Summary
In nature, bacteria reside in biofilms– multicellular differentiated communities held together by an extracellular matrix. This work identified a novel subpopulation—mineral-forming cells—that is essential for biofilm formation in Bacillus subtilis biofilms. This subpopulation contains an intracellular calcium-accumulating niche, in which the formation of a calcium carbonate mineral is initiated. As the biofilm colony develops, this mineral grows in a controlled manner, forming a functional macrostructure that serves the entire community. Consistently, biofilm development is prevented by the inhibition of calcium uptake. Our results provide a clear demonstration of the orchestrated production of calcite exoskeleton, critical to morphogenesis in simple prokaryotes.
Subject areas: Microbiology, Microbiofilms, cell biology
Graphical abstract
Highlights
-
•
The orchestrated formation of calcite scaffolds supports the morphogenesis of microbial biofilms
-
•
A novel subpopulation—mineral-forming cells—is essential for biofilm formation
-
•
This subpopulation contains an intracellular calcium-accumulating niche, supporting the formation of calcium carbonate
-
•
Intracellular calcium homeostasis and calcium export are associated with a functional biofilm macrostructure
Microbiology; Microbiofilms; Cell biology
Introduction
Although bacteria have been historically studied in homogeneous monocultures, they are typically found in natural ecosystems and clinical settings in interface-associated multicellular communities called biofilms (Kolter and Greenberg, 2006). It was recently indicated that in soils, oceans, the deep subsurface, phyllosphere, and animal microbiomes, 40–80% of bacteria reside in biofilms (Flemming and Wuertz, 2019). Bacterial biofilms are differentiated communities in which the cells are held together by an extracellular matrix. The formation of a biofilm is frequently favored over a free-living planktonic lifestyle, as it increases the fitness of member cells promoting their survival, such as better attachment to hosts, division of metabolic labor, and protection from the environment.
A hallmark of biofilms is their ability to form robust and complex 3D structures, and the distinctive spatial organization provides the residing bacteria with several benefits. The biofilm architecture was suggested to relieve metabolic stress. For example, channels formed below the ridges and wrinkles within the colony may facilitate the diffusion of fluids, nutrients, and oxygen (Bloom-Ackermann et al., 2016; Dietrich et al., 2013; Kolodkin-Gal et al., 2013; Wilking et al., 2013). Furthermore, cells located in different areas of the colony are exposed to varying levels of oxygen, nutrients, and quorum sensing molecules, which affect the genetic programs they express, promoting differentiation (Asally et al., 2012; Hassanov et al., 2018; Liu et al., 2015; Monds and O'Toole, 2009; Serra and Hengge, 2014; Stewart and Franklin, 2008). Thus, community structure allows the genetically identical biofilm cells to display phenotypic heterogeneity and endure stressful environments. A biofilm colony model often enables the study of microbial microenvironments, reflecting different aspects of this complex system. In a biofilm colony growing on agar plates, nutrients come from the medium located below, oxygen comes from the surrounding atmosphere, and carbon dioxide comes from microbial respiration and the surrounding atmosphere (Povolotsky et al., 2021).
Owing to physical protection offered by the biofilm biomass, differentiation, and physiological adaptations, cells residing deep within the biofilm are protected from environmental assaults, such as antibiotics.
Until recently, the ability of biofilm-forming bacteria to generate complex architectures was attributed exclusively to self-produced organic extracellular matrix (ECM) (Dragos and Kovacs, 2017; Reichhardt and Parsek, 2019; Steinberg and Kolodkin-Gal, 2015), composed of carbohydrate-rich polymers (i.e., lipids or exopolysaccharides), proteins, and nucleic acids (Branda et al., 2005). However, we and others have recently shown that microbial biofilms contain an organized internal mineral structure composed of crystalline calcium carbonate (calcite) that also contributes to their 3D morphology (Keren-Paz et al., 2018; Keren-Paz and Kolodkin-Gal, 2020; Li et al., 2015). The formation of minerals is induced in bacterial biofilms through passive surface-mediated processes promoted by by-products of bacterial metabolism. Biologically induced minerals associated with bacteria include oxides of Fe, Mn, and other metals; metal sulfates and sulfites; phosphates and carbonates; and Fe and Fe-Al silicates (Frankel and Bazylinski, 2003). Of all examples of biomineralization, microbial-induced calcium carbonate precipitation (MICCP) is most frequently associated with microbial communities (Weiner and Dove, 2003; Douglas and Beveridge, 1998). In geological settings, bacterial metabolic processes, such as urea hydrolysis by urease-positive bacteria, increase the local concentration of bicarbonate and elevate pH. When enough environmental calcium is present, those changes promote spontaneous precipitation of calcium carbonate, with bacterial envelopes serving as nucleation sites for the growing mineral, leading to the formation of crystalline calcium carbonate (Dhami et al., 2013). Similar processes were also suggested to promote biofilm-associated calcification of Proteus mirabilis on catheters (Morris and Stickler, 1998) and during dual-species biofilm formation (Li et al., 2016a) and to contribute to Pseudomonas aeruginosa virulence in an insect host (Lotlikar et al., 2019). In clinical and environmental scenarios, calcium deposition in biofilms mainly was seen as an uncontrolled and unintentional by-product of bacterial metabolic activity.
Recently, intracellular amorphous calcium carbonate (ACC) granules were detected in some species of Gram-negative autotrophic bacteria (Blondeau et al., 2018). Those intracellular deposits were suggested to contribute to single-cell energy metabolism by promoting photosynthesis (Blondeau et al., 2018) or chemolithoautotrophy (Monteil et al., 2020). Although the precise molecular mechanisms regulating this process are still unresolved, the presence of a microcompartment dedicated to mineralization raises the possibility that some bacteria might be capable of controlling the formation of the biogenic mineral.
Our recent discovery of precisely organized mineral macrostructures within biofilms suggests that biofilm biomineralization is tightly controlled. These calcite scaffolds contributed to the fitness of biofilm colonies in two unrelated soil bacteria: Bacillus subtilis and Mycobacterium smegmatis (Keren-Paz et al., 2018; Oppenheimer-Shaanan et al., 2016). For both, the mineral structure acted as a structural scaffold supporting the 3D architecture of the colony and as a diffusion barrier preventing the penetration of solutes into the biofilms. The formation of controlled calcium carbonate macrostructures in phylogenetically distinct heterotrophic biofilms raises the possibility that the function of calcium carbonate is not limited to specific species utilizing it for intracellular chemical energy but is instead a general phenomenon in the bacterial kingdom.
Those reports challenge the current view of biofilm development as a process depending solely on organic ECM production. They raise several fundamental questions regarding the developmental role of mineral scaffold formation within a bacterial community and the relation between structure and function in microbial biofilms. If mineralization is a controlled process resulting in a structure crucial to biofilm development and function, this is a novel aspect of basic biofilm biology. What is the role of mineral structure in the development of a differentiated biofilm community? Are there dedicated cells initiating mineral production? What are the fitness advantages that regulated mineralization confers to the community as a whole?
In this work, we discovered that the calcium-dependent 3D organization of the biofilm colony leads to transcriptional reprogramming of the bacterial community and is essential for biofilm development. Calcite formation and the assembly of a functional mineralized scaffold supporting the 3D morphology of a biofilm colony were associated with defined intracellular calcium-rich deposits present in a distinct subpopulation of biofilm cells, differing from flagellin and organic ECM producers.
Our results identify a previously overlooked process essential for B. subtilis biofilm development — the tightly regulated formation of mineral scaffolds by dedicated cells. We expect future research exploring this newly discovered process to shed further light on mechanisms of bacterial development.
Results
Calcium regulation of B. subtilis biofilms
In previous work, we demonstrated that B. subtilis biofilm morphology is calcium-dependent (Oppenheimer-Shaanan et al., 2016). Removing calcium acetate from the undefined biofilm-promoting B4 medium prevented the development of the complex 3D architecture characteristic of a biofilm colony without affecting bacterial growth. This 3D structure required calcium (as no equivalent cation could replace it and as various calcium salts induced it) and metabolically produced carbonate (as it was eliminated under anaerobic conditions and increased in a CO2-enriched environment). The mineral component was identified as calcite by FTIR and XRD (Oppenheimer-Shaanan et al., 2016). It spanned the entire colony and was spatially co-localized with colony wrinkles, suggesting it acted as a “scaffold” supporting them (Oppenheimer-Shaanan et al., 2016). The mineral structure developed and increased over time as the colony grew (Keren-Paz et al., 2018). Finally, as the biofilm colonies aged and their 3D spatial structure diminished, the internal and organized mineral deteriorated (potentially because of fermentation and acidification of the medium by oxygen-depleted cells within the inner mass of the aging biofilm), and only large nonstructural crystals were left in the colony periphery because of a passive growth of calcite crystals (Oppenheimer-Shaanan et al., 2016). During biomineralization, the formation of exopolysaccharides and the amyloid protein TasA was also essential for complex morphology formation, and interactions between the organic and inorganic matrix were evident (Oppenheimer-Shaanan et al., 2016).
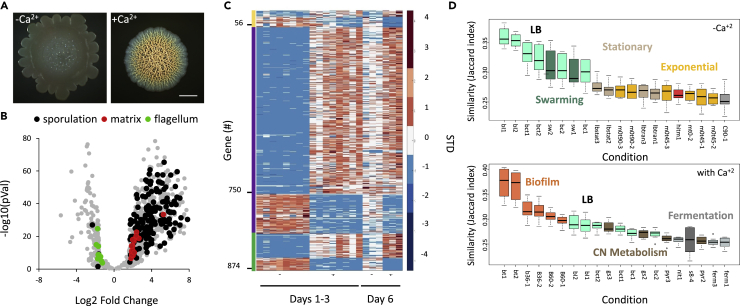
To better understand the molecular mechanisms underlying the formation of functional mineral macrostructures, we examined the effect of calcium on gene expression in a B. subtilis biofilm colony by sequencing the transcriptome of colonies grown either with or without excess calcium (Figure 1A). The addition of calcium resulted in a dramatic reprogramming of the transcriptional profile of the colony, with 20% of the genome (n = 875) significantly changing between the two conditions at all time-points tested (File S1). Specific developmental processes defining the biofilm state, such as induction of ECM production and sporulation, and the repression of motility, were observed (Figure 1B). These fundamental changes were mostly sustained over time (Figure 1C). We next compared the two transcriptional signatures to a collection of published data containing 269 mRNA profiles of B. subtilis grown under over 100 different conditions (Nicolas et al., 2012) (Figure 1D). The transcriptome of cells grown with excess calcium was similar to that of previously analyzed biofilm cells, regardless of the biofilm-promoting medium used. On the other hand, the transcription profile of colonies grown without added calcium was similar to that of planktonic cells experiencing nutrient starvation but was not identified as biofilm in this unbiased analysis.
Figure 1.
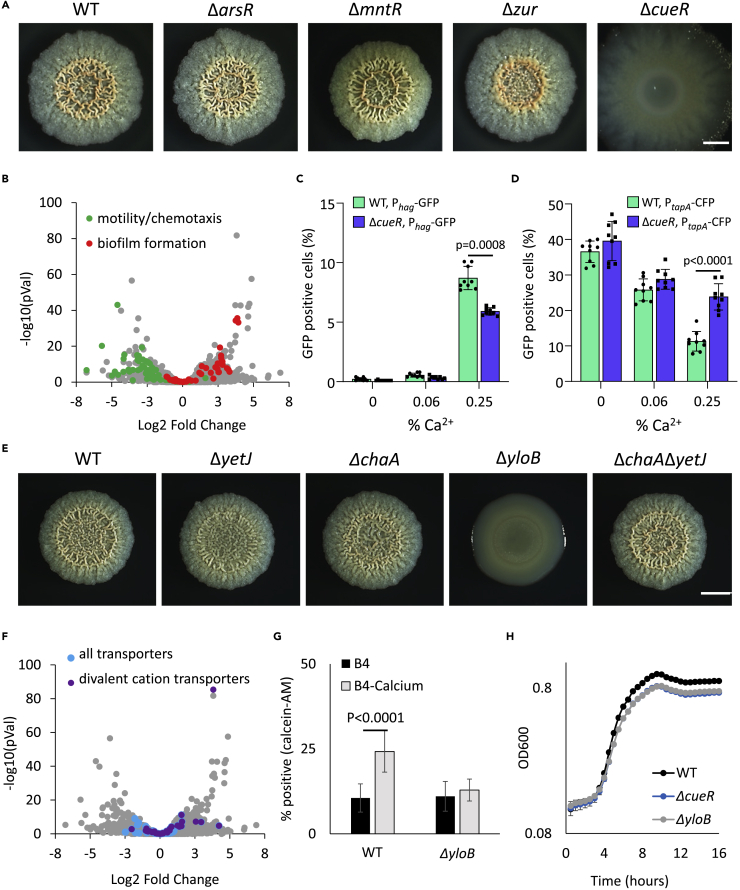
Calcium is necessary for normal biofilm development and structure
(A) Light microscopy images of 6-day-old B. subtilis NCIB 3610 biofilm colonies grown on B4 agar, showing the effect of addition of calcium (0.25% v/v calcium acetate) on colony architecture. Scale bar – 2 mm. A representative image (out of n = 3 experiments) is shown.
(B) Volcano-plot depicting calcium-dependent changes in the transcriptome in B. subtilis biofilm colonies. Significance (p values) vs difference (fold changes) values are plotted. Functional categories were determined by DAVID analysis.
(C) A heatmap with all differentially expressed genes (n = 876), scaled to the mean expression level of each gene. The map is ordered by age of biofilm colonies as shown by the left colored bar, where green are genes that are differentially expressed in days 1–3, and day 6; purple genes that are differentially expressed in days 1–3 and yellow genes that are differentially expressed in day 6.
(D) The top 20 conditions (Nicolas et al., 2012), showing the highest similarity (expressed as Jaccard index) to the transcriptome of B. subtilis 6-day-old biofilm colonies grown with or without calcium.
Visualizing the extracellular calcium carbonate in B. subtilis biofilms
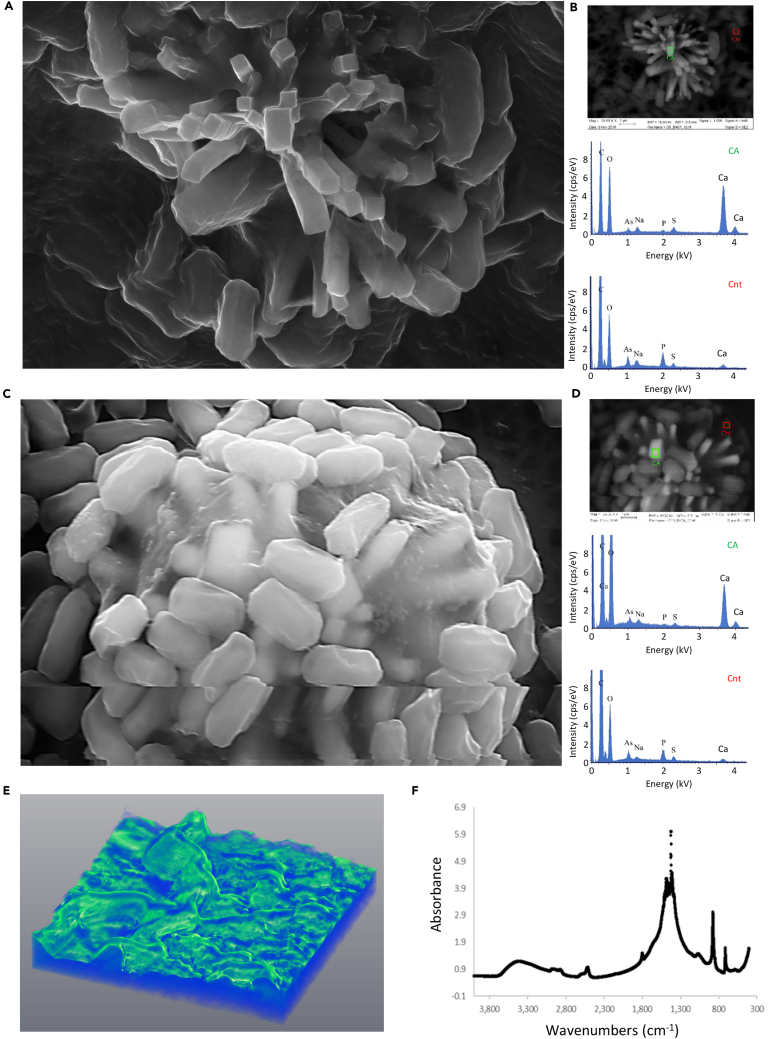
These findings highlight the central role of calcium in biofilm development and suggest an intimate connection between biofilm structure and function, raising the possibility that microbial cells are actively regulating calcium carbonate biomineralization. Indeed, when a biofilm colony was visualized by scanning electron microscopy (SEM), we observed a highly mineralized subpopulation of cells tightly associated with mineral crystals (Figures 2A–2D). Consistent with our previous characterization of bleached biogenic calcite, the unbleached crystals had rough faces, a smooth and flat crystal face, and displayed elongated prismatic morphology instead of the rhombohedral morphology of calcite grown in pure solution (Oppenheimer-Shaanan et al., 2016).
Figure 2.
Calcite mineralization occurs in distinct areas within the biofilms
(A–D). Biofilm cells closely associated with prismatic mineral structures, as visualized by Scanning Electron Microscopy (SEM) image of 8 (A and B) and 10-day-old (C and D) B. subtilis biofilm colonies grown with calcium. (A and C) - secondary mode; (B and D) upper panel - backscattering mode. Magnification – X25000, scale bar – 1 μm. A representative field (out of n = 5 fields, from six experiments) is shown. (B and D) Lower panel - EDX analysis of boxed areas, indicating that mineral structures are rich in calcium. Ca – calcium-rich, Cnt – control.
(E) 3D reconstruction of microCT X-ray analysis revealing 3D mineral distribution in a 5-day old B. subtilis colony grown with calcium. Color indicates intensity, with green representing the densest mineral. Scale bar – 0.2 mm. A representative image (out of n = 3 experiments) is shown.
(F) FTIR spectra of bleached B. subtilis biofilm colonies, displaying vibrations characteristic of calcite (crystalline calcium carbonate).
The crystals were consistent with biogenic calcite generated by B. subtilis (Oppenheimer-Shaanan et al., 2016) as judged by shape, backscatter mode, and energy dispersive X-ray spectroscopy (EDX) (Figures 2B and 2D). In the absence of added calcium, mineral crystals were rarely detected (Figure S1).
To create the 3D reconstruction, a whole, unfixed bacterial colony was transferred to a plastic slide for a microCT scan and rotated between the X-ray source and the detector positioned at optimal distances for a voxel size of 0.87 μm. 2D projections were taken at different angles until a full rotation (360°) was completed. The full set of images was then used to reconstruct the whole volume of the sample by back-projection algorithm, and thus a high-resolution 3D image was generated (see Videos S1 and S2). This microCT X-ray scan confirmed that the crystalline calcium is organized in a macrostructure, developing a nonuniform continuous layer throughout the wrinkles (Figure 2E). Consistently with our previous observations (Oppenheimer-Shaanan et al., 2016), the accumulated mineral was calcite (Figure 2F).
B. subtilis colony. The intact colony was visualized under X-ray, and the obtained 2D images were used to generate high-resolution 3D images. Rotation of 360° was taken around an axis perpendicular to the biofilm surface. Results are of a representative experiment out of three independent repeats
intact colony was visualized as in supplementary movie 1
Intracellular calcium carbonate is detected in a subpopulation of biofilm cells
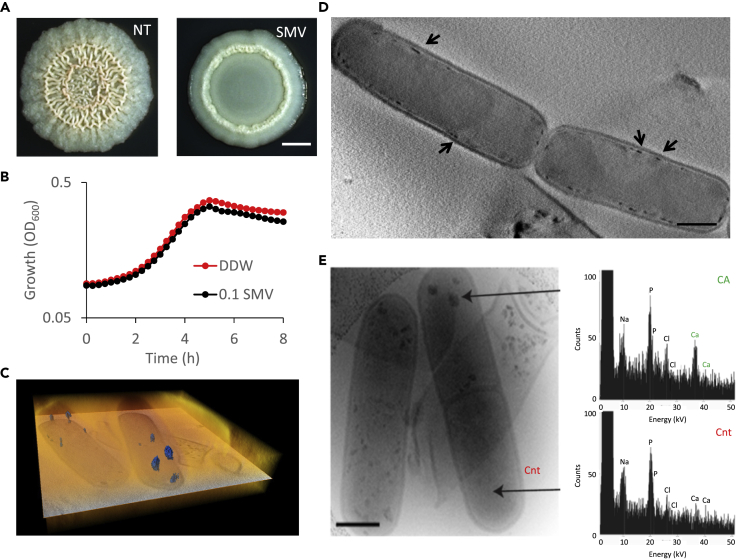
The tight association of calcium carbonate minerals with the bacterial cells led us to examine the role of intracellular calcium. Disruption of intracellular calcium homeostasis by a chemical inhibitor of P-type ATPases sodium metavanadate (SMV) (Clausen et al., 2016; Guragain et al., 2013; Kuhlbrandt, 2004) prevented the formation of a complex 3D structure even in the presence of excess calcium (Figure 3A). This morphological defect was not because of inhibition of planktonic growth, which was only mildly affected by SMV (Figure 3B). This need for controlled intracellular calcium levels raised the possibility that carbonate mineralization starts intracellularly. One possible mechanism linking calcium mineralization and colony structure is that, in the presence of calcium, some kind of specialized cells located in specific regions of the colony would serve as nucleation sites and promote localized mineral formation. Therefore, we next examined single cells from biofilms grown with excess calcium.
Figure 3.
Calcium storage in biofilm cells
(A) Light microscopy images of 5-day-old B. subtilis biofilm colonies grown with calcium, and supplemented with calcium uptake inhibitor (0.1 mg/mL sodium metavanadate (SMV)), as indicated. NT indicated untreated culture. Scale bar – 2 mm. A representative image (out of n = 3 experiments) is shown.
(B) The effect of inhibiting calcium uptake on planktonic growth. B. subtilis was grown shaking in liquid B4 medium with calcium, supplemented with 0.1 mg/mL SMV, as indicated. Results are averages of nine wells. Bars represent standard deviations. A representative experiment (out of n = 3 experiments) is shown.
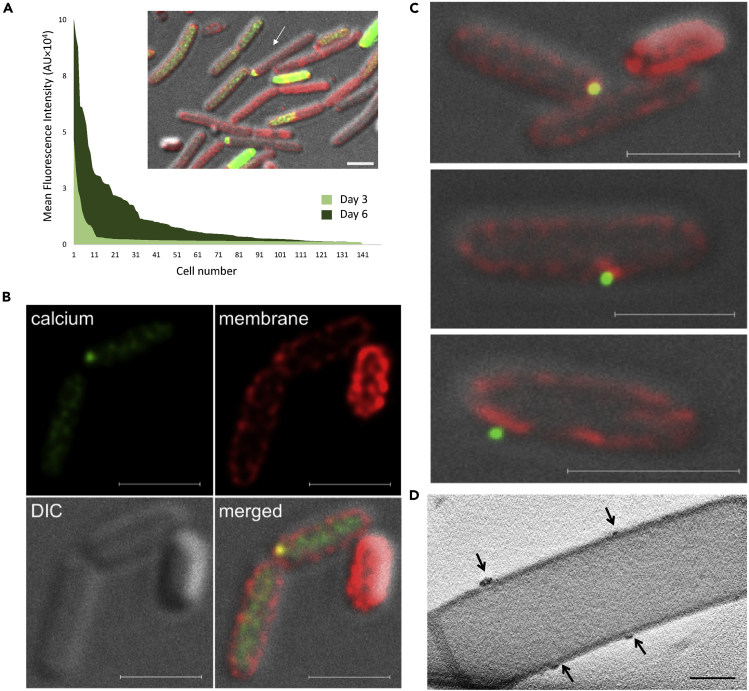
(C) Volume rendering (orange) and a single orthoslice (greyscale) through the center of the volume, from a 3D CSTET reconstruction. The calcium-rich deposits are artificially colored (blue).
(D) A 30-nm thick virtual slice through a CSTET 3D reconstruction of B. subtilis cells showing intracellular calcium-rich deposits (arrows). Scale bar – 400 nm. A representative field (out of n = 20 fields, from three experiments) is shown.
(E) Left panel - Bright-field STEM image of representative B. subtilis biofilm cells showing cellular calcium deposits, from a colony grown 10 days on B4-Ca2+ agar. The black arrow indicates the cell used for EDX analysis in panels (B-C). Right panel – EDX analysis of the calcium deposits. A representative mineralizing cell imaged (out of n = 50 cells, from four independent experiments performed with EDX is shown.
3D reconstructions of cryo-fixed bacteria with scanning transmission electron tomography (CSTET) (Wolf et al., 2014) (Figure 3C) revealed calcium dense foci inside the cells associated with the membrane. CryoSTEM imaging and cryo-EDX measurements verified excess calcium in these deposits (Figures 3D and 3E). The appearance of these deposits was calcium-dependent, as we could not observe them in cells grown without added calcium (Figure S2). Next, we examined the distribution of cellular calcium levels within the biofilm community. Although the cytoplasm of most cells was stained evenly, a small fraction of cells (1.6% out of 300 cells examined) displayed concrete, membrane-localized calcium deposits (Figure S3), comparable in their frequency, distribution, and localization with calcium deposits identified by CSTET (Figures 3C–3E). Although the mineral-producing subpopulation was small, it was enriched in the structured zones of the colony (Figure S3).
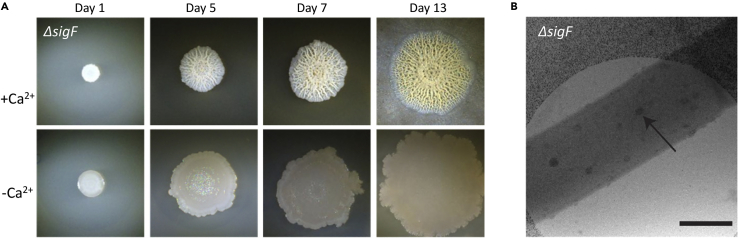
Biomineralization can take place in the absence of sporulating cells
The observed induction of sporulation regulon transcription by calcium (Figure 1B) raised the possibility that spores could serve this function, possibly because of the unique membrane properties of the spore. However, sporulation does not seem to be involved in cellular calcium deposition, consistent with our previous observation that a mutant strain lacking sporulation sigma factor SigF (de Hoon et al., 2010), which does not form spores, was able to create highly structured colonies in a calcium-dependent manner (Figure 4A), suggesting that biomineralization promotes biofilm development and sporulation, and not vice versa. Moreover, ΔsigF cells contained calcium-rich foci similar to those of wild type (Figure 4B). Finally, as this mutant strain is blocked in the early stages of sporulation and as cells that accumulate calcium granules did not exhibit an asymmetric septum, mineral producers are considered as a different subpopulation from sporulating cells.
Figure 4.
Biofilm mineralization is sporulation independent
(A) Light microscopy images of ΔsigF mutant strain. Biofilm colonies were grown on B4-Ca2+ agar for the indicated time at 30°C. The experiment was repeated 3 times, in a technical quadruplicate – and representative images are shown.
(B) Cryo-STEM bright-field images of representative biofilm cells from a ΔsigF colony grown for 10 days on B4-Ca2+ agar. Black arrows point to aggregates with edge-on views (left panel) and top views (right panel). Scale bars 500 nm. A representative field imaged (out of n = 30 cells), from four independent experiments is shown.
Identifying the regulators of calcium homeostasis in B. subtilis biofilms
In B. subtilis, the transcriptional regulation of calcium uptake is poorly characterized. Therefore, we examined the potential involvement of several general transcriptional regulators of cation uptake. Non-ferric metal homeostasis in bacteria is regulated by the MntR, MerR, and ArsR families of transcriptional regulators (Moore and Helmann, 2005). Strains lacking arsR, mntR, and zur developed robust and structured biofilm colonies. In contrast, ΔcueR mutants had a severely altered biofilm phenotype and formed completely flat colonies (Figure 5A). CueR/YhdQ is a poorly characterized transcriptional regulator belonging to the metal-responsive MerR family (Moore and Helmann, 2005). CueR/YhdQ binds but does not regulate the promoter of copper transporting P-type ATPase CopA (Smaldone and Helmann, 2020). Consistantly, the deletion of copA has no effect on biofilm morphology (Figure S4). Those observations suggest that CueR/YhdQ regulates additional, yet undiscovered, P-type ATPase involved in calcium uptake. To characterize the regulon of CueR/YhdQ we repeated the transcriptome analysis of the wild-type strain and ΔcueR mutant, both grown in the presence of calcium. Compared to the wild-type colony, the expression of matrix genes was upregulated, and motility was inhibited in the ΔcueR mutant, suggesting a general dysregulation of biofilm transcriptional profile (Figure 5B and File S2). Biofilm formation is a tightly regulated developmental process. Both inhibition and overexpression of key components cause severe defects in biofilm development (for example, both deletion and over-activation of the biofilm regulator Spo0A prevent normal biofilm development (Veening et al., 2006). High ECM gene expression levels further indicate that the featureless morphology of the ΔcueR mutant is not a result of the lack of organic ECM components. The calcium-dependent repression of motility and induction of organic ECM was also demonstrated in the WT but not its ΔcueR mutant derivative by flow cytometry (Figures 5C and 5D). As the defect of ΔcueR is not because of a regulatory role of the ECM, we suspected it could alter calcium homeostasis by regulating cation influx.
Figure 5.
Calcium homeostasis supports biofilm development
(A) Light microscopy images of 3-day-old biofilm colonies of wild-type B. subtilis and its mutant derivatives (see text for details) grown with calcium. Scale bar – 2 mm. A representative experiment (out of n = 3 experiments) is shown.
(B) Volcano-plot depictingCueR/YhdQ -dependent changes in expression of biofilm genes in 3-day old B. subtilis biofilm colony. Significance (p values) vs difference (fold changes) values are plotted. Functional categories were determined by DAVID analysis.
(C and D) A dual reporter of WT B. subtilis strain harboring Phag-GFP (C) and PtapA-CFP (D) and its ΔcueR deletion mutant were inoculated on B4 plates with either no calcium (no Ca), 0.0625% Ca or 0.25% Ca. Data were collected from 24 h post inoculation, 100,000 cells were counted. Y axis represents the % of cells expressing the reporters, graphs represent mean ± SD from three independent experiments (n = 9). Statistical significance was determined by two-way ANOVA with Sidak’s multiple comparison test. p < 0.05 was considered statistically significant.
(E) Light microscopy images of 2-day-old biofilm colonies of wild-type B. subtilis and its mutant derivatives (see text for details) grown with calcium. Scale bar – 2 mm. A representative experiment (out of n = 3 experiments) is shown.
(F) Volcano-plot depicting CueR/YhdQ -dependent changes in expression of transporter genes in 3-day old B. subtilis biofilm colony. Significance (p values) vs difference (fold changes) values are plotted. Functional categories were determined by DAVID analysis.
(G) Intracellular calcium levels of bacterial cells of indicated strains (n > 200) were stained with calcein-AM, a calcium-specific fluorescent dye. Cells were isolated from B. subtilis biofilm colonies (day 3) grown with or without calcium. % Calcein-AM positive cells were determined by ImageJ (Schneider et al., 2012). Results represent average and SD of a representative experiment (out of n = 3 experiments) performed with five technical repeats. Statistical significance was determined by two tailed Student’s t- test. p < 0.05 was considered statistically significant.
(H) The effect of inhibiting calcium uptake on planktonic growth. Indicated strains were grown shaking in liquid B4 medium with calcium. Results are averages of five wells, bars represent standard deviation. A representative experiment (out of n = 3 experiments) is shown.
Previous studies suggested a role for CueR/YhdQ in the homeostasis of magnesium (Pi et al., 2020; Wendel et al., 2022). However,the subsequent analysis of the expression of all divalent cation transporters revealed that their expression is significantly altered in ΔcueR mutant, whereas other transporters are less affected (Figure 5F). Among those, we identified three calcium transporters with transcriptional levels significantly changed in the mutant: yloB (ATP-driven Ca2+ pump) (Gupta et al., 2017), chaA (H+/Ca2+ exchanger) (Fujisawa et al., 2009), and yetJ (pH-dependent calcium leak channel) (Chang et al., 2014) (File S2). Deletion of yloB was sufficient to prevent 3D colony morphology formation (Figure 5E). On biomineralization medium B4, magnesium does not induce the formation of complex morphologies in Bacillus subtilis (Oppenheimer-Shaanan et al., 2016), but magnesium transporters are regulated by CueR/YhdQ under our conditions (Supporting File 2). Collectively these results suggest that CueR/ YhdQ is involved in the coordination of Mg/Ca homeostasis.Onlthough magnesium transporters are also regulated by CueR/YhdQ under our conditions (File S2). Collectively these results suggest that CueR/ YhdQ has involved in the coordination of Mg/Ca homeostasis.
Notably, calcium homeostasis and internal calcium (as judged by cueR mutation) affect the transcriptome differently than external calcium (Figure 1D and (Oppenheimer-Shaanan et al., 2016)). The expression of extracellular matrix genes increases with the increase in extracellular calcium but is also increased in the absence of CueR/YhdQ (Figure 5C). Interestingly, the effect of calcium on ECM expression cannot be explained by the response of the transcriptome to the loss of complex morphology: wild-type biofilms grown in the absence of extracellular calcium and cueR mutants (in the presence and absence of calcium) are both featureless, but their effects on the expression of the ECM genes are inverse.
Calcium storage and export are detected in a subpopulation of biofilm cells
To better comprehend the dynamic between intracellular and extracellular calcium in living biofilm cells, we stained biofilm cells with calcein-AM – a dye that can be used to visualize intracellular calcium in live cells (Hale et al., 2000). This lipophilic stain can freely enter the cells but only binds calcium and becomes fluorescent after cellular esterases remove the acetoxymethyl (AM) groups. Indeed ΔyloB biofilm cells reduced calcium uptake (Figures 5G and S5), indicating the importance of calcium uptake and homeostasis. Although colony biofilm formation was blocked altogether in both cueR and yloB mutants, only a very slight defect was observed in their planktonic growth (Figure 5H). The deletion of yetJ and chaA, alternate calcium channels regulated by CueR/YhdQ had little or no effect on biofilm morphology (Figure 5E) and did not enhance the effect of yloB deletion (Figure S6).
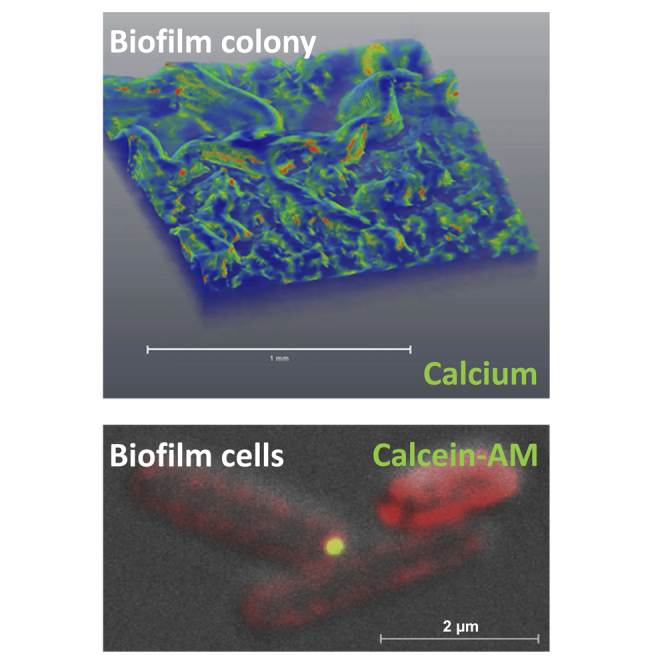
To better comprehend the dynamic between intracellular and extracellular calcium in living biofilm cells, we stained live biofilm cells with calcein-AM. We relied on Gated-STED fluorescent microscopy to image intracellular calcium with high resolution. This analysis revealed heterogeneity in cellular calcium levels within the biofilm colony, with most cells containing low calcium levels, whereas a smaller fraction had very high levels (Figure 6A). As the biofilm developed and the calcified structures matured, the number of cells showing a strong calcein signal increased. Furthermore, we could detect calcium-rich deposits associated with the cell envelope (Figure 6B)—on the cytoplasm side, embedded within the membrane, and closely associated with the cell from the outside (Figure 6C)—suggesting different stages of export. Similar calcium deposits were also observed by CSTET in close association with the outer membrane of bacterial cells (Figure 6D). These findings support the existence of a calcium-concentrating compartment/niche in a subpopulation of heterotrophic biofilm cells, similar to that recently discovered in autotrophic bacteria (Blondeau et al., 2018), and that the mechanism for its formation involves an active and regulated calcium uptake as well as the export of the stored calcium. Exporting calcium deposits may be essential for them to grow into functional mineral scaffolds via interactions with the dense extracellular matrix (Keren-Paz and Kolodkin-Gal, 2020).
Figure 6.
The dynamics of intracellular calcium storage in biofilm cells and its subsequent secretion
(A) Intracellular calcium levels of bacterial cells (n = 150) stained with calcein-AM, a calcium-specific fluorescent dye. Cells were isolated from B. subtilis biofilm colonies (day 3 and 6) grown with calcium. Inset – a representative image of stained cells isolated from a 6-day-old biofilm. Green – calcein-AM, red – NileRed membrane stain, gray – DIC. Scale bar – 2 μm.
(B and C) G-STED images B. subtilis cells isolated from 6-day-old biofilm colonies grown with calcium. Green – calcein-AM, red – NileRed membrane stain, gray – DIC. Scale bar – 2 μm.
(D) 30-nm thick virtual slice through a CSTET 3D reconstruction. Arrows indicate calcium-rich deposits associated with a membrane from the outside of the cell. Scale bar – 400 nm.
These observations suggest that the first critical step in the formation of calcite minerals is initiated in saturated mineral precursor microenvironments within the cell. After the initial nucleation, the precursor seems to be exported out of the bacterial cell without hampering the membrane integrity of its producer and to further grow on organic ECM templates in the highly complex biofilm microenvironment until an organized functional mineral macrostructure is formed.
Discussion
Bacterial biofilms are widespread in nature, forming multicellular colonies on various surfaces. The molecular mechanisms guiding the complex network of events that transition from a free-living planktonic bacterium to a differentiated community are well characterized. Much is known about the production of the organic matrix, the division of labor between the different cell types, the metabolic adaptations, and the methods of communication between the community members; besides, the fitness advantages of such a communal lifestyle are evident.
In addition, bacterial biofilms were extensively shown to promote biomineralization of metals (such as gold and iron), phosphate, and carbonate (Konhauser et al., 2008). This work uncovers a previously overlooked but fundamental connection between the highly regulated biofilm development and the seemingly random biomineralization occurring within biofilms. We propose that bacterial communities actively control the formation of mineral macrostructures and that those “exoskeletons” play an important role in biofilm development and fitness.
The first clues to the presence of a structural mineral component in a biofilm came when we demonstrated that a mature 3D structure of biofilm colonies of B. subtilis depends on their ability to precipitate precisely organized patterns of calcium carbonate (Oppenheimer-Shaanan et al., 2016). Controlled deposition of a calcium carbonate scaffold structurally supports the complex architecture of biofilm colonies and protects them from the environment by limiting solute diffusion (Keren-Paz et al., 2018).
This work builds on those preliminary observations to show that crystalline calcium carbonate production is a controlled process fundamental to biofilm development. Only when calcification occurs, B. subtilis can successfully activate and maintain the characteristic biofilm transcriptional program, activating matrix production and sporulation while repressing motility (Figure 1). As observed by us previously, the transcription of ECM genes is induced by the extracellular calcium [(Oppenheimer-Shaanan et al., 2016), File S1]; however, mutation in cueR enhances the transcription of ECM genes, potentially as a nonspecific response to disturbane of calcium homeostasis, aimed to overcome the defect in biofilm development.
Mechanistically, we suggest that in the presence of sufficient calcium, the formation of mineral macrostructures is initiated within intracellular compartment/dedicated cellular niches within a small subpopulation of cells, which secrete small calcium-rich granules, while remaining intact (Figures 3 and 6). The process is likely actively controlled, as the membrane integrity of the exporting cells is not compromised. Initiating mineral formation within the controlled cellular environment allows the organism to regulate the composition (e.g., the purity and identity of the crystalline mineral), structure, and physical properties of the mineral, thereby controlling the final function (Benzerara et al., 2014; Komeili, 2012; Kovacs et al., 2021). In contrast, the lack of biological regulation is expected to result in heterogeneous mineral compositions with poor crystallinity, including large size variations, poorly defined crystal morphologies, and the inclusion of impurities (Jiang et al., 2020; Konhauser et al., 2008). Consistently with the existence of a biological regulation, the structured biomineralization in B. subtilis biofilms cannot be explained by passive calcium saturation (Figure S7).
The best known example of bacteria producing functional minerals formed in a defined intracellular compartment/niche by a highly regulated cellular pathway is the magnetotactic bacteria, forming invagination of membranes to generate magnetosomes (Komeili et al., 2006). Recently, intracellular calcium carbonate mineralization was reported in cyanobacteria (Blondeau et al., 2018) and in a subgroup of poorly characterized magnetotactic bacteria (Monteil et al., 2020). Unlike the aforementioned examples, which occur in a specific group of phylogenetically related planktonic aquatic bacteria, the formation of mineralized macrostructures in biofilms is conserved across the microbial domain, highlighting their potential importance for general microbial fitness, unrelated to specific adaptations of single-cell microbial metabolism.
We found here that in a subpopulation of cells, calcium uptake and functionally conserved pathways of carbonate production alter the intracellular microenvironment. Therefore, the intracellular calcium homeostasis is involved in mineral nucleation and the formation of a mineral macrostructure. We found that calcium accumulation in biofilm cells is heterogeneous (Figure 6A) probably because of differential uptake of calcium, but the accumulation of calcium alone is insufficient to explain dense calcium foci, occurring in a subset of calcium storing cells (Figure 6A). The subpopulation that is capable of storing mineralized calcium (Figures 3 and 6) may differ in their internal membrane properties (as suggested by the observed membrane invagination around the calcium deposits), their intracellular pH, promoting mineralization or the presence of a protein nucleator (Bazylinski et al., 1995).
Numerous studies of B. subtilis, P. aeruginosa, and additional biofilm formers, established that complex colony architecture is dependent on the organic extracellular matrix (Branda et al., 2005; Colvin et al., 2012; Jones and Wozniak, 2017; Kobayashi and Iwano, 2012; Romero et al., 2010; Vlamakis et al., 2013), and thus it was hypothesized that the presence of the organic ECM components is sufficient for bacterial biofilm morphogenesis. We found that nonorganic ECM components are also necessary for the development of structured bacterial communities. Although our results indicate that mineral formation initiates within the cells and requires a controlled intracellular environment, the final mineral macrostructure assembly has to occur in the extracellular microenvironment (requiring export), guided by complex local interactions with the extracellular organic matrix (Keren-Paz and Kolodkin-Gal, 2020). The evidence for these interactions are consistent with our observations with scanning electron microscopy (Figures 2A and 2B and FTIR analysis (Oppenheimer-Shaanan et al., 2016)). Therefore, both organic ECM components and biogenic minerals are essential but not sufficient for the formation of complex structures. Furthermore, their interaction and timely export are probably imperative to generate the functional biominerals in microbial biofilms.
Although in our settings bacteria relied on calcite to generate functional scaffolds, other minerals may serve similar functions. For example, deposition of calcium phosphate crystals in P. mirabilis, P. vulgaris, and Providencia rettgeri biofilms blocks catheters in infected patients (Broomfield et al., 2009; Mathur et al., 2006; Tan et al., 2018). In addition, urease-dependent calcium phosphate mineralization was shown to increase P. mirabilis resistance to ciprofloxacin (Li et al., 2016b) and increase its survival in dual-species biofilms (Li et al., 2016a). Importantly, the physiological role and regulation of calcium phosphate deposits within these medical biofilms remain to be determined.
Our work suggests that it is time to reconsider the old definition of bacterial mineralization solely as a passive, unintended byproduct of bacterial environmental activity. Instead, here we provide evidence that it can also be a regulated developmental process originating from a dedicated subcellular environment, conferring clear benefits to biofilm-residing bacteria. Biomineralization is highly conserved and thus is of enormous clinical significance, as it can yield completely novel classes of broad-spectrum drugs to combat emerging biofilm infections.
Limitations of the study
Although this study has demonstrated the role of intracellular calcium carbonate in biofilm formation in B. subtilis, we have not provided direct evidence that calcium carbonate (either crystalline or amorphous) is actively secreted to interact with the organic extracellular matrix. The suggested roles of the P-type ATPases YloB and the regulator CueR in this dynamic process are pending additional research. Lastly, the importance of intracellular and extracellular calcium for biofilm formation in the rhizosphere and soil remains to be determined.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Bacillus subtilis 168 | Lab collection | N/A |
| Bacillus subtilis NCIB3610 | Lab collection | N/A |
| Bacilus subtilis deletion mutants (See Method details) | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| sodium metavanadate (SMV) | Sigma-Aldrich | Cat. #590088 |
| Plasmids | ||
| pAH54 for specR | Lab collection | N/A |
| pAH52 for ermR | Lab collection | N/A |
| Deposited data | ||
| RNA seq - with vs without calcium | This study | GSE199052 |
| RNA seq - cueR deletion mutant vs wild type | This study | GSE199052 |
| Oligonucleotides | ||
| Oligonucleotides (See File S3) | This study | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ilana Kolodkin-Gal (ilana.kolodkin@mail.huji.ac.il)
Materials availability
Bacterial strains generated in this study are available from the lead contact upon request.
Experimental model and subject details
Bacterial strains
B. subtilis 168, NCIB3610 and their derivate mutant strains were maintained in standard LB medium at 30°C, unless specified otherwise.
A list of strains used in this study can be found in Key resources table below.
Method details
Bacterial strains
Deletions of arsR, mntR, zur, cueR, copA and chaA were generated by transforming B. subtilis NCIB 3610 (Branda et al., 2001) with genomic DNA isolated from B. subtilis 168 deletion library (Koo et al., 2017) (Addgene, Cat#1000000115), or when indicated 3610 derivative (Steinberg et al., 2020) and verified by PCR. Deletion of yetJ and yloB was done by long flanking homology (LFH-PCR). Briefly, a plasmid with an antibiotic cassette (pAH54 for specR, pAH52 for ermR was used as a template to generate of a long DNA fragment containing an antibiotic-resistance gene flanked by 1-kb-long regions directly upstream and downstream to the target gene to be deleted. The resulting linear PCR product was used to transform a wild type B. subtilis strain, transformants were selected for the presence of the resistance cassette (by plating on the appropriate antibiotic plates), and the absence of the target gene was verified by PCR. The list of primers can be found in Key resources table below. Double and triple mutants were generated by natural transformation with genomic DNA, and verified by PCR.
Biofilm assays
B. subtilis biofilms were grown on B4 biofilm-promoting solid medium (0.4% yeast extract, 0.5% glucose, and 1.5% agar) (Barabesi et al., 2007) supplemented with calcium acetate at 0.25% v/v, incubated at 30°C, in a sealed box with a lit candle for enriched CO2 environment (candle jar method (Oppenheimer-Shaanan et al., 2016).
When indicated, the medium was supplemented with sodium metavanadate (SMV) (Sigma-Aldrich, Cat. #590088). The concentrations used are indicated in the text.
Phase microscopy
Biofilm colonies were observed using a Nikon D3 camera or a Stereo Discovery V20″ microscope (Tochigi, Japan) with objectives Plan Apo S ×0.5 FWD 134 mm or Apo S × 1.0 FWD 60 mm (Zeiss, Goettingen, Germany) attached to Axiocam camera, as required. Data were captured using Axiovision suite software (Zeiss).
RNA extraction and library preparation
Biofilm colonies were grown on biofilm-promoting B4 solid medium with and without calcium for 1, 2, 3 and 6 days. Three independent experiments were conducted, with three colonies from each treatment combined for RNA extraction in each experiment. The samples were frozen in liquid nitrogen and stored until extraction. Frozen bacterial pellets were lysed using the Fastprep homogenizer (MP Biomedicals) and RNA was extracted with the FastRNA PROT blue kit (MP Biomedicals, 116025050) according to the manufacturer’s instructions. RNA levels and integrity were determined by Qubit RNA BR Assay Kit (Life Technologies, Q10210) and TapeStation, respectively. All RNA samples were treated with TURBO DNase (Life Technologies, AM2238).
A total of five ug RNA from each sample was subjected to rRNA depletion using the Illumina Ribo-Zero rRNA Removal Kit (Bacteria, MRZB12424), according to the manufacturers’ protocols. RNA quantity and quality post-depletion was assessed as above. RNA-seq libraries were contracted with NEBNext® Ultra™ Directional RNA Library Prep Kit (NEB, E7420) according to the manufacturer’s instructions. Libraries concentrations and sizes were evaluated as above, and were sequenced as multiplex indexes in one lane using the Illumina HighSeq2500 platform.
RNAseq processing
Reads were trimmed from their adapter with cutadapt and aligned to the B. subtilis genome (subsp. subtilis str. NCIB 3610, NZ_CM000488.1) with Bowtie2 version 2.3.4.1 (Langmead and Salzberg, 2012). The number of uniquely mapped reads per gene was calculated with HT-seq (Anders et al., 2015). Normalization and testing for differential expression was performed with DESeq2 v1.16. A gene was considered to be differentially expressed using the following criteria: normalized mean read count ≥30, fold change ≥3, and adjusted p value <0.05. First, we tested for differential expression between samples grown with and without calcium separately for each time point; however, since the results for days 1, 2 and 3 were very similar, we joined days 1–3. The crude read count, normalized read count, and the result of the differentially expression tests are available in File S1.
Comparison between growth conditions
We compared our RNAseq expression data to publically available transcriptomes representing 269 different growth conditions (Nicolas et al., 2012). Because that study used microarray platform and not RNAseq, the comparison was performed using the top 10% genes with the highest expression level of every condition and every replicate (383 genes per sample). We than used Jaccard index to measure the overlap between the conditions of the two platforms (i.e. the current study and (Nicolas et al., 2012)). Prior to the analysis, we removed 152 genes that appear among the top 10% in more than 80% of the conditions.
Scanning electron microscopy and EDX
Biofilm colonies were grown for 1, 3, 6, 10 and 15 days at 30°C on biofilm-promoting B4 solid medium, with or without calcium. The colonies were fixed overnight at 4°C with 2% glutaraldehyde, 3% paraformaldehyde, 0.1 M sodium cacodylate (pH 7.4) and 5 mM CaCl2, dehydrated and dried (Bucher et al., 2015, 2016).
Mounted samples were coated with 15 nm thick carbon layer in carbon coater (EDWARDS). The imaging by secondary electron (SE) or back scattered electron (BSE) detectors and the Energy Dispersive X-ray Spectroscopy (EDS, Bruker) were performed using Carl Zeiss Ultra 55 or Supra scanning electron microscopes.
MicroCT X-Ray analysis
Images of indicated magnification were taken using a Zeiss micro XCT 400 instrument (Pleasanton, CA, USA). Tomography was carried out using a micro-focused source set at 20 kV and 100 μA. 1200 separate 2D images were taken with a pixel size of 0.87 mm over 1800, exposure time of 30 s. Image analysis was carried out with Avizo software (VSG, Hillsboro, OR, USA).
FTIR spectrophotometer analysis
Calcite was collected as described by Mahamid et al., with some modifications (Mahamid et al., 2008): agar samples were slightly bleached with 6% sodium hypochlorite for 1 min to remove organic matter, washed with Milli-Q water twice and dehydrated in acetone.
FTIR spectra of the bleached samples were acquired in KBr pellets by using a NICOLET iS5 spectrometer (Thermo Scientific, Pittsburgh, PA, USA). The samples were homogenized in an agate mortar and pestle with about 40 mg of KBr, and pressed into a 7 mm pellet using a manual hydraulic press (Specac, Orpington, UK). Infrared spectra were obtained at 4 cm−1 resolution for 32 scans.
The infrared calcite spectrum has three characteristic peaks, designated v2, v3, and v4. Calcium carbonate ν3 peak is expected at 1425 cm−1 for calcite, 1490 cm−1 for vaterite and 1475 cm-1 for aragonite. The calcium carbonate ν2 peak is expected at 875 cm−1 for calcite, 850 cm−1 for vaterite and 855 cm−1 for aragonite. Finally, the calcium carbonate ν4 peak is expected at 713 cm−1 for calcite, 750 cm−1 for vaterite and 715 cm−1 for aragonite (Politi et al., 2004).
Planktonic growth assays
All strains were grown from a single colony isolated over lysogeny broth (LB) plates to a mid-logarithmic phase of growth (4 h at 37°C with shaking). Cells were diluted 1:100 in 150 μL liquid B4 medium in 96-well microplate (Thermo Scientific). Cells were grown at 30°C for 20 h in a microplate reader (Synergy 2, BioTek), and the optical density at 600 nm (OD600) was measured every 15 min. Three independent experiments were conducted, with three technical repeats per plate.
Cryo-STEM analysis
Bacterial colonies grown as described were suspended in PBS buffer. Quantifoil TEM grids were glow-discharged with an Evactron Combi-Clean glow-discharge device, and 5 μL of suspended cells were deposited onto the glow-discharged grids. Ten nm-diameter gold fiducials (Duchesne et al., 2008) were applied before blotting and verification using a Leica EM-GP automated plunging device (Leica). No chemical fixation was used to ovoid artifacts
Vitrified samples were observed with a Tecnai F20 S/TEM instrument (Thermo Fisher Scientific) at 200 kV, with Gatan 805 brightfield and Fischione HAADF detectors. Microscope conditions: extraction voltage = 4300 V, gun lens = 3 or 6, and spot size = 5 or 6 with 10 μm condenser apertures, yielding probe diameters of 1–2 nm and semi-convergence angles of ∼1.3–2.7 mrad. Images of 2048 × 2048 pixels were recorded with probe dwell times of 8–18 μs. Spatial sampling was set between 1 and 4 nm/pixel. Electron doses were 1–3 electrons/A2 per image. Single-axis tilt series were recorded using SerialEM (Kremer et al., 1996). EDX was performed in STEM mode on vitrified cell samples with the same electron microscope set-up as used for STEM imaging, using a liquid N2 cooled Si(Li) detector (EDAX).
Tomography reconstructions and visualization
The CSTET tomographic tilt series were aligned using fiducial markers and reconstructed using weighted back projection (Frangakis and Hegerl, 2001) as implemented in the IMOD software suite (Kremer et al., 1996). Reconstructions are displayed after non-linear anisotropic diffusion filtering within IMOD. Segmentation and volume rendering were performed using Amira 6.3 (FEI Visualization Sciences Group).
Quantification of cells with mineral deposits using STEM
Vitrified grids contacting 100–200 cells per grid were automatically imaged on a Talos Arctica (Thermo Fisher Scientific) microscope in STEM mode, and mapped at intermediate magnifications (10–13k) using SerialEM software to obtain large fields of view. The quantification was done by manually counting the cells with or without dense deposits.
Flow cytometry
Starter cultures of indicated strains were inoculated on B4 plates with varying calcium concentrations. The plates were incubated at 30°C and biofilms colonies were harvested at 24 h post inoculation. The biofilms were suspended in PBS and sonicated using BRANSON digital sonicator, at an amplitude of 10% for 10 s. After sonication samples were diluted in PBS and flow cytometry was performed using LSR-II 597 cytometer (Becton Dickinson, San Jose, CA, USA). Fluorescence of GFP positive cells was measured using laser excitation of 488 nm, coupled with 505 599 LP and 525/50 sequential filters, while CFP fluorescence was measured using a solid state violet 407 nm laser, coupled with 450/50 sequential filters. A total of 100,000 cells were counted and flow cytometry analyses was performed using FACS Diva (BD biosciences).
STED image acquisition
Cells were isolated from 3 to 6 day old biofilm colonies and stained with Calcein-AM (20 μM) and NileRed (100 μg/mL). Immediately after the staining procedure, the cells were mounted on a coverslip and immobilized by an agarose pad (1% agarose, S750 minimal media). STED microscopy was performed using a Leica SP8 confocal microscope with a 100x oil immersion objective (NA: 1.4). A 552 nm laser line was used for confocal detection of NileRed, a 494 nm excitation laser and a 592 nm depletion laser were used for G-STED detection of Calcein-AM. Channels were acquired with 200 Hz (4 x line averaging) and the appropriately set HyD hybrid detectors. Images processing was performed using Leica LAS X and the deconvolution of the respective channels was performed using the Huygens algorithm.
Quantification and statistical analysis
All details of quantification methods, statistical analysis and significance calculations are discussed in each figure legend.
Acknowledgments
This research was supported by the Israel Science Foundation grant number 119/16, Israel Foundation grant number JSPS 184.20, Kamin grant by Israel Chief Scientist no. 67459, Israel Ministry of Science, Technology & Space (grant no. 713454), Ministry of Health (grant no. 713645), Angel-Fiavovich fund for ecological research, Dr. Barry Sherman Institute for Medicinal Chemistry, Kekst Family Institute for Medical Genetics and by the Helen and Milton A. Kimmelman Center for Biomolecular Structure & Assembly. The electron microscopy studies were partially supported by the Irving and Cherna Moskowitz Center for Nano and BioNano Imaging (Weizmann Institute of Science). We thank Prof. Ehud Banin and to Itzhak Zander (Bar-Ilan University) for PA strains and E.B. for helpful discussions.
Author contributions
I.K.G. designed the study. A.K.P., S.G.W., I.K., i.k.g A.G., H.M., S.K., V.B., E.K., and S.D. designed and performed experiments. T.O. analyzed the transcriptome data. I.K.G. and P.G. mentored. a.k.p and I.K.G. wrote the paper.
Declaration of interest
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104308.
Supplemental information
Bacterial strains used in this study and their construction details
Differentially expressed genes – wild type B. subtilis, with vs without excess calcium
Differentially expressed genes – wild type B. subtilis vs cueR deletion mutant
Data and code availability
-
•
RNA-seq data have been deposited at NCBI GEO: GSE199052 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All data reported in this paper, including microscope images, will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asally M., Kittisopikul M., Rue P., Du Y., Hu Z., Cagatay T., Robinson A.B., Lu H., Garcia-Ojalvo J., Suel G.M. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl. Acad. Sci. U S A. 2012;109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabesi C., Galizzi A., Mastromei G., Rossi M., Tamburini E., Perito B. Bacillus subtilis gene cluster involved in calcium carbonate biomineralization. J. Bacteriol. 2007;189:228–235. doi: 10.1128/jb.01450-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazylinski D.A., Frankel R.B., Heywood B.R., Mann S., King J.W., Donaghay P.L., Hanson A.K. Controlled biomineralization of magnetite (Fe(inf3)O(inf4)) and Greigite (Fe(inf3)S(inf4)) in a magnetotactic bacterium. Appl. Environ. Microbiol. 1995;61:3232–3239. doi: 10.1128/aem.61.9.3232-3239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzerara K., Skouri-Panet F., Li J., Ferard C., Gugger M., Laurent T., Couradeau E., Ragon M., Cosmidis J., Menguy N., et al. Intracellular Ca-carbonate biomineralization is widespread in cyanobacteria. Proc. Natl. Acad. Sci. U S A. 2014;111:10933–10938. doi: 10.1073/pnas.1403510111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau M., Sachse M., Boulogne C., Gillet C., Guigner J.M., Skouri-Panet F., Poinsot M., Ferard C., Miot J., Benzerara K. Amorphous calcium carbonate granules form within an intracellular compartment in calcifying cyanobacteria. Front. Microbiol. 2018;9:1768. doi: 10.3389/fmicb.2018.01768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom-Ackermann Z., Steinberg N., Rosenberg G., Oppenheimer-Shaanan Y., Pollack D., Ely S., Storzi N., Levy A., Kolodkin-Gal I. Toxin-Antitoxin systems eliminate defective cells and preserve symmetry in Bacillus subtilis biofilms. Environ. Microbiol. 2016;18:5032–5047. doi: 10.1111/1462-2920.13471. [DOI] [PubMed] [Google Scholar]
- Branda S.S., Gonzalez-Pastor J.E., Ben-Yehuda S., Losick R., Kolter R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda S.S., Vik S., Friedman L., Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Broomfield R.J., Morgan S.D., Khan A., Stickler D.J. Crystalline bacterial biofilm formation on urinary catheters by urease-producing urinary tract pathogens: a simple method of control. J. Med. Microbiol. 2009;58:1367–1375. doi: 10.1099/jmm.0.012419-0. [DOI] [PubMed] [Google Scholar]
- Bucher T., Oppenheimer-Shaanan Y., Savidor A., Bloom-Ackermann Z., Kolodkin-Gal I. Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ. Microbiol. Rep. 2015;7:990–1004. doi: 10.1111/1758-2229.12346. [DOI] [PubMed] [Google Scholar]
- Bucher T., Kartvelishvily E., Kolodkin-Gal I. Methodologies for studying B. Subtilis biofilms as a model for characterizing small molecule biofilm inhibitors. J. Vis. Exp. 2016:54612. doi: 10.3791/54612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Bruni R., Kloss B., Assur Z., Kloppmann E., Rost B., Hendrickson W.A., Liu Q. Structural basis for a pH-sensitive calcium leak across membranes. Science. 2014;344:1131–1135. doi: 10.1126/science.1252043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen J.D., Bublitz M., Arnou B., Olesen C., Andersen J.P., Moller J.V., Nissen P. Crystal structure of the vanadate-inhibited Ca2+-ATPase. Structure. 2016;24:617–623. doi: 10.1016/j.str.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Colvin K.M., Irie Y., Tart C.S., Urbano R., Whitney J.C., Ryder C., Howell P.L., Wozniak D.J., Parsek M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M.J., Eichenberger P., Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 2010;20:R735–R745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhami N.K., Reddy M.S., Mukherjee A. Biomineralization of calcium carbonate polymorphs by the bacterial strains isolated from calcareous sites. J. Microbiol. Biotechnol. 2013;23:707–714. doi: 10.4014/jmb.1212.11087. [DOI] [PubMed] [Google Scholar]
- Dietrich L.E.P., Okegbe C., Price-Whelan A., Sakhtah H., Hunter R.C., Newman D.K. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J. Bacteriol. 2013;195:1371–1380. doi: 10.1128/jb.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S., Beveridge T.J. Mineral formation by bacteria in natural microbial communities. Fems Microbiol. Ecol. 1998;26:79–88. doi: 10.1111/j.1574-6941.1998.tb00494.x. [DOI] [Google Scholar]
- Dragos A., Kovacs A.T. The peculiar functions of the bacterial extracellular matrix. Trends Microbiol. 2017;25:257–266. doi: 10.1016/j.tim.2016.12.010. [DOI] [PubMed] [Google Scholar]
- Duchesne L., Gentili D., Comes-Franchini M., Fernig D.G. Robust ligand shells for biological applications of gold nanoparticles. Langmuir. 2008;24:13572–13580. doi: 10.1021/la802876u. [DOI] [PubMed] [Google Scholar]
- Flemming H.C., Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- Frangakis A.S., Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Frankel R.B., Bazylinski D.A. 4. Biologically induced mineralization by bacteria. Rev. Mineral. Geochem. 2003;54:95–114. doi: 10.1515/9781501509346-009. [DOI] [Google Scholar]
- Fujisawa M., Wada Y., Tsuchiya T., Ito M. Characterization of Bacillus subtilis YfkE (ChaA): a calcium-specific Ca2+/H+ antiporter of the CaCA family. Arch. Microbiol. 2009;191:649–657. doi: 10.1007/s00203-009-0494-7. [DOI] [PubMed] [Google Scholar]
- Gupta H.K., Shrivastava S., Sharma R. A novel calcium uptake transporter of uncharacterized P-type ATPase family supplies calcium for cell surface integrity in Mycobacterium smegmatis. mBio. 2017;8 doi: 10.1128/mbio.01388-17. e01388–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guragain M., Lenaburg D.L., Moore F.S., Reutlinger I., Patrauchan M.A. Calcium homeostasis in Pseudomonas aeruginosa requires multiple transporters and modulates swarming motility. Cell Calcium. 2013;54:350–361. doi: 10.1016/j.ceca.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale L.V., Ma Y.F., Santerre R.F. Semi-quantitative fluorescence analysis of calcein binding as a measurement of in vitro mineralization. Calcified Tissue Int. 2000;67:80–84. doi: 10.1007/s00223001101. [DOI] [PubMed] [Google Scholar]
- Hassanov T., Karunker I., Steinberg N., Erez A., Kolodkin-Gal I. Novel antibiofilm chemotherapies target nitrogen from glutamate and glutamine. Scientific Rep. 2018;8:7097. doi: 10.1038/s41598-018-25401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Liu X., Yin H., Liang Y., Liu H., Miao B., Peng Q., Meng D., Wang S., Yang J., Guo Z. The utilization of biomineralization technique based on microbial induced phosphate precipitation in remediation of potentially toxic ions contaminated soil: a mini review. Ecotoxicol. Environ. Saf. 2020;191:110009. doi: 10.1016/j.ecoenv.2019.110009. [DOI] [PubMed] [Google Scholar]
- Jones C.J., Wozniak D.J. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. mBio. 2017;8 doi: 10.1128/mbio.00864-17. e00864-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Paz A., Brumfeld V., Oppenheimer-Shaanan Y., Kolodkin-Gal I. Micro-CT X-ray imaging exposes structured diffusion barriers within biofilms. NPJ Biofilms Microbiomes. 2018;4:8. doi: 10.1038/s41522-018-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Paz A., Kolodkin-Gal I. A brick in the wall: discovering a novel mineral component of the biofilm extracellular matrix. N. Biotechnol. 2020;56:9–15. doi: 10.1016/j.nbt.2019.11.002. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol. Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I., Elsholz A.K., Muth C., Girguis P.R., Kolter R., Losick R. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 2013;27:887–899. doi: 10.1101/gad.215244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Greenberg E.P. Microbial sciences: the superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol. Rev. 2012;36:232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., Li Z., Newman D.K., Jensen G.J. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Konhauser K.O., Lalonde S.V., Phoenix V.R. Bacterial biomineralization: where to from here? Geobiology. 2008;6:298–302. doi: 10.1111/j.1472-4669.2008.00151.x. [DOI] [PubMed] [Google Scholar]
- Koo B.M., Kritikos G., Farelli J.D., Todor H., Tong K., Kimsey H., Wapinski I., Galardini M., Cabal A., Peters J.M., et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 2017;4:291–305.e7. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs C.S., Chaussain C., Osdoby P., Brandi M.L., Clarke B., Thakker R.V. The role of biomineralization in disorders of skeletal development and tooth formation. Nat. Rev. Endocrinol. 2021;17:336–349. doi: 10.1038/s41574-021-00488-z. [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chopp D.L., Russin W.A., Brannon P.T., Parsek M.R., Packman A.I. Spatial patterns of carbonate biomineralization in biofilms. Appl. Environ. Microbiol. 2015;81:7403–7410. doi: 10.1128/aem.01585-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lu N., Brady H.R., Packman A.I. Biomineralization strongly modulates the formation of Proteus mirabilis and Pseudomonas aeruginosa dual-species biofilms. FEMS Microbiol. Ecol. 2016;92:fiw189. doi: 10.1093/femsec/fiw189. [DOI] [PubMed] [Google Scholar]
- Li X., Lu N., Brady H.R., Packman A.I. Ureolytic biomineralization reduces Proteus mirabilis biofilm susceptibility to ciprofloxacin. Antimicrob. Agents Chemother. 2016;60:2993–3000. doi: 10.1128/aac.00203-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Prindle A., Humphries J., Gabalda-Sagarra M., Asally M., Lee D.Y.D., Ly S., Garcia-Ojalvo J., Suel G.M. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature. 2015;523:550–554. doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotlikar S.R., Kayastha B.B., Vullo D., Khanam S.S., Braga R.E., Murray A.B., McKenna R., Supuran C.T., Patrauchan M.A. Pseudomonas aeruginosa beta-carbonic anhydrase, psCA1, is required for calcium deposition and contributes to virulence. Cell Calcium. 2019;84:102080. doi: 10.1016/j.ceca.2019.102080. [DOI] [PubMed] [Google Scholar]
- Mahamid J., Sharir A., Addadi L., Weiner S. Amorphous calcium phosphate is a major component of the forming fin bones of zebrafish: indications for an amorphous precursor phase. Proc. Natl. Acad. Sci. U S A. 2008;105:12748–12753. doi: 10.1073/pnas.0803354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S., Suller M.T.E., Stickler D.J., Feneley R.C.L. Factors affecting crystal precipitation from urine in individuals with long-term urinary catheters colonized with urease-positive bacterial species. Urol. Res. 2006;34:173–177. doi: 10.1007/s00240-006-0036-6. [DOI] [PubMed] [Google Scholar]
- Monds R.D., O'Toole G.A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Monteil C.L., Benzerara K., Menguy N., Bidaud C.C., Michot-Achdjian E., Bolzoni R., Mathon F.P., Coutaud M., Alonso B., Garau C., et al. Intracellular amorphous Ca-carbonate and magnetite biomineralization by a magnetotactic bacterium affiliated to the Alphaproteobacteria. ISME J. 2020;15:1–18. doi: 10.1038/s41396-020-00747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.M., Helmann J.D. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 2005;8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Morris N.S., Stickler D.J. Encrustation of indwelling urethral catheters by Proteus mirabilis biofilms growing in human urine. J. Hosp. Infect. 1998;39:227–234. doi: 10.1016/s0195-6701(98)90262-6. [DOI] [PubMed] [Google Scholar]
- Nicolas P., Mader U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M., Aymerich S., et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y., Sibony-Nevo O., Bloom-Ackermann Z., Suissa R., Steinberg N., Kartvelishvily E., Brumfeld V., Kolodkin-Gal I. Spatio-temporal assembly of functional mineral scaffolds within microbial biofilms. NPJ Biofilms Microbiomes. 2016;2:15031. doi: 10.1038/npjbiofilms.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H., Wendel B.M., Helmann J.D. Dysregulation of Magnesium Transport Protects Bacillus subtilis against Manganese and Cobalt Intoxication. J Bacteriol. 2020;202:e00711–e00719. doi: 10.1128/JB.00711-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi Y., Arad T., Klein E., Weiner S., Addadi L. Sea urchin spine calcite forms via a transient amorphous calcium carbonate phase. Science. 2004;306:1161–1164. doi: 10.1126/science.1102289. [DOI] [PubMed] [Google Scholar]
- Povolotsky T.L., Keren-Paz A., Kolodkin-Gal I. Metabolic microenvironments drive microbial differentiation and antibiotic resistance. Trends Genet. 2021;37:4–8. doi: 10.1016/j.tig.2020.10.007. [DOI] [PubMed] [Google Scholar]
- Reichhardt C., Parsek M.R. Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front. Microbiol. 2019;10:677. doi: 10.3389/fmicb.2019.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D., Aguilar C., Losick R., Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra D.O., Hengge R. Stress responses go three dimensional - the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 2014;16:1455–1471. doi: 10.1111/1462-2920.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaldone G.T., Helmann J.D. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology. 2020;153:4123–4128. doi: 10.1099/mic.0.2007/011742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg N., Keren-Paz A., Hou Q., Doron S., Yanuka-Golub K., Olender T., Hadar R., Rosenberg G., Jain R., Camara-Almiron J., et al. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aaw8905. [DOI] [PubMed] [Google Scholar]
- Steinberg N., Kolodkin-Gal I. The matrix reloaded: probing the extracellular matrix synchronizes bacterial communities. J. Bacteriol. 2015;197:2092–2103. doi: 10.1128/JB.02516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P.S., Franklin M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Tan Y., Leonhard M., Moser D., Ma S., Schneider-Stickler B. Inhibitory effect of probiotic lactobacilli supernatants on single and mixed non-albicans Candida species biofilm. Arch. Oral Biol. 2018;85:40–45. doi: 10.1016/j.archoralbio.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Veening J.W., Kuipers O.P., Brul S., Hellingwerf K.J., Kort R. Effects of phosphorelay perturbations on architecture, sporulation, and spore resistance in biofilms of Bacillus subtilis. J. Bacteriol. 2006;188:3099–3109. doi: 10.1128/jb.188.8.3099-3109.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner S., Dove P.M. 1. An overview of biomineralization processes and the problem of the vital effect. Rev. Mineral. Geochem. 2003;54:1–29. doi: 10.1515/9781501509346-006. [DOI] [Google Scholar]
- Wendel B.M., Pi H., Krüger L., Herzberg C., Stülke J., Helmann J.D. A central role for magnesium homeostasis during adaptation to osmotic stress. mBio. 2022;13 doi: 10.1128/mbio.00092-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking J.N., Zaburdaev V., De Volder M., Losick R., Brenner M.P., Weitz D.A. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U S A. 2013;110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S.G., Houben L., Elbaum M. Cryo-scanning transmission electron tomography of vitrified cells. Nat. Methods. 2014;11:423–428. doi: 10.1038/nmeth.2842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
B. subtilis colony. The intact colony was visualized under X-ray, and the obtained 2D images were used to generate high-resolution 3D images. Rotation of 360° was taken around an axis perpendicular to the biofilm surface. Results are of a representative experiment out of three independent repeats
intact colony was visualized as in supplementary movie 1
Bacterial strains used in this study and their construction details
Differentially expressed genes – wild type B. subtilis, with vs without excess calcium
Differentially expressed genes – wild type B. subtilis vs cueR deletion mutant
Data Availability Statement
-
•
RNA-seq data have been deposited at NCBI GEO: GSE199052 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All data reported in this paper, including microscope images, will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.