Abstract
Introduction
In a diploid organism, two alleles from a single genetic locus are expressed to generate a normal phenotype. Heterozygous deleterious mutation causes a reduction of functional proteins to a half dose and insufficient amounts of functional proteins can occur to generate an in–normal phenotype, namely haploinsufficiency. Heterozygous deleterious mutation of microRNAs (miRs), non-coding RNAs that regulate the expression level of target transcripts, is still not well understood. The hsa-miR-302/367 cluster is the most abundant and specifically up-regulated miR cluster in human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) and plays an important role in the maintenance of pluripotency.
Methods
We targeted the hsa-miR-302/367 region via a Cas9 nuclease complex with guide RNA and replaced that region with green fluorescent protein (GFP). Using a homologous donor, consisting of left and right arms and GFP, we confirmed deletion of the hsa-miR-302/367 cluster by homologous recombination without cellular destruction by microscopy. We sub-cloned GFP-positive colonies and checked the genotype of each sub-clone by genomic PCR. We then analyzed the pluripotency of heterozygous knockout cells with a hsa-miR-302/367 cluster by assessing cell proliferation ratio, morphology, and undifferentiated marker gene expression. We also used an embryoid body formation assay and transplanted wild-type and heterozygous knockout cells into immune-deficient mice. Furthermore, to analyze the lineage-specific differentiation potential of heterozygous knockout cells, we differentiated both wild-type and heterozygous knockout cells into neural stem cells.
Results
Here, we show that the half dose of mature miRs from the hsa-miR-302/367 cluster loci was sufficient for the continued self-renewal of hiPSCs. All GFP-positive clones were revealed to be heterozygous knockout cells, suggesting hsa-miR-302/367 cluster homozygous knockout cells were not maintained. The cell proliferation ratio, morphology, and expression of undifferentiated marker genes were comparable between wild-type and heterozygous knockout of undifferentiated human iPSCs. In addition, we found that heterozygous knockout human iPSCs have the capacity to differentiate into three germ layers, including neural stem cells.
Conclusions
Taken together, a single allele of the hsa-miR-302/367 cluster expresses a sufficient amount of miRs to maintain the pluripotent properties of human stem cells.
Keywords: microRNAs, iPS cells, miR-302, CRISPR/Cas9
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; EGFP, enhanced green fluorescent protein; hetKO, hsa-miR-302/367 cluster heterozygous knockout; LHA, left homologous arm; miRs, microRNAs; Poly(A) signal, polyadenylation signal; PSC, pluripotent stem cell; RHA, right homologous arm; TALE, transcription activator-like effector; NSC, neural stem cell; EB, embryoid body; RFP, red fluorescent protein; tRFP, TurboRFP
Highlights
-
•
hsa-miR-302/367 cluster was deleted with CRISPR/Cas9 in human pluripotent stem cells.
-
•
Homozygous hsa-miR-302/367 knockout cell was not generated.
1. Introduction
In a diploid organism, two alleles of a single genetic locus are expressed to generate a normal phenotype. Heterozygous deleterious mutation reduces the level of encoded proteins by half. For most gene loci, a single functional allele provides sufficient activity such that a standard phenotype is maintained. In some cases, insufficient amounts of functional proteins can still generate an in–normal phenotype, that is haploinsufficiency [1]. MicroRNAs (miRs) are small, non–protein-coding RNAs, 21–25 nucleotides in length, that negatively regulate the post-transcriptional expression of targeted mRNAs [2]. MicroRNA sequences are dispersed throughout the genomes of from Caenorhabditis elegans [3], [4] to humans [5] and have either RNA polymerase II or III regulatory elements in their own promoters [6]. Since a single miR can recognize and consequently regulate the expression of more than 100 different transcripts, miRs were estimated to be able to regulate up to 30% of the protein-coding genes in the human genome [7],[8] As a result, miRs received widespread attention for their potential roles in complicated biological processes and multifactorial diseases. However, the impact of heterozygous deleterious mutations of miRs to such processes and disease remains to be elucidated.
Recent studies have implicated miRs in the self-renewal and differentiation of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) [[9], [10], [11], [12]]. Compared to differentiated cells, particular miR families are specifically upregulated in human pluripotent stem cells (PSCs) [[13], [14], [15], [16]], including Homo sapiens (hsa)-miR-302/367 [[17], [18], [19]], hsa-miR-372 [19], hsa-miR-17-92 [20], and hsa-miR-200 families [21]. However, the functional roles of human PSC–specific miRs and the genes they regulate remain largely unknown. DICER1 deletion led to increased cell death by receptor–mediated apoptosis and a failure of self-renewal in human ESCs [22]. Also, knockout (KO) of the hsa-miR-302/367 cluster in primary fibroblasts completely blocked iPSC generation [23]. Furthermore, study of knockdown of the hsa-miR-302/367 cluster by employing transcription activator-like effector (TALE)-based transcriptional repressors revealed that these miRs dually regulated the cell cycle and apoptosis in human ESCs [24]. The miRs cannot be functionally knocked out by simple insertions and deletions like protein-coding sequences, so exchanging the whole region, including all target miRs, may be the first option in investigating miR functions [25]. To the best of our knowledge, hsa-miR-302/367 KO PSCs have not been established.
In this study, we investigated the functional roles of hsa-miR-302/367 in human PSCs by targeting the hsa-miR-302/367 region via a CRISPR/Cas9 genome editing system. Using a donor plasmid with homologous arms of hsa-miR-302/367 flanking regions and a green fluorescent protein (GFP)-polyadenylation tail, we isolated hsa-miR-302/367 region–deleted clones detected by fluorescence protein expression under the microscope. All fluorescent-positive clones were shown to be heterozygous knockouts by genomic PCR, suggesting that the hsa-miR-302/67 cluster is essential for maintaining human iPSCs. Next, we investigated the pluripotency of heterozygous knockout human iPSCs and found that characteristics were properly maintained by a half dose of the mature form of hsa-miR-302/367. Previously, we demonstrated that overexpression of DAZAP2 protein, one of the target genes of hsa-miR-302a/b/c/d, disrupted neural differentiation [26]. To observe ectodermal differentiation properties in more detail, we applied more lineage-specific differentiation methods such as differentiation into neural stem cells (NSCs) and found that neural differentiation was comparable between wild-type and heterozygous knockouts. Our study revealed that the hsa-miR-302/367 cluster was essential for self-renewal. Also, similar to most protein-coding genes, a single functional allele of the hsa-miR-302/367 cluster provided sufficient amounts of miRs such that a standard phenotype was maintained in human iPSCs.
2. Materials and methods
2.1. Cell culture
Human menstrual blood (Edom22) cell lines were established in our laboratory under a protocol approved by the Institutional Review Board of the National Research Institute for Child Health and Development of Japan [27]. We generated human iPSCs in our laboratory [26] and maintained these on irradiated mouse embryonic fibroblast feeder layers in human iPSC medium, which contained KODMEM (Thermo Fisher Scientific, Waltham, MA, USA), 20% KO serum replacement, GlutaMAX, sodium pyruvate, and non-essential amino acids (Thermo Fisher Scientific) supplemented with 10 ng/mL recombinant human basic fibroblast growth factor (Wako Pure Chemical Industries, Ltd., Osaka, Japan) or on iMatrix-511 (Nippi. Inc., Tokyo, Japan) in StemFit medium (Reprocell Inc., Kanagawa, Japan). The medium was changed every other day and cells were passaged approximately once per week using enzymatic (TrypLE Select Enzyme; Thermo Fisher Scientific) or by mechanical methods.
2.2. Generation of fluorescence protein knock-in human PSCs
Oligo DNAs for guide (g)RNAs targeted to the hsa-miR-302 cluster region were cloned into a PX459 ver.2.0 plasmid (Addgene, Cambridge, MA, USA) using FastDigest BpiI (Thermo Fisher Scientific). The primer sequences of oligo DNAs are listed in the Supplementary Table. For donor vector construction, we first amplified a 2.3-kb region that included hsa-miR-302/367 using GoTaq MasterMix (Promega Corp., Madison, WI, USA) with primers as a donor vector listed in the Supplementary Table and cloned this into a pGEM-T easy vector (Promega Corp., Madison, WI, USA). To remove the hsa-miR-302/367 region, SpeI and HindIII (NEB Inc., Ipswich, MA, USA) were used and then DNA fragments containing GFP or RFP (red fluorescent protein) with a polyadenylation signal were ligated by a Gibson assembly kit (NEB Inc., Ipswich, MA, USA). Constructed vectors were extracted using a QIAGEN Plasmid Plus Midi Kit (QIAGEN GmbH, Venlo, Netherlands) and electroporated using a NEPA21 Super Electroporator (Nepa Gene Co., Ltd., Chiba, Japan) following the manufacturer's protocol.
2.3. EB formation
On the day of embryoid body (EB) formation, cells were grown to 60–80% confluence. To form EBs, human iPSC colonies were washed using Dulbecco's phosphate-buffered saline (DPBS; Thermo Fisher Scientific). Cells were then detached to become single cells or small aggregates after 10 min incubation with TrypLE Select Enzyme (Thermo Fisher Scientific), and transferred to a 15-mL conical tube. Cell culture medium was added to the tube and the cell suspension pipetted to separate cells. The cell number was counted, and cells spun down at 1000 rpm for 5 min. We re-suspended the cells in StemFit medium with ROCK inhibitor (Y-27632; FUJIFILM Wako Pure Chemicals Corporation, Osaka, Japan), and seeded 2–3 × 103 cells in a total volume of 100 μL into each well of Nunclon Sphera 96-well round (U) bottom plates (Thermo Fisher Scientific). StemFit medium was added on days 2 and 4. The resulting EB cultures were maintained for 7 days and collected for microarray assay.
2.4. miRNA extraction and qRT–PCR of mature microRNA
Total RNA (including small RNA) was isolated using TRIzol LS reagent (Thermo Fisher Scientific), following the manufacturer's protocol. Complementary DNA templates for quantitative reverse transcription polymerase chain reaction (qRT–PCR) analysis of miRNA expression was prepared using a TaqMan MicroRNA Reverse Transcription Kit and miR-specific RT Primers. Mature miRNA expression was analyzed using a TaqMan miRNA assay (Life Technologies, Waltham, CA, USA) and TaqMan Universal Master Mix II with UNG (Life Technologies) following the manufacturer's protocol.
2.5. Differentiation of iPSCs into neural stem cells
Neural stem cells were differentiated from wild type (wt) and hsa-miR-302/367 cluster heterozygous knockout (hetKO) cells following the protocol of Chambers et al., 2009 with a slight modification [28]. Briefly, human iPS cells were plated on iMatrics-511 in StemFit medium with ROCK inhibitor. Human iPS cells were allowed to expand until from 50 to 60% confluent. We then changed the medium to differentiation medium containing 10 nM SB431542 (FUJIFILM Wako Pure Chemicals Corporation) and 500 ng/mL Noggin (PEPROTECH, Cranbury, NJ, USA) in StemFit. After two days, SB432542 was withdrawn and increasing amounts of N-2 medium (25%, 50% and 75%) was gradually added to the StemFit medium every two days while maintaining 500 ng/mL Noggin. N2 medium contained DMEM/F12 (Thermo Fisher Scientific) and 1% N-2 supplement (Thermo Fisher Scientific).
2.6. Immunofluorescence staining
Human iPSCs were grown on 35-mm glass coverslips (AGC TECHNO GLASS, Shizuoka, Japan), fixed in 4% paraformaldehyde, and permeabilized in phosphate buffered saline (PBS) containing 0.5% Triton X-100. Subsequently, cells were blocked with PBS containing 5% goat normal serum appropriate for each antibody in preparation for immunohistochemistry studies. Cells were then incubated overnight at 4 °C with primary antibodies against POU5F1 (sc-5792; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), NANOG (RCAB0003P; ReproCELL, Yokohama, Japan), and stage-specific embryonic antigen 4 (SSEA4; RCAB0003P; ReproCELL). Following overnight incubation, cells were washed with PBS containing 5% normal serum. Next, cells were incubated in the dark for 1 h at room temperature in PBS containing 0.1% bovine serum albumin and appropriate secondary anti-mouse, anti-rabbit, or anti-goat antibodies coupled with Alexa 546 (BD Biosciences, Franklin Lakes, NJ, USA), as well as 4’, 6-diamidino-2-phenylindole (DAPI) for nuclear staining. In immunofluorescence assays, we showed that endogenous pluripotent markers, such as POU5F1 and NANOG proteins, were expressed in wt and hetKO cells. We also found that both wt and hetKO cells expressed human ESC-specific surface antigens, such as SSEA-4.
2.7. Teratoma formation assay
The animal use protocol was approved by the Institutional Animal Care and Use Committee of the National Research Institute for Child Health and Development (NRICHD, Permit Number: A2003-002). All experiments with mice were subject to 3 R (refine, reduce, and replace) consideration, and all efforts were made to minimize animal suffering and to reduce the number of animals used. In vivo pluripotency was assessed by teratoma formation in severe combined immunodeficient nude mice (BALB/cAJcl-nu/nu) purchased from CLEA (Tokyo, Japan). A 60-mm plate of undifferentiated hESCs was washed with DPBS, and the cells were harvested with a cell scraper. The cell suspension was collected into a 15-mL conical tube and centrifuged at 1000 rpm for 4 min. The cell pellet was resuspended in hESC culture medium and Matrigel (BD Biosciences, Franklin Lakes, NJ, USA; #356234) to a final total volume of 400 μL. Approximately 2–5 × 106 cells in 200 μL were injected subcutaneously into the dorsolateral area on both sides. Mice were sacrificed after 8–10 weeks. Tumors were then excised surgically, fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Hematoxylin and eosin–stained paraffin-embedded sections were histologically examined for the presence of differentiated human tissue derived from all three embryonic germ layers.
2.8. Quantitative RT–PCR analysis
Total RNA was isolated using the RNeasy Plus Mini Kit (QIAGEN GmbH). Single-stranded cDNA was synthesized from 0.1 to 2 μg of total RNA in 20 μL reactions containing random primers using a Superscript III or IV First Strand cDNA Synthesis System (Thermo Fisher Scientific). For qRT–PCR, we used SYBR Green–based assays (Luna Universal qPCR Master Mix; NEB). Transcript levels were determined using a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific). All qRT–PCR reactions using SYBR Green were conducted in triplicate or quadruplicate, and relative quantification was performed using GAPDH as a reference gene. In addition, we ran multiple gene expression assays with EBs derived from wild-type and hsa-miR-302/367 heterozygous knockout iPSCs by qRT–PCR, using TaqMan™ hPSC Scorecard™ Panel (Thermo Fisher Scientific).
2.9. Statistical analysis
The values shown in the graphs are expressed as mean ± standard deviation. Statistical analysis for comparison between groups was performed using the paired Student t-test. Statistical significance was set as P < 0.05.
3. Results
3.1. Hsa-miR-302/367 cluster is essential for human iPSCs
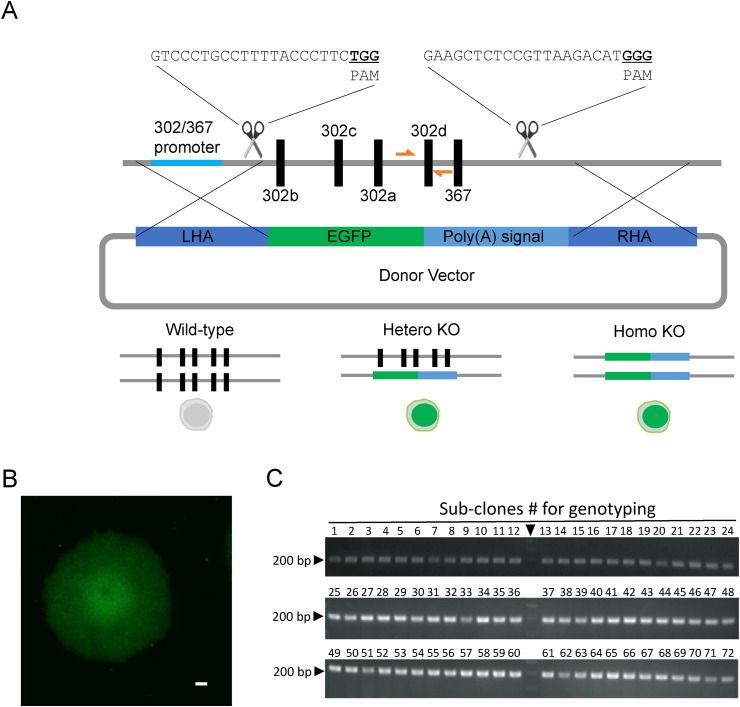
Recently, we analyzed miR expression profiles in human PSCs such as ESCs and iPSCs. We identified the hsa-miR-302/367 cluster as being the most abundant and specific. Furthermore, DAZAP2 overexpression was found to have an impact on neural differentiation of human iPSCs [26]. To further investigate a functional role for the hsa-miR-302/367 cluster in human PSCs we designed two gRNAs that were located upstream and downstream of the hsa-miR-302/367 cluster (Fig. 1A). A flanking sequence of the hsa-miR-302/367 cluster was cloned as a homologous arm of a homology-directed repair donor for the recombination of GFP with a polyadenylation signal sequence on the hsa-miR-302/367 cluster region (Fig. 1A). We transfected two gRNAs and a donor vector into human Edom_iPSCs [26]. In case homologous recombination (HR) occurred, GFP expression would be driven by an endogenous promoter of the hsa-miR-302/367 cluster so that HR would be observable by fluorescence microscopy. One week after transfection, we found several GFP-positive cells. After a few passages, we successfully sub-cloned GFP-positive cells (Fig. 1B) and genotyped them by genomic PCR. Unexpectedly, we did not obtain any homozygous KO human iPSC lines after genotyping more than a hundred clones by repeated experiments of transfection (Fig. 1C). Next, we confirmed that GFP was inserted in the hsa-miR-302/367 cluster region in a subclone of heterozygous knockout Edom_iPSCs by genomic PCR (Supplemental figure 1). We also checked the locus of potential off-target sites by COSMID (https://crispr.bme.gatech.edu). Among the top 10 potential off-target sites, only one is the site on protein-coding region. We amplified this region (GPRIN3 gene coding region) by genomic PCR and sequenced it by sanger sequencing. As a result, we confirmed no obvious off-target effect using these gRNAs (data not shown). These results suggest that the hsa-miR-302/367 cluster is essential for human PSC survival and that miR-deficient human PSCs have self-renewal defects. Thereafter, we analyzed the properties of heterozygous knockout human iPSCs to investigate the impact of heterozygous deleterious mutation of miRs.
Fig. 1.
Generation of hsa-miR-302/367 cluster knockout by CRISPR/Cas9. (A) Schematic image of Homo sapiens (hsa)-miR-302/367 cluster knockout (KO) by CRISPR/Cas9 technology. Two guide (g)RNAs were designed in immediately before and after miR cluster coding loci. Donor vectors for homologous recombination were designed to observe green fluorescent protein (GFP) signals in case recombination occurred. (B) Images of GFP-positive transfected clones of Edom_iPSCs. Scale bars are 200 μm. (C) PCR analysis of genotyping using genomic DNA of sub-clones. An orange primer pair was used to amplify the endogenous hsa-miR-302/367 cluster coding loci. Markers are indicated by an arrowhead. EGFP, enhanced green fluorescent protein; iPSC, induced pluripotent stem cell; LHA, left homologous arm; RHA, right homologous arm.
3.2. Heterozygous knockout of the hsa-miR-302/367 cluster does not affect human iPSC renewal
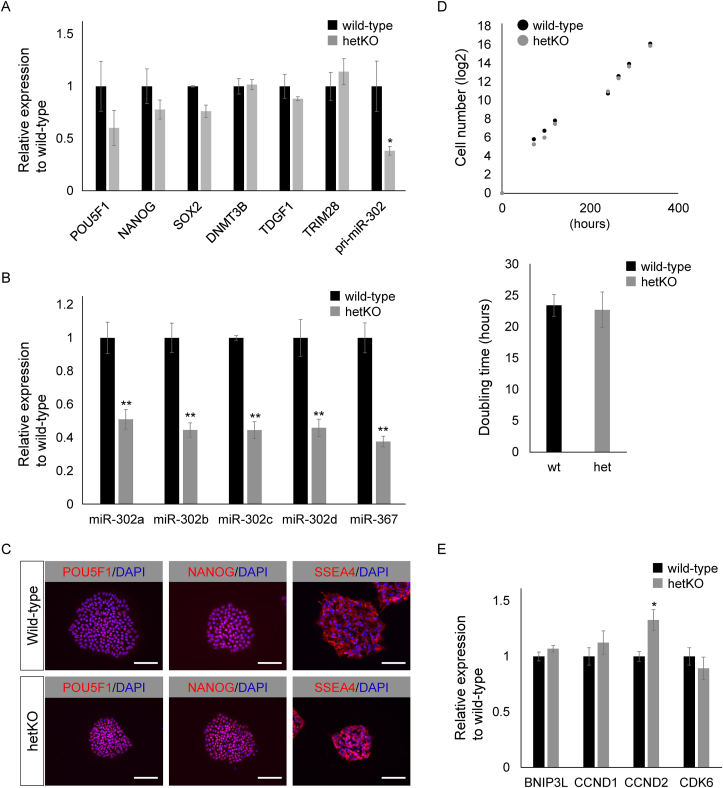
Human Edom_iPSCs with heterozygous knockout of the hsa-miR-302/367 cluster (hetKO) showed a normal morphology and were comparable to wild-type isogenic lines. At the time of writing, the hetKO had been maintained for more than 30 passages. Since the hsa-miR-302/367 cluster plays a role in pluripotent gene signaling networks, we first examined the expression level of pluripotent marker genes, pri-miR-302, and five mature hsa-miR-302/367 cluster members in the hetKO by qRT–PCR. Pluripotent marker genes, such as POU5F1, NANOG, SOX2, DNMT3B, TDGF1, and TRIM28, were expressed at high levels in the hetKO comparable to those in the control isogenic line (Fig. 2A). As expected, expression of the precursor of mature miR-302a/b/c/d/367 (pri-miR-302) in the hetKO was significantly reduced by half when compared with the control isogenic cell line (Fig. 2A). Also, the expression of five mature miRs from hsa-miR-302/367 cluster in the hetKO were significantly reduced by 50% when compared with the control isogenic cell line (Fig. 2B). We also tested whether mature miR expression levels exerted effects on pluripotent markers at the protein level. We found that the expression level and subcellular localization of POU5F1, NANOG, and SSEA-4 proteins were comparable with that in the control isogenic cell line (Fig. 2C). Taken together, despite the importance of hsa-miR-302/367 members in the maintenance of human iPSCs, a reduction of a half dose of miRs did not have severe effects on marker gene expression in human iPSCs.
Fig. 2.
Pluripotent marker gene expression in hetKO iPSCs was comparable with isogenic control line. (A) The expression of undifferentiated marker genes, including POU5F1, NANOG, SOX2, DNMT3B, TDGF1, and TRIM28, was comparable between control isogenic line and Homo sapiens (hsa)-miR-302/367 cluster heterozygous knockout Edom_iPSCs. Primary microRNA (miR) of hsa-miR-302/367 (pri-miR-302) was reduced by half in heterozygous knockout Edom_iPSCs. (B) The expression of mature miRs from the hsa-miR-302/367 cluster was analyzed by a TaqMan microRNA assay system in heterozygous knockout Edom_iPSCs and isogenic control lines. Mature miR-302a/b/c/d/and miR-367 were reduced by half in heterozygous knockout Edom_iPSCs. (C) Undifferentiated markers, including POU5F1, NANOG, and stage-specific embryonic antigen 4 (SSEA4), were normally expressed and localized in heterozygous knockout Edom_iPSCs and isogenic control lines. 4′,6-diamino-2-phenylindole (DAPI) staining is shown in blue. Scale bars are 100 μm. (D) The cell proliferation rate was comparable between the control isogenic line and hsa-miR-302/367 cluster heterozygous knockout Edom_iPSCs. (E) Apoptosis and cell-cycle related marker gene expression was comparable between the control isogenic line and hsa-miR-302/367 cluster heterozygous knockout Edom_iPSCs. hetKO, hsa-miR-302/367 cluster heterozygous knockout; iPSC, induced pluripotent stem cell; wt, wild-type. ∗P < 0.05. ∗∗P < 0.01.
Previous studies reported that hsa-miR-302/367 cluster members regulated cell-cycle–related genes. Also, Zhang and colleagues reported that the miR-302/367 cluster dually regulated the cell cycle and apoptosis in human ESCs by employing TALE-based transcriptional repressors of such miRs [24]. Thus, we investigated the expression of the genes that play a role in either apoptosis or the cell cycle. First, to investigate the impacts of the heterozygous knockout of the hsa-miR-302/367 cluster on the self-renewal of human iPSCs, we assessed the cell growth rate in the hetKO line. Unexpectedly, the growth rate did not significantly differ between the hetKO and control isogenic line (Fig. 2D). BNIP3L that contained a BH3 domain with homology to the BH3-only pro-apoptotic members of the Bcl-2 family of proteins is a bifunctional mitochondrial protein that can induce cell death. The up-regulation of this gene was not observed in hetKO cells (Fig. 2E). Using qRT–PCR, we examined the expression of cell-cycle regulators that were down-regulated in hsa-miR-302/367 knockdown cells in the previous study. We found that CCND1, CCND2, and CDK6 were unchanged in hetKO cells (Fig. 2E), which is consistent with the result in Fig. 2D. Next, we investigated the expression level of genes that were previously reported as targets for the hsa-miR-302/367 cluster: DAZAP2, KLF4, MBD2, MBLN2, and TGFBR2; we found that they were comparable with levels in the control isogenic cell line. Since the hsa-miR-302/367 cluster is located in the intron of the LARP7 gene, we also investigated the expression level of LARP7 and found that this was comparable with the level in the control isogenic cell line (Supplemental figure 2). Taken together, we concluded that despite the importance of the hsa-miR-302/367 cluster in human iPSCs, a reduction to a half dose of mature hsa-miR302a/b/c/d and hsa-miR 367 induced by heterozygous knockout did not affect the self-renewal of human iPSCs.
3.3. Heterozygous knockout of hsa-miR-302/367 cluster does not modulate iPSC differentiation
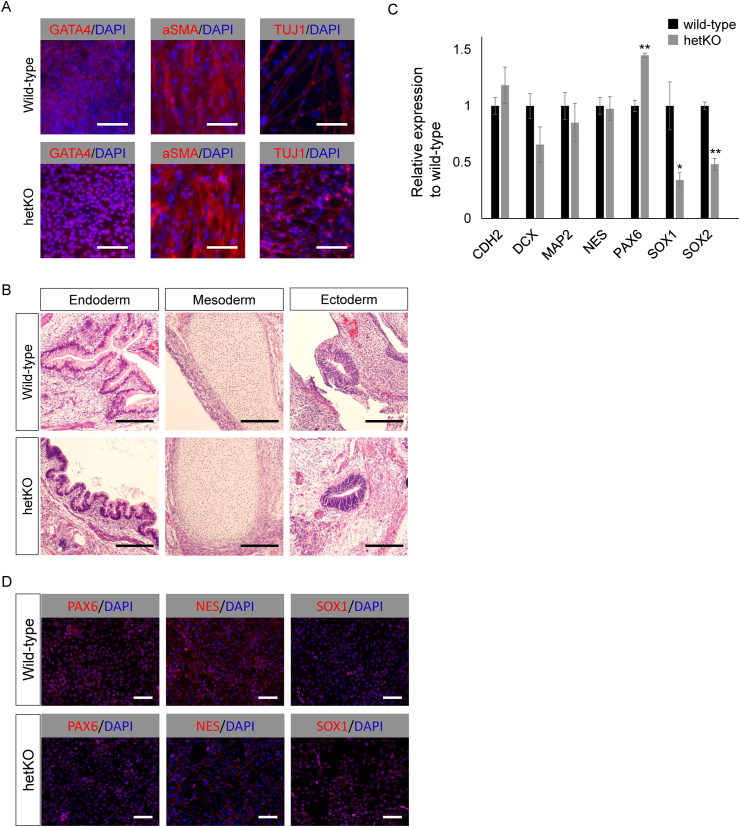
To investigate the effect of heterozygous knockout of the hsa-miR-302/367 cluster on differentiation potencies, we generated embryoid bodies (EBs) and compared differentiation properties between hetKO and the control isogenic line according to lineage-specific marker gene expression. First, the spontaneous differentiation of human iPSCs via EB formation for seven days was used to observe differentiation properties by TaqMan qPCR array analysis (Supplemental figure 3). PAX6, one of the ectodermal marker genes, was up-regulated in hetKO cells (Supplemental figure 3); endodermal and mesodermal marker gene expression was comparable between hetKO and control isogenic lines (Supplemental figure 3). To further evaluate whether hetKO cells maintained their pluripotency in vitro, we performed EB adhesion culture assays. Embryoid bodies that had differentiated on Matrigel-coated glass dishes from hetKO cells and control isogenic cell lines expressed markers associated with the three major germ layers: TUJ1 (ectoderm), αSMA (mesoderm), and HNF4α (endoderm; Fig. 3A). Additionally, in an in vivo teratoma formation assay, structures from all three germ layers were detected, including neural tissues and pigmented epithelium (ectoderm), cartilage (mesoderm), and intestinal epithelial structure (endoderm; Fig. 3B).
Fig. 3.
Both control isogenic line and hsa-miR-302/367 cluster heterozygous knockout Edom_iPSCs can differentiate into three germ layers. (A) Both the control isogenic line and Homo sapiens (hsa)-miR-302/367 cluster heterozygous knockout Edom_iPSCs differentiated in vitro via EB adhesion culture. Both cell lines expressed markers of primary germ layers. Immunohistochemical analyses of markers of ectoderm (TUJ1), mesoderm (αSMA), and endoderm (GATA4) layers are shown. Scale bars are 100 μm. (B) Both the control isogenic line and hsa-miR-302/367 cluster heterozygous knockout Edom_iPSCs differentiated in vivo via teratoma formation. Hematoxylin and eosin staining revealed germ layer derivatives, such as neural tissues (ectoderm; right panels), cartilage (mesoderm; middle panels), and gut epithelial tissues (endoderm; left panels). Scale bars are 100 μm. (C) Neural stem cell (NSC) marker genes were analyzed in NSCs derived from both a control isogenic line and hsa-miR-302/367 cluster heterozygous knockout Edom_iPSCs. CDH2, DCX, MAP2, and NES expression were comparable between heterozygous knockout and the control isogenic line. PAX6 was relatively high in the heterozygous knockout line, while SOX1 and SOX2 were relatively low. (D) Neural stem cell markers, including PAX6, NES, and SOX1, were normally expressed and localized in the heterozygous knockout Edom_iPSC and isogenic control lines. Scale bars are 100 μm. DAPI staining is shown in blue. hetKO, hsa-miR-302/367 cluster heterozygous knockout. ∗P < 0.05. ∗∗P < 0.01.
To observe ectodermal differentiation properties in more detail, we applied more lineage-specific differentiation methods such as differentiation into NSCs. As expected, expression of the NSC marker, PAX6, was significantly higher in the hetKO compared to control isogenic cell line (Fig. 3C). However, the expression of other NSC markers, such as SOX1, and SOX2, was lower in the hetKO compared to control isogenic cell line (Fig. 3C). Also, the expression of CDH2, DCX, MAP2, and NES were comparable between hetKO and control isogenic cell lines. Immunostaining of NSCs revealed that the expression of neural marker proteins, such as PAX6, NES, and SOX1, was comparable between hetKO and control isogenic cell lines (Fig. 3D). Taken together, a single allele of the functional hsa-miR-302/367 cluster allows the activity of mature miRs such that standard differentiation properties are maintained.
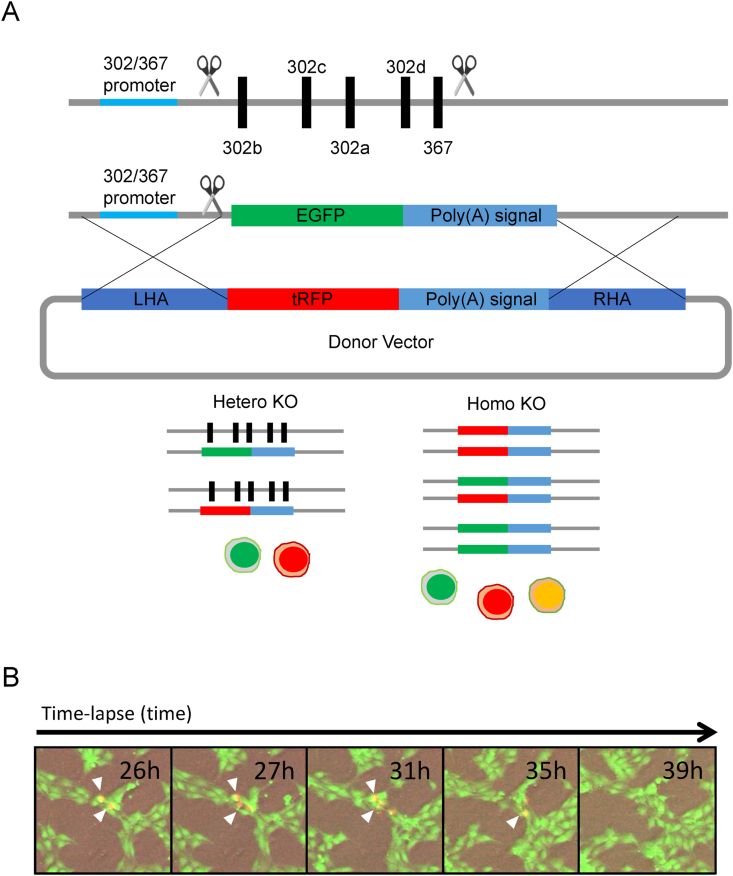
3.4. Visualization of hsa-miR-302/367 cluster homozygous knockout cells
To visualize whether we could generate homozygous knockouts of the hsa-miR-302/367 cluster, we cloned turboRFP (tRFP) instead of GFP in the donor vector (Fig. 4A). When we transfected the same two gRNAs and donor vector with tRFP into GFP positive hetKO cells. A successful homozygous knockout of the hsa-miR-302/367 cluster would be detected as GFP/tRFP double-positive cells. A day after transfection, we observed several GFP/tRFP double-positive cells, suggesting that hsa-miR-302/367 homozygous KO cells were generated. However, three days after transfection we rarely observed double-positive cells. By a week after transfection, we did not detect any double-positive cells. Next, we acquired sequential cell image data 24 h after transfection (Fig. 4B). We observed GFP/tRFP double-positive cells 26 h after transfection (Fig. 4B left panel). The GFP/tRFP double-positive cells started to shrink by 31 h after transfection (Fig. 4B middle panel). By 39 h after transfection, GFP/tRFP positive cells had disappeared (Fig. 4B right panel). These results further indicated that the hsa-miR-302 cluster is essential for human PSC survival and that miR-deficient human PSCs have self-renewal defects.
Fig. 4.
Visualization of homozygous hsa-miR-302/367 knockout cells. (A) Schematic image of Homo sapiens (hsa)-miR-302/367 cluster knockout (KO) by CRISPR/Cas9 technology. Donor vectors for homologous recombination were designed to show green and red fluorescent signals in case recombination occurred homozygously. (B) Time-lapse imaging after tRFP donor plasmid transfection into a green fluorescent protein (GFP)-positive heterozygous hsa-miR-302/367 knockout Edom_iPSCs. White arrowhead indicates a tRFP/GFP doble-positive cell. is EGFP, enhanced green fluorescent protein; hetero KO, heterozygous knockout; homo KO, homozygous knockout; iPSC, induced pluripotent stem cell; LHA, left homologous arm; RHA, right homologous arm; tRFP, turbo red florescent protein.
4. Discussion
An miR-302/367 cluster consists of five members, including miR-302b, miR-302c, miR-302a, miR-302d, and miR-367, from a 5′–3′ direction and is highly conserved in higher vertebrates such as mammals and birds [29], [30]. Such a cluster is involved in various important biological processes, such as cell proliferation, cell differentiation, cell reprogramming, tumorigenesis, organ development, autoimmunity, and maintenance of pluripotency in pluripotent stem cells [[31], [32], [33]] However, the biological functions of an miR-302/367 cluster remains to be elucidated. In this study, we investigated the biological function of hsa-miR-302/367 by target deletion in human PSCs using a CRISPR/Cas9 system. We subsequently found that we could not generate homozygous KOs of hsa-miR-302/367, indicating that these miRs were essential for the maintenance of human PSCs. Next, we investigated the functional properties of heterozygous KO cells. We did not observe any clear morphological and functional changes because of a reduction to a half dose of mature hsa-miR-302a/b/c/d and −367. Therefore, a single allele of the functional hsa-miR-302 cluster provides sufficient miR activities such that standard pluripotent properties are maintained.
In a diploid organism, two alleles of a single genetic locus are expressed to produce a normal phenotype. For most genes, however, a single copy is enough to support normal growth and development of diploid organisms despite the heterozygous deleterious mutation reducing the amount of functional protein by half. A small subset of genes exhibits sensitivity to decreased gene dosage, namely haploinsufficiency. MicroRNAs, small non-coding RNAs, can recognize and consequently regulate the expression of more than 100 different transcripts such that a reduction in the amount of mature miRs might affect the expression of over a hundred genes and functional properties of cells. A reduction of miRs by deleterious mutations was reported to cause metabolic abnormalities in a cell. Starczynowski et al. reported that the deletion of chromosome 5q resulted in the loss of two miRNAs that are abundant in hematopoietic stem/progenitor cells: miR-145 and miR-146a. Loss of miR-145 and miR-146a results in hematopoietic abnormalities reminiscent of 5q– syndrome, including thrombocytosis, characteristic dysmegakaryopoiesis, and variable neutropenia [34]. In our study, heterozygous KO of the hsa-miR-302/367 cluster did not have an impact on the self-renewal and differentiation of human PSCs despite the expression level of mature miRs decreasing by half. Although heterozygous KO cells have the potency to differentiate into all three germ layers, the differentiation properties of heterozygous KO cells may differ from those of the isogenic control line. Further study is needed to fully elucidate the effects of the loss of a hsa-miR-302/367 cluster.
CRISPR-Cas9 induces double-strand breaks at target sites, resulting in their repair and generating random insertions, deletions, and changes in the DNA sequence. Gene knockout by CRISPR/Cas9 is a methodology widely utilized to investigate the functional role of protein-coding genes. When targeting CRISPR/Cas9 to the hsa-miR-302/367 cluster, a single site of gene editing is not sufficient to disrupt function since several miRs, including those of the same family, share a seed sequence that is critical for the recognition of target genes. To the best of our knowledge, hsa-miR-302/367 KO human PSCs have not been established. To overcome this limitation, paired gRNAs targeting both 5′ and 3’ of the precursor miR region with a homologous donor vector have been shown to result in larger deletions. The efficiency of CRISPR/Cas9 technology still forces us to spend time genotyping and subcloning to obtain the desired cells. Here, we employed a donor plasmid with fluorescent protein expression genes to visualize HR clones without cell destruction, allowing us to easily pick and expand KO cells. Since Dicer or DGCR8 KO mouse ESCs could self-renew but showed a differentiation deficit, we expected similar results in human PSCs [35], [12]. Unexpectedly, we observed the disappearance of GFP/tRFP double-positive homozygous KO cells within 1 week and could not generate stable human PSCs from the homozygous KO of a hsa-miR-302/367 cluster. Consistent with our results, Teijeiro and colleagues reported the generation of DICER1-KO human ESCs, and found that DICER and small RNAs were essential for self-renewal [22]. In addition, deletion of the miR-302 cluster in primary human fibroblasts completely blocked iPSC generation [23]. The inconsistency between human and mouse cells could be explained by the complementary function of Mus musculus (mmu)-miR-290-295 and the mmu-miR-302 cluster in mouse ESCs [[36], [37], [38]]. Consistent with this speculation, hsa-miR-372 and -373, which share a seed sequence with hsa-miR 302 members, rescued the phenotype produced by Dicer depletion. Our miR profiling data indicated that the expression of hsa-miR-371, -372, and −373 was much less compared to that of the hsa-miR-302 cluster in our human PSCs (data not shown), suggesting that the hsa-miR-302 cluster was essential for human PSC survival. However, the mechanisms behind why hsa-miR-302 cluster fully depleted cells had disappeared remains to be elucidated. Zhang and colleagues reported that the miR-302 cluster dually regulated the cell cycle and apoptosis in human ESCs by employing TALE-based transcriptional repressors of such miRs [24]. The identification of hsa-miR-302/367 cluster targets will be needed to understand molecular mechanisms for cell death by hsa-miR-302/367 depletion.
In summary, our study revealed that a single allele of the hsa-miR-302/367 cluster maintained the self-renewal and differentiation potencies of human PSCs. Further studies will be needed to investigate which hsa-miR-302/367 target gene plays a role in apoptosis, cell-cycle regulation or differentiation.
Authors’ contributions
TS: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. TK: collection and/or assembly of data, data analysis and interpretation. YK: collection and/or assembly of data, data analysis and interpretation. AU: financial support, administrative support, provision of study material. HA: conception and design, financial support, administrative support, provision of study material, data analysis and interpretation, manuscript writing, final approval of manuscript.
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Acknowledgments
We thank Masakazu Machida for providing technical assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to HA, AU; Grant-in-aids for Scientific Research (21390456), (26293364) and (25670710) to HA; and a Grant-in-aid for Scientific Research (22770233) to TS. This research was supported by the Japan Agency for Medical Research and Development (AMED) under Grant Number 20bm0704023h0002 to TS and 20bk0104089h0002 to HA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.reth.2022.05.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplemental figure 1 Schematic image of Homo sapiens (hsa)-microRNA (miR)-302/367 cluster knockout (KO) by CRISPR/Cas9 technology. Primer positions for genotyping heterozygous knockout cells are shown (a to d). PCR analysis of genotyping using genomic DNA of sub-clones. An orange primer pair was used to amplify endogenous hsa-miR-302/367 cluster coding loci. A blue primer pair was used to confirm the insertion of enhanced green fluorescent protein (EGFP) into endogenous hsa-miR-302/367 cluster coding loci. Markers are indicated by arrowheads. hetKO, hsa-miR-302/367 cluster heterozygous knockout; LHA, left homologous arm; RHA, right homologous arm; wild-type, Edom_iPSCs.
Supplemental figure 2Homo sapiens (hsa)-miR-302/367 target gene expression, including DAZAP2, MBD2, MBLN2, and TGFBR2, was comparable between control isogenic line and hsa-miR-302/367 cluster heterozygous knockout Edom_iPSCs. An miR-hsa-miR-302/367 cluster was located in the intron of the LARP7 gene and the expression level of LARP7 was comparable between heterozygous knockout and isogenic control lines. iPSC, induced pluripotent stem cell; miR, microRNA; wild-type, Edom_iPSCs.
Supplemental figure 3 Heat map and score box plot from scorecard analysis for differentiation potential. The samples were embryoid bodies (EBs) differentiated from the iPSC lines (wild-type and hetKO). The EBs were analyzed using a TaqMan hPSC Scorecard Assay (Thermo Fisher Scientific) according to the manufacturer's protocol. The resulting expression data set was analyzed using cloud-based analysis software that compares the expression pattern against a reference standard composed of multiple functionally validated ESC and iPSC lines. (A) Heat map of expression of self-renewal and three germ layer markers by the EBs differentiated from the wild-type and the hetKO, respectively. (B) Score box plot represents comparison of two samples. Black, and gray dots indicate EBs from wild-type iPSCs and hetKO iPSCs, respectively. ESC, embryonic stem cell; hetKO, hsa-miR-302/367 cluster heterozygous knockout; iPSC, induced pluripotent stem cell; fc, fold change; wild-type, Edom_iPSCs.
References
- 1.Morrill S.A., Amon A. Why haploinsufficiency persists. Proc Natl Acad Sci U S A. 2019;116:11866–11871. doi: 10.1073/pnas.1900437116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger C.J., Rougvie A.E., et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Hutvágner G., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 6.Schanen B.C., Li X. Transcriptional regulation of mammalian miRNA genes. Genomics. 2011;97:1–6. doi: 10.1016/j.ygeno.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallanna S.K., Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiscornia G., Izpisúa Belmonte J.C. MicroRNAs in embryonic stem cell function and fate. Genes Dev. 2010;24:2732–2741. doi: 10.1101/gad.1982910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar M., Wyman S.K., Fritz B.R., Qi J., Garg K.S., Parkin R.K., et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cell. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakshmipathy U., Love B., Goff L.A., Jörnsten R., Graichen R., Hart R.P., et al. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cell Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Wilson K.D., Venkatasubrahmanyam S., Jia F., Sun N., Butte A.J., Wu J.C. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cell Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barroso-delJesus A., Romero-López C., Lucena-Aguilar G., Melen G.J., Sanchez L., Ligero G., et al. Embryonic stem cell-specific miR 302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R., et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 22.Teijeiro V., Yang D., Majumdar S., González F., Rickert R.W., Xu C., et al. DICER1 is essential for self-renewal of human embryonic stem cells. Stem Cell Rep. 2018;11:616–625. doi: 10.1016/j.stemcr.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., Xiang D., Heriyanto F., Gao Y., Qian Z., Wu W.S. Dissecting the roles of miR-302/367 cluster in cellular reprogramming using TALE-based repressor and TALEN. Stem Cell Rep. 2013;1:218–225. doi: 10.1016/j.stemcr.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Hong Y., Xiang D., Zhu P., Wu E., Li W., et al. MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Rep. 2015;4:645–657. doi: 10.1016/j.stemcr.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang H., Yi B., Ma R., Zhang X., Zhao H., Xi Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep. 2016;6:22312. doi: 10.1038/srep22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugawara T., Miura T., Kawasaki T., Umezawa A., Akutsu H. The hsa-miR-302 cluster controls ectodermal differentiation of human pluripotent stem cell via repression of DAZAP2. Regen Ther. 2020;15:1–9. doi: 10.1016/j.reth.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui C.H., Uyama T., Miyado K., Terai M., Kyo S., Kiyono T., et al. Menstrual blood-derived cells confer human dystrophin expression in the murine model of Duchenne muscular dystrophy via cell fusion and myogenic transdifferentiation. Mol Biol Cell. 2007;18:1586–1594. doi: 10.1091/mbc.E06-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Z., Zhu X., Dou Y. The miR-302/367 cluster: a comprehensive update on its evolution and functions. Open Biol. 2015;5:150138. doi: 10.1098/rsob.150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosa A., Spagnoli F.M., Brivanlou A.H. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Hu S., Wilson K.D., Ghosh Z., Han L., Wang Y., Lan F., et al. MicroRNA-302 increases reprogramming efficiency via repression of NR2F2. Stem Cell. 2013;31:259–268. doi: 10.1002/stem.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosa A., Brivanlou A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipchina I., Elkabetz Y., Hafner M., Sheridan R., Mihailovic A., Tuschl T., et al. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starczynowski D.T., Kuchenbauer F., Argiropoulos B., Sung S., Morin R., Muranyi A., et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 35.Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medeiros L.A., Dennis L.M., Gill M.E., Houbaviy H., Markoulaki S., Fu D., et al. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc Natl Acad Sci U S A. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parchem R.J., Moore N., Fish J.L., Parchem J.G., Braga T.T., Shenoy A., et al. miR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 2015;12:760–773. doi: 10.1016/j.celrep.2015.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S.L., Yang M., Herrlinger S., Liang C., Lai F., Chen J.F. MiR-302/367 regulate neural progenitor proliferation, differentiation timing, and survival in neurulation. Dev Biol. 2015;408:140–150. doi: 10.1016/j.ydbio.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.