Significance
Efforts to improve tuberculosis therapy include optimizing multidrug regimens to take advantage of drug–drug synergies. However, the complex host environment has a profound effect on bacterial metabolic state and drug activity, making predictions of optimal drug combinations difficult. In this study, we leverage a newly developed library of conditional knockdown Mycobacterium tuberculosis mutants in which genetic depletion of essential genes mimics the effect of drug therapy. This tractable system allowed us to assess the effect of growth condition on predicted drug–drug interactions. We found that these interactions can be differentially sensitive to the metabolic state, and select in vitro–defined interactions can be leveraged to accelerate bacterial killing during infection. These findings suggest strategies for optimizing tuberculosis therapy.
Keywords: Mycobacterium tuberculosis, chemical–genetic interactions, carbon metabolism, antibiotics
Abstract
Current chemotherapy against Mycobacterium tuberculosis (Mtb), an important human pathogen, requires a multidrug regimen lasting several months. While efforts have been made to optimize therapy by exploiting drug–drug synergies, testing new drug combinations in relevant host environments remains arduous. In particular, host environments profoundly affect the bacterial metabolic state and drug efficacy, limiting the accuracy of predictions based on in vitro assays alone. In this study, we utilized conditional Mtb knockdown mutants of essential genes as an experimentally tractable surrogate for drug treatment and probe the relationship between Mtb carbon metabolism and chemical–genetic interactions (CGIs). We examined the antitubercular drugs isoniazid, rifampicin, and moxifloxacin and found that CGIs are differentially responsive to the metabolic state, defining both environment-independent and -dependent interactions. Specifically, growth on the in vivo–relevant carbon source, cholesterol, reduced rifampicin efficacy by altering mycobacterial cell surface lipid composition. We report that a variety of perturbations in cell wall synthesis pathways restore rifampicin efficacy during growth on cholesterol, and that both environment-independent and cholesterol-dependent in vitro CGIs could be leveraged to enhance bacterial clearance in the mouse infection model. Our findings present an atlas of chemical–genetic–environmental interactions that can be used to optimize drug–drug interactions, as well as provide a framework for understanding in vitro correlates of in vivo efficacy.
The current chemotherapeutic regimen for tuberculosis (TB) is the product of many decades of basic and clinical research. Since the first trials of streptomycin monotherapy in 1948 were rapidly followed by the emergence of antibiotic-resistant clones (1), multidrug regimens to both suppress resistance and accelerate bacterial killing have become standard. The current regimen used against drug-sensitive strains of Mycobacterium tuberculosis (Mtb) consists of four antibiotics, isoniazid (INH), rifampicin (RIF), pyrazinamide (PZA), and ethambutol (EMB), and was optimized in a series of clinical trials in the 1970s (2, 3). While this “short-course regimen” has been credited with curing over 50 million patients, its delivery is complicated by the need for 6 to 9 mo of drug administration (4). Furthermore, even in clinical trial settings where the delivery of this extended regimen is assured, 5 to 10% of patients fail therapy (5). The frequent transmission of antibiotic-resistant Mtb strains has further complicated TB treatment options, requiring the use of less optimized drug combinations that are administered for even longer periods (6).
The factors that necessitate prolonged therapy are complex and specific to the infection environment. While antibiotics such as INH and RIF cause rapid cell death in vitro, their antimicrobial activities are much slower during infection (7). Limited drug penetration into Mtb-containing tissue lesions may reduce the efficacy of some drugs. For example, the intralesional RIF concentrations are significantly lower than what is achieved in the plasma (8, 9), and clinical studies suggest that increased RIF dosing improves bacterial clearance (10, 11). However, drug penetration is unlikely to fully account for the reduced antibiotic efficacy in host tissue, as this complex and stressful environment has also been shown to alter the physiology of the pathogen to induce a drug-tolerant state (12–14). While a number of host immune-related stresses may be involved in this process, simple changes in macronutrient availability can have important consequences. Mtb has access to a mixture of glycolytic carbon sources, fatty acids, and cholesterol in host tissue (15–17), and altering the availability of these carbon sources in vitro can change the efficacy of antitubercular compounds (18–23). A role for differential carbon catabolism in determining drug efficacy is also supported by the identification of natural genetic variants in clinical Mtb isolates that enhance drug tolerance by altering either glycerol, lipid, or sterol catabolism (24, 25). The profound effect of the host environment on bacterial metabolic state and drug activity makes it difficult to predict the ultimate efficacy of an antibiotic regimen based on in vitro assays alone.
The most advanced efforts to accelerate TB therapy involve optimizing multidrug regimens to take advantage of drug–drug synergies. These pharmacological interactions can improve therapy by increasing drug exposure, decreasing minimum inhibitory concentration (MIC), or enhancing the maximal effect of the treatment (26). However, despite the demonstrated benefits of synergistic regimens in many therapeutic realms, the combinatorial burden of testing all potential multidrug combinations remains cumbersome. While recent advances to more efficiently predict synergies in vitro have proven valuable (27, 28), it remains unclear whether the environmental influences that alter the efficacy of individual drugs will also influence drug–drug interactions (29, 30).
In this study, we leveraged a newly developed genetic resource to understand the effect of growth conditions on potential drug–drug interactions. This “hypomorph library” consists of individual DNA barcoded strains in which individual essential genes are tagged with the DAS+4 sequence that targets proteins for degradation upon inducible induction of the SspB adapter (31). Graded sspB expression produces different degrees of depletion to model the effect of partial chemical inhibition (32). This approach allowed for highly parallel assessment of changes in TB drug sensitivity of individual hypomorphic mutants under different growth conditions, creating a drug-specific chemical–genetic interaction (CGI) profile (33). We report that drugs of distinct classes produce interactions that are differentially sensitive to the environment, and that both condition-independent and -dependent CGIs can be identified for all drugs tested. In particular, RIF efficacy was impaired as a result of cell surface lipid alterations that occur during growth in cholesterol, and the cell could be resensitized through a condition-specific interaction with the cell wall synthetic machinery both in vitro and in the mouse lung. These observations provide a compendium of chemical–genetic interactions that can be exploited to enhance therapy and begin to define in vitro correlates of in vivo efficacy.
Results
Genetic Strategy to Define Essential Bacterial Functions that Alter Drug Efficacy In Vitro.
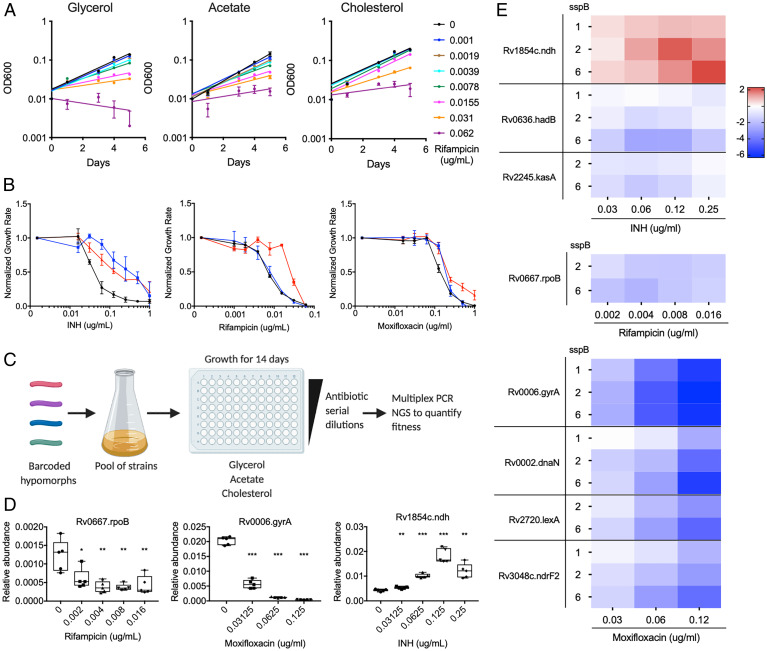
Host environmental factors alter antibiotic efficacy against TB (12, 24, 34, 35). To determine whether carbon source availability specifically affects antibiotic efficacy, we used the Mtb strain H37Rv in minimal medium with glycerol, acetate, or cholesterol as sole carbon sources in a growth inhibition assay with the TB drugs INH, RIF, and MOX. As these carbon sources support different growth rates, endpoint-based antibiotic activity measurements such as MIC assays can be difficult to interpret. Therefore we quantified growth rates in each carbon source over multiple concentrations of antibiotics to determine the concentration that decreased the growth rate by 50% (GR50) (24, 36). Growth of individual strains was monitored during exponential phase (OD600 0.20 to 0.25) and the growth rate was determined by fitting an exponential curve (Fig. 1A). We found different carbon sources to alter the GR50 of these drugs in distinct ways. For INH, glycerol-dependent growth increased efficacy, relative to the nonglycolytic carbon sources. In contrast, growth in cholesterol was found to decrease RIF efficacy, relative to the other conditions. Media composition had a more modest effect on MOX, altering GR50 by only approximately twofold (Fig. 1B). These findings show that carbon metabolism plays an important role in antibiotic efficacy and has distinct effects on different drugs.
Fig. 1.
Genetic strategy to define essential bacterial functions that alter drug efficacy in vitro. (A) Growth of H37Rv in minimal media with glycerol, acetate, and cholesterol as the sole carbon source and increasing concentrations of RIF. Results shown as mean from three biological replicates with SDs. (B) Normalized growth inhibition of WT Mtb across increasing concentrations of INH, RIF, and MOX in minimal media with glycerol (black), acetate (blue), and cholesterol (red) as sole carbon sources. Results shown as means from three biological replicates with SDs. (C) Barcoded hypomorph mutants were pooled and grown in 96-well plates containing minimal media with glycerol, acetate, or cholesterol as the sole carbon source for 14 d. Antibiotics were added to individual wells as well as untreated controls. Chromosomal barcodes were PCR amplified and pooled for Illumina NGS. Barcodes were analyzed to quantify changes in fitness of individual strains from different conditions. (D) Boxplot representing changes in relative abundances of rpoB, gyrA, and ndh mutants grown in glycerol during RIF, MOX, and INH treatment, respectively. The highest drug concentrations tested for RIF, MOX, and INH represent the concentrations at which most of the strains in the library were inhibited. Data represent five biological replicates. Significance was calculated using unpaired t test and compared to untreated conditions, *P < 0.05, **P < 0.01, ***P < 0.001. (E) Heatmap representing changes in fitness of individual mutants, shown as log2 fold change, during INH, RIF, and MOX treatment in glycerol growth conditions. Mutants were chosen from previous association with respective antibiotic. sspB numbers denote sspB expression level of mutant. Results shown as means from five biological replicates.
To understand the mechanisms linking drug efficacy and metabolism, we utilized a barcoded-hypomorph library consisting of 465 essential genes to identify CGI with these drugs (32). The library was grown in minimal media with glycerol, acetate, or cholesterol, and antibiotics were added at concentrations ranging from 0.05× to 1× GR50 for each. Bacteria were subjected to these conditions in 96-well plates for 14 d, at which point the relative abundance of individual mutants was assessed through multiplex PCR and next-generation sequencing (NGS) (Fig. 1C and Dataset S1). Relative abundances of individual mutants were calculated by normalizing the mutant’s barcode count to the total barcode count and comparing the relative abundance of each mutant in different drug concentrations with the untreated control samples.
We initially investigated the validity of this dataset by determining whether genetic inhibition of each drug’s target produced the expected CGI with that compound in standard glycerol-dependent growth conditions. RIF and MOX inhibit the RNA polymerase subunit, RpoB, or the DNA gyrase subunit, GyrA, respectively. As expected, rpoB and gyrA hypomorphic mutants were hypersensitive to RIF or MOX, and the CGI increased with drug concentration (Fig. 1D). The major target of INH, InhA, was not present in our mutant pool. However, the library did contain a ndh mutant. Deficiency in this nicotinamide adenine dinucleotide (NADH)-dehydrogenase is known to decrease INH efficacy, likely due to the inefficient formation of the active INH–NAD adduct (37). Consistent with these observations, we found that the ndh hypomorphic mutant showed an INH dose–dependent increase in abundance in our dataset (Fig. 1D).
The hypomorph library was constructed to contain up to five different versions of each DAS+4-tagged mutant, which express different levels of sspB and therefore produce graded levels of protein depletion (31, 38). To investigate the relationship between target protein abundance and phenotype, we correlated the strength of sspB expression with drug sensitivity for a number of genes that were expected to alter antibiotic efficacy (Fig. 1E). For RIF, we concentrated on rpoB. For INH, we examined ndh as well as genes associated with mycolate biosynthesis and found kasA and hadB mutants to be significantly hypersensitive to increasing INH concentrations. For MOX, in addition to gyrA, depletion of proteins associated with DNA integrity or deoxynucleotide production, dnaN, lexA, and ndrF2, showed significantly decreased fitness over increasing MOX concentrations. For some mutants, particularly those involved in DNA metabolism, we noted a dose-dependent effect of sspB expression, with larger phenotypes corresponding to greater degrees of depletion. However, this was not a universal phenomenon, and we often found that independent mutants expressing different sspB levels produced consistent phenotypic effects. The observed differences in sensitivity to protein depletion likely reflect the unique biochemistry of each pathway (39). As a result, we considered each mutant corresponding to a target protein independently in the following analyses.
CGI Profiles Are Determined by the Combination of Carbon Source and Drug.
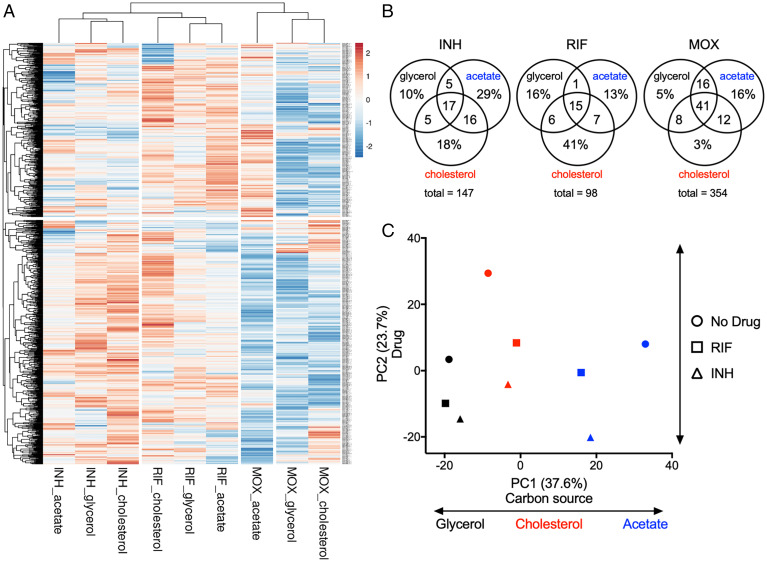
To understand the relative importance of carbon metabolism and drug on bacterial physiology, we compared the “fitness profiles” of each condition, which represented the relative fitness of each mutant strain. We first selected the fold change in abundance (log2FC) values of every mutant at 0.65× GR50 drug concentration that showed a statistically significant fitness difference when at least one treated sample was compared to the untreated control (918 mutants, Q < 0.05). The drug concentration 0.65× GR50 displayed the largest number of mutants with a statistically significant fitness difference across most conditions. Upon hierarchical clustering of both genes and conditions, we found three major condition clusters that corresponded to each antibiotic (Fig. 2A). These condition clusters were differentiated by two distinct clusters of mutants, one large gene set differentiated MOX from the other drugs (Fig. 2 A, Bottom cluster), and differential fitness effects on a second smaller gene set differentiated INH and RIF (Fig. 2 A, Top cluster). Within each condition cluster, fitness effects due to carbon source were apparent.
Fig. 2.
Carbon sources mediate antibiotic–genetic interactions of essential bacterial functions. (A) Heatmap of hypomorph mutants shown as log2 fold change of 0.65× GR50 of INH, RIF, and MOX against untreated controls. Results shown as means from five biological replicates. Each antibiotic (INH, RIF, and MOX) in a single carbon source (glycerol, acetate, and cholesterol) conditions were compared using hierarchical clustering methods. (B) Venn diagrams showing the total number of genes with log2FC > −1 and P < 0.05 at 0.65× GR50 of INH, RIF, and MOX, and the % of genes associated with specific conditions. (C) Relative abundance datasets of the hypomorph libraries from glycerol (black), acetate (blue), and cholesterol (red) growth conditions with 0.65× GR50 INH (triangle) and RIF (square) treatment as well as untreated controls (circle) were examined using principal component analysis. Units shown on axes are arbitrary values in principal component space.
To catalog the genes that differentiate these conditions, we compared each drug-treated sample (0.65× GR50) with the corresponding untreated control and chose genes with log2FC < −1 and P < 0.05 for each carbon source. For all antibiotics tested, there were mutants that showed decreased fitness across all carbon sources as well as those associated with a single carbon source (Fig. 2B). Notably, MOX produced the largest number of mutants with decreased fitness in a condition-independent manner, while INH and RIF produced the largest number of mutants with decreased fitness during acetate- and cholesterol-dependent growth, respectively. To more quantitatively assess the relative contribution of carbon source and drug, we used principal component analysis (PCA) to examine the amount of variance contributed by each, concentrating only on the INH and RIF conditions, which produced the most condition-specific effects. PC1 aligned to distinguish the carbon sources, while PC2 differentiated among drug treatments (Fig. 2C). The first and second principal components accounted for 37.6% and 23.7%, respectively, of the overall variance across the nine datasets, indicating that drug and carbon source played a relatively equal role in shaping bacterial physiology.
Condition-Independent CGI Can Be Reproduced with Individual Mutants.
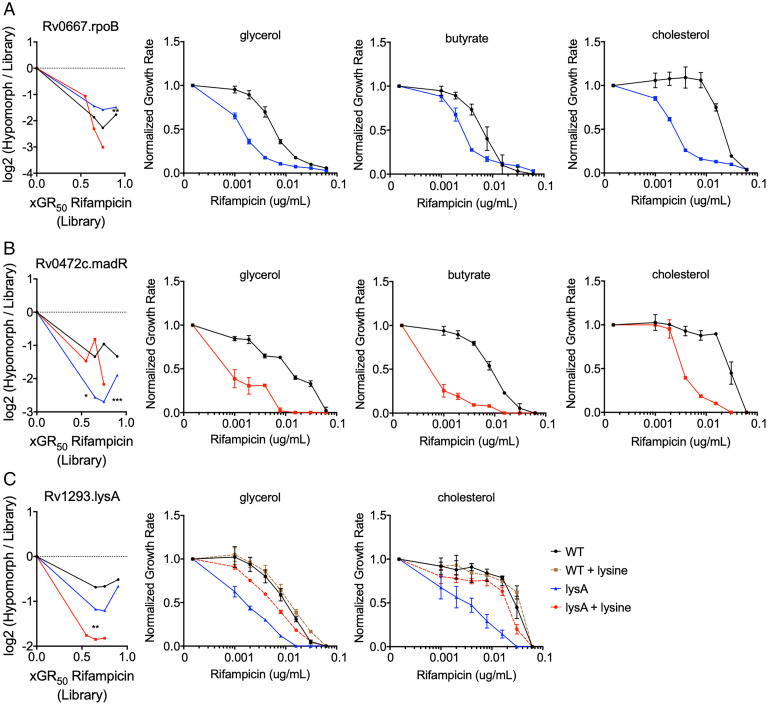
Genes found to alter antibiotic efficacy from the high-throughput hypomorph analysis were validated using individual mutant strains. For initial validation studies, we concentrated on mutants predicted to show drug hypersensitivity under all media conditions. We also replaced acetate with butyrate, as butyrate provided relatively enhanced growth that allowed for robust validation. As these hypomorph mutants target essential genes that may alter growth, we compared the relative growth rates of individual mutants to WT in different carbon sources (SI Appendix, Fig. S1). Variability in growth rates of mutants in different growth conditions led to the utilization of the growth rate inhibition method described above to determine the RIF MIC. The repression of a drug’s target is thought to increase sensitivity to inhibition by reducing the fraction of the target that needs to be inhibited to reduce growth (40). As such, we anticipated that drug target inhibition would result in condition-independent effects. This prediction was verified using a rpoB hypomorphic mutant, which our screening data predicted to increase RIF efficacy in all carbon sources (Fig. 3A). As such, we verified that this degree of RpoB depletion decreased the RIF MIC by 4 to 16-fold in each of the three media conditions (Fig. 3A). In addition to known drug targets, a number of additional genes were predicted to enhance RIF efficacy in a carbon source–independent manner, including the transcriptional regulator, Rv0472c. This gene has been named, mycolic acid desaturase regulator (madR), based on its specific effect on the mycolate-modifying desA1 and desA2 genes (41). As we found for RpoB, MadR depletion sensitized the bacterium to RIF in all tested carbon sources in the single strain assay (Fig. 3B). Finally, to ensure the specificity of these CGIs, we investigated whether they could be reversed by specific metabolite complementation. As predicted by the screen, depletion of the lysine biosynthetic enzyme, LysA, produced RIF hypersensitivity in both glycerol and cholesterol media, and we found this phenotype was largely reversed by supplementation with 3 mM lysine (Fig. 3C). These data support the predictions of the primary screen and demonstrate that certain CGIs are relatively unaffected by carbon source.
Fig. 3.
Condition-independent chemical–genetic interactions can be reproduced with individual mutants. (A) Leftmost panel: RpoB depletion shows decreased fitness compared to the library over increasing concentrations of RIF for glycerol (black), acetate (blue), and cholesterol (red). Remaining panels: Normalized growth inhibition assay shows rpoB hypomorph mutant (blue) has decreased RIF MIC compared to WT (black) across multiple carbon conditions. (B) Leftmost panel: MadR depletion shows decreased fitness compared to the library over increasing concentrations of rifampicin for glycerol (black), acetate (blue), and cholesterol (red). Remaining panels: Normalized growth inhibition assay shows madR hypomorph mutant (red) has decreased RIF MIC compared to WT (black) across multiple carbon conditions. (C) Leftmost panel: LysA depletion shows decreased fitness compared to the library over increasing concentrations of rifampicin for glycerol (black), acetate (blue), and cholesterol (red). Remaining panels: Normalized growth inhibition assay shows lysA hypomorph mutant (blue) has decreased RIF MIC compared to WT (black), which can be reversed by 3 mM lysine supplementation (red). Library results in leftmost panels are shown as means from five biological replicates, and significance was calculated using unpaired t test with Benjamini–Hochberg multiple testing correction, *q < 0.05, **q < 0.01, ***q < 0.001. Normalized growth inhibition results in remaining panels are shown as means from three biological replicates with SDs.
Depletion of Cell Wall Biosynthetic Genes Produces Condition-Specific RIF Interactions.
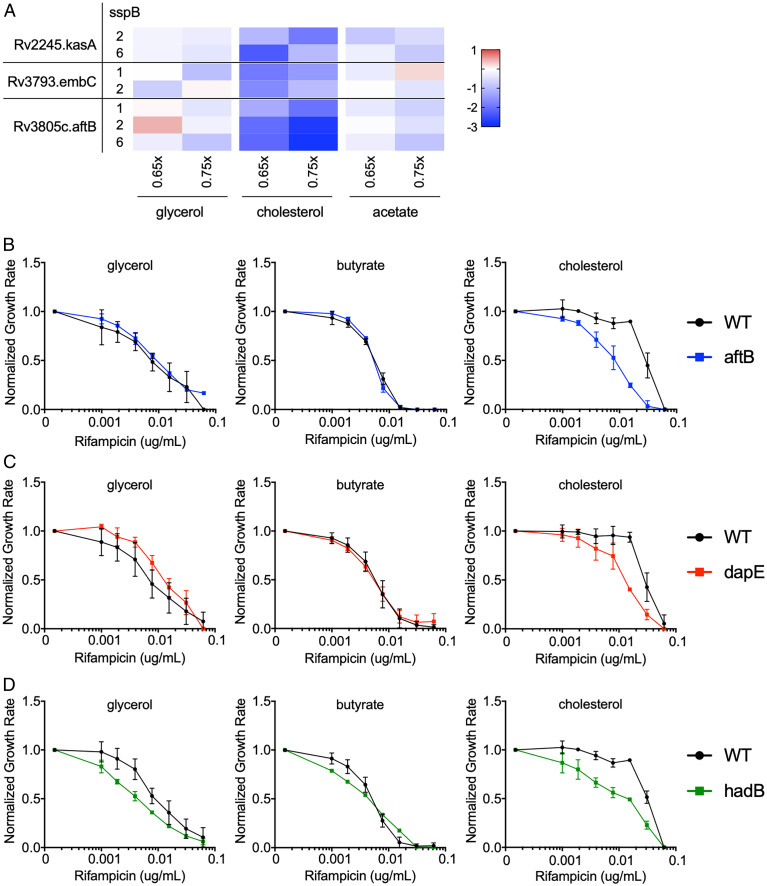
As cholesterol represents a primary carbon source for Mtb during infection (16, 42), and RIF efficacy is thought to be limited by drug exposure (43–45), we probed the hypomorph fitness profiles for clues to the mechanism underlying the cholesterol-dependent increase in RIF MIC. We found that inhibition of genes associated with different components of the mycobacterial cell wall, including arabinogalactan (embC and aftB) and mycolic acid (kasA) (46, 47), preferentially sensitized the bacterium to RIF during cholesterol growth (Fig. 4A). These mutants were significantly underrepresented by up to eightfold upon RIF treatment predominantly in cholesterol growth conditions across multiple sspB doses and drug concentrations. To further explore the effect of cell wall inhibition, we tested individual mutants with defects in peptidoglycan (dapE), arabinogalactan (aftB), and mycolic acid (hadB) synthesis (46, 48–50). We first compared the relative growth rates of individual mutants to wild type (WT) in different carbon sources and found no significant differences for dapE and hadB hypomorph mutants across all conditions, while aftB hypomorph mutant showed a modest growth rate increase in glycerol (SI Appendix, Fig. S2). We then found that perturbation in each of these cell wall layers produced only relatively modest effects in glycerol or butyrate media, but all consistently sensitized the bacterium to RIF in cholesterol growth conditions (Fig. 4 B–D). These observations suggest that alterations in cell wall structure might underlie the relationship between cholesterol catabolism and reduced RIF efficacy.
Fig. 4.
Cell wall biosynthesis hypomorph mutants show altered RIF efficacy during cholesterol growth conditions. (A) Heatmap showing hypomorph mutants of select mycobacterial cell wall biosynthesis genes and pathways with significant changes in fitness during cholesterol growth, shown as log2 fold change at 0.65× and 0.75× GR50 of RIF against untreated controls. Results shown as means from five biological replicates. Classification of mutants was done using Mycobrowser. (B–D) Normalized growth inhibition of WT and select mutants across increasing concentrations of RIF in minimal media with sole carbon sources. Depletion of AftB (blue), DapE (red), and HadB (green) show decreased RIF MIC compared to WT (black) only during cholesterol growth conditions. Results shown as means from three biological replicates with SDs.
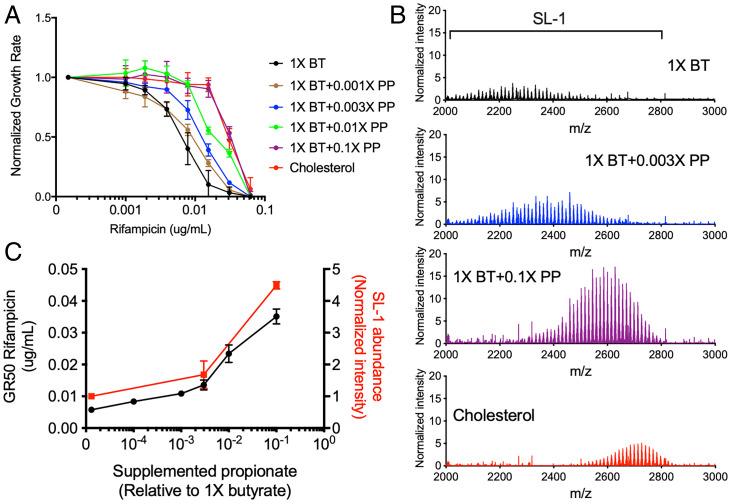
RIF Efficacy Correlates with Propionate Catabolism and Branched Chain Lipid Abundance.
Mtb catabolizes cholesterol to pyruvate, propionyl-CoA, and acetyl-CoA (17, 51). Propionyl-CoA is a precursor of branched chain lipid synthesis, and catabolism of either cholesterol or propionate increases cellular propionyl-CoA levels, causing an increase in both the abundance and chain length of cell wall lipids, such as diacyltrehalose (DAT), polyacyltrehalose (PAT), and sulfolipid-1 (SL-1) (51–54). To understand whether these alterations in cell wall lipids could be responsible for altering RIF efficacy, we compared RIF GR50 values in cells grown in cholesterol to those grown in butyrate media supplemented with increasing concentrations of propionate. Growth rate in cholesterol showed no significant difference compared to both butyrate and butyrate supplemented with propionate (SI Appendix, Fig. S3). We observed that GR50 values increased up to eightfold with increasing propionate levels, and the addition of 0.1× propionate, reflecting a 1:10 ratio of propionate to butyrate (weight:weight), mimicked the effect of cholesterol (Fig. 5A). To more quantitatively associate branched chain lipid abundance with drug efficacy, we performed relative quantification of the abundance of SL-1 during growth in butyrate, butyrate and propionate, and cholesterol using mass spectrometry. As expected, we found that growth in either propionate or cholesterol increased both the m/z range of SL-1 and its total abundance, and the effect of propionate was dose dependent in the tested range (Fig. 5B). This dose dependency allowed us to correlate changes in sulfolipid abundance with changes in rifampicin efficacy. At multiple propionate concentrations, we calculated the GR50 for RIF and the total abundance of SL-1. We found a correlation between these values, as 0.003× propionate supplementation had no effect on either SL-1 abundance or RIF GR50, and 0.1× increased both values (Fig. 5C). These findings implied that the propionyl-CoA derived from cholesterol alters rifampicin efficacy through modification of mycobacterial surface lipids.
Fig. 5.
Propionate and cholesterol catabolism decreases RIF efficacy in correlation with increased branched chain lipid length and abundance. (A) Normalized growth inhibition of WT across increasing concentrations of RIF in minimal media with 1× butyrate (BT) and corresponding propionate (PP) supplementations as well as cholesterol. Results shown as means from three biological replicates with SDs. (B) Mass spectra of total lipid extract from WT grown in minimal media with 1× BT and corresponding PP supplementations as well as cholesterol. MS spectra intensities were normalized using GM2 ganglioside internal standard and cell density. Results shown as representative from two independent experiments. Mass range characteristic of sulfolipid-1 is shown. (C) Plotted GR50 values (black) and relative sulfolipid abundances (red) across increasing propionate levels. Relative sulfolipid abundances were determined by the combined normalized mass spectra intensities from m/z 2,000 to 2,800 for each growth condition. Results shown as means with SDs.
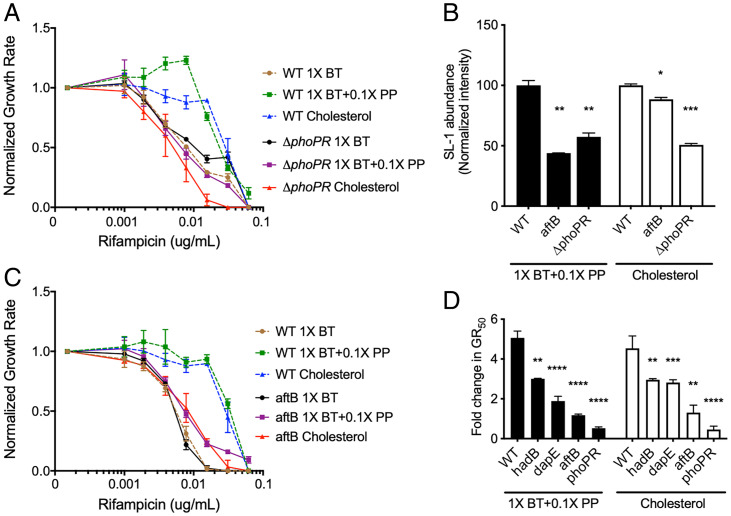
The different branched chain lipid species of Mtb are produced by distinct biosynthetic pathways, and inhibiting the synthesis of one lipid can produce a compensatory increase in others (53). Therefore, to assess whether increased lipid abundance was causally related to RIF efficacy, we employed an Mtb mutant lacking the PhoPR regulatory system, which is required for the synthesis of multiple branched chain lipid species (55–57). We first compared the relative growth rates of ΔphoPR deletion mutant to WT in different carbon sources and found that while ΔphoPR deletion mutant showed a modest growth rate decrease in butyrate, no significant differences were seen in butyrate supplemented with propionate and cholesterol conditions (SI Appendix, Fig. S3). In contrast to wild-type Mtb, a ΔphoPR deletion mutant showed little change in rifampicin efficacy with propionate supplementation or cholesterol compared to butyrate growth conditions and possessed significantly decreased SL-1 levels compared to WT (Fig. 6 A and B). Similar to the effects seen with the ΔphoPR mutant, cell wall defective hypomorphs (aftB, hadB, and dapE) also maintained a relatively consistent RIF GR50 in all media conditions, compared to wild type (Fig. 6 C and D). These findings suggest that cholesterol catabolism reduces RIF efficacy via a propionyl-CoA–driven increase in synthesis of cell surface lipids, and that this effect can be reversed by perturbing the structure of the cell envelope. We note that while depletion of AftB sensitized Mtb to RIF and significantly decreased SL-1 abundance in both cholesterol- and propionate-containing media, the correlation between these two effects was not absolute. AtfB depletion decreased sulfolipid levels to differing degrees under these conditions (Fig. 6B). This implies that cell wall perturbation might be able to reverse the cholesterol- and lipid-driven RIF resistance phenotype without directly altering lipid abundance.
Fig. 6.
Disruption of the cell envelope reverses propionate and cholesterol catabolism dependent decrease in RIF efficacy. (A) Normalized growth inhibition of ΔphoPR mutant across increasing concentrations of RIF in minimal media compared to WT. Results shown as means from three biological replicates. (B) Relative sulfolipid abundances of WT, aftB mutant, and ΔphoPR mutant grown in minimal media with 1× butyrate and 0.1× propionate (1× BT+0.1× PP), and cholesterol. Relative sulfolipid abundances were determined by the combined normalized mass spectra intensities from m/z 2,000 to 2,800 for each growth condition. MS spectra intensities were normalized using GM2 ganglioside internal standard and cell density. Results shown as means with SDs from two independent experiments. Significance was calculated using unpaired t test, *P < 0.05, **P < 0.01. (C) Normalized growth inhibition of aftB mutant across increasing concentrations of RIF in minimal media compared to WT. Results shown as means from three biological replicates. (D) Calculated fold change in GR50 values of 1× butyrate and 0.1× propionate (1× BT+0.1× PP) and cholesterol conditions against 1× butyrate alone for WT and mutant strains. Significance was calculated using unpaired t test, **P < 0.01, ***P < 0.001, ****P < 0.0001.
In Vitro CGIs Predict Drug Efficacy In Vivo.
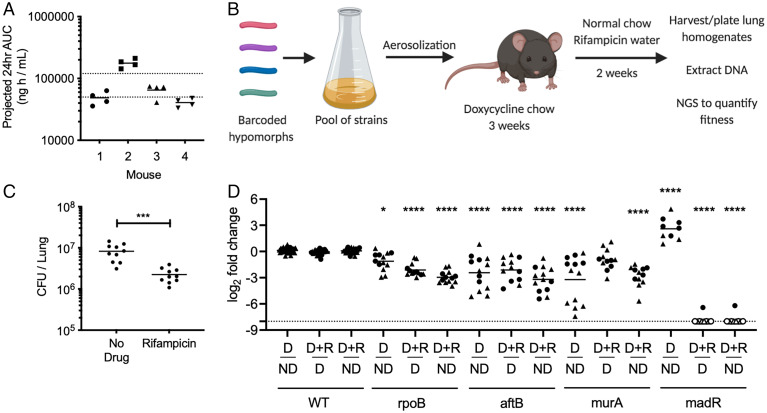
To determine whether CGIs identified in vitro predict strategies to accelerate bacterial killing in the lung, we infected C57BL/6J mice via the aerosol route with a pooled culture consisting of three barcoded WT strains and a number of select hypomorph mutants. We concentrated on genes that either displayed carbon source–independent increase in RIF efficacy (rpoB and madR), or mutants that disrupt the peptidoglycan (murA) or arabinogalactan (aftB) layer of the cells wall and would be expected to reverse the effect of host-derived cholesterol catabolism (Figs. 3 A and B and 4 B and C and SI Appendix, Fig. S4). To ensure that the animals were treated with a relevant dose of antibiotic, we measured the plasma concentration of RIF in mice and adjusted the dosing to match the 24-h exposure observed during clinical TB therapy (Fig. 7A) (58). The mice were fed doxycycline chow starting 3 d before infection to repress sspB expression in the mutants upon infection and allow them to grow normally. At 3 wk postinfection, mice were split into three groups. One group maintained doxycycline for the remainder of the infection without RIF treatment (ND, nondepleted). A second group had doxycycline withdrawn to initiate protein degradation for the remainder of the infection without RIF treatment (D, depleted). The third group had doxycycline withdrawn and 2 wk of RIF treatment was administered (D+R, depleted and RIF treated) (Fig. 7B) (59). The antibiotic regimen effectively decreased the total number of lung colony-forming units (CFU) (Fig. 7C). Lung homogenates were washed and plated on 7H10 plates with supplementation to recover viable clones. The relative abundance of individual mutants was normalized to the average abundance of three barcoded WT strains for each mouse and fitness changes were calculated by comparing between conditions. Comparing groups D and ND shows the relative fitness effects of depletion alone on individual hypomorphs, while examining groups D+R and D shows the relative change in fitness due to RIF treatment across depleted samples. Groups ND and D+R were compared to investigate the combined impact of both depletion and RIF treatment.
Fig. 7.
Essential bacterial functions that alter drug efficacy in vivo. (A) RIF plasma concentrations were measured in mice over a 12-h period, 24 h post-RIF administration (0.1 g/L). Results shown as area under the concentration (AUC) time profile. Dotted lines indicate RIF plasma range observed during clinical TB therapy (58). (B) C57BL/6J mice were infected through the aerosol route with a pooled culture of individual hypomorph mutants and barcoded WT strains. Mice were fed doxycycline chow starting 3 d before infection to 3 wk postinfection. Mice were subsequently switched to normal chow and water with or without RIF (0.1 g/L) for 2 additional weeks. Lungs were harvested, homogenized, and plated for Mtb outgrowth. Upon DNA extraction of grown colonies, chromosomal barcodes were PCR amplified and pooled for Illumina NGS. Barcode abundances of individual mutants were normalized to WT strains and analyzed to quantify changes in fitness during RIF treatment. (C) RIF-treated mice show significant decrease in Mtb CFU in lungs compared to untreated controls. Each dot represents a single mouse. Significance was calculated using unpaired t test, ***P < 0.001. (D) Change in fitness of individual mutants was determined by comparing normalized counts from nondepleted (ND), depleted (D), as well as depleted and RIF-treated (D+R) conditions. Depletion of RpoB, AftB, and MurA display significant decrease in fitness, shown as log2 fold change, compared to nondepleted controls. Depletion of RpoB, AftB, and MadR show significant decrease in fitness during RIF treatment compared to depleted but untreated controls. Results shown as two independent experiments (circle and triangle) with each dot representing a single mouse. Empty dot indicates that the relative abundance was below the limit of detection (dotted line). Significance was calculated using Sidak’s multiple comparison test, *P < 0.05, ****P < 0.0001.
RpoB, AftB, and MurA depletion decreased fitness compared to nondepleted controls, while MadR depletion increased relative fitness in the mouse model. Upon RIF treatment, we found that AftB depletion increased bacterial clearance to the same degree as RpoB depletion. Depleting MadR had an even greater effect, and the abundance of this mutant was reduced below our limit of detection in 11 out of 12 animals (Fig. 7D). The effect of MurA depletion was more variable and its effect on RIF efficacy was not statistically significant. In sum, the reproducible effects of RpoB, AtfB, and MadR on RIF efficacy show that CGI identified in vitro can accelerate bacterial clearance during infection and they highlight the importance of the environment for defining relevant interactions.
Discussion
Optimizing combination therapy is critical to improving TB treatment. While efficient strategies for predicting and quantifying drug–drug interactions under in vitro conditions have been developed, the impact of bacterial environment on these interactions has not been determined (27, 28). As a result, it remains unclear how well these in vitro data predict the effects of combination therapy in the infection environment. In this study, we modeled drug–drug interactions using a newly developed genetic hypomorph library to assess the impact of bacterial carbon metabolism on CGI. This work revealed that CGIs with distinct drugs are differentially sensitive to the environment, suggesting tailored approaches to optimization.
While global analysis of our chemical–genetic–environmental dataset suggested that drug mechanism and environment both play critical roles in shaping chemical–genetic interactions, all drugs were not equally affected by condition. In particular, the individual effect of the fluoroquinolone, MOX, was unaffected by carbon source, and this relative indifference to the environment was also observed for CGI with MOX. These observations contrast with RIF and INH, where carbon source plays an important role in determining the MIC of the drug alone, as well as shaping chemical–genetic interaction profiles. While we can only speculate on the mechanistic basis for these differences between drugs, we found that the cholesterol-dependent decrease in RIF sensitivity is due to the effect of carbon source on cell envelope structure that is likely to alter RIF penetration. Similarly, the activation of the prodrug, INH, is affected by the redox state of the cell (60), another process that is impacted by carbon catabolism. In contrast, MOX is not a prodrug, and we speculate that it may not be limited by cellular penetration as strongly as RIF. More practically, these observations suggest that drug–drug interactions with MOX may be particularly robust to changes in condition, and therefore may translate well between in vitro and in vivo conditions.
Complex drug–drug interactions may both increase or decrease sensitivity. Our dataset found potential antagonistic interactions that may arise with different TB antibiotics. For example, the inhibition of Ndh, a NADH-dehydrogenase, has previously been shown to decrease INH efficacy and was also identified in our dataset (37). Antagonistic interactions have important implications for TB therapeutics due to the reliance on multidrug regimens, and our study provides a global assessment of these effects.
While RIF interactions were complex and condition dependent compared to MOX, we found that the cholesterol-specific effects translate well to the infection environment and may be particularly relevant to treatment. As reduced RIF exposure limits the efficacy of this critical component of our standard regimen, even small increases in MIC during infection could have a significant effect on bacterial killing. A number of observations indicate that RIF efficacy is limited by similar mechanisms in vivo and in cholesterol-containing media. The increase in branched chain lipid abundance that is associated with reducing RIF efficacy in cholesterol- or propionate-containing media has also been observed during infection, where similar carbon sources are utilized (16, 53). Furthermore, genetic perturbation of the same cell wall synthetic enzymes sensitized Mtb to RIF in both conditions. This effect may underlie the increased cell-associated RIF upon simultaneous INH and ethambutol treatment in vitro (61–63). While the importance of cell wall architecture and cell envelope lipid production suggests that cellular permeability to RIF may be altered during growth on cholesterol, we note that lipid synthesis also has important effects on central carbon metabolic pathways that could influence antibiotic susceptibility (15). In addition, the mixture of carbon sources available in different host microenvironments remains ill defined, and cholesterol may not be the only macronutrient capable of limiting RIF efficacy. Additional enzymes identified in this study may also be attractive targets for further drug development, such as AftB, a arabinofuranosyltransferase distinct from EmbA and EmbB that are targets of ethambutol (47). Similarly, enzymes in the diaminopimelic acid (DAP) biosynthesis pathway produce precursors for the synthesis of peptidoglycan and lysine (64), and the loss of either sensitizes the cell to RIF.
Using a chemical–genetic system, we determined that cellular environment plays an important role in shaping CGIs, and that even the use of relatively simple in vitro culture conditions can identify interactions that are relevant during infection. These studies provide a tractable system that can incorporate more complex culture systems, or even cellular models, to identify additional in vivo relevant interactions. Expanding this CGI atlas promises to elucidate the processes limiting drug efficacy during infection and guide drug development and regiment optimization efforts.
Materials and Methods
Bacterial Cultures.
Mtb (H37Rv) cells were cultured in Middlebrook 7H9 medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.5% glycerol, and 0.05% Tween-80, or in minimal medium supplemented with 0.1% tyloxapol and variable levels of glycerol, acetate, butyrate, propionate, or cholesterol up to 0.1%. Minimal medium was made as previously described but with ferric chloride (100 μM) replacing ferric ammonium citrate (51).
Cells were also cultured on 7H10 agar medium supplemented with 10% OADC and 0.5% glycerol. Streptomycin (20 μg/mL) was supplemented when necessary. Anhydrotetracycline (ATC, 500 ng/mL) was supplemented periodically until cultures reached exponential growth. ATC was removed from cultures prior to antibiotic growth inhibition assays by washing in phosphate-buffered saline (PBS) supplemented with 0.1% tyloxapol. ΔphoPR mutant was generated as described previously (65).
Multiplexed Library Screening.
Library pools of hypomorph mutants were prepared as described previously (32). Pools were initially cultured in Middlebrook 7H9 medium supplemented with 10% OADC, 0.5% glycerol, and 0.05% Tween-80. Cultures were supplemented with ATC (500 ng/mL) periodically until cultures reached exponential growth. ATC was removed from cultures by washing in PBS supplemented with 0.1% tyloxapol prior to inoculation in 96-well plates to allow induction of SspB. Pools were grown in minimal medium with 0.1% glycerol, 0.1% acetate, or 0.1% cholesterol on 96-well plates for 2 wk. Isoniazid and moxifloxacin were used at 1 μg/mL and rifampicin was used at 0.062 μg/mL. Antibiotics were added to select wells and serially diluted in a twofold manner prior to inoculation. An untreated growth condition was included for each study. Samples were inoculated at OD600 0.01 and growth was monitored using OD600. Upon completion, 96-well plates were heat-inactivated at 85 °C for 2 h. Barcodes were PCR (polymerase chain reaction)-amplified as described previously (32). Individual libraries were mixed with 1:1 20% dimethyl sulfoxide (DMSO) and heated for 10 min at 98° prior to PCR. Amplified barcodes were purified using SPRI (solid phase reversible immobilization)-based purification methods and sequenced using next-generation sequencing methods. Sequence alignment and analysis were conducted using the Bowtie software package with index mismatch set to 2 bases and barcode mismatch set to 1 base. Relative abundance of every mutant was calculated as mean ratio of barcode abundance of mutant relative to total barcode abundance of library. Log2FC of a mutant was calculated as the log2 of the ratio of the mean relative abundances in a given antibiotic condition relative to its untreated control. Hierarchical clustering and principal component analysis were conducted using ClustVis (66). Hierarchical clustering was applied to vectors of log2FC of each gene across all conditions. PCA was performed on relative abundances across all conditions. Statistical significance was determined by unpaired t test with Benjamini–Hochberg multiple testing correction.
Antibiotic Growth Inhibition Assay.
Mtb cells were grown in minimal medium with 0.1% glycerol, 0.01% acetate, 0.01% butyrate, 0.01% propionate, or 0.01% cholesterol on 96-well plates. Isoniazid and moxifloxacin were used at 1 μg/mL and rifampicin was used at 0.062 μg/mL. Antibiotics were added to select wells and serially diluted in a twofold manner prior to inoculation. Untreated condition was included for each study. Cells were inoculated at OD600 0.01 and growth was monitored using OD600 until exponential phase (OD600 0.20 to 0.25). Antibiotic efficacy was determined using growth rate inhibition as done previously (24). The exponential growth constant (k) value was calculated by plotting an exponential curve over all growth points. The k value of each antibiotic concentration was normalized to the k value of the no-drug control. The GR50 value was defined as the concentration of antibiotic that resulted in 50% decrease in growth rate. L-lysine (Sigma) was added to growth conditions to a final concentration of 3 mM.
Lipid Extraction and Mass Spectrometry.
Mtb cells were grown in minimal medium with 0.01% butyrate, propionate, or cholesterol. Cells were inoculated at OD600 0.1 and grown to final OD600 of 0.7. Cells were pelleted and heat inactivated at 85 °C for 45 min. Cells were washed in 10% glycerol to remove residual detergents from growth media. Lipid extraction was conducted with methyl tert-butyl ether (MTBE) as described previously (67). A total of 1.5 mL methanol and 5 mL MTBE were mixed with the cell pellet and placed into a glass tube with a Teflon-lined cap. The mixture was incubated at room temperature for 5 h while shaking. Phase separation was achieved by adding 0.75 mL water to the tubes, incubating for 10 min at room temperature, then centrifuging at 1,000 × g for 10 min. The 2.5 mL of the top phase was collected and transferred to a fresh glass tube with a Teflon-lined cap. Samples were dried using a nitrogen evaporator and stored in −20 °C prior to injection to the mass spectrometer. After evaporation of MTBE, 300 μL of 2:1 methanol:chloroform was added to each tube followed by vigorous vortex. Solubilized lipids were then transferred to a new, preweighed tube, dried using a nitrogen evaporator, and then tubes were reweighed to determine the lipid mass in each sample. Then 1.5 μL of 100 ng/μL N-omega-CD3-octadecanoyl monosialoganglioside GM2 (Matreya) was added to each sample for normalization and samples were reconstituted in 300 μL of 2:1 methanol:chloroform for mass spectrometry analysis.
Samples were analyzed by direct infusion on a syringe pump to an Orbitrap Velos Pro (Thermo Fisher Scientific) mass spectrometer operating in the negative electrospray ionization mode. Mass spectra were acquired at a flow rate of 10 μL/min for 2 min in two different mass ranges, m/z 200 to 2,000 and m/z 300 to 3,000, acquiring two replicates for each range using a resolution of 30,000 (m/z 200), an automatic gain control (AGC) target population of 5e5 and a maximum ion injection time of 100 ms. Data were analyzed in Xcalibur 2.2 (Thermo Scientific). Briefly, mass spectra were averaged over the entire acquisition range (0 to 2 min), producing one averaged spectrum for each analysis. Peak lists were exported in Excel and intensity data were normalized to the intensity of the GM2 ganglioside spike.
Plasma Pharmacokinetic Analysis.
C57BL/6J mice were purchased from The Jackson Laboratory. Housing and experimentation were done in accordance with the guidelines set by the Department of Animal Medicine of University of Massachusetts Medical School and Institutional Animal Care and Use Committee and adhered to the laws of the United States and regulations of the Department of Agriculture. Eight- to 12-wk mice were administered 0.1 g/L RIF through drinking water for 3 d. Blood was collected during a 12-h period 24 h post-RIF administration, kept on ice, centrifuged at ∼2,500 × g for 5 min. After centrifugation, plasma was collected and stored at −80 °C until analysis.
Next 1 mg/mL DMSO stocks for RIF were serial diluted in 50/50 acetonitrile/water to create neat spiking stocks. Standards and quality control (QC) samples were created by adding 10 μL of spiking stock to 90 μL of drug-free plasma. Ten microliters of control, standard, QC, or study sample were added to 100 μL of acetonitrile/methanol 50/50 protein precipitation solvent containing 20 ng/mL RIF-d8. Extracts were vortexed for 5 min and centrifuged at 4,000 RPM for 5 min. A total of 100 μL of supernatant was combined with 5 μL of 75 mg/mL ascorbic acid to stabilize RIF. A total of 100 μL of mixture was combined with 100 μL of Milli-Q water prior to high-performance liquid chromatography tandem mass spectrometry (HPLC-MS/MS) analysis. Mouse control plasma (potassium ethylenediaminetetraacetic acid) was sourced from Bioreclamation. Mouse control lungs were collected in house. RIF was sourced from Sigma-Aldrich and RIF-d8 was purchased from Toronto Research Chemicals.
LC-MS/MS analysis was performed on a Sciex Applied Biosystems Qtrap 6500+ triple-quadrupole mass spectrometer coupled to a Shimadzu Nexera ×2 UHPLC system to quantify each drug in plasma. Chromatography was performed on a Agilent SB-C8 (2.1 × 30 mm; particle size, 3.5 μm) using a reverse phase gradient. Milli-Q deionized water with 0.1% formic acid was used for the aqueous mobile phase and 0.1% formic acid in acetonitrile for the organic mobile phase. Multiple-reaction monitoring of precursor/product transitions in electrospray positive-ionization mode was used to quantify the analytes. Sample analysis was accepted if the concentrations of the quality-control samples were within 20% of the nominal concentration. The compounds were ionized using electrospray ionization (ESI) positive mode ionization and monitored using masses of RIF (823.50/791.60) and RIF-d8 (831.50/799.60). Data processing was performed using Analyst software (version 1.6.2; Applied Biosystems Sciex).
In Vivo Antibiotic Susceptibility.
C57BL/6J mice were purchased from The Jackson Laboratory. Housing and experimentation were done in accordance with the guidelines set by the Department of Animal Medicine of the University of Massachusetts Medical School and Institutional Animal Care and Use Committee and adhered to the laws of the United States and regulations of the Department of Agriculture. Eight- to 12-wk mice were infected with pools of strains at equal ratios through the aerosol route (500 to 1,000 CFU/mouse). Mice were fed doxycycline-containing chow (Purina 5001 with 2,000 ppm doxycycline, Research Diets C11300-2000i) starting 3 d preinfection to 21 d postinfection. At 21 d postinfection, 0.1g/L rifampicin was administered through drinking water for 14 d. At 35 d postinfection, mice were euthanized, spleen and lungs were isolated and homogenized, and CFU was determined by plating dilutions on 7H10 agar with 50 μg/mL streptomycin and 500 ng/mL ATC. For library recovery, ∼1 million CFU per mouse were plated on 7H10 agar with 50 μg/mL streptomycin and 500 ng/mL ATC. Genomic DNA was extracted and normalized as done previously (35) and sequenced using multiplex PCR methods. Sequence alignment and analysis were conducted using the Bowtie software package as described above. Relative abundance of every mutant was calculated as mean ratio of barcode abundance of mutant relative to the average barcode abundance of three barcoded H37Rv strains. Fitness changes were calculated by comparing normalized counts between conditions. Statistical significance was determined using Sidak's multiple comparison test.
Supplementary Material
Acknowledgments
We are thankful to the members of the C.M.S. laboratory for both technical assistance and helpful discussion. We thank Sovie Lavalette-Levy and Curtis Engelhart for technical help. This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Program, Focused Program Award, under Award W81XWH-17-1-0692. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense. The work was additionally supported by the NIH Grant AI095208. Model figures were created with BioRender.com.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201632119/-/DCSupplemental.
Data Availability
All study data neccessary to interpret or repeat analyses are included in the article and/or supporting information.
References
- 1.Streptomycin Treatment of Pulmonary Tuberculosis, Streptomycin treatment of pulmonary tuberculosis. BMJ 2, 769–782 (1948). [PMC free article] [PubMed] [Google Scholar]
- 2.Fox W., Mitchison D. A., Short-course chemotherapy for pulmonary tuberculosis. Am. Rev. Respir. Dis. 111, 325–353 (1975). [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization, Guidelines for Treatment of Tuberculosis (World Health Organization, ed. 4, 2010). [Google Scholar]
- 4.World Health Organization, Global Tuberculosis Report 2020 (World Health Organization, 2020). [Google Scholar]
- 5.Gillespie S. H., et al. , Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N. Engl. J. Med. 371:1577–1587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nahid P., et al. , Treatment of drug-resistant tuberculosis. An official ATS/CDC/ERS/IDSA clinical practice guideline. Am. J. Respir. Crit. Care Med. 200, e93–e142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez J. E., McKinney J. D., M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb.) 84, 29–44 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Ordonez A. A., et al. , Dynamic imaging in patients with tuberculosis reveals heterogeneous drug exposures in pulmonary lesions. Nat. Med. 26, 529–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kjellsson M. C., et al. , Pharmacokinetic evaluation of the penetration of antituberculosis agents in rabbit pulmonary lesions. Antimicrob. Agents Chemother. 56, 446–457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svensson R. J., et al. , Greater early bactericidal activity at higher rifampicin doses revealed by modeling and clinical trial simulations. J. Infect. Dis. 218, 991–999 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Boeree M. J., et al. ; PanACEA Consortium, High-dose rifampicin, moxifloxacin, and SQ109 for treating tuberculosis: A multi-arm, multi-stage randomised controlled trial. Lancet Infect. Dis. 17, 39–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baek S.-H., Li A. H., Sassetti C. M., Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 9, e1001065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams K. N., et al. , Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., et al. , Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J. Exp. Med. 213, 809–825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovewell R. R., Sassetti C. M., VanderVen B. C., Chewing the fat: Lipid metabolism and homeostasis during M. tuberculosis infection. Curr. Opin. Microbiol. 29, 30–36 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Pandey A. K., Sassetti C. M., Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baughn A. D., Rhee K. Y., Metabolomics of central carbon metabolism in Mycobacterium tuberculosis. Microbiol. Spectr. 2, 2.3.02 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Aguilar-Ayala D. A., et al. , Antimicrobial activity against Mycobacterium tuberculosis under in vitro lipid-rich dormancy conditions. J. Med. Microbiol. 67, 282–285 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Baker J. J., Abramovitch R. B., Genetic and metabolic regulation of Mycobacterium tuberculosis acid growth arrest. Sci. Rep. 8, 4168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzblau S. G., et al. , Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis (Edinb.) 92, 453–488 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Pethe K., et al. , A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat. Commun. 1, 57 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopal P., et al. , Pyrazinamide resistance is caused by two distinct mechanisms: Prevention of coenzyme a depletion and loss of virulence factor synthesis. ACS Infect. Dis. 2, 616–626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalia N. P., et al. , Carbon metabolism modulates the efficacy of drugs targeting the cytochrome bc1:aa3 in Mycobacterium tuberculosis. Sci. Rep. 9, 8608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellerose M. M., et al. , Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. MBio 10, e00663-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicks N. D., et al. , Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3, 1032–1042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann G. R., Lehár J., Keith C. T., Multi-target therapeutics: When the whole is greater than the sum of the parts. Drug Discov. Today 12, 34–42 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Cokol M., Kuru N., Bicak E., Larkins-Ford J., Aldridge B. B., Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis. Sci. Adv. 3, e1701881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma S., et al. , Transcriptomic signatures predict regulators of drug synergy and clinical regimen efficacy against tuberculosis. MBio 10, e02627-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasipanodya J., Gumbo T., An oracle: Antituberculosis pharmacokinetics-pharmacodynamics, clinical correlation, and clinical trial simulations to predict the future. Antimicrob. Agents Chemother. 55, 24–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuermberger E., Sizemore C., Romero K., Hanna D., Toward an evidence-based nonclinical road map for evaluating the efficacy of new tuberculosis (TB) drug regimens: Proceedings of a critical path to TB drug regimens-National Institute of Allergy and Infectious Diseases In Vivo Pharmacology Workshop for TB Drug Development. Antimicrob. Agents Chemother. 60, 1177–1182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.-H., et al. , Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic Acids Res. 39, 2210–2220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson E. O., et al. , Large-scale chemical-genetics yields new M. tuberculosis inhibitor classes. Nature 571, 72–78 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Parsons A. B., et al. , Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126, 611–625 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Stokes J. M., Lopatkin A. J., Lobritz M. A., Collins J. J., Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251–259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellerose M. M., et al. , Distinct bacterial pathways influence the efficacy of antibiotics against Mycobacterium tuberculosis. mSystems 5, e00396-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hafner M., Niepel M., Chung M., Sorger P. K., Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods 13, 521–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee A. S. G., Teo A. S. M., Wong S.-Y., Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 45, 2157–2159 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.-H., et al. , A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc. Natl. Acad. Sci. U.S.A. 110, 19095–19100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei J.-R., et al. , Depletion of antibiotic targets has widely varying effects on growth. Proc. Natl. Acad. Sci. U.S.A. 108, 4176–4181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giaever G., et al. , Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 21, 278–283 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Peterson E. J., et al. , Path-seq identifies an essential mycolate remodeling program for mycobacterial host adaptation. Mol. Syst. Biol. 15, e8584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Geize R., et al. , A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diacon A. H., et al. , Early bactericidal activity of high-dose rifampin in patients with pulmonary tuberculosis evidenced by positive sputum smears. Antimicrob. Agents Chemother. 51, 2994–2996 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chigutsa E., et al. , Impact of nonlinear interactions of pharmacokinetics and MICs on sputum bacillary kill rates as a marker of sterilizing effect in tuberculosis. Antimicrob. Agents Chemother. 59, 38–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasipanodya J. G., et al. , Serum drug concentrations predictive of pulmonary tuberculosis outcomes. J. Infect. Dis. 208, 1464–1473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jankute M., Cox J. A. G., Harrison J., Besra G. S., Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Seidel M., et al. , Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 282, 14729–14740 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Usha V., et al. , Reconstruction of diaminopimelic acid biosynthesis allows characterisation of Mycobacterium tuberculosis N-succinyl-L,L-diaminopimelic acid desuccinylase. Sci. Rep. 6, 23191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jankute M., et al. , Disruption of mycobacterial AftB results in complete loss of terminal β(1 → 2) arabinofuranose residues of lipoarabinomannan. ACS Chem. Biol. 12, 183–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefebvre C., et al. , HadD, a novel fatty acid synthase type II protein, is essential for alpha- and epoxy-mycolic acid biosynthesis and mycobacterial fitness. Sci. Rep. 8, 6034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griffin J. E., et al. , Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson M., Stadthagen G., Gicquel B., Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: Biosynthesis, transport, regulation and biological activities. Tuberculosis (Edinb.) 87, 78–86 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Jain M., et al. , Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc. Natl. Acad. Sci. U.S.A. 104, 5133–5138 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X., Nesbitt N. M., Dubnau E., Smith I., Sampson N. S., Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry 48, 3819–3821 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walters S. B., et al. , The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60, 312–330 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Gonzalo Asensio J., et al. , The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J. Biol. Chem. 281, 1313–1316 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Baker J. J., Johnson B. K., Abramovitch R. B., Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol. Microbiol. 94, 56–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stott K. E., et al. , Pharmacokinetics of rifampicin in adult TB patients and healthy volunteers: A systematic review and meta-analysis. J. Antimicrob. Chemother. 73, 2305–2313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gengenbacher M., et al. , Tissue distribution of doxycycline in animal models of tuberculosis. Antimicrob. Agents Chemother. 64, e02479-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wengenack N. L., Hoard H. M., Rusnak F., Isoniazid oxidation by Mycobacterium tuberculosis KatG: A role for superoxide which correlates with isoniazid susceptibility. J. Am. Chem. Soc. 121, 9748–9749 (1999). [Google Scholar]
- 61.McNeil M. B., Chettiar S., Awasthi D., Parish T., Cell wall inhibitors increase the accumulation of rifampicin in Mycobacterium tuberculosis. Access Microbiol. 1, e000006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piddock L. J. V., Williams K. J., Ricci V., Accumulation of rifampicin by Mycobacterium aurum, Mycobacterium smegmatis and Mycobacterium tuberculosis. J. Antimicrob. Chemother. 45, 159–165 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Bhusal Y., Shiohira C. M., Yamane N., Determination of in vitro synergy when three antimicrobial agents are combined against Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 26, 292–297 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Pavelka M. S. Jr., Jacobs W. R. Jr., Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J. Bacteriol. 178, 6496–6507 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J., Repasy T., Papavinasasundaram K., Sassetti C., Kornfeld H., Mycobacterium tuberculosis induces an atypical cell death mode to escape from infected macrophages. PLoS One 6, e18367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Metsalu T., Vilo J., ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43 (W1), W566–W570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., Schwudke D., Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data neccessary to interpret or repeat analyses are included in the article and/or supporting information.