Significance
Skeletal muscle is one of the largest tissues in the body and can regenerate when damaged through a population of resident muscle stem cells. A type of muscle trauma called volumetric muscle loss overwhelms the regenerative capacity of muscle stem cells and engenders fibrotic supplantation. A comparison of muscle injuries resulting in regeneration or fibrosis revealed that intercellular communication between neutrophils and natural killer cells impacts muscle stem cell-mediated repair. Perturbation of neutrophil–natural killer cell interactions resulted in a variation of healing outcomes and suggested that immunomodulatory interventions can be effective to prevent aberrant healing outcomes.
Keywords: muscle stem cells, inflammation, skeletal muscle, single-cell RNA sequencing
Abstract
Volumetric muscle loss (VML) overwhelms the innate regenerative capacity of mammalian skeletal muscle (SkM), leading to numerous disabilities and reduced quality of life. Immune cells are critical responders to muscle injury and guide tissue resident stem cell– and progenitor-mediated myogenic repair. However, how immune cell infiltration and intercellular communication networks with muscle stem cells are altered following VML and drive pathological outcomes remains underexplored. Herein, we contrast the cellular and molecular mechanisms of VML injuries that result in the fibrotic degeneration or regeneration of SkM. Following degenerative VML injuries, we observed the heightened infiltration of natural killer (NK) cells as well as the persistence of neutrophils beyond 2 wk postinjury. Functional validation of NK cells revealed an antagonistic role in neutrophil accumulation in part via inducing apoptosis and CCR1-mediated chemotaxis. The persistent infiltration of neutrophils in degenerative VML injuries was found to contribute to impairments in muscle stem cell regenerative function, which was also attenuated by transforming growth factor beta 1 (TGFβ1). Blocking TGFβ signaling reduced neutrophil accumulation and fibrosis and improved muscle-specific force. Collectively, these results enhance our understanding of immune cell–stem cell cross talk that drives regenerative dysfunction and provide further insight into possible avenues for fibrotic therapy exploration.
The traumatic or surgical loss of a critical volume of skeletal muscle (SkM) that results in a permanent functional impairment (1), known as volumetric muscle loss (VML), is responsible for more than 90% of all muscle conditions that lead to long-term disability and is the principal cause of nearly 10% of all medical retirements from the military (2). VML results in reduced quality of life through pronounced disabilities (3) ranging from reduced muscle function and atrophy to aggressive development of osteoarthritis (4). An increasing prevalence of extremity traumas during combat since the turn of the century has incentivized increased therapeutic development for VML (5). Toward this end, research groups have developed and tested strategies ranging from acellular biological scaffolds (6–8) to stem cell transplants (9), drug therapies (3, 10), and rehabilitation (11), as well as various combinations of each (12–14). There is considerable variation in the success of such strategies (13), and further improvements in these results may be inhibited due to the loss of architectural units required for proper tissue reconstruction (15) as well as the prolonged inflammation and inhibitive microenvironment produced by the fibrotic scar (13, 16, 17). However, little information exists for why VML permanently overwhelms the regenerative capacity of SkM, resulting in pathological fibrosis and degeneration.
The regenerative capacity of SkM is dependent upon a pool of resident muscle stem cells (MuSCs), also known as satellite cells (18). Upon muscle injury, quiescent MuSCs are triggered to proliferate, differentiate into muscle progenitor cells, and fuse to form new or repair existing myofibers (18). The immune system plays a complex role in communicating with MuSCs during the regenerative response and guides healing outcomes (19, 20). Following SkM damage, a wave of proinflammatory cells, including neutrophils, macrophages, and conventional T lymphocytes, infiltrate the injured site and encourage MuSC proliferation through proinflammatory cytokine secretion, while regulatory T cells (21, 22) balance the magnitude and duration of the inflammatory response. Once the debris has been cleared and the progenitor pool has appropriately expanded, the immune microenvironment transitions to an anti-inflammatory state to coordinate matrix rebuilding and the formation of new myotubes. As part of this transition, macrophages undergo a phenotypic switch to support myogenic progenitor differentiation and myofiber growth (23, 24). However, after VML, macrophages persist in the defect for weeks to months (13) and their polarization switch is impaired (25), resulting in an intermediate phenotype (26, 27). Moreover, VML triggers an increase in T helper and cytotoxic T lymphocytes (28), which persist as long as 4 wk postinjury (27) compared to returning to preinjury levels within 7 to 10 d, as occurs in models of successful muscle regeneration (29). Comprehensive etiological assessment after VML and how dysregulated immune signaling networks converge to influence MuSC functions remain enigmatic. Elucidating these mechanisms stands to potentiate the development of regenerative therapies.
Herein, we characterized the cellular and molecular mechanisms driving responses of SkM by comparing VML injuries that regenerate to those that degenerate and result in fibrosis. Using immunohistofluorescent analyses, single-cell RNA sequencing (scRNA-Seq), lineage-tracing mouse models, cellular transplants, and small-molecule inhibition, we provide a resource that elucidates cellular and molecular players after VML. We observed and validated that degenerative VML injuries result in the persistent infiltration of inflammatory cells, such as neutrophils, which exert lasting consequences on the myogenic capacity of resident MuSCs. Further, we identified and characterized an intercellular communication circuit between neutrophils and cytolytic natural killer (NK) cells, which combat neutrophil accumulation via a CCL5/CCR1 axis. The inhibition of CCR1 exacerbated neutrophil accumulation in degenerative defects, while NK transplants significantly reduced neutrophil populations and enhanced healing. Finally, a cell–cell communication network prediction from scRNA-Seq data suggested TGFβ-conferred MuSC impairments. Small-molecule inhibition of TGFβ signaling in vivo improved tissue morphology and strength at late time points following degenerative VML injuries. Together, these findings enhance our understanding of cellular communication dynamics governing muscle healing outcomes.
RESULTS
Volumetric Muscle Loss Induces Muscle Degeneration and Fibrotic Supplantation.
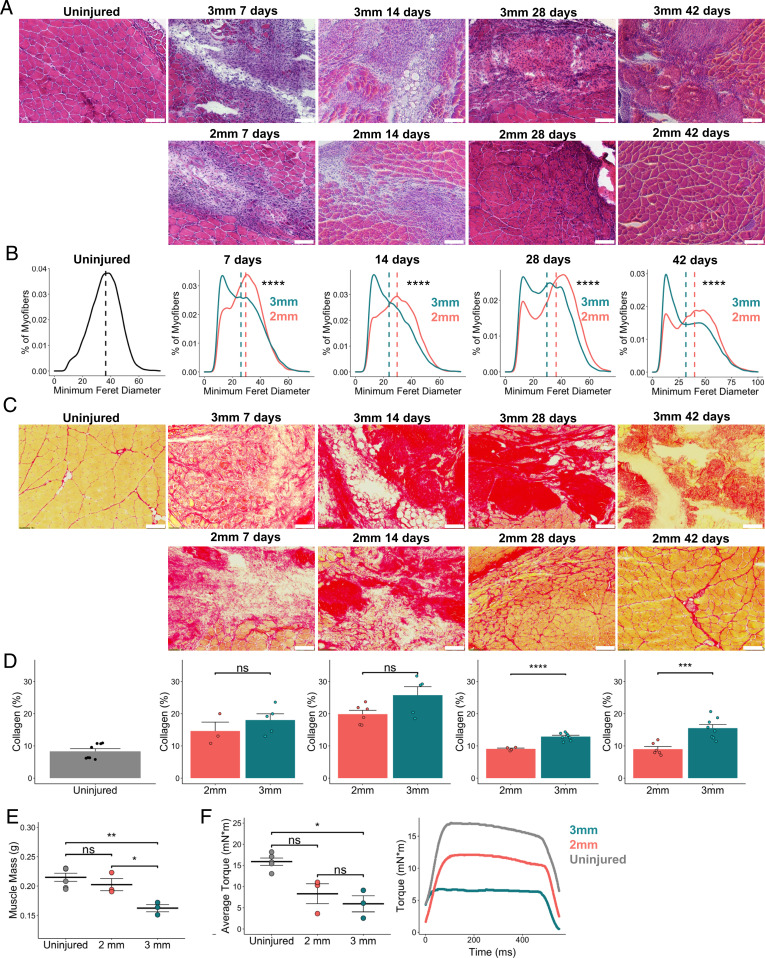
To model regenerative and degenerative muscle healing outcomes, we administered bilateral full-thickness 2-mm and 3-mm punch biopsy defects to the rectus femoris of adult C57BL6/J mice (30). Cross-sections of mouse quadriceps were stained with hematoxylin and eosin (H&E) and picrosirius red (PSR) and immunohistochemically stained for laminin at 7, 14, 28, and 42 d postinjury (dpi) to assess muscle degeneration and fibrosis compared to uninjured controls. Consistent with previous literature (30), the 3-mm muscle defects failed to resolve by 42 dpi (subsequently termed degenerative defects), whereas regenerating myofibers filled the 2-mm defects (subsequently termed regenerative defects) by 28 dpi (Fig. 1A and SI Appendix, Fig. S1). Compared to regenerative defects, degenerative defects were further characterized by higher proportions of smaller fibers (Fig. 1B; n = 5 to 7 tissues from 3 to 4 mice), significantly increased collagen deposition at 28 and 42 dpi (Fig. 1 C and D and SI Appendix, Fig. S1; n = 3 to 7 tissues from 3 to 4 mice, unpaired), and muscle atrophy at 28 dpi (Fig. 1E; n = 3 to 5 tissues from 3 to 5 mice, unpaired). Degenerative defects also exhibited significant reductions in isometric torque at 28 dpi compared to uninjured controls (Fig. 1F; n = 3 to 5 tissues from 3 to 5 mice, unpaired). Combining these results shows that VML defects of different sizes beget distinct regenerative trajectories.
Fig. 1.
Degenerative VML defects exhibit increased fibrosis and reduced myofiber diameters. (A) Representative H&E stains of uninjured quadriceps along with 2-mm and 3-mm defects harvested 7, 14, 28, and 42 dpi. (Scale bars, 200 μm.) Single windows of the defect area are shown for detail. Representative full-section images are shown in SI Appendix, Fig. S1. (B) Portion of myofibers stratified by minimum Feret diameter shows smaller fiber size distribution following 3-mm defect. ****P < 0.0001 by two-sample, two-sided Kolmogorov–Smirnov test for equal distributions. Feret diameters were calculated for 5 to 7 full-section images each from a distinct defect, per group, and pooled for comparison of distributions. Dashed lines indicate mean minimum Feret diameters. (C) Representative PSR stains of 2-mm and 3-mm defects throughout the time course. (D) Quantification of collagen content showing a return to preinjury levels following 2-mm defects by 28 dpi but a significant increase and persistence following 3-mm defects. (Scale bars, 200 μm.) Bars show mean ± SEM; ns denotes not significant (P > 0.05). ****P < 0.0001, ***P < 0.001 by two-sided t test. n = 3 to 7 tissues per group. Cohen’s d = 0.7566, 1.2678, 2.2536, and 2.4264 for 7, 14, 28, and 42 dpi, respectively. Quantifications were performed on full section stitched images (representative images shown in SI Appendix, Fig. S1). Single windows of the defect area are shown for detail. (E) Muscle mass is significantly reduced among limbs that received 3-mm defects at 28 dpi compared to uninjured limbs and those that received 2-mm defects. **P < 0.01, *P < 0.05, ns denotes P > 0.05 by one-way ANOVA and posthoc analysis. n = 3 to 5 tissues per group. Bars show mean ± SEM. (F) Average isometric torque (Left) and representative force curves (Right) of uninjured quadriceps or quadriceps 28 d following 2-mm or 3-mm injuries. *P < 0.05, ns denotes P > 0.05 by one-way ANOVA and posthoc analysis. n = 3 to 5 tissues per group. Bars show mean ± SEM.
scRNA-Seq Reveals Alterations in Cellular Ecosystem in Response to Degenerative Muscle Defects.
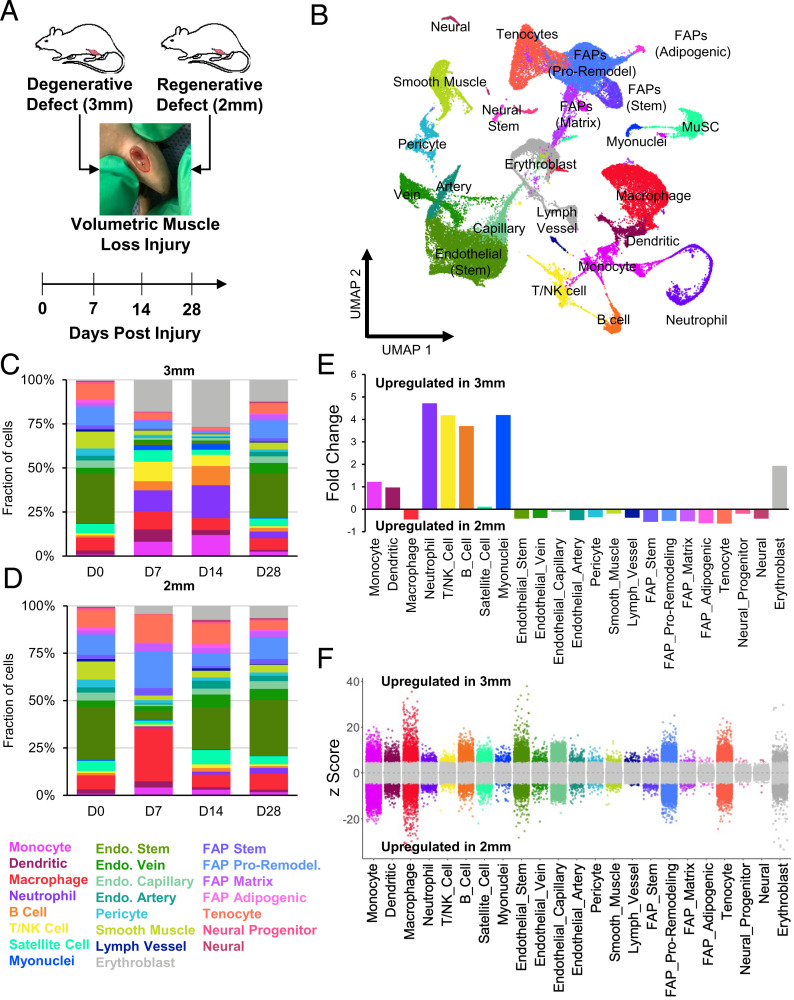
To probe the cellular and molecular drivers that contribute to the regenerative versus degenerative responses, we performed droplet-based scRNA-Seq on viable mononucleated cells isolated from uninjured quadriceps (0 dpi, 7 dpi, 14 dpi, and 28 dpi) (Fig. 2A). We generated 58,405 high-quality scRNA-Seq libraries encompassing on average 1,875 genes per cell with an average read depth of 7,489 unique molecular identifiers per cell (SI Appendix, Fig. S2 A and B). Technical variations between batches were regressed out using Seurat version 3 (31), followed by dimensionality reduction using Uniform Manifold Approximation and Projection (UMAP) (32) (SI Appendix, Fig. S2C). Unsupervised Louvain clustering revealed 23 cell types (Fig. 2B). Cluster-based cell-type annotation was performed by matching cluster marker genes to a muscle-specific cell taxonomy reference database using the single-cell Cluster-based Annotation Toolkit for Cellular Heterogeneity (scCATCH) (33) as well as by overlaying marker gene expression with previously published reference datasets (34–36) (SI Appendix, Fig. S2D). We observed variation in the proportion and type of captured cells among degenerative defects compared to regenerative defects, whereby increases in neutrophils and lymphocytes were observed in degenerative defects at 7 and 14 dpi compared to cells sequenced from regenerative defects at the same time points (Fig. 2 C and D). When we merged all time points, the scRNA-Seq results suggested a four- to fivefold increase in lymphocytes and neutrophils among degenerative defects (Fig. 2E). Further, substantial differences in gene expression were observed among most cell populations isolated from degenerative versus regenerative defects using the MAST toolkit (37, 38) (Fig. 2F). A protein annotation through evolutionary relationship (PANTHER) (39) pathway analysis of the top 150 differentially expressed genes (DEGs) up-regulated among degenerative defects identified increased chemokine- and cytokine-mediated inflammation (Il1b, Ccl5, Ccr7, Ccr1, Rac2, Acta1, Actb, Actc1, Actg1, Arpc3, and Alox5ap), T and B lymphocyte activation (Trbc2, MHCII antigens, Cd3d, Cd79a and b, Ighm, and Rac2), glycolysis (Aldoa, Gapdh, Pgam2, and Bpgm), and apoptosis signaling (Ltb, heat shock proteins, and Cycs). Most of these genes were expressed among dendritic cells (Il1b, Ccr7, Rac2, Alox5ap), neutrophils (Il1b, Ltb, Ccr1, Alox5ap), T/NK cells (Ccl5, Ltb, Rac2, Cd3d, Trbc2, and heat shock proteins), and B cells (Rac2, Ccr7, Ltb, Cd79, Ighm). Moreover, GO term analysis on up-regulated genes from regenerative defects showed enrichment in terms associated with extracellular matrix (ECM) organization and cell migration (SI Appendix, Fig. S2E). Finally, we validated the proportions of T cells and NK cells using flow cytometry from regenerative and degenerative defects after 7 dpi and observed cell abundances largely similar to our scRNA-Seq observations (SI Appendix, Fig. S3 A and B; n = 6 muscles from 3 mice, unpaired). These results suggest that degenerative VML defects induce stronger and sustained inflammatory responses compared to VML injuries that heal.
Fig. 2.
scRNA-Seq of regenerative and degenerative muscle defects show exacerbated and persistent inflammation in injuries that do not heal. (A) Schematic of experiment, whereby adult (10 to 12 wk) mice were administered 2-mm or 3-mm biopsy punches to their rectus femoris and humanely euthanized before injury, or 7, 14, or 28 dpi for scRNA-Seq analysis. (B) Dimensional reduction and unsupervised clustering of mononucleated cells isolated from uninjured quadriceps as well as injured quadriceps at 7, 14, and 28 dpi showing 23 different recovered cell types according to marker gene overlays and scCATCH cluster annotation. Two mice were pooled and sequenced for each defect size at each time point, and between 2,337 and 7,500 high-quality libraries were generated for each condition. Quantification of cell abundances at each time point sequenced following (C) 3-mm and (D) 2-mm VML defects shows increased and persistent inflammation among 3-mm defects. (E) Fold changes in cell abundance following 3-mm defects in comparison to 2-mm defects merged across all time points shows nearly fivefold increase in neutrophils, B cells, and T and NK cells. (F) Differential gene expression among each cell type merged across time points and normalized to 2-mm defects. Gray region indicates adjusted P value less than 0.05. Z-scores and P values were calculated for each gene using MAST.
Degenerative VML Injuries Result in Variations of Inflammatory Cell Infiltration.
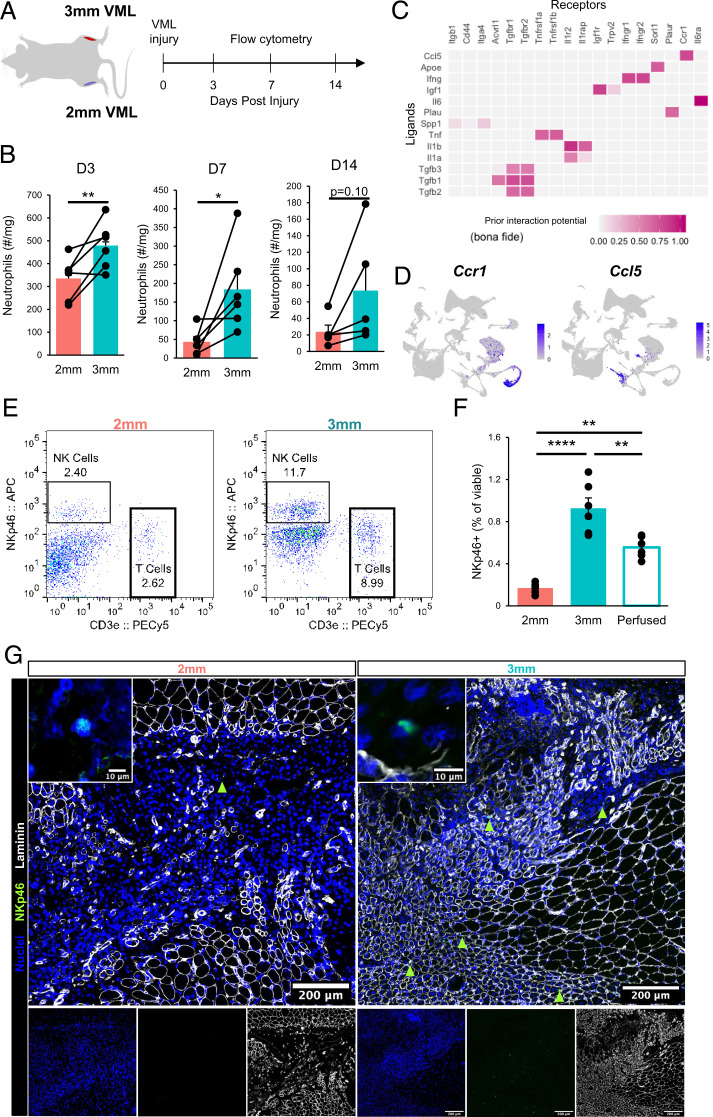
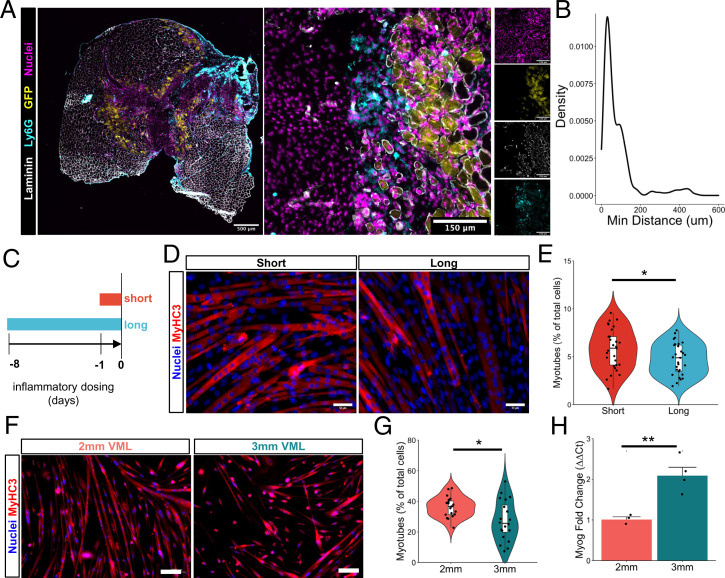
Our scRNA-Seq results predicted increases in neutrophils in degenerative injuries, which play a primary role in the clearance of tissue debris after injury and secrete a myriad of proinflammatory cytokines and chemokines to recruit and activate inflammatory macrophages and T lymphocytes before being rapidly cleared (20, 40). To validate these predictions, we isolated neutrophil populations (CD11b+Ly6G+) after multiple time points following regenerative and degenerative injuries and enumerated their fractions using flow cytometry (Fig. 3A). Consistent with our scRNA-Seq observations, we observed significant increases in neutrophil counts for degenerative VML injuries at 3 and 7 dpi and a trend toward increased neutrophils at 14 dpi (Fig. 3B and SI Appendix, Fig. S3C, n = 5 to 6 tissues from 5 to 6 mice, paired). To understand the mechanisms governing neutrophil accumulation following degenerative VML injuries, we employed the recently developed NicheNet (41) algorithm. NicheNet integrates prior knowledge on ligand-target pathways with transcriptomic expression data to predict influential ligands, match them with target genes (which we defined according to differential expression across defects), and identify possible signaling mediators. NicheNet predicted that one of the top ligand–receptor interactions driving neutrophil responses in 3-mm defects was CCL5/CCR1 signaling (Fig. 3C). CCL5 was expressed almost exclusively in NK and T cells, while CCR1 was most highly expressed in neutrophils (Fig. 3D). While more than fourfold increases in all lymphocytes were observed in degenerative defects from our scRNA-Seq data, one of the largest lymphocyte populations, and the primary source of CCL5, was NK cells (Fig. 3D and SI Appendix, Fig. S4A and B). To confirm the increased influx of NK cells into degenerative defects, we performed flow cytometry on degenerative and regenerative muscle defects at 7 dpi (Fig. 3 E and F; n = 6 tissues from 3 mice, unpaired) and observed approximately 4 times as many NK cells in degenerative defects compared with regenerative defects, which only slightly decreased after saline perfusion. Moreover, qualitative immunofluorescent stains revealed that NK cells are situated at the periphery of the VML defects (Fig. 3G; n = 6 tissues from 3 mice, unpaired).
Fig. 3.
Perturbations in inflammatory cell signaling after degenerative muscle injury. (A) Schematic of experiment design whereby mice received 3-mm defects on one limb and 2-mm defects on the contralateral limb, then neutrophils were quantified by flow cytometry (PI−CD11b+Ly6g+) at 3, 7, and 14 dpi. (B) Time course of neutrophil infiltration in 2-mm and 3-mm defects as quantified by flow cytometry. Bars show mean ± SEM. n = 5 to 6 defects from 5 to 6 mice per time point. *P < 0.05, **P < 0.01 by two-sided, paired t test. Cohen’s d = 2.1, 1.17, and 0.94 for 3, 7, and 14 dpi, respectively. (C) Top-ranked NicheNet ligand-receptor pairs based on prior literature and their prediction of downstream target gene expression. Ccl5 is one of the top ligands predicted to influence neutrophils following 3-mm defects. (D) UMAP overlays of Ccr1 and Ccl5 expression showing neutrophils with the highest Ccr1 mRNA expression and T and NK cells with the highest Ccl5 mRNA expression. (E) Representative flow cytometry scatter plots showing T cell and NK cell abundance in 2-mm vs. 3-mm VML defects at 7 dpi. (F) Flow cytometry quantification of NK cell abundance in 2-mm and 3-mm defects 7 dpi and in 3-mm defects 7 dpi following whole-body saline perfusion prior to dissection. Graph shows mean ± SEM. **P < 0.001, ****P < 0.0001 by one-way ANOVA and Bonferroni posthoc analysis. n = 6 injuries per group. n = 9 mice. Effect size = 2.21. (G) Immunofluorescent stains qualitatively show NK cell localization primarily within the defects and increased numbers in 3-mm defects. (Scale bars, 200 μm; Inset, 10 μm.)
NK cells rapidly produce and secrete proinflammatory cytokines (42) and have previously been reported in SkM (22, 43, 44), though their roles remain undefined. In addition to numerous NK cell marker genes (Ncr1, Nkg7, Klre1, Klra8, Klra8) (45), differential gene expression analysis revealed elevated expression of proinflammatory molecules (Gzma, Gzmb, Fcer1g) and chemokines (Ccl3, Ccl4, Ccl5) among NK cells compared with the recovered T cells (SI Appendix, Fig. S4C). Additionally, NK cells were observed to display a strong expression of genes associated with activation and cytolytic function (Ncr1, Klrk1, Gzma, Gzmb, Fasl) and cytokine secretion (Klrc1, Ifng, Ltb) (SI Appendix, Fig. S4D). Summing these results showed that NK cells and neutrophils display enhanced infiltration following degenerative VML injuries and may communicate via a CCL5-CCR1 circuit.
NK Cells Interact with Neutrophils in Degenerative VML Injuries.
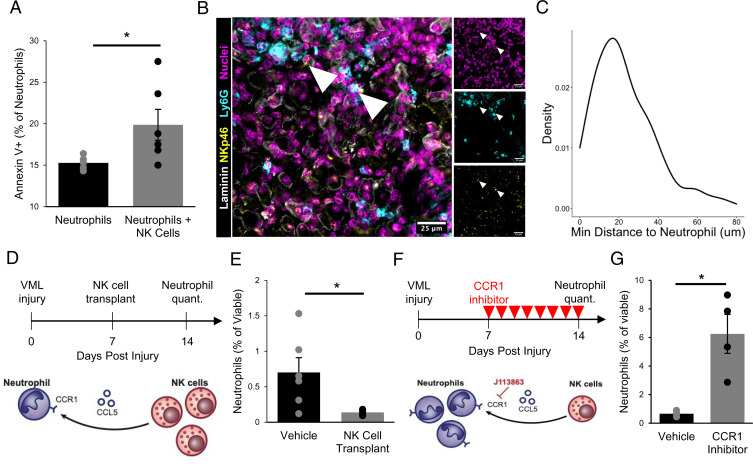
To elucidate the impact of NK cells on neutrophils in VML injuries, we first cocultured neutrophils isolated from degenerative VML defects at 7 dpi with activated NK cells. Consistent with studies showing that human NK cells induce contact-dependent neutrophil apoptosis (46), we observed significantly increased neutrophil apoptosis after 4 h (Fig. 4A and SI Appendix, Fig. S5A; n = 6 wells using neutrophils from 4 mice, unpaired). To determine whether such communication could be occurring in vivo, we next evaluated the colocalization of NK cells and neutrophils within degenerative VML defects at 7 dpi using immunofluorescence. More than 90% of NK cells were within 50 μm of the nearest neutrophil, which is within the previously reported maximum cytokine propagation distances up to 250 μm depending on the density of receptors (47, 48) (Fig. 4 B and C; n = 159 NK cells and 397 neutrophils, from 4 defects, unpaired). These results suggest that NK cells induce neutrophil apoptosis and are proximal to neutrophils after degenerative VML injuries.
Fig. 4.
NK cells interact with neutrophils in VML injuries. (A) NK cells induce neutrophil apoptosis in coculture based on increased annexin V+ cells, as analyzed by flow cytometry. *P < 0.05. n = 5 to 6 wells. Each well contains a pool of neutrophils that are FACS-enriched from four 3-mm VML defects 7 dpi. NK cells were MACS-enriched from one spleen and activated in vitro for 48 h. Bars show mean ± SEM. (B) Immunofluorescent stains for DAPI, NKp46, Ly6G, and laminin in VML defects 7 dpi show colocalization of neutrophils and NK cells. (Scale bar, 25 μm.) (C) Distributions of minimum Euclidean distances between NK cells; 92.5% of NK cells are within 50 μm of the nearest neutrophil. n = 159 NK cells and 397 neutrophils from nine 60× images of four defects. (D) Schematic whereby 50,000 in vitro activated NK cells were transplanted into degenerative (3-mm) defects at 7 dpi. Contralateral control limbs received PBS injections. Neutrophils were quantified at 14 dpi using flow cytometry (CD11b+Ly6g+). (E) NK cell transplant significantly reduced neutrophil abundance. *P < 0.05 by two-sided, two-sample t test. Cohen’s d = 1.56. n = 5 to 6 muscles (n = 6 mice), repeated twice. Graph shows mean ± SEM. (F) Schematic of experiment design whereby a cohort of mice received degenerative (3-mm) defects followed by daily intraperitoneal injection of CCR1 inhibitor (J113863) between 7 and 14 dpi. Neutrophil populations were quantified by flow cytometry (CD11b+Ly6g+) at 14 dpi. (G) CCR1 inhibition significantly increased neutrophil abundance in 3-mm defects, to a similar extent as the NK cell transplant reduced neutrophil populations, suggesting that NK cells may locally recruit neutrophils via CCL5 secretion to induce contact-dependent apoptosis. *P < 0.05 by two-sided, two-sample t test. n = 3 to 4 mice per group, repeated twice. Cohen’s d = 2.64. Graph shows mean ± SEM.
Given our results that NK cells had transcriptional profiles aligning with a cytolytic phenotype and induced neutrophil apoptosis in vitro, we reasoned that NK cell transplantation into VML defects would reduce neutrophil abundance (Fig. 4D). We transplanted 50,000 in vitro activated NK cells into degenerative (3-mm) and regenerative (2-mm) VML injuries at 7 dpi and quantified the neutrophils and NK cells at 14 dpi using flow cytometry (Fig. 4D and SI Appendix, Fig. S5 B and C). Though the transplanted NK cells had largely cleared by 14 dpi (SI Appendix, Fig. S5D; n = 3 tissues from 3 mice, paired), we observed a significant reduction of neutrophils in response to NK cell transplantation following both degenerative (Fig. 4E; n = 5 to 6 tissues from 6 mice, unpaired) and regenerative injury (SI Appendix, Fig. S5E, n = 6 tissues from 6 mice, paired).
To determine whether NK–neutrophil communication occurs through CCR1 signaling, we administered a small-molecule CCR1 inhibitor, J-113863, daily starting 7 d after degenerative VML injury (Fig. 4F). At 14 dpi, we observed a substantial increase in neutrophil abundance (Fig. 4G; n = 4 tissues from 4 mice, unpaired) without significant changes to overall immune cell (CD45+) abundances (SI Appendix, Fig. S5F; n = 4 tissues from 4 mice, unpaired). Taken together, these results suggest that after VML injury, NK cells reduce neutrophil abundance through contact-mediated apoptosis and that this effect is in part regulated or reinforced by CCR1 signaling.
Degenerative VML Injuries Engender Impairments in Muscle Progenitor Fusion.
The persistence of neutrophils after injury can have deleterious consequences for tissue repair, and failure to appropriately clear these cells from the injury site can lead to secondary damage as well as inefficient regeneration (20, 40, 49). To determine whether prolonged infiltration of neutrophils observed in degenerative VML injuries were colocated with MuSCs or their progeny, we administered degenerative VML defects (3 mm) to adult Pax7cre × Rosa26mTmG mice, which express membrane-tagged green fluorescent protein (GFP) in all Pax7+ cells and their progeny following tamoxifen induction. To capture both regenerating fibers and neutrophil persistence, we collected the quadriceps at 14 dpi and stained for GFP, neutrophils (Ly6G), laminin, and nuclei using immunofluorescence (Fig. 5A). We did not detect MuSCs or GFP+ myofibers within the defect but did observe that ∼75% of GFP+ myofibers were within 250 μm of the edge of the defect. In contrast, a large majority of Ly6G+ cells were observed within the defect (∼85%) and were interspersed among or adjacent to GFP+-regenerating myofibers at the defect edge (Fig. 5B; n = 382 neutrophils and 410 GFP+ fibers from 20× images of 4 defects; SI Appendix, Fig. S6 A and B; n = 5 full section stitched images from 5 tissues). These results suggest that MuSCs and their progeny are exposed to neutrophil signaling for extended periods in degenerative VML injuries.
Fig. 5.
Neutrophil secretome impairs myoblast fusion in accordance with reduced muscle stem cell regenerative capacity following critical-sized defects. (A) Representative image showing interspersion of neutrophils (cyan) within GFP+-regenerating myofibers (yellow) at 14 dpi. (B) Distributions of minimum Euclidean distances between neutrophils and GFP+-regenerating fibers. More than 93% of neutrophil cells are within 250 μm of the nearest regenerating fiber. n = 382 neutrophils and 410 GFP+ fibers from 20× images of four defects at 14 dpi. (C) Schematic of experiment design, whereby C2C12s are cultured in neutrophil-conditioned myoblast media for 8 (long) or 1 (short) d before addition of differentiation medium without conditioning. Fusion was assessed 72 h later. (D) Representative immunofluorescent images of C2C12s cultured for long or short durations in neutrophil-conditioned medium, then induced to differentiate for 72 h in low-serum media. Blue indicates nuclei (DAPI) and red indicates embryonic myosin heavy chain (MYH3). (Scale bar, 50 mm.) (E) Quantification of number of myotubes as percentage of total cells after short or long exposure to neutrophil-conditioned medium. *P < 0.05 by two-sided, two-sample t test after removal of one outlier. n = 29 10× images taken from three culture wells. (F) Representative immunofluorescent images of primary myoblasts harvested 28 dpi following regenerative (2-mm) and degenerative (3-mm) defects and cultured in differentiation medium for 72 h. Blue indicates nuclei (DAPI) and red indicates embryonic myosin heavy chain (MYH3). (Scale bar, 100 mm.) (G) Quantification of fusion index according to the fraction of nuclei located in myofibers containing at least two nuclei, calculated manually. *P value < 0.05 by two-sided, two-sample t test. n = 20 10× images per defect from three mice per defect size, repeated twice. Cohen’s d = 0.86. (H) Fold change in Myog expression measured by qRT-PCR following in vitro differentiation of MuSCs isolated from 2-mm and 3-mm defects at 28 dpi. Bars show mean ± SEM. **P < 0.01 by two-sided, two-sample t test. n = 3 to 4. Replicates with Cq values > 30 were excluded from further analysis.
To understand the potential consequences of the prolonged exposure of myogenic progenitors to the neutrophil secretome in the context of degenerative VML injuries, we cultured immortalized myoblasts (C2C12s) with neutrophil-conditioned growth medium for a short (1 d) or long (8 d) period, then induced differentiation and fusion (Fig. 5C and SI Appendix, Fig. S6C). We reasoned that the short exposure would mimic the quick inflammatory burst that follows regenerative defects, while the long exposure would mimic the observed significant increase in neutrophil presence beyond 7 dpi among degenerative defects. As a result of prolonged exposure to the neutrophil secretome, we observed a reduction in myoblast fusion (Fig. 5D and E; n = 30 images from 3 culture wells, unpaired), which is consistent with recently published findings (50). Based on the observed impairments in fusion, we examined whether MuSCs isolated from degenerative and regenerative defects that had been exposed to neutrophil signaling for different periods also displayed fusion defects. We isolated MuSCs at 28 dpi after inflammation had largely resolved and evaluated their ability to differentiate and fuse. Consistent with our previous findings, we observed reductions in fusion (Fig. 5F and G; n = 20 images from 5 culture wells, unpaired) among MuSCs enriched from degenerative defects compared to those enriched from regenerative defects. The negative changes in fusion were not results of differentiation impairments, as Myog expression was higher among MuSCs from degenerative defects compared to regenerative defects (Fig. 5H; n = 3 to 4 culture wells, unpaired). Integrating these results suggests that MuSC differentiation is not impaired following VML, but fusion may be attenuated, and this effect may be partially conferred by extended exposure to the neutrophil secretome.
To determine the drivers of impairments to MuSC fusion between degenerative and regenerative defects, we reclustered the myogenic scRNA-Seq clusters and performed trajectory and differential expression analyses (SI Appendix, Fig. S7A–C). The largest number of DEGs across all time points and both defects were observed among MuSCs harvested from degenerative defects at 7 dpi (455 genes, log-fold change > 1.5, adjusted P value < 0.05). Among degenerative defects, the most up-regulated genes (ordered according to adjusted P values) were associated with myogenic progenitor activation [Itm2a, Cdkn1c, Ckb (51), Atp2a1, Cebpb (52, 53)], metabolism [Mt1 and Mt2 (54), Gamt, Socs3), fibrosis (Sparc) (55)], stress response, and inflammation [Fos, Fosb, Jun, S100a8, S100a9, Prdx5, Mif (56)] (SI Appendix, Fig. S7D). A PANTHER (39) pathway analysis displayed enriched terms for the regulation of apoptosis (17.0% of up-regulated genes), increased the regulation of metabolism (35.7% of up-regulated genes), and elevated a stress response (24.9% of up-regulated genes). The expression of many genes associated with myoblast fusion (GO:0007520) was reduced among MuSCs in degenerative defects compared to those in regenerative defects at the same time point, and, consistent with our previous results, MuSCs from degenerative defects were observed to strongly express Myog compared to MuSCs from regenerative defects at the same time points (SI Appendix, Fig. S7E). We did not observe differences in MuSC numbers or apoptosis at 14 dpi (SI Appendix, Fig. S7 F and G; n = 6 tissues from 6 mice, paired). However, there was a significant difference in β1 integrin expression in MuSCs isolated from degenerative defects by flow cytometry (SI Appendix, Fig. S7H; n = 6 tissues from 6 mice, paired). Together, these results suggest that the inflammatory milieu in degenerative defects induces variations in stress response, metabolism, and matrix attachment that may contribute to reductions in fusion and myogenic repair.
Targeting Predicted Intercellular Interactions Alleviates Muscle Fibrosis by Helping to Resolve MuSC Impairments.
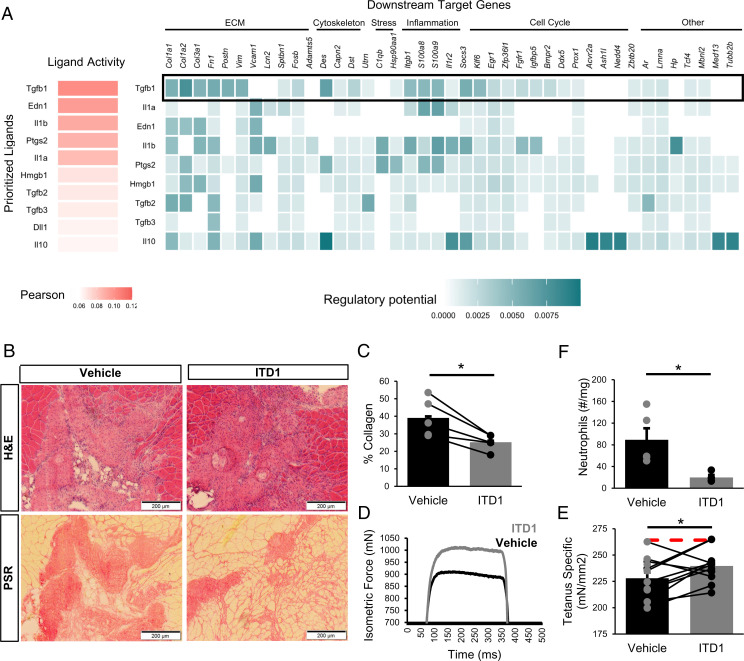
To elucidate which intercellular communication networks drive the observed differential gene expression patterns among MuSCs and impairments in fusogenic behavior, we again employed NicheNet. We observed increases in interleukin 1 (IL-1) and TGFβ1 among degenerative defects as potential drivers of the altered MuSC response (Fig. 6A), influencing the transcription of genes associated with attachment (Vcam1, Itgb1), stress response (Fos, Fosb, Jun, C1qb, S100a8, S100a9, Hsp90aa1), and ECM synthesis (Col1a1, Col3a1, Fn1) (SI Appendix, Fig. S8A). Many of the gene targets of IL-1 and TGFβ1 signaling were identified by RNA velocity dynamical modeling as putative drivers governing myogenic cell fate transitions (SI Appendix, Fig. S8B), further supporting the influence of IL-1 and TGFβ1 signaling on altered MuSC regenerative kinetics. Il1b was highly expressed among neutrophils and dendritic cells. Tgfb1 was expressed in various cell types, though the most drastic up-regulation across defect sizes was observed among macrophages and monocytes (SI Appendix, Fig. S8C). TGFβ1 has previously been shown to impair MuSC fusion (57, 58) but not differentiation (59), consistent with our in vitro observations (Fig. 5) and scRNA-Seq datasets following degenerative muscle defects (SI Appendix, Fig. S7). Moreover, TGFβ1 is one of the most potent chemokines for neutrophils (60) and was predicted by NicheNet to be a top regulator of neutrophil populations in this system (Fig. 2C). Thus, to evaluate the impact of TGFβ signaling on regeneration, we locally blocked the TGFβ signaling axis following degenerative VML injury with a TGFβ receptor II inhibitor (ITD1) through intramuscular injection. At 28 dpi, there was a significant reduction in collagen deposition (Fig. 6 B and C; n = 5 tissues from 5 mice, paired). Moreover, ITD1 treatment yielded increases in maximal tetanic force, with and without normalization to the muscle cross-sectional area, suggesting functional improvements (Fig. 6D and E and SI Appendix, Fig. S8 D–I; n = 10 tissues from 10 mice, paired). Since TGFβ1 is chemotactic for neutrophils and was among the ligands that NicheNet predicted to influence neutrophils, we evaluated whether ITD1 treatment reduced neutrophil accumulation in degenerative defects using flow cytometry. In accordance with predictions, significantly fewer neutrophils were observed in ITD1-treated defects (Fig. 6F; n = 4 tissues from 2 mice, unpaired). Cumulatively, our results support a network driving VML-induced muscle degeneration whereby neutrophils infiltrate and persist in degenerative defects, and with elevated TGFβ signaling, impair fusion of MuSCs (52, 61).
Fig. 6.
Inhibition of TGFβ signaling partially restores muscle regeneration and reduces fibrosis after VML injury. (A) NicheNet ranking of the ligands that best predict MuSC differential gene expression at 7 dpi. Pearson correlation coefficient indicates the ability of each ligand to predict the expression of differentially expressed genes. TGFβ1 is predicted to be a key contributor to MuSC dysfunction. (B) Representative H&E and PSR images from 3-mm defects 28 dpi following intramuscular treatment with ITD1 (a TGFβ-signaling inhibitor) or vehicle every 3 d until 15 dpi. (C) Defects treated with ITD1 showed reduced collagen deposition. Collagen was calculated as a percentage of the 10× magnification image of the defect that was stained red by PSR. n = 4 tissues per condition (n = 4 mice), where contralateral limbs were treated with vehicle. *P < 0.05 by two-sided, paired t test. Bars show mean ± SEM. Cohen’s d = 1.83. (D) Representative force curves for muscles treated with ITD1 or vehicle. (E) Specific force following muscle stimulation of injured TA muscles 28 dpi improved by ITD1 treatment. Bars show mean ± SEM, and red dashed line indicates specific force of uninjured, untreated TA muscles. *P = 0.023 by two-sided, paired t test. n = 10 muscles (n = 10 mice), Cohen’s d = 1.1. (F) Neutrophil abundance following ITD1 treatment, as quantified by flow cytometry 14 dpi, shows that TGFβ signaling inhibition reduces neutrophil recruitment. Boxplots show mean ± SEM. n = 4 defects (n = 4 mice). *P = 0.025 by two-sided, two-sample t test. Cohen’s d = 1.91.
DISCUSSION
Skeletal muscle displays a remarkable ability to regenerate following injuries through the coordinative actions of MuSCs (62). Yet the dysregulated cellular and molecular mechanisms that develop after VML and prevent MuSC-based regeneration remain elusive, limiting therapeutic efficacy and restoration of function. Understanding the drivers of this pathological behavior is critical to glean the effects of new therapeutic interventions targeting these circuits as well as improve existing therapeutic modalities. Herein, we use scRNA-Seq to characterize injuries that result in degeneration and fibrosis and show that degenerative VML displays exacerbated and prolonged inflammation that negatively influences the regenerative capacity of resident MuSCs. These datasets and results offer a rich resource to further improve our understanding of the VML etiology as well as the restorative benefits of therapies.
Regeneration of skeletal muscle after injury is critically dependent on multiple types of immune cells that remove debris, condition the injury site with inflammation to prevent infection, and signal to resident stromal and stem cells to guide repair (40). In accordance with other muscle-regenerative injury models (40), neutrophil abundance in regenerative VML defects peaked after injury and returned to baseline before 7 d. However, in degenerative VML injuries, neutrophils persisted beyond 1 wk post injury and were colocated with regenerating myofibers. Collateral skeletal muscle damage caused by neutrophil-derived oxidants, as well as the ability to attenuate such damage by blocking neutrophil activity, has been extensively studied in myopathies, age-associated muscle decline, and various models of muscle injury and fibrosis (63). In line with these observations (64, 65), we observed that sustained exposure to neutrophil secretomes in vitro resulted in decreases in fusion of MuSCs and correlations between reduced neutrophil abundance and improved functional recovery in vivo. As such, targeting neutrophils and impacting the timeline of neutrophil clearance after VML may enhance the efficacy of regenerative therapies and alter fibrotic versus regenerative outcomes.
A corollary of increased and sustained neutrophil infiltration in degenerative VML injury was increases in a compensatory population of NK cells. We did not observe NK cells at later time points (>14 d) after degenerative injury, suggesting that these cells are not targeting fibrotic cells but rather play an immunoregulatory role by targeting neutrophil populations. The enhancements in apoptosis in vitro as well as the timing of NK cell infiltration, gene expression profile, and colocalization with neutrophils suggest that NK cells function to attenuate the negative inflammation induced from VML by locally recruiting and inducing neutrophil apoptosis (46). This is consistent with previous reports demonstrating an anti-inflammatory role of NK cells following autoimmune myocarditis (66). While we cannot exclude that additional factors and other cell types may also interact with NK cells (67), cross talk between neutrophils and NK cells has also previously been observed in cancer and chronic infections (68), but their interactions in muscle have not been elucidated. A seminal question is why degenerative VML injury results in sustained inflammation and fibrosis despite the presence of NK cells that act to induce neutrophil apoptosis. A clue to the mechanism of this behavior was increased TGFβ signaling in degenerative defects, which inhibits NK cell cytotoxicity (69). Further exploration of the capacity of NK cells to control neutrophil-based inflammation in the pathological microenvironment of VML injuries is warranted, as well as how this cross talk contributes to the behavior of T cells and macrophages.
The role of elevated TGFβ in muscle degeneration following VML most likely results from a complex network of macrophage and fibro-adipogenic progenitor (FAP) interactions (70). FAPs are muscle resident cells that function as important regulators of ECM deposition. In response to exacerbated or chronic inflammation, FAPs act as pathological drivers of muscle fibrosis, intramuscular fatty infiltration, and heterotopic ossification (71). TGFβ signaling is a critical determinant of FAP activity and response (72), and previous efforts to reduce TGFβ after VML have shown reductions in fibrosis (72, 73). Beyond reported influences on ECM secretion by mesenchymal cells and negative impacts on MuSC activation, differentiation (3), and fusion (58, 59), TGFβ may also contribute to persistent neutrophil infiltration (60, 74). Moreover, it is possible that TGFβ contributes to a feed-forward degenerative loop, whereby neutrophil secretion of IL-1β increases macrophage secretion of TGFβ1 (75), which then recruits more neutrophils (60, 74). This cascade has potent effects on mesenchymal stem cells and fibroblasts to synthesize collagen and tissue inhibitor of metalloproteinase 1(TIMP1) (75), which in turn impairs MuSC regenerative capacity by manipulating activation, migration, and differentiation (76, 77). In line with this degenerative loop, we observed differential gene expression patterns between regenerative and degenerative MuSCs and alterations in attachment through β1 integrin (78). These results suggest that aberrant cortical tension (79) from exacerbated ECM deposition and composition (80, 81) may alter membrane remodeling and the fusogenic behavior of MuSCs (82), (83). Consistent with this and other previous findings (59), a blockade of TGFβ signaling following degenerative VML injury reduced collagen deposition and helped recover muscle strength, though treatment duration and and/or delivery modality (systemic versus local delivery) may stimulate variations in the level of function restored (84). Because the macrophage secretome, including TGFβ1, is so critical to MuSC-mediated repair and sensitive to the local milieu (85), additional exploration into how macrophage-MuSC cross talk networks are dysregulated as a result of VML-induced microenvironmental changes is needed.
The cellular and molecular mechanisms driving pathological remodeling and degeneration following VML injuries remains underexamined. The approaches used in the present study begin to elucidate inter- and intracellular signaling networks that are dysregulated as a result of critical injuries, contributing to muscle atrophy and fibrotic development. We envision that these insights will provide a valuable resource for further exploration into mechanisms preventing healing after VML and therapeutics targeting those mechanisms.
Materials and Methods
Animals.
Adult (3 to 4 mo) female and male mice were obtained from Jackson Laboratories or a breeding colony at the University of Michigan (UM). Pax7CreER/+;Rosa26mTmG/+ (mTmG) and Pax7CreER/+;Rosa26TdTomato/+ mice were administered 5 daily 100 μL intraperitoneal injections of 20 mg/mL tamoxifen in corn oil. All mice were fed normal chow ad libitum and housed on a 12:12-h light–dark cycle under UM veterinary staff supervision. All procedures were approved by the University Committee on the Use and Care of Animals at UM and the Institutional Animal Care and Committee and were in accordance with the NIH.
Injury Model.
Adult mice (3 to 4 mo) were anesthetized and administered buprenorphine. Hair was removed from the hindlimbs and the surgical area was wiped with iodine and ethanol to sterilize. A 1-cm incision was made in the skin on the anterior side of each quadriceps, followed by the removal of a 2-mm or 3-mm full-depth muscle section from the middle of the rectus femoris muscle. The skin was sutured closed.
Histology.
Tissue sectioning.
Quadriceps muscles from both hindlimbs were embedded in an optical cutting temperature compound and frozen in isopentane cooled with liquid nitrogen and then stored at −80 °C. Serial cross-sections were cut using a cryotome at the suture delineating the midpoint of the injury and collected on positively charged glass slides. Duplicate or triplicate cross-sections from each time point and wound size were placed on each slide.
H&E.
Slides were submerged in hematoxylin for 2 min followed by quick immersions in distilled water and Scott’s tap water, then incubated in 80% ethanol. Slides were next incubated in eosin for 1 min, followed by quick immersions in 95% and 100% ethanol. Finally, slides were submerged in SafeClear II, and 2 drops of Permount were added before placing the coverslip on top. Brightfield images were taken using a motorized Olympus IX83 microscope at 10× magnification and stitched using the Olympus CellSense software to obtain an image of the complete tissue section.
PSR.
Slides were fixed in 4% paraformaldehyde for 15 min at room temperature (RT), washed three times in phosphate-buffered saline (PBS) and two times in deionized water, allowed to dry, and incubated in Direct Red 80 solubilized in picric acid for 1 h at RT. Next, slides were washed twice with acidified water followed by two washes in deionized water. Tissue samples were then dehydrated in a series of ethanol washes, incubated in xylene twice, and mounted with Permount. Brightfield images were taken as described above. Collagen fraction was calculated using ImageJ. Two or three sections per tissue were stained and quantified, then averaged to get the percentage of collagen for each tissue.
Immunohistofluorescence.
Immunofluorescence staining was performed as previously reported (85). Images were acquired with a Nikon A1 confocal microscope and pseudo-colored using ImageJ. Myofiber area and Feret diameters were quantified based on stitched images of laminin stains using MuscleJ (86).
Flow Cytometry.
Mouse quadriceps were extracted and weighed. Using sterile surgical scissors, muscles were minced and collagenase type II (0.2%) and dispase II (2.5 U/mL) were added to 10 mL Dulbecco's Modified Eagle Medium (DMEM) per quadriceps. Samples were placed on rocker in a 37 °C incubator for 1 h. The enzymes within the slurry were inactivated by the addition of 20% heat-inactivated fetal bovine serum (FBS) in Ham’s F-10 media. The solution was passed through a 70 μm cell strainer, centrifuged, washed, and counted. The cell pellet was resuspended in staining buffer (PBS with 2% bovine serum albumin [BSA], 2 mM ethylenediaminetetraacetic acid, and 0.01% sodium azide), then plated into round-bottom 96-well plates for staining. Cells were centrifuged at 250× for 2.5 min. The supernatant was discarded and cells were resuspended in a primary antibody mixture, incubated for 30 min on ice, washed with staining buffer, then resuspended in staining buffer containing propidium iodide (PI) for 1 min at RT in the dark, centrifuged, and resuspended in staining buffer for flow cytometry analysis. Prior to acquisition, cells were filtered through 40 μm cell strainers. UltraComp eBeads compensation beads were used for single-color controls. Samples were acquired within 1 h, and the data were processed using FlowJo.
qRT-PCR.
RNA was extracted using the Qiagen miRNeasy Micro Kit. Purity and concentration were measured using a NanoDrop and Qubit RNA HS Assay. cDNAs were synthesized using the SuperScript III cDNA Synthesis Kit. DNA quality and concentration were determined using a NanoDrop. We plated 80 to 100 μg cDNA template in triplicate along with SYBR Green PCR MasterMix and 500 nM PCR primer, then it was cycled 40 times starting at 95 °C for 10 s followed by 60 °C for 30 s on a CFX96 Real-Time thermocycler. Gene expression was quantified using the ΔΔCt method.
NK Cell and Neutrophil Coculture.
Murine IL15 super agonist (mIL15SA) was complexed as previously described (87). Splenic NK cells were magnetically enriched from WT mice using a mouse NK Cell Isolation Kit and activated for 48 h in Roswell Park Memorial Institute (RPMI) containing 10% FBS, 60 ng/mL mIL15SA, and antibiotics. NKp46+ NK cell purity following magnetic enrichment was > 80% measured by flow cytometry (SI Appendix, Fig. S4B). Neutrophils were fluorescence-activated cell sorting (FACS)–enriched from mouse skeletal muscle at 7 d after 3-mm VML injury. NK cells and neutrophils were cocultured at a 10:1 ratio for 4 h at 37 °C in RPMI with 10% FBS and antibiotics. Following incubation, cells were stained with annexin V and PI, then immediately analyzed by flow cytometry. Data were analyzed using FlowJo.
NK Cell Transplants.
Splenic NK cells were obtained and activated in vitro as described above. Activated NK cells were washed and resuspended in sterile PBS (50,000 cells/10 μL) and transplanted into 3-mm and 2-mm VML defects 7 dpi. Contralateral limbs received PBS. At 14 dpi, mice were humanely euthanized, and neutrophil abundance was quantified by flow cytometry.
CCR1 Inhibition.
Mice were administered bilateral 3-mm VML defects. Starting 7 dpi, mice received daily intraperitoneal injections of the CCR1 inhibitor J-113863 (8 mg/kg) solubilized in sterile PBS with 5% dimethyl sulfoxide (DMSO) and 5% CremophorEL. Neutrophil abundance was quantified by flow cytometry at 14 dpi.
TGFβRII Inhibition.
Mice were administered bilateral 3-mm VML defects to the quadriceps or 2-mm VML defects to the tibialis anterior (TA) for force testing. Starting 3 dpi, mice received intramuscular injections of ITD1 (200 pg) in PBS with 0.01% DMSO every third day until 15 dpi. Contralateral limbs were injected with 0.01% DMSO in PBS. Neutrophil abundance was quantified by flow cytometry at 14 dpi, and healing was assessed histologically at 28 dpi.
Muscle Force Testing.
Quadriceps.
After isolating the posterior branch of the femoral nerve with a nerve cuff, the anesthetized mouse was restrained such that the knee was at a 90° angle and the ankle was connected to a force transducer. The nerve was stimulated using a stimulus generator to induce the contraction of the quadriceps femoris. The pulse duration, frequency, and train duration of the stimulator were set to 0.2 ms, 175 Hz, and 500 ms, respectively. The nerve was stimulated with progressively increasing voltages until the peak isometric tetanic torque was reached. Maximal torque was recorded three times in a row for consistent force production.
TA.
Force testing was performed as previously reported by Dellorusso et al. with some modifications (88). Mice were anesthetized, hindlimb fur was removed, and the TA muscle was exposed. The distal TA tendon was isolated, and the distal half of the TA was freed from adjacent muscles. A suture was tied around the distal tendon, and the tendon was severed. A 25-gauge needle was driven through the knee and immobilized to prevent the knee from moving. The tendon was tied securely to the lever arm of a servomotor via the suture ends. The TA muscle was initially stimulated with 0.2 ms pulses via the peroneal nerve using platinum electrodes. Stimulation voltage and muscle length were adjusted for maximum isometric twitch force. While held at optimal muscle length (Lo), the muscle was stimulated at increasing frequencies until a maximum force (Po) was reached, with a 1 min rest period between each tetanic contraction. Subsequently, the same procedure was repeated, but rather than activating the muscle via the peroneal nerve, a cuff electrode was placed around the muscle for stimulation. Muscle length was measured with calipers, based on well-defined anatomical landmarks near the knee and the ankle. Optimum fiber length (Lf) was determined by multiplying Lo by the TA Lf/Lo ratio of 0.6 (89). After the evaluation of isometric force, the TA muscle was removed from the mouse and the muscle was weighed. The total muscle fiber cross-sectional area (CSA) of TA muscles was calculated by dividing muscle mass by the product of Lf and 1.06 mg/mm3, the density of mammalian skeletal muscle (90). Specific Po was calculated by dividing Po by CSA.
Muscle Progenitor Cell Differentiation.
Primary cells.
After obtaining a single cell suspension, MuSCs were magnetically enriched using a satellite cell isolation kit, then seeded onto a matrigel-coated dish in 8 mL myoblast medium (91). Cells were expanded with fresh myoblast media added every other day. Following expansion, 50,000 cells were seeded into a matrigel-coated 12-well plate and expanded until 90 to 100% confluency in myoblast media. Myoblast media was then replaced with differentiation media (DMEM with 5% horse serum and 1% penicillin/streptomycin), and cells were allowed to differentiate and fuse for 72 h.
C2C12s.
Neutrophils were magnetically enriched from peripheral blood mononuclear cell using a mouse neutrophil isolation kit, then incubated for 48 h in C2C12 growth media (DMEM with 10% FBS, basic fibroblast growth factor, and antibiotics). Conditioned media was collected and filtered to remove cell debris. C2C12s were expanded in a 96-well plate for either 8 d or 1 d in C2C12 growth media containing conditioned media. Media was replenished every 48 h. Following expansion, media was replaced with differentiation media and cells were incubated for 72 h.
Immunofluorescence staining.
After 72 h, myotubes were rinsed with PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.1% TritonX-100, then blocked with 1% BSA, 0.1% Tween-20, and 22.52 mg/mL glycine in PBS. After blocking, cells were incubated with rabbit anti-mouse MYH3 followed by AF555 anti-rabbit for 1 h at RT. Nuclei were stained with Hoechst. Cells were imaged on an epifluorescent microscope. The fusion index was calculated as the ratio of nuclei within myofibers containing more than 2 nuclei divided by the total number of nuclei per image. Myonuclei as a percentage of total cells were calculated as the number of MyH3+ myotubes with more than 2 nuclei divided by the total number of cells.
scRNA-Seq.
Sample preparation and sequencing.
Freshly isolated cells were briefly stained with PI and FACS sorted to remove dead cells and debris. An equal number of viable cells were pooled from 2 mice, 10,000 to 16,000 cells were loaded into the 10× Genomics chromium single-cell controller, and single cells were captured into nL-scale gel bead-in-emulsions. Complementary DNAs were prepared using the single-cell 3′ Protocol as per the manufacturer’s instructions and sequenced on a NovaSeq 6000.
Data processing and analysis.
A 10× CellRanger was run to generate HDF5 matrix files, which were imported into R using Seurat version 3 (92). Low-quality libraries were removed. Following log normalization, identification of variable features, and scaling using default parameters, datasets were integrated using Seurat’s FindIntegrationAnchors and IntegrateData functions. Dimensional reduction was performed in Seurat using principal component analysis, then UMAP (32) followed by community detection using the Louvain algorithm. Cluster marker genes were determined with both Seurat and scCATCH to annotate cell types. Defect-associated differential gene expression among cell types was calculated using MAST (37, 38). NicheNet (41) interaction predictions were generated using the Seurat Wrapper to compare the 3-mm defects to the 2-mm defects. Trajectory analysis was performed on the myogenic cell clusters using scVelo (93). Loom files were generated using velocyto (94). Seurat and ggplot2 were used for data visualization.
Statistics.
Experiments were repeated at least twice, apart from scRNA-Seq. Statistical analysis was performed in MATLAB or R. All statistical tests performed were two-sided. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank James Markworth and Eric Buras for assistance with histological and immunohistochemical analyses and the University of Michigan DNA Sequencing Core for assistance with single-cell sequencing library preparation. Research reported in this publication was partially supported by the Defense Advanced Research Projects Agency Bioelectronics for Tissue Regeneration Program through cooperative agreement (D20AC0002), awarded by the U.S. Department of the Interior, the Interior Business Center, the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH under award number P30 AR069620 (C.A.A.), the 3M Foundation (C.A.A.), the American Federation for Aging Research Grant for Junior Faculty (C.A.A.), the Department of Defense and Congressionally Directed Medical Research Program (W81XWH2010336 and W81XWH2110491; C.A.A.), the University of Michigan Geriatrics Center and National Institute of Aging under award number P30 AG024824 (C.A.A.), the University of Michigan Rackham Graduate School, and the NSF Graduate Research Fellowship Program under grant number DGE 1256260 (J.A.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, the NSF, or the Department of Defense, or the policy of the government, and no official endorsement should be inferred.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111445119/-/DCSupplemental.
Data Availability
High-throughput sequencing data have been deposited in Gene Expression Omnibus (GSE163376) (95).
References
- 1.Grogan B. F., Hsu J. R.; Skeletal Trauma Research Consortium, Volumetric muscle loss. J. Am. Acad. Orthop. Surg. 19 (suppl. 1), S35–S37 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Corona B. T., Rivera J. C., Owens J. G., Wenke J. C., Rathbone C. R., Volumetric muscle loss leads to permanent disability following extremity trauma. J. Rehabil. Res. Dev. 52, 785–792 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Garg K., Corona B. T., Walters T. J., Therapeutic strategies for preventing skeletal muscle fibrosis after injury. Front. Pharmacol. 6, 87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg K., et al. , Volumetric muscle loss: Persistent functional deficits beyond frank loss of tissue. J. Orthop. Res. 33, 40–46 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Owens B. D., Kragh J. F. Jr., Macaitis J., Svoboda S. J., Wenke J. C., Characterization of extremity wounds in Operation Iraqi Freedom and Operation Enduring Freedom in Journal of Orthopaedic Trauma. J. Orthop. Trauma 21, 254–257 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Dziki J., et al. , An acellular biologic scaffold treatment for volumetric muscle loss: Results of a 13-patient cohort study. npj. Regen. Med. 1, 16008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sicari B. M., et al. , An acellular biologic scaffold promotes skeletal muscle formation in mice and humans with volumetric muscle loss. Sci. Transl. Med. 6, 234ra58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grasman J. M., Zayas M. J., Page R. L., Pins G. D., Biomimetic scaffolds for regeneration of volumetric muscle loss in skeletal muscle injuries. Acta Biomater. 25, 2–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shayan M., Huang N. F., Pre-clinical cell therapeutic approaches for repair of volumetric muscle loss. Bioengineering (Basel) 7, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corona B. T., Rivera J. C., Dalske K. A., Wenke J. C., Greising S. M., Pharmacological mitigation of fibrosis in a porcine model of volumetric muscle loss injury. Tissue Eng. Part A 26, 636–646 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Greising S. M., et al. , Early rehabilitation for volumetric muscle loss injury augments endogenous regenerative aspects of muscle strength and oxidative capacity. BMC Musculoskelet. Disord. 19, 173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greising S. M., Corona B. T., McGann C., Frankum J. K., Warren G. L., Therapeutic approaches for volumetric muscle loss injury: A systematic review and meta-analysis. Tissue Eng. Part B Rev. 25, 510–525 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Aguilar C. A., et al. , Multiscale analysis of a regenerative therapy for treatment of volumetric muscle loss injury. Cell Death Discov. 4, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S., et al. , Pre-innervated tissue-engineered muscle promotes a pro-regenerative microenvironment following volumetric muscle loss. Commun. Biol. 3, 330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webster M. T., Manor U., Lippincott-Schwartz J., Fan C. M., Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell 18, 243–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber R. L., Ward S. R., Cellular mechanisms of tissue fibrosis. 4. Structural and functional consequences of skeletal muscle fibrosis. Am. J. Physiol. Cell Physiol. 305, C241–C252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larouche J., Greising S. M., Corona B. T., Aguilar C. A., Robust inflammatory and fibrotic signaling following volumetric muscle loss: A barrier to muscle regeneration. Cell Death Dis. 9, 409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin H., Price F., Rudnicki M. A., Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard E. E., Pasiakos S. M., Blesso C. N., Fussell M. A., Rodriguez N. R., Divergent roles of inflammation in skeletal muscle recovery from injury. Front. Physiol. 11, 87 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wosczyna M. N., Rando T. A., A muscle stem cell support group: Coordinated cellular responses in muscle regeneration. Dev. Cell 46, 135–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burzyn D., et al. , A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panduro M., Benoist C., Mathis D., Treg cells limit IFN-γ production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc. Natl. Acad. Sci. U.S.A. 115, E2585–E2593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold L., et al. , Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dort J., Fabre P., Molina T., Dumont N. A., Macrophages are key regulators of stem cells during skeletal muscle regeneration and diseases. Stem Cells Int. 2019, 4761427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommerfeld S. D., et al. , Interleukin-36γ-producing macrophages drive IL-17-mediated fibrosis. Sci. Immunol. 4, eaax4783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak M. L., Weinheimer-Haus E. M., Koh T. J., Macrophage activation and skeletal muscle healing following traumatic injury. J. Pathol. 232, 344–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurtgen B. J., et al. , Severe muscle trauma triggers heightened and prolonged local musculoskeletal inflammation and impairs adjacent tibia fracture healing. J. Musculoskelet. Neuronal Interact. 16, 122–134 (2016). [PMC free article] [PubMed] [Google Scholar]
- 28.Sadtler K., et al. , Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science 352, 366–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deyhle M. R., Hyldahl R. D., The role of T lymphocytes in skeletal muscle repair from traumatic and contraction-induced injury. Front. Physiol. 9, 768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson S. E., et al. , Determination of a critical size threshold for volumetric muscle loss in the mouse quadriceps. Tissue Eng. Part C Methods 25, 59–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becht E., et al. , Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–47 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Shao X., et al. , scCATCH: Automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience 23, 100882 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabula Muris Consortium, Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larouche J. A., et al. , Murine muscle stem cell response to perturbations of the neuromuscular junction are attenuated with aging. eLife 10, e66749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giordani L., et al. , High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell 74, 609–621.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Finak G., et al. , MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dulken B. W., et al. , Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas P. D., et al. , PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tidball J. G., Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 17, 165–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browaeys R., Saelens W., Saeys Y., NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Terabe M., Berzofsky J. A., Tissue-specific roles of NKT cells in tumor immunity. Front. Immunol. 9, 1838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capote J., et al. , Osteopontin ablation ameliorates muscular dystrophy by shifting macrophages to a pro-regenerative phenotype. J. Cell Biol. 213, 275–288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vetrone S. A., et al. , Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J. Clin. Invest. 119, 1583–1594 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bezman N. A., et al. ; Immunological Genome Project Consortium, Molecular definition of the identity and activation of natural killer cells. Nat. Immunol. 13, 1000–1009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorén F. B., et al. , Human NK cells induce neutrophil apoptosis via an NKp46- and Fas-dependent mechanism. J. Immunol. 188, 1668–1674 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Francis K., Palsson B. O., Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proc. Natl. Acad. Sci. U.S.A. 94, 12258–12262 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oyler-Yaniv A., et al. , A tunable diffusion-consumption mechanism of cytokine propagation enables plasticity in cell-to-cell communication in the immune system. Immunity 46, 609–620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Neutrophils in tissue injury and repair. Cell Tissue Res. 371, 531–539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo B. R., et al. , Skeletal muscle regeneration with robotic actuation-mediated clearance of neutrophils. Sci. Transl. Med. 13, eabe8868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simionescu-Bankston A., et al. , Creatine kinase B is necessary to limit myoblast fusion during myogenesis. Am. J. Physiol. Cell Physiol. 308, C919–C931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchildon F., Lamarche É., Lala-Tabbert N., St-Louis C., Wiper-Bergeron N., Expression of CCAAT/enhancer binding protein beta in muscle satellite cells inhibits myogenesis in cancer cachexia. PLoS One 10, e0145583 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.AlSudais H., Lala-Tabbert N., Wiper-Bergeron N., CCAAT/enhancer binding protein β inhibits myogenic differentiation via ID3. Sci. Rep. 8, 16613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Summermatter S., et al. , Blockade of metallothioneins 1 and 2 increases skeletal muscle mass and strength. Mol. Cell. Biol. 37, e00305-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jørgensen L. H., et al. , Secreted protein acidic and rich in cysteine (SPARC) in human skeletal muscle. J. Histochem. Cytochem. 57, 29–39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reimann J., et al. , Macrophage migration inhibitory factor in normal human skeletal muscle and inflammatory myopathies. J. Neuropathol. Exp. Neurol. 69, 654–662 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Petrany M. J., Millay D. P., Cell fusion: Merging membranes and making muscle. Trends Cell Biol. 29, 964–973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melendez J., et al. , TGFβ signalling acts as a molecular brake of myoblast fusion. Nat. Commun. 12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girardi F., et al. , TGFβ signaling curbs cell fusion and muscle regeneration. Nat. Commun. 12, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brandes M. E., Mai U. E., Ohura K., Wahl S. M., Type I transforming growth factor-beta receptors on neutrophils mediate chemotaxis to transforming growth factor-beta. J. Immunol. 147, 1600–1606 (1991). [PubMed] [Google Scholar]
- 61.Cohen T. V., et al. , Upregulated IL-1β in dysferlin-deficient muscle attenuates regeneration by blunting the response to pro-inflammatory macrophages. Skelet. Muscle 5, 24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aguilar C. A., et al. , Transcriptional and chromatin dynamics of muscle regeneration after severe trauma. Stem Cell Reports 7, 983–997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziemkiewicz N., Hilliard G., Pullen N. A., Garg K., The role of innate and adaptive immune cells in skeletal muscle regeneration. Int. J. Mol. Sci. 22, 3265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pizza F. X., Peterson J. M., Baas J. H., Koh T. J., Neutrophils contribute to muscle injury and impair its resolution after lengthening contractions in mice. J. Physiol. 562, 899–913 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walden D. L., et al. , Neutrophils accumulate and contribute to skeletal muscle dysfunction after ischemia-reperfusion. Am. J. Physiol. 259, H1809–H1812 (1990). [DOI] [PubMed] [Google Scholar]
- 66.Ong S., et al. , Natural killer cells limit cardiac inflammation and fibrosis by halting eosinophil infiltration. Am. J. Pathol. 185, 847–861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruscetti M., et al. , NK cell–mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 362, 1416–1422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costantini C., Cassatella M. A., The defensive alliance between neutrophils and NK cells as a novel arm of innate immunity. J. Leukoc. Biol. 89, 221–233 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Ghiringhelli F., et al. , CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-β-dependent manner. J. Exp. Med. 202, 1075–1085 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stepien D. M., et al. , Tuning macrophage phenotype to mitigate skeletal muscle fibrosis. J. Immunol. 204, 2203–2215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Theret M., Rossi F. M. V., Contreras O., Evolving roles of muscle-resident fibro-adipogenic progenitors in health, regeneration, neuromuscular disorders, and aging. Front. Physiol. 12, 673404 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mázala D. A. G., et al. , TGF-β-driven muscle degeneration and failed regeneration underlie disease onset in a DMD mouse model. JCI Insight 5, e135703 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garg K., Boppart M. D., Influence of exercise and aging on extracellular matrix composition in the skeletal muscle stem cell niche. J. Appl. Physiol. (1985) 121, 1053–1058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reibman J., et al. , Transforming growth factor β 1, a potent chemoattractant for human neutrophils, bypasses classic signal-transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 88, 6805–6809 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidal B., et al. , Fibrinogen drives dystrophic muscle fibrosis via a TGFbeta/alternative macrophage activation pathway. Genes Dev. 22, 1747–1752 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamada M., et al. , Matrix metalloproteinases are involved in mechanical stretch-induced activation of skeletal muscle satellite cells. Muscle Nerve 34, 313–319 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Chen X., Li Y., Role of matrix metalloproteinases in skeletal muscle: Migration, differentiation, regeneration and fibrosis. Cell Adhes. Migr. 3, 337–341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rozo M., Li L., Fan C. M., Targeting β1-integrin signaling enhances regeneration in aged and dystrophic muscle in mice. Nat. Med. 22, 889–896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J. H., et al. , Mechanical tension drives cell membrane fusion. Dev. Cell 32, 561–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bentzinger C. F., et al. , Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell 12, 75–87 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lukjanenko L., et al. , Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 22, 897–905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gilbert P. M., et al. , Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tierney M. T., et al. , Autonomous extracellular matrix remodeling controls a progressive adaptation in muscle stem cell regenerative capacity during development. Cell Rep. 14, 1940–1952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garg K., Corona B. T., Walters T. J., Losartan administration reduces fibrosis but hinders functional recovery after volumetric muscle loss injury. J. Appl. Physiol. (1985) 117, 1120–1131 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Markworth J. F., et al. , Resolvin D1 supports skeletal myofiber regeneration via actions on myeloid and muscle stem cells. JCI Insight 5, e137713 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mayeuf-Louchart A., et al. , MuscleJ: A high-content analysis method to study skeletal muscle with a new Fiji tool. Skelet. Muscle 8, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo Y., et al. , IL-15 superagonist-mediated immunotoxicity: Role of NK cells and IFN-γ. J. Immunol. 195, 2353–2364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dellorusso C., Crawford R. W., Chamberlain J. S., Brooks S. V., Tibialis anterior muscles in mdx mice are highly susceptible to contraction-induced injury. J. Muscle Res. Cell Motil. 22, 467–475 (2001). [DOI] [PubMed] [Google Scholar]
- 89.Burkholder T. J., Fingado B., Baron S., Lieber R. L., Relationship between muscle fiber types and sizes and muscle architectural properties in the mouse hindlimb. J. Morphol. 221, 177–190 (1994). [DOI] [PubMed] [Google Scholar]
- 90.Mendez J., Keys A., Density and composition of mammalian muscle. Metabolism 9, 184–188 (1960). [Google Scholar]
- 91.Motohashi N., Asakura Y., Asakura A., Isolation, culture, and transplantation of muscle satellite cells. 10.3791/50846 J. Vis. Exp., (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bergen V., Lange M., Peidli S., Wolf F. A., Theis F. J., Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 94.La Manno G., et al. , RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.J. A. Larouche, S. J. Kurpiers, C. A. Aguilar, Single cell deconstruction of murine volumetric muscle loss reveals inflammatory imbalances preventing muscle stem cell mediated regeneration. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163376. Accessed 16 December 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data have been deposited in Gene Expression Omnibus (GSE163376) (95).