Abstract
Background
Leptospirosis, caused by Leptospira bacteria, is a common zoonosis worldwide, especially in the tropics. Reservoir species and risk factors have been identified but surveys for environmental sources are rare. Furthermore, understanding of environmental Leptospira containing virulence associated genes and possibly capable of causing disease is incomplete, which may convolute leptospirosis diagnosis, prevention, and epidemiology.
Methodology/Principal findings
We collected environmental samples from 22 sites in Puerto Rico during three sampling periods over 14-months (Dec 2018-Feb 2020); 10 water and 10 soil samples were collected at each site. Samples were screened for DNA from potentially pathogenic Leptospira using the lipL32 PCR assay and positive samples were sequenced to assess genetic diversity. One urban site in San Juan was sampled three times over 14 months to assess persistence in soil; live leptospires were obtained during the last sampling period. Isolates were whole genome sequenced and LipL32 expression was assessed in vitro.
We detected pathogenic Leptospira DNA at 15/22 sites; both soil and water were positive at 5/15 sites. We recovered lipL32 sequences from 83/86 positive samples (15/15 positive sites) and secY sequences from 32/86 (10/15 sites); multiple genotypes were identified at 12 sites. These sequences revealed significant diversity across samples, including four novel lipL32 phylogenetic clades within the pathogenic P1 group. Most samples from the serially sampled site were lipL32 positive at each time point. We sequenced the genomes of six saprophytic and two pathogenic Leptospira isolates; the latter represent a novel pathogenic Leptospira species likely belonging to a new serogroup.
Conclusions/Significance
Diverse and novel pathogenic Leptospira are widespread in the environment in Puerto Rico. The disease potential of these lineages is unknown but several were consistently detected for >1 year in soil, which could contaminate water. This work increases understanding of environmental Leptospira diversity and should improve leptospirosis surveillance and diagnostics.
Author summary
Leptospirosis is a common zoonotic disease worldwide, but more prevalent in the tropics. Cases are more common following severe weather events, possibly due to flooding, which may more readily distribute soil and/or water contaminated with Leptospira spp., the disease agents. Human cases increased following the 2017 hurricanes that ravaged Puerto Rico (Maria and Irma), prompting environmental sampling of soil and water to assess the presence, abundance, and possible persistence of pathogenic leptospires in these environments. The goal was to better understand these potential reservoirs of human and animal disease. Diverse and novel groups of pathogenic Leptospira were abundant and widespread in soil and water in Puerto Rico and sometimes were consistently detected in these environments for >1 year. However, most groups we identified have not previously been described from humans and/or other animals, so the disease potential of these novel organisms is unknown. The results of this study reveal a tremendous amount of previously uncharacterized Leptospira diversity in soil and water in Puerto Rico. The description and characterization of these novel types improves our understanding of the genus Leptospira, and will aid in the development of improved diagnostics and preventative tools to advance public health outcomes.
Introduction
Leptospirosis is caused by pathogenic bacteria in the genus Leptospira. The three types of leptospires within this genus are categorized using 1) phylogenetic placement based upon single genes (e.g. secY, 16S rRNA) or whole genome sequences, 2) in vitro phenotypes, and 3) actual or presumed pathogenicity (i.e., the presence of the lipL32 gene, which is conserved in all pathogenic leptospires) [1, 2]. To date, there are 17 described species in the pathogenic group (group P1), 21 species in the intermediate group (P2), and 26 in the two saprophytic groups (S1 and S2) [2]. The genomes of leptospires are comprised of two circular chromosomes that are larger than those of other spirochetes and more diverse than those of many other bacterial genera [3] due to significant horizontal gene transfer and gene duplication [4]. This potentially facilitates an increased ability to survive in a variety of hosts (e.g., humans, domestic and wild animals), environmental conditions (e.g., soil and water), and climates (e.g., tropical, temperate, etc.) [3, 5].
Leptospirosis is the most widespread global bacterial zoonosis [6, 7] and disproprotionately affects resource-poor populations [8]; recent estimates of global human disease cases are >1 million annually, including >58,000 deaths [8], with 73% of cases occuring in the tropics [9]. In the United States (US), leptospirosis is considered endemic with most human cases arising in Puerto Rico and Hawaii [9]. It was first reported in Puerto Rico in 1942 [10] and is common in other parts of the Caribbean: 12,475 human cases were reported by nine Caribbean countries/territories between 1980 and 2005 [11]. Notwithstanding this, it was removed from the US reportable disease list in 1995 but later reinstated in 2014. Despite the lack of requirement to report this disease, 759 cases of leptospirosis were reported in Puerto Rico during that intervening time; 570 of these were confirmed, including 92 deaths [10]. Heavy rains and floods have been identified as major risk factors for human leptospirosis, particularly in tropical regions and on islands [12]. In just the three months following the 2017 hurricanes Irma and Maria, which caused major damage to infrastructure across Puerto Rico [13], the US Centers for Disease Control and Prevention (CDC) identified 85 leptospirosis cases, including 14 deaths, which was greater than the reported annual cases for any of the three previous years (2014–2016; 45–73 annual cases) (CDC personal communication).
Much current thinking suggest pathogenic leptospires are unable to multiply in the environment [14, 15] and are only maintained in animal reservoir hosts, such as rodents, livestock, and domestic dogs [16, 17]. Leptospires colonize the proximal renal tubules of infected animals and these reservoir hosts then excrete leptospires in their urine, thereby contaminating the environment wherein leptospires can transiently survive under moist conditions [18] and then infect other susceptible hosts [2, 19]. Transmission of leptospirosis to humans typically occurs via direct contact with infected animal urine or, more frequently, indirectly via exposure to contaminated soil and water [19]. Because indirect contamination via the environment is the most frequent source of human infection [15], leptospirosis is considered an environmentally-borne infection [20].
In recent years the use of the term “environmental reservoirs” for leptospirosis has increased, particularly when referring to moist soil in endemic regions [15], and the discovery of novel, pathogenic leptospires likely persisting in these environments (this study and [2, 21, 22]) supports the adoption of this term. Although moist soil conditions seem to be associated with environmental persistence of leptospires [23–25] little is known about the duration of survival in the environment [15, 26]. Environmental surveys are thus of critical importance toward our understanding of these environmental reservoirs and their role in the persistence and proliferation of pathogenic leptospires, as well as their potential role in disease transmission. To this end, we conducted environmental surveys in Puerto Rico after the two 2017 hurricanes to: 1) characterize the prevalence and geographic distribution of pathogenic Leptospira spp. in soil and water, 2) understand the diversity of pathogenic strains that occupy these environments, 3) investigate the possible persistence of pathogenic Leptospira spp. in moist soil, and 4) isolate and characterize pathogenic Leptospira spp. occuring in soil in Puerto Rico.
Methods
Ethics statement
All animal experimental procedures were performed in accordance with relevant guidelines and regulations, and as approved by United States Department of Agriculture institutional guidelines.
Sample collection
The focus of our environmental sampling efforts, conducted over three time periods (Dec 3–14, 2018; Feb 9–23, 2019; and Feb 13–14, 2020), were 12 municipalities in Puerto Rico (Fig 1) in which there is a potentially higher leptospirosis risk to humans based on previous human and canine leptospirosis cases and environmental factors, such as areas prone to flooding and poor sanitation [10]. We collected 440 samples (10 water and 10 soil per site) from 22 field sites within these municipalities (S1 Table); we targeted urban rivers and canals as well as runoff sites and/or flood zones near livestock farms where urine contamination from livestock was likely. At each site we established five short (<3m) linear transects and collected two soil samples (~50g each) and two water samples (>150mL each) per transect (sampling strategy illustrated in S1 Fig). For each transect, the first soil sample was collected from the edge of the water and the second soil sample was 1m back and directly perpendicular to the edge of the water. Likewise, the first water sample was collected at the edge of the water and the second water sample was collected 1m beyond and perpendicular to the edge of the water (S1 Fig). Soil samples were collected ≤10cm below the surface and placed into 50mL capacity conical tubes and, when possible, water samples were collected from stagnate or less turbulent areas of the subsurface water (10-30cm deep) into a Whirl-Pak. For site 22, collection in transects was not possible due to access constraints at this urban canal. At that site we collected 10 water and 10 soil samples within ≤1m of the edge of the water. For soil samples at all sites we recorded approximate elevation of the samples relative to the water, in meters. Soil elevation was categorized into four groupings: 0–0.33m, >0.33–0.66m, >0.66-1m, and >1m (S2 Table). Finally, we revisited one pathogenic Leptospira spp. positive site in San Juan (site 16) that was identified during collection period 1 (Dec 3–14) twice more, once in February 2019 and again in February 2020 (Fig 1). In February 2019 we collected 10 additional soil samples at this site in a linear transect along the edge of the water in the same location where the previous positive soil samples were collected. In February 2020, for the purpose of culturing attempts to obtain live leptospires, we collected 20 soil samples at this site clustered at the edge of the water (two clusters of 10 samples) where we detected a high abundance of pathogenic Leptospira DNA from the previous two collection periods.
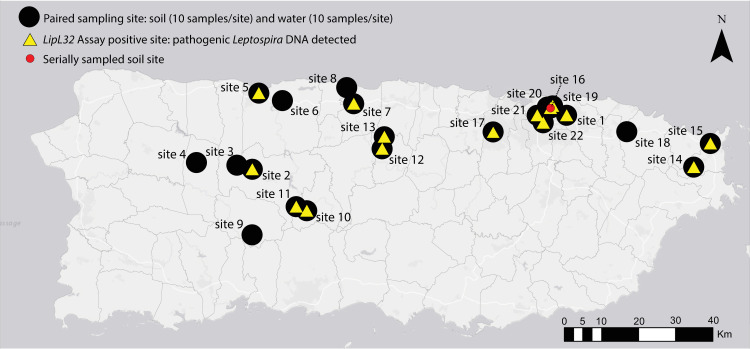
Fig 1. Map of Puerto Rico indicating sites where environmental samples were collected for the detection of pathogenic Leptospira spp. DNA from 2018–2020.
Black dots represent paired water and soil sites (n = 22); 20 samples were collected from each site (10 soil and 10 water). Yellow triangles indicate sites where pathogenic Leptospira DNA was detected (15/22 sites). One site in San Juan (site 16, red circle) was serially sampled three times over a 14-month period to assess persistence of pathogenic Leptospira in soil. This map was created using ArcGIS software by Esri. ArcGIS and ArcMap are the intellectual property of Esri and are used herein under license. Copyright Esri. All rights reserved. For more information about Esri software, please visit www.esri.com. Basemap: http://goto.arcgisonline.com/maps/Canvas/World_Light_Gray_Base.
Soil and water processing
Water samples (150mL) were filtered through a 250mL capacity 0.22μm nitrocellulose filter as previously describe [27]; afterward the filter was cut in half using sterile scissors and half of the filter was placed in a 5mL DNeasy PowerWater DNA bead tube (Qiagen, Valencia, CA, USA) and refrigerated at 4°C until shipment to the laboratory. Prior to filtering, 1mL of each homogenous water sample was placed in a 2mL capacity screw-cap tube for downstream pH testing. We collected pH values directly from the water sample using a 6mm glass pH probe (Cole-Parmer, Vernon Hills, IL, USA, Part # EW-55510) and from soil using methods outlined by Hall et al., 2019 [27] (S2 Table).
DNA extractions
DNA extractions were conducted in a class II A2 biosafety cabinet (BSC) as required by our USDA-APHIS-PPQ soil transport permit. DNA was isolated from water filters using DNeasy PowerWater DNA extraction kits (Qiagen, Valencia, CA, USA) following the recommendations of the manufacturer, and from ~0.5g of soil using Qiagen PowerSoil DNA extraction kits (Qiagen, Valencia, CA, USA) following previously published conditions [28] with one modification: after the addition of solution C3, the sample was incubated at 4°C overnight. DNA extractions were assessed for overall quality and bacterial abundance using a universal 16S real-time PCR assay as previously described [28]; this quality control step is necessary prior to conducting a pathogen detection PCR assay because it provides confidence that a negative result truly reflects the absence of pathogen DNA and not a technical failure associated with poor quality or low yield DNA extracts.
Leptospira qPCR detection and direct sequencing
All DNA extracts were screened in triplicate 10μL PCRs to detect the presence of DNA from pathogenic Leptospira spp. using the lipL32 TaqMan real-time PCR assay [1] containing the following: 1x TaqMan Environmental PCR master mix (Applied Biosystems, Foster City, CA, USA), 0.9μM of each primer, 0.45μM of the MGB probe, and 1μl undiluted DNA template. Quantitative PCRs were run interchangeably on Applied Biosystems QuantStudio (QS) QS7 and QS12 Real-Time PCR Systems with QS 12K Flex or QS Real-Time PCR software, as appropriate, under the following conditions: 50°C for 2 minutes, 95°C for 10 minutes, and 45 cycles of 95°C for 15 seconds and 58°C for 1 minute. Positive (L. interrogans strain Fiocruz L1-130) and non template controls were included on all runs. LipL32 amplicons from all positive samples were subjected to direct Sanger sequencing to confirm that the amplified product was Leptospira, using the same forward and reverse primers for the PCR (all replicates from a single sample were sequenced independently). Treatment and sequencing conditions are described below.
secY 203bp amplification
All samples that yielded a positive PCR result with the lipL32 assay (defined as amplification of at least 1 of 3 replicates with a Ct <45) were also subjected to PCR amplification of the secY gene; we amplified a 203bp fragment using the “SecYIVF” and “SecYIVR” primers designed to amplify pathogenic Leptospira spp. as described in Ahmed et al., 2009 [29]. PCRs were carried out in 10μL volumes containing the following reagents (given in final concentrations): 1μL of 1/25 diluted DNA template (none of the PowerWater extractions were diluted), 1x SYBR Green Universal master mix (Applied Biosystems, Foster City, CA, USA), 1.2M betaine, 0.2U Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) to improve efficiency, and 0.4μM of each primer. The assay was run on an Applied Biosystems 7500 Fast Real-Time PCR System with SDS 7500 software v2.0.6 under the following conditions: 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 57°C for 1 minute. Positive and non template controls were included on all runs. Due to the low abundance of Leptospira DNA in many of these samples, triplicate reactions were employed to increase the number of secY sequences generated; frequently only a single reaction (out of three) would yield an amplicon.
secY 549bp amplification
We also attempted to sequence a 549bp region of the secY gene for all lipL32 positive samples to gain more phylogenetic resolution for certain samples and increase the number of secY sequences generated for this dataset. We used primer pair F-ATGCCGATCATTTTTGCTTC and R-CCGTCCCTTAATTTTAGACTTCTTC, which were designed to amplify pathogenic Leptospira spp. as previously described [30]. Duplicate PCRs were carried out in 10μL volumes containing the following reagents (given in final concentrations): 1μL of 1/25 diluted DNA template (undiluted for all PowerWater extractions), 1x PCR buffer, 2.5mM MgCl2, 0.2mM dNTPs, 0.8U Platinum Taq DNA polymerase, and 0.4μM of each primer. PCRs were thermocycled according to following conditions: 95°C for 10 minutes to release the polymerase antibody, followed by 38 cycles of 94°C for 60 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, and a final extension step of 72°C for 10 minutes to ensure completion of the fragments. Positive and non template controls were included on all runs.
Sanger sequencing
For all three amplicons (lipl32-242bp, secY-203bp, and secY-549bp), PCR products were visualized on a 2% agarose gel to ensure that the products were of the expected size and treated with 1μL of ExoSAP-IT (Affymetrix, Santa Clara, CA, USA) added to 7μL of PCR product under the following conditions: 37°C for 15 minutes followed by 80°C for 15 minutes. Treated products were then diluted (based on amplicon intensity) and sequenced in both directions using the same forward and reverse primers from the PCR in a BigDye Terminator v3.1 Ready Reaction Mix (Applied Biosystems, Foster City, CA, USA). We used 10μL volumes for sequencing reactions containing the following reagents (given in final concentrations): 5x Sequencing Buffer, 1μL BigDye Terminator v3.1 Ready Reaction Mix, 1μM primer, and 5μL diluted PCR product. The following thermocycling conditions were used: 96°C for 20 seconds, followed by 30 cycles of 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 4 minutes. Sanger sequences were assembled and primers were manually removed using SeqMan Pro (DNASTAR Lasergene, Madison, WI, USA), resulting in sequence lengths of 202bp (lipL32-242bp), 162bp (secY-203bp), and 504bp (secY-549bp). FASTA database files for each gene (lipL32 and secY) were generated in BioEdit [31]; sequences from both secY amplicons (162 and 504bp) were combined into a single FASTA file.
lipL32 high fidelity amplicon sequencing
Sanger sequences from a subset of samples displayed multiple genotypes from replicate amplicons and/or heterogeneous nucleotide calls in a single sequence, suggesting multiple Leptospira spp. genotypes (i.e., mixtures). Also, within some lipL32 positive samples the concentration of Leptospira spp. DNA was too low to sequence using traditional Sanger sequencing methods. Thus, to recover sequences from these low-level positives and identify and characterize individual genotypes within these mixtures, we employed a high fidelity amplicon sequencing approach (AmpSeq) that enables differentiation of alleles without the need for traditional cloning and sequencing of the PCR amplicon while providing confidence in the detection and sequencing accuracy of low level positives [32]. We amplified the same 242bp fragment of the lipL32 gene described above in triplicate reactions using modified primers that incorporated universal tails [32]. The PCR setup and conditions for this modified lipl32 AmpSeq assay were the same as described above. Replicates were pooled and sequence libraries were prepared for all positive samples as previously described [32], except a 1x Agencourt AMPure XP (Beckman Coulter, Indianapolis, IN, USA) bead cleanup was used. Uniquely indexed sample libraries were pooled together in equimolar amounts and sequenced on an Illumina MiSeq instrument using a 500 cycle (2 x 250) MiSeq Reagent Kit v2 Nano with PhiX control (Illumina, San Diego, CA, USA, part# MS-103-1003). We sequenced all lipL32 positive soil and water samples (n = 86) in parallel on a single run.
Illumina sequence processing and lipL32 phylogenetic analysis
Paired-end Illumina reads were processed within QIIME 2 (v2019.7 and 2020.6) [33]; primers were trimmed with cutadapt [34]. The DADA2 [35] QIIME 2 plugin was used for quality trimming (--p-trunc-len-f 115 --p-trunc-len-r 110), denoising, dereplication of sequences, and chimera removal. LipL32 sequences were identified with BLASTN [36] searches of representative sequences against the NCBI nr/nt database (NCBI Resource Coordinators 2016) and BLAST databases consisting of lipL32 Sanger sequences generated from Leptospira positive samples as part of this study. LipL32 Illumina and Sanger sequences were combined with 538 publically available lipL32 sequences that were extracted from whole genomes (GenBank accession numbers in S3 Table) using BLASTN v2.9.0+ [37] (lipL32 sequence from L. interrogans strain Fiocruz L1-130 was used as a reference) and aligned with MUSCLE (v3.8.31) [38]. Phylogenetic analysis was conducted using MEGA version 7 [39] and a maximum likelihood phylogeny was inferred using the Tamura-Nei model; bootstrap values were calculated using 1,000 replicates.
secY phylogenetic analysis
Near full-length secY gene sequences (~1,300bp) were extracted from publicly available genome assemblies (S3 Table) with BLASTN (v2.2.29) [40]. These nucleotide sequences were combined with Sanger sequences generated from environmental samples in this study (162bp and 504bp amplicons) and aligned with MUSCLE (v3.8.31) [38]. A maximum likelihood phylogeny was inferred with IQ-TREE (v1.6.12) [41] using the best-fit model (TIM+F+I+G4) identified by ModelFinder [42] and the UFBoot2 ultrafast bootstrapping [43] and SH-aLRT [44] options.
Statistical analyses
We tested for associations between environmental and sampling variables and the detection of pathogenic Leptospira spp. in soil and water (S2 Table). Sampling variables were not available for 13 soil samples and site 22 was removed entirely from this analysis because the sampling strategy for this site was different from the other sites (see above). Sample sizes used for each test are listed in S4 Table. For categorical variables (sample type, location, and elevation), we used chi-square tests of independence, whereas for pH (a continuous variable) we used Wilcoxon Rank Sum (W) test because these data were not normally distributed. Categories for each environmental and sampling variable are described above under “Sample collection” and “Soil and water processing” and listed in S2 Table; p-values of <0.05 were considered significant. All analyses were conducted in R studio v3.6.1 with tidyverse and ggplot2 packages [45].
Soil culturing
Ten grams of soil samples were resuspended in 20mL of sterile water in 50mL conical tubes, vigorously vortexed, and allowed to settle for 15 minutes for sedimentation of solid matter. Supernatants were filtered through a 0.45μm filter as a selection step to remove larger bacteria, and 2mL of filtrate was used to inoculate either a 2x HAN [46] or 2x EMJH media (Difco, BD, Franklin Lakes, NJ, USA), both containing STAFF [47]. EMJH cultures were grown at 29°C and HAN cultures were prepared in duplicate for growth at both 29 and 37°C. Each culture was assessed daily by dark-field microscopy and, upon reaching high densities of growth (>108 spirochetes/mL), an aliquot was removed for flourescent antibody testing prior to storage in liquid nitrogen for downstream analysis.
Fluorescent antibody test (FAT)
Ten microliters of cultured samples were placed on a glass slide with a 7mm well as previously described [48]. Briefly, slides were air dried overnight and fixed in acetone for 15 minutes and then placed in a humid chamber; 50μL of rabbit anti-LipL32 sera (1:250) was added to each spot and then incubated for 1 hour at 37°C. Next, slides were washed for 10 minutes in PBS with gentle rocking and incubated with secondary Alexa Fluor 488 F(ab’)2 goat anti-rabbit IgG (Invitrogen, Waltham, MA, USA) for 1 hour at 37°C in the dark. After extensive washing, slides were dried and counterstained for 15 seconds with Flazo Orange (1:50, National Veterinary Services Laboratory). Slides were then rinsed with PBS and mounted using ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher, Waltham, MA, USA). Microscopic examination was done using a Nikon Eclipse E800 microscope and B2-A filter (excitation, 450–490 nm; emission, 520 nm) at 400× magnification [49].
ELISA
Cultured spirochetes from soil were recovered from liquid media (4,000 x g for 15 minutes), washed twice with PBS, and resuspended in 500μL of PBS. Cells were counted by dark field microscopy, suspensions were brought to a concentration of 5x107 spirochetes/mL, and 100μL (~5x106) was used to coat individual wells on ELISA plates in triplicate (Thermo Fisher, Waltham, MA, USA) for 16 hours at room temperature. Plates were then washed three times with PBS and blocked with PBS-containing 1% bovine serum albumin (BSA) for 1 hour at 29°C. For detection of pathogenic leptospires within the sample population, wells were incubated for 1 hour with polyclonal rabbit anti-LipL32 (1:1,000 in blocking buffer) followed by secondary horseradish peroxidase-conjugated goat anti-rabbit IgG (1:3,000). Wells were washed six times and reactivity was revealed by the addition of 100μL of TMB substrate (SeraCare, Milford, MA, USA) for 15 minutes. Reactions were stopped with 50μL of TMB STOP Solution and optical densities were taken at 620nm in a 96-well plate reader. Sample analysis was performed in triplicate. Positive and negative controls were comprised of wells coated with 100μL (containing 5x106, 2.5x106, 1.25x106, or 6.25x105) of L. interrogans serovar Copenhageni (LipL32 positive) and 100μL (5x106) L. biflexa serovar Patoc (LipL32 negative), respectively.
Selection and culture of pathogenic leptospires
ELISA and FAT-positive mixed cultures were enumerated by dark field microscopy and leptospires were diluted in HAN media to 104 cells/mL; 100μL was spread onto HAN agar plates supplemented with 0.4% rabbit serum [49] and incubated at 37°C until colonies were visible. Individual colonies were then harvested from the plates, vigorously homogenized in 100μL of HAN for microscopic visualization and, after confirmation of motile leptospires, 10μL of each suspension was used for coating FAT slides for LipL32 detection, as described above. The remaining sample was maintained at 29°C and, in the event of a positive FAT with anti-LipL32, used to inoculate fresh HAN media. Cultures were grown at 37°C, harvested at mid-log phase, and were further confirmed as pathogens by PCR with the lipL32 and secY-549bp primer sets described above. SecY amplicons were Sanger sequenced (as above) to identify and differentiate individual colonies.
WGS sequencing of isolated leptospires
Genomic DNA was extracted from isolated colonies using DNeasy kits (Qiagen, Valencia, CA, USA) according to the recommendations of the manufacturer, except the buffer AL incubation step occurred at 80°C for 1 hour. The gDNA was assessed for quality and quantity on a 0.7% agarose gel using λ DNA-HindIII Digest (New England Biolabs, Ipswich, MA, USA). WGS library construction was performed using the KAPA Hyper Prep Kits for Illumina NGS platforms per the manufacturer’s protocol with double-sided size-selection performed after sonication (KAPA Biosystems, Woburn, MA, USA, part# KK8504). The adapters and 8bp index oligos purchased from IDT (Integrated DNA Technologies, San Diego, CA, USA), based on Kozarewa and Turner, 2011 [50], were used in place of those supplied in the KAPA preparation kit. The final libraries were quantified on an Applied Biosystems QuantStudio 7 Flex Real-Time PCR System using the KAPA SYBR FAST ROX Low qPCR Master Mix for Illumina platforms (part# KK4873). The libraries were then pooled together at equimolar concentrations and quality was assessed with a Bioanalyzer High Sensitivity DNA kit (Agilent Technologies, Santa Clara, CA, USA, part# 5067–1504). Final quantification by qPCR preceded sequencing of the final library. The samples were sequenced on an Illumina MiSeq using the 600-cycle v3 kit (part# MS-102-3003) with the standard Illumina procedure. The appropriate sequencing primers were added to the MiSeq kit as previously described [50].
WGS phylogenetic analysis
External genomes
Publically available Leptospira genomes were downloaded from the GenBank assembly database [51] with the ncbi-genome-download tool (https://github.com/kblin/ncbi-genome-download) on September 9th, 2021. Genomes were renamed to include the assembly accession and taxonomic information. The dataset resulted in a set of 802 reference genomes that included pathogenic, intermediate, and saprophytic leptospires (S3 Table).
Genome assembly
Raw reads were trimmed with bbduk.sh (v38.92) (https://sourceforge.net/projects/bbmap/) and assembled with SPAdes (v3.13.0) [52]. Reads were mapped back against contigs with minimap2 (v2.22) [53] and the depth of coverage was calculated with Samtools (v1.13) [54]. Two hundred nucleotides from each contig were aligned against the GenBank nt database with BLASTN (v.2.11.0) [37] and the taxonomy of the top hit was recorded. Contigs with an anomalously low depth of coverage or aligned against contaminants were manually removed.
Leptospira dendrogram
Pairwise MASH distances [55] were calculated on all Leptospira genomes and a dendrogram was generated with mashPy (https://gist.github.com/jasonsahl/24c7cb0fb78b4769521752193a43b219), a tool that incorporates SciPy [56] and Skbio (http://scikit-bio.org).
Average nucleotide identity
Average nucleotide identity (ANI) between the novel Leptospira spp. described herein and L. yasudae strain 201601115 was calculated using the ANIb stat in PYANI (v0.2.11) as described previously [57].
SNP and indel discovery
Genome assemblies were annotated with Prokka (v1.14.6) [58]. Reads were aligned against each reference and SNPs and indels were called with Snippy (v4.6.0) (https://github.com/tseemann/snippy) using default parameters.
Core genome SNP phylogenies
For pathogenic genomes, assemblies were aligned against Leptospira kmetyi strain LS-001/16 (GCA_003722295.1) with NUCmer (v3.1) [59] and SNPs were called with NASP (v1.2.0) [60]; SNPs that fell within duplicated regions, based on a reference self-alignment with NUCmer, were filtered from downstream analyses. A maximum likelihood phylogeny was inferred on the concatenated SNP alignment with IQ-TREE (v2.0.3) [61] in conjunction with ModelFinder [42] and rooted with Leptospira interrogans FDAARGOS 203 (GCA_002073495.2). The reference genome, Leptospira montravelensis strain 201800278 (GCA_004770045.1), was used for the phylogeny including saprophytic genomes, which was rooted by Leptospira noumeaensis strain 201800287 (GCA_004770765.1).
Hamster infection with isolated colonies
To assess virulence of the newly identified pathogenic isolate described herein, Golden Syrian hamsters were inoculated by intraperitoneal (IP) injection with 108 leptospires in 1mL as previously described [62]. Negative control hamsters received 1mL of media alone. Animals were assessed for acute disease through day 11 post infection and euthanized on day 21 to provide ample time for renal carriage to develop.
Microscopic agglutination test (MAT)
The MAT was performed according to OIE guidelines [63] at two-fold dilutions from an initial dilution of 1:50 to 1:6400 using 18 serovars of Leptospira spp. listed in S5 Table and the newly identified pathogenic isolates described herein.
Culture
At three weeks post-infection, kidneys were harvested using aseptic techniques for culture of leptospires in T80/40/LH semi-solid media incubated at 29°C [64] and HAN semi-solid media incubated at both 29 and 37°C [46].
Results
Pathogenic Leptospira detection
Pathogenic Leptospira spp. were common and distributed widely throughout Puerto Rico. Using lipL32 PCR, we detected pathogenic Leptospira spp. DNA at 15 of 22 (68.2%) paired soil and water exploratory sites (Fig 1); 8/15 sites yielded positive soil samples only, 2/15 positive water samples only, and 5/15 both positive soil and water samples (S2 Fig). From these exploratory sites, 63 samples (out of 440 total) were positive, including 15 water and 48 soil samples. At the serially sampled soil site (site 16), an additional 23 (out of 30 total) soil samples collected during the second and third visits to this site also were positive. In total, 86 positive samples from Puerto Rico were detected. At five paired soil and water sites (sites 7, 12, 17, 21, and 22) we detected pathogenic Leptospira spp. in both sample types (Table 1 and S2 Fig). At four of these five sites (12, 17, 21, and 22) a higher proportion of soil samples were positive than water samples and the phylogenetic clades represented in the water samples were a subset of those identified in the corresponding soil samples (Table 1). This observation was not possible at site 7 because only a single soil and a single water sample were lipL32 positive and lipL32 sequence was not recovered from the positive soil sample (S1 Table).
Table 1. All positive sites in Puerto Rico illustrating the relationship between lipL32 genotypes found in soil and water.
In total, 15 sites were positive for pathogenic Leptospira DNA; eight sites wherein only soil was positive, two sites wherein only water was positive, and five sites wherein both soil and water were positive (highlighted in gray). At four of the latter sites, the clades present in water were a subset of the clades identified in soil (bolded text), suggesting that soil reservoirs may lead to the contamination of water.
| Site ID | Site Type | Proportion Positive | Clades |
|---|---|---|---|
| 1 | Water | 0.1 | 4b |
| 2 | Soil | 0.2 | 4a |
| 5 | Water | 0.7 | 1, 6 |
| 7 | Water | 0.1 | 6 |
| 7 | Soil | 0.1 | unknown |
| 10 | Soil | 0.1 | 4a |
| 11 | Soil | 0.2 | 4a |
| 12 | Water | 0.1 | 4b |
| 12 | Soil | 0.6 | 3, 4a, 4b, 6, 9 |
| 13 | Soil | 0.1 | 6 |
| 14 | Soil | 0.4 | 3, 4a, 4b, 9 |
| 15 | Soil | 0.3 | 4b |
| 16 | Soil | 0.7 | 4b, 6 |
| 17 | Water | 0.1 | 6 |
| 17 | Soil | 0.4 | 3, 4b, 6 |
| 19 | Soil | 0.3 | 4b, 6 |
| 21 | Water | 0.1 | 6 |
| 21 | Soil | 0.9 | 4a, 4b, 6, 9 |
| 22 | Water | 0.3 | 4b, 6 |
| 22 | Soil | 0.5 | 4b, 6, 9 |
Gene sequencing lipL32
Sequencing of the lipL32 amplicons revealed extensive and previously undescribed diversity in pathogenic Leptospira spp. at this locus. Using AmpSeq and Sanger sequencing methods, we generated 147 lipL32 sequences from 83 of the 86 positive samples representing all 15 positive sites (S1 Table) to: 1) confirm that the generated amplicons were from Leptospira DNA, and 2) assess genetic diversity of pathogenic Leptospira spp. within and across sites and samples; information on generated nucleotide sequences is provided in S6 Table. Thirty-four alleles were identified in this dataset from Puerto Rico of which 32 were novel (i.e., were not a 100% nucleotide match to any available sequences in GenBank; the remaining two alleles were a 100% match to L. gomenensis and L. yasudae). We assigned either a high or moderate level of confidence to each newly identified allele. If a sequence was observed in more than one sample or was generated via Sanger sequencing methods, this allele was assigned a high level of confidence; 20 of 34 alleles met this criterion. The remaining 14 alleles were only observed in a single sample, only in the Illumina dataset, and/or with a low depth of coverage (3–42 paired end reads) and thus were assigned a moderate level of confidence. As a quality control step, we also translated all the lipL32 sequences in frame to confirm that they resulted in functional amino acid sequences. Although the dual indexing and paired end analysis approach described above provides additional confidence in the accuracy of these sequences, we acknowledge that these putative alleles could benefit from further validation. As such, no major phylogenetic conclusions are inferred from these 14 lower confidence alleles and none of these alleles define the novel lipL32 clades described herein. However, all 34 alleles were used to illustrate the diversity of pathogenic Leptospira spp. genotypes present in Puerto Rico soil and water.
Gene sequencing secY
Unlike lipL32, which has only been documented in pathogenic leptospires, the secY gene is present in both pathogenic (P1 and P2) and saprophytic (S1 and S2) Leptospira groups [2, 65]. It is also commonly used to obtain Leptospira spp. identification from clinical samples. Thus, to compliment the lipL32 sequence data and provide support for phylogenetic assignment to the group P1 pathogenic clade and determine the relationship to other pathogenic leptospires, we generated secY sequences for these environmental samples. This was an important quality control step because diverse alleles that have not previously been described from the lipL32 gene were observed in these samples. Twenty-one secY-203bp sequences, representing 13 alleles, from 16 samples (3 water and 13 soil), and 29 secY-549bp sequences, representing 19 alleles, from 23 samples (2 water and 21 soils), were generated (S1 Table). In combination, secY sequences were generated from 32/86 positive samples representing 10/15 sites. All sequences fell within the pathogenic group of the secY phylogeny (S3 Fig), thereby confirming the lipL32 results; nucleotide sequences are provided in S7 Table.
Mixed pathogenic genotypes within samples
Sequences generated for the lipL32 and secY genes revealed mixtures (multiple pathogenic genotypes within a single sample) in 39 soil and two water samples from 12 sites; up to five unique lipL32 genotypes in a single sample were identified (S1 and S6 Tables).
Phylogenetics
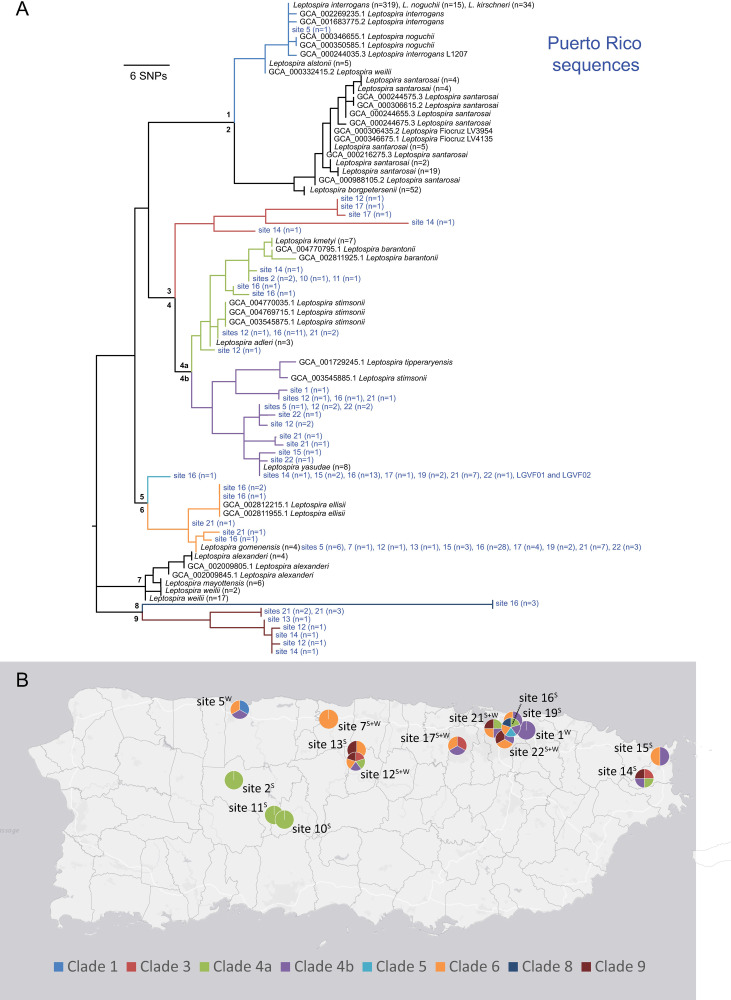
Phylogenetic analyses of lipL32 sequences revealed a high level of genetic diversity among these samples, including four previously undescribed pathogenic Leptospira phylogenetic clades (3, 5, 8, and 9; Figs 2 and S4); 18 sequences obtained from the environment in Puerto Rico assigned to these four clades. Clades 4 and 6 contain 128 lipL32 sequences from Puerto Rico as well as environmentally acquired isolates from other locations [2]. The two most common lipL32 genotypes from soil and water in Puerto Rico were a perfect match to L. gomenensis in clade 6 (n = 56) or a close match to L. yasudae (previously L. dzianensis) in clade 4b (n = 28) (Figs 2 and S4). Interestingly the L. gomenensis genotype was recently described in New Caledonia and L. yasudae (L. dzianensis) in Mayotte [2], suggesting these environmental Leptospira spp. are distributed globally. The secY phylogeny provided additional confidence in the phylogenetic placement of these novel types because those genotypes were also a close match to L. yasudae and L. gomenensis (S3 Fig). Interestingly, there also was sequence similarity to L. santarosai for some secY genotypes, including those from the newly acquired LGVF01 and LGVF02 isolates that are described herein (S3 Fig). Clades 1, 2, and 7 of the lipL32 phylogeny (Figs 2 and S4) contain described pathogenic leptospires that are commonly found in human and animal infections: L. interrogans, L. noguchii, and L. kirschneri (clade 1); L. borgpetersenii and L. santarosai (clade 2); and L. alexanderi, L. weilii, and L. mayottensis (clade 7). A single sequence from site 5 assigned to clade 1 (Figs 2 and S4) and no sequences from Puerto Rico assigned to clades 2 and 7.
Fig 2. Genetically diverse and previously undescribed lineages of pathogenic Leptospira are widespread in Puerto Rico.
A) Maximum likelihood phylogeny constructed using a 202bp segment of the lipL32 gene, including sequences obtained from soil and water samples collected in this study (blue text) and those available in public databases (black text). Nine major genetic clades are represented, seven of which were present in Puerto Rico, including four not previously described. Clades are labelled 1 through 9 on the tree and color coded. B) Sites where each major clade was collected, with colors within the pie charts corresponding to those in the phylogeny; superscript indicates if soil (S) and/or water (W) samples were positive at each site. Fifteen positive sites are included on this map. This map was created using ArcGIS software by Esri. ArcGIS and ArcMap are the intellectual property of Esri and are used herein under license. Copyright Esri. All rights reserved. For more information about Esri software, please visit www.esri.com. Basemap: http://goto.arcgisonline.com/maps/Canvas/World_Light_Gray_Base.
Detection statistics
We observed a significant association between the distribution (median) of pH values for samples collected from water that were positive (median pH = 8.4) and negative (median pH = 8.2) for pathogenic Leptospira spp. (W = 550, p = 0.0015), but not from soil, where positive and negative median pH values were ~7.5 regardless of pathogenic Leptospira status (W = 3057, p = 0.9399). We also detected pathogenic Leptospira spp. in soil more commonly than in water: 48/210 (22.9%) of soil samples compared to 15/210 (7.1%) of water samples (x2 = 15.087, p<0.001). No other significant associations were observed among environmental/sampling variables and the detection of pathogenic Leptospira spp. (S4 Table).
Persistence in soil
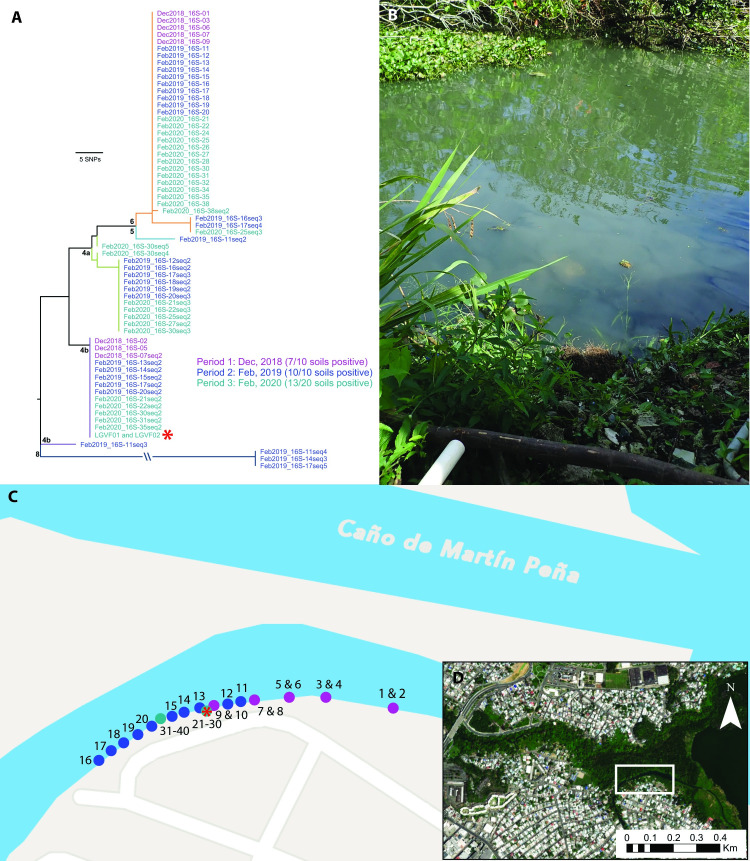
During our first sampling period (December 2018) we identified one exploratory site in an urban district of San Juan where seven soil samples (out of ten) tested positive for the presence of pathogenic Leptospira spp. DNA (S1 Table), the highest single site detection rate from this first period. We revisited the site two months later (February 2019) and collected an additional ten soil samples in a linear transect along the edge of the water to test the hypothesis that pathogenic Leptospira spp. persist in water-soaked soil at this site; all ten of the new soil samples tested positive for pathogenic Leptospira spp. DNA (S1 Table). We revisited this site once again 14 months after the initial sampling period (February 2020) and collected 20 samples clustered along the edge of the water to use for live Leptospira spp. recovery and isolation attempts, as well as persistence analysis; 13 of these 20 soil samples tested positive for pathogenic Leptospira spp. DNA (S1 Table). Thus, at all three time points the majority of soil samples collected at this site were positive for pathogenic Leptospira spp. DNA: 7/10, 10/10, and 13/20 of samples, respectively (Fig 3). Among these positive samples, identical LipL32 sequences from clades 4b and 6 were identified across all three time points (Fig 3A), suggesting these genotypes persisted in soil at this site.
Fig 3. Site 16 in Caño Martín Peña, San Juan, Puerto Rico where sampling was conducted at three separate timepoints to assess environmental persistence of pathogenic Leptospira in soil near the edge of the water.
A) Maximum likelihood phylogeny of all lipL32 sequences obtained from this soil site over the course of three sampling periods (color coded by date of sampling period); two of five clades detected at this site were identified at all three timepoints, suggesting environmental persistence. Clade colors correspond to Fig 2. Two isolates representing a novel pathogenic Leptospira species were obtained from soil sample 27 (red asterisk). B) Photo of exact location where samples 21–30 were collected and the isolates were obtained. C) Transect where 40 soil samples were collected over three sampling periods. Circles are color coded to indicate sampling period as in the phylogeny (panel A) and the red asterisk denotes the location where the Leptospira isolates originated. D) Location of site 16 in Caño Martín Peña. The maps in panels B and C were created using ArcGIS software by Esri. ArcGIS and ArcMap are the intellectual property of Esri and are used herein under license. Copyright Esri. All rights reserved. For more information about Esri software, please visit www.esri.com. Basemap for panel C: Community, https://www.arcgis.com/home/item.html?id=273bf8d5c8ac400183fc24e109d20bcf. Basemap for panel D: World Imagery (WGS84), https://www.arcgis.com/home/item.html?id=898f58f2ee824b3c97bae0698563a4b3.
Quantification of pathogenic Leptospira spp. in soil cultures
Single aliquots of culture from FAT-positive soil sample #27 and FAT-negative soil sample #40 (both from site 16) were defrosted in a water bath at 37°C and used for a confirmative FAT and for coating onto ELISA plates. As expected, sample #27 presented with a high proportion of LipL32-reactive leptospires (S5A Fig), which was quantified by ELISA (S5B Fig); approximately 50% of the mixed population was pathogens. In contrast, only basal reactivity was obtained when wells were coated with cells from sample #40, consistent with a small number, or absence of, pathogenic leptospires. Cultures were seeded onto HAN plates and incubated at 37°C in a 3% CO2 atmosphere; colonies were typically observed in five days. Colonies were picked from both plates (27 colonies from sample #27 and 54 from sample #40), vigorously homogenized in liquid HAN, and the suspensions used to coat FAT slides. In strong agreement with ELISA results, 14 out of 27 colonies from sample #27 were FAT positive (51.9%), whereas none of the 54 colonies from sample #40 showed significant LipL32 reactivity. Representative FAT images of positive (colony 9 and 17) and negative (colony 8) clonal suspensions from sample #27 are presented in S5C Fig. FAT-positive cells were inoculated into liquid HAN and harvested at mid-log phase for PCR confirmation, wherein colony 8 was also employed as a negative control. All 14 FAT-positive cultures were also PCR-positive for both lipL32 and secY and, as expected, no bands were observed when cells from colony 8 were used as a template (S5D Fig).
Sequence identification of pathogenic colonies
All secY amplicons were sequenced and the assembled sequences (446bp in length after trimming) were submitted to nucleotide alignment by BLASTN [36]. The 14 clones presented an average identity of 95% to L. santarosai serovar Princestown strain TRVL 112499. We identified two clones (colony 9 and 18) that were slightly divergent from the other 12 and contained several SNPs within the secY sequence (S3 Fig and S7 Table). One representative isolate of each genotype, designated as strain LGVF01 and strain LGVF02, respectively, were re-grown in liquid HAN for DNA extraction and whole genome sequencing. As a quality control step we also sequenced the lipL32 gene from these isolates and confirmed that they matched sequences we obtained from the originating soil samples.
Isolation of other leptospires
A total of 40 saprophytic isolates also were obtained during the purification process for pathogenic colonies. We sequenced near full length 16S rRNA fragments (1,330bp) for these isolates using universal primers [66] and identified two genotypes among them that differed by four SNPs. One genotype (16S-01) was a perfect match to L. mtsangambouensis, L. jelokensis, L. noumeaensis, L. congkakensis, L. levettii, and L. macculloughii and the other (16S-02) was a perfect match to L. bandrabouensis, L. kemamanensis, L. bouyouniensis, and L. meyeri; three representatives of each genotype were whole genome sequenced (S8 Table).
WGS
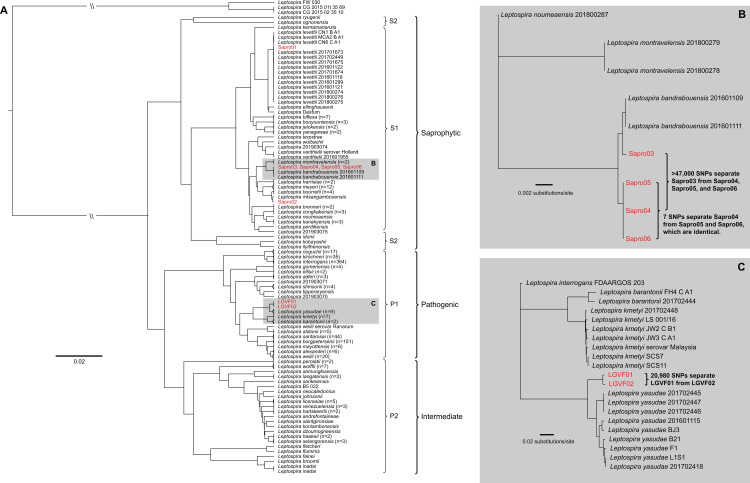
In total, we generated whole genome sequences for two pathogenic and six saprophytic isolates obtained from a single soil sample (#27) at the serially sampled site 16 (Fig 3); the former are two distinct strains of a novel pathogenic Leptospira species that differ from each other by 20,980 pairwise SNPs (4,982 non-synonymous) (Fig 4B). This novel pathogen species falls within clade 4b of the lipL32 phylogeny (Figs 2 and S4) and is most closely related to, but is clearly distinct from, L. yasudae; the ANI between these species is 92.8%. Samples from five other positive soil sites and one water site from eastern Puerto Rico (sites 14, 15, 17, 19, 21, and 22) share the same lipL32 genotype found in this novel pathogenic species, suggesting it may be fairly common and widespread in Puerto Rico (Fig 2). The six saprophytic isolates represent three previously known species of Leptospira: they were genomically identified as L. levettii (n = 1), L. bandrabouensis (n = 4), and L. mtsangambouensis (n = 1) (Fig 4). The 16S-01 genotype was conserved among the L. levettii, L. mtsangambouensis, and one of the L. bandrabouensis isolates, whereas the 16S-02 genotype was conserved among the remaining three L. bandrabouensis isolates (S8 Table). The L. bandrabouensis isolate (Sapro03) that contained the 16S-01 genotype differed from the other three L. bandrabouensis isolates by >47,000 SNPs (Fig 4B). The genome assembly details for all eight isolates are provided in S9 Table.
Fig 4. Whole genome dendrogram of 802 known pathogenic, intermediate, and saprophytic Leptospira spp. isolates, including six saprophytic and two pathogenic isolates obtained in this study from a single soil sample from site 16.
A) Pairwise genomic distance dendrogram contextualizing the relationship among all known saprophytic (S1 and S2), intermediate (P2), and pathogenic (P1) Leptospira spp. reveals the pathogenic isolates LGVF01 and LGVF02 represent two genotypes of the same previously undescribed pathogenic species, whereas the saprophytes belong to three known Leptospira spp. B) Detailed view of the S1 clade that contains four saprophytic isolates obtained during this study. This maximum likelihood phylogeny reveals significant diversity among isolates that fall within the L. bandrabouensis clade. C) Detailed view of the P1 clade that contains two pathogenic isolates obtained during this study. This maximum likelihood phylogeny reveals two genotypes of a novel pathogenic Leptospira spp. isolated from soil in Puerto Rico.
Hamster infection and MAT response
MAT reactivity was detected when sera from hamsters infected with the new pathogenic isolate LGVF02 were tested against the challenge isolate (LGVF02) at a 1:1600 dilution. However, no experimentally infected hamsters showed clinical signs of infection or weight loss after inoculation (S10 Table) and at three weeks post-infection all hamster kidneys were culture negative. In addition, at three weeks post-infection all hamsters were seronegative for the 18 serovars, representative of 15 serogroups, listed in S5 Table.
Discussion
This study provides new insights into the diversity of pathogenic leptospires detectable in the environment in Puerto Rico. It raises important questions regarding persistence in the environment, host/environmental adaptation, genomic exchange, the evolution or acquisition/loss of virulence associated genes and pathogenicity in the genus Leptospira, and the disease potential of environmental leptospires. Our results also demonstrate the need for updated diagnostics to detect and identify these novel and diverse types to better understand their potential role in disease.
Our survey for pathogenic Leptospira spp. in the environment in Puerto Rico revealed that diverse and novel pathogenic types that assigned to the P1 phylogenetic group are widespread in soil and water across the island, and more commonly detected in soil than water. Thirty-two previously undescribed lipL32 alleles were identified, which suggests a vast amount of uncharacterized diversity within the pathogenic P1 group of this genus that is present in the environment in Puerto Rico. However, we only identified a single sequence from one water sample that shared a lipL32 genotype with Leptospira spp. from clade 1 of the P1 group (Figs 2 and S4); species from clade 1 (e.g., L. interrogans, L. kirschneri, and L. noguchii) are commonly identified from human leptospirosis infections. This might suggest that clade 1 types of the P1 pathogenic group are not well adapted for survival in soil and water. Indeed, host adaptation among these commonly identified pathogenic types has been suggested [4, 67], which could explain the limited detection of these pathogenic types in soil and water in this study, even though they previously have been reported from human cases in Puerto Rico [68]. This is in line with the described transmission dynamics of human leptospirosis in which infecting clade 1 types are maintained in the kidneys and renal tubules of animal reservoir hosts and excreted through urine whereby only transient contamination of soil and water can occur but still lead to human infections. Alternatively, the limited detection of clade 1 pathogenic types in soil and water also could be due to their relative rarity in these sample types.
In contrast, we identified 33 less common or novel lipL32 alleles in these environmental samples whose disease potential is unknown. Although the pathogenicity of many of these novel types is yet to be determined, the presence of the lipL32 gene along with phylogenetic placement in the pathogenic P1 group using lipL32, secY, and whole-genome sequences suggest pathogenic potential in humans and animals. Of course it is also possible that the presence of the lipL32 gene in these strains is not indicative of pathogenicity but perhaps an artifact of genetic exchange and recombination and/or serves some other purpose in these complex environments. Indeed it has been shown that the LipL32 protein is not required for infection with L. interrogans [69], and furthermore, the presence of LipL32 in intermediate P2 group leptospires [65] that are only occasionally associated with disease [70] highlights the need to use caution when inferring pathogenicity based upon the presence of LipL32 alone. Either way, the abundance and possible persistence of these types in soil and water suggests adaptation for survival, and potentially proliferation, in these environments.
In support of this hypothesis, we found evidence for lipL32 positive Leptospira spp. in water-soaked soil near the edge of the water for more than one year at an urban site in San Juan, Puerto Rico. Two of the five pathogenic (P1) genetic clades found in soil at this site were resampled again at two months and 14 months after the initial sampling date. Although persistent survival and proliferation in this environment seems the most likely explanation, possibly via formation of biofilms [71, 72], it is also possible that this site is continually contaminated from some other source. This same urban site was described in a 2015 study wherein the research team evaluated Leptospira seroprevalence in humans and carriage in rodents [73]. It was determined in that study that Leptospira exposure was high in humans in this urban community (seroprevalence of 27.2%) but the species/serotypes identified in humans and the species identified and/or recovered from rodents in that study were not a match to those found in our study but, rather, were the commonly identified human and animal pathogens L. borgpetersenii and L. interrogans. However, it is important to point out that seroprevalence testing for this other study employed a MAT panel containing only common human and animal pathogenic serogroups (27 serovars represented), so if exposure from these novel soil dwelling types had occurred they may not have been detected with this method. It is interesting to speculate that exposure to these environmental types, if in fact they are low-virulent, could provide some primed immunity to known pathogenic types like L. interrogans and L. borgpetersenii; a question that potentially could be addressed in this urban community with updated diagnostics that include the novel types.
Our discovery of many novel pathogenic genotypes at this urban site (n = 8) and in Puerto Rico as a whole (n = 32) highlights the need to address the limited scope of leptospirosis diagnostics (e.g., an existing MAT panel that does not include all local strains), as it is possible that these novel pathogenic types also are associated with human and animal disease in Puerto Rico but remain undetected in the clinic. Indeed, it is thought that many cases of leptospirosis go undetected or are misdiagnosed as other febrile illnesses due to challenges with implementation and/or limitations of the diagnostic tools themselves [74, 75]. If it were shown that these novel types also contribute to human and animal disease and proliferation in the environment is experimentally confirmed, it would contradict the current dogma that all pathogenic leptospires require an animal reservoir host for maintenance and proliferation and, thus, suggest a much more complicated scenario of leptospirosis transmission that includes infection by soil dwelling leptospires.
We generated whole genome sequences for two pathogenic and six saprophytic Leptospira isolates obtained from a single soil sample collected from serially sampled site 16. The whole genome dendrogram contextualized the phylogenetic relationships of these isolates to all other known Leptospira spp. and revealed that the two pathogenic isolates, LGVF01 and LGVF02, were unique strains representing a single, novel pathogenic species. We classified these isolates as pathogenic based upon the presence of the lipL32 gene in their genomes and the expression of the LipL32 protein, as well as their whole-genome phylogenetic relationships to other pathogenic species. The saprophytic isolates represent three known species, of which two (L. bandrabouensis and L. mtsangambouensis) have previously only been identified from the island of Mayotte in the Indian ocean, and the third (L. levettii) has previously been identified only in Mayotte, Malaysia, and New Caledonia. In other words, these three species have not, to our knowledge, previously been identified in Puerto Rico, the Caribbean, or the Western Hemisphere. In total, four Leptospira species were isolated from a single 10g soil sample from site 16 in Puerto Rico, and the lipL32 analysis from this same site identified four additional phylogenetic clades that were not represented by the isolates obtained from this sample (Fig 3A), suggesting the presence of even more pathogenic species in soil from this site. Finally, the SNP diversity identified among isolates within the same species from a single soil sample is considerable (20,980 SNPs between LGVF01 and LGVF02 and >47,000 SNPs differentiating two subgroups of L. bandrabouensis). Together, these findings reveal a tremendous amount of unrecognized Leptospira spp. diversity within the environment in Puerto Rico, and very likely many other geographic locations, that have yet to be characterized.
The novel pathogenic Leptospira spp. we isolated in San Juan, represented by strain LGVF02, was confirmed to express LipL32 in vitro. Sera from infected Golden Syrian hamsters were positive by MAT when using this challenge strain, although no signs of acute disease were observed. Interestingly, sera from challenged hamsters were MAT negative when tested against a panel of 18 commonly tested strains representing 15 serogroups; as such, we suspect that this isolate represents an entirely novel serogroup and, thus, would not be detected using a MAT panel of common isolates. Importantly, the absence of disease symptoms in challenged hamsters does not negate its classification as a pathogen; other pathogenic Leptospira spp., including L. borgpetersenii, L. mayottensis, and L. tipperaryensis, have also failed to cause acute disease in Golden Syrian hamsters [76, 77]. LGVF02 was also subjected to LipL32 gene silencing via CRISPRi as part of another study [49] and the results of that study (specifically, the detection of the LipL32 and LipL41 proteins) further support the pathogenic classification of this novel leptospire. The species description of LGVF02 will be treated in a separate publication in which we will investigate whether it represents an entirely new serogroup. Either way, the ability of strain LGVF02 to evade detection with MAT based on common isolates again highlights the need to include regionally relevant isolates when conducting MAT in the clinic to ensure detection of these lesser known types and better understand their potential role in human and animal disease.
Soil may serve as a long-term reservoir for contamination of waterways with pathogenic Leptospira spp. in Puerto Rico. At four of the five sites where leptospires from the pathogenic P1 group were detected in both soil and water, the Leptospira clade(s) identified in water were a subset of those identified in soil and a higher proportion of soil samples were positive when compared to water samples at these sites. These observations suggest soil may serve as a reservoir for Leptospira from the pathogenic P1 group that then leads to the contamination of waterways in Puerto Rico, which could occur when the soil is disturbed by animal or human activity, or during weather events such as heavy rains and floods. This is in line with the dynamics of Leptospira environmental contamination reviewed elsewhere [15]. However, our findings also provide support for the hypothesis that some Leptospira from the pathogenic P1 group are maintained in the soil long term, rather than just transiently being present due to urine contamination from an infected reservoir host. If this is the case, the potential for genetic exchange among pathogenic isolates that could lead to the emergence of novel outbreak strains is something that needs to be explored further. Although speculative, a possible example of this may be the documented outbreak between 1999–2003 in Thailand that was associated with a novel type of L. interrogans that emerged, caused illness in humans for several years, and then inexplicably seemed to fade out [78]. It is possible this type of outbreak is due to the exchange of genetic material among transiently present (via urine contamination) and soil dwelling pathogenic leptospires that result in increased pathogenic potential of certain strains in these complex environments. Although we did detect pathogenic Leptospira from clade 1 of the pathogenic P1 group at a water site (site 5) that also contained two novel pathogenic types, a genomic analyses among these types was not possible because no isolates were obtained. However, a comprehensive comparative genomics study conducted by Xu et al., 2016 [4] revealed that horizontal gene transfer events resulting in the acquisition of novel genes in Leptospira are fairly common. It has also been suggested that loss of virulence may be a feature of environmentally adapted leptospires after excretion into the environment from reservoir hosts [21].
Overall, this study provides evidence for the existence of diverse lineages of soil dwelling Leptospira from the pathogenic P1 group in Puerto Rico; the disease potential of these novel types in humans and animals is unknown. The genetic diversity within this genus found in the environment in Puerto Rico is vast and, thus, there is a need to characterize this diversity to improve our understanding of the disease agents of leptospirosis. Many cases of leptospirosis are thought to go undetected, or are misdiagnosed as other febrile illnesses, and it is possible that these novel types play a role in such cases. It is also interesting to consider that immunity could be provided by exposure to low-virulent types. A follow up study in Puerto Rico to investigate the possibility that these novel types are found in human and animal samples would be highly valuable. Furthermore, environmental surveys aimed at the discovery of these novel types in all areas where leptospirosis is a public health concern will be of critical importance towards improving diagnostics, our understanding of leptospirosis disease agents, and public health outcomes.
Supporting information
Black and blue stars represent soil and water samples, respectively. Transects were a minimum of 2.5 meters apart. This sampling design was also applied in a linear fashion for the sampling of rivers and streams.
(PDF)
The yellow triangles indicate sites where pathogenic Leptospira DNA was detected. Eight soil sites were positive for pathogenic Leptospira DNA, compared to two water sites; both soil and water were positive at five sites. Site 16, which was used to assess environmental persistence in soil is indicated with a red circle. This map was created using ArcGIS software by Esri. ArcGIS and ArcMap are the intellectual property of Esri and are used herein under license. Copyright Esri. All rights reserved. For more information about Esri software, please visit www.esri.com. Basemap: http://goto.arcgisonline.com/maps/Canvas/World_Light_Gray_Base.
(PDF)
Sequences generated from soil and water samples collected in Puerto Rico are in blue or pink text, the novel pathogenic Puerto Rico isolates from soil are also included; reference sequences for pathogenic, intermediate, and saprophytic Leptospira are in black. Bootstrap/aLRT support values are indicated on branch nodes.
(PDF)
Sequences generated from soil (blue text) and water (pink text) samples collected in Puerto Rico are displayed. Reference sequences representing all other known pathogenic Leptospira spp. are in black. Nine major clades are represented of which seven were identified in Puerto Rico and four of those (3, 5, 8, and 9) have not been described previously. Identical genotypes were present in soil and water from Puerto Rico at two sites (17 and 21), with both occurrences representing a single genotype from clade 6 (in orange). Bootstrap values are indicated on each branch. Major clades are numbered 1 through 9 and color-coding matches Fig 2.
(PDF)
LipL32 PCR positive (Soil #27) and negative (Soil #40) samples were subjected to FAT (Panel A) and ELISA (Panel B) along with positive and negative controls (L. interrogans and L. biflexa, respectively) to assess expression of the LipL32 pathogenicity protein. Individual colonies from Soil #27 were subjected to additional FAT testing (Panel C) and confirmatory PCR (Panel D) to verify that the obtained isolates were pathogenic.
(PDF)
Summary of detection and sequencing results for environmental samples from Puerto Rico, 2018–2020.
(XLSX)
Sample and site ID, location, elevation, and pH.
(XLSX)
GenBank accession numbers for all sequences used to build the lipL32 and secY phylogenies and the whole genome dendrogram.
(XLSX)
Statistical tests used to assess associations between the detection of pathogenic Leptospira spp. and environmental or sampling variables.
(XLSX)
Antigens used in the microscopic agglutination test (MAT) that were unreactive when tested against sera from hamsters infected with the novel pathogenic Leptospira spp. isolated from Puerto Rico soil.
(XLSX)
All lipL32 sequences generated during this study using Sanger and AmpSeq methods.
(XLSX)
All secY sequences generated during this study for two secY amplicons (203 and 549bp).
(XLSX)
Near full length 16S sequences (1330bp) generated for six saprophytic isolates.
(XLSX)
Genome assembly details including assembly size, number of contigs, and NCBI assembly accession numbers.
(XLSX)
The weight of each hamster was recorded daily, beginning two days preinfection.
(XLSX)
Acknowledgments
We thank Ryelan McDonough and Amber Jones for laboratory assistance preparing samples for whole genome sequencing and Karen LeCount for performing MAT.
USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture or the Centers for Disease Control and Prevention.
Data Availability
All WGS reads and assemblies generated during this study have been deposited in NCBI BioProject database under accession # PRJNA766613. The associated BioSample accession numbers for these isolates are sequentially assigned beginning with # SAMN21855208 and ending with # SAMN21855215. Likewise, SRA accession numbers for these isolates are sequentially assigned beginning with # SRR16134453 and ending with # SRR16134460. Partial gene sequences for lipL32 (S6 Table) and secY (S7 Table) have been deposited in NCBI (accession numbers sequentially assigned beginning with # OK345072 and ending with # OK345268).
Funding Statement
This work was funded in part by the United States Department of Health and Human Services Centers for Disease Control and Prevention (Award# 6NU1ROT000006-01-04 to DMW). The Brazilian agency FAPESP also financially supported this work; LGVF is funded with a fellowship from FAPESP (2017/06731-8 and 2019/20302-8). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. 2009;64(3):247–55. doi: 10.1016/j.diagmicrobio.2009.03.014 . [DOI] [PubMed] [Google Scholar]
- 2.Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. 2019;13(5):e0007270. doi: 10.1371/journal.pntd.0007270 ; PubMed Central PMCID: PMC6532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, et al. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One. 2008;3(2):e1607. doi: 10.1371/journal.pone.0001607 ; PubMed Central PMCID: PMC2229662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y, Zhu Y, Wang Y, Chang YF, Zhang Y, Jiang X, et al. Whole genome sequencing revealed host adaptation-focused genomic plasticity of pathogenic Leptospira. Sci Rep. 2016;6:20020. doi: 10.1038/srep20020 ; PubMed Central PMCID: PMC4735792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3(12):757–71. doi: 10.1016/s1473-3099(03)00830-2 . [DOI] [PubMed] [Google Scholar]
- 6.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14(2):296–326. doi: 10.1128/CMR.14.2.296-326.2001 ; PubMed Central PMCID: PMC88975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler B, de la Pena Moctezuma A. Leptospira and leptospirosis. Vet Microbiol. 2010;140(3–4):287–96. doi: 10.1016/j.vetmic.2009.03.012 . [DOI] [PubMed] [Google Scholar]
- 8.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl Trop Dis. 2015;9(9):e0003898. doi: 10.1371/journal.pntd.0003898 ; PubMed Central PMCID: PMC4574773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marinova-Petkova A, Guendel I, Strysko JP, Ekpo LL, Galloway R, Yoder J, et al. First Reported Human Cases of Leptospirosis in the United States Virgin Islands in the Aftermath of Hurricanes Irma and Maria, September-November 2017. Open Forum Infect Dis. 2019;6(7):ofz261. doi: 10.1093/ofid/ofz261 ; PubMed Central PMCID: PMC6602892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos LJS, Lazaro PM, Barbosa JS, Cardona CP. Spatial and Epidemiological Analysis of Human Leptospirosis in Puerto Rico, 1996 to 2014. Geofocus-Rev Int Cie. 2018;(21):227–51. doi: 10.21138/Gf.592 PubMed PMID: WOS:000440383300013. [DOI] [Google Scholar]
- 11.Peters A, Vokaty A, Portch R, Gebre Y. Leptospirosis in the Caribbean: a literature review. Rev Panam Salud Publica. 2017;41:e166. doi: 10.26633/RPSP.2017.166 ; PubMed Central PMCID: PMC6650629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mwachui MA, Crump L, Hartskeerl R, Zinsstag J, Hattendorf J. Environmental and Behavioural Determinants of Leptospirosis Transmission: A Systematic Review. PLoS Negl Trop Dis. 2015;9(9):e0003843. doi: 10.1371/journal.pntd.0003843 ; PubMed Central PMCID: PMC4574979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nedelman M. Suspected leptospirosis cases increasing in Puerto Rico after hurricane Maria. <https://wwwcnncom/2017/10/24/health/leptospirosis-puerto-rico/indexhtml>. 2017.
- 14.Casanovas-Massana A, Pedra GG, Wunder EA Jr., Diggle PJ, Begon M, Ko AI. Quantification of Leptospira interrogans Survival in Soil and Water Microcosms. Appl Environ Microbiol. 2018;84(13). doi: 10.1128/AEM.00507-18 ; PubMed Central PMCID: PMC6007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bierque E, Thibeaux R, Girault D, Soupe-Gilbert ME, Goarant C. A systematic review of Leptospira in water and soil environments. PLoS One. 2020;15(1):e0227055. doi: 10.1371/journal.pone.0227055 ; PubMed Central PMCID: PMC6984726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picardeau M. Diagnosis and epidemiology of leptospirosis. Med Mal Infect. 2013;43(1):1–9. doi: 10.1016/j.medmal.2012.11.005 . [DOI] [PubMed] [Google Scholar]
- 17.Putz EJ, Nally JE. Investigating the Immunological and Biological Equilibrium of Reservoir Hosts and Pathogenic Leptospira: Balancing the Solution to an Acute Problem? Front Microbiol. 2020;11:2005. doi: 10.3389/fmicb.2020.02005 ; PubMed Central PMCID: PMC7456838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monahan AM, Callanan JJ, Nally JE. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect Immun. 2008;76(11):4952–8. doi: 10.1128/IAI.00511-08 ; PubMed Central PMCID: PMC2573331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haake DA, Levett PN. Leptospirosis in humans. Curr Top Microbiol Immunol. 2015;387:65–97. doi: 10.1007/978-3-662-45059-8_5 ; PubMed Central PMCID: PMC4442676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarantola A, Goarant C. Leptospirosis in French Historical Medical Literature: Weil’s Disease or Kelsch’s Disease? Am J Trop Med Hyg. 2018;99(6):1366–8. doi: 10.4269/ajtmh.18-0629 ; PubMed Central PMCID: PMC6283489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito M, Villanueva SY, Chakraborty A, Miyahara S, Segawa T, Asoh T, et al. Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl Environ Microbiol. 2013;79(2):601–9. Epub 20121109. doi: 10.1128/AEM.02728-12 ; PubMed Central PMCID: PMC3553789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito M, Miyahara S, Villanueva SY, Aramaki N, Ikejiri M, Kobayashi Y, et al. PCR and culture identification of pathogenic Leptospira spp. from coastal soil in Leyte, Philippines, after a storm surge during Super Typhoon Haiyan (Yolanda). Appl Environ Microbiol. 2014;80(22):6926–32. Epub 20140829. doi: 10.1128/AEM.02568-14 ; PubMed Central PMCID: PMC4248998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller E, Barragan V, Chiriboga J, Weddell C, Luna L, Jimenez DJ, et al. Leptospira in river and soil in a highly endemic area of Ecuador. BMC Microbiol. 2021;21(1):17. doi: 10.1186/s12866-020-02069-y ; PubMed Central PMCID: PMC7792295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz-Zanzi C, Mason MR, Encina C, Astroza A, Romero A. Leptospira contamination in household and environmental water in rural communities in southern Chile. Int J Environ Res Public Health. 2014;11(7):6666–80. doi: 10.3390/ijerph110706666 ; PubMed Central PMCID: PMC4113836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores B, Escobar K, Muzquiz JL, Sheleby-Elias J, Mora B, Roque E, et al. Detection of Pathogenic Leptospires in Water and Soil in Areas Endemic to Leptospirosis in Nicaragua. Trop Med Infect Dis. 2020;5(3). doi: 10.3390/tropicalmed5030149 ; PubMed Central PMCID: PMC7559144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barragan V, Olivas S, Keim P, Pearson T. Critical Knowledge Gaps in Our Understanding of Environmental Cycling and Transmission of Leptospira spp. Appl Environ Microbiol. 2017;83(19). doi: 10.1128/AEM.01190-17 ; PubMed Central PMCID: PMC5601346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall CM, Jaramillo S, Jimenez R, Stone NE, Centner H, Busch JD, et al. Burkholderia pseudomallei, the causative agent of melioidosis, is rare but ecologically established and widely dispersed in the environment in Puerto Rico. PLoS Negl Trop Dis. 2019;13(9):e0007727. doi: 10.1371/journal.pntd.0007727 ; PubMed Central PMCID: PMC6748447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stone NE, Sidak-Loftis LC, Sahl JW, Vazquez AJ, Wiggins KB, Gillece JD, et al. More than 50% of Clostridium difficile Isolates from Pet Dogs in Flagstaff, USA, Carry Toxigenic Genotypes. PLoS One. 2016;11(10):e0164504. doi: 10.1371/journal.pone.0164504 ; PubMed Central PMCID: PMC5056695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS One. 2009;4(9):e7093. doi: 10.1371/journal.pone.0007093 ; PubMed Central PMCID: PMC2740861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed N, Devi SM, Valverde Mde L, Vijayachari P, Machang’u RS, Ellis WA, et al. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob. 2006;5:28. doi: 10.1186/1476-0711-5-28 ; PubMed Central PMCID: PMC1664579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;(41):95–8.10780396 [Google Scholar]
- 32.Colman RE, Schupp JM, Hicks ND, Smith DE, Buchhagen JL, Valafar F, et al. Detection of Low-Level Mixed-Population Drug Resistance in Mycobacterium tuberculosis Using High Fidelity Amplicon Sequencing. Plos One. 2015;10(5). doi: ARTN e0126626 doi: 10.1371/journal.pone.0126626 PubMed PMID: WOS:000354544200122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–7. doi: 10.1038/s41587-019-0209-9 ; PubMed Central PMCID: PMC7015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. M. Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnetjournal. 2011;17(1):10–2. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 35.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–3. doi: 10.1038/nmeth.3869 ; PubMed Central PMCID: PMC4927377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2 . [DOI] [PubMed] [Google Scholar]
- 37.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421 ; PubMed Central PMCID: PMC2803857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340 ; PubMed Central PMCID: PMC390337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2007;30(12):2725–9. Epub 2013/10/16. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389 ; PubMed Central PMCID: PMC146917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular biology and evolution. 2015;32(1):268–74. doi: 10.1093/molbev/msu300 ; PubMed Central PMCID: PMC4271533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9. doi: 10.1038/nmeth.4285 ; PubMed Central PMCID: PMC5453245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular biology and evolution. 2018;35(2):518–22. doi: 10.1093/molbev/msx281 ; PubMed Central PMCID: PMC5850222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. doi: 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- 45.Team R. RStudio: Integrated Development for R. PBC, Boston, MA: RStudio; 2020.
- 46.Hornsby RL, Alt DP, Nally JE. Isolation and propagation of leptospires at 37 degrees C directly from the mammalian host. Sci Rep-Uk. 2020;10(1). doi: ARTN 9620 10.1038/s41598-020-66526-4. PubMed PMID: WOS:000544936100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakraborty A, Miyahara S, Villanueva SY, Saito M, Gloriani NG, Yoshida S. A novel combination of selective agents for isolation of Leptospira species. Microbiol Immunol. 2011;55(7):494–501. doi: 10.1111/j.1348-0421.2011.00347.x . [DOI] [PubMed] [Google Scholar]
- 48.Zuerner RL. Laboratory maintenance of pathogenic Leptospira. Curr Protoc Microbiol. 2005;Chapter 12:Unit 12E 1. doi: 10.1002/9780471729259.mc12e01s00 . [DOI] [PubMed] [Google Scholar]
- 49.Fernandes LGV, Hornsby RL, Nascimento A, Nally JE. Genetic manipulation of pathogenic Leptospira: CRISPR interference (CRISPRi)-mediated gene silencing and rapid mutant recovery at 37 degrees C. Sci Rep. 2021;11(1):1768. doi: 10.1038/s41598-021-81400-7 ; PubMed Central PMCID: PMC7815788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozarewa I, Turner DJ. 96-plex molecular barcoding for the Illumina Genome Analyzer. Methods Mol Biol. 2011;733:279–98. doi: 10.1007/978-1-61779-089-8_20 . [DOI] [PubMed] [Google Scholar]
- 51.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40(Database issue):D48–53. doi: 10.1093/nar/gkr1202 ; PubMed Central PMCID: PMC3245039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021 ; PubMed Central PMCID: PMC3342519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100. doi: 10.1093/bioinformatics/bty191 ; PubMed Central PMCID: PMC6137996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352 ; PubMed Central PMCID: PMC2723002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17(1):132. doi: 10.1186/s13059-016-0997-x ; PubMed Central PMCID: PMC4915045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17(3):261–72. doi: 10.1038/s41592-019-0686-2 ; PubMed Central PMCID: PMC7056644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods-Uk. 2016;8(1):12–24. doi: 10.1039/c5ay02550h PubMed PMID: WOS:000366905700001. [DOI] [Google Scholar]
- 58.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. doi: 10.1093/bioinformatics/btu153 . [DOI] [PubMed] [Google Scholar]
- 59.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30(11):2478–83. doi: 10.1093/nar/30.11.2478 ; PubMed Central PMCID: PMC117189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom. 2016;2(8):e000074. doi: 10.1099/mgen.0.000074 ; PubMed Central PMCID: PMC5320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular biology and evolution. 2020;37(5):1530–4. doi: 10.1093/molbev/msaa015 ; PubMed Central PMCID: PMC7182206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nally JE, Hornsby RL, Alt DP, Bayles D, Wilson-Welder JH, Palmquist DE, et al. Isolation and characterization of pathogenic leptospires associated with cattle. Vet Microbiol. 2018;218:25–30. doi: 10.1016/j.vetmic.2018.03.023 . [DOI] [PubMed] [Google Scholar]
- 63.Cole JR Jr., Sulzer CR, Pursell AR. Improved microtechnique for the leptospiral microscopic agglutination test. Appl Microbiol. 1973;25(6):976–80. doi: 10.1128/am.25.6.976-980.1973 ; PubMed Central PMCID: PMC380950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellis WA, Montgomery J, Cassells JA. Dihydrostreptomycin treatment of bovine carriers of Leptospira interrogans serovar hardjo. Res Vet Sci. 1985;39(3):292–5. . [PubMed] [Google Scholar]
- 65.Picardeau M. Virulence of the zoonotic agent of leptospirosis: still terra incognita? Nat Rev Microbiol. 2017;15(5):297–307. doi: 10.1038/nrmicro.2017.5 . [DOI] [PubMed] [Google Scholar]
- 66.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55(3):541–55. doi: 10.1016/j.mimet.2003.08.009 . [DOI] [PubMed] [Google Scholar]
- 67.Thibeaux R, Iraola G, Ferres I, Bierque E, Girault D, Soupe-Gilbert ME, et al. Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb Genom. 2018;4(1). Epub 20180103. doi: 10.1099/mgen.0.000144 ; PubMed Central PMCID: PMC5857368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorbea H, Garcia-Rivera EJ, Torres H, Lorenzi OD, Rivera A, Galloway RL, et al. Leptospirosis Cases Infected with Uncommon Serogroups, Puerto Rico, 2013–2015. Am J Trop Med Hyg. 2018;98(1):258–61. Epub 20180101. doi: 10.4269/ajtmh.17-0538 ; PubMed Central PMCID: PMC5928731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murray GL, Srikram A, Hoke DE, Wunder EA Jr., Henry R, Lo M, et al. Major surface protein LipL32 is not required for either acute or chronic infection with Leptospira interrogans. Infect Immun. 2009;77(3):952–8. Epub 20081222. doi: 10.1128/IAI.01370-08 ; PubMed Central PMCID: PMC2643616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsuboi M, Koizumi N, Hayakawa K, Kanagawa S, Ohmagari N, Kato Y. Imported Leptospira licerasiae Infection in Traveler Returning to Japan from Brazil. Emerg Infect Dis. 2017;23(3):548–9. doi: 10.3201/eid2303.161262 ; PubMed Central PMCID: PMC5382744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thibeaux R, Soupe-Gilbert ME, Kainiu M, Girault D, Bierque E, Fernandes J, et al. The zoonotic pathogen Leptospira interrogans mitigates environmental stress through cyclic-di-GMP-controlled biofilm production. NPJ Biofilms Microbiomes. 2020;6(1):24. doi: 10.1038/s41522-020-0134-1 ; PubMed Central PMCID: PMC7293261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ristow P, Bourhy P, Kerneis S, Schmitt C, Prevost MC, Lilenbaum W, et al. Biofilm formation by saprophytic and pathogenic leptospires. Microbiol-Sgm. 2008;154:1309–17. doi: 10.1099/mic.0.2007/014746-0 PubMed PMID: WOS:000256113900005. [DOI] [PubMed] [Google Scholar]
- 73.Briskin EA, Casanovas-Massana A, Ryff KR, Morales-Estrada S, Hamond C, Perez-Rodriguez NM, et al. Seroprevalence, Risk Factors, and Rodent Reservoirs of Leptospirosis in an Urban Community of Puerto Rico, 2015. J Infect Dis. 2019;220(9):1489–97. doi: 10.1093/infdis/jiz339 ; PubMed Central PMCID: PMC6761939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Izurieta R, Galwankar S, Clem A. Leptospirosis: The "mysterious" mimic. J Emerg Trauma Shock. 2008;1(1):21–33. doi: 10.4103/0974-2700.40573 ; PubMed Central PMCID: PMC2700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Musso D, La Scola B. Laboratory diagnosis of leptospirosis: a challenge. J Microbiol Immunol Infect. 2013;46(4):245–52. doi: 10.1016/j.jmii.2013.03.001 . [DOI] [PubMed] [Google Scholar]
- 76.Zuerner RL, Alt DP, Palmer MV. Development of chronic and acute golden Syrian hamster infection models with Leptospira borgpetersenii serovar Hardjo. Vet Pathol. 2012;49(2):403–11. doi: 10.1177/0300985811409252 . [DOI] [PubMed] [Google Scholar]
- 77.Cordonin C, Turpin M, Bascands JL, Dellagi K, Mavingui P, Tortosa P, et al. Three Leptospira Strains From Western Indian Ocean Wildlife Show Highly Distinct Virulence Phenotypes Through Hamster Experimental Infection. Frontiers in Microbiology. 2019;10. doi: ARTN 382 doi: 10.3389/fmicb.2019.00382 PubMed PMID: WOS:000460773800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1(1):e56. doi: 10.1371/journal.pntd.0000056 ; PubMed Central PMCID: PMC2041815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Black and blue stars represent soil and water samples, respectively. Transects were a minimum of 2.5 meters apart. This sampling design was also applied in a linear fashion for the sampling of rivers and streams.
(PDF)
The yellow triangles indicate sites where pathogenic Leptospira DNA was detected. Eight soil sites were positive for pathogenic Leptospira DNA, compared to two water sites; both soil and water were positive at five sites. Site 16, which was used to assess environmental persistence in soil is indicated with a red circle. This map was created using ArcGIS software by Esri. ArcGIS and ArcMap are the intellectual property of Esri and are used herein under license. Copyright Esri. All rights reserved. For more information about Esri software, please visit www.esri.com. Basemap: http://goto.arcgisonline.com/maps/Canvas/World_Light_Gray_Base.
(PDF)
Sequences generated from soil and water samples collected in Puerto Rico are in blue or pink text, the novel pathogenic Puerto Rico isolates from soil are also included; reference sequences for pathogenic, intermediate, and saprophytic Leptospira are in black. Bootstrap/aLRT support values are indicated on branch nodes.
(PDF)
Sequences generated from soil (blue text) and water (pink text) samples collected in Puerto Rico are displayed. Reference sequences representing all other known pathogenic Leptospira spp. are in black. Nine major clades are represented of which seven were identified in Puerto Rico and four of those (3, 5, 8, and 9) have not been described previously. Identical genotypes were present in soil and water from Puerto Rico at two sites (17 and 21), with both occurrences representing a single genotype from clade 6 (in orange). Bootstrap values are indicated on each branch. Major clades are numbered 1 through 9 and color-coding matches Fig 2.
(PDF)
LipL32 PCR positive (Soil #27) and negative (Soil #40) samples were subjected to FAT (Panel A) and ELISA (Panel B) along with positive and negative controls (L. interrogans and L. biflexa, respectively) to assess expression of the LipL32 pathogenicity protein. Individual colonies from Soil #27 were subjected to additional FAT testing (Panel C) and confirmatory PCR (Panel D) to verify that the obtained isolates were pathogenic.
(PDF)
Summary of detection and sequencing results for environmental samples from Puerto Rico, 2018–2020.
(XLSX)
Sample and site ID, location, elevation, and pH.
(XLSX)
GenBank accession numbers for all sequences used to build the lipL32 and secY phylogenies and the whole genome dendrogram.
(XLSX)
Statistical tests used to assess associations between the detection of pathogenic Leptospira spp. and environmental or sampling variables.
(XLSX)
Antigens used in the microscopic agglutination test (MAT) that were unreactive when tested against sera from hamsters infected with the novel pathogenic Leptospira spp. isolated from Puerto Rico soil.
(XLSX)
All lipL32 sequences generated during this study using Sanger and AmpSeq methods.
(XLSX)
All secY sequences generated during this study for two secY amplicons (203 and 549bp).
(XLSX)
Near full length 16S sequences (1330bp) generated for six saprophytic isolates.
(XLSX)
Genome assembly details including assembly size, number of contigs, and NCBI assembly accession numbers.
(XLSX)
The weight of each hamster was recorded daily, beginning two days preinfection.
(XLSX)
Data Availability Statement
All WGS reads and assemblies generated during this study have been deposited in NCBI BioProject database under accession # PRJNA766613. The associated BioSample accession numbers for these isolates are sequentially assigned beginning with # SAMN21855208 and ending with # SAMN21855215. Likewise, SRA accession numbers for these isolates are sequentially assigned beginning with # SRR16134453 and ending with # SRR16134460. Partial gene sequences for lipL32 (S6 Table) and secY (S7 Table) have been deposited in NCBI (accession numbers sequentially assigned beginning with # OK345072 and ending with # OK345268).