Abstract
Tomato brown rugose fruit virus (ToBRFV) is an emerging virus of the genus Tobamovirus. ToBRFV overcomes the tobamovirus resistance gene Tm-22 and is rapidly spreading worldwide. Genetic resources for ToBRFV resistance are urgently needed. Here, we show that clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9)-mediated targeted mutagenesis of four tomato (Solanum lycopersicum) homologs of TOBAMOVIRUS MULTIPLICATION1 (TOM1), an Arabidopsis (Arabidopsis thaliana) gene essential for tobamovirus multiplication, confers resistance to ToBRFV in tomato plants. Quadruple-mutant plants did not show detectable ToBRFV coat protein (CP) accumulation or obvious defects in growth or fruit production. When any three of the four TOM1 homologs were disrupted, ToBRFV CP accumulation was detectable but greatly reduced. In the triple mutant, in which ToBRFV CP accumulation was most strongly suppressed, mutant viruses capable of more efficient multiplication in the mutant plants emerged. However, these mutant viruses did not infect the quadruple-mutant plants, suggesting that the resistance of the quadruple-mutant plants is highly durable. The quadruple-mutant plants also showed resistance to three other tobamovirus species. Therefore, tomato plants with strong resistance to tobamoviruses, including ToBRFV, can be generated by CRISPR/Cas9-mediated multiplexed genome editing. The genome-edited plants could facilitate ToBRFV-resistant tomato breeding.
Editing of host susceptibility genes in tomato confers strong resistance against an emerging virus capable of overcoming currently available resistance genes.
Introduction
Tomato mosaic virus (ToMV), a member of the genus Tobamovirus, was a major threat to the production of tomato (Solanum lycopersicum) (Broadbent, 1976), but ToMV infection has been controlled for >40 years since the introduction of durable resistance gene Tm-22 into commercial tomato cultivars (Pelham, 1966; Hall, 1980). However, a new tobamovirus, tomato brown rugose fruit virus (ToBRFV), which is insensitive to Tm-22, emerged in the Middle East in 2014 (Salem et al., 2016; Luria et al., 2017). ToBRFV rapidly spread worldwide and threatens global tomato production (van de Vossenberg et al., 2021). Tobamovirus virions can survive for many years in soil. These viruses are transmitted mechanically during agricultural activities, or via seeds (Broadbent, 1976; Dombrovsky et al., 2017). Therefore, effective and durable resistance genes to control ToBRFV and other tobamoviruses are needed. Tomato cultivars showing tolerance to ToBRFV, in which ToBRFV multiplies but disease symptoms are not evident, and others showing resistance to ToBRFV, in which ToBRFV multiplication is suppressed, have been reported (Zinger et al., 2021). Genetic analyses revealed that the tolerance trait is controlled by a single recessive gene, whereas the resistance trait is controlled at least by the gene conferring tolerance and another gene located near or at the ToMV resistance gene Tm-1 (Zinger et al., 2021). The genes controlling these phenotypes have not been identified and the durability of these traits has not been described.
Many resistance genes used to protect crops from virus infection are genetically dominant. Most, such as Tm-22, encode nucleotide binding site-leucine-rich repeat proteins (Kang et al., 2005; de Ronde et al., 2014). These dominant resistance gene products recognize viral factors to elicit host defense reactions. In general, viral mutations that promote escape from recognition can easily occur, and if they do not affect viral fitness, the resistance is overcome. Other virus resistance genes are genetically recessive. They represent loss-of-function alleles of genes that encode the host factors necessary for efficient multiplication of viruses (Kang et al., 2005; Hashimoto et al., 2016). Interestingly, most natural recessive resistance genes identified to date are to potyviruses that encode eukaryotic translation initiation factor 4E (eIF4E) or its isoform eIF(iso)4E (Kang et al., 2005; Sanfaçon, 2015). Mutations in the eIF4E or eIF(iso)4E genes confer resistance to specific classes of potyviruses (Piron et al., 2010; Chandrasekaran et al., 2016; Gauffier et al., 2016; Pyott et al., 2016). Knockout of the host genes necessary for virus multiplication could confer virus resistance, but successful examples are still limited to several cases (Chandrasekaran et al., 2016; Pyott et al., 2016; Gomez et al., 2019; Atarashi et al., 2020; Pramanik et al., 2021), possibly because the loss-of-function of endogenous genes also affects host growth and development (Gauffier et al., 2016).
Previously, we identified TOBAMOVIRUS MULTIPLICATION1 (TOM1) in Arabidopsis (Arabidopsis thaliana) (Ishikawa et al., 1991). TOM1 is necessary for efficient multiplication of tobamoviruses and encodes a seven-pass transmembrane protein that interacts with tobamovirus-encoded replication proteins (Yamanaka et al., 2000). The TOM1 protein plays roles in the formation of, and is a component of active tobamovirus replication complexes (Nishikiori et al., 2011). Simultaneous loss-of-function mutations in TOM1 and its putative paralog TOM3 resulted in near-complete inhibition of tobamovirus multiplication (Yamanaka et al., 2002). Importantly, the tom1 tom3 double mutant and a triple mutant with an additional T-DNA insertion in TOM THREE HOMOLOG1, another putative TOM1 paralog in Arabidopsis, do not exhibit substantial defects in plant growth and development (Fujisaki et al., 2006). Although the function of TOM1, other than supporting tobamovirus multiplication, is unknown, TOM1 homologs are found in numerous plant species. In tobacco (Nicotiana tabacum), Nicotiana benthamiana, and tomato, knockdown of TOM1 homologs by RNA interference (RNAi) suppresses tobamovirus multiplication (Asano et al., 2005; Chen et al., 2007; Ali et al., 2018). However, RNAi plants fall under GMO legislation, hampering their use in agriculture. Thus, we aimed here to confer ToBRFV resistance to tomato by targeted mutagenesis of TOM1 homologs.
Results
Knockout of TOM1 gene homologs in tomato
We used the clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) to knock out TOM1 genes in tomato to confer ToBRFV resistance. A BLAST search of a tomato genome sequence database (ITAG release 4.0) using the Arabidopsis TOM1 amino acid sequence as a query identified five genes, named here SlTOM1a–e (Table 1). SlTOM1a, b, and c were previously identified and named LeTH1, LeTH2, and LeTH3, respectively (Ali et al., 2018). Following the change of the scientific name of tomato, we propose gene names starting with “Sl” (S. lycopersicum) instead of “Le” (Lycopersicon esculentum). We targeted these three genes (SlTOM1a–c) and another expressed homolog, SlTOM1d. Because the expression level of SlTOM1e was extremely low in the public data (Tomato functional genomics database; http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi), it was not analyzed here. SlTOM1a–d proteins showed amino acid sequence identity >50% with one another (Supplemental Figure S1). Chromosomal locations of these genes were different from those of previously mapped tobamovirus resistance genes (Tm-1 and Tm-2) or a tolerance locus (Zinger et al., 2021).
Table 1.
TOM1 homologs in tomato
| Proposed Gene Name | SGNa Gene ID | SGNa BLASTb to Arabidopsis TOM1 |
Former Gene Name | GenBank ID | NCBIc Reference Sequence ID | Length (Amino Acids) | Normalized Expressiond (RPKM) | |
|---|---|---|---|---|---|---|---|---|
| Identity (%) | E-value | |||||||
| SlTOM1a | Solyc04g008540 | 60 | 6.00E−102 | LeTH1 | AB193041 | NP_001234096.1 | 295 | 16.14 |
| SlTOM1b | Solyc01g105270 | 67 | 1.00E−112 | LeTH2 | AB193042 | NP_001234100.1 | 297 | 3.93 |
| SlTOM1c | Solyc02g080370 | 78 | 4.00E−153 | LeTH3 | AB193043 | NP_001234306.1 | 288 | 26.96 |
| SlTOM1d | Solyc01g007900 | 61 | 4.00E−101 | XP_010315372.1 | 295 | 7.76 | ||
| SlTOM1e | Solyc09g005240 | 44 | 4.00E−62 | 246 | 0 | |||
Solanaceae Genomics Network (https://solgenomics.net/).
SGN blastn (https://solgenomics.net/tools/blast/), database: tomato genome coding sequences (ITAG release 4.0), query: Arabidopsis TOM1 amino acid sequence.
National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/).
Tomato functional genomics database (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi), Experiment D004: Transcriptome analysis of various tissues in tomato cultivar Heinz and the wild relative Solanum pimpinellifolium; Data S0593: Heinz leaves.
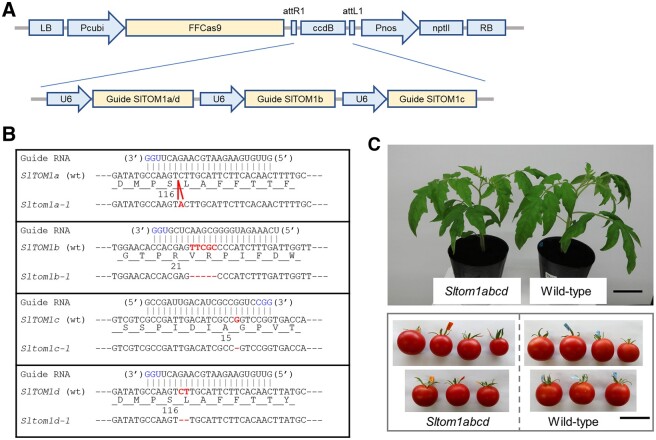
For targeted mutagenesis of SlTOM1a–d genes by the CRISPR/Cas9 system, guide RNA sequences were designed using the CRISPR-P program (http://cbi.hzau.edu.cn/crispr/; Lei et al., 2014). Because of the high nucleotide sequence identity, we used a common guide RNA sequence for the SlTOM1a and d genes. Therefore, we constructed a vector to express Streptococcus pyogenes Cas9 and three single guide RNA (sgRNA) sequences to target the SlTOM1a–d genes (Figure 1A). This gene cassette was introduced to tomato cultivar Craigella GCR26, which is susceptible to ToMV and ToBRFV, by Agrobacterium-mediated transformation. Among the progenies of the transformants, we identified individuals that carried frameshift mutations in the SlTOM1a–d genes but lost the Cas9-sgRNA cassette (Figure 1B). By crossing these plants with each other or with wild-type (wt) plants, we established single-, double-, triple-, and quadruple-mutant lines in which homozygous mutations were introduced in any of SlTOM1a–d. No obvious differences in growth or fruit production were observed between wt plants and the mutant plants including the quadruple mutant (Figure 1C). In triple- or quadruple-mutant plants, the accumulation levels of SlTOM1a, c, or d mRNA from the mutant genes were lower than those from the nonmutated genes in triple mutants or from wt plants, probably due to nonsense-mediated mRNA decay (Supplemental Figure S2). Expression of SlTOM1e was undetectable in the triple or quadruple mutant plants or in the wt plants.
Figure 1.
CRISPR/Cas9 mutagenesis of SlTOM1 genes. A, CRISPR/Cas9 constructs for disruption of SlTOM1 genes. Pcubi, ubiquitin gene (Ubi4-2) promoter from Petroselinum crispum; FFCas9, S. pyogenes Cas9 gene codon-optimized for Arabidopsis; Pnos, Nopaline synthase gene promoter from Rhizobium radiobacter; nptII, neomycin phosphotransferase II; U6, Arabidopsis U6-26 promoter. B, CRISPR/Cas9-induced mutations in SlTOM1 genes. The protospacer adjacent motif sequences and inserted or deleted nucleotide residues are indicated by blue and red letters, respectively. C, wt and Sltom1 quadruple-mutant plants (29 d after imbibition) and fruits. Scale bars represent 5 cm.
Effects of tom1 mutations on virus multiplication in tomato
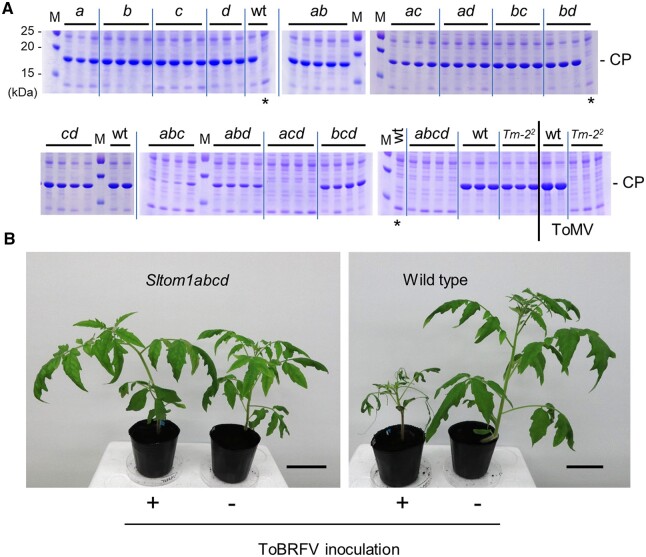
We inoculated ToBRFV onto cotyledons of Sltom1 mutant plants and examined viral coat protein (CP) accumulation in the inoculated leaves at 7 d post-inoculation (dpi) by SDS-PAGE followed by Coomassie blue staining (Figure 2A andSupplemental Figure S3, A and B) or immunoblotting (Supplemental Figure S3C). In Sltom1 single or double mutants, ToBRFV CP accumulated to nearly wt levels. In some double mutants, including Sltom1ac, ToBRFV CP accumulation was slightly reduced compared with wt plants (Figure 2A andSupplemental Figure S3). After longer incubation, ToBRFV-inoculated single and double mutants systemically accumulated viral CP and showed disease symptoms. In Sltom1 triple mutants, ToBRFV CP accumulation was reduced compared with that in wt plants. The level of CP accumulation at 7 dpi was in the order wt > Sltom1bcd ≥ Sltom1abd > Sltom1abc > Sltom1acd (Figure 2A andSupplemental Figure S3). This suggests that the contribution of SlTOM1 genes to ToBRFV multiplication is in the order SlTOM1a ≥ SlTOM1c > SlTOM1d > SlTOM1b. The contribution of SlTOM1 genes to ToBRFV multiplication showed weak association with relative mRNA accumulation levels of respective SlTOM1 genes (Supplemental Figure S2). Because ToBRFV CP accumulation was higher in the Sltom1ac double mutant than the Sltom1abc triple mutant (Figure 2A andSupplemental Figure S3), the contribution of SlTOM1b to ToBRFV multiplication is nonnegligible. In ToBRFV-inoculated Sltom1acd mutant plants, CP was barely detectable at 7 dpi, but after longer (e.g. 21 d) incubation, a low level of ToBRFV CP was detected not only in inoculated leaves, but also in systemic ones, and disease symptoms were observed in some plants. The other triple mutants (Sltom1bcd, abd, and abc) systemically accumulated viral CP and showed disease symptoms. In Sltom1abcd quadruple-mutant plants, neither ToBRFV CP accumulation nor disease symptoms were observed at ≥21 dpi (Figure 2, A and B and Supplemental Figure S3).
Figure 2.
ToBRFV multiplication in wt and Sltom1 mutant plants. A, ToBRFV CP accumulation in wt, Sltom1 single- (a, b, c, and d), double- (ab, ac, ad, bc, bd, and cd), triple- (abc, abd, acd, and bcd), and quadruple- (abcd) mutant and Tm-22 plants. Inoculated cotyledons were harvested at 7 dpi and CP accumulation was examined by SDS-PAGE (NuPAGE; Invitrogen, 12%) and Coomassie blue staining. Each lane corresponds to an individual plant. The rightmost five plants in the lower panel were inoculated with ToMV. Samples prepared in parallel from noninoculated plants were analyzed for comparison and are indicated by asterisks. M: Protein Marker Precision Plus Protein Dual Color Standards (Bio-Rad, Hercules, CA, USA). Positions of ToBRFV and ToMV CP bands are indicated by “CP.” Numbers represent approximate molecular masses of the marker proteins in kilodalton. Only the relevant parts of the gels with the CP bands were shown in this figure and whole gel images are provided in Supplemental Figure S3. B, ToBRFV-inoculated wt and Sltom1 quadruple-mutant plants. Uninoculated plants grown side-by-side are shown for comparison. ToBRFV-inoculated and uninoculated plants are shown by “+” and “-,” respectively. Photographs were taken at 21 dpi. Scale bars represent 5 cm.
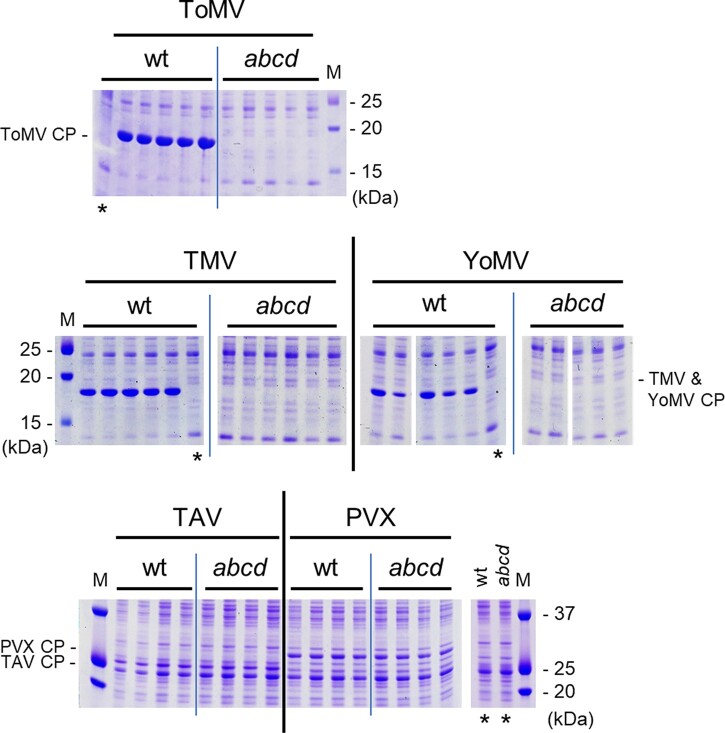
Similar results were obtained when ToMV was inoculated onto Sltom1 mutant plants (Supplemental Figure S4). Importantly, ToMV CP accumulation was not detected in Sltom1 quadruple-mutant plants (Figure 3 and Supplemental Figure S4, A and B). The Sltom1 quadruple-mutant plants did not show CP accumulation when inoculated with other tobamoviruses; that is, tobacco mosaic virus (TMV) (strain OM) or youcai mosaic virus (YoMV, or TMV-Cg) (Figure 3 and Supplemental Figure S5). Viruses taxonomically distinct from tobamoviruses—tomato aspermy virus (TAV; genus Cucumovirus) and potato virus X (TVX; genus Potexvirus)—multiplied and accumulated CPs in Sltom1 quadruple-mutant plants to levels similar to those in wt tomato plants (Figure 3 and Supplemental Figure S5). These results indicated that simultaneous loss-of-function mutations in SlTOM1a–d genes confer tobamovirus resistance to tomato.
Figure 3.
CP accumulation of ToMV, TMV, YoMV, TAV, and PVX in wt and Sltom1 quadruple mutant plants. Virus-inoculated cotyledons of wt plants were harvested at 7 dpi (ToMV, TAV, and PVX) or 6 dpi (TMV and YoMV), and those of Sltom1 quadruple-mutant plants (abcd) were harvested at 7 dpi (ToMV, TAV, and PVX) or 10 dpi (TMV and YoMV). CP accumulation was examined and presented as described in Figure 2A. Positions of viral CP bands are indicated by “CP” with virus names. Only the relevant parts of the gels with the CP bands are shown in this figure and whole gel images are provided in Supplemental Figures S4 (ToMV) and S5 (TMV, YoMV, TAV, and PVX).
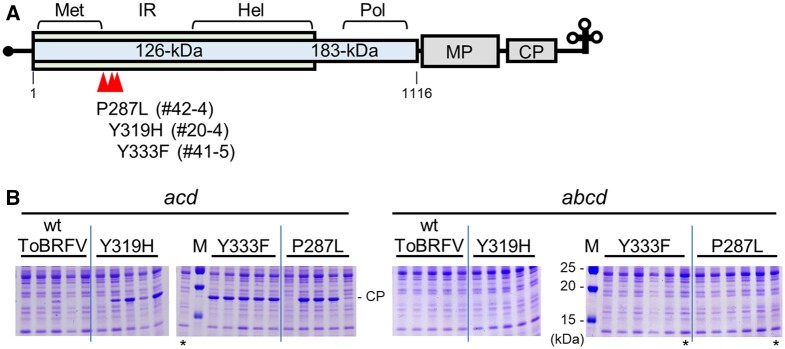
CP accumulation was delayed in ToBRFV-inoculated Sltom1acd mutant plants. To examine whether ToBRFV multiplied slowly with or without mutating, we inoculated ToBRFV to 42 Sltom1acd plants. About half (22/42) of the plants accumulated CP in systemic leaves at 18 dpi (Supplemental Figure S6A). Virions were separately recovered from plants #20, 41, and 42, in which CP accumulated to high levels (Supplemental Figure S6A), and separately passaged in Sltom1acd mutant plants. After the passage, we purified virions from one plant per inoculum (#20-4, 41-5, and 42-4). In the Sltom1acd triple-mutant plants inoculated with virions recovered from plants #20-4, 41-5, and 42-4, CP accumulation was higher and more frequent, and disease symptoms were observed more frequently, than in those inoculated with original ToBRFV (Supplemental Figure S6B), suggesting that the recovered virions contain mutants that can multiply in Sltom1acd plants more efficiently. We next sequenced the replication protein-coding region of genomic RNA of the mutants. The viruses carried missense mutations in close but distinct codons (Figure 4A). We introduced these mutations back into the wt ToBRFV infectious cDNA clone and confirmed that the infectious clone-derived mutant viruses accumulated more CP and caused disease symptoms in Sltom1acd triple-mutant plants more frequently than wt ToBRFV (Figure 4B andSupplemental Figure S7, left two panels). Also, the introduced mutations were maintained and no additional mutation occurred after multiplication of the cDNA-derived mutant viruses in Sltom1acd triple-mutant plants. These results suggest that ToBRFV multiplies with low efficiency using SlTOM1b in Sltom1acd triple-mutant plants, and mutants with increased infectivity and multiplication efficiency in Sltom1acd triple-mutant plants emerged. The ToBRFV mutants may have evolved their replication proteins to have higher affinity to SlTOM1b.
Figure 4.
ToBRFV mutants with increased infectivity to Sltom1acd triple-mutant plants. A, Mutation sites in the ToBRFV genome. Boxes represent the coding regions for the 126-kDa and 183-kDa replication proteins, movement protein (MP), and CP. Regions for methyltransferase-like (Met), helicase-like (Hel), and polymerase-like (Pol) domains, and intervening region of the replication proteins are shown. Amino acid changes from original ToBRFV to the mutants are shown along with the ID numbers of plants from which the mutant ToBRFV was recovered. B, ToBRFV CP accumulation in Sltom1 quadruple- (abcd) and Sltom1acd triple- (acd) mutant plants. Tomato plants were cotyledon-inoculated with leaf homogenates that were prepared from N. benthamiana leaves to which capped RNA synthesized from wt or mutant ToBRFV infectious clone plasmids had been inoculated. Upper uninoculated leaves were harvested at 21 dpi (Sltom1acd) or 20 dpi (Sltom1abcd) and CP accumulation was examined and presented as described in Figure 2A. Only the relevant parts of the gels with the CP bands are shown in this figure and whole gel images are provided in Supplemental Figure S7.
Importantly, no CP accumulation was observed after inoculation of these mutant viruses to Sltom1 quadruple-mutant plants (Figure 4B andSupplemental Figure S7, right two panels). Because loss of TOM1 function inhibits tobamovirus RNA replication (more specifically, negative-strand RNA synthesis (Nishikiori et al., 2011)), ToBRFV RNA replication in initially infected Sltom1 quadruple-mutant cells would hardly occur, minimizing the risk of emergence of mutant viruses that can overcome the resistance. In fact, no emergence of such a mutant virus in the quadruple mutant has been observed to date.
Discussion
In this study, we demonstrated that simultaneous knockout of the four TOM1 homologs in tomato confers strong resistance to ToBRFV. Single mutations in TOM1 homologs do not affect ToBRFV or ToMV CP accumulations, indicating that SlTOM1a–d are functionally redundant. Such a redundancy, frequently observed in plant genes, has been an obstacle to breeding. CRISPR/Cas9-mediated genome editing enables the introduction of multiple loss-of-function mutations in plant genes, which accelerates crop improvement if appropriate target genes to confer beneficial traits are available. Host genes necessary for the multiplication of pathogens (so-called “susceptibility genes”) are suitable for creation of beneficial traits using the CRISPR/Cas9 system, because simple loss-of-function mutations lead to resistance to pathogens. The functions of TOM1 family genes in plants are unknown but knockout-mutant Arabidopsis and tomato plants did not show obvious growth defects under greenhouse conditions. Therefore, knockout of TOM1 homologs is expected to have no apparent negative side effects, and to be useful to confer tobamovirus resistance to other plant species. In pepper (Capsicum annuum L.), tobamovirus resistance genes L1–L4 have been used to protect plants from pepper mild mottle virus but virus strains that can overcome this resistance have emerged (Genda et al., 2007). ToBRFV is also an emerging disease agent in pepper (Salem et al., 2020). In cucurbit crops, effective natural resistance genes against tobamoviruses are not available and cucurbit production is damaged by tobamoviruses (Dombrovsky et al., 2017). Because conferring tobamovirus resistance by knockout of TOM1 homologs does not rely on natural resistance resources, the strategy is applicable to other tobamovirus-sensitive crops for which genome editing or other targeted mutagenesis methods are available. Also in these cases, it is preferable to completely knockout all functional TOM1 homologs because low rate of viral accumulation can lead to the emergence and spread of resistance-breaking mutants as observed in Sltom1acd triple mutant plants.
Materials and methods
Targeted gene disruption in tomato
The CRISPR vector pDe-Cas9 (Km) carries gene cassettes expressing codon-optimized S. pyogenes Cas9 and NPTII and a gateway recombination cassette (R1–R2) to clone sgRNA modules (Fauser et al., 2014; Ritter et al., 2017). To combine three sgRNA modules, we used the entry clones pMR203, pMR204, and pMR205, which are suitable for MultiSite Gateway LR cloning to pDe-Cas9 (Km). These plasmids were constructed and provided by Drs. Mily Ron and Anne Britt. Briefly, to construct pMR203, pMR204, and pMR205, primers with appropriate attB/attBr flanking sites (B1–B4, B4r–B3r, and B3–B2, respectively) were used to amplify the sgRNA-expression cassette from pEn_Chimera (L1–L2) (Fauser et al., 2014). The amplified fragment was cloned into the corresponding pDONR221 vector (pDONR221 P1-P4, P4r-P3r, and P3-P2, respectively) by a Gateway BP reaction. An additional BbsI site in the pDONR backbone was eliminated by site-directed mutagenesis via In-Fusion reaction with the primers noBbsI_F and noBbsI_R (Ritter et al., 2017). The CRISPR vectors were obtained via Drs. Masaki Endo and Seiichi Toki.
For each sgRNA sequence, two complementary 23–24-nucleotide 5′-phosphorylated synthetic DNA oligos (Supplemental Table S1) were annealed and inserted at the BbsI site of the entry clones. These entry clones were mixed with pDe-Cas9 (Km) and subjected to a Gateway LR reaction to obtain the final expression plasmid.
The expression plasmid was introduced to Agrobacterium tumefaciens GV3101 (pMP90) and used for transformation of cotyledon explants of the tomato (S. lycopersicum) cultivar Craigella GCR26, as described previously (Sun et al., 2006).
Viruses
A gene cassette containing the T7 RNA polymerase promoter, complete cDNA of ToBRFV genomic RNA (GenBank: KT383474.1, nucleotides 6–9 were deleted), and a unique AgeI restriction site were chemically synthesized, assembled, and cloned in pUC57 (pToBRFV-A2, prepared by GenScript). ToBRFV was propagated by inoculating capped in vitro transcript synthesized from AgeI-linearized pToBRFV-A2 onto tomato (GCR26) or N. benthamiana leaves. The other tobamoviruses—TMV (strain OM) (Asano et al., 2005), ToMV (strain L) (Ishikawa et al., 1991), and YoMV (Ishikawa et al., 1991)—were described previously. TAV and PVX were obtained from the NARO Genebank (MAFF260123 and MAFF715063, respectively).
Virus multiplication assay
Tomato plants were grown at 25°C under a 16-h light/8-h dark cycle in a growth chamber. Viruses or viral infectious transcripts were rub-inoculated onto cotyledons 10 d after imbibition using carborundum. Viral CPs were analyzed by SDS-PAGE (NuPAGE 12% Bis–Tris gel; Thermo Fisher Scientific, Beverly, MA, USA) and Coomassie blue staining. Total RNA was purified from tomato leaves using RNAiso Plus (TaKaRa, Shiga, Japan) and analyzed.
Accession numbers
Accession numbers of the SlTOM1 genes and proteins are shown in Table 1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of amino acid sequences of SlTOM1 proteins.
Supplemental Figure S2. Expression of TOM1 homologs in wt, Sltom1 quadruple-, and triple-mutant tomato plants.
Supplemental Figure S3. ToBRFV CP accumulation in wt, Sltom1 single-, double-, triple-, and quadruple-mutant and Tm-22 plants.
Supplemental Figure S4. ToMV CP accumulation in wt, Sltom1 single-, double-, triple-, and quadruple-mutant plants.
Supplemental Figure S5. TMV, YoMV, TAV, and PVX CP accumulation in wt and Sltom1 quadruple-mutant plants.
Supplemental Figure S6. Emergence of ToBRFV mutants in tomato Sltom1acd triple-mutant plants.
Supplemental Figure S7. CP accumulation of ToBRFV mutants in Sltom1acd triple-mutant and Sltom1 quadruple-mutant plants.
Supplemental Table S1. DNA oligonucleotides used to construct the sgRNA sequence-containing entry clones.
Supplementary Material
Acknowledgments
We thank Drs. Holger Puchta, Friedrich Fauser, Simon Schiml, Mily Ron, Anne Britt, Masaki Endo, and Seiichi Toki for providing CRISPR/Cas9-related plasmids. We also thank Dr. Manabu Yoshikawa for discussion and Dr. Miao Shu and Ms. Hiroe Hatomi for technical assistance.
Funding
This research was supported in part by the Research Program on Development of Innovative Technology Grants from the Project of the Bio-oriented Technology Research Advancement Institution (BRAIN).
Conflict of interest statement. A.K. is employed by Takii and Company, Limited. M.I., A.K., and K.I. are named as inventors on a patent application related to this work (JP6810946B1).
M.I., A.K., and K.I. conceived the idea and designed the experiments. M.I., T.Y., M.M., and K.I. conducted the experiments. M.I., T.Y., M.M., Y.K., A.K., and K.I. analyzed the data. M.I. and K.I. drafted the manuscript. All the authors read and approved the final manuscript. K.I. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Kazuhiro Ishibashi (bashi@affrc.go.jp).
References
- Ali ME, Ishii Y, Taniguchi JI, Waliullah S, Kobayashi K, Yaeno T, Yamaoka N, Nishiguchi M (2018) Conferring virus resistance in tomato by independent RNA silencing of three tomato homologs of Arabidopsis TOM1. Arch Virol 163:1357–1362 [DOI] [PubMed] [Google Scholar]
- Asano M, Satoh R, Mochizuki A, Tsuda S, Yamanaka T, Nishiguchi M, Hirai K, Meshi T, Naito S, Ishikawa M (2005) Tobamovirus-resistant tobacco generated by RNA interference directed against host genes. FEBS Lett 579:4479–4484 [DOI] [PubMed] [Google Scholar]
- Atarashi H, Jayasinghe WH, Kwon J, Kim H, Taninaka Y, Igarashi M, Ito K, Yamada T, Masuta C, Nakahara KS (2020) Artificially edited alleles of the eukaryotic translation initiation factor 4E1 gene differentially reduce susceptibility to cucumber mosaic virus and potato virus Y in tomato. Front Microbiol 11:564310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent L (1976) Epidemiology and control of tomato mosaic virus. Annu Rev Phytopathol 14:75–96 [Google Scholar]
- Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17:1140–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Jiang JH, Zhou XP (2007) A TOM1 homologue is required for multiplication of tobacco mosaic virus in Nicotiana benthamiana. J Zhejiang Univ Sci B 8:256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ronde D, Butterbach P, Kormelink R (2014) Dominant resistance against plant viruses. Front Plant Sci 5:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrovsky A, Tran-Nguyen LTT, Jones RAC (2017) Cucumber green mottle mosaic virus: Rapidly increasing global distribution, etiology, epidemiology, and management. Annu Rev Phytopathol 55:231–256 [DOI] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359 [DOI] [PubMed] [Google Scholar]
- Fujisaki K, Ravelo GB, Naito S, Ishikawa M (2006) Involvement of THH1, an Arabidopsis thaliana homologue of the TOM1 gene, in tobamovirus multiplication. J Gen Virol 87:2397–2401 [DOI] [PubMed] [Google Scholar]
- Gauffier C, Lebaron C, Moretti A, Constant C, Moquet F, Bonnet G, Caranta C, Gallois J-L (2016) A TILLING approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J 85: 717–729 [DOI] [PubMed] [Google Scholar]
- Genda Y, Kanda A, Hamada H, Sato K, Ohnishi J, Tsuda S (2007) Two amino acid substitutions in the coat protein of pepper mild mottle virus are responsible for overcoming the L4 gene-mediated resistance in Capsicum spp. Phytopathology 97:787–793 [DOI] [PubMed] [Google Scholar]
- Gomez MA, Lin ZD, Moll T, Chauhan RD, Hayden L, Renninger K, Beyene G, Taylor NJ, Carrington JC, Staskawicz BJ, et al. (2019) Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol J 17:421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TJ (1980) Resistance at the Tm-2 locus in the tomato to tomato mosaic virus. Euphytica 29:189–197 [Google Scholar]
- Hashimoto M, Neriya Y, Yamaji Y, Namba S (2016) Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front Microbiol 7:1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Obata F, Kumagai T, Ohno T (1991) Isolation of mutants of Arabidopsis thaliana in which accumulation of tobacco mosaic virus coat protein is reduced to low levels. Mol Gen Genet 230:33–38 [DOI] [PubMed] [Google Scholar]
- Kang BC, Yeam I, Jahn MM (2005) Genetics of plant virus resistance. Annu Rev Phytopathol 43:581–621 [DOI] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7:1494–1496 [DOI] [PubMed] [Google Scholar]
- Luria N, Smith E, Reingold V, Bekelman I, Lapidot M, Levin I, Elad N, Tam Y, Sela N, Abu-Ras A, et al. (2017) A new Israeli tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PLoS ONE 12:e0170429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikiori M, Mori M, Dohi K, Okamura H, Katoh E, Naito S, Meshi T, Ishikawa M (2011) A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication. PLoS Pathog 7:e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham J (1966) Resistance in tomato to tobacco mosaic virus. Euphytica 15:258–267 [Google Scholar]
- Piron F, Nicolaï M, Minoïa S, Piednoir E, Moretti A, Salgues A, Zamir D, Caranta C, Bendahmane A (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 5:e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik D, Shelake RM, Park J, Kim MJ, Hwang I, Park Y, Kim JY (2021) CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int J Mol Sci 22:1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyott DE, Sheehan E, Molnar A (2016) Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol Plant Pathol 17:1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter A, Iñigo S, Fernández-Calvo P, Heyndrickx KS, Dhondt S, Shi H, De Milde L, Vanden Bossche R, De Clercq R, Eeckhout D, et al. (2017) The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat Commun 8:15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem N, Mansour A, Ciuffo M, Falk BW, Turina M (2016) A new tobamovirus infecting tomato crops in Jordan. Arch Virol 161:503–506 [DOI] [PubMed] [Google Scholar]
- Salem NM, Cao MJ, Odeh S, Turina M, Tahzima R (2020) First report of tobacco mild green mosaic virus and tomato brown rugose fruit virus infecting Capsicum annuum in Jordan. Plant Disease 104:601 [Google Scholar]
- Sanfaçon H (2015) Plant translation factors and virus resistance. Viruses 7:3392–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47:426–431 [DOI] [PubMed] [Google Scholar]
- van de Vossenberg BTLH,, Dawood T,, Woźny M,, Botermans M (2021) First Expansion of the Public Tomato Brown Rugose Fruit Virus (ToBRFV) Nextstrain Build; Inclusion of New Genomic and Epidemiological Data. PhytoFrontiers™ 1: 359–363 [Google Scholar]
- Yamanaka T, Imai T, Satoh R, Kawashima A, Takahashi M, Tomita K, Kubota K, Meshi T, Naito S, Ishikawa M (2002) Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J Virol 76:2491–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Ohta T, Takahashi M, Meshi T, Schmidt R, Dean C, Naito S, Ishikawa M (2000) TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA 97:10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinger A, Lapidot M, Harel A, Doron-Faigenboim A, Gelbart D, Levin I (2021) Identification and mapping of tomato genome loci controlling tolerance and resistance to tomato brown rugose fruit virus. Plants (Basel) 10:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.