Abstract
A novel cell surface display system was developed by employing Escherichia coli outer membrane protein C (OmpC) as an anchoring motif. Polyhistidine peptides consisting of up to 162 amino acids could be successfully displayed on the seventh exposed loop of OmpC. Recombinant cells displaying polyhistidine could adsorb up to 32.0 μmol of Cd2+ per g (dry weight) of cells.
Cell surface display aims to display proteins or peptides on the surfaces of prokaryotic or eukaryotic cells, especially bacterial and yeast cells (6, 26). The possible applications of this technique include live vaccine development (11, 15), antibody production (3, 13), peptide library screening (2, 5), environmental bioadsorbent development (24, 25), and whole-cell biocatalyst development (14, 21). A variety of surface displaying systems have been developed, and these systems can be divided into the following three groups on the basis of their recombinant profiles: C-terminal fusion, N-terminal fusion, and sandwich fusion. If a native surface protein has a discrete localization signal within its N-terminal portion, a C-terminal fusion strategy may be used to conjugate foreign peptides to the C terminus of that functional portion (4, 5, 21). Similarly, N-terminal fusion systems have been developed by using Staphylococcus aureus protein A (8, 23), fibronectin binding protein B (28), Streptococcus pyogenes fibrillar M protein (18), and Saccharomyces cerevisiae α-agglutinin (14), all of which contain C-terminal sorting signals, to target foreign proteins to the cell wall. However, many surface proteins, such as outer membrane proteins (OMPs) and extracellular appendage subunits, do not have anchoring regions, and the whole structure is required for assembly. In this situation, sandwich fusion is the only choice. Escherichia coli PhoE (1), FimH (17), FliC (12), and PapA (27) have been shown to be good sandwich carriers for small peptides.
In this study, our goal was to develop a novel cell surface display system by using E. coli OmpC, one of the most abundant OMPs in E. coli cells (up to 105 molecules per cell may be present). This protein is one of the three classical porins of E. coli K-12 strains (the other two are OmpF and PhoE) and consists of 367 amino acids, including a signal peptide consisting of 21 amino acids. Three OmpC molecules form a pore structure on the outer membrane of an E. coli cell, which allows small hydrophilic molecules to pass through. One OmpC molecule consists of 16 transmembrane, antiparallel β-strands, which produce a β-barrel structure surrounding a large channel and are connected by seven internal loops and eight external loops (16). In general, the amino acid sequences of the external loops are less conservative, and thus these loops may be relatively tolerant to insertion and deletion. Therefore, we decided to use one of the external loops as the point of insertion for foreign peptides for cell surface display. Polyhistidine peptides (containing several hexahistidine [6His] linkers) were used as model inserts for the following two main reasons: (i) they are good chelators for divalent metal ions, such as Cd2+, Cu2+, Zn2+, Ni2+, and Pb2+, and therefore may be used as biosorbents for heavy metal removal; and (ii) the permissive size limit of the polypeptide to be fused to the external loops of OmpC can be easily examined by inserting different numbers of copies of 6His linkers.
Bacterial strains and growth conditions.
E. coli K-12 strain MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR] (ATCC 35695) was used as a host strain for all of the plasmids used in this study. All recombinant strains were cultivated in Luria-Bertani medium supplemented with 50 μg of ampicillin per ml. Recombinant cells were induced at an optical density at 600 nm of 0.6 by adding isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 10 μM. Unless indicated otherwise, strains harboring pTCdP, pTCHP1, pTCHP2, pTCHP3, or pTCHP6 were cultivated at 30°C and were grown for an additional 2 h after induction, and strains harboring pTCHP12 or pTCHP18 were cultivated at 25°C and were grown for an additional 4 h after induction.
Plasmids and DNA manipulation.
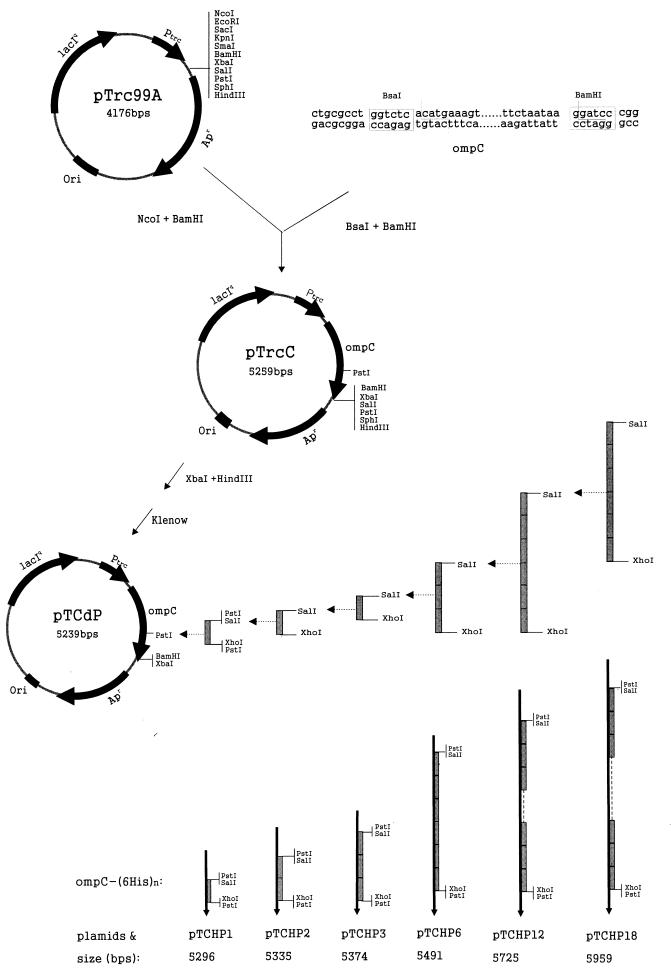
The plasmids used in this study are shown in Fig. 1. Plasmid pTrc99A was purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). One 6His linker was obtained by performing overlapping PCR with the following primers: 5′-GATAGATATCCTGCAGGTCGACCCAAGCGGACATCACCATCATCACCAT-3′ and 5′-CCAAC TG CAG GATATCC TCGAGACCAGAATG G TGATGATGGTGATG-3′. This 6His linker was designed so that it contained PstI-SalI sites at the 5′ end and XhoI-PstI sites at the 3′ end. The PstI sites were used to introduce the first copy of 6His into the ompC gene, and the SalI and XhoI sites were used to insert 6His linkers in tandem. The E. coli ompC gene was cloned from K-12 strain MC4100 by performing PCR with the following primers: 5′-CTGCGCCTGGTCTCACATGAAAGTTAAAGTACTG-3′ and 5′-CCGGGATCCTTATTAGAACTGGTAAACCAG-3′. All DNA manipulations were carried out by using the procedures described by Sambrook et al. (22). Restriction enzymes and modifying enzymes were purchased from New England Biolabs (Beverly, Mass.) and were used as recommended by the manufacturer.
FIG. 1.
Construction of the plasmids used in this study. The grey rectangles represent sets of 6His linkers.
Construction of OmpC-(6His)n fusion vectors.
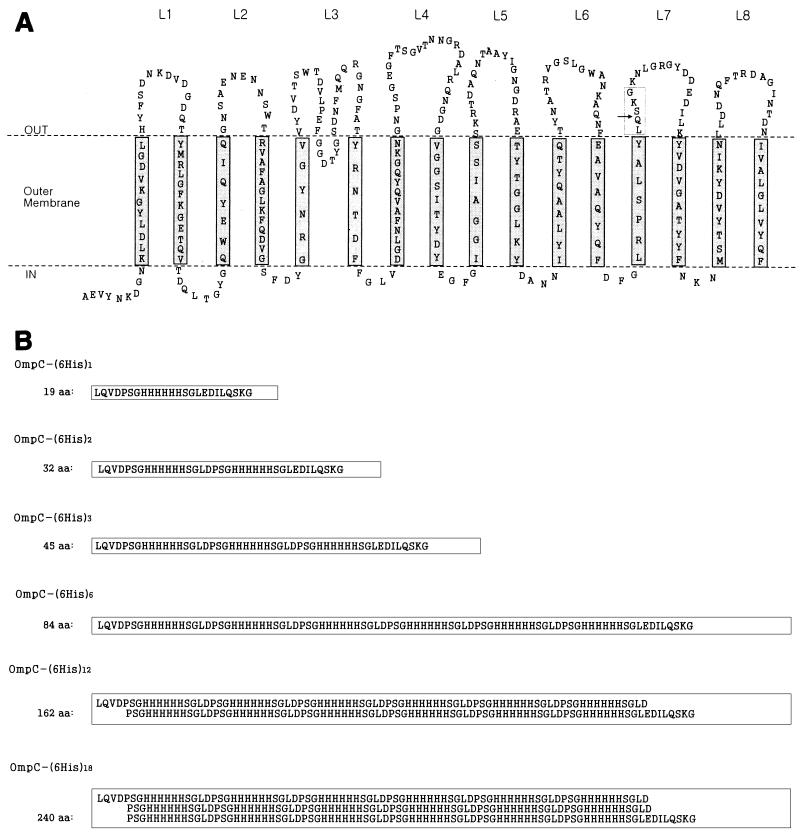
Based on homology studies (9, 20) and information concerning the Salmonella typhi OmpC structure (19), we suggested the presumed structure of E. coli OmpC shown in Fig. 2A. Loop 7 of E. coli OmpC, the second external loop from the C terminus, was selected as the fusion point. The PstI site loop 7 was used for inserting 6His linkers.
FIG. 2.
Structure of OmpC and amino acid sequences of polyhistidine inserts. (A) Predicted structure of E. coli OmpC, based on the findings of Jeanteur et al. (9) and Puente et al. (19, 20). (B) Sequences of polyhistidine inserts. The arrow indicates the site of insertion. The boxes in panel B correspond to the box in panel A. aa, amino acids; L1, loop 1.
The entire ompC gene, including the signal sequence, was inserted into the NcoI-BamHI site of pTrc99A downstream of the trc promoter in order to make pTrcC (Fig. 1). Therefore, expression of ompC and its derivatives carrying (6His)n could be induced by IPTG. Plasmid pTrcC was digested with HindIII and XbaI and filled in by using the Klenow enzyme, and the linear fragment was self-ligated to make pTCdP. In this way the PstI site present in the multiple cloning site was deleted. The following pTCHP series plasmids bearing different ompC-(6His)n fusion genes were constructed as shown in Fig. 1: pTCHP1, pTCHP2, pTCHP3, pTCHP6, pTCHP12, and pTCHP18, which contained 1, 2, 3, 6, 12, and 18 sets of 6His linkers, respectively. The sequences of the resulting peptides are shown in Fig. 2B.
Expression of OmpC-(6His)n fusion proteins.
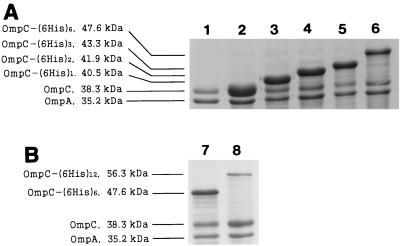
First, all recombinant strains were cultivated at 30°C and were incubated for an additional 2 h after IPTG induction. Outer membrane fraction samples were prepared from the induced cultures and were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3A). OMPs were prepared as described by Puente et al. (19), except that 0.5% (wt/vol) Sarkosyl was used instead of Triton X-100 and the samples were incubated on ice. Protein samples were analyzed by electrophoresis (7) on a 12% (wt/vol) SDS-PAGE gel as described by Laemmli (10). The apparent molecular mass of OmpC-(6His)n increased in proportion to n, as we intended. The results suggested that polyhistidine peptides up to 84 amino acids long could be targeted efficiently to the E. coli outer membrane by OmpC protein (Fig. 3A). The expression level of OmpC-(6His)n decreased as the number of inserted 6His linkers increased, as follows: 34.7, 33.1, 32.8, and 26.7% of the total OMPs for OmpC-(6His)1, OmpC-(6His)2, OmpC-(6His)3, and OmpC-(6His)6, respectively. Expression of OmpC-(6His)12 and OmpC-(6His)18 could not be detected in this experiment (data not shown). However, when the recombinant strains were cultivated at 25°C and were incubated for an additional 4 h, OmpC-(6His)12 could be targeted to the outer membrane quite efficiently (up to 10.2% of the total OMPs) (Fig. 3B). Under these conditions, the amount of expressed OmpC-(6His)6 increased to 29.9% of the total OMPs (Fig. 3B). However, OmpC-(6His)18 was still not detected under the same conditions or even after induction with a higher concentration of IPTG (100 or 500 μM or 1 mM) (data not shown).
FIG. 3.
SDS-PAGE of OMPs from recombinant strains that were cultivated at 30°C and were grown for an additional 2 h after induction (A) and recombinant strains that were cultivated at 24°C and were grown for an additional 4 h after induction (B). Lane 1, MC4100; lane 2, MC4100(pTCdP); lane 3, MC4100(pTCHP1); lane 4, MC4100(pTCHP2); lane 5, MC4100(pTCHP3); lanes 6 and 7, MC4100(pTCHP6); lane 8, MC4100(pTCHP12).
Cells began to display less OmpC-(6His)n fusion protein when the insert became larger. There are two possible reasons for this. First, large inserts interfere with the folding of OmpC-(6His)n peptide into the correct conformation much more severely than small inserts do. Second, cells may encounter a shortage of aminoacyl-tRNAHis when they have to synthesize large inserts containing many histidines. The same explanation can be proposed for the failure to display OmpC-(6His)18.
Whole-cell adhesion to Ni-NTA-agarose beads.
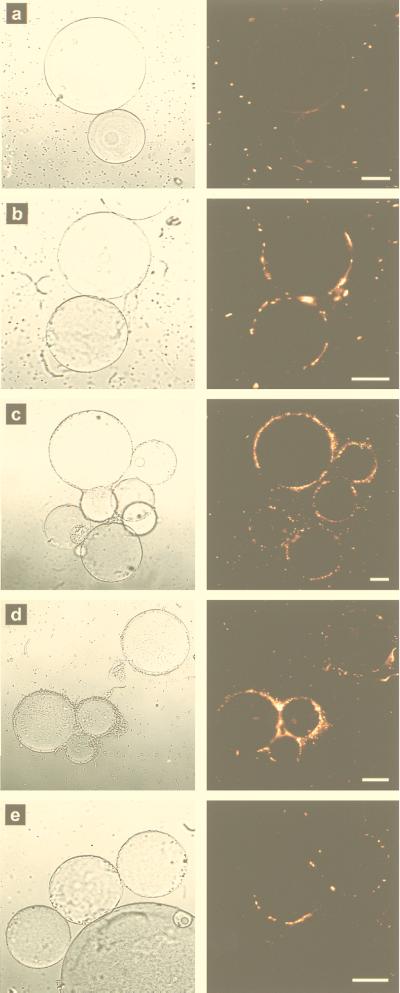
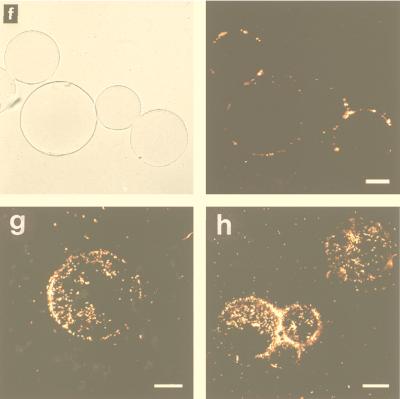
Adhesion of recombinant E. coli cells to nickel-nitrilotriacetic acid (NTA)-agarose (Qiagen GmbH, Hilden, Germany) beads was examined as described by Sousa et al. (24). Ni-NTA-agarose is precharged with nickel ions, and six pairs of electrons are necessary to form a stable coordination sphere around each ion. Four of the pairs are available from NTA, and the other two should come from neighboring histidines. Figure 4 shows recombinant E. coli cells expressing OmpC-(6His)n fusion proteins bound to Ni-NTA-agarose beads. MC4100(pTCdP) did not bind to the beads, and therefore, the beads were invisible under fluorescent light (Fig. 4a). With MC4100(pTCHP) strains, the outlines of the beads were clearly visible due to the attached recombinant E. coli cells. Since only the exposed (6His)n was accessible to nickel ions on the agarose beads, the micrographs indicate that OmpC-(6His)n fusion proteins were successfully exposed outside the recombinant cells. Among the MC4100(pTCHP) strains, MC4100(pTCHP3) attached to the beads most efficiently (Fig. 4d and h), followed by MC4100(pTCHP2) (Fig. 4c and g), MC4100(pTCHP1) (Fig. 4b), and finally MC4100(pTCHP6) (Fig. 4e) and MC4100(pTCHP12) (Fig. 4f). Again, MC4100(pTCHP18) gave no indication of adhesion (data not shown).
FIG. 4.
Adhesion of recombinant strains to Ni-NTA-agarose beads. (a to f) Transmission micrographs (left) and matching fluorescent micrographs (right). (a) MC4100(pTCdP). (b) MC4100(pTCHP1). (c) MC4100(pTCHP2). (d) MC4100(pTCHP3). (e) MC4100(pTCHP6). (f) MC4100(pTCHP12). (g and h) Fluorescent micrographs. (g), MC4100(pTCHP2). (h) MC4100(pTCHP3). The fluorescent images in panels a to f were taken by focusing on the outlines of agarose beads. The images in panels g and h were taken by focusing on the surface-bound cells. The transmission images in panels a to f are background reference images. The large circles are nickel-precharged agarose beads, which did not fluoresce by themselves. The bright dots in the fluorescence images are stained E. coli cells. Bars = 30 μm.
Theoretically, stronger binding should occur with cells expressing more 6His units. This was true for MC4100(pTCHP1), MC4100(pTCHP2), and MC4100(pTCHP3). Just one set of 6His linkers was enough to make cells attach to the beads, while two and three sets were much better (Fig. 4g and h). The decreased binding efficiencies observed when the number of 6His sets was increased to 6 or 12 seemed to be due to long protruding loops that became obstacles for cells trying to access restricted nickel ions. The relatively lower level of expression might be another reason. No attachment was observed when the number of 6His linkers was increased to 18, which is consistent with the finding that no protein band was visible in the SDS-PAGE gel.
Adsorption of Cd2+.
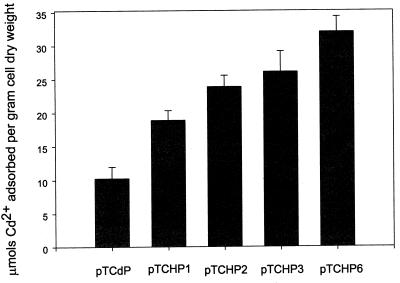
Induced cells were washed twice with 0.85% (wt/vol) NaCl and were resuspended in 0.85% (wt/vol) NaCl (pH 5.8) to an optical density at 600 nm of 5.0. An equal volume of CdCl2 (50 ppm in 0.85% [wt/vol] NaCl [pH 5.8]) was added, and the mixtures were incubated for 24 h with shaking at 25°C. The cells were pelleted, washed twice with 0.85% (wt/vol) NaCl, and then digested with 70% (wt/vol) nitric acid overnight at room temperature. Samples were analyzed with an atomic analysis system (model 3300; Perkin-Elmer, Norwalk, Conn.) by using an air-acetylene flame and a hollow cathode lamp. The wavelength and slit width were 228.8 and 0.7 nm, respectively. Recombinant E. coli cells displaying OmpC-(6His)n fusion proteins containing up to six copies of 6His were examined to determine their abilities to adsorb Cd2+. Cells harboring pTCdP, pTCHP1, pTCHP2, pTCHP3, and pTCHP6 could absorb 10.3, 18.9, 23.9, 26.1, and 32.0 μmol of Cd2+ per g (dry weight) of cells, respectively (Fig. 5). The ability to adsorb Cd2+ increased as the number of 6His units increased, a result which was different from the results of the Ni-NTA-agarose binding test. The reason for this was probably the different status of metal ions (free versus restricted). In the bead binding test, Ni2+ ions are confined to the surfaces of agarose beads, and therefore, cells have to overcome steric hindrance to access Ni2+ ions. In the Cd2+ biosorption test, however, all Cd2+ ions exist as free ions and thus are free to be adsorbed. The greatest Cd2+ removal capacity observed in this study was 32 μmol per g (dry weight) of cells, which was twice the value obtained with the LamB–polyhistidine displaying motif (24) and also was slightly greater than values obtained with the recently developed LamB systems displaying metallothioneins (25).
FIG. 5.
Adsorption of Cd2+ by recombinant strains expressing OmpC-(6His)n fusion proteins.
In conclusion, we developed a novel cell surface displaying system in which E. coli OmpC was used as an anchoring motif. A large polyhistidine peptide up to 162 amino acids long could be successfully displayed on the E. coli outer membrane, which is not consistent with the size limit for peptides to be inserted previously suggested for sandwich fusion motifs (6, 26). Furthermore, the recombinant strains developed in this study could adsorb large amounts of Cd2+, suggesting that they may be useful as bioadsorbents.
Acknowledgments
We thank M.-Y. Lee and K. Beak for help with the atomic adsorption analysis and Z.-W. Lee and K.-S. Ha of the Korea Basic Science Institute for help with confocal microscopy.
This work was supported by the First Young Scientist’s Award to S.Y.L. from the President of Korea and by the Korea Academy of Science and Technology. Z.X. was a visiting student from Huazhong Agricultural University, Wuhan, People’s Republic of China, and was supported in part by the BioProcess Engineering Research Center.
REFERENCES
- 1.Agterberg M, Adriaanse H, van Bruggen A, Karperien M, Tommassen J. Outer-membrane PhoE protein of Escherichia coli K-12 as an exposure vector: possibilities and limitations. Gene. 1990;88:37–45. doi: 10.1016/0378-1119(90)90057-x. [DOI] [PubMed] [Google Scholar]
- 2.Boder E T, Wittrup K D. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 3.Charbit A, Molla A, Saurin W, Hofnung M. Versatility of a vector for expressing foreign polypeptides at the surface of Gram-negative bacteria. Gene. 1988;70:181–189. doi: 10.1016/0378-1119(88)90116-3. [DOI] [PubMed] [Google Scholar]
- 4.Francisco J A, Earhart C F, Georgiou G. Transport and anchoring of β-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:2713–2717. doi: 10.1073/pnas.89.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francisco J A, Campbell R, Iverson B L, Georgiou G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proc Natl Acad Sci USA. 1993;90:10444–10448. doi: 10.1073/pnas.90.22.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgiou G, Stathopoulos C, Daugherty P S, Nayak A R, Iverson B L, Curtiss R., III Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol. 1997;15:29–34. doi: 10.1038/nbt0197-29. [DOI] [PubMed] [Google Scholar]
- 7.Georgiou G, Shuler M L, Wilson D B. Release of periplasmic enzymes and other physiological effects of β-lactamase overproduction in Escherichia coli. Biotechnol Bioeng. 1988;32:741–748. doi: 10.1002/bit.260320603. [DOI] [PubMed] [Google Scholar]
- 8.Gunneriusson E, Samuelson P, Uhlen M, Nygren P-A, Stahl S. Surface display of a functional single-chain Fv antibody on staphylococci. J Bacteriol. 1996;178:1341–1346. doi: 10.1128/jb.178.5.1341-1346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Liljeqvist S, Samuelson P, Hansson M, Nguyen T N, Binz H, Stahl S. Surface display of the cholera toxin B subunit on Staphylococcus xylosus and Staphylococcus carnosus. Appl Environ Microbiol. 1997;63:2481–2488. doi: 10.1128/aem.63.7.2481-2488.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z, Murray K S, Cleave V V, LaVallie E R, Stahl M L, McCoy J M. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagellin: a system designed for exploring protein-protein interactions. Bio/Technology. 1995;13:366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 13.Martineau P, Charbit A, Leclerc C, Werts C, O’Callaghan D, Hofnung M. A genetic system to elicit and monitor anti-peptide antibodies without peptide synthesis. Bio/Technology. 1991;9:170–172. doi: 10.1038/nbt0291-170. [DOI] [PubMed] [Google Scholar]
- 14.Murai T, Ueda M, Kawaguchi T, Arai M, Tanaka A. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing β-glucosidase and carboxymethylcellulase from Aspergillus aculeatus. Appl Environ Microbiol. 1998;64:4857–4861. doi: 10.1128/aem.64.12.4857-4861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen T N, Hansson M, Stahl S, Bachi T, Robert A, Domzig W, Binz H, Uhlen M. Cell-surface display of heterologous epitopes on Staphylococcus xylosus as a potential delivery system for oral vaccination. Gene. 1993;128:89–94. doi: 10.1016/0378-1119(93)90158-y. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido H. Outer membrane. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 29–47. [Google Scholar]
- 17.Pallesen L, Poulsen L K, Christiansen G, Klemm P. Chimeric FimH adhesin of type 1 fimbriae: a bacterial surface display system for heterologous sequences. Microbiology. 1995;141:2839–2848. doi: 10.1099/13500872-141-11-2839. [DOI] [PubMed] [Google Scholar]
- 18.Pozzi G, Contorni M, Oggioni M R, Manganelli R, Tommasino M, Cavalieri F, Fischetti V A. Delivery and expression of a heterologous antigen on the surface of streptococci. Infect Immun. 1992;60:1902–1907. doi: 10.1128/iai.60.5.1902-1907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puente J L, Juarez D, Bobadilla M, Arisa C F, Calva E. The Salmonella ompC gene: structure and use as a carrier for heterologous sequences. Gene. 1995;156:1–9. doi: 10.1016/0378-1119(94)00883-t. [DOI] [PubMed] [Google Scholar]
- 20.Puente J L, Alvarez-Scherer V, Gosset G, Calva E. Comparative analysis of the Salmonella typhi and Escherichia coli ompC genes. Gene. 1989;83:197–206. doi: 10.1016/0378-1119(89)90105-4. [DOI] [PubMed] [Google Scholar]
- 21.Richins R D, Kaneva I, Mulchandani A, Chen W. Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nat Biotechnol. 1997;15:984–987. doi: 10.1038/nbt1097-984. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Samuelson P, Hansson M, Ahlborg N, Andreoni C, Gotz F, Bachi T, Nguyen T N, Binz H, Uhlen M, Stahl S. Cell surface display of recombinant proteins on Staphylococcus carnosus. J Bacteriol. 1995;177:1470–1476. doi: 10.1128/jb.177.6.1470-1476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sousa C, Cebolla A, de Lorenzo V. Enhanced metalloadsorption of bacterial cells displaying poly-His peptides. Nat Biotechnol. 1996;14:1017–1020. doi: 10.1038/nbt0896-1017. [DOI] [PubMed] [Google Scholar]
- 25.Sousa C, Kotrba P, Ruml T, Cebolla A, de Lorenzo V. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J Bacteriol. 1998;180:2280–2284. doi: 10.1128/jb.180.9.2280-2284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl S, Uhlen M. Bacterial surface display: trends and progress. Trends Biotechnol. 1997;15:185–192. doi: 10.1016/S0167-7799(97)01034-2. [DOI] [PubMed] [Google Scholar]
- 27.Steidler L, Remaut E, Fiers W. Pap pili as a vector system for surface exposition of an immunoglobulin G-binding domain of protein A of Staphylococcus aureus in Escherichia coli. J Bacteriol. 1993;175:7639–7643. doi: 10.1128/jb.175.23.7639-7643.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss A, Gotz F. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol. 1996;21:491–500. doi: 10.1111/j.1365-2958.1996.tb02558.x. [DOI] [PubMed] [Google Scholar]