Significance
CD8 T cell exhaustion is a key underlying factor limiting immunity in chronic infections and cancer. Persistent antigen exposure antagonizes formation of functional memory CD8 T cells that provide long-term protection and, instead, drives the development of exhausted CD8 T cells (TEX). Improving TEX persistence and function is a major goal for reinvigorating immune responses against chronic infections and tumors. Here, we identify miR-29a as a molecule that attenuates exhaustion and enhances persistence and function of TEX. Enforced expression of miR-29a alters TEX transcriptome, resulting in robust changes in molecular pathways governed by fundamental transcription factors and epigenetic modulators. Thus, enforced miR-29a expression enhances TEX responses, attenuates exhaustion, and represents a target for improving the outcome of immunotherapy.
Keywords: CD8 T cells, microRNA, exhaustion
Abstract
CD8 T cells mediate protection against intracellular pathogens and tumors. However, persistent antigen during chronic infections or cancer leads to T cell exhaustion, suboptimal functionality, and reduced protective capacity. Despite considerable work interrogating the transcriptional regulation of exhausted CD8 T cells (TEX), the posttranscriptional control of TEX remains poorly understood. Here, we interrogated the role of microRNAs (miRs) in CD8 T cells responding to acutely resolved or chronic viral infection and identified miR-29a as a key regulator of TEX. Enforced expression of miR-29a improved CD8 T cell responses during chronic viral infection and antagonized exhaustion. miR-29a inhibited exhaustion-driving transcriptional pathways, including inflammatory and T cell receptor signaling, and regulated ribosomal biogenesis. As a result, miR-29a fostered a memory-like CD8 T cell differentiation state during chronic infection. Thus, we identify miR-29a as a key regulator of TEX and define mechanisms by which miR-29a can divert exhaustion toward a more beneficial memory-like CD8 T cell differentiation state.
CD8 T cells are key mediators of immunity to intracellular pathogens and tumors. During a CD8 T cell response to an acutely resolved infection, naïve CD8 T cells (TN) undergo clonal expansion and differentiate into effector CD8 T cells (TEFF). Upon antigen elimination, antigen-experienced CD8 T cells differentiate into long-lasting memory CD8 T cells (TMEM) that provide protection upon subsequent reinfection (1, 2). In contrast, during chronic infections, persistent antigen stimulation prevents the generation of optimal TMEM and results in T cell exhaustion (3, 4). Exhausted CD8 T cells (TEX) produce limited cytokines and fail to protect upon secondary antigen challenge. Transcriptional profiling identified TEX characteristics, including high expression of inhibitory receptors, changes in signaling pathways, altered expression and use of transcription factors, and bioenergetic alterations, including reduced expression of ribosomal subunit genes (5–7). Moreover, TEX have a distinct open chromatin landscape compared with TEFF and TMEM, which identifies TEX as a distinct branch of mature CD8 T cell differentiation (8–10). The distinct transcriptional and epigenetic features of TEX are found not only in chronic infections but also in tumor-infiltrating CD8 T cells in mice and humans, suggesting that common pathways underlie TEX differentiation in different disease settings.
Despite considerable previous work on the phenotypic, transcriptional, and epigenetic characteristics of TEX, the posttranscriptional circuits involved in TEX differentiation remain relatively unexplored. Noncoding RNAs constitute ∼40% of the human genome (11) and have the potential to regulate diverse areas of cellular biology. Specifically, microRNAs (miRs) are a class of short, double-stranded, noncoding RNAs that repress complementary mRNA targets (12). miRs can exert powerful regulatory effects on a specific biological pathway by simultaneously targeting several mRNAs in the same pathway. As a result, miRs have the potential to exert substantial posttranscriptional control over cell state, differentiation, and function (13, 14). Our current understanding of the role of miRs in CD8 T cell differentiation is mostly restricted to the differentiation of TEFF and TMEM (15–24). However, how miRs regulate CD8 T cell differentiation in chronic infections, and specifically the biology of TEX, remains poorly understood.
Persistent T cell receptor (TCR) stimulation is a key factor leading to CD8 T cell exhaustion. Thus, two miRs induced upon TCR signaling, miR-31 and miR-155, were shown to regulate TEX. miR-31 promotes exhaustion by increasing CD8 T cell sensitivity to type I interferon (25). Instead, although miR-155 also fosters exhaustion during chronic viral infection, this miR also enhances long-term TEX persistence by targeting the AP-1 transcription factor Fosl2 (26). This latter effect of miR-155 may represent an adaptation that allows TEX to withstand the stress of constant TCR stimulation and persist over the long term, contributing to partial disease containment. Several studies have investigated the potential miR regulation of inhibitory receptor expression (27–29). However, a comprehensive understanding of how miRs may regulate CD8 T cell differentiation, especially in the context of exhaustion, remains limited.
Here, we compared the expression of miRs in virus-specific CD8 T cells responding to acutely resolved or chronic viral infection and defined miR expression patterns in TEFF, TMEM, or TEX CD8 T cells. These studies revealed miR-29a as a key TMEM associated miR. Enforced expression of miR-29a enhanced virus-specific CD8 T cell responses to acutely resolved and chronic viral infection, antagonized development of exhaustion, and promoted TMEM-like patterns of differentiation even during chronic infection. Mechanistically, miR-29a attenuated inflammatory and TCR signaling in TEX, targeted the transcription factor Eomes, and altered key transcriptional pathways associated with exhaustion, including Tox and AP-1. Thus, we identify miR-29a as a key regulator of TMEM versus TEX biology, demonstrate underlying miR-29a targeted mechanisms, and reveal a potential therapeutic opportunity through manipulation of miR-29a to improve TEX responses during chronic infection and cancer.
Results
MicroRNAs Are Differentially Regulated in CD8 T Cells Responding to Acutely Resolved or Chronic Viral Infection.
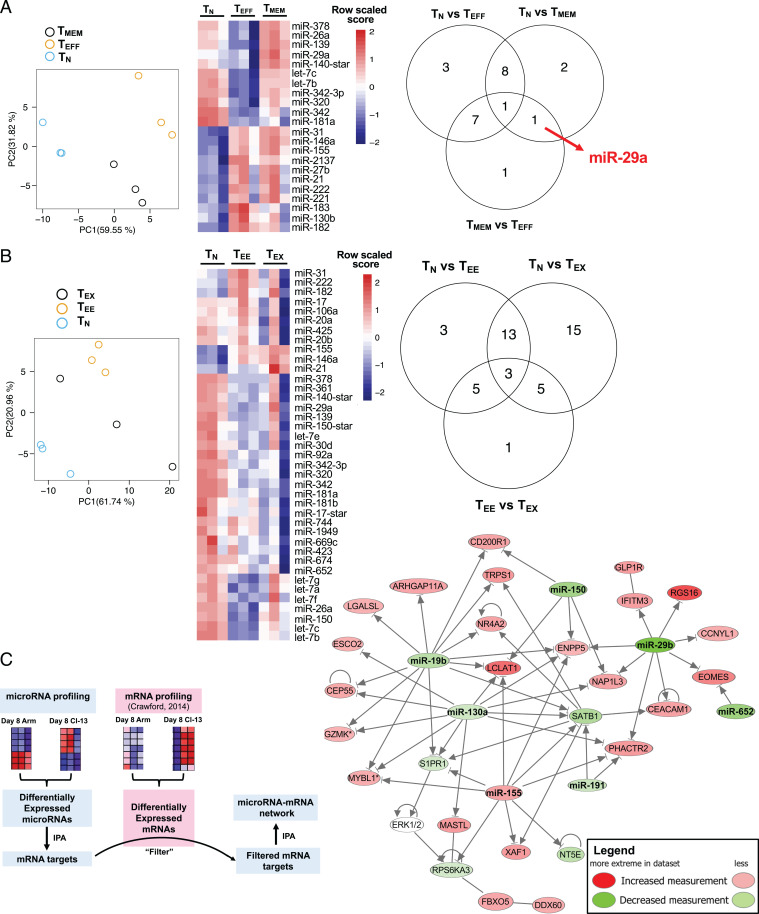
We investigated global miR expression profiles in virus-specific CD8 T cells following acutely resolved or chronic infection of mice with the Armstrong (Arm) or clone 13 strains of lymphocytic choriomeningitis virus (LCMV), respectively. LCMV DbGP33–41–TEFF and TMEM were isolated from mice infected with LCMV Arm at day 8 (d8) or d30 postinfection (p.i.), respectively. As a control, TN were isolated from uninfected mice. We examined the miR expression of isolated LCMV DbGP33–41–specific CD8 T cells using Affymetrix miR arrays. Principal component analysis demonstrated that miR expression patterns were distinct in TN, TEFF, and TMEM (Fig. 1A). Among differentially expressed miRs (DEMs; false discovery rate [FDR] < 0.05) among TN, TEFF, and TMEM were several previously shown to regulate T cell differentiation, including miR-155, miR-146a, let-7b, and let-7c (16, 18, 22, 30) (Fig. 1A and SI Appendix, Fig. S1 A and B). We asked whether any individual miRs were distinctly expressed by TMEM. Indeed, miR-29a was the only miR uniquely expressed by TMEM (i.e., differential expression [DE] in TMEM versus TN [1.3-fold] and TMEM versus TEFF [1.6-fold]) but not in TN versus TEFF; Fig. 1A and SI Appendix, Fig. S1 A, B, and F). These data suggested a potential role for miR-29a in the biology of TMEM.
Fig. 1.
miR-29a is a key memory CD8 T cell–specific miR dysregulated during exhaustion. C57/BL6 mice were infected with LCMV Arm (acute) or LCMV clone 13 (chronic). At d8 and d30 p.i., LCMV Db gp-33–specific CD8 T cells were purified from spleens and their miR profile was examined. As a control, TN were purified. (A) Principal component analysis (PCA) among TN, TEFF, and TMEM. Heat map and Venn diagram showing the DE miRs with FDR < 0.05. (B) PCA among TN, TEE, and TMEM. Heat map and Venn diagram showing the DE miRs with FDR < 0.05. (C) The DE miRs between CD8 T cells responding to acute and chronic infection were used to create a list of predicted mRNA targets using Ingenuity Pathway Analysis. A list of DE mRNAs between CD8 T cells responding to acute and chronic infection was created from ref. 31. The DE mRNA list was used to filter the miRNA target list and select only the miRNA targets that were DE during the same time point but in the opposite direction of the miRNA. The filtered miRNA target list, together with the list of DE miRs, was then used to create a network of miRs and their predicted targets that were DE between acute and chronic infection at d30 p.i.
During chronic LCMV infection, optimal TEFF and TMEM do not develop; instead, virus-specific CD8 T cells become exhausted. Therefore, we next examined miR expression in virus-specific CD8 T cells at d8 (early exhausted CD8 T cells at d8 [TEE]) and at d30 (TEX) p.i. with LCMV clone 13. Similar to acute infection, PCA revealed distinct miR profiles in virus-specific CD8 T cells during chronic infection (Fig. 1B). Comparison of miR expression between virus-specific CD8 T cells from acute versus chronic infection identified 12 DEMs at d8 p.i. (TEFF versus TEE) and 46 DEM at d30 p.i. (TMEM versus TEX), indicating that miR expression patterns diverge as CD8 T cell differentiation patterns become more distinct over time following acutely resolved versus chronic viral infection (SI Appendix, Fig. S1 C and D). All DEMs at d30 p.i., including miR-29a, were down-regulated in TEX compared with TMEM, suggesting that failure to up-regulate or sustain specific miR expression during chronic infection may contribute to CD8 T cell exhaustion. We further confirmed the microarray data with quantitative RT-PCR. CD45.1+ P14 CD8 T cells were adoptively transferred to CD45.2+ recipient mice that were infected with LCMV Arm or LCMV clone 13. At d30 p.i., miR-29a was 1.9-fold increased in TMEM P14 versus TEX P14, confirming the microarray data. Thus, these analyses identified distinct patterns of miR expression in CD8 T cells and revealed miR-29a as a TMEM–specific miR that was down-regulated in TEX.
We hypothesized that the effects of miRs in CD8 T cell differentiation were due to effects on complementary target mRNAs. Therefore, we performed an integrated analysis of miR expression patterns with transcriptional profiles of mRNA for these cell types from ref. 31. We used miR expression patterns (i.e., DEMs; P < 0.05) between acute and chronic infection at d8 and d30 p.i. to generate a list of predicted miR target mRNAs at each time point. We then cross-referenced this list with differentially expressed genes (DEGs) at these time points (P < 0.05). Since miRs function typically by inducing mRNA degradation, we examined DEGs that were expressed in the opposite direction of their predicted targeting miR. A network constructed using these miR–mRNA data revealed several miR nodes regulating key genes, including some previously shown to regulate CD8 T cell biology (namely, miR-150 and miR-155) but also identified several other miRs, including miR-29a, miR-19b, miR-130a, and associated mRNA targets (Fig. 1C and SI Appendix, Fig. S2A).
The transcription factor Eomes has been implicated in the biology of TEX (3, 4) and has been shown to be directly targeted by miR-29a (21). Indeed, higher Eomes expression in CD8 T cells from LCMV clone 13 infection correlated with lower amounts of miR-29a, compared with CD8 T cells from acute LCMV Arm infection that expressed less Eomes and more miR-29a (SI Appendix, Fig. S2B). Upstream regulators of the miR–mRNA network were central to the cellular response to inflammation (i.e., STATs, IRFs, NFKB1) and TCR signaling (i.e., NFATC2, NR4A1; SI Appendix, Fig. S2C), suggesting that this set of miRs may function as a rheostat, limiting CD8 T cell responses to inflammatory and/or antigen signaling and that lower expression of these miRs in TEX may contribute to chronic overstimulation and exhaustion.
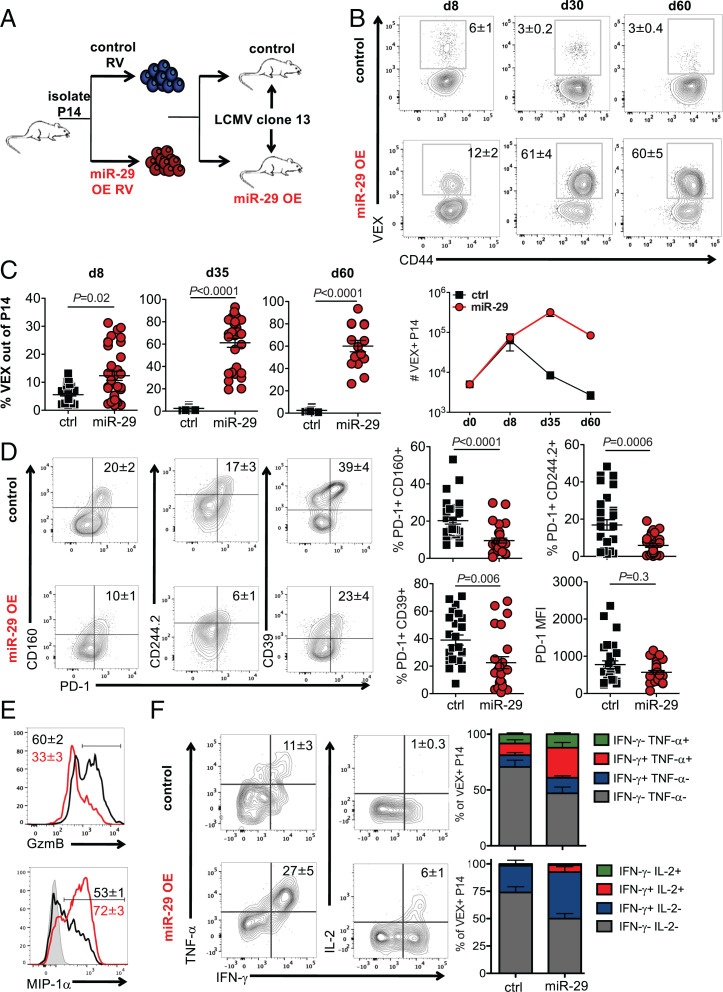
TEX are characterized by a distinct transcriptional and epigenetic profile (3). We hypothesized that miR expression patterns contribute to the TEX-associated mRNA expression profiles, specifically due to the absence in TEX of miRs that are up-regulated in TMEM. miR-29a was a prime candidate as it was the only TMEM-specific miR in our analysis (Fig. 1A) and this miR was down-regulated in CD8 T cells during chronic infection (Fig. 1 and SI Appendix, Fig. S1). We, therefore, hypothesized that enforced expression of miR-29a might improve CD8 T cell responses during chronic infection by fostering TMEM-like differentiation. To test this idea, we used retrovirus (RV) transduction to enforce expression of miR-29a in TCR transgenic CD8 T cells (P14) that recognize the LCMV DbGP33–41 epitope. P14 cells were transduced with miR-29a RV or control RV and adoptively transferred into congenically distinct LCMV clone 13–infected recipient mice at d1 p.i. (Fig. 2A). Control (empty) RV and miR-29a RV resulted in equal transduction efficiency (∼30%; SI Appendix, Fig. S2D), which corresponded to a 7 ± 2-fold increase of miR-29a expression in miR-29a VEX+ compared with control VEX+ cells, based on quantitative RT-PCR. Transduction with miR-29a expressing RV increased the frequency and number of responding P14 cells (Fig. 2 B and C) with an increasing advantage of the miR29a RV–transduced P14 cells at 1 and 2 mo p.i.
Fig. 2.
miR-29a attenuates CD8 T cell exhaustion. CD45.1+ P14 CD8 T cells were transduced with either control empty-VEX RV (control [ctrl]) or miR-29a OE-VEX RV (miR) and adoptively transferred to CD45.2+ recipient mice that were infected with LCMV clone 13 24 h earlier. (A) Experimental design. (B and C) Frequency and number of donor VEX+ P14 cells in spleens (mean+/-SEM). Fluorescence-activated cell-sorting (FACS) plots are gated on total CD45.1+ P14 CD8 T cells. (D) Expression of inhibitory receptors on VEX+ P14 cells at d30 p.i. (E) MIP-1α and GzmB production by VEX+ P14 cells at d30 p.i. (F) Cytokine production by VEX+ P14 cells at d30 p.i. FACS plots in D–F are gated on VEX+ CD45.1+ P14 cells. Each data point represents an independent mouse. Representative results of at least three independent experiments are reported with at least 11 mice per group.
The numerical increase and enhanced persistence of miR-29a overexpression (OE) P14 cells in chronic infection suggested that miR-29a may antagonize CD8 T cell exhaustion. Indeed, although miR-29a OE did not have a dramatic impact on expression of PD-1, expression of other inhibitory receptors was decreased, resulting in substantial reduction in inhibitory-receptor coexpression, a key feature of TEX (Fig. 2D). These effects of miR-29a were mostly observed during the later stages of TEX differentiation (d30 p.i.); no significant differences were observed at d8 p.i. (SI Appendix, Fig. S2E). To exclude any potential effects of the empty RV on CD8 T cell differentiation, we also transduced P14 cells with an RV expressing a scrambled sequence of miR-29a. Compared with cells transduced with the RV expressing the scrambled version of miR-29a, wild-type miR-29a OE also increased the number of transduced P14 cells (SI Appendix, Fig. S2F), decreased expression of inhibitory receptors, and promoted expression of memory-associated markers CD127 and Ly108 (SI Appendix, Fig. S2G). This effect of miR29a OE on inhibitory receptor coexpression was not likely due to changes in viral load, because the number of P14 cells initially adoptively transferred does not impact viral replication, according to results reported from previous studies (32, 33), and viral load in serum and kidney at d30 p.i. was similar between the miR-29a OE and the control RV group (SI Appendix, Fig. S3A). Moreover, inhibitory receptor expression by the nontransduced (VEX−) P14 cells in each group was indistinguishable (SI Appendix, Fig. S3B), consistent with a cell-intrinsic role for miR29a.
To confirm the role of miR-29a in antagonizing exhaustion, we adoptively transferred miR-29a–deficient CD8 T cells (miR-29ab1fl/fl CD4 Cre±) into congenically marked recipient mice that were then infected with LCMV clone 13. miR-29a–deficient CD8 T cells did not up-regulate CD127 and had increased expression of PD-1 and CD160 compared with wild-type CD8 T cells (SI Appendix, Fig. S3C), consistent with a role of miR-29a in antagonizing exhaustion in chronic infection. The miR-29ab1fl/fl CD4 Cre± CD8 cells were deficient in both miR-29a and miR-29b1; however, miR-29b1 was not differentially expressed during CD8 T cell differentiation in acute or chronic LCMV infection (Fig. 1). In addition, our OE data suggest a specific role for miR-29a, as only miR-29a was OE and not miR-29b1. Therefore, we suggest that the observed phenotypes can be attributed to miR-29a, despite the fact that miR-29b1 contains the same seed sequence and could have other roles in T cell biology. TEX maintain expression of granzyme B but have reduced cytokine production upon stimulation, in contrast to TMEM (3). miR-29a–OE P14 cells expressed less granzyme B (Fig. 2E) but had increased cytokine and chemokine production (Fig. 2 E and F). Thus, miR29a OE promoted robust CD8 T cell expansion and persistence during chronic viral infection and antagonized key features of exhaustion.
To begin to dissect the molecular mechanisms by which miR-29a antagonized CD8 T cell exhaustion, we investigated direct mRNA targeting. The transcription factor Eomes regulates TEX differentiation (3, 4), is highly expressed by TEX compared with TMEM (SI Appendix, Fig. S2B), and plays a key role in exhaustion (3, 4). miR-29a has been shown to directly target Eomes in CD4 T cells (21). Thus, we hypothesized that the effect of miR-29a in attenuating exhaustion could be mediated by direct targeting of Eomes. We used a 3′ untranslated region (3′UTR) sensor construct (34) containing the 3′UTR of Eomes downstream of GFP and transduced primary mouse CD8 T cells together with miR-29a OE or control RV expressing the VEX reporter. CD8 T cells transduced with the Eomes 3′UTR in the presence of miR-29a–OE RV expressed less GFP than did cells transduced with Eomes 3′UTR in the presence of control RV (SI Appendix, Fig. S3D). These data indicate that miR-29a directly targets Eomes at the 3′UTR in CD8 T cells.
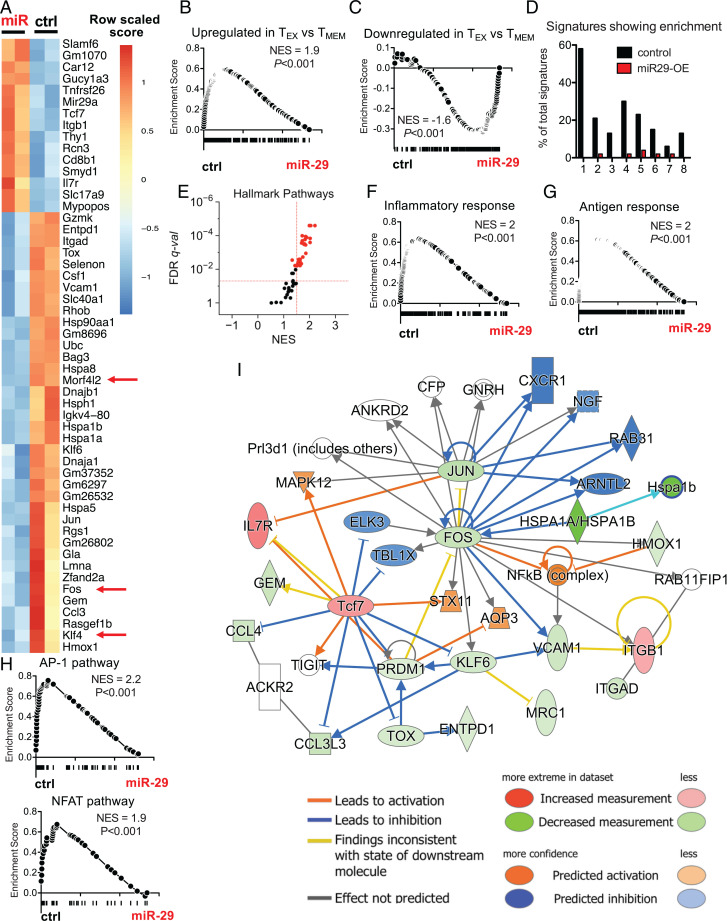
To investigate the downstream molecular mechanisms by which miR-29a antagonized CD8 T cell exhaustion, we analyzed the transcriptional program of miR-29a–OE and control RV–transduced P14 cells at d30 p.i. miR-29a was up-regulated in miR-29a–OE cells, confirming stable transduction and overexpression (SI Appendix, Table S1 and Fig. S3E). We noted that 61 transcripts were significantly changed (FDR < 0.05) due to miR-29a OE (Fig. 3A). The majority of these transcripts (72%) were down-regulated upon miR-29a OE (Fig. 3A). Predicted miR-29a target genes were significantly enriched in the control RV group compared with miR-29a–OE P14 cells (SI Appendix, Fig. S3F). Of the 452 predicted miR-29a targets, 145 showed significant enrichment in control versus miR-29a–OE cells, resulting in 32% of predicted targets being enriched. The AP-1 transcription factor Fos was the third most enriched miR-29a predicted target in control versus miR-29a–OE cells (SI Appendix, Fig. S3G). Down-regulated genes in miR-29a–OE P14 cells included three predicted miR-29a targets (Klf4, Fos, and Morfl2), as well as transcription factors implicated in TEX differentiation, such as Jun and Tox (Fig. 3A). Among the few transcripts that were up-regulated upon miR-29a OE were IL-7Ra and Tcf7, the latter of which is a key TMEM promoting transcription factor. These transcriptional data are consistent with the cellular and functional data reported earlier in Results and support the notion that miR-29a can antagonize exhaustion.
Fig. 3.
miR-29a instructs a memory-like CD8 T cell transcriptional profile during chronic infection. P14 cells were transduced with miR-29a OE (miR) or control empty (ctrl) RV and adoptively transferred as described in Fig. 2A. At d30 p.i., VEX+ P14 cells were sorted and RNA-seq was performed. (A) Heat map shows DE transcripts with FDR < 0.05. Red arrows highlight predicted targets of miR-29a. (B and C) GSEA was performed for gene signatures obtained from MSigDB (data set: GSEA 9650). (D) The percentage of pathways from each MSigDB database enriched (with FDR < 0.05) in ctrl (black) or miR-29a–OE cells (red). Databases are numbered as follows on the x-axis: 1: Hallmark; 2: Kyoto Encyclopedia of Genes and Genomes; 3: BioCarta; 4: Gene Ontology (GO) Molecular Process; 5: GO Cellular Component; 6: GO Molecular Function; 7: Gene Transcription Regulation Database; 8: miR predicted targets. (E) Hallmark pathways enriched in ctrl versus miR-29a–OE P14 CD8 T cells. (F–H) GSEA plots for the following data sets: (F) Inflammatory response (Hallmark); (G) antigen response (Goldrath); (H) AP-1 (PID) and NFAT (PID). (I) Network analysis for genes DE between miR-29a OE and ctrl with FDR < 0.05. NES, normalized enrichment score; q-val, q value.
Since miR-29a was strongly associated with TMEM, we next asked whether miR-29a OE promoted a more global pattern of TMEM-like differentiation during chronic infection. Indeed, Gene Set Enrichment Analysis (GSEA) revealed substantially reduced enrichment of TEX-associated genes in the miR-29a–OE P14 cells (Fig. 3B). Similarly, genes that were down-regulated in TEX, compared to TMEM, were strongly enriched in miR-29a–OE P14 cells compared with control RV–transduced P14 cells (Fig. 3C). Furthermore, genes up-regulated in TEFF, versus TMEM, were enriched in control versus miR-29a–OE P14 cells, suggesting that miR-29a fostered differentiation of P14 cells toward TMEM rather than TEFF (SI Appendix, Fig. S3H). One of the few miRs implicated in TEX is miR-155. However, unlike miR-29a, miR-155 promotes durability, but not reversal, of exhaustion. Therefore, we asked whether miR-29a antagonized exhaustion by antagonizing the effects of miR-155. Indeed, the gene signature associated with miR-155 OE was enriched in control versus miR-29a–OE P14 cells (SI Appendix, Fig. S3I), suggesting that miR-29a antagonizes the effect of miR-155. The subset of enriched genes included inhibitory receptors (CD244, CD200R1, CD200R2, CD200R4), suggesting that miR-29a may antagonize the effect of miR-155 by antagonizing the expression of surface inhibitory receptors and exhaustion markers, consistent with the observation that OE of either miR-155 or miR-29a enhances CD8 T cell persistence but has opposing effects on the phenotype of CD8 T cells in chronic infection. Thus, enforced miR-29a expression in virus-specific CD8 T cells antagonizes a transcriptional profile associated with TEX and fosters transcriptional features associated with TMEM.
To further interrogate the underlying mechanisms by which miR-29a fosters TMEM-like transcriptional, phenotypic, and functional features during chronic viral infection, we examined the biological pathways and transcriptional circuits regulated by miR-29a. Only a small number of Hallmark, Kyoto Encyclopedia of Genes and Genomes, BioCarta, or Gene Ontology–term biological pathways were enriched in miR-29a–OE P14 cells at d30 p.i. (Fig. 3 D and E), consistent with global mRNA down-regulation as the major transcriptional effect of miR-29a OE (Fig. 3A). Among the few biological pathways induced by miR-29a were several related to ribosome biogenesis and protein translation (SI Appendix, Table S2). Regulation of the translational machinery is critical for CD8 T cell differentiation (35), and down-regulation of genes encoding ribosomal subunits is a prominent feature of TEX (7) that may be associated with poor bioenergetics (6). A reversal of this feature of TEX may contribute to better expression of effector molecules by miR-29a–OE P14 cells. Several cytokine signaling and inflammatory pathways were also down-regulated upon miR-29a OE in P14 cells during chronic infection (Fig. 3F and SI Appendix, Table S3), suggesting that miR-29a may attenuate the response to inflammatory cytokines and, thus, abrogate the deleterious effect of chronic inflammation on TMEM differentiation (36). Moreover, a transcriptional signature of antigen stimulation was enriched in control versus miR-29a–OE P14 cells (Fig. 3G), suggesting a potential role for miR-29a in limiting overstimulation of antigen-specific CD8 T cells that drives exhaustion during chronic infection. Thus, miR-29a may antagonize exhaustion and promote TMEM-like differentiation by regulating responses to both antigen and inflammation.
We next investigated the impact of miR-29a OE on transcription factors, the downstream mediators of changes in inflammatory or TCR signaling pathways. We used the Pathway Interaction Database (PID), a collection of cellular signaling pathways and intracellular molecular interactions. Although none of the 180 gene sets from the PID database enriched in miR-29a–OE P14 cells, 41 PID pathway gene sets enriched in control RV–transduced P14 cells, suggesting down-regulation of these pathways by miR-29a OE. These pathways included key transcription-factor pathways, such as AP-1, c-Myb, and NFAT, as well as the TCR_CALCIUM pathway (Fig. 3H and SI Appendix, Table S4). Indeed, a transcriptional network involving Fos and Jun, as well as the exhaustion-related transcription factors Prdm1 and Tox was significantly affected by miR-29a OE (Fig. 3I). This network also included differential expression of Klf4 and Tcf7, two transcription factors implicated in TMEM differentiation. Collectively, these results suggest miR-29a has a role as a central regulator of key transcriptional networks in CD8 T cells, acting as a rheostat between central exhaustion pathways (Tox/AP-1) and memory-associated pathways (Tcf7).
To further investigate whether miR-29a OE antagonized the effect of these central exhaustion pathways directed by Tox, Fos, and Jun, we used a double RV OE system to enforce expression of miR-29a (with GFP reporter) together with Tox, Fos, or Jun (with VEX reporter) in P14 cells. Upon adoptive transfer into LCMV clone 13–infected mice, double-transduced P14 cells expressing miR-29a and Tox, Fos, or Jun were identified as GFP+VEX+ P14 cells. OE of Fos or Tox abrogated the effect of miR-29a OE on inhibitory receptor expression, suggesting that miR-29a antagonizes the effects of these central exhaustion pathways (SI Appendix, Fig. S4A). Although Tox and Jun are not predicted to be directly targeted by miR-29a, and their effect in abrogating the miR-29a effect is likely indirect, we identified these key transcription factors with functional relevance in overcoming the miR-29a effect in antagonizing exhaustion.
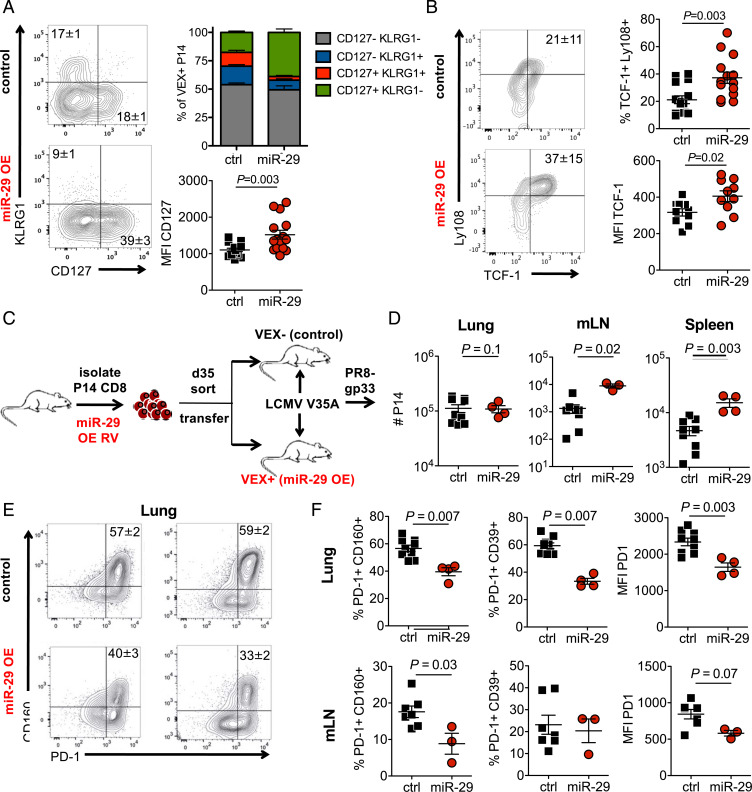
These data provoke the hypothesis that enforced miR-29a expression may foster TMEM-like differentiation and function in virus-specific CD8 T cells during chronic infection. We, therefore, examined how miR-29a OE affected development of other phenotypic and functional TMEM properties during chronic viral infection. Indeed, miR-29a OE enhanced expression of the memory-associated molecule IL-7Rα by P14 cells in chronic LCMV infection (Fig. 4A). Expression of the TEFF marker KLRG1 by TEX is typically low, but miR29a OE further reduced expression of this molecule consistent with a shift toward TMEM or a memory precursor cell (IL-7Rα+ KLRG1−) differentiation state (Fig. 4A). This effect of miR-29a OE on P14 cell differentiation in chronic infection promoted a pattern of IL-7Rα and KLRG1 expression similar to what was observed during acutely resolved infection (SI Appendix, Fig. S4 B–D). The transcription factor TCF-1 plays a key role in both long-term TMEM following acutely resolved infections and also in TEX progenitor cells during chronic infections and cancer (37–39). Enforced expression of miR-29a enhanced TCF-1 expression in P14 cells during chronic infection (Fig. 4B), consistent with a shift toward progenitor or TMEM-like differentiation.
Fig. 4.
miR-29a promotes memory-like CD8 T cell responses in chronic infection. (A and B) CD45.1+ P14 CD8 T cells were transduced with either control empty-VEX RV (ctrl) or miR-29a OE-VEX RV (miR) and adoptively transferred as shown in Fig. 2A. (A) Percentages of terminal effector and memory precursor P14 cells (gated on VEX+ P14 cells) at d30 p.i. (B) Intracellular expression of TCF-1 and surface Ly108 at d30 p.i. (C–F) At d34 p.i., transduced VEX+ and nontransduced VEX- P14 cells were sorted from the spleens of donor mice. A total of 50,000 sorted VEX+ or VEX− P14 cells were separately adoptively transferred to congenic recipient mice that were infected with LCMV V35A at 35 d prior. Recipient mice were then challenged with influenza virus PR8-gp33 2 d later. (D) Secondary expansion of transferred P14 cells was analyzed on 9 d after PR8-gp33 infection. (E and F) The phenotype of transferred P14 cells was analyzed on d9 after PR8-gp33 infection. Fluorescence-activated cell-sorting plots are gated on CD45.1+ P14 cells. mLN, mediastinal lymph nodes.
A canonical property that distinguishes TMEM from TEFF or TEX is the ability of TMEM to mount robust recall responses upon reinfection. We therefore tested whether miR-29a OE in chronic infection improved recall responses. We purified miR-29a OE and nontransduced P14 cells from chronically infected mice at d30 p.i. Equal numbers of control or miR29a–OE P14 cells were then adoptively transferred to new congenic recipient mice. To avoid the potential caveat of infecting these secondary recipients with LCMV clone 13 associated with the adoptively transferred P14 cells, we used recipient mice previously infected with LCMV V35A, a variant of LCMV that lacks the gp33 epitope (40). These secondary recipients were then challenged intranasally 2 d later with influenza PR8 expressing the GP33–41 epitope (PR8-GP33) (Fig. 4C). The P14 cells with enforced expression of miR-29 mounted a more robust recall response upon reinfection than did the control P14 populations (Fig. 4D). In addition, miR-29a–OE cells showed reduced expression of inhibitory receptors upon reinfection (Fig. 4 E and F). These data are consistent with the transcriptional and phenotypic changes driven by miR-29a OE in chronic infection and suggest that miR-29a can foster changes that allow improved recall responses to be preserved despite the persistent antigen stimulation of chronic infection. Together, these results suggest miR-29a as a potential therapeutic target for enhancing TEX function and diverting TEX differentiation toward more TMEM-like differentiation in cancer and chronic infections.
Discussion
We identified miR-29a as a molecule that attenuates exhaustion and enhances persistence and function of CD8 T cells during chronic viral infection. Mechanistically, we identified a role for miR-29a as a rheostat between exhaustion-related (AP-1, NFAT, Tox) and memory-related (TCF-1) transcriptional pathways that are implicated in TMEM versus TEX differentiation. Moreover, our data suggest that miR-29a functions by attenuating TCR and/or inflammatory signaling pathways that feed into these key transcriptional circuits, consistent with the known importance of overstimulation driving T cell exhaustion. Together, these studies suggest that enhanced expression of miR-29a may be a strategy to foster more functional, durable TMEM-like differentiation in the context of persistent antigen stimulation, such as chronic infections and cancer.
A major gap in our understanding of T cell exhaustion has been defining the roles of noncoding RNAs, including miRs. miRs can simultaneously target several mRNAs and, therefore, modulating expression of a single miR could have broader biological impact than modulating expression of individual mRNAs. Although some work has identified roles for miR-31 (25) or miR-155 (26) in TEX, little other information exists. Our global miR profiling here revealed patterns of miR expression in TEFF, TMEM, and TEX in vivo and, although we focused on miR-29a, these data also highlighted many other miRs that warrant investigation in the future for regulating the biology of TEFF, TMEM, and TEX.
TEX reinvigoration by checkpoint blockade has had remarkable clinical success (41, 42). Despite these successes, many patients do not benefit from durable clinical responses (42), and recent data suggest that the immunological response to checkpoint blockade may be transient (8, 43). In other words, PD-1–pathway blockade may not induce long-term TEX reinvigoration or TMEM-like differentiation. Optimal immunotherapies aimed at reversing or preventing exhaustion may, therefore, need to address issues related to acquisition of TMEM-like properties to optimally enhance durability, persistence, and recall capacity. The effects of miR-29a on quantitively and qualitatively improving TEX responses and inducing phenotypic, functional, and transcriptional changes are consistent with TMEM-like differentiation and suggest changes in central pathways involved in the dichotomous TMEM versus TEX differentiation states. Indeed, miR-29a OE resulted in lower expression of Tox, the epigenetic inducer of TEX differentiation (44–48). miR29a OE also up-regulated expression of Tcf7, the key transcription factor that governs TEX progenitor cells during chronic infection and is necessary for responses to anti-PD1 therapy (37–39). However, TCF-1 (encoded by Tcf7) is also a major regulator of long-term, quiescent, central memory CD8 T cells (49, 50) potentially connecting this effect of miR29a OE to the improved memory-like properties observed. Thus, we demonstrate miR-29a is a key player of CD8 T cell differentiation by regulating transcriptional pathways central to TMEM versus TEX differentiation.
Our data on miR-29a promoting persistence of CD8 T cells during chronic infection complement our understanding of how another miR, miR-155, functions to promote long-term persistence of TEX (26). The underlying mechanisms of how these two miRs alter TEX differentiation are different yet complementary. miR-155 enhances long-term persistence of exhausted CD8 T cells by increasing surface inhibitory-receptor expression and, therefore, rendering the cells less susceptible to the deleterious effects of persistent TCR and inflammatory signals. On the contrary, miR-29a enhances long-term persistence by directing the cells to a TMEM-like phenotype and altering expression of transcription-factor pathways downstream of TCR, such as Jun, Fos, NF-κB, Tox, and NFAT. In fact, miR-29a antagonized the effect of miR-155, suggesting opposing mechanisms by which two individual miRs regulate TEX differentiation. Whereas miR-155 inhibits the responsiveness to external stimuli by increasing inhibitory receptor expression, miR-29a affects downstream molecular pathways. In both cases, TCR and inflammatory signaling are inhibited, in line with the known role of TCR and inflammatory signaling in driving exhaustion. However, whereas miR155 allows exhausted CD8 T cells to withstand the stress of overstimulation and persist despite this chronic activation, miR29a prevents CD8 T cells from entering into the state of full exhaustion by limiting pathways driving the primary overstimulation signal. The mechanistically different roles of miR-29a and miR-155 in regulating TEX lead us to ask if there is a potential synergistic effect in CD8 T cell persistence and differentiation. Furthermore, this notion of two miRs affecting the same problem of overstimulation that leads to exhaustion by employing two distinct mechanisms suggests new opportunities to prevent and/or reverse exhaustion by controlling antigen and inflammatory signaling. It will be interesting to dissect how miR-29a affects the induction and/or stability of the epigenetic landscape of TEX and determine whether de novo expression of miR29a, once exhaustion has been established, affects reversal of exhaustion or reprogramming of TEX.
In conclusion, we have identified a major role for miR-29a in regulating TEX differentiation, promoting long-term persistence and fostering a TMEM-like differentiation state in CD8 T cells responding to chronic viral infection that would otherwise become exhausted. Thus, we suggest that miR-29a might represent an immunotherapeutic target to promote long-term, functional CD8 T cell responses in chronic infections and cancers, including for cellular therapies.
Materials and Methods
Mice.
We purchased 6- to 8-week-old C57BL/6 Ly5.2CR (CD45.1) and C57BL/6 (CD45.2) mice from the US National Cancer Institute. miR-29ab1fl/fl CD4 Cre+ mice (51) were obtained from K.M. Ansel (University of California, San Francisco). Both male and female mice were used. P14 TCR transgenic mice expressing a TCR specific for the LCMV Dbgp33-41 epitope were bred in house. All mice were used in accordance with Institutional Animal Care and Use Committee guidelines for the University of Pennsylvania.
Viral Infections.
Mice were infected intraperitoneally with 2 × 105 plaque-forming units (PFU) of LCMV Arm or intravenously via tail-vein injection with 4 × 106 PFU of LCMV Cl-13 or 2 × 104 PFU of LCMV V35A. Recombinant influenza virus (H1N1) expressing the LCMV gp33-41 epitope (PR8-GP33) was obtained from Dr. Richard J. Webby (St. Jude Children's Research Hospital, Memphis, TN). Plaque assay was performed as previously described (8, 26, 31).
RV Experiments.
The miR-29a (MI0000576) cDNA clone was obtained from OriGene. miR-29a cDNA was cloned into the MSCV-IRES-VEX plasmid and MIGR1-IRES-GFP plasmid. Scrambled miR-29a (5′-UUCUCCGAACGUGUCACGUTT-3′) (52) was synthesized and inserted into MIGR1-GFP plasmid by Alta Biotech. For Tox, Fos, and Jun OE, only the coding regions (not including the 3′UTR) were cloned into the MSCV-IRES-VEX plasmid. The 3′UTR sensor GFP construct was obtained from Dr. Alejandro Villarino, University of Miami, Florida (33). RV was produced in 293T cells with MSCV and pCL-Eco plasmids using Lipofectamine 3000. RV transduction was performed as described (53). Briefly, purified CD8+ T cells were stimulated with 100 U/mL recombinant human IL-2, 1 μg/mL anti-mouse CD3ε, and 0.5 μg/mL anti-mouse CD28. After 18–24 h of stimulation, cells were transduced in the presence of polybrene (0.5 μg/mL) during spin infection (2,000g for 60 min at 32 °C) following incubation at 37 °C for 6 h. Cells were then washed and counted, and equal numbers were transferred immediately to clone 13–infected recipients.
Cell Preparation, Flow Cytometry, and Cell Sorting.
Spleens were mechanically disrupted onto a 70-μM cell strainer and red blood cells were lysed with ACK buffer (Gibco). Cells were stained with extracellular antibodies for 30 min on ice. For transcription-factor detection, cells were fixed and permeabilized using the Foxp3 Transcription Factor buffer set (Thermo Fisher Scientific). Samples were acquired on an LSR II and analyzed with FlowJo, version 10 software (Tree Star Inc). For cell sorting, CD8+ T cells were enriched using the EasySep CD8+ T Cell Isolation Kit (StemCell) and VEX+ cells were sorted based on CD8, CD45.1, CD45.2, and VEX on a BD FACSARIA (BD Bioscience) using a 70-μm nozzle.
Intracellular Cytokine Staining.
Splenocytes (1–2 × 106) were restimulated in vitro for 5 h at 37 °C in Roswell Park Memorial Institute medium supplemented with GolgiStop (1/250; BD Bioscience), GolgiPlug (1/500; BD Bioscience), gp33–41 peptide (0.4 μg/mL; National Institutes of Health [NIH]), and CD107a antibodies (1/500). Cells were then washed and stained using the BD Fixation/Permeabilization Kit (BD Bioscience).
Microarray Processing and Analysis.
Sorted LCMV Db gp-33–specific T cells and TN were resuspended in TRIzol (Thermo Fisher Scientific). RNA was isolated using the RNeasy Micro Kit (Qiagen). Microarray microRNA 2.0 (Affymetrix) was performed at the Penn Microarray Facility. CEL files from the microarrays were read in R using the ReadAffy function from the affy package, and the counts were quantile normalized using the NormiR function from ExiMiR package (https://www.rdocumentation.org/packages/ExiMiR/versions/2.14.0/topics/NormiR). The R package limma was used to fit the counts data to a model based on groups.
RNA Isolation, Quantitative RT-PCR, and Sequencing.
RNA was isolated using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. For quantitative RT-PCR, reverse transcription was performed using the TaqMan Advanced miRNA cDNA Synthesis Kit (for miRNAs) and TaqMan SuperScript VILO cDNA Synthesis Kit (for mRNAs). miRNAs were detected using TaqMan Advanced miRNA Assays, and mRNAs were detected using TaqMan gene expression assays, according to the manufacturer’s instructions. RT-PCR was performed using a StepOne Plus Real-Time PCR system (Applied Biosystems).
For RNA sequencing (RNA-seq), quality-control analysis, library generation, and RNA-seq were carried out by the Oncogenomics Core Facility at the University of Miami. RNA-seq libraries were prepared using Roche Kapa RNA HyperPrep with Riboerase. The RNA-sample RNA integrity number was equal to 10. Input amounts were split into two batches: low input (11.2 ng) and standard input (30 ng). Library amplification cycles were 14 for low input and 12 for standard input. Libraries were cleaned using standard AMPure bead protocols and balanced using fragment analysis (Agilent 5200) and DNA quantitation (Qubit). The library pool was sequenced on an Illumina NovaSeq 6000 on an S2 flow cell as 2× 150-bp reads. Basecalling and demultiplexing was performed in BaseSpace using default bcl2fastq parameters. Raw paired-ended FASTQ data were assessed for quality with FastQC (version11.5) (54). Trimmomatic (version 0.32) was then used to remove adapters, platform-specific sequences, and low-quality leading and trailing bases from reads (55). Then STAR (version 2.5.0) was used to map reads to the reference genome GRCm38 (56). The mapped data were assigned genomic features with featureCounts, version1.5.0 (57). Fold changes of differential expression were estimated through DESeq2 (58,59,60).
Network Analysis and GSEA.
Network analysis was performed with Ingenuity Pathway Analysis. Differential miRNAs were used as input to the MicroRNA Target Filter in IPA (Qiagen, https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/features/microrna-target-filter/) along with differential mRNAs from a previous study (5) to examine the miR-mRNA pairings. GSEA was performed using MSigDb (version 5.1) from the Broad Institute (https://www.broadinstitute.org/gsea/index.jsp).
Statistical Analysis.
Samples were tested for normal distribution using the D’Agostino and Pearson omnibus normality test. For samples that passed the normality test, statistical significance was calculated using an unpaired two-tailed Student t test (for n = 2) or one-way ANOVA with Bonferroni multiple comparisons posttest (for n > 2). For samples that did not pass normal distribution, statistical significance was calculated using the nonparametric Mann–Whitney test (for n = 2) or Kruskal–Wallis test with Dunn multiple comparisons posttest (for n > 2). Statistical significance was calculated by Prism 5 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank members of the E.J.W. laboratory who provided feedback. We thank the Flow Cytometry Core Facility at the University of Pennsylvania, the Onco-Genomics Shared Resource at Sylvester Comprehensive Cancer Center at the University of Miami, and the Biostatistics and Bioinformatics Shared Resource of the Sylvester Comprehensive Cancer Center, University of Miami. This work was supported by NIH grants AI105343, AI112521, AI082630, AI201085, and AI117950 to E.J.W.; NIH grants HL109102 and HL107202 to K.M.A.; and NIH grant 5-R21-AI-144732-02 to M.S.J. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award P30CA240139 to the Onco-Genomics Shared Resource at Sylvester Comprehensive Cancer Center. E.J.W. is also supported by the Parker Institute for Cancer Immunotherapy, which supports the cancer immunology program at the University of Pennsylvania.
Footnotes
Competing interest statement: E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Marengo, Janssen, Related Sciences, Synthekine, and Surface Oncology. E.J.W. is a founder of Surface Oncology, Danger Bio, and Arsenal Biosciences. E.J.W. has a patent licensing agreement on the PD-1 pathway with Roche/Genentech.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106083119/-/DCSupplemental.
Data Availability
RNASeq and microarray data have been deposited in Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) (data related to Fig. 1 [GEO], accession no. GSE196616; data related to Fig. 3 [SRA], accession no PRJNA811256).
References
- 1.Kaech S. M., et al. , Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLane L. M., Abdel-Hakeem M. S., Wherry E. J., CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doering T. A., et al. , Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bengsch B., et al. , Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity 45, 358–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wherry E. J., et al. , Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Pauken K. E., et al. , Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philip M., et al. , Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen D. R., et al. , The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel D. P., MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 13.O’Connell R. M., Rao D. S., Chaudhuri A. A., Baltimore D., Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122 (2010). [DOI] [PubMed] [Google Scholar]
- 14.O’Connell R. M., Rao D. S., Baltimore D., microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30, 295–312 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Chen Z., et al. , miR-150 regulates memory CD8 T cell differentiation via c-Myb. Cell Rep. 20, 2584–2597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gracias D. T., et al. , The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat. Immunol. 14, 593–602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hope J. L., et al. , The transcription factor T-bet is regulated by microRNA-155 in murine anti-viral CD8+ T cells via SHIP-1. Front. Immunol. 8, 1696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lind E. F., Elford A. R., Ohashi P. S., micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J. Immunol. 190, 1210–1216 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Ma F., et al. , The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 12, 861–869 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Smith N. L., Wissink E. M., Grimson A., Rudd B. D., miR-150 regulates differentiation and cytolytic effector function in CD8+ T cells. Sci. Rep. 5, 16399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steiner D. F., et al. , microRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35, 169–181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells A. C., et al. , Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. eLife 6, e26398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells A. C., Pobezinskaya E. L., Pobezinsky L. A., Non-coding RNAs in CD8 T cell biology. Mol. Immunol. 120, 67–73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H., et al. , miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One 2, e1020 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moffett H. F., et al. , The microRNA miR-31 inhibits CD8+ T cell function in chronic viral infection. Nat. Immunol. 18, 791–799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stelekati E., et al. , Long-term persistence of exhausted CD8 T cells in chronic infection is regulated by microRNA-155. Cell Rep. 23, 2142–2156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q., et al. , miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget 7, 53735–53750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., et al. , Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 27, 443–452 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Wei J., et al. , miR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. 18, 639–648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., et al. , microRNA-146a feedback suppresses T cell immune function by targeting Stat1 in patients with chronic hepatitis B. J. Immunol. 191, 293–301 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Crawford A., et al. , Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity 40, 289–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z., et al. , TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51, 840–855.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z., et al. , In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell 184, 1262–1280.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villarino A. V., et al. , Posttranscriptional silencing of effector cytokine mRNA underlies the anergic phenotype of self-reactive T cells. Immunity 34, 50–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araki K., et al. , Translation is actively regulated during the differentiation of CD8+ effector T cells. Nat. Immunol. 18, 1046–1057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stelekati E., et al. , Bystander chronic infection negatively impacts development of CD8(+) T cell memory. Immunity 40, 801–813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Im S. J., et al. , Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Utzschneider D. T., et al. , T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Wu T., et al. , The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin H., Blackburn S. D., Blattman J. N., Wherry E. J., Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J. Exp. Med. 204, 941–949 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callahan M. K., Postow M. A., Wolchok J. D., Targeting T cell co-receptors for cancer therapy. Immunity 44, 1069–1078 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Topalian S. L., Drake C. G., Pardoll D. M., Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang A. C., et al. , A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat. Med. 25, 454–461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alfei F., et al. , TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Scott A. C., et al. , TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo H., et al. , TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. U.S.A. 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan O., et al. , TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao C., et al. , Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeannet G., et al. , Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl. Acad. Sci. U.S.A. 107, 9777–9782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X., et al. , Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hines M. J., et al. , miR-29 sustains B cell survival and controls terminal differentiation via regulation of PI3K signaling. Cell Rep. 33, 108436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei W., et al. , miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 4, e668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurachi M., et al. , Optimized retroviral transduction of mouse T cells for in vivo assessment of gene function. Nat. Protoc. 12, 1980–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.S. Andrews, FastQC: A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 20 May 2020. [Google Scholar]
- 55.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.E. Stelekati, E. J. Wherry, MicroRNA expression data from LCMV infected CD8 T cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196616. Deposited 11 February 2022. [Google Scholar]
- 60.Y. Ban, MicroRNA-29a attenuates CD8 T cell exhaustion and induces memory-like CD8 T cells during chronic infection. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA811256. Accessed 28 February 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNASeq and microarray data have been deposited in Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) (data related to Fig. 1 [GEO], accession no. GSE196616; data related to Fig. 3 [SRA], accession no PRJNA811256).