Abstract
DNA methyltransferase (DNMT) 1 is an enzyme that functions as a maintenance methyltransferase during DNA replication, and depletion of this enzyme from cells is considered to be a rational goal in DNA methylation–dependent disorders. Two DNMT1-depleting agents 5-aza-2′-deoxycytidine (aza-dCyd, decitabine) and 5-aza-cytidine (aza-Cyd, azacitidine) are currently used for the treatment of myelodysplastic syndromes and acute myeloid leukemia and have also been investigated for nononcology indications, such as sickle cell disease. However, these agents have several off-target activities leading to significant toxicities that limit dosing and duration of treatment. Development of more selective inhibitors of DNMT1 could therefore afford treatment of long durations at effective doses. We have discovered that 5-aza-4′-thio-2′-deoxycytidine (aza-T-dCyd) is as effective as aza-dCyd in depleting DNMT1 in mouse tumor models but with markedly low toxicity. In this review we describe the preclinical studies that led to the development of aza-T-dCyd as a superior DNMT1-depleting agent with respect to aza-dCyd and will describe its pharmacology, metabolism, and mechanism of action. In an effort to understand why aza-T-dCyd is a more selective DNMT1 depleting agent than aza-dCyd, we will also compare and contrast the activities of these two agents.
SIGNIFICANCE STATEMENT
Aza-T-dCyd is a potent DNMT1-depleting agent. Although similar in structure to decitabine (aza-dCyd), its metabolism and mechanism of action is different than that of aza-dCyd, resulting in less off-target activity and less toxicity. The larger therapeutic index of aza-T-dCyd (DNMT1 depletion vs. toxicity) in mice suggests that it would be a better clinical candidate to selectively deplete DNMT1 from target cells and determine whether or not depletion of DNMT1 is an effective target for various diseases.
Introduction: DNA Hypomethylation Using DNA Methyltransferase 1–Depleting Nucleosides
Methylation of cytosines in DNA occurs mainly in the context of CG dinucleotides that are distributed throughout the genome, including within repetitive sequences, enhancers, promoters, and coding regions (Ramsahoye et al., 2000; Robertson, 2005; Du et al., 2015). Cytosine methylation is mediated by a family of enzymes known as the DNA methyltransferases (DNMTs), which catalyze the transfer of a methyl group from S-adenosylmethionine to the 5 position of cytosine residues in DNA. DNMT1, which is the most abundant DNMT, is a maintenance methyltransferase that methylates the newly replicated strand and preserves the existing methylation pattern in DNA, whereas DNMT3A and DNMT3B largely establish methylation patterns de novo on previously unmethylated DNA (Du et al., 2015). DNA methylation of DNA results in transcriptional silencing by regulating formation of heterochromatin (Bachman et al., 2001) and influencing the accessibility of transcription factors to DNA. Methylated CGs can regulate transcription factor binding and also affect transcription by recruiting methyl-CG binding proteins that affect chromatin structure through interactions with other proteins, such as those that modify histones (Hendrich and Bird, 2000; Della Ragione et al., 2016).
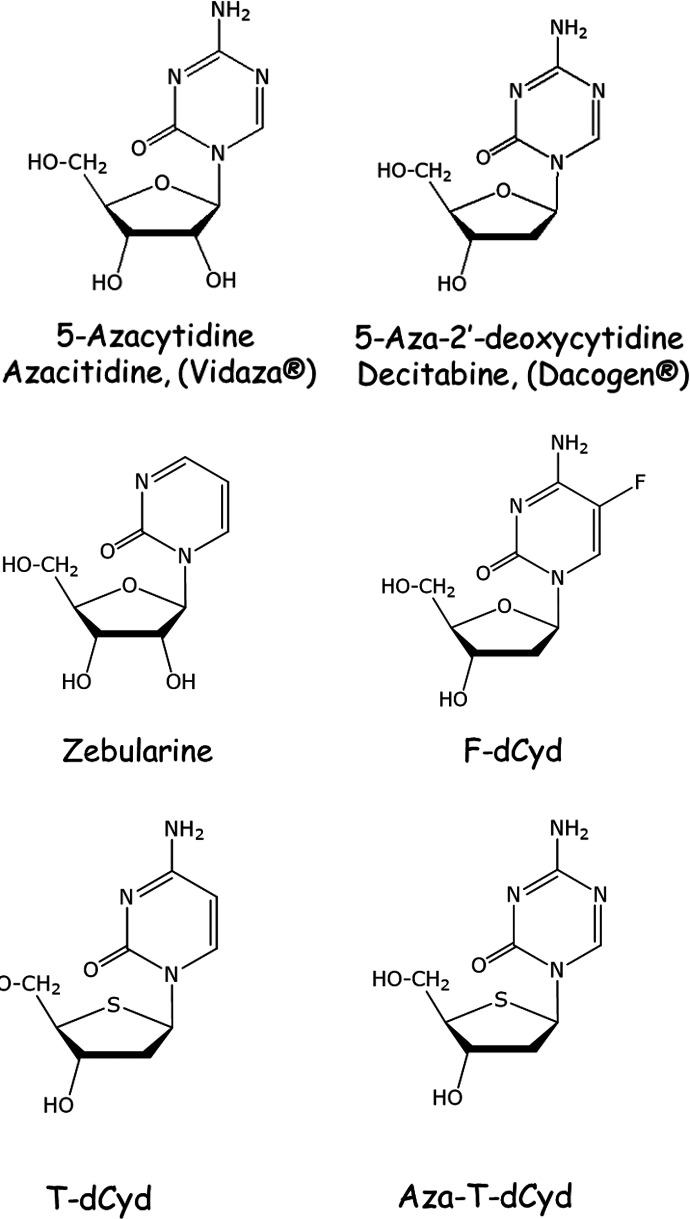
DNA methylation must be actively maintained in every replication cycle by DNMT1 and can therefore be reversed by inhibition or depletion of the enzyme. Several DNA-hypomethylating nucleosides have been evaluated in the clinic, and two of these agents 5-aza-2′-deoxycytidine (aza-dCyd, decitabine) and 5-aza-cytidine (aza-Cyd, azacitidine) (Fig. 1) have been approved by the Food and Drug Administration for the treatment of myelodysplastic syndromes and chronic myelomonocytic leukemia. Treatment with either of these agents results in the incorporation of aza-cytosine in place of cytosine in DNA. DNMT1 gets covalently trapped when it attempts to methylate aza-cytosines that are diagonally opposite a 5-methylcytosine in the template strand. The covalent complex thus effectively reduces the availability of active DNMT1 and also induces proteasomal degradation of the free enzyme (Santi et al., 1984; Ghoshal et al., 2005; Patel et al., 2010). Although active against heme malignancies, these agents are much less effective in solid tumors. Both agents have poor oral bioavailability, poor stability due to hydrolytic cleavage of the triazine ring, and considerable toxicity.
Fig. 1.
Structures of aza-Cyd, aza-dCyd, zebularine, F-dCyd, T-dCyd, and aza-T-dCyd.
5-Fluoro-2′-deoxycytidine (F-dCyd, Fig. 1), a stable nucleoside, was shown in early studies to be active in solid tumor models as well as in leukemias (Eidinoff et al., 1959; Mukherjee and Heidelberger, 1962). Like aza-dCyd, F-dCyd is activated via 2′-deoxycytidine (dCyd) kinase to F-dCTP , which is readily incorporated into DNA. It was later recognized to inhibit methylation in cells and was shown to induce expression of tumor suppressor genes, such as p16/CDKN2A (Jones and Taylor, 1980; Kaysen et al., 1986; Osterman et al., 1988; Coyne et al., 2020). Because of the fluorine in the 5-position of the pyrimidine ring, DNMT1 forms a covalent complex with DNA, which causes its subsequent depletion. However, F-dCyd has a serious toxicity problem because it is readily converted via both cytidine (Cyd) deaminase and dCMP deaminase (Fig. 2) (Newman and Santi, 1982; Boothman et al., 1985) to 5-F-2′-deoxyuridine-5′-phosphate, a potent inhibitor of thymidylate synthase (Parker and Cheng, 1990). Therefore, F-dCyd is not a selective DNMT1-depleting agent. F-dCyd was tested in patients in combination with tetrahydrouridine (THU), a potent inhibitor of Cyd deaminase, in an effort to enhance formation of F-dCMP (Beumer et al., 2008a,b; Holleran et al., 2015). However, recently reported results in different solid tumor patient cohorts have demonstrated very low response rates (Coyne et al., 2020).
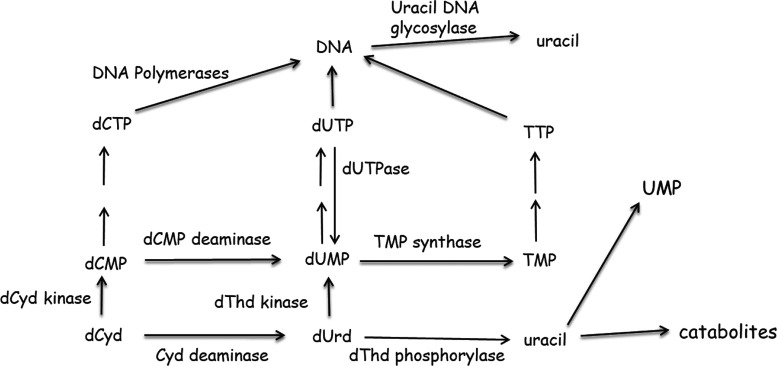
Fig. 2.
Metabolism of dCyd in human cells.
Zebularine ([1-(β-d-ribofuranosyl)-1,2-dihydropyrimidin-2-one], Fig. 1) is another stable cytidine analog that is an inhibitor of DNA methylation with improved oral bioavailability. Once incorporated into DNA, this agent also forms covalent complexes with DNMT1 resulting in its depletion, the reactivation of previously hypermethylated tumor suppressor genes, and antitumor activity in various mouse models (Yoo et al., 2008). However, it is a riboside analog and was found to have poor potency against DNMT1 owing to its limited incorporation into DNA (Ben-Kasus et al., 2005). Zebularine’s weak conversion to 2′-deoxynucleotide metabolites and the inability of 2′-deoxyzebularine to be phosphorylated to its active metabolite resulted in the discontinuation of this line of investigation.
Recently other DNMT1-depleting nucleosides, such as guadecitabine (SGI-110 or S-110) and NUC013, have also entered development. Guadecitabine is a dinucleotide of aza-dCyd linked via a phosphodiester to deoxyguanosine, which was developed as a stable prodrug of aza-dCyd that is also less vulnerable to deamination by Cyd deaminase (Roboz et al., 2018). However, there have been two recent phase III failures with this drug when it was tested in myelodysplastic syndrome, acute myeloid leukemia, and chronic myelomonocytic leukemia (NCT02920008, NCT02907359). NUC013 (5-aza-2′,2′-difluoro-2′-deoxycytidine) is another aza-nucleoside that decreases DNMT1 levels and has shown efficacy in xenograft models of human leukemia and colon cancer (Daifuku et al., 2017). However, since this compound is an analog of gemcitabine (2′,2′-difluoro-2′-deoxycytidine), it is expected to induce chain termination during replication resulting in off-target activities. NUC013 and its prodrug NUC041 have therefore not moved beyond preclinical development stages (Daifuku et al., 2017, 2018).
DNMT1 Depletion and Treatment of Solid Tumors
Diseases, such as cancer, wherein hypermethylation of CG regions plays a role in their development, could theoretically be treated with agents that induce the proteolytic degradation of DNMT1. Since cells with reduced or absent DNMT1 are able to complete replication, hypermethylated tumor suppressor genes are demethylated and re-expressed in subsequent generations, which leads to eventual growth inhibition and apoptosis in cancer cells within 1–2 replication cycles. Several preclinical studies in solid tumors have shown that treatment with DNA-hypomethylating agents can synergize with or restore sensitivity to chemotherapies, targeted agents (Anzai et al.,1992; Soengas et al., 2001; Hou et al., 2019), and very recently even immune checkpoint agents (Yu et al., 2019; Huang et al., 2020; Gonda et al., 2020). The biologic basis of these effects was consistent with a number of reports that had demonstrated 1) reversal of hypermethylation of CG islands in tumor suppressor genes, 2) epigenetic immunomodulation by these drugs, and 3) the role of DNMT1 overexpression in tumorigenesis (El-Deiry et al., 1991; Merlo et al., 1995; Coral et al., 1999). The importance of DNMT1 overexpression has not only been recognized in early studies in which it was shown to increase proliferation and induce transformation (Wu et al., 1993) but was further strengthened by the observation that it correlates well with metastasis, poor prognosis, and aberrant methylation in several solid tumor patient cohorts (Saito et al., 2003; Peng et al., 2006; Zhao et al., 2011; Yuan et al., 2016). Consistent with these studies, several reports have also demonstrated that depletion of DNMT1 by siRNA approaches can inhibit growth of tumor cells both in vitro and in vivo (Milutinovic et al., 2000, 2004; Xiang et al., 2014). A systematic meta-analysis of 58 clinical studies of DNA-hypomethylating drugs concluded that although these drugs improved outcome and altered methylation status in patients with solid tumor, overall response was limited (Linnekamp et al., 2017). Despite the plethora of clinical trials that have investigated DNA hypomethylators, none of these drugs have been approved for solid tumors. One major reason that has been suggested is the limited number of treatment cycles in these studies with these drugs often due to toxicities, and given that delayed responses were seen in a number of patients, long treatment durations would be necessary to achieve clinical benefit.
DNMT1 Depletion and Hemoglobinopathies
Initial empirical observations have demonstrated that antileukemic effects of aza-dCyd were accompanied by irreversible hemoglobinization and morphologic differentiation in erythroleukemic cells (Pinto A et al., 1983). These observations coupled with the knowledge that the expression of γ-globin [fetal hemoglobin (HbF)] is associated with the status of its CG methylation in the adult (van der Ploeg and Flavell, 1980, Mavilio et al., 1983) spurred the extension of aza-nucleoside studies into hemoglobinopathies, such as sickle cell disease and thalassemia (Desimone et al., 1982; Charache et al., 1983). DNMT1 by interacting with transcriptional regulators like Myb and Bcl11A has been shown to be required to maintain HbF silencing (Xu et al., 2013). The role of DNMT1 also appears to involve methylation of promoters of other HbF gene modifiers (e.g., ZBTB7 ) (Roosjen et al., 2014; Chondrou et al., 2018). Inhibition of DNMT1 activity by aza-dCyd, aza-Cyd, or guadecitabine has been shown to enhance HbF expression in primary erythroid cells, nonhuman primates, and humans (Desimone et al., 1982; Humphries et al.,1985; Akpan et al., 2010; Lavelle et al., 2018; Stomper and Lubbert, 2019). This led to clinical trials wherein aza-dCyd was shown to successfully induce HbF in patients with sickle cell disease intolerant to hydroxyurea and in β-thalassemia (Saunthararajah et al., 2003; Olivieri et al., 2011; Saunthararajah, 2019). Although this provided clinical proof of concept that subcutaneous administration of low-dose aza-dCyd increases HbF, Molokie et al. (2017) decided to provide an oral route of administration by combining the drug with THU to inhibit intestinal and liver Cyd deaminase. This small, randomized study concluded that twice-weekly low-dose aza-dCyd (5 mg/m2 vs. the approved i.v. 5-day dose of 20 mg/m2) when combined orally with THU achieved the pharmacokinetic objectives of low Cmax with long half-life/Tmax and the pharmacodynamic objectives of DNMT1 depletion, DNA hypomethylation, increase in HbF, and an increase in numbers of healthy red blood cells. Since this was a first-in-human study of this combination with a small number of patients for only 8 weeks, it is unknown whether this will translate into long-term clinical benefits in terms of reducing veno-occlusive events, associated morbidities, and low life spans. However, an oral toxicity study in mice that evaluated the THU and aza-dCyd combination did show increased toxicity as compared with aza-dCyd alone, which suggests that the trade-off for improved oral pharmacokinetics might be increased adverse events (Terse et al., 2014).
Off-Target Activities of DNA-Hypomethylating Nucleosides
What is the evidence that clinically used DNA-hypomethylating nucleosides could be exerting their pharmacological and toxic effects at least in part by employing alternative mechanisms? First and foremost, DNMT1 inhibition per se is relatively nontoxic to cells. An orally bioavailable non-nucleoside DNMT1 inhibitor has been shown to induce HbF levels in primary human erythroid cells equivalent to that induced by aza-dCyd but with little cellular growth inhibition and had good tolerability in transgenic sickle cell mice (Gilmartin et al., 2021). In contrast, as described in greater detail below, aza-dCyd is a known cytotoxic nucleoside analog. Aza-dCyd induces genome-wide DNA damage and cytotoxicity arising from the DNA-DNMT1 crosslinks that can block DNA synthesis and induce directly or indirectly DNA double-strand breaks, eventually leading to cell death (Juttermann et al., 1994; Palii et al., 2008). Aza-dCyd has also been reported to induce point mutations and genome rearrangements most likely due to protein-DNA crosslinks (Jackson-Grusby et al., 1997; Maslov et al., 2012). It is well established that aza-dCyd treatment induces the activation of a specific DNA damage response that includes the phosphorylation of histone H2AX (Palii et al., 2008; Wilsker et al., 2019). Recently, it has also been proposed that base excision repair might initiate the repair of aza-dCyd–induced DNA base lesions, although the precise mechanisms and the DNA glycosylases involved remain unknown (Orta et al., 2014). Lastly, an alternative mechanism for aza-dCyd in which its metabolism ultimately leads to the genotoxic accumulation of dUTP in the cellular pool has also been demonstrated (Requena et al., 2016). In the case of aza-Cyd, in addition to toxicities associated with its metabolism to aza-2′-deoxynucleotides, it is also impaired by those stemming from its incorporation into RNA (with the RNA:DNA incorporation ratio being 65:35) (Hollenbach et al., 2010; Cheng et al., 2018). Given the above considerations with approved DNA-hypomethylating drugs and the potential for leveraging the benefits of long-term depletion of DNMT1, it is imperative to identify newer and more selective DNMT1-depleting agents that exhibit less toxicity.
Design of Novel 4′-Thio-2′deoxycytidine Agents
The design of new nucleoside analogs targeting DNMT1 must recognize that sequential intracellular steps that lead to DNA incorporation and strand elongation need to be efficient: In other words, modifications in the nucleoside should not 1) impede activation to the triphosphate and incorporation into DNA, 2) inhibit extension of the DNA chain after incorporation, or 3) prevent extrusion of the base from the double helix and binding to the active site of DNMT1. In addition, any new agent (or its metabolites) should not inhibit other enzymes that are involved in nucleotide metabolism (Fig. 2). In our studies (Parker et al., 2000; Thottassery et al., 2014), we have evaluated 4′-thio-2′-deoxycytidine (T-dCyd) and its 5-aza analog [5-aza-4′-thio-2′-deoxycytidine (aza-T-dCyd)] (Fig. 1) and found that they fulfill many of the requirements described above. T-dCyd is readily phosphorylated and incorporated into DNA, and the 4′-thio modification to 2′-deoxycytidine (dCyd) alters metabolism in a positive manner resulting in relatively little production of 2′-deoxyuridine (dUrd) or thymidine (dThd) nucleotide metabolites in human cells.
In this work we review the development and rationale for use of aza-T-dCyd as a DNMT1-depleting agent, which we have discovered is as effective as aza-dCyd in depleting DNMT1 but with much less toxicity. In addition, we will compare and contrast its biochemical pharmacology and mechanism of action with aza-dCyd in an effort to understand why aza-T-dCyd is a more selective DNMT1-depleting agent. For the purposes of this review, we have restricted the comparison of aza-T-dCyd to aza-dCyd since aza-Cyd is a ribonucleoside and its off-target activities result from its incorporation into RNA in addition to its effects after conversion to deoxynucleotide metabolites.
Metabolism and Depletion of DNMT1 by T-dCyd
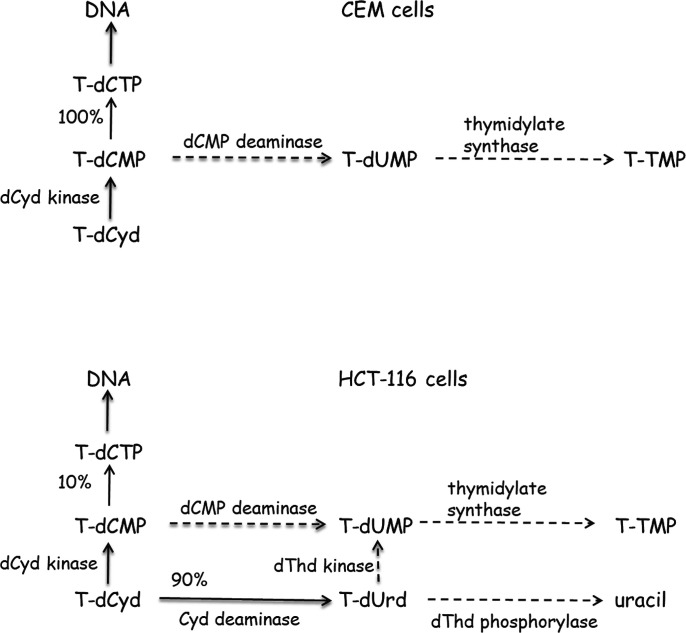
Previously we have thoroughly evaluated the metabolism of T-dCyd in cells in culture (Parker et al., 2000; Thottassery et al., 2014), which provides insight into the effect of a 4′-thio group on the metabolism of dCyd and dUrd nucleosides and nucleotides (Fig. 3). In CCRF-CEM cells (a cell line that does not express Cyd deaminase) T-dCyd was readily routed through dCyd kinase but was not converted to dThd nucleotides since T-dCMP is at best a poor substrate for dCMP deaminase (Parker et al., 2000). In stark contrast to the predominant conversion of dCMP to dUMP (Fig. 2) in these cells when incubated with dCyd, no T-dUMP was formed from T-dCyd (Fig. 3). The T-dCTP formed from T-dCyd was readily incorporated into DNA without inhibition of DNA replication (Parker et al., 2000). Therefore, its cytotoxicity in CEM cells is due to some disruption of DNA function once incorporated into the DNA chain. 4′-Thio-thymidine (T-dThd) is also a potent cytotoxic compound that, like T-dCyd, is readily phosphorylated in cells and incorporated into DNA without inhibiting DNA replication (Parker et al., 1995). Therefore, the cytotoxicity resulting from treatment with either of these 4′-thio-deoxynucleosides is due to some disruption of DNA function once incorporated into the DNA chain.
Fig. 3.
Metabolism of T-dCyd in CEM and HCT-116 cells. Dashed lines indicate little if any enzymatic activity.
In HCT-116 cells (a human colon carcinoma cell line that expresses high levels of cytidine deaminase) T-dCyd was mostly converted to 4′-thio-2′-deoxyuridine (T-dUrd) (Thottassery et al., 2014), which indicated that T-dCyd was a good substrate for Cyd deaminase (Fig. 3). The addition of Cyd deaminase inhibitor THU to cells treated with T-dCyd totally inhibited the formation of T-dUrd but in these short-term experiments had very little effect on the incorporation of T-dCyd into DNA or the level of T-dCTP. The catalytic efficiency of T-dCyd with Cyd deaminase in crude cell extracts from HCT-116 cells was approximately 50% that of dCyd with a Km that was ∼5 times that of dCyd (110 vs. 23 μM) and a Vmax that was ∼3 times that of dCyd (37 vs. 13 nmol/mg-min). Although most T-dCyd was converted to T-dUrd, there was little evidence that T-dUrd was phosphorylated by dThd kinase and then converted to T-TMP via thymidylate synthase: 12% of dCyd was converted to dThd nucleotides in HCT116 cells, however, only 0.2% of T-dCyd was converted to T-dThd nucleotides. This observation supports previous studies (Secrist et al., 1991; Verri et al., 2000) that indicated T-dUrd was a poor substrate for dThd kinase. In addition, T-dUrd was not a substrate for dThd phosphorylase in crude cell extracts of HCT-116 cells [unpublished observation].
Pharmacokinetic studies in mice support the observation that T-dUrd is at best a poor substrate for dThd phosphorylase. After intraperitoneal administration, T-dCyd was primarily converted to T-dUrd, and a major metabolite detected in mice treated with dCyd (presumably uracil and its catabolites) was not detected in the plasma after administration of T-dCyd (Thottassery et al., 2014). T-dUrd was also the primary circulating metabolite in pharmacokinetic studies conducted by investigators at the NCI (Kinders et al., 2014). The half-life of T-dCyd after intravenous or intraperitoneal administration in mice was on the order of 10–15 minutes, whereas the plasma concentration of T-dUrd was stable for up to 4 hours at levels similar to those of the peak concentration of T-dCyd. A small amount of T-dThd was also detected in plasma (500-fold less than T-dUrd), which indicated that T-dUrd could be phosphorylated and converted to T-TMP via thymidylate synthase.
In summary, the metabolic studies described above indicated that T-dCyd metabolites (T-dCMP and T-dUrd) are at best poor substrates for dCMP deaminase, dThd kinase, and dThd phosphorylase (Fig. 3), which significantly reduces the amount of dThd metabolites created in human cells after treatment with T-dCyd. The understanding that T-dCTP and T-TTP do not inhibit DNA replication (Parker et al., 1995, 2000) and structural studies (Kumar et al., 1997), which indicated that T-dCyd could inhibit methylation by Hhal methyltransferase, led us to consider T-dCyd as a novel agent for DNMT1 depletion. Since dUrd and dThd metabolites of T-dCyd (T-dUMP, T-TTP, others) would not contribute to the inhibition of DNMT1 activity and could only contribute to toxicity of the compounds, we hypothesized that T-dCyd could be a much more selective agent in its ability to deplete DNMT1 in human cells.
Although T-dCyd was able to deplete DNMT1 from tumor cells in vivo and in vitro (Thottassery et al., 2014), its ability to do so was not universal in vitro and was only effective in mice at high concentrations, which were near its maximally tolerated dose. In addition, because T-dCyd is a good substrate for Cyd deaminase and a poor substrate for dThd phosphorylase, the primary circulating metabolite of T-dCyd in mice was T-dUrd (Kinders et al., 2014). Moreover, the detection of T-dThd in the plasma of mice indicated that T-dUMP is a substrate for thymidylate synthase and that T-dThd nucleotides were formed in mouse tissues. Although only a small amount of T-dThd was detected in plasma, our previous studies with this compound (Parker et al., 1995) indicated that it is readily incorporated into DNA and is not removed by proofreading enzymes. Therefore, the bulk of T-dThd nucleotides formed in cells would be sequestered in the DNA in the time frame of the pharmacokinetic studies. These results suggest that because of the long plasma half-life of T-dUrd, the primary target responsible for in vivo antitumor activity of T-dCyd was the incorporation of T-TTP into DNA, which would not contribute to DNMT1 depletion. Toxicity associated with the incorporation of T-dThd into DNA would likely interfere with T-dCyd depletion of DNMT1. In support of this conclusion, studies done by the NCI (unpublished observation) indicated that treatment with T-dUrd resulted in excellent antitumor activity in the NCI-H23 mouse xenograft model, and that addition of THU to T-dCyd treatment resulted in decreased antitumor activity. This result suggests that the antitumor activity seen with T-dCyd is primarily due to metabolites of T-dUrd.
T-dCyd in First-in-Human Clinical Trial
After successful demonstration of efficacy in preclinical tumor xenografts in mice, the NCI developed the analytical assays for T-dCyd (Liu et al., 2016), prepared an Investigational New Drug application, and began a first-in-human open-label phase I trial with T-dCyd (O’Sullivan Coyne et al., 2016). Subjects were patients with solid tumors whose disease had progressed on standard therapy. The primary objective was to establish the safety, tolerability, maximal tolerated dose, and recommended phase II dose. The study had entered dose level 5 enrollment, but recruitment has been suspended, presumably due to lack of clinical activity at nontoxic doses.
Development of Aza-T-dCyd
Because of the above considerations with T-dCyd, we turned our attention to aza-T-dCyd, which is structurally similar to aza-dCyd (Fig. 1). The in vitro cytotoxicity of aza-T-dCyd was 3- to 4-fold more potent than T-dCyd in various human tumor cells (Teicher et al., 2019) and was also 10-fold more potent than aza-dCyd in HCT-116 cells (Laranjeira et al., 2017). Experiments were conducted to compare the cytotoxicity of aza-T-dCyd and aza-dCyd in HCT116 parental cells and an isogenic HCT116 cell line in which the DNMT1 gene had been knocked-out (Laranjeira et al., 2017). The DNMT1 knockout HCT116 cells were 300-fold less sensitive to aza-T-dCyd (96 hours of drug exposure), whereas these cells were only 10-fold less sensitive to aza-dCyd. These results are consistent with the suggestion that off-target activities of aza-dCyd contribute more to its cytotoxic activity than they do to the cytotoxic activity of aza-T-dCyd.
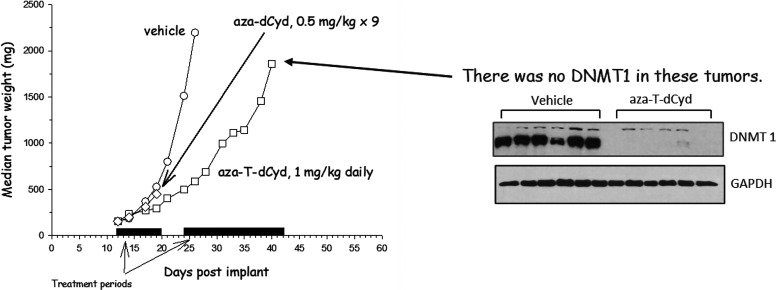
Treatment of mice bearing CEM xenografts with either aza-T-dCyd or aza-dCyd resulted in the depletion of DNMT1 from the tumor cells (and supposedly any host cell that was replicating during this period). However, aza-dCyd was much more toxic to the mice than aza-T-dCyd (Table 1). In another experiment (Fig. 4) mice bearing subcutaneous CEM tumor xenografts were treated with either 1 mg/kg aza-T-dCyd or 0.5 mg/kg aza-dCyd daily for 9 days to determine what effect these compounds had on tumor growth. Aza-dCyd had no effect on tumor growth during the 9-day treatment period and resulted in the death of all mice. Aza-T-dCyd did inhibit tumor growth modestly but importantly did not result in any weight loss or death of the mice. Therefore, after a 3-day rest period treatment was resumed for another 20 days also without weight loss or death of mice. As seen above, DNMT1 was undetectable in tumor cells at the end of the 20-day treatment. This result demonstrates that aza-T-dCyd effectively depletes DNMT1 in tumor cells in vivo at doses that were very well tolerated by the mice.
TABLE 1.
Toxicity and DNMT1 depletion in CEM tumor xenografts in mice
post-treatment with either aza-T-dCyd or aza-dCyd
Mice bearing CEM tumor xenografts were treated daily for 9 days with either aza-T-dCyd or aza-dCyd (0.3–5 mg/kg per dose). At death of animal or day 10, tumors were removed and evaluated for DNMT1 (Thottassery et al., 2014).
| Dose (mg/kg × 9) |
Aza-T-dCyd | Aza-dCyd | ||
|---|---|---|---|---|
| DNMTI | Death | DNMTI | Death | |
| 5 | ND | No | ND | Yes |
| 2.5 | ND | No | ND | Yes |
| 1.2 | ND | No | ND | Yes |
| 0.6 | ND | No | ND | Yes |
| 0.3 | ± | No | ± | No |
| ND, DNMT1 not detected; ±, DNMT1 barely detected | ||||
Fig. 4.
Effect of aza-T-dCyd and aza-dCyd on tumor growth and DMNT1 levels in CEM tumor xenografts. Mice bearing CEM tumor xenografts were treated daily for 9 days with either 1 mg/kg aza-T-dCyd or 0.5 mg/kg aza-dCyd. Treatment with 0.5 mg/kg aza-dCyd daily for 9 days resulted in the death of all mice. After a 3-day rest period, aza-T-dCyd treatment was resumed for another 20 days. At the end of the 20-day treatment period the mice were sacrificed, and DNMT1 levels in tumor were determined.
In a subsequent experiment (unpublished) mice were treated with either 1 or 2 mg/kg aza-T-dCyd daily for 20 days. The low dose again had a small impact on tumor growth, but the high dose resulted in excellent antitumor activity, although at this dose there were three deaths (out of eight treated mice) that may have resulted from the therapy (all deaths occurred shortly after treatment ended). In theory, depletion of DNMT1 should not be acutely cytotoxic to tumor cells but should instead slow tumor growth due to re-expression of tumor suppressor genes. In support of this we have demonstrated that the tumor suppressor genes p15/CDKN2B (Thottassery et al., 2014) and p16/CDKN2A (unpublished) are re-expressed in aza-TdCyd–treated cells. Therefore, it is likely that the antitumor activity seen with high doses is due to off-target activity of aza-T-dCyd unlike that seen at low doses. Regardless, the experiments suggest that depletion of DNMT1 in host tissues is not toxic and that any off-target activities and toxicities of aza-T-dCyd occur at doses that are well above those needed to deplete DNMT1. Because the therapeutic index of aza-T-dCyd (DNMT1 depletion vs. toxicity) is large, our in vivo results in mouse models of cancer indicate that mice can be treated with aza-T-dCyd on an extended schedule at nontoxic, DNMT1-depleting doses, which would allow for treatment of all target cells as they enter S phase.
Antitumor Activity of Aza-T-dCyd
We demonstrated that aza-T-dCyd was very effective in inducing growth inhibition in NCI-H23 lung tumor xenografts (Thottassery et al., 2014). Studies conducted at the NCI (unpublished observation) showed that unlike T-dCyd addition of THU to aza-T-dCyd treatment enhanced the antitumor activity of aza-T-dCyd, suggesting that deamination of this compound resulted in inactive metabolites. In HL-60 leukemia xenografts, treatment with either aza-T-dCyd alone or aza-T-dCyd in combination with THU resulted in significant tumor growth inhibition (Nguyen et al., 2016), which was associated with simultaneous inhibition of DNMT1 and DNMT3B. Interestingly, the tumor growth delay in mice treated with aza-T-dCyd plus THU was less than that seen in mice treated with only aza-T-dCyd. Treatment with aza-dCyd did not result in tumor growth delay in HL-60 leukemia xenografts even though both DNMT3A and DNMT1 levels were decreased. Treatment of mice with aza-dCyd or aza-T-dCyd along with isotopically labeled methionine resulted in a reduction in the enrichment of methyl-deoxycytidine in DNA by 20%–70% in tumor tissue, bone marrow, and intestine (Anderson et al., 2017), which supported the observation that treatment with these compounds results in the depletion of DNMT1 in various tissues.
Aza-T-dCyd was also evaluated in comparison with T-dCyd in pediatric acute lymphoblastic leukemia (ALL) patient-derived xenograft models to assess their potential in hematologic malignancies. Treatment with aza-T-dCyd showed impressive remission-inducing activity for ALL xenografts and was, in general, more active than T-dCyd (Teicher et al., 2018, 2019). Profound responses were observed across a range of ALL biology, including B-ALL (ALL-2, ALL-7, ALL-19), T-ALL (ALL-8, ALL-31), early T-cell phenotype ALL (early T-cell phenotype-1), mixed-lineage leukemia (MLL)-rearranged infant ALL (MLL -7), and Ph-like ALL (PAKHZT). T-dCyd and aza-T-dCyd were also tested in patient-derived xenograft models of Ewing sarcoma, rhabdoid tumor, rhabdomyosarcoma, and osteosarcoma. At tolerable doses, the anticancer activity of both T-dCyd and aza-T-dCyd were found to be modest in the Ewing sarcoma (four) and rhabdomyosarcoma (four) models. The activity of both compounds was also modest in five out of six osteosarcoma models. However, the median survival of mice bearing the OS-9 osteosarcoma doubled (42 days) upon treatment with T-dCyd and increased nearly 4-fold (84 days) in mice treated with aza-T-dCyd (Teicher et al., 2018, 2019). Neither aza-T-dCyd nor T-dCyd were active against pediatric brain tumor xenografts. In these studies, there was moderate correlation between expression of DNMT1 and the antitumor activity of aza-T-dCyd.
Although toxicity was observed, treatment with aza-T-dCyd (1–2 mg/kg daily × 5 repeated weekly for 3 or 4 weeks) resulted in impressive remissions in acute lymphocytic leukemia xenografts (Teicher et al., 2018). Aza-T-dCyd was not active against solid tumor xenografts, although it did prevent growth of two sarcoma xenografts. T-dCyd was not active in these tumor models. Treatment with T-dCyd but not aza-T-dCyd resulted in significantly advanced overall survival associated with hematologic improvement in an in vivo model of myelodysplastic syndrome (Khawaja et al., 2018), although aza-T-dCyd was toxic at the selected dose (4 mg/kg/day on weekdays for 2 weeks repeated after 21-day rest).
Recently Morris et al. (2021) have described the antitumor activity of 2′-fluoro-5-aza-4′-thio-2′-deoxy-arabinofuranosyl cytosine (F-aza-T-dCyd) and compared its in vitro and in vivo activity with aza-T-dCyd. Interestingly, there was a low COMPARE correlation between these two compounds in the 2-day NCI-60 assay, which suggested that the mechanism of action of these two compounds is different. Studies with clofarabine (2-chloro-2′-fluoro-2′-deoxy-arabinofuranosyl adenine, a molecule that like F-aza-T-dCyd has a fluorine atom at the 2′ position) indicate that addition of F at this position in an arabinose configuration results in potent inhibition of DNA polymerase once converted to the triphosphate (Parker et al., 1991, 1999). Therefore, it is likely that the mechanism of action of F-aza-T-dCyd involves inhibition of DNA replication more so than aza-T-dCyd.
Pharmacokinetics of Aza-T-dCyd
Studies conducted at the NCI indicate that the oral bioavailability of aza-T-dCyd in mice (2 mg/kg) was 80% of that seen after intravenous administration (unpublished observation). Studies by Momparler (1985) indicated that the oral bioavailability of aza-dCyd was only 9%. The low oral bioavailability of aza-dCyd in nonhuman primates could be improved by pretreatment with THU, which indicated that Cyd deaminase activity contributed to its poor bioavailability (Lavelle et al., 2012), and there are now oral formulations for administering aza-dCyd to humans. The high oral bioavailability of aza-T-dCyd suggests that this compound may be a poor substrate for Cyd deaminase. It is also possible that the rapid decomposition of aza-dCyd contributes to its poor oral bioavailability (Momparler, 1985). We have shown that the half-life of aza-T-dCyd in phosphate buffer at pH 6.2 is approximately 5-fold that of decitabine (Thottassery et al., 2014), which suggests that the increased chemical stability of this compound could contribute to its good oral bioavailability.
Aza-T-dCyd in First-in-Human Clinical Trial
Based on the good antitumor activity of aza-T-dCyd observed in mouse xenografts, the NCI initiated a phase I clinical trial, which is ongoing (Monge et al., 2019, Nguyen et al., 2021). In this trial aza-T-dCyd is administered orally once a day for 5 days of each week for 2 weeks, with 1 week off, in 21-day cycles. As of January 2021 (Nguyen et al., 2021), a total of 18 patients with solid tumors have been enrolled in the trial, and a maximal tolerated dose of 32 mg has been determined. Of 14 patients that were evaluable for response, 11 had a best response of stable disease, and 3 had a best response of progressive disease. The median cycles of therapy was four, and one patient with clear cell ovarian carcinoma has been on study for 10+ cycles (30+ weeks). The authors concluded that aza-T-dCyd is safe and well tolerated with a toxicity profile similar to currently approved hypomethylating agents. Global DNA methylation profiling, RNAseq, and DNMT immunohistochemical analyses of tumor biopsies are planned for the currently accruing dose expansion cohort.
Comparison of the Metabolism and Activity of Aza-T-dCyd and Aza-dCyd
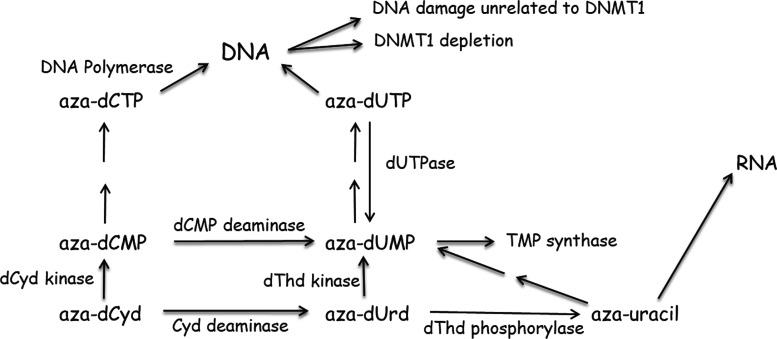
Although aza-dCyd has been studied for many years (Momparler, 2005), there is still much that is not known about its mechanism of action, and a brief review of its activity may be useful to understand how aza-T-dCyd could differ from aza-dCyd and possibly suggest reasons for their different toxicity profiles. The enzymes involved in the metabolism and activity of aza-dCyd are shown in Fig. 5. In in vitro studies it is clear that aza-dCyd is readily activated to aza-dCTP , which is incorporated into DNA, resulting in the depletion of DNMT1. Aza-dCyd is a good substrate for dCyd kinase (Momparler and Derse, 1979; Cihak et al., 1980; Vesely and Cihak, 1980; Vesely, 1987), which is the rate-limiting enzyme in the activation of most deoxynucleoside analogs. It has also been shown to be a good substrate for DNA polymerase α (Vesely and Cihak, 1977; Bouchard and Momparler, 1983), one of the primary DNA polymerases responsible for DNA replication, without causing chain termination.
Fig. 5.
Metabolism of aza-dCyd in human cells.
It is known that aza-dCyd is a reasonably good substrate for Cyd deaminase (Chabot et al., 1983; Laliberte et al., 1992), and most of aza-dCyd that is administered to an animal is converted to 5-aza-2′-deoxyuridine (aza-dUrd) (Lavelle et al., 2012; Ebrahem et al., 2012). The Km for aza-dCyd was 10- to 20-fold that of dCyd, but its Vmax was similar. Aza-dCMP is also a substrate for dCMP deaminase (Momparler et al., 1984, 1986), which creates aza-dUMP . Although in this case, the Km of aza-dCMP was similar to that of dCMP, but its Vmax was only 1% that seen with dCMP. In vivo studies (Terse et al., 2014) showed that treatment of mice with THU plus aza-dCyd was more toxic to mice than treatment with aza-dCyd alone and that the increased toxicity correlated with increased aza-dCyd plasma levels. These results suggest that the toxicity of aza-dCyd in mice is primarily due to its conversion to aza-dCMP. However, THU does not inhibit dCMP deaminase activity, and deamination of aza-dCMP could still produce considerable amounts of aza-dUMP in cells. Our studies with CEM cells (Parker et al., 2000) indicate that 90% of dCMP formed in cells is deaminated to form dUMP and it is possible that much of the aza-dCMP would be converted to aza-dUMP. However very little is known about the activities of aza-dUMP and its metabolites.
Thymidylate synthase methylates the 5-carbon of dUMP, and inhibition of this enzyme by 5-fluoro-2′-deoxyuridine-5′-monophosphate results in cytotoxicity due to depression of TTP levels and incorporation of uracil and 5-fluorouracil into DNA (Parker and Cheng, 1990). Therefore, it is possible that the 5-aza-moiety could inhibit the methylation of aza-dUMP by thymidylate synthase in a manner similar to that of aza-cytosine and DNMT1. Recently a compound screen of 11,000 compounds in intact cells using a cellular thermal shift assay identified aza-dUMP as an inhibitor of thymidylate synthase (Almqvist et al., 2016). In other studies (Requena et al., 2016) the toxicity of aza-dCyd was enhanced in cells deficient in dUTPase activity, which was correlated with increased intracellular levels of dUTP (and aza-dUTP ) and incorporation of uracil into DNA. More recently in in vitro and in vivo experiments, treatment with aza-dCyd resulted in the depletion of thymidylate synthase (Gu et al., 2021). These three studies suggest that sufficient aza-dUMP is produced in cells and that this compound inhibits thymidylate synthase resulting in the decrease of TTP levels and accumulation of dUTP and aza-dUTP. The removal of uracil or aza-uracil from DNA by uracil DNA glycosylase results in a futile cycle of incorporation and removal that overwhelms DNA repair mechanisms.
Early studies have shown that aza-dCyd can cause alkali-labile breaks in DNA (D’Incalci et al., 1985; Covey et al., 1986), which are caused by a saturable (enzymatic) DNA repair mechanism (Limonta et al., 1993). It was proposed in this work that the toxicity of aza-dCyd was due to the removal of 5-azacytosine from DNA by DNA glycosylases leading to the formation of apyrimidinic sites, and the excessive incorporation and removal of aza-dCyd sets up a futile cycle that overwhelms the DNA repair process causing DNA damage. These studies suggest that there is a balance between the amount of aza-dCyd that is incorporated into DNA that is necessary for depletion of DNMT1 and the amount incorporated that results in toxicity. Based on the more recent studies (Almqvist et al., 2016; Requena et al., 2016; Gu et al., 2021), it is possible that the DNA damage reported in these studies was due to the incorporation of dUTP or aza-dUTP.
No studies have been reported that evaluate aza-dUrd as a substrate for dThd kinase, so it is not known whether aza-dUMP can be formed from aza-dUrd. However, aza-dUrd is a substrate for dThd phosphorylase (Cihak and Vesely, 1978; Cihak, 1978), which is expressed throughout the body, and, therefore, a considerable amount of 5-aza-uracil should also circulate after exposure to aza-dCyd. Early studies with aza-dCyd indicated that 5-aza-uracil is activated and incorporated into RNA (Cihak and Sorm, 1972), which could also contribute to the toxicity of aza-dCyd. Although generation of aza-uracil may not contribute to aza-dCyd activity in vitro, it may be very important in vivo since most of the administered drug is deaminated to aza-dUrd.
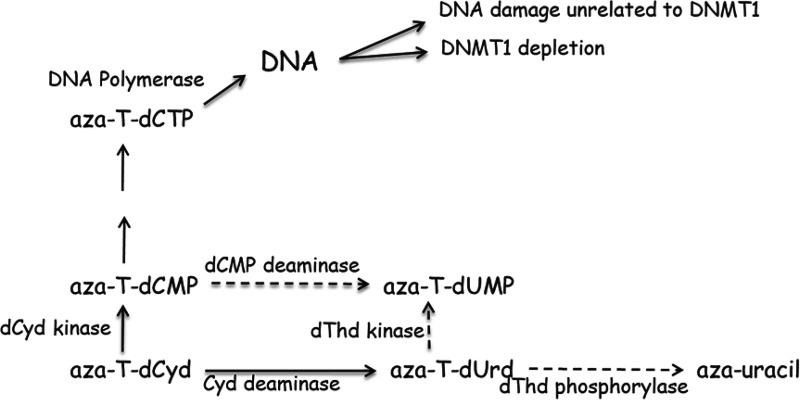
Unfortunately, no studies have been conducted to evaluate the metabolism of aza-T-dCyd. However, since treatment with aza-T-dCyd results in the depletion of DNMT1, it is clear that it is readily converted to aza-T-dCTP and incorporated into DNA. Furthermore, if one assumes that the 4′-thio moiety of aza-T-dCyd affects its metabolism in a similar manner as that seen with T-dCyd, then one would predict that the metabolism of aza-T-dCyd (Fig. 6) would be very different than that of aza-dCyd (Fig. 5) and that little aza-T-dCyd would be converted to either aza-T-dUMP or aza-uracil. If true, then this difference in metabolism could explain the large therapeutic index of aza-T-dCyd (depletion of DNMT1 vs. toxicity) relative to aza-dCyd (Table 1). Because of a potential difference in activity of dCMP deaminase with these two agents, it is possible that the primary intracellular metabolite of aza-dCyd in cells after activation by dCyd kinase is aza-dUMP, whereas for aza-T-dCyd it is aza-T-dCTP . Differences in how these molecules interact with DNA polymerases and influence DNA function could also explain the difference observed in their therapeutic indices. As discussed above, some studies have indicated that the toxicity of aza-dCyd is due to DNA damage caused by the incorporation of aza-dCyd (Limonta et al., 1993) or aza-dUrd (Requena et al., 2016) and their subsequent removal by DNA glycosylases. Others have shown that 4′-thio-2′-deoxyuridine is not able to inhibit uracil DNA glycosylase (Verri et al., 2000), which suggests that 4’-thio nucleosides in DNA would not be substrates for DNA glycosylases. Therefore, it is possible that aza-T-dCyd residues in DNA are not recognized by DNA glycosylases and that a futile cycle of incorporation/removal does not occur with aza-T-dCyd, resulting in less DNA damage.
Fig. 6.
Potential metabolism and mechanism of action of aza-T-dCyd in human cells.
Conclusion
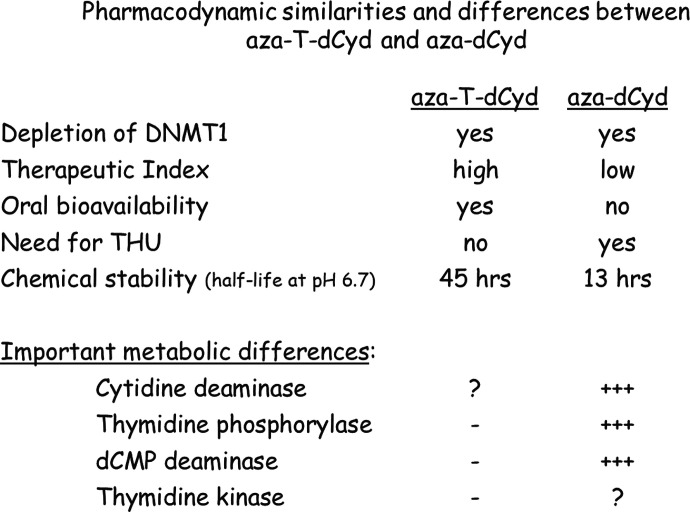
Aza-T-dCyd has two important attributes (oral bioavailability and lower toxicity) that distinguish it from aza-dCyd and argue strongly for its evaluation in various disease settings wherein selective depletion of DNMT1 is considered to be a rational strategy (Fig. 7). The reason for these differences is not known at present. Without a complete understanding of how these molecules are metabolized in cells as well as the effect of their metabolites on various enzymatic targets, it is not productive to speculate too much about how the mechanism of action of these two compounds differs. It will be necessary to determine the kinetic constants of aza-T-dCyd and aza-dCyd with dCyd kinase and Cyd deaminase as well as how their metabolites interact with various enzymes involved in nucleotide metabolism, such as DNA polymerases, dCMP deaminase, and thymidylate synthase. That said, knowledge from studies with other 4′-thio molecules suggests significant differences in the metabolism of these two compounds could explain the relatively low toxicity of aza-T-dCyd. In addition, the fact that aza-T-dCyd has high oral bioavailability and does not need THU as a combination partner suggests that aza-T-dCyd may be a poor substrate for cytidine deaminase.
Fig. 7.
Pharmacodynamic similarities and differences between aza-T-dCyd and aza-dCyd. “-” indicates little if any enzymatic activity. “+++” indicates good enzymatic activity. “?” indicates enzymatic activity unknown.
Although both aza-Cyd and aza-dCyd are approved for the treatment of some diseases, there is no strong evidence that depletion of DNMT1 is primarily responsible for their clinical activity. Both aza-dCyd and aza-Cyd are currently administered for short periods with significant toxicity followed by long recovery period. Because depletion of DNMT1 is not expected to cause acute toxicity, it is likely that their clinical activity is due to inhibition of enzymes other than DNMT1. Regardless, the therapeutic index for aza-dCyd seems to be very narrow, and the larger therapeutic index of aza-T-dCyd in mice suggests that it would be a better candidate to selectively deplete DNMT1 from cells and determine in humans whether depletion of DNMT1 is an effective target for these various diseases. Our in vivo results in murine models of cancer indicate that mice can be treated on an extended schedule with aza-T-dCyd at an effective DNMT1-depleting dose without toxicity, which would allow for treatment of all target cells as they enter S phase.
Abbreviations
- ALL
acute lymphoblastic leukemia
- aza-Cyd
5-aza-cytidine (azacitidine)
- aza-dCyd
5-aza-2′-deoxycytidine (decitabine)
- aza-T-dCyd
5-aza-4′-thio-2′-deoxycytidine
- aza-dUrd
5-aza-2′-deoxyuridine
- Cyd
cytidine
- dCyd
2′-deoxycytidine
- DNMT
DNA methyltransferase
- dThd
thymidine
- dUrd
2′-deoxyuridine
- F-aza-T-dCyd
2′-fluoro-5-aza-4′-thio-2′-deoxy-arabinofuranosylcytosine
- F-dCyd
5-fluoro-2′-deoxycytidine
- HbF
fetal hemoglobin
- NCI
National Cancer Institute
- T-dCyd
4′-thio-2′-deoxycytidine
- T-dThd
4′-thio-thymidine
- T-dUrd
4′-thio-2′-deoxyuridine
- THU
tetrahydrouridine
Authorship Contributions
Participated in research design: Parker, Thottassery.
Conducted experiments: Parker, Thottassery.
Performed data analysis: Parker, Thottassery.
Wrote or contributed to the writing of the manuscript: Parker, Thottassery.
Footnotes
This work was supported by the Alabama Drug Discovery alliance and the University of Alabama Center for Clinical and Translational Science [Grant 5UL1 RR025777] and National Institutes of Health National Cancer Institute [Grant CA34200 and Contract N01-CO-12400]. This research was supported in part by the Intramural Research Program of National Institutes of Health (National Cancer Institute).
No author has an actual or perceived conflict of interest with the contents of this article
References
- Almqvist HAxelsson HJafari RDan CMateus AHaraldsson MLarsson AMartinez Molina DArtursson PLundbäck T, et al. (2016) CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil. Nat Commun 7:11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Majerova E, Hill KD, Carter J, Stottlemyer J, Stotler H, Hollingshead MG, Collins JM (2017) Effects of epigenetic agents on methylation of DNA in vitro and in vivo, as measured with stable isotopically labeled methionine. Cancer Res 77: (13 supplement) Abstract # 4351. [Google Scholar]
- Anzai H, Frost P, Abbruzzese JL (1992) Synergistic cytotoxicity with 2′-deoxy-5-azacytidine and topotecan in vitro and in vivo. Cancer Res 52:2180–2185. [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB (2001) Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem 276:32282–32287. [DOI] [PubMed] [Google Scholar]
- Ben-Kasus T, Ben-Zvi Z, Marquez VE, Kelley JA, Agbaria R (2005) Metabolic activation of zebularine, a novel DNA methylation inhibitor, in human bladder carcinoma cells. [DOI] [PubMed] [Google Scholar]
- (•••). Biochem Pharmacol 70:121–133.15885659 [Google Scholar]
- Beumer JH, Eiseman JL, Parise RA, Florian JA Jr, Joseph E, D’Argenio DZ, Parker RS, Kay B, Covey JM, Egorin MJ (2008a) Plasma pharmacokinetics and oral bioavailability of 3,4,5,6-tetrahydrouridine, a cytidine deaminase inhibitor, in mice. Cancer Chemother Pharmacol 62:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer JH, Parise RA, Newman EM, Doroshow JH, Synold TW, Lenz HJ, Egorin MJ (2008b) Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU). Cancer Chemother Pharmacol 62:363–368. [DOI] [PubMed] [Google Scholar]
- Boothman DA, Briggle TV, Greer S (1985) Metabolic channeling of 5-fluoro-2′-deoxycytidine utilizing inhibitors of its deamination in cell culture. Mol Pharmacol 27:584–594. [PubMed] [Google Scholar]
- Bouchard J, Momparler RL (1983) Incorporation of 5-aza-2′-deoxycytidine-5′-triphosphate into DNA. Interactions with mammalian DNA polymerase α and DNA methylase. Mol Pharmacol 24:109–114. [PubMed] [Google Scholar]
- Chabot GG, Bouchard J, Momparler RL (1983) Kinetics of deamination of 5-aza-2′-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside. Biochem Pharmacol 32:1327–1328. [DOI] [PubMed] [Google Scholar]
- Charache S, Dover G, Smith K, Talbot CC Jr, Moyer M, Boyer S (1983) Treatment of sickle cell anemia with 5-azacytidine results in increased fetal hemoglobin production and is associated with nonrandom hypomethylation of DNA around the gamma-delta-beta-globin gene complex. Proc Natl Acad Sci USA 80:4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JXChen LLi YCloe AYue MWei JWatanabe KAShammo JMAnastasi JShen QJ, et al. (2018) RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun 9:1163–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondrou VStavrou EFMarkopoulos GKouraklis-Symeonidis AFotopoulos VSymeonidis AVlachaki EChalkia PPatrinos GPPapachatzopoulou A, et al. (2018) Impact of ZBTB7A hypomethylation and expression patterns on treatment response to hydroxyurea. Hum Genomics 12:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cihak A, Sorm F (1972) Metabolic transformations of 5-aza-uracil and 5-azaorotic acid in mouse liver and Escherichia coli. Biochem Pharmacol 21:607–617. [DOI] [PubMed] [Google Scholar]
- Cihák A, Veselý J (1978) Effects of 5-aza-2′-deoxycytidine on DNA synthesis in mouse lymphatic tissues. Neoplasma 25:385–393. [PubMed] [Google Scholar]
- Cihák A (1978) Transformation of 5-aza-2′-deoxycytidine-3H and its incorporation in different systems of rapidly proliferating cells. Eur J Cancer 14:117–124. [DOI] [PubMed] [Google Scholar]
- Cihak A, Vesely J, Hynie S (1980) Transformation and metabolic effects of 5-aza-2′-deoxycytidine in mice. Biochem Pharmacol 29:2929–2932. [DOI] [PubMed] [Google Scholar]
- Coral S, Sigalotti L, Gasparollo A, Cattarossi I, Visintin A, Cattelan A, Altomonte M, Maio M (1999) Prolonged upregulation of the expression of HLA class I antigens and costimulatory molecules on melanoma cells treated with 5-aza-2′-deoxycytidine (5-AZA-CdR). J Immunother 22:16–24. [DOI] [PubMed] [Google Scholar]
- Covey JM, D’Incalci M, Tilchen EJ, Zaharko DS, Kohn KW (1986) Differences in DNA damage produced by incorporation of 5-aza-2′-deoxycytidine or 5,6-dihydro-5-azacytidine into DNA of mammalian cells. Cancer Res 46:5511–5517. [PubMed] [Google Scholar]
- Daifuku R, Hu Z, Saunthararajah Y (2017) 5-Aza-2′,2′-difluoro deoxycytidine (NUC013): a novel nucleoside DNA methyl transferase inhibitor and ribonucleotide reductase inhibitor for the treatment of cancer. Pharmaceuticals (Basel) 10:65 Basel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daifuku R, Grimes S, Stackhouse M (2018) NUC041, a prodrug of the DNA methytransferase inhibitor 5-aza-2′,2′-difluoro deoxycytidine (NUC013), leads to tumor regression in a model of non-small cell lung cancer. Pharmaceuticals (Basel) 11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone J, Heller P, Hall L, Zwiers D (1982) 5-Azacytidine stimulates fetal hemoglobin synthesis in anemic baboons. Proc Natl Acad Sci USA 79:4428–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Incalci M, Covey JM, Zaharko DS, Kohn KW (1985) DNA alkali-labile sites induced by incorporation of 5-aza-2′-deoxycytidine into DNA of mouse leukemia L1210 cells. Cancer Res 45:3197–3202. [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, Patel DJ (2015) DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahem Q, Mahfouz RZ, Ng KP, Saunthararajah Y (2012) High cytidine deaminase expression in the liver provides sanctuary for cancer cells from decitabine treatment effects. Oncotarget 3:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidinoff ML, Rich MA, Perez AG (1959) Growth inhibition of a human tumor cell strain by 5-fluorocytidine and 5-fluoro-2′-deoxycytidine: reversal studies. Cancer Res 19:638–642. [PubMed] [Google Scholar]
- el-Deiry WS, Nelkin BD, Celano P, Yen RW, Falco JP, Hamilton SR, Baylin SB (1991) High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc Natl Acad Sci USA 88:3470–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST (2005) 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol 25:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gilmartin AGGroy AGore ERAtkins CLong ERMontoute MNWu ZHalsey WMcNulty DEEnnulat D, et al. (2021) In vitro and in vivo induction of fetal hemoglobin with a reversible and selective DNMT1 inhibitor. Haematologica 106:1979–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda TAFang JSalas MDo CHsu EZhukovskaya ASiegel ATakahashi RLopez-Bujanda ZADrake CG, et al. (2020) A DNA hypomethylating drug alters the tumor microenvironment and improves the effectiveness of immune checkpoint inhibitors in a mouse model of pancreatic cancer. Cancer Res 80:4754–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XTohme RTomlinson BSakre NHasipek MDurkin LSchuerger CGrabowski DZidan AMRadivoyevitch T, et al. (2021) Decitabine- and 5-azacytidine resistance emerges from adaptive responses of the pyrimidine metabolism network. Leukemia 35:1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A (2000) Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation. Curr Top Microbiol Immunol 249:55–74. [DOI] [PubMed] [Google Scholar]
- Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C, MacBeth KJ (2010) A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One 5:e9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran JL, Beumer JH, McCormick DL, Johnson WD, Newman EM, Doroshow JH, Kummar S, Covey JM, Davis M, Eiseman JL (2015) Oral and intravenous pharmacokinetics of 5-fluoro-2′-deoxycytidine and THU in cynomolgus monkeys and humans. Cancer Chemother Pharmacol 76:803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Ma J, Hu C, Zou F, Jiang S, Wang Y, Han C, Zhang Y (2019) Decitabine reverses gefitinib resistance in PC9 lung adenocarcinoma cells by demethylation of RASSF1A and GADD45β promoter. Int J Clin Exp Pathol 12:4002–4010. [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Chiang SF, Chen WT, Chen TW, Hu CH, Yang PC, Ke TW, Chao KSC (2020) Decitabine augments chemotherapy-induced PD-L1 upregulation for PD-L1 blockade in colorectal cancer. Cancers (Basel) 12:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries RK, Dover G, Young NS, Moore JG, Charache S, Ley T, Nienhuis AW (1985) 5-Azacytidine acts directly on both erythroid precursors and progenitors to increase production of fetal hemoglobin. J Clin Invest 75:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R (1997) Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci USA 94:4681–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM (1980) Cellular differentiation, cytidine analogs and DNA methylation. Cell 20:85–93. [DOI] [PubMed] [Google Scholar]
- Jüttermann R, Li E, Jaenisch R (1994) Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA 91:11797–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaysen J, Spriggs D, Kufe D (1986) Incorporation of 5-fluorodeoxycytidine and metabolites into nucleic acids of human MCF-7 breast carcinoma cells. Cancer Res 46:4534–4538. [PubMed] [Google Scholar]
- Khawaja G, Chung YJ, Park E, Difilippantonio M, Doroshow JH, Aplan PD(2018) A novel DNA methyltransferase inhibitor is effective in an in vivo model of myelodysplastic syndrome. Annual meeting American Society of Hematology Abstract # 1804. 10.1182/blood-2018-99-119265. [Google Scholar]
- Kinders R, Hollingshead M, Thottassery JV, Parker WB, Pfister T, Morris J, Anderson L, Tomaszewski J, Collins J, Doroshow J (2014) Pre-clinical Development of 4’-thio-2’-deoxycytidine (TdCyd) as a DNA-demethylating Agent for Use in Treating Solid Tissue Tumors. Cancer Res 74: (19 supplement) Abstract # 2306. [Google Scholar]
- Kumar S, Horton JR, Jones GD, Walker RT, Roberts RJ, Cheng X (1997) DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res 25:2773–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberté J, Marquez VE, Momparler RL (1992) Potent inhibitors for the deamination of cytosine arabinoside and 5-aza-2′-deoxycytidine by human cytidine deaminase. Cancer Chemother Pharmacol 30:7–11. [DOI] [PubMed] [Google Scholar]
- Laranjeira AB, Nguyen D, Huang E, Doroshow JH, Yang SX (2017) Disruption of DNA methyltransferase (DNMT) 1 confers resistance to DNMT inhibitors in human colorectal cancer cells. Cancer Res 77: (13 supplement) Abstract # 5081. [Google Scholar]
- Akpan I, Banzon V, Ibanez V, Vaitkus K, DeSimone J, Lavelle D (2010) Decitabine increases fetal hemoglobin in Papio anubis by increasing γ-globin gene transcription. Exp Hematol 38:989–993.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle DVaitkus KLing YRuiz MAMahfouz RNg KPNegrotto SSmith NTerse PEngelke KJ, et al. (2012) Effects of tetrahydrouridine on pharmacokinetics and pharmacodynamics of oral decitabine. Blood 119:1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle D, Engel JD, Saunthararajah Y (2018) Fetal hemoglobin induction by epigenetic drugs. Semin Hematol 55:60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limonta MColombo TDamia GCatapano CVConter VGervasoni MMasera GLiso VSpecchia GGiudici G, et al. (1993) Cytotoxic activity and mechanism of action of 5-aza-2′-deoxycytidine in human CML cells. Leuk Res 17:977–982. [DOI] [PubMed] [Google Scholar]
- Linnekamp JF, Butter R, Spijker R, Medema JP, van Laarhoven HWM (2017) Clinical and biological effects of demethylating agents on solid tumours - a systematic review. Cancer Treat Rev 54:10–23. [DOI] [PubMed] [Google Scholar]
- Liu M, Wang J, Liu P (2016) HPLC method development, validation, and impurity characterization of a potent antitumor nucleoside, T-dCyd (NSC 764276). J Pharm Biomed Anal 131:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov AY, Lee M, Gundry M, Gravina S, Strogonova N, Tazearslan C, Bendebury A, Suh Y, Vijg J (2012) 5-Aza-2′-deoxycytidine-induced genome rearrangements are mediated by DNMT1. Oncogene 31:5172–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio F, Giampaolo A, Carè A, Migliaccio G, Calandrini M, Russo G, Pagliardi GL, Mastroberardino G, Marinucci M, Peschle C (1983) Molecular mechanisms of human hemoglobin switching: selective undermethylation and expression of globin genes in embryonic, fetal, and adult erythroblasts. Proc Natl Acad Sci USA 80:6907–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D (1995) 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1:686–692. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, Knox JD, Szyf M (2000) DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAF1/CIP1/sdi1). J Biol Chem 275:6353–6359. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, Brown SE, Zhuang Q, Szyf M (2004) DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem 279:27915–27927. [DOI] [PubMed] [Google Scholar]
- Molokie RLavelle DGowhari MPacini MKrauz LHassan JIbanez VRuiz MANg KPWoost P, et al. (2017) Oral tetrahydrouridine and decitabine for non-cytotoxic epigenetic gene regulation in sickle cell disease: a randomized phase 1 study. PLoS Med 14:e1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momparler RL, Derse D (1979) Kinetics of phosphorylation of 5-aza-2′-deoxyycytidine by deoxycytidine kinase. Biochem Pharmacol 28:1443–1444. [DOI] [PubMed] [Google Scholar]
- Momparler RL, Rossi M, Bouchard J, Vaccaro C, Momparler LF, Bartolucci S (1984) Kinetic interaction of 5-AZA-2′-deoxycytidine-5′-monophosphate and its 5′-triphosphate with deoxycytidylate deaminase. Mol Pharmacol 25:436–440. [PubMed] [Google Scholar]
- Momparler RL (1985) Molecular, cellular and animal pharmacology of 5-aza-2′-deoxycytidine. Pharmacol Ther 30:287–299. [DOI] [PubMed] [Google Scholar]
- Momparler RL, Rossi M, Bouchard J, Bartolucci S, Momparler LF, Raia CA, Nucci R, Vaccaro C, Sepe S (1986) 5-Aza-2’-deoxycytidine synergistic action with thymidine on leukemic cells and interaction of 5-aza-dCMP with dCMP deaminase. Adv Exp Med Biol 195: ptB, 157-163. [DOI] [PubMed] [Google Scholar]
- Momparler RL (2005) Pharmacology of 5-Aza-2′-deoxycytidine (decitabine). Semin Hematol 42(3, Suppl 2)S9–S16. [DOI] [PubMed] [Google Scholar]
- Monge C, O’Sullivan Coyne G, Piekarz R, Takebe N, Bruns A, Mittra A, Khan SS, Collins JM, Anderson L, Juwara L, Miller B, Kinders RJ, Rubinstein L, Chen AP, Doroshow JH (2019) Trial in progress abstract phase 1 trial of 5-aza-4’-thio-2’-deoxycytidine (aza-TdC) in patients with advanced solid tumors. J Clin Oncol 37: (Suppl; abstract TPS3148). [Google Scholar]
- Morris JWishka DGLopez ODRudchenko VHuang GHoffman SNBorgel SGeorgius KCarter JStotler H, et al. (2021) F-aza-T-dCyd (NSC801845), a novel cytidine analog, in comparative cell culture and xenograft studies with the clinical candidates T-dCyd, F-T-d Cyd, and aza-T-dCyd. Mol Cancer Ther 20:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee KL, Heidelberger C (1962) Studies of fluorinated pyrimidines. XV. Inhibition of the incorporation of formate-C14 into DNA thymine of Ehrlich ascites carcinoma cells by 5-fluoro-2′-deoxyuridine-5′-monophosphate and related compounds. Cancer Res 22:815–822. [PubMed] [Google Scholar]
- Newman EM, Santi DV (1982) Metabolism and mechanism of action of 5-fluorodeoxycytidine. Proc Natl Acad Sci USA 79:6419–6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Hollingshead M, Rubinstein L, Doroshow J, Yang SX (2016) Selective inhibition of DNA methyltransferase and efficacy of novel DNMT inhibitors in leukemia xenografts. Cancer Res 76: (14 supplement) Abstract # 4722. [Google Scholar]
- Nguyen J, O’Sullivan Coyne H, Takebe N, Naqash AR, Mukherjee J, Bruns A, Piekarz R, Collins JM, Anderson L, Miller BL, Parchment RE, Rubinstein L, Kummar S, Sharon E, Streicher H, Chen AP, Doroshow JH (2021) Phase I trial of 5-aza-4’-thio-2’-deoxycytidine (aza-TdC) in patients with advanced solid tumors. J. Clin. Oncol. 39: (Suppl 15; abstract 3088). [Google Scholar]
- Olivieri NF, Saunthararajah Y, Thayalasuthan V, Kwiatkowski J, Ware RE, Kuypers FA, Kim H-Y, Trachtenberg FL, Vichinsky EP; Thalassemia Clinical Research Network (2011) A pilot study of subcutaneous decitabine in β-thalassemia intermedia. Blood 118:2708–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman DG, DePillis GD, Wu JC, Matsuda A, Santi DV (1988) 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry 27:5204–5210. [DOI] [PubMed] [Google Scholar]
- O’Sullivan Coyne GChen AJummar SCollins JMMeehan RSSuto MRubinstein LKinders RMoore NParchment R, et al. (2016) First-in-human trial of 4′-thio-2′-deoxycytidine (TdCyd) in patients with advanced solid tumors. Ann Oncol 27 (Supplement 6):vi114–vi135 10.1093/annonc/mdw368.54. [Google Scholar]
- Coyne GOWang LZlott JJuwara LCovey JMBeumer JHCristea MCNewman EMKoehler SNieva JJ, et al. (2020) Intravenous 5-fluoro-2′-deoxycytidine administered with tetrahydrouridine increases the proportion of p16-expressing circulating tumor cells in patients with advanced solid tumors. Cancer Chemother Pharmacol 85:979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orta ML, Höglund A, Calderón-Montaño JM, Domínguez I, Burgos-Morón E, Visnes T, Pastor N, Ström C, López-lázaro M, Helleday T (2014) The PARP inhibitor Olaparib disrupts base excision repair of 5-aza-2′-deoxycytidine lesions. Nucleic Acids Res 42:9108–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD (2008) DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol 28:752–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker WB, Cheng YC (1990) Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther 48:381–395. [DOI] [PubMed] [Google Scholar]
- Parker WB, Shaddix SC, Chang CH, White EL, Rose LM, Brockman RW, Shortnacy AT, Montgomery JA, Secrist JA 3rd, Bennett LL Jr (1991) Effects of 2-chloro-9-(2-deoxy-2-fluoro-β-D-arabinofuranosyl)adenine on K562 cellular metabolism and the inhibition of human ribonucleotide reductase and DNA polymerases by its 5′-triphosphate. Cancer Res 51:2386–2394. [PubMed] [Google Scholar]
- Parker WB, Shaddix SC, Rose LM, Tiwari KN, Montogmery JA, Secrist JA 3rd, Bennett LL Jr (1995) Metabolism and metabolic actions of 4′-thiothymidine in L1210 cells. Biochem Pharmacol 50:687–695. [DOI] [PubMed] [Google Scholar]
- Parker WB, Shaddix SC, Rose LM, Shewach DS, Hertel LW, Secrist JA 3rd, Montgomery JA, Bennett LL Jr (1999) Comparison of the mechanism of cytotoxicity of 2-chloro-9-(2-deoxy-2- fluoro-β-D-arabinofuranosyl)adenine, 2-chloro-9-(2-deoxy-2-fluoro- β-D-ribofuranosyl)adenine, and 2-chloro-9-(2-deoxy-2,2-difluoro- β-D-ribofuranosyl)adenine in CEM cells. Mol Pharmacol 55:515–520. [PubMed] [Google Scholar]
- Parker WB, Shaddix SC, Rose LM, Waud WR, Shewach DS, Tiwari KN, Secrist JA 3rd (2000) Metabolism of 4′-thio-beta-D-arabinofuranosylcytosine in CEM cells. Biochem Pharmacol 60:1925–1932. [DOI] [PubMed] [Google Scholar]
- Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B (2010) Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res 38:4313–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D-F, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, Hirohashi S(2006) DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis; 27:1160–1168. [DOI] [PubMed] [Google Scholar]
- Pinto A, Attadia V, Di Fiore PP, Fusco A, Spada OA, Vecchio G (1983) 2'-Deoxy-5-aza-cytydine induces functional and morphological differentiation of a human erythroleukemic cell line (K562). Rich MA (Ed.), Leukemia Reviews International, vol 1, Marcel Dekker, New York. [Google Scholar]
- Della Ragione F, Vacca M, Fioriniello S, Pepe G, D’Esposito M (2016) MECP2, a multi-talented modulator of chromatin architecture. Brief Funct Genomics 15:420–431. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA 97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena CE, Pérez-Moreno G, Horváth A, Vértessy BG, Ruiz-Pérez LM, González-Pacanowska D, Vidal AE (2016) The nucleotidohydrolases DCTPP1 and dUTPase are involved in the cellular response to decitabine. Biochem J 473:2635–2643. [DOI] [PubMed] [Google Scholar]
- Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6:597–610. [DOI] [PubMed] [Google Scholar]
- Roboz GJKantarjian HMYee KWLKropf PLO’Connell CLGriffiths EAStock WDaver NGJabbour ERitchie EK, et al. (2018) Dose, schedule, safety, and efficacy of guadecitabine in relapsed or refractory acute myeloid leukemia. Cancer 124:325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosjen M, McColl B, Kao B, Gearing LJ, Blewitt ME, Vadolas J (2014) Transcriptional regulators Myb and BCL11A interplay with DNA methyltransferase 1 in developmental silencing of embryonic and fetal β-like globin genes. FASEB J 28:1610–1620. [DOI] [PubMed] [Google Scholar]
- Santi DV, Norment A, Garrett CE (1984) Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA 81:6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, Hirohashi S (2003) Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer 105:527–532. [DOI] [PubMed] [Google Scholar]
- Secrist JA 3rd, Tiwari KN, Riordan JM, Montgomery JA (1991) Synthesis and biological activity of 2′-deoxy-4′-thio pyrimidine nucleosides. J Med Chem 34:2361–2366. [DOI] [PubMed] [Google Scholar]
- Saunthararajah Y, Hillery CA, Lavelle D, Molokie R, Dorn L, Bressler L, Gavazova S, Chen YH, Hoffman R, DeSimone J (2003) Effects of 5-aza-2′-deoxycytidine on fetal hemoglobin levels, red cell adhesion, and hematopoietic differentiation in patients with sickle cell disease. Blood 102:3865–3870. [DOI] [PubMed] [Google Scholar]
- Saunthararajah Y (2019) Targeting sickle cell disease root-cause pathophysiology with small molecules. Haematologica 104:1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas MSCapodieci PPolsky DMora JEsteller MOpitz-Araya XMcCombie RHerman JGGerald WLLazebnik YA, et al. (2001) Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 409:207–211. [DOI] [PubMed] [Google Scholar]
- Stomper J, Lübbert M (2019) Can we predict responsiveness to hypomethylating agents in AML? Semin Hematol 56:118–124. [DOI] [PubMed] [Google Scholar]
- Teicher BA, Lock RB, Collins JM, Gorlick R, Kolb EA, Houghton PJ, Kumasheva RT, Li XN, Erickson SW, Guo Y, Evans K, Qi L, Smith MA (2018) Pediatric preclinical testing consortium evaluation of 4’-thio-2’-deoxycytidine (T-dCyd) and 5-aza-4’-thio-2’-deoxycytidine (aza-T-dCyd). Mol Cancer Ther 17 (1 Suppl): Abstract nr LB-B12. [Google Scholar]
- Teicher BA, Lock RB, Evans K, Houghton PJ, Kurmasheva RT, Gorlicki R, Erickson S, Wishka D, Morris J, Difilippantonio M, Collins JE, Smith MA, Doroshow JH (2019) Comparison of thio-deoxy-cytidine (T-dCyd) and aza-thio-deoxy-cytidine (Aza-TdCyd) in solid and liquid tumor cell lines and PPTC pediatric xenografts. Cancer Res 79: (13 supplement) Abstract # 3839. [Google Scholar]
- Terse P, Engelke K, Chan K, Ling Y, Sharpnack D, Saunthararajah Y, Covey JM (2014) Subchronic oral toxicity study of decitabine in combination with tetrahydrouridine in CD-1 mice. Int J Toxicol 33:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thottassery JV, Sambandam V, Allan PW, Maddry JA, Maxuitenko YY, Tiwari K, Hollingshead M, Parker WB (2014) Novel DNA methyltransferase-1 (DNMT1) depleting anticancer nucleosides, 4′-thio-2′-deoxycytidine and 5-aza-4′-thio-2′-deoxycytidine. Cancer Chemother Pharmacol 74:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verri AFocher FDuncombe RJBasnak IWalker RTCoe PLde Clercq EAndrei GSnoeck RBalzarini J, et al. (2000) Anti-(herpes simplex virus) activity of 4′-thio-2′-deoxyuridines: a biochemical investigation for viral and cellular target enzymes. Biochem J 351:319–326. [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg LH, Flavell RA (1980) DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell 19:947–958. [DOI] [PubMed] [Google Scholar]
- Veselý J, Cihák A (1977) Incorporation of a potent antileukemic agent, 5-aza-2′-deoxycytidine, into DNA of cells from leukemic mice. Cancer Res 37:3684–3689. [PubMed] [Google Scholar]
- Veselý J, Cihák A (1980) Kinetics of 5-aza-2′-deoxycytidine phosphorylation in mouse spleen and L1210 leukemic cell extracts. Neoplasma 27:121–127. [PubMed] [Google Scholar]
- Veselý J (1987) High degree of resistance to 5-aza-2′-deoxycytidine in L1210 cells in vitro associated with almost complete loss of deoxycytidine kinase activity. Neoplasma 34:713–720. [PubMed] [Google Scholar]
- Wilsker DF, Barrett AM, Dull AB, Lawrence SM, Hollingshead MG, Chen A, Kummar S, Parchment RE, Doroshow JH, Kinders RJ (2019) Evaluation of pharmacodynamic responses to cancer therapeutic agents using DNA damage markers. Clin Cancer Res 25:3084–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Issa JP, Herman J, Bassett DE Jr, Nelkin BD, Baylin SB (1993) Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci USA 90:8891–8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Luo F, Chen Y, Zhu F, Wang J (2014) si-DNMT1 restore tumor suppressor genes expression through the reversal of DNA hypermethylation in cholangiocarcinoma. Clin Res Hepatol Gastroenterol 38:181–189. [DOI] [PubMed] [Google Scholar]
- Xu J, Bauer DE, Kerenyi MA, Vo TD, Hou S, Hsu YJ, Yao H, Trowbridge JJ, Mandel G, Orkin SH (2013) Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci USA 110:6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CB, Chuang JC, Byun HM, Egger G, Yang AS, Dubeau L, Long T, Laird PW, Marquez VE, Jones PA (2008) Long-term epigenetic therapy with oral zebularine has minimal side effects and prevents intestinal tumors in mice. Cancer Prev Res (Phila) 1:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wu Y, Wang W, Xu J, Lv X, Cao X, Wan T (2019) Low-dose decitabine enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by re-modulating the tumor microenvironment. Cell Mol Immunol 16:401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZSánchez Claros CSuzuki MMaggi ECKaner JDKinstlinger NGorecka JQuinn TJGeha RCorn A, et al. (2016) Loss of MEN1 activates DNMT1 implicating DNA hypermethylation as a driver of MEN1 tumorigenesis. Oncotarget 7:12633–12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SL, Zhu ST, Hao X, Li P, Zhang ST (2011) Effects of DNA methyltransferase 1 inhibition on esophageal squamous cell carcinoma. Dis Esophagus 24:601–610. [DOI] [PubMed] [Google Scholar]