Abstract
The expression of the reverse transsulfuration enzyme cystathionine-β-synthase (CBS) is markedly increased in many forms of cancer, including colorectal, ovarian, lung, breast and kidney, while in other cancers (liver cancer and glioma) it becomes downregulated. According to the clinical database data in high-CBS-expressor cancers (e.g. colon or ovarian cancer), high CBS expression typically predicts lower survival, while in the low-CBS-expressor cancers (e.g. liver cancer), low CBS expression is associated with lower survival. In the high-CBS expressing tumor cells, CBS, and its product hydrogen sulfide (H2S) serves as a bioenergetic, proliferative, cytoprotective and stemness factor; it also supports angiogenesis and epithelial-to-mesenchymal transition in the cancer microenvironment. The current article reviews the various tumor-cell-supporting roles of the CBS/H2S axis in high-CBS expressor cancers and overviews the anticancer effects of CBS silencing and pharmacological CBS inhibition in various cancer models in vitro and in vivo; it also outlines potential approaches for biomarker identification, to support future targeted cancer therapies based on pharmacological CBS inhibition.
Keywords: Hydrogen sulfide, Mitochondria, Angiogenesis, Signalling, Stemness, Colon cancer
List of abbreviations
- ACLY
ATP citrate lyase
- Akt
protein kinase B = PKB
- ALT
glutamate pyruvate transaminase = GPT
- AMPK
AMP-activated protein kinase
- AOAA
aminooxyacetic acid
- AP-1
activating protein-1
- ATF
Activating Transcription Factor
- ATP
adenosine triphosphate
- BPH1
a benign prostatic hyperplasia line
- BRCA
breast cancer gene
- CaMKK2
calcium/calmodulin-activated protein kinase 2
- cAMP
cyclic adenosine monophosphate
- CAT
cysteine aminotransferase
- CBS
cystathionine β-synthase
- cGMP
cyclic guanosine monophosphate
- CNS
central nervous system
- CO
carbon monoxide
- COAD
Colon Adenocarcinoma Dataset
- CREB
cAMP-response element binding protein
- CSE
cystathionine γ-lyase
- CXCR
CXC chemokine receptor 4
- DNA
deoxyribonucleic acid
- DOPA
dihydroxyphenylalanine
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- eNOS
endothelial nitric oxide synthase
- EREG
epiregulin
- ERK
extracellular signal-regulated protein kinase
- ETHE1
persulfide dioxygenase 1 (=ethylmalonic encephalopathy protein 1)
- EXOG
5′-exonuclease
- G6PD
glucose-6-phosphate dehydrogenase
- GABA
gamma-aminobutyric acid
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GCL-C
glutamate cysteine ligase (C subunit)
- GEPIA
interactive web application for gene expression analysis
- GI
growth inhibition
- GOT
aspartate aminotransferase
- GPT2
glutamic pyruvate transaminase 2
- GSH
glutathione
- GSR
glutathione reductase
- H2S
hydrogen sulfide
- H2Sn
polysulfide
- HER2
human epidermal growth factor receptor 2
- HIF-1α
hypoxia-inducible factor-1α
- HMEC
human mammary epithelial cells
- HMPSNE
2-[(4-hydroxy-6-methylpyrimidin-2-yl)sulfanyl]-1-(naphthalen-1-yl)ethan-1-one
- IC50
half maximal inhibitory concentration
- kDa
kilodaltons
- KIRC
kidney renal clear cell carcinoma
- LDH
lactate dehydrogenase
- LKB1
liver kinase B1
- MET
mesenchymal-to-epithelial transition
- MDR-1
multidrug resistance protein-1
- MFN2
mitofusin 2
- MMP-2
matrix metalloproteinase-2
- mRNA
messenger ribonucleic acid
- 3-MST
3-mercaptopyruvate sulfurtransferase
- mTOR
mechanistic target of rapamycin
- NAD
nicotinamide adenine dinucleotide
- NAMPT
nicotinamide phoshophoribosyltransferase
- NANOG
homeobox transcription factor nanog
- NCI
National Cancer Institute
- NK
natural killer cells
- NO
nitric oxide
- Nrf2
NF-E2 related factor-2
- OCT4
octamer-binding transcription factor
- p38MAPK
p38 mitogen-activated protein kinase
- PAG
propargylglycine
- PCR
polymerase chain reaction
- PDE2A
cGMP-dependent 3′,5′-cyclic phosphodiesterase
- P-gp
P-glycoprotein
- PKC
protein kinase C
- PKM2
pyruvate kinase
- PLP
pyridoxal-5′-phosphate
- PTEN
phosphatase and tensin homolog
- RNA
ribonucleic acid
- ROS
reactive oxygen species
- rpL3
ribosomal protein L3
- SAM
S-adenosyl methionine
- SIRT1
sirtuin 1
- STAT3
signal transducer and activator of transcription 3
- TCGA
The Cancer Genome Atlas
- TFAM
mitochondrial transcription factor A
- TNBC
triple negative breast cancer
- TYMS
thymidylate synthetase
- VCAM
cytokine-induced endothelial adhesion molecule
- VEGF
vascular endothelial growth factor
- VHL
Von Hippel-Lindau
1. Introduction
Hydrogen sulfide (H2S), for centuries, has been viewed as a poisonous gas and environmental toxin. However, studies conducted over the last 25 years demonstrate that when H2S is produced endogenously by mammalian cells and tissues, it acts as a regulatory gasotransmitter. The physiological roles of H2S and the transition of the H2S field from environmental toxicology to mammalian biology, physiology and pathophysiology have been covered in multiple recent reviews [[1], [2], [3], [4], [5]]. Mammalian cells generate H2S in a regulated fashion by cystathionine γ-lyase (CSE), cystathionine β-synthase (CBS), and 3-mercaptopyruvate sulfurtransferase (3-MST). H2S, in turn, acts as an autocrine cellular regulator in the same cell where it was generated, but it can also cross cell membranes and affect neighboring cells to serve as a paracrine modulator [[1], [2], [3], [4], [5]].
The current article focuses on the emerging roles of CBS – one of the 3 mammalian H2S-producing enzymes – in cancer. This rapidly-growing area of basic and translational research was initiated in 2013 by the demonstration of increased CBS expression and H2S generation in surgically excised tissues of colon cancer patients (compared to non-cancerous surrounding tissues), and the demonstration of the functional tumor-supportive bioenergetic and pro-angiogenic roles of CBS-derived H2S in various cell-based and animal models of colon cancer [6]. A follow-up report, in ovarian cancer, demonstrated a similar upregulation of CBS in clinical ovarian cancer specimens and described various tumor-supporting roles of CBS-derived H2S in various ovarian cancer models in vitro and in vivo [7]. Over the subsequent decade, over 50 papers have appeared implicating the various tumor-supporting roles of tumor-cell-derived H2S, and demonstrated the anticancer effects of inhibition of H2S biosynthesis in many forms of cancer. Depending on the cancer type and the species, the source of H2S generation in various cancer cells can be CBS, and/or CSE and/or 3-MST. However, several forms of cancer were also identified where CBS expression and H2S production was found to be lower than in the surrounding normal tissue.
The current article overviews this emerging field, primarily focusing on the role of CBS in high-CBS expressor tumors, and the effects of silencing or pharmacological inhibition of this enzyme in cancer cells and in tumor-bearing mice. However, in the final sections of the article, the broader tumor-supporting roles of H2S (generated by various enzymatic sources) will also be covered. The article will start out with the clinical data, showing CBS expression in patient-derived materials, and the presence of various clinical correlations (e.g. survival or drug responsiveness) with CBS expression, followed by preclinical studies in cancer cells and tumor-bearing mouse models focusing on the effect of genetic or pharmacological modulation of CBS in cancer.
2. CBS
CBS is a well-characterized mammalian enzyme, which is a central constituent of the reverse transsulfuration pathway. When narrowly defined, transsulfuration refers to the transfer of sulfur from homocysteine to cysteine, and serves the physiological role of elimination of homocysteine, a cytotoxic metabolite and cardiovascular risk factor. Several of the biochemical pathways that CBS catalyzes generate H2S. This phenomenon was already observed over 75 years, but the physiological importance of CBS-derived H2S generation has only been recognized over the last two decades, and the role of this pathway in cancer only been recognized in 2013 [[4], [5], [6]].
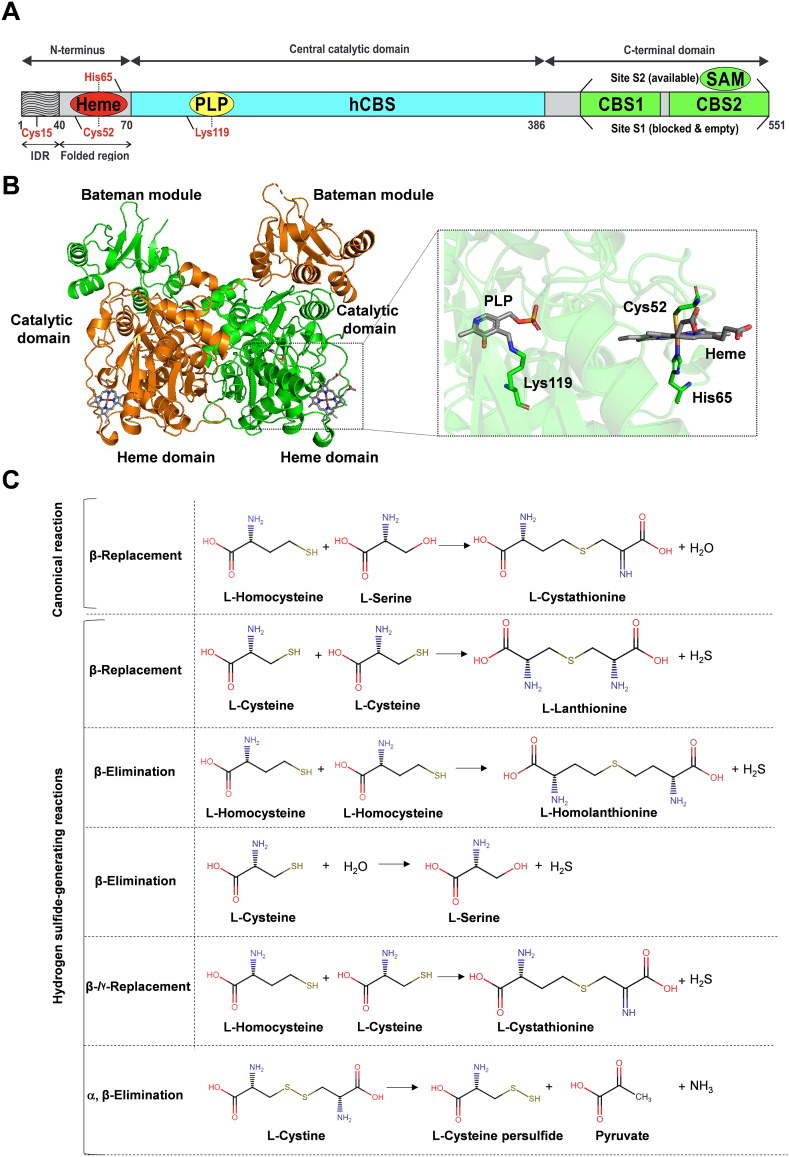
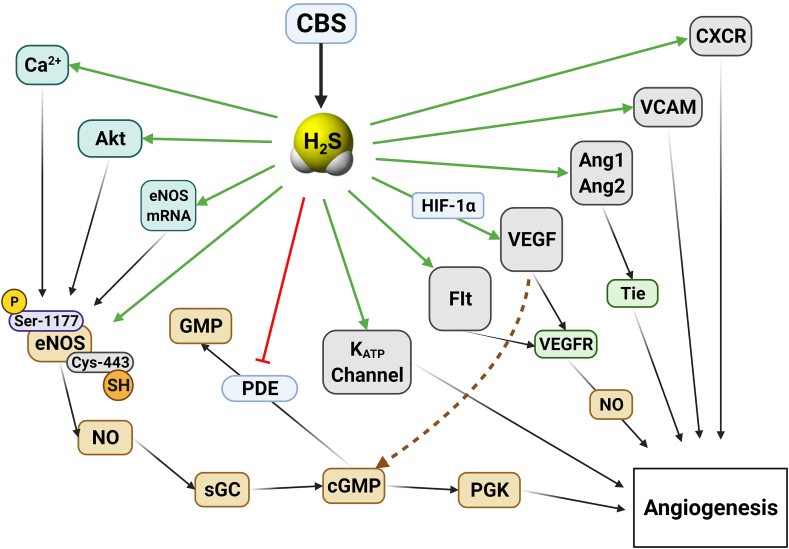
The organization and the biochemistry of CBS are covered in specialized review articles [[8], [9], [10], [11], [12]]. Human CBS is a tetramer comprising of 551 amino acids with a subunit molecular weight of 63 kDa (Fig. 1A and B). Each subunit binds its various substrates (see below) as well as three additional ligands: pyridoxal-5′-phosphate (PLP, a co-factor required for its enzymatic activity) and heme, the function of which is incompletely understood. From the multiple reactions that CBS catalyzes, the canonical reaction is a β-replacement reaction of l-serine with l-homocysteine forming l-cystathionine and water (i.e. this reaction does not yield H2S). CBS catalyzes multiple H2S-yielding reactions, including the condensation of l-cysteine and l-homocysteine to form l-cystathionine and H2S, the condensation of two l-cysteine molecules to form l-lanthionine and H2S, the condensation of two l-homocysteine molecules to form l-homolanthionine and H2S, the conversion of l-cysteine and water to l-serine and H2S, the β-γ replacement reaction of l-homocysteine with l-cysteine forming l-cystathionine and H2S and the formation of l-cysteine persulfide from l-cystine – where l-cysteine persulfide, in subsequent steps, releases H2S in the presence of reductants or transfers the sulfane sulfur moiety to acceptor proteins (Fig. 1C). In the cellular environment, the production of H2S is clearly detectable in cells and tissues that express CBS; when CBS is pharmacologically inhibited or silenced, the cellular H2S generation is diminished; when CBS overexpression is achieved in a cell, the result is increased H2S generation (see below). In the body, under physiological conditions, the liver and the brain are major CBS-expressor organs; the H2S generated by these organs is likely to contribute to the systemic (circulating) H2S ‘pools’ in the body [3,12]. In the current section, we will focus on the aspects of CBS that are most relevant in the context of CBS cancer biology, such as (a) its transcriptional regulation; (b) its posttranscriptional regulation including its proteolytic degradation; and (c) its intracellular distribution.
Fig. 1.
Structure and function of CBS. A) The organization of CBS. B) Crystal structure of the Δ516-525 human CBS homodimer (PDB# 4COO). Human CBS is architecturally organized in three regions: the Bateman module, the catalytic domain and the heme-binding domain. The engineered hCBS Δ516-525 is catalytically identical to the full-length native enzyme even if it lacks a loop consisting of 10 amino acid residues from the C-terminal regulatory domain. hCBS Δ516-525 forms dimers, rather then tetramers or higher order oligomers typical of the full-length CBS, that are colored in green and orange, respectively. The PLP and the heme cofactors are shown in sticks. The inset represents a zoom-in view into the catalytic (PLP) and regulatory (heme) sites. The PLP forms an internal aldimine intermediate via the Schiff base bond with the amino group of Lys119, while the heme is coordinated by Cys52 and His65. Figures were generated with PyMol 2.5. C) Key biochemical reactions catalyzed by hCBS. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The CBS gene is located on human chromosome 21 in the subtelomeric region q.22.3, with 23 exons, 15 of which are coding for the enzyme. CBS has two principal promoters -1a and -1b. Both promoters are rich in GC and contain numerous putative binding sites for transcription factors and an estrogen receptor binding site. CBS basal transcription is regulated by specific protein Sp1 and Sp3, upstream stimulatory factor 1 and nuclear factor-Y on the -1b promoter [12,13]. CBS mRNA induction has been shown to occur in response to the transcription factor Nrf2 [14], in response to several hormones including 1,25-dihydroxyvitamin D3 (the biologically active form of vitamin D) [15], various estrogen receptor ligands [16] and glucocorticoids [17]. Certain physiological conditions can also upregulate CBS expression in cell culture models. These conditions include cell growth and cell proliferation itself – at least in part due to the stimulatory effect of serum and/or various growth factors in the culture medium [18] and hypoxia – at least in part due to the action of hypoxia-inducible factor α and hypoxia-inducible factor β on the hypoxia-response element in the CBS promoter [19]. Importantly, CBS expression is also known to be regulated by the methylation status of CpG islands in its promoter [[20], [21], [22], [23], [24], [25], [26]]. Some of the mechanisms discussed above – alone or in combination – may contribute to the upregulation of CBS mRNA and CBS protein in various cancers (see below). Interestingly, the chemotherapeutic agent 5-fluorouracil, at relatively high concentrations, has also been demonstrated to suppress the expression of CBS; the underlying mechanism involves the induction of the proapoptotic factor ribosomal protein L3 (rpL3), and consequent suppression of the binding of Sp1 to the CBS promoter [27].
Importantly, the levels of CBS protein are not only regulated via transcriptional processes, but also post-transcriptionally. One of these mechanisms is ubiquitination, a common post-translational modification of cellular proteins, which is intrinsically linked to proteosomal protein degradation. Lys72 and Lys481 are two significant ubiquitination sites on human CBS [28]. The role of ubiquitination and proteosomal protein degradation in the regulation of CBS levels is highlighted by studies of the Kruger group who demonstrated that pharmacological inhibition of proteosomal activity can increase cellular CBS protein levels [29]. CBS is also subject to various posttranscriptional modifications – such as phosphorylation, glutathionylation and SUMOylation. From these modifications, SUMOylation may affect the stability of CBS [12,30]. CBS stability is also affected by a mitochondrial class of proteases, called Lon proteases, which regulate CBS degradation in an O2-dependent manner [31]. One of the implications of significant post-transcriptional regulation of CBS for the field of cancer is that correlation analyses based on cancer transcriptome databases (which are based on mRNA and not protein levels) may not always or not fully reflect the actual CBS protein levels in cancers. This topic will be further discussed in the subsequent sections.
Proteolytic modification is another, relatively poorly studied, but potentially important regulatory mechanism of CBS, with implications for various pathophysiological conditions, including cancer. In 1984 Skovby, Kraus, and Rosenberg have noticed that —in addition to the regular 63 kDa Mw form of CBS – liver tissue also contains a 48 kDa form of CBS, which appears to be the product of limited proteolysis [32]. This form of the enzyme has been designated early on as the “evolutionarily conserved active core of CBS” [[33], [34], [35]]. Subsequent studies demonstrated the appearance of this truncated form in cells and animals exposed to pro-inflammatory conditions [36] or to hypoxia [37]. The 45 kDa CBS is also present in various cancer cells [38] and in several brain regions of rats in a Down syndrome model that contains a triplicated (extra) copy of CBS [39]. The truncated form of CBS no longer contains its allosteric regulatory domain and is in a constitutively active (“hyperactive”) form capable of higher rate of H2S generation than the normal isoform [33,34]. It is conceivable that CBS cleavage is relevant for the biology of cancer cells – which are often exposed to local inflammatory conditions.
CBS is primarily a cytosolic protein, but it has been also found in various other intracellular components, and can undergo intracellular redistribution under certain conditions. For instance, in response to hypoxia or ischemia, CBS can translocate into the mitochondria, at least in part due to the consequence of the regulation of mitochondrial CBS stability by Lon proteases [31]. CBS has also been shown to enter the nucleus, perhaps as a consequence of its SUMOylation [30]. In cancer cells, both cytosolic and mitochondrial CBS has been observed already in the initial studies published in 2013 [6,7] and confirmed subsequently by multiple studies (see below). Since H2S in mitochondria stimulates a multitude of processes that can support cancer cell bioenergetics and viability [[40], [41], [42], [43], [44], [45], [46], [47]] – the mitochondrial translocation of CBS may be important in the regulation of cancer cell bioenergetics and survival.
3. CBS expression in various human cancers: correlation with clinical outcomes
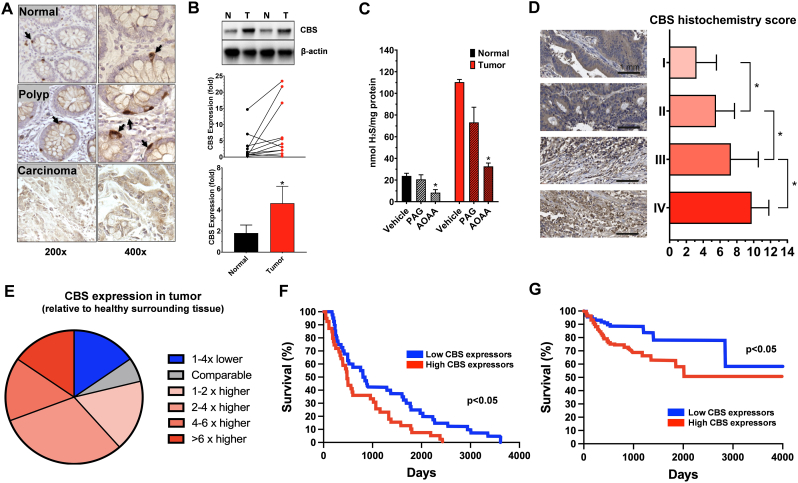
Colon cancer. Increased CBS expression in cancer was first demonstrated, in a limited number of clinical specimens obtained from patients undergoing colorectal tumor resection (n = 3). Western blotting studies demonstrated higher expression of CBS (but not CSE or 3-MST) in the tumor tissue, as compared to the surrounding nominally “normal” tissue which was also removed during surgery [6].
Studies focusing on the evolution of CBS expression in the context of colorectal polyp formation and carcinogenesis revealed (once again, only in a limited number of clinical samples) that CBS protein expression – quantified by immunohistochemistry as well as Western blotting – is low in healthy colonic mucosa, but gradually increases as the epithelial cells are transformed into polyps, hyperplastic polyps, tubular adenoma (dysplasia), and adenocarcinoma (in situ) (Fig. 2A) [48].
Fig. 2.
CBS is upregulated in human colon cancer and correlates with worse clinical prognosis. A) Formalin-fixed paraffin-embedded sections of normal colonic mucosa, hyperplastic polyp and adenocarcinoma, stained with CBS antibodies showing a gradual increase in CBS expression corresponding to the severity of the disease. The panel was adapted from data published in Ref. [48]). B). CBS expression in human colon cancer biopsies compared to normal surrounding tissue. Representative western blots and summary of expression data are shown (mean ± SEM). Arbitrary relative densitometry units were normalized with β-actin using image analysis software. ∗p < 0.05 T (tumor) vs. N (normal surrounding tissue); n = 15. The panel was redrawn from data presented in Ref. [49]. C) Effect of the CBS/CSE inhibitor AOAA (1 mM) and the CSE inhibitor PAG (3 mM) on H2S production in homogenates of a colorectal cancer and patient-matched normal colonic tissue. Data are presented as mean ± SEM of 3 independent experiments. ∗p < 0.05 shows significant inhibition of H2S production. The panel was redrawn from data presented in Ref. [6]. D) Representative images of CBS immunohistochemical staining in different-stage tumor tissues (n = 90) in the microarray. Immunohistochemical score for CBS in different-stage colon tumor tissues in the microarray. (stage 1 = 28 cases; stage 2 = 28 cases; stage 3 = 21 cases; stage 4 = 13 cases). Data are presented as mean ± SEM of at least 3 independent experiments. ∗p < 0.05 shows significantly higher expression of CBS in tumors, compared to adjacent normal tissues. The panel was redrawn from data presented in Ref. [52]. E) CBS mRNA expression in 52 colon cancer tissues and paired adjacent normal colon tissues showing that CBS in tumor tissue is higher than in the surrounding normal tissue in the majority of the patients studied. The panel was redrawn from data presented in Ref. [50]. F) Survival curve showing the impact of CBS expression on overall survival in colon cancer from COAD dataset. p < 0.05 reflects higher survival rate in low-CBS-expressor patients. The panel was redrawn from data presented in Ref. [55]. G) Survival curve showing the impact of CBS expression on overall survival in colon cancer from TCGA-OV dataset. ∗p < 0.05 reflects higher survival rate in low-CBS-expressor patients. The panel was redrawn from data presented in Ref. [52].
In a larger, commercially available collection of primary human tumor tissues and surrounding normal tissues (n = 15), once again, CBS expression was found significantly higher than normal surrounding tissue, while no statistically significant differences in CSE or 3-MST were detected [49]. Out of the 15 tumor/normal tissue pair studied, the tumor tissue showed high CBS expression in 8 cases, while in the other samples both tumor CBS and surrounding normal CBS protein levels were low. There were 2 patients in whom the surrounding normal tissue CBS levels were relatively high; in one of these patients, the tumor CBS was even higher, while in the other one the tumor CBS was lower than the peritumor CBS levels [49] (Fig. 2B). Unfortunately, no clinical or genetic information was reported on these ‘outliers’ vs. the rest of the cohort.
Homogenates of the tumor tissue produce significantly higher amounts of H2S than healthy control tissue homogenates; H2S production can be suppressed by the combined CBS + CSE inhibitor aminooxyacetic acid (AOAA), but is only slightly inhibited by the CSE inhibitor propargylglycine (PAG) (Fig. 2C) [6]. (The effect of pharmacological inhibitors of various H2S producing enzymes in cancer cells and cancer models, and the selectivity and limitations of these agents will be covered in a subsequent section).
In a subsequent, larger patient cohort, immunohistochemistry analysis on a tissue microarray composed of 90 colorectal cancer and paired adjacent normal tissues further confirmed the increased expression of CBS in colorectal cancer tissues, with CBS mainly expressed in the colon cancer cells – as opposed to interstitial cells or tumor-infiltrating immune cells [50]. In another study, 6 primary colorectal tumor samples were analyzed by Western blotting for CBS expression; 1 of these samples exhibited very high CBS levels, 2 samples medium levels, 1 sample low levels and 1 sample undetectable levels [51]. This analysis – although very limited in terms of case number – is important because the samples analyzed for CBS expression were also implanted into nude mice, and their growth rate and responsiveness to the antiproliferative effect to in vivo pharmacological CBS inhibition showed a good correlation with CBS expression levels (see below).
In a recent study, CBS expression (quantified by immunohistochemical analysis) was analyzed in a larger patient cohort; CBS protein levels in human colon cancer specimens closely correlated with the severity/tumor stage: more advanced tumors were found to express higher levels of CBS (Fig. 2D); in addition, higher CBS levels showed a positive correlation with higher expression of VEGF in the tumor tissue [52].
Similar to CBS protein, the degree of CBS mRNA expression in the clinical colon cancer materials is also higher than CBS expression in the surrounding tissue. Chen and colleagues have performed real-time PCR in 52 colon cancer tissues and paired adjacent normal colon tissues and found that the expression of CBS mRNA is higher in colon cancer tissues compared to the patient-matched healthy surrounding tissues in approximately 80% of the cases, while in 20% of the cases the opposite was seen. CBS mRNA expression was at least 2-fold higher in the tumor tissues than in the surrounding normal tissues in approximately 50% of the patients analyzed (Fig. 2E) [50]. Chen and colleagues have also re-analyzed the cancer mRNA databases of Gaedcke and colleagues (rectal adenocarcinoma, n = 65) [53] and Graudens and colleagues (colorectal carcinoma, n = 12–18) [54]. In the larger dataset of Gaedcke, CBS mRNA levels were higher in the tumor tissue than in the normal controls [50]. However, in a recent report by Silver and colleagues, using data curated from the GEPIA dataset, CBS mRNA levels were compared in normal colon (n = 349) versus colon adenocarcinoma (n = 275). In this comparison (which, however, did not compare tumor tissue with matching surrounding tissue, but, rather, compared two different sets of tissues), no significant difference in CBS mRNA levels was reported [55].
The regulation of CBS mRNA levels in cancer appears to be, at least in part, related to the methylation status of the CBS promoter, although the existing body of literature is conflicting. In one report, hypermethylation of the CBS promoter was reported in colon cancer clinical specimens; the degree of hypermethylation correlated with tumor stage, metastasis frequency, tumor recurrence rate and overall mortality [56]. In contrast, in another report, hypomethylation of the CBS promoter was reported; this was associated with the upregulation of CBS mRNA and protein in the tumors and was more pronounced in patients with low circulating folate levels than in patients with high folate levels [22]. In the same study, circulating tumor DNA was also analyzed for the methylation of the CBS promoter, and once again, DNA hypomethylation was detected, which was associated with worse clinical outcomes (increased recurrence rates and lower survival) [22]. According to most reports, many cancers, including colorectal, are associated with a CpG island methylator phenotype, which is characterized by aberrant, pervasive, and genome-wide DNA hypermethylation of CpG islands and subsequent global transcriptional alterations [[57], [58], [59], [60]]. In this context, the hypomethylation of the CBS promoter appears to be atypical.
Taken together, it appears that measurements of CBS mRNA levels are less informative than direct CBS protein analysis to assess CBS expression in colon cancer, a finding which is likely related to patient and database heterogeneity, but may also be due to the fact that CBS levels are determined not only by mRNA levels but also at the level of stability/degradation (see above). Moreover, the role of CBS promoter methylation in the regulation of CBS expression in colon cancer remains to be further investigated.
Do, then, intratumoral CBS levels correlate with various clinical parameters and with clinical outcomes in colorectal cancer? In colon adenocarcinoma, according to analysis of “The Cancer Genome Atlas Colon Adenocarcinoma Dataset” (TCGA COAD), where median gene expression was used to differentiate between n = 40 high and 41 low CBS mRNA expressing patients, high CBS mRNA in the primary tumor is associated with a significantly lower patient survival (p = 0.0128); 75% of the patients succumbed to the disease at a median of approx. 1200 vs. 2000 days in the high and low CBS expressor groups, respectively (Fig. 2F) [55]. A more recent report also reported a similar difference, with patients expressing CBS mRNA at a lower level associated with better clinical outcomes (Fig. 2G) [52]. The best working hypothesis that can be formulated from the above data – as well as from the variety of preclinical data discussed in a subsequent section – is that in colon cancer, CBS mRNA and protein levels are increased in a significant proportion of patients: higher intratumoral CBS serves tumor-cell-supporting roles, which, in turn, leads to worse clinical outcomes.
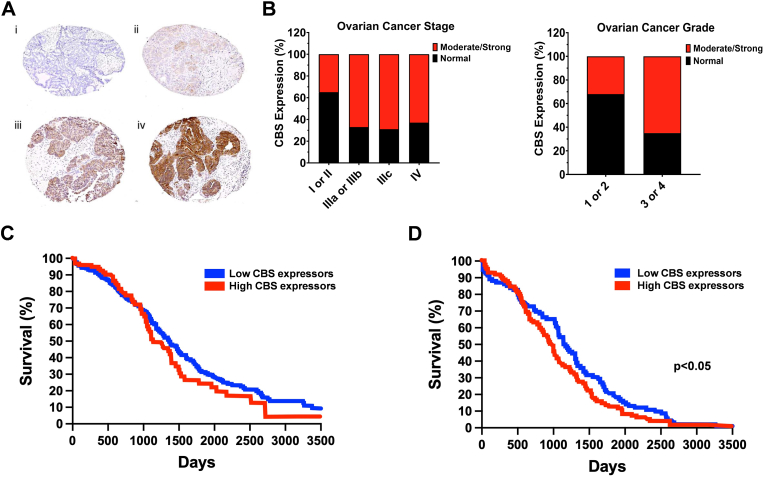
Ovarian cancer. The second form of cancer where the most clinical information is currently available with respect to CBS is ovarian cancer. Bhattacharyya and colleagues have analyzed 210 tissue microarrays constructed from primary epithelial ovarian cancers for CBS protein expression using immunohistochemical analysis. CBS expression was detected in the cytosol of primary ovarian tumors, particularly in serous carcinoma, the most common histologic variant. CBS expression was highest in ovarian cancers exhibiting serous histology and in higher stage and grade cancers (Fig. 3A and B). Nevertheless, CBS expression was also present in 35% of the early-stage tumors analyzed indicating that upregulation of CBS is a relatively early characteristic of serous ovarian cancers [7].
Fig. 3.
CBS is upregulated in human ovarian cancer and correlates with worse clinical prognosis. A) Immunohistochemical staining of a tissue microarray of epithelial ovarian cancer samples. Representative images are shown of none (i), weak (ii), moderate (iii), and (iv) strong staining. The panel was redrawn from data presented in Ref. [7]. B) CBS overexpression in the late stages and grades of ovarian cancer. The panel was redrawn from data presented in Ref. [7]. C) Survival curve showing the impact of CBS expression in ovarian cancer on overall survival from Atlas database (https://www.proteinatlas.org). The panel was redrawn from data presented in Ref. [61]. D) Survival curve showing the impact of CBS expression in ovarian serous cystic adenocarcinoma on overall survival from OV dataset. ∗p < 0.05 reflects higher survival rate in low-CBS-expressor patients. The panel was redrawn from data presented in Ref. [55].
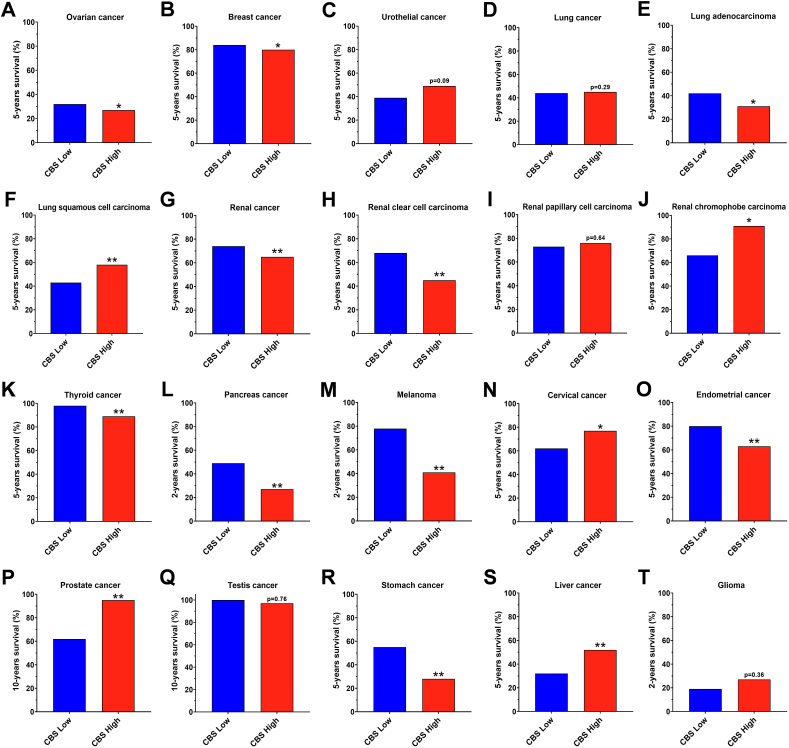
As opposed to the CBS protein data, the CBS mRNA data do not show consistent signs of upregulation in ovarian cancer. In a recent report by Silver and colleagues, data curated and downloaded from the GEPIA dataset, CBS mRNA levels were compared in normal ovary (n = 88) versus ovary serous cystadenocarcinoma (n = 275). In this comparison (which, similarly to the analysis related to colon carcinoma discussed earlier, did not compare tumor tissue with matching surrounding tissue, but, rather, compared two different sets of tissues), no difference in CBS mRNA was reported [56]. Chakraborty has published Kaplan-Meier overall survival curves in ovarian cancer patients stratified for low- and high CBS mRNA expression via analysis of the OvCa cases from The Cancer Genome Atlas (Gene Expression Omnibus) database [61]. The percent probability of survival was plotted vs. time since diagnosis in months. Higher CBS mRNA expression was associated with a trend towards worse survival (Fig. 3C); the difference was statistically significant (p < 0.05) in an analysis conducted by Silver and colleagues from another cancer mRNA database comparing 92 low-CBS mRNA expressor and 93 high-CBS mRNA expressor ovarian serous cyst adenocarcinoma patients (Fig. 3D) [55]. The data in the Human Protein Atlas also tended to be associated with better 5-year survival of low-CBS mRNA expressor ovarian cancer patients, compared to high CBS-expressors (Fig. 4A).
Fig. 4.
Patient survival rates, as a function of CBS mRNA levels in the primary tumor, in several types of cancer. A-T) Data were obtained from the Atlas database (https://www.proteinatlas.org). ∗p < 0.05, ∗∗p < 0.01, reflect significant differences in patient survival between the high- and low-CBS expressor groups.
Since preclinical data indicated that CBS-derived H2S stimulates the expression of mitofusin 2 (MFN2, a mitochondrial membrane protein that plays a central role in regulating mitochondrial fusion and cell metabolism) function, Chakraborty and colleagues cross-referenced CBS mRNA expression data with MFN2 mRNA expression, found a statistically significant correlation [7].
Taken together, the clinical data in ovarian cancer (similar to colon cancer) predict a pathogenetic role of CBS expression.
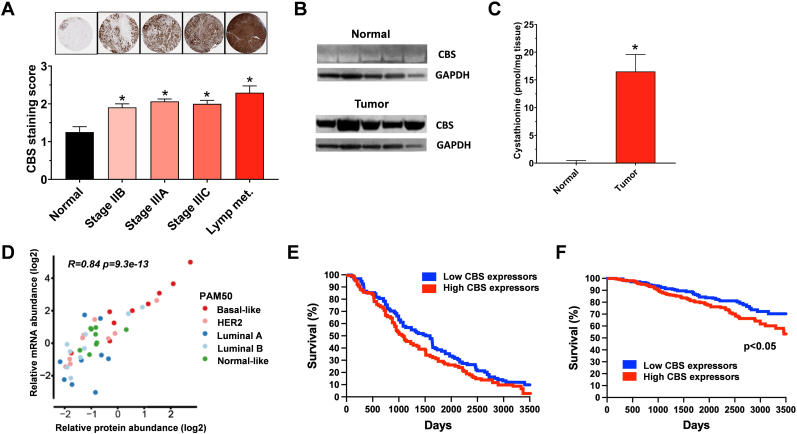
Breast cancer. In 2015, Sen and colleagues have conducted an immunohistochemical analysis of 60 commercially available human breast cancer tissue arrays for CBS expression [62]. Significantly higher CBS expression was observed in the breast cancer tissue compared with adjacent normal controls; CBS levels increased with the progression of the disease, with most pronounced CBS expression (up to 3-fold over healthy tissue) detected in the lymph node metastatic tissue (Fig. 5A). CBS expression was independent of the estrogen receptor, progesterone receptor, HER2 and p53 status [62]. The high CBS expression was also confirmed by Western blotting in a limited number (n = 5) of human breast cancer tissue homogenates (Fig. 5B); the tissue homogenates also produced high levels of the CBS biochemical product cystathionine (Fig. 5C): in fact, cystathionine was designated as an “oncometabolite” by the authors [63]. Analysis of in-depth quantitative proteomics data of 45 human breast cancer tumors from the Oslo 2 study cohort was subsequently conducted by the group of Nagy in 2021 [64]. Elevated CBS protein levels were reported in the luminal, HER2-negative and “normal-like” breast cancer subtypes, with a relatively rare subgroup of breast cancer, the basal-like cancer showing the highest CBS expression levels (Fig. 5D). CBS expression positively correlated with proliferation-related gene sets and negatively correlated with estrogen response–related genes [64].
Fig. 5.
CBS is upregulated in human breast cancer and correlates with worse clinical prognosis. A) Immunohistochemical staining of a tissue microarray of 60 human breast cancer samples. Data are presented as mean ± SD of CBS staining. ∗p < 0.05 shows significantly higher tumoral CBS levels compared to normal tissue. The panel was redrawn from data presented in Ref. [62]. B) Representative western blot, detecting CBS in-patient derived breast cancer tissues and matched normal breast tissues (n = 5). The panel was redrawn from data presented in Ref. [63]. C) Cystathionine levels in patient derived breast tumor and normal breast tissues (n = 5). Data are represented as mean ± SEM from three independent experiments, ∗p < 0.05. The panel was redrawn from data presented in Ref. [63]. D) CBS overexpression in basal-like breast cancer. Correlation of CBS mRNA and protein levels in 45 tumors from the Oslo 2 cohort. The panel was redrawn from data presented in Ref. [64]. E) Survival curve showing the impact of CBS expression on overall survival in invasive breast carcinoma from the BRCA dataset showing a trend for better survival in low-CBS-expressing patients. The panel was redrawn from data presented in Ref. [55]. F) Survival curves showing the impact of CBS expression on overall survival in breast cancer from Atlas database (https://www.proteinatlas.org). ∗p < 0.05 shows significantly better survival in low-CBS-expressing patients.
CBS mRNA upregulation in human breast cancer clinical specimens was subsequently demonstrated in 2021 by two independent groups. The group of Gad has analyzed tumor biopsies from a cohort of 80 Egyptian women, diagnosed with breast cancer [65]. Tumors from the patients showed significantly higher CBS mRNA levels than histologically normal breast tissues isolated from the same mastectomy sample. The degree of CBS mRNA upregulation was comparable in the various subtypes of breast cancer (Luminal A, luminal B, HER2 enriched and TNBC) (Fig. 5D) [64]. Serum H2S levels – detected by a microplate based colorimetric method – were approximately 45% higher in breast cancer patients than in healthy gender and age matched controls [65]. When stratified by tumor proliferation index (ki-67), rapidly proliferating tumors expressed approximately 40-times higher CBS mRNA levels than slow-proliferating ones [65]. The study conducted by the group of Nagy – which has already demonstrated high CBS protein expression in breast cancer clinical specimens (see prior paragraph) – has also found a significant upregulation of CBS mRNA in several breast cancer clinical databases (Oslo 2, TCGA, NeoAva), with particularly high CBS mRNA levels in basal like breast cancer [64]. There was a good correlation between CBS mRNA and CBS protein expression in human breast cancer tissues [65], suggesting that the mechanism of CBS upregulation has a substantial transcriptional component in breast cancer cells.
In contrast to the above data, in a recent report by Silver and colleagues, data curated and downloaded from The Cancer Genome Atlas (TCGA), CBS mRNA levels did not find differences in mRNA expression between normal breast tissue (n = 291) versus breast cancer tissue (n = 1085) [55]. This comparison – similarly to the case in colon and ovarian cancer analysis from the same TCGA dataset where it failed to reveal differences – did not compare tumor tissue with matching surrounding tissue, but, rather, compared two different sets of tissues, in this case, with very different patient group size. We conclude that data derived from the TCGA dataset should be interpreted with caution, and studies using direct comparisons of tumor tissue and healthy surrounding tissue from matching subjects (and, when available, direct information on CBS protein, rather than mRNA) is significantly more informative, even when the analyzed patient group sizes are smaller.
Do, then, intratumoral CBS levels correlate with various clinical parameters and clinical outcomes in breast cancer? According to analysis of the “BRCA Dataset” from TCGA, where median gene expression was used to differentiate between n = 99 high and 99 low CBS mRNA expressing patients, a statistically non-significant trend was noted for worse clinical outcomes in invasive breast carcinoma patients with higher CBS mRNA expression (Fig. 5E) [55]. However, according to data in the Human Protein Atlas, high CBS mRNA expression tended to predict worse survival probability (Fig. 4B).
According to the analysis of Nagy's group, CBS expression may affect treatment response–related behavior in breast cancer patients [64]. In samples from the NeoAva trial, patients with complete responses had higher CBS mRNA expression levels than noncomplete responders. CBS levels, in complete responders, and, to a lesser extent, also in noncomplete responders, decreased significantly by 12 weeks of treatment and remained low at 25 weeks after treatment. One possible interpretation of these findings may be that breast cancer chemotherapy causes the downregulation or degradation of CBS in the tumor tissue. These data are in contrast to the analysis of gastric cancer patient outcomes by Zhao and colleagues [66] (discussed below), which found that low-CBS mRNA expressor gastric cancer patients respond better to adjuvant chemotherapy than high-CBS mRNA expressors.
Other cancers exhibiting increased CBS expression. In human samples of urothelial cell carcinoma of the bladder, increased CBS expression was reported by immunohistochemical analysis; moreover, the H2S producing ability of cancer tissue was 3-4-fold higher than the normal tissue. Higher-grade tumors showed higher CBS expression than low-grade tumors [67,68]. There are no peer-reviewed publications with respect to a potential correlation of CBS expression with clinical outcomes in urothelial cell carcinoma; according to the Human Protein Atlas (93 high and 313 low CBS expressors) low-CBS expressors show a trend for better clinical prognosis (p = 0.09) (Fig. 4C).
In human non-small cell lung adenocarcinoma Western blotting analysis of the tumor tissue vs. the normal adjacent lung tissues (n = 20) showed significantly higher, approximately 5-fold more CBS protein in the tumor homogenates; these tissues also produced about 2-times more H2S than the surrounding normal tissue [69]. Increased CBS protein or mRNA levels have also been reported subsequently in two different lung cancer clinical specimen collections [70,71]. According to the Human Protein Atlas, no overall difference can be found in 5-year survival in lung cancer, when all forms of lung cancer are grouped together (Fig. 4D). However, interestingly, in lung adenocarcinomas, high CBS expression is associated with significantly lower 5-year survival (Fig. 4E) while in lung squamous cell carcinomas the opposite is the case (Fig. 4F). Thus – similar to the case of breast cancer – in lung cancer, the association of CBS with clinical outcomes shows significant subtype differences.
Renal oncocytoma, renal urothelial carcinoma, and renal clear cell carcinoma clinical samples also showed increased CBS expression, as evidenced by immunohistochemical analysis [72,73]. In an earlier report, in renal clear cell carcinoma samples a clear increase in CBS expression was seen as a function of increasing tumor stage; Fuhrman IV samples had over 3-times more CBS than Fuhrman I stage samples [72]. However, in a subsequent report, CBS enzyme expression patterns in renal clear cell carcinoma versus normal tissue did not correlate with nuclear grade, stage, histological type or cancer recurrence/metastasis [73]. According to analysis of the renal clear cell adenocarcinoma; KIRC Dataset, the level of CBS expression does not affect clinical prognosis [55]. However, according to the data contained in the Human Protein Atlas Database, high CBS mRNA levels in renal cancer (when all subgroups are analyzed together) is associated with significantly worse survival (p < 0.01) (Fig. 4G), and this difference is driven primarily by the most common subgroup, the renal clear cell carcinoma (Fig. 4H), but not by renal papillary cell carcinoma (Fig. 4I) or chromophobe carcinoma (Fig. 4J).
CBS expression and H2S generation of tumor homogenates were also increased in oral cavity and esophageal squamous cell carcinomas [[74], [75], [76]], in various thyroid malignancies [77], in extrahepatic cholangiocarcinoma and in gallbladder carcinomas [78,79]. Increased CBS expression was also reported in esophageal squamous cell carcinoma [79], biliary tract carcinoma [80] and multiple myeloma [81,82]. To our knowledge, no published studies reported any correlation of CBS expression with clinical outcomes in the above cancers, with the exception of squamous cell/adenosquamous carcinomas, where high CBS predicts worse survival [76]. According to the Human Protein Atlas, high CBS expression is associated with lower survival probability in thyroid cancer (Fig. 4K), pancreatic cancer (Fig. 4L), melanoma (Fig. 4M), and endometrial cancer (Fig. 4O), while in cervical cancer and prostate cancer, the opposite is the case: high CBS predicts better survival (Fig. 4N, P).
In gastric cancer, CBS mRNA and protein levels were found to be relatively low in clinical samples; nevertheless, in a limited analysis (n = 10), CBS in the gastric cancer homogenates appeared to be higher than in the surrounding healthy tissues [83]. Zhao and colleagues have analyzed a large patient cohort (GC cohort from the Cancer Genome Atlas) for CBS mRNA expression and clinical outcomes [66]. The study demonstrated that relatively higher CBS mRNA levels are associated with worse clinical outcomes and similar data are also contained in the Human Protein Atlas database (Fig. 4R). In addition, patients with higher CBS mRNA levels in their tumors respond worse to adjuvant chemotherapy than patients with low CBS expression [66]. Based on these findings it was concluded that CBS expression in the cancer tissues may be a marker to predict adjuvant chemotherapy responsiveness.
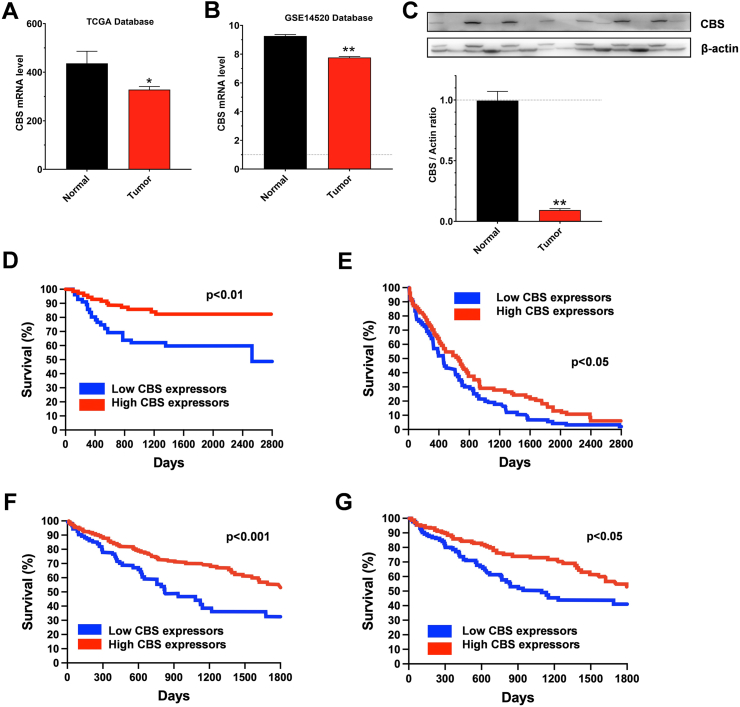
Cancers exhibiting decreased CBS expression. Hepatic carcinoma appears to be one outlier from the above discussed, rather extensive list of carcinomas where CBS is upregulated, and this upregulation is frequently associated with worse clinical outcomes. The downregulation of CBS in liver cancer was first reported by Kim and colleagues in 2009. When comparing hepatocellular carcinoma tissues with corresponding non-cancerous hepatic tissues obtained from 120 patients who underwent curative hepatectomy for primary hepatocellular carcinoma, a significant downregulation of CBS mRNA was noted; the degree of CBS mRNA downregulation was more pronounced in higher-grade tumors, in patients with higher plasma alfa-fetoprotein level and in older patients [84]. Multivariate Cox regression analysis for overall survival among patients with low alfa-fetoprotein levels showed that the downregulation of CBS expression predicts better overall survival; in fact, is appears to be a better predictor for overall survival than even tumor grade [84]. Similarly, in a large clinical database of clinical materials, CBS mRNA in hepatocellular carcinoma tissue (n = 369) was found to be lower than in healthy human tissues (n = 160) [55]. The stage-dependent downregulation of CBS in hepatocellular carcinoma was also demonstrated recently in a third, independent patient dataset [85]. In hepatocellular carcinoma – once again, in contrast to the other tumors discussed in the previous sections – high CBS mRNA expression is associated with better clinical outcomes, as shown by multiple independent analyses of large clinical datasets [55,80,86,87]. According to the Human Protein Atlas (264 high and 101 low CBS expressor patients), a marked difference can be found in 5-year survival in liver cancer, once again, high CBS predicting significantly better (p < 0.01) survival (Fig. 4S). Direct measurements of H2S generation in human liver cancer homogenates also confirmed that these tissues produce less H2S than the surrounding normal tissue [87]. The clinical findings and correlations in liver cancer (some of which are summarized in Fig. 6) are in striking contrast with the majority of findings in many other types of cancer (discussed above), where high CBS expression correlates with worse survival (see above) and where CBS inhibition or silencing exerts anticancer effects (see below).
Fig. 6.
CBS is downregulated in human hepatocellular carcinoma and correlates with better clinical prognosis. A, B) Relative CBS expression in hepatocellular carcinoma tumor versus peritumor tissues from TCGA and GSE14520 database, respectively. Data are shown as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01 showing better survival in high-CBS expressor patients. The panel was redrawn from data presented in Ref. [87]. C) Western blot analysis of CBS protein levels in 28 hepatocellular carcinoma tumor tissues and paired peritumor tissues. Data, shown as mean ± SEM, show a significant downregulation of CBS in hepatocellular carcinoma ∗∗p < 0.01. The panel was redrawn from data presented in Ref. [87]. D) Survival curve showing the impact of CBS expression on overall survival in hepatocellular carcinoma. ∗∗p < 0.01 reflects better patient survival in high-CBS-expressing patients. The panel was redrawn from data presented in Ref. [84]. E) Survival curve showing the impact of CBS expression on overall survival in hepatocellular carcinoma from the LIHC dataset. ∗p < 0.05 reflects better patient survival in high-CBS-expressing patients. The panel was redrawn from data presented in Ref. [55]. F) Survival curve showing the impact of CBS expression on overall survival in hepatocellular carcinoma from Atlas database (https://www.proteinatlas.org). ∗∗∗p < 0.001 reflects better patient survival in high-CBS-expressing patients. G) Survival curve showing the impact of CBS expression on overall survival in hepatocellular carcinoma from TCGA database. p < 0.05 reflects the patient survival statistics. The panel was redrawn from data presented in Ref. [87].
Another clear outlier tumor type – with respect to the role of CBS – is glioma. In this tumor type, a downregulation, rather than an upregulation of CBS expression has been reported in multiple studies using clinical materials or human cell lines. Among the various forms of glioma, patients with glioblastoma multiforme present the highest degree of CBS downregulation, compared to patients with other tumor types within the glioma family [55,88]. Preclinical studies show that in glioma – as opposed to the vast majority of cancers discussed in the previous paragraphs – it is the loss of CBS (as opposed to its upregulation) that plays tumor-supporting roles (see below). According to the Human Protein Atlas (64 high and 89 low CBS expressors), higher CBS expression tends to predict better 2-year survival in glioma, but the difference in this small clinical data set is not statistically significant (Fig. 4T).
The reason why the role of the CBS pathway in liver cancer and glioma is different from many other types of tumors is currently unclear. Nevertheless, it is important to keep in mind that the two organs with the highest physiological (baseline) CBS expression (and, consequently, H2S production) are the liver and the brain. (In the liver, CBS is a key effector of transsulfuration, and an essential enzyme in the degradation of homocysteine, while in the brain H2S plays key roles as a neuromodulator and neuroprotectant [12].) Thus, it stands to reason that the role of CBS in the pathophysiology of cancers that strive in a high-baseline-H2S environment is different from those that occur and develop on the background of low H2S levels in their environment.
4. CBS expression in various cancer cell lines
A study from 2005, focusing on the expression profiling of “homocysteine junction enzymes” used the NCI60 panel of human cancer cell lines to assess the expression of CBS (as well as methionine synthase and methionine sulfoxide reductase) using Western blotting [89]. Expression of CBS in different cancer cell lines was quantified relative to its levels in HepG2 cells (which is a cell type, which expresses fairly high levels of CBS; see also below). Thus, it is not surprising that in many of the studied cell types, CBS expression was characterized, in relative terms, as “fairly low”; when considering all cancer cell lines studied, it was approximately 50% of the levels detected in HepG2 cells. The only group of cancers where CBS was found to be higher than the HepG2 levels was the group of breast cancer lines. Relatively high CBS expression was also reported in the group of renal cancer lines, ovarian cancer lines, prostate cancer lines [89]. When the measured CBS protein expression data were compared with CBS mRNA levels – obtained from a microarray database at the NCI website –a good correlation was observed. The same study also attempted to correlate CBS expression with drug sensitivity – assessed by GI50 (50% growth inhibition level of effect, as specified by the NCI Developmental Therapeutics Program database), no correlations were noted. Likewise, the study did not find any correlation between CBS expression and the cells’ methionine dependence.
While the above study was pioneering in its overall approach, it has several deficiencies: (a) first, it did not compare the expression levels of the cancer cells to non-transformed control cells and (b) it did not assess the functional consequence of CBS inhibition in the cancer cells. However, many of the subsequent studies focusing on CBS expression in various human cancer cell lines have been conducted with the above considerations in mind. In colon cancer cell lines, in 2013, CBS expression in various colon cancer cell lines was compared to the expression levels in a normal cell comparator. In this study, NCM356 cells were used, which were considered as a “non-transformed epithelial cell line”. Compared to NCM356 cells, several colon cancer cell lines (HCT116, LoVo, HT-29) exhibited higher CBS expression [6]. (Although the NCM356 cells, for a long time, were considered non-transformed, later Next Generation sequencing studies demonstrated the existence of a mutant TP53 gene in this cell line, and identified additional functional mutations in the KRAS and APC genes, suggesting that this NCM356 cell line, in fact, exhibit the characteristics of a late adenoma [53].) Several independent studies have confirmed CBS protein expression in the above colon cancer lines, and extended the findings to additional colon cancer cell lines (e.g. DLD-1 and SW480) as well [27,38,49,51,52,[90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100]].
Alix-Panabières and colleagues compared the expression of various genes in CTC-MCC-41 cells, a colon cancer cell line isolated from the blood of a patient with colon cancer with the gene expression profile of HT-29 cells [100]. CTC-MCC-41 cells (a metastasis-competent cell line that exhibits intermediate epithelial-mesenchymal phenotype and stem cell–like properties) exhibited differential expression of multiple genes, including genes that regulate energy metabolism, DNA repair and stemness. The genes which exhibited higher expression in CTC-MCC-41 cells than in HT-29 cells also included CBS, which, therefore, was classified as a “stemness gene” [100].
In ovarian cancer, OV202, SKOV3, A2780, A2780/CP-70, CP20, OV90 and OVCA429 cells are typically used for studies elucidating the role of CBS – without a healthy non-transformed cell comparator; all of these cells express high levels of CBS protein and generate H2S [7,61,101,102].
In breast cancer studies, human cell lines that have been used include the MDA-MB-231, MDA-MB-435S, MDA-MB-468, MCF-7, MCF-10A and Hs 578T lines (many of which are triple negative – i.e. lacking estrogen or progesterone receptors and HER2), and, more recently, two basal-like subtype breast cancer cell lines, Cal51 and HCC1143 [[62], [63], [64], [65],[103], [104], [105]]. In some studies, human mammary epithelial cells (HMEC) were also included as healthy control comparators. While these latter cells express low levels of CBS protein, most of the breast cancer cell lines (with the exception of MCF-10A) express high CBS levels; with particularly high CBS expression reported in the Cal51 and HCC1143 lines [[62], [63], [64], [65],[103], [104], [105]].
To demonstrate the increased expression of CBS in bladder urothelial cell carcinoma cell lines, RT4, SW780, 5637, EJ, T24 and UM-UC-3 cells were used; in some experiments SV-HUC-1 cells were employed as a non-transformed (and low CBS-expressor) comparator [67,68]. Most of these cells (except RT4 cells, which express relatively less CBS) showed significant CBS expression – higher levels than the non-transformed control cells.
In lung cancer, the three published studies that examined the functional role of CBS, utilized the lung adenocarcinoma cell lines A549, H522 and H1944, or A549 and 95D cells, or Calu-6 cells, respectively; CBS expression in the cancer lines was significantly higher than CBS expression in the non-transformed lung epithelial cell line Beas2B comparator [[69], [70], [71]].
The only currently available study aimed to examine the role of CBS in renal carcinoma cell lines utilized Caki-1 (a Von Hippel-Lindau [VHL] tumor suppressor wild-type human renal cell carcinoma cell line), and two of its VHL-deficient subclones (786-O and 769-P). CBS expression and functional parameters in these cells were compared to the normal human renal epithelial cell line HK-2. Although all cells expressed CBS, the cancer cell lines did not express it to higher levels than the control comparator line [106].
Examples of other human cancer cell lines that exhibited high expression of CBS and have been used to study the functional role of CBS in other forms of cancer [67,76,80,81,[103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113]] are listed in Table 1. Importantly, the various hepatoma and glioma lines studied in various in vitro studies and tumor-bearing mice xenograft models contain significant amounts of CBS, in some cases higher than the corresponding non-transformed cell control [106] – despite of the clinical data (see prior section) demonstrating that liver cancer and glioma tissues express lower levels of CBS than surrounding normal liver and brain tissue, respectively. In case of a few cell lines, the published reports are conflicting; for instance, different groups reported high or low CBS expression in the BEL-7404 hepatoma cell line [108,109].
Table 1.
Expression and role of CBS in various cancers.
| Colorectal cancer | Ovarian cancer | Breast cancer | Urothelial cell carcinoma of bladder | Lung adeno-carcinoma | Renal cancers | Oral and esophageal carcinoma | Extrahepatic cholangio-carcinoma | Gallbladder adenocarcinoma | Gastric cancer | Multiple myeloma | Liver cancer | Glioma, glioblastoma | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CBS protein expression in clinical tumor tissue relative to surrounding normal | Higher | Higher | Higher | Higher | Higher | Higher | Higher | Higher | Higher | Higher | Not yet assessed | Lower | Lower |
| Correlation of CBS protein expression in clinical tumor specimen with disease stage | Correlation confirmed both in terms of colon cancer stage and also in terms of progression from polyp formation to colon adenocarcinoma formation | Correlation confirmed | Correlation confirmed for basal-like cancer | Not yet assessed | Not yet assessed | Inconclusive data | Not yet assessed | Correlation confirmed | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed |
| CBS mRNA expression in clinical tumor tissue relative to surrounding normal | Higher | Higher | Higher | Not yet assessed | Higher | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Higher | Higher | Lower | Lower |
| Correlation of CBS mRNA expression in clinical tumor tissues with clinical prognosis | Higher CBS mRNA is associated with worse clinical outcomes in several (but not all) published analyses | Higher CBS mRNA is associated with worse clinical outcomes | Higher CBS mRNA is associated with worse clinical outcomes | Higher CBS mRNA tends to be associated with better clinical outcomes. | Higher CBS mRNA is associated with worse clinical outcomes. | Higher CBS mRNA is associated with worse clinical outcomes (especially in renal clear cell carcinoma) | Not yet assessed | Higher CBS mRNA is associated with worse clinical outcomes. | Not yet assessed | Higher CBS mRNA is associated with worse clinical outcomes and poorer chemotherapy responses | Not yet assessed | Higher CBS mRNA is associated with better clinical outcomes. | Higher CBS mRNA tends to be associated with better clinical outcomes. |
| Expression in human cancer cell lines | High expression: HCT116, LoVo, HT-29, DLD-1, SW480, CTC-MCC-41 | High expression: OV202, SKOV3, A2780, A2780/CP-70, CP20, OV90, OVCAR3, OVCAR4, OVCAR5, OVCA429 |
High expression: MDA-MB-231 MDA-MB-435S, MDA-MB-468, MCF-7, Hs578T, Cal51, HCC1143 | High expression: RT4, SW780, 5637, T24, EJ, UM-UC-3 | High expression in A549, H522, H1944, 95D, Calu-6 | Average expression: Caki-1, 786-O, 769-P is comparable to non-transformed cell controls | Variable expression ranging from non-detectable to high: KYSE510, KYSE150, KYSE140, Eca109, EC9706, TE-13, KYSE70, KYSE450 | Not yet assessed | Not yet assessed | Average expression: BGC-823, SGC-7901 | Average/low expression: U266 | Relatively high CBS expression: HepG2, SMMC-7721, BEL-7402, BEL-7404 | Relatively low CBS expression: U87-MG, hGBM 23, hGBM 124, hGBM 3691, BT142, TS603, NCH1681 |
| Effect of CBS silencing in human cancer cell lines in vitro | Inhibition of tumor growth, potentiation of the effect of chemotherapy | Inhibition of tumor growth, potentiation of the effect of chemotherapy | Inhibition of tumor growth, potentiation of the effect of immune cell mediated tumor killing | Not yet assessed | Inhibition of tumor growth, potentiation of the effect of chemotherapy | Not yet assessed | Inhibition of tumor growth | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Variable effects: either pro- or antiproliferative and either cytotoxic or cytoprotective actions in various hepatoma cell lines | Stimulation of tumor growth |
| Effect of CBS silencing in human cancer cell lines on tumor growth in tumor-bearing mice | Inhibition of tumor growth | Inhibition of tumor growth, potentiation of the effect of chemotherapy | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Not yet assessed | Enhancement of tumor growth | Enhancement of tumor growth |
Regarding the potential role of CBS in prostate cancer the published information is minimal. There are some data comparing survival probabilities in high vs. low CBS expressor patients in the Human Protein Atlas (see above), but in peer-reviewed publications, the only available information relates to prostate cancer, where high CBS protein levels measured by Western blotting in two androgen-dependent prostate cancer lines (LNCaP cells and DU145 cells); lower, but detectable levels in BPH1 cells (a benign prostatic hyperplasia line), while CBS was not detected in the benign prostatic epithelial cell lines RWPE-1 and WPMY1 [114,115]. Some of the above data suggest that the expression of CBS becomes higher with the progression of prostate cancer; however, the fact that CBS expression was low in PC-3 cells (a cell line derived from a metastasis of prostatic adenocarcinoma) does not fully support this notion [115].
The form(s) of CBS expressed in the various cancer cell lines has not been investigated extensively. In HCT116 cells, both the full-length and the proteolytically cleaved, constitutively active truncated form of the enzyme have been detected [38]. In many other reports, the Western blots published only show the canonical 63 kDa band, and it is unclear if the lower protein band was also present in the homogenate. Clearly, further work needs to be conducted to characterize the cleavage process (e.g. the enzymes involved, the relative specific activity of the two forms of CBS, and the stability and degradation of the two enzymes). The subcellular distribution of CBS has also not been explored in most published studies; there is some evidence that hypoxic conditions can induce CBS translocation into the mitochondria, and, indeed, in HCT116 cells and in A2780 cells, CBS was detected both in the cytosolic and mitochondrial fractions [6,7], but subcellular distribution of CBS in most studies published to date has not been studied. One rare exception is the study of Sen and colleagues, where, in human breast cancer cells, CBS was localized to the plasma membrane (in addition to its normal cytoplasmic localization), but not to the mitochondria [62]. Likewise, potential post-transcriptional modifications (e.g. SUMOylation) have not yet been investigated in cancer cell lines. Finally, the potential changes in the various co-factors and substrates of CBS remain to be explored in the future.
The regulation of CBS expression in cancer cells, however, has been investigated in several studies. Already in the early 2000's, Kraus and colleagues have suggested that increased cell proliferation, on its own, is a driver of CBS induction through a redox-sensitive mechanism [18]. However, clearly, high CBS expression has not been observed in all rapidly proliferating cell types studied, so proliferation, by itself, can not be the sole driving factor. Nevertheless, in some cases, a close correlation has been found between CBS upregulation and increased cell proliferation; for instance, in human colon cancer cell lines treated with increasing concentrations of the polyamine N12-diacetylspermine [98]. The potential role of hypoxia as a CBS-inducing or enhancing factor, has been explored by multiple investigators, and yielded variable results. In U87-MG glioma cells, hypoxia was identified as a marked inducer of CBS [19,88]. However, in renal cell carcinoma cells, and in Cal51 breast cancer cells, hypoxia did not have any marked effect on CBS expression [64,106]. In A549 lung cancer cells, chemical hypoxia (cobalt chloride treatment) caused a slight upregulation of CBS mRNA and protein levels [70]. Finally, in HepG2 cells, hypoxia was reported to induce an upregulation of CBS protein expression [116]. In the context of hypoxia and H2S biology it should be also mentioned that hypoxic conditions, on their own, are known to suppress the degradation of H2S to elevate H2S cell and tissue levels [3], and, therefore, hypoxia – even if the expression or activity of the various H2S-producing enzymes remain unchanged – is expected to enhance the biological effect of the H2S produced by the tumor cells.
In various prostate cancer cells, there is conflicting evidence for an android-hormone-dependent regulation of CBS: in LNCaP cells, incubation with dihydrotestosterone was reported to upregulate, while incubation with testosterone was reported to downregulate CBS protein levels [114,115]. Interestingly, exogenous administration of a pharmacological H2S donor compound was also reported to induce the upregulation of CBS protein in thyroid carcinoma cells in a bell-shaped fashion (i.e. only noted with intermediate concentrations of the donor) [117]; it is currently unclear if such a positive feedback cycle between H2S generation and CBS expression is physiologically relevant, and if so, if it is a general feature of cancer, or only relevant for specific cell types.
Finally, a marked down-regulation of CBS occurs in glioma under conditions of high-fat diet [55]. The role of CBS in glioma is completely different from its role in many other cell types, in that it is cancer-suppressive, as opposed to cancer promoting (see above and below). In this context, the additional downregulation of CBS in glioma during obesity may be a potential mechanism by which obesity accelerates the development of this type of cancer [55].
Sanokawa-Akakura and colleagues applied glucose deprivation, hypoxia and hydrogen peroxide treatment to various hepatoma and breast cancer cells to produce damage-recovered cells. These cells – which became resistant to subsequent injury – exhibited an upregulation of CBS; it appears that oxidative stress is the most significant factor which contributed to the upregulation of CBS in this experimental model [103,104]. When the cells assumed the damage-recovered phenotype, a variety of stemness-associated genes were upregulated in them. Similarly, an association between stemness and CBS upregulation was suggested by comparison of CTC-MCC-41 cells (a metastasis-competent circulating colon epithelial cell line) with less aggressive colon cancer lines [100]. Moreover, after exposing HepG2 cells to ionizing radiation, CBS upregulation has been demonstrated [110,116]. HCT116 cells, as they develop multidrug-resistance as a result of prolonged culturing in a 5-fluorouracil-containing culture medium, also exhibit increased expression of CBS – together with the induction of various other enzymes, including the drug-metabolizing cytochrome P450 enzymes CYP1A2 and CYP2A6 [93]. Similarly, doxorubicin-resistant MCF-7 cells exhibit significantly higher expression of CBS than normal MCF-7 cells [118]. Only some of these studies (discussed below) examined directly if there is a casual relationship between CBS upregulation, or H2S production and the increase in stemness in the surviving dedifferentiated and resistant cells.
5. Effect of “forced CBS expression” on various cellular functions in the context of cancer biology
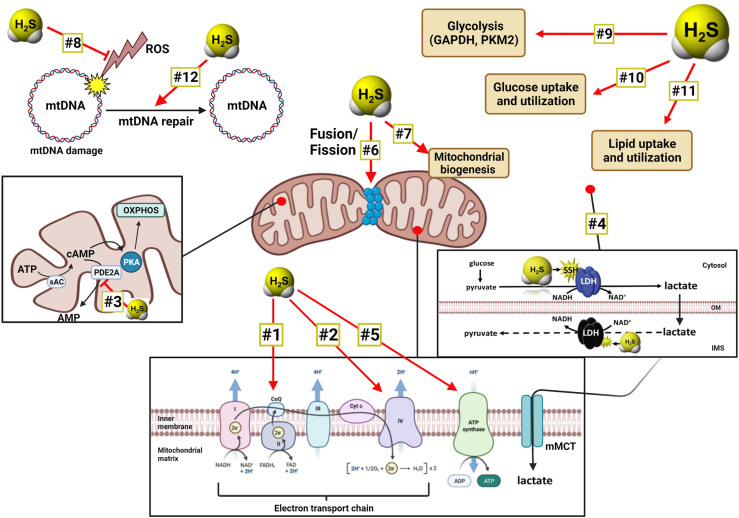
One highly instructive way to investigate the functional role of CBS in cancer cells is to induce the over-expression of this enzyme in non-transformed cells that don't normally express it, or express it only at low levels. One of the most comprehensive analyses, focusing on this approach, has been conducted in NCM356 cells, a low-CBS expressor, slowly proliferating human epithelial cell line, which – as mentioned earlier – according to Next Generation sequencing, does exhibit several mutations that make it resemble a late adenoma, rather than a ‘normal epithelial cell’. (In fact, no ‘cell line’ which can be propagated in cell culture conditions will completely resemble the character of freshly isolated primary cells.) NCM356 cells with forced overexpression of CBS produced increased amounts of H2S, and exhibited a faster proliferation rate than the parental control cell line, both in vitro [6,48] and in vivo when xenografted onto nude mice [48]. This increased proliferation could be suppressed by pharmacological inhibition of CBS [48]. The CBS overexpressor cells also exhibited increased migration and invasiveness, and colony formation in vitro.
Metabolomic profiling of the cells demonstrated the expected shifts in the transsulfuration pathway – including an increase in the CBS enzymatic byproduct lanthionine in vitro and in vivo [48,51] –, but also increased flux through both the pentose phosphate and glycolytic pathways. Accordingly, pharmacological inhibition of the pentose phosphate pathway suppressed the proliferation rate of these cells [48]. There was also a significant increase in the cellular respiration and bioenergetic parameters of these cells, indicating that CBS expression, on its own, can drive up the metabolism of colonic epithelial cells, in line of the well-established ability of cancer-cell-generated H2S to stimulate mitochondrial electron transport and ATP generation [47]. Part of the enhanced bioenergetics of the CBS-overexpressor cells may be related to their upregulated citrate synthase activity, enhanced lipid biosynthesis and induction of the hexose phosphate pathway [48].
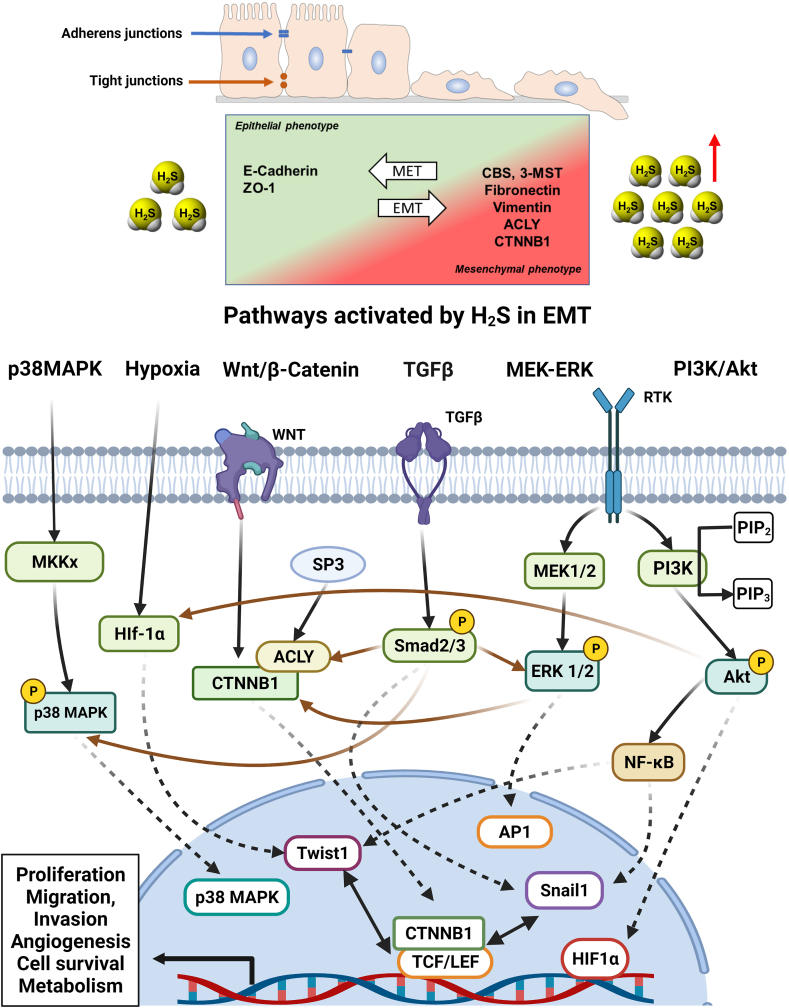
Whole transcriptome RNA-seq revealed that forced expression of CBS in NCM356 cells induces the upregulation of 243 genes and the downregulation of 108 genes. GSEA gene family categorization revealed the regulation of many transcriptions factor genes (20 up- and 10 downregulated), cytokines and growth factors (18 up and 3 down), protein kinases (7 up and 3 down), and oncogenes (ASPSCR1, PER1, PRDM16, HOXD11, CCND2 up while LCP1 and DDX10 down). The upregulated genes significantly overlapped with gene sets related to glycolysis, hypoxia, and a colon cancer cell phenotype including NF-κB, KRAS, and p53 signaling, apical junction gene set, and Wnt pathway genes. Other upregulated genes due to forced CBS overexpression have been shown to enhance hypoxia responses (PER1, APLN), promote cytokines, growth factors and inflammation-mediated responses (ETS2, GDF15, FGF19, S100A6, TNC, FGF11, TNSF15, KITLG, IGF2, TNFSF9, MDK, INHBB, SEMA6B, ADM, INHBE, VEGFA, ADM2, GDF15, APLN, FGF3), stimulate cell proliferation (CCND2, MAP3K5, PTK7, RPS6K2, E2F2, FGF19, MDK, KIT, JUNB, CXCR1, ELF3, FOS), promote cell migration and invasion (CDC42BPG, DDR1, ELF3, HOXD11, PTK7, KIT) and induce angiogenesis (ADM, ADM2, VEGFA, CXCR1, FOS) [48]. Finally, there was significant overlap with genes related to increased extracellular matrix (ECM) and ECM-related proteins, cell adhesion molecules, and epithelial-to-mesenchymal transition (EMT), suggesting that the presence of CBS, on its own may be able to shift cells into a migratory and invasive phenotype [48]. This latter property of CBS has been directly confirmed by the fact that CBS-overexpressor NCM356 cells exhibit increased invasive and migratory properties, and it is also supported by multiple lines of data that suggest that CBS silencing or CBS inhibition in cancer cells suppresses invasion and migration, and reverses EMT (discussed in the subsequent sections). Importantly, however, no secondary NCM356 cell loci (i.e. metastases) were found in the nude mice bearing xenografted CBS-overexpressor NCM356 cells – at least in the timeframe of the experiments conducted (up to 30 days) [51]. Thus, CBS overexpression, on its own, may not be sufficient to induce a metastatic tumor phenotype, although this question remains to be further investigated, for instance in longer-term models or using intracecal, as opposed to subcutaneous implantation of the cells.
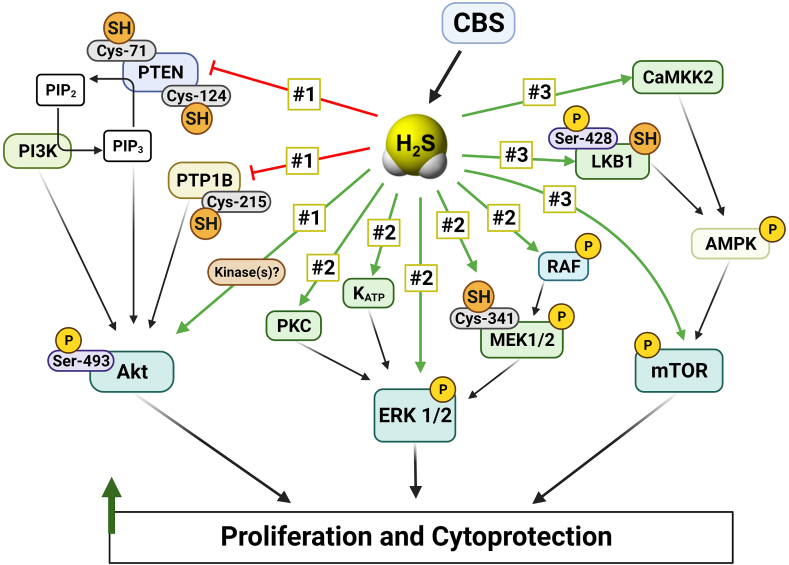
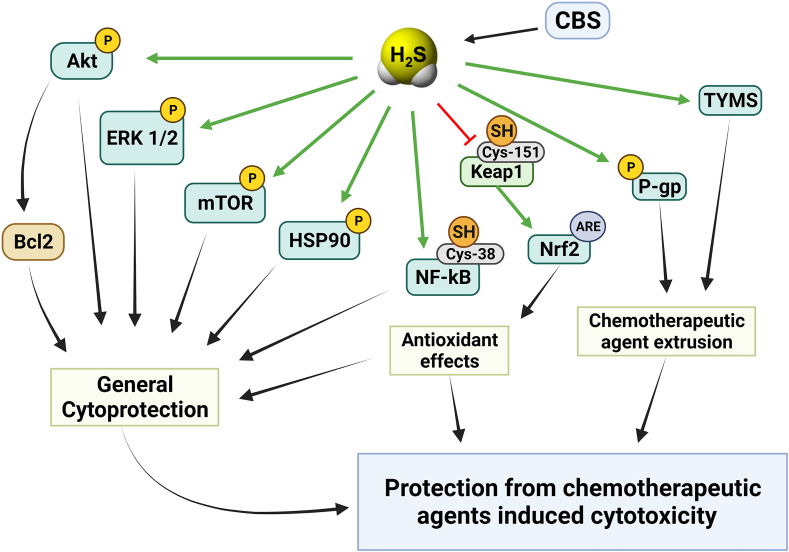
The other published studies utilizing CBS overexpression approaches mainly focused on functional parameters, such as cell proliferation. One study utilized BEAS-2B cells, a non-transformed human epithelial cell line; in these cells, CBS overexpression caused a slight increase in cell proliferation rate and increased the migration and invasion capacity of the cells [69]. Another study utilized BEL-7404 cells, a human hepatoma line, which – in contrast to most hepatoma cells studied, including HepG2 – exhibits very low CBS basal expression. In these cells, forced expression of CBS increased cell proliferation and migration, and protected the cells from the antiproliferative effect of doxorubicin, and from the antiproliferative effect of the tyrosine kinase inhibitor chemotherapeutic agent sunitinib [109]. In ovarian cancer cells, forced CBS overexpression conferred resistance to erastin (a small molecule that induces ferroptosis – an iron-dependent, lipid peroxide-mediated cell death) [102]. These findings, taken together, indicate that CBS upregulation (at least in part via its enzymatic product H2S) can “reprogram” cancer cells into a multi-drug resistant phenotype. CBS overexpression also induced the upregulation of P-glycoprotein [109], a key effector in the development of multidrug resistance. This membrane protein (also termed multidrug resistance protein 1; MDR-1) exerts its effect by accelerating the extrusion of chemotherapeutic agents from cancer cells; in fact, the lower concentration of doxorubicin in CBS-overexpressor cells, when compared to wild-type cells, was confirmed by direct measurements [109]. (Indeed, there are several independent studies demonstrating or suggesting that H2S, on its own, is able to confer chemotherapeutic resistance to various organisms ranging from bacteria to cancer cells [[119], [120], [121]].) Additional changes in the CBS-overexpressing BEL-7404 cells included an increased STAT3 phosphorylation and Akt activation, increased intracellular level of Bcl-2 and increased MMP-2 expression [109]. These effects – many of which have been previously demonstrated in cells exposed to various classes of H2S donors [114,[122], [123], [124], [125], [126], [127], [128], [129], [130]] – are likely contribute to the increased proliferative, invasive, and drug-resistant character of the CBS overexpressor cells.
Similar to the above-discussed studies, forced CBS expression into Eca109 cells (a human esophageal squamous carcinoma cell line with low basal expression of CBS) induced an increase in cell proliferation, migration and improved the viability of these cells; these effects were associated with – and may be mediated, at least in part by – the induction of SIRT1 and Hes1 and the suppression of Notch1 expression [76]. Moreover, in co-culture experiments, Eca109 cells with CBS overexpression stimulated more angiogenesis and lymphangiogenesis than the responses induced by wild-type Eca109 cells, an effect which occurred, at least in part, via the upregulation of VEGF protein expression [76]. These findings are consistent with multiple sets of studies focusing on the pro-angiogenic role of H2S; it has been well established that (a) pharmacological H2S donators stimulate angiogenesis; (b) certain endogenous pro-angiogenic hormones, including VEGF, utilize the cellular biosynthesis of H2S to exert their pro-angiogenic action and (c) H2S can upregulate various elements of multiple angiogenic pathways, including VEGF, VEGF receptor and several other pro-angiogenic factors and mediators [52,64,70,[122], [123], [124], [125], [126],[131], [132], [133], [134], [135], [136], [137], [138], [139]].
Overexpression of CBS into MCF-10A cells (a human breast cancer cell line, which expresses relatively low levels of CBS under baseline conditions) conferred resistance against activated macrophage-induced cell killing in an in vitro co-culture experiment [62].
Finally, in two liver cancer cell lines that express low-to-medium levels of CBS (MHCC97H and Hepa1-6), forced overexpression of CBS was reported to suppress, rather than stimulate proliferation [87]. These findings are in contrast with the observations made in multiple other cell types but are clearly in line with the clinical data showing that in liver cancer (in contrast to most other forms of cancer), high CBS expression correlates with better outcomes (see above).
Taken together, the above findings demonstrate that when CBS expression is “forced” into various cell types that are expressing it at low baseline levels, multiple transcriptional and posttranscriptional effects occur. In any case, the CBS-overexpressing cells assume a more highly metabolic, more proliferative, more invasive, more dedifferentiated/stem-like state and chemotherapy- and immune-cell-resistant phenotype. The CBS-induced cellular “reprogramming effects” most likely are mediated via its enzymatic product, H2S – although additional roles of CBS, for example via protein-protein interactions, may also play a role. Importantly, in many experiments, a “mirror-image” biological response (i.e. inhibition of cell proliferation and invasion, a suppression of cellular bioenergetics, etc.) can be obtained in high-CBS expressor tumor cells with CBS silencing or CBS inhibition (see subsequent sections). Importantly, however, the results obtained with CBS overexpression in with low-basal-CBS-expressor cells are markedly different from the results with cells that are expressing high baseline levels of CBS. In addition, liver cancer cells are a clear outlier: in these cells, CBS overexpression decreases, rather than increases cell proliferation.
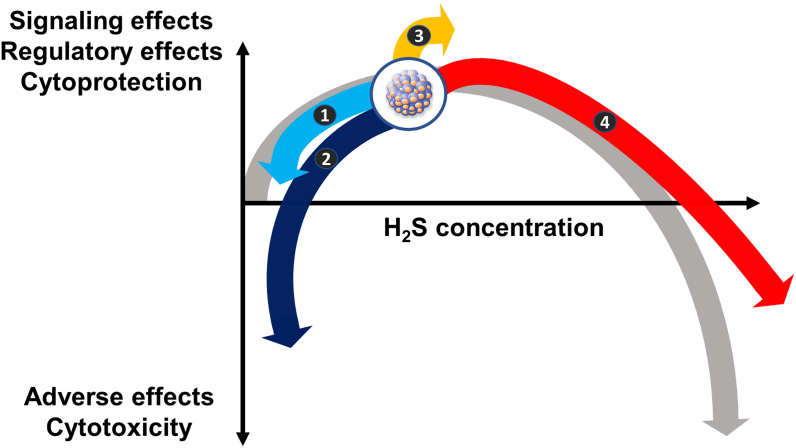
In a recent paper a CBS-overexpressor subclone of the high-CBS expressor HT-29 cells (a rapidly proliferating human colon cancer cell line) has been created; these CBS-overexpressor cells, in fact, exhibited decreased cell viability, slower cell proliferation in vitro and slower tumor growth and less metastasis formation after xenografting into nude mice in vivo [99]. The phenotype of CBS-overexpressor HT-29 cells most likely reflects the bell-shaped (biphasic, hormetic) nature of H2S – elevation of H2S concentrations over and beyond an optimal range suppresses cell metabolism, impairs proliferation and can lead to cytotoxic effects [140,141]. (This topic will be discussed in further detail at the final section of the current article.)
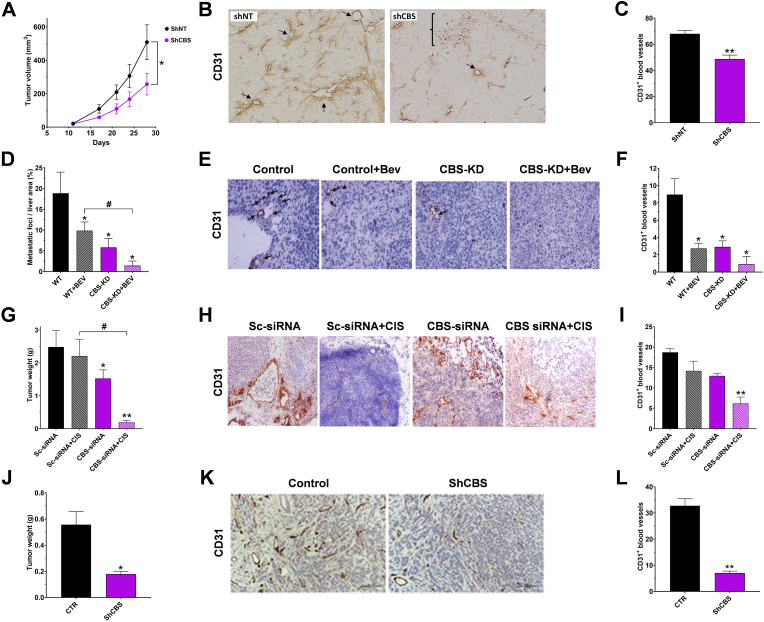
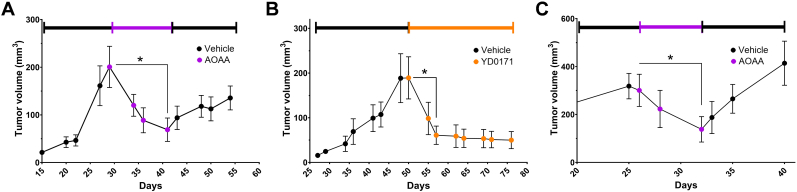
6. Effects of CBS silencing in cancer cells and tumor-bearing mouse models
The most elegant and straight-forward way to elucidate the functional role of CBS in cancer cells is to downregulate its levels via various transient and permanent silencing and knock-down approaches. There are some caveats to this approach, which are related to several issues: (a) First of all, the degree of CBS silencing, in many instances, is only partial (the degree of silencing efficiency depends on the technique used as well as the cell type). Thus, CBS may play a more significant role in the studied process that revealed by this approach. (b) Second, the absence of CBS protein does not equal to the lack of CBS catalytic activity – i.e. lack of CBS-derived H2S production; with CBS silencing/knockdown, the ”scaffolding” roles of CBS – its role in various protein-protein interactions – is also eliminated. This may be one of the reasons why pharmacological CBS inhibition and CBS silencing/knockdown do not always produce the same cellular responses (see also below). (c) Third, CBS silencing/knockdown is a long-term process; time gives an opportunity to the cell to initiate various compensatory responses, e.g. the upregulation of other H2S-generating enzymes. This possibility has not been investigated in most of the published studies.
Even with the above caveats, studies using CBS silencing/knockdown approaches have revealed many important regulatory roles of CBS in various cancer cells. The first study that utilized a silencing approach to delineate the role of CBS in the proliferation of cancer cells was published in 2013 and used a lentiviral shRNA to produce a fairly robust (approx. 80%, as quantified by Western blotting) suppression of the CBS protein in these cells [6]. HCT116 cells with CBS silencing proliferated approximately 50% slower than wild-type cells, and exhibited a reduction in their mitochondrial respiration and calculated aerobic ATP production, as well as in their glycolytic rate, suggesting that CBS (most likely via its product H2S) supports both the aerobic and the anaerobic bioenergetic pathways in these cells [6]. When subcutaneously implanted into nude mice, shCBS HCT116 cells exhibited an approximately 50% slower growth rate than wild-type cells; peritumor angiogenesis (evidenced by CD31 staining) was also reduced [6] (Fig. 7A, B, 7C).
Fig. 7.
CBS knockdown suppresses tumor growth and tumor angiogenesis in colon and ovarian cancer. Effects of CBS knockdown on colon cancer xenografts: A) tumor growth rate from shNT (nontargeting shRNA control) and CBS knockdown (shCBS) groups. Data are shown as mean ± SEM from n = 6 mice, ∗p < 0.05 compared to shRNA control. The panel was redrawn from data presented in Ref. [6]. B) Photomicrographs of representative sections (10 μm) from shNT and shCBS xenografts showing CD31-positive blood vessels (brown). Arrows indicate larger vessels and bracket indicates areas of necrosis with shCBS xenograft. The panel was redrawn from data presented in Ref. [6]. C) CD31-positive blood vessel density quantification in HCT116 tumor xenografts. Data are shown as mean ± SEM from n = 6 mice, ∗∗p < 0.01 compared to shRNA control. The panel was redrawn from data presented in Ref. [6]. D) Average area of metastatic foci in liver sections from control, control + bevacizumab (control + Bev), CBS knockdown (CBS-KD), CBS knockdown + bevacizumab (CBS-KD + Bev) groups. Data are shown as mean ± SEM from n = 5 mice, ∗p < 0.05 vs. the control group, #p < 0.05 vs. the group of mice implanted with WT cells and treated with Bev. The panel was redrawn from data presented in Ref. [52]. E) Representative images of CD31 immunohistochemical staining in liver tissue sections from control, control + Bev, CBS-KD, CBS-KD + Bev groups. The panel was redrawn from data presented in Ref. [52]. F) CD31-positive blood vessel density quantification in liver tissue sections from control, control + bevacizumab (control + Bev), CBS knockdown (CBS-KD), CBS knockdown + bevacizumab (CBS-KD + Bev) groups. Data are shown as mean ± SEM from n = 5 mice, ∗p < 0.05 vs. the control group. The panel was redrawn from data presented in Ref. [52]. Effect of CBS knockdown on chemoresistant ovarian cancer xenografts: G) tumor growth rate from Sc-siRNA, Sc-siRNA + cisplatin (Sc-siRNA + CIS), CBS siRNA, CBS siRNA + cisplatin (CBS siRNA + CIS) groups. Data are shown as mean ± SD from n = 10 mice, ∗p < 0.05, ∗∗p < 0.01 compared to siRNA control, #p < 0.05 vs. the group of mice implanted with siRNA control cells and treated with cisplatin. The panel was redrawn from data presented in Ref. [7]. H) Representative images of CD31 immunohistochemical staining in tumor tissue sections from Sc-siRNA, Sc-siRNA + CIS, CBS siRNA, CBS siRNA + CIS groups. The panel was redrawn from data presented in Ref. [7]. I) CD31-positive blood vessel density quantification in tumor tissue sections from Sc-siRNA, Sc-siRNA + CIS, CBS siRNA, CBS siRNA + CIS groups. Data are shown as mean ± SD from n = 4 mice, ∗∗p < 0.01 compared to siRNA control. The panel was redrawn from data presented in Ref. [7]. Effects of CBS knockdown on basal-like breast cancer xenografts: J) tumor growth rate from Cal51 control and shCBS groups. Data are shown as mean ± SD from n = 8 mice, ∗p < 0.05, compared to control. The panel was redrawn from data presented in Ref. [64]. K) Representative images of CD31 immunohistochemical staining in tumor tissue sections from HCC1143 control and shCBS groups. The panel was redrawn from data presented in Ref. [64]. L) CD31-positive blood vessel density quantification in tumor tissue sections from HCC1143 control and shCBS groups. The values were determined by Image J analysis from the figures shown in K). Data are shown as mean ± SEM, ∗∗p < 0.01 compared to control. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The above data, suggesting a proliferation-supportive, bioenergetic stimulatory and pro-angiogenic role of CBS and H2S in colon cancer, have subsequently been confirmed and extended in various colon cancer models. In 2022, using the CRISPR/Cas9 system to knock down CBS gene expression in SW480 and DLD1 colon cancer cell lines, the above findings have been confirmed and extended. Knocking down CBS in these cells suppressed their proliferation by approximately 30–40% and suppressed their migratory and invasive capacity by approximately 50–80% [52]. In a dorsal skinfold model, xenografting of the colon cancer cells induced an angiogenic response, which was suppressed by approximately 50% after CBS silencing [52]. CBS silencing suppressed the expression of AP-1 mRNA and inhibited the expression of VEGF by regulating its promoter [52]. In a model of intrahepatic injection of tumor cells, CBS deficient SW480 cells produced approximately 70% less metastatic foci and nodules than wild-type controls; CBS deficiency also enhanced the effect of the angiogenesis inhibitor bevacizumab [52] (Fig. 7D, E, 7F).
Additional effects of CBS silencing in colon cancer cells include an enhancement of their sensitivity to the cytotoxic action of 5-fluorouracil; the effect of CBS silencing was evident both in wild-type cells and in chemotherapy-resistant cells. This effect of CBS knockdown was associated with an increased expression of the micro-RNA miR-215-5p and a consequent decrease the expression of both epiregulin (EREG) and thymidylate synthetase (TYMS) [50]. Via these actions, CBS induces or enhances the EREG/EGFR pathway, which is considered an important effector of tumor progression and multidrug resistance in cancer [142].
In wild-type HCT116 cells, the allosteric CBS activator SAM stimulated cell proliferation and cellular bioenergetics at lower concentrations while it had exerted an inhibitory effect at higher concentrations. In CBS-silenced cells, these effects of SAM were markedly attenuated. These data (also confirmed by Western blot studies) indicate that in HCT116 cells, both a full-length and a truncated, constitutively active version of CBS co-exists; the former (but not the latter) is responsive to allosteric stimulation [38]. These data also indicate that when CBS activity (and consequent H2S generation) in cancer cells is raised beyond a certain “optimum-point”, the cells no longer benefit from it, and a cytotoxic effect and a suppression of cell proliferation occurs. This conclusion is consistent with the findings with CBS overexpression in HT-29 cells ([140], discussed above) and with the general concept of the bell-shaped H2S concentration-response (discussed in more detail below).
Ovarian cancer is another cancer type, where multiple studies have been published using CBS silencing approaches, confirming multiple tumor-supportive roles of this enzyme. In the first report, Bhattacharyya and colleagues demonstrated that CBS silencing – using an efficient siRNA method, producing approximately 80% suppression of CBS protein – in OV202, SKOV3, A2780 and A2780/CP-70 cells reduced the cells’ proliferative, migration and invasive capacity by 70–90%; CBS silencing also resulted in a 40–50% suppression of cellular O2 consumption and cell viability [7,101,102] and sensitized the cancer cells to the cytotoxic effect of the chemotherapeutic agent cisplatin [7] and the ferroptosis inducer erastin [102]. The underlying mechanisms include (a) a mitochondrial/bioenergetic effect; i.e. inhibition of mitochondrial respiration and of cellular ATP generation, similar to what has also been observed in colon cancer cells (see above) [7,61]; (b) inhibition of mitochondrial morphogenesis, leading to mitochondrial fragmentation, i.e. increased individual mitochondria numbers and overall smaller mitochondrial networks – [61]; (c) a pro-oxidant effect; depletion of cellular glutathione levels and increased cellular ROS generation [7,61,101]; and (d) a suppression of NF-κB activation, which likely affects multiple signalling pathways in the ovarian cancer cells [7]. The mitochondrial fragmentation and the increased intracellular oxidative stress after CBS silencing are functionally interlinked: when CBS silencing increases oxidative stress, this leads to the degradation of mitofusin 2 (MFN2 is a key regulator of mitochondrial morphogenesis) by the ubiquitin-proteasome system [61]. The functional role of suppressed NF-κB activation after CBS silencing remains to be directly confirmed in cancer cells but in immortalized human adipose-derived mesenchymal stem cells, CBS silencing has been shown to induce the suppression of various cytokine mRNAs [143], which is the expected consequence of the suppression of NF-κB.
Part of the reason CBS plays a vital role in the bioenergetics of ovarian cancer cells is that it supports the metabolic transition of the cells into a lipid-dependent biochemical pathway via the induction of the lipogenic transcription factors SREBP1 and SREBP2 and the upregulation of several key enzymes involved in lipid synthesis (e.g. FASN and ACC1) [101] (The upregulation of FASN gene expression after CBS silencing was also observed in other models, e.g. in immortalized human adipose-derived mesenchymal stem cells [143].). The net result is that ovarian cancer cells enhance their lipid uptake and increase their lipid content and utilize the metabolism of these lipids to support their bioenergetics and their function; CBS silencing markedly suppresses all of these responses [101]. These roles of CBS are, at least in part, mediated by modulation of Sp1 activation: CBS silencing suppressed the nuclear translocation of this transcription factor [101]. Importantly, several studies – in experimental models other than cancer) – show that Sp1 is target of sulfhydration by H2S [133,144,145]. A potential working hypothesis is that in ovarian cancer CBS-derived H2S promotes Sp1 sulfhydration, which, in turn, increases its stability and/or facilitates its nuclear translocation, which promotes the expression of various lipogenic genes.
The functional importance of CBS in ovarian cancer is also highlighted by in vivo studies, demonstrating that silencing of CBS in a model of intraperitoneal administration of A2780/CP20 cells into mice markedly reduces tumor growth, tumor nodule numbers and the proliferative activity (assessed by Ki67 staining) and suppresses the tumor's angiogenic potential (assessed by CD31 staining). In line with the in vitro studies showing a synergistic effect of CBS silencing with cisplatin (see above), a marked synergy of CBS silencing with cisplatin therapy was observed [7] (Fig. 7G, H, 7I).
Similar to colon and ovarian cancer cell lines, CBS silencing also exerts significant functional effects in various human breast cancer cell lines, although there are some significant differences in the published literature. In MCF7 and MDA-MB-468 cells, CBS silencing, on its own, did not affect proliferation rates or cell viability in vitro over 5 days in the studies of Sen and colleagues [62], while in the same cell line (MCF7) and a closely related one (MDA-MB-231), under slightly different experimental conditions, Youness and colleagues have observed an antiproliferative and anti-migratory effect of CBS silencing, as well as a lower colony-forming capacity in vitro [65]. Most recently, once again, antiproliferative effects and diminished cell viability were also observed after CBS silencing in the basal-like breast cancer cell lines Cal51 and HCC1143 [64]. All three groups working in the area of breast cancer and CBS agree, however, that CBS silencing markedly sensitizes breast cancer cells to the effect of various external cytotoxic/cytostatic interventions, including various pro-oxidative conditions such as exposure to 4-hydroxynonenal, a cytotoxic and genotoxic product of lipid peroxidation [64], exposure to hydrogen peroxide, a cytotoxic oxidant [64], and the pharmacological depletion of intracellular glutathione [62,64]. Moreover, CBS depletion in various breast cancer cell lines sensitizes to the antiproliferative and cytotoxic effect of activated macrophages [62], activated NK cells [65] or activated T-cells [65] in various co-culture conditions.
The relationship of CBS to the regulation of various intracellular redox processes has been investigated, in various breast cancer cell lines, to some extent, by Kawahara in 2017 [105], and subsequently, in significant detail, by Nagy's group in 2021 [64]. Kawahara has demonstrated in MCF7, MDA-MB-468, and Hs 578T cells that CBS silencing suppresses intracellular glutathione levels, GSH/GSSG ratio, together with increased intracellular ROS generation [105]. These changes, at least in part, were attributed to the fact that CBS silencing decreases the expression of genes that promote GSH biosynthesis and regeneration such as Nrf2, G6PD, GCLC and GSR [105]. Subsequent studies, in Cal51 and HCC1143 cells, however, have observed increased, rather than decreased intracellular GSH levels after CBS silencing, suggesting that the increased vulnerability of shCBS cells to oxidative stress is not due to impaired GSH levels.
In Cal51 cells CBS knockdown sensitized to CySSCy deprivation–induced lipid peroxidation and cell death; when the cells were grown in reduced CySSCy-containing medium, shCBS cells became more sensitive to the cytotoxic effect of oxidative stress. Taken together, Cal51 and HCC1143 cells appear to have a “CySSCy-addicted” phenotype and in this regard the upregulated transsulfuration pathway in them serves to maintain their cysteine need [64]. A potential future anticancer approach that follows from these observations may be the “starving” of the cancer cells from their special cysteine “need”, either by inhibiting relevant intracellular pathways and/or by suppressing their ability to take up cysteine from the extracellular space (see also below).
Importantly, in vivo, breast cancer cells with CBS silencing grow substantially slower than wild-type cells, as shown with the subcutaneous implantation of MCF7 cells into nude mice where an approximately 90% suppression of tumor growth was observed [62], or HCC1143 or Cal51 cells, where an approximately 60–80% suppression of tumor growth was observed, together with an enhancement of intratumor necrosis and an inhibition of peritumor angiogenesis (Fig. 7J, K, 7L) [64].
A similar role of CBS was also demonstrated, using various CBS silencing approaches, in A549 lung cancer cells where CBS silencing was found to suppress cancer cell proliferation, migration, and invasion; it also delayed the repair of mitochondrial DNA after oxidative cell injury and potentiated the cytotoxic effect of the anticancer therapeutic agent camptothecin [69].
Although from the analysis of clinical samples it is clear that in liver cancer tissues, CBS becomes downregulated, rather than upregulated, and in liver cancer high CBS predicts better, rather than worse outcomes (see above), in various in vitro and preclinical in vivo studies, the effect of CBS inhibition in various liver cancer models is not drastically different from the effects observed in the cancer types discussed earlier (colon, ovarian, breast and lung). This may be due to the fact that in liver cancer cell lines – even after some degree of CBS downregulation compared to healthy tissue – sufficient levels of CBS remain in these cells, which may provide tumor-cell-supporting roles, especially in an in vitro system where the cells are not grown on the background of a (basally high-CBS-expressor and high-H2S-producer) normal healthy liver tissue. The main findings in various liver cell lines with CBS silencing include variable effects on proliferation, i.e. either pro- or antiproliferative and either cytotoxic or cytoprotective actions in various hepatoma cell lines [87,108,109]; inhibition of the expression of certain EMT/MET markers [110] and enhancement of the radiation-induced cell killing (reported in HepG2 cells) [110]. In Hep3B cells, – as opposed to many cell types discussed earlier – CBS silencing exerted no significant effect on any of the bioenergetic parameters (oxidative phosphorylation or glycolysis) [87]. Importantly, in vivo, in nude mice bearing subcutaneous Hep3B xenografts, silencing of CBS in these cells resulted in an increased tumor growth [87]. This increase was unrelated to any changes in peritumor angiogenesis, but were linked to the regulation of tumor-infiltrating immune cells (Tregs) [87] (see also below). These findings in hepatoma cells, once again, are in contrast with the observations made in multiple other cell types but are clearly in line with the human clinical correlations showing that high CBS expression in liver cancer tissue, correlates with better prognosis (see above).
Regarding other forms of cancer – e.g. various forms of renal or gastric carcinoma – where clinical data or pharmacological inhibitor studies indicate a role of the CBS pathway (see above and below), no published studies are available using CBS silencing approaches. The only exemption is esophageal squamous cell carcinoma, where the functional effect of CBS silencing has been examined using KYSE450 cells. In these cells, viability, proliferation and migration were suppressed in the absence of CBS, and the percentage of apoptotic cell proliferation increased. In in vitro tumor-endothelial cell co-culture studies, CBS silencing suppressed angiogenic and lymphangiogenic responses, at least in part via suppression of VEGF expression [76]. In vivo, when CBS-knockdown KYSE450 cells were injected subcutaneously into the right flank of nude mice, tumor growth was reduced by approximately 70% and lymph node metastasis was also significantly suppressed [76]. With regards to the mode of action, in KYSE450 cells, the mechanism of CBS silencing was, at least in part, linked to the suppression of SIRT1 induction, and to the stimulation of the Notch and Hes1 signalling pathways [76]. In many respects, CBS silencing produced an exact “mirror image” of the responses seen with CBS expression in a low-baseline CBS-expressor cell line Eca109 (see above).
Similar to the clinical data where role of CBS in glioma represents an “outlier” (with high CBS expression correlating with better prognosis, see above), also in cell-based and animal-based studies, the role of CBS in glioma models is the exact opposite of what has been observed in many other cancer models. In U87-MG cells, CBS silencing increased (rather than decreased) cell proliferation and invasion in vitro and promoted (rather than suppressed) tumor growth in a xenograft model in vivo; these effects were associated with an increase (rather than decrease) of tumor angiogenesis [88]. The mechanism of action of CBS silencing in this experimental system/in this tumor type has not yet been fully characterized but they were found to be related, at least in part to increases in HIF2α expression, VEGF and angiopoietin 2 and 4 expression and anchorage-independent cell growth; there are also more recent studies confirming and extending these findings and showing that the growth of glioma is further enhanced under conditions of high-fat diet [55]. Clearly, the role of CBS in the pathophysiology of glioma/glioblastoma is markedly different from all other cancers [55,88,113].
Finally – and in contrast to the multiple lines of additional studies showing the effect of CBS silencing and CBS inhibition in many forms of cancer – in a paper by Yamamoto and colleagues, in 2014, CBS silencing in HCT116 cells was found to increase, rather than decrease, proliferation rate and tumor formation in metastatic tumors of the liver [146]. The reason for the discrepancy is currently unclear; in the subsequent 8 years since this study was published the Yamamoto group, to our knowledge, has not followed up this isolated report and this study has not since been reproduced or confirmed by any other group.
Some key studies demonstrating the functional effect of CBS silencing in various cancer cell lines are shown in Fig. 7. The vast majority of the published literature using CBS silencing in multiple cancer models (except in hepatoma and glioma models) supports the notion that when CBS expression is high, cancer cells assume a hypermetabolic, hyperproliferative, hyperinvasive, dedifferentiated/stem-like phenotype and they become resistant to oxidative stress, chemotherapy and immune cell mediated killing. This notion is also indirectly supported by CBS silencing studies conducted in non-transformed cells. For instance, when human dermal fibroblasts are placed in a 3D culture, a dedifferentiation-type reprogramming response takes place, which is driven, at least in part, by the upregulation of the pluripotency factors such as OCT4, and Nanog in these cells. Importantly, this dedifferentiation process has been functionally linked to the upregulation of CBS: CBS silencing suppressed the expression of OCT4, and Nanog [147].
Taken together, the body of literature describing the effects of CBS overexpression (previous section), CBS silencing (current section) and pharmacological inhibition (next section) supports the view that CBS is tumor-supportive factor. Thus – with the important exception of liver cancer and glioma – CBS, in many cancers, can be characterized a preclinically validated anticancer drug target.
7. Pharmacological inhibition of CBS in cancer cells and cancer models
When considering the translation of the CBS/H2S/cancer concept into future clinical, therapeutic applications, pharmacological CBS inhibition becomes a crucial topic. CBS inhibition has been subject of multiple recent reviews [3,12]; in the current section we will focus on those CBS inhibitors which have been tested in cancer models (Table 2).
Table 2.
Summary of the effects of various pharmacological CBS inhibitors in cancer models.
| CBS inhibitor compound | Name | hCBS IC50 | Characteristics | Effects in cancer models |
|---|---|---|---|---|
 |
AOAA | 8 μM | PLP-dependent covalent inhibitor; inhibits multiple other enzymes as well | Efficacy in a cellular model in vitro and in murine models in vivo |
| AOAA methyl ester AOAA isopropyl ester |
YD0171YD0251 | 8 μM (with better cell uptake) | After hydrolysis by esterases, it yields AOAA and exerts similar effects as AOAA at lower concentrations/doses. | Efficacy in a cellular model in vitro and in murine models in vivo |
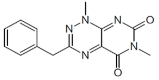 |
CH004 | 2 μM | Competitive inhibitor, identified by HTS, has known or likely off-target effects | Efficacy in a cellular model in vitro and in murine models in vivo |
 |
Sikokianin C | 3 μM | Competitive inhibitor, identified by HTS, has known off-target effects, not druggable | Efficacy in a cellular model in vitro and in a murine model in vivo |
 |
Benserazide | 30 μM | PLP-dependent covalent inhibitor; known clinically used drug – may be a possible candidate for drug repurposing | Efficacy in a cellular model in vitro and in murine models in vivo |
 |
Disulfiram | 1 μM (after conversion to CuDCC) | Covalent inhibitor via copper; known clinically used drug with multiple MOA's – may be a possible candidate for drug repurposing; currently in several cancer clinical trials (but the proposed mechanism of action that is unrelated to CBS) | Efficacy in a cellular model in vitro and in murine models in vivo |
| Various polyphenols | [Multiple] | 5–30 μM | Multiple targets, most of them natural compounds | No information |
 |
NSC11041 NSC67078 |
4–10 μM | Partially characterized compounds from various HTS campaigns; CBS selectivity and cell uptake insufficiently characterized | No information |
Although CBS is an enzyme that has been studied for many decades, pharmacological inhibitors of this enzyme remain to be improved: all of the currently known CBS inhibitors have some significant problems in terms of potency, selectivity, and/or cell permeability. While they are useful as experimental tools (with many caveats), they are not suitable for clinical translation.
Aminooxyacetic acid. Aminooxyacetic acid (AOAA, also known as carboxymethoxyamine), is commonly referred to in the literature as a “CBS inhibitor” – and in some cases even as a “potent CBS inhibitor”. In fact, AOAA is an inhibitor of several (but not all) PLP-dependent enzymes, including a second H2S-producing enzyme, CSE, as well as several different transaminases [12,148]. On the various non-CBS enzymes, the inhibitory potency of AOAA is variable. The mammalian enzymes which are inhibited by AOAA with the highest potency (IC50 < 10 μM) are – in addition to CBS and CSE – (a) the various isoforms of alanine transaminase (ALT, also known as glutamate pyruvate transaminase or GPT); (b) glutamate decarboxylase; and (c) GABA transaminase (GABA-T). The latter two enzymes are predominantly expressed in the CNS, and they are not considered significant actors in the pathobiology of most cancers. Thus, in the context of cancer biology, the principal secondary (or, in the context of H2S biology, “off-target”) effect of AOAA is inhibition of ALT/GPT. Indeed, silencing studies have demonstrated that several subtypes of these transaminases play significant bioenergetic and other regulatory roles in various cancer cells. In fact, in several cancer papers AOAA is referred to as a “GOT inhibitor” – or more specifically, an inhibitor of GOT2, an isoform with specific roles in cancer – or as an inhibitor of glutamic pyruvate transaminase 2 (GPT2), another transaminase with significant cancer cell supporting functions [[149], [150], [151], [152], [153], [154]].
Inhibition of CBS by AOAA is generally viewed as pharmacologically irreversible; it is based on the covalent interaction of AOAA with the PLP prosthetic group in the active site of CBS [12]. The off-target actions of AOAA have been extensively discussed recently [3,12,41,42]. Even though AOAA exhibits the above problems of selectivity, it remains a frequently used compound, due to the fact that the alternative pharmacological tools are rather limited, and due to the fact that it is, in fact, a fairly potent inhibitor of CBS (depending on the assay conditions its IC50 ranges between 1 and 10 μM). The multiple off-target effects of AOAA must be considered when interpreting the anticancer effects of this compound.
In many of the studies discussed in the previous section that employed CBS silencing approaches, AOAA has also been used to confirm and extend the findings. In colon cancer cells, AOAA phenocopied the effect of CBS silencing in terms of inhibition of cell proliferation, suppression of mitochondrial electron transport and ATP generation, and suppression of cancer cell invasion and tumor angiogenesis [6,50]. In addition – and in line with multiple studies demonstrating a similar effect of CBS silencing – AOAA was found to enhance the cytotoxic effect of 5-fluorouracil in several different human colon cancer cell lines in vitro [50]. AOAA also exerted effects in HCT116 cells that are consistent with inhibition of EMT [97], but in this cell type, it has been not assessed if CBS silencing exerts similar effects. However, in lung cancer cell lines, both CBS silencing and AOAA were found to exert comparable effects in terms of inhibition of EMT (see below).
AOAA, in some experiments, especially at higher concentrations, exerts direct cytotoxic (apoptosis-inducing) effects to cancer cells – which may be due to the combination of inhibition of H2S biosynthesis, as well as additional inhibitory effects on various transaminases, as well as possible non-specific toxic effects of the compound at higher concentrations [6,7,95]. In most studies, however, AOAA did not have major effects on the distribution of the cells in various phases of the cell cycle – AOAA definitely does not induce the type of cell cycle arrest that is characteristic to many chemotherapeutic drugs [6,50,51,155]. Importantly – and in contrast to the effect of CBS silencing – AOAA does not, however, appear to be particularly effective in reversing the acquired resistance of cancer cells to chemotherapeutic agents [50,93].
In vivo, AOAA was also found to suppress the growth and metastatic capacity of xenografted human colon cancer cell lines [6]. AOAA treatment of nude mice bearing NCM356 cells with CBS overexpression resulted in the regression of the tumor growth (Fig. 8A) [48]. AOAA also suppressed the growth of patient-derived colon cancer xenografts, and this effect was independent of the patients’ KRAS status [51,155,156]. Importantly, AOAA potentiated the inhibitory effect of 5-fluorouracil in a colon cancer-bearing nude mouse model [50]. Moreover, role of CBS-derived H2S in the promotion of peritumor angiogenesis – as demonstrated using CBS silencing and CBS overexpressor approaches (see above) – has also been confirmed in multiple studies using AOAA [6,50].
Fig. 8.
Pharmacological inhibition of CBS causes the regression of established tumors in tumor-bearing mouse models. A) Effect of CBS inhibitor (AOAA) in tumor growth rate of NCM356 cell line overexpressing CBS xenografts. Data are shown as mean ± SEM of n = 10 animals per group; ∗p < 0.05 shows significant difference between tumor size between the indicated two time points. The figure was redrawn from data presented in Ref. [48]. B) Effect of CBS inhibitor (YD0171) in tumor growth rate of HCT116 xenografts. Data are shown as mean ± SEM of n = 8 animals per group; ∗p < 0.05 shows significant difference between tumor size between the indicated two time points. The figure was redrawn from data presented in Ref. [155]. C) Effect of CBS inhibitor (AOAA) in tumor growth rate of A549 xenografts. Data are shown as mean ± SEM of n = 5 animals per group; ∗p < 0.05 shows significant difference between tumor size between the indicated two time points. The figure was redrawn from data presented in Ref. [69].
In cell-based studies, and especially in studies with tumor-bearing mice, one of the problems with AOAA is its limited cell uptake, which necessitates the use of high micromolar, in some cases low millimolar concentrations of this compound in cancer cells. By creating ester prodrugs of AOAA, more potent compounds, such as the AOAA ethyl ester YD0171 and the AOAA isopropyl ester YD0251 have been created and tested in models of colon cancer [51,155]. These compounds exert more potent anticancer effects than AOAA, but – since after cleavage by esterases, they generate AOAA intracellularly – the prodrug approach cannot solve the selectivity or specificity issues of AOAA itself. Nevertheless, using one of these prodrug molecules, the regression of established colon cancer tumors has been demonstrated in a nude mouse model (Fig. 8B) [155]. The efficacy of another one of these prodrug molecules (YD0251) has also been demonstrated that high-CBS expressor PDTX-bearing nude mice [51].
AOAA has also been extensively utilized in ovarian cancer models, mostly to confirm and extend the findings obtained with CBS silencing. In various human ovarian cancer cell lines, AOAA phenocopied the effects of CBS silencing in terms of suppression of cell proliferation, migration, invasion and metabolism [7,101,102]. AOAA also phenocopied many of the signalling and gene expression alterations that were noted with CBS silencing [101,102].
In breast cancer studies, most of the recent literature utilizes CBS silencing, rather than CBS inhibitors. However, already a report from 2008 has reported the inhibitory effect of AOAA on the growth of human breast cancer cells in a murine model [149].
Similar to colon and ovarian cancer cell lines, AOAA also phenocopied many of the CBS silencing effects in various human lung cancer cell lines, including the antiproliferative effects, the suppression of mitochondrial activity and the inhibitory effect on mitochondrial DNA repair [69,70]. Using AOAA, the mechanism underlying this latter response has been identified: it relates to a mechanism by which CBS-derived H2S, via sulfhydration of the mitochondrial DNA repair enzyme EXOG, which, after sulfhydration, is more effective in catalyzing the formation of a mitochondrial DNA repair complex consisting of EXOG, APE1, PolG and Lig3 [69]. In various lung cancer cell lines in vitro, as well as in a tumor-bearing mouse model in vivo, AOAA not only exerted antiproliferative effects but also potentiated the anticancer effect of irinotecan, oxaliplatin and camptothecin [69]. In A549 lung cancer cells AOAA also phenocopied the effect of CBS silencing in its ability to decrease vimentin expression, indicating an inhibitory effect against epithelial-to-mesenchymal transition [70]. Importantly, treatment of mice bearing A549 xenografts with AOAA markedly suppressed tumor growth [69,70]; moreover, delayed treatment with this agent resulted in tumor growth arrest/regression [69] (Fig. 8C); so far, tumor regression in response to CBS inhibition has only been reported in this model and in some of the colon cancer models discussed in other paragraphs of the current section.
Similar to CBS silencing (see above), AOAA has also been shown to act as a radiosensitizer in various cancer cells in vitro [116,157]. Additional effects of AOAA in various cancer cells include inhibition of cell proliferation, suppression of cell viability and inhibition of tumor growth [82,108,112,[158], [159], [160], [161]]. However, in several of the studies cited here, the mode of action of AOAA has been attributed to mechanisms other than CBS inhibition: most commonly to inhibition transaminases, glutaminolysis and/or the inhibition of the malate-aspartate shuttle.
In line with the clinical observations in liver cancer and in line with studies of CBS silencing in various liver cancer cell models, in several liver cancer cell lines, AOAA was found to enhance cell proliferation in vitro; it also stimulated tumor growth in vivo [87]. We would like to reiterate that the pathomechanistic role of CBS in liver cancer is markedly different from its role in multiple other tumor models and this distinction is supported not only by genetic but also by pharmacological experiments.
Hydroxylamine. Another, non-selective compound which inhibits CBS (as well as a variety of additional enzymes) is hydroxylamine. From the 1950's, in the cancer literature, many reports can be found showing its anticancer effects in various models [[162], [163], [164], [165]], but the mechanism of its action is difficult to determine. Fortunately, nowadays this compound is only very rarely being used experimentally; there are only one published study, in renal cancer cell lines where it has been discussed in the context of CBS and cancer [106].
Benserazide. Various screening campaigns have been conducted to determine if any of the clinically used drugs have a secondary, CBS inhibitory property, which may be, in turn, used in the context of drug repurposing. These efforts have identified benserazide, which is one component of a clinically used two-component anti-Parkinson drug. The mode of benserazide's action, in the clinical context, is inhibition of DOPA decarboxylase (which is also a PLP-dependent enzyme). Benserazide inhibits CBS through a covalent inhibition via PLP that is similar to the action of AOAA [166,167]. Its potential anticancer effects have not been characterized extensively, but in one study it has been shown a partial inhibitory effect on colon cancer cell growth in mice at high doses (up to 600 mg/kg/day in mice) – even though the proposed mechanism of its action was attributed to hexokinase inhibition in this report [168]. Recently, benserazide has also been characterized as a pharmacological inhibitor of pyruvate kinase (PKM2); and in this role, it has been tested in melanoma models, where it reduced cell proliferation, migration, and invasion in the concentration range of 10–50 μM and inhibited tumor growth in melanoma-bearing nude mice in the dose range of 40–80 mg/kg/day [169]. Benserazide appears to be fairly well tolerated in vivo in the above animal studies, even at the high doses used. Prior observations, demonstrating that benserazide increases circulating homocysteine levels in experimental animals and in patients [[170], [171], [172]], suggests that the compound does, indeed, exert a CBS inhibitory effect in vivo. Nevertheless, it is unclear whether clinical repurposing is a viable translational option for this compound.
Disulfiram. Another clinically used drug which exhibits CBS inhibitory effects – both in cell-based systems and in vivo – is disulfiram. This compound has been originally identified as a CBS inhibitor in a yeast-based screen in the context of a CBS/Down syndrome project, and, indeed, treatment of Down syndrome mice with this compound yielded neurobehavioral responses that phenocopied the functional effect of CBS silencing in this model [173]. The mechanism of its CBS inhibitory effect involves its reduction to diethyldithiocarbamate (which is considered the major metabolite of disulfiram), followed by the chelation of intracellular copper (II) and the “delivery” of copper to the CBS molecule, resulting in an inhibitory effect [96]. (In fact, on a molar basis copper (II) ions are probably the most potent known inhibitors of CBS, with an IC50 of approximately 300 nM [167,174,175]). Disulfiram has been shown to exert anticancer effects in various preclinical models, and several targets have been proposed to explain this effect [[176], [177], [178], [179]]. In fact, disulfiram (alone, or in various combinations, including a combination with copper) is currently in several clinical cancer trials [[176], [177], [178], [179]]. Thus, it is possible that disulfiram may be ultimately repurposed for cancer therapy, although the extent to which a CBS inhibitory effect contribute to its clinical anticancer action remains unclear.
CH004. CH004 (3-benzyl-1,6-dimethylpyrimido(5,4-e)(1,2,4) triazine-5,7(1H,6H)- dione or 3-benzyl-toxoflavin) was first introduced into the scientific literature in 2018 through a paper by Wang and colleagues [111] who have reported it as a CBS inhibitor with high potency (IC50: 1 μM in human enzyme) and some selectivity for CBS over CSE [111]. This paper refers back to an earlier report, a high-throughput screen conducted in 2013 by Zhou and colleagues, which identified several novel classes of CBS inhibitors [180]. However, in this latter paper, only the core structure of CH-004 (i.e. 1,6-dimethylpyrimido(5,4-e)(1,2,4) triazine-5,7(1H,6H)-dione) has been reported; this compound (also known as toxoflavin or NSC67078) also inhibits CBS-derived H2S generation in the single μM IC50 range [167,180]. However, the first mention of both of these compounds (toxoflavin and its 3-benzyl derivative), appears in the patent literature, with its priority dating back to 2008, in the context of CBS-derived H2S overproduction in Down syndrome [181].
Toxoflavin is, in fact, a natural compound; it is the prosthetic group of a yellow pigment formed by Burkholderia gladioli. Closer look at the pharmacological action of toxoflavin (and, likely, its derivative including CH004) reveals several problematic issues. First of all, toxoflavin not only inhibits CBS-derived H2S signals, but, with almost comparable potency, it also inhibits H2S signals from chemical H2S generators (“H2S donors”) suggesting a combined CBS inhibitory and H2S decomposing/scavenging mode of action [167]. Second, toxoflavin is known to have several molecular targets unrelated to CBS or H2S, including inhibition of sirtuins, inhibition of KDM4A (lysine demethylase 4A), inhibition of the protein-protein interaction between T cell factor 4 (Tcf4) and β-catenin, inhibition of protein disulfide isomerase, inhibition of tyrosyl-DNA phosphodiesterase II, as well as a significant redox activity that can yield the generation of various pro-oxidant species in biological systems [[182], [183], [184], [185], [186], [187], [188], [189]]. Third, toxoflavin – most likely due to the combination of the above actions – is a known cytotoxic compound, which inhibits the mitochondrial electron transport chain, suppresses cell respiration and exerts antiproliferative and cytotoxic effects in various cell types including several cancer cell lines [111,167,[182], [183], [184], [185], [186], [187], [188], [189]]. Thus, there is no doubt that toxoflavin and CH004, are, indeed “anticancer compounds”. However, these anticancer effects – for instance as reported in HepG2 cells in the study of Wang and colleagues [111] – are likely to involve several mechanisms in addition to CBS inhibition. Nevertheless, the anticancer effect of CH004 was tested in several cell lines that express high levels of CBS protein (HepG2, HEK293T, Huh7, H22, Panc-28, HCT116, and MDA-MB-231) [111,167]. In all cases, the molecule inhibited cell proliferation with IC50 values in the 10–20 μM range. In the same concentration range, cellular H2S generation was inhibited as well. In HEK293T cells after CBS silencing was achieved using an siRNA approach, the pharmacological action of CH004 diminished but was not absent, once again, reinforcing the idea that the “toxoflavin class” of compounds have multiple cellular and molecular targets.
Sikokianin C. The CBS inhibitory effect of several different flavonoids emerged from various small molecule screening campaigns seeking to identify novel CBS inhibitors [166,180,190]. The CBS inhibitory effect of Sikokianin C, a natural biflavonoid compound (originally isolated from the roots of Wikstroemia indica), was first reported by Niu and colleagues who have identified it through a high-throughput screening, employing a fluorescent thiol to capture the CBS-catalyzed production of methanethiol from the artificial substrate methylcysteine [191]. In a subsequent report, Niu and colleagues followed up on the pharmacological effects of sikokianin C in biochemical models (molecular docking with CBS) and in cell-based and animal models of colon cancer [92]. The molecular docking highlighted five amino acid residues in CBS (His203, Tyr308, Tyr223, Asn194 and Thr193) which interact with the molecule via hydrogen bonds.
Follow-up studies demonstrated the inhibitory effect of sikokianin C on the proliferation of the human colon cancer line HT29 cells in vitro and the suppression of HT29 growth in tumor-bearing mice in vivo [52,92]. Its antiproliferative effect was attenuated (but was not abolished) after siRNA-mediated CBS silencing in HT29 cells [92]. In a murine model of liver metastasis of colon cancer cells, sikokianin C enhanced the antitumor effect of the VEGF-A inhibitor bevacizumab [52]. These data suggest that – similar to the case of CH004 – the mode of sikokianin C's action involves multiple mechanisms in addition to the inhibition of CBS. In fact, sikokianin C is known to have several pharmacological effects including antimalarial effects, anti-inflammatory effects, and an inhibitory effect on the expression of the inducible nitric oxide synthase [[192], [193], [194]].
8. Potential roles of host CBS in cancer
CBS is expressed in many mammalian cells and tissues. As mentioned earlier, the brain and the liver contain particularly high levels of CBS, and, according to our working hypothesis (see above) the fact that CBS has a markedly different functional role in liver cancer and glioma than in most other cancer types may be related to the fact that these cancers are developing on the background of particularly high CBS/H2S environment. CBS is also present in many other organ systems (e.g. kidney, lung, gastrointestinal system, cardiovascular system) and – perhaps most importantly for the purpose of the current article – various immune cells [12,195]. Moreover, part of the intestinal microbiome consists of H2S-producing bacteria, which produce H2S by several different enzymes, including a bacterial CBS-equivalent [[196], [197], [198], [199]]; although much of the H2S generated in the intestinal system becomes neutralized by the intestinal epithelial cells, some of it crosses into the circulation and contributes to the systemic H2S levels [199,200]. Thus, cancer cells – in addition to their internally generated H2S – are also exposed to “external” H2S – produced by various cell types in the cancer microenvironment and/or delivered to them via the blood supply – which may, in turn, regulate their function via a variety of mechanisms.
CBS and carcinogenesis. From the standpoint of intestinal carcinogenesis, there are multiple lines of emerging data that a dysregulation of H2S homeostasis in the intestinal microbiome may contribute to the pathogenesis of colitis, which, in turn, may contribute to the pathogenesis of intestinal polyp formation and carcinogenesis. This field of research is covered in recent review articles [[198], [199], [200], [201], [202]]. As far as the host (mammalian) CBS and carcinogenesis goes, the body of literature is minimal. There is some clinical correlative indication that single nucleotide polymorphisms in CBS shows an association with postmenopausal breast cancer incidence [203]. Moreover, individuals who are heterozygotes for 68-basepair insertion in the exon 8 coding region of CBS are associated with a reduced colorectal cancer risk [204,205]. In animal studies, the results are conflicting. In a model of azoxymethane-induced aberrant crypt foci formation in mice, CBS heterozygous (CBS+/-) mice developed approximately 50% less foci than wild-type control mice [48]. In contrast, in model of diethylnitrosamine-induced liver cancer in mice, only the CBS heterozygous (CBS+/-) mice developed micro-tumor lesions after 4 months (1/3 of the animals), while none of the wild-type controls have developed lesions [87]. In light of the contrasting role of CBS in other aspects of colon cancer vs. liver cancer (see above), the discrepancy between these two sets of findings is not particularly surprising. Nevertheless, the underlying molecular mechanisms remain uncharacterized. When taken together, the body of data regarding the role of host CBS and carcinogenesis is minimal and remains to be explored in the future.
CBS and tumor immunity. CBS is expressed in various immune cells and has been shown to affect immune cell development and differentiation in multiple ways [195,206]. H2S, at low concentrations, can increases T cell activation, and IL-2 production, while T cell activation and proliferation is significantly inhibited when CBS expression is suppressed by siRNA. H2S also increases the capacity of T cells to create immunological synapses [[207], [208], [209]]. Interestingly, CBS expression is markedly increased during T-cell activation [210]. Moreover, when Treg cells are polarized to Th1, Th2 or Th17, Treg cells from CBS−/- mice tend to be directed into the Th1 and Th17, but not Th2 lineage [211,212].
T regulatory cells (Tregs, which are known to play an especially important role in immunological homeostasis, self-tolerance and antitumor responses) also express high levels of CBS; inhibition of CBS in mice decreases the relative proportion of FoxP3+ Tregs, indicating that CBS plays a role in the T cell polarization and/or Treg maintenance [211,212]. CBS knockout mice have fewer Tregs, and the reduction of Tregs cells is linked to immune cell infiltration and higher autoantibody production [211,212]. Elevation of intracellular H2S levels in T cells (achieved by CSE overexpression) was recently shown to enhance the antitumor effect of these cells in various model systems in vivo: importantly, administration of CSE-overexpressing CD8+ T cells to melanoma-bearing mice suppressed tumor growth and improved survival to a greater extent than infusion of wild-type CD8+ T cells [213]. Interestingly, this effect was not attributed to increased proliferation or higher antitumor cytotoxicity of these T cells, but likely to be related to an altered metabolic milieu of these cells [213].
Although the roles of H2S and CBS in various immune cell subsets are incompletely understood, one would predict that CBS deficiency or CBS inhibition may influence several components of antitumor immunity and may affect the growth of tumors in vivo. However, the data published in this regard are very limited. The only publication which directly examines the role of the host CBS in tumor-bearing mice is a recent study by Zhou and colleagues who have compared the growth of human hepatoma xenografts in wild-type vs. CBS ± mice and reported that tumor growth is increased in the CBS ± mice; this is associated with an increased infiltration of Tregs into the tumor tissue [87]. These data indicate that host CBS, in this model, has a tumor-suppressive role. It should be pointed out, nevertheless, that the above study focused on the pathogenesis of liver cancer, and, in terms of the role of CBS, this form of cancer is a clear outlier compared to most other forms of cancer studied (see above). Thus, the role of host CBS in other forms of cancer (e.g. colon cancer, ovarian cancer, breast cancer, lung cancer etc.) remains to be investigated in the future. One problem that is likely to arise during the course of future studies that when using immunocompetent models (fully immunocompetent mice bearing murine tumors), the relative contribution of the various H2S-producing enzymes in murine tumors will be often different from the relative contribution of these enzymes in human cancers (see also below).
9. Roles of CSE and 3-MST in cancer; interactions with CBS
As already mentioned in the Introduction, mammalian cells produce H2S from three principal enzymes, CBS, CSE and 3-MST. The current review's focus is CBS. Nevertheless, there are multiple lines of data that CSE and 3-MST can also play tumor-supporting roles in various forms of cancer. There are also examples where CSE and 3-MST may exert additive or cooperative roles with CBS to support the bioenergetic and proliferative function of various cancer cells. The following section provides a brief summary of these findings – with CBS used as a reference point and comparator.
CSE. The clinical correlations with respect to intratumor CSE expression and patient outcomes are less prominent than in the case of CBS. Silver and colleagues have not reported any differences in clinical outcomes between high and low CSE expressors in most cancers (colon, ovarian, breast, lung, kidney, pancreas), while in liver cancer – similar to CBS – high CSE expression was associated with better survival [55]. In clinical colon and ovarian cancer specimens, most studies have not noticed higher CSE expression than in normal surrounding tissue and CSE inhibition or silencing was not found to exert significant functional effects [6,7,49,102], except in one study in SW480 human colon cancer cells, where CSE inhibition or CSE silencing exerted a partial, but statistically significant inhibitory effect on cancer growth and suppressed the development of EMT [214].
In breast cancer, lung cancer, renal cancer, oral cancer, bladder cancer and to a smaller extent in gastric cancer, however, higher CSE expression was noted in the tumor tissue relative to the surrounding tissue [[62], [63], [64], [65],67,69,70,73,75,83]. Examples of cancers that show CSE overexpression include bladder, melanoma, papillary thyroid cancer, testicular cancer and prostate cancer [68,[215], [216], [217], [218], [219]]. Silencing or pharmacological inhibition of CSE has been shown to exert antiproliferative or cytotoxic effects in some of these cancers; in several cases the efficacy of CBS inhibition was less than the efficacy of CSE inhibition [64,70,106,107,112,[215], [216], [217], [218], [219]]. However, in melanoma and in prostate cancer CSE (and not CBS) appears to be the primary driver [215]. In addition, several lines of recently emerging data underline the importance of CSE in various aspects of breast cancer biology including growth, metastasis and the development of multidrug resistance [[220], [221], [222], [223], [224], [225]]. Finally, recent studies demonstrate the inhibitory effect of CSE inhibition and/or silencing in models of Ewing sarcoma [226] and glioma [227].
In studies utilizing HepG2 liver cancer cells subjected to irradiation it was demonstrated that as the cells that recover from irradiation and undergo EMT, CSE is significantly upregulated and appears to have a more important functional role than CBS; CSE inhibition enhances radiation responses and suppresses tumor growth [110,228]. Importantly, Nagy's group demonstrated in basal-like breast cancer cell models that after CBS silencing, the residual CSE activity is able to support the growth and proliferation of the cancer cells to some extent: the most pronounced anticancer response was achieved through the combination of CBS silencing and pharmacological CSE inhibition [64].
From the body of data presented above, it appears that inhibition of CSE may be a potential therapeutic direction in several forms of cancers, perhaps breast cancer being the most supported one. One important fact, however, should be re-emphasized in the context of CSE inhibition and cancer: the pharmacological tools used to inhibit this enzyme are, for most part, relatively weak in terms of potency and selectivity. Moreover – as discussed above – AOAA is a combined inhibitor of CBS and CSE [3,148] and this should be kept in mind when interpreting the antitumor efficacy of this compound. Other CBS inhibitors (e.g. CH004) are also only semi-selective to CBS over CSE [111]. In addition, the selectivity of I157172 and I194496, which are claimed as a novel CSE inhibitors [[222], [223], [224]] remains to be assessed on the other H2S producing enzymes CBS and 3-MST and their effects in cancer models remain to be tested in the future.
3-MST. 3-MST is a constitutive cytoplasmic and mitochondrial enzyme, which is highly expressed in most parenchymal cells under physiological conditions, and is not subject to significant transcriptional or posttranscriptional regulation [3,5,[229], [230], [231], [232]]. It produces H2S, but it is also a key mammalian source of polysulfides (H2Sn), a class of reactive species that have significantly different signalling and regulatory roles than H2S [232]. 3-MST can be localized in most tumor cells and tumor tissues [233,234]. As far as primary tumor tissues, it has been detected in human brain gliomas, where its expression tended to decrease with higher grades of the malignancy [235]. Accordingly, glioblastoma-bearing ipsilateral hemispheres contain greater amounts of H2Sn than the glioblastoma-free control hemispheres [236]. In human melanoma specimens, the expression of 3-MST was detectable in 25–50% of the sections analyzed [216]. 3-MST was also detected in human colon cancer resections [49], in human lung carcinoma resections, human bladder urothelial cell carcinoma resections, human oral squamous cell carcinoma resections and in oral adenoid cystic carcinoma resections [49,67,69,75,237]; in some instances its levels were significantly higher or tended to be higher than its levels in the surrounding normal tissues. Paired analysis of colon cancer specimen 3-MST expression suggests that there may be a subgroup of colon cancer patients where colon cancer 3-MST levels are substantially higher than surrounding tissue 3-MST [49], but this possibility remains to be further assessed in larger datasets. In contrast to all of these tumors, in liver cancer (similar to the downregulation of CBS, discussed above), mRNA and protein expression of 3-MST was significantly downregulated when compared to their paired nontumor counterparts [238].
No differences in clinical outcomes have been reported between high and low 3-MST expressors in most cancers (ovarian, breast, lung, kidney and pancreas cancers and melanoma) [55]. However, in colon cancer, high 3-MST expression – similar to high CBS expression – was associated with worse survival [55]. In contrast, in liver cancer – once again, similar to the case with CBS – high 3-MST expression was associated with better prognosis [238].
Similar to most primary tumor tissues, 3-MST is also highly expressed in various human and murine tumor cell lines, including several different colon cancer lines, lung cancer lines and others [49,97,[234], [235], [236], [237], [238], [239], [240], [241], [242], [243], [244]]. Interestingly, in several murine colon cancer lines, H2S generation is primarily driven by 3-MST, with CBS only playing a secondary role [241,242]. 3-MST expression (similar to CBS expression) is significantly increased when cancer cell lines assume multidrug-resistant and/or exhibit stem-like properties, for instance during recovery from a stressful/cytotoxic stimulus [93,104]. As far as the functional role of 3-MST in cancer cells, in most models, it plays tumor-supporting roles similar to those described for CBS: stimulation of cellular bioenergetics, maintenance of cell proliferation and viability, promotion of EMT [49,97,[234], [235], [236], [237], [238], [239], [240], [241], [242], [243], [244]] and perhaps a pro-angiogenic role in the tumor microenvironment [238,245]. Its activity relies on the catalytic activity of cysteine aminotransferase (CAT), which produces its substrate, 3-mercaptopyruvate [246,247]. Similar to the tumor-supportive role of CBS, which is fairly uniform except for glioma and liver cancer, also for 3-MST, a notable exception is liver cancer, where 3-MST silencing has been demonstrated to stimulate, rather than suppress cell proliferation and tumor growth [238].
The field of 3-MST biology has been substantially held back by the limited number of potent and selective 3-MST inhibitors. In addition, so far, no studies have investigated a potential cooperative relationship between the 3-MST system and CBS or CSE. Also, there are currently no published studies on the role of the 3-MST in carcinogenesis, nor has 3-MST been examined with respect to the immune aspects of tumor biology.
10. Role of CBS in cancer: an integrated concept
Taken together, for the majority of cancers where the role of CBS has been investigated (with the exception of liver cancer and glioma, which will be discussed separately), an integrated concept will be presented in the subsequent paragraphs. This concept is based, in part, on experimental data involving CBS overexpression or CBS silencing, and, in some part, on experimental data using various pharmacological agents, such as CBS inhibitors – which, as, discussed above, must be interpreted with caution. In some cases the concept will also utilize data that were generated in experimental models or cell types other than cancer, and in other cases it will also be extended to incorporate data generated with external pharmacological H2S donors, which also must be interpreted with caution due to the well-known bell-shaped pharmacological character of H2S.
Overall, it appears that many cancer cells upregulate the production of H2S and benefit from it. In many cancers, the increase in H2S is due to the increased expression of CBS – in some cases in combination with the other H2S- or H2Sn-generating enzymes CSE and 3-MST. The mechanism(s) underlying the upregulation of CBS may be transcriptional, i.e. related to increased CBS mRNA expression and transcription, as well as post-transcriptional, i.e. decreased CBS degradation. The factors driving these events may be functional (i.e. hypoxia or oxidative stress), but various hormonal, receptor-mediated mechanisms may also contribute. It is likely that CBS upregulation is influenced by interactions of the cancer cells with the tumor microenvironment, although these mechanisms remain to be explored. The upregulation of CBS is generally further increased in more advanced cancers – i.e. as the cancer cells assume multi-drug resistant, stem-like and metastasis-prone phenotypes.
The “purpose” (in the evolutionary sense) of a H2S and polysulfide-rich cellular environment is to help the cancer cells thrive by induction of an internal functional ‘remodelling’ of the cell (e.g. metabolic and signalling processes) and an external functional ‘remodelling’ of the tumor microenvironment (e.g. by promoting tumor angiogenesis). H2S can also help the cancer cell to withstand the hostile environment created by the host immune system and/or by chemotherapeutic agents. Given the evolutionarily conserved roles of H2S that can be traced back to several billions of years, the creation of a H2S-rich environment by the cancer cell may be viewed as another example of a “regressive step” that may, in part, recreate certain aspects of the primordial environment and the associated biochemical mechanisms [248,249]. In this respect, the fact that many of the regulatory roles of H2S in cancer cells relate to various bioenergetic and mitochondrial functions may be related to the fact that mitochondria have a bacterial evolutionary origin, and bacteria (both present-day and ancestral) utilize fundamental biochemical mechanisms that involve H2S. H2S production by cancer cells may also be conceptually viewed as an attempt to mobilize yet another biochemical mechanism that may support its aggressive growth and invasion “needs” in the short- and mid-term – even at the price of potential longer-term disadvantages.
H2S, as a diffusible, multifunctional autocrine and paracrine mediator and modulator, is an ideal “vehicle” to serve as a metabolic and signalling “reprogrammer”. Through its interactions with multiple cellular targets (metal centers, proteins, other diffusible/reactive species), it can simultaneously affect a wide range of essential processes in the cancer cell. The pluripotent nature of the action of H2S is well illustrated in the studies where the effect of CBS overexpression has been evaluated on various transcriptomic and metabolomic parameters (see above). It is likely that a significant part of these actions is related to protein sulfhydration; in various cell types it has been established that thousands of proteins are subject to H2S/polysulfide-mediated posttranscriptional modifications (sulfhydration) [[250], [251], [252], [253], [254], [255]]. However, “cancer cell sulfhydromes” remain to be comprehensively defined in the future.
H2S reprograms cancer cell metabolism and stimulates cellular bioenergetics. What, then, are the main bioenergetic benefits that cancer cell can gain from boosting their H2S/polysulfide levels? First of all, H2S can stimulate, perhaps maximize, the cancer cell's cellular bioenergetic capacity, to meet the high ATP demand of its rapid proliferation, growth, and movement (migration, invasion, metastasis, etc.). These effects can involve both aerobic and anaerobic energetic pathways. Although the “Warburg hypothesis” (i.e. the concept that cancer cells switch to glycolysis instead of oxidative phosphorylation) remains a popular concept, the reality is that most cancer cells maximize their ATP generation by upregulating both anaerobic (i.e. glycolysis) and aerobic (mitochondrial electron transport, ATP generation) processes; they also expand on the utilization of nutrient sources (in addition to glucose, glutamine, aspartate and others; at least in part by inducing the upregulation of transporters that support the uptake of various ‘fuels’ from the extracellular space (“opportunistic modes of nutrient acquisition”) through reprogramming the intracellular metabolic pathways that support various metabolic interconversions [[256], [257], [258], [259]].
H2S can, indeed, support many of the above processes in cancer cells. For instance, H2S can serve as a direct electron donor into the mitochondrial electron transport chain; primarily at the level of Complex II via SQR (Fig. 9, Pathway #1) [[40], [41], [42],243,260], but also, at low concentrations, it can serve as an electron donor at Complex IV (Fig. 9, Pathway #2) [[261], [262], [263], [264]] – in stark contrast to its (perhaps better-known) inhibitory effect on Complex IV at higher (toxicological) concentrations [41,42,262,263]. This latter, inhibitory action not relevant in the context of cancer cell biochemistry but it is relevant in the context of H2S intoxications and in certain CNS diseases, e.g. Down syndrome and ETHE1 deficiency [3,49]. The stimulatory actions of H2S on mitochondrial electron transport cannot support mitochondrial ATP generation in the complete absence of the ‘regular’ electron donors, but H2S can balance and maximize the flow of electrons, especially under conditions where the flux of electrons from the traditional Krebs-cycle-derived electron donors is submaximal [41,42,243,260]. Additionally, H2S can inhibit mitochondrial cAMP phosphodiesterases (as PDE2A), and thereby stimulate intramitochondrial cAMP-dependent protein kinases, which can phosphorylate and thus further activate the electron transport chain (Fig. 9, Pathway #3) [265]. In addition, H2S can sulfhydrate LDH-A, resulting in its activation (Fig. 9, Pathway #4) [91,266]. Activation of LDH-A (which is primary cytosolic), increases cytosolic lactate, followed by an increased flux of lactate into the mitochondria through the intracellular lactate shuttle, providing additional support to mitochondrial electron transport and ATP generation [91]. Moreover, H2S can also directly stimulate the activity of ATP synthase via the sulfhydration of specific cysteines [267,268] (Fig. 9, Pathway #5).
Fig. 9.
Roles of H2S in the stimulation of cellular bioenergetics in cancer cells. H2S can serve as a direct electron donor into the mitochondrial electron transport chain at the level of Complex II via SQR (Pathway #1), but also, at low concentrations, it can serve as an electron donor at Complex IV (Pathway #2). H2S can inhibit mitochondrial cAMP phosphodiesterases (as PDE2A), and thereby stimulate intramitochondrial cAMP-dependent protein kinases, which can phosphorylate and thus further activate the electron transport chain (Pathway #3). In addition, H2S can sulfhydrate LDH-A, resulting in its activation (Pathway #4). H2S can also directly stimulate the activity of ATP synthase via the sulfhydration of specific cysteines (Pathway #5). H2S can regulate mitochondrial dynamics (fusion/fission) to maintain the mitochondrial pool in its most effective state (Pathway #6). H2S can also increase mitochondrial mass via the stimulation of mitochondrial biogenesis (Pathway #7). Additional mechanisms underlying mitochondrial “stabilization” may be simply related to the general antioxidant role of H2S (Pathway #8). Another mitochondrial protective mechanism is related to the stimulation of mitochondrial DNA repair (Pathway #12). H2S may also support the cancer cell metabolism via the stimulation of glycolysis, in part through GAPDH and PKM2 sulfhydration and their consequent activation (Pathway #9). H2S may also stimulate the uptake of glucose and its utilization (Pathway #10), and lipid uptake and its utilization (Pathway #11) into the cells.
In addition, H2S can regulate mitochondrial dynamics (fusion/fission) to maintain the mitochondrial pool in its most effective state. Part of these mitochondrial regulatory roles occur via specific regulatory actions (e.g. via effects on mitofusin) [61,269]; others are related to other signalling pathways, e.g. the Drp1/ERK1/2 signaling pathway [270] (Fig. 9, Pathway #6).
H2S can also increase mitochondrial mass via the stimulation of mitochondrial biogenesis [[271], [272], [273], [274]] (Fig. 9, Pathway #7), although the experimental data supporting these actions are typically not derived from cancer biology studies and such actions remain to be directly assessed for cancer cells. H2S can enhance the binding of IRF-1 at the Dnmta promoter, leading to a suppression of its expression, which leads to demethylation of TFAM promoter leading to increased expression of TFAM and mitochondrial biogenesis [271]. Other mechanisms by which H2S can stimulate mitochondrial biogenesis involve the AMPK pathway as well as PGC-1α and PPRC signaling [125,272].
Additional mechanisms underlying mitochondrial “stabilization” may be simply related to the general antioxidant role of H2S (Fig. 9, Pathway #8), which – as demonstrated in several cell types, both transformed and non-transformed – also helps to maintain mitochondrial functions [[275], [276], [277], [278], [279], [280]]. The underlying mechanisms may be direct redox reactions between H2S and various reactive species, although these reactions typically have fairly slow reaction rates; additional antioxidant responses may the consequences of Nrf2 and p66Shc activation and secondary upregulation of various intracellular antioxidant systems. (These mechanisms are further discussed in the next paragraphs in the context of cytoprotective roles of H2S). Some of the above-listed mechanisms have been demonstrated in cancer cells; others have been shown in other biological systems, and yet others in reductionist biochemical assays only. Nevertheless, the end result, which is that CBS silencing or CBS inhibition suppresses mitochondrial electron transport and mitochondrial ATP generation, has been clearly shown in multiple cancer cells (best characterized, so far, in colon cancer cells and ovarian cancer cells) [6,7,61,101,241]. Another mitochondrial protective mechanism is related to the stimulation of mitochondrial DNA repair (see below for further discussion) (Fig. 9, Pathway #12). Mitochondrial DNA integrity would be expected to maintain proper regulation of mitochondrial proteins, including key electron transport chain protein components encoded locally within the mitochondrial DNA, which, ultimately, would help mitochondrial function and cellular bioenergetics.
H2S may also support the cancer cell metabolism via the stimulation of glycolysis (Fig. 9, Pathway #9). Indeed, in a variety of experimental models, H2S donation has been shown to increase glycolytic activity of the cells [103,[281], [282], [283], [284]]; also in several cancer cells, CBS inhibition and/or CBS silencing was shown to suppress glycolytic activity [6,7]. The underlying mechanisms are likely multiple. First of all, H2S can sulfhydrate and activate GAPDH, a key glycolytic enzyme [281]. Second, aerobic glycolysis can be stimulated via a H2S-induced reductive shift in the mitochondrial NAD+/NADH pool [283]. A third, recently suggested mechanism may be a H2S-mediated metabolic switch related to the sulfhydration of PKM2 (an alternatively spliced form of pyruvate kinase), which enhances aerobic glycolysis and is known to provide tumor growth advantage [253].
H2S may also stimulate the uptake of glucose into the cells [137,266,280] (Fig. 9, Pathway #10), - and possibly the uptake of other metabolic fuels, as well. H2S in cancer cells may also stimulate metabolic functions via modulating the activity of various Krebs cycle enzymes, leading to the activation of the cycle and the increased supply of electron donors into the mitochondria. Indeed, multiple studies demonstrate that H2S, either via transcriptional actions, and/or via posttranscriptional mechanisms (sulfhydration) can regulate a variety of pathways including central carbon metabolism, amino acid biosynthesis and interconversion, lipid and steroid biosynthesis and metabolism and others [253,285,286]. These actions occur via actions on a variety of enzymes including pyruvate kinases 1 and 2, isocitrate dehydrogenase, oxoglutarate dehydrogenase, succinate dehydrogenase, pyruvate dehydrogenase, aconitase 1, phosphoglycerate mutase 1, TP53 induced glycolysis regulatory phosphatase, UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, carbohydrate sulfotransferase 4 and many others [253,285,286]. One prominent example by which CBS reprograms metabolism – discussed in an earlier section – occurs via the upregulation of lipid uptake and lipid utilization in ovarian cancer [101] (Fig. 9, Pathway #11).
The various roles of H2S in the stimulation of cellular bioenergetics in cancer cells are summarized in Fig. 9. The translational consequence of the above mechanisms is that by inhibiting CBS (or, more broadly, H2S generation) the ‘energy charge’ of the cancer cells may be lowered, which, in turn, will suppress cell growth, proliferation and may sensitize the cells to chemotherapeutic agents or immune-cell-mediated attack.
H2S can stimulate proliferative cell signalling. According to the classical ‘cancer hallmark’ concept, one of the hallmarks of cancer cells is that they stimulate their own growth and proliferation independently from external signals (e.g. various growth factors). This switch from external-signal-dependent to intrinsic/perpetual proliferative signaling is often the consequence of key oncogenic mutations of the cancer cell that induce specific growth suppressor pathways and/or delete various tumor suppressors. Classical pathways that are considered essential for cancer cell proliferation include the PI3K/Akt, the Ras/Erk, and the mTOR pathways [[287], [288], [289], [290], [291]].
With respect to the PI3K/Akt pathway, multiple studies demonstrate that administration of exogenous H2S to various cell types can stimulate this pathway [[292], [293], [294], [295], [296], [297], [298], [299]]; the role of this mechanism has been extensively investigated, for instance in the context of H2S-induced vascular regulation [122,124,126,135]. As reviewed previously [135], H2S-induced Akt activation is not a direct effect of H2S on PI3K, but may be related to (a) stimulation of Akt via stimulation of the phosphorylation of its activating site (Ser493) (probably via the activation of intermediary kinases); (b) inhibition of Phosphatase and Tensin homolog (PTEN) - an essential counterregulatory enzyme of the PI3K pathway - via sulfhydration of Cys124 or Cys71 and (c) inhibition of the activity of PTP1B (another counterregulatory enzyme in the PI3K pathway) via sulfhydration at Cys215 (Fig. 10, Pathway #1). All of these actions result in increased PIP3 levels. When PI3K/Akt is activated, the result is the activation of both proliferative and cytoprotective downstream pathways [[287], [288], [289], [290], [291]]. Exogenous H2S or polysulfide administration can, indeed, stimulate the Akt pathway in cancer cells – as shown, for instance in colon cancer cells [90] and neuroblastoma cells [298,299], multiple myeloma cells [128], hepatocellular carcinoma cells [129,300] and thyroid carcinoma cells [117]. Whether endogenously produced, CBS-derived H2S contributes to the stimulation of the PI3K/Akt pathway, is less clear and may be cell-type dependent. In HCT116 colon cancer cells, Akt phosphorylation was unaffected by treatment of the cells with AOAA under certain conditions [49]. Under other experimental conditions in the same cell type, AOAA decreased total Akt1 (but not Akt2) levels, but increased the phosphorylated proportion [97]. However, in MCF7 and MDA-MB-231 breast cancer cells Akt phosphorylation was suppressed by CBS silencing [65].
Fig. 10.
The various roles of H2S in the stimulation of cells signaling in cancer cells. H2S can stimulate the PI3K/Akt pathway through: (a) stimulation of Akt via induction of the phosphorylation of its active site (Ser493) (probably via the activation of intermediary kinases); (b) inhibition of Phosphatase and Tensin homolog (PTEN) - an essential counterregulatory enzyme of the PI3K pathway - via sulfhydration of Cys124 or Cys71 and (c) inhibition of the activity of PTP1B (another counterregulatory enzyme in the PI3K pathway) via sulfhydration at Cys215 (Pathway #1). All of these actions result in increased PIP3 levels. H2S can stimulate ERK1/2 pathway through: ERK1/2 phosphorylation; RAF phosphorylation that, in turn, can phosphorylate MEK1; sulfhydration of MEK1 (at Cys341). MEK1 activation, in turn, leads to the phosphorylation of ERK1/2 and translocation of ERK1/2 into the nucleus to stimulate ERK1/2 mediated downstream signaling; KATP channels opening and PKC activation resulting in downstream ERK1/2 activation (Pathway #2). H2S can stimulate mTOR pathway through: mTOR phosphorylation; AMPK phosphorylation via sulfhydration of liver kinase B1 (LKB1) leading to phosphorylation at Ser428 and/or to the activation of calcium/calmodulin-activated protein kinase 2 (CaMKK2), the phosphorylate AMPK can, in turn, phosphorylate mTOR resulting in downstream mTOR activation (Pathway #3). The PI3K/Akt, ERK1/2 and mTOR pathways activation result in the activation of both proliferative and cytoprotective downstream pathways.
With respect to the Erk and mTOR pathways, once again, multiple studies demonstrate that administration of exogenous H2S to various cell types, including cancer cells, can activate these pathways – primarily via stimulation of Erk1/2 phosphorylation and via enhancement of AMPK phosphorylation, respectively (Fig. 10, Pathway #2 and #3) [90,117,129,270,272,297,300,301]. The activation of this pathway by H2S is initiated by sulfhydration of MEK1 (at Cys341), which, in turn, leads to the phosphorylation of ERK1/2 and translocation of ERK1/2 into the nucleus to stimulate ERK1/2 mediated downstream signalling. ERK1/2 can also be activated through KATP channels opening and PKC activation [292]. The typical biphasic H2S concentration-response is also relevant in this context; for instance, in mammary epithelial cells, low concentrations of exogenously administered H2S stimulated mTOR phosphorylation and activated the mTOR pathway (thereby increasing cell proliferation), while at higher concentrations, the opposite effects were found [302]. The mechanism by which H2S stimulates AMPK phosphorylation may be related, at least in part, to the sulfhydration of liver kinase B1 (LKB1) leading to phosphorylation at Ser428 and/or to the activation of calcium/calmodulin-activated protein kinase 2 (CaMKK2) (Fig. 10, Pathway #3) [303,304]. The body of information regarding the capacity of endogenous H2S to regulate these pathways in cancer cells remains to be further investigated.
Many additional signalling pathways have been shown to be modulated by H2S in various experimental systems in vitro and in vivo in a cell-type and context-dependent manner [[1], [2], [3],305,306]. CBS overexpression in various cancer cell types has been also demonstrated to activate a variety of signalling pathways (see above), although the functional consequence of many of them remains to be further determined. Signalling pathways with relevance to cytoprotection, cancer cell stemness and regulation of the EMT/MET transitions will be discussed in subsequent sections. Taken together, it is likely that H2S contributes to the induction of various signalling pathways in cancer cells in a cell-type and context-dependent fashion, thereby potentially conferring a more proliferative and invasive phenotype to the cells, as well as protecting them from various external noxious stimuli aimed at their elimination (e.g. the immune system and chemotherapeutic agents).
The various roles of H2S in the stimulation of cells signalling in cancer cells are summarized in Fig. 10. The translational consequence of the above mechanisms is that by inhibiting CBS (or, more broadly, H2S generation), the growth and proliferation of the cancer cells may be inhibited.
H2S serves as a cytoprotective factor in cancer cells. As already discussed, elevated intracellular H2S levels can act as antioxidants to protect the mitochondria. This protection, in fact, extends to other cellular compartments as well; thus, the upregulation of CBS and the elevation of cellular H2S levels can be viewed as an attempt by the cancer cell to create a ‘general, primordial cytoprotective shield’. Indeed, the origins of the ability of H2S to protect organisms from various forms can be traced back to the bacterial kingdom; H2S production in bacteria serves as a generic mechanism to protect against antibiotic-mediated killing, as well as against immune-cell-mediated elimination [[119], [120], [121],[307], [308], [309], [310], [311]]. Likewise – although the quantified reaction rate constants between H2S and various oxygen-derived reactive species are fairly low [[312], [313], [314]] – a large number of studies, conducted in various mammalian cell types, demonstrate that H2S supplementation can protect various mammalian cells from oxidative and nitrosative stress [41,42,[315], [316], [317], [318], [319], [320], [321], [322], [323], [324], [325], [326], [327]]. In addition to the direct antioxidant/cytoprotective effects, H2S can lead to the upregulation of various antioxidant systems; for instance via the induction of the Nrf2/ARE pathway [14,102,125]. The underlying molecular mechanism include the H2S-mediated sulfhydration of Keap1 (at Cys151) [328]. Keap1, in turn, undergoes a conformational change which leads to the dissociation of Nrf2 from the Keap1-Cul3-RBX1 E3 ligase complex. Subsequently, the free Nrf2 translocates into the nucleus to induce a global change in gene expression, which includes the upregulation of a host of antioxidant genes and enzyme systems [329]. Several other H2S-activated cytoprotective pathways have also been implicated, including Akt, ERK, AMPK/mTOR and others. Cytoprotective effects may also be the consequence of some of the pathways discussed in the previous section (the PI3K/Akt pathway, the Erk pathway and the mTOR pathway). In addition, H2S upregulation in cancer cells may also induce the upregulation of the NF-κB pathway via sulfhydration at Cys-38; the activation of this pathway can also confer cytoprotective effects to the cells [7,48,330]. Moreover, H2S may also upregulate various heat-shock protein pathways in cancer cells; this process can also confer cytoprotective effects [7,48].
In the context of the mechanisms discussed above, it is not surprising that CBS upregulation can protect the cancer cell from the anticancer effects of various chemotherapeutic agents, from radiation-induced cell killing – as well as from immune-cell-mediated elimination (see below) – since many of the underlying processes are linked to the generation of various reactive species (e.g. superoxide, hypochlorous acid, hydrogen peroxide, hydroxyl radical, etc.) [[331], [332], [333], [334], [335], [336], [337], [338]]. However, in the context of chemotherapeutic agents, additional, more specific mechanisms may also be involved in the protection provided by H2S. As discussed in prior sections, CBS-derived H2S can, for instance, induce the STAT3/Akt/Bcl-2 pathway [109]; via promoting Akt activation (and, possibly, via additional signalling pathways) H2S can also promote the upregulation of P-glycoprotein (P-gp) [109] as well as of epiregulin (EREG) and thymidylate synthetase (TYMS) [50] – all of which can confer specific chemotherapeutic resistance.
The various roles of H2S as a cytoprotective factor in cancer cells are summarized in Fig. 11. The translational consequence of the above mechanisms is that by inhibiting CBS (or, more broadly, H2S generation) the cancer cells can be sensitized to the cytotoxic effect of various chemotherapeutic agents or to radiation therapy.
Fig. 11.
Various roles of H2S as a cytoprotective factor in cancer cells. H2S can lead to the upregulation of various antioxidant systems. H2S can stimulate Nrf2/ARE pathway through: sulfhydration of Keap1 (at Cys151), that in turn, undergoes a conformational change which leads to the dissociation of Nrf2 from the Keap1-Cul3-RBX1 E3 ligase complex. Subsequently, the free Nrf2 translocates into the nucleus to induce a global change in gene expression, which includes the upregulation of a host of antioxidant genes and enzyme systems. H2S can stimulate PI3K/Akt, ERK1/2 and mTOR pathways, already discussed in Fig. 10. H2S can stimulate NF-kB pathway through: sulfhydration of NF-kB at Cys38. H2S can stimulate HSP90 pathway. The activation of these pathways confers general cytoprotection and cancer cell resistance to chemotherapeutic agents induced cytotoxicity. However, in the context of chemotherapeutic agents, additional, more specific mechanisms may also be involved in the protection provided by H2S. H2S can stimulate STAT3/Akt/Bcl-2 pathway through Akt activation. H2S can also promote the upregulation of P-glycoprotein (P-gp) and thymidylate synthetase (TYMS).
H2S stimulates DNA repair in cancer cells. Although the earliest publications in relation to H2S and DNA integrity have suggested that H2S is a genotoxic agent [[339], [340], [341]], now it is clear that these studies have utilized high exogenous concentrations of H2S, and they are only relevant in a toxicological context. In the physiological context, however, the role of H2S in regulating DNA integrity is markedly different, and H2S can, in fact, promote various pathways of DNA repair, both in the nuclear DNA and in the mitochondrial DNA. With respect to nuclear DNA repair, the underlying mechanisms may include the activation of ataxia telangiectasia-mutated and Rad3-related (ATR) serine/threonine kinase as well as the regulation of MEK1, PARP1 and ATM [325,[342], [343], [344]]. (Please note that these mechanisms have been characterized in a variety of cell types, not only in cancer cells, and the H2S used was often exogenously provided and not endogenously produced.)
With respect to mitochondrial DNA repair (as discussed above), the mechanism involves sulfhydration of EXOG and the promotion of the formation of a mitochondrial DNA repair complex [69] and the induction of DNA methyltransferase 3A [271]. Whether these mechanisms apply to cancer cells remains to be determined in future studies. Nevertheless, we can hypothesize that one of the roles of CBS upregulation in cancer may be to promote DNA repair of the cancer cell. In this respect, stimulation of DNA repair can be viewed as one sub-category of the broadly discussed cytoprotective role of H2S in the cancer cell.
The translational consequence of the above mechanisms is that by inhibiting CBS (or, more broadly, H2S generation), the cancer cells can be sensitized to DNA-damaging agents (e.g. certain chemotherapeutic agents or radiation).
H2S stimulates epithelial-to-mesenchymal transition (EMT). In epithelial cancers (e.g. colon adenocarcinoma, squamous cell carcinoma, basal cell carcinoma, most ovarian cancers), the progression from normal (epithelial) to transformed (mesenchymal) state is termed EMT. In this process, epithelial cells lose their cell-to-cell adhesion capacity and gain mesenchymal characteristics. Via EMT, the cells develop increased migratory capabilities and gain a higher invasive and metastatic potential [[345], [346], [347], [348]]. EMT is also closely associated with ‘tumor budding’, a process whereby isolated single cancer cells or small clusters of cancer cells appear at the invasive tumor front [349,350]. EMT is typically detected by its markers, e.g. increased N-cadherin expression and loss of membranous E-cadherin. The reversal of EMT is MET (i.e. mesenchymal-to-epithelial transition); pharmacological induction of MET has significant experimental therapeutic potential [351,352]. While the EMT/MET transitions are regulated by multiple signalling events, one key regulator is the Wnt/β-catenin signaling pathway. β-catenin is constitutively bound to a multiprotein “destruction complex”, whose fate is to be degraded by the proteasome. In the presence of Wnt ligands such as Wnt3, the formation of the destruction complex is prevented, followed by the β-catenin translocation in the nucleus, where it stimulates the expression of genes, such as Snail1 and Twist1, involved in cell proliferation, migration and invasion [[346], [347], [348], [349], [350], [351], [352], [353], [354], [355], [356], [357], [358], [359]].
The role of H2S in the regulation of EMT has been investigated in several models, including lung fibrosis (another disease condition where EMT/MET transitions are pathophysiologically relevant) and in various forms of cancer. The effect of exogenously administered H2S appears to be context-dependent; in some models (e.g. cultured lung epithelial cells, renal tubular epithelial cells or cultured breast cancer cells challenged with TGFβ), H2S suppresses this process [[353], [354], [355], [356], [357], [358], [359]]. However, more importantly, endogenously produced H2S appears to have a different role. Indeed, forced expression of CBS into human colon epithelial cells is sufficient to upregulate several elements of this pathway, at least on the level of mRNA [48]. Moreover, multiple studies demonstrate that inhibition of H2S biogenesis – either by CSE or CBS silencing [70,110,214,228] or by CBS or 3-MST inhibition [97] – in fact, suppresses the process of EMT: for instance in a model of radiation induced EMT in hepatocellular carcinoma [110,228], and in non-small cell lung cancer cells [70] and in cultured human colon cancer cells in vitro [97].
The stimulatory effect of endogenously produced H2S on the Wnt/β-catenin signaling pathway has also been demonstrated in experimental models other than cancer, for instance during the differentiation of human periodontal ligament cells [360] and in mesenchymal stem cells [361]; in the latter cell type, endogenous, CBS-derived H2S maintains calcium influx, which, in turn, induces PKC phosphorylation, which, in turns, stimulates the Wnt/β-catenin signaling pathway. The pathways affected by endogenously produced H2S in the process of EMT promotion in colon cancer cells include the induction of the Sp3 transcription factor, a subsequent induction of ACLY mRNA transcription and ACLY protein expression and a consequent interaction of ACLY with β-catenin to block β-catenin ubiquitination leading to its accumulation in the cytoplasm. In turn, β-catenin translocates into the nucleus and activates Twist1 and Snail1 transcription factors to induce the ‘classical’ pathway of EMT [97]. Another pathway of EMT, stimulated by endogenous H2S generation in lung cancer cells, involves the PI3K/Akt pathway, which upregulates HIF1α, thereby inducing or maintaining EMT [70]. In irradiated hepatic carcinoma cells – similar to colon cancer cells – CBS (or, more prominently CSE) appears to promote the induction of Snail1 to induce or maintain EMT [110].
The various roles of H2S in the stimulation of EMT in cancer cells are summarized in Fig. 12. The translational consequence of the above mechanisms is that by inhibiting CBS (or, more broadly, H2S generation), a pharmacological MET induction may be facilitated or achieved, which would be expected to reduce the metastatic potential of the cancer cells.
Fig. 12.
The various roles of H2S in the stimulation of EMT in cancer cells. H2S can stimulate Wnt/β-catenin pathway through: Sp3 transcription factor upregulation that, in turn, upregulate ACLY. ACLY interacts with β-catenin and may block β-catenin ubiquitination leading to its accumulation in the cytoplasm which, in turn, also translocates into the nucleus. In the nucleus, β-catenin binds to LEF and activates Twist1 and Snail1 transcription factors. H2S can stimulate PI3K/Akt pathway, which upregulates HIF1α, or induces Snail1 transcription factors expression through NF-κB. H2S can also stimulate the MEK/ERK pathway, leading to the activation of the transcription factor AP1. H2S can stimulate TGFβ pathway through: Smad2/3 activation, which, in turn, activates the transcription factor Snail1. H2S can stimulate p38MAPK pathway, either by activation of the upstream kinases MKKx which consequently phosphorylate p38MAPK or by the autophosphorylation of p38MAPK. Under hypoxic conditions H2S can induce HIF-1α expression and further up-regulate VEGFA and the transcription factor Twist1. Some of these pathways are interconnected and can be activated at the same time. Possible crosstalks between these pathways are indicated by brown arrows. The bell-shaped effect H2S should be emphasized in the above processes, meaning that these pathways can be upregulated during EMT in cancer cells, however this process can be reverted if additional H2S is added to the cells or if the H2S biosynthesis is inhibited in cancer cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
H2S promotes dedifferentiation and maintains cancer cell stemness. As discussed in the previous sections, the development of stem-like properties of cancer cells (e.g. after surviving a non-lethal insult, such as irradiation, oxidative stress, hypoxia or chemotherapeutic agents) often coincides with the upregulation of H2S-producing enzymes, and an increase in H2S biogenesis [100,103,104,147]. CBS, via its product H2S, in fact, may be a factor that contributes to the transition of the cells into the stem-like state, either by promoting the induction of specific stemness genes – as shown, for the wnt pathway, for the stemness gene CXCR4 and for various pluripotency genes, e.g. OCT4 and NANOG in various experimental systems in vitro [70,97,130,147,[360], [361], [362], [363]] –, and/or perhaps it serves as a cytoprotective mediator (as discussed in an earlier section), which helps with the maintenance and ultimate selection of stress-resistant cancer cell clones. The induction of OCT4 by H2S may be related to the induction of STAT3 [109,220,221]. The upregulation of intracellular NAD + levels by H2S may be an additional mechanism, which can affect both stemness/pluripotency genes and cell viability/survival. The underlying mechanisms may either be related to the induction by H2S of NAMPT (a key enzyme recovering NAD+ from nicotinamide and generating most of the NAD+ cellular pools) and/or via the regulation of various sirtuin systems [103,104,147,364,365].
The translational consequence of the above mechanisms is that by inhibiting CBS (or, more broadly, H2S generation), cancer stem cells may be sensitized to elimination by chemotherapeutic agents or by the immune system.
H2S promotes tumor angiogenesis. The role of H2S in the promotion of angiogenesis in the cardiovascular system has been subject to multiple recent reviews [3,132,134,135] and will not be discussed here in detail. What is important for the current article is that endogenously produced H2S, generated by CBS (on its own, or in combination with CSE or 3-MST) acts as a promoter of tumor angiogenesis. This is well established by various in vitro studies (e.g. cancer cell/endothelial cell co-cultures) [6,7,52,64,76,106] and by in vivo studies demonstrating that tumor-bearing mice subjected to implantation of various tumor cell lines develop less peri- and intratumor vasculature when the H2S-producing ability of the implanted tumor cells is suppressed (Fig. 7) [6,52,64,70,101]. The signalling mechanisms by which H2S promotes angiogenesis are multiple and are summarized in Fig. 13. One of the key mechanisms is calcium mobilization, which, in turn, can activate various pro-angiogenic signalling pathways, including the activation of eNOS and the production of NO [124,132,135]. Another mechanism relates to the ability of H2S to open KATP channels on the endothelial cell membrane; another mechanism involves a cooperative interaction between H2S and NO whereby H2S upregulates eNOS, directly activates eNOS via sulfhydration, dimerization and stabilization and also activates the eNOS system via Akt activation and phosphorylation [124,132,135]. An additional pro-angiogenic mechanism may involve the cooperative interaction between H2S and eNOS-derived NO in the activation of cGMP signalling; in this system NO serves as the primary activator of this system, while H2S prolongs the action of cGMP by inhibiting cGMP phosphodiesterase [126,132,135]. Further mechanisms may include the upregulation of VEGF and its receptor [2,48,64,70,76,[122], [123], [124], [125], [126],[131], [132], [133], [134], [135], [136], [137], [138], [139]] and the consequent activation of this system (possibly, in part, also via Flt induction [64]), the induction of the angiopoietin/tie system, possibly via the upregulation of Ang1 and Ang2 system [64] as well as though the upregulation of Tie [64] and, possibly, the induction of the adrenomedullin system [48] and the induction of various additional endothelial cell receptors with pro-angiogenic functions (CXCR1, VCAM1) [48,64]. Finally, a further mechanism may be simply related to the fact that tumors are often hypoxic, and hypoxia prolongs the biological half-life of H2S, which, in turn, may be more effective in exerting its various pro-angiogenic actions. Please note that some of these mechanisms have been demonstrated in reductionist models that did not utilize cancer cells. In particular, the cooperative interaction between the NO and the H2S pathway – which appears to be important in the context of physiological angiogenesis – remains to be explored in the future in the specific context of tumor angiogenesis.
Fig. 13.
Various roles of H2S in the stimulation of angiogenesis. H2S can stimulate intracellular Ca2+ mobilization in endothelial cells, which stimulates eNOS via the calcium-calmodulin system. H2S can activate PI3K/Akt pathway resulting in eNOS activation through phosphorylation at Ser1177. H2S can also induce a direct posttranslational modification of eNOS via sulfhydration on Cys443. This action, in turn, can further activate and stabilizes eNOS by promoting its dimerization. H2S can also stimulate the expression of eNOS mRNA, increasing eNOS protein levels. Further downstream, NO produced by eNOS binds to soluble guanylate cyclase (sGC) and induces the formation of cGMP. H2S can inhibit PDE, suppressing the degradation of cGMP and increasing cGMP levels enhancing the activation of its downstream protein kinase, PKG. H2S can open KATP channels on the endothelial cell membrane. H2S can upregulate VEGF and its receptor through HI–F1α upregulation but also through Flt induction. H2S can stimulate angiopoietin/tie system, possibly via the upregulation of Ang1, Ang2 and Tie. H2S can also stimulate various endothelial cell receptors with pro-angiogenic roles, including VEGFR, CXCR1 and VCAM.
Although the underlying mechanisms are incompletely understood, it has been observed in multiple studies that tumors transplanted into various host animals develop less peri- and intratumor vasculature if CBS in the tumor has been silenced [6,7,52,64,70] (Fig. 7). The translational consequence of the above mechanisms is that by inhibiting CBS (or, more broadly, H2S generation), the growing tumor tissue may be deprived of its blood vessel network, which would be expected to be therapeutically relevant.
H2S may protect the tumor cell from immune-mediated elimination. The question whether tumor-cell-derived H2S may protect against immune-activation mediated elimination is a very important one. Based on the ‘generic’ cytoprotective effect of H2S (see above), such a role is expected. However, to date, only a very limited number of studies have directly investigated this matter; in co-cultures of breast cancer cells and activated macrophages, CBS silencing in the breast cancer cells markedly accelerated the killing efficiency of the macrophages [62] and in co-cultures of breast cancer cells with activated NK cells [65] or activated T-cells [65], a similar enhancement was demonstrated. As discussed in a previous section, the role of the host H2S production (in the tumor microenvironment in general and especially in the host immune cells) in the antitumor immune responses remains to be further explored.
The translational consequence of the above mechanisms would be very important. Inhibiting CBS – or, more broadly, inhibition of H2S generation in the tumor cells – may be a way to enhance the efficacy of the host's immune response in eliminating tumor cells – either basally, or perhaps in conjunction with immunostimulatory therapies.
All of the above roles of CBS/H2S in cancer, and the expected effects of CBS inhibition are summarized in Fig. 14. It should be emphasized that the above mechanisms do not apply to liver cancer and glioma, where the role of CBS/H2S is markedly different – it acts as a tumor suppressor, rather than a tumor promoter. Also, the mechanisms discussed in the current section are not operative simultaneously in all cancer cells. Rather, they represent possible mechanisms, the relative contribution of which is likely be dependent on the type and stage of cancer.
Fig. 14.
Various tumor-supporting roles of H2S in high-CBS-expressing tumors; expected effects of CBS inhibition on these processes. Please note that these mechanisms are not present simultaneously and do not apply to all cancer types.
11. The path to clinical translation
The need for a selective, potent, clinically useful CBS inhibitor. Given the fact that CBS inhibition has been shown to beneficially affect multiple hallmarks of cancer (Fig. 14), the clinical translation of this concept is warranted for several forms of cancer – with colon cancer, ovarian cancer, lung cancer and certain subtypes of breast cancer being the most supported by clinical correlations and preclinical data. In contrast, based on the majority of published data, CBS inhibition is contraindicated in liver cancer and glioma (Table 1).
Inhibition of CBS, in a clinical setting, would require a potent and selective pharmacological inhibitor suitable for human use. As discussed earlier, none of the inhibitors used in the preclinical studies have the necessary selectivity and potency for clinical translation, and the potentially repurposable compounds (e.g. disulfiram and benserazide) are less than optimal, due to their low potency, limited selectivity and less than ideal therapeutic index. Thus, the best approach, to identify new CBS inhibitors would require the discovery of new CBS inhibitory scaffolds, either through physical screening or through in silico approaches. A competitive inhibitor of CBS that would prevent the binding of its substrates and would suppress H2S generation, would probably be an ideal small molecule for cancer therapy; covalent inhibitors at the PLP site (similar to the mode of AOAA's action) may also be clinically translatable, although such molecules may also inhibit other PLP-dependent enzymes, which may, in turn, could lead to various side effects.
Although selective targeting of a CBS inhibitor to the tumor cells, in theory, would be ideal, such approaches are beyond the current state-of-the art. What, then, would be the side effects of a systemically administered CBS inhibitor in a cancer patient? Given the central role of CBS in transsulfuration, the main side effect of a CBS inhibitor would be the inhibition of homocysteine degradation, i.e. an increase in circulating homocysteine levels. Homocysteine is considered a reactive, cytotoxic molecule, which is particularly deleterious to the integrity of the vascular endothelium [366,367]; hyperhomocysteinemia is viewed as a cardiovascular risk factor, primarily for thrombotic vascular disease and atherosclerosis [[368], [369], [370], [371], [372]]. However, the topic is complicated. First of all, a temporary elevation in homocysteine (e.g. in conjunction with a 3-month cancer therapy) would not carry the same risk as a year-long or life-long hyperhomocysteinemia. Second, the correlation of cardiovascular risk with increased plasma homocysteine levels is not a linear one; slight-to-median elevations do not carry a substantial additional risk [369]. Third, the causal link between elevated homocysteine levels and the cardiovascular adverse events has been put into question by the results of recent trials, where homocysteine-lowering therapy did not produce the expected reduction in cardiovascular adverse events, especially in the mild-to-moderate hyperhomocysteinemia group, suggesting that – in this group of patients – homocysteine may only be a marker, rather than an active contributor to cardiovascular disease [373].
Thus, the question that would need to be addressed is the following: how severe increase in homocysteine can be expected with a therapeutically efficacious dosing of a CBS inhibitor in a cancer patient? One way to answer this question is through preclinical studies. The majority of the publications utilizing various CBS inhibitors in cancer models have not measured circulating homocysteine levels. However, in a mouse model, the AOAA prodrug YD0171, at the therapeutically efficacious dose where it produced a >80% suppression of HCT116 colon cancer growth, induced a 36% elevation of plasma homocysteine [155]. If a similar increase would occur in patients, this would be considered a mild elevation (unlikely to be associated with increased cardiovascular risk). Mice that lack CBS (homozygote CBS−/- animals) exhibit significant elevations in circulating homocysteine levels (up to 500 μM); these animals also exhibit severe developmental and functional defects and early lethality [[374], [375], [376], [377]]. However, CBS ± mice are clinically healthy and don't exhibit marked plasma homocysteine levels (<20 μM) [[374], [375], [376], [377]]. With a partial normalization of circulating homocysteine levels (when levels remain in the range of 80–100 μM), achieved by CBS enzyme therapy of CBS−/- mice, normal development and phenotype can be readily achieved [377].
Additional potential side effects of systemic CBS inhibition may also include the loss of systemic (circulating, vascular or intra-tissue) H2S, which has various cytoprotective and anti-inflammatory roles as well as a vasodilatory effect [3]. However, these roles of H2S are likely compensated, at least in part, by the remaining presence of the other two H2S-producing enzymes, CSE and 3-MST. Also – although H2S is considered a physiological vasodilator – there is no strong indication, based on studies in isolated vascular preparations or measurements of blood pressure in CBS deficient mice [[378], [379], [380], [381], [382], [383], [384]] that CBS inhibition would produce a clinically significant hypertensive side effect.
Taken together, we do not believe that a modest, temporary inhibition of CBS – along with a temporary reduction in circulating H2S levels and a temporary elevation of homocysteine levels – during a course of anticancer therapy will be a factor that could limit the clinical translation of the CBS inhibition concept. On the other hand, measurement of plasma homocysteine levels may be used as a marker to confirm “target engagement”.
In light of the role of additional H2S generating enzymes, CSE and 3-MST in various cancers (including those cancers where CBS appears to be the primary driver), the question arises as to whether it may be desirable (and feasible) to seek simultaneous inhibition of multiple H2S producing enzymes. Based on the stand-alone effect of CBS silencing or the effect of the various CBS inhibitors (even with each having its own sets of caveats), inhibition of CBS, on its own, is likely to exert significant effects. If one seeks to target several of the H2S-producing enzymes together, a PLP-interaction-based CBS/CSE inhibitor (similar to AOAA) may be a possibility – although the risk of such a compound may be an inhibition of additional PLP-dependent enzymes. Since 3-MST has a markedly different structure and enzymatic mechanism, a combined CBS/3-MST inhibitor would be a very challenging medicinal chemistry task.
Another ‘broader’ approach to inhibit cancer cell H2S generation may be based on the restriction of the cancer cell of its substrates, cysteine and homocysteine, by blocking the respective uptake mechanisms such as the cystine/glutamate antiporter system Xc (xCT) [[385], [386], [387], [388], [389]], and/or by producing the degradation of circulating substrates, by systemically administering an extracellular enzyme that utilizes these substrates [[390], [391], [392]]. All of these approaches have been assessed preclinically; in each case there is evidence for their preclinical efficacy, but each of these approaches would also come with their own significant set of challenges, which are discussed elsewhere [[385], [386], [387], [388], [389], [390], [391], [392]].
The need for a biomarker of cancer cell CBS activation. Any future therapeutic modality based on CBS would be more efficacious if it was applied in a personalized manner. This notion is supported by recent data indicate that in patient-derived xenografts, CBS inhibition is significantly more efficacious in suppressing the growth of the tumors when the tumors contain high levels of CBS [51]. How, then, could one prospectively select “likely responder” high-CBS-expressor patients? Of course, the direct way to assess this matter is to quantify CBS expression in the primary tumor tissue after surgical resection; this quantification can be performed, for instance, by Western blotting or by immunohistochemistry. Naturally, this approach, can only guide postoperative (i.e. adjuvant) therapy, e.g. to suppress tumor relapse. If one wishes to prospectively identify high-CBS-expressors, a biomarker-based approach would need to be developed. H2S is a gaseous mediator, which is present in the circulation and exits the body, in part, via the exhaled air [393]; indeed, exhaled H2S measurements have been attempted to be used as disease markers, most commonly of various pulmonary diseases [[394], [395], [396]]. Interestingly, various animal species – from dogs to ants – can be ‘trained’ to recognize cancer patients [[397], [398], [399], [400]]; it is likely that the volatile biomarkers in the patient's exhaled breath or urine include H2S; olfactory receptors are extremely highly sensitive to H2S (in the parts per billion range). In fact, there are several studies showing that cancer patients exhale higher levels of H2S than healthy controls [394,401]. Detection methods based on this principle (“artificial noses” [[402], [403], [404]]) are in various stages of development. The exhaled breath-based approach, however, could not distinguish between H2S produced in the airways vs. systemically produced H2S which is upregulated during various inflammatory conditions [405,406] vs. H2S generated by the tumor tissue. Nor could this approach identify the enzymatic source of the H2S detected.
Thiosulfate is one of the principal stable metabolites of H2S – it can be detected in the circulation, and it is excreted via the urine [407,408]. This metabolite is easier to quantify than exhaled H2S, but, once again, any disease that induces the upregulation of any of the H2S-producing enzymes would be expected to increase its levels. Currently, there is no clear indication that urinary thiosulfate is elevated in cancer; an early report indicated that this may be the case in prostate cancer [409], but the preliminary findings could not be confirmed in a subsequent multicenter study [410].
Increased H2S production in vivo may also be detected by various bio-imaging approaches – e.g. using various chemiluminescent probes [[411], [412], [413], [414]] or using 99mTc-labeled gluconate or alpha-hydroxy acids [415,416]; one of these probes, in fact, shows that systemic treatment of HCT116-bearing mice with the allosteric CBS activator SAM increases the H2S signal in the tumor tissue [414]. These methods are currently only in preclinical stage and are not readily implementable in clinical trials.
If one looks for specific and non-invasive ways to identify CBS expression or activity in cancer patients, several biochemical and several molecular possibilities can be considered. On the biochemical side, measurement of various side-products of CBS enzymatic activity, such as lanthionine or cystathionine may be possible. Both of these mediators have been proposed as potential cancer biomarkers; cystathionine levels as well as lanthionine levels were found to be elevated in the supernatant of high-CBS-expressor cancer cells [51,63]. Increased lanthionine levels could be measured in the urine of mice bearing CBS-overexpressing epithelial cell tumors, compared to normal epithelial cells which express CBS only at low levels [51]. However, so far, no attempts have been made to measure these markers in the circulation of cancer patients.
Yet another potential approach may be to detect CBS-related markers in the circulating tumor cells or the circulating tumor DNA of cancer patients. There is some evidence that circulating tumor cells may express higher levels of CBS than resident ones [100], and also, generally, circulating cells (which have undergone EMT and/or stemness genes) are likely to be higher CBS-expressors, based on the data demonstrating that CBS expression increases as cancer cells undergo dedifferentiation (see above). However, the expression of CBS in circulating tumor cells has not yet been investigated in clinical studies. A related approach may be the assessment of the CBS promoter's methylation status in circulating cell-free tumor DNA (cfDNA), in light of the fact the methylation of the CBS promoter regulates the expression of CBS [[20], [21], [22], [23], [24], [25], [26]]. However – as discussed earlier – the clinical data published so far are conflicting with respect to the role of hypo-vs hypermethylation of this promoter in various forms cancer [22,56]. Thus, additional basic research, followed by clinical translation, remains to be conducted to expand the body of knowledge in this direction. The isolation of circulating tumor cells, and the separation of various cell populations (e.g. stem cells) as well as the isolation of circulating tumor DNA now feasible in clinical settings; multiple studies suggest its predictive potential in terms of clinical prognosis [e.g. Refs. [[417], [418], [419], [420], [421], [422], [423], [424], [425]]. Thus, a similar detection of CBS is not beyond the realms of possibilities. Obviously, direct detection of CBS protein in circulating tumor cells would be a more direct approach than studying the CBS promoter, given the fact that CBS levels are regulated at multiple levels (transcriptionally and post-transcriptionally), and it is currently not known, how tight is the correlation between CBS promoter activation in the circulation vs. CBS protein levels in the tumor tissue.
The concept of H2S donation. The bell-shaped character of H2S in the modulation of cancer cell viability is summarized in Fig. 15. As discussed in the previous pages, by inhibiting CBS (or, more generally, H2S production in tumor cells), we deprive the cancer cell from an endogenous tumor-promoting, cytoprotective and pro-angiogenic factor.
Fig. 15.
The bell-shaped concentration-response of H2S in cancer. H2S in the low-to-medium concentration range serves signaling functions, stimulates cellular bioenergetics, exerts cytoprotective effects, while high concentrations of H2S are generally cytotoxic. High-CBS-expressor cancer cells upregulate their H2S generation so that they are near the top of the bell-shaped H2S concentration-response curve, so that they best benefit from the supporting actions of H2S. This is beneficial for the cancer cell, but detrimental to the tumor-bearing host. Arrow #1: Inhibition of CBS-mediated H2S production in cancer cells can “take away” the stimulatory effects of H2S on cellular signaling and bioenergetics and can suppress cancer cell proliferation; this intervention, on its own, is typically not directly cytotoxic. However, CBS inhibition can synergize with the effect of chemotherapeutic agents (Arrow #2). Addition of lower concentrations of H2S to cancer cells may, in some conditions, induce a further proliferative or cytoprotective effect, by moving the cells to the “top” of the H2S concentration-response curve (Arrow #3), but donation of H2S, at higher concentrations, moves the curve to the right, where the suppressive bioenergetic and cytotoxic effects of H2S prevail (Arrow #4).
However, at higher concentrations, H2S becomes a cytotoxic molecule, which can reduce the viability (or kill) various cell types, including cancer cells. Consequently – in parallel with the development of the anticancer concept based on inhibition of H2S biosynthesis discussed in the current article – an entirely different concept, based on the anticancer effect of H2S donation – has also been advanced. The existence of this distinct body of literature – which is often confusing for scientists entering the field of H2S biology – does not invalidate the concepts and ideas discussed in the current article, but highlights an entirely different mechanism and therapeutic opportunity. The data supporting this concept – as well as the challenges related to this approach – e.g. the selective targeting of H2S donors to the tumors in vivo, as opposed to the surrounding normal tissues – are discussed in separate review articles [140,141,[426], [427], [428], [429], [430], [431]] and will not be covered, in detail, in the current review.
Conclusions and outlook. Taken together, the preclinical and clinical data presented in the current article support the view that CBS plays a significant pathogenetic role in multiple forms of cancer. Clinical correlations and the various in vitro and in vivo studies support the view that pharmacological inhibition of CBS may exert antitumor effects in a clinical setting, and supports the concept of further basic, as well as clinical/translational work in this area. The cancers currently best supported by consistent bodies of data are colon (or colorectal) cancer, ovarian cancer, and certain forms of breast and lung cancer – while the role of CBS in several other cancers (e.g. liver cancer and glioma) is markedly different, and in the therapy of these cancers CBS inhibition is unlikely to be a useful approach (in fact, the opposite approach, i.e. upregulation or induction of CBS or H2S donation may be considered).
Based on the role of CBS in cancer cells, and based on the multiple lines of data demonstrating the link of this enzyme to stemness, EMT and multidrug resistance, one possible translational direction may be combination therapy with chemotherapeutic agents, in various (curative, palliative or preventative, i.e. neoadjuvant or adjuvant) settings to enhance chemotherapeutic efficacy and/or to prevent cancer relapse. We predict that CBS inhibitors, when given to patients in reasonable durations, would be well tolerated, with safety profiles likely superior to the profile of standard chemotherapeutic agents. There is no indication in the literature to suggest that CBS inhibitors would exacerbate the side effects of chemotherapeutic agents – although this topic remains to be further investigated. Taken together, we hope that the current article will serve as a reference point and possible guide for the future clinical translation of the CBS inhibitor concept.
Author contributions
KA: literature review, manuscript writing, figure preparation; CS: literature review, manuscript writing, figure preparation.
Declaration of competing interest
None.
Acknowledgements
This work was supported by the Swiss Krebsliga (KLS 4504-08-2018R) to C.S.
References
- 1.Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol. Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 2.Huang C.W., Moore P.K. H2S synthesizing enzymes: biochemistry and molecular aspects. Handb. Exp. Pharmacol. 2015;230:3–25. doi: 10.1007/978-3-319-18144-8_1. [DOI] [PubMed] [Google Scholar]
- 3.Szabo C., Papapetropoulos A. International union of basic and clinical pharmacology. CII: pharmacological modulation of H2S levels: H2S donors and H2S biosynthesis inhibitors. Pharmacol. Rev. 2017;69:497–564. doi: 10.1124/pr.117.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szabo C. A timeline of hydrogen sulfide (H2S) research: from environmental toxin to biological mediator. Biochem. Pharmacol. 2018;149:5–19. doi: 10.1016/j.bcp.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura H. Hydrogen sulfide (H2S) and polysulfide (H2Sn) signaling: the first 25 years. Biomolecules. 2021;11:896. doi: 10.3390/biom11060896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo C., Coletta C., Chao C., Modis K., Szczesny B., Papapetropoulos A., Hellmich M.R. Tumor-derived hydrogen sulfide, produced by cystathionine-beta-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya S., Saha S., Giri K., Lanza I.R., Nair K.S., Jennings N.B., Rodriguez-Aguayo C., Lopez-Berestein G., Basal E., Weaver A.L., Visscher D.W., Cliby W., Sood A.K., Bhattacharya R., Mukherjee P. Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee R., Zou C.G. Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor protein. Arch. Biochem. Biophys. 2005;433:144–156. doi: 10.1016/j.abb.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 9.Singh S., Banerjee R. PLP-dependent H2S biogenesis. Biochim. Biophys. Acta. 2011;1814:1518–1527. doi: 10.1016/j.bbapap.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vicente J.B., Malagrinò F., Arese M., Forte E., Sarti P., Giuffrè A. Bioenergetic relevance of hydrogen sulfide and the interplay between gasotransmitters at human cystathionine β-synthase. Biochim. Biophys. Acta. 2016;1857:1127–1138. doi: 10.1016/j.bbabio.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Majtan T., Pey A.L., Gimenez-Mascarell P., Martínez-Cruz L.A., Szabo C., Kožich V., Kraus J.P. Potential pharmacological chaperones for cystathionine beta-synthase-deficient homocystinuria. Handb. Exp. Pharmacol. 2018;245:345–383. doi: 10.1007/164_2017_72. [DOI] [PubMed] [Google Scholar]
- 12.Zuhra K., Augsburger F., Majtan T., Szabo C. Cystathionine-beta-synthase: molecular regulation and pharmacological inhibition. Biomolecules. 2020;10:697. doi: 10.3390/biom10050697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge Y., Konrad M.A., Matherly L.H., Taub J.W. Transcriptional regulation of the human cystathionine beta-synthase -1b basal promoter: synergistic transactivation by transcription factors NF-Y and Sp1/Sp3. Biochem. J. 2001;357:97–105. doi: 10.1042/0264-6021:3570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hourihan J.M., Kenna G., Hayes J. The gasotransmitter hydrogen sulfide induces nrf2-target genes by inactivating the keap1 ubiquitin ligase substrate adaptor through formation of a disulfide bond between cys-226 and cys-613. Antioxidants Redox Signal. 2013;19:465–481. doi: 10.1089/ars.2012.4944. [DOI] [PubMed] [Google Scholar]
- 15.Kriebitzsch C., Verlinden L., Eelen G., van Schoor N.M., Swart K., Lips P., Meyer M.B., Pike J.W., Boonen S., Carlberg C., Vitvitsky V., Bouillon R., Banerjee R., Verstuyf A. 1,25-dihydroxyvitamin D3 influences cellular homocysteine levels in murine preosteoblastic MC3T3-E1 cells by direct regulation of cystathionine β-synthase. J. Bone Miner. Res. 2011;26:2991–3000. doi: 10.1002/jbmr.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechuga T.J., Qi Q.R., Kim T., Magness R.R., Chen D.B. E2β stimulates ovine uterine artery endothelial cell H2S production in vitro by estrogen receptor-dependent upregulation of cystathionine β-synthase and cystathionine γ-lyase expression. Biol. Reprod. 2019;100:514–522. doi: 10.1093/biolre/ioy207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratnam S., Maclean K.N., Jacobs R.L., Brosnan M.E., Kraus J.P., Brosnan J.T. Hormonal regulation of cystathionine beta-synthase expression in liver. J. Biol. Chem. 2002;277:42912–42918. doi: 10.1074/jbc.M206588200. [DOI] [PubMed] [Google Scholar]
- 18.Maclean K.N., Janosík M., Kraus E., Kozich V., Allen R.H., Raab B.K., Kraus J.P. Cystathionine beta-synthase is coordinately regulated with proliferation through a redox-sensitive mechanism in cultured human cells and Saccharomyces cerevisiae. J. Cell. Physiol. 2002;192:81–92. doi: 10.1002/jcp.10118. [DOI] [PubMed] [Google Scholar]
- 19.Takano N., Peng Y.J., Kumar G.K., Luo W., Hu H., Shimoda L.A., Suematsu M., Prabhakar N.R., Semenza G.L. Hypoxia-inducible factors regulate human and rat cystathionine β-synthase gene expression. Biochem. J. 2014;458:203–211. doi: 10.1042/BJ20131350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi F., Zhou Y., Xiao Y., Tao J., Gu J., Jiang X., Xu G.Y. Promoter demethylation of cystathionine-β-synthetase gene contributes to inflammatory pain in rats. Pain. 2013;154:34–45. doi: 10.1016/j.pain.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H.H., Hu J., Zhou Y.L., Hu S., Wang Y.M., Chen W., Xiao Y., Huang L.Y., Jiang X., Xu G.Y. Promoter interaction of nuclear factor-kappaB with demethylated cystathionine-beta-synthetase gene contributes to gastric hypersensitivity in diabetic rats. J. Neurosci. 2013;33:9028–9038. doi: 10.1523/JNEUROSCI.1068-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue G., Lu C.J., Pan S.J., Zhang Y.L., Miao H., Shan S., Zhu X.T., Zhang Y. DNA hypomethylation of CBS promoter induced by folate deficiency is a potential noninvasive circulating biomarker for colorectal adenocarcinomas. Oncotarget. 2017;8:51387–51401. doi: 10.18632/oncotarget.17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C., Xu G., Wen Q., Peng X., Chen H., Zhang J., Xu S., Zhang C., Zhang M., Ma J., Hui Z., Wu G., Ma M. CBS promoter hypermethylation increases the risk of hypertension and stroke. Clinics (Sao Paulo) 2019;74 doi: 10.6061/clinics/2019/e630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S., Zuo S., Zhu J., Yue T., Bu D., Wang X., Wang P., Pan Y., Liu Y. Decreased expression of cystathionine beta-synthase exacerbates intestinal barrier injury in ulcerative colitis. J. Crohns Colitis. 2019;13:1067–1080. doi: 10.1093/ecco-jcc/jjz027. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Q., Zhang C., Li D., Huang X., Ren B., Yue L., Du B., Godfrey O., Zhang W. CBS gene polymorphism and promoter methylation-mediating effects on the efficacy of folate therapy in patients with hyperhomocysteinemia. J. Gene Med. 2020;22:e3156. doi: 10.1002/jgm.3156. [DOI] [PubMed] [Google Scholar]
- 26.Huang X., Zhao Q., Li D., Ren B., Yue L., Shi F., Wang X., Zheng C., Chen X., Zhang C., Zhang W. Association between gene promoter methylation of the one-carbon metabolism pathway and serum folate among patients with hyperhomocysteinemia. Eur. J. Clin. Nutr. 2020;74:1677–1684. doi: 10.1038/s41430-020-0657-9. [DOI] [PubMed] [Google Scholar]
- 27.Pagliara V., Saide A., Mitidieri E., d'Emmanuele di Villa Bianca R., Sorrentino R., Russo G., Russo A. 5-FU targets rpL3 to induce mitochondrial apoptosis via cystathionine-β-synthase in colon cancer cells lacking p53. Oncotarget. 2016;7:50333–50348. doi: 10.18632/oncotarget.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akimov V., Barrio-Hernandez I., Hansen S.V.F., Hallenborg P., Pedersen A.K., Bekker-Jensen D.B., Puglia M., Christensen S.D.K., Vanselow J.T., Nielsen M.M., Kratchmarova I., Kelstrup C.D., Olsen J.V., Blagoev B. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018;25:631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- 29.Gupta S., Wang L., Anderl J., Slifker M.J., Kirk C., Kruger W.D. Correction of cystathionine β-synthase deficiency in mice by treatment with proteasome inhibitors. Hum. Mutat. 2013;34:1085–1093. doi: 10.1002/humu.22335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabil O., Zhou Y., Banerjee R. Human cystathionine β-synthase is a target for sumoylation. Biochemistry. 2006;45:13528–13536. doi: 10.1021/bi0615644. [DOI] [PubMed] [Google Scholar]
- 31.Teng H., Wu B., Zhao K., Yang G., Wu L., Wang R. Oxygen-sensitive mitochondrial accumulation of cystathionine β-synthase mediated by Lon protease. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12679–12684. doi: 10.1073/pnas.1308487110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skovby F., Kraus J.P., Rosenberg L.E. Biosynthesis and proteolytic activation of cystathionine beta-synthase in rat liver. J. Biol. Chem. 1984;259:588–593. [PubMed] [Google Scholar]
- 33.Kéry V., Poneleit L., Kraus J.P. Trypsin cleavage of human cystathionine β-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch. Biochem. Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 34.Bruno S., Schiaretti F., Burkhard P., Kraus J.P., Janosik M., Mozzarelli A. Functional properties of the active core of human cystathionine β-synthase crystals. J. Biol. Chem. 2000;276:16–19. doi: 10.1074/jbc.C000588200. [DOI] [PubMed] [Google Scholar]
- 35.Pey A.L., Martínez-Cruz L.A., Kraus J.P., Majtan T. Oligomeric status of human cystathionine beta-synthase modulates AdoMet binding. FEBS Lett. 2016;590:4461–4471. doi: 10.1002/1873-3468.12488. [DOI] [PubMed] [Google Scholar]
- 36.Zou C.G., Banerjee R. Tumor necrosis factor-alpha-induced targeted proteolysis of cystathionine beta-synthase modulates redox homeostasis. J. Biol. Chem. 2003;278:16802–16808. doi: 10.1074/jbc.M212376200. [DOI] [PubMed] [Google Scholar]
- 37.Chan S.J., Chai C., Lim T.W., Yamamoto M., Lo E.H., Lai M.K., Wong P.T. Cystathionine beta-synthase inhibition is a potential therapeutic approach to treatment of ischemic injury. ASN Neuro. 2015;7 doi: 10.1177/1759091415578711. 1759091415578711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Módis K., Coletta C., Asimakopoulou A., Szczesny B., Chao C., Papapetropoulos A., Hellmich M.R., Szabo C. Effect of S-adenosyl-L-methionine (SAM), an allosteric activator of cystathionine-β-synthase (CBS) on colorectal cancer cell proliferation and bioenergetics in vitro. Nitric Oxide. 2014;41:146–156. doi: 10.1016/j.niox.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panagaki T., Lozano-Montes L., Janickova L., Zuhra K., Szabo M.P., Majtan T., Rainer G., Maréchal D., Herault Y., Szabo C. Overproduction of hydrogen sulfide, generated by cystathionine β-synthase, disrupts brain wave patterns and contributes to neurobehavioral dysfunction in a rat model of Down syndrome. Redox Biol. 2022;51 doi: 10.1016/j.redox.2022.102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goubern M., Andriamihaja M., Nübel T., Blachier F., Bouillaud F. Sulfide, the first inorganic substrate for human cells. Faseb. J. 2007;21:1699–1706. doi: 10.1096/fj.06-7407com. [DOI] [PubMed] [Google Scholar]
- 41.Szabo C., Ransy C., Módis K., Andriamihaja M., Murghes B., Coletta C., Olah G., Yanagi K., Bouillaud F. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br. J. Pharmacol. 2014;171:2099–2122. doi: 10.1111/bph.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Módis K., Bos E.M., Calzia E., van Goor H., Coletta C., Papapetropoulos A., Hellmich M.R., Radermacher P., Bouillaud F., Szabo C. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part II. Pathophysiological and therapeutic aspects. Br. J. Pharmacol. 2014;171:2123–2146. doi: 10.1111/bph.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy B., Bhattacharya R., Mukherjee P. Hydrogen sulfide signaling in mitochondria and disease. Faseb. J. 2019;33:13098–13125. doi: 10.1096/fj.201901304R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul B.D., Snyder S.H., Kashfi K. Effects of hydrogen sulfide on mitochondrial function and cellular bioenergetics. Redox Biol. 2021;38 doi: 10.1016/j.redox.2020.101772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borisov V.B., Forte E. Impact of hydrogen sulfide on mitochondrial and bacterial bioenergetics. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222312688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng W., Guo W., Zheng Y. Recent development of hydrogen sulfide therapeutics in the treatment of cardiovascular diseases. Curr. Top. Med. Chem. 2021;21:2230–2242. doi: 10.2174/1568026621666210831163817. [DOI] [PubMed] [Google Scholar]
- 47.Szabo C. Hydrogen sulfide, an endogenous stimulator of mitochondrial function in cancer cells. Cells. 2021;10:220. doi: 10.3390/cells10020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips C.M., Zatarain J.R., Nicholls M.E., Porter C., Widen S.G., Thanki K., Johnson P., Jawad M.U., Moyer M.P., Randall J.W., Hellmich J.L., Maskey M., Qiu S., Wood T.G., Druzhyna N., Szczesny B., Modis K., Szabo C., Chao C., Hellmich M.R. Upregulation of cystathionine-beta-synthase in colonic epithelia reprograms metabolism and promotes carcinogenesis. Cancer Res. 2017;77:5741–5754. doi: 10.1158/0008-5472.CAN-16-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olah G., Modis K., Toro G., Hellmich M.R., Szczesny B., Szabo C. Role of endogenous and exogenous nitric oxide, carbon monoxide and hydrogen sulfide in HCT116 colon cancer cell proliferation. Biochem. Pharmacol. 2018;149:186–204. doi: 10.1016/j.bcp.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen S., Yue T., Huang Z., Zhu J., Bu D., Wang X., Pan Y., Liu Y., Wang P. Inhibition of hydrogen sulfide synthesis reverses acquired resistance to 5-FU through miR-215-5p-EREG/TYMS axis in colon cancer cells. Cancer Lett. 2019;466:49–60. doi: 10.1016/j.canlet.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Hellmich M.R., Chao C., Módis K., Ding Y., Zatarain J.R., Thanki K., Maskey M., Druzhyna N., Untereiner A.A., Ahmad A., Xue Y., Chen H., Russell W.K., Wang J., Zhou J., Szabo C. Efficacy of novel aminooxyacetic acid prodrugs in colon cancer models: towards clinical translation of the cystathionine beta-synthase inhibition concept. Biomolecules. 2021;11:1073. doi: 10.3390/biom11081073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo S., Li J., Huang Z., Yue T., Zhu J., Wang X., Liu Y., Wang P., Chen S. The CBS-H2S axis promotes liver metastasis of colon cancer by upregulating VEGF through AP-1 activation. Br. J. Cancer. 2022;126:1055–1066. doi: 10.1038/s41416-021-01681-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaedcke J., Grade M., Jung K., Camps J., Jo P., Emons G., Gehoff A., Sax U., Schirmer M., Becker H., Beissbarth T., Ried T., Ghadimi B.M. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 2010;49:1024–1034. doi: 10.1002/gcc.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graudens E., Boulanger V., Mollard C., Mariage-Samson R., Barlet X., Grémy G., Couillault C., Lajémi M., Piatier-Tonneau D., Zaborski P., Eveno E., Auffray C., Imbeaud S. Deciphering cellular states of innate tumor drug responses. Genome Biol. 2006;7:R19. doi: 10.1186/gb-2006-7-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silver D.J., Roversi G.A., Bithi N., Wang S.Z., Troike K.M., Neumann C.K., Ahuja G.K., Reizes O., Brown J.M., Hine C., Lathia J.D. Severe consequences of a high-lipid diet include hydrogen sulfide dysfunction and enhanced aggression in glioblastoma. J. Clin. Invest. 2021;131 doi: 10.1172/JCI138276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., He Y., Tu X., Huang S., Chen Z., Wang L., Song J. Mapping of DNA hypermethylation and hypomethylation induced by folate deficiency in sporadic colorectal cancer and clinical implication analysis of hypermethylation pattern in CBS promoter. Clin. Lab. 2017;63:733–748. doi: 10.7754/Clin.Lab.2016.161018. [DOI] [PubMed] [Google Scholar]
- 57.Nazemalhosseini Mojarad E., Kuppen P.J., Aghdaei H.A., Zali M.R. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol. Hepatol. Bed Bench. 2013;6:120–128. [PMC free article] [PubMed] [Google Scholar]
- 58.Mármol I., Sánchez-de-Diego C., Pradilla Dieste A., Cerrada E., Rodriguez Yoldi M.J. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 2017;18:197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X., Zhang W., Cao P. Advances in CpG island methylator phenotype colorectal cancer therapies. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.629390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Padmanabhan N., Kyon H.K., Boot A., Lim K., Srivastava S., Chen S., Wu Z., Lee H.O., Mukundan V.T., Chan C., Chan Y.K., Xuewen O., Pitt J.J., Isa Z.F.A., Xing M., Lee M.H., Tan A.L.K., Ting S.H.W., Luftig M.A., Kappei D., Kruger W.D., Bian J., Ho Y.S., Teh M., Rozen S.G., Tan P. Highly recurrent CBS epimutations in gastric cancer CpG island methylator phenotypes and inflammation. Genome Biol. 2021;22:167. doi: 10.1186/s13059-021-02375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakraborty P.K., Murphy B., Mustafi S.B., Dey A., Xiong X., Rao G., Naz S., Zhang M., Yang D., Dhanasekaran D.N., Bhattacharya R., Mukherjee P. Cystathionine beta-synthase regulates mitochondrial morphogenesis in ovarian cancer. Faseb. J. 2018;32:4145–4157. doi: 10.1096/fj.201701095R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sen S., Kawahara B., Gupta D., Tsai R., Khachatryan M., Roy-Chowdhuri S., Bose S., Yoon A., Faull K., Farias-Eisner R., Chaudhuri G. Role of cystathionine beta-synthase in human breast cancer. Free Radic. Biol. Med. 2015;86:228–238. doi: 10.1016/j.freeradbiomed.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 63.Sen S., Kawahara B., Mahata S.K., Tsai R., Yoon A., Hwang L., Hu-Moore K., Villanueva C., Vajihuddin A., Parameshwar P., You M., Bhaskar D.L., Gomez O., Faull K.F., Farias-Eisner R., Chaudhuri G., Cystathionine A novel oncometabolite in human breast cancer. Arch. Biochem. Biophys. 2016;604:95–102. doi: 10.1016/j.abb.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 64.Erdélyi K., Ditrói T., Johansson H.J., Czikora Á., Balog N., Silwal-Pandit L., Ida T., Olasz J., Hajdú D., Mátrai Z., Csuka O., Uchida K., Tóvári J., Engebraten O., Akaike T., Børresen Dale A.L., Kásler M., Lehtiö J., Nagy P. Reprogrammed transsulfuration promotes basal-like breast tumor progression via realigning cellular cysteine persulfidation. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2100050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Youness R.A., Gad A.Z., Sanber K., Ahn Y.J., Lee G.J., Khallaf E., Hafez H.M., Motaal A.A., Ahmed N., Gad M.Z. Targeting hydrogen sulphide signaling in breast cancer. J. Adv. Res. 2021;27:177–190. doi: 10.1016/j.jare.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao J., Zhao Y., Ding S., Liu T., Meng F. Low CBS expression can identify patients who benefit from adjuvant chemotherapy in gastric cancer. Expert Rev. Anticancer Ther. 2021;21:1287–1298. doi: 10.1080/14737140.2021.1962298. [DOI] [PubMed] [Google Scholar]
- 67.Gai J.W., Qin W., Liu M., Wang H.F., Zhang M., Li M., Zhou W.H., Ma Q.T., Liu G.M., Song W.H., Jin J., Ma H.S. Expression profile of hydrogen sulfide and its synthases correlates with tumor stage and grade in urothelial cell carcinoma of bladder. Urol. Oncol. 2016;34:166. doi: 10.1016/j.urolonc.2015.06.020. e15–20. [DOI] [PubMed] [Google Scholar]
- 68.Wahafu W., Gai J., Song L., Ping H., Wang M., Yang F., Niu Y., Xing N. Increased H2S and its synthases in urothelial cell carcinoma of the bladder, and enhanced cisplatin-induced apoptosis following H2S inhibition in EJ cells. Oncol. Lett. 2018;15:8484–8490. doi: 10.3892/ol.2018.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szczesny B., Marcatti M., Zatarain J.R., Druzhyna N., Wiktorowicz J.E., Nagy P., Hellmich M.R., Szabo C. Inhibition of hydrogen sulfide biosynthesis sensitizes lung adenocarcinoma to chemotherapeutic drugs by inhibiting mitochondrial DNA repair and suppressing cellular bioenergetics. Sci. Rep. 2016;6:36125. doi: 10.1038/srep36125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang M., Yan J., Cao X., Hua P., Li Z. Hydrogen sulfide modulates epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via HIF-1alpha activation. Biochem. Pharmacol. 2020;172 doi: 10.1016/j.bcp.2019.113775. [DOI] [PubMed] [Google Scholar]
- 71.Russo A., Saide A., Cagliani R., Cantile M., Botti G., Russo G. rpL3 promotes the apoptosis of p53 mutated lung cancer cells by down-regulating CBS and NFkappaB upon 5-FU treatment. Sci. Rep. 2016;6:38369. doi: 10.1038/srep38369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shackelford R.E., Abdulsattar J., Wei E.X., Cotelingam J., Coppola D., Herrera G.A. Increased nicotinamide phosphoribosyltransferase and cystathionine-beta-synthase in renal oncocytomas, renal urothelial carcinoma, and renal clear cell carcinoma. Anticancer Res. 2017;37:3423–3427. doi: 10.21873/anticanres.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sogutdelen E., Pacoli K., Juriasingani S., Akbari M., Gabril M., Sener A. Patterns of expression of H2S-producing enzyme in human renal cell carcinoma specimens: potential avenue for future therapeutics. In Vivo. 2020;34:2775–2781. doi: 10.21873/invivo.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patel S., Ansari J., Meram A., Abdulsattar J., Cotelingam J., Coppola D., Ghali G., Shackelford R. Increased nicotinamide phosphoribosyltransferase and cystathionine-beta-synthase in oral cavity squamous cell carcinomas. Int. J. Clin. Exp. Pathol. 2017;10:702–707. [Google Scholar]
- 75.Meram A.T., Chen J., Patel S., Kim D.D., Shirley B., Covello P., Coppola D., Wei E.X., Ghali G., Kevil C.G., Shackelford R.E. Hydrogen sulfide is increased in oral squamous cell carcinoma compared to adjacent benign oral mucosae. Anticancer Res. 2018;38:3843–3852. doi: 10.21873/anticanres.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Y., Pan L., Li Y., Deng Y., Han X., Fu H., Wang T. Cystathionine-beta-synthase (CBS) / H2S system promotes lymph node metastasis of esophageal squamous cell carcinoma by activating SIRT1. Carcinogenesis. 2022;43:382–392. doi: 10.1093/carcin/bgac002. [DOI] [PubMed] [Google Scholar]
- 77.Turbat-Herrera E.A., Kilpatrick M.J., Chen J., Meram A.T., Cotelingam J., Ghali G., Kevil C.G., Coppola D., Shackelford R.E. CBS is increased in thyroid malignancies. Anticancer Res. 2018;38:6085–6090. doi: 10.21873/anticanres.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L., Yang Z., Wu Z., He J., Xu S., Li D., Zou Q., Yuan Y. Increased expression of cystathionine beta-synthase and chemokine ligand 21 is closely associated with poor prognosis in extrahepatic cholangiocarcinoma. Medicine (Baltim.) 2020;99 doi: 10.1097/MD.0000000000022255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li D., Yang Z., Liu Z., Zou Q., Yuan Y. Clinical significance of CBS and CCL21 in gallbladder adenocarcinomas and squamous cell/adenosquamous carcinomas. Appl. Immunohistochem. Mol. Morphol. 2020;28:103–110. doi: 10.1097/PAI.0000000000000705. [DOI] [PubMed] [Google Scholar]
- 80.Hansel D.E., Rahman A., Hidalgo M., Thuluvath P.J., Lillemoe K.D., Schulick R., Ku J.L., Park J.G., Miyazaki K., Ashfaq R., Wistuba I.I., Varma R., Hawthorne L., Geradts J., Argani P., Maitra A. Identification of novel cellular targets in biliary tract cancers using global gene expression technology. Am. J. Pathol. 2003;163:217–229. doi: 10.1016/S0002-9440(10)63645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Vos J., Thykjær T., Tarte K., Ensslen M., Raynaud P., Requirand G., Pellet F., Pantesco V., Rème T., Jourdan M., Rossi J.F., Ørntoft T., Klein B. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene. 2002;21:6848–6857. doi: 10.1038/sj.onc.1205868. [DOI] [PubMed] [Google Scholar]
- 82.Zhang M., Li J., Huang B., Kuang L., Xiao F., Zheng D. Cystathionine beta synthase/hydrogen sulfide signaling in multiple myeloma regulates cell proliferation and apoptosis. J. Environ. Pathol. Toxicol. Oncol. 2020;39:281–290. doi: 10.1615/JEnvironPatholToxicolOncol.2020034851. [DOI] [PubMed] [Google Scholar]
- 83.Zhang L., Qi Q., Yang J., Sun D., Li C., Xue Y., Jiang Q., Tian Y., Xu C., Wang R. An anticancer role of hydrogen sulfide in human gastric cancer cells. Oxid. Med. Cell. Longev. 2015;2015 doi: 10.1155/2015/636410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim J., Hong S.J., Park J.H., Park S.Y., Kim S.W., Cho E.Y., Do I.G., Joh J.W., Kim D.S. Expression of cystathionine beta-synthase is downregulated in hepatocellular carcinoma and associated with poor prognosis. Oncol. Rep. 2009;21:1449–1454. doi: 10.3892/or_00000373. [DOI] [PubMed] [Google Scholar]
- 85.Cai F.F., Song Y.N., Lu Y.Y., Zhang Y., Hu Y.Y., Su S.B. Analysis of plasma metabolic profile, characteristics and enzymes in the progression from chronic hepatitis B to hepatocellular carcinoma. Aging (Albany NY) 2020;12:14949–14965. doi: 10.18632/aging.103554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang X., Chen M., Ni X., Wang Y., Zheng X., Zhang H., Xu S., Yang C.T. Metabolic reprogramming of sulfur in hepatocellular carcinoma and sulfane sulfur-triggered anti-cancer strategy. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.571143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou Y.F., Song S.S., Tian M.X., Tang Z., Wang H., Fang Y., Qu W.F., Jiang X.F., Tao C.Y., Huang R., Zhou P.Y., Zhu S.G., Zhou J., Fan J., Liu W.R., Shi Y.H. Cystathionine β-synthase mediated PRRX2/IL-6/STAT3 inactivation suppresses Tregs infiltration and induces apoptosis to inhibit HCC carcinogenesis. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takano N., Sarfraz Y., Gilkes D.M., Chaturvedi P., Xiang L., Suematsu M., Zagzag D., Semenza G.L. Decreased expression of cystathionine beta-synthase promotes glioma tumorigenesis. Mol. Cancer Res. 2014;12:1398–1406. doi: 10.1158/1541-7786.MCR-14-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang W., Braun A., Bauman Z., Olteanu H., Madzelan P., Banerjee R. Expression profiling of homocysteine junction enzymes in the NCI60 panel of human cancer cell lines. Cancer Res. 2005;65:1554–1560. doi: 10.1158/0008-5472.CAN-04-1554. [DOI] [PubMed] [Google Scholar]
- 90.Cai W.J., Wang M.J., Ju L.H., Wang C., Zhu Y.C. Hydrogen sulfide induces human colon cancer cell proliferation: role of Akt, ERK and p21, Cell. Biol. Int. 2010;34:565–572. doi: 10.1042/CBI20090368. [DOI] [PubMed] [Google Scholar]
- 91.Untereiner A.A., Oláh G., Módis K., Hellmich M.R., Szabo C. H2S-induced S-sulfhydration of lactate dehydrogenase a (LDHA) stimulates cellular bioenergetics in HCT116 colon cancer cells. Biochem. Pharmacol. 2017;136:86–98. doi: 10.1016/j.bcp.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Niu W., Chen F., Wang J., Qian J., Yan S. Antitumor effect of sikokianin C, a selective cystathionine beta-synthase inhibitor, against human colon cancer in vitro and in vivo. Medchemcomm. 2017;9:113–120. doi: 10.1039/c7md00484b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Untereiner A.A., Pavlidou A., Druzhyna N., Papapetropoulos A., Hellmich M.R., Szabo C. Drug resistance induces the upregulation of H2S-producing enzymes in HCT116 colon cancer cells. Biochem. Pharmacol. 2018;149:174–185. doi: 10.1016/j.bcp.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bobba K.N., Saranya G., Sujai P.T., Joseph M.M., Velusamy N., Podder A., Maiti K.K., Bhuniya S. Endogenous H2S-assisted cancer-cell-specific activation of theranostics with emission readout. ACS Appl. Bio Mater. 2019;2:1322–1330. doi: 10.1021/acsabm.9b00019. [DOI] [PubMed] [Google Scholar]
- 95.Yue T., Zuo S., Bu D., Zhu J., Chen S., Ma Y., Ma J., Guo S., Wen L., Zhang X., Hu J., Wang Y., Yao Z., Chen G., Wang X., Pan Y., Wang P., Liu Y. Aminooxyacetic acid (AOAA) sensitizes colon cancer cells to oxaliplatin via exaggerating apoptosis induced by ROS. J. Cancer. 2020;11:1828–1838. doi: 10.7150/jca.35375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zuhra K., Panagaki T., Randi E.B., Augsburger F., Blondel M., Friocourt G., Herault Y., Szabo C. Mechanism of cystathionine-β-synthase inhibition by disulfiram: the role of bis(N,N-diethyldithiocarbamate)-copper(II) Biochem. Pharmacol. 2020;182 doi: 10.1016/j.bcp.2020.114267. [DOI] [PubMed] [Google Scholar]
- 97.Ascencao K., Dilek N., Augsburger F., Panagaki T., Zuhra K., Szabo C. Pharmacological induction of mesenchymal-epithelial transition via inhibition of H2S biosynthesis and consequent suppression of ACLY activity in colon cancer cells. Pharmacol. Res. 2021;165 doi: 10.1016/j.phrs.2020.105393. [DOI] [PubMed] [Google Scholar]
- 98.Mu T., Chu T., Li W., Dong Q., Liu Y. N1, N12-diacetylspermine is elevated in colorectal cancer and promotes proliferation through the miR-559/CBS axis in cancer cell lines. JAMA Oncol. 2021;2021 doi: 10.1155/2021/6665704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y., Chen S., Zhu J., Guo S., Yue T., Xu H., Hu J., Huang Z., Chen Z., Wang P., Liu Y. Overexpression of CBS/H2S inhibits proliferation and metastasis of colon cancer cells through downregulation of CD44. Cancer Cell Int. 2022;22:85. doi: 10.1186/s12935-022-02512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alix-Panabières C., Cayrefourcq L., Mazard T., Maudelonde T., Assenat E., Assou S. Molecular portrait of metastasis-competent circulating tumor cells in colon cancer reveals the crucial role of genes regulating energy metabolism and DNA repair. Clin. Chem. 2017;63:700–713. doi: 10.1373/clinchem.2016.263582. [DOI] [PubMed] [Google Scholar]
- 101.Chakraborty P.K., Xiong X., Mustafi S.B., Saha S., Dhanasekaran D., Mandal N.A., McMeekin S., Bhattacharya R., Mukherjee P. Role of cystathionine beta synthase in lipid metabolism in ovarian cancer. Oncotarget. 2015;6:37367–37384. doi: 10.18632/oncotarget.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu N., Lin X., Huang C. Activation of the reverse transsulfuration pathway through NRF2/CBS confers erastin-induced ferroptosis resistance. Br. J. Cancer. 2020;122:279–292. doi: 10.1038/s41416-019-0660-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sanokawa-Akakura R., Ostrakhovitch E.A., Akakura S., Goodwin S., Tabibzadeh S. A H2S-Nampt dependent energetic circuit is critical to survival and cytoprotection from damage in cancer cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ostrakhovitch E.A., Akakura S., Sanokawa-Akakura R., Goodwin S., Tabibzadeh S. Dedifferentiation of cancer cells following recovery from a potentially lethal damage is mediated by H2S-Nampt. Exp. Cell Res. 2015;330:135–150. doi: 10.1016/j.yexcr.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 105.Kawahara B., Moller T., Hu-Moore K., Carrington S., Faull K.F., Sen S., Mascharak P.K. Attenuation of antioxidant capacity in human breast cancer cells by carbon monoxide through inhibition of CBS activity: implications in chemotherapeutic drug sensitivity. J. Med. Chem. 2017;60:8000–8010. doi: 10.1021/acs.jmedchem.7b00476. [DOI] [PubMed] [Google Scholar]
- 106.Sonke E., Verrydt M., Postenka C.O., Pardhan S., Willie C.J., Mazzola C.R., Hammers M.D., Pluth M.D., Lobb I., Power N.E., Chambers A.F., Leong H.S., Sener A. Inhibition of endogenous hydrogen sulfide production in clear-cell renal cell carcinoma cell lines and xenografts restricts their growth, survival and angiogenic potential. Nitric Oxide. 2015;49:26–39. doi: 10.1016/j.niox.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sekiguchi F., Sekimoto T., Ogura A., Kawabata A. Endogenous hydrogen sulfide enhances cell proliferation of human gastric cancer AGS cells. Biol. Pharm. Bull. 2016;39:887–890. doi: 10.1248/bpb.b15-01015. [DOI] [PubMed] [Google Scholar]
- 108.Jia H., Ye J., You J., Shi X., Kang W., Wang T. Role of the cystathionine β-synthase/H2S system in liver cancer cells and the inhibitory effect of quinolone-indolone conjugate QIC2 on the system. Oncol. Rep. 2017;37:3001–3009. doi: 10.3892/or.2017.5513. [DOI] [PubMed] [Google Scholar]
- 109.Wang L., Han H., Liu Y., Zhang X., Shi X., Wang T. Cystathionine beta-synthase induces multidrug resistance and metastasis in hepatocellular carcinoma. Curr. Mol. Med. 2018;18:496–506. doi: 10.2174/1566524019666181211162754. [DOI] [PubMed] [Google Scholar]
- 110.Zhang H., Song Y., Zhou C., Bai Y., Yuan D., Pan Y., Shao C. Blocking endogenous H2S signaling attenuated radiation-induced long-term metastasis of residual HepG2 cells through inhibition of EMT. Radiat. Res. 2018;190:374–384. doi: 10.1667/RR15074.1. [DOI] [PubMed] [Google Scholar]
- 111.Wang L., Cai H., Hu Y., Liu F., Huang S., Zhou Y., Yu J., Xu J., Wu F. A pharmacological probe identifies cystathionine beta-synthase as a new negative regulator for ferroptosis. Cell Death Dis. 2018;9:1005. doi: 10.1038/s41419-018-1063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ye F., Li X., Sun K., Xu W., Shi H., Bian J., Lu R., Ye Y. Inhibition of endogenous hydrogen sulfide biosynthesis enhances the anti-cancer effect of 3,3'-diindolylmethane in human gastric cancer cells. Life Sci. 2020;261 doi: 10.1016/j.lfs.2020.118348. [DOI] [PubMed] [Google Scholar]
- 113.Ruiz-Rodado V., Dowdy T., Lita A., Kramp T., Zhang M., Jung J., Dios-Esponera A., Zhang L., Herold-Mende C.C., Camphausen K., Gilbert M.R., Larion M. Cysteine is a limiting factor for glioma proliferation and survival. Mol. Oncol. 2022;16:1777–1794. doi: 10.1002/1878-0261.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prudova A., Albin M., Bauman Z., Lin A., Vitvitsky V., Banerjee R. Testosterone regulation of homocysteine metabolism modulates redox status in human prostate cancer cells. Antioxidants Redox Signal. 2007;9:1875–1881. doi: 10.1089/ars.2007.1712. [DOI] [PubMed] [Google Scholar]
- 115.Guo H., Gai J.W., Wang Y., Jin H.F., Du J.B., Jin J. Characterization of hydrogen sulfide and its synthases, cystathionine β-synthase and cystathionine γ-lyase, in human prostatic tissue and cells. Urology. 2012;79:483. doi: 10.1016/j.urology.2011.10.013. e1–5. [DOI] [PubMed] [Google Scholar]
- 116.Zhang J., Xie Y., Xu Y., Pan Y., Shao C. Hydrogen sulfide contributes to hypoxia-induced radioresistance on hepatoma cells. J. Radiat. Res. 2011;52:622–628. doi: 10.1269/jrr.11004. [DOI] [PubMed] [Google Scholar]
- 117.Wu D., Li J., Zhang Q., Tian W., Zhong P., Liu Z., Wang H., Wang H., Ji A., Li Y. Exogenous hydrogen sulfide regulates the growth of human thyroid carcinoma cells. Oxid. Med. Cell. Longev. 2019 doi: 10.1155/2019/6927298. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ryu C.S., Kwak H.C., Lee K.S., Kang K.W., Oh S.J., Lee K.H., Kim H.M., Ma J.Y., Kim S.K. Sulfur amino acid metabolism in doxorubicin-resistant breast cancer cells. Toxicol. Appl. Pharmacol. 2011;255:94–102. doi: 10.1016/j.taap.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 119.Shukla P., Khodade V.S., Sharath Chandra M., Chauhan P., Mishra S., Siddaramappa S., Pradeep B.E., Singh A., Chakrapani H. On demand" redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem. Sci. 2017;8:4967–4972. doi: 10.1039/c7sc00873b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xuan G., Lü C., Xu H., Chen Z., Li K., Liu H., Liu H., Xia Y., Xun L. Sulfane sulfur is an intrinsic signal activating MexR-regulated antibiotic resistance in Pseudomonas aeruginosa. Mol. Microbiol. 2020;114:1038–1048. doi: 10.1111/mmi.14593. [DOI] [PubMed] [Google Scholar]
- 121.Mao Z., Yang X., Mizutani S., Huang Y., Zhang Z., Shinmori H., Gao K., Yao J. Hydrogen sulfide mediates tumor cell resistance to thioredoxin inhibitor. Front. Oncol. 2020;10:252. doi: 10.3389/fonc.2020.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cai W.J., Wang M.J., Moore P.K., Jin H.M., Yao T., Zhu Y.C. The novel proangiogenic effect of hydrogen sulfide is dependent on Akt phosphorylation. Cardiovasc. Res. 2007;76:29–40. doi: 10.1016/j.cardiores.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 123.Jha S., Calvert J.W., Duranski M.R., Ramachandran A., Lefer D.J. Hydrogen sulfide attenuates hepatic ischemia-reperfusion injury: role of antioxidant and antiapoptotic signaling. Am. J. Physiol. Heart Circ. Physiol. 2008;295:H801–806. doi: 10.1152/ajpheart.00377.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Papapetropoulos A., Pyriochou A., Altaany Z., Yang G., Marazioti A., Zhou Z., Jeschke M.G., Branski L.K., Herndon D.N., Wang R., Szabó C. Hydrogen sulfide is an endogenous stimulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21972–21977. doi: 10.1073/pnas.0908047106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Calvert J.W., Jha S., Gundewar S., Elrod J.W., Ramachandran A., Pattillo C.B., Kevil C.G., Lefer D.J. Hydrogen sulfide mediates cardioprotection through Nrf2 signaling. Circ. Res. 2009;105:365–374. doi: 10.1161/CIRCRESAHA.109.199919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Coletta C., Papapetropoulos A., Erdelyi K., Olah G., Módis K., Panopoulos P., Asimakopoulou A., Gerö D., Sharina I., Martin E., Szabo C. Hydrogen sulfide and nitric oxide are mutually dependent in the regulation of angiogenesis and endothelium-dependent vasorelaxation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9161–9166. doi: 10.1073/pnas.1202916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Majid A.S., Majid A.M., Yin Z.Q., Ji D. Slow regulated release of H2S inhibits oxidative stress induced cell death by influencing certain key signaling molecules. Neurochem. Res. 2013;38:1375–1393. doi: 10.1007/s11064-013-1034-z. [DOI] [PubMed] [Google Scholar]
- 128.Zheng D., Chen Z., Chen J., Zhuang X., Feng J., Li J. Exogenous hydrogen sulfide exerts proliferation, anti-apoptosis, migration effects and accelerates cell cycle progression in multiple myeloma cells via activating the Akt pathway. Oncol. Rep. 2016;36:1909–1916. doi: 10.3892/or.2016.5014. [DOI] [PubMed] [Google Scholar]
- 129.Wu D., Li M., Tian W., Wang S., Cui L., Li H., Wang H., Ji A., Li Y. Hydrogen sulfide acts as a double-edged sword in human hepatocellular carcinoma cells through EGFR/ERK/MMP-2 and PTEN/AKT signaling pathways. Sci. Rep. 2017;7:5134. doi: 10.1038/s41598-017-05457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lei Y.Y., Feng Y.F., Zeng B., Zhang W., Xu Q., Cheng F., Lan J., Luo H.H., Zou J.Y., Chen Z.G., Su C.H., Zhen Y.L., Chen J.F. Exogenous H2S promotes cancer progression by activating JAK2/STAT3 signaling pathway in esophageal EC109 cells. Int. J. Clin. Exp. Pathol. 2018;11:3247–3256. [PMC free article] [PubMed] [Google Scholar]
- 131.Tao B.B., Liu S.Y., Zhang C.C., Fu W., Cai W.J., Wang Y., Shen Q., Wang M.J., Chen Y., Zhang L.J., Zhu Y.Z., Zhu Y.C. VEGFR2 functions as an H2S-targeting receptor protein kinase with its novel Cys1045-Cys1024 disulfide bond serving as a specific molecular switch for hydrogen sulfide actions in vascular endothelial cells. Antioxidants Redox Signal. 2013;19:448–464. doi: 10.1089/ars.2012.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Katsouda A., Bibli S.I., Pyriochou A., Szabo C., Papapetropoulos A. Regulation and role of endogenously produced hydrogen sulfide in angiogenesis. Pharmacol. Res. 2016;113:175–185. doi: 10.1016/j.phrs.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saha S., Chakraborty P.K., Xiong X., Dwivedi S.K., Mustafi S.B., Leigh N.R., Ramchandran R., Mukherjee P., Bhattacharya R. Cystathionine beta-synthase regulates endothelial function via protein S-sulfhydration. Faseb. J. 2016;30:441–456. doi: 10.1096/fj.15-278648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kanagy N.L., Szabo C., Papapetropoulos A. Vascular biology of hydrogen sulfide. Am. J. Physiol. Cell Physiol. 2017;312:C537–549. doi: 10.1152/ajpcell.00329.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Szabo C. Hydrogen sulfide, an enhancer of vascular nitric oxide signaling: mechanisms and implications. Am. J. Physiol. Cell Physiol. 2017;312 doi: 10.1152/ajpcell.00282.2016. C3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Majumder A., Singh M., George A.K., Behera J., Tyagi N., Tyagi S.C. Hydrogen sulfide improves postischemic neoangiogenesis in the hind limb of cystathionine-beta-synthase mutant mice via PPAR-gamma/VEGF axis. Phys. Rep. 2018;6 doi: 10.14814/phy2.13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Longchamp A., Mirabella T., Arduini A., MacArthur M.R., Das A., Treviño-Villarreal J.H., Hine C., Ben-Sahra I., Knudsen N.H., Brace L.E., Reynolds J., Mejia P., Tao M., Sharma G., Wang R., Corpataux J.M., Haefliger J.A., Ahn K.H., Lee C.H., Manning B.D., Sinclair D.A., Chen C.S., Ozaki C.K., Mitchell J.R. Amino acid restriction triggers angiogenesis via GCN2/ATF4 regulation of VEGF and H2S production. Cell. 2018;173:117–129. doi: 10.1016/j.cell.2018.03.001. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abdollahi Govar A., Törő G., Szaniszlo P., Pavlidou A., Bibli S.I., Thanki K., Resto V.A., Chao C., Hellmich M.R., Szabo C., Papapetropoulos A., Módis K. 3-Mercaptopyruvate sulfurtransferase supports endothelial cell angiogenesis and bioenergetics. Br. J. Pharmacol. 2020;177:866–883. doi: 10.1111/bph.14574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhu M.L., Zhao F.R., Zhu T.T., Wang Q.Q., Wu Z.Q., Song P., Xu J., Wan G.R., Yin Y.L., Li P. The antihypertension effect of hydrogen sulfide (H2S) is induced by activating VEGFR2 signaling pathway. Life Sci. 2021;267 doi: 10.1016/j.lfs.2020.118831. [DOI] [PubMed] [Google Scholar]
- 140.Szabo C. Gasotransmitters in cancer: from pathophysiology to experimental therapy. Nat. Rev. Drug Discov. 2016;15:185–203. doi: 10.1038/nrd.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cao X., Ding L., Xie Z.Z., Yang Y., Whiteman M., Moore P.K., Bian J.S. A review of hydrogen sulfide synthesis, metabolism, and measurement: is modulation of hydrogen sulfide a novel therapeutic for cancer? Antioxidants Redox Signal. 2019;31:1–38. doi: 10.1089/ars.2017.7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cheng W.L., Feng P.H., Lee K.Y., Chen K.Y., Sun W.L., Van Hiep N., Luo C.S., Wu S.M. The role of EREG/EGFR pathway in tumor progression. Int. J. Mol. Sci. 2021;22:12828. doi: 10.3390/ijms222312828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Comas F., Latorre J., Ortega F., Oliveras-Cañellas N., Lluch A., Ricart W., Fernández-Real J.M., Moreno-Navarrete J.M. Permanent cystathionine-β-synthase gene knockdown promotes inflammation and oxidative stress in immortalized human adipose-derived mesenchymal stem cells, enhancing their adipogenic capacity. Redox Biol. 2021;42 doi: 10.1016/j.redox.2020.101668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dasgupta P., Sengupta S.B. Role of diallyl disulfide-mediated cleavage of c-Myc and Sp-1 in the regulation of telomerase activity in human lymphoma cell line U937. Nutrition. 2015;31:1031–1037. doi: 10.1016/j.nut.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 145.Meng G., Xiao Y., Ma Y., Tang X., Xie L., Liu J., Gu Y., Yu Y., Park C.M., Xian M., Wang X., Ferro A., Wang R., Moore P.K., Zhang Z., Wang H., Han Y., Ji Y. Hydrogen sulfide regulates Krüppel-like factor 5 transcription activity via specificity protein 1 S-sulfhydration at Cys664 to prevent myocardial hypertrophy. J. Am. Heart Assoc. 2016;5 doi: 10.1161/JAHA.116.004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yamamoto T., Takano N., Ishiwata K., Ohmura M., Nagahata Y., Matsuura T., Kamata A., Sakamoto K., Nakanishi T., Kubo A., Hishiki T., Suematsu M. Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat. Commun. 2014;5:3480. doi: 10.1038/ncomms4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ostrakhovitch E.A., Akakura S., Tabibzadeh S. Hydrogen sulfide facilitates reprogramming and trans-differentiation in 3D dermal fibroblast. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Asimakopoulou A., Panopoulos P., Chasapis C.T., Coletta C., Zhou Z.M., Cirino G., Giannis A., Szabo C., Spyroulias G.A., Papapetropoulos A. Selectivity of commonly used pharmacological inhibitors for cystathionine synthase (CBS) and cystathionine lyase (CSE), Brit. J. Pharmacol. 2013;169:922–932. doi: 10.1111/bph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Thornburg J.M., Nelson K.K., Clem B.F., Lane A.N., Arumugam S., Simmons A., Eaton J.W., Telang S., Chesney J. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008;10:R84. doi: 10.1186/bcr2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mohrenz I.V., Antonietti P., Pusch S., Capper D., Balss J., Voigt S., Weissert S., Mukrowsky A., Frank J., Senft C., Seifert V., von Deimling A., Kögel D. Isocitrate dehydrogenase 1 mutant R132H sensitizes glioma cells to BCNU-induced oxidative stress and cell death. Apoptosis. 2013;18:1416–1425. doi: 10.1007/s10495-013-0877-8. [DOI] [PubMed] [Google Scholar]
- 151.Wang C., Chen H., Zhang M., Zhang J., Wei X., Ying W. Malate-aspartate shuttle inhibitor aminooxyacetic acid leads to decreased intracellular ATP levels and altered cell cycle of C6 glioma cells by inhibiting glycolysis. Cancer Lett. 2016;378:1–7. doi: 10.1016/j.canlet.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 152.Korangath P., Teo W.W., Sadik H., Han L., Mori N., Huijts C.M., Wildes F., Bharti S., Zhang Z., Santa-Maria C.A., Tsai H., Dang C.V., Stearns V., Bhujwalla Z.M., Sukumar S. Targeting glutamine metabolism in breast cancer with aminooxyacetate. Clin. Cancer Res. 2015;21:3263–3273. doi: 10.1158/1078-0432.CCR-14-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hao Y., Samuels Y., Li Q., Krokowski D., Guan B.J., Wang C., Jin Z., Dong B., Cao B., Feng X., Xiang M., Xu C., Fink S., Meropol N.J., Xu Y., Conlon R.A., Markowitz S., Kinzler K.W., Velculescu V.E., Brunengraber H., Willis J.E., LaFramboise T., Hatzoglou M., Zhang G.F., Vogelstein B., Wang Z. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat. Commun. 2016;7:11971. doi: 10.1038/ncomms11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moreno-Sánchez R., Marín-Hernández Á., Del Mazo-Monsalvo I., Saavedra E., Rodríguez-Enríquez S. Assessment of the low inhibitory specificity of oxamate, aminooxyacetate and dichloroacetate on cancer energy metabolism. Biochim. Biophys. Acta Gen. Subj. 2017;1861:3221–3236. doi: 10.1016/j.bbagen.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 155.Chao C., Zatarain J.R., Ding Y., Coletta C., Mrazek A.A., Druzhyna N., Johnson P., Chen H., Hellmich J.L., Asimakopoulou A., Yanagi K., Olah G., Szoleczky P., Törö G., Bohanon F.J., Cheema M., Lewis R., Eckelbarger D., Ahmad A., Módis K., Untereiner A., Szczesny B., Papapetropoulos A., Zhou J., Hellmich M.R., Szabo C. Cystathionine-beta-synthase inhibition for colon cancer: enhancement of the efficacy of aminooxyacetic acid via the prodrug approach. Mol. Med. 2016;22:361–379. doi: 10.2119/molmed.2016.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hellmich M.R., Coletta C., Chao C., Szabo C. The therapeutic potential of cystathionine β-synthetase/hydrogen sulfide inhibition in cancer. Antioxidants Redox Signal. 2015;22:424–448. doi: 10.1089/ars.2014.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hunter A.J., Blekkenhorst G.H. The effects of 2-deoxyglucose and amino-oxyacetic acid on the radiation response of mammalian cells in vitro. Int. J. Radiat. Biol. 1998;73:311–324. doi: 10.1080/095530098142419. [DOI] [PubMed] [Google Scholar]
- 158.Sun L., Yin Y., Clark L.H., Sun W., Sullivan S.A., Tran A.Q., Han J., Zhang L., Guo H., Madugu E., Pan T., Jackson A.L., Kilgore J., Jones H.M., Gilliam T.P., Zhou C., Bae-Jump V.L. Dual inhibition of glycolysis and glutaminolysis as a therapeutic strategy in the treatment of ovarian cancer. Oncotarget. 2017;8:63551–63561. doi: 10.18632/oncotarget.18854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Muthu M., Kumar R., Syed Khaja A.S., Gilthorpe J.D., Persson J.L., Nordström A. GLUL ablation can confer drug resistance to cancer cells via a malate-aspartate shuttle-mediated mechanism. Cancers (Basel) 2019;11:1945. doi: 10.3390/cancers11121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kariagina A., Lunt S.Y., McCormick J.J. Genomic and metabolomic analysis of step-wise malignant transformation in human skin fibroblasts. Carcinogenesis. 2020;41:656–665. doi: 10.1093/carcin/bgz126. [DOI] [PubMed] [Google Scholar]
- 161.Xiang J., Chen C., Liu R., Gou D., Chang L., Deng H., Gao Q., Zhang W., Tuo L., Pan X., Liang L., Xia J., Huang L., Yao K., Wang B., Hu Z., Huang A., Wang K., Tang N. Gluconeogenic enzyme PCK1 deficiency promotes CHK2 O-GlcNAcylation and hepatocellular carcinoma growth upon glucose deprivation. J. Clin. Invest. 2021;131 doi: 10.1172/JCI144703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cudkowitz G. Effect of hydroxylamine and sodium azide on the growth of various transplantable tumors. Tumori. 1955;41:181–185. doi: 10.1177/030089165504100204. [DOI] [PubMed] [Google Scholar]
- 163.Tarnowski G.S., Kreis W., Schmid F.A., Cappuccino J.G., Burchenal J.H. Effect of hydroxylamine (NSC-26250) and related compounds on growth of transplanted animal tumors. Cancer Res. 1966;26:1279–1301. [PubMed] [Google Scholar]
- 164.Vítek J.A. Hydroxylamine-caused mitotic inhibition of a human neoplastic cell line. Folia Biol. (Praha) 1974;20:286–288. [PubMed] [Google Scholar]
- 165.Stenbäck F., Weisburger J., Williams G.M. Hydroxylamine effects on cryptogenic neoplasm development in C3H mice. Cancer Lett. 1987;38:73–85. doi: 10.1016/0304-3835(87)90202-3. [DOI] [PubMed] [Google Scholar]
- 166.Thorson M.K., Majtan T., Kraus J.P., Barrios A.M. Identification of cystathionine β-synthase inhibitors using a hydrogen sulfide selective probe. Angew Chem. Int. Ed. Engl. 2013;52:4641–4644. doi: 10.1002/anie.201300841. [DOI] [PubMed] [Google Scholar]
- 167.Druzhyna N., Szczesny B., Olah G., Módis K., Asimakopoulou A., Pavlidou A., Szoleczky P., Gerö D., Yanagi K., Törö G., López-García I., Myrianthopoulos V., Mikros E., Zatarain J.R., Chao C., Papapetropoulos A., Hellmich M.R., Szabo C. Screening of a composite library of clinically used drugs and well-characterized pharmacological compounds for cystathionine beta-synthase inhibition identifies benserazide as a drug potentially suitable for repurposing for the experimental therapy of colon cancer. Pharmacol. Res. 2016;113:18–37. doi: 10.1016/j.phrs.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li W., Zheng M., Wu S., Gao S., Yang M., Li Z., Min Q., Sun W., Chen L., Xiang G., Li H. Benserazide, a dopadecarboxylase inhibitor, suppresses tumor growth by targeting hexokinase 2. J. Exp. Clin. Cancer Res. 2017;36:58. doi: 10.1186/s13046-017-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou Y., Huang Z., Su J., Li J., Zhao S., Wu L., Zhang J., He Y., Zhang G., Tao J., Zhou J., Chen X., Peng C. Benserazide is a novel inhibitor targeting PKM2 for melanoma treatment. Int. J. Cancer. 2020;147:139–151. doi: 10.1002/ijc.32756. [DOI] [PubMed] [Google Scholar]
- 170.Muller T., Kuhn W. Homocysteine levels after acute levodopa intake in patients with Parkinson's disease. Mov. Disord. 2009;24:1339–1343. doi: 10.1002/mds.22607. [DOI] [PubMed] [Google Scholar]
- 171.Alirezaei M. Betaine protects cerebellum from oxidative stress following levodopa and benserazide administration in rats. Iran J. Basic Med. Sci. 2015;18:950–957. [PMC free article] [PubMed] [Google Scholar]
- 172.Guo G., Xu S., Cao L.D., Wu Q.Y. The effect of levodopa benserazide hydrochloride on homocysteinemia levels in patients with Parkinson's disease and treatment of hyperhomocysteinemia. Eur. Rev. Med. Pharmacol. Sci. 2016;20:2409–2412. [PubMed] [Google Scholar]
- 173.Marechal D., Brault V., Leon A., Martin D., Lopes Pereira P., Loaëc N., Birling M.C., Friocourt G., Blondel M., Herault Y. CBS overdosage is necessary and sufficient to induce cognitive phenotypes in mouse models of Down syndrome and interacts genetically with Dyrk1a. Hum. Mol. Genet. 2019;28:1561–1577. doi: 10.1093/hmg/ddy447. [DOI] [PubMed] [Google Scholar]
- 174.Matsuo Y., Greenberg D.M. A crystalline enzyme that cleaves homoserine and cystathionine. II. Prosthetic group. J. Biol. Chem. 1958;230:561–571. [PubMed] [Google Scholar]
- 175.Bar-Or D., Rael L., Thomas G., Kraus J. Inhibitory effect of copper on cystathionine - synthase activity: protective effect of an analog of the human albumin N-terminus. Protein Pept. Lett. 2005;12:271–273. doi: 10.2174/0929866053587048. [DOI] [PubMed] [Google Scholar]
- 176.Li H., Wang J., Wu C., Wang L., Chen Z.S., Cui W. The combination of disulfiram and copper for cancer treatment. Drug Discov. Today. 2020;25:1099–1108. doi: 10.1016/j.drudis.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 177.Lu C., Li X., Ren Y., Zhang X. Disulfiram: a novel repurposed drug for cancer therapy, Cancer Chemother. Pharmacol. 2021;87:159–172. doi: 10.1007/s00280-020-04216-8. [DOI] [PubMed] [Google Scholar]
- 178.Kannappan V., Ali M., Small B., Rajendran G., Elzhenni S., Taj H., Wang W., Dou Q.P. Recent advances in repurposing disulfiram and disulfiram derivatives as copper-dependent anticancer agents. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.741316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lu Y., Pan Q., Gao W., Pu Y., Luo K., He B., Gu Z. Leveraging disulfiram to treat cancer: mechanisms of action, delivery strategies, and treatment regimens. Biomaterials. 2022;281 doi: 10.1016/j.biomaterials.2021.121335. [DOI] [PubMed] [Google Scholar]
- 180.Zhou Y., Yu J., Lei X., Wu J., Niu Q., Zhang Y., Liu H., Christen P., Gehring H., Wu F. High-throughput tandem-microwell assay identifies inhibitors of the hydrogen sulfide signaling pathway. Chem. Commun. (Camb). 2013;49:11782–11784. doi: 10.1039/c3cc46719h. [DOI] [PubMed] [Google Scholar]
- 181.D. Charre, H. Blehaut, F. Bellamy, Inhibitors of Cystathionine Beta Synthase to Reduce the Neurotoxic Overproduction of Endogenous Hydrogen Sulfide, WO/2013/068592, Patent publication date: 16.05.2013.
- 182.Stern K.G. Oxidation-reduction potentials of toxoflavin. Biochem. J. 1935;29:500–508. doi: 10.1042/bj0290500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Latuasan H.E., Berends W. On the origin of the toxicity of toxoflavin. Biochim. Biophys. Acta. 1961;52:502–508. doi: 10.1016/0006-3002(61)90408-5. [DOI] [PubMed] [Google Scholar]
- 184.Akimenko V.K., Golovchenko N.P., Medentsev A.G., Esipov S.E. Effect of xanthothricin on the respiratory chain. FEBS Lett. 1974;46:23–28. doi: 10.1016/0014-5793(74)80326-1. [DOI] [PubMed] [Google Scholar]
- 185.Choi G., Lee J., Ji J.Y., Woo J., Kang N.S., Cho S.Y., Kim H.R., Ha J.D., Han S.Y. Discovery of a potent small molecule SIRT1/2 inhibitor with anticancer effects. Int. J. Oncol. 2013;43:1205–1211. doi: 10.3892/ijo.2013.2035. [DOI] [PubMed] [Google Scholar]
- 186.Antony L., van der Schoor F., Dalrymple S.L., Isaacs J.T. Androgen receptor (AR) suppresses normal human prostate epithelial cell proliferation via AR/β-catenin/TCF-4 complex inhibition of c-MYC transcription. Prostate. 2014;74:1118–1131. doi: 10.1002/pros.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kyani A., Tamura S., Yang S., Shergalis A., Samanta S., Kuang Y., Ljungman M., Neamati N. Discovery and mechanistic elucidation of a class of protein disulfide isomerase inhibitors for the treatment of glioblastoma. ChemMedChem. 2018;13:164–177. doi: 10.1002/cmdc.201700629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Letfus V., Jelić D., Bokulić A., Petrinić Grba A., Koštrun S. Rational design, synthesis and biological profiling of new KDM4C inhibitors. Bioorg. Med. Chem. 2020;28 doi: 10.1016/j.bmc.2019.115128. [DOI] [PubMed] [Google Scholar]
- 189.Wang S., Liu D., Zhang X., Li S., Sun Y., Li J., Zhou Y., Zhang L. Study on glycosylated prodrugs of toxoflavins for antibody-directed enzyme tumor therapy. Carbohydr. Res. 2007;342:1254–1260. doi: 10.1016/j.carres.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 190.Thorson M.K., Van Wagoner R.M., Harper M.K., Ireland C.M., Majtan T., Kraus J.P., Barrios A.M. Marine natural products as inhibitors of cystathionine beta-synthase activity. Bioorg. Med. Chem. Lett. 2015;25:1064–1066. doi: 10.1016/j.bmcl.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 191.Niu W., Wu P., Chen F., Wang J., Shang X., Xu C. Discovery of selective cystathionine β-synthase inhibitors by high-throughput screening with a fluorescent thiol probe. MedChemComm. 2016;8:198–201. doi: 10.1039/c6md00493h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nunome S., Ishiyama A., Kobayashi M., Otoguro K., Kiyohara H., Yamada H., Omura S. In vitro antimalarial activity of biflavonoids from Wikstroemia indica. Planta Med. 2004;70:76–78. doi: 10.1055/s-2004-815462. [DOI] [PubMed] [Google Scholar]
- 193.Wang L.Y., Unehara T., Kitanaka S. Anti-inflammatory activity of new guaiane type sesquiterpene from Wikstroemia indica. Chem. Pharm. Bull. 2005;53:137–139. doi: 10.1248/cpb.53.137. [DOI] [PubMed] [Google Scholar]
- 194.Aitken S.M., Kirsch J.F. Kinetics of the yeast cystathionine β-synthase forward and reverse reactions: continuous assays and the equilibrium constant for the reaction. Biochemistry. 2003;42:571–578. doi: 10.1021/bi026681n. [DOI] [PubMed] [Google Scholar]
- 195.Dilek N., Papapetropoulos A., Toliver-Kinsky T., Szabo C. Hydrogen sulfide: an endogenous regulator of the immune system. Pharmacol. Res. 2020;161 doi: 10.1016/j.phrs.2020.105119. [DOI] [PubMed] [Google Scholar]
- 196.Carbonero F., Benefiel A.C., Alizadeh-Ghamsari A.H., Gaskins H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012;3:448. doi: 10.3389/fphys.2012.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Blachier F., Beaumont M., Kim E. Cysteine-derived hydrogen sulfide and gut health: a matter of endogenous or bacterial origin. Curr. Opin. Clin. Nutr. Metab. Care. 2019;22:68–75. doi: 10.1097/MCO.0000000000000526. [DOI] [PubMed] [Google Scholar]
- 198.Blachier F., Andriamihaja M., Larraufie P., Ahn E., Lan A., Kim E. Production of hydrogen sulfide by the intestinal microbiota and epithelial cells and consequences for the colonic and rectal mucosa. Am. J. Physiol. Gastrointest. Liver Physiol. 2021;320:G125–135. doi: 10.1152/ajpgi.00261.2020. [DOI] [PubMed] [Google Scholar]
- 199.Gui D.D., Luo W., Yan B.J., Ren Z., Tang Z.H., Liu L.S., Zhang J.F., Jiang Z.S. Effects of gut microbiota on atherosclerosis through hydrogen sulfide. Eur. J. Pharmacol. 2021;896 doi: 10.1016/j.ejphar.2021.173916. [DOI] [PubMed] [Google Scholar]
- 200.Shen X., Carlström M., Borniquel S., Jädert C., Kevil C.G., Lundberg J.O. Microbial regulation of host hydrogen sulfide bioavailability and metabolism. Free Radic. Biol. Med. 2013;60:195–200. doi: 10.1016/j.freeradbiomed.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fang Y., Yan C., Zhao Q., Xu J., Liu Z., Gao J., Zhu H., Dai Z., Wang D., Tang D. The roles of microbial products in the development of colorectal cancer: a review. Bioengineered. 2021;12:720–735. doi: 10.1080/21655979.2021.1889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Buret A.G., Allain T., Motta J.P., Wallace J.L. Effects of hydrogen sulfide on the microbiome: from toxicity to therapy. Antioxidants Redox Signal. 2022;36:211–219. doi: 10.1089/ars.2021.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Stevens V.L., McCullough M.L., Pavluck A.L., Talbot J.T., Feigelson H.S., Thun M.J., Calle E.E. Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol. Biomarkers Prev. 2007;16:1140–1147. doi: 10.1158/1055-9965.EPI-06-1037. [DOI] [PubMed] [Google Scholar]
- 204.Le Marchand L., Donlon T., Hankin J.H., Kolonel L.N., Wilkens L.R., Seifried A. B-vitamin intake, metabolic genes, and colorectal cancer risk (United States) Cancer Causes Control. 2002;13:239–248. doi: 10.1023/a:1015057614870. [DOI] [PubMed] [Google Scholar]
- 205.Sharp L., Little J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: a HuGE review. Am. J. Epidemiol. 2004;159:423–443. doi: 10.1093/aje/kwh066. [DOI] [PubMed] [Google Scholar]
- 206.Pozzi G., Gobbi G., Masselli E., Carubbi C., Presta V., Ambrosini L., Vitale M., Mirandola P. Buffering adaptive immunity by hydrogen sulfide. Cells. 2022;11:325. doi: 10.3390/cells11030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Miller T.W., Wang E.A., Gould S., Stein E.V., Kaur S., Lim L., Amarnath S., Fowler D.H., Roberts D.D. Hydrogen sulfide is an endogenous potentiator of T cell activation. J. Biol. Chem. 2012;287:4211–4221. doi: 10.1074/jbc.M111.307819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Miller T.W., Kaur S., Ivins-O'Keefe K., Roberts D.D. Thrombospondin-1 is a CD47-dependent endogenous inhibitor of hydrogen sulfide signaling in T cell activation. Matrix Biol. 2013;32:316–324. doi: 10.1016/j.matbio.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kaur S., Schwartz A.L., Miller T.W., Roberts D.D. CD47-dependent regulation of H2S biosynthesis and signaling in T cells. Methods Enzymol. 2015;555:145–168. doi: 10.1016/bs.mie.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lund R., Aittokallio T., Nevalainen O., Lahesmaa R. Identification of novel genes regulated by IL-12, IL-4, or TGF-beta during the early polarization of CD4+ lymphocytes. J. Immunol. 2003;171:5328–5336. doi: 10.4049/jimmunol.171.10.5328. [DOI] [PubMed] [Google Scholar]
- 211.Yang R., Qu C., Zhou Y., Konkel J.E., Shi S., Liu Y., Chen C., Liu S., Liu D., Chen Y., Zandi E., Chen W., Zhou Y., Shi S. Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity. 2015;43:251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Oh S.A., Li M.O. TETs link hydrogen sulfide to immune tolerance. Immunity. 2015;43:211–213. doi: 10.1016/j.immuni.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 213.Lancien M., Gueno L., Salle S., Merieau E., Beriou G., Nguyen T.H., Abidi A., Dilek N., Solomon P., Poschmann J., Michielin O., Vuillefroy de Silly R., Vanhove B., Louvet C. Cystathionine-gamma-lyase overexpression in T cells enhances antitumor effect independently of cysteine autonomy. Cancer Sci. 2021;112:1723–1734. doi: 10.1111/cas.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fan K., Li N., Qi J., Yin P., Zhao C., Wang L., Li Z., Zha X. Wnt/beta-catenin signaling induces the transcription of cystathionine-gamma-lyase, a stimulator of tumor in colon cancer. Cell. Signal. 2014;26:2801–2808. doi: 10.1016/j.cellsig.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 215.Leikam C., Hufnagel A., Walz S., Kneitz S., Fekete A., Müller M.J., Eilers M., Schartl M., Meierjohann S. Cystathionase mediates senescence evasion in melanocytes and melanoma cells. Oncogene. 2014;33:771–782. doi: 10.1038/onc.2012.641. [DOI] [PubMed] [Google Scholar]
- 216.Panza E., De Cicco P., Armogida C., Scognamiglio G., Gigantino V., Botti G., Germano D., Napolitano M., Papapetropoulos A., Bucci M., Cirino G., Ianaro A. Role of the cystathionine gamma lyase/hydrogen sulfide pathway in human melanoma progression. Pigment Cell Melanoma Res. 2015;28:61–72. doi: 10.1111/pcmr.12312. [DOI] [PubMed] [Google Scholar]
- 217.Xu Y., Ma N., Wei P., Zeng Z., Meng J. Expression of hydrogen sulfide synthases and Hh signaling pathway components correlate with the clinicopathological characteristics of papillary thyroid cancer patients. Int. J. Clin. Exp. Pathol. 2018;11:1818–1824. [PMC free article] [PubMed] [Google Scholar]
- 218.Ozluk E., Patel S., Coppola D., Ghali G., Cotelingam J.D., Kevil C.G., Shackelford R.E. Cystathionine gamma-lyase is increased in testicular seminomas, embryonal, and yolk sac tumors. Anticancer Res. 2021;41:4211–4214. doi: 10.21873/anticanres.15225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang Y.H., Huang J.T., Chen W.L., Wang R.H., Kao M.C., Pan Y.R., Chan S.H., Tsai K.W., Kung H.J., Lin K.T., Wang L.H. Dysregulation of cystathionine γ-lyase promotes prostate cancer progression and metastasis. EMBO Rep. 2019;20 doi: 10.15252/embr.201845986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.You J., Shi X., Liang H., Ye J., Wang L., Han H., Fang H., Kang W., Wang T. Cystathionine- gamma-lyase promotes process of breast cancer in association with STAT3 signaling pathway. Oncotarget. 2017;8:65677–65686. doi: 10.18632/oncotarget.20057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang L., Shi H., Zhang X., Zhang X., Liu Y., Kang W., Shi X., Wang T. I157172, a novel inhibitor of cystathionine γ-lyase, inhibits growth and migration of breast cancer cells via SIRT1-mediated deacetylation of STAT3. Oncol. Rep. 2019;41:427–436. doi: 10.3892/or.2018.6798. [DOI] [PubMed] [Google Scholar]
- 222.Wang L., Shi H., Liu Y., Zhang W., Duan X., Li M., Shi X., Wang T. Cystathionine-γ-lyase promotes the metastasis of breast cancer via the VEGF signaling pathway. Int. J. Oncol. 2019;55:473–487. doi: 10.3892/ijo.2019.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Tan Z., Ge C., Feng D., Xu C., Cao B., Xie Y., Zhou H., Wang G., Aa J. The interleukin-6/signal transducer and activator of transcription-3/cystathionine gamma-lyase axis deciphers the transformation between the sensitive and resistant phenotypes of breast cancer cells. Drug Metab. Dispos. 2021;49:985–994. doi: 10.1124/dmd.121.000571. [DOI] [PubMed] [Google Scholar]
- 224.Yamaga T., Suehiro J., Wada Y., Sakurai H. Induction of CTH expression in response to amino acid starvation confers resistance to anti-LAT1 therapy in MDA-MB-231 cells. Sci. Rep. 2022;12:1021. doi: 10.1038/s41598-022-04987-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Liu Y., Wang L., Zhang X., Deng Y., Pan L., Li H., Shi X., Wang T. A novel cystathionine gamma-lyase inhibitor, I194496, inhibits the growth and metastasis of human TNBC via downregulating multiple signaling pathways. Sci. Rep. 2021;11:8963. doi: 10.1038/s41598-021-88355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang H.F., Hughes C.S., Li W., He J.Z., Surdez D., El-Naggar A.M., Cheng H., Prudova A., Delaidelli A., Negri G.L., Li X., Ørum-Madsen M.S., Lizardo M.M., Oo H.Z., Colborne S., Shyp T., Scopim-Ribeiro R., Hammond C.A., Dhez A.C., Langman S., Lim J.K.M., Kung S.H.Y., Li A., Steino A., Daugaard M., Parker S.J., Geltink R.I.K., Orentas R.J., Xu L.Y., Morin G.B., Delattre O., Dimitrov D.S., Sorensen P.H. Proteomic screens for suppressors of anoikis identify IL1RAP as a promising surface target in Ewing sarcoma. Cancer Discov. 2021;11:2884–2903. doi: 10.1158/2159-8290.CD-20-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cano-Galiano A., Oudin A., Fack F., Allega M.F., Sumpton D., Martinez-Garcia E., Dittmar G., Hau A.C., De Falco A., Herold-Mende C., Bjerkvig R., Meiser J., Tardito S., Niclou S.P. Cystathionine-γ-lyase drives antioxidant defense in cysteine-restricted IDH1-mutant astrocytomas. Neurooncol. Adv. 2021;3:vdab057. doi: 10.1093/noajnl/vdab057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pan Y., Zhou C., Yuan D., Zhang J., Shao C. Radiation exposure promotes hepatocarcinoma cell invasion through epithelial mesenchymal transition mediated by H2S/CSE pathway. Radiat. Res. 2016;185:96–105. doi: 10.1667/RR14177.1. [DOI] [PubMed] [Google Scholar]
- 229.Nagahara N. Regulation of mercaptopyruvate sulfurtransferase activity via intrasubunit and intersubunit redox-sensing switches. Antioxidants Redox Signal. 2013;19:1792–1802. doi: 10.1089/ars.2012.5031. [DOI] [PubMed] [Google Scholar]
- 230.Nagahara N. Multiple role of 3-mercaptopyruvate sulfurtransferase: antioxidative function, H2S and polysulfide production and possible SO(x) production. Br. J. Pharmacol. 2018;175:577–589. doi: 10.1111/bph.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pedre B., Dick T.P. 3-mercaptopyruvate sulfurtransferase: an enzyme at the crossroads of sulfane sulfur trafficking. Biol. Chem. 2020;402:223–237. doi: 10.1515/hsz-2020-0249. [DOI] [PubMed] [Google Scholar]
- 232.Kimura H. Signalling by hydrogen sulfide and polysulfides via protein S-sulfuration. Br. J. Pharmacol. 2020;177:720–733. doi: 10.1111/bph.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Augsburger F., Szabo C. Potential role of the 3-mercaptopyruvate sulfurtransferase (3-MST)-hydrogen sulfide (H2S) pathway in cancer cells. Pharmacol. Res. 2020;154 doi: 10.1016/j.phrs.2018.11.034. [DOI] [PubMed] [Google Scholar]
- 234.Kaczor-Kamińska M., Kaminski K., Wróbel M. The expression and activity of rhodanese, 3-mercaptopyruvate sulfurtransferase, cystathionine γ-lyase in the most frequently chosen cellular research models. Biomolecules. 2021;11:1859. doi: 10.3390/biom11121859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wrobel M., Czubak J., Bronowicka-Adamska P., Jurkowska H., Adamek D., Papla B. Is development of high-grade gliomas sulfur-dependent? Molecules. 2014;19:21350–21362. doi: 10.3390/molecules191221350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shiota M., Naya M., Yamamoto T., Hishiki T., Tani T., Takahashi H., Kubo A., Koike D., Itoh M., Ohmura M., Kabe Y., Sugiura Y., Hiraoka N., Morikawa T., Takubo K., Suina K., Nagashima H., Sampetrean O., Nagano O., Saya H., Yamazoe S., Watanabe H., Suematsu M. Gold-nanofève surface-enhanced Raman spectroscopy visualizes hypotaurine as a robust anti-oxidant consumed in cancer survival. Nat. Commun. 2018;9:1561. doi: 10.1038/s41467-018-03899-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Dongsoo K., Chen J., Wei E., Ansari J., Meram A., Patel S., Ghali G., Kevil C., Shackelford R.E. Hydrogen sulfide and hydrogen sulfide-synthesizing enzymes are altered in a case of oral adenoid cystic carcinoma. Case Rep. Oncol. 2018;11:585–590. doi: 10.1159/000492464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Li M., Song X., Jin Q., Chen Y., Zhang J., Gao J., Cen L., Lin Y., Xu C., He X., Li Y., Yu C. 3-mercaptopyruvate sulfurtransferase represses tumor progression and predicts prognosis in hepatocellular carcinoma. Liver Int. 2022;42:1173–1184. doi: 10.1111/liv.15228. [DOI] [PubMed] [Google Scholar]
- 239.Jurkowska H., Placha W., Nagahara N., Wróbel M. The expression and activity of cystathionine-gamma-lyase and 3-mercaptopyruvate sulfurtransferase in human neoplastic cell lines. Amino Acids. 2011;41:151–158. doi: 10.1007/s00726-010-0606-3. [DOI] [PubMed] [Google Scholar]
- 240.Zuhra K., Tomé C.S., Masi L., Giardina G., Paulini G., Malagrinò F., Forte E., Vicente J.B., Giuffrè A. N-acetylcysteine serves as substrate of 3-mercaptopyruvate sulfurtransferase and stimulates sulfide metabolism in colon cancer cells. Cells. 2019;8:828. doi: 10.3390/cells8080828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Augsburger F., Randi E.B., Jendly M., Ascencao K., Dilek N., Szabo C. Role of 3-mercaptopyruvate sulfurtransferase in the regulation of proliferation, migration, and bioenergetics in murine colon cancer cells. Biomolecules. 2020;10:447. doi: 10.3390/biom10030447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bantzi M., Augsburger F., Loup J., Berset Y., Vasilakaki S., Myrianthopoulos V., Mikros E., Szabo C., Bochet C.G. Novel aryl-substituted pyrimidones as inhibitors of 3-mercaptopyruvate sulfurtransferase with antiproliferative efficacy in colon cancer. J. Med. Chem. 2021;64:6221–6240. doi: 10.1021/acs.jmedchem.1c00260. [DOI] [PubMed] [Google Scholar]
- 243.Módis K., Coletta C., Erdélyi K., Papapetropoulos A., Szabo C. Intramitochondrial hydrogen sulfide production by 3-mercaptopyruvate sulfurtransferase maintains mitochondrial electron flow and supports cellular bioenergetics. Faseb. J. 2013;27:601–611. doi: 10.1096/fj.12-216507. [DOI] [PubMed] [Google Scholar]
- 244.Módis K., Asimakopoulou A., Coletta C., Papapetropoulos A., Szabo C. Oxidative stress suppresses the cellular bioenergetic effect of the 3-mercaptopyruvate sulfurtransferase/hydrogen sulfide pathway. Biochem. Biophys. Res. Commun. 2013;433:401–407. doi: 10.1016/j.bbrc.2013.02.131. [DOI] [PubMed] [Google Scholar]
- 245.Tao B., Wang R., Sun C., Zhu Y. 3-mercaptopyruvate sulfurtransferase, not cystathionine beta-synthase nor cystathionine gamma-lyase, mediates hypoxia-induced migration of vascular endothelial cells. Front. Pharmacol. 2017;8:657. doi: 10.3389/fphar.2017.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kimura H. Signaling molecules: hydrogen sulfide and polysulfide. Antioxidants Redox Signal. 2015;22:362–376. doi: 10.1089/ars.2014.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hipólito A., Nunes S.C., Vicente J.B., Serpa J. Cysteine aminotransferase (CAT): a pivotal sponsor in metabolic remodeling and an ally of 3-mercaptopyruvate sulfurtransferase (MST) in cancer. Molecules. 2020;25:3984. doi: 10.3390/molecules25173984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Farber E., Rubin H. Cellular adaptation in the origin and development of cancer. Cancer Res. 1991;51:2751–2761. [PubMed] [Google Scholar]
- 249.Poljsak B., Kovac V., Dahmane R., Levec T., Starc A. Cancer etiology: a metabolic disease originating from life's major evolutionary transition? Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/7831952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Doka E., Pader I., Biro A., Johansson K., Cheng Q., Ballago K., Prigge J.R., Pastor-Flores D., Dick T.P., Schmidt E.E., Arner E.S., Nagy P. A novel persulfide detection method reveals protein persulfide and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Gao X.H., Krokowski D., Guan B.J., Bederman I., Majumder M., Parisien M., Diatchenko L., Kabil O., Willard B., Banerjee R., Wang B., Bebek G., Evans C.R., Fox P.L., Gerson S.L., Hoppel C.L., Liu M., Arvan P., Hatzoglou M. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife. 2015;4 doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Filipovic M.R., Zivanovic J., Alvarez B., Banerjee R. Chemical biology of H2S signaling through persulfidation. Chem. Rev. 2018;118:1253–1337. doi: 10.1021/acs.chemrev.7b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gao X.H., Li L., Parisien M., Wu J., Bederman I., Gao Z., Krokowski D., Chirieleison S.M., Abbott D., Wang B., Arvan P., Cameron M., Chance M., Willard B., Hatzoglou M. Discovery of a redox thiol switch: implications for cellular energy metabolism. Mol. Cell. Proteomics. 2020;19:852–870. doi: 10.1074/mcp.RA119.001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zivanovic J., Kouroussis E., Kohl J.B., Adhikari B., Bursac B., Schott-Roux S., Petrovic D., Miljkovic J.L., Thomas-Lopez D., Jung Y., Miler M., Mitchell S., Milosevic V., Gomes J.E., Benhar M., Gonzalez-Zorn B., Ivanovic-Burmazovic I., Torregrossa R., Mitchell J.R., Whiteman M., Schwarz G., Snyder S.H., Paul B.D., Carroll K.S., Filipovic M.R. Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-sulfhydration. Cell Metabol. 2020;31:207. doi: 10.1016/j.cmet.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Bibli S.I., Hu J., Looso M., Weigert A., Ratiu C., Wittig J., Drekolia M.K., Tombor L., Randriamboavonjy V., Leisegang M.S., Goymann P., Delgado Lagos F., Fisslthaler B., Zukunft S., Kyselova A., Justo A.F.O., Heidler J., Tsilimigras D., Brandes R.P., Dimmeler S., Papapetropoulos A., Knapp S., Offermanns S., Wittig I., Nishimura S.L., Sigala F., Fleming I. Mapping the endothelial cell S-sulfhydrome highlights the crucial role of integrin sulfhydration in vascular function. Circulation. 2021;143:935–948. doi: 10.1161/CIRCULATIONAHA.120.051877. [DOI] [PubMed] [Google Scholar]
- 256.Vander Heiden M.G. Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 257.Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 258.Cantor J.R., Sabatini D.M. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Pavlova N.N., Zhu J., Thompson C.B. The hallmarks of cancer metabolism: still emerging. Cell Metabol. 2022;34:355–377. doi: 10.1016/j.cmet.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lagoutte E., Mimoun S., Andriamihaja M., Chaumontet C., Blachier F., Bouillaud F. Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim. Biophys. Acta. 2010;1797:1500–1511. doi: 10.1016/j.bbabio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 261.Landry A.P., Ballou D.P., Banerjee R. Hydrogen sulfide oxidation by sulfide quinone oxidoreductase. Chembiochem. 2021;22:949–960. doi: 10.1002/cbic.202000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Nicholls P., Kim J.K. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can. J. Biochem. 1982;60:613–623. doi: 10.1139/o82-076. [DOI] [PubMed] [Google Scholar]
- 263.Nicholls P., Marshall D.C., Cooper C.E., Wilson M.T. Sulfide inhibition of and metabolism by cytochrome c oxidase. Biochem. Soc. Trans. 2013;41:1312–1316. doi: 10.1042/BST20130070. [DOI] [PubMed] [Google Scholar]
- 264.Vitvitsky V., Miljkovic J.L., Bostelaar T., Adhikari B., Yadav P.K., Steiger A.K., Torregrossa R., Pluth M.D., Whiteman M., Banerjee R., Filipovic M.R. Cytochrome c reduction by H2S potentiates sulfide signaling. ACS Chem. Biol. 2018;13:2300–2307. doi: 10.1021/acschembio.8b00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Módis K., Panopoulos P., Coletta C., Papapetropoulos A., Szabo C. Hydrogen sulfide-mediated stimulation of mitochondrial electron transport involves inhibition of the mitochondrial phosphodiesterase 2A, elevation of cAMP and activation of protein kinase A. Biochem. Pharmacol. 2013;86:1311–1319. doi: 10.1016/j.bcp.2013.08.064. [DOI] [PubMed] [Google Scholar]
- 266.Liang M., Jin S., Wu D.D., Wang M.J., Zhu Y.C. Hydrogen sulfide improves glucose metabolism and prevents hypertrophy in cardiomyocytes. Nitric Oxide. 2015;46:114–122. doi: 10.1016/j.niox.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 267.Módis K., Ju Y., Ahmad A., Untereiner A.A., Altaany Z., Wu L., Szabo C., Wang R. S-sulfhydration of ATP synthase by hydrogen sulfide stimulates mitochondrial bioenergetics. Pharmacol. Res. 2016;113:116–124. doi: 10.1016/j.phrs.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Wang C., Du J., Du S., Liu Y., Li D., Zhu X., Ni X. Endogenous H2S resists mitochondria-mediated apoptosis in the adrenal glands via ATP5A1 S-sulfhydration in male mice. Mol. Cell. Endocrinol. 2018;474:65–73. doi: 10.1016/j.mce.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 269.Rao G., Murphy B., Dey A., Dwivedi S.K.D., Zhang Y., Roy R.V., Chakraborty P., Bhattacharya R., Mukherjee P. Cystathionine beta synthase regulates mitochondrial dynamics and function in endothelial cells. Faseb. J. 2020;34:9372–9392. doi: 10.1096/fj.202000173R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Qiao P., Zhao F., Liu M., Gao D., Zhang H., Yan Y. Hydrogen sulfide inhibits mitochondrial fission in neuroblastoma N2a cells through the Drp1/ERK1/2 signaling pathway. Mol. Med. Rep. 2017;16:971–977. doi: 10.3892/mmr.2017.6627. [DOI] [PubMed] [Google Scholar]
- 271.Li S., Yang G. Hydrogen sulfide maintains mitochondrial DNA replication via demethylation of TFAM. Antioxidants Redox Signal. 2015;23:630–642. doi: 10.1089/ars.2014.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Untereiner A.A., Fu M., Módis K., Wang R., Ju Y., Wu L. Stimulatory effect of CSE-generated H2S on hepatic mitochondrial biogenesis and the underlying mechanisms. Nitric Oxide. 2016;58:67–76. doi: 10.1016/j.niox.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 273.Shimizu Y., Polavarapu R., Eskla K.L., Nicholson C.K., Koczor C.A., Wang R., Lewis W., Shiva S., Lefer D.J., Calvert J.W. Hydrogen sulfide regulates cardiac mitochondrial biogenesis via the activation of AMPK. J. Mol. Cell. Cardiol. 2018;116:29–40. doi: 10.1016/j.yjmcc.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Sun F., Luo J.H., Yue T.T., Wang F.X., Yang C.L., Zhang S., Wang X.Q., Wang C.Y. The role of hydrogen sulphide signalling in macrophage activation. Immunology. 2021;162:3–10. doi: 10.1111/imm.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kimura Y., Goto Y., Kimura H. Hydrogen sulfide increases glutathione production and suppresses oxidative stress in mitochondria. Antioxidants Redox Signal. 2010;12:1–13. doi: 10.1089/ars.2008.2282. [DOI] [PubMed] [Google Scholar]
- 276.Noda M., Fujita K., Lee C.H., Yoshioka T. The principle and the potential approach to ROS-dependent cytotoxicity by non-pharmaceutical therapies: optimal use of medical gases with antioxidant properties. Curr. Pharmaceut. Des. 2011;17:2253–2263. doi: 10.2174/138161211797052600. [DOI] [PubMed] [Google Scholar]
- 277.Suzuki K., Olah G., Modis K., Coletta C., Kulp G., Gerö D., Szoleczky P., Chang T., Zhou Z., Wu L., Wang R., Papapetropoulos A., Szabo C. Hydrogen sulfide replacement therapy protects the vascular endothelium in hyperglycemia by preserving mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13829–13834. doi: 10.1073/pnas.1105121108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Coletta C., Módis K., Szczesny B., Brunyánszki A., Oláh G., Rios E.C., Yanagi K., Ahmad A., Papapetropoulos A., Szabo C. Regulation of vascular tone, angiogenesis and cellular bioenergetics by the 3-mercaptopyruvate sulfurtransferase/H2S pathway: functional impairment by hyperglycemia and restoration by DL-alpha-lipoic acid. Mol. Med. 2015;21:1–14. doi: 10.2119/molmed.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Xie Z.Z., Shi M.M., Xie L., Wu Z.Y., Li G., Hua F., Bian J.S. Sulfhydration of p66Shc at cysteine59 mediates the antioxidant effect of hydrogen sulfide. Antioxidants Redox Signal. 2014;21:2531–2542. doi: 10.1089/ars.2013.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhang H., Huang Y., Chen S., Tang C., Wang G., Du J., Jin H. Hydrogen sulfide regulates insulin secretion and insulin resistance in diabetes mellitus, a new promising target for diabetes mellitus treatment? A review. J. Adv. Res. 2021;27:19–30. doi: 10.1016/j.jare.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Mustafa A.K., Gadalla M.M., Sen N., Kim S., Mu W., Gazi S.K., Barrow R.K., Yang G., Wang R., Snyder S.H. H2S signals through protein S-sulfhydration. Sci. Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wondimu E.T., Zhang Q., Jin Z., Fu M., Torregrossa R., Whiteman M., Yang G., Wu L., Wang R. Effect of hydrogen sulfide on glycolysis-based energy production in mouse erythrocytes. J. Cell. Physiol. 2022;237:763–773. doi: 10.1002/jcp.30544. [DOI] [PubMed] [Google Scholar]
- 283.Vitvitsky V., Kumar R., Libiad M., Maebius A., Landry A.P., Banerjee R. The mitochondrial NADH pool is involved in hydrogen sulfide signaling and stimulation of aerobic glycolysis. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Parsanathan R., Jain S.K. Hydrogen sulfide increases glutathione biosynthesis, and glucose uptake and utilisation in C2C12 mouse myotubes. Free Radic. Res. 2018;52:288–303. doi: 10.1080/10715762.2018.1431626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Libiad M., Vitvitsky V., Bostelaar T., Bak D.W., Lee H.J., Sakamoto N., Fearon E., Lyssiotis C.A., Weerapana E., Banerjee R. Hydrogen sulfide perturbs mitochondrial bioenergetics and triggers metabolic reprogramming in colon cells. J. Biol. Chem. 2019;294:12077–12090. doi: 10.1074/jbc.RA119.009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sun Q., Zhong H., Yue Y., Xiong F., Chen L., Peng X., Chen P., Wan C., Yao Z., Zeng Y. Endogenous hydrogen sulfide promotes human preimplantation embryonic development by regulating metabolism related gene expression. Nitric Oxide. 2022;120:9–15. doi: 10.1016/j.niox.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 287.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 288.Rad E., Murray J.T., Tee A.R. Oncogenic signalling through mechanistic target of rapamycin (mTOR): a driver of metabolic transformation and cancer progression. Cancers (Basel) 2018;10:5. doi: 10.3390/cancers10010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zou Z., Tao T., Li H., Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020;10:31. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Hon K.W., Zainal Abidin S.A., Othman I., Naidu R. The crosstalk between signaling pathways and cancer metabolism in colorectal cancer. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.768861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Hu Y., Chen X., Pan T.T., Neo K.L., Lee S.W., Khin E.S., Moore P.K., Bian J.S. Cardioprotection induced by hydrogen sulfide preconditioning involves activation of ERK and PI3K/Akt pathways. Pflügers Archiv. 2008;455:607–616. doi: 10.1007/s00424-007-0321-4. [DOI] [PubMed] [Google Scholar]
- 293.Manna P., Jain S.K. Hydrogen sulfide and L-cysteine increase phosphatidylinositol 3,4,5-trisphosphate (PIP3) and glucose utilization by inhibiting phosphatase and tensin homolog (PTEN) protein and activating phosphoinositide 3-kinase (PI3K)/serine/threonine protein kinase (AKT)/protein kinase Czeta/lambda (PKCzeta/lambda) in 3T3l1 adipocytes. J. Biol. Chem. 2011;286:39848–39859. doi: 10.1074/jbc.M111.270884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Li H., Wang Y., Wei C., Bai S., Zhao Y., Li H., Wu B., Wang R., Wu L., Xu C. Mediation of exogenous hydrogen sulfide in recovery of ischemic post-conditioning-induced cardioprotection via down-regulating oxidative stress and up-regulating PI3K/Akt/GSK-3beta pathway in isolated aging rat hearts. Cell Biosci. 2015;5:11. doi: 10.1186/s13578-015-0003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.He X.L., Yan N., Chen X.S., Qi Y.W., Yan Y., Cai Z. Hydrogen sulfide down-regulates BACE1 and PS1 via activating PI3K/Akt pathway in the brain of APP/PS1 transgenic mouse. Pharmacol. Rep. 2016;68:975–982. doi: 10.1016/j.pharep.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 296.Xu D., Jin H., Wen J., Chen J., Chen D., Cai N., Wang Y., Wang J., Chen Y., Zhang X., Wang X. Hydrogen sulfide protects against endoplasmic reticulum stress and mitochondrial injury in nucleus pulposus cells and ameliorates intervertebral disc degeneration. Pharmacol. Res. 2017;117:357–369. doi: 10.1016/j.phrs.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 297.Ma J., Du D., Liu J., Guo L., Li Y., Chen A., Ye T. Hydrogen sulphide promotes osteoclastogenesis by inhibiting autophagy through the PI3K/AKT/mTOR pathway. J. Drug Target. 2020;28:176–185. doi: 10.1080/1061186X.2019.1624969. [DOI] [PubMed] [Google Scholar]
- 298.Koike S., Ogasawara Y., Shibuya N., Kimura H., Ishii K. Polysulfide exerts a protective effect against cytotoxicity caused by t-buthylhydroperoxide through Nrf2 signaling in neuroblastoma cells. FEBS Lett. 2013;587:3548–3555. doi: 10.1016/j.febslet.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 299.Zhang H., Gao Y., Zhao F.L., Qiao P.F., Yan Y. Hydrogen sulfide-induced processing of the amyloid precursor protein in SH-SY5Y human neuroblastoma cells involves the PI3-K/Akt signaling pathway. Cell. Mol. Neurobiol. 2015;35:265–272. doi: 10.1007/s10571-014-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wang S.S., Chen Y.H., Chen N., Wang L.J., Chen D.X., Weng H.L., Dooley S., Ding H.G. Hydrogen sulfide promotes autophagy of hepatocellular carcinoma cells through the PI3K/Akt/mTOR signaling pathway. Cell Death Dis. 2017;8:e2688. doi: 10.1038/cddis.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Cao Q., Zhang L., Yang G., Xu C., Wang R. Butyrate-stimulated H2S production in colon cancer cells, Antioxid. Redox Signal. 2010;12:1101–1109. doi: 10.1089/ars.2009.2915. [DOI] [PubMed] [Google Scholar]
- 302.Zhang J., Ye J., Yuan C., Fu Q., Zhang F., Zhu X., Wang L., Gao P., Shu G., Jiang Q., Wang S. Exogenous H2S exerts biphasic effects on porcine mammary epithelial cells proliferation through PI3K/Akt-mTOR signaling pathway. J. Cell. Physiol. 2018;233:7071–7081. doi: 10.1002/jcp.26630. [DOI] [PubMed] [Google Scholar]
- 303.Cui C., Fan J., Zeng Q., Cai J., Chen Y., Chen Z., Wang W., Li S.Y., Cui Q., Yang J., Tang C., Xu G., Cai J., Geng B. CD4+ T-cell endogenous cystathionine γ lyase-hydrogen sulfide attenuates hypertension by sulfhydrating liver kinase B1 to promote T regulatory cell differentiation and proliferation. Circulation. 2020;142:1752–1769. doi: 10.1161/CIRCULATIONAHA.119.045344. [DOI] [PubMed] [Google Scholar]
- 304.Zhou X., Cao Y., Ao G., Hu L., Liu H., Wu J., Wang X., Jin M., Zheng S., Zhen X., Alkayed N.J., Jia J., Cheng J. CaMKKβ-dependent activation of AMP-activated protein kinase is critical to suppressive effects of hydrogen sulfide on neuroinflammation. Antioxidants Redox Signal. 2014;21:1741–1758. doi: 10.1089/ars.2013.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Li L., Rose P., Moore P.K. Hydrogen sulfide and cell signaling. Annu. Rev. Pharmacol. Toxicol. 2011;51:169–187. doi: 10.1146/annurev-pharmtox-010510-100505. [DOI] [PubMed] [Google Scholar]
- 306.Giuffrè A., Tomé C.S., Fernandes D.G.F., Zuhra K., Vicente J.B. Hydrogen sulfide metabolism and signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 2020;1219:335–353. doi: 10.1007/978-3-030-34025-4_17. [DOI] [PubMed] [Google Scholar]
- 307.Shatalin K., Shatalina E., Mironov A., Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 308.Mironov A., Seregina T., Nagornykh M., Luhachack L.G., Korolkova N., Lopes L.E., Kotova V., Zavilgelsky G., Shakulov R., Shatalin K., Nudler E. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2017;114:6022–6027. doi: 10.1073/pnas.1703576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Toliver-Kinsky T., Cui W., Törö G., Lee S.J., Shatalin K., Nudler E., Szabo C. H2S, a bacterial defense mechanism against the host immune response. Infect. Immun. 2018;87 doi: 10.1128/IAI.00272-18. e00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Croppi G., Zhou Y., Yang R., Bian Y., Zhao M., Hu Y., Ruan B.H., Yu J., Wu F. Discovery of an inhibitor for bacterial 3-mercaptopyruvate sulfurtransferase that synergistically controls bacterial survival. Cell. Chem. Biol. 2020;27:1483–1499. doi: 10.1016/j.chembiol.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 311.Shatalin K., Nuthanakanti A., Kaushik A., Shishov D., Peselis A., Shamovsky I., Pani B., Lechpammer M., Vasilyev N., Shatalina E., Rebatchouk D., Mironov A., Fedichev P., Serganov A., Nudler E. Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science. 2021;372:1169–1175. doi: 10.1126/science.abd8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kolluru G.K., Shen X., Bir S.C., Kevil C.G. Hydrogen sulfide chemical biology: pathophysiological roles and detection. Nitric Oxide. 2013;35:5–20. doi: 10.1016/j.niox.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ono K., Akaike T., Sawa T., Kumagai Y., Wink D.A., Tantillo D.J., Hobbs A.J., Nagy P., Xian M., Lin J., Fukuto J.M. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic. Biol. Med. 2014;77:82–94. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Cuevasanta E., Möller M.N., Alvarez B. Biological chemistry of hydrogen sulfide and persulfides. Arch. Biochem. Biophys. 2017;617:9–25. doi: 10.1016/j.abb.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 315.Elrod J.W., Calvert J.W., Morrison J., Doeller J.E., Kraus D.W., Tao L., Jiao X., Scalia R., Kiss L., Szabo C., Kimura H., Chow C.W., Lefer D.J. Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15560–15565. doi: 10.1073/pnas.0705891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Szabó G., Veres G., Radovits T., Gero D., Módis K., Miesel-Gröschel C., Horkay F., Karck M., Szabó C. Cardioprotective effects of hydrogen sulfide. Nitric Oxide. 2011;25:201–210. doi: 10.1016/j.niox.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Dong X.B., Yang C.T., Zheng D.D., Mo L.Q., Wang X.Y., Lan A.P., Hu F., Chen P.X., Feng J.Q., Zhang M.F., Liao X.X. Inhibition of ROS-activated ERK1/2 pathway contributes to the protection of H2S against chemical hypoxia-induced injury in H9c2 cells. Mol. Cell. Biochem. 2012;362:149–157. doi: 10.1007/s11010-011-1137-2. [DOI] [PubMed] [Google Scholar]
- 318.Wen Y.D., Wang H., Kho S.H., Rinkiko S., Sheng X., Shen H.M., Zhu Y.Z. Hydrogen sulfide protects HUVECs against hydrogen peroxide induced mitochondrial dysfunction and oxidative stress. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Szczesny B., Módis K., Yanagi K., Coletta C., Le Trionnaire S., Perry A., Wood M.E., Whiteman M., Szabo C. AP39, a novel mitochondria-targeted hydrogen sulfide donor, stimulates cellular bioenergetics, exerts cytoprotective effects and protects against the loss of mitochondrial DNA integrity in oxidatively stressed endothelial cells in vitro. Nitric Oxide. 2014;41:120–130. doi: 10.1016/j.niox.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Yang M., Huang Y., Chen J., Chen Y.L., Ma J.J., Shi P.H. Activation of AMPK participates hydrogen sulfide-induced cyto-protective effect against dexamethasone in osteoblastic MC3T3-E1 cells. Biochem. Biophys. Res. Commun. 2014;454:42–47. doi: 10.1016/j.bbrc.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 321.Ahmad A., Olah G., Szczesny B., Wood M.E., Whiteman M., Szabo C. AP39, a mitochondrially targeted hydrogen sulfide donor, exerts protective effects in renal epithelial cells subjected to oxidative stress in vitro and in acute renal injury in vivo. Shock. 2016;45:88–97. doi: 10.1097/SHK.0000000000000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Yan X., Wu H., Wu Z., Hua F., Liang D., Sun H., Yang Y., Huang D., Bian J.S. The new synthetic H2S-releasing SDSS protects MC3T3-E1 osteoblasts against H2O2-induced apoptosis by suppressing oxidative stress, inhibiting MAPKs, and activating the PI3K/Akt pathway. Front. Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00007. 07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Lin J., Chen M., Liu D., Guo R., Lin K., Deng H., Zhi X., Zhang W., Feng J., Wu W. Exogenous hydrogen sulfide protects human umbilical vein endothelial cells against high glucose-induced injury by inhibiting the necroptosis pathway. Int. J. Mol. Med. 2018;41:1477–1486. doi: 10.3892/ijmm.2017.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Zhang Y.E., Huang G.Q., Wu B., Lin X.D., Yang W.Z., Ke Z.Y., Liu J. Hydrogen sulfide protects H9c2 cardiomyoblasts against H2O2-induced apoptosis. Braz. J. Med. Biol. Res. 2019;52 doi: 10.1590/1414-431X20187626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Chen J., Shen X., Pardue S., Meram A.T., Rajendran S., Ghali G.E., Kevil C.G., Shackelford R.E. The ataxia telangiectasia-mutated and Rad3-related protein kinase regulates cellular hydrogen sulfide concentrations. DNA Repair (Amst.) 2019;73:55–63. doi: 10.1016/j.dnarep.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Lohakul J., Jeayeng S., Chaiprasongsuk A., Torregrossa R., Wood M.E., Saelim M., Thangboonjit W., Whiteman M., Panich U. Mitochondria-targeted hydrogen sulfide delivery molecules protect against UVA-induced photoaging in human dermal fibroblasts, and in mouse skin in vivo. Antioxidants Redox Signal. 2022 doi: 10.1089/ars.2020.8255. In press. [DOI] [PubMed] [Google Scholar]
- 327.Lin J., Li X., Lin Y., Huang Z., Wu W. Exogenous sodium hydrosulfide protects against high glucose-induced injury and inflammation in human umbilical vein endothelial cells by inhibiting necroptosis via the p38 MAPK signaling pathway. Mol. Med. Rep. 2021;23:67. doi: 10.3892/mmr.2020.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Xie L., Gu Y., Wen M., Zhao S., Wang W., Ma Y., Meng G., Han Y., Wang Y., Liu G., Moore P.K., Wang X., Wang H., Zhang Z., Yu Y., Ferro A., Huang Z., Ji Y. Hydrogen sulfide induces Keap1 S-sulfhydration and suppresses diabetes-accelerated atherosclerosis via Nrf2 activation. Diabetes. 2016;65:3171–3184. doi: 10.2337/db16-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 2020;40 doi: 10.1128/MCB.00099-20. e00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Sen N., Paul B.D., Gadalla M.M., Mustafa A.K., Sen T., Xu R., Kim S., Snyder S.H. Hydrogen sulfide-linked sulfhydration of NF-κB mediates its antiapoptotic actions. Mol. Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Caputo F., Vegliante R., Ghibelli L. Redox modulation of the DNA damage response. Biochem. Pharmacol. 2012;84:1292–1306. doi: 10.1016/j.bcp.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 332.Saeidnia S., Abdollahi M. Antioxidants: friends or foe in prevention or treatment of cancer: the debate of the century. Toxicol. Appl. Pharmacol. 2013;271:49–63. doi: 10.1016/j.taap.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 333.Zou Z., Chang H., Li H., Wang S. Induction of reactive oxygen species: an emerging approach for cancer therapy. Apoptosis. 2017;22:1321–1335. doi: 10.1007/s10495-017-1424-9. [DOI] [PubMed] [Google Scholar]
- 334.Jiang H., Wang H., De Ridder M. Targeting antioxidant enzymes as a radiosensitizing strategy. Cancer Lett. 2018;438:154–164. doi: 10.1016/j.canlet.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 335.Du S., Huang Y., Jin H., Wang T. Protective mechanism of hydrogen sulfide against chemotherapy-induced cardiotoxicity. Front. Pharmacol. 2018;9:32. doi: 10.3389/fphar.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Tang J.Y., Ou-Yang F., Hou M.F., Huang H.W., Wang H.R., Li K.T., Fayyaz S., Shu C.W., Chang H.W. Oxidative stress-modulating drugs have preferential anticancer effects - involving the regulation of apoptosis, DNA damage, endoplasmic reticulum stress, autophagy, metabolism, and migration. Semin. Cancer Biol. 2019;58:109–117. doi: 10.1016/j.semcancer.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 337.Srinivas U.S., Tan B.W.Q., Vellayappan B.A., Jeyasekharan A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019;25 doi: 10.1016/j.redox.2018.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Wang J., Liu N., Jiang H., Li Q., Xing D. Reactive oxygen species in anticancer immunity: a double-edged sword. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.784612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Attene-Ramos M.S., Wagner E.D., Plewa M.J., Gaskins H.R. Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 340.Attene-Ramos M.S., Wagner E.D., Gaskins H.R., Plewa M.J. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 341.Attene-Ramos M.S., Nava G.M., Muellner M.G., Wagner E.D., Plewa M.J., Gaskins H.R. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen. 2010;51:304–314. doi: 10.1002/em.20546. [DOI] [PubMed] [Google Scholar]
- 342.Shackelford R., Ozluk E., Islam M.Z., Hopper B., Meram A., Ghali G., Kevil C.G. Hydrogen sulfide and DNA repair, Redox. Biol. 2021;38 doi: 10.1016/j.redox.2020.101675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Jiang X., MacArthur M.R., Treviño-Villarreal J.H., Kip P., Ozaki C.K., Mitchell S.J., Mitchell J.R. Intracellular H2S production is an autophagy-dependent adaptive response to DNA damage. Cell. Chem. Biol. 2021;28:1669–1678. doi: 10.1016/j.chembiol.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Shackelford R.E., Li Y., Ghali G.E., Kevil C.G. Bad smells and broken DNA: a tale of sulfur-nucleic acid cooperation. Antioxidants (Basel) 2021;10:1820. doi: 10.3390/antiox10111820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Vella L.J. The emerging role of exosomes in epithelial-mesenchymal-transition in cancer. Front. Oncol. 2014;4:361. doi: 10.3389/fonc.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Cao H., Xu E., Liu H., Wan L., Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: a system review. Pathol. Res. Pract. 2015;211:557–569. doi: 10.1016/j.prp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 347.Buyuk B., Jin S., Ye K. Epithelial-to-mesenchymal transition signaling pathways responsible for breast cancer metastasis. Cell. Mol. Bioeng. 2021;15:1–13. doi: 10.1007/s12195-021-00694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Jayachandran J., Srinivasan H., Mani K.P. Molecular mechanism involved in epithelial to mesenchymal transition. Arch. Biochem. Biophys. 2021;710 doi: 10.1016/j.abb.2021.108984. [DOI] [PubMed] [Google Scholar]
- 349.Grigore A.D., Jolly M.K., Jia D., Farach-Carson M.C., Levine H. Tumor budding: the name is EMT. Partial EMT. J. Clin. Med. 2016;5:51. doi: 10.3390/jcm5050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Lugli A., Zlobec I., Berger M.D., Kirsch R., Nagtegaal I.D. Tumour budding in solid cancers. Nat. Rev. Clin. Oncol. 2021;18:101–115. doi: 10.1038/s41571-020-0422-y. [DOI] [PubMed] [Google Scholar]
- 351.Sipos F., Galamb O. Epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions in the colon. World J. Gastroenterol. 2012;18:601–608. doi: 10.3748/wjg.v18.i7.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Brabletz S., Schuhwerk H., Brabletz T., Stemmler M.P. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 2021;40 doi: 10.15252/embj.2021108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Fang L.P., Lin Q., Tang C.S., Liu X.M. Hydrogen sulfide attenuates epithelial-mesenchymal transition of human alveolar epithelial cells. Pharmacol. Res. 2010;61:298–305. doi: 10.1016/j.phrs.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 354.Lv M., Li Y., Ji M.H., Zhuang M., Tang J.H. Inhibition of invasion and epithelial-mesenchymal transition of human breast cancer cells by hydrogen sulfide through decreased phospho-p38 expression. Mol. Med. Rep. 2014;10:341–346. doi: 10.3892/mmr.2014.2161. [DOI] [PubMed] [Google Scholar]
- 355.Huang J., Yang B., Xiang T., Peng W., Qiu Z., Wan J., Zhang L., Li H., Li H., Ren G. Diallyl disulfide inhibits growth and metastatic potential of human triple-negative breast cancer cells through inactivation of the β-catenin signaling pathway. Mol. Nutr. Food Res. 2015;59:1063–1075. doi: 10.1002/mnfr.201400668. [DOI] [PubMed] [Google Scholar]
- 356.Guo L., Peng W., Tao J., Lan Z., Hei H., Tian L., Pan W., Wang L., Zhang X. Hydrogen sulfide inhibits transforming growth factor-beta1-induced EMT via Wnt/catenin pathway. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Cheng S., Lu Y., Li Y., Gao L., Shen H., Song K. Hydrogen sulfide inhibits epithelial-mesenchymal transition in peritoneal mesothelial cells. Sci. Rep. 2018;8:5863. doi: 10.1038/s41598-018-21807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Bai Y.W., Ye M.J., Yang D.L., Yu M.P., Zhou C.F., Shen T. Hydrogen sulfide attenuates paraquat-induced epithelial-mesenchymal transition of human alveolar epithelial cells through regulating transforming growth factor-beta1/Smad2/3 signaling pathway. J. Appl. Toxicol. 2019;39:432–440. doi: 10.1002/jat.3734. [DOI] [PubMed] [Google Scholar]
- 359.Sun H.J., Xiong S.P., Cao X., Cao L., Zhu M.Y., Wu Z.Y., Bian J.S. Polysulfide-mediated sulfhydration of SIRT1 prevents diabetic nephropathy by suppressing phosphorylation and acetylation of p65 NF-κB and STAT3. Redox Biol. 2021;38 doi: 10.1016/j.redox.2020.101813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Cen S.D., Yu W.B., Ren M.M., Chen L.J., Sun C.F., Ye Z.L., Deng H., Hu R.D. Endogenous hydrogen sulfide is involved in osteogenic differentiation in human periodontal ligament cells. Arch. Oral Biol. 2016;68:1–8. doi: 10.1016/j.archoralbio.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 361.Liu Y., Yang R., Liu X., Zhou Y., Qu C., Kikuiri T., Wang S., Zandi E., Du J., Ambudkar I.S., Shi S. Hydrogen sulfide maintains mesenchymal stem cell function and bone homeostasis via regulation of Ca2+ channel sulfhydration. Cell Stem Cell. 2014;15:66–78. doi: 10.1016/j.stem.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Khanna A., Indracanti N., Chakrabarti R., Indraganti P.K. Short-term ex-vivo exposure to hydrogen sulfide enhances murine hematopoietic stem and progenitor cell migration, homing, and proliferation. Cell Adhes. Migrat. 2020;14:214–226. doi: 10.1080/19336918.2020.1842131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Yang R., Liu Y., Shi S. Hydrogen sulfide regulates homeostasis of mesenchymal stem cells and regulatory T cells. J. Dent. Res. 2016;95:1445–1451. doi: 10.1177/0022034516659041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Corsello T., Komaravelli N., Casola A. Role of hydrogen sulfide in NRF2- and sirtuin-dependent maintenance of cellular redox balance. Antioxidants (Basel) 2018;7:129. doi: 10.3390/antiox7100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Navas L.E., Carnero A. NAD+ metabolism, stemness, the immune response, and cancer. Signal Transduct. Targeted Ther. 2021;6:2. doi: 10.1038/s41392-020-00354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Esse R., Barroso M., Tavares de Almeida I., Castro R. The contribution of homocysteine metabolism disruption to endothelial dysfunction: state-of-the-art. Int. J. Mol. Sci. 2019;20:867. doi: 10.3390/ijms20040867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Balint B., Jepchumba V.K., Guéant J.L., Guéant-Rodriguez R.M. Mechanisms of homocysteine-induced damage to the endothelial, medial and adventitial layers of the arterial wall. Biochimie. 2020;173:100–106. doi: 10.1016/j.biochi.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 368.Chrysant S.G., Chrysant G.S. The current status of homocysteine as a risk factor for cardiovascular disease: a mini review. Expert Rev. Cardiovasc Ther. 2018;16:559–565. doi: 10.1080/14779072.2018.1497974. [DOI] [PubMed] [Google Scholar]
- 369.Kang S.S., Rosenson R.S. Analytic approaches for the treatment of hyperhomocysteinemia and its impact on vascular disease. Cardiovasc. Drugs Ther. 2018;32:233–240. doi: 10.1007/s10557-018-6790-1. [DOI] [PubMed] [Google Scholar]
- 370.Al Mutairi F. Hyperhomocysteinemia: clinical insights. J. Cent. Nerv. Syst. Dis. 2020;12 doi: 10.1177/1179573520962230. 1179573520962230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Stabler S.P. Alterations in sulfur amino acids as biomarkers of disease. J. Nutr. 2020;150:S2532–2537. doi: 10.1093/jn/nxaa118. [DOI] [PubMed] [Google Scholar]
- 372.Guieu R., Ruf J., Mottola G. Hyperhomocysteinemia and cardiovascular diseases. Ann. Biol. Clin. 2022;80 doi: 10.1684/abc.2021.1694. 7–14. [DOI] [PubMed] [Google Scholar]
- 373.Martí-Carvajal A.J., Solà I., Lathyris D., Dayer M. Homocysteine-lowering interventions for preventing cardiovascular events. Cochrane Database Syst. Rev. 2017;8:CD006612. doi: 10.1002/14651858.CD006612.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Gupta S., Kühnisch J., Mustafa A., Lhotak S., Schlachterman A., Slifker M.J., Klein- Szanto A., High K.A., Austin R.C., Kruger W.D. Mouse models of cystathionine β-synthase deficiency reveal significant threshold effects of hyperhomocysteinemia. Faseb. J. 2009;23:883–893. doi: 10.1096/fj.08-120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Maclean K.N., Sikora J., Kozich V., Jiang H., Greiner L.S., Kraus E., Krijt J., Overdier K.H., Collard R., Brodsky G.L., Meltesen L., Crnic L.S., Allen R.H., Stabler S.P., Elleder M., Rozen R., Patterson D., Kraus J.P. A novel transgenic mouse model of CBS-deficient homocystinuria does not incur hepatic steatosis or fibrosis and exhibits a hypercoagulative phenotype that is ameliorated by betaine treatment. Mol. Genet. Metabol. 2010;101:153–162. doi: 10.1016/j.ymgme.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Kruger W.D. Cystathionine beta-synthase deficiency: of mice and men. Mol. Genet. Metabol. 2017;121:199–205. doi: 10.1016/j.ymgme.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Majtan T., Park I., Cox A., Branchford B.R., di Paola J., Bublil E.M., Kraus J.P. Behavior, body composition, and vascular phenotype of homocystinuric mice on methionine-restricted diet or enzyme replacement therapy. Faseb. J. 2019;33:12477–12486. doi: 10.1096/fj.201901203R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Carnicer R., Navarro M.A., Guillén N., Arbonés-Mainar J.M., Surra J.C., Acín S., Osada J. Simvastatin reverses the hypertension of heterozygous mice lacking cystathionine beta-synthase and apolipoprotein A-I. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2008;377:35–43. doi: 10.1007/s00210-007-0247-6. [DOI] [PubMed] [Google Scholar]
- 379.Munjal C., Givvimani S., Qipshidze N., Tyagi N., Falcone J.C., Tyagi S.C. Mesenteric vascular remodeling in hyperhomocysteinemia. Mol. Cell. Biochem. 2011;348:99–108. doi: 10.1007/s11010-010-0643-y. [DOI] [PubMed] [Google Scholar]
- 380.Roy A., Khan A.H., Islam M.T., Prieto M.C., Majid D.S. Interdependency of cystathione gamma-lyase and cystathione beta-synthase in hydrogen sulfide-induced blood pressure regulation in rats. Am. J. Hypertens. 2012;25:74–81. doi: 10.1038/ajh.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Greaney J.L., Kutz J.L., Shank S.W., Jandu S., Santhanam L., Alexander L.M. Impaired hydrogen sulfide-mediated vasodilation contributes to microvascular endothelial dysfunction in hypertensive adults. Hypertension. 2017;69:902–909. doi: 10.1161/HYPERTENSIONAHA.116.08964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Winchester L.J., Veeranki S., Pushpakumar S., Tyagi S.C. Exercise mitigates the effects of hyperhomocysteinemia on adverse muscle remodeling. Phys. Rep. 2018;6 doi: 10.14814/phy2.13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.George A.K., Homme R.P., Majumder A., Laha A., Metreveli N., Sandhu H.S., Tyagi S.C., Singh M. Hydrogen sulfide intervention in cystathionine-β-synthase mutant mouse helps restore ocular homeostasis. Int. J. Ophthalmol. 2019;12:754–764. doi: 10.18240/ijo.2019.05.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Ghalwash M., Elmasry A., Omar N.M.A. Possible cardioprotective role of NaHS on ECG and oxidative stress markers in an unpredictable chronic mild stress model in rats. Can. J. Physiol. Pharmacol. 2021;99:321–327. doi: 10.1139/cjpp-2019-0646. [DOI] [PubMed] [Google Scholar]
- 385.Santos I., Ramos C., Mendes C., Sequeira C.O., Tomé C.S., Fernandes D.G.H., Mota P., Pires R.F., Urso D., Hipólito A., Antunes A.M.M., Vicente J.B., Pereira S.A., Bonifácio V.D.B., Nunes S.C., Serpa J. Targeting glutathione and cystathionine beta-synthase in ovarian cancer treatment by selenium-chrysin polyurea dendrimer nanoformulation. Nutrients. 2019;11:2523. doi: 10.3390/nu11102523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Combs J.A., DeNicola G.M. The non-essential amino acid cysteine becomes essential for tumor proliferation and survival. Cancers (Basel) 2019;11:678. doi: 10.3390/cancers11050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Tarragó-Celada J., Foguet C., Tarrado-Castellarnau M., Marin S., Hernández-Alias X., Perarnau J., Morrish F., Hockenbery D., Gomis R.R., Ruppin E., Yuneva M., Atauri P., Cascante M. Cysteine and folate metabolism are targetable vulnerabilities of metastatic colorectal cancer. Cancers (Basel) 2021;13:425. doi: 10.3390/cancers13030425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Nunes S.C., Ramos C., Santos I., Mendes C., Silva F., Vicente J.B., Pereira S.A., Félix A., Gonçalves L.G., Serpa J. Cysteine boosts fitness under hypoxia-mimicked conditions in ovarian cancer by metabolic reprogramming. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.722412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Jyotsana N., Ta K.T., DelGiorno K.E. The role of cystine/glutamate antiporter SLC7A11/xCT in the pathophysiology of cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.858462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Cramer S.L., Saha A., Liu J., Tadi S., Tiziani S., Yan W., Triplett K., Lamb C., Alters S.E., Rowlinson S., Zhang Y.J., Keating M.J., Huang P., DiGiovanni J., Georgiou G., Stone E. Systemic depletion of L-cyst(e)ine with cyst(e)inase increases reactive oxygen species and suppresses tumor growth. Nat. Med. 2017;23:120–127. doi: 10.1038/nm.4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Lu W.C., Saha A., Yan W., Garrison K., Lamb C., Pandey R., Irani S., Lodi A., Lu X., Tiziani S., Zhang Y.J., Georgiou G., DiGiovanni J., Stone E. Enzyme-mediated depletion of serum l-Met abrogates prostate cancer growth via multiple mechanisms without evidence of systemic toxicity. Proc. Natl. Acad. Sci. U.S.A. 2020;117:13000–13011. doi: 10.1073/pnas.1917362117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Badgley M.A., Kremer D.M., Maurer H.C., DelGiorno K.E., Lee H.J., Purohit V., Sagalovskiy I.R., Ma A., Kapilian J., Firl C.E.M., Decker A.R., Sastra S.A., Palermo C.F., Andrade L.R., Sajjakulnukit P., Zhang L., Tolstyka Z.P., Hirschhorn T., Lamb C., Liu T., Gu W., Seeley E.S., Stone E., Georgiou G., Manor U., Iuga A., Wahl G.M., Stockwell B.R., Lyssiotis C.A., Olive K.P. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science. 2020;368:85–89. doi: 10.1126/science.aaw9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Toombs C.F., Insko M.A., Wintner E.A., Deckwerth T.L., Usansky H., Jamil K., Goldstein B., Cooreman M., Szabo C. Detection of exhaled hydrogen sulphide gas in healthy human volunteers during intravenous administration of sodium sulphide. Br. J. Clin. Pharmacol. 2010;69:626–636. doi: 10.1111/j.1365-2125.2010.03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Liu N., Tseng Y., Zhang H., Chen J. The role of exhaled hydrogen sulfide in the diagnosis of colorectal adenoma, Can. J. Infect. Dis. Med. Microbiol. 2021;2021 doi: 10.1155/2021/8046368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Li L., Liu Y., Wang Q., Wang Z., Cui L., Xu Y., Guan K. Levels of nasal exhaled hydrogen sulfide in the general population and allergic rhinitis patients. J. Clin. Lab. Anal. 2021;35 doi: 10.1002/jcla.23678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Suzuki Y., Saito J., Munakata M., Shibata Y. Hydrogen sulfide as a novel biomarker of asthma and chronic obstructive pulmonary disease. Allergol. Int. 2021;70:181–189. doi: 10.1016/j.alit.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 397.Palmieri B., Nardo B., Lippi G., Palmieri L., Vadalà M., Laurino C. Dogs'olfactory diagnostics applied on human species: state of the art and clinical perspectives. Clin. Ter. 2016;167:e78–84. doi: 10.7417/CT.2016.1943. [DOI] [PubMed] [Google Scholar]
- 398.Biehl W., Hattesohl A., Jörres R.A., Duell T., Althöhn U., Koczulla A.R., Schmetzer H. VOC pattern recognition of lung cancer: a comparative evaluation of different dog- and eNose-based strategies using different sampling materials. Acta Oncol. 2019;58:1216–1224. doi: 10.1080/0284186X.2019.1634284. [DOI] [PubMed] [Google Scholar]
- 399.Junqueira H., Quinn T.A., Biringer R., Hussein M., Smeriglio C., Barrueto L., Finizio J., Huang X.Y.M. Accuracy of canine scent detection of non-small cell lung cancer in blood serum. J. Am. Osteopath. Assoc. 2019 doi: 10.7556/jaoa.2019.077. In press. [DOI] [PubMed] [Google Scholar]
- 400.Piqueret B., Bourachot B., Leroy C., Devienne P., Mechta-Grigoriou F., d'Ettorre P., Sandoz J.C. Ants detect cancer cells through volatile organic compounds. iScience. 2022;25 doi: 10.1016/j.isci.2022.103959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Yamagishi K., Onuma K., Chiba Y., Yagi S., Aoki S., Sato T., Sugawara Y., Hosoya N., Saeki Y., Takahashi M., Fuji M., Ohsaka T., Okajima T., Akita K., Suzuki T., Senawongse P., Urushiyama A., Kawai K., Shoun H., Ishii Y., Ishikawa H., Sugiyama S., Nakajima M., Tsuboi M., Yamanaka T. Generation of gaseous sulfur-containing compounds in tumour tissue and suppression of gas diffusion as an antitumour treatment. Gut. 2012;61:554–561. doi: 10.1136/gutjnl-2011-300721. [DOI] [PubMed] [Google Scholar]
- 402.Moon H.G., Jung Y., Han S.D., Shim Y.S., Shin B., Lee T., Kim J.S., Lee S., Jun S.C., Park H.H., Kim C., Kang C.Y. Chemiresistive electronic nose toward detection of biomarkers in exhaled breath. ACS Appl. Mater. Interfaces. 2016;8:20969–20976. doi: 10.1021/acsami.6b03256. [DOI] [PubMed] [Google Scholar]
- 403.Yoon J.W., Lee J.H. Toward breath analysis on a chip for disease diagnosis using semiconductor-based chemiresistors: recent progress and future perspectives. Lab Chip. 2017;17:3537–3557. doi: 10.1039/c7lc00810d. [DOI] [PubMed] [Google Scholar]
- 404.Vajhadin F., Mazloum-Ardakani M., Amini A. Metal oxide-based gas sensors for detection of exhaled breath markers. Med. Devices Sens. 2020;14 doi: 10.1002/mds3.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Banik G.D., De A., Som S., Jana S., Daschakraborty S.B., Chaudhuri S., Pradhan M. Hydrogen sulphide in exhaled breath: a potential biomarker for small intestinal bacterial overgrowth in IBS. J. Breath Res. 2016;10 doi: 10.1088/1752-7155/10/2/026010. [DOI] [PubMed] [Google Scholar]
- 406.Bee N., White R., Petros A.J. Hydrogen sulfide in exhaled gases from ventilated septic neonates and children: a preliminary report. Pediatr. Crit. Care Med. 2017;18:e327–332. doi: 10.1097/PCC.0000000000001223. [DOI] [PubMed] [Google Scholar]
- 407.Milby T.H., Baselt R.C. Hydrogen sulfide poisoning: clarification of some controversial issues. Am. J. Ind. Med. 1999;35:192–195. doi: 10.1002/(sici)1097-0274(199902)35:2<192::aid-ajim11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 408.Zhang M.Y., Dugbartey G.J., Juriasingani S., Sener A. Hydrogen sulfide metabolite, sodium thiosulfate: clinical applications and underlying molecular mechanisms. Int. J. Mol. Sci. 2021;22:6452. doi: 10.3390/ijms22126452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Chwatko G., Forma E., Wilkosz J., Głowacki R., Jóźwiak P., Różański W., Bryś M., Krześlak A. Thiosulfate in urine as a facilitator in the diagnosis of prostate cancer for patients with prostate-specific antigen less or equal 10 ng/mL. Clin. Chem. Lab. Med. 2013;51:1825–1831. doi: 10.1515/cclm-2013-0069. [DOI] [PubMed] [Google Scholar]
- 410.Stephan C., Wilkosz J., Różański W., Ecke T.H., Lein M., Bryś M., Krześlak A., Chwatko G., Jung K. Urinary thiosulfate as failed prostate cancer biomarker - an exemplary multicenter re-evaluation study. Clin. Chem. Lab. Med. 2015;53:477–483. doi: 10.1515/cclm-2014-0729. [DOI] [PubMed] [Google Scholar]
- 411.Wu H., Krishnakumar S., Yu J., Liang D., Qi H., Lee Z.W., Deng L.W., Huang D. Highly selective and sensitive near-infrared-fluorescent probes for the detection of cellular hydrogen sulfide and the imaging of H2S in mice. Chem. Asian J. 2014;9:3604–3611. doi: 10.1002/asia.201402860. [DOI] [PubMed] [Google Scholar]
- 412.Cao J., Lopez R., Thacker J.M., Moon J.Y., Jiang C., Morris S.N., Bauer J.H., Tao P., Mason R.P., Lippert A.R. Chemiluminescent probes for imaging H2S in living animals. Chem. Sci. 2015;6:1979–1985. doi: 10.1039/c4sc03516j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Zhang L., Liu H., Wu C., Zheng Y., Kai X., Xue Y. A near-infrared fluorescent probe that can image endogenous hydrogen polysulfides in vivo in tumour-bearing mice. Org. Biomol. Chem. 2021;19:911–919. doi: 10.1039/d0ob02253e. [DOI] [PubMed] [Google Scholar]
- 414.Xu G., Yan Q., Lv X., Zhu Y., Xin K., Shi B., Wang R., Chen J., Gao W., Shi P., Fan C., Zhao C., Tian H. Imaging of colorectal cancers using activatable nanoprobes with second near-infrared window emission. Angew Chem. Int. Ed. Engl. 2018;57:3626–3630. doi: 10.1002/anie.201712528. [DOI] [PubMed] [Google Scholar]
- 415.Kweon Y., Park J.Y., Kim Y.J., Lee Y.S., Jeong J.M. Imaging hydrogen sulfide in hypoxic tissue with [(99m)Tc]Tc-gluconate. Molecules. 2020;26:96. doi: 10.3390/molecules26010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Park J.Y., Kim Y.J., Lee J.Y., Lee Y.S., Jeong J.M. Imaging of the third gasotransmitter hydrogen sulfide using 99mTc-labeled alpha-hydroxy acids. Nucl. Med. Biol. 2019;76–77:28–35. doi: 10.1016/j.nucmedbio.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 417.Malara N., Trunzo V., Foresta U., Amodio N., De Vitis S., Roveda L., Fava M., Coluccio M., Macrì R., Di Vito A., Costa N., Mignogna C., Britti D., Palma E., Mollace V. Ex-vivo characterization of circulating colon cancer cells distinguished in stem and differentiated subset provides useful biomarker for personalized metastatic risk assessment. J. Transl. Med. 2016;14:133. doi: 10.1186/s12967-016-0876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Zhang B., Niu X., Zhang Q., Wang C., Liu B., Yue D., Li C., Giaccone G., Li S., Gao L., Zhang H., Wang J., Yang H., Wu R., Ni P., Wang C., Ye M., Liu W. Circulating tumor DNA detection is correlated to histologic types in patients with early-stage non-small-cell lung cancer. Lung Cancer. 2019;134:108–116. doi: 10.1016/j.lungcan.2019.05.034. [DOI] [PubMed] [Google Scholar]
- 419.Wang Z.L., Zhang P., Li H.C., Yang X.J., Zhang Y.P., Li Z.L., Xue L., Xue Y.Q., Li H.L., Chen Q., Chong T. Dynamic changes of different phenotypic and genetic circulating tumor cells as a biomarker for evaluating the prognosis of RCC. Cancer Biol. Ther. 2019;20:505–512. doi: 10.1080/15384047.2018.1537576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Chen J., Ye C., Dong J., Cao S., Hu Y., Situ B., Xi X., Qin S., Xu J., Cai Z., Zheng L., Wang Q. Metabolic classification of circulating tumor cells as a biomarker for metastasis and prognosis in breast cancer. J. Transl. Med. 2020;18:59. doi: 10.1186/s12967-020-02237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Alizadeh-Sedigh M., Fazeli M.S., Mahmoodzadeh H., Sharif S.B., Teimoori-Toolabi L. Cancer Biomark; 2021. Methylation of FBN1, SPG20, ITF2, RUNX3, SNCA, MLH1, and SEPT9 Genes in Circulating Cell-free DNA as Biomarkers of Colorectal Cancer. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Wang D., O'Rourke D., Sanchez-Garcia J.F., Cai T., Scheuenpflug J., Feng Z. Development of a liquid biopsy based purely quantitative digital droplet PCR assay for detection of MLH1 promoter methylation in colorectal cancer patients. BMC Cancer. 2021;21:797. doi: 10.1186/s12885-021-08497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Jiang J.H., Gao J., Chen C.Y., Ao Y.Q., Li J., Lu Y., Fang W., Wang H.K., de Castro D.G., Santarpia M., Hashimoto M., Yuan Y.F., Ding J.Y. Circulating tumor cell methylation profiles reveal the classification and evolution of non-small cell lung cancer. Transl. Lung Cancer Res. 2022;11:224–237. doi: 10.21037/tlcr-22-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Richter F., Röder C., Möller T., Egberts J.H., Becker T., Sebens S. Detection of circulating and disseminated tumor cells and their prognostic value under the influence of neoadjuvant therapy in esophageal cancer patients. Cancers (Basel) 2022;14:1279. doi: 10.3390/cancers14051279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Gerratana L., Pierga J.Y., Reuben J.M., Davis A.A., Wehbe F.H., Dirix L., Fehm T., Nolé F., Gisbert-Criado R., Mavroudis D., Grisanti S., Garcia-Saenz J.A., Stebbing J., Caldas C., Gazzaniga P., Manso L., Zamarchi R., Bonotto M., Fernandez de Lascoiti A., De Mattos-Arruda L., Ignatiadis M., Sandri M.T., Generali D., De Angelis C., Dawson S.J., Janni W., Carañana V., Riethdorf S., Solomayer E.F., Puglisi F., Giuliano M., Pantel K., Bidard F.C., Cristofanilli M. Modeling the prognostic impact of circulating tumor cells enumeration in metastatic breast cancer for clinical trial design simulation. Oncol. 2022 doi: 10.1093/oncolo/oyac045. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Yang C.T., Chen L., Xu S., Day J.J., Li X., Xian M. Recent development of hydrogen sulfide releasing/stimulating reagents and their potential applications in cancer and glycometabolic disorders. Front. Pharmacol. 2017;8:664. doi: 10.3389/fphar.2017.00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Li H., Xu F., Gao G., Gao X., Wu B., Zheng C., Wang P., Li Z., Hua H., Li D. Hydrogen sulfide and its donors: novel antitumor and antimetastatic therapies for triple-negative breast cancer, Redox. Biol. 2020;34 doi: 10.1016/j.redox.2020.101564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Ngowi E.E., Afzal A., Sarfraz M., Khattak S., Zaman S.U., Khan N.H., Li T., Jiang Q.Y., Zhang X., Duan S.F., Ji X.Y., Wu D.D. Role of hydrogen sulfide donors in cancer development and progression. Int. J. Biol. Sci. 2021;17:73–88. doi: 10.7150/ijbs.47850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Wang R.H., Chu Y.H., Lin K.T. The hidden role of hydrogen sulfide metabolism in cancer. Int. J. Mol. Sci. 2021;22:6562. doi: 10.3390/ijms22126562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Li M., Liu Y., Deng Y., Pan L., Fu H., Han X., Li Y., Shi H., Wang T. Therapeutic potential of endogenous hydrogen sulfide inhibition in breast cancer (Review) Oncol. Rep. 2021;45:68. doi: 10.3892/or.2021.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Hu X., Xiao Y., Sun J., Ji B., Luo S., Wu B., Zheng C., Wang P., Xu F., Cheng K., Hua H., Li D. New possible silver lining for pancreatic cancer therapy: hydrogen sulfide and its donors. Acta Pharm. Sin. B. 2021;11:1148–1157. doi: 10.1016/j.apsb.2020.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]