Abstract
Clostridium thermocellum is actively being developed as a microbial platform to produce biofuels and chemicals from renewable plant biomass. An attractive feature of this bacterium is its ability to efficiently degrade lignocellulose using surface-displayed cellulosomes, large multi-protein complexes that house different types of cellulase enzymes. C. thermocellum tailors the enzyme composition of its cellulosome using nine membrane-embedded anti-σ factors (RsgI1-9), which are thought to sense different types of extracellular carbohydrates and then elicit distinct gene expression programs via cytoplasmic σ factors. Here we show that the RsgI9 anti-σ factor interacts with cellulose via a C-terminal bi-domain unit. A 2.0 Å crystal structure reveals that the unit is constructed from S1C peptidase and NTF2-like protein domains that contain a potential binding site for cellulose. Small angle X-ray scattering experiments of the intact ectodomain indicate that it adopts a bi-lobed, elongated conformation. In the structure a Conserved RsgI Extracellular (CRE) domain is connected to the bi-domain via a proline-rich linker, which is expected to project the carbohydrate binding unit ~160 Å from the cell surface. The CRE and proline-rich elements are conserved in several other C. thermocellum anti-σ factors, suggesting that they will also form extended structures that sense carbohydrates.
Keywords: Biofuels, biomass, cellulosomes, anti-sigma factor, crystallography, small angle X-ray scattering
INTRODUCTION
Concerns about climate change and declining petroleum supplies have created a pressing need for renewable transportation fuels1. Bioethanol produced from lignocellulosic plant biomass (called cellulosic ethanol) is potentially an attractive solution to this problem, as plant biomass is highly abundant and renewable. Typically, cellulosic ethanol is produced by first degrading lignocellulose into its component sugars using purified cellulase enzymes, followed by fermentation of the sugars into ethanol2. However, the high costs of the cellulase cocktails used in this process are prohibitive, such that relatively little cellulosic ethanol is produced industrially3,4. To increase the cost-efficiency of bioethanol production, much attention has been devoted to developing a consolidated bioprocessing (CBP) platform that does not require the use of purified cellulases. In CBP, a single organism would both produce the cellulases that hydrolyze the cellulose and hemicellulose polymers in plant biomass as well as ferment the resultant sugars into ethanol, or other biofuels or materials5,6. Clostridium thermocellum (also renamed as Ruminiclostridium thermocellum, Hungateiclostridium thermocellum, and Acetivibrio thermocellus) is actively being developed as a CBP platform due to its high native cellulolytic activity, ethanologenic potential, and preference to grow under thermophilic conditions7. The ability of C. thermocellum to degrade lignocellulose is particularly notable, as it is one of the most cellulolytic organisms thus far discovered8,9. Current research is focused toward uncovering fundamental genome-encoded properties that confer potent, tunable cellulolytic activity in C. thermocellum, and are an important step toward developing cost-effective microbial approaches to produce biofuels and chemicals from plant biomass.
C. thermocellum and other anaerobic bacteria within the bacterial orders Clostridiales and Bacteroidales degrade lignocellulose using cellulosomes, multi-enzyme complexes that house an array of enzymes with different substrate specificities (cellulases, hemicellulases, pectinases, esterases etc.)10. Cellulosome-displaying bacteria degrade lignocellulose more efficiently than microbes that simply secrete cellulases, because enzyme colocalization within the cellulosome promotes enzyme-enzyme synergy, enzyme-proximity enhancement, and cellulose-enzyme-microbe interactions11,12. Cellulosomes adopt elaborate structures that are built using a series of surface-displayed scaffoldin proteins that coordinate the binding of the enzyme machinery via dockerin-cohesin domain interactions. They are assembled in modular fashion to create massive polycellulosomal structures that are readily visible by electron microscopy and can harbor >140 distinct dockerin-borne enzymes in some bacterial species10,13. The primary scaffoldin in the C. thermocellum cellulosome, CipA, contains nine type-I cohesin modules that bind to cellulases harboring type-I dockerin modules7. The scaffoldin also contains a carbohydrate-binding module (CBM) that tethers the cellulosome complex to its cellulose substrate, as well as a C-terminal type-II dockerin module that anchors the complex to the cell by interacting with surface displayed proteins that contain type-II cohesin domains14,15.
To degrade various types forms of plant biomass, C. thermocellum tailors the enzyme composition of its cellulosome using membrane embedded anti-σ factors16,17. Work by the Lamed and Bayer groups has shown that ten membrane-associated anti-σ factors may control the composition of the cellulosome, nine of which share homology to the B. subtilis RsgI anti-σ factor (named RsgI1 to RsgI9)18. Based on studies of SigI1 and RsgI1, each anti-σ factor is thought to bind to a different type of biomass-derived carbohydrate on the extracellular surface, and then trigger distinct gene expression programs by releasing a cognate extracytoplasmic function (ECF) σ factor (SigI1 to SigI8) that confers promoter specificity for the RNA polymerase17-19. With the notable exception of the RsgI9, genes expressing each anti-σ factor are located on the same operon as a gene encoding an ECF σ factor, suggesting that their protein products are functionally linked; for example, the rsgI1 and sigI1 genes are expressed from the same operon suggesting Rsg1 and SigI1 proteins form a functional anti-σ/σ pair. However, our understanding of how these regulatory systems sense carbohydrates and alter gene expression remains limited. In vitro binding studies have experimentally identified carbohydrates that interact with the RsgI1, RsgI2, RsgI3, RsgI4, and RsgI6 anti-σ factors, but the specific genes whose expression is ultimately regulated by these binding events is generally not known18,20-22. This is because at present only partial regulons have only been identified for SigI1, SigI3, and SigI6 using bioinformatics analyses and verified experimentally17. Finally, the molecular mechanism through which extracellular carbohydrate binding to each anti-σ factor triggers gene expression by releasing its cognate ECF σ factor remains to be determined.
The function of the RsgI9 anti-σ factor is poorly understood because it is an ‘orphan’; the gene encoding RsgI9 is not located within an operon that also contains a gene that encodes for an ECF σ factor. Moreover, it is not known if RsgI9 is capable of sensing biomass as its primary sequence does not encode for a known carbohydrate-binding module (CBM) or carbohydrate-active enzyme. Herein, we report the first characterization of RsgI9’s ectodomain to gain insight into its function. Using X-ray crystallography, we show that RsgI9 contains a unique C-terminal bi-domain unit that is formed from S1C peptidase and NTF2-like domains. Biochemical experiments indicate that the NTF2-like domain interacts with cellulose, while the S1C peptidase domain is not enzymatically active. In the ectodomain of RsgI9, the C-terminal bi-domain module is connected to a Conserved RsgI Extracellular (CRE) domain via a proline-rich linker. Using Small Angle X-ray Scattering (SAXS), we show that RsgI9’s intact ectodomain primarily adopts an extended conformation that can be expected to project the bi-domain module up to 160 Å from the cell surface. Inspection of other RsgI proteins reveals that they also contain CRE domains and proline-rich linkers, indicating they will also adopt extended structures to sense different types of biomass.
MATERIALS AND METHODS
Cloning and expression of RsgI9.
The nucleotide sequence encoding the C-terminal region of the C. thermocellum (DSM 1313) RsgI9 ectodomain (residues 387-702, RsgI9S1C-CTD) was purchased from Twist Bioscience (San Francisco, CA). The pET-29b expression plasmid includes an N-terminal hexahistidine (6xHis) tag followed by a TEV protease recognition site. The plasmid was transformed into E. coli BL21 (DE3) cells (New England Biolabs) and grown at 37°C in LB media in the presence of 50 μg/mL kanamycin until OD600 of 0.6-0.8 was reached, followed by induction with 1 mM IPTG and overnight (12-18 hours) protein expression at 17°C. Expression pellets were resuspended in lysis buffer containing 50 mM sodium phosphate, 300 mM NaCl, 2 mM DTT, pH 8.0, and lysed via sonication in the presence of 1 mg/mL egg white lysozyme, 400 μL of protease inhibitor cocktail (Calbiochem), 2 mM phenylmethanesulfonyl fluoride (PMSF, Sigma), and 0.5 mg S. marcescens nuclease23. Lysates were clarified by centrifugation at 15,000 rpm for 45 minutes. The supernatant was applied to HisPur cobalt resin (Thermo Fisher Scientific) equilibrated in lysis buffer, and unbound proteins were washed by several column volumes of lysis buffer followed by a wash buffer (lysis buffer + 10 mM imidazole). The bound protein was eluted using an elution buffer composed of the lysis buffer with 200 mM imidazole. TEV protease (purified in-house) was added to the eluted sample followed by dialysis against imidazole-free lysis buffer. The sample was then loaded back onto cobalt resin and the cleaved protein was collected by rinsing with lysis buffer. The resulting protein was further purified by gel-filtration chromatography with a Superdex S75 size-exclusion column on an Akta FPLC system (GE Healthcare Life Sciences). Protein was concentrated via Amicon Ultra centrifugal filters (EMD Millipore), with purity confirmed to be >98% via SDS-PAGE.
The nucleotide sequence encoding the full extracellular region of RsgI9 (residues 167-707, RsgI9ecto) was cloned out of C. thermocellum (ATCC 27405) genomic DNA and inserted into a pET-28b-based vector containing N-terminal 6xHis and small ubiquitin-like modifier (SUMO) fusion tags using Gibson assembly. Growth and expression in E. coli BL21 (DE3) cells proceeded as described above. Purification was also carried out identically, but with ULP1 protease (purified in-house) used to cleave the 6xHis-SUMO tag from RsgI9. Expression plasmids producing the isolated Conserved RsgI Extracellular (CRE) portion of the ectodomain (residues 167-343, RsgI9CRE) and the S1C domain (residues 387-592, RsgI9S1C) were also prepared by cloning the relevant portions of the gene, with expression and purification carried out as above. Site-directed mutagenesis was used to create a T535S variant of the protease domain, RsgI9S1C,T535S.
Crystallization, data collection, and crystal structure determination.
RsgI9S1C-CTD was concentrated to 10 mg/mL, and spontaneously formed small protein crystals in a solution of 20 mM HEPES, 2 mM DTT, pH 8.0. However, these crystals were too small to be suitable for diffraction. The small crystals were dissolved in the same buffer at a concentration of 8 mg/mL and used to screen in 24-well hanging drop format with increasing concentrations of glycerol and PEG chain additives, and larger crystals were grown after a week in 20 mM HEPES, 2 mM DTT, pH 7.5, and 10% PEG-3350. For data collection, the crystals were cryoprotected with reservoir solution containing 30% glycerol. Diffraction datasets were collected at the Advanced Photon Source (Argonne National Laboratory) on beamline 24-ID-C equipped with a Dectris EIGER2 X 16M detector at 100K. Data was collected at a wavelength of 0.97903 Å, detector distance of 280 mm, and 0.25° oscillations. The crystals diffracted X-rays to 2.0 Å resolution. XDS/XSCALE was used to index, integrate, and scale data in the P21 space group24. The asymmetric unit of the crystal contained two molecules, resulting in a Matthews coefficient of 2.68 Å3/Da and 54.15% solvent content in the unit cell25-27. The MRage automated suite in Phenix was used to perform molecular replacement with templates of S1C domain homologs using its built-in NCBI BLAST search28-30. The top 5 hits each resulted in excellent solutions (LLG > 130), with the top solution (PDB 5B6L, LLG of 162.5) used to continue building a complete structure using the AutoBuild function31,32. The structure was iteratively improved by manual rebuilding in Coot and automatic refinement in BUSTER, with NCS restraints enabled and TLS groups defined for the separate domains33-36. Complete refinement and structure statistics are reported in Table 1. Coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 7SJY. Figure images were generated using PyMOL 2.4.1 (Schrodinger, LLC).
Table 1.
Crystal data collection and structure refinement statistics
| RsgI9S1C-CTD | |
|---|---|
| Data collection | |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 63.63, 78.06, 81.21 |
| α, β, γ (°) | 90.00, 112.98, 90.00 |
| Resolution (Å) | 74.77-2.00 (2.05-2.00) |
| Wavelength (Å) | 0.97903 |
| Total observations | 246720 (17477) |
| Unique reflections | 48797 (3594) |
| Rmerge (%) | 4.9 (61.8) |
| I / σI | 16.51 (2.60) |
| CC1/2 | 99.9 (83.9) |
| Completeness (%) | 98.4 (98.2) |
| Multiplicity | 5.1 (4.9) |
| Wilson B-factor (Å2) | 40.84 |
| Refinement | |
| Resolution (Å) | 74.77-2.00 |
| No. of reflections | 48797 |
| Rwork / Rfree (%) | 19.7/21.9 |
| No. atoms | 5058 |
| Protein | 4788 |
| Ligand/ion | 78 |
| Water | 192 |
| B-factors (Å2) (all atoms) | 51.5 |
| Protein | 51.4 |
| Ligand/ion | 64.1 |
| Water | 48.1 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.49 |
| Ramachandran favored (%) | 99.03 |
| Ramachandran allowed (%) | 0.97 |
| Ramachandran outliers (%) | 0.00 |
| PDB ID | 7SJY |
Values in parentheses are for the highest-resolution shell.
Nuclear magnetic resonance and small-angle X-ray scattering (SAXS) experiments.
A 15N-labeled sample of RsgI9S1C-CTD was produced by following the expression and purification methods above, but with expression induced after exchanging the cultures into minimal media supplemented with 15NH4Cl (Cambridge Isotope Laboratories, Tewksbury, MA)37. Spectra were acquired using a 0.4 mM sample in NMR buffer (50 mM sodium phosphate, 200 mM NaCl, 0.03% sodium azide, pH 7.8) at 298 K on a Bruker Avance III HD 600 MHz (14.1 T) spectrometer equipped with a triple resonance cryogenic probe. A 2-D 15N-1H TROSY-HSQC spectrum showed a well-folded protein. The rotational correlation time (tc) of RsgI9S1C-CTD was estimated from a series of 1-D 15N-TRACT experiments, with 2,048 complex points, 32 transients, 100 experiments for each spin state, and the relevant delay incremented by 4 ms38. The decrease in integrated backbone amide intensity was fitted to an exponential decay function, resulting in a measured transverse cross-correlated relaxation rate (ηxy) of 22.9 Hz, which corresponds to a tc value of ~22.1 ns, calculated via algebraic solutions39.
Size-exclusion chromatography small-angle X-ray scattering (SEC-SAXS) was performed at the SIBYLS beamline (Advanced Light Source beamline 12.3.1, Lawrence Berkeley National Laboratory)40. Samples were injected at 5 mg/mL on an Agilent 1290 HPLC with a Shodex KW-803 column equilibrated in size exclusion buffer (50 mM sodium phosphate, 300 mM NaCl, pH 7.0) at a flow rate of 0.65 mL/min. X-ray scattering of eluent was continuously collected on a Dectris PILATUS3 X 2M detector in two-second frames, with X-ray wavelength at 1.216 Å and a sample-to-detector distance of 2.0 m. Multi-angle light scattering (MALS), quasi-elastic light scattering (QELS), and refractometry data were also collected on this eluent and processed using the ASTRA software package (Wyatt) to estimate molecular weight. SAXS frames from before and after the protein elution peak were used to subtract the scattering contribution of the buffer alone, and frames corresponding to protein were merged using ScÅtter IV (www.bioisis.net). SAXS analysis (Guinier analysis and distance distribution calculations) to determine Rg and p(r) function was performed using BioXTAS RAW and the ATSAS Suite41,42. Molecular weights were estimated using the Bayesian inference approach43. The experimental SAXS data of RsgI9S1C-CTD was compared to the theoretical scattering curve from the crystal structure (with missing N-terminal residues added using MODELLER) using the FoXS server44-46. For RsgI9CRE, RsgI9S1C-CTD, and RsgI9ecto, ab initio bead modelling was performed on the output of GNOM using DAMMIF to generate 15 models in slow mode, following by averaging and refinement with DAMMIN47,48. DENSS was used to reconstruct electron density of the same constructs in slow mode to generate 20 density maps, followed by averaging and refinement49. Models were fit into maps using UCSF ChimeraX, which was also used to generate images50.
Cellulose-binding and peptidase activity assays.
Binding to the microcrystalline cellulose substrate (Avicel, Sigma-Aldrich) was qualitatively assayed via SDS-PAGE pull-down assays modified from previous protocols20,22. Recombinant protein and control samples were prepared using 50 μg of protein in 200 μL of lysis buffer (50 mM Tris-HCl, 300 mM NaCl, pH 8.0) and mixed with 15 μg of microcrystalline cellulose (Avicel). Each sample was thoroughly vortexed and then incubated at room temperature with gentle rotation for 60 min. After incubation, samples were separated by centrifugation at 12,000 x g for 10 min. 10 μL of the supernatant containing unbound protein was taken for SDS-PAGE analysis. The remaining sedimented polysaccharide was washed five times with 200 μL aliquots of lysis buffer with 0.1% Triton-200 detergent to mitigate nonspecific protein binding. After the final centrifugation, the sedimented polysaccharide and bound protein was resuspended in 200 μL SDS loading buffer and boiled for 5 min before loading for SDS-PAGE. Each assay was repeated at least three times.
The proteolytic activity of the separate RsgI9 domains was quantitatively measured with UV spectroscopy in triplicate by monitoring the hydrolysis of the modified nonspecific protease substrate azocasein (Sigma-Aldrich). 200 μM protein samples were dissolved in lysis buffer and combined in equal volume with 1% azocasein dissolved in glycine-NaOH buffer (100 mM, pH 10.0 or pH 8.0) for a final reaction volume of 500 μL. A sample containing no enzyme was used as a blank and additional control. Samples were incubated at either 37°C or 50°C with gentle rotation for 30 min. Each reaction was terminated via the addition of 750 μL of 10% trifluoroacetic acid (TFA) and subsequently chilled on ice for 30 min. Samples were centrifuged at 10,000 g for 10 min to separate precipitated enzyme after TFA addition. 125 μL samples were plated into a 96-well plate and the absorbance at 440 nm (A440) was measured using a SpectraMax iD3 Multi-Mode Microplate spectrophotometer (Molecular Devices).
RESULTS AND DISCUSSION
The ectodomain in the RsgI9 anti-σ factor contains a C-terminal bi-domain module.
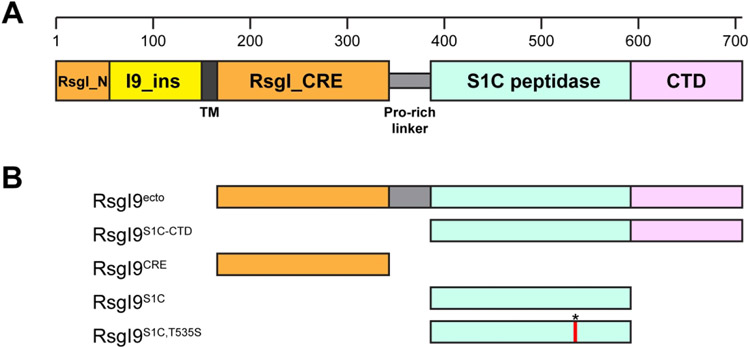
We inspected the primary sequence of the RsgI9 anti-σ factor to gain insight into its function. An alignment of the amino acid sequences of the nine RsgI anti-σ factors in C. thermocellum (RsgI1-9) reveals that they possess the same basic architecture (Fig. 1A). Each contains a cytoplasmic N-terminal region that is followed by a transmembrane helix and a larger C-terminal ectodomain that in some of the anti-σ factors has been shown to bind to carbohydrates17. Several regions are highly conserved amongst the proteins (Fig. 1A, shaded orange). They include a ~40 amino acid N-terminal segment annotated RsgI_N (Pfam: PF12791) that binds to cognate σ factors in the cytoplasm, and a ~170 residue segment that immediately follows the transmembrane helix and is presumably located on the extracellular surface, hereafter called the Conserved RsgI Extracellular (CRE) domain51. NMR studies indicate the CRE domain is autonomously folded, but its structure has not been determined52. In RsgI9 and several other RsgI proteins a proline-rich segment connects the CRE region to a C-terminal region that is varied amongst the anti-σ factors. In RsgI9, the C-terminal region contains amino acids that share homology with trypsin-like S1C domains (residues I396-H578). Residues following this segment do not display sequence homology to any protein of known structure. However, they are classified by the program DISOPRED3 as being structurally ordered and are hereafter referred to as the C-terminal domain (CTD)53. The DISOPRED3 prediction suggests that the S1C and CTD are closely linked, raising the possibility that they form an ordered bi-domain unit. The module may be involved in biomass sensing, as a search of microbial genomes reveals that it is only present in a few bacterial species within the genus Acetivibrio, several of which exhibit cellulolytic activity similar to C. thermocellum (Fig. S1)54.
Figure 1.
RsgI9 contains both unique and RsgI-conserved domains. (A) RsgI9 contains conserved domains that are found in other RsgI-family proteins (orange). These include an intracellular anti-σ factor domain (RsgI_N, residues 1-55), a transmembrane helix (TM, dark gray) and an extracellular domain of unknown function (CRE or RsgI_CRE, residues 167-343). RsgI9 and many other RsgI proteins also contain a Pro-rich linker segment (light gray) that connects the RsgI_CRE to a variable C-terminal region that differs in each type of anti-σ factor. In RsgI9, the C-terminal region contains a domain that is homologous to S1C peptidase domains (mint, residues 396-578) and a C-terminal domain (CTD) of unknown structure (purple, residues 579-707). RsgI9 also contains a unique insertion immediately following RsgI_N (I9_ins, yellow) that is located in the cytoplasm. (B) Depiction of the RsgI9 polypeptide constructs used in this study. In RsgI9S1C,T535S an asterisk and red line indicates the location of the mutation.
The bi-domain module forms a structurally ordered unit that contains peptidase- and NTF2-like domains.
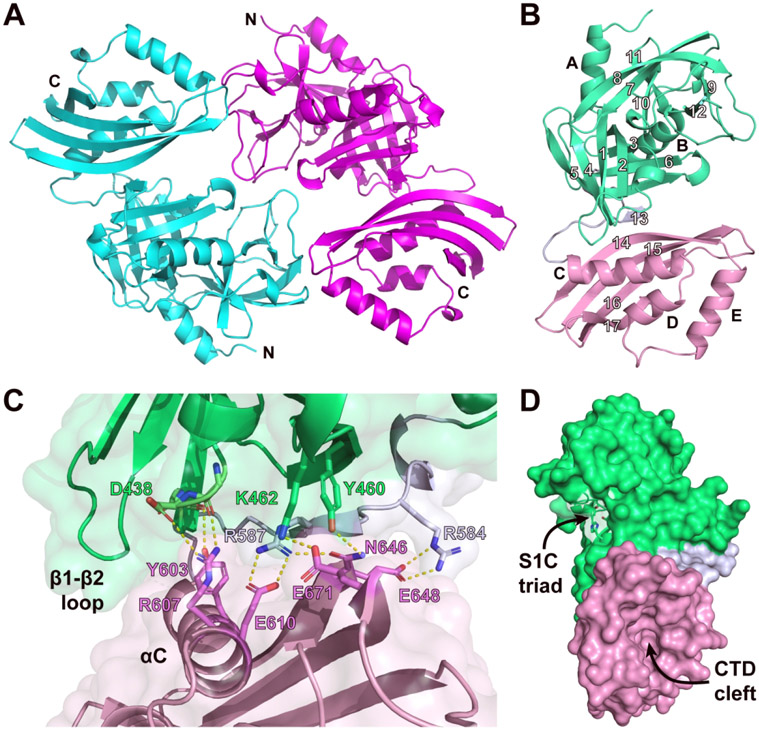
To shed light onto the structure and function of the bi-domain module we determined the 2.0 Å crystal structure of a polypeptide containing the predicted S1C and CTD domains (RsgI9S1C-CTD, residues A387-K702). RsgI9S1C-CTD crystallized in the P21 space group and its structure was determined by molecular replacement using the coordinates of the S1C peptidase domain within the HhoA protein as a search model (PDB: 5B6L, 34% sequence identity), followed by iterative model building of the CTD coordinates (Fig. 2A)31. Continuous electron density allowed modeling of the entirety of the protein, except for amino acid G502 in the β7-β8 loop. Two RsgI9S1C-CTD molecules are present in the asymmetric unit and are arranged in head-to-tail manner. The proteins are related by two-fold non-crystallographic symmetry and adopt similar atomic structures (backbone RMSD < 0.5 Å). Dimerization buries 907 Å2 of protein surface area, but based on a PDBePISA analysis this interface is not biologically relevant55. This is consistent with size-exclusion chromatography with multi-angle light scattering detection (SEC-MALS) analyses that show that RsgI9S1C-CTD is monomeric; the SEC-MALS derived molecular weight is 35.3 ± 0.4 kDa, as compared to a predicted value of 34.5 kDa. Complete data collection and structure refinement statistics are shown in Table 1.
Figure 2.
Structure of RsgI9’s bi-domain unit, RsgI9S1C-CTD. (A) Cartoon representation of the crystal structure of RsgI9S1C-CTD showing the two proteins in the asymmetric unit (N- and C-termini are labeled). (B) Ribbon drawing of a single molecule of RsgI9S1C-CTD with its secondary structural elements labeled. The S1C and CTD domains are colored mint and pink, respectively, while the tether is colored blue-gray. (C) Close-up view of the interface between the S1C and CTD domains, coloring as in the previous panel. Hydrogen-bonding and salt bridge interactions are indicated by yellow dashed lines. (D). Surface representation of RsgI9S1C-CTD showing the locations of the recessed cleft in the CTD and a pocket within the S1C domain that in homologous proteins forms a peptidase active site.
The bi-domain unit adopts a rigid structure that houses potential ligand binding sites within S1C peptidase and NTF2-like domains (Fig. 2A, B). The N-terminal S1C domain is composed of residues P389-H578 and shares structural homology with the Deg/HtrA family of serine proteases. Its structure is formed by two α-helices (αA, αB) that flank a pair of β-barrels (β1-β6 and β7-β12). In Deg/HtrA proteases these β-barrels surround a pocket housing a His-Asp-Ser active site triad that mediates catalysis56. However, in RsgI9 the serine residue in the triad is replaced with threonine (H434-D465-T535, RsgI9 numbering). The CTD is formed by residues F596-K702 and contains three helices (αC, αD, and αE) that are placed over a four-stranded β-sheet (β14-β17). Based on DALI and PDBeFold analyses it adopts a NTF2-like fold28,57, which was originally identified in the Nuclear Transport Factor 2 (NTF2) protein58. However, the CTD shares less than 20% sequence identity with NTF2 or its structural homologs. NTF2-like folds are broadly distributed in biology and typically form cone-like structures that harbor a recessed hydrophobic cavity that mediates ligand binding59. In the CTD, residues that would enclose this pocket are replaced by a short loop between the αD and αE helices, generating a more open cleft that might serve as a ligand binding site (Fig. 2D, S2). A network of hydrogen-bonding and salt-bridging interactions between the S1C and NTF2 domains buries ~680 Å2 of solvent exposed surface area (Fig. 2C). The short tether that links the primary sequences of the domains is at the heart of the interface (residues S579-D595). It forms strand β13 that connects the β-sheets of each module by hydrogen bonding; strand β13 pairs with strands β5 and β14 in the S1C and CTD folds, respectively. Additionally, the β1/β2 loop in the S1C domain, known as loop LA in serine protease nomenclature, packs over the αC helix of the CTD60. These inter-domain interactions appear to be important for the stability of the CTD, as attempts to express a polypeptide containing only this domain were unsuccessful. NMR and SAXS analyses reveal that packing of the S1C and CTD domains causes them to adopt a rigid structure in solution. In particular, the experimental molecular correlation time (tc) of RsgI9S1C-CTD was determined by NMR to be 22.1 ns, which agrees with the theoretical value for a 34.5 kDa globular protein (~20.9 ns)61,62 (Fig. S3). SAXS experiments described below also indicate that the domains are immobilized with respect to one another. Thus, we conclude that the phylogenetically conserved bi-domain within RsgI9 forms a structurally ordered unit that houses potential ligand binding sites within its S1C and CTD domains.
The bi-domain unit interacts with cellulose.
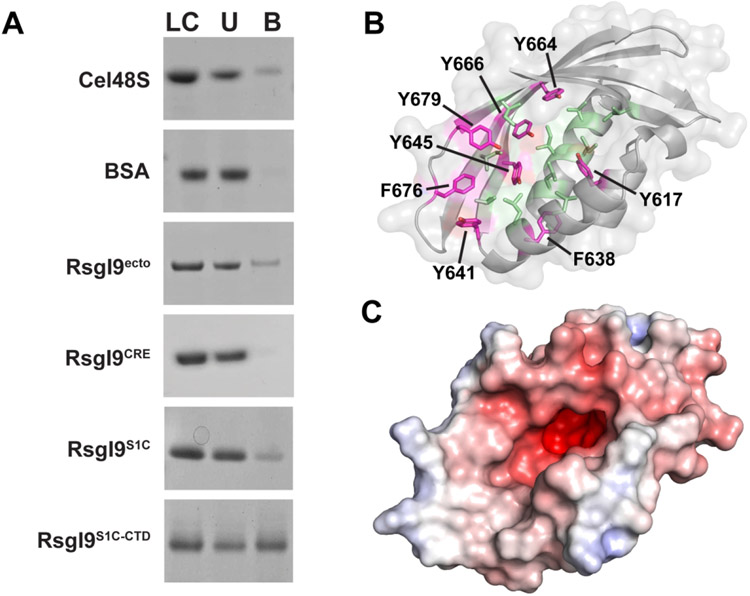
The RsgI1, RsgI2, RsgI3, RsgI4, and RsgI6 anti-σ factors in C. thermocellum bind to carbohydrates and are thought to serve as biomass sensors20-22. We therefore probed whether RsgI9 could interact with cellulose using an established pull-down assay20,22. In these experiments polypeptides containing either the entire ectodomain (RsgI9ecto), the bi-domain unit (RsgI9S1C-CTD), the S1C domain (RsgI9S1C), or the CRE domain (RsgI9CRE) were tested. Only peptides containing the bi-domain unit domains interact with microcrystalline cellulose (Avicel, Fig. 3A), as the CRE domain did not interact with any of the carbohydrates that were tested. Control experiments confirm specificity, as a known cellulase Cel48S binds to cellulose in these experiments, whereas no binding was detected for bovine serum albumin (BSA). Interestingly, significantly more RsgI9S1C-CTD protein binds to Avicel as compared to RsgI9S1C, suggesting that the majority of affinity for cellulose originates from the CTD. Although the molecular basis of cellulose binding remains to be determined, it is possible that it is mediated by interactions with CTD’s cleft. This is because the walls of this groove contain a series of surface exposed aromatic and non-polar amino acid side chains (Fig. 3B, C), which in other carbohydrate-binding proteins form π-stacking interactions with bound sugars63. Notably, the C. thermocellum RsgI1, RsgI2, and RsgI4 anti-σ factors each contain CBM3 carbohydrate binding domains that employ a linear strip of aromatic amino acids to interact with the pyranoside rings within cellulose22,64. Although the CTD and CBM3 adopt distinct folds, the CTD cleft also contains a linear strip of aromatic residues in strands β14-β16 (Y641, Y645, Y664, Y666, F676, Y679) suggesting that it may form similar interactions. A more deeply recessed anionic patch is located immediately adjacent to the cleft and occupied by a glycerol molecule in the structure of RsgI9S1C-CTD. Although its function remains unknown, similar anionic pockets are present in carbohydrate-binding proteins and often interact with metal cations (e.g. Ca2+)65-67.
Figure 3.
The RsgI9 ectodomain binds to crystalline cellulose. (A) Pull-down assay that tests protein binding to microcrystalline cellulose (Avicel). In each experiment purified protein samples were incubated with Avicel, and then centrifuged to assess binding to this insoluble substrate. Lanes are labeled: LC, loading control; U, unbound protein; B, bound protein. (B) Potential carbohydrate recognition groove in the CTD exposes aromatic (magenta) and non-polar residues (light green). The aromatic sidechains form a continuous strip on one side from Y641 to Y664. (C) Solvent-excluded surface of this domain colored by surface electrostatics (from red to blue, −7.5 to +7.5 e) indicates a negatively charged patch at the base of the groove.
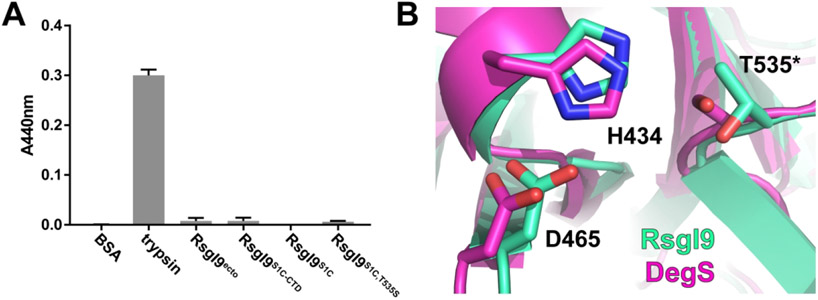
Despite exhibiting structural homology with members of the Deg/HtrA protease family, the S1C domain in RsgI9 is enzymatically inactive in vitro. In marked contrast to the trypsin control, neither the intact ectodomain nor the bi-domain unit exhibits proteolytic activity when assayed using azocasein as a substrate (Fig. 4A). The structure of RsgI9’s S1C domain is similar to the E. coli DegS protease, which mediates the heat shock response by degrading the RseA anti-σ factor 68. Superimposing the coordinates of their S1C domains reveals striking similarities in the positioning of residues that form the active site in DegS. The side chains that construct the His-Asp-Ser catalytic triad in DegS overlay well with residues in the RsgI9 S1C domain, except that the serine within the triad is replaced with a threonine in RsgI9 (Fig. 4B). Interestingly, a T535S variant of RsgI9 is also enzymatically inactive even though it contains a full complement of triad residues. This suggests that the pocket housing the triad in RsgI9 lacks other essential features that are needed for catalysis69. Indeed, a detailed comparison with the structure of DegS reveals that the oxyanion hole used to stabilize reaction intermediates may not be properly formed in RsgI9 (Fig. S4)70. Interestingly, DegS and the bi-domain unit also have similar domain architectures. In DegS the S1C domain packs against a smaller C-terminal PDZ domain that regulates proteolytic activity in response to binding to unfolded peptides, while in RsgI9 the S1C domain is packed against a NTF2-like module that may bind to carbohydrates. As the S1C domain in RsgI9 is inactive, it may simply function as a spacer that properly positions the CTD and/or its pocket containing the His-Asp-Thr triad may be used to bind to a ligand whose identity remains to be determined.
Figure 4.
Functional studies of the protease-like domain in RsgI9 and its structural homology with DegS. (A) Protease activity assay results. RsgI9 constructs were all inactive against a promiscuous proteolytic substrate, azocasein. (B) The catalytic triad residues as they appear in RsgI9’s S1C domain (mint), including a threonine in place of the conserved serine (T535*), aligned with active form of structural homolog DegS (magenta, PDB: 4RQZ). Residues are numbered as they appear in the RsgI9 primary sequence.
The full-length ectodomain adopts a rod-like conformation that may extend from the cell surface.
To gain insight into the solution conformation of the ectodomain, size-exclusion chromatography coupled with small-angle X-ray scattering (SEC-SAXS) data was collected for polypeptides containing the isolated bi-domain unit and the intact ectodomain. The SEC-SAXS data for RsgI9S1C-CTD indicates that the bi-domain unit adopts a rigid structure, consistent with its crystal structure and NMR measurements of its molecular correlation time. This is evident from the dimensionless Kratky plot, which shows the presence of a peak near q*Rg = √3 and normalized intensity = 1.1 for RsgI9S1C-CTD (Fig. 5A, red)71. Furthermore, the scattering data also fits very well to the FoXS predicted scattering curve for the RsgI9S1C-CTD crystal structure (χ2 = 1.23) (Fig. S5A),44 and the calculated P(r) function for RsgI9S1C-CTD is also consistent with the crystal structure (Dmax of 82 Å versus 80 Å in the crystal structure) (Fig. 5B, red). Finally, ab initio bead model and electron density reconstructions using DAMMIN/F and DENSS, respectively, fit well to a single polypeptide from the crystal structure (Fig. S5B)47,48.
Figure 5.
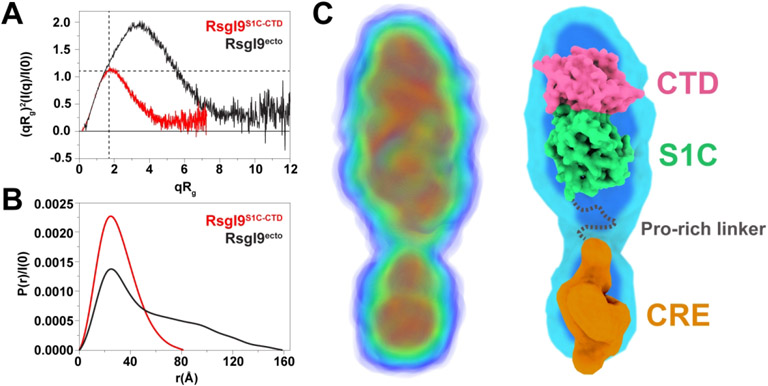
SAXS analyses show the RsgI9 ectodomain forms an extended structure. (A) Dimensionless Kratky plots of both the RsgI9S1C-CTD bi-domain unit (red) and intact ectodomain, RsgI9ecto (black). The two dashed lines indicate the theoretical peak position for a typical globular protein. The bi-domain unit behaves as a compact globular protein, while the intact ectodomain forms an extended rod-like structure. (B) Distance distribution function P(r) for both RsgI9S1C-CTD (red, Dmax 82 Å) and RsgI9ecto (black, Dmax 160 Å). (C) SAXS derived electron density reconstruction of the ectodomain obtained using the program DENSS49. The volume of electron density is colored with a gradient from blue to red to indicate regions of highest density (left). Coordinates of the crystal structure of RsgI9S1C-CTD fitted into the electron density (right). A low-resolution model of the CRE domain that was obtained using the program DENSS and RsgI9CRE SAXS data is also fitted into the density.
The SAXS analysis indicates that the intact ectodomain adopts a bi-lobed, rod-shaped structure in solution. RsgI9ecto eluted from the size exclusion chromatography step as a monomer; the theoretical molecular weight of monomeric RsgI9ecto is 59.7 kDa, compatible with estimates from MALS and SAXS of 62.7 ± 0.2 kDa and 58.1 kDa, respectively. Notably, the scattering profile of RsgI9ecto is indicative of an extended protein as evidenced by the up- and right-ward shift of its peak in the dimensionless Kratky plot (Fig. 5A, black). Furthermore, the P(r) distance distribution function of RsgI9ecto shows a Dmax at 160 Å with a Rg value of 45 Å, much larger than expected for a compact spherical protein of the same molecular weight (Fig. 5B, black)71. The peak centered at ~25 Å in the p(r) function of RsgI9S1C-CTD is also present in the corresponding plot of RsgI9ecto, but the broad and nearly linear decrease toward its large Dmax is indicative of a rod-like extension72. Ab initio electron density reconstruction using DENSS reveals that the ectodomain adopts an elongated, bi-lobed structure (Fig. 5C)49. The coordinates of the crystal structure of the bi-domain unit fit well into the major lobe of the density, while the small lobe presumably houses the CRE domain that adopts a globular structure. Density in between the lobes corresponds to the proline-rich linker that connects the CRE and S1C domains, but is less well-defined. In totality, the SAXS data strongly support the notion that the ectodomain predominantly adopts an extended configuration in which the bi-domain and CRE units do not interact with one another. Residues in the intervening proline-rich linker (residues 344-386) presumably cause RsgI9 to be elongated. Interestingly, the proline-rich linker is also present in six other C. thermocellum anti-σ factors (RsgI1 to RsgI6) and varies in length from 75 to 262 residues (Fig. S6). Thus, it appears that like RsgI9, these anti-σ factors will also adopt elongated structures that project carbohydrate-binding modules away from the cell surface.
CONCLUSIONS
The results of our studies provide new insight into how RsgI9 and other anti-σ factors enable C. thermocellum to sense different types of biomass. RsgI9’s ectodomain adopts an elongated structure in which two folded domains are connected by a proline-rich linker. A highly conserved CRE domain is presumably located near the cell membrane and is joined by the extended linker to an unusual bi-domain unit that interacts with cellulose. This basic architecture is likely conserved in other C. thermocellum anti-σ factors, as the primary sequences of RsgI1 to RsgI6 encode for CRE domains that are connected by proline-rich linkers to carbohydrate binding modules (Fig. S5). In order to promote new transcriptional programs, the RsgI-type anti-σ factors presumably transduce carbohydrate binding signals received on the extracellular surface to the cytoplasm, a process that releases the cognate ECF σ factor needed for RNA polymerase targeting to specific promoters. Recent structural studies have provided insight into how the intracellular RsgI_N domain engages its cognate sigma factor19, but how these interactions are disrupted in response to carbohydrate binding on the cell surface remains unknown. The common architecture adopted by the ectodomains in C. thermocellum and the fact that each contains a highly conserved (CRE) domain near the membrane surface suggests that they may employ a similar mechanism to promote signaling.
Supplementary Material
FUNDING INFORMATION
This material is based upon work supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research program under Award Number DE-FC02-02ER63421, which also provides support for resources at the UCLA-DOE Institute’s X-ray and EM Structure Determination Core and Macromolecular NMR Core. We also acknowledge NIH equipment grants S10OD025073 and S10OD016336 for partial support of the NMR core facilities, as well as NIH grant R01-AI052217 (R.T.C). The X-ray diffraction experiments were performed at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). The research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract number DE-AC02-06CH11357. SAXS data was collected at SIBYLS which is supported by the DOE-BER IDAT DE-AC02-05CH11231 and NIGMS ALS-ENABLE (P30 GM124169 and S10OD018483).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
DATA AVAILABILITY
The coordinates and structure factors of the RsgI9S1C-CTD structure have been deposited in the Protein Data Bank under accession code 7SJY.
REFERENCES
- 1.Watts N, Amann M, Ayeb-Karlsson S, et al. The Lancet Countdown on health and climate change: from 25 years of inaction to a global transformation for public health.The Lancet. 2018;391(10120):581–630. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Cruz-Morales P, Zargar A, et al. Biofuels for a sustainable future. Cell.2021; 184(6): 1636–1647. [DOI] [PubMed] [Google Scholar]
- 3.Sinitsyn AP, Sinitsyna OA. Bioconversion of Renewable Plant Biomass. Second-Generation Biofuels: Raw Materials, Biomass Pretreatment, Enzymes, Processes, and Cost Analysis. Biochemistry (Moscow). 2021;86(S1):S166–S195. [DOI] [PubMed] [Google Scholar]
- 4.Viikari L, Vehmaanpera J, Koivula A. Lignocellulosic ethanol: From science to industry. Biomass and Bioenergy. 2012;46:13–24. [Google Scholar]
- 5.Olson DG, McBride JE, Joe Shaw A, Lynd LR. Recent progress in consolidated bioprocessing. Current Opinion in Biotechnology. 2012;23(3):396–405. [DOI] [PubMed] [Google Scholar]
- 6.La Grange DC, Den Haan R, Van Zyl WH. Engineering cellulolytic ability into bioprocessing organisms. Applied Microbiology and Biotechnology. 2010;87(4):1195–1208. [DOI] [PubMed] [Google Scholar]
- 7.Akinosho H, Yee K, Close D, Ragauskas A. The emergence of Clostridium thermocellum as a high utility candidate for consolidated bioprocessing applications. Front Chem. 2014;2:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson EA, Sakajoh M, Halliwell G, Madia A, Demain AL. Saccharification of Complex Cellulosic Substrates by the Cellulase System from Clostridium thermocellum. Appl Environ Microbiol. 1982;43(5):1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Zhang YH, Lynd LR. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc Natl Acad Sci U S A. 2006; 103(44): 16165–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alves VD, Fontes CMGA, Bule P. Cellulosomes: Highly Efficient Cellulolytic Complexes. In: Springer International Publishing; 2021:323–354. [DOI] [PubMed] [Google Scholar]
- 11.Hu BB, Zhu MJ. Reconstitution of cellulosome: Research progress and its application in biorefinery. Biotechnology and Applied Biochemistry. 2019;66(5):720–730. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Zhang X, Yuan H, et al. Research progress and the biotechnological applications of multienzyme complex. Applied Microbiology and Biotechnology. 2021; 105(5): 1759–1777. [DOI] [PubMed] [Google Scholar]
- 13.Lamed R, Bayer EA. The Cellulosome of Clostridium thermocellum. In: Laskin AI, ed. Advances in Applied Microbiology. Vol 33. Academic Press; 1988:1–46. [Google Scholar]
- 14.Leibovitz E, Ohayon H, Gounon P, Beguin P. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J Bacteriol. 1997; 179(8):2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Currie MA, Adams JJ, Faucher F, Bayer EA, Jia Z, Smith SP. Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome. Journal of Biological Chemistry. 2012;287(32):26953–26961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoav S, Barak Y, Shamshoum M, et al. How does cellulosome composition influence deconstruction of lignocellulosic substrates in Clostridium (Ruminiclostridium) thermocellum DSM 1313? Biotechnology for Biofuels. 2017; 10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataf Y, Bahari L, Kahel-Raifer H, et al. Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proceedings of the National Academy of Sciences. 2010;107(43):18646–18651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahel-Raifer H, Jindou S, Bahari L, et al. The unique set of putative membrane-associated anti-σ factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiology Letters. 2010;308(1):84–93. [DOI] [PubMed] [Google Scholar]
- 19.Wei Z, Chen C, Liu Y-J, et al. Alternative σI/anti-σI factors represent a unique form of bacterial σ/anti-σ complex. Nucleic Acids Research. 2019;47(11):5988–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grinberg IR, Yaniv O, Ora LO, et al. Distinctive ligand-binding specificities of tandem PA14 biomass-sensory elements from Clostridium thermocellum and Clostridium clariflavum. Proteins: Structure, Function, and Bioinformatics. 2019;87(11):917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahari L, Gilad Y, Borovok I, et al. Glycoside hydrolases as components of putative carbohydrate biosensor proteins in Clostridium thermocellum. Journal of Industrial Microbiology & Biotechnology. 2011;38(7): 825–832. [DOI] [PubMed] [Google Scholar]
- 22.Yaniv O, Fichman G, Borovok I, et al. Fine-structural variance of family 3 carbohydrate-binding modules as extracellular biomass-sensing components of Clostridium thermocellum anti-sigmal factors. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 2):522–534. [DOI] [PubMed] [Google Scholar]
- 23.Friedhoff P, Gimadutdinow O, Ruter T, et al. A procedure for renaturation and purification of the extracellular Serratia marcescens nuclease from genetically engineered Escherichia coli. Protein Expr Purif. 1994;5(1):37–43. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallographica Section D Biological Crystallography. 2010;66(2): 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weichenberger CX, Rupp B. Ten years of probabilistic estimates of biocrystal solvent content: new insights via nonparametric kernel density estimate. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 6): 1579–1588. [DOI] [PubMed] [Google Scholar]
- 26.Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Science. 2003; 12(9): 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33(2):491–497. [DOI] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallographica Section D Biological Crystallography. 2004;60(12):2256–2268. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40(4):658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liebschner D, Afonine PV, Baker ML, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallographica Section D Structural Biology. 2019;75(10):861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong W, Wang J, Niu G, Zhao S, Liu L. Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803. FEBS Letters. 2016;590(19):3435–3442. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallographica Section D Biological Crystallography. 2008;64(1):61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smart OS, Womack TO, Flensburg C, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallographica Section D Biological Crystallography. 2012;68(4):368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D Biological Crystallography. 2010;66(4):486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bricogne G, Blanc E, Brandl M, et al. BUSTER version 2.10.4 Global Phasing Ltd, Cambridge, UK. 2021. [Google Scholar]
- 37.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. Journal of Biomolecular NMR. 2001;20(1):71–75. [DOI] [PubMed] [Google Scholar]
- 38.Lee D, Hilty C, Wider G, Wuthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. Journal of Magnetic Resonance. 2006;178(1):72–76. [DOI] [PubMed] [Google Scholar]
- 39.Robson SA, Dağ Ç, Wu H, Ziarek JJ. TRACT revisited: an algebraic solution for determining overall rotational correlation times from cross-correlated relaxation rates. Journal of Biomolecular NMR. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Classen S, Hura GL, Holton JM, et al. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. Journal of Applied Crystallography. 2013;46(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hopkins JB, Gillilan RE, Skou S. BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. Journal of Applied Crystallography. 2017;50(5): 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manalastas-Cantos K, Konarev PV, Hajizadeh NR, et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. Journal of Applied Crystallography. 2021;54(1):343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hajizadeh NR, Franke D, Jeffries CM, Svergun DI. Consensus Bayesian assessment of protein molecular mass from solution X-ray scattering data. Scientific Reports. 2018;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Research. 2016;44(W1):W424–W429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneidman-Duhovny D, Hammel M, John, Sali A. Accurate SAXS Profile Computation and its Assessment by Contrast Variation Experiments. Biophysical Journal. 2013;105(4):962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Current Protocols in Bioinformatics. 2016;54(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.σDl Svergun. Restoring Low Resolution Structure of Biological Macromolecules from Solution Scattering Using Simulated Annealing. Biophysical Journal. 1999;76(6):2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42(2):342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grant TD. Ab initio electron density determination directly from solution scattering data. Nature Methods. 2018;15(3):191–193. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX : Structure visualization for researchers, educators, and developers. Protein Science. 2021;30(1):70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021. Nucleic Acids Research. 2021;49(D1):D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding X, Chen C, Cui Q, Li W, Feng Y. Resonance assignments of the periplasmic domain of a cellulose-sensing trans-membrane anti-sigma factor from Clostridium thermocellum. Biomolecular NMR Assignments. 2015;9(2):321–324. [DOI] [PubMed] [Google Scholar]
- 53.Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31(6):857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Research. 2018;46(W1):W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. Journal of Molecular Biology. 2007;372(3):774–797. [DOI] [PubMed] [Google Scholar]
- 56.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416(6879):455–459. [DOI] [PubMed] [Google Scholar]
- 57.Holm L Using Dali for Protein Structure Comparison. In: Springer US; 2020:29–42. [DOI] [PubMed] [Google Scholar]
- 58.Bullock TL, Clarkson WD, Kent HM, Stewart M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2). J Mol Biol. 1996;260(3):422–431. [DOI] [PubMed] [Google Scholar]
- 59.Eberhardt RY, Chang Y, Bateman A, et al. Filling out the structural map of the NTF2-like superfamily. BMC Bioinformatics. 2013;14(1):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perona JJ, Craik CS. Evolutionary Divergence of Substrate Specificity within the Chymotrypsin-like Serine Protease Fold. Journal of Biological Chemistry. 1997;272(48):29987–29990. [DOI] [PubMed] [Google Scholar]
- 61.Cavanagh J Protein NMR spectroscopy : principles and practice. 2nd ed. Amsterdam ; Boston: Academic Press; 2007. [Google Scholar]
- 62.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by nitrogen-15 relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31(23):5269–5278. [DOI] [PubMed] [Google Scholar]
- 63.Hudson KL, Bartlett GJ, Diehl RC, et al. Carbohydrate–Aromatic Interactions in Proteins. Journal of the American Chemical Society. 2015;137(48):15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tormo J, Lamed R, Chirino AJ, et al. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15(21):5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 65.Garron ML, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20(12):1547–1573. [DOI] [PubMed] [Google Scholar]
- 66.Yang L, Connaris H, Potter JA, Taylor GL. Structural characterization of the carbohydrate-binding module of NanA sialidase, a pneumococcal virulence factor. BMC Structural Biology. 2015;15(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding N, Zhao B, Ban X, et al. Carbohydrate-Binding Module and Linker Allow Cold Adaptation and Salt Tolerance of Maltopentaose-Forming Amylase From Marine Bacterium Saccharophagus degradans 2-40 (T). Front Microbiol. 2021;12:708480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alba BM. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma E-dependent extracytoplasmic stress response. Genes & Development. 2002;16(16):2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Research. 2018;46(D1):D624–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sohn J, Grant RA, Sauer RT. Allosteric Activation of DegS, a Stress Sensor PDZ Protease. Cell. 2007;131(3):572–583. [DOI] [PubMed] [Google Scholar]
- 71.Durand D, Vives C, Cannella D, et al. NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J Struct Biol. 2010;169(1):45–53. [DOI] [PubMed] [Google Scholar]
- 72.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Reports on Progress in Physics. 2003;66(10):1735–1782. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors of the RsgI9S1C-CTD structure have been deposited in the Protein Data Bank under accession code 7SJY.