Abstract
Inflammation and dysfunction of the extrahepatic biliary tree are common causes of human pathology, including gallstones and cholangiocarcinoma. Despite this, we know little about the local regulation of biliary inflammation. Tuft cells, rare sensory epithelial cells, are particularly prevalent in the mucosa of the gallbladder and extrahepatic bile ducts. Here, we show biliary tuft cells express a core genetic tuft cell program in addition to a tissue-specific gene signature, and, in contrast to small intestinal tuft cells, decreased postnatally, coincident with maturation of bile acid production. Manipulation of enterohepatic bile acid recirculation revealed that tuft cell abundance is negatively regulated by bile acids, including in a model of obstructive cholestasis in which inflammatory infiltration of the biliary tree correlated with loss of tuft cells. Unexpectedly, tuft cell-deficient mice spontaneously displayed an increased gallbladder epithelial inflammatory gene signature accompanied by neutrophil infiltration that was modulated by the microbiome. We propose that biliary tuft cells function as bile-acid sensitive negative regulators of inflammation in biliary tissues, and serve to limit inflammation under homeostatic conditions.
One Sentence Summary:
Using bulk and single-cell RNA sequencing paired with novel lineage-tracking and -ablating tools, we describe the phenotype and bile-sensitive nature of biliary tuft cells, uncovering a novel immunomodulatory role for these cells in regulating the inflammatory tone of the extrahepatic biliary tree.
INTRODUCTION
Bile acids (BAs) are required for uptake of fats/fat-soluble vitamins, and serve as signaling molecules (1). Regulated excretion and concentration of bile is enabled by the gallbladder (GB) and extrahepatic bile ducts (EHBDs), a key physiologic feature in mammals with diurnal cycles of fasting and feeding (2, 3). Chronic inflammation of the biliary tree promotes liver and extrahepatic biliary tissue damage (4–6), and inflammatory/obstructive biliary disease is a leading cause of liver transplant (7). Despite this, biliary inflammation is poorly characterized. Understanding epithelial – immune interactions in the extrahepatic biliary tree could provide important insights into common biliary pathologies.
Tuft cells are rare chemosensory epithelial cells found in many tissues (8) including GB/EHBDs (9–14). Tuft cells are best characterized in the mouse small intestine, where they relay luminal signals from intestinal helminths and protists to activate lamina propria Group 2 innate lymphoid cells (ILC2s), promoting differentiation of goblet cells and tuft cells themselves and adaptation of the small intestine (10, 15–18). In other organs, tuft cells have been shown to play roles in response to allergens and bacteria (19–21) and induction of innate-like lymphocytes (22–24). GB/EHBD tuft cells express canonical tuft cell markers, including transient receptor potential cation channel subfamily M member 5 (TRPM5), interleukin (IL)-25, doublecortin-like kinase 1 (DCLK1) and enzymes supporting eicosanoid biosynthesis (8, 16), but their physiologic role remains unknown.
We set out to characterize the tuft cell compartment of the extrahepatic biliary tree. Using bulk and single-cell RNA sequencing (scRNA-seq), together with flow cytometry and novel lineage-tracking and -ablating tools, we describe the BA-sensitive nature of biliary tuft cells and uncover an unanticipated role for these cells in modulating the microbiome-dependent infiltration of activated neutrophils and biliary inflammatory tone.
RESULTS
Biliary tuft cells express tuft cell- and tissue-specific gene signatures
We first optimized methods in the mouse for isolation of viable epithelial, immune and stromal cells from digested GB/EHBDs (Fig S1A). Using IL-25 reporter mice (Flare25) to identify tuft cells by flow cytometry (10), we found that tuft cells represented a greater percentage of epithelial cells in the biliary tree compared to small intestine (Fig 1A,B). As previously noted (9, 14), tuft cells were present in both the GB and EHBDs by imaging of DCLK1 (8) (Fig S1B) and Flare25 (Fig 1C), but absent from intrahepatic bile ducts (Fig S1C).
Fig. 1. Biliary tuft cells express tuft cell-specific and tissue-specific gene signature and are not dependent on type 2 cytokines.
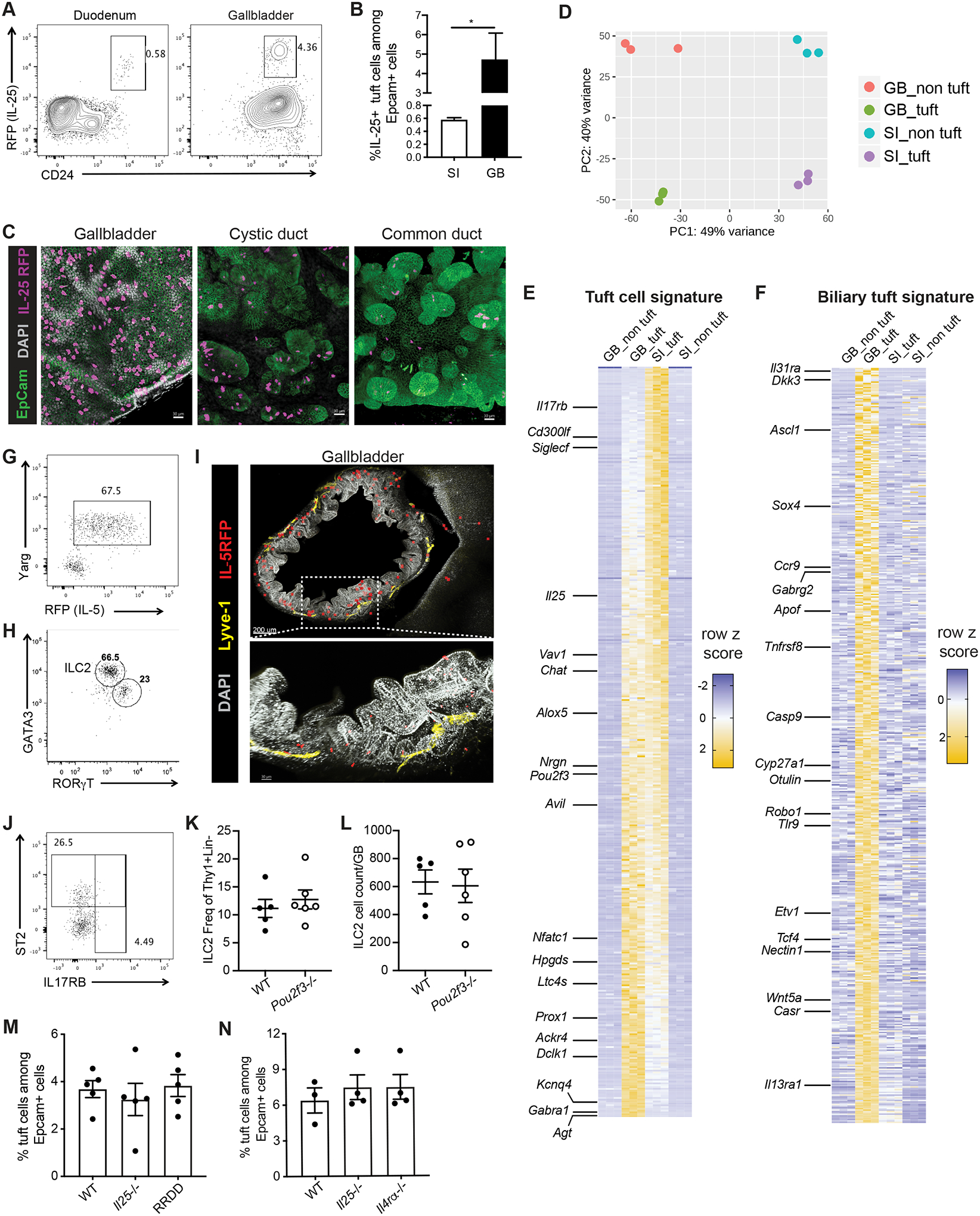
A,B) Epithelial cells from duodenum (SI) or gallbladder (GB) of IL-25 reporter (Flare25) mice were examined by flow cytometry. A) Representative flow plot. Previously gated on live singlets, FSC-A × SSC-A, EpCam+CD45-. B) IL-25+ cells as frequency of EpCam+ epithelial cells from SI and GB, 3 mice, *p<.05 by two-tailed paired T test. Representative of >3 experiments. C). Whole mount confocal imaging of Flare25 mouse GB, cystic duct, and common bile duct. D) Tuft and non-tuft epithelial cells were sorted from duodenum or GB of Flare25 mice and analyzed by RNA sequencing. Unsupervised PCA of top 500 genes by variance: biliary tuft (GB_tuft), biliary non-tuft (GB_non tuft), small intestinal tuft (SI_tuft), and small intestinal non-tuft (SI_non tuft). E,F) Row z score for tuft cell-specific transcripts (E) and (F) biliary tuft cell-specific transcripts. G,H) Representative flow plots. ILC2s identified in total GB/EHBD digests from Arginase1 (YARG) and IL-5 (Red5) reporter mice (G) and by transcription factor staining (H). Previously gated on live singlets, FSC-A × SSC-A, CD45+lineage-Thy1+,TCR-NKp46-NK1.1-. I) Imaging of GB and liver from RosaAi14-RFP × Red5 mice, stained for RFP (ILC2s, red), EpCam (green), LYVE-1 (yellow), and DAPI (gray). J) As in (G,H), staining for ST2 and IL17RB on ILC2s in total GB/EHBD digest. Previously gated on live singlets, FSC-A × SSC-A, CD45+lineage-Thy1+,TCR-NKp46-NK1.1-.YARG+. K,L) Relative frequency (K) and count (L) of ILC2s per GB/EHBD from WT and Pou2f3−/− YARG/Red5 reporter mice, as assessed by flow cytometry. M,N) Frequency of tuft cells among biliary epithelial cells from Il25−/−, ILC2 deficient (RRDD), and Il4Rα−/− mice determined by flow cytometry for DCLK1.
To assess similarity of biliary and small intestinal tuft cells, we sorted biliary and duodenal tuft cells from Flare25 mice for bulk RNA sequencing. PCA analysis indicated that cells clustered by tissue as well as tuft cell identity (Fig 1D). Analysis of the top 1000 genes by variance across all samples (Fig S1D, Table S1) revealed that biliary and small intestinal tuft cells share a core gene program, including transcripts for Il25, Dclk1, Chat, Alox5 and Pou2f3 (Fig 1E) (8). Biliary tuft cells expressed a unique signature, distinct from duodenal tuft cells and non-tuft epithelial cells (Fig 1F). Gene ontology analysis showed biliary tuft cell-associated transcripts were enriched in genes related to axon guidance and neural development (e.g., Robo1, Ascl1, Sema3a), immune-related genes (e.g., Tlr9, Il31ra, Il13ra1), cholesterol metabolism (e.g., Cyp27a1, Apof), and WNT signaling (e.g.: Wnt5a, Dkk3) (Supp Fig 1E).
Biliary tuft cell and ILC2 abundance are regulated independently
Small intestinal tuft cells make IL-25 and cysteinyl leukotrienes which drive lamina propria ILC2 function (10, 25, 26). We identified biliary resident ILC2s by flow cytometry using IL-5 (Red5; IL-5tdtomato-Cre (6)) and Arginase-1 (Yarg (27)) reporter mice (Fig 1G, Fig S1F), and by staining for the transcription factor GATA3 (Fig 1H). Intravenous anti-CD45 labelling confirmed tissue residency of CD45+ immune cells isolated from GB/EHBDs (Fig S1G). We corroborated biliary ILC2 identity by flow cytometry analysis of Rag1‒/‒ mice (Fig S1H) and by imaging of IL-5 lineage tracker mice (IL-5tdtomato-Cre; Rosa26RFP-Ai14) (Fig 1I) (28). IL-5+ lymphocytes were observed in the GB and liver, while tuft cells were exclusive to the extrahepatic epithelium (Supp Fig S1I). In contrast to small intestine ILC2s, biliary ILC2s expressed ST2, a component of the IL-33 receptor, but not IL17RB, the IL-25 receptor (25) (Fig 1J). Abundance of biliary ILC2s was comparable in in Pou2f3−/− mice, which lack tuft cells (Fig 1K,L) (29). Consistent with this, biliary tuft cell abundance was not significantly different in mice lacking IL-25, ILC2s, or IL-4Rα compared to controls (Fig 1M,N), in contrast to small intestinal tuft cells, which are reduced in these strains (10). Together, these data suggest that factors other than type 2 cytokine signals regulate biliary tuft cells.
Biliary tuft cells are long-lived and replenish slowly in adult tissue
Biliary epithelial cells are considered quiescent, with proliferation activated by injury (30, 31), but the longevity of biliary tuft cells has not been examined. We observed strong concordance between DCLK1 and IL-25/Siglec F expression among biliary tuft cells and loss of DCLK1 staining in Pou2f3−/− mice (Fig 2A). This indicated that DCLK1 is a faithful tuft cell marker in the biliary tree, allowing us to use Dclk1ERT2/+;R26YFPDclk1ERT2/+;R26YFP (32) mice for tuft cell labelling. In tamoxifen-treated Dclk1ERT2/+ ;R26YFP mice we observed robust labeling and minimal decay over six months, suggesting biliary tuft cells are long-lived (Fig 2B).
Fig. 2. Biliary tuft cells exhibit developmental regulation.
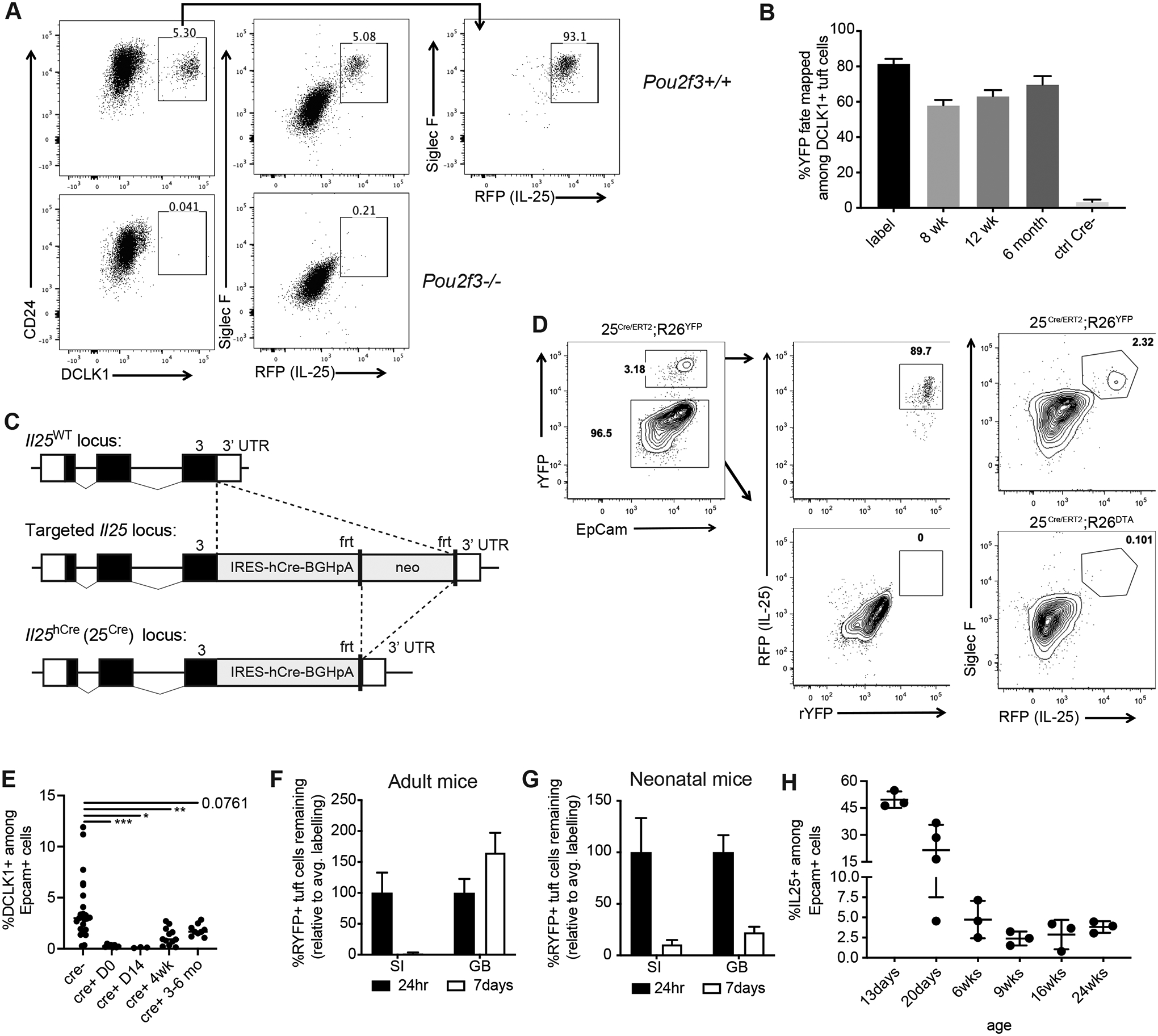
A) Total GB/EHBD digests from Flare25 reporter WT or Pou2f3−/− mice examined by flow cytometry for tuft cell surface markers (CD24 and Siglec F), expression of DCLK1 (intracellular staining), and endogenous RFP (IL-25). Previously gated on live singlets, FSC-A × SSC-A, EpCam+. B) Dclk1ERT2/+ ;R26YFP mice received two wks tamoxifen. Percent YFP expression among DCLK1+ epithelial cells was examined in total biliary GB/EHBD digests by flow cytometry after tamoxifen removal (label) and for indicated chase periods. C) Targeting strategy for IL-25 humanized cre mice. D) Co-expression of IL-25 reporter and 25cre-driven YFP expression in 25Cre/ERT2;R26YFP/+ and loss of RFP+ tuft cells in 25Cre/ERT2;R26DTA/+ mice were determined by flow cytometry. Representative flow plots. E) Tuft cell frequency in biliary epithelial prep from DT-injected 25Cre/+;R26iDTR/iDTR mice analyzed one day after DT (D0) and for indicated chase periods by intracellular staining for DCLK1. Significance determined by one-way ANOVA, *p<.05, **p<.01, ***p<.001. F,G) 25ERT/ERT2;R26YFP/YFP adult (G) or neonatal (p10-p12, H) mice were injected with tamoxifen; YFP expression among IL-25+ tuft cells was quantified in biliary epithelial prep by flow cytometry 24 hr and seven after the second injection. Data shown as percentage loss of YFP labelling relative to average 24 hr YFP expression in IL-25+ tuft cells. H). IL-25+ tuft cells in biliary epithelial prep were quantified by flow cytometry from Flare25 mice of the indicated ages.
To assess the recovery capacity of biliary tuft cells, we developed two Il25 locus knock-in mouse strains, one expressing humanized Cre recombinase (“25Cre”, Fig 2C) and the other expressing tdTomato-2A-CreERT2 (“25ERT”, Fig S2A). Both retain Il25 expression, with the latter also serving as an IL-25 reporter. To validate 25Cre specificity, we generated homozygous 25ERT2/ERT2;R26YFP/YFP mice and intercrossed this strain with 25Cre/Cre mice (25Cre/ERT2;R26YFP/+), generating tuft cells that co-expressed the tdTomato IL-25 reporter and were lineage traced with YFP under control of the 25Cre driver (Fig S2B). The majority of IL-25+ tuft cells co-expressed YFP, while IL-25- biliary epithelial cells were YFP-negative, indicating good efficacy and specificity (Fig 2D). Biliary tuft cells were ablated when 25Cre/ERT2 mice were intercrossed with ROSA26-diphtheria toxin A mice (25Cre/ERT2; R26DTA). We crossed 25Cre/+ mice to R26 inducible diphtheria toxin (DT) receptor mice (25Cre/+;R26iDTR/iDTR) to assess recovery of biliary tuft cells after inducible DT-mediated ablation. We observed near complete depletion of biliary tuft cells, which was sustained for 3–6 months after DT treatment (Fig 2E). Thus, biliary tuft cells are long-lived and replenish slowly in adult mice.
The extrahepatic biliary epithelium undergoes significant postnatal expansion (33–35) (Fig S2C). To compare biliary tuft cell turnover in neonatal (actively proliferating) and adult (quiescent) extrahepatic epithelium, we treated adult or P10 neonatal 25ERT2/ERT2;R26YFP/YFP mice with tamoxifen. We assessed induction of YFP in IL-25+ tuft cells 24 hrs after tamoxifen injection and presence of YFP+ tuft cells seven days later. Labeling efficiency was less robust than in Dclk1ERT2/+;R26YFP mice (Fig S2D,E), but was stably retained in adult biliary tuft cells, in contrast to rapid loss from small intestinal tuft cells (Fig 2F). YFP label was poorly retained by both biliary and small intestinal IL-25+ tuft cells in neonates (Fig 2G). IL-25+ tuft cells were highly abundant pre-weaning and declined to adult levels around 6 wks of age (Fig 2H), further supporting distinct biliary tuft cell regulation in adult vs. neonatal mice.
Biliary tuft cell abundance is modulated by bile acids
Weaning promotes maturation of enterohepatic recirculation (Fig 3A), dependent on the transition from milk to solid foods and acquisition of a stable intestinal microbiota (33). As this coincides with the postnatal reduction of biliary tuft cell frequency, we assessed the effects of enterohepatic recirculation/BAs on tuft cells by feeding mice a 2% cholestyramine diet to sequester intestinal BAs. As expected, cholestyramine reduced small intestinal expression of the FXR gene product Fgf15, which negatively regulates hepatic BA synthesis, thereby resulting in increased liver Cyp7a1 expression (Fig S3A). Biliary tuft cells were almost completely eliminated in cholestyramine-fed mice compared to controls (Fig 3B). Since cholestyramine acts indirectly to increase hepatic BA output, we tested whether the effect of cholestyramine on biliary tuft cells could be blocked by systemic activation of FXR, which would directly inhibit hepatic BA synthesis. Treatment of cholestyramine-fed mice with the FXR agonist GW4064 (36) sustained biliary tuft cells at normal levels (Fig 3C). In cholestyramine-induced tuft cell loss we observed no evidence for trans-differentiation to non-tuft epithelial cells using our fate-mapping 25CreERT/Cre;R26YFP/+ mice (Fig S3B). These data suggest a role for liver BA production in the regulation of biliary tuft cell frequency.
Fig. 3. Biliary tuft cell abundance is modulated by bile acids.
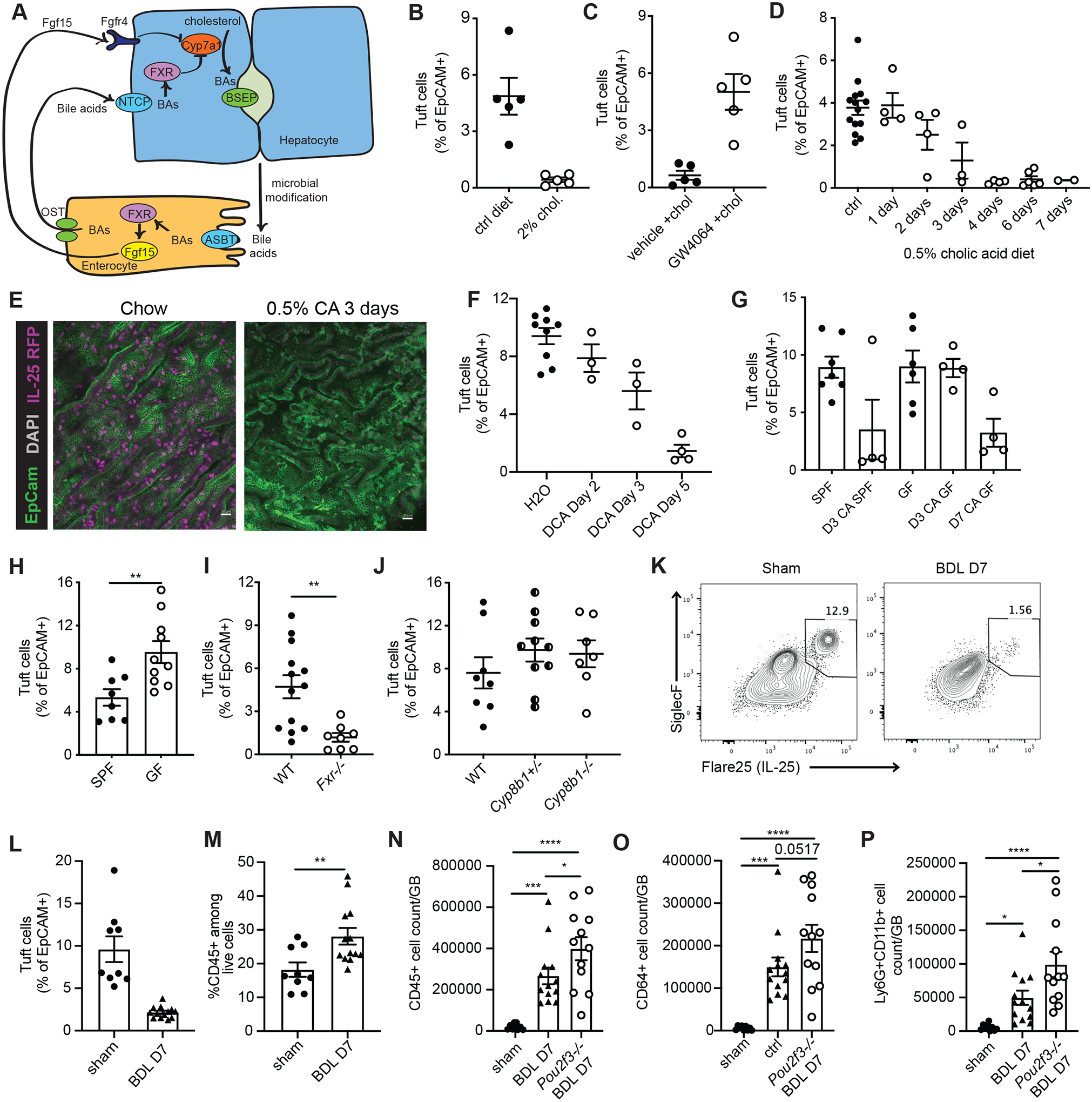
A) Schematic of BA regulation via enterohepatic recirculation. B) Flare25 mice were fed control (ctrl) diet or 2% cholestyramine (2% chol) diet for two wks; IL-25+ tuft cell frequency in biliary epithelial prep was determined by flow cytometry. Representative of three experiments. C) Flare25 mice fed 2% cholestyramine diet for ten days were injected IP every other day with GW4064 (GW4064 + chol) or vehicle (veh + chol) starting on day 1 of diet. Representative of three experiments. D) Flare25 mice were fed 0.5% cholic acid (CA) diet for the indicated time periods; IL-25+ tuft cell frequency in biliary epithelial prep was determined by flow cytometry. E) Wholemount confocal imaging of Flare25 GB from mice fed chow or 0.5% CA diet for three days. F) Germfree (GF) were fed irradiated 0.5% CA diet in sterile isolators. Frequency of DCLK1+ tuft cells among epithelial cells in total GB/EHBD digest was examined by flow cytometry at indicated timepoints and compared to SPF mice fed irradiated 0.5% CA diet. G) Flare25 mice received 0.2% sodium deoxycholate in drinking water ad libitum. Tuft cell frequency among epithelial cells from total GB/EHBD digest was determined by flow cytometry. H,I) Frequency of DCLK1+ tuft cells among epithelial cells from total GB/EHBD digests was analyzed by flow cytometry in age- and sex-matched H) SPF mice from the UCSF vivarium compared to GF mice and I) Fxr −/− and WT controls. J) The frequency of IL-25+ tuft cells among biliary epithelial cells from total GB/EHBD digests was examined by flow cytometry in Flare25 reporter Cyp8b1−/−, Cyp8b1+/−, and Cyp8b1+/+ mice. K-M) Flare25 mice were subjected to bile duct ligation (BDL) or sham surgery. Seven days later total GB/EHBD digests were analyzed by flow cytometry for frequency of IL-25+ tuft cells among epithelial cells (K,L) and frequency of CD45+ immune cells (M). N-P) Total GB/EHBD digests from Pou2f3−/− and littermate controls were analyzed by flow cytometry seven days after BDL or sham surgery. Total immune cells (CD45+, N), macrophages (CD64+CD11b+Ly6G-, O), and neutrophils (Ly6G+CD11b+CD64-, P) were enumerated. Significance determined by unpaired two-tailed T test (H,I,M) or one-way ANOVA (N-P), *p<.05, **p<.01, ***p<.001, ****p<.0001.
Cholic acid (CA) is the primary BA in mice and humans (1), and its production is increased after cholestyramine treatment and reduced by FXR activation. Feeding 0.5% CA led to an abundance of CA in bile (Fig S3C), and resulted in rapid and near complete elimination of tuft cells in the biliary tree (Fig 3D,E) but not in the small intestine (Fig S3D). Deoxycholic acid (DCA), generated by bacterial dehydroxylation of CA (37, 38), was also elevated in bile after 0.5% CA feeding (Fig S3E). Supplementation of drinking water with 0.2% DCA in drinking water was sufficient to reduce biliary tuft cell frequency (Fig 3F), suggesting a role for the microbiota in BA-induced tuft cell loss. Consistent with this, tuft cell reduction was delayed in GF mice fed 0.5% CA (Fig SG), and untreated GF mice had significantly more biliary tuft cells compared to age- and sex-matched SPF controls (Fig 3H).
Further implicating BAs as negative regulators of biliary tuft cell abundance, we found that frequency of tuft cells in the biliary epithelium was greatly reduced in FXR-deficient mice, which lack negative feedback on CA production in the liver (39) (Fig 3I). Conversely, tuft cell frequency was unchanged in mice with CRISPR-mediated deletion of Cyp8b1 (Fig 3J), which is required for liver CA production (40) (Fig S3F,G). Cyp27a1−/− mice, in which only CA and its derivative are present (41), were similar to Fxr−/− mice in their diminished biliary tuft cell population (Fig S3H). Taken together, these results suggest that CA and its metabolites negatively regulate GB and EHBD tuft cell abundance.
Inflammatory cell infiltration induced by bile duct ligation increases in the absence of tuft cells
Bile duct ligation (BDL) is an established model in which altered BA levels, including increased CA (42, 43), lead to cholestatic liver injury and liver fibrosis (44). We observed significantly reduced biliary tuft cell frequency seven days after BDL as compared to sham surgery controls (Fig 3K,L).
In contrast to genetic and dietary BA manipulations, BDL induces a robust inflammatory response (Fig 3M). Therefore, we asked whether biliary tuft cells could play a role in cholestatic biliary inflammation. One week post-surgery, Pou2f3−/− mice had modestly increased mortality (Fig S3I) and weight loss (Fig S3J), and we observed a trend towards increased serum bilirubin (Fig S3K) as compared to littermate controls. Neither genotype developed liver fibrosis one week post-surgery as assessed by quantification of liver hydroxyproline and Col6a1 expression (Fig S3L,M). Pou2f3−/− BDL mice had significantly more biliary-infiltrating CD45+ cells (Fig 3N), with a trend towards increased macrophages (Fig 3O) and a significant increase in neutrophils (Fig 3P), as compared to tuft cell-sufficient BDL mice. Extrahepatic epithelial cell hyperplasia after BDL was comparable between genotypes (Fig S3N), suggesting that increased tissue hyperplasia did not explain the increased immune cells. Hepatic ductal reaction was evident one week after BDL but was unaffected by loss of tuft cells (Fig S3O,P). There was no evidence of intrahepatic IL-25+ tuft cells in liver from Flare25 mice subjected to BDL (Fig S3P). Taken together, the loss of tuft cells was correlated with enhanced myeloid cell infiltration and a trend towards increased morbidity and mortality after BDL.
Tuft cell-deficient Pou2f3−/− mice develop spontaneous biliary neutrophil infiltration
The altered myeloid cell infiltration in Pou2f3−/− mice subjected to BDL prompted us to examine the biliary myeloid compartment in Pou2f3−/− mice at homeostasis. We found spontaneous accumulation of CD45+ cells, predominantly neutrophils, in tuft cell-deficient mice compared to age- and sex-control mice (Fig 4A,B), and replicated this finding in littermate Pou2f3−/− and control mice (Fig 4C–E). Neutrophilia was not evident in spleen, small intestine lamina propria, or peripheral blood of Pou2f3−/− mice relative to littermate controls (Fig S4A,B).
Fig. 4. Pou2f3−/− mice have increased biliary neutrophil infiltration under homeostatic conditions.
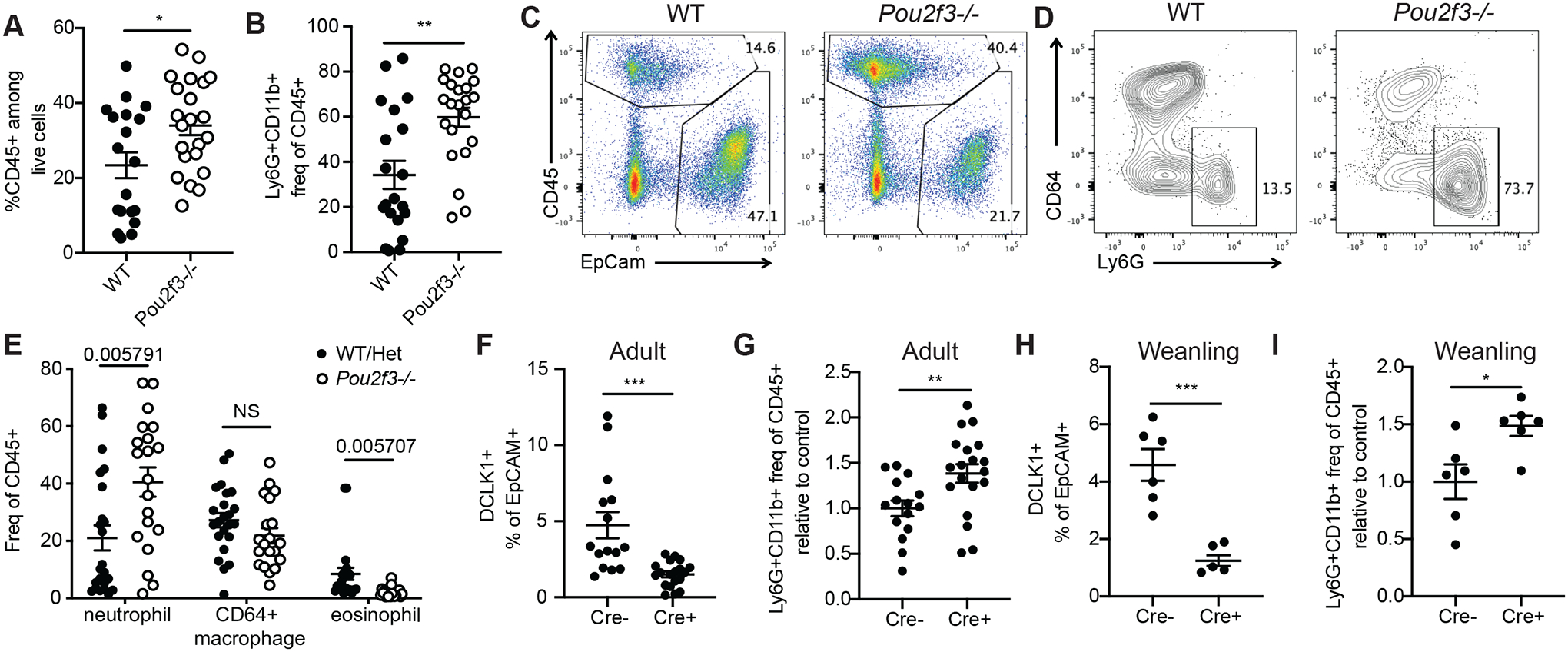
A-B) Relative frequency of CD45+ cells (among live cells, A) and neutrophils (Ly6G+CD11b+CD64- among CD45+ cells, B) as assessed by flow cytometry on total GB/EHBD digests from non-littermate age- and sex-matched Pou2f3−/− and WT mice. Statistical significance determined by unpaired two-tailed T test. C-E) Total GB/EHBD digests from littermate Pou2f3−/− and control mice were analyzed by flow cytometry for presence of CD45+ cells (representative flow plot previously gated on live singlets, FSC-A × SSC-A, C) and frequency of neutrophils (Ly6G+CD11b+CD64-), macrophages (CD64+CD11b+Siglec F-Ly6G-) and eosinophils (Siglec F+CD11b+Ly6G-CD64) among CD45+ cells (D,E). P values calculated by one-way ANOVA (E). F-I) 25cre/+;RosaiDTR or 25+/+;RosaiDTR littermate mice received two retroorbital injections of diphtheria toxin. Frequency of tuft cells among epithelial cells (F,H) and neutrophils among CD45+ cells (G, I) in total GB/EHBD digests were determined by flow cytometry. F,G) Adult mice were injected at 8 wks of age and chased 1–6 months. Data pooled from four experiments. H,I) Mice were injected at weaning and analyzed 4–6 wks later. Data pooled from two experiments. G,I) Neutrophil frequencies were normalized to the average of control cre- mice per experiment. Statistical significance determined by unpaired two-tailed T test (F-I). *p<.05, **p<.01, ***p<.001, ****p<.0001.
We next tested whether neutrophil infiltration could be induced by acute tuft cell depletion. IL-25+ tuft cells were depleted in DT-treated 25Cre/+;R26iDTR/iDTR mice in all tissues, but our recovery timecourse indicated long-term suppression of biliary tuft cells. Adult DT-injected 25Cre/+; R26iDTR/iDTR mice analyzed after 4 wks to 6 months had reduced tuft cell frequency (Fig 4F), and increased biliary neutrophils as compared to DT-injected 25+/+; R26iDTR/iDTR littermate controls (Fig 4G, Fig S4C). Similar increases in biliary neutrophils occurred in mice subjected to DT-mediated tuft cell depletion shortly after weaning (4 wks) (Fig 4H,I; Fig S4D,E).
We next asked whether non-inflammatory BA-induced tuft cell loss would promote biliary neutrophil influx. When we quantified biliary neutrophils in Cyp27a1−/− and Fxr−/− mice relative to controls we found reduced frequency of biliary neutrophils (Fig S4F,G). To further examine the impact of increased BA on biliary neutrophils, we fed Pou2f3−/− and littermate control mice 0.5% CA for 21 days, which in tuft cell-sufficient mice led to sustained tuft cell depletion (Fig S4H). Prolonged elevated dietary CA reduced immune cell infiltration in Pou2f3−/− mice, largely eliminating neutrophil influx (Fig S4I), in contrast to what we observed with tuft cell deletion under normal BA production and in cholestasis (BDL).
Loss of tuft cells drives accumulation of activated neutrophils and altered biliary epithelium
To understand how loss of biliary tuft cells impacts the extrahepatic biliary immune niche, we performed scRNA-seq on total GB/EHBD digests from Pou2f3−/− or littermate control mice (Fig S5A). We identified all major cellular types by canonical gene expression in both genotypes (Fig 5A,B; Fig S5B).
Fig. 5. Total biliary scRNA-seq reveals heightened inflammatory tone of biliary tissue in the absence of tuft cells.
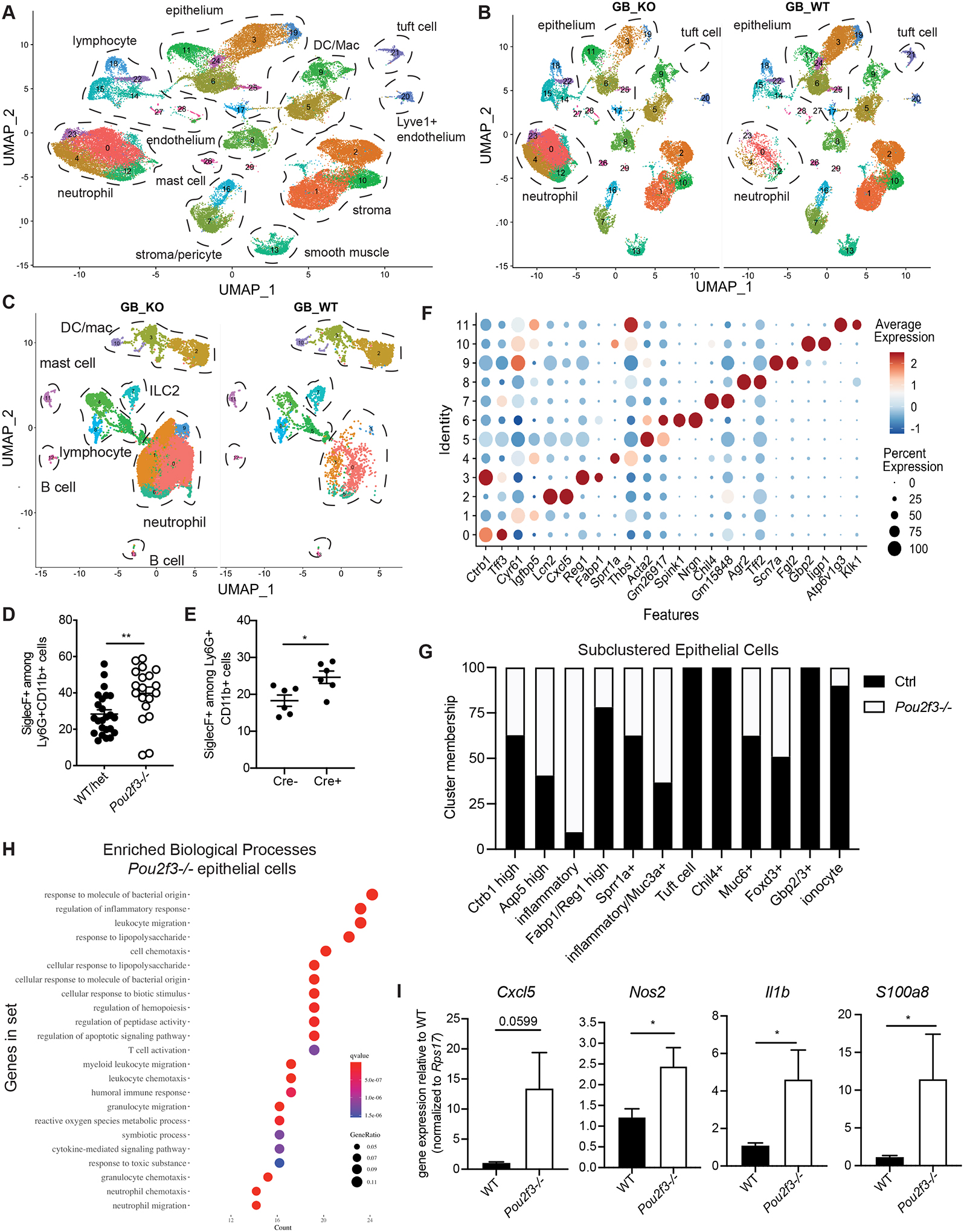
A-B) Live cells were sorted from Pou2f3−/− and littermate controls total GB/EHBD digests and subjected to single cell sequencing using the 10X platform. UMAP shows major cell types, determined by canonical gene expression. C) Subclustered CD45+ cells split by genotype: Pou2f3−/− (GB_KO), littermate controls (GB_WT) . D,E) Quantification of Siglec F+ neutrophils from littermate Pou2f3−/− and littermate controls (D) and 25cre/+;R26iDTR or 25+/+;R26iDTR/iDTR mice injected with diphtheria toxin at 4 wks of age and analyzed 4–6 wks later (E) as determined by flow cytometry on total GB/EHBD digests. F) Dotplot of top two most differentially expressed genes (DEGs) after subclustering of biliary epithelial cells. G) Epithelial cell cluster membership after normalizing to the total number of epithelial cells sequenced per genotype. H) Gene ontology biological processes enrichment for genes upregulated in epithelial cells from Pou2f3−/− mice compared to controls. I) Total GB/EHBD from non-littermate Pou2f3−/− (n=17) and WT (n=15) mice was analyzed by qPCR for the indicated target genes relative to the housekeeping gene Rps17. Data pooled from three experiments, Pou2f3−/− RQ normalized to average of WT mice in each experiment. Statistical significance determined by unpaired student’s T-test.
As expected, neutrophils were the dominant immune population in Pou2f3−/− mice (Fig 5C). Subclustering CD45+ immune cells and assessing the cluster representation of each genotype revealed multiple neutrophil clusters, all of which were increased in Pou2f3−/− mice (Fig 5C; Fig S5C). Biliary neutrophils in Pou2f3−/− mice had a more activated phenotype, characterized by upregulation of chemotaxis/cytokine response genes by GO term enrichment analysis (Fig S5D). We also observed increased expression of Siglecf and Ffar2 (Fig S5E), aligning with recently described gene profiles of tissue-infiltrating neutrophils (45, 46). We confirmed the higher frequency of biliary Siglec F+ neutrophils in Pou2f3−/− mice as compared to littermate controls using flow cytometry (Fig 5D, Fig S5F). We also observed a significant increase in Siglec F+ neutrophils when tuft cells were depleted by DT injection in 4-week old 25Cre/+; R26iDTR/iDTR mice as compared to littermate controls (Fig 5E).
We next analyzed the sequenced epithelial cells. Subclustering revealed highly distinct extrahepatic biliary epithelial cell gene expression programs (Fig 5F), suggesting distinct biliary epithelial cell identities. We found major shifts in epithelial cell cluster representation in mice lacking tuft cells (Fig 5G, Fig S5G). Pou2f3−/− mice, which lacked tuft cells as expected, had an increase in cells in cluster 2, which expressed genes such as Cxcl5 and Lcn2, and loss of two epithelial clusters of unknown function—clusters 7 (Chil4+) and 10 (Cd247+) (Fig 5F,G).
Previous single cell analyses have demonstrated heterogeneous gene expression among tuft cells (47, 48). While all biliary tuft cells expressed canonical tuft cell genes including Avil and Trpm5, subclustering revealed four distinct gene expression programs (Fig S5H–J) which did not align with previously described “tuft-1” and “tuft-2” subsets (Fig S5J,K).
Enrichment analysis of upregulated genes among all Pou2f3−/− epithelial cells relative to control cells suggested increased inflammatory/defense pathways, including those related to neutrophil chemotaxis and response to bacteria (Fig 5H). To verify this, we isolated RNA from extrahepatic biliary tissue (bile, GB, cystic and common duct) and examined expression of top DEGs upregulated in cluster 2 epithelial cells from Pou2f3−/− mice (Fig S5L). We observed upregulation of the genes Cxcl5, Nos2, S100a8, and Il1b in biliary tissue from Pou2f3−/− mice relative to controls (Fig 5I).
Biliary inflammatory tone and neutrophil influx is driven by host microbiota
This inflammatory tissue signature, coupled with the enrichment in GO terms related to bacterial responses driven by the increased prevalence of cluster 2 inflammatory epithelial cells in Pou2f3−/− mice (Fig S6A), prompted us to examine whether biliary neutrophil infiltration could be affected by the microbiome. We examined the frequency of immune cells in GB/EHBDs from UCSF-housed mice as compared to mice from The Jackson Laboratory (Jax) and GF mice and found increased frequency of total CD45+ immune cells and Ly6G+ neutrophils in UCSF mice compared to Jax and GF mice; neutrophils were near-absent in GF mice (Fig 6A,B). Consistent with this, biliary tissue from Jax mice had lower expression of Cxcl5, Nos2, S100a8, and Il1b in biliary tissues from Jax mice as compared to UCSF mice (Fig S6B).
Fig. 6. Biliary neutrophilia is driven by microbiota.
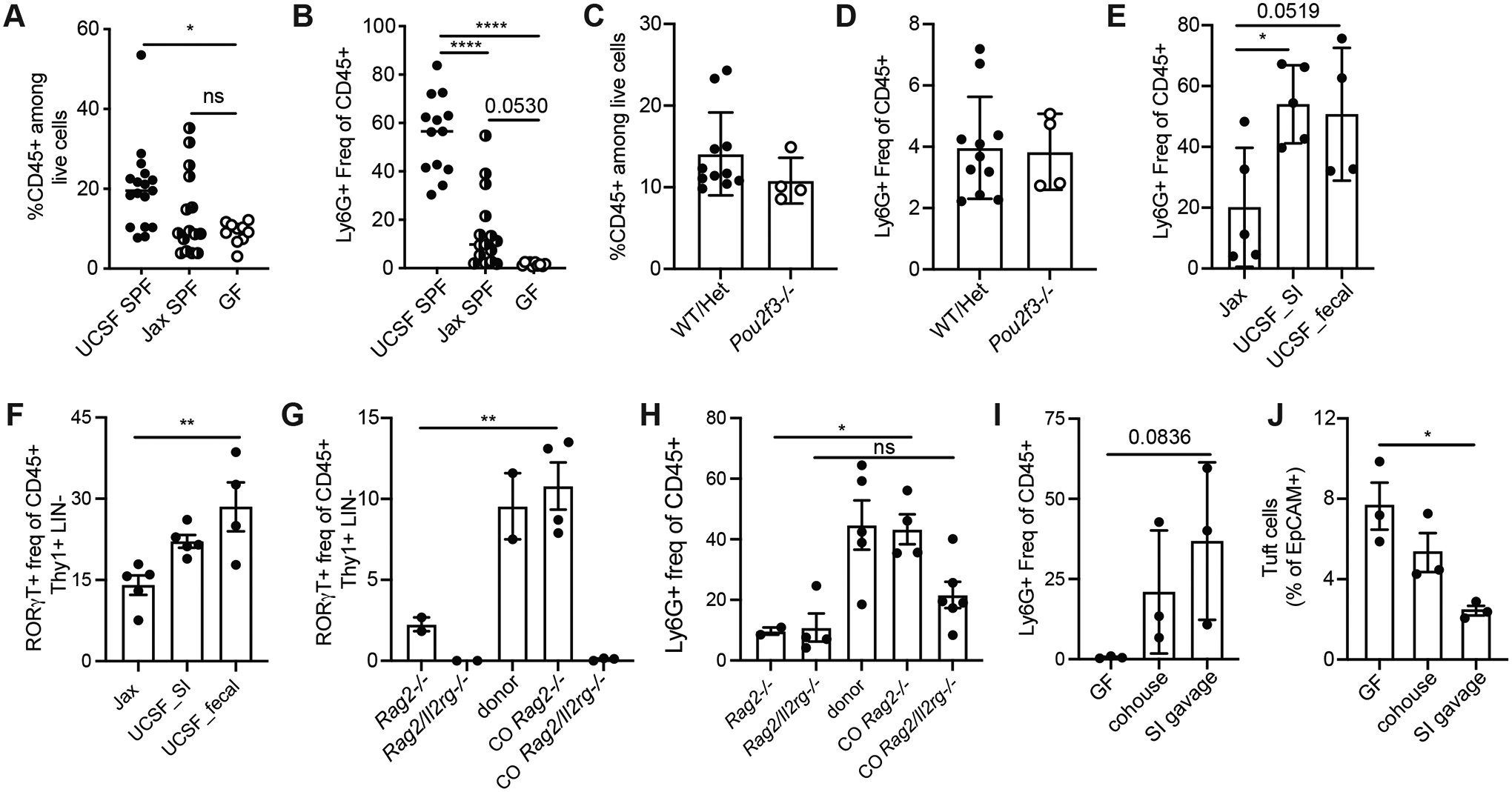
A,B) Frequency CD45+ cells among live cells (B), and frequency of Ly6G+CD11b+ neutrophils among CD45+ cells (B) in total GB/EHBD digests from UCSF-housed SPF mice compared to Jax SPF mice and germfree (GF) mice analyzed by flow cytometry. C,D) Frequency CD45+ cells among live cells (C), and frequency of Ly6G+CD11b+ neutrophils among CD45+ cells (D) in total GB/EHBD digests from 3.5 wk old Pou2f3−/− mice and littermate controls analyzed by flow cytometry. E,F) Jax mice received fecal or small intestinal contents from UCSF donor mice by oral gavage and were analyzed 6 wks later by flow cytometry on total GB/EHBDs for frequency of Ly6G+CD11b+ neutrophils (E) or RORγT+ lymphocytes (F) as identified by intracellular staining. G, H) Rag2−/− or Rag2/Il2rg−/− mice from Taconic were cohoused with UCSF donor mice for 6 wks. RORγT+ lymphocytes and neutrophils were quantified by flow cytometry on total GB/EHBDs. I,J) GF mice received small intestinal contents from UCSF donor mice by oral gavage or were cohoused with UCSF donors. 6 wks later mice were compared to GF controls by flow cytometry on total GB/EHBDs analyzing frequency of Ly6G+CD11b+ neutrophils (I) or tuft cells (J) by intracellular staining. P values calculated by one-way ANOVA. *p<.05, **p<.01, ****p<.0001. All data shown +/− SEM.
In UCSF-housed WT mice we found that the reduced tuft cell frequency post-weaning correlated with expansion of CD45+ infiltrating immune cells (Fig S6C,D), suggesting that the post-weaning microbiome is required for biliary neutrophil influx. Indeed, we observed no differences in biliary immune cell infiltration and a paucity of biliary neutrophils in both Pou2f3−/− and littermate controls analyzed at weaning (Fig 6C,D).
To determine whether transfer of microbiota from UCSF-housed WT mice could induce biliary neutrophil influx, we colonized Jax mice with fecal or small intestinal content from UCSF donor mice and examined biliary immune cells six weeks later. Mice that received microbiome transfer had higher levels of biliary infiltrating neutrophils compared to uncolonized Jax mice housed in the UCSF vivarium (Fig 6E) while tuft cell frequency was unchanged (Fig S6E). We also observed an increase in the frequency of lineage-negative RORγT+ lymphocytes, but not ILC2s, in colonized mice (Fig 6F, Fig S6F). Analysis of RORγT+ lymphocytes in Pou2f3−/− and control mice suggested this was not tuft cell dependent (Fig S6G).
This prompted us to explore the role of adaptive vs. innate lymphoid cells in microbiome-dependent neutrophil influx. When Rag2−/− or Rag2/Il2rg−/− mice, which lack most adaptive and innate lymphocytes, were cohoused with UCSF donors, Rag2−/− recipients had robust induction of RORγT+ lymphocytes; these cells were not present in Rag2/Il2rg−/− mice (Fig 6G). Compared to uncolonized controls, cohoused Rag2−/− mice had significantly increased biliary neutrophils (Fig 6H); the frequency of biliary neutrophils in colonized Rag2/Il2rg−/− mice was not significantly increased relative to controls (Fig 6H).
Having established the transferability of neutrophil-promoting microbiota, we asked whether this could impact tuft cell frequency in GF mice, which have significantly more tuft cells than SPF mice. We compared GF controls to GF mice cohoused with UCSF donors or GF mice colonized with small intestinal content. Colonization with small intestinal contents from UCSF-raised donors induced biliary neutrophilia (Fig 6I), and a reduction in biliary tuft cells (Fig 6J) in GF mice.
DISCUSSION
Though critical to understanding human disease, immune responses in the extrahepatic biliary tree are poorly characterized. Here we show that tuft cells, which are highly abundant in GB/EHBDs, impact the immune balance in this tissue. Biliary tuft cells express the core tuft cell gene expression program and a tissue-specific gene signature. In contrast to small intestinal tuft cells, biliary tuft cells are highly sensitive to BAs and not regulated by ILC2 cytokines. The reduction in biliary tuft cells in the BDL model of cholestasis, which superimposes inflammatory injury with elevated BA levels, correlates with increased immune cells, with the complete lack of tuft cells driving accumulation of neutrophils. Similarly, loss of tuft cells leads to spontaneous accumulation of activated neutrophils and an increase in inflammatory gene expression in unperturbed mice. Biliary neutrophils infiltration is dependent on the microbiota, which additionally impacts accumulation of RORγT+ lymphocytes and can promote tuft cell reduction. Our findings add to the understanding of biliary immune cells and inflammation, suggesting unappreciated mechanisms for affecting immune balance in the biliary tree that could be therapeutically targeted.
The GB/EHBDs undergo significant postnatal development in response to stabilization of diet, microbiome, and enterohepatic recirculation; this development is synchronous with substantial liver growth and increasing metabolic demand (1, 33, 35). Biliary tuft cells are prevalent in the pre-weaning period and decrease to adult levels around 6–8 wks, after the period of growth and metabolic consolidation in which BA synthesis increases (33). Although this developmental reduction is consistent with the negative relationship between CA/CA metabolites and biliary tuft cells that we established through manipulation of the enterohepatic circulation, genetic knockouts and dietary CA, the precise mechanism driving the inverse relationship of CA and biliary tuft cell abundance remains unclear and a focus of future investigation.
Loss of tuft cells led to increased biliary infiltration of neutrophils with a transcriptomic signature similar to that seen in neutrophils in other inflammatory settings (45, 46, 49). Accumulation of inflammatory neutrophils was not observed in other tissues in Pou2f3−/− mice, suggesting a biliary-specific consequence for tuft cell loss, which could be induced by 25Cre-mediated tuft cell deletion. Tuft cell loss induced by elevated levels of BAs in the absence of other inflammatory insult did not induce neutrophil influx; rather, overabundance of CA induced by dietary manipulation or genetic deficiency repressed immune cell infiltration, even in tuft cell deficient mice. Given the robust link we identified between microbiota and neutrophil influx, it is plausible that impacts of altered BAs on the microbiome itself could explain the disconnect between BA-induced loss of tuft cells and inducible or constitutive loss of tuft cells in driving neutrophil influx and inflammatory tissue tone.
Our single-cell and total tissue analysis suggests that loss of tuft cells promotes an anti-microbial gene expression program in the biliary epithelium (e.g.: Cxcl5 (50), Lcn2 (51) and S100a8/a9 (52)). This tissue gene signature, and corresponding neutrophil influx, can be modulated by the microbiome. Whether the epithelium itself induces neutrophil recruitment directly (through chemotactic factors such as CXCL5) remains to be tested.
Our GB and EHBD atlas of epithelial, immune and stromal cells extends previous efforts analyzing this tissue in mouse (53), and complements recent analysis of human biliary epithelium (54). We document extensive, previously undescribed heterogeneity of biliary epithelial and immune cells. A deeper analysis of these populations, and their role in both biliary disease and normal function is clearly warranted. Notably, this sequencing was performed on “total” biliary digests (GB and ducts). Therefore, distinct epithelial cell transcriptional profiles and immune cell phenotypes could be regional. This could be the case for tuft cells themselves—the majority of tuft cells from the single cell analysis overlap with the bulk RNAseq data from GB tuft cells only, while a minority show a distinct gene expression program and could be ductal.
The inflammatory biliary tissue gene signature observed in the absence of tuft cells, coupled with infiltration of activated neutrophils, shares similarities with hallmarks of the inflamed mucosa in inflammatory bowel disease and in intestinal response to pathogenic bacteria (55–58). However, the existence of a biliary microbiome is controversial in health (59, 60). Microbiome-dependent effects on the biliary tree could be regional (common duct vs. GB), related to bacterial infiltration from the small intestine, or induced from microbiome-dependent metabolites or microbial byproducts that could directly engage biliary tuft cells.
Understanding the complex interplay between biliary immunity, bile acids, tuft cells and the microbiome has great potential to improve understanding of biliary inflammation in health and disease. Our observations raise the intriguing possibility that biliary tuft cells may be key to limiting inflammatory responses to microbial exposures, and that this may condition biliary immune tone in a way that impacts progression of, or predisposition to, common biliary maladies.
STUDY DESIGN
The aim of this study was to understand the phenotype/ function of IL-25+ tuft cells in the extrahepatic biliary tree. To this end, we optimized methods for isolation of live cells from murine GB/EHBDs, developed new genetically modified mice, and used dietary, pharmacological, genetic, and surgical perturbation of BA recirculation. We performed scRNA-seq, reverse transcription polymerase chain reaction (RT-PCR), flow cytometry, and histology. All experiments used randomly assigned mice without investigator blinding. All data points and n values reflect biological replicates.
MATERIALS AND METHODS
Mice
Mice were housed in the University of California San Francisco specific pathogen–free animal facility in accordance with Institutional Animal Care and Use Committee guidelines. Mice were ≥ six wks old, unless otherwise noted. Experiments were performed using sex- and age- matched mice, or littermate controls as indicated. Fxr−/− (Nr1h4tm1Gonz/J) were purchased from the Jackson Laboratory and maintained in-house. Rag2−/− and Rag2−/−Il2rg−/− mice were purchased from Taconic. Cyp27a1−/− (B6.129-Cyp27a1tm1Elt/J) were provided by the Cyster Lab at UCSF. Pou2f3−/− (C57BL/6N-Pou2f3tm1.1(KOMP)Vlcg) were provided by the Anderson Lab at UCSF (22). IL-25 reporter mice (Flare25), Arginase 1 reporter mice (Yarg, B6.129S4-Arg1tm1Lky/J), IL-5 reporter/Cre KO/KI mice (Red5, B6(C)-Il5tm1.1(icre)Lky/J), and IL-13 (Smart13, B6.129S4(C)-Il13tm2.1Lky/J) reporter mice have been described (6, 10, 27, 61). IL-5 deleter mice were generated by intercrossing Red5 mice with Rosa-DTA mice (B6.129P2-Gt(ROSA)26Sortm1(DTA)Lky/J) to generate homozygous RRDD mice. IL4ra−/− mice (62), were provided by F. Brombacher, and backcrossed ≥ 8 generations to C57BL/6J. Il25−/− mice (63), were provided by A. McKenzie and backcrossed ≥ 8 generations to C57BL/6J. Dclk1 CreERT2 (32) from M. Buchert at the Olivia Newton John Cancer Research Institute, Australia were intercrossed with YFP Gt(ROSA)26STOP-flox, provided by O. Klein at UCSF.
Generation of new mouse strains
Il25-driven Cre and CreERT2
The 25cre and 25ERT mice were generated as described for the IL-25 reporter allele Flare25 (10), with the following modifications:
Removal of the 5’ loxP site in front of the last exon of Il25 from the 5’ homologous arm.
A humanized (eukaryotic codon-optimized) Cre (for 25Cre) or tandem RFP and tamoxifeninducible Cre recombinase (64) (for 25ERT2) was used to replace the tandem RFP fluorescent protein sequence in the original Flare 25.
Cyp8b1−/− mice
Two sgRNAs targeting the Cyp8b1 gene main exon (GGCATCCCACAACCGTGAGC and GATCCGTCGCGGAGATAAGG) and recombinant Cas9 (PNA Bio) were simultaneously injected as RNPs into the zygotes of C57BL/6J homozygous reporter mice (Flare25, Yarg, Smart13; Gladstone Institutes, San Francisco CA). Founders were screened by PCR using primers AGCCTGCTGGAGCCTAGCCATGACG and AGCTGGGGAGAGGAAGGAGTGCCTC to identify the 1.2 kb deletion event resulted from the end-joining event of two double-stranded breaks at the sgRNA-targeted sites. Individual positive founder was backcrossed with wild-type homozygous reporter mice to segregate the alleles before intercrossing to obtain homozygous Cyp8b1 knockouts.
Germfree mice
Germfree mice were from the UCSF Gnotobiotic Core Facility. Feeding experiments were performed in gnotobiotic isolators. Fecal pellet DNA was examined by qPCR to verify absence of bacterial colonization.
Preparation of single-cell suspensions
GB, cystic duct and common duct (at the duodenum) were resected, hepatocytes/pancreas were removed by scraping, GB was fileted. and tissue was rinsed in cold HBSS (Ca2+/Mg2+ free, Life Technologies). Tissue was incubated at 37 C for ten min in 1 mL HBSS (Ca2+/Mg2+ free) with 2% fetal calf serum (FCS, company), 10 mM HEPES (Sigma) and 5 mM DTT (Sigma). Supernatants were discarded and tissue was incubated at 37 C for 20 min in 1 mL HBSS (Ca2+/Mg2+ free) with 2% FCS and 5 mM EDTA (Teknova). Supernatant was collected into FACS wash buffer (FWB: PBS, 3% FCS, 0.05% NaN3) and kept on ice. This step was repeated. After collection of the EDTA supernatant, cells were pelleted and resuspended in FWB. For epithelial prep, cells were then stained for flow cytometry. For total GB/EHBD remaining tissue was incubated at 37 C for 5 min in 1 mL HBSS (with Ca2+/Mg2+) with 3% FCS and 10 mM HEPES, supernata was discaeded and tissue was minced and digested at 37 C for 20–25 min in 4 mL HBSS (with Ca2+/Mg2+) supplemented with 3% FCS, 10 mM HEPES, 100 μg/ml Liberase TM (Roche) and 30 μg/ml DNase I (Roche). Incubations were performed with gentle rocking. Tissue was dissociated in GentleMACS C tubes (Miltenyi Biotec) using program m_intestine_01. The epithelial fraction in cold FWB was to digest, passed through a 100 μm filter, and washed in cold FWB. Small intestine epithelium and lamina propria were processed as described (15, 65).
In vivo manipulations
0.5% (w/w) CA in Teklad 7001 or Teklad 2019 base diet was from Envigo. 2% (w/w) cholestyramine (Sigma) diet on AIN93G was made by Research Diets. 0.2% (w/v) sodium deoxycholic acid (Sigma) was dissolved in sterile water. Tamoxifen (Sigma) treatment was performed by intraperitoneal (IP) injection or given in chow (Envigo TD.130858) (64), as indicated in legends. Tamoxifen was dissolved in ethanol/cremaphore at 20 mg/ml, diluted 1:1 in sterile PBS and mice received 1 mg in 100 ul. Tamoxifen chow was provided for 2–4 wks. Labelling was assessed 24 hrs after last injection/removal of diet. Diphtheria toxin (Sigma) was injected retro-orbitally, 500 ng/100 ul sterile PBS/mouse. Two injections were given 48 hrs apart, and mice were analyzed for deletion after 24 hrs. GW4064 (Selleck Chem) was dissolved in DMSO, resuspended in sterile methycellulose and given IP at 100 mg/kg body weight.
Microbiota transfer
GF/Jax mice were cohoused for a minimum of 6 wks with adult UCSF-raised WT females with known biliary neutrophil infiltration. For microbiota transfer, the full length of the small intestine from affected UCSF-housed WT donor mice was fileted and flushed with sterile PBS, followed by gentle scraping with curved forceps (into a total volume of 5–7 mls), or fecal pellets were homogenized in PBS (two donors, 5 ml volume). Slurry was filtered through a 100 uM filter. Recipient mice received 150 ul by oral gavage.
Bile acid quantification
Mice were fasted overnight with access to water. Following euthanasia, GB/EHBDs were resected as described above; intact GB (containing bile) was snap-frozen in liquid nitrogen and stored at −80 C. Tauro-cholic acid (TCA) and tauro-deoxycholic acid (TDCA) were quantified by GC-MS (West Coast Metabolomics, UC Davis).
Total tissue RNA extraction and qPCR
GB with bile, cystic and common duct or liver were snap-frozen in liquid nitrogen or on dry ice and stored at −80 C. Samples were dissociated in 1 ml RNAzol (Molecular Research Center, Inc) using Miltenyi M tubes and the gentleMACS tissue dissociator (Miltenyi Biotec) RNA1 program. RNA was isolated following manufacturer’s protocol with additional BAN phase separation. RNA was subjected to reverse transcription using the SuperScript Vilo Master Mix (Life Technologies). cDNA was used as template for quantitative PCR (qPCR) with the Power SYBR Green reagent on a StepOnePlus cycler (Applied Biosystems). Transcripts were normalized to Rps17 (40S ribosomal protein S17) expression and relative expression shown as 2-ΔΔCt compared to the average ΔCt of experiment-matched controls using the following primers:
Rps17: 5′- CGCCATTATCCCCAGCAAG-3′, 5′-TGTCGGGATCCACCTCAATG-3′
Fgf15: 5’- GAGGACCAAAACGAACGAAATT-3’, 5’- ACG TCC TTG ATG GCA ATC G-3’
Cyp7a1: 5’- GGGATTGCTGTGGTAGTGAGC-3’; 5’- GGTATGGAATCAACCCGTTGTC-3’ (PrimerBank 31542445a1)
Nos2: 5’- GAATCTTGGAGCGAGTTGTGG-3’, 5’- CAGGAAGTAGGTGAGGGCTTG-3’
Il1b: 5’-GCAACTGTTCCTGAACTCAACT-3’, 5’- ATCTTTTGGGGTCCGTCAACT-3’
S100a8: 5’- AAATCACCATGCCCTCTACAAG-3’, 5’- CCCACTTTTATCACCATCGCAA-3’ (PrimerBank 7305453a1)
Col1a1: 5’- CCAAGAAGACATCCCTGAAGTCA-3’, 5’-TGCACGTCATCGCACACA-3’
Epcam: 5’- GCGGCTCAGAGAGACTGT-3’, 5’-CCAAGCATTTAGACGCCAGTTT-3’
Cxcl5: 5’- TCCAGCTCGCCATTCATGC-3’, 5’- TTGCGGCTATGACTGAGGAAG-3’ (PrimerBank 6677887a1)
Cyp8b1: 5’-TGTTTCTGGGTCCTCTTATTCCTGC-3’; 5’-ACTCTCCTCCATCACGCTGTCCAAC-3’
Bile duct ligation
Mice were anesthetized in an aseptic surgical field by IP ketamine/xylazine (80–100 mg/kg Ketamine + 5–10 mg/kg Xylazine) followed by inhalative isoflurane and subcutaneous buprenorphine analgesia (0.05–0.1 mg/kg). The abdomen was opened by midline laparotomy approximately 2 cm in length and peritoneum cut open along the linea alba. The operation area was spread using a Colibri retractor in the peritoneal cavity, and lifted with a saline-moisturized cotton swab to reveal the hilum. The bile duct was exposed and separated from the portal vein and hepatic artery using a micro-serrations forceps. 7–0 suture was placed around the bile duct and secured with two surgical knots. A second cranial ligation was added in the same manner. Abdominal layers (peritoneum and cutis plus facia) were closed with running 6–0 sutures with absorbable suture material. Mice were weighed daily and monitored for humane endpoint.
Wholemount imaging
GB/EHBDs were resected, fileted to drain bile, and rinsed in cold PBS before fixation in 4% paraformaldehyde (PFA, Thermo Scientific, catalog# 28906) for four hrs RT or overnight at 4 C. Tissue was rinsed in PBS three times, blocked with 5% serum (secondary antibody host species) in 0.1% TritonX PBS for one hr, stained with primary unconjugated antibody for 2–4 hrs RT or overnight at 4C, washed three times, and stained with secondary antibody and conjugated primary antibody for 1–2 hrs at room temperature. Samples were washed three times and incubated in DAPI/PBS 1:1000 for ten min. Samples were washed four times and mounted on glass slides with glass coverslips in Vectashield (Vector Labs). Amplification of Flare25 RFP was not necessary. Samples were scanned using a NikonA1R laser scanning confocal (405, 488, 561, and 650 laser lines) with 20x air objective. High-resolution images were acquired in Galvano mode with pixel sizes of 300 nm. Bitplane Imaris v9.5 software package (Andor Technology PLC, Belfast, N. Ireland) was used for image analysis.
Thick section liver staining
Animals were perfused with PBS. Liver/GB was harvested and fixed in 4% PFA overnight at 4°C. Fixed tissues were washed with PBS, cut into 200 μm sections using a vibratome (Leica VT1000S), permeabilized with 0.2% triton X-100/0.3M glycine in PBS for 3–4 hrs at RT, and blocked with 0.3% triton X-100, 5% FBS, 0.5% BSA, and 5% serum (host species of the secondary antibody) in PBS overnight at 4°C. Samples were incubated with primary antibodies in 0.15% triton X-100, 7.5% FBS, 0.75% BSA, 3% serum in PBS at 4°C overnight, washed in 0.2% triton X-100 in PBS for 30 min 3–4 times, then incubated with secondary antibodies diluted in 0.15% triton X-100, 7.5% FBS, 0.75% BSA, 3% serum in PBS at 4°C overnight. Samples were washed in 0.2% triton X-100 in PBS for 30 min, 3–4 times followed by dehydration in ethanol gradation (20, 30, 50, 70, 95, 100%) for 5 min each. Samples were cleared by soaking in methyl salicylate for at least 10 min. Samples were scanned using NikonA1R laser scanning confocal (405, 488, 561, and 650 laser lines) and 16X/0.8 water immersion objective. Acquisition and analysis were performed as described above. For localization of IL-5+ lymphocytes in the liver and gall bladder a channel was generated and surfaced.
Imaging antibodies
Primary antibodies included Living Colors Rabbit anti-dsRed Polyclonal Pan Antibody (1:200; Takara), Goat anti-Type 1 Collagen Polyclonal Antibody (1:200, Southern Biotech), Rat anti-CD326/EpCam Monoclonal Antibody (1:200, clone G8.8, BD Pharmingen), and Rabbit anti-DCAMKL1 Monoclonal Antibody (1:300, clone EPR6085, Abcam). The following species-specific secondary antibodies were used at 1:300 dilution: Alexa Fluor 488 donkey anti-goat IgG (H+L) cross-adsorbed (Thermo Fisher Scientific), Alexa Fluor 488 donkey anti-rat IgG (H+L) cross-adsorbed (Thermo Fisher Scientific), Alexa Fluor 555 donkey anti-rabbit IgG (H+L) cross adsorbed (Thermo Fisher Scientific), Alexa Fluor 647 donkey anti-goat IgG (H+L) cross adsorbed (Thermo Fisher Scientific), and Alexa Fluor 647 donkey anti-rabbit IgG (H+L) cross adsorbed (Thermo Fisher Scientific). The directly conjugated eFluor 660 anti-Lyve1 monoclonal antibody (clone ALY7, eBioscience) was used at 1:200 dilution. The directly conjugated AF488 anti-CD326/EpCam monoclonal antibody (clone G8.8, Biolegend) was used at 1:200 dilution.
Flow cytometry
Single-cell suspensions were incubated with Fc Block (Bio X Cell, 2.4G2) and stained with antibodies to surface markers diluted in FWB. Cells were washed in FWB and resuspended in FWB containing DAPI (Roche) and flow cytometric counting beads (CountBright Absolute; Life Technologies) for live cell gating and counts. Cells for intracellular staining were washed in PBS and stained with Violet Live/Dead fixable stain (Life Technologies) according to manufacturer’s instructions. Transcription factor staining was done using the FoxP3/Transcription Factor Staining Buffer Set (eBiosciences) according to manufacturer’s instructions. Cells for DCLK1 staining were fixed in 4% PFA for 2–5 min, washed in FWB, and stained with rabbit anti-DCLK1 (Abcam; ab31704; 1:4000) antibody in perm/wash buffer from the FoxP3 staining buffer kit, followed by F(ab’)2 donkey anti-rabbit IgG-PE (Life Technologies; 1:1000) or goat anti-Rabbit AF488 (Invitrogen; 1:4000) for 10 min. Staining was done on ice. For co-staining of DCLK1 and endogenous IL-25 RFP, PFA fixation was performed for 30 seconds on ice. Samples were analyzed on a LSRFortessa (BD Biosciences) with five lasers (355 nm, 405 nm, 488 nm, 561 nm and 640 nm). Samples were FSC-A/SSC-A gated to exclude debris, FSC-H/FSC-A gated to select single cells, and gated to exclude dead cells. Data were analyzed with FlowJo 9/10 (Treestar/Flowjo).
Flow cytometry antibodies
The following flow cytometry antibodies were purchased from BioLegend: EpCam PerCypCy5.5 and BV711 (clone G8.8), Ly6G APCCy7 (1A8), CD11b BV711 (M1/70), CD45 BV785 (30-F11), Thy1 BV786 (30-H12), TCRgd PerCPCy5.5 (GL3), NK1.1 PECy7 (PK136), NKp46/CD335 PECy7 (29A1.4). Lineage-positive cells were excluded in ILC staining using the antibodies in Pacific Blue, from BioLegend: CD11c (N418), F4/80 (BM8), CD11b (M1/70), FCeR1 (MAR-1), CD19 (6D5), B220 (RA3–6B2), Ter-119, and GR-1. T1/ST2 Fitc/PE (DJ8) was purchased from MD Biosciences. The following antibodies were from eBioscience/Invitrogen: GATA3 ef660 (TWAJ), TCRb APC-ef780 (H57–597). The following antibodies were purchased from BD: CD64a/b AF647 (X54–5/7.1), RORgT PE (B2D), CD45 BUV395 (30-F11), Siglec-F BB515 (E50–2440), Thy1 BUV395 (3-H12)
RNAseq analysis
Tuft (live, RFP+ Epcam+) and non-tuft (live, RFP- Epcam+) cells from single-cell suspensions of GB and duodenal epithelium were sorted from three Flare25 mice into Dynabead mRNA Direct Micro Purification kit lysis buffer (Thermofisher); mRNA was isolated according to the manufacturer’s protocol. Amplified cDNA was prepared using the NuGen Ovation RNA-Seq system V2 kit according to the manufacturer’s protocol (NuGen Technologies). Sequencing libraries were generated using the Nextera XT library preparation kit with multiplexing primers according to manufacturer’s protocol (Illumina). Library fragment size distributions were assessed using the Bioanalyzer 2100 and the DNA high-sensitivity chip (Agilent Technologies). Library sequence quality was assessed by sequencing single-end 50 bp reads using the Illumina MiSeq platform and pooled for high-throughput sequencing on the Illumina HiSeq 4000 by using equal numbers of uniquely mapped reads (Illumina). Twelve samples per lane were multiplexed. Sequencing yielded a median read depth of 89.2 million reads per sample. The analytic pipeline included de-multiplexing raw sequencing results, trimming adapter sequences, and aligning to the reference genome. Sequence alignment and splice junction estimation was performed using STAR software (2.7.2b). For differential expression testing, the genomic alignments were restricted to unique Ensembl IDs (all protein coding mRNAs, coding and noncoding RNAs). STAR aggregated mappings on a per-gene basis were used as raw input for normalization by DESeq2. Differential expression analysis was performed in the R version 36.1 using DESeq2 version 1.26. Significance thresholds were FDR<0.05 and log fold change >1 or <−1. Unsupervised PCA was performed on the top 500 genes by variance.
Shared tuft cell genes were defined as genes in SI/GB tuft cells >2 log2FC relative to non-tuft cells from the same tissue, with an FDR <0.01, and a sum normalized read count >500 in SI or GB tuft cells (413 total genes). GB tuft cell exclusive gene signature was defined as genes with FDR<0.05 relative to SI tuft cells and GB non-tuft cells. Genes were further filtered to include those with a sum normalized read count >150 in GB tuft cells (555). GO Term and pathway analysis was performed using the ClusterProfiler package in R (66).
Single cell sequencing
GB/EHBDs from 5 mice per genotype were prepared using the total digest described above. Single-cell suspensions were stained with DAPI, and live cells (DAPI negative, FSC-A × SSC-A) were sorted using a MoFlo XDP (Beckman Coulter). Concentration was determined using CountBright beads on an LSRFortessa. Single-cell libraries from 37000 cells per sample were prepared with the Chromium Single Cell 3ʹ GEM, Library & Gel Bead Kit v3 (10 × Genomics PN-1000075) following the manufacturer’s protocol. The libraries were sequenced on the NovaSeq 6000 at the UCSF Institute for Human Genetics with the following number of cycles for each read:
| Read 1 | 28 |
| I7 Index | 8 |
| I5 Index | 0 |
| Read 2 | 91 |
Data were analyzed in Seurat V3 using SCTransform normalization: min.cells =3, min.features=200; nFeature_RNA >= 300; nFeature_RNA< 5000; nCount_RNA < 20000; percent mitochondrial reads < 15; percent hemoglobin reads 0.1. Percent mitochondrial reads was regressed out of clustering/aggregation. After clustering of all cells at resolution 0.6, epithelial or immune clusters were identified by canonical markers and subclustered at resolution 0.3. Contaminating doublets containing were removed and the populations were clustered again before DEG analysis for cluster-defining genes and differential expression between genotypes. Tuft cells were subclustered at resolution 0.3 and were only found in cells from control mice. GO Term/pathway analysis was performed using the ClusterProfiler package in R (66).
Statistical analysis
Statistical analysis was performed using Prism 6/8 (GraphPad Software). Figures display mean ± S.E.M. p values were calculated using unpaired two-tailed student’s T test for comparison between two groups or one-way ANOVA for multiple comparisons. Intragroup variation was not assessed. Transcriptomic data was analyzed and visualized in R, using the DESeq2 and Seurat packages for calculation of differential gene expression as described above.
Supplementary Material
Acknowledgments:
The authors thank Jimmy Ye and members of the Institute for Human Genetics for assistance with 10X sequencing. Embryo injections were performed at the Gladstone Institutes. The UCSF Gnotobiotic Core Facility and Director Jessie Turnbaugh provided GF mice and performed GF experiments. The West Coast Metabolomics Core performed bile acid analysis. Z. Wang, M. Ji, and M. Consengco provided technical assistance. The authors acknowledge Drs. Clifford Lowell and Averil Ma at UCSF for their critical commentary.
Funding:
National Institutes of Health grant R01AI026918 (RML)
National Institutes of Health grant R01HL128903 (RML)
National Institutes of Health grant F32 DK121476 (CEO)
National Institutes of Health grant T32 DK007007 (CEO)
National Institutes of Health grant F32HL140868 (MEK)
National Institutes of Health grant T32HL007185 (MEK),
The Howard Hughes Medical Institute (RML)
The A.P. Giannini Foundation (MEK)
The Diabetes Research Center (RML)
Sandler Asthma Basic Research Center (RML)
Swiss National Science Foundation P300PA_171591 (CS)
Swiss National Science Foundation P4P4PM_180832 (CS)
Peter Hans Hofschneider Professorship for Molecular Medicine (CS)
Footnotes
Competing interests:
The authors have no competing interests to declare.
List of Supplementary Materials
Fig. S1. Biliary tuft cells express tuft cell- and tissue-specific gene signature and are not dependent on type 2 cytokines.
Table S1. RNAseq top 1000 genes by variance
Fig. S2. Biliary tuft cells exhibit developmental regulation.
Fig. S3. Biliary tuft cell abundance is modulated by bile acids.
Fig. S4. Pou2f3−/− mice have increased biliary neutrophil infiltration under homeostatic conditions.
Fig. S5. Total biliary scRNA-seq reveals activated state of biliary neutrophils and epithelium in the absence of tuft cells
Fig. S6. The microbiome drives biliary neutrophil influx.
Data and materials availability:
RNAseq and single cell sequencing data will be uploaded to GEO. All other data are available in the main text or supplementary materials. Requests for reagents should be directed to RML.
References
- 1.Wahlstrom A, Sayin SI, Marschall HU, Backhed F, Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab 24, 41–50 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Gorham F. W. a. I., A. C., General function of the gall bladder from the evolutionary standpoint. American Journal of Digestive Diseases and Nutrition 22, (1938). [Google Scholar]
- 3.Higashiyama H, Uemura M, Igarashi H, Kurohmaru M, Kanai-Azuma M, Kanai Y, Anatomy and development of the extrahepatic biliary system in mouse and rat: a perspective on the evolutionary loss of the gallbladder. Journal of anatomy 232, 134–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia W, Xie G, Jia W, Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 15, 111–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen PL, Ghallab A, Vartak N, Reif R, Schaap FG, Hampe J, Hengstler JG, The ascending pathophysiology of cholestatic liver disease. Hepatology (Baltimore, Md.) 65, 722–738 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang HE, Locksley RM, Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabris L, Spirli C, Cadamuro M, Fiorotto R, Strazzabosco M, Emerging concepts in biliary repair and fibrosis. American journal of physiology. Gastrointestinal and liver physiology 313, G102–g116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Leary CE, Schneider C, Locksley RM, Tuft Cells-Systemically Dispersed Sensory Epithelia Integrating Immune and Neural Circuitry. Annual review of immunology, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schutz B, Jurastow I, Bader S, Ringer C, von Engelhardt J, Chubanov V, Gudermann T, Diener M, Kummer W, Krasteva-Christ G, Weihe E, Chemical coding and chemosensory properties of cholinergic brush cells in the mouse gastrointestinal and biliary tract. Frontiers in physiology 6, 87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Moltke J, Ji M, Liang HE, Locksley RM, Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luciano L, Reale E, Presence of brush cells in the mouse gallbladder. Microscopy research and technique 38, 598–608 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Luciano L, Castellucci M, Reale E, The brush cells of the common bile duct of the rat. This section, freeze-fracture and scanning electron microscopy. Cell and tissue research 218, 403–420 (1981). [DOI] [PubMed] [Google Scholar]
- 13.Nevalainen TJ, Ultrastructural characteristics of tuft cells in mouse gallbladder epithelium. Acta anatomica 98, 210–220 (1977). [DOI] [PubMed] [Google Scholar]
- 14.Schutz B, Ruppert AL, Strobel O, Lazarus M, Urade Y, Buchler MW, Weihe E, Distribution pattern and molecular signature of cholinergic tuft cells in human gastrointestinal and pancreatic-biliary tract. Scientific reports 9, 17466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider C, O’Leary CE, von Moltke J, Liang HE, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, Locksley RM, A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 174, 271–284.e214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, Erle DJ, Anderson MS, Locksley RM, Raftery D, von Moltke J, Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 49, 33–41.e37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P, Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, Artis D, Garrett WS, Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science (New York, N.Y.) 351, 1329–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ualiyeva S, Hallen N, Kanaoka Y, Ledderose C, Matsumoto I, Junger WG, Barrett NA, Bankova LG, Airway brush cells generate cysteinyl leukotrienes through the ATP sensor P2Y2. Science immunology 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perniss A, Liu S, Boonen B, Keshavarz M, Ruppert AL, Timm T, Pfeil U, Soultanova A, Kusumakshi S, Delventhal L, Aydin Ö, Pyrski M, Deckmann K, Hain T, Schmidt N, Ewers C, Günther A, Lochnit G, Chubanov V, Gudermann T, Oberwinkler J, Klein J, Mikoshiba K, Leinders-Zufall T, Offermanns S, Schütz B, Boehm U, Zufall F, Bufe B, Kummer W, Chemosensory Cell-Derived Acetylcholine Drives Tracheal Mucociliary Clearance in Response to Virulence-Associated Formyl Peptides. Immunity 52, 683–699.e611 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Bankova LG, Dwyer DF, Yoshimoto E, Ualiyeva S, McGinty JW, Raff H, von Moltke J, Kanaoka Y, Frank Austen K, Barrett NA, The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Science immunology 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, Fries AC, Lwin WW, Wigton EJ, Parent AV, Kyewski B, Erle DJ, Hogquist KA, Steinmetz LM, Locksley RM, Anderson MS, Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Toth B, Goldberg O, Itzkovitz S, Taylor N, Jay P, Zimmermann VS, Abramson J, Amit I, Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature, (2018). [DOI] [PubMed] [Google Scholar]
- 24.Lucas B, White AJ, Cosway EJ, Parnell SM, James KD, Jones ND, Ohigashi I, Takahama Y, Jenkinson WE, Anderson G, Diversity in medullary thymic epithelial cells controls the activity and availability of iNKT cells. Nature communications 11, 2198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, Locksley RM, Tissue signals imprint ILC2 identity with anticipatory function. Nature immunology 19, 1093–1099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinty JW, Ting HA, Billipp TE, Nadjsombati MS, Khan DM, Barrett NA, Liang HE, Matsumoto I, von Moltke J, Tuft-Cell-Derived Leukotrienes Drive Rapid Anti-helminth Immunity in the Small Intestine but Are Dispensable for Anti-protist Immunity. Immunity 52, 528–541.e527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, Locksley RM, Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447, 92–96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlgren MW, Jones SW, Cautivo KM, Dubinin A, Ortiz-Carpena JF, Farhat S, Yu KS, Lee K, Wang C, Molofsky AV, Tward AD, Krummel MF, Peng T, Molofsky A, Adventitial Stromal Cells Define Group 2 Innate Lymphoid Cell Tissue Niches. Immunity 50, 707–722.e706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita J, Ohmoto M, Yamaguchi T, Matsumoto I, Hirota J, Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PloS one 12, e0189340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvaro D, Mancino MG, Glaser S, Gaudio E, Marzioni M, Francis H, Alpini G, Proliferating cholangiocytes: a neuroendocrine compartment in the diseased liver. Gastroenterology 132, 415–431 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Lamote J, Willems G, DNA synthesis, cell proliferation index in normal and abnormal gallbladder epithelium. Microscopy research and technique 38, 609–615 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, Brandtner A, Setlik W, Remotti H, Muley A, Chen X, May R, Houchen CW, Fox JG, Gershon MD, Quante M, Wang TC, Long-lived intestinal tuft cells serve as colon cancer-initiating cells. The Journal of clinical investigation 124, 1283–1295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui JY, Aleksunes LM, Tanaka Y, Fu ZD, Guo Y, Guo GL, Lu H, Zhong XB, Klaassen CD, Bile acids via FXR initiate the expression of major transporters involved in the enterohepatic circulation of bile acids in newborn mice. American journal of physiology. Gastrointestinal and liver physiology 302, G979–996 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khandekar G, Llewellyn J, Kriegermeier A, Waisbourd-Zinman O, Johnson N, Du Y, Giwa R, Liu X, Kisseleva T, Russo PA, Theise ND, Wells RG, Coordinated development of the mouse extrahepatic bile duct: Implications for neonatal susceptibility to biliary injury. J Hepatol 72, 135–145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Best N, Rolle-Kampczyk U, Schaap FG, Basic M, Olde Damink SWM, Bleich A, Savelkoul PHM, von Bergen M, Penders J, Hornef MW, Bile acids drive the newborn’s gut microbiota maturation. Nature communications 11, 3692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Binz J, Numerick MJ, Dennis S, Luo G, Desai B, MacKenzie KI, Mansfield TA, Kliewer SA, Goodwin B, Jones SA, Hepatoprotection by the farnesoid X receptor agonist GW4064 in rat models of intra- and extrahepatic cholestasis. The Journal of clinical investigation 112, 1678–1687 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Funabashi M, Grove TL, Wang M, Varma Y, McFadden ME, Brown LC, Guo C, Higginbottom S, Almo SC, Fischbach MA, A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White BA, Lipsky RL, Fricke RJ, Hylemon PB, Bile acid induction specificity of 7 alpha-dehydroxylase activity in an intestinal Eubacterium species. Steroids 35, 103–109 (1980). [DOI] [PubMed] [Google Scholar]
- 39.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM, Bile acids: natural ligands for an orphan nuclear receptor. Science (New York, N.Y.) 284, 1365–1368 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Li-Hawkins J, Gåfvels M, Olin M, Lund EG, Andersson U, Schuster G, Björkhem I, Russell DW, Eggertsen G, Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. The Journal of clinical investigation 110, 1191–1200 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosen H, Reshef A, Maeda N, Lippoldt A, Shpizen S, Triger L, Eggertsen G, Bjorkhem I, Leitersdorf E, Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. The Journal of biological chemistry 273, 14805–14812 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Hong JY, Rockwell CE, Copple BL, Jaeschke H, Klaassen CD, Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int 32, 58–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuzaki Y, Bouscarel B, Ikegami T, Honda A, Doy M, Ceryak S, Fukushima S, Yoshida S, Shoda J, Tanaka N, Selective inhibition of CYP27A1 and of chenodeoxycholic acid synthesis in cholestatic hamster liver. Biochimica et biophysica acta 1588, 139–148 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Mariotti V, Strazzabosco M, Fabris L, Calvisi DF, Animal models of biliary injury and altered bile acid metabolism. Biochimica et biophysica acta, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engblom C, Pfirschke C, Zilionis R, Da Silva Martins J, Bos SA, Courties G, Rickelt S, Severe N, Baryawno N, Faget J, Savova V, Zemmour D, Kline J, Siwicki M, Garris C, Pucci F, Liao HW, Lin YJ, Newton A, Yaghi OK, Iwamoto Y, Tricot B, Wojtkiewicz GR, Nahrendorf M, Cortez-Retamozo V, Meylan E, Hynes RO, Demay M, Klein A, Bredella MA, Scadden DT, Weissleder R, Pittet MJ, Osteoblasts remotely supply lung tumors with cancer-promoting SiglecF(high) neutrophils. Science (New York, N.Y.) 358, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfirschke C, Engblom C, Gungabeesoon J, Lin Y, Rickelt S, Zilionis R, Messemaker M, Siwicki M, Gerhard GM, Kohl A, Meylan E, Weissleder R, Klein AM, Pittet MJ, Tumor-Promoting Ly-6G(+) SiglecF(high) Cells Are Mature and Long-Lived Neutrophils. Cell reports 32, 108164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, Rogel N, Burgin G, Tsankov AM, Waghray A, Slyper M, Waldman J, Nguyen L, Dionne D, Rozenblatt-Rosen O, Tata PR, Mou H, Shivaraju M, Bihler H, Mense M, Tearney GJ, Rowe SM, Engelhardt JF, Regev A, Rajagopal J, A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A, A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fachi JL, Sécca C, Rodrigues PB, Mato FCP, Di Luccia B, Felipe JS, Pral LP, Rungue M, Rocha VM, Sato FT, Sampaio U, Clerici M, Rodrigues HG, Câmara NOS, Consonni SR, Vieira AT, Oliveira SC, Mackay CR, Layden BT, Bortoluci KR, Colonna M, Vinolo MAR, Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. The Journal of experimental medicine 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, Guttentag SH, Kolls JK, Oliver PM, Bushman FD, Worthen GS, Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. The Journal of clinical investigation 122, 974–986 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moschen AR, Adolph TE, Gerner RR, Wieser V, Tilg H, Lipocalin-2: A Master Mediator of Intestinal and Metabolic Inflammation. Trends in endocrinology and metabolism: TEM 28, 388–397 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA, Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. Journal of immunology (Baltimore, Md. : 1950) 170, 3233–3242 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Peters AL, Luo Z, Li J, Mourya R, Wang Y, Dexheimer P, Shivakumar P, Aronow B, Bezerra JA, Single cell RNA sequencing reveals regional heterogeneity of hepatobiliary innate lymphoid cells in a tissue-enriched fashion. PloS one 14, e0215481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sampaziotis F, Muraro D, Tysoe OC, Sawiak S, Beach TE, Godfrey EM, Upponi SS, Brevini T, Wesley BT, Garcia-Bernardo J, Mahbubani K, Canu G, Gieseck R 3rd, Berntsen NL, Mulcahy VL, Crick K, Fear C, Robinson S, Swift L, Gambardella L, Bargehr J, Ortmann D, Brown SE, Osnato A, Murphy MP, Corbett G, Gelson WTH, Mells GF, Humphreys P, Davies SE, Amin I, Gibbs P, Sinha S, Teichmann SA, Butler AJ, See TC, Melum E, Watson CJE, Saeb-Parsy K, Vallier L, Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science (New York, N.Y.) 371, 839–846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eckmann L, Kagnoff MF, Intestinal mucosal responses to microbial infection. Springer Semin Immunopathol 27, 181–196 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nuñez G, Camp JG, Hattori M, Umesaki Y, Honda K, Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salzman AL, Eaves-Pyles T, Linn SC, Denenberg AG, Szabó C, Bacterial induction of inducible nitric oxide synthase in cultured human intestinal epithelial cells. Gastroenterology 114, 93–102 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Kolios G, Rooney N, Murphy CT, Robertson DA, Westwick J, Expression of inducible nitric oxide synthase activity in human colon epithelial cells: modulation by T lymphocyte derived cytokines. Gut 43, 56–63 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molinero N, Ruiz L, Milani C, Gutiérrez-Díaz I, Sánchez B, Mangifesta M, Segura J, Cambero I, Campelo AB, García-Bernardo CM, Cabrera A, Rodríguez JI, González S, Rodríguez JM, Ventura M, Delgado S, Margolles A, The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 7, 100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye F, Shen H, Li Z, Meng F, Li L, Yang J, Chen Y, Bo X, Zhang X, Ni M, Influence of the Biliary System on Biliary Bacteria Revealed by Bacterial Communities of the Human Biliary and Upper Digestive Tracts. PloS one 11, e0150519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM, Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nature immunology 13, 58–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohrs M, Ledermann B, Köhler G, Dorfmüller A, Gessner A, Brombacher F, Differences between IL-4- and IL-4 receptor alpha-deficient mice in chronic leishmaniasis reveal a protective role for IL-13 receptor signaling. Journal of immunology (Baltimore, Md. : 1950) 162, 7302–7308 (1999). [PubMed] [Google Scholar]
- 63.Fallon PG, Ballantyne SJ, Mangan NE, Barlow JL, Dasvarma A, Hewett DR, McIlgorm A, Jolin HE, McKenzie AN, Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. The Journal of experimental medicine 203, 1105–1116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang HE, Locksley RM, Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity 50, 1425–1438.e1425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Leary CE, Feng X, Cortez VS, Locksley RM, Schneider C, Interrogating the Small Intestine Tuft Cell-ILC2 Circuit Using In Vivo Manipulations. Curr Protoc 1, e77 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu G, Wang LG, Han Y, He QY, clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq and single cell sequencing data will be uploaded to GEO. All other data are available in the main text or supplementary materials. Requests for reagents should be directed to RML.


