Significance
Carotenoid pigments accumulate in specific patterns in vertebrate tissues and play important roles as colorants, chromophores, and hormone precursors. However, proteins that facilitate transportation of these lipophilic pigments within cells have not been identified. We provide evidence that Aster proteins are key components for this process and show that they bind the pigments with high affinity. We observed in mice that carotenoids accumulate in tissues that express Aster-B and this accumulation can be prevented by enzymatic turnover by the BCO2 protein. Accordingly, we found opposing expression patterns of the Aster-B protein and BCO2 in the human retina that seemingly contribute to the unique carotenoid concentration in the macula lutea.
Keywords: GRAMD1, carotenoids, cholesterol, BCO2, retina
Abstract
Some mammalian tissues uniquely concentrate carotenoids, but the underlying biochemical mechanism for this accumulation has not been fully elucidated. For instance, the central retina of the primate eyes displays high levels of the carotenoids, lutein, and zeaxanthin, whereas the pigments are largely absent in rodent retinas. We previously identified the scavenger receptor class B type 1 and the enzyme β-carotene-oxygenase-2 (BCO2) as key components that determine carotenoid concentration in tissues. We now provide evidence that Aster (GRAM-domain-containing) proteins, recently recognized for their role in nonvesicular cholesterol transport, engage in carotenoid metabolism. Our analyses revealed that the StART-like lipid binding domain of Aster proteins can accommodate the bulky pigments and bind them with high affinity. We further showed that carotenoids and cholesterol compete for the same binding site. We established a bacterial test system to demonstrate that the StART-like domains of mouse and human Aster proteins can extract carotenoids from biological membranes. Mice deficient for the carotenoid catabolizing enzyme BCO2 concentrated carotenoids in Aster-B protein-expressing tissues such as the adrenal glands. Remarkably, Aster-B was expressed in the human but not in the mouse retina. Within the retina, Aster-B and BCO2 showed opposite expression patterns in central versus peripheral parts. Together, our study unravels the biochemical basis for intracellular carotenoid transport and implicates Aster-B in the pathway for macula pigment concentration in the human retina.
Carotenoids are a familiar sight as yellow, orange, and red colors in the natural world and are beneficial for human health (1). These organic compounds retain an extended polyene chromophore with distinct physical and chemical properties (2). The pigments act as antioxidants in lipophilic environments and as filters of short wavelength light (3). Many animals, including humans, concentrate carotenoids in specific cell types and tissues (4). For instance, the macula lutea in the primate retina owes its yellow color to the accumulation of the carotenoids lutein and zeaxanthin (5). These macula pigments reduce chromatic aberration and light scattering in vision and protect the photoreceptors against abiotic stress (6). Additionally, carotenoids such as β-carotene are the metabolic precursors of vitamin A (all-trans-retinol) (7) that plays crucial roles in mammalian physiology. The vitamin A metabolites retinaldehyde and retinoic acid act as chromophore of G protein coupled visual receptors (rhodopsin) and as ligand of nuclear hormone receptors (8), respectively.
Carotenoids are produced by plants, bacteria, and fungi, and animals acquire them almost exclusively through their diets (9). Upon absorption in enterocytes of the intestine, carotenoids are packaged in lipoproteins and delivered to target tissues (10). Once in cells, carotenoids can move from the plasma membrane to the endoplasmic reticulum (ER) and to the outer and inner mitochondrial membranes (11). In mitochondria, carotenoids are converted to apocarotenoids by the enzyme β-carotene-oxygenase-2 (BCO2) (12). However, little is known about mechanisms that facilitate the intracellular transport of these lipids that contribute to either accumulation or catabolic turnover in cells.
The scavenger receptor class 1 type 2 (SR-B1) mediates the bidirectional flux of cholesterol between high density lipoproteins and cells (13) and also facilitates the uptake of other terpenoids, including carotenoids (14). Recent research revealed that members of the Aster protein family, also known as GRAM domain-containing (GRAMD), act downstream of SR-B1 and engage in nonvesicular cholesterol transport in cells (15). Mammalian genomes encode three closely related family members Aster-A (GRAMD1A), Aster-B (GRAMD1B), and Aster-C (GRAMD1C) (SI Appendix, Fig. S1) (16). Aster proteins are anchored with their C-terminal transmembrane domain in the ER whereas the bulk of the protein is exposed to the cytoplasm space. The central part of Aster proteins contains a StART-like domain that binds sterols in its lipophilic cleft (15). The N-terminal GRAM domain of the Aster proteins interacts with phosphatidylserine residues of the inner leaflet of the plasma membrane (SI Appendix, Fig. S1). The domain-like structure allows Aster proteins to form bridges between the ER and plasma membrane and to facilitate the exchange of cholesterol (15). Studies in cell lines and mouse models revealed that Aster proteins play a critical role in cellular cholesterol homeostasis and steroidogenesis by mobilizing cholesterol from the plasma membrane to ER (17) and mitochondria (18).
The chemical similarities between cholesterol and carotenoids led us to speculate that Aster proteins mediate the movement of carotenoids between cellular compartments, including their transportation to mitochondria. To test this hypothesis, we established a bacterial test system to determine whether the StART-like domain of Aster proteins can extract carotenoids from membranes. By structural modeling and binding assays, we demonstrate that this domain is large enough to accommodate the bulky pigments. In BCO2-deficient mice, we showed that carotenoids accumulated in Aster-B-expressing tissues such as the adrenal glands. Furthermore, we provide evidence that Aster-B is expressed at high levels in the human but not in the mouse retina. Consistent with the spatial distribution of carotenoids, the Aster-B and the carotenoid catabolizing enzyme BCO2 displayed opposite expression patterns in peripheral versus central parts of the human retina.
Results
The StART Domain of Aster-B Extracts Carotenoids from Membranes.
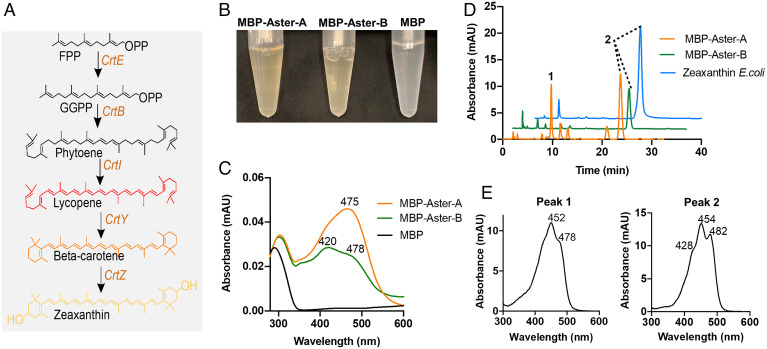
We cloned the cDNA encoding the StART-like domains from Gramd1A (Aster-A) and Gramd1B (Aster-B) from mouse liver (SI Appendix, Fig. S1). We then expressed the MBP-Aster-A and MBP-Aster-B fusion proteins in Escherichia coli cells equipped with genes for zeaxanthin synthesis. This synthesis takes place at bacterial membranes via the intermediates phytoene, lycopene, β-carotene, and β-cryptoxanthin (19) (Fig. 1A). The bacterial expression system allowed testing whether the StART-like domain of Aster proteins can extract the membrane-born pigments and form carotenoprotein complexes. As a control, we expressed maltose binding protein (MBP) alone in this bacterial strain. The eluate of the affinity chromatography purified recombinant MBP-Aster fusion proteins showed a characteristic yellow hue whereas the MBP eluate was colorless (Fig. 1B). We recorded ultraviolet/visible (UV/Vis) spectra of the putative carotenoprotein complexes of the MBP-Aster fusion proteins and compared it to purified MBP (Fig. 1C). The MBP-Aster-A fusion protein displayed a peak maximum at 475 nm and the MBP-Aster-B fusion protein at 420 nm with an additional peak shoulder at 478 nm (Fig. 1C). To demonstrate that the spectral properties of the purified fusion proteins resulted from carotenoid binding, we denatured the complexes in organic solvent and extracted the bound pigments. The organic phases were concentrated and subjected to high-performance liquid chromatography (HPLC) analysis as described in the experimental section. In lipid extracts from purified MBP-Aster-A, we detected β-cryptoxanthin and zeaxanthin (Fig. 1 D and E). Lipid extracts of purified MBP-Aster-B contained zeaxanthin as sole carotenoid (Fig. 1 D and E). MBP lipid extracts did not contain any carotenoids. Thus, we established a bacterial test system for the characterization of mammalian carotenoid binding proteins and observed that the StART-like domains of Aster-A and B can extract carotenoids from bacterial membranes.
Fig. 1.
A bacterial test system for carotenoid binding proteins. (A) Bacterial zeaxanthin biosynthesis pathway. (B) Color comparison of the eluates of purified recombinant MBP-Aster-A, MBP-Aster-B, and MBP. (C) UV/Visible absorption spectra of recombinant murine MBP-Aster-A (orange trace), MBP-Aster-B (green trace), and MBP (black trace) (D) HPLC traces at 460 nm of organic extracts from the zeaxanthin biosynthesizing E.coli cells (blue trace) and purified MBP-Aster-A (orange trace) and MBP-Aster-B (green trace). (E) UV/Vis absorption spectra for the indicated peak 1 (β-cryptoxanthin) and peak 2 (zeaxanthin) in the chromatograms. The absorption maximum of each peak is indicated in the panels.
Characterization of the Interaction of Aster Proteins with Carotenoids.
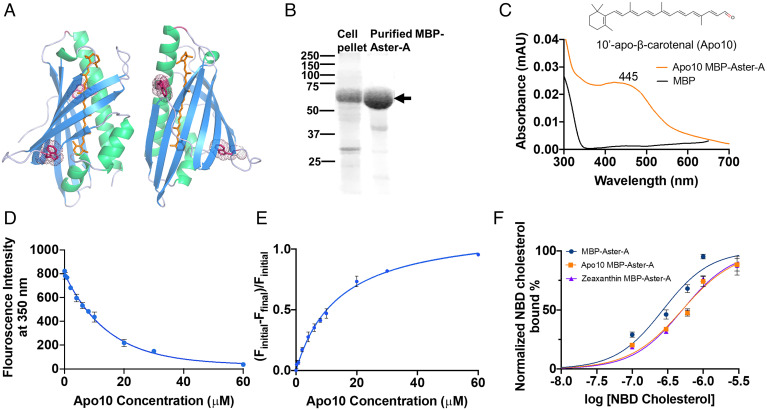
Recent structural analysis of the StART-like domains of mouse Aster-A (PDB ID: 6GQF) and Aster-C (PDB ID: 7AZN) revealed that in addition to cholesterol (C27), the binding cavity accommodated a glycerol molecule (SI Appendix, Fig. S1) (15). This finding suggested that the dimensions of the binding cavity are large enough to accommodate the rigid and elongated structure of a carotenoid (C40). In silico analysis revealed that a carotenoid can be modeled into the binding cavity of mouse Aster A without steric clashes (Fig. 2A). To further characterize the binding of carotenoids, we focused on MBP-Aster-A because we were able to express significant amounts of the fusion protein in soluble form in E. coli BL21 and to purify the apo-protein by affinity chromatography (Fig. 2B). To establish an in vitro binding assay, we used 10′-apo-β-carotenal (Apo10). Apo10 is a carotenoid metabolite that can interact with recombinant proteins when dissolved in Triton X-100 detergent solution. Previously, we used this in vitro system to characterize the interaction of Apo10 and other apo-β-carotenoids with the enzyme BCO2 (20). Thus, we incubated MBP-Aster-A with detergent solubilized Apo10 and purified the resulting carotenoprotein complexes by affinity chromatography. The purified proteins displayed characteristic spectral properties with a maximum of absorption at 445 nm (Fig. 2C). To further examine the interaction between MBP-Aster-A and Apo10, we took advantage of two tryptophan residues in the lipid binding cavity of the StART-like domain of Aster A (Fig. 2A). We observed in the test tube that the binding of Apo10 to MBP-Aster-A led to a dose-dependent quenching of the tryptophan fluorescence of these residues (SI Appendix, Fig. S2). By recording the quenching at different concentrations of Apo10 in a fluorimeter, we generated binding curves with MBP-Aster-A protein (Fig. 2D). Plotting the ratio of the bound over free fraction with increasing Apo10 concentrations revealed that the binding approached a maximum as indicated by the hyperbolic binding curve (Fig. 2E). Using a curve fitting program, we determined the dissociation constant of the Apo10 carotenoprotein complex to be in the micromolar range.
Fig. 2.
Recombinant MBP-Aster-A binds carotenoids and sterols. (A) Two-side view (in 180° angle) of the structure of the StART-domain of murine Aster A (PDB ID- 6GQF) with a bound carotenoid (orange) modeled into the binding cavity. Tryptophan residues in close vicinity to binding cavity are shown as dark-pink color sticks. (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of the protein purification of recombinant MBP-Aster-A. Cell pellet (insoluble) fraction and affinity purified MBP-Aster-A were resolved by SDS-PAGE. The black arrow indicates the size of the MBP-Aster-A protein (67 kDa). (C) UV/Vis absorption spectrum of the β-apo-10′-carotenal (Apo10) bound to recombinant MBP-Aster-A and MBP. (D) Tryptophan fluorescence quenching of MBP-Aster-A in the presence of increasing concentrations of Apo10. (E) Bound over free fraction of binding assays with MBP-Aster-A and Apo10. (F) Competition assays with MBP-Aster-A with NBD cholesterol and carotenoids. MBP-Aster-A was incubated in the presence of increasing amounts of NBD cholesterol (black curve); the purified Apo10-MBP-Aster-A caroteneoprotein complex was incubated in the presence of increasing amounts of NBD cholesterol (orange curve). The purified zeaxanthin-MBP-Aster A caroteneoprotein complex was incubated in the presence of increasing amounts of NBD cholesterol (purple curve).
We next wished to confirm that carotenoids and sterols bind to the same binding site of Aster-A. In competition assays for protein binding, we used 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol (NBD cholesterol) as ligand. NBD cholesterol emits a characteristic fluorescence upon protein binding. Thus, we incubated apo-MBP-Aster-A in the presence of increasing amount of NBD cholesterol and plotted the data in a semi logarithmic dose–response curve (Fig. 2F). The binding curve displayed a sigmoidal shape with an estimated dissociation constant of 0.27 µM. To determine whether carotenoids compete with NBD cholesterol for the same binding site, we purified MBP-Aster-A with bound zeaxanthin and Apo10, respectively. We then incubated the carotenoprotein complexes in the presence of increasing amounts of NBD cholesterol and monitored the fluorescence in a fluorimeter. The binding curves showed a significant right shift when compared to the NBD cholesterol binding curve with apo-MBP-Aster-A. With increasing concentration, NBD cholesterol replaced the carotenoids from the carotenoprotein complexes and reached maximum binding. The increased dissociation constant and the shape of the curve indicated a competition between the sterol and the carotenoids for the same binding site.
Carotenoids Accumulate in GRAMD1B-Expressing Mouse Tissues.
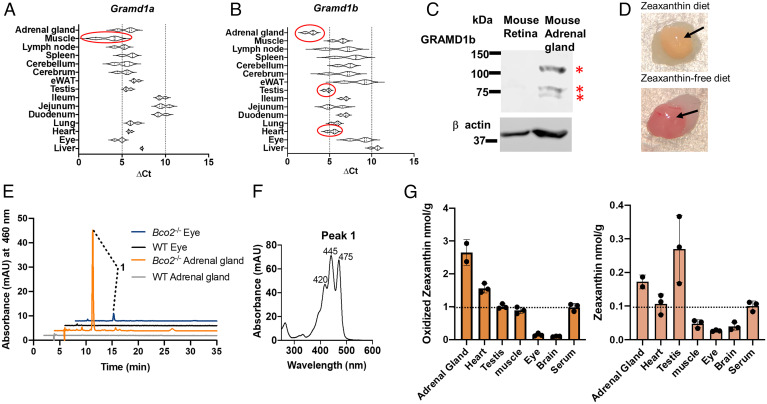
We performed quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis for Gramd1A and Gramd1B mRNA expression in dissected mouse tissues using β-actin as housekeeping gene. Plots of the ΔCT values revealed that Gramd1A did not display a pronounced tissue-specific expression pattern (Fig. 3A). By contrast, Gramd1b mRNA expression was particularly abundant in the adrenal glands, testis, and heart (Fig. 3B) as previously reported by others (15). Western blot analyses confirmed high expression of Aster-B protein in the adrenal gland (Fig. 3C and SI Appendix, Table S1). In contrast, Aster-B protein was not detectable in the murine retina consistent with its far lower mRNA expression (Fig. 3 B and C).
Fig. 3.
Carotenoids accumulate in Gramd1-expressing tissues of Bco2−/− mice. (A and B) qRT-PCR analysis of mRNA expression of (A) Gramd1a gene (encoding Aster A) and (B) Gramd1b (Aster-B) in tissues of wild-type mice. The ΔCt values are normalized to the β-actin housekeeping gene (lower ΔCt values indicate higher mRNA expression levels). The tissues with the highest mRNA expression are highlighted by red circles. (C) Western blot with protein extracts (20 μg per lane) of mouse retina and mouse adrenal gland with anti-GRAMD1b antibody. β-actin was used as the loading control. Three different isoforms (red asterisks) were detected in the adrenal gland. (D) Comparison of the color difference between freshly dissected adrenal glands collected from Bco2−/− mice fed with the zeaxanthin diet (upper photograph) and with zeaxanthin-free diet (lower photograph). The glandular parts are indicated with an arrow. (E) HPLC traces at 460 nm of lipid extracts of adrenal glands and eyes of wild-type (WT) and Bco2−/− mice subjected to feeding with zeaxanthin-rich diet. (F) UV/Vis spectra of the eluted oxidized zeaxanthin (peak 1). (G) The contents of oxidized zeaxanthin (Left) and zeaxanthin (Right) of different tissues of Bco2−/− mice (n = 3) fed with zeaxanthin. The dashed lines indicate the serum concentration of the carotenoids.
In adrenal glands, the scavenger receptor SR-B1 acquires cholesterol from circulating high density proteins for steroidogenesis (13, 21). We and others showed that similar to cholesterol, carotenoids are transported in high density lipoproteins (11) and acquired via SR-B1 by cells (14). In cells, carotenoids such as zeaxanthin are transported to mitochondria where they can be catabolized by the enzyme BCO2 (11, 12). Notably, Aster-B contains an N-terminal mitochondrial target sequence and facilitates uptake of cholesterol into mitochondria for steroidogenesis (18). We now assumed that similar to cholesterol, carotenoids will be transported to mitochondria in an Aster-B-dependent manner. In fact, the adrenal glands of zeaxanthin supplemented Bco2−/− mice showed a characteristic yellow color when compared to glands of nonsupplemented mice (Fig. 3D). HPLC analysis of extracted lipids revealed that zeaxanthin accumulated as oxidized (β,β-carotene-3,3′-di-one) metabolite in the gland as seen by the shift in the retention time and the characteristic spectral properties of the compound (Fig. 3 E and F). The concentration of the zeaxanthin metabolite in the adrenal gland exceeded the plasma concentration by threefold (Fig. 3G). The heart also displayed a higher concentration of the oxidized zeaxanthin metabolite than the serum, consistent with high expression levels of Aster-B in this organ (Fig. 3 B and G). The highest concentration of unmodified zeaxanthin was found in the testis whereas the concentration of oxidized zeaxanthin in this tissue was comparable to the plasma concentration. Notably, neuronal tissues including the retina with low Aster-B mRNA and protein expression (Fig. 3 B and C) showed concentrations that were well below the serum concentration of zeaxanthin and its oxidized metabolite (Fig. 3G). Thus, we observed in Bco2−/− mice that zeaxanthin was concentrated in tissues with high Aster-B expression.
GRAMD1B and BCO2 Display Distinct Expression Patterns in the Human Retina.
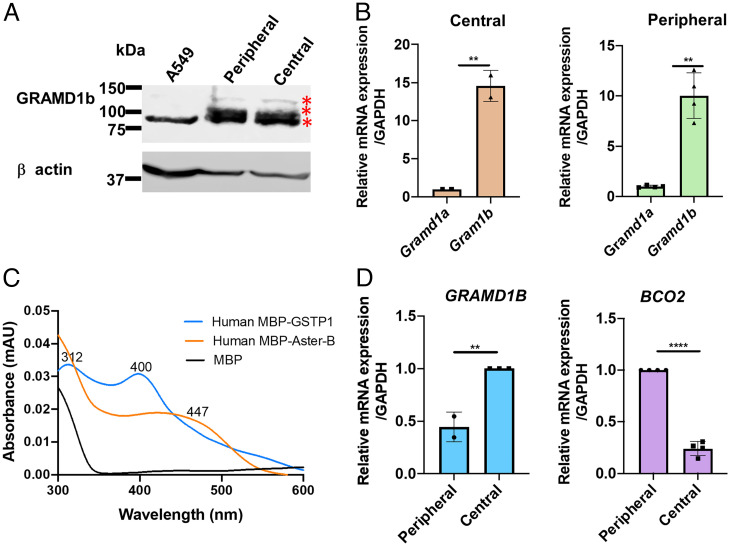
Rodents do not concentrate carotenoids to a large extent in the retina even in the absence of BCO2 (22). In contrast, the human retina displays a high concentration of the pigments, particularly in the macula lutea (5). Therefore, we wondered whether this difference is associated with Aster protein expression. To analyze GRAMD1 expression in the human retina, we performed Western blot and qRT-PCR analysis with human donor eyes. In Western blot analysis, we used the human lung cancer cell line A549 with high GRAMD1B expression as a control. In the human retina, we detected three different isoforms of GRAMD1B protein in peripheral and central parts (Fig. 4A, asterisks). In qRT-PCR analysis, we observed more than tenfold higher mRNA expression of GRAMD1B when compared to GRAMD1A both in peripheral and central parts of the retina of human donor eyes (Fig. 4B). Thus, we concluded that GRAMD1B is expressed at high levels in the human retina.
Fig. 4.
Opposite expression patterns of GRAMD1B and BCO2 in the human retina. (A) Western blot for GRAMD1B with protein extracts of A549 cells (positive control), and human peripheral and central retina. 20 μg of protein was separated in each lane. β-actin was used as loading control. (B) Relative mRNA expression of GRAMD1A and GRAMD1B in central and peripheral parts of human retinas (n = 3 individual donor retinas). (C) UV-Vis absorption spectrum of purified recombinant purified human MBP-ASTER-B and MBP-GSTP1 as well as MBP expressed in zeaxanthin synthesizing E. coli cells. (D) Relative mRNA expression of GRAMD1B and BCO2 in peripheral and central parts of the human retinas (n = 3 individual donor retinas). In (B) and (D), GAPDH was used to normalize the expression.
To demonstrate that the StART-like domain of human Aster-B binds carotenoids, we cloned its cDNA. Additionally, we cloned the cDNA of the GSTP1 (pi isoform) protein that has been implicated as zeaxanthin binding protein of the primate eyes by others (23). We expressed both proteins as MBP fusion proteins in zeaxanthin producing E. coli and purified the recombinant proteins (SI Appendix, Fig. S3 A and B). Spectral analysis of the eluate of the purified recombinant proteins showed that human MBP-Aster-B displayed an absorption maximum at 447 nm (Fig. 4C). The MBP-GSTP1 showed different spectral properties with maxima at lower wavelength (400 nm and 312 nm) that likely resulted from bound ferrous iron (Fig. 4C) (24). To confirm carotenoid binding, we denatured the purified protein complexes in organic solvent and extracted lipids. HPLC analysis confirmed that zeaxanthin was present in purified human MBP-Aster-B solution (SI Appendix, Fig. S3 C and D), whereas the pigment was largely absent in organic extracts of purified human MBP-GSTP1 fusion protein preparations. Thus, recombinant human Aster-B, but not GSTP1, was capable to extracting the pigment from membranes in the bacterial test system.
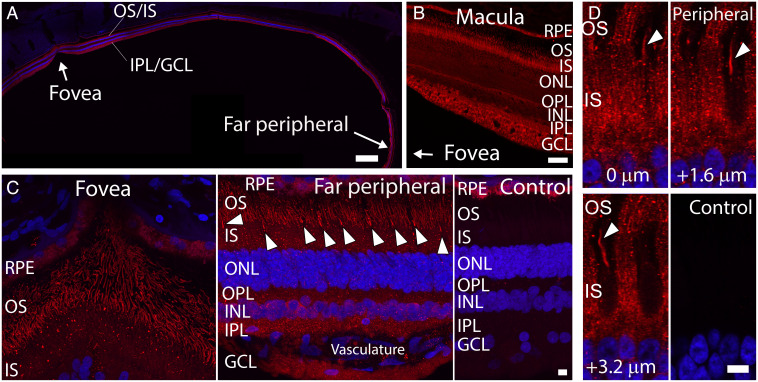
We next performed immunohistochemistry to determine the spatial distribution of Aster-B expression in the human retina. Consistent with the expression pattern observed by qRT-PCR (Fig. 4B), GRAMD1B was expressed in the central and peripheral retina with similar localization patterns (Fig. 5). In the foveal region (Fig. 5A, arrow) that is occupied with cone photoreceptors, their outer segments were immunopositive for anti-GRAMD1B (Fig. 5 B and C). Both in central (Fig. 5B) and peripheral regions (Fig. 5A), anti-GRAMD1B antibody demonstrated particularly strong labels in photoreceptor outer segments, inner plexiform layer, and ganglion cell layer (Fig. 5 A and C, Left). Weaker signals were observed in photoreceptor inner segments, outer nuclear layer, inner nuclear layer, as well as outer plexiform layer. We also detected Aster-B in cone outer segments of the fovea and the peripheral retina (Fig. 5C, Fovea and far peripheral, arrowheads). Cone outer segment demonstrated signal intensities similar to those of rod outer segments. In the outer segment layer, matrices surrounding cone outer segments (cone sheathes) were negative to GRAMD1B antibody, consistent with the idea that GRAMD1B is an intracellular protein and not secreted by cones (Fig. 5D). As negative control, purified rabbit immunoglobulin G (IgG) from unimmunized animals did not demonstrate significant staining in the human retina (Fig. 5 C and D, Control). Therefore, we concluded that GRAMD1B is expressed in cones of both central and peripheral retina, as well as rods throughout the retina. Macular regions, including the fovea, are known to demonstrate photoreceptor and ganglion cell densities and ratios higher than those in the periphery (25). These high cell densities might have contributed to the high expression of GRAMD1B in the central region (Figs. 4D and 5 B and C, Left). The distribution of GRAMD1B in different retinal cell types is in line with its putative role as carotenoid transport protein and is consistent with the presence of zeaxanthin in different cellular layers of the human retina (26).
Fig. 5.
Immunofluorescence localization of GRAMD1B (Aster B) in the human retina. (A) Low magnification image of human retina labeled with anti-GRAMD1B antibody (red). Foveal and far peripheral regions are indicated by arrows. (B) High magnification image of macula region labeled with anti-GRAMD1B antibody (red). The direction of the foveal region is indicated by an arrow. (C) High magnification images of the foveal region (Left) and far peripheral retina (Middle). Cones in the far peripheral region are indicated by arrowheads. As control, IgG from unimmunized rabbits was applied to human retina (Right). Significant immunofluorescence signals were not detected throughout the retina. RPE demonstrated autofluorescence. (D) In the outer segment layer of the peripheral retina, anti-GRAMD1B antibody labeled rod and cone (arrowhead) outer segments (Top Row and Bottom Left). Images at three different z-heights (relative positions of 0, 1.6, and 3.2 µm) are shown. Control rabbit IgG does not demonstrate significant signal within the photoreceptor inner and outer segments (Bottom Right). In (A), (C), and (D), nuclei were labeled with hoechst33342 dye (blue). Scale bars are 600 µm for (A), 60 µm for (B), and 6 µm for (C and D). Abbreviations: RPE, retinal pigment epithelium; OS, photoreceptor outer segments; IS, photoreceptor inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer, INL, inner nuclear layer; IPL, outer plexiform layer; GCL, ganglion cell layer.
Carotenoids exist in different cell types but are disproportionately concentrated in the retina. They exist in low concentration in the peripheral and high concentration in the central foveal parts (27, 28). In lizard skin, the expression pattern of the carotenoid catabolizing enzyme BCO2 determines coloration patterns (29). Thus, we compared the expression of BCO2 and GRAMD1B mRNA in peripheral and central parts of human donors. This analysis showed that BCO2 mRNA levels were tenfold higher in peripheral than in central parts of the retina (Fig. 4D). The opposite picture emerged when we analyzed GRAMD1B mRNA expression that was higher in central than in peripheral parts of the retina. These findings implicated that a differential expression of BCO2 and GRAMD1B exists in the human retina (Fig. 4D). In central parts, low expression of BCO2 would allow for GRAMD1B facilitated accumulation of the pigments whereas in peripheral parts carotenoids would be catabolized by BCO2 when transferred to mitochondria (30).
Discussion
Carotenoids are responsible for some of the most spectacular colors in animals (4) and play important roles as chromophores and hormone precursors (8). In humans, high tissue carotenoid concentrations have been associated with beneficial health effects and the prevention of chronic diseases (31). Almost exclusively acquired from the diet, carotenoids must be transported, distributed through the circulation, taken up by cells, and metabolically modified or converted to apocarotenoids, including retinoids (8). Identifying the genes underlying this metabolism is an essential step in clarifying carotenoid phenotypes and their association with disease states. Based on the work on blind Drosophila mutants, members of carotenoid cleavage oxygenase family and class B scavenger receptors were identified as key components for animal carotenoid metabolism (32, 33). Studies from various laboratories confirmed that these genes account for many carotenoid phenotypes including the plumage coloration of birds (34, 35) and the coloration of vertebrate skin (29, 36). Moreover, mutations and polymorphisms in these genes are associated with macular pigment density of the retina in the human eyes (37), vitamin A deficiency (38), and retinitis pigmentosa (39).
Previously, we and others observed that carotenoid metabolism shares similar features with mammalian cholesterol metabolism, including the transport in HDL and its cellular uptake by SR-B1. Recently, the Aster protein family was shown to act downstream of SR-B1 and to move excessive amounts of cholesterol from the plasma membrane to the ER (15, 17) and mitochondria (18). Here, we speculated that Aster proteins also facilitate intracellular carotenoid transport. We established a bacterial test system to demonstrate that the StART-domain of mouse and human Aster proteins can extract carotenoids from membranes. The bacterial test system for carotenoid binding proteins allowed us to isolate carotenoprotein complexes with specific spectral proprieties. Our analysis in this test system indicated that GRAMD1 proteins prefer zeaxanthin and β-cryptoxanthin over other intermediates produced in the carotenoid synthesis path in the bacteria. In Bco2−/− mice, we observed that zeaxanthin accumulated in the adrenal glands that express high levels of SR-B1 and Aster-B. This finding indicated that carotenoids and cholesterol use the same transport protein machinery and accumulate in the same tissues when the BCO2 function is inactivated by genetic means. The association of carotenoid accumulation with Aster-B expression is remarkable. Aster-B contains a mitochondrial target sequence that is not present in other Aster protein family members. Aster-B facilitates the transport of cholesterol and fatty acids to mitochondria to support steroidogenesis (18). Similar to cholesterol, carotenoids are transported to mitochondria where they accumulate or are metabolically converted to oxidized metabolites by BCO2 (11, 12). Currently, we lack direct genetic proof that carotenoid transport in mitochondria is impaired in Aster-B protein deficiency. Thus, the consequences of loss-of-function mutations in the Gramd1b gene in mice should be further studied to firmly establish the interplay between Aster-B, cholesterol, and carotenoid metabolism.
Molecular modeling revealed that the StART-like domain of Aster proteins can accommodate the rigid and elongated structure of a carotenoid without steric clashes. Our in vitro binding assays revealed a dissociation constant in the lower micromolar range for the model carotenoid Apo10 and provided evidence that carotenoids and cholesterol compete for the same binding cavity of Aster-A. We also compared the carotenoid binding properties of the StART-like domain of the Aster proteins with human GSTP1. GSTP1 is a small globular protein with a single glutathione-binding site located within the thioredoxin-like domain. GSTP1 has been associated with xanthophylls in the fovea and was proposed as putative zeaxanthin-binding protein (23). However, the GSTP1-MBP fusion protein was not able to extract significant amounts of carotenoids from membranes in the bacterial test system.
GSTP1 is expressed both in the human and also in the mouse retina, although rodents do not accumulate the pigments (6). In contrast, GRAMD1B showed a species-specific expression pattern and is expressed at high levels in the human but not in the mouse retina. The expression patterns of GRAMD1B in different retinal layers corresponded to the distribution of carotenoids in the human retina that is not restricted to a single neuronal cell type (26, 28). For instance, expression in photoreceptor outer segments suggests that GRAMD1B acts downstream of interphotoreceptor binding protein that has been implicated in intercellular transport of carotenoids from the retina pigment epithelium to photoreceptor cells (40). Expression of GRAMD1B in ganglion cells suggests that carotenoids are not only delivered via the retinal pigment epithelium and interphotoreceptor retinoid-binding protein (41) but also taken up at the inner blood–retinal barrier. SR-B1 has been implicated in carotenoid uptake at the outer blood-retinal barrier in the RPE (42). It remains to be investigated whether SR-B1 is expressed at the inner retinal blood barrier and facilitates carotenoid uptake from circulating lipoproteins. In all retinal cell types, however, GRAMD1B would facilitate nonvesicular transport of carotenoids and cholesterol between cellular membranes.
The concentration of carotenoids in the fovea region of the macula exceeds 100-fold the concentration in the peripheral retina (43). Consistently, we observed that GRAMD1B is expressed at higher levels in the central than in the peripheral retina. However, this expression difference alone would likely not explain the high concentration of carotenoids in the fovea. Differential expression of BCO2 establishes distinct patterns of carotenoids in tissues of other vertebrates, including lizards (36). In fact, we observed that BCO2 is expressed at high levels in the peripheral and significantly lower levels in the central human retina. This finding confirmed previous transcriptomic studies by Voigt et al. (44) and has significant implication on the distribution of carotenoids in the retina.
Photoreceptors largely rely on aerobic glycolysis for energy production (45). Nevertheless, they are packed with clusters of mitochondria. Our findings suggest that mitochondria contribute to carotenoid metabolism in these specialized neurons. High expression of BCO2 in the peripheral retina would accelerate carotenoid turnover in mitochondria whereas low expression of BCO2 in central parts would assure carotenoid concentration in mitochondria. Notably, histological analysis of the fovea of a patient suffering from macular telangiectasia (MacTel) type 2 revealed damaged mitochondria with disrupted inner membranes (46). Mitochondrial inner membranes are the place of carotenoid accumulation (11) and MacTel type 2 is characterized by dramatically decreased macula pigment density (47).
In conclusion, we provide biochemical and genetic evidence that Aster proteins can bind carotenoids. Among these proteins, Aster-B protein is highly expressed in carotenoid-rich tissues of human and mice, suggesting its critical role in carotenoid transfer and retention. Thus, our study ends the quest for carotenoid transport proteins in mammals and highlights the close relationship between the metabolism of different isoprenoid compounds. It remains to be investigated whether Aster proteins also engage in other aspects of carotenoid metabolism and function. It will be fascinating to further study the role of the Aster protein family in the metabolism of the various dietary isoprenoids, including carotenoids and fat-soluble vitamins.
Materials and Methods
All materials used in the study, including descriptions of the plasmids, antibodies, oligonucleotide primers, and chemical can be found in the SI Appendix. The protocols for protein production (e.g., MBP-Aster-A, MBP-Aster-B, and MBP-GSTP1), the description of their purification and spectroscopic analysis can be found in the SI Appendix. The qualitative and quantitative HPLC analysis of carotenoids is described in the SI Appendix. We used a zeaxanthin-producing E. coli strain (19) to establish an in vivo carotenoid binding assay which is described in the SI Appendix. The in vitro binding assays (APO10), tryptophan fluorescence assays, and the NBD-cholesterol competition assays with murine MBP-Aster-A protein are described in the SI Appendix. The preparation of protein and RNA samples from mouse tissues and from retina of human donor eyes are outlined in the SI Appendix. The molecular modeling approach for determining carotenoid binding to the StART domain of Aster A (PDB ID 6gqf), the qRT-PCR protocols, and the statistical analyses are described in the SI Appendix. Last, immunohistochemistry and microscopic analysis of the ocular tissue were conducted as previously described (48), and the modifications of this protocol are outlined in the SI Appendix.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Eye Institute (EY028121 and EY020551 to J.v.L. and EY029680 and EY028884 to Y.I.). L.D.T. was supported by the T32 Visual Science Training Grant (EY007157), and we received support through the P30 Core grant EY011373. This work was also supported in part by a Challenge Grant from Research to Prevent Blindness to the Department of Ophthalmology, Indiana University School of Medicine. Research activities at Eversight are supported by funding from LC Industries (Durham, NC), Eye Bank Association of America, Connecticut Lions Eye Research Foundation, Blue Cross Blue Shield of Michigan Foundation, The Louise H. and David S. Ingalls Foundation (Cleveland, OH), Lowell F. Johnson Foundation (Pittsburgh, PA), and William G. and Helen C. Hoffman Foundation (Las Vegas, NV).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200068119/-/DCSupplemental.
Data Availability
All study data are included in the main text and the SI Appendix.
References
- 1.von Lintig J., Colors with functions: Elucidating the biochemical and molecular basis of carotenoid metabolism. Annu. Rev. Nutr. 30, 35–56 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Álvarez R., Vaz B., Gronemeyer H., de Lera A. R., Functions, therapeutic applications, and synthesis of retinoids and carotenoids. Chem. Rev. 114, 1–125 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Demmig-Adams B., W. W. Adams, 3rd, Antioxidants in photosynthesis and human nutrition. Science 298, 2149–2153 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Toews D. P. L., Hofmeister N. R., Taylor S. A., The evolution and genetics of carotenoid processing in animals. Trends Genet. 33, 171–182 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Bone R. A., Landrum J. T., Tarsis S. L., Preliminary identification of the human macular pigment. Vision Res. 25, 1531–1535 (1985). [DOI] [PubMed] [Google Scholar]
- 6.Bernstein P. S., et al. , Lutein, zeaxanthin, and meso-zeaxanthin: The basic and clinical science underlying carotenoid-based nutritional interventions against ocular disease. Prog. Retin. Eye Res. 50, 34–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Lintig J., Wyss A., Molecular analysis of vitamin A formation: Cloning and characterization of beta-carotene 15,15′-dioxygenases. Arch. Biochem. Biophys. 385, 47–52 (2001). [DOI] [PubMed] [Google Scholar]
- 8.von Lintig J., Moon J., Babino D., Molecular components affecting ocular carotenoid and retinoid homeostasis. Prog. Retin. Eye Res. 80, 100864 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Lintig J., Moon J., Lee J., Ramkumar S., Carotenoid metabolism at the intestinal barrier. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison E. H., Mechanisms of transport and delivery of vitamin a and carotenoids to the retinal pigment epithelium. Mol. Nutr. Food Res. 63, e1801046 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Palczewski G., Amengual J., Hoppel C. L., von Lintig J., Evidence for compartmentalization of mammalian carotenoid metabolism. FASEB J. 28, 4457–4469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amengual J., et al. , A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 25, 948–959 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acton S., et al. , Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271, 518–520 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Widjaja-Adhi M. A., Lobo G. P., Golczak M., Von Lintig J., A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 24, 3206–3219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandhu J., et al. , Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mammalian cells. Cell 175, 514–529.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naito T., Saheki Y., GRAMD1-mediated accessible cholesterol sensing and transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1866, 158957 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Ferrari A., et al. , Aster proteins regulate the accessible cholesterol pool in the plasma membrane. Mol. Cell. Biol. 40, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen J. P., et al. , Aster-B coordinates with Arf1 to regulate mitochondrial cholesterol transport. Mol. Metab. 42, 101055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misawa N., et al. , Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172, 6704–6712 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandara S., et al. , The structural and biochemical basis of apocarotenoid processing by β-carotene oxygenase-2. ACS Chem. Biol. 16, 480–490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigotti A., Miettinen H. E., Krieger M., The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocr. Rev. 24, 357–387 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Babino D., et al. , Characterization of the role of β-carotene 9,10-dioxygenase in macular pigment metabolism. J. Biol. Chem. 290, 24844–24857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhosale P., et al. , Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. J. Biol. Chem. 279, 49447–49454 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Vasieva O., The many faces of glutathione transferase pi. Curr. Mol. Med. 11, 129–139 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Curcio C. A., Allen K. A., Topography of ganglion cells in human retina. J. Comp. Neurol. 300, 5–25 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Li B., et al. , Imaging lutein and zeaxanthin in the human retina with confocal resonance Raman microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 12352–12358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bone R. A., et al. , Macular pigment in donor eyes with and without AMD: A case-control study. Invest. Ophthalmol. Vis. Sci. 42, 235–240 (2001). [PubMed] [Google Scholar]
- 28.Bone R. A., et al. , Distribution of lutein and zeaxanthin stereoisomers in the human retina. Exp. Eye Res. 64, 211–218 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Andrade P., et al. , Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl. Acad. Sci. U.S.A. 116, 5633–5642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas L. D., et al. , The human mitochondrial enzyme BCO2 exhibits catalytic activity toward carotenoids and apocarotenoids. J. Biol. Chem. 295, 15553–15565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eggersdorfer M., Wyss A., Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 652, 18–26 (2018). [DOI] [PubMed] [Google Scholar]
- 32.von Lintig J., Dreher A., Kiefer C., Wernet M. F., Vogt K., Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation invivo. Proc. Natl. Acad. Sci. U.S.A. 98, 1130–1135 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiefer C., Sumser E., Wernet M. F., Von Lintig J., A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 99, 10581–10586 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toomey M. B., et al. , High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. U.S.A. 114, 5219–5224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gazda M. A., et al. , A genetic mechanism for sexual dichromatism in birds. Science 368, 1270–1274 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Eriksson J., et al. , Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4, e1000010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohn T., et al. , Mechanistic aspects of carotenoid health benefits - where are we now? Nutr. Res. Rev. 34, 276–302 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Lindqvist A., Sharvill J., Sharvill D. E., Andersson S., Loss-of-function mutation in carotenoid 15,15′-monooxygenase identified in a patient with hypercarotenemia and hypovitaminosis A. J. Nutr. 137, 2346–2350 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Marlhens F., et al. , Mutations in RPE65 cause Leber’s congenital amaurosis. Nat. Genet. 17, 139–141 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Vachali P. P., Besch B. M., Gonzalez-Fernandez F., Bernstein P. S., Carotenoids as possible interphotoreceptor retinoid-binding protein (IRBP) ligands: A surface plasmon resonance (SPR) based study. Arch. Biochem. Biophys. 539, 181–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shyam R., Vachali P., Gorusupudi A., Nelson K., Bernstein P. S., All three human scavenger receptor class B proteins can bind and transport all three macular xanthophyll carotenoids. Arch. Biochem. Biophys. 634, 21–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas S. E., Harrison E. H., Mechanisms of selective delivery of xanthophylls to retinal pigment epithelial cells by human lipoproteins. J. Lipid Res. 57, 1865–1878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bone R. A., Landrum J. T., Dixon Z., Chen Y., Llerena C. M., Lutein and zeaxanthin in the eyes, serum and diet of human subjects. Exp. Eye Res. 71, 239–245 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Voigt A. P., et al. , Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res. 184, 234–242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurley J. B., Retina metabolism and metabolism in the pigmented epithelium: A busy intersection. Annu. Rev. Vis. Sci. 7, 665–692 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zucker C. L., Bernstein P. S., Schalek R. L., Lichtman J. W., Dowling J. E., A connectomics approach to understanding a retinal disease. Proc. Natl. Acad. Sci. U.S.A. 117, 18780–18787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charbel Issa P., et al. , Quantification of reduced macular pigment optical density in the central retina in macular telangiectasia type 2. Exp. Eye Res. 89, 25–31 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Imanishi Y., et al. , Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur. J. Neurosci. 15, 63–78 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and the SI Appendix.