Abstract
The extent to which persisting species may fill the functional role of extirpated or declining species has profound implications for the structure of biological communities and ecosystem functioning. In North America, arthropodivorous bats are threatened on a continent‐wide scale by the spread of white‐nose syndrome (WNS), a disease caused by the fungus Pseudogymnoascus destructans. We tested whether bat species that display lower mortality from this disease can partially fill the functional role of other bat species experiencing population declines. Specifically, we performed high‐throughput amplicon sequencing of guano from two generalist predators: the little brown bat (Myotis lucifugus) and big brown bat (Eptesicus fuscus). We then compared changes in prey consumption before versus after population declines related to WNS. Dietary niches contracted for both species after large and abrupt declines in little brown bats and smaller declines in big brown bats, but interspecific dietary overlap did not change. Furthermore, the incidence and taxonomic richness of agricultural pest taxa detected in diet samples decreased following bat population declines. Our results suggest that persisting generalist predators do not necessarily expand their dietary niches following population declines in other predators, providing further evidence that the functional roles of different generalist predators are ecologically distinct.
Keywords: bat diet, community science, Eptesicus fuscus, high‐throughput sequencing, metabarcoding, Myotis lucifugus, white‐nose syndrome (WNS)
While many bat species are known to function as suppressors of arthropod abundance, including several economically important agricultural pest taxa, the broader ecological consequences of disease‐related bat declines have not been fully assessed in many regions. In our study, we tested whether big brown bats (Eptesicus fuscus), which demonstrate lower mortality from white‐nose syndrome, could potentially fill the ecological role formerly occupied by the more severely affected little brown bat (Myotis lucifugus). Overall, we found that big brown bats did not adopt more of the prey formerly consumed by little brown bats and that the dietary niche overlap between the two bat species did not change over time.

1. INTRODUCTION
Species tend to diverge and specialize over long periods of time, but generalist habits can be an asset in eras dominated by change. On an evolutionary scale, specialization can lead to speciation, yet the persistence of generalism may offer a buffer against extinction (Dennis et al., 2011; Loxdale et al., 2011). Flexible resource requirements are often considered advantageous in the rapidly changing environments of the Anthropocene (Boyles & Storm, 2007; Colles et al., 2009; Purvis et al., 2000). As certain species decline, the question of whether persisting sympatric species can serve as ecological replacements to maintain interaction networks becomes increasingly important (Parker et al., 2010; Rubenstein et al., 2006; Tylianakis et al., 2010). In some examples, such as the reintroduction of Galápagos tortoises with saddle‐backed phenotypes (Chelonoidis spp.) as replacements for saddle‐backed giant tortoises (Chelonoidis abingdonii, Hunter et al., 2013), intentional introductions of similar species with the purpose of restoration have been successful. Other examples, such as American mink (Neogale vison) populations increasing following the decline in red foxes (Vulpes vulpes, Carlsson et al., 2010) and grey squirrel (Sciurus carolinensis) populations increasing following the decline in red squirrels (S. vulgaris, Tompkins et al., 2003), demonstrate unintentional introductions or range expansions that involve one species adopting the vacant trophic niches left behind by another. Considering global predator declines and recent emphases on the role of predators in conservation biology (e.g., Estes et al., 2011; Ritchie et al., 2012), the question of whether and to what extent surviving predators may be capable of filling the functional roles of extirpated predators remains largely unanswered. Assessing the foraging flexibility of different taxa therefore represents an important step for developing conservation strategies focused on preserving ecosystem function in the era of species extinctions.
Whether extant persisting taxa can compensate for the functional roles of other declining populations is generally unknown in aerial arthropodivores, largely due to methodological challenges associated with characterizing the consumption of diverse prey communities prior to population declines. Anatomical and physiological constraints often limit the flexibility of generalist predator foraging. For example, among arthropodivorous bats, body size, flight agility and maneuverability, bite force, and echolocation call frequencies influence the type of prey that can be captured (Aguirre et al., 2003; Barclay & Brigham, 1991). Some bat species are capable of consuming prey larger than their own body size and will occasionally land and consume the preferred parts of the prey while discarding the rest (O'Shea & Vaughan, 1977; Santana & Cheung, 2016). Since most arthropodivorous bats rely on echolocation while hunting, certain types of echolocation are also adapted for specific prey and may play a role as a factor‐limiting dietary niche breadth and flexibility (Arbour et al., 2019). For example, while echolocation frequency is related to the habitats in which different bats forage, the diets of bats that use lower‐frequency echolocation are also more likely to include larger prey, while the diets of bats that use higher‐frequency echolocation are more likely to include smaller prey (Denzinger & Schnitzler, 2013; Jones & Holderied, 2007). Such physiological constraints are generally considered to be more important determinants of bat trophic niches than direct competition with other bats in similar guilds (Schoeman & Jacobs, 2011). As such, the morphological constraints that shape bat foraging strategies call into question the potential for ecological equivalency among taxa.
In North America, many hibernating bat species have experienced rapid and precipitous declines due to white‐nose syndrome (WNS), a disease caused by the fungus Pseudogymnoascus destructans (Frick et al., 2016; Lorch et al., 2011). Bat population declines from WNS present a unique circumstance under which basic ecological questions about the role of arthropodivorous bats as predators may be answered. In the eastern region of North America where WNS has been present for over a decade, little brown bats (Myotis lucifugus, Leconte 1831) have declined by more than 90% (Frick et al., 2016). Comparatively, other common bat species such as big brown bats (Eptesicus fuscus, Palisot de Beauvois 1796) experience infection from WNS yet have declined with much less severity (Frank et al., 2014; Frick et al., 2010). Additional studies have shown that some overlap in prey resource usage occurs between these two species in this study region, with molecular methods indicating a 21.3% overlap in OTUs detected in diets (Wray et al., 2021) and stable isotope methods indicating a 45% overlap between dietary profiles (Wray & Peery, 2022). These observed patterns, combined with the apparent lesser severity of big brown bat mortality due to WNS, raise the question of the extent to which persisting taxa may further expand their dietary profiles to include prey resources formerly consumed by other arthropodivorous bats that experience more severe population declines.
In this study, we quantified changes in dietary composition and niche overlap in two generalist arthropodivorous bat species following the rapid, WNS‐induced decline in one species (little brown bats) and the persistence of another (big brown bats). Specifically, we tested whether declines in little brown bats would lead to dietary niche expansion among big brown bats by comparing changes in prey consumption, including agricultural pests, as measured using high‐throughput amplicon sequencing methods.
2. METHODS
2.1. Study species
Little brown and big brown bats are among the most common bat species in North America (Fenton, 1980; Kurta & Baker, 1990). Little brown bats are higher‐frequency echolocators that tend to be generalist in their foraging habits, mostly consuming aquatic insects—particularly those with swarming behaviors (such as chironomid midges)—although they also consume terrestrial prey including moths, true bugs, beetles, and spiders (Clare, Symondson, Broders, et al., 2014; Whitaker & Lawhead, 1992; Wray et al., 2021). In contrast, big brown bats are lower‐frequency echolocators and are often speciously referred to as “beetle specialists” but are well known to consume a variety of other arthropods such as flies, caddisflies, true bugs, and moths (Agosta, 2002; Clare et al., 2014; Wray et al., 2021). We selected little brown bats as a focal study species because, prior to WNS‐related declines, they were the most abundant bat species in the study region (Huebschman, 2019). We selected big brown bats as a second focal study species because they are also abundant in the region but were expected to decline less from WNS (based on previous population trends observed in the eastern region of North America, e.g., Cheng et al., 2021), thus allowing for a comparative analysis of the effects of disease‐related bat declines in differentially afflicted species. During the breeding season, both little brown and big brown bats also frequently form large maternity roosts within human‐built structures (Voigt et al., 2016), which allows for ease of detection and monitoring between years.
2.2. Bat guano collection and detection of arthropod DNA
We collected roost‐level bat guano samples (e.g., multiple pellets from multiple individuals at the same location) weekly at five little brown and five big brown bat roosts in Southern Wisconsin, USA, including one site with a little brown bat colony present in a bat house and a big brown bat colony present in a nearby barn (Figure 1a). We collected guano from each roost by placing a clean plastic sheet under each roost for 1 week, with samples collected weekly at all sites for the duration of the summer from 2015 to 2018 (late May to late August, ordinal weeks 20–36). Temporal differences in sample collection were minimized as much as possible, with a median ordinal collection week of 30 for the first 2 years (interquartile range [IQR] = 27, 32) and 29 for the second 2 years (IQR = 26, 32). Following collection, samples were stored on wet ice during transport and subsequently kept at –80°C for long‐term storage. Additional samples were collected in the same manner by community scientists at three or four time periods during the summer from 2015 to 2018, yielding an additional two little brown and two big brown bat roost sites (Figure 1a). These samples were initially stored at –20°C, then shipped overnight with a frozen ice pack and kept at –80°C for long‐term storage. This sample collection effort builds upon a previous study (Wray et al., 2021), with two additional years of data and five additional roost sites (n = 296 additional samples collected). The identity of bat species was confirmed by directly observing bat appearance (roosting bats were visible at all sites) and by comparing relative guano pellet size at the time of collection. The number of bats per roost was also estimated via emergence counts conducted as part of the Wisconsin Bat Program's Great Wisconsin Bat Count. These counts occurred at least twice per year in the early and late summer, which roughly corresponds to the pre‐ and post‐volancy reproductive periods. Approximately 30 minutes before sunset, volunteers positioned themselves near bat roosts and counted bats as they emerged. In 2015, little brown bat colonies had an average of 210 bats per roost (ranging from 94 to 409 bats), while big brown bat colonies had an average of 137 bats per roost (ranging from 21 to 428 bats). By 2018, little brown bat colonies declined to an average of 19 bats per roost (ranging from 0 to 50 bats), while big brown bat colonies declined to an average of 76 bats per roost (ranging from 26 to 180 bats). Overall, the average number of little brown bats in total per year was 1016 in the first 2 years, and 120 in the second 2 years, representing a decline of –88%. The average number of big brown bats in total per year was 929 in the first 2 years, and 542 in the second 2 years, representing a decline of –42%. All sample collection and animal observation methods were carried out in accordance with the guidelines of the Wisconsin Department of Natural Resources and the American Society of Mammalogists (Sikes, 2016). Experimental protocols were approved by the Wisconsin Natural History Inventory Program and the University of Wisconsin Animal Care and Use Committee.
FIGURE 1.
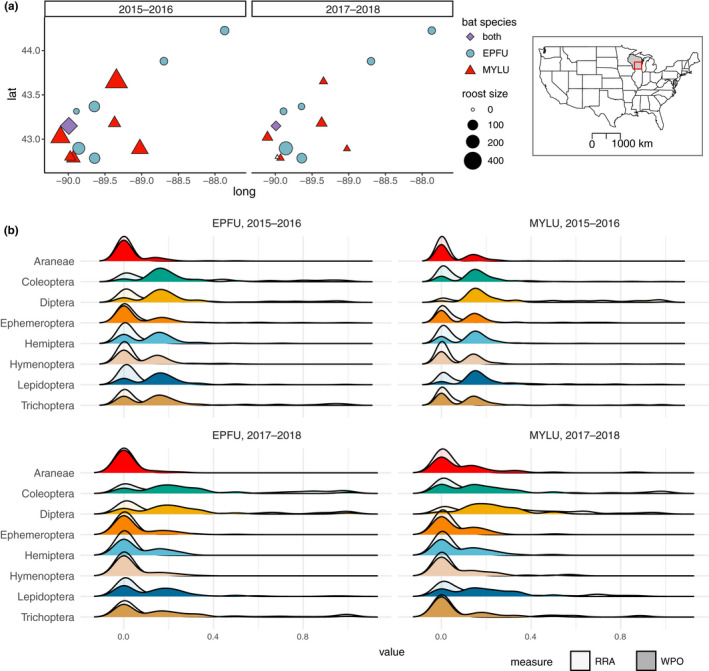
Characterization of study sites and bat dietary composition. (a) Map of study locations with points indicating the relative size of roosts. Map inset indicates the location of study sites within the continental United States. (b) Density plots of ordinal level dietary composition between time periods for each bat species. Solid regions represent weighted percent occurrence (wPO) values, and translucent regions indicate RRA (relative read abundance) values. EPFU = big brown bat (Eptesicus fuscus), MYLU = little brown bat (Myotis lucifugus)
2.3. Sequencing arthropod COI isolated from bat guano
DNA extraction, PCR, and high‐throughput amplicon sequencing followed Jusino et al. (2019) with the same modifications presented in Wray et al. (2021). Briefly, DNA was extracted from an 80mg subsample (~8 pellets) using a Qiagen DNA Stool mini kit (Qiagen Inc.), with a 180‐bp region of the COI subunit c amplified using PCR with ANML primers (Jusino et al., 2019). Thermocycler parameters followed Hebert et al. (2003), with the exception of the final extension at 72°C increased from 5 to 7 min. A single‐copy mock community of 34 known arthropod constituents was also amplified under the same conditions as a positive control (Jusino et al., 2019). Negative controls were included during each extraction batch and for each set of PCR reactions, which were visualized on a 2% agarose gel and did not yield any visible bands. Similarly, positive controls were also included for each set of PCR reactions, which did yield visible bands. PCR products were purified using a Zymo Select‐a‐Size Clean & Concentrator kit (Zymo Research), and then quantified using a Qubit 2.0 fluorometer (Invitrogen) with a high‐sensitivity dsDNA kit. Following quantification, the purified PCR products were then equilibrated. Five total equimolar libraries were then constructed, each with approximately 72 samples per library. Samples were processed in a randomized order to reduce potential batch processing biases. Sequencing was performed on an Ion Torrent Personal Genome Machine platform (PGM; ThermoFisher Scientific Inc.) according to the manufacturer's recommendations with an Ion PGM 318v2 chip. Raw sequence data were then processed using AMPtk v1.4.2 (Palmer et al., 2018). This data processing procedure includes de‐multiplexing using unique barcode index sequences, stripping of forward and reverse primers, and quality filtering and denoising with the DADA2 algorithm (Callahan et al., 2016). The resulting amplicon sequence variants (ASVs) were then clustered at 97% similarity using the UCLUST algorithm employed in VSEARCH to generate operational taxonomic units (OTUs; Jusino et al., 2019). Demultiplexed sequences were mapped back to these OTUs to generate an OTU table. Taxonomy was then assigned using the built‐in COI database in AMPtk. Finally, we removed all OTUs that were not identified as insects or arachnids, as well ectoparasites including mites (Mesostigmata and Trombidiformes), acari (Sarcoptiformes), and fleas (Siphonatera), which do not represent typical prey items.
2.4. Data cleaning and statistical analyses
Post‐processing, OTU tables were converted into weighted percent occurrence (wPO), a presence‐based metric, and relative read abundance (RRA), a read‐based metric, following Deagle et al. (2019). Since reads do not necessarily reflect abundance or biomass, occurrence‐based metrics are often considered more conservative, yet can be sensitive to overinflating the influence of rare taxa (Deagle et al., 2013, 2019). For this reason, we present both read‐based and occurrence‐based metrics for visual and qualitative comparison but use wPO for statistical analyses. Within major arthropod orders (Araneae, Coleoptera, Diptera, Ephemeroptera, Hemiptera, Hymenoptera, Lepidoptera, and Trichoptera), we compared mean values of wPO from samples collected between 2015 and 2016 with samples collected from 2017 to 2018 using separate Wilcoxon rank‐sum tests for each bat species. To account for the effects of multiple comparisons, we used a Bonferroni correction to adjust p‐values. We then aggregated OTU tables into presence/absence matrices at the family level for the estimation of niche metrics. We calculated niche breadth as Levin's adjusted niche breadth, Ba , and calculated niche overlap as Pianka's measure of niche overlap, Ojk , which provides a symmetrical estimate of the niche overlap between two species (Hurlbert, 1978; Levins, 1968; MacArthur & Levins, 1967). To visualize diet communities in multivariate space, we performed non‐metric multidimensional scaling (NMDS) using the metaMDS function in the R package “vegan” (Oksanen et al., 2013) with a modification of the raupcrick function as described by Chase et al. (2011), which was performed separately for presence/absence matrices at the OTU and family levels. For both taxonomic levels, we excluded any taxon groups that were not detected at least five times in order to reduce the influences of infrequently detected diet items. To focus on general trends in community composition, outliers were also sequentially removed following a visual inspection of NMDS plots (n = 12 samples removed). To assess whether the bat diet communities differed by species, time period, collection site, and ordinal week, we used non‐parametric PERMANOVA tests (Anderson, 2001) that were performed by the “adonis” function with 999 replicates and assessed the influences of multivariate dispersion (Anderson, 2006) using the “betadisper” function to separately test each predictor variable. We also searched taxonomy tables for certain arthropod taxa that are known agricultural pests in the area. To compare the incidence and taxonomic richness of pest taxa detected in samples collected between 2015 and 2016 with samples collected between 2017 and 2018, we used chi‐squared tests and Welch's t‐tests, respectively, which were conducted separately for each bat species. All post‐OTU table data cleaning and analyses were conducted in R version 4.1.0 (R Core Team, 2021). Additional packages used for data processing and visualization include “dplyr”, “ggplot2”, “tidyverse”, “wesanderson”, and “reshape2” (Ram & Wickham, 2018; Wickham, 2007, 2016; Wickham et al., 2019).
3. RESULTS
A total of 350 samples were successfully amplified of 558 samples collected (62.7%), yielding 20,990,143 sequencing reads. Following filtering and bioinformatic processing, 326 of the amplified samples (93.1%) were retained, which yielded a total of 2274 valid OTUs with 16,777,947 sequencing reads. All members of the insect mock community were recovered. After excluding sample duplicates (n = 11), the final sample set included 173 little brown and 142 big brown bat samples (n = 315 total samples). All OTUs were identified to phylum, class, and order, with 87.4% identified to family, 74.8% identified to genus, and 56.9% identified to species. After removing non‐arthropod OTUs and taxa that were unlikely prey items (i.e., ectoparasites, n = 611 OTUs removed), 1663 arthropod prey OTUs remained (73% of the total valid OTUs), representing 19 orders, 221 families, 703 genera, and 891 species. Samples had a median of 41,666 reads per sample (IQR = 22,080–54,122) and a median of 17 OTUs per sample (IQR = 9–27). A total of 1334 OTUs were found among little brown bats and 865 OTUs were found among big brown bats, of which 536 OTUs were shared between both. The most detected prey families for little brown bats, as measured by incidence, were Diptera: Chironomidae, followed by Lepidoptera: Tortricidae and Diptera: Limoniidae. For big brown bats, the most detected prey families were Coleoptera: Elateridae, Diptera: Limoniidae, and unidentified Coleoptera (Table 1). The most common prey, as measured by wPO, and relative reads were generally consistent, except for big brown bats where Trichoptera: Hydropsychidae had a much higher RRA in comparison to wPO (Table 1).
TABLE 1.
Twenty most commonly detected prey families for each bat species, ranked by incidence
| Little brown bat (Myotis lucifugus) | Big brown bat (Eptesicus fuscus) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Order | Family | Incidence | Mean RRA | Mean wPO | Order | Family | Incidence | Mean RRA | Mean wPO |
| Diptera | Chironomidae | 138 | 0.213 | 0.077 | Coleoptera | Elateridae | 83 | 0.160 | 0.061 |
| Lepidoptera | Tortricidae | 93 | 0.037 | 0.041 | Diptera | Limoniidae | 71 | 0.100 | 0.055 |
| Diptera | Limoniidae | 78 | 0.036 | 0.031 | Coleoptera | unidentified | 70 | 0.010 | 0.045 |
| Coleoptera | Elateridae | 75 | 0.057 | 0.036 | Coleoptera | Carabidae | 67 | 0.018 | 0.047 |
| Diptera | unidentified | 71 | 0.013 | 0.028 | Trichoptera | Hydropsychidae | 65 | 0.196 | 0.070 |
| Lepidoptera | Gelechiidae | 58 | 0.015 | 0.028 | Coleoptera | Scarabaeidae | 56 | 0.075 | 0.049 |
| Ephemeroptera | Caenidae | 53 | 0.044 | 0.022 | Lepidoptera | Tortricidae | 54 | 0.030 | 0.037 |
| Hemiptera | Miridae | 51 | 0.011 | 0.022 | Diptera | Chironomidae | 45 | 0.063 | 0.036 |
| Lepidoptera | Depressariidae | 50 | 0.049 | 0.020 | Coleoptera | Pyrochroidae | 43 | 0.020 | 0.028 |
| Trichoptera | Hydropsychidae | 49 | 0.083 | 0.028 | Diptera | Tipulidae | 42 | 0.004 | 0.023 |
| Diptera | Culicidae | 46 | 0.028 | 0.021 | Coleoptera | Cerambycidae | 36 | 0.007 | 0.022 |
| Diptera | Tipulidae | 44 | 0.003 | 0.019 | Hemiptera | Miridae | 35 | 0.010 | 0.020 |
| Hemiptera | Corixidae | 41 | 0.021 | 0.017 | Coleoptera | Hydrophilidae | 33 | 0.026 | 0.021 |
| Trichoptera | Leptoceridae | 40 | 0.016 | 0.017 | Ephemeroptera | Heptageniidae | 32 | 0.044 | 0.025 |
| Ephemeroptera | Heptageniidae | 39 | 0.017 | 0.018 | Megaloptera | Corydalidae | 30 | 0.058 | 0.029 |
| Diptera | Psychodidae | 37 | 0.028 | 0.015 | Hymenoptera | Ichneumonidae | 27 | 0.004 | 0.015 |
| Hymenoptera | Ichneumonidae | 34 | 0.002 | 0.013 | Diptera | unidentified | 26 | 0.005 | 0.022 |
| Coleoptera | unidentified | 32 | 0.002 | 0.014 | Coleoptera | Tenebrionidae | 21 | 0.003 | 0.013 |
| Coleoptera | Carabidae | 31 | 0.013 | 0.015 | Hemiptera | Cicadellidae | 21 | 0.000 | 0.013 |
| Diptera | Ceratopogonidae | 30 | 0.001 | 0.010 | Coleoptera | Silphidae | 21 | 0.001 | 0.010 |
Abbreviations: RRA, relative read abundance; wPO, weighted percent occurrence.
Overall, we found that little brown and big brown bat dietary composition was distinct, and intraspecific differences in dietary composition did not change substantially between pre‐ and post‐WNS time periods for either bat species. Little brown bats diets contained a higher richness of Diptera, Lepidoptera, and Hemiptera, and big brown bat diets contained a higher richness of Coleoptera. The mean wPO within arthropod orders differed by bat species but remained similar between time periods within each bat species (Figure 1b; Table 2). Hemiptera were less common in both little brown and big brown bat guano samples in the later time periods, and for little brown bats, other prey groups did not change (Figure 1b; Table 2). In the later time period, Hymenoptera were marginally less common in big brown bat guano samples (Figure 1b; Table 2). The arthropod families most commonly detected for each bat species also remained similar between time periods (Table 3).
TABLE 2.
Changes in mean weighted percent occurrence (wPO) of prey orders between time periods
| Order | Big brown bat (Eptesicus fuscus) | Little brown bat (Myotis lucifugus) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean, 2015–2016 | Mean, 2017–2018 | p | p, adjusted | Mean, 2015–2016 | Mean, 2017–2018 | p | p, adjusted | |
| Araneae | 0.033 | 0.020 | .229 | 1.000 | 0.059 | 0.091 | .341 | 1.000 |
| Coleoptera | 0.180 | 0.231 | .102 | .814 | 0.141 | 0.153 | .991 | 1.000 |
| Diptera | 0.155 | 0.220 | .037 | .295 | 0.185 | 0.222 | .032 | .260 |
| Ephemeroptera | 0.067 | 0.050 | .339 | 1.000 | 0.088 | 0.058 | .067 | .537 |
| Hemiptera | 0.118 | 0.067 | .004 | .035 | 0.124 | 0.079 | .005 | .041 |
| Hymenoptera | 0.078 | 0.041 | .007 | .056 | 0.084 | 0.067 | .098 | .786 |
| Lepidoptera | 0.147 | 0.129 | .370 | 1.000 | 0.159 | 0.172 | .828 | 1.000 |
| Trichoptera | 0.122 | 0.136 | .636 | 1.000 | 0.092 | 0.069 | .082 | .657 |
Adjusted p‐values below .05 are highlighted in bold.
TABLE 3.
Top 20 family‐level prey items detected in big brown and little brown bat guano samples, ranked by weighted percent occurrence (wPO). Changes in top family‐level prey items between time periods are highlighted in bold
| Big brown bat (Eptesicus fuscus), 2015–2016 | Big brown bat (Eptesicus fuscus), 2017–2018 | ||||
|---|---|---|---|---|---|
| Order | Family | Mean wPO | Order | Family | Mean wPO |
| Coleoptera | Elateridae | 0.0527 | Trichoptera | Hydropsychidae | 0.089 |
| Trichoptera | Hydropsychidae | 0.0524 | Coleoptera | Scarabaeidae | 0.070 |
| Coleoptera | Carabidae | 0.0498 | Coleoptera | Elateridae | 0.070 |
| Coleoptera | unidentified | 0.0469 | Diptera | Limoniidae | 0.064 |
| Diptera | Limoniidae | 0.0459 | Coleoptera | Carabidae | 0.044 |
| Lepidoptera | Tortricidae | 0.0376 | Coleoptera | unidentified | 0.044 |
| Ephemeroptera | Heptageniidae | 0.0366 | Megaloptera | Corydalidae | 0.043 |
| Diptera | Chironomidae | 0.0330 | Diptera | Chironomidae | 0.040 |
| Coleoptera | Scarabaeidae | 0.0305 | Lepidoptera | Tortricidae | 0.036 |
| Coleoptera | Pyrochroidae | 0.0269 | Coleoptera | Pyrochroidae | 0.030 |
| Coleoptera | Cerambycidae | 0.0262 | Diptera | unidentified | 0.024 |
| Hemiptera | Miridae | 0.0241 | Diptera | Tipulidae | 0.023 |
| Diptera | Tipulidae | 0.0222 | Coleoptera | Hydrophilidae | 0.021 |
| Coleoptera | Hydrophilidae | 0.0213 | Hemiptera | Cicadellidae | 0.019 |
| Diptera | unidentified | 0.0196 | Coleoptera | Cerambycidae | 0.017 |
| Hymenoptera | Ichneumonidae | 0.0180 | Hemiptera | Miridae | 0.015 |
| Megaloptera | Corydalidae | 0.0167 | Diptera | Culicidae | 0.014 |
| Lepidoptera | Tineidae | 0.0148 | Coleoptera | Tenebrionidae | 0.014 |
| Trichoptera | Leptoceridae | 0.0135 | Ephemeroptera | Heptageniidae | 0.012 |
| Coleoptera | Tenebrionidae | 0.0128 | Hymenoptera | Ichneumonidae | 0.012 |
| Little brown bat (Myotis lucifugus), 2015–2016 | Little brown bat (Myotis lucifugus), 2017–2018 | ||||
| Diptera | Chironomidae | 0.0744 | Diptera | Chironomidae | 0.086 |
| Lepidoptera | Tortricidae | 0.0387 | Lepidoptera | Tortricidae | 0.049 |
| Diptera | Limoniidae | 0.0337 | Coleoptera | Elateridae | 0.044 |
| Coleoptera | Elateridae | 0.0337 | Trichoptera | Hydropsychidae | 0.043 |
| Diptera | unidentified | 0.0285 | Diptera | Culicidae | 0.032 |
| Lepidoptera | Gelechiidae | 0.0268 | Lepidoptera | Gelechiidae | 0.031 |
| Ephemeroptera | Caenidae | 0.0259 | Lepidoptera | Depressariidae | 0.031 |
| Hemiptera | Miridae | 0.0251 | Diptera | unidentified | 0.028 |
| Trichoptera | Hydropsychidae | 0.0240 | Araneae | unidentified | 0.025 |
| Trichoptera | Leptoceridae | 0.0207 | Coleoptera | Scarabaeidae | 0.025 |
| Hemiptera | Corixidae | 0.0193 | Coleoptera | Dermestidae | 0.024 |
| Diptera | Tipulidae | 0.0184 | Diptera | Tipulidae | 0.023 |
| Ephemeroptera | Heptageniidae | 0.0182 | Araneae | Theridiidae | 0.021 |
| Lepidoptera | Depressariidae | 0.0176 | Coleoptera | unidentified | 0.021 |
| Diptera | Culicidae | 0.0175 | Diptera | Limoniidae | 0.019 |
| Diptera | Psychodidae | 0.0166 | Diptera | Chaoboridae | 0.019 |
| Hymenoptera | Ichneumonidae | 0.0155 | Ephemeroptera | Heptageniidae | 0.018 |
| Diptera | Cecidomyiidae | 0.0154 | Coleoptera | Carabidae | 0.018 |
| Lepidoptera | Tineidae | 0.0148 | Diptera | Tachinidae | 0.017 |
| Coleoptera | Carabidae | 0.0140 | Lepidoptera | Crambidae | 0.015 |
In general, little brown and big brown bats showed interspecific differences in family‐level dietary niche breadth, which did not change substantially between the pre‐ and post‐WNS time periods. Little brown bats displayed higher niche breadth in comparison to big brown bats, and total interspecific niche overlap was 0.281 (Table 4). For both bat species, dietary niche breadth from samples collected in 2015–2016 was higher than dietary niche breadth from samples collected in 2017–2018. Little brown bat dietary niche breadth decreased by 24.2% between time periods, while big brown bat dietary niche breadth decreased by 34.6% between time periods (Table 4). Interspecific dietary niche overlap was similar between time periods, increasing by only 2.3% from 0.281 to 0.288 (Table 4). NMDS plots indicated visually that diet composition differed more between species than between time periods (Figure 2a). PERMANOVA demonstrated that species was the best predictor of variation at the family and OTU levels (Figure 2a; Table 5). At both the family and OTU levels, ordinal week was also a significant predictor of variation, while collection site and time period were also significant predictors at the OTU level (Table 5). At both the family and OTU levels, there were significant differences in multivariate dispersion between bat species and between collection sites, and at the OTU level, there were also significant differences in multivariate dispersion between time periods and ordinal week (Figure 2a; Table 5). These results suggest that while bat species was the most important factor in determining diet community structure and dispersion, other factors were also somewhat influential, particularly at the OTU level.
TABLE 4.
Family‐level dietary niche breadth (B, Levin's measure of niche breadth and Ba , Levin's adjusted niche breadth) and overlap Ojk (Pianka's measure of symmetrical niche overlap) for little brown and big brown bats
| Time | B, MYLU | Ba , MYLU | B, EPFU | Ba , EPFU | Overlap, Ojk |
|---|---|---|---|---|---|
| 2015–2018 | 51.3252 | 0.2207 | 35.8268 | 0.1527 | 0.2813 |
| 2015–2016 | 52.1028 | 0.2241 | 42.0538 | 0.1801 | 0.2811 |
| 2017–2018 | 39.7123 | 0.1698 | 27.8405 | 0.1177 | 0.2876 |
Abbreviations: EPFU, big brown bat (Eptesicus fuscus);MYLU, little brown bat (Myotis lucifugus).
FIGURE 2.
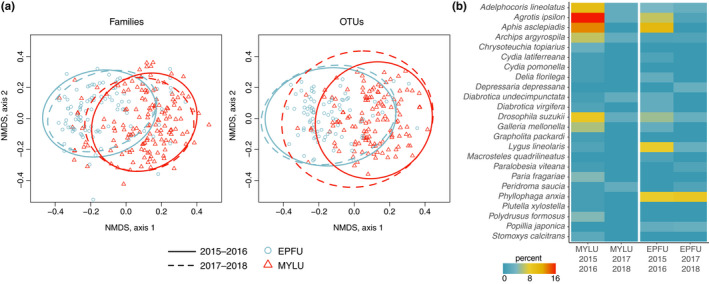
Changes in bat dietary composition over time. (a) NMDS plot of family‐level and OTU‐level dietary communities with 80% confidence interval ellipses. Solid lines indicate the first time period (2015–2016), while dashed lines indicate the second time period (2017–2018). Shapes indicate samples from each bat species. (b) Heatmap of agricultural pest taxa detected in bat guano samples. Values indicate the percentage of samples for which each agricultural pest taxa were detected. EPFU = big brown bat (Eptesicus fuscus), MYLU = little brown bat (Myotis lucifugus)
TABLE 5.
PERMANOVA and Betadisper test results
| Term | PERMANOVA | Betadisper | ||||||
|---|---|---|---|---|---|---|---|---|
| df | Sums of Sqs | Mean Sqs | F | R 2 | p | F | p | |
| Family level | ||||||||
| Species | 1 | 2.42 | 2.416 | 13.97 | 0.052 | .01 | 15.89 | <.001 |
| Site | 12 | 2.83 | 0.236 | 1.36 | 0.061 | .14 | 1.89 | .04 |
| Time | 1 | 0.29 | 0.286 | 1.65 | 0.006 | .24 | 0.01 | .92 |
| Week | 1 | 0.68 | 0.682 | 3.94 | 0.015 | .03 | 0.99 | .47 |
| Residuals | 233 | 40.30 | 0.173 | 0.866 | ||||
| Total | 248 | 46.52 | 1.000 | |||||
| OTU level | ||||||||
| Species | 1 | 10.12 | 10.124 | 62.14 | 0.117 | .01 | 16.40 | <.001 |
| Site | 13 | 32.25 | 2.481 | 15.22 | 0.371 | .01 | 3.17 | <.001 |
| Time | 1 | 2.20 | 2.202 | 13.51 | 0.025 | .01 | 3.98 | .047 |
| Week | 1 | 2.00 | 2.004 | 12.30 | 0.023 | .01 | 2.90 | <.001 |
| Residuals | 247 | 40.25 | 0.163 | 0.464 | ||||
| Total | 263 | 86.82 | 1.000 | |||||
P values below .05 are highlighted in bold.
Several agricultural pests were detected in bat guano samples, which were most common in little brown bat samples collected prior to WNS‐related declines. Cumulatively, we detected at least one agricultural pest taxon in 45.1% of little brown bat samples and in 33.8% of big brown samples. For little brown bats, this percentage decreased from 53.3% to 15.8% between the two time periods, while for big brown bats this percentage decreased from 43.2% to 23.6% between the two time periods. The most common agricultural pest taxon in little brown bat guano samples was Agrotis ipsilon (black cutworm), which was detected in 13% of all samples, while the most common agricultural pest taxon for big brown bats was Phyllophaga anxia (cranberry white grub), which was detected in 8% of all samples. For both bat species, the incidence of most individual pest taxa also declined between time periods (Figure 2b). The difference in the proportion of samples with at least one agricultural pest taxon present between time periods was statistically significant for little brown bats (χ 2 = 15.4, df = 1, p < .001) and for big brown bats (χ 2 = 5.31, df = 1, p = .021). Little brown bats had an average of 0.54 pest taxa per sample (ranging from 0 to 4 pest taxa per sample), while big brown bats had an average of 0.45 pest taxa per sample (ranging from 0 to 3 pest taxa per sample). For little brown bat guano samples, the average richness of pest taxa per sample was significantly higher in the first time period (x̅ = 0.89 ± 0.17) in comparison to the second time period (x̅ = 0.18 ± 0.15, t 136.61 = 6.19, p < .001). For big brown bat guano samples, the average richness of pest taxa per sample was also significantly higher in the first time period (x̅ = 0.61 ± 0.19) in comparison to the second time period (x̅ = 0.29 ± 0.15, t 133.18 = 2.61, p = .01).
4. DISCUSSION
Overall, our findings indicate that big brown bats do not immediately expand their dietary profiles after rapid declines in little brown bat populations, and are therefore unlikely to function as ecological replacements. In this study, we tested whether the effects of bat population declines influenced changes in dietary composition, interspecific niche overlap, and the amount of agricultural pest taxa consumed. Little brown bat roost sizes declined to a much greater extent than big brown bats, in accordance with our a priori assumptions. After the observed declines in little brown bat populations, intraspecific dietary composition did not change substantially, nor did overlap increase, although niche breadth decreased for both bat species. These results indicate that while little brown and big brown bats provide some complementarity as predators with different dietary niches, they already maintained some dietary niche overlap prior to WNS‐related population declines, which is also consistent with a long‐term study using stable isotope analyses (Wray & Peery, 2022). The observed decrease in niche breath following declines in roost sizes suggests that both bat species may display some degree of individual specialization (Bolnick et al., 2002, 2003), with the individuals within a population consuming different prey resources that ultimately contribute to a broader population‐level dietary niche. The decreasing prevalence of agricultural pest arthropods in bat diets following WNS‐related declines may be an artefact of overall decreasing dietary niche breadth but could also suggest that individuals select for other prey items, potentially as a result of reduced inter‐ or intraspecific competition. These results are consistent with one previous study which also suggested that big brown bat dietary niche breadth may be driven by individual specialization (Cryan et al., 2012), although the contributions of individuals to population‐level dietary breadth have been seldom explored in arthropodivorous bats despite evidence of within‐population variation in several other taxa (Johnston & Fenton, 2001).
While niche breadth decreased for both bat species, niche overlap did not change, but rather, some niche overlap (0.281) was observed prior to declines in bat populations. One previous study, which compared pre‐ and post‐WNS dietary composition based on stomach contents of bat carcasses, found that overlap in dietary composition did increase following WNS‐related bat declines and suggested that this may indicate increasing competition (Morningstar et al., 2019). Considering the observed variation between sites and between weeks, as well as the known high spatial and temporal turnover in the diets of both bat species (Wray et al., 2021), the comparison of pre‐ and post‐WNS bat diets may not be appropriate for individual bats collected at different sites or during different seasonal time periods. Interannual variation in diets may also influence conclusions for studies with limited or uneven year‐to‐year sampling schemes. Separate studies also suggested that pre‐ and post‐WNS changes in bat acoustic activity could indicate shifts in temporal and spatial niche partitioning (Jachowski et al., 2014) or reduced interspecific competition (Mayberry et al., 2020). However, just as co‐occurrence does not necessarily imply the presence of ecological interactions (Blanchet et al., 2020), decreasing habitat or dietary niche overlap does not necessarily imply decreasing competition. The principle of competition relies on the supposition that resources shared by two species must be limiting for competition to occur, and coexistence has been shown to persist under many cases where the assumptions of competitive exclusion are not met (Chase et al., 2002; Holt, 1977). As such, the results of our study suggest that WNS‐related declines in little brown bats likely do not lead to increases in niche overlap in this study area, but rather demonstrates that interspecific niche overlap between little brown and big brown bats has remained fairly consistent in recent time.
The results of this study suggest that while big brown bats share some prey resources with little brown bats, they do not readily shift their diets to include more prey resources following little brown bat declines. These findings are consistent with previous research and suggest that big brown bat foraging may be limited by other factors such as body size. Indeed, little brown and big brown bats are estimated to have diverged from each other more than 30 million years ago (Amador et al., 2018; Lack & Bussche, 2010), and have developed adaptations for foraging on different prey types. In this study, we did not quantify the influences of prey availability, although other studies have detected declining arthropod abundance in many regions (Hallmann et al., 2017; Sánchez‐Bayo & Wyckhuys, 2019; Seibold et al., 2019). A previous study in this region showed that these bat species generally maintain strong prey preferences independently of changing local prey availability (Wray et al., 2021). Nonetheless, other factors, such as habitat or roost availability, could represent limiting resources where the larger body size of big brown bats could make for a better competitor (Agosta, 2002). We observed complete roost abandonment at two little brown bat roost sites by 2018, and if these roosts were later adopted by big brown bats, it may be unlikely that little brown bats could reoccupy them upon population recovery. As such, further exploration into the potential competition between little brown and big brown bats for roost space or other habitat requirements may be warranted and are potentially more important than limitations due to food resources.
We characterized the functional role of bats as predators with the goal of assessing the extent to which widespread, flexible, and comparatively successful sympatric species have the potential to serve as ecological replacements for other declining species. While big brown bats likely cannot fully fill the trophic role of little brown bats, possibly due to morphological or other physiological constraints, other bat species may be more ecologically similar to little brown bats. However, most of these species also experience severe declines due to WNS, and non‐affected species such as migratory bats do not cluster in large colonies in this study region (Huebschman, 2019), and as such probably do not influence prey communities in the same manner. While other studies have demonstrated the successful reintroductions of extirpated predators leading to restoration ecosystem functioning (e.g., Mittelbach et al., 1995; Ripple & Beschta, 2012), such efforts often rely on the possibility of conservation strategies such as captive breeding or translocation—none of which have successfully been implemented for little brown bats or other bat species severely affected by WNS (Davy & Whitear, 2016). As such, it is unlikely that the functional role of the little brown bat can be restored either naturally or through management strategies, and several bat species severely impacted by WNS are expected to face extirpation in many regions (Frick et al., 2010; Thogmartin et al., 2013). The growing body of evidence regarding the function of arthropodivorous bats as ecologically important predators thus raises serious concerns regarding potential top‐down consequences of WNS‐related bat declines. These findings highlight the importance of continuing to support little brown bat population recovery, while also emphasizing the need for conservation of bats and other aerial arthropodivores in general due to the probability that each unique species cannot necessarily be replaced by another.
AUTHOR CONTRIBUTIONS
Amy Wray: Data curation (lead); Formal analysis (lead); Investigation (lead); Visualization (lead); Writing—original draft (lead); Writing—review & editing (lead). Claudio Gratton: Conceptualization (equal); Funding acquisition (equal); Writing—review & editing (supporting). Michelle Jusino: Data curation (supporting); Investigation (supporting); Methodology (lead); Resources (supporting); Writing—review & editing (supporting). Jing Jamie Wang: Investigation (supporting); Writing—review & editing (supporting). Jade Kochanski: Investigation (supporting); Writing—review & editing (supporting). Jonathan M Palmer: Data curation (supporting); Methodology (supporting); Software (supporting); Writing—review & editing (supporting). Mark Banik: Methodology (supporting); Resources (supporting); Writing—review & editing (supporting). Daniel L. Lindner: Conceptualization (equal); Resources (supporting); Writing—review & editing (supporting). Zach Peery: Conceptualization (equal); Funding acquisition (equal); Resources (lead); Writing—review & editing (supporting).
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
The authors thank Pen & Lynn Ackley, Mike & Jane Balch, Rhonda Enge, Marci & Jim Hess, Elaine Horn, Gary Jansen, Ron & Phillip Kerl, Emma Pelton, Liese Pfeifer, Cathy Prescott, Tom & Linda Ross, Will Sandvold, Elaine Swanson, and Mimi Wuest for their contributions to sample collection for this project. We thank John Arthur, Kathleen Gruentzel, and Darren Marsh for assistance in coordinating sampling at state and county parks. We thank Heather Kaarakka, J. Paul White, and the many volunteers who participated in the Great Wisconsin Bat Count for collecting and providing bat emergence count data. Finally, we thank two anonymous reviewers for their helpful suggestions for improving an earlier version of this manuscript. This study was funded by USDA Hatch Formula Fund WIS01841.
Wray, A. K. , Gratton, C. , Jusino, M. A. , Wang, J. J. , Kochanski, J. M. , Palmer, J. M. , Banik, M. T. , Lindner, D. L. , & Peery, M. Z. (2022). Disease‐related population declines in bats demonstrate non‐exchangeability in generalist predators. Ecology and Evolution, 12, e8978. 10.1002/ece3.8978
DATA AVAILABILITY STATEMENT
The final OTU table and sample metadata are permanently archived online and can be accessed at https://figshare.com/articles/dataset/ch4_OTU_table_taxonomy_with_metadata/17267150. Raw sequence data are available at SRA accession number PRJNA668526.
REFERENCES
- Agosta, S. J. (2002). Habitat use, diet and roost selection by the big brown bat (Eptesicus fuscus) in North America: A case for conserving an abundant species. Mammal Review, 32, 179–198. 10.1046/j.1365-2907.2002.00103.x [DOI] [Google Scholar]
- Aguirre, L. , Herrel, A. , Van Damme, R. , & Matthysen, E. (2003). The implications of food hardness for diet in bats. Functional Ecology, 17, 201–212. 10.1046/j.1365-2435.2003.00721.x [DOI] [Google Scholar]
- Amador, L. I. , Arévalo, R. L. M. , Almeida, F. C. , Catalano, S. A. , & Giannini, N. P. (2018). Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. Journal of Mammalian Evolution, 25, 37–70. 10.1007/s10914-016-9363-8 [DOI] [Google Scholar]
- Anderson, M. J. (2001). A new method for non‐parametric multivariate analysis of variance. Austral Ecology, 26, 32–46. [Google Scholar]
- Anderson, M. J. (2006). Distance‐based tests for homogeneity of multivariate dispersions. Biometrics, 62, 245–253. 10.1111/j.1541-0420.2005.00440.x [DOI] [PubMed] [Google Scholar]
- Arbour, J. H. , Curtis, A. A. , & Santana, S. E. (2019). Signatures of echolocation and dietary ecology in the adaptive evolution of skull shape in bats. Nature Communications, 10, 1–13. 10.1038/s41467-019-09951-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay, R. M. R. , & Brigham, R. M. (1991). Prey detection, dietary niche breadth, and body size in bats ‐ why are aerial insectivorous bats so small. American Naturalist, 137, 693–703. 10.1086/285188 [DOI] [Google Scholar]
- Blanchet, F. G. , Cazelles, K. , & Gravel, D. (2020). Co‐occurrence is not evidence of ecological interactions. . Ecology Letters, 23(7), 1050–1063. 10.1111/ele.13525 [DOI] [PubMed] [Google Scholar]
- Bolnick, D. I. , Svanbäck, R. , Fordyce, J. A. , Yang, L. H. , Davis, J. M. , Hulsey, C. D. , & Forister, M. L. (2003). The ecology of individuals: Incidence and implications of individual specialization. The American Naturalist, 161, 1–28. 10.1086/343878 [DOI] [PubMed] [Google Scholar]
- Bolnick, D. I. , Yang, L. H. , Fordyce, J. A. , Davis, J. M. , & Svanbäck, R. (2002). Measuring individual‐level resource specialization. Ecology, 83, 2936–2941. [Google Scholar]
- Boyles, J. G. , & Storm, J. J. (2007). The perils of picky eating: Dietary breadth is related to extinction risk in insectivorous bats. PLoS One, 2, e672. 10.1371/journal.pone.0000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B. J. , McMurdie, P. J. , Rosen, M. J. , Han, A. W. , Johnson, A. J. A. , & Holmes, S. P. (2016). DADA2: High‐resolution sample inference from Illumina amplicon data. Nature Methods, 13, 581. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, N. O. , Jeschke, J. M. , Holmqvist, N. , & Kindberg, J. (2010). Long‐term data on invaders: When the fox is away, the mink will play. Biological Invasions, 12, 633–641. 10.1007/s10530-009-9470-z [DOI] [Google Scholar]
- Chase, J. M. , Abrams, P. A. , Grover, J. P. , Diehl, S. , Chesson, P. , Holt, R. D. , Richards, S. A. , Nisbet, R. M. , & Case, T. J. (2002). The interaction between predation and competition: A review and synthesis. Ecology Letters, 5, 302–315. 10.1046/j.1461-0248.2002.00315.x [DOI] [Google Scholar]
- Chase, J. M. , Kraft, N. J. , Smith, K. G. , Vellend, M. , & Inouye, B. D. (2011). Using null models to disentangle variation in community dissimilarity from variation in α‐diversity. Ecosphere, 2, 1–11. 10.1890/ES10-00117.1 [DOI] [Google Scholar]
- Cheng, T. L. , Reichard, J. D. , Coleman, J. T. , Weller, T. J. , Thogmartin, W. E. , Reichert, B. E. , Bennett, A. B. , Broders, H. G. , Campbell, J. , Etchison, K. , & Feller, D. J. (2021). The scope and severity of white‐nose syndrome on hibernating bats in North America. Conservation Biology, 35, 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare, E. L. , Symondson, W. O. , Broders, H. , Fabianek, F. , Fraser, E. E. , MacKenzie, A. , Boughen, A. , Hamilton, R. , Willis, C. K. , & Martinez‐Nuñez, F. (2014). The diet of Myotis lucifugus across Canada: Assessing foraging quality and diet variability. Molecular Ecology, 23, 3618–3632. [DOI] [PubMed] [Google Scholar]
- Clare, E. L. , Symondson, W. O. , & Fenton, M. B. (2014). An inordinate fondness for beetles? Variation in seasonal dietary preferences of night‐roosting big brown bats (Eptesicus fuscus). Molecular Ecology, 23, 3633–3647. [DOI] [PubMed] [Google Scholar]
- Colles, A. , Liow, L. H. , & Prinzing, A. (2009). Are specialists at risk under environmental change? Neoecological, paleoecological and phylogenetic approaches. Ecology Letters, 12, 849–863. 10.1111/j.1461-0248.2009.01336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan, P. M. , Stricker, C. A. , & Wunder, M. B. (2012). Evidence of cryptic individual specialization in an opportunistic insectivorous bat. Journal of Mammalogy, 93, 381–389. 10.1644/11-MAMM-S-162.1 [DOI] [Google Scholar]
- Davy, C. , & Whitear, A. (2016). Feasibility and pitfalls of ex situ management to mitigate the effects of an environmentally persistent pathogen. Animal Conservation, 19, 539–547. [Google Scholar]
- Deagle, B. E. , Thomas, A. C. , McInnes, J. C. , Clarke, L. J. , Vesterinen, E. J. , Clare, E. L. , Kartzinel, T. R. , & Eveson, J. P. (2019). Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Molecular Ecology, 28, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deagle, B. E. , Thomas, A. C. , Shaffer, A. K. , Trites, A. W. , & Jarman, S. N. (2013). Quantifying sequence proportions in a DNA‐based diet study using Ion Torrent amplicon sequencing: Which counts count? Molecular Ecology Resources, 13(4), 620–633. [DOI] [PubMed] [Google Scholar]
- Dennis, R. L. , Dapporto, L. , Fattorini, S. , & Cook, L. M. (2011). The generalism–specialism debate: The role of generalists in the life and death of species. Biological Journal of the Linnean Society, 104, 725–737. 10.1111/j.1095-8312.2011.01789.x [DOI] [Google Scholar]
- Denzinger, A. , & Schnitzler, H.‐U. (2013). Bat guilds, a concept to classify the highly diverse foraging and echolocation behaviors of microchiropteran bats. Frontiers in Physiology, 4, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes, J. A. , Terborgh, J. , Brashares, J. S. , Power, M. E. , Berger, J. , Bond, W. J. , Carpenter, S. R. , Essington, T. E. , Holt, R. D. , Jackson, J. B. C. , Marquis, R. J. , Oksanen, L. , Oksanen, T. , Paine, R. T. , Pikitch, E. K. , Ripple, W. J. , Sandin, S. A. , Scheffer, M. , Schoener, T. W. , … Wardle, D. A. (2011). Trophic downgrading of planet Earth. Science, 333, 301–306. 10.1126/science.1205106 [DOI] [PubMed] [Google Scholar]
- Fenton, M. B. , & Barclay, R. M. R. (1980). Myotis lucifugus . Mammalian Species, 142, 1–8. [Google Scholar]
- Frank, C. L. , Michalski, A. , McDonough, A. A. , Rahimian, M. , Rudd, R. J. , & Herzog, C. (2014). The resistance of a North American bat species (Eptesicus fuscus) to white‐nose syndrome (WNS). PLoS One, 9, e113958. 10.1371/journal.pone.0113958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick, W. F. , Pollock, J. F. , Hicks, A. C. , Langwig, K. E. , Reynolds, D. S. , Turner, G. G. , Butchkoski, C. M. , & Kunz, T. H. (2010). An emerging disease causes regional population collapse of a common North American bat species. Science, 329, 679–682. 10.1126/science.1188594 [DOI] [PubMed] [Google Scholar]
- Frick, W. F. , Puechmaille, S. J. , & Willis, C. K. R. (2016). White‐nose syndrome in bats. In Voigt C. C. & Kingston T. (Eds.), Bats in the Anthropocene: Conservation of bats in a changing world (pp. 245–262). Springer. [Google Scholar]
- Hallmann, C. A. , Sorg, M. , Jongejans, E. , Siepel, H. , Hofland, N. , Schwan, H. , Stenmans, W. , Müller, A. , Sumser, H. , Hörren, T. , Goulson, D. , & de Kroon, H. (2017). More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PLoS One, 12, e0185809. 10.1371/journal.pone.0185809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, P. D. N. , Cywinska, A. , Ball, S. L. , & de Waard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, 270, 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, R. D. (1977). Predation, apparent competition, and the structure of prey communities. Theoretical Population Biology, 12, 197–229. 10.1016/0040-5809(77)90042-9 [DOI] [PubMed] [Google Scholar]
- Huebschman, J. J. (2019). Bats in southwest Wisconsin during the era of white‐nose syndrome. Northeastern Naturalist, 26, 168–182. 10.1656/045.026.0115 [DOI] [Google Scholar]
- Hunter, E. A. , Gibbs, J. P. , Cayot, L. J. , & Tapia, W. (2013). Equivalency of Galápagos giant tortoises used as ecological replacement species to restore ecosystem functions. Conservation Biology, 27, 701–709. 10.1111/cobi.12038 [DOI] [PubMed] [Google Scholar]
- Hurlbert, S. H. (1978). The measurement of niche overlap and some relatives. Ecology, 59, 67–77. 10.2307/1936632 [DOI] [Google Scholar]
- Jachowski, D. S. , Dobony, C. A. , Coleman, L. S. , Ford, W. M. , Britzke, E. R. , & Rodrigue, J. L. (2014). Disease and community structure: White‐nose syndrome alters spatial and temporal niche partitioning in sympatric bat species. Diversity and Distributions, 20, 1002–1015. [Google Scholar]
- Johnston, D. S. , & Fenton, M. B. (2001). Individual and population‐level variability in diets of pallid bats (Antrozous pallidus). Journal of Mammalogy, 82, 362–373. [Google Scholar]
- Jones, G. , & Holderied, M. W. (2007). Bat echolocation calls: Adaptation and convergent evolution. Proceedings of the Royal Society B: Biological Sciences, 274, 905–912. 10.1098/rspb.2006.0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusino, M. A. , Banik, M. T. , Palmer, J. M. , Wray, A. K. , Xiao, L. , Pelton, E. , Barber, J. R. , Kawahara, A. Y. , Gratton, C. , Peery, M. Z. , & Lindner, D. L. (2019). An improved method for utilizing high‐throughput amplicon sequencing to determine the diets of insectivorous animals. Molecular Ecology Resources, 19, 176–190. 10.1111/1755-0998.12951 [DOI] [PubMed] [Google Scholar]
- Kurta, A. , & Baker, R. H. (1990). Eptesicus fuscus . Mammalian Species, 356, 1–10. 10.2307/3504258 [DOI] [Google Scholar]
- Lack, J. B. , & Van Den Bussche, R. A. (2010). Identifying the confounding factors in resolving phylogenetic relationships in Vespertilionidae. Journal of Mammalogy, 91, 1435–1448. 10.1644/09-MAMM-A-354.1 [DOI] [Google Scholar]
- Levins, R. (1968). Evolution in changing environments: Some theoretical explorations. Princeton University Press. [Google Scholar]
- Lorch, J. M. , Meteyer, C. U. , Behr, M. J. , Boyles, J. G. , Cryan, P. M. , Hicks, A. C. , Ballmann, A. E. , Coleman, J. T. H. , Redell, D. N. , Reeder, D. A. M. , & Blehert, D. S. (2011). Experimental infection of bats with Geomyces destructans causes white‐nose syndrome. Nature, 480, 376–378. 10.1038/nature10590 [DOI] [PubMed] [Google Scholar]
- Loxdale, H. D. , Lushai, G. , & Harvey, J. A. (2011). The evolutionary improbability of ‘generalism’ in nature, with special reference to insects. Biological Journal of the Linnean Society, 103, 1–18. 10.1111/j.1095-8312.2011.01627.x [DOI] [Google Scholar]
- MacArthur, R. , & Levins, R. (1967). The limiting similarity, convergence, and divergence of coexisting species. The American Naturalist, 101, 377–385. 10.1086/282505 [DOI] [Google Scholar]
- Mayberry, H. W. , McMillan, M. R. , Chochinov, A. V. , Hinds, J. C. , & Ratcliffe, J. M. (2020). Potential foraging niche release in insectivorous bat species relatively unaffected by white‐nose syndrome? Canadian Journal of Zoology, 98(10), 667–680. [Google Scholar]
- Mittelbach, G. G. , Turner, A. M. , Hall, D. J. , Rettig, J. E. , & Osenberg, C. W. (1995). Perturbation and resilience: A long‐term, whole‐lake study of predator extinction and reintroduction. Ecology, 76, 2347–2360. 10.2307/2265812 [DOI] [Google Scholar]
- Morningstar, D. E. , Robinson, C. V. , Shokralla, S. , & Hajibabaei, M. (2019). Interspecific competition in bats and diet shifts in response to white‐nose syndrome. Ecosphere, 10, e02916. 10.1002/ecs2.2916 [DOI] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Kindt, R. , Legendre, P. , Minchin, P. R. , Ohara, R. , Simpson, G. L. , Solymos, P. , Stevens, M. H. H. , & Wagner, H. (2013). Package ‘vegan’. Community ecology package, version 2.5‐6. https://CRAN.R‐project.org/package=vegan
- O'Shea, T. J. , & Vaughan, T. A. (1977). Nocturnal and seasonal activities of the pallid bat, Antrozous pallidus . Journal of Mammalogy, 58, 269–284. 10.2307/1379326 [DOI] [Google Scholar]
- Palmer, J. M. , Jusino, M. A. , Banik, M. T. , & Lindner, D. L. (2018). Non‐biological synthetic spike‐in controls and the AMPtk software pipeline improve mycobiome data. PeerJ, 6, e4925. 10.7717/peerj.4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, K. A. , Seabrook‐Davison, M. , & Ewen, J. G. (2010). Opportunities for nonnative ecological replacements in ecosystem restoration. Restoration Ecology, 18, 269–273. 10.1111/j.1526-100X.2010.00676.x [DOI] [Google Scholar]
- Purvis, A. , Gittleman, J. L. , Cowlishaw, G. , & Mace, G. M. (2000). Predicting extinction risk in declining species. Proceedings of the Royal Society of London. Series B: Biological Sciences, 267, 1947–1952. 10.1098/rspb.2000.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R‐project.org/ [Google Scholar]
- Ram, K. , & Wickham, H. (2018). wesanderson: A Wes Anderson Palette Generator. R package version 0.3.6. https://CRAN.R‐project.org/package=wesanderson
- Ripple, W. J. , & Beschta, R. L. (2012). Trophic cascades in Yellowstone: The first 15 years after wolf reintroduction. Biological Conservation, 145, 205–213. 10.1016/j.biocon.2011.11.005 [DOI] [Google Scholar]
- Ritchie, E. G. , Elmhagen, B. , Glen, A. S. , Letnic, M. , Ludwig, G. , & McDonald, R. A. (2012). Ecosystem restoration with teeth: What role for predators? Trends in Ecology & Evolution, 27, 265–271. 10.1016/j.tree.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Rubenstein, D. R. , Rubenstein, D. I. , Sherman, P. W. , & Gavin, T. A. (2006). Pleistocene Park: Does re‐wilding North America represent sound conservation for the 21st century? Biological Conservation, 132, 232–238. 10.1016/j.biocon.2006.04.003 [DOI] [Google Scholar]
- Sánchez‐Bayo, F. , & Wyckhuys, K. A. (2019). Worldwide decline of the entomofauna: A review of its drivers. Biological Conservation, 232, 8–27. 10.1016/j.biocon.2019.01.020 [DOI] [Google Scholar]
- Santana, S. E. , & Cheung, E. (2016). Go big or go fish: Morphological specializations in carnivorous bats. Proceedings of the Royal Society B: Biological Sciences, 283(1830), 20160615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman, M. C. , & Jacobs, D. S. (2011). The relative influence of competition and prey defences on the trophic structure of animalivorous bat ensembles. Oecologia, 166, 493–506. 10.1007/s00442-010-1854-3 [DOI] [PubMed] [Google Scholar]
- Seibold, S. , Gossner, M. M. , Simons, N. K. , Blüthgen, N. , Müller, J. , Ambarlı, D. , Ammer, C. , Bauhus, J. , Fischer, M. , Habel, J. C. , Linsenmair, K. E. , Nauss, T. , Penone, C. , Prati, D. , Schall, P. , Schulze, E.‐D. , Vogt, J. , Wöllauer, S. , & Weisser, W. W. (2019). Arthropod decline in grasslands and forests is associated with landscape‐level drivers. Nature, 574, 671–674. 10.1038/s41586-019-1684-3 [DOI] [PubMed] [Google Scholar]
- Sikes, R. S. (2016). 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. Journal of Mammalogy, 97, 663–688. 10.1093/jmammal/gyw078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thogmartin, W. E. , Sanders‐Reed, C. A. , Szymanski, J. A. , McKann, P. C. , Pruitt, L. , King, R. A. , Runge, M. C. , & Russell, R. E. (2013). White‐nose syndrome is likely to extirpate the endangered Indiana bat over large parts of its range. Biological Conservation, 160, 162–172. 10.1016/j.biocon.2013.01.010 [DOI] [Google Scholar]
- Tompkins, D. , White, A. R. , & Boots, M. (2003). Ecological replacement of native red squirrels by invasive greys driven by disease. Ecology Letters, 6, 189–196. 10.1046/j.1461-0248.2003.00417.x [DOI] [Google Scholar]
- Tylianakis, J. M. , Laliberté, E. , Nielsen, A. , & Bascompte, J. (2010). Conservation of species interaction networks. Biological Conservation, 143, 2270–2279. 10.1016/j.biocon.2009.12.004 [DOI] [Google Scholar]
- Voigt, C. C. , Phelps, K. L. , Aguirre, L. F. , Schoeman, M. C. , Vanitharani, J. , & Zubaid, A. (2016). Bats and buildings: The conservation of synanthropic bats. In Voigt C. C. & Kingston T. (Eds.), Bats in the Anthropocene: Conservation of bats in a changing world (pp. 427–462). Springer. [Google Scholar]
- Whitaker, J. O. , & Lawhead, B. (1992). Foods of Myotis lucifugus in a maternity colony in central Alaska. Journal of Mammalogy, 646–648. 10.2307/1382037 [DOI] [Google Scholar]
- Wickham, H. (2007). Reshaping data with the reshape Package. Journal of Statistical Software, 21, 1–20. [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant graphics for data analysis. Springer‐Verlag. [Google Scholar]
- Wickham, H. , Averick, M. , Bryan, J. , Chang, W. , McGowan, L. , François, R. , Grolemund, G. , Hayes, A. , Henry, L. , Hester, J. , Kuhn, M. , Pedersen, T. , Miller, E. , Bache, S. , Müller, K. , Ooms, J. , Robinson, D. , Seidel, D. , Spinu, V. , … Yutani, H. (2019). Welcome to the Tidyverse. Journal of Open Source Software, 4, 1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- Wray, A. K. , & Peery, M. Z. (2022). Comparing little brown and big brown bat isotopic niches over the past century in an agriculturally‐dominated landscape. Journal of Mammalogy, in press. 10.1093/jmammal/gyac018 [DOI] [Google Scholar]
- Wray, A. K. , Peery, M. Z. , Jusino, M. A. , Kochanski, J. M. , Banik, M. T. , Palmer, J. M. , Lindner, D. L. , & Gratton, C. (2021). Predator preferences shape the diets of arthropodivorous bats more than quantitative local prey abundance. Molecular Ecology, 30, 855–873. 10.1111/mec.15769 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The final OTU table and sample metadata are permanently archived online and can be accessed at https://figshare.com/articles/dataset/ch4_OTU_table_taxonomy_with_metadata/17267150. Raw sequence data are available at SRA accession number PRJNA668526.


