Abstract
The mouse visual system serves as an accessible model to understand mammalian circuit wiring. Despite rich knowledge in retinal circuits, the long-range connectivity map from distinct retinal ganglion cell (RGC) types to diverse brain neuron types remains unknown. Here we developed an integrated approach, named Trans-Seq, to map RGC to superior collicular (SC) circuits. Trans-Seq combines a fluorescent anterograde transsynaptic tracer, consisting of codon-optimized wheat germ agglutinin fused to mCherry, with single-cell RNA Sequencing. We used Trans-Seq to classify SC neuron types innervated by genetically-defined RGC types and predicted a neuronal pair from αRGCs to Nephronectin-positive wide-field neurons (NPWFs). We validated this connection using genetic labeling, electrophysiology, and retrograde tracing. We then utilized transcriptomic data from Trans-Seq to identify Nephronectin as a determinant for selective synaptic choice from αRGC to NPWFs via binding to Integrin-α8β1. The Trans-Seq approach can be broadly applied for postsynaptic circuit discovery from genetically-defined presynaptic neurons.
Introduction
Precise neural circuit assembly is necessary for the brain to process complex sensory information and produce proper behaviors. While significant progress has been made on classifying mammalian neuronal types, there is a lack of systematic methods to map long-range synaptic connections and decode the underlying molecular determinants of such connectivity. Efforts to comprehensively map neural circuits have been fruitful in invertebrate systems through cataloging neuronal cell types and then leveraging cell identity to trace circuits1. Similar approaches to understanding mammalian brain circuit assembly remain challenging. The development of neuron type-specific Cre driver mouse lines2 offers direct genetic access to neuronal types of interests. The critical next step is to utilize these tools to understand how diverse neuronal types are interconnected and to elucidate cellular and molecular mechanisms underlying synaptic partner choice3. The mouse visual pathway represents one of the most established models to understand the wiring principles of the mammalian circuits. There is close-to-complete molecular profiling and genetic marking for RGCs, representing distinct neuronal morphologies and visual features4. The integration of genetic labeling of RGC types with optogenetic reagents and high-resolution imaging approaches5 has led to significant insights into the collective input from diverse interneurons onto individual RGC types6. In contrast, although retinotectal connections represent one of the most conserved circuits7, little is known regarding the connectivity map from distinct RGC types onto SC cell types. The enriched genetic access to RGC types provides knowledge on axonal projection patterns8; it is also suitable to develop a genetically encoded anterograde tracing method to map retinotectal connectivity at single-neuron type resolution. Using such a system, we established a system to understand what the targets of a given neuron type are; and how connections are determined in the mammalian brain.
One of the major challenges in mammalian circuit mapping is to efficiently decode the complexity of circuit connectivity over long distances. To this end, viral tracing tools have enabled direct access to distinct neuron types and allowed functional manipulation of neurons along with each relay of a given neural circuit. While pseudo-typed monosynaptic rabies viruses have proven invaluable for retrograde tracing9, efficient anterograde transsynaptic tracers remain limited. Modern anterograde tracers include H129-HSV10 and adeno-associated-virus (AAV) serotype 1 encoding a Cre recombinase11. However, each possesses limitations: neuronal cytotoxicity from H129-HSV complicates functional studies, such as electrophysiological characterizations, and the large HSV-genome size also makes it difficult to work with. In contrast, AAV1 has a smaller genome to handle but is not compatible with Cre-driver line-based circuit tracing. A high-resolution anterograde tracing system is desired from genetically or regionally defined presynaptic neurons, with the following features: (1) to be single-component and can be conveniently delivered using standard viral vectors; (2) to be conjugated to a fluorescent protein so that connected neurons can be characterized via live-imaging; (3) to specifically and efficiently label monosynaptically connected postsynaptic partners; and (4) to allow isolation and subsequent profiling of the connected neurons.
We established a system named Trans-Seq, which offers a generalizable framework to conduct anterograde transsynaptic circuit mapping at the neuron-type resolution, and a step-wise protocol to decode molecular cues governing neuronal partner choice in parallel. Trans-Seq integrates the following components. First, we screened for a fluorescent transsynaptic tracer in vivo based on wheat germ agglutinin (WGA), and obtained an optimized configuration, mWGA-mCherry (mWmC) meeting the criteria above. mWmC allowed efficient and specific monosynaptic anterograde tracing in multiple retinal and brain circuits. The mWmC tracer marks postsynaptic neurons with perisomatic fluorescence, enabling direct electrophysiology validations of connectivity, representing a significant improvement over conventional WGA-based tracers12,13. Second, the faithful anterograde transsynaptic properties of mWmC enable isolation and transcriptional profiling of individual postsynaptic partners. We applied Trans-Seq to investigate mouse retinotectal connections at single-neuron type resolution7. By applying mWmC to all retinal ganglion cells (RGCs), Trans-Seq allowed the classification of retinorecipient neuron types in the SC. Third, we identified distinct input patterns onto these SC recipient neurons by conducting differential Trans-Seq from two distinct RGC subclasses14. We identified a specific synaptic connection from αRGCs to Nephronectin-positive wide-field neurons (NPWFs) through such characterizations and comparisons. This synaptic pair was subsequently validated using genetic labeling, electrophysiology, and retrograde tracing. Last, we revealed, using Trans-Seq transcriptomics data, that the synaptic pair is formed via interactions between presynaptic Integrin-α8β115 and postsynaptic Nephronectin. Nephronectin is the first cue discovered in mammalian SC to confer retinal axon sublaminar choice. These results demonstrate how Trans-Seq efficiently maps long-range synaptic connections at the neuron-type resolution and simultaneously captures molecular determinants for synaptic specificity, offering a new avenue to decode mammalian circuits.
Results
A genetically encoded fluorescent tracer for anterograde neuron mapping
We sought to re-engineer WGA12 as a genetically encoded reagent delivered through adeno-associated-viruses (AAVs). WGA and its closely related lectins exhibit anterograde transsynaptic transfer16 with the caveat that the plant-derived original WGA tracer, when applied as a protein, exhibits retrograde transfer, likely through axonal uptake, and the anterograde transfer can be polysynaptic12,17. We took a stepwise approach to re-engineer a monosynaptic anterograde tracer and tested its anterograde and monosynaptic properties in vivo.
First, we synthesized a mammalian codon-optimized cDNA (mWGA) to improve WGA expression in mammalian neurons. Second, we examined whether the fusion of a fluorescent protein onto either the N- or C- terminus of mWGA allows direct visualization of connected neurons (Extended Data Fig. 1a, b). Third, to maximize compatibility with existing GFP lines for RGC types4 and identify efficient fluorescent protein fusion configurations, we generated mWGA constructs fused to red fluorescent protein (RFP) variants, including mCherry, mRuby3, and tdTomato (Extended Data Fig. 1a, c). We then tested the anterograde transferring efficiencies. AAV2 vectors expressing mWGA-RFPs were delivered into the left eyes (Fig. 1a), and the transduction of major RGC subclasses was confirmed (Fig. 1c, d, and Extended Data Fig. 1m–q). AAV2 demonstrates its tropism towards RGCs and excludes infection of inner retinal neurons. We evaluated anterograde transfer efficiency by assessing the number of RFP+ neurons on the contralateral SC using native fluorescence (Fig. 1e, f, Extended Data Fig. 1e–h). In acute SC slices, we found that the mWGA-mCherry (mWmC) construct resulted in higher anterograde transfer efficiency than mCherry-mWGA under the same experimental conditions (Extended Data Fig. 1a–b). Furthermore, among the panel of C-terminal fused RFP variants tested, mWmC resulted in a multi-fold higher efficiency than other RFP variants (Extended Data Fig. 1a, c–h). mWmC anterograde transfer predominantly labeled SC neurons and few astrocytes in retinorecipient regions (Extended Data Fig. 1v–x). The live-fluorescent signals were evident in retinorecipient regions, such as the dorsal lateral geniculate nucleus (Extended Data Fig. 1r, s) in addition to the SC (Extended Data Fig. 1t, u). In contrast, such signals were absent in di-synaptic regions from RGCs, such as the primary visual cortex (V1) and the lateral posterior thalamic nucleus (LP) (Extended Data Fig. 1u, s). The restricted fluorescent signals to retinorecipient regions suggest that mWmC transfer primarily occurs via monosynaptic transfer.
Fig. 1. Engineering a genetically encoded fluorescent anterograde transsynaptic tracer (mWGA-mCherry) to map postsynaptic neurons.

a, Schematic diagram of intraocular injections of anterograde transsynaptic red-fluorescent tracers to label postsynaptic retinorecipient neurons in the superior colliculus (SC). b, Optimized adeno-associated virus (AAV) vector design encoding mWGA-mCherry (mWmC). mCherry is fused at the C-terminal of a codon-optimized mWGA. c, Retina wholemount showing mWmC coverage to the ganglion cell layer (GCL). d, magnified view of inset of (c). Both images were stained with anti-RFP. Scale bars: (c, 2mm; d, 200 μm). 3 times each experiment was repeated independently with similar results. e, Live red fluorescent labeling of the contralateral SC after acute brain slice preparation, indicating transsynaptic transfer onto the recipient neurons enriched in stratum griseum superficiale (SGS) and stratum opticum (SO), but not in stratum griseum intermedium (SGI). f, Magnified view of inset from e. showing individual neurons labeled with bright red fluorescent protein from RGC anterograde transfer without signal amplification through immunostaining. Scale bars: (e, 500μm; f, 50μm). 3 times each experiment was repeated independently with similar results. g, Schematic drawing of mWmC-mediated monosynaptic anterograde transsynaptic tracing using electrophysiology recording after co-injection of AAV-mWmC (red) and AAV-ChR2-YFP (green). h, A sample image of patched mWmC-positive red fluorescent neurons under DIC camera (top) and epi-fluorescence scope (bottom). Scale bar: 25 μm. At least 10 times each experiment was repeated independently with similar results. i, Whole-cell recordings of mWmC-positive fluorescent neurons, indicating that excitatory postsynaptic currents (EPSCs) in response to a short pulse (2ms, blue line) of blue-light (475nm) excitation possess an onset latency of <5ms, suggesting mWmC+ SC neurons receive monosynaptic inputs from RGCs. j to m. Evoked currents of the mWmC+ SC neurons are glutamatergic and monosynaptic. j, Postsynaptic currents persisted in TTX (1μM) and 4-AP (100μM) (black) and were blocked by CNQX (10μM) and APV (50μM) (red). k. Average ESPC amplitudes in control conditions (black) and the presence of CNQX and APV (red). Both conditions had bath-applied (TTX+4-AP). n=20 cells ****, p<0.0001, two-sided Student’s t-test. l, Distributions of the response latency of the evoked EPSCs to the short pulse blue-light, with the average latency at 2.2±0.2ms. n=27 cells m, EPSCs measured from randomly patched mCherry-positive (red, n=5 animals) and mCherry-negative (black, n=4 animals) neurons under the same electrophysiological recording conditions and subject to the same LED stimulations. In total, 28 out 30 (93%) of mCherry-positive neurons received monosynaptic inputs from the retina. In comparison, only 1 out of 20 (5%) mCherry-negative neurons showed monosynaptic inputs. ****, p<0.0001, two-sided Student’s t-test. All data in this figure are presented as mean ± SEM.
Monosynaptic connections revealed by mWmC coupled with functional testing
We utilized whole-cell recordings to test whether mWmC-positive SC neurons receive direct synaptic inputs from RGCs. We co-administered two AAVs, encoding ChR2-YFP and mWmC into the left eye through intraocular injections (Fig. 1g) and identified bright perisomatic mWmC (red) signals transfer in the SC at four weeks post-injection (wpi) (Fig. 1h). Using red fluorescence as a guide for targeting connected cells, we detected excitatory postsynaptic currents (EPSCs) in response to 2ms blue-light excitation, with an onset latency of 2.2±0.2ms, suggesting these neurons receive monosynaptic RGC inputs (Fig. 1i, l). The evoked EPSCs persisted in the presence of TTX and 4-AP but were blocked by CNQX and APV, confirming that the synaptic connections were monosynaptic and glutamatergic (Fig. 1j, k). To test the efficiency and specificity of the anterograde transfer, we recorded from mWmC-positive neurons and compared their properties with neighboring mWmC-negative neurons. We found that 93% of mWmC-positive neurons received monosynaptic inputs, but only 5% of mWmC-negative neurons did so (Fig. 1m). These mWmC-positive SC neurons exhibited similar intrinsic properties as their mWmC-negative neighboring neurons, such as resting membrane potentials and firing properties at 4wpi (Extended Data Fig. 1i–l). In summary, the mWmC tracer allows for direct detection of SC neurons postsynaptic to RGC starters, with low cytotoxicity in vivo.
Lysosomal mWmC-enrichment for efficient anterograde transsynaptic labeling
mWmC yielded the brightest fluorescent labeling of recipient cells in a perisomatic fashion (Fig. 1f, h). In comparison, prior tracer configurations based on WGA exhibit signal diffusivity and label regions outside of the cell soma. A proposed explanation for the enriched perisomatic fluorescence in mWmC is mCherry’s resistance to degradation: mCherry possesses a low pKa that retains its fluorescence properties under the acidic pH in the late endosomes and lysosomes (unlike the higher pKa of other RFPs in Extended Data Fig 1a). Moreover, mCherry accumulates in lysosomes because it resists lysosomal protease degradation18. To test this hypothesis, we compared AAV-mWmC to WGA configurations previously used in literature for transsynaptic tracing: WGA-protein conjugated with Alexa-555, and AAV-encoding C-terminus truncated WGA (ctt-WGA) cDNA. We injected these tracers into the retina and imaged postsynaptic cells in the SC. We observed significantly higher enrichment of mWmC within lysosomes (Lamp1) but not in prior WGA configurations. The enrichment of mCherry in the lysosomes of the recipient neurons (Fig. 2e–g) led to signal enhancement of mWmC, thereby serving as a high-threshold filter for detection of mono-synaptic recipient neurons. Similarly, the lysosomal enrichment of mWmC in primary recipient neurons will not escape into the next neuronal relay, such as from the SC onto the thalamic LP neurons (Extended Data Fig. 1s). This distinct lysosomal enrichment serves as a hallmark for mWmC, ensuring efficient and specific detection of monosynaptically connected neurons.
Fig. 2. Lysosomal enrichment of mWmC leads to stable red-fluorescent labeling of postsynaptic neurons, facilitating electrophysiology and FACS identification of postsynaptic neurons in vivo.

a-f, mWmC-mediated anterograde trans-synaptic labeling (e, f) leads to a selective enhancement of tracer signals in the lysosomes (Lamp1) of postsynaptic neurons, compared to ctt-WGA tracer12 (c, d) or WGA-protein conjugated with Alexa555 (a, b). Transferred mWmC signals within recipient neurons were observed to aggregate in a perisomatic manner, facilitating the identification of labeled neurons (as in Figs. 1&3). The mWmC aggregates highly overlap with lysosomes (f, marked by Lamp1), suggesting that the perisomatic structures are formed through mWmC enrichment within the lysosomes of postsynaptic neurons (Green, Lamp1, lysosomal marker; Red, mWmC fluorescence). 4 times each experiment was repeated independently with similar results. Scale bars: (a, c, e, 20μm; b, d, e, 5μm). g, The percentages of WGA-protein-conjugates, ctt-WGA, and mWmC overlapping with Lamp1 were quantified, showing the unique properties of mWmC in live. ****, p<0.0001, two-sided Student’s t-test. n=7 biologically independent samples. Data in this figure are presented as mean ± SEM.
Genetically encoded mWmC with a minimal retrograde spread
We next investigated the specificity and directionality of mWmC transfer using well-characterized driver lines to mark individual neuronal subsets, such as direction-selective circuits (Fig. 3a). Following the introduction of mWmC into Starburst Amacrine Cells (SACs), we detected robust anterograde and monosynaptic transfer into On-Off Direction Selective Ganglion Cells (ooDSGCs) (Fig. 3a, b, g). We did not detect retrograde transfer of mWmC onto the inner nuclear layer (INL) cells (Fig. 3c, g). When we applied the mWmC tracer onto ooDSGCs as starter neurons (Fig. 3d, e). We did not detect significant retrograde spread of mWmC from RGCs onto SACs or other interneurons (Fig. 3f, Extended Data Fig. 2a–g). The restriction of mWmC transfer from SACs to ooDSGCs is notable as the synaptic contacts between SACs and ooDSGCs are highly mixed with other neurons in the same neuropil19. Next, we applied mWmC to SC neurons, postsynaptic to RGCs and presynaptic to neurons in the thalamic LP. We compared AAV-mWmC to tracer configurations based on WGA previously used in literature for transsynaptic tracing: WGA-protein conjugated with Alexa-55520 and AAV-encoding ctt-WGA cDNA12. Purified WGA isolectins exhibit bidirectional transfer. Each tracer was injected into the SC as starter cells (Extended Data Fig. 2h–k)12,13,17. All tracers exhibited efficient anterograde transfer from the SC to downstream targets, such as LP of the thalamus. In contrast, each tracer displayed different patterns of retrograde labeling back to the retina. WGA-conjugated dye led to bright RGC labeling, likely due to axonal uptake of the protein by RGCs (Extended Data Fig. 2i). Similarly, injecting AAV expressing ctt-WGA cDNA or mWmC into the SC resulted in intense labeling of RGCs, likely due to WGA expression from internalized AAV following uptake by RGC axon terminals (Extended Data Fig. 2j,k). We detected weaker WGA-stained RGCs at the lower-level expression in addition to bright signals from primary axonal uptake, suggesting retrograde transfer of protein from SC to RGCs (Extended Data Fig. 2j, k). Importantly, such a low level of retrograde transfer was significantly reduced in the AAV-mWmC configuration (Extended Data Fig 2l, m). Furthermore, when we introduced Cre-dependent mWmC into excitatory SC neurons (vGlut2-Cre positive) as starter neurons (Fig. 3h, j), the Cre-dependent genetic control of mWmC expression in starter neurons greatly reduced ectopic expression due to retrograde axonal uptake of AAVs (Extended Data Fig. 2n). We detected limited if any retrograde spread of mWmC from the SC to RGCs (Fig. 3i). In the same experimental preparation, we detected robust SC anterograde transfer onto the LP of the thalamus (Fig. 3k). The normalized ratios of anterograde versus retrograde transfer of each tracer were quantified in the RGC-SC-LP circuit (Fig. 3l). In summary, mWmC exhibited primarily anterograde transfer but limited, if any, retrograde spread.
Fig. 3. Anterograde but not retrograde transfer of mWmC in retinal and brain circuits.
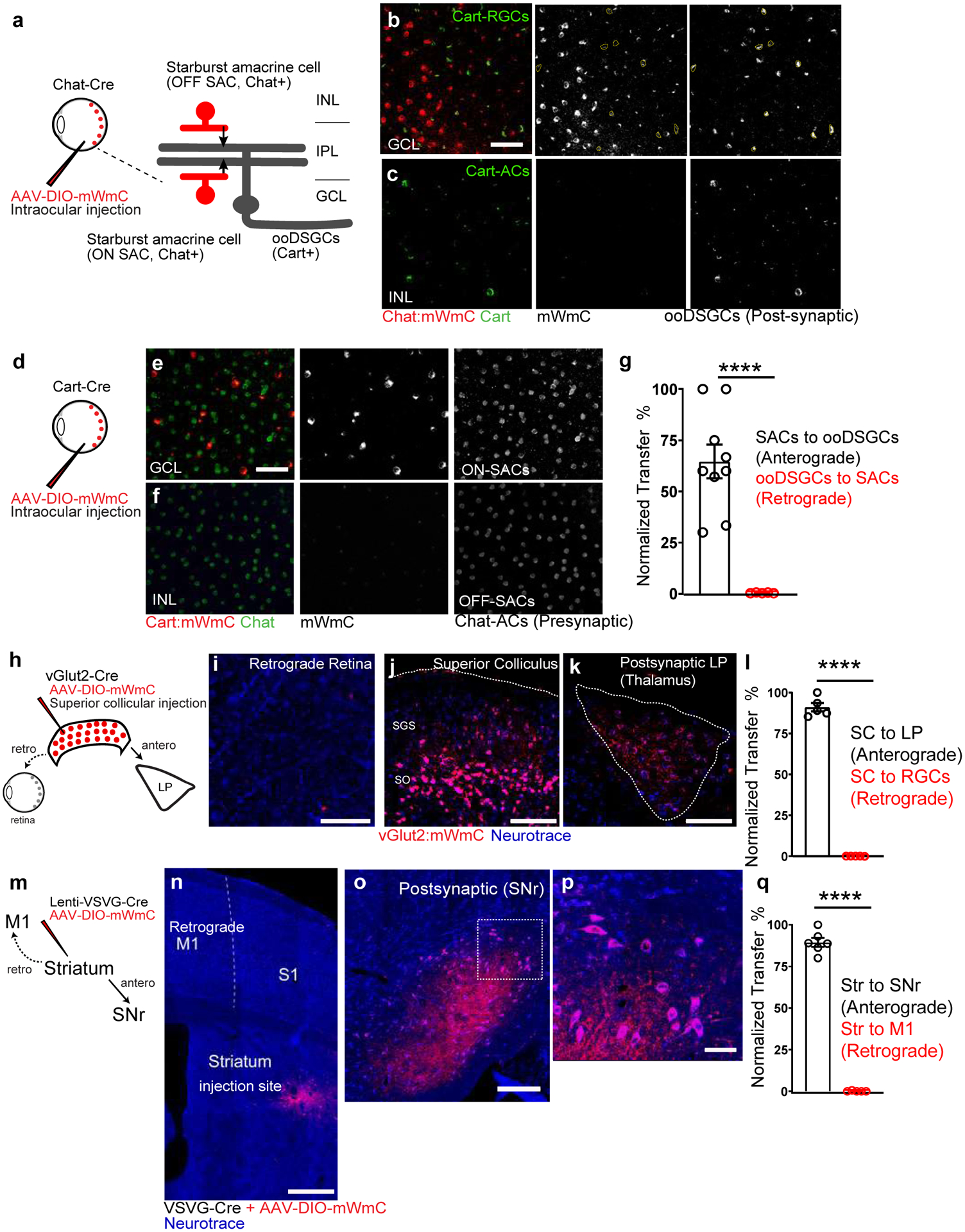
a-c, (a) Intraocular injections of mWmC into Chat-Cre led to (b) expression of mWmC within Starburst Amacrine Cells (SACs, red) within the GCL as starter cells. ooDSGCs (Cart-positive, green), which receive direct input from SACs, were labeled by mWmC through anterograde transfer (mWmC double-positive cells labeled with yellow-dotted circles). No signal was detected in the INL, indicating that bipolar cells were not labeled from SACs retrogradely (c), 8 times each experiment was repeated independently with similar results. d-f, (d) Intraocular injections of mWmC into Cart-Cre led to (e) Efficient transduction of ooDSGCs as starter cells for anterograde tracing into the brain (Fig. 5). No retrograde spread was seen into SACs (Chat-positive) or bipolar cells in either the GCL or INL (f), 5 times each experiment was repeated independently with similar results. Scale bars: b, c, e, f, 50μm. g, Quantifications of the anterograde transfer ratio from SACs to ooDSGCs (black) n=9 biologically independent samples, versus retrograde transfer ratio from ooDSGCs to SACs (red), n=5 biologically independent samples. ****, p<0.0001, two-sided Student’s t-test. Within each retina region, 43±6% of the Cart-positive RGCs received mWmC transfer from SACs. h-l, (h) Introduction of mWmC into excitatory SC neurons (vGlut2-positive) for anterograde and retrograde transfer tests. (j) Efficient start neurons at the SC demonstrated very few retrograde spread back to the retina (i), but specific and efficient anterograde transfer onto the LP of the thalamus (k). Scale bars: (i, 50μm, j, 200μm, k, 100μm). l. Quantifications of the anterograde transfer ratio from SC to LP (black) versus retrograde transfer ratio from SC to RGCs (red), n=5 animals. ****, p<0.0001, two-sided Student’s t-test. Within each SC starting region, 81±6% of the NeuN-positive SC neurons were infected with mWmC; within the LP recipient neuron regions, 86±4% received mWmC transfer from the SC. m-q, (m) Introduction of Cre-dependent mWmC with VSVG-lentivirus expressing Cre into the dorsal lateral striatal neurons for anterograde and retrograde transfer tests. (n) Efficient start neurons at the striatum demonstrated no retrograde transfer back to M1 on the same brain slice. In contrast, mWmC displayed specific anterograde transfer onto the Substantia Nigra (o, SNr) as zoomed-in (p). Notably, axons are also filled by mWmC, in addition to somata filling. Scale bars: (n, 500μm, o, 100μm, p, 50μm). q. Quantifications of the anterograde transfer ratio from the striatum to SNr (black) versus retrograde transfer ratio from the Striatum to M1 (red), mWmC-positive neurons, n=6 animals ****, p<0.0001, two-sided Student’s t-test. Within the SNr recipient neuron regions, 89±2% received mWmC transfer from the Striatum. All data in this figure are presented as mean ± SEM.
mWmC-mediated anterograde tracing across different brain regions
Beyond the visual pathways, we performed anterograde transsynaptic tracing from several brain regions as starter neurons, including the vibrassal motor cortex (vM1) (Fig. 4a). We observed extensive anterograde mWmC transfer in the dorsal striatum (Fig. 4b, c); thalamus; the barrel Cortex (S1) (Fig. 4d); anterior medial (AM), ventral anterior-lateral (VAL), and ventral medial (VM) thalamic nuclei (Fig. 4e); and intermediate reticular (IRt) and ventral medullary reticular (MdV) nuclei in the brainstem area (Fig. 4f, g)21. Similarly, we confirmed anterograde transsynaptic tracing from the barrel cortex (S1) (Fig. 4h–n). Functionally, we tested the cortical-striatum transfer using electrophysiology by co-administrating ChR2-YFP and mWmC into vM1. We observed bright perisomatic labeling by mWmC in the striatum (Fig. 4c). We detected evoked EPSCs from mWmC-positive striatum neurons in response to blue-light stimulation of M1 axons in the presence of TTX and 4-AP, which were reduced by CNQX and APV (Fig. 4o, p), indicating that mWmC-positive striatal neurons receive monosynaptic vM1 inputs. We also applied mWmC to trace striatal projections. To bypass the retrograde axonal uptake of AAV-expressing mWmC and restrict mWmC only in the striatum, we injected Cre-dependent mWmC AAV with a VSVG-lentiviruses expressing Cre (Fig. 3m). Axons rarely take up lentiviruses; therefore, the mixture locally restricted mWmC expression to the injection site, where a focal population of dorsal-lateral striatal neurons serves as starter neurons (Fig. 3n). We detected no retrograde transfer of mWmC back M1 (Fig. 3n, Extended Data Fig. 2q). In contrast, we detected mWmC anterograde transfer from the striatum onto both the Substantia Nigra (SNr) and Globus Pallidus (GPe) (Fig. 3o, p, Extended Data Fig. 2p). Collectively, these data suggest that mWmC can be applied for circuit mapping within the brain as an anterograde transsynaptic tracer from regionally defined neurons.
Fig. 4. Utilizing mWGA-mCherry (mWmC) for downstream neuron discovery and circuit mapping in various brain regions.

a-g. Anterograde transsynaptically labeled neurons (red) in vM1 projection areas. Presynaptic starter neuron axons are labeled by the YFP signal (green). a, The injection site in the vibrissal primary motor cortex (vM1). b, The dorsal striatum with a magnified view c. d, The thalamus, and S1. e, Magnified view of the boxed area in d. (AM: anterior medial, VAL: ventral anterior-lateral, and VM: ventral medial thalamic nuclei). f, The intermediate reticular (IRt) and ventral medullary reticular (MdV) nuclei in the brainstem. g, Magnified view of the boxed area in f. Spinal trigeminal nucleus caudalis, SpVc; dorsal medullary reticular nucleus dorsal, MdD; Hypoglossal motor nucleus, 12N. Scale bars: (a, d, 1mm; b, e, f, 200μm; g, 100μm; c, 20μm), n=4 animals. h-n, Anterograde transsynaptic tracing from barrel (somatosensory) cortex (S1, h) via stereotaxic injections into the designated cortical areas as starters cells. We used a YFP marker that labels the axons of starter neurons in addition to mWmC to separate presynaptic axons from postsynaptic mWmC-positive cells. From S1 (h), we detected mWmC-transfer signals in known recipient areas, including contralateral S1, M1, thalamus, dorsal lateral ventral posterior medial (VPM), and principal trigeminal sensory nucleus (PrV) as transsynaptically labeled neurons (red) in S1 projection areas. Presynaptic starter axons are labeled by the YFP signal (green). h, The injection site in the barrel field of the primary somatosensory cortex (S1). i, Contralateral S1. Magnified view of the boxed area in h. j, M1. k, The thalamus. l, Dorsal lateral ventral posterior medial (VPM). Magnified view of the boxed area in k. m, The principal trigeminal sensory nucleus (PrV). n, Magnified view of the boxed area in m. Scale bars: (h, 1mm; k, 500μm; i, j, m, 200μm; l, 100μm; n, 20μm), 5 times each experiment was repeated independently with similar results.. Abbreviations: Posterior (PO), ventral lateral (VL), and ventral posterior lateral (VPL) nuclei of the thalamus. o to p, Monosynaptic connectivity test from the motor cortex (M1) to the striatum using mWmC-mediated anterograde transsynaptic transfer. The recordings were all done in ACSF with bath applied TTX (1μM) and 4-AP (100μM). o, An average EPSC from a sample mWmC-positive neuron responds to a 2ms light pulse (blue dot). p, All 32 cells showed EPSCs, and such responses were drastically reduced by CNQX (10μM) and APV (50μM) (N= 2 animals, 32 neurons). ***, p<0.005, two-sided Student’s t-test. Data in this figure are presented as mean ± SEM.
Establishing Trans-Seq platform to define RGC downstream neuron type diversity in the SC
We next sought to couple mWmC anterograde tracing with single-cell RNA-Sequencing (scRNA-Seq). This integration helps generate transcriptomic connectivity maps from genetically or regionally defined starter cells in a high-throughput manner. We named the mWmC-based tracing and sequencing system, Trans-Seq. We first established the Trans-Seq protocol by generating a molecular atlas for retinorecipient SC neurons, as defined expressing mWmC in most RGC subtypes (pan-RGC). Following unilateral eye injections, we collected ~39,000 mWmC-positive retinorecipient cells via tissue dissociation and FACS at 4wpi (Fig. 5a, Extended Data Fig. 3a–h). Due to low adult neuronal survival, we recovered scRNA-Seq data from ~1000 adult SC recipient neurons among three replicates (Fig. 5b; and Extended Data Fig. 3i–m) using the standard 10X Genomics protocol for scRNA-Seq. We obtained eight neuronal clusters representing a molecular map of retinorecipient neurons connected with pan-RGCs, but not other SC neurons without retinal inputs. We clustered the cells into three excitatory SC types (ESC1–3) and five inhibitory SC types (ISC1–5) (Fig. 5b, c, Extended Data Fig. 4). A distinct and unbiased set of marker genes were identified to define the neuron type identities (Fig. 5c), including mutiple candidates based on SC subset expressions22 (Extended Data Fig. 4e–g) but without known cell type information.
Fig. 5. Trans-Seq integrates mWGA-mCherry-mediated anterograde transsynaptic tracing and scRNA-Seq to categorize the RGC connected neurons in the SC.
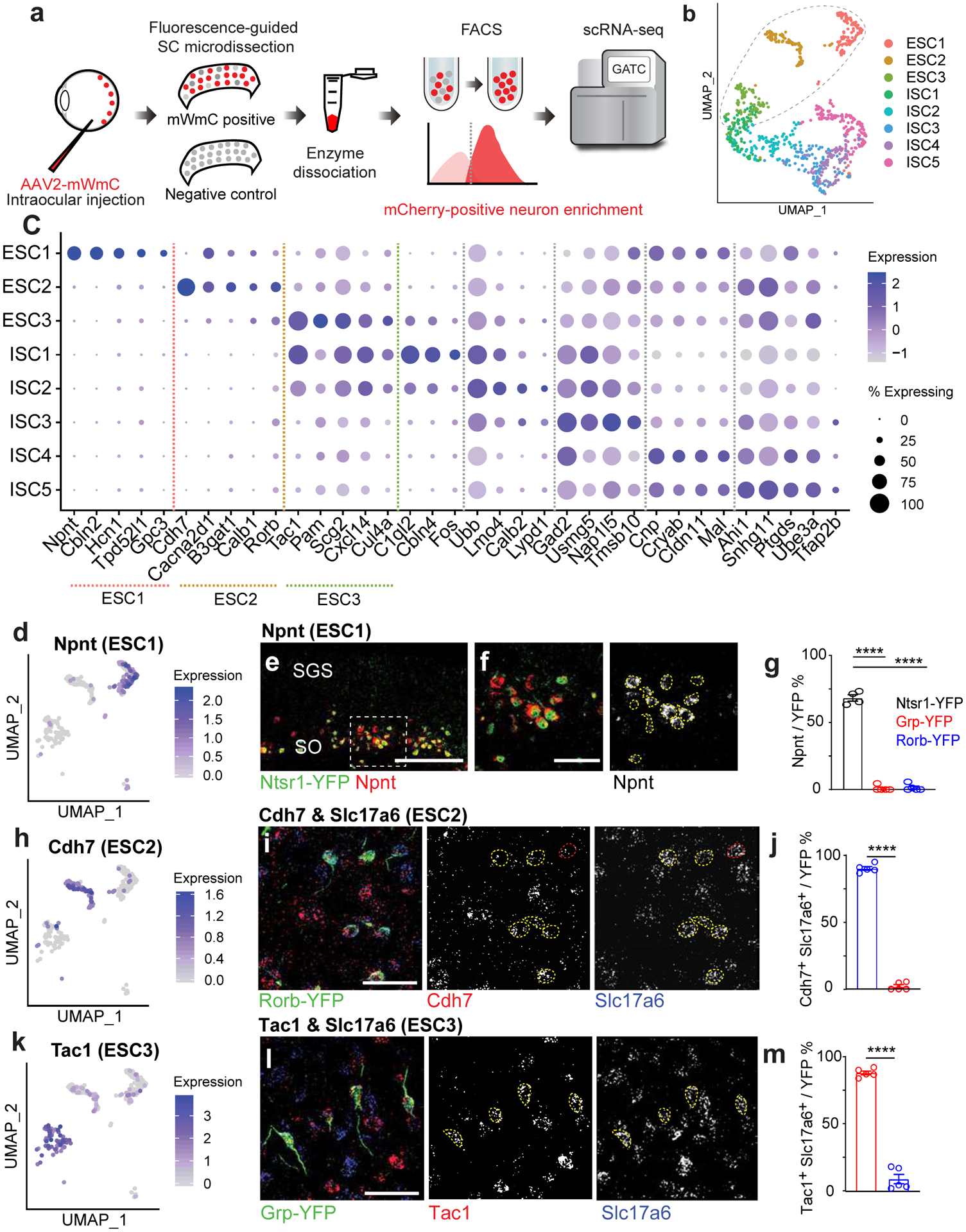
a, To generate a molecular atlas for all RGC-connected SC neurons, the following workflow was used to dissociate the SC and isolate mCherry-positive recipient neurons by FACS. The workflow starts with the injection of mWmC into the left retinas of mice, followed by right SC slicing, fluorescence-based microdissection, enzyme dissociation, and FACS for red fluorescence from mWmC. The scRNA-Seq library preparation and subsequent data analysis using the standard 10XGenomics platform and Seurat Packages. b, scRNA-Seq libraries were generated using the 10XGenomics platform. From three replicates, we recovered scRNA-Seq data from 898 adult SC recipient neurons, identified through neuronal markers, thereby generating a molecularly defined connectivity map from pan-RGC anterograde tracing. UMAP plots of the Trans-Seq data from all RGCs (Pan-RGC) show three excitatory neuron clusters (ESCs) and five inhibitory neuron clusters (ISCs). Among all retinorecipient SC neurons subtypes, there were 307 excitatory neurons (within the dotted circle) and 591 inhibitory neurons. c, Dot plot of top marker genes. ESC1-ESC3 are represented on the left of the plot for top feature genes, while ISC1–5 are represented on the right of the plot for top feature genes. (d, h, k) UMAP plots for primary marker genes for each ESC, d, Npnt for ESC1, h, Cdh7 for ESC2, and k, Tac1 for ESC3. e, f, Validation of Trans-Seq marker gene expression from ESC1 using RNA-Scope in situ hybridization. e, Npnt (red) is enriched in Ntsr1-GN209-YFP (green) transgenic for wide-field excitatory neurons as quantified in g. f, Higher magnification of boxed area highlighted in e. n=4 animals, Scale bars: (e, 250μm; f, 50μm). i, Validation of Trans-Seq marker gene expression from ESC2 using RNA-Scope in situ hybridization. Cdh7 (red) and Slc17a6 (blue) double-positive neurons are enriched in the Rorb-YFP line primarily for SC stellate cells as quantified in j, n=5 animals. l, Validation of Trans-Seq marker gene expression from ESC3 using RNA-Scope in situ hybridization. Tac1 (red) and Slc17a6 (blue) double-positive neurons are enriched in the Grp-KH288-YFP transgenic line for narrow-field excitatory neurons as quantified in m. n=5 animals, Scale bars: (i, l, 50μm). Dotted yellow circles (f, i, l) indicate overlapped in situ signals with GFP staining, and red circles in i indicate in situ signals non-overlapping with GFP staining. Quantification plots ****, p<0.0001, two-sided Student’s t-test. (g, j, m). All data in this figure are presented as mean ± SEM.
First, we validated marker gene expression of ESC1–3 (Fig. 5d, h, k) using in situ hybridization onto established transgenic lines23. These lines mark three major excitatory neuron types in the superficial SC characterized by morphology: Wide-field neurons (Ntsr1-GN209) (Fig. 5e), Stellate neurons (Rorb-Cre knock-in marks these neurons in addition to other SC neuron types) (Fig. 5i), and Narrow-field neurons (Grp-KH288) (Fig. 5l), facilitating electrophysiological and morphological analyses. We started by defining the molecular identities of these neurons and registering transcriptomic-defined types, ESC1–3, onto these functionally defined GFP transgenic lines. We validated two top marker genes from ESC1: Nephronectin (Npnt)15 and Cerebellin2 (Cbln2)24 (Fig. 5c). Npnt and Cbln2 are enriched in wide-field neurons (Fig. 5d, Extended Data Fig. 5h). High Npnt expression is restricted within the stratum opticum (SO), marking 88% of Ntsr1-GN209-YFP neurons (Fig. 5e, f, g). Npnt was not detected in inhibitory neurons (Extended Data Fig. 5a–c). We also validated an Npnt antibody showing that Npnt is restrictively expressed by Ntsr1-GN209-YFP neurons in the SO within the deep sublamina of the retinorecipient area (Extended Data Fig. 5d). In contrast, Npnt protein is absent from Narrow-field neurons and Stellate neurons (Extended Data Fig. 5e, f). Importantly, we found near-absent Ntsr1 expression from the scRNA-Seq analysis. Thus, Ntsr1-GN209-YFP expression may not reflect the endogenous Ntsr1 gene expression. As Npnt serves a bona fide marker for ESC1. We therefore defined ESC1 as Npnt-positive wide-field neurons (NPWFs). We also validated markers for ESC2 and ESC3 (Fig. 5c). These included Cdh7 (Fig. 5h) for ESC2 and Tac1 (Substance-P, Fig. 5k) for ESC3, in addition to Slc17a6 (vGluT2), a secondary marker for excitatory neurons (Extended Data Fig. 4c). We found that Cdh7/Slc17a6 ESC2 neurons are putative stellate cells marked by the Rorb-YFP cells in the upper SGS (Fig. 5i, j); whereas, Tac1/Slc17a6 double expression in ESC3s matches with narrow-field neurons marked by the Grp-KH288-YFP cells (Fig. 5l, m).
Second, we examined whether SC clusters identified through Trans-Seq receive retinal inputs functionally. We inquired whether mWmC tracing and subsequent Trans-Seq revealed such synaptic specificity beyond just laminar localizations within the SC. We focused on the SO, where NPWF (ESC1) somata localize. We characterized one neuronal cluster, absent in Trans-Seq. Traditional in situ hybridizations characterized a population of SC neurons expressing a high level of Etv1 (Er81) in the SO layer22 (Fig. 6a, b), which did not uptake mWmC (Fig. 6c). We applied optogenetics-assisted circuit mapping to validate this finding, as established above (Fig. 1g), we rarely detected monosynaptic retinal inputs onto Etv1-positive SO neurons (Fig. 6e, f), which was in contrast to the strong retinal monosynaptic inputs onto the Ntsr1-GN209-YFP SO neurons (ESC1, Fig. 6d, f). The tracing data and functional validations indicated that mWmC-mediated tracing reveals precise connectivity based on synaptic specificity beyond just sublaminar positions.
Fig. 6. Etv1-positive neurons in the SO do not uptake mWmC from the retina and do not receive retinal monosynaptic inputs.

a, in situ hybridization of Etv1 (red, RNA-probe) showed no overlap with ESC1 neurons (green), even though the somata of both populations reside in the same sublamina (SO) at the SC. Scale bars: 100 μm. b, Genetically labeled Etv1-positive neurons (Etv1-CreER; LSL-YFP) were detected as a subset of neurons within the SO, above the vAChT-positive bands marking the SGI (red). Scale bar: 100 μm. The dotted line marks the pial surface. c Etv1-positive neurons were not labeled by mWmC following retinal injections of AAV2-WmC. In contrast, neighboring cells within the SO were labeled by mWmC. Red dotted circles mark cells that were not labeled by mWmC. Scale bar: 20um. d-e, Whole-cell recordings showed Etv1-positive SC neurons do not receive retinal inputs (e), while the ESC1s (Ntsr1-GN209-YFP) receive direct retinal inputs (d). The stimulation paradigms were established in Fig. 1. The blue lines indicate the onset of blue light to active ChR2.Postsynaptic currents persisted in TTX (1μM) and 4-AP (100μM). F, Average ESPC amplitudes in ESC1s (black) and Etv1-positive neurons (red). n= 5animals, ****, p<0.0001, two-sided Student’s t-test. Data in this figure are presented as mean ± SEM.
Third, to cross-compare the identified marker genes from Trans-Seq data, we examined the gene expression correlations from ESC1, ESC2, and ESC3 to MEGLU6, MEGLU5, and MEGLU4 using a published scRNA-seq adult brain database25. Npnt and Cdh7 qualified as top markers based on this orthogonal dataset25 (Extended Data Fig. 6a). Trans-Seq data identified three among six collicular excitatory neuronal clusters (Extended Data Fig. 6a–e)25. Importantly, Trans-Seq recapitulated the specific markers for each of these three neuronal types, separating the collicular ESCs into retinorecipient versus non-recipient neurons. Taking the MEGLU3 marker as an example, we found that this cluster represented a population of SO neurons expressing Pou4f2, distinct from ESC1s (Extended Data Fig. 6f, i). These Pou4f2-positive neurons did not uptake mWmC from the retina (Extended Data Fig. 6g, h). Thus, transcriptomic data from Trans-Seq offers direct insights regarding the connectivity of these distinct neuronal clusters (MEGLU3 versus MEGLU6/ESC1). Such information is unavailable from standard single-cell RNA-Seq experiments using whole tissue dissection and dissociation. We could distinguish retinorecipient SC neuron clusters from non-retinorecipient clusters for their differential roles in local SC computational units7. Together these validations using genetics, immunohistochemistry, and electrophysiology demonstrate the fidelity of transcriptional neuronal classification by Trans-Seq and its ability to define circuit connectivity simultaneously.
RGC type-specific downstream SC neuronal clusters revealed by differential Trans-Seq
We applied Trans-Seq to map the differential wiring paradigms. We chose two RGC subclasses as starter neurons, αRGCs, and ooDSGCs, for differential Trans-Seq, to directly compare the downstream SC targets. Using Kcng4-Cre for αRGC subclass and Cart-Cre for ooDSGC subclass (Fig. 7a) as starter neurons, Cre-dependent mWmC tracer (Extended Data Fig. 7a–e) was selectively expressed within each RGC subclass. The fluorescence intensity of mWmC within cells across the SC followed a bimodal fluorescence intensity distribution (Extended Data Fig. 7f), which ensured a clear definition of connected neurons. Their positions could then be marked to determine their differential distribution along with the depth of the SC (Extended Data Fig. 7g–i). This bright fluorescent labeling was furthermore critical for the success of recovery of mCherry+ by FACS (Extended Data Fig. 3g). We isolated and profiled mWmC-positive SC neurons from the two different starter neurons. After aligning the two datasets with the pan-RGC Trans-Seq dataset (Fig. 7b), we found different proportions of 3 ESCs were represented in each dataset, indicating differential connectivity: ESC1 (NPWFs) were present among αRGC- but not ooDSGC-connected neurons (Fig. 7c), ESC2s were enriched among ooDSGC-connected neurons, and ESC3s were enriched among αRGC-connected neurons (Fig. 6c). These data generated a prediction of selective synaptic wiring from αRGCs, but not ooDSGCs to NPWFs (ESC1). We established an Npnt-FlpO knock-in mouse line to mark and manipulate NPWFs (Fig. 7g, h, Extended Data Fig. 8a–c) in conjunction with existing Cre drivers to mark RGCs. NPWFs elaborated two or three primary dendrites from the soma and extended their distal dendrites to the pia surface of the SC (Fig. 7h, Extended Data Fig. 8d–h), matching the known morphological and physiological features of classic SC wide-field neurons23. We demonstrated that mWmC was anterogradely transferred to a high fraction of NPWFs (Extended Data Fig. 8k, l) from αRGCs, but not from ooDSGCs (Extended Data Fig. 8k, m). We also functionally validated the selective αRGC to NPWF connection (Fig. 7i). We delivered Cre-dependent ChR2-YFP within the left eyes to activate presynaptic RGC subclasses. Td-Tomato-positive NPWF neurons in SC slices were recorded at 4wpi. We detected monosynaptic contacts from αRGCs (Kcng4: ChR2YFP) to NPWFs (Fig. 7j, l, Extended Data Fig. 9a, b), but few NPWFs were found to receive inputs from ooDSGCs (Cart: ChR2YFP)(Fig. 7k, l, Extended Data Fig. 9c).
Fig. 7. Comparative analysis of the Trans-Seq data predicted a selective synapse from αRGCs, but not ooDSGCs to Npnt-positive Wide-field neurons (NPWFs), confirmed experimentally.
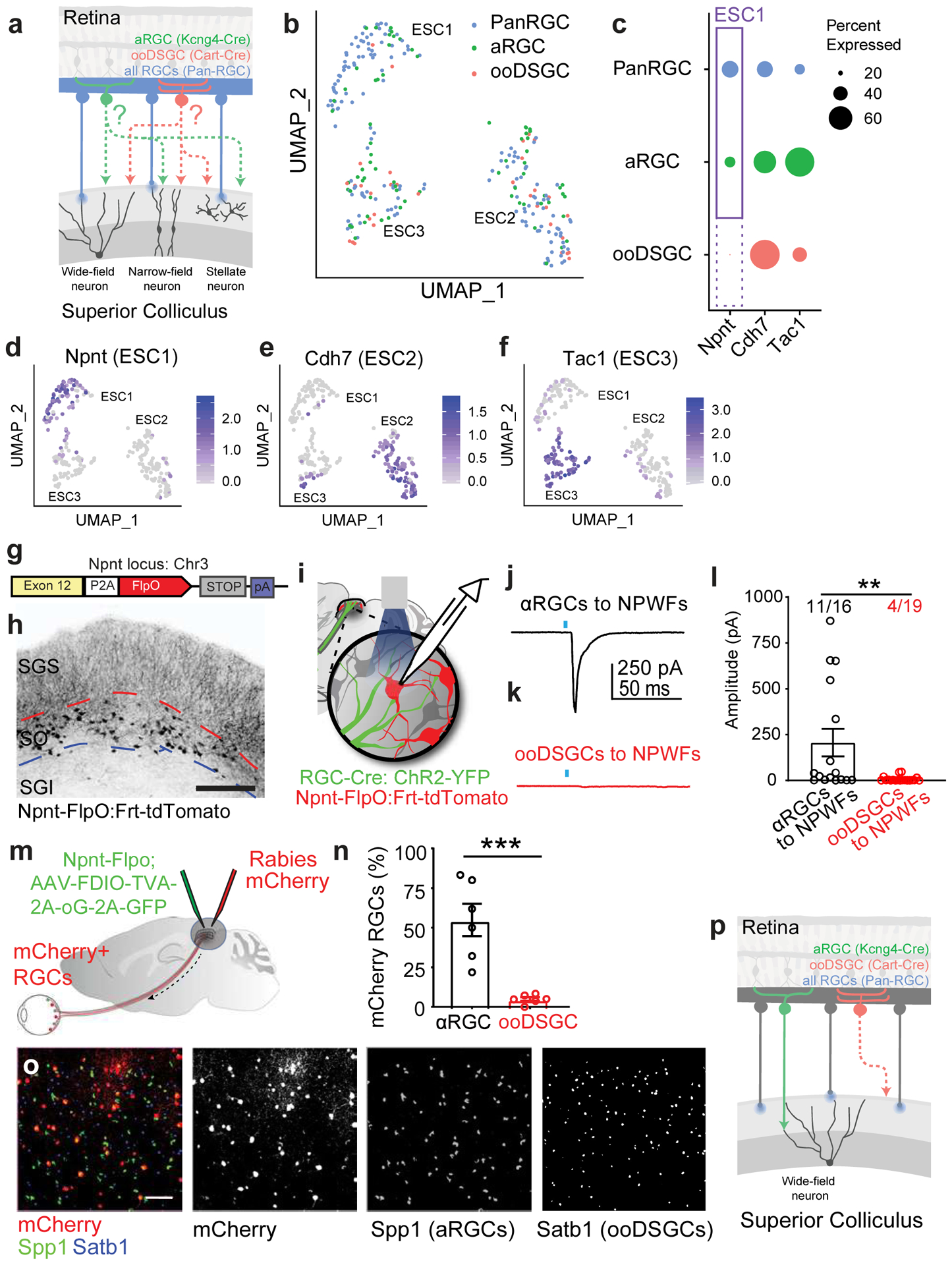
a, Schematic drawing for a comparative Trans-Seq between the downstream SC neuron types of αRGCs (green) and ooDSGCs (red), in addition to the existing pan-RGC (blue). Tracing dataset, with three replicates for each. The goal is to identify selective retinotectal circuits from RGC types to SC neuron types. RGC subclass-specific Cre-drivers include Kcng4-Cre for αRGCs and Cart-Cre for ooDSGCs. b, UMAP plots generated after aligning the pan-RGC (blue), αRGC (green), and ooDSGC (red) tracing datasets and clustering 268 excitatory neurons into the three ESCs established in the pan-RGC mapping (Fig. 4). Notably, very few red dots from ooDSGC tracing are present in ESC1, indicating limited ooDSGCs innervation of ESC1s (NPWFs); By contrast, the αRGC tracing dataset (green) contain significant ESC1 [ESC1, 67 blue, 18 green, 2 red; ESC2, 62 blue, 29 green, 18 red; ESC3 32 blue, 25 green, 15 red). c, Dot-plot of three ESCs showing differential gene expression of validated marker genes among three different tracing datasets from pan-RGCs (blue), αRGCs (green), and ooDSGCs (red). Confirmed marker genes were established in Fig. 3, including Npnt for ESC1, Cdh7 for ESC2, and Tac1 for ESC3. The sizes of the dots encode the percentages of cells expressing each marker gene within each RGC tracing dataset. The presence of ESC1 (Npnt+) in αRGC tracing datasets (solid-line frame), but the absence of ESC1 (Npnt+) in ooDSGC tracing datasets (dotted-line frame), indicate that ESC1s receive selective inputs from αRGCs but not ooDSGCs. d-f, UMAP plots validating the same set of ESC markers in the combined tracing datasets of pan-RGCs, αRGCs, and ooDSGCs. d, Enriched Npnt expression in ESC1 (NPWF neurons), e, Cdh7 in ESC2, and f, Tac1 in ESC3. The normalized log expression of each gene is presented here. g, Design of Npnt-FlpO targeting vector to mark and manipulate Nephronectin-positive wide-field neurons (NPWFs) using the endogenous Npnt locus on mouse Chr3. h, Npnt-FlpO; Frt-Td-Tomato specifically labels ESC1 as a unique neuronal population in the SO but not in SGI. Scale bar: 250 μm. i, Schematic drawing of the binary genetic strategy to examine selective connectivity from specific RGC-subclasses (RGC-Cre; AAV-FLEX-ChR2-YFP, green) to NPWF neurons (Npnt-FlpO; Frt-TdTomato, red), using optogenetics-mediated electrophysiology. Kcng4-Cre; Npnt-Flp and Cart-Cre; Npnt-Flp crosses were compared. j, Average of five trials of evoked EPSCs recorded from one NPWF neuron, driven by ChR2-YFP expressed in αRGC (Kcng4-Cre; AAV-DIO-ChR2-YFP). The blue dot indicates the 2-ms blue LED stimulation, followed by a monosynaptic evoked current, n=7animals. k, Sample EPSC trace recorded from NPWF neurons, driven by ooDSGCs (Cart-Cre; AAV-DIO-ChR2-YFP), n=4animals. l, Average EPSC amplitudes were quantified. Percentages of connectivity were compared. Significantly higher ChR2-mediated synaptic currents onto Npnt-positive ESC1 neurons were detected from αRGCs (black). In contrast, very small currents can be detected in a few ooDSGCs (red), which can be further blocked by TTX (1μM) and 4-AP (100μM)(Extended Data Fig. 13c). **, p<0.001, two-sided Student’s t-test. m, Schematic drawing of Npnt-FlpO-dependent retrograde tracing from NPWFs to the retina at neuronal type resolution, using two viral components (Flp-dependent EGFP-2a-TVA-2a-oG, green; and RdGV-mCherry, red) that infect SC neurons in green and red, and retrogradely labeled RGCs in red only. n, Percentage of αRGCs (Spp1, black) and ooDSGCs (Satb1, red) RGCs among all mCherry-positive RGCs were quantified, n= 6 animals. ***, p<0.005, two-sided Student’s t-test. o, Retina wholemount images showing that retrogradely labeled RGCs (mCherry-positive, red) highly overlap with αRGCs (SPP1-positive, green), but not ooDSGCs (Satb1-positive, blue). Scale bar: 100 μm. p, Schematic drawing for the finding based on Trans-Seq prediction and experimental validations shows a selective retinotectal synapse from αRGCs (green), but not ooDSGCs (red) to NPWFs in the SC. All data in this figure are presented as mean ± SEM.
We also verified the finding using retrograde tracing system. We introduced the rabies-helper cassette (FDIO-EGFP-2A-TVA-2a-oG) into the right SC of Npnt-FlpO mice, followed by a second injection of EnvA-dG-Rabies-mCherry (Fig. 7m, Extended Data Fig. 9d)26. We evaluated the RGC subclass composition of Rabies-mCherry retrogradely labeled RGCs in the contralateral (left) eyes. NPWFs primarily traced back to αRGCs, as well as a few other RGC subsets, but not to ooDSGCs labeled by Satb127 (Fig. 6n, o, Extended Data Fig. 9e–k). The fidelity of the Npnt-FlpO line for NPWFs combined with the optimized retrograde tracing system26 allows comprehensive determination of connected RGC types. Altogether, using the retinotectal projection as the model system, we demonstrated how Trans-Seq could be utilized to define neuronal clusters, identify morphologically and functionally characterized recipient neurons of genetically defined presynaptic cells, and ultimately reveal a previously uncharacterized and selective neuronal pair from the retina to the brain (Fig. 7p).
Npnt instructs the assembly of the selective retinotectal connection via Itga8 recognition
We reasoned that transcriptomic data of Trans-Seq offer a rational strategy to identify molecular candidates for selective circuit wiring based on differential expression of molecules within the connectivity map. To understand the molecular mechanisms underlying the synaptic choice from αRGCs to NPWFs, we asked whether Nephronectin (Npnt), one of the top marker genes from transcriptomes of ESC1 (Fig. 5c, d), may regulate wiring specificity. Npnt is a member of the EGF-like superfamily of extracellular matrix glycoproteins and has been shown to mediate heterophilic cell-cell adhesion between Npnt-expressing cells and Itgα8β1-expressing cells in skin and kidney28,29. The restricted sublaminar expression and cell-adhesion properties of Npnt made it a strong candidate for mediating selective synapse formation from RGCs to the SC.
αRGCs and ooDSGCs send their axons to lower and upper SGS lamina, respectively (Extended Data Fig. 9m, n). Therefore, we used used RGC axon lamination patterns as a proxy for altered RGC-SC connectivity. First, we examined axon lamination patterns following selective elimination of NPWFs30 (Extended Data Fig. 10a–c). NPWF neuron elimination resulted in αRGC axon mistargeting into the upper SGS sublamina, reaching as far as the pia (Fig. 8b, c), while control αRGC axons arborize within a restricted lamina in the lower SGS31 (Fig. 8a). NPWF elimination did not perturb ooDSGC axons (Fig. 8d–f). Second, we tested whether selective Npnt gene knockout from the SC phenocopies NPWF elimination, using SC-specific Npnt knockout approach (Extended Data Fig. 10f–h)29. We found that αRGC axons were no longer confined to the lower SGS, with a large fraction of their axons sprouting into the upper SGS (Fig. 8g–i). In contrast, ooDSGC axons were unaffected (Fig. 8j–l). Third, functionally, we generated Npnt-FlpO; H11-Cas932 mice to simultaneously visualize NPWFs and enables efficient CRISPR-mediated Npnt deletion in vivo (Extended Data Fig. 10o–r). The Cas9/sgRNA strategy for Npnt deletion also resulted in αRGC axonal mistargeting phenotypes into the upper SC lamina, consistent with the Npnt knockout analysis (Fig. 8g–i). Functionally, we observed a significant decrease in the percentages of connected cells and the average ESPC amplitudes in the Npnt deletion group (Fig. 8u, v) in terms of αRGCs to NPWFs synaptic connections. Biochemical studies have shown that Npnt possesses the highest binding affinity for α8β1 integrin over other RGD-binding integrin receptors33. Since the α8 subunit solely heterodimerizes with the β1 subunit to form α8β1, expression of Integrin α8 directly reflects that of α8β134. We found that 74.6% αRGCs produce high levels of α8, while ooDSGCs have minimal α8 (3.4%, Fig. 8s, t). Following an in utero embryonic retinal injection mediated CRISPR/Cas9 to knockdown Itga8 specifically (Extended Data Fig. 10k–n), αRGC axons displayed mistargeting axon phenotypes (Fig. 8m–o), phenocopying the deficits in Npnt knockout. In contrast, the ooDSGC axons remained unaffected (Fig. 8p–r). Collectively, our current data reveal a unique role for Itgα8β1 in αRGCs and Npnt in NPWFs in guiding the selective axonal sublaminar choice, affecting subsequent synaptic recognition (Fig. 7w).
Fig. 8. Npnt instructs the selective retinotectal circuit wiring from αRGCs to NPWFs.
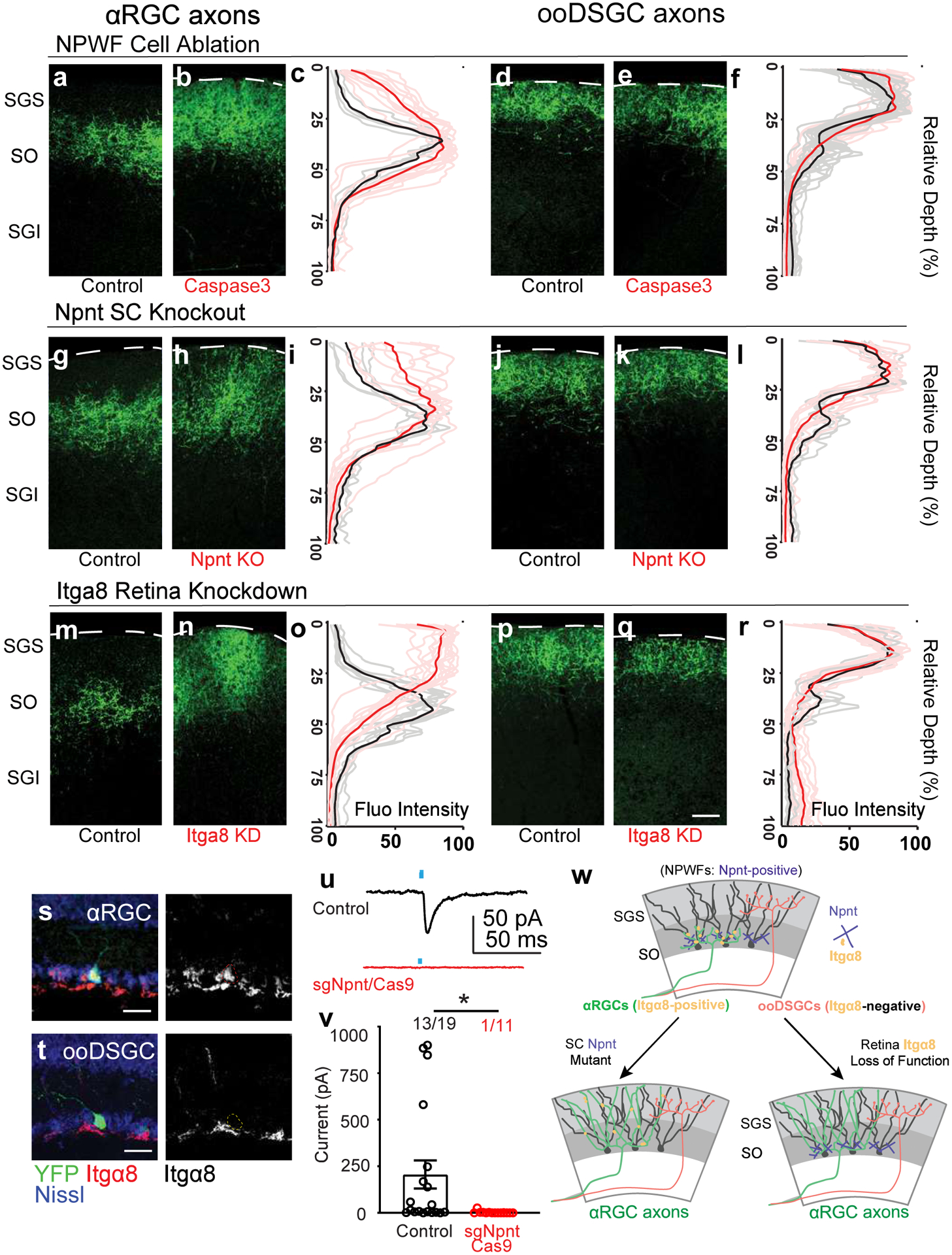
a, b, Sample images of αRGC axons (Kcng4-YFP) in the SC, subject to NPWF neuron elimination mediated by (Npnt-FlpO; AAV-FDIO-Caspase3-Tev) b and control a. c, Normalized GFP axon fluorescence intensity of line scans drawn perpendicular to the SC of control (black line is mean intensity curve, gray lines are each example) and after NPWF elimination (red line is mean intensity curve, light red lines are each example). The dotted line marks the pial surface. d, e, Sample images of ooDSGC axons in the SC, subject to NPWF neuron elimination mediated by (Npnt-FlpO; AAV-FDIO-Caspase3-Tev) e and control d. f, Quantification as in c for ooDSGC axons. (n >=20 axons per animal, n=4 animals, per genotype). g to l, Sample images of αRGC axons in the SC subject to SC-specific Npnt knockout mediated by AAV-Cre injection, leading to axonal sublamination deficits h and control g. i as in c, Quantification shows the ectopic sublaminar distribution of αRGC axons in the upper SC, phenocopying NPWF-elimination c. j, k, Sample images of ooDSGC axons in the SGS, subject to SC-specific Npnt knockout mediated by AAV-Cre injection, k and control j. l, Quantification as in c showing that ooDSGC axons are not perturbed subject to Npnt knockout. (n >=20 axons per animal, n=4 animals, per genotype). m to r, sgRNA/Cas9 targeting Itga8 was delivered at embryonic (E) day 14.5 through in utero intraocular injections. m, n, Sample images of αRGC axons in the SC, subject to retina-specific Itga8 knockout mediated by sgRNA-Itga8/Cas9, leading to axonal sublamination deficits n and control m. o, Quantification shows the ectopic sublaminar distribution of αRGC axons in the upper SGS, phenocopying Npnt mutants h. Sample images of ooDSGC axons in the SC, subject to retina-specific Itga8 knockout mediated by sgRNA-Itga8/Cas9 q and control p. r, Quantification shows that ooDSGC axons are not perturbed subject to Itga8 knockout. (n>=20 axons per animal, n=4 animals, per genotype). Scale bars: (a-q, 50 μm). s, t, Sample images show selectively enriched expression of Itga8 in αRGCs (s, 74.6±8.3%, n=5 animals) but very limited if any expression in ooDSGCs (t, 3.4±1.2%, N= 5 animals). Scale bar: 25 μm. u, v AAV-sgNpnt-FDIO-mCherry was injected into the right SC hemisphere (Kcng4-Cre; Npnt-FlpO; H11Cas9) mice, followed by injections of AAV-DIO-ChR2-YFP to induce ChR2-YFP expression in αRGC cells. Kcng4-Cre; Npnt-FlpO; H11Cas9 mice injected with a non-cutter sgRNA-mCherry construct in the SC served as controls. mCherry+ NPWFs were targeted for whole-cell patch-clamp recordings. u, Sample evoked ESPCs traces showing the loss of specific connectivity subject to Npnt-loss-of-function (red), compared to controls (black). Blue dots mark the onsets of blue-light stimulations. v, The average ESPC amplitudes and percent connectivity from αRGCs to NPWFs were assessed by ChR2 activation in αRGCs. Both amplitudes and percentages connectivity decrease after Npnt-knockout (red, n=8 animals), compared to controls (black, n=8 animals). *, p<0.05, two-sided Student’s t-test. w, Model summarizing how Npnt-Integrin interactions guide RGC axons and specify the specific retinotectal synapse. Npnt acts as an anchor for axons from α8 integrin-positive αRGCs to laminate within the lower SGS and forms synapses onto NPWFs. ooDSGCs, which do not express Itgα8, do not bind Npnt and do not synapse onto NPWFs. Data in this figure are presented as mean ± SEM.
Discussion
In summary, Trans-Seq establishes a new technical avenue for genetics-based circuit studies in mammalian systems. It directly couples neuron taxonomy and functional connectivity testing, facilitating discovery by identifying specific neuronal pairs that can be further validated using electrophysiology. Through this means of converting neuronal transcriptomes to digital connectomes and identifying connections, the Trans-Seq platform represents a generalizable anterograde tracing and sequencing approach. Furthermore, leveraging the insights from the single-cell transcriptomic data of Trans-Seq, we identified an extracellular matrix molecule, Nephronectin, required for selective retinotectal sublaminar choice in vivo.
mWGA-mCherry is a broadly applicable anterograde transsynaptic tracer
In developing the mWGA-mCherry (mWmC) tracer for anterograde transsynaptic tracing, we sought to design a genetically encoded single-component tracer targeting specific cell types. We showed that the re-engineered mWmC tracer ensures efficient and specific targeting to monosynaptically connected neurons across diverse brain regions, allowing sensitive and efficient detection of long-range connectivity using native fluorescence signal. We showed that mWmC transfer carries bright red fluorescence and is compatible with live studies, including electrophysiology recordings and FACS (Figs. 1&4). Our side-by-side comparison showed that mWmC possesses significantly higher transfer efficiency into postsynaptic cells than other tested RFP fusion constructs (Extended Data Fig. 1a–c). The unique enrichment of lysosomal mWmC also raises an interesting possibility that the inefficiency of mWmC to escape from the endo-lysosome compartment results in the accumulation of mCherry-fluorescence within the cell soma, which is likely the key to producing high signal-to-noise in recipient neurons. Additional studies into the chemical properties and cell biology of the mWmC construct will help test these possibilities35–37. We also show the degree of anterograde transfer is markedly improved over prior WGA-based tracers. Of the retrogradely labeled cells, many were as bright as starter cells in the injection area, suggesting that labeling likely resulted from retrograde uptake of AAV through axons. The restriction of WGA to starter cells at the injection site is of key importance to reducing retrograde labeling. mWmC-based fluorescence detection further enhanced the signal versus noise ratios in the circuits that we examined. WGA-protein-based transsynaptic tracing greatly suffered from retrograde labeling, mainly due to the uptake of WGA protein by axons projecting to the injection site. Prior studies of WGA also support the idea that dominant retrograde WGA expression is largely due to axon uptake rather than retrograde transport of WGA from starter cells to their presynaptic cells. For example, retrograde labeling was rare in a transgenic mouse line in which WGA expression was induced by Cre-mediated recombination17. Additionally, we established a workflow to test neuronal pairs using electrophysiology. The current study demonstrated such a capacity in retinal and brain circuits (Figs. 1&4). The power of mWmC to enable circuit discovery from genetically defined presynaptic neurons, high transfer efficiency, and suitability for live imaging and recording. Complementary electrophysiology, anatomy, and retrograde tracing methods can substantiate the connectome discovery in other brain areas.
Trans-Seq in advancing the scRNA-Seq approach for circuit reconstruction
Traditionally, efforts towards understanding how a given neuron chooses its postsynaptic partners rely on labor-intensive gain-of-function or loss-of-function studies at the individual gene level, including our past work in the retina circuits38. Trans-Seq focuses on the connected neurons at single neuron-type resolution and offers an orthogonal view of the single-cell RNA-Seq data from conventional whole tissue preparations. For the retinotectal transsynaptic tracing, mWmC excluded non-connected neurons in the deeper SC laminae, which are hard to distinguish based solely on anatomical dissections. Trans-Seq differs from other established tracing and sequencing techniques, such as a Sindbis virus-based barcoding method, termed MAP-Seq39. While MAP-seq, an axon-projection-based single-cell RNA-Seq tracing method, provides a high-throughput mapping of neuron projection patterns (“projectomes”), it does not provide any information regarding the identity of their postsynaptic targets (“connectomes”). All subsequent procedures utilized commercial resources, including FACS and 10XGenomics library kit, followed by standard data analysis. Of note, neuronal loss during dissociation and FACS-enrichment is a known problematic issue for scRNA-Seq of adult neurons. Trans-Seq on retinotectal circuits is reflected in the low recovery rates from FACS to scRNA-Seq library preparation. This shortcoming may be mitigated by complementing single-nuclei RNA-Seq of the targeted area, or database inquiries. Nonetheless, current protocols allowed clear comparisons of the differential connectivity from distinct presynaptic neurons and offers insights into how a given neuron chooses its postsynaptic partners based on its transcriptomics data.
Molecular mechanisms regulating retinotectal circuit wiring
Past studies using the retina projection onto the optic tectum or SC yielded significant insights into the cellular and molecular mechanisms regulating circuit formation. The selective synaptic choices consist of multiple steps. The mouse retina sends its axons to the SC by the neonatal stage. By the second postnatal week, the retinotectal map reaches its adult precision. The axonal innervation pattern is a highly coordinated cellular process in which RGC axons are instructed by molecular guidance cues and structured activity to find their correct sublaminar locations40–42. The increased availability of transgenic mouse reporters has further highlighted the importance of laminar specificity in instructing synaptic specificity from the retina to the brain. Before our work, the molecular cues that mediate RGC axonal sublaminar targeting in mammals were largely unknown43. This gap in knowledge contrasts greatly with the amount of knowledge regarding the molecular cues44–50 or activity-dependent mechanisms51,52 for retinotopic map development in the SC. While Ephrins have been well characterized as essential molecular cues for mediating retinal axon guidance and topographic mapping within the SC, they do not play a role in RGC laminar choice46. By contrast, molecular cues in zebrafish and chick tectum have shown that extracellular matrix (ECM) proteins pre-pattern lamina to guide axons into their target layers or restrict them from neighboring layers; for example, basement-membrane bound Slit1 is important for laminar RGC axon targeting in zebrafish53. Furthermore, ECM proteins such as Tenascin-R54, Versican55, and Nel56 are laminarly expressed within zebrafish or chick tectum and function to inhibit retinal axon outgrowth in vitro.
Utilizing the Trans-Seq platform for biological discoveries, we identified molecular cues that mediate RGC axon sublaminar choice in mammals43. The neuronal type definition of NPWFs and genetic manipulation of Npnt provide critical ideas to understand how diverse RGC axons choose their respective SC sublamina in mammals. Npnt belongs to the EGF-like superfamily of extracellular glycoproteins, including factors that influence axonal growth and targeting, such as tenascins, laminins, and thrombospondins57. Our molecular and genetic work utilized a circuit-tracing-based approach and identified Npnt-Integrin α8β1 signaling as a unique mechanism to guide RGC sublaminar choice and wire up a selective retinotectal circuit from αRGCs to NPWFs. Thus, our current data identified Npnt-integrin signaling as a molecular determinant for RGC to SC sublaminar choice. The anatomical and electrophysiological measurements revealed molecular mechanisms for synaptic choices from αRGCs to NPWFs. However, we know that the αRGCs also synapse onto other SC neuron subtypes (Fig. 6); conversely, other RGCs also converge onto NPWF neurons. The Npnt-α8β1 recognition does not fully account for the assembly of all parallel retinotectal connections. Cell-cell recognition molecules, such as Cdhs14, or Cblns24 may provide additional signaling mechanisms for parallel circuit wiring in vivo. Continued genetic constructions and Trans-Seq data mining of the retinotectal circuits and beyond will help establish a tractable model in the mammalian central nervous system to study long-range circuit assembly across diverse neuron types, mirroring the molecular and functional studies in invertebrate model systems58. We envision the broad application of Tran-Seq and the mWmC tracer in conjunction with genetic and functional studies will offer generalized principles for mammalian circuit assembly across the brain.
METHODS
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committees at UCSF and MIT. Mice were maintained under regular housing conditions with standard access to drink and food in a pathogen-free facility. Immunohistochemistry experiments were carried out using Postnatal (P) P28-P56 mice unless indicated otherwise. Slice physiological recordings were carried out in young adults (6–10 weeks old). The FACS of the superior colliculi and single-cell RNA sequencing experiments were performed at P28–56. Male and female mice were both used in roughly equal numbers; no sexual dimorphisms were observed in retinotectal connectivity, and all ages and numbers were documented. Genotypes were determined by PCR from toe biopsy. Specifically, the following mouse lines were used in the following categories:
Kcng4-Cre and Cart-Cre mouse lines were previously established for studies of retinal ganglion cell (RGC) subclasses. Kcng4-Cre marks all αRGC subtypes, and Cart-Cre marks all ON-OFF direction-selective Ganglion Cell (ooDSGC) subtypes. Both Kcng4-Cre and Cart-Cre are knock-in alleles but are viable and fertile. They show no outward abnormality or retina phenotypes. Both Kcng4-Cre14 and Cart: Cre8,59 lines have been studied in the retina. The dendritic and axonal projection patterns of both lines have also been comprehensively characterized8.
Hb9-GFP transgenic mice express EGFP in ventral-ooDSGCs60, serving as a defined marking line for ooDSGC axons.
Gad2-Cre, Rorb-Cre, Ntsr1–GN209-Cre, and Grp–KH288 –Cre were previously used to label horizontal, stellate, wide-field, and narrow-field neurons in the superior colliculus61. Ntsr1-GN209-Cre and Grp-KH288-Cre were generous gifts of Charles Gerfen (NIMH)62.
vGlut2-Cre (Jax: 016963)63 was previously characterized to label all excitatory neurons in the superior colliculus11.
ChAT- Cre (Jax: 006410) was characterized to mark and manipulate starburst amacrine cells (SACs)38.
Etv1-CreER (Jax: 013048)64 was made for cortical studies, labeling Layer 5 pyramidal cells. Induction of Cre expression was driven by administering 100ug tamoxifen/g bodyweight two times on postnatal days P13 and P15.
Npnt-FlpO (Jax: 034305)65 was a generous gift of J. Ngai (UC Berkeley). In brief, this line was generated through CRISPR/Cas-9-mediated homologous recombination in ES cells.
Thy1-stop-YFP Line #15 transgenic mice express EYFP driven by Cre-recombinase in many neuronal populations66, including most retinal ganglion cells and projection neurons in the brain and spinal cord. We crossed this line with the superior collicular neuron-marking Cre driver lines listed above to visualize neuronal morphology.
Ai65F (Rosa-CAG- Frt-LSL-Frt- TdTomato) express TdTomato driven by FlpO-recombinase67. Ai65F mice were crossed to Npnt-FlpO to label Nephronectin-positive wide-field neurons and visualize somata and dendrites.
Npntfl/fl mice were generated by targeting the first exon of Npnt with flanking loxP sites as previously characterized68. Mutant knockout efficacies were confirmed through immunostaining with a specific antibody targeting Npnt, generated from the same study, established by D. K. Marciano (UT-Southwestern, co-author).
H11-Cas9 knock-in mice were a generous gift from J. Weissman and D. Yang (UCSF and Whitehead Institute). H11-Cas9 mice possess constitutive Cas9 expression through the insertion of Cas9 in the H11 intergenic region on chromosome 11, which drives high-level global gene expression using the CAG promoter32. H11Cas9 mice require only delivery of a specific single-guide RNA (sgRNA) for generating single or multiple simultaneous mutations. We delivered AAV-sgRNAs against Npnt and confirmed the knockout/knockout efficiencies with the same set of antibodies onto the SC tissues. The high-efficacy sgRNA sequences were ATACTTGAGCAGGACCCGTC (#1) and GACATCGACGAGTGCTCTCT (#2), both independently confirmed. We used sgRNA (Non-Cutter): 5′-AACGACTAGTTAGGCGTGTA-3′ targeting Gal4 sequence69 as a control.
Histology
Retina histology: Eyes were collected and fixed in 4% PFA/PBS on ice for 30 minutes, followed by retina dissection, post-fixation for 30 min, and rinsing with PBS. Retinas were analyzed as cryosections and wholemounts as previously described6. Wholemount retina samples were incubated with blocking buffer (5% normal donkey serum, 0.5% Triton-X-100 in PBS) overnight, then incubated for 2–4 days at 4°C with primary antibodies. For sectioning, fixed mouse retinas were incubated with 30% sucrose in PBS for 2 hours, then quickly frozen in OCT as blocks and sectioned at 20μm. Vertical sections were incubated with 0.3% Triton X-100, 3% donkey serum in PBS for 1 hour, and then with primary antibodies overnight at 4°C. Secondary antibodies were applied for 2 hours at room temperature. Retinas or sections were mounted onto glass slides using Vectashield (Vector Lab) or Prolong Gold Antifade medium (Life Technologies).
Brain section histology: 4 weeks after the intraocular injection, animals were deeply anesthetized and transcardially perfused with 10% sucrose in Milli-Q water, followed by ice-cold 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. After dissection, the brains were post-fixed in the same fixative solution overnight at 4°C and cryoprotected in 30% sucrose in phosphate buffer saline (PBS) at 4°C until they sank. The brains were either embedded in OCT compound (Tissue-Tek) and frozen in dry‐ice‐cooled ethanol or directly frozen on a sliding microtome stage cooled with dry-ice. 40–80μm free-floating coronal sections were made using a cryostat or sliding microtome (Leica). For mWGA-mCherry immunostaining, sections were incubated in a blocking buffer for 1 hour at room temperature. Sections were then incubated with antibodies in the blocking buffer overnight at 4°C. After washing in PBST, sections were incubated with antibodies in the blocking buffer overnight at 4°C. After washing in PBST, sections were mounted on slide glasses.
Antibodies used were as follows: rabbit and chicken anti-GFP (1:1000, Millipore; 1:500, Abcam); rabbit anti-RFP (1:500 Rockland); goat anti-WGA (1:500, Vector Labs); mouse anti-NeuN (1: 500, Millipore); mouse anti-GFAP (1:500, Sigma); rabbit anti-Satb1 (1:1000, Abcam); goat anti-choline acetyltransferase (ChAT) (1:500, Millipore); rabbit anti-Cart (1:2500, Phoenix Pharmaceuticals); rabbit anti-Melanopsin (1:5000, Thermo Scientific); guinea-pig anti-RBPMS (1:1000, PhosphoSolutions); goat anti-Osteopontin/Spp1 (1:500, R&D); rabbit anti-Nephronectin (1:100, produced in68); goat anti-Itga8 (1:100, R&D); rabbit anti-Lamp1 (1:500, abcam); mouse anti-Brn3b/Pou4f2 (1:500, Santa Cruz Biotechnology). Nuclei were labeled with NeuroTrace Nissl 435/455 (1:500, Molecular Probes). Secondary antibodies were conjugated to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647 (Jackson ImmunoResearch) and used at 1:500.
RNA-Scope in situ hybridization
In situ hybridization was performed using RNA-Scope Fluorescent Multiplex Kit (Advanced Cell Diagnostics) following the manufacturer’s instructions. In brief, sections were fixed in 4% PFA as regular brain sections described above in the histology section, washed in PBS, dehydrated in a series of ethanol washes, and dried. A hydrophobic barrier was drawn around the sections with an ImmEdge pen (Vector Lab, H-4000). The sections were treated with Protease IV, incubated with target probes, treated with a series of amplification reagents, and developed with TSA fluorophores. Slides were mounted using Vectorshield (Vector Lab). RNA-Scope Probes included the following ones: RNAscope® Probe- Mm-Cbln2 (Cat No. 428551); RNAscope® Probe- Mm-Npnt (Cat No. 316771); RNAscope® Probe- Mm-Cdh7 (Cat No. 520761); RNAscope® Probe- Mm-Tac1 (Cat No. 410351); RNAscope® Probe- Mm-ETV1 (Cat No. 557891).
AAV vectors
-
mWGA-mCherry: Codon-optimized WGA (mWGA) was synthesized at Genewiz Inc and fused with red fluorescent proteins (RFPs). We used mCherry (p-mCherry-N1, p-mCherry-C1, Clontech) to tag the N- or C-terminus of mWGA and then cloned the mWGA-mCherry or mCherry-mWGA fusion fragment into an AAV-CAG-overexpression-WPRE vector. In the Cre-dependent cassette (Flex-switch), we inserted the fusion protein (mWGA-mCherry) in the 3’ to 5’ orientation to achieve Cre-dependent control. These vectors are available at Addgene for requests. The reference sequence for mWGA-mCherry cDNA (Genewiz) was optimized as below.
ATGGAGACCGACACCCTGCTGCTGTGGGTGCTTCTGCTGTGGGTCCCTGGCAGCACTGGCGATGGGCCTGTGATGACCGCCCAAGCTCAGAGGTGCGGCGAGCAAGGCAGCAACATGGAGTGCCCTAATAACTTGTGCTGCTCTCAGTACGGCTATTGCGGCATGGGTGGCGACTACTGCGGCAAGGGCTGTCAGAACGGCGCCTGCTGGACTAGCAAGAGGTGCGGCTCCCAAGCCGGCGGTGCCACCTGCCCTAACAATCACTGTTGCTCACAGTACGGTCACTGCGGCTTCGGCGCCGAGTACTGTGGGGCTGGTTGCCAAGGCGGCCCTTGTAGGGCCGATATCAAGTGCGGCAGTCAAAGCGGCGGCAAATTGTGCCCTAACAACCTGTGCTGCTCTCAGTGGGGTTTCTGCGGACTGGGAAGCGAGTTTTGCGGCGGCGGGTGTCAATCCGGCGCTTGTAGCACCGACAAGCCTTGCGGCAAGGACGCCGGCGGAAGGGTGTGCACCAACAACTACTGCTGCAGCAAATGGGGATCGTGTGGCATAGGCCCTGGCTACTGCGGCGCTGGGTGTCAGTCGGGCGGCTGCGACGCCGCTAGGGACCCTCCTGTGGCAAGCGCCACCATGGTGAGCAAGGGCGAGGAGGACAACATGGCCATCATCAAGGAGTTCATGAGGTTCAAGGTGCACATGGAGGGCAGCGTGAACGGCCACGAGTTCGAGATCGAGGGCGAGGGCGAGGGAAGGCCTTACGAGGGCACACAGACCGCCAAGCTGAAGGTGACCAAGGGCGGCCCTCTGCCTTTCGCCTGGGACATCCTGAGCCCTCAGTTCATGTACGGCAGCAAGGCCTACGTGAAGCACCCTGCCGACATCCCTGACTACCTGAAGCTGAGCTTCCCTGAGGGCTTCAAGTGGGAGAGGGTGATGAACTTCGAGGACGGCGGCGTGGTGACCGTGACCCAAGACAGCAGCCTGCAAGACGGCGAGTTCATCTACAAGGTGAAGCTGAGGGGCACCAACTTCCCTAGCGACGGCCCTGTGATGCAGAAGAAGACCATGGGCTGGGAGGCAAGCAGCGAGAGGATGTACCCTGAGGACGGCGCCCTGAAGGGCGAGATCAAGCAGAGGCTGAAGCTGAAGGACGGCGGCCACTACGACGCCGAGGTGAAGACCACCTACAAGGCCAAGAAGCCTGTGCAGCTGCCTGGCGCCTACAACGTGAACATCAAGCTGGACATCACAAGCCACAACGAGGACTACACCATCGTGGAGCAGTACGAGAGGGCCGAGGGAAGGCACAGCACCGGCGGCATGGACGAGCTGTACAAGTGA
sgRNA Constructs: To make AAV-U6sgRNA-EF1-FDIO-mCherry (sgRNA-Npnt and control), or Lentiviral-U6-sgRNA-Ef1aCas9–2a-Cherry (sgRNA-Itga8 and control), the following sequences were used: sgRNA- Npnt: ATACTTGAGCAGGACCCGTC (#1) and GACATCGACGAGTGCTCTCT (#2), #1 sgRNA-for Npnt was validated with high potencies and primarily used in this study.
sgRNA-Itga8: CCCCTTCTACGATATCCGGC (#1 for Itga8) and GCTCCCCCTTCTACGATATC (#2 for Itga8). Both sgRNA showed high potencies and were applied in the current study using the CRISPR-V2-Cherry system (Addgene) using high titer lentiviral vectors. sgRNA-Non-Cutter Control: 5′-AACGACTAGTTAGGCGTGTA-3′ (targets Gal4 sequence) as published by69 was adopted for both lentiviral CRISPRV2-Cherry vector, and subsequently in AAV-sgRNA-FDIO-mCherry vector.
These oligonucleotides were separately amplified and cloned into the lentiviral CRISPRV2-Cas9-2a-Cherry vector. For AAV applications, U6-sgRNA fragments were further amplified by PCR and subcloned into an AAV-U6-sgRNA-FDIO-mCherry vector. sgRNA for Npnt knockout efficacy was determined by immunostaining against Npnt in the collicular tissues subject to neonatal AAV injection, compared to controls; sgRNA for Itga8 knockout efficacy was determined by immunostaining against Itga8 in the retinas subject to embryonic lentiviral injection. Rabbit anti-Npnt (1:1000, D. Marciano Lab, UT-Southwestern) or goat anti-Itga8 (1:100, R&D) antibodies were used immunohistochemistry.
AAV and Lentivirus production
All AAVs were made at the Boston Children’s Hospital Viral Core at the titer of (2×10E13 GC/Unit) unless stated otherwise. All experiments were carried out using AAV Serotype 2. AAVs used in the current study included:
AAV2-CAG-mWGA-mCherry-WPRE; AAV2-CAG-DIO-mWGA-mCherry-WPRE; AAV2-CAG-mWGA-Ruby3-WPRE; AAV2-CAG-mWGA-TdTmt-WPRE; AAV2-CAG-mCherry-mWGA-WPRE; AAV2-CAG-WGA(original)-WPRE; AAV2-CAG-mWGA-WPRE; AAV2-EF1a-FDIO-EGFP-WPRE; AAV2-sgRNA-EF1a-FDIO-mCherry-WPRE (For both Npnt knockout and Non-cutter controls); AAV2-CAG-ChR2-YFP-WPRE; AAV2-CAG-DIO-ChR2(H134R)-YFP; AAV2-EF1a-DIO-EGFP; AAV2-CAG-Cre-WPRE. AAV2-EF1a-FDIO-Caspase3-2a-TEV and AAV8-hSyn-FDIO- EGFP-2a-TVA-2a-oG were gifts of X. Chen and G. Nachtrab, Stanford. Pseudo-del-G Rabies Virus-(RV-mCherry) was purchased from Salk Institute Viral Core. Lentiviruses (VSVG, CRISPR-V2-mCherry, Addgene; hSyn-Cre-WPRE, Addgene) were produced using standard triple transfection protocol followed by ultra-centrifugation at the titer of 1E7 Unit70.
In utero intraocular injection
In utero injection of lentiviruses encoding sgRNA/Cas9 was carried out as previously described71. Timed pregnant E14.5 CD1 females were deeply anesthetized with ketamine. The incision area was shaved and cleaned with 70% ethanol and betadine. A midline incision was used to expose the embryos, then removed from the abdominal cavity for injection. Sharpened glass pipettes containing lentiviruses (CRISPV2-Cas9-2a-mCherry, sgRNA-control/Cas9 or sgItga8/Cas9 viruses) mixed with fast green dye were injected into the eyes of targeted embryos by Femto-Jet (Eppendorf). Embryos were then gently placed back inside the abdominal cavity and irrigated with warm, sterile saline. Finally, the abdominal wall and skin were sutured, and the mouse was placed on a warming pad for recovery. After confirmation of genotype after birth, tissue from embryonic transduction experiments was collected and processed at P28. Retinal coverage was assessed by screening for lentiviral mCherry coverage. Only animals with high coverage for mCherry were processed for subsequent analysis.
Postnatal intraocular injection
Intraocular injection of AAVs was performed as previously described14,31.
For WGA tracing experiments, AAV encoding constitutive or Cre-dependent WGA were intravitreally injected into wild-type, Kcng4-Cre, or Cart-Cre mice with 0.5–1μl AAV (in 1X DPBS, AAV was standardized to the same titer of 2×10E13/GC). AAVs were injected into the left eyes using a sharpened glass pipette as previously described8,72. Animals were processed for analysis 4 weeks post-injection (wpi).
For axonal tracing and ChR2YFP stimulations during development, neonatal mice (P2-P4) were anesthetized on ice, and 0.5μl AAV (in 1X DPBS) encoding for AAV-EF1a-DIO-YFP (for axon tracing experiments), or AAV2-EF1a-DIO-ChR2YFP (for optogenetic stimulations) was injected into the left eyes using a sharpened glass pipette.
Optogenetics and anterograde transfer co-injections, AAV2-CAG-ChR2-FYP-WPRE, and AAV2-CAG-mWGA-mCherry-WPRE, were mixed as (1:3) based on titer calculations. All AAVs (Serotype 2 targeting RGCs) were produced at Boston Children’s Hospital Viral Core (AAVs were standardized to the same titer of 2×10E13/GC). Animals were euthanized, and retinas and brains were harvested ~4 wpi.
For the in vivo screen of WGA-RFP tracers (AAV2-CAG-mWGA-mCherry-WPRE; AAV2-CAG-mWGA-Ruby3-WPRE; AAV2-CAG-mWGA-TdTmt-WPRE; AAV2-CAG-mCherry-mWGA-WPRE) 1μl AAV (in 1X DPBS, AAV was standardized to the same titer of 2×10E13/GC). AAVs were injected into the left eyes using a sharpened glass pipette as previously described8,72. Animals were processed for analysis 4 weeks post-injection (wpi). High retinal coverage (> 90% of the regions) of tracers was first confirmed, followed by SC examination using live fluorescent imaging. The number of fluorescent positive SC neurons was quantified under a DIC/Fluorescent setup that is used for electrophysiology experiments. For tissues with histology experiments, animals were further perfused following the protocols listed below.
For the re-engineering of the WGA-based tracers, WGA-Alexa Fluor555 conjugate (Life technology/Molecular Probes; 1.0mg/mL), AAV2-CAG-ctt-WGA-WPRE; and AAV2-CAG-mWGA-mCherry-WPRE were tested and compared using the same intraocular injection methods. WGA-Alexa dye injected animals were analyzed at 1 wpi64,73, while the rest of the AAV-encoded tracer were analyzed at 4wpi. High retinal coverage (> 90% of the regions) of tracers were first evaluated using histology approaches, followed by brain immunohistochemistry experiments.
Neonatal intracranial injection
Superior collicular neonatal injections were carried out as previously described74,75. Neonatal pups (P0–1) were anesthetized with ice. 0.5 ul AAVs were injected using sharpened glass pipettes targeting the right superior collicular hemisphere, visible as a triangular region posterior to the cortex. The lack of further resistance indicates the penetration of the tip past the skull after piercing through the skull.
Stereotaxic injections
For Pseudo-rabies-mediated retrograde tracing, mice were anesthetized with 2% isoflurane and fixed in a stereotaxic apparatus (Model 940, David Kopf Instruments). Meloxicam (5mg/kg) was used for analgesia before the surgery and one day after the surgery by intraperitoneal injection. The viruses were loaded into a pulled-glass pipette connected with a syringe (7634–0, Hamilton) by a dual ferrule adaptor (55750–0, Hamilton). The injection speed and volume were controlled by a Microinjection Syringe Pump (UMP3T-1, WPI). AAV8-hSyn-FDIO-EGFP-2a-TVA-2a-oG viruses (1μl, Titer 1×10E13/GC, Gift of G. Nachtrab and X. Chen, Stanford) were injected into the right hemisphere of the Npnt-FlpO mice by neonatal superior colliculus injections at P0–1 as described above. Four weeks later, the pseudo-typed rabies-mCherry virus (400nl/site, Units/2×10E8) was injected into the right hemisphere at a constant speed (40nl/min). Two sites were injected on the right hemisphere in order to cover the whole superior colliculus. One site was 3.4mm anterior, 0.7mm lateral to the Bregma, and 1.4mm below the skull. The other site was 4.2mm posterior, 0.7 mm lateral to the Bregma, and 1.2mm below the skull. 6–7 days after pseudo-typed rabies injection, the mice were sacrificed with transcardial perfusion. The brains and the left retinas were collected for immunohistochemical analysis.
For adult cortical and collicular tracing and scRNA-seq based tracing, animals were anesthetized with isoflurane during the surgery (4% for induction and 1.5%–3% for maintenance). Meloxicam (5mg/kg) was administered subcutaneously for analgesia before the surgery. The body temperature was maintained at 37°C with a homeothermic blanket (Harvard Apparatus). The animals were fixed in the stereotaxic apparatus (Model 963, David Kopf Instruments) and stereotaxically injected with viruses using a pulled glass pipette connected with a microsyringe pump (WPI). The viruses, animal lines, and injection sites used are as follows: For Figure 1M, a mixture of AAV2-CAG-WGA-mCherry (400nl) and AAV2-CAG-ChR2-EYFP (200nl) (30nl/min) was injected into the vibrissal motor cortex of C57BL/6 mice, 1mm anterior, 1mm lateral to the Bregma, and 0.75mm below the brain surface the vibrissal motor cortex, or the barrel field of the primary somatosensory cortex of C57BL/6 mice, 1.5mm posterior, 3.5 mm lateral to the Bregma, and 0.6mm below the brain surface.
For SC stereotaxic injection and traces comparison, AAV2-CAG-DIO-WGA-mCherry (400nl, 40nl/min) were introduced into the superior colliculus of vGlut2-Cre mice. The coordinate was 3.6mm posterior, 0.7 mm lateral to the Bregma, and 1.0 mm below the skull. To compare the re-engineered mWmC with traditional WGA-based tracers, WGA-Alexa Fluor555 conjugate (Life technology/Molecular Probes; 1.0mg/mL), AAV2-CAG-cttWGA-WPRE; AAV2-CAG-mWGA-WPRE and AAV2-CAG-mWGA-mCherry-WPRE were introduced into the superficial SC. WGA-Alexa dye injected animals were analyzed at 1wpi, while the rest of the AAV-encoded tracer were analyzed at 4wpi. The mice were sacrificed with transcardial perfusion. The brains and the left retinas were collected for immunohistochemical analysis.
To restrict the starter neurons within the region of interest, and to avoid axonal uptake by Cre-expressing neurons in the presynaptic side, we mixed lentivirus-hSyn-Cre and AAV2-CAG-DIO-mWmC at 1:3 ratio and introduced them onto either the superficial SC or the dorsal lateral striatum the coordinate is 0.6mm anterior, 3.0 mm lateral to the Bregma, and 2.8 mm below the skull. Similar to the experiments established above, we traced presynaptic sites to the SC or the Striatum or postsynaptic sites to them using histology approaches as established above.
Slice physiology and optogenetics
For retinotectal connectivity mapping, SC neurons were labeled in mCherry (mWmC-positive for connected neurons; or Npnt-FlpO-RFP positive for genetically labeled neurons), while the presynaptic axons were excited with RGC subtype-specific ChR2-YFP as previously described76. We crossed presynaptic RGC Cre drivers such as Kcng4-Cre or Cart-Cre to postsynaptic Npnt-FlpO mice. ChR2 expression was induced through the injection of AAV2-DIO-ChR2-YFP into the left eye. Labeled NPWF-mCherry or mWmC neurons were identified from the right (and contralateral) SC hemisphere and targeted whole-cell patch-clamp recording. We used blue light to excite RGC axon terminals while recording from SC retinorecipient neurons. Mice were anesthetized with ketamine and xylazine (100 mg/kg, 12.5 mg/kg) and transcardially perfused with ice-cold cutting solution (78.3 mM NaCl, 2.3 mM KCl, 33.8 mM Choline-Cl, 0.45 mM CaCl2, 6.4 mM MgCl2, 1.1 mM NaH2PO4, 23 mM NaHCO3, 20 mM D-Glucose, 0.5 mM L-glutamine, pH 7.4) at 4 wpi. The brains were then dissected, and coronal brain sections were prepared in ice-cold and oxygenated cutting solution by vibratome sectioning (VT1200S, Leica). Mouse superior colliculus slices (250μm in thickness) were collected and transferred to an incubation chamber filled with the cutting solution. The incubation was maintained at 32.5 °C for 30 minutes. After incubation, the brain slices were transferred to room temperature ACSF (125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 1 mM MgCl2.6H2O, 2 mM CaCl2, 20 mM D-Glucose, pH 7.4) before recording. All solutions were constantly bubbled with 5% CO2,95% O2 (Carbogen, Airgas).
Viral coverage on the retina and the SC were examined under a fluorescence microscope (BX51WI, Olympus) to identify a red cell surround by green axons. Targeted whole-cell recordings were made under IR-DIC visualization. Images of the targeted cell were taken by a CCD camera (IR-2000, DAGE-MTI). The recording glass pipettes (BF150-86-10, Sutter Instrument) were pulled by a micropipette puller (P1000, Sutter Instrument) into a recording electrode (3–5 MΩ). The recording electrode was filled with potassium-based internal solution (135mM K-Gluconate, 8 mM KCl, 10 mM HEPES, 0.25 mM EGTA, 2 mM MgATP, 0.3 mM Na2GTP, 7 mM Phosphocreatine, pH 7.3).
The electrophysiology data were acquired with a MultiClamp 700B Amplifier and 1440A Digi-data (Molecular Devices). The traces were digitized at 10kHz and low-pass filtered at 2kHz. The data were analyzed by Clampfit 10.7 (Molecular Devices). Photo-stimulation was performed using a 473 nm LED (Cool LED, pE300). Two light pulses (2ms, 15mW/mm2) were given every trial, five trials per cell. After breaking into the neurons, the cells were held at −65mV. Voltage-clamp experiments were carried out to record EPSCs, and current clamp tests were performed to record action potentials. Tetrodotoxin (TTX, 1 μM) and 4-aminopyridine (4-AP, 100 μM) were added into ACSF perfusions to confirm that postsynaptic currents were monosynaptic. CNQX (10 μM) and APV (50 μM) were added to block the glutamatergic components.
For cortical-striatum connectivity mapping, four weeks after viral injection of AAV2-CAG-WGA-mCherry (2×10E12/GC, Boston Children’s Viral Core) into the primary motor cortex of wildtype C57BL/6 male mice, mice were anesthetized with isofluorane and transcardially perfused in ice-cold slicing solution (2.5mM KCl, 1.25mM NaH2PO4, 25mM NaHCO3, 7mM MgCl2, 0.5mM CaCl2, 7mM dextrose, 210 mM sucrose, 3 mM sodium pyruvate, 1.3 mM ascorbic acid, bubbled with 5% CO2/95% O2). The brain was then extracted, and striatum-containing slices (250μm in thickness) were cut coronally with a Leica microtome (VT-1000S, Leica) and immediately transferred to an incubation beaker filled with an aerated holding solution: 125mM NaCl, 2.5mM KCl, 1.25mM NaH2PO4, 25mM NaHCO3, 2mM MgCl2, 2mM CaCl2, 12.5mM dextrose, 3mM sodium pyruvate, 1.3mM ascorbic acid. The incubator was preset to 34.5 °C to help brain recovery. After about 60-min incubation, we transferred slices to a submerged chamber perfused with aerated normal ACSF containing: 124 mM NaCl, 2.5mM KCl, 1.25mM NaH2PO4, 26mM NaHCO3, 2mM MgSO4, 2.5mM CaCl2, 10mM dextrose (315 mOsm, pH 7.4) and visualized by infrared differential interference contrast and fluorescence video microscopy (Examiner D1, Zeiss). To confirm whether postsynaptic currents were monosynaptic, tetrodotoxin (TTX, 1μM) and 4-aminopyridine (4-AP, 100μM) was bath applied in ACSF. The patch-clamp electrode (4–6 MΩ) was filled with an intracellular solution containing 130 mM D-gluconic acid, 130 mM CsOH, 5 mM NaCl, 10 mM HEPES, 12 mM phosphocreatine, 3mM MgATP, 0.2mM Na2GTP and 1mM EGTA. Photostimulation was performed using a 473nm LED (CoolLED) controlled by Spike2 software (Cambridge Electronic Design). Light intensity was set to trigger the light responses averagely within a range of 100–200 pA with a pulse length of 1 ms once every 5 seconds and was fixed for all cells. To verify the properties of light-evoked synaptic responses on mWGA-mCherry-positive striatum neurons, we applied a combination of CNQX (10μM) and APV (50μM) to block the glutamatergic components. We employed a MultiClamp 700B amplifier (Molecular Devices) for patch-clamp recording and Spike2 software (Cambridge Electronic Design) for data acquisition. The amplitudes of light-evoked responses were analyzed offline with custom-written MATLAB code (MathWorks) by averaging 25 consecutive traces.
Neuronal morphology reconstruction and dye filling
Biocytin (0.5%, B4261–100mg, Sigma) was added to the internal solution for electrophysiology measurement. After recording, the brain slices were fixed with 4% PFA overnight at 4°C, then blocked with blocking solution (PBS with 0.3% TritonX-100 and 5% normal donkey serum) for 2 hours at room temperature. Then the tissues were incubated with Streptavidin-488 (1:1000, Invitrogen) for 2 hours at room temperature. The slices were mounted on glass slides in order and covered with glass slips. The images were acquired using a Leica (SP8) confocal microscope and reconstructed by IMARIS software (Oxford Instruments) or Fiji (NIH).
Neuronal preparation and FACS enrichment of the SC neurons
The contralateral superior colliculi corresponding to four high-coverage eyes without damage were prepared through an electrophysiology slice protocol. Each mouse was transcardially perfused with ice-cold, carbogenated cutting solution (78.3 mM NaCl, 2.3 mM KCl, 33.8 mM Choline-Cl, 0.45 mM CaCl2, 6.4 mM MgCl2, 1.1 mM NaH2PO4, 23 mM NaHCO3, 20 mM D-Glucose, 0.5 mM L-glutamine, pH 7.4). Coronal slices of 500μm thick were cut with a vibratome (Leica VT1200S) in ice-cold carbogenated cutting solution and then immediately transferred into ice-cold, carbogenated ACSF (125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 1 mM MgCl2.6H2O, 2 mM CaCl2, 20 mM D-Glucose, pH 7.4). Slices containing the superior colliculus were transferred to a Petri-dish with ice-cold carbogenated ACSF. The mCherry-positive superficial superior colliculus hemisphere was microdissected under a fluorescence dissection microscope. The dissected regions were transferred into resuspension buffer (Hibernate-A supplemented with B27, 25% glucose, 0.01% BSA, 1% Glutamax, and 0.1mg/mL DNaseI). The mCherry negative SC hemisphere was dissected and processed in parallel to prepare an unstained sample (negative control) for FACS gating. Resuspension buffer was then exchanged with papain solution (2mg/mL papain, 0.1mg/mL DNaseI in Hibernate-Calcium), and the dissected tissue was digested in papain for 25min at 37°C. Following incubation, papain solution was exchanged with trituration buffer (Hibernate-A supplemented with B27, 25% glucose, 0.1% BSA). Tissue pieces were subsequently gently triturated using a 200ul pipette tip to obtain a single-cell suspension. Tissue debris was allowed to settle for 1 min at room temperature, and the supernatant was transferred to a new microcentrifuge tube and centrifuged at 300g for 5 min at 4°C. The supernatant was then exchanged with 1 ml resuspension buffer. The resulting cell suspension was gently pipetted and filtered with a 70μm filter to remove cell clumps.
FACS sorting was performed on a BD FACSAriaII Cell Sorter with a 100 μm nozzle. DAPI was added before sorting at a concentration of 3ng/mL. FACS was used to select cells with low DAPI fluorescence and high mCherry fluorescence. The native (mCherry-negative, without injections) sample was used to establish gating for mCherry. Sorted cells were immediately processed for cDNA library preparation. Cell concentration was confirmed using a Countess hemocytometer.
10XGenomics library preparation and neuronal clustering analysis
Library preparation was performed by the UCSF Institute of Human Genetics Core Facility, using 10X Chromium Single 3Prime Reagent Kits according to the manufacturer’s protocols. Library length and concentration were quantified using a Bioanalyzer (Agilent). A sequencing depth of 50,000 read pairs/cell was targeted for library preparation as recommended by 10x Genomics. Paired-end sequencing was performed on an Illumina NovaSeq. The Cell Ranger pipeline was used to generate FASTQ files, perform an alignment with the mm10 mouse genome assembly, and generate feature-barcode matrixes. The UCSF Institute of Human Genetics Computational Core ran the Cell Ranger pipeline. Downstream analysis was carried out using Seurat V377. Cells with (1) expression of >400 genes; (2) mitochondrial count <30%; (3) expression of neuronal gene signatures Snap25 or Syt1 >0 were retained after filtering. SCTransform, which uses regularized negative binomial regression, was used to normalize UMI count data and regress out mitochondrial genes. The principal component analysis was run using the top 30 principal components (PCs). To integrate datasets, PrepSCTIntegration was run, and integration was run on SCT-transformed data. Cells with the expression of Gad2, Gad1, or Slc17a6>0 were retained. Analysis of one replicate led to the identification of Npnt, Cbln2, Cdh7, and Tac1 as marker genes; therefore, supervised PCA analysis with Gad2, Slc17a6, Tac1, Cdh7, Npnt, Cbln2 as features were implemented on the integrated data. A KNN graph was then constructed using the first 15 PCs. Louvain-based modularity optimization was used to cluster cells with resolution = 0.8 to generate UMAP plots for cluster visualization78. The FindAllMarker function was run to identify top marker genes for each cluster.
To compare outputomes, datasets obtained through αRGC, ooDSGC, and pan-RGC tracing were aligned using SCTransform in the same manner as above. Clusters expressing Slc17a6 and either Npnt, Cdh7, or Tac1 were retained for analysis. The percentage of cell types specified by their molecular markers was quantified in each dataset.
Statistics and Reproducibility
For data acquisition for confocal images: Confocal imaging and subsequent data analysis were performed double-blindly. To analyze RGC axons in the superior colliculus, coronal brain sections were immunostained and imaged on a Leica SP8 confocal microscope equipped with 10X air and 40X and 63X oil immersion objectives. Confocal stacks of 20um thickness were acquired and processed to obtain maximum intensity projections. Using ImageJ, each right SC hemisphere was divided into five equal sections. The fluorescence profile of axon arbors was measured by taking 200μm-thick line scans perpendicular to each section’s pial surface to a depth of 500 μm below the pial surface. Signal intensity values were normalized to the maximum intensity of the curve in across images and plotted. The lowess (Locally Weighted Scatterplot Smoothing) function in Matlab was used for curve smoothing. ImageJ (NIH) software was used to analyze confocal stacks and generate maximum intensity projections. To quantify axon depth across the SC, the shortest distance marking the upper and lower axonal arbors’ boundaries to the pial surface was measured within each section. A Student’s t-test was used to assess significance. At least three animals, taking all SC slices, were used for each experimental condition. No statistical method was used to predetermine sample size.
To quantitatively evaluate the anterograde transfer efficiencies under live conditions for different tracers, a 20X water objective on an electrophysiology rig was used to acquire ten images within the SC. Neurons were counted for visible cellular labeling 4 wpi. To calculate the anterograde transfer efficiencies across different conditions, neurons within a field of view were counted, and the counts across different conditions were normalized to the condition with the highest number of counted neurons. Subsequently, the ratios and statistics were calculated across different experimental animals within the same group, as well as different virus conditions (includinf WGA-protein dye; AAV2-ctt-WGA, and AAV2-mWmC). AAV2-mWmC demonstrated the highest ratio of number of LP neuron/SC neuron labelled among all conditions, and therefore was set to possess an anterograde efficiency of 100%. All other configurations were subsequently normalized to this value. Individual neuron numbers at each relay were documented, and the ratios were calculated and reported across animals for each different tracer configuration. Similarly, the retrograde transfer efficiencies were calculated and normalized. The WGA-555 dye possessed the highest ratio of retrograde transfer among all tested configurations. Thus, it was set as 100%, and subsequently, all retrograde ratios (presynaptic/postsynaptic neuron numbers) were quantified and compared across animals for each different configuration. Statistical methods and the number of animals tested with each configuration are documented for each experiment under the figure legends.
To quantify the signal intensity of mCherry signal within cells, Neurotrace signal was used to generate cell masks. This was done by thresholding the Neurotrace signal within ImageJ, followed by the “fill holes” function, and two erode steps followed by two dilate steps to smooth the cell masks and remove the extraneous signal. Each cell mask was then added as a unique ROI within ImageJ. A custom script was used to determine the mean mCherry intensity within each cell mask and rank all cell ROIs according to decreasing mean intensity. The resulting mean intensities were then plotted in a histogram to visualize a bimodal distribution of mWmC intensity. The intensity corresponding to the valley of the bimodal distribution was used to set the threshold for marking a cell as positive for mWmC. The depth of the centroid of each cell ROI from the pial surface was then measured using ImageJ and plotted. Each trace represents an SC slice from a similar anatomical position taken from separate animals. The traces were normalized by dividing by to the total number of WGA-positive cells in each slice.
To quantify the subcellular overlap of LAMP1 with WGA signals, images were acquired with a 63X objective with a digital 2X zoom for a resulting 126X magnification. Confocal stacks were acquired to capture the entire thickness of a cell of interest. To analyze the enrichment of mWmC in lysosomes, object-based colocalization within ImageJ was used (Clinicaltrials.gov), which is an established method for analyzing colocalization of two signals. LAMP1 and WGA signals were manually thresholded using the Watershed image filter to obtain binary masks. The percentage area overlap of WGA within LAMP1 masks was then calculated for each image. This provides a measure of co-occurrence, or the extent to which mWmC signal overlapped with lysosomes marked by Lamp1. A Student’s t-test was used to assess significance. At least three animals were included, taking all SC slices for each experimental condition. No statistical method was used to predetermine sample size.
For data acquired in electrophysiology: Data collection and analysis were not performed blind to the conditions of the experiments. All whole-cell recording data were acquired by Camplex 10.7 (Molecular Devices) and analyzed in MATLAB (MathWorks) or Clampfit 10.7 (Molecular Devices). Optogenetically-evoked currents were plotted as an average of five trials. The peak amplitudes of evoked currents were measured by Clampfit. The intrinsic properties of recorded neurons were analyzed by MATLAB. All statistics were calculated by GraphPad Prism 8. Comparisons of the peak currents between two groups were made using a two-tailed t-test, and multiple samples were compared using a one-way analysis of variance.
Extended Data
Extended Data Fig. 1. Multiple configurations of mWGA-based tracer were examined for anterograde synaptic transfer efficiency in the retinotectal synapses.
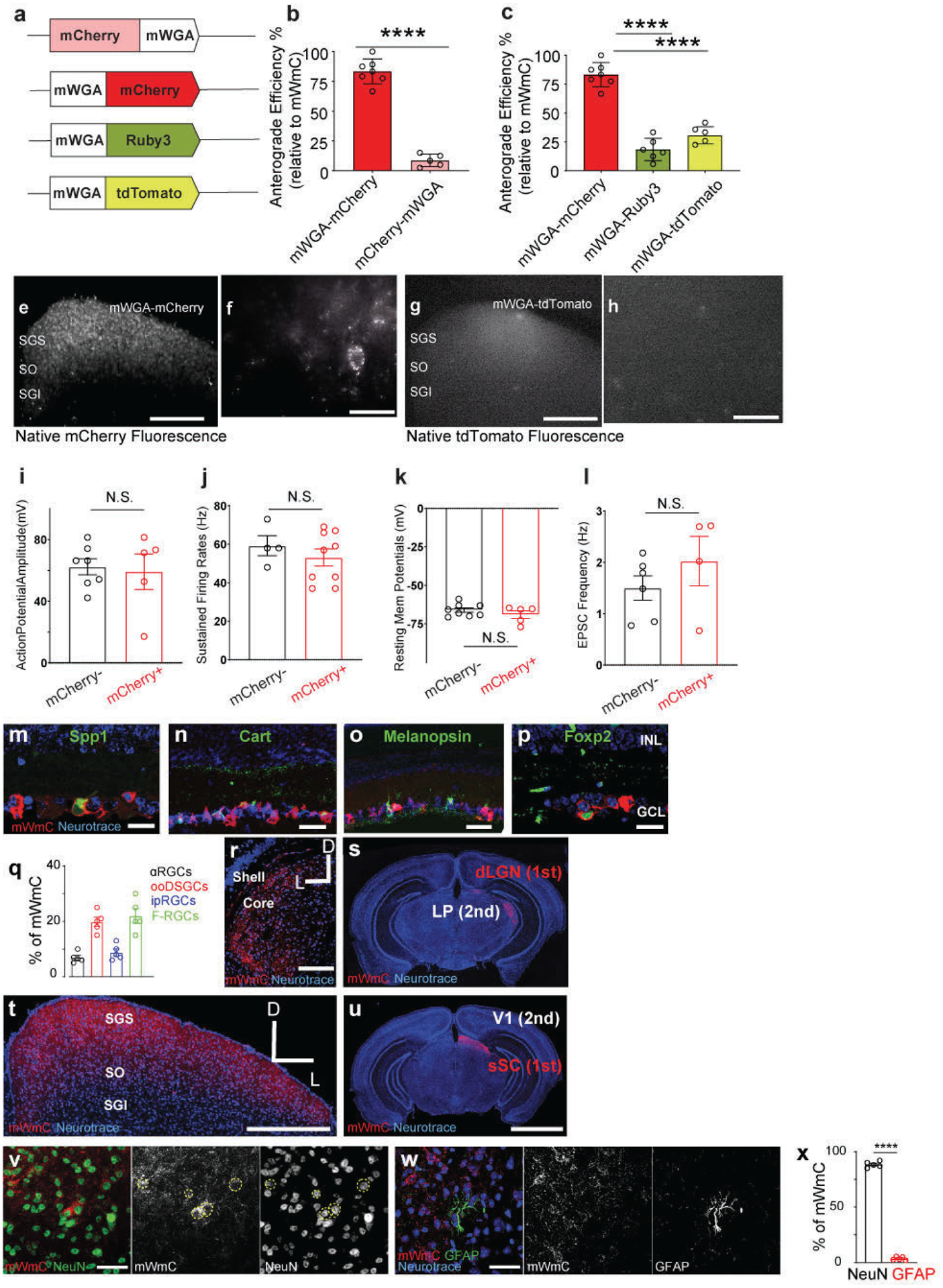
The numbers of fluorescence-positive neurons in the SGS from different mWGA-RFP configurations were counted. The percentages were normalized to the highest number as (100%) set by the mWGA-mCherry configuration, which possessed the highest efficiency. The anterograde transfer number was quantified by counting the number of RFP-positive neurons in acute SC slices on the contralateral side (right SC) without fluorescence amplification through immunostaining. We focused on the contralateral (right) SC, as 90% of RGCs project contralaterally31. We examined RGC coverage across the retina to ensure there was no retinotopy bias in SC analysis. mWGA-fusion protein configurations were compared under consistent conditions by delivering AAV2 expressing the fusion proteins into the left eyes (Fig. 1a). We used standard AAV Serotype-2 followed by 4 weeks post-injection (wpi) sampling, a commonly established window to examine RGC axonal projections and carry out the optogenetic measurement8. a, Several WGA-fusion proteins were compared side-by-side using an AAV-mediated in vivo screen for highly efficient anterograde transfer from the retina to the brain. b, Comparisons between N-terminal (mCherry-mWGA) and C-terminal fusion (mWGA-mCherry) transfer efficiencies were quantified. c, Transfer efficiencies of different RFP C-terminal fusions, including mWGA-mCherry, mWGA-Ruby3, mWGA-tdTmt, were quantified, n=4 animals for each condition. Statistics for b, two-sided Student’s t-test, ****, P<0.0001; c, one-way ANOVA test. ****, P<0.0001. e-f, (e) Additional samples showing live red-fluorescent labeling of the contralateral SC after acute brain slice preparation 4 weeks post-injection (wpi), indicating transsynaptic transfer onto the recipient neurons enriched in stratum griseum superficiale (SGS) and stratum opticum (SO), but not in stratum griseum intermedium (SGI). f, Magnified view of inset from e showing individual neurons labeled with bright red fluorescent protein from RGC anterograde transfer without signal amplification through immunostaining. The ability to detect native fluorescence is unique to mWGA-mCherry (mWmC) but absent in other mWGA-RFP configurations, such as mWGA-TdTomato shown here (g, h). Scale bars: (e, g, 5 mm; f, h, 50μm). i-l, Intrinsic electrophysiological properties of mCherry-positive recipient neurons (red, n=5 animals) and neighboring mCherry-negative neurons (black, n=4 animals) are similar as showed in Fig. 1m. i, Action potential amplitudes, j, sustained firing rates, k, resting membrane potentials, and l, EPSC frequencies are shown. These intrinsic properties were unperturbed by mWmC-transfer. N.S. not significant. two-sided Student’s t-test. m-p, Retinal vertical sections to show high mWmC coverage across major RGC types, including (m) Spp1 for αRGCs, (n) Cart for ooDSGCs, (o) Melanopsin for ipRGCs, and (p) Foxp2 for F-RGCs. Scale bar: 20 μm. INL: inner nuclear layer; GCL: ganglion cell layer. The percentages of each RGC subclasses were quantified in q, representing a similar fraction of RGC subclasses among all RGCs14, n=5 animals. f-i, Intraocular injections of mWmC lead to efficient monosynaptic transfer to connected neurons in multiple retino-recipient areas, including SC (t,u) and LGN (r, s). By contrast, the secondary relay neurons in V1 (u) or those in the lateral posterior nucleus of the thalamus (LP) (s) do not show mWmC transfer. Scale bars: (r, t, 2mm; s, u, 20mm). v, Immunostaining for RFP (mWmC) indicates high efficiency of anterograde transfer onto the recipient neurons in stratum griseum superficiale (SGS) and stratum opticum (SO). mWmC-positive cells are largely NeuN-positive. Dotted-yellow circles indicate mCherry and NeuN double-positive neurons from RGC anterograde transsynaptic transfer. w, mWmC-positive cells are largely GFAP-negative, which were quantified in x. n=5 animals for each condition. ****, p<0.0001, two-sided Student’s t-test. Scale bars: (v, w, 50μm). All data in this figure are presented as mean ± SEM.
Extended Data Fig. 2. Comparison of different versions of WGA tracer for their anterograde and retrograde transfer capacities in the retinotectal synapses.
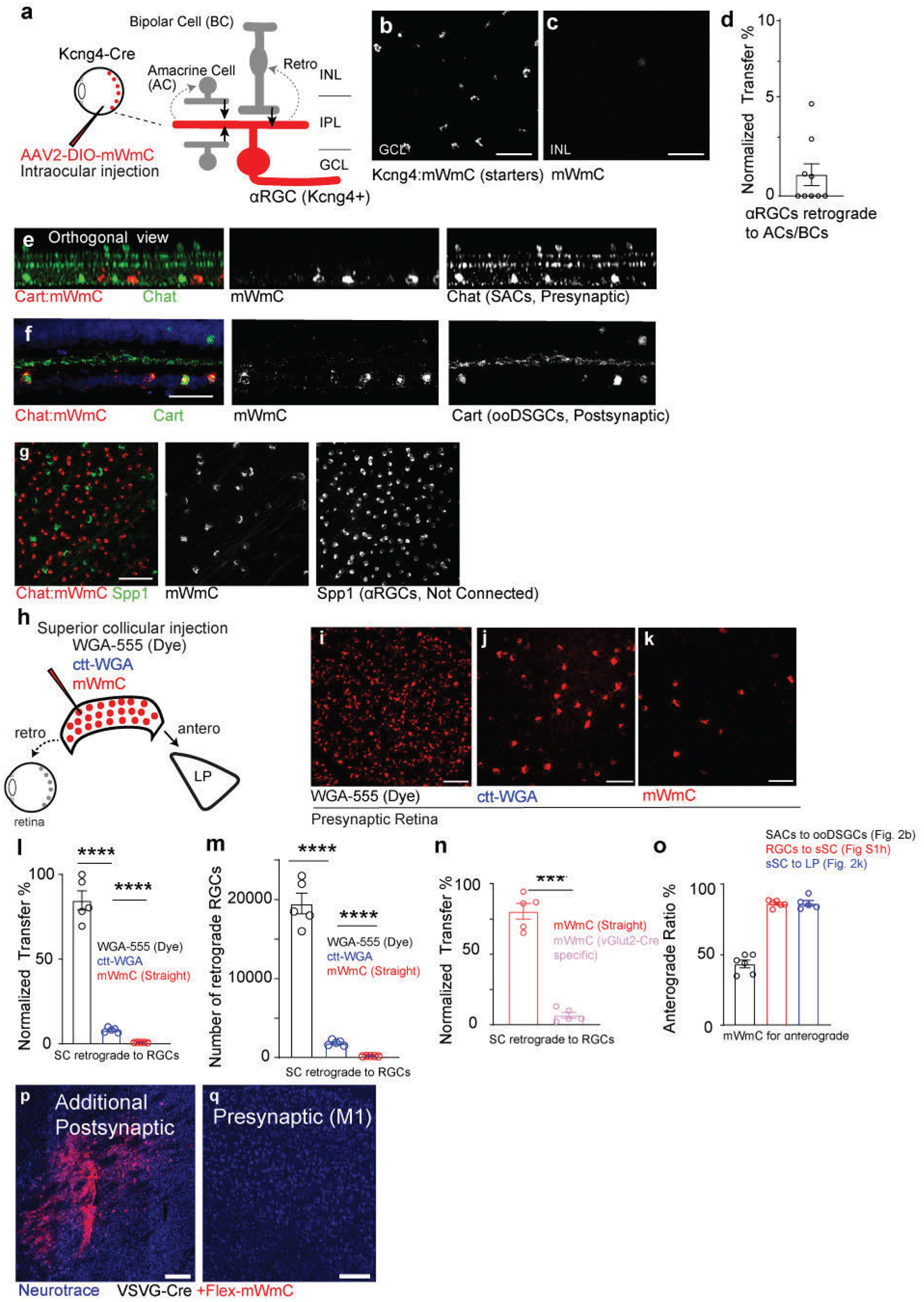
a-d, a, a schematic drawing of the injection within the retinal circuit. b, Retinal wholemount view to show mWmC coverage from RGC starter neurons (Kcng4-Cre for αRGC subclass neurons) in the ganglion cell layer (GCL, b), with limited retrograde transfer to the inner plexiform layer (INL, c). Scale bar: 50μm, with the retrograde transfer ratios quantified in (d). n=9 biologically independent samples. e-g, Orthogonal view (e) of RGC labeling with mWmC (red) without any retrograde transfer onto SACs (green, Chat). Intraocular injections of mWmC into Chat-Cre led to labeling of SACs (red) for anterograde and monosynaptic tracing onto ooDSGCs (Cart-positive, green; mWmC, red double-positive) in (f). Scale bar: 50μm (e, f). Additionally, Chat-mWmC labeling does not lead transfer mWmC to SPP1-positive neurons (αRGCs, g), indicating the specificity of anterograde transsynaptic transfer as opposed to trans-cellular transfer within a local area. Scale bar: 50μm(g) 0% Spp1-positive, n=4 animals. h, Injections of WGA-protein conjugated dye, AAV-expressing ctt-WGA cDNA, and AAV-expressing mWmC into SC lead to efficient anterograde transfer to LP. However, they displayed different levels of retrograde labeling, including bright axonal uptake and faint retrograde transfer. mWmC (j) displayed with very limited retrograde spread onto the retina. This is a significant improvement in limiting retrograde transfer compared to WGA-Alexa555 dye (i) or AAV-expressing ctt-WGA (k). n=4 animals in each condition. Scale bars: (i, j, k, 50 μm). l, Retrograde transfer ratios were normalized across three configurations of the WGA, using the highest numbers from WGA-555 as 100%. n=5 biologically independent samples. ****, p<0.0001, two-sided Student’s t-test. m, Average numbers of retinal ganglion cells per retina were quantified and compared across the three conditions. n=5 biologically independent samples. ****, p<0.0001, two-sided Student’s t-test. n. Additionally, restricting the starter neurons genetically in SC vGlut2-Cre cells further eliminated the axonal retrograde uptake (compared to global mWmC expression). n=5 biologically independent samples. ****, p<0.0001, two-sided Student’s t-test. o, Retrograde transfer ratios were normalized across two conditions, using the highest numbers from straight mWmC expression as 100%. n=5 biologically independent samples. ****, p<0.0001, two-sided Student’s t-test. p, GPe, another downstream target of the striatum injection (Fig. 2m, n) was traced here; q, Magnified view of M1 in Fig. 3n from the striatum retrograde tracing showed no retrograde spread from the Striatum. Scale bar p-q, 100μm. All data in this figure are presented as mean ± SEM.
Extended Data Fig. 3. The Trans-Seq platform’s workflow ensures quality control and generates a bioinformatic prediction for neuronal clustering.

To improve adult cell isolation and survival throughout the Trans-Seq protocol, we adopted well-controlled physiological conditions to preserve adult neurons79 and optimized slice preparation and dissociation conditions. a, Freshly prepared brain slice for contralateral SC dissociations at 4 weeks post-injection (wpi). Bright red fluorescence without immunostaining indicates a high-efficiency anterograde transsynaptic transfer from the retina onto contralateral SC recipient neurons. Fluorescence-activated cell sorting (FACS) enriched mCherry-positive neurons and excluded dead cells, debris, and doublets. n=4 animals each preparation. b-h, FACS gating parameters to isolate mWmC+ neurons from freshly prepared SC slices reflecting the bimodal distribution of the mCherry signals enriched by mWmC tracer, feasible for FACS. b, First, cell debris was excluded using forward scatter area (FSC-A) vs. side scatter area (SSC-A) profiles. Second, cell doublets were excluded using double gating composed from c forward scatter height (FSC-W) vs. forward scatter area (FSC-A) and d side scatter height (SSC-W) vs. side scatter area (SSC-A) profiles. e, Third, gating for DAPI-negative live cells was subsequently performed, followed by f the last gating for mCherry-positive cells. g, Sample gatings for both DAPI and mCherry were performed by establishing gating parameters on unstained control SC suspensions and backgating isolated cells. h, mCherry-gating results from mWmC-positive samples, summarizing the fraction of cells passing each of the FACS parameters listed above b-g. 25000–50000 mWmC-positive cells were collected each preparation. Using the scRNA-Seq data of the connected neurons from Pan-RGC tracing, we obtained eight neuronal clusters, including three excitatory SC clusters (ESC1–3) and five inhibitory SC clusters (ISC1–5). i, UMAP plot showing the alignment of all three replicates of Pan-RGC Tran-Seq experiments, showing the even distribution of all neuronal clusters across experimental procedures. j, Contribution of each preparation from i to individual neuronal clusters to show the batch effects across three experiments. These results included all three ESCs and five ISCs, indicating no neuronal clusters were biased across the triplicates. (Red, 1st; Green, 2nd; Blue, 3rd preparation). k-m, QC plots of the sorted mCherry-positive neurons for each neuronal cluster, including ESCs and ISCs, examining the number of genes expressed k, counts of transcripts l, and the mitochondria counts m.
Extended Data Fig. 4. Data analysis of the Pan-RGC Trans-Seq data.

a-d, UMAP plots of Pan-RGC Trans-Seq data show major neuronal markers, including (a) Snap25 and (b) Syt1 as pan-neuronal markers, (c) Slc17a6 (vGluT2) for excitatory neurons including three ESCs, and (d) Gad2 for inhibitory neurons including five ISCs. These markers defined neuronal populations and separated the clusters among three ESCs (Slc17a6-positive, c) and five ISCs (Gad2-positive, d). e-g, We queried previously identified marker genes expressed in SC neuronal subsets (f)22,80. Additionally, several cell-cell adhesion molecules, such as Type II Cdhs (g) and neuropeptides and their putative receptors (e), appeared as good molecular markers for neuronal subsets in the mammalian nervous system81,82. Notably, when we queried the gene expression of Ntsr1 in the Trans-Seq dataset, we found no Ntsr1 expression in any SC neuron cluster (e). Dot-plots as bioinformatics predictions of (e) Neuropeptides and receptors, and (g) Cdhs for SC subset expression.
Extended Data Fig. 5. Additional histology data to validate Trans-Seq data in the SC.
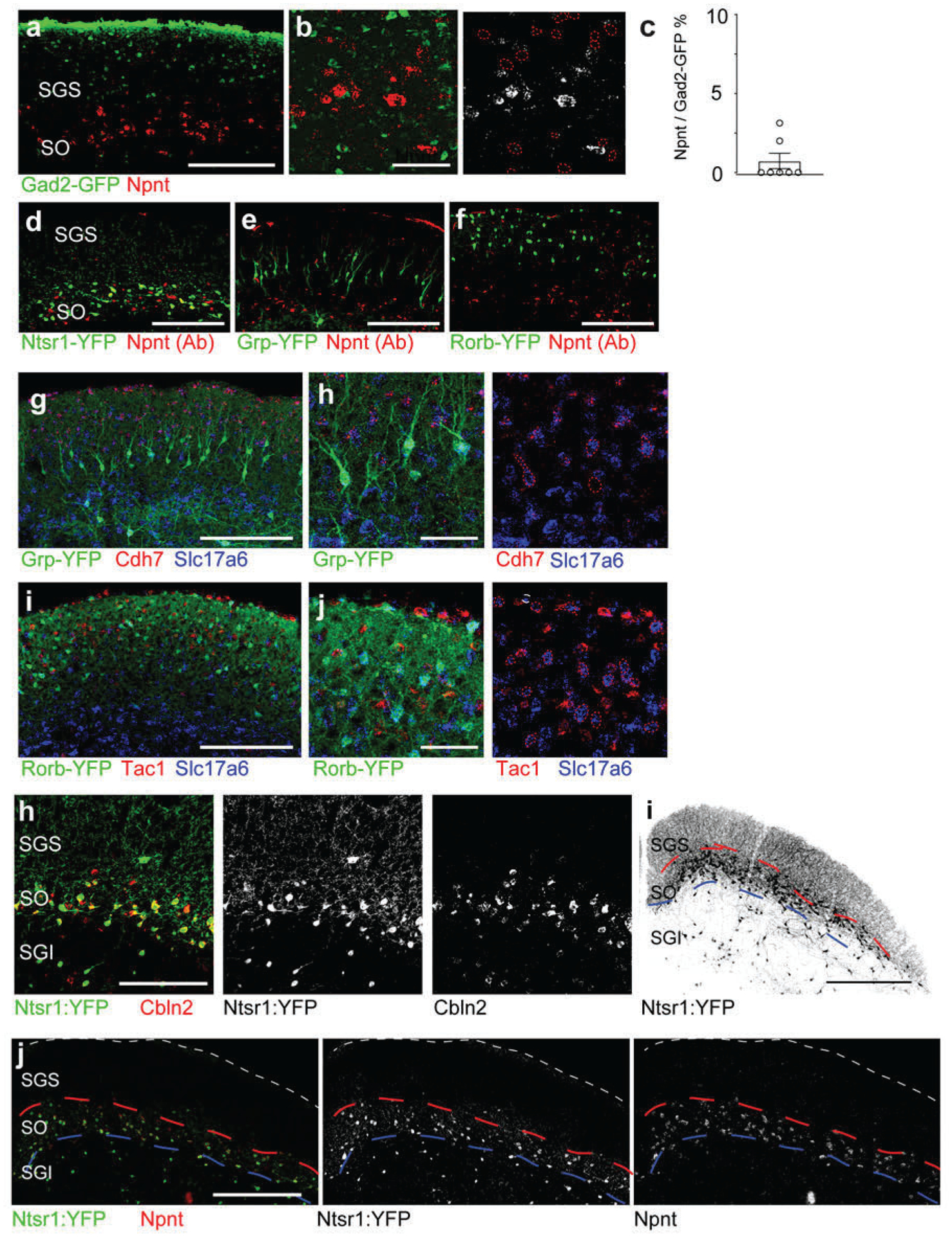
a-c, Validation of Trans-Seq marker gene expression from ESC1 using RNA-Scope in situ hybridization. Npnt (red) is not found in inhibitory neurons marked by Gad2-GFP (a). b, Higher magnification of the boxed area in a. Scale bars: (a, 250μm; b, 50μm), quantified in (c). n=4 animals, per genotype. Data in this figure are presented as mean ± SEM. d-f, Immunohistochemistry using Npnt antibody onto SC excitatory-marking lines. d, Npnt antibody (red) co-localizes with Ntsr1-GN209-YFP (green) for wide-field excitatory neurons. e, f, By contrast, the Npnt antibody (red) does not co-localize with the Grp-KH288-YFP line (green) for narrow-field neurons (e) or the Rorb-YFP line (green) for stellate cells (f). Scale bars: (d-f, 250μm). g-h, Validation of Trans-Seq marker gene expression from ESC2 using RNA-Scope in situ hybridization. (g) Cdh7 (red)/Slc17a6 (blue) double in situs do not label neurons in the Grp-KH288-YFP transgenic line for narrow-field excitatory neurons at higher magnification (h), n= 4 animals. i-j, Validation of Trans-Seq marker gene expression from ESC3 using RNA-Scope in situ hybridization. (i) Tac1 (red)/Slc17a6 (blue) double in situs do not label neurons in the Rorb-YFP knock-in line primarily for stellate excitatory neurons, at higher magnification (j), n=4 animals. Red dotted circles in (b, h, j) indicate the absence of in situ signals with GFP staining. Scale bars (h-j, 50μm). h, Validation of Trans-Seq top marker gene expression from ESC1 using RNA-Scope for in situ hybridization shows that Cbln2 RNA probes (red) are enriched in the Ntsr1-GN209-YFP transgenic line labeling wide-field excitatory neurons (green). Scale bar: 200 μm. n=3 animals. i, Neuronal distribution of Ntsr1-GN209-YFP transgenic line, including wide-field neurons present in SO and a significant fraction of neurons in the SGI and other deep SC layers. The wide distribution of YFP-positive neurons beyond SO indicates that Ntsr1-GN209-YFP cannot be used to manipulate SO neurons within the SC selectively. Red and blue dotted lines mark the borders of the three zones (SGS, SO, and SGI) within the superficial SC. j, Only wide-field neurons in the SO are Npnt-positive (between red and blue dotted lines), but not the rest of Ntsr1-YFP positive neurons in the SC’s SGI and deep layers. These data further suggest that Npnt is a faithful molecular marker for these neurons within SO. n=3 animals. Scale bars: (i, 2mm; j, 500μm).
Extended Data Fig. 6. Alignment of ESC1–3 clusters from Trans-Seq to a public database of whole-brain excitatory neuron clustering.
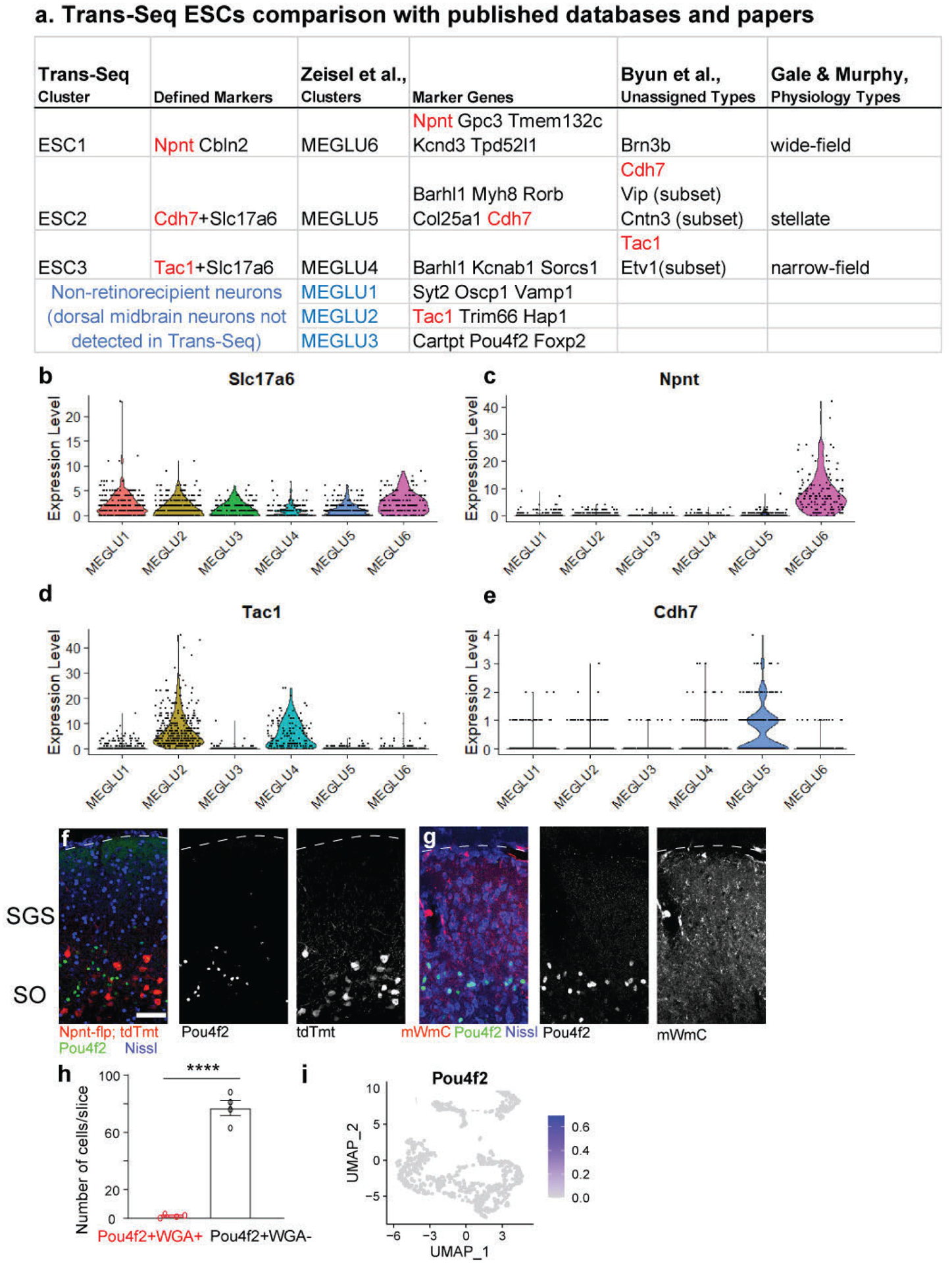
a, A summary of the three excitatory neuron clusters (ESCs) characterized in Trans-Seq. ESC1–3 were first correlated to a published bioinformatics database for all excitatory neuronal clusters throughout the brain25, indicating that ESC1–3 are characterized as retinorecipient neurons among six excitatory neuron clusters (MEGLU1–6) annotated from the dorsal midbrain. These six clusters were classified as putative SC excitatory clusters among all 11 collicular clusters. ESC1–3 were compared to an immunohistochemical study22 that identified laminar markers but did not link to cell types. ESC1–3 were matched to cell types defined by morphology and physiology23. b-e, Violin plots of candidate marker genes (b, Slc17a6, pan-excitatory neurons), (c, Npnt, ESC1), (d, Tac1, ESC3), and (e, Cdh7, ESC2) among MEGLU1–6. f, Immunohistochemistry of Pou4f2 (green, antibodies) showed no overlap with ESC1 neurons (Npnt-TdTomato, red). Pou4f2 was revealed as an excitatory neuron subset marker for MEGLU3 from a public scRNA-Seq database. A defined Pou4f2-positive population of neurons resides in the SGI. g, Pou4f2-positive SGI neurons did not uptake mWmC following 4wpi of mWmC infection of the RGCs, corroborating the findings from Trans-Seq in defining specific clusters of retino-recipient neurons. Scale bar: 100 μm. h. Quantifications of cell number for Pou4f2-positive and mWmC-positive neuron (red), versus all Pou4f2-positive neurons (black), n=3 animals, ****, p<0.0001, two-sided Student’s t-test. Data in this figure are presented as mean ± SEM. i. A UMAP Plot shows the absence of the MEGLU3 marker (Pou4f2) within Trans-Seq data (Fig. 5).
Extended Data Fig. 7. Cre-dependent mMmC for RGC type-specific circuit mapping.
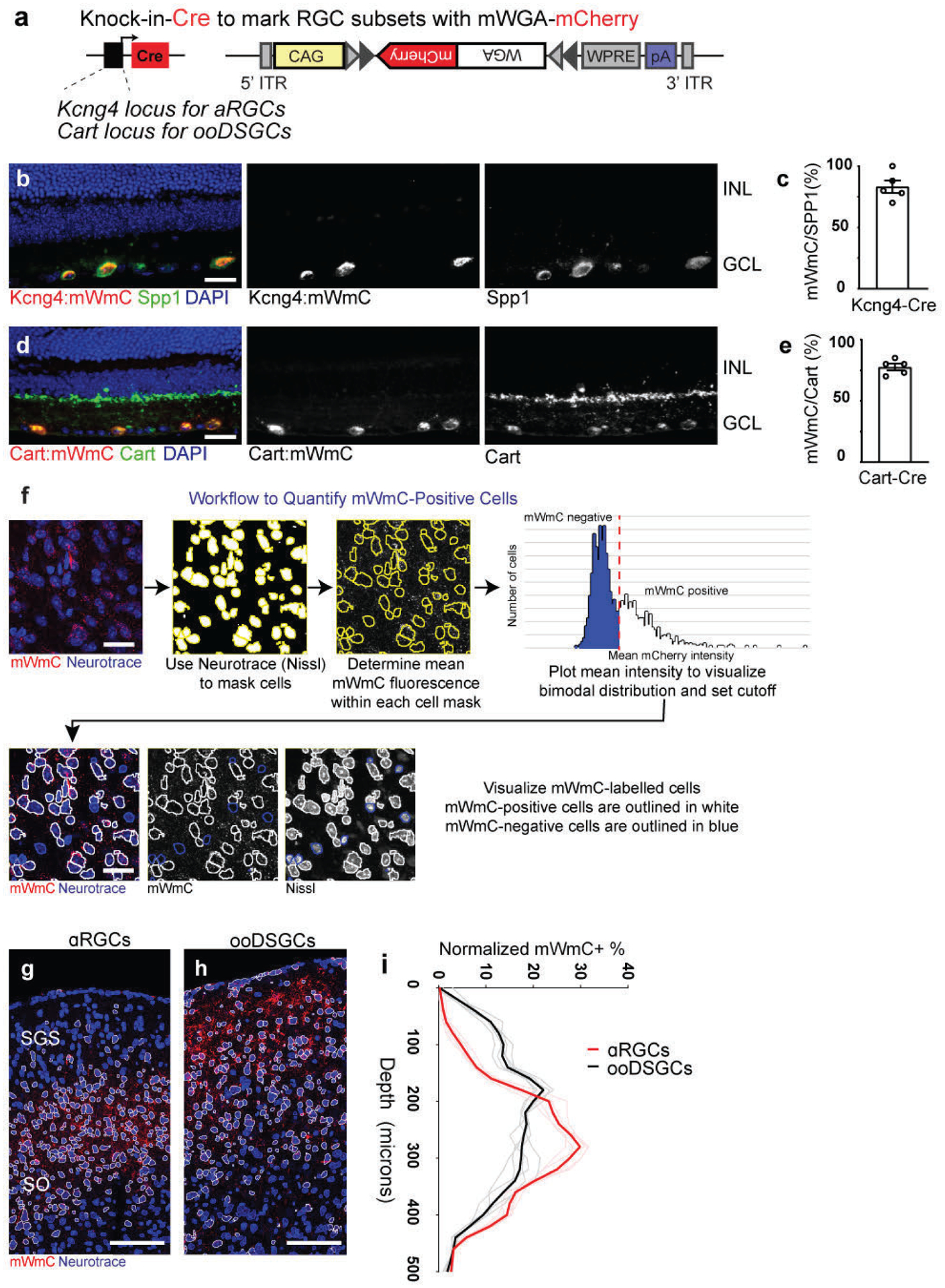
a, Genetic design of AAV vector for Cre-dependent mWmC (AAV2-CAG-DIO-mWmC-WPRE) expression to achieve neuronal subtype-specific anterograde transsynaptic tracing. b-e, mWmC infection can achieve efficient and restricted expression in RGC starter neuron subclasses: Kcng4-Cre is a driver for αRGC subtypes (Spp1-positive, b); and Cart-Cre is a driver for all ooDSGC subtypes (Cart-positive, d). Notably, these Cre-dependent mWmC expressions are restricted to the starter neurons without retrograde spread into the inner retina (i.e., no bipolar cells and amacrine cells uptake mWmC), scale bars: 20μm. The specificities of the Cre-drivers were quantified in c (Kcng4-Cre) and e (Cart-Cre), respectively. n=5 animals, per genotype. Data in this figure are presented as mean ± SEM. f, mWmC demonstrated a bimodal distribution of red fluorescence in recipient neurons. Thresholding for the high fluorescence intensity population allowed neurons across tissue slices to be classified as transferred or un-transferred. g-i, RGC-subclass specific mWmC anterograde transfer properties can be quantified in the postsynaptic neurons. Anterograde transsynaptic tracing from different starter retinal ganglion cell (RGC) types was compared (g, αRGCs; h, ooDSGCs as Fig. 6) and quantified using this workflow. This quantification (i) demonstrated the differential distribution of SC neurons receiving mWmC transfer from RGC starter cells, supporting the electrophysiology and genetic data from Fig. 5. n=5 animals, per genotype Scale bar: 100μm. (Black line is mean intensity curve, gray lines are each example. Red line is mean intensity curve, light red lines are each example).
Extended Data Fig. 8. Characterizations of Nephronectin-positive-wide-field neurons (NPWFs) and their differential inputs from αRGCs versus ooDSGCs.
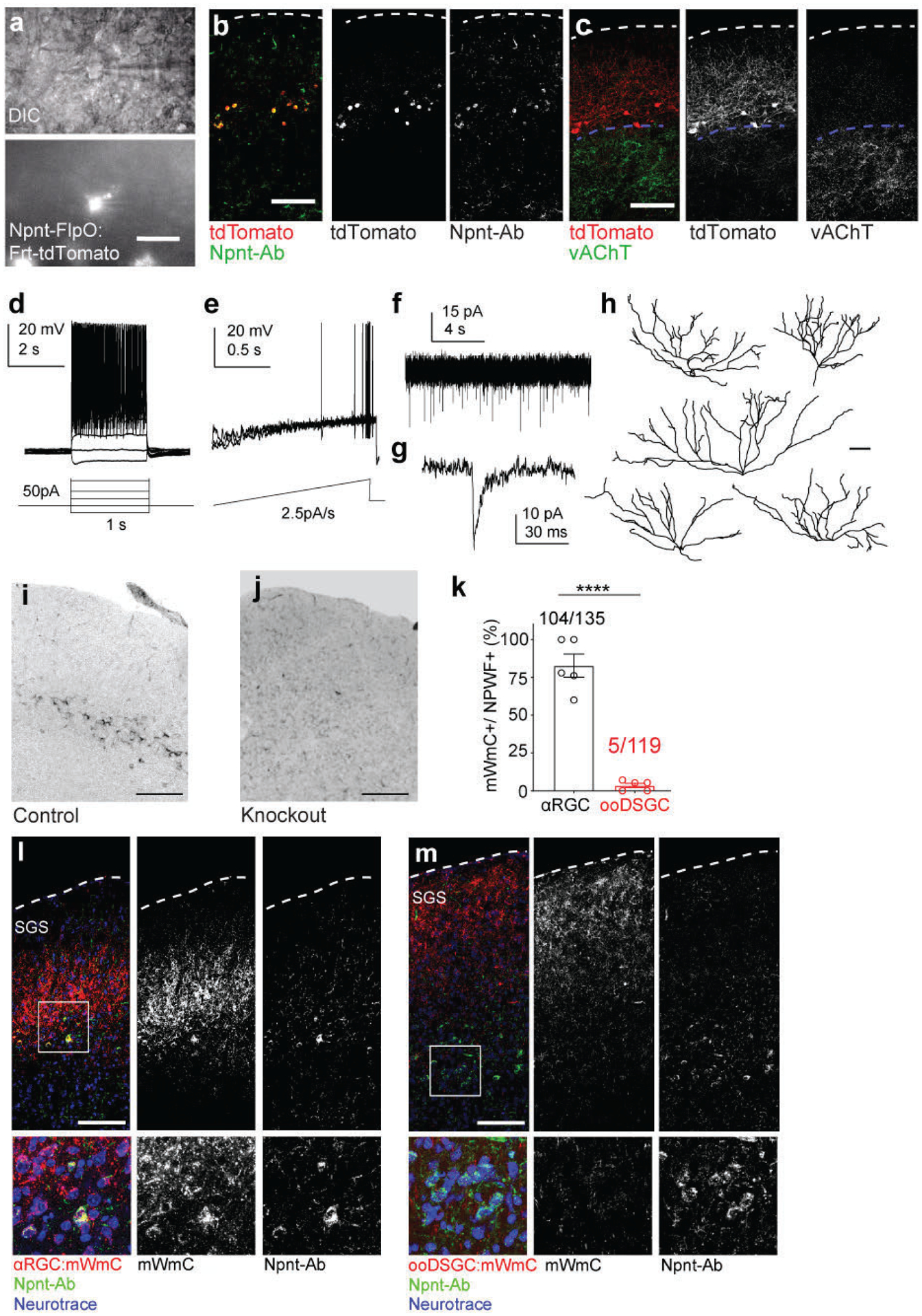
a, Sample DIC and fluorescent microscope images showing the targeted postsynaptic Npnt-FlpO-TdTomato neurons for connectivity tests. At least 30 times each experiment was repeated independently with similar results. Scale bar: 20 μm. b, The majority of the labeled Npnt-FlpO-TdTomato neurons are positive for Npnt antibody immunostaining (96.5±2.5% double-positive neurons among all TdTomato-positive neurons in b, n= 3 animals). The dotted line marks the pial surface. c, Npnt-FlpO-TdTomatao neurons have their somata positioning right above the top boundary of the SGI (dotted lines) marked by the vAChT. Scale bars: (b and c, 100 μm). d-g, Current-clamp recordings of NPWFs in response to (d) step-stimulation, 50pA per step for 1second, 10 steps per sweep, and (e) ramp-stimulation, 2.5pA/s for 20 seconds per trail, 3 trails per sweep. f, Voltage-clamp recordings of spontaneous EPSCs. g, A sample spontaneous ESPC trace from f. h, Single-cell morphologies of NPWFs (ESC1) were reconstructed after physiological recording and intracellular dye filling showing the dendritic complexity. Scale bar: 100 μm. i, j, SC-specific Npnt expression using immunohistochemistry (i) was absent in the SC-specific conditional knockout (j). Scale bar: 200μm. k-m, Sample immunohistochemistry images showing differential mWmC transfer from distinct RGC starter neurons onto retinorecipient neurons in the SC (mWmC, mCherry signal in red). k, Percentages of Npnt-positive wide-field neurons (NPWFs, ESC1) within connected neurons of the αRGCs and ooDSGCs. Data in this figure are presented as mean ± SEM. NPWFs were enriched in αRGC tracing datasets (77.0±8.2% double-positive neurons among all Npnt-positive cells in l, n=5 animals) but largely absent in ooDSGC tracing datasets (4.5±2.1% double-positive cells among all Npnt-positive neurons in m, n=5 animals). ****, p<0.0001, two-sided Student’s t-test. l, αRGC ouputomes labeled with Kcng4:mWmC. ESC1 neurons were immunostained for Npnt. m, ooDSGC tracing datasets in SC labeled with Cart:mWmC. ESC1 neurons stained with Npnt. Bottom panels are magnified view of inset from l and m labeled by solid square line.
Extended Data Fig. 9. Physiological and histological characterization of the selective RGC synapses onto NPWF neurons (ESC1).

a, Sample EPSCs from NPWF neurons driven by αRGCs with bath-applied TTX+4-AP (blue trace) and after bath perfusion of CNQX+APV (red trace). Blue dots indicate 2ms of the blue LED light. b, Quantification of the average amplitudes before and after adding pharmacological blockers in αRGC-NPWF connected pairs. Current amplitudes before adding any drugs are shown in black dots and bars; blue dots and bars show current amplitude after adding TTX+4-AP; red dots and bars show current amplitude after adding CNQX+APV. n=8 animals, N.S., no significance, *, p<0.05, two-sided Student’s t-test. The dots are amplitudes of individual cells, and the bar is the average of all cells. c, Among all the ooDSGCs and NPWF, connected pairs, only four cells showed a very small evoked current (black dots and bars). These small currents were blocked by bath-applied TTX and 4-AP (blue dots and bars), suggesting that the evoked currents are not monosynaptic inputs from ooDSGCs. The dots are amplitudes of individual cells, and the bar is the average of the three cells. n=4 cells, *, p<0.05, two-sided Student’s t-test. d, Sample images of NPWFs in the SC showing high efficiency of Flp-dependent EGFP-2a-TVA-2a-oG (green) and RdGV: Rabies-Cherry (red) overlap. Scale bar: 100 μm. e–j, Vertical sections of contralateral retinas following rabies tracing indicating αRGC subtypes (e and h) OFF-transient αRGCs, (f and i) OFF-sustained αRGCs, and (g and j) On-sustained αRGCs, were retrogradely labeled. Scale bar: 50μm. (e–g),vAChT staining (green) shows RGC dendritic sublaminations within the retina; (h–j), SPP1 staining (green) to confirm cell identities as αRGCs. k, Quantifications of different RGC subclasses among mCherry+ neurons including αRGCs, ooDSGCs, and other RGC types. l, Magnified view of Fig. 6o to show retrogradely labeled RGCs (red) highly overlap with αRGCs (SPP1-positive, green) in filled arrowheads, but not ooDSGCs (Satb1-positive, blue) in open arrowheads, as quantified in Fig. 6n. n=6 animals, ***, p<0.005, two-sided Student’s t-test. Scale bar: 100 μm. m, n, Sample images with axons of m, αRGCs, and n, ooDSGCs labeled by YFP (green) in relative position to the dendrites of NPWF neurons (Npnt-RFPs). n=4 animals each genotype, Scale bar: 100μm. All data in this figure are presented as mean ± SEM.
Extended Data Fig. 10. Validation of genetic and molecular reagents for Npnt mutant analysis.
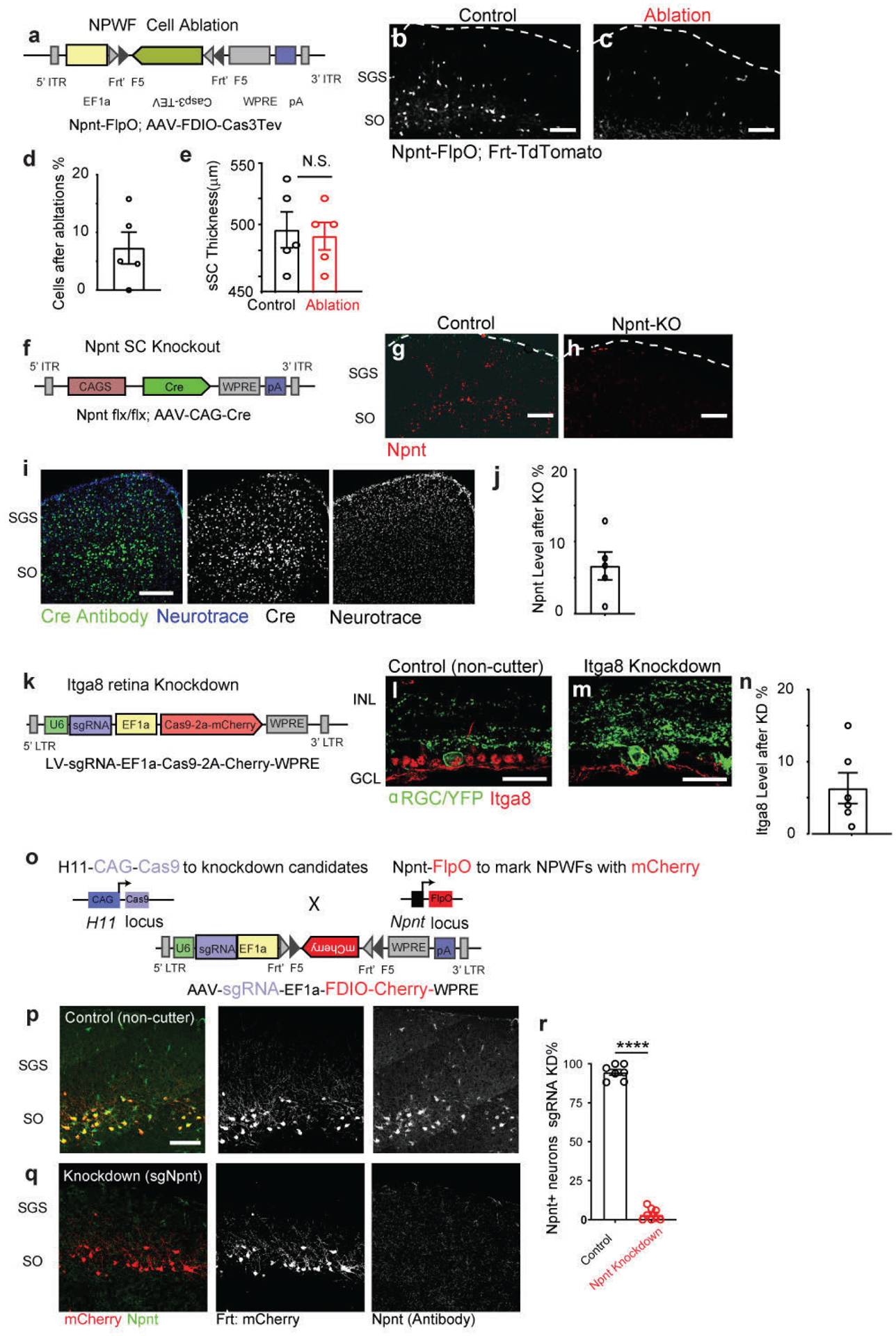
a, The AAV vector design for FlpO-dependent Caspase3-TEV expression, modified from (Addgene #45580)30. b, c, Confirmation of successfully eliminating ESC1s using AAV-fDIO-Caspase3 in Npnt-FlpO by comparing control b and Cas3-TEV injections c. The dotted line marks the pial surface. Scale bar: 100μm. d, Quantifications of Cas3-deletion of NPWFs efficiencies, compared to control side. N=5 animals. e, Quantifications of the superficial SC thickness subject to NPWF eliminations. n=5 animals, per genotype, N.S., no significance, two-sided Student’s t-test. f, The AAV vector design for Cre overexpression. g-h, Confirmation of SC-specific knockout of Npnt using Npnt f/f conditional mutants by comparing controls (g) and mutants (h), n=3 animals. Scale bar: 100μm. No obvious cell-autonomous migration or morphological changes of labeled wide-field neurons were observed after Npnt knockout. i. Sample images showing comprehensive Cre coverage across different sublaminae of SC subject to neonatal AAV injections. Anti-Cre staining was displayed here in the green channel. j, Quantifications of AAV-Cre mediated Npnt knockout efficiency, compared to the control side. n=5 animals. k, The lentiviral vector design for Itga8 knockout based on CRISPR-V2-system to simultaneously express sgRNA/Cas9-2a-Cherry (Addgene Plasmid #99154). l, m, Confirmation of the lentivirus-mediated Itga8 knockout in the retina (m) compared with control (l). Scale bar: 50μm. n, Quantifications of sgRNA-mediated retinal Itga8 knockdown efficacies, compared to control side. n=6 animals, Several sgRNAs were evaluated, and an efficient sgRNA targeting the fourth exon of Npnt was selected (See Methods). Cas9/sgRNA-mediated Npnt deletion was validated by immunostaining with Npnt antibody. We performed neonatal injections of AAV-encoding for sgRNA targeting Npnt or control sgRNA-non-cutter and Flp-dependent RFP expression (AAV-sgNpnt-FDIO-mCherry; or AAV-sg-Noncutter-FDIO-mCherry;) into Kcng4-YFP; Npnt-FlpO; H11Cas9 mice. o, The AAV vector is carrying sgRNAs and Frt-dependent mCherry. This vector can simultaneously knock down endogenous Npnt while labeling the NPWF neurons using Npnt-FlpO dependent RFP reporter. The Cas9 was genetically harbored in the H11-Cas9 line via a genetic cross to Npnt-FlpO. p, q, Npnt in the SC was knocked down (q) compared to sg-Non-cutter control (p). Scale bar: 100μm. The percentages of Npnt knockdown were quantified in (r), n=4 animals, per genotype, ****. p<0.0001, two-sided Student’s T-test. All data in this figure are presented as mean ± SEM.
Acknowledgments:
We thank J. Ngai for the Npnt-FlpO line; G. Nachtrab, Q. Wang and X. Chen for FlpO-dependent rabies tracing system; C. Gerfen for Ntsr1-GN209 and Grp-KH288 (GENSAT). We acknowledge A. Basbaum, L. Jan, X. Jin, B. Huang, L. Liang, J. Sanes, M. Scanziani, M. Stryker, and X. Wei for their advice of the manuscript. We acknowledge (NEI P30EY002162), the funding from the RPB unrestricted fund to UCSF-Ophthalmology; from NEI (F30EY033201) to N.Y.T.; from NIDDK (RO1DK118032) to D. K. M; from NIMH (R01MH08188) to J.L.R.; from NINDS (R01NS077986) to Fan. W.; from RBP-CDA, Klingenstein-Simons Neuroscience Fellowship, Whitehall Foundation, E. M. Ziegler Funds for Blindness, Glaucoma Research Foundation (CFC3), and NIH (R01EY030138; R01NS123912) to X.D.
Footnotes
Competing interests: Authors declare that they have no competing interests.
Materials & Correspondence: Raw scRNA-seq data reported in this paper is available through Sequence Read Archive PRJNA715507. GEO Accession Number for the scRNA-Seq Data is GSE202257. Scripts and R-markdown files for data analysis are available through https://github.com/duanxlab/Trans-Seq. All other data are available in the main manuscript or supplementary materials. Materials requests should be directed to X.D.
Data and Code availabilities
Raw scRNA-seq data reported in this paper is available through GEO Acession Number: PRJNA715507. GEO Accession Number for the scRNA-Seq Data is GSE202257. Scripts and R-markdown files for data analysis are available through https://github.com/duanxlab/Trans-Seq. All other data are available in the main manuscript or supplementary materials. AAV plasmids are available at Addgene associated with this paper. Materials requests should be directed to X.D.
REFERENCES
- 1.Yemini E et al. NeuroPAL: A Multicolor Atlas for Whole-Brain Neuronal Identification in C. elegans. Cell, doi: 10.1016/j.cell.2020.12.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He M & Huang ZJ Genetic approaches to access cell types in mammalian nervous systems. Curr Opin Neurobiol 50, 109–118, doi: 10.1016/j.conb.2018.02.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanes JR & Zipursky SL Synaptic Specificity, Recognition Molecules, and Assembly of Neural Circuits. Cell 181, 1434–1435, doi: 10.1016/j.cell.2020.05.046 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Siegert S et al. Genetic address book for retinal cell types. Nature neuroscience 12, 1197–1204, doi: 10.1038/nn.2370 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Yonehara K et al. The first stage of cardinal direction selectivity is localized to the dendrites of retinal ganglion cells. Neuron 79, 1078–1085, doi: 10.1016/j.neuron.2013.08.005 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Duan X, Krishnaswamy A, De la Huerta I & Sanes JR Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158, 793–807, doi: 10.1016/j.cell.2014.06.047 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Cang J, Savier E, Barchini J & Liu X Visual Function, Organization, and Development of the Mouse Superior Colliculus. Annual review of vision science 4, 239–262, doi: 10.1146/annurev-vision-091517-034142 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Martersteck EM et al. Diverse Central Projection Patterns of Retinal Ganglion Cells. Cell Rep 18, 2058–2072, doi: 10.1016/j.celrep.2017.01.075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callaway EM & Luo L Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J Neurosci 35, 8979–8985, doi: 10.1523/JNEUROSCI.0409-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo L & Anderson DJ A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72, 938–950, doi: 10.1016/j.neuron.2011.12.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zingg B et al. AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron 93, 33–47, doi: 10.1016/j.neuron.2016.11.045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshihara Y et al. A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron 22, 33–41 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Basbaum AI & Menetrey D Wheat germ agglutinin-apoHRP gold: a new retrograde tracer for light- and electron-microscopic single- and double-label studies. J Comp Neurol 261, 306–318, doi: 10.1002/cne.902610211 (1987). [DOI] [PubMed] [Google Scholar]
- 14.Duan X et al. Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron 85, 1244–1256, doi: 10.1016/j.neuron.2015.02.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandenberger R et al. Identification and characterization of a novel extracellular matrix protein nephronectin that is associated with integrin alpha8beta1 in the embryonic kidney. J Cell Biol 154, 447–458, doi: 10.1083/jcb.200103069 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braz JM & Basbaum AI Genetically expressed transneuronal tracer reveals direct and indirect serotonergic descending control circuits. J Comp Neurol 507, 1990–2003, doi: 10.1002/cne.21665 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braz JM, Rico B & Basbaum AI Transneuronal tracing of diverse CNS circuits by Cre-mediated induction of wheat germ agglutinin in transgenic mice. Proc Natl Acad Sci U S A 99, 15148–15153, doi: 10.1073/pnas.222546999 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama H, Yamamoto A, Mizushima N, Yoshimori T & Miyawaki A GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct 33, 1–12, doi: 10.1247/csf.07011 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Morrie RD & Feller MB Development of synaptic connectivity in the retinal direction selective circuit. Curr Opin Neurobiol 40, 45–52, doi: 10.1016/j.conb.2016.06.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuriel S, Gudes S, Draft RW, Binshtok AM & Lichtman JW Multispectral labeling technique to map many neighboring axonal projections in the same tissue. Nat Methods 12, 547–552, doi: 10.1038/nmeth.3367 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Takatoh J et al. New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron 77, 346–360, doi: 10.1016/j.neuron.2012.11.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byun H et al. Molecular features distinguish ten neuronal types in the mouse superficial superior colliculus. J Comp Neurol 524, 2300–2321, doi: 10.1002/cne.23952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gale SD & Murphy GJ Distinct representation and distribution of visual information by specific cell types in mouse superficial superior colliculus. J Neurosci 34, 13458–13471, doi: 10.1523/JNEUROSCI.2768-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seigneur E & Sudhof TC Cerebellins are differentially expressed in selective subsets of neurons throughout the brain. The Journal of comparative neurology 525, 3286–3311, doi: 10.1002/cne.24278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeisel A et al. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014 e1022, doi: 10.1016/j.cell.2018.06.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beier KT et al. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell 162, 622–634, doi: 10.1016/j.cell.2015.07.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng YR et al. Satb1 Regulates Contactin 5 to Pattern Dendrites of a Mammalian Retinal Ganglion Cell. Neuron 95, 869–883 e866, doi: 10.1016/j.neuron.2017.07.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linton JM, Martin GR & Reichardt LF The ECM protein nephronectin promotes kidney development via integrin alpha8beta1-mediated stimulation of Gdnf expression. Development 134, 2501–2509, doi: 10.1242/dev.005033 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiwara H et al. The basement membrane of hair follicle stem cells is a muscle cell niche. Cell 144, 577–589, doi: 10.1016/j.cell.2011.01.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang CF et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell 153, 896–909, doi: 10.1016/j.cell.2013.04.017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong YK, Kim IJ & Sanes JR Stereotyped axonal arbors of retinal ganglion cell subsets in the mouse superior colliculus. J Comp Neurol 519, 1691–1711, doi: 10.1002/cne.22595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiou SH et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes & development 29, 1576–1585, doi: 10.1101/gad.264861.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato Y et al. Molecular basis of the recognition of nephronectin by integrin alpha8beta1. The Journal of biological chemistry 284, 14524–14536, doi: 10.1074/jbc.M900200200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphries JD, Byron A & Humphries MJ Integrin ligands at a glance. J Cell Sci 119, 3901–3903, doi: 10.1242/jcs.03098 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epp AL et al. A novel motif in the proximal C-terminus of Pannexin 1 regulates cell surface localization. Sci Rep 9, 9721, doi: 10.1038/s41598-019-46144-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright CS Structural comparison of the two distinct sugar binding sites in wheat germ agglutinin isolectin II. J Mol Biol 178, 91–104, doi: 10.1016/0022-2836(84)90232-8 (1984). [DOI] [PubMed] [Google Scholar]
- 37.Linhoff MW et al. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron 61, 734–749, doi: 10.1016/j.neuron.2009.01.017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnaswamy A, Yamagata M, Duan X, Hong YK & Sanes JR Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature 524, 466–470, doi: 10.1038/nature14682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kebschull JM et al. High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 91, 975–987, doi: 10.1016/j.neuron.2016.07.036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim IJ, Zhang Y, Meister M & Sanes JR Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 1452–1462, doi: 10.1523/JNEUROSCI.4779-09.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng TW et al. Emergence of lamina-specific retinal ganglion cell connectivity by axon arbor retraction and synapse elimination. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 16376–16382, doi: 10.1523/JNEUROSCI.3455-10.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osterhout JA, El-Danaf RN, Nguyen PL & Huberman AD Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell reports 8, 1006–1017, doi: 10.1016/j.celrep.2014.06.063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varadarajan SG & Huberman AD Assembly and repair of eye-to-brain connections. Curr Opin Neurobiol 53, 198–209, doi: 10.1016/j.conb.2018.10.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frisen J et al. Ephrin-A5 (AL-1/RAGS) is essential for proper retinal axon guidance and topographic mapping in the mammalian visual system. Neuron 20, 235–243, doi: 10.1016/s0896-6273(00)80452-3 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Feldheim DA et al. Genetic analysis of ephrin-A2 and ephrin-A5 shows their requirement in multiple aspects of retinocollicular mapping. Neuron 25, 563–574, doi: 10.1016/s0896-6273(00)81060-0 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Sweeney NT, James KN, Sales EC & Feldheim DA Ephrin-As are required for the topographic mapping but not laminar choice of physiologically distinct RGC types. Dev Neurobiol 75, 584–593, doi: 10.1002/dneu.22265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito S & Feldheim DA The Mouse Superior Colliculus: An Emerging Model for Studying Circuit Formation and Function. Front Neural Circuits 12, 10, doi: 10.3389/fncir.2018.00010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triplett JW Molecular guidance of retinotopic map development in the midbrain. Curr Opin Neurobiol 24, 7–12, doi: 10.1016/j.conb.2013.07.006 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Cang J, Wang L, Stryker MP & Feldheim DA Roles of ephrin-as and structured activity in the development of functional maps in the superior colliculus. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 11015–11023, doi: 10.1523/JNEUROSCI.2478-08.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knoll B et al. Graded expression patterns of ephrin-As in the superior colliculus after lesion of the adult mouse optic nerve. Mech Dev 106, 119–127, doi: 10.1016/s0925-4773(01)00431-2 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Ackman JB & Crair MC Role of emergent neural activity in visual map development. Curr Opin Neurobiol 24, 166–175, doi: 10.1016/j.conb.2013.11.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLaughlin T, Torborg CL, Feller MB & O’Leary DD Retinotopic map refinement requires spontaneous retinal waves during a brief critical period of development. Neuron 40, 1147–1160, doi: 10.1016/s0896-6273(03)00790-6 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Xiao T et al. Assembly of lamina-specific neuronal connections by slit bound to type IV collagen. Cell 146, 164–176, doi: 10.1016/j.cell.2011.06.016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker CG, Schweitzer J, Feldner J, Schachner M & Becker T Tenascin-R as a repellent guidance molecule for newly growing and regenerating optic axons in adult zebrafish. Mol Cell Neurosci 26, 376–389, doi: 10.1016/j.mcn.2004.03.003 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Yamagata M & Sanes JR Versican in the developing brain: lamina-specific expression in interneuronal subsets and role in presynaptic maturation. J Neurosci 25, 8457–8467, doi: 10.1523/JNEUROSCI.1976-05.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang Y et al. In vitro guidance of retinal axons by a tectal lamina-specific glycoprotein Nel. Mol Cell Neurosci 41, 113–119, doi: 10.1016/j.mcn.2009.02.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grahovac J & Wells A Matrikine and matricellular regulators of EGF receptor signaling on cancer cell migration and invasion. Lab Invest 94, 31–40, doi: 10.1038/labinvest.2013.132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanes JR & Zipursky SL Design principles of insect and vertebrate visual systems. Neuron 66, 15–36, doi: 10.1016/j.neuron.2010.01.018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kay JN et al. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci 31, 7753–7762, doi: 10.1523/JNEUROSCI.0907-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trenholm S, Johnson K, Li X, Smith RG & Awatramani GB Parallel mechanisms encode direction in the retina. Neuron 71, 683–694, doi: 10.1016/j.neuron.2011.06.020 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gale SD & Murphy GJ Distinct cell types in the superficial superior colliculus project to the dorsal lateral geniculate and lateral posterior thalamic nuclei. J Neurophysiol 120, 1286–1292, doi: 10.1152/jn.00248.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gerfen CR, Paletzki R & Heintz N GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80, 1368–1383, doi: 10.1016/j.neuron.2013.10.016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vong L et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154, doi: 10.1016/j.neuron.2011.05.028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taniguchi H et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013, doi: 10.1016/j.neuron.2011.07.026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Network, B. I. C. C. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102, doi: 10.1038/s41586-021-03950-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buffelli M et al. Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature 424, 430–434, doi: 10.1038/nature01844 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Madisen L et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958, doi: 10.1016/j.neuron.2015.02.022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimmerman SE et al. Nephronectin Regulates Mesangial Cell Adhesion and Behavior in Glomeruli. Journal of the American Society of Nephrology : JASN 29, 1128–1140, doi: 10.1681/ASN.2017070752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staahl BT et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol 35, 431–434, doi: 10.1038/nbt.3806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duan X et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130, 1146–1158, doi: 10.1016/j.cell.2007.07.010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng YR et al. Binary Fate Choice between Closely Related Interneuronal Types Is Determined by a Fezf1-Dependent Postmitotic Transcriptional Switch. Neuron 105, 464–474 e466, doi: 10.1016/j.neuron.2019.11.002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duan X et al. Cadherin Combinations Recruit Dendrites of Distinct Retinal Neurons to a Shared Interneuronal Scaffold. Neuron 99, 1145–1154 e1146, doi: 10.1016/j.neuron.2018.08.019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bai L et al. Genetic Identification of Vagal Sensory Neurons That Control Feeding. Cell 179, 1129–1143 e1123, doi: 10.1016/j.cell.2019.10.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim JY et al. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. The European journal of neuroscience 37, 1203–1220, doi: 10.1111/ejn.12126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Breunig JJ et al. Rapid genetic targeting of pial surface neural progenitors and immature neurons by neonatal electroporation. Neural development 7, 26, doi: 10.1186/1749-8104-7-26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bei F et al. Restoration of Visual Function by Enhancing Conduction in Regenerated Axons. Cell 164, 219–232, doi: 10.1016/j.cell.2015.11.036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502, doi: 10.1038/nbt.3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol, doi: 10.1038/nbt.4314 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Wu YE, Pan L, Zuo Y, Li X & Hong W Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329 e316, doi: 10.1016/j.neuron.2017.09.026 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Shang C et al. BRAIN CIRCUITS. A parvalbumin-positive excitatory visual pathway to trigger fear responses in mice. Science 348, 1472–1477, doi: 10.1126/science.aaa8694 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Shrestha BR et al. Sensory Neuron Diversity in the Inner Ear Is Shaped by Activity. Cell 174, 1229–1246 e1217, doi: 10.1016/j.cell.2018.07.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abraira VE et al. The Cellular and Synaptic Architecture of the Mechanosensory Dorsal Horn. Cell 168, 295–310 e219, doi: 10.1016/j.cell.2016.12.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw scRNA-seq data reported in this paper is available through GEO Acession Number: PRJNA715507. GEO Accession Number for the scRNA-Seq Data is GSE202257. Scripts and R-markdown files for data analysis are available through https://github.com/duanxlab/Trans-Seq. All other data are available in the main manuscript or supplementary materials. AAV plasmids are available at Addgene associated with this paper. Materials requests should be directed to X.D.


