Summary
Purpose
The emergence of antimicrobial resistance is a major global health challenge and becoming an urgent priority for policymakers. There is a paucity of scientific studies presenting the multidrug resistance pattern from one health perspective in Ethiopia. Therefore, a systematic review and meta-analysis aimed to determine the pooled prevalence of multidrug resistance in bacteria from human, animal, food, and environmental sources.
Methods
In this systematic review and meta-analysis, an electronic search was made in PubMed & Google scholar using different keywords. The studies conducted in all areas of Ethiopia, published from 2015 to 2020 in peer-reviewed journals, English full-length papers were included. The meta-analysis was done on STATA version 14. The pooled prevalence of multidrug resistance for each bacterium was analysed using the random-effects model; Cochran Q statistics and the I2 statistic was used to analyse heterogeneity and considered significant at p < 0.01.
Results
81 studies were included in the systematic review and meta-analysis; 53 human studies, eight animal studies, and 16 environments/food studies. The meta-analysis included six species from gram-positive bacteria and 13 from gram-negative bacteria. S. aureus 53% (95%CI: 42–64%), Coagulase negative Staphylococci 68%(95%CI:53–82), Pseudomonas spp. 73%(95%CI:48–93%), E. coli 70% (95%CI:61–78%), Citrobacter spp. 71%(95%CI:54–87%), Klebsiella spp. 68% (54–80%), Enterobacter spp. 67% (48–83%) and Salmonella spp. 65% (95%CI:48–81%) were the common multidrug-resistant species of bacteria from two or more sources.
Conclusion
In Ethiopia, the pooled prevalence of MDR is high in most bacterial species from humans, animals, food, and environmental sources. Staphylococcus, most members of the Enterobacteriaceae and Pseudomonas, are the standard MDR bacterial population involving all sources. Therefore, integrated policy and intervention measures should be implemented to reduce the emergence and spread of MDR bacteria for better animal and human health outcomes.
Keywords: Antimicrobial resistance, Multidrug resistance, bacteria, One health, Ethiopia
Abbreviations: CI, Confidence interval; CoNS, Coagulase-negative Staphylococci; ES, Estimate; MDR, Multidrug resistance
1. Introduction
The emergence of antimicrobial resistance (AMR) is a major global health challenge and becoming an urgent priority for policymakers [1]. Despite the natural factors, repeated exposure to antimicrobial agents in human and veterinary medicine is driving the emergence of antimicrobial resistance [2]. This widespread availability and misuse of antimicrobials and escalating AMR jeopardise the modern health care system [1,3]. Besides, there is a lack of new drug products due to economic shortcomings and challenges related to the regulatory requirements [4]. Because antimicrobials used to treat human infections are often used in animals, and exposed organisms have been transferred to humans through food and released into the environment through human and animal excretions, livestock and the ecosystem play a significant role in changing the resistance profile of microorganism [3]. Furthermore, the global mobility of people, food, and animals adds to the problem [5].
Infection due to resistant organisms leads to increased morbidity, hospital stay, and mortality [6]. Antimicrobial resistance organisms affect the outcome of patients due to enhanced virulence and delay in the administration of antimicrobial therapies [7]. The changes in regimens may also lead to two-fold higher adverse drug effects, increased disease severity, and unintended surgical procedures [7,8]. The prescription is syndrome-biased and rarely correlated with the specific etiologic agent [3]. Hence, this misleads physicians for inappropriate prescription of antimicrobials, ultimately ending with sub-inhibitory drug exposure and genetic rearrangement within the pathogen; the genetic rearrangement leads to altered gene expression and increased virulence spread of antimicrobial resistance [9].
Antimicrobial resistance also has a significant impact on the health care system. Infections and admissions associated with drug-resistant organisms lead to much higher health care costs; these increased costs are associated with complicated conditions that demand increased resource utilisation and therapeutic alterations [8]. The inflated health care costs may also be attributed to the cost of isolation, infection control, and wastage due to cancelled procedures [5].
In developing countries, the burden of antimicrobial resistance is much higher due to several reasons. The common causes include inadequate patient education, inappropriate medications or unregulated dispensing, limited facilities, and poor regulatory mechanisms [10]. The increased burden of infectious disease, shortfalls in infection control, and public health accelerate the rate of antimicrobial resistance [11]. Overall, poverty was the driving factor for multidrug resistance problems in developing countries [4].
Antibiotic-resistant bacteria infections are more complicated and, in some cases, impossible to treat with currently available medications. Such conditions result in increased morbidity and mortality and a significant increase in healthcare costs [12]. As a result of evolutionary pressure or horizontal gene transfer, microorganisms have become resistant to two or more kinds of antibiotics in recent years. Several studies have been done on the antimicrobial resistance of common pathogens such as Escherichia coli, Staphylococcus aureus, Klebsiella species, P. aeruginosa, Acinetobacter baumannii, and Enterobacter species [13]. Antibiotic resistance trends among widely encountered bacterial etiologies in Ethiopia have previously been characterised [14]. Antibiotic resistance in bacteria is also recognised to be a dynamic phenomenon. Because of the close association between antibiotic efficiency and antibiotic resistance, resistance patterns reported in the past may not accurately reflect the current situation. As a result, understanding the dynamic and trend of resistance requires knowledge about bacteria's current antibiotic resistance pattern. Therefore, the systematic review and meta-analysis aimed to determine the pooled prevalence of multidrug resistance in bacteria from human, animal, food, and environmental sources.
2. Methods
2.1. Search strategy
This systematic review and meta-analysis did an electronic search in PubMed &Google scholar. The search strategy on PubMed was made using different keywords and corresponding Mesh words, including “Food Microbiology” or “Environmental Microbiology”, “Microbial sensitivity” or “Drug resistance”, and “Ethiopia”. Furthermore, the searching process was filtered by year of publication to include full-length research articles published from 2015 to 2020. According to our search, we accessed a total of 546 studies.
2.2. Inclusion criteria and screening strategy
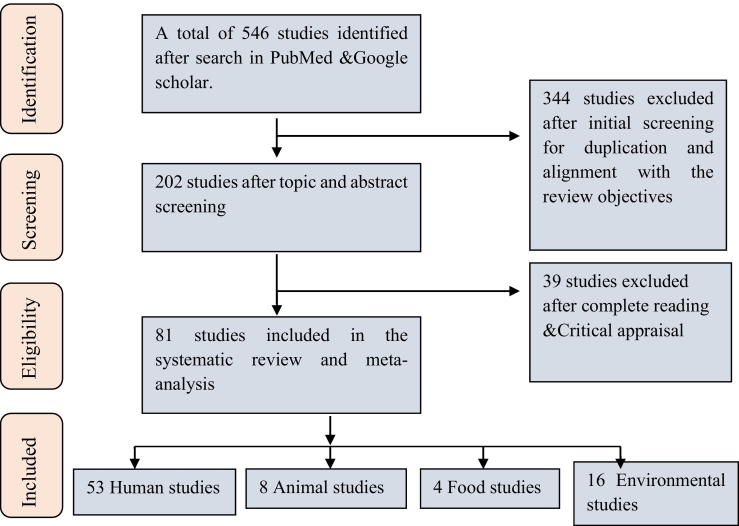
The studies conducted in all areas of Ethiopia, published from 2015-to 2020 he peer-reviewed journals, English full-length papers were included in this study. Studies that use human clinical samples and samples from animals, food, and environmental sources were included in the review. Studies isolated and tested for antimicrobial susceptibility using standard microbiological methods were included; the studies that did not indicate the prevalence of MDR isolates based on the commonly accepted MDR definition have been excluded from the systematic review and meta-analysis. Two members made the screening process independently with the fixed inclusion criteria to ensure double-checking. The Joanna Briggs Institute (JBI) checklist for critical appraisal was used for quality assessment and to enrol each article in this systematic review and meta-analysis [[15], [16], [17]]. The selection process is presented based on the preferred reporting items for systematic reviews and meta-analysis (PRISMA) guidelines [18] (Fig. 1). Mycobacterium tuberculosis has been excluded from systematic review and meta-analysis because it is extensively addressed in a recent study [19].
Fig. 1.
Prisma flow chart showing the selection process of articles on antimicrobial resistance in Ethiopia.
2.3. Data extraction process
The data extraction was made by a carefully designed excel data extraction sheet. The data extraction sheet recorded the author, year of publication, study area, study design, source (human source, animal source, food, or environment), and the type of specimen taken. Additionally, we have extracted the kind of bacterial isolates, the number of each isolate, and the number of multidrug-resistant isolates. We have also extracted the type of infection or type of anatomical colonization considered in each study for human studies.
2.4. Statistical analysis and synthesis
The data extracted was cleaned for any legibility errors, and the meta-analysis was done on STATA version 14. The pooled prevalence of MDR for each bacteria combination was analysed using the random-effects model; Cochran Q statistics and the I2 statistic was used to analyse heterogeneity and considered significant at p < 0.01 [18,20]. A forest plot was done for each bacterial and MDR combination. The results were narrated in words and plotted in terms of tables and forest plots that best suit readers. For the studies on Animals and the environment, MDR estimates were pooled if reported in at least three studies, whereas, in the case of Human sources, the point estimates were pooled if the bacterium-MDR combinations were reported in at least four studies [18]. The Begg and Mazumdar rank correlation (P < 0.05) was used to test publication bias.
3. Results
3.1. Description of the included studies
In this review, 81 studies were included in the systematic review and meta-analysis; 81 of the studies were cross-sectional studies, and one study was retrospective. The different features of the included studies are summarised below in Table 1, Table 2, Table 3. The studies were grouped into regions based on the study area; the higher percentage of the studies were from the Amhara region (25.9%), followed by the Addis Ababa area (23.5%) area (Table 1). On the other hand, few studies were included from eastern Ethiopia due to limited published articles on MDR.
Table 1.
Description of included studies based on the study area: clustered into regions.
| Description | Number of studies | Percentage | |
|---|---|---|---|
| Study area | Addis Ababa area | 19 | 23.5 |
| Eastern Ethiopia | 5 | 6.5 | |
| Southern Ethiopia | 11 | 13.6 | |
| Oromia region | 13 | 16 | |
| Amhara region | 21 | 25.9 | |
| Tigray region | 12 | 14.8 | |
| Total | 81 | 100 | |
N: indicates the number of studies.
Table 2.
Description of human studies (n = 53).
| Disease condition (n) | Type of sample taken (n) |
|---|---|
| Multiple infections (14) | Multiple clinical samples (14) |
| Nasal/nasopharyngeal Colonization (11) | Nasal/nasopharyngeal swab (12) |
| Skin colonization (1) | Skin swab (1) |
| Intestinal infection/colonization (11) | Stool (14) |
| Sepsis (4), Enteric fever (2) | Blood (4) |
| Genital colonization/infection (6) | Viginal swab &rectal swab/urethral discharge/cervical swab (6) |
| UTI (5) | Urine (5) |
| Ocular infection (5) | Eye discharge or swab (5) |
| Ear infection (2) | Ear discharge (2) |
| Nosocomial infection (2) | Multiple clinical samples (2) |
N: indicates the number of studies.
Table 3.
Description of Animal, food, and environmental studies.
| Animal studies (8) | |
|---|---|
| Type of animal (n) | Type of sample taken (n) |
| Cattle (3) | Feces (1), Milk (1), Feces &surface swab (1) |
| Chicken (1) | Visceral organ samples (1) |
| Dog (1) | Rectal swab (1) |
| Goat (1) | Different clinical samples (1) |
| Cockroaches (2) | External and internal body samples (2) |
| Food & Environment studies (n = 16) | |
|---|---|
| Type of food/Environment (n) | Type of sample taken (n) |
| Meat (3), Milk (2), Egg (1) | Meat or carcass swabs (3) |
| Raw milk (2), Egg (1) | |
| Hospital environment (6) | Surface swab (4), Air (2), wastewater (1) |
| Wastewater from urban rivers (2) | Water sample (2) |
| Abattoir/butcher shops/retailer houses (5) | A swab from meat & surfaces (4) |
| Poultry industry (1) | Surface swab (2) |
| Dairy farm (1) | Carcass swab & excretes (1) |
| Transport buses (1) | A swab from hand surfaces (1) |
| Mobile phones (1) | Surface swab (1) |
N: indicates the number of studies associated with that particular description.
3.2. Characteristics of studies that involved humans
In this data extraction, we have considered studies involving various medical conditions; multiple infections, anatomical colonization, hospital-acquired (nosocomial) infections, UTIs, and other types of diseases described in Table 2. Out of the 53 included studies on humans, 14 studies have isolated bacteria from patients with multiple infections and tested organisms isolated from various clinical samples. The nasopharyngeal area, intestine, genitalia, and skin were the commonly studied anatomical sites for the presence of drug-resistant bacteria isolates (Table 2).
3.3. Characteristics of studies on animals, food, and environment
Eight studies on different animals have reported MDR bacteria, and three out of the eight animal studies have isolated MDR bacteria from dairy cattle. Hospital environment, abattoir, and meat establishments were the most frequent categories of the environment associated with MDR isolates (Table 3).
3.4. The pooled proportion of MDR for commonly reported bacterial isolates
This review extracts the type of bacterial isolate, the number of isolates tested for antimicrobial resistance, and the number of isolates reported as MDR. A total of 6 species of gram-positive bacteria and 13 species of gram-negative bacteria were included in the meta-analysis (Table 4). Among all, different studies found that S. aureus, E. coli, and Klebsiella spp. were the top three frequently reported species. The pooled proportion of MDR was computed for each bacterium from all categories (animal studies, human studies, environment, and food) as summarised below (Table 5, Table 6). Organisms were included in the meta-analysis if reported by at least four human studies and at least three animal studies, food &environmental studies.
Table 4.
The type of bacterial isolates considered for the meta-analysis.
| Type of bacteria | Number of studies from each source |
Reference | |||
|---|---|---|---|---|---|
| Humans | Animals | Environment/Food | Total | ||
| S. aureus | 25 | 3 | 9 | 37 | [[21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56]] |
| CoNS | 12 | 1 | 7 | 8 | [[21], [22], [23],[25], [26], [27], [28],31,[33], [34], [35],38,[42], [43], [44],[46], [47], [48],52,53] |
| S. agalactae | 5 | – | – | 5 | [40,53,[57], [58], [59]] |
| Streptococcus pyogenes | 8 | – | – | 8 | [33,38,40,42,43,52,53,56] |
| Streptococcus pneumoniae | 8 | – | – | 8 | [20,35,40,42,53,56,60,61] |
| Enterococcus spp. | 6 | – | – | 6 | [35,38,41,[62], [63], [64]] |
| E. coli | 19 | 4 | 14 | 37 | [21,22,[24], [25], [26], [27], [28], [29], [30], [31],[33], [34], [35],38,40,[42], [43], [44], [45], [46],48,52,53,[65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78]] |
| Klebsiella spp. | 18 | 1 | 7 | 26 | [21,[26], [27], [28],31,[33], [34], [35],38,40,[42], [43], [44], [45], [46], [47], [48],52,53,[67], [68], [69],74,[76], [77], [78]] |
| Enterobacter spp. | 15 | 1 | 3 | 19 | [24,31,[33], [34], [35],38,40,42,43,[45], [46], [47],53,67,69,74,[76], [77], [78]] |
| Citrobacter spp. | 14 | 1 | 5 | 20 | [25,26,31,[33], [34], [35],38,40,42,43,45,47,53,[67], [68], [69],74,[76], [77], [78]] |
| Salmonella spp. | 12 | 5 | 6 | 23 | [21,30,31,35,38,40,53,69,75,77,[79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91]] |
| Shigella spp. | 11 | 2 | – | 13 | [30,31,35,40,53,75,[87], [88], [89],[91], [92], [93]] |
| Proteus spp. | 11 | 1 | 2 | 14 | [21,28,31,[33], [34], [35],38,40,42,44,47,53,74,76] |
| Serratia spp. | 7 | 1 | 1 | 9 | [28,31,33,38,43,45,52,53,77] |
| Acinetobacter spp. | 7 | – | 1 | 8 | [22,35,36,38,44,45,48,77] |
| Pseudomonas spp. | 11 | – | 5 | 16 | [21,22,[26], [27], [28],35,36,38,40,42,45,47,52,53,76,77] |
| N. gonorrhea | 4 | – | – | 4 | [35,38,40,94] |
| Neisseria meningitidis | 4 | – | – | 4 | [35,40,53,95] |
| Haemophilus influenzae | 4 | – | – | 4 | [38,42,52,53] |
Table 5.
A summary of the pooled proportion of MDR for each gram-positive bacterium included in the meta-analysis, 2015–2020.
| Type of bacteria | Estimate (95%CI) for bacterial isolates from each source |
Overall pooled proportion | Heterogeneity between groups | ||
|---|---|---|---|---|---|
| Humans | Animals | Environment/Food | |||
| S. aureus | 0.57 (0.44, 0.70) | 0.32(0.00, 0.97) | 0.49 (0.31, 0.68) | 0.53 (0.42, 0.64), I2 = 96.10% p = 0.000 | P = 0.677 |
| CoNS | 0.78(0.62–0.91) | – | 0.51(0.28–0.74) | 0.68 (0.53–0.82), I2 = 93.35% p = 0.00 | P = 0.062 |
| Streptococcus pyogenes | 0.42(0.07–0.80) | – | – | I2 = 83.07% p = 0.00 | NA |
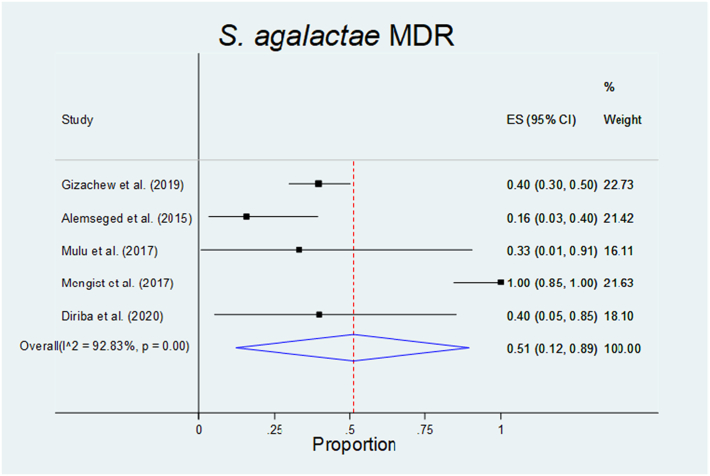
| S. agalactae | 0.51(0.15–0.89) | – | – | I2 = 92.83% p = 0.00 | NA |
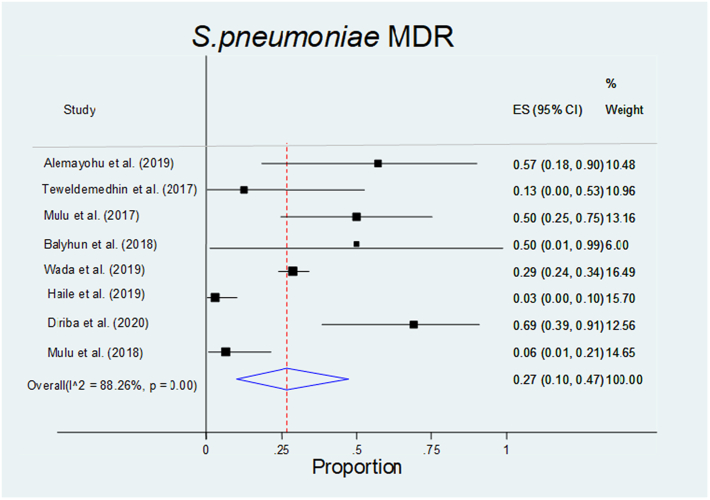
| S. pneumonia | 0.27(0.10–0.47) | – | – | I2 = 88.26% p = 0.01 | NA |
| Enterococcus spp. | 0.65(0.41–0.85) | – | – | I2 = 88.69% p = 0.000 | NA |
Table 6.
A summary of the pooled proportion of MDR for each gram-negative bacteria included in the meta-analysis, 2015–2020.
| Type of bacteria | Estimate (95%CI) for bacterial isolates from each source |
Overall pooled proportion: ES (95%CI), I2 = % p = value | Heterogeneity between groups | ||
|---|---|---|---|---|---|
| Humans | Animals | Environment/Food | |||
| E. coli | 0.81(0.70–090) | 0.72(0.57–0.86) | 0.53(0.35–0.70) | 0.70(0.61–0.78) I2 = 89.4% p = 0.000 |
No, p = 0.035 |
| Klebsiella spp. | 0.81(0.70–0.90) | – | 0.37(0.20–0.57) | 0.68(0.54–0.80) I2 = 89.92% p = 0.00 |
Yes, P = 0.0000 |
| Enterobacter spp. | 0.64(0.43–0.84) | – | 0.73(0.43–0.98) | 0.67(0.48–0.83) I2 = 69.95% p = 0.00 |
No, P = 0.622 |
| Citrobacter spp. | 0.73(0.54–0.90) | – | 0.69(0.32–0.97) | 0.71(0.54–0.87) | No, P = 0.849 |
| Salmonella spp. | 0.70(0.50–0.87) | 0.64(0.43–0.83) | 0.56(0.13–0.94) | 0.65(0.48–0.81) I2 = 91.88% p = 0.000 |
No,0.068 |
| Shigella spp. | 0.69(0.54–0.82) | – | – | I2 = 58.13% p = 0.01 | NA |
| Proteus spp. | 0.54(0.34–0.75) | – | – | I2 = 57.50% p = 0.01 | NA |
| Serratia spp. | 0.42(0.12–0.74) | – | – | I2 = 25.00% p = 0.24 | NA |
| Acinetobacter spp. | 0.86(0.46–1.00) | – | – | I2 = 77.69% p = 0.00 | NA |
| Pseudomonas spp. | 0.82(0.52–1.00) | – | 0.53(0.13–0.92) | 0.73 (0.48–0.93) I2 = 89.18% p = 0.00 |
No, P = 0.312 |
| N. gonorrhoae | 0.74(0.51–0.92) | – | – | I2 = 20.80% p = 0.29 | NA |
| N. Meningitidis | 0.44(0.00–0.95) | – | – | I2 = 81.29% p = 0.00 | NA |
| Haemophilus influenzae | 0.10(0.00–0.39) | – | – | I2 = 0.00% p = 0.99 | NA |
NA: not applicable.
3.5. Multidrug resistance in gram-positive bacteria
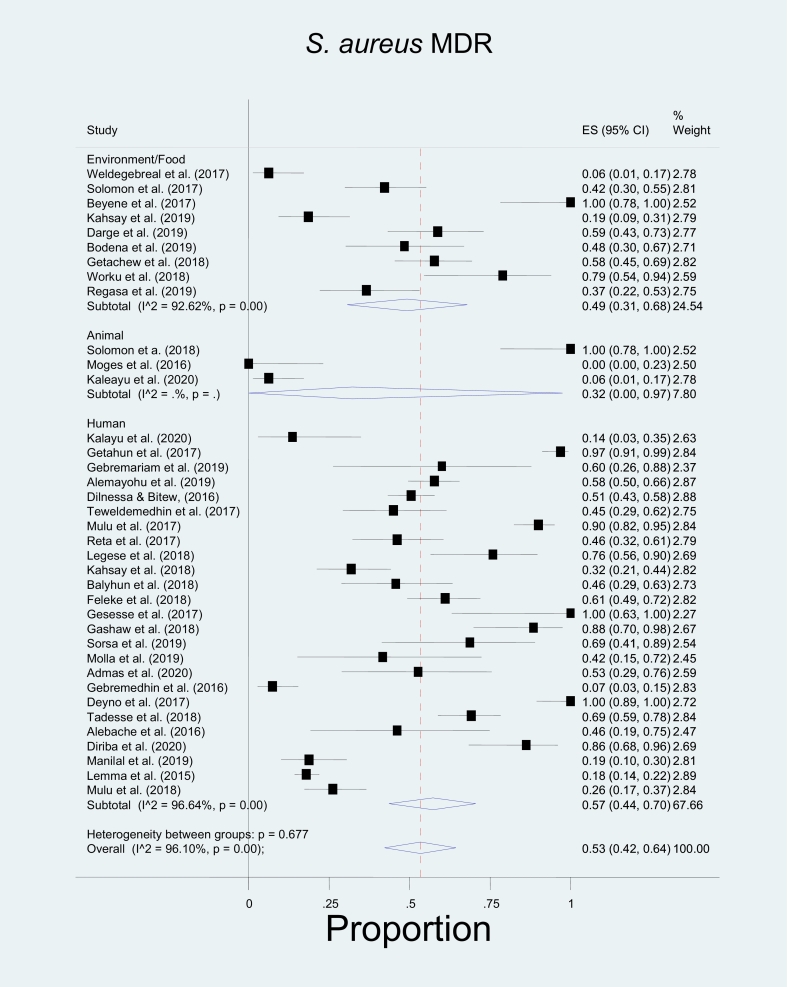
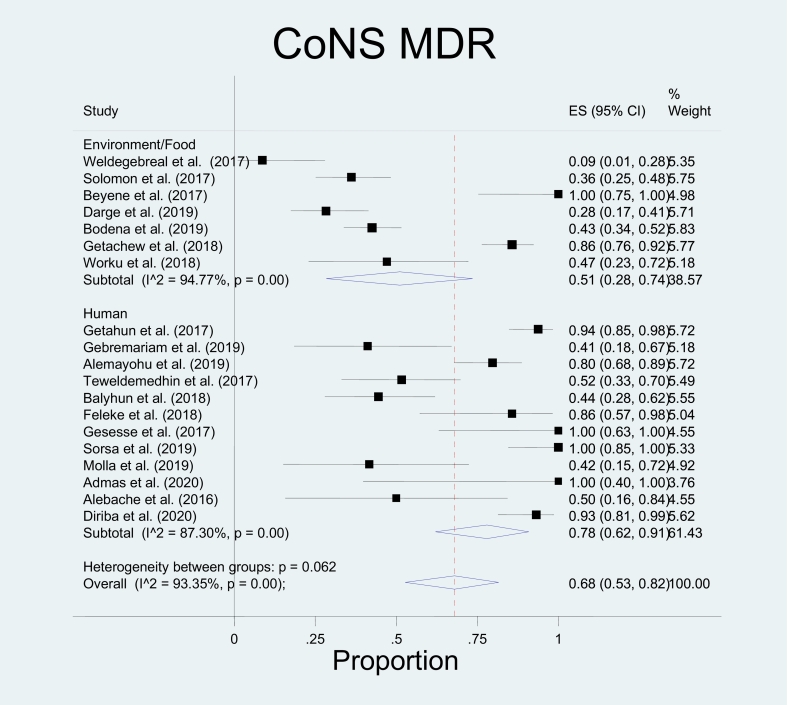
In this study, bacterial isolates that belong to three genera (genus Staphylococcus, genus Streptococcus, and Enterococci) have been included in the meta-analysis because reports from other genera did not fulfil the criteria for meta-analysis. Among the included species, the pooled proportion of MDR for S. aureus was found to be 0.57(0.44, 0.70) for isolates from humans, 0.32(0.00, 0.97) in animal studies, and 0.49(0.31, 0.68) for isolates from the environment or food (Table 5). This study has yielded an overall estimated MDR of 53%. The meta-analysis indicated no significant heterogeneity between the three sources; p = 0.677 (Fig. 2). In the case of Coagulase-negative Staphylococci (CoNS), the pooled proportion of MDR was 0.78(0.62–0.91) in human studies, 0.51(0.28–0.74) in environment or food with an overall estimated MDR of 68%. There was no significant heterogeneity between the two sources (Fig. 3). However, group heterogeneity has been explained by study area and year in both species. Concerning CoNS, a single animal study reported the proportion of MDR CoNS as 0.29 [31].
Fig. 2.
The Forest plot shows the pooled prevalence of multidrug-resistant (MDR) S. aureus in humans was 0.57(0.44, 0.70), 0.32(0.00, 0.97) in animal studies, and 0.49(0.31, 0.68) for isolates from the environment or food. There was no significant heterogeneity between the three sources; p = 0.677.
Fig. 3.
Forest plot of the pooled proportion of Coagulase-negative Staphylococci (CoNS) Multidrug-resistant (MDR) was 0.78(0.62–0.91) in human studies, 0.51(0.28–0.74) in environment or food with an overall estimated MDR of 68% and has no significant heterogeneity between the two sources.
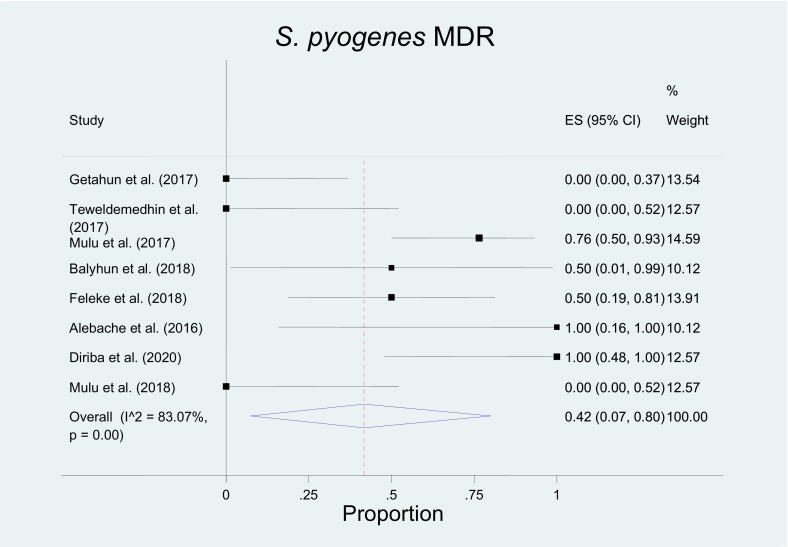
All reported isolates of S. pyogens, S. agalactae, Streptococcus pneumoniae, and Enterococcus species were from human sources. Hence, the pooled proportion of MDR S. pyogens and GBS in humans were 0.42(0.07–0.80), I2 = 83.07% p = 0.00 and 0.51(0.15–0.89), I2 = 92.83% p = 0.00 respectively (Fig. 3, Fig. 4, Fig. 5, Fig. 6; Table 5).
Fig. 4.
The pooled prevalence of Multidrug-resistant (MDR) S. pyogens in humans was 0.42(0.07–0.80), I2 = 83.07%, and p = 0.
Fig. 5.
The pooled prevalence of multidrug- resistant (MDR) Streptococcus pneumoniae in humans was 0.27 (0.10–0.47), I2 = 88.26%, and p = 0.
Fig. 6.
The pooled prevalence of multidrug-resistant (MDR) S. agalactae in humans was 0.51(0.12–0.89), I2 = 92.83%, and p = 0.
3.6. Multidrug resistance in gram-negative bacteria
As summarised in Table 6, pooled MDR was computed for 13 species of gram-negative bacteria. Accordingly, the highest pooled MDR in human sources was for Acinetobacter species 86% (95% CI: 46–100%) followed by Pseudomonas species 82% (95% CI: 52–100%), E. coli 81% (95% CI: 70–90%) and Klebsiella species 70% (95% CI: 50–87%). In the case of animal sources, E. coli and Salmonella species were frequently reported with a pooled MDR proportion of 0.72 (0.57–0.86) and 0.64(0.43–0.83), respectively.
Among the isolates from environment or food sources, the highest pool of MDR was found in Enterobacter species (0.73(0.43–0.98)), Citrobacter species (0.69 (0.32–0.97)), and Salmonella species (0.56(0.13–0.94) (Table 6). Overall, for the species that involved all sources, there was no significant heterogeneity between the subgroups (Haman source, Animal source, Environment, or food sources) except for Klebsiella species (Table 6). The pooled proportion of MDR and group differences can be compared from the forest plots for each bacterium.
4. Discussion
Multidrug-resistant organisms pose a significant burden worldwide [96]. Mitigation of the problem needs to determine its magnitude from the perspective of human infections and the animals, food, and the environment from a health perspective. Hence, this systematic review and meta-analysis determined the prevalence of MDR for commonly reported bacteria from humans, animals, environment, and food studies in Ethiopia.
From the perspective of human studies, CoNS (78%), Enterococcus spp. (65%) and S. aureus (57%) had the highest pooled percentage of MDR among the gram-positive bacteria. In gram-negative bacteria, Acinetobacter spp. (86%), Pseudomonas spp., E. coli and Klebsiella spp. (81%) were the top MDR bacteria. Various mechanisms contribute to the emergence of drug resistance in human pathogens. One mechanism is the carriage state in various anatomical sites, as Staphylococcus, Enterococci, and E. coli results in frequent suboptimal exposure to antimicrobials used for the treatment and prophylactic measures. The other scenario is prolonged hospitalisation [97], prolonged use of prophylaxis, and predispositions to hospital-acquired infections, a common scenario in S. aureus, E. coli, Klebsiella, and Pseudomonas spp. [98]. These MDR organisms could pose a triple burden because humans take the highest share of releasing MDR organisms to the environment and animals through direct contact, faecal pollution, mucosal secretion, and recreational activities. Compared to other studies, the pooled prevalence of MDR for Klebsiella spp. and E. coli is higher than the findings in Cameroon through the consistent finding was found for Proteus spp. and Staphylococcus spp. [18].
Out of the eight species of bacteria extracted from environmental or food studies, the pooled prevalence of MDR was above 50% for seven of the species, specifically, Enterobacter spp. (73%), Citrobacter spp. (69%), Salmonella spp. (56%), E. coli (53%) and Pseudomonas spp. (53%) were the top MDR bacteria. Almost half of the staphylococcal isolates from environment/food studies were also MDR. As stated in the results section, most of the included studies from the environment domain were from the hospital environment and wastewater. Hence, the selective pressure of nosocomial pathogens such as Pseudomonas, E. coli, Enterobacter, Citrobacter species, and S. aureus might have contributed to the high degree of MDR because the stress response, adaptation, and gene transfer events in the hospital environment favour the emergence of multidrug resistance foci within the bacterial genome [99]. Furthermore, the mixed release of organisms and antimicrobials in the health care wastes and urban rivers may contribute to the development and acquisition of drug resistance [100], which may sound specifically for common water contaminants such as E. coli and Enterococci [101]. Hence, the released drug-resistant bacteria pollute drinking water sources, which may affect human health [102].
On the other hand, many studies were from the abattoir, butcher shops, meat chains, and milk chains. In this case, firstly, there might be contamination with skin and mucosal floras such as MDR Staphylococcus from the food handlers; secondly, there might also be surface contamination with faecal matter, animal excreta, and water sources which may also be true for E. coli, Enterococci, and Salmonella spp. It may allow the transmission of drug resistance determining genes between bacterial populations from raw meat, carcasses, surfaces/soil, water, and humans [100]. Hence, the habitual consumption of raw meat and raw milk may be a potential risk of these MDR bacteria.
In the case of animal studies, E. coli (72%), Salmonella spp. (64%) and S. aureus (32%) were the common MDR bacteria. Animals have direct and indirect contributions to the spread of MDR bacteria to the environment and humans [103]. This may be explained by direct contact with the environment (soil, water), faecal contamination, and manure associated spread of antibiotic resistance genes in agricultural fields [104]. The frequent contact between humans, dairy cattle, and poultry may also be a good opportunity for the bidirectional transmission of MDR bacteria such as S. aureus and E. coli. Lastly, small animals such as cockroaches may contribute to the spread of MDR bacteria between humans, animals, and the environment [72]. Overall, the spread of multidrug-resistant bacteria is a cyclic process between the environment, animals, and the environment. It is also evidenced that MDR bacteria are associated with excess morbidity, mortality, and cost [96]. Therefore, understanding the nature of the problem from one health perspective has paramount importance.
Various environmental routes are essential avenues for human exposure to resistant bacteria and their genes coming from animal and plant reservoirs and opportunities for improved antimicrobial resistance regulation [105]. The transmission of bacteria and resistance genes from agricultural sources is largely foodborne. In the developed countries with good sewage and drinking water treatment, most people have little to no direct contact with food-producing animals. Bacteria and resistance genes are transmitted directly from contaminated meat and poultry during slaughter and processing or indirectly from contaminated fruit and vegetables from manure or irrigation water [106]. In countries with inadequate sewage and water treatment, drinking water is expected to significantly transmit resistant germs and genes from animals [107,108].
Furthermore, poor sanitation allows for indirect person–person waterborne transmission of enteric germs among residents and international tourists, who return home colonised with resistant bacteria [109]. Globalised trade in animals and food, and long-distance migratory patterns of wildlife, are used to spread antimicrobial-resistant bacteria worldwide. Among the broad actions to prevent antibiotic resistance in the wider environment are improved pollution controls from industrial, residential, and agricultural sources. More research and environmental monitoring and risk assessment are needed to understand better the role of the environment in the selection and spread of antimicrobial resistance and create more specialised approaches to combat resistance in this area [110]. The WHO Global Plan is supported by five pillars: 1. Reduce the Incidence of Infection via Effective Sanitation, Hygiene, and Infection Prevention Measures; 2. Strengthen the Knowledge and Evidence Base through Surveillance and Research; 3. Optimise Antimicrobial Medicine Use in Human and Animal Health; 4. Develop an Economic Case for Long-Term Investment.
The One Health approach of the WHO Global Action Plan is appropriate and consistent with remarks made in other international and national action plans [111]. However, until a fully integrated One health plan to combat antibiotic resistance is adopted at the national and global levels, there is still a long way to go. Multiple sectors (including animals, humans, and the environment) and organisations have competing interests, and there are gaps in antimicrobial resistance surveillance, antimicrobial use regulation, and infection control in many regions of the world. History has proven that it is impossible to clearly divide antimicrobial classes into those used solely in the human or non-human sectors, except for novel antimicrobial types. As long as there are few or no options, they should probably be reserved for use in humans. On the other hand, most classes will be offered in both sectors, and One health's goal would be to ensure their utilisation is efficient collectively. It is more likely to happen if antimicrobials used in both industries are only used for therapy, never for prophylaxis, not for improving plant growth, and if we effectively manage the types and amounts of antimicrobials we allow into the environment, as well as the frequency of resistant bacteria.
There are several obstacles to enhancing antibiotic management in One health, including a lack of knowledge, limited awareness, antibiotic misuse, and insufficient regulatory approaches in various countries. Excessive antibiotic usage in food-producing animals has been shown to have hostile public and animal health consequences. However, the poor world has been sluggish to integrate scientific development and accept the findings. Advanced molecular approaches, including whole genome sequencing, metagenomics, metadata analysis, and phylogenetic research, are required for a deeper understanding of global antibiotic resistance. These cutting-edge tools will help us better understand how resistant superbugs and antibiotic resistance spread among humans, animals, insects, plants, water, and soil. One health is a topic that improves health via multidisciplinary collaboration by gathering data from multiple areas of One health to understand how antibiotic resistance spreads at the human-animal-environment interface. Microbiome study of the various regions of One health will be broad and challenging in the future. Plans for antibiotic stewardship must be feasible to reduce antibiotic usage.
5. Conclusion
In Ethiopia, the pooled prevalence of MDR is high in most bacterial species from humans, animals, food, and environmental sources. Staphylococcus species, most members of the Enterobacteriaceae, and Pseudomonas species are the standard MDR bacterial population involving all sources. To reduce the human, animal, and environmental health hazards, the global emergence and spread of antibiotic resistance need the implementation of a harmonised and multidisciplinary One Health approach. The proliferation of antibiotic resistance infections worldwide due to antibiotic abuse, poor sanitation, and ineffective control mechanisms has a significant influence on global public health. Therefore, this demands integrated policy and intervention measures should be implemented to reduce the emergence and spread of MDR bacteria for better animal and human health outcomes.
Funding
This research did not receive any specific funding.
Ethical approval
Not applicable.
CRediT authorship contribution statement
Mebrahtu Tweldemedhin: Writing – original draft, Conceptualization. Saravanan Muthupandian: Supervision. Tsega Kahsay Gebremeskel: Data curation, Investigation. Kibrti Mehari: Formal analysis. Getahun Kahsay Abay: Methodology, Validation. Teklay Gebrecherkos Teklu: Resources. Ranjithkumar Dhandapani: Visualization, Validation. Ragul Paramasivam: Software, Formal analysis. Tsehaye Asmelash: Validation.
Declaration of Competing Interest
The author reports no conflicts of interest in this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2022.100390.
Appendix A. Supplementary data
Supplementary material
References
- 1.Amann R.I., Ludwig W., Schleifer K.-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faine S., Organization W.H. World Health Organization; 1982. Guidelines for the Control of Leptospirosis. [Google Scholar]
- 3.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3(2):1–8. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pond S.K., Wadhawan S., Chiaromonte F., Ananda G., Chung W.-Y., Taylor J., Nekrutenko A., Team G. Windshield splatter analysis with the galaxy metagenomic pipeline. Genome Res. 2009;19(11):2144–2153. doi: 10.1101/gr.094508.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stach E., Bull A.T. Estimating and comparing the diversity of marine actinobacteria. Antonie Van Leeuwenhoek. 2005;87(1):3–9. doi: 10.1007/s10482-004-6524-1. [DOI] [PubMed] [Google Scholar]
- 6.Cowan D.A. Microbial genomes–the untapped resource. Trends Biotechnol. 2000;18(1):14–16. doi: 10.1016/s0167-7799(99)01395-5. [DOI] [PubMed] [Google Scholar]
- 7.Daniel R. The metagenomics of soil. Nat. Rev. Microbiol. 2005;3(6):470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- 8.Prasanna R., Nain L., Pandey A.K., Saxena A.K. Microbial diversity and multidimensional interactions in the rice ecosystem. Arch. Agron. Soil Sci. 2012;58(7):723–744. [Google Scholar]
- 9.Rondon M.R., August P.R., Bettermann A.D., Brady S.F., Grossman T.H., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000;66(6):2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunin V., Copeland A., Lapidus A., Mavromatis K., Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008;72(4):557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kollef K.E., Schramm G.E., Wills A.R., Reichley R.M., Micek S.T., Kollef M.H. Predictors of 30-day mortality and hospital costs in patients with ventilator-associated pneumonia attributed to potentially antibiotic-resistant gram-negative bacteria. Chest. 2008;134(2):281–287. doi: 10.1378/chest.08-1116. [DOI] [PubMed] [Google Scholar]
- 13.Finley R.L., Collignon P., Larsson D.J., McEwen S.A., Li X.-Z., Gaze W.H., Reid-Smith R., Timinouni M., Graham D.W., Topp E. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 2013;57(5):704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 14.Abera B., Kibret M., Mulu W. Extended-Spectrum beta (β)-lactamases and Antibiogram in Enterobacteriaceae from clinical and drinking water Sources from Bahir Dar City, Ethiopia. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0166519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M., Eisen J.A. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9(10):1–11. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause L., Diaz N.N., Goesmann A., Kelley S., Nattkemper T.W., Rohwer F., Edwards R.A., Stoye J. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36(7):2230–2239. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teeling H., Meyerdierks A., Bauer M., Amann R., Glöckner F.O. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 2004;6(9):938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- 18.Von Mering C., Hugenholtz P., Raes J., Tringe S., Doerks T., Jensen L., Ward N., Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315(5815):1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- 19.McHardy A.C., Martín H.G., Tsirigos A., Hugenholtz P., Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods. 2007;4(1):63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 20.Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9(1):1–8. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goecks J., Nekrutenko A., Taylor J., Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;25:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlach W., Jünemann S., Tille F., Goesmann A., Stoye J. WebCARMA: a web application for the functional and taxonomic classification of unassembled metagenomic reads. BMC Bioinformatics. 2009;10(1):1–10. doi: 10.1186/1471-2105-10-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.-Y., Cohoon M., de Crécy-Lagard V., Diaz N., Disz T., Edwards R. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seshadri R., Kravitz S.A., Smarr L., Gilna P., Frazier M. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5(3) doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fieller E.C., Hartley H.O., Pearson E.S. Tests for rank correlation coefficients. I. Biometrika. 1957;44(3/4):470–481. [Google Scholar]
- 26.Rosen G.L., Essinger S.D. Comparison of statistical methods to classify environmental genomic fragments. IEEE Trans. Nanobiosci. 2010;9(4):310–316. doi: 10.1109/TNB.2010.2081375. [DOI] [PubMed] [Google Scholar]
- 27.Stackebrandt E., Murray R., Trüper H. Proteobacteria classis nov., a name for the phylogenetic taxon that includes the “purple bacteria and their relatives”. Int. J. Syst. Evol. Microbiol. 1988;38(3):321–325. [Google Scholar]
- 28.Vijayakumar R., Muthukumar C., Thajuddin N., Panneerselvam A., Saravanamuthu R. Studies on the diversity of actinomycetes in the Palk Strait region of Bay of Bengal, India. Actinomycetologica. 2007;21(2):59–65. [Google Scholar]
- 29.Le Ouay B., Stellacci F. Antibacterial activity of silver nanoparticles: a surface science insight. Nano Today. 2015;10(3):339–354. [Google Scholar]
- 30.Solomon F., Kibru G., Ali S. Multidrug-resistant pattern of food borne illness associated bacteria isolated from cockroaches in meal serving facilities, Jimma, Ethiopia. Afr. Health Sci. 2018;18(1):32–40. doi: 10.4314/ahs.v18i1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnusson M.H., Deppert K., Malm J.-O., Bovin J.-O., Samuelson L. Gold nanoparticles: production, reshaping, and thermal charging. J. Nanopart. Res. 1999;1(2):243–251. [Google Scholar]
- 32.Abdelghany T., Al-Rajhi A.M., Al Abboud M.A., Alawlaqi M., Magdah A.G., Helmy E.A., Mabrouk A.S. Recent advances in green synthesis of silver nanoparticles and their applications: about future directions. A review. BioNanoScience. 2018;8(1):5–16. [Google Scholar]
- 33.Venkateswaran P., Millman I., Blumberg B.S. Effects of an extract from Phyllanthus niruri on hepatitis B and woodchuck hepatitis viruses: in vitro and in vivo studies. Proc. Natl. Acad. Sci. 1987;84(1):274–278. doi: 10.1073/pnas.84.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W., Qiao X., Chen J. Synthesis of silver nanoparticles—effects of concerned parameters in water/oil microemulsion. Mater. Sci. Eng. B. 2007;142(1):1–15. [Google Scholar]
- 35.Kalikar M., Thawani V., Varadpande U., Sontakke S., Singh R., Khiyani R. Immunomodulatory effect of Tinospora cordifolia extract in human immuno-deficiency virus positive patients. Indian J. Pharm. 2008;40(3):107. doi: 10.4103/0253-7613.42302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma H., Patil P., Kolhapure R., Gopalkrishna V. Antiviral activity of the Indian medicinal plant extract, Swertia chirata against herpes simplex viruses: A study by in-vitro and molecular approach. Indian J. Med. Microbiol. 2008;26(4):322. [PubMed] [Google Scholar]
- 37.Lee S.H., Tang Y.Q., Rathkrishnan A., Wang S.M., Ong K.C., Manikam R., Payne B.J., Jaganath I.B., Sekaran S.D. Effects of cocktail of four local Malaysian medicinal plants (Phyllanthus spp.) against dengue virus 2. BMC Complement. Altern. Med. 2013;13(1):1–13. doi: 10.1186/1472-6882-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Husen A., Siddiqi K.S. Phytosynthesis of nanoparticles: concept, controversy and application. Nanoscale Res. Lett. 2014;9(1):1–24. doi: 10.1186/1556-276X-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manikandan V., Yi P.-I., Velmurugan P., Jayanthi P., Hong S.-C., Jang S.-H., Suh J.-M., Sivakumar S. Production, optimisation and characterisation of silver oxide nanoparticles using Artocarpus heterophyllus rind extract and their antifungal activity. Afr. J. Biotechnol. 2017;16(36):1819–1825. [Google Scholar]
- 40.Sundeep D., Kumar T.V., Rao P.S., Ravikumar R., Krishna A.G. Green synthesis and characterisation of Ag nanoparticles from Mangifera indica leaves for dental restoration and antibacterial applications. Prog. Biomater. 2017;6(1):57–66. doi: 10.1007/s40204-017-0067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rai M., Yadav A., Gade A. CRC 675—current trends in phytosynthesis of metal nanoparticles. Crit. Rev. Biotechnol. 2008;28(4):277–284. doi: 10.1080/07388550802368903. [DOI] [PubMed] [Google Scholar]
- 42.Karuppiah M., Rajmohan R. Green synthesis of silver nanoparticles using Ixora coccinea leaves extract. Mater. Lett. 2013;97:141–143. [Google Scholar]
- 43.Bar H., Bhui D.K., Sahoo G.P., Sarkar P., De S.P., Misra A. Green synthesis of silver nanoparticles using latex of Jatropha curcas. Colloids Surf. A. 2009;339(1–3):134–139. [Google Scholar]
- 44.Prakash P., Gnanaprakasam P., Emmanuel R., Arokiyaraj S., Saravanan M. Green synthesis of silver nanoparticles from leaf extract of Mimusops elengi, Linn. for enhanced antibacterial activity against multi drug resistant clinical isolates. Colloids Surf. B: Biointerfaces. 2013;108:255–259. doi: 10.1016/j.colsurfb.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Bagherzade G., Tavakoli M.M., Namaei M.H. Green synthesis of silver nanoparticles using aqueous extract of saffron (Crocus sativus L.) wastages and its antibacterial activity against six bacteria. Asian Pac. J. Trop. Biomed. 2017;7(3):227–233. [Google Scholar]
- 46.Edison T.J.I., Sethuraman M. Instant green synthesis of silver nanoparticles using Terminalia chebula fruit extract and evaluation of their catalytic activity on reduction of methylene blue. Process Biochem. 2012;47(9):1351–1357. [Google Scholar]
- 47.Das J., Das M.P., Velusamy P. Sesbania grandiflora leaf extract mediated green synthesis of antibacterial silver nanoparticles against selected human pathogens. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013;104:265–270. doi: 10.1016/j.saa.2012.11.075. [DOI] [PubMed] [Google Scholar]
- 48.Klasen H. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns. 2000;26(2):131–138. doi: 10.1016/s0305-4179(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 49.Morones J., Elechiguerra J., Camacho A., Holt K., Kouri J.B., Ramirez J.T., Yacaman M.J. Nanotechnology. 2005;16:2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 50.Kvitek D.J., Will J.L., Gasch A.P. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 2008;4(10) doi: 10.1371/journal.pgen.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monteiro D.R., Gorup L.F., Takamiya A.S., Ruvollo-Filho A.C., de Camargo E.R., Barbosa D.B. The growing importance of materials that prevent microbial adhesion: antimicrobial effect of medical devices containing silver. Int. J. Antimicrob. Agents. 2009;34(2):103–110. doi: 10.1016/j.ijantimicag.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 52.Kumar P., Selvi S.S., Praba A., Selvaraj M., Rani L.M., Suganthi P., Devi B.S., Govindaraju M. Antibacterial activity and in-vitro cytotoxicity assay against brine shrimp using silver nanoparticles synthesised from Sargassum ilicifolium. Digest J. Nanomater. Biostruct. 2012;7(4):1447–1455. [Google Scholar]
- 53.Lundberg D.S., Lebeis S.L., Paredes S.H., Yourstone S., Gehring J., Malfatti S., Tremblay J., Engelbrektson A., Kunin V., Del Rio T.G. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peiffer J.A., Spor A., Koren O., Jin Z., Tringe S.G., Dangl J.L., Buckler E.S., Ley R.E. Diversity and heritability of the maise rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. 2013;110(16):6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fact sheet . Global HIV & AIDS Statistics – 2020 Fact Sheet; 2020. Latest Global and Regional Statistics on the Status of the AIDS Epidemic. [Google Scholar]
- 56.Malve H. Exploring the ocean for new drug developments: marine pharmacology. J. Pharm. Bioallied Sci. 2016;8(2):83. doi: 10.4103/0975-7406.171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thakkar K.N., Mhatre S.S., Parikh R.Y. Biological synthesis of metallic nanoparticles. Nanomedicine. 2010;6(2):257–262. doi: 10.1016/j.nano.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 58.Kalishwaralal K., Deepak V., Pandian S.R.K., Kottaisamy M., BarathManiKanth S., Kartikeyan B., Gurunathan S. Biosynthesis of silver and gold nanoparticles using Brevibacterium casei. Colloids Surf. B: Biointerfaces. 2010;77(2):257–262. doi: 10.1016/j.colsurfb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 59.Kumar P.V., Pammi S., Kollu P., Satyanarayana K., Shameem U. Green synthesis and characterisation of silver nanoparticles using Boerhaavia diffusa plant extract and their anti bacterial activity. Ind. Crop. Prod. 2014;52:562–566. [Google Scholar]
- 60.Mallikarjuna K., Narasimha G., Dillip G., Praveen B., Shreedhar B., Lakshmi C.S., Reddy B., Raju B.D.P. Green synthesis of silver nanoparticles using Ocimum leaf extract and their characterisation. Digest J. Nanomater. Biostruct. 2011;6(1):181–186. [Google Scholar]
- 61.Amaladhas T.P., Sivagami S., Devi T.A., Ananthi N., Velammal S.P. Biogenic synthesis of silver nanoparticles by leaf extract of Cassia angustifolia. Adv. Nat. Sci. Nanosci. Nanotechnol. 2012;3(4) [Google Scholar]
- 62.Vaidyanathan R., Kalishwaralal K., Gopalram S., Gurunathan S. RETRACTED: nanosilver—the burgeoning therapeutic molecule and its green synthesis. Biotechnol. Adv. 2009;27(6):924–937. doi: 10.1016/j.biotechadv.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 63.Ramesh P., Kokila T., Geetha D. Plant mediated green synthesis and antibacterial activity of silver nanoparticles using Emblica officinalis fruit extract. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015;142:339–343. doi: 10.1016/j.saa.2015.01.062. [DOI] [PubMed] [Google Scholar]
- 64.Meyer F., Bagchi S., Chaterji S., Gerlach W., Grama A., Harrison T., Paczian T., Trimble W.L., Wilke A. MG-RAST version 4—lessons learned from a decade of low-budget ultra-high-throughput metagenome analysis. Brief. Bioinform. 2017;20(4):1151–1159. doi: 10.1093/bib/bbx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson A.S., Wellington E. The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 2001;51(3):797–814. doi: 10.1099/00207713-51-3-797. [DOI] [PubMed] [Google Scholar]
- 66.Chin K.-J., Janssen P.H. Propionate formation by Opitutus terrae in pure culture and in mixed culture with a hydrogenotrophic methanogen and implications for carbon fluxes in anoxic rice paddy soil. Appl. Environ. Microbiol. 2002;68(4):2089–2092. doi: 10.1128/AEM.68.4.2089-2092.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Janssen P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noll M., Matthies D., Frenzel P., Derakshani M., Liesack W. Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 2005;7(3):382–395. doi: 10.1111/j.1462-2920.2005.00700.x. [DOI] [PubMed] [Google Scholar]
- 69.Pukall R., Lapidus A., Del Rio T.G., Copeland A., Tice H., Cheng J.-F., Lucas S., Chen F., Nolan M., Bruce D. Complete genome sequence of Conexibacter woesei type strain (ID131577 T) Stand. Genomic Sci. 2010;2(2):212–219. doi: 10.4056/sigs.751339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hardin J., Mitani A., Hicks L., VanKoten B. A robust measure of correlation between two genes on a microarray. BMC Bioinformatics. 2007;8(1):1–13. doi: 10.1186/1471-2105-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahmed S., Ahmad M., Swami B.L., Ikram S. A review on plants extract mediated synthesis of silver nanoparticles for antimicrobial applications: a green expertise. J. Adv. Res. 2016;7(1):17–28. doi: 10.1016/j.jare.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iravani S., Korbekandi H., Mirmohammadi S.V., Zolfaghari B. Synthesis of silver nanoparticles: chemical, physical and biological methods. Res. Pharm. Sci. 2014;9(6):385. [PMC free article] [PubMed] [Google Scholar]
- 73.Goodsell D.S. John Wiley & Sons; 2004. Bionanotechnology: Lessons from Nature. [Google Scholar]
- 74.Kapil A. The challenge of antibiotic resistance: need to contemplate. Indian J. Med. Res. 2005;121(2):83. [PubMed] [Google Scholar]
- 75.Oei J.D., Zhao W.W., Chu L., DeSilva M.N., Ghimire A., Rawls H.R., Whang K. Antimicrobial acrylic materials with in situ generated silver nanoparticles. J Biomed Mater Res B Appl Biomater. 2012;100(2):409–415. doi: 10.1002/jbm.b.31963. [DOI] [PubMed] [Google Scholar]
- 76.Song H., Ko K., Oh L., Lee B. Fabrication of silver nanoparticles and their antimicrobial mechanisms. Eur. Cells Mater. 2006;11(Suppl. 1):58. [Google Scholar]
- 77.Tziveleka L.-A., Vagias C., Roussis V. Natural products with anti-HIV activity from marine organisms. Curr. Top. Med. Chem. 2003;3(13):1512–1535. doi: 10.2174/1568026033451790. [DOI] [PubMed] [Google Scholar]
- 78.Gupta A., Bonde S.R., Gaikwad S., Ingle A., Gade A.K., Rai M. Lawsonia inermis-mediated synthesis of silver nanoparticles: activity against human pathogenic fungi and bacteria with special reference to formulation of an antimicrobial nanogel. IET Nanobiotechnol. 2013;8(3):172–178. doi: 10.1049/iet-nbt.2013.0015. [DOI] [PubMed] [Google Scholar]
- 79.Bonett D.G., Wright T.A. Sample size requirements for estimating Pearson, Kendall and Spearman correlations. Psychometrika. 2000;65(1):23–28. [Google Scholar]
- 80.Hugenholtz P., Goebel B.M., Pace N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998;180(18):4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kruis F.E., Fissan H., Rellinghaus B. Sintering and evaporation characteristics of gas-phase synthesis of size-selected PbS nanoparticles. Mater. Sci. Eng. B. 2000;69:329–334. [Google Scholar]
- 82.Wintachai P., Kaur P., Lee R.C.H., Ramphan S., Kuadkitkan A., Wikan N., Ubol S., Roytrakul S., Chu J.J.H., Smith D.R. Activity of andrographolide against chikungunya virus infection. Sci. Rep. 2015;5(1):1–14. doi: 10.1038/srep14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sandin S.A., Smith J.E., DeMartini E.E., Dinsdale E.A., Donner S.D., Friedlander A.M., Konotchick T., Malay M., Maragos J.E., Obura D. Baselines and degradation of coral reefs in the northern line Islands. PLoS One. 2008;3(2) doi: 10.1371/journal.pone.0001548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zandi K., Teoh B.-T., Sam S.-S., Wong P.-F., Mustafa M.R., AbuBakar S. Novel antiviral activity of baicalein against dengue virus. BMC Complement. Altern. Med. 2012;12(1):1–9. doi: 10.1186/1472-6882-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sinha S., Pan I., Chanda P., Sen S.K. Nanoparticles fabrication using ambient biological resources. J. Appl. Biosci. 2009;19:1113–1130. [Google Scholar]
- 86.Sharma V.K., Yngard R.A., Lin Y. Silver nanoparticles: green synthesis and their antimicrobial activities. Adv. Colloid Interf. Sci. 2009;145(1–2):83–96. doi: 10.1016/j.cis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Sadeghi B., Gholamhoseinpoor F. A study on the stability and green synthesis of silver nanoparticles using Ziziphora tenuior (Zt) extract at room temperature. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015;134:310–315. doi: 10.1016/j.saa.2014.06.046. [DOI] [PubMed] [Google Scholar]
- 88.Perumal V., Manickam T., Bang K.-S., Velmurugan P., Oh B.-T. Antidiabetic potential of bioactive molecules coated chitosan nanoparticles in experimental rats. Int. J. Biol. Macromol. 2016;92:63–69. doi: 10.1016/j.ijbiomac.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 89.Sudhakar C., Selvam K., Govarthanan M., Senthilkumar B., Sengottaiyan A., Stalin M., Selvankumar T. Acorus calamus rhizome extract mediated biosynthesis of silver nanoparticles and their bactericidal activity against human pathogens. J. Genet. Eng. Biotechnol. 2015;13(2):93–99. doi: 10.1016/j.jgeb.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rastogi L., Arunachalam J. Sunlight based irradiation strategy for rapid green synthesis of highly stable silver nanoparticles using aqueous garlic (Allium sativum) extract and their antibacterial potential. Mater. Chem. Phys. 2011;129(1–2):558–563. [Google Scholar]
- 91.Kumarasamyraja D., Jeganathan N. 2013. Green Synthesis of Silver Nanoparticles Using Aqueous Extract of Acalypha indica and Its Antimicrobial Activity. [Google Scholar]
- 92.Durán N., Marcato P.D., Alves O.L., De Souza G.I., Esposito E. Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J. Nanobiotechnol. 2005;3(1):1–7. doi: 10.1186/1477-3155-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hamouda T., Baker J., Jr. Antimicrobial mechanism of action of surfactant lipid preparations in enteric gram-negative bacilli. J. Appl. Microbiol. 2000;89(3):397–403. doi: 10.1046/j.1365-2672.2000.01127.x. [DOI] [PubMed] [Google Scholar]
- 94.Niraimathi K., Sudha V., Lavanya R., Brindha P. Biosynthesis of silver nanoparticles using Alternanthera sessilis (Linn.) extract and their antimicrobial, antioxidant activities. Colloids Surf. B: Biointerfaces. 2013;102:288–291. doi: 10.1016/j.colsurfb.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 95.Mayer A., Guerrero A.J., Rodríguez A.D., Taglialatela-Scafati O., Nakamura F., Fusetani N. Marine pharmacology in 2014–2015: marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, antiviral, and anthelmintic activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Marine Drugs. 2020;18(1):5. doi: 10.3390/md18010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dhanasekaran D., Thangaraj R. Evaluation of larvicidal activity of biogenic nanoparticles against filariasis causing Culex mosquito vector. Asian Pacific J. Trop. Dis. 2013;3(3):174–179. [Google Scholar]
- 97.Sahayaraj K., Rajesh S. 2022. Bionanoparticles: Synthesis and Antimicrobial Applications. [Google Scholar]
- 98.Logaranjan K., Raiza A.J., Gopinath S.C., Chen Y., Pandian K. Shape-and size-controlled synthesis of silver nanoparticles using Aloe vera plant extract and their antimicrobial activity. Nanoscale Res. Lett. 2016;11(1):1–9. doi: 10.1186/s11671-016-1725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rajaram K., Aiswarya D., Sureshkumar P. Green synthesis of silver nanoparticle using Tephrosia tinctoria and its antidiabetic activity. Mater. Lett. 2015;138:251–254. [Google Scholar]
- 100.Nezamdoost T., Bagherieh-Najjar M., Aghdasi M. Biogenic synthesis of stable bioactive silver chloride nanoparticles using Onosma dichroantha Boiss. Root extract. Mater. Lett. 2014;137:225–228. [Google Scholar]
- 101.Ahmed M.J., Murtaza G., Mehmood A., Bhatti T.M. Green synthesis of silver nanoparticles using leaves extract of Skimmia laureola: characterisation and antibacterial activity. Mater. Lett. 2015;153:10–13. [Google Scholar]
- 102.Ravindran D., Ramanathan S., Arunachalam K., Jeyaraj G., Shunmugiah K., Arumugam V. Phytosynthesized silver nanoparticles as antiquorum sensing and antibiofilm agent against the nosocomial pathogen Serratia marcescens: an in vitro study. J. Appl. Microbiol. 2018;124(6):1425–1440. doi: 10.1111/jam.13728. [DOI] [PubMed] [Google Scholar]
- 103.Srinivasan R., Vigneshwari L., Rajavel T., Durgadevi R., Kannappan A., Balamurugan K., Devi K.P., Ravi A.V. Biogenic synthesis of silver nanoparticles using Piper betle aqueous extract and evaluation of its anti-quorum sensing and antibiofilm potential against uropathogens with cytotoxic effects: an in vitro and in vivo approach. Environ. Sci. Pollut. Res. 2018;25(11):10538–10554. doi: 10.1007/s11356-017-1049-0. [DOI] [PubMed] [Google Scholar]
- 104.Amin M., Anwar F., Janjua M.R.S.A., Iqbal M.A., Rashid U. Green synthesis of silver nanoparticles through reduction with Solanum xanthocarpum L. berry extract: characterisation, antimicrobial and urease inhibitory activities against helicobacter pylori. Int. J. Mol. Sci. 2012;13(8):9923–9941. doi: 10.3390/ijms13089923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mollenkopf D.F., Stull J.W., Mathys D.A., Bowman A.S., Feicht S.M., Grooters S.V., Daniels J.B., Wittum T.E. Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob. Agents Chemother. 2017;61(2) doi: 10.1128/AAC.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nelson J.M., Chiller T.M., Powers J.H., Angulo F.J. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin. Infect. Dis. 2007;44(7):977–980. doi: 10.1086/512369. [DOI] [PubMed] [Google Scholar]
- 107.Kennedy K., Collignon P. Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur. J. Clin. Microbiol. Infect. Dis. 12, 2010;29:1501–1506. doi: 10.1007/s10096-010-1031-y. [DOI] [PubMed] [Google Scholar]
- 108.Aubertheau E., Stalder T., Mondamert L., Ploy M.-C., Dagot C., Labanowski J. Impact of wastewater treatment plant discharge on the contamination of river biofilms by pharmaceuticals and antibiotic resistance. Sci. Total Environ. 2017;579:1387–1398. doi: 10.1016/j.scitotenv.2016.11.136. [DOI] [PubMed] [Google Scholar]
- 109.Walsh T.R., Weeks J., Livermore D.M., Toleman M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 2011;11(5):355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 110.Ashbolt N.J., Amézquita A., Backhaus T., Borriello P., Brandt K.K., Collignon P., Coors A., Finley R., Gaze W.H., Heberer T. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013;121(9):993–1001. doi: 10.1289/ehp.1206316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Collignon P.J., McEwen S.A. One health—its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019;4(1):22. doi: 10.3390/tropicalmed4010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material