Significance
Pluripotency and development can be governed at the level of epigenetics. Growing lines of evidence underscore the importance of the Set1A/complex of proteins associated with SET1 (COMPASS) histone 3 lysine 4 methyltransferase complex in embryogenesis and neurodevelopment. We show that the catalytic SET domain of Set1A is essential for mouse embryogenesis; however, having this domain extends the viability of embryos compared to complete loss of Set1A. We additionally show that Ing5, a core component of several complexes involved in histone acetylation, can functionally interact with Set1AΔSET in regulating ESC viability and developmental gene expression. Insights into their physiological activity and regulation will assist our understanding of their dysfunction in disease and ultimately facilitate the discovery of new targets for future therapy.
Keywords: COMPASS, H3K4 methylation, ING5, Set1A, pluripotency
Abstract
Embryonic stem cells (ESCs) are defined by their ability to self-renew and the potential to differentiate into all tissues of the developing organism. We previously demonstrated that deleting the catalytic SET domain of the Set1A/complex of proteins associated with SET1 histone methyltransferase (Set1A/COMPASS) in mouse ESCs does not impair their viability or ability to self-renew; however, it leads to defects in differentiation. The precise mechanisms by which Set1A executes these functions remain to be elucidated. In this study, we demonstrate that mice lacking the SET domain of Set1A are embryonic lethal at a stage that is unique from null alleles. To gain insight into Set1A function in regulating pluripotency, we conducted a CRISPR/Cas9-mediated dropout screen and identified the MOZ/MORF (monocytic leukaemia zinc finger protein/monocytic leukaemia zinc finger protein-related factor) and HBO1 (HAT bound to ORC1) acetyltransferase complex member ING5 as a synthetic perturbation to Set1A. The loss of Ing5 in Set1AΔSET mouse ESCs decreases the fitness of these cells, and the simultaneous loss of ING5 and in Set1AΔSET leads to up-regulation of differentiation-associated genes. Taken together, our results point toward Set1A/COMPASS and ING5 as potential coregulators of the self-renewal and differentiation status of ESCs.
Embryonic stem cells (ESCs) have two major defining features: 1) the extensive ability to self-renew and 2) the potential to differentiate into all cell lineages, known as pluripotency. These unique characteristics bestow ESCs the fortune to serve as an effective platform for disease modeling and drug discovery research. In addition to a core network of transcription factors, self-renewing and pluripotent states are mediated by various epigenetic factors that encompass both chromatin remodelers and histone modifiers that promote a transcriptionally permissive environment (1–3). For that reason, it is necessary to tightly regulate the expression and interactions of numerous proteins to prevent untimely differentiating events. However, how these proteins impact one another and the transcriptional program to jointly control ESC pluripotency remains to be thoroughly understood.
The complex of proteins associated with SET1 (COMPASS) family was first identified in the budding yeast Saccharomyces cerevisiae. SET1 forms a multiprotein complex in budding yeast that implements mono-, di-, and trimethylation of the fourth lysine residue of histone H3 (H3K4). In addition to genome-wide H3K4me2 and H3K4me3, the COMPASS family deposits H3K4me1 at enhancers and gene-specific H3K4me2 and H3K4me3 at developmental promoters (4–8). These functions are carried out by six different COMPASS/COMPASS-like families in mammals. SET1A/COMPASS and SET1B/COMPASS are responsible for bulk H3K4 methylation of the mammalian genome (9–11).
Among the COMPASS family of six H3K4 methyltransferases identified in mammals, Set1A is the only member whose full genetic knockout has been shown to be essential for ESC proliferation and self-renewal (12, 13). However, significant loss of genome-wide H3K4me3 only occurs when both SET1A and SET1B are removed (12, 14). There is an increasing amount of evidence in the field regarding catalytic-independent functions of COMPASS and how they play a crucial role in development and disease (reviewed in ref. 11). We previously demonstrated that although removing the catalytic SET domain of Set1A via CRISPR/Cas9 resulted in defective ESC differentiation, Set1AΔSET ESCs remained viable and could undergo proper self-renewal (15). It is likely that Set1A coordinates with other proteins (in addition to Set1B) and/or downstream effectors to regulate pluripotency. These additional functional interactors are currently undefined.
By leveraging a CRISPR/Cas9-based dropout screen approach, we sought to identify these factors that genetically interact with Set1AΔSET in an unbiased manner. Our genome-wide screen revealed several candidates where their perturbations are synthetic lethal with Set1AΔSET in ESCs, one of which is Ing5. ING5 is a core subunit of three histone acetyltransferase (HAT) complexes (MOZ versus MORF versus HBO1) responsible for lysine acetylation on H3 and H4 (16). To date, only a handful of studies have implicated the role of ING5 in preserving self-renewal, although all three HAT complexes show context-specific functional relevance in maintaining stem cell self-renewal. In this study, we identify ING5 and Set1A as potential coregulators of ESC function. Our findings provide additional insight into the intricacy underlying epigenetic regulation of ESC pluripotency, illustrating that the interplay among chromatin regulators is composed of a complex chromatin language that guides transcriptional regulation.
Results
The Catalytic SET Domain of Set1A Is Required for Embryonic Development in Mice.
Previously, we had shown that the CRISPR/Cas9-mediated deletion of the catalytic SET domain of Set1A in V6.5 mouse ESCs does not affect their self-renewal and proliferation but impairs their differentiation (15). Furthermore, Set1A has been shown to be required for gastrulation; Set1A-knockout (KO) embryos implant but cannot progress past the epiblast stage, leading to early embryonic lethality at embryonic day 7.5 (E7.5) (12). These findings warranted further investigation into the role of Set1A’s catalytic activity in development. To define the in vivo consequences of deleting the SET domain of Set1A, we generated mutant mice harboring a Set1A SET domain mutation via pronuclear injection of previously published (15) single guide RNAs (sgRNAs) flanking the SET domain (SI Appendix, Fig. S1A). Next-generation sequencing (NGS) analysis of filial (F)0 founder mice using uniquely barcoded primers around the sgRNA cut site in exon 17 of Set1A showed a two-nucleotide insertion at the start of the SET domain of Set1A, resulting in an early stop codon that removes the majority of the SET domain (SI Appendix, Fig. 1 A and B). This allele was designated as SETless to differentiate it from the previously published Set1AΔSET ESCs.
Heterozygous intercrosses between F1 mice harboring the 2-base pair (bp) insertion (hereafter referred to as SETless-HET mice) demonstrated that F2 SETless-HET mice were healthy and indistinguishable from wild-type (WT) mice both in terms of phenotype and gene expression (Fig. 1B and SI Appendix, Fig. 1C). However, the homozygous loss of the Set1A SET domain (SETless-KO) proved to be incompatible with postnatal life. To determine the stage at which SETless-KO is lethal, we performed timed mating and dissected embryos at multiple developmental timepoints. E8.5 was chosen as the first timepoint because Set1A-KO mice are E7.5 lethal (12), and we predicted that the loss of the catalytic domain (as opposed to the entire protein) might delay lethality. Indeed, surviving embryos were observed up to E12.5 (Fig. 1E). However, beginning at E8.5, gross developmental defects and a general delay in development were observed in the SETless-KO embryos (Fig. 1A).
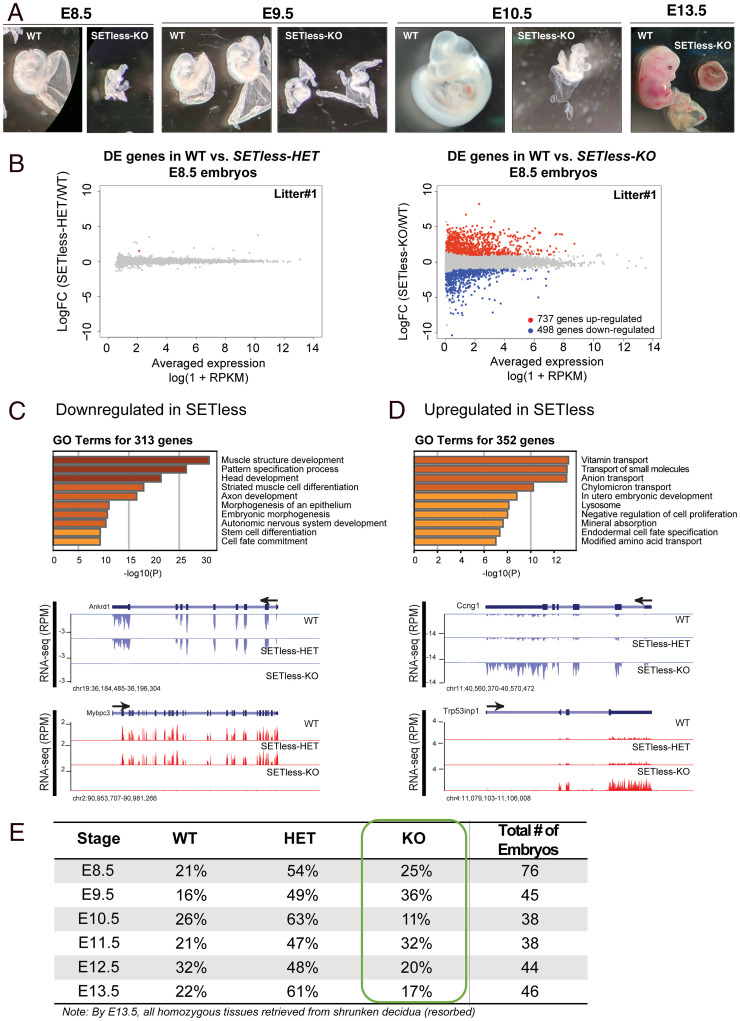
Fig. 1.
The deletion of the SET domain of Set1A (SETless) is embryonic lethal at a later stage than Set1A-KO and leads to gross developmental defects. (A) Representative images of dissected littermates WT versus homozygous mutants (SETless-KO) at E8.5, E9.5, E10.5, and E13.5. Note the severe delay in development of the homozygous mutant compared to its WT littermate. The yolk sacs were left attached to preserve the integrity of the embryos. (B) MA plots (M [log ratio] and A [mean average] scale) comparing the transcriptomes between WT and heterozygous littermates (Left) and WT and homozygous littermates (Right). Three individual litters of E8.5 embryos were analyzed, and one is shown in this figure. Criteria used to determine DE genes include 1) logFC > |1|, 2) log(average counts per million) > 1, and 3) Benjamini–Hochberg adjusted P values < 0.01; RPKM, reads per kilobase of transcript per million mapped reads. (C) GO analysis for 313 down-regulated genes identified by RNA-seq with genome browser track examples of 2 down-regulated genes in the homozygous mutants compared to WT and heterozygous littermates. (D) GO analysis for 352 up-regulated genes identified by RNA-seq and genome browser track examples of 2 up-regulated genes in the homozygous mutants compared to WT and heterozygous littermates. GO analysis was carried out using the Metascape software. Enriched pathways that are statistically significant are shown with corresponding −log10 (P) values. (E) Genotypes of resulting embryos for each of the specified stages during embryogenesis. A few cases of resorption in which decidua were very small and extremely minute amounts of embryo material were recovered for genotyping were included in the total number. These cases were determined to be homozygous mutants from PCR genotyping.
To better understand the phenotype of the SETless-KO embryos, we investigated gene expression changes in the mutant embryos by extracting RNA from three separate litters of SETless-KO, SETless-HET, and WT whole E8.5 embryos (Fig. 1 C and D and SI Appendix, Fig. 1 C and D). While there were no changes in gene expression between WT and SETless-HET embryos (SI Appendix, Fig. 1C), significant differences were observed between SETless-KO and WT embryos for all three litters analyzed (Fig. 1B and SI Appendix, Fig. 1D). Overlapping the significantly differentially expressed (DE) genes from the three litters revealed 313 down-regulated genes, whereas 352 genes were up-regulated (SI Appendix, Fig. 1E). Gene ontology (GO) analysis demonstrated that many of the down-regulated genes were important for differentiation, cell fate commitment, and muscle development (Fig. 1C), whereas up-regulated genes were enriched for negative regulation of cell proliferation and transport of nutrients and small molecules (Fig. 1D). This is an intriguing finding, as our previously published data do not show significant differences in gene expression between Set1AΔSET and WT ESCs (15), and suggests that context-dependent gene regulatory functions for Set1A exist.
Since the SETless mouse harbors a premature truncation of Set1A instead of an exclusive catalytic domain deletion, there is a small possibility that the remainder of the C terminus accounts for the phenotype. The C terminus of the protein, designated the post-SET domain, is required for zinc (Zn) binding. It is intrinsically disordered in the absence of Zn but forms a knot-like structure close to the catalytic domain when Zn is present (17). In Neurospora crassa, this structuring is necessary for S-adenosyl-methionine and histone tail interactions and mutation studies in which cysteines are replaced by serines show a loss of catalytic activity (18). These findings suggest that the post-SET domain is important in the context of the protein’s catalytic activity, and, as such, the premature termination ought to have the same effect as exclusively deleting the catalytic domain.
A Genome-Wide CRISPR Dropout Screen Identifies Genetic Dependencies of Set1AΔSET ESCs.
Both our previous studies in ESCs and our assessment of the SETless mice underscore the importance of Set1A and its catalytic-dependent and catalytic-independent functions in pluripotency and development. Because ESC proliferation and viability are not perturbed by the loss of the SET domain, we decided that further investigation into the pathways regulated by Set1A and its potential coregulators is warranted to better understand how it functions in self-renewal. Therefore, we performed a genome-wide CRISPR/Cas9 dropout screen to identify genes that are required for cell viability of Set1AΔSET ESCs (Fig. 2A).
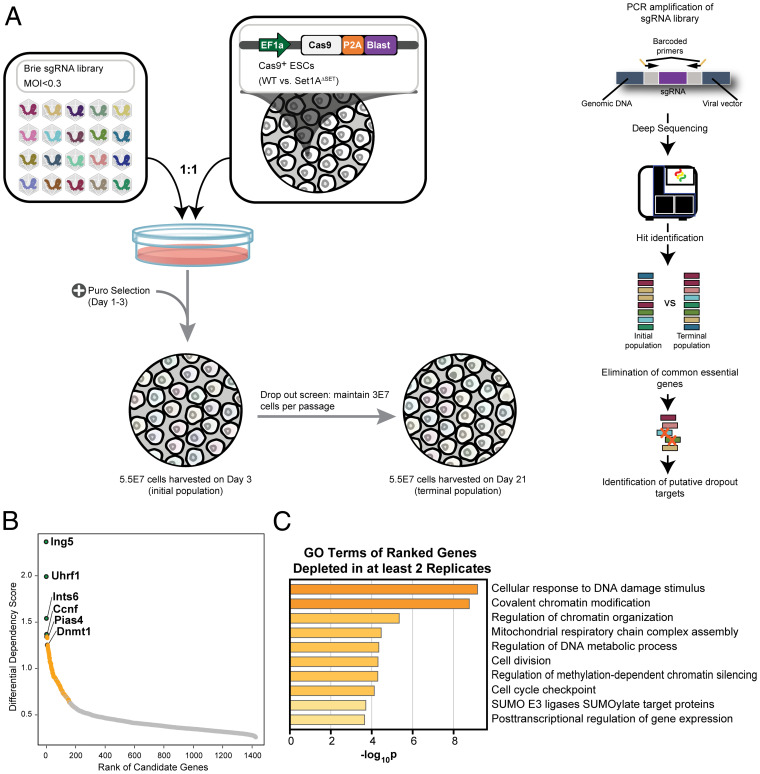
Fig. 2.
A genome-wide CRISPR dropout screen identifies genetic dependencies of Set1AΔSET ESCs. (A) Pooled Cas9-expressing WT versus Set1AΔSET ESCs were transduced with Brie sgRNA library at an MOI of <0.3. Under puromycin selection, cells were passaged every 2 to 3 d and maintained at 3 × 107 cells per passage to ensure sufficient sgRNA representation. Cells (5.5 × 107) were harvested at day 3 (initial population) and day 21 (terminal population) after transduction. Depleted candidate targets were identified following library amplification, Illumina sequencing, and elimination of common essential genes. (B) Dropout targets (1,425) were ranked by their assigned DDS, which encompasses both the magnitude and reproducibility of dropout depletion across four replicates of the dropout screen. Ranked targets had at least two sgRNAs depleted out of four sgRNAs. Genes in green were depleted in at least three out of four replicates, and genes in orange were depleted in at least two out of four replicates. The six labeled targets correspond to the genes in green with their ranking as determined by the DDS score in parentheses. (C) GO analysis for the 1,425 dropout targets in B using the Metascape software. Enriched pathways that are statistically significant are shown with corresponding −log10 (P) values.
To retrieve the list of putative candidates that synthetically perturb Set1AΔSET, we focused on genes that were depleted only in Set1AΔSET cells. An initial analysis revealed 1,425 targets that had at least two sgRNAs depleted out of the four sgRNAs represented in the Brie library (Fig. 2B). GO analysis of these genes revealed the most significant enrichment in pathways involved in DNA damage response and repair and chromatin regulation (Fig. 2C). To identify the most essential genetic dependencies of Set1AΔSET ESCs, each target candidate was assigned a “differential dependency score” (DDS), which reflects both the degree and consistency of dropout across multiple replicates of the 21-d screen, and was subsequently ranked. Six targets were found to be depleted in at least three of four replicates (Fig. 2B). These targets, Ing5, Uhrf1, Ints6, Ccnf, Pias4, and Dnmt1, are all known to play a role in chromatin regulation and/or DNA damage response. As a result, we decided to focus our subsequent target validation efforts on the genes in the top two significant GO term categories (Fig. 2C).
For our secondary analysis, the 39 selected genes shown in SI Appendix, Fig. 2A, were individually targeted using one sgRNA per gene (chosen from the Brie library) in WT-Cas9 and Set1AΔSET-Cas9 ESCs and selected with puromycin for 10 d. Subsequently, alkaline-phosphatase staining was performed to assess viability and colony formation (SI Appendix, Fig. 2B). In each assay plate, an sgRNA against Pcna and a nontarget control (NTC) sgRNA were included as positive and negative controls, respectively. Multiple targets, including ING5, conferred a visible growth disadvantage to Set1AΔSET ESCs compared to their WT counterpart. These qualitative observations were subsequently confirmed by measurement of colony number and areas in ImageJ (SI Appendix, Fig. 2B). Targets selected for further validation were those that consistently showed a decreased colony number between the two replicates.
Upon closer scrutiny of the 39 selected candidates (SI Appendix, Fig. 2A), we noticed two known interactors of ING5: BRPF1 and JADE1. ING5 is a core component of two different HAT complexes: MOZ/MORF or HBO1 complexes (also known as the KAT6A/6B and KAT7 complexes) (19). These complexes include alternate scaffold proteins BRPF1/2/3 or JADE1/2/3 that help dictate acetylation specificity on the H3 or H4 tail (16, 20). The identification of these additional complex members increased the level of confidence of ING5 being a genetic dependency to Set1A in ESC maintenance and viability. As such, both BRPF1 and JADE1 were included in subsequent validation assays.
The MOZ/MORF and HBO1 Complex Subunit ING5 Shows Synthetic Perturbation with Set1AΔSET.
For a more robust validation of our screen results, we selected Ing5, Brpf1, and Jade1 as well as the catalytic subunits of the MOZ/MORF and HBO1 complexes (MOZ/KAT6A, MORF/KAT6B, and HBO1/KAT7) as targets for a CRISPR/Cas9-based cellular competitive growth assay (Fig. 3). In brief, WT-Cas9 and Set1AΔSET-Cas9 were respectively labeled with mCherry and enhanced green fluorescent protein (eGFP) by stable lentiviral expression, and then these two lines (hereafter referred to as WT-mCherry and Set1AΔSET-eGFP) were plated in the same well at a 1:1 ratio (Fig. 3A). The next day, the mixed population was transduced with individual sgRNAs against the above genes; two to four sgRNAs were used per target. Depletion was confirmed at the protein level for ING5, JADE1, and HBO1 for these sgRNAs (SI Appendix, Fig. 3A). The rationale behind this assay is that if depletion of any of the candidates confers an additional growth disadvantage to Set1AΔSET-eGFP cells, the WT-mCherry cells should eventually outcompete the Set1AΔSET-eGFP cells over successive passages. The cells were subcultured over the course of 21 d, and the mCherry+ and eGFP+ populations were counted every 2 to 3 d by flow cytometry (Fig. 3A). As expected, under ING5 depletion conditions, the WT-mCherry population steadily overtook the Set1AΔSET-eGFP population over time (SI Appendix, Fig. 3B).
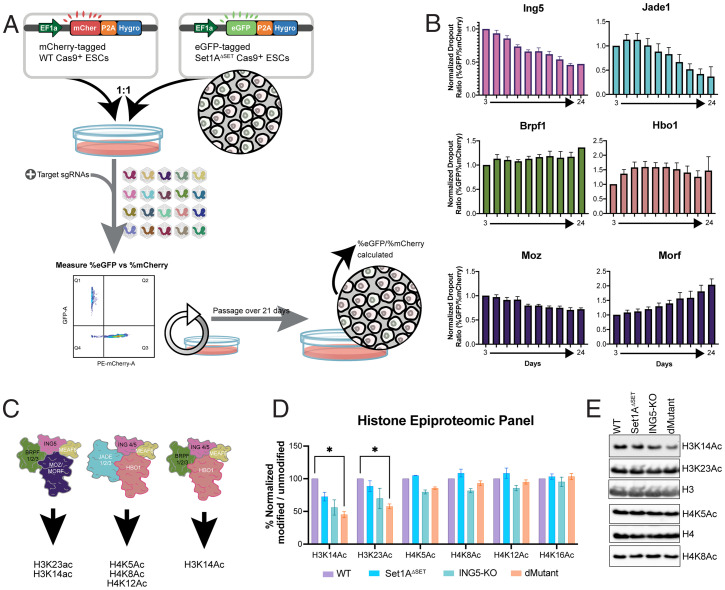
Fig. 3.
The MOZ/MORF and HBO1 complex subunit ING5 shows synthetic perturbation with Set1A. (A) A competitive growth assay was used to validate screen targets. WT-Cas9 and Set1AΔSET-Cas9 cells were labeled with mCherry and eGFP, respectively, and mixed at a 1:1 ratio before sgRNA lentivirus transduction the next day. During the 21-d screen, fractions of eGFP+ versus mCherry+ were measured by flow cytometry. (B) Dropout ratios (% eGFP/% mCherry, corresponding to % Set1AΔSET-Cas9/% WT-Cas9) were calculated after flow cytometry every 2 d and normalized to ratios for day 3 and NTC (lentiGuide-Puro). Data are shown as mean ± SEM (n = 3). (C) The top screen hit ING5 is known to be a part of the MOZ/MORF and HBO1 complexes, which are involved in acetylation at H3 and/or H4. MOZ, MORF, and HBO1 are the catalytic subunits of the complexes. (D) Histone modification mass spectrometry analysis was performed by acid extraction of histones from WT, Set1AΔSET, ING5-KO, and dMutant mouse ESC nuclear extracts and derivatization via propionylation reaction trypsin digestion. Percent relative abundances of three technical replicates were normalized to the abundance of unmodified residues to show data as percent normalized values (mean ± SEM); *, P < 0.05. (E) Representative image of a Western blot performed to confirm mass spectrometry results. Total H3 and H4 were used as loading controls.
Dropout ratios were calculated by measuring the percentage of Set1AΔSET-GFP/WT-mCherry and normalizing these to the baseline timepoint (day 3) and the NTC (lentiGuide-Puro; Fig. 3B). ING5 and JADE1 depletion resulted in a significant growth disadvantage for Set1AΔSET-GFP cells, whereas BRPF1 depletion did not lead to visible changes in Set1AΔSET-GFP ESC growth relative to WT-mCherry (Fig. 3B and SI Appendix, Fig. 3 B and C). The catalytic subunit MOZ gave a small but significant growth disadvantage to Set1AΔSET-GFP cells. Interestingly, MORF depletion gave a significant growth advantage to Set1AΔSET-GFP cells (Fig. 3B). These results were reproducible across a minimum of three replicates with at least two different sgRNAs (SI Appendix, Fig. 3C).
The MOZ/MORF and HBO1 complexes are members of the MYST family of acetyltransferases and are responsible for a variety of histone modifications (Fig. 3C), including H3K14/K23 acetylation by MOZ/MORF complexes (19). To make an unbiased assessment of the histone modification profile of WT and Set1AΔSET ESCs with or without loss of ING5, we generated ING5-KO and ING5-KO;Set1AΔSET (hereafter referred to as dMutant) ESCs using CRISPR/Cas9-mediated deletion that led to the loss of the entire Ing5 transcript (Fig. 4A).
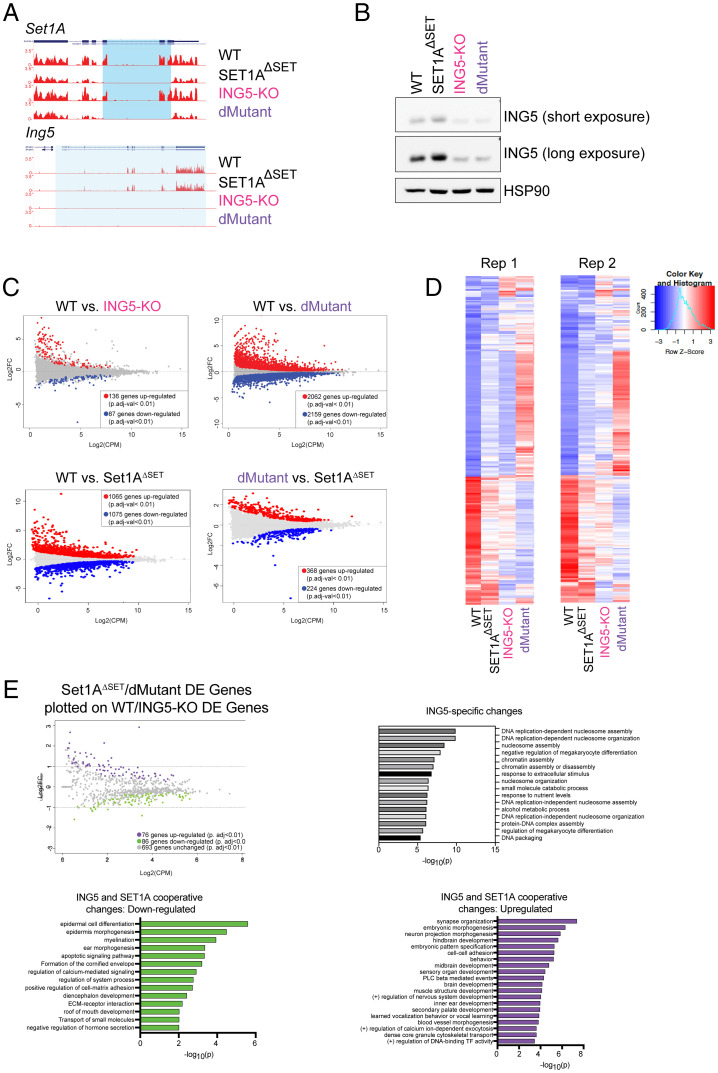
Fig. 4.
Loss of ING5 results in global gene expression changes and dysregulation of chromatin assembly and nucleosome organization changes both alone and in combination with Set1AΔSET. (A) UCSC genome browser tracks showing the loss of the deletion of the SET domain of Set1A (Top) and the loss of the ING5 transcript (Bottom). The deleted regions are highlighted in light blue. (B) Western blotting for ING5 shows loss of ING5 protein in ING5-KO and dMutant (Set1AΔSET, ING5-KO) cells. The faint band detected in the KO cells was attributed to the cross-reaction of the ING5 antibody with ING4. (C) The log2FC changes in gene expression from RNA-seq data indicate that while loss of ING5 alone does not affect gene expression significantly, the joint perturbation of ING5 and Set1A together lead to major changes in gene expression. (D) Heat maps of RNA-seq data were generated by plotting z scores of log2FC gene expression changes. Three independent genetic clones for both ING5-KO and dMutant were analyzed, and the experiment was repeated twice. (E) To investigate ING5-specific gene expression changes, dMutant/Set1AΔSET DE genes were plotted on ING5-KO/WT DE genes. Gray genes indicate ING5-specific DE genes. GO analysis of ING5-specific gene expression changes with a logFC of <−1 and P value of <0.01 show that chromatin assembly and nucleosome organization genes are down-regulated with loss of ING5. ING5/Set1A cooperative up-regulated genes are plotted in green, and down-regulated genes are plotted in purple. GO analysis was performed using the Metascape software. Enriched pathways that are statistically significant are shown with corresponding −log10(P) values.
Interestingly, loss of Ing5 function in conjunction with Set1AΔSET did not impair survival. dMutant cells grew slightly slower than the other three genotypes (WT, Set1AΔSET, and ING5-KO) but were still viable. Thus, we hesitate to denote the interaction between ING5 and Set1A as synthetic lethal based on our screen and have instead opted for the term “synthetic perturbation”. The most likely explanation for the blunting of the phenotype is the redundancy between ING4 and ING5 compensating for the loss of ING5. ING4 and ING5 can form heterodimers (21), and several studies suggest that the two proteins can function redundantly (19). For instance, both ING4 and ING5 bind P300 HAT and the tumor suppressor P53 to modulate the function of P53 (22).
Following the confirmation of ING5 loss (Fig. 4 A and B), epiproteomic histone modification panel mass spectrometry (23–25) was conducted in WT, Set1AΔSET, ING5-KO, and dMutant ESCs. This analysis did not reveal a significant loss of modifications implemented by the MOZ/MORF and HBO1 complexes (H3K14ac, H3K23ac, H4K5ac, H4K8ac, and H4K12ac), aside from a 30% decrease in H3K14ac and 20% decrease in H3K23ac when comparing the dMutant to the WT (P < 0.05, Fig. 3D). Western blotting analysis confirmed these results (Fig. 3E). This suggests that while the catalytic activity of MOZ/MORF and HBO1 complexes could be important for the genetic dependency between Set1A and ING5, the small effect of the loss of the aforementioned histone acetylations could also point toward a MOZ/MORF/HBO1 complex–independent role for ING5 in the context of its genetic dependency with Set1A.
Loss of ING5 Causes Global Gene Expression Changes Alone and in Combination with Set1AΔSET.
To account for clonal effects, gene expression of three independent genetic clones for both ING5-KO and dMutant ESCs were assessed. Analysis of the transcriptomic data showed that loss of ING5 alone did not lead to a major change in gene expression profile, whereas the combination of Ing5 and Set1AΔSET lead to a large number of gene expression changes (Fig. 4C, Top). In our previously published results, WT and Set1AΔSET ESCs had an overall similar gene expression profile (15). In this study, we did observe a difference in gene expression profile between WT and Set1AΔSET ESCs (Fig. 4C, Bottom Left). We attribute this to a single Set1AΔSET line being used for this RNA sequencing (RNA-seq) as opposed to two independent lines in the original study, as clonal effects can contribute to changes in gene expression profile. The difference between dMutant and Set1AΔSET was negligible (Fig. 4C, Bottom Right). Overall, the ING5-KO ESC gene expression profile differed slightly from that of WT and Set1AΔSET ESCs; however, the dMutant ESCs showed the most divergent gene expression pattern from both WT and Set1AΔSET (Fig. 4D).
To understand which of the gene expression changes were specific to Ing5 loss, we took our list of DE genes between Set1AΔSET versus dMutant and plotted it against the list of WT versus ING5-KO DE genes. In this setting, the genes that are no longer significantly changed when the two lists are overlapped are likely changes exclusively conferred by the deletion of Ing5 (Fig. 4E). These genes are indicated in gray, whereas the genes that still show statistically significant up- or down-regulation are indicated in purple and green, respectively. GO analysis of these Ing5-specific (gray) genes showed that the list is significantly enriched for genes associated with nucleosome assembly and organization and DNA replication (Fig. 4E). We additionally examined the genes that remained up-regulated (purple) or down-regulated (green) in the overlay, with the rationale that they could indicate ING5–Set1A cooperative differential expression (Fig. 4E, Bottom). Interestingly, the up-regulated DE genes were strongly enriched for developmental pathways, suggesting that ING5 and Set1A could be regulating the state of differentiation in ESCs.
The Simultaneous Loss of Ing5 and the SET Domain of Set1A Lead to Up-Regulation of Differentiation-Associated Genes at Regions Cooccupied by ING5 and Set1A.
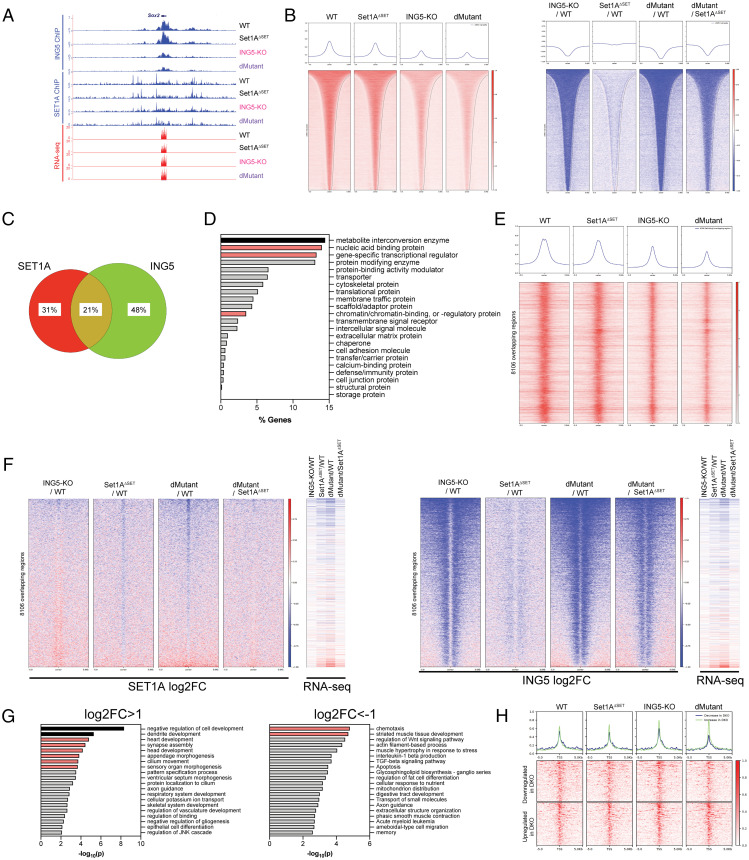
To better understand the role of ING5 and Set1A in ESC function at the chromatin level, we performed chromatin immunoprecipitation with sequencing (ChIP-seq) for ING5 and Set1A in our WT, Set1AΔSET, ING5-KO, and dMutant ESC lines. As anticipated, the deletion of ING5 led to a visible signal loss in ING5 ChIPs (Fig. 5 A and B). There was residual ING5 signal at several promoter regions, including SOX2 (Fig. 5A), which we attribute to the antibody potentially cross-reacting with ING4, similar to what we observed for total protein levels by Western blotting (Fig. 4B). Interestingly, while there was overlap in Set1A and ING5 signal, we observed a difference in the binding pattern of the two, whereas Set1A signal appeared as sharp, narrow peaks primarily at gene promoters and the transcription start site (TSS); ING5 signal tended to be broad and spanned the promoter, TSS, and gene bodies (Fig. 5 A and B).
Fig. 5.
The simultaneous loss of ING5 and the SET domain of Set1A lead to up-regulation of differentiation-associated genes at regions cooccupied by ING5 and Set1A. (A) ING5 and Set1A ChIP-seq was performed in WT, ING5-KO, Set1AΔSET, and dMutant cells. Representative UCSC genome browser tracks depict a loss of ING5 signal in the ING5-KO and dMutant cells at the Sox2 locus. (B) Global occupancy (Left) and log2FC (Right) analyses also confirm loss of ING5 across the genome. (C) Peak calling was performed by MACS2 for ING5 and Set1A, and a 20% overlap was observed, indicating that one-fifth of the identified regions are cooccupied by Set1A and ING5. (D) PantherDB protein class analysis was performed on the genes nearest to the Set1A/ING5 cooccupied regions. The top three categories identified were metabolic genes, nucleic acid–binding proteins, and transcriptional regulators. (E) Genome-wide occupancy of ING5 of Set1A/ING5 cooccupied regions was mapped. (F) log2FC analysis of Set1A/ING5 cooccupied regions and corresponding RNA-seq log2FC indicate a loss of gene expression at ING5/Set1A cooccupied regions when ING5 is deleted with or without the deletion of the SET domain of Set1A. (G) GO analysis of genes annotated by the nearest TSS to ING5/Set1A cooccupied regions shows that differentiation-related genes are significantly up-regulated, whereas stress response genes are down-regulated in dMutant cells. Analysis was performed by Metascape software, and a cutoff of |log2FC| > 1 and P < 0.01 was used for the inclusion of genes in the analysis. Enriched pathways that are statistically significant are shown with corresponding −log10(P) values. (H) ING5 occupancy grouped by DE genes in dMutant versus WT. Clusters respectively show down-regulated versus up-regulated genes, and peaks are centered at the TSS.
ING5 contains a Zn-finger PHD domain, which is responsible for its DNA binding (26). We reasoned that ING5 could be responsible for recruiting Set1A/COMPASS to appropriate developmental gene promoters. However, global analysis of Set1A ChIP-seq data did not show a significant loss of Set1A signal in the ING5-KO or dMutant (SI Appendix, Fig. 4C). We additionally looked at Set1A signal at ING5 peaks as well as ING5 signal at Set1A peaks but did not observe any change in the binding of one transcription factor at the other’s binding site(s) (SI Appendix, Fig. 4 D and E).
Taking a different approach to understand how Set1A and ING5 work together, we performed peak calling for both Set1A and ING5 and looked at the overlapping regions for both transcription factors. A total of 8,106 Set1A/ING5 cooccupying regions were identified, and there was an overall overlap of 20% between Set1A and ING5 ChIP-seq peaks (Fig. 5C). We bioinformatically identified the nearest TSS for these regions and performed pantherDB protein classification analysis on these genes. A significant number of them were nucleic acid–binding proteins, transcriptional regulators, or chromatin-binding proteins (Fig. 5D), suggesting that ING5 and Set1A play an important role in transcriptional regulation of developmental gene expression in ESCs. Indeed, when we ordered our RNA-seq log fold change (logFC) data by the Set1A/ING5 cooccupying coordinates, the loss of both ING5 and Set1A signal correlated strongly with down-regulation of gene expression, and signal loss was the lowest at the promoters of up-regulated genes (Fig. 5 E and F). Most importantly, we observed that the up-regulated genes were primarily enriched for development and morphogenesis (Fig. 5G). Finally, we divided the DE genes in the dMutant into two clusters, up-regulated (dMutant-UP) and down-regulated (dMutant-DN), and assessed Set1A occupancy at these regions (Fig. 5H). As expected, the dMutant-UP genes had the highest Set1A signal in the dMutant, and the dMutant-DN genes had the lowest Set1A signal. Interestingly, the highest Set1A occupancy at dMutant-DN genes was observed in the ING5-KO.
Differentiation-Associated Genes Are Up-Regulated in the dMutant at H3K4me3-Enriched ING5 Clusters.
The C-terminal PHD domain of ING5 is known to interact with H3K4me3 (26). Therefore, we performed H3K4me3 ChIP-seq on WT, Set1AΔSET, ING5-KO, and dMutant cells to investigate the effect the loss of ING5, alone and in combination with Set1AΔSET, has on H3K4me3. While loss of ING5 lead to a decrease in H3K4me3, this decrease in H3K4 methylation was not striking (SI Appendix, Fig. 5 A and B). To evaluate H3K4me3 signal in the context of ING5, we partitioned ING5 ChIP-seq peaks from WT, Set1AΔSET, ING5-KO, and dMutant ESCs into three independent clusters by unbiased k-means clustering (SI Appendix, Fig. 5 C and D). ING5 signal did not vary significantly between the three clusters (SI Appendix, Fig. 5C). However, when the H3K4me3 signal was sorted by the ING5 clusters, we observed that cluster 1 had the lowest H3K4me3 occupancy, while cluster 3 had the highest (SI Appendix, Fig. 5D).
To understand how this correlated with gene expression, we ordered the RNA-seq log2FC data comparing ING5-KO versus WT and dMutant versus WT ESCs according to the three ING5 clusters. We observed that clusters 1 and 2 had primarily down-regulated genes, whereas cluster 3 contained a higher number of up-regulated genes (SI Appendix, Fig. 5D, Right). GO analysis of these three clusters indicated that clusters 2 and 3 were enriched for tissue-specific differentiation-associated genes (SI Appendix, Fig. 5E). In addition, we observed that H3K4me3 and ING5 had similar binding patterns, and >50% of ING5 and H3K4me3 peaks showed overlap (SI Appendix, Fig. 5 F and G). Evidence from the literature indicates that ING5 binding occurs primarily at TSSs of transcribed genes and correlates positively with mRNA expression in epidermal stem cells (27), which is in line with the colocalization with H3K4me3 signal.
Finally, we investigated whether any histone modification changes occurred secondary to the loss of ING5 alone and in combination with Set1AΔSET by performing ChIP-seq for H3K14ac, H3K23ac, H4K5ac, H4K8ac, H4K12ac, and H4K16ac, which are the histone marks known to be deposited by the MOZ/MORF and HBO1 complexes. The epiproteomics data suggested that H3K14ac and H3K23ac levels could be affected by the joint perturbation of SET1A and ING5, with only minimal changes in H4K5ac, H4K8ac, and H4K12ac (Fig. 3D). The ChIP-seq data demonstrated that no significant changes in the global levels were observed for H3K14ac, H4K5ac, or H4K8ac (SI Appendix, Fig. 6). A minimal loss of H4K12ac and H4K16ac was observed. A strong decrease in H3K23ac in the ING5-KO and dMutant was observed by ChIP-seq (SI Appendix, Fig. 6). While changes in these histone modifications could be helpful in identifying whether ING5’s role in SET1A-mediated regulation of pluripotency is driven through MOZ/MORF and HBO1, the modest changes in the majority of the histone modifications suggest that other enzyme-independent functions are also possible for ING5 in the context of SET1A.
ING5-KO ESCs Can Be Efficiently Differentiated into Embryoid Bodies (EBs).
To functionally assess the effect of knocking out ING5 on ESC differentiation, we performed EB differentiation assays with WT, Set1AΔSET, ING5-KO, and dMutant ESCs as previously described (15). To account for clonal effects, two independent genetic lines were assayed for each genotype. As published previously, Set1AΔSET ESCs had defective differentiation and formed smaller, deformed EBs. Conversely, the ING5-KO EBs were significantly larger (27%, P < 0.0001) than their WT counterparts. dMutant EBs were phenotypically similar to Set1AΔSET EBs, suggesting that the Set1AΔSET has the dominant effect in the dMutant (SI Appendix, Fig. 7 A and B).
When differentiation is induced, stem cells down-regulate the four pluripotency factors Oct4, Sox2, Nanog, and Klf4 (28, 29). We had previously shown that this down-regulation is not observed in the Set1AΔSET EBs. We observed that ING5-KO EBs further suppressed these pluripotency factors compared to WT (SI Appendix, Fig. 7C). Accordingly, GO analyses of up- and down-regulated genes in the ING5-KO versus WT showed that the up-regulated genes were primarily associated with differentiation into terminal lineages, whereas the down-regulated genes were associated with pluripotency (SI Appendix, Fig. 7D).
Discussion
The Set1/COMPASS family plays key roles in development. The mouse models for Set1A and Set1B clearly show discrete developmental roles; Set1A is required during the epiblast stage, whereas Set1B becomes essential later for gastrulation (12). In ESCs, we see some redundancy between SET1A/B as far as catalytic functions go. SET1AΔSET is not sufficient to reduce bulk H3K4me3, but SET1AΔSET,SET1B-KO leads to visible loss of bulk H3K4me3 (14). It is clear that there is still much we do not know about the role of not just Set1A but also the other COMPASS family members’ H3K4 methylation–independent functions governed by other domains of the proteins. Some structure–function studies have been conducted with yeast Set1, which might hint at what happens in mammalian systems, although none of these implicate the catalytic domain specifically. The N terminus of yeast SET1 appears to be required for RNA polymerase II (RNAP II) C-terminal domain binding and H3K4 methylation (30), and loss of the N-terminal RNA-recognition motif domain leads to loss of H3K4 methylation (31). Further domain-mapping studies and unbiased approaches are necessary to elucidate the role of the catalytic domain in pluripotency.
As such, using a genome-wide CRISPR/Cas9 negative selection screen, we have identified several plausible genetic dependencies to Set1AΔSET in maintaining ESC viability, among which ING5 was our primary hit. Only a handful of studies to date have implicated the role of Ing5 in various stem cell populations with minimal understanding of the underlying mechanisms pertaining to its function: 1) overexpression of ING5 in glioblastoma stem cells promotes their self-renewal, while ING5 knockdown results in a higher fraction of differentiated cells (32); 2) Ing5 was identified via a focused genetic screen as part of an epigenetic network controlling epidermal stem cell maintenance (27); and 3) ING5–HAT complexes (elaborated further below) have been shown to maintain pluripotency and proliferation of ESCs (33), hematopoietic stem cells (34, 35), and adult neural stem cells (36).
Our study suggests that ING5 may cooperate with Set1A to jointly control ESC viability and self-renewal (Fig. 6). The loss in the fitness of Set1AΔSET ESCs with the depletion of ING5 (Fig. 3) strongly supports this claim. However, the lack of evidence for a functional relationship between the two makes it difficult to dissect the precise mechanism by which this occurs. Below, we discuss possible consequences of the Set1AΔSET-ING5 synthetic perturbation and consider potential roles for ING5 in this relationship.
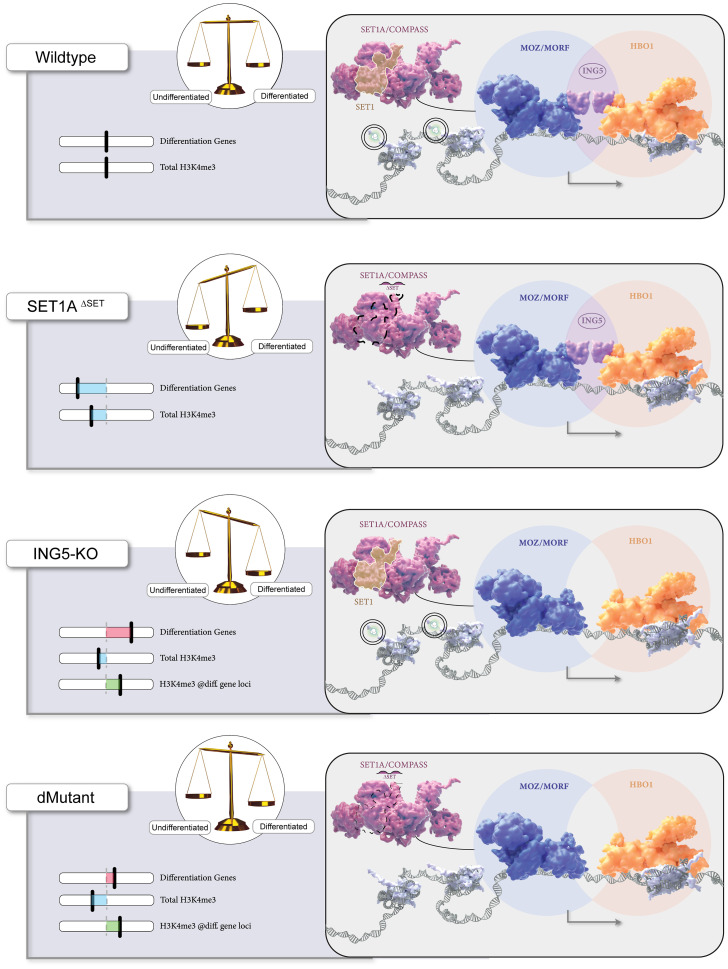
Fig. 6.
Working model. ING5 is a synthetic perturbation of Set1AΔSET and a potential coregulator of ESC function. Precise gene regulatory networks maintain the pluripotent ESC state, directing cells to self-renew or differentiate. The loss of H3K4 methylation due to the deletion of Set1A’s SET domain impairs differentiation of ESCs, whereas the genetic deletion of ING5 alone or in the background of Set1AΔSET leads to up-regulation of differentiation-associated genes with increased H3K4me3 implementation at the promoters of these genes. Taken together, our findings suggest that ING5, via its adapter function in the MOZ/MORF and HBO1 complexes, maintains ESC fitness and self-renewal by cooperating with Set1A in mouse ESCs.
ING5 as a Regulator of Developmental Gene Expression.
Several studies in the literature, including our own, highlight the importance of Set1A in developmental gene expression. Loss of Set1A is incompatible with survival and development; mice harboring the global deletion of it cannot progress past the epiblast stage, and Set1A-KO ESCs cannot be generated (12). The catalytic domain is required for differentiation and activation of the developmental gene expression program; monolayer neuronal differentiation is impaired in Set1AΔSET ESCs, and they are unable to extinguish pluripotency-associated factors and up-regulate mesoderm differentiation genes upon induction of EB differentiation (14).
Our results reported in this study suggest a role for ING5 in the neuronal gene expression program. This is intriguing in the context of a recent publication showing a requirement for Set1A in a neural signature–driven prometastatic memory in oral carcinomas (37). Our findings also show an up-regulation of neural genes upon loss of ING5, and our data suggest that this effect could be occurring synergistically with loss of the Set1A SET domain (Fig. 4E). The extent of this potential cooperativity is difficult to dissect, as ING5 alone also leads to up-regulation of neuronal genes and improved spheroid formation in EB differentiation. Elucidating the molecular mechanisms of this could lead to a better understanding of the many neurodevelopmental disorders associated with Set1A.
We sought to isolate Ing5-specific transcriptomic changes by comparing the DE gene profile of dMutant/Set1AΔSET DEs and that of ING5-KO/WT (Fig. 4 C–E). Ing5-specific DE genes were enriched for chromatin assembly and organization and DNA replication, which precisely fit the functional profile of Ing5 (38, 39). However, since loss of Ing5 by itself does not cause dramatic phenotypic and gene expression changes in ESCs, it is unlikely for Ing5 alone to explain the gene expression and chromatin-binding profile alterations we observe in our datasets. The transcriptomic profile comparison in Fig. 4E also demonstrates the DE genes observed in dMutant/Set1AΔSET but not in ING5-KO/WT. This implies that the genes that are up-regulated/down-regulated are the consequence of the perturbation of both Set1A and Ing5.
Set1A/ING5 cooccupying regions (Fig. 5) and gene expression changes associated with the loss of binding at these regions were assessed to further investigate the joint function of Set1A and ING5. Interestingly, when Set1A signal was ordered by Set1A/ING5 cooccupying regions, the up-regulated genes showed an increase in Set1A signal in ING5-KO versus WT (Fig. 5 F and G). GO analysis of these genes showed that the up-regulated genes were strongly represented in developmental gene categories. These findings suggest that a potential mistargeting of Set1A occurs with the loss of ING5, and this leads to aberrant up-regulation of developmental genes. This effect is not strong enough to cause ING5-KO or dMutant ESCs to spontaneously differentiate. However, ING5-KO ESCs do form larger EBs and suppress the four pluripotency factors robustly (SI Appendix, Fig. 7). Together with the up-regulation of developmental genes in the dMutant cells, these indicate a possible role for ING5 in suppressing differentiation.
ING5 and Histone Acetylation via the MYST Acetyltransferases.
As stated earlier, ING5 is a core component of the MOZ/MORF and HBO1 complexes. MOZ/MORF complexes are believed to acetylate H3K9/K14/K23 at transcriptionally active promoters (38, 40–45), while the HBO1 complex reportedly acetylates H3K14 at promoters and gene bodies (34, 46–49) and H4K5/K8/K12 (20, 38, 50). Native HBO1 complexes can contain either JADE1/2/3 or BRPF1/2/3 scaffold proteins to acetylate either H4 or H3, respectively. It is conceivable that there may be additional HAT subcomplexes to be unveiled. Our findings from the competitive growth assay suggest a possible “MOZ-Jade” complex in regulating ESC viability alongside Set1A (Fig. 3 and SI Appendix, Fig. 3). The reduction in H3K23ac, a mark primarily deposited by the MOZ complex, supports this (SI Appendix, Fig. 6).
Despite residing in similar multimeric complexes, each HAT has a unique role in development as reflected by the different phenotypes observed in mutant mice. Hbo1-null mice are embryonic lethal at E10.5; deleting Hbo1 appears to adversely affect embryonic patterning and organogenesis, especially the development of blood vessels and somites (47). Determining the consequent effects on histone modifications upon targeting Moz (and/or Morf and Hbo1) in Set1AΔSET ESCs may also explain the underlying mechanism of epigenetic regulation. Our preliminary analyses in sgHbo1 and Set1AΔSET sgHbo1 ESCs point toward a cooperation between Set1A and HBO1, as the loss in H3K14ac and H4 acetylation, the primary acetylations implemented by the HBO1 complex, are augmented when the catalytic subunit HBO1 is depleted in the background of Set1AΔSET (SI Appendix, Fig. 8). Hence, both MOZ and HBO1 can be implicated as playing roles in the synthetic perturbation of ING5 in Set1AΔSET ESCs. In either case, detailed biochemical analysis of Set1A, ING5-containing complexes, and their interactors is necessary to understand the precise molecular mechanisms of these pathways and is among the future directions of this study.
ING5 in DNA Damage Response and Proliferation.
Since ESCs propagate quickly and indefinitely, it becomes imperative that they maintain genomic integrity to protect self-renewal and that continual DNA replication does not induce spurious differentiation (51, 52). Recently, several studies have demonstrated Set1A’s role in orchestrating the DNA damage response and repair pathway to prevent genome instability, especially during replicative stress (53–56). ING5 and its associated complexes also participate in DNA replication. In fact, HBO1 was first discovered via its interaction with ORC1 and MCM helicases, which are involved in the prereplication complex (57, 58). Additional studies have linked the function of ING5 and its associated complexes to p53 signaling, including physically interacting with p53 to activate p53-downstream targets (e.g., p21) in response to DNA damage (22, 59, 60). ING5 has also been reported to regulate cell proliferation in a p53-independent manner (61). Given the current literature, it is reasonable to extrapolate that Set1A/COMPASS cooperates with ING5-associated complexes to maintain ESC self-renewal via mechanisms involving regulating the DNA damage response. DNA damage response was also shown to be the primarily enriched pathway in the GO analysis of the initial CRISPR screen (Fig. 2). Furthermore, cross-talk between the two families of chromatin-modifying complexes have been previously described; Mll1/COMPASS and MOZ cooperate to regulate Hox genes in human cord blood cells (62), and recruitment of HBO1 by Mll1 regulates the HoxA gene cluster in leukemic stem cells (LSCs) (34). Interestingly, two independent studies have also implicated the involvement of HBO1 in transcriptional elongation (34, 63); in particular, H3K14ac deposition by HBO1 may facilitate RNAP II processivity throughout the coding regions of LSC genes (34). Since studies have shown that yeast Set1, homolog to mammalian Set1A and Set1B, could be recruited to chromatin by associating with elongating RNAP II (64–66), it is also therefore possible that the connection between Set1A/COMPASS and Ing5-related complexes is via the RNAP II elongation complex.
In conclusion, we present here initial findings and speculation on a previously unrecognized potential functional interaction between ING5 and Set1A/COMPASS in ESCs, showcasing a sophisticated relationship among different families of epigenetic modifiers in mediating self-renewal. Loss of ING5 in ESCs leads to up-regulation of differentiation-associated genes but is not sufficient to induce differentiation. The loss of H3K23ac in the ING5-KO and dMutant as well as the decrease in fitness of MOZ-depleted Set1AΔSET-GFP cells point toward the MOZ complex as a potential downstream effector. However, this requires validation, as does the role of ING5 in DNA damage response in the context of stem cells. Of course, these are not mutually exclusive, and all require further investigation to understand the relationship between Set1A and ING5. Clearly, much remains to be investigated about these underlying epigenetic mechanisms in governing ESC pluripotency. Such molecular insights would be highly applicable for understanding the behavior of cancer stem cells, given their shared characteristics with ESCs as well as stem cell reprogramming, which would greatly advance the field of regenerative medicine.
Materials and Methods
Generation of SETless Mice, Screening, and Genotyping.
SETless mutant C57BL/6 mice were generated via pronuclear injection of CRISPR sgRNAs to the SET domain of Set1A (sgRNA sequences are provided in SI Appendix, Table 1) with the assistance of the Northwestern University Transgenic and Targeted Mutagenesis Laboratory (TTML). The resulting F0 founder mice were genotyped using PCR and NGS to identify the F0 mice harboring the SETless mutation. In brief, genomic DNA was extracted from tail snips (provided by TTML) of resulting F0 mice. We designed NGS primers that amplify the intended sgRNA target region of Set1A to include Illumina adaptor sequences, a staggering length sequence, and an 8-bp barcode for multiplexing of different F0 samples: forward primer, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT (1- to 9-bp staggering length sequence) GGAAGAAGAAACTCCGATTTGG-3′, and reverse primer, 5′-CAAGCAGAAGACGGCATACGAGAT (unique 8-bp barcode) GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT ACCATCTCATCAGCGGCAAT-3′.
For each F0 sample, we used Phusion high-fidelity DNA polymerase (NEB, M0530) for PCR amplification (35 cycles) and then combined the resulting amplicons across samples. Pooled PCR reactions were precipitated using isopropanol, gel extracted, and sequenced using a NextSeq 500 sequencing platform (Illumina). Raw BCL (basecall) output files were converted into fastq files using bcl2fastq (Illumina, version 2.17.1.14), followed by quality trimming using Trimmomatic (67). Trimmed reads were then aligned to the mouse genome (University of California Santa Cruz [UCSC] mm9) using Burrows–Wheeler aligner (68). Output binary alignment map files were converted into SAM (sequence alignment map) files using SAMtools (69), from which CIGAR strings were retrieved for each F0 sample and subsequently analyzed to determine which F0 mouse harbored an intended mutation in the Set1A SET domain. One F0 mouse harboring a two-nucleotide insertion at the guide RNA (gRNA) cut site located at the start of the SET domain of Set1A was identified and subsequently crossed with WT female C57BL/6 mice, and the resulting offspring validated the germline transmission of the mutant allele. Heterozygous breeding was used to establish and maintain the mouse colony, and heterozygous intercrosses were carried out to obtain progeny with homozygous Set1AΔSET mutation. Developmentally staged embryos from heterozygous intercrosses were dissected, genotyped, and characterized for developmental deformities. For mouse genotyping after colony establishment, mice were ear notched, and the ear notch biopsies were used for genotyping following a previously published protocol (70). Genotyping primers are listed in SI Appendix, Table 1.
Genome-Wide CRISPR/Cas9 Dropout Screen.
The Brie mouse library (71) was purchased from Addgene. This library targets each of the 19,674 mouse genes with ∼4 sgRNAs per gene plus 1,000 NTC sgRNAs (71). Brie library amplification, lentiviral production, multiplicity of infection (MOI) determination, and transduction were performed as previously described (71). In brief, 3 × 107 WT V6.5 and Set1AΔSET ESCs stably expressing Cas9 (denoted as WT-Cas9 and Set1AΔSET-Cas9, respectively) were transduced with the Brie library at an MOI of <0.3. Twenty-four hours after transduction, infected cells were treated with puromycin (2 µg/mL, Life Technologies), and 5.5 × 107 cells were pelleted and snap-frozen 2 d later (day 3). Remaining cells were passaged every 2 d for an additional 18 d, during which at least 3 × 107 cells were maintained per passage to ensure adequate sgRNA representation. On day 21, 5.5 × 107 cells were pelleted and snap-frozen. Four replicates of this 21-d dropout screen were performed. As described previously (72), genomic DNA was extracted from pelleted cells collected on day 3 and day 21, which serve as the initial and terminal populations of transduced cells, respectively, and the sgRNA library was amplified from the extracted DNA by PCR with primers containing adaptors for Illumina sequencing. Deep sequencing on a NextSeq 500 sequencing system followed by statistical analyses were used to analyze sgRNA library composition in the initial and terminal populations.
For the analysis of the CRISPR screen data, count tables and sgRNA rankings were generated using the MAGeCK algorithm, as previously described (73). Following this, we performed additional filtering by fold change. Each sgRNA with a log2FC of >−1 in the terminal population compared to the initial population was considered as depleted, and the total number of depleted sgRNAs was computed. Essential genes, such as Pcna and Ctcf, were depleted in both WT and Set1AΔSET cells and were filtered out and excluded from further dropout candidate identification analyses. An initial analysis revealed 1,425 targets depleted only in Set1AΔSET cells that had at least two sgRNAs depleted out of the four sgRNAs represented in the Brie library (Fig. 2B). To identify the most essential genetic dependencies of Set1AΔSET ESCs, each target candidate was assigned a DDS, which reflects both the degree and consistency of dropout across multiple replicates of the 21-d screen, and was subsequently ranked. The DDS score was calculated as follows:
where Wi = 1 if there are at least two sgRNAs of a gene that meet the criteria −log2FCSet1AΔSET > 1 and −log2FCWT < 1; otherwise, Wi = 0.
For the final ranking, the DDS score was averaged across all four replicates. A cutoff of 0.25 was used for the average DDS in order to designate a target as depleted. Based on this, six targets were found to be depleted in at least three out of four replicates (Fig. 2C). Of the 1,425 ranked targets, 139 genes had at least three out of four sgRNAs depleted in a minimum of two replicates, from which 39 genes were selected for further validation based on their role in DNA damage response and/or chromatin modification (SI Appendix, Fig. 2).
Competitive Growth Assay for Secondary Validation of Screen Hits.
Select targets resulting in perturbed Set1AΔSET proliferation determined by alkaline-phosphatase staining were subjected to further validation using a cell competition assay. For the cell competition assay, we first labeled WT-Cas9 and Set1AΔSET-Cas9 ESCs with mCherry and eGFP. The plasmid for expressing mCherry in ESCs was purchased from Addgene (120426) (74), into which we cloned the eGFP transgene in place of mCherry to generate the eGFP-expressing plasmid. mCherry-labeled WT-Cas9 cells and eGFP-labeled Set1AΔSET-Cas9 cells were mixed at a 1:1 ratio and seeded 12 h before gRNA lentivirus transduction. Twenty-four hours after transduction (day 1), infected cells were selected with puromycin. The percentages of mCherry+ versus eGFP+ cells per gRNA perturbation were measured between day 3 and day 21 after transduction every 2 d via flow cytometry using the BD FACSAria II. At least two replicates of the 21-d cell competition assay were performed as part of the target validation process. At least two sgRNAs were used per target. Flow cytometry analyses, including gating, were performed on FlowJo v10.6.2.
Mass Spectrometry.
Mass spectrometry was performed in collaboration with the Northwestern University Excellence in Proteomics Core. Three replicates were submitted per genotype. For each sample, 5 × 106 ESCs were harvested, snap frozen in liquid nitrogen, and stored at −80 °C until submission. Upon submission to the Proteomics Core, nuclei were isolated from 100% of the material provided. Histones were acid extracted from 100% of nuclei, and 100% of each sample was derivatized via propionylation reaction and digested with trypsin as previously described (75). Each sample was resuspended in 50 µL of 0.1% TFA/mH2O (trifluoracetic acid) and 3 µL was injected with three technical replicates. For data analysis, the percent abundance values were calculated for each histone modification and normalized to the percent abundance of its unmodified form and the nontarget sgRNA control (AAVS1) for all three replicates. An ordinary two-way ANOVA test with main effects only was performed for significance testing. For comparisons between individual groups, Tukey hypothesis testing for multiple comparisons was conducted, and the multiplicity adjusted P value was reported for each comparison. All statistical analyses were performed using GraphPad Prism 9.0.1 software.
Supplementary Material
Acknowledgments
Research in the A.S. laboratory is supported by NIH grant R35CA197569 to A.S. C.C.S. was supported by the NIH/National Cancer Institute (NCI) Predoctoral to Postdoctoral Transition Award (Grant 5F99CA234945-02). K.C. was supported by the NIH Pathway to Independence Award from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Grant K99HD094906). We thank Dr. Lihua Zou for the initial analysis of the CRISPR screen data and designing the parameters for differential dependency score calculation. We would also like to thank Oliver Vickman for technical assistance. We would additionally like to thank Stacy Marshall for the preparation and running of NGS samples. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center, instrumentation award (S10OD025194) from NIH Office of Director, and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. We would additionally like to thank Drs. Jeannie Camarillo and Neil Kelleher for their assistance in the design and analysis of the mass spectrometry experiments. The genetically engineered mice were generated with the assistance of Northwestern University TTML. The Northwestern University TTML is partially supported by NIH grant CA60553 to the Robert H. Lurie Comprehensive Cancer Center at Northwestern University. We thank Nicole Ethen for providing the illustrations in Figs. 2A and 3A and Brianna Monroe for providing the illustrations in Fig. 6. Finally, we would like to acknowledge Drs. Marc Morgan and Edwin Smith and all the members of the A.S. laboratory for their insights and suggestions on improving our study and manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118385119/-/DCSupplemental.
Data Availability
NGS data have been deposited to the Gene Expression Omnibus database under accession number GSE196946 and are publicly available. All other study data are included in the article and/or SI Appendix.
References
- 1.Boland M. J., Nazor K. L., Loring J. F., Epigenetic regulation of pluripotency and differentiation. Circ. Res. 115, 311–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Los Angeles A., et al. , Hallmarks of pluripotency. Nature 525, 469–478 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Fisher C. L., Fisher A. G., Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr. Opin. Genet. Dev. 21, 140–146 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Krogan N. J., et al. , COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277, 10753–10755 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Miller T., et al. , COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U.S.A. 98, 12902–12907 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roguev A., et al. , The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137–7148 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider J., et al. , Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 19, 849–856 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Wood A., et al. , Ctk complex-mediated regulation of histone methylation by COMPASS. Mol. Cell. Biol. 27, 709–720 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J. H., Skalnik D. G., CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280, 41725–41731 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Lee J. H., Tate C. M., You J. S., Skalnik D. G., Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 282, 13419–13428 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Cenik B. K., Shilatifard A., COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 22, 38–58 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Bledau A. S., et al. , The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 141, 1022–1035 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Fang L., et al. , H3K4 methyltransferase Set1a is a key Oct4 coactivator essential for generation of Oct4 positive inner cell mass. Stem Cells 34, 565–580 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Sze C. C., et al. , Coordinated regulation of cellular identity-associated H3K4me3 breadth by the COMPASS family. Sci. Adv. 6, eaaz4764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sze C. C., et al. , Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev. 31, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lalonde M. E., Cheng X., Côté J., Histone target selection within chromatin: An exemplary case of teamwork. Genes Dev. 28, 1029–1041 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X., et al. , Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 12, 177–185 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., et al. , Structure of the neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111, 117–127 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avvakumov N., Côté J., The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 26, 5395–5407 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Lalonde M. E., et al. , Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 27, 2009–2024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ormaza G., et al. , The tumor suppressor ING5 is a dimeric, bivalent recognition molecule of the histone H3K4me3 mark. J. Mol. Biol. 431, 2298–2319 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Shiseki M., et al. , p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 63, 2373–2378 (2003). [PubMed] [Google Scholar]
- 23.LaFave L. M., et al. , Loss of BAP1 function leads to EZH2-dependent transformation. Nat. Med. 21, 1344–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y., et al. , Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc. Natl. Acad. Sci. U.S.A. 109, 13549–13554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y., Thomas P. M., Kelleher N. L., Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat. Commun. 4, 2203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Champagne K. S., et al. , The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins 72, 1371–1376 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder K. W., et al. , Diverse epigenetic strategies interact to control epidermal differentiation. Nat. Cell Biol. 14, 753–763 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Ying Q. L., et al. , The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Bae H. J., et al. , The Set1 N-terminal domain and Swd2 interact with RNA polymerase II CTD to recruit COMPASS. Nat. Commun. 11, 2181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fingerman I. M., Wu C. L., Wilson B. D., Briggs S. D., Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J. Biol. Chem. 280, 28761–28765 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F., et al. , ING5 activity in self-renewal of glioblastoma stem cells via calcium and follicle stimulating hormone pathways. Oncogene 37, 286–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M. S., et al. , The histone acetyltransferase Myst2 regulates Nanog expression, and is involved in maintaining pluripotency and self-renewal of embryonic stem cells. FEBS Lett. 589, 941–950 (2015). [DOI] [PubMed] [Google Scholar]
- 34.MacPherson L., et al. , HBO1 is required for the maintenance of leukaemia stem cells. Nature 577, 266–270 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Perez-Campo F. M., Borrow J., Kouskoff V., Lacaud G., The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood 113, 4866–4874 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merson T. D., et al. , The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 26, 11359–11370 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascual G., et al. , Dietary palmitic acid promotes a prometastatic memory via Schwann cells. Nature 599, 485–490 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Doyon Y., et al. , ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21, 51–64 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Russell M., Berardi P., Gong W., Riabowol K., Grow-ING, age-ING and die-ING: ING proteins link cancer, senescence and apoptosis. Exp. Cell Res. 312, 951–961 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Klein B. J., et al. , Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nat. Commun. 10, 4724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lv D., et al. , Histone acetyltransferase KAT6A upregulates PI3K/AKT signaling through TRIM24 binding. Cancer Res. 77, 6190–6201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Y., et al. , Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 26, 1376–1391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simó-Riudalbas L., et al. , KAT6B is a tumor suppressor histone H3 lysine 23 acetyltransferase undergoing genomic loss in small cell lung cancer. Cancer Res. 75, 3936–3945 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Voss A. K., Collin C., Dixon M. P., Thomas T., Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev. Cell 17, 674–686 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Voss A. K., et al. , MOZ regulates the Tbx1 locus, and Moz mutation partially phenocopies DiGeorge syndrome. Dev. Cell 23, 652–663 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y., et al. , BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 35, 176–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kueh A. J., Dixon M. P., Voss A. K., Thomas T., HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 31, 845–860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishima Y., et al. , The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 118, 2443–2453 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Newman D. M., Voss A. K., Thomas T., Allan R. S., Essential role for the histone acetyltransferase KAT7 in T cell development, fitness, and survival. J. Leukoc. Biol. 101, 887–892 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Foy R. L., et al. , Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J. Biol. Chem. 283, 28817–28826 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su J., et al. , Genomic integrity safeguards self-renewal in embryonic stem cells. Cell Rep. 28, 1400–1409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vitale I., Manic G., De Maria R., Kroemer G., Galluzzi L., DNA damage in stem cells. Mol. Cell 66, 306–319 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Arndt K., et al. , SETD1A protects HSCs from activation-induced functional decline in vivo. Blood 131, 1311–1324 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Higgs M. R., et al. , Histone methylation by SETD1A protects nascent DNA through the nucleosome chaperone activity of FANCD2. Mol. Cell 71, 25–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoshii T., et al. , A non-catalytic function of SETD1A regulates cyclin K and the DNA damage response. Cell 172, 1007–1021.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tajima K., et al. , SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat. Commun. 6, 8257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burke T. W., Cook J. G., Asano M., Nevins J. R., Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276, 15397–15408 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Iizuka M., Stillman B., Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274, 23027–23034 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Avvakumov N., et al. , Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol. Cell. Biol. 32, 689–703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu N., et al. , ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 73, 3749–3760 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Linzen U., et al. , ING5 is phosphorylated by CDK2 and controls cell proliferation independently of p53. PLoS One 10, e0123736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paggetti J., et al. , Crosstalk between leukemia-associated proteins MOZ and MLL regulates HOX gene expression in human cord blood CD34+ cells. Oncogene 29, 5019–5031 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Saksouk N., et al. , HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 33, 257–265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krogan N. J., et al. , The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Lee J. H., Skalnik D. G., Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol. Cell. Biol. 28, 609–618 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng H. H., Robert F., Young R. A., Struhl K., Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H., et al. , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Truett G. E., et al. , Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29, 52, 54 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joung J., et al. , Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W., et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parekh U., et al. , Mapping cellular reprogramming via pooled overexpression screens with paired fitness and single-cell RNA-sequencing readout. Cell Syst. 7, 548–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia B. A., et al. , Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2, 933–938 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data have been deposited to the Gene Expression Omnibus database under accession number GSE196946 and are publicly available. All other study data are included in the article and/or SI Appendix.