Abstract
Selective metal coordination is central to the functions of metalloproteins1,2: each metalloprotein must pair with its cognate metallocofactor to fulfill its biological role3. However, achieving metal selectivity solely through a three-dimensional protein structure is a great challenge, because there is a limited set of metal-coordinating amino acid functionalities and proteins are inherently flexible, impeding steric selection of metals3,4. In fact, metal binding affinities of natural proteins are primarily dictated by the electronic properties of metal ions and universally follow the Irving-Williams (IW) series (MnII<FeII<CoII<NiII<CuII>ZnII)5 with few exceptions6,7. Accordingly, metalloproteins overwhelmingly bind CuII and ZnII in isolation, regardless of the nature of their active sites and their cognate metal ions1,3,8. This led organisms to evolve complex homeostatic machinery and non-equilibrium strategies to achieve correct metal speciation1,3,8-10. Here, we report an artificial dimeric protein, (AB)2, that thermodynamically overcomes the IW restrictions in vitro and cellulo, favoring binding of lower-IW transition metals over CuII, the most dominant ion in the IW series. Counter to the convention in molecular design to achieve specificity through structural preorganization, (AB)2 was deliberately designed to be flexible. This flexibility enabled (AB)2 to adopt mutually exclusive, metal-dependent conformational states, leading to the discovery of structurally coupled coordination sites that disfavor CuII ions by enforcing an unfavorable coordination geometry. Aside from highlighting flexibility as a valuable protein design element, our results illustrate design principles for constructing selective metal sequestration agents.
Several strategies have been developed for building artificial metalloproteins11-15, including the de novo design of new protein folds/complexes16-18, repurposing of proteins for incorporation of metal coordination sites19,20, and metal-templated construction of supramolecular protein assemblies21,22. These approaches have yielded many examples of functional metalloproteins that are assembled in vitro (i.e., in homogeneous solutions). However, building from scratch a protein that selectively binds a transition metal ion over others in a heterogeneous environment represents a formidable task. Rare successes in this arena are either based on the redesign of the interiors of pre-existing proteins23,24 or do not display complete counter-IW selectivity25. This difficulty stems from the fact that de novo protein design approaches have yet to provide the requisite structural precision (well below 0.5 Å) needed to sterically discriminate between transition metal ions, which is exacerbated by the challenge of computationally modeling metal-protein interactions26,27. Furthermore, our predictive understanding of how protein structure and metal coordination influence one another is limited, which poses a fundamental problem for any deterministic metalloprotein design campaign with a “single structure-single function” goal.
Against this backdrop, we set out to pursue an alternative, probabilistic metalloprotein design approach, which relies on a flexible protein-protein interface that is replete with metal-binding residues. Our inspiration came from natural multi-metallic proteins (e.g., metallothionein28, calmodulins29, calprotectin30) with inherent flexibilities that enable structural coupling between different metal coordination sites. We were particularly intrigued by the selective MnII-over-FeII binding in the dinuclear sites of class Ib ribonucleotide reductases and the suggestion that such counter-IW selectivity may be achieved by the cooperativity between the two sites6,7. Accordingly, we envisioned that structural coupling between multiple metal sites within a flexible protein scaffold may engender enhanced binding affinities or selectivities.
Design and Characterization of (AB)2.
We recently introduced a strategy termed MASCoT (Metal Active Sites through Covalent Tethering)31. In MASCoT, a protein-protein interface is simply created by the disulfide-mediated linking of two copies of a monomeric protein, and metal-binding motifs are subsequently incorporated near the disulfide bond to generate interfacial metal coordination sites. The disulfide “pivot” furnishes a malleable assembly that possesses a shallow free-energy landscape and is capable of exploring different conformations. Yet, the assembly is also structurally constrained by the disulfide bond, and, upon metal binding, forms a well-ordered interface. In proof-of-principle experiments, cytochrome cb562 (a monomeric, four-helix bundle hemeprotein32) was tailored with a single Cys (C96) and a proximal tris-His (3His) motif comprising H67, H71, and H97 (Fig. 1a). The resulting dimer, (Cys3His)2, was observed to coordinate all divalent, mid-to-late first row transition metal ions in an identical, distorted square-pyramidal geometry, consisting of five of the available six His residues (H67/H67’, H71/H71’ and H97). The MII-(Cys3His)2 dissociation constants (Kd) ranged from 20 μM for MnII to 400 fM for CuII, following the IW series.31
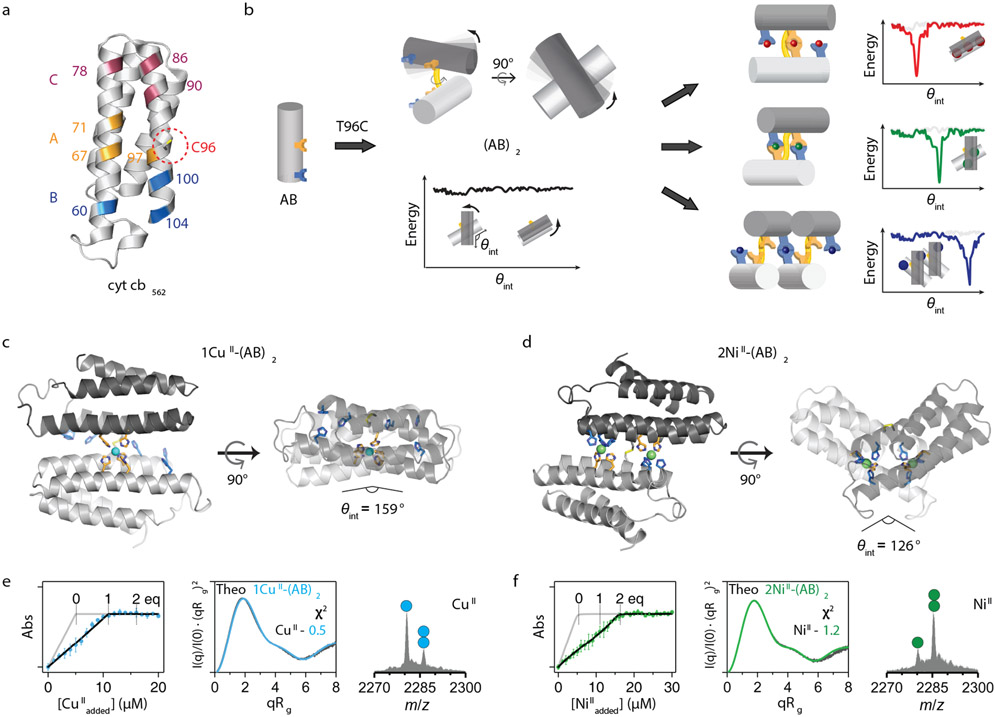
Figure 1 ∣. Design and characterization of the (AB)2 scaffold.
a, Locations of metal coordination motifs (A, B, C) and Cys96 on the cytochrome cb562 surface. b, Design of disulfide-linked (AB)2 with a malleable dimer interface, along with cartoon representations of possible metal-free/bound conformations and corresponding free-energy landscapes. Crystal structures of c, 1CuII-(AB)2 and d, 2NiII-(AB)2. CuII and NiII ions are represented as cyan and green spheres, respectively. X-ray data collection and refinement statistics are listed in Extended Data Table 2. Characterization of e, 1CuII-(AB)2 and f, 2NiII-(AB)2 complexes in solution using competitive metal titrations, solution SAXS, and ESI-MS. Log-scale SAXS plots are shown in Extended Data Figs. 2j-m. Changes in Fura-2 absorbance at 335 nm (left) are plotted with theoretical metal-binding isotherms (grey) in the absence of (AB)2. Experimental data points and error bars are presented as mean and standard deviation of three independent measurements. Theoretical SAXS profiles (middle) derived from the crystal structures of 1CuII-(AB)2 (cyan) and 2NiII-(AB)2 (green) are plotted with experimental SAXS profiles (black) and corresponding χ2 values. Circles in ESI-MS spectra (right) represent the number of NiII (green) and CuII (cyan) ions bound to (AB)2.
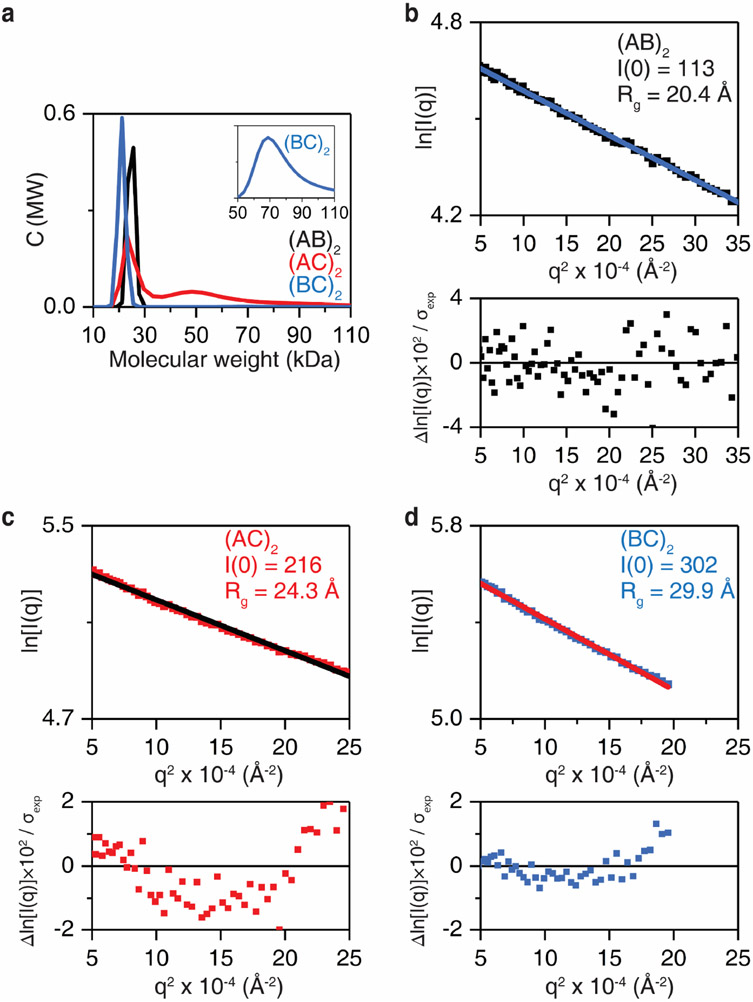
Here, to access alternative dimer conformations with multiple metal binding sites, we engineered three new cytochrome cb562 variants (AB, AC and BC) bearing two 3His motifs surrounding the C96 pivot. These variants combine two of following three 3His sites in addition to the native H63: Motif A (H67/H71/H76), Motif B (H60/H100/H104), and Motif C (H78/H86/H90). These His-enriched variants were isolated in high yields from bacterial cultures and formed stable disulfide-linked dimers upon oxidation. Analytical ultracentrifugation and solution small-angle X-ray scattering (SAXS) experiments indicated that the metal-free AB dimer was monodisperse at all concentrations tested, but AC and BC variants appeared to form some higher-order oligomers/aggregates and were therefore not studied further (Extended Data Fig. 1).
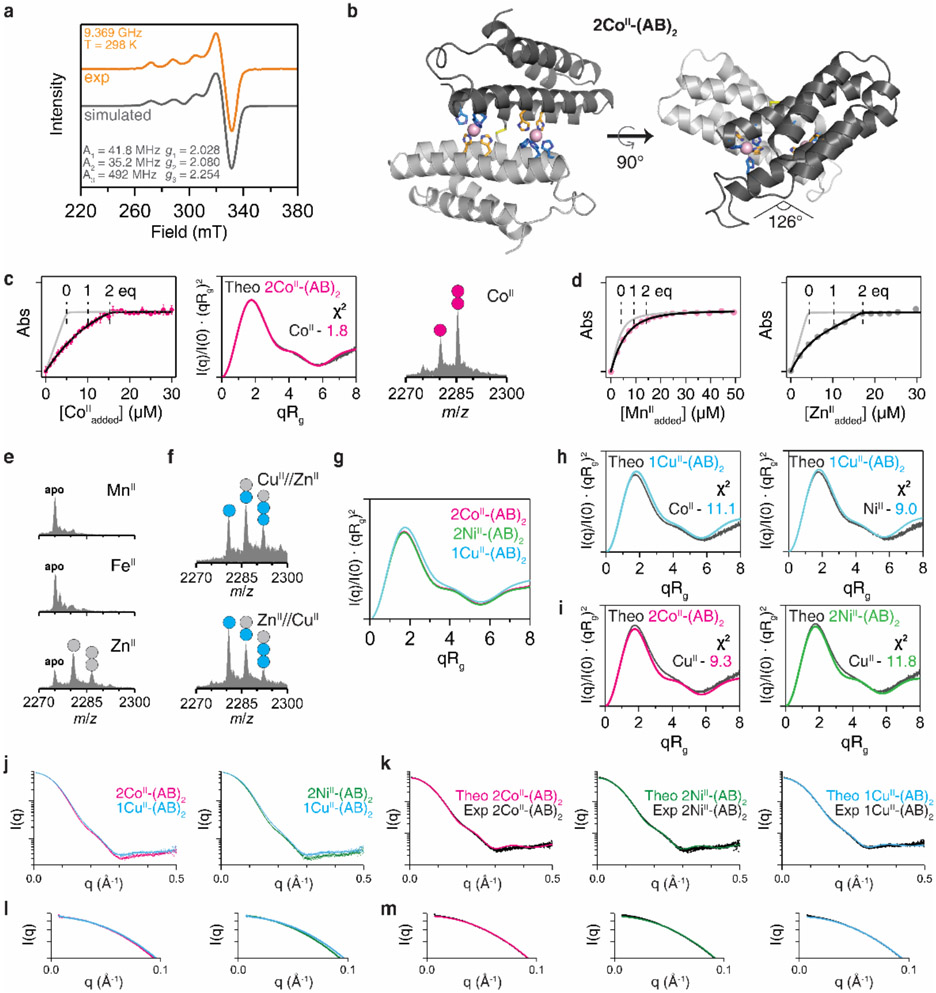
We anticipated that the flexible AB dimer, (AB)2, could form several different metal-dependent conformations (Fig. 1b). To explore these possibilities, we first sought to crystallize (AB)2 in the presence of first-row transition metal ions (MnII, FeII, CoII, NiII, CuII, ZnII). Of these, we determined the crystal structures of the CoII-, NiII- and CuII-complexes at high resolution (<2.0 Å). The CuII-(AB)2 adduct was isostructural with the previously described MII-(Cys3His)2 complexes (RMSD over all Cα’s ≈ 0.5 Å), featuring a flat, antiparallel dimer interface with an intermonomer angle θint = 159° (Fig. 1c). We refer to this dimer geometry as Conformation-1. In Conformation-1, only a single, central 5His coordination site consisting of two A motifs participated in CuII coordination (electron paramagnetic spectrum shown in Extended Data Fig. 2a), and the two B motifs were surprisingly unoccupied. In contrast, CoII- and NiII-(AB)2 complexes possessed two symmetrical metal coordination sites on either side of the C96-C96 linkage, each composed of A and B motifs in a square-pyramidal 5His geometry, with H60/H100/H104 from one monomer and H67’/H71’ from the opposing monomer (Fig. 1d, Extended Data Fig. 2b). To accommodate this new metal-binding configuration, the monomers in 2CoII- and 2NiII-(AB)2 complexes (RMSD = 0.27 Å with respect to one another) rotate by more than 30° about the C96-C96 linkage relative to the CuII-(AB)2 structure, yielding a kinked architecture with θint = 126° (Conformation-2).
To examine whether the crystal structures of MII-(AB)2 complexes were reflective of their solution structures, we carried out metal-binding assays and SAXS studies (Figs. 1e-f, Extended Data Fig. 2c, Extended Data Table 1). Titrations using Fura-2 as a competitive indicator confirmed that (AB)2 tightly bound a single equivalent of CuII and two equivalents of CoII or NiII, with fM and nM Kd’s, respectively. MnII and FeII (which lie low in the IW series) or ZnII (which typically disfavors all-His, high-coordination-number geometries) did not display appreciable binding affinities or selectivities over CuII (Extended Data Figs. 2d-f). Given that the (AB)2 adducts of these ions also did not yield crystal structures, we did not consider them further. For SAXS analyses, we used the crystal structures of the CuII- and 2CoII/2NiII-(AB)2 complexes to calculate theoretical scattering profiles. The calculated SAXS pattern of CuII-(AB)2 (Conformation-1) is quite distinct from those of 2CoII- and 2NiII-(AB)2 (Conformation-2), as expected from their different crystal structures (Extended Data Fig. 2f). The experimental SAXS profiles for each species (CuII vs. NiII or CoII) matched their corresponding theoretical profiles closely (χ2 = 0.5–1.8) but deviated from those of the other species (χ2 = 9.3–14.1), indicating that the crystallographically observed conformations of (AB)2 complexes were populated in solution (Extended Data Figs. 2g-h). These observations established that (AB)2 formed two distinct and mutually exclusive metal-dependent conformations in solution.
Counter-IW Selectivity of (AB)2.
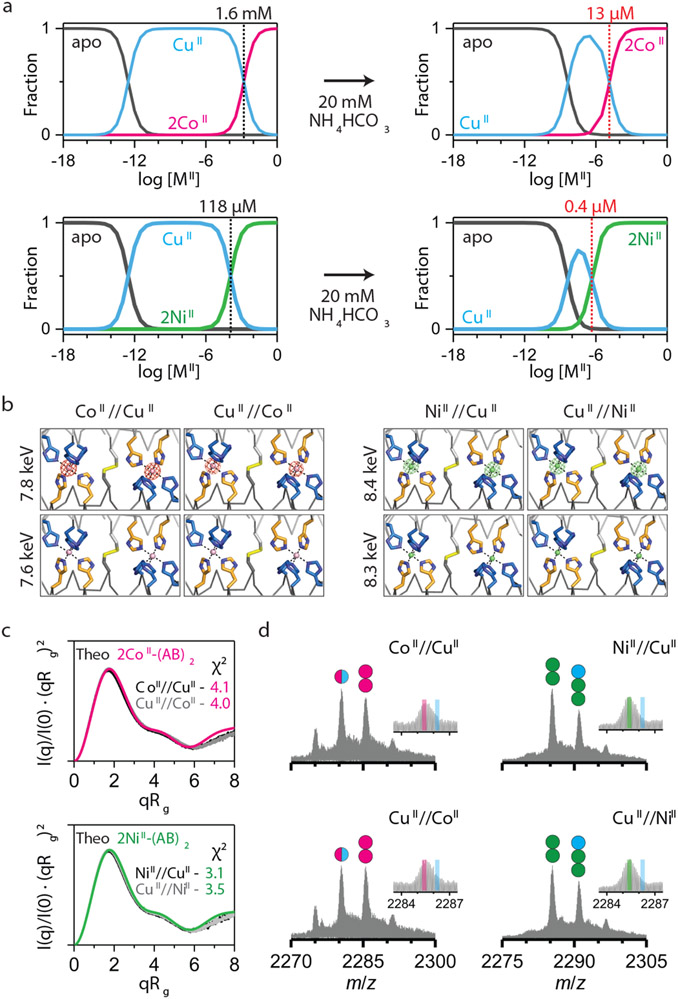
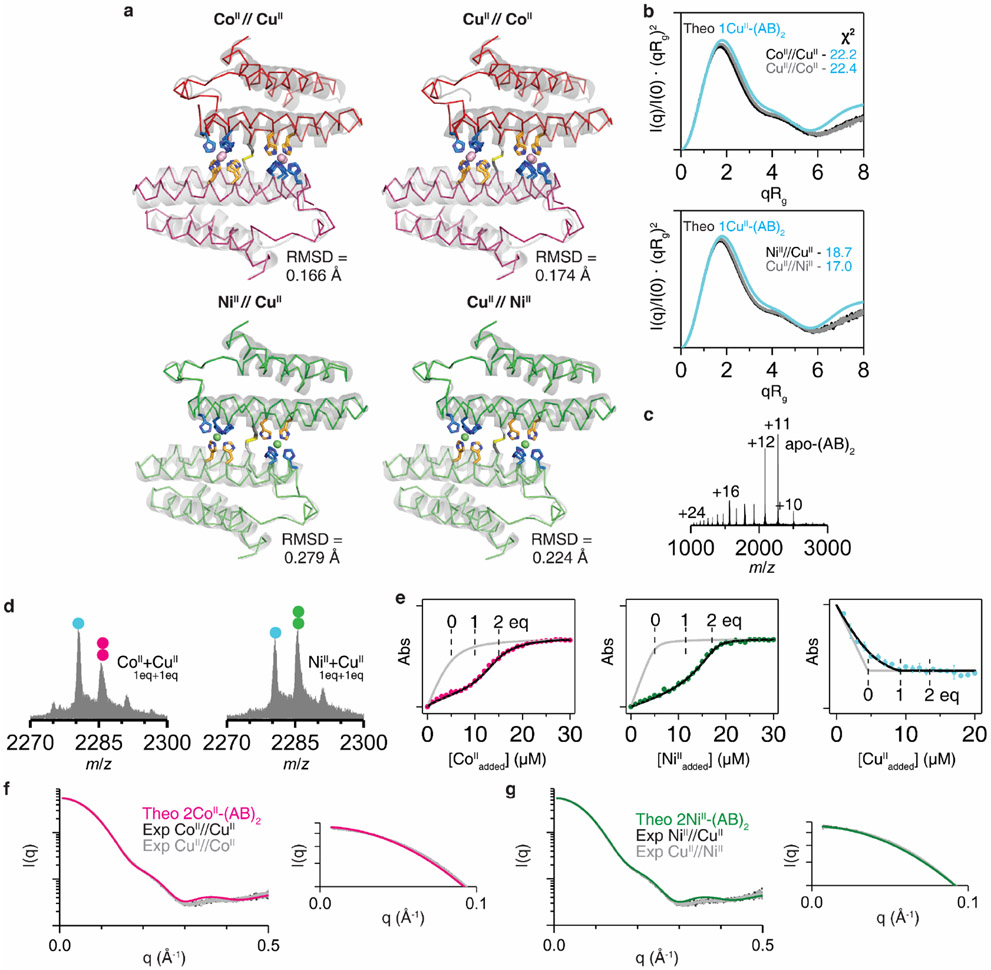
Although the per-site CuII binding affinity of (AB)2 is >10,000-fold higher than that for CoII and NiII, we envisioned that CoII and NiII might outcompete CuII given their higher valency and potential cooperativity. Equilibrium calculations based on the experimentally determined MII-(AB)2 Kd’s indicated that that population of 2CoII- and 2NiII-(AB)2 complexes should indeed begin to dominate the CuII-(AB)2 species at [MII]total > 10−3–10−4 M (Fig. 2a, Supplementary Information). Given that these concentrations are typical of crystallization conditions, we first examined the metal selectivity of (AB)2 in the crystalline state. (AB)2 (2 mM) was crystallized in a mixture of 4 mM CuII and 4 mM CoII or NiII. In these experiments, the protein solution was incubated first with either CuII or with CoII or NiII for at least 1 h, after which the second metal ion was added and the resulting mixture was left to equilibrate for 1 day. Under all four conditions (CoII//CuII, CuII//CoII, NiII//CuII, CuII//NiII), the observed crystal structures were identical to those of 2CoII- and 2NiII-(AB)2 (i.e., Conformation-2) with two peripheral metal coordination sites (Extended Data Fig. 3a). Anomalous X-ray diffraction experiments confirmed that these sites were populated exclusively by Co or Ni (Fig. 2b).
Figure 2 ∣. CoII- and NiII-selectivity of (AB)2.
a, Theoretical fractions of metal-free/bound (AB)2 species calculated from Kd values and buffered metal concentrations in 20 mM MOPS and 150 mM NaCl (left) and 20 mM NH4HCO3 (right). b, Anomalous scattering densities of interfacial metal sites of (AB)2. Anomalous densities contoured at 5.0 σ were collected around K-edge of Co (~7.7 keV) and Ni (~8.3 keV). X-ray data collection and refinement statistics are listed in Extended Data Table 3. c, Experimental Kratky plots of (AB)2 (black/grey) compared with theoretical plots of 2CoII-(AB)2 (magenta) and 2NiII-(AB)2 (green) structures (log-scale plots are shown in Extended Data Figs. 3f-g). d, ESI-MS spectra of (AB)2 under CoII//CuII, CuII//CoII, NiII//CuII, and CuII//NiII competition conditions. Circles in ESI-MS spectra represent the number of CoII (magenta), NiII (green), and CuII (cyan) ions bound to (AB)2. Half circles (magenta/cyan) indicate a mixture of CoII and CuII complexes. Insets represent expanded m/z ranges of 2MII-(AB)2 complexes with magenta, green, and cyan lines corresponding to theoretical m/z values of 2CoII, 2NiII, and 2CuII-(AB)2 complexes, respectively.
In parallel, we prepared similar samples for solution SAXS experiments. The Kratky plots of these samples showed close correspondence with the theoretical scattering patterns of 2CoII- and 2NiII-(AB)2 structures in Conformation-2 (χ2 = 3.5–4.1) (Fig. 2c), but not with that of CuII-(AB)2 in Conformation-1 (χ2 = 17.0–22.4) (Extended Data Fig. 3b), confirming that CoII and NiII fully outcompete CuII in solution under thermodynamic control.
To investigate whether the counter-IW selectivity of (AB)2 was operative under lower protein and metal concentrations, we turned to native electrospray ionization mass spectrometry (ESI-MS)33. ESI-MS of 5 μM (AB)2 in 20 mM NH4HCO3 (pH 7.8) without metal ions showed a broad charge-state distribution from +10 to +24 (Extended Data Fig. 3c). We chose the most abundant, +11 state for further analyses of MII-(AB)2 complexes, whereby two molar equivalents of MII (10 μM) were added to (AB)2. Consistent with crystal structures and competitive titrations, we observed 1CuII-(AB)2, 2CoII-(AB)2 and 2NiII-(AB)2 as the predominant species with >80% abundance (Figs. 1e-f, Extended Data Fig. 2c).
For ESI-MS under competitive binding conditions, we prepared similar CoII//CuII, CuII//CoII, NiII//CuII, CuII//NiII samples, using micromolar (AB)2 and MII (Fig. 2d). In CoII//CuII and CuII//CoII samples, we observed approximately equal amounts of 2CoII-(AB)2 and 1MII-(AB)2 species, with the latter attributed to a mixture of 1CoII-(AB)2 and 1CuII-(AB)2. These results indicated that CoII could still compete with CuII for binding (AB)2 at μM concentrations. Importantly, in the NiII//CuII and CuII//NiII samples, 2NiII-(AB)2 was the dominant species, with no evidence for a 1CuII-(AB)2 complex. The analysis of a sample containing 5 μM (AB)2 and substoichiometric amounts of CuII (5 μM) and NiII or CoII (5 μM) revealed roughly equal populations of 1CuII-(AB)2 and 2NiII-(AB)2 or 2CoII-(AB)2 complexes with no indication of heterobimetallic species (Extended Data Fig. 3d). This observation supports our structural findings that two equivalents of NiII or CoII cooperatively bind to the peripheral sites of Conformation-2 in an all-or-nothing manner, whereas CuII only coordinates Conformation-1 in one equivalent.
Curiously, the equilibrium fractionation profiles shown in Fig. 2a suggested that NiII and CoII should not be able to outcompete CuII for (AB)2 binding at μM concentrations. Yet, we considered that the ESI-MS samples also contained 20 mM NH4HCO3 as a volatile buffer and the observed MII-(AB)2 populations must also be influenced by the metal-NH3 binding equilibria: complexation with NH3 would be expected to lower the concentration of free CuII in solution to a greater extent compared to CoII and NiII owing to the considerably higher formation constants of CuII-ammine complexes34. Indeed, competitive binding titrations of (AB)2 carried out in 20 mM NH4HCO3 showed that the apparent Kd for the CuII-(AB)2 complex decreased by ~15,000-fold, whereas those for the CoII and NiII complexes decreased by less than 10-fold (Extended Data Fig. 3e, Extended Data Table 1). The revised equilibrium fractionation profiles (calculated using the apparent Kd’s in 20 mM NH4HCO3) show that CoII and NiII begin dominating CuII binding at ~13 μM and ~0.4 μM concentrations, respectively, consistent with ESI-MS results (Fig. 2a). These results illustrate that competing equilibria involving exchange-labile ligands in solution can amplify the metal selectivity of a protein scaffold. Indeed, this strategy is similar to what is observed in cells, which contain a surplus of competing ligands that buffer intracellular metals to the inverse of the Irving-Williams series35,36.
Basis for Counter-IW Selectivity.
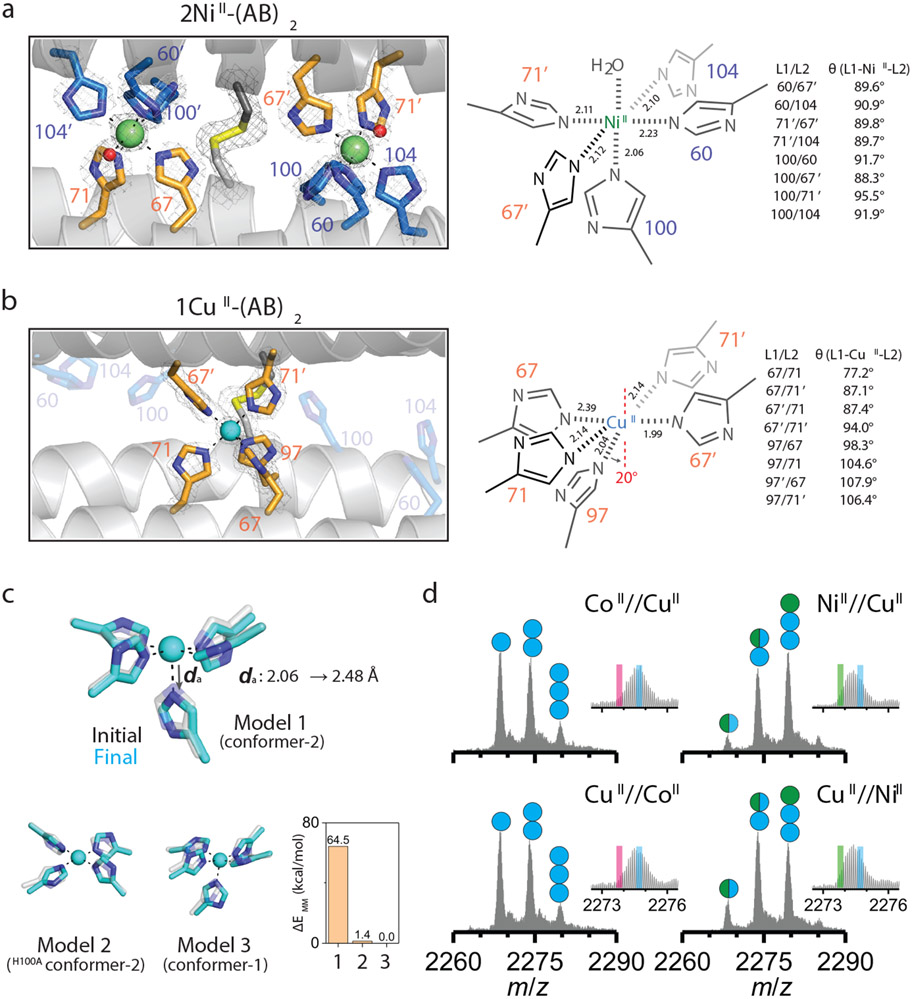
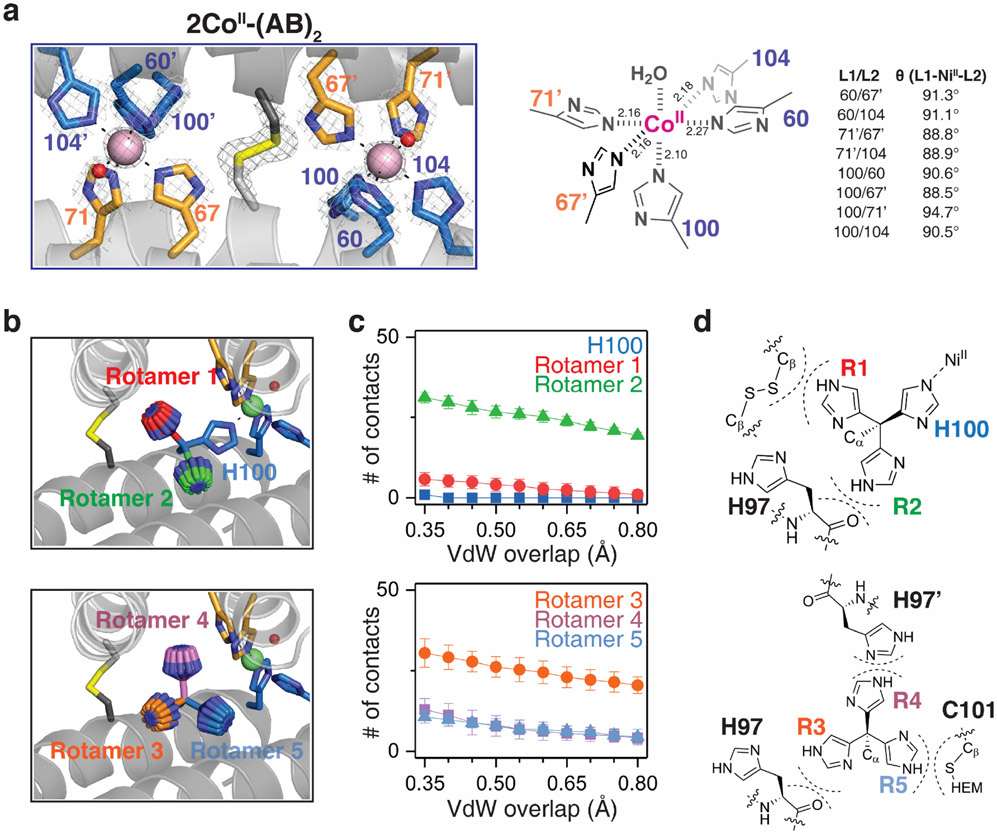
Having established (AB)2 as a unique protein scaffold that defies the IW series, we next sought to understand the structural basis for its selectivity. It should be noted at the outset that the CoII and NiII selectivity cannot arise from their higher valency of binding compared to CuII, because the two peripheral, 5His coordination motifs in Conformation-2 are also available in solution for two CuII ions to bind. The fact that: a) CuII binds singly to the central 5His site in Conformation-1 and b) CoII and NiII exclusively occupy the peripheral 5His sites over CuII in Conformation-2 mandates that each peripheral site must individually disfavor CuII binding (i.e., each site binds CoII and NiII more strongly than CuII). Because both the central and peripheral sites are 5His motifs, we surmised that any selectivity must arise from steric effects, which prompted us to closely inspect the coordination geometries. The peripheral sites possess an almost-perfect octahedral geometry, with nearly equidistant MII-ligand bonds (2.17±0.06 Å for CoII and 2.12±0.06 Å for NiII) and minimal deviation from ideal octahedral bond angles (90.5±2.0° and 90.9±2.2°) (Fig. 3a, Extended Data Fig. 4a). All His sidechains are positioned for optimal bonding between their Nε atoms and the metal, including the axial H100 ligand. In contrast, the geometry of the central site is distorted from an octahedron with a broad distribution of bond lengths (2.16±0.16 Å for CuII) and angles (95.3±11.9°). Notably, the imidazole group of the axial H97 ligand is slanted by nearly 20° with respect to the equatorial plane normal (Fig. 3b), presenting a less than ideal σ-donation geometry to CuII.
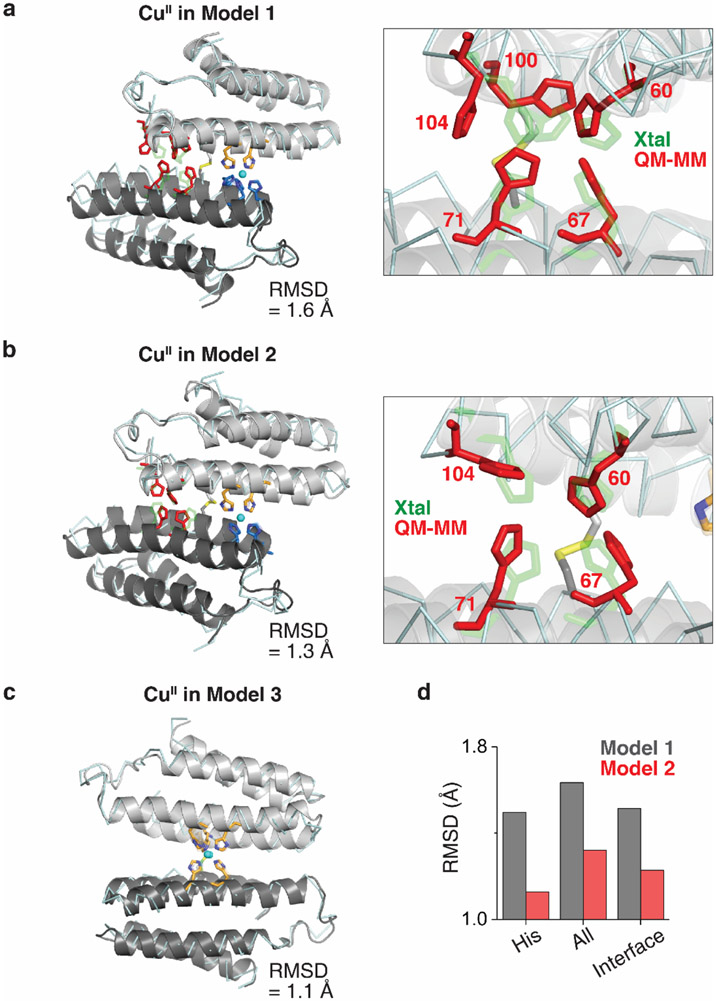
Figure 3 ∣. Primary metal coordination spheres of central and peripheral sites in (AB)2.
Close-up view of 5His-coordination of a, NiII and b, CuII with the 2mFo-DFc electron density map (grey mesh) contoured at 5.0 σ (metal) or 1.5 σ (ligand). Coordination distances and angles between MII and ligands are shown on the right. c, QM-MM optimized CuII coordination in Model 1 (CuII in peripheral site of (AB)2), Model 2 (CuII in the peripheral site of H100A(AB)2), and Model 3 (CuII in the central site of (AB)2). The initial (crystal structures) and final (QM-MM optimized structures) conformations of Models 1–3 are represented as grey and cyan, respectively. Relative MM energies of (AB)2 conformations resulting from the optimized Models 1–3 are presented in the bar graph. d, ESI-MS spectra of H100A(AB)2 under competitive metal binding conditions. Circles in ESI-MS spectra represent the number of NiII (green) and CuII (cyan) ions bound to (AB)2. Half circles (green/cyan) indicate the mixture of NiII and CuII complexes. Insets show expanded m/z ranges for 2MII-(AB)2 complexes with magenta, green, and cyan lines corresponding to theoretical m/z values of 2CoII, 2NiII, and 2CuII_H100A(AB)2 complexes, respectively.
Thus, we hypothesized that the peripheral sites in Conformation-2 might disfavor CuII binding by rigidly enforcing an ideal octahedral coordination geometry (featuring the strongly σ-donating axial H100 ligand), thereby preventing the energetically favored, tetragonal distortion of the CuII center. To probe this hypothesis, we first examined the protein environment around H100 and found that the imidazole sidechain had no alternative rotameric configurations that could be adopted without sterically clashing with nearby sidechains and protein backbone (Extended Data Figs. 4b-d). Importantly, any tetragonal distortion in one peripheral site would necessitate a conformational change in the entire dimeric assembly, thereby deforming the second peripheral site. This expectation was borne out by quantum mechanics/molecular mechanics (QM/MM) calculations (Fig. 3c), in which we considered three structural models of (AB)2 as initial states for simulations: Model 1: Conformation-2 with a single CuII ion substituted into one of the peripheral metal binding sites based on the crystal structure of 2NiII-(AB)2; Model 2: same as Model 1 but with a H100A mutation to eliminate axial coordination; Model 3: the crystal structure of CuII-(AB)2 (Conformation-1). In the case of Model 1, QM (B3LYP) and MM (AMBER) optimization led to a drastic distortion of CuII coordination with an increase in the axial H100-CuII distance from 2.06 to 2.48 Å. This distortion was accompanied by a large conformational change in the dimer structure, eliminating the other peripheral binding site and leading to an increase in the overall energy of the system (Fig. 3c, Extended Data Fig. 5). In contrast, Model 2 and Model 3 did not undergo significant changes in the CuII coordination geometries and overall (AB)2 conformations, and both were considerably more stable compared to Model 1 (Extended Data Fig. 5).
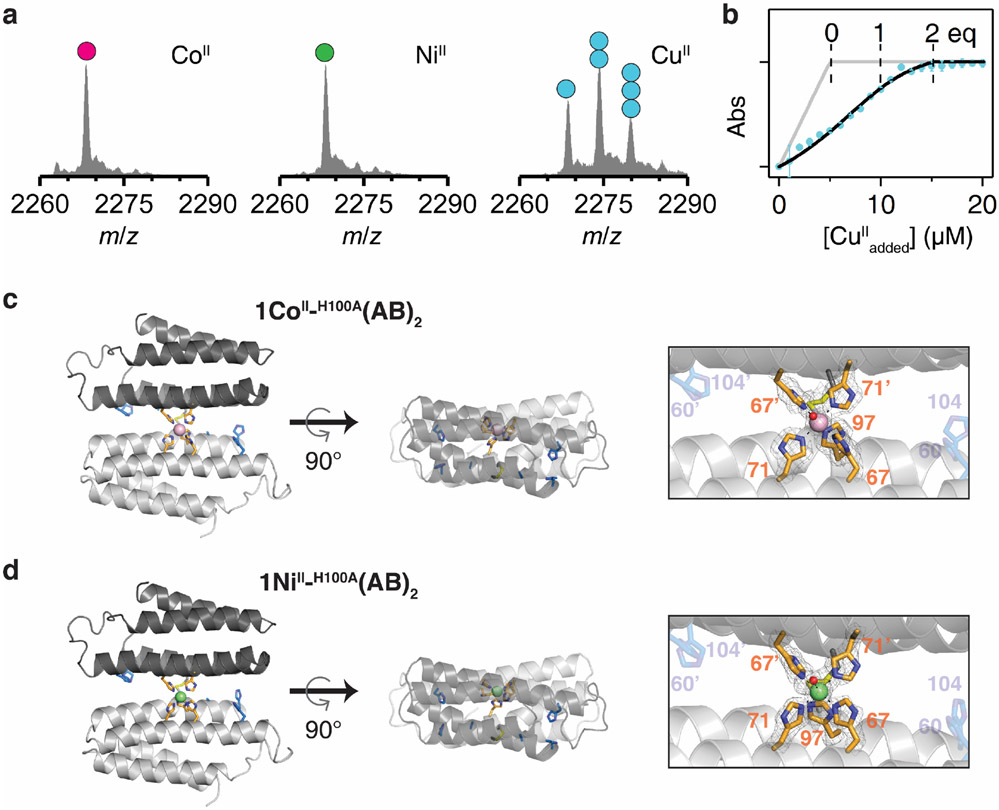
Next, we experimentally evaluated the effects of the H100A mutation. In a complete reversal of the metal binding properties of (AB)2, ESI-MS experiments indicated the H100A(AB)2 dimer bound two equivalents of CuII, but only one equivalent of CoII or NiII (Extended Data Fig. 6a). Fura-2 competition assays further indicated that H100A(AB)2 coordinated two equivalents of CuII (Extended Data Fig. 6b). Crystal structures of the CoII- and NiII_H100A(AB)2 complexes revealed that they adopted Conformation-1 with a single metal ion in the central 5His site (Extended Data Figs. 6c-d). Competition experiments showed that the H100A mutation completely abolished CoII and NiII selectivity over CuII, with the latter ion dominating binding under all conditions tested (Fig. 3d). Collectively, our results confirmed the counterintuitive mechanism by which (AB)2 destabilizes CuII binding through an increased coordination number and enforced octahedral geometry, thereby achieving counter-IW selectivity.
In vivo Metal Selectivity of (AB)2.
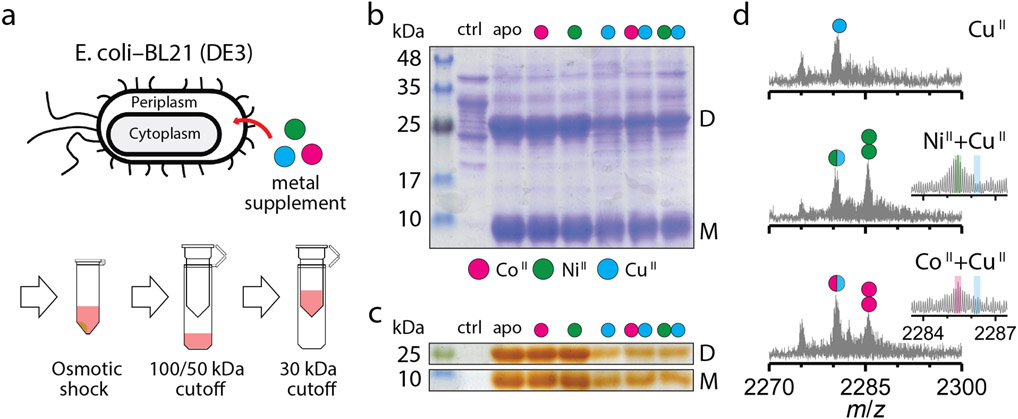
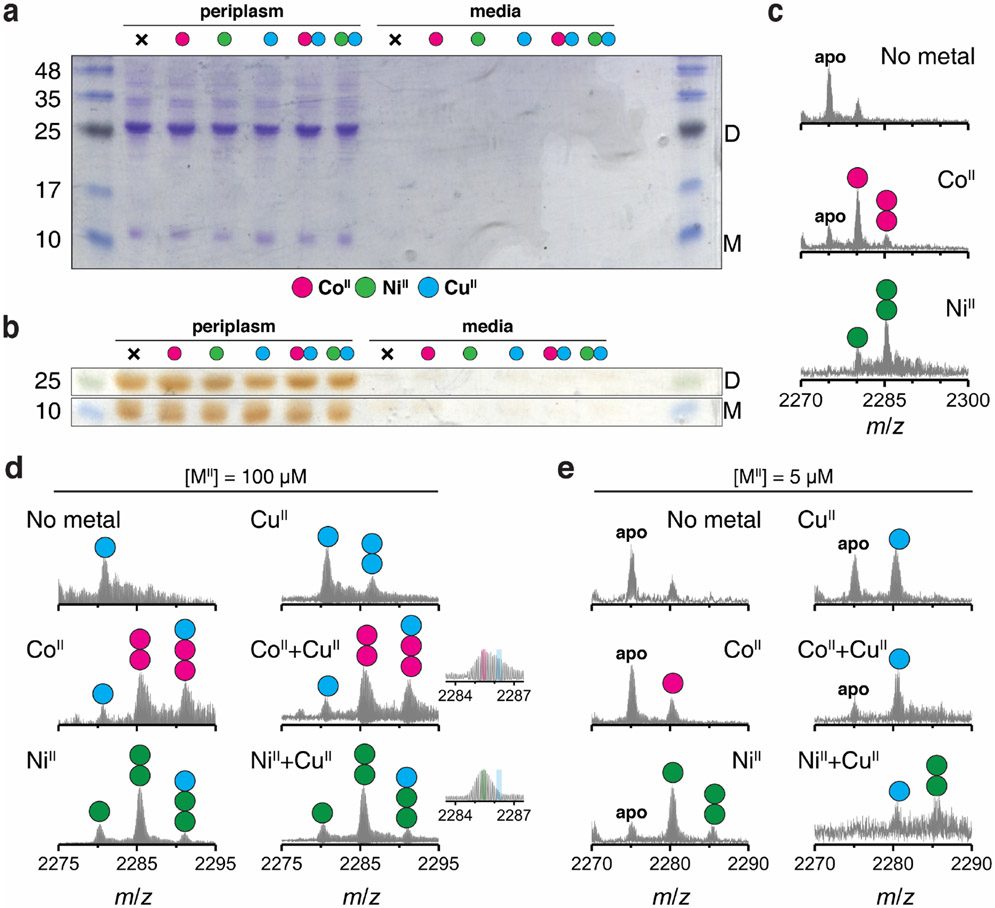
Selective incorporation of metals in the cellular environment is essential for the development of artificial metalloproteins/metalloenzymes with in vivo activities21,37. With such synthetic biological applications in mind, we investigated whether (AB)2 displayed metal selectivity in the periplasm of bacterial cells. AB is equipped with an N-terminal leader sequence for translocation to the periplasm whose oxidizing environment would allow the formation of the disulfide-mediated (AB)2 dimer21. We expressed AB in E. coli (BL21) cells, followed by the incubation of the intact cells in a growth medium supplemented by 20 μM of CoII, NiII, CuII or a mixture CoII + CuII and NiII + CuII (Fig. 4a). The cells were harvested and subjected to osmotic shock for the periplasmic extraction of protein components, which were analyzed for metal content. Gel electrophoretic analyses indicated that AB exists as a mixture of roughly equal amounts of dimers and monomers (Figs. 4b-c) and is trapped in periplasm without secretion to the external medium (Extended Data Figs. 7a-b). ESI-MS measurements of the extracts from CoII- and NiII-supplemented cells indicated both singly and doubly MII-bound (AB)2 complexes (Extended Data Fig. 7c) whereas the (AB)2 complexes extracted from CuII-supplemented cells contained one CuII equivalent (Fig. 4d), paralleling findings of in vitro studies. Under competitive conditions with a mixture of NiII + CuII, 2NiII-(AB)2 was detected as the major species (~60%) with a smaller population (~40%) of singly-MII bound (AB)2 complex (Fig. 4d). The population of the doubly-bound, 2CoII-(AB)2 species was reduced (~35%) compared to singly-bound species (~65%), likely owing to the lower CoII affinity of (AB)2. Consistent with a metal-exchange equilibrium between the periplasm and the external medium, when the metal concentrations in the LB medium were raised to 100 μM under competition conditions, the periplasmic fractions of 2CoII- and 2NiII-(AB)2 increased to >80% (Extended Data Fig. 7d). These observations illustrate that the in vitro, metal selectivity of (AB)2 for CoII and NiII over CuII is also operative in the heterogeneous environment of the periplasm and controllable through metal concentrations in the external milieu.
Figure 4 ∣. Selective CoII- and NiII-binding of (AB)2 in the E. coli periplasmic space.
a, Extraction and purification scheme of periplasmic AB species. SDS-PAGE of periplasmic extracts stained with b, Coomassie Blue for all proteinaceous contents and c, o-dianisidine for heme-proteins: supplemented metal ions are represented as magenta (CoII), green (NiII), and cyan (CuII). D and M refer to the (AB)2 dimer and the AB monomer, respectively. Reproducibility of SDS-PAGE was tested using three independently extracted sample sets. ESI-MS spectra of (AB)2 extracted from periplasm of the cells grown in d, CuII, CoII+CuII, or NiII+CuII conditions. Circles in ESI-MS spectra represent the number of CoII (magenta), NiII (green), and CuII (cyan) ions bound to (AB)2. Half circles (green/cyan and magenta/cyan) indicate the mixture of metal-bound (AB)2. Insets show expanded m/z ranges for 2MII-(AB)2 complexes with magenta, green, and cyan lines corresponding to theoretical m/z values of 2CoII, 2NiII, and 2CuII-(AB)2 complexes.
Conclusions.
Metal selectivity by natural proteins relies on a precise arrangement of amino acids that is frequently associated with preorganized architectures. For instance, the rigid selectivity filter in the potassium ion channel can distinguish between K+ and Na+ ions whose ionic radii differ by only 0.36 Å38. Yet, there are also prominent examples such as calmodulin29 or metalloregulatory39 proteins that are inherently flexible but become well-structured upon binding their cognate metals to reach the same level of sub-Å precision. These features are realized in the design of (AB)2, whose “acquired rigidity” is driven by direct structural coupling between symmetrical coordination sites in de novo interfaces. (AB)2 was not designed with the specific goal of selecting against CuII binding at the outset. However, the disulfide-bridged (AB)2 interface was deliberately constructed to be flexible, yet conducive to yielding structurally well-ordered states, which was essential for the discovery of a new design principle for selective metal coordination. This mode of construction is highly reminiscent of natural protein folds that simultaneously possess structural order and flexibility to enable the emergence of new functions40 and cooperativity/allosterism41,42. We believe that such flexible, minimally tethered, and evolutionarily unconstrained protein-protein interfaces provide an ideal platform for the design and directed-evolution of selective binding sites for metal ions as well as other substrates.
Our findings also show that it is possible to thermodynamically overcome the near-universal dominance of Irving-Williams series solely through the design of a 3D protein architecture. However, achieving selectivity over CuII comes at the cost of a rigidified protein structure and a highly restrictive metal coordination geometry, leaving little room for developing higher-order metal-based functions such as catalysis, electron transfer and signaling that rely on diverse metal coordination environments. By extension, this implies that the bioinorganic complexity of a cell could not have evolved solely by relying on the 3D structures of proteins to selectively bind metal ions under thermodynamic control, therefore necessitating actively driven metal homeostasis mechanisms for the emergence of complex organisms.
Methods
Protein mutagenesis, expression, and purification.
A pET20b(+)/Cys(His)3 vector with ampicillin resistance was utilized for site-directed mutagenesis of the original Cys(His)3 variant into (AB)2 variants31. PCR products generated from site-directed mutagenesis were transformed to chemically competent XL-1 blue cells (Agilent) and plated onto LB-agar plates with 100 mg/L ampicillin at 37 °C for 24 h. Plasmids were purified using Express plasmid Miniprep kit (Biomiga) and sequenced (Eton Bioscience). Sequenced plasmids were transformed to chemically competent BL21(DE3) cells containing a ccm (cytochrome c maturation) cassette with chloramphenicol resistance and plated onto LB-agar plates containing 100 mg/L ampicillin and 33 mg/L chloramphenicol at 37 °C for 24 h32,43. Isolated colonies were transferred to 5 mL LB media with 100 mg/L ampicillin and 33 mg/L chloramphenicol and grown for 12–14 h. These cultures were inoculated into 2.8-L glass flasks containing 1.3 L of LB media with 100 mg/L ampicillin and 33 mg/L chloramphenicol. Flasks were shaken at 100 rpm for 16–24 h at 37 °C. Pink-colored cells were harvested using centrifugation at 6,000×g for 7 min, and the cell pellet was stored at −80 °C. Frozen cell pellets were suspended in a 10 mM sodium acetate buffer solution (50–100 mL, pH 4.5) with vigorous stirring for 1 h. The pellets were lysed by repeated cycles of sonication in an ice bath. The pH of the lysate was adjusted to 10 and lowered to 4.5 to precipitate cell components other than cyt cb562 variants. After centrifugation (10,000 rpm for 10 min), red-colored supernatant was filtered using 0.45 μm syringe disk-filter. The clear supernatant was diluted with a 10 mM sodium acetate buffer solution (0.8 L, pH 4.5) and applied to a CM Sepharose Fast Flow resin pre-equilibrated with the same buffer. The column was washed with additional buffer (0.2 L) and eluted using a step-gradient from 0–1000 mM NaCl. Fractions with A415/A280 values > 4 were pooled, concentrated, and exchanged into a 10 mM sodium acetate buffer solution (pH 4.5). The protein was then purified on a High-S cartridge column using a step-gradient of 0–1000 mM NaCl using a Biologic DuoFlow workstation (BioRad). Fractions with A415/A280 values > 6 were pooled, concentrated, and exchanged into a 20 mM MOPS buffer solution (pH 7.4) using a Vivaspin-6 3kDa-centrifugal filter (Sartorious). CuCl2 (0.5× protein monomer concentration) was added to catalyze the formation of disulfide bond. The mixture of protein and CuCl2 was incubated at 37 °C for 8 h. The protein was treated with 10-fold excess EDTA overnight at 4 °C to prepare metal-free protein. EDTA and monomeric protein were removed using a Chelex100-treated, 20 mM MOPS buffer solution (pH 7.4) in a Vivaspin-6 10 kDa-centrifugal filter (Sartorius). The purity of the protein was confirmed by SDS-PAGE and A415/A280 values44. Concentrated proteins (~10 mM) were kept at 4 °C and used within a month.
Solution small angle X-ray scattering (SAXS).
SAXS scattering profiles of (AB)2 were obtained at Beamline 4-2 of the Stanford Synchrotron Radiation Laboratory (SSRL). Sample-to-detector distance was set to 1.7 m, and the temperature was maintained at 20 °C. Protein concentrations were set to 20 mg/mL (800 μM dimer) in a 20 mM MOPS buffer solution (pH 7.4) containing 150 mM NaCl solution. All samples were transferred to 8×PCR strip tubes and placed in the BL4-2 autosampler45. The autosampler injected 25 μL of the solution into a quartz capillary tube with a diameter of 1.5 mm equipped in the lab-made sample holder. SAXS profiles of the samples were recorded five times (1 pattern/sec) with the interval of 2 s to monitor the structural transition induced by the incident X-ray, and no X-ray induced change occurred in (AB)2 variants. Then, SAXS scattering patterns of two independently prepared protein solutions were collected using Blu-Ice 5 software. All measurements were averaged and background-subtracted in SAXSPipe. Experimental SAXS profiles were also reproducible in identical conditions after 6 months. Further data analysis of SAXS profiles was performed using ATSAS software package46. Intensities of all merged scattering profiles were normalized in SAS data analysis. Guinier analysis of (AB)2, (AC)2, and (BC)2 variants was performed using the equation below in SAS data analysis software of ATSAS package46.
where q and I(q) are the scattering vector and the scattering intensity. Theoretical scattering profiles of 2CoII-(AB)2, 2NiII-(AB)2, and 1CuII-(AB)2 structures were generated in CRYSOL47 using default parameters and were compared with experimental scattering profiles. Chi-square (χ2) values were evaluated from differences between experimental scattering profiles and theoretical scattering profiles using the equation below:
where m is the total number of points in scattering profiles, and σexp is the standard deviation of experimental scattering profiles. aexp values were obtained using repeat exposures of the same sample. Experimental scattering profiles were converted to normalized Kratky plots using the function below to emphasize q-range from 0.1 to 0.3 Å−1, which corresponds to the change of dimer orientation.
X-ray crystallography.
Red-colored crystals of (AB)2 variants were obtained by sitting-drop vapor diffusion at room temperature. Crystallization conditions are listed in Supplementary Table 1. Protein solutions were preincubated with the metal ion of interest for 1 h. For crystallization under metal competition conditions, protein solutions were incubated with the first metal ion for 1 h, and then the second metal ion was added and incubated for an additional 24 h. Sitting drop contained 1–2 μL each of the protein and precipitant solutions and the reservoir contained 500 μL of the precipitant solution. Crystals typically appeared within a week and were harvested after 2–8 weeks. Experimental crystal growth conditions are tabulated in Supplementary Table 1. Concentrations of dimer and metal ions were 2 mM and 4 mM, respectively. Perfluoro polyether (Hampton) was utilized for cryoprotection prior to crystal freezing in liquid N2. Diffraction data were collected at ALS Beamline 5.0.2, and SSRL Beamline 9-2 using multi-wavelength radiation (Blu-Ice 5 software) or at UCSD crystallography facility using a Bruker APEX II Cu-edge source diffractometer at 100 K (APEX3 software). Diffraction patterns of 3–5 protein crystals in 1–2 screening conditions were confirmed without significant changes in overall protein structures, and the highest resolution data set was chosen for further analysis. Diffraction data were processed using iMosflm and Scala48. Molecular replacement was performed using Phaser-MR with monomeric cyt cb562 (PDB: 2BC5) as a search model. Model refinement using phenix.refine and metal/water placement using COOT49 were performed to generate final models. Electron density maps were generated using Phenix50 and all graphics related to structures and electron densities were produced using PYMOL51. X-ray data collection and refinement statistics are listed in Extended Data Tables 2 and 3.
Analytical ultracentrifugation (AUC).
To confirm the oligomeric state of initially designed (AB)2, (AC)2, and (BC)2 variants, AUC analysis was performed at 25 μM dimer concentration in a buffer solution containing 20 mM MOPS and 150 mM NaCl (pH 7.4). Sedimentation velocities were measured on a Beckman XL-A instrument using an An-60 Ti rotor. For each sample, 250-600 scans were obtained at 41,000 RPM and 25 °C for 12–16 h using ProteomLab software. All data were processed using SEDFIT software52 with the following fixed parameters: dbuffer = 1.0049 g/mL; ηbuffer = 0.010214 poise; Vbar = 0.73084.
Electrospray ionization mass spectrometry (ESI-MS).
ESI-MS experiments were performed using Elite hybrid linear ion trap-orbitrap mass spectrometer equipped with a heated-electrospray ionization (HESI) source (Thermo). All solutions for ESI-MS samples were prepared using LC-MS grade water (JT Baker). The concentrations of (AB)2 dimers were adjusted to 5 μM in 20 mM NH4HCO3. Protein solutions were exchanged using 20 mM to remove non-specifically bound metal ions. The capillary voltage was adjusted to 2.5 kV, the flow rate was set to 3–10 μL/min, the source temperature was set to 100 °C, and the gas flow rates (sheath and aux) were set to 12. For each sample, 200 spectra were obtained for 3–4 min and averaged for further analysis using Xcalibur software. To prevent cross-contamination of metal ions, peek tubing and steel capillary were flushed out using 0.1% formic acid and 20 mM NH4HCO3 in each measurement. High mass mode at 240,000 resolution was applied to obtain the best resolution limit. Estimated resolution (m/Δm) of the mass spectrometer was ~80,000 around +11 charge state of (AB)2. To assure reproducibility of ESI-MS results, two independent experiments were performed with the interval of 1–2 weeks. Theoretical and experimental m/z values of (AB)2 and H100A(AB)2 complexes are tabulated in Supplementary Tables 2-5.
Periplasmic extraction of (AB)2.
BL21(DE3) colonies with pET20b(+) vector encoding (AB)2 were inoculated to 25 mL LB media in 50 mL Falcon tubes and shaken at 200 rpm for 24 h at 37 °C. Then, metal ions were supplemented to LB media and incubated at 200 rpm for additional 8 h at 37 °C. Cells were centrifuged at 8,000×g for 10 min at 4 °C. The periplasmic extraction protocol was adopted from references21,53 with the following modifications. The cell pellets were washed with 4 mL ice-cold 1× phosphate buffered saline (PBS). No CoII, NiII, and CuII contaminant was observed in the ICP-MS analysis of the fresh 1× PBS solution. Only negligible amounts (<10 nM) of CoII, NiII, and CuII were detected in the 1× PBS washing step, indicating that nearly all non-intracellular metal ions were removed by the cell pelleting and the washing step. The pellet was resuspended in 1.5 mL of a 20 mM NH4HCO3 solution with 20% (w/v) sucrose and transferred to 2 mL tubes. The suspension was incubated for 20 mins at room temperature and centrifuged at 17,000×g for 1 min. The pellet was then resuspended with 0.75 mL of ice-cold 20 mM NH4HCO3 solution and incubated for 20 mins at room temperature to induce the lysis of the outer membrane, followed by centrifugation at 17,000 × g for 2 min. Freshly prepared solutions of 20 mM NH4HCO3 (with or without 20% sucrose) were also free of CoII, NiII, and CuII contaminants, as determined by ICP-MS analysis. The red-colored supernatant was diluted to 2 mL using 20 mM NH4HCO3 and filtered using a Vivaspin-6 100 kDa-centrifugal filter (Sartorius) to remove cell debris and large molecular weight components. The extract was filtered using an Amicon ultra-0.5 50kDa-centrifugal filter (Millipore). The filtrate was concentrated with an Amicon ultra-0.5 30kDa-centrifugal filter (Millipore) and buffer-exchanged using a 20 mM NH4HCO3 solution 4-6 times to remove low molecular weight species. The A415/A280 ratio was in the range of 2–4. ESI-MS spectra of purified extracts were obtained under the same parameters as above. ESI-MS experiments of the extracts were also performed in duplicate using two independent samples to examine reproducibility of metal-selectivity in the periplasm.
QM-MM calculations.
To evaluate the conformational stabilities of CuII-bound (AB)2 structures, Quantum Mechanics/Molecular Mechanics (QM/MM) calculations of CuII-bound (AB)2 were performed using two-layer ONIOM (Our own n-layered integrated molecular orbital and molecular mechanics) approach54,55. Using Chimera 1.13.1, water boxes with 1 nm × 1 nm × 1 nm dimension were created around the structures of Model 1 (CuII in the peripheral site of (AB)2), Model 2 (CuII in the peripheral site of H100A(AB)2), and Model 3 (CuII in the central site of (AB)2). These structures were converted to Gaussian input files using TAO package56. 5His/4His metal coordination spheres were designated as the QM region and the remaining parts of the model structures were set as the MM region. QM calculations were performed using B3LYP57,58 function with 6-31G+ (d,p) basis set59-61 and MM calculations were performed using the AMBER62,63 force field equipped in Gaussian 09. The interactions between QM and MM parts were treated using mechanical embedding64,65. Force field for the heme group is not defined in AMBER, but even without the heme group, the cyt cb562 scaffold was highly stable in a pilot test of MM optimization. Thus, all input files were prepared without the heme group. The QM region was released during optimization to find the most stable CuII-bound 5His/4His coordination sites whereby the MM region is optimized. After QM-MM optimization of Models 1–3, water molecules were removed and single point energy calculation was performed to evaluate relative stabilities of the optimized structures. The MM energy [EMM (model)] of the QM (model) part was subtracted from the MM energy [EMM (real)] of the whole (real) system to correct the energy of the MM region. Because the total number of atoms in the MM region of Models 1–3 is identical, EMM (real) - EMM (model) values were utilized to compare conformational stabilities of (AB)2 in Models 1–3. Model 3 showed the lowest energy value in the MM region, so Model 3 was set as zero point to compare relative stabilities of Models 1–3.
Fura-2 titration.
Equilibration dissociation constants of (AB)2 with metal ions were obtained by competitive binding titrations using Fura-2, Mag-Fura-2, and Newport Green DCF25,31,66. Trace amounts of metal ions were removed from buffer solutions containing 20 mM MOPS and 150 mM NaCl (pH 7.4) using a Chelex100 resin (Sigma-Aldrich). To a 1-mL buffer solution, 5 μM of metal-free dye and 5 μM of metal-free (AB)2 were added, followed by the addition of aliquots of 2 mM MnCl2, CoCl2, NiCl2, CuCl2 or ZnCl2 stock solutions. Concentrations of all metal solutions were determined using a PAR assay67. Competitive binding of metal ions, dyes, and (AB)2 was monitored on an Agilent 8453 UV-vis spectrophotometer. Complete equilibration was achieved within 5 minutes following the addition of each metal aliquot. Changes in the absorbance at 335 nm (Fura-2), 324 nm (Mag-Fura-2), and 511 nm (Newport Green DCF) were used to measure the concentration of metal-bound indicators. Addition of metal aliquots were sequentially performed until the absorbances of the dyes were fully saturated (i.e., no further changes were observed). All titrations were done in triplicate and the titration curves were fit to a one- or a two-site binding model for protein using Dynafit68, taking into account the total indicator concentration and the metal concentration at which the absorbance for the metal-bound indicator reached the saturation point.
Electron Paramagnetic Resonance Measurements.
The EPR sample (5 mM CuII-(AB)2) was prepared in a buffer solution containing 20 mM MOPS (pH 7.4) and 150 mM NaCl and loaded into a Wilmad Q-1.0X1.2 quartz capillary tube. The capillary tube was then placed in a 4mm OD Wilmad quartz tube (707-SQ-250M). Continuous wave (CW) EPR data for CuII-(AB)2 were collected at X-band frequency (9–9.8 GHz) on a Bruker EMXplus EPR spectrometer equipped with a high-sensitivity resonator (ER4119HS) at UCSD. The CW-EPR spectrum was collected at 298 K using Xenon software and fit using Easyspin software69.
Mathematical calculation of metal-free and metal-bound (AB)2 in competitive conditions with CuII.
Fractions of metal-free and metal-bound (AB)2 species were calculated using Kd values of 1CuII-(AB)2, 2NiII-(AB)2, and 2CoII-(AB)2 as a function of metal concentration. Mathematical model of the (AB)2 fractions is available in Supplementary Methods.
Extended Data
Extended Data Figure 1 ∣. Analysis of the oligomerization states of (AB)2, (AC)2, and (BC)2 variants.
a, Sedimentation velocity/AUC analysis of the oligomerization state of (AB)2 (black), (AC)2 (red), and (BC)2 (blue) at 25 μM dimer concentration. The inset shows a higher-order oligomer population of (BC)2. Guinier analysis of b, AB (black), c, AC, and d, BC constructs measured by solution small-angle X-ray scattering (SAXS) at 0.8 mM dimer concentration. Larger I(0) values in (AC)2 and (BC)2 indicate higher oligomeric states compared to (AB)2. Radius of gyration (Rg) value of (AB)2 corresponds to approximately 25 kDa in Mw–Rg relationship of globular proteins72, indicating a dimeric state for (AB)2.
Extended Data Figure 2 ∣. Structural characterization and metal-binding analysis of MII-(AB)2 complexes.
a, X-band EPR spectrum of CuII-(AB)2 (orange) and simulated pattern (black line), along with the fit parameters consistent with a rhombic coordination geometry. Conditions: 2.5 mM CuII, 20 mM MOPS (pH 7.4) and 150 mM NaCl, 298 K. b, Crystal structure of 2CoII-(AB)2. CoII ions are represented as magenta spheres. c, Solution characterization of 2CoII-(AB)2 complex by competitive Fura-2 titration (left), SAXS (middle), and ESI-MS (right). Circles (magenta) in ESI-MS spectrum represent the number of CoII ions bound to (AB)2. Experimental data points and error bars in the Fura-2 titration are presented as mean and standard deviation of three independent measurements. d, Competitive metal-binding titrations (AB)2 for MnII (pink) and ZnII (grey) binding. Mag-Fura-2 and Fura-2 were used for MnII and ZnII titrations, respectively. e, Investigation of MII-(AB)2 complexation using ESI-MS: MnII (top), FeII (middle), and ZnII (bottom). MnII- and FeII-(AB)2 complexes were not observed owing to the low metal binding affinities. f, ESI-MS spectra of (AB)2 collected under CuII//ZnII and ZnII//CuII competition conditions. CuII outcompetes ZnII or forms heterometallic complexes of (AB)2. g, Theoretical SAXS scattering profiles of MII-(AB)2 generated by CRYSOL. Experimental SAXS profiles of h, 2CoII- and 2NiII-(AB)2 complexes compared with theoretical scattering profile of 1CuII-(AB)2 (cyan), and i, 1CuII-(AB)2 complex compared with theoretical scattering profiles of 2CoII- (magenta) and 2NiII-(AB)2 (green). Log-scale plots of Fig. 1e-f to compare j, experimental scattering profiles between 2CoII/2NiII-(AB)2 and 1CuII-(AB)2 and k, experimental scattering profiles of 2CoII- (left), 2NiII- (middle), and 1CuII-(AB)2 (right) with theoretical scattering profiles. Expanded low q-ranges of the scattering plots in l, Extended Data Fig. 2j and m, Fig. 2k.
Extended Data Figure 3 ∣. Structural characterization and metal-binding analysis of MII-(AB)2 complexes under competitive conditions with CuII.
a, Crystal structures (ribbon models) of (AB)2 formed under CoII//CuII (PDB:7RRB), CuII//CoII (PDB:7RRR), NiII//CuII (PDB:7LR5), and CuII//NiII (PDB:7LRA) conditions. Root mean square deviation (RMSD) values were determined in comparison with the crystal structures of 2CoII-(AB)2 and 2NiII-(AB)2 (grey cartoon models). b, Experimental SAXS profile of (AB)2 (0.8 mM) in CoII//CuII, CuII//CoII, NiII//CuII, and CuII//NiII conditions ([MII] = 1.6 mM) compared with the theoretical SAXS profile of 1CuII-(AB)2. c, ESI-MS spectrum of (AB)2 (5 μM) without metal ions. d, ESI-MS spectrum of (AB)2 (5 μM) with sub-stoichiometric amounts of CuII (5 μM) and CoII or NiII (5 μM). Circles in ESI-MS spectra represent the number of CoII (magenta), NiII (green), and CuII (cyan) ions bound to (AB)2. e, Competitive metal-binding titration of (AB)2 with CoII (magenta), NiII (green), and CuII (cyan) in 20 mM NH4HCO3 (pH 7.8). Mag-Fura-2 (5 μM) was used for competitive CoII and NiII titration, and Newport green DCF (5 μM) was used for competitive CuII titration. Experimental data points and error bars are presented as mean and standard deviation of three independent measurements. Log-scale plots (left) of Fig. 2c to compare experimental scattering profiles of f, CoII//CuII and CuII//CoII, and g, NiII//CuII and CuII//NiII with theoretical scattering profiles. Right panels present expanded low q-ranges of the scattering plots (left).
Extended Data Figure 4 ∣. Peripheral 5His coordination site of 2CoII-(AB)2 and rotamer analysis of the H100 side chain.
a, CoII-coordination in the dimer interface of (AB)2 with the 2mFo-DFc electron density map (grey mesh) contoured at 5.0 σ (metal) and 1.5 σ (ligand). Coordination distances and angles between CoII and ligands are shown on the right. b, Possible rotamer orientations of H100 residue in the crystal structure of 2NiII-(AB)2. Rotamers were generated with the combination of χ1 (torsion angle of Cα-Cβ = 60–300°, 60° interval) and χ2 (torsion angle of Cβ-Cγ = 0–330°, 30° interval). c, Averaged number of atomic contacts of H100 rotamers as a function of van der waals (VdW) overlap. Error bars reflect the standard deviations of the atomic contacts in rotamers with fixed χ1 and variable χ2. Compared to the original conformation of H100, all possible rotamers show significant clashes with proximal residues. d, Location of proximal residues overlapped with H100 rotamers.
Extended Data Figure 5 ∣. QM/MM calculations of Models 1–3.
QM/MM-optimized structures of a, Model 1: CuII in the peripheral site of (AB)2 b, Model 2: CuII in the peripheral site of H100A(AB)2, and c, Model 3: CuII in the central site of (AB)2. Conformations of the opposite peripheral binding site (red) from the modeled CuII coordination site are shown in the panel on the right in a and b. d, RMSDs of Cα positions between QM/MM-optimized models and the crystal structure (His: Cα’s of 5His or 4His residues in the opposite peripheral site, All: all Cα’s in (AB)2 or H100A(AB)2, and Interface: Cα’s of residues in the dimer interface.
Extended Data Figure 6 ∣. Characterization of metal-bound H100A(AB)2 complexes.
a, ESI-MS spectra of H100A(AB)2 (5 μM) complexed with metal ions. Circles in ESI-MS spectra represent the number of CoII (magenta), NiII (green), and CuII (cyan) ions bound to H100A(AB)2. Metal concentrations were 10 μM. b, Fura-2 competitive metal titration assay of H100A(AB)2 with CuII. Changes in Fura-2 absorbance at 335 nm (cyan) are plotted with theoretical metal-binding isotherms in the absence (grey) and the presence (black) of (AB)2. Experimental data points and error bars in the Fura-2 titration are presented as mean and standard deviation of three independent measurements. Crystal structure of c, CoII_H100A(AB)2 (PDB:7N4G) and d, NiII_H100A(AB)2 (PDB:7N4F). CoII and NiII ions are represented as magenta and green. Atomic details of each metal coordination site are shown in the right panels, with the 2mFo-DFc electron density map (grey mesh) contoured at 5.0 σ (metal) and 1.5 σ (ligand).
Extended Data Figure 7 ∣. Characterization of periplasmic extracts using SDS-PAGE and ESI-MS.
SDS-PAGE of periplasmic extracts and media stained with a, Coomassie Blue for all proteinaceous contents and b, o-dianisidine for heme proteins. Regardless of supplemented metal ions, no significant amount of (AB)2 dimer and (AB) monomer was observed in media. c, ESI-MS spectra of (AB)2 extracted from cells grown without metal supplement (top) and with CoII (middle) or NiII (bottom) supplement (20 μM) in LB media. ESI-MS spectra of (AB)2 extracted from cells grown with d, 100 μM and e, 5 μM metal ions in LB media. Inlet spectra represent expanded m/z ranges for 2MII-(AB)2 complexes with magenta, green, and cyan lines corresponding to theoretical m/z values of 2CoII, 2NiII, and 2CuII-(AB)2 complexes. Supplemented metal ions are represented as magenta (CoII), green (NiII), and cyan (CuII). Filled circles indicate the number of metal ions bound to (AB)2. D and M in gel pictures refer to the dimer and the monomer, respectively.
Extended Data Table 1 ∣. Equilibrium dissociation constants (Kd) of MII-(AB)2 complexes.
Metal-competition assays were performed using Fura-2 in Chelex100-treated buffer solution (pH 7.4) containing 20 mM MOPS and 150 mM NaCl. Kd values in 20 mM NH4HCO3 were determined using Mag-Fura-2 (CoII and NiII) and Newport Green DCF (CuII) as metal-binding competitor. Errors are derived from non-linear least square fitting analyzed in Dynafit.
| (AB)2 | (AB)2 (20 mM NH4HCO3) |
||
|---|---|---|---|
| Mn II | K d1 | 2.7 (±0.4) × 10−6 | |
| K d2 | 4.3 (±1.1) × 10−6 | ||
| Co II | K d1 | 1.1 (±0.2) × 10−8 | 5.2 (±1.8) × 10−8 |
| K d2 | 4.1 (±0.7) × 10−8 | 1.1 (±0.3) × 10−7 | |
| Ni II | K d1 | 4.5 (±1.1) × 10−9 | 3.0 (±0.7) × 10−8 |
| K d2 | 7.6 (±1.0) × 10−9 | 6.1 (±0.7) × 10−8 | |
| Cu II | K d1 | 2.9 (±0.5) × 10−13 | 4.3 (±1.4) × 10−9 |
| K d2 | - | - | |
| Zn II | K d1 | 1.4 (±0.3) × 10−8 | |
| K d2 | 1.3 (±0.4) × 10−8 |
Extended Data Table 2 ∣. X-ray data collection, processing and refinement statistics.
Numbers in parentheses correspond to statistics of the highest resolution shell.
| 2CoII-(AB)2 | 2NiII-(AB)2 | 1CuII-(AB)2 | 1CoII_H100A(AB)2 | 1NiII_H100A(AB)2 | |
|---|---|---|---|---|---|
| PDB ID | 7MK4 | 7LRV | 7LV1 | 7N4G | 7N4F |
| Data collection | |||||
| Resol. range (Å) | 34.42 - 1.269 (1.315 - 1.269) | 41.61 - 1.4 (1.45 - 1.4) | 37.88 - 1.91 (1.978 - 1.91) | 34.6 - 1.93 (1.999 - 1.93) | 35.03 - 1.8 (1.864 - 1.8) |
| Space group | P 21 21 21 | P 21 21 21 | P 1 21 1 | P 1 21 1 | P 1 21 1 |
| Unit cell (Å, °) | 54.53 61.08 64.57 90 90 90 |
54.91 60.72 63.77 90 90 90 |
34.17 83.21 38.57 90 100.83 90 |
35.21 78.27 39 90 100.68 90 |
34.47 84.65 39.14 90 100.51 90 |
| Total reflections | 416194 (26635) | 1371309 (118524) | 131503 (8872) | 217256 (21702) | 242223 (24300) |
| Unique reflections | 57551 (5614) | 42610 (4185) | 16289 (1530) | 15651 (1535) | 20432 (1985) |
| Multiplicity | 7.2 (4.7) | 32.2 (28.3) | 8.07 (5.8) | 13.9 (14.0) | 11.9 (11.8) |
| Completeness (%) | 99.79 (98.86) | 99.80 (99.12) | 98.82 (93.62) | 98.89 (98.65) | 96.31 (96.60) |
| Mean I/sigma(I) | 10.0 (1.5) | 152.76 (9.62) | 20.9 (5.8) | 159.03 (6.55) | 271.29 (19.40) |
| Wilson B-factor | 14 | 16.52 | 15.15 | 36.53 | 25.88 |
| R-merge | 0.067 (0.847) | 0.744 (0.9591) | 0.063 (0.214) | 0.5892 (0.8141) | 0.5845 (0.5606) |
| R-meas | 0.072 (0.953) | 0.7563 (0.9731) | 0.067 (0.237) | 0.6126 (0.8456) | 0.6132 (0.5889) |
| R-pim | 0.024 (0.429) | 0.1333 (0.1631) | 0.022 (0.099) | 0.1643 (0.2242) | 0.1812 (0.1773) |
| CC1/2 | 0.999 (0.765) | 0.738 (0.804) | 0.999 (0.978) | 0.82 (0.83) | 0.749 (0.817) |
| Refinement | |||||
| Reflections used in refinement | 57550 (5612) | 42605 (4183) | 16289 (1511) | 15519 (1533) | 19738 (1986) |
| Reflections used for R-free | 2002 (191) | 1995 (193) | 1625 (150) | 1549 (153) | 1974 (199) |
| R-work | 0.1960 (0.3160) | 0.1901 (0.2097) | 0.1908 (0.2724) | 0.2640 (0.3957) | 0.2078 (0.2966) |
| R-free | 0.2135 (0.3582) | 0.2161 (0.2557) | 0.2371 (0.2766) | 0.3245 (0.4594) | 0.2537 (0.3489) |
| Number of non-hydrogen atoms | 2123 | 1975 | 1927 | 1757 | 1801 |
| macromolecules | 1671 | 1636 | 1670 | 1651 | 1640 |
| ligands | 90 | 88 | 91 | 87 | 87 |
| solvent | 362 | 251 | 166 | 19 | 74 |
| Protein residues | 214 | 214 | 214 | 214 | 214 |
| RMS(bonds) | 0.006 | 0.006 | 0.008 | 0.009 | 0.008 |
| RMS(angles) | 0.81 | 0.78 | 0.84 | 1.19 | 0.93 |
| Ramachandran favored (%) | 100 | 99.04 | 99.52 | 96.63 | 99.52 |
| Ramachandran allowed (%) | 0 | 0.96 | 0.48 | 3.37 | 0.48 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 | 0 |
| Rotamer outliers (%) | 0 | 0 | 0 | 0 | 0.61 |
| Clashscore | 1.76 | 1.8 | 3.79 | 12.11 | 4.48 |
| Average B-factor | 19.01 | 23.24 | 18.1 | 49.13 | 32.97 |
| macromolecules | 17.84 | 22.77 | 17.93 | 49.45 | 33.31 |
| ligands | 15.22 | 21.96 | 13.72 | 46.2 | 26.74 |
| solvent | 25.36 | 26.73 | 22.24 | 34.22 | 32.78 |
Extended Data Table 3 ∣. X-ray data collection, processing and refinement statistics.
Numbers in parentheses correspond to statistics of the highest resolution shell.
| 2NiII-(AB)2 (NiII/CuII) |
2NiII-(AB)2 (CuII/NiII) |
2CoII-(AB)2 (CoII//CuII) |
2CoII-(AB)2 (CuII//CoII) |
|
|---|---|---|---|---|
| PDB ID | 7LR5 | 7LRA | 7LRB | 7LRR |
| Data collection | ||||
| Resol. range (Å) | 41.32 - 1.7 (1.761 - 1.7) | 41.65 - 1.7 (1.761 - 1.7) | 41.63 - 1.78 (1.844 - 1.78) | 44.27 - 1.89 (1.958 - 1.89) |
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Unit cell (Å, °) | 54.63 60.92 63.16 90 90 90 |
54.76 61 64.16 90 90 90 |
54.63 61.05 64.30 90 90 90 |
54.51 61.15 64.17 90 90 90 |
| Total reflections | 919338 (85881) | 686593 (63701) | 541511 (41580) | 531273 (31757) |
| Unique reflections | 23765 (2334) | 24213 (2371) | 20861 (1911) | 17578 (1611) |
| Multiplicity | 38.7 (36.7) | 28.4 (26.8) | 26.0 (21.6) | 30.2 (19.4) |
| Completeness (%) | 99.50 (99.62) | 99.54 (99.37) | 97.65 (91.70) | 98.54 (92.48) |
| Mean I/sigma(I) | 35.10 (6.50) | 49.46 (8.63) | 31.17 (6.16) | 14.77 (3.30) |
| Wilson B-factor | 17.96 | 17.98 | 17.17 | 16.99 |
| R-merge | 0.7238 (0.9307) | 0.7754 (0.8503) | 0.7359 (0.7766) | 0.8529 (0.9726) |
| R-meas | 0.7335 (0.9442) | 0.79 (0.8665) | 0.7509 (0.7961) | 0.8678 (0.9993) |
| R-pim | 0.1161 (0.156) | 0.1475 (0.1641) | 0.1447 (0.1692) | 0.156 (0.223) |
| CC1/2 | 0.878 (0.727) | 0.73 (0.771) | 0.676 (0.788) | 0.883 (0.417) |
| Refinement | ||||
| Reflections used in refinement | 23704 (2333) | 24165 (2371) | 20721 (1911) | 17488 (1611) |
| Reflections used for R-free | 2001 (196) | 2010 (198) | 1995 (180) | 1762 (158) |
| R-work | 0.1893 (0.2192) | 0.1802 (0.2215) | 0.1774 (0.2232) | 0.1902 (0.2544) |
| R-free | 0.2296 (0.2790) | 0.2141 (0.2508) | 0.2112 (0.2791) | 0.2294 (0.3165) |
| Number of non-hydrogen atoms | 1899 | 1968 | 1990 | 1940 |
| macromolecules | 1637 | 1635 | 1680 | 1667 |
| ligands | 88 | 88 | 90 | 90 |
| solvent | 174 | 245 | 220 | 183 |
| Protein residues | 214 | 214 | 214 | 214 |
| RMS(bonds) | 0.007 | 0.006 | 0.006 | 0.007 |
| RMS(angles) | 0.8 | 0.78 | 0.86 | 0.82 |
| Ramachandran favored (%) | 99.52 | 99.52 | 100 | 100 |
| Ramachandran allowed (%) | 0.48 | 0.48 | 0 | 0 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 |
| Rotamer outliers (%) | 0 | 0 | 0.58 | 0 |
| Clashscore | 3 | 4.51 | 2.91 | 2.64 |
| Average B-factor | 20.56 | 21.82 | 19.86 | 19.43 |
| macromolecules | 20.34 | 21.23 | 19.41 | 19.26 |
| ligands | 19.44 | 20.83 | 17.83 | 17.48 |
| solvent | 23.22 | 26.13 | 24.08 | 21.93 |
Supplementary Material
Acknowledgements
We thank members of the Tezcan Group, J. Rittle, H. Gray, S. Cohen and M. Green for helpful discussions. We also thank A. Kakkis and C.-J. Yu, for AUC and EPR measurements, respectively. This work was funded primarily by the NIH (R01-GM138884) and by NASA (80NSSC18M0093; ENIGMA: Evolution of Nanomachines in Geospheres and Microbial Ancestors (NASA Astrobiology Institute Cycle 8)). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (supported by the DOE, Office of Basic Energy Sciences contract DE-AC02-76SF00515 and NIH P30-GM133894) and the Advanced Light Source (supported by the DOE, Office of Basic Energy Sciences contract DE-AC02-05CH11231and NIH P30-GM124169-01).
Footnotes
Supplementary information is available for this paper at https://doi.org/
Competing interests
The authors declare no competing interests.
Data Availability
The principal data supporting the findings of this work are available within the figures and the Supplementary Information. Additional data that support the findings of this study are available from the corresponding author on request. Crystallographic data for protein structures (coordinates and structure factors) have been deposited into the RCSB data bank under the following accession codes: 7MK4, 7LRV, 7LV1, 7N4G, 7N4F, 7LR5, 7LRA, 7LRB and 7LRR. The model structure used for molecular replacement of processed X-ray diffraction data is also available in the RCSB data bank (code: 2BC5).
References
- 1.Waldron KJ, Rutherford JC, Ford D & Robinson NJ Metalloproteins and metal sensing. Nature 460, 823–830 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Gray HB, Stiefel EI, Valentine JS & Bertini I Biological Inorganic Chemistry: Structure and Reactivity (University Science Books, Sausalito, 2007). [Google Scholar]
- 3.Waldron KJ & Robinson NJ How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol 7, 25–35 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Dudev T & Lim C Competition among Metal Ions for Protein Binding Sites: Determinants of Metal Ion Selectivity in Proteins. Chem. Rev 114, 538–556 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Frausto da Silva JJR & Williams RJP The biological chemistry of the elements. (Oxford University Press, 2001). [Google Scholar]
- 6.Kisgeropoulos EC et al. Key Structural Motifs Balance Metal Binding and Oxidative Reactivity in a Heterobimetallic Mn/Fe Protein. J. Am. Chem. Soc 142, 5338–5354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grāve K, Griese JJ, Berggren G, Bennett MD & Högbom M The Bacillus anthracis class Ib ribonucleotide reductase subunit NrdF intrinsically selects manganese over iron. J. Biol. Inorg. Chem 25, 571–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes-Caballero H, Campanello GC & Giedroc DP Metalloregulatory proteins: Metal selectivity and allosteric switching. Biophys. Chem 156, 103–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Halloran TV & Culotta VC Metallochaperones, an Intracellular Shuttle Service for Metal Ions. J. Biol. Chem 275, 25057–25060 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Tottey S et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455, 1138–1142 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Lombardi A, Pirro F, Maglio O, Chino M & DeGrado WF De Novo Design of Four-Helix Bundle Metalloproteins: One Scaffold, Diverse Reactivities. Acc. Chem. Res 52, 1148–1159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y, Yeung N, Sieracki N & Marshall NM Design of functional metalloproteins. Nature 460, 855–862 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu F et al. Protein design: toward functional metalloenzymes. Chem. Rev 114, 3495–3578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwizer F et al. Artificial Metalloenzymes: Reaction Scope and Optimization Strategies. Chem. Rev 118, 142–231 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Churchfield LA & Tezcan FA Design and Construction of Functional Supramolecular Metalloprotein Assemblies. Acc. Chem. Res 52, 345–355 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Faiella M et al. An artificial di-iron oxo-protein with phenol oxidase activity. Nat. Chem. Biol 5, 882–884 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zastrow ML, Peacock FA, Stuckey JA & Pecoraro VL Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat. Chem 4, 118–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studer S et al. Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science 362, 1285–1288 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Khare SD et al. Computational redesign of a mononuclear zinc metalloenzyme for organophosphate hydrolysis. Nat. Chem. Biol 8, 294–300 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung N et al. Rational design of a structural and functional nitric oxide reductase. Nature 462, 1079–1082 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song WJ & Tezcan FA A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346, 1525–1528 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Churchfield LA, Medina-Morales A, Brodin JD, Perez A & Tezcan FA De Novo Design of an Allosteric Metalloprotein Assembly with Strained Disulfide Bonds. J. Am. Chem. Soc 138, 13163–13166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L et al. A protein engineered to bind uranyl selectively and with femtomolar affinity. Nat. Chem 6, 236–241 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Wegner SV, Boyaci H, Chen H, Jensen MP & He C Engineering A Uranyl-Specific Binding Protein from NikR. Angew. Chem. Int. Ed. Engl 48, 2339–2341 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Brodin JD et al. Evolution of Metal Selectivity in Templated Protein Interfaces. J. Am. Chem. Soc 132, 8610–8617 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guffy SL, Der BS & Kuhlman B Probing the minimal determinants of zinc binding with computational protein design. Protein Eng. Des. Sel 29, 327–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akcapinar Gunseli B. & Sezerman Osman U. Computational approaches for de novo design and redesign of metal-binding sites on proteins. Biosci. Rep 37, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byrd J & Winge DR Cooperative cluster formation in metallothionein. Arch. Biochem. Biophys 250, 233–237 (1986). [DOI] [PubMed] [Google Scholar]
- 29.Halling DB, Liebeskind BJ, Hall AW & Aldrich RW Conserved properties of individual Ca2+-binding sites in calmodulin. Proc. Natl. Acad. Sci. USA 113, E1216–E1225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zygiel EM & Nolan EM Transition Metal Sequestration by the Host-Defense Protein Calprotectin. Annu. Rev. Biochem 87, 621–643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rittle J, Field MJ, Green MT & Tezcan FA An efficient, step-economical strategy for the design of functional metalloproteins. Nat. Chem 11, 434–441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faraone-Mennella J, Tezcan FA, Gray HB & Winkler JR Stability and Folding Kinetics of Structurally Characterized Cytochrome c-b562. Biochemistry 45, 10504–10511 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Choi TS, Lee HJ, Han JY, Lim MH & Kim HI Molecular Insights into Human Serum Albumin as a Receptor of Amyloid-β in the Extracellular Region. J. Am. Chem. Soc 139, 15437–15445 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Burgot J-L Ionic Equilibria in Analytical Chemistry (Springer, 2012). [Google Scholar]
- 35.Osman D et al. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol 15, 241–249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young TR et al. Calculating metalation in cells reveals CobW acquires CoII for vitamin B12 biosynthesis while related proteins prefer ZnII. Nat. Commun 12, 1195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeschek M et al. Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature 537, 661 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Thompson AN et al. Mechanism of potassium-channel selectivity revealed by Na+ and Li+ binding sites within the KcsA pore. Nat. Struct. Mol. Biol 16, 1317–1324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capdevila DA, Braymer JJ, Edmonds KA, Wu H & Giedroc DP Entropy redistribution controls allostery in a metalloregulatory protein. Proc. Natl. Acad. Sci. USA 114, 4424–4429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokuriki N & Tawfik DS Protein Dynamism and Evolvability. Science 324, 203–207 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Motlagh HN, Wrabl JO, Li J & Hilser VJ The ensemble nature of allostery. Nature 508, 331–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papaleo E et al. The Role of Protein Loops and Linkers in Conformational Dynamics and Allostery. Chem. Rev 116, 6391–6423 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Arslan E, Schulz H, Zufferey R, Künzler P & Thöny-Meyer L Overproduction of the Bradyrhizobium japonicum c-Type Cytochrome Subunits of the cbb3 Oxidase in Escherichia coli. Biochem. Biophys. Res. Commun 251, 744–747 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Bailey JB, Subramanian RH, Churchfield LA & Tezcan FA in Method. Enzy Vol. 580 (ed Pecoraro Vincent L.) 223–250 (Academic Press, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martel A, Liu P, Weiss TM, Niebuhr M & Tsuruta H An integrated high-throughput data acquisition system for biological solution X-ray scattering studies. J. Synchrotron Radiat 19, 431–434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manalastas-Cantos K et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr 54, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svergun D, Barberato C & Koch MHJ CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr 28, 768–773 (1995). [Google Scholar]
- 48.Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8 (2015).
- 52.Schuck P Size-Distribution Analysis of Macromolecules by Sedimentation Velocity Ultracentrifugation and Lamm Equation Modeling. Biophys. J 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manoil C & Beckwith J A genetic approach to analyzing membrane protein topology. Science 233, 1403–1408 (1986). [DOI] [PubMed] [Google Scholar]
- 54.Dapprich S, Komáromi I, Byun KS, Morokuma K & Frisch MJ A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct.THEOCHEM 461–462, 1–21 (1999). [Google Scholar]
- 55.Vreven T, Morokuma K, Farkas Ü, Schlegel HB & Frisch MJ Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J. Comput. Chem 24, 760–769 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Tao P et al. Matrix Metalloproteinase 2 Inhibition: Combined Quantum Mechanics and Molecular Mechanics Studies of the Inhibition Mechanism of (4-Phenoxyphenylsulfonyl)methylthiirane and Its Oxirane Analogue. Biochemistry 48, 9839–9847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becke AD Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys 98, 5648–5652 (1993). [Google Scholar]
- 58.Lee C, Yang W & Parr RG Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785–789 (1988). [DOI] [PubMed] [Google Scholar]
- 59.Hariharan PC & Pople JA The effect of d-functions on molecular orbital energies for hydrocarbons. Chem. Phys. Lett 16, 217–219 (1972). [Google Scholar]
- 60.Rassolov VA, Pople JA, Ratner MA & Windus TL 6-31G* basis set for atoms K through Zn. J. Chem. Phys 109, 1223–1229 (1998). [Google Scholar]
- 61.Rassolov VA, Ratner MA, Pople JA, Redfern PC & Curtiss LA 6-31G* basis set for third-row atoms. J. Comput. Chem 22, 976–984 (2001). [Google Scholar]
- 62.Freindorf M, Shao Y, Furlani TR & Kong J Lennard–Jones parameters for the combined QM/MM method using the B3LYP/6-31G*/AMBER potential. J. Comput. Chem 26, 1270–1278 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Case DA et al. The Amber biomolecular simulation programs. J. Comput. Chem 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakowies D & Thiel W Hybrid Models for Combined Quantum Mechanical and Molecular Mechanical Approaches. J. Phys, Chem 100, 10580–10594 (1996). [Google Scholar]
- 65.Weiner SJ, Singh UC & Kollman PA Simulation of formamide hydrolysis by hydroxide ion in the gas phase and in aqueous solution. J. Am. Chem. Soc 107, 2219–2229 (1985). [Google Scholar]
- 66.Kakkis A, Gagnon D, Esselborn J, Britt RD & Tezcan FA Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites. Angew. Chem. Int. Ed 59, 21940–21944 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kocyła A, Pomorski A & Krężel A Molar absorption coefficients and stability constants of metal complexes of 4-(2-pyridylazo)resorcinol (PAR): Revisiting common chelating probe for the study of metalloproteins. J. Inorg. Biochem 152, 82–92 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Kuzmič P Program DYNAFIT for the Analysis of Enzyme Kinetic Data: Application to HIV Proteinase. Anal. Biochem 237, 260–273 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Stoll S & Schweiger A EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson 178, 42–55 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Smilgies D-M & Folta-Stogniew E Molecular weight-gyration radius relation of globular proteins: a comparison of light scattering, small-angle X-ray scattering and structure-based data. J. Appl. Crystallogr 48, 1604–1606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The principal data supporting the findings of this work are available within the figures and the Supplementary Information. Additional data that support the findings of this study are available from the corresponding author on request. Crystallographic data for protein structures (coordinates and structure factors) have been deposited into the RCSB data bank under the following accession codes: 7MK4, 7LRV, 7LV1, 7N4G, 7N4F, 7LR5, 7LRA, 7LRB and 7LRR. The model structure used for molecular replacement of processed X-ray diffraction data is also available in the RCSB data bank (code: 2BC5).