Abstract

Drug metabolite identification is a bottleneck of drug metabolism studies due to the need for time-consuming chromatographic separation and structural confirmation. Ion mobility-mass spectrometry (IM-MS), on the other hand, separates analytes on a rapid (millisecond) time scale and enables the measurement of collision cross section (CCS), a unique physical property related to an ion’s gas-phase size and shape, which can be used as an additional parameter for identification of unknowns. A current limitation to the application of IM-MS to the identification of drug metabolites is the lack of reference CCS values. In this work, we assembled a large-scale database of drug and drug metabolite CCS values using high-throughput in vitro drug metabolite generation and a rapid IM-MS analysis with automated data processing. Subsequently, we used this database to train a machine learning-based CCS prediction model, employing a combination of conventional 2D molecular descriptors and novel 3D descriptors, achieving high prediction accuracies (0.8–2.2% median relative error on test set data). The inclusion of 3D information in the prediction model enables the prediction of different CCS values for different protomers, conformers, and positional isomers, which is not possible using conventional 2D descriptors. The prediction models, dmCCS, are available at https://CCSbase.net/dmccs_predictions.
Introduction
Drug metabolism studies are a critical component of the drug development process. Metabolites can inform metabolic soft spots and may be pharmacologically active and/or elicit unexpected toxicity or other off-target effects, making knowledge of their structures essential.1,2 Current and conventional approaches to drug metabolite structural determination have typically involved a combination of liquid chromatography (LC), coupled with UV–vis spectroscopy and/or mass spectrometry (MS), and nuclear magnetic resonance spectroscopy (NMR).3−5 LC–MS and LC–UV benefit from low sample requirements and fast analysis time, but identification of unknowns can be limited when relying upon UV spectra or MS fragmentation data alone. In contrast, NMR allows for definitive assignment of chemical structures, but it requires large amounts of materials and is relatively low throughput.
Ion mobility spectrometry (IMS) is an analytical technique that rapidly separates ions based on differences in their gas-phase size and shape, which is orthogonal to polarity-based LC separation and partially orthogonal to mass.6−10 In time-dispersive ion mobility (IM) separations, ions are driven through a neutral buffer gas under the influence of an electric field. Ions are differentially impeded as they interact with the buffer gas molecules, and as a result, they traverse the mobility cell in different amounts of time (i.e., drift time). An ion’s drift time can be converted into collision cross section (CCS), a unique physical property reflecting its gas-phase size and shape, using appropriate experimental measurements and/or calibration. Excellent reproducibility has been demonstrated for CCS measured across different instrumentation and laboratories,11−14 making it a robust parameter for compound identification. CCS also provides useful information related to shape, conformation, and polarity. When IM is coupled with MS (IM-MS), an additional dimension of separation is achieved without adversely affecting analytical throughput.
The use of IM-MS for the determination of drugs and their metabolites has gained significant traction in recent years,15 but inadequate reference CCS databases remains a significant limitation to the application of CCS to identifying unknown metabolites. Large CCS databases covering drug and drug-like compounds have been presented in the literature,16−19 but due to the vastness and complexity of small molecule chemical space, many unknowns may not be represented. This issue of chemical representation is even more pronounced for drug metabolites, for which no such large-scale CCS database exists. Theory-based computation methods are generally very time-consuming and resource-demanding.15,20−22 ISiCLE is a higher throughput computational workflow, but this method still requires large amounts of computational resources and multiple steps of computational setup, and thus, the throughput is still not ideal.23 This problem can be addressed by leveraging structural trends in existing CCS databases to predict CCS for unknowns that are not in experimental databases, and this approach has been demonstrated by multiple groups, including us.12,18,19,24−29 An important consideration in this approach, however, is the dependence of CCS prediction performance on the quality and coverage of chemical space in the data used to train the model.27 Therefore, a drug metabolite-specific CCS database is needed for accurate prediction of CCS for drug metabolites. Another limitation in current machine learning (ML)-based CCS prediction models is that the 2D features used in previous work (e.g., molecular quantum numbers, MQNs) do not adequately capture more complex IM behavior arising from the presence of different protomers, conformers, or positional isomers that are common among drugs and drug metabolites.15 Previous work by Soper-Hopper et al. compared the performance of 2D vs 3D molecular descriptors using partial least-squares linear multivariate regression, but this previous work generated 3D molecular descriptors using the structures of the neutral parent molecules and did not demonstrate the capability to predict CCS values for different conformers and isomers.30
Here, we present (1) the generation of a high-quality drug and drug metabolite CCS database through the use of high-throughput in vitro drug metabolite generation and rapid IM-MS analysis with automated data processing and (2) the training of drug- and drug metabolite-specific CCS prediction models using ML with novel 3D molecular descriptors, which enables the prediction of CCS values for protomers, conformers, and positional isomers with high accuracy and throughput.
Experimental Section
High-Throughput In vitro Drug Metabolite Generation
Drug metabolites were generated in vitro using pooled subcellular fractions (S9 and microsomes) derived from human liver following a protocol from our previous work,31 adapted to a high-throughput 384-well plate format with all sample preparation performed using automated sample handling systems at the Quellos High-Throughput Screening Core at the University of Washington. Briefly, HLM/S9 stock (5 mM GSH, 5 mM MgCl, 0.01 mg/mL alamethicin, 0.2 mg of protein/mL pooled HLM, 0.2 mg of protein/mL S9, 100 mM potassium phosphate buffer at pH 7.4) was prepared and allowed to stand on ice for 15 min (alamethicin pretreatment to enhance UGT activity). A 90 μL sample of the HLM/S9 stock was dispensed into each well of 14 384-well plates, and then 0.5 μL of each drug stock (50 mM in DMSO) from the MicroSource Spectrum Discovery Collection (seven plates) was dispensed into the plates in duplicate. Ten microliters of a cofactor-containing activation mixture (10 mM NADPH, 50 mM UDPGA, 100 mM potassium phosphate buffer at pH 7.4) or potassium phosphate buffer without cofactors (as control) were then added to the duplicate plates, initiating the drug metabolism reactions for plates containing activation mixture. All plates were incubated at room temperature for 90 min before being quenched with 100 μL of ice-cold acetonitrile (with 10 μM lysophosphatidylethanolamine 13:0 as an internal standard). After quenching, all plates were stored at 4 °C for at least 15 min to promote precipitation of proteins. Each plate was centrifuged at 3500G for 15 min at 4 °C to sediment the precipitated proteins, and then 150 μL of the supernatant was transferred to fresh plates. All plates were stored at −80 °C until IM-MS analysis.
High-Throughput Ion Mobility-Mass Spectrometry
Samples (5 μL) were injected and separated using a Waters Acquity FTN UPLC coupled to a reversed-phase column (Phenomenex Kinetex, 2.6 μm, polar C18, 100 Å, 30 × 21 mm), eluting with a gradient of water with 0.1% formic acid (A) and methanol with 0.1% formic acid (B) at 0.5 mL/min: 0.00–0.20 min, 100% A; 0.20–0.30 min, 100 → 25% A; 0.30–0.75 min, 25 → 0% A; 0.75–1.05 min, 0% A; 1.05–1.10 min, 0 → 100% A. The total analysis time for each sample, factoring in acquisition and autosampler operations, was just under 2 min. For each injection, the first 0.20 min of eluent was diverted to waste in order to avoid buildup of salt on the ESI source, and after that, the flow was automatically diverted back to the instrument via an electronically controlled switching valve. TWIM-MS analysis was performed on a Waters Synapt G2-Si mass spectrometer (Waters Corp., Milford, MA) equipped with an ESI source and using nitrogen as the drift gas. ESI conditions were as follows: capillary, +2.3 kV; sampling cone, 40 V; source temperature, 130 °C; desolvation temperature, 350 °C; cone gas, 90 L/h; and desolvation gas, 600 L/h.
Mass calibration was performed using sodium formate for the range of m/z 50–1200. IM separations were performed at a traveling wave velocity of 650 m/s and a height of 24.9 V. For post-IM fragmentation analyses, collision energy was added to the transfer region using a ramp from 30 to 50 eV. Data was acquired from 0.20 to 1.15 min with a 1 s scan time over m/z 50–1200, which resulted in approximately 57 scans across the acquired elution region (individual peaks typically spanned ∼0.05 min for roughly three scans per peak). The 384-well plates were analyzed on three separate occasions over two months.
TWIM CCS Calibration
A series of singly charged polyalanines (n = 2–14) and a mixture of druglike compounds were used for calibration of TWIM drift times into CCS (TWIMCCSN2) using their drift tube CCS values in nitrogen (DTCCSN2), as described previously.16,31 Briefly, arrival time distributions (ATDs) for CCS calibrants were extracted from the raw data (acquired multiple times throughout acquisition of each plate) using accurate mass with a window of ±0.01 Da, and a CCS calibration curve was constructed from reference CCS values in an automated fashion using a Python script developed in-house.31 Drift times for each calibrant were obtained as the mean from a least-squares fit of a Gaussian function on the ATD and were corrected for mass-dependent flight time outside the mobility region to give the corrected drift times (td′), and reference CCS values were corrected for the ion charge state (Z) and reduced mass with the drift gas to give the corrected CCS (CCS′).32 A calibration curve was generated by fitting these corrected values with the function CCS′ = A(td′ + t0)B, where A, t0, and B were the fitted parameters.11,33 A calibration curve displaying randomly distributed fit residuals with a maximal absolute error of less than 3% was considered acceptable. CCS calibrant data was acquired 3–5 times over the course of analysis of each plate, and all of this calibrant data was used to construct a combined CCS calibration curve to account for any variation that occurred over the course of data acquisition. The measurement and report of the CCS values are consistent with the recommendations in the field.34
Ion Mobility-Mass Spectrometry Data Processing
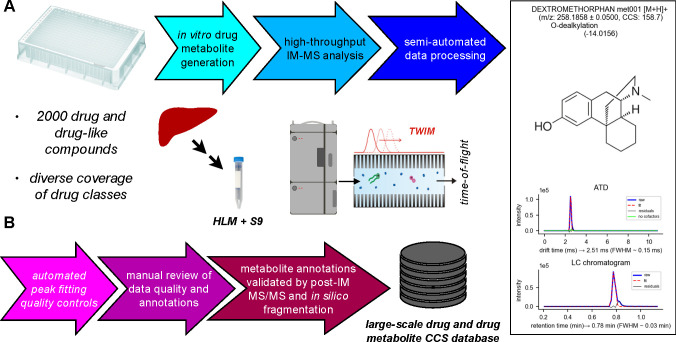
The raw IM-MS data was processed in a number of steps to extract, annotate, and validate CCS values for drugs and putative metabolites (Figure 1B) and was performed separately for each batch of data acquired on the same day (two plates were analyzed each day). The first set of data processing steps was completely automated using Python scripts developed in-house and were applied only to the first technical replicates. First, a target list was assembled for the parent drugs based on the known plate contents. For each parent compound, m/z-selected arrival time distributions (ATDs) were extracted for common ionized species with a tolerance of 0.05 Da. ATDs were fit with a Gaussian function to obtain drift time, and the fitted drift time was used to calculate calibrated CCS. If an ATD was not able to be fit or the fitted peak did not meet empirically determined rough quality cutoffs (intensity >1000, peak width between 0.06 and 1.77 ms), the corresponding ionized species was not processed any further. Upon successful ATD peak fitting, a drift time-selected LC chromatogram was also extracted, and an attempt was made to fit for retention time. All data and metadata were stored in custom Python data structures for subsequent processing. Putative metabolites were generated using BioTransformer,35 with the “allHuman” setting and up to two metabolism steps. Putative metabolites were filtered to exclude isobaric metabolites, metabolites with the same neutral mass as the parent compound, and metabolites resulting from the breakdown of secondary metabolites (i.e., free glucuronic acid or glutathione), then their corresponding ATDs were extracted from the raw data and fitted as described above. Successfully fitted ATDs were stored along with metadata (including putative metabolite annotation) in custom Python data structures for subsequent processing. Plots containing compound/putative metabolite structures, m/z, metabolism reaction information, CCS, and ATDs with fits were generated and stored (see Figure 1A) for subsequent manual review.
Figure 1.
(A) Workflow for high-throughput in vitro drug metabolite generation and IM-MS analysis. Drug metabolites were generated from the MicroSource Spectrum Discovery Collection, containing ∼2000 drug and drug-like compounds, in a high-throughput 384-well plate format using subcellular fractions (microsomes and S9) pooled from 200 human livers. Samples were analyzed using a rapid IM-MS protocol, including semiautomated data processing, including extraction and fitting of drift times from ATDs, calibration of CCS, prediction of metabolites, and establishment of cofactor dependence for oxidative metabolites. (B) Semiautomated data processing with multiple steps of automated and manual quality controls, including automated quality controls on peak fitting, manual review of extracted data quality and metabolite annotations, and validation of metabolite annotations with MS/MS data. The processed data was finally compiled into a SQLite3 database for use in CCS prediction by ML.
The resulting initial data set (>11k analytes) was next subjected to a manual review process. Each of the generated plots described above was manually inspected for general quality of ATD peak fitting (clean ATD fit without secondary peaks) and cofactor dependence for oxidative and glucuronide metabolites and then accepted or rejected accordingly. The results of this manual review process were used to curate an analyte m/z target list for automated data extraction from the second and third replicates. Data extraction from the second and third replicates followed the same automated workflow described above, except that the curated target list was used to search for putative metabolites rather than through in silico metabolite prediction. All extracted data and metadata from the second and third replicates were stored in custom Python data structures for subsequent processing.
The final step in data processing was validating compound annotations, which was performed using a semiautomated process. The identities of the parent compounds were known from the plate contents, so further validation was not required. To validate the annotations of the putative metabolites, known metabolites of the parent compounds were manually searched for in the DrugBank database.36 A list of potential metabolites and associated metadata were compiled from these searches and later matched to metabolites (superseding the original putative metabolite annotation from BioTransformer) on the basis of their neutral mass (within 50 ppm was considered a match). Finally, all metabolite annotations were subjected to filtering based on postmobility MS/MS data that were acquired for the first replicate. Drift time-selected MS/MS spectra were extracted and scored against in silico fragmentation spectra using MetFrag,37 and all annotations with a fragmenter score above the empirical cutoff of 100 were accepted. This empirical MetFrag scoring cutoff was determined by a rank test using the known identities of the parent compounds as follows. The drift time-selected MS/MS spectrum for each parent compound was compared to the in silico fragmentation spectra of all parent compounds, resulting in ranked identifications with corresponding fragmenter scores. The rank and score of the known identity were recorded for each compound, and the empirical scoring cutoff was determined by looking at the distribution of scores for parent compounds with true identities ranking in the top 500 (Figure S5A). Ultimately, this cutoff represents a rough way of ruling out unlikely annotations given their corresponding MS/MS spectrum, and in total 861 putative metabolite annotations were removed based on this criterion (587 annotations without scores +274 with scores <100).
Assembly of a Drug and Metabolite CCS Database
A SQLite3 database was used to store experimental data, associated metadata, annotations, 3D structures, and computed molecular descriptors for all of the drugs and metabolites observed in this study. The overall database architecture is summarized in Figure S6. The database has separate tables for CCS measurement data and metadata (plate_N), MS2 spectra (plate_N_ms2), compound annotations (plate_N_id), 2D molecular descriptors (plate_N_mqn), 3D structures (plate_N_3d), and 3D molecular descriptors (plate_N_md3d). All of the experimental plates (seven in total) have their own set of corresponding tables for consistency with the organization of the experimental source data. The database was constructed in a stepwise, automated fashion using a series of Python build scripts developed in-house. Briefly, the database was first initialized with all of the empty tables, and then the measured data were added to the plate_N tables according to plate number. Next, compound annotations (names and SMILES structures) were added to the plate_N_id tables. Parent drug annotations were already known from the plate contents, but metabolite annotations were assigned via a combination of automated and manual processes as discussed above. Drift time-selected MS2 spectra were added to the plate_N_ms2 tables, with each entry in the measured data tables having a corresponding MS2 spectrum. Next, MQNs were computed using SMILES structures (see below for detail) from the annotation tables and added to the plate_N_mqn tables. 3D structures (in plain text format) were generated (see below for detail) and added to the plate_N_3d tables, and corresponding 3D molecular descriptors were computed for each structure (vide infra) and added to the plate_N_md3d tables. The plate_N, plate_N_ms2, and plate_N_id tables are all related by a unique (across all seven sets of plates) text identifier, dmim_id. All of the annotations in the plate_N_id tables have an additional unique integer identifier, ann_id, relating them to entries in the plate_N_mqn and plate_N_3d tables. The plate_N_3d tables have an additional unique integer identifier, str_id, relating their entries to the plate_N_md3d tables.
Generation of 3-Dimensional Structures for Ionized Drugs and Metabolites
3-Dimensional structures were computed from SMILES structures for experimentally observed ionized (protonated and Na+/K+ adducts) drug and metabolite species using a series of scripts developed in-house employing a combination of molecular mechanics and semiempirical methods. Briefly, initial 3D structures were generated by a Monte Carlo conformer search followed by steepest descent energy minimization using the MMFF94 force field in the OpenBabel38 software package. The initial 3D structures were then further optimized at the PM7 semiempirical theory level in Gaussian16.39 Finally, the optimized atom positions, masses, and partial charges were stored along with relevant metadata for the measured species. This process was repeated 3 times for each individual ion species to increase the chances that a minimum energy structure would be sampled in this nonextensive modeling protocol.
Generation of 3D structures for protonated species followed the same protocol, but with the inclusion of additional steps to account for multiple potential sites of protonation within a molecule. First, potential protomers were determined by presence of ionizable groups, and the SMILES structures were modified to reflect each protomer. 3D structure generation was performed using each of the protomer SMILES structures as described above, but with additional thermodynamic calculations specified in the semiempirical optimization step. After the 3D structures had been produced for all potential protomers, the structures having the lowest energy and highest partial charge located on the protonation site (if different from lowest energy structure) were selected and stored. The above protocol was repeated three times for each species, resulting in three to six structures for each protonated species.
The 3D structure generation protocol resulted in the production of three to six structures for each ionized species in an attempt to capture multiple energetically similar conformers; however, for most compounds, many or all of the produced structures were virtually the same. To avoid undue influence in predictive model training from such duplications, all structures for a given compound were subjected to RMSD filtering. Briefly, for each compound, a mass-weighted RMSD matrix was computed between all predicted structures, and only those differing by more than 0.01 Å were retained. The RMSD cutoff of 0.01 Å was determined empirically by computing the distribution of RMSD values for all structures in the database (Figure S7A), in addition to manual inspection of a handful of compound structures. All of the filtered 3D structures were added as a separate table to the drug and metabolite CCS database.
Multivariate Analysis of Drug and Metabolite CCS Database
PCA and PLS-RA are implemented in Scikit-Learn, a free and open-source machine learning library for Python (sklearn.decomposition.PCA and sklearn.cross_decomposition.PLSRegression, respectively).40 PCA and PLS-RA are dimensionality reduction techniques that work by determining successive orthogonal axes within a high-dimensional data set that contain maximal variance. PLS-RA differs from PCA in that the first axis is chosen such that it corresponds to the direction of maximal variance in an external target variable (in this case CCS), making it a targeted analysis.
Prediction of CCS Using Machine Learning
Prior to model training, the data were processed in a stepwise fashion. First, the data set was randomly (seeded for deterministic results) split into training and test sets in proportions of 80% and 20%, respectively, and the test set was held aside during model training. Rough stratification based on distribution of CCS was used during data set splitting to ensure comparability between the training and test sets. The training data were centered and scaled such that each feature would have a mean of 0 and unit variance in order to avoid undue emphasis of features on the basis of their magnitudes. A support vector regression (SVR) model with radial basis function kernel was used for CCS prediction (sklearn.svm.SVR). The model hyperparameters (C and gamma) were optimized using a grid search with 5-fold cross validation (sklearn.optimize.GridSearchCV) on the training data. The model trained using the optimal hyperparameters was then used to compute performance metrics (vide infra) from predictions made on the training and test data sets.
CCS Prediction Performance Metrics
A standard set of metrics was used to determine the bulk performance of CCS prediction using ML and other methods as described previously.27 Briefly, these include R2, mean and median absolute error (MAE and MDAE, respectively, Å2), mean and median relative error (MRE and MDRE, respectively,%), root mean squared error (RMSE, Å2), and cumulative error distribution at 1, 3, 5 and 10% levels (CE135A,%).
Calculation of PA/EHS CCS
Theoretical CCS values were calculated for all 3D structures in the drug and metabolite CCS database by the projection approximation (PA) and exact hard-sphere scattering (EHS) methods using MobCal.21,22
Calculation of Metabolite Compaction Factors
Gas-phase compaction factors of metabolites relative to parents were computed using the equation
as described previously.31 Briefly, since the CCS at a given mass is analogous to a gas-phase density, a change in mass (i.e., due to metabolic modification) is expected to produce a monotonic change in CCS, and that change is isotropic if the density does not change (C = 1). If C > 1, then the metabolite is denser than expected under isotropic growth, while C < 1 indicates the metabolite is less dense.
LC-IM-MS Analysis of Pooled Drug Metabolism Incubations
Four pooled samples were prepared from drug metabolism incubations containing different parent compounds (five parent compounds per pooled sample) taken from the first plate of the MicroSource Spectrum Discovery Collection. Pooled incubations (5 μL) were injected and separated using a Waters Acquity FTN UPLC coupled to a reversed-phase column (Thermo Hypersil GOLD, 1.9 μm, C18, 100 × 2.1 mm), eluting with a gradient of water with 0.1% formic acid and 2 mM ammonium formate (A) and acetonitrile (B) at 0.4 mL/min: 0.0–15.0 min, 85 → 10% A; 15.0–16.0 min, 10% A; 16.0–16.1 min, 10 → 85% A; 16.1–20.0 min, 85% A. TWIM-MS analysis was performed on a Waters Synapt G2-XS mass spectrometer (Waters Corp., Milford, MA) equipped with an ESI source and using nitrogen as the drift gas. ESI conditions were as follows: capillary, + 2.5 kV; sampling cone, 30 V; source temperature, 150 °C; desolvation temperature, 500 °C; cone gas, 50 L/h; and desolvation gas, 1000 L/h. Mass calibration was performed using sodium formate for the range of m/z 50–1200. IM separations were performed at a traveling wave velocity of 500 m/s and a height of 40 V. CCS was calibrated using a mixture of polyalanines and drug standards as described above. Automated peak picking was performed in DriftScope (Waters Corp., Milford, MA).
Code and Data Availability
All code for generating the database and prediction models and for data processing are available on GitHub (https://github.com/dylanhross/dmccs). Raw mass spectrometry data is available at MassIVE under MSV000088549 (doi:10.25345/C5CZ90).
Results and Discussion
High-Throughput Measurement of Drug and Drug Metabolite CCS
To obtain a large collection of drug metabolites, we first carried out high-throughput drug metabolism reactions in 384-well plates using human liver microsomes and S9 fraction on 2000 drug and drug-like compounds in the MicroSource Discovery Systems’ Spectrum Collection, containing 50% approved drugs, 30% natural products, and 20% bioactive compounds (Figure 1A). This compound collection has broad coverage of small molecule chemical space and contains drugs spanning a range of bioactivities (e.g., antibacterial, anti-inflammatory, antineoplastic, antihypertensive, analgesic, etc.), which was described in detail previously.16 Reactions catalyzed by the Phase-I enzymes, such as cytochromes P450 (CYPs), flavin-containing monooxygenases, and reductases, and Phase-II enzymes, such as glutathione S-transferases (GSTs) and UDP-glucuronosyltransferases (UGTs), were probed. The drug metabolism reactions were carried out with or without enzyme cofactors, such as NADPH (cofactor of CYPs), glutathione (GSH, cosubstrate of GSTs), UDP-glucuronic acid (UDPGA, cosubstrate of UGTs), and alamethicin41 (enabling access of substrates to UGTs). Plates incubated with HLM + S9, but in the absence of enzyme cofactors, served as controls. After the reactions and sample processing, we carried out rapid IM-MS analysis using a 30 mm reversed-phase column, resulting in just under 2 min per run (Figure 1A). Measuring the roughly 2000 compound collection in triplicate with or without enzyme cofactors/cosubstrates resulted in >8900 samples analyzed. This large and complex set of raw data was analyzed using a stepwise approach with a high degree of automation (Figure 1B), including extraction of arrival time distributions (ATDs), Gaussian fitting, CCS calibration, and calculation of CCS of observed ATD peaks. The CCS values of parent compounds were obtained by extracting the ATDs of the exact masses of various adducts. For metabolites, we first generated a theoretical list of potential metabolites using Biotransformer35 and then extracted the ATDs of these potential metabolites (see the Experimental Section for details). Only ATD peaks meeting the criteria of intensity >1000 and peak width between 0.06 and 1.77 ms (roughly 1–30 drift time bins) were retained. This approach ultimately led to the assembly of a large CCS database specific to drugs and drug metabolites with high reproducibility in replicate CCS values (0.39 and 0.15% mean and median RSD, respectively). Figure 2A summarizes the composition of the drug and metabolite CCS database. The database contained 6245 measured CCS values from 3286 different compounds, of which 1333 were from parent drugs (3675 CCS values) and 1953 were from metabolites (2570 CCS values). The measured CCS values corresponded to a number of ionization states commonly observed in positive mode ESI including [M + H]+ (1936), [M + Na]+ (1656), [M + K]+ (1235), [M + H – H2O]+ (1299), and [M]+ (119).
Figure 2.
(A) Composition of the assembled CCS database for drugs and drug metabolites (dmCCS). Metabolic modification abbreviations: GSH, glutathione conjugated metabolites; Glc, glucuronide metabolites; Ox, oxidative metabolites (e.g., −2H, +O, −Me). (B) PCA projections of the dmCCS database (color) from a PCA computed using the CCSbase database (gray), colored by CCS. (C) PCA projections of parent compounds (blue) and metabolites (red) from the dmCCS database from a PCA computed using the CCSbase database (gray). (D) CCS vs m/z of parent compounds (blue) and metabolites (red) from the dmCCS database overlaid on the CCSbase database (gray). Dotted lines represent individual power fits for parent (chartreuse) and metabolite (orange) data, and residual CCS from these fits are included below the main plot. (E) PCA projections of dmCCS database computed using MQNs as molecular descriptors, colored by CCS. (F) PCA projections of dmCCS database computed using MD3Ds as molecular descriptors, colored by CCS. (G) PCA projections of dmCCS database computed using the combination of MQNs and MD3Ds as molecular descriptors, colored by CCS. (H) Individual feature loadings for principal component 1 from PCA computed on dmCCS using MQNs as molecular descriptors. (I) Individual feature loadings for principal component 1 from PCA computed on dmCCS using MD3Ds as molecular descriptors. (J) Individual feature loadings for principal component 1 from PCA computed on dmCCS using the combination of MQNs and MD3Ds as molecular descriptors.
To validate the identity of the potential metabolites, we first carried out a thorough search of DrugBank36 for reported metabolites of known drugs in our collection and matched our observed metabolites with those previously reported. For those without reported metabolites, we matched the experimental MS/MS spectra obtained from post-IM fragmentation against an in-silico generated MS/MS spectra of potential metabolite structures using MetFrag,37 and ruled out low-scoring metabolite annotations. The validation process is discussed in greater detail in the Experimental Section. We evaluated the potential for multiple isobaric metabolites to be predicted for different metabolic modifications (see Figure S11) and found that this potential was much higher for oxygenation reactions than for dealkylated and conjugated metabolites. This analysis indicates that, in general, the dealkylated and conjugated metabolite identifications have a higher likelihood of being accurate (or at least being inconsequential with respect to CCS predictions) than the oxygenated metabolites. However, oxygenated metabolites undergo smaller MQN feature changes relative to the parent compounds when compared with more significant metabolic modifications, and thus, the impact of a potential mis-assignment on CCS prediction would be small. After this process, 4408 of the measured CCS values were retained with an annotation, corresponding to all parent drugs with a CCS value (1333 compounds and 3675 values) and 29.3% of metabolite CCS values (572 compounds and 744 values). Among these CCS values, roughly 2876 of the parent values and all of the metabolite values are new.16 Overall, the 3675 parent CCS values (83.4% of the database) are confidently assigned, and 396 CCS values (9.0%) correspond to only one possible isomer for the metabolites, which together represents 92.4% of the entire database. The entries with multiple potential isomers were further narrowed down using MetFrag as described above. The 799 parent CCS values observed previously displayed excellent agreement with the prior measurements, having mean and median errors of only 1.16 and 0.71%, respectively. The annotated metabolite CCS values represent a range of metabolic modifications, including glutathione adducts (22), glucuronide conjugates (200), and oxidative metabolites (166). The coverage of glutathione and glucuronide conjugates is particularly important for this work because such Phase-II metabolism introduces a large mass and structural modification to a parent drug.15,31 2D molecular descriptors, i.e., molecular quantum numbers (MQNs, 42 features, see Table S1),27,42 were generated for all annotated species as described previously and in the Supporting Information. Furthermore, novel 3D molecular descriptors (Table S2), including principal moments of inertia (PMI) and radial mass distributions (RMD) (eight features, see the Supporting Information), were generated to better capture the relationship between conformation and CCS during machine learning as discussed below. Briefly, we attempted to generate 3D structures for all annotated [M + H]+, [M + Na]+, and [M + K]+ species at a low level of theory (MMFF94 and PM7, see the Experimental Section), resulting in a total of 9813 modeled structures (4074, 3172, and 2567 for each ionized species, respectively). 3D molecular descriptors were generated from 3D structures using in-house developed Python scripts as described in the Supporting Information (Figure S6 and related text). In total, 7652 and 2161 3D structures with 3D molecular descriptors were generated for parent drugs and metabolites, respectively.
Characteristics of the Drug and Metabolite CCS Database
Principal components analysis (PCA) was used to probe the chemical characteristics (as captured by 2D or 3D molecular descriptors) that contribute to variance in the drug and metabolite CCS database. We had previously characterized a comprehensive collection of compounds from a diverse set of chemical classes using MQNs as features (which capture compositional and topological information about chemical structures),42 so we first computed a PCA using this comprehensive database (CCSbase) to serve as the chemical space background. We then projected the new drug and metabolite CCS database (dmCCS) into this PCA to examine the chemical space that dmCCS spans within the context of CCSbase. Parts B and C of Figure 2 show the PCA projections of compounds from dmCCS (color) overlaid over compounds from CCSbase (gray). In Figure 2B, it can be seen that CCS values of the compounds from dmCCS generally increase along the direction of PC2, indicating that the strongest sources of variance in CCSbase do not correspond with sources of variance in dmCCS that relate to CCS. Figure 2C shows where the parent compounds and metabolites from dmCCS group fall within the chemical space defined by CCSbase, which indicates that the dmCCS occupies a broad region corresponding roughly to “small molecules”. The metabolites occupy a subspace within the chemical space occupied by the parent compounds. Figure 2D shows where the parent compounds and metabolites from dmCCS (color) map into the IM-MS conformational space (i.e., CCS vs m/z), compared to the compounds from CCSbase (gray), with individual power fits for parent compounds and metabolites (dashed lines). Generally, the compounds from dmCCS occupy the low m/z region of this space and span a wide range of CCS values. Interestingly, the metabolites seem to occupy a slightly narrower CCS envelope with mostly similar average CCS values to those of the parent compounds. Even in the context of the large chemical space of CCSbase, the compounds from dmCCS represent considerable structural diversity. We also looked into the structural changes that were induced by groups of metabolic modifications (see Figure S9) and found that some modifications, such as Phase-II metabolites, led to similar shifts in CCS regardless of the parent compound, while other modifications, such as oxygenation and dealkylation, led to CCS changes that were highly dependent upon the structure of the parent compound.
Separate PCAs were computed on dmCCS using the 2D (MQN) and 3D (MD3D) molecular descriptors to determine how each set of descriptors reflected the chemical space covered by this database. Parts E and F of Figure 2 show the PCA projections from the 2D and 3D features, respectively. For both feature sets, PC1 correlates well with variation in CCS, indicating that among these compounds the primary sources of variance are related to CCS. PC1 and PC2 of the PCA computed on the 2D feature set captured 19.3% and 13.4% of the overall variance, respectively, compared to 58.8% and 18.0% for the 3D feature set, indicating that a high degree of variance orthogonal to CCS in the 2D feature set is not present in the 3D feature set. Indeed, the PCA computed on the 2D features required 24 components to capture 95% of the variance in the data set, in contrast to only 5 components needed for the 3D feature set. Figure 2H,I show the 2D and 3D feature loadings, respectively, for PC1 in each PCA, both of which correlate well with CCS. The strongest contributors to separation along the first principal component (Figure 2H) for the 2D features were counts of atoms (hac: heavy atoms, ao: acyclic oxygens, c: carbons), bonds (adb: acyclic double bonds, atb: acyclic triple bonds), and topological features (asv: acyclic monovalent nodes, ctv: cyclic trivalent nodes). All of the 3D features contributed similarly to the separation along the PC1 (Figure 2I), and interestingly, the second and third PMI had slightly larger contributions than the first. Together, these results demonstrate that both the 2D and 3D features capture the important characteristics of this set of compounds that relate to CCS, but the 3D features contain somewhat less extraneous information.
We next examined the degree to which the 2D and 3D feature sets (MQN and MD3D, respectively) offered complementary information by computing a PCA on dmCCS using a combination of both feature sets (COMB) (Figure 2G). The PCA projections overall appear quite similar to those from the 2D feature set alone. PC1 captured 23.6% of the total variance, while 27 total components were required to capture 95% of the variance in the data set. The top features contributing to separation along PC1 (Figure 2J) consist of a combination of those identified from the 2D and 3D feature sets.
We also performed a set of analogous analyses using partial least-squares regression analysis computed on the 2D, 3D, and combined feature sets with CCS as the target variable (Figure S1). The results from these analyses largely mirrored those discussed above, which is expected given the alignment of CCS with PC1 in all three PCAs.
Training Drug and Drug Metabolite-Specific CCS Prediction Models
The 2D and 3D feature sets were used to train individual ML models for CCS prediction on dmCCS. Despite the different sizes (42 features vs 8) and characteristics of the 2D and 3D feature sets, the MQN and MD3D predictive models achieved very similar performance in CCS prediction by multiple metrics, with robust performance between training and test set data (Figure 3). We next sought to test the degree to which the two feature sets provided orthogonal information by training a ML model on the combined 2D and 3D feature sets (COMB). Although the COMB model achieved significantly improved predictive performance relative to models trained on either feature set (Figure 3), there was a significant lapse in performance between the training and test set data, indicating model overfitting likely attributable to the presence of redundant and/or superfluous features.
Figure 3.

CCS prediction performance comparison for ML models trained on dmCCS using MQN, MD3D, a combination of MQN and MD3D (COMB), or a minimal feature set (MIN) as molecular descriptors.
To address potential overfitting in the COMB model, a set of feature ranking and successive feature removal trials including PLS-RA, gradient boosting regression (GBR), and a permutation feature importance function in Scikit-Learn (PER) were run in order to select a minimal feature set combining the most influential features from the 2D and 3D feature sets while avoiding overfitting by removing extraneous features (see the Supporting Information and Figure S2 for details). Molecular descriptors retained by at least two of the feature removal methods were kept as the minimal feature set (MIN), which consisted of only 11 descriptors from both the 2D and 3D feature sets: hac, c, asv, adb, ctv, hbam (H-bond acceptor sites), hbd (H-bond donor atoms), pmi1, pmi2, pmi3, and rmd02. A new ML model was trained using this feature set. Although there was still an appreciable degree of correlation between the features (Figure S3), the MIN model achieved an intermediate increase in performance relative to the models trained on the 2D or 3D features alone (Figure 3), and importantly, this performance was better maintained between the training and test set data.
The MIN model was also used to compare with fast theory-based CCS prediction methods (e.g., projection approximation, PA, and exact hard-sphere scattering, EHS),21 which again showed superior performance in terms of accuracy and precision (see Figure S4 and related text).
Metabolite annotations were based on predictions from BioTransformer35 and validated by scoring in MetFrag37 and as such are subject to the limitations of both tools. For example, BioTransformer may not cover all possible metabolic transformers, and the empirical score cutoff based on the parent compounds may not be equally applicable to the metabolites. As a result, there is a potential for misidentified metabolite species, which could reduce the accuracy of the CCS predictions if the structural characteristics of the misidentified species differ greatly from the true identities. As described above, we characterized the degree to which various metabolic modifications produce multiple possible annotations for single metabolites in our workflow and found that the oxidative metabolites displayed the greatest propensity for such uncertainty. In order to examine whether these metabolite annotations were increasing the uncertainty of our model’s CCS predictions, we retrained the MIN model using a data set with all oxidative metabolites removed and characterized the CCS prediction accuracy (Figure S13). We found that exclusion of the oxidative metabolites from the training data did not significantly change the accuracy of the model’s predictions; in fact, the accuracy decreased very slightly. This result indicates that to the extent that there are misidentified metabolites within this data set, their presence does not seem to significantly detract from the accuracy of this CCS prediction model. To increase transparency and allow researchers in the field to better utilize the experimental data, we have added a column for the metabolite data to specify whether potential multiple isomers are present based on BioTransformer.
Application of CCS Prediction to Compounds with Multimodal ATDs
Multimodal ATDs can arise from a number of circumstances, such as constitutional isomers (e.g., positional isomers of metabolites or protomers formed in the ESI process) and conformers.15,16,31 However, previous CCS prediction models based on 2D molecular descriptors generally do not allow the differentiation of such isomers or conformers. Inclusion of 3D features in CCS prediction could in theory capture such multimodal differences for given 3D structures, so we sought to evaluate some known examples of multimodal distributions using CCS prediction models trained with different feature sets.
The oxygenated (+O) metabolite of terfenadine displays a more compact conformation relative to the parent, likely attributable to the introduction of an intramolecular polar–polar interaction (Figure 4B).31Figure 4A compares the experimentally measured CCS values of terfenadine and its +O metabolites to values predicted using the different models discussed above. The CCS values predicted using the MQN and MD3D models are lower than the experimental values, and the + O metabolite has a larger CCS than the parent, indicating that these feature sets do not adequately capture the structural differences between these compounds. The MIN model produced the closest predictions and, importantly, reproduced the decreased CCS of the metabolite relative to the parent with a compaction factor (see the Experimental Section) of 1.030 (compared to 1.042 in the experimental values).
Figure 4.
(A) Comparison of measured (hatched) and predicted (solid) CCS for terfenadine and its +O metabolite. Each plot presents CCS values predicted using ML models trained on dmCCS using different feature sets. (B) Representative structures of terfenadine and its +O metabolite demonstrating the gas-phase compaction of metabolite relative to the parent. (C) Comparison of measured (hatched) and predicted (solid) CCS for two protomers of cefpodoxime proxetil. Each plot presents CCS values predicted using ML models trained on dmCCS using different feature sets. (D) Representative structures of the two protomers of cefpodoxime proxetil. (E) Comparison of measured (dashed lines) and predicted (solid bars) CCS for the positional isomers of quercetin glucuronide. Each plot presents CCS values predicted using ML models trained on dmCCS using different feature sets. (F) Representative structures of the positional isomers of quercetin glucuronide.
Cefpodoxime proxetil is a β-lactam antibiotic that has previously been shown to form two protomers in ESI with distinct CCS values (Figure 4D).16 As seen in Figure 4C, the MQN model was unable to distinguish between the different protomers (likely due to them being constitutional isomers), and the predicted CCS values were significantly smaller than the experimental values. The MD3D model produced predictions that differed between the two protomers, but their rank order was reversed relative to the experimental values. The MIN model produced CCS predictions close to the experimental values while preserving the experimentally observed rank-order.
Quercetin is a flavonoid compound with multiple hydroxyl (−OH) groups available for glucuronidation (Figure 4F).43 Glucuronidation of quercetin has previously been observed to produce a bimodal CCS distribution, likely attributable to glucuronidation at different positions.31 Again, the MQN model failed to capture any CCS differences between the positional isomers, while both MD3D and MIN models were able to distinguish between the different positional isomers and the assignment of isomers to the two experimental values was largely in agreement with previous results (Figure 4E).31 Prediction results from both MD3D and MIN models suggest that the larger CCS values likely have contributions from the isomers at the 3-, 5-, and/or 7-positions, while the smaller CCS value likely arises from 3′- and/or 4′-isomers, which is mostly consistent with previous results using high-level computation methods.31
We also examined CCS prediction performance using existing CCS prediction models trained using 2D descriptors (AllCCS, CCSbase),27,29 and neither were able to replicate the multimodal CCS associated with the protomers of cefpodoxime proxetil or the positional isomers of quercetin glucuronide (see Figure S8).
The 2D MQN model and the 2D/3D MIN model are now available at CCSbase.net. Predictions can be made easily with an input of a SMILES structure of an ion and a 3D structure in .mol2 format.
Demonstration of Metabolite Identification Using Predicted CCS
In order to demonstrate the benefit of CCS prediction for identification of metabolites from complex samples, we analyzed pooled drug metabolism incubations from a total of 20 parent compounds using LC-IM-MS (see the Experimental Section). Putative candidate metabolites were generated using BioTransformer as described above, and CCS values were predicted using the MQN CCS prediction model, yielding a metabolite target list of 2114 species. The peak-picked LC-IM-MS data was annotated from this target list, with matching based on either m/z or m/z and predicted CCS (m/z tolerance: 0.05 Da, CCS tolerance: 3%). Out of a total of 24358 cofactor-dependent LC-IM-MS peaks, 9949 peaks were annotated using only m/z, while 4815 were annotated using m/z and predicted CCS, a reduction of 5134 peaks. Because of the untargeted nature of the experiment and the complexity of the samples, there were many cases in which multiple annotations were assigned to a single peak or the same annotation was assigned to multiple peaks (see Figure S10). The mean number of annotations assigned to a single peak was reduced from 5.8 to 4.9 with the inclusion of predicted CCS, and the mean number of peaks with the same annotation was reduced from 3.2 to 2.4 with the inclusion of predicted CCS. Taken together, these results demonstrate that inclusion of predicted CCS decreases the total number of annotated peaks and increases the confidence of putative annotations by reducing both the number of annotations per peak and peaks per annotation. The reduction in number and increase in confidence of putative annotations saves time in downstream analysis and interpretation of complex drug metabolism studies.
Conclusion
This work addresses several major gaps in applying IM-MS to drug metabolite identification and building ML-based CCS prediction models. First, there is a lack of large-scale experimental CCS database for drug metabolites, which was accomplished here with a high-throughput in vitro drug metabolite generation system, followed by high-throughput IM-MS analysis and automated data processing. This large database provided the basis for building ML-based CCS prediction models for drug metabolites. Second, previous ML approaches for the prediction of CCS values rely on 2D molecular descriptors,12,18,19,24−27 which cannot differentiate protomers, positional isomers, and conformers. By incorporating novel 3D molecular descriptors, such as PMIs and RMDs, our ML model using minimum combined 2D and 3D features successfully overcame these limitations. Third, our approach represents a hybridization of data- and theory-driven CCS prediction, which showed superior performance than fast theory-based computation approaches in terms of accuracy and precision. Although our ML-based model cannot replace high-level theoretical CCS calculation as small differences captured by the high-level computation methods may not be readily captured by the low-level methods used to generate the training 3D structures in this work, the time-efficiency and accuracy of our approach (a few seconds vs hours using high-level computation) makes it easily integrated into drug development processes. To summarize, the ML approach reported in this work enables high-accuracy and high-throughput generation of CCS values for drugs and drug metabolites with sufficient precision to differentiate isomers and conformers. The dmCCS prediction models are available to the public at CCSbase.
Acknowledgments
This work was supported by the University of Washington (UW) CoMotion Innovation Gap Fund to L.X. We thank Prof. Matthew F. Bush at UW Chemistry for helpful discussions on CCS calculation using MobCal.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.2c00111.
Additional experimental details and results; additional figures related to characterization of the CCS database, training of the machine learning models, feature selection trials, description of 3D molecular descriptors, comparison of the performance of the dmCCS prediction model with PA/EHS, AllCCS, and CCSbase, application of drug metabolite identification using a pooled drug metabolism incubation, characterization of Mass and CCS Shifts for Metabolites, analysis of isobaric metabolites predicted by BioTransformer, performance of MIN model using all or reduced metabolite data sets; supplemental tables listing all MQN and 3D molecular descriptors (PDF)
entire experimental database as an Excel file (XLSX)
Author Present Address
# Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352
Author Contributions
D.H.R. and L.X. designed the study; D.H.R. and R.P.S. developed in vitro drug metabolite generation methodology; D.H.R. developed and performed IM-MS analysis; D.H.R. designed and conducted semiautomated data analysis; D.H.R. performed ML experiments with contributions from A.K.; D.H.R. and L.X. prepared the manuscript with contributions from R.P.S. All authors approved the submitted version of this manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Fura A.; Shu Y. Z.; Zhu M.; Hanson R. L.; Roongta V.; Humphreys W. G. Discovering drugs through biological transformation: role of pharmacologically active metabolites in drug discovery. J. Med. Chem. 2004, 47, 4339–4351. 10.1021/jm040066v. [DOI] [PubMed] [Google Scholar]
- Park B. K.; Kitteringham N. R.; Maggs J. L.; Pirmohamed M.; Williams D. P. The role of metabolic activation in drug-induced hepatotoxicity. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 177–202. 10.1146/annurev.pharmtox.45.120403.100058. [DOI] [PubMed] [Google Scholar]
- Prakash C.; Shaffer C. L.; Nedderman A. Analytical strategies for identifying drug metabolites. Mass Spectrom. Rev. 2007, 26, 340–369. 10.1002/mas.20128. [DOI] [PubMed] [Google Scholar]
- Zhu M.; Zhang H.; Humphreys W. G. Drug metabolite profiling and identification by high-resolution mass spectrometry. J. Biol. Chem. 2011, 286, 25419–25425. 10.1074/jbc.R110.200055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B.; Zhu M. Applications of mass spectrometry in drug metabolism: 50 years of progress. Drug Metab. Rev. 2015, 47, 71–87. 10.3109/03602532.2014.1001029. [DOI] [PubMed] [Google Scholar]
- Clemmer D. E.; Hudgins R. R.; Jarrold M. F. Naked Protein Conformations: Cytochrome c in the Gas Phase. J. Am. Chem. Soc. 1995, 117, 10141–10142. 10.1021/ja00145a037. [DOI] [Google Scholar]
- von Helden G.; Wyttenbach T.; Bowers M. T. Conformation of macromolecules in the gas phase: use of matrix-assisted laser desorption methods in ion chromatography. Science 1995, 267, 1483–1485. 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- McLean J. A.; Ruotolo B. T.; Gillig K. J.; Russell D. H. Ion mobility-mass spectrometry: a new paradigm for proteomics. Int. J. Mass Spectrom. 2005, 240, 301–315. 10.1016/j.ijms.2004.10.003. [DOI] [Google Scholar]
- Kanu A. B.; Dwivedi P.; Tam M.; Matz L.; Hill H. H. Jr. Ion mobility-mass spectrometry. J. Mass Spectrom. 2008, 43, 1–22. 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- Fenn L. S.; Kliman M.; Mahsut A.; Zhao S. R.; McLean J. A. Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal. Bioanal. Chem. 2009, 394, 235–244. 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines K. M.; May J. C.; McLean J. A.; Xu L. Evaluation of Collision Cross Section Calibrants for Structural Analysis of Lipids by Traveling Wave Ion Mobility-Mass Spectrometry. Anal. Chem. 2016, 88, 7329–7336. 10.1021/acs.analchem.6b01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Shen X.; Tu J.; Zhu Z. J. Large-Scale Prediction of Collision Cross-Section Values for Metabolites in Ion Mobility-Mass Spectrometry. Anal. Chem. 2016, 88, 11084–11091. 10.1021/acs.analchem.6b03091. [DOI] [PubMed] [Google Scholar]
- Stow S. M.; Causon T. J.; Zheng X.; Kurulugama R. T.; Mairinger T.; May J. C.; Rennie E. E.; Baker E. S.; Smith R. D.; McLean J. A.; Hann S.; Fjeldsted J. C. An Interlaboratory Evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section Measurements. Anal. Chem. 2017, 89, 9048–9055. 10.1021/acs.analchem.7b01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Mesa M.; D’Atri V.; Barknowitz G.; Fanuel M.; Pezzatti J.; Dreolin N.; Ropartz D.; Monteau F.; Vigneau E.; Rudaz S.; Stead S.; Rogniaux H.; Guillarme D.; Dervilly G.; Le Bizec B. Interlaboratory and Interplatform Study of Steroids Collision Cross Section by Traveling Wave Ion Mobility Spectrometry. Anal. Chem. 2020, 92, 5013–5022. 10.1021/acs.analchem.9b05247. [DOI] [PubMed] [Google Scholar]
- Ross D. H.; Xu L. Determination of drugs and drug metabolites by ion mobility-mass spectrometry: A review. Anal. Chim. Acta 2021, 1154, 338270. 10.1016/j.aca.2021.338270. [DOI] [PubMed] [Google Scholar]
- Hines K. M.; Ross D. H.; Davidson K. L.; Bush M. F.; Xu L. Large-Scale Structural Characterization of Drug and Drug-Like Compounds by High-Throughput Ion Mobility-Mass Spectrometry. Anal. Chem. 2017, 89, 9023–9030. 10.1021/acs.analchem.7b01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejada-Casado C.; Hernandez-Mesa M.; Monteau F.; Lara F. J.; Olmo-Iruela M. D.; Garcia-Campana A. M.; Le Bizec B.; Dervilly-Pinel G. Collision cross section (CCS) as a complementary parameter to characterize human and veterinary drugs. Anal. Chim. Acta 2018, 1043, 52–63. 10.1016/j.aca.2018.09.065. [DOI] [PubMed] [Google Scholar]
- Bijlsma L.; Bade R.; Celma A.; Mullin L.; Cleland G.; Stead S.; Hernandez F.; Sancho J. V. Prediction of Collision Cross-Section Values for Small Molecules: Application to Pesticide Residue Analysis. Anal. Chem. 2017, 89, 6583–6589. 10.1021/acs.analchem.7b00741. [DOI] [PubMed] [Google Scholar]
- Mollerup C. B.; Mardal M.; Dalsgaard P. W.; Linnet K.; Barron L. P. Prediction of collision cross section and retention time for broad scope screening in gradient reversed-phase liquid chromatography-ion mobility-high resolution accurate mass spectrometry. J. Chromatogr., A 2018, 1542, 82–88. 10.1016/j.chroma.2018.02.025. [DOI] [PubMed] [Google Scholar]
- Mesleh M. F.; Hunter J. M.; Shvartsburg A. A.; Schatz G. C.; Jarrold M. F. Structural information from ion mobility measurements: Effects of the long-range potential. J. Phys. Chem-Us 1996, 100, 16082–16086. 10.1021/jp961623v. [DOI] [Google Scholar]
- Shvartsburg A. A.; Jarrold M. F. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem. Phys. Lett. 1996, 261, 86–91. 10.1016/0009-2614(96)00941-4. [DOI] [Google Scholar]
- Campuzano I.; Bush M. F.; Robinson C. V.; Beaumont C.; Richardson K.; Kim H.; Kim H. I. Structural characterization of drug-like compounds by ion mobility mass spectrometry: comparison of theoretical and experimentally derived nitrogen collision cross sections. Anal. Chem. 2012, 84, 1026–1033. 10.1021/ac202625t. [DOI] [PubMed] [Google Scholar]
- Colby S. M.; Thomas D. G.; Nunez J. R.; Baxter D. J.; Glaesemann K. R.; Brown J. M.; Pirrung M. A.; Govind N.; Teeguarden J. G.; Metz T. O.; Renslow R. S. ISiCLE: A Quantum Chemistry Pipeline for Establishing in Silico Collision Cross Section Libraries. Anal. Chem. 2019, 91, 4346–4356. 10.1021/acs.analchem.8b04567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Tu J.; Xiong X.; Shen X.; Zhu Z. J. LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision To Support Ion Mobility-Mass Spectrometry-Based Lipidomics. Anal. Chem. 2017, 89, 9559–9566. 10.1021/acs.analchem.7b02625. [DOI] [PubMed] [Google Scholar]
- Soper-Hopper M. T.; Petrov A. S.; Howard J. N.; Yu S. S.; Forsythe J. G.; Grover M. A.; Fernandez F. M. Collision cross section predictions using 2-dimensional molecular descriptors. Chem. Commun. (Camb) 2017, 53, 7624–7627. 10.1039/C7CC04257D. [DOI] [PubMed] [Google Scholar]
- Plante P. L.; Francovic-Fontaine E.; May J. C.; McLean J. A.; Baker E. S.; Laviolette F.; Marchand M.; Corbeil J. Predicting Ion Mobility Collision Cross-Sections Using a Deep Neural Network: DeepCCS. Anal. Chem. 2019, 91, 5191–5199. 10.1021/acs.analchem.8b05821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. H.; Cho J. H.; Xu L. Breaking Down Structural Diversity for Comprehensive Prediction of Ion-Neutral Collision Cross Sections. Anal. Chem. 2020, 92, 4548–4557. 10.1021/acs.analchem.9b05772. [DOI] [PubMed] [Google Scholar]
- Tsugawa H.; Ikeda K.; Takahashi M.; Satoh A.; Mori Y.; Uchino H.; Okahashi N.; Yamada Y.; Tada I.; Bonini P.; Higashi Y.; Okazaki Y.; Zhou Z.; Zhu Z.-J.; Koelmel J.; Cajka T.; Fiehn O.; Saito K.; Arita M.; Arita M. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159. 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Luo M.; Chen X.; Yin Y.; Xiong X.; Wang R.; Zhu Z. J. Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nat. Commun. 2020, 11, 4334. 10.1038/s41467-020-18171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper-Hopper M. T.; Vandegrift J.; Baker E. S.; Fernández F. M. Metabolite collision cross section prediction without energy-minimized structures. Analyst 2020, 145, 5414–5418. 10.1039/D0AN00198H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. H.; Seguin R. P.; Xu L. Characterization of the Impact of Drug Metabolism on the Gas-Phase Structures of Drugs Using Ion Mobility-Mass Spectrometry. Anal. Chem. 2019, 91, 14498–14507. 10.1021/acs.analchem.9b03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruotolo B. T.; Benesch J. L.; Sandercock A. M.; Hyung S. J.; Robinson C. V. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008, 3, 1139–1152. 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- Forsythe J. G.; Petrov A. S.; Walker C. A.; Allen S. J.; Pellissier J. S.; Bush M. F.; Hud N. V.; Fernandez F. M. Collision cross section calibrants for negative ion mode traveling wave ion mobility-mass spectrometry. Analyst 2015, 140, 6853–6861. 10.1039/C5AN00946D. [DOI] [PubMed] [Google Scholar]
- Gabelica V.; Shvartsburg A. A.; Afonso C.; Barran P.; Benesch J. L. P.; Bleiholder C.; Bowers M. T.; Bilbao A.; Bush M. F.; Campbell J. L.; Campuzano I. D. G.; Causon T.; Clowers B. H.; Creaser C. S.; De Pauw E.; Far J.; Fernandez-Lima F.; Fjeldsted J. C.; Giles K.; Groessl M.; Hogan C. J. Jr.; Hann S.; Kim H. I.; Kurulugama R. T.; May J. C.; McLean J. A.; Pagel K.; Richardson K.; Ridgeway M. E.; Rosu F.; Sobott F.; Thalassinos K.; Valentine S. J.; Wyttenbach T. Recommendations for reporting ion mobility Mass Spectrometry measurements. Mass Spectrom. Rev. 2019, 38, 291–320. 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou-Feunang Y.; Fiamoncini J.; Gil-de-la-Fuente A.; Greiner R.; Manach C.; Wishart D. S. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J. Cheminform 2019, 11, 2. 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Knox C.; Guo A. C.; Shrivastava S.; Hassanali M.; Stothard P.; Chang Z.; Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–672. 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttkies C.; Schymanski E. L.; Wolf S.; Hollender J.; Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform 2016, 8, 3. 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: An open chemical toolbox. J. Cheminform 2011, 3, 33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A. V.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams D. F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery Jr J. A.; Peralta J. E.; Ogliaro F.; Bearpark M. J.; Heyd J. J.; Brothers E. N.; Kudin K. N.; Staroverov V. N.; Keith T. A.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A. P.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 16, Rev. C.01; Gaussian, 2016. [Google Scholar]
- Pedregosa F.; Varoquaux G.; Gramfort A.; Michel V.; Thirion B.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Dubourg V.; Vanderplas J.; Passos A.; Cournapeau D.; Brucher M.; Perrot M.; Duchesnay E. Scikit-learn: Machine Learning in Python. J. Mach Learn Res. 2011, 12, 2825–2830. [Google Scholar]
- Walsky R. L.; Bauman J. N.; Bourcier K.; Giddens G.; Lapham K.; Negahban A.; Ryder T. F.; Obach R. S.; Hyland R.; Goosen T. C. Optimized assays for human UDP-glucuronosyltransferase (UGT) activities: altered alamethicin concentration and utility to screen for UGT inhibitors. Drug Metab. Dispos. 2012, 40, 1051–1065. 10.1124/dmd.111.043117. [DOI] [PubMed] [Google Scholar]
- Nguyen K. T.; Blum L. C.; van Deursen R.; Reymond J. L. Classification of organic molecules by molecular quantum numbers. ChemMedChem. 2009, 4, 1803–1805. 10.1002/cmdc.200900317. [DOI] [PubMed] [Google Scholar]
- Boersma M. G.; van der Woude H.; Bogaards J.; Boeren S.; Vervoort J.; Cnubben N. H.; van Iersel M. L.; van Bladeren P. J.; Rietjens I. M. Regioselectivity of phase II metabolism of luteolin and quercetin by UDP-glucuronosyl transferases. Chem. Res. Toxicol. 2002, 15, 662–670. 10.1021/tx0101705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code for generating the database and prediction models and for data processing are available on GitHub (https://github.com/dylanhross/dmccs). Raw mass spectrometry data is available at MassIVE under MSV000088549 (doi:10.25345/C5CZ90).