Significance
Nitric oxide is a multifaceted signaling molecule that affects multiple pathways and cellular systems. Here, we report that inducible nitric oxide synthase expression, which is strongly correlated with inflammation and poor outcomes in patients with cancer, substantially alters DNA methylation to regulate cellular plasticity. Our data connect inflammation mechanistically with stress signals, DNA demethylation, and genotoxic retrotransposon expression. Passive DNA demethylation occurs during conditions of reduced DNA (cytosine-5)–methyltransferase 1 (DNMT1) activity; however, the cellular pathways that control passive demethylation are not clear. Our results show that sustained cellular stress signals result in DNMT1 protein loss and DNA hypomethylation, similar to DNMT1 inhibition by 5-azacytidine. This implies that chronic inflammation drives cellular transformation via DNA hypomethylation and retrotransposon activation.
Keywords: nitric oxide, NOS2, DNA methylation, retrotransposon, S-nitrosation
Abstract
Inducible nitric oxide synthase (NOS2) produces high local concentrations of nitric oxide (NO), and its expression is associated with inflammation, cellular stress signals, and cellular transformation. Additionally, NOS2 expression results in aggressive cancer cell phenotypes and is correlated with poor outcomes in patients with breast cancer. DNA hypomethylation, especially of noncoding repeat elements, is an early event in carcinogenesis and is a common feature of cancer cells. In addition to altered gene expression, DNA hypomethylation results in genomic instability via retrotransposon activation. Here, we show that NOS2 expression and associated NO signaling results in substantial DNA hypomethylation in human cell lines by inducing the degradation of DNA (cytosine-5)–methyltransferase 1 (DNMT1) protein. Similarly, NOS2 expression levels were correlated with decreased DNA methylation in human breast tumors. NOS2 expression and NO signaling also resulted in long interspersed noncoding element 1 (LINE-1) retrotransposon hypomethylation, expression, and DNA damage. DNMT1 degradation was mediated by an NO/p38-MAPK/lysine acetyltransferase 5–dependent mechanism. Furthermore, we show that this mechanism is required for NO-mediated epithelial transformation. Therefore, we conclude that NOS2 and NO signaling results in DNA damage and malignant cellular transformation via an epigenetic mechanism.
Nitric oxide (NO) is an endogenously produced signaling molecule with a broad array of biological functions, ranging from neurotransmission to viral immune response (1–3). Inducible nitric oxide synthase (NOS2) is an inflammatory enzyme that produces high NO levels and initiates S-nitrosothiol (SNO) signaling, a term that we use herein to refer to signaling induced by NO-dependent cellular thiol oxidation (4, 5). SNO signaling results in altered protein function and signaling pathways (6–8), in addition to protein disulfide bond formation (9). NOS2 expression activates multiple oncogenic signaling pathways via SNO signaling (10, 11) and modulates the tumor immune response (12). Furthermore, NOS2 expression is associated with poor outcomes in patients with cancer and aggressive cancer cell phenotypes (13–18), while NOS2 inhibition reduces tumor burden and metastasis (19, 20).
DNA methylation is frequently altered in human cancer cells compared with normal cells, as cancer cell genomes are predominantly hypomethylated with hypermethylated promoter regions of tumor suppressor genes (21–23). In addition to altered gene expression, global DNA hypomethylation results in increased DNA mutation rates (24), endogenous retrotransposon and retrovirus expression, genomic and microsatellite instability, and insertional mutagenesis (25–27). The most abundant retrotransposon in the human genome is the long interspersed noncoding element 1 (LINE-1), which is normally suppressed by promoter methylation but commonly is expressed in many human tumors. (26, 28). LINE-1 expression results in multiple genomic alterations, including indel mutations, transcriptional interference, and DNA strand breaks (29). Notably, both genome-wide hypomethylation and LINE-1 expression are observed in precancerous lesions, implicating DNA demethylation as an early event in tumorigenesis (30–32). Furthermore, the extent of DNA hypomethylation and LINE-1 expression also correlates with tumor stage (33, 34). Therefore, DNA hypomethylation and LINE-1 expression appear to be involved in both tumor initiation and progression.
DNA (cytosine-5)–methyltransferase 1 (DNMT1) is the critical epigenetic writer responsible for maintaining DNA methylation during replication (35), as well as de novo methyltransferase activity at retrotransposons (36). Two molecular processes achieve DNA demethylation: TET dioxygenase-mediated “active” and replication-dependent “passive” demethylation (37). While active demethylation is, in part, regulated by TET dioxygenase expression and cofactor availability, passive demethylation occurs during DNA replication in the absence of DNMT1 (37). DNMT1 protein stability is governed by multiple posttranslational modifications, such as acetylation, methylation, and phosphorylation (38, 39). Reduced DNMT1 expression results in global hypomethylation, loss of genomic imprinting (40), and spontaneous tumors in mice (41). Furthermore, DNMT1 mutations that result in the deletion of the catalytic domain can be found in human colorectal cancers, implicating loss of DNMT1 activity during tumorigenesis (42). Therefore, decreased DNMT1 expression and activity are associated with tumorigenesis.
As tumor DNA hypomethylation and NOS2 expression are both associated with aggressive cancer phenotypes and poor survival rates among patients, we investigated the role of NOS2 on DNMT1 activity, expression and DNA methylation. Here, we show that NOS2 expression and SNO signaling caused the proteasomal degradation of DNMT1 and global DNA hypomethylation. Mechanistic studies revealed that NO-mediated DNMT1 degradation and epithelial cell transformation was p38-MAPK dependent. Consistent with DNA hypomethylation, we also show that NOS2 expression resulted in LINE-1 expression and DNA damage. These data indicate that NOS2 activity, via SNO signaling, dramatically alters cellular DNA methylation profiles to promote genomic instability and cellular transformation.
Results
NOS2 and SNO Signaling Induces Passive DNA Demethylation.
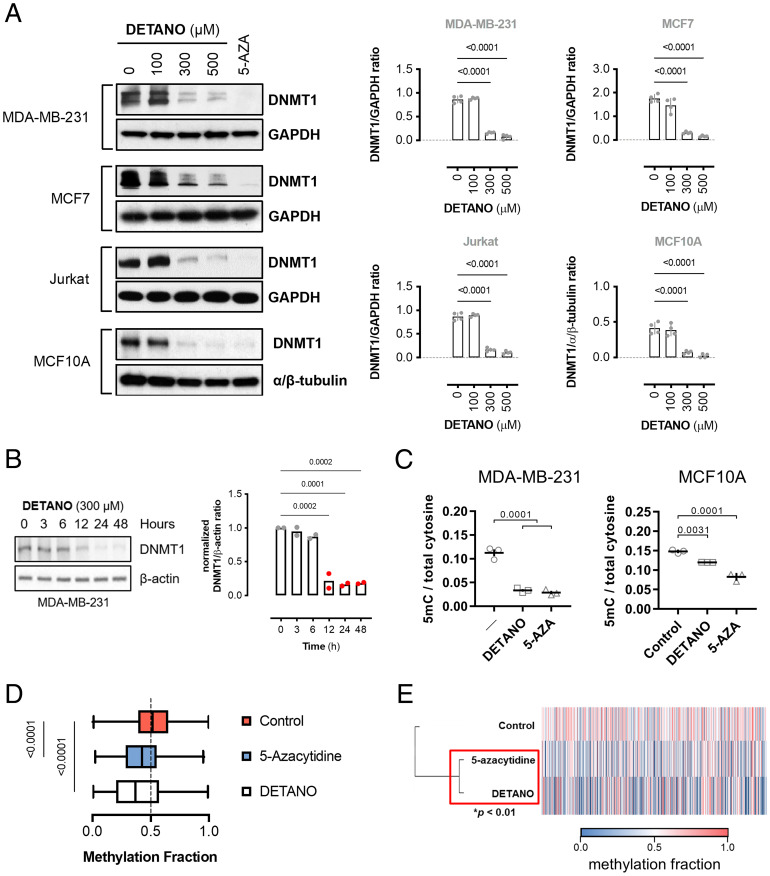
Cancer cell NOS2 expression is correlated with a basal-like breast cancer gene signature in estrogen receptor–negative (ER−) breast tumors (14). However, a majority of genes up-regulated in NOS2high compared with NOS2low ER− breast tumors have a repressive CpG island located at the transcriptional start site (SI Appendix, Table S1), which led us to hypothesize that NOS2 activity may alter DNA methylation levels. DNMT1 catalysis requires a cysteine at the active site and is, therefore, potentially inhibited by SNO signaling (5). To determine if methyltransferase activity is inhibited by SNO bond formation, recombinant human DNMT1 protein was incubated with the rapid-releasing, NO-donor 1,1-diethyl-2-hydroxy-2-nitroso-hydrazine sodium (DEANO; half-life [t1/2] = 2 min). NO significantly reduced rhDNMT1 activity and resulted in DNMT1 S-nitrosation (SI Appendix, Fig. S1A). DEANO-treated isolated nuclei also exhibited reduced (cytosine-5)-methyltransferase activity (SI Appendix, Fig. S1B). However, in a whole-cell context using the slow-releasing NO-donor 2,2′-(hydroxynitroso-hydrazono)bis-ethanimine (DETANO; t1/2 = 22 h), a seminal and intriguing observation in multiple human cell lines was made, namely that physiologically relevant concentrations of NO led to a significant loss of DNMT1 protein (Fig. 1A and SI Appendix, Fig. S1 C–F). DETANO reduced DNMT1 levels similar to the clinically used DNMT1 inhibitor, 5-azacytidine, which also results in DNMT1 protein loss (43). NO-mediated loss of DNMT1 has profound implications for cell lineage survival and identity, as we describe later, and occurs without significantly reducing cell proliferation or viability (SI Appendix, Fig. S2A). To examine the temporal regulation of DNMT1 by NO, cells were exposed to DETANO for the indicated times and DNMT1 protein was measured by immunoblotting. DETANO exposure decreased DNMT1 expression between 6 and 12 h and remained low during NO exposure (Fig. 1B). DNMT3A protein levels were not affected by NO signaling (SI Appendix, Fig S2B). Similar to DNMT1 protein levels, total DNA methylation levels measured by liquid chromatography–mass spectrometry (Fig. 1C) and Illumina BeadChip arrays (Fig. 1 D and E and SI Appendix, Fig. S2C and Dataset S1) were significantly reduced in NO-treated cells compared with control cells and were statistically similar to results after 5-azacytidine treatment. The decrease of DNMT1 expression followed an NO threshold–concentration effect, similar to the previously described SNO-mediated oncogenic pathway activation (10). NO-mediated loss of DNMT1 protein was not sensitive to soluble guanylyl cyclase inhibition (SI Appendix, Fig. S2D), and a stable cGMP analog did not significantly alter DNMT1 protein levels (SI Appendix, Fig. S2E). These data indicate that canonical NO/cGMP signaling does not contribute to reduced DNMT1 protein expression and suggest that NO mediates DNA demethylation by SNO signaling.
Fig. 1.
SNO signaling induces passive DNA demethylation. (A) Immunoblot analysis of relative DNMT1 expression from cells treated with DETANO or 5-azacytidine (AZA; 1 μM) for 48 h. Graphs represent mean normalized DNMT1 levels (±SEM; n = 3–6). Significance was calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (B) Immunoblot of DNMT1 expression in MDA-MB-231 cells treated with DETANO for the indicated times. Graph shows mean DNMT1 to actin ratios (n = 2). (C) Total DNA cytosine methylation quantification from cells treated with either DETANO (300 μM) or 5-azacytidine (5 μM) for 48 h. Graphs represent mean 5mC to total cytosine ratios (±SEM; n = 3). Significance was calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (D) Box-and-whisker plot of mean methylation fraction from DNA isolated from MDA-MB-231 cells treated with DETANO and with 5-azacytidine (5 μM) for 48 h and compared with untreated controls. Graph represents 6,070 loci with Δβ ≥ 0.2 of control mean methylation. Statistical significance was calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (E) Heat map of mean methylation values of loci with Δβ ≥ 0.2 of control. Statistical significance was calculated by hierarchical cluster analysis with multiscale bootstrap resampling.
In addition to passive demethylation, TET iron dioxygenases catalyze the oxidation of 5mC during active DNA demethylation (37). However, NO treatment decreased TET expression, activity, and 5hmC content compared with vehicle-treated cells (SI Appendix, Fig. S2 F–I). As NO is a pleiotropic signaling molecule that affects multiple pathways (5), it is conceivable that NO results in decreased DNA 5mC by an alternative mechanism that is independent of DNMT1 protein status and TET activity. However, the effect of NO-mediated DNMT1 protein degradation and DNA demethylation was rescued by DNMT1 overexpression (SI Appendix, Fig. S2J), indicating that SNO signaling caused the shift in global 5mC content by decreasing DNMT1 protein levels and inducing passive demethylation.
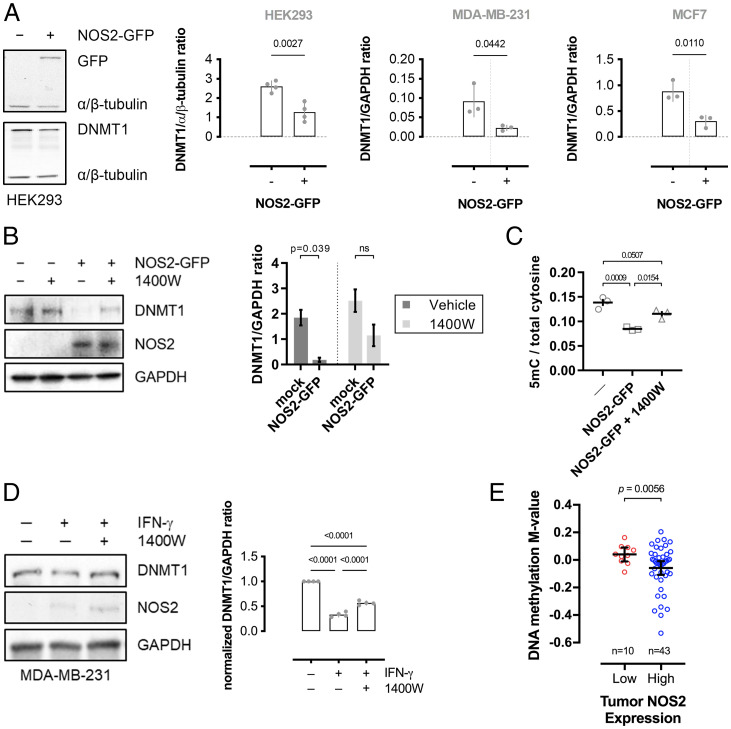
Human NOS2 transgene expression in NOS2-null breast cancer cell lines (MDA-MB-231 and MCF7) or a noncancerous cell line (HEK293) resulted in decreased DNMT1 protein levels (Fig. 2A and SI Appendix, Fig. S2K), which was partially reversed by NOS2 inhibition (Fig. 2B), indicating that NOS2 activity and subsequent SNO signaling decrease DNMT1 protein expression. NOS2 expression also resulted in a significant decrease in 5mC content, which was attenuated by NOS2 inhibition (Fig. 2C). Additionally, IFN-γ treatment resulted in NOS2 expression and significant DNMT1 downregulation, which was partially reversed by the NOS2-selective inhibitor, 1400W (Fig. 2D). Similarly, high NOS2 expression in human breast tumors is associated with decreased global DNA methylation levels compared with tumors with low NOS2 expression (Fig. 2E and SI Appendix, Table S2). Therefore, NOS2 expression and activity are associated with decreased DNA methylation.
Fig. 2.
NOS2 expression decreases DNA methylation. (A) Immunoblot and densitometric analyses of NOS2 and DNMT1 expression in cells 48 h after mock or NOS2-GFP transient transfection. Graphs represent mean relative DNMT1 densitometry (±SEM; n = 4). Significance was calculated by an unpaired, two-tailed t test. (B) Immunoblot and densitometric analyses of NOS2 and DNMT1 expression in HEK293 cells 48 h after mock or NOS2-GFP transient transfection with or without NOS2 inhibitor 1400W (10 μM) added 24 h after transfection. Graphs represent mean relative DNMT1 densitometry (±SEM; n = 3). Significance was calculated by two-way ANOVA with Tukey’s multiple comparisons test. (C) Total DNA cytosine methylation from NOS2-GFP expressing HEK293 cells treated with or without 1400W for 24 h. Graphs represent mean 5mC to total cytosine ratios (±SEM; n = 3). Significance was calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (D) Immunoblot showing relative DNMT1 and NOS2 expression in MDA-MB-231 cells stimulated with IFN-γ and 1400W. Graph shows mean DNMT1 to GAPDH ratios (±SEM; n = 4). Significance was calculated by one-way ANOVA with Dunnett’s test. (E) DNA methylation M-values from human breast tumors expressing either low or high levels of NOS2. Horizontal line represents the mean and 95% CI M-value of each genome. Significance was determined by two-tailed, unpaired t test with Welch’s correction. Data were obtained from Illumina 450K methylation BeadChip arrays and NOS2 expression levels were determined by immunohistochemistry.
SNO Signaling Induces DNMT1 Degradation.
To account for the reduced DNMT1 protein expression in response to SNO signaling, DNMT1 transcript expression was measured in MDA-MB-231 and MCF7 cell lines exposed to vehicle or 500 μM DETANO. In both cell lines, SNO signaling resulted in increased DNMT1 mRNA levels, indicating that SNO signaling does not acutely regulate DNMT1 protein at the transcriptional level (SI Appendix, Fig. S3A). However, DNMT1 protein levels did not change in response to SNO signaling in cells treated with the proteasome inhibitor MG-132 (SI Appendix, Fig. S3B), indicating that SNO signaling induces the degradation of DNMT1. To determine if SNO signaling resulted in DNMT1 ubiquitination (Ub), HEK293 cells coexpressing Myc-DNMT1 and HA-ubiquitin were treated with DETANO or vehicle. SNO signaling resulted in increased DNMT1-Ub compared with control cells (SI Appendix, Fig. S3C), indicating that SNO signaling reduces DNMT1 protein expression by inducing its proteasomal degradation.
p38-MAPK Activity Is Required for SNO-Mediated DNMT1 Degradation.
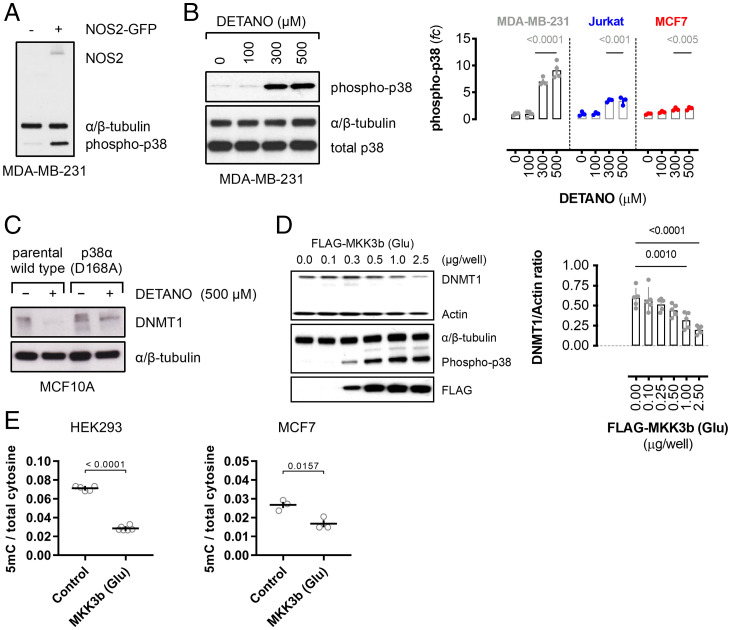
SNO signaling activates multiple intracellular pathways such as Src, Akt, and β-catenin (5, 10). Gene set enrichment analysis of the NOS2-associated signature from patients with ER− breast cancer (14) revealed an enrichment of p38-MAPK–regulated transcription factors present in the NOS2 up-regulated genes using the TRANSFAC database (SI Appendix, Fig. S4A and Table S3), and similar results were obtained from oPOSSUM transcription factor binding-site cluster analysis (SI Appendix, Fig. S4 B and C and Table S4) (44, 45). These data indicate that p38-MAPK activity is a dominant driver of NOS2-associated gene expression in human ER− breast tumors; therefore, the role of p38 in regulating DNMT1 protein levels was examined. NOS2 transgene expression and DETANO, at concentrations that reduce DNMT1 protein levels, resulted in significant p38 phosphorylation (Fig. 3 A and B and SI Appendix, Fig. S4 D and E). SNO signaling did not reduce DNMT1 protein levels in either CRISPR-mutated, kinase-inactive p38 (D168A) cells (Fig. 3C), p38-inhibited or p38 kinase-dead mutant-expressing cells (SI Appendix, Fig. S4 F and G). These data strongly indicate that p38 activity is critical for SNO-signaling-induced DNMT1 degradation.
Fig. 3.
p38-MAPK mediates DNMT1 protein degradation by SNO signaling. (A) Immunoblot of phospho-p38 (Thr-180/Tyr-182) and NOS2 expression in control and NOS2-GFP–transfected MDA-MB-231 cells. (B) Immunoblot of phospho-p38 expression in MDA-MB-231 cells cultured with vehicle or DETANO for 24 h. Graphs show normalized relative phospho-p38 expression as fold change (fc) compared with vehicle controls in human cell lines (±SEM; n = 3–4). Significance was calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (C) Immunoblot showing DNMT1 expression in vehicle- or DETANO-treated, p38-inactive, CRISPR-edited MCF10A cells compared with parental cells. (D) Immunoblot of DNMT1, phospho-p38, and FLAG expression in HEK293 cells transfected with increasing amounts of FLAG-MKK3b (Glu) plasmid. Graph shows mean DNMT1 expression normalized to actin (±SEM; n = 3). Significance calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (E) DNA 5mC content normalized to total cytosine in cells expressing FLAG-MKK3b (Glu). Data shown are mean ratios (±SEM; HEK293, n = 6; MCF7, n = 3). Significance was calculated by an unpaired, two-tailed t test.
The requirement of p38-MAPK activity for SNO-mediated DNMT1 loss prompted us to examine if activation of this kinase is sufficient for DNMT1 loss and passive DNA demethylation. Direct p38 activation by expression of constitutively active MKK3b (Ser218Glu/Thr222Glu) double mutant (Glu) resulted in a plasmid concentration–dependent decrease of DNMT1 protein expression (Fig. 3D and SI Appendix, Fig. S5A). MKK3b (Glu) expression failed to reduce DNMT1 levels in HEK293 cells coexpressing a kinase-dead p38 mutant (SI Appendix, Fig. S5B). Similar to SNO signaling, MKK3b (Glu) expression resulted in increased DNMT1 mRNA expression, DNMT1-Ub, and loss of DNMT1 protein that was blocked by proteasome inhibition (SI Appendix, Fig. S5 C–E). MKK3b (Glu) expression in HEK293 and MCF7 cells resulted in decreased DNA 5mC content, compared with control cells (Fig. 3E). DNA demethylation by MKK3b (Glu) was blocked in HEK293 cells expressing a kinase-dead p38 mutant, indicating that p38 activity is required for both DNMT1 degradation and DNA demethylation (SI Appendix, Fig. S5F). MKK3b (Glu) expression resulted in replication-dependent passive DNA demethylation, as the DNA polymerase inhibitor aphidicolin blocked the ability of MKK3b (Glu) expression to reduce DNA 5mC content (SI Appendix, Fig. S4G). MKK3b (Glu) expression did not alter TET activity or 5hmC levels (SI Appendix, Fig. S5 H and I), and TET2 protein expression was not significantly altered (SI Appendix, Fig. S5J), demonstrating that p38 signaling does not induce active DNA demethylation. Transient p38 activation achieved by CD3/CD28 activation of Jurkat cells or EGF stimulation of breast cancer cells did not alter DNMT1 protein levels (SI Appendix, Fig. S5 K and L). Thus, we conclude that prolonged p38 activation is sufficient to induce passive DNA demethylation.
p38 Signaling Induces Passive DNA Demethylation Via KAT5 Activation.
DNMT1 protein stability is partially controlled by lysine acetylation (38) and p38 activates lysine acetyltransferase 5 (KAT5) to drive cellular senescence and p53-PUMA signaling (46, 47). To determine if KAT5 mediates DNMT1 degradation, cells were transiently transfected with either wild-type (WT) KAT5 (isoform 3) or a (T191A) mutant that cannot be phospho-activated by p38. Coexpression of MKK3b (Glu) with KAT5-WT in HEK293 cells resulted in decreased DNMT1 protein levels; however, MKK3b (Glu) expression did not alter DNMT1 protein levels in cells coexpressing mutant KAT5 (T191A) (SI Appendix, Fig. S6A). MKK3b (Glu) caused an increase in Myc-DNMT1 lysine acetylation in KAT5-WT–expressing cells, but not in KAT5 (T191A)–expressing cells (SI Appendix, Fig. S6B). Furthermore, MKK3b (Glu) expression significantly increased KAT5 activity toward rhDNMT1 in an in vitro acetyltransferase assay but did not increase KAT5 (T191A) activity (SI Appendix, Fig. S6C). Additionally, the KAT5-specific inhibitor TH1834 (48) blocked DNMT1 degradation by SNO signaling or MKK3b (Glu) expression (SI Appendix, Fig. S6 D and E). These data indicate that p38-mediated KAT5 phosphorylation and activation are required for DNMT1 acetylation and subsequent degradation.
SNO/p38 Signaling Induces LINE-1 Retrotransposon Expression and Double-Strand Breaks.
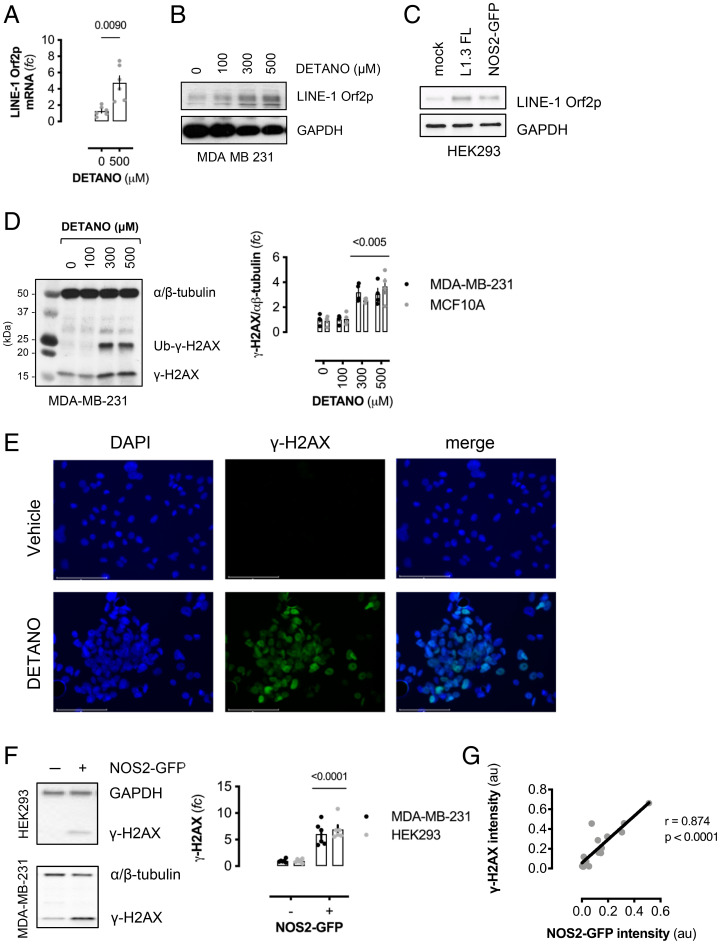
A consequence of global DNA demethylation is the activation of LINE-1 retrotransposons, which are normally suppressed by promoter methylation (29). LINE-1 repression is alleviated during global DNA demethylation events, and LINE-1 expression is commonly observed in human cancer cells (49). SNO signaling induced LINE-1 promoter demethylation similar to 5-azacytidine (SI Appendix, Fig. S7A). Consistent with L1 promoter demethylation, DETANO treatment resulted in LINE-1 Orf2p mRNA expression (Fig. 4A), and SNO signaling caused a concentration threshold effect on LINE-1 orf2p expression, similar to DNMT1 degradation (Fig. 4B and SI Appendix, Fig. S7B). Additionally, NOS2 expression in HEK293 cells resulted in LINE-1 orf2p expression, compared with mock transfected cells, and similar to cells expressing full-length L1.3 element (Fig. 4C). These data indicate that NOS2-mediated SNO signaling induces LINE-1–promoter demethylation and expression.
Fig. 4.
NOS2 and SNO signaling induce LINE-1 expression and DNA damage. (A) Relative LINE-1 Orf2p mRNA in DETANO-treated cells compared with vehicle controls. Data shown are mean ratios normalized to HPRT (±SEM; n = 6). Significance was calculated by two-tailed, unpaired t test with Welch's correction. (B) Immunoblot of LINE-1 Orf2p and GAPDH expression in MDA-MB-231 cells cultured with DETANO for 48 h. (C) Immunoblot of LINE-1 Orf2p and GAPDH expression in HEK293 cells transiently transfected with either full-length human L1.3 element or NOS2-GFP expression plasmids and compared with mock transfected cells. (D) Immunoblot of γ-H2AX expression in DETANO-treated MDA-MB-231 cells. Graph represents mean densitometric analyses of γ-H2AX normalized to an α/β-tubulin from DETANO-treated MDA-MB-231 and MCF10A cell lines (±SEM; n = 3–5). Significance from vehicle controls was calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (E) Immunofluorescent images of γ-H2AX expression in MDA-MB-231 cells treated with DETANO or vehicle for 48 h. γ-H2AX was visualized with Alexa488-conjugated secondary, and cells were counterstained with DAPI. (F) Immunoblot of γ-H2AX expression in mock or NOS-GFP–transfected HEK293 or MDA-MB-231 cells. Graph represents γ-H2AX densitometric analysis as fold change (fc) of control (±SEM; n = 6). Significance was calculated by two-tailed, unpaired t tests. (G) Pearson correlation analysis of proximal NOS2 and γ-H2AX expression in NOS2-GFP–transfected HEK293 cells. Regions of interest (ROI) from immunofluorescent images (SI Appendix, Fig. S7E) were analyzed for green and red channel intensities, normalized to mock-transfected ROI. Graph shows ROI signal intensities and linear correlation with calculated Pearson coefficient and two-tailed P value inset.
L1 expression results in DNA double-strand breaks (DSBs) (50); therefore, the effect of SNO signaling on DSBs was examined by γ-H2AX expression. Similar to DNMT1 degradation and L1 Orf2p expression, SNO-signaling levels of DETANO resulted in significantly increased γ-H2AX expression (Fig. 4D and SI Appendix, Fig. S7C) and punctate nuclear staining (Fig. 4E). NOS2 expression resulted in significant γ-H2AX expression (Fig. 4 F and G) and was similar to cells expressing full-length L1.3 element (SI Appendix, Fig. S7D). Furthermore, DNA damage was strongly associated with proximal NOS2-GFP expression. Immunofluorescent imaging shows NOS2 and γ-H2AX expression had a positive Pearson correlation coefficient (r = 0.874; P < 0.0001), suggesting that NOS2 expression is causally linked to γ-H2AX expression (Fig. 4G and SI Appendix, Fig. S7E).
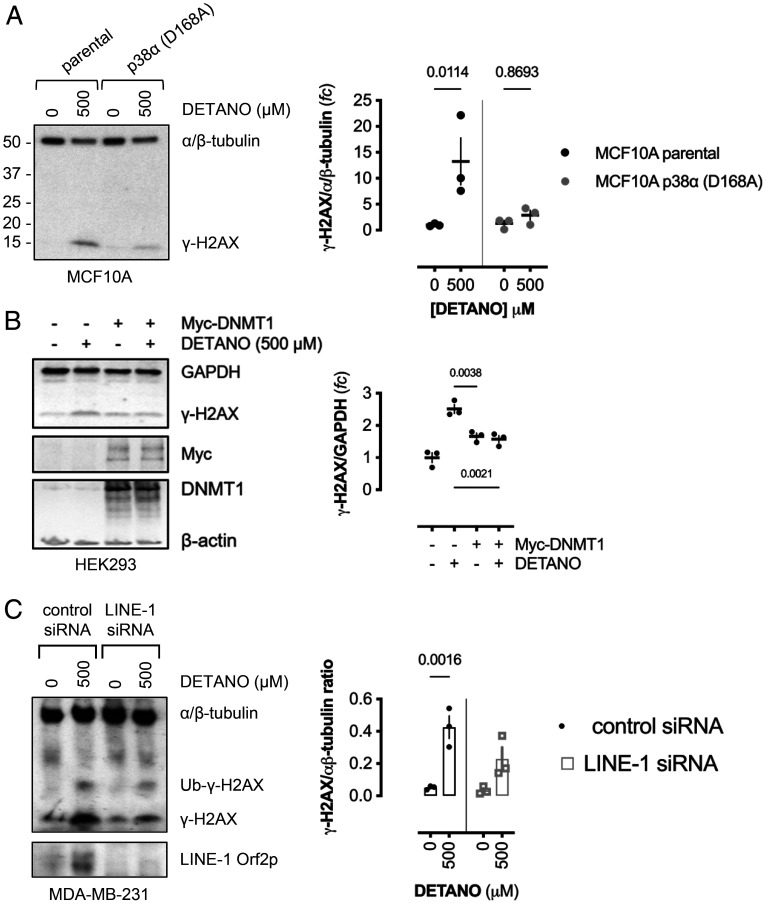
To determine if p38 signaling is required for SNO-mediated DNA damage, CRISPR-mutated kinase-dead p38 (D168A)–expressing MCF10A cells were treated with DETANO or vehicle and compared with parental MCF10A breast epithelial cells. SNO signaling failed to induce γ-H2AX expression in p38 (D168A) mutant cells, in contrast to significant γ-H2AX expression in parental cells (Fig. 5A).
Fig. 5.
NO induces DNA damage via a p38/DNMT1/LINE-1 mechanism. (A) Immunoblot of γ-H2AX expression in DETANO-treated parental or p38α-inactive mutant MCF10A cells compared with vehicle controls. Graph shows normalized mean γ-H2AX expression as fold change (fc) of vehicle controls (±SEM; n = 3). Significance was calculated by two-way ANOVA with Šídák's multiple comparisons test. (B) Immunoblot of γ-H2AX expression in control or Myc-DNMT1 overexpressing HEK293 cells treated with vehicle or DETANO for 24 h. Graph shows normalized mean γ-H2AX densitometric ratios as fold change of control (±SEM; n = 3). Significance was calculated by one-way ANOVA with Dunnett's multiple comparisons test. (C) Immunoblot of γ-H2AX expression in MDA-MB-231 cells transfected with either control or LINE-1 silencing RNA and cultured with vehicle or DETANO for 24 h. Graph shows the mean densitometric analyses of normalized γ-H2AX expression (±SEM; n = 3). Significance was calculated by two-way ANOVA with Tukey's multiple comparisons test. siRNA, small interfering RNA.
DNMT1 overexpression blocked SNO-mediated demethylation. To examine if DNMT1 overexpression rescues NO-induced DNA damage, Myc-tagged DNMT1-overexpressing cells were treated with DETANO and compared with mock transfected cells. While DETANO induced γ-H2AX expression in control cells, compared with vehicle-treated control cells, SNO signaling did not alter γ-H2AX expression in DNMT1-overexpressing cells (Fig. 5B), indicating SNO signaling induced DNA damage via DNMT1 protein degradation. Additionally, SNO-induced γ-H2AX expression was significantly attenuated in transiently LINE-1–silenced cells (Fig. 5C), indicating that SNO signaling and reactive nitrogen species induce DNA cleavage by an epigenetic-retrotransposition mechanism.
Chronic SNO Signaling Transforms Breast Epithelial Cells Via a p38/DNMT1 Mechanism.
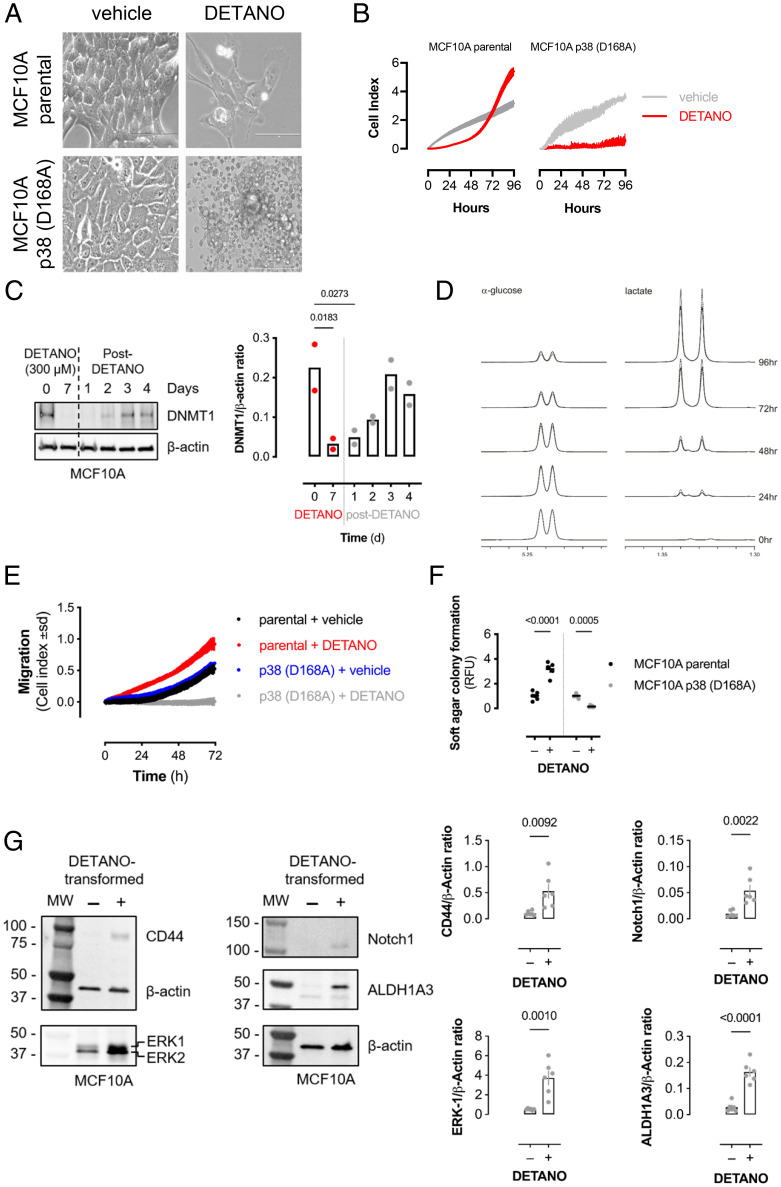
To determine if SNO signaling induced cellular transformation via DNA demethylation, a normal, human breast epithelial cell line (MCF10A) was chronically treated with DETANO or vehicle. Control cells retained a cuboidal, epithelial structure, whereas DETANO-treated cells exhibited an altered cellular morphology similar to a mesenchymal phenotype (Fig. 6A). However, in MCF10A p38 inactive-kinase cells, long-term exposure of DETANO resulted in cellular toxicity, while vehicle-treated mutant cells retained a cuboidal epithelial morphology. The toxicity observed in DETANO-treated p38-inactive cells is similar to the induction of apoptosis by hydrogen peroxide in p38-null cells (51).
Fig. 6.
p38-MAPK is required for epithelial transformation by chronic SNO signaling. (A) Brightfield images of MCF10A parental or p38α (D168A)–mutant MCF10A cells cultured for 7 d with vehicle or DETANO (500 μM). Images were recoded at ×40 magnification. Scale bars: 100 μm. (B) Cellular proliferation as measured by electric impedance (cell index) of MCF10A parental or mutant p38α (D168A) cells cultured with vehicle or DETANO for 7 d prior to 0 h time point. Data shown are mean cell index (±SD; n = 2–3). (C) DNMT1 protein expression from MCF10A cells treated with or recovering from DETANO for the indicated times. Bar graph shows mean DNMT1 expression values (n = 2). (D) Proton NMR spectra of α-glucose and lactate in conditioned media from MCF10A cells chronically treated with DETANO (dashed line) or vehicle (solid line) for 7 d prior to 0 h time point. Media were collected at indicated times after culture in fresh media without DETANO. (E) Cellular migration as measured by electric impedance (cell index) of MCF10A parental or mutant p38α (D168A) cells cultured with vehicle or DETANO for 7 d prior to 0 h time point. Data shown are mean cell index (±SD; n = 3–4). (F) Soft-agar colony growth ability of cells described in A. Colony formation was quantified by fluorescent dye incorporation. Graph shows mean relative fluorescence unit (RFU) values normalized to control (±SEM; n = 6). Significance was calculated by two-way ANOVA with Šídák’s multiple comparisons test. (G) Immunoblots showing CD44, ERK, Notch1, and ALDH1A3 protein expression in control MCF10A or DETANO-transformed cells. Graphs show mean relative protein expression (±SEM; n = 6) and significance was calculated by unpaired, two-way t test.
After chronic exposure to DETANO, cellular proliferation was measured in real time via electrical impedance, which showed that DETANO-treated cells initially showed reduced proliferation compared with control cells (Fig. 6B). However, after approximately 60 h of recovery, DETANO-pretreated cells showed rapid proliferation compared with control cells, whereas inactive p38-mutated cells exposed to DETANO showed minimal growth over 4 d in culture. DNMT1 expression remained low after NO stress and returned to normal levels 3 d after removing DETANO from the culture medium, and this correlated with increased growth rates at 72 and 96 h after NO treatment (Fig. 6C). In parallel experiments, glucose and lactate levels were measured after 96 h by 1H-NMR (52). DETANO-treated cells showed increased glucose consumption and elevated lactate production compared with control cells (Fig. 6D and SI Appendix, Fig. S7F). Long-term DETANO- and vehicle-treated cells were cultured for 72 h before assessing cellular migration. Consistent with a mesenchymal phenotype and cellular transformation, WT cells exposed to long-term SNO signaling exhibited increased chemotactic migration compared with vehicle-treated controls. In contrast, CRISPR-mutated inactive p38 kinase–expressing breast epithelial cells exposed to long-term SNO signaling showed decreased migration compared with vehicle-treated p38 inactive cells (Fig. 6E). To measure cellular transformation, vehicle- or DETANO-pretreated MCF10A cells were grown in a soft agar matrix. Consistent with altered metabolism and growth kinetics, DETANO pretreatment resulted in increased colony formation compared with vehicle-treated cells, whereas SNO signaling decreased the colony-forming ability of p38 inactive cells (Fig. 6F). NO resulted in the hypomethylation of cancer stem cell markers (e.g., Notch, ALDH1) and progrowth genes (e.g., ERK1) in a breast cancer cell line (Dataset S1). Thus, the expression of breast cancer stem cell markers was examined in NO-transformed breast epithelial cells. NO-transformed breast epithelial cells had significantly higher expression of breast cancer stem cell markers (CD44, Notch1, and ALDH1A3) and ERK1, a finding that correlates with increased growth rates and metabolic activity (Fig. 6G). Therefore, these results indicate that sustained SNO signaling resulted in cellular transformation via a p38-dependent, epigenetic mechanism.
Discussion
NO has numerous cellular signaling effects, some of which are due to protein SNO bond formation. Here, we tested the hypothesis that SNO signaling would inhibit DNMT1 activity to reduce DNA methylation levels. While S-nitrosation of the active-site cysteine residue resulted in inhibited DNMT1 activity, the cellular response of coordinated DNMT1 degradation was unexpected. Our results are consistent with recent reports that cellular oxidants inhibit S-adenosylmethionine formation to limit DNA methylation (53). Therefore, SNO signaling limits DNMT1 cofactor availability while simultaneously degrading and inhibiting this enzyme required for copying the methylome to the nascent DNA strand during replication.
NO is a potent driver of gene expression and can alter the transcriptome by modulating histone methylation (54). Here, we extend the known epigenetic functions of NO to include the regulation of DNA methylation. However, the effect of NO on DNA methylation has a detrimental outcome, as LINE-1 demethylation and expression induced by NO resulted in DNA damage, consistent with known retrotransposon biology (50). Interestingly, NOS2 appears to be a unique stimulator of LINE-1 expression, as high NOS2 levels in tumor cells are correlated with both decreased DNA methylation (Fig. 2) and the upregulation of transcription factor families required for LINE-1 expression (nkx-2.5, Sry, Runx) (SI Appendix, Tables S3 and S4). Furthermore, these transcription-factor families are p38 regulated, indicating that the NOS2/p38 signaling axis described here drives LINE-1 expression. Additionally, NO signaling activates p53 (55) and RUNX3 transcriptional activity (56), which result in LINE-1 expression (57, 58). SNO signaling also reduces the expression of BRCA1 (59), a tumor suppressor protein that represses LINE-1 retrotransposition (60). Therefore, NOS2 expression in humans, which is normally tightly regulated, may be a significant driver of LINE-1 retrotransposition and may partially explain its mutagenic and tumorigenic properties.
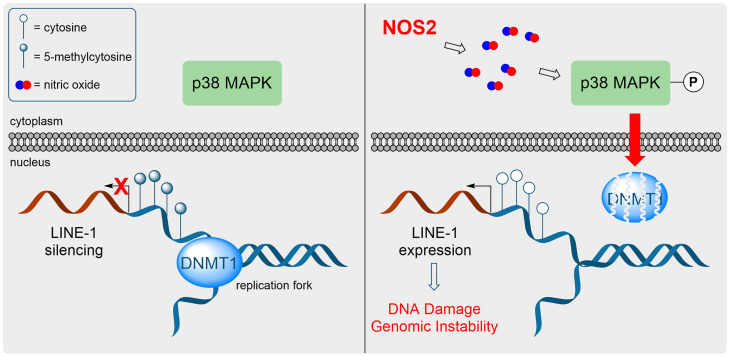
NO forms potent oxidants capable of DNA damage and mutagenesis (61, 62). However, NO-derived oxidants, such as NO2, are rapidly quenched by abundant cytosolic thiols such as glutathione. Furthermore, S-nitrosation of cellular thiols is generally resolved to disulfides (9). Therefore, it is not clear how NO-derived oxidants can penetrate the reductive capacity of the cytosol to oxidize DNA. The p38-dependent DNMT1-degradation mechanism is an elegant way to chemically bypass the cellular reducing capacity of cytosolic glutathione. Furthermore, we conclude that DNMT1-SNO formation is not required as direct p38 activation via mutant MKK3b also resulted in DNMT1 degradation (Fig. 3 D and E); therefore, the loss of DNMT1 is an indirect effect caused by NOS2 and SNO signaling. In the mechanism proposed here, the nitrosative signal is sensed by p38 and/or upstream activator(s) and is transduced via a series of PTMs (i.e., KAT5 phosphorylation and DNMT1 acetylation, Ub) to degrade DNMT1 protein, allowing for passive DNA demethylation and the expression of LINE-1 retrotransposons that causes genomic alterations and DNA DSBs (Fig. 7). In addition to SNO signaling, reactive oxygen species, nickel, and IL-6 also result in LINE-1 expression (26, 63). Therefore, oxidants such as NO may drive DNA damage and genomic instability via an epigenetic and molecular mechanism instead of direct chemical oxidation of DNA bases.
Fig. 7.
NOS2 promotes passive DNA demethylation and LINE-1 expression. (Left) Under normal physiological conditions, DNMT1 methylates the nascent DNA strand during replication and LINE-1 retrotransposons are silenced by promoter CpG methylation. (Right) NOS2 expression results in sustained p38-MAPK activation, which leads to DNMT1 protein degradation. Replication in the absence of DNMT1 activity results in passive DNA demethylation, LINE-1 expression, and genomic instability.
Our data indicate that NOS2 expression and associated SNO signaling promote aggressive tumor phenotypes by driving DNA hypomethylation and genomic instability via LINE-1 retrotransposon expression. This is consistent with the link between tumors with high NOS2 expression and epigenomic instability in breast cancer with higher tumor grades and poor outcomes in patients (14, 64). While NOS2 inhibition has therapeutic efficacy in breast cancer models (19), our mechanism points to multiple targets for cancer chemoprevention or intervention, as p38, KAT5, and protein acetylation can be pharmacologically manipulated. Studies will be required to elucidate if these targets limit tumorigenesis.
The loss of DNMT1 protein expression and passive DNA demethylation caused by SNO signaling was dependent on p38-MAPK activity. In addition to controlling cell cycle and differentiation, p38 has both tumor suppressor and oncogene functions. Interestingly, cellular differentiation and tumorigenesis both require DNA demethylation events, and the duration of p38 activation appears to modulate its demethylation function, as transient p38 activation did not alter DNMT1 levels. This is consistent with differential signaling effects depending on whether cells encounter chronic stress or acute mitogenic signals (65), suggesting that p38 signaling can be distinguished by source and duration of stimulation. Given that cellular stress and chronic p38 signaling is induced by carcinogens (66), it is plausible that p38-mediated DNA demethylation may be a common event in tumorigenesis, as we have linked the chronic cellular stress response to epigenetic changes associated with tumor initiation.
A potential limitation of this study is the physiological relevance of NO levels from NOS2 transgene expression or a NO-donor compound. While both scenarios resulted in p38-MAPK activation and DNMT1 protein degradation, it is difficult to compare the nitrosative environments of each scenario. NOS2 activity produces high, local, intracellular NO concentrations, while DETANO releases NO into the bulk media, where it autoxidizes to form nitrosating species. However, IFN-γ–induced NOS2 expression also resulted in decreased DNMT1 protein expression, indicating that this effect is physiologically relevant. This is further supported by DNA methylation levels being significantly decreased in breast tumors with high NOS2 expression compared with those with low NOS2 expression. While the use of NOS2 or DETANO may have disadvantages, the results demonstrate that DNMT1 is a target of nitrosative signaling in human cells.
To conclude, our data reveal an epigenetic function for NOS2 and cellular nitrosative signaling. NOS2 activity and SNO signaling induced passive DNA demethylation by chronically activating a p38/KAT5-signaling pathway to degrade DNMT1 protein. Consistent with genome-wide DNA hypomethylation, NOS2 and SNO signaling also resulted in LINE-1 retrotransposon expression. We conclude that NOS2 promotes cellular transformation and an aggressive cancer cell phenotype by inducing genomic hypomethylation, retrotransposition, and instability.
Materials and Methods
Cell Lines and Cell Culture.
MDA-MB-231, MCF7, Jurkat E6.1 (European Collection of Authenticated Cell Cultures), HEK293, HL-60, and RAMOS (ATCC) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), penicillin (50 units/mL), and streptomycin (50 µg/mL) in a humidified incubator at 37 °C with 5% CO2. MCF10A breast epithelial cell line (ATCC) was cultured in mammary epithelial cell growth medium (MEGM) with MEGM SingleQuot Supplements (hydrocortisone, rhEGF, insulin, and bovine pituitary extract) as instructed (Lonza) and 100 ng/mL cholera toxin (Sigma). Cell lines were passaged by 0.05% trypsin and 0.5 mM EDTA (Gibco) and quenched with serum-containing medium. MCF10A cells were trypsinized and quenched with Defined Trypsin Inhibitor (Thermo Fisher Scientific). Cell viability was measured by trypan blue exclusion assay. Briefly, 10 µL of cell suspension was mixed with 10 µL of 4% trypan blue solution and total and trypan-stained cells were quantified by Countess II Automated Cell Counter (Thermo Fisher Scientific).
Chemicals and Reagents.
DEANO and DETANO were obtained from Sigma and stock solutions were prepared in 10 mM NaOH. NO-donor purity was confirmed by measuring ultraviolet absorbance, ensuring a single symmetrical peak at 250 nm without a shoulder or nitrite peak at approximately 225 nm. Concentration was calculated using Beer’s law using 250-nm absorbance values (ε250 = 8000 M-1•cm-1). DETANO treatment of cells was done in RPMI + 10% FBS. We prepared stock solutions of 1400W (Sigma), 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one (Cayman Chemical), 5-azacytidine (Cambridge Bioscience), MG132 (Fisher), and SB203580 (Cambridge Bioscience) in DMSO. We dissolved 8-(4-chlorophenylthio)-guanosine 3′,5′-cyclic monophosphate sodium salt (8-pCPT-cGMP) (Sigma) and TH1834 (Axon Medchem) in water. Aphidicolin, Ready Made Solution from Nigrospora sphaerica, was obtained from Sigma. Recombinant human IFN-γ was obtained from R&D Systems and used as instructed; final IFN-γ concentrations were 200 ng/mL
Plasmids and Transfections.
The following constructs were obtained from Addgene: pcDNA3/Myc-DNMT1 (plasmid 36939, from A. Riggs), pRK5-HA-Ubiquitin-WT (plasmid 17608, from T. Dawson), pCDNA3 Flag MKK3b(Glu) (plasmid 50449, from R. Davis), and pcDNA3 Flag HA (plasmid 10792, from W. Sellers). We obtained pCMV6-AC-GFP-NOS2 (NOS2-GFP) from OriGene (RG211819), and pcDNA3.1-HA-KAT5 (isoform 3) and pcDNA3.1-HA-KAT5 (isoform 3; T191A) were synthesized by GenScript. Cells (0.25 × 106 in 3 mL of medium) were seeded into 6-well plates and cultured overnight. Plasmids were diluted in Opti-MEM medium (125 µL/well) and supplemented with P3000 (5 µL/well) and added to Lipofectamine 3000 (5 µL/well) diluted in Opti-MEM medium (125 µL/well). DNA complexes were incubated for 15 min at room temperature and added dropwise to cells. Mock-transfected cells were treated with Lipofectamine and P3000 diluted in Opti-MEM as described. Cells were then incubated for the times indicated before analysis.
DNMT Activity and S-Nitrosation.
Full-length human recombinant DNMT1 (Reaction Biology Corp.) was desalted using a Zeba micro spin column and was immediately diluted to 1.5 µM in 20 mM Tris HCl at pH 7.5, aliquoted into microcentrifuge tubes, and incubated with 1,1-diethyl-2-hydroxy-2-nitroso-hydrazine sodium (DEANO) or vehicle for 30 min at 37 °C in a 100-µL reaction. DNMT1 activity was then measured using EpiQuik DNMT1 Activity/Inhibitor Screening Assay Core Kit (EpiGentek) as instructed using a final DNMT1 concentration of 100 nM. Formation of SNO bond was determined by a biotin-switch method, as previously described (9). HL-60 nuclear fractions were prepared using Subcellular Protein Fractionation Kit for Cultured Cells (Thermo) as instructed, and protein concentrations were measured using bicinchoninic acid (BCA) assay. Nuclear fractions were then incubated with DEANO for 30 min at 37 °C, and total nuclear DNMT activity was measured using 1 µg of nuclear protein, as described.
Immunoblotting.
Cells were lysed with 2× RIPA buffer containing complete protease inhibitor mixture and phosphatase inhibitor mixture 2 (Sigma). Lysates were denatured with 4× SDS sample buffer supplemented with 5% (vol/vol) 2-mercaptoethanol and incubated at 85 °C for 5 min. Lysates were then separated on 4 to 20% or 4 to 15% sodium dodecyl-sulfate polyacrylamide gel electrophoresis gels and transferred onto polyvinylidene fluoride or polyvinylidene difluoride membranes using the Bio-Rad Trans-Blot Turbo system. Membranes were blocked with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBS-Tween) and incubated overnight at 4 °C with primary antibodies in 2% BSA in PBS-Tween. Primary antibodies and dilutions used are listed in SI Appendix, Table S5. After washing, membranes were incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies in 2% BSA PBS-Tween for 1 h at room temperature. After washing with PBS-Tween, membranes were exposed to Pierce ECL reagent and imaged with Amersham Hyperfilm ECL or iBright FL1500. Immunoblots were quantified with Gel-Pro Analyzer v3.1 software.
Flow Cytometry Analysis.
Cells (1.0 × 106 in 15 mL of medium) were seeded into T75 flasks and cultured overnight before vehicle, DETANO (500 µM), or 5-azacytidine (1 µM) was added and cultured for 48 h. Cells were then trypsinizsed and quenched in defined trypsin inhibitor and washed in PBS. Cells were then fixed in 4% formaldehyde for 15 min at room temperature and washed in PBS. Cells were permeablized with 90% cold methanol for 30 min and washed in PBS, followed by resuspension in PBS + 0.5% BSA. IgG-PE or anti–DNMT1-PE (3 µg/100 µL) was added to cells and incubated for 1 h in the dark at room temperature. Cells were then washed in PBS + 0.5% BSA before analysis on a BD FACSCanto II analyzer. Data were then further analyzed using FlowJo software.
qPCR Analysis.
RNA was isolated from cells using RNeasy Mini Kit (Qiagen) and quantified on a P200 Picodrop spectrophotometer. Complementary DNA (cDNA) was prepared using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) as instructed. qPCR assays for DNMT1 and HRPT utilized the Universal ProbeLibrary System (Roche), and primers were designed using the Universal ProbeLibrary (UPL) Assay Design Center. Human LINE-1 was measured using iTaq Universal SYBR Green Supermix (Bio-Rad). UPL probes and primers used are listed in SI Appendix, Table S6. qPCR reactions were carried out using 5.1 ng of cDNA, 10 µM left and right primers, 10 µM UPL probe, and TaqMan Universal Master Mix II, with Uracil-N-glycosylase in a 20-µL reaction volume. qPCR reactions were performed on an Applied Biosystems StepOnePlus Real-Time PCR system. Relative expression normalized to HPRT was calculated using the comparative cycle threshold method.
DNA Methylation.
DNA was isolated from cells using DNeasy Blood & Tissue Kit (Qiagen) and 1 µg of DNA was digested with DNA Degradase Plus (Zymo Research) for 4 h at 37 °C in a 25-µL reaction. After digestion, 0.1% formic acid was added to each sample and stored at −20 °C until liquid chromatography–tandem mass spectrometry analysis. Genomic 5mC, 5hmC, and total cytosine content was measured on an Agilent 1100 LC system interfaced with a Waters Quattro LC triple quadrupole mass spectrometer as previously described (67). Genomic methylation was measured with MethylationEPIC BeadChip, using the manufacturer’s standard protocol (Illumina, Inc.). Genomic DNA was extracted from cells using DNeasy Blood & Tissue Kit and bisulfite converted with EZ-96 DNA Methylation Kit (Zymo Research Corp.). The converted DNA was processed by the Biomedical Research Centre based at Guy’s and St Thomas’ National Health Service Foundation Trust and King’s College London. Genomic methylation data were analyzed using Partek Genomics Suite and only differentially methylated positions with a Δβ = ±0.2 relative to control samples were included for some analyses, as indicated in figure legends.
Human Tumor NOS2 and DNA Methylation Analysis.
DNA methylation data (Illumina 450K BeadChip arrays) were obtained from a publicly available data set (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37754) and corresponding NOS2 immunohistochemistry and patient data were generously provided by Dr. Stefan Ambs (National Cancer Institute, NIH, Bethesda, MD) (68). Mean M-values for each genome were plotted according to NOS2 expression level. Corresponding patient characteristics are shown in SI Appendix Table S3.
TET Activity Assay.
HEK293 cells (0.25 × 106/well in 3 mL of RPMI 1640 + 10% FBS) were seeded into 6-well plates and incubated overnight. Cells were then transiently transfected with FLAG-MKK3b (Glu) expression plasmid or FLAG-HA control plasmid, as described previously. Nuclear fractions were isolated 24 h after transfection and protein concentrations were determined by BCA assay. TET activity was measured using the Epigenase 5mC-Hydroxylase TET Activity/Inhibition Assay Kit as instructed and normalized to the protein assessed.
KAT5 Activity Assay.
HEK293 cells were transiently transfected with HA-KAT5 (WT) or HA-KAT5 (T191A) and FLAG-MKK3b (Glu), and cells were lysed after 24 h with RIPA buffer containing protease and phosphatase inhibitors. Lysates were then centrifuged to remove debris and supernatants were incubated overnight with anti–HA-tag–conjugated magnetic beads (Cell Signaling Technology, 11846) at 4 °C. Beads were then isolated in a magnetic field and washed three times with acetyltransferase activity buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 100 μM dithiothreitol). Recombinant human DNMT1 (10 ng/reaction) and acetyl-CoA (50 µM; Sigma) were added to immuno-captured KAT5 and incubated at 37 °C for 30 min. Sample buffer (4×) was added to terminate reactions, and samples were analyzed by immunoblotting as described.
Immunofluorescence and Brightfield Microscopy.
γ-H2AX expression was visualized by immunofluorescent imaging. Cells were grown on sterile glass coverslips and treated with DETANO or vehicle or transiently transfected with NOS2-GFP plasmid for 48 h. Cells were fixed with PBS containing 4% formaldehyde and permeabilized with PBS containing 0.3% Triton X-100 and blocked with 1% BSA in PBS containing 0.3% Triton X-100 for 1 h at room temperature. Coverslips were then incubated overnight at 4 °C with anti–γ-H2AX (Cell Signaling, #9718; 1:500 dilution) in blocking buffer. Coverslips were then washed with PBS containing 0.3% Triton X-100 and incubated with Alexa488- or Alexa594-conjugated anti-rabbit secondary antibody for 1 h at room temperature, washed, and coverslips attached to glass slides with DAPI-containing mounting medium. Fluorescence and bright field images were recorded on an EVOS M7000 Imaging System (ThermoFisher Scientific).
LINE-1 Promoter Methylation Assay.
LINE-1 methylation was measured using the Global DNA Methylation LINE-1 Kit (Active Motif) as instructed. Briefly, genomic DNA isolated from cells using DNeasy Blood & Tissue Kit (Qiagen) was digested with MseI, annealed to biotinylated LINE-1 promoter oligonucleotides and bound to a streptavidin-coated, 96-well plate. Immobilized LINE-1 promoters were then incubated with 5mC antibody, washed, incubated with an HRP-conjugated secondary antibody, and developed with a colorimetric substrate. Data are reported as the mean of the percentage of 5mC per total cytosines in the LINE-1 consensus promoter.
LINE-1 Silencing.
The effect of LINE-1 silencing on DNA damage was assessed by transfecting MDA-MB-231 cells with scrambled control or endo 453 (5′-UGUUUACCUAAGCAAGCCUGGG-3′) small interfering RNA using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen) as instructed. Medium was removed after 24 h and replaced with medium containing vehicle or DETANO and cultured for 48 h before analysis.
Proliferation and Migration.
Cellular proliferation and migration were assessed by electrical impedance using an Agilent xCELLigence Real-Time Cell Analysis dual-purpose instrument. Proliferation was measured by seeding a glass E-Plate 16 with 2.0 × 104 cells, while migration was measured by seeding a RTCA CIM plate 16 (Agilent) with 3.5 × 104 cells in 200 µL medium, and cell index (electrical impedance) was recorded every 30 min for 96 h at 37 °C in a humidified, 5% CO2 atmosphere. Data were baseline corrected to medium-only–containing wells.
Glucose and Lactate Measurements.
Cellular glucose consumption and lactate production were monitored by 1H-NMR, as previously described (52). Briefly, cells were seeded into 24-well plates and incubated for indicated times. Conditioned medium was passed through 0.22-μm filters and stored at −80C until NMR analysis, and cells were assayed for protein content using the BCA assay with a BSA standard curve. Proton peaks corresponding to glucose and lactate in conditioned medium were measured and normalized to protein content of the well. Control (0 h) conditions were aliquots of fresh medium placed into 24-well plates and assayed as described.
Soft Agar Assay.
Soft-agar colony formation was assessed using the CytoSelect 96-Well Cell Transformation Assay (Cell Biolabs, Inc.). Briefly, 1.0 × 104 cells were seeded between agar layers and incubated for 8 d with MEGM. The agar was solubilized, and cells stained with a fluorescent dye and quantified on a plate reader. Relative fluorescence units were normalized to untreated parental MCF10A cells, and data are reported as fold change relative to this group.
Statistical Analysis.
All statistical analyses were calculated using Prism GraphPad 9 unless otherwise stated. Comparisons between treatment groups and control were determined by one-way ANOVA with Dunnett’s post hoc test. Comparisons between two conditions were determined by two-way unpaired t test, while comparisons between two conditions and two variables were determined by two-way ANOVA with Sidak’s post hoc test. The effect of tumor NOS2 levels on DNA methylation was analyzed by two-tailed Welch’s t test (low NOS2 levels, n = 10; high NOS2 levels, n = 43). Hierarchical clustering was analyzed in RStudio running the pvclust package, an implementation of multiscale bootstrap resampling to calculate an approximately unbiased P value. Pearson correlation between NOS2-GFP and H2AX intensity was calculated with a two-tailed P value. Statistical significance was defined as P < 0.05.
Supplementary Material
Acknowledgments
C.H.S. is supported by British Heart Foundation Project Grant PG/19/33/34385. P.E. is supported by The Barts Charity Cardiovascular Programme Award G00913 and by program grants from the British Heart Foundation and the Medical Research Council. T.R.E. acknowledges support from the National Institute of Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ National Health Service Foundation Trust and King’s College London; the Centre of Excellence in Medical Engineering, funded by the Wellcome Trust and Engineering and Physical Sciences Research Council grant (WT 203148/Z/16/Z); and the British Heart Foundation Centre of Research Excellence grant (RE/18/2/34213). The Centre for Biomolecular Spectroscopy is funded by grants from the Wellcome Trust (202767/Z/16/Z) and British Heart Foundation (IG/16/2/32273). P.L. was supported by Medical Research Council Grant G1100238/1. We thank the BRC Genomics Platform at the NIHR Biomedical Research Centre at Guy’s and St. Thomas’ Hospitals, London. We thank Miguel Branco (Blizzard Institute, Queen Mary University of London) for LINE-1 primers and plasmids and helpful discussions regarding the manuscript. We thank A. Riggs, T. Dawson, R. Davis, and W. Sellers for providing plasmids. We also thank Stefan Ambs (National Cancer Institute, NIH) for providing tumor NOS2 expression annotations and David Wink and Robert Cheng (National Cancer Institute, NIH) for sharing the NOS2 TRANSFAC data set.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200022119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Ignarro L. J., Nitric Oxide: Biology and Pathobiology (Academic Press, 2000). [Google Scholar]
- 2.Akaike T., Maeda H., Nitric oxide and virus infection. Immunology 101, 300–308 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bult H., et al. , Nitric oxide as an inhibitory non-adrenergic non-cholinergic neurotransmitter. Nature 345, 346–347 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Thomas D. D., et al. , The chemical biology of nitric oxide: Implications in cellular signaling. Free Radic. Biol. Med. 45, 18–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas D. D., et al. , Signaling and stress: The redox landscape in NOS2 biology. Free Radic. Biol. Med. 87, 204–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaston B. M., Carver J., Doctor A., Palmer L. A., S-nitrosylation signaling in cell biology. Mol. Interv. 3, 253–263 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Sha Y., Marshall H. E., S-nitrosylation in the regulation of gene transcription. Biochim. Biophys. Acta 1820, 701–711 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marozkina N. V., Gaston B., S-nitrosylation signaling regulates cellular protein interactions. Biochim. Biophys. Acta 1820, 722–729 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolhuter K., et al. , Evidence against stable protein S-nitrosylation as a widespread mechanism of post-translational regulation. Mol. Cell 69, 438–450.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Switzer C. H., et al. , S-nitrosylation of EGFR and Src activates an oncogenic signaling network in human basal-like breast cancer. Mol. Cancer Res. 10, 1203–1215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aranda E., López-Pedrera C., De La Haba-Rodriguez J. R., Rodriguez-Ariza A., Nitric oxide and cancer: The emerging role of S-nitrosylation. Curr. Mol. Med. 12, 50–67 (2012). [DOI] [PubMed] [Google Scholar]
- 12.McGinity C. L., et al. , Nitric oxide modulates metabolic processes in the tumor immune microenvironment. Int. J. Mol. Sci. 22, 7068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas D. D., Wink D. A., NOS2 as an emergent player in progression of cancer. Antioxid. Redox Signal. 26, 963–965 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Glynn S. A., et al. , Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J. Clin. Invest. 120, 3843–3854 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyler C. E., et al. , Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell 146, 53–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J., et al. , Inducible nitric oxide synthase enhances disease aggressiveness in pancreatic cancer. Oncotarget 7, 52993–53004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okayama H., et al. , NOS2 enhances KRAS-induced lung carcinogenesis, inflammation and microRNA-21 expression. Int. J. Cancer 132, 9–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomsen L. L., et al. , Nitric oxide synthase activity in human breast cancer. Br. J. Cancer 72, 41–44 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granados-Principal S., et al. , Inhibition of iNOS as a novel effective targeted therapy against triple-negative breast cancer. Breast Cancer Res. 17, Article 25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dávila-González D., et al. , Pharmacological inhibition of NOS activates ASK1/JNK pathway augmenting docetaxel-mediated apoptosis in triple-negative breast cancer. Clin. Cancer Res. 24, 1152–1162 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Baylin S. B., DNA methylation and gene silencing in cancer. Nat. Rev. Clin. Oncol. 2, S4–S11 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Feinberg A. P., Vogelstein B., Hypomethylation of ras oncogenes in primary human cancers. Biochem. Biophys. Res. Commun. 111, 47–54 (1983). [DOI] [PubMed] [Google Scholar]
- 23.Gama-Sosa M. A., et al. , The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 11, 6883–6894 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R. Z., Pettersson U., Beard C., Jackson-Grusby L., Jaenisch R., DNA hypomethylation leads to elevated mutation rates. Nature 395, 89–93 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Kemp J. R., Longworth M. S., Crossing the LINE toward genomic instability: LINE-1 Retrotransposition in cancer. Front Chem. 3, Article 68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodić N., Burns K. H., Long interspersed element-1 (LINE-1): Passenger or driver in human neoplasms? PLoS Genet. 9, e1003402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson A. S., Power B. E., Molloy P. L., DNA hypomethylation and human diseases. Biochim. Biophys. Acta 1775, 138–162 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Kazazian H. H. Jr., Moran J. V., Mobile DNA in health and disease. N. Engl. J. Med. 377, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazazian H. H. Jr., Goodier J. L., LINE drive. Retrotransposition and genome instability. Cell 110, 277–280 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Miousse I. R., Koturbash I., The Fine LINE: Methylation drawing the cancer landscape. BioMed Res. Int. 2015, 131547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki K., et al. , Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 9, 199–207 (2006). [DOI] [PubMed] [Google Scholar]
- 32.van Veldhoven K., et al. , Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin. Epigenetics 7, Article 67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soares J., et al. , Global DNA hypomethylation in breast carcinoma: Correlation with prognostic factors and tumor progression. Cancer 85, 112–118 (1999). [PubMed] [Google Scholar]
- 34.Bernardino J., et al. , DNA hypomethylation in breast cancer: An independent parameter of tumor progression? Cancer Genet. Cytogenet. 97, 83–89 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Hermann A., Goyal R., Jeltsch A., The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 279, 48350–48359 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Haggerty C., et al. , Dnmt1 has de novo activity targeted to transposable elements. Nat. Struct. Mol. Biol. 28, 594–603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S. C., Zhang Y., Active DNA demethylation: Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Z., et al. , DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 3, Article ra80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott A., Song J., Ewing R., Wang Z., Regulation of protein stability of DNA methyltransferase 1 by post-translational modifications. Acta Biochim. Biophys. Sin. (Shanghai) 46, 199–203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holm T. M., et al. , Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8, 275–285 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Gaudet F., et al. , Induction of tumors in mice by genomic hypomethylation. Science 300, 489–492 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Kanai Y., Ushijima S., Nakanishi Y., Sakamoto M., Hirohashi S., Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett. 192, 75–82 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Ghoshal K., et al. , 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 25, 4727–4741 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Kwon A. T., Arenillas D. J., Worsley Hunt R., Wasserman W. W., oPOSSUM-3: Advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2, 987–1002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matys V., et al. , TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y., Liao R., Li N., Xiang R., Sun P., Phosphorylation of Tip60 by p38α regulates p53-mediated PUMA induction and apoptosis in response to DNA damage. Oncotarget 5, 12555–12572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng H., et al. , A posttranslational modification cascade involving p38, Tip60, and PRAK mediates oncogene-induced senescence. Mol. Cell 50, 699–710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao C., et al. , Rational design and validation of a Tip60 histone acetyltransferase inhibitor. Sci. Rep. 4, Article 5372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bae J. M., et al. , ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int. J. Cancer 131, 1323–1331 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Gasior S. L., Wakeman T. P., Xu B., Deininger P. L., The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutiérrez-Uzquiza Á., Arechederra M., Bragado P., Aguirre-Ghiso J. A., Porras A., p38α mediates cell survival in response to oxidative stress via induction of antioxidant genes: Effect on the p70S6K pathway. J. Biol. Chem. 287, 2632–2642 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hill D. K., et al. , 1H NMR and hyperpolarized 13C NMR assays of pyruvate-lactate: A comparative study. NMR Biomed. 26, 1321–1325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Connor K. M., Das A. B., Winterbourn C. C., Hampton M. B., Inhibition of DNA methylation in proliferating human lymphoma cells by immune cell oxidants. J. Biol. Chem. 295, 7839–7848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hickok J. R., Vasudevan D., Antholine W. E., Thomas D. D., Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. J. Biol. Chem. 288, 16004–16015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofseth L. J., et al. , Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proc. Natl. Acad. Sci. U.S.A. 100, 143–148 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L., et al. , NO• /RUNX3/kynurenine metabolic signaling enhances disease aggressiveness in pancreatic cancer. Int. J. Cancer 146, 3160–3169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang N., Zhang L., Zhang Y., Kazazian H. H. Jr., An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 31, 4929–4940 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris C. R., et al. , p53 responsive elements in human retrotransposons. Oncogene 28, 3857–3865 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yakovlev V. A., Nitric oxide-dependent downregulation of BRCA1 expression promotes genetic instability. Cancer Res. 73, 706–715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mita P., et al. , BRCA1 and S phase DNA repair pathways restrict LINE-1 retrotransposition in human cells. Nat. Struct. Mol. Biol. 27, 179–191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keefer L. K., Wink D. A., DNA damage and nitric oxide. Adv. Exp. Med. Biol. 387, 177–185 (1996). [DOI] [PubMed] [Google Scholar]
- 62.Tamir S., Burney S., Tannenbaum S. R., DNA damage by nitric oxide. Chem. Res. Toxicol. 9, 821–827 (1996). [DOI] [PubMed] [Google Scholar]
- 63.El-Sawy M., et al. , Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J. Mol. Biol. 354, 246–257 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Batra R. N., et al. , DNA methylation landscapes of 1538 breast cancers reveal a replication-linked clock, epigenomic instability and cis-regulation. Nat. Commun. 12, Article 5406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faust D., et al. , Differential p38-dependent signalling in response to cellular stress and mitogenic stimulation in fibroblasts. Cell Commun. Signal. 10, Article 6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kyriakis J. M., Avruch J., Mammalian MAPK signal transduction pathways activated by stress and inflammation: A 10-year update. Physiol. Rev. 92, 689–737 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Burr S., et al. , Oxygen gradients can determine epigenetic asymmetry and cellular differentiation via differential regulation of Tet activity in embryonic stem cells. Nucleic Acids Res. 46, 1210–1226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terunuma A., et al. , MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J. Clin. Invest. 124, 398–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.