Abstract
Background
Treatment options have been historically limited for cisplatin-ineligible patients with advanced urothelial carcinoma (UC). Given the need for alternatives to platinum-based chemotherapy, including non-chemotherapy regimens for patients with both impaired renal function and borderline functional status, in 2010 (prior to the immune checkpoint blockade era in metastatic UC), we initiated a phase II trial to test the activity of everolimus or everolimus plus paclitaxel in the cisplatin-ineligible setting.
Methods
This was an open-label phase II trial conducted within the US-based Hoosier Cancer Research Network (ClinicalTrials.gov number: NCT01215136). Patients who were cisplatin-ineligible with previously untreated advanced UC were enrolled. Patients with both impaired renal function and poor performance status were enrolled into cohort 1; patients with either were enrolled into cohort 2. Patients received everolimus 10 mg daily alone (cohort 1) or with paclitaxel 80 mg/m2 on days 1, 8, and 15 of each 28-day cycle (cohort 2). The primary outcome was clinical benefit at 4 months. Secondary outcomes were adverse events, progression-free survival (PFS), and 1-year overall survival (OS). Exploratory endpoints included genomic correlates of outcomes. The trial was not designed for comparison between cohorts.
Results
A total of 36 patients were enrolled from 2010 to 2018 (cohort 1, N = 7; cohort 2, N = 29); the trial was terminated due to slow accrual. Clinical benefit at 4 months was attained by 0 (0%, 95% confidence interval [CI] 0-41.0%) patients in cohort 1 and 11 patients (37.9%, 95% CI 20.7-57.7%) in cohort 2. Median PFS was 2.33 (95% CI 1.81-Inf) months in cohort 1 and 5.85 (95% CI 2.99-8.61) months in cohort 2. Treatment was discontinued due to adverse events for 2 patients (29%) in cohort 1 and 11 patients (38%) in cohort 2. Molecular alterations in microtubule associated genes may be associated with treatment benefit but this requires further testing.
Conclusion
Everolimus plus paclitaxel demonstrates clinical activity in cisplatin-ineligible patients with metastatic UC, although the specific contribution of everolimus cannot be delineated. Patients with both impaired renal function and borderline functional status may be difficult to enroll to prospective trials. (ClinicalTrials.gov Identifier NCT01215136).
Keywords: cisplatin-ineligible, everolimus, genomic, paclitaxel, urothelial cancer
Treatment options for chemotherapy ineligible patients are needed. This phase II trial tested the activity of everolimus or everolimus plus paclitaxel in the cisplatin-ineligible setting.
Lessons Learned.
Everolimus plus paclitaxel demonstrates clinical activity in cisplatin-ineligible patients with metastatic urothelial cancer, although the contribution of everolimus is unclear.
There is a need for treatment options for “chemotherapy-ineligible” patients, but these patients are challenging to enroll in prospective trials.
Discussion
Cisplatin remains the backbone of treatment for advanced UC. However, many patients are not eligible for cisplatin due to performance status or comorbidities. The subgroup of cisplatin-ineligible patients with both poor performance status and poor renal function experience increased toxicity and reduced benefit from carboplatin-based regimens necessitating novel treatment approaches. We initiated a phase II trial to test the activity of everolimus or everolimus plus paclitaxel in the cisplatin-ineligible setting shortly prior to a new era in drug development in metastatic UC. The shifting landscape, coupled with pragmatic considerations related to cohort 1, contributed to early closure due to poor accrual. Nonetheless, this trial has generated insights that may inform future treatment strategies.
There was a 4-month clinical benefit rate of 37.9% associated with everolimus and paclitaxel (EVP) among patients with either poor performance status or poor renal function (cohort 2). This benefit was most likely driven by paclitaxel, which has demonstrated efficacy in this context both as a single-agent and in combinations. The EVP combination has also been studied in patients with UC progressing despite platinum-based chemotherapy with an objective response rate of 13%, similar to the response rate with paclitaxel alone, suggesting limited benefit by adding everolimus, although the specific contribution of each agent cannot be defined here.
We initiated our trial in 2010 prior to the immune checkpoint blockade era and the subsequent shifts in the metastatic UC treatment landscape. Current standard first-line treatment for cisplatin-ineligible patients with metastatic UC includes carboplatin-based chemotherapy followed by switch maintenance immune checkpoint blockade or single agent immune checkpoint in patients with tumors harboring high levels of PD-L1 expression or patients who are “chemotherapy ineligible” (eg, those with poor functional status and renal function). The current trial, although performed in an earlier era and with a treatment without substantial activity, highlights the potential challenges of enrolling “chemotherapy ineligible” patients to prospective clinical trials; the median OS of patients in cohort 1 was only 4.5 months.
We examined genomic data from 17 patients in cohort 2 to identify possible biomarkers of response to EVP. There were no significant associations between somatic mutations, copy number variants, or mutational signatures and response. However, power was limited. One notable, albeit non-significant, observation was the high response rates to EVP among those with mutations in either of the microtubule-associated genes MACF1 or FRY (100%; Fisher’s exact P = .24 two sided; P = .14 one sided). To our knowledge, there have not been in vitro or in vivo experiments testing the relationship between mutations in these genes and paclitaxel sensitivity. Though the use of taxanes in latter lines of therapy for metastatic UC is decreasing in the context of new treatment options, treatment selection biomarkers for these newer treatments are still lacking and biomarkers of paclitaxel benefit could still impact clinical treatment strategies and warrant further testing.
Trial Information
| Disease | bladder cancer |
| Stage of disease/treatment | metastatic/advanced |
| Prior therapy | none |
| Type of study | phase II |
| Primary endpoint | clinical benefit rate at 4 months from treatment initiation |
| Secondary endpoint | toxicity, safety, correlative endpoint, other |
| Investigator’s analysis | active but results overtaken by other developments |
Additional Details of Endpoints or Study Design
Exploratory endpoints included genomic correlates of outcomes.
The study included two parallel cohorts and was not designed for statistical comparison of the cohorts. Each cohort used a separate Simon’s two-stage minimax design, with one-sided α 0.05 and power 0.8. For cohort 1, the minimal activity threshold was a 4-month clinical benefit rate (CBR) of ≤10% while the substantial activity threshold was a CBR ≥30%. For cohort 2, the minimal activity threshold was a CBR ≤25% while the substantial activity threshold was a CBR ≥45%.
Based on these parameters, we planned to accrue 15 evaluable patients in the first stage for cohort 1, and an additional 10 patients in the second stage. For cohort 2, we planned to enroll 17 patients in the first stage, and an additional 19 patients in the second stage. Anticipating a 10% dropout rate, the target accrual was 68 patients: 28 in cohort 1 and 40 in cohort 2. The trial opened in 2010 but was closed in 2018 due to slow accrual after having enrolled 36 patients: 7 in cohort 1 and 29 in cohort 2. All patients who received at least one dose of the trial medication were included in the final analyses for efficacy and safety.
Drug Information for Cohort 1
| Everolimus | |
|---|---|
| Generic/Working name | Everolimus |
| Drug Type | Small molecule |
| Drug Class | m-TOR |
| Dose | 10 mg per flat dose |
| Route | oral (p.o.) |
| Schedule of administration | daily |
Drug Information for Cohort 2
| Everolimus | |
|---|---|
| Generic/working name | everolimus |
| Drug type | small molecule |
| Drug class | m-TOR |
| Dose | 10 mg per flat dose |
| Route | oral (p.o.) |
| Schedule of administration | daily |
| Paclitaxel | |
| Generic/working name | paclitaxel |
| Drug type | chemotherapy |
| Drug class | taxane |
| Dose | 80 mg/m² |
| Route | i.v. |
| Schedule of administration: days 1, 8, and 15 of each 28-day cycle |
Patient Characteristics for Cohort 1
| Number of patients, male | 5 |
| Number of patients, female | 2 |
| Age | Median (range): 79 (59-90) years |
| Number of prior systemic therapies | Median (range): 0 |
| Performance status: ECOG | 0–0 |
| 1–0 | |
| 2–0 | |
| 3–0 | |
| Unknown–0 | |
| Other | Karnofsky performance status, Median (range) |
| 60 (60-70) | |
| Calculated creatinine clearance, median (range) | |
| 36.03 (10.54-60) |
Patient Characteristics for Cohort 2
| Number of patients, male | 22 |
| Number of patients, female | 7 |
| Age | Median (range): 72 (54-88 years) |
| Number of prior systemic therapies | |
| Performance status: ECOG | 0–0 |
| 1–0 | |
| 2–0 | |
| 3–0 | |
| Unknown–0 | |
| Other | Karnofsky performance status, median (range): |
| 80 (60-100) | |
| Calculated creatinine clearance, median (range): | |
| 51.3 (22-96) |
Primary Assessment Method for Cohort 1
| Title | Response at 4 months |
|---|---|
| Number of patients screened | 0 |
| Number of patients enrolled | 7 |
| Number of patients evaluable for toxicity | 7 |
| Number of patients evaluated for efficacy | 4 |
| Evaluation method | RECIST 1.1 |
| Response assessment CR | n = 0 (0%) |
| Response assessment PR | n = 0 (0%) |
| Response assessment SD | n = 0 (0%) |
| Response assessment PD | n = 4 (57.1%) |
| Response assessment other | n = 3 (42.9%) |
Primary Assessment Method for Cohort 2
| Title | Radiographic response at 4 months |
|---|---|
| Number of patients screened | 0 |
| Number of patients enrolled | 29 |
| Number of patients evaluable for toxicity | 29 |
| Number of patients evaluated for efficacy | 20 |
| Evaluation method | RECIST 1.1 |
| Response assessment CR | n = 0 (0%) |
| Response assessment PR | n = 8 (27.6%) |
| Response assessment SD | n = 3 (10.3%) |
| Response assessment PD | n = 9 (31%) |
| Response assessment other | n = 9 (31%) |
Outcome Notes
Table 2 shows additional details of study outcome.
Table 2.
Radiographic outcomes.
| Cohort 1 (N = 7) | Cohort 2 (N = 29) | Overall (N = 36) | |
|---|---|---|---|
| Response at 4 months | |||
| Complete response, N (%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Partial response, N (%) | 0 (0%) | 8 (27.6%) | 8 (22.2%) |
| Stable disease, N (%) | 0 (0%) | 3 (10.3%) | 3 (8.3%) |
| Progressive disease, N (%) | 4 (57.1%) | 9 (31%) | 13 (36.1%) |
| Not evaluable, N (%) | 3 (42.9%) | 9 (31%) | 12 (33.3%) |
| Best response | |||
| Complete response, N (%) | 0 (0%) | 1 (3.4%) | 1 (2.8%) |
| Partial response, N (%) | 0 (0%) | 13 (44.8%) | 13 (36.1%) |
| Stable disease, N (%) | 4 (57.1%) | 6 (20.7%) | 10 (27.8%) |
| Prog. disease, N (%) | 2 (28.6%) | 4 (13.8%) | 6 (16.7%) |
| Not evaluable, N (%) | 1 (14.3%) | 5 (17.2%) | 6 (16.7%) |
Adverse Events: Cohort 1, All Cycles
| Name | *NC/NA | 1 | 2 | 3 | 4 | 5 | All grades |
|---|---|---|---|---|---|---|---|
| Anemia | 29% | 14% | 0% | 57% | 0% | 0% | 71% |
| Anorexia | 57% | 0% | 43% | 0% | 0% | 0% | 43% |
| Cholesterol high | 57% | 43% | 0% | 0% | 0% | 0% | 43% |
| Constipation | 57% | 43% | 0% | 0% | 0% | 0% | 43% |
| Diarrhea | 57% | 29% | 0% | 14% | 0% | 0% | 43% |
| Dysgeusia | 57% | 43% | 0% | 0% | 0% | 0% | 43% |
| Dyspnea | 57% | 14% | 14% | 14% | 0% | 0% | 43% |
| Fatigue | 0% | 57% | 29% | 14% | 0% | 0% | 100% |
| Hypertension | 57% | 0% | 43% | 0% | 0% | 0% | 43% |
| Nausea | 57% | 14% | 29% | 0% | 0% | 0% | 43% |
| Rash acneiform | 57% | 0% | 43% | 0% | 0% | 0% | 43% |
| Urinary tract infection | 43% | 0% | 43% | 14% | 0% | 0% | 57% |
Data shown here are the AEs observed in at least 40% of patients. Table 3 shows a detailed listing.
Table 3.
Treatment-emergent adverse events occurring in at least 5% of patients, sorted alphabetically.
| Cohort 1 (N = 7) | Cohort 2 (N = 29) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adverse event | Grade 1 | Grade 2 | Grade 3 | Grade 4 | Any | Grade 1 | Grade 2 | Grade 3 | Grade 4 | Any | ||||||||||
| No.% | No.% | No.% | No.% | No.% | No.% | No.% | No.% | No.% | No.% | |||||||||||
| Any adverse event | 7 | 100 | 6 | 86 | 5 | 71 | 2 | 29 | 7 | 100 | 29 | 100 | 28 | 97 | 26 | 90 | 6 | 21 | 29 | 100 |
| Abdominal pain | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 9 | 31 | 3 | 10 | 2 | 7 | 0 | — | 12 | 41 |
| Acute kidney injury | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 3 | 10 | 0 | — | 0 | — | 3 | 10 |
| Alanine aminotransferase increased | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 1 | 3 | 1 | 3 | 0 | — | 2 | 7 |
| Alopecia | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 10 | 34 | 7 | 24 | 0 | — | 0 | — | 13 | 45 |
| Anemia | 3 | 43 | 4 | 57 | 4 | 57 | 0 | — | 5 | 71 | 17 | 59 | 16 | 55 | 9 | 31 | 0 | — | 22 | 76 |
| Anorexia | 2 | 29 | 3 | 43 | 0 | — | 0 | — | 3 | 43 | 11 | 38 | 6 | 21 | 1 | 3 | 0 | — | 15 | 52 |
| Anxiety | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 6 | 21 | 2 | 7 | 0 | — | 0 | — | 6 | 21 |
| Aspartate aminotransferase increased | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 1 | 3 | 0 | — | 2 | 7 |
| Back pain | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 3 | 10 | 2 | 7 | 0 | — | 0 | — | 4 | 14 |
| Blood and lymphatic system disorders - Other | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 0 | — | 0 | — | 2 | 7 |
| Cataract | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Chills | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 4 | 14 | 0 | — | 0 | — | 0 | — | 4 | 14 |
| Cholesterol high | 3 | 43 | 0 | — | 0 | — | 0 | — | 3 | 43 | 8 | 28 | 0 | — | 0 | — | 0 | — | 8 | 28 |
| Chronic kidney disease | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 2 | 7 | 1 | 3 | 3 | 10 |
| Constipation | 3 | 43 | 0 | — | 0 | — | 0 | — | 3 | 43 | 14 | 48 | 5 | 17 | 0 | — | 0 | — | 16 | 55 |
| Cough | 2 | 29 | 0 | — | 0 | — | 0 | — | 2 | 29 | 11 | 38 | 1 | 3 | 0 | — | 0 | — | 11 | 38 |
| Creatinine increased | 1 | 14 | 1 | 14 | 1 | 14 | 0 | — | 2 | 29 | 2 | 7 | 3 | 10 | 0 | — | 0 | — | 5 | 17 |
| Dehydration | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 3 | 10 | 3 | 10 | 0 | — | 5 | 17 |
| Depression | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 7 | 24 | 2 | 7 | 0 | — | 0 | — | 7 | 24 |
| Diarrhea | 2 | 29 | 0 | — | 1 | 14 | 0 | — | 3 | 43 | 16 | 55 | 5 | 17 | 4 | 14 | 0 | — | 18 | 62 |
| Dizziness | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 5 | 17 | 0 | — | 0 | — | 0 | — | 5 | 17 |
| Dry skin | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Dysgeusia | 3 | 43 | 0 | — | 0 | — | 0 | — | 3 | 43 | 6 | 21 | 3 | 10 | 0 | — | 0 | — | 7 | 24 |
| Dyspepsia | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 7 | 24 | 0 | — | 0 | — | 0 | — | 7 | 24 |
| Dyspnea | 3 | 43 | 1 | 14 | 1 | 14 | 0 | — | 3 | 43 | 12 | 41 | 2 | 7 | 4 | 14 | 0 | — | 15 | 52 |
| Edema limbs | 1 | 14 | 1 | 14 | 0 | — | 0 | — | 2 | 29 | 12 | 41 | 8 | 28 | 0 | — | 0 | — | 14 | 48 |
| Epistaxis | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 5 | 17 | 0 | — | 0 | — | 0 | — | 5 | 17 |
| Fatigue | 7 | 100 | 2 | 29 | 1 | 14 | 0 | — | 7 | 100 | 25 | 86 | 10 | 34 | 2 | 7 | 0 | — | 25 | 86 |
| Fever | 2 | 29 | 0 | — | 0 | — | 0 | — | 2 | 29 | 11 | 38 | 3 | 10 | 0 | — | 0 | — | 12 | 41 |
| Flatulence | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Fracture | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 0 | — | 0 | — | 2 | 7 |
| Gastrointestinal disorders - other | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 2 | 7 | 0 | — | 0 | — | 4 | 14 |
| Generalized muscle weakness | 0 | — | 1 | 14 | 0 | — | 0 | — | 1 | 14 | 2 | 7 | 0 | — | 1 | 3 | 0 | — | 2 | 7 |
| Hallucinations | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Headache | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 4 | 14 | 0 | — | 0 | — | 0 | — | 4 | 14 |
| Hematuria | 1 | 14 | 1 | 14 | 1 | 14 | 0 | — | 2 | 29 | 1 | 3 | 1 | 3 | 0 | — | 0 | — | 2 | 7 |
| Hyperglycemia | 0 | — | 1 | 14 | 1 | 14 | 0 | — | 2 | 29 | 3 | 10 | 1 | 3 | 1 | 3 | 0 | — | 3 | 10 |
| Hyperhidrosis | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Hypertension | 0 | — | 3 | 43 | 0 | — | 0 | — | 3 | 43 | 5 | 17 | 7 | 24 | 4 | 14 | 0 | — | 12 | 41 |
| Hypertriglyceridemia | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 8 | 28 | 3 | 10 | 0 | — | 0 | — | 8 | 28 |
| Hypoalbuminemia | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 3 | 10 | 1 | 3 | 0 | — | 0 | — | 3 | 10 |
| Hypocalcemia | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 1 | 3 | 1 | 3 | 1 | 3 | 2 | 7 |
| Hypokalemia | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 3 | 10 | 4 | 14 | 2 | 7 | 0 | — | 6 | 21 |
| Hypomagnesemia | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 4 | 14 | 1 | 3 | 0 | — | 0 | — | 4 | 14 |
| Hyponatremia | 1 | 14 | 1 | 14 | 0 | — | 0 | — | 1 | 14 | 3 | 10 | 0 | — | 1 | 3 | 0 | — | 3 | 10 |
| Hypophosphatemia | 1 | 14 | 1 | 14 | 0 | — | 0 | — | 2 | 29 | 1 | 3 | 1 | 3 | 2 | 7 | 0 | — | 2 | 7 |
| Infections and infestations - other | 0 | — | 1 | 14 | 1 | 14 | 0 | - | 2 | 29 | 1 | 3 | 3 | 10 | 3 | 10 | 0 | - | 5 | 17 |
| Insomnia | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 14 | 48 | 0 | — | 0 | — | 0 | — | 14 | 48 |
| Lung infection | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 0 | — | 2 | 7 |
| Mucositis oral | 2 | 29 | 2 | 29 | 1 | 14 | 0 | — | 2 | 29 | 10 | 34 | 8 | 28 | 0 | — | 0 | — | 14 | 48 |
| Myocardial infarction | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 2 | 7 |
| Nail infection | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 1 | 3 | 0 | — | 2 | 7 |
| Nasal congestion | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 0 | — | 0 | — | 2 | 7 |
| Nausea | 1 | 14 | 2 | 29 | 0 | — | 0 | — | 3 | 43 | 14 | 48 | 3 | 10 | 2 | 7 | 0 | — | 14 | 48 |
| Neck pain | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Nervous system disorders - other | 0 | — | 1 | 14 | 1 | 14 | 0 | — | 1 | 14 | 0 | — | 2 | 7 | 0 | — | 0 | — | 2 | 7 |
| Neutrophil count decreased | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 4 | 14 | 4 | 14 | 5 | 17 | 1 | 3 | 11 | 38 |
| Non-cardiac chest pain | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 0 | — | 0 | — | 2 | 7 |
| Pain | 3 | 43 | 0 | — | 0 | — | 0 | — | 3 | 43 | 17 | 59 | 7 | 24 | 2 | 7 | 0 | — | 18 | 62 |
| Pain in extremity | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 6 | 21 | 0 | — | 0 | — | 0 | — | 6 | 21 |
| Peripheral motor neuropathy | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 1 | 3 | 0 | — | 0 | — | 2 | 7 |
| Peripheral sensory neuropathy | 2 | 29 | 0 | — | 0 | — | 0 | — | 2 | 29 | 12 | 41 | 4 | 14 | 0 | — | 0 | — | 14 | 48 |
| Platelet count decreased | 2 | 29 | 1 | 14 | 0 | — | 0 | — | 2 | 29 | 6 | 21 | 2 | 7 | 1 | 3 | 0 | — | 6 | 21 |
| Pneumonitis | 0 | — | 0 | — | 1 | 14 | 0 | — | 1 | 14 | 0 | — | 3 | 10 | 1 | 3 | 0 | — | 4 | 14 |
| Pruritus | 1 | 14 | 1 | 14 | 0 | — | 0 | — | 2 | 29 | 4 | 14 | 2 | 7 | 0 | — | 0 | — | 6 | 21 |
| Rash acneiform | 1 | 14 | 3 | 43 | 0 | — | 0 | — | 3 | 43 | 8 | 28 | 4 | 14 | 1 | 3 | 0 | — | 9 | 31 |
| Rash maculo-papular | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 3 | 10 | 1 | 3 | 0 | — | 0 | — | 3 | 10 |
| Renal and urinary disorders - Other, specify | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 0 | — | 0 | — | 1 | 3 | 2 | 7 |
| Respiratory failure | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 2 | 7 |
| Sepsis | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 2 | 7 |
| Sinus disorder | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Sore throat | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 0 | — | 2 | 7 |
| Supraventricular tachycardia | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 1 | 3 | 1 | 3 | 0 | — | 2 | 7 |
| Thromboembolic event | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 3 | 10 | 0 | — | 0 | — | 4 | 14 |
| Tooth infection | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 2 | 7 | 0 | — | 0 | — | 2 | 7 |
| Urinary frequency | 0 | — | 0 | — | 0 | — | 0 | — | 0 | — | 4 | 14 | 3 | 10 | 0 | — | 0 | — | 6 | 21 |
| Urinary tract infection | 0 | — | 4 | 57 | 1 | 14 | 0 | — | 4 | 57 | 0 | — | 11 | 38 | 6 | 21 | 0 | — | 13 | 45 |
| Urinary tract pain | 1 | 14 | 1 | 14 | 0 | — | 0 | — | 2 | 29 | 2 | 7 | 1 | 3 | 0 | — | 0 | — | 3 | 10 |
| Vomiting | 2 | 29 | 0 | — | 0 | — | 0 | — | 2 | 29 | 9 | 31 | 1 | 3 | 2 | 7 | 0 | — | 12 | 41 |
| Weight loss | 1 | 14 | 0 | — | 0 | — | 0 | — | 1 | 14 | 3 | 10 | 2 | 7 | 0 | — | 0 | — | 4 | 14 |
Highest grade treatment-emergent adverse events occurring in at least 5% of patients, sorted alphabetically. Treatment was discontinued due to adverse events for 2 patients (29%) in cohort 1 and 11 patients (38%) in cohort 2. Treatment-emergent grades 3-4 adverse events developed in 5 patients (71%) in cohort 1 and 26 patients (90%) in cohort 2 (Table 3). The most common grades 3-4 adverse event in both cohorts was anemia (cohort 1, N = 4; cohort 2, N = 9). The most common adverse events of any grade in cohort 1 were fatigue (N = 7, 100%), anemia (N = 5, 71%), and urinary tract infections (N = 4, 57%). The most common adverse events of any grade in cohort 2 were fatigue (N = 25, 86%), anemia (N = 22, 76%), pain (N = 18, 62%), dyspnea (N = 15, 52%), and gastrointestinal symptoms eg, diarrhea (N = 18, 62%), constipation (N = 16, 55%), anorexia (N = 15, 52%), and nausea (N = 14, 48%).
Adverse Events: Cohort 2, All Cycles
| Name | *NC/NA | 1 | 2 | 3 | 4 | 5 | All grades |
|---|---|---|---|---|---|---|---|
| Abdominal pain | 59% | 24% | 10% | 7% | 0% | 0% | 41% |
| Alopecia | 55% | 21% | 24% | 0% | 0% | 0% | 45% |
| Anemia | 24% | 17% | 28% | 31% | 0% | 0% | 76% |
| Anorexia | 48% | 28% | 21% | 3% | 0% | 0% | 52% |
| Constipation | 45% | 38% | 17% | 0% | 0% | 0% | 55% |
| Diarrhea | 38% | 38% | 10% | 14% | 0% | 0% | 62% |
| Dyspnea | 48% | 31% | 7% | 14% | 0% | 0% | 52% |
| Fatigue | 14% | 52% | 28% | 7% | 0% | 0% | 86% |
| Fever | 59% | 31% | 10% | 0% | 0% | 0% | 41% |
| Hypertension | 59% | 14% | 14% | 14% | 0% | 0% | 41% |
| Insomnia | 52% | 48% | 0% | 0% | 0% | 0% | 48% |
| Mucositis oral | 52% | 21% | 28% | 0% | 0% | 0% | 48% |
| Nausea | 52% | 34% | 7% | 7% | 0% | 0% | 48% |
| Peripheral sensory neuropathy | 52% | 34% | 14% | 0% | 0% | 0% | 48% |
| Urinary tract infection | 55% | 0% | 24% | 21% | 0% | 0% | 45% |
| Vomiting | 59% | 31% | 3% | 7% | 0% | 0% | 41% |
Data shown here are AEs occurring in at least 40% of patients. Table 3 shows details.
Assessment, Analysis, and Discussion
| Completion | did not fully accrue |
| Investigator’s assessment | active but results overtaken by other developments |
Introduction
The standard treatment for metastatic or unresectable urothelial carcinoma (UC) is cisplatin-based chemotherapy. However, a large subset of patients with UC are considered ineligible for cisplatin due to comorbidities such as chronic renal insufficiency.1,2
Treatment options for cisplatin-ineligible patients are limited. The EORTC 30986 trial compared the combination of gemcitabine plus carboplatin (GCa) versus methotrexate, carboplatin, plus vinblastine (M-CAVI) and demonstrated severe acute toxicity in 9.3% of patients receiving GCa and 21.2% of patients receiving M-CAVI.3 Patients with both impaired renal function and borderline functional status experienced even higher rates of severe acute toxicity, questioning the role of platinum-based regimens in this context.
Overexpression of the mTOR pathway has been observed in invasive UC and inactivation of endogenous mTOR inhibitors, such as PTEN, has been linked to UC progression.4,5 The mTOR inhibitor everolimus has demonstrated single-agent antitumor activity in patients with tumors harboring somatic alterations associated with mTOR pathway activation.6 Paclitaxel has single-agent activity in UC and has demonstrated safety in the treatment of cisplatin-ineligible patients with advanced UC.7 In model systems of cancer, PI3K/AKT/mTOR pathway upregulation is associated with taxane resistance and mTOR pathway inhibition has been shown to synergize with paclitaxel.8-10 The combination of everolimus and paclitaxel (EVP) has also demonstrated safety and activity across a variety of tumor types.11-13
Given the need for alternatives to platinum-based chemotherapy, including nonchemotherapy regimens for patients with both impaired renal function and borderline functional status, in 2010 (prior to the immune checkpoint blockade era in metastatic UC), we initiated a phase II trial to test the activity of everolimus or everolimus plus paclitaxel in the cisplatin-ineligible setting.
Patients and Methods
Participants
Adult patients (aged 18 or older) with histologically proven UC who were ineligible for cisplatin and who had not been previously treated for metastatic disease were eligible for this study. Upper tract disease and mixed histology (with a UC component) were allowed. Cisplatin ineligibility was based on one of two criteria: (1) calculated creatinine clearance (by the Cockroft-Gault formula) <60 mL/minute, (2) Karnofsky performance status 60-70%. Patients meeting both criteria were assigned to cohort 1 while patients meeting only one criterion were assigned to cohort 2. Key exclusion criteria included active brain metastases and lack of measurable disease (per RECIST14). Patients were enrolled from treatment centers within the US-based Hoosier Cancer Research Network.
Trial Oversight
The protocol was approved by the Institutional Review Board of each participating institution. Written informed consent was obtained from all participants prior to enrollment. The study was performed in accordance with ethical principles originating from the Declaration of Helsinki, which are consistent with ICH/Good Clinical Practice, and applicable regulatory requirements.
Interventions
Patients in cohort 1 were assigned to take everolimus alone (EVE) at a dose of 10 mg by mouth daily, without interruption. Medications were dispensed on an outpatient basis on day 1 of each 28-day cycle. Patients in cohort 2 were assigned to a combination of EVP. Everolimus was prescribed at the same dose and schedule as for cohort 1. Paclitaxel 80 mg/m2 was given as a 1-hour intravenous infusion on days 1, 8, and 15 of each 28-day cycle.
Dose reductions were permitted in accordance with a schedule specified in the protocol. Paclitaxel could be reduced to 60 mg/m2 and everolimus could be reduced to a minimum of 5mg every other day. The study drugs were discontinued if further dose reductions were required or if treatment was interrupted for greater than 4 weeks.
Treatment was continued until radiographic progression (by RECIST criteria), unacceptable toxicity, death, or discontinuation for any other reason. Cross-sectional imaging was obtained every 2 cycles until disease progression.
Outcomes
The primary objective was to evaluate CBR at 4 months from treatment initiation. Clinical benefit was defined as complete response (CR), partial response (PR), or stable disease (SD) per RECIST criteria. Secondary objectives were to evaluate the safety of EVE and EVP in this population, and to determine progression-free survival (PFS) and 1-year overall survival (OS). Exploratory objectives included identifying genomic correlates of outcomes using whole-exome and transcriptome sequencing data from archived tumor samples.
Genomic Analyses
Formalin-fixed paraffin-embedded tumor and paired blood normal samples (N = 17) were submitted for whole-exome sequencing (WES). Exome capture and sequencing library preparation were performed using the SureSelect Human All Exon V7, no UTR hybridization capture kit from Agilent (Santa Clara, CA). Libraries were sequenced on an Illumina HiSeq 4000 instrument with 100-bp paired-end reads. An in-house GATK4-based pipeline (TIGRIS) was used to analyze the WES profiles. Somatic variants with a general allelic fraction (AF) or ethnic-specific AF ≥ 0.5% in the gnomAD database were removed from analysis. Copy number variant (CNV) segmentation profiles were called using saasCNV,15 then fed into GISTIC 2.016 across the entire cohort to look for significant CNV regions. Mutational signature analysis was done via R package quadprog,17 and only samples with SNVs in exome region ≥50 at AF ≥ 5% were included, resulting a total of 14 samples. The signature fitting step was conducted using a reference catalog consisting of bladder cancer specific COSMIC v2 mutational signatures 1, 2, 5, 10, and 13.18
RNA sequencing was performed on 8 of the 17 WES samples using SureSelect RNADirect (Agilent, Santa Clara, CA). An in-house RNaseq data processing pipeline (EUPHRATES) was used to analyze the data. UCSC’s hg19 genome build was used as the standard reference genome for all analyses. Gene annotations were derived from UCSC’s refGene table. Briefly, STAR (v2.6.1.d)19 was used for read alignment, and featureCounts (v1.4.4)20 was used to measure abundance of genomic features. Differential gene expression analysis was then performed using the DESeq221 package using read counts from the previous step. Gene fusion events were also screened for using open source fusion calling tools FusionCatcher (v1.0)22 and FusionInspector(v2.1.0).23
Statistical Considerations
The study included two parallel cohorts and was not designed for statistical comparison of the cohorts. Each cohort used a separate Simon’s two-stage minimax design, with one-sided α 0.05 and power 0.8. For cohort 1, the minimal activity threshold was a 4-month CBR of ≤10% while the substantial activity threshold was a CBR ≥30%. For cohort 2, the minimal activity threshold was a CBR ≤25% while the substantial activity threshold was a CBR ≥45%.
Based on these parameters, we planned to accrue 15 evaluable patients in the first stage for cohort 1, and an additional 10 patients in the second stage. For cohort 2, we planned to enroll 17 patients in the first stage, and an additional 19 patients in the second stage. Anticipating a 10% dropout rate, the target accrual was 68 patients: 28 in cohort 1 and 40 in cohort 2. The trial opened in 2010 but was closed in 2018 due to slow accrual after having enrolled 36 patients: 7 in cohort 1 and 29 in cohort 2. All patients who received at least one dose of the trial medication were included in the final analyses for efficacy and safety.
Descriptive statistics were summarized using medians and ranges for continuous variables and counts and proportions for categorical variables. The primary outcome of 4-month CBR was calculated as the number of patients achieving clinical benefit at 4 months divided by the total number of patients in the cohort. 95% confidence intervals for proportions were calculated using the Clopper-Pearson exact method. Survival outcomes were estimated using the Kaplan-Meier method.
Statistical analyses were conducted in R statistical software, version 4.0.0.
Results
Patient Characteristics
Of the 36 patients enrolled, the majority (75%, N = 27) were men. Seven patients with both impaired renal function and poor performance status were assigned to EVE in cohort 1. Twenty-nine patients with either impaired renal function or poor performance status were assigned to EVP in cohort 2. The median Karnofsky performance status (60% vs 80%, P < .001) and calculated creatinine clearance (36.03 vs 51.3 mL/minute, P = .12) were both numerically lower in cohort 1 compared with cohort 2 (Table 1).
Table 1.
Baseline characteristics.
| Cohort 1 (N = 7) | Cohort 2 (N = 29) | Overall (N = 36) | |
|---|---|---|---|
| Age, median (range) | 79 (59-90) | 72 (54-88) | 73 (54-90) |
| Male, N (%) | 5 (71.4%) | 22 (75.9%) | 27 (75%) |
| White, N (%) | 5 (71.4%) | 27 (93.1%) | 32 (88.9%) |
| Black, N (%) | 2 (28.6%) | 2 (6.9%) | 4 (11.1%) |
| Non-Hispanic, N (%) | 7 (100%) | 28 (96.6%) | 35 (97.2%) |
| Karnofsky performance status, median (range) | 60 (60-70) | 80 (60-100) | 80 (60-100) |
| Calculated creatinine clearance, median (range) | 36.03 (10.54-60) | 51.3 (22-96) | 50.35 (10.54-96) |
Of the 36 patients enrolled, the majority (75%, N = 27) were men. Seven patients with both impaired renal function and poor performance status were assigned to EVE in cohort 1. Twenty-nine patients with either impaired renal function or poor performance status were assigned to EVP in cohort 2. The median Karnofsky performance status (60% vs 80%, P < .001) and calculated creatinine clearance (36.03 vs 51.3 mL/minute, P = .12) were both numerically lower in cohort 1 compared with cohort 2.
Efficacy
No patients (0%, 95% confidence interval [CI] 0-41.0%) in cohort 1 attained the primary outcome of clinical benefit at 4 months; 11 patients (37.9%, 95% CI 20.7-57.7%) in cohort 2 attained the primary outcome (Table 2). Twelve patients who were not evaluable for the primary outcome (due to lack of imaging) were included in this intent to treat analysis. Three patients in cohort 1 died prior to the 4-month evaluation. Nine patients in cohort 2 were not evaluable at 4 months; 4 had died prior to that time point, while the remaining 5 did not have 4-month imaging.
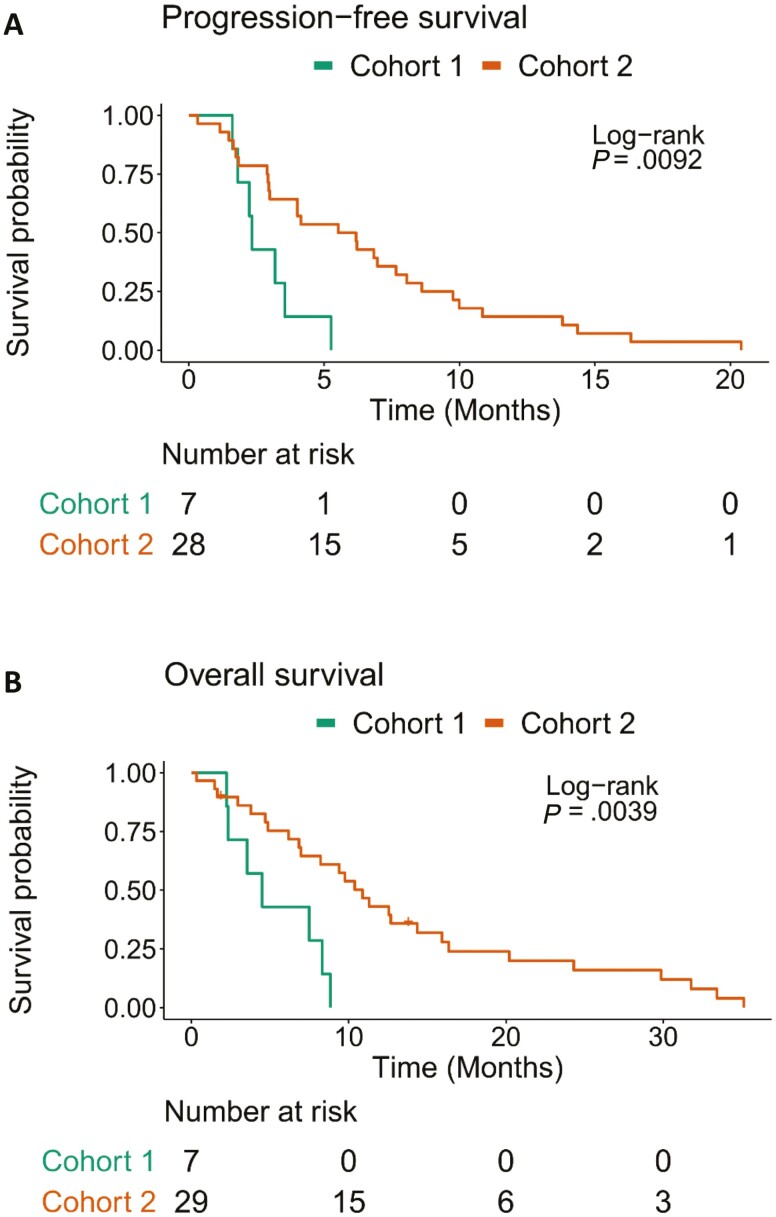
The median PFS was 2.33 (95% CI 1.81-Inf) months in cohort 1 and 5.85 (95% CI 2.99-8.61) months in cohort 2 (Figure 1A). Median OS was 4.5 (95% CI 2.33-Inf) months in cohort 1 and 10.9 (95% CI 6.97-16.4) months in cohort 2 (Figure 1B). Overall survival at 1 year was not estimable for cohort 1 and was 43% (95% CI 28.1-65.9) in cohort 2.
Figure 1.
Kaplan-Meier curves for (A) progression-free survival and (B) overall survival, stratified by cohort. The median progression-free survival was 2.33 (95% CI 1.81-Inf) months in cohort 1 and 5.85 (95% CI 2.99-8.61) months in cohort 2. Median overall survival was 4.5 (95% CI 2.33-Inf) months in cohort 1 and 10.9 (95% CI 6.97-16.4) months in cohort 2.
Safety and Tolerability
The median duration of exposure to EVE in cohort 1 was 1.87 months (range 0.84-5); the median duration of exposure to EVP in cohort 2 was 2.83 months (range 0.23-20). Treatment was discontinued due to adverse events for 2 patients (29%) in cohort 1 and 11 patients (38%) in cohort 2. Treatment-emergent grades 3-4 adverse events developed in 5 patients (71%) in cohort 1 and 26 patients (90%) in cohort 2 (Table 3). The most common grades 3-4 adverse event in both cohorts was anemia (cohort 1, N = 4; cohort 2, N = 9).
The most common adverse events of any grade in cohort 1 were fatigue (N = 7, 100%), anemia (N = 5, 71%), and urinary tract infections (N = 4, 57%). The most common adverse events of any grade in cohort 2 were fatigue (N = 25, 86%), anemia (N = 22, 76%), pain (N = 18, 62%), dyspnea (N = 15, 52%), and gastrointestinal symptoms, eg, diarrhea (N = 18, 62%), constipation (N = 16, 55%), anorexia (N = 15, 52%), and nausea (N = 14, 48%).
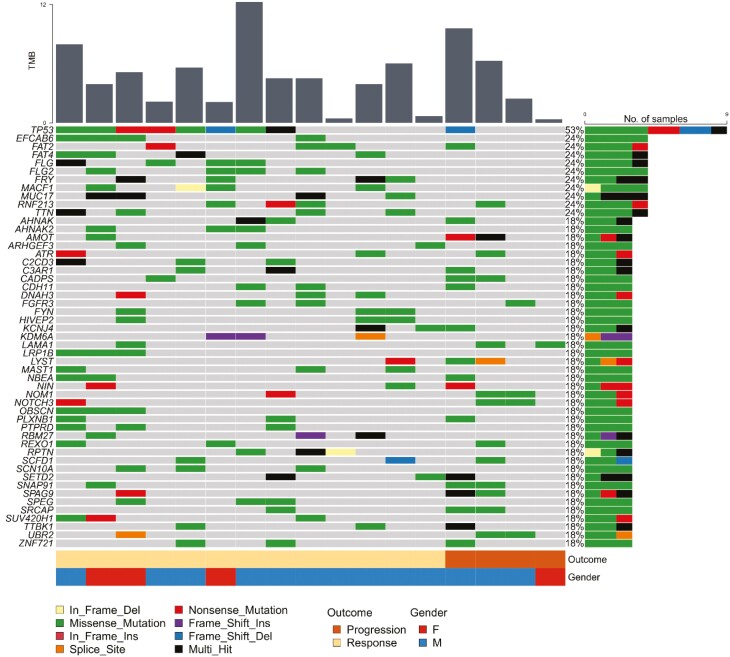
Genomic Alterations Associated with Response
Whole-exome sequencing was performed on baseline biopsy samples from 19 patients in cohort 2 (Figure 2). Of these, 17 patients were evaluable for radiographic response at 4 months. We classified these patients into 13 responders and 4 non-responders, with response defined as CR, PR, or SD with a PFS of at least 120 days. The most commonly mutated gene in the cohort was TP53 (N = 9); 8 of 9 patients with TP53 mutations were responders (Fisher’s exact P = .29). Other notable recurrent mutations included the microtubule-related genes MACF1 (N = 4) and FRY (N = 4). All 6 patients with mutations in either MACF1 or FRY were responders, although the association was not statistically significant (Fisher’s exact P = .24 two sided; P = .14 one sided). TSC1 mutations have previously been linked with everolimus sensitivity in mUC.6 One patient had a TSC1 mutation (p.Pro17fs) and was a responder.
Figure 2.
Mutational landscape of whole-exome sequencing cohort (N = 17). Genes mutated in at least three samples in the cohort are listed. Each column represents one sample. The vertical bar plot depicts tumor mutation burden in each sample. The horizontal bar plot summarizes the number and type of mutations (by color) for each gene. The tracks along the bottom provide additional clinical context, color coding each sample according to the patient’s gender and clinical outcome. The most commonly mutated gene in the cohort was TP53 (N = 9); 8 of 9 patients with TP53 mutations were responders (Fisher’s exact P = .29). Other notable recurrent mutations included the microtubule-related genes MACF1 (N = 4) and FRY (N = 4). All 6 patients with mutations in either MACF1 or FRY were responders, although the association was not statistically significant (Fisher’s exact P =.24 two sided; P =.14 one sided).
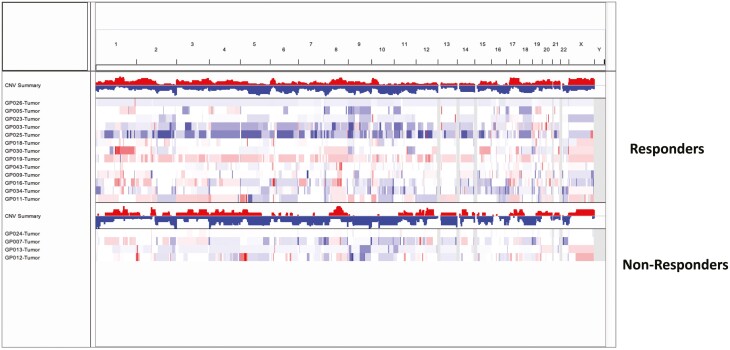
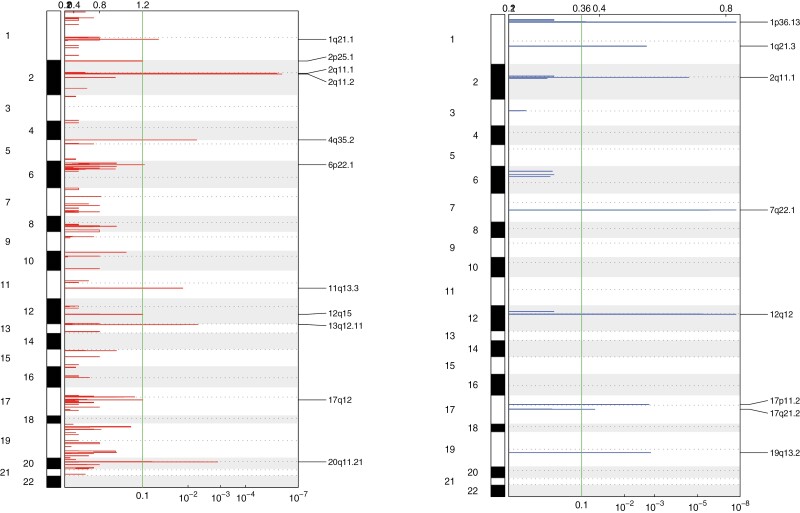
Copy number segmentation profiles were qualitatively similar between responders and non-responders (Figure 3). Due to the small number of non-responders, the two cohorts were combined to identify significantly enriched CNVs by GISTIC 2.0 (Tables 4-6). The most significant regions included 2q11.2, 2q11.1 for gains and 1p36.13, 7q22.1, 12q12, 2q11.1 for losses (Figure 4). The significantly amplified regions included several genes involved in fibroblast growth factor signaling, upstream of the PI3K/AKT/mTOR pathway: FGF3, FGF4, FGF9, FGF19, and FRS2.
Figure 3.
A heatmap of the genome-wide copy number variation (CNV) profiles based on median log2ratio, stratified by response. Individual patient samples are shown along the y-axis with amplification events in red and loss events in blue. The number of CNV events at each genomic locus for responders and nonresponders are summarized as bar plots at the top for both responders and non-responders. Copy number segmentation profiles were qualitatively similar between responders and non-responders.
Table 4.
Significant CNV gain regions called by GISTIC across all 17 WES T/N samples submitted for whole exome sequencing (WES). GISTIC is an algorithm that identified regions of the genome that are gained or lost more than expected by chance across a set of samples.
| Cytoband | 2q11.2 | 2q11.1 | 20q11.21 | 13q12.11 | 4q35.2 | 11q13.3 | 1q21.1 | 6p22.1 | 2p25.1 | 12q15 | 17q12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| q value | 1.20E−06 | 2.34E−06 | 0.0012368 | 0.0053277 | 0.0057245 | 0.013732 | 0.049551 | 0.092026 | 0.098716 | 0.098716 | 0.098716 |
| Residual q value | 1.82E−06 | 4.00E−06 | 0.0012368 | 0.0053277 | 0.0057245 | 0.013732 | 0.049551 | 0.092026 | 0.098716 | 0.098716 | 0.098716 |
| Wide peak boundaries | chr2:97820512-97828928 | chr2:96604806-96610328 | chr20:29628342-29632831 | chr13:22255168-23253312 | chr4:189022421-190878451 | chr11:68512544-69949326 | chr1:144852237-145293666 | chr6:29692730-29911259 | chr2:8943044-10729896 | chr12:66990486-70918140 | chr17:37557600-38062138 |
| Genes in wide peak | ANKRD36 | [LOC729234] | FRG1B | FGF9 | FRG1 | hsa-mir-3164 | SEC22B | HLA-A | hsa-mir-4261 | hsa-mir-1279 | ERBB2 |
| HSP90AA4P | CCND1 | PDE4DIP | HLA-F | HPCAL1 | CPM | GRB7 | |||||
| TRIML2 | CPT1A | NOTCH2NL | HLA-G | ODC1 | IFNG | NEUROD2 | |||||
| TRIML1 | FGF3 | NBPF10 | HLA-H | RRM2 | LYZ | PNMT | |||||
| LOC401164 | FGF4 | HCG4 | ADAM17 | MDM2 | MED1 | ||||||
| IGHMBP2 | HCG4B | KLF11 | CNOT2 | TCAP | |||||||
| MTL5 | HLA-F-AS1 | ASAP2 | PTPRB | STARD3 | |||||||
| FGF19 | IFITM4P | TAF1B | RAP1B | IKZF3 | |||||||
| MYEOV | LOC554223 | ITGB1BP1 | YEATS4 | CDK12 | |||||||
| ANO1 | YWHAQ | DYRK2 | GSDMB | ||||||||
| MRGPRD | GRHL1 | CCT2 | PPP1R1B | ||||||||
| MRGPRF | CPSF3 | FRS2 | MIEN1 | ||||||||
| MRPL21 | KIDINS220 | CPSF6 | FBXL20 | ||||||||
| TPCN2 | NOL10 | GRIP1 | PGAP3 | ||||||||
| ORAOV1 | MBOAT2 | KCNMB4 | ZPBP2 | ||||||||
| CYS1 | IL22 | MIR4728 | |||||||||
| IAH1 | SLC35E3 | ||||||||||
| C2orf48 | IL26 | ||||||||||
| SNORA80B | CAND1 | ||||||||||
| MIR4261 | MDM1 | ||||||||||
| NUP107 | |||||||||||
| RAB3IP | |||||||||||
| BEST3 | |||||||||||
| LRRC10 | |||||||||||
| MIR1279 | |||||||||||
| SNORA70G | |||||||||||
| MIR3913-2 | |||||||||||
| MIR3913-1 | |||||||||||
| LOC100507250 | |||||||||||
Table 6.
GISTIC output describing significantly amplified and deleted regions across the samples. The columns represent individual patient samples and indicate the regions gained or deleted in these samples.
| Unique name | Descriptor | Wide peak limits | Peak limits | Region limits | q values | Residual q values after removing segments shared with higher peaks | Amplitude threshold | GP034-tumor | GP019-tumor | GP003-tumor | GP018-tumor | GP023-tumor | GP043-tumor | GP016-tumor | GP030-tumor | GP011-tumor | GP007-tumor | GP026-tumor | GP013-tumor | GP009-tumor | GP024-tumor | GP005-tumor | GP012-tumor | GP025-tumor |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amplification peak 1 | 1q21.1 | chr1:144852237-145293666(probes 61189:64677) | chr1:145109662-145115657(probes 63931:64118) | chr1:145109662-145115809(probes 63931:64121) | 0.049551 | 0.049551 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Amplification peak 2 | 2p25.1 | chr2:8943044-10729896(probes 112423:113242) | chr2:8946569-10729896(probes 112424:113241) | chr2:8946569-10729896(probes 112424:113244) | 0.098716 | 0.098716 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 |
| Amplification Peak 3 | 2q11.1 | chr2:96604806-96610328(probes 140471:140516) | chr2:96604811-96607047(probes 140472:140515) | chr2:96604811-96610758(probes 140472:140589) | 2.34E−06 | 4.00E−06 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 2 | 0 |
| Amplification Peak 4 | 2q11.2 | chr2:97820512-97828928(probes 142142:142325) | chr2:97820635-97827942(probes 142143:142295) | chr2:97817579-97829924(probes 142113:142399) | 1.20E−06 | 1.82E−06 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 0 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 2 | 0 |
| Amplification Peak 5 | 4q35.2 | chr4:189022421-190878451(probes 290591:290876) | chr4:189022508-190878451(probes 290592:290875) | chr4:189022508-190878451(probes 290592:290878) | 0.0057245 | 0.0057245 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 0 | 2 | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Amplification Peak 6 | 6p22.1 | chr6:29692730-29911259(probes 346349:346874) | chr6:29910483-29910864(probes 346455:346697) | chr6:29910483-29910985(probes 346455:346700) | 0.092026 | 0.092026 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 2 |
| Amplification Peak 7 | 11q13.3 | chr11:68512544-69949326(probes 624468:624919) | chr11:68527601-69949326(probes 624469:624918) | chr11:68527601-69949326(probes 624469:624921) | 0.013732 | 0.013732 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Amplification Peak 8 | 12q15 | chr12:66990486-70918140(probes 683150:683997) | chr12:69048132-70672148(probes 683373:683974) | chr12:69048132-70672148(probes 683373:683977) | 0.098716 | 0.098716 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Amplification Peak 9 | 13q12.11 | chr13:22255168-23253312(probes 706758:706956) | chr13:22255302-23243644(probes 706759:706876) | chr13:22255302-23253391(probes 706759:706958) | 0.0053277 | 0.0053277 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t>0.9 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| Amplification Peak 10 | 17q12 | chr17:37557600-38062138(probes 876351:876621) | chr17:37558370-38061277(probes 876352:876620) | chr17:37558370-38062195(probes 876352:876623) | 0.098716 | 0.098716 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Amplification Peak 11 | 20q11.21 | chr20:29628342-29632831(probes 1016094:1016357) | chr20:29632520-29632826(probes 1016171:1016345) | chr20:29449510-30142483(probes 1015484:1016377) | 0.0012368 | 0.0012368 | 0: t < 0.85; 1: 0.85 < t < 0.9; 2: t > 0.9 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 |
| Deletion Peak 1 | 1p36.13 | chr1:16903823-16913583(probes 17147:17600) | chr1:16905719-16913583(probes 17152:17599) | chr1:16890673-16913583(probes 16646:17602) | 2.21E−08 | 2.21E−08 | 0: t > −0.74; 1: −0.74>t> −1.3; 2: t < −1.3 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| Deletion Peak 2 | 1q21.3 | chr1:151862662-152285860(probes 70611:71079) | chr1:152187589-152285860(probes 70612:71078) | chr1:152187589-152285929(probes 70612:71081) | 0.0019967 | 0.0019967 | 0: t>−0.74; 1: −0.74>t> −1.3; 2: t < −1.3 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| Deletion Peak 3 | 2q11.1 | chr2:96604592-96606984(probes 140356:140472) | chr2:96604731-96606943(probes 140376:140471) | chr2:96604592-96606984(probes 140356:140474) | 2.95E−05 | 2.95E−05 | 0: t>−0.74; 1: −0.74>t> −1.3; 2: t < −1.3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| Deletion Peak 4 | 7q22.1 | chr7:100638857-100642454(probes 435615:435646) | chr7:100639055-100642454(probes 435616:435645) | chr7:100638857-100647874(probes 435615:435933) | 2.21E−08 | 2.21E−08 | 0: t>−0.74; 1: −0.74>t> −1.3; 2: t < −1.3 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 2 | 1 | 2 | 2 | 0 | 0 | 2 | 2 |
| Deletion Peak 5 | 12q12 | chr12:40879121-40897247(probes 667577:667888) | chr12:40879178-40897247(probes 667578:667887) | chr12:40837332-40897247(probes 667398:667890) | 2.21E−08 | 2.21E−08 | 0: t>−0.74; 1: −0.74>t> -1.3; 2: t < -1.3 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 |
| Deletion Peak 6 | 17p11.2 | chr17:21217587-21318587(probes 868540:868629) | chr17:21217597-21318585(probes 868541:868628) | chr17:21217597-21318587(probes 868541:868631) | 0.0016159 | 0.0016159 | 0: t>−0.74; 1: −0.74>t> -1.3; 2: t < -1.3 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 0 |
| Deletion Peak 7 | 17q21.2 | chr17:39253750-39432561(probes 878741:879305) | chr17:39383074-39406408(probes 879136:879249) | chr17:39383074-39406408(probes 879136:879252) | 0.054262 | 0.054262 | 0: t>−0.74; 1: −0.74>t> -1.3; 2: t < -1.3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Deletion Peak 8 | 19q13.2 | chr19:40368281-40373890(probes 975747:975836) | chr19:40368331-40373888(probes 975748:975826) | chr19:40368331-40373890(probes 975748:975838) | 0.001411 | 0.001411 | 0: t>−0.74; 1: −0.74>t> −1.3; 2: t < −1.3 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| Amplification Peak 1 - CN values | 1q21.1 | chr1:144852237-145293666(probes 61189:64677) | chr1:145109662-145115657(probes 63931:64118) | chr1:145109662-145115809(probes 63931:64121) | 0.049551 | 0.049551 | Actual copy change given | 4.5797 | 0.69103 | 0.7953 | 0.53731 | 0.095471 | 0.20289 | 2.3826 | 3.6975 | 0.47619 | 0.88043 | 0.0027178 | -0.079543 | 1.1278 | 0.12261 | 0.40286 | 0.82495 | 0 |
| Amplification Peak 2 - CN values | 2p25.1 | chr2:8943044-10729896(probes 112423:113242) | chr2:8946569-10729896(probes 112424:113241) | chr2:8946569-10729896(probes 112424:113244) | 0.098716 | 0.098716 | Actual copy change given | −0.029333 | 0.042857 | 0 | 0.40765 | −0.059497 | 0.094427 | 0.86 | 0.062812 | 0.51852 | 0.21129 | −0.01085 | 6 | 0.1783 | −0.0046285 | 0 | 2.6817 | −0.12104 |
| Amplification Peak 3 - CN values | 2q11.1 | chr2:96604806-96610328(probes 140471:140516) | chr2:96604811-96607047(probes 140472:140515) | chr2:96604811-96610758(probes 140472:140589) | 2.34E−06 | 4.00E−06 | Actual copy change given | 1.6485 | 3.6735 | 0.96643 | 2.3769 | 1.1326 | 0.35596 | 1.2282 | 2.6182 | 0.35639 | 0.59483 | 0.30052 | 5.0601 | −0.80329 | 1.2419 | 0 | 1.6588 | 0.031842 |
| Amplification Peak 4 - CN values | 2q11.2 | chr2:97820512-97828928(probes 142142:142325) | chr2:97820635-97827942(probes 142143:142295) | chr2:97817579-97829924(probes 142113:142399) | 1.20E−06 | 1.82E−06 | Actual copy change given | 0.7777 | 3.8889 | 0.96643 | 2.5124 | 1.0965 | 0.018477 | 1.0605 | 2.5096 | 2.6595 | 4.4545 | 0.30052 | 0.072293 | -1.1464 | 1.0502 | 4.101 | 0.97825 | 0.031842 |
| Amplification Peak 5 - CN values | 4q35.2 | chr4:189022421-190878451(probes 290591:290876) | chr4:189022508-190878451(probes 290592:290875) | chr4:189022508-190878451(probes 290592:290878) | 0.0057245 | 0.0057245 | Actual copy change given | 0.33263 | 2.9254 | 3.1022 | 2.3359 | −0.0051105 | −0.15262 | 2.0461 | 3.2015 | 0.8205 | 0 | −0.040664 | −0.10707 | −0.13909 | 0.18688 | 0.48085 | 0.97708 | −0.42871 |
| Amplification Peak 6 - CN values | 6p22.1 | chr6:29692730-29911259(probes 346349:346874) | chr6:29910483-29910864(probes 346455:346697) | chr6:29910483-29910985(probes 346455:346700) | 0.092026 | 0.092026 | Actual copy change given | −0.80453 | −0.19355 | 3.8034 | −0.20754 | 0.08222 | 0.20604 | 2.8335 | −0.46171 | 0.020576 | −0.50571 | −0.019879 | 1.7421 | 0.024452 | −0.21015 | 1.1299 | 0.19199 | 2.5654 |
| Amplification Peak 7 - CN values | 11q13.3 | chr11:68512544-69949326(probes 624468:624919) | chr11:68527601-69949326(probes 624469:624918) | chr11:68527601-69949326(probes 624469:624921) | 0.013732 | 0.013732 | Actual copy change given | −0.78822 | −0.30548 | 0.19049 | −0.10101 | 6 | 0.043358 | −0.43478 | −0.022714 | 0.49808 | 0.027778 | 0.0092321 | 6 | 0.0037594 | −0.079159 | −0.13152 | 0.66311 | 0.65257 |
| Amplification Peak 8 - CN values | 12q15 | chr12:66990486-70918140(probes 683150:683997) | chr12:69048132-70672148(probes 683373:683974) | chr12:69048132-70672148(probes 683373:683977) | 0.098716 | 0.098716 | Actual copy change given | 0.39584 | 3.8839 | 0.31016 | 0.24606 | 0.059919 | −0.19368 | 0.1612 | 0.023994 | −0.47619 | 1.366 | 0.012851 | −0.082443 | 0.12666 | 0.21429 | −0.021505 | 0.23653 | 4.0643 |
| Amplification Peak 9 - CN values | 13q12.11 | chr13:22255168-23253312(probes 706758:706956) | chr13:22255302-23243644(probes 706759:706876) | chr13:22255302-23253391(probes 706759:706958) | 0.0053277 | 0.0053277 | Actual copy change given | 0.41427 | 0.30303 | 6 | 0.1941 | 0.048364 | 1.547 | −0.15 | −0.028577 | 0.53333 | −0.096774 | 0.004384 | −0.06446 | −0.095616 | 0.16703 | 6 | −0.17063 | 0.42759 |
| Amplification Peak 10 - CN values | 17q12 | chr17:37557600-38062138(probes 876351:876621) | chr17:37558370-38061277(probes 876352:876620) | chr17:37558370-38062195(probes 876352:876623) | 0.098716 | 0.098716 | Actual copy change given | 2.7139 | 0.17476 | 0.48521 | 6 | 0.53333 | −0.056568 | 0.38103 | 0.1334 | 0.72741 | 0.44949 | 0.0087288 | 0.16865 | 0.41472 | −0.0072883 | 0.10496 | 1.2383 | 0.15732 |
| Amplification Peak 11 - CN values | 20q11.21 | chr20:29628342-29632831(probes 1016094:1016357) | chr20:29632520-29632826(probes 1016171:1016345) | chr20:29449510-30142483(probes 1015484:1016377) | 0.0012368 | 0.0012368 | Actual copy change given | 1.4558 | 3.3422 | 3.0339 | 1.2699 | 1.2105 | 0.16143 | 1.38 | 2.3559 | 1.358 | 0.31044 | 0.034156 | 0.73932 | 0.34586 | 0.71369 | 2.336 | 1.9477 | 0.030591 |
| Deletion Peak 1 - CN values | 1p36.13 | chr1:16903823-16913583(probes 17147:17600) | chr1:16905719-16913583(probes 17152:17599) | chr1:16890673-16913583(probes 16646:17602) | 2.21E−08 | 2.21E−08 | Actual copy change given | −0.63903 | −0.44118 | −0.80967 | -1.2147 | -1.3043 | −0.37043 | −0.41538 | −0.76366 | −0.38567 | −0.85185 | −0.62946 | -1.0867 | -1.1864 | −0.76243 | −0.6107 | −0.013426 | −0.77209 |
| Deletion Peak 2 - CN values | 1q21.3 | chr1:151862662-152285860(probes 70611:71079) | chr1:152187589-152285860(probes 70612:71078) | chr1:152187589-152285929(probes 70612:71081) | 0.0019967 | 0.0019967 | Actual copy change given | 0.61226 | -1.1672 | 0.7953 | 0.13654 | −0.7693 | 0.58767 | 1.004 | −0.10414 | −0.66347 | 0.36364 | 0.0027178 | -1.2965 | −0.91416 | −0.058036 | 0.52776 | 0.29752 | 0.74062 |
| Deletion Peak 3 - CN values | 2q11.1 | chr2:96604592- 96606984(probes 140356:140472) | chr2:96604731- 96606943(probes 140376:140471) | chr2:96604592- 96606984(probes 140356:140474) | 2.95E−05 | 2.95E−05 | Actual copy change given | −0.0115 | −0.53153 | 0.96643 | −0.95155 | −0.059497 | −0.67375 | −0.62034 | -1.1527 | 0.35639 | 0.59483 | −0.83263 | -1.0643 | −0.70605 | −0.23196 | 0 | -1.197 | 0.031842 |
| Deletion Peak 4 - CN values | 7q22.1 | chr7:100638857-100642454(probes 435615:435646) | chr7:100639055-100642454(probes 435616:435645) | chr7:100638857-100647874(probes 435615:435933) | 2.21E−08 | 2.21E−08 | Actual copy change given | 0.24656 | -1.1321 | −0.69576 | 0.21496 | -1.5 | −0.069574 | −0.26788 | −0.80776 | -1.101 | -1.3636 | -1.2313 | -1.5 | -1.5 | −0.60593 | −0.04892 | -1.5 | -1.3881 |
| Deletion Peak 5 - CN values | 12q12 | chr12:40879121-40897247(probes 667577:667888) | chr12:40879178-40897247(probes 667578:667887) | chr12:40837332-40897247(probes 667398:667890) | 2.21E−08 | 2.21E−08 | Actual copy change given | 0.34613 | -1.458 | −0.30748 | −0.97993 | 0.059919 | 0.13048 | -1.2019 | -1.173 | -1.2852 | −0.5011 | 0.012851 | 0.053358 | -1.5 | −0.64087 | −0.021505 | −0.84181 | 0.025247 |
| Deletion Peak 6 - CN values | 17p11.2 | chr17:21217587-21318587(probes 868540:868629) | chr17:21217597-21318585(probes 868541:868628) | chr17:21217597-21318587(probes 868541:868631) | 0.0016159 | 0.0016159 | Actual copy change given | -1.5 | -1.1805 | 0.9029 | -1.0398 | −0.95596 | −0.24475 | −0.5611 | −0.80132 | −0.91292 | -1.0783 | -1.1403 | -1.3682 | -1.5 | -1.0251 | -1.2051 | −0.97453 | −0.30211 |
| Deletion Peak 7 - CN values | 17q21.2 | chr17:39253750-39432561(probes 878741:879305) | chr17:39383074-39406408(probes 879136:879249) | chr17:39383074-39406408(probes 879136:879252) | 0.054262 | 0.054262 | Actual copy change given | −0.1487 | −0.40741 | 2.2584 | −0.809 | 0.53333 | −0.056568 | 0.38103 | -1.0423 | 0.72741 | 0.44949 | 0.0087288 | 0.16865 | -1.215 | −0.0072883 | 0.10496 | −0.83901 | 2.2532 |
| Deletion Peak 8 - CN values | 19q13.2 | chr19:40368281-40373890(probes 975747:975836) | chr19:40368331-40373888(probes 975748:975826) | chr19:40368331-40373890(probes 975748:975838) | 0.001411 | 0.001411 | Actual copy change Given | −0.48534 | -1.0282 | 1.5499 | 0.13636 | −0.68163 | −0.88589 | 0.28891 | −0.45791 | −0.89906 | −0.075668 | -1.1579 | -1.2162 | −0.19894 | −0.20003 | 0.84998 | -1.1049 | −0.38052 |
Figure 4.
Significantly enriched amplification (red) and deletion (blue) events in the overall cohort of 17 samples, using GISTIC 2.0. Annotated cytobands indicate significant calls (FDR < 0.1) with the peaks corresponding to the significance value on the x-axis. The most significant regions included 2q11.2, 2q11.1 for gains and 1p36.13, 7q22.1, 12q12, 2q11.1 for losses. The significantly amplified regions included several genes involved in fibroblast growth factor signaling, upstream of the PI3K/AKT/mTOR pathway: FGF3, FGF4, FGF9, FGF19, and FRS2.
Table 5.
Significant CNV loss regions called by GISTIC across all 17 samples submitted for whole-exome sequencing. GISTIC is an algorithm which identifies regions of the genome that are gained or lost more than expected by chance across a set of samples.
| Cytoband | 1p36.13 | 7q22.1 | 12q12 | 2q11.1 | 19q13.2 | 17p11.2 | 1q21.3 | 17q21.2 |
|---|---|---|---|---|---|---|---|---|
| q value | 2.21E-08 | 2.21E-08 | 2.21E-08 | 2.95E-05 | 0.001411 | 0.0016159 | 0.0019967 | 0.054262 |
| Residual q value | 2.21E-08 | 2.21E-08 | 2.21E-08 | 2.95E-05 | 0.001411 | 0.0016159 | 0.0019967 | 0.054262 |
| Wide peak boundaries | chr1:16903823-16913583 | chr7:100638857-100642454 | chr12:40879121-40897247 | chr2:96604592-96606984 | chr19:40368281-40373890 | chr17:21217587-21318587 | chr1:151862662-152285860 | chr17:39253750-39432561 |
| Genes in wide peak | NBPF1 | MUC12 | [LRRK2] | [LOC729234] | FCGBP | KCNJ12 | FLG | KRTAP9-9 |
| MAP2K3 | S100A10 | KRTAP4-6 | ||||||
| KCNJ18 | S100A11 | KRTAP4-12 | ||||||
| TCHH | KRTAP9-2 | |||||||
| THEM4 | KRTAP9-3 | |||||||
| TCHHL1 | KRTAP9-8 | |||||||
| RPTN | KRTAP4-4 | |||||||
| HRNR | KRTAP9-4 | |||||||
| KRTAP4-1 | ||||||||
| KRTAP4-5 | ||||||||
| KRTAP4-3 | ||||||||
| KRTAP4-2 | ||||||||
| KRTAP4-11 | ||||||||
| KRTAP4-8 | ||||||||
| KRTAP9-1 | ||||||||
| KRTAP4-9 |
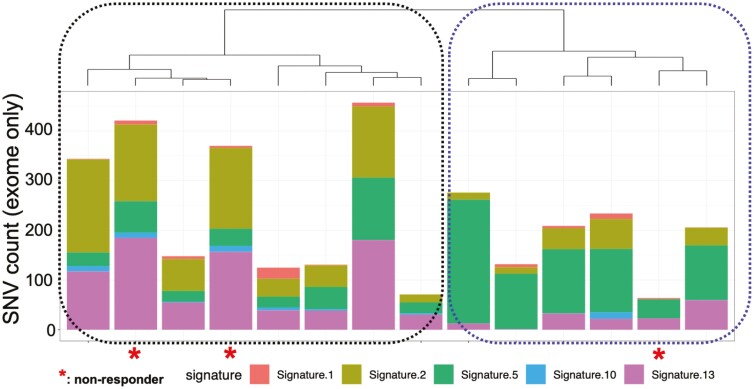
Mutational signature analysis revealed two clusters of samples based on their signature decomposition results (Figure 5, Table 7). Cluster 1 (N = 8) was dominated by signatures 2 and 13, which are associated with activity of APOBEC cytidine deaminases.18 Cluster 2 (N = 6) was characterized by the dominance of signature 5, which has been associated with ERCC2 mutations.24 Mutational signature clusters were not associated with response (Fisher’s exact P = 1 two sided).
Figure 5.
Mutational signature analysis revealed that samples fell into two clusters with distinct patterns of single-nucleotide alterations throughout the genome. Cluster 1 (N = 8) was dominated by COSMIC v2 signatures 2 and 13, which are associated with APOBEC cytidine deaminases. Cluster 2 (N = 6) was characterized by dominance of signature 5, which has been associated with ERCC2 mutations. Mutational signature clusters were not associated with response (Fisher’s exact P = 1 two-sided).
Table 7.
Mutational signatures in each sample (N = 15 samples from cohort 2 with whole-exome sequencing), based on bladder-specific signatures in the COSMIC v2 database. Mutational signatures represent global patterns in the types of single-nucleotide changes throughout the genome and are thought to reflect distinct underlying mutational processes. The numbers indicate the percent of mutations attributed to each signature within each sample.
| Sample | Signature.1 | Signature.2 | Signature.5 | Signature.10 | Signature.13 |
|---|---|---|---|---|---|
| GP012 | 0.01943257 | 0.36660802 | 0.149879 | 0.02486539 | 0.43921502 |
| GP043 | 0.01095983 | 0.33135712 | 0.34225969 | 0.02093894 | 0.29448443 |
| GP003 | 3.28E-18 | 0.05226691 | 0.9015513 | 0 | 0.04618179 |
| GP025 | 0.0391949 | 0.43356456 | 0.14695689 | 0.00935119 | 0.37093247 |
| GP023 | 0.17884431 | 0.28888586 | 0.17462784 | 0.04510776 | 0.31253424 |
| GP011 | 0.01690564 | 0.31356415 | 0.27458071 | 0 | 0.39494949 |
| GP016 | 0.04755421 | 0.09875741 | 0.84644402 | 0 | 0.00724437 |
| GP034 | 0.02233931 | 0.202452 | 0.61821754 | -8.67E-19 | 0.15699115 |
| GP007 | 0.01140029 | 0.43952321 | 0.09374151 | 0.03198736 | 0.42334762 |
| GP018 | 0.00392937 | 0.54435374 | 0.07943001 | 0.03159905 | 0.34068783 |
| GP032 | 0.09088163 | 0.05547802 | 0.76846216 | 0 | 0.08517819 |
| GP013 | 0.03954119 | 0.00325378 | 0.59572065 | 0 | 0.36148437 |
| GP005 | 0.00370014 | 0.1725524 | 0.53444981 | 0 | 0.28929765 |
| GP030 | 0.04876071 | 0.25637049 | 0.54555192 | 0.05520981 | 0.09410708 |
| GP009 | 0 | 0.22447006 | 0.309945 | 0.03486223 | 0.4307227 |
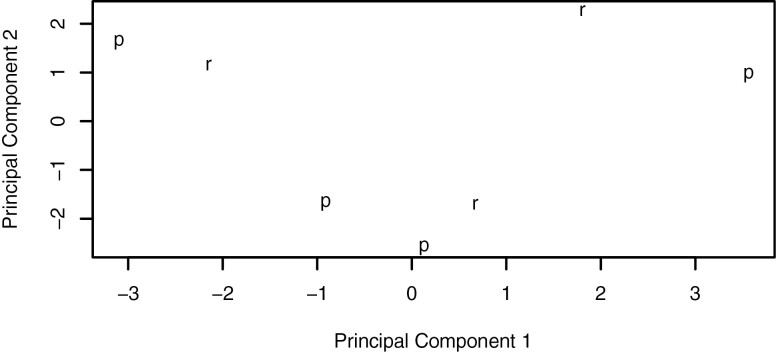
RNA sequencing was performed on 8 of the WES samples; 1 patient was not evaluable for response, leaving 7 evaluable patients with RNA expression data. Clustering of transcriptomic profiles showed no clear separation between responders (N = 3) versus non-responders (N = 4; Figure 6). Differential expression analyses identified 9 differentially expressed genes between responders versus non-responders (at adj. P cutoff of .05). None of these were associated with mTOR signaling or microtubule function (Table 8).
Figure 6.
Clustering of samples by transcriptomic profile using principal components analysis failed to identify separate clusters based on paclitaxel treatment responsiveness. R: responders; P: nonresponders.
Table 8.
Differentially expressed genes between responders (N = 3) and nonresponders (N = 4) (including only those with successful RNA sequencing). Negative log2 foldchange values indicate decreased expression in responders compared with nonresponders, while positive log2 foldchange values indicate increased expression. All patients for this analysis were from Cohort 2.
| Gene ID | Approved Symbol | Log2 foldchange | Adj P |
|---|---|---|---|
| HGNC:19133 | HS6ST2 | −4.176714 | .008258125 |
| HGNC:1047 | BHMT | −5.999643 | .008258125 |
| HGNC:21226 | LRFN2 | −7.754684 | .008258125 |
| HGNC:26731 | C8orf31 | −4.328898 | .0206424 |
| HGNC:7423 | MTCP1 | −2.920986 | .032091609 |
| HGNC:21923 | STEAP4 | −2.754969 | .03327653 |
| HGNC:32406 | IQCJ | −6.218168 | .038737815 |
| HGNC:23596 | KRTAP5-1 | 4.892141 | .038737815 |
| HGNC:4020 | FUT9 | −6.352472 | .040669222 |
No recurrent gene fusions were identified after false positives were eliminated by manual review (Tables 9 and 10).
Table 9.
Gene fusions in responders.
| Fusion name | Patient count |
|---|---|
| ZNF137P:ZNF83 | 1 |
| ADGRE5:ADGRE2 (FALSE POSITIVE) | 3 |
| ADGRE2:ADGRE5 (FALSE POSITIVE) | 3 |
| SMG1:NPIPB5 | 1 |
| KANSL1:ARL17A | 1 |
| KANSL1:ARL17B | 1 |
| SCNN1A:TNFRSF1A | 1 |
| PSMD14:ZNF638 | 1 |
| ANK2:CAMK2D | 1 |
| PIP4K2A:RAB18 | 1 |
| STX16:NPEPL1 | 1 |
| STX16:STX16-NPEPL1 | 1 |
| ACLY:DNAJC7 | 1 |
| ZNF486:GATAD2A | 1 |
| TBCEL:TECTA | 1 |
| STX16-NPEPL1:NPEPL1 | 1 |
| STX16-NPEPL1:STX16-NPEPL1 | 1 |
| CYTIP:ERMN | 1 |
Table 10.
Gene fusions in non-responders.
| Fusion name | Patient count |
|---|---|
| ADGRE5:ADGRE2 (FALSE POSITIVE) | 3 |
| ADGRE2:ADGRE5 (FALSE POSITIVE) | 2 |
| SMG1:NPIPB5 | 1 |
| NAIP:OCLN | 1 |
| CLTC:VMP1 | 1 |
| KANSL1:ARL17A | 1 |
| KANSL1:ARL17B | 1 |
| EIF3K:ACTN4 | 1 |
| PTPN1:PPTC7 | 1 |
Discussion
Cisplatin remains the backbone of treatment for advanced UC. However, many patients are not eligible for cisplatin due to performance status or comorbidities. The subgroup of cisplatin-ineligible patients with both poor performance status and poor renal function experience increased toxicity and reduced benefit from carboplatin-based regimens necessitating novel treatment approaches. We initiated a phase II trial to test the activity of everolimus or everolimus plus paclitaxel in the cisplatin-ineligible setting shortly prior to a new era in drug development in metastatic UC. Novel regimens such as enfortumab vedotin alone or combined with pembrolizumab have demonstrated promising activity in cisplatin-ineligible patients.25,26 The shifting landscape, coupled with pragmatic considerations related to cohort 1, contributed to early closure due to poor accrual. Nonetheless, this trial has generated insights that may inform future treatment strategies.
We observed a 4-month CBR of 37.9% associated with EVP among patients with either poor performance states or poor renal function (cohort 2). This benefit was most likely driven by paclitaxel, which has demonstrated efficacy in this context both as a single-agent and in combinations.7,27,28 This degree of activity is similar to that observed with carboplatin-based regimens in the phase II/III EORTC 30986 trial3 and reinforces the value of single-agent paclitaxel in the cisplatin-ineligible setting. However, treatment was still associated with a notable adverse event burden in this population. The EVP combination has also been studied in patients with UC progressing despite platinum-based chemotherapy with an objective response rate of 13%,29 similar to the response rate with paclitaxel, suggesting limited benefit by adding everolimus although the specific contribution of each agent cannot be defined here. Notably, everolimus monotherapy was disappointing across different solid tumors selected for genomic alterations predicted to confer vulnerability, despite previous promising case reports.6,30,31
We initiated our trial in 2010 prior to the immune checkpoint blockade era and the subsequent shifts in the metastatic UC treatment landscape.32 Current standard first-line treatment for cisplatin-ineligible patients with metastatic UC includes carboplatin-based chemotherapy followed by switch maintenance immune checkpoint blockade or single agent immune checkpoint in patients with tumors harboring high levels of PD-L1 expression or patients who are “chemotherapy ineligible” (in certain regions of the world). Notably, such regimens have the potential for durable disease control in a subset of patients not observed in the current study. Patients for whom the risks of any platinum-based chemotherapy outweigh the potential benefits of treatment are complicated to define in both clinical care and for the purposes of trial design. However, the results of EORTC 30986 suggest that patients with both impaired renal function and borderline functional status suffer excessive toxicity from carboplatin-based chemotherapy and this definition of “chemotherapy ineligibility” was reinforced in a recent survey of oncologists.33 The current trial, although performed in an earlier era and with a treatment without substantial activity, highlights the potential challenges of enrolling “chemotherapy ineligible” patients to prospective clinical trials; the median OS of patients in Cohort 1 was only 4.5 months.
We examined genomic data from 17 patients in cohort 2 to identify possible biomarkers of response to EVP. There were no significant associations between somatic mutations, copy number variants, or mutational signatures and response. However, power was limited by the number of samples and an imbalance of responders (N = 13) and nonresponders (N = 4). One notable, albeit non-significant, observation was the high response rates to EVP among those with mutations in either of the microtubule-associated genes MACF1 or FRY (100%; Fisher’s exact P = .24 two sided; P = .14 one sided). MACF1 is a microtubule binding protein that bridges cytoskeletal elements and has roles in cellular migration, adhesion, and intracellular transport.34,35FRY also binds microtubules and regulates the mitotic spindle during cell division.36 To our knowledge, there have not been in vitro or in vivo experiments testing the relationship between mutations in these genes and paclitaxel sensitivity. Though the use of taxanes in latter lines of therapy for metastatic UC is decreasing in the context of new treatment options, treatment selection biomarkers for these newer treatments are still lacking and biomarkers that might define patients deriving most benefit from paclitaxel could still impact clinical treatment strategies and warrant further testing.
Conclusions
Paclitaxel demonstrated activity comparable to historical reports of carboplatin-based regimens in cisplatin-ineligible patients with metastatic UC in this small cohort, although is not associated with durable responses that occur in a subset of patients treated with modern regimens. Everolimus did not demonstrate obvious additivity in this combination regimen. Outcomes in “chemotherapy ineligible” patients remain suboptimal and enrollment of such patients to prospective trials is challenging.
Contributor Information
Tomi Jun, Sema4, Stamford, CT, USA; The Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Noah M Hahn, Indiana University Melvin and Bren Simon Cancer Center, Indianapolis, IN Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD, USA.
Guru Sonpavde, University of Alambama at Birmingham, Birmingham, AL Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA.
Constantine Albany, Indiana University Melvin and Bren Simon Cancer Center, Indianapolis, IN, USA.
Gary R MacVicar, Robert H. Lurie Comprehensive Cancer Center, Northwestern University Feinberg School of Medicine, Chicago, IL Illinois CancerCare, Peoria, IL, USA.
Ralph Hauke, Nebraska Cancer Specialists/ Nebraska Methodist Hospital, Omaha, NE, USA.
Mark Fleming, Virginia Oncology Associates, Norfolk, VA, USA.
Theodore Gourdin, Medical University of South Carolina Hollings Cancer Center, Charleston, SC, USA.
Bagi Jana, University of Texas Medical Branch at Galveston, Galveston, TX, USA.
William K Oh, Sema4, Stamford, CT, USA; The Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Patricia Taik, Sema4, Stamford, CT, USA.
Huan Wang, Sema4, Stamford, CT, USA.
Ajay Ramakrishnan Varadarajan, Sema4, Stamford, CT, USA.
Andrew Uzilov, Sema4, Stamford, CT, USA.
Matthew D Galsky, The Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Acknowledgments
This study was sponsored by the Hoosier Cancer Research Network.
Funding
The study was funded by Novartis Pharmaceuticals Corporation.
Conflict of Interest
Tomi Jun: Sema4 (E, OI); Noah M. Hahn: Merck, Genentech, GlaxoSmithKline, Ferring, Champions Oncology, Health Advances, Keyquest Health, Guidepoint Global, Seattle Genetics, Mirati, Incyte, TransMed, CicloMed, Janssen, Pfizer, Boehringer Ingelheim, Pfizer, EMD Serono (C/A), HTG Molecular Diagnostics, AstraZeneca, Bristol-Myers Squibb, Genentech, Seattle Genetics, OncoGenex, Pieris, Inovio, Principia Biopharm (RF-institutional); Creative Educational Concepts, Large Urology Group Practice Association (H); Guru Sonpavde: Bristol-Meyers Squibb, Genentech, EMD Serono, Merck, Sanofi, Seattle Genetics/Astellas, AstraZeneca, Exelixis, Janssen, Bicycle Therapeutics, Pfizer, Immunomedics/Gilead, Scholar Rock, G1 Therapeutics (SAB), Sanofi, AstraZeneca, Immunomedics/Gilead, QED, Predicine, Bristol-Meyers Squibb (RF–institutional), Bristol-Meyers Squibb, Bavarian Nordic, Seattle Genetics, QED, G1 Therapeutics (steering committees, unpaid), AstraZeneca, EMD Serono, Debiopharm (steering committees, paid), Mereo (data safety monitoring committee), Bristol-Meyers Squbbi, AstraZeneca (travel costs), Up-to-Date, Editor of Elsevier Practice Update Bladder Cancer Center of Excellence (writing/editor fees), Physicians Education Resource (PER), Onclive, Research to Practice, Medscape (speaking fees, all educational); Bagi R. Jana: Genzyme (SAB); William K. Oh: Astellas, AstraZeneca, Bayer, Janssen, Pfizer, Sanofi (C/A), Sema4 (Chief Medical Science Officer); Patricia Taik: Sema4 (E); Huan Wang: Sema4 (E, OI); Ajay Ramakrishnan Varadarajan: Sema4 (E, OI); Andrew Uzilov: Sema4 (E, OI); Matthew D. Galsky: BioMotiv, Janssen, Dendreon, Merck, GlaxoSmithKline, Lilly, Astellas Pharma, Genentech, Bristol-Myers Squibb, Novartis, Pfizer, EMD Serono, AstraZeneca, Seattle Genetics, Incyte, Aileron Therapeutics, Dracen, Inovio Pharmaceuticals, NuMab, Dragonfly Therapeutics, Basilea, Urogen, Infinity Pharmaceuticals, Gilead (C/A), Janssen Oncology, Dendreon, Novartis, Bristol-Myers Squibb, Merck, AstraZeneca, Genetech/Roche (RF–institutional), Rappta Therapeutics (OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) research funding; (E) employment; (ET) expert testimony; (H) honoraria received; (OI) ownership interests; (IP) intellectual property rights/inventor/patent holder; (SAB) scientific advisory board.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Dash A, Galsky MD, Vickers AJ, et al. . Impact of renal impairment on eligibility for adjuvant cisplatin-based chemotherapy in patients with urothelial carcinoma of the bladder. Cancer. 2006;107(3):506-513. [DOI] [PubMed] [Google Scholar]
- 2. Galsky MD, Hahn NM, Rosenberg J, et al. . Treatment of patients with metastatic urothelial cancer “unfit” for Cisplatin-based chemotherapy. J Clin Oncol. 2011;29(17):2432-2438. [DOI] [PubMed] [Google Scholar]
- 3. De Santis M, Bellmunt J, Mead G, et al. . Randomized phase II/III trial assessing gemcitabine/ carboplatin and methotrexate/carboplatin/vinblastine in patients with advanced urothelial cancer “unfit” for cisplatin-based chemotherapy: phase II–results of EORTC study 30986. J Clin Oncol. 2009;27(33):5634-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tickoo SK, Milowsky MI, Dhar N, et al. . Hypoxia-inducible factor and mammalian target of rapamycin pathway markers in urothelial carcinoma of the bladder: possible therapeutic implications. BJU Int. 2011;107(5):844-849. [DOI] [PubMed] [Google Scholar]
- 5. Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, et al. . Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23(6):675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iyer G, Hanrahan AJ, Milowsky MI, et al. . Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338(6104):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dreicer R, Gustin DM, See WA, Williams RD.. Paclitaxel in advanced urothelial carcinoma: its role in patients with renal insufficiency and as salvage therapy. J Urol. 1996;156(5):1606-1608. [DOI] [PubMed] [Google Scholar]
- 8. Hu L, Hofmann J, Lu Y, Mills GB, Jaffe RB.. Inhibition of phosphatidylinositol 3’-kinase increases efficacy of paclitaxel in in vitro and in vivo ovarian cancer models. Cancer Res. 2002;62(4):1087-1092. [PubMed] [Google Scholar]
- 9. Faried LS, Faried A, Kanuma T, et al. . Inhibition of the mammalian target of rapamycin (mTOR) by rapamycin increases chemosensitivity of CaSki cells to paclitaxel. Eur J Cancer. 2006;42(7):934-947. [DOI] [PubMed] [Google Scholar]
- 10. Liu Z, Zhu G, Getzenberg RH, Veltri RW.. The upregulation of PI3K/Akt and MAP kinase pathways is associated with resistance of microtubule-targeting drugs in prostate cancer. J Cell Biochem. 2015;116(7):1341-1349. [DOI] [PubMed] [Google Scholar]
- 11. Campone M, Levy V, Bourbouloux E, et al. . Safety and pharmacokinetics of paclitaxel and the oral mTOR inhibitor everolimus in advanced solid tumours. Br J Cancer. 2009;100(2):315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hurvitz SA, Dalenc F, Campone M, et al. . A phase 2 study of everolimus combined with trastuzumab and paclitaxel in patients with HER2-overexpressing advanced breast cancer that progressed during prior trastuzumab and taxane therapy. Breast Cancer Res Treat. 2013;141(3):437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurvitz SA, Andre F, Jiang Z, et al. . Combination of everolimus with trastuzumab plus paclitaxel as first-line treatment for patients with HER2-positive advanced breast cancer (BOLERO-1): a phase 3, randomised, double-blind, multicentre trial. Lancet Oncol. 2015;16(7):816-829. [DOI] [PubMed] [Google Scholar]
- 14. Eisenhauer EA, Therasse P, Bogaerts J, et al. . New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228-247. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z, Hao K.. SAAS-CNV: a joint segmentation approach on aggregated and allele specific signals for the identification of somatic copy number alterations with next-generation sequencing data. PLoS Comput Biol. 2015;11(11):e1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G.. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lynch AG. Decomposition of mutational context signatures using quadratic programming methods. F1000Res. 2016;5:1253. [Google Scholar]
- 18. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobin A, Davis CA, Schlesinger F, et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao Y, Smyth GK, Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923-930. [DOI] [PubMed] [Google Scholar]
- 21. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicorici D, Şatalan M, Edgren H, et al. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014. doi:10.1101/011650 [Google Scholar]
- 23. Haas BJ, Dobin A, Stransky N, et al. STAR-fusion: fast and accurate fusion transcript detection from RNA-Seq. bioRxiv. 2017. doi:10.1101/120295 [Google Scholar]
- 24. Kim J, Mouw KW, Polak P, et al. . Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48(6):600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu EY, Petrylak DP, O’Donnell PH, et al. . Enfortumab vedotin after PD-1 or PD-L1 inhibitors in cisplatin-ineligible patients with advanced urothelial carcinoma (EV-201): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2021;22(6):872-882. [DOI] [PubMed] [Google Scholar]
- 26. Rosenberg JE, Flaig TW, Friedlander TW, et al. Study EV-103: durability results of enfortumab vedotin plus pembrolizumab for locally advanced or metastatic urothelial carcinoma. JCO. 2020;38(15_suppl):5044. [Google Scholar]
- 27. Siefker-Radtke AO, Campbell MT, Munsell MF, Harris DR, Carolla RL, Pagliaro LC.. Front-line treatment with gemcitabine, paclitaxel, and doxorubicin for patients with unresectable or metastatic urothelial cancer and poor renal function: final results from a phase II study. Urology. 2016;89:83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narayanan S, Lam A, Vaishampayan U, et al. . Phase II study of pazopanib and paclitaxel in patients with refractory urothelial cancer. Clin Genitourin Cancer. 2016;14(5):432-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niegisch G, Retz M, Thalgott M, et al. . Second-line treatment of advanced urothelial cancer with paclitaxel and everolimus in a German phase II Trial (AUO Trial AB 35/09). Oncology. 2015;89(2):70-78. [DOI] [PubMed] [Google Scholar]
- 30. Adib E, Klonowska K, Giannikou K, et al. Phase II clinical trial of everolimus in a pan-cancer cohort of patients with mTOR pathway alterations. Clin Cancer Res. 2021;27(14):3845-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bellmunt J, Lalani AA, Jacobus S, et al. . Everolimus and pazopanib (E/P) benefit genomically selected patients with metastatic urothelial carcinoma. Br J Cancer. 2018;119(6):707-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patel VG, Oh WK, Galsky MD.. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70(5):404-423. [DOI] [PubMed] [Google Scholar]
- 33. Gupta S, Sonpavde G, Grivas P, et al. Defining “platinum-ineligible” patients with metastatic urothelial cancer (mUC). JCO. 2019;37(7_suppl):451. [Google Scholar]
- 34. Kakinuma T, Ichikawa H, Tsukada Y, Nakamura T, Toh BH.. Interaction between p230 and MACF1 is associated with transport of a glycosyl phosphatidyl inositol-anchored protein from the Golgi to the cell periphery. Exp Cell Res. 2004;298(2):388-398. [DOI] [PubMed] [Google Scholar]
- 35. Ning W, Yu Y, Xu H, et al. . The CAMSAP3-ACF7 complex couples noncentrosomal microtubules with actin filaments to coordinate their dynamics. Dev Cell. 2016;39(1):61-74. [DOI] [PubMed] [Google Scholar]
- 36. Nagai T, Ikeda M, Chiba S, Kanno S, Mizuno K.. Furry promotes acetylation of microtubules in the mitotic spindle by inhibition of SIRT2 tubulin deacetylase. J Cell Sci. 2013;126(Pt 19):4369-4380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.