Abstract
The nymphalid butterfly genus Junonia has remarkable dispersal abilities. Occurring on every continent except Europe and Antarctica, Junonia are often among the only butterflies on remote oceanic islands. The biogeography of Junonia has been controversial, plagued by taxonomic disputes, small phylogenetic datasets, incomplete taxon sampling, and shared interspecific mitochondrial haplotypes. Junonia originated in Africa but its route into the New World remains unknown. Presented here is, to our knowledge, the most comprehensive Junonia phylogeny to date, using full mitogenomes and nuclear ribosomal RNA repeats from 40 of 47 described species. Junonia is monophyletic and the genus Salamis is its probable sister clade. Genetic exchange between Indo-Pacific Junonia villida and New World Junonia vestina is evident, suggesting a trans-Pacific route into the New World. However, in both phylogenies, the sister clades to most New World Junonia contain both African and Asian species. Multiple trans-Atlantic or trans-Pacificinvasions could have contributed to New World diversification. Hybridization and lateral transfer of mitogenomes, already well-documented in New World Junonia, also occurs in at least two Old World lineages (Junonia orithya/Junonia hierta and Junonia iphita/Junonia hedonia). Variation associated with reticulate evolution creates challenges for phylogenetic reconstruction, but also may have contributed to patterns of speciation and diversification in this genus.
Keywords: Junonia, mitogenomics, Junoniini, phylogenomics, reticulate evolution, evolutionary radiation
1. Introduction
The balance between immigration, extinction and diversification determines species richness in geographical localities [1]. Most immigrants fail to establish populations, and the majority that colonize successfully undergo little or no diversification [2]. Only rare immigrant lineages show high diversification rates and adaptive radiation [2]. Understanding factors that make immigrant lineages successful in new habitats is important to the disciplines of biogeography, invasion biology, evolution and conservation. The butterfly genus Junonia has extraordinary abilities for dispersal and diversification. Junonia originated 15–27 million years ago (Ma) [3] and are often among the few butterfly species present on remote oceanic islands [4,5], suggesting that they can survive crossing thousands of kilometers of open water to colonize and diversify in new habitats. Some Old World Junonia (28 extant species) appear to have crossed an ocean basin 2–4 Ma to establish a radiation of 18 New World species [3]. This unparalleled ability to disperse may have fomented diversification in Junonia, but the lack of effective geographical barriers to gene flow also may have stymied previous attempts to resolve its phylogenetic and biogeographic history.
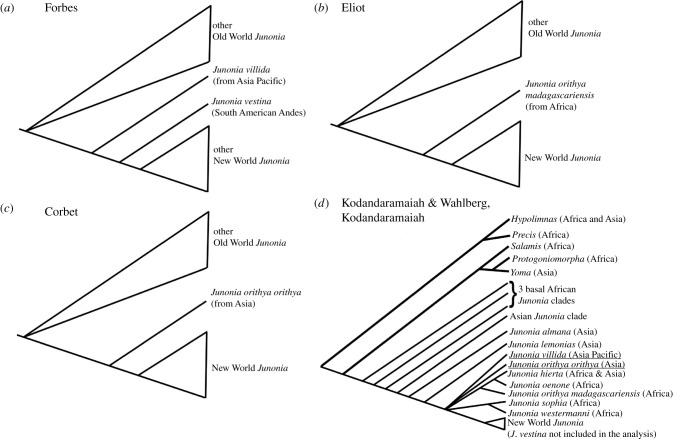
Junonia has a convoluted taxonomic history, especially for New World species. This is attributable to conflation of the generic names Precis (restricted to a related African clade) and Junonia (distributed nearly worldwide) by some authors, misidentifications owing to intraspecific seasonal and geographical variation, loss or absence of type specimens, and the failure of many authors to reference taxonomic authorities used for specimen identification [6–8]. Consequently, creating a robust phylogeny for the genus Junonia has been difficult. Several morphology-based phylogenetic hypotheses are mutually contradictory (figure 1a–c; [11–15]). They also differ from molecular phylogenies based on small mitochondrial and nuclear sequence datasets (COI, wingless and EF1-alpha) (figure 1d; [3,9,16–19]). Finally, New World mitochondrial DNA haplotypes are often shared among all sympatric species in any given locality, resulting in identical heterospecific mitochondrial genome sequences [6,17–20].
Figure 1.
Hypotheses for the phylogeny of Junonia with proposed sister groups to the New World Junonia: (a) Junonia villida by Forbes [14,15], (b) African Junonia orithya madagascariensis by Eliot [12,13] and (c) Asian Junonia orithya orithya by Corbet [11] based on morphology; (d) presents the molecular phylogeny of Kodandaramaiah & Wahlberg [3] and Kodandaramaiah [9] as interpreted by McCullagh [10].
Morphology-based taxonomic assignments have become easier owing to recent clarification of New World species taxonomy [6,7,17,20–23], but remain challenging for reasons described above, and owing to poorly described, undescribed and cryptic Junonia species [20,23]. In general, Junonia phylogenetic studies have either contained very few (≤3) New World species [9,24,25], or (≤2) Old World species [19,23]. Further, some phylogenetic studies of New World taxa have included GenBank sequences labelled as either Junonia evarete or Junonia genoveva, but in the absence of museum vouchers it is unclear if the source-specimens were correctly identified or if they belong to one of six other potential Junonia species [6,16–19]. The use of DNA barcoding [3,17–19] and later, full mitochondrial genome sequencing [5,26–28], confirmed that Junonia is a monophyletic clade. Although still missing species, these studies improved phylogenetic resolution over earlier studies.
The invasion of the New World by Junonia has been the subject of decades of speculation. Based on molecular phylogenetic and biogeographic analyses, the consensus is that Junonia originated in Africa [3,19], but which lineages invaded the New World remains unanswered. Four taxa have been suggested as sister to the New World Junonia: Junonia villida (Indo-Pacific) [15], Junonia orithya madagascariensis (Africa) [12,13], Junonia orithya orithya (Asia) [11] and Junonia lemonias (Asia) [5,10]. Also unknown is whether the occupation of the New World by Junonia was a unique event [3,8,12–15,19] or caused by multiple invasions involving one or more Old World species [10,18]. Recent full mitochondrial genome sequence data suggests a trans-Pacific route and that the same lineage (J. villida) crossed the Pacific more than once to reach the New World [5,10].
Individual Old World Junonia species have been considered to be monophyletic based on morphology and molecular characters [3,9,24,29]. By contrast, New World Junonia species do not form monophyletic clades based on most molecular studies [5,6,10,17–20,23,26]. Instead, all New World Junonia species tend to share mitochondrial haplotypes and sympatric populations of Junonia species typically show the same mitochondrial haplotype group frequencies [18,30,31]. Many New World Junonia species have the capacity to interbreed, so hybridization and mitochondrial introgression events are possible [20,23,32,33]. Consequently, mitochondrial gene-based phylogenies are not useful for New World species delimitation [3,5,6,9,17–20,23], but if species determination is first done using other characteristics, the study of mitochondrial haplotype distributions permits characterization of biogeographic patterns of genetic variation [18,20,30].
(a) . DNA barcodes, short fragment nuclear DNA and haplotype groups
Three primary mitochondrial haplotype groups are shared among New World Junonia species [3,6,9,16–20,23]. This pattern was first discussed by Pfeiler et al. [19] based on the 658 base pair cytochrome oxidase I DNA barcoding fragment used to delimit many animal species [34,35]. Pfeiler et al. [19] suggested that shared mitochondrial haplotypes may be owing to the recent invasion of Junonia into the New World (approx. 2–4 Ma) [3], such that lineage sorting is incomplete and hybridization between species is ongoing. Haplotype group A is common in South American and Caribbean Junonia, while haplotype group B is common in North and Central America, and these two monophyletic groups diverged approximately 2.2 Ma [19]. Pfeiler et al. [19] found that Junonia vestina, a high-elevation South American species, had a distinct sequence (called A1) within haplotype group A and considered all other species to carry subgroup A2. Based on outgroup selection, Pfeiler et al. [19] speculated that the sister taxon to New World Junonia may have been an African lineage related to J. orithya or Junonia westermanni, although this was based on very limited sampling of Old World species.
Later, Gemmell & Marcus [18] mapped the New World distribution patterns of haplotype groups and confirmed that Californian Junonia grisea specimens carried a distinct variant (BCA) of haplotype group B. Haplotype BCA also occurs at a low frequency in four other American southwest Junonia species [20]. Gemmell & Marcus [18] suggested that mitochondrial DNA sequences from Indo-Pacific J. villida are more similar to the New World Junonia than sequences from J. orithya, consistent with some previous hypotheses based on morphology and geographical distributions [4,14,36]. They proposed two hypotheses for the origin of haplotype B: either it evolved from haplotype group A mitochondria within the New World, or that two separate New World invasions carried haplotype groups A and B, followed by hybridization between species descended from the two invasion events [18].
(b) . The use of full mitochondrial DNA genomes
Mitochondrial genome (mitogenome) sequences are a rich source of genetic variation for improved taxonomic resolution in phylogenetic studies [37–41]. The first two full approximately 15.2 kb Junonia mitochondrial genome sequences were reported from Old World species [40,41]. In 2016, 14 New World Junonia mitogenomes and four additional Old World mitogenomes were used to reconstruct patterns of diversification [10,42]. Subfamily Nymphalinae and genus Junonia formed monophyletic groups, but like previous findings using COI sequences, New World Junonia showed a lack of monophyly, consistent with earlier proposals of multiple Junonia invasions of the New World [10,18]. Separate invasions by different Old World species may have created each of the New World haplotype groups [5,10]. The haplotype groups A and B in Junonia were estimated to have diverged 2.31 ± 0.42 Ma, consistent with previous molecular clock estimates [3,19]. Divergence times for all other New World haplotypes were estimated: divergence of A1 and A2 1.52 ± 0.31 Ma and the divergence of B1 from the rest of haplotype B 1.19 ± 0.29 Ma [10]. Peters & Marcus [5] recognized the distinctiveness of New World haplotype group C in high-elevation J. vestina, (more closely related to J. lemonias from Asia than it is to other New World haplotypes) and was estimated to have diverged 1.16 ± 0.32 Ma [5]. Consistent with previous studies, haplotypes A and B each formed monophyletic groups [10,16–19], but the placement of haplotype group C makes the mitochondria of the New World Junonia paraphyletic [5].
Recently, Cong et al. [23] analysed next-generation sequencing libraries from 11 New World Junonia species to describe two new species (Junonia pacoma; Pacific mangrove buckeye and Junonia stemosa; South Texas dark buckeye). Fixed diagnostic characters (morphological or molecular) that allow for the consistent separation of J. stemosa from the morphologically nearly identical Junonia nigrosuffusa could not be identified, so we will treat this form as a subspecies J. nigrosuffusa stemosa nov. stat. [23]. To date, the largest mitogenome-based Junonia phylogeny consists of 28 mitogenomes (15 Junonia species (eight New World, seven Old World), and one from each of the five other Junoniini genera) [26]. This analysis concluded that Junonia was monophyletic, haplotype groups A and B form monophyletic clades, but the New World Junonia are not monophyletic. The most likely sister clade to genus Junonia (though with weak bootstrap support) contains both African Protogoniomorpha and Asian Yoma, all consistent with a recent analysis of mitochondrial DNA barcodes from Old World Junonia species [25].
Many mitogenomes have now been published for Junonia (18 of 47 species) and other genera from tribe Junoniini (one species each from the five other genera) through GenBank (Dataverse electronic supplementary material, table S1). Here, we build upon this pre-existing knowledge by assembling additional full Junonia mitochondrial genomes (22 new species: 40 of the 47 described species), tribe Junoniini (16 additional species across five genera) and additional outgroup species into a phylogenetic analysis. To complement the mitogenomes, we conducted a phylogenetic analysis based on the nuclear ribosomal RNA (rRNA) repeat sequence (an 8–10 kb sequence containing three rRNA repeat subunits (2.8S, 18S and 28S), two internal transcribed spacers (ITS1 and ITS2), and 5′ and 3′ non-transcribed spacers) [43] from the same group of samples. Creating phylogenies using both types of sequences will aid in reconstructing patterns of reticulate evolution and describing biogeographic distributions for a rapidly diversifying clade.
2. Material and methods
(a) . Specimen collection, preparation and sequence generation
Ninety-seven specimens were analysed, consisting of 64 Junonia (40 of 47 described species), 21 additional specimens from tribe Junoniini and 11 outgroup species within subfamily Nymphalinae (Dataverse electronic supplementary material, table S1). We generated most data ourselves, and supplemented with additional sequences from GenBank. Specimens were identified based on morphology. DNA was extracted from a single leg per specimen using a Qiagen DNEasy Blood and Tissue Kit (Qiagen, Düsseldorf, Germany) either manually as previously described [18] or using the animal tissue DNA program on a Qiagen QiAcube. Samples were stored at −20°C before sequencing.
Sequence data were obtained using Ion Torrent (ThermoFisher Scientific, Waltham, Massachusetts, USA), Illumina MiSeq (San Diego, California, USA) or Illumina NovaSeq6000 sequencing (mean size of sequence libraries approximately 3.42 Gigabase pairs) [26]. Sequences were assembled and annotated to previously published reference sequences with Geneious 10.2.6 (Dataverse electronic supplementary material, table S1).
(b) . Mitogenome phylogeny
Phylogenetic reconstruction employed 97 mitogenome sequences from 64 Junonia specimens from 40 species, 21 specimens from other genera in tribe Junoniini and 12 outgroup specimens within subfamily Nymphalinae (Dataverse electronic supplementary material, table S1). Mitogenome sequences (Dataverse electronic supplementary material, file S1) were aligned in ClustalX 2.1 [44,45] and analysed using Bayesian inference with the GTR + I + G model (model selected by jModeltest 2.1.1 [46]) in MrBayes version 3.2.7 [47,48] for 10 million Markov chain Monte Carlo iterations, sampling every 1000 generations, with the first 25% of iterations discarded as burn-in. The trees produced were rendered in FigTree version 1.4.3 [49] and illustrated using Canvas X Draw.
(c) . Nuclear ribosomal RNA and internal transcribed spacer repeat phylogeny
Phylogenetic reconstruction employed 90 nuclear rRNA repeat sequences from 62 Junonia specimens, 19 specimens from other genera in tribe Junoniini and nine outgroup specimens from subfamily Nymphalinae (Dataverse electronic supplementary material, table S1). Our laboratory generated all of the nuclear rRNA repeat sequences, some of which were published previously [26–28,50–57], but are analysed in concert here for the first time, to our knowledge. When samples in the mitogenome dataset lacked GenBank Sequence Read Archives (SRAs) containing sufficient raw sequence data to assemble a nuclear rRNA repeat, they were excluded. Sequences were aligned (Dataverse electronic supplementary material, file S2) and analysed as described above.
3. Results
(a) . Junonia mitogenome phylogeny
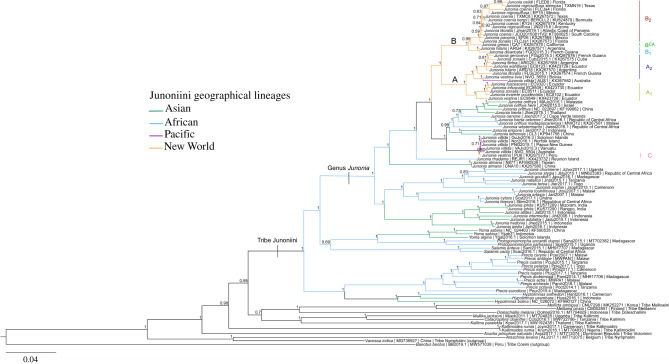
The mitogenome phylogeny was constructed using Bayesian inference with a GTR + I + G model with a best state likelihood of −170 867 and a final average deviation of split frequencies of 0.002109 (figure 2). Tribe Junoniini was monophyletic. The oldest node within Junoniini defines the divergence of Precis and Hypolimnas species from the rest of the tribe with a Bayesian posterior probability of 1 (figure 2). Sister to Junonia was a clade containing the genera Protogoniomorpha, Yoma and Salamis. Salamis diverges at the oldest node in the clade with low (0.69) Bayesian posterior probability, making the placement of this genus tentative. New World Junonia species resolve into a single large clade, with the exception of a single J. villida specimen from Australia grouping with New World sequences, and a single South American J. vestina specimen with Old World taxa. Individual New World species are not monophyletic in this analysis and interspecific relationships in the New World are unresolved (figure 2). Haplotype group A and subgroup A2 remain most prevalent in South American Junonia populations. Haplotype subgroup A1 originally thought to only occur in high-elevation J. vestina populations is shared by some Junonia fuscescens, Junonia infuscata, Junonia zonalis and J. evarete populations in Ecuador.
Figure 2.
Junonia mitogenome Bayesian inference phylogeny (GTR + I + G model, best state likelihood = −170 867 and a deviation of split frequencies = 0.002109). Sixty-four Junonia mitogenomes, 21 other tribe Junoniini mitogenomes and 12 mitogenomes from other Nymphalinae tribes. The MrBayes Bayesian posterior probability values are given at each node. (Online version in colour.)
Haplotype group B is most prevalent in North and Central America, and Bermuda, and occurs with haplotype A in the Caribbean. Haplotype subgroup B2 is the most recently diverged within group B (figure 2) and is the predominant haplotype in the Western Hemisphere from Panama northwards. The BCA haplotype group from J. grisea and other American Southwest Junonia is sister to the New World B2 subgroup. Subgroup B1 is the most divergent B haplotype lineage and only occurs in South American Junonia populations. The oldest nodes in both haplotype groups A and B show divergence of South American lineages (figure 2). Haplotype group C is restricted to J. vestina in Peru and is unique in that it does not cluster with other New World haplotype groups but instead with a J. villida mitogenome clade (Old World species with an Indo-Pacific distribution). Similarly, a single J. villida mitogenome is sister to the New World haplotype A2 clade, suggesting that there may be recent or historical gene flow between populations of New World J. vestina and Indo-Pacific J. villida populations.
In contrast with the New World, prior studies concluded that individual Old World Junonia species were monophyletic based on limited sampling of species and populations. The current analysis of complete mitochondrial genomes suggests a lack of monophyly in at least some Old World Junonia species (figure 2). For example, J. orithya and Junonia hierta form two separate lineages, one in Asia and one in Africa, which are more closely related to the sympatric lineage of the other species than they are to conspecific allopatric lineages. This suggests that there has either been remarkable parallel morphological and colour pattern evolution in these lineages, or that there may be lateral transfer and introgression of mitochondrial haplotypes between them. Another species pair that may be experiencing introgression and ongoing geneflow are Junonia iphita and Junonia hedonia. The J. iphita mitogenome sequence from Indonesia forms a clade with sympatric sequences from J. hedonia rather than with J. iphita sequences from elsewhere in Asia. This suggests that lateral transfer events and introgression is more widespread in Old World Junonia than previously appreciated.
(b) . Junonia nuclear ribosomal RNA repeat phylogeny
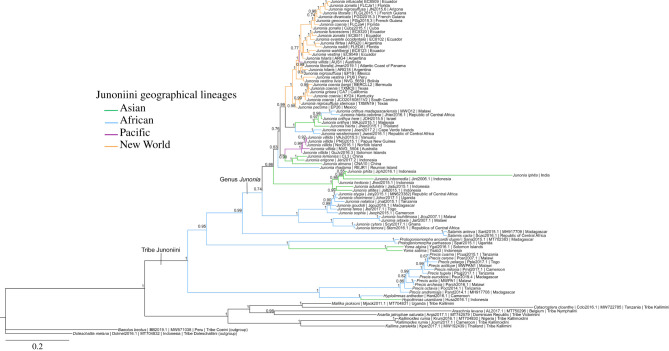
The nuclear rRNA repeat phylogeny was constructed using Bayesian inference with a GTR + I + G model with a best state likelihood of −119 000 and a final average deviation of split frequencies of 0.003566 (figure 3). Tribe Junoniini and its component genera are monophyletic. Nearly all of the New World Junonia nuclear rRNA repeats form two distinct clades. One clade is restricted to North America and Bermuda. The other clade contains specimens from South, Central, and parts of North America, and the Caribbean. Some New World Junonia species include individuals assigned to both clades. As a whole, the New World Junonia are monophyletic with only a single exception: the same Australian J. villida specimen that grouped with New World mitogenomes also clusters with New World nuclear rRNA repeats. The J. vestina specimen with a distinct mitogenome (haplotype group C) that had grouped with the Old World J. villida clade in the previous analysis (figure 2) is the sister taxon to another South American J. vestina sample within the New World clade in the nuclear rRNA repeat phylogeny (figure 3).
Figure 3.
Junonia complete rRNA repeat Bayesian inference phylogeny (GTR + I + G model, best state likelihood = −119 000 and an average deviation of split frequencies = 0.003566) based on 62 Junonia specimens, 19 other sequences from tribe Junoniini and nine outgroup species from tribes within subfamily Nymphalinae. The Bayesian posterior probability values determined by MrBayes are given at each node. (Online version in colour.)
The nuclear rRNA repeat phylogeny for Old World Junonia taxa is consistent with the mitogenome phylogeny with two major differences. The J. villida clade, for which a J. vestina sequence from the New World was a sister clade in the mitogenome tree, is more closely associated with a group of Asian Junonia species (J. lemonias, Junonia erigone and Junonia almana) in the nuclear rRNA repeat phylogeny. The second is the sister clade to this Asian clade in the mitogenome tree, is the sister clade to the New World Junonia in the rRNA repeat phylogeny (figure 3). This clade, which includes both African and Asian lineages, contains J. orithya and J. hierta. The African lineages of J. orithya and J. hierta form a clade together and Asian lineages from these two species and are interspersed with one another, similar to the results of the mitogenome analysis. By contrast, the nuclear rRNA repeats from J. iphita form a monophyletic group and do not show the affinity with J. hedonia sequences as observed using mitogenomes (figure 2).
The relationships within tribe Junoniini based on rRNA repeats are consistent with the mitogenome analysis except for the placement of Salamis, which is placed as sister taxon to Junonia with a very high probability value (0.99). The relationships of the outgroup species are consistent with the mitogenome analysis, with the exception of tribe Kallimini, which is paraphyletic in the nuclear rRNA repeat analysis. Mallika jacksoni and Catacroptera cloanthe are assigned to tribe Kallimini as expected, but Kallima paralekta clusters with tribe Kallimoidini within subfamily Nymphalinae (figure 3).
4. Discussion
(a) . Tribe Junoniini, the sister clade of Junonia and the paraphyly of Kallimini
The clade defined by the oldest node within the Junoniini (figures 2 and 3) contains the monophyletic genera Precis and Hypolimnas, as reported previously [3,24,25]. The sister clade to Junonia differs between mitochondrial and nuclear analyses. The mitogenome sister clade contains the genera Protogoniomorpha, Yoma and Salamis, with Salamis as the outgroup to the other two genera, consistent with some prior phylogenetic analyses [3,24–26]. Pyrcz et al. [25] found genus Salamis embedded within Junonia. Our rRNA repeat phylogeny shows Salamis as sister to genus Junonia (posterior probability value of 0.99), with the next further outgroup containing the genera Yoma and Protogoniomorpha.
Kallima (tribe Kallimini) originally served as a catch-all genus for Asian and African nymphalid butterflies masquerading as leaf mimics [58]. Based on genitalia and behavioural characteristics, it was later determined that genus Kallima is restricted to Asia, and African leaf mimics and were reassigned to three other Nymphalinae genera (Junonia (Junoniini), Mallika (Kallimini) or Kallimoides (Kallimoidini)) [29,59,60]. Tribe Kallimini comprised the genera Kallima, Mallika and Catacroptera and was viewed as the sister clade to the Junoniini, since most Kallimini and the basal character state within Junoniini is leaf mimicry [59]. Molecular phylogenetics has consistently placed tribe Melitaeini as the sister to the Junoniini, with tribe Kallimini placed as a farther outgroup [29,61]. While the mitogenome tree (figure 2) is consistent with these results, phylogenetic reconstruction using the rRNA repeat region (figure 3) shows genera Mallika and Catacroptera (both African genera) forming a monophyletic clade. Genus Kallima falls outside this grouping and as sister to Kallimoides, with a Bayesian probability value of 1 (figure 3). These differences could be attributable to limited taxon sampling among outgroups within the rRNA repeat phylogeny (figure 3). Both the Junoniini and Kallimini show close molecular phylogenetic associations with the Melitaeini in our mitogenome reconstruction (figure 2) and in previous studies [29,62,63], but nuclear rRNA repeat sequences, helpful for inferring higher level taxonomic relationships within the Nymphalinae, are not yet available from the Melitaeini.
(b) . New World Junonia
Species-level relationships in the New World Junonia were unresolved by mitogenomes (figure 2), consistent with earlier DNA barcode and mitogenome studies [3,5,16–19,23]. Many New World species include individuals that carry both A and B haplotype groups. Similarly, most New World Junonia species do not form monophyletic groups based on nuclear rRNA repeats. This is indicative of gene flow between New World Junonia species and is consistent with prior observations of hybridization between many of these species [20,23,30,32]. Based on the mitogenome phylogeny (figure 2), the New World Junonia is monophyletic with two exceptions. The first is a J. vestina sample that possesses haplotype group C and is most closely related to a lineage of J. villida from the Indo-Pacific. This finding differs from that of Peters & Marcus [5], which placed J. vestina as the sister taxon to J. lemonias. Peters & Marcus [5] only included single representatives of most Junonia species, including J. villida, so this discrepancy can be attributed to limited sampling. In both the current study and Peters & Marcus [5], a single J. villida sample from Australia is most closely related to the haplotype group A2 in the New World. These findings signify that long-range dispersal and gene flow across the Pacific may be occurring and is of importance to understanding the relationships between the Old World and New World species, as well as the origins of the New World Junonia.
Complete nuclear rRNA repeats occur at high copy number in the genome and are easily recovered from the same genome skimming datasets used for assembling whole mitogenomes (figure 3; [43]). Phylogenetic analysis of Junonia nuclear rRNA repeats did not recover the same clades found through analyses of mitogenomes, nor do rRNA repeats resolve New World species-level relationships. Like the mitogenome, there is an apparent geographical signal, with most North American specimens forming one rRNA repeat lineage, while a second is made up of Central and South American, Caribbean and a few specimens from southern portions of North America. Based on nuclear rRNA repeat sequences, the New World Junonia are monophyletic, except for an Australian J. villida sample that also groups with the New World in the mitogenome phylogeny. Unlike the mitogenome phylogeny, the nuclear rRNA repeats of all J. vestina samples fall within the New World rRNA clade. This further supports the hypothesis that gene flow between New World J. vestina and the Indo-Pacific J. villida by means of long-distance dispersal across the Pacific, followed by hybridization, may be ongoing since individual specimens do not share the same level of molecular affinity with one another, with some forming clades with the other taxon.
(c) . Old World Junonia
Previous phylogenetic studies of Old World Junonia could not test for monophyly or make strong statements about species-level relationships, because only a single sample of each species was used [9,16–18,24–26,29]. Kodandaramaiah & Wahlberg [3] was exceptional in including multiple specimens from some Old World species comparable with this analysis. First, specimens from Asian and African populations of J. hierta and J. orithya form clades based on geography rather than taxonomic species assignment here (figures 2 and 3) and in Kodandaramaiah & Wahlberg [3], though they did not comment on the pattern in the text of their paper. Also consistent with Kodandaramaiah & Wahlberg [3] is the placement of this clade as sister to the New World Junonia, although our mitogenome data includes J. villida within the clade. Further, mitogenomes from J. iphita pair with either J. hedonia or Junonia atlites (figure 2). By contrast, in the analysis of rRNA nuclear repeats (figure 3), the J. iphita sequences form a monophyletic clade and none are sister to J. hedonia. A recent lateral transfer of the J. hedonia mitogenome to J. iphita in Indonesia seems likely as the sequences are nearly identical, but additional sampling and sequencing from these and other species in this lineage would clarify patterns of organelle capture. What seems increasingly clear is that while lateral transfer events may be most frequent in the New World Junonia, it also occurs in some Old World lineages, contributing to reticulate evolution of the genus as a whole.
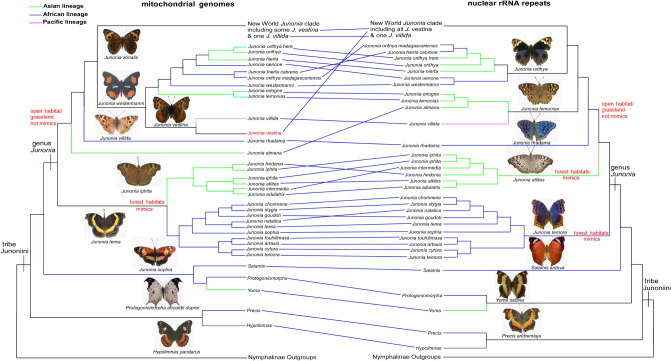
Despite instances of apparent lateral transfer, the Old World Junonia form distinct molecular clades made up of species that share phenotypic features (figure 4). Although there are small differences between the two different phylogenetic analyses in respect to species placement, all Old World species can be grouped based on geography, habitat type and mimicry strategy. Some clades defined by the oldest Junonia nodes in the mitogenome phylogeny (figure 4) consist of two lineages of forest-dwelling species. The first is an Asian forest-dwelling butterfly lineage containing species (J. iphita, J. atlites, Junonia intermedia, Junonia adulatrix and J. hedonia) that at rest have closed wings that masquerade imprecisely as leaves [58]. The second lineage includes African species that are also forest-dwelling but most are considered to be very good leaf mimics [24] (figure 4). There is one exception, Junonia sophia, which is thought to be a Batesian mimic of the false diadem butterfly (Pseudacraea lucretia) [59]. Within this African Junonia lineage, there are two subclades that cluster based on similar coloration of the dorsal wing surfaces (either blue or brown).
Figure 4.
Modified molecular phylogeny of the Old World Junonia. Interpretations based on mitogenome and rRNA repeat phylogenetic reconstructions from figures 2 and 3. Geographical origins, habitat preferences and the display of mimicry of lineages are indicated. (Online version in colour.)
The next large Junonia clade in the mitogenome phylogeny includes all New World Junonia species as well as some Asian (J. orithya, J. hierta, J. lemonias, J. villida, J. erigone and J. almana) and African species (J. orithya, J. hierta, Junonia oenone, J. westermanni and Junonia rhadama). These are grassland or open habitat specialists, and none are considered to be mimics (figure 4, [59,64]). Consistent with prior studies, this lineage originated in Africa, dispersed to Asia, and then returned to Africa, perhaps several times [3]. Later, this lineage established the New World Junonia radiation. The rRNA repeat phylogeny yields similar results to the mitogenome analysis, with phylogenetic trends based on habitat type, mimicry and geography, but differences do exist. The clade defined by the oldest node within Junonia recovered by this analysis supports an African origin for this genus, as all species in this clade are forest-dwelling and restricted to Africa (figure 4), reinforcing mitogenome results and an earlier three-gene analysis [3]. Like the mitogenome phylogeny, all species within this clade are mimics. The next clade consists of Asian and Indo-Pacific Junonia, with one exception, J. rhadama that occupies Madagascar and several other Indian Ocean islands. The first major split in this clade is by habitat type with forest-dwelling imperfect leaf mimics and grassland/open habitat lineages. Forest-dwelling species include the J. iphita and J. hedonia complex discussed above in the mitogenome phylogeny and the same pattern is observed here. Open habitat species include J. rhadama. The remaining open habitat species are more similar morphologically to New World taxa than they are to other Old World species. A key difference in the composition of this clade exists between our phylogenetic analyses. In the mitogenome analysis, J. villida is the earliest diverging species and sister to the New World Junonia (figure 4), while in the nuclear rRNA repeat analysis, the sister clade is limited to African J. westermanni, and J. oenone and African/Asian J. orithya and J. hierta, and the remaining taxa (Asian: J. villida, J. lemonias, J. erigone and J. almana) are transferred into a separate clade (figure 4). This difference between the mitogenomic and nuclear rRNA repeat phylogenies raises the intriguing possibility that the New World Junonia were established with contributions from two Old World Junonia lineages: one containing J. villida which crossed the Pacific Ocean and a second containing J. orithya and J. hierta, which could have crossed either the Atlantic or Pacific Ocean to reach the New World. Thus, several of the early hypotheses for the origin of the New World Junonia may not have been mutually exclusive after all and instead may turn out to be simultaneously correct [12,13,15].
5. Conclusion
Compared with many other butterfly taxa, Junonia have particularly effective dispersal abilities, allowing them to colonize remote new habitats, perhaps in some cases repeatedly. This creates scenarios where resident and new immigrant Junonia are sympatric and lateral transfer is possible through hybridization and reticulate evolution. This appears to have taken place both in the Old World and New World. The large number of New World Junonia (18 species) that have evolved in a very short period of time (2–4 Myr) suggest that speciation in this group has been accelerated compared to the older lineages in the Old World (28 species) which have diverged since the origins of the genus 15–27 Ma [3]. The variety of new ecological niches (especially larval host plant associations) presented by new habitats, in combination with frequent lateral transfer of adaptive genes and traits, and reticulate evolution may have contributed to the substantially greater rate of speciation in the New World, making this system similar to the explosive species radiation events such as the Lake Victoria cichlids [65] or the Hawaiian Drosophila [66].
To address the challenges of species delimitation in Junonia in the face of reticulate evolution, one recent study successfully employed complete Z chromosome sequences [23]. Although their dataset only included specimens from some New World Junonia taxa, they were able to resolve the species into monophyletic clades. Accumulating complete Z chromosome sequences from the rest of the genus will provide an interesting comparison to results from analyses of mitochondrial genomes and nuclear rRNA repeats, and together this may permit better delimitation of the remaining species, further resolve species-level phylogenetic relationships, and should be considered for future phylogenetic studies of this genus.
Acknowledgements
We sincerely thank prior laboratory members Bonnie McCullagh, Melissa Peters, Rayna Hamilton, Josephine Payment and Mackenzie Alexiuk for their work which laid the foundations for this project. Thanks also to the Next Generation Sequencing Platform at the Manitoba Institute of Child Health, and Genome Quebec for library preparation and sequencing.
Data accessibility
All sequences and SRAs have been made submitted to be publically available through GenBank and accession numbers for all data used in the analyses can be found in the Dataverse electronic supplementary material, table S1 (https://doi.org/10.34990/FK2/O9UKCE/LQDEC8) which has been deposited through the online data repository Dataverse. Sequence alignment data is available as electronic supplementary files through the online data repository Dataverse for both the full mitochondrial genomes (https://doi.org/10.34990/FK2/O9UKCE/5S8ZJ8) and rRNA repeats (https://doi.org/10.34990/FK2/O9UKCE/YYM2ZT).
Authors' contributions
M.M.L.L.: conceptualization, data curation, formal analysis, investigation, methodology, software, validation, visualization, writing—original draft, writing—review and editing; J.M.M.: conceptualization, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing—review and editing.
Both authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work received support from NSERC under grant nos. RGPIN386337-2011 and RGPIN-2016-06012 and from the University of Manitoba under the University Research Grants Program.
References
- 1.Lomolino MV. 2016. The unifying, fundamental principles of biogeography: understanding Island Life. Front. Biogeogr. 8, e29920. ( 10.21425/F58229920) [DOI] [Google Scholar]
- 2.Patiño J, et al. 2017. A roadmap for island biology: 50 fundamental questions after 50 years of The Theory of Island Biogeography. J. Biogeogr. 44, 963-983. ( 10.1111/jbi.12986) [DOI] [Google Scholar]
- 3.Kodandaramaiah U, Wahlberg N. 2007. Out-of-Africa origin and dispersal-mediated diversification of the butterfly genus Junonia (Nymphalidae: Nymphalinae). J. Evol. Biol. 20, 2181-2191. ( 10.1111/j.1420-9101.2007.01425.x) [DOI] [PubMed] [Google Scholar]
- 4.Vane-Wright RI, Tennent WJ. 2011. Colour and size variation in Junonia villida (Lepidoptera, Nymphalidae): subspecies or phenotypic plasticity? Syst. Biodivers. 9, 289-305. ( 10.1080/14772000.2011.640497) [DOI] [Google Scholar]
- 5.Peters MJ, Marcus JM. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst. Ent. 42, 288-300. ( 10.1111/syen.12214) [DOI] [Google Scholar]
- 6.Lalonde MML, McCullagh BS, Marcus JM. 2018. The taxonomy and population structure of the buckeye butterflies (genus Junonia, Nymphalidae: Nymphalini) of Florida, USA. J. Lepid Soc. 72, 97-115. ( 10.18473/lepi.v72i2.a2) [DOI] [Google Scholar]
- 7.Brévignon L, Brévignon C. 2012. Nouvelles observations sur le genre Junonia en Guyane Française. Lepidoptera: Nymphalidae)(Seconde partie). Lépidoptères de Guyane 7, 8-35. [Google Scholar]
- 8.Neild AFE. 2008. The butterflies of Venezuela, part 2: Nymphalidae II (Acraeinae, Libytheinae, Nymphalinae, Ithomiinae, Morphinae). London, UK: Meridian. [Google Scholar]
- 9.Kodandaramaiah U. 2009. Eyespot evolution: phylogenetic insights from Junonia and related butterfly genera (Nymphalidae: Junoniini). Evol. Dev. 11, 489-497. ( 10.1111/j.1525-142X.2009.00357.x) [DOI] [PubMed] [Google Scholar]
- 10.McCullagh, B. S. 2016. Sequence evolution among divergent mitochondrial haplotypes within species of Junonia butterflies. Winnipeg, Canada: University of Manitoba. (http://hdl.handle.net/1993/31105) [Google Scholar]
- 11.Corbet AS. 1948. Papers on Malaysian Rhopalocera. V. The conspecificity of the American Precis lavinia (Cramer) with the Oriental Precis orithya (Cramer). Entomologist 81, 54-56. [Google Scholar]
- 12.Eliot N. 1946. Continental drift and Precis lavinia. Entomologist 79, 225-228. [Google Scholar]
- 13.Eliot N. 1947. More on continental drift. Precis lavinia Hb. and P. villida F. Entomologist 80, 230-234. [Google Scholar]
- 14.Forbes WTM. 1928. Variation in Junonia lavinia (Lepidoptera, Nymphalidae). J. N. Y. Entomol. Soc. 36, 306-321. [Google Scholar]
- 15.Forbes WTM. 1947. Buckeyes and Wegener. Entomologist 80, 56-58. [Google Scholar]
- 16.Borchers TE, Marcus JM. 2014. Genetic population structure of buckeye butterflies (Junonia) from Argentina. Syst. Ent. 39, 242-255. ( 10.1111/syen.12053) [DOI] [Google Scholar]
- 17.Gemmell AP, Borchers TE, Marcus JM. 2014. Molecular population structure of buckeye butterflies (Junonia) from French Guiana, Martinique, and Guadeloupe. Psyche 2014, 1-21. ( 10.1155/2014/897596) [DOI] [Google Scholar]
- 18.Gemmell AP, Marcus JM. 2015. A tale of two haplotype groups: the origin and distribution of divergent New World Junonia COI haplotypes. Syst. Ent. 40, 532-546. ( 10.1111/syen.12120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeiler E, Johnson S, Markow TA. 2012. DNA barcodes and insights into the relationships and systematics of buckeye butterflies (Nymphalidae: Nymphalinae: Junonia) from the Americas. J. Lepid Soc. 66, 185-198. ( 10.18473/lepi.v66i4.a1) [DOI] [Google Scholar]
- 20.Lalonde MML, Marcus JM. 2019. Getting Western: biogeographical analysis of morphological variation, mitochondrial haplotypes, and nuclear markers reveals cryptic species and hybrid zones in the Junonia butterflies of the American Southwest and Mexico. Syst. Ent. 44, 465-489. ( 10.1111/syen.12335) [DOI] [Google Scholar]
- 21.Brévignon C. 2004. Description de deux nouvelle sous-espèces Guadeloupeennes du genere Junonia Hübner, 1819 (Lepidoptera, Nymphalidae, Nymphalinae). Lambillionea CIV, 72-80. [Google Scholar]
- 22.Brévignon C. 2009. Nouvelles observations sur le genre Junonia en Guyane Française. (Lepidoptera: Nymphalidae) Première Partie. Lambillionea 109, 3-7. [Google Scholar]
- 23.Cong Q, Zhang J, Shen J, Cao X, Brévignon C, Grishin NV. 2020. Speciation in North American Junonia from a genomic perspective. Syst. Ent. 45, 803-837. ( 10.1111/syen.12428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke JW. 2017. Evolutionary trends in phenotypic elements of seasonal forms of the tribe Junoniini (Lepidoptera: Nymphalidae). In Diversity and evolution of butterfly wing patterns: an integrative approach (eds Sekimura T, Nijhout HF), pp. 239-253. Singapore: Springer Singapore. [Google Scholar]
- 25.Pyrcz TW, Florczyk K, Collins S, Safian S, Mahecha-J O, Lachowska-Cierlik D. 2021. Alpha-taxonomy and phylogeny of African Junoniini butterflies based on morphological data, with an emphasis on genitalia, and COI barcode (Lepidoptera: Nymphalidae). Zootaxa 4991, 401-433. ( 10.11646/zootaxa.4991.3.1) [DOI] [PubMed] [Google Scholar]
- 26.Lalonde MML, Marcus JM. 2020. The complete mitochondrial genome of the Malagasy clouded mother-of-pearl butterfly Protogoniomorpha ancardii duprei (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 5, 3261-3263. ( 10.1080/23802359.2020.1810156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalonde MML, Marcus JM. 2019. The complete mitochondrial genome of the Madagascar banded commodore butterfly Precis andremiaja (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 4, 277-279. ( 10.1080/23802359.2018.1541721) [DOI] [Google Scholar]
- 28.Living Prairie Mitogenomics Consortium. 2020. The complete mitochondrial genome of the brown pansy butterfly, Junonia stygia (Aurivillius, 1894), (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 5, 41-43. ( 10.1080/23802359.2019.1693921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahlberg N, Brower AVZ, Nylin S. 2005. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol. J. Linn. Soc. Lond. 86, 227-251. ( 10.1111/j.1095-8312.2005.00531.x) [DOI] [Google Scholar]
- 30.Lalonde M, Marcus J. 2020. Back to the future: an update on the invasion history of Junonia butterflies in Florida, USA and an assessment of the origins of early 20th century insect specimens attributed to Chokoloskee. J. Lepid. Soc. 74, 83-94. ( 10.18473/lepi.74i2.a3) [DOI] [Google Scholar]
- 31.Lalonde MML, Marcus JM. 2020. How old can we go? Evaluating the age limit for effective DNA recovery from historical insect specimens. Syst. Ent. 45, 505-515. ( 10.1111/syen.12411) [DOI] [Google Scholar]
- 32.Hafernik JE. 1982. Phenetics and ecology of hybridization in buckeye butterflies (Lepidoptera: Nymphalidae). Univers. Calif. Publ. Entomol. 96, 1-109. ( 10.5962/p.266775) [DOI] [Google Scholar]
- 33.Paulsen SM. 1996. Quantitative genetics of the wing color pattern in the buckeye butterfly (Precis coenia and Precis evarete). Evolution 50, 1585-1597. ( 10.1111/j.1558-5646.1996.tb03931.x) [DOI] [PubMed] [Google Scholar]
- 34.Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc. R. Soc. B 270, 313-321. ( 10.1098/rspb.2002.2218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hebert PDN, Ratnasingham S, deWaard JR. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B 270, S96-S99. ( 10.1098/rsbl.2003.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seitz A. 1914. Die Gross Schmetterlinge der Erde. Stuttgart, Germany: Alfred Kernen. [Google Scholar]
- 37.Hao J, Sun M, Shi Q, Sun X, Shao L, Yang Q. 2013. Complete mitogenomes of Euploea mulciber (Nymphalidae: Danainae) and Libythea celtis (Nymphalidae: Libytheinae) and their phylogenetic implications. ISRN Genomics 2013, 1-14. ( 10.1155/2013/491636) [DOI] [Google Scholar]
- 38.Gillett CPDT, Crampton-Platt A, Timmermans MJTN, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk De Novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol. Biol. Evol. 31, 2223-2237. ( 10.1093/molbev/msu154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timmermans MJTN, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol. Phylogenet. Evol. 79, 169-178. ( 10.1016/j.ympev.2014.05.031) [DOI] [PubMed] [Google Scholar]
- 40.Wu LW, Lin LH, Lees DC, Hsu YF. 2014. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics 15, 468. ( 10.1186/1471-2164-15-468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Q, Huang D, Wang Y, Hao J. 2015. The complete mitochondrial genome of blue pansy, Junonia orithya (Lepidoptera: Nymphalidae: Nymphalinae). Mitochond. DNA 26, 245-246. ( 10.3109/19401736.2013.823182) [DOI] [PubMed] [Google Scholar]
- 42.McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of lemon pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J. Asia-Pacific Ent. 18, 749-755. ( 10.1016/j.aspen.2015.08.006) [DOI] [Google Scholar]
- 43.Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 5, 1-23. ( 10.3934/genet.2018.1.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947-2948. ( 10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- 45.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windown interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876-4882. ( 10.1093/nar/25.24.4876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574. ( 10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 48.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539-542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambaut A. 2016. FigTree, version 1.4.3, Computer program distributed by the author, website: See http://tree.bio.ed.ac.uk/software/figtree/.
- 50.Alexiuk MR, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome and phylogenetic analysis of the European map butterfly Araschnia levana (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 5, 3264-3266. ( 10.1080/23802359.2020.1810163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alexiuk MR, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome of the Jackson's leaf butterfly Mallika jacksoni (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 5, 3316-3318. ( 10.1080/23802359.2020.1814885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamilton RV, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome of the black dead leaf butterfly Doleschallia melana (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 5, 3306-3308. ( 10.1080/23802359.2020.1814172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lalonde MML. 2021. Phylogenetic analysis of the complete mitochondrial genome of the graphic beauty butterfly Baeotus beotus (Doubleday 1849) (Lepidoptera: Nymphalidae: Nymphalinae: Coeini). Mitochondr. DNA B Resour. 6, 1516-1518. ( 10.1080/23802359.2021.1914526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lalonde MML. 2022. The complete mitochondrial genome of the pirate butterfly Catacroptera cloanthe (Stoll 1781)(Insecta: Lepidoptera: Nymphalidae: Kallimini). Mitochondr. DNA B Resour. 7, 306-308. ( 10.1080/23802359.2022.2030818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lalonde MML, Marcus JM. 2019. The complete mitochondrial genome of Madagascar mother-of-pearl butterfly Salamis anteva (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 4, 296-298. ( 10.1080/23802359.2018.1542989) [DOI] [Google Scholar]
- 56.Payment JE, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome of the African leaf butterfly Kallimoides rumia (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 5, 3415-3417. ( 10.1080/23802359.2020.1823261) [DOI] [Google Scholar]
- 57.Payment JE, Marcus JM, Lalonde MML. 2020. Phylogenetic analysis of the complete mitochondrial genome of the white peacock butterfly Anartia jatrophae saturata (Insecta: Lepidoptera: Nymphalidae). Mitochondr. DNA B Resour. 5, 3708-3710. ( 10.1080/23802359.2020.1832929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skelhorn J. 2015. Masquerade. Curr. Biol. 25, R643-R664. ( 10.1016/j.cub.2015.02.069) [DOI] [PubMed] [Google Scholar]
- 59.Larsen T. 1999. Butterflies of West Africa - origins, natural history, diversity, and conservation. Stenstrup, Denmark: Apollo Books. [Google Scholar]
- 60.Shirôzu T, Nakanishi A. 1984. A revision of the genus Kallima DOUBLEDAY (Lepidoptera, Nymphalidae): I. Generic Classification. Tyô to Ga 34, 97-110. ( 10.18984/lepid.34.3_97) [DOI] [Google Scholar]
- 61.Su C, Shi Q, Sun X, Ma J, Li C, Hao J, Yang Q. 2017. Dated phylogeny and dispersal history of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae). Sci. Rep. 7, 8799. ( 10.1038/s41598-017-08993-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wahlberg N, Weingartner E, Nylin S. 2003. Towards a better understanding of the higher systematics of Nymphalidae (Lepidoptera: Papilionoidea). Mol. Phylogenet. Evol. 28, 473-484. ( 10.1016/S1055-7903(03)00052-6) [DOI] [PubMed] [Google Scholar]
- 63.Freitas AVL, Brown KS Jr. 2004. Phylogeny of the Nymphalidae (Lepidoptera). Syst. Biol. 53, 363-383. ( 10.1080/10635150490445670) [DOI] [PubMed] [Google Scholar]
- 64.Aoki T, Yamaguchi S, Uemura Y. 1982. Butterflies of the south east Asian islands III: Satyridae, Libytheidae. Tokyo, Japan: Plapac Co., Ltd. [Google Scholar]
- 65.Keller I, Wagner CE, Greuter L, Mwaiko S, Selz OM, Sivasundar A, Wittwer S, Seehausen O. 2013. Population genomic signatures of divergent adaptation, gene flow and hybrid speciation in the rapid radiation of Lake Victoria cichlid fishes. Mol. Ecol. 22, 2848-2863. ( 10.1111/mec.12083) [DOI] [PubMed] [Google Scholar]
- 66.Price DK, Muir C. 2008. Conservation implications of hybridization in Hawaiian picture-winged Drosophila. Mol. Phylogenet. Evol. 47, 1217-1226. ( 10.1016/j.ympev.2007.12.003) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences and SRAs have been made submitted to be publically available through GenBank and accession numbers for all data used in the analyses can be found in the Dataverse electronic supplementary material, table S1 (https://doi.org/10.34990/FK2/O9UKCE/LQDEC8) which has been deposited through the online data repository Dataverse. Sequence alignment data is available as electronic supplementary files through the online data repository Dataverse for both the full mitochondrial genomes (https://doi.org/10.34990/FK2/O9UKCE/5S8ZJ8) and rRNA repeats (https://doi.org/10.34990/FK2/O9UKCE/YYM2ZT).