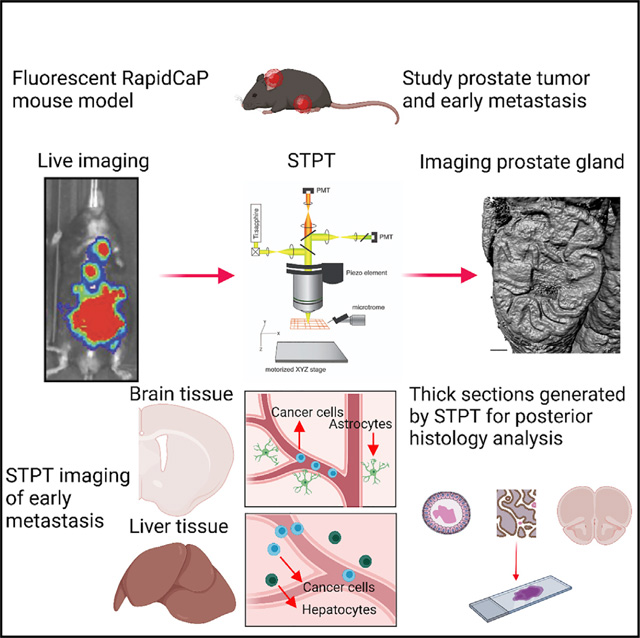
SUMMARY
Early steps of cancer initiation and metastasis, while critical for understanding disease mechanisms, are difficult to visualize and study. Here, we describe an approach to study the processes of initiation, progression, and metastasis of prostate cancer (PC) in a genetically engineered RapidCaP mouse model, which combines whole-organ imaging by serial two-photon tomography (STPT) and post hoc thick-section immunofluorescent (IF) analysis. STPT enables the detection of single tumor-initiating cells within the entire prostate, and consequent IF analysis reveals a transition from normal to transformed epithelial tissue and cell escape from the tumor focus. STPT imaging of the liver and brain reveal the distribution of multiple metastatic foci in the liver and an early-stage metastatic cell invasion in the brain. This imaging and data analysis pipeline can be readily applied to other mouse models of cancer, offering a highly versatile whole-organ platform to study in situ mechanisms of cancer initiation and progression.
In brief
Scientists have long known that tumors are initiated by few cells. The detection of these cells with high resolution is a challenge due to the microscopic dimensions of organs. Taranda et al. use STP tomography combined with traditional histology to describe these events in prostate cancer and its metastasis.
Graphical abstract
INTRODUCTION
Among men in the United States, prostate cancer (PC) is the most commonly diagnosed cancer and the second leading cause of cancer-related deaths (Siegel et al., 2020). Since the disease is clinically identified only when large tumors comprising hundreds of millions of cancer cells are present, studies in faithful animal models applying sensitive whole-organ imaging and tissue analysis methods are needed to understand PC initiation and early progression, including early metastatic spread to distant tissues.
Advances in whole-organ microscopy, such as block-face serial two-photon tomography (STPT) introduced by our laboratory to study brain anatomy and pathology (Hyun et al., 2021; Kim et al., 2017; Mandelbaum et al., 2019; Ragan et al., 2012; Taranda and Turcan, 2021; Turcan et al., 2018) and light-sheet fluorescence microscopy (LSFM) used with chemically cleared tissues (Keller and Dodt, 2012; Susaki et al., 2014), have brought new opportunities for imaging cancer at high spatial resolution across entire organs, promising visualization of early disease stages comprising only a few cancer cells, which would be difficult to detect by traditional tissue-section-based analyses. Recently, LSFM methods have indeed been applied to visualize cancer cells in dissected mouse tissues (Cuccarese et al., 2017) and even at the whole-animal level (Cai et al., 2019; Kubota et al., 2017; Pan et al., 2019; Park et al., 2018). However, the chemical delipidation needed for tissue clearing in LSFM was shown to cause damage to the cellular membrane (Kubota et al., 2017), suggesting that this may, at least to some extent, limit the reliability of the use of the LSFM methods in combination with traditional immunostaining and cytology methods used in the study of cancer pathology. The need for such combined whole-organ imaging and traditional cellular analyses is exemplified in prostate adenocarcinoma, the focus of our study, where cancer cells disrupt the cytoarchitecture of the epithelial gland, including the basal layer, where disseminated invasive cells are found (Shen and Abate-Shen, 2010).
Here, we describe a novel approach to the study of cancer in animal models, one that combines single-cell resolution imaging of entire organs by STPT, computational quantification of putative cancer cell distribution across the imaged organs, and immunostaining of thick tissue sections generated during STPT imaging to validate and further characterize the combination of molecular and morphological pathology. We demonstrate these methods in the study of PC in a fluorescent version of the Rapid-CaP model, a genetically engineered mouse (GEM) model for native metastatic PC, which is based on a focal deletion of Pten and Trp53 genes by lentivirus-based delivery of Cre recombinase directly into the prostate gland (Cho et al., 2014, 2015). We show how our approach addresses one of the main challenges of studying natively growing PC in GEM models—finding the needle in the haystack; while large tumors are easy to detect and analyze, studying early disease evolution requires finding and visualizing very few cells within entire organs. First, we establish the application of STPT to post-mortem high-resolution prostate imaging following in vivo bioluminescence detection of PC in the RapidCaP model. Second, we quantitate the time course of disease progression in the prostate from 1 to 50 days post-initiation. Third, we use prostate tissue sections generated during STPT imaging for immunofluorescent (IF) analysis of cytoskeletal rearrangements during disease progression. And fourth, we use STPT to detect and study micro-metastasis in the liver and the brain, identifying and quantifying the spatial distribution of multiple disease foci.
In summary, the STPT platform, comprising automated organ imaging, 3D organ reconstruction, computational analysis of primary and metastatic cancer cell distribution, and IF analysis of cellular pathological features, provides a high-resolution quantitative assay for the analysis of PC initiation and progression, including mapping of metastatic spread. Importantly, the same method pipeline can be readily applied to other tissues, organs, and cancer mouse models.
RESULTS
PC imaging by STPT: Visualization of transformed epithelial cells in a fluorescent RapidCaP mouse model
STPT is an automated block-face microscopy that operates by iterative steps of two-photon imaging of the top of the tissue (typically at 1 × 1 μm xy resolution) and vibratome-based sectioning and removal of the imaged top 50-μm-thick section to generate a digital serial section dataset that can be readily reconstructed in 3D (Figures 1 and 2A–2C) (Ragan et al., 2012). Importantly, the tissue preparation for STPT imaging includes only fixation by 4% paraformaldehyde (PFA) and embedded in a 6% agar block (Ragan et al., 2012), without the need for disruptive chemical clearing treatments used in LSFM imaging. Therefore, the 50-μm tissue section generated during STPT can be collected and further processed by traditional immuno- or histo-staining for different protein and cellular markers, as done here in determining pathological features of PC cells in the RapidCaP mouse model.
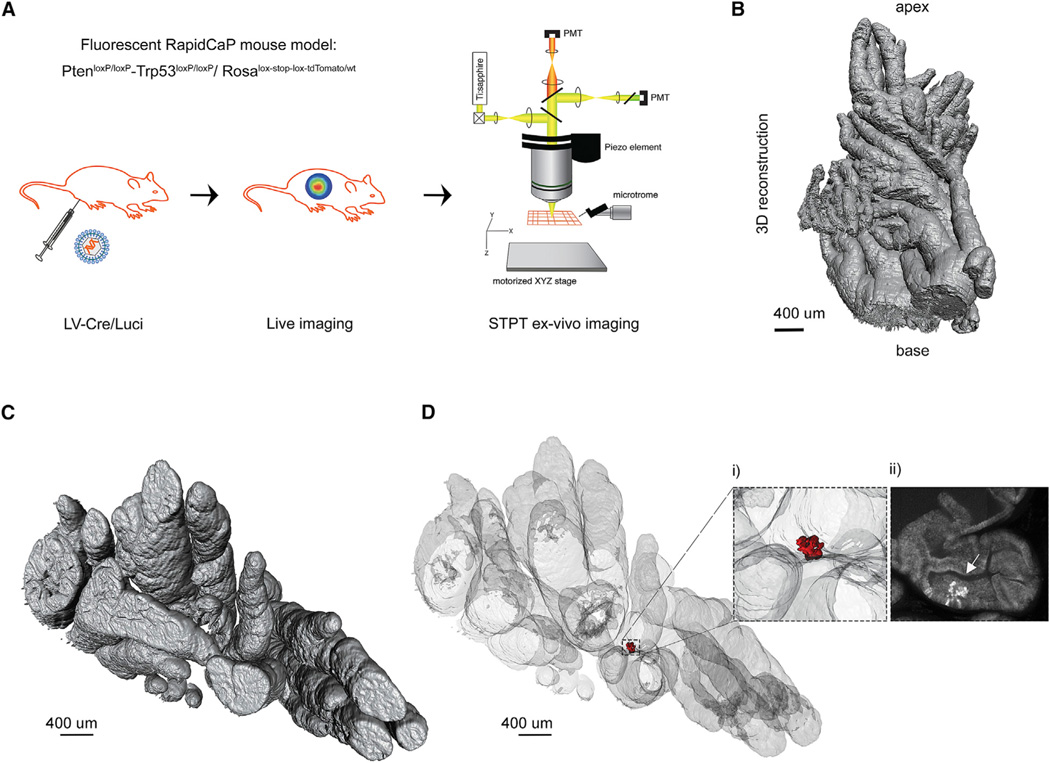
Figure 1. High-resolution 3D imaging of RapidCaP mouse prostate by STPT.
(A) The RapidCaP eTOM mouse: Surgical delivery of Cre/Luciferase lentivirus (LV-Cre/Luci) into the right anterior prostate in triple-knockin PtenloxP/loxP-Trp53loxP/loxP-Ai9(RCL-tdT) mice. The Pten-Trp53 deletion can be visualized in vivo by bioluminescence imaging before ex vivo imaging of the prostate by STPT.
(B) 3D reconstruction of the imaged prostate (4 mm × 1.6 mm × 7 mm) from a RapidCaP mouse at 10 days post-injection.
(C) Top view of the same mouse prostate reconstruction.
(D) 3D transparent prostate reconstruction and detection of a small cluster of epithelial tdT+ cells (shown in red). (i) Enlarged region with tdT+ epithelial cells. (ii) 2D transverse section from the same area shown in (i), where the arrow points to tdT+ epithelial cells (see also Figure S1). This example is from a dataset of seven mice analyzed at 10 days post-injection (see also Figure 2).
Figure 2. Time course of PC initiation and proliferation in the eTOM RapidCaP.
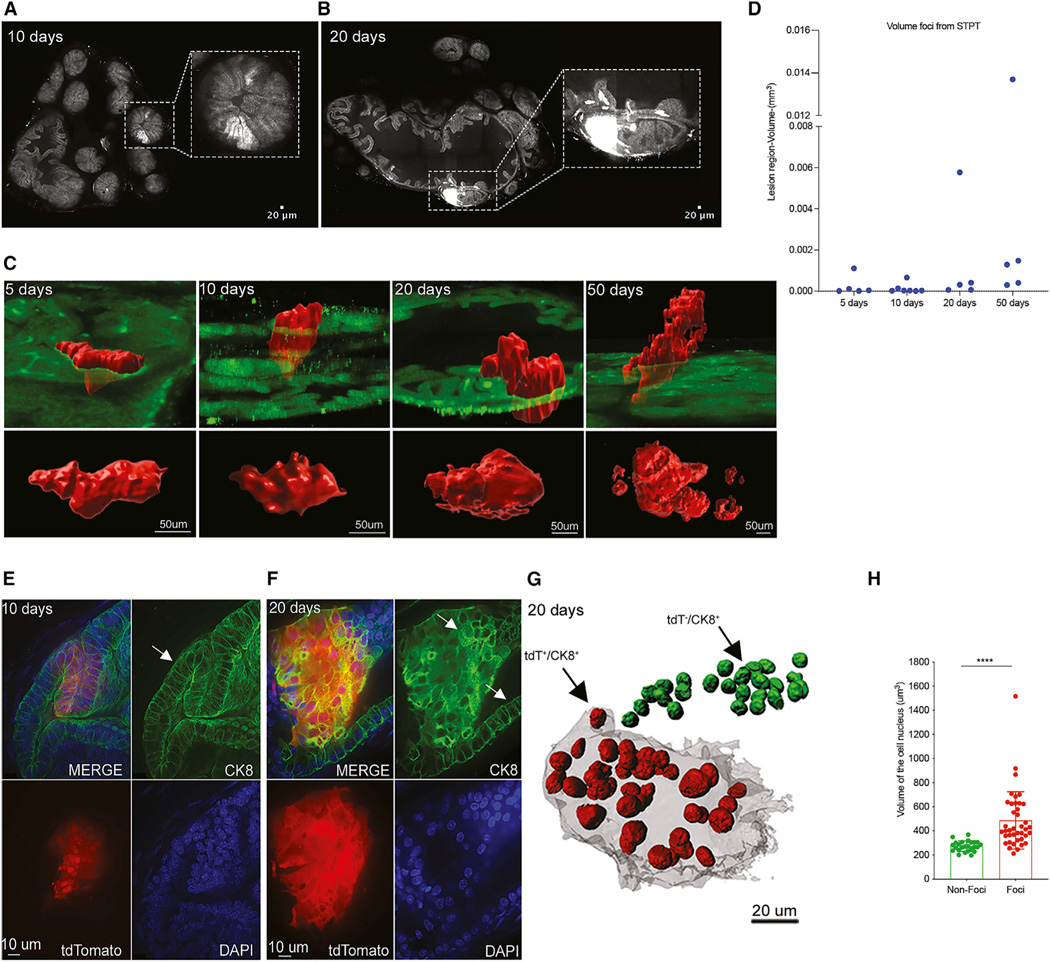
(A) Transverse section of prostate at 10 days post-injection reveals a focus of tdT+ epithelial cells with normal epithelial architecture (inset) (see also Figure S3A).
(B) In contrast, a transverse section of prostate at 20 days post-injection reveals a focus of tdT+ cells that appears to lose normal epithelial architecture (see also Figure S3B).
(C) Reconstruction and volume quantification of tdT+ foci in prostate at 5, 10, 20, and 50 days post-injection. 3D view of the foci is shown in red (see also Figures S3A–S3C and S4A).
(D) Quantification of the complete volume of the tdT+ focal regions (mm3) in prostate at 5, 10, 20, and 50 days post-injection, including five, seven, five, and five mice analyzed at days 5, 10, 20, and 50 post-injection, respectively (see also Table S1).
(E and F) Immunofluorescence (IF) with cytokeratin 8 (CK8) antibody in STPT sections confirms normal epithelial architecture at 10 days (E) but disrupted hyperplastic-like changes at 20 days post-injection (see also Figures S6 and S7).
(G) Visualization of DAPI-labeled nuclei in prostate at 20 days post-injection showing tdT+ cells in red and tdT− in green. (H) Quantification of the increase in nuclear size of tdT+ compared to nearby tdT− epithelial cells (mean ± SD): 487 ± 237.3 μm3 (n = 40) and 273 ± 42.57 μm3 (n = 25); p < 0.001, unpaired t tests (see also Figures S6 and S7 and Table S2) (n = 2 mice).
The fluorescent RapidCaP GEM model is based on crossing conditional double-knockout PtenloxP/loxP and Trp53loxP/loxP mice with mice carrying the Cre-activatable “lox-stop-lox” red fluorescent protein (RFP) variant tdTomato in the Rosa26 locus (Ai9(RCL-tdT)) (Madisen et al., 2010), generating triple transgenic animals which we termed eTOM mice. In this model, surgical delivery of lentiviruses co-expressing Cre recombinase and firefly luciferase (LV-Cre/Luci) (Cho et al., 2014) to the prostate tissue results in a focal deletion of Pten/Trp53 genes and co-expression of luciferase and RFP tdTomato reporters in the same cells (Figure 1A). The luciferase expression can then be used for bioluminescence-based monitoring of PC progression in live cohorts (Figure S1A), while the tdTomato expression enables the detection of individual infected cells by STPT (Figure 1). As shown in Figure 1B–1D, STPT imaging of eTOM prostate at 10 days after lentivirus injection was indeed able to identify a small cluster of transformed tdTomato-positive (tdT+) cells, determining their precise position within the 3D volume of the right anterior prostate lobe (see more below). Visualization of a single STPT optical section revealed the exact localization of those cells within the targeted prostate epithelial layer (Figure 1D, inset ii, and Figure S1).
STPT enables early detection and morphological characterization of epithelial PC cell expansion
Having established conditions for STPT prostate imaging, we next analyzed the progression of prostate epithelial cell transformation in a time course of 1, 5, 10, 20, and 50 days following the surgical delivery of the LV-Cre/Luci into the right anterior part of the prostate. This analysis revealed the following findings. First, STPT imaging was able to identify individual few tdT+ cells present in the entire gland at day 1 post-injection, demonstrating the power of this approach to detect the earliest sites of LV-infected single cells (Figure S2A). Second, STPT allowed us to also map the size of the tdT+ region in each gland, revealing a gradual time-dependent increase in the volume comprising tdT+ cells, and this was seen at the latest after day 20 post-injection (Figures 2C and 2D; Table S1). And third, while the cellular morphology of the tdT+ regions in tissues taken at days 1, 5, and 10 retained normal columnar epithelial architecture, these foci appeared to lose normal tissue organization in glands taken at 20 and 50 days post-injection (Figures 2, S2B, and S3). As a control for the specificity of these morphological changes on the deletion of the Pten and Trp53 genes, we also injected the LV-Cre/Luci lentivirus into a single transgenic Ai9(RCL-tdT) reporter mice. As expected, we did not detect any time-dependent gross morphological changes in the lsl-tdTomato-reporter mice (Ai9, RCL-tdT) harboring no conditional Pten/Trp53 knockout genes, confirming that these events are contingent on the loss of Pten/Trp53 expression in the RapidCaP mice (Figure S4). As a second control for the specificity of the deletion of Pten and Trp53 genes in the prostate, we imaged three animals at 10 days post-injection to exclude the possibility that LV particles were disseminated by bloodstream into the liver and brain, inducing direct recombinations in these distant tissues. Imaging of the prostate, liver, and brain in each mouse detected the primary foci in the prostate as described in Figure 2D and did not detect tdT+ cells in the liver and the brain at this time point (data not shown). This supports our conclusion of metastatic spread of the primary PC tumor, which is further examined below (Figures 3 and 4).
Figure 3. Mapping 3D distribution of PC metastatic cells in the liver.
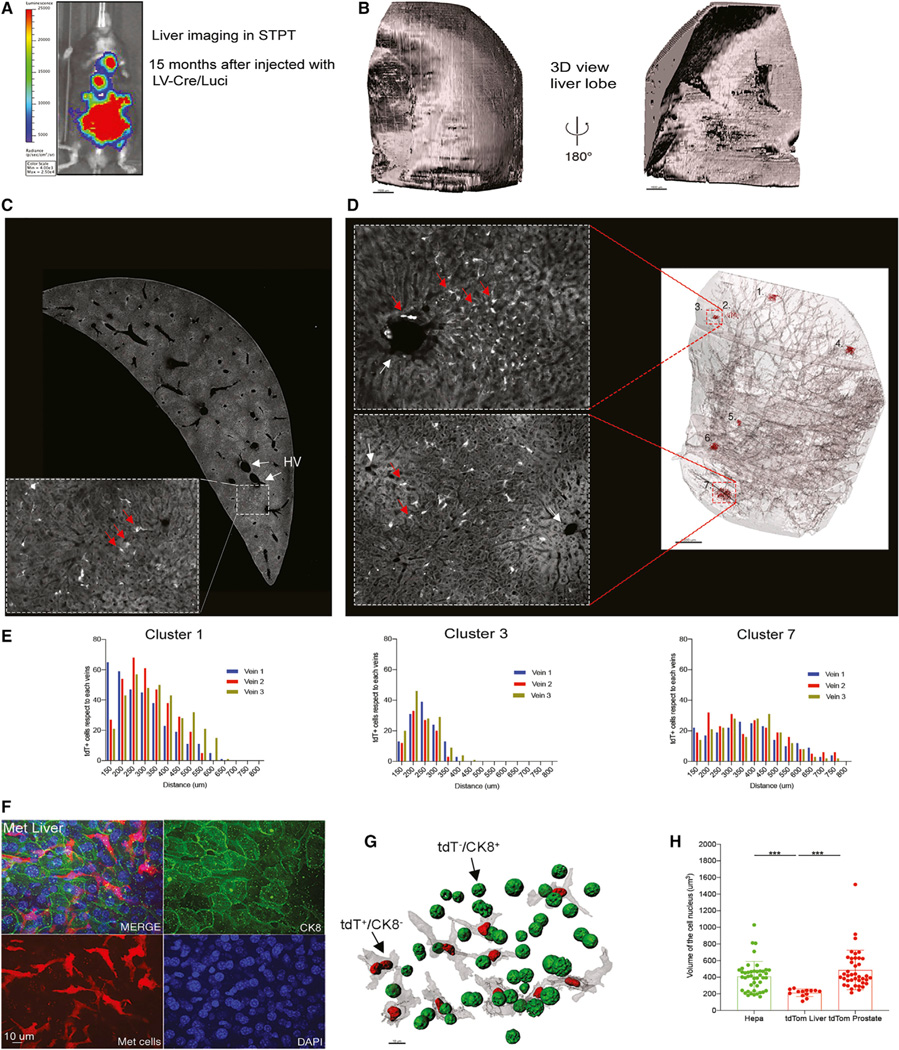
(A) Bioluminescence signal reveals metastatic disease spread across the body at 15 months post-injection.
(B) 3D reconstruction of the right liver lobe imaged by STPT.
(C) A cross-section of the imaged liver and an enlarged zoom-in view with tdT+ cells (red arrows) close to the two hepatic veins (HVs) (white arrows).
(D) 3D view of the transparent-rendered liver highlighted seven tdT+ foci (shown in red) distributed among main blood vessels. Enlarged images show two of the foci detected (tdT+ cells, red arrows. Each cluster has an ID number, and they were quantified).
(E) Histograms of the distribution of clusters 1, 3, and 7 representing the distance between tdT+ cells and the closest veins to each cluster (see also Figure S9).
(F) Anti-CK8 IF labeling reveals that the metastatic tdT+ cells in the liver are CK8−.
(G) Visualization of DAPI-labeled tdT+/CK8− and tdT−/CK8+ nuclei in the liver.
(H) Quantification of the nuclear size of tdT+/CK8− and tdT−/CK8+ nuclei in the liver (mean ± SD): 213 ± 44 um3 (n = 13) and tdT+/CK8+ prostate 409 ± 179.0 um3 (n = 44); p < 0.001, unpaired t tests (n = 2 mice) (see also Table S3).
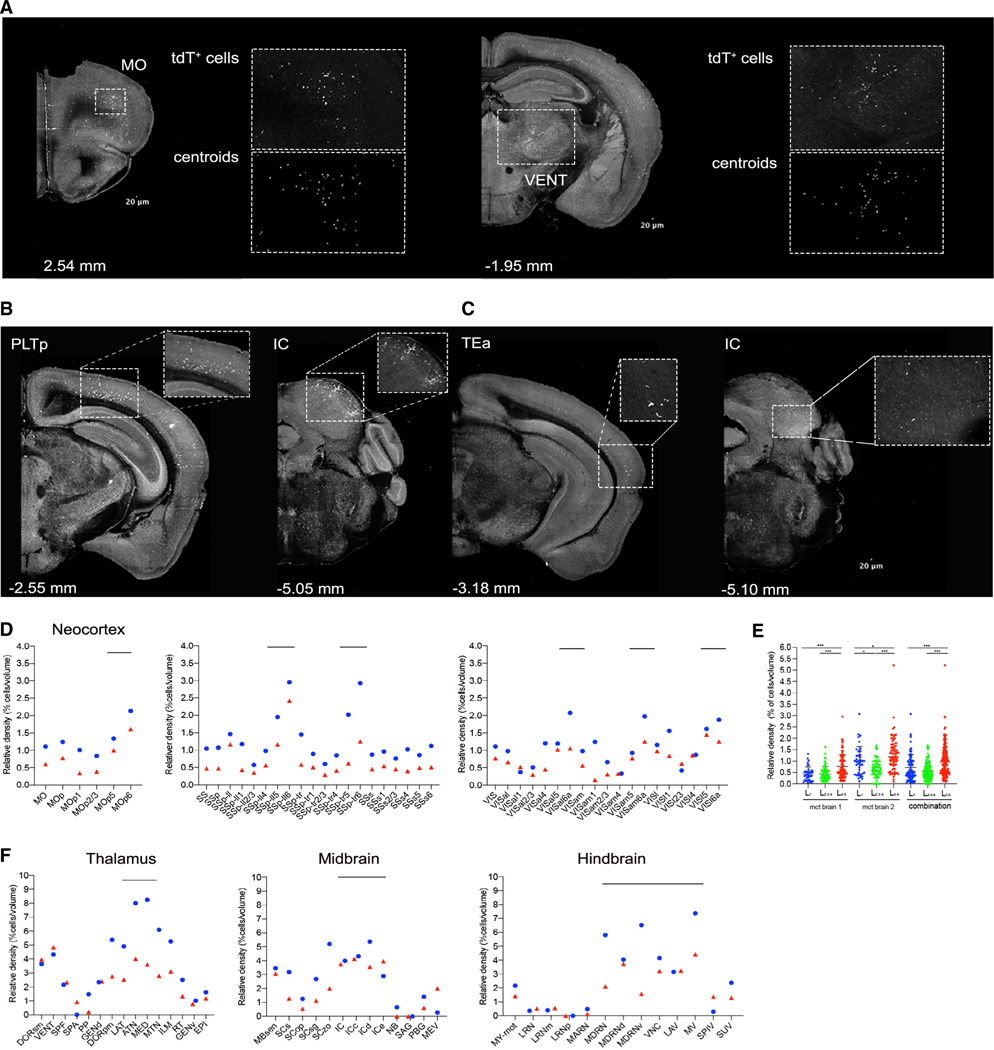
Figure 4. Detection of PC metastatic cells in the brain.
(A) Examples of coronal brain sections from STPT imaging showing metastatic tdT+ cells in the motor cortex (MO) and a ventral group of the dorsal thalamus (VENT). Enlarged inset views show tdT+ cells (top) and their detection as centroids using machine learning (bottom) (Kim et al., 2015).
(B and C) Additional examples of STPT coronal brain sections with tdT+ cells in the posterior parietal association area (PLTp) area, inferior colliculus (IC), and temporal association area (TEa) (see also Figure S10).
(D) The distribution of metastatic tdT+ cells in the neocortex shows enrichment in deep cortical layers, mainly the deepest layer 6 (the circle [blue] and triangles [red] represent measurements from two different animals). Examples are from MO, somatosensory (SS), and visual (VIS) cortex.
(E) Comparison of layer-specific cortical distribution confirms deep-layer tdT+ cell enrichment in two brains, One-factor (mean ± SD) ANOVA; ***p < 0.001 (see also Table S4).
(F) Quantification of the distribution of the metastatic tdT+ cells in thalamus, midbrain, and hindbrain across two brains (n = 2 mice).
Next, we employed the unique aspect of our platform—combining whole-prostate STPT imaging with traditional immunostaining and cytological assays of the imaged 50-μm-thick prostate sections comprising the tdT+ foci—to further classify the morphological changes following PC initiation.
Immunostaining with antibodies against the cytoskeletal protein cytokeratin 8 (CK8) and the surface protein E-cadherin allowed us to characterize the transition from normal to neoplastic cellular pathology that the above STPT data predicted to occur between days 10 and 20 post-injection. This assay revealed that at 10 days post-injection, there was a detectable expansion of the cell number of the tdT+ cells in columnar epithelia, consistent with Pten/Trp53 loss and phosphatidylinositol 3-kinase (PI3K) pathway activation as shown in Figures S5A and S5B, but without other cellular morphological changes (Figures 2E, S6B, and 6C; see also Figures S5C and S5D for normal reference epithelium). We also inferred a doubling time of ~1.9 days (Figure S6A). In contrast, the sites captured at 20 days post-injection showed overt disruption of the lateral cytokeratin and E-cadherin patterns (Figures 2F and S6D and quantified in Figures S7A and S7B) and irregular cell sizes and shapes of the tdT+ cells (Figures 2F and S7C). 3D reconstruction of agarose thick-section immunostaining on 20 days and 50 days lesions revealed invasive cells that have escaped from the tumor into the stroma (Figure S8). Convexity analysis (Figure S8B) revealed that the tumor-stroma interface is significantly less convex (spherical) in the 20-day lesion than in the 10-days lesion (Figures 2E, 2F, and S8C), consistent with an irregular shaped invasive front. In addition, cytological DAPI staining enabled us to compare nuclei of tdT+ and tdT− epithelial cells in the same tissue sections, revealing a significant increase in nuclear size in the transformed tdT+ cells compared to wild-type tdT− epithelial cells at 20 days post-injection (Figures 2G and 2H; Table S2).
STPT detection and quantification of micro-metastases in the liver
Liver metastasis is a typical adverse event in late-stage human PC (Siegel et al., 2020) and also a terminal event in the RapidCaP mouse model (Cho et al., 2015; Cho et al., 2014). Here, we used STPT to map the spatial distribution of the metastatic cells across the liver in three eTOM mice with a significant metastatic spread of the disease to liver, as determined by longitudinal live bioluminescence imaging (Figure 3A). As shown in Figures 3B–3D, STPT imaging of a liver right medial lobe (1 cm × 0.43 cm × 1.43 cm [xyz]) enabled us to generate a 3D reconstruction of the liver tissue and visualize numerous metastatic tdT+ cells across a total of seven metastatic foci. The metastatic cells were seen distributed in small clusters near hepatic veins (Figures 3C and 3D). This morphology was confirmed in classic formalin-fixed paraffin-embedded (FFPE) tissue-based analysis of an eTOM RapidCaP animal with advanced liver metastasis (Figure S9A). To further analyze the early disseminated tumor cell position in relation to the liver vasculature across the organ, we quantified the distance between individual tdT+ cells (converted to a centroid position) of the seven detected cell clusters and nearby hepatic veins (Figures 3D and 3E). This analysis revealed a distance distribution between at least 28–71 μm and at most 400–850 μm from the most proximal blood vessel for all clusters, suggesting a comparable tissue entry and spread with the tissue parenchyma for all metastatic cells detected (Figures 3E, S9B, and S9C).
Finally, we also used the 50-μm liver sections generated by STPT imaging for additional analyses by immunostaining and cytology. Immunostaining with anti-CK8 antibodies confirmed our previous finding that metastatic the tdT+ cells are CK8− and can be visualized among CK8+ hepatocytes (Figures 3F and S9A) (Cho et al., 2015; Cho et al., 2014). This also allowed us to quantify DAPI-labeled nuclei of the metastatic cells and nearby hepatocytes, revealing that the metastatic tdT+/CK8− cells have significantly smaller nuclei than the tdT−/CK8+ hepatocytes (Figures 3G and 3H; Table S3). Of note, bulk proliferation markers like Ki-67 yielded ambiguous results on these sparse cells in STPT agarose slices. Thus, it remains to be seen whether the early sparse metastatic tumor cells in liver are actively proliferating.
STPT detection and quantification of micro-metastases in the brain
Metastatic prostate cells can also reach the brain, as metastases to the dura mater (a connective tissue surrounding the brain) and brain parenchyma have been reported in 1%–2% of patients with metastatic PC (Benjamin, 2002; Chang et al., 2017). This phenomenon, however, is yet to be detected in mouse GEMs of PC. Given the sensitivity of our platform for imaging whole organs, we next chose the three RapidCaP animals analyzed for liver metastasis above to also screen for brain micro-metastases. Notably, two of the three mice indeed revealed tdT+ meta static cells in different brain areas (Figures 4A–4C), which we quantified using our computational single-cell detection pipeline for the brain (Kim et al., 2017). This analysis revealed a similar distribution profile of tdT+ cell between the two brains, comprising deep cortical layers of the motor, somatosensory, and visual cortical areas (Figures 4D and 4E), as well as subcortical areas of the ventral anterior-lateral nucleus of the thalamus, inferior colliculus, and sensory and motor-related medulla (Figures 4D and S10; Table S4). This suggest a similar route of entry into the brain parenchyma in both animals, which may be explored in future studies focused on the underlying mechanisms.
In the final set of experiments, we again used the 50-μm tissue sections generated by the STPT imaging and analyzed the presence of tdT+ cells in the brain environment. Immunolabeling with anti-CD31 antibodies to visualize brain vasculature revealed that the tdT+ cells were located in the lumen of brain vessels, either as single cells or small groups of ~3–5 cells (Figures S11, S13B, and S13D), with some tdT+ cells appearing to expand the blood vessel walls that surrounded them (Figure S11B, white arrows). These data thus suggest that the tdT+ cells are part of the earliest step of brain metastasis cascade defined as “intravascular cell arrest” (Feinauer et al., 2021; Kienast et al., 2010). Next, anti-GFAP immunolabeling of astrocytes revealed a modest reactive astrocytic activation near the metastatic cells (Lorger and Felding-Habermann, 2010) (Figures S12, S13A, and S13B). Finally, DAPI labeling of nuclei of the metastatic cells and nearby neurons revealed that the metastatic tdT+/NeuN− cells have significantly larger nuclei than tdT−/NeuN neurons, with a considerable variability in their nucleus volumes (Figures S13C–S13E; Table S5).
DISCUSSION
The STPT-based approach to imaging the different stages of PC in the RapidCaP model uniquely combines automated high-resolution whole-organ imaging and analysis of PC initiation, progression, and metastasis at cellular resolution with traditional IF analysis of cytoskeletal protein markers to further examine cellular features of PC cells in the prostate and other metastatic tissues.
STPT imaging and computational methods for analysis of the 3D reconstructed tissues, which until now have been used to study mouse brain anatomy (Hou et al., 2016; Kim et al., 2017; Mandelbaum et al., 2019; Matho et al., 2020; Muñoz-Castaneda et al., 2020; Ragan et al., 2012), provide several key advantages for analysis of cancer in mouse models. First, during STPT, the tissue is imaged as an intact whole mount, which allows for a complete 3D digital reconstruction of the imaged tissue without any spatial distortions. This in turn can be used for building averaged tissue volumes onto which multiple datasets can be precisely registered for detailed spatial analysis, as done here for the analysis of the distribution of single metastatic cells in an entire liver lobe relative to its vasculature and in the brain registered to our reference brain template and the Allen Mouse Brain Atlas (Kim et al., 2017; Ragan et al., 2012). Second, the use of two-photon microscopy in STPT allows for a high versatility in the spatial resolution at which the tissue can be imaged. Here we selected x-y resolution of 1 μm at z spacing of 25–50 μm, but this can be further increased, for example to an x-y resolution of 0.3 μm and a z resolution of 1.5 μm (Ragan et al., 2012). Such high spatial resolution could reveal further details in local 3D cell distribution, such as in the migration of metastatic cells from the cancer primary site. Third, STPT imaging requires only traditional tissue fixation (e.g., by formaldehyde) so that the serial tissue sections generated during STPT are therefore immediately compatible with traditional immunostaining methods used for analysis of cellular morphology, cytoskeleton, and other markers of the cancer and its native microenvironment (Turcan et al., 2018). Importantly, while the current application has focused on the RapidCaP PC mouse model, the same methods can be readily applied to other cancer mouse models, promising to advance our understanding of cellular events of cancer initiation and progression within intact tissues and organs.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contacts
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts Dr. Lloyd Trotman (trotman@cshl.edu) and Dr. Pavel Osten (osten@cshl.edu).
Materials availability
Viruses and transgenic animals generated for this study (see key resources table) have been archived and maintained in the Trotman laboratory (trotman@cshl.edu) at Cold Spring Harbor Laboratory.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit polyclonal cytokeratin CK8 | Abcam | Cat# ab59400; RRID: AB_942041 |
| Recombinant Alexa Flour 488 anti-Cytokeratin 8 | Abcam | Cat# 192467: RRID: AB_2864346 |
| Recombinant Alexa Flour 647 anti-Cytokeratin 5 | Abcam | Cat# 193895; RRID: AB_2728796 |
| Rabbit monoclonal E-Cadherin (24E10) | Cell Signaling technology | Cat# 3195T |
| Donkey anti-rabbit IgG secondary antibody Alexa 488 conjugate. | Thermo-Fisher Scientific | Cat# A21206; RRID: AB_2535792 |
| NeuN | Millipore | Cat# MAB377; RRID: AB_2298772 |
| GFAP | Agilent DAKO | Cat# Z033429-2; RRID: AB_10013382 |
| CD31 | R&D systems | Cat# AF3608; RRID: AB_2169833 |
| Ki67 | Spring Bioscience | Cat# M3062; RRID: AB_11219741 |
| RFP | Rockland | Cat# 600-401-379; RRID: AB_2209751 |
| Phospho-S6 | Cell Signaling technology | Cat# 5364; RRID: AB_10694233 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Luc/Cre lentiviral plasmid | Cho et al., 2014, 2015 | Addgene plasmid 20905 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Dapi | Thermo Fisher Scientific | Cat# D1306; RRID: AB_2629482 |
| Normal Donkey Serum | Sigma | D9663-10ML; RRID: AB_2810235 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# 19210 |
| Sodium Periodate | Sigma | Cat# S1878 |
| Sodium borohydrate | Sigma | Cat# 452882 |
| Sucrose | Sigma | Cat# S8501 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed data | This paper | Tables S1, S2, S3, S4, and S5 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J | Jackson Laboratory | Cat # 000664 |
| PtenloxP/loxP Trp53loxP/loxP | Cho et al., 2014 | PMID: 24444712 |
| RosaCAG-tdTomato-loxP-stop-loxP | Jackson Laboratory | Cat # 007909 |
|
| ||
| Oligonucleotides | ||
|
| ||
| PtenloxP/loxP, F: 5′-TGTTTTTGACCAATTAAAGTAGGCTGTG-3′ | This paper | N/A |
| PtenloxP/loxP, R:5′AAAAGTTCCCCTGCTGATGATTTGT-3′ | This paper | N/A |
| Trp53loxP/loxP, F: 5′-CACAAAAACAGGTTAAACCCAG-3′ | This paper | N/A |
| Trp53loxP/loxP, R: 5′-AGCACATAGGAGGCAGAGAC-3′ | This paper | N/A |
| Wt tdTomato, F: 5′-AAGGGAGCTGCAGTGGAG TA-3′ | This paper | N/A |
| Wt tdTomato, R: 5′-CCGAAAATCTGTGGGAAGTC-3′ | This paper | N/A |
| Mut tdTomato F: 5′-GGCATTAAAGCAGCGTATCC-3′ | This paper | N/A |
| Mut tdTomato R: 5′-CTGTTCCTGTACGGCATG 3′ | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FIJI software | NIH | https://imagej.net/software/fiji, RRID: SCR_00228 |
| MATLAB software | MathWorks | https://www.mathworks.com/products/matlab.html?s_tid=hp_products_matlab; RRID: SCR_001622 |
| GraphPad Prism 9 software | GraphPad Software | https://www.graphpad.com/; RRID: SCR_002798 |
| Imaris software | Version 9.2 Bitplane | http://www.bitplane.com/imaris/imaris; RRID: SCR_007370 |
| Code for STPT | (Kim et al., 2015) | https://www.ncbi.nlm.nih.gov/pubmed/25558063 |
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
The codes were published in (Hyun et al., 2021; Kim et al., 2017) and are available upon request to Dr. Pavel Osten.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
PtenloxP/loxP, Trp53loxP/loxP, and wild-type C57BL/6J mice were used in this study. All the animals used were males between 2 to 6 months and animals of 15 months old for distant metastasis analysis. Animals were kept on a 12:12 light/dark cycle or a reversed cycle under standard housing conditions. All protocols for mouse experiments were performed following the institutional guidelines protocols and were approved by the Institutional Animal Care and Use Committee (IACUC). PtenloxP/loxP, Trp53loxP/loxP transgenic mice were generated by crossing PtenloxP/loxP with Trp53loxP/loxP (Jonkers et al., 2001). Then, the F1 was crossed with Cre-dependent tdTomato reporter mice (Rosa lox-stop-lox-tdTomato+/WT, Jackson Labs # 007905) (Madisen et al., 2010). For genotyping, tail DNA was subjected to PCR analysis with the following primers. For PtenloxP/loxP, primer 1 (5′-TGTTTTTGACCAATTAAAGTAGGCTGTG-3′) and primer 2 (5′AAAAGTTCCCCTGCTGATGATTTGT-3′) were used. For Trp53loxP/loxP, primer 1 (5′-CACAAAAACAGGTTAAACCCAG-3′) and primer 2 (5′-AGCACATAGGAGGCAGAGAC-3′) were used. For the tdTomato primers wild-type forward (5′-AAGGGAGCTGCA GTGGAG TA-3′), wild-type reverse (5′-CCGAAAATCTGTGGGAAGTC-3′), mutant reverse (5′-GGC ATTAAAGCAGCGTATCC-3′), mutant forward (5′-CTGTTCCTGTACGGCATG 3′) were used.
METHOD DETAILS
Viral constructs
Lentiviral constructs-the Luc. Cre lentiviral plasmid (Tyler Jacks, Addgene plasmid 20905), pMD2.G, and psPAX2-were purchased (University of Iowa Viral Vector core).
Lentivirus productions and infections
Lentivirus Productions, Infections, and Injections Lentiviruses were produced by calcium phosphate transfection. 293FT cells were plated for transfection density 8 Å~10 6 cells per10-cm plate. Target plasmid (10 μg) was combined with helper constructs, 8.5 μg of pMD2.G, and 3.5 μg of psPAX2, for transfection. It was harvested 24, 36, 48, and 60 hours after transfection and centrifuged (4,500 rpm, 15 minutes) before filtering through 0.45-μm pore cellulose acetate filters. Viral supernatant was concentrated by ultracentrifugation (2 hours at 20,000 Å ~g), then an in vitro infection test for each batch of viruses was conducted in advance.
Intra-prostate injection
After exposure to anesthesia (isoflurane, 2%), the lower half of the abdomen was shaved, and the mouse was placed in a surgery hood. The mouse was exposed continuously to isoflurane via a nose cone for the entire duration of the 10-minute surgery. The shaved region was cleaned with betadine, followed by 70% ethanol three times. A 0.5-inch incision in both the skin and peritoneum was made along the lower abdominal midline to allow the right anterior prostate to be positioned for injection on sterile support. Typically, 20 μL of the concentrated virus was injected into the right anterior prostate. The incision was then sutured, and the skin was stapled shut using two to three stainless steel EZ Clip wound closures. After animals were observed for complete recovery from anesthesia, they were warmed under a heating lamp to regain the ability to maintain sternal recumbence and given DietGel.
Bioluminescence Imaging
In vivo and ex-vivo, bioluminescence imaging was performed using a Xenogen IVIS Spectrum imager, which uses a highly sensitive, cooled CCD camera mounted in a light-tight camera box. For in vivo imaging, animals received intraperitoneal injections of 200 mg/kg luciferin 10 minutes before imaging. The animals were then anesthetized using 2% isoflurane and placed onto the warmed stage inside the camera box. The animals received continuous exposure to 2% isoflurane to sustain sedation for 3 minutes of imaging. For quantification, regions of interest were measured with standardized rectangular regions covering the mouse trunk and extremities.
Imaging acquisition in STPT
Animals were perfused transcardially with ice-cold PBS followed by 4% PFA. After a 24-hour post-fix in 4% PFA, the prostate, liver, and brains were kept in a 0.7% glycine solution for 48 hours. The prostate, liver, and brain were stored in 0.05 M phosphate buffer at 4 degrees. Then, were embedded in 6% agarose for prostate and 4% agarose for liver and brain in 0.05M PB. The agarose block was cross-linked in a 0.2% sodium borohydride solution by 5–8 hours, depending on the tissue. After imaging with a high-speed multiphoton microscope with integrated vibratome sectioning employing x-y resolution of 1 μm, z-step of 50 μm, or adding an optical section of 25 um in the z axis (TissueCyte 1000, TissueVision) as described before (Ragan et al., 2012). The raw image files were corrected for illumination, stitched in 2D, and aligned in 3D.
Immunofluorescence
Fixed sections 50 um thickness from the prostate and liver were immediately collected from the STPT bath and kept in cryoprotectant solution in 12 well plates at —20 degrees until using them. Employing an inverted fluorescent microscope (Zeiss Observer), we scanned the sections with foci that were detected previously in the 3D images from STPT. Foci sections and their controls were gently washed in 1X PBS and used for immunolabeling. The sections were incubated in 50mM glycine for 15 mins and washed with 1X PBS. Next, the sections were blocked for 2 hours with a blocking buffer comprising 0.6% Triton X-100 and 10% goat serum. Post blocking, the sections were stained overnight at room temperature with primary antibodies at 1:100 dilution for CK8 and 1:100 for E-Cad. Following three 10-minutes washes with PBS, sections were incubated with Alexa Fluor 488 donkey anti-rabbit (1:500; ThermoFisher) antibody for 5 hours at room temperature for visualization. Sections were incubated with a 10 μg/mL solution of 4′,6-diamidino-2-phenylindole (DAPI) for 5 mins at room temperature to stain the cell nuclei. Sections were rinsed in PBS and distilled water serially, and finally mounted with Mounting Medium (ProLong gold antifade mountant; ThermoFisher). Stained slides were imaged and analyzed using both the Ultraview VoX Spinning disk confocal microscope (PerkinElmer).
Immunohistochemistry
After running the samples in STPT, fixed brain sections 50 um thickness were immediately collected from the STPT bath and kept in cryoprotectant solution in 12 well plates at −20 degrees until using them. Employing an inverted fluorescent microscope (Zeiss Observer), we scanned the sections with tdT+ cells detected previously in the 3D images from STPT. We processed the 50um sections between histology sponges and embedded them in paraffin. Then, we sliced at 5 um serial sections thickness in the microtome. IHC slides were stained in Discovery Ultra automatic IHC stainer (Roche) following standard protocols. Briefly, after deparaffinization and rehydration, slides were subjected to antigen retrieval (Benchmark Ultra CC1, Roche) at 96◦C for 1hr; primary Ab incubation was performed at 37◦C for 1hr (RFP-1:200, Ki-67–1:250) and we employed the Discovery multimer detection system (Discovery OmniMap HRP, Discovery DAB) to detect immuno-signals. Additionally, we processed coronal brain sections with H&E staining.
QUANTIFICATION AND STATISTICAL ANALYSIS
3D images reconstruction
Employing the STPT dataset with full resolution from prostate and liver, 3D rendering was done using Imaris version 7.6.5 (Bitplane). STPT images resolution for the liver and the prostate was 1 μm-1 μm in x-y and 50 μm. Also, Imaris software developed transparent prostate or liver surfaces and allowed visualized the tdTomato positive cells in both cases.
Quantification of the volume foci
To quantify the prostate focus, we use the start and the end cross-sections that covert the focus from the STPT 2D images. Then with Imaris software, we calculate the volume of each focus employing the tdTomato+/− fluorescent and as control the green channel as an internal control to cover the whole surface of the focus. The resolution for prostate focus was 1 μm-1 μm in x-y and 25 μm.
Volume quantification of the cell nucleus using DAPI labeling
In 50 um prostate and liver sections containing focus at 20 days and metastasis liver, we quantify the volume of the DAPI signal. Then, using Imaris, we applied a threshold to the DAPI signal to epithelial cells located into the focus and outside the focus, simultaneously getting the corresponding volume for several cells. In case the metastasis cells in the liver, we did calculate the nucleus volume for hepatocytes and the metastatic tdTomato+/− cells.
Metastatic tdT+ cells quantification, and analysis of the distance of them with vasculature structure in the liver
Centroids of all structures (metastatic cells, portal veins, central veins) are calculated as follows. First, all images are manually annotated (Fiji-plugins Labkit). Every pixel of each structure is given a unique intensity corresponding to its structure type (e.g., all pixels that are part of any portal vein are given an intensity of 2). Annotations are done in such a way that independent structures will not have annotated pixels that are adjacent to each other. Next, a connected component labeling using 8-connectivity is done on each image so that the annotated pixels of each independent structure can be grouped. Then, for each independent structure, the centroid of a structure is calculated by taking the mean of the coordinates of all pixels that belong to that structure.
STPT cell count image analysis
Raw images were corrected for non-uniform illumination and stitched in 2D, and stacked in 3D. To improve the signal to noise ratio, we applied subtracted channels between the green channel to the red channel (fluorescent cells). Then, a convolutional network trained to recognize nuclear labeling that automatically detects cell body neurons was applied (Turuga et al., 2010). The 3D stack was then registered to a 3D reference brain (built using 40 STPT imaged brains) based on the ABA (Allen Brain Atlas) (Sunkin et al., 2013; Kim et al., 2015) by 3D affine registration followed by a 3D B-spline registration using the software Elastix (Klein et al., 2010). The density of total tdTomato+/− cells in the brain areas was obtained by the percentage of tdTomato+/− positive cells normalized for the volume in the specific brain regions (Isocortex, Thalamus, Midbrain, and Hindbrain).
Brain volume quantification
To measure the volume of anatomical regions, the average reference brain was aligned to the ABA. Segmentation areas were registered onto each brain using the B-spline registration procedure described above (i.e., the ABA segmentation was registered onto each brain sample). The number of voxels belonging to each region in the transformed ABA segmentation was counted and multiplied by 0.02 × 0.02 × 0.05 mm3 (the dimensions of an anatomical voxel spacing unit), resulting in the total volume of each region.
Convexity analysis
The Convexity morphological feature algorithm was developed to quantify shape variation of the tumor border. Convexity is the relative amount that an object differs from an ideal convex (spherical) object of convexity = 1.0. A measure of convexity can be obtained by forming the ratio of the perimeter of an object’s convex hull to the perimeter of the object itself. A planar tumor region was generated using the Imaris software (Bitplane AG) whose convex hull and perimeter were computed using a custom Python 3.7 script from: https://github.com/Ironhorse1618/Python3.7-Imaris-XTensions
General
Data points are stated and plotted as mean values ± SD. Samples sizes (n) represent the numbers of animals unless otherwise specified. All experiments are plotted, showing all the data points. p values are represented by symbols using the following code: * for 0.01 < p < 0.05, ** for 0.001 < p < 0.01, and *** for p < 0.001. Exact p values and statistical tests are stated in figure legends. Data were analyzed one-way analyses of variance (ANOVAs) with repeated-measures followed by Tukey’s multiple comparison test. Two-tailed P values of < 0.05 were considered the cutoff for statistical significance.
Supplementary Material
Highlights.
STP tomography has the resolution to detect tumor initiation and its progression in PC
Description of the micro-metastasis in mouse liver with whole-organ reconstruction
Whole-brain imaging and quantification of metastatic cells from primary prostate cancer
This technology allows the use of post hoc thick sections for traditional histology
ACKNOWLEDGMENTS
We thank members of Dr. Lloyd C. Trotman’s laboratory and Dr. Pavel Osten’s laboratory for helpful discussions. We thank the LAR facility at CSHL, especially to Eileen Earl, supervisor of lab animal resources. We thank Ramesh Palaniswamy and Dr. Arun Narasimhan for help with STPT. This work was supported by grants from the Simons Foundation (to P.O. and L.C.T.), the NIH (U01 MH105971 to P.O. and R01CA237154-01 to L.C.T.), and The Gertrude and Louis Feil Family Trust (P.O.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110027.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Benjamin R. (2002). Neurologic complications of prostate cancer. Am. Fam. Physician 65, 1834–1840. [PubMed] [Google Scholar]
- Cai R, Pan C, Ghasemigharagoz A, Todorov MI, Förstera B, Zhao S, Bhatia HS, Parra-Damas A, Mrowka L, Theodorou D, et al. (2019). Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci. 22, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Kwan B, Panjwani N, Villanueva N, Diamond S, Wong-Sefidan I, and Muchmore E. (2017). Prostate adenocarcinoma metastases to the testis and brain: case report and review of the literature. Oxf. Med. Case Rep. 2017, omx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Herzka T, Zheng W, Qi J, Wilkinson JE, Bradner JE, Robinson BD, Castillo-Martin M, Cordon-Cardo C, and Trotman LC (2014). Rapid-CaP, a novel GEM model for metastatic prostate cancer analysis and therapy, reveals myc as a driver of Pten-mutant metastasis. Cancer Discov. 4, 318–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Herzka T, Stahlhut C, Watrud K, Robinson BD, and Trotman LC (2015). Rapid in vivo validation of candidate drivers derived from the PTEN-mutant prostate metastasis genome. Methods 77-78, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccarese MF, Dubach JM, Pfirschke C, Engblom C, Garris C, Miller MA, Pittet MJ, and Weissleder R. (2017). Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nat. Commun. 8, 14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinauer MJ, Schneider SW, Berghoff AS, Robador JR, Tehranian C, Karreman MA, Venkataramani V, Solecki G, Grosch JK, Gunkel K, et al. (2021). Local blood coagulation drives cancer cell arrest and brain metastasis in a mouse model. Blood 137, 1219–1232. [DOI] [PubMed] [Google Scholar]
- Hou XH, Hyun M, Taranda J, Huang KW, Todd E, Feng D, Atwater E, Croney D, Zeidel ML, Osten P, and Sabatini BL (2016). Central Control Circuit for Context-Dependent Micturition. Cell 167, 73–86.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun M, Taranda J, Radeljic G, Miner L, Wang W, Ochandarena N, Huang KW, Osten P, and Sabatini BL (2021). Social isolation uncovers a circuit underlying context-dependent territory-covering micturition. Proc. Natl. Acad. Sci. USA 118, e2018078118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, and Berns A. (2001). Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425. [DOI] [PubMed] [Google Scholar]
- Keller PJ, and Dodt HU (2012). Light sheet microscopy of living or cleared specimens. Curr. Opin. Neurobiol. 22, 138–143. [DOI] [PubMed] [Google Scholar]
- Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, and Winkler F. (2010). Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122. [DOI] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. (2015). Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 10, 292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yang GR, Pradhan K, Venkataraju KU, Bota M, García Del Molino LC, Fitzgerald G, Ram K, He M, Levine JM, et al. (2017). Brain-wide Maps Reveal Stereotyped Cell-Type-Based Cortical Architecture and Subcortical Sexual Dimorphism. Cell 171, 456–469.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA, and Pluim JPW (2010). elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging, 196–205. 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- Kubota SI, Takahashi K, Nishida J, Morishita Y, Ehata S, Tainaka K, Miyazono K, and Ueda HR (2017). Whole-Body Profiling of Cancer Metastasis with Single-Cell Resolution. Cell Rep. 20, 236–250. [DOI] [PubMed] [Google Scholar]
- Lorger M, and Felding-Habermann B. (2010). Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis. Am. J. Pathol. 176, 2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbaum G, Taranda J, Haynes TM, Hochbaum DR, Huang KW, Hyun M, Umadevi Venkataraju K, Straub C, Wang W, Robertson K, et al. (2019). Distinct Cortical-Thalamic-Striatal Circuits through the Parafascicular Nucleus. Neuron 102, 636–652.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matho KS, Huilgol D, Galbavy W, Kim G, He M, An X, Lu J, Wu P, Di Bella DJ, Shetty AS, et al. (2020). Genetic dissection of glutamatergic neuron subpopulations and developmental trajectories in the cerebral cortex. bioRxiv, 2020.2004.2022.054064. [Google Scholar]
- Muñoz-Castaneda R, Zingg B, Matho KS, Wang Q, Chen X, Foster NN, Narasimhan A, Li A, Hirokawa KE, Huo B, et al. (2020). Cellular Anatomy of the Mouse Primary Motor Cortex. bioRxiv, 2020.2010. 2002.323154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Schoppe O, Parra-Damas A, Cai R, Todorov MI, Gondi G, von Neubeck B, Bogurcu-Seidel N, Seidel S, Sleiman K, et al. (2019). Deep Learning Reveals Cancer Metastasis and Therapeutic Antibody Targeting in the Entire Body. Cell 179, 1661–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YG, Sohn CH, Chen R, McCue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W, et al. (2018). Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. Published online December 17, 2018. 10.1038/nbt.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, and Osten P. (2012). Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, and Abate-Shen C. (2010). Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 24, 1967–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30. [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, and Dang C. (2013). Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res, D996–D1008. 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. (2014). Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739. [DOI] [PubMed] [Google Scholar]
- Taranda J, and Turcan S. (2021). 3D Whole-Brain Imaging Approaches to Study Brain Tumors. Cancers (Basel) 13, 1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Makarov V, Taranda J, Wang Y, Fabius AWM, Wu W, Zheng Y, El-Amine N, Haddock S, Nanjangud G, et al. (2018). Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence. Nat. Genet. 50, 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turuga S, Murray JF, Jain V, Roth F, Helmstaedter M, Briggman K, Denk W, and Seung HS (2010). Convolutional networks can learn to generate affinity graphs for image segmentation. Neural Comput. 22, 511–538. 10.1162/neco.2009.10-08-881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
The codes were published in (Hyun et al., 2021; Kim et al., 2017) and are available upon request to Dr. Pavel Osten.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.