Abstract
Innate immune cells participate in the detection of tumor cells via complex signaling pathways mediated by pattern-recognition receptors, such as Toll-like receptors and nucleotide-binding and oligomerization domain–like receptors. These pathways are finely tuned via multiple mechanisms, including epigenetic regulation. It is well established that hematopoietic progenitors generate innate immune cells that can regulate cancer cell behavior, and the disruption of normal hematopoiesis in pathologic states may lead to altered immunity and the development of cancer. In this review, we discuss the epigenetic and transcriptional mechanisms that underlie the initiation and amplification of innate immune signaling in cancer. We also discuss new targeting possibilities for cancer control that exploit innate immune cells and signaling molecules, potentially heralding the next generation of immunotherapy.
Introduction
Host immunity can be classified into innate immunity, which is rapid to develop but less specific, and adaptive immunity, which is slower to develop but more specific. Innate immunity plays an important role in host defense against infection and cancer, recognizing various antigens via pattern recognition receptors (PRR). Innate immune cells comprise a wide range of myeloid and lymphoid cell types that share common hematopoietic origin (1). Two major conceptual advances have highlighted our rapidly evolving understanding of innate immunity. First, bone marrow hematopoietic stem and progenitor cells (HSPC) can sense systemic inflammation and adapt by increasing their proliferation rate and skewing differentiation toward myeloid cells. Such HSPC adaptations are beneficial in eliminating pathogens during the acute phase of infection. However, they may contribute to chronic inflammation, and to HSPC malfunction and exhaustion when sustained (2). Second, innate immune cells and HSPCs can exhibit adaptive characteristics, termed trained immunity; previous exposure to a pathogen leads to an enhanced innate immune response upon rechallenge (3). In this review, we focus on the epigenetic and transcriptional regulation of innate immune cells and signaling in different cancers. Also, we summarize how targeting innate immunity can spur the development of next generation cancer immunotherapies.
Regulation of the Genesis of Innate Immune Cells
HSPCs
HSPCs generate a variety of cells that participate in innate immunity. IFNs play an important role in the response of HSPCs to inflammation. Activation of the type 1 IFN signaling pathway, mediated by IFNα/β receptor IFNAR, drives the proliferation of dormant HSCs (4) while its inhibition, such as by the negative regulator, Irf2, promotes hematopoietic stem cell (HSC) quiescence (5). Additional mechanisms against IFNα-induced HSC dysfunction include retinoic acid signaling (6), and the circular RNA cia-cGAS that antagonizes cyclic GMP-AMP (cGAMP) synthase cGAS-mediated virus DNA sensing (7). The effects of IFNγ on HSCs are context-dependent. For example, HSC proliferation is promoted by IFNγ-STAT1 signaling in response to mycobacterium infection (8). However, upon lymphocytic choriomeningitis virus infection, IFNγ inhibits the proliferation of HSCs by reducing STAT5 phosphorylation (9). IFNγ also induces myeloid differentiation dependent on Batf2 activity (10). TNF induces HSC proliferation and myeloid lineage differentiation by upregulating PU.1 (11). IL3, produced by innate response activator B cells, induces myelopoiesis (12). The IL6/IL12 cytokine family members, including IL27, also act on HSPCs to promote emergency myelopoiesis (13, 14). IL1 directly accelerates cell division and myeloid differentiation by activating a PU.1-dependent gene program (15). This allows for rapid myeloid recovery following acute marrow injury; however, chronic IL1 exposure compromises HSC self-renewal and restricts HSC lineage output (15).
Neutrophils
The generation of neutrophils from HSCs is regulated by a number of transcription factors (TF), including CAAT/enhancer binding protein (C/EBP), GATA1, and PU.1. C/EBP-α induces early myeloid precursors to differentiate by negatively regulating the expression of c-Myc, via an E2F binding site in the c-Myc promoter (16). Then, the acetylation of C/EBP-ε at K121 and K198, together with the absence of GATA-1, triggers CMPs to commit to terminal neutrophil differentiation (17). PU.1 recruits histone deacetylase 1 (HDAC1) to inhibit the accessibility of AP-1 binding motifs, thereby preventing the hyperactivation of neutrophils (18). AP-1 is a heterodimeric TF that is activated by inflammatory cytokines, growth factors, and infection, which activate kinases that modulate its transcriptional activity (19). AP-1 cooperates with other TFs, including NF-κB and IRF to stimulate the expression of type I IFNs and proinflammatory cytokines (20).
Our understanding of the roles that neutrophils play in cancer is growing. The prognostic value of circulating neutrophils and tumor-associated neutrophils have been assessed in various cancers (21). Antitumor (N1) and protumor (N2) subpopulations of neutrophils have been identified (22). N1 are characterized by an immunostimulatory profile (TNFαhigh, CCL3high, ICAM-1high, Arginaselow) and cytotoxic activity towards tumor cells, whereas N2 exhibit upregulation of chemokine production (CCL2, 3, 4, 8, 12, and 17, and CXCL1, 2, 8 and 16; ref. 23). The fate switch between N1 and N2 is controlled by TGFβ, which promotes the N2 phenotype (24), and IFNβ, which promotes the N1 phenotype (25). TGFβ signaling is transduced via SMAD proteins that regulate chromatin remodeling and transcription, either by direct DNA binding or indirectly through other TFs (26). IFNβ signaling is mediated by the activator of transcription (STAT) family of TFs (27). However, the epigenetic mechanisms that regulate cytokine and chemokine gene expression in neutrophils, in response to TGFβ or IFNβ, remains to be determined.
Macrophages
Macrophages phagocytize microorganisms and apoptotic cells, and produce inflammatory cytokines (28). Tissue-resident macrophages are established during embryonic and fetal hematopoiesis, but they can also arise from circulating monocytes after local macrophage depletion, inflammation, and normal aging (29). Regardless of their cell of origin, the major regulator of the macrophage lineage is the colony stimulating factor (CSF) 1 receptor (30). Expression of MafB and c-Maf also play a role in driving terminal macrophage differentiation (31). Moreover, the NAD-dependent lysine deacetylases, Sirtuins 1 and 2, play a critical role in macrophage differentiation via a direct interaction with DNMT3B (32). Notably, tumor-associated macrophages (TAM) are mostly protumorigenic in solid tumors, functioning to promote carcinogenesis, neoangiogenesis, immune-suppressive tumor microenvironment, chemoresistance, and metastasis. Reprogramming of immune-suppressive TAMs by pharmacologic approaches has gathered much interest in recent years to improve cancer therapies (33).
Macrophages can be classified into M1 (classically activated macrophages, or kill-type macrophages) that are primed by Th1 cytokines such as IFNγ and bacterial products, and M2 (alternatively activated macrophages, or repair-type macrophages) that are primed by Th2 cytokines such as IL4 and IL13 (34). IFNγ plays a pivotal role in promoting immunity against cancer (35). IFNγ triggers the receptor association of the JAK1 and JAK2 tyrosine kinases, which then induce the phosphorylation of STAT1 and STAT2. This promotes the binding of STAT1 homodimers to consensus DNA sequences termed GAS elements, triggering the expression of IFN-stimulated genes (36). In contrast, IFNα leads to phosphorylation of STAT1 and STAT2, which heterodimerize and bind to an IFN-stimulated response element (ISRE) in conjunction with IRF9 (37). M1 macrophages elicit rapid proinflammatory responses to infection and tissue damage by sensing lipopolysaccharide and damage-associated molecular patterns, respectively, while M2 macrophages possess anti-inflammatory activities that enable these cells to resolve inflammation and promote tissue repair (38).
Epigenetic mechanisms that control macrophage polarization are being uncovered. M1 polarization, which can be driven by LPS and IFNγ, requires dynamic metabolic reprogramming and a two-stage remodeling of the tricarboxylic acid (TCA) cycle, including the transient accumulation and subsequent decrease in metabolites such as succinate and itaconate (39). The tumor environment provides signals such as PGE2 or TGFβ that inhibit M1 activation, thus M2 macrophages predominate in most human cancers, where they produce growth-promoting molecules for tumors (40). Jumonji domain–containing protein D3 (Jmjd3), a key H3K27 demethylase, whose activity is dependent on glutamine metabolism, controls M2 macrophage activation (41, 42); the production of α-ketoglutarate via glutaminolysis promotes M2 macrophage activation via fatty-acid oxidation and the Jmjd3-dependent epigenetic reprogramming of M2 genes (43). IL4-induced M2 polarization of liver macrophages is dependent on the activation of STAT6-JMJD3 signaling and suppression of TLR4–NF-κB signaling (44). Macrophage heterogeneity is not fully represented by a dichotomy between M1 and M2. Macrophages also exhibit intermediate phenotypes and are in fact a continuum of polarization states from the two ends marked by M1 and M2. Heterogeneous subpopulations of macrophages take on a variety of roles depending on tissue type and the specific pathology (45). Macrophages even show plasticity after polarization (46). Thus, altering macrophage polarization dynamics, such as triggering an M2 to M1 macrophage transition, could slow or stop cancer growth, a strategy that form the basis for novel cancer immunotherapy (40).
Myeloid-derived suppressor cells
Myeloid-derived suppressor cells (MDSC) are cells of myeloid origin with potent immune-suppressive functions (47). MDSC generation occurs in two phases in cancer, an expansion phase driven mainly by tumor-derived growth factors that promote the accumulation of immature myeloid cells, and an activation phase driven mainly by tumor stroma-derived proinflammatory cytokines, which convert immature myeloid cells into MDSCs (48). A complex network of extracellular signals, chromatin modulators, and TFs is involved in the regulation of MDSCs. For example, the increased production of IL6 in mouse myeloid cells results in STAT3 activation and MDSC expansion (49). Hypoxia-inducible factor 1α (HIF1α) positively regulates the V-domain Ig suppressor of T-cell activation (VISTA) promoter, increasing VISTA expression on myeloid cells and facilitating MDSC-mediated suppression of T-cell activity (50). DNMT3A downregulation erases MDSC-specific hypermethylation and abolishes their immunosuppressive capacity in cancer (51). Moreover, NLRP3 is expressed in MDSCs (52); Nlrp3-deficient mice exhibit decreased MDSCs at the tumor site, implicating NLRP3 in MDSC expansion and/or recruitment (53).
The epigenetic regulation of the generation of different MDSC subsets is being defined. STAT3 signaling, induced by various soluble mediators, is required for monocytic MDSC generation (54). The IFNγ-STAT1-IRF1 axis appears to be specifically crucial for monocytic MDSC suppressive activity (55). In contrast, the generation of human granulocytic MDSCs is less clear. These cells are morphologically heterogeneous, ranging in appearance from immature to mature neutrophils (56), and recent studies suggest that immunosuppressive granulocytic MDSCs can be derived from mature neutrophils (57). STAT3 can also trigger polymorphonuclear (PMN)-MDSC accumulation by increasing the expression of several components of the NADPH complex, such as S100A9, that leads to increased reactive oxygen species (ROS) production (58). IRF8 limits MDSC generation, particularly the PMN-MDSC subset, in mouse mammary tumor models (59). The biology and regulatory mechanisms of MDSC subsets need further characterization.
Innate lymphoid cells and dendritic cells
Innate lymphoid cells (ILC) and dendritic cells (DC) also contribute to the innate arm of the immune system. ILCs are a heterogeneous group of cells that derive from common lymphoid progenitors but lack rearranged antigen receptor genes. Five classes of ILCs [natural killer (NK) cells, ILC1, ILC2, ILC3, and lymphoid tissue-inducer cells] have been defined based on differences in TF expression and their cytokine production. We refer readers to two recent reviews that comprehensively cover the roles of NK cells and other ILCs in cancer (60, 61).
DCs represent a heterogeneous family of immune cells that bridge innate and adaptive immunity, as impaired DC activation, licensing, or maturation also compromises antigen-specific T-cell immunity. The importance of DC biology in antitumor immunity has gained attention recently. DC-based antitumor vaccines have been FDA-approved for treating prostate cancer, while similar and other approaches are begin studied and assessed in clinical trials. Given space constraints, we refer readers to two recent reviews that cover this growing field (62, 63).
Innate Immune Signaling in Cancer
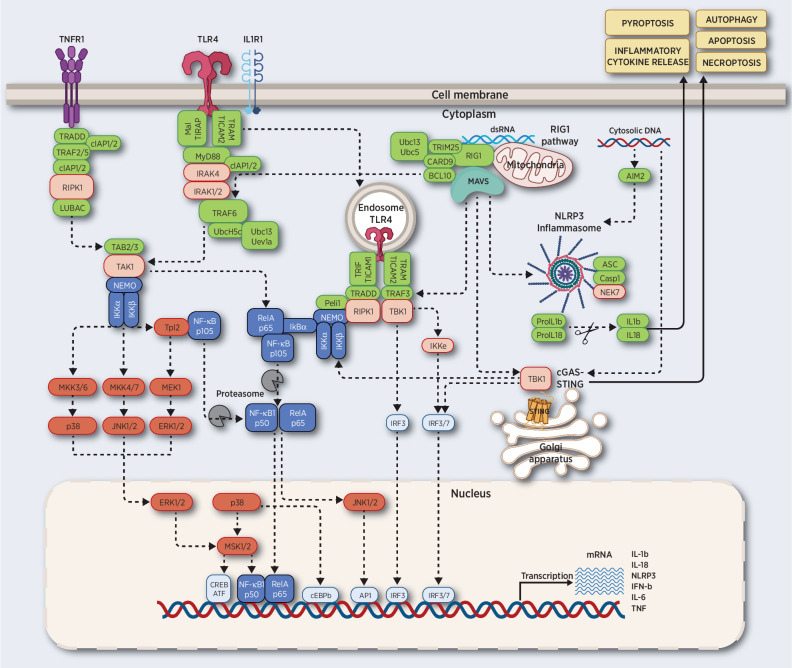
PRRs mediate innate immune signaling and can be classified based on their subcellular localizations: membrane-bound receptors, including Toll-like receptors (TLR) and C-type lectin receptors (CLR), and intracellular receptors, including nucleotide-binding and oligomerization domain (NOD)–like receptors (NLR), AIM2-like receptors (ALR), and RIG-I–like receptors (RLR; ref. 64). Upon recognition of “nonself” antigens, the innate immune system responds by producing cytokines, chemokines and IFNs, and by activating phagocytosis, autophagy, and cell death (Fig. 1).
Figure 1.
Innate immune signaling pathways. TNF binding to TNFR1 triggers the assembly of LUBAC complex, and the activation of TAK1 and subsequently IKK. TLR4, or IL1R1 triggers the interaction of the MyD88-IRAK complex, which engages TRAF6. Activated TLR4 can also be endocytosed and signaled via RIPK1. RIG1 binding to dsRNA promotes its association to MAVS complex, which converges on TRAF6. MAVS also interacts with TRAF3, TBK1, and STING to activate IRF3 and IRF7. NLRP3 triggers secretion of IL1b and IL18.
TLRs
TLRs are expressed in antigen-presenting cells (APC) and other immune cell types including mast cells, NK cells, regulatory T cells, monocytes, neutrophils, and basophils (65). TLRs are also expressed by tumor cells, where they play anti- or protumor roles depending on cell context (66). Upon ligand binding, the TLRs dimerize and signal through different sets of adaptor proteins, including (i) the Toll/IL1 receptor (TIR)-containing adaptor protein (TIRAP) and the protein myeloid differentiation primary response 88 (MyD88; TIRAP/MyD88), (ii) TRAM/TRIF, the TIR domain–containing adaptor-inducing IFNβ (TRIF) and the TRIF-related adaptor molecule (TRAM; TRAM/TRIF), (iii) the mitochondrial antiviral signaling protein (MAVS), and (iv) Apoptosis-associated speck-like proteins containing a caspase recruitment domain (CARD; ASC), which allow for different transcriptional outputs (64).
Intriguingly, TLRs are also expressed by HSPCs. TLR ligand binding blocks HSPC expansion (67) and induces myeloid differentiation in a MyD88-dependent manner (68). The activation of TLRs in short-term HSCs and MPPs also results in substantial cytokine production via activation of NF-κB. Loss of TLR signaling enhances HSC repopulating capacity, as HSCs from Tlr2−/−, Tlr4−/−, Tlr9−/−, or MyD88−/− mice show an advantage over normal HSCs (69, 70). Similarly, TLR activation leads to compromised self-renewal and HSPC exhaustion, mediated by TRIF, rather than MyD88, with the production of ROS and activation of the MAPK p38, leading to replication stress (71, 72). These findings highlight the role of TLR signaling in regulating the behavior of HSPCs, cancer cells, and host defense.
Aberrant TLR signaling is also linked to malignant hematopoiesis. Overexpression of TLR genes in HSCs may contribute to the pathogenesis of myelodysplastic syndrome (MDS; ref. 73). A recent study of 149 patients with MDS showed that TLR1, TLR2, and TLR6 and multiple TLR downstream genes are significantly overexpressed in CD34+ MDS bone marrow cells (74). TLR signaling is promoted by several bromodomain and extraterminal (BET) proteins, which bind to acetylated proteins including histones H3 and H4, and promote tumor growth in several lymphoma models (75). BET inhibitors suppress the expression of TLR2/4 and the transcription of IL1β, IL6, and TNFα and thus may have antitumor activity (76).
NLRs and the inflammasome
NLRs are cytoplasmic receptors that mediate the innate immune response (77). Members of the NLR family of PRRs possess a C-terminal leucine-rich repeat (LRR) domain, a central nucleotide binding domain (NBD), and a distinct N-terminal domain that differs between subfamilies (78). The two most prominent NLR subfamilies have either a pyrin domain (PYD) at their N-terminus, or one or more CARDs (79). Upon sensing pathogens or host derived proteins, NLRs oligomerize and assemble the inflammasome, which serves as a caspase-1–activating scaffold to activate the proinflammatory IL1 family of cytokines, IL1β and IL18, triggering a specific type of inflammatory cell death, termed pyroptosis (80). NLR-inflammasome pathways have been linked to both solid tumors and hematologic malignancies (81, 82).
Priming signals are required for the expression of inflammasome components, which can be induced by TLR ligands, or via the TNF or IL1β signaling pathways that lead to NF-κB activation (83). Hypomethylation of CpG sites within other inflammasome genes such as NLRC4 and NLRP12, and IL1β has been associated with upregulation of their expression in Kawasaki disease (84). Furthermore, increased expression of C-terminal–binding proteins (CtBP), together with p300 and AP-1, activates NLRP3 expression, which aggravates the inflammatory response in osteoarthritis (85). HDAC6, inhibits the activation of the NLRP3 inflammasome in mouse bone marrow–derived macrophages by directly associating with ubiquitinated NLRP3 through its ubiquitin-binding domain (86). Inflammasomes also regulate hematopoiesis and the generation of innate immune cells; loss of inflammasome components or caspase-1 inhibition inhibits myelopoiesis, in a GATA1-dependent manner (87). The impact of cross-talk between epigenetic modifiers and the various NLR-inflammasome pathways is largely unknown in specific cancers.
CLRs, RLRs, and AIM2-like receptors
CLRs, characterized by C-type lectin-like domains, form a large heterogeneous group of transmembrane and soluble receptors (88). The roles of CLRs in immunity and cancer are being delineated and were recently reviewed (89, 90). However, little is known about the epigenetic regulation of CLR generation or function. The TF NFATc2 activates the expression of specific cytokines and chemokines in DCs in response to CLR dectin-1 stimulation, and induces the H3K4 trimethylation that is associated with enhanced gene expression (91). Manipulation, i.e., a decrease in the activity of the histone lysine-methyltransferase Ezh2 increases the generation of IL15R(+) CD122(+) NK precursors and mature NK progeny from mouse and human HSPCs (92). The enhanced NK cell expansion and cytotoxicity against tumor cells are associated with the upregulation of both CD122 and the CLR NKG2D (92).
RLRs induce the transcription of type I IFNs and other genes by sensing viral and host-derived RNAs. The regulatory mechanisms that control the generation of RLRs, their roles in viral infection, autoimmunity, and cancer, and their therapeutic potential, have recently been reviewed (93). ALRs represent a newly recognized class of PRRs that function in cytosolic and nuclear pathogen DNA sensing. ALRs recruit ASC and caspase-1 to form inflammasomes, which elicit inflammatory responses by producing IL1β and IL18, and by triggering pyroptosis (94). The epigenetic regulation of ALRs is yet to be studied.
Trained Immunity in Cancer
Transcriptional and epigenetic reprogramming
The molecular basis of trained immunity is only partially understood, but it is clearly regulated by transcriptional and epigenetic reprogramming, involving chromatin organization at the level of the topologically associated domains (TAD), long noncoding RNAs, and reprogramming of cellular metabolism (Fig. 2; refs. 95, 96). Trained immunity occurs in part via the epigenetic regulation of the monocyte-to-macrophage differentiation transition, where roughly equal numbers of promoters are turned on or off (97). Furthermore, DNA methylation patterns in peripheral blood mononuclear cells can also reflect their capability of undergoing trained immunity (98, 99). During adaptive NK cell responses, specific TFs promote permissive histone modifications and chromatin accessibility, including RUNX family members, STAT family members, IRF8, IRF9, KLF12, and T-box TFs (100–105).
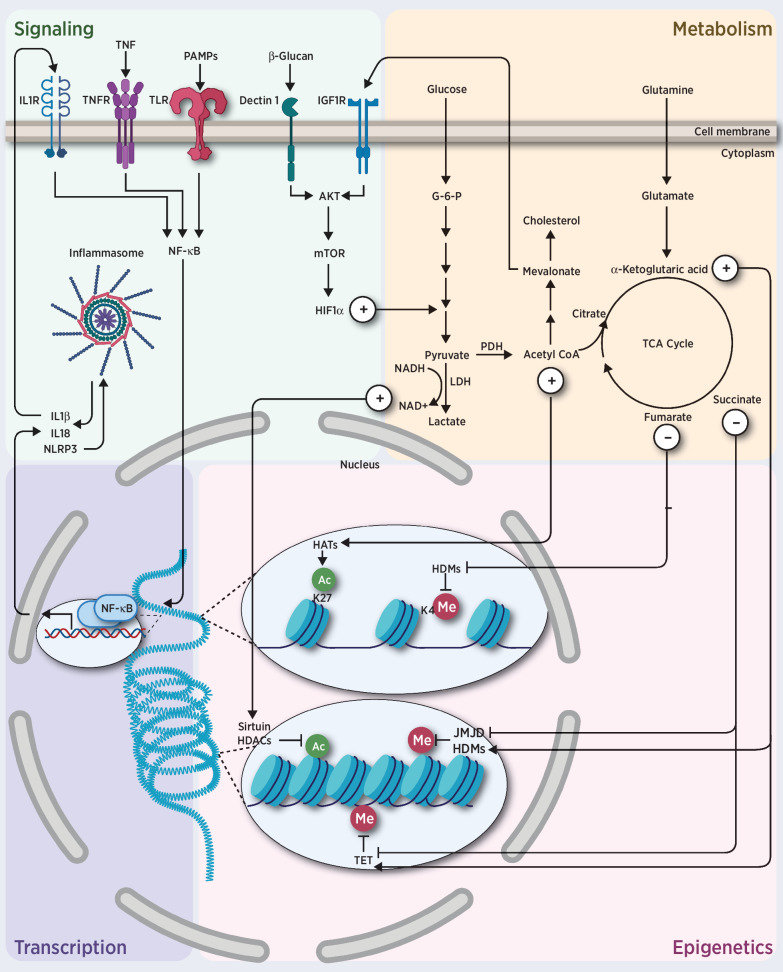
Figure 2.
Trained immunity. Challenge of innate immune cells and HSPCs with training stimuli induces changes in cell signaling and metabolism. Specific TFs, such as NF-κB or AP-1, and epigenetic enzymes, induce chromatin and DNA modifications, and regulate gene transcription. Expression of these genes in turn feeds the machinery of innate immunity and promotes cytokine secretion.
Trained immunity also occurs within HSPCs. For example, exposure to β-glucans, which are fungal cell wall polysaccharides, promotes the expansion of myeloid progenitors and increases innate immune signaling in mice (106). Likewise, Bacille Calmette-Guérin (BCG), an attenuated version of Mycobacterium bovis, increases the chromatin openness of specific TADs in mouse HSCs (107). Intriguingly, inflammasomes promote trained immunity at the level of HSPCs. Transcriptomic and epigenomic reprogramming induced by a high-fat diet is dependent on NLRP3 (108). Recently, trained immunity that occurs in HSPCs is termed central trained immunity, which in part explains the long-lasting phenotype of trained immunity (96).
Immunometabolism and inflammaging
Metabolic intermediates can function as signaling nodes, substrates, cofactors, or inhibitors for chromatin-modifying enzymes (109). Trained monocytes show increased glycolysis, which is dependent on activation of mTOR through the dectin-1/Akt/HIF1α pathway (110). Subsequent studies show that glycolysis, glutaminolysis, and the cholesterol synthesis pathway are essential for the induction of trained immunity in monocytes (111, 112). Accumulation of fumarate induces the epigenetic reprogramming of monocytes by inhibiting KDM5 histone demethylases (111), while mevalonate, an intermediate in the cholesterol synthesis pathway, contributes to the training of human monocytes, via activation of IGF1R and mTOR and via subsequent histone modifications in inflammatory pathways (113). These findings indicate that rewiring of cellular metabolism toward aerobic glycolysis and cholesterol synthesis may be integral to trained immunity.
How metabolic changes affect trained immunity in cancer is not well characterized. However, chronic, sterile, low-grade inflammation, termed inflammaging, has been studied in age-related diseases, including cancer (114). Clonal hematopoiesis of indeterminate potential (CHIP), more common in the elderly, is associated with an increased risk of developing myeloid malignancies and atherosclerosis (115, 116). Individuals with CHIP frequently have mutations in epigenetic modifiers such as DNMT3A, TET2, or ASXL1. Intriguingly, Tet2-deficient HSPCs demonstrate clonal expansion and accelerate atherosclerosis in mice, in a NLRP3 inflammasome/IL1β pathway-dependent manner (117). Circulating monocytes of patients with atherosclerosis exhibit enhanced cytokine production and glycolytic metabolism, with epigenetic reprogramming at the level of histone methylation (118, 119). These findings highlight the role of metabolic and epigenetic changes in aging and age-related diseases. Further studies are needed to dissect the links between metabolic and epigenetic remodeling, trained immunity, aging, and cancer.
Targeting Innate Immunity: Next Generation of Cancer Immunotherapy
The current, FDA-approved immunotherapies for cancer rely largely on the adaptive immune system. However, advances in our understanding of innate immunity provide a novel framework for targeting this aspect of immune homeostasis to maintain health and prevent disease (Table 1). β-glucans and BCG have been evaluated in a variety of cancers, including neuroblastoma, bladder cancer, and lung cancer (120, 121). The protective effect of BCG vaccine relies on trained immunity, specifically the epigenetic reprogramming of monocytes at the level of H3K4m3 (122). Similarly, β-glucans induce trained immunity at the HSPC level and promote myeloid differentiation, innate immune signaling, and metabolic adaptations (106). β-glucans, given in combination with cetuximab in patients with KRAS-mutant colorectal cancer, have promising clinical activity in a phase II trial (123). While increases in the histone marks H3K4me3 and H3K9me3 underlie both BCG-induced and β-glucan–induced trained immunity (111, 112, 124), how this control trained immunity is unclear.
Table 1.
Immunotherapies in cancer.
| Drugs | Targets | Effect on epigenetics or biology |
|---|---|---|
| Targeting trained immunity | ||
| BCG vaccine | Mycobacterium tuberculosis | H3K4 trimethylation of monocytes |
| B-glucans | Dectin-1 and complement receptor 3 (CR3) | H3K4 and H3K9 trimethylation |
| MDP | NOD2 | Activate NF-κB pathway |
| Statins | Mevalonate | Change DNA methylation and prevent trained immunity induction |
| B-hydroxybutyrate and MCC950 | NLRP3 | Inhibit NLRP3 inflammasome-mediated trained immunity |
| Targeting innate immune signaling | ||
| ODNs | TLR9 | Active innate immune responses by producing proinflammation cytokines and Th1 cells |
| Imiquimod | TLR7 | Reverse local immunosuppression and induce tumor cell specific apoptosis |
| poly(I:C) | TLR9 | Induce stable maturation of functionally active dendritic cells |
| TLR3 | Induce cancer cell apoptosis | |
| Flagellin-protein fusions | TLR5 | Induces inflammatory responses through the activation of APCs |
| IRAK1/4 inhibitor I | IRAK1/4 | Suppresses solid tumor growth in several distinct combination therapies |
| NCGC1481 | FLT3-IRAK1/4 | Eliminates adaptive resistance of FLT3-mutant AML cells |
| NG25 | TAK1 | Inhibits colorectal cancer cell proliferation, especially for KRAS-mutant cells |
| 5Z-7-oxozeaenol | TAK1 | Enhances the sensitivity of glioblastoma cells to chemotherapy |
| Suppresses triple-negative breast cancer metastasis by altering TAK1-p38 signaling | ||
| C-178, C-176 | STING | Inhibits STING palmitoylation and attenuates autoinflammatory features in mice |
| NCGC00138783, Pep-20, etc. | CD47/SIRPα | Small-molecule inhibitors targeting macrophage checkpoints that induce phagocytosis |
Abbreviations: MDP, muramyl dipeptide; ODN, CpG oligo-deoxynucleotides.
The synthetic peptide conjugate muramyl dipeptide (MDP), a peptidoglycan minimal bioactive motif common to all bacteria, activates innate immune cells through NOD2, activating NF-κB and inducing epigenetic rewiring and trained immunity (125). The ketone metabolite β-hydroxybutyrate and the small-molecule inhibitor MCC950 can inhibit NLRP3 inflammasome-mediated trained immunity (108, 126, 127). Combinatorial use of epigenetic drugs with immunotherapy is also being investigated. For example, DNMT inhibitor 5-azacytidine triggers immune response via double-stranded RNA (dsRNA) sensing pathway, sensitizing melanoma cells to anti-CTLA4 therapy (128).
TLR agonists are being exploited as adjuvants for cancer vaccines as well as direct cancer therapeutics. Imiquimod binds to TLR7 to reverse local immunosuppression and induce skin cancer cell apoptosis (129). Flagellin fusion proteins can induce specific immune responses, mediated by TLR5 activation on target APCs (130). CpG oligo-deoxynucleotides, which are TLR9 agonists, have shown promising results as vaccine adjuvants and when used in combination with cancer immunotherapy (131). Polyriboinosinic-polyribocytidylic acid [poly(I:C)], which targets TLR3 and TLR9, can boost immune system activation and promote anticancer effects (132).
Given the established track record of kinase inhibitors being approved to treat cancer, and other disorders, inhibitors of the kinases involved in innate immunity are being studied. IRAK functions downstream of TLRs and IL1R to regulate the expression of inflammatory molecules. These kinases play a protumor roles in several cancers (133): For example, inhibition of IRAK1/4 synergizes with sorafenib in suppressing the growth of hepatocellular carcinoma in a xenograft mouse model (134), while the combination of IRAK1/4 inhibition and lenvatinib decreases tumor volume in a mouse anaplastic thyroid cancer model, better than either therapy alone (135). A multikinase FLT3-IRAK1/4 inhibitor displays superior efficacy, compared with current FLT3-targeted therapies to eliminate adaptive resistance of FLT3-mutant acute myeloid leukemia (AML; ref. 136). Other kinase inhibitors, such as those targeting TAK1 kinase, which is downstream of TLR and TNFR pathways, also show therapeutic efficacy in various cancer models (137–139). Taken together, kinase inhibitors and other drugs that target aspects of innate immunity, such as the cGAS-STING pathway or macrophage checkpoints (CD47/SIRPα axis) show important promise. Several of these approaches have recently been reviewed (140, 141).
Perspectives
The study of innate immunity in cancer is fast growing and drugs that target protumorigenic cellular infiltration or inflammation are being tested preclinically and clinically. However, multiple challenges exist, given the complexity of the interactions between tumor cells and their environment and the importance of targeting only certain aspects of the immune system, without impairing others. On the molecular level, a better understanding of the mechanistic details underlying the intricate interactions between cancer biology, innate immunity signaling, and inflammation is needed. Clearly, the effects of feedback and compensatory pathways on tumor growth are hard to predict or control, despite the research advances we have outlined above. On the cellular level, the heterogeneity and plasticity of the immune cells that infiltrate tumors must be taken into account in cancer to target the biology of a specific immune cell type or the interactions between cell types. Also, the roles that aging or the microbiome have on the outcome of anti-inflammatory therapies remain to be characterized (142).
Notably, trained immunity provides a compelling layer of control on myeloid cell function by integrating epigenetic, transcriptional, and metabolic regulations, although the precise mechanisms are just beginning to be discovered. The presence of persistent epigenetic marks in trained innate immune cells, generated following a pathologic process or during aging, could underlie an increased susceptibility to certain cancer events. Innate immunity can be therapeutically manipulated, at the level of epigenetic modifiers, TFs, cellular metabolism, and signaling pathways. Moreover, mediators of innate immunity such as IFNs and cytokines can enhance adaptive immunity-based therapy by sensitizing tumor cells (143, 144). Mechanistic studies of innate immunity regulation in cancer are underway, which will help lay the groundwork for the development of innate immunity-based mono- or combination therapies.
Authors' Disclosures
No disclosures were reported.
References
- 1. Gasteiger G, D'Osualdo A, Schubert DA, Weber A, Bruscia EM, Hartl D. Cellular innate immunity: an old game with new players. J Innate Immun 2017;9:111–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nahrendorf M. Myeloid cell contributions to cardiovascular health and disease. Nat Med 2018;24:711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe 2011;9:355–61. [DOI] [PubMed] [Google Scholar]
- 4. Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009;458:904–8. [DOI] [PubMed] [Google Scholar]
- 5. Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med 2009;15:696–700. [DOI] [PubMed] [Google Scholar]
- 6. Cabezas-Wallscheid N, Buettner F, Sommerkamp P, Klimmeck D, Ladel L, Thalheimer FB, et al. Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell 2017;169:807–23. [DOI] [PubMed] [Google Scholar]
- 7. Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 2018;48:688–701. [DOI] [PubMed] [Google Scholar]
- 8. Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 2010;465:793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Bruin AM, Demirel O, Hooibrink B, Brandts CH, Nolte MA. Interferon-gamma impairs proliferation of hematopoietic stem cells in mice. Blood 2013;121:3578–85. [DOI] [PubMed] [Google Scholar]
- 10. Matatall KA, Jeong M, Chen S, Sun D, Chen F, Mo Q, et al. Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep 2016;17:2584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Etzrodt M, Ahmed N, Hoppe PS, Loeffler D, Skylaki S, Hilsenbeck O, et al. Inflammatory signals directly instruct PU.1 in HSCs via TNF. Blood 2019;133:816–9. [DOI] [PubMed] [Google Scholar]
- 12. Weber GF, Chousterman BG, He S, Fenn AM, Nairz M, Anzai A, et al. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science 2015;347:1260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao JL, Ma C, O'Connell RM, Mehta A, DiLoreto R, Heath JR, et al. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell 2014;14:445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furusawa J, Mizoguchi I, Chiba Y, Hisada M, Kobayashi F, Yoshida H, et al. Promotion of expansion and differentiation of hematopoietic stem cells by interleukin-27 into myeloid progenitors to control infection in emergency myelopoiesis. PLoS Pathog 2016;12:e1005507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol 2016;18:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR, et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol 2001;21:3789–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bartels M, Govers AM, Fleskens V, Lourenço AR, Pals CE, Vervoort SJ, et al. Acetylation of C/EBPε is a prerequisite for terminal neutrophil differentiation. Blood 2015;125:1782–92. [DOI] [PubMed] [Google Scholar]
- 18. Fischer J, Walter C, Tönges A, Aleth H, Jordão MJC, Leddin M, et al. Safeguard function of PU.1 shapes the inflammatory epigenome of neutrophils. Nat Immunol 2019;20:546–58. [DOI] [PubMed] [Google Scholar]
- 19. Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003;3:859–68. [DOI] [PubMed] [Google Scholar]
- 20. Kawamura T, Ogawa Y, Aoki R, Shimada S. Innate and intrinsic antiviral immunity in skin. J Dermatol Sci 2014;75:159–66. [DOI] [PubMed] [Google Scholar]
- 21. Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol 2019;16:601–20. [DOI] [PubMed] [Google Scholar]
- 22. Sionov RV, Fridlender ZG, Granot Z. The multifaceted roles neutrophils play in the tumor microenvironment. Cancer Microenviron 2015;8:125–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fridlender ZG, Albelda SM. Tumor-associated neutrophils: friend or foe? Carcinogenesis 2012;33:949–55. [DOI] [PubMed] [Google Scholar]
- 24. Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell 2009;16:183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jablonska J, Leschner S, Westphal K, Lienenklaus S, Weiss S. Neutrophils responsive to endogenous IFN-beta regulate tumor angiogenesis and growth in a mouse tumor model. J Clin Invest 2010;120:1151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol 2012;13:616–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mogensen TH. IRF and STAT transcription factors - from basic biology to roles in infection, protective immunity, and primary immunodeficiencies. Front Immunol 2018;9:3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496:445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 2016;44:755–68. [DOI] [PubMed] [Google Scholar]
- 30. Hume DA. Macrophages as APC and the dendritic cell myth. J Immunol 2008;181:5829–35. [DOI] [PubMed] [Google Scholar]
- 31. Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science 2009;326:867–71. [DOI] [PubMed] [Google Scholar]
- 32. Li T, Garcia-Gomez A, Morante-Palacios O, Ciudad L, Ozkaramehmet S, Van Dijck E, et al. SIRT1/2 orchestrate acquisition of DNA methylation and loss of histone H3 activating marks to prevent premature activation of inflammatory genes in macrophages. Nucleic Acids Res 2020;48:665–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang N, Wang S, Wang X, Zheng Y, Yang B, Zhang J, et al. Research trends in pharmacological modulation of tumor-associated macrophages. Clin Transl Med 2021;11:e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012;122:787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001;410:1107–11. [DOI] [PubMed] [Google Scholar]
- 36. Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 2002;3:651–62. [DOI] [PubMed] [Google Scholar]
- 37. Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls type I and type II IFN responses. Front Immunol 2018;9:1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014;41:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seim GL, Britt EC, John SV, Yeo FJ, Johnson AR, Eisenstein RS, et al. Two-stage metabolic remodelling in macrophages in response to lipopolysaccharide and interferon-gamma stimulation. Nat Metab 2019;1:731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mills CD, Lenz LL, Harris RA. A breakthrough: macrophage-directed cancer immunotherapy. Cancer Res 2016;76:513–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 2009;114:3244–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010;11:936–44. [DOI] [PubMed] [Google Scholar]
- 43. Liu PS, Wang H, Li X, Chao T, Teav T, Christen S, et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 2017;18:985–94. [DOI] [PubMed] [Google Scholar]
- 44. Deng M, Wang J, Wu H, Wang M, Cao D, Li J, et al. IL-4 alleviates ischaemia-reperfusion injury by inducing kupffer cells M2 polarization via STAT6-JMJD3 pathway after rat liver transplantation. Biomed Res Int 2020;2020:2953068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008;8:958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol 2016;17:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, et al. The terminology issue for myeloid-derived suppressor cells. Cancer Res 2007;67:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Condamine T, Mastio J, Gabrilovich DI. Transcriptional regulation of myeloid-derived suppressor cells. J Leukoc Biol 2015;98:913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu L, Yan C, Czader M, Foreman O, Blum JS, Kapur R, et al. Inhibition of PPARγ in myeloid-lineage cells induces systemic inflammation, immunosuppression, and tumorigenesis. Blood 2012;119:115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deng J, Li J, Sarde A, Lines JL, Lee YC, Qian DC, et al. Hypoxia-induced VISTA promotes the suppressive function of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Immunol Res 2019;7:1079–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rodriguez-Ubreva J, Catala-Moll F, Obermajer N, Alvarez-Errico D, Ramirez RN, Company C, et al. Prostaglandin E2 leads to the acquisition of DNMT3A-dependent tolerogenic functions in human myeloid-derived suppressor cells. Cell Rep 2017;21:154–67. [DOI] [PubMed] [Google Scholar]
- 52. van Deventer HW, Burgents JE, Wu QP, Woodford RM, Brickey WJ, Allen IC, et al. The inflammasome component NLRP3 impairs antitumor vaccine by enhancing the accumulation of tumor-associated myeloid-derived suppressor cells. Cancer Res 2010;70:10161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chow MT, Sceneay J, Paget C, Wong CS, Duret H, Tschopp J, et al. NLRP3 suppresses NK cell-mediated responses to carcinogen-induced tumors and metastases. Cancer Res 2012;72:5721–32. [DOI] [PubMed] [Google Scholar]
- 54. Millrud CR, Bergenfelz C, Leandersson K. On the origin of myeloid-derived suppressor cells. Oncotarget 2017;8:3649–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schouppe E, Mommer C, Movahedi K, Laoui D, Morias Y, Gysemans C, et al. Tumor-induced myeloid-derived suppressor cell subsets exert either inhibitory or stimulatory effects on distinct CD8+ T-cell activation events. Eur J Immunol 2013;43:2930–42. [DOI] [PubMed] [Google Scholar]
- 56. Bergenfelz C, Leandersson K. The generation and identity of human myeloid-derived suppressor cells. Front Oncol 2020;10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Singel KL, Emmons TR, Khan ANH, Mayor PC, Shen S, Wong JT, et al. Mature neutrophils suppress T cell immunity in ovarian cancer microenvironment. JCI Insight 2019;4:e122311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med 2008;205:2235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Netherby CS, Messmer MN, Burkard-Mandel L, Colligan S, Miller A, Cortes Gomez E, et al. The granulocyte progenitor stage is a key target of IRF8-mediated regulation of myeloid-derived suppressor cell production. J Immunol 2017;198:4129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huntington ND, Cursons J, Rautela J. The cancer-natural killer cell immunity cycle. Nat Rev Cancer 2020;20:437–54. [DOI] [PubMed] [Google Scholar]
- 61. Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol 2018;18:671–88. [DOI] [PubMed] [Google Scholar]
- 62. Marciscano AE, Anandasabapathy N. The role of dendritic cells in cancer and anti-tumor immunity. Semin Immunol 2021;52:101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mastelic-Gavillet B, Balint K, Boudousquie C, Gannon PO, Kandalaft LE. Personalized dendritic cell vaccines-recent breakthroughs and encouraging clinical results. Front Immunol 2019;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 2015;33:257–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hopkins PA, Sriskandan S. Mammalian Toll-like receptors: to immunity and beyond. Clin Exp Immunol 2005;140:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dajon M, Iribarren K, Cremer I. Toll-like receptor stimulation in cancer: a pro- and anti-tumor double-edged sword. Immunobiology 2017;222:89–100. [DOI] [PubMed] [Google Scholar]
- 67. Rodriguez S, Chora A, Goumnerov B, Mumaw C, Goebel WS, Fernandez L, et al. Dysfunctional expansion of hematopoietic stem cells and block of myeloid differentiation in lethal sepsis. Blood 2009;114:4064–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, Akira S, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity 2006;24:801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ichii M, Shimazu T, Welner RS, Garrett KP, Zhang Q, Esplin BL, et al. Functional diversity of stem and progenitor cells with B-lymphopoietic potential. Immunol Rev 2010;237:10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schuettpelz LG, Borgerding JN, Christopher MJ, Gopalan PK, Romine MP, Herman AC, et al. G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia 2014;28:1851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang H, Rodriguez S, Wang L, Wang S, Serezani H, Kapur R, et al. Sepsis induces hematopoietic stem cell exhaustion and myelosuppression through distinct contributions of TRIF and MYD88. Stem Cell Reports 2016;6:940–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Takizawa H, Fritsch K, Kovtonyuk LV, Saito Y, Yakkala C, Jacobs K, et al. Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell 2017;21:225–40. [DOI] [PubMed] [Google Scholar]
- 73. Varney ME, Melgar K, Niederkorn M, Smith M, Barreyro L, Starczynowski DT. Deconstructing innate immune signaling in myelodysplastic syndromes. Exp Hematol 2015;43:587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wei Y, Dimicoli S, Bueso-Ramos C, Chen R, Yang H, Neuberg D, et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia 2013;27:1832–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bernasconi E, Gaudio E, Lejeune P, Tarantelli C, Cascione L, Kwee I, et al. Preclinical evaluation of the BET bromodomain inhibitor BAY 1238097 for the treatment of lymphoma. Br J Haematol 2017;178:936–48. [DOI] [PubMed] [Google Scholar]
- 76. Meng S, Zhang L, Tang Y, Tu Q, Zheng L, Yu L, et al. BET inhibitor JQ1 blocks inflammation and bone destruction. J Dent Res 2014;93:657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010;140:805–20. [DOI] [PubMed] [Google Scholar]
- 78. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell 2014;157:1013–22. [DOI] [PubMed] [Google Scholar]
- 79. Platnich JM, Muruve DA. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys 2019;670:4–14. [DOI] [PubMed] [Google Scholar]
- 80. Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev 2015;265:6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Karki R, Man SM, Kanneganti TD. Inflammasomes and cancer. Cancer Immunol Res 2017;5:94–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ratajczak MZ, Bujko K, Cymer M, Thapa A, Adamiak M, Ratajczak J, et al. The Nlrp3 inflammasome as a "rising star" in studies of normal and malignant hematopoiesis. Leukemia 2020;34:1512–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 2019;19:477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Huang YH, Lo MH, Cai XY, Kuo HC. Epigenetic hypomethylation and upregulation of NLRC4 and NLRP12 in Kawasaki disease. Oncotarget 2018;9:18939–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sun X, Xiao L, Chen J, Chen X, Chen X, Yao S, et al. DNA methylation is involved in the pathogenesis of osteoarthritis by regulating CtBP expression and CtBP-mediated signaling. Int J Biol Sci 2020;16:994–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hwang I, Lee E, Jeon SA, Yu JW. Histone deacetylase 6 negatively regulates NLRP3 inflammasome activation. Biochem Biophys Res Commun 2015;467:973–8. [DOI] [PubMed] [Google Scholar]
- 87. Tyrkalska SD, Perez-Oliva AB, Rodriguez-Ruiz L, Martinez-Morcillo FJ, Alcaraz-Perez F, Martinez-Navarro FJ, et al. Inflammasome regulates hematopoiesis through cleavage of the master erythroid transcription factor GATA1. Immunity 2019;51:50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J 2005;272:6179–217. [DOI] [PubMed] [Google Scholar]
- 89. Ganguly K, Kishore U, Madan T. Interplay between C-type lectin receptors and microRNAs in cellular homeostasis and immune response. Febs j 2021;288:4210–29. [DOI] [PubMed] [Google Scholar]
- 90. Brown GD, Willment JA, Whitehead L. C-type lectins in immunity and homeostasis. Nat Rev Immunol 2018;18:374–89. [DOI] [PubMed] [Google Scholar]
- 91. Yu HB, Yurieva M, Balachander A, Foo I, Leong X, Zelante T, et al. NFATc2 mediates epigenetic modification of dendritic cell cytokine and chemokine responses to dectin-1 stimulation. Nucleic Acids Res 2015;43:836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yin J, Leavenworth JW, Li Y, Luo Q, Xie H, Liu X, et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci U S A 2015;112:15988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 2020;20:537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Caneparo V, Landolfo S, Gariglio M, De Andrea M. The absent in melanoma 2-like receptor IFN-inducible protein 16 as an inflammasome regulator in systemic lupus erythematosus: the dark side of sensing microbes. Front Immunol 2018;9:1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, et al. Trained immunity: a program of innate immune memory in health and disease. Science 2016;352:aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Netea MG, Dominguez-Andres J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol 2020;20:375–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014;345:1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Verma D, Parasa VR, Raffetseder J, Martis M, Mehta RB, Netea M, et al. Anti-mycobacterial activity correlates with altered DNA methylation pattern in immune cells from BCG-vaccinated subjects. Sci Rep 2017;7:12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Das J, Verma D, Gustafsson M, Lerm M. Identification of DNA methylation patterns predisposing for an efficient response to BCG vaccination in healthy BCG-naive subjects. Epigenetics 2019;14:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, Sun JC. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J Exp Med 2016;213:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rapp M, Lau CM, Adams NM, Weizman OE, O'Sullivan TE, Geary CD, et al. Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Sci Immunol 2017;2:eaan3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Geary CD, Krishna C, Lau CM, Adams NM, Gearty SV, Pritykin Y, et al. Non-redundant ISGF3 components promote NK cell survival in an auto-regulatory manner during viral infection. Cell Rep 2018;24:1949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lam VC, Folkersen L, Aguilar OA, Lanier LL. KLF12 regulates mouse NK cell proliferation. J Immunol 2019;203:981–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Adams NM, Lau CM, Fan X, Rapp M, Geary CD, Weizman OE, et al. Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity 2018;48:1172–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Madera S, Geary CD, Lau CM, Pikovskaya O, Reiner SL, Sun JC. Cutting edge: divergent requirement of T-box transcription factors in effector and memory NK cells. J Immunol 2018;200:1977–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Mitroulis I, Ruppova K, Wang B, Chen LS, Grzybek M, Grinenko T, et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 2018;172:147–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kaufmann E, Sanz J, Dunn JL, Khan N, Mendonca LE, Pacis A, et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 2018;172:176–90. [DOI] [PubMed] [Google Scholar]
- 108. Christ A, Gunther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, et al. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 2018;172:162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Donohoe DR, Bultman SJ. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J Cell Physiol 2012;227:3169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014;345:1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Arts RJ, Novakovic B, Ter Horst R, Carvalho A, Bekkering S, Lachmandas E, et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab 2016;24:807–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Arts RJW, Carvalho A, La Rocca C, Palma C, Rodrigues F, Silvestre R, et al. Immunometabolic pathways in BCG-induced trained immunity. Cell Rep 2016;17:2562–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden C, Li Y, et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell 2018;172:135–46. [DOI] [PubMed] [Google Scholar]
- 114. Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 2018;14:576–90. [DOI] [PubMed] [Google Scholar]
- 115. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017;355:842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bekkering S, van den Munckhof I, Nielen T, Lamfers E, Dinarello C, Rutten J, et al. Innate immune cell activation and epigenetic remodeling in symptomatic and asymptomatic atherosclerosis in humans in vivo. Atherosclerosis 2016;254:228–36. [DOI] [PubMed] [Google Scholar]
- 119. Shirai T, Nazarewicz RR, Wallis BB, Yanes RE, Watanabe R, Hilhorst M, et al. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J Exp Med 2016;213:337–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pettenati C, Ingersoll MA. Mechanisms of BCG immunotherapy and its outlook for bladder cancer. Nat Rev Urol 2018;15:615–25. [DOI] [PubMed] [Google Scholar]
- 121. Mulder WJM, Ochando J, Joosten LAB, Fayad ZA, Netea MG. Therapeutic targeting of trained immunity. Nat Rev Drug Discov 2019;18:553–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Buffen K, Oosting M, Quintin J, Ng A, Kleinnijenhuis J, Kumar V, et al. Autophagy controls BCG-induced trained immunity and the response to intravesical BCG therapy for bladder cancer. PLoS Pathog 2014;10:e1004485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Segal NH, Gada P, Senzer N, Gargano MA, Patchen ML, Saltz LB. A phase II efficacy and safety, open-label, multicenter study of imprime PGG injection in combination with cetuximab in patients with stage IV KRAS-mutant colorectal cancer. Clin Colorectal Cancer 2016;15:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A 2012;109:17537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 2003;278:8869–72. [DOI] [PubMed] [Google Scholar]
- 126. Youm YH, Nguyen KY, Grant RW, Goldberg EL, Bodogai M, Kim D, et al. The ketone metabolite beta-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat Med 2015;21:263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med 2015;21:248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015;162:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Schön M, Bong AB, Drewniok C, Herz J, Geilen CC, Reifenberger J, et al. Tumor-selective induction of apoptosis and the small-molecule immune response modifier imiquimod. J Natl Cancer Inst 2003;95:1138–49. [DOI] [PubMed] [Google Scholar]
- 130. Cuadros C, Lopez-Hernandez FJ, Dominguez AL, McClelland M, Lustgarten J. Flagellin fusion proteins as adjuvants or vaccines induce specific immune responses. Infect Immun 2004;72:2810–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000;408:740–5. [DOI] [PubMed] [Google Scholar]
- 132. Bianchi F, Pretto S, Tagliabue E, Balsari A, Sfondrini L. Exploiting poly(I:C) to induce cancer cell apoptosis. Cancer Biol Ther 2017;18:747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jain A, Kaczanowska S, Davila E. IL-1 receptor-associated kinase signaling and its role in inflammation, cancer progression, and therapy resistance. Front Immunol 2014;5:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cheng BY, Lau EY, Leung HW, Leung CO, Ho NP, Gurung S, et al. IRAK1 augments cancer stemness and drug resistance via the AP-1/AKR1B10 signaling cascade in hepatocellular carcinoma. Cancer Res 2018;78:2332–42. [DOI] [PubMed] [Google Scholar]
- 135. Kawamura Y, Saijo K, Imai H, Ishioka C. Inhibition of IRAK1/4 enhances the antitumor effect of lenvatinib in anaplastic thyroid cancer cells. Cancer Sci 2021;112:4711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Melgar K, Walker MM, Jones LM, Bolanos LC, Hueneman K, Wunderlich M, et al. Overcoming adaptive therapy resistance in AML by targeting immune response pathways. Sci Transl Med 2019;11:eaaw8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ma Q, Gu L, Liao S, Zheng Y, Zhang S, Cao Y, et al. NG25, a novel inhibitor of TAK1, suppresses KRAS-mutant colorectal cancer growth in vitro and in vivo. Apoptosis 2019;24:83–94. [DOI] [PubMed] [Google Scholar]
- 138. Campolo M, Lanza M, Casili G, Paterniti I, Filippone A, Caffo M, et al. TAK1 inhibitor enhances the therapeutic treatment for glioblastoma. Cancers 2020;13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Iriondo O, Liu Y, Lee G, Elhodaky M, Jimenez C, Li L, et al. TAK1 mediates microenvironment-triggered autocrine signals and promotes triple-negative breast cancer lung metastasis. Nat Commun 2018;9:1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jiang M, Chen P, Wang L, Li W, Chen B, Liu Y, et al. cGAS-STING, an important pathway in cancer immunotherapy. J Hematol Oncol 2020;13:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yu WB, Ye ZH, Chen X, Shi JJ, Lu JJ. The development of small-molecule inhibitors targeting CD47. Drug Discov Today 2021;26:561–8. [DOI] [PubMed] [Google Scholar]
- 142. Hou J, Karin M, Sun B. Targeting cancer-promoting inflammation - have anti-inflammatory therapies come of age? Nat Rev Clin Oncol 2021;18:261–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 2017;547:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019;565:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]