Summary
Background
The epilepsies are highly heritable conditions that commonly follow complex inheritance. While monogenic causes have been identified in rare familial epilepsies, most familial epilepsies remain unsolved. We aimed to determine (1) whether common genetic variation contributes to familial epilepsy risk, and (2) whether that genetic risk is enriched in familial compared with non-familial (sporadic) epilepsies.
Methods
Using common variants derived from the largest epilepsy genome-wide association study, we calculated polygenic risk scores (PRS) for patients with familial epilepsy (n = 1,818 from 1,181 families), their unaffected relatives (n = 771), sporadic patients (n = 1,182), and population controls (n = 15,929). We also calculated separate PRS for genetic generalised epilepsy (GGE) and focal epilepsy. Statistical analyses used mixed-effects regression models to account for familial relatedness, sex, and ancestry.
Findings
Patients with familial epilepsies had higher epilepsy PRS compared to population controls (OR 1·20, padj = 5×10−9), sporadic patients (OR 1·11, padj = 0.008), and their own unaffected relatives (OR 1·12, padj = 0.01). The top 1% of the PRS distribution was enriched 3.8-fold for individuals with familial epilepsy when compared to the lowest decile (padj = 5×10−11). Familial PRS enrichment was consistent across epilepsy type; overall, polygenic risk was greatest for the GGE clinical group. There was no significant PRS difference in familial cases with established rare variant genetic etiologies compared to unsolved familial cases.
Interpretation
The aggregate effects of common genetic variants, measured as polygenic risk scores, play an important role in explaining why some families develop epilepsy, why specific family members are affected while their relatives are not, and why families manifest specific epilepsy types. Polygenic risk contributes to the complex inheritance of the epilepsies, including in individuals with a known genetic etiology.
Funding
National Health and Medical Research Council of Australia, National Institutes of Health, American Academy of Neurology, Thomas B and Jeannette E Laws McCabe Fund, Mirowski Family Foundation.
Keywords: Epilepsy genetics, Familial epilepsies, Common genetic variation, Polygenic risk scores
Research in context.
Evidence before this study
The study of multiplex families with epilepsy led to the first monogenic epilepsy gene being discovered in 1995. However, monogenic families are rare, and epidemiological studies have suggested complex (rather than monogenic) inheritance in most families with epilepsy. We searched PubMed with the terms “complex genetics” and “familial epilepsy” for reports published before 01 June, 2021, with no language restrictions. Although the contribution of rare epilepsy-associated microdeletions and enrichment of ultra-rare variants in dominant epilepsy-associated genes have been described in familial epilepsies, there were no reports describing the role of common genetic variation. Whilst genome-wide association studies (GWAS) have determined that common genetic variants contribute to the complex inheritance of epilepsy, these have all been performed without regard to familial status. Polygenic risk scores (PRS) derived from these GWAS have shown that individuals (again, unselected for familial status) with epilepsy have higher PRS than population controls. These findings indicate a role for common variants in the overall risk of epilepsy in the population, however, the contribution of common variation to familial epilepsies has not been investigated.
Added value of this study
This study shows that common genetic variation is enriched in a large collection of 1,181 independent families with epilepsy. Until now, the study of families with epilepsy have been limited in scope to the contribution of rare genetic variation. Our results demonstrate that common epilepsy risk variants can help explain why some family members develop epilepsy while others do not, as well as why families manifest specific epilepsy types. Extending our analysis to capture rare variant status enabled us to discuss the relative contribution of rare and common genetic variants in familial epilepsies, with evidence now clearly supporting a complex inheritance role for both.
Implications of all the available evidence
The present study provides three key findings towards our understanding of familial epilepsy genetics. First, we show that common genetic variation explains a significant portion of familial epilepsy risk, significantly greater than that seen in sporadic epilepsy. Second, our results support an important role for the degree of positive family history with the highest polygenic risk observed in individuals from multiplex families with ≥3 affected relatives. Finally, with regards to the magnitude of common variant risk captured, we demonstrate a significant 3.8-fold enrichment for individuals with familial epilepsy compared to controls in the top 1% highest epilepsy PRS distribution; this is comparable to several established epilepsy-associated microdeletions. Collectively, these results all have important genetic counselling implications for people with epilepsy, and their family members, particularly in the case of a strong positive family history for epilepsy, and regardless of rare variant status.
Alt-text: Unlabelled box
Introduction
Families with epilepsy have played a critical role in unravelling the genetic basis of the epilepsies. Early twin and family-based studies demonstrated familial aggregation and provided evidence for high heritability.1,2 Rare multi-generational pedigrees, consistent with monogenic inheritance, then led to the first discoveries of epilepsy genes.3 Since then, the past decade has seen tremendous progress in gene discovery, with hundreds of genes now established as monogenic causes of epilepsy.4 However, this recent gene discovery has predominantly occurred in the most severe group of epilepsies, the developmental and epileptic encephalopathies (DEEs).5 As these pathogenic variants have often arisen de novo and these affected individuals seldom have children, paradoxically these gene discoveries have not explained most of epilepsy's heritability. Indeed, most families with epilepsy do not segregate a single monogenic gene.
The underlying genetic architecture of common epilepsies and most familial epilepsies is not well understood. Epidemiological studies have suggested complex (rather than monogenic) inheritance in most families,1,6 yet the contributory genetic factors remain largely unknown. More recently, genome-wide association studies have demonstrated the contribution of common variants to epilepsy genetic risk.7 Polygenic risk scores (PRS) are individual-level risk estimates based on the aggregate effects of common variants derived from genome-wide association studies. PRS provide meaningful risk measures for the epilepsies overall,8 and for families with other complex neurological disorders such as migraine, bipolar disorder and late-onset Alzheimer's disease.9, 10, 11
We hypothesised that the accumulation of common variants with small individual effect size contributes to familial aggregation of epilepsy. We aimed to determine whether PRS contribute to familial epilepsy risk, and whether that genetic risk is enriched in familial compared with non-familial (sporadic) epilepsies. We also compared the effects of separate risk scores for generalised and focal epilepsies. Finally, we compared PRS in families with, versus those without, an established monogenic etiology or rare epilepsy-associated microdeletion.
Methods
Cohorts
Individuals with and without epilepsy from families with epilepsy were ascertained from the Epi4K Consortium12 and The University of Melbourne's Epilepsy Genetics Program. Singleton patients with epilepsy were also ascertained from the Epi4K Consortium and from tertiary hospitals in Australia, New Zealand and the US. Control data were obtained from the QSkin Sun and Health Study, a prospective cohort study of men and women, randomly sampled from the Australian state of Queensland13 (Supplementary Table 1).
Ethics statement
This study was approved by the Austin Health Human Research Ethics Committee (H2007/02961). All participants, or their legal guardians, provided signed informed consent according to local IRB requirements.
Clinical classifications
People with epilepsy were classified as familial if there was any reference to a positive family history for epilepsy recorded clinically. People with epilepsy who did not have a known family history of epilepsy were classified as sporadic. We further divided our familial group into individuals from multiplex families with ≥3 affected relatives (as defined previously)12 versus individuals from non-multiplex families (with only two affected relatives) or a positive family history with unknown number of affected relatives.
Patients were diagnosed with epilepsy and classified into epilepsy types according to the International League Against Epilepsy (ILAE) classification criteria.14 Relatives diagnosed with febrile seizures, but not epilepsy, were classified as affected. Finally, to identify any families or individuals that carry established pathogenic or epilepsy risk variants, we cross-referenced our case identifiers with results from previous exome sequencing,15,16 copy number variant analyses17 and the University of Melbourne's epilepsy genetics database. Individuals were coded as rare variant carriers if one or more of their variants was 1) a pathogenic/likely pathogenic rare variant according to ACMG guidelines,18 or 2) one of three most common recurrent epilepsy risk microdeletions (15q13.3, 15q11.2, 16p13.11).19,20 While the risk conferred by these three microdeletions differs, we included these genetic etiologies given that they have been historically included in many epilepsy genetic studies.
Data processing
Case and control SNP genotyping was performed on one of four Illumina arrays: Global Screening Array (GSA), HumanCore (HC), Human Multi-Ethnic (HME) or Multi-Ethnic Global Array (MEGA) (Supplementary Table 1, Supplementary Figure 1).
We performed standard quality control (QC) measures on each array dataset separately (Supplementary Figure 2). Using PLINK v1.9,21 we excluded SNPs that exhibited high ‘missingness’ rates (>2%), low minor allele frequency (MAF<0·5%), or failed a test of Hardy-Weinberg equilibrium (p<10−6). We also excluded samples with high (>0·2) or low (< -0·2) rates of heterozygosity, a high proportion of missing genotypes (>2%), and those with missing, ambiguous, or sex mismatch between X-chromosome genotype and reported gender. Finally, we merged each dataset with 1000 Genomes data for ancestry PCA using GCTA.22 Samples that did not cluster with the European super population were excluded.
We then merged the HME and MEGA datasets, given their 98·2% SNP concordance, and repeated the above quality control steps. Furthermore, we performed a pseudo-association analysis between samples from each dataset to identify markers with significantly different MAFs for removal (p<10−5).
After these QC measures, three datasets (GSA, HC, HME/MEGA) required imputation before merging. Imputation to the HRC r1.1 2016 (GRCh37/hg19) reference panel was performed using Minimac4 as implemented on the Michigan Imputation Server23 with pre-imputation phasing using Eagle v2.4.24
Post-imputation, we selected high-quality imputed and genotyped SNPs with Minimac4 imputation quality scores R2>0·9 and repeated the standard quality control measures for the three datasets separately. We then merged the imputed datasets one by one, repeating the standard QC steps each time. We also excluded any markers that failed a test of differential missingness (p<10−5) between the merged datasets. PCA for ancestry was performed on the final merged cohort using PC-Air to account for related samples25 (Supplementary Figure 3).
Finally, we generated pairwise identity-by-descent (IBD) estimates with PLINK v1·921 to identify any duplicate or cryptically related samples across the merged datasets. In the case of duplicate samples, we kept the sample with the least missing data. In the case of cryptically related samples (proportion IBD > 0·125), if they were control samples, we again kept the sample with the least missing data. However, if the two samples were individuals with epilepsy, we kept both and linked their family identifiers (FID). We also used these results to check that the pedigree relationships within our multiplex families were as expected and removed any problem samples (Supplementary Figure 4).
Polygenic risk scores
We calculated polygenic risk scores (PRS) for all case and control individuals using the SNP effect sizes estimated from the largest GWAS of epilepsy conducted thus far.7 Our cohorts did not contribute to the underlying source GWAS that was used to generate the weights (betas) for the PRS.7
PRS for epilepsy, genetic generalised epilepsy (GGE), and focal epilepsy were generated using PRSice-226 for each sample that survived pre- and post- imputation quality control measures. We used a significance threshold of p<0·5 for SNP inclusion in our primary analyses, the optimal threshold determined by a previous epilepsy PRS study.8 Other thresholds were also investigated. As a negative control experiment, we also calculated PRS for asthma, based on publicly available summary statistics from an independent GWAS study, thresholded at p<0·5.27 All PRS values were normalised to a standard normal distribution with a mean of zero and standard deviation of one.
Statistical analyses
Because our dataset included related individuals within families, we used mixed-effects regression analysis to account for clustered data. For most analyses, we applied the logistic mixed-effects regression models implemented in GMMAT28 that treated epilepsy phenotype as a categorical (binary) outcome variable, PRS as the exposure variable, sex, and the first five ancestry principal components as fixed-effects covariates, and the genetic relatedness matrix (GRM) as a random-effects covariate. The GRM was generated based on genetic variants using PLINK v1·9.21 Results of the logistic mixed-effects models are expressed as odds ratios, representing the increase in odds of being in the test group versus the control group, for every 1-standard deviation unit increase in polygenic risk score, after adjusting for covariates and genetic relatedness. P-values were derived from Wald tests.
To assess the impact of polygenic risk for individuals carrying the highest burden of risk alleles, we stratified the PRS into deciles based on the distribution of PRS in population controls. We then assigned epilepsy patients and unaffected relatives to these deciles based on their PRS values. These decile categorizations then replaced quantitative PRS values in the logistic mixed-effects regression models as a predictor of the phenotype outcome.
We corrected for multiple comparisons using the Benjamini-Hochberg procedure to control the false discovery rate at 0.05, a method robust to multiple dependent tests,29 and report adjusted p-values. Analyses were performed using the statistical software R version 4.0.2.
Role of funders
The funders played no role in study design, data collection, data analyses, interpretation, or writing of this manuscript.
Results
Cohort
The cohort characteristics are shown in Table 1. The 1,818 patients with familial epilepsy belonged to 1,181 families; in 270 families, more than one affected individual was genotyped (Supplementary Figure 5). The 771 unaffected relatives of familial cases came from 212 families. The 1,182 patients with sporadic epilepsy included 719 individuals with a documented negative family history and 463 individuals with unknown family history. Overall, 31% of individuals with epilepsy had GGE, 42% focal epilepsies, and 27% other epilepsies comprised largely of DEEs, febrile seizures, or unclassified epilepsy (demographic and phenotypic information in Supplementary Tables 2 and 3).
Table 1.
Study cohort.
| Group | N | GGE, n (%) | Focal, n (%) | Other Epilepsies, n (%) |
|---|---|---|---|---|
| Epilepsy Cases | 3,000 | 922 (31%) | 1,276 (42%) | 802 (27%) |
| - Familial | 1,818 | 678 (37%) | 601 (33%) | 539 (30%) |
| - Sporadic | 1,182 | 244 (21%) | 675 (57%) | 263 (22%) |
| Unaffected Relativesa | 771 | •• | •• | •• |
| Controls | 15,929 | •• | •• | •• |
Unaffected Relatives are relatives of Familial Epilepsy cases only.
Abbreviations: GGE, genetic generalised epilepsies.
Epilepsy polygenic risk is enriched in familial cases
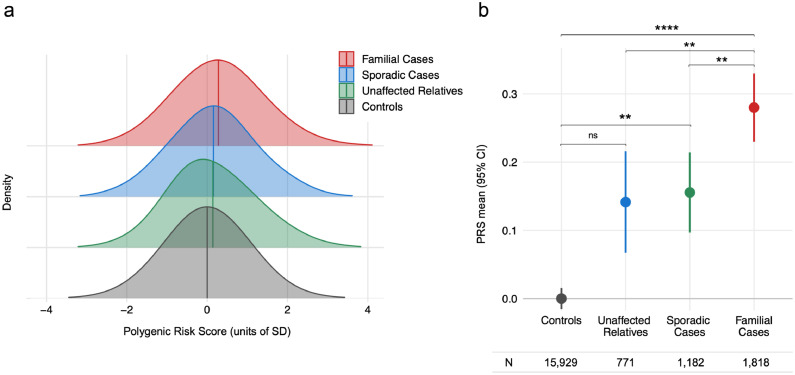
Using the epilepsy polygenic risk model (Figure 1), we found that polygenic risk was higher in familial cases compared to population controls (OR 1·20, 95% CI 1·13, 1·27, Wald test padj = 5×10−9). For every 1-standard deviation increase in PRS, the odds of an individual being a familial epilepsy case increased by 20%. Polygenic risk was also higher in familial cases compared to sporadic cases (OR 1·11, 95% CI 1·02, 1·20, padj = 0.008) and compared to unaffected relatives of familial cases (OR 1·12, 95% CI 1·03, 1·21, padj = 0.01).
Figure 1.
Epilepsy polygenic risk is enriched in familial cases.
Polygenic risk scores (PRS) for all epilepsies were higher in patients with familial epilepsy than in population controls, sporadic epilepsy patients, and unaffected relatives of familial cases. (a) Normalised distributions of epilepsy PRS (b) Mean epilepsy PRS values. Logistic mixed-effects regression models used PRS as exposure variable, sex and the first five ancestry principal components as fixed-effects covariates, and genetic relatedness matrix as random-effects covariate. P-values from Wald tests were corrected for multiple comparisons. ns Padj ≥ 0.05, * Padj ≤ 0.05, ** Padj ≤ 0.01, *** Padj ≤ 0.001, **** Padj ≤ 0.0001. Abbreviations: PRS, polygenic risk score; CI, confidence interval.
Polygenic risk was higher in sporadic cases compared to population controls (OR 1·09, 95% CI 1·02, 1·16, padj = 0·01). Polygenic risk in unaffected relatives of familial cases was not different from population controls after adjusting for multiple comparisons (OR 1·07, 95% CI 0.98, 1.17, padj = 0.22).
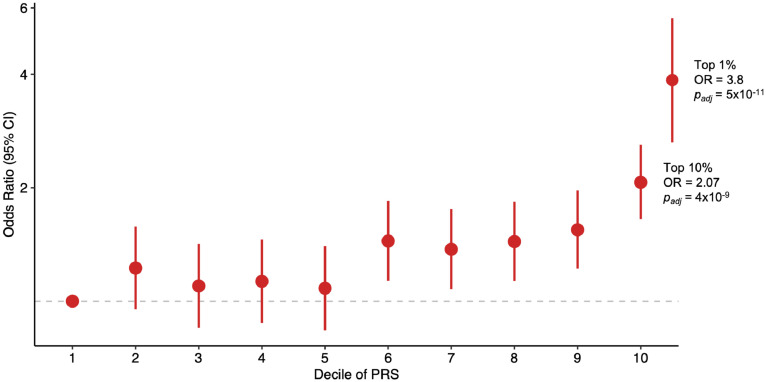
Using the lowest decile of polygenic risk as the referent, the highest decile of polygenic risk was enriched in familial epilepsy cases compared to controls (Figure 2; OR 2·07, 95% CI 1·65, 2·60, padj = 4×10−9). The top 1% of polygenic risk was even further enriched in familial epilepsy cases (OR 3·80, 95% CI 1·92, 5·69, padj = 5×10−11). A similar trend was observed in sporadic epilepsy cases and unaffected relatives (Supplementary Table 4).
Figure 2.
Familial epilepsy cases by decile of Epilepsy PRS.
Familial epilepsy cases were assigned to PRS deciles derived from the population control distribution. Odds ratios are the odds of being an affected case relative to the lowest decile (denoted by dashed grey line).
In our primary analysis, the 1,818 familial cases were those with any family history of epilepsy, regardless of the number or degree of relatedness of the affected relatives. We further stratified familial patients into those from multiplex families, defined as three or more affected relatives per family (n = 807), and non-multiplex families, defined as two affected relatives per family or a positive family history with unknown number of affected relatives (n = 1,011). Enrichment of epilepsy polygenic risk compared to population controls was greatest in the multiplex families (OR 1·31, 95% CI 1·18, 1·44, padj = 4×10−7, Supplementary Figure 6). This trend was present within the GGE, Focal, and Other Epilepsy subgroups (data not shown), suggesting it was not caused by imbalances in epilepsy types between multiplex and non-multiplex families.
Our primary analyses used a polygenic risk model that included all risk alleles below the threshold of p < 0·5, based on prior studies.8 We also tested the effects of polygenic risk models that used more stringent thresholds that included fewer variants in calculating risk scores. Consistent with prior studies, the trends remained robust across thresholds, with the more lenient thresholds (p<0·5 and p<0·1) providing the greatest power to distinguish cases from controls (Supplementary Table 5).
To confirm that the differences in polygenic risk between cases and controls were specific to epilepsy, and not due to a systematic bias between cases and controls, we performed a negative control experiment. We calculated PRS for asthma, a non-neurological disorder that is not co-morbid with epilepsy nor expected to share genetic risk. There was no significant difference in asthma PRS between our epilepsy cohorts and population controls (Supplementary Table 6).
Polygenic enrichment stratified by epilepsy phenotype
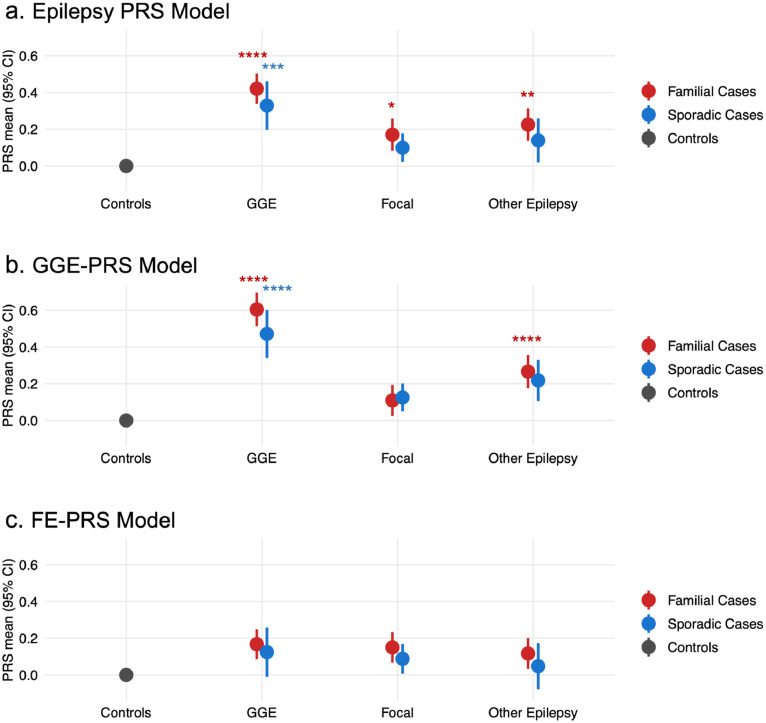
To determine whether the familial epilepsy PRS enrichment was being driven by a particular type of epilepsy, we further stratified cases according to epilepsy type. The magnitude of enrichment for familial compared to sporadic cases was consistent across each of the three broad epilepsy groups (Figure 3a; Supplementary Table 7a).
Figure 3.
Polygenic risk stratified by epilepsy type.
(a) The Epilepsy PRS model was highest in familial cases across all epilepsy phenotype groups. (b) The polygenic risk scores (PRS) model for risk of genetic generalised epilepsy (GGE) was highest in individuals with familial GGE. (c) The PRS model for risk of focal epilepsy was not different from controls in any of the phenotypic groups. Logistic mixed-effects regression models used PRS as exposure variable, sex and the first five ancestry principal components as fixed-effects covariates, and genetic relatedness matrix as random-effects covariate. Stars indicate statistically significant comparisons to the control group, corrected for multiple comparisons (* Padj ≤ 0.05, ** Padj ≤ 0.01, *** Padj ≤ 0.001, **** Padj ≤ 0.0001); all other comparisons were non-significant (Padj > 0.05). Full results are available in Supplementary Table 7.
We next generated PRS models specific to genetic generalised epilepsies (GGE-PRS) and focal epilepsies (FE-PRS), using summary statistics from the GWAS of each phenotype.7 We applied these PRS models to our patients with epilepsy, again stratified by epilepsy type and familial status (Figure 3b, 3c).
For the GGE-PRS model, enrichment of polygenic risk compared to population controls was greatest in familial GGE cases (OR 1·58, 95% CI 1·45, 1·72, padj = 6×10−25). GGE-PRS was enriched to a lesser extent in sporadic GGE cases (OR 1·46, 95% CI 1·28, 1·65, padj = 5×10−8), however, the difference between familial and sporadic GGE cases was not significant after adjustment (OR 1·11, 95% CI 0·97, 1·28, padj = 0.21). GGE-PRS values in individuals with focal epilepsies did not differ from population controls (Supplementary Table 7b). Individuals with other epilepsies and a family history of epilepsy had higher GGE-PRS compared to population controls (OR 1·25, 95% CI 1·14, 1·37, padj = 4×10−6).
For the FE-PRS model, no epilepsy group was significantly different from population controls (Supplementary Table 7c).
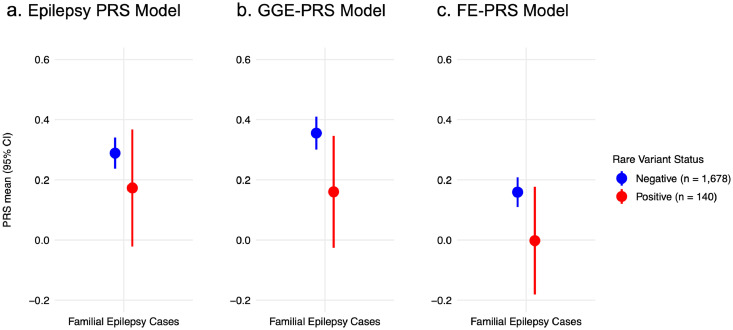
Polygenic risk in rare epilepsy variant carriers and non-carriers
In our familial epilepsy cohort, 140/1,818 individuals (7·4%) from 92 families carried an epilepsy-associated rare pathogenic variant (pathogenic single nucleotide variant or recurrent microdeletion); Supplementary Table 8 shows their phenotypes. Across all three PRS models, there was a non-significant trend towards lower PRS values in rare pathogenic variant carriers compared to non-carriers (Figure 4; Supplementary Table 9a). No trend was evident in individuals with sporadic epilepsy who carried rare pathogenic epilepsy variants (Supplementary Table 9b; Supplementary Figure 7).
Figure 4.
PRS in familial cases carrying rare epilepsy variants no different to non-carriers.
Polygenic risk score (PRS) models specific to (a) all epilepsy, (b) genetic generalised epilepsies (GGE) and (c) focal epilepsies in individuals with familial epilepsy who had known rare pathogenic variants (red; n = 140) versus individuals without rare pathogenic variants (blue; n = 1,678). Statistical comparisons were non-significant (Padj > 0.05). Full results are available in Supplementary Table 9.
Discussion
We examined the contribution of polygenic risk scores to familial epilepsies, using a large cohort of 1,181 epilepsy families genotyped for common genetic variations. We found that individuals with familial epilepsy had higher PRS than population controls, their unaffected relatives, and individuals with sporadic epilepsy. This effect was greatest for patients from multiplex families comprising ≥3 affected relatives. For people in the top percentile of the epilepsy PRS distribution, the odds of having familial epilepsy was increased approximately 4-fold; this is comparable to several established epilepsy-associated microdeletions.19 These findings provide strong evidence that the aggregate effect of common variants, each conferring small amounts of risk, plays an important role in familial epilepsy and, by extension, in the overall genetic contribution to epilepsy within families.
Prior studies showed that unselected persons with epilepsy (not stratified by family history) have higher PRS than population controls.8 Additionally, GWAS identified common variant risk alleles for the epilepsies (without regard to familial status), although the number of genome-wide significant loci was small.7 These findings supported a role for common variants in the overall risk of epilepsy in the population. We extend these findings by showing enrichment of common polygenic risk in people with epilepsy who have affected relatives compared to those without affected relatives. A role for such complex, non-Mendelian genetic factors in epilepsy has been hypothesised from epidemiological data, for example, the observation that most family pedigrees are not consistent with a monogenic (dominant or recessive) mechanism.6,12,30
Similar familial polygenic enrichment has been observed in other complex neurological disorders. For example, in both migraine9 and bipolar disorder,11 significantly higher PRS burden in familial cases versus non-familial cases and unaffected relatives has also been observed. Interestingly, the unaffected relatives from our multiplex families with epilepsy showed no statistically significant difference in epilepsy PRS burden compared to unrelated controls; this result differs from both the migraine and bipolar disorder studies.9,11 As our smallest cohort, it may be that our unaffected relative group was underpowered as opposed to there being no true difference from controls.
Epilepsy is a clinically heterogeneous disorder. Clinical stratification of our cohort into GGE, focal and other epilepsy types, revealed that the degree of familial polygenic enrichment was consistent across clinical groups. This indicates that the epilepsy PRS model is capturing greatest risk in familial cases, regardless of phenotype. Overall, the GGE group showed greatest epilepsy PRS enrichment. As expected, enrichment for the GGE-PRS model was even stronger with highest mean GGE-PRS reserved for individuals with familial GGE. No significant enrichment was achieved for the FE-PRS model even when applied to the focal epilepsy group.
That the generalised and focal epilepsy polygenic models and clinical groups yielded different results is not surprising. Our findings are consistent with the observation that GGE is both more heritable overall1,2 and has a larger proportion of heritability explained by common variants compared to focal epilepsy (approximately 32% vs 9%).7 They are also consistent with epidemiological studies of familial epilepsies that have found evidence for distinct genetic factors underlying different epilepsy phenotypes.31 Our findings help in part to explain why families, including the majority without a known variant of large effect, tend to manifest concordant epilepsy types, particularly the GGEs. Conversely, families that segregate a monogenic variant often show phenotypic heterogeneity (e.g., genetic epilepsy with febrile seizures plus [GEFS+] families).32 We hypothesise that this phenotypic variability could be due in part to differences in the background distribution of common variants between individuals; a hypothesis that should be tested by future research.
An important question is the relative role of rare and common variants within families. Our cohort included a relatively small sample of familial cases with known rare pathogenic variants or microdeletions, and our ability to draw strong conclusions is limited. Rare epilepsy variant carriers had a trend toward lower PRS than non-carriers, suggesting a diminished role of common variants in the presence of stronger risk alleles as has been recently shown in other complex diseases such as type 2 diabetes.33 However, our result for epilepsy was not significant and, although this may be due to a lack of statistical power, we cannot confidently infer that a difference exists. Future studies will require larger sample sizes to investigate the role of common variants in families with identified rare genetic variants. It could be that an inverse relationship exists—that is, in the most extreme case, a family is either monogenic or polygenic. Alternatively, common variants may play a role even in monogenic families, perhaps explaining phenomena such as phenotypic variability and incomplete penetrance.34, 35, 36, 37
As familial epilepsies are also enriched for ultra-rare variants in dominant epilepsy-associated genes,15 it is reasonable to assume that the polygenic background may involve a complex combination of both rare and common variants. Recent studies of the DEEs, and severe neurodevelopmental disorders more broadly, found enrichment of damaging ultra-rare variants38 and common genetic variation,39,40 that was present in cases both with and without de novo monogenic variants. An additive effect of polygenic risk and de novo pathogenic variants has also been observed in autism spectrum disorder.41 Our results suggest that the presence of a family history may indicate that common variants are making an important contribution to the genetic background, even in families with an established (or presumed) monogenic etiology. Finally, it is known that the risk of epilepsy following acquired injury42 is increased in people with no prior seizures, but a family history of epilepsy. We hypothesise that polygenic background also may underlie this observation.
Our study had several limitations. As with all PRS analyses, the predictive power will improve with increasing GWAS sample sizes that will permit more reliable estimation of common variant effect sizes. This is particularly relevant for the FE-PRS with its lower heritability and thus greater requirement for larger GWAS sample sizes. Furthermore, we know that rare risk alleles for epilepsy exist (e.g., recurrent microdeletions), yet our PRS models are currently limited to the common variants captured by GWAS. Certainly, there would also be additional clinical features associated with risk of developing epilepsy not captured, but we were limited by both these data not being available for our control cohort and for some of our epilepsy cases (particularly those from retrospective consortium collections). We do not, however, feel that the absence of additional covariates undermines our findings or conclusions. We defined sporadic epilepsy as individuals without a known family history of epilepsy. Some of these individuals may have a family history of epilepsy that was not reported, although such misclassification would make it harder to detect a significant difference between the groups. Further, our cohort included a wide range of epilepsy types and subtypes, which may have different genetic architectures. Finally, our study was limited to individuals of European descent because our polygenic risk models were derived from European-based GWAS. In epilepsy8 and other disorders, it has been shown that PRS models do not generalise well across different populations. Future large-scale GWAS studies in diverse populations will be essential to help address this important disparity.
In conclusion, we show that the combined effects of common variants, measured here as PRS, play an important role in familial epilepsies, and helps to explain the complex genetic basis of the epilepsies which is incompletely explained by known monogenic genes.
Contributors
Co-wrote first draft: KLO, CAE; Study design: SFB, MB, KLO, CAE, MPE, RO, CL; Patient phenotyping: IES, LGS, SFB, IH, CAE, Epi4K; Data access and curation: KLO, CAE, RVH, ELH, HCM, SG, SWC, CL, Epi4K; Control data: DCW; Data processing and generation: KLO; Statistical analysis: CAE, AJB, MPE; Manuscript revisions: IES, SG, CL, LGS, ELH, HCM, AJB, SWC, RVH, Epi4K, DCW, IH, RO, MPE, MB, SFB; Verified underlying data: KLO, CAE, MB, SFB. All authors read and approved the final version of the manuscript.
Data sharing statement
Genetic data used in this study is not available due to patient privacy and ethical restrictions.
Declaration of interests
Samuel Berkovic has received grants from UCB Pharma, Eisai, SciGen; consulting fees from Praxis Precision Medicines, Sequiris; honoraria from Eisai; has a patent for SCN1A testing held by Bionomics Inc licensed to Athena Diagnostics and Genetics Technologies Ltd. Ingrid Scheffer has served on scientific advisory boards for UCB, Eisai, GlaxoSmithKline, BioMarin, Nutricia, Rogcon, Chiesi, Encoded Therapeutics, Knopp Biosciences and Xenon Pharmaceuticals; has received speaker honoraria from GlaxoSmithKline, UCB, BioMarin, Biocodex, Chiesi, Liva Nova and Eisai; has received funding for travel from UCB, Biocodex, GlaxoSmithKline, Biomarin and Eisai; has served as an investigator for Zogenix, Zynerba, Ultragenyx, GW Pharma, UCB, Eisai, Xenon Pharmaceuticals, Anavex Life Sciences, Ovid Therapeutics, Epigenyx, Encoded Therapeutics and Marinus; and has consulted for Zynerba Pharmaceuticals, Atheneum Partners, Ovid Therapeutics, Care Beyond Diagnosis, Epilepsy Consortium and UCB; and is a Non-Executive Director of Bellberry Ltd. She may accrue future revenue on pending patent WO61/010176 (filed: 2008): Therapeutic Compound; has a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies; has a patent molecular diagnostic/theranostic target for benign familial infantile epilepsy (BFIE) [PRRT2] 2011904493 & 2012900190 and PCT/AU2012/001321 (TECH ID:2012-009). David Whiteman has received speaker fees from Pierre Fabre. Melanie Bahlo has received payment for thesis examination from University of Melbourne, University of Sydney, University of Western Australia; has received support for attending conferences: Genemappers, International Conference on Familial Cortical Myoclonic Tremor Epilepsy and Repeat Expansion Diseases, Lorne Genome, Bioinforsummer, Australian Academy of Science Australia-China symposium on precision medicine; has served as an advisor for ALADIN, Kinghorn Sequencing Center, Murdoch Children's Research Institute, Viertel Foundation; is a committee member for Australian Academy of Health and Medical Sciences, American Epilepsy Society Basic Sciences, Gen V Bioresource Genetics. Costin Leu received conference travel support from the International Epilepsy Congress/ILAE. Ingo Helbig received travel support from the Americal Epilepsy Society. Heather Mefford has received support from CURE Epilepsy and sits on the American Society of Human Genetics Board of Directors. Lynette Sadleir has received consulting fees from the Epilepsy Consortium, Zynerba; is an advisor to Eisai; and is treasurer for the New Zealand League Against Epilepsy.
Acknowledgements
We thank the patients, their families, and the participants in the QSKIN Study for contributing to this research.
Karen L Oliver is supported by an Australian Government Research Training Program Scholarship [APP533086] provided by the Australian Commonwealth Government and the University of Melbourne. Colin A Ellis is supported by the National Institute Of Neurological Disorders And Stroke of the National Institutes of Health Award Number K23NS121520; by the American Academy of Neurology Susan S Spencer Clinical Research Training Scholarship; by the Thomas B and Jeannette E Laws McCabe Fund at the University of Pennsylvania; and by the Mirowski Family Foundation. Ingrid Scheffer and Samuel Berkovic are supported by a National Health and Medical Research Council (NHMRC) of Australia Program Grant [1091593], and Ingrid Scheffer is further supported by a NHMRC Practitioner Fellowship [1006110] and Senior Investigator grant [1172897], the Medical Research Future Fund, Australian Epilepsy Research Fund, Shenzhen Sanming Development Grant, Einstein Foundation Berlin and the Health Research Council of New Zealand. Melanie Bahlo is supported by an NHMRC Investigator grant [APP1195236]. David Whiteman is supported by a Research Fellowship [APP1155413] from the NHMRC. Samuel Berkovic and Heather Mefford are supported by the National Institute Of Neurological Disorders And Stroke of the National Institutes of Health. Lynette Sadleir is supported by the Health Research Council of New Zealand and Cure Kids New Zealand. Ingo Helbig is supported by the German Research Foundation (HE5415/6-1, DFG/FNR INTER Research Unit FOR2715 HE5415/7-1 and HE5415/7-2); The Hartwell Foundation; The National Institute for Neurological Disorders and Stroke (K02 NS112600); The Center Without Walls on the Ion channel function in Epilepsy (U54 NS108874); Through the Intellectual and Developmental Disabilities Research Center (IDDRC); The University of Pennsylvania (U54 HD086984); The National Center for Advancing Translational Sciences of the National Institutes of Health (Award Number UL1TR001878); The Institute for Translational Medicine Therapeutics’ Transdisciplinary Program (ITMAT); The Epilepsy NeuroGenetics Initiative (ENGIN); EuroEpinomics-Rare Epilepsy Syndrome (RES); The International League Against Epilepsy (ILAE). The QSKIN Study is supported by Grants [APP1185416, APP1073898, APP1063061] from the NHMRC. This work was also supported by the Victorian Government's Operational Infrastructure Support Program and the NHMRC Independent Research Institute Infrastructure Support Scheme (IRIISS).
Genomic data for participants from tertiary hospitals in the AUS, NZ, and the US that contributed to this study were provided by the Epi25 Collaborative (www.epi-25.org).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104079.
Appendix. Supplementary materials
References
- 1.Annegers J.F., Hauser W.A., Anderson V.E., Kurland LT. The risks of seizure disorders among relatives of patients with childhood onset epilepsy. Neurology. 1982;32:174–179. doi: 10.1212/wnl.32.2.174. [DOI] [PubMed] [Google Scholar]
- 2.Lennox W.G. The heredity of epilepsy as told by relatives and twins. J Am Med Assoc. 1951;146:529–536. doi: 10.1001/jama.1951.03670060005002. [DOI] [PubMed] [Google Scholar]
- 3.Steinlein O.K., Mulley J.C., Propping P., et al. A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 1995;11:201–203. doi: 10.1038/ng1095-201. [DOI] [PubMed] [Google Scholar]
- 4.Perucca P., Bahlo M., Berkovic S.F. The genetics of epilepsy. Annu Rev Genom Hum Genet. 2020;21:205–230. doi: 10.1146/annurev-genom-120219-074937. [DOI] [PubMed] [Google Scholar]
- 5.Guerrini R., Balestrini S., Wirrell E.C., Walker M.C. Monogenic epilepsies: disease mechanisms, clinical phenotypes, and targeted therapies. Neurology. 2021;97(17):817–831. doi: 10.1212/WNL.0000000000012744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peljto A.L., Barker-Cummings C., Vasoli V.M., et al. Familial risk of epilepsy: a population-based study. Brain. 2014;137:795–805. doi: 10.1093/brain/awt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International League Against Epilepsy Consortium on Complex Epilepsies Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. 2018;9:5269. doi: 10.1038/s41467-018-07524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leu C., Stevelink R., Smith A.W., et al. Polygenic burden in focal and generalized epilepsies. Brain. 2019;142:3473–3481. doi: 10.1093/brain/awz292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gormley P., Kurki M.I., Hiekkala M.E., et al. Common variant burden contributes to the familial aggregation of migraine in 1,589 families. Neuron. 2018;99:1098. doi: 10.1016/j.neuron.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 10.Tosto G., Bird T.D., Tsuang D., et al. Polygenic risk scores in familial Alzheimer disease. Neurology. 2017;88:1180–1186. doi: 10.1212/WNL.0000000000003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andlauer T.F.M., Guzman-Parra J., Streit F., et al. Bipolar multiplex families have an increased burden of common risk variants for psychiatric disorders. Mol Psychiatry. 2021;26:1286–1298. doi: 10.1038/s41380-019-0558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epi4K Consortium Phenotypic analysis of 303 multiplex families with common epilepsies. Brain. 2017;140:2144–2156. doi: 10.1093/brain/awx129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen C.M., Green A.C., Neale R.E., et al. Cohort profile: the QSkin Sun and Health Study. Int J Epidemiol. 2012;41:929. doi: 10.1093/ije/dys107. -i. [DOI] [PubMed] [Google Scholar]
- 14.Scheffer I.E., Berkovic S., Capovilla G., et al. ILAE classification of the epilepsies: position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epi4K Consortium, Epilepsy Phenome/Genome Project Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017;16:135–143. doi: 10.1016/S1474-4422(16)30359-3. [DOI] [PubMed] [Google Scholar]
- 16.Epi4K Consortium, Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epilepsy Phenome/Genome Project, Epi4K Consortium Copy number variant analysis from exome data in 349 patients with epileptic encephalopathy. Ann Neurol. 2015;78:323–328. doi: 10.1002/ana.24457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Kovel C.G., Trucks H., Helbig I., et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helbig I., Mefford H.C., Sharp A.J., et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S., Forer L., Schonherr S., et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh P.R., Danecek P., Palamara P.F., et al. Reference-based phasing using the haplotype reference consortium panel. Nat Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi S.W., O’Reilly P.F. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8(7) doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demenais F., Margaritte-Jeannin P., Barnes K.C., et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H., Wang C., Conomos M.P., et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 30.Ottman R., Hauser W.A., Barker-Cummings C., Lee J.H., Risch N. Segregation analysis of cryptogenic epilepsy and an empirical test of the validity of the results. Am J Hum Genet. 1997;60:667–675. [PMC free article] [PubMed] [Google Scholar]
- 31.Winawer M.R., Rabinowitz D., Barker-Cummings C., et al. Evidence for distinct genetic influences on generalized and localization-related epilepsy. Epilepsia. 2003;44:1176–1182. doi: 10.1046/j.1528-1157.2003.58902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheffer I.E., Berkovic S.F. Generalized epilepsy with febrile seizures plus. A genetic disorder with heterogeneous clinical phenotypes. Brain. 1997;120(Pt 3):479–490. doi: 10.1093/brain/120.3.479. [DOI] [PubMed] [Google Scholar]
- 33.Lu T., Forgetta V., Richards J.B., Greenwood C.M.T. Polygenic risk score as a possible tool for identifying familial monogenic causes of complex diseases. Genet Med. 2022 doi: 10.1016/j.gim.2022.03.022. In press. [DOI] [PubMed] [Google Scholar]
- 34.Fahed A.C., Wang M., Homburger J.R., et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun. 2020;11:3635. doi: 10.1038/s41467-020-17374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassanin E., May P., Aldisi R., et al. Breast and prostate cancer risk: the interplay of polygenic risk, rare pathogenic germline variants, and family history. Genet Med. 2022;24:576–585. doi: 10.1016/j.gim.2021.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Goodrich J.K., Singer-Berk M., Son R., et al. Determinants of penetrance and variable expressivity in monogenic metabolic conditions across 77,184 exomes. Nat Commun. 2021;12:3505. doi: 10.1038/s41467-021-23556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mars N., Widen E., Kerminen S., et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat Commun. 2020;11:6383. doi: 10.1038/s41467-020-19966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takata A., Nakashima M., Saitsu H., et al. Comprehensive analysis of coding variants highlights genetic complexity in developmental and epileptic encephalopathy. Nat Commun. 2019;10:2506. doi: 10.1038/s41467-019-10482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell C., Leu C., Feng Y.C.A., et al. The role of common genetic variation in presumed monogenic epilepsies. eBioMedicine. 2022 doi: 10.1016/j.ebiom.2022.104098. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niemi M.E.K., Martin H.C., Rice D.L., et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562:268–271. doi: 10.1038/s41586-018-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiner D.J., Wigdor E.M., Ripke S., et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet. 2017;49:978–985. doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen J., Pedersen M.G., Pedersen C.B., Sidenius P., Olsen J., Vestergaard M. Long-term risk of epilepsy after traumatic brain injury in children and young adults: a population-based cohort study. Lancet. 2009;373:1105–1110. doi: 10.1016/S0140-6736(09)60214-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.