Abstract
The infection rate of 60 pigs (10 pigs from each of six farms) by Helicobacter species was studied by two techniques. Histological examination of the cardiac area of the stomach yielded a 58% positive result versus an 80% positive result by PCR with genus-specific primers. Eighty percent of the 16S rRNA gene was amplified, classified in four groups by PCR-restriction fragment length polymorphism, and sequenced. Isolates from all farms except one (farm C) were identified as Helicobacter heilmannii type 1, while those from farm C were identified as H. heilmannii type 2. Attempts to culture this organism in vitro failed. Helicobacter pylori was not found in these animals.
Since the description of Helicobacter pylori (33), many studies have been conducted to determine its importance in human diseases (17, 20). The discovery of H. pylori has led to a renewed interest in the prevalence and significance of mucus-associated spiral bacteria in the gastrointestinal tract of animals. A possible link between such bacteria and gastric diseases of pigs has been evoked (3, 4, 26). Such diseases constitute an important problem in veterinary medicine leading to significant economic consequences, with up to 2.5% of mortality in pigs due to gastrointestinal hemorrhaging (34). Ulceration of the gastric pars esophagea is also a common problem in intensive pig production. Several factors have been suspected in the pathogenesis of these lesions (11, 14, 22), but the etiology remains unknown.
In 1990, a bacterium from the gastric mucosa of a pig was described, included in the new genus “Gastrospirillum” proposed by McNulty et al. (16), and named Gastrospirillum suis (19). This bacterium is now considered to be a member of the Helicobacter genus and has been renamed Helicobacter heilmannii type 1 (formerly Gastrospirillum hominis type 1) (28). Only a few studies have been performed on the prevalence of H. heilmannii infection in swine, with controversial results linked to the different methods used (3, 4, 9, 25, 26, 29); molecular identification has rarely been performed (30, 31). Furthermore, because piglets constitute one of the rare animal models of H. pylori infection (6) and a laboratory pig was found to be infected with H. pylori (12), it has been suggested that these animals may be a source of infection for human beings.
The aim of the present study was to study the prevalence of infection by Helicobacter species in fattening pigs randomly selected from six farms in the west of France and to identify the species by 16S rDNA sequencing.
Sixty fattening pigs, 5 months old and apparently healthy, weighing about 100 kg and originating from one of six farms (designated A, B, C, D, E, and F) situated in western France, were studied. At each sampling, 10 pigs from the same farm were randomly selected at the slaughterhouse after the carcasses were eviscerated. The farms were also selected at random.
The stomachs were excised along the greater curvature, and the contents were discarded. The stomachs were then washed gently in tap water, taking care to remove only food particles. The mucosal surfaces were examined and assessed for the presence of macroscopic lesions. Biopsy samples were taken from the cardiac (site 1), fundic (site 2), and pyloric (site 3) areas of the stomach. For each site, a fraction of the biopsy was immersed in 10% neutral buffered formalin for histological examination, another fraction was introduced into a transport medium (Portagerm pylori; bioMérieux, Marcy l’Etoile, France) for culture, and the remaining fraction was frozen at −80°C in order to perform molecular studies.
Biopsy specimens were processed for histological examination according to a standard procedure. Formalin-fixed fractions from the cardiac area, the fundus, and the pylorus were paraffin embedded and cut in 5-μm sections. They were stained with hematoxylin and eosin and modified Giemsa stains. Tissue sections were examined by light microscopy for the presence and localization of spiral bacteria.
Macroscopic examination of the 60 stomach specimens showed the presence of ulcerous lesions in six pigs (10%). These ulcers were small in size (<5 mm in diameter), unique, and essentially localized in the fundic area (five cases), with only one found in the cardiac area. Upon examination of the histological preparations of the three sites, spiral bacteria in samples from 39 pigs (65%) were observed (Table 1). In all cases, their morphology corresponded to that of H. heilmannii or Helicobacter felis, i.e., a diameter of 7 to 10 μm and spires of limited amplitude. Bacteria with a morphology similar to that of H. pylori were not observed.
TABLE 1.
Comparison of the results obtained by histology and HS1-HS2 PCR
| Farm | No. of pigs positive for spiral bacteria bya:
|
||||
|---|---|---|---|---|---|
| Histology
|
PCR with primer pair HS1-HS2 (site 1) | ||||
| Site 1 | Site 2 | Site 3 | Total | ||
| A | 7 | 6 | 6 | 7 | 9 |
| B | 8 | 6 | 8 | 8 | 9 |
| C | 3 | 3 | 4 | 6b | 2 |
| D | 6 | 2 | 3 | 6 | 10 |
| E | 6 | 4 | 3 | 6 | 9 |
| F | 5 | 5 | 6 | 6 | 9 |
| Total | 35 | 26 | 30 | 39 | 48 |
Sites 1, 2, and 3 correspond to the cardiac, fundic, and pyloric areas, respectively.
Pig C3 was only positive in site 2, and pigs C7 and C9 were only positive in site 3.
Gastric biopsy specimens were cultured after being ground for 2 to 3 s with an electric tissue homogenizer followed by inoculation onto selective and nonselective in-house media and incubation under microaerobic conditions at 37°C for 12 days. A drop of each suspension obtained was also used for a Gram stain. No bacteria which could be identified as members of the Helicobacter genus grew from the 180 biopsy specimens tested, even when such bacteria were present in the corresponding histological preparations and the suspensions were used for culture.
The DNA was extracted by the QIAamp tissue method (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. For PCR analysis, 5 μl of the extracted DNA was added to a 50-μl reaction mixture volume containing 1 U of Taq DNA polymerase (EurobioTaq; Eurobio, Les Ulis, France), 25 pmol of each primer, 1.5 mM of MgCl2, 200 μM of deoxyribonucleoside triphosphate, and 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl). Amplification was performed in a DNA thermal cycler (model 480; Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). PCR products were resolved by electrophoresis in a 1% (wt/vol) agarose gel containing 0.5 μg of ethidium bromide per ml. Several primers were used for the PCR (Table 2). Primers HS1 and HS2 are specific to the Helicobacter genus (8), AL1 and AL2 to H. pylori (13), and primer pairs FH1-FH2 and IS-IR were chosen on the basis of the 16S rRNA gene sequence of H. heilmannii types 1 and 2.
TABLE 2.
Primers used for PCR and sequencing
| Primer designation | Gene | Nucleotide sequence (5′→3′) | Position (nt) | Type of analysis | Cycle temp (°C), time (min) |
|---|---|---|---|---|---|
| HS1a | 16S rRNA | AAC GAT GAA GCT TCT AGC TTG CTA G | 65–89 | PCR and sequencing | 94, 1 |
| 60, 1.5 | |||||
| HS2a | GTG CTT ATT CGT TAG ATA CCG TCA T | 464–439 | 72, 1 | ||
| FH1b | 16S rRNA | CGC TAA AGG ATT GGT CTA TGT CCT ATC | 173–199 | PCR and sequencing | 94, 1 |
| 65, 1 | |||||
| FH2b | TAT TCA CCG CAA CAT RGC TGA TTT G | 1306–1282 | 72, 1 | ||
| ISb | 16S rRNA | TTA CCT AGG CTT GAC ATT GAA | 909–929 | Sequencing | |
| IRb | TTC AAT GTC AAG CCT AGG TAA | 929–909 | |||
| AL1c | glmM | GGA TAA GCT TTT AGG GGT GTT AGG GG | 1289–1314 | PCR | 94, 1 |
| 56, 1 | |||||
| AL2c | GCT TAC TTT CTA ACA CTA ACG CGC | 1584–1561 | 72, 1 |
Primer numbering is based upon 16S rRNA gene sequence of H. pylori MC123 (GenBank accession no. UO1328).
Primer sequence and numbering are based upon 16S rRNA gene sequence of G. hominis G1A1 (GenBank accession no. L10079).
Primer sequence and numbering are based upon glmM gene sequence of H. pylori taxon 210 (GenBank accession no. M60398).
Utilization of the HS1 and HS2 primers allowed the amplification of a 400-bp fragment of the expected size. Detection was carried out first on the three specimen collection sites of the pigs from farm A. Site 1 (the cardiac area) being more frequently colonized than the other sites, it was therefore the only site used for further detection in animals originating from the other farms. PCR analysis indicated that 80% of the pigs (48 of 60) were infected at this site with bacteria from the Helicobacter genus. Positive samples were detected in pigs from all six farms (Table 1). PCR was more sensitive than histological examination, which detected bacteria in only 35 of 60 specimens (58%) from the cardiac area. However, for farm C samples, PCR did not detect a positive case which was observed microscopically and, in addition, three other cases were detected microscopically only at site 2 or 3. Primers specific for H. pylori (AL1 and AL2) were also used, but none of the samples led to a positive reaction (data not shown).
In order to determine the Helicobacter species present, sequencing of the HS1-HS2 PCR products was performed on both strands without preliminary cloning, with a commercially available kit (Taq Dye Deoxy Terminator Cycle sequencing kit, Applied Biosystems, Inc., Foster City, Calif.) (27). The sequence analyses were resolved with the ABI PRISM collection program (Perkin-Elmer). When these sequences were compared to those present in databases by using BLAST software (1), 99% homology with the 16S rDNA of H. heilmannii type 1 (G. hominis type 1, GenBank accession no. L10079) was found for samples from farms A, B, D, E, and F; 100% homology with the 16S rDNA of H. heilmannii type 2 (G. hominis type 2, GenBank accession no. L10080) was found for the samples from farm C.
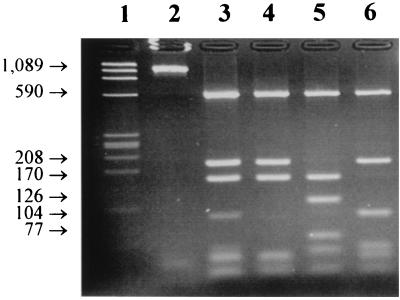
In order to determine more exactly the species present in the pig samples, a fragment of 1,089 bp overlapping the HS1-HS2 fragment was amplified with primers FH1 and FH2 from all the cases where the first PCR results were positive. The PCR products showing a single band of the expected size (1,089 bp) were submitted to restriction fragment length polymorphism (RFLP) analysis with AluI (Roche Diagnostic, Meylan, France) in order to screen for the different types of fragments. Four AluI profiles were obtained (Fig. 1, lanes 3 to 6). Profile I was the most common (85%). It was found in specimens from all of the farms except farm C. Profile II was present only in farm C. Profiles III and IV were found in farms B (specimens B1 and B8) and D (D1, D7, and D9), respectively. It should be noted that profile III may result from the combination of profiles I and IV (Fig. 1, lanes 3, 4, and 6). Mixed infections (as in cats) may explain this result, as described by Dieterich et al. (5). For each RFLP profile, one amplified FH1-FH2 amplicon was sequenced except for profile I, which was the most common and for which four amplicons (A2, B6, E1, and F8) were sequenced. The amplified products corresponding to profile I showed sequences that were identical except for three bases. The amplified products corresponding to profile II (C2) differed from profile I by 53 bases.
FIG. 1.
PCR-RFLP of FH1 and FH2 products with AluI. Lane 1, marker X174 restriction fragment DNA HaeIII digest; lane 2, FH1-FH2 product before restriction; lanes 3 to 6, restriction profiles III, I, II, and IV, respectively. The sizes are indicated in base pairs.
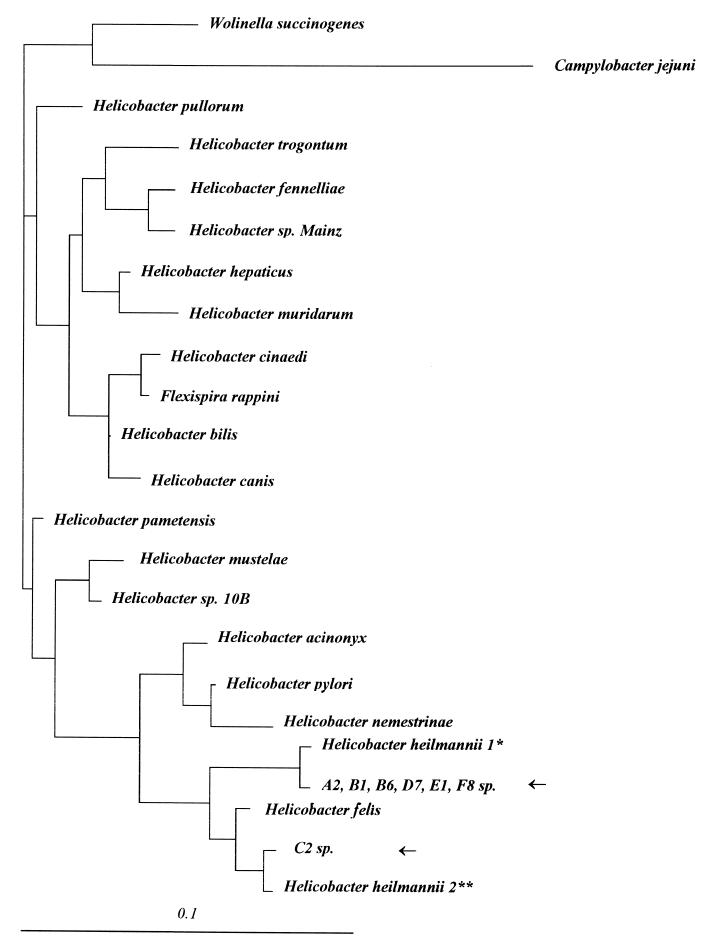
A total of 1,226 bp (HS1 to FH2) were sequenced for seven strains, i.e., more than 80% of the 16S rRNA gene, and were compared to the sequences present in the database. In six cases (A2, B1, B6, D7, E1, and F8), a homology of 98.3% with the 16S rDNA of H. heilmannii type 1 was found, and in one case (C2), a homology of 98.9% with the 16S rDNA of H. heilmannii type 2 was found. Using the fast DNA ml program in the Ribosomal Database Project site (15) and Treeview software (24), a phylogenetic tree was constructed (Fig. 2) showing the relationship between the Helicobacter species found in pigs A2, B1, B6, D7, E1, and F8 with H. heilmannii type 1 and that found in pig C2 with H. heilmannii type 2.
FIG. 2.
Phylogenetic tree showing relationship between A2, B1, B6, D7, E1, F8, and C2 species (shown on tree with arrows) and other Helicobacter and related species. The scale represents a 1% difference in nucleotide sequence. Vertical distance has no meaning. The tree was rooted with Wolinella succinogenes and Campylobacter jejuni as outgroups. ∗, formerly known as G. hominis type 1; ∗∗, formerly known as G. hominis type 2.
It is well known that H. pylori and H. heilmannii can colonize the stomachs of pigs (6, 9, 18, 25, 26). The model of H. pylori-infected piglets has been used as an argument in favor of the possibility that farmed pigs may naturally carry H. pylori (6). This argument was supported by the isolation of H. pylori in a laboratory pig (12) and the observation of H. pylori in pigs by immunofluorescence (32). However, according to Grasso et al. (9), Mendes et al. (18, 19), and Queiroz et al. (25), as well as this study, H. pylori was not detected in pig stomach.
This study presents data on the prevalence of H. heilmannii infection in fattening pigs and particularly on the type of strains present. To avoid bias, we did not study consecutive pigs which could have originated from the same farm, but we randomly chose six different farms and, subsequently, 10 pigs from each farm. The farms produced 1,200 to 3,600 pigs per year. In contrast to the studies of Queiroz et al. (25) and Grasso et al. (9), in which 10.8 and 9.4% of the animals respectively, were colonized, 86.6% of the pigs in this study were H. heilmannii positive when the results of PCR analysis and histology were combined. This difference can be explained partly by the fact that in the studies mentioned only histology was used, a less sensitive technique than PCR. Nonetheless, even with histology, we found that 65% of the pigs were infected, indicating that other unknown reasons for infection may exist. A similarly high rate of infection in swine was also reported in another French study carried out on 60 sows, in which 63% were positive for Helicobacter species (30).
We carried out an extensive molecular investigation and H. heilmannii type 1 was the most frequently identified isolate in our samples, as in the study of Thiberge et al. (30). Confusion still exists about the exact nature of the tightly coiled spiral bacteria present in animals and sometimes in humans. This is essentially because these bacteria cannot be cultured, with a few exceptions (2, 10), and sequencing of more than 80% of the 16S rDNA, the only method to reach a precise identification, is rarely performed.
There was no correlation between the presence of ulcers and the presence of H. heilmannii on the gastric mucosa. However, this phenomenon is also found in humans: ulcers do not occur in all human subjects infected with H. pylori. Only a small proportion will develop ulcers, depending on the presence of environmental and host factors (17). The situation is most likely the same with swine infected with H. heilmannii. All the pigs with ulcerous lesions were, however, infected by H. heilmannii. Lymphoid aggregates or lymphoid follicles in the gastric mucosa of bacterium-positive pigs were observed more frequently than in that of bacterium-negative pigs (data not shown), in agreement with the observations made by Queiroz et al. (26).
In this study of gastric colonization, a greater degree of homogeneity was found among Helicobacter species in pigs than in animals such as cats and dogs (5, 7, 21, 23). It remains to be proven whether this homogeneity results from an exquisite adaptation of H. heilmannii to this particular niche (as is seen for H. pylori in humans) or from the process of selection of fattening pigs. Over the years, the pigs may have been selected with their Helicobacter flora.
Several factors may influence the prevalence rate of infection in pigs, such as the geographical zone and the type of breeding. In previous studies (9, 25, 30), no information about the breeding of the pigs analyzed was given. Indeed, in our study, we pointed out differences in the prevalence of infection by H. heilmannii in fattening pigs between the six different farms that ranged from 30 to 80% with the histology technique performed on site 1.
Follow-up and intervention studies should be carried out to address the question of the relationship between H. heilmannii infection and ulcers in pigs, as already initiated by Queiroz et al. (26), and to identify the risk factors in the contamination of the pigs.
Nucleotide sequence accession numbers.
The nucleotide sequences were deposited with GenBank under accession nos. AF142146, AF142147, AF142148, AF142149, AF142150, AF142151, and AF142152 for the Helicobacter species found in pigs A2, B1, B6, D7, E1, F8, and C2, respectively.
Acknowledgments
We are grateful to industrial partners for generously supplying animals and assistance. We thank Florence Jugiau and Florence Rama for technical assistance.
This study was supported by grants from the Ministère Français de l’Agriculture et de la Pêche (Direction Générale de l’Enseignement et de la Recherche) as well as by the Institut National de la Recherche Agronomique and the Université Victor Ségalen Bordeaux 2.
REFERENCES
- 1.Altshul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersen L P, Norgaard A, Holck S, Blom J, Elsborg L. Isolation of a “Helicobacter heilmannii”-like organism from the human stomach. Eur J Clin Microbiol Infect Dis. 1996;15:95–96. doi: 10.1007/BF01586196. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa A J A, Silva J C, Nogueira A M, Paulino E, Jr, Miranda C R. Higher incidence of Gastrospirillum sp. in swine with gastric ulcer of the pars oesophagea. Vet Pathol. 1995;32:134–139. doi: 10.1177/030098589503200206. [DOI] [PubMed] [Google Scholar]
- 4.Bedel A, Pichard F, Wuscher N, Labigne A, Huerre M. Prevalence of Helicobacter infection and gastric-associated pathologies in swine originating from pork producers in the west of France. Gut. 1997;41(Suppl. 1):A124. [Google Scholar]
- 5.Dieterich C, Wiesel P, Neiger R, Blum A, Corthésy-Theulaz I. Presence of multiple “Helicobacter heilmannii” strains in an individual suffering from ulcers and in his two cats. J Clin Microbiol. 1998;36:1366–1370. doi: 10.1128/jcm.36.5.1366-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton K A, Morgan D R, Krakowka S. Persistence of Helicobacter pylori in conventionalized piglets. J Infect Dis. 1990;161:1299–1301. doi: 10.1093/infdis/161.6.1299. [DOI] [PubMed] [Google Scholar]
- 7.Eaton K A, Dewhirst F E, Paster B J, Tzellas N, Coleman B E, Paola J, Sherding R. Prevalence and varieties of Helicobacter species in dogs from random sources and pet dogs: animal and public health implications. J Clin Microbiol. 1996;34:3165–3170. doi: 10.1128/jcm.34.12.3165-3170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Germani Y, Dauga C, Duval P, Huerre M, Levy M, Pialoux G, Sansonetti P, Grimond P A D. Strategy for the detection of Helicobacter species by amplification of 16S rRNA genes and identification of H. felis in a human gastric biopsy. Res Microbiol. 1997;148:315–326. doi: 10.1016/S0923-2508(97)81587-2. [DOI] [PubMed] [Google Scholar]
- 9.Grasso G M, Ripabelli G, Sammarco M L, Ruberto A, Iannitto G. Prevalence of Helicobacter-like organisms in porcine gastric mucosa: a study of swine slaughtered in Italy. Comp Immunol Microbiol Infect Dis. 1996;19:213–217. doi: 10.1016/0147-9571(96)00007-0. [DOI] [PubMed] [Google Scholar]
- 10.Hänninen M L, Jalava K, Saari S, Happonen I, Westermarck F. Culture of “Gastrospirillum” from gastric biopsies of dogs. Eur J Clin Microbiol Infect Dis. 1995;14:145–146. doi: 10.1007/BF02111876. [DOI] [PubMed] [Google Scholar]
- 11.Huber W G, Wallin R F. Pathogenesis of porcine gastric ulcers. Am J Vet Res. 1967;28:1455–1459. [PubMed] [Google Scholar]
- 12.Jones D M, Elridge J. Gastric campylobacter-like organisms from man (C. pyloridis) compared with GCLO strains from the pig, baboon and ferret. In: Kaijser B, Falsen E, editors. Campylobacter IV. Göteborg, Sweden: University of Göteborg; 1988. p. 44. [Google Scholar]
- 13.Kansau I, Raymond J, Bingen E, Courcoux P, Kalach N, Bergeret M, Braimi N, Dupont C, Labigne A. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res Microbiol. 1996;147:661–669. doi: 10.1016/0923-2508(96)84023-x. [DOI] [PubMed] [Google Scholar]
- 14.Kowalczyk T. Etiologic factors of gastric ulcers in swine. Am J Vet Res. 1969;30:393–399. [PubMed] [Google Scholar]
- 15.Maidack B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Databank Project) Nucleic Acids Res. 1997;205:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNulty C A, Dent J C, Curry A, Uff J S, Ford G A, Gear M W, Wilkinson S P. New spiral bacterium in gastric mucosa. J Clin Pathol. 1989;42:585–591. doi: 10.1136/jcp.42.6.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mégraud F, Lamouliatte H. Helicobacter pylori and duodenal ulcer. Evidence suggesting causation. Dig Dis Sci. 1992;37:769–772. doi: 10.1007/BF01296437. [DOI] [PubMed] [Google Scholar]
- 18.Mendes E N, Queiroz D M M, Dewhirst F E, Paster B J, Rocha G A, Fox J B. Are pigs a reservoir host for human Helicobacter infection? Am J Gastroenterol. 1994;89:1296. [Google Scholar]
- 19.Mendes E N, Queiroz D M M, Rocha G A, Moura S B, Leite V H, Fonseca M E. Ultrastructure of a spiral micro-organism from pig gastric mucosa (“Gastrospirillum suis”) J Med Microbiol. 1990;33:61–66. doi: 10.1099/00222615-33-1-61. [DOI] [PubMed] [Google Scholar]
- 20.NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. Helicobacter pylori in peptic ulcer disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- 21.Norris C R, Marks S L, Eaton K A, Torabian S Z, Munn R J, Solnick J V. Healthy cats are commonly colonized with “Helicobacter heilmannii” that is associated with minimal gastritis. J Clin Microbiol. 1999;37:189–94. doi: 10.1128/jcm.37.1.189-194.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien J J. Gastric ulceration (of the pars esophagea) in the pig; a review. Vet Bull. 1969;39:75–82. [Google Scholar]
- 23.Otto G, Hazel S H, Fox J G, Howlett C R, Murphy J C, O’Rourke J L, Lee A. Animal and public health implications of gastric colonization of cats by Helicobacter-like organisms. J Clin Microbiol. 1994;32:1043–1049. doi: 10.1128/jcm.32.4.1043-1049.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page R D M. Treeview, an application to display phylogenetic trees on personal computers. CABIOS. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 25.Queiroz D M, Rocha G A, Mendes E N, Lage A P, Carvalho A C, Barbosa A J. A spiral microorganism in the stomach of pigs. Vet Microbiol. 1990;24:199–204. doi: 10.1016/0378-1135(90)90067-6. [DOI] [PubMed] [Google Scholar]
- 26.Queiroz D M M, Rocha G A, Mendes E N, Moura S B, Oliveira A M R, Miranda D. Association between Helicobacter and gastric ulcer disease of the pars esophagea in swine. Gastroenterology. 1996;111:19–27. doi: 10.1053/gast.1996.v111.pm8698198. [DOI] [PubMed] [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain reaction-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solnick J V, O’Rourke J, Lee A, Paster B J, Dewhirst F E, Tompkins L S. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 29.Suarez D L, Wesley I V, Larson D J. Detection of Arcobacter species in gastric samples from swine. Vet Microbiol. 1997;57:325–336. doi: 10.1016/s0378-1135(97)00107-7. [DOI] [PubMed] [Google Scholar]
- 30.Thiberge J M, Bedel A, Huerre M, Pichard F, Labigne A. Comparison of several diagnostic tests for the detection of Helicobacter infection in swine. Gut. 1997;41(Suppl. 1):A125. [Google Scholar]
- 31.Utriainen M, Hänninen M L. Detection of Helicobacter-like bacteria in porcine gastric biopsy samples by amplification of 16S rRNA, ureB, vacA and cagA genes by PCR. Vet Res Commun. 1998;22:373–383. doi: 10.1023/a:1006141211452. [DOI] [PubMed] [Google Scholar]
- 32.Vaira D, Ferron P, Negrini R, Cavazzini L, Holton J, Ainley C, Londei M, Vergura M, Dei R, Colecchia A, et al. Detection of Helicobacter pylori-like organisms in stomach of some food-source animals using a monoclonal antibody. Ital J Gastroenterol. 1992;24:181–184. [PubMed] [Google Scholar]
- 33.Warren J R, Marshall B J. Unidentified curved bacilli on gastric epithelium in active gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 34.Yeomans N D. Helicobacter heilmannii (formerly Gastrospirillum): association with pig and human gastric pathology. Gastroenterology. 1996;111:244–259. doi: 10.1053/gast.1996.v111.agast961110244. [DOI] [PubMed] [Google Scholar]