Abstract
Background
Multiple lines of evidence have demonstrated that circular RNAs (circRNAs) play oncogenic or tumor-suppressive roles in various human cancers. Nevertheless, the biological functions of circRNAs in small cell lung cancer (SCLC) are still elusive.
Methods
CircVAPA (annotated as hsa_circ_0006990) was identified by mining the circRNA profiling dataset of six paired SCLC tissues and the RNA-seq data of serum samples from 36 SCLC patients and 118 healthy controls. The circVAPA expression level was evaluated using quantitative real-time PCR in SCLC cells and tissues. Cell viability, colony formation, cell cycle and apoptosis analysis assays and in vivo tumorigenesis were used to reveal the biological roles of circVAPA. The underlying mechanism of circVAPA was investigated by Western blot, RNA pulldown, RNA immunoprecipitation, dual-luciferase reporter assay and rescue experiments.
Results
We revealed that circVAPA, derived from exons 2-4 of the vesicle-associated membrane protein-associated protein A (VAPA) gene, exhibited higher expression levels in SCLC cell lines, clinical tissues, and serum from SCLC patients than the controls, and facilitated SCLC progression in vitro and in vivo. Mechanistically, circVAPA activated the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway by modulating the miR-377-3p and miR-494-3p/insulin-like growth factor 1 receptor (IGF1R) axis to accelerate SCLC progression. Furthermore, circVAPA depletion markedly enhanced the inhibitory effects of BMS-536924, an IGF1R kinase inhibitor in cellular and xenograft mouse models.
Conclusions
CircVAPA promotes SCLC progression via the miR-377-3p and miR-494-3p/IGF1R/AKT axis. We hope to develop clinical protocols of combinations of circVAPA inhibition and BMS-536924 addition for treating SCLC with circVAPA upregulation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-022-01595-9.
Keywords: CircVAPA, SCLC, Progression, miR-377-3p, miR-494-3p, IGF1R, AKT, BMS-536924
Background
Small cell lung cancer (SCLC) is a highly aggressive and lethal subtype of lung cancer, characterized by bi-allelic inactivation of tumor suppressors RB1 and p53 [1–3]. Despite the significant advances achieved in chemotherapy, SCLC patients almost recur within one year, resulting in a quite poor prognosis and its 5-year survival rate less than 7% [4–7]. Therefore, it is urgent to elucidate the underlying mechanisms of SCLC progression and identify the potential biomarkers and therapeutic targets in SCLC.
Circular RNAs (circRNAs) are back-spliced from precursor mRNA (pre-mRNA) formed with exons or introns, in which a downstream splice site is joined with an upstream splice site [8–12]. Reverse complementary sequences such as Alu elements in the flanking region of circularized exons and RNA binding proteins (RBPs) are the major factors contributing to circRNA biogenesis [13–17]. Due to its covalent closed loop structure and lack of exposed terminal ends, circRNAs are resistant to the degradation by exonucleases and highly stable in blood and other body fluids [10, 18]. Accumulating evidence has demonstrated that the dysregulated circRNAs are associated with specific hallmarks of cancer, including sustaining proliferative signaling, evading growth suppressors, activating invasion, and metastasis [10, 18–25]. For instance, circPTPRA acts as a competitive endogenous RNA (ceRNA) against miR-96-5p to erase the repressive role of RASSF8, consequently suppressing epithelial-mesenchymal transition and metastasis of non-small cell lung carcinoma (NSCLC) [22]. CircURI1 behaves as a decoy of heterogeneous nuclear ribonucleoprotein M (hnRNPM) to modulate alternative splicing of VEGFA, thereby inhibiting gastric cancer metastasis [20]. Several circRNAs with translational effects are also involved in tumorigenesis [10, 18, 25]. For example, circular RNA E-cadherin undergoes translation to encode a unique E-cadherin protein variant (C-E-Cad), promoting glioblastoma tumorigenicity by activating the EGFR–STAT3 signaling [25]. However, the biological functions and underlying mechanisms of circRNAs in SCLC occurrence and development remain unclear.
The insulin-like growth factor 1 receptor (IGF1R) exerts essential functions in transmitting signals through the IGF1/IGF1R signaling axis-mediated pathway, which belongs to the tyrosine kinase receptor family [26–28]. Aberrant regulation of the IGF1R signaling pathway has been recognized as a well-established therapeutic target in a variety of human malignancies, including SCLC [29, 30]. BMS-536924, an ATP-competitive IGF1R inhibitor, can suppress the IGF1R-induced phosphorylation of phosphoinositide 3-kinase (PI3K)/ protein kinase B (AKT), consequently repressing cancer cell proliferation and differentiation [31–34].
To clarify the functions of circRNAs in SCLC tumorigenesis, bioinformatics analysis of differentially expressed circRNAs in SCLC tissues and serum from SCLC patients identified circVAPA as an up-regulated circular RNA. Loss- and gain- of function experiments revealed that circVAPA accelerated cell cycle progression, cell proliferation, and colony formation in SCLC. Mechanically, we found that circVAPA served as a ceRNA against miR-377-3p and miR-494-3p to weaken their inhibitory effects on IGF1R mRNA expression, thus promoting SCLC progression by activating the PI3K/AKT pathway.
Materials and methods
Clinical SCLC samples
All SCLC clinical samples and the corresponding non-tumor tissues were collected from Anhui Provincial Hospital. Written informed consent was obtained from all patients for this study. All fresh samples were immediately frozen in liquid nitrogen after removing from the operation and stored in liquid nitrogen for further investigation.
Cell lines and reagents
All human SCLC cell lines (H69, DMS79, H82, DMS273, H446, and H526) and non-small cell lung cancer (NSCLC) cell lines (HCC827 and PC9) were maintained under standard culture conditions with RPMI-1640 media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PS) at 37 °C in a humidified atmosphere with 5% CO2. BMS-536924 was purchased from Glpbio (Cat NO. GC17773) and kept as a stock solution of 100 nM in dimethyl sulfoxide (DMSO).
RNA preparation and quantitative real-time PCR
The nuclear and cytoplasmic fractions were isolated as described previously [20]. Total RNA from whole-cell lysates or the cytoplasmic and nuclear fractions were extracted using TRIzol (Thermo Scientific) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche). 18S rRNA was used for miRNAs template normalization, and β-actin was used as an internal standard for circVAPA and mRNAs. Real-time quantitative PCR (RT-qPCR) was performed using ChamQ SYBR qPCR Master Mix (Vazyme) on a Roche Light Cycler 96 Real-Time PCR System. Oligonucleotide sequences for primers using in RT-qPCR were listed in Table S5.
Fluorescence in situ hybridization (FISH)
The Cy3-labeled probe against the back-spliced junction in circVAPA was synthesized by RiboBio (Guangzhou, China). FISH was performed using a FISH kit (RiboBio) following the manufacturer's guidelines. The images were visualized using an Olympus SpinSR10 confocal microscope.
Quantification of RNA copy number per cell
Quantification of RNA copy number per cell was carried out as previously described with some modifications [35]. The DNA fragments corresponding to circVAPA, IGF1R, miR-377-3p and miR-494-3p were amplified with cDNA and the amount of the purified product were used to plot standard curves through real-time PCR. R squares for Spearman’s correlation coefficient, and P values were calculated by Spearman’s correlation test. Total RNA was extracted from 105 DMS273 and H82 cells, respectively, and cDNA was subsequently synthesized. The copy numbers per cell in each cell line were calculated based on the specific number of cells and the Ct value using the standard curve.
Plasmid construction and cell transfection
The short hairpin RNA (shRNA) oligonucleotides targeting the junction site of circVAPA were inserted into the pLKO.1 vector (Sigma). Then the constructs were packaged into lentivirus, which were used to infect SCLC cells. The cells were subsequently selected using puromycin resistance for one week. The surviving cells were regarded as stable circVAPA knockdown cells. For circVAPA overexpression, the second, third and fourth exons of VAPA gene and the endogenous flanking sequence including the complementary Alu element pairs were inserted into the backbone vector of pcDNA3. Transfection was carried out using Effectene Transfection Reagent (QIAGEN) according to the manufacturer’s protocol. The circVAPA level was assessed using RT-qPCR. Oligonucleotide sequences for primers used in plasmid construction, short interfering RNAs (siRNAs), and shRNA were listed in Table S5. The pGL-3 basic luciferase reporter vector and pRL-TK renilla luciferase vector were purchased from Promega.
Cell viability assay
SCLC cells (3 × 103/well) were seeded into 96-well plates. After transfection or drug treatment for 48 h, these cells were analyzed using the CellTiter-Glo luminescent assay according to the manufacturer’s instructions [36]. The multi-label plate reader (Envision PerkinElmer) was used to detect the luminescence signals.
Cell cycle and apoptosis analysis
SCLC cells were cultured in 6-well plates at a concentration of 2 × 105 cells per well. For cell cycle analysis, 48 h after transfection, SCLC cells were fixed in 80% ethanol at -20 °C overnight, followed by the staining with PI/RNase staining buffer (BD Biosciences). Cells were measured for cell cycle distribution by flow cytometry, and the cell-cycle profiles were further analyzed using ModFit software (Verity Software House). For cell apoptosis assay, apoptotic cells were determined as previously described [36]. Cells were stained with FITC Annexin V and PI using the FITC Annexin V Apoptosis Detection Kit (BD Pharmingen) according to the manufacturer’s instructions.
Cell colony formation assay
For adherent cells, 1.5 × 103 DMS273 cells 48 h after transfection were plated into 6-well plates in triplicates for each condition and then cultured for 3 weeks. Then the colonies were fixed with methanol for 30 min, followed by staining with 1.5% crystal violet for 10 min at room temperature. For suspension cells, 48 h after transfection, 103 cells in 1 ml of RPMI1640 containing 10% (v/v) FBS and 0.33% (w/v) agarose were overlaid onto bottom agar consisting of 1 ml of RPMI-1640 containing 10% (v/v) FBS and 0.5% (w/v) agarose in a 6-well culture plate. Then the cells were cultured for 3 weeks. The colonies were photographed and analyzed by Image J.
Western blot analysis and antibodies
Western blot analysis was performed as described previously [36]. The following antibodies were used in this study: anti-AKT (CST, #9272), anti-phosphorylated-AKT (Ser-473) (CST, #9271), anti-IGF1R (Proteintech, #20,254-1-AP), anti-phosphorylated-S6 Ribosomal Protein (Ser235/236) (CST, #4858), anti-S6 Ribosomal Protein (5G10) (CST, #2217), anti-PARP (CST, #9542), anti-p21 (Proteintech, 10,355-1-AP), anti-β-actin (TransGen, HC201).
RNA pull-down assay with a biotinylated circVAPA probe
RNA pull-down was conducted as previously described [35]. Briefly, we designed a biotin-labeled 30nt probe against the back-spliced junction of circVAPA to specifically pull down circVAPA and its intracellular RNA-RNA complex. A biotin-labeled probe with scrambled sequence was set as a negative control. 107 cells were cross-linked in ice-cold PBS buffer with 1% formaldehyde for 10 min. Upon PBS buffer removal, these cells were lysed in RNA immunoprecipitation (RIP) buffer on ice for 30 min. After sonication, the cell supernatant was harvested and divided into two equal parts for subsequent RNA pull-down after centrifugation. The biotin-labeled and control probes were incubated with the respective cell lysate for 4 h at 4 °C with gentle rotation. Identical blocked M280 Streptavidin magnetic Dynabeads (Invitrogen) were added to the above lysates and further rotated for 4 h at 4 °C. After washing with RIP buffer and RIP buffer supplemented with 500 mM NaCl, the bound RNA was isolated using TRIzol and used for RNA detection by RT-qPCR assay.
RNA immunoprecipitation
107 cells were cross-linked in ice-cold PBS buffer with 1% formaldehyde for 10 min. Then they were harvested and lysed in RIP lysis buffer, incubated with Dynabeads protein G (Invitrogen) conjugated with anti-IgG (CST, #2729) or anti-AGO2 (Sigma, SAB4200085), and rotated at 4 °C overnight. The immunoprecipitated RNAs were extracted by TRIzol reagent and further detected by RT-qPCR with specific primers.
Dual-luciferase reporter assay
The recombinant luciferase reporter plasmids were inserted with the potential miR-377-3p and miR-494-3p binding site sequences in circVAPA and the 3’ UTR of IGF1R. When the 293T cells grew to 80% confluency, the cells were co-transfected with miR-377-3p and miR-494-3p mimics or inhibitors, luciferase reporter plasmids (circVAPA-WT, circVAPA-Mut, IGF1R-WT, IGF1R-Mut), and pRL-TK renilla luciferase vector. Activities of firefly luciferase (FL) and renilla luciferase (RL) were measured after transfection for 48 h using the Dual-Luciferase® Reporter Assay System, and the relative ratio of the FL/RL was used to reveal the interactions between miR-377-3p, miR-494-3p and circVAPA, IGF1R.
Small cell lung cancer xenograft mouse models
Animal experiments were carried out according to a protocol approved by the Ethics Committee of Hefei Institutes of Physical Science, China Academy of Sciences [36]. Four-week-old nude mice were injected subcutaneously with 107 stably transfected sh-circVAPA or sh-NC in DMS273 cells suspended in an equal volume of Matrigel on the both left and right flanks (n = 5 per group). When tumors volume reached 100-200 mm3, 0.5% carboxymethyl cellulose sodium (CMC-Na) or BMS-536924 (100 mg/kg) was administered daily by gavage for 16 consecutive days. The width and length of the tumor were measured every day for 5 weeks, and the tumor size was calculated according to the formula: volume (mm3) = (length × width2)/2. Thirty days after injection, mice were sacrificed, and tumors were harvested.
Histological and immunohistochemical analyses
3 paired clinical SCLC tissues and harvested xenograft tumor tissues were fixed in 4% paraformaldehyde for 24 h and embedded in paraffin. The embedded tissues were cut into 4 μm thick sections and then processed for hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) staining (anti-IGF1R, CST, #14,534; anti-phosphorylated-AKT, Affinity, #AF0016; anti-Ki67, CST, #12,202).
CircRNA RNase R and Actinomycin D treatments
For RNase R treatment, 2 μg total RNA was incubated with or without 20 U RNase R at 37 °C for 30 min. For actinomycin D treatment, 2 mg/ml actinomycin D was added to the culture medium to block RNA transcription at indicated time points. After treatment, RT-qPCR was used to assess the expression levels of circVAPA and VAPA mRNA.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism 6.0.1 (GraphPad Software). Data were listed mean ± SD of at least three independent experiments. Differences between groups were analyzed using Student’s t-test. A P-value of < 0.05 was considered to be significant.
Results
Identification of circVAPA in SCLC
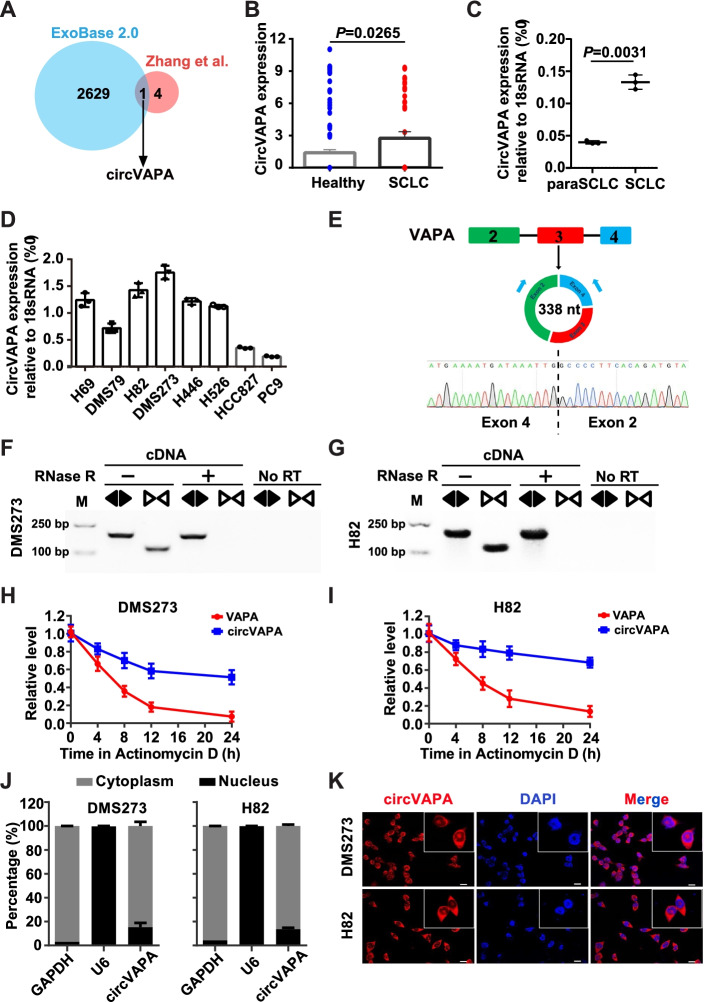
Integrative analysis of the previously reported upregulated circRNAs based on circRNA profiling of six paired SCLC tissues and the RNA-seq data of serum samples from 36 SCLC patients and 118 healthy controls [37, 38] identified circVAPA as a significantly upregulated circRNA (Fig. 1A, B and Table S1, S2, S3). Then, we analyzed the expression of circVAPA in both lung cancer cell lines and human primary SCLC tissues. The endogenous circVAPA was significantly elevated in 3-paired SCLC tissues compared to the corresponding non-tumor controls (paraSCLC) (Fig. 1C). Similarly, circVAPA exhibited remarkably higher expressions in SCLC cell lines than NSCLC cell lines (Fig. 1D), suggesting that circVAPA is a significantly upregulated circRNA in SCLC meriting further investigation. Two SCLC cell lines (DMS273 and H82) were chosen for subsequent experiments due to the highest circVAPA expression levels among six SCLC cell lines (Fig. 1D). CircVAPA (annotated as hsa_circ_0006990 in circBase (http://www.circbase.org/) with 338 nucleotides (nt) in length, was back-spliced from exon 2-4 of the VAMP-associated protein A (VAPA) gene (Fig. 1E), which is located on human chromosome 18p11.22 [39, 40]. The putative back-spliced junction fragment of circVAPA was verified by PCR amplification with divergent primers from complementary DNA (cDNA) of SCLC cell lines and confirmed by Sanger sequencing (Fig. 1E). RNase R exonuclease assay examined by RT-qPCR verified that circVAPA was resistant to digestion (Fig. 1F, G, and Fig. S1A), consistent with the characteristics of circRNAs [41, 42]. To further evaluate the stability of circVAPA, actinomycin D (an inhibitor of transcription) treatment assay revealed that circVAPA was more stable than VAPA mRNA in SCLC cells (Fig. 1H, I). The function of non-coding RNA, including circRNA, is tightly and closely related to its subcellular location pattern [9, 11, 43]. FISH assay with a probe targeting the back-spliced junction of circVAPA and RT-qPCR analysis of nuclear and cytoplasmic RNAs revealed the predominately cytoplasmic enrichment of circVAPA in both DMS273 and H82 cells (Fig. 1J, K). Finally, ~ 770 and ~ 500 circVAPA copies per cell were determined in DMS273 and H82 cells, respectively (Fig. S1B, C). Taking above results together, our findings demonstrated that circVAPA was a cytoplasmic circRNA and upregulated in SCLC.
Fig. 1.
Expression and characterization of circVAPA in SCLC. a Venn diagram revealing the overlap of upregulated circRNAs based on circRNA profiling of SCLC tissues from Zhang et al. [38] and the RNA-seq data of serum samples of SCLC from exoRBase 2.0. b Box plots illustrating circVAPA expression in 36 serum samples from SCLC patients versus 118 serum samples from healthy controls. c The levels of circVAPA expression in 3 paired SCLC and matched adjacent normal tissues were detected by RT-qPCR. d RT-qPCR analysis of relative expression of circVAPA in six SCLC cell lines (H69, DMS79, H82, DMS273, H446, and H526) and two NSCLC cell lines (HCC827 and PC9). e The upper schematic illustration demonstrated the circularization of exons 2-4 of VAPA gene forms circVAPA by “head-to-tail” junction. The presence of circVAPA was validated by RT-PCR followed by Sanger sequencing, and the black dotted line indicates the putative back-splice site. f and g PCR products with divergent primers (circVAPA) or convergent primers (VAPA mRNA) in cDNA of DMS273 cells (f) and H82 cells (g), treated with or without RNase R, as assessed by the agarose gel electrophoresis. No RT, no reverse transcription. h and i The relative expression of circVAPA and VAPA mRNA in SCLC cells were measured after actinomycin D treatment for 0 h, 4 h, 8 h, 12 h, 16 h, 20 h, and 24 h. j RT-qPCR analysis of circVAPA in the nuclear and cytoplasmic fractions of SCLC cells. (GAPDH is mainly expressed in the cytoplasm and U6 in the nucleus). k The subcellular localization of circVAPA in DMS273 and H82 cells performed with FISH. Nuclei was stained blue (DAPI) and circVAPA was stained red (Cy3). Scale bar, 20 μm. (All data are presented as the mean ± SD; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays
CircVAPA promotes SCLC progression in vitro
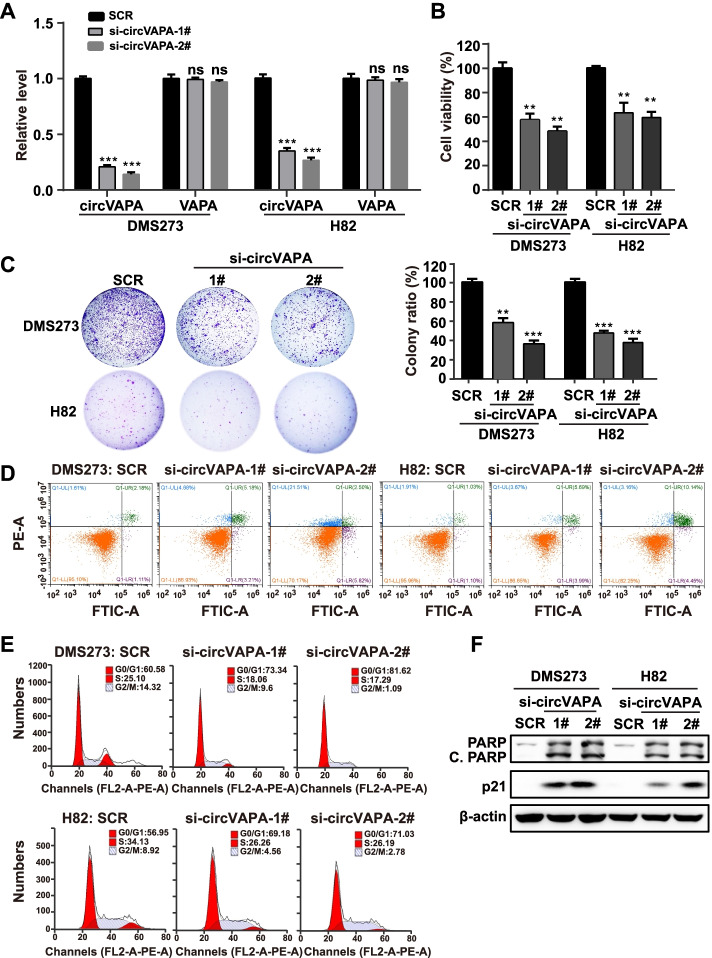
The aberrant expression of circVAPA in SCLC tissues prompted further research into the role of circVAPA in SCLC progression. RNA interference is a practical approach to investigating the biological functions of non-coding RNA (ncRNA) of interest. Two independent siRNAs targeting the back-spliced junction site of circVAPA resulted in the effective silence of circVAPA in SCLC cells, whereas no significant changes on VAPA mRNA (Fig. 2A and Fig. S2A). The CellTiter-Glo luminescent assay revealed that circVAPA knockdown with either siRNA decreased the SCLC cell viability (Fig. 2B). Subsequently, siRNA-mediated circVAPA inhibition resulted in the reduction in colony formation of SCLC cells (Fig. 2C). Flow cytometry analysis demonstrated that the depletion of circVAPA led to cell cycle G0/G1 arrest in SCLC cells and increased the proportion of apoptotic SCLC cells (Fig. 2D, E and Fig. S2B). Meanwhile, the p21 and cleaved PARP protein levels examined by western blot were robustly elevated upon circVAPA knockdown in SCLC cells (Fig. 2F).
Fig. 2.
Silencing circVAPA suppresses cell viability, induces apoptosis of SCLC cells, and inhibits cell cycle progression in vitro. a RT-qPCR analysis of circVAPA and VAPA mRNA expression in SCLC cells treated with the corresponding siRNA. SCR, siRNA with scrambled sequences; si-circVAPA 1# and si-circVAPA 2#, two siRNAs specifically against the junction site of circVAPA. b Cell viability was assessed by the CellTiter-Glo assay. c Colony formation assay (DMS273) and soft agar colony formation assay (H82) were used to assess cell survival in SCLC cells transfected with the indicated siRNAs. d The apoptosis rate was analyzed by flow cytometry after depleting circVAPA in SCLC cells. e Effects on cell cycle progression analyzed by flow cytometry after downregulation of circVAPA. f The expressions of apoptosis-related protein (cleaved-PARP) and cycle-related protein (p21) were detected in SCLC cells transfected with the indicated siRNA by Western blot. β-actin was used as an internal reference. C.PARP, cleaved PARP. (All data are presented as the mean ± SD, ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays
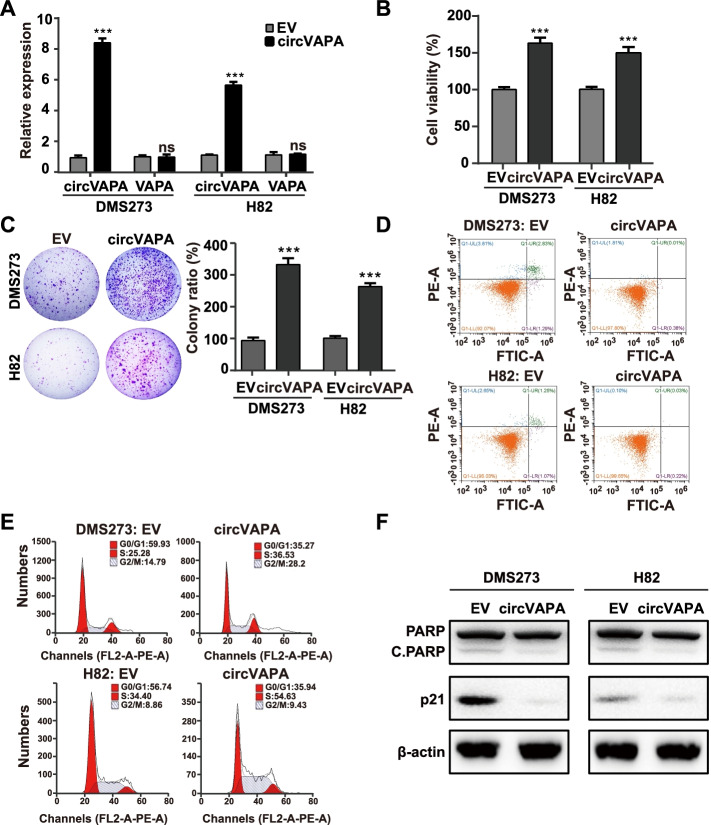
Furthermore, we generated the construction of overexpressing circVAPA (OE-circVAPA) with its endogenous flanking sequences including complementary Alu element pairs to gain the function of the ectopic circVAPA. As depicted in Fig. 3A, OE-circVAPA dramatically increased the circVAPA expressions and unchanged the VAPA mRNA levels in SCLC cells. In contrast to the effect of circVAPA silencing, overexpression of circVAPA contributed to the increase in cell viability, colony formation, and cell cycle progression, and the decrease in the proportion of apoptotic cells and the protein levels of p21 and cleaved PARP in SCLC cells (Fig. 3B-F, and Fig. S2C). Collectively, our results concluded that circVAPA promoted SCLC progression in vitro.
Fig. 3.
Overexpressing circVAPA elevates cell viability, reduces apoptosis of SCLC cells, and accelerates cell cycle progression in vitro. a RT-qPCR analysis of circVAPA and VAPA mRNA expression upon circVAPA overexpression. EV, the empty vector; circVAPA, the circVAPA overexpression plasmid. b Cell viability was assessed by the CellTiter-Glo assay. c Colony formation assay (DMS273) and soft agar colony formation assay (H82) were used to assess cell survival in SCLC cells transfected with indicated plasmid. d The apoptosis rate was analyzed by flow cytometry after overexpressing circVAPA in SCLC cells. e Effects on cell cycle progression analyzed by flow cytometry after circVAPA overexpression. f The expressions of apoptosis-related protein (cleaved-PARP) and cycle-related protein (p21) were detected by Western blot. β-actin was used as an internal reference. C.PARP, cleaved PARP. (All data are presented as the mean ± SD; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays
CircVAPA functions as a sponge for miR-377-3p and miR-494-3p
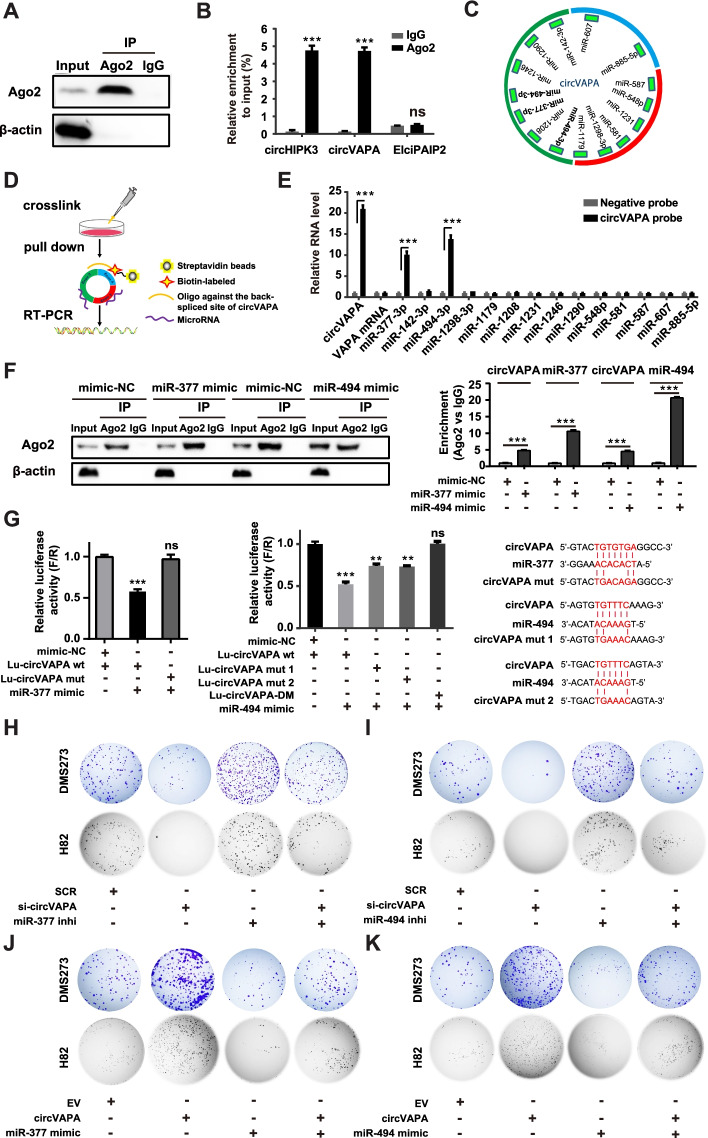
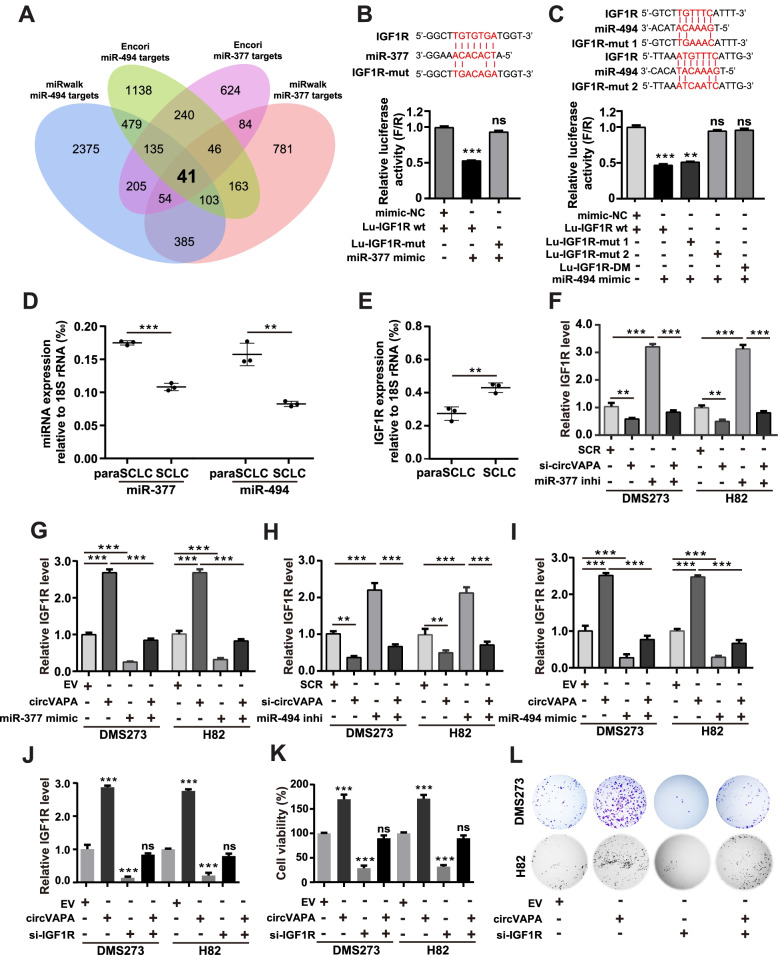
Up to now, the well-characterized mechanism for cytoplasmic circRNAs is to sequester miRNA to regulate target gene expression [11, 44]. Considering that circVAPA is preferentially localized within the cytoplasm, we speculated that circVAPA participated in SCLC progression through a ceRNA mechanism. Since Ago2 is an essential mediator of circRNA-miRNA interaction [44, 45], Ago2 RNA immunoprecipitation (RIP) was conducted to validate the binding of circVAPA in DMS273 cells (Fig. 4A). RIP assay confirmed the direct binding of Ago2 to circVAPA and circHIPK3 (a positive control), but not EIciPAIP2 (a negative control) (Fig. 4B) [46, 47]. We then employed CircInteractome (https://circinteractome.nia.nih.gov/), a web tool for exploring the interaction between circRNAs and miRNAs [48], to predict the putative miRNA binding sites of circVAPA. As illustrated in Fig. 4C, 14 putative miRNA binding sites were predicted in circVAPA sequences, among which there are two potential binding sites for miR-494-3p. Afterward, a biotin-labeled oligonucleotide probe antisense to the junction site of circVAPA was synthesized and applied to perform RNA pull-down assay to further evidence the possible miRNA-circVAPA interactions (Fig. 4D). The antisense probe was able to effectively and precisely capture the endogenous circVAPA, as well as co-pulldown miR-377-3p and miR-494-3p compared to the control probe, but no other 12 predicted miRNAs (Fig. 4E). Moreover, RT-qPCR analysis of Ago2 RIP demonstrated that circVAPA was much more enriched after the overexpression of either miR-377-3p or miR-494-3p with the corresponding mimic in DMS273 cells (Fig. 4F). Furthermore, we constructed luciferase reporter gene plasmids where either the linear sequence or the sequence with the mutation of the putative binding sites for miR-377-3p or miR-494-3p of circVAPA was fused to the 3’ UTR of luciferase. Dual-luciferase reporter assay verified the direct binding of circVAPA to miR-377-3p/miR-494-3p in 293T cells (Fig. 4G). Notably, two putative miR-494-3p binding sites in circVAPA are both required for their interactions (Fig. 4G). Upon siRNA-mediated circVAPA knockdown, the expression levels for both miR-377-3p and miR-494-3p were markedly increased in DMS273 and H82 cells, while circVAPA overexpression caused the decreased levels of miR-377-3p and miR-494-3p (Fig. S3A). Importantly, we showed that suppressing miR-377-3p or miR-494-3p in circVAPA-depleted cells could rescue the inhibitory roles of circVAPA knockdown on cell viability and colony formation in DMS273 and H82 cells (Fig. 4H, I and Fig. S3G, H). On the contrary, miR-377-3p or miR-494-3p overexpression eliminated the promotive effects of circVAPA overexpression on cell viability and colony formation of DMS273 and H82 cells (Fig. 4J, K and Fig. S3I, J). These results indicated that circVAPA served as a molecular sponge for miR-377-3p and miR-494-3p in SCLC cells.
Fig. 4.
circVAPA functions as a sponge for miR-377-3p and miR-494-3p. a Anti-Ago2 RNA IP assay for detecting circVAPA in DMS273 cells. Anti-IgG was used as a negative control. b Anti-Ago2 RIP was performed in DMS273 cells. CircHIPK3 was a positive control, while EIciPAIP2 was a negative control. c Predicted putative binding miRNAs of circVAPA with CircInteractome. d Illustration of the experimental procedure for the circVAPA pull-down assay with biotinylated antisense oligonucleotides. e RNA pull-down efficiency of circVAPA in DMS273 cells. The enrichment of the circVAPA probe or negative probe was detected by RT-qPCR assay to analyze potential miRNAs associated with circVAPA. Negative probe, a biotin-labeled oligonucleotide with scrambled sequences; circVAPA probe, a biotin-labeled oligonucleotide with antisense sequences targeting the circVAPA junction site. f Anti-Ago2 RIP was performed in DMS273 cells transiently overexpressing mimics NC/miR-377-3p/miR-494-3p to detect RNA enrichment in IP complexes. Anti-IgG was used as a negative control. The expression of circVAPA and miR-377-3p/miR-494-3p were detected by RT-qPCR. g The relative luciferase activities were detected in 293T cells after co-transfection with Lu-circVAPA-WT or Lu-circVAPA-Mut and NC or miR-377-3p/miR-494-3p mimics, respectively (left). Lu-circVAPA-DM represents Lu-circVAPA-Mut 1 & 2, which contains two mutation sites. The firefly luciferase activities were measured and normalized to renilla luciferase activities (F/R). Wild-type (WT) and mutated-type (MuT) sequences of the putative binding sites between circVAPA and miR-377-3p/miR-494-3p were listed (right). h–k Colony formation assay of DMS273 cells and H82 cells transiently transfected with siRNAs, plasmids, miRNA inhibitors or mimics as indicated. miR-377 inhi, miR-377 inhibitor; miR-494 inhi, miR-494 inhibitor. SCR, siRNA with scrambled sequences; si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; EV, the empty vector; circVAPA, the circVAPA overexpression plasmid; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively. (All data are presented as the mean ± SD; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. *** miR-377/miR-494 in this article represents miR-377-3p/miR-494-3p, respectively
IGF1R is a functional target of miR-377-3p and miR-494-3p
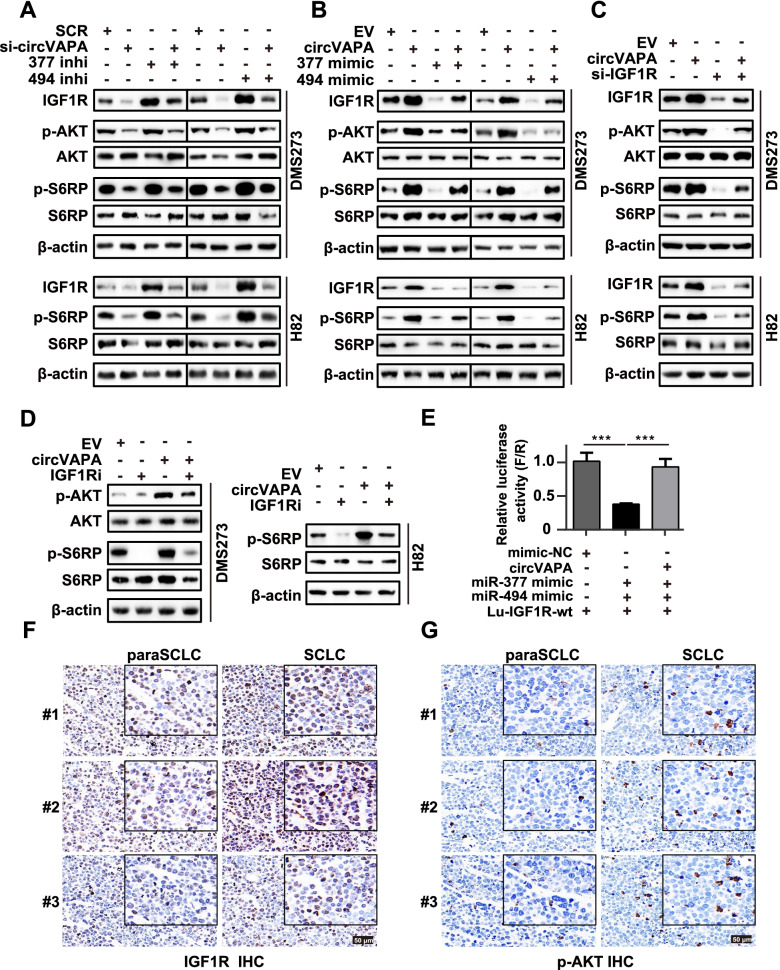
Since circVAPA could behave as a ceRNA against miR-377-3p and miR-494-3p, we set out to identify the downstream targets of miRNAs. The potential targets of miR-377-3p and miR-494-3p were predicted based on two databases (miRWalk (http://mirwalk.umm.uni-heidelberg.de/) and ENCORI (https://rna.sysu.edu.cn/encori/rriPathways.php)) [49, 50], revealing that miR-377-3p and miR-494-3p shared 41 common candidate genes (Fig. 5A, Table S4). Integrative analysis of significantly upregulated target genes in SCLC from the dataset (GSE149507) and the 41 common candidates ultimately focused on ten potential target genes. IGF1R, as an indispensable player in SCLC [28–30], was further confirmed as the only common target of miR-377-3p and miR-494-3p, evidenced by that the IGF1R mRNA level was significantly decreased or increased upon the transfection of miR-377-3p/miR-494-3p mimics or inhibitors, respectively (Fig. S4). We further unveiled that miR-377-3p/miR-494-3p combined with IGF1R mRNA wild type at the molecular level, but not with IGF1R mRNA carrying mutations in the putative binding sites for miR-377-3p/miR-494-3p in 293T cells by dual-luciferase reporter assay (Fig. 5B, C). Notably, there are 2 putative binding sites (Positions 2199–2204, 5648–5654) for miR-494-3p in the 3’ UTR of IGF1R, and only the site 2 rescued miR-494-3p-induced inhibitory effect on luciferase activity (Fig. 5C). Next, we examined the miR-377-3p/miR-494-3p and IGF1R expression in SCLC clinical samples and found that miR-377-3p/miR-494-3p were both down-regulated in SCLC compared to paraSCLC, whereas the IGF1R mRNA showed the opposite trend (Fig. 5D, E). Furthermore, silencing miR-377-3p/miR-494-3p with their corresponding inhibitors robustly increased the IGF1R expression at mRNA and protein levels in DMS273 and H82 cells (Fig. 5F, H and Fig. 6A and Fig. S5B). Conversely, overexpression of miR-377-3p and miR-494-3p with their mimics significantly reduced the mRNA and protein levels of IGF1R in SCLC cells (Fig. 5G, I and Fig. 6B and Fig. S5B). These results together indicated that IGF1R was the direct and functional target of miR-377-3p and miR-494-3p.
Fig. 5.
circVAPA facilitates SCLC cell viability via the miR-377-3p & miR-494-3p/IGF1R/AKT axis. a Two databases (miRwalk and ENCORI) were used to predict the potential target mRNAs of miR-377-3p and miR-494-3p. The Venn diagram shows the number of overlapping miRNAs. b and c Dual-luciferase reporter assay was performed to validate the interaction of IGF1R and miR-377-3p (b) or miR-494-3p (c). 293T cells were co-transfected with a dual-luciferase reporter vector containing the putative binding sites/ mutated sequences for IGF1R and miR-377-3p/miR-494-3p, miR-377-3p/miR-494-3p mimics or negative control mimics (mimic-NC), respectively. Lu-IGF1R-DM represents Lu-IGF1R-Mut 1 & 2, which contains two mutation sites. d-e RT-qPCR analysis of miR-377-3p/miR-494-3p (d) and IGF1R (e) in 3-paired SCLC and paraSCLC samples. 18S rRNA was used as the internal control. f-j RT-qPCR analysis of IGF1R expression in SCLC cells transiently transfected with siRNAs or plasmids as indicated. k and l Cell viability (k) and colony formation (l) assays of SCLC cells transiently transfected with siRNAs, plasmids, miRNA inhibitors or mimics as indicated. SCR, siRNA with scrambled sequences; si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; EV, the empty vector; circVAPA, the circVAPA overexpression plasmid; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively; si-IGF1R, an independent siRNA targeting IGF1R. (All data are presented as the mean ± SD; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. *** miR-377/miR-494 represents miR-377-3p/miR-494-3p, respectively
Fig. 6.
circVAPA promotes the progression of SCLC through the miR-377-3p and miR-494-3p/IGF1R/AKT axis. a and b Western blot analysis of the effects of circVAPA or miR-377-3p/miR-494-3p on IGF1R, AKT, and its downstream protein expression in SCLC cells. 377 inhi, miR-377 inhibitor, 494 inhi, miR-494 inhibitor; 377 mimic, miR-377 mimic; 494 mimic, miR-494 mimic. c Western blot analysis of the effect of overexpressing circVAPA or silencing IGF1R on AKT and its downstream protein expression in SCLC cells. d Western blot analysis of the effect of overexpressing circVAPA or IGF1R inhibitor (drug BMS-536924) on AKT and its downstream protein expression in SCLC cells. e The relative luciferase activities were detected in 293T cells after co-transfection with Lu-IGF1R-WT and mimic-NC or the circVAPA overexpression plasmid or miR-377-3p/miR-494-3p mimics, respectively. The firefly luciferase activities were measured and normalized to renilla luciferase activities (F/R). f-g Representative images of immunohistochemistry analysis of IGF1R (f) and p-AKT (g) in three independent SCLC cases. Scale bar, 50 μm. SCR, siRNA with scrambled sequences; si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; EV, the empty vector; circVAPA, the circVAPA overexpression plasmid; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively; si-IGF1R, an independent siRNA targeting IGF1R; IGF1Ri, the addition of IGF1R inhibitor (drug BMS-536924). (All data are presented as the mean ± SD; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. *** miR-377/miR-494 in this article represents miR-377-3p/miR-494-3p, respectively
CircVAPA facilitates SCLC cell viability via the miR-377-3p & miR-494-3p/IGF1R/AKT axis
To test whether circVAPA behaved as a ceRNA targeting miR-377-3p and miR-494-3p to regulate the expression level of IGF1R, dual-luciferase reporter assay was carried out to assess the interaction between circVAPA, miR-377-3p & miR-494-3p, and IGF1R. The ectopic circVAPA overexpression reversed the suppression of luciferase activities caused by miR-377-3p and miR-494-3p overexpression, demonstrating that circVAPA served as a ceRNA against miR-377-3p and miR-494-3p to relieve the inhibition of IGF1R expression in 293T cells (Fig. 6E). We also evaluated the copies per cell for miR-377-3p, miR-494-3p and IGF1R mRNA in DMS273 and H82 cells, and found that the average molecular ratio is approximately 4.5:1:9 (Fig. S5C-F). IGF1R has been reported to activate the PI3K/AKT signaling [51]. We wondered whether circVAPA regulated the PI3K/AKT signaling pathway via the miR-377-3p & miR-494-3p/IGF1R axis to promote SCLC progression. First, we showed that the miR-377-3p or miR-494-3p inhibitors could rescue the decrease in the mRNA and protein levels of IGF1R and its downstream targets, phosphorylated AKT (p-AKT) and phosphorylated S6 ribosomal protein (p-S6RP) expression upon circVAPA silencing in SCLC cells (Fig. 5F, H and Fig. 6A). Notably, the p-AKT level was too low to be detected by western blot in H82 cells according to our previously reported study [36]. Conversely, the promotive effects of circVAPA on the IGF1R mRNA, IGF1R protein, p-AKT, and p-S6RP protein expression could be abolished by the miR-377-3p or miR-494-3p mimics (Fig. 5G, I and Fig. 6B). Importantly, IGF1R has been demonstrated as a potential target, and inhibition of IGF1R in SCLC cells displays a promising antitumor effect [29, 30, 51]. IGF1R inhibition with either siRNA or BMS-536924, an ATP-competitive IGF1R inhibitor, could recover the circVAPA-induced promoting phenotype of cell viability, colony formation, IGF1R mRNA, and the protein levels of IGF1R, p-AKT and p-S6RP (Fig. 5J-L, Fig. 6C, D and Fig. S6A-D). The IGF1R and p-AKT protein levels examined by IHC staining, are both upregulated in SCLC compared to paraSCLC (Fig. 6F, G). Taking these results together, circVAPA facilitated SCLC cell viability and colony formation by activating the IGF1R/AKT axis by sequestering miR-377-3p and miR-494-3p.
CircVAPA facilitates SCLC proliferation through regulating IGF1R in vivo and in vitro
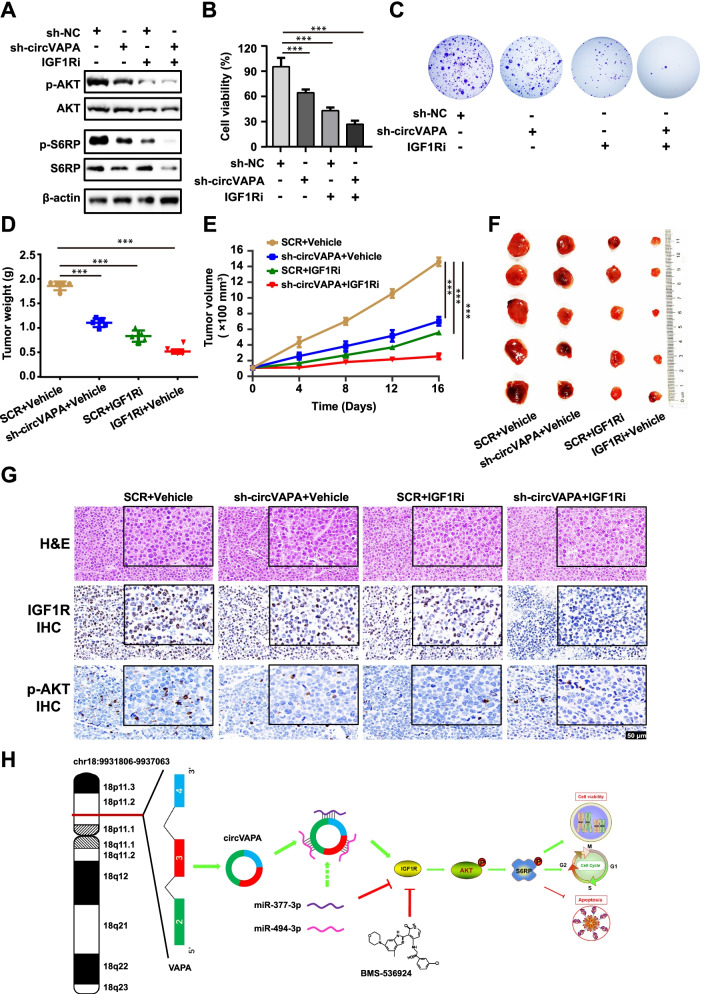
BMS-536924, a small molecule inhibitor targeting IGF1R, has been confirmed to suppress IGF1R phosphorylation and block IGF1R-mediated activation of AKT signaling cascades [33]. Addition of BMS-536924 could attenuate the expression of p-AKT and p-S6RP protein, but this negative regulation of AKT signaling cascade was diminished upon circVAPA overexpression (Fig. 6D). Moreover, we established a DMS273 stable cell line with lentivirus shRNA to silence circVAPA and confirmed the effective knockdown efficiency of circVAPA (Fig. S6E). Treatment with BMS-536924 addition or circVAPA inhibition displayed a moderate impact on the reduction of AKT signaling cascade, whereas a combination treatment with both exhibited the maximal repressive effect (Fig. 7A and Fig. S6B). In support of the western blot results, the BMS-536924 addition alone or circVAPA silencing displayed an appreciable effect on blocking the cell viability and colony formation of DMS273 and H82 (Fig. 7B, C and Fig. S6C, D). However, the combination of BMS-536924 treatment and circVAPA depletion achieved the maximal inhibitory effects on cell viability and colony formation in DMS273 and H82 (Fig. 7B, C and Fig. S6C, D).
Fig. 7.
CircVAPA facilitates SCLC proliferation through regulating IGF1R in vivo and in vitro. a Western blot analysis of the effect of SCLC stable cell line with circVAPA knockdown or the control with or without IGF1R inhibitor (drug BMS-536924) on AKT and its downstream protein expression. b-c Cell viability (b) and colony formation (c) assays of the SCLC stable cell line with circVAPA knockdown or the control with or without IGF1R inhibitor (drug BMS-536924). d-f Therapeutic efficacy of circVAPA depletion and IGF1R inhibitor (drug BMS-536924) as single-agents or in combination in vivo (n = 5 for each group). Tumor weights (d), tumor volume curves (e), and tumor photos (f) of xenograft tumors treated with circVAPA depletion and IGF1R inhibitor alone or in combination. g Immunohistochemistry analysis of IGF1R and p-AKT in tumors. Scale bar, 50 μm. h Model patterns of circVAPA/miR-377-3p & miR-494-3p/IGF1R/AKT axis. Vehicle, negative control cells for silencing circVAPA; sh-circVAPA, stable cell line with lentivirus shRNA to knockdown circVAPA; IGF1Ri, the addition of IGF1R inhibitor (drug BMS-536924). (All data are presented as the mean ± SD; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays
To explore the biological functions of circVAPA in SCLC in vivo, we then performed the subcutaneous injection of circVAPA knockdown and control DMS273 cells into nude mice to investigate the in vivo roles of circVAPA. Cells with stable circVAPA knockdown formed significantly smaller tumors’ size, volume, and weight than the control (Fig. 7D, E, F and Fig. S6E). IHC staining demonstrated that the levels of Ki67, IGF1R, p-AKT were significantly decreased in the tumors derived from cells with stable circVAPA knockdown compared to the control (Fig. 7G and Fig. S6F). The IGF1R inhibitor BMS-536924 exhibited the enhancement in the circVAPA-silencing-caused suppressive effects on cell viability, colony formation, p-AKT, and p-S6RP in vitro and tumors’ size, volume, and weight in vivo, suggesting that BMS-536924 and circVAPA depletion might achieve a potential synergistic effect on the treatment of SCLC (Fig. 7A-G and Fig. S6A-D, F). These results indicated that circVAPA promoted SCLC proliferation by targeting IGF1R in vivo.
Discussion
SCLC is an aggressive malignancy with high mortality and poor prognosis [2, 6]. Even though a significant improvement in chemotherapy efficacy, the clinical outcome of SCLC patients remains poor, mainly due to recurrence and drug resistance [1, 2, 6]. Therefore, it is essential to identify novel biomarkers and effective therapeutic targets for SCLC. Emerging shreds of evidence uncover that circRNAs are dysregulated in diverse human cancers, and these aberrantly expressed circRNAs may be associated with the oncogenesis and progression of multiple cancers [10, 11]. Nevertheless, researches on the role and molecular mechanism of circRNAs in SCLC are still in its infancy.
Previous studies have revealed that circVAPA played an oncogenic role in colorectal and breast cancer [52, 53]. Li et al. discovered that circVAPA facilitated colorectal cancer progression by sponging miR-101 [52]. Additionally, we have also found that RNA pull-down with the probe against the back-spliced junction of circVAPA displayed the enrichment of miR-101 and miR-101 did not affect the IGF1R/PI3K/AKT signaling pathway in SCLC (Fig. S3D-F). Zhou’s team reported that miR-130a-5p suppressed breast cancer cell migration and invasion, and circVAPA served as a sponge for miR-130a-5p [53]. However, no detailed studies on the role of circVAPA in SCLC were performed. With a series of molecular, cellular and biochemical experiments, we propose a working model (Fig. 7H) that circVAPA promotes SCLC progression in vitro and in vivo by modulating the miR-377-3p and miR-494-3p/IGF1R/AKT axis, expanding the knowledge about circRNAs in SCLC.
It has been widely accepted that most circRNAs could act as a ceRNA to regulate mRNA expression via competitively adsorbing miRNAs [23, 44, 46]. Herein, we verified that circVAPA could be enriched in the Ago2 RIP fraction, which is necessary for circVAPA to act as a miRNA sponge. We subsequently predicted that potential miRNAs might bind to circVAPA and confirmed the interaction between circVAPA and miR-377-3p/miR-494-3p by various approaches, such as RNA pull-down assay, Ago2 RIP assay in circVAPA over-expressed SCLC cells, and dual-luciferase reporter assay.
As miRNAs exert their regulatory functions by targeting downstream mRNAs, we explored miR-377-3p/ miR-494-3p downstream mRNA. miR-377 and miR-494 have been found to be related to human cancer and exerted important roles in human cancer by their respective target mRNAs [54, 55]. For example, Li found that miR-377 expression was significantly downregulated in esophageal squamous cell carcinoma (ESCC), and miR-377 expression was positively correlated with ESCC patient survival [54]. Moreover, miR-377 inhibits the initiation and progression of esophageal squamous cell carcinoma through the negative regulation of CD133 and VEGF [54]. Additionally, miR-494 suppresses gastrointestinal stromal tumor (GIST) cell proliferation via targeting KIT, a critical regulatory protein in the development and progression of GIST [55]. In this study, we have explored the common downstream targets of miR-377-3p/miR-494-3p using the miRWalk and ENCORI prediction tools [49, 50]. Of note, IGF1R was predicted to be a potential mRNA target in the downstream pathway of miR-377-3p/miR-494-3p. Then we utilized dual-luciferase reporter assay based on the putative binding sites of miR-377-3p/miR-494-3p on IGF1R, which verified that IGF1R was the common target of miR-377-3p/miR-494-3p.
Numerous circRNAs are significantly associated with clinicopathological characteristics of cancer by regulating the PI3K/AKT signaling pathway [51]. Given that IGF1R plays vital roles in PI3K-AKT signaling cascades [29, 30, 51], we speculated that the mechanism of action of circVAPA might affect the PI3K-AKT signaling cascades. As a result, we aimed to investigate whether IGF1R and its downstream PI3K-AKT signaling cascade could be activated by circVAPA altered. The effect of circVAPA knockdown on IGF1R and the PI3K-AKT signaling cascades in SCLC cells could be reversed by co-transfection of miR-377-3p/miR-494-3p inhibitors. On the contrary, the impact of circVAPA over-expression on IGF1R and the PI3K-AKT signaling cascades in SCLC cells could be rescued by co-transfection of miR-377-3p/miR-494-3p mimics or IGF1R inhibition. These in vitro experiments revealed that circVAPA might act as a molecular sponge to relieve the suppressive effects of miR-377-3p/miR-494-3p on their downstream target IGF1R.
The IGF1/IGF1R signaling axis-mediated pathway has been implicated in the tumorigenesis and development of multiple malignancies, and IGF1R inhibitor emerged as a potential anticancer agent [27, 28]. We revealed that overexpressing circVAPA could recover the reduction in IGF1R activity and eliminate the PI3K-AKT signaling cascades caused by BMS-536924 stimulation in SCLC. Moreover, BMS-536924 could block IGF1R activity and downstream signaling cascades, and this negative regulation could be further enhanced by knocking down circVAPA in vitro. Furthermore, the combination of circVAPA inhibition and BMS-536924 addition exhibited a better therapeutic efficacy in vivo than circVAPA silencing or BMS-536924 alone.
In conclusion, our study demonstrated that circVAPA might serve as an oncogenic circRNA and promote the progression of SCLC. Mechanistically, circVAPA acted as a sponge for miR-377-3p/miR-494-3p to elevate the IGF1R expression to activate the PI3K/AKT signaling pathway. Additionally, the combination of circVAPA inhibition and BMS-536924 displayed a more potent antitumor effect in SCLC. We hope to develop clinical protocols of combinations of circVAPA inhibition and BMS-536924 addition for treating SCLC with circVAPA upregulation.
Conclusions
In summary, our work may provide novel insights into the mechanisms involved in SCLC progression, as well as a promising biomarker for SCLC. We advocate that the circVAPA/miR-377-3p and miR-494-3p/IGF1R/AKT axis may serve as a potent therapeutic target in SCLC.
Supplementary Information
Additional file 1: Figure S1. (a) The relative expression of circVAPA, CircHIPK3 and 18S rRNA in SCLC cells were detected by RT-qPCR after RNase R treatment. CircHIPK3 was a positive control, while 18S rRNA was a negative control. (b-c) Quantification of circVAPA copy numbers in DMS273 and H82 cell lines. The red and blue dots indicate the Ct value and the amount of RNA in DMS273 and H82 SCLC cells, respectively. More experimental details are provided in the Methods section. (d) Prediction of the potential translation ability of circVAPA. An interaction model shows that circVAPA harbors potential internal ribosome entry site (IRES), but not open reading frame (ORF), which is analyzed by the circRNADb and ORFfinder databases. (All data are presented as the mean ± SD; ns, no significance; ***P < 0.001 by two-tailed Student’s t-test). Figure S2. (a) Schematic diagram of knocking down circVAPA using two independent siRNAs target circVAPA junction. (b-c) The apoptosis rate was analyzed by flow cytometry after downregulation (b) or overexpression (c) of circVAPA in SCLC cells. (All data are presented as the mean ± SD; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S3. (a) The effect of circVAPA on miR-377-3p and miR-494-3p expression levels in SCLC cells were detected by RT-qPCR. (b and c) RT-qPCR analysis of miR-377-3p (b) and miR-494-3p (c) expression in SCLC cells transiently transfected with the corresponding mimic, respectively. (d) After RNA pull-down by biotin-labeled circVAPA probe or negative probe, the miR-101 enrichment level in DMS273 cells was detected by RT-qPCR. (e) RT-qPCR analysis of miR-101 expression in SCLC cells transiently transfected with the corresponding mimic. (f) Western blot analysis of the effect of miR-101 on IGF1R, AKT, and its downstream protein expression in DMS273 (left) and H82 (right) SCLC cells. β-actin was used as an internal reference. (g-j) Cell viability analysis of SCLC cells transiently transfected with siRNAs, plasmids, miRNA inhibitors or mimics as indicated. SCR, siRNA with scrambled sequences; si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; EV, the empty vector; circVAPA, the circVAPA overexpression plasmid; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively. (All data are presented as the mean ± SD; N.S., not significant; *P < 0.05; **P < 0.01; ***P <0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S4. (a-d) RT-qPCR analysis of common candidate targets of miR-377-3p & miR-494-3p in SCLC cells transiently transfected with the corresponding mimics or inhibitors. The common targets of miR-377-3p and miR-494-3p were predicted by miRWalk and ENCORI, and they were further in comparison with upregulated genes in the GEO dataset GSE149507. Overlapped genes matching the condition where |fold change| > 1 and p-value < 0.001 were selected. NC, negative control cells for overexpressing or suppressing miR-377-3p/miR-494-3p; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively. (All data are presented as the mean ± SD; **P < 0.01; ***P <0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S5. (a) RT-qPCR analysis of miR-377-3p/miR-494-3p expression in DMS273 cells transiently transfecting the indicated mimics. (b) RT-qPCR analysis of the effects of miR-377-3p/miR-494-3p on IGF1R in SCLC cells. (c-f) Quantification of miR-377-3p, miR-494-3p, and IGF1R copy numbers in DMS273 and H82 cell lines. The red and blue dots indicate the Ct value and the amount of RNA in DMS273 and H82 SCLC cells, respectively. More experimental details are provided in the Methods section. NC, negative control cells for overexpressing or suppressing miR-377-3p/miR-494-3p; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively. (All data are presented as the mean ± SD; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S6. (a) Cell viability analysis of the effect of circVAPA overexpression or IGF1R inhibitor (drug BMS-536924) on SCLC cells. (b) Western blot analysis of the effects of circVAPA depletion or IGF1R inhibitor (drug BMS-536924) addition on S6RP and p-S6RP protein expression in H82 cells. (c and d) Cell viability (c) and colony formation (d) analysis of the H82 cells with circVAPA knockdown or the control with or without IGF1R inhibitor. (e) RT-qPCR analysis of circVAPA and VAPA mRNA expression in stable DMS273 cells transfected with the lentivirus-shRNA. Vector was the negative control for knocking down circVAPA. (f) Immunohistochemistry analysis of ki67 in tumors. Scale bar, 50 μm. si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; IGF1Ri, the addition of IGF1R inhibitor (drug BMS-536924). (All data are presented as the mean ± SD; ns, no significance; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays.
Additional file 2: Table S1. The differentially expressed circRNAs between serum samples from 36 SCLC patients and that from 118 healthy controls.
Additional file 3: Table S2. The upregulated circRNAs previously reported based on circRNA profiling of six paired SCLC tissues.
Additional file 4: Table S3. The expression of circVAPA in serum samples.
Additional file 5: Table S4. Common candidate targets of miR-377-3p and miR-494-3p.
Additonal file 6: Table S5. Oligos used in the study.
Acknowledgements
Not applicable.
Abbreviations
- AKT
Protein kinase B
- cDNA
Complementary DNA
- CeRNA
Competitive endogenous RNA
- CircRNA
Circular RNA
- IGF1R
Insulin-like growth factor 1 receptor
- ncRNA
Non-coding RNA
- NSCLC
Non-small cell lung cancer
- OE-circVAPA
Overexpressing circVAPA
- PI3K
Phosphoinositide 3-kinase
- Pre-mRNA
Precursor mRNA
- RBP
RNA binding protein
- RT-qPCR
Real-time quantitative PCR
- S6RP
S6 ribosomal protein
- SCLC
Small cell lung cancer
- shRNA
Short hairpin RNA
- siRNA
Short interfering RNA
- VAPA
VAMP (Vesicle-associated membrane protein)-associated protein A
Authors’ contributions
W.L., X.W., and J.H. designed this study. J.H., L.M., G.C., and S.Z. conducted the RT-qPCR, western blot, FACS, cell transfection, RNA immunoprecipitation and RNA pull down experiments. X.W. and J.L. preformed plasmid construction and luciferase reporter assay experiments. X.W. performed bioinformatics analyses. J.H., L.M. and S.Z. conducted animal experiments, histological and immunohistochemical analyses. J.H., X.W. and W.L. wrote this manuscript. The author(s) read and approved the final manuscript.
Funding
This study was supported by National Natural Science Foundation of China (81972191 and 81672647), Science and Technology Major Project of Anhui Province (18030801140), and the 100-Talent Program of Chinese Academy of Sciences. A portion of this work was supported by the High Magnetic Field Laboratory of Anhui Province.
Availability of data and materials
Quantitative data that support this study are available within this article and its supplementary files. All other data that support the findings of this study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Ethics statements: The studies involving human participants were reviewed and approved by Ethics Committee of Hefei Institutes of Physical Science, Chinese Academy of Sciences. The patients/participants provided their written informed consent to participate in this study.
Consent for publication
We have obtained consents to publish this paper from all the participants of this study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jinghan Hua and Xiaolin Wang contributed equally to this work.
References:
- 1.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers. 2021;7:3. doi: 10.1038/s41572-020-00235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 4.Blackhall F, Frese KK, Simpson K, Kilgour E, Brady G, Dive C. Will liquid biopsies improve outcomes for patients with small-cell lung cancer? Lancet Oncol. 2018;19:e470–e481. doi: 10.1016/S1470-2045(18)30455-8. [DOI] [PubMed] [Google Scholar]
- 5.Chan JM, Quintanal-Villalonga A, Gao VR, Xie Y, Allaj V, Chaudhary O, et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. 2021;39:1479–1496. doi: 10.1016/j.ccell.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol. 2017;14:549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet. 2011;378:1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Hua J, Li J, Zhang J, Dzakah EE, Cao G, et al. Mechanisms of non-coding RNA-modulated alternative splicing in cancer. RNA Biol. 2022;19:541–547. doi: 10.1080/15476286.2022.2062846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 10.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Huang C, Wang X, Shan G. Circular RNAs in Eukaryotic Cells. Curr Genomics. 2015;16:312–318. doi: 10.2174/1389202916666150707161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 13.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci USA. 2017;114:E5207–E5215. doi: 10.1073/pnas.1612235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Huang C, Shan G. Circular RNAs in physiology and non-immunological diseases. Trends Biochem Sci. 2022;47:250–264. doi: 10.1016/j.tibs.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 19.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869–881. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Li J, Bian X, Wu C, Hua J, Chang S, et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc Natl Acad Sci USA. 2021;118:e2012881118. doi: 10.1073/pnas.2012881118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu YZ, Lv DJ, Wang C, Song XL, Xie T, Wang T, et al. Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. Mol Cancer. 2022;21:12. doi: 10.1186/s12943-021-01480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi: 10.1016/j.ebiom.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Ma XX, Luo J, Chung HK, Kwon MS, Yu TX, et al. Circular RNA CircHIPK3 Promotes Homeostasis of the Intestinal Epithelium by Reducing MicroRNA 29b Function. Gastroenterol. 2021;161:1303–1317. doi: 10.1053/j.gastro.2021.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang T, Wang H, Liu L, Song H, Zhang Y, Wang J, et al. CircIL4R activates the PI3K/AKT signaling pathway via the miR-761/TRIM29/PHLPP1 axis and promotes proliferation and metastasis in colorectal cancer. Mol Cancer. 2021;20:167. doi: 10.1186/s12943-021-01474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X, et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol. 2021;23:278–291. doi: 10.1038/s41556-021-00639-4. [DOI] [PubMed] [Google Scholar]
- 26.Nagao H, Cai W, Albrechtsen NJW, Steger M, Batista TM, Pan H, et al. Distinct signaling by insulin and IGF-1 receptors and their extra- and intracellular domains. Proc Natl Acad Sci USA. 2021;118:e2019474118. doi: 10.1073/pnas.2019474118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guha M. Anticancer IGF1R classes take more knocks. Nat Rev Drug Discov. 2013;12:250. doi: 10.1038/nrd3992. [DOI] [PubMed] [Google Scholar]
- 28.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 29.Warshamana-Greene GS, Litz J, Buchdunger E, García-Echeverría C, Hofmann F, Krystal GW. The insulin-like growth factor-I receptor kinase inhibitor, NVP-ADW742, sensitizes small cell lung cancer cell lines to the effects of chemotherapy. Clin Cancer Res. 2005;11:1563–1571. doi: 10.1158/1078-0432.CCR-04-1544. [DOI] [PubMed] [Google Scholar]
- 30.Ferte C, Loriot Y, Clémenson C, Commo F, Gombos A, Bibault JE, et al. IGF-1R targeting increases the antitumor effects of DNA-damaging agents in SCLC model: an opportunity to increase the efficacy of standard therapy. Mol Cancer Ther. 2013;12:1213–1222. doi: 10.1158/1535-7163.MCT-12-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrickson AEW, Haluska P, Schneider PA, Loegering DA, Peterson KL, Attar R, et al. Expression of Insulin Receptor Isoform A and Insulin-like Growth Factor-1 Receptor in Human Acute Myelogenous Leukemia: Effect of the Dual-Receptor Inhibitor BMS-536924 In vitro. Cancer Res. 2009;69:7635–7643. doi: 10.1158/0008-5472.CAN-09-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou X, Huang F, Carboni JM, Flatten K, Asmann YW, Eyck CT, et al. Drug Efflux by Breast Cancer Resistance Protein Is a Mechanism of Resistance to the Benzimidazole Insulin-Like Growth Factor Receptor/Insulin Receptor Inhibitor, BMS-536924. Mol Cancer Ther. 2011;10:117–125. doi: 10.1158/1535-7163.MCT-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litzenburger BC, Kim HJ, Kuiatse I, Carboni JM, Attar RM, Gottardis MM, et al. BMS-536924 Reverses IGF-IR-Induced Transformation of Mammary Epithelial Cells and Causes Growth Inhibition and Polarization of MCF7 Cells. Clin Cancer Res. 2009;15:226–237. doi: 10.1158/1078-0432.CCR-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tognon CE, Martin MJ, Moradian A, Trigo G, Rotblat B, Cheng SWG, et al. A tripartite complex composed of ETV6-NTRK3, IRS1 and IGF1R is required for ETV6-NTRK3-mediated membrane localization and transformation. Oncogene. 2012;31:1334–1340. doi: 10.1038/onc.2011.323. [DOI] [PubMed] [Google Scholar]
- 35.Hu S, Wang X, Shan G. Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat Struct Mol Biol. 2016;23:1011–1019. doi: 10.1038/nsmb.3302. [DOI] [PubMed] [Google Scholar]
- 36.Ma L, Bian X, Lin W. The dual HDAC-PI3K inhibitor CUDC-907 displays single-agent activity and synergizes with PARP inhibitor olaparib in small cell lung cancer. J Exp Clin Cancer Res. 2020;39:219. doi: 10.1186/s13046-020-01728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Zhang B, Yuan B, Chen C, Zhou Y, Zhang Y, et al. RNA-Seq profiling of circular RNAs in human small cell lung cancer. Epigenomics. 2020;12:685–700. doi: 10.2217/epi-2019-0382. [DOI] [PubMed] [Google Scholar]
- 39.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 44.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 45.Hauptmann J, Meister G. Argonaute regulation: two roads to the same destination. Dev Cell. 2013;25:553–554. doi: 10.1016/j.devcel.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 48.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue C, Li G, Lu J, Li L. Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression. Signal Transduct Target Ther. 2021;6:400. doi: 10.1038/s41392-021-00788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother. 2019;112:108611. doi: 10.1016/j.biopha.2019.108611. [DOI] [PubMed] [Google Scholar]
- 53.Zhou SY, Chen W, Yang SJ, Li J, Zhang JY, Zhang HD, et al. Circular RNA circVAPA regulates breast cancer cell migration and invasion via sponging miR-130a-5p. Epigenomics. 2020;12:303–317. doi: 10.2217/epi-2019-0124. [DOI] [PubMed] [Google Scholar]
- 54.Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee NPY, et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986–4000. doi: 10.1038/onc.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim WK, Park M, Kim YK, Tae YK, Yang HK, Lee JM, et al. MicroRNA-494 downregulates KIT and inhibits gastrointestinal stromal tumor cell proliferation. Clin Cancer Res. 2011;17:7584–7594. doi: 10.1158/1078-0432.CCR-11-0166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. (a) The relative expression of circVAPA, CircHIPK3 and 18S rRNA in SCLC cells were detected by RT-qPCR after RNase R treatment. CircHIPK3 was a positive control, while 18S rRNA was a negative control. (b-c) Quantification of circVAPA copy numbers in DMS273 and H82 cell lines. The red and blue dots indicate the Ct value and the amount of RNA in DMS273 and H82 SCLC cells, respectively. More experimental details are provided in the Methods section. (d) Prediction of the potential translation ability of circVAPA. An interaction model shows that circVAPA harbors potential internal ribosome entry site (IRES), but not open reading frame (ORF), which is analyzed by the circRNADb and ORFfinder databases. (All data are presented as the mean ± SD; ns, no significance; ***P < 0.001 by two-tailed Student’s t-test). Figure S2. (a) Schematic diagram of knocking down circVAPA using two independent siRNAs target circVAPA junction. (b-c) The apoptosis rate was analyzed by flow cytometry after downregulation (b) or overexpression (c) of circVAPA in SCLC cells. (All data are presented as the mean ± SD; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S3. (a) The effect of circVAPA on miR-377-3p and miR-494-3p expression levels in SCLC cells were detected by RT-qPCR. (b and c) RT-qPCR analysis of miR-377-3p (b) and miR-494-3p (c) expression in SCLC cells transiently transfected with the corresponding mimic, respectively. (d) After RNA pull-down by biotin-labeled circVAPA probe or negative probe, the miR-101 enrichment level in DMS273 cells was detected by RT-qPCR. (e) RT-qPCR analysis of miR-101 expression in SCLC cells transiently transfected with the corresponding mimic. (f) Western blot analysis of the effect of miR-101 on IGF1R, AKT, and its downstream protein expression in DMS273 (left) and H82 (right) SCLC cells. β-actin was used as an internal reference. (g-j) Cell viability analysis of SCLC cells transiently transfected with siRNAs, plasmids, miRNA inhibitors or mimics as indicated. SCR, siRNA with scrambled sequences; si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; EV, the empty vector; circVAPA, the circVAPA overexpression plasmid; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively. (All data are presented as the mean ± SD; N.S., not significant; *P < 0.05; **P < 0.01; ***P <0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S4. (a-d) RT-qPCR analysis of common candidate targets of miR-377-3p & miR-494-3p in SCLC cells transiently transfected with the corresponding mimics or inhibitors. The common targets of miR-377-3p and miR-494-3p were predicted by miRWalk and ENCORI, and they were further in comparison with upregulated genes in the GEO dataset GSE149507. Overlapped genes matching the condition where |fold change| > 1 and p-value < 0.001 were selected. NC, negative control cells for overexpressing or suppressing miR-377-3p/miR-494-3p; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively. (All data are presented as the mean ± SD; **P < 0.01; ***P <0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S5. (a) RT-qPCR analysis of miR-377-3p/miR-494-3p expression in DMS273 cells transiently transfecting the indicated mimics. (b) RT-qPCR analysis of the effects of miR-377-3p/miR-494-3p on IGF1R in SCLC cells. (c-f) Quantification of miR-377-3p, miR-494-3p, and IGF1R copy numbers in DMS273 and H82 cell lines. The red and blue dots indicate the Ct value and the amount of RNA in DMS273 and H82 SCLC cells, respectively. More experimental details are provided in the Methods section. NC, negative control cells for overexpressing or suppressing miR-377-3p/miR-494-3p; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively. (All data are presented as the mean ± SD; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. Figure S6. (a) Cell viability analysis of the effect of circVAPA overexpression or IGF1R inhibitor (drug BMS-536924) on SCLC cells. (b) Western blot analysis of the effects of circVAPA depletion or IGF1R inhibitor (drug BMS-536924) addition on S6RP and p-S6RP protein expression in H82 cells. (c and d) Cell viability (c) and colony formation (d) analysis of the H82 cells with circVAPA knockdown or the control with or without IGF1R inhibitor. (e) RT-qPCR analysis of circVAPA and VAPA mRNA expression in stable DMS273 cells transfected with the lentivirus-shRNA. Vector was the negative control for knocking down circVAPA. (f) Immunohistochemistry analysis of ki67 in tumors. Scale bar, 50 μm. si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; IGF1Ri, the addition of IGF1R inhibitor (drug BMS-536924). (All data are presented as the mean ± SD; ns, no significance; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays.
Additional file 2: Table S1. The differentially expressed circRNAs between serum samples from 36 SCLC patients and that from 118 healthy controls.
Additional file 3: Table S2. The upregulated circRNAs previously reported based on circRNA profiling of six paired SCLC tissues.
Additional file 4: Table S3. The expression of circVAPA in serum samples.
Additional file 5: Table S4. Common candidate targets of miR-377-3p and miR-494-3p.
Additonal file 6: Table S5. Oligos used in the study.
Data Availability Statement
Quantitative data that support this study are available within this article and its supplementary files. All other data that support the findings of this study are available from the corresponding author on reasonable request.