Abstract
Three-dimensional (3D) chromatin structure plays a critical role in development, gene regulation, and cellular identity. Alterations to this structure can have profound effects on cellular phenotypes and have been associated with a variety of diseases including multiple types of cancer. One of several forces that help shape 3D chromatin structure is liquid-liquid phase separation (LLPS), a form of self-association between biomolecules that can sequester regions of chromatin into sub-nuclear droplets or even membraneless organelles like nucleoli. This review focuses on a class of oncogenic fusion proteins that appear to exert their oncogenic function via phase-separation driven alterations to 3D chromatin structure. Here we review what is known about the mechanisms by which these oncogenic fusion proteins phase separate in the nucleus and their role in shaping the 3D chromatin structure. We discuss the potential for this phenomenon to be a more widespread mechanism of oncogenesis.
Introduction
Three-dimensional (3D) chromatin structure plays a critical role in gene regulation by connecting distant regulatory elements to gene promoters and modulating gene expression. Modifications to this structure have been associated with the development of human diseases including cancer. Several molecular forces have been identified for establishing and/or modulating the 3D chromatin structure, such as those directed by CTCF and multi-subunit cohesin complexes, and liquid-liquid phase separation (LLPS), a form of self-association between biomolecules that can sequester regions of chromatin fibers into sub-nuclear droplets or even membraneless organelles like nucleoli. This review focuses on an emerging class of oncogenic fusion proteins that appear to exert their oncogenic function at least partly, via phase-separation-driven alterations to 3D chromatin structure. Despite the relatively small number of such fusions that have been characterized, this phenomenon is potentially a more widespread mechanism of oncogenesis and further identification and characterization of such fusions could have important implications for diagnostic and therapeutic development.
3D chromatin organization
3D chromatin structure plays a fundamental role in gene regulation and cellular identity by rewiring contacts between regulatory loci and gene promoters. Current theories suggest that this organization is driven largely by two coexisting, dynamic, yet sometimes opposing forces: compartmentalization and loop extrusion [1–6]. Although the exact mechanisms driving compartmentalization are still under investigation, this phenomenon is thought to be largely mediated by affinity interactions between genomic regions with similar epigenetic marks and levels of transcription. In contrast, loop extrusion is mediated by cohesin, a ring-like protein complex that extrudes DNA until it encounters CTCF proteins bound in a convergent orientation, at which point it stabilizes point-to-point interactions called chromatin loops. The combined forces of loop extrusion and compartmentalization produce contact domains, also known as “topologically associated domains” (TADs), which are broad regions of self-interacting chromatin. The interplay of these forces in shaping 3D chromatin architecture is nicely discussed by Rowley and Corces [7].
Increasing evidence suggests that LLPS—long known for forming membraneless nuclear bodies such as nucleoli, nuclear speckles, and Cajal bodies—may play a broader role in 3D chromatin organization than previously suspected. Indeed, the association of chromatin-bound proteins in these nuclear bodies is thought to play a role in shaping 3D chromatin architecture [8]. LLPS is mostly driven by a collection of weak, multivalent interactions between proteins that contain intrinsically disordered regions (IDRs) [9,10]. Transcription factors (TF) and co-regulators implicated in chromatin remodeling and gene regulation, such as Mediator and RNA polymerase II (RNA Pol II), are enriched with IDRs and are thought to exercise their function in part via phase separation [11–16]. Additionally, although not the focus of this review, RNA has been proposed to have a prominent role in chromatin organization in a way that is compatible with the concept of LLPS [17–20].
Despite mounting evidence supporting the role of LLPS in 3D chromatin organization, it remains unclear exactly what types of chromatin structures LLPS can generate and in which biological contexts. One hypothesis that has gained support in recent years proposes LLPS as one of the driving forces for chromatin compartmentalization [19,21–25]. Moreover, while DNA loops are thought to be formed largely via the ATP-dependent process of extrusion [3,26,27], recent evidence suggests that cohesin itself may undergo LLPS [28]. Finally, a model proposing phase separation as a general mechanism driving super-enhancer-mediated gene regulation has also gained extensive support [29,30], extending the potential role of phase separation to all levels of chromatin organization.
3D chromatin structure, phase separation, and cancer
Alterations in both 3D chromatin structure via a variety of different mechanisms has been linked to the development of various cancers. Mutations in cohesin, which are among the most common mutations found in cancer, have been shown to result in misregulation of intra-chromosomal DNA looping, affecting genome organization and gene expression (recently reviewed by Waldman [31]). Point mutations and somatic structural variations found in multiple cancer types have been shown to disrupt TADs and alter transcription levels of the surrounding genes [32]. One example of this phenomenon is the IDH gain-of-function mutation in gliomas, which alters TAD boundaries, resulting in the induced expression of the PDGFRA oncogene [33].
LLPS has also been associated with cancer, independently from 3D chromatin conformation. For example, several oncogenes and tumor suppressor genes exert their normal activity via LLPS which when disrupted—via alterations of either gene expression or LLPS capability—can promote cancer development. For example, p53 binding protein 1 (53BP1), uses LLPS to recruit DNA repair complexes to sites of DNA damage. Underexpression of 53BP1 in various contexts promotes DNA instability and the development of cancer [34]. In another example, mutations disrupting the phase separation capacity of the IDR in the histone H3K27 demethylase UTX (also known as KDM6A) abolish its tumor suppression capabilities [35]. In contrast, a mutation in the histone acetylation reader ENL, commonly found in pediatric kidney cancer, confers novel phase separation capacity and has been linked to alterations in transcription and oncogenesis [36].
IDR-DNA binding fusion proteins in cancer
In addition to the observations mentioned above that independently link phase separation and 3D chromatin structure to cancer, several recent studies describe a novel paradigm in which phase separation of cancer-related fusion proteins promotes oncogenesis by directly inducing changes to 3D chromatin structure. Chromosomal translocations, a common feature of cancers, can produce chimeric genes that encode fusion proteins, which have been shown to influence diverse functions including cellular cycle, cell shape, cell mobility and RNA metabolism, amongst others [37]. Intriguingly, IDRs are enriched in cancer-associated fusion proteins (43.3% vs. 20.7% in all human proteins) [38] and a subset of these fuse the IDR of one protein to a chromatin-associated domain of another (Figure 1A). This unique combination of domains provides, in theory, the potential to alter 3D chromatin structure by pulling a set of bound regions into condensates formed via IDR LLPS, and as a result of this process, alter the expression of surrounding genes (Figure 1A).
Figure 1. IDR/chromatin-associating fusion proteins in cancer.
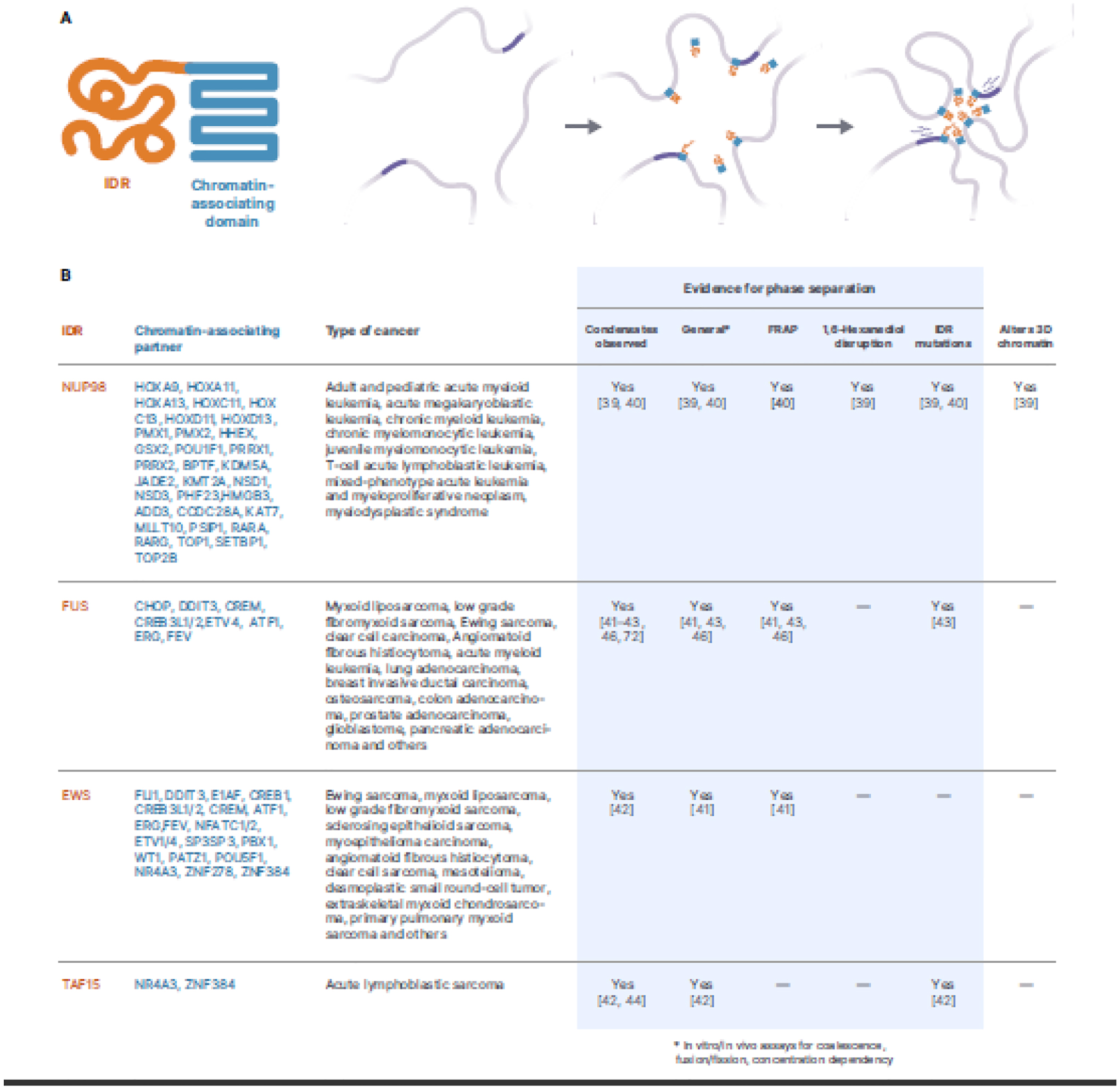
A. The proposed mechanism of action of IDR/chromatin-associating fusion proteins in cancer. Fusion proteins bind chromatin at enhancers and gene promoters and recruit bound loci into phase separation dependent condensates activating oncogenic transcriptional profiles. B. A list of IDR/DNA-binding fusion proteins frequently found in cancer that have been described to phase separate in the nucleus. *In vitro/in vivo assays for coalescence, fusion/fission, concentration dependency. FRAP: Fluorescence recovery after photobleaching.
We recently showed that a cancer-associated fusion between an IDR and a DNA binding domain could induce oncogenic transcription programs at least partly via de novo loop formation [39]**. This study focused on the NUP98–HOXA9, a classic example for a set of transcription factor (TF) fusion oncoproteins found recurrently in human haematological malignancies such as acute myeloid leukemia. The two components of this chimera are: (1) NUP98, a constitutively expressed nucleoporin protein that contains a phenylalanine-glycine(FG)-repeat-rich IDR, and (2) HOXA9, a TF that contains a DNA binding homeodomain regulating gene expression essential for proper cell proliferation and differentiation during embryogenesis. Expression of this fusion in primary hematopoietic stem/progenitor cells (HSPCs) is sufficient to transform cells and activate leukemic gene-expression profiles. In contrast, a phase-separation-incompetent mutant of NUP98–HOXA9, or one harboring a shorter IDR incapable of inducing LLPS, were unable to transform HSPCs, highlighting a critical requirement of IDR-mediated phase separation for cancerous transformation. ChIP-seq and Hi-C analysis revealed that NUP98–HOXA9-expressing cells formed DNA loops whose anchors were bound by NUP98–HOXA9, but not by CTCF, supporting a model that proposes that phase separation but not loop extrusion is the driving force for the formation of de novo chromatin contacts in the context of this fusion. Genes whose promoters overlap the anchors of the NUP98–HOXA9-bound loops were upregulated, including proto-oncogenes such as PBX3 and HOX cluster genes. These findings suggest phase separation as the driver of aberrant chromatin looping and the resulting changes in gene expression in leukemias harboring this fusion. Some of these results were confirmed by Chandra et al., who demonstrated the phase separation capabilities of NUP98-HOXA9 and other leukemia-related fusion oncoproteins [40]. It is important to mention that NUP98-HOXA9-induced alterations to 3D chromatin structure have only been investigated in HEK 293T cells and moving forward it will be important to confirm these changes in leukemia-relevant cell types.
Several other cancer-associated fusion proteins that have a similar composition —including an IDR and a chromatin interaction domain— are likely to impart oncogenic properties via similar alterations to 3D chromatin structure (Figure 1B). One group of likely candidates are the FUS/EWS/TAF15 (FET) fusion oncoproteins. These FET oncogenic fusions form multiple large nuclear condensates in cells similar to the ones observed when the aforementioned oncogenic fusions are present. FET fusion proteins are essential oncogenic drivers in various cancers including myxoid liposarcoma, Ewing sarcoma, and adenocarcinomas. They were shown to undergo phase separation at target binding loci and form phase-separated transcriptional condensates recruiting RNA Pol II and other co-factors (e.g. BRD4), which results in the increase in gene expression [41–43]*. Chong et al. also identified IDR-dependent nuclear hubs of FET fusion oncoproteins and demonstrated their role in transcriptional regulation; however, they propose that such hubs are formed via a non LLPS-driven mechanism [44]. Finally, several studies have reported that the SWI/SNF chromatin-remodeling complex, also known as BAF (BRG1-associated factors), can be recruited into FET fusion condensates. This process is likely to occur via heterotypic interactions among prion-like domains commonly present in both FET fusions and components of the SWI/SNF (BAF) complex, resulting in cancer-specific chromatin alterations favorable for gene activation [45–47].
Other nuclear phase separating fusion proteins in cancer
Several other fusions have been described that also act via phase separation driven changes to 3D chromatin organization, albeit by mechanisms distinct from the one described above (Table 1). Rosencrance et. al. found that BRD4-NUT, a BRD4 fusion oncoprotein found in midline carcinoma, alters 3D chromatin at the compartment scale in a manner that is consistent with phase separation [48]**. In this case, the fusion protein BRD4-NUT produces changes observed at a larger scale, resulting in the formation of aberrant chromatin sub-compartments referred to as subcompartments “M”. These subcompartments “M” encompass broad stretches of active chromatin that exhibit heightened interactions both within and between different chromosomes. These modifications in interaction occur due to the ability of BRD4-NUT to drive massive changes in histone acetylation. The BRD4 portion of the fusion protein binds acetylated histones, while the NUT portion recruits the p300 histone acetyltransferase, driving a proposed feed-forward loop of histone acetylation which results in exceptionally broad linear (100 kb to 2Mb) hyperacetylated chromatin areas called “megadomains”[49]. These megadomains interact with one another even between chromosomes, are associated with increased transcription of Myc and other important oncogenes for NUT-carcinomas, and appear as large nuclear puncta [48]. Although no experiments were performed to directly test for BRD4-NUT’s LLPS capabilities, previous studies have shown that it is clearly present in nuclear puncta, consistent with an LLPS-driven mechanism [49,50] and evidence for phase separation of the short isoform of BRD4 has been reported [51]; also, LLPS of the acetylated chromatin in combination with the multi-bromodomain of BRD4 as it has been previously proposed [24]. Interestingly, a subclass of NUP98 fusions that carry the plant homeodomain (PHD), a H3K4me2/3 reader motif, were shown to exhibit comparable large nuclear condensates [52]. Moreover, similar to the BRD4-NUT fusion, we have observed that condensates of NUP98-PHD fusions are enriched for H3K4me2/3, to which the PHD domain of the fusion binds, as well as MLL1, an enzyme that catalyzes H3K4 methylations (unpublished). This suggests that a similar feed-forward loop mechanism could underlie the formation of broad domains of inter/intrachromosomal interactions in leukemia cells.
Table 1. Other nuclear phase-separating fusion proteins in cancer.
A list of phase-separating fusion proteins frequently found in cancer that combine an IDR/chromatin-associating-containing proteins and a fusion partner that alters the distribution and/or role of the first component. *In vitro/in vivo assays for coalescence, fusion/fission, concentration dependency. FRAP: Fluorescence recovery after photobleaching.
| Fusion proteins | Type of cancer | Evidence for phase separation | |||||
|---|---|---|---|---|---|---|---|
| Condensates observed | General* | FRAP | 1,6-Hexanediol disruption | IDR mutations | Alters 3D chromatin | ||
| BRD4-NUT | Squamous cell carcinoma, midline carcinoma | Yes[48–50] | - | - | - | - | Yes[48] |
| SS18-SSX1/2 | Synovial sarcoma | Yes[53,55] | Yes [53] | Yes [53] | Yes [53] | Yes [53] | - |
In vitro/in vivo assays for coalescence, fusion/fission, concentration dependency
Similarly, a fusion protein typically found in synovial sarcoma between the SWI/SNF (BAF) complex member SS18 and one of several SSX genes is another example of a fusion onco-protein containing an unstructured region that can associate with specific chromatin areas and alter gene transcription. SS18 contains an IDR-rich C-terminal and has been shown to mediate the BAF complex assembly via LLPS [53]. When SSX is fused to SS18, this fusion evicts another member of the BAF complex and retargets BAF from enhancer regions to broad polycomb domains in the chromatin. This new BAF complex occupancy opposes PRC2-mediated chromatin repression and results in activation of bivalent genes [54]. The SS18-SSX fusion forms multiple large nuclear condensates and exhibits dense and broad binding patterns at target loci, reflecting typical characteristics of phase-separated molecules [55]. Despite the broadly known capacity of these fusion proteins to phase separate and cause gene mis-regulation, their potential roles in inducing alterations in the 3D chromatin structure have not yet been fully explored. Conversely, additional research needs to be performed to uncover whether other fusion onco-proteins that alter 3D chromatin structure do so via a LLPS mechanism. One such fusion is PAX3-FOXO1, a recurrent mutation found in rhabdomyosarcoma that is associated with altered 3D chromatin structure and has the capacity to form de novo super enhancers and recruit BRD4 and other master transcription factors [56,57].
Future Directions: Fusions, Mechanisms, and Interventions
These studies provide the first glimpse into a cancer-driving mechanism in which fusions between IDRs and chromatin-associated domains drive oncogenic transcription via alterations in the 3D chromatin structure. However, it is currently unclear how broad this mechanism is across various cancer subtypes and other diseases. One ongoing challenge will be to identify and characterize other cancer fusions that operate via alterations to chromatin interaction frequencies. Ever expanding databases of disease-associated fusion proteins (e.g chimerDB [58] and COSMIC [59]), intrinsically disordered domains (e.g. MobiDB [60], DisProt [61], and IDEAL [62]), and phase-separating proteins (e.g. LLPSDB [63], PhaSePro [64], PhaSepDB [65]) provide rich resources to mine for such possibilities. Improving and assessing our ability to detect IDRs and phase-separation-competent regions should accelerate discovery further. Necci et al. [66] suggest that deep learning-based approaches show the most promise here, highlighting one way in which advances in artificial intelligence may contribute to this field moving forward.
Further mechanistic studies are required to better understand how these 3D structures are formed as well as their impact on transcription. While DNA loops formed via NUP98-HOXA9 are not anchored by CTCF [39], their dependence on cohesin and/or ATP is unclear. Another open question involves how the oncogenic condensates interact with other nuclear condensates and microenvironments. For example, it has been shown that RNA Pol II forms phase-separated condensates [12–14] and in some cases is recruited into condensates formed by fusion onco-proteins to activate gene transcription [41,48]. Further development of experimental approaches to simultaneously monitor and visualize multiple classes of nuclear condensates will enable a deeper understanding of the interplay between them.
It is important to note that several researchers have expressed concern regarding the widespread attribution of LLPS to explain IDR-driven nuclear structures [67–69]. The main concerns refer to studies employing in vitro assays or ectopic overexpression models, which might fail to fully reflect physiological conditions (i.e. local concentrations, microenvironment, and interaction partners) that are fundamental for the occurrence of LLPS. Moving forward, the use of approaches that involve targeting of endogenous IDR-containing proteins and single molecule tracking might enable more precise mechanistic interrogations.
Finally, the discovery of 3D chromatin alterations produced by IDR-containing fusion oncoproteins as a cancer-driving mechanism may reveal new avenues for therapeutic intervention. As reviewed by Wheeler [70], such interventions may look quite different from the classical enzyme lock-and-key or structured binding site approaches, since they would take into account the physicochemical properties of nuclear condensates and drug molecules for their mechanism of action. For example, a recent study showed how the partitioning, concentration, and activity of some onco-drugs were highly influenced by the physicochemical properties of nuclear condensates [71]. Tailoring drugs for either inclusion or exclusion from specific nuclear condensates could potentially improve specificity and/or efficacy. Another study demonstrated that small molecule induced degradation of BRD4-NUT eliminates its effect on chromatin compartmentalization, pointing to the potential of altering 3D chromatin structure as a novel approach to treat disease [48].
Conclusions
Recent studies have illuminated how aberrant gene fusion events seen in cancer can create chimeric proteins capable of binding DNA, altering 3D chromatin structure, and driving oncogenic transcription. While only a handful of such chimeras have been characterized, the enrichment of fusion proteins and IDRs in tumors suggests that this is a much broader mechanism to promote oncogenesis. Future research is required to determine the extent to which this mechanism explains other cancer-related fusions, the mechanisms through which these 3D chromatin structures form and impact gene transcription, and how such structures can be targeted to improve therapeutic interventions.
Acknowledgements
We would like to acknowledge Erika Deoudes for illustrations and graphical design. This work was supported by NIH grants (R01-CA215284 and R01-CA218600 to G.G.W.; R35-GM128645 to D.H.P.), and UNC Lineberger Stimulus Awards (to D.H.P.). IYQ is supported by a BrightFocus Foundation postdoctoral fellowship 911831. G.G.W. is a Leukemia & Lymphoma Society Scholar and an American Cancer Society Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest
none
References
- 1.Barrington C, Finn R, Hadjur S: Cohesin biology meets the loop extrusion model. Chromosome Res 2017, 25:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. : Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA: Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A 2018, 115:E6697–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng H, Xie W: The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 2019, 20:535–550. [DOI] [PubMed] [Google Scholar]
- 5.Mirny LA, Imakaev M, Abdennur N: Two major mechanisms of chromosome organization. Curr Opin Cell Biol 2019, 58:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spracklin G, Abdennur N, Imakaev M, Chowdhury N, Pradhan S, Mirny L, Dekker J: Heterochromatin diversity modulates genome compartmentalization and loop extrusion barriers [preprint]. 2021, doi: 10.1101/2021.08.05.455340. [DOI] [PMC free article] [PubMed]
- 7.Rowley MJ, Corces VG: Organizational principles of 3D genome architecture. Nat Rev Genet 2018, 19:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirny L, Dekker J: Mechanisms of Chromosome Folding and Nuclear Organization: Their Interplay and Open Questions. Cold Spring Harb Perspect Biol 2021, doi: 10.1101/cshperspect.a040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung HYJ, Birol M, Rhoades E: IDPs in macromolecular complexes: the roles of multivalent interactions in diverse assemblies. Curr Opin Struct Biol 2018, 49:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin EW, Holehouse AS: Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack thereof. Emerg Top Life Sci 2020, 4:307–329. [DOI] [PubMed] [Google Scholar]
- 11.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. : Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175:1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladouceur A-M, Parmar BS, Biedzinski S, Wall J, Tope SG, Cohn D, Kim A, Soubry N, Reyes-Lamothe R, Weber SC: Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc Natl Acad Sci U S A 2020, 117:18540–18549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, Zhou Q: Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558:318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, et al. : RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol 2018, 25:833–840. [DOI] [PubMed] [Google Scholar]
- 15.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. : Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II: Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakur J, Henikoff S: Architectural RNA in chromatin organization. Biochem Soc Trans 2020, 48:1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinodoz SA, Jachowicz JW, Bhat P, Ollikainen N, Banerjee AK, Goronzy IN, Blanco MR, Chovanec P, Chow A, Markaki Y, et al. : RNA promotes the formation of spatial compartments in the nucleus. Cell 2021, 184:5775–5790.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nozawa R-S, Yamamoto T, Takahashi M, Tachiwana H, Maruyama R, Hirota T, Saitoh N: Nuclear microenvironment in cancer: Control through liquid-liquid phase separation. Cancer Sci 2020, 111:3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh HJ, Aguilar R, Kesner B, Lee H-G, Kriz AJ, Chu H-P, Lee JT: Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell 2021, 184:6157–6173.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erdel F, Rippe K: Formation of Chromatin Subcompartments by Phase Separation. Biophys J 2018, 114:2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrand EM, Dekker J: Mechanisms and Functions of Chromosome Compartmentalization. Trends Biochem Sci 2020, 45:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rippe K: Liquid-Liquid Phase Separation in Chromatin. Cold Spring Harb Perspect Biol 2021, doi: 10.1101/cshperspect.a040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK: Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179:470–484.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulianov SV, Velichko AK, Magnitov MD, Luzhin AV, Golov AK, Ovsyannikova N, Kireev II, Gavrikov AS, Mishin AS, Garaev AK, et al. : Suppression of liquid-liquid phase separation by 1,6-hexanediol partially compromises the 3D genome organization in living cells. Nucleic Acids Res 2021, 49:10524–10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA: Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 2016, 15:2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson IF, Peters J-M: Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Biol 2021, 22:445–464. [DOI] [PubMed] [Google Scholar]
- 28.Ryu J-K, Bouchoux C, Liu HW, Kim E, Minamino M, de Groot R, Katan AJ, Bonato A, Marenduzzo D, Michieletto D, et al. : Bridging-induced phase separation induced by cohesin SMC protein complexes. Sci Adv 2021, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA: A Phase Separation Model for Transcriptional Control. Cell 2017, 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng L, Li E-M, Xu L-Y: From start to end: Phase separation and transcriptional regulation. Biochim Biophys Acta Gene Regul Mech 2020, 1863:194641. [DOI] [PubMed] [Google Scholar]
- 31.Waldman T: Emerging themes in cohesin cancer biology. Nat Rev Cancer 2020, 20:504–515. [DOI] [PubMed] [Google Scholar]
- 32.Akdemir KC, Le VT, Chandran S, Li Y, Verhaak RG, Beroukhim R, Campbell PJ, Chin L, Dixon JR, Futreal PA, et al. : Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat Genet 2020, 52:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE: Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016, 529:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M: Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J 2019, 38:e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi B, Li W, Song Y, Wang Z, Ju R, Ulman A, Hu J, Palomba F, Zhao Y, Le JP, et al. : UTX condensation underlies its tumour-suppressive activity. Nature 2021, 597:726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, Carroll TS, Li Y, Feng L, Chen G, et al. : Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 2020, 577:121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latysheva NS, Babu MM: Molecular Signatures of Fusion Proteins in Cancer. ACS Pharmacol Transl Sci 2019, 2:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegyi H, Buday L, Tompa P: Intrinsic structural disorder confers cellular viability on oncogenic fusion proteins. PLoS Comput Biol 2009, 5:e1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, Li J, Storey AJ, Tsai Y-H, Keeley DP, et al. : Phase separation drives aberrant chromatin looping and cancer development. Nature 2021, 595:591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper describes the capacity of the cancer-associated fusion NUP98-HOXA9 to bind DNA, form super-enhancer-like loci, undergo LLPS, induce aberrant chromatin loops, and activate an oncogenic transcriptional program. The authors use in vitro assays, cell lines, and mouse models to demonstrate the requirement of FG repeats in the NUP98 IDR for phase separation, chromatin loop formation, and capacity for oncogenic transformation. Then they use multiple genomics techniques (ChIP-seq, RNA-seq, and Hi-C) to identify novel chromatin loops formed between NUP98-HOXA9 binding sites. The anchors of those de novo loops were bound by NUP98–HOXA9, but not by CTCF, supporting a model that proposes that phase separation but not loop extrusion is the driving force for the formation of these de novo chromatin contacts. The anchors of these loops overlap the promoters of many leukemia-associated genes.
- 40.Chandra B, Michmerhuizen NL, Shirnekhi HK, Tripathi S, Pioso BJ, Baggett DW, Mitrea DM, Iacobucci I, White MR, Chen J, et al. : Phase Separation mediates NUP98 Fusion Oncoprotein Leukemic Transformation. Cancer Discov 2021, doi: 10.1158/2159-8290.CD-21-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuo L, Zhang G, Massett M, Cheng J, Guo Z, Wang L, Gao Y, Li R, Huang X, Li P, et al. : Loci-specific phase separation of FET fusion oncoproteins promotes gene transcription. Nat Commun 2021, 12:1491. [DOI] [PMC free article] [PubMed] [Google Scholar]; *In this article the authors use a high-throughput single-molecule technique called DNA Curtains to demonstrate that FET fusion proteins undergo phase separation at target binding loci and that the phase separated condensates recruit RNA polymerase II and enhance gene transcription. Furthermore, they determine a threshold number of fusion-binding DNA elements that can enhance the formation of FET fusion protein condensates.
- 42.Wei M-T, Chang Y-C, Shimobayashi SF, Shin Y, Strom AR, Brangwynne CP: Nucleated transcriptional condensates amplify gene expression. Nat Cell Biol 2020, 22:1187–1196. [DOI] [PubMed] [Google Scholar]; *In this paper, the authors use the optoDroplet system to show that the IDR of the FET-family transcriptional regulators exhibit a strong tendency to phase separate within living cells and drive localized RNA transcription. They found that TAF15 has a unique charge distribution among the FET family members that enhances its interactions with the C-terminal domain of RNA polymerase II. Nascent C-terminal domain clusters at primed genomic loci lower the energetic barrier for nucleation of TAF15 condensates, which in turn further recruit RNA polymerase II to drive transcriptional output.
- 43.Owen I, Yee D, Wyne H, Perdikari TM, Johnson V, Smyth J, Kortum R, Fawzi NL, Shewmaker F: The oncogenic transcription factor FUS-CHOP can undergo nuclear liquid-liquid phase separation. J Cell Sci 2021, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This article characterizes the FET fusion FUS-CHOP in Myxoid liposarcoma cell lines and in vitro, describing novel phase-separating properties relative to unmodified CHOP. Furthermore, the data indicates that FUS-CHOP forms phase-separated condensates that colocalize with BRD4, a marker of super enhancer condensates.
- 44.Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. : Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindén M, Thomsen C, Grundevik P, Jonasson E, Andersson D, Runnberg R, Dolatabadi S, Vannas C, Luna Santamarίa M, Fagman H, et al. : FET family fusion oncoproteins target the SWI/SNF chromatin remodeling complex. EMBO Rep 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis RB, Kaur T, Moosa MM, Banerjee PR: FUS oncofusion protein condensates recruit mSWI/SNF chromatin remodeler via heterotypic interactions between prion-like domains. Protein Sci 2021, 30:1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. : Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171:163–178.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosencrance CD, Ammouri HN, Yu Q, Ge T, Rendleman EJ, Marshall SA, Eagen KP: Chromatin Hyperacetylation Impacts Chromosome Folding by Forming a Nuclear Subcompartment. Mol Cell 2020, 78:112–126.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper describes how the fusion oncoprotein BRD4-NUT found in midline carcinoma, alters 3D chromatin at the compartment scale in a manner that is consistent with phase separation forming aberrant chromatin sub-compartments referred as “M compartments” that exhibit heightened interactions both within and between different chromosomes. These modifications in interaction occur due to the ability of BRD4-NUT to drive massive changes in histone acetylation. The BRD4 portion of the fusion protein binds acetylated histones, while the NUT portion recruits the p300 histone acetyltransferase, driving a proposed feed-forward loop of histone acetylation which results in exceptionally broad linear hyperacetylated chromatin megadomains. The M compartments were conserved even in the absence of RNA Pol II transcription.
- 49.Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, Kuroda MI, French CA: The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev 2015, 29:1507–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, Caron C, Kimura H, Rousseaux S, Cole PA, et al. : Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J 2010, 29:2943–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han X, Yu D, Gu R, Jia Y, Wang Q, Jaganathan A, Yang X, Yu M, Babault N, Zhao C, et al. : Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat Struct Mol Biol 2020, 27:333–341. [DOI] [PubMed] [Google Scholar]
- 52.Fahrenkrog B, Martinelli V, Nilles N, Fruhmann G, Chatel G, Juge S, Sauder U, Di Giacomo D, Mecucci C, Schwaller J: Expression of Leukemia-Associated Nup98 Fusion Proteins Generates an Aberrant Nuclear Envelope Phenotype. PLoS One 2016, 11:e0152321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuang J, Zhai Z, Li P, Shi R, Guo W, Yao Y, Guo J, Zhao G, He J, Xu S, et al. : SS18 regulates pluripotent-somatic transition through phase separation. Nat Commun 2021, 12:4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McBride MJ, Pulice JL, Beird HC, Ingram DR, D’Avino AR, Shern JF, Charville GW, Hornick JL, Nakayama RT, Garcia-Rivera EM, et al. : The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 2018, 33:1128–1141.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoneda Y, Ito S, Kunisada T, Morimoto Y, Kanzaki H, Yoshida A, Shimizu K, Ozaki T, Ouchida M: Truncated SSX protein suppresses synovial sarcoma cell proliferation by inhibiting the localization of SS18-SSX fusion protein. PLoS One 2013, 8:e77564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gryder BE, Pomella S, Sayers C, Wu XS, Song Y, Chiarella AM, Bagchi S, Chou H-C, Sinniah RS, Walton A, et al. : Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet 2019, 51:1714–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gryder BE, Yohe ME, Chou H-C, Zhang X, Marques J, Wachtel M, Schaefer B, Sen N, Song Y, Gualtieri A, et al. : PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov 2017, 7:884–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang YE, Jang I, Kim S, Cho S, Kim D, Kim K, Kim J, Hwang J, Kim S, Kim J, et al. : ChimerDB 4.0: an updated and expanded database of fusion genes. Nucleic Acids Res 2020, 48:D817–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA: The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018, 18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piovesan D, Necci M, Escobedo N, Monzon AM, Hatos A, Mičetić I, Quaglia F, Paladin L, Ramasamy P, Dosztányi Z, et al. : MobiDB: intrinsically disordered proteins in 2021. Nucleic Acids Res 2021, 49:D361–D367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quaglia F, Mészáros B, Salladini E, Hatos A, Pancsa R, Chemes LB, Pajkos M, Lazar T, Peña-Díaz S, Santos J, et al. : DisProt in 2022: improved quality and accessibility of protein intrinsic disorder annotation. Nucleic Acids Res 2022, 50:D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukuchi S, Amemiya T, Sakamoto S, Nobe Y, Hosoda K, Kado Y, Murakami SD, Koike R, Hiroaki H, Ota M: IDEAL in 2014 illustrates interaction networks composed of intrinsically disordered proteins and their binding partners. Nucleic Acids Res 2014, 42:D320–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q, Peng X, Li Y, Tang W, Zhu J ‘an, Huang J, Qi Y, Zhang Z: LLPSDB: a database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res 2020, 48:D320–D327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mészáros B, Erdős G, Szabó B, Schád É, Tantos Á, Abukhairan R, Horváth T, Murvai N, Kovács OP, Kovács M, et al. : PhaSePro: the database of proteins driving liquid-liquid phase separation. Nucleic Acids Res 2020, 48:D360–D367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You K, Huang Q, Yu C, Shen B, Sevilla C, Shi M, Hermjakob H, Chen Y, Li T: PhaSepDB: a database of liquid-liquid phase separation related proteins. Nucleic Acids Res 2020, 48:D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Necci M, Piovesan D, CAID Predictors, DisProt Curators, Tosatto SCE: Critical assessment of protein intrinsic disorder prediction. Nat Methods 2021, 18:472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narlikar GJ, Myong S, Larson D, Maeshima K, Francis N, Rippe K, Sabari B, Strader L, Tjian R: Is transcriptional regulation just going through a phase? Mol Cell 2021, 81:1579–1585. [DOI] [PubMed] [Google Scholar]
- 68.McSwiggen DT, Mir M, Darzacq X, Tjian R: Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 2019, 33:1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McSwiggen DT, Hansen AS, Teves SS, Marie-Nelly H, Hao Y, Heckert AB, Umemoto KK, Dugast-Darzacq C, Tjian R, Darzacq X: Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wheeler RJ: Therapeutics-how to treat phase separation-associated diseases. Emerg Top Life Sci 2020, 4:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall’Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, et al. : Partitioning of cancer therapeutics in nuclear condensates. Science 2020, 368:1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thelin-Järnum S, Göransson M, Burguete AS, Olofsson A, Aman P: The myxoid liposarcoma specific TLS-CHOP fusion protein localizes to nuclear structures distinct from PML nuclear bodies. Int J Cancer 2002, 97:446–450. [DOI] [PubMed] [Google Scholar]


