Abstract
Background
One possible transmission route for nosocomial pathogens is contaminated medical devices. Formation of biofilms can exacerbate the problem. We report on a carbapenemase-producing Klebsiella pneumoniae that had caused an outbreak linked to contaminated duodenoscopes. To determine whether increased tolerance to disinfectants may have contributed to the outbreak, we investigated the susceptibility of the outbreak strain to disinfectants commonly used for duodenoscope reprocessing. Disinfection efficacy was tested on planktonic bacteria and on biofilm.
Methods
Disinfectant efficacy testing was performed for planktonic bacteria according to EN standards 13727 and 14561 and for biofilm using the Bead Assay for Biofilms. Disinfection was defined as ≥ 5log10 reduction in recoverable colony forming units (CFU).
Results
The outbreak strain was an OXA-48 carbapenemase-producing K. pneumoniae of sequence type 101. We found a slightly increased tolerance of the outbreak strain in planktonic form to peracetic acid (PAA), but not to other disinfectants tested. Since PAA was the disinfectant used for duodenoscope reprocessing, we investigated the effect of PAA on biofilm of the outbreak strain. Remarkably, disinfection of biofilm of the outbreak strain could not be achieved by the standard PAA concentration used for duodenoscope reprocessing at the time of outbreak. An increased tolerance to PAA was not observed in a K. pneumoniae type strain tested in parallel.
Conclusions
Biofilm of the K. pneumoniae outbreak strain was tolerant to standard disinfection during duodenoscope reprocessing. This study establishes for the first time a direct link between biofilm formation, increased tolerance to disinfectants, reprocessing failure of duodenoscopes and nosocomial transmission of carbapenem-resistant K. pneumoniae.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13756-022-01112-z.
Keywords: Outbreak, Antimicrobial resistance, Disinfection, Peracetic acid, Duodenoscope, Reprocessing, Gram-negative, Carbapenemase, OXA-48
Background
One of the most prominent fast-spreading, multidrug-resistant nosocomial pathogens is Klebsiella pneumoniae. In Europe, carbapenemase-producing K. pneumoniae (CRKP) have been estimated to show the highest increase in the number of infections and attributable deaths between 2007 and 2015 compared to other antibiotic-resistant bacteria [1]. The epidemiology of CRKP shows a fast spread of highly successful, well-adapted strains in healthcare settings [2].
An important contributing factor to the successful nosocomial spread of CRKP is its ability to form biofilms [3]. In general, biofilms may resist cleaning and disinfection measures to a higher degree than planktonic bacteria and may thus serve as a reservoir for subsequent spread. K. pneumoniae biofilms have been found on hospital surfaces [4], sinks and drains [5] and, importantly, on medical devices such as endoscopes [6].
In the past decade, several outbreaks by CRKP were reported in which transmission was linked to contaminated duodenoscopes [7–9]. Duodenoscopes are flexible, complex instruments that are difficult to clean and made of sensitive materials that do not allow thermal disinfection [10–13]. Thorough cleaning and disinfection are critical to prevent transmission of microorganisms. Yet, in spite of strict adherence to cleaning and disinfection protocols, reprocessing failure of endoscopes has been frequently found to be involved in nosocomial transmissions and outbreaks of CRKP [13]. However, due to the lack of systematic monitoring and the limited sensitivity of diagnostic tools, the true extent of this problem remains unknown [14].
We investigated a CRKP strain which had been causative of an outbreak affecting thirteen patients in a hospital in Berlin in 2014. The endoscopy unit and a distinct type of duodenoscope were identified as the source. The outbreak investigation revealed that duodenoscope reprocessing had been performed by strictly following the manufacturer´s instructions. In this study we assessed the susceptibility of the outbreak strain to disinfectants commonly used for duodenoscope reprocessing to test whether increased tolerance to disinfectants may have contributed to the outbreak.
Currently, disinfection recommendations are based on experimental data obtained from reference type strains that are tested as planktonic cells in suspension or attached to a surface according to the international standards EN 13727 and EN 14561. The application of disinfectant efficacy testing to bacteria in biofilm has not been established yet.
We here investigated the efficacy of disinfection on planktonic bacteria and on biofilm, hypothesizing that the latter might display a higher, and thus clinically relevant tolerance to disinfectants. Efficacy testing was performed on: (1) planktonic cells in suspension, (2) planktonic cells fixed to a surface, and (3) cells embedded in biofilm.
For the latter part we used the Bead Assay for Biofilms which has been developed in our laboratory and has proven to be a reliable and robust method for testing the efficacy of disinfectants on biofilm-embedded bacteria [15]. We tested several disinfectants that are commonly used for disinfection of duodenoscopes in Germany, including glutaraldehyde (GA) and peracetic acid (PAA) [16, 17]. Our objective was to establish a direct link between biofilm formation, decreased susceptibility to disinfectants, and inadequate disinfection during reprocessing leading to a nosocomial outbreak.
Description of the outbreak
Between May and November 2014, an outbreak of CRKP occurred in a hospital in Berlin affecting 13 patients. The outbreak strain was recovered from clinical specimens in eight patients (including blood cultures, wound swabs and tracheal aspirates), rectal screening cultures in five patients, two duodenoscopes and one gastroscope (16 isolates in total). Both duodenoscopes belonged to a brand that had previously been involved in other CRKP outbreaks [6, 7], and which was retracted by the manufacturer and redesigned thereafter. The reprocessing recommendations were also subsequently updated by the manufacturer. During the outbreak, duodenoscope reprocessing in the affected unit was performed according to the manufacturers’ instructions using a commercially available product containing PAA, with a working concentration of 0.15% (w/w) PAA and 10 min exposure time at 25 °C in an automated washer-disinfector. In response to this outbreak, parts of the duodenoscopes were replaced with newly designed elements by the manufacturer and the duodenoscope reprocessing procedure was changed to a GA-based disinfection (10 min exposure time, 55 °C). No further cases occurred after these changes.
Molecular characterization of the outbreak strain
Pulsed-field gel electrophoresis (PFGE)-based typing confirmed that the CRKP isolates obtained during the outbreak belonged to the same strain (Additional file 1: Fig. S1A). As a representative of the outbreak strain, the K. pneumoniae isolate 886/14 obtained from a clinical sample of a patient was further analysed. Antimicrobial susceptibility testing revealed that the isolate was resistant to cephalosporins and carbapenems and susceptible only to amikacin, colistin and tigecycline (Additional file 2: Table S1). Carbapenemase production was proven by modified Hodge test and was encoded by the blaOXA48 gene located on a self-conjugable IncL/M megaplasmid, determined by PCR-based Sequencing as described previously [18] (Additional file 1: Fig. S1B). The outbreak strain was subjected to whole genome sequencing. Multilocus Sequence Typing (MLST) using the MLST tool revealed that the outbreak strain belonged to the sequence type (ST)101 [19]. The complete antimicrobial susceptibility profile and detailed molecular characterization are provided in Additional file 2: Material 1 and Table S2.
Efficacy testing of disinfectants on the outbreak strain
We investigated the susceptibility of the outbreak strain by initially testing four disinfectants that are commonly used for the reprocessing of duodenoscopes: hydrogen peroxide (H2O2), GA, isopropanol and PAA. For comparison, a K. pneumoniae type strain (ATCC 13883) was tested in parallel. Disinfection was defined as ≥ 5log10 reduction in recoverable mean CFUs. Experimental conditions, disinfectants and neutralizers are listed in Additional file 2: Table S3.
Testing of planktonic cells in suspension
As a first step, efficacy testing on planktonic cells in suspension was performed using the suspension test according to EN 13727 [20]. For H2O2, GA and isopropanol we found similar results for disinfection of the outbreak strain and the type strain (Table 1). While the latter three substances completely inactivated both strains at similar concentrations, we found a slight difference between the outbreak strain and the type strain with regard to the efficacy of PAA. The PAA concentration needed for disinfection of the outbreak strain was six-fold higher than the concentration needed for the type strain (0.003% versus ≤ 0.0005% (w/v) PAA) (Table 1). Still, both strains were sensitive to PAA in the suspension test. Since PAA was the disinfectant used in the respective hospital for endoscope reprocessing at the time of the outbreak the effect of PAA was further investigated using two additional tests, as described below.
Table 1.
Results of efficacy testing by suspension test of four disinfectants
| Disinfectant | Exposure time (min) | K. pneumoniae ATCC 13883 | K. pneumoniae outbreak strain |
|---|---|---|---|
| H2O2 | 5 | 5 % (w/v) | 5 % (w/v) |
| Glutaraldehyde | 10 | 0.04 % (w/v) | 0.04 % (w/v) |
| Isopropanol | 1 | 30 % (w/v) | 30 % (w/v) |
| Peracetic acid | 10 | ≤ 0.0005 % (w/v) | 0.003 % (w/v) |
Conditions resulted in ≥ 5log10 reduction in CFU of K. pneumoniae strains are presented
Testing of surface-fixed planktonic cells
Efficacy testing for PAA on surface-fixed bacteria was performed using the quantitative carrier test for evaluation of bactericidal activity for medical instruments (instrument disinfection) according to EN 14561 [21]. At 10 min exposure time, higher concentrations were required for the inactivation of both strains as compared to the suspension test (Fig. 1). For surface-fixed bacteria, PAA concentrations of 0.01% and 0.15% were required for disinfection of the type strain and the outbreak strain, respectively. Still, complete inactivation was achieved for both strains in this assay at the PAA concentration used in duodenoscope reprocessing, i.e. 0.15%.
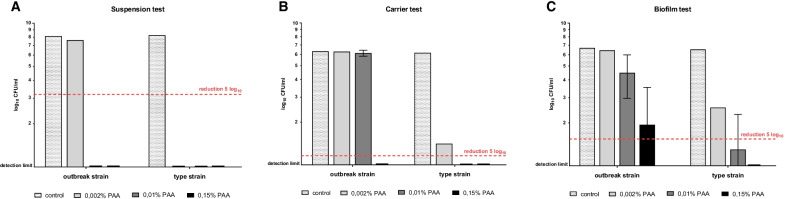
Fig. 1.
Efficacy of peracetic acid (PAA) using three different disinfectant testing methods. Bars show mean recovered CFUs after 10 min exposure to different concentrations of PAA of the K. pneumoniae outbreak strain and the type strain that were tested as: (a) planktonic cells (suspension test, EN 13727) (b) surface-fixed planktonic cells (carrier test, EN 14561) and (c) biofilm (Bead Assay for Biofilms). Disinfection was defined as ≥ 5log10 reduction in mean recoverable CFUs and is marked by a dashed red line in the respective assays. At comparable PAA concentrations, the outbreak strain shows a higher number of recovered CFUs than the type strain in all three models. Experiments were performed in triplicates
Testing bacteria in biofilm
We next assessed the efficacy of PAA on 24-h old biofilm of the outbreak strain and the type strain using the Bead Assay for Biofilms [15]. An even higher concentration of PAA was required to achieve disinfection of both strains (Fig. 1). Only at a PAA concentration of 1%, which is markedly higher than the concentration used for duodenoscope reprocessing (0.15%), did we observe a ≥ 5log10 CFU reduction of the outbreak strain in all biological replicates (Fig. 2). In contrast, the disinfection of the type strain was achieved at a lower concentration, i.e. 0.15% PAA.
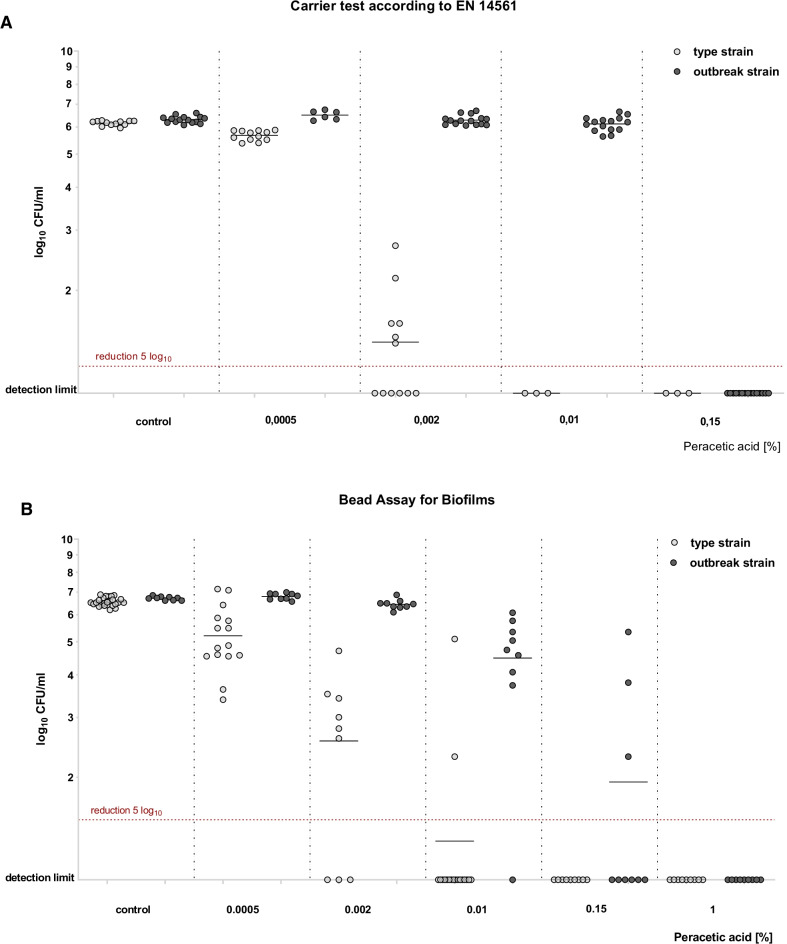
Fig. 2.
Effect of PAA on K. pneumoniae strains as surface-fixed cells (a) and biofilm (b). Scatter Plot with each dot representing recovered CFUs for a replicate after 10 min exposure to PAA. Disinfection was defined as ≥ 5log10 reduction in mean recoverable CFUs (dashed red line). The horizontal lines show the mean value for the respective replicates. In the biofilm model, 0.15% PAA, the concentration used in duodenoscope reprocessing, was not sufficient to achieve disinfection of the outbreak strain
Discussion
We investigated a CRKP strain responsible for an outbreak in a duodenoscopy unit and found a clinically relevant increased tolerance towards PAA-based disinfection. Since PAA was used for duodenoscopes reprocessing at the time of the outbreak, this factor may be a key feature contributing to the nosocomial transmission of the outbreak strain to several patients, even though the reprocessing unit strictly followed standard disinfection protocols. Of note, the outbreak strain displayed tolerance to PAA, which was used for reprocessing, but not to the other disinfectants included in the initial testing, suggesting that the enhanced tolerance to PAA confers a specific advantage to the outbreak strain.
Bacteria in biofilm have previously been shown to be less susceptible to disinfectants than surface-fixed planktonic bacteria [15, 22]. Our data confirm these observations and show the clinical impact of this feature, meaning that biofilm of the outbreak strain could not be inactivated by the PAA concentration deemed as safe for routine reprocessing. It should be noted that biofilm of the type strain was successfully inactivated by the PAA concentration used in duodenoscope reprocessing, suggesting that this trait may be a unique feature of the outbreak strain.
The CRKP outbreak strain characterized in this study belongs to the epidemic ST101, a highly successful clonal lineage that often comprises carbapenemase-producing isolates [2, 23, 24]. Future research is needed to evaluate additional isolates of the ST101 lineage and maybe other epidemic CRKP lineages for tolerance to disinfectants.
Reprocessing of complex medical instruments such as duodenoscopes is a critical issue in infection prevention and control (IPC). It has been hypothesized that transmissions of pathogens by contaminated endoscopes may be severely underreported [14]. As a disinfectant, PAA is valued for its efficacy and is widely accepted by personnel due to safe handling [25]. Still, in spite of strict adherence to best practice, several CRKP outbreaks have been linked to thermolabile endoscopes [7–9]. Interestingly, an outbreak similar to the one described here involving a K. pneumoniae ST101 (OXA-48, CTX-M-15) strain associated with duodenoscopes occurred in another hospital in Berlin in 2012 [6], indicating that problems with CRKP and contaminated endoscopes are more common than generally assumed. K. pneumoniae and other biofilm producing species such as Pseudomonas aeruginosa and Acinetobacter baumannii were recently shown to survive the PAA-based duodenoscope reprocessing, with increasing colony counts after each reprocessing cycle over the course of one working day [26]. Our study shows that even correct reprocessing of endoscopes may result in a residual risk for nosocomial transmission of pathogens. Higher PAA concentrations that could efficiently inactivate biofilms such as those formed by the outbreak strain described here would inflict damage on the sensitive materials of duodenoscopes. The formation of miniscule cracks, for example in the duodenoscopes mantle, would provide additional niches for biofilm formation and persistence.
Our study has some limitations. First, we assessed only one outbreak isolate and compared it with a type strain. Future investigation of additional clinical CRKP isolates, including highly successful clonal lineages, is needed to assess whether the decreased susceptibility to disinfectants observed in the outbreak strains biofilm is a unique feature or might represent a general adaptive step of nosocomial strains to withstand continued exposure to disinfectants. Second, since the outbreak ended as both the duodenoscope model used and the disinfection procedure applied were changed, we cannot say which step was the most effective one in terminating the outbreak.
Conclusion
In conclusion, we found that biofilm of the K. pneumoniae outbreak strain was tolerant to standard disinfection conditions that are currently established for duodenoscopes reprocessing. We consider the markedly increased tolerance of the outbreak strains' biofilm to PAA to be a major factor contributing to the outbreak. This study establishes for the first time a direct link between biofilm formation, increased tolerance to disinfectants, reprocessing failure of duodenoscopes and nosocomial transmission of K. pneumoniae. Bacterial biofilms must be considered systematically to avoid underexposure of bacterial pathogens to disinfectants, resulting in selective pressure towards development of tolerant strains.
Supplementary Information
Additional file 2. Supplemental Material.
Acknowledgements
The authors are grateful to Prof. Dr. Martin Mielke for the initiating scientific investigation of the outbreak and Dr. Martin Eisenblätter for providing the outbreak strain. We thank Katja Levin and Dóra Csertö for excellent technical assistance.
Author contributions
MB, KK and MA conceptualized the project and the manuscript. KK, LB, BP, YP, IS, and CS conducted the investigation. MB wrote the initial draft. MB, KK and MA revised the manuscript. MA supervised the project. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. The authors declare no funding.
Availability of data and materials
The outbreak strain may be provided upon request to M.A. through a material transfer agreement. The authors will share data on request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Melanie S. Brunke and Katharina Konrat contributed equally to this work.
References
- 1.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL; Burden of AMR Collaborative Group. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed]
- 2.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandeira M, Carvalho PA, Duarte A, Jordao L. Exploring dangerous connections between Klebsiella pneumoniae biofilms and healthcare-associated infections. Pathogens (Basel, Switzerland) 2014;3(3):720–731. doi: 10.3390/pathogens3030720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa DM, Johani K, Melo DS, Lopes LKO, Lopes Lima LKO, Tipple AFV, et al. Biofilm contamination of high-touched surfaces in intensive care units: epidemiology and potential impacts. Lett Appl Microbiol. 2019;68(4):269–276. doi: 10.1111/lam.13127. [DOI] [PubMed] [Google Scholar]
- 5.Kizny Gordon AE, Mathers AJ, Cheong EYL, Gottlieb T, Kotay S, Walker AS, et al. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—a systematic review of the literature. Clin Infect Dis. 2017;64(10):1435–1444. doi: 10.1093/cid/cix132. [DOI] [PubMed] [Google Scholar]
- 6.Kola A, Piening B, Pape UF, Veltzke-Schlieker W, Kaase M, Geffers C, et al. An outbreak of carbapenem-resistant OXA-48—producing Klebsiella pneumoniae associated to duodenoscopy. Antimicrob Resist Infect Control. 2015;4:8. doi: 10.1186/s13756-015-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aumeran C, Poincloux L, Souweine B, Robin F, Laurichesse H, Baud O, et al. Multidrug-resistant Klebsiella pneumoniae outbreak after endoscopic retrograde cholangiopancreatography. Endoscopy. 2010;42(11):895–899. doi: 10.1055/s-0030-1255647. [DOI] [PubMed] [Google Scholar]
- 8.Bourigault C, Le Gallou F, Bodet N, Musquer N, Juvin ME, Corvec S, et al. Duodenoscopy: an amplifier of cross-transmission during a carbapenemase-producing Enterobacteriaceae outbreak in a gastroenterology pathway. J Hosp Infect. 2018;99(4):422–426. doi: 10.1016/j.jhin.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Kovaleva J, Peters FT, van der Mei HC, Degener JE. Transmission of infection by flexible gastrointestinal endoscopy and bronchoscopy. Clin Microbiol Rev. 2013;26(2):231–254. doi: 10.1128/CMR.00085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCafferty CE, Aghajani MJ, Abi-Hanna D, Gosbell IB, Jensen SO. An update on gastrointestinal endoscopy-associated infections and their contributing factors. Ann Clin Microbiol Antimicrob. 2018;17(1):36. doi: 10.1186/s12941-018-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutala WA, Kanamori H, Gergen MF, Knelson LP, Sickbert-Bennett EE, Chen LF, et al. Enhanced disinfection leads to reduction of microbial contamination and a decrease in patient colonization and infection. Infect Control Hosp Epidemiol. 2018;39(9):1118–1121. doi: 10.1017/ice.2018.165. [DOI] [PubMed] [Google Scholar]
- 12.Muscarella LF. Risk of transmission of carbapenem-resistant Enterobacteriaceae and related "superbugs" during gastrointestinal endoscopy. World J Gastrointest Endosc. 2014;6(10):457–474. doi: 10.4253/wjge.v6.i10.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dirlam Langlay AM, Ofstead CL, Mueller NJ, Tosh PK, Baron TH, Wetzler HP. Reported gastrointestinal endoscope reprocessing lapses: the tip of the iceberg. Am J Infect Control. 2013;41(12):1188–1194. doi: 10.1016/j.ajic.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Konrat K, Schwebke I, Laue M, Dittmann C, Levin K, Andrich R, et al. The bead assay for biofilms: a quick, easy and robust method for testing disinfectants. PLoS ONE. 2016;11(6):e0157663. doi: 10.1371/journal.pone.0157663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kommission für Krankenhaushygiene und Infektionsprävention beim Robert Koch-Institut (KRINKO), Bundesinstitut für Arzneimittel und Medizinprodukte (BfArM). Anforderungen an die Hygiene bei der Aufbereitung von Medizinprodukten. Empfehlung der KRINKO und des BfArM. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55(10):1244–310. [DOI] [PubMed]
- 16.Kommission für Krankenhaushygiene und Infektionsprävention beim Robert Koch-Institut (KRINKO), Bundesinstitut für Arzneimittel und Medizinprodukte (BfArM). Mitteilung der KRINKO sowie des BfArM: Kommentar zur Anlage 8 „Anforderungen an die Hygiene bei der Aufbereitung flexibler Endoskope und endoskopischen Zusatzinstrumentariums“ der Empfehlung „Anforderungen an die Hygiene bei der Aufbereitung von Medizinprodukten“. Epidemiol Bull. 2013;(28):253–5.
- 17.Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother. 2012;56(1):559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DIN EN 13727, Chemical disinfectants and antiseptics—quantitative suspension test for the evaluation of bactericidal activity in the medical area—test method and requirements (phase 2, step 1); German version EN 13727:2012+A2:2015.
- 20.DIN EN 14561. Chemical disinfectants and antiseptics—quantitative carrier test for the evaluation of bactericidal activity for instruments used in the medical area - Test method and requirements (phase 2, step 2); German version EN 14561:2006. 2006-08.
- 21.Bridier A, Briandet R, Thomas V, Dubois-Brissonnet F. Resistance of bacterial biofilms to disinfectants: a review. Biofouling. 2011;27(9):1017–1032. doi: 10.1080/08927014.2011.626899. [DOI] [PubMed] [Google Scholar]
- 22.Becker L, Kaase M, Pfeifer Y, Fuchs S, Reuss A, von Laer A, et al. Genome-based analysis of Carbapenemase-producing Klebsiella pneumoniae isolates from German hospital patients, 2008–2014. Antimicrob Resist Infect Control. 2018;7:62. doi: 10.1186/s13756-018-0352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roe CC, Vazquez AJ, Esposito EP, Zarrilli R, Sahl JW. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 Lineage. Front Microbiol. 2019;10:542. doi: 10.3389/fmicb.2019.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otterspoor S, Farrell J. An evaluation of buffered peracetic acid as an alternative to chlorine and hydrogen peroxide based disinfectants. Infect Dis Health. 2019;24:240–243. doi: 10.1016/j.idh.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Cristina ML, Sartini M, Schinca E, Ottria G, Dupont C, Bova P, et al. Is post-reprocessing microbiological surveillance of duodenoscopes effective in reducing the potential risk in transmitting pathogens? Int J Environ Res Public Health. 2019;17(1):140. doi: 10.3390/ijerph17010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 27.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7(2):88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 28.Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226(2):235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 29.Wetzker W, Pfeifer Y, Wolke S, Haselbeck A, Leistner R, Kola A, et al. Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from flies in the urban center of Berlin, Germany. Int J Environ Res Public Health. 2019;16(9):1530. doi: 10.3390/ijerph16091530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2. Supplemental Material.
Data Availability Statement
The outbreak strain may be provided upon request to M.A. through a material transfer agreement. The authors will share data on request.