Summary
We previously reported the recovery of five ST11 carbapenem resistant hypervirulent Klebsiella pneumoniae (CR-HvKP) strains that harbored pLVPK-like virulence plasmids, yet molecular mechanisms underlying acquisition of virulence plasmid by ST11 K. pneumoniae have not been characterized. In this study, we showed that virulence plasmids in these CR-HvKP strains could be transferred to Escherichia coli strain EC600 via conjugation. Transmission of the virulence plasmids was found to involve formation of fusion plasmids with an Incl1 type conjugative plasmid and a small ColRNAI plasmid through homologous recombination and by insertion sequences IS26 and IS903B. The conjugative fusion event would transform different ST types of K. pneumoniae, in particular, the clinically prevalent ST11 or ST258 CRKP into CR-HvKP. Clinical factors that promote or suppress the occurrence of this fusion process should be further investigated to devise new approaches to halt such bacterial evolution trends.
Subject areas: Microbiology, Molecular microbiology, Clinical microbiology
Graphical abstract
Highlights
-
•
The pLVPK-like virulence plasmid could be transferred to Escherichia coli via conjugation
-
•
Transmission of the virulence plasmids was found to involve formation of fusion plasmids
-
•
The fusion processes involved homologous recombination or insertion sequences IS26/IS903B
Microbiology; Molecular microbiology; Clinical microbiology.
Introduction
Klebsiella pneumoniae is an opportunistic pathogen that can cause serious hospital infections, especially among patients with a compromised immune system (Paczosa and Mecsas, 2016). A subset of hypervirulent K. pneumoniae (HvKP), which may cause life-threatening community-acquired infections in healthy individuals, has become prevalent since early 1980s (Shon and Russo, 2012). HvKP isolates are highly associated with several biomarkers including peg-344 (putative transporter), iroB (salmochelin siderophore biosynthesis), iucA (aerobactin siderophore biosynthesis), and plasmid-borne rmpA and rmpA2 (regulators of the mucoid phenotype), the latter four being commonly presented in a large virulence plasmid (Russo et al., 2018). HvKP strains are generally susceptible to commonly used antibiotics (Wyres et al., 2019). However, convergence of hypervirulence and antibiotic resistance encoding genetic elements harbored by HvKP and multidrug resistant (MDR) K. pneumoniae strains has occurred, generating highly virulent strains that are also difficult to eliminate during antimicrobial treatments (Lam et al., 2019; Shen et al., 2019; Turton et al., 2019; Yang et al., 2021). Furthermore, hypervirulence-encoding genes have been found to disseminate extensively to MDR Klebsiella strains through various plasmid-mediated conjugation mechanisms (Li et al., 2020; Xie et al., 2021). A recent study discovered that the pLVPK-like virulence plasmid could be transferred from HvKP strains to ST11 CRKP and Escherichia coli strains with the help of a self-transferable IncF plasmid in various modes (Xu et al., 2021).
Previously, we reported a fatal outbreak of ST11 CR-HvKP (carbapenem resistant HvKP) strains in a Chinese hospital and revealed the complete genome sequences of five CR-HvKP strains, namely CR-HvKP1∼5 (Dong et al., 2018; Gu et al., 2018). Five plasmids were recovered from each isolate, including a 178.2-kbp (kilobase pair) IncHI1B/FIB pLVPK-like virulence plasmid in which certain sequences were deleted, a 177.5-kbp IncFII/R self-transferable blaKPC-2-bearing MDR plasmid, a 99.7-kbp Incl1 plasmid and two ColRNAI-type plasmids of 11.9 and 5.6-kbp, designated as pVir-CR-HvKP, pKPC-CR-HvKP, p3-CR-HvKP, p4-CR-HvKP, and p5-CR-HvKP, respectively. However, how these ST11 CRKP strains acquired the virulence plasmid and became HvKP remained unclear. In this work, we found that the virulence plasmids in the five CR-HvKP strains could be transferred to E. coli strain EC600 via conjugation. Transmission of the virulence plasmid was demonstrated to be mediated by a 99.7-kbp Incl1 helper plasmid, which should be the mechanism underlying the acquisition of the virulence plasmid by these ST11 K. pneumoniae strains. Furthermore, we identified eight modes of virulence plasmid mobilization, including transfer with or without the 11.9-kbp ColRNAI-type plasmid or the Incl1-type conjugative plasmid, fusion with the 11.9-kbp ColRNAI-type plasmid via homologous recombination, fusion with the Incl1 plasmid via homologous recombination, or insertion sequences from the virulence plasmid or the helper plasmid. Our findings provide a new insight into mechanisms of dissemination of virulence plasmid in Klebsiella species.
Results and discussion
Virulence plasmids in the five CR-HvKP strains were transmissible by conjugation
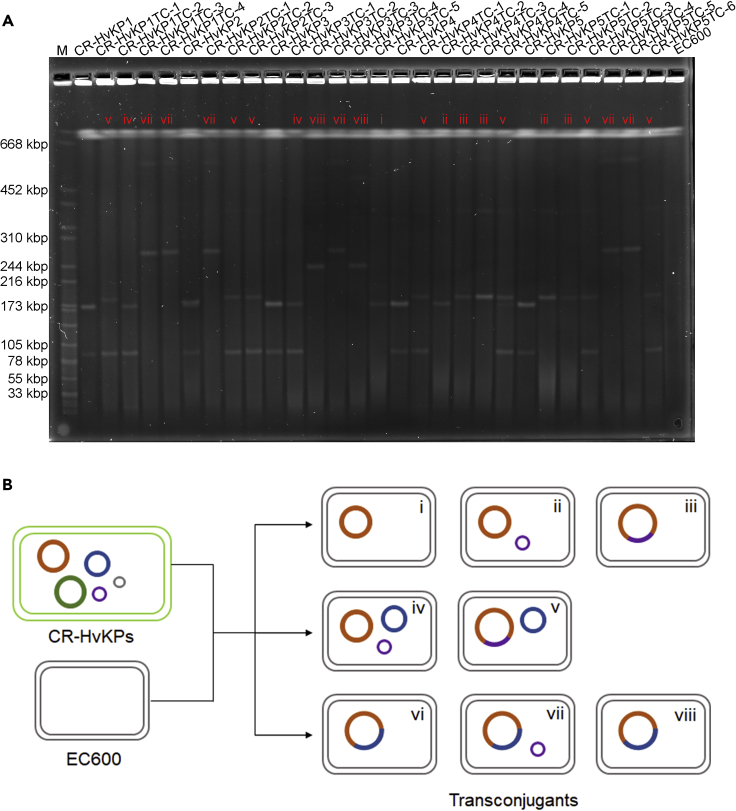
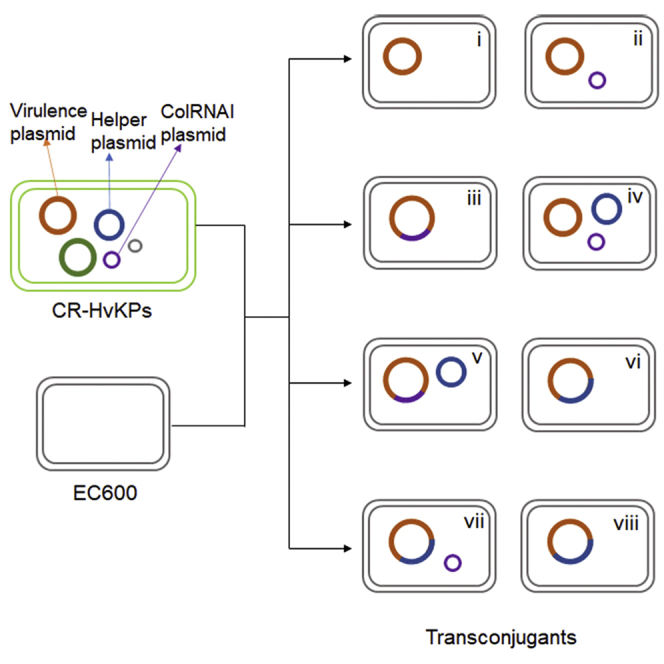
The five CR-HvKP strains were subjected to a conjugation experiment using approaches developed in our laboratory, with E. coli strain EC600 as recipient. Surprisingly, our results showed that each of the five virulence plasmids could be transferred to E. coli EC600 via conjugation, with efficiency ranging from 8.52E-07 to 3.13E-06 (Table 1). The conjugation experiments were repeated twice for each strain and a total of 23 transconjugants were randomly selected for further analysis. Transconjugants of each strain were picked up and subjected to S1-PFGE analysis. Interestingly, their transconjugants exhibited different profiles of plasmids according to the S1-PFGE results (Figure 1A). In brief, the virulence plasmid was found to transfer to E. coli strain EC600 in eight modes: i) alone; ii) with the 11.9-kbp ColRNAI plasmid, individually; iii) fusion with the 11.9-kbp ColRNAI plasmid; iv) with both the 99.7-kbp Incl1 plasmid and the 11.9-kbp ColRNAI plasmid, individually; v) with the 99.7-kbp Incl1 plasmid in the form of a fusion plasmid with the 11.9-kbp ColRNAI plasmid; vi) fusion with the 99.7-kbp Incl1 plasmid; vii) with the 11.9-kbp ColRNAI plasmid in the form of a fusion plasmid with the 99.7-kbp Incl1 plasmid; and viii) fusion with the 99.7-kbp Incl1 plasmid with deletion (Figure 1B). These different profiles of the transconjugants indicated that various genetic recombination events might have occurred during the conjugation process. However, there might be additional modes that were not identified as limited transconjugants were subjected for analysis. The five CR-HvKP strains also harbored a self-transmissible blaKPC-2-bearing plasmid, whereas fusion of the virulence plasmid and the blaKPC-2-bearing plasmid was not observable in this study. The blaKPC-2-bearing plasmid was found to transfer from these CR-HvKP strains to E. coli strain under selection by meropenem in our previous study (Gu et al., 2018). However, no fusion of these two plasmids was observable either.
Table 1.
Phenotypic and genotypic characteristics of the five CR-HvKP strains and their corresponding transconjugants
| Strain ID | Bacterial Species | MIC (μg/mL)a |
rmpA2 | Conjugation Efficiency | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ATM | CTX | CAZ | MEM | AMK | CIP | CLS | TE | ||||
| CR-HvKP1 | K. pneumoniae | >128 | >128 | >128 | >128 | >128 | 64 | 128 | >128 | + | NAb |
| CR-HvKP2 | K. pneumoniae | >128 | >128 | >128 | >128 | >128 | 64 | 2 | >128 | + | NA |
| CR-HvKP3 | K. pneumoniae | >128 | >128 | >128 | >128 | >128 | 64 | 2 | >128 | + | NA |
| CR-HvKP4 | K. pneumoniae | >128 | >128 | >128 | >128 | >128 | 128 | 2 | >128 | + | NA |
| CR-HvKP5 | K. pneumoniae | >128 | >128 | >128 | >128 | >128 | 64 | 128 | >128 | + | NA |
| EC600 | E. coli | <1 | <1 | <1 | <1 | 2 | <1 | <1 | <1 | – | NA |
| CR-HvKP1TC | E. coli | <1 | <1 | <1 | <1 | 2 | <1 | <1 | 128 | + | 1.88E-06 |
| CR-HvKP2TC | E. coli | <1 | <1 | <1 | <1 | 2 | <1 | <1 | >128 | + | 8.52E-07 |
| CR-HvKP3TC | E. coli | <1 | <1 | <1 | <1 | 2 | <1 | <1 | >128 | + | 1.22E-06 |
| CR-HvKP4TC | E. coli | <1 | <1 | <1 | <1 | 2 | <1 | <1 | >128 | + | 2.22E-06 |
| CR-HvKP5TC | E. coli | <1 | <1 | <1 | <1 | 2 | <1 | <1 | >128 | + | 3.13E-06 |
ATM, aztreonam; CTX, cefotaxime; CAZ, ceftazidime; MEM, meropenem; AMK, amikacin; CIP, ciprofloxacin; CLS, colistin; TE, tellurite. All tests were performed in duplicate, and each test included three biological replicates.
NA, not available.
Figure 1.
Transmission of virulence plasmids from CR-HvKP strains to E. coli EC600 by conjugation
(A) S1-PFGE of the five CR-HvKP strains and their corresponding transconjugants. Each transconjugant was indicated with a specific pattern.
(B) Schematic diagram of different modes of the transfer of the virulence plasmid. Plasmids pVir-CR-HvKP, pKPC-CR-HvKP, p3-CR-HvKP, p4-CR-HvKP, and p5-CR-HvKP were indicated by orange, green, blue, purple, and gray, respectively.
Genetic basis of transmission of virulence plasmids in CR-HvKP strains
To depict the mechanism underlying the transmission of virulence plasmids, transconjugants of these CR-HvKP strains were then subjected to whole genome sequencing. As the plasmid content of the five strains was almost the same, plasmid sequences of a representative transconjugant of each mode were present (Table 2). The transconjugant CR-HvKP3TC-5 which showed one band with the size of 180-kbp in the S1-PFGE analysis was found to harbor only one plasmid with the size of 178,159 bp, identical to plasmid pVir-CR-HvKP3, which was defined as mode i (Table 2). The transconjugant CR-HvKP4TC-2 that showed two bands with the size of 180-kbp and 10-kbp, respectively, in the S1-PFGE analysis was found to harbor two plasmids with the size of 178,159 bp and 11,970 bp, respectively, identical to plasmids pVir-CR-HvKP4 and p4-CR-HvKP4, which was defined as mode ii.
Table 2.
Characterization of different modes of transconjugants of the five CR-HvKP strains
| Modes | Transconjugants | Plasmid(s) | Size (bp) | tra genes | GenBank No. |
|---|---|---|---|---|---|
| i) alone | CR-HvKP3TC-5 | pCR-HvKP3TC-5_Vir | 178,159 | - | GenBank: OM001476.1 |
| ii) with the 11.9-kbp ColRNAI plasmid | CR-HvKP4TC-2 | pCR-HvKP4TC-2_Vir | 178,159 | - | GenBank: OM001478.1 |
| pCR-HvKP4TC-2_p4 | 11,970 | - | GenBank: OM001477.1 | ||
| iii) fusion with the 11.9-kbp ColRNAI plasmid | CR-HvKP4TC-3 CR-HvKP4TC-4 CR-HvKP5TC-1 CR-HvKP5TC-2 |
pCR-HvKP4TC-3_Vir-p4 | 190,129 | - | GenBank: MW598247.1 |
| iv) with both the 99.7-kbp Incl1 and the 11.9-kbp ColRNAI plasmid independently | CR-HvKP1TC-2 CR-HvKP3TC-1 |
pCR-HvKP3TC-1_Vir | 178,159 | - | GenBank: MW598242.1 |
| pCR-HvKP3TC-1_p3 | 99,764 | + | GenBank: MW598243.1 | ||
| pCR-HvKP3TC-1_p4 | 11,970 | - | GenBank: MW598244.1 | ||
| v) with the 99.7-kbp Incl1 plasmid in the form of a fusion plasmid with the 11.9-kbp ColRNAI plasmid | CR-HvKP1TC-1 CR-HvKP2TC-2 CR-HvKP2TC-3 CR-HvKP4TC-1 CR-HvKP4TC-5 CR-HvKP5TC-3 CR-HvKP5TC-6 |
pCR-HvKP1TC-1_Vir-p4 | 190,129 | - | GenBank: MW598240.1 |
| pCR-HvKP1TC-1_p3 | 99,717 | + | GenBank: MW598239.1 | ||
| vi) fusion with the 99.7-kbp Incl1 plasmid | CR-HvKP2TC-1 | pCR-HvKP2TC-1_Vir-p3 | 277,911 | + | GenBank: MW598241.1 |
| vii) with the 11.9-kbp ColRNAI plasmid in the form of a fusion plasmid with the 99.7-kbp Incl1 plasmid | vii-1) CR-HvKP1TC-3 CR-HvKP1TC-4 CR-HvKP3TC-3 CR-HvKP5TC-4 |
pCR-HvKP1TC-3_Vir-p3 | 277,876 | + | GenBank: OM001474.1 |
| pCR-HvKP1TC-3_p4 | 11,970 | - | GenBank: OM001473.1 | ||
| vii-2) CR-HvKP5TC-5 |
pCR-HvKP5TC-5_Vir-p3 | 278,709 | + | GenBank: OM001480.1 | |
| pCR-HvKP5TC-5_p4 | 11,970 | - | GenBank: OM001479.1 | ||
| viii) fusion with the 99.7-kbp Incl1 plasmid with deletion | CR-HvKP3TC-2 CR-HvKP3TC-4 |
pCR-HvKP3TC-2_Vir-p3 | 244,787 | + | GenBank: OM001475.1 |
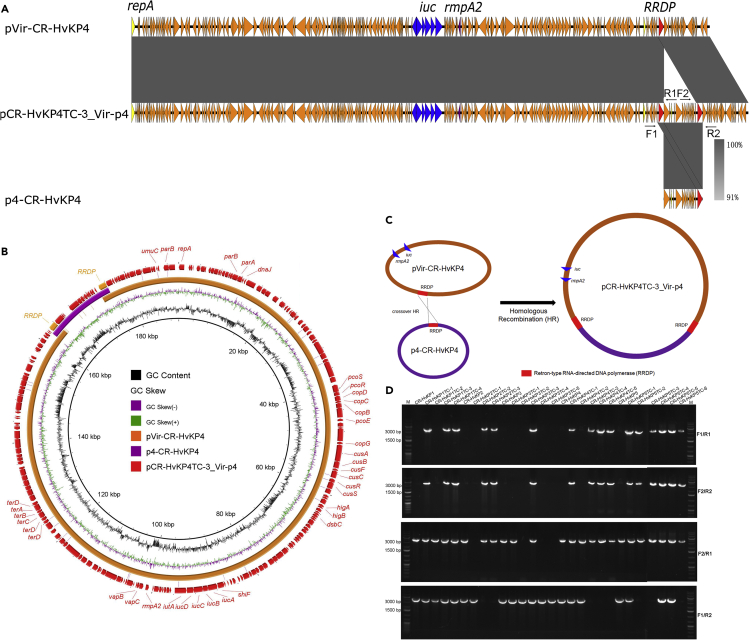
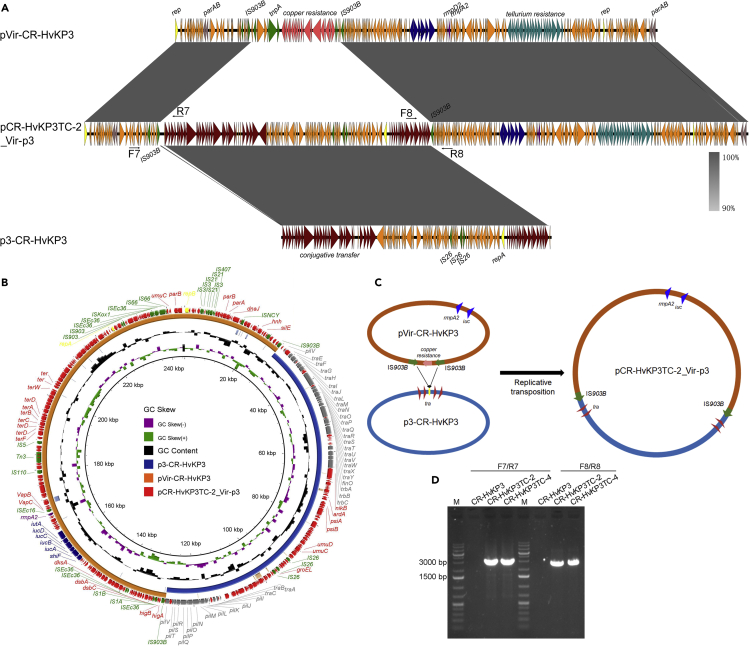
The transconjugants CR-HvKP4TC-3, CR-HvKP4TC-4, CR-HvKP5TC-1, and CR-HvKP5TC-2, which showed one band with the size of 190-kbp in the S1-PFGE analysis were found to harbor only one plasmid with the size of 190,129 bp, which was defined as mode iii. The 190,129 bp plasmid in transconjugant CR-HvKP4TC-3 was found to be a fusion plasmid of pVir-CR-HvKP4 and p4-CR-HvKP4 and was designated as pCR-HvKP4TC-3_Vir-p4 (Figure 2). As fusion product of the virulence plasmid and the ColRNAI plasmid frequently occurred in the transconjugants, we further analyzed the underlying mechanism of its generation. The 11.9-kbp ColRNAI plasmid p4-CR-HvKP4 harbored 13 ORFs, which encoded colicin E3, a mobilization protein and various hypothetical proteins. Further sequence analysis identified a homologous region which encoded the Retron-type RNA-directed DNA polymerase in both pVir-CR-HvKP4 and p4-CR-HvKP4 (Figures 2A, 2B, and S1). A crossover homologous recombination event then took place in this homologous region, leading to integration of these two plasmids and formation of the 190,129 bp fusion plasmid (Figure 2C). As quite a lot of the transconjugants harbored this fusion plasmid, we designed two pairs of primers to amplify sequences that span across both ends of the integration sites of plasmid pCR-HvKP4TC-3_Vir-p4 and performed PCR using all donor strains and transconjugants as templates to check the presence of this kind of fusion plasmid (Figure 2A and Table S1). PCR products could be generated only in transconjugants CR-HvKP1TC-1, CR-HvKP1TC-3, CR-HvKP1TC-4, CR-HvKP2TC-2, CR-HvKP2TC-3, CR-HvKP3TC-3, CR-HvKP4TC-1, CR-HvKP4TC-3, CR-HvKP4TC-4, CR-HvKP4TC-5, CR-HvKP5TC-1, CR-HvKP5TC-2, CR-HvKP5TC-3, CR-HvKP5TC-4, CR-HvKP5TC-5, and CR-HvKP5TC-6, indicating that these two plasmids were not fused in the donor strains and other transconjugants (Figure 2D). We also used the primers F1/R2 and F2/R1 to detect the unfused virulence plasmid and ColRNAI plasmid in the transconjugants. Interestingly, some transconjugants such as CR-HvKP1TC-1, which harbored the fusion plasmid, also harbored unfused plasmids. However, some transconjugants such as CR-HvKP2TC-2 only harbored the unfused ColRNAI plasmid rather than the unfused virulence plasmid. These data indicated that the fusion product of the virulence plasmid and the ColRNAI plasmid could be disintegrated to release the independent virulence and ColRNAI plasmid.
Figure 2.
Fusion of pVir-CR-HvKP4 and p4-CR-HvKP4 through homologous recombination
(A) alignment of plasmid pCR-HvKP4TC-3_Vir-p4 with plasmids pVir-CR-HvKP4 and p4-CR-HvKP4 by easyfig, a colour gradient indicating the identities of the BLAST hits was present.
(B) alignment of plasmid pCR-HvKP4TC-3_Vir-p4 with plasmids pVir-CR-HvKP4 and p4-CR-HvKP4 by BRIG.
(C) Illustration of the process of fusion by pVir-CR-HvKP4 and p4-CR-HvKP4.
(D) PCR assays to confirm the presence of the hybrid plasmid pCR-HvKP4TC-3_Vir-p4 and individual plasmids in CR-HvKP donor strains and their transconjugants.
The transconjugant strain CR-HvKP3TC-1 was found to harbor three plasmids with the size of 178,159 bp, 99,764 bp, and 11,970, respectively, consistent with the S1-PFGE result, which was defined as mode iv. These three plasmids were almost identical to the plasmids pVir-CR-HvKP3, p3-CR-HvKP3, and p4-CR-HvKP3 harbored by the donor strain, indicating that these three plasmids might be transferred to the recipient strain simultaneously. Transconjugant strain CR-HvKP1TC-2 also exhibited the same mode. However, fusion plasmids derived from these three plasmids were not detectable by both S1-PFGE and sequencing, indicating any fusion plasmids might have disintegrated after the conjugation process. The transconjugant strain CR-HvKP1TC-1 was found to harbor two plasmids with the size of 190,129 bp and 99,717 bp, respectively, which was defined as mode v. The 99,717 bp plasmid was found to be identical to plasmid p3-CR-HvKP1 and was designated as pCR-HvKP1TC-1_p3. The 190,129 bp plasmid was found to be a fusion plasmid of pVir-CR-HvKP1 and p4-CR-HvKP1 and was designated as pCR-HvKP1TC-1_Vir-p4, similar to plasmid pCR-HvKP4TC-3_Vir-p4. Transconjugants CR-HvKP2TC-2, CR-HvKP2TC-3, CR-HvKP4TC-1, CR-HvKP4TC-5, CR-HvKP5TC-3, and CR-HvKP5TC-6 also exhibited similar modes. The virulence plasmid or the fusion plasmid with the 11.9-kbp ColRNAI plasmid contained no conjugative genes and was thought to be non-transmissible. Transmissibility of these two plasmids were verified by conjugation using transconjugant strains CR-HvKP3TC-5 and CR-HvKP4TC-3 as donors, respectively, and a K. pneumoniae strain as recipient. No transconjugant was obtained, indicating that the virulence plasmid or the ColRNAI plasmid only was non-transmissible. Because another 99.7-kbp Incl1 conjugative plasmid was also detected in the transconjugants of modes iv and v, we speculated that this plasmid might mediate the conjugation of the virulence plasmid, even though fusion plasmid of these two plasmids was not observed by both PFGE and sequencing in modes iv and v. Nevertheless, plasmids with similar functional role have been reported in a previous study (Chen et al., 2019). However, the conjugative plasmid was not found in the transconjugants of modes i, ii, and iii, indicating that it could be lost among the offsprings of the transconjugants. The reason why the conjugative plasmid was not observable in these transconjugants remains unknown; therefore, further studies are required to depict this phenomenon.
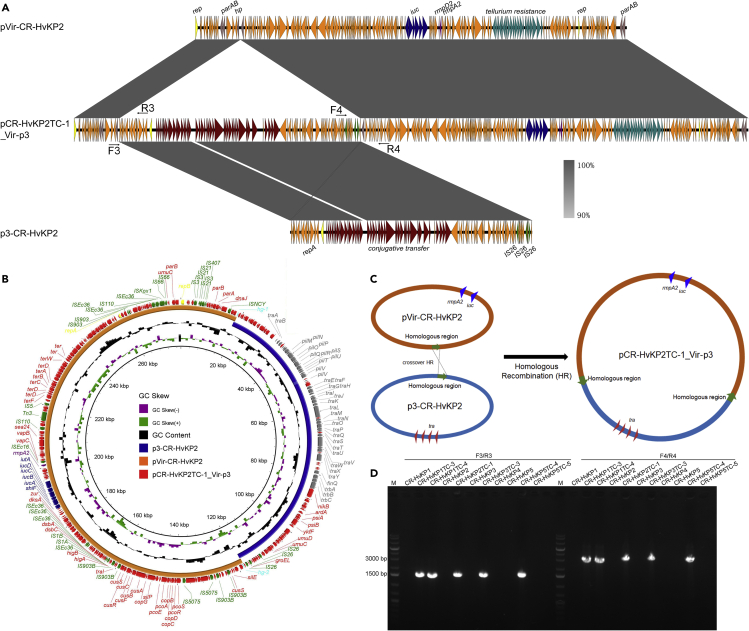
Only one plasmid with a size of 277,911 bp was found in transconjugant CR-HvKP2TC-1, which was defined as mode vi. This plasmid was found to be a fusion plasmid of pVir-CR-HvKP2 and p3-CR-HvKP2 and was designated as pCR-HvKP2TC-1_Vir-p3 (Table 2). Further sequence analysis identified a 259 bp homologous region in both plasmids pVir-CR-HvKP2 and p3-CR-HvKP2, which shared 70% identity (Figures 3A, 3B, and S2). This region was a noncoding region that is located between two hypothetical proteins. A crossover homologous recombination event then took place in this homologous region, leading to integration of these two plasmids and formation of the 277,911 bp fusion plasmid (Figure 3C). These data further confirmed that the 99.7-kbp Incl1 plasmid could mediate the transmission of the virulence plasmid through formation of fusion plasmid. Similarly, we designed two pairs of primers to amplify the region that spans both ends of the integration sites of fusion plasmid pCR-HvKP2TC-1_Vir-p3 and performed PCR on all the donor strains and transconjugants harbored a 280-kbp plasmid (Figures 3A and Table S1). Transconjugants CR-HvKP1TC-3, CR-HvKP1TC-4, CR-HvKP2TC-1, CR-HvKP3TC-3, and CR-HvKP5TC-4 all showed positive bands, whereas CR-HvKP donor strains and transconjugant CR-HvKP5TC-5 were negative for this fusion plasmid (Figure 3D). These findings showed that there might be different modes of plasmid fusion between the virulence plasmid and the conjugative helper plasmid during the conjugation process. A similar fusion plasmid was also identified in transconjugants CR-HvKP1TC-3, CR-HvKP1TC-4, CR-HvKP3TC-3, and CR-HvKP5TC-4. But these four transconjugants also harbored the 11,970 bp ColRNAI plasmid, which was defined as mode vii-1.
Figure 3.
Fusion of pVir-CR-HvKP2 and p3-CR-HvKP2 through a 259 bp homologous region
(A) alignment of plasmid pCR-HvKP2TC-1_Vir-p3 with plasmid pVir-CR-HvKP2 and p3-CR-HvKP2 by easyfig, a colour gradient indicating the identities of the BLAST hits was present.
(B) alignment of plasmid pCR-HvKP2TC-1_Vir-p3 with plasmid pVir-CR-HvKP2 and p3-CR-HvKP2 by BRIG.
(C) Illustration of the process of fusion by pVir-CR-HvKP2 and p3-CR-HvKP2 through a 259 bp homologous region.
(D) PCR assays to confirm the presence of the hybrid plasmid, pCR-HvKP2TC-1_Vir-p3 in CR-HvKP donor strains and their transconjugants.
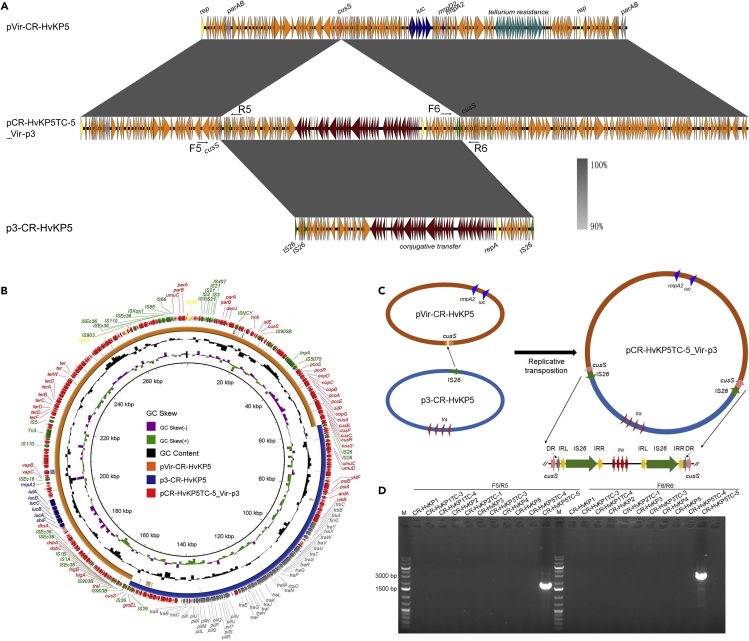
Two plasmids were identified in transconjugant CR-HvKP5TC-5 with the size of 278,709 and 11,970 bp, respectively, defined as mode vii-2 (Table 2). The 11970 bp plasmid was identical to plasmid p4-CR-HvKP5, whereas the 278,709 bp plasmid was found to be a fusion plasmid of pVir-CR-HvKP5 and p3-CR-HvKP5, designated as pCR-HvKP5TC-5_Vir-p3 (Figures 4A, 4B, and S3). Plasmid pCR-HvKP5TC-5_Vir-p3 was not fused as the way of pCR-HvKP2TC-1_Vir-p3. Further analysis indicated that the fusion plasmid was generated by the insertion sequence IS26. Two IS26 were identified at the insertion site of plasmid pCR-HvKP5TC-5_Vir-p3. The two IS26 were both flanked by the 14 bp terminal inverted repeat sequences (IR) and the 8 bp direct target repeats were identified outside each IS26 (Figure 4C). Thus, the IS26 in plasmid p4-CR-HvKP5 targeted the cusS gene in the virulence plasmid, lead to the conjugative plasmid fuse into the virulence plasmid and generate a duplicated IS26 and the 8 bp direct target repeats, resulting in 278,709 bp fusion plasmids (Figure 4C). Similarly, we designed two pairs of primers to amplify the region that spans both ends of the integration sites of fusion plasmid pCR-HvKP5TC-5_Vir-p3 and performed PCR on all the donor strains, and transconjugants harbored a 280-kbp plasmid (Figures 4A and Table S1). Only transconjugant CR-HvKP5TC-5 was tested positive, whereas CR-HvKP donor strains and other transconjugants were negative for this fusion plasmid (Figure 4D).
Figure 4.
Fusion of pVir-CR-HvKP5 and p3-CR-HvKP5 by IS26
(A) alignment of plasmid pCR-HvKP5TC-5_Vir-p3 with plasmid pVir-CR-HvKP5 and p3-CR-HvKP5 by easyfig, a colour gradient indicating the identities of the BLAST hits was present.
(B) alignment of plasmid pCR-HvKP5TC-5_Vir-p3 with plasmid pVir-CR-HvKP5 and p3-CR-HvKP5 by BRIG.
(C) Illustration of the process of fusion by pVir-CR-HvKP5 and p3-CR-HvKP5 by IS26. The structure of the insertion sequences was indicated. Left (IRL, GGCACTGTTGCAAA) and right (IRR, TTTGCAACAGTGCC) terminal 14 bp IRs were shown as yellow filled arrows. The 8 bp direct target repeats (CTTCGTAA) were shown as gray filled arrows.
(D) PCR assays to confirm the presence of the hybrid plasmid, pCR-HvKP5TC-5_Vir-p3 in CR-HvKP donor strains and their transconjugants.
Interestingly, two plasmids were identified in transconjugant CR-HvKP3TC-2 with the size of 244,787 and 11,970 bp, respectively, defined as mode viii (Table 2). The 11,970 bp plasmid was identical to plasmid p4-CR-HvKP3, whereas the 244,787 bp plasmid was found to be a fusion plasmid of pVir-CR-HvKP3 and p3-CR-HvKP3, designated as pCR-HvKP3TC-2_Vir-p3 (Figures 5A, 5B, and S4). Plasmid pCR-HvKP3TC-2_Vir-p3 was shorter than fusion plasmids generated by modes vii-1 and vii-2. Further analysis indicated that the two IS903B that flanked a region encoding the copper resistance in the virulence plasmid mediated the fusion. Briefly, the IS903B targeted the tra gene locus in the plasmid p3-CR-HvKP3. Then a recombination occurred in this region, leading to integration of these two plasmids, excision of the copper resistance encoding region, and formation of the 244,787 bp fusion plasmid (Figure 5C). Similarly, we designed two pairs of primers to amplify the region that spans both ends of the integration sites of fusion plasmid pCR-HvKP3TC-2_Vir-p3 and performed PCR on the donor strain CR-HvKP3 and transconjugants CR-HvKP3TC-2 and CR-HvKP3TC-4, which harbored a 240-kbp plasmid (Figures 5A and Table S1). Both transconjugants CR-HvKP3TC-2 and CR-HvKP3TC-4 were tested positive, while CR-HvKP donor strain was negative for this fusion plasmid (Figure 5D).
Figure 5.
Fusion of pVir-CR-HvKP3 and p3-CR-HvKP3 by IS903B
(A) alignment of plasmid pCR-HvKP3TC-2_Vir-p3 with plasmid pVir-CR-HvKP3 and p3-CR-HvKP3 by easyfig, a colour gradient indicating the identities of the BLAST hits was present.
(B) alignment of plasmid pCR-HvKP3TC-2_Vir-p3 with plasmid pVir-CR-HvKP3 and p3-CR-HvKP3 by BRIG.
(C) Illustration of the process of fusion by pVir-CR-HvKP3 and p3-CR-HvKP3 by IS903B.
(D) PCR assays to confirm the presence of the hybrid plasmid, pCR-HvKP3TC-2_Vir-p3 in donor strain CR-HvKP3 and transconjugants CR-HvKP3TC-2 and CR-HvKP3TC-4.
Plasmid fusion has increasingly been shown to be a major mechanism by which bacteria transfer an originally nonconjugative plasmid to organisms of different types or species, disseminating virulence and antimicrobial resistance determinants in pathogens (Li et al., 2019; Xu et al., 2021; Yang et al., 2019). Homologous recombination and insertion sequences play important roles in these plasmid fusion events (He et al., 2015; Yang et al., 2019). Our data identified several plasmid fusion events through homologous recombination and insertion sequences, which could have occurred in the five CR-HvKP strains, all carrying genetic elements involved in the recombination events.
Prevalence of p3-CR-HvKP1-like conjugative helper plasmids
To investigate the prevalence of p3-CR-HvKP1-like conjugative helper plasmids in different bacterial pathogens, we searched the NCBI database and a total of 85 strains carrying similar conjugative helper plasmids were identified, among which four were K. pneumoniae strains (Table S2 and Figure S5). However, this plasmid was detected much more frequently in E. coli and Salmonella enterica strains from both livestock and homo sapiens (Table S2 and Figure S5). Interestingly, a similar plasmid pSa64-96 from an S. enterica subsp. enterica serovar Derby strain was reported as a conjugative helper plasmid, which could fuse with a nonconjugative ciprofloxacin resistance-encoding plasmid to form a hybrid conjugative ciprofloxacin resistance-encoding plasmid (Chen et al., 2019). Among the four K. pneumoniae strains, three of them also contained the pLVPK-like virulence plasmid (Table S2 and Figure S6). All the three K. pneumoniae strains belonged to ST11 and recovered from homo sapiens, whereas the rest belonged to ST1 and recovered from livestock (Table S2). Furthermore, we screened clinical samples collected in the hospital in which the five CR-HvKP strains were isolated and identified 27 K. pneumoniae strains that harbored both the virulence plasmid and the conjugative helper plasmid (Table S2, Figures S5, and S6), indicating that transmission of virulence plasmid via the help of p3-CR-HvKP1-like helper plasmid has already occurred.
Conclusion
In this work, we showed that the virulence plasmids harbored by our previously reported CR-HvKP strains could be transferred to E. coli strain EC600 via conjugation. Transmission of these virulence plasmids was found to be mediated by events that resulted in formation of fusion plasmids between the virulence plasmid, an Incl1 type conjugative helper plasmid and a small ColRNAI plasmid through homologous recombination and by insertion sequences IS26 and IS903B. Fusion events will lead to generation of new virulence elements in K. pneumoniae and enhanced dissemination of such elements, posing grave threat to human health. Therefore, our findings provide new insight into genetic mechanisms underlying the dissemination of virulence plasmid in Klebsiella species.
Limitations of the study
Various genetic recombination events might have occurred between the plasmids during the conjugation process. However, there might be additional fusion events which were not identified as limited transconjugants were subjected for analysis. Another limitation is that the 99.7-kbp Incl1 conjugative plasmid was demonstrated as a helper plasmid, whereas the role of the ColRNAI plasmid remains unknown.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| K. pneumoniae CR-HvKP1-5 | Patients in our previous studies (Gu et al., 2018; Dong et al., 2018) | |
| E. coli EC600 | Laboratory stock | |
| Deposited Data | ||
| CR-HvKP1 | Dong et al. (2018) | GenBank: CP040534.1-CP040538.1 |
| CR-HvKP2 | This study | GenBank: MW598231.1-MW598233.1; GenBank: MW598248.1-MW598249.1 |
| CR-HvKP3 | This study | GenBank: MW598234.1-MW598238.1 |
| CR-HvKP4 | Dong et al. (2018) | GenBank: CP040540.1-CP040544.1 |
| CR-HvKP5 | Dong et al. (2018) | GenBank: CP040546.1-CP040550.1 |
| CR-HvKP3TC-5 | This study | GenBank: OM001476.1 |
| CR-HvKP4TC-2 | This study | GenBank: OM001477.1-OM001478.1 |
| CR-HvKP4TC-3 | This study | GenBank: MW598247.1 |
| CR-HvKP3TC-1 | This study | GenBank: MW598242.1-MW598244.1 |
| CR-HvKP1TC-1 | This study | GenBank: MW598239.1-MW598240.1 |
| CR-HvKP2TC-1 | This study | GenBank: MW598241.1 |
| CR-HvKP1TC-3 | This study | GenBank: OM001474.1 |
| CR-HvKP5TC-5 | This study | GenBank: OM001479.1-OM001480.1 |
| CR-HvKP3TC-2 | This study | GenBank: OM001475.1 |
| Oligonucleotides | ||
| See Table S1 | ||
| Software and Algorithms | ||
| Unicycler | Wick et al. (2017) | Wick, R.R. |
| RAST | Brettin et al. (2015) | Brettin, T. |
| BLAST Ring Image Generator (BRIG) | Alikhan et al. (2011) | Alikhan, N.F. |
| Easyfig | Sullivan et al. (2011) | Sullivan, M.J. |
| Isfinder&ISsaga | Siguier et al. (2006) | Siguier, P. |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Sheng CHEN (shechen@cityu.edu.hk).
Materials availability
This study did not generate new unique reagent.
Experimental model and subject details
Bacterial strains
E. coli and K. pneumoniae strains were cultured in Luria-Bertani (LB) broth at 37°C with shaking.
Method details
Conjugation experiments
To detect the transmissibility the virulence plasmid, conjugation experiments were performed using the five CR-HvKP strains as donors and rifampin-resistant E. coli strain EC600 as recipient. Both donor and recipient strains were cultured to logarithmic phase (OD∼0.6) at 37°C in LB broth. Then 100 μL culture of the donor cells and 400 μL culture of the recipient cells were mixed and added carefully onto a 0.45 μm membrane which was placed on an LB agar plate. After incubation at 37°C overnight, bacteria on the membrane were collected, resuspended in saline and serially diluted. The virulence plasmids carried the tellurium resistance genes, so transconjugants were selected on China Blue agar plates containing 2 μg/mL potassium tellurite (K2TeO3) and 600 μg/mL rifampin. The presence of the rmpA2 gene as a marker gene of the virulence plasmid in transconjugants was determined by PCR. Antimicrobial sensitive test (AST) was performed by the microdilution method on donor, recipient and transconjugant strains to verify them. Antimicrobials and compounds aztreonam, cefotaxime, ceftazidime, meropenem, amikacin, ciprofloxacin, colistin and potassium tellurite were included for AST. S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) was performed to confirm the transfer of the plasmids. The conjugation efficiency was calculated as the number of transconjugants cells divided by the number of recipient cells.
Whole genome sequencing and bioinformatics analysis
Genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen, USA) according to the manufacturer’s instructions. The extracted DNA was then subjected to library preparation by NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, USA) and sequenced via the 150-bp paired-end NextSeq 500 platform (Illumina, San Diego, CA). Genomic DNA was also subjected to the long-read MinION platform (Oxford Nanopore Technologies, Oxford, United Kingdom). MinION libraries were prepared using the SQK-RBK004 nanopore sequencing kit according to the manufacturer’s instructions. The library was then added to a MinION flow cell (R9. 4.1) and sequenced. Both short and long reads were de novo hybrid assembled using Unicycler v0.4.8 (Wick et al., 2017). Assembled genome sequences were annotated with RAST v2.0 (Brettin et al., 2015). Alignment of plasmids with similar structures were generated by BLAST Ring Image Generator (BRIG) version 0.95.22 (Alikhan et al., 2011) and Easyfig_win_2.1 (Sullivan et al., 2011). Virulence genes were identified by searching against the BIGSdb Klebsiella genome database (Jolley et al., 2018). The BLAST command lines, with an 80% coverage and identity cutoff, were used to map genome sequences against plasmid replicons and insertion sequences (ISs). The plasmid replicons databases were obtained from the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/). The insertion sequences (ISs) were identified using Isfinder and ISsaga (Siguier et al., 2006).
Detection of fusion plasmids by polymerase chain reaction
To detect the presence of the fusion plasmids pCR-HvKP1TC-1_Vir-p4, pCR-HvKP2TC-1_Vir-p3, pCR-HvKP5TC-5-Vir_p3 and pCR-HvKP3TC-2-Vir_p3, we designed two pairs of primers to amplify sequences at each end of the integration sites, respectively. The primer sequences and the expected sizes of products were listed in Table S1. The positions of these primers were shown in Figures 2A, 3A, 4A, and 5A.
Quantification and statistical analysis
The conjugation efficiency was calculated as the number of transconjugants cells divided by the number of recipient cells.
Acknowledgments
This work was supported by the Guangdong Major Project of Basic and Applied Basic Research (2020B0301030005) .
Author contributions
X.M.Y. performed the experiment and drafted the manuscript. L.W.Y. performed the DNA sequencing. M.M.X., Q.X., C.Y., and N.D. helped with the experiment. E.W.C.C. edited the manuscript and contributed to experimental design. R.Z. and S.C. designed and supervised the study and interpreted the data. S.C. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104428.
Supplemental information
Data and code availability
Complete sequences of plasmids of strains CR-HvKP1-5 and transconjugants of different modes have been deposited in the GenBank database and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
References
- Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Dong N., Chan E.W., Chen S. Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microbes Infect. 2019;8:857–865. doi: 10.1080/22221751.2019.1626197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Yang X., Zhang R., Chan E.W., Chen S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg. Microbes Infect. 2018;7:146. doi: 10.1038/s41426-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., Chan E.W.C., Shu L., Yu J., Zhang R., Chen S. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- He S., Hickman A.B., Varani A.M., Siguier P., Chandler M., Dekker J.P., Dyda F. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio. 2015;6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M.M.C., Wyres K.L., Wick R.R., Judd L.M., Fostervold A., Holt K.E., Lohr I.H. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 2019;74:1218–1222. doi: 10.1093/jac/dkz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Cheng J., Dong H., Li L., Liu W., Zhang C., Feng X., Qin S. Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int. J. Antimicrob. Agents. 2020;55:105952. doi: 10.1016/j.ijantimicag.2020.105952. [DOI] [PubMed] [Google Scholar]
- Li R., Xie M., Dong N., Lin D., Yang X., Wong M.H.Y., Chan E.W.-C., Chen S. Erratum to: efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. GigaScience. 2019;8:giz031. doi: 10.1093/gigascience/giz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczosa M.K., Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016;80:629–661. doi: 10.1128/mmbr.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T.A., Olson R., Fang C.T., Stoesser N., Miller M., MacDonald U., Hutson A., Barker J.H., La Hoz R.M., Johnson J.R., et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 2018;56 doi: 10.1128/jcm.00776-18. e00776–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Ma G., Li C., Jia X., Qin C., Yang T., Wang L., Jiang X., Ding N., Zhang X., et al. Emergence of a multidrug-resistant hypervirulent Klebsiella pneumoniae sequence type 23 strain with a rare bla CTX-M-24-harboring virulence plasmid. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/aac.02273-18. e02273–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon A.S., Russo T.A. Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol. 2012;7:669–671. doi: 10.2217/fmb.12.43. [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton J., Davies F., Turton J., Perry C., Payne Z., Pike R. Hybrid resistance and virulence plasmids in "High-Risk" clones of Klebsiella pneumoniae, including those carrying blaNDM-5. Microorganisms. 2019;7:326. doi: 10.3390/microorganisms7090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres K.L., Wick R.R., Judd L.M., Froumine R., Tokolyi A., Gorrie C.L., Lam M.M.C., Duchene S., Jenney A., Holt K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:e1008114. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Yang X., Xu Q., Ye L., Chen K., Zheng Z., Dong N., Sun Q., Shu L., Gu D., et al. Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun Biol. 2021;4:650. doi: 10.1038/s42003-021-02148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhang J., Wang M., Liu M., Liu G., Qu H., Liu J., Deng Z., Sun J., Ou H.-Y., Qu J. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 2021;13:119. doi: 10.1186/s13073-021-00936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Dong N., Chan E.W.C., Zhang R., Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29:65–83. doi: 10.1016/j.tim.2020.04.012. [DOI] [PubMed] [Google Scholar]
- Yang X., Wai-Chi Chan E., Zhang R., Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. 2019;4:2039–2043. doi: 10.1038/s41564-019-0566-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete sequences of plasmids of strains CR-HvKP1-5 and transconjugants of different modes have been deposited in the GenBank database and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.