Abstract
Severe acute respiratory syndrome coronavirus 2 was originally identified in Wuhan city of China in December 2019 and it spread rapidly throughout the globe, causing a threat to human life. Since targeted therapies are deficient, scientists all over the world have an opportunity to develop novel drug therapies to combat COVID-19. After the declaration of a global medical emergency, it was established that the Food and Drug Administration (FDA) could permit the use of emergency testing, treatments, and vaccines to decrease suffering, and loss of life, and restore the nation's health and security. The FDA has approved the use of remdesivir and its analogs as an antiviral medication, to treat COVID-19. The primary protease of SARS-CoV-2, which has the potential to regulate coronavirus proliferation, has been a viable target for the discovery of medicines against SARS-CoV-2. The present research deals with the in silico technique to screen phytocompounds from a traditional medicinal plant, Bauhinia variegata for potential inhibitors of the SARS-CoV-2 main protease. Dried leaves of the plant B. variegata were used to prepare aqueous and methanol extract and the constituents were analyzed using the GC-MS technique. A total of 57 compounds were retrieved from the aqueous and methanol extract analysis. Among these, three lead compounds (2,5 dimethyl 1-H Pyrrole, 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide, and Benzonitrile m phenethyl) were shown to have the highest binding affinity (−5.719 to −5.580 kcal/mol) towards SARS-CoV-2 Mpro. The post MD simulation results also revealed the favorable confirmation and stability of the selected lead compounds with Mpro as per trajectory analysis. The Prime MM/GBSA binding free energy supports this finding, the top lead compound 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide showed high binding free energy (−64.377 ± 5.24 kcal/mol) towards Mpro which reflects the binding stability of the molecule with Mpro. The binding free energy of the complexes was strongly influenced by His, Gln, and Glu residues. All of the molecules chosen are found to have strong pharmacokinetic characteristics and show drug-likeness properties. The lead compounds present acute toxicity (LD50) values ranging from 670 mg/kg to 2500 mg/kg; with toxicity classifications of 4 and 5 classes. Thus, these compounds could behave as probable lead candidates for treatment against SARS-CoV-2. However further in vitro and in vivo studies are required for the development of medication against SARS-CoV-2.
Keywords: Bauhinia variegata; SARS-CoV-2 main protease; Molecular docking; Secondary metabolites; 2,5 dimethyl 1-H Pyrrole; Dynamic simulation; Prime MM/GBSA free Energy calculations
Graphical abstract
1. Introduction
The World Health Organization (WHO) has acknowledged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection as a universal emergency in January 2020. This disease i.e. COVID-19 originated in Wuhan, China and it merely acquired four months to become a pandemic [1,2]. COVID-19 has affected more than 332,617,707 patients, including 5,551,314 deaths, due to a lack of treatment possibilities and vaccines, particularly in middle and low-income countries [3]. The disease's etiological agent from the Coronaviridae family is a single-stranded RNA virus. The virus has been named SARS-CoV-2 because it is phylogenetically related to the SARS-CoV-2 that was materialized in 2002 [4]. COVID-19 is categorized by severe infection in the lungs called pneumonia and, in severe cases results in respiratory failure [5]. SARS-CoV-2 normally fits the beta-CoVs class with a diameter between 60 and 140 nm with a circular or ovoid shape [6]. SARS-CoV-2 is an RNA virus with positive strands, that encrypts 16 proteins without structural format (Nsp1-16), RNA synthesis constituents, viral RNA dependent RNA polymerase, and a pair of bulky non-structural polyproteins in part of its genome. The remaining portion of the genetic makeup encrypts tetrad structural proteins which include Spike protein (S), an Envelope protein (E), Membrane protein (M), Nucleocapsid (N), and the supplementary proteins like nonstructural proteins (Nsp1-16). Among these, Nsp3 represents papain-like protease (PLpro), Nsp5: 3C-like protease (3CLpro), and Nsp12: RNA-dependent RNA polymerase (RdRp) [[7], [8], [9]]. Furthermore, SARS-CoV-2 encodes its own miRNAs, which can enter host cells and be undetected by the immune system, causing diseases by targeting host function genes [10,11].
After the declaration of a global medical emergency, the Food and Drug Administration (FDA) has permitted the use of emergency testing, treatments, and vaccines to decrease suffering, and loss of life, and restore the nation's health and security. The FDA has approved the use of Remdesivir and its analogs as an antiviral medication, to treat COVID-19. Additional emergency treatments, including convalescent plasma, monoclonal antibodies, and other therapeutic combinations, have been approved by the FDA. Through the hopeful outcome of various vaccines has been established around the globe, it is widely accredited that supplementary modifications are of prime and foremost requirement due to the unforeseen behavior of the virus due to genetic mutations [12]. It could be a long time before Covid-19 is completely eradicated from all corners of the globe. The human immune response to the virus is not completely understood, and its experimental exhibitions are not entirely implicit either [13,14]. Therefore, in the current global epidemic scenario, there is an immediate necessity to explore bioactive compounds with the potential to combat this viral infection as well as provide our organs with resistance and strength to battle against the disease.
According to our ancient science, ‘Ayurveda’, Indian medicinal herbs have been utilized for the development of medications and to toughen our immune systems for a variety of diseases. Indigenous herbal plants have a number of biochemical and bioactive ingredients. These bioactive substances could be used to minimize the risk of getting a variety of dangerous viral infections by targeting specific viral receptors. Medicinal plants and natural products have been studied as prophylactics, treatments, and other helpful applications in recent years, depending on healing traditions or scientific evidence [15]. Herbal medicine has plentiful compensations, including the fact that it is naturally accessible, helps with hormones and metabolism, anti-inflammatory, and boosts the immune system [16]. For example, Sulphated polysaccharides have immunomodulatory and bioactive characteristics. Consumption of Tinospora cordifolia leaves could boost IgG antibody production in the body, which leads to macrophage activation. It improves humoral immunity [17].
B. variegata can be found in the tropical and temperate Indian subcontinent, Southeast Asia, and China. According to Ayurveda, the stem bark and blooms of this plant are frequently utilized as medication in treatment in pharmaceutical formulations; it has been used in the folklore remedy for diverse purposes such as; Galaganda (Goiter), Gandamala (Lymphadenopathy) [18], Ashthila which is also called as Benign Prostatic Hyperplasia (BPH)], Kapha-Pitta Dosha diseases, and Arbuda means tumor. These are all conditions where Kachnar bark is recommended. The flowers, on the other hand, have Rakta Pradaraghna i.e. for treating the dysfunctional uterine hemorrhage, Pittaghna i.e. for curing Pitta Dosha, Kshyaghna i.e. for treatment of tuberculosis, and Kaasghna i.e. utilized in the management of cough. However, it is also used to treat a variety of cancers [19]. Mohamed Shaheen et al. studied various extracts of B. variegata for antiviral activity. After the application of extracts to infected cells, the aqueous extract was found to be more effective against rotavirus infection, decreasing virus infectivity by 11.5% when compared to virus control. This finding suggested that the B. variegata extract could reduce virus infectivity by interfering with one or more steps of the viral replication cycle following entry into the host cell [20].
According to a review, irrespective of the fact that Bauhinia variegata's phytochemical constituents have a wide range of therapeutic applications, there has been inadequate research conducted to substantiate the beneficial claim in a systematic manner. Plants involved in the genus Bauhinia are a rich source of flavonoids, steroids, alkaloids, terpenes, and phenolic compounds [21]. The present research work deals with the objective to discover the therapeutic potential of B. variegata against the SARS-CoV-2 main protease by implementing computational methods including docking of molecules and molecular dynamics simulation to discover the probable medication for COVID-19. The results are very promising and show that B. variegata has the competence of being effective in the management of SARS-CoV-2. The molecular docking process helps to forecast the binding affinity with help of scoring functions amongst protein and ligand [21]. Many computational studies have revealed possible inhibitors of SARS-CoV-2 target proteins from various sources [[22], [23], [24], [25]]. In order to screen possible targets in a short amount of time, computational methodologies could be used [[26], [27], [28], [29]]. Since many of the SARS-CoV-2 proteins have high-resolution structures, we were able to discover lead therapeutic candidates for SARS-CoV-2 quickly using in silico screening methods. It allocates and utilizes a range of conformation search strategies to receptors and ligands, as well as anticipates potential binding ability by establishing chemical interactions between them [30]. While performing simulations in molecular dynamics, there is the amalgamation of Newton's laws of motion for producing sequential conformations of the evolving system which offers trajectories that stipulate the velocities and locations of the elements over time [31].
2. Materials and methods
2.1. Plant collection, processing, and soxhlet extraction
Leaves of B. variegata plant were collected, shade dried and pulverized in the Pune region Maharashtra, India in the month of January 2020. These powdered leaves were used for further Soxhlet extraction using organic solvents.100 gm powdered leaves were subjected to a continuous Soxhlet extraction unit with various organic solvents one by one as per polarities such as petroleum ether (60–80 °C), chloroform, and methanol and water respectively. The process is continued until the crude leaves powder has been completely exhausted, at which time a solvent pouring from the extraction chamber leaves no residue. This approach works well with plant materials that are partially soluble in the solvent of choice, as well as plant materials that contain insoluble contaminants. The requirement of solvent in this process is less as compared to maceration [32,33]. The obtained methanol and aqueous extract were dried and further used for Gas chromatography and mass spectrometry analysis. (GC-MS).
2.2. GC-MS analysis
In recent years FTIR, GC-MS, and LC-MS have frequently used techniques for the identification of functional groups and secondary metabolites of respective medicinal plant extracts. Some recent databases like MMEASE, NOREVA and ANPELA are also being used to identify the compound from the natural resources [[34], [35], [36], [37], [38]]. A gas chromatography along with a mass spectrometry (GC-MS) system was used to conduct the analysis of B. variegata aqueous as well as methanol leaves extract (model; QP2010 Ultra, Shimadzu Corporation, Kyoto, Japan). A capillary column of 30 m long, 0.25 mm i.d. coated with a 0.25 m film thickness stationary phase-separated the sample components (Rtx-5MS, Restek Corporation, Bellefonte PA, U.S.A). By keeping the linear velocity constant, of 36.3 cm/s, helium gas (99.999%) was used as the carrier gas. The AOC-20i + s auto-injector was used to inject a 1 μl sample volume. In split-less mode, at 260 °C the injection port was set. The temperature of the GC oven was set at 50 °C for initial 5 min, then increased to 300 °C at 2 °C per minute and kept for 10 min [39,40].
Ion source temperature was set to 240 °C in mass spectroscopy, whereas the interface was set to 240 °C. For the m/z range of 40–800, a total ion chromatogram (TIC) was generated. The mass spectra of GC peaks were linked to the National Institute of Standards and Technology database to identify them (NIST). By comparing the peak area of each constituent to the overall area of peaks in the chromatogram, the relative percentage amount of each component was computed.
2.3. Molecular docking calculations
2.3.1. Retrieval of secondary metabolites of B. variegata and their preparation
The 57 natural secondary metabolites (19 common compounds from aqueous and methanol extract and 57 from methanol extract) of B. variegata (Kanchnar) were identified from Gas chromatography-mass spectroscopic analysis (aqueous and methanol extract), and utilized as ligand dataset for the present study (Supplementary Table S1). Further, the 3-D structures of these identified phytochemicals were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and saved in SDF file format. Before docking, all compounds were energy minimized using the Ligprep module of Schrodinger with OPLS3e force field till an energetically stable conformation was achieved. The preparation of ligands was done by keeping the pH to 7.0 ± 2.0.
2.3.2. Absorption, distribution, metabolism, excretion and toxicity (ADMET) profiling of the phytoconstituents
The retrieved 57 phytocompounds from B. variegata leaves extract were evaluated for the determination of ADME properties, and drug-likeness with the QikProp module of Maestro Schrodinger designed by Professor William L. Jorgensen. The QikProp module of Schrodinger is a simple, precise, and easy-to-use tool for absorption, distribution, metabolism, and excretion forecast that generates specific ADME characteristics. It predicts physicochemically important descriptors as well as pharmacokinetically important features based on Lipinski's rule of five, which indicates the drug-like action of ligand molecules. Rule of five proposed by Lipinski [41,42], which is frequently used to examine possible correlations amongst proposed leads and targeted biomolecules, analyses a compound's tendency to have a pharmacological or biological influence. The rule is worthwhile for screening promising therapeutic agents/drugs as a filter from the commencement of the project, slashing production costs, plummeting the expense of clinical drug development exercises, as well as, to a significant part, preventing late-stage clinical failures [43,44]. These secondary metabolites have the potential to act as lead compounds against SARS-CoV-2. Compared to existing computational models, the ProTox-II is a freely available computational toxicity web server that offers significant advantages. The ProTox web software is based on machine learning algorithms that act on both chemical and molecular targets. The ProTox-II website is unique in that it categorizes the prediction scheme into distinct categories of toxicity, such as oral toxicity, organ toxicity (hepatotoxicity), and toxicological endpoints (such as mutagenicity, carcinogenicity, cytotoxicity, and immunotoxicity) [45,46]. The LD50 values for selected lead compounds are provided via the ProTox-II servers. After being exposed to a material, the LD50 is the dose at which 50% of test participants die [[47], [48], [49]].
2.3.3. Crystal structure of SARS-CoV-2's main protease (Mpro)
The X-ray crystal structure of the SARS-CoV-2 main protease (Mpro) complexed with inhibitor N3 has been retrieved from the Protein Data Bank with PDB ID: 6LU7 [50]. Chains A and B comprise the homodimeric Mpro. In this study, chain A has been selected which was prepared using Schrodinger's Protein Preparation Wizard and utilized for next-level investigation. Input data of the receptor must be compiled by determining the receptor atoms, calculating the essential hydrogen atom's position, and defining and specifying the atoms of the binding site [51,52].
2.3.4. Receptor grid generation and molecular docking
The area of interaction between the protein and the ligand is defined by the Receptor grid. The tool in maestro 13 named the receptor grid generation tool was used to characterize the area around the active site in terms of coordinates x, y, and z. The determination of the grid box was done by using a co-crystallized peptide inhibitor i.e. N3 of 6LU7, to ensure that the center of each docked ligand had the identical binding box dimensions of the crystal ligand. Amino acids present in Mpro's binding site are, His41, Gln192 Cys44, Met49, Glu166, His172, Phe181, Pro52, Leu141, Gly143, His164, Thr24, Thr26, Leu27, Ser139, Phe140, Gln189, Thr190, and Asn142. Also, these binding site residues were confirmed with a literature survey [53,54].
Interaction between compounds of B. variegata and Mpro was achieved through molecular docking simulation. Glide's (v93137, Release: 2021-4) standard precision (SP) mode was used to conduct a rigid receptor docking process based on the OPLS3e force field [[55], [56], [57]]. During the docking simulations, the prepared structure of Mpro is reserved as rigid; whereas the ligands were flexible during the docking process. All molecules were docked into the binding site of the SARS-CoV-2 Mpro and used to estimate binding poses for the extracted compounds [58,59]. The ligand interaction tool was used to see how the ligands interacted with the residues in the target protein's active site. Also, a detailed interaction analysis of protein-ligand complexes has been performed using the PDBsum server (www.ebi.ac.uk/pdbsum/) [60] by providing docked complexes of lead compounds with SARS-CoV-2 Mpro protein.
Anti-COVID-19 medicines such as Remdesivir (antiviral), Ivermectin (anti-parasitic), Favipiravir (anti-influenza), Hydroxychloroquine (anti-malarial and anti-rheumatic diseases), and arbidol are now being studied (anti-influenza) by various researchers [[61], [62], [63], [64]]. FDA-approved drugs (commercially available) acting on SARS-CoV-2 Mpro target proteins like Ritonavir, Ribavirin, Remdesivir, Lopinavir, Ivermectin, Imatinib, Flavipiravir, Azithromycin, Baricitinib and Aspirin were downloaded from DrugBank database (https://go.drugbank.com/). [65]. These target proteins were compared with lead compounds using Glide SP of Schrödinger suite 2021-4.
2.4. Molecular dynamics simulation
The Desmond module of the Schrödinger suite was implemented for Molecular Dynamics (MD) simulation on a Linux system. The SARS-CoV-2 Mpro complexes with the topmost three ranked compounds were subjected to MD Simulation analysis for 100 ns (ns) to determine the docked complexes’ stability [56]. The Desmond had been used to examine the complexes in the specified solvent system with the OPLS-2005 force field. For a 10 Å buffer region, the solvation of the molecular system was done with explicit water (TIP3P) molecules by keeping under cubic periodic boundary conditions.
Neutralization of the system was completed by adding Na + as counter ions, which removed the overlying water molecules. To keep the temperature at (300 K) and pressure at (1 bar) of the systems constant, an ensemble (NPT) of Nose–Hoover thermostat and barostat was used [58]. The researchers used a hybrid energy minimization approach which involved 1000 steps of steepest descent which are followed by coupled gradient algorithms. Furthermore, RMSD and RMSF values were calculated for the assessment of the stability and dynamic properties of these complexes [66,67].
The backbone RMSDs of the SARS-CoV-2 Mpro structure were used to investigate the structural and dynamic characteristics of the protein-ligand complexes across a simulation period of 100 ns. To measure the flexibility of each amino acid residue in the complexes of protein-ligand, the root-mean-square fluctuations (RMSF) were determined and plotted. For all of the complexes, the RMSF of the protein-ligand composites represented the least variation.
Energy minimization experiments after molecular dynamic simulation were conducted utilizing Prime (v3.000, Release 2021-4) molecular mechanics-generalized Born surface area (MM-GB/SA) to forecast the free energy of binding for a group of ligands in a complex with a receptor [68,69]. Trajectories were captured at intervals of 10ns from the 1000 snapshots generated during 100ns of MD simulation along with initial conformation (docking pose) for free energy calculations and provided cumulative average energies with standard deviations [[70], [71], [72], [73]]. These postures were used as inputs for minimizing the energy of protein-ligand complexes (Ecomplex), free proteins (Eprotein), and free ligand (Eligand). Using the OPLS3e force field and the generalized born/surface area (GB/SA) continuum VSGB 2.0 solvent model, the energy for a minimal glide docked position of the ligand-receptor complex was computed [[74], [75], [76]]. The binding free energy Gbind was calculated using the following formula:
The Gcomplex represents complex energy, Gprotein is the receptor energy and Gligand is the unbound ligand energy [[77], [78], [79]].
3. Results and discussion
3.1. Characterization of phytoconstituents from B. variegata
3.1.1. GC-MS analysis
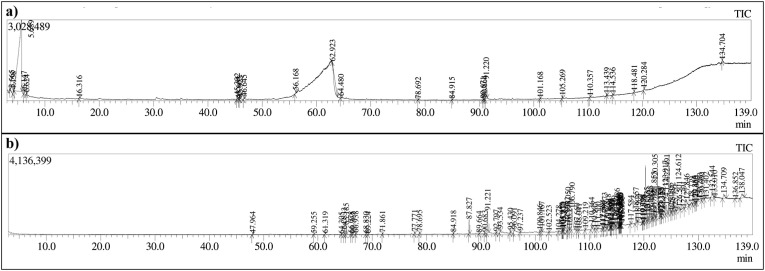
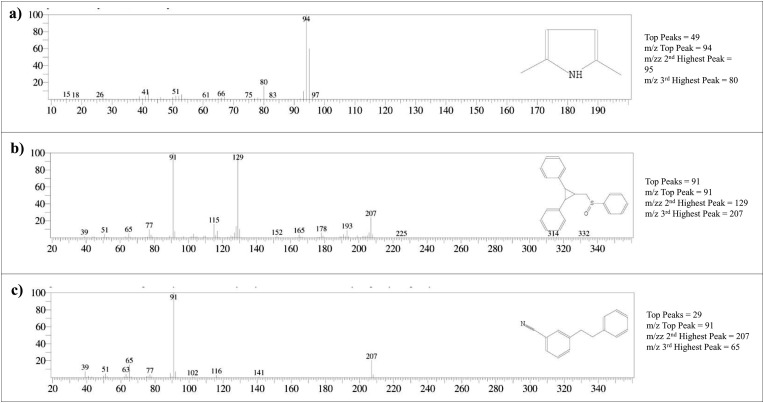
The Gas chromatography–mass spectrometry analysis of B. variegata leaf extracts in distilled water and methanol revealed a total of 19 and 57 peaks respectively corresponding to phytoconstituents which were identified by comparing peak retention time, peak area (percent), height (percent), and mass spectral fragmentation patterns to those of known compounds in the National Institute of Standards and Technology (NIST) library (Fig. 1 ). As the complex and challenging samples like plant extracts are analyzed with GC-MS analysis over the temperature range 40–800 °C m/z peaks of top docked compounds i.e 2,5 dimethylpyrrole,2,3 diphenyl cyclopropyl, methyl phenyl sulphoxide, and benzonitrile m phenethyl was matched as per NIST library and illustrated in Fig. 2 . The horizontal axis of a mass spectrum is expressed in units of m/z, which represents mass divided by charge number. Because z is virtually always 1 in GCMS, the m/z number is frequently taken as the mass. Peak percentage areas covered by 2,5 dimethylpyrrole is 0.52% with retention time 6.140–6.150 min and 4.92% for both 2,3 diphenyl cyclopropyl, methyl phenyl sulphoxide, and benzonitrile m phenethyl with retention time in between 91.215 and 91.225 min.
Fig. 1.
Total Ionic chromatogram (TIC) of (a) aqueous and (b) methanol extract of B. variegata leaves by GCMS.
Fig. 2.
Total Ionic chromatogram (TIC) of (a) 2,5 Dimethyl 1-H-Pyrrole, (b) 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide, (c) and Benzonitrile m phenethyl.
3.2. Catalytic position of the SARS-CoV-2 Mpro
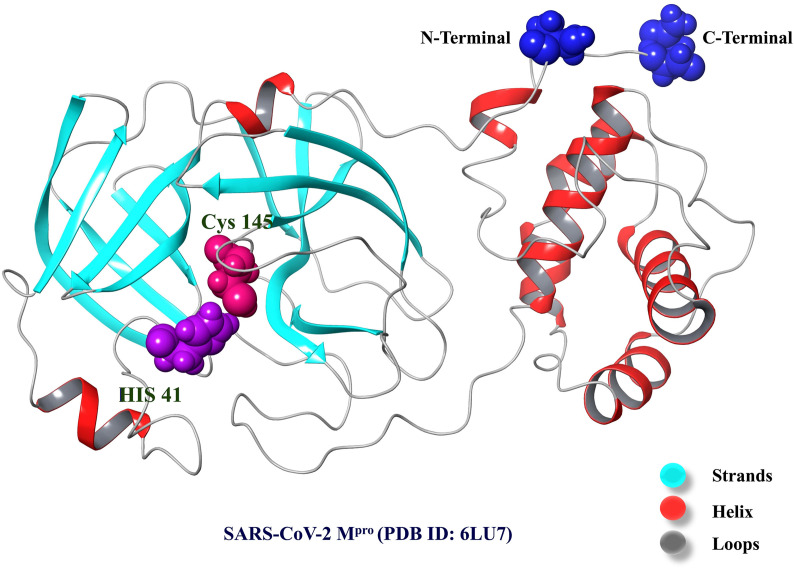
SARS-CoV-2 Mpro relates to the MEROPS peptidase C30 class, with Cys145 and His41 creating an active or catalytic dyad in the catalytic domain. As per the conserved domain search, Mpro comprises three functional domains, the first two of which include anti-parallel beta barrels and the third of which is made up of alpha-helices. The catalytic residues are situated between the first two domains in a cleft (Fig. 3 ). His41 is vital for the deprotonation of the catalytic Cys145 in order to bind the glutamine backbone at the peptide substrate's P1 site. As a result, His41 was replaced with aspartate and glutamate, which were thought to serve as generic bases for the deprotonation of Cys145 in SARS-CoV-2 Mpro. Additionally, Cys145 was replaced with serine, as in the catalytic triad of serine proteases, where serine serves as a nucleophile throughout catalysis. The catalytic activity of SARS-CoV-2 Mpro was not affected by serine substitution of Cys145. SARS-CoV-2 Mpro was inactivated by all amino acid changes analyzed [[77], [78], [79], [80]].
Fig. 3.
The crystal structure of SARS-CoV-2 Mpro (PDB ID: 6LU7). Residues (His41 and Cys145) involved in the catalytic dyad of the SARS-CoV-2 Mpro are shown in CPK representation.
3.3. Molecular docking
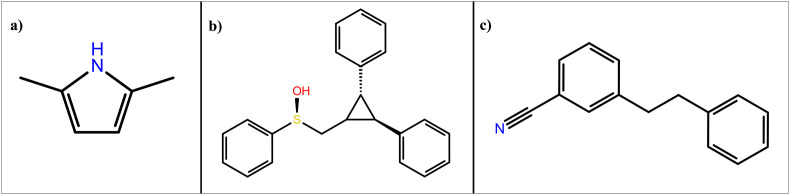
The compounds identified by GC-MS analysis of aqueous and methanol extracts of B. variegata leaves were docked against SARS-CoV-2 main protease (PDB ID 6LU7) using advanced molecular docking software i.e., the Glide module of the Schrodinger Suite. Binding affinity was represented by a lower docking score and Glide score (Table 1 and Table 2 ). Based on the docking score, the best three compounds 2,5 Dimethyl 1-H-Pyrrole (CID12265) (Fig. 4 (a)), 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide (CID562543) (Fig. 4 (b)) and Benzonitrile m phenethyl (CID141877) (Fig. 4 (c)) from B. variegata were screened whose energy value was ranged from −5.719 kcal/mol to −5.589 kcal/mol−1. The successfully docked other compounds with their docking score are provided in Supplementary Table S2. The bioactive phytocompound 2,5 dimethyl 1-H Pyrrole showed the maximum binding affinity for Mpro (6LU7), with a glide score of −5.719 kcal/mol. The results of the molecular docking studies showed the presence of non-bonded interactions between receptors and ligands which included hydrogen bonds, electrostatic bonds, and hydrophobic bonds. These three compounds were selected for further evaluation for their thermodynamic stability. The known FDA-approved drugs Favipiravir [81,82], Remdesivir [83], and Ritonavir [84], were also docked to protease enzyme. These drugs showed good interaction with the enzyme with a binding score of −5.560 kcal/mol with Favipiravir, −5.260 with remdesivir, and −5.153 with ritonavir. Favipiravir showed the best binding with the protease followed by Remdesivir and then Ritonavir. A similar observation was also reported by Tiago da silva et al. [84] when they tried to study their interaction. The lead compounds (Table 1) of B. variegata showed better binding interaction with the SARS-CoV-2 Mpro as compared to the FDA-approved drugs (Table 2).
Table 1.
The binding affinities and detailed interaction studies of lead compounds from B. variegata with SARS-CoV-2 Mpro.
| Compound | Binding Affinity (kcal/mol) | Interacting Amino Acid | Type of Intermolecular Bond |
Bond Distance (Å) |
|---|---|---|---|---|
| 2,5 Dimethyl 1-H-Pyrrole | −5.719 | His41 | Pi-Pi Stacking | 4.05 |
| His164 | Hydrogen bond | 1.86 | ||
| 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide | −5.690 | Gln189 | Hydrogen bond | 2.05 |
| His164 | Hydrogen bond | 2.65 | ||
| His164 | Aromatic HB | 2.21 | ||
| His41 | Pi-Pi Stacking | 4.09 | ||
| Glu189 | Aromatic HB | 2.69 | ||
| Benzonitrile-m Phenethyl | −5.580 | His41 | Pi-Pi Stacking | 5.03 |
| His163 | Hydrogen bond | 2.00 | ||
| N3 | −5.444 | Glu189 | Hydrogen Bond | 2.04 |
| Glu189 | Hydrogen Bond | 2.93 | ||
| Phe140 | Hydrogen Bond | 1.70 |
Table 2.
The binding affinities and detailed interaction studies of FDA Approved Drugs with SARS-CoV-2 Mpro.
| Compound | Binding Affinity (kcal/mol) | Interacting Amino Acid | Type of Intermolecular Bond |
Bond Distance (Å) |
|---|---|---|---|---|
| Favipiravir | −5.560 | Glu166 | Hydrogen bond | 2.27 |
| Phe140 | Hydrogen bond | 2.40 | ||
| Hip163 | Hydrogen bond | 2.18 | ||
| Hip163 | Hydrogen bond | 2.25 | ||
| Remdesivir | −5.262 | Gly 143 | Hydrogen bond | 2.31 |
| Asn142 | Hydrogen bond | 2.30 | ||
| Ritonavir | −5.153 | Asn142 | Hydrogen bond | 2.10 |
| Gly143 | Hydrogen bond | 2.43 | ||
| Glu166 | Hydrogen bond | 2.64 | ||
| His41 | Pi-Pi Stacking | 4.76 | ||
| Aspirin | −4.512 | Glu166 | Hydrogen bond | 2.18 |
| Glu166 | Hydrogen bond | 1.83 | ||
| His163 | Hydrogen bond | 2.12 | ||
| Lopinavir | −4.203 | His41 | Pi-Pi Stacking | 5.38 |
| Asn142 | Hydrogen bond | 2.15 | ||
| Gln189 | Hydrogen bond | 2.26 | ||
| Imatinib | −3.820 | His41 | Hydrogen bond | 1.97 |
Fig. 4.
The 2D structures of top 3 lead compounds: (a) 2,5 Dimethyl 1-H-Pyrrole (CID12265), (b) 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide (CID562543), (c) and Benzonitrile m phenethyl (CID141877).
3.3.1. Interaction analysis of lead compounds with SARS-CoV-2 Mpro
Maestro was used to examine the interactive prototype and binding mechanism of these docked lead compounds with the Mpro's crystalline structure. Table 1 shows the comprehensive docking analysis with their interaction pattern. It is reported that the novel series of compound 2,5 dimethyl pyrrole and its derivatives are used as anticancer agents against the MDA-MB-435 cell line of melanoma having good projected oral bioavailability, [77]. Derivatives of this compound showed considerable antiviral activity as HIV fusion inhibitors targeting gp41 [85,86] In present research work, 2,5 dimethylpyrrole had proved an effective lead compound to hinder SARS-CoV-2 main protease as it demonstrated superior binding affinity compared to other molecules.
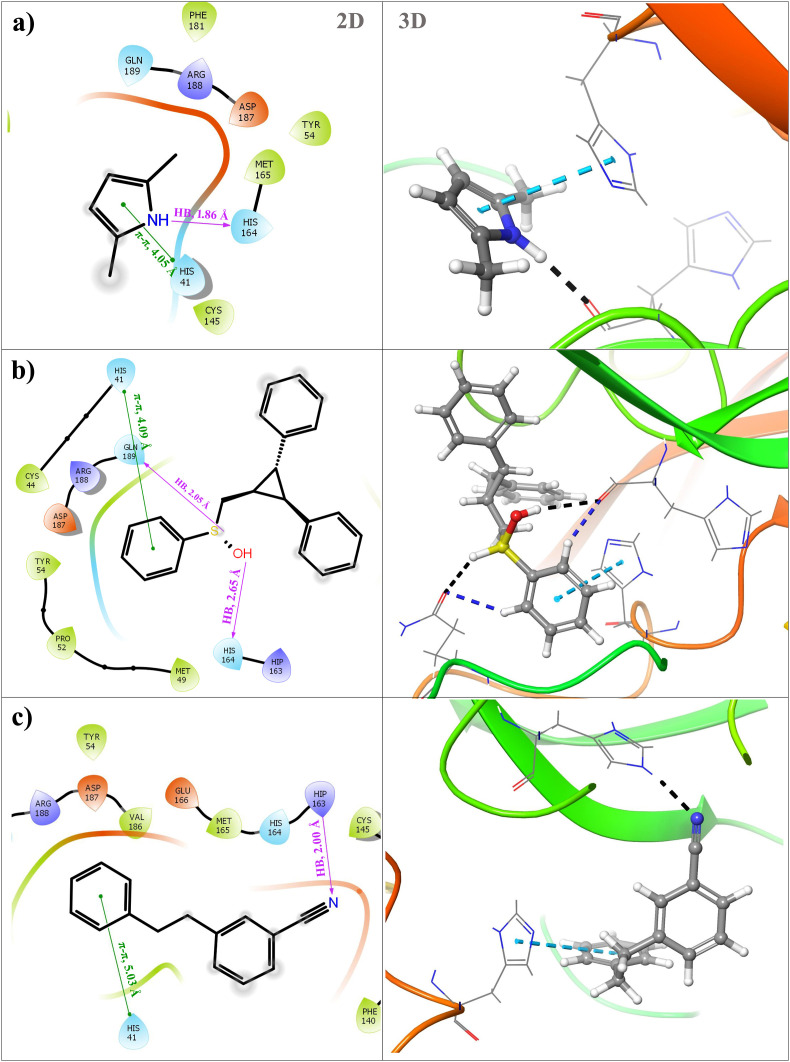
2,5 dimethylpyrrole showed the docking score of - 5.719 kcal/mol which advocates strong binding of this molecule to the inhibitory site of the Mpro crystal structure. The binding pose of the 2,5 dimethylpyrrole in the inhibitory site of Mpro has been exposed in Fig. 5 (a). 2,5 dimethylpyrrole formulate one hydrogen bond and one pi-pi stacking with main protease. The hydrogen bond is a force between two molecules holding two or more molecules together. The basic amino acid histidine (His164), as well as (HIS 41) are involved in the formation of hydrogen bonding and pi-pi stacking with 2,5 dimethylpyrrole. The HIS 164 forms a hydrogen bond with 2,5 dimethylpyrrole with a bond distance of 1.86 Å, whereas HIS 41 forms pi-pi stacking with a bond distance of 4.05 Å. The detailed PDBsum interaction analysis of 2,5 dimethylpyrrole interacted with SARS-CoV2 Mpro via one hydrogen bond along with 17 non-bonded contacts (Supplementary Fig. S1 and Supplementary Table S3).
Fig. 5.
Intermolecular interactions between (a) 2,5 dimethyl 1-H Pyrrole with SARS-CoV-2 Mpro, (b) 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide with SARS-CoV-2 Mpro, (c) Benzonitrile-m Phenethyl with SARS-CoV-2 Mpro.
Fig. 5 (b) demonstrates the intermolecular interactions of second docked lead compound i.e. 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide with the crystalline structure of SARS-CoV-2 Mpro. This compound is from a class of stilbenoids. Stilbenoids are formed when one or more hydroxyl groups are added to a phenyl ring. These molecules are structurally linked since they all have the same stilbene backbone, but the substituent groups on the rings are different with regard to type and positions. Stilbene-based compounds have been intensively investigated in recent years due to their various biological effects on human beings [86,87]. 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide is proven to act as a promising potential therapeutic agent against ovarian cancer stem cell lines [88,89]. The compound 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide forms two hydrogen bonds with acidic amino acid glutamate (Gln189) and basic amino acid histidine (His141) with bond distances of 2.05 Å and 2.65 Å respectively. With histidine (His141) and Glutamate (Gln189) this compound also formed aromatic hydrogen bonds at distances of 2.21 Å and 2.69 Å. Likewise, the pi-pi stacking was detected with interacting amino acid His41 with a bond distance of 4.09 Å. The docking score of 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide is −5.69 kcal which recommends that this molecule is also capable of obstructing SARS-CoV-2 Mpro. A study also revealed a similar kind of interaction between Favipiravir and protease enzymes. It showed interaction with HIS 164 and HIS 41. In our docking study also similar interactions were observed through binding energy. The detailed PDBsum interaction analysis of 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide with SARS-CoV-2 Mpro shows one hydrogen bond along with 84 non-bonded contacts (Supplementary Fig. S2 and Supplementary Table S4).
Fig. 5 (c) illustrates the intermolecular binding interactions of the third top docked compound i.e. Benzonitrile m phenethyl with the crystalline structure of Mpro. In a dose-dependent manner, derivatives of benzonitrile m phenethyl showed a considerable effect on duplication of zika virus ribonucleic acid and expression of virus protein at small micromolar concentrations [90]. The derivatives of this compound also combat the rhinovirus and enteroviruses [91]. Benzonitrile m phenethyl formed a hydrogen bond with a bond distance of 2.00 Å and pi-pi stacking with a bond distance of 5.03 Å. Amino acids which are responsible for interactions are HIS163 and HIS41 respectively. The detailed PDBsum interaction analysis of Benzonitrile m phenethyl with SARS-CoV-2 Mpro shows 37 non-bonded contacts (Supplementary Fig. S3 and Supplementary Table S5).
In the present docking research, it has been discovered that the diamino-monocarboxylic amino acid histidine (His164) and (His41) is a critical residue for holding and building a firm complex of testified lead compounds by generating hydrogen bonds and pi-pi stacking. In addition to that, polar charged acidic amino acid i.e. glutamic acid (Gln189) has also played an essential role in the formation of a stable complex.
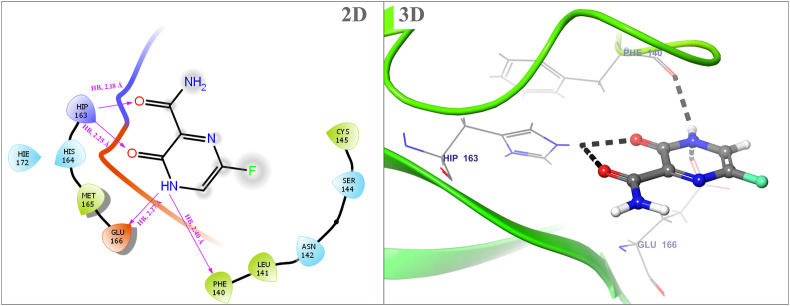
Fig. 6 demonstrates the intermolecular interactions of the top FDA-approved drug i.e. Favipiravir with the crystalline structure of SARS-CoV-2 Mpro. Favipiravir (6-fluoro-3-hydroxy-2-pyrazinecarboxamide), also known as T-705, was developed as a structural analog of 3-hydroxy-2-pyrazinecarboxamide (T-1105). Favipiravir selectively inhibits the RNA-dependent RNA polymerase (RdRp) of RNA viruses and induces lethal RNA transversion mutations, with the help of the production of a nonviable virus phenotype [92]. Because of its distinct antiviral properties, Favipiravir could be a promising treatment option for RNA viral infections that are currently untreatable. In current in silico docking analysis, Favipiravir forms four hydrogen bonds with SARS-CoV-2 Mpro with bond distances 2.27 Å, 2.40 Å, 2.18 Å, and 2.25 Å. Amino acids which are crucial for these bond formation are Glutamate (Glu166), Phenylalanine (Phe140), and Histidine (His163). The Co-crystal ligand i.e., N3 gives −5.444 kcal/mol binding affinity towards SARS-CoV-2 Mpro. Three hydrogen bonds formed between N3 and Mpro. The residue Glu189 forms two hydrogen bonds and Phe140 is involved in one hydrogen bond with Mpro at bond distances of 2.04 Å, 1.93 Å, and 1,72 Å, respectively. It was observed that the selected lead compounds form a conformation very similar to co-crystal ligand i.e. N3 within the binding pocket of SARS-CoV-2 Mpro.
Fig. 6.
Intermolecular interactions between Favipiravir and SARS-CoV-2 Mpro.
The data regarding the biological activity of some selected FDA-approved drugs with SARS-CoV-2 Mpro has been collected and correlated with our predicted docking score to show the consistency between the calculated binding score and the reported target activities. The activities of Remdesivir (IC50 = 10.11 μM/L, pIC50 = 4.995 μM/L) [93,94], Ritonavir: (IC50 = 13.7 μM/L, pIC50 = 4.863 μM/L) [95]; Favipiravir (IC50 = 9.60, μM/L, pIC50 = 5.017, μM/L) [96], and Imatinib: (IC50 = 3.24, μM/, pIC50 = 5.489 μM/L) [97] were compared with the Glide docking score. The resulted coefficient correlation between docking score and biological activity of these drugs has been obtained. (R2 = 0.81) (Supplementary Fig. S4) It suggests a significant predictive ability of the Glide docking technique used in this study.
3.4. Calculation of RMSD and RMSF
The rigid crystal conformation of SARS-CoV-2 Mpro was further utilized for molecular dynamic simulation. The behavior of the receptor-ligand complex was studied by means of molecular dynamics simulation to interrogate the consistency of linked conformation upon binding of lead molecules inside the main protease cavity. The values of the Root Mean Square Fluctuation and Root Mean Square Deviation of the C-α atoms in the main protease evaluation were utilized to determine thermodynamic complex stability during a 100-ns timeframe. Additionally, using the Simulation Event Analysis (SEA) panel of Schrodinger, 1000 conformational structures captured during the molecular dynamics simulation were collected and structurally aligned on the crystal structure of SARS-CoV-2 Mpro which was initially used in ".dat" format.
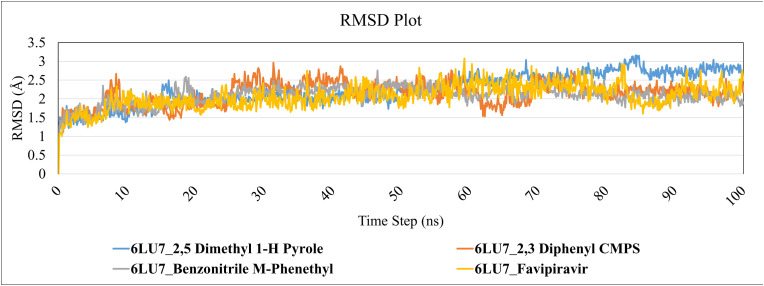
RMSD of the protein-ligand complex was observed to be in the range between 1 and 3 Å which is perfectly acceptable. If the RMSD value is greater than 3 Å, the protein structure has undergone a significant conformational shift. The simulation was performed on Desmond systems up to 100 ns individually to explore the structural stability and variations of protein-ligand complexes. In this investigation, 100 ns simulation was performed, which is enough for the Cα atoms of SARS-CoV-2 main protease to rearrange in complexes with selected lead compounds. The RMSD plot (Fig. 7 ) reveals that these three lead molecules i.e. 2,5 dimethylpyrrole, 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide, and Benzonitrile m phenethyl are in the acceptable range of RMSD. The obtained averaged RMSD value is 2.16 Å, specifying that all the testified lead molecules are bound firmly inside the Mpro cavity. After examining the RMSD graph (Fig. 7) all the complexes reached the equilibrium from 30 ns to 100 ns in the RMSD range of 2.09 Å to 2.26 Å resulting from the averaged RMSD value of 2.16 Å.
Fig. 7.
Root Mean Square Deviation (RMSD) Plot of SARS-CoV-2 Mpro structure (C-α atoms) when complexed with lead compounds during 100ns MD simulation.
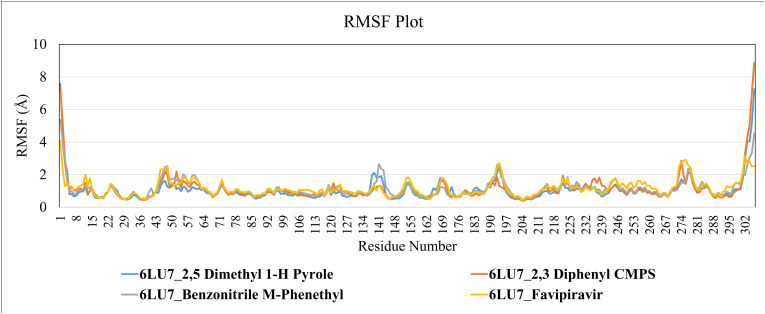
RMSF values for C-α atoms of all the amino acid residues were calculated to scrutinize the binding affinity of 2,5 dimethyl 1 H pyrrole (CID12265), 2,3-Diphenyl cyclopropyl methyl phenyl sulphoxide (CID562543) and Benzonitrile m phenethyl (CID141877) with SARS-CoV-2 Mpro, based on 100 ns MD Simulation data. The average RMSFs measured for Mpro upon binding of lead compounds is 1.11 Å. Since all of the residues oscillated in the RMSF range of 1.0 Å to 3.0 Å (Fig. 8 ), the SARS-CoV-2 Mpro exhibited the least fluctuation and relative secondary conformational stability when bound to the reported lead compounds. MD trials resulted, with lead 2,5 dimethylpyrrole, 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide and Benzonitrile m phenethyl have proven to be promising lead compounds for novel class drug design and development against SARS-CoV-2 Mpro.
Fig. 8.
Root Mean Square Fluctuation (RMSF) Plot of SARS-CoV-2 Mpro structure (C-α atoms) when complexed with lead compounds during 100ns MD simulation.
3.5. Hydrogen bonds
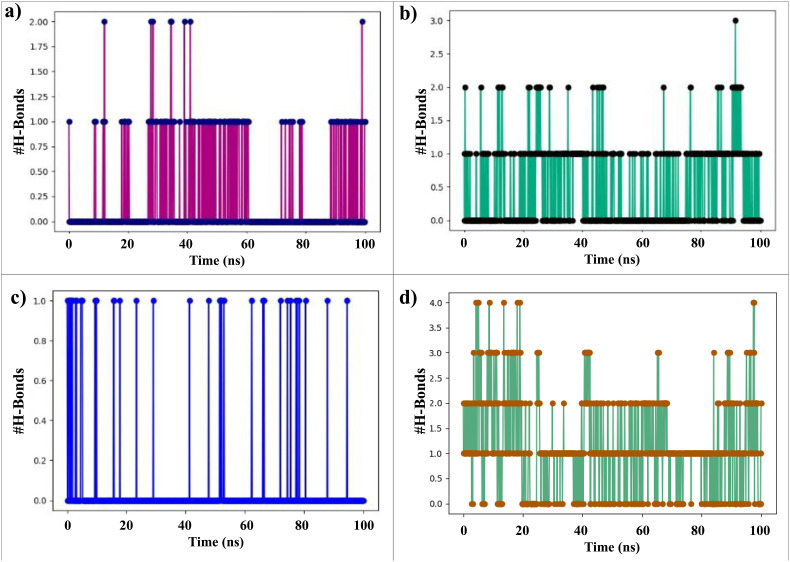
Drugs' binding affinity to the catalytic position of the desired protein relies heavily on hydrogen bonding. The number of hydrogen bonds between protein-ligand complexes has a major role to maintain the stability of the ligand within the binding cavity of the receptor. Hence, the number of hydrogen bonds formed by the selected docked compounds i.e. 2,5 dimethylpyrrole, 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide, and Benzonitrile m phenethyl with the Mpro was calculated for complex systems by examining conformations after every 100 ps during 100ns of MD simulation. Lead compounds formed a maximum of three hydrogen bonds until 100ns simulation time.
The hydrogen bond profile of SARS-CoV-2 Mpro-Benzonitrile-m Phenethyl complex indicates 1–2 hydrogen bonds formed between 100ns MD simulations (Fig. 9 (a)). In Fig. 9 (b), the SARS-CoV-2 Mpro-2,3 diphenyl cyclopropyl methyl phenyl sulphoxide complex formed 1–2 hydrogen bonds during 100ns MD simulations, also at 93ns, there are 3 hydrogen bonds formed between the 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide and SARS-CoV-2 Mpro. Fig. 9 (c) shows only one hydrogen bond formation between 2,5 dimethyl 1-H Pyrrole and SARS-CoV-2 Mpro as the molecular structure of 2,5 dimethyl 1-H Pyrrole didn't have any branched functional group over the pyrrole ring. Therefore, these compounds improve the strength and stability of the ligands–receptor interaction considerably. Fig. 9 (d) shows the hydrogen bond profile of the SARS-CoV-2 Mpro-Favipiravir complex. It formed 4 hydrogen bonds initially from 0ns to 20ns and thereafter 2 hydrogen bonds formation occurred up to 70ns. During the 70s to 90ns, a single hydrogen bond between protein and ligand complex was formed. From 90ns to 100ns again 2 hydrogen bonds are formed during MD simulations. The consistency of H-bonds was not observed during MD simulation.
Fig. 9.
Intermolecular hydrogen bond profile between lead compounds with SARS-CoV-2 Mpro during 100ns MD simulation. (a) SARS-CoV-2 Mpro-Benzonitrile-m Phenethyl complex, (b) SARS-CoV-2 Mpro-2,3 diphenyl cyclopropyl methyl phenyl sulphoxide complex, (c) SARS-CoV-2 Mpro-2,5 dimethyl 1-H Pyrrole complex, (d) SARS-CoV-2 Mpro-Favipiravir Pyrrole complex.
3.6. Prime MM/GBSA energies of lead compounds with SARS-CoV-2 Mpro
Determination of MM/GBSA is the most appropriate approach for computing free binding energies (ΔG Bind), which articulates the outcomes in terms of hydrophobic, van Der Waals forces, and solvation components. The screened ligands (2,5 dimethylpyrrole, 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide, and Benzonitrile m phenethyl) were exposed to an ensemble-averaged Prime MM/GBSA system for an extensive period of MD simulation. Table 3 expresses the computed binding free energies of three complexes utilizing ensemble-averaged MM/GBSA. The top docked receptor-ligand complex's stability is directly proportional to the negative calculated values of binding free energies [98].
Table 3.
Binding free energies between SARS-CoV-2 Mpro and lead compounds using Prime.
| Complex | ΔG Bind | ΔG Bind Coulomb | ΔG Bind Covalent | ΔG Bind Hbond | ΔG Bind Lipo | ΔG Bind vdW | Receptor Energy |
|---|---|---|---|---|---|---|---|
| 6LU7_2,3 diphenyl cyclopropyl methyl phenyl sulphoxide | −64.377 ±5.24 |
−9.722 ±2.15 |
1.630 ±0.54 |
−0.111 ±0.10 |
−34.481 ±2.84 |
−35.821 ±1.93 |
−9945.677 ±56.53 |
| 6LU7_Benzonitrile M Phenethyl | −31.254 ±3.57 |
−4.585 ±2.69 |
0.715 ±0.56 |
−0.174 ±0.06 |
−16.55 ±3.335 |
−21.183 ±3.68 |
−9970.835 ±55.68 |
| 6LU7_2_5-Dimethyl Pyrrole | −16.900 ±3.848 |
−4.115 ±2.31 |
0.130 ±0.11 |
−0.158 ±0.07 |
−8.69 ±2.364 |
−10.808 ±2.54 |
−9909.906 ±63.84 |
The maximum negative binding free energy obtained from the data is characterized to have the value of −64.377 kcal/mol, which deliberates the binding stability of the 2,3 diphenyl cyclopropyl methyl phenyl sulphoxide to be higher towards SARS-CoV-2 Mpro. However, other compounds i.e. Benzonitrile m phenethyl and 2,5 dimethylpyrrole also expressed favorable binding free energy with SARS-CoV-2 Mpro. These MM/GBSA conclusions indicate that the obtained lead molecules are capable of forming a stable complex with SARS-CoV-2 Mpro using the prime MM/GBSA technique and that all of the predictions of the energies produced by the prime module MM/GBSA are thermodynamically stable.
3.7. ADME profile of the top 10 compounds
The pharmacokinetics of all lead compounds were studied, as well as the ADME features, and the rules of five (RO5) of future drug molecules stated by Lipinski were justified with the QikProp application of Schrodinger software. All of the lead compounds chosen have strong pharmacokinetic characteristics and maintain the RO5. This is significant while assessing the efficacy of the lead molecule to be drug molecules. The parameters such as drug-likeness, dipole moment, molecular weight, hydrogen bond donor, acceptor traits, solvent accessible surface area, octanol/water coefficient, water-solubility, number of probable metabolic reactions, binding affinity to serum albumin, coefficient of brain/blood barrier and oral absorption of the drug molecule. The toxicity properties of the drug with good ADME properties were used to determine the adverse effect on people or animals [44]. The selected lead compounds were proven to have no or little toxicity after toxicity testing (Table 4 ). Also, ADME profile ADME profiles have been provided for all the compounds obtained from B. variegata in Supplementary Table S6. The test metabolites utilized in this research are expected to have a significant solubility and intestinal permeability, allowing them to penetrate the bloodstream or the intestinal wall for further action [99]. The lead compounds showed acute toxicity (LD50) values ranging from 670 mg/kg to 2500 mg/kg; with toxicity classifications of 4 and 5 classes (Supplementary Table S7). Toxicity classifications are determined using a globally harmonized system of chemical classification and labeling (GHS). The lead molecules have minimum toxicity profiles, as predicted by these studies.
Table 4.
The ADME profile of the top 10 screened compounds from in silico study.
| Compound Name | Mol MW | QP log S | QPlog HERG | QP logBB | QP logKp | Human Oral Absorption | PSA | Rule of Five | Rule of Three |
|---|---|---|---|---|---|---|---|---|---|
| 2_3-Diphenyl-cyclopropyl_methyl_phynyl_sulphoxide | 334.475 | −6.383 | −7.067 | −0.098 | −0.015 | 1 | 22.198 | 1 | 1 |
| 2_5-Dimethyl_1_H_Pyrole | 95.144 | −1.294 | −3.272 | 0.409 | −1.531 | 3 | 13.376 | 0 | 0 |
| Benzonitrile m-phenethyl_ | 207.274 | −4.377 | −5.625 | −0.338 | −1.199 | 3 | 25.794 | 0 | 0 |
| 1_2_Benzenedicarboxylic_acid_butyl_2_methyl_propyl_ester_ | 278.347 | −3.72 | −4.639 | −0.509 | −1.627 | 3 | 59.025 | 0 | 0 |
| 1_2_propandiol_3_Benzyloxy-1_2-diacetyl | 266.293 | −3.48 | −5.666 | −1.05 | −2.278 | 3 | 82.474 | 0 | 0 |
| 1_5_Anhydroglucitol | 452.885 | −9.082 | −3.767 | −0.665 | 0.425 | 1 | 29.45 | 0 | 1 |
| 1-Tetradecene | 198.391 | −8.903 | −4.774 | 1.39 | 3.531 | 1 | 0 | 1 | 1 |
| 2-_Aziridinylethil_amine | 86.136 | 0.773 | −3.361 | 0.19 | −4.713 | 3 | 36.217 | 0 | 0 |
| 2_2_Dimethoxybutane | 118.175 | −1.593 | −2.49 | 0.168 | −1.581 | 3 | 14.033 | 0 | 0 |
| 2_3-Butanediol | 90.122 | 0.357 | −2.401 | −0.198 | −2.669 | 3 | 40.098 | 0 | 0 |
4. Conclusion
There is currently no effective medication against SARS-CoV-2. Because of their severe side effects, synthetic medications are not always appropriate for human health. As a result, there is an urgent need for the discovery of a non-toxic, novel, and cost-effective treatment to battle this fatal virus is crucial. Drugs from natural origin have the potential to fight in this situation; as of their minimum toxicity. Further, they have a vast array of applications in human health and disease without creating any negative adverse effects.
In the present situation, inventors from all around the world have been researching hard to identify prospective lead molecules from natural sources along with medicinal plants and natural resources which are effective against SARS-CoV-2 [100,101]. Till now no fixed treatment for SARS-CoV-2 is followed; and all parts of our experimental plant B. variegata were known to be used in folklore medicine for treating various disorders like bronchitis, leprosy, and tumors [102]. B. variegata extracts on oral administration with a specific time interval with tumor inoculation exhibited a significant decrease in the tumor volume [103]. B. variegata methanolic extract was also effective against rotavirus-induced gastroenteritis which proved that the plant is not having any toxic side effects. This phytochemical constituent has a number of well-established pharmacological actions that could help with the overall treatment of COVID-19, with antiviral activities being the most important [104]. As a result, the present research aims to use computational drug design methodologies to discover novel natural Mpro inhibitors in order to combat the existing disastrous scenario caused by SARS-CoV-2.
In the present research, it has been hypothesized that phytoconstituents from the B. variegata plant could be capable of inhibiting the main protease of SARS-CoV-2. For the present investigation, a molecular docking technique was done against the main protease of SARS-CoV-2. Binding affinity and free energies between lead compounds and the Mpro enzyme recommended that all three compounds showed good interaction with the binding site of this Mpro. The energetic behavior of SARS-CoV-2 Mpro on phytocompounds from B. variegata binding was examined using comparative molecular dynamic simulations. It was observed that the values obtained from the root mean square deviation for the SARS-CoV-2 Mpro C-α atoms are in 2.5 Å suggests that the reported lead compounds 2,5 dimethylpyrrole,2,3 diphenyl cyclopropyl methyl phenyl sulphoxide and Benzonitrile m phenethyl could form a strong complex with SARS-CoV-2 Mpro, as per molecular dynamics calculations. Furthermore, the observed RMSF values for binding site residues indicate that when lead compounds bound to the binding pocket SARS-CoV-2 Mpro, the least fluctuations occurred. The calculated Prime MM/GBSA binding free energies are found to be thermodynamically favorable. The binding free energy of the complexes was strongly influenced by His, Gln, and Glu residues. These compounds possess comparatively good ADMET parameters therefore further could be considered for formulation and clinical trials. Thus it could be concluded that the top three compounds 2,5 dimethylpyrrole,2,3 diphenyl cyclopropyl methyl phenyl sulphoxide and Benzonitrile m phenethyl have the potential to prevent the activity of SARS-CoV-2 Mpro and result in obstructing the duplication of SARS-CoV-2 into the human host cytosol. Further studies could be helpful in finding a potent molecule or formulation that could be promising to treat the disease with less adverse effects.
Funding
No external funding is received for this research.
Statement of availability of data
All docking structures are accessible upon demand from the corresponding author.
Declaration of competing interest
There is no conflict of interest declared by all authors.
Acknowledgments
The authors are thankful to the School of Pharmacy, MIT World Peace University for providing research lab facilities and Dr. D. Y. Patil Biotechnology and Bioinformatics Institute, Dr. D. Y. Patil Vidyapeeth, Pune, for allowing to work at Bioinformatics Research Laboratory. The authors also acknowledge the DST-SERB, Govt. of India, New Delhi, (File Number: YSS/2015/002035) for Optimized Supercomputer facility for dynamics calculations. Mr. Kiran Bharat Lokhande acknowledges the ICMR (Indian Council of Medical Research), New Delhi, India for Senior Research Fellowship (Project ID: 2019–3458; file: ISRM/11(54)/2019). The authors also would like to acknowledge the support of Schrodinger, and the team for providing an Evaluation License.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105679.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gostin L.O. COVID-19 reveals urgent need to strengthen the world health organization. JAMA. 2020;323(23):2361–2362. doi: 10.1001/jama.2020.8486. [DOI] [PubMed] [Google Scholar]
- 2.Pettersson H. 2020. Tracking Covid-19's Global Spread. [Google Scholar]
- 3.https://covid19.who.int/accesed on 20th Jan 2022.
- 4.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K.…Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakib M., Nishat A.A., Islam M.T., Raihan Uddin M.A., Iqbal M.S., Bin Hossen F.F., Ahmed M.I., Bashir M.S., Hossain T., Tohura U.S., Saif S.I., Jui N.R., Alam M., Islam M.A., Hasan M.M., Sufian M.A., Ali M.A., Islam R., Hossain M.A., Halim M.A. Computational screening of 645 antiviral peptides against the receptor-binding domain of the spike protein in SARS-CoV-2. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nag A., Paul S., Banerjee R., Kundu R. In silico study of some selective phytochemicals against a hypothetical SARS-CoV-2 spike RBD using molecular docking tools. Comput. Biol. Med. 2021;137 doi: 10.1016/j.compbiomed.2021.104818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S., Amahong K., Zhang C., Li F., Gao J., Qiu Y., Zhu F. RNA-RNA interactions between SARS-CoV-2 and host benefit viral development and evolution during COVID-19 infection. Briefings Bioinf. 2022;23(1):bbab397. doi: 10.1093/bib/bbab397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S., Amahong K., Sun X., Lian X., Liu J., Sun H., Lou Y., Zhu F., Qiu Y. The miRNA: a small but powerful RNA for COVID-19. Briefings Bioinf. 2021;22(2):1137–1149. doi: 10.1093/bib/bbab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calina D., Docea A.O., Petrakis D., Egorov A.M., Ishmukhametov A.A., Gabibov A.G., Shtilman M.I., Kostoff R., Carvalho F., Vinceti M., Spandidos D.A., Tsatsakis A. Towards effective COVID-19 vaccines: updates, perspectives and challenges (Review) Int. J. Mol. Med. 2020;46(1):3–16. doi: 10.3892/ijmm.2020.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abduljalil J.M., Abduljalil B.M. Epidemiology, genome, and clinical features of the pandemic SARS-CoV-2: a recent view. N. Microb. N. Infect. 2020;35 doi: 10.1016/j.nmni.2020.100672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lokhande K.B., Banerjee T., Swamy K.V., Ghosh P., Deshpande M. An in silico scientific basis for LL-37 as a therapeutic for Covid-19. Proteins. 2021 doi: 10.1002/prot.26198. 10.1002/prot.26198. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhama K., Karthik K., Khandia R., Munjal A., Tiwari R., Rana R., Khurana S.K., Ullah Sana, Khan R.U., Alagawany M., Farag M.R., Dadar M., Joshi S.K. Medicinal and therapeutic potential of herbs and plant metabolites/extracts countering viral pathogens - current knowledge and future prospects. Curr. Drug Metabol. 2018;19(3):236–263. doi: 10.2174/1389200219666180129145252. [DOI] [PubMed] [Google Scholar]
- 16.Ministry of Ayush . 2020. Ayurveda's Immunity Boosting Measures for Self Care during COVID 19 Crisis. [WWW Document]. https://www. ayush.gov.in/docs/123.pdf. [Google Scholar]
- 17.Llivisaca-Contreras S.A., Naranjo-Morán J., Pino-Acosta A., Pieters L., Vanden Berghe W., Manzano P., Vargas-Pérez J., León-Tamariz F., Cevallos-Cevallos J.M. Plants and natural products with activity against various types of coronaviruses: a review with focus on SARS-CoV-2. Molecules. 2021;26(13):4099. doi: 10.3390/molecules26134099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manoj A.F., Kumar A., Yunus S.M. Screening of hepatoprotective activity of ethanolic extractof stem bark of Bauhinia variegatain rats. Int. J. Pharm. Pharmaceut. Sci. 2013;5(2):624–628. [Google Scholar]
- 19.Sharma Kartik, Kumar Vikas, Kumar Satish, Sharma Rakesh, Mehta Chandra. Bauhinia variegata: a comprehensive review on bioactive compounds, health benefits and utilization. Orient. Pharm. Exp. Med. 2020 doi: 10.1007/s13596-020-00472-4. [DOI] [Google Scholar]
- 20.Shaheen M., El-Gamal M., Mousa A., Mostafa S., El-Esnawy N. Antiviral activity of Bauhinia variegata extracts against rotavirus in vitro. Curr. Sci. Int. 2014;3(3):172–178. [Google Scholar]
- 21.Sharma N., Sharma A., Bhatia G., Landi M., Brestic M., Singh B., Singh J., Kaur S., Bhardwaj R. Isolation of phytochemicals from Bauhinia variegata L. Bark and their in vitro antioxidant and cytotoxic potential. Antioxidants. 2019;8(10):492. doi: 10.3390/antiox8100492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh R., Bhardwaj V.K., Sharma J., Purohit R., Kumar S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. Tradit. Compl Med. 2022;12(1):35–43. doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346 doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dynam. 2021;39(10):3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R., Bhardwaj V.K., Purohit R. Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: an in-silico approach. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R., Bhardwaj V.K., Sharma J., Kumar D., Purohit R. Identification of potential plant bioactive as SARS-CoV-2 Spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Bioactive molecules of tea as potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2. Front. Med. 2021;8 doi: 10.3389/fmed.2021.684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talukdar R., Padhi S., Rai A.K., Masi M., Evidente A., Jha D.K., Cimmino A., Tayung K. Isolation and characterization of an endophytic fungus colletotrichum coccodes producing tyrosol from houttuynia cordata thunb. Using ITS2 RNA secondary structure and molecular docking study. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.650247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G., Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Med. Chem. 2016;8(14):1707–1710. doi: 10.4155/fmc-2016-0143. [DOI] [PubMed] [Google Scholar]
- 32.Ingle K.P., Deshmukh A.G., Padole D.A., Dudhare M.S., Moharil M.P., Khelurkar V.C. Phytochemicals: extraction methods, identification and detection of bioactive compounds from plant extracts. J. Pharmacogn. Phytochem. 2017;6(1):32–36. [Google Scholar]
- 33.Azwanida N.N. A review on the extraction methods use in medicinal plants, principle, strength and limitation. Med. Aromatic Plants. 2015;4(196) 2167-0412. [Google Scholar]
- 34.Fu J., Zhang Y., Wang Y., Zhang H., Liu J., Tang J., Yang Q., Sun H., Qiu W., Ma Y., Li Z., Zheng M., Zhu F. Optimization of metabolomic data processing using NOREVA. Nat. Protoc. 2022;17(1) doi: 10.1038/s41596-021-00636-9. 129–151. [DOI] [PubMed] [Google Scholar]
- 35.Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017;45(W1):W162–W170. doi: 10.1093/nar/gkx449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Q., Wang Y., Zhang Y., Li F., Xia W., Zhou Y., Qiu Y., Li H., Zhu F. NOREVA: enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Res. 2020;48(W1):W436–W448. doi: 10.1093/nar/gkaa258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang J., Fu J., Wang Y., Li B., Li Y., Yang Q., Cui X., Hong J., Li X., Chen Y., Xue W., Zhu F. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Briefings Bioinf. 2020;21(2):621–636. doi: 10.1093/bib/bby127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Q., Li B., Chen S., Tang J., Li Y., Li Y., Zhang S., Shi C., Zhang Y., Mou M., Xue W., Zhu F. MMEASE: online meta-analysis of metabolomic data by enhanced metabolite annotation, marker selection and enrichment analysis. J. Proteonomics. 2021;232 doi: 10.1016/j.jprot.2020.104023. [DOI] [PubMed] [Google Scholar]
- 39.Satapute P., Paidi M.K., Kurjogi M., Jogaiah S. Physiological adaptation and spectral annotation of Arsenic and Cadmium heavy metal-resistant and susceptible strain Pseudomonas taiwanensis. Environ. Pollut. 2019;251:555–563. doi: 10.1016/j.envpol.2019.05.054. 1987. [DOI] [PubMed] [Google Scholar]
- 40.Fan S., Chang J., Zong Y., Hu G., Jia J. GC-MS analysis of the composition of the essential oil from dendranthema indicum var. Aromaticum using three extraction methods and two columns. Molecules. 2018;23(3):576. doi: 10.3390/molecules23030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrödinger Release 2021-4. LigPrep, Schrödinger, LLC; New York, NY: 2021. [Google Scholar]
- 42.De Vivo M., Masetti M., Bottegoni G., Cavalli A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 2016;59(9):4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- 43.Jayaram B., Singh T., Mukherjee G., Mathur A., Shekhar S., Shekhar V. Sanjeevini: a freely accessible web-server for target directed lead molecule discovery. BMC Bioinf. 2012;13(Suppl 17):S7. doi: 10.1186/1471-2105-13-S17-S7. Suppl 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Niveshika, Singh S., Verma E., et al. In silico molecular docking analysis of cancer biomarkers with GC/MS identified compounds of Scytonema sp. Netw. Model. Anal. Inf. Bioinf. 2020;9:30. doi: 10.1007/s13721-020-00235-w. [DOI] [Google Scholar]
- 46.Banerjee Priyanka, et al. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257–W263. doi: 10.1093/nar/gky318. W1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabie A.M. Discovery of Taroxaz-104: the first potent antidote of SARS-CoV-2 VOC-202012/01 strain. J. Mol. Struct. 2021;1246 doi: 10.1016/j.molstruc.2021.131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabie A.M. Potent inhibitory activities of the adenosine analogue cordycepin on SARS-CoV-2 replication. ACS Omega. 2022;7(3):2960–2969. doi: 10.1021/acsomega.1c05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabie A.M. Two antioxidant 2, 5-disubstituted-1, 3, 4-oxadiazoles (CoViTris2020 and ChloViD2020): successful repurposing against COVID-19 as the first potent multitarget anti-SARS-CoV-2 drugs. New J. Chem. 2021;45(2):761–771. doi: 10.1039/D0NJ03708G. [DOI] [Google Scholar]
- 50.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X.…Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 51.Lokhande K.B., Doiphode S., Vyas R., Swamy K.V. Molecular docking and simulation studies on SARS-CoV-2 Mpro reveals Mitoxantrone, Leucovorin, Birinapant, and Dynasore as potent drugs against COVID-19. J. Biomol. Struct. Dynam. 2021;39(18):7294–7305. doi: 10.1080/07391102.2020.1805019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lokhande K.B., Pawar S.V., Madkaiker S., Nawani N., Venkateswara S.K., Ghosh P. High throughput virtual screening and molecular dynamics simulation analysis of phytomolecules against BfmR of Acinetobacter baumannii: anti-virulent drug development campaign. J. Biomol. Struct. Dynam. 2022:1–15. doi: 10.1080/07391102.2022.2038271. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y.W., Yiu C.B., Wong K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research. 2020;9:129. doi: 10.12688/f1000research.22457.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kollar J., Frecer V. How accurate is the description of ligand-protein interactions by a hybrid QM/MM approach? J. Mol. Model. 2017;24(1):11. doi: 10.1007/s00894-017-3537-z. [DOI] [PubMed] [Google Scholar]
- 55.Schrödinger Release 2020–1: Glide. Schrödinger, LLC; New York, NY: 2020. [Google Scholar]
- 56.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 57.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 58.Schrödinger Release 2020–1: Protein Preparation Wizard. Schrödinger, LLC; New York, NY: 2020. Epik, Schrödinger, LLC, New York, NY, 2020; Impact, Schrödinger, LLC, New York, NY, 2020; Prime. [Google Scholar]
- 59.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 60.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: structural summaries of PDB entries. Protein Sci. : Publ. Protein Soc. 2018;27(1):129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabie A.M. CoViTris2020 and ChloViD2020: a striking new hope in COVID-19 therapy. Mol. Divers. 2021;25(3):1839–1854. doi: 10.1007/s11030-020-10169-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Rabie A.M. Cyanorona-20: the first potent anti-SARS-CoV-2 agent. Int. Immunopharm. 2021;98 doi: 10.1016/j.intimp.2021.107831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rabie A.M. 2021. Discovery of (E)-N-(4-cyanobenzylidene)- 6-fluoro- 3-hydroxypyrazine-2 -carboxamide (Cyanorona-20): the First Potent and Specific Anti-COVID-19 Drug. Chemicke Zvesti; pp. 1–17. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Rabie A.M. Teriflunomide: a possible effective drug for the comprehensive treatment of COVID-19. Curr. Res. Pharmacol. Drug. Discov. 2021;2 doi: 10.1016/j.crphar.2021.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A.…Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lokhande K.B., Apte G.R., Shrivastava A., Singh A., Pal J.K., Venkateswara Swamy K., Gupta R.K. Sensing the interactions between carbohydrate-binding agents and N-linked glycans of SARS-CoV-2 spike glycoprotein using molecular docking and simulation studies. J. Biomol. Struct. Dynam. 2020:1–19. doi: 10.1080/07391102.2020.1851303. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gupta R.K., Apte G.R., Lokhande K.B., Mishra S., Pal J.K. Carbohydrate-binding agents: potential of repurposing for COVID-19 therapy. Curr. Protein Pept. Sci. 2020;21(11):1085–1096. doi: 10.2174/1389203721666200918153717. [DOI] [PubMed] [Google Scholar]
- 68.Jacobson M.P., Pincus D.L., Rapp C.S., Day T.J., Honig B., Shaw D.E., Friesner R.A. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55(2):351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 69.Schrödinger Release 2020–1: Prime. Schrödinger, LLC; New York, NY: 2020. [Google Scholar]
- 70.Fu T., Zheng G., Tu G., Yang F., Chen Y., Yao X., Li X., Xue W., Zhu F. Exploring the binding mechanism of metabotropic glutamate receptor 5 negative allosteric modulators in clinical trials by molecular dynamics simulations. ACS Chem. Neurosci. 2018;9(6):1492–1502. doi: 10.1021/acschemneuro.8b00059. [DOI] [PubMed] [Google Scholar]
- 71.Shen L., Liu F., Huang L., Liu G., Zhou L., Peng L. VDA-RWLRLS: an anti-SARS-CoV-2 drug prioritizing framework combining an unbalanced bi-random walk and Laplacian regularized least squares. Comput. Biol. Med. 2021;140 doi: 10.1016/j.compbiomed.2021.105119. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xue W., Yang F., Wang P., Zheng G., Chen Y., Yao X., Zhu F. What contributes to serotonin-norepinephrine reuptake inhibitors' dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 2018;9(5):1128–1140. doi: 10.1021/acschemneuro.7b00490. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y., Ying J.B., Hong J.J., Li F.C., Fu T.T., Yang F.Y., Zheng G.X., Yao X.J., Lou Y., Qiu Y., Xue W.W., Zhu F. How does chirality determine the selective inhibition of histone deacetylase 6? A lesson from trichostatin A enantiomers based on molecular dynamics. ACS Chem. Neurosci. 2019;10(5):2467–2480. doi: 10.1021/acschemneuro.8b00729. [DOI] [PubMed] [Google Scholar]
- 74.Hayes J.M., Archontis G. MM-GB (PB) SA calculations of protein-ligand binding free energies. Mol. Dynam. Stud. Synth. Biolog. Macromol. 2012:171–190. [Google Scholar]
- 75.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 76.Li J., Abel R., Zhu K., Cao Y., Zhao S., Friesner R.A. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins. 2011;79(10):2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hao G.F., Zhu X.L., Ji F.Q., Zhang L., Yang G.F., Zhan C.G. Understanding the mechanism of drug resistance due to a codon deletion in protoporphyrinogen oxidase through computational modeling. J. Phys. Chem. B. 2009;113(14):4865–4875. doi: 10.1021/jp807442n. [DOI] [PubMed] [Google Scholar]
- 78.Pan Y., Gao D., Zhan C.G. Modeling the catalysis of anti-cocaine catalytic antibody: competing reaction pathways and free energy barriers. J. Am. Chem. Soc. 2008;130(15):5140–5149. doi: 10.1021/ja077972s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferreira J.C., Fadl S., Villanueva A.J., Rabeh W.M. Catalytic dyad residues His41 and Cys145 impact the catalytic activity and overall conformational fold of the main SARS-CoV-2 protease 3-chymotrypsin-like protease. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.692168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yadav P., Rana M., Chowdhury P. DFT and MD simulation investigation of favipiravir as an emerging antiviral option against viral protease (3CLpro) of SARS-CoV-2. J. Mol. Struct. 2021;1246 doi: 10.1016/j.molstruc.2021.131253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aggarwal B., Verma S.K. Favipiravir may acts as covid-19 main protease pdb id 6lu7 inhibitor: docking analysis. Biointerface Res. Appl. Chem.. 2020:6821–6828. [Google Scholar]
- 83.Narkhede R.R., Cheke R.S., Ambhore J.P., Shinde S.D. The molecular docking study of potential drug candidates showing anti-COVID-19 activity by exploring of therapeutic targets of SARS-CoV-2. Eurasian J. Med. Oncol. 2020;4(3):185–195. [Google Scholar]
- 84.Silva Arouche T.D., Reis A.F., Martins A.Y., S Costa J.F., Carvalho Junior R.N., J C Neto A.M. Interactions between remdesivir, Ribavirin, favipiravir, Galidesivir, hydroxychloroquine and chloroquine with fragment molecular of the COVID-19 main protease with inhibitor N3 complex (PDB ID:6LU7) using molecular docking. J. Nanosci. Nanotechnol. 2020;20(12):7311–7323. doi: 10.1166/jnn.2020.18955. [DOI] [PubMed] [Google Scholar]
- 85.Rasal N.K., Sonawane R.B., Jagtap S.V. Potential 2,4-dimethyl-1H-pyrrole-3-carboxamide bearing benzimidazole template: design, synthesis, in vitro anticancer and in silico ADME study. Bioorg. Chem. 2020;97 doi: 10.1016/j.bioorg.2020.103660. [DOI] [PubMed] [Google Scholar]
- 86.He X.Y., Zou P., Qiu J., Hou L., Jiang S., Liu S., Xie L. Design, synthesis and biological evaluation of 3-substituted 2,5-dimethyl-N-(3-(1H-tetrazol-5-yl)phenyl)pyrroles as novel potential HIV-1 gp41 inhibitors. Bioorg. Med. Chem. 2011;19(22):6726–6734. doi: 10.1016/j.bmc.2011.09.047. [DOI] [PubMed] [Google Scholar]
- 87.Weiskirchen S., Weiskirchen R. Resveratrol: how much wine do you have to drink to stay healthy? Adv. Nutr. 2016;7(4):706–718. doi: 10.3945/an.115.011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mattio L.M., Catinella G., Pinto A., Dallavalle S. Natural and nature-inspired stilbenoids as antiviral agents. Eur. J. Med. Chem. 2020;202 doi: 10.1016/j.ejmech.2020.112541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vermeersch K.A., Wang L., McDonald J.F., Styczynski M.P. Distinct metabolic responses of an ovarian cancer stem cell line. BMC Syst. Biol. 2014;8:134. doi: 10.1186/s12918-014-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y., Zhou R., Quan Y., Chen S., Shi X., Li Y., Cen S. Design, synthesis, and evaluation of novel 4-amino-2-(4-benzylpiperazin-1-yl)methylbenzonitrile compounds as Zika inhibitors. Bioorg. Med. Chem. Lett. 2020;30(4) doi: 10.1016/j.bmcl.2019.126906. [DOI] [PubMed] [Google Scholar]
- 91.Egorova A., Ekins S., Schmidtke M., Makarov V. Back to the future: advances in development of broad-spectrum capsid-binding inhibitors of enteroviruses. Eur. J. Med. Chem. 2019;178:606–622. doi: 10.1016/j.ejmech.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.ElNaggar M.H., Abdelwahab G.M., Kutkat O., GabAllah M., Ali M.A., El-Metwally M., Sayed A.M., Abdelmohsen U.R., Khalil A.T. Aurasperone A inhibits SARS CoV-2 in vitro: an integrated in vitro and in silico study. Mar. Drugs. 2022;20(3):179. doi: 10.3390/md20030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou Y., Zhang Y., Lian X., Li F., Wang C., Zhu F., Qiu Y., Chen Y. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;50(D1):D1398–D1407. doi: 10.1093/nar/gkab953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mahdi M., Mótyán J.A., Szojka Z.I., Golda M., Miczi M., Tőzsér J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2's main protease. Virol. J. 2020;17(1):190. doi: 10.1186/s12985-020-01457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Purwati, Miatmoko A., Nasronudin, Hendrianto E., Karsari D., Dinaryanti A., Ertanti N., Ihsan I.S., Purnama D.S., Asmarawati T.P., Marfiani E., Yulistiani, Rosyid A.N., Wulaningrum P.A., Setiawan H.W., Siswanto I., Tri Puspaningsih N.N. An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0252302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weston S., Coleman C.M., Haupt R., Logue J., Matthews K., Li Y., Reyes H.M., Weiss S.R., Frieman M.B. Broad anti-coronavirus activity of Food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J. Virol. 2020;94(21):e01218–e01220. doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dynam. 2021;39(10):3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Padhi S., Masi M., Chourasia R., Rajashekar Y., Rai A.K., Evidente A. ADMET profile and virtual screening of plant and microbial natural metabolites as SARS-CoV-2 S1 glycoprotein receptor binding domain and main protease inhibitors. Eur. J. Pharmacol. 2021;890 doi: 10.1016/j.ejphar.2020.173648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tahir Ul Qamar M., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharmaceut. Anal. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rakib A., Paul A., Chy M., Sami S.A., Baral S.K., Majumder M., Tareq A.M., Amin M.N., Shahriar A., Uddin M.Z., Dutta M., Tallei T.E., Emran T.B., Simal-Gandara J. Biochemical and computational approach of selected phytocompounds from Tinospora crispa in the management of COVID-19. Molecules. 2020;25(17):3936. doi: 10.3390/molecules25173936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pandey S. Ethno-pharmacological review of Bauhinia variegata: a potential herbal drug. Res. Rev.: J. Herb. Sci.. 2013;2(2):6–11. [Google Scholar]
- 103.Pandey S. In vivo antitumor potential of extracts from different parts of Bauhinia variegata linn. Against b16f10 melanoma tumour model in c57bl/6 mice. Appl. Cancer Res. 2017;37(1):1–14. [Google Scholar]
- 104.Shaheen M., El-Gamal M., Mousa A., Mostafa S., El-Esnawy N. Antiviral activity of Bauhinia variegata extracts against rotavirus in vitro. Curr. Sci. Int. 2014;3(3):172–178. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.