Abstract
Prokaryotes adapt to challenges from mobile genetic elements by acquiring foreign DNA-derived spacers into the CRISPR array1. Spacer insertion is carried out by the Cas1-Cas2 integrase complex2-4. A significant fraction of CRISPR-Cas systems further utilize an Fe-S cluster containing nuclease Cas4 to ensure spacers are acquired from a DNA flanked by a protospacer adjacent motif (PAM)5,6 and inserted into the CRISPR array unidirectionally, so that the transcribed CRISPR RNA can guide target-searching in PAM-dependent fashion. Focusing on Type I-G CRISPR in Geobacter sulfurreducens where Cas4 is naturally fused with Cas1, here we provide a high-resolution mechanistic explanation for the Cas4-assisted PAM-selection, spacer biogenesis and directional integration. During biogenesis, only DNA duplexes possessing a PAM-embedded 3′-overhang trigger Cas4/Cas1-Cas2 assembly. Importantly, during this process the PAM-overhang is specifically recognized, sequestered, but not cleaved by Cas4. This “molecular constipation” prevents the PAM-side prespacer from participating in integration. Lacking such sequestration, the non-PAM overhang is trimmed by host nucleases and integrated to the leader-side CRISPR repeat. Half-integration subsequently triggers PAM cleavage and Cas4 dissociation, allowing spacer-side integration. Overall, the intricate molecular interaction between Cas4 and Cas1-Cas2 selects PAM-containing prespacers for integration, and couples the timing of PAM processing with the stepwise integration to establish directionality.
Prokaryotes have a unique ability to acquire immunological memories against mobile genetic elements by integrating short fragments of DNA (i.e. spacers) between CRISPR repeats. The array of repeat-spacers is transcribed to generate guide RNAs that direct CRISPR effector complexes to find and cleave DNA or RNA targets. DNA-targeting CRISPR-Cas systems further require the spacers to be acquired adjacent from PAM. PAM helps crRNA-guided complexes distinguish true targets from spacers in the CRISPR array, and thereby prevents lethal self-targeting. PAM also speeds up the target-searching process, by reducing the total number of candidate sites dramatically7. To ensure CRISPR spacers are only derived from PAM-flanking sequences, both Class I (type I-A, I-B, I-C, I-D, I-G) and Class II (type II-B, V-A, V-B) CRISPR-Cas systems further encode a dedicated CRISPR adaptation protein, Cas4, that works in conjunction with the core spacer acquisition machinery consisting of Cas1 and Cas22-4,8-13. While early studies mainly showed that deletion of cas4 impaired spacer acquisition in type I-B systems in Haloarcula hispanica14 and I-A in Sulfolobus islandicus15, recent studies using type I-A in Pyrococcus furiosus16, I-D in Synechocystis sp.17 and I-G (previously I-U) in Geobacter sulfurreducens18 established a critical role for Cas4 in acquiring spacers with a functional PAM. Cas4 protein was found to harbour an Fe-S cluster and to catalyse various exo- and endonuclease activities19-21. Only recently did it become clear from work in I-C Bacillus halodurans that Cas4 uses its nuclease activity to cleave PAM sequences in spacer precursors just before integration into the CRISPR array22,23. Follow-up work showed that Cas4 forms a complex with a dimer of Cas1 and associates with Cas2 upon prespacer binding22,23.
Results
Cas4 is a PAM-cleaving endonuclease
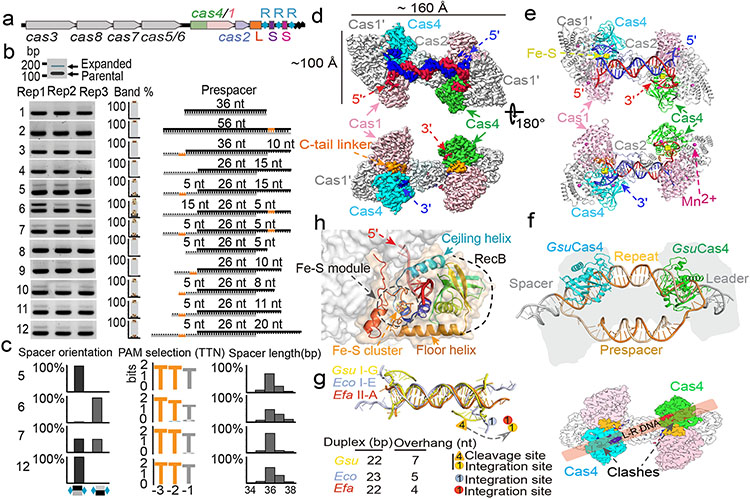
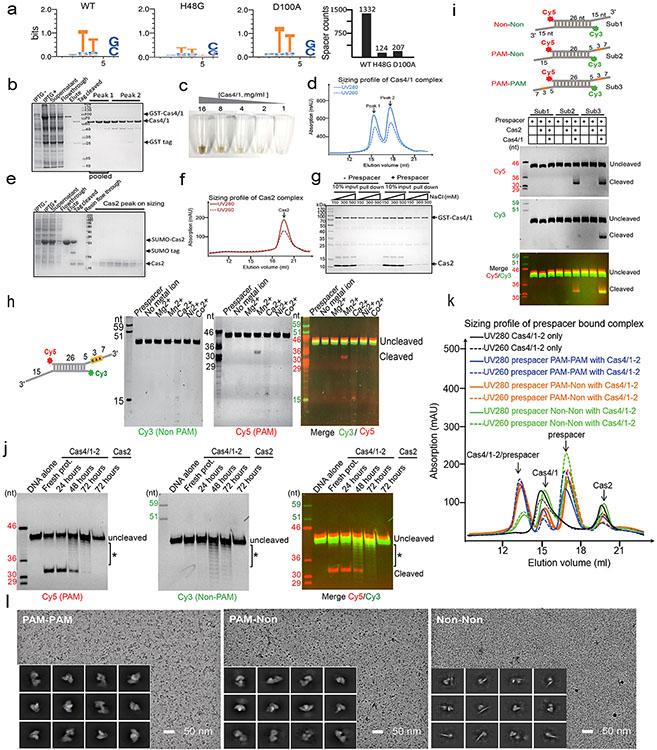
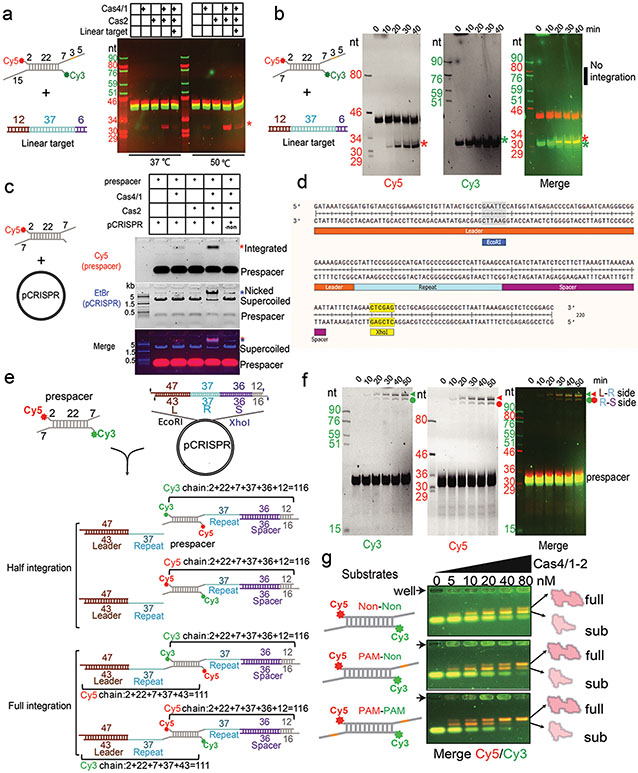
Geobacter sulfurreducens I-G CRISPR-Cas contains a highly active spacer acquisition module, in which Cas4 is naturally fused with Cas1 (Fig. 1a)18. This module acquires 34-40 base pair (bp)-long spacers into the CRISPR locus in a PAM-dependent manner (5′-TTN)18. To understand the prespacer processing and integration mechanisms, we electroporated prespacers of various sequence and structure compositions into E. coli cells containing a G.sul cas4/cas1-cas2/CRISPR genomic locus and analyzed cells for newly acquired spacers using PCR and deep sequencing methods (Fig. 1b-c, Extended Data Fig. 1a). It was hypothesized that GsuCas4/Cas1-Cas2 may preferentially integrate prespacers containing a 26-bp mid-duplex and 5-nt 3′-overhangs18,22. Such prespacers were indeed robustly integrated in a single-stranded PAM (ss-PAM) dependent fashion; prespacers lacking ss-PAM were not integrated (Fig. 1b). The context surrounding PAM also influenced the integration outcome. Whereas a ss-PAM 5-nt away from the mid-duplex were efficiently integrated, the same ss-PAM immediately adjacent to the mid-duplex, or a ds-PAM in the middle of a duplex, did not enable spacer integration (Fig. 1b). Dual-PAM containing prespacers were integrated with scrambled directionality but a precise length distribution, whereas the single-PAM containing prespacers were integrated directionally but with a 2-3 nt length distribution (Fig. 1c). These data suggest that GsuCas4/Cas1-Cas2 preferentially recognizes prespacers containing a correctly spaced PAM in the 3′-overhang of a DNA duplex.
Figure 1. PAM-dependent spacer acquisition and the dual-PAM prespacer bound GsuCas4/Cas1-Cas2 structure.
a. Organization of the G. sul. CRISPR-Cas operon. b. PAM dependency analyzed using in vivo spacer acquisition assay. N=3 biologically independent assays detected by PCR are shown, as well as relative percentages of expanded and non-expanded bands. Data is presented as mean ± s.e.m. PAM is represented in orange. c. Deep-sequencing analysis of spacer orientation, length and PAM code for selected prespacers in b. PAM-1 appears conserved because a single prespacer was assayed. d. Cryo-EM density and e. structure of the dual-PAM bound GsuCas4/1-2 complex, f. Superposition with EfaCas1-Cas2 structure in full-integration state. Cas4 binding is incompatible with repeat-spacer docking into Cas1 integrase. g. Comparison of the 3′-overhang status among three prespacer-bound Cas1-Cas2 structures. The overhang is guided away from Cas1 and sequestered by Cas4 in GsuCas4/1-2. h. Organization of Cas4 structural elements around the PAM-containing 3′-overhang.
In biochemical reconstitutions (Extended Data Figs. 1b-k), the PAM-containing 3′-overhang of the prespacer was found to be specifically cleaved by recombinant GsuCas4/Cas1-Cas2 complex; the non-PAM 3′-overhang remained intact (Extended Data Fig. 1i). Cleavage was Mn2+-dependent and took place precisely, if inefficiently, after PAM (3′-A−3A−2G−1↓; Extended Data Fig. 1h-i). Only ~5% of the PAM-containing overhang was processed after 1 hr of incubation in 37 °C, in 50-fold excess of GsuCas4/Cas1-Cas2 (Extended Data Figs. 1h). The underlining mechanism for the attenuated PAM processing only became clear after structural analysis. Interestingly, extended exposure to air induced promiscuous DNA cleavage (Extended Data Fig. 1j), likely due to the oxidation of the Fe-S cluster in Cas4. The various level of oxidation may explain the spectrum of reported endo and exonuclease activities for Cas4 in the literature19-23.
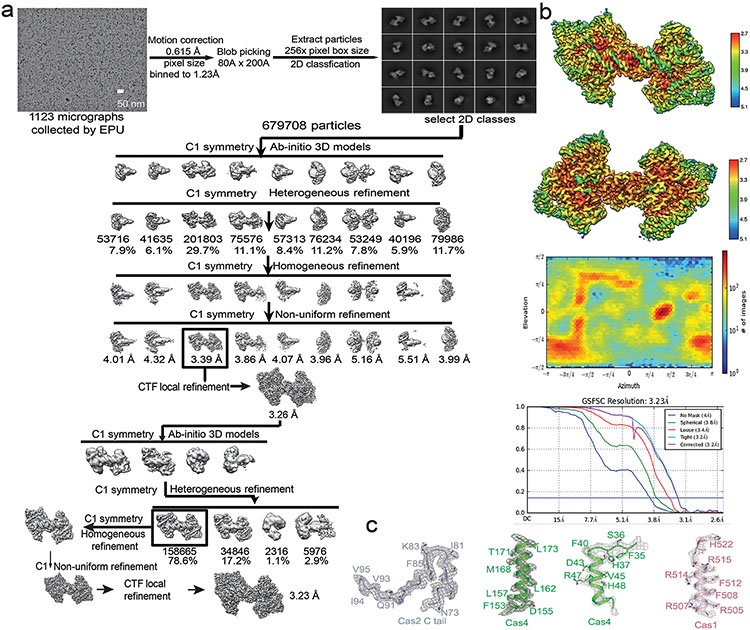
Dual-PAM prespacer/Cas4/1-2 structure
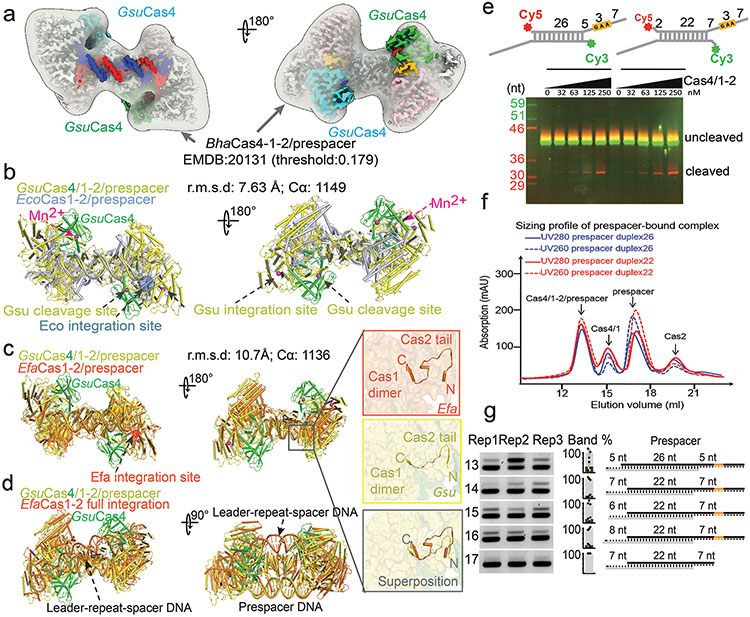
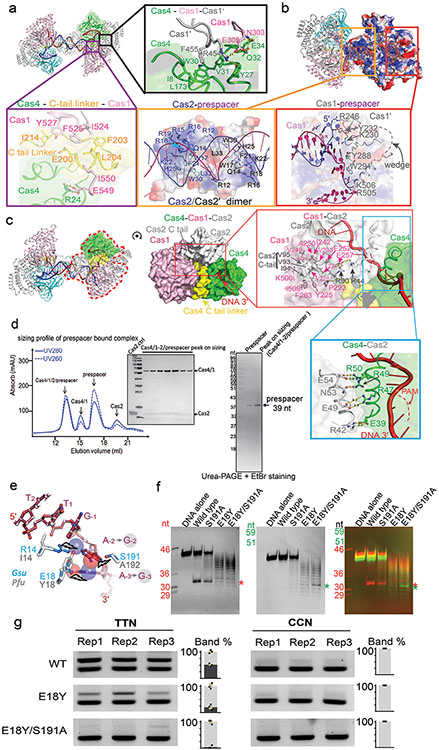
Whereas weak interaction could be detected between GsuCas4/Cas1 and GsuCas2, functional complex formation required prespacer presence. A dual- or single-PAM containing prespacer led to stable higher-order complex formation, as revealed by size-exclusion chromatography (SEC) and electron microscopy (EM) analyses; a PAM-less prespacer was inefficient at complex formation (Extended Data Figs. 1g, 1k-l). The dual-PAM prespacer bound GsuCas4/Cas1-Cas2 complex was especially homogeneous, and its single particle reconstruction reached 3.23 Å in resolution, which revealed significant more structural details than previous studies22 (Extended Data Figs. 2-3). The Cas14-Cas22 integrase core assumes its characteristic dumbbell shape - the Cas2 dimer constitute the central handle, and two Cas1 dimers constitute the two distal weights (Fig. 1d-e). In each dimer, only one Cas1 participates in spacer integration, the other plays structural roles. The architecture and interfaces are more consistent with Enterococcus faecalis rather than E. coli Cas14-Cas2210,12 (Extended Data Fig. 2b-d). Surprisingly, Cas1-Cas2 was found to specify a 22-bp rather than 26-bp mid-duplex as defined by the integration assay; an additional two base-pairs are unwound from each end. Indeed, prespacers containing a 22-bp mid-duplex integrated as efficiently as the 26-bp version in various assays (Extended Data Fig. 2b-f). We predict that Cas1-Cas2 among different CRISPR systems likely share a preference for a 22-bp-long mid-duplex but specify idiosyncratic 3′-overhang length in prespacer11,12 (Fig. 1g).
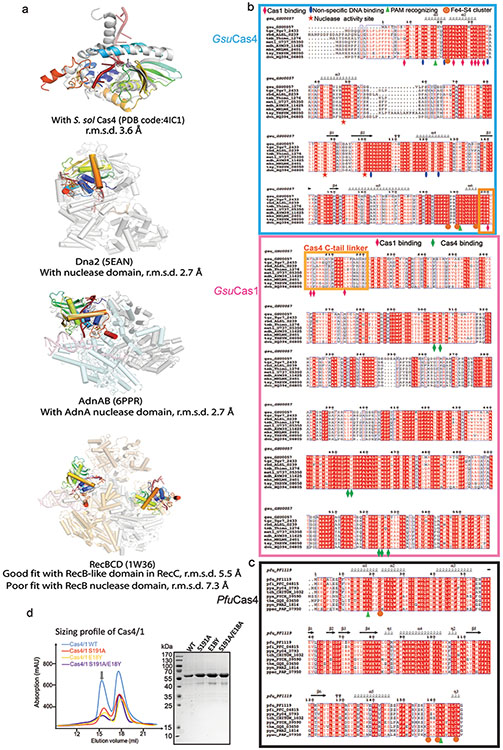
Among the four fused Cas4s, only the two PAM-engaging ones were resolved in the EM density; the other two Cas4s fused to the catalytic Cas1s were presumably too mobile (Fig. 1d-e). Since the Cas4/1 fusion does not alter the dynamic nature of the Cas4-Cas1-Cas2 interaction, the mechanistic insights from this study would apply to all Cas4 systems. This Cas4 structure aligns well with those of the stand-alone Cas4s19,20 and the nuclease domains in helicase-nuclease fusion proteins AddAB/AdnAB, RecBCD, and eukaryotic Dna2 (Extended Data Fig. 5). Cas4 organizes its structural modules to form a narrow passage for the PAM-containing 3′-overhang. Its N-terminal α-helical floor connects to the ceiling helix on the top, which reaches overhead to the RecB nuclease center on the opposite side, which then weaves back through the floor helix, and the remaining C-terminal region assembles with the N-terminal helical region to form the Fe-S cluster module, a hallmark to all Cas4 nucleases (Fig. 1h).
Importantly, the Cas4 interface on Cas1-Cas2 overlaps with that of the leader-repeat DNA for spacer integration (Fig. 1f)10,12. Cas4-binding therefore sterically blocks integration at the PAM-side Cas1. Cas4 contacts Cas1s through an extensive interface; many interface residues are conserved (Extended Data Figs. 4a-c, 5b). However, it is difficult to identify key interface residues that are universally conserved across all Cas4 branches. There may exist evolutionary pressure to maintain idiosyncratic Cas4 and Cas1-Cas2 interactions in order to avoid crosstalk among coexisting CRISPR systems. If true, this scheme would be analogous to the highly selective binding relationship between Cas3 and Cascade24.
Cas4-mediated PAM recognition explained
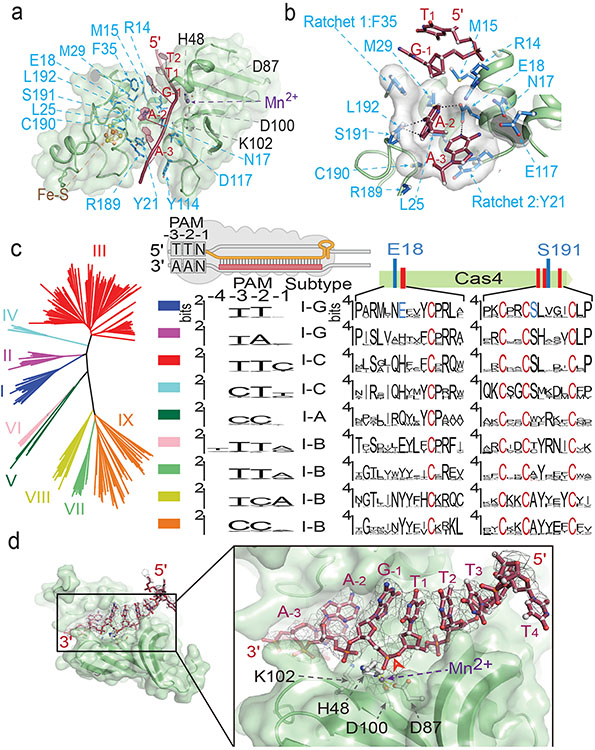
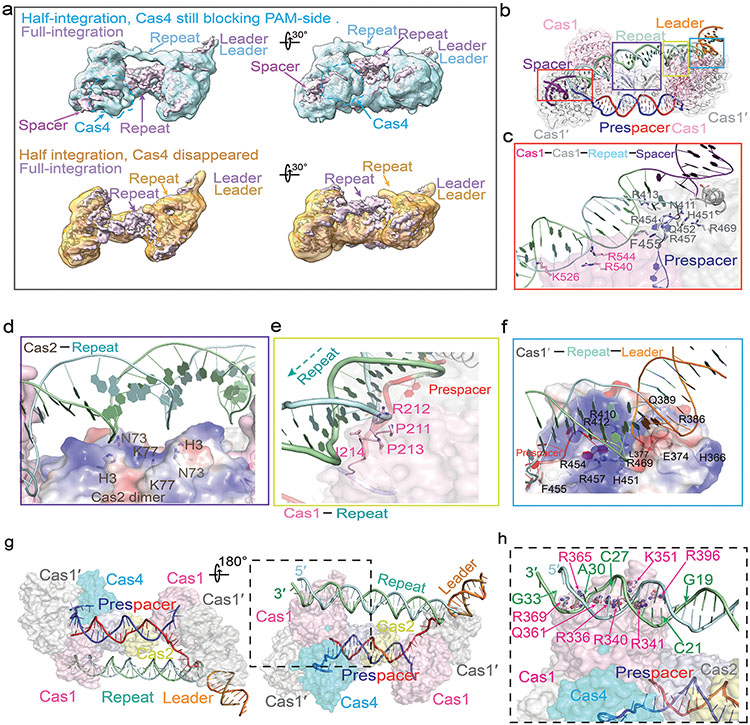
Despite extensive studies, the PAM recognition and cleavage mechanisms inside Cas4-Cas1-Cas2 remain unresolved. This EM structure brings such mechanisms into focus. The substrate-binding groove in Cas4 aligns with that in Cas1 to form a continuous 3′-overhang-binding groove. The 11-nt 3′-overhang (5′-dA7C6T5T4T3T2T1G−1A−2A−3 T−4) travels deep inside, protected from random nuclease cleavage. The nucleotides 1-4 travel along the previously described path towards Cas1 active site8,10,12. However, nucleotides 5-11 detour into Cas4 (Fig. 1d-e). They travel through the RecB nuclease module and into a narrow passage, where PAM recognition takes place (Fig. 2a). Two hydrophobic residues F35 and Y21 interdigitates into the ssDNA before and after the narrow passage, forming molecular ratchets that cage the di-deoxyadenosine PAM (3′-A−3A−2) inside (Fig. 2b). They likely enforce a ratcheting motion to slowly thread the 3′-overhang through. Inside the narrow passage, the edges of A−2 and A−3 are surrounded by hydrophobic or long side chain residues (R14, M29, L25, L192, E117, N17, C190) that probe nucleotides for shape complementarity. Deoxyguanosines would not fit in the same cage because their exocyclic N2 amines would cause steric clash; whereas the smaller-sized pyrimidines may slip through without a chance to establish favorable contacts. Two Cas4 residues establish polar contacts with PAM: E18 makes bidentate hydrogen-bonding interactions with A−2 and A−3, and S191 forms a hydrogen bond with A−2 (Fig. 2b). They likely contribute significantly to the PAM specificity. Consistent with the in vivo data 18, there is no sequence-specific recognition to the first residue of PAM, G−1. This nucleotide is excluded from the PAM-recognition box and points to the solvent (Fig. 2a-b).
Figure 2. Cas4-mediated PAM recognition delays overhang cleavage.
a. Cross-section of the narrow passage that sequesters the PAM-containing 3’-overhang. b. PAM (A−3A−3) is surrounded by Van der Waals interactions that probe for shape complementarity, and by sequence-specific hydrogen-bonding interactions from E18 and S191. c. Correlations between PAM code in Cas4-containing CRISPR systems and the recognition motif consensus in Cas4. d. Arrangement in the Cas4 nuclease center. Cryo-EM density suggests the PAM-containing overhang is not cleaved. Red arrow: labile bond.
Because Cas4 is responsible for PAM selection in a large fraction of CRISPR systems, we attempted to rationalize the PAM code in other CRISPR systems. A structure-guided mutagenesis was carried out to switch the PAM specificity of GsuCas4 to that of Pyrococcus furiosus Cas4. PfuCas4 share 17% sequence identity with GsuCas4 and specifies a 5′-CCN PAM (3′-GGN in the overhang). We substituted the two sequence-specific PAM contacting residues in GsuCas4 to their counterparts in PfuCas4. In single substitutions, S191A retained Gsu-PAM specificity; cleavage activity was slightly compromised. E18Y lost sequence-specific cleavage activity on both PAMs, and cleaved ssDNA distributively. Interestingly, the double substitution led to a cleavage preference for Pfu-PAM on a distributive cleavage background. These results suggest E18 plays a more important role than S191 in PAM recognition (Extended Data Fig. 4e-f) . However, this partial success in switching PAM specificity did not further extend into in vivo spacer acquisition assays, which put further demand on complex stability and PAM cleavage timing. While E18Y/S191A Cas4 showed compromised Gsu-PAM-prespacer integration, it was unable to support Pfu-PAM-prespacers integration (Extended Data Fig. 4g). These results suggest that while the hydrogen-bonding interactions are important, a significant portion of the PAM specificity is likely conferred by the peripheral residues mediating hydrophobic interactions.
Next, we used bioinformatics to establish a correlation between structural features in Cas4 and PAM sequence variations. A phylogenetic tree (Fig. 2c) was generated based on the alignment of Cas4s for which we could reliably couple PAM code with clades of Cas4s25. We expected that residues crucial for PAM selection would be conserved within the clades, but differ between groups selecting a different PAM (Fig. 2c). Structure-defined E18 is one such discriminant residue because it is highly conserved among Type I-G Cas4s specifying TTN PAMs and among Type I-B Cas4s specifying a TTA or TTG PAM. S191 is not a discriminant residue as it was also found in Type I-G Cas4s specifying TAN PAMs. However, the highly conserved neighboring residue, L192, was exclusively found in Cas4 groups specifying T−2 in PAM, including the distant Type I-C Cas4s that specify either TTC or CTT. Therefore, the presence of L192 in Cas4 is a good predictor of PAM-T−2. Similarly, informatics identified R14 and L25 as good predictors of T−2. The reverse argument is not necessarily true, as there is likely more than one evolutionary solution for Cas4 to specify a particular PAM.
PAM recognition prevents integration
The most important mechanistic insight from the dual-PAM structure is the observation that the PAM-containing 3′-overhang is recognized, sequestered, but not cleaved by Cas4 (Fig. 2c). The labile phosphate of G−1 is correctly positioned into the active site, which consists a DEK motif (D87, D100, K102) and a histidine residue (H48), all of which are highly conserved among Cas4 and RecB family of nucleases. These residues coordinate a catalytic metal ion, presumably Mn2+, which is shown by the EM density to be tightly coordinated to the scissile phosphate. In the AdnAB structure, such active site configuration was shown to cleave DNA efficiently26. However, here the EM density clearly argues for an intact DNA substrate at the active site (Fig. 2c). which was subsequently confirmed by denaturing PAGE (Extended Data Fig. 4d). The exact cleavage inhibition mechanism in Cas4 will require a more focused analysis in the future. Among the many mechanistic possibilities, we speculate that it might be caused by the sub-optimally placed K102 residue, an essential catalytic residue in the DEK motif18. Rather than pointing towards the labile phosphate, K102 is twisted away by the residing β-strand. A minor conformational change in Cas4 may reorient K102 to participate in PAM cleavage. Without PAM cleavage, Cas4 is trapped in place and the adjacent integration center is blocked. This structural observation agrees with the spacer directionality requirement in Type I CRISPR systems.
Directional spacer integration reconstituted
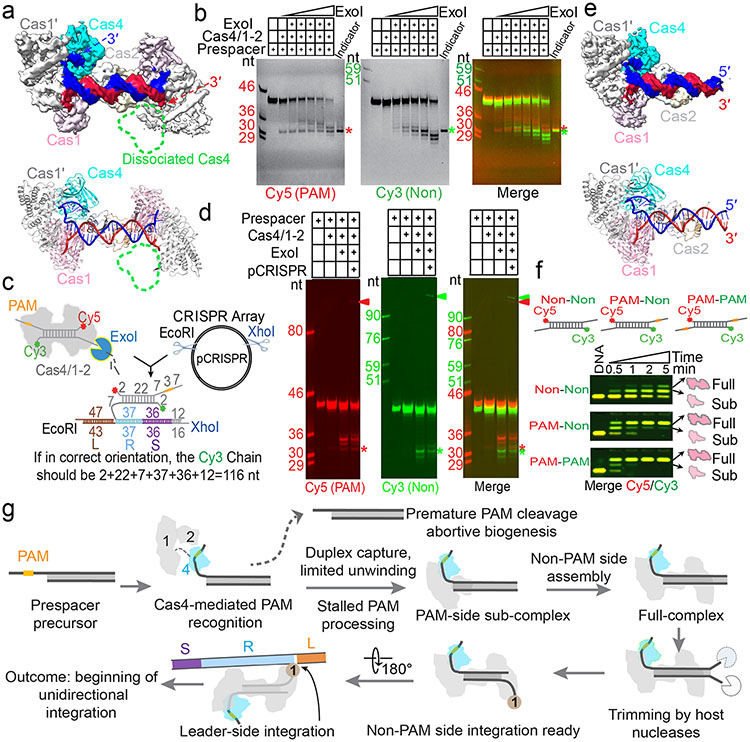
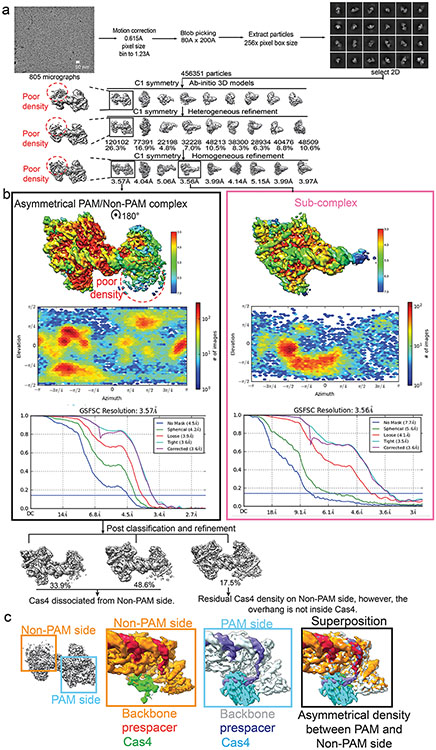
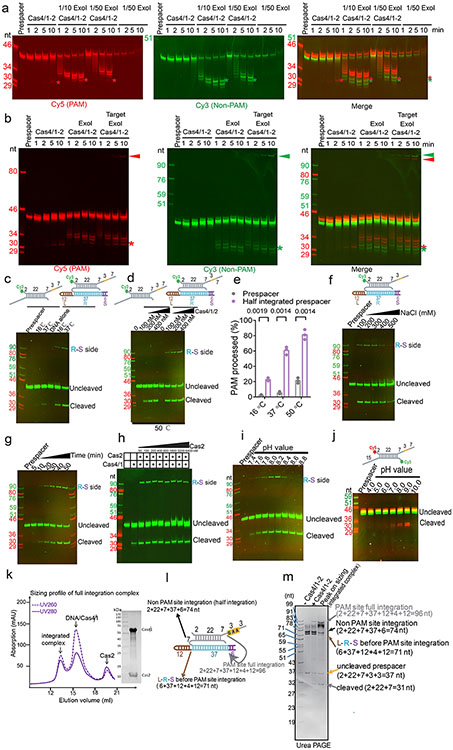
Next, to investigate the status of the non-PAM 3′-overhang, we determined the cryo-EM structure of the GsuCas4/Cas1-Cas2 complex programmed with a single-PAM containing prespacer. This led to an asymmetric full complex structure at 3.57 Å resolution, and a 3.56 Å assemble intermediate that will be discussed later (Fig. 3). Whereas a Cas4 docks onto the PAM-side of GsuCas4/Cas1-Cas2, 82.5% of the particles do not contain a docked Cas4 at the non-PAM side (Fig. 3a; Extended Data Fig. 6); 17.5% contain a docked Cas4 evidenced by the weak densities, however, the non-PAM overhang is not captured inside (Extended Data Fig. 6c). In both cases, the non-PAM side Cas4/1 dimer density is weaker than the PAM-side counterpart, due to a hinge motion around the non-catalytic Cas1. Only the first four nucleotides of the non-PAM 3′-overhang can be traced in the density, along a similar path as in the PAM-side (Extended Data Fig. 6c). Because the non-PAM overhang lacks Cas4 protection, we reasoned that it may be trimmed to the optimal overhang length by certain host nucleases, then captured by the nearby Cas1 and preferentially integrated to the leader-repeat DNA. This host nuclease-assisted integration mechanism would lead to a fixed spacer directionality that is consistent with the CRISPR biology. We directly tested this mechanistic model. Indeed, E. coli SbcB (ExoI) protein could trim the non-PAM 3′-overhang to the preferred length of ~7-nt, (Fig. 4b). Even the distributive cleavage pattern was categorically consistent with the spacer length distribution in the G. sul CRISPR systems (Fig. 1c)18. In the same reaction, the PAM-side 3′-overhang was protected by the footprint of Cas4 (Fig. 3b). Next, we established an in vitro integration assay to test whether the ExoI-trimmed prespacer can be integrated unidirectionally. An obstacle to this effort is that although GsuCas4/Cas1-Cas2 readily integrated prespacers into a negatively supercoiled leader-repeat-containing plasmid, it failed to do so on a linear dsDNA (Extended Data Figs. 7a-d). This behavior is similar to that of E. coli Cas1-Cas2, which was later shown to rely on the host integration factor (IHF) to integrate into a linear target27. Given the limitation, in order to resolve the integration directionality, we first integrated the fluorescently labeled prespacer into plasmid, then restriction-digested out the leader-repeat region to determine the integration directionality based on the product size on denaturing polyacrylamide gel (Extended Data Figs. 7c-f). In control experiments, we verified GsuCas4/Cas1-Cas2’s preference to integrate first into the leader-proximal side (Extended Data Figs. 7e-f). We went on to demonstrate that ExoI-trimming enabled the non-PAM side of the prespacer to specifically integrate into the leader-proximal side of the repeat (Fig. 3c-d). This pattern is in agreement with the observed spacer directionality in the G. sul CRISPR array.
Figure 3. Mechanistic insights from the single-PAM prespacer bound GsuCas4/Cas1-Cas2 structure.
a. Cryo-EM density and structure of the single-PAM prespacer bound GsuCas4/1-2 complex. Cas4 is absent from the non-PAM side. b. ExoI is capable of trimming the non-PAM overhang to the optimal length for integration; PAM-side is protected by Cas4. c. In vitro integration assay setup and the expected readout. d. Non-PAM overhang is unidirectionally integrated to the leader-proximal end of the CRISPR repeat after ExoI trimming. e. Cryo-EM density and structure of a sub-complex. Cas4/1 dimer is missing from the non-PAM side. f. EMSA showing Cas4/1-2 is assembled sequentially, and preferentially on PAM-containing prespacers. g. Mechanistic model explaining Cas4-dependent prespacer biogenesis and directional integration.
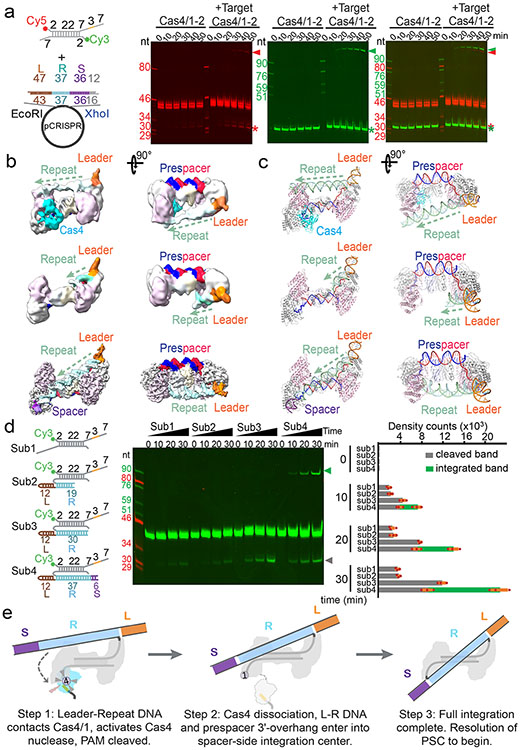
Figure 4. Structural basis for integration-coupled PAM cleavage by Cas4.
a. Time-course experiments showing half-integration stimulates PAM cleavage by Cas4, enabling full integration. b. Three cryo-EM reconstructions and c. corresponding structures captured from Cas4/1-2 incubated with half-integration mimicking substrate. They depict the initial blockage of spacer-side integration by PAM-engaging Cas4 (top), the Cas4 dissociation after PAM cleavage (middle), and the full-integration state (bottom). Resolutions are 5.83, 5.76, and 3.81 Å, respectively. d. Time-course experiments showing that PAM cleavage is stimulated when leader-repeat DNA contacts the spacer-side Cas4/1. Left: substrate design; middle: urea-PAGE; right: quantification of PAM cleavage and integration bands; Data was collected from N=3 biologically independent experiments and presented as mean ± s.e.m. e. Mechanistic model depicting the coupling between half-integration, PAM cleavage, Cas4 dissociation, and full-integration.
Structural basis for prespacer biogenesis
The single-PAM cryo-EM reconstruction further captured an important functional state that corresponds to a prespacer biogenesis intermediate. In this 3.6 Å structure, the PAM-side arrangement is essentially the same and the mid-duplex is protected by a Cas2 dimer, however, the non-PAM side lacks the (Cas4/Cas1)2 protection (Fig 3e; Extended Data Fig. 6). This structure raises the mechanistic possibility that components of the integration complex assemble onto prespacer in a stepwise fashion. Indeed, in time-course and concentration-titration based electrophoretic mobility shift assays (EMSA), the GsuCas4/Cas1-Cas2 integrase was found to assemble in a stepwise fashion, and the PAM-containing overhang strongly promoted the assembly of the full-complex (Fig 3f; Extended Data Fig. 7g). Collectively, these structural snapshots provide the much-needed temporal resolution for prespacer biogenesis. We conclude that the (Cas4/Cas1)2-Cas22 sub-complex is capable of scouting for precursor DNA with a PAM-containing 3′-overhang. Binding of such precursor triggers enzymatic stalling in Cas4 and recruits a second (Cas4/Cas1)2 complex to the opposite side, leading to the formation of an integration-competent (Cas4/Cas1)4-Cas22 full complex. The stepwise assembly process provides a quality-control mechanism to selectively recruit PAM-containing precursors for further processing and integration (Fig. 3g; Supplementary Information Video 1). The length of the precursor duplex is likely longer than the preferred duplex length by Cas14-Cas22. In a previous study we explored this scenario and found that the host nucleases are capable of trimming the duplex and overhangs to optimal prespacer specifications as defined by the Cas14-Cas22 footprint11.
Half-integration triggers PAM cleavage
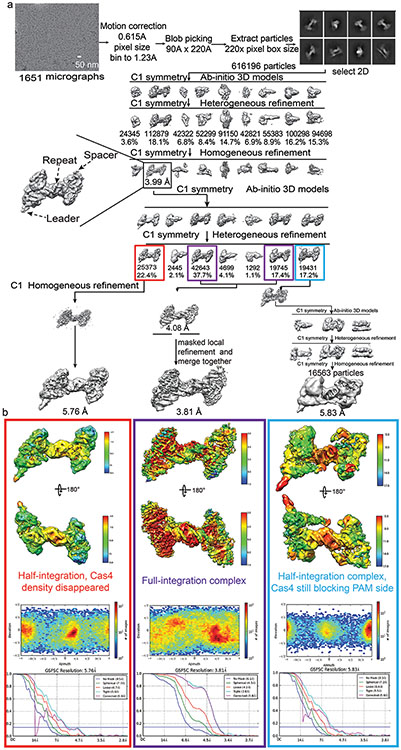
Having established that Cas4 defines the spacer directionality by blocking the PAM-side integrase center before integration, we next probed into the mechanism that relieves this blockage after half-integration, since the PAM-side prespacer needs to be processed and integrated to the opposite side of the CRISPR repeat to complete full integration. What serves as the molecular switch? We hypothesized that the half-integration itself may stimulate PAM cleavage and Cas4 dissociation. To test this, we programmed GsuCas4/Cas1-Cas2 to the half-integrated state using an annealed prespacer and leader-repeat DNA that mimics the half-integration product10, and monitored the extent of PAM processing and half-to-full integration transition at different conditions (Extended Data Fig. 8a-j). Indeed, half-integration led to faster and higher extent of PAM cleavage, and full integration quickly followed (Fig. 4a; Extended Data Fig. 8b). As controls, PAM cleavage was much slower and weaker when the leader-repeat DNA was absent (Fig. 4a).
To understand the structural basis for the observed mechanistic coupling, we snap-froze the reacted sample (Extended Data Figs. 8k-m) for cryo-EM analysis. We were able to capture three conformational states from the single-particle reconstruction, each depicts a distinct functional state during the half-to-full integration transition (Extended Data Figs. 9). The three states differ significantly in their spacer-side contacts and in Cas4 and integration status. In the 5.83 Å early state reconstruction, density clearly reveals that Cas4 still blocks the PAM-side integration site and the PAM-containing 3′-overhang is still sequestered in Cas4. Unable to dock into the integration site, the CRISPR repeat reaches over from the leader-side Cas1 directly to the spacer-side counterpart, without contacting the Cas2 dimer in the middle. The spacer-side CRISPR repeat contacts a positively-charged region on Cas1, near Cas4 (Fig. 4b-c; Extended Data Fig. 10g). The DNA density is weak, suggesting that it samples multiple conformations. In the 5.76 Å intermediate state, the Cas4 density disappears and the CRISPR repeat DNA points towards the spacer-side integration center, however, its density is too weak for model building at the spacer-side (Fig. 4b-c; Extended Data Fig. 10a). This suggests that even with Cas4 dissociation, the spacer-side CRISPR DNA capture and integration is inefficient due to the lack of favorable leader-sequence contacts11. Lastly, in the 3.81 Å full-integration state reconstruction, densities clearly reveal that the CRISPR repeat DNA has been accommodated into the spacer-side integration center, and full-integration has taken place (Fig. 4b-c). This snapshot is architecturally similar to the E. fae post-integration structure12, however, the G. sul leader-repeat DNA is not as sharply kinked at the Cas2 binding site as the E. fae counterpart (Extended Data Fig. 10). Connecting the dots together, the three snapshots define the order of molecular events and support a strong mechanistic coupling between the leader-half integration and the Cas4-mediated PAM processing, which enables PAM-specific spacer-side integration.
How does the leader-side integration activate spacer-side PAM cleavage remotely? There are at least two mechanistic possibilities: 1) the leader-half integration may trigger a global conformational change that allosterically activates Cas4; 2) the physical contacts by the integrated leader-repeat DNA somehow activates Cas4. Since no significant conformational change in Cas1-Cas2 was observed among apo, half- and full-integration structures, we ruled out the allosteric activation model and probed deeper into the role of the leader-repeat DNA contact on Cas4 activation. The leader-repeat DNA in the integration assay was systematically shortened (Fig. 4d). A strong correlation was observed. When the leader-repeat was too short to reach spacer-side Cas4/1 (Sub2: 19-bp CRISPR repeat), the extent of PAM cleavage was indistinguishable from the prespacer-only control. When the leader-repeat was long enough to reach the spacer-side Cas4/1 but still too short to allow spacer-side integration (Sub3: 30-bp CRISPR repeat), the PAM cleavage was significantly enhanced, approaching the extent in the positive control (Sub4) (Fig. 4d). We therefore conclude that contacts by the half-integrated DNA efficiently stimulates the PAM cleavage activity of Cas4. PAM cleavage leads to Cas4 dissociation, which exposes the spacer-side integrase center and allows full integration (Fig. 4e; Supplementary Information Video 2).
Discussion
We provide a comprehensive set of mechanism to explain the PAM-dependent spacer acquisition process in Cas4-containing CRISPR systems. Our study firmly establishes that Cas4 is a dedicated PAM-cleaving endonuclease that is tightly regulated. In the context of the Cas1-Cas2 integrase complex, Cas4 specifically recognizes but refrains from cleaving the PAM-containing 3′-overhang in a prespacer. This unexpected “molecular constipation” is the cornerstone for productive prespacer biogenesis and functional spacer integration in Types I and V CRISPR systems. We provide direct evidence that PAM recognition and the subsequent “molecular constipation” takes place early during prespacer biogenesis. In essence Cas4 serves as a gatekeeper to only channel productive precursors into the biogenesis pathway. We further show that host nucleases can assist the further processing of these precursors, and this eventually leads to a directional integration to the leader-side CRISPR repeat. Moreover, we reveal that the leader-side integration efficiently activates the PAM cleavage activity of Cas4, which causes Cas4 dissociation and allows the half-to-full integration transition. Exactly how spacer directionality is established in Cas4-less CRISPR systems requires further investigation. In Type I-E CRISPR, such mechanism has been shown to involve Cas1-mediated PAM sequestration and integration-dependent desequestration13,28. Therefore, the PAM-dependent blockage/activation of the two integration centers in Cas1-Cas2 may be a universal theme to achieve directional spacer integration.
The structural similarity of Cas4 to the nuclease domains of AddAB/AdnAB and a structural domain in the equivalent location in RecBCD sheds light on the ancient function of Cas4 in spacer acquisition. These helicase-nuclease machines not only play essential roles in homology-directed repair, but also provide innate immunity for bacteria by preferentially degrading linear DNA lacking χ sites, which are more likely of foreign origin. Functional interactions between RecBCD/AddAB and Cas1-Cas2 mediated spacer acquisition have been noted in previous studies29,30. Certain traits in the AdnA nuclease (and its structural equivalent in RecBCD) may have made them particularly desirable by Cas1-Cas2. For example, the subtle sequence preference and occasional enzymatic pausing may have been exploited by Cas1-Cas2 to establish PAM-dependent directional integration. This dramatically increased the productive spacer acquisition in the ancient CRISPR systems. It is possible that the ancient Cas1-Cas2 relied on RecBCD or AddAB for spacer precursors so heavily, that it started to establish a physical interaction with the nuclease domain to facilitate the process. It eventually led to the hijacking of this host nuclease domain into the cas operon as cas4.
Methods
PAM prediction
221,089 unique spacers along with genome source, cas gene information31,32, and repeat sequence were obtained from CRISPRCasDb33 in February 2020. These spacers were blasted against our own sequence database containing all sequences from the NCBI nucleotide database34,35, environmental nucleotide database36, PHASTER37, Mgnify38, IMG/M39, IMG/Vr40, HuVirDb41, HMP database42, and data from Pasolli et al.43. All databases were accessed in February 2020.
Hits between spacers and sequences from the aforementioned nucleotide databases were obtained using the BLASTN program44 version 2.10.0, which was run with parameters word_size 10, gap open 10, penalty 1 and an e-value cutoff of 1. Hits inside CRISPR arrays were detected and filtered out by aligning the repeat sequence of the spacer to the flanking regions of the spacer hit (23 nucleotides on both sides). To minimize the number of false positive hits, we further filtered hits based on the fraction of spacer nucleotides that hit the target sequence. In a first step, only hits with this fraction >90% were kept. To find targets for even more spacers while keeping the number of false positives low, we included a second step where hits with a matching percentage >80% were kept if another spacer from the same phylogenetic genus hit the same sequence in the stringent first round. Finally, we removed spacers that were shorter than 27 nt.
Highly similar repeat sequences of the same length were clustered using CD-HIT45 with a 90% identity threshold. To increase the number of aligned sequences for PAM5,46,47 determination, we hypothesized that similar repeat sequences would be used in the same orientation and would correspond to the same PAM sequences, as coevolution of PAM, repeat and Cas1 sequences has been shown previously48,49. The PAM for each aligned repeat cluster was then determined by aligning the flanking regions of the spacer hits in each cluster. To equally weigh each spacer within the repeat cluster, irrespective of the number of blast hits, consensus flanks were obtained per spacer. These consensus flanks contained the most frequent nucleotide per position of the flanking regions. From the alignment of consensus flanks (for clusters with at least 10 unique spacer hits) the nucleotide conservation in each flank was calculated. Conserved nucleotides were considered part of the PAM in case nucleotide conservation was higher than 0.5 bit score, and the bit score in that position was at least 5 times higher than the median bit score of the two 23-nt flanks. This PAM database was manually curated to fix PAMs determined incompletely when nucleotides that were slightly below the threshold did occur in other repeat clusters of the same subtype. The orientation of the PAM was set to match the overall orientations of experimentally determined PAMs in literature for different systems (upstream of 5′-end of the protospacer in Type I systems and downstream of 3′ of the protospacer in Type II systems).
Cas4 phylogenomics
Cas4 sequences were retrieved from each Cas4-containing genome in the PAM database. Cas4 sequences were discarded in case multiple Cas4 sequences of that subtype (subtypes defined by CRISPRCasdb) were present in a single genome, or when Cas4 belonged to a different subtype than the predicted subtype of the repeat cluster. The tree was generated with PhyML50 from a MAFFT alignment of all Cas4 sequences51. The sequence logos were generated with Berkeley weblogo52 and were performed on each group of Cas4 sequences with a similar PAM, where redundant sequences were removed by CD-hit (threshold 0.9). For groups with a small amount of nonredundant sequences (I-G TTN, I-G TAN and I-C CTT), additional Cas4 sequences were retrieved by BLAST search of repeat sequences of predetermined PAM repeat clusters and retrieving adjacent Cas4 sequences in the NCBI nucleotide database.
Bacterial strains
See Supplementary Information Table 1 for plasmids and their corresponding selection markers.
Plasmid construction
Plasmids used in this work are listed in Supplementary Information Table 1. The type IG CRISPR-Cas acquisition module from G. sulfurreducens DSMZ 12127 was amplified by PCR using the Q5 High-Fidelity Polymerase (New England Biolabs) and primers BN462 and BN1196 (Supplementary Information Table 2). The amplicon was cloned into the p13S-S ligation-independent (LIC) cloning vector (http://qb3.berkeley.edu/macrolab/addgene-plasmids/) by TA cloning, generating plasmid pCas4/1-2. For plasmid pCRISPR, a synthetic construct composed of T7 terminator, a CRISPR array (leader-repeat-spacer1-repeat), the mCherry gene, and flanking 20-bp homology regions to the vector, was introduced into pET cloning vector 2A-T amplified with primers BN1247 and BN1650 by Gibson assembly. E18Y mutant of Cas41 (pCas4/1-2-E18Y) was generated by mutagenesis using pCas4/1-2 as a template with primers BN3392 and BN3393. Double mutant E18Y/S191A (pCas4/1-2-E18Y/S191A) was generated by mutagenesis using pCas4/1-2-E18Y as a template with primers BN3394 and BN3395. All plasmids were verified by Sanger sequencing.
Spacer acquisition assay
Escherichia coli BL21-AI was co-transformed with pCas4/1-2, pCas4/1-2-E18Y, or pCas4/1-2-E18Y/S191A and pCRISPR. Colonies were grown in 5 ml of LB supplemented with spectinomycin and ampicillin at 37 °C with shaking. After 2.5 h of growth, the expression of cas genes was induced with IPTG and L-arabinose, and the cultures were incubated for additional 2 h. Cells were made electrocompetent and transformed with 5 μl of each 50 μM prespacer prepared by mixing primers (Supplementary Information Table 2) at 1:1 from the 100 μM stock. Cells were recovered in LB for 1h at 37 °C, 180 rpm, and then grown overnight in 10 ml of LB supplemented with spectinomycin and ampicillin at 37 °C with shaking. Plasmids were extracted from the overnight cultures (Thermo Scientific GeneJet Plasmid Extraction Kit) and digested with EcoRI and NcoI to avoid amplification of larger products from the plasmid backbone. Digested plasmids were used to detect spacer acquisition by PCR using OneTaq 2x MasterMix (New England Biolabs) and a mix of three degenerate primers with different 3′ nucleotides (BN464, BN465, and BN1314) and primer BN170817. Samples were run on 2% agarose gels and visualization for spacer acquisition using SYBR Safe. Unexpanded and expanded band percentages were determined using the Analysis Tool Box of ImageLab software using unmodified images. The expanded CRISPR DNA band was purified by automated size selection and submitted to a second round of PCR using the degenerate primers and the internal reverse primer BN175417,53.
Expanded CRISPR array sequencing
PCR amplicons of the expanded CRISPR arrays were purified using the GeneJET PCR Purification kit (Thermo Fisher Scientific) and the DNA concentration was measured using Qubit Fluorometric Quantification (Invitrogen). Samples were prepared for sequencing using the NEB Next Ultra II DNA Library Prep Kit for Illumina and each library was individually barcoded with the NEBNext Multiplex Oligos for Illumina (Index Primers Set1 and Set2). Sample size and concentration were then assessed using the Agilent 2200 TapeStation D100 high sensitivity kit, and samples were pooled with equal molarity. Pooled samples were denatured and diluted as recommended by Illumina and spiked with 15% of PhiX174 control DNA (Illumina). Sequencing was performed on a Nano flowcell (2 × 250 base paired-end) with an Illumina MiSeq. Image analysis, base calling, de-multiplexing, and data quality assessments were performed on the MiSeq instrument. Resulting FASTQ files were analyzed by pairing and merging the reads using Geneious 9.0.5. Acquired spacers were extracted and analyzed as described previously17.
Cloning, expression and purification
Gsu_cas4/1 (Gsu0057 in KEGG) gene was cloned into pET28a -His6-Twin-Strep-SUMO vector or pGEX-41-T-His6-Flag-GST, between BamHI and XhoI sites and expressed in E. coli BL21 (DE3) star cells. A 6-L cell culture was grown in LB medium at 37 °C until OD600 reached 0.5. Expression was induced with 0.5 mM IPTG, 0.2 mg/mL ferrous sulfate and 0.4 mg/mL L-cysteine at 16 °C overnight. Harvested cells were resuspended in 100 mL buffer A containing 50 mM HEPES pH 7.5, and 500 mM NaCl, 10% glycerol, and 5 mM TCEP, lysed by sonication, and centrifuged at 17,000 g for 50 min at 4 °C. The supernatant was transferred into anaerobic conditioned glove box and applied onto the pre-equilibrated 4 mL Ni-NTA column (SUMO tagged expression) or 5 mL GST column (GST tagged expression). After washing with 100 ml of buffer A, the protein was eluted with 20 ml buffer A plus 300 mM imidazole for SUMO-tagged purification and buffer A plus 15 mM reduced GSH for GST tagged purification, then incubated with SUMO-protease or 3C protease at 4 °C for 2 h. 2 mL of concentrated eluate was loaded onto a Superdex 200 16/60 size-exclusion column (GE Healthcare) equilibrated with buffer C (10 mM HEPES pH 7.5, 500 mM NaCl, and 5 mM TCEP), the peak fractions were pooled and snap-frozen in liquid nitrogen for later usage.
Gsu_cas2 (Gsu0058 in KEGG) gene was cloned into His6-Twin-Strep-SUMO-pET28a vectors (KanR) between BamHI and XhoI sites. Protein expression, Ni-NTA purification, and SUMO-tag cleavage were carried out in similar conditions as for the His-SUMO-Cas4/1. Cas2 after tag-cleavage was purified on Superdex 200 16/60. The peak fractions were pooled and snap-frozen in liquid nitrogen for later usage.
Affinity pull-down assay
15 μg GST-tagged Cas4/1 and 30 μg untagged Cas2 were mixed and incubated with 10 μL GST resin at 4 °C for 30 min in different salt concentration buffer (50 mM HEPES pH7.5, 10% glycerol, 5 mM TCEP, and 150/300/500 mM NaCl) in presence or absence of prespacer, in a total assay volume of 50 μL. The GST resin was pelleted by centrifugation at ~100 g for 30 seconds, washed 3 times with 200 μL of the corresponding binding buffer, then eluted with 70 μL elution buffer (50 mM HEPES pH7.5, 500 mM NaCl, 5 mM TCEP, and 15 mM reduced GSH). Eluted proteins were separated on 12% SDS-PAGE and stained by Coomassie blue.
Fluorescently labeled prespacer substrate preparation
Fluorescent DNA oligos (Supplementary Information Table 2) for biochemistry were synthesized (Integrated DNA Technologies) with either a /5AmMC6/ or /3AmMO/ label, fluorescently-labeled in-house, and annealed at equimolar amount, and native PAGE purified to remove unannealed ssDNA.
Prespacer cleavage assays
Prespacer cleavage assays were set up in 20 μL reactions containing 10 nM final concentration of labelled prespacer, 500 nM Cas4/1, 250 nM Cas2 in a cleavage buffer containing 50 mM Tris pH 8.0, 100 mM KCl, 10% glycerol, 5 mM TCEP, and 5 mM metal ion MnCl2 or different metal ions in Extended Data Fig. 1h. After 37 °C incubation for 1 h, reactions were quenched by vortexing with 20 μL of phenol-chloroform. The extracted aqueous phases were mixed with equi-volume of 100% formamide and separated on 13% urea-PAGE. Signals from each fluorescent dye were recorded using ChemiDoc (Bio-Rad). The KMnO4 foot printing assay was carried out following previously published protocols12.
Reconstitution of prespacer bound/integration Cas4/1-2 complex
Complex was formed by mixing Cas42/Cas12, Cas2, and prespacer (or half-integration mimicking substrate) at a final concentration of 30 μM, 60 μM, and 60 μM respectively in 500 μL total volume with a reconstitute buffer containing 25 mM Tris pH 8.0, 300 mM NaCl, 5 mM TCEP and 5 mM MnCl2. After 37 °C incubation for 30 min, the complex was separated on Superdex 200 16/30 column equilibrated in the same buffer. The full-complex peak was pooled and concentrated to appropriate concentration and snap-frozen in liquid nitrogen for long-term storage.
Integration assays
The in vitro integration assays were set up as follows. 10 nM of prespacer were incubated with 250 nM Cas4/1-2 complex in the integration buffer containing 50 mM Tris pH 8.0, 100 mM KCl, 5 mM TCEP and 5 mM MnCl2 in 20 μL reaction volume. After an initial incubation at 37 °C for 5 min, 300 ng of pCRISPR plasmid was introduced into the reaction. Integration was allowed at 37 °C for 1 h, after which 0.5 μL of EcoRI and XhoI restriction enzymes (NEB) were introduced for 10 min more at 37 °C to digest out the leader-repeat region of the plasmid, together with the integrated prespacer. Reactions were quenched by vortexing with 20 μL phenol-chloroform solution. The extracted aqueous phase was mixed with equi-volume of formamide, separated on 13% urea-PAGE, and scanned on ChemiDoc imaging system.
ExoI trimming and follow-up integration assays
10 nM of prespacer were pre-incubated with 250 nM of Cas4/1-2 complex at 37 °C for 5 min in 20 μL containing the trimming buffer (50 mM Tris pH 8.0, 100 mM KCl, 10% glycerol, 5 mM TCEP, 5 mM MnCl2 and 10 mM MgCl2). The 2-fold ExoI dilution series in Fig. 3b was prepared by dilution E. coli ExoI (NEB, 20 U/μL) to a final concentration of 0.2, 0.1, 0.05, 0.025, 0.0125 U/μL in each reaction. The 1/10 and 1/50 ExoI concentrations in the Extended Data Fig. 9a correspond to 0.1, 0.02 U/μL. The ExoI concentration in the Extended Data Fig. 8b was 0.1 U/μL across. In reactions where the trimming and integration were coupled, 300 ng of pCRISPR plasmid (~5 nM final concentration) was introduced at the same time with ExoI into the reaction. After incubation, the reaction was quenched by mixing with equi-volume of a buffer containing 95% formamide, 10 mM EDTA and 0.2% SDS, phenol-extracted, then separated on 13% urea-PAGE, and scanned on ChemiDoc imaging system (Bio-Rad), as described above.
Electrophoretic mobility shift assay
2 nM final concentration of fluorescently-labeled prespacer DNA was incubated with an increasing concentration of Cas4/1-2 complex for 15 min (in concentration titration experiments), or with 50 nM Cas4/1-2 complex for 0.5, 1, 2, 5 min (in time-course experiments) at 4 °C in a total 20 μL system containing 50 mM Tris pH 8.0, 100 mM KCl, 5 mM TCEP, 5 mM MnCl2 and 10% glycerol. After incubation, 15 μL of each sample was loaded onto 1% agarose gel equilibrated in 1x TG buffer (20 mM Tris pH 8.0, 200 mM glycine) immediately. Electrophoresis was performed at 60 V for 40 min. The fluorescent signals from the gel were recorded using a ChemiDoc imaging system (Bio-Rad).
Negative-stain electron microscopy
4 μL of 0.01 mg/mL prespacer-bound Cas4/1-2 complex was applied to a glow-discharged copper 400-mesh continuous carbon grid. After a 30-sec incubation, the grid was blotted on a filter paper, immediately transferred carbon-face down on top of a 2% (w/v) uranyl acetate solution for 1 min. The grid was then blotted on a filter paper again to remove residual stain, then air-dried on bench for 5 min. The grid was examined under a Morgagni transmission electron microscope operated at 100 keV with a direct magnification of x140000 (3.2 A ° pixel size) by AMT camera system. Each image was acquired using an 800 ms exposure time and −1 to −2 mm defocus setting. Data processing and 2D classification were performed on CyoSPARC software.
Cryo-EM data acquisition
4 μL of 0.6 mg/mL SEC-purified prespacer-bound or half-integration mimicking substrate-bound Cas4/1-2 complexes were applied to a Quantifoil holey carbon grid (1.2/1.3, 400 mesh) which had been glow-discharged for 30 sec. Grids were blotted for 4 sec at 6 °C, 100% humidity and plunge-frozen in liquid ethane using a Mark IV FEI/Thermo Fisher Vitrobot. Cryo-EM images were collected on a 200 kV Talos Arctica transmission microscope (Thermo Fisher) equipped with a K3 Summit direct electron detector (Gatan). The total exposure time of each movie stack was ~ 3.5 sec, leading to a total accumulated dose of 50 electrons per A ° which fractionated into 50 frames. Dose fractionated super-resolution movie stacks collected from the K3 Summit direct electron detector were 1x binned to a pixel size of 1.234 Å. The defocus value was set between −1.5 μm to −3.5 μm.
Cryo-EM data processing
Motion correction, CTF-estimation, blob particle picking, 2D classification, 3D classification and non-uniform 3D refinement were performed in cryoSPARC v.254. Refinements followed the standard procedure, a series of 2D and 3D classifications with C1 symmetry were performed as shown in Extended Data Fig. 4a, Extended Data Fig. 7 and Extended Data Fig. 10a, to generate the final maps. A solvent mask was generated and was used for all subsequent refinement steps. CTF post refinement was conducted to refine the beam-induced motion of the particle set, resulting in the final maps. The final map ‘CTF Post-refinement was used to estimate resolution based on the Fourier shell correlation (FSC) = 0.143 criterion after correcting for the effects of a soft shape mask using high-resolution noise substitution. We noticed that the map of the full-integration complex was not homogeneous in both sides, so we divided the map into two half parts from the middle site by Chimera UCSF. Then imported two half maps into Relion 3.055 to make a mask for next masked local refinement respectively. Finally imported these two masks into cryoSPARC again and did a local refinement to get two half local refined maps and merged two maps to a final map in Extended Data Fig. 10. The detailed data processing and refinement statistics for all cryo-EM structures are summarized in Extended Data figures and Extended Data Table 1.
Data availability
The cryo-EM density maps that support the findings of this study have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers of EMD-23839 (PAM/PAM prespacer bound), EMD-23840 (PAM/Non-PAM prespacer bound), EMD-23843 (full integration complex), EMD-23845(half integration, Cas4 still blocking the PAM side), EMD-23849 (half integration, Cas4 dissociated), and EMD-23847 (sub-complex). The coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers of 7MI4 (PAM/PAM prespacer-bound), 7MI5 (PAM/non-PAM prespacer-bound), 7MI9 (full integration), 7MIB (half integration, Cas4 still blocking the PAM side), 7MID (sub-complex). MiSeq sequencing data that support analysis of in vivo prespacer integration have been deposited in the European Nucleotide Archive (ENA) under accession number PRJEB41616. Plasmids used in this study are available upon request.
Statistics and Reproducibility
We typically drew biochemistry conclusions based on the best quality gels. Such gels typically were repeated multiple times during the optimization stage to ensure reproducibility, albeit they may not have been repeated in the exact same format or loading sequence. When a conclusion was drawn based on the band intensity changes/differences in a gel, we typically carried out n=3 biologically independent assays to ensure reproducibility and statistical significance (i.e. Fig. 4d; ED Figs. 8e). In vivo assays were carried out in n=3 biologically independent assays for quantification. All data points were displayed on the figure panels.
Extended Data
Extended Data Figure 1. Reconstitution and characterization of the GsuCas4/Cas1-Cas2 complex.
a. Active site substitution in Cas4 nuclease center (H48G, D100A) reduced in vivo spacer acquisition efficiency dramatically. Left three panels display the WebLogo of PAM code from spacers integrated by each Cas4/1-2 variant. Rightmost panel displays the number of deep-sequencing reads that confirm spacer integration. b-d. GsuCas4/1 purification analyzed by SDS-PAGE, coloring from the Fe-S cluster, and SEC profile, respectively. e,f. Affinity purification of GsuCas2, SDS-PAGE, and SEC analysis, respectively. g. GST pull-down experiments revealing the physical interaction between GsuCas4/1 and GsuCas2, with or without prespacer present. h. Metal ion dependency in PAM cleavage reaction. i. Biochemistry showing Cas4/1-2 specifically cleaves the PAM-embedded 3′-overhang in prespacer. j. PAM-cleavage specificity is lost over time, presumably due to Fe-S oxidation in Cas4. k. SEC profile of GsuCas4/Cas1-Cas2, alone or programmed with different prespacer substrates. PAM-containing prespacers drive high-order complex formation. l. Cryo-electron micrographs of three different complexes, with corresponding preliminary 2D averages to investigate sample quality.
Extended Data Figure 2. In-depth analysis of the dual-PAM prespacer bound GsuCas4/Cas1-Cas2 structure.
a. Comparison between the current 3.2 Å cryo-EM reconstruction with the previous negative staining reconstruction of the B. hal Cas4/1-2 complex (EMDB 20131)22. b-d. Pairwise alignment between GsuCas4/Cas1-Cas2/prespacer and EcoCas1-Cas2/prespacer8,31 (PDB 5DS4), EfaCas1-Cas2/prespacer12 (PDB 5XVN), and EfaCas1-Cas2/full-integration12 (PDB 5XVO), respectively. Alignments details are noted on the figure panel. Inset: the C-terminal tail of Cas2 plays similar roles in G. sul and E. fae structures in mediating edge-stacking with both Cas2 and Cas1. e. PAM was processed similarly in 22-bp or 26-bp mid-duplex containing prespacer by GsuCas4/Cas1-Cas2. f. SEC profile was similar when the two different prespacers were used to assemble the complex. g. Validation that prespacers containing a 22-bp mid-duplex are actively acquired in vivo. N=3 biologically independent assays were evaluated by PCR detection as shown, as well as relative percentages of expanded and non-expanded bands. Data presented as mean ± s.e.m.
Extended Data Figure 3. Flow-chart of the cryo-EM single particle reconstruction of the dual-PAM prespacer bound GsuCas4/Cas1-Cas2.
a. Cryo-EM reconstruction workflow for the dual-PAM prespacer bound Cas4/1-2 complex. b. Cryo-EM density of the dual-PAM prespacer bound Cas4/1-2 complex, colored according to local resolution (top). The viewing direction distribution plot (middle) and FSC curves (bottom) for data processing. c. Representative EM densities for Cas2, Cas4, and Cas1, superimposed with their corresponding structural model.
Extended Data Figure 4. In-depth GsuCas4/Cas1-Cas2 interface analysis and structure-guided mutagenesis attempt to switch PAM specificity.
a. Overall dual-PAM structure. Insets: zoom-ins of interface between Cas4 and the two neighboring Cas1s. Cas4 connects to the non-catalytic Cas1 through a 20-amino acid fusion linker (colored in yellow), which mediates the dynamic docking and dissociation of Cas4. b. Surface electrostatic potential. Left inset: Cas2 contacts to the mid-duplex; Right inset: Cas1 end-stacking to the mid-duplex. Residues responsible for guiding the 3’-overhang are also shown. Cas1-Cas2 was found to specify a 22-bp mid-duplex rather than a 26-bp mid-duplex as defined by the integration assay; an additional two base-pairs are unwound from each end, and the mid-duplex is end-stacked by the N-terminal domain of the catalytic Cas1s on opposite ends. The 22-bp specification and the limited end-unwinding activity was previously observed in EfaCas1-Cas2 11,12. c. Cas1-Cas2 and Cas4-Cas2 interfaces. Top inset: the highly conserved C-terminus of Cas2 inserting into a hydrophobic pocket in Cas1, stabilizing complex formation. Bottom inset: the ceiling helix of Cas4 (aa 39-50) makes extensive polar contacts with a helix in Cas2 (aa 42-53). d. SEC, SDS-PAGE, and urea-PAGE analyses of the prespacer-bound complex used in cryo-EM analysis. They reveal the molecular weight, protein integrity, and prespacer integrity, respectively. For example, urea-PAGE reveals the PAM-overhang is not cleaved inside the Cas4/1-2 complex. e. Modeling the impact on PAM recognition by introducing the equivalent residues of E18 and S191 in P. fur CasA into G. sul Cas4 (E18Y and S191A substitutions). Specific atom changes in A-to-G switching (N6O substitution and N2 amine addition) are highlighted in colored balls. The steric clashes (lightening arrows) to PfuPAM (3’-GGN in the 3’-overhang) are expected to be partially relieved when substitutions are in place. f. Impact of E18Y and S191A substitutions on PAM cleavage activity. g. In vivo spacer acquisition assay results for the wild type and PAM-specificity Cas4 mutants. While E18Y/S191A Cas4 showed compromised Gsu-PAM (TTN) prespacer integration, it was able to support integration of Pfu-PAM (CCN) containing prespacers in vivo. N=3 biological independent assays were analyzed by PCR and the band quantification revealed integration efficiency. Data presented as mean ± s.e.m.
Extended Data Figure 5. In-depth analysis of the structure and sequence conservation in Cas4.
a. Superposition of GsuCas4 with a standalone Cas419,20, and the nuclease domains in helicase-nuclease fusion proteins AddAB32, AdnAB26, RecBCD33, and eukaryotic Dna234. The caging of the ssDNA substrate and the arrangement of the Fe-S cluster and the catalytic triad are conserved themes. Interestingly, the Cas4 structure aligns poorly with the RecB nuclease in RecBCD; it agrees better with the RecB-like fold in RecC instead, b, c. Sequence alignment of GsuCas4, GsuCas1, and PfuCas4 with their close homologs. Based on the structural analysis, we marked the residues important for subunit interaction, substrate binding, catalysis and Fe-S cluster formation, d. Quality of the purified GsuCas4 mutants that carry the PAM-recognition residues from PfuCas4. These mutants were used in the structure-guided PAM-switching experiments in Extended Data Fig. 4.
Extended Data Figure 6. Cryo-EM single particle reconstruction of the single-PAM prespacer bound GsuCas4/Cas1-Cas2.
a. Flow-chart of the cryo-EM single particle reconstruction process that led to the reconstruction of two major snapshots. Left: Asymmetrical PAM/Non-PAM prespacer bound Cas4/1-2 complex. Right: That of the sub complex lacking (Cas4/1)2 on the non-PAM side. b. Cryo-EM density of the two reconstructions colored according to local resolution (top); viewing direction distribution plot (middle); and FSC curves (bottom). c. Superposition of the PAM side and non-PAM side densities showing that Cas4 density is largely missing at the non-PAM side, and the non-PAM 3’-overhang is largely disordered. Only the first four nucleotides of the non-PAM 3′-overhang can be traced in the density, along a similar path as in the PAM-side.
Extended Data Figure 7. In vitro assays to distinguish integration directionality.
a, b. Biochemistry showing that GsuCas4/1-2 is unable to integrate prespacer into the linear form of leader-repeat DNA. c. Successful prespacer integration into a leader-repeat containing plasmid by Cas4/1-2. d. The leader-repeat sequence cloned into the plasmid. We cleaved the leader-repeat sequence via the EcoRI and XhoI sites after the integration assay to further resolve the integration directionality on urea-PAGE. e. Schematic diagram explaining how the integration directionality can be resolved based on the fluorescent ssDNA sizes. f. Integration profile in urea-PAGE when both overhangs are integration-ready (7-nt long). Results showed that from the leader-repeat point of view, integration preferentially initiates from the leader-side, as the spacer-side integration trails after the leader-side integration in the time-course experiment. From the prespacer point of view, the integration directionality is scrambled. Each integration band contains two overlapping fluorescent signals. g. Native PAGE showing that in the concentration-gradient experiment, complex formation between Cas4/1-2 and prespacer takes place in a stepwise and PAM-dependent fashion.
Extended Data Figure 8. In-depth analysis of the mechanistic coupling between half-integration and PAM cleavage by Cas4.
a. Time-course experiment showing ExoI trims PAM and non-PAM overhangs differently. The non-PAM 3′-overhang was trimmed to within one nucleotide of the preferred length, 7 nt. The PAM-side 3′-overhang was protected by the footprint of Cas4 in the same reaction. b. Time-course experiment resolving the order of events from prespacer processing to full integration. Using the Cas4/1-2 (left set) and Cas4/1-2 plus ExoI (middle set) lanes as controls, the right set of experiment shows ExoI trimming triggers the integration of the non-PAM overhang into the leader-proximal target DNA. This is followed by the stimulation of PAM cleavage, and then the full integration from PAM-overhang to spacer-side target. c. Temperature-dependency of PAM cleavage and spacer-side integration. d. Side-by-side comparison of PAM cleavage at 50 °C, prespacer alone or programmed to the half-integrated state. e. Quantification of the cleaved band in c. and d. revealing the elevated PAM cleavage and full integration when leader-side integration already took place. Data were collected from N=3 biologically independent experiments and presented with mean ± s.e.m.. Statistical significance was assessed by two-tailed t-test, with the exact P values displayed. f. Salt-dependency of PAM cleavage and full integration. g-i. Optimization of full integration reaction by defining its time course, Cas2-dependency, and pH-dependency, respectively. j. Defining pH-dependency of PAM cleavage by Cas4. k. SEC analysis of the Cas4/1-2 complex programmed with the half-integration product mimic. Samples in the integrated complex peak was used for cryo-EM data collection and single particle reconstruction. l, Schematics of the half-integration product mimic annealed from oligonucleotides. m. Urea-PAGE analysis of the SEC peak in k. revealing that Cas4/1-2 further catalyzed the full-integration reaction after binding to the half-integration mimic.
Extended Data Figure 9. Cryo-EM single particle reconstruction of GsuCas4/Cas1-Cas2 programmed with a half-integration mimic.
a. Workflow of cryo-EM data processing. b. Overall cryo-EM density showing resolution distribution, viewing direction distribution plot, and FSC curves of three different snapshots. Left: half-integration, Cas4 disappeared; Middle: full-integration; Right: half-integration, Cas4 still blocking PAM-side.
Extended Data Figure 10. In-depth analysis of the three snapshots captured from GsuCas4/Cas1-Cas2 programmed with a half-integration mimic.
a. Superposition of cryo-EM reconstructions to reveal the structural differences among three functional states. b. Orientation view of the full integration snapshot for additional interface analysis. The entire leader- repeat DNA is contacted in a quasi-symmetric fashion at the following four regions. c. Contacts from the two Cas1 subunits to the spacer-repeat DNA. The spacer-side DNA density is degenerate and DNA bending is not significant. The leader-recognition α-helix in the catalytic Cas1 is not inserted into the minor groove of the spacer-side DNA. d. The backbone of the central dyad of CRISPR repeat is contacted by the positive charges and a proline-rich motif on the ridge of the Cas2 dimer. e. Immediately adjacent to the catalytic loop, the linker connecting Cas4 to Cas1 is involved in DNA contact. A conserved PRPI motif is exposed upon Cas4 dissociation and is involved in DNA minor groove contact. f. The 4-bp leader region immediately upstream of the CRISPR repeat is favorably recognized and significantly bent upwards by the DNA minor groove insertion of a glycine-rich α-helix in Cas1. As previously revealed, this recognition leads to strong leader-proximal preference at the first half-integration reaction10-12. A pair of inverted repeats is found at the border region of the CRISPR repeat. This inverted repeat is recognized at the major groove region by the catalytic Histidine-containing loop in Cas1 12. g. Overall structure of the “Half-integration, Cas4 still blocking PAM-side” snapshot. This represents an early state, when Cas4 is still engaged in PAM recognition and the spacer-side leader-repeat is not allowed to enter into the integration site. h. The low-resolution EM density defines that the leader-repeat DNA preferentially contact a positively charged patch in Cas1. It should be noted that we are not able to define which specific DNA contact activates Cas4. This will require even higher temporal and spatial resolutions to resolve.
Extended Data Table 1.
Cryo-EM data collection, refinement and validation statistics.
| PAM/PAM bound complex (EMDB-23839) (PDB 7MI4) |
PAM/non-PAM bound complex (EMDB-23840) (PDB 7MI5) |
Sub-complex (EMDB-23847) (PDB 7MID) |
Full integration (EMDB-23843) (PDB 7MI9) |
Half-int, PAM intact, Cas4 remains. (EMDB-23845) (PDB 7MIB) |
Half-int, PAM cleaved, Cas4 dissociated. (EMDB-23849) (PDB N/A) |
|
|---|---|---|---|---|---|---|
| Data collection and processing | ||||||
| Magnification | 63,000 | 63,000 | 63,000 | 63,000 | 63,000 | 63,000 |
| Voltage (kV) | 200 | 200 | 200 | 200 | 200 | 200 |
| Electron exposure (e–/υ2) | 50 | 50 | 50 | 50 | 50 | 50 |
| Defocus range (μm) | 1.5~3.5 | 1.5~3.5 | 1.5~3.5 | 1.5~3.5 | 1.5~3.5 | 1.5~3.5 |
| Pixel size (Å) | 1.23 | 1.23 | 1.42 | 1.31 | 2.18 | 1.32 |
| Symmetry imposed | C1 | C1 | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 1214203 | 896858 | 896858 | 1711962 | 1711962 | 1711962 |
| Final particle images (no.) | 158665 | 120102 | 32228 | 62074 | 16563 | 25373 |
| Map resolution (Å) | 3.2 | 3.6 | 3.6 | 3.8 | 5.8 | 5.8 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 20-2.8 | 20-3.0 | 20-3.2 | 20-3.5 | 20-5.0 | 20-5.0 |
| Refinement | ||||||
| Initial model used (PDB code) | N/A | N/A | N/A | N/A | N/A | N/A |
| Model resolution (Å) | 3.2 | 3.6 | 3.6 | 3.8 | 5.8 | - |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | ||
| Model resolution range (Å) | 20-3.1 | 20-3.6 | 20-3.6 | 20-3.8 | 20-5.8 | - |
| Map sharpening B factor (Å2) | −50 | −50 | −50 | −50 | − | − |
| Model composition | ||||||
| Non-hydrogen atoms | 17162 | 15469 | 9789 | 15924 | 17216 | - |
| Protein residues | 2048 | 1852 | 1137 | 1706 | 1922 | - |
| Ligands | 10 | 8 | 6 | 4 | 6 | - |
| DNA base | 70 | 60 | 57 | 170 | 121 | - |
| B factors (Å2) | ||||||
| Protein | 71.98 | 146.39 | 71.96 | 54.24 | 54.24 | - |
| Ligand | ||||||
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.009 | 0.009 | 0.009 | 0.006 | 0.008 | - |
| Bond angles (°) | 0.952 | 0.932 | 0.945 | 0.840 | 0.899 | - |
| Validation | ||||||
| MolProbity score | 2.7 | 2.8 | 2.7 | 2.9 | 2.8 | - |
| Clashscore | 13 | 33 | 14 | 30 | 57 | - |
| Poor rotamers (%) | 5.8 | 8.1 | 5.7 | 2.08 | 0.81 | - |
| Ramachandran plot | - | |||||
| Favored (%) | 91.45 | 90.47 | 91.32 | 86.9 | 87.54 | - |
| Allowed (%) | 8.44 | 9.42 | 8.68 | 12.97 | 12.45 | - |
| Disallowed (%) | 0.1 | 0.1 | 0.1 | 0.13 | 0.1 | - |
Supplementary Material
Acknowledgements
This work is supported by the Netherlands Organization for Scientific Research (NWO) VICI grant [VI.C.182.027] to S.J.J.B. and the National Institutes of Health (NIH) grant [GM118174] to A.K. This work made use of the Cornell Center for Materials Research Shared Facilities which are supported through the NSF MRSEC program (DMR-1719875). We thank S.N. Kieper, R. Miojevic, M. Ramos, G. Schuler and K. Spoth for helpful discussions, advice and technical assistance.
Footnotes
Competing Interests Statement
The authors declare no competing financial interests.
Reprints and permissions information are available upon request.
Main text references
- 1.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712, doi: 10.1126/science.1138140 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Yosef I, Goren MG & Qimron U Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic acids research 40, 5569–5576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuñez JK et al. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nature structural & molecular biology 21, 528–534 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nuñez JK, Lee AS, Engelman A & Doudna JA Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J & Almendros C Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740, doi: 10.1099/mic.0.023960-0 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Marraffini LA & Sontheimer EJ Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571, doi: 10.1038/nature08703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vink JNA et al. Direct Visualization of Native CRISPR Target Search in Live Bacteria Reveals Cascade DNA Surveillance Mechanism. Mol Cell 77, 39–50 e10, doi: 10.1016/j.molcel.2019.10.021 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Nuñez JK, Harrington LB, Kranzusch PJ, Engelman AN & Doudna JA Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature 527, 535–538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright AV & Doudna JA Protecting genome integrity during CRISPR immune adaptation. Nature Structural & Molecular Biology (2016). [DOI] [PubMed] [Google Scholar]
- 10.Wright AV et al. Structures of the CRISPR genome integration complex. Science 357, 1113–1118, doi: 10.1126/science.aao0679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budhathoki JB et al. Real-time observation of CRISPR spacer acquisition by Cas1–Cas2 integrase. Nat Struct Mol Biol 27, 489–499, doi: 10.1038/s41594-020-0415-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao Y, Ng S, Nam KH & Ke A How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature 550, 137–141, doi: 10.1038/nature24020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S et al. Selective loading and processing of prespacers for precise CRISPR adaptation. Nature 579, 141-+, doi: 10.1038/s41586-020-2018-1 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Li M, Wang R, Zhao D & Xiang H Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res 42, 2483–2492, doi: 10.1093/nar/gkt1154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T et al. Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res 45, 8978–8992, doi: 10.1093/nar/gkx612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiimori M, Garrett SC, Graveley BR & Terns MP Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci. Molecular Cell 70, 814-+, doi: 10.1016/j.molcel.2018.05.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieper SN et al. Cas4 Facilitates PAM-Compatible Spacer Selection during CRISPR Adaptation. Cell Reports 22, 3377–3384, doi: 10.1016/j.celrep.2018.02.103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almendros C, Nobrega FL, McKenzie RE & Brouns SJJ Cas4-Cas1 fusions drive efficient PAM selection and control CRISPR adaptation. Nucleic Acids Res 47, 5223–5230, doi: 10.1093/nar/gkz217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemak S et al. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. Journal of the American Chemical Society 135, 17476–17487, doi: 10.1021/ja408729b (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemak S et al. The CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis contains a [2Fe-2S] cluster: crystal structure and nuclease activity. Nucleic Acids Res 42, 11144–11155, doi: 10.1093/nar/gku797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Kasciukovic T & White MF The CRISPR Associated Protein Cas4 Is a 5 ' to 3 ' DNA Exonuclease with an Iron-Sulfur Cluster. Plos One 7, doi:ARTN e47232 10.1371/journal.pone.0047232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H, Dhingra Y & Sashital DG The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation. Elife 8, doi:ARTN e44248 10.7554/eLife.44248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H, Zhou Y, Taylor DW & Sashital DG Cas4-Dependent Prespacer Processing Ensures High-Fidelity Programming of CRISPR Arrays. Molecular Cell 70, 48-+, doi: 10.1016/j.molcel.2018.03.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao Y, Luo M, Dolan AE, Liao M & Ke A Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 361, doi: 10.1126/science.aat0839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah SA, Erdmann S, Mojica FJ & Garrett RA Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol 10, 891–899, doi: 10.4161/rna.23764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia N et al. Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection. Proceedings of the National Academy of Sciences of the United States of America 116, 24507–24516, doi: 10.1073/pnas.1913546116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuñez JK, Bai L, Harrington LB, Hinder TL & Doudna JA CRISPR immunological memory requires a host factor for specificity. Molecular cell 62, 824–833 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Ramachandran A, Summerville L, Learn BA, DeBell L & Bailey S Processing and integration of functionally oriented prespacers in the Escherichia coli CRISPR system depends on bacterial host exonucleases. The Journal of biological chemistry 295, 3403–3414, doi: 10.1074/jbc.RA119.012196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy A et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505-+, doi: 10.1038/nature14302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modell JW, Jiang W & Marraffini LA CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544, 101–104, doi: 10.1038/nature21719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 31.Makarova KS et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83, doi: 10.1038/s41579-019-0299-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudaiberdiev S et al. Phylogenomics of Cas4 family nucleases. Bmc Evol Biol 17, doi:ARTN 232 10.1186/s12862-017-1081-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pourcel C et al. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res 48, D535–D544, doi: 10.1093/nar/gkz915 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruitt KD, Tatusova T & Maglott DR NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33, D501–504, doi: 10.1093/nar/gki025 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson DA et al. GenBank. Nucleic Acids Res 46, D41–D47, doi: 10.1093/nar/gkx1094 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayers EW et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 37, D5–15, doi: 10.1093/nar/gkn741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arndt D et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44, W16–21, doi: 10.1093/nar/gkw387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell AL et al. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48, D570–D578, doi: 10.1093/nar/gkz1035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen IA et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 45, D507–D516, doi: 10.1093/nar/gkw929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paez-Espino D et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res 47, D678–D686, doi: 10.1093/nar/gky1127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soto-Perez P et al. CRISPR-Cas System of a Prevalent Human Gut Bacterium Reveals Hyper-targeting against Phages in a Human Virome Catalog. Cell Host Microbe 26, 325–335 e325, doi: 10.1016/j.chom.2019.08.008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Group NHW et al. The NIH Human Microbiome Project. Genome Res 19, 2317–2323, doi: 10.1101/gr.096651.109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasolli E et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 176, 649–662 e620, doi: 10.1016/j.cell.2019.01.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. Journal of molecular biology 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
- 45.Fu L, Niu B, Zhu Z, Wu S & Li W CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152, doi: 10.1093/bioinformatics/bts565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deveau H et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190, 1390–1400, doi: 10.1128/JB.01412-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almendros C, Guzman NM, Diez-Villasenor C, Garcia-Martinez J & Mojica FJ Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli. PLoS One 7, e50797, doi: 10.1371/journal.pone.0050797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange SJ, Alkhnbashi OS, Rose D, Will S & Backofen R CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res 41, 8034–8044, doi: 10.1093/nar/gkt606 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alkhnbashi OS et al. CRISPRstrand: predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 30, i489–496, doi: 10.1093/bioinformatics/btu459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guindon S et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59, 307–321, doi: 10.1093/sysbio/syq010 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Katoh K & Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780, doi: 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: a sequence logo generator. Genome Res 14, 1188–1190, doi: 10.1101/gr.849004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenzie RE, Almendros C, Vink JNA & Brouns SJJ Using CAPTURE to detect spacer acquisition in native CRISPR arrays. Nat Protoc 14, 976–990, doi: 10.1038/s41596-018-0123-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296, doi: 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Xu K, Zang X, Peng M, Zhao Q & Lin B Magnesium Lithospermate B Downregulates the Levels of Blood Pressure, Inflammation, and Oxidative Stress in Pregnant Rats with Hypertension. Int J Hypertens 2020, 6250425, doi: 10.1155/2020/6250425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps that support the findings of this study have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers of EMD-23839 (PAM/PAM prespacer bound), EMD-23840 (PAM/Non-PAM prespacer bound), EMD-23843 (full integration complex), EMD-23845(half integration, Cas4 still blocking the PAM side), EMD-23849 (half integration, Cas4 dissociated), and EMD-23847 (sub-complex). The coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers of 7MI4 (PAM/PAM prespacer-bound), 7MI5 (PAM/non-PAM prespacer-bound), 7MI9 (full integration), 7MIB (half integration, Cas4 still blocking the PAM side), 7MID (sub-complex). MiSeq sequencing data that support analysis of in vivo prespacer integration have been deposited in the European Nucleotide Archive (ENA) under accession number PRJEB41616. Plasmids used in this study are available upon request.