Abstract
Kidney Precision Medicine Project (KPMP) is building a spatially specified human kidney tissue atlas in health and disease with single-cell resolution. Here, we describe the construction of an integrated reference map of cells, pathways, and genes using unaffected regions of nephrectomy tissues and undiseased human biopsies from 56 adult subjects. We use single-cell/nucleus transcriptomics, subsegmental laser microdissection transcriptomics and proteomics, near-single-cell proteomics, 3D and CODEX imaging, and spatial metabolomics to hierarchically identify genes, pathways, and cells. Integrated data from these different technologies coherently identify cell types/subtypes within different nephron segments and the interstitium. These profiles describe cell-level functional organization of the kidney following its physiological functions and link cell subtypes to genes, proteins, metabolites, and pathways. They further show that messenger RNA levels along the nephron are congruent with the subsegmental physiological activity. This reference atlas provides a framework for the classification of kidney disease when multiple molecular mechanisms underlie convergent clinical phenotypes.
Pathway-based integration of multiomics and imaging assays maps physiological function of the human kidney at the single-cell level.
INTRODUCTION
The kidney has one of the most diverse cellular populations and spatial organization in the human body, and it is critical in maintaining the physiological homeostasis by regulating fluid and electrolyte balance, osmolarity, and pH. The basic unit of organization in the kidney is the nephron embedded in the interstitium; the human kidney has between 210,000 and 2.7 million nephrons. There are multiple cell types in the nephron and the interstitium including those that comprise the blood vessels and capillaries (such as endothelial cells and vascular smooth muscle cells) and many types of immune cells.
For decades, there has been a sustained effort to develop a detailed understanding of structure-function-endophenotype relationships within the kidney tissue to understand its physiology and pathophysiology (1). Over the past decade, with the advent of single-cell (sc) RNA sequencing (RNAseq) technologies, substantial advances have been made in enumerating the different cell types in the human and mouse kidney (2–11). Computational analyses and modeling of sc transcriptomic data and other omics data are starting to provide rich and deep insight into different kidney disease processes, including kidney cancers (11) and fibrosis (6). These studies demonstrate the power of omics technologies in developing atlases that map structure-function relationships at the single-cell level within tissues.
Datasets from different omics technologies provide an unparalleled opportunity to understand how the diversity of cell types and their constituents underlie physiological functions and how they are altered in different disease states. The Kidney Precision Medicine Project (KPMP) is a consortium funded by the National Institute of Diabetes and Digestive and Kidney Diseases. Using kidney biopsies that are ethically and safely obtained from participants with kidney disease, KPMP aims to create a kidney atlas in health and disease. Such an atlas can identify critical cells, pathways, and targets for novel therapies and preventive strategies (12, 13). To identify and understand disease states, it is necessary to have a detailed atlas of tissues that do not show disease phenotype by standard clinical histopathological evaluation. We call such an atlas “a reference atlas.” Using multiple reference kidney sources, different groups in the consortium have generated diverse types of data. Among these are single-nucleus (sn) (14) and sc (15) transcriptomics, regional bulk transcriptomics, proteomics, and metabolomics, as well as multiple complementary types of imaging methods. We have analyzed and integrated these different data types obtained from reference kidney tissue specimens, as evaluated by standard pathology analysis, from 56 adult human subjects and analyzed 80,289 single cells. For quality control, we also assessed 24,282 additional single cells from public sn RNAseq data from the literature (16, 17). We constructed maps of the different cell types in the kidney and the molecular entities and functional pathways within these cell types to develop the first version of a reference human kidney atlas. To determine whether the molecular details in the atlas enable previously unknown insights into physiological activity, we compare our data to the transcellular sodium reabsorption along the nephron, which is essential for maintaining normal blood pressure in individuals with normo- and hypertension (18). We find strong congruence between physiological activity and the mRNA expression signatures of different sodium transporters, indicating that such a detailed atlas can provide a deeper understanding of the molecular and cellular principles underlying physiological processes. All data presented in this study are available for download, and as an interactive cellular atlas (atlas.kpmp.org/explorer) to serve as a starting point from which data of disease states can be projected onto a functional context to drive new molecular classification of kidney disease.
RESULTS
The KPMP Consortium conducted different types of omics assays and low-throughput imaging experiments at different sites for these reference atlas studies. Although it is impossible to definitively characterize tissue as healthy, clinical pathologists adjudicated that the specimens used in this study show no structural signs of disease manifestations. Nevertheless, since we use unaffected tissue regions from nephrectomies and biopsies from both living donors and transplant recipients (i.e., surveillance biopsies), we use the general term reference tissue (table S1). In future studies, these can be compared to diseased tissue specimens.
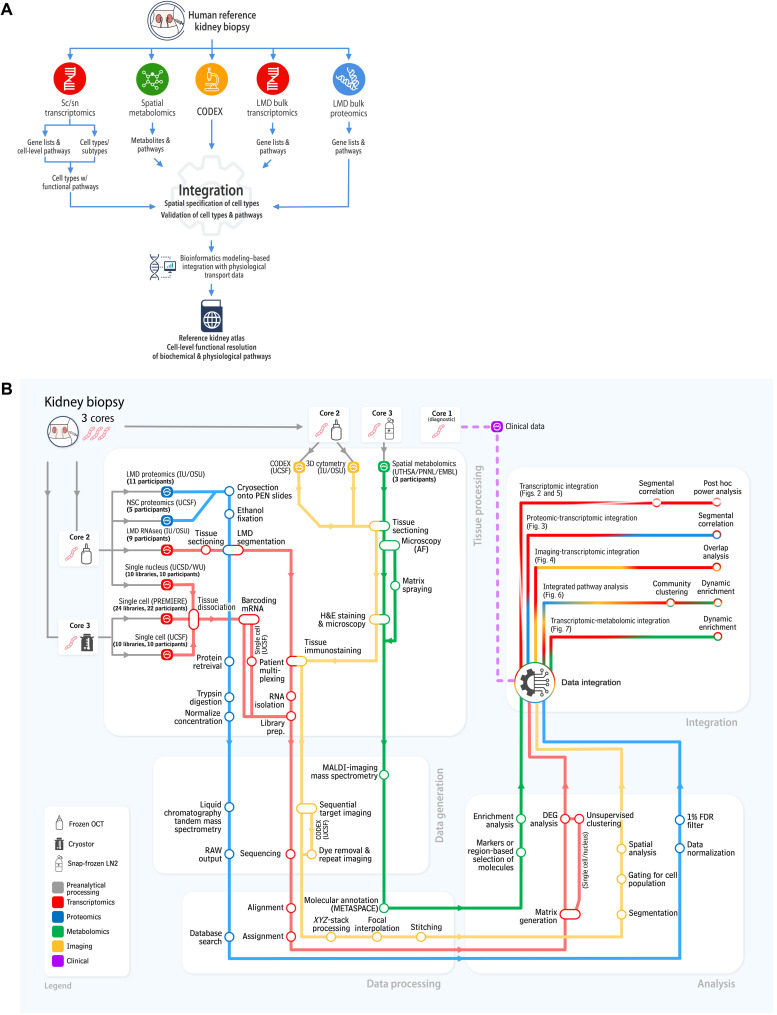
There were four transcriptomic, two proteomic, two imaging-based, and one spatial metabolomics tissue interrogation assays deployed on the shared tissue samples (Fig. 1A). These assays yielded 3 to 48 different datasets obtained from 3 to 22 subjects per assay for a total of 56 different adult human subjects (table S1). The assays and their detailed tissue preanalytical, tissue processing, data acquisition, and analytical data processing pipelines are schematically depicted as a flowchart in Fig. 1B. We also summarize, in the integration segment of our flowchart, the steps by which the datasets from different assays were integrated and harmonized. This is shown on the upper right side of the descriptive map.
Fig. 1. Graphic outline of KPMP data integration and harmonization procedures.
The “subway map” representation of the experimental and analytical protocols used within KPMP is shown in operational flow from kidney biopsy to the integrated multimodal data represented in this manuscript. The kidney biopsy, which is processed through three different tissue processing methods, is shared among tissue interrogation sites (TISs) that generate the data. Four key modalities of molecular data are generated: transcriptomic (red), proteomic (blue), imaging (yellow), and metabolomic (green). Biopsy cores 2 and 3 are used for the molecular analysis; biopsy core 1 (not depicted) is used for histological analysis.
Integration of multiple transcriptomic interrogation techniques shows agreement and technological synergy between assays
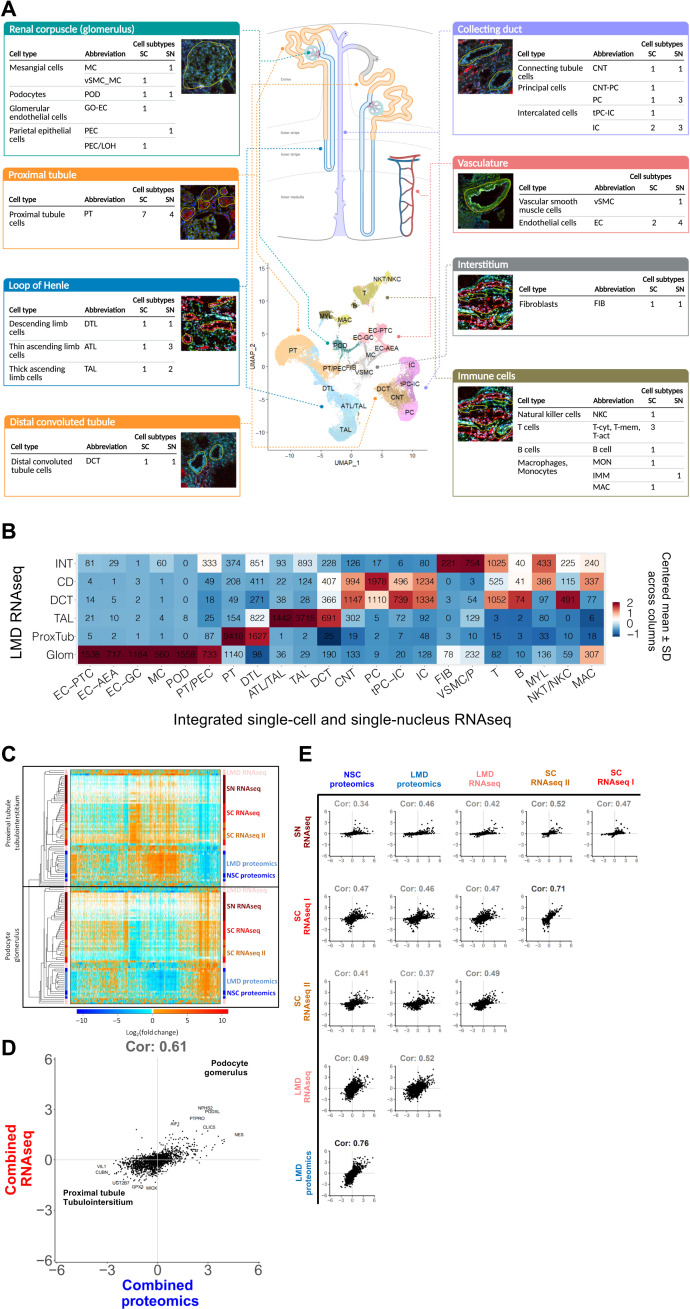
Separate and integrated analysis of sc, multiplexed sc, and sn transcriptomic datasets confirmed all known major kidney tissue cell types of the nephron and multiple immune cells (Fig. 2A). Clustering algorithms used to analyze the sc and sn RNAseq data separately identified multiple subtypes for several cells. We observed differences between the numbers of subtypes in the sc versus sn data as different cutoffs were used in the initial analyses (14, 15). Nevertheless, when sc and sn RNAseq data were analyzed in an integrated manner, all major cell types were identified, as shown in the central panel in Fig. 2A, yielding 16 main clusters (note that some clusters contain multiple closely related subtypes). These clusters were annotated to 14 cell types based on cluster-specific marker gene expression (fig. S1G). Almost every cluster contained cells and nuclei from every dataset, documenting consistency of our transcriptomic datasets (fig. S1E). For quality control measures see fig. S1 (A to G).
Fig. 2. Integration of transcriptomic, proteomic, and image-based assays documents concordance across different omics technologies.
(A) Scheme showing the major nephron segments as identified in our datasets. Sc and sn datasets were either analyzed separately or combined. Uniform manifold approximation and projection documents the results of the combined analysis. Cell subtype counts were obtained from the separated analyses (fig. S4, A and B). The corresponding LMD segments shown include the markers used to identify each subsegment: phalloidin, fluorescein isothiocyanate–labeled phalloidin for dissection of glomeruli and other structures; LRP2, megalin with Alexa Fluor 568 secondary (red); UMOD, directly conjugated Alexa Fluor 546 antibody to uromodulin (red); fluorescein-labeled PNA, peanut agglutinin labels collecting ducts (green); 4′,6-Diamidino-2-phenylindole was included for nuclei (blue). (B) We used Pearson correlation analysis of gene expression data to identify the closed subsegment in the LMD RNAseq data for each cell or nucleus in the combined transcriptomic analysis. Numbers document the number of cells/nuclei of each cluster mapped to each segment. (C) We calculated log2 fold changes between podocyte (or glomerulus) and PT cells (or tubulointerstitium) for each subject based on each assay. Pairwise correlation coefficients between all log2 fold changes were determined and used for hierarchical clustering. The variation in the axial ranges represents the divergences in the dynamic range of different assays as the axes are non-normalized. (D) Log2 fold changes obtained by the same assay were averaged across all subjects, followed by averaging of the results across all four transcriptomics and two proteomics assays. Positive or negative log2 fold changes indicate podocyte/glomerular or PT/tubulointerstitial expression. (E) Pairwise correlations between the sc/sn RNAseq and proteomic datasets document highest concordance between both proteomic and single-cell assays. Positive and negative log2 fold changes indicate podocyte/glomerular and PT/tubulointerstitial expression, respectively.
To provide spatial context with respect to different regions of the nephron, we compared the sc and sn transcriptomic datasets with nephron segment–specific bulk transcriptomic datasets that were obtained after laser microdissection (LMD) of kidney segments (table S2) (19). Cross-assay Pearson correlation analysis shows that there is strong concordance across the data obtained by the different technologies, whereby most of the cells and nuclei from each cluster were assigned to the correct corresponding LMD subsegment in an unbiased manner (Fig. 2B). For example, 83% proximal tubule (PT) cells were assigned to the PT subsegment, while 99.5% podocytes were assigned to the glomerular subsegment. As the total numbers of cells analyzed are modest compared to some of the recent reports, we evaluated whether other independent technologies improve the overall atlas framework using integration of orthogonal omics assays and post hoc power analyses.
Proteomic and transcriptomic assays produce biologically complementary descriptions of subsegmental molecular composition
In addition to transcriptomic profiles, we present subsegment-specific protein expression profiles using two different proteomic assays. These assays identify protein expression in the glomerulus and the tubulointerstitium (LMD proteomics) or PT [near-single-cell (NSC) proteomics] (tables S3 and S4, respectively). For an unbiased cross-platform comparison, we focused on podocyte/glomerular and PT cells and subsegments in the four transcriptomic datasets. Pairwise correlation of logarithmic ratios of gene or protein expression values for the glomerular versus tubular cell types or subsegments (fig. S2A) followed by hierarchical clustering resulted in grouping of the datasets by appropriate regions of the kidney (Fig. 2C; see fig. S2B for quantitative information about the clustering heights in the dendrogram). Within this broad classification, the subgroupings by different assays could be readily identified and are shown (right-side labels in Fig. 2C). From this clustering, we conclude that irrespective of the assay, we can readily identify groups of genes or proteins associated with the appropriate anatomical region (i.e., glomerulus versus tubulointerstitium). This pattern is observed with or without the removal of genes or proteins that are not identified by all technologies (fig. S2C). In contrast, if we cluster by absolute expression values, the clustering is primarily driven by the assay used rather than the anatomical region. This is irrespective of whether we use datasets with and without removing genes or proteins not detected by all technologies (fig. S2, D and E, respectively). These results suggest that rather than the absolute presence or absence of the different genes or proteins, the relative expression levels are more indicative of the corresponding anatomical region of the kidney. Correlation analysis of averaged log2 fold changes between all combined RNAseq and proteomic datasets suggests that similar entities are identified by different assays (Fig. 2D). The 0.61 correlation value that we obtain agrees with the canonical value across mammalian tissues as described in (20). However, our comparison is based on fold changes and not absolute mRNA and protein abundances. Hence, integration of multiple datasets increases the accuracy of the results since integrated RNAseq and proteomic datasets show a higher correlation than any individual RNAseq and proteomic datasets. Nevertheless, correlation coefficients are high between technologies where assays were conducted at different sites (Fig. 2E).
Imaging-based molecular data and nonspatial proteomic and transcriptomic assays together produce spatial marker expression signatures
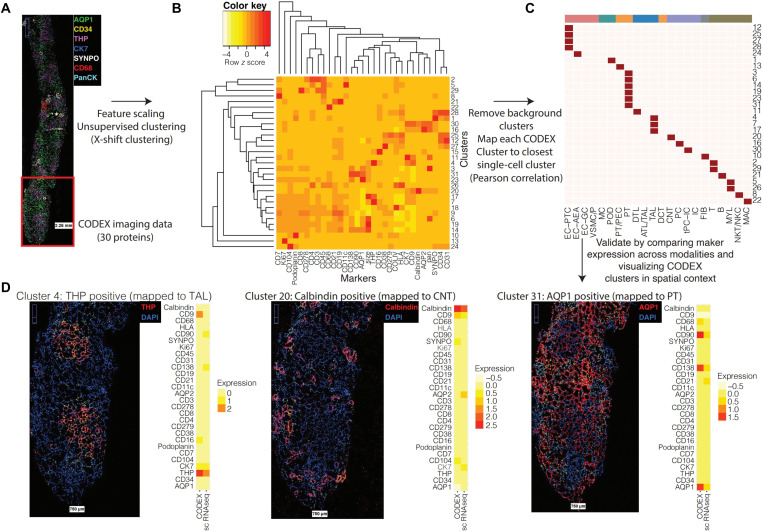
Imaging assays can provide the spatial specification of omics data, such as bulk proteomics (21), and help characterize the contextual framework for cell types inferred through dissociated cell transcriptomics; those with well-characterized markers can identify the spatial localization of individual cells, which can be independently identified from gene expression patterns. By analyzing the relationship between cells identified from sc/sn sequencing technologies and co-detection by indexing (CODEX) imaging of canonical markers across 27,236 cells, we establish the concordance between the assay types for independently identifying cell types and inferring molecular profiles for spatially localized cells (Fig. 3). We find a strong correspondence between the molecular profiles of cell types across data modalities (Fig. 3 and fig. S3). By mapping sc/sn expression profiles to either LMD segments or the independent imaging assays, we were able to further assign dissociated transcriptomic signatures to anatomical regions within the kidney. Using these comparisons, we can arrange different cell subtypes along the nephron and in the interstitium, allowing documentation of the order by which they encounter the glomerular ultrafiltrate in the reference state.
Fig. 3. Integration of single-cell and nucleus transcriptomic data with CODEX imaging data.
(A) CODEX imaging provides spatial localization for 30 proteins across 27,236 cells (spatial distribution of selected proteins visualized). (B) CODEX data were clustered using X-shift clustering to identify groups of cells expressing common subsets of protein markers. (C) Each CODEX cluster was mapped to the most similar transcriptomic cluster based on the Pearson correlation between the average scaled expression profiles. (D) Visualization of CODEX clusters in spatial context. Yellow dots indicate cells mapped to each cluster, and the side-by-side average expression profiles of the CODEX cluster and corresponding mapped transcriptomic cluster are shown.
Integrated pathway enrichment analysis enables identification of functional capabilities of different cell types of the kidney
After establishing the consistency between transcriptomic, proteomic, and imaging datasets, we used these integrated data to identify the cell type–specific functional pathways and network modules. We started by individual analyses of the sc/sn RNAseq datasets and by identifying the pathways inferred from the expressed genes (14, 15). Individualized analyses of sc/sn transcriptomic data ensure that these two related technologies do not computationally influence each other. In contrast to our integrated analysis of these datasets described above, the individual analyses used more relaxed quality control cutoffs, such as allowing up to 50% mitochondrial gene expression, so the cell subtype–specific gene expression obtained by the sc RNAseq dataset was based on 22,264 cells instead of 17,529 cells after quality control. The number of nuclei after quality control was the same with 17,659 nuclei. These dissociated cell analyses also allowed us to ascertain that all cell types could be observed independently of the method by which the reference tissue was obtained. We find that all major kidney cell types can be identified in nephrectomy, living donor biopsy, and transplant surveillance biopsy tissues based on the individual sc and sn RNAseq datasets (fig. S4, A and B, respectively). An exception to this finding is that immune cells were mostly identified only within the sc RNAseq dataset, while only one cluster of the sn RNAseq dataset that contained less than 1% of all nuclei was annotated to an immune cell type, i.e., macrophage (fig. S4B). Almost all cells identified in sc or sn RNAseq datasets in the individualized analyses were annotated to the same cell type as in the combined analysis (fig. S4, C and D, respectively), expressed cell type–specific marker genes (fig. S4, E and F, respectively), and mapped to the appropriate LMD segment as well (fig. S4, G and H, respectively).
Post hoc power analysis documents consistent cell type detection and affirms adequate sampling
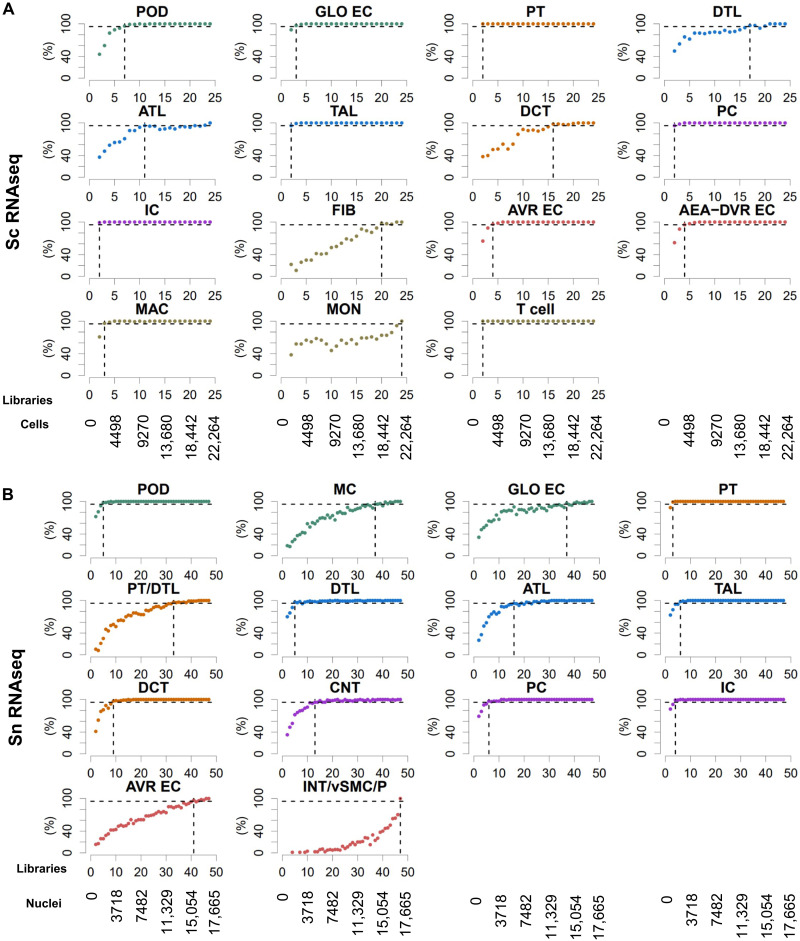
Before focusing on cell type–specific functions that we predicted from pathway enrichment analysis and module mappings, we evaluated how many reference subject samples need to be processed to obtain consistently reproducible results. Twenty-four and 47 libraries obtained from 22 and 15 subjects were subjected to sc (15) and sn (14) RNAseq (table S1), yielding 22,264 cells and 17,655 nuclei after quality control, respectively. We separately subjected both RNAseq datasets to a standardized sc and sn RNAseq analysis pipeline with and without random and progressive removal of libraries (fig. S5A). Our analysis pipeline automatically screens for an optimal clustering resolution to identify as many major kidney cells with high confidence as possible. Results obtained for the downsampled datasets were compared to those obtained for the complete datasets (fig. S5B). Our results indicate that for a consistent detection of podocytes (i.e., in at least 95% of all downsampled datasets with the same library counts), at least seven (6403 cells) or five libraries (1921 nuclei) are needed if subjected to sc RNAseq (Fig. 4A) or sn RNAseq (Fig. 4B), respectively. The observed higher identification rate by the sn RNAseq assay agrees with a previous report that compared sn and sc RNAseq results obtained from mouse kidneys (10). PT cells, thick ascending limb (TAL) cells, principal cells (PC), intercalated cells (IC), and T cells were always detected in the downsampled sc RNAseq datasets. Macrophages (MAC), glomerular endothelial cells (GO-EC), and two other endothelial cell (EC) types were consistently detected if three (2843 cells), three, or four libraries (3817 cells), respectively, were analyzed. In the sn RNAseq datasets, we consistently detected PT cells, TAL cells, distal convoluted tubule cells (DTL), connecting tubule (CNT) cells, principal cells, and intercalated cells in 3, 6, 9, 13, 6, and 4 libraries (1043, 2235, 3395, 4930, 2235, and 1464 nuclei), respectively. Descending limb cells were first detected in 95% of all downsampled datasets at a library count of five (1921 nuclei). For additional cell types, see Fig. 4 (A and B). In addition, our results suggest that the accuracy of sc or sn assignments to the selected cell types is relatively stable, as documented by the low number of cells that are assigned as different cell types or mapped to an unrelated tissue subsegment in the downsampled sc and sn datasets (fig. S5, C and D, respectively). Similarly, Pearson correlation between cell type–specific differentially expressed genes (DEGs) in the downsampled and complete datasets follow the same trend. These analyses establish the rigor with which we are able to assign pathways and physiological functions to the different cell types.
Fig. 4. Single-cell/nucleus transcriptomic post hoc power analyses show that nine libraries are sufficient to identify most major kidney cell types.
Subject libraries (or samples) were randomly and progressively removed from (A) the sc (24 libraries) and (B) sn (47 libraries) RNAseq to generate at maximum 100 non-overlapping random groups for the remaining samples. Sc and sn datasets were subjected to an automated data analysis pipeline (fig. S5A). To assign cell types to the identified clusters, we compared cluster-specific markers of each analysis with literature curated cell type–specific genes (fig. S5B). We counted how many analyses based on the same number of remaining libraries that have identified a particular cell type. Horizontal dashed lines mark the 95% plateau; vertical dashed lines indicate the lowest library quantity that allowed identification of a given cell type with a probability of 95%. See fig. S5 for complete post hoc power analysis results. See Fig. 2A for cell type abbreviations.
Pathway enrichment analysis and module identification helps understand molecular basis of physiological functions with cellular resolution
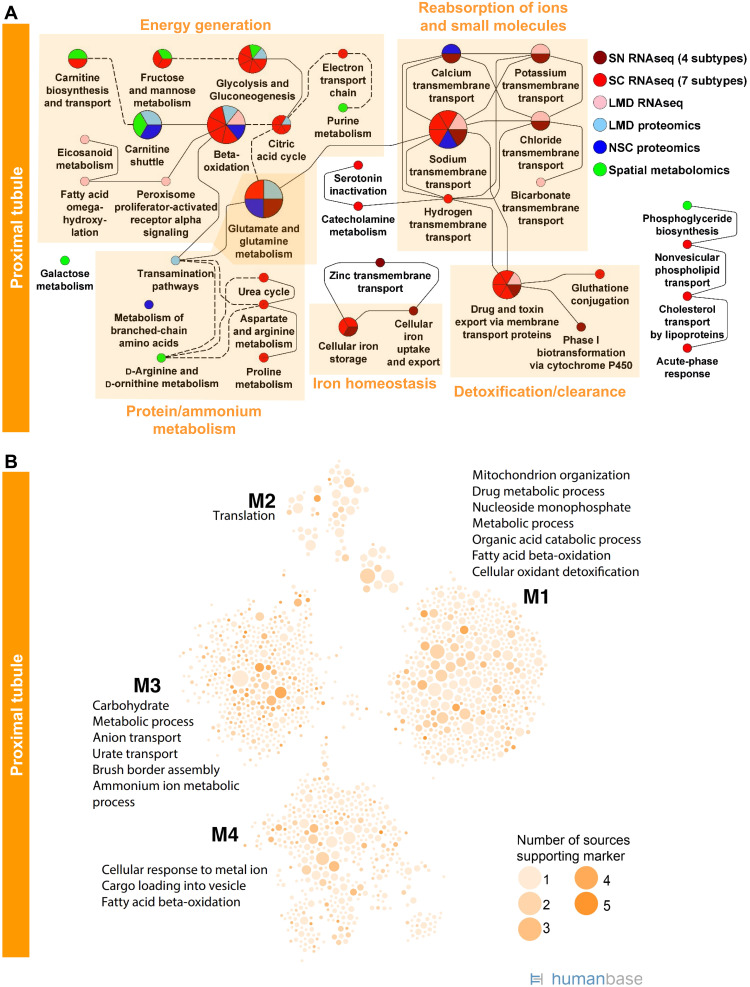
The top 300 significant gene and protein markers of each cell type or subtype and subsegment (table S5) were subjected to dynamic enrichment analysis using the Molecular Biology of the Cell Ontology (MBCO; table S7) (22). In many cases, less than 300 markers were significant (table S6), and we consequently used only those for our downstream analysis. In contrast to standard enrichment analysis, dynamic enrichment analysis is not limited to a single process but instead determines whether a set of experimental genes maps to multiple functionally related subcellular processes (SCPs). Resulting SCP networks can give rise to whole-cell physiological function. Nonglomerular and glomerular metabolites (table S8) were subjected to pathway enrichment analysis using MetaboAnalyst (fig. S6, A and B, respectively) (23) and predicted pathways mapped to MBCO SCPs. Similarly, we subjected significant mRNA and protein markers to community clustering in a kidney-specific functional network using HumanBase (table S9) (24, 25), which was constructed by integrating thousands of public genomic datasets using a regularized Bayesian framework to predict the probability that every pair of genes in the genome is related in a specific tissue context. Combining both network approaches, we created a detailed map of pathway activities in all major cell types in the kidney, which is described and discussed in detail within the “Cells of the kidney” section in Supplementary Information. As documented by the high metabolic, reabsorption and detoxification activity predicted for the PT cells (Fig. 5, A and B), our approach identified many well-known cellular activities for most kidney cell types and numerous previously unknown functions (figs. S7 and S16).
Fig. 5. Enrichment analysis of markers for PT and glomerular cells and segments predicts well-known cell functions.
(A) Marker genes and proteins of each PT cell subtype or subsegment were subjected to dynamic enrichment analysis using the MBCO. SCPs that were among the top seven predictions were connected by dashed lines, if their interaction was part of the top 25% inferred MBCO SCP interactions, and by dotted lines, if their functional relationship was curated from the literature. Figure S8 shows additional predicted SCPs involved in cell adhesion and translation. Metabolites associated with nonglomerular compartments were subjected to MetaboAnalyst enrichment analysis (fig. S6). Any pathway among the top eight predicted pathways that was predicted on the basis of metabolites specifically for that pathway was mapped to MBCO SCPs, if possible, and integrated into the PT SCP network. MBCO SCPs carnitine shuttle and carnitine biosynthesis and transport were added to the predicted MetaboAnalyst pathways since four and two involved metabolites were among the nonglomerular metabolites (see Methods for details). (B) HumanBase analysis of PT marker genes and proteins.
Using a similar approach to our post hoc power analysis, we investigated the robustness of the SCP-identified cell biological functions by randomly downsampling libraries from the sc/sn datasets. Investigation of the ranks that were obtained for downsampled SCPs allows estimation of SCPs that are consistently predicted and are probably describing core biological functions. In case of the PT cells, most of the consistently identified SCPs by the sc (fig. S17A) or sn RNAseq data (fig. S17H) are related to cellular metabolism and energy generation, reabsorption, and detoxification. In the case of the podocytes, the consistently identified SCPs are involved in cell-cell and cell-matrix adhesion. Among other cell adhesion pathways, we consistently identify the two SCPs “tight junction organization” and “adherens junction organization” based on both the sc (fig. S17B) and sn RNAseq assay (fig. S17I). These results document the central importance of the glomerular slit diaphragm that is described as a specialized form of both tight junctions (26) and adherens junctions (27). Figure S17 also shows the results obtained for the other cell types.
Comparison of variation of oxygen supply and inferred levels of energy metabolism help identify sites vulnerable for kidney injury
To identify energy generation pathways in the different cells along the renal tubule of the nephron, we generated a focused ontology of metabolic pathways (fig. S18A). Using this ontology for enrichment analysis allowed us to distinguish between aerobic and anaerobic as well as catabolic and anabolic pathways (fig. S18B). To rigorously define the groups, we used a rule-based analysis that ignores predicted parent pathways if a given child that contains the reactions specific for the parent pathway was not among the predictions. For example, the pathway anaerobic glycolysis was only considered if the pathway lactate dehydrogenase was predicted as well. See Methods for details.
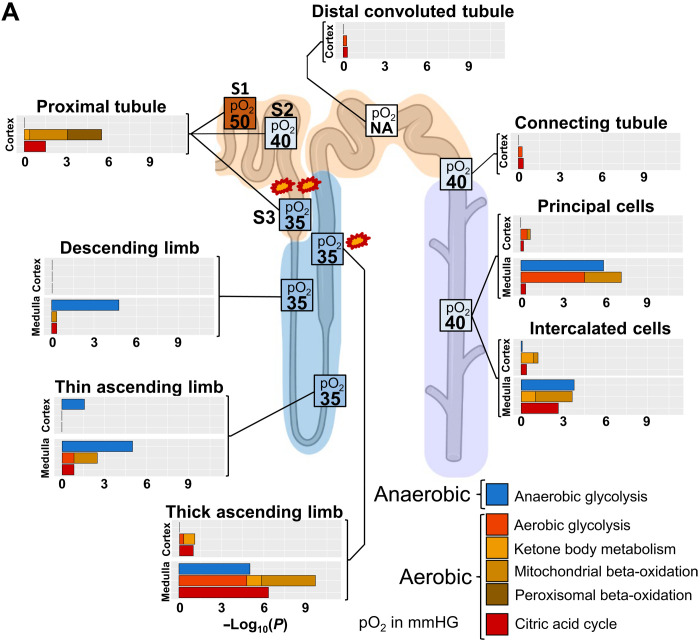
Expression patterns of different pathways involved in aerobic and anaerobic energy generation (Fig. 6 and table S11) were mapped to the varying levels of oxygen availability in different regions of the nephron (28). This comparison helped us identify regions with high susceptibility toward hypoxia-induced kidney injury. Missing capability for anaerobic energy generation as predicted from the human transcriptomics data (Fig. 6 and fig. S18B) and observed in animal experiments (29) combined with already low pO2 in the healthy kidney suggests S3 segment of the PT as a primary site for hypoxia-based injury. This reasoning agrees with experimental observations (28). High levels of capacity for aerobic energy generation activity in the medullary TAL (mTAL), a region with low oxygen supply, are complemented by high capacity for anaerobic energy generation, as also documented in animal experiments (29). When the output of the anaerobic energy generation is depleted, mTAL can become a second site of kidney injury. Our conclusion agrees with the experimental observation that mTAL injury during hypoxia depends on epithelial transport activity (28). It can be readily seen that molecular profiles of metabolic pathways in our atlas provide a basis for understanding and predicting kidney injury due to hypoxia in the clinical setting.
Fig. 6. Aerobic and anaerobic energy generation profiles and oxygen supply accurately highlights sites of hypoxia-induced injury.
To compare energy generation profiles with experimentally determined oxygen supply in the different nephron regions, we generated an ontology that allows the separation of aerobic and anaerobic pathways involved in energy generation. Enrichment analysis of cell type, subtype, and subsegment marker genes with this ontology predicts high dependency of PT cells on aerobic energy generation, suggesting S3 as a primary injury site during hypoxia (marked by two explosions) because of its low oxygen supply under basal conditions. Enrichment results predict a high aerobic energy generation activity for the mTAL that can be compensated by anaerobic energy generation. In combination with the already low oxygen saturation in that segment under normal conditions, our results suggest that mTAL is the second, although less likely, injury site during hypoxia (marked by one explosion). Enrichment results are combined from those shown in fig. S18B. Numbers in boxes indicate pO2 in mmHg taken from (28). NA, not available.
Predicted reabsorption capacities agree with experimental reabsorption profiles and provide a molecular basis for differential physiological activity along the nephron
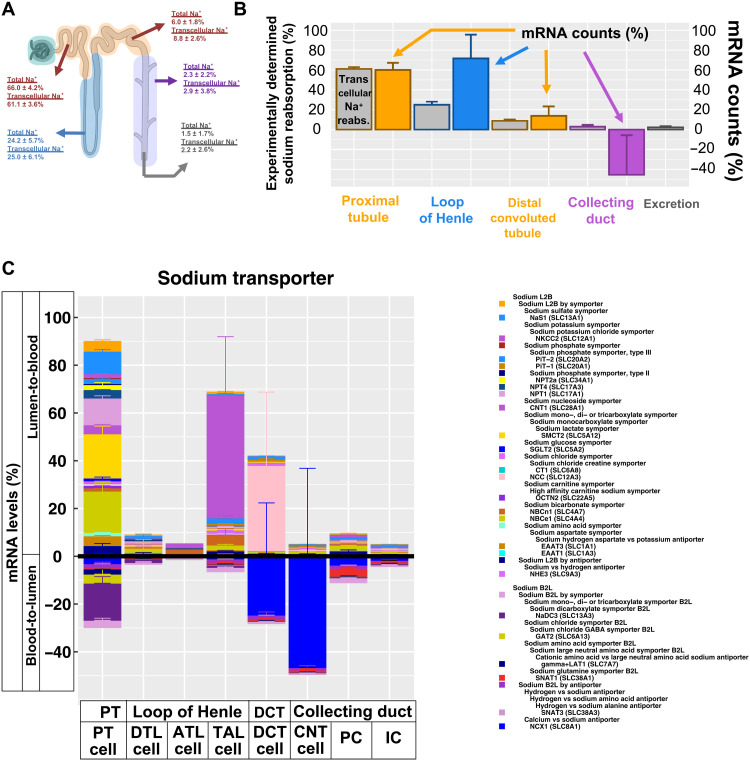
Physiological experiments allow us to determine what percentage of a filtered ion or small molecule is reabsorbed in a particular nephron segment or excreted into the kidney pelvis and ureter. Sodium is the central ion for all reabsorption activities; hence, we focused on sodium reabsorption as an example of how a tissue atlas with cellular resolution can help us understand physiological homeostasis. We averaged sodium reabsorption profiles obtained from four different standard physiology and school textbooks (30–33) (Fig. 7A). Since we estimated that 37% (34) and 30% (35) of the total sodium in the PT and loop of Henle are respectively reabsorbed by passive paracellular mechanisms, we removed those percentages from the reabsorption profiles. Modified experimental values were renormalized (Fig. 7A), and an ontology for sodium transport mechanisms across the plasma membrane was generated (fig. S19, A to C) to predict sodium reabsorption capacities from transcriptomic data. Using our sn RNAseq dataset (14) and two additional sets from the literature (16, 17), we calculated the sum of all mRNA levels mapping to sodium transporter genes across all cells in each segment. Averaged results compared well with the experimentally measured reabsorption profiles both without (fig. S19D) and with (Fig. 7B) the removal of paracellular reabsorption. As the spare capacity for sodium reabsorption is most likely to be within the loop of Henle (36–39), our data show substantial agreement between calculated capacities and the experimentally measured reabsorption profiles.
Fig. 7. Predicted sodium transport capacities match with experimentally determined reabsorption profiles.
(A) Estimated transcellular sodium reabsorption before and after removal of estimated paracellular sodium reabsorption from experimentally determined total sodium reabsorption profiles. (B) Using our and two other sn RNAseq datasets, we calculated the sum of all mRNA counts that mapped to genes involved in sodium lumen-to-blood (L2B) and blood-to-lumen (B2L) transport for each segment of the renal tubule. Net reabsorption capacities for sodium (colored bars) were determined by subtracting both sums and compared to experimentally determined transcellular sodium reabsorption (gray bars). (C) L2B and B2L cell type–specific transport mechanisms for sodium are visualized above and below the abscissa, respectively. Error bars document SEs. Parent-child relationships are documented in the legend, where children SCPs are written below their parent SCPs and shifted to the right. To prevent double counting, we removed any mRNA levels from each parent SCP that are already visualized as part of its child SCPs. Parent SCPs missing in the diagram were added to the legend next to an uncolored box for a proper documentation of the SCP hierarchy. In case of multiple parent SCPs, we only show one parent. Stacked bar diagram colors are in the same or reverse order as in the legend for L2B and B2L, respectively.
Some nephron segments, such as the loop of Henle and the collecting duct, contain multiple cell types with different reabsorption mechanisms (36, 40); hence, we mostly focus on cell type–specific transport mechanisms (Fig. 7C). We define mRNA levels mapping to transporters involved in blood-to-lumen transport (table S12) as negative to account for the opposite direction when compared to lumen-to-blood transport. Adding all transporter mRNA levels of each segment gives the net transport capacities described above (Fig. 7B). Regarding individual transport mechanisms, we highlight two details here (Fig. 7C): In the PT, we find high capacity for sodium-coupled transport of multiple molecules, such as amino acids, organic anions, and sugars. In agreement, sodium reabsorption in the PT is the driver for the absorption of many metabolites (30–33). mRNA encoding NKCC2 is the predominant species of sodium transporter in the TAL cells, in agreement with the large contribution of NKCC2 to sodium reabsorption in the TAL (36). We acknowledge that the epithelial Na channel ENaC (SCNN1) plays a major role in sodium absorption in the connecting tubule and collecting duct, where it reabsorbs ~5% of the filtered load of sodium (18), but our analysis focuses only on transporters and excludes channels. This probably explains our prediction of a higher blood-to-lumen versus lumen-to-blood sodium transport in the collecting duct. Calculation of the reabsorption capacities after inclusion of our sc RNAseq dataset along with sn RNAseq data slightly decreases the match with the physiological reabsorption profiles and mRNA levels (fig. S19E) but still gives similarly favorable results (fig. S19F). The decrease is mainly due to the high mRNA levels associated with the basolateral amino acid transporter LAT1 that exports cationic amino acids into the blood in exchange for large neutral amino acids and sodium (fig. S19F) (41).
In summary, the cell-level atlas provides a detailed picture of sodium reabsorption that agrees with experimental reabsorption profiles and was up to now not attainable. Similarly to sodium, we find that glucose transport along the nephron agrees with glucose transporter mRNAs (fig. S19, G and I) and is mainly mediated by SGLT2 (fig. S19, H and J), the glucose symporter responsible for over 80% of filtered glucose reabsorption in the PT (42).
DISCUSSION
The integration of multiple types of omics data allows us to describe in depth multiple SCPs and pathways at cell-level resolution. High cross-correlation coefficients for independent technology pairs and the successful unbiased clustering of segments affirm the high quality of our data, as technological bias can be overcome by a relatively simple algorithm. From such an integrated description, we can hypothesize key functions that can help define disease states when perturbed. These disease states could have convergent clinical phenotypes, although the underlying molecular changes are different. Thus, our detailed characterization of the reference state described here can provide a new framework for molecular classification of kidney diseases. For example, identification of both mitochondrial and peroxisomal β-oxidation and carnitine transport and local biosynthesis pathway in PT cells suggests how individual variations in any of these SCPs can contribute to the effects of kidney injury, including fibrosis (6). Thus, a convergent clinical phenotype can arise from very different molecular changes related to energy metabolism. Mapping these changes in individual patients may allow for better classification of disease states.
Integrated view of kidney cellular functions
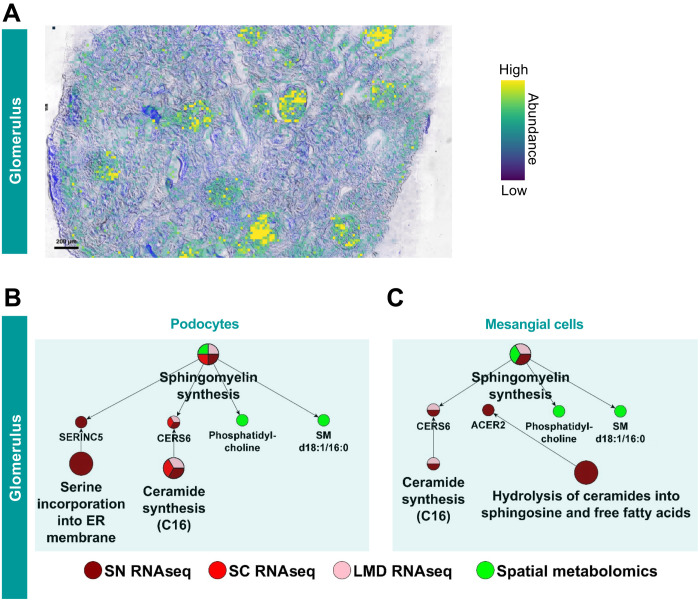
One advantage of the presented multiomics data integration strategy is the ability to infer how different classes of biomolecules may enable complex multicellular functions leading to potentially predictive biomarkers. Spatial metabolomics identifies N-palmitoylsphingomyelin (SM d18:1/16:0) as a spatial correlate of glomerular kidney segments (correlation coefficient > 0.9; Fig. 8A), as described previously (43, 44). To identify the cell types involved in its synthesis, we screened all glomerular cell types for expression of genes involved in ceramide, sphingomyelin, and sphingosine metabolism (45, 46). Transcriptomics identify SERINC5 (serine incorporator 5) and CERS6 (ceramide synthetase 6) as specifically expressed in podocytes or in podocytes and mesangial cells, respectively (Fig. 8B). SERINC5 incorporates serine into the membrane of the endoplasmic reticulum, making it available for ceramide and phosphatidylserine synthesis (45). CERS6 is identified by all transcriptomic assays in the podocyte. CERS6 is one of six ceramide synthases that converts sphingosine and acyl–coenzyme A (acyl-CoA) into ceramide. In contrast to the other five ceramide synthases, it has a high substrate specificity toward palmitoyl-CoA (C16:0) (46), thereby generating ceramides with the correct acyl chain length to be converted into SM d18:1/16:0. Only one technology, sn RNAseq, shows CERS6 to be expressed in mesangial cells as well, albeit at a lower level of significance (rank 293 in mesangial cells versus 116 and 126 in podocytes; fig. S20).
Fig. 8. Podocytes are the synthesis site for glomerular SM d18:1/16:0.
(A) Matrix-assisted laser desorption/ionization mass spectrometry imaging reveals that the ion distribution of SM d18:1/16:0, [M + Na]+, correlates with the glomerular kidney regions. (B) Podocytes express two genes involved in sphingomyelin synthesis including the genes CERS6 that is identified by both sn and sc RNAseq datasets and the LMD RNAseq dataset. CERS6 specifically generates C16 ceramides, the direct precursor for SM d18:1/16:0. (C) CERS6 is also expressed in mesangial cells, although only detected by the sn RNAseq dataset. Glomerular expression of the gene SERINC2 is detected by the LMD RNAseq assay.
Transcriptomic datasets also predict the expression of enzymes involved in sphingomyelin synthesis in nonglomerular cells. In general, only one specific enzyme of this pathway is expressed per cell type. Consequently, podocytes (and mesangial cells) are the most likely synthesis site(s) for this particular sphingomyelin, as demonstrated by the spatial metabolomics data. Altered metabolism of multiple sphingolipids including sphingomyelin and its metabolites is observed in several glomerular diseases (47). Cellular sphingomyelin is predominantly localized at membranes derived from the trans-Golgi and plasma membrane (48). It is involved in multiple functions, including cell signaling, lipid rafts formation (49), caveolar endocytosis (48), and apoptosis (50). In addition, it has long been known that sphingomyelin (as part of lipid rafts) is enriched in desmosomes (49, 51) and tight junctions (52). Given the central importance of foot process interactions between neighboring podocytes, the potential importance of this metabolite in making different types of cell-cell contacts and the enzymes involved in its biosynthesis in podocytes can be readily appreciated. Five technologies that focus on genes and proteins identify cell-cell/cell-matrix adhesion as podocyte key functions, and the sixth technology identifies a specific metabolite that we consequently predict would be involved in the same key functions, thus providing integrated support for the role of sphingomyelins in podocyte cell-cell interactions. Sphingomyelin with very long (C24) acyl chains, but not palmitoyl (C16) sphingomyelin, is the predominant species associated with tight junctions (52). Tight and adherens junctions are morphologically only observed in developing or diseased podocytes (53), while healthy podocytes form the glomerular slit diaphragm (53). These observations indicate that we may be able to use decreased levels of C16 SM and increased long-chain SM as a predictive biomarker for disease progression even before changes in glomerular filtration rates.
The distribution of mRNAs for the various transporters mapped in this study also agrees well with the known levels of reabsorption activities identified in physiological experiments, suggesting that the transporter mRNA levels along the nephron can provide an indication of transcellular transport capacity for individual cell subtypes. While our results are confirmatory of known physiology, we note that only through our atlas, one will now have the framework to investigate the molecular mechanisms of renal dysfunction in the context of individual cell subtypes and to generate hypotheses that can be directly tested using physiological approaches.
The value of a reference tissue atlas
There have been several valuable studies focused on sc transcriptomic analyses of human kidney tissue (6, 7, 9, 11) in the context of different diseases. Although each of these studies have provided substantial insight into disease processes, their mapping of reference kidney tissue has often been limited to a single source of tissue and focused on cell types relevant to the disease of interest. In contrast, in this study, we have studied only human kidney specimens without disease; we use multiple omics technologies, including regional and sc/sn transcriptomics, proteomics, and spatial metabolomics in conjunction with imaging assays to obtain an extensive, near-comprehensive spatial map of the human kidney at the single-cell resolution. Our experiments identify all known major cell types in the kidney and recapitulate several known subtypes. In addition, we can identify different types of endothelial cells, vascular smooth muscle cells, fibroblasts, and different circulating immune cells. Together, these different cell types and subtypes provide a detailed picture of the cellular and molecular composition of the human kidney.
Here, we have extended our bioinformatics analyses beyond ranked lists of genes (table S13) and associated pathways to identify coherent networks of pathways that give rise to function. We have developed our model in a systematic manner such that we identify key functions for each cell type and subtypes. These physiological roles identified through pathways and marker gene lists enable the development of a multiscale atlas that connects expression patterns to whole-cell and tissue-level physiological functions. The proteomics and metabolomics and the spatial imaging data from CODEX allows for the mapping of sc/sn RNAseq–based cell type identification to canonical cell type markers and appropriate spatial regions. This exercise provides independent orthogonal validation of both the cell type and the spatial localization within the nephron (Figs. 2A and 7).
Limitations and future refinements
Several limitations of our study should be noted. Not all cell types are identified with the same certainty and depth, although our cell types agree well with other published studies (6, 8, 11, 17). In addition, our sc/sn RNAseq assays contain a relatively low number of cells compared to some of the contemporary studies. However, this is compensated for by use of multiple omics and other technologies, all of which provide convergent conclusions in the identification of cell types.
As seen in our three-dimensional (3D) imaging assays, the spatial distribution of different cell types within the kidney varies substantially; therefore, it is expected that the composition of sc/sn RNAseq signatures also varies. When fewer cells are detected, typically, we also identify a lesser number of marker genes and SCPs. Currently, we do not know whether the relative number of cells that we detect in the sc/sn RNAseq assays reflect the proportions in situ. Further sampling within the KPMP will likely resolve this issue. It is also likely that further sampling will also enable mapping of SCPs in every single-cell type with similar certainties. Nevertheless, for all the major cell types of the kidney, post hoc power analyses indicate that we have sufficient power to map the core cell-level functions from SCPs generated from orthogonal assays. In addition, the number and functional identity of major cell subtypes need to be further studied. Currently, subtype identification is based on statistical reasoning used for the clustering algorithms. How many subtypes there are for a given kidney cell and whether these subtypes exist in all individuals require further studies using confirmatory spatial imaging technologies. Studies in other tissues, such as brain (54) and heart, have identified multiple subtypes of paraventricular interneurons and ventricular myocytes. Hence, it is likely that kidney cell types may also contain major subtypes. Despite these limitations, this study provides a detailed functional view of a kidney map at the single-cell resolution, which can be used to understand major aspects of kidney physiology, as demonstrated by the two examples described above.
Information becomes knowledge only when it is deliberately and systematically cataloged such that new cohesive insights can readily be drawn, as shown above for sphingomyelin-related functions in podocytes. Ontological classification could enable the generation of new insights, especially when it involves multiscale relationships between molecules, cell types and their subtypes and tissue level physiological function. In addition to the integrated analytics presented here, KPMP is also building a community-based Kidney Tissue Atlas Ontology (55) that may aid such relationship building. Together, the final knowledge environment and the kidney tissue atlas constructed by KPMP, which is available at www.atlas.kpmp.org, should be able to help molecularly characterize cellular types and subtypes in the kidney, improve patient care by providing new disease classifications, and may ultimately lead to new patient-specific therapeutic approaches.
METHODS
Omics and imaging assays used within KPMP target different types of molecular components with different resolution, sensitivity, and precision. An important function of the KPMP Central Hub is to integrate the different types of data using a set of analytical techniques. This process is summarized in Fig. 1. Throughout the paper, we consistently use the same continuous color code to identify different assays or cell types. The experimental assays that generate the raw data and all their technical details including standard operating procedures are detailed under “Data generation and initial analysis” and publicly released with all their technical details and version-controlled release dates on the KPMP protocols.io page (www.protocols.io/workspaces/kpmp/publications).
Identification of DEGs, proteins, and metabolites
We analyzed data from four types of transcriptomic, two proteomic, one imaging-based, and one metabolomic tissue interrogation assays. The pilot data presented for each assay comprises 3 to 48 different datasets that are obtained from 3 to 22 subjects (table S1). Kidney tissue was procured from a spectrum of tissue resources including from unaffected parts of tumor nephrectomy specimen (n = 38), living donor preperfusion biopsies (n = 3), diseased donor nephrectomies (n = 5), and normal surveillance transplant (n = 5) and native kidney biopsies (n = 4). Single-cell and sn transcriptomics clusters were obtained from previous analyses (14, 15). Within each assay, we generated lists of DEGs, differentially expressed proteins (DEPs), and metabolites that describe those genes, proteins, or metabolites that are up-regulated or enriched in a particular sc cluster, sn cluster or kidney subsegment, if compared to all other clusters or subsegments.
For pathway enrichment analysis and module identification, cluster-specific DEGs were obtained from published analyses from PREMIERE Tissue Interrogation Site (TIS) (Michigan, Princeton, Broad) sc RNAseq (15) and UCSD/WU TIS sn RNAseq (14) datasets. We excluded the clusters PT cells 3 and principal cells 2 from the sn RNAseq dataset since these clusters showed an inflammatory stress response. Similarly, we excluded the cluster “Unk” from the sn and the clusters “Pax8positivecells” and “LOH/DCT/IC” from the sc RNAseq assays. LMD RNAseq and proteomics (OSUIU), NSC proteomics (UCSF) and spatial metabolomics (UTHSA-PNNL-EMBL) datasets were individually processed as described in the Supplementary Materials. Only DEGs and DEPs that indicate genes and proteins that are higher expressed in a particular cell subtype, type, or segment were considered for all analyses.
Ranking of DEGs and DEPs
In the case of the DEGs and DEPs that were used for dynamic enrichment analysis, (22) module identification, (25) and post hoc power analysis, sn and sc DEGs were first ranked by adjusted P value and then by decreasing fold changes (i.e., fold changes were used as a tiebreaker). Top ranked 300 entities with a maximum adjusted P value of 0.05 were subjected to downstream analysis. Similarly, DEGs and DEPs obtained for each kidney subsegment based on LMD bulk RNAseq (19) or LMD and NSC proteomics were ranked first by P value and decreasing fold changes, and the top ranked 300 DEGs and DEPs with maximum nominal P value of 0.05 were subjected to pathway enrichment analysis or module detection (see below).
Dynamic enrichment analysis
Top DEGs and DEPs for each podocyte cluster/glomerulus, PT cell cluster/tubulointerstitium, and principal cell cluster/collecting duct subsegment were separately subjected to dynamic enrichment analysis using the MBCO (version 1.1) level-3 SCPs (22) that can be found at github.com/SBCNY/Molecular-Biology-of-the-Cell and www.mbc-ontology.org. The annotated interconnected hierarchy of MBCO is enriched using a unique algorithm that infers weighted relationships between functionally related SCPs. For all analyses, we considered the top 25% weighted relationships. Dynamic enrichment analysis uses the top relationships to generate context-specific higher-level processes by merging functionally related SCPs that contain at least one DEG or DEP. The context-specific higher-level SCPs contain all annotated genes of the original SCPs and are added to the annotated ontology to generate a context-specific ontology. The context specific ontology at this point contains single and merged SCPs. This list is then used for enrichment analysis of the DEPs or DEGs using Fisher’s exact test. All SCPs that are among the first seven predictions are connected on the basis of the top inferred relationships using solid lines. All networks for a particular cell type and the corresponding segment were merged, and each SCP was color-coded according to the source assay(s) that initiated its dynamic enrichment. SCPs predicted by multiple assays contain multiple slices that are color-coded accordingly. SCP size is determined by the number of assays that identified a particular SCP. Multiple subtypes or a particular cell type (e.g., PT cells) are all color-coded by the same assay specific color. If an SCP was predicted for more than one subtype, it contains multiple slices colored with the assay specific color. SCPs predicted by different assays for the same cell type or corresponding segment were connected on the basis of the top 25% inferred MBCO relationships using solid lines. Additional well-known functionally related SCPs were connected using dashed lines.
We used the right-tailed Fisher’s exact test to calculate the likelihood of obtaining the observed or a higher overlap between a list of DEGs/DEPs and a list of genes/proteins annotated to a particular SCP. To calculate this likelihood, we consider which genes or proteins have a chance to be identified as differentially expressed. Only genes/proteins that are detected by a particular assay and are statistically analyzed for differential expression can be identified as DEGs/DEPs. Consequently, only these genes/proteins are considered as the experimental background set for the Fisher’s exact test. Similarly, the ontology background set only contains genes that have a chance to be assigned to a given SCP. In the case of the sc (15) and sn (14) RNAseq datasets, all genes that are part of the UMI (unique molecular identifier) read count matrices comprise the experimental background genes. In the case of the LMD bulk RNAseq and the LMD and NSC proteomics datasets, the experimental background genes/proteins were all genes/proteins that were statistically analyzed for differential expression (tables S2 to S4, respectively). MBCO contains an SCP that is labeled “Background genes” and contains all genes that were identified during its population via text mining. The intersection of the experimental and ontological background genes/proteins is called background genes/proteins and is different for every assay and ontology combination. For additional statistical accuracy, we removed all genes and proteins that were not part of the background genes/proteins from the lists of DEGs, DEPs, and SCP genes before each enrichment analysis.
Module detection
In parallel to enrichment analyses, we also performed another network-based pathway enrichment technique, identifying modules of cell type–specific marker genes within the kidney-specific functional network using the HumanBase interface (hb.flatironinstitute.org). For each cell type, module detection was performed using all cell type–specific DEGs detected by sc and sn RNAseq (adjusted P value < 0.01) and segment-specific DEGs and DEPs detected by the other four technologies (nominal P value < 0.01). Module detection is a network-based approach described by Krishnan et al. (25), and construction of the functional networks is described by Greene et al. (24). In contrast to the prior knowledge-based MBCO networks, the kidney-specific functional network is constructed using a data-driven regularized Bayesian framework based on the information in thousands of datasets, which include coexpression, transcription factor binding, protein-protein interactions, and other data types. Modules are detected using a community clustering algorithm based on connectivity between genes in the kidney-specific functional network, and enrichment analysis is subsequently performed to identify functional enrichments in each module.
Enrichment analysis for metabolites
All glomerular and nonglomerular metabolites that were identified for the three subjects were merged and subjected to pathway enrichment analysis using MetaboAnalyst (23) with the selections: hypergeometric test, relative-betweenness centrality, Homo sapiens (Kyoto Encyclopedia of Genes and Genomes), website version 3/22/2021. We analyzed which metabolites were part of the top eight predicted metabolic pathways. We removed those pathways among the top eight predictions that were predicted on the basis of the metabolites that are shared substrates in multiple pathways and consequently unspecific for the identified pathway [i.e., we ignored the glomerular pathways “linoleic acid metabolism,” “alpha-linoleic acid metabolism,” “glycosylphosphatidylinositol-anchor synthesis,” and “arachidonic acid metabolism” that were predicted on the basis of the lipids “phosphatidylethanolamine” and “phosphatidylcholine” and the pathway “phenylalanine, tyrosine, and tryptophan biosynthesis” that was predicted on the basis of the central precursor “3-(4-hydroxyphenyl)pyruvate”]. We mapped the kept MetaboAnalyst pathways onto MBCO pathways whenever possible; if those pathways did not have a corresponding MBCO pathway, then the original pathway names were preserved. Since the nonglomerular metabolites contained multiple carnitine derivates, we added the MBCO pathways “carnitine shuttle” (based on l-acetylcarnitine, malonyl-carnitine, l-palmitoylcarnitine, and l-carnitine) and “carnitine biosynthesis and transport” (based on l-carnitine and 3-dehydroxycarnitine) to the pathways predicted from spatial metabolomics, assigning the ranks 9 and 10, respectively.
Integration of single-cell/single-nucleus transcriptomics
In contrast to bulk mRNA sequencing, where the gene expression measurements reflect an average across all captured cell types, sc or sn mRNA sequencing allows the measurement and comparison of comprehensive gene sets obtained from individual cells. This approach enables mapping of cellular heterogeneity with high throughput. In the first phase of the project, three KPMP tissue interrogation sites (TISs) performed this approach to generate sc/sn expression data from normal adult kidney tissue. In addition to locally acquired kidney tissue samples, each TIS also used a set of common KPMP pilot tumor nephrectomy tissue samples to generate the expression data. Sc transcriptomic data was produced by PREMIERE (24 libraries from 22 subjects) (15) and UCSF (10 libraries from 10 subjects), whereas the sn data were made by UCSD (47 libraries from 15 subjects) (14). Following is a brief description of the integration of the data from the three sites.
Data from each site were first processed using the Seurat 3.0 R package (56). As a quality control step, nuclei/cells with less than 500 and more than 5000 features and more than 20% mitochondrial genes were removed, yielding 17,529 and 13,130 cells along with 17,657 nuclei. The processing steps included normalization and identification of highly variable genes. We then removed potential doublets using DoubletFinder (57) from each dataset. Next, we used the integration algorithm embedded in the Seurat R package to perform combined analysis of sc/sn transcriptomic data. The integration algorithm first identified a set of anchor genes in each processed dataset. These anchor genes were then used to harmonize the datasets. The downstream process included scaling, principal components analysis, batch integration using harmony, dimensionality reduction using uniform manifold approximation and projection, and unsupervised clustering. The clustering was performed at low resolutions (clustering granularity of 0.5). Enriched genes for each cluster compared to all other clusters were identified using the Wilcoxon rank-sum test.
Integration of single-cell, single-nucleus, and laser capture microdissection bulk transcriptomics
To integrate sc sequencing, sn sequencing, and LMD bulk transcriptomic datasets, we first determined the overlap between genes identified both in the LMD dataset and in the corresponding single-cell transcriptomic dataset. From this set of shared genes, we restricted further analyses to a subset of genes showing variable expression in the single-cell dataset. We then computed the Pearson correlation between each individual cell in a scaled single cell/single nucleus dataset and the LMD transcriptomic dataset. For this correlation, we used the logarithmized mean fold change that was obtained by dividing the average expression of each gene within a subsegment by the average expression of the same gene within all other subsegments. Using this approach, we can assign each cell to the appropriate LMD segment that shows the highest correlation value. To evaluate the overall segment assignments for individual cell clusters, we examine the normalized distribution of cells assigned to each LMD segment within a given single-cell cluster and present this as a normalized heatmap that represents overlap between different transcriptomic assays.
Proteomic-transcriptomic coexpression analysis
LMD and NSC proteomic datasets identified protein expression in two kidney subsegments: glomeruli and tubulointerstitium for LMD and glomeruli and PT for NSC. Here, we did not combine sequencing and proteomic results of multiple subjects to generate DEGs and DEPs but compared the results obtained for each individual person. Because only one dataset per segment was generated from each individual person by the LMD and NSC technologies, we could not calculate P values in this analysis. Furthermore, both proteomic technologies only generated results for two subsegments, i.e., the glomerular and PT segments for NSC proteomics and the glomerular and tubulointerstitial subsegments for the LMD proteomics. Consequently, we collectively calculated the fold changes between podocyte/glomeruli and PT/tubulointerstitial cells or subsegments for each individual subject.
For the sc and sn transcriptomic datasets, we identified technology-, subject- and cell type–specific gene expression, using the “Average Expression” functionality embedded in the Seurat R package (RNA assay and counts slot) on the integrated single cell and nucleus RNAseq data analysis described above (fig. S2). For each technology, we characterized all genes/proteins that were identified in at least one cell type or subsegment of at least one subject and defined these genes/proteins as a technology-specific background set. The intersection of all background sets was defined as the set of common genes/proteins. Subject-specific podocyte or glomerular gene and protein expression was calculated by dividing gene and protein expression in podocytes, or glomeruli, by gene and protein expression in PT cells or PT/tubulointerstitial subsegments, after adding 1 to prevent division by 0. Ratios were inverted to describe PT/tubulointerstitial-specific gene expression. Log10 absolute expression values and log2 ratios of all genes/proteins or all common genes/proteins were subjected to pairwise correlation, followed by hierarchical clustering. Log2 ratios were averaged over each subject within each technology and pairwise Pearson correlation coefficients were determined between the different technologies using the set of common genes. Mean log2 ratios were averaged across the four RNAseq platforms and the two proteomic platforms, followed by determination of the Pearson correlation coefficient using the set of common genes.
Comparison of cell type–specific imaging and transcriptomic expression data
Clustering of the CODEX data was performed using 31 features (size and 30 protein markers: AQP1,CD34,THP,CK7,CD104,CD7,podoplanin,CD16, CD38, CD279, CD4, CD8, CD278, CD3, COLIV, AQP2, CD11c, CD21, CD19, CD138, CD31, CD45, Ki67, SYNPO, CD90, HLA-DR, CD68, CD9, pan-CK, and calbindin) using the most recent version of the VorTeX software (26 April 2018 release). Before clustering, cells were filtered to only those with QC parameter of 1, and each feature was scaled to have mean 0 and SD of 1 across all remaining cells. X-shift clustering was performed using parameters as specified by Black et al. (58). The clustering with K (number of neighbors for the density estimate) of 60 (31 clusters) was selected using the elbow point method. The normalized intensities of each marker were averaged within each cluster to generate per-cluster profiles and three clusters corresponding to background or noisy signal were removed. To map between the CODEX clusters and the integrated sc/sn dataset, all features were used except for size, pan-CK, and COLIV. Integrated sc and sn data were normalized and scaled, and the average expression value of each marker was determined for each sc cluster. Genes in the sc data were subsetted to genes matching CODEX features (AQP1, CD34, UMOD, KRT7, ITGB4, CD7, PDPN, FCGR3A, CD38, PDCD1, CD4, CD8A, ICOS, CD3D, AQP2, ITGAX, CR2, CD19, SDC1, PECAM1, PTPRC, MKI67, SYNPO, THY1, HLA-DRA, CD68, CD9, and CALB1). The Pearson correlation between each CODEX cluster and each single-cell cluster was computed, and each CODEX cluster was mapped to the single-cell cluster with the maximum Pearson correlation.
Post hoc power analysis
The sc and sn RNAseq datasets were obtained from 22 and 15 subjects, respectively, whose samples were sequenced in 24 and 47 libraries (table S1). We used these datasets to assess the reproducibility and reliability of both assays in a post hoc power analysis. This analysis compares results by the complete datasets with the results by down-sampled datasets where libraries are randomly and progressively removed from the complete data.
Both complete datasets were separately subjected to a standardized Seurat pipeline for the identification of sc or sn clusters and DEGs (fig. S5A). Nuclei and cells with less than 400 or 500 features, respectively, and more than 5000 features as well as more than 50% mitochondrial genes were removed. “SCTransform” was used for data normalization and scaling (based on top 2000 features), followed by principal components analysis. The first 30 principal components were used for dimensionality reduction before identifying cell neighborhoods using the Seurat functionality “FindNeighbors.”
Our analysis pipeline now searched for an optimal resolution that identified as many predefined cell types as possible. The predefined cell types included all major kidney cell types for both sc and sn RNAseq datasets, while we excluded the immune cell types in case of the single nucleus datasets. Our pipeline identified clusters using the Seurat functionality “FindClusters” and an initial resolution of 0.5, followed by annotation of cell types to each cluster. Briefly, our pipeline calculates cluster-specific marker genes using the Seurat functionality “FindAllMarkers” and an adjusted P value cutoff of 0.05. If more than 300 marker genes were identified, we only used the top 300 most significant genes for cell type annotation. Marker genes of each cluster were compared with literature-curated cell type–specific essential genes (table S10) using Fisher’s exact test. For each cluster we generated a list of cell type assignments that can be ranked by significance P values. If the quotient of the first and the second most significant P values was smaller than 0.05, our pipeline assigned the cell type associated with the first P value to that particular cluster. If not, it investigated if the quotient between the second and the third most significant P values is smaller than 0.05. In this case, it assigned both the first and second cell type to that particular cluster. This way our algorithm acknowledges that closely related cell types cannot always be distinguished from each other based on sc/sn RNAseq datasets (e.g., PT and descending limb cells). If the quotient between the second and third P values is higher than 0.05, our pipeline left that cluster unassigned. Our pipeline now analyzed whether all predefined cell types were identified (ignoring double cell type assignments). If not, it increased the resolution by 0.1 and repeated all steps leading to cluster identification and cell type annotation, until all predefined cell types were identified or a maximum resolution of 2.0 was achieved. The final selected resolution was that resolution that lead to the identification of the highest number of predefined cell types, under the requirement that all cell types that were identified at lower resolutions needed to be identified at this resolution as well. If this was not the case, we ignored the resolution completely. If multiple resolutions identified the same number of cell types, we selected that resolution that was associated with the lowest number of cells belonging to clusters with no cell type annotation. If there were still multiple candidate resolutions, we selected the lowest one. Clusters annotated to the same cell types were merged and cell type–specific marker genes were calculated (adjusted P value = 0.05).
Nuclei and cells that were assigned to a particular cell type and map or did not map to the corresponding LMD tissue subsegment were counted, using the subsegmental correlation analysis described above. Cell type–specific marker genes were subjected to dynamic enrichment analysis using MBCO. If more than 300 marker genes were identified, we only subjected the top 300 genes. All SCPs among the top seven predictions were further investigated.
We progressively and randomly removed libraries from the complete (reference) datasets to generate 100 non-overlapping downsampled datasets for each number of remaining libraries. Down-sampled data were subjected to the same analysis pipeline and results were compared with the reference results. We calculated the percentage of downsampled datasets for each number of remaining libraries that identified a particular cell type. If a particular cell type was identified in a down-sampled dataset, we counted how many of its nuclei/cells were assigned to the same or to a different cell type in the reference analysis. To visualize both counting results in the same plot, we defined those cell counts that mapped to a different cell type to be negative, so these counts are plotted below the abscissa. Similarly, we counted how many nuclei/cells of a particular cell type mapped and did not map to a particular tissue subsegment. Here, we also defined those cell counts that mapped to a different subsegment to be negative. We calculated the Pearson correlation between the DEGs of each cell type in the downsampled datasets and the reference datasets based on log2 fold changes.
Pathway enrichment analysis normally involves identification of the most significant pathways irrespective of their P values. To document the reliability of the SCPs that were among the top seven predictions in the complete dataset, we identified the ranks of these SCPs in each downsampled dataset as well. Ranks were averaged for each SCP and number of analyzed libraries.
Documentation of cellular metabolism
We generated a small ontology that contains the major metabolic pathways involved in energy generation and sphingomyelin synthesis. We defined parent-child relationships, where child pathways described subfunctions of their parent pathways (fig. S17A). Pathways were populated with genes curated from the literature, parent pathways also populated with the genes of their child pathways. The ontology is publicly available at github.com/SBCNY/Molecular-Biology-of-the-Cell and mbc-ontology.org. Before enrichment analysis, we added all genes of the “Gene_ontology_biological_process_2018” library that we downloaded from the enrichR website (59) as background genes of the ontology.
We subjected the top 500 significant marker genes and proteins (sc/sn RNAseq: adjusted P value 0.05, LMD RNAseq, LMD/NSC proteomics: nominal P value 0.05) to enrichment analysis using this ontology and Fisher’s exact test. Investigation of the predicted pathways that are specific for a particular reaction allowed to decide in which reaction(s) those enzymes participate that are shared by multiple pathways. Child pathways that specifically describe the function of their parent pathways are visualized in the same color in fig. S17A. If only pathways that contained the shared reactions of multiple parent pathways were predicted, we assumed that they participated in the default parent pathways “glycolysis,” “ketone body catabolism” or “aerobic glycolysis.”
Because the sn RNAseq data were derived from cortical, medullary, and mixed samples (fig. S4B), we distinguished between medullary (DTL, ATL1-3, TAL-1, PC-3, and IC-A2, for abbreviations see Fig. 2A) and cortical cell types (all other cell types of the renal tubule of the nephron). All other datasets were assigned as cortical. Enrichment result negative log10 P values were first averaged across the different cell subtypes of the same cell type and then across the different transcriptomic datasets. In case of the sc and sn RNAseq assays, we considered the number of cells assigned to each subtype of a particular cell type. The averaged negative log10 P values is representative of the cell counts of each cluster.
Because marker genes characterize those genes that are higher expressed in a particular cell type, subtype, or subsegment if compared to all other cell types or subsegments, missing identification of a particular pathway does not mean that there is no pathway activity at all. For example, we did not identify any podocyte marker genes involved in any of the analyzed energy generation pathways, although podocytes can generate energy by anaerobic glycolysis (60). Here, our analysis simply documents that the levels for those genes are lower in podocytes than in most of the other kidney cells.
Comparison of experimental reabsorption and gene expression profiles
Experimentally determined reabsorption capacity profiles that describe what percentage of a filtered sodium or glucose is reabsorbed in a particular nephron segment were curated from standard medical and physiological text books (30, 31, 33, 61), followed by averaging of the curated numbers for each ion or molecule. As these are widely used medical school textbooks, we assumed that the information is correct and did not further track down the values given in these books to the primary papers from which these values were obtained. In addition, we assumed conversation of physiological processes across mammalian species, and we did not ascertain whether all values were derived from the same species or more than one species.
Generation of a transmembrane transport ontology
Because human single cell and single nucleus RNAseq datasets (14–17) contain gene expression profiles in all major nephron cell types, we reasoned we could compare segment specific gene expression levels of the transporter or channel of interest with these physiologically measured reabsorption profiles. Using gene ontology MBCO (22), Wikipedia articles, and selected reviews as sources (36, 40), we generated a comprehensive ontology of transmembrane transporter at the plasma membrane (fig. S19A). Within gene ontology, we focused on all biological processes and molecular functions that were children of “sodium ion transport,” “sodium ion transmembrane transporter activity” as well as “glucose transport,” “glucose transmembrane transporter activity,” as defined by the “is_a” and “part_of” relationships. From MBCO, we added all genes assigned to the SCP “sodium transmembrane transport.” The initial list of transporter candidates was manually investigated to validate their transporter activity. True positives were assigned to de novo SCPs that describe the movement mechanism (i.e., transport via symporter or antiporter), the movement direction (i.e., lumen-to-blood or blood-to-lumen) and all ions or molecules that are transported by that mechanism. In case of antiporters, we specified which ions or molecules move in opposing directions by separating them with the term “vs.” If the protein translated from a particular gene had a unique name that is commonly used and is different from the official National Center for Biotechnology Information (NCBI) gene symbol, we assigned the gene to that particular protein name (e.g., SLC12A1 and SCL12A3 were assigned to NKCC2 and NCC, respectively). Here, we did not describe the activity mediated by that protein (e.g., “sodium potassium chloride transport by the symporter NKCC2”), because this would create unnecessarily long names in our figure legends. Nevertheless, in all analyses, these proteins were processed as if they were SCPs. Consequently, whenever we use the term SCP in the manuscript, we refer to these proteins as well. Each SCP-gene association was supported by at least one reference that could be the NCBI gene summary, UniProt gene summary or a PubMed ID for a supporting article (table S12). To allow systematic analysis and grouping of transmembrane movements, we integrated all SCPs into a SCP hierarchy of parent and children SCPs (fig. S19B) using a strategy we have described for the MBCO ontology (22). This hierarchy converges children on parent SCPs that describe more generalized shared transport mechanisms. For example, the SCP “sodium potassium chloride symporter” (that is the parent of the SCP “NKCC”) is the child of the two parent SCPs “sodium chloride symporter” and “sodium potassium symporter.” We left out the SCP “potassium chloride symporter,” because here, we focused on sodium and glucose transmembrane transport. These SCPs are then connected to the higher-level SCP “sodium lumen-to-blood transport by symporter.” For both sodium and glucose all SCPs finally converge on either one of two different overall parent SCPs describing transcellular lumen-toward-blood transport and transcellular blood-toward-lumen transport. Discussed example of SCP relationships and two additional examples are shown in fig. S19B. Figure S19C shows the hierarchical organization of all SCPs involved in sodium lumen-to-blood and blood-to-lumen transport. All parent SCPs were populated with the genes of all of their children SCPs. Last, we kept only those genes in the ontology that localize to the plasma membrane based on the jensenlab human compartment ontology with a minimum confidence score of 4 (out of maximal 5) (www.compartments.jensenlab.org).
Calculation of predicted reabsorption capacities
Besides our own sc and sn RNAseq dataset (14, 15), we used two different sn RNA seq datasets that were generated from undiseased tissue as well (16, 17). All datasets document how many mRNA molecules are transcribed from each gene in each individual cell. These numbers are described as UMI counts (62), but in this study, we use the term mRNA counts or levels to indicate that it is a quantitative measure of mRNA levels of a certain species. The cells and nuclei in the sc and sn RNAseq datasets were previously grouped into clusters using standard software packages, followed by identification of cluster-specific marker genes and cell type and subtype annotation (14, 15, 17). We analyzed the raw UMI matrix (GSE114156) (16) using the Seurat package (as outlined in fig. S5A) and annotated kidney cell types based on cell type–specific gene expression (table S10).
We assumed that that mRNA molecule counts (i.e., UMI counts) of each transporter or channel in each cell reflect the capacity of that particular cell for transmembrane movement of that particular ion or molecule. The following explanation of how we predicted movement capacities from those mRNA levels is summarized in fig. S19A. We initially processed all four datasets, i.e., one sc RNAseq and three sn RNAseq datasets, independently. For each dataset and SCP of our transmembrane movement ontology we summed up all mRNA molecules that are expressed in all cells of a particular cell type or nephron segment and map to genes involved in that SCP. It should be noted that we documented total and not mean capacities, because we did not divide the mRNA count sums by the number of cells in each particular cell type or segment. If a particular cell type or nephron segment contains more cells, it is assumed to contribute more to the reabsorption of a particular ion or molecule, if the appropriate transporter is present. Measured physiological reabsorption profiles describe net lumen-to-blood transport values in each segment. To account for the different transport directions predicted from SCPs that are involved in lumen-to-blood and blood-to-lumen transport, we defined all mRNA levels mapping to blood-to-lumen transport SCPs as negative. This allowed the calculation of net lumen-to-blood transport capacities by adding up all mRNA counts involved in lumen-to-blood transport and all (negative) mRNA counts involved in blood-to-lumen transport of each ion or molecule. Because the physiological profiles document how much percent of a particular ion or molecule is reabsorbed in a particular nephron segment, we expressed all SCP capacities in percent of the net lumen-to-blood transport capacities of the corresponding ions or molecules. Consequently, the sum of all predicted transport capacities along the nephron is 100% for both sodium and glucose (mRNA levels assigned to blood-to-lumen transport are still defined as negative), allowing the direct comparison of mRNA levels and reabsorption profiles. Any SCPs that mediate the transport of multiple ions or molecules were normalized independently for each ion and molecule to calculate the relative contribution of that SCP to the total reabsorption capacity of each ion or molecule. Final percentages of the same SCPs predicted by the three sn RNAseq datasets or all four datasets were averaged.
Identification of cell type markers
Marker genes and proteins of each cell subtype or segment were ranked as described above. If multiple cell subtypes were identified on the basis of the single cell or single nucleus RNAseq datasets, we calculated the average rank for that cell type. Original or averaged ranks were averaged again over the different assays for each cell type and selected segment, followed by reranking. The top five reranked genes/proteins were selected as cell type markers in table S12. To identify combinations of markers that best define each cell type, we also used a machine learning approach (63) to find marker combinations that optimally separate each cell type from all other cell types for sc, sn, and integrated sc/sn RNA-seq datasets.
Generation of nephron schema
We used BioRender.com to create the nephron schema in Fig. 7.
Data generation and initial analysis
Seven different RNAseq, proteomics, metabolomics, and imaging datasets were generated and analyzed by five different TISs. The PREMIERE TIS (composed of Michigan, Princeton, Broad) generated sc RNAseq data, the USCD/WashU TIS generated sn data, the UCSF TIS generated sc RNAseq, NSC proteomics and Codex imaging data, the IU/OSU TIS generated LMD RNAseq and LMD proteomics data, and the UTHSA-PNNL-EMBL TIS generated spatial metabolomics data.
Sn RNAseq (UCSD/WashU) and single-cell RNAseq (PREMIERE)
UMI count matrixes and list of DEGs were downloaded from published analyses for the PREMIERE TIS (composed of Michigan, Princeton, Broad) single-cell RNAseq (15) and UCSD/WashU TIS Single-nucleus RNAseq (14) datasets. We excluded the PT cells-3 and principal cells-2 clusters from the single-nucleus RNAseq dataset, because these clusters showed an inflammatory or a stress response.
Subsegmental LMD transcriptomics (IU/OSU)
A comprehensive LMD protocol is published on protocols.io (www.protocols.io/view/laser-microdissection-8rkhv4w). Briefly, 12-μm frozen sections are obtained from an optimal cutting temperature (OCT) preserved tissue block and adhered to LMD membrane slides (Leica, Buffalo Grove, IL). The tissue undergoes a rapid staining protocol involving acetone fixation, washes with ribonuclease (RNAse)–free phosphate-buffered saline, and antibody incubation in 10% bovine serum albumin. The slides undergo dissection with a Leica LMD6500 system with pulsed ultraviolet laser. After collecting a minimum tissue area of 500,000 μm2 in an RNAse-free microcentrifuge tube, the RNA is isolated using the PicoPure RNA Isolation Kit according to the manufacturer’s instructions (Applied Biosystems, catalog no. KIT0204). The RNA quality is assessed by Bioanalyzer; ribosomal RNA is depleted, and cDNA libraries are prepared using the SMARTer Universal Low-Input RNA Kit (Takara, no. 634938). Sequencing was conducted on an Illumina HiSeq 4000. Mapping was performed using STAR (v2.5.2b), and read counts were quantified with featureCounts (subread v.1.5.0). Total read counts mapping to each gene were generated with edgeR, normalized, and converted to expression ratios.
Segment-specific gene expression was compared to the gene expression in all other subsegments using an unpaired t test with equal variance. Subsegment-specific gene expression ratios were calculated similarly.
Subsegmental LMD proteomics (IU/OSU)
A comprehensive LMD proteomics protocol is published on protocols.io (www.protocols.io/view/laser-microdissection-for-regional-transcriptomics-8rkhv4w?version_warning=no). Our LMD proteomic methods have also been previously published in detail (64, 65). Briefly, 10-μm frozen sections are obtained from an OCT-preserved tissue block and adhered to polyethylene naphthalate membrane slides for LMD. Frozen sections are fixed in 70% ethanol, incubated in H2O to remove OCT, briefly stained with hematoxylin, and dehydrated in ethanol. LMD is performed and glomeruli and tubulointerstitial samples are collected separately in 0.5% RapiGest/50 mm NH3HCO3 solution. The collected samples are then boiled for 20 min for protein retrieval and digested overnight with trypsin. Peptides are dried, resuspended in acetonitrile/formic acid, and analyzed using liquid chromatography tandem–mass spectrometry analysis using an Easy-nLC 1000 HPLC coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific, Waltham, MA). Data are searched using Proteome Discoverer 2.1 (Thermo Scientific) and searched against a human UniProt database (version 05/26/18). Data are analyzed following global normalization of spectral counts.
Glomerular gene expression was compared to the tubulointerstitial gene expression using an unpaired t test with equal variance. Glomerular to tubular-specific gene expression ratios were calculated similarly.
3D immunofluorescence imaging and tissue cytometry (IU/OSU)
The entire 3D fluorescence imaging and tissue cytometry protocol is published on protocols.io (dx.doi.org/10.17504/protocols.io.9avh2e6). Briefly, frozen cores are sectioned at 50 μm using a cryostat and fixed using 4% paraformaldehyde. A panel of up to eight antibodies was incubated to identify renal and immune cell types. Images were acquired in up to eight channels using a Leica SP8 confocal microscope. Volume stacks spanning the whole thickness of the tissue were taken using a 20× numerical aperture (NA) 0.75 or 40× NA 1.3 objectives with 0.5- to 1.0-μm spacing. Large-scale confocal imaging of overlapping volumes was performed with an automated stage and stitched using Leica LASX software (Germany). A 3D image rendering was done using Voxx v2.09d. The 3D tissue cytometry was performed on image volumes using VTEA, which was developed as a plugin for ImageJ/Fiji as previously described (66).
CODEX imaging (UCSF)
The CODEX technology comprises of immunofluorescence staining with antibodies conjugated to unique oligonucleotide sequence targeting markers of interest probed with complementary oligonucleotide reporters tagged to fluorophores (67). CODEX fluidics instrument is used for iterative cycles of imaging, dye, and reporter removal allowing high plexity. 4′,6-Diamidino-2-phenylindole for nuclear signal along with complementary oligonucleotide probes tagged to Alexa Fluor 488, Atto 550, and Cy5 fluorophores are revealed in each cycle. CODEX Software Suite allows raw image processing for subtraction of background noise, deconvolution to eliminate out-of-focus light, collapsing z-stacks into single best focus, shading correction, and cycle alignment in addition to image segmentation based on watershed algorithm. Flow cytometry standard expression data and raw images as TIFF files are generated and can be visualized in external software for gating, clustering, and phenotyping.
Transplant normal biopsy was stained with 29-plex panel of CODEX antibodies, followed by immunofluorescent staining of calbindin and imaged at 20× resolution on a Keyence BZ-X710 microscope. Raw data were transferred and processed with CODEX Software Suite comprising CODEX Instrument Manager and processor. Processed data were visualized in the CODEX Multiple Analysis Viewer (MAV), an ImageJ plugin to eliminate artifacts, assess cell segmentation, gating, annotation, and generation of subpopulation for downstream analyses. Clustered population were visualized in MAV by overlaying with corresponding markers and annotated population to assess cell type enrichment.
Spatial metabolomics (UTHSA-PNNL-EMBL)
Ten-micrometer-thick renal cortical tissues were sectioned using a cryostat (Leica Microsystems), thaw-mounted on indium tin oxide–coated slides (Bruker Daltonics), and prepared for matrix-assisted laser deposition/ionization mass spectrometry (MALDI-MSI) by spraying with 2,5-dihydroxybenzoic acid (40 mg/ml in 50% MeOH:H2O) using the TM-Sprayer automated spraying robot (HTX Technology). The following spraying parameters were used: 80°C nozzle temperature, a flow rate of 0.05 ml/min, 10 passes, a N2 pressure of 10 psi, a track spacing of 3 mm, and a 40-mm distance between the nozzle, and sample was maintained. MALDI-MSI was performed using a MALDI-Fourier-transform ion cyclotron resonance (FTICR) imaging mass spectrometer (Bruker Daltonics) set at a 120,000 resolving power at mass/charge ratio (m/z) 400 or a MALDI-Orbitrap mass spectrometer (Thermo Scientific) set at the 120,000 resolving power at m/z 200. The data were inspected following the quality control guidelines as developed within KPMP and converted into the imzML centroided format using the SCiLS software (Bruker Daltonics) or ImageInsight software (Spectroglyph LLC), followed by the submission to METASPACE and annotation against the SwissLipids and Human Metabolome Database (HMDB) molecular databases with the false discovery rate of 20%, as described in (68).
We have developed an approach to find glomeruli markers in MALDI-MSI data by using METASPACE and colocalization analysis. First, we have selected a template marker that was localized within the glomerular regions, as confirmed by the histology. This ion was annotated by METASPACE as ceramide phosphate CerP(d34:1) (68). Then, we performed a spatial colocalization analysis by calculating for all other detected metabolites and lipids their spatial correlation with CerP(d34:1) using the cosine score. The molecules with the correlation above 0.2 were considered and manually curated to show the colocalization with the glomeruli regions by overlaying every ion image with the histological image. The resulting 30 markers were uploaded to the KPMP DataLake and were used for the multiomics integration analysis.
Acknowledgments
KPMP acknowledges all the participants, patients, and the scientific officers from the National Institute of Diabetes and Digestive and Kidney Diseases. KPMP was supported by NIH grants UH3 DK114923, UH3 DK114920, UH3 DK114933, UH3 DK114937, UH3 DK114907, and U2C DK114886. A complete list of all KPMP members can be found at kpmp.org. We thank J. Goldfarb for critically reading of the manuscript.
Author contributions; Integrated analysis and interpretation: J.Ha., R.S., R.M., J.C.H., J.Hi., L.M.S., O.G. T., M.K., R.I., and E.U.A.. Pilot tissue procurement, data coordination, and metadata curation: B.S., A.S.N., J.B.H., M.K., E.U.A.. Sc/sn RNAseq data generation and processing: R.M., B.B.L., J.Ha., E.A.O., J.B.H., L.B., M.M.S., K.Z., M.T.V., S.J., M.K., and E.U.A. LMD transcriptomics data generation and processing: M.T.E., D.B., and P.C.D. LMD and NSC proteomic data generation and processing: S.P., J.P.S., T.K.S., P.R., M.M.S., and B.R. Imaging data generation and processing: K.A., D.D., M.R., S.W., T.M.E-A., and Z.L. Spatial metabolomics data generation and processing: T.A., D.V., C.R.A., G.Z., A.P., and K.S.. Manuscript preparation: J.Ha., R.S., R.M., M.P.R., Y.X., Y.H., I.H.d.B., L.M.S., O.G.T., M.K., R.I., and E.U.A. All authors commented and edited the manuscript and assisted in the assembly of the final version.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All raw and processed data described in this manuscript are available through the KPMP Data Portal at kpmp.org (DOI: 10.48698/z30t-0a62) and Zenodo (DOI: 10.5281/zenodo.6410326). In addition, all version-controlled quality control metrics and annotated hierarchical metadata standards can be viewed along with the primary data in the interactive cellular kidney atlas section at atlas.kpmp.org/explorer. The raw transcriptomics data are also available on Gene Expression Omnibus with the accession IDs GSE163603, GSE121862, and GSE140989. Raw proteomics data are publicly available on MassIVE repository with the accession ID MSV000089251.
Code availability: Fully annotated scripts and readme files that can be used to generate all the main figures in this manuscript are freely available at the KPMP GitHub portal (https://github.com/KPMP/Reference-Tissue-Cell-Atlas-Manuscript-2022). The containerized code can also be obtained from Docker as Windows (for C# programs, https://hub.docker.com/r/iyengarlab/kpmp-cs-apps) or Linux (for R and Python programs, https://hub.docker.com/r/iyengarlab/kpmp-rp-apps) containers.
Supplementary Materials
This PDF file includes:
Figs. S1 to S20
Supplemental Information
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S13
REFERENCES AND NOTES
- 1.Park J., Liu C. L., Kim J., Susztak K., Understanding the kidney one cell at a time. Kidney Int. 96, 862–870 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L., Chou C. L., Knepper M. A., Targeted single-cell RNA-seq identifies minority cell types of kidney distal nephron. J. Am. Soc. Nephrol. 32, 886–896 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conway B. R., O’Sullivan E. D., Cairns C., O’Sullivan J., Simpson D. J., Salzano A., Connor K., Ding P., Humphries D., Stewart K., Teenan O., Pius R., Henderson N. C., Bénézech C., Ramachandran P., Ferenbach D., Hughes J., Chandra T., Denby L., Kidney single-cell atlas reveals myeloid heterogeneity in progression and regression of kidney disease. J. Am. Soc. Nephrol. 31, 2833–2854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He B., Chen P., Zambrano S., Dabaghie D., Hu Y., Möller-Hackbarth K., Unnersjö-Jess D., Korkut G. G., Charrin E., Jeansson M., Bintanel-Morcillo M., Witasp A., Wennberg L., Wernerson A., Schermer B., Benzing T., Ernfors P., Betsholtz C., Lal M., Sandberg R., Patrakka J., Single-cell RNA sequencing reveals the mesangial identity and species diversity of glomerular cell transcriptomes. Nat. Commun. 12, 2141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L., Liao J., He J., Pan S., Zhang H., Yang X., Cheng J., Chen Y., Mo Z., Single-cell profiling reveals sex diversity in human renal PTs. Gene 752, 144790 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kuppe C., Ibrahim M. M., Kranz J., Zhang X., Ziegler S., Perales-Patón J., Jansen J., Reimer K. C., Smith J. R., Dobie R., Wilson-Kanamori J. R., Halder M., Xu Y., Kabgani N., Kaesler N., Klaus M., Gernhold L., Puelles V. G., Huber T. B., Boor P., Menzel S., Hoogenboezem R. M., Bindels E. M. J., Steffens J., Floege J., Schneider R. K., Saez-Rodriguez J., Henderson N. C., Kramann R., Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Suszták K., Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart B. J., Ferdinand J. R., Young M. D., Mitchell T. J., Loudon K. W., Riding A. M., Richoz N., Frazer G. L., Staniforth J. U. L., Vieira Braga F. A., Botting R. A., Popescu D. M., Vento-Tormo R., Stephenson E., Cagan A., Farndon S. J., Polanski K., Efremova M., Green K., del Castillo Velasco-Herrera M., Guzzo C., Collord G., Mamanova L., Aho T., Armitage J. N., Riddick A. C. P., Mushtaq I., Farrell S., Rampling D., Nicholson J., Filby A., Burge J., Lisgo S., Lindsay S., Bajenoff M., Warren A. Y., Stewart G. D., Sebire N., Coleman N., Haniffa M., Teichmann S. A., Behjati S., Clatworthy M. R., Spatiotemporal immune zonation of the human kidney. Science 365, 1461–1466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson P. C., Wu H., Kirita Y., Uchimura K., Ledru N., Rennke H. G., Welling P. A., Waikar S. S., Humphreys B. D., The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. U.S.A. 116, 19619–19625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H., Kirita Y., Donnelly E. L., Humphreys B. D., Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 30, 23–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young M. D., Mitchell T. J., Vieira Braga F. A., Tran M. G. B., Stewart B. J., Ferdinand J. R., Collord G., Botting R. A., Popescu D. M., Loudon K. W., Vento-Tormo R., Stephenson E., Cagan A., Farndon S. J., del Castillo Velasco-Herrera M., Guzzo C., Richoz N., Mamanova L., Aho T., Armitage J. N., Riddick A. C. P., Mushtaq I., Farrell S., Rampling D., Nicholson J., Filby A., Burge J., Lisgo S., Maxwell P. H., Lindsay S., Warren A. Y., Stewart G. D., Sebire N., Coleman N., Haniffa M., Teichmann S. A., Clatworthy M., Behjati S., Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer I. H., Alpers C. E., Azeloglu E. U., Balis U. G. J., Barasch J. M., Barisoni L., Blank K. N., Bomback A. S., Brown K., Dagher P. C., Dighe A. L., Eadon M. T., el-Achkar T. M., Gaut J. P., Hacohen N., He Y., Hodgin J. B., Jain S., Kellum J. A., Kiryluk K., Knight R., Laszik Z. G., Lienczewski C., Mariani L. H., McClelland R. L., Menez S., Moledina D. G., Mooney S. D., O’Toole J. F., Palevsky P. M., Parikh C. R., Poggio E. D., Rosas S. E., Rosengart M. R., Sarwal M. M., Schaub J. A., Sedor J. R., Sharma K., Steck B., Toto R. D., Troyanskaya O. G., Tuttle K. R., Vazquez M. A., Waikar S. S., Williams K., Wilson F. P., Zhang K., Iyengar R., Kretzler M., Himmelfarb J., Knight R., Lecker S., Stillman I., Waikar S., Mcmahon G., Weins A., Short S., Hacohen N., Hoover P., Aulisio M., Cooperman L., Herlitz L., O’Toole J., Poggio E., Sedor J., Jolly S., Appelbaum P., Balderes O., Barasch J., Bomback A., Canetta P. A., d’Agati V. D., Kiryluk K., Kudose S., Mehl K., Radhakrishnan J., Weng C., Barisoni L., Alexandrov T., Ashkar T., Barwinska D., Dagher P., Dunn K., Eadon M., Ferkowicz M., Kelly K., Sutton T., Winfree S., Menez S., Parikh C., Rosenberg A., Villalobos P., Malik R., Fine D., Atta M., Monroy Trujillo J. M., Slack A., Rosas S., Williams M., Azeloglu E., He C. (J.), Iyengar R., Hansen J., Parikh S., Rovin B., Anderton C., Pasa-Tolic L., Velickovic D., Lukowski J., Oliver G. (H.), Ardayfio J., Bebiak J., Brown K., Campbell T., Campbell C., Hayashi L., Jefferson N., Koewler R., Roberts G., Saul J., Shpigel A., Stutzke E. C., Wright L., Miegs L., Pinkeney R., Sealfon R., Troyanskaya O., Tuttle K., Dobi D., Goltsev Y., Lake B., Zhang K., Joanes M., Laszik Z., Schroeder A., Sarwal M., Sigdel T., Balis U., Blanc V., He O., Hodgin J., Kretzler M., Mariani L., Menon R., Otto E., Schaub J., Steck B., Lienczewski C., Eddy S., Elder M., Hall D., Kellum J., Kruth M., Murugan R., Palevsky P., Randhawa P., Rosengart M., Sims-Lucas S., Stefanick M., Stull S., Tublin M., Alpers C., de Boer I., Dighe A., Himmelfarb J., Mcclelland R., Mooney S., Shankland S., Williams K., Blank K., Carson J., Dowd F., Drager Z., Park C., Sharma K., Zhang G., Bansal S., Venkatachalam M., Kermani A., Lee S., Lu C., Miller T., Moe O., Park H., Sambandam K., Sanchez F., Torrealba J., Robert T., Vazquez M., Wang N., Gaut J., Jain S., Vijayan A., Luciano R., Moledina D., Ugochukwu U., Wilson F. P., Alfano S., Rationale and design of the Kidney Precision Medicine Project. Kidney Int. 99, 498–510 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Achkar T. M., Eadon M. T., Menon R., Lake B. B., Sigdel T. K., Alexandrov T., Parikh S., Zhang G., Dobi D., Dunn K. W., Otto E. A., Anderton C. R., Carson J. M., Luo J., Park C., Hamidi H., Zhou J., Hoover P., Schroeder A., Joanes M., Azeloglu E. U., Sealfon R., Winfree S., Steck B., He Y., D’Agati V., Iyengar R., Troyanskaya O. G., Barisoni L., Gaut J., Zhang K., Laszik Z., Rovin B. H., Dagher P. C., Sharma K., Sarwal M. M., Hodgin J. B., Alpers C. E., Kretzler M., Jain S., A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: Guidelines from the Kidney Precision Medicine Project. Physiol. Genomics 53, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lake B. B., Chen S., Hoshi M., Plongthongkum N., Salamon D., Knoten A., Vijayan A., Venkatesh R., Kim E. H., Gao D., Gaut J., Zhang K., Jain S., A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 10, 2832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menon R., Otto E. A., Hoover P., Eddy S., Mariani L., Godfrey B., Berthier C. C., Eichinger F., Subramanian L., Harder J., Ju W., Nair V., Larkina M., Naik A. S., Luo J., Jain S., Sealfon R., Troyanskaya O., Hacohen N., Hodgin J. B., Kretzler M., Kpmp K. P. M. P.; Nephrotic Syndrome Study Network (NEPTUNE) , Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight 5, e133267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H., Malone A. F., Donnelly E. L., Kirita Y., Uchimura K., Ramakrishnan S. M., Gaut J. P., Humphreys B. D., Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J. Am. Soc. Nephrol. 29, 2069–2080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muto Y., Wilson P. C., Ledru N., Wu H., Dimke H., Waikar S. S., Humphreys B. D., Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat. Commun. 12, 2190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer L. G., Schnermann J., Integrated control of Na transport along the nephron. Clin. J. Am. Soc. Nephrol. 10, 676–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barwinska D., el-Achkar T. M., Melo Ferreira R., Syed F., Cheng Y. H., Winfree S., Ferkowicz M. J., Hato T., Collins K. S., Dunn K. W., Kelly K. J., Sutton T. A., Rovin B. H., Parikh S. V., Phillips C. L., Dagher P. C., Eadon M. T.; for the Kidney Precision Medicine Project , Molecular characterization of the human kidney interstitium in health and disease. Sci. Adv. 7, eabd3359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buccitelli C., Selbach M., mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 21, 630–644 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Calizo R. C., Bhattacharya S., van Hasselt J. G. C., Wei C., Wong J. S., Wiener R. J., Ge X., Wong N. J., Lee J. J., Cuttitta C. M., Jayaraman G., Au V. H., Janssen W., Liu T., Li H., Salem F., Jaimes E. A., Murphy B., Campbell K. N., Azeloglu E. U., Disruption of podocyte cytoskeletal biomechanics by dasatinib leads to nephrotoxicity. Nat. Commun. 10, 2061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen J., Meretzky D., Woldesenbet S., Stolovitzky G., Iyengar R., A flexible ontology for inference of emergent whole cell function from relationships between subcellular processes. Sci. Rep. 7, 17689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia J., Psychogios N., Young N., Wishart D. S., MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene C. S., Krishnan A., Wong A. K., Ricciotti E., Zelaya R. A., Himmelstein D. S., Zhang R., Hartmann B. M., Zaslavsky E., Sealfon S. C., Chasman D. I., FitzGerald G. A., Dolinski K., Grosser T., Troyanskaya O. G., Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan A., Zhang R., Yao V., Theesfeld C. L., Wong A. K., Tadych A., Volfovsky N., Packer A., Lash A., Troyanskaya O. G., Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat. Neurosci. 19, 1454–1462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukasawa H., Bornheimer S., Kudlicka K., Farquhar M. G., Slit diaphragms contain tight junction proteins. J. Am. Soc. Nephrol. 20, 1491–1503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiser J., Kriz W., Kretzler M., Mundel P., The glomerular slit diaphragm is a modified adherens junction. J. Am. Soc. Nephrol. 11, 1–8 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Scholz H., Boivin F. J., Schmidt-Ott K. M., Bachmann S., Eckardt K. U., Scholl U. I., Persson P. B., Kidney physiology and susceptibility to acute kidney injury: Implications for renoprotection. Nat. Rev. Nephrol. 17, 335–349 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Bagnasco S., Good D., Balaban R., Burg M., Lactate production in isolated segments of the rat nephron. Am. J. Physiol. 248, F522–F526 (1985). [DOI] [PubMed] [Google Scholar]
- 30.R. M. Berne, M. N. Levy, Physiology (Mosby-Year Book, ed. 3, 1993), chap. 42. [Google Scholar]
- 31.L. S. Costanzo, Physiology (Elsevier, ed. 4, 2010), chap. 6. [Google Scholar]
- 32.R. Brandes, F. Lang, R. F. Schmidt, Physiologie des Menschen (Springer, 2010). [Google Scholar]
- 33.K. Rainer, S. Silbernagl, Lehrbuch der Physiologie (Georg Thieme, ed. 2, 1996), chap. 12. [Google Scholar]
- 34.Muto S., Hata M., Taniguchi J., Tsuruoka S., Moriwaki K., Saitou M., Furuse K., Sasaki H., Fujimura A., Imai M., Kusano E., Tsukita S., Furuse M., Claudin-2-deficient mice are defective in the leaky and cation-selective paracellular permeability properties of renal PTs. Proc. Natl. Acad. Sci. U.S.A. 107, 8011–8016 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu A. S. L., Paracellular transport and energy utilization in the renal tubule. Curr. Opin. Nephrol. Hypertens. 26, 398–404 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Mount D. B., Thick ascending limb of the loop of Henle. Clin. J. Am. Soc. Nephrol. 9, 1974–1986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ares G. R., Ortiz P. A., Constitutive endocytosis and recycling of NKCC2 in rat thick ascending limbs. Am. J. Physiol. Renal Physiol. 299, F1193–F1202 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caceres P. S., Ares G. R., Ortiz P. A., cAMP stimulates apical exocytosis of the renal Na(+)-K(+)-2Cl(−) cotransporter NKCC2 in the thick ascending limb: Role of protein kinase A. J. Biol. Chem. 284, 24965–24971 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giménez I., Forbush B., Short-term stimulation of the renal Na-K-Cl cotransporter (NKCC2) by vasopressin involves phosphorylation and membrane translocation of the protein. J. Biol. Chem. 278, 26946–26951 (2003). [DOI] [PubMed] [Google Scholar]
- 40.van der Wijst J., Belge H., Bindels R. J. M., Devuyst O., Learning physiology from inherited kidney disorders. Physiol. Rev. 99, 1575–1653 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Rotoli B. M., Barilli A., Visigalli R., Ferrari F., Dall’Asta V., y+LAT1 and y+LAT2 contribution to arginine uptake in different human cell models: Implications in the pathophysiology of lysinuric protein intolerance. J. Cell Mol. Med. 24, 921–929 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sano R., Shinozaki Y., Ohta T., Sodium-glucose cotransporters: Functional properties and pharmaceutical potential. J Diabetes Investig. 11, 770–782 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukowski J. K., Pamreddy A., Velickovic D., Zhang G., Pasa-Tolic L., Alexandrov T., Sharma K., Anderton C. R.; the Kidney Precision Medicine Project , Storage conditions of human kidney tissue sections affect spatial lipidomics analysis reproducibility. J. Am. Soc. Mass Spectrom. 31, 2538–2546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velickovic D., Zhang G., Bezbradica D., Bhattacharjee A., Paša-Tolić L., Sharma K., Alexandrov T., Anderton C. R.; KPMP Consortium , Response surface methodology as a new approach for finding optimal MALDI matrix spraying parameters for mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 31, 508–516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inuzuka M., Hayakawa M., Ingi T., Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J. Biol. Chem. 280, 35776–35783 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Merrill A. H. Jr., De novo sphingolipid biosynthesis: A necessary, but dangerous, pathway. J. Biol. Chem. 277, 25843–25846 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Merscher S., Fornoni A., Podocyte pathology and nephropathy–Sphingolipids in glomerular diseases. Front Endocrinol. 5, 127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slotte J. P., Biological functions of sphingomyelins. Prog. Lipid Res. 52, 424–437 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Hullin-Matsuda F., Taguchi T., Greimel P., Kobayashi T., Lipid compartmentalization in the endosome system. Semin. Cell Dev. Biol. 31, 48–56 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Green D. R., Apoptosis and sphingomyelin hydrolysis. The flip side. J. Cell Biol. 150, F5–F8 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drochmans P., Freudenstein C., Wanson J. C., Laurent L., Keenan T. W., Stadler J., Leloup R., Franke W. W., Structure and biochemical composition of desmosomes and tonofilaments isolated from calf muzzle epidermis. J. Cell Biol. 79, 427–443 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shigetomi K., Ono Y., Inai T., Ikenouchi J., Adherens junctions influence tight junction formation via changes in membrane lipid composition. J. Cell Biol. 217, 2373–2381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee D. B., Huang E., Ward H. J., Tight junction biology and kidney dysfunction. Am. J. Physiol. Renal Physiol. 290, F20–F34 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Mickelsen L. E., Bolisetty M., Chimileski B. R., Fujita A., Beltrami E. J., Costanzo J. T., Naparstek J. R., Robson P., Jackson A. C., Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nat. Neurosci. 22, 642–656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ong E., Wang L. L., Schaub J., O’Toole J. F., Steck B., Rosenberg A. Z., Dowd F., Hansen J., Barisoni L., Jain S., de Boer I. H., Valerius M. T., Waikar S. S., Park C., Crawford D. C., Alexandrov T., Anderton C. R., Stoeckert C., Weng C., Diehl A. D., Mungall C. J., Haendel M., Robinson P. N., Himmelfarb J., Iyengar R., Kretzler M., Mooney S., He Y.; Kidney Precision Medicine Project , Modelling kidney disease using ontology: Insights from the Kidney Precision Medicine Project. Nat. Rev. Nephrol. 16, 686–696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Black S., Phillips D., Hickey J. W., Kennedy-Darling J., Venkataraaman V. G., Samusik N., Goltsev Y., Schürch C. M., Nolan G. P., CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., Ma’ayan A., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brinkkoetter P. T., Bork T., Salou S., Liang W., Mizi A., Özel C., Koehler S., Hagmann H. H., Ising C., Kuczkowski A., Schnyder S., Abed A., Schermer B., Benzing T., Kretz O., Puelles V. G., Lagies S., Schlimpert M., Kammerer B., Handschin C., Schell C., Huber T. B., Anaerobic glycolysis maintains the glomerular filtration barrier independent of mitochondrial metabolism and dynamics. Cell Rep 27, 1551–1566.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R. F. Schmidt, G. Thews, Physiologie des Menschen (Springer, ed. 27, 1997), chap. 35. [Google Scholar]
- 62.Islam S., Zeisel A., Joost S., la Manno G., Zajac P., Kasper M., Lönnerberg P., Linnarsson S., Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Aevermann B., Zhang Y., Novotny M., Keshk M., Bakken T., Miller J., Hodge R., Lelieveldt B., Lein E., Scheuermann R. H., A machine learning method for the discovery of minimum marker gene combinations for cell type identification from single-cell RNA sequencing. Genome Res. 31, 1767–1780 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Satoskar A. A., Shapiro J. P., Bott C. N., Song H., Nadasdy G. M., Brodsky S. V., Hebert L. A., Birmingham D. J., Nadasdy T., Freitas M. A., Rovin B. H., Characterization of glomerular diseases using proteomic analysis of laser capture microdissected glomeruli. Mod. Pathol. 25, 709–721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shapiro J. P., Biswas S., Merchant A. S., Satoskar A., Taslim C., Lin S., Rovin B. H., Sen C. K., Roy S., Freitas M. A., A quantitative proteomic workflow for characterization of frozen clinical biopsies: Laser capture microdissection coupled with label-free mass spectrometry. J. Proteomics 77, 433–440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winfree S., Khan S., Micanovic R., Eadon M. T., Kelly K. J., Sutton T. A., Phillips C. L., Dunn K. W., el-Achkar T. M., Quantitative three-dimensional tissue cytometry to study kidney tissue and resident immune cells. J. Am. Soc. Nephrol. 28, 2108–2118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G. P., Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palmer A., Phapale P., Chernyavsky I., Lavigne R., Fay D., Tarasov A., Kovalev V., Fuchser J., Nikolenko S., Pineau C., Becker M., Alexandrov T., FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat. Methods 14, 57–60 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Bhargava P., Schnellmann R. G., Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 13, 629–646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang K., Kestenbaum B., Proximal tubular secretory clearance: A neglected partner of kidney function. Clin. J. Am. Soc. Nephrol. 13, 1291–1296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Swelm R. P. L., Wetzels J. F. M., Swinkels D. W., The multifaceted role of iron in renal health and disease. Nat. Rev. Nephrol. 16, 77–98 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Zarjou A., Bolisetty S., Joseph R., Traylor A., Apostolov E. O., Arosio P., Balla J., Verlander J., Darshan D., Kuhn L. C., Agarwal A., Proximal tubule H-ferritin mediates iron trafficking in acute kidney injury. J. Clin. Invest. 123, 4423–4434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Legouis D., Faivre A., Cippa P. E., de Seigneux S., Renal gluconeogenesis: An underestimated role of the kidney in systemic glucose metabolism. Nephrol. Dial. Transplant. 2020, gfaa302 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Nakagawa T., Johnson R. J., Andres-Hernando A., Roncal-Jimenez C., Sanchez-Lozada L. G., Tolan D. R., Lanaspa M. A., Fructose production and metabolism in the kidney. J. Am. Soc. Nephrol. 31, 898–906 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer C., Nadkarni V., Stumvoll M., Gerich J., Human kidney free fatty acid and glucose uptake: Evidence for a renal glucose-fatty acid cycle. Am. J. Physiol. 273, E650–E654 (1997). [DOI] [PubMed] [Google Scholar]
- 76.Minami S., Yamamoto T., Takabatake Y., Takahashi A., Namba T., Matsuda J., Kimura T., Kaimori J. Y., Matsui I., Hamano T., Takeda H., Takahashi M., Izumi Y., Bamba T., Matsusaka T., Niimura F., Isaka Y., Lipophagy maintains energy homeostasis in the kidney proximal tubule during prolonged starvation. Autophagy 13, 1629–1647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wanders R. J., Waterham H. R., Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Houten S. M., Violante S., Ventura F. V., Wanders R. J., The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu. Rev. Physiol. 78, 23–44 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Kang H. M., Ahn S. H., Choi P., Ko Y. A., Han S. H., Chinga F., Park A. S. D., Tao J., Sharma K., Pullman J., Bottinger E. P., Goldberg I. J., Susztak K., Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 21, 37–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strijbis K., Vaz F. M., Distel B., Enzymology of the carnitine biosynthesis pathway. IUBMB Life 62, 357–362 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Faul C., Asanuma K., Yanagida-Asanuma E., Kim K., Mundel P., Actin up: Regulation of podocyte structure and function by components of the actin cytoskeleton. Trends Cell Biol. 17, 428–437 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Márquez M. G., Favale N. O., Leocata Nieto F., Pescio L. G., Sterin-Speziale N., Changes in membrane lipid composition cause alterations in epithelial cell-cell adhesion structures in renal papillary collecting duct cells. Biochim. Biophys. Acta 1818, 491–501 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Schunk S. J., Floege J., Fliser D., Speer T., WNT–β-catenin signalling—A versatile player in kidney injury and repair. Nat. Rev. Nephrol. 17, 172–184 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Mallipattu S. K., He J. C., The beneficial role of retinoids in glomerular disease. Front. Med. 2, 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Y., Li Q., Ouyang Q., Wu L., Chen X., Activated mesangial cells acquire the function of antigen presentation. Cell. Immunol. 361, 104279 (2021). [DOI] [PubMed] [Google Scholar]
- 86.Zuo L., Kuo W. T., Turner J. R., Tight junctions as targets and effectors of mucosal immune homeostasis. Cell. Mol. Gastroenterol. Hepatol. 10, 327–340 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.G. Gamba, W. Wang, L. Schild, Sodium chloride transport in the loop of Henle, distal convoluted tubule, and collecting duct, in Seldin and Giebisch’s The Kidney, R. J. Alpern, O. W. Moe, M. Caplan, Eds. (Academic Press, ed. 5, 2013), pp. 1143–1179. [Google Scholar]
- 88.A. Guillermo, A. Luis Reuss, in Seldin and Giebisch’s The Kidney, R. J. Alpern, O. W. Moe, M. Caplan, Eds. (Academic Press, ed. 5, 2013), pp. 95–120. [Google Scholar]
- 89.Nielsen S., Maunsbach A. B., Ecelbarger C. A., Knepper M. A., Ultrastructural localization of Na-K-2Cl cotransporter in thick ascending limb and macula densa of rat kidney. Am. J. Physiol. 275, F885–F893 (1998). [DOI] [PubMed] [Google Scholar]
- 90.Maeoka Y., McCormick J. A., NaCl cotransporter activity and Mg2+ handling by the distal convoluted tubule. Am. J. Physiol. Renal Physiol. 319, F1043–F1053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.A. S. L. Yu, F. Hanner, J. Peti-Peterdi, in Seldin and Giebisch’s The Kidney, R. J. Alpern, O. W. Moe, M. Caplan, Eds. (Academic Press, ed. 5, 2013), pp. 347–368. [Google Scholar]
- 92.Gao C., Higgins P. J., Zhang W., AQP2: Mutations associated with congenital nephrogenic diabetes insipidus and regulation by post-translational modifications and protein-protein interactions. Cell 9, 2172 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fuller P. J., Young M. J., Mechanisms of mineralocorticoid action. Hypertension 46, 1227–1235 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Hamm L. L., Nakhoul N., Hering-Smith K. S., Acid-base homeostasis. Clin. J. Am. Soc. Nephrol. 10, 2232–2242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saxena V., Gao H., Arregui S., Zollman A., Kamocka M. M., Xuei X., McGuire P., Hutchens M., Hato T., Hains D. S., Schwaderer A. L., Kidney intercalated cells are phagocytic and acidify internalized uropathogenic Escherichia coli. Nat. Commun. 12, 2405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klasen C., Ohl K., Sternkopf M., Shachar I., Schmitz C., Heussen N., Hobeika E., Levit-Zerdoun E., Tenbrock K., Reth M., Bernhagen J., el Bounkari O., MIF promotes B cell chemotaxis through the receptors CXCR4 and CD74 and ZAP-70 signaling. J. Immunol. 192, 5273–5284 (2014). [DOI] [PubMed] [Google Scholar]
- 97.Fan H., Hall P., Santos L. L., Gregory J. L., Fingerle-Rowson G., Bucala R., Morand E. F., Hickey M. J., Macrophage migration inhibitory factor and CD74 regulate macrophage chemotactic responses via MAPK and Rho GTPase. J. Immunol. 186, 4915–4924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zang Z. S., Xu Y. M., Lau A. T. Y., Molecular and pathophysiological aspects of metal ion uptake by the zinc transporter ZIP8 (SLC39A8). Toxicol Res 5, 987–1002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nielsen M. J., Moller H. J., Moestrup S. K., Hemoglobin and heme scavenger receptors. Antioxid. Redox Signal. 12, 261–273 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S20
Supplemental Information
References
Tables S1 to S13