity. Immunofluorescence analyses suggested that each treatment targeted overlapping, but not identical subsets of cells within the patient’s tumor. Future discovery studies should aim to further dissect how potential recurrence-driving or drug-resistant populations can be identified using single-cell approaches. Additionally, which targeted combinations are tolerable should be explored, as the combination of erlotinib and rapamycin was well tolerated, while the combination of trametinib and rapamycin had increased toxicity.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Ihrie and Irish Labs at Vanderbilt and the Pediatric Neuro-Oncology Tumor Board at VUMC for thoughtful discussions. Research was supported by the following funding resources: NIH T32 GM007628 (Laura C. Geben), NIH R01 NS096238 (Rebecca A. Ihrie), DODW81XWH-16-1-0171/TS150037 (Rebecca A. Ihrie), the Michael David Greene Brain Cancer Fund (Rebecca A. Ihrie), and the Eric T. Klindt Fund for Brain Cancer Research (Rebecca A. Ihrie).
Funding information
National Institute of General Medical Sciences, Grant/Award Number: T32GM007628; U.S. Department of Defense, Grant/Award Numbers: TS150037, W81XWH-16-1-0171; National Institute of Neurological Disorders and Stroke, Grant/Award Number: R01 NS096238; Michael David Greene Brain Cancer Fund; Eric T. Klindt Fund for Brain Cancer Research
Abbreviations:
- EGFR
epidermal growth factor receptor
- GFAP
glial fibrillary acidic protein
- HPF
high-power field
- MEK (MAP2K)
mitogen-activated protein kinase kinase
- mTOR
mechanistic target of rapamycin
Footnotes
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Preliminary data for this manuscript were previously presented in poster form at the ISPNO 2018 annual meeting.
DATA AVAILABILITY STATEMENT
Data are available on request from the authors.
REFERENCES
- 1.Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: the Rosetta Stone of therapy resistance. Cancer Cell. 2020;37(4):471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson A, Severson E, Gay L, et al. Comprehensive genomic profiling of 282 pediatric low- and high-grade gliomas reveals genomic drivers, tumor mutational burden, and hypermutation signatures. Oncologist. 2017;22(12):1478–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salloum R, McConechy MK, Mikael LG, et al. Characterizing temporal genomic heterogeneity in pediatric high-grade gliomas. Acta Neuropathol Commun. 2017;5(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aihara K, Mukasa A, Nagae G, et al. Genetic and epigenetic stability of oligodendrogliomas at recurrence. Acta Neuropathol Commun. 2017;5(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yalon M, Rood B, MacDonald TJ, et al. A feasibility and efficacy study of rapamycin and erlotinib for recurrent pediatric low-grade glioma (LGG). Pediatr Blood Cancer. 2013;60(1):71–76. [DOI] [PubMed] [Google Scholar]
- 7.Gerdes MJ, Sevinsky CJ, Sood A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110(29):11982–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin JR, Izar B, Wang S, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife. 2018;7:e31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten M, Barbacid M. Targeting the MAPK pathway in KRAS-driven tumors. Cancer Cell. 2020;37(4):543–550. [DOI] [PubMed] [Google Scholar]
- 10.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request from the authors.