Abstract
A reverse transcription (RT)-PCR technique was developed to analyze global gene regulation in Escherichia coli. A novel combination of primers designed specifically for the start and stop regions of E. coli genes (based on the findings of Fislage et al. [R. Fislage, M. Berceanu, Y. Humboldt, M. Wendt, and H. Oberender, Nucleic Acids Res. 25:1830–1835, 1997]) was used as an alternative to the poly(T) primers often used in eukaryotic RT-PCR. The validity of the technique was demonstrated by applying it to heat shock analysis. Specifically, RT-PCR-amplified total RNA from heat-shocked and non-heat-shocked cells were hybridized with slot blots of the Kohara set (U. Kohara, K. Akiyama, and K. Isono, Cell 50:495–508, 1987; S. Chuang, D. Daniels, and F. Blattner, J. Bacteriol. 175:2026–2036, 1993). The signals obtained for heat-shocked and control cultures of each clone were compared, and differences in intensity were evaluated by calculating induction ratios. Clones that were considered significantly induced were subsequently mapped by the Southern blot technique in order to determine specific gene upregulation. Also, for several genes, Northern blotting and total RNA dot blotting were performed to confirm that the transcript levels in the original RNA samples were different. This technique extended previously described methods for studying global gene regulation in E. coli by incorporating a PCR amplification step in which global, mRNA-specific primers were used. In addition, the method employed here can be easily extended to study E. coli global gene regulation in response to additional environmental stimuli.
The first comprehensive studies of cellular response resulted from the development of two-dimensional gel electrophoresis and a complementary method for quantitatively measuring protein levels (15, 16). In parallel, Casadaban (2) created transcriptional fusion proteins to assist in the study of gene regulation, and these proteins have been particularly useful for analyzing genes whose products are difficult to characterize. These techniques have been refined (3, 12), and detection of transcriptional regulation has been simplified due to bioluminescent reporter proteins such as luciferase (18, 20) and green fluorescent protein (9).
Chuang et al. (4) demonstrated that global gene regulation in Escherichia coli could be analyzed by using single-stranded reverse-transcribed cDNA and a technique in which radiolabeled cDNA was hybridized with the Kohara set of overlapping λ bacteriophage clones. These clones contain the entire E. coli genome and were used to map the locations of cDNA homologs. By performing follow-up Southern blotting (19), Chuang and Blattner identified 26 new heat shock genes (5). This technique was a significant improvement over the methods used previously (two-dimensional electrophoresis and transcriptional fusion) to analyze global genetic regulation; however, the ratio of mRNA signal to total RNA noise remained small (2, 15, 16). Wong and McClelland (23) used a random arbitrarily primed PCR amplification step, after reverse transcription (RT) of total RNA, to detect a stress-induced gene in Salmonella typhimurium. While this technique improved the mRNA signal level, the signal-to-noise ratio did not change due to random amplification of RNA templates. In addition, because sequencing gels were used for transcript identification, this technique did not allow quantification at the genomic level. An innovative refinement of the two methods mentioned above was described by de Saizieu et al. (6); in the refined technique nonradioactively labeled total prokaryotic RNA was hybridized directly to an oligonucleotide array that was synthesized on and bonded to a silicon chip. This method, which allowed quantification of a large subset of transcribed genes, required scanning confocal microscopy as RNA levels were detected as unamplified transcripts.
Conversely, there have been rapid advances in differential display techniques based on PCR amplification in eukaryotic systems due to mRNA polyadenlyation. The RT-PCR (14) and random arbitrarily primed PCR (RAP-PCR) (22) techniques were developed to enhance the signal-to-noise ratio in differential display experiments. While these techniques have provided methods for acquiring vast amounts of regulatory information concerning eukaryotic systems, differential display has not been widely utilized with prokaryotic systems due to the absence of polyadenylation in prokaryotic mRNA. Other developments in eukaryote-based RT-RAP-PCR include the use of random arbitrary primers or motif primers (7, 14, 22). Based on the results obtained, we suspected that an RT-RAP-PCR-based technique in which primers designed specifically for prokaryotic mRNA were used would improve differential display analysis of E. coli.
Fislage et al. (8) performed a detailed statistical evaluation of the coding regions extracted from bacterial genetic databases and designed 10 RT primers for the 3′ end of prokaryotic mRNA and 10 PCR primers for the 5′ end of prokaryotic mRNA. These primers exhibited increased specificity in the 3′ and 5′ regions surrounding E. coli genes and decreased specificity for rRNA or other abundant small RNA species, so that mRNA were preferentially transcribed. In their RT-PCR analysis, Fislage et al. used one RT primer in combination with a single PCR primer for an RT-PCR, and the process was subsequently repeated for each primer set so that 100 different amplification experiments were performed for every sample. Following amplification, each sample was analyzed by performing gel electrophoresis, reamplification, and sequencing.
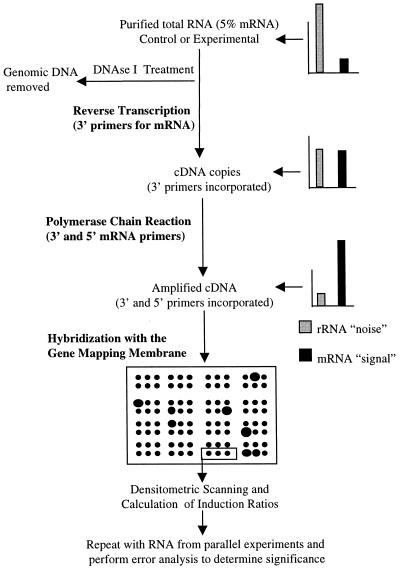
In this paper, we describe an E. coli RT-PCR technique in which a novel combination of the primers of Fislage et al. (Fig. 1) is used in a degenerate fashion for mRNA-specific amplification. A single amplification experiment was required for each RNA sample, and repeated amplifications of the same RNA under identical conditions allowed us to analyze experimental error. This technique was made possible by the availability of an E. coli gene mapping membrane (4) which could be used for global analysis of amplified RNA. Specifically, by avoiding the use of sequencing gels, we were able to avoid performing multiple amplification experiments with the same RNA sample. The RT-PCR method described here was validated by comparing quantified Kohara clone intensities determined with amplified RNA from heat-shocked E. coli cells and non-heat-shocked E. coli cells. Since Kohara clones contain more than one E. coli gene, restriction enzyme-digested Kohara clones were hybridized with RT-PCR products to confirm that amplification of specific heat shock genes occurred. In addition, Northern blotting and total RNA dot blotting were performed to confirm the differentially displayed transcript levels.
FIG. 1.
Schematic diagram of the E. coli RT-PCR method used. Purified total RNA was treated with DNase I to remove chromosomal DNA and then repurified. RT and PCR were then performed sequentially in the presence of mRNA-specific primers. Amplified cDNA was end labeled and hybridized with an E. coli gene mapping membrane. Finally, the signal levels for repeated control and stressed samples were quantified, and the induction ratio and standard error were calculated as described in the text.
MATERIALS AND METHODS
Microorganisms.
E. coli W3110(pIL2) [F− λ− IN(rrnD-rrnE)] was used exclusively in this work. W3110 was also utilized by Kohara et al. (13) to create the Kohara miniset.
Fermentation and media.
Organisms were grown in Luria-Bertani media (17) supplemented with 0.1 mg of ampicillin per ml. The growth temperature was 37°C, and experiments were performed in 250-ml shake flasks. Shake flask experiments were performed by using either an air shaker (New Brunswick Scientific, Edison, N.J.) at 250 rpm or a reciprocating water bath shaker (New Brunswick Scientific) at 100 rpm.
Induction of stress response and sample preparation.
Heat shock was induced in the mid-exponential phase of growth. All samples were taken immediately before the stress and 15 min after the stress. The heat shock was initiated by transferring shake flask cultures grown at 37°C to a 42°C water bath. Samples (25 ml) were removed from the shake flasks and immediately mixed with an equal volume of crushed ice in 50-ml centrifuge tubes. The samples were centrifuged at 5,000 × g for 15 min at 4°C, washed once in ice-cold 50 mM Tris buffer (pH 7.5), and then pelleted by centrifugation under the same conditions before total RNA was isolated.
RNA and DNA purification.
RNA was purified by using an RNAqueous total RNA isolation kit (Ambion Inc., Austin, Tex.). This kit typically purifies 10 μg of total RNA from every 1 ml of E. coli culture at an optical density of 1.0. Purified RNA was incubated with 50 U of DNase I (Boehringer Mannheim, Indianapolis, Ind.) per ml at 37°C for 30 min and then was repurified by using an RNAqueous kit. This procedure has been shown to produce RNA that is pure enough to be used in RT-PCR mixtures (Ambion, Inc.). E. coli and RT-PCR product DNA were purified by ethanol-sodium acetate precipitation by using the protocol recommended by Boehringer Mannheim. λ DNA was purified by using a λ DNA purification kit (Promega, Madison, Wis.).
RT-PCR primers.
The specific combinations of primers (based on the findings of Fislage et al. [8]) utilized for RT and PCR are shown in Table 1. Mixtures containing equimolar quantities of RT and PCR primers were utilized for the RT reactions and PCR, respectively. PCR primers PCR1, PCR3, and PCR5 were also added to the RT reaction mixtures at equimolar concentrations.
TABLE 1.
RT-PCR primersa
| RT primers
|
PCR primers
|
||
|---|---|---|---|
| Designation | Sequence (5′-3′) | Designation | Sequence (5′-3′) |
| RT1 | TTTTATCCAGC | PCR1 | GCTGGAAAAA |
| RT2 | ACTTTACGCAG | PCR2 | GCTGCTGGCG |
| RT3 | TTTATCCAGCG | PCR3 | GAAGTGCTGG |
| RT4 | TCAGCGTTTTA | PCR4 | TGGCGGCGGC |
| RT5 | TTTCAGCGCCT | PCR5 | AACTGGCGAA |
| RT6 | TTTTTTCAGCA | PCR6 | ATGCGCTGGC |
| RT7 | TCTTTTTTACC | PCR7 | TGCCGATGAA |
| RT8 | ATCATCCAGCA | PCR8 | CTGGAAGAAG |
| RT9 | TTTTACCCAGC | PCR9 | ATGGCGCTGG |
| RT10 | TTCAGCCAGCG | PCR10 | ATGGCGATGA |
Adapted from reference 8 with permission from the publisher.
RT.
The RT reaction was performed with a Peltier thermal cycler (MJ Research, Watertown, Mass.). Each reaction mixture contained avian myeoblastosis virus reverse transcriptase (AMVRT) reaction buffer (50 mM Tris HCl [pH 8.3], 30 mM KCl, 8 mM MgCl2)(Boehringer Mannheim), each deoxynucleotide (Boehringer Mannheim) at a concentration of 0.5 mM, 0.8 U of RNase inhibitor (Boehringer Mannheim) per μl, 0.5 U of AMVRT (Boehringer Mannheim) per μl, each RT primer (Life Technologies, Gaithersburg, Md.) at a concentration of 0.5 μM, 15 μg of RNA sample, and enough autoclaved deionized water to bring the total volume to 60 μl. The RNA was denatured at 70°C for 10 min before it was mixed with the reaction solution. The RNA templates were allowed to anneal to the primers for 10 min at 25°C before the elongation reaction was started. The elongation reaction lasted for 3 h at 42°C. Final denaturation was carried out by heating the preparation at 95°C for 10 min, and cDNA was stored at −20°C.
PCR.
The PCR was performed with a Peltier thermal cycler (MJ Research). Each reaction mixture contained Taq DNA polymerase reaction buffer (10 mM Tris HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2)(Boehringer Mannheim), 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dTTP, 0.5 mM dGTP (all deoxynucleoside triphosphates were obtained from Boehringer Mannheim), each RT primer (Life Technologies) at a concentration of 0.5 μM, each PCR primer (Life Technologies) at a concentration of 0.5 μM, 0.05 U of Taq DNA polymerase (Boehringer Mannheim) per μl, 5 μl of cDNA, and enough autoclaved deionized water to bring the total volume to 60 μl. The PCR cycle consisted of initial denaturation at 94°C for 4 min, followed by 40 cycles consisting of denaturation at 94°C for 1 min, annealing at 40°C for 1 min, and extension at 72°C for 1.5 min. A final extension step consisting of 72°C for 5 min was included. PCR products were quantified by determining absorbance at 260 nm.
3′ end labeling of probe DNA with DIG-dUTP.
The 3′-end-labeling reaction mixture contained 25 μg of probe DNA, 50 U of terminal transferase (Boehringer Mannheim), terminal transferase reaction preparation (Boehringer Mannheim), 20 μM dATP (Boehringer Mannheim), 3 μM digoxygenin-labeled dUTP (DIG-dUTP)(Boehringer Mannheim), and CoCl2 (Boehringer Mannheim). This reaction mixture was placed in a 37°C water bath and incubated for 45 min. DIG-labeled probe was purified by ethanol-sodium acetate precipitation by using the protocol recommended by Boehringer Mannheim and was quantified by determining the absorbance at 260 nm.
We evaluated the signal strength resulting from end labeling and found that the signal was sufficient for chemiluminescent detection with gene mapping membrane (thus, the use of radioisotopes was avoided). We investigated the following two factors to make this determination: the ability of terminal transferase to 3′ end label DNA of the size expected for RT-PCR products (200 to 800 bp) and the ability of DIG-labeled probes to allow visualization of the Kohara clones on the gene-mapping membranes. The efficiency of end labeling of the RT-PCR products was evaluated by dot blotting, which clearly revealed a strong signal from end-labeled RT-PCR products. To determine the stringency required for detection of clones on the gene mapping membranes, hybridization of 3′ DIG-end-labeled λ DNA (restriction digested to sizes similar to those of RT-PCR products) was performed. Each Kohara clone was visible on a gene mapping membrane, which validated the detection method.
Hybridization and detection of amplified mRNA.
The E. coli gene mapping membranes used consisted of the Kohara set bound to nylon membranes in an ordered matrix (Panvera Inc., Milwaukee, Wis.). The E. coli gene-mapping membranes were prehybridized for 3 h in 15 ml of prehybridization-hybridization solution (Boehringer Mannheim). The prehybridization solution was decanted, and 5 ml of fresh prehybridization-hybridization solution was added. DNA was added to the hybridization mixtures at concentrations between 200 and 1,500 ng/ml. Hybridization occurred during incubation overnight at 40 to 65°C, and the membranes were developed by using the DIG development protocols recommended by Boehringer Mannheim, anti-DIG alkaline phosphatase (Boehringer Mannheim), and the CSPD chemiluminescent substrate (Boehringer Mannheim). Developed membranes were incubated at 37°C for 30 min and then exposed to X-ray film for 2 h. A schematic diagram of the overall RT-PCR and gene mapping technique is shown in Fig. 1.
Southern blot analysis.
Kohara clones were amplified, and λ DNA was purified by using a λ DNA purification kit (Promega). The clones were restriction digested as described below before they were loaded onto an agarose gel. DNA that was separated by agarose gel electrophoresis was denatured by submersion in denaturation solution (0.5 M NaOH, 1.5 M NaCl) for 30 min. The gel was neutralized with neutralization solution (0.5 M Tris-HCl [pH 7.5], 3 M NaCl) for 30 min prior to blotting. The gel was blotted overnight onto a nylon membrane (Boehringer Mannheim) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7). DNA was fixed to the membrane by using UV irradiation-induced cross-linking, and the membrane was stored at −20°C. The procedures used for hybridization with RT-PCR products and the DIG development procedures were identical to the procedures used for the gene mapping membrane.
Northern blotting.
Samples of total RNA (10 μg/well) were separated by electrophoresing a 1% agarose denaturing gel (17% formaldehyde) at 75 V for approximately 2 h. Unused regions of the gel were removed, and blotting onto a nylon membrane (Boehringer Mannheim) was performed at 4°C overnight in 20× SSC. Total-RNA dot blots were prepared by pipetting 1 to 5 μg of total RNA (in 50% formamide–6.5% formaldehyde–1 × SSC) directly onto nylon membranes by using a microsample filtration manifold (Schleicher and Schuell, Keene, N.H.). All of the nylon membranes were fixed by UV-induced cross-linking. The procedures used for hybridization with DIG-labeled Northern probes and the DIG development procedures were identical to the procedures used for the gene-mapping membrane. The Northern probes were prepared by using a PCR DIG probe synthesis kit (Boehringer Mannheim) and were approximately 400 bp long. A detailed standard curve for serially diluted DIG-labeled λ DNA was utilized to quantify Northern blots or total-RNA dot blots. The dilutions used spanned a 50-fold difference in DNA mass per dot, and the resulting standard curve had an r2 of 0.9. Northern blotting was performed by using ibpA and groEL to correlate with dot blot data and to check for probe sensitivity. The following primer sequences were used to prepare probes: clpA, 5′ ATGCTCAATCAAGAACT 3′ and 5′ AAAGTTCACCACATCGA 3′; ibpA, 5′ ATGCGTAACTTTGATTT 3′ and 5′ TTAGTTGATTTCGATAC 3′; groEL, 5′ ATGGCAGCTAAAGACGT 3′ and 5′ CTTTGTCCATCGCTTCA 3′; and degP, 5′ TTTAATGACCGTCGCGT 3′ and 5′CCACATTAGCACTGAGT 3′.
Quantification of RT-PCR product signal.
Densitometric scanning was performed to analyze RT-PCR product signals. Images of exposed film were acquired with an Eagle Eye II image acquisition system (Stratagene, La Jolla, Calif.). Each image was quantified by using Scion Image analysis software (Scion, Inc., Frederick, Md.) to measure the area under a curve of the scanned image minus the background of the film surrounding the image. Images were analyzed by determining the densities of each Kohara clone that contained a known heat shock gene and the two adjacent Kohara clones. For example, clones 101, 201, and 301 were evaluated due to the presence of the dnaKJ gene on clone 101 (clones 201 and 301, which are clones that do not contain a heat shock gene, were directly adjacent). The three Kohara clones were scanned simultaneously to minimize the quantification error. Since the technique for integrating the density of a scanned image directly affected the value obtained, the method used for quantification was performed in the same way for all of the film samples analyzed.
To obtain an induction ratio, the following set of calculations was performed. First, the quantified signal intensity for each clone was divided by the average signal intensity for all of the clones evaluated on the same film. This accounted for film-to-film variation in intensity and served as an internal control. Note that dividing by the average film signal intensity made visual interpretation of the films inaccurate. Second, the signal intensity of a clone derived from heat-shocked cells was divided by the signal intensity of the non-heat-shocked control. Third, to account for RT-PCR amplification error, the induction ratios from parallel experiments were averaged for each clone. In each case a minimum of three parallel experiments were included. Finally, for convenience, the data was normalized to an average induction ratio of 1 by dividing each clone value by the average induction ratio for all clones. As a result, any systematic error in RT, PCR, or hybridization that occurred between experiments or membranes was eliminated. An error analysis was performed to determine the level of significance. Specifically, the average normalized intensity for each clone for repeated experiments was determined, and the standard error was calculated by determining the standard deviation of the mean divided by the clone’s average intensity. To be considered “significantly induced,” a clone had to have an induction ratio greater than 1 plus the standard deviation of the error for that clone. To be considered “significantly repressed,” a clone had to have a value less than 1 minus the standard deviation of the error for that clone. All other clones were classified as “stable.” The induction ratio (IR) was calculated as follows:
IR = [(densitykc)/(densityfilm avg)]test/[(densitykc)/(densityfilm avg)]control, where densitykc is the density of the Kohara clone and densityfilm avg is the average density of the 105 clones analyzed on each film.
RESULTS
Evaluation of RT-PCR primers.
The ability of the RT-PCR primers described by Fislage et al. (8) (Table 1) to hybridize in a degenerate fashion was compared to the use of random hexamers for differential display analysis (data not shown). In these experiments, the RT reaction mixtures were incubated for 2 or 4 h, and this was followed by 10 or 30 PCR cycles. The best results, which yielded the greatest number of bands on agarose gels, were obtained when random hexamers were used for the RT reaction and an equimolar mixture of the 10 PCR primers of Fislage et al. (8) was used for the PCR. These primers, however, do not preferentially amplify mRNA so that a large amount of rRNA which confounds later analysis can be amplified (14). When RT primers were used in combination with the PCR primers, mRNA was preferentially targeted (8), but in our experiments the lowest number of bands was observed. Consequently, we sought a new combination of RT-PCR primers so that significant levels of mRNA would be amplified.
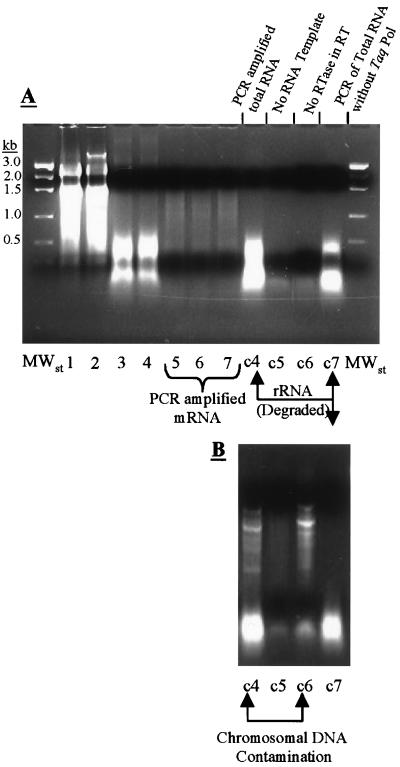
After a systematic analysis of several primer combinations and after several new primers were examined, one combination appeared to be most effective. Primers PCR1, PCR3, and PCR5 of Fislage et al. (8) were combined with the 10 RT primers for the RT reaction (a total of 13 primers were used), and all 20 primers of Fislage et al. were used for the PCR. When this combination was used, multiple bands were present at high concentrations, as determined by agarose gel electrophoresis (Fig. 2A). Lanes 5 through 7 in Fig. 2A contained a broad ethidium bromide smear corresponding to RT-PCR-amplified cDNA. Lanes c4 through c7 contained controls for contaminating chromosomal DNA (lane c4), degraded rRNA (lanes c6 and c7), and contaminated RT-PCR components (lane c5). Specifically, lane c4 contained PCR-amplified total RNA which was not subjected to the RT reaction (to determine if the purified RNA was contaminated with DNA), lane c5 contained RT-PCR products obtained in the absence of any RNA template (to determine whether the RT and PCR solutions were contaminated), lane c6 contained RT-PCR-amplified total RNA obtained without AMVRT (to ensure that total RNA degradation occurred during RT-PCR; otherwise, total RNA might have appeared on the gel), and lane c7 contained the PCR products obtained from total RNA without Taq polymerase (to assess total RNA degradation during PCR). Therefore, a comparison of lanes c4, c6, and c7 allowed us to determine if products amplified from purified RNA or contaminating DNA were present. Because lanes c4 and c7 were the same, RT-PCR amplification of RNA occurred; however, because lanes c4 and c6 were not the same, the purified RNA was not contaminated with DNA. Figure 2B shows an identical control agarose gel in which RT-PCR was performed in the presence of contaminating chromosomal DNA. When RNA was added to the PCR mixture (with no RT step), amplification resulted in multiple bands of product cDNA between 0.2 and 1 kbp (Fig. 2B, lanes c4 and c6). In Fig. 2A, there are no distinct contamination bands in this region in lanes c4 and c6; therefore, there was no contaminating chromosomal DNA (the chromosomal DNA was removed by DNase I treatment). The results obtained with the RNA-only control (Fig. 2A, lane c4) closely resembled the results obtained when RNA was added to the PCR mixture in the absence Taq DNA polymerase (Fig. 2A, lane c7). In this case, the visualized bands corresponded to RNA fragments degraded during the PCR. Therefore, the higher-molecular-weight bands in lanes 5, 6, and 7 correspond to RT-PCR-amplified RNA, not degraded RNA or amplified chromosomal DNA.
FIG. 2.
(A) Agarose (2%) gel containing RT-PCR samples from heat-shocked and control cells. Lanes 1 and 2 contained control and heat-shocked RNA, respectively. Lanes 3 and 4 contained the RT products in lanes 1 and 2, respectively. Lane 5 contained the products resulting from PCR amplification of the material in lane 3. Lanes 6 and 7 contained parallel PCR mixtures from lane 4. Lanes c4 through c7 contained controls. Lane c4 contained the product resulting from PCR amplification of purified RNA (lanes 1 and 2). Lane c5 contained the product resulting from a complete RT-PCR performed with no initial template RNA. Lane c6 contained the RT-PCR product obtained without AMVRT, and lane c7 contained the PCR product resulting from RNA obtained without DNA Taq polymerase. (B) Agarose (2%) gel containing new RNA samples loaded in the same manner as those in panel A. However, the purified RNA samples were contaminated with DNA, as shown by the multiple bands in control lanes c4 and c6.
The RT-PCR procedure was utilized to amplify mRNA from total RNA of both heat-shocked and control (non-heat-shocked) cells. The total-RNA, cDNA, and RT-PCR products are shown in Fig. 2A, lanes 1 through 7. We observed distinct bands in the RT-PCR product lanes and different levels of bands of similar sizes between the RT-PCR products obtained from control and heat-shocked cells (data not shown). In addition, the lanes containing control samples revealed that the RT-PCR products were not degraded RNA (lane c7), amplified full-length rRNA (lanes 1 and 2), or amplified E. coli chromosomal DNA (lanes c4 and c6) or the result of contamination of RT reaction or PCR components (lane c5). However, the results did not reveal whether the amplification data indicated of the relative amounts of mRNA transcripts. To determine this, we evaluated RT-PCR products for a well-characterized stress response, the heat shock response, by using a gene-mapping membrane.
Hybridization to the gene-mapping membrane.
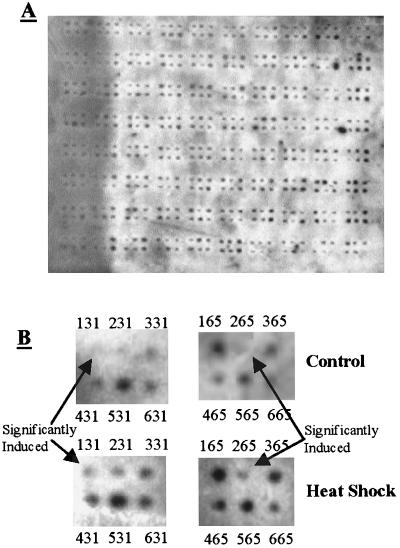
In order to quantify and map RT-PCR products, the end-labeled RT-PCR products were hybridized to gene-mapping membranes (Fig. 3A). All of the Kohara clones were clearly visible on all of the film samples analyzed; this indicated that amplified mRNA fragments representing the entire E. coli genome hybridized and were detected on the membranes. The signal-to-noise ratio was high enough to quantify the signal at each clone location. In fact, the signals for all 630 clones evaluated were the same within 1%. In addition, the signals for the clones containing rRNA genes were not the maximum signals for all of the clones analyzed, which reaffirmed the ability of the RT-PCR primers to preferentially amplify non-rRNA molecules.
FIG. 3.
(A) Hybridization of the RT-PCR product obtained from heat-shocked cells to an E. coli gene-mapping membrane. (B) Gene mapping membrane results obtained for control and heat-shocked films for the regions containing clones 131 and 265. Significantly induced clones 131 and 265 are visible in the heat shock films but are not visible in the control films. In addition, after quantification and analysis of repeated measurements, the induction ratios minus the standard errors were all greater than 1.
We compared the quantified signals for each clone obtained with heat-shocked and control cells. Table 2 shows the clones that were significantly induced or repressed by the heat shock treatment. The induction ratio was determined as described above. Briefly, clone signals were quantified by using Scion Image analysis software and were normalized by using the average signal for all of the quantified clones on the film being analyzed. The induction ratio was calculated by dividing the average heat shock signal by the average control signal, and the standard error was calculated by adding the heat shock and control errors for each clone. Hence, each clone had an associated standard error. Significance was then determined based on the individual clone errors rather than on the average error for all of the clones analyzed. To be significantly induced, the clone-specific induction ratio minus the clone-specific standard error had to be greater than 1. Figure 3B shows the results obtained for two significantly induced clones (clones 131 and 265).
TABLE 2.
Significantly induced and repressed clones and their induction ratios
| Induced clones
|
Repressed clones
|
||
|---|---|---|---|
| Designation | Induction ratio | Designation | Induction ratio |
| 131 | 4.8 ± 0.6 | 258 (pspA) | 0.23 ± 0.1 |
| 566 (ibpAB) | 4.6 ± 2.5 | 675 | 0.40 ± 0.1 |
| 117 (degP) | 3.9 ± 1.3 | 232 (hslD) | 0.40 ± 0.2 |
| 212 (clpA) | 3.3 ± 0.2 | 639 | 0.45 ± 0.2 |
| 152 (htpG) | 2.1 ± 0.2 | 449 | 0.46 ± 0.2 |
| 265 (hslIJ) | 1.8 ± 0.2 | 549 | 0.48 ± 0.3 |
| 248 | 1.8 ± 0.6 | 547 | 0.49 ± 0.4 |
| 537 | 1.7 ± 0.3 | 160 | 0.49 ± 0.1 |
| 334 (hslK) | 1.7 ± 0.6 | 410 | 0.50 ± 0.1 |
| 648 (groESL, hslW) | 1.6 ± 0.1 | 638 | 0.51 ± 0.3 |
| 448 | 1.5 ± 0.3 | 575 | 0.51 ± 0.2 |
| 234 | 1.3 ± 0.2 | 509 (rpoD) | 0.52 ± 0.2 |
| 652 (hslXYZ) | 1.2 ± 0.1 | 302 | 0.52 ± 0.4 |
| 233 (hslD) | 1.2 ± 0.1 | 132 | 0.56 ± 0.3 |
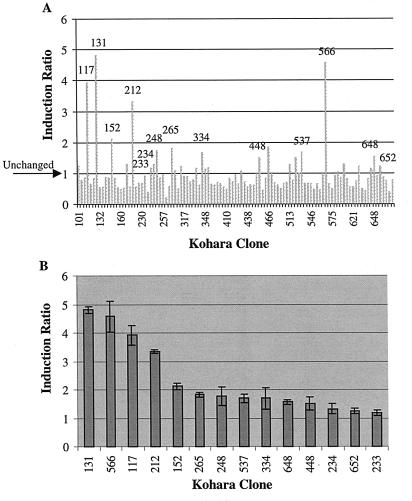
Our results indicated that 9 of the 14 significantly induced clones contained known heat shock genes and that several contained multiple heat shock genes. For example, clone 566 contained the ibpA and ibpB genes, and clone 648 contained the groES, groEL, and hslW genes. A histogram analysis revealed that 71% of the significantly induced clones contained heat shock genes. This was in contrast to the significantly repressed clones, only 28% of which contained known heat shock genes.
Figure 4A shows the induction ratio for each clone after heat shock treatment and RT-PCR, calculated as described above. Importantly, 87% of the clones evaluated had either stable or significantly repressed induction ratios. This was expected during the heat shock response due to RNA polymerase sequestering by heat shock sigma factor ς32 (11). Figure 4B shows the induction ratio and percent error for each of the significantly induced clones. As Fig. 4B shows, each significantly induced clone had an induction ratio minus standard error that was greater than 1. Also, the benefit of using clone-specific error analysis was revealed. For example, clone 233 had a induction ratio of only 1.2; however, the error was relatively small, which allowed us to classify this clone as significantly induced. Based on the initial differential display results, a number of the significantly induced clones were evaluated further by performing Southern blotting with restriction enzyme-digested Kohara clones and Northern blotting with mapped genes.
FIG. 4.
(A) Induction ratios for all of the clones analyzed. The clones identified on the figure were determined to be significantly induced. An induction ratio of 1 indicated that the clone signal did not change after the heat shock treatment. (B) Induction ratios and standard errors for all clones determined to be significantly induced. Note that the error bars for significantly induced clones do not go below 1.
Southern blotting and Northern blotting for identification of specific genes.
To ensure that amplified Kohara clone signals were the result of heat shock genes and not other genes in each clone, Southern blotting with restriction enzyme-digested Kohara clones and Northern blot and total-RNA dot blot experiments were performed.
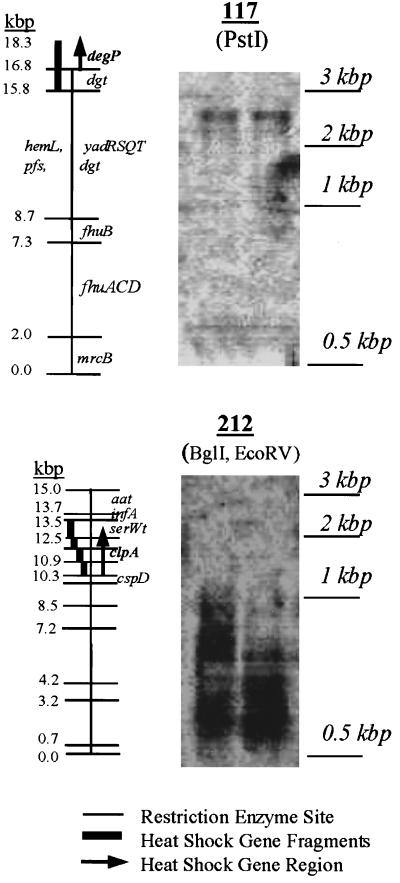
A subset of the significantly induced Kohara clones were restriction digested either to isolate previously identified heat shock genes or to provide DNA fragments less than 3 kbp long or both. The results obtained for clones 212 and 117 are shown in Fig. 5. The restriction maps for each clone are shown next to the Southern blot results. For example, Kohara clone 212 contained the heat shock clpA gene, which was located on two 0.6-kbp fragments and two 1.0-kbp fragments after digestion. Therefore, the signal detected was a broad smear with two distinct band ends between 0.6 and 1.0 kbp corresponding to hybridization of the RT-PCR heat shock product and the clpA gene of E. coli. Kohara clone 117 has been reported to contain the heat shock degP gene (1); however, the E. coli linkage map indicates that degP is located on clones 118 and 119. Therefore, the DNA band (which was obtained from highly induced clone 117) located at 2.5 kbp could be either degP or a DNA fragment containing a potentially new heat shock gene. In addition, Southern blotting was performed with clones 152 and 265, and the fragments identified mapped correctly to heat shock gene regions (5). On the other hand, clone 131 contained the argF gene and multiple putative genes whose functions are not known (determined from a database search of Entrez [14a]). Southern blotting mapped the RT-PCR product to yagP; however, Northern blotting did not confirm differential display. This was an example of a false positive.
FIG. 5.
Southern blots obtained by using restriction enzyme-digested Kohara clones 117 and 212 and by probing with RT-PCR-amplified RNA from heat-shocked E. coli. Each clone was electrophoresed twice, in adjacent lanes of the same gel. Restriction maps for the clones are shown next to the Southern blots. Restriction sites are indicated by horizontal lines in the restriction maps. The restriction enzymes used are indicated below the clone numbers. The genes and locations of the genes on the restriction fragments are indicated. The vertical bars on the restriction maps indicate the fragments detected by Southern blotting. For clone 117 a single 2.5-kbp fragment was detected, and for clone 212 a smear of fragments corresponding to two 0.6-kbp fragments and two 1.0-kbp fragments was visible.
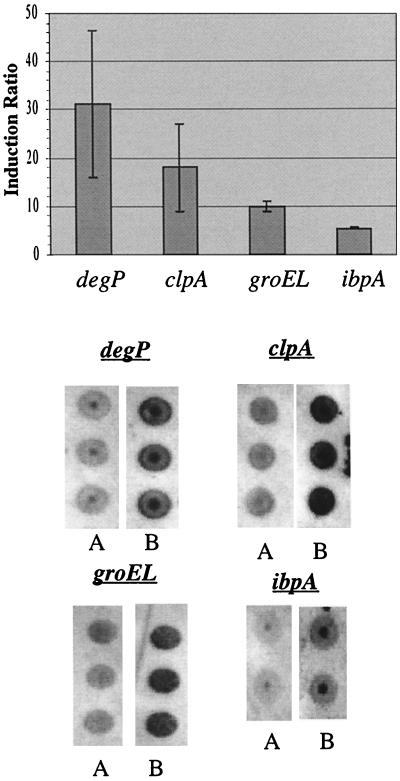
Northern dot blot experiments were performed with clpA, degP, ibpA, and groEL in order to confirm that different transcript levels were present in the heat-shocked and control samples (Fig. 6). In each case the level of the transcript increased significantly after the heat shock. The level of degP increased 31-fold, the level of clpA increased 18-fold, the level of groEL increased 10-fold, and the level of ibpA increased 5-fold. All of the clones which contained these genes, clones 117 (degP), 212 (clpA), 648 (groEL), and 566 (ibpA), were also significantly induced. Note that the induction ratios calculated by using dot blots were not the same as the induction ratios calculated by using RT-PCR mapping, although this result should not have been expected as the RT-PCR mapping technique is not gene specific. It was interesting to note, however, that the relative levels of induction were similar for the two analytical approaches. To confirm that the RNA probes were specific and that total RNA dot blot data was quantifiable, a Northern blot analysis was performed with ibpA and groEL, and a reliable standard curve was created by using prelabeled λ DNA (see above) (data not shown). In summary, the combination of the Southern blot, Northern blot, and total-RNA dot blot data confirmed that RT-PCR amplification and Kohara clone-based analysis performed in this study indicated the relative abundance of mRNA in the original total-RNA sample.
FIG. 6.
Total RNA dot blots for degP, clpA, ibpA, and groEL. The data in the bar graph are data obtained from dot blots of heat-shocked samples collected in triplicate under identical conditions. The signal intensity levels for each dot were quantified and averaged for each sample. The induction ratios were then calculated by dividing the average signal intensity for each heat-shocked sample by the corresponding average control signal intensity. For each dot blot, A was the control and B was the preparation obtained from cells that were heat shocked (15 min, 42°C). Note that the dark dot in each of the dot blots resulted from the apparatus used in the experiments.
DISCUSSION
To prove that the technique described here is an experimentally viable tool, we established that the RT-PCR product was the result of amplified mRNA (not contaminating DNA or total RNA) and that the amplified products indicated the relative mRNA amounts at the times that the samples were obtained. The first criterion was established by judiciously using experimental controls (Fig. 2). The second criterion, which was much more difficult to determine, was established by using a known stress response and by comparing our observed results to expected results. In addition, we utilized Southern blot-based mapping to ensure that the results observed corresponded to heat shock genes. Finally, Northern blotting and total-RNA dot blotting were performed to confirm the different levels of specific genes in our RNA samples.
RT-PCR primers and use of the gene-mapping membrane.
The RT-PCR method, in which we used the combination of RT-PCR primers described here, allowed us to quantify induced and repressed Kohara clones containing differentially displayed genes in response to heat shock. Our results confirmed that using the primers of Fislage et al. (8), modified as described above, resulted in significant quantities of RT-PCR product cDNA that were representative of the relative amounts of mRNA before and after the heat shock.
We used several experimental controls in this study which allowed us to quantify the results with statistical significance. Specifically, we evaluated PCR performed with purified RNA without an RT step, degradation of RNA by RT and PCR (with no AMVRT or Taq DNA polymerase added), RT-PCR of components with no RNA or cDNA templates added, and parallel RT or PCR performed with identical RNA or cDNA. These controls accounted for contamination by E. coli chromosomal DNA, erroneous bands of degraded rRNA, DNA contamination of reaction components, and variations in RT-PCR amplification, respectively. The control products obtained from PCR-amplified RNA in the presence and in the absence of DNA Taq polymerase were identical. Therefore, the ethidium bromide smear in control lanes contained degraded RNA, not amplified contaminating DNA. This result was supported by the results of agarose gel electrophoresis performed before the DNase I treatment and double RNA purification steps were used (Fig. 2B). When chromosomal DNA was present, the PCR-amplified RNA lanes contained multiple, distinct bands. These lanes contained different band patterns than the RT-PCR product lanes. We believe that this was the result of primer incorporation during the RT reaction and perfect match amplification during PCR.
The use of an internal control was very important when we attempted to quantify results of differential display experiments. It has been shown that minute pipetting errors or gradients in the thermal cycler can lead to differences in parallel differential display experiments (10). One common method for controlling for these variations is to add a known amount of template DNA to the reaction mixture. After amplification, the results obtained for the amplified control template in parallel reactions can be compared. We utilized the average signal from each film as our internal control. By dividing by the average clone signal, we accounted for differences in pipetting and thermal cycling conditions; this was similar to adding a known amount of template. In addition, the signal from the same clone on the same membrane was compared after hybridization with heat-shocked or control samples. Therefore, clone-to-clone and membrane-to-membrane errors were eliminated.
The gene-mapping membrane was the greatest source of error throughout this study. The major problems associated with the gene-mapping membrane were not due to inconsistent readings at individual clone locations; rather, individual membranes probed under identical conditions at times did not produce detectable or quantifiable signals. In fact, the intermembrane variability was almost 20%. This was consistent with the findings of Chuang and Blattner (5), who reported that repeatability was significantly improved when the same membrane was utilized. We independently evaluated the effects of poor membrane stripping, probe precipitation, and nonspecific hybridization (data not shown). Specifically, the stripping procedure was evaluated by hybridizing unlabeled probe and developing the membrane in the standard manner. The resulting films were perfectly clean; therefore, the signal obtained was consistently the result of labeled probe alone. Due to the highly ordered nature of the gene-mapping membrane, precipitation of a probe was easily detected as miscellaneous dots in random order. Because precipitation was detected easily and relatively infrequently, it was eliminated from the analysis. Importantly, 71% of the significantly induced clones contained heat shock genes, which reflected the ability of our RT-PCR method to amplify mRNA based on the relative abundance of a transcript. In summary, the signals from each Kohara clone location were determined to be the result of hybridization with RT-PCR-amplified mRNA present in the cells at the time of harvest.
RT-PCR as a screening method for differentially displayed genes.
The RT-PCR technique used here to analyze differentially displayed genes in E. coli is useful as a screening technique for rapidly identifying Kohara clones containing genes that respond to certain environmental stresses. Since Kohara clones contain more than one E. coli gene, this technique must be followed by additional techniques for identifying specific genes that respond to previously uncharacterized stress responses. Specifically, Northern blotting with gene-specific probes or Southern blotting (19) with restriction enzyme-digested Kohara clones (5) can be used to identify differentially displayed genes. We found that Northern blotting with specific genes produced much better results given the amount of experimental work required. In addition, given the wealth of information about the E. coli genome, educated guesses can be made about differentially displayed genes in lieu of the somewhat lengthy Southern blotting steps.
Our comparison of heat shock clones to control clones provided the strongest evidence in support of the use of this technique as a differential display screening tool. This was possible only because of the abundance of data pertaining to the heat shock response (11). As an alternative to mapping an unknown regulatory response, we sought to validate this technique by using a well-known, repeatedly validated stress response. This allowed us not only to develop a sensitive method for determining significance (based on clone-specific error) but also to achieve a true positive ratio of close to 71%.
Using the 2.5-day procedure described here, we isolated stressed and control cellular total RNA, RT-PCR-amplified the mRNA, and identified amplified clones nonradioactively with relatively inexpensive gene-mapping membranes. Nine clones containing 15 previously identified heat shock genes were found to have significant induction ratios. Finally, Southern blotting performed with RT-PCR product probes and restriction enzyme-digested Kohara clone DNA and gene-specific Northern blotting were utilized to ensure that the levels of the RT-PCR products were representative of the levels of mRNA in the original total-RNA samples.
The average induction ratio for clones containing heat shock genes was 44% greater than the induction ratio for clones that did not contain heat shock genes. In addition, clones containing heat shock genes were generally not represented in the group of clones that were significantly repressed, as would have been expected for random, nonsense amplification and hybridization. The true hit rate for differential display experiments has been reported to be approximately 50% (21). Therefore, the true hit rate of 71% reported here represents a significant improvement over identification by electrophoresis and subsequent reamplification and sequencing.
The Kohara set was originally developed not only to map the location of E. coli genes but also to “map and clone the gene or genes that is (are) induced in response to a certain external or internal signal(s) … It was with these goals in mind that we initiated this work” (13). Chuang et al. (4) responded to the research of Kohara et al. with their work performed with single-stranded reverse-transcribed cDNA. The present work represents a natural extension of the work of Chuang et al., in which we included a PCR amplification step that was made possible by the availability of the prokaryotic mRNA-specific primers described by Fislage et al. (8). It will be interesting to apply this RT-PCR technique to uncharacterized E. coli responses in order to better understand a global genetic regulatory response. In order to carry out this research, the technique described here should be complemented with subcloning performed with restriction enzyme-digested Kohara clones and/or Northern blotting. At present, such techniques are being utilized to study additional E. coli stress responses in our laboratory.
ACKNOWLEDGMENTS
Funds for this project were provided by grant DAAM01-96-0037 from the U.S. Army Engineering, Research, and Development Center, Edgewood, Md.
We thank Yuji Kohara for graciously supplying the Kohara miniset of overlapping λ clones.
REFERENCES
- 1.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- 2.Casadaban M J. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976;104:557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang S, Daniels D, Blattner F. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuang S, Blattner F R. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nature Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 7.Donohue P, Hsu D, Winkles J. Differential display methods and protocols. Totowa, N.J: Humana Press; 1997. Differential display using random hexamer-primed cDNA, motif primers, and agarose gel electrophoresis; pp. 25–35. [DOI] [PubMed] [Google Scholar]
- 8.Fislage R, Berceanu M, Humboldt Y, Wendt M, Oberender H. Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res. 1997;25:1830–1835. doi: 10.1093/nar/25.9.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill R T, Cha H, Jain A, Rao G, Bentley W E. Physiological effects of DTT addition to E. coli including growth rate, specific oxygen uptake, heat shock protein expression, and specific activity of recombinant protein. Biotechnol Bioeng. 1998;59:248–259. [PubMed] [Google Scholar]
- 10.Gilliland G, Perrin S, Bunn F. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. Competitive PCR for quantitation of mRNA; pp. 60–69. [Google Scholar]
- 11.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 12.Kenyon C J, Walker G C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci USA. 1980;177:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohara U, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 14.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 14a.National Center for Biotechnology Information. 13 October 1999, posting date. [Online.] National Institutes of Health. http://www.ncbi.nlm.nih.gov. [27 October 1999, last date accessed.]
- 15.O’Farell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 16.Pederson S, Bloch P L, Reeh S, Neidhardt F C. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978;14:179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez R L, Tait R E. Recombinant DNA techniques: an introduction. Menlo Park, Calif: Benjamin/Cummings; 1983. [Google Scholar]
- 18.Rupani S, Gu M B, Konstantinov K B, Dhurjati P S, Van Dyk T K, LaRossa R A. Characterization of the stress response of a bioluminescent biological sensor in batch and continuous cultures. Biotechnol Prog. 1996;12:387–392. doi: 10.1021/bp960015u. [DOI] [PubMed] [Google Scholar]
- 19.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 20.Van Dyk T K, Majarian W R, Konstantinov K B, Young R M, Dhurjati P S, Larossa R A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan J, Erlander M. Differential display methods and protocols. Totowa, N.J: Humana Press; 1997. Cloning differentially expressed genes by using differential display and subtractive hybridization; pp. 45–68. [DOI] [PubMed] [Google Scholar]
- 22.Welsh J, Chada K, Dalal S, Cheng R, Ralph D, McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992;20:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong K K, McClelland M. Stress-inducible gene of Salmonella typhimurium identified by arbitrarily primed PCR of RNA. Proc Natl Acad Sci USA. 1994;91:639–643. doi: 10.1073/pnas.91.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]