Abstract
Tris (1,3-dichloro-2-propyl) phosphate (TDCIPP) is an organophosphate ester-based flame retardant widely used within the United States. Within zebrafish, initiation of TDCIPP exposure at 0.75 h post-fertilization (hpf) reliably disrupts cytosine methylation from cleavage (2 hpf) through early-gastrulation (6 hpf). Therefore, the objective of this study was to determine whether TDCIPP-induced effects on cytosine methylation persist beyond 6 hpf. First, we exposed embryos to vehicle or TDCIPP from 0.75 hpf to 6, 24, or 48 hpf, and then conducted bisulfite amplicon sequencing of a target locus (lmo7b) using genomic DNA derived from whole embryos. Within both vehicle- and TDCIPP-treated embryos, CpG methylation was similar at 6 hpf and CHG/CHH methylation were similar at 24 and 48 hpf (relative to 6 hpf). However, relative to 6 hpf within the same treatment, CpG methylation was lower within vehicle-treated embryos at 48 hpf and TDCIPP-treated embryos at 24 and 48 hpf – an effect that was driven by acceleration of CpG hypomethylation. Similar to our previous findings with DNA methyltransferase, we found that, even at high μM concentrations, TDCIPP had no effect on zebrafish and human thymine DNA glycosylase activity (a key enzyme that decreases CpG methylation), suggesting that TDCIPP-induced effects on CpG methylation are not driven by direct interaction with thymine DNA glycosylase. Finally, using 5-methylcytosine (5-mC)-specific whole-mount immunochemistry and automated imaging, we found that exposure to TDCIPP increased 5-mC abundance within the yolk of blastula-stage embryos, suggesting that TDCIPP may impact cytosine methylation of maternally loaded mRNAs during the maternal-to-zygotic transition. Overall, our findings suggest that TDCIPP disrupts the trajectory of cytosine methylation during zebrafish embryogenesis, effects which do not appear to be driven by direct interaction of TDCIPP with key enzymes that regulate cytosine methylation.
Keywords: Zebrafish, Embryo, TDCIPP, Cytosine methylation
1. Introduction
Tris (1,3-dichloro-2-propyl) phosphate (TDCIPP) is an organophosphate ester-based additive flame retardant and semi-volatile organic compound (SVOC) widely used within the United States (Dishaw et al., 2014; Doherty et al., 2019; Fu et al., 2013; Liu et al., 2016; McGee et al., 2012; Tran et al., 2021). As an SVOC, TDCIPP migrates from end-use products (e.g., polyurethane foam) into the air and partitions into environmental media such as indoor dust. For example, longer commutes are associated with increased human exposure to TDCIPP (Brandsma et al., 2014; Reddam et al., 2020), a finding that may be due to migration of TDCIPP from interior parts of vehicles such as car seats. In a cross-sectional European human biomonitoring study, TDCIPP and bis(1,3-dichloro-2-propyl) phosphate (BDCIPP, the primary metabolite of TDCIPP) were detected within 93% of hair follicle samples from mother-child pairs (Kucharska et al., 2015), suggesting that human exposure to TDCIPP is ubiquitous.
Previous studies using albino mice found that exposure to 10 μM TDCIPP decreased blastocyst formation and exposure to 100 μM TDCIPP was embryonic lethal, indicating that TDCIPP disrupted development of early mouse embryos in a dose-dependent manner (Yin et al., 2019). Within Chinese rare minnows, TDCIPP exposure induced apoptosis and altered multiple pathways related to DNA damage, effects that were due to TDCIPP-induced oxidative DNA lesions (Chen et al., 2019). Moreover, using zebrafish as a model, prior studies within our lab found that initiation of TDCIPP exposure at 0.75 h post fertilization (hpf) reliably disrupted DNA methylation from cleavage (2 hpf) through early-gastrulation (6 hpf) as well as induced epiboly delay or arrest during gastrulation (Dasgupta et al., 2017, 2018, 2019; Kupsco et al., 2017; McGee et al., 2012). However, it’s unclear whether TDCIPP-induced effects on cytosine methylation occur during later stages of embryonic development.
Epigenetic reprogramming occurs during early embryonic development (fertilization through blastocyst formation) and is highly conserved across multiple model organisms (Breton-Larrivée et al., 2019; Efimova et al., 2020; Jessop et al., 2018). Following fertilization, three processes occur during epigenetic reprogramming: 1) demethylation of paternal DNA; 2) degradation of maternally-loaded mRNA; and 3) remethylation of the zygotic genome (Potok et al., 2013; Tadros and Lipshitz, 2009). Regulation of cytosine methylation is critical during the maternal-to-zygotic transition (MZT) in order to activate the zygotic genome (Potok et al., 2013). During epigenetic reprograming, an embryo progresses through a series of methylation and demethylation steps that drive key processes of development such as cell fate, tissue formation, organogenesis, and neuronal development. Once epigenetic reprogramming is complete, zygotic DNA transcription commences and the zygote decreases reliance on maternal mRNA for cellular maintenance and function (Tadros and Lipshitz, 2009). Therefore, epigenetic reprogramming is critical for proper embryonic development, as key developmental landmarks (e.g., organogenesis) are tightly regulated and coordinated by the timing of genome-wide transcription.
Cytosine methylation is highly conserved across mammalian species in long stretches of cytosine-guanine pairs, or CpG islands. Approximately 2–5% of all cytosines are methylated in the mammalian genome, and methylated cytosines are found across the genome in a CpG, CHG, or CHH context (Fang et al., 2013). During epigenetic reprogramming, active methylation and demethylation are driven by enzymatic modification of cytosine into 5-methylcytosine (5-mC) by DNA methyltransferase (DNMT) as well as oxidation of 5-mC into 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine by ten-eleven translocation (TET) enzymes. In addition, demethylase and thymine DNA glycosylase (TDG, coupled with base excision repair) are involved in decreasing 5-mC levels (Jessop et al., 2018). During early stages of development, regulation of cytosine methylation is necessary for the recruitment of transcriptional machinery and ultimately gene expression. Alterations in the normal trajectory of cytosine methylation may disrupt zygotic DNA transcription which, in turn, may adversely affect normal cell adherence, migration, viability, and division within developing embryos (Jones and Takai, 2001).
Using zebrafish as a model, the objective of this study was to determine whether TDCIPP-induced effects on DNA methylation persist beyond 6 hpf as well as whether TDCIPP directly affects zebrafish and human TDG activity. In order to accomplish this objective, we relied on 1) bisulfite amplicon sequencing to test whether TDCIPP-induced alterations in cytosine methylation within a previously identified locus (lmo7b) persisted later in embryonic development; 2) thymine DNA glycosylase (TDG) activity assays to determine whether TDCIPP directly affects zebrafish and human TDG activity; and 3) 5-mC-specific whole-mount immunochemistry and automated imaging to investigate whether TDCIPP affected the abundance of 5-mC in situ within intact embryos.
2. Materials and methods
2.1. Animals
Adult wild-type (5D strain) zebrafish were raised and maintained and bred on a recirculating system using previously described procedures (Mitchell et al., 2018). Adult breeders were handled and treated in accordance with an Institutional Animal Care and Use Committee (IACUC)-approved animal use protocol (#20180063) at the University of California, Riverside.
2.2. Chemicals
TDCIPP (99% purity) was purchased from Chem Service, Inc. (West Chester, PA). Stock solutions of TDCIPP were prepared in high-performance liquid chromatography (HPLC)-grade dimethyl sulfoxide (DMSO) and stored within 2-mL amber glass vials with polytetrafluoroethylene-lined caps. Working solutions (1:1000 dilution of stock solutions) of vehicle (0.1% DMSO) and TDCIPP were prepared in particulate-free water from our recirculating system (pH and conductivity of ~7.2 and ~950 μS, respectively) immediately prior to each experiment.
2.3. Bisulfite amplicon sequencing (BSAS)
Based on our previous findings (Kupsco et al., 2017), we selected a concentration of TDCIPP (0.78 μM) that only induced a ~1.3-h epiboly delay at 6 h post-fertilization (hpf) in the absence of effects on embryo survival. To test whether TDCIPP-induced alterations in cytosine methylation within a previously identified locus (lmo7b) (Dasgupta et al., 2019; Kupsco et al., 2017) persisted later in embryonic development, embryos (50 per replicate dish) were treated with 10 mL of vehicle (0.1% DMSO) or 0.78 μM TDCIPP from 0.75 hpf to 6 hpf, 0.75 hpf to 24 hpf, or 0.75 hpf to 48 hpf in glass petri dishes (eight replicate dishes per treatment). Exposures were conducted under static conditions at 28 °C within a temperature-controlled incubator under a 14-h:10-h light:dark cycle. At 6, 24, and 48 hpf, 100 surviving embryos were pooled from two replicate dishes (4 replicate pools per group), snap-frozen in liquid nitrogen, and stored at −80 °C. .
Genomic DNA (gDNA) was extracted, purified, and bisulfite-treated following previously described protocols (Kupsco et al., 2017). The region of interest (lmo7b) was PCR-amplified and amplicons were purified using previously described protocols (Kupsco et al., 2017). DNA quality was confirmed using an Agilent 2100 Bioanalyzer System (DNA 1000 and High Sensitivity DNA Kits for amplicons and libraries, respectively), and all amplicon and library concentrations were quantified using a Qubit 4.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Amplicons were pooled and libraries were prepared using a Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA), and all libraries were paired-end sequenced on our Illumina MiniSeq Sequencing System using a 300-cycle High-Output Reagent Kit. Raw Illumina (fastq.gz) sequencing files (24 files) are available via NCBI’s BioProject database under BioProject ID PRJNA780909, and a summary of sequencing run metrics is provided within Table S1 (>88.51% of reads were ≥Q30 across all runs). Using previously described protocols (Kupsco et al., 2017), downstream analysis of quality control and methylation differences were identified within Illumina’s BaseSpace. MethylSeq was used to align reads and remove duplicate reads, and MethylKit was used for positions with ≥10X CpG coverage to identify significant methylation differences relative to time-matched vehicle controls; q < 0.01 was used to minimize false positives.
2.4. Integration and analysis of BSAS data
Including this study, we have utilized BSAS across three separate studies to investigate methylation of lmo7b within 2-, 4-, 6-, 24-, or 48-hpf zebrafish embryos following exposure to vehicle (0.1% DMSO), 0.78 μM TDCIPP, or 3.12 μM TDCIPP. Raw Illumina (fastq.gz) sequencing files for all three studies are available via NCBI’s BioProject database under BioProject IDs PRJNA395080, PRJNA553577, and PRJNA780909. Cytosine report files containing methylation metrics and cytosine context (CpG, CHG, or CHH) data at base-pair resolution were obtained from MethylSeq output files for the current study as well as our two prior studies (Dasgupta et al., 2019; Kupsco et al., 2017). For each treatment replicate and timepoint, the <Sample>.CX_report.txt.gz report was downloaded, and raw text files were extracted, indexed, and, depending on the size of the file, trimmed manually or via MATLAB to focus only on positions containing cytosines mapped to Chr1. Trimmed files were then imported into R where package deployer and writexl were used to 1) remove negative strand data, 2) calculate the total number of methylated and unmethylated cytosines (see below for equation), 3) aggregate data by cytosine context, 4) filter out positions where the summation of count methylated and unmethylated were equal to 0, and 5) export output data into Excel files. Data were then combined into a single Excel file and compiled relative to stage (hpf) to be further trimmed by creating a column where the summation between the count methylated and unmethylated could be used as a filter. Only positions with ≥10X coverage were used for statistical analysis.
2.5. Thymine-DNA glycosylase assay
We determined whether TDCIPP directly affects zebrafish and human TDG activity by 1) extracting nuclear proteins from untreated zebrafish embryos and HepG2 cells and 2) quantifying TDG activity in vitro in the presence or absence of TDCIPP. Immediately after spawning, newly fertilized zebrafish eggs were collected and placed into groups of approximately 100 per glass Petri dish within a light- and temperature-controlled incubator. Zebrafish embryos (100 per replicate) were collected at 24 hpf and stored at −80 °C. HepG2 cells were purchased from American Type Culture Collection (Manassas, VA, USA) and grown within T75 culture flasks (Millipore Sigma, St. Louis, MO, USA) using previously described methods (Cheng et al., 2021). HepG2 cells were grown to 75% confluency then collected after 24 h, lysed, and stored in the lysate at −80 °C. Nuclear proteins were extracted from whole zebrafish embryos and HepG2 cells using an EpiQuick Nuclear Extraction kit (Epigentek Group, Farmingdale, NY). Nuclear extracts were aliquoted and stored at −80 °C following the manufacturer’s instructions. Aliquots from each replicate were thawed on ice, pooled, and immediately analyzed using a BCA Protein Assay (Pierce Biotechnology, Rockford, IL, USA) to quantify protein concentrations. Absorbance was quantified using a Promega GloMax Multiplus Plate Reader/Luminometer, and total protein was quantified using a standard curve generated from bovine serum albumin (BSA).
TDG inhibition was quantified using an Epigenase Thymine DNA Glycosylase (TDG) Activity/Inhibition Assay Colorimetric Kit (Epigentek Group, Farmingdale, NY, USA). TDG activity within nuclear extracts (5 μg of protein per reaction) derived from 24-hpf zebrafish embryos and HepG2 cells was quantified in the presence of vehicle (0.1% DMSO) or TDCIPP (3.91, 7.81, 15.63, 31.25, and 62.5 μM). Four replicate reactions were conducted per treatment group. Absorbance was measured using a Promega GloMax Multiplus Plate Reader/Luminometer, and data were corrected for background and reported as relative fluorescence units. TDG activity was calculated using the following equation:
2.6. 5-mC-specific whole-mount immunochemistry and automated imaging
To investigate whether TDCIPP affected the abundance of 5-mC in situ, embryos were exposed to vehicle (0.1% DMSO) or TDCIPP (0.78 or 1.56 μM) from 0.75- to 10-hpf using procedures described above. Based on our previous findings (Kupsco et al., 2017), we selected 0.78 and 1.56 μM TDCIPP since these concentrations only induced a ~1.3- and ~1.7-h epiboly delay, respectively, at 6 hpf. At exposure termination, embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4 °C. Fixed embryos were then manually dechorionated in a glass petri dish under a Leica MZ10 F stereomicroscope prior to storage in 1X phosphate-buffered saline (PBS) at 4 °C. Intact dechorionated embryos were then incubated with a 1:100 dilution of monoclonal mouse anti-5-mC antibody (Sigma-Aldrich, St. Louis, MO, USA) for 16 h at 4 °C, washed in 1X PBS +0.1% Tween-20 (1X PBST) three times for 15 min, and then incubated with a 1:500 dilution of AlexaFluor 488-conjugated goat anti-mouse IgG antibody for 16 h at 4 °C (Thermo Fisher Scientific, Waltham, MA, USA).
Using a Leica MZ10 F stereomicroscope, intact embryos were identified and transferred to clear polystyrene 96-well plates (Thermo Fisher Scientific, Waltham, MA, USA) containing 250 μL 1X PBS per well, centrifuged for 3 min at 130 rpm, and then imaged (at 2X magnification) under transmitted light and FITC using our ImageXpress Micro XLS Widefield High-Content Screening System. Within MetaXpress 6.March 0, 1658, each embryo was analyzed for total fluorescence area and integrated fluorescence intensity using custom automated image analysis procedures. After exporting data from MetaXpress into Excel files, R coding along with packages deployer and writexl were used to sort, summate data points for each well, filter by well number, and export summated data output from MetaXpress into an Excel file. A summary of our optimized protocol for labeling and imaging 5-mC is provided within Figure S1.
2.7. Statistical analyses
A general linear model (GLM) analysis of variance (ANOVA) (α = 0.05) and Tukey-based multiple comparisons were performed using SPSS Statistics 24 for detecting significant within-treatment and/or within-stage differences in context-specific cytosine methylation data, TDG assay data, and 5-mC IHC data. For BSAS-derived data from this study, percent methylation differences were grouped by hierarchical clustering within Morpheus (Broad Institute, Cambridge, MA, USA) using Euclidean distance with a complete linkage method.
3. results
3.1. TDCIPP-induced impacts on CpG methylation persist into later stages of development
We relied on BSAS to quantify alterations in cytosine methylation within a previously identified locus (lmo7b) that was susceptible to TDCIPP exposure. Relative to embryos exposed to vehicle (0.1% DMSO) from 0.75 to 6 hpf, CpG methylation of lmo7b was significantly decreased following exposure to the vehicle (0.1% DMSO) from 0.75 to 48 hpf (but not 0.75 to 24 hpf) (Fig. 1; Table S2), whereas CHG and CHH methylation of lmo7b were not significantly different following exposure to the vehicle (0.1% DMSO) from 0.75 to 24 hpf nor 0.75 to 48 hpf (Fig. 1; Tables S2). Relative to embryos exposed to 0.78 μM TDCIPP from 0.75 to 6 hpf, CpG methylation of lmo7b was significantly decreased following exposure to 0.78 μM TDCIPP from 0.75 to 24 hpf and 0.75 to 48 hpf (Fig. 1; Table S2), whereas CHG and CHH methylation of lmo7b was not significantly different following exposure to 0.78 μM TDCIPP from 0.75 to 24 hpf nor 0.75 to 48 hpf (Fig. 1; Tables S2). Overall, these data suggest that 1) exposure to 0.78 μM TDCIPP may accelerate CpG hypomethylation of lmo7b by at least 24 h and 2) effects of TDCIPP on CpG methylation persist until at least 48 hpf.
Fig. 1.

Mean (± standard deviation) percent cytosine methylation of lmo7b in a CpG (A), CHG (B), or CHH (C) context following exposure of zebrafish embryos to vehicle (0.1% DMSO) or 0.78 μM TDCIPP from 0.75 hpf to 6 hpf, 0.75 hpf to 24 hpf, or 0.75 hpf to 48 hpf. Asterisk (*) denotes significant within-treatment difference (p < 0.05) relative 6 hpf; N = 4 independent replicates of genomic DNA per group.
Within-stage and within-treatment effects on CpG methylation were then assessed on four positions on lmo7b (Chr1), all of which were localized to a single amplicon that was sequenced using BSAS. Based on this analysis, we found that the magnitude of within-treatment effects (i.e., comparing 24 hpf or 48 hpf relative to 6 hpf within the same treatment) was higher than the magnitude of within-stage effects (i.e., comparing vehicle vs. TDCIPP within the same stage) (Fig. 2; Table S3), suggesting that, similar to overall CpG methylation (Fig. 1), CpG methylation at the position-level is strongly stage-dependent within both vehicle- and TDCIPP-exposed embryos. Moreover, we found that positions 33,664,419 and 33,664,494 were more susceptible to TDCIPP-induced acceleration of CpG hypomethylation relative to positions 33,664,391 and 33,664,274 (Fig. 2; Table S3), suggesting that the effects of TDCIPP on CpG methylation within lmo7b may also be position-dependent (e.g., within regions that are CG-rich).
Fig. 2.

Differences in percent CpG methylation on four lmo7b positions following exposure of zebrafish embryos to vehicle (0.1% DMSO) or 0.78 μM TDCIPP from 0.75 hpf to 6 hpf, 0.75 hpf to 24 hpf, or 0.75 hpf to 48 hpf. Data are presented as within-stage (relative to vehicle) and within-treatment (relative to 6 hpf) percent CpG methylation differences. Percent methylation differences were grouped by hierarchical clustering within Morpheus using Euclidean distance with a complete linkage method. N = 4 independent replicates of genomic DNA per group; ns = not significant. Blue circles denote a significant percent methylation difference less than zero (hypomethylation), whereas red circles denote a significant percent methylation difference greater than zero (hypermethylation). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. TDCIPP disrupts the normal trajectory of CHH methylation within lmo7b
Including this study, we have utilized BSAS across three separate studies to investigate methylation of lmo7b within 2-, 4-, 6-, 24-, or 48-hpf zebrafish embryos following exposure to vehicle (0.1% DMSO), 0.78 μM TDCIPP, or 3.12 μM TDCIPP. However, in our prior studies, we did not analyze raw percent methylation data at base-pair resolution by cytosine context (CpG, CHG, or CHH). Therefore, we downloaded cytosine report files to integrate lmo7b-specific data from all three studies and identify potential cytosine context-specific effects as a function of stage and/or TDCIPP concentration. As expected, we found that, across all stages and treatments, percent CpG methylation (>95%) was substantially higher than percent CHG and CHH methylation (<10% and <1%, respectively) (Fig. 3; Table S4). There were more cytosines on a single amplicon of lmo7b within a CHH context (52 total) relative to positions within a CpG context (6 total) and CHG context (19 total). Therefore, we detected significant differences in CHH methylation at 4-, 6-, 24-, and 48-hpf (relative to 2 hpf) following initiation of exposure to vehicle (0.1% DMSO) at 0.75 hpf (Fig. 3; Table S4). However, this normal stage-dependent effect on CHH methylation was absent following exposure to 0.78 or 3.12 μM TDICPP, suggesting that, similar to CpG methylation, TDCIPP disrupts the normal trajectory of CHH methylation within developing zebrafish embryos (Fig. 3; Table S4).
Fig. 3.
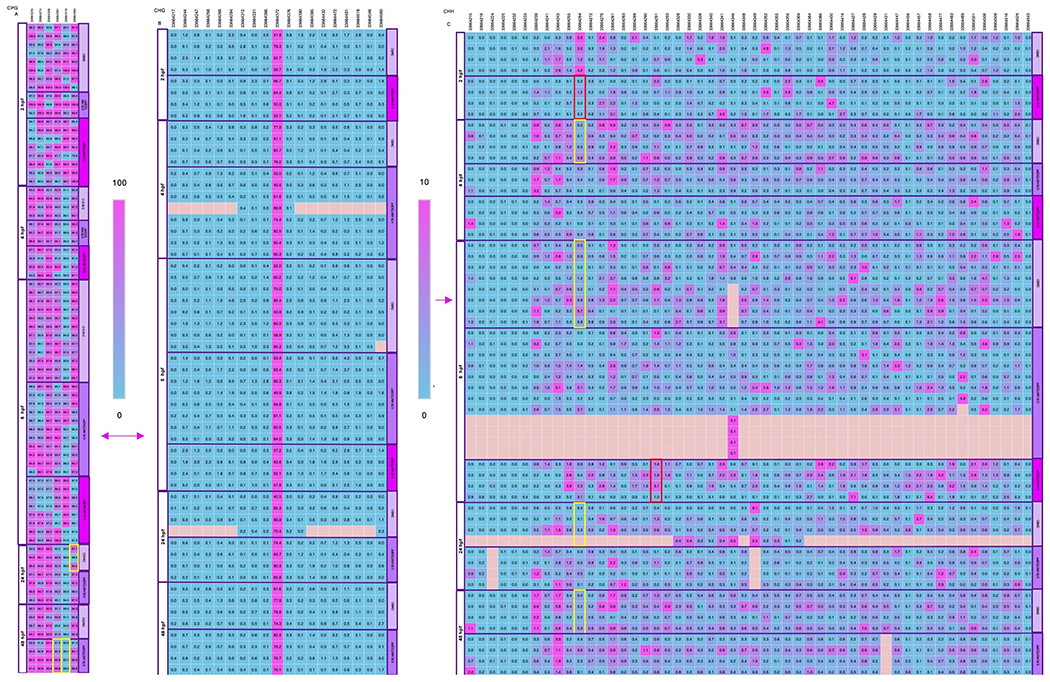
Percent cytosine methylation of lmo7b at base-pair resolution in a CpG (A), CHG (B), or CHH (C) context within 2-, 4-, 6-, 24-, or 48-hpf zebrafish embryos following exposure to vehicle (0.1% DMSO), 0.78 μM TDCIPP, or 3.12 μM TDCIPP from 0.75 to 48 hpf. Squares denote percent cytosine methylation by position for each replicate. Blue denotes 0% cytosine methylation, whereas pink denotes 100% cytosine methylation. Significant within-stage or within-treatment positions (p < 0.05) are outlined with a red or yellow box, respectively. N = 4 independent replicates of genomic DNA per group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. TDCIPP does not affect zebrafish nor human TDG activity in vitro
TDCIPP concentrations ranging from 3.91 to 62.5 μM (the highest concentration tested) had no effect on zebrafish nor human TDG activity (Fig. 4), suggesting that TDCIPP-induced effects on cytosine methylation are not driven by direct interaction with thymine DNA glycosylase.
Fig. 4.

Mean (± standard deviation) thymine DNA glycosylase (TDG) activity within nuclear extracts derived from 24-hpf zebrafish embryos and HepG2 cells following incubation in the presence of vehicle (0.1% DMSO) or TDCIPP (3.91, 7.81, 15,63, 31,25, and 62.5 μM). Numbers above bars denote number of replicate reactions conducted per treatment group.
3.4. TDCIPP increases 5-mC abundance within the yolk of blastula-stage embryos
We relied on 5-mC-specific whole-mount IHC and automated imaging to monitor TDCIPP-induced effects on cytosine methylation in situ as a function of stage and TDCIPP concentration. At 2 and 4 hpf, the vast majority of 5-mC was detected within the yolk of developing embryos (Fig. 5; Table S5), suggesting that the anti-5-mC antibody we used also labeled yolk-localized maternal mRNA containing methylated cytosines. However, at 6, 8 and 10 hpf, 5-mC was increasingly detected within portions of the cell mass, suggesting that cytosine methylation within the cell mass increases during gastrulation. Interestingly, using two different fluorescence-based measurements (total area or integrated intensity) as endpoints, we found that initiation of exposure at 0.75 hpf to 0.78 μM or 1.56 μM TDICPP resulted in a significant increase in yolk-localized cytosine methylation at 2 hpf and/or 4 hpf relative to vehicle-exposed, phenotypically identical embryos at the same stage (Fig. 5; Table S5), suggesting that the downstream effects on TDCIPP on epiboly during gastrula (5.25–10 hpf) may be driven by disruption of yolk-localized cytosine methylation within the first 4 h of development.
Fig. 5.

TDCIPP-induced effects on cytosine methylation in situ as a function of developmental stage and TDCIPP concentration. Zebrafish embryos were exposed to vehicle (0.1% DMSO) or TDCIPP (0.78 or 1.56 μM) from 0.75- to 10-hpf and then processed for 5-mC-specific whole-mount immunochemistry. Intact embryos were imaged under transmitted light and FITC, and then analyzed using our ImageXpress Micro XLS Widefield High-Content Screening System. Representative images of 4-hpf embryos are shown in Panel A. Within MetaXpress 6.March 0, 1658, each embryo was analyzed for total area (B) and total integrated intensity (C) of fluorescence using custom automated image analysis procedures. Asterisk (*) denotes significant within-stage difference (p < 0.05) relative to vehicle controls. Numbers above bars denote number of intact embryos imaged per group. cm = cell mass; ys = yolk sac.
4. Discussion
This study builds upon our previous research demonstrating that TDCIPP impacts cytosine methylation within developing zebrafish embryos (Dasgupta et al., 2019; Kupsco et al., 2017; McGee et al., 2012; Volz et al., 2016). While we acknowledge that the nominal concentrations of TDCIPP (0.78 and 1.56 μM) used within this study may be higher than concentrations detected within blood and/or placental tissues, our study suggests that TDCIPP – if elevated in utero – has the potential to impact the normal trajectory of cytosine methylation during early stages of embryonic development. Human exposure to TDCIPP continues to be ubiquitous around the world, and the concentration of BDCIPP – a primary metabolite of TDCIPP detected in urine – within humans has increased over the past 15 years (Hoffman et al., 2017). As the dynamics of cytosine methylation are highly conserved among mammalian species and critical for regulating cell differentiation (Martin and Fry, 2018; Shen et al., 2007), our findings suggest that TDCIPP may impact cytosine methylation within mammalian embryos in addition to zebrafish embryos.
Interestingly, we found that TDCIPP-induced impacts on CpG methylation within lmo7b persisted into later stages of development, and the severity of this effect was dependent on the developmental stage, TDCIPP concentration, and chromosomal position. Although lmo7b was used as a readout for TDCIPP-induced effects on cytosine methylation in zebrafish embryos, we suspect that, similar to lmo7b, there may be genome-wide effects of TDCIPP on CpG methylation (Volz et al., 2016). Moreover, the magnitude of TDCIPP-induced effects on CpG methylation across the genome may be associated with downstream effects during blastulation and gastrulation (e.g., epiboly delays, etc.), as regulation of 5-mC is necessary for signaling processes that specify cell tissue fate across multiple species (Bartels et al., 2018; Jones, 2012). Indeed, similar to our findings in zebrafish, prior studies have reported that TDCIPP exposure disrupts early mouse embryonic development by inducing uneven blastomeres and abnormal blastocyst formation (Yin et al., 2019) – effects that may be a result of aberrant CpG methylation following fertilization.
Similar to CpG methylation, our analysis across a total of three BSAS studies (including this study) conducted within our lab revealed that TDCIPP also disrupts the normal trajectory of CHH methylation of lmo7b. To our knowledge, little is known about the role of CHH methylation in embryonic development, including within plants. In cotyledons, normal CHH methylation increases from 6% to 11% in earlier to later stages of development, respectively (An et al., 2017). In Arabadopsis, normal CHH methylation increases to 25% in normal mature embryos during development followed by a decrease in CHH methylation during germination (Gutzat et al., 2020). In zebrafish, benzo [a]pyrene (a known human carcinogen) induces changes in CHH methylation within the first few hours of development (Corrales et al., 2014), suggesting that other environmental chemicals may have an impact on CHH methylation during zebrafish embryogenesis. Overall, little is known about the physiological importance and impacts of alterations in CHH methylation, underscoring the importance of future investigations into how these alterations impact epigenetic reprogramming and developmental landmarks such as the MZT.
Our prior study demonstrated that TDCIPP does not affect DNMT activity in vitro at concentrations as high as 500 μM (the highest concentration tested) (Volz et al., 2016). Similarly, in this study we found that TDCIPP did not affect zebrafish nor human TDG activity in vitro at concentrations as high as 62.5 μM (the highest concentration tested). As a result, our studies to date suggest that the impacts of TDCIPP on cytosine methylation do not appear to be driven by direct interaction of TDCIPP with key enzymes (such as DNMT and TDG) that regulate cytosine methylation. However, additional research is needed to confirm whether TDCIPP directly affects the activity of TET and demethylase, as both enzymes are, similar to TDG, involved in decreasing 5-mC levels.
5-mC is a direct and stable biomarker for detecting alterations in epigenetic modifications (Efimova et al., 2020). Although 5-mC is one of the most important RNA modifications and TET proteins are responsible for 5-mC oxidation, little is known about the biological significance of methylated RNA beyond potential functions in control/regulation of gene transcription and protein translation (Zhang et al., 2016). To our knowledge, this is the first study to successfully detect and quantify 5-mC levels in situ within zebrafish embryos during early development. Surprisingly, we found that exposure to TDCIPP increased 5-mC abundance within the yolk of blastula-stage embryos, suggesting that, in addition to impacts on DNA methylation within the cell mass, TDCIPP may affect cytosine methylation of yolk-localized maternal mRNAs during the MZT. During early zebrafish development, the translation and subsequent degradation of maternal mRNAs are essential for zygotic genome activation (Tadros and Lipshitz, 2009) – a process that is tightly regulated by zygotically-derived miR-430 that deadenylates and targets several hundred maternal mRNAs for clearance (Giraldez et al., 2006). Moreover, 5-mC-modifed maternal mRNAs are more stable than non-5-mC-modifed maternal mRNAs during early zebrafish embryogenesis (Yang et al., 2019), suggesting that an abnormal increase in 5-mC-modifed maternal mRNAs may stall maternal mRNA decay and lead to a disruption in the timing of the MZT as well as developmental landmarks such as epiboly.
5. Conclusions
In conclusion, our findings show that TDCIPP disrupts the normal trajectory of cytosine methylation within developing zebrafish embryos. Specifically, we found that 1) TDCIPP-induced impacts on CpG methylation persist into later stages of development; 2) TDCIPP disrupts the normal trajectory of CHH methylation within lmo7b; 3) TDCIPP does not affect zebrafish nor human TDG activity in vitro; and 4) TDCIPP increases 5-mC abundance within the yolk of blastula-stage embryos. Overall, our findings suggest that TDCIPP disrupts cytosine methylation of zygotic DNA within the cell mass as well as maternal mRNA within the yolk – effects that appear to be independent of direct interaction of TDCIPP with enzymes that regulate cytosine methylation. Importantly, TDCIPP-induced disruption of cytosine methylation during epigenetic reprogramming has the potential to lead to downstream effects on embryonic development as well as adverse outcomes into adulthood such as cancer. Indeed, TDCIPP was added to California’s Proposition 65 (Prop 65) list in 2011 based on its potential to cause cancer within humans.
Interestingly, TDCIPP-induced impacts on methylation do not appear to be driven by 1) direct interaction of TDCIPP with key enzymes (such as DNMT and TDG) that regulate cytosine methylation (Volz et al., 2016) nor 2) effects on methyl donor concentrations (Kupsco et al., 2017). Therefore, our findings to date point to an alternative mechanism underlying aberrant methylation such as TDCIPP-induced oxidative stress. TDCIPP is known to induce reactive oxygen species (ROS) production within human neuroblastoma cells (Li et al., 2017), and the role of oxidative stress in modifying the epigenome has been recognized as an important mechanism driving carcinogenesis (Wu and Ni, 2015). Therefore, additional research is needed in order to explore alternative mechanisms (such as oxidative stress) underlying aberrant methylation as well as understand the potential impacts of TDCIPP-induced effects on 5-mC-specific methylation and stability of maternally loaded mRNAs during early embryonic development within zebrafish and other human-relevant models. In addition, more research is needed to assess the potential association of TDCIPP exposure and altered DNA methylation status within human populations that experience chronic and ubiquitous exposure.
Supplementary Material
Acknowledgements
Research support was provided by a UCR Graduate Division Fellowship to SAB and JLW, a NRSA T32 Training Program Fellowship (T32ES018827) to VC and SAB, and a National Institutes of Health grant (R01ES027576) and USDA National Institute of Food and Agriculture Hatch Project (1009609) to DCV.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A.: Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2022.113078.
References
- An Y-QC, Goettel W, Han Q, Bartels A, Liu Z, Xiao W, 2017. Dynamic changes of genome-wide DNA methylation during soybean seed development. Sci. Rep 7, 12263. 10.1038/s41598-017-12510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels A, Han Q, Nair P, Stacey L, Gaynier H, Mosley M, Huang QQ, Pearson JK, Hsieh T-F, An Y-QC, Xiao W, 2018. Dynamic DNA methylation in plant growth and development. Int. J. Mol. Sci 19 10.3390/ijms19072144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma SH, de Boer J, van Velzen MJM, Leonards PEG, 2014. Organophosphorus flame retardants (PFRs) and plasticizers in house and car dust and the influence of electronic equipment. Chemosphere 116, 3–9. 10.1016/j.chemosphere.2014.02.036. [DOI] [PubMed] [Google Scholar]
- Breton-Larrivée M, Elder E, McGraw S, 2019. DNA methylation, environmental exposures and early embryo development. Anim. Reprod 16, 465–474. 10.21451/1984-3143-AR2019-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hou R, Hong X, Yan S, Zha J, 2019. Organophosphate flame retardants (OPFRs) induce genotoxicity in vivo: a survey on apoptosis, DNA methylation, DNA oxidative damage, liver metabolites, and transcriptomics. Environ. Int 130, 104914. 10.1016/j.envint.2019.104914. [DOI] [PubMed] [Google Scholar]
- Cheng V, Reddam A, Bhatia A, Hur M, Kirkwood JS, Volz DC, 2021. Utilizing systems biology to reveal cellular responses to peroxisome proliferator-activated receptor γ ligand exposure. Curr Res Toxicol 2, 169–178. 10.1016/j.crtox.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales J, Fang X, Thornton C, Mei W, Barbazuk WB, Duke M, Scheffler BE, Willett KL, 2014. Effects on specific promoter DNA methylation in zebrafish embryos and larvae following benzo[a]pyrene exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol 163, 37–46. 10.1016/j.cbpc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Cheng V, Vliet SMF, Mitchell CA, Volz DC, 2018. Tris(1,3-dichloro-2-propyl) phosphate exposure during the early-blastula stage alters the normal trajectory of zebrafish embryogenesis. Environ. Sci. Technol 52, 10820–10828. 10.1021/acs.est.8b03730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Vliet SM, Kupsco A, Leet JK, Altomare D, Volz DC, 2017. Tris(1,3-dichloro-2-propyl) phosphate disrupts dorsoventral patterning in zebrafish embryos. PeerJ 5, e4156. 10.7717/peerj.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Vliet SMF, Cheng V, Mitchell CA, Kirkwood J, Vollaro A, Hur M, Mehdizadeh C, Volz DC, 2019. Complex interplay among nuclear receptor ligands, cytosine methylation, and the metabolome in driving tris(1,3-dichloro-2-propyl)phosphate-induced epiboly defects in zebrafish. Environ. Sci. Technol 53, 10497–10505. 10.1021/acs.est.9b04127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw LV, Hunter DL, Padnos B, Padilla S, Stapleton HM, 2014. Developmental exposure to organophosphate flame retardants elicits overt toxicity and alters behavior in early life stage zebrafish (Danio rerio). Toxicol. Sci 142, 445–454. 10.1093/toxsci/kfu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty BT, Hammel SC, Daniels JL, Stapleton HM, Hoffman K, 2019. Organophosphate esters: are these flame retardants and plasticizers affecting children’s Health? Curr Environ Health Rep 6, 201–213. 10.1007/s40572-019-00258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimova OA, Koltsova AS, Krapivin MI, Tikhonov AV, Pendina AA, 2020. Environmental epigenetics and genome flexibility: focus on 5-Hydroxymethylcytosine. Int. J. Mol. Sci 21 10.3390/ijms21093223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Corrales J, Thornton C, Scheffler BE, Willett KL, 2013. Global and gene specific DNA methylation changes during zebrafish development. Comp. Biochem. Physiol. B Biochem. Mol. Biol 166, 99–108. 10.1016/j.cbpb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Han J, Zhou B, Gong Z, Santos EM, Huo X, Zheng W, Liu H, Yu H, Liu C, 2013. Toxicogenomic responses of zebrafish embryos/larvae to tris(1,3-dichloro-2-propyl) phosphate (TDCPP) reveal possible molecular mechanisms of developmental toxicity. Environ. Sci. Technol 47, 10574–10582. 10.1021/es401265q. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF, 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Gutzat R, Rembart K, Nussbaumer T, Hofmann F, Pisupati R, Bradamante G, Daubel N, Gaidora A, Lettner N, Donà M, Nordborg M, Nodine M, Mittelsten Scheid O, 2020. Arabidopsis shoot stem cells display dynamic transcription and DNA methylation patterns. EMBO J. 39, e103667 10.15252/embj.2019103667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman K, Butt CM, Webster TF, Preston EV, Hammel SC, Makey C, Lorenzo AM, Cooper EM, Carignan C, Meeker JD, Hauser R, Soubry A, Murphy SK, Price TM, Hoyo C, Mendelsohn E, Congleton J, Daniels JL, Stapleton HM, 2017. Temporal trends in exposure to organophosphate flame retardants in the United States. Environ. Sci. Technol. Lett 4, 112–118. 10.1021/acs.estlett.6b00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop P, Ruzov A, Gering M, 2018. Developmental functions of the dynamic DNA methylome and hydroxymethylome in the mouse and zebrafish: similarities and differences. Front. Cell Dev. Biol 6, 27. 10.3389/fcell.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet 13, 484–492. 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jones PA, Takai D, 2001. The role of DNA methylation in mammalian epigenetics. Science 293, 1068–1070. 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kucharska A, Cequier E, Thomsen C, Becher G, Covaci A, Voorspoels S, 2015. Assessment of human hair as an indicator of exposure to organophosphate flame retardants. Case study on a Norwegian mother-child cohort. Environ. Int 83, 50–57. 10.1016/j.envint.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Kupsco A, Dasgupta S, Nguyen C, Volz DC, 2017. Dynamic alterations in DNA methylation precede tris(1,3-dichloro-2-propyl)phosphate-induced delays in zebrafish epiboly. Environ. Sci. Technol. Lett 4, 367–373. 10.1021/acs.estlett.7b00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhou P, Guo Y, Lee J-S, Zhou B, 2017. Tris (1,3-dichloro-2-propyl) phosphate-induced apoptotic signaling pathways in SH-SY5Y neuroblastoma cells. Neurotoxicology 58, 1–10. 10.1016/j.neuro.2016.10.018. [DOI] [PubMed] [Google Scholar]
- Liu C, Su G, Giesy JP, Letcher RJ, Li G, Agrawal I, Li J, Yu L, Wang J, Gong Z, 2016. Acute exposure to tris(1,3-dichloro-2-propyl) phosphate (TDCIPP) causes hepatic inflammation and leads to hepatotoxicity in zebrafish. Sci. Rep 6, 19045. 10.1038/srep19045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EM, Fry RC, 2018. Environmental influences on the epigenome: exposure-associated DNA methylation in Human populations. Annu. Rev. Publ. Health 39, 309–333. 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- McGee SP, Cooper EM, Stapleton HM, Volz DC, 2012. Early zebrafish embryogenesis is susceptible to developmental TDCPP exposure. Environ. Health Perspect 120, 1585–1591. 10.1289/ehp.1205316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CA, Dasgupta S, Zhang S, Stapleton HM, Volz DC, 2018. Disruption of nuclear receptor signaling alters triphenyl phosphate-induced cardiotoxicity in zebrafish embryos. Toxicol. Sci 163, 307–318. 10.1093/toxsci/kfy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, Nix DA, Parnell TJ, Cairns BR, 2013. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772. 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddam A, Tait G, Herkert N, Hammel SC, Stapleton HM, Volz DC, 2020. Longer commutes are associated with increased human exposure to tris(1,3-dichloro-2-propyl) phosphate. Environ. Int 136, 105499. 10.1016/j.envint.2020.105499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa J-PJ, 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 3, 2023–2036. 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD, 2009. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Tran CM, Lee H, Lee B, Ra J-S, Kim K-T, 2021. Effects of the chorion on the developmental toxicity of organophosphate esters in zebrafish embryos. J. Hazard Mater 401, 123389. 10.1016/j.jhazmat.2020.123389. [DOI] [PubMed] [Google Scholar]
- Volz DC, Leet JK, Chen A, Stapleton HM, Katiyar N, Kaundal R, Yu Y, Wang Y, 2016. Tris(1,3-dichloro-2-propyl)phosphate induces genome-wide Hypomethylation within early zebrafish embryos. Environ. Sci. Technol 50, 10255–10263. 10.1021/acs.est.6b03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Ni X, 2015. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr. Drug Targets 16, 13–19. 10.2174/1389450116666150113121054. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhao Y, Li M, Du M, Li X, Li Y, 2019. A review of a class of emerging contaminants: the classification, distribution, intensity of consumption, synthesis routes, environmental effects and expectation of pollution abatement to organophosphate flame retardants (OPFRs). Int. J. Mol. Sci 20 10.3390/ijms20122874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S-Y, Chen L, Wu D-Y, Wang T, Huo L-J, Zhao S, Zhou J, Zhang X, Miao Y-L, 2019. Tris(1,3-dichloro-2-propyl) phosphate disturbs mouse embryonic development by inducing apoptosis and abnormal DNA methylation. Environ. Mol. Mutagen 60, 807–815. 10.1002/em.22322. [DOI] [PubMed] [Google Scholar]
- Zhang H-Y, Xiong J, Qi B-L, Feng Y-Q, Yuan B-F, 2016. The existence of 5-hydroxymethylcytosine and 5-formylcytosine in both DNA and RNA in mammals. Chem. Commun 52, 737–740. 10.1039/c5cc07354e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


