Abstract
The aryl hydrocarbon receptor (AHR) regulates the expression of numerous genes in response to activation by agonists including xenobiotics. Although it is well appreciated that environmental signals and cell intrinsic features may modulate this transcriptional response, how it is mechanistically achieved remains poorly understood. We show that hexokinase 2 (HK2) a metabolic enzyme fuelling cancer cell growth, is a transcriptional target of AHR as well as a modulator of its activity. Expression of HK2 is positively regulated by AHR upon exposure to agonists both in human cells and in mice lung tissues. Conversely, over-expression of HK2 regulates the abundance of many proteins involved in the regulation of AHR signalling and these changes are linked with altered AHR expression levels and transcriptional activity. HK2 expression also shows a negative correlation with AHR promoter methylation in tumours, and these tumours with high HK2 expression and low AHR methylation are associated with a worse overall survival in patients. In sum, our study provides novel insights into how AHR signalling is regulated which may help our understanding of the context-specific effects of this pathway and may have implications in cancer.
INTRODUCTION
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor belonging to the basic helix-loop-helix/Per-ARNT-SIM (bHLH-PAS) family (1,2). According to classical models, AHR is retained by a chaperone complex in the cytoplasm and upon binding of its ligand, AHR translocates to the nucleus to form a heterodimer with its most common partner, the AHR nuclear translocator (ARNT, also known as hypoxia-inducible factor 1β – HIF1β) (1–3). This new complex then regulates the expression of different set of genes containing consensus AHR-responsive elements in their promoter such as prototypical target genes coding proteins of the cytochrome P-450 family of monooxygenases (e.g. CYP1A1 and CYP1B1) and aldehyde dehydrogenases (e.g. ALDH3A1) (3–7). AHR plays a critical function in the activation of genes involved in the metabolism of xenobiotics and in additional processes including cell proliferation, cell differentiation, immune response and in cancer processes (8–10). AHR is also emerging as a promising therapeutic target in a number of human diseases, including cancer and immune diseases (11–16). It is thus important to better understand how AHR signalling is regulated by environmental signals and intrinsic features to control such diverse functions and transcriptional programs.
A first level of regulation is the activity of AHR promoter itself, and hence AHR expression. AHR is expressed in most tissues, although levels are quite variable across tissues and cell types, with high levels in epithelia (e.g. lung, intestine) and immune cells (17). Several studies have shown that AHR promoter is regulated by epigenetic mechanisms including DNA methylation and chromatin marks (18–21). For instance, in human cells AHR promoter is susceptible to DNA methylation and heavy DNA methylation of the promoter is correlated with a lower expression of the gene (19).
AHR transcriptional activity is also modulated by a wide variety of exogenous and endogenous ligands. This includes halogenated aromatic hydrocarbon compounds (HAHs) (e.g. 2,3,7,8-tetrachlorodibenzodioxin, also known as TCDD or dioxin), polycyclic aromatic hydrocarbons (PAHs) (e.g. benzo(a)pyrene (BaP) and 3-methylcholanthrene (3-MC)), and tryptophan derivatives (e.g. formylindolo[3,2-b]carbazole (FICZ) and kynurenic acid) (11,12). These ligands exhibit quite different impacts on AHR activity due to their chemical properties and variable affinity for AHR (11,12). For instance, TCDD causes long lasting effects as it persists in tissues following exposure, while FICZ only transiently enhances AHR transcriptional activity due to its rapid metabolism and inactivation in cells (7,12,22).
Finally, AHR activity is regulated by numerous cofactors including proteins that promote its activity such as transcriptional coactivators (e.g. EP300, NCOA3, SRC) (1). On the contrary AHR activity is negatively regulated by repressors such as AHR repressor (AHRR) and TCDD-inducible poly(ADP-ribose) polymerase (TIPARP) (1,23–25). These latter two, in addition to repress AHR signalling, are also induced at the transcriptional level by AHR upon agonists exposure, establishing negative feedback loops that attenuate transcriptional activation of AHR targets (23–25).
AHR is thus regulated at several levels. In this prospect, it is important to characterize additional proteins involved in the regulation of AHR signalling to better understand how it leads to different transcriptional outcomes and target these key regulatory links in disease. In this study, we report a molecular link between hexokinase 2 (HK2) and AHR signalling pathway. HK2 is one of the five hexokinases encoded in the human genome which phosphorylates glucose to produce glucose-6-phosphate, the first rate-limiting step of glycolysis (26). Its over-expression is extensively described to promote metabolic alterations supporting tumour growth and aggressiveness, and its knock-out markedly reduces the growth of genetically-induced tumours in mouse models of lung and breast cancers (26–28). We showed that AHR binds to HK2 gene in human cancer cells and promotes its expression in response to exposure to a variety of AHR agonists in different cell types. We further demonstrated that over-expression of HK2 alters the abundance of numerous proteins involved in the AHR network, including regulatory proteins and targets of AHR, and that HK2 also regulates AHR gene expression. These regulations are linked with gene-specific changes in AHR transcriptional outcomes. Thus, HK2 is a target as well as a regulator of AHR signalling, and we showed that the expression of both genes is positively correlated in diverse tumour types suggesting that this regulatory link might play an important function in cancer.
MATERIALS AND METHODS
ChIP-sequencing analyses of AHR binding sites in human cells
AHR ChIP-sequencing data were retrieved from NCBI GEO and fastq files downloaded from EMBL-EBI ENA website (https://www.ebi.ac.uk/ena) on the Galaxy web-interface (29). We used Bowtie2 to map sequencing reads against human reference genome hg19 and produce bam files and ChIP-sequencing peaks (30).
ChIP-sequencing datasets were previously published and prepared as follows. AHR ChIPs in MCF-7 breast cancer cells were performed using an AHR antibody (Santa Cruz, H-211) in triplicate, starting from MCF-7 treated with 10 nM TCDD (2,3,7,8-tetrachlorodibenzodioxin) or DMSO (dimethylsulfoxide) for 45 min and 24 h (GSE90550) (5). AHR ChIPs in GM17212 EBV-immortalised lymphocytes (GSE116638) were performed using an AHR antibody (Cell Signaling Technology (CST); D5S6H) in duplicate, starting from GM17212 cells treated with 1 μM 3-MC (3-methylcholanthrene) or DMSO for 24 h (6). AHR ChIPs in HepG2 hepatocarcinoma cells were produced using an antibody directed against Flag-Tag (Sigma-Aldrich) from cells engineered to express an AHR flag-tagged protein (GSE127649; ENCSR412ZDC, released on 10 February 2018) (31).
Gene classification and gene-ontology (GO) enrichment were performed using the PANTHER interface (version 16.0 released 2020–12-01) using Fisher's exact t-test and false discovery rate (32).
The UCSC Genome Browser and Integrative Genome Viewer (IGV_2.3.92) were used to visualize the ChIP-sequencing data as well as to prepare the different panels in the manuscript (33). The UCSC CpG Islands, GeneHancer and RefSeq gene tracks were used to locate promoters and enhancers as well as genes in the human genome (reference hg19) (33). ENCODE datasets, including transcription factors ChIP-sequencing data and DNAseI sites, are freely usable and were retrieved from the ENCODE download portal (31). Histone ChIP-sequencing data are freely usable and were retrieved from the ChIP-Atlas database (https://chip-atlas.org/) (34).
A search for classical recognition motif of the AHR/ARNT complex (5′-GCGTG-3′; 3′-CACGC-5′ and its longer version 5′-TNGCGTG-3′; 3′-CACGCNA-5′) was performed manually on the sequence of HK2 gene (coordinates: chromosome 2:75059782–75120481 on human reference genome hg19).
Cell lines, culture and treatments
U2OS human osteosarcoma and HCT116 human colon cancer cells were authenticated at the start of the project by DNA/STR profiling (Eurofins Genomics). 143B osteosarcoma cell line was a gift from Dr Olivia Fromigué (INSERM UMR981, Institut Gustave Roussy, Villejuif, France). All cell lines were routinely tested for mycoplasma contamination by conventional PCR using the mycoplasma detection kit Venor®GeM Classic (Minerva Biolabs). Authentication certificates and mycoplasma-test results are available upon request. 143B, U2OS and HCT116 cell lines encode a wild-type AHR protein (35).
U2OS, 143B and HCT116 were cultured at 37°C in a humid chamber with 5% CO2. U2OS and 143B cells were cultured in DMEM (GlutaMAX, glucose 4.5 g/l and pyruvate) supplemented with 10% foetal bovine serum and 1% penicillin-streptomycin. HCT116 cells were cultured in RPMI-1640 with 10% foetal bovine serum and 1% penicillin-streptomycin. Cells were split every 2 to 3 days.
U2OS-GFP, U2OS-GFP-HK2, 143B-GFP and 143B-GFP-HK2 were maintained in DMEM (GlutaMAX, glucose 4.5 g/l and pyruvate) supplemented with 10% foetal bovine serum, 1% penicillin-streptomycin and experimentally-defined concentrations of neomycin/G418 (Thermo Fisher Scientific; 10131027) as a selection agent (see section ‘Plasmids’ below).
Cells were seeded at a density of 7800 cells/cm2 in six-well plates and after cell attachment to the culture plates, AHR agonists were added to the culture media. TCDD solution in toluene (Supelco; 48599) was added at final concentration of 1 or 10 nM, and toluene (Sigma-Aldrich; 244511) was used as control vehicle treatment. BaP solution (Supelco; 40071) was diluted in DMSO and used at final concentration of 0.5–2 μM. FICZ (Sigma-Aldrich; SML1489) was resuspended at 1 mM in DMSO and used at final concentration of 1 nM to 10 μM. CH-223191 (Sigma-Aldrich; C8124) resuspended in DMSO was used at final concentrations of 0.001–10 μM and added alone or at the same time as TCDD or BaP for 48 h. 5-aza-2′-deoxycytidine (A3656; Sigma-Aldrich) diluted in DMSO was used at final concentration of 10 μM for 48 h and cell culture media was changed every 24 h as previously described (36). DMSO was used as a control for FICZ, BaP, CH-223191 and 5-aza-2′-deoxycytidine exposure experiments. 2-Deoxy-d-glucose (Sigma-Aldrich; D6134) was resuspended in water and used at final concentrations of 2 mM alone or in combination with BaP for 48 h.
ChIP-qPCR analysis of AHR binding
The chromatin immunoprecipitation (ChIP) was performed using a human AHR specific antibody (Cell Signaling Technology; 13790) following a procedure previously described (37). qPCR analysis was performed with the kit LightCycler® 480 SYBR Green I Master (Roche Diagnostics; 04887352001) according to manufacturer recommendations on a Roche LightCycler® 480 system (software version 1.5.1.62 SP3). ChIP and input DNAs were amplified and data are presented as percentage of input DNA recovered in each ChIPs.
qPCR primers were as follows: CYP1A1 promoter 5′-CCT GGG ATC ACA AGG ATC AGG-3′ and 5′-CGT ACA AGC CCG CCT ATA AA-3′, HK2 cis-regulatory region A 5′- CCA CTA CCA GGG AAG GCT CA-3′ and 5′-TCC TGC CCA GTG ACT AGA GG-3′, HK2 cis-regulatory region B 5′-CAG GGA GCT GGT CAG ATG TG-3′ and 5′-AGT GAA GCG GAA TGG GTC AG-3′, HK2 cis-regulatory region C 5′-GAG GTA GTC GGC TCT CAG GA-3′ and 5′-TCC AGG TTG CTA CGA ATG CC-3′, HK2 cis-regulatory region D 5′-CAT GCT GGG GTT GGA GAA GG-3′ and 5′-TTG GTG CAG GCA TAG GAG TG-3′, HK2 promoter 5′-CAA CAT CGT GTC ACC CAG CT-3′ and 5′-GCT AAC TTC GGC CAC AGG AT-3′, region 2-kilobases upstream of HK2 promoter 5′-CCC GGC ATC CCT TGA ATT CT-3′ and 5′-TCC AGG CCT GTC TCC AAC TC-3′, region 2-kilobases downstream of HK2 promoter 5′-ATG TAG TGA TGG CGC GTG AA-3′ and 5′-CAG AGC CAC ATC CCA GGA ATT-3′.
RT-qPCR analysis
RNAs were isolated using the TRIzol™ reagent (Thermo Fisher Scientific; 15596026) followed by DNAse treatment and removal (Thermo Fisher Scientific; AM1906) according to a standard protocol previously described (36) or the RNeasy mini-kit according to manufacturer recommendations (QIAGEN; 74106). RNA concentration and purity were assessed using NanoDrop™ 2000 (Thermo Fisher Scientific). Reverse transcription was performed using the Superscript™ II reverse transcriptase enzyme with reaction conditions recommended by the manufacturer (Thermo Fisher Scientific; 18064) starting with 500 ng of total RNA preparation and using random hexamers (Thermo Fisher Scientific; N8080127). qPCR analysis was performed using the kit LightCycler® 480 SYBR Green I Master (Roche Diagnostics; 04887352001) according to manufacturer recommendations on a Roche LightCycler® 480 system (software version 1.5.1.62 SP3).
Primers used for qPCR analyses were as follows for human genes: HK2 5′-GAG CCA CCA CTC ACC CTA CT-3′ and 5′-CCA GGC ATT CGG CAA TGT G-3′, AHR 5′-ACA TCA CCT ACG CCA GTC G-3′ and 5′-CGC TTG GAA GGA TTT GAC TTG A-3′ (used for Figure 2), AHR 5′-GCC GGT GCA GAA AAC AGT AAA-3′ and 5′-AGC CAA ACG GTC CAA CTC TG-3′ (used for Figure 4), CYP1A1 5′-TCT TGA GGC CCT GAT TAC CCA-3′ and 5′-TTC GGC CAC GGA GTT TCT TC-3′, CYP1B1 5′-AAC GTA CCG GCC ACT ATC AC-3′ and 5′-GCA CTC GAG TCT GCA CAT CA-3′, ALDH3A1 5′-TGA TCC AGG AGC AGG AGC A-3′ and 5′-CCT CTA GGA CGT ACA CCA CC-3′. Gene expression values were normalised using the DeltaCq method (2–ΔΔCq) against reference MCM3 gene expression in Figures 4A–F, 5A, B, F, G and Supplementary Figure S5 (5′-GCT CCT CTG GAG TGG GTC TG -3′ and 5′-TCC TGT TTC CTG GTC TGT GGT-3′) and in all other figures against the average value of expression of two reference genes: B2M 5′-GGC TAT CCA CGT ACT CCA AA-3′ and 5′-CGG CAG GCA TAC TCA TCT TTT T-3′ and MAPK14 5′-TGC CGA AGA TGA ACT TTG CGA-3′ and 5′-TCA TAG GTC AGG CTT TTC CAC T-3′. Cq is the quantification cycle corresponding to the Cp value on Roche LightCycler® 480 system.
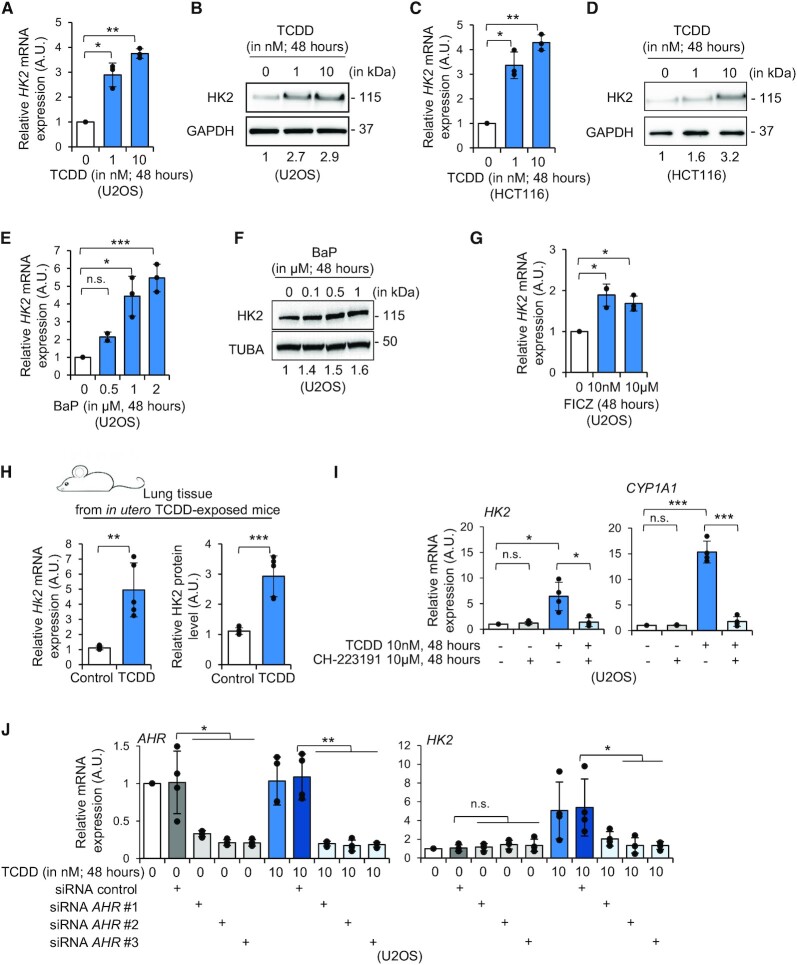
Figure 2.
AHR regulates HK2 mRNA expression. (A) RT-qPCR analysis of HK2 expression in U2OS cells treated with vehicle (i.e. toluene) or TCDD (1 and 10 nM) for 48 h (n = 3). A.U., arbitrary unit. (B) Western blot analysis of HK2 and GAPDH levels in U2OS cells treated with vehicle (i.e. toluene) or TCDD (1 and 10 nM) for 48 h. Relative quantifications are indicated below the blots (versus GAPDH control and relative to vehicle condition). (C) RT-qPCR analysis of HK2 expression in HCT116 human colon cancer cells treated with vehicle (i.e. toluene) or TCDD (1 and 10 nM) for 48 h (n = 3). A.U., arbitrary unit. (D) Western blot analysis of HK2 and GAPDH levels in HCT116 cells treated with vehicle (i.e. toluene) or TCDD (1 and 10 nM) for 48 h. Relative quantifications are indicated below the blots (versus GAPDH control and relative to vehicle condition). (E) RT-qPCR analysis of HK2 expression in U2OS cells treated with vehicle (i.e. DMSO) or BaP (0.1 μM up to 2 μM) for 48 h (n = 3). A.U., arbitrary unit. (F) Western blot analysis of HK2 and TUBA levels in U2OS cells treated with vehicle (i.e. DMSO) or BaP (0.1 μM up to 1 μM) for 48 h. Relative quantifications are indicated below the blots (versus TUBA control and relative to vehicle condition). (G) RT-qPCR analysis of HK2 expression in U2OS cells treated with vehicle (i.e. DMSO) or FICZ (10 nM and 10 μM) for 48 h (n = 3). A.U., arbitrary unit. (H) RT-qPCR (left graph) and relative quantification of western blots (right graph) of Hk2 expression and protein levels in lung tissues from adult male mice exposed to TCDD or vehicle (i.e. n-nonane) in utero and during lactation (n = 5). A.U., arbitrary unit. (I) RT-qPCR analysis of HK2 (left graph) and CYP1A1 (right graph) expression in U2OS cells treated with vehicle (i.e. toluene), TCDD (10 nM), CH-223191, an AHR antagonist (10 μM) or with TCDD + CH-223191 (n = 4). A.U., arbitrary unit. (J) RT-qPCR analysis of AHR (left graph) and HK2 (right graph) expression in U2OS cells transfected with control or AHR siRNAs and further exposed to vehicle (i.e. toluene) or TCDD (10 nM) for 48 h (n = 4). A.U., arbitrary unit. Paired Student's t-test was used to calculate P-value in panels A, C, E, G and I, and unpaired Student's t-test for panels H and J. n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
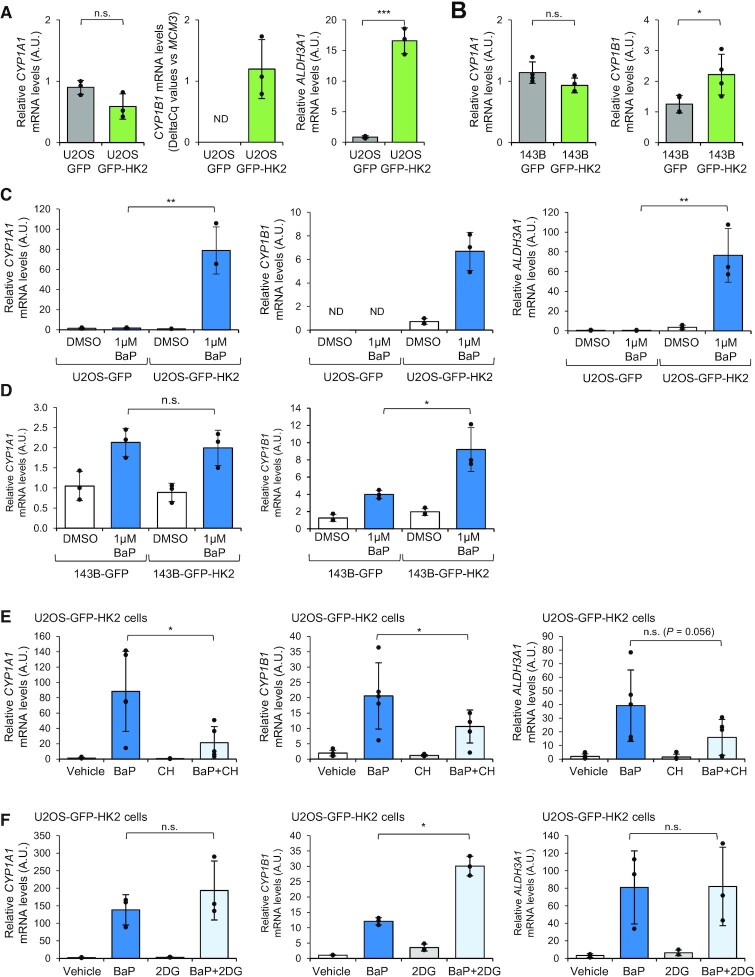
Figure 4.
Over-expression of HK2 enhances a subset of AHR transcriptional target genes expression and potentiates the AHR transcriptional response to BaP exposure. (A) RT-qPCR analysis of CYP1A1 (left graph), CYP1B1 (middle graph) and ALDH3A1 (right graph) expression in U2OS cells expressing GFP or GFP-HK2 (n = 3). A.U., arbitrary unit. ND, not detected by RT-qPCR. (B) RT-qPCR analysis of CYP1A1 (left graph) and CYP1B1 (right graph) expression in 143B-GFP and 143B-GFP-HK2 cells (n = 4). A.U., arbitrary unit. Of note, ALDH3A1 is not detected in these cell lines. (C) RT-qPCR analysis of CYP1A1 (left graph), CYP1B1 (middle graph) and ALDH3A1 (right graph) expression in U2OS cells expressing GFP and GFP-HK2 and treated with BaP 1 μM or vehicle (i.e. DMSO) for 48 h (n = 3). ND, not detected by RT-qPCR. (D) RT-qPCR analysis of CYP1A1 (left graph), CYP1B1 (right graph) expression in 143B cells expressing GFP and GFP-HK2 and treated with BaP 1 μM or vehicle (i.e. DMSO) for 48 h (n = 3). (E) RT-qPCR analysis of CYP1A1 (left graph), CYP1B1 (middle graph) and ALDH3A1 (right graph) expression in U2OS-GFP-HK2 cells treated with vehicle (i.e. DMSO), BaP (1 μM), AHR antagonist CH-223191 (10 μM) or BaP + CH-223191 for 48 h (n = 5). A.U., arbitrary unit. (F) RT-qPCR analysis of CYP1A1 (left graph), CYP1B1 (middle graph) and ALDH3A1 (right graph) mRNA levels in U2OS-GFP-HK2 cells treated with vehicle (i.e. DMSO), BaP (1 μM), hexokinase inhibitor 2-DG (2mM) or BaP + 2-DG for 48 h (n = 3). A.U., arbitrary unit. P-value was calculated using unpaired Student's t-test for panels A, B, C, D, and paired Student's t-test for panels E and F. n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
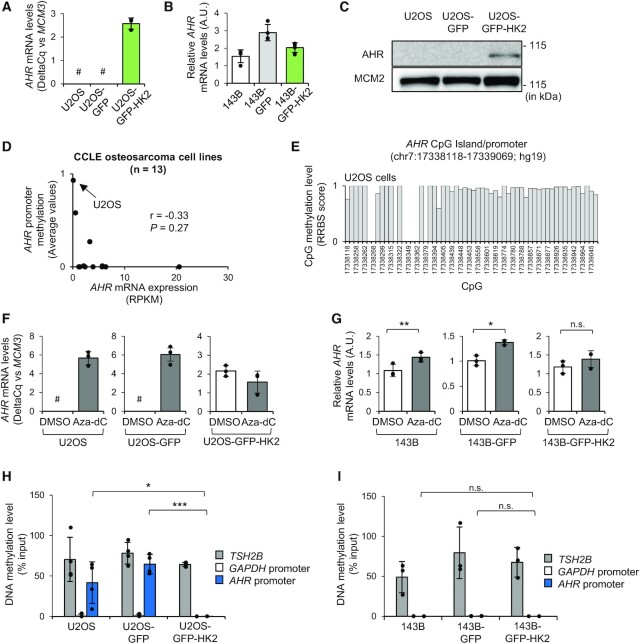
Figure 5.
Over-expression of HK2 enhances AHR expression and it correlates with the DNA demethylation of AHR promoter in U2OS cells. (A) RT-qPCR analysis of AHR expression in U2OS, U2OS-GFP, U2OS-GFP-HK2. Data are presented as DeltaCq values versus MCM3 for U2OS cell lines (n = 3; #, high Cq values for AHR (>38)). (B) RT-qPCR analysis of AHR expression in 143B, 143B-GFP and 143B-GFP-HK2 cells (n = 4). A.U., arbitrary unit. (C) Western blot analysis of AHR and MCM2 levels in U2OS, U2OS-GFP and U2OS-GFP-HK2 cells. MCM2 is used as loading control as it is not differentially expressed between U2OS and U2OS-GFP-HK2 cells according to the proteomic data. (D) Graph showing the correlation between AHR promoter methylation status and AHR expression from the CCLE osteosarcoma cell lines repository. DNA methylation are presented as average beta-value for AHR promoter (0: no methylation; 1: complete methylation). (E) Individual CpG methylation levels at the AHR CpG island promoter in U2OS cells defined by RRBS score (0: no methylation; 1: complete methylation). (F) RT-qPCR analysis of AHR expression in U2OS, U2OS-GFP and U2OS-GFP-HK2 cells after treatment with vehicle (i.e. DMSO) or with the DNA demethylating agent 5-aza-2′-deoxycytidine (10 μM; Aza-dC) for 48 h. Data are presented as ‘DeltaCq’ values versus MCM3 (n = 3). A.U., arbitrary unit. #, high Cq values for AHR (>38). (G) RT-qPCR analysis of AHR expression in 143B, 143B-GFP and 143B-GFP-HK2 cells after treatment with vehicle (i.e. DMSO) or with the DNA demethylating agent 5-aza-2′-deoxycytidine (10 μM; Aza-dC) for 48 h (n = 3). A.U., arbitrary unit. (H, I) MeDIP-qPCR analysis of the methylation level of AHR promoter in (H) U2OS, U2OS-GFP and U2OS-GFP-HK2 cells (n = 4) and (I) 143B, 143B-GFP and 143B-GFP-HK2 cells (n = 3). TSH2B region is used as reference of heavily methylated sequence, while GAPDH region is used as a reference of unmethylated DNA. Paired and unpaired Student's t-test were used to calculate P-value respectively in panels G and H–I. n.s., not significant; *P < 0.05; ***P < 0.001.
For Hk2 murine transcript we used the following primers: 5′-ATG ATC GCC TGC TTA TTC ACG-3′ and 5′-CGC CTA GAA ATC TCC AGA AGG G-3′. We normalised Hk2 expression using the DeltaCq (2–ΔΔCq) method and the average value of expression of two reference genes: Actb 5′-CAA TAG TGA TGA CCT GGC CGT-3′ and 5′-AGA GGG AAA TCG TGC GTG AC-3′ and Mapk14 5′-TGA CCC TTA TGA CCA GTC CTT T-3′ and 5′-GTC AGG CTC TTC CAC TCA TCT AT-3′.
Western blot analysis
Methods for protein extraction, separation and detection were previously described (36). Proteins of interest were revealed using the following antibodies: AHR (13790), HK1 (C35C4), HK2 (C64G5), GAPDH (D16H11) and LDHA (C4B5) purchased from Cell Signaling Technology, NQO1 (A180) purchased from Novus Biologicals and CHK2 (ab109413) and Histone H3 (ab1791) purchased from Abcam. α-Tubulin (T6199-100UL), β-Actin (PA1-183) and MCM2 (A300-191A) were purchased from Sigma-Aldrich, Thermo Fisher Scientific and Bethyl Laboratories respectively. Rabbit homemade antiserum against JUN was a gift from Dr Frédérique Verdier (Institut Cochin, INSERM U1016, Paris, France).
Exposition of mouse pups to TCDD
C57BL/6J mice were provided by the CDTA (Center for animal Distribution, Typing and Archiving, CNRS, Orléans, France). Experiments on animals were performed at the animal facility of CDTA and subsequently at the animal core facility of BioMed Tech facilities (Campus Saint Germain des Prés, INSERM US36, CNRS UMS2009, Université Paris Cité, Paris, France). The European Communities Council directive 2010/63/EU on the protection of the animals were followed for the experiments using animals. Animals were treated humanely and with regard for alleviation of suffering. All procedures were given approval by the ethical committees for animal research of CNRS (Orléans, France) and ‘Université Paris Cité’ (Paris, France) (CEEA34.XC.049.12).
C57BL/6J mice were housed in a temperature-controlled room (22 ± 1°C) with a relative humidity of 55 ± 5% and a 12-h light/dark cycle. Water and food (Safe® D30) were provided ad libitum. Pregnant CD57Bl/6J mice were randomly administered either oral doses of TCDD in vehicle (1 ng TCDD/g body weight; LGC Standards) or n-nonane/corn oil vehicle alone (1/24 v/v; Sigma-Aldrich) on embryonic days E7.5, E14.5 and post-natal days P0.5, P7.5, P14.5, P21 in the CDTA animal facility by using a curved gavage probe fitted to the mouse mounted on a 1 ml syringe. The oral route of exposure was used to mimic the major route of exposure in humans. Each administration corresponded to 1 ng/g body weight of TCDD in n-nonane (corresponding to 0.2 μl/g body weight) or n-nonane alone (0.2 μl/g body weight) diluted in corn oil (10 μl/g body weight). Pups were weaned at 3 weeks of age and allowed to acclimate to the BioMed Tech animal facility from 5 to 9 weeks of age. Five male pups from these two groups of dams were analysed at 9 weeks of age. Their lung tissues were collected and analysed by RT-qPCR and western blot.
The Comparative Toxicogenomics Database (CTD)
The Comparative Toxicogenomics Database (CTD) was interrogated on 1 September 2021 on an updated version of the database including 14 036 unique chemicals (38). Using the search engine ‘chemical-gene interaction’ tool, 2284 chemical interactions with AHR in human (taxon:9606) were retrieved among which 748 interactions modulate AHR activity (either ‘increase’, ‘decrease’ or ‘affect with degree unspecified’). Using a similar approach, we identified 180 chemical interactions with HK2 and 158 chemical interactions regulating HK2 expression in human. We manually curated the lists (to remove duplications) and then compared them to identify the chemicals common to the regulation of HK2 expression and AHR activity, and we assessed the resulting overlap by hypergeometric test.
Electroporation of small-interfering RNAs (siRNAs)
Transfection of control and AHR siRNA duplexes was performed with Neon Transfection System according to the manufacturer's instructions (Thermo Fisher Scientific). Functional AHR duplexes were purchased from Thermo Fisher Scientific under references: HSS100337, HSS100336 and HSS100338. Control silencing RNA was also purchased from Thermo Fisher Scientific under reference: 465377. siRNA-electroporated cells were cultured in six-well plates at 37°C under 5% CO2 in a humid chamber for 24 h prior to treatment.
Plasmids
The following mammalian plasmids pCMV6-AC-GFP and pCMV6-AC-HK2-GFP were purchased from ORIGENE with references PS100010 and RG209482 respectively. pCMV6-AC-HK2-GFP encodes a functional full-length human HK2 protein fused to TurboGFP in its C-terminus. The plasmid backbone further encodes a G418/neomycin selection cassette.
To establish the U2OS-GFP and U2OS-GFP-HK2 cell lines, U2OS cells were transfected with pCMV6-AC-GFP and pCMV6-AC-HK2-GFP respectively, with Neon Transfection System according to the manufacturer's instructions (Thermo Fisher Scientific). Electroporated cells were exposed to a range of G418 from 0.3 to 0.6 mg/ml (Thermo Fisher Scientific, 10131027) and the appropriate concentration empirically determined. 0.6 mg/ml was deemed appropriate to maintain U2OS-GFP-HK2 cells, and 0.4 mg/ml for U2OS-GFP cells.
A similar approach was utilised to produce 143B osteosarcoma cells with stable expression of GFP and GFP-HK2.
pH measurement
The level of media acidification reached by the U2OS-GFP and U2OS-GFP-HK2 cell lines were monitored using a FiveEasy™ pH meter (Mettler Toledo). One million cells were seeded in regular media and allowed to attach for 6 h to the plate. Then, the cell culture media were replaced with fresh media and the pH of the cells’ supernatant measured 4 h later (n = 6).
Cell proliferation assay
Fifty thousand cells were seeded in six-well plates. Cells were then numbered 7 days later with a Countess™ II FL automated cell counter (Thermo Fisher Scientific). Trypan blue (Amresco, K940) staining was used to discriminate living and dead cells in six independent experiments.
Proteomic analysis
Digestion
U2OS-GFP-HK2 and U2OS cell pellets (four samples per group) were lysed during 5 min at 95°C in 100 mM Tris/HCl pH8.5, 2% SDS. Protein concentration was determined using SDS PAGE of an aliquot and Imagelab software (Bio-Rad Laboratories). 50 μg of proteins from each lysate were reduced and alkylated with 10 mM tris(2-carboxyethyl)phosphine (TCEP) and 50 mM chloroacetamide for 5 min at 95°C. After cooling to room temperature, extracts were diluted with 300 μl 8 M urea, 50mM Tris/HCl pH 8.5, transferred onto 30 kDa centrifugal filters and prepared for FASP digestion as described previously (39). Proteins were digested overnight at 37°C with 1μg trypsin (V511A; Promega).
Peptide desalting
Peptides were desalted on C18 StageTips, manufactured by stacking six layers of C18 reverse-phase from a disk of 3M Empore Octadecyl C18 High Performance Extraction Disk into a 200 μl micropipet tip.
Peptide fractionation
Peptides were then separated in five fractions using strong cation exchange (SCX) resin (40). Briefly, peptides were loaded into pipette-tip columns made by stacking six layers of a 3M Empore cation extraction disk into a 200 μl micropipet tip. Column conditioning was performed using acetonitrile (ACN). We used 0.1% Trifluoroacetic acid (TFA) for column equilibration. Samples acidified with TFA were loaded on the column and washed with 0.1% TFA. Peptides were finally successively eluted using 20% ACN, 0.05% formic acid, ammonium acetate at 75, 125, 200, 300 mM. The 5th fraction was eluted in 1.4% NH4OH, 80% ACN.
LC–MS/MS
After speed-vacuum drying, fractions were solubilized in 10 μl of 0.1% TFA, 10% ACN. Liquid chromatography and mass spectrometry analyses were performed on an U3000 RSLC nanoflow-HPLC system coupled to a Q-Exactive Orbitrap mass spectrometer (both from Thermo Fisher Scientific). 1 μl of each fraction were concentrated and washed on a C18 reverse-phase precolumn (3 μm particle size, 100 Å pore size, 75 μm inner diameter, 2 cm length, Thermo Fischer Scientific), then separated using a C18 reverse-phase analytical column (2 μm particle size, 100 Å pore size, 75 μm inner diameter, 25 cm length from Thermo Fischer Scientific) with a 3 h gradient starting from 99% of solvent A (0.1% formic acid) to 55% of solvent B (80% ACN and 0.085% formic acid). The mass spectrometer acquired data throughout the elution process and operated in a data-dependent scheme with full MS scans acquired, followed by up to 10 successive MS/MS HCD-fragmentations on the most abundant ions detected. Settings for Q-Exactive were: full MS automated gain control (AGC) target 1.106 with 60 ms maximum ion injection time (MIIT) and resolution of 70 000. The MS scans spanned from 350 to 1500 Th. Precursor selection window was set at 2 Th. HCD normalized collision energy (NCE) was set at 27% and MS/MS scan resolution was set at 17 500 with AGC target 1.105 within 60ms MIIT. Dynamic exclusion time was set to 30 s and spectra were recorded in profile mode.
Identification and quantification
The mass spectrometry data were analyzed using Maxquant version 1.6.2.6 (41). The database used was a concatenation of human sequences from the Uniprot-Swissprot database (Uniprot, release 2018-06) and an incremented list of contaminants. The enzyme specificity was trypsin. The precursor mass tolerance was set to 4.5 ppm and the fragment mass tolerance to 20 ppm. Carbamidomethylation of cysteins was set as constant modification and acetylation of protein N-terminus and oxidation of methionines were set as variable modifications. Second peptide search was allowed and minimal length of peptides was set at seven amino acids. False discovery rate (FDR) was kept below 1% on both peptides and proteins. Label-free protein quantification (LFQ) was done using both unique and razor peptides. At least two ratio counts were required for LFQ. All experiments were analyzed simultaneously with the ‘match between runs’ option with a match time window of 0.7 min and an alignment time window of 20 min.
Proteome data analysis
Using Perseus software (version 1.6.2.3) (42) false proteins discovery were filtered out, to wit proteins that match with contaminant, to the reverse database and proteins identified only with modified peptide, leading to a matrix of 7278 proteins. LFQ intensity data were transformed into log2 and a Student's t-test was performed using the Benjamini–Hochberg false discovery rate to identify the differentially expressed proteins. Proteins present in at least 3/4 replicates only in U2OS or U2OS-GFP-HK2 cells were manually given the scores: P-value = 0 and |log ratio| = 10 (i.e. log2(mean LFQ intensity in U2OS) – log2(mean LFQ intensity in U2OS-GFP-HK2)). Then Canonical Pathways and Upstream Regulator analyses were generated through the use of Ingenuity Pathway Analysis software IPA (QIAGEN, https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) version 65367011, considering proteins with a cut-off P-value <0.001 and an absolute |log ratio| > 1.
Cytoscape visualization
The list including the differentially regulated proteins belonging to the AHR signalling pathway, the Upstream Regulators predicted in this pathway and their target proteins was submitted into Cytoscape software (v.3.8.2) for network visualisation (43). Proteins of the ‘AHR signalling’ were manually highlighted by bold lines. Proteins were colour coded according to their log ratio between U2OS and U2OS-GFP-HK2 cells.
Tryptophan and other metabolites quantification
Metabolites from 2 millions of thawed cells were extracted using a methanol/chloroform extraction method with minor modifications (44). Cold methanol/chloroform (2:1, v/v; 1.5 ml) was added to cell pellets and homogenized on ice. The sample tube was centrifuged at 15 000g for 10 min at 4°C, and the supernatant was transferred to a new sample tube through a 70-mm cell strainer. Ice-cold water (0.6 ml) was added, and the sample tube was vortexed and centrifuged (15 000g, 5 min, 4°C) to obtain phase separation. The upper and lower phases were separately collected in fresh sample tubes with a syringe, taking care not to disturb the interface. The polar (upper) phase (500 ml) was evaporated to dryness in a SpeedVac concentrator (Savant), and then was reconstituted in 100 μl of methanol/water (1:1, v/v). Extracted metabolites were stored at –80°C until analysis. Tryptophan (TRP) and TRP metabolites were measured via HPLC using a coulometric electrode array (ESA Coultronics, ESA Laboratories) and fluorometric detection. Separation of TRP metabolites were done by reversed-phase liquid chromatography using a 20 mM NaH2PO4 buffer (not pH adjusted) with 5.0% acetonitrile. The mobile phase was delivered by an HPLC pump (Shimadzu) through a Supelcosil™ LC-18 column (250 mm × 4.6 mm, 5 μm, Supelco) at a rate of 1 ml/min. Quantifications were performed by referencing calibration curves obtained with internal standards.
Cancer Cell Line Encyclopedia (CCLE) data
RNA sequencing data were downloaded from the authors’ manuscript through the cBioPortal web interface (https://www.cbioportal.org) and retrieved in reads per kilobase per million mapped reads (RPKM) (35,45). Illumina 450K methylation data and reduced-representation bisulfite sequencing (RRBS) data of the AHR promoter in osteosarcoma cell lines were retrieved using the CellMiner Cross-Database (CDB) web interface (https://discover.nci.nih.gov/cellminercdb/) (46). Only CpG located in the AHR CpG islands with coverage >5 were exported for RRBS data.
Methylated-DNA immunoprecipitation (MeDIP)
Genomic DNA were extracted and purified using a standard phenol/chloroform procedure (36). Methylated DNA immunoprecipitation (MeDIP) was performed with the MagMeDIP kit (Diagenode; C02010021) according to the supplier recommendations starting from 1.2 μg of genomic DNA sonicated to 200–400 bp using a Bioruptor® PLUS sonicator (Diagenode). qPCR analysis was performed using the kit LightCycler® 480 SYBR Green I Master (Roche Diagnostics; 04887352001) according to manufacturer recommendations on a Roche LightCycler® 480 system (software version 1.5.1.62 SP3). Quality of the MeDIP procedure was monitored using control primers for methylated DNA (TSH2B gene) and for unmethylated DNA (GAPDH promoter) provided by the MagMeDIP kit manufacturer (Diagenode). AHR promoter methylation was assessed using the following primers: 5′-GAC CGC CAG CTC AGA ACA G-3′ and 5′-CTC CCA GCT TCC GTT CGG-3′ (coordinates chr7:17338323–17338373 on hg19) and data are presented as percentage of input DNA recovered after MeDIP (i.e. DNA methylation level).
The Cancer Genome Atlas (TCGA) data
Gene expression, DNA methylation and clinical data in 33 cancer types were obtained from the TCGA and cBioPortal data portal (45,47,48). Correlations were evaluated with Pearson coefficient values. High and low HK2 expression groups were defined using the median expression value of HK2 in each TCGA cancer type. High and low AHR expression and promoter methylation level groups were defined using the same criteria. All data used in this study were freely available, freely re-usable and patients’ information anonymized. Adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS) and uveal melanoma (UVM).
Osteosarcoma patients’ data
Gene expression values were retrieved from the National Cancer Institute (NCI) Therapeutically Applicable Research to Generate Effective Treatments (TARGET) Osteosarcoma (OS) project (accession studies phs000218 and phs000468) and from GSE21257 (49). TARGET Osteosarcoma datasets are available without restrictions on their use in publications since 2019.
Statistical analysis
All the data points as well as the different statistical tests are available in Supplementary Table S5. All error bars represent standard deviation. Student's t-tests and hypergeometric tests were performed. P < 0.05 was considered significant. *P < 0.05; **P < 0.01 and ***P < 0.001. n.s., not significant.
RESULTS
A genomic analysis of AHR binding sites identifies Hexokinase 2, HK2, as a potential transcriptional target of AHR
To identify key AHR signalling targets in human cells, we retrieved AHR binding sites identified by chromatin immunoprecipitation followed by deep-sequencing (ChIP-sequencing) in three human cancer cell types. We compared the lists of AHR targets identified from human hepatocellular carcinoma cells (HepG2), from human breast cancer cells (MCF-7) exposed to 10 nM TCDD for 45 min or 24 h or vehicle alone, and from human EBV-immortalised lymphocytes (GM17212) treated with 3-methylcholanthrene (3-MC) at 1 μM for 24 h or vehicle alone (5,6,31). As previously described and discussed by the authors, we observed that most AHR binding sites are detectable in non-treated conditions in these cell lines, possibly due to the chronic activation of AHR by endogenous ligands (5,6). We identified 1314 AHR-bound genes common between the different ChIP-sequencing datasets and experimental conditions (Figure 1A). We performed a functional annotation of this list of genes using the PANTHER GO-Slim Biological Process database (32) and observed that a large proportion of AHR target genes encode proteins involved in metabolic pathways and processes (Figure 1B and Supplementary Table S1).
Figure 1.
HK2 is a genomic target of AHR. (A) Venn diagram showing the intersection of AHR-bound genes between AHR ChIP-sequencing analyses in MCF-7 breast cancer cells (vehicle and TCDD-treated for 45 min and 24 h), in GM17212 lymphoblastoid cells exposed to vehicle and 3-MC (24 h) and in HepG2 hepatocellular carcinoma cells. Data were retrieved from publicly available datasets deposited at NCBI GEO under accession numbers GSE90550, GSE116638 and GSE127649. (B) Functional classification in biological processes of common AHR targets (n = 1314) using the PANTHER GO-Slim Biological Process database. (C–E) Snapshot of ChIP-sequencing data showing AHR binding sites at the HK2 locus (C) in MCF-7 cells treated with TCDD (45 min) compared to vehicle (DMSO) treatment, (D) in GM17212 cells exposed to 3-MC for 24 h or DMSO and (E) in HepG2 cells. The location of CpG island (in green), promoters (in red) as well as enhancers (in grey) from the GeneHancer database are indicated. (F) ChIP-qPCR characterisation of AHR binding sites at the CYP1A1 promoter and the HK2 locus in U2OS cells treated with vehicle (i.e. toluene) or 10 nM TCDD for 1 h and 24 h (n = 3). P-value as determined by paired Student's t-test (vehicle versus TCDD). *P < 0.05; ***P < 0.001.
We notably observed that HK2, a key cancer gene, is an AHR target in all experimental conditions of the three studies which led us to further explore this AHR/HK2 axis. In MCF-7 cells, AHR binding is detectable in a cluster of cis-regulatory elements in the vicinity of HK2 exon 3 (i.e. introns 2 and 3; that we called A, B, C and D cis-regulatory elements hereafter) and the binding is enhanced after 45 min of TCDD exposure (Figure 1C). In GM17212 and HepG2 cells, AHR ChIP-sequencing profile at the HK2 loci is different: AHR binds at the HK2 promoter and no binding is detectable around exon 3 in all conditions (Figure 1D, E). Consistent with these ChIP-sequencing data, a search for AHR DNA binding consensus sequences confirmed the presence of putative AHR-response elements in the promoter of HK2 and in the cis-regulatory regions A and D near exon 3 (Supplementary Figure S1A). In addition, profiling of ChIP- and DNAseI-sequencing data in MCF-7 indicates that cis-regulatory regions A–D are located in an enhancer region of HK2 as illustrated by the presence of DNAseI hyper-sensitive sites, enrichment for enhancer-associated histone marks and binding sites for transcription factors including FOXA1 and cMYC, which are transcriptional co-factors of AHR (50,51) (Figure 1C and Supplementary Figure S1B). These observations support the hypothesis that these AHR-bound genomic sites are functional cis-regulatory elements of HK2.
To confirm that HK2 is a bona-fide target of AHR, we treated U2OS human osteosarcoma cells with 10 nM TCDD or vehicle for 1 h and 24 h before assessing the binding of AHR at genomic sites nearby exon 3 (i.e. regions A, B, C and D), at the HK2 promoter as well as at negative control regions located ±2 kb from the promoter. As an additional control we also monitored AHR binding at the CYP1A1 promoter, commonly assessed as a read-out of AHR transcriptional activity (3). Importantly, using histone marks ChIP-sequencing data produced in U2OS cells, we could confirm the location of HK2 enhancers, although only AHR-binding sites A, B and C are enriched for enhancer-associated histone marks (Supplementary Figure S1C and D). In U2OS cells treated with vehicle, we did not observe significant AHR binding at the CYP1A1 and HK2 promoters, nor at the other regions tested (Figure 1F). By contrast, in TCDD-treated cells we observed a significant binding of AHR at the CYP1A1 promoter 1 h and 24 h after TCDD treatment as well as a significant enrichment of AHR at the HK2 promoter and nearby exon 3 in cis-regulatory region C 1 h post TCDD-treatment, and in regions B and C 24 h post TCDD-treatment (Figure 1F). No AHR ChIP-qPCR enrichment is detected at negative control sites (i.e. ±2 kb from HK2 promoter) monitored in our assays, nor at regions A and D, at either time points (Figure 1F). These results demonstrate that in U2OS cells, AHR binds to the HK2 promoter and several cis-regulatory elements near exon 3 in response to TCDD exposure.
Collectively, our data demonstrate that the transcription factor AHR binds to HK2 gene in different human cell types.
Agonists of AHR promote HK2 expression in human cells and mice lung tissue
We next investigated whether HK2 expression was regulated in response to AHR agonists exposure. To do so, we treated human U2OS cancer cells with TCDD and monitored by RT-qPCR and western blot the expression level of HK2. We observed that HK2 mRNA and protein levels are significantly increased at 48-h post-treatment with 1 and 10 nM TCDD compared to control cells exposed to vehicle alone (Figure 2A, B). In addition, we observed that HK2 expression was up-regulated as soon as 24 h post-TCDD (10 nM) treatment (Supplementary Figure S2A). We also assessed HK2 expression level upon TCDD treatment in the colon cancer cells HCT116 and we observed that TCDD exposure significantly increases HK2 expression at mRNA and protein levels (Figure 2C, D). We next assessed whether other AHR agonists could promote HK2 expression. We treated U2OS cells with benzo[a]pyrene (BaP) or with 6-formylindolo[3,2-b]carbazole (FICZ) at different concentrations and for different durations. We observed that HK2 expression was up-regulated in a time- and concentration-dependent manner upon BaP treatment at mRNA and protein levels (Figure 2E, F, Supplementary Figure S2B). Upon FICZ treatment, HK2 mRNA expression was slightly up-regulated but no difference was detectable at protein level compared to vehicle-treated cells (Figure 2G and Supplementary Figure S2C, D). Altogether, these data indicate that different agonists of AHR induce HK2 expression in human cells in a time- and concentration-dependent manner, although the kinetic and amplitude of induction are quite variable.
We also investigated whether HK2 expression was regulated upon TCDD exposure in vivo, in tissues expressing high levels of AHR, such as the lung (18). To do that, we studied Hk2 expression by RT-qPCR and western blot in lung tissues from adult male mice exposed to TCDD in utero and during lactation. We observed significant higher Hk2 mRNA and protein levels compared to vehicle-treated mice tissues (Figure 2H). Hk2 is thus also regulated by TCDD exposure in vivo.
Finally, to gain a broader insight into the association between modulators of AHR activity and the expression of HK2, we interrogated the Comparative Toxicogenomics Database (CTD) (38). Among the 266 chemicals (or mixtures) interactions reported to modulate AHR activity in human cells, we identified 18 compounds that also regulate HK2 expression (fold enrichment: 8.05; P(X = 18) = 8.06 × 10–12) (Supplementary Figure S2E and Table S2). Among these associations, we retrieved complex chemical mixtures such as cigarette smoke as well as several HAH and PAH compounds. This data mining analysis further supports that HK2 is transcriptionally regulated by several modulators of AHR pathway in human cells, although it appears context (agonist- and cell type-) dependent.
AHR regulates HK2 mRNA level upon activation by TCDD
We next investigated whether AHR activity was involved in the regulation of HK2 expression upon AHR agonists exposure. We first used a competitive inhibitor of AHR called CH-223191 (52). We tested the effect of different concentrations of CH-223191 in U2OS cells on HK2 expression. We did not observe any change in HK2 expression at the mRNA and protein levels with concentrations up to 10 μM for 48 h (Supplementary Figure S2F–G). We then treated U2OS cells with CH-223191, at the time of TCDD treatment, and monitored HK2 and CYP1A1 expression. We observed that both genes were induced by TCDD as expected, and that co-treatment with CH-223191 abolishes their induction (Figure 2I).
To strengthen these observations, we next transfected U2OS cells with AHR small interfering RNAs (siRNAs) and 24 h later treated the cells with 10 nM TCDD for 48 h. RT-qPCR analyses indicated low levels of AHR expression in U2OS cells and confirmed the knock-down of AHR expression (Figure 2J). We also observed that depletion of AHR prevents HK2 mRNA up-regulation in response to TCDD treatment, while it has no detectable effect on HK2 mRNA expression in control treated cells (Figure 2J). All these results show that AHR regulates HK2 expression in response to its activation by TCDD.
Over-expression of HK2 promotes an imbalance in AHR signalling pathway proteins abundance
We then investigated the consequences of HK2 over-expression in human cells by establishing U2OS cells stably expressing either GFP-HK2 (U2OS-GFP-HK2 cells) or GFP alone as control (U2OS-GFP cells). By western blot, we observed that 48 h post-transfection, GFP-HK2 is expressed at its expected size in U2OS-GFP-HK2 cells (Supplementary Figure S3A). Numerous studies have previously reported, notably in osteosarcoma cells (53,54), that the stable over-expression of HK2 increases culture media acidification (due to increased release of lactate, an end-product of glycolysis) and cell proliferation. We thus monitored these two parameters. We confirmed that the pH of the culture medium of the U2OS-GFP-HK2 cells is lower than that of U2OS-GFP cells (Supplementary Figure S3B) and that expression of GFP-HK2 promotes cell proliferation compared to GFP alone (Supplementary Figure S3C). These results validate that GFP-HK2 is a functional protein in our experimental conditions.
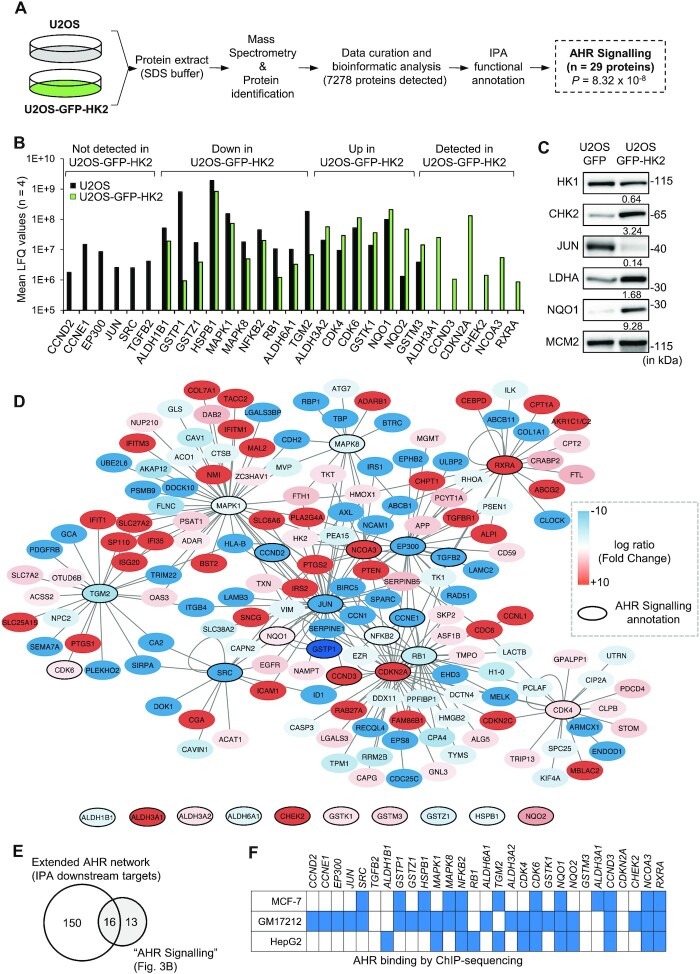
We next wanted to get deeper insight into the signalling and molecular pathways regulated by HK2 over-expression. We used a quantitative proteomic approach to characterise and compare the proteome of U2OS and U2OS-GFP-HK2 cells (Figure 3A). We detected 7278 proteins by mass spectrometry and as anticipated, we observed that the amount of HK2 is significantly higher in U2OS-GFP-HK2 compared to U2OS cells (P < 0.001) (Supplementary Figure S3D). We also confirmed previous reports (27) showing that HK2 over-expression promotes an imbalance in glycolytic enzymes abundance with for instance lower levels of HK1 (P < 0.001) and GAPDH (P < 0.001), and heightened level of LDHA (P < 0.001) (Supplementary Figure S3E). Following downstream statistical analyses, we identified the differentially expressed proteins (|log ratio| > 1 and P-value < 0.001) between the two cell lines. A functional annotation of this list using the QIAGEN Ingenuity Pathway Analysis (IPA) software revealed several pathways deregulated upon HK2 over-expression, among which ‘AHR signalling’ (P = 8.32 × 10–8) with 29 associated proteins. This list includes proteins involved in phase-I and phase-II detoxification mechanisms (ALDH1B1, ALDH3A1, ALDH3A2, ALDH6A1, GSTK1, GSTM3, GSTP1, GSTZ1, NQO1 and NQO2); cell cycle regulators (CCND2, CCND3, CCNE1, CDK4, CDK6, CDKN2A, CHK2 and RB); chromatin factors (EP300, JUN, NCOA3 and NFKB2) and signalling proteins (HSPB1, MAPK1, MAPK8, RXRA, SRC, TGFB2 and TGM2) (Figure 3B). Of note, additional proteins of the AHR network were detected in our proteomic analysis although not robustly enough to be considered by IPA, including most of the CYP enzymes and AHR itself (Supplementary Table S3). As an orthogonal validation of the proteomic data, we performed western blot analyses. We validated that HK2 over-expression leads to the down-regulation of HK1 and JUN and the up-regulation of LDHA, CHK2 and NQO1 at protein levels in U2OS-GFP-HK2 compared to U2OS-GFP cells (Figure 3C).
Figure 3.
Over-expression of HK2 alters the abundance of a subset of proteins involved in the AHR network. (A) Schema of the protocol used to characterise U2OS and U2OS-GFP-HK2 proteomes. Four samples of each cell lines were analysed by mass spectrometry. (B) Mean of label free quantification (LFQ) values of proteins annotated by Ingenuity Pathway Analysis (IPA) software as belonging to the ‘AHR signalling pathway’ and identified in the proteomic approach with significant changes in abundance between U2OS and U2OS-GFP-HK2 cells (n = 4). Targets are grouped in four classes according to their abundance levels in U2OS and U2OS-GFP-HK2 cells. (C) Western blot analysis of a subset of proteins identified in the proteomic analysis and belonging to the ‘AHR signalling pathway’. Relative quantifications between cell lines are indicated below the blots (relative to MCM2 levels which is not differentially expressed between U2OS and U2OS-GFP-HK2 cells according to the proteomic data). (D) Cytoscape visualization of proteins deregulated in U2OS-GFP-HK2 compared to U2OS cells and annotated as ‘AHR signalling pathway’ or ‘downstream targets’ (i.e targets of predicted Upstream Regulators) using the IPA software. Proteins are colour-coded with up-regulated proteins in U2OS-GFP-HK2 depicted in red and proteins down-regulated depicted in blue. Proteins annotated as ‘AHR signalling pathway’ are highlighted by bold lines. (E) Venn diagram showing the overlap between ‘AHR signalling pathway’ proteins and ‘downstream targets’. (F) Genes encoding proteins of the HK2/AHR network identified as AHR genomic targets in MCF-7, GM17212 and HepG2 cells (either in presence or absence of agonist). Blue indicates the presence of an AHR binding sites in the vicinity of the promoter and/or in the coding sequence of the gene.
Our analysis, using the IPA software, also indicated that 12 of these 29 proteins are predicted as ‘upstream regulators’, suggesting that they potentially regulate additional proteins identified as mis-regulated by HK2 over-expression (Supplementary Table S4). For instance, CDKN2A is predicted to regulate 32 proteins regulated by HK2 over-expression in our analysis (P = 1.52 × 10–8). This complementary analysis led us to identify 166 downstream proteins including 16 which are annotated as belonging to ‘AHR signalling’ (Figure 3D, E). We also observed that 24 of the 29 genes coding the proteins of the HK2/AHR signalling were previously identified as AHR ChIP targets in other cell lines such as MCF-7, HepG2 or GM17212 cells (Figure 3F). These correlative associations suggest that HK2 might actually regulate AHR signalling at different levels and that several proteins of the HK2/AHR network might cross-regulate each other functions.
Over-expression of HK2 enhances a subset of AHR transcriptional target genes expression
Based on our proteomics findings, we tested whether AHR transcriptional activity was altered in U2OS-GFP-HK2 cells. To do so, we compared by RT-qPCR the expression level of prototypical AHR target genes CYP1A1, CYP1B1 and ALDH3A1 between U2OS-GFP and U2OS-GFP-HK2 cells. We observed higher expression levels of CYP1B1 and ALDH3A1 in U2OS-GFP-HK2 cells compared to U2OS-GFP, while the level of expression of CYP1A1 was similar between the two cell lines (Figure 4A). We repeated such analyses in another osteosarcoma cell line, 143B, stably expressing GFP or GFP-HK2. We could confirm the expression of the GFP-HK2 fusion protein in these cells (Supplementary Figure S3F) and observed again gene-specific effects, with increased expression of CYP1B1 in 143B-GFP-HK2 cells compared to 143B-GFP control cells, while ALDH3A1 expression was not detected in both cell lines and CYP1A1 expression was similar between the two cell lines (Figure 4B). These analyses revealed that over-expression of HK2 and subsequent perturbation of the AHR network alters the expression levels of a subset of AHR target genes in both cell lines.
We wondered whether these changes in gene expression could be a consequence of a change in the intracellular abundance of known endogenous ligands of AHR such as tryptophan (TRP) derivatives (11,12). We observed no major difference in the levels of 3-OH-TRP, TRP, kynurenic acid and 3-OH-kynurenine between U2OS-GFP and U2OS-GFP-HK2 cells, arguing against such a scenario (Supplementary Figure S4).
Over-expression of HK2 promotes a better response to AHR agonists in specific cell lines
We then investigated whether HK2 over-expression might also alter the response to AHR agonists. To do so, we treated cells over-expressing GFP and GFP-HK2 with BaP or FICZ and monitored by RT-qPCR the expression level of CYP1A1, CYP1B1 and ALDH3A1. We observed that the induction of each gene was markedly higher in U2OS-GFP-HK2 cells compared to U2OS-GFP cells upon BaP or FICZ treatment (Figure 4C and Supplementary Figure S5A). HK2 over-expression is thus potentiating AHR transcriptional activity in U2OS cells, in response to agonists. Intriguingly, we did not observe a similar effect in the 143B cell line. We observed that the induction of CYP1A1 mRNA expression in response to BaP and FICZ treatments is similar in 143B-GFP and in 143B-GFP-HK2 (Figure 4D and Supplementary Figure S5B). In the case of CYP1B1, the gene was induced by BaP and FICZ and levels of expression were much higher in 143B-GFP-HK2 cells upon treatments compared to 143B-GFP cells. Nonetheless, since the CYP1B1 gene was already expressed at higher levels in untreated 143B-GFP-HK2 cells, the amplitude of induction was actually similar in the two cell lines (Figure 4D and Supplementary Figure S5B).
To confirm that the up-regulation of these AHR prototypical target genes is mediated by AHR, we co-treated U2OS-GFP-HK2 cells with BaP and CH-223191. We observed that CH-223191 limits the induction of CYP1A1, CYP1B1 and ALDH3A1 upon BaP treatment, consistent with AHR involvement (Figure 4E). We also tested whether inhibition of hexokinase activity would affect the expression of these AHR targets upon BaP treatment in U2OS-GFP-HK2 cells. We co-treated these cells with BaP and with 2-deoxy-d-glucose (2-DG), an allosteric inhibitor of hexokinase activity (55). In BaP + 2-DG co-treated cells, we did not observe lower expression levels of CYP1A1, CYP1B1 and ALDH3A1 compared to BaP treated cells (Figure 4F). These data indicate that 2-DG treatment does not block the induction of AHR target genes upon BaP exposure.
All these data show that over-expression of HK2 amplifies the transcriptional levels of AHR target genes in response to AHR agonists, and that this response is AHR dependent and not prevented by 2-DG treatment.
Over-expression of HK2 is associated with the DNA demethylation of AHR promoter and higher AHR expression in U2OS cells
We hypothesize that the cellular context, including AHR expression level, might explain the differential regulation of AHR activity in the two osteosarcoma cell lines. We thus monitored AHR expression by RT-qPCR in the different U2OS and 143B cell lines we established (Figure 5A, B). We detected higher AHR mRNA levels in U2OS-GFP-HK2 compared to respective control cells while AHR levels were quite similar between 143B, 143B-GFP and 143B-GFP-HK2 cells (Figure 5A, B). This observation suggests that a dramatic change in AHR transcriptional status between U2OS/U2OS-GFP and U2OS-GFP-HK2 cells is linked to the dramatic change in AHR transcriptional outcomes in response to agonists exposure. Importantly, by western blot, we confirmed higher levels of AHR protein in U2OS-GFP-HK2 cells compared to control cells (Figure 5C).
As AHR promoter contains CpG sequences susceptible to DNA methylation (19), we investigated whether DNA methylation was involved in the regulation of AHR expression. We first assessed the methylation status and the expression level of AHR in a panel of osteosarcoma cells using the Cancer Cell Line Encyclopedia (CCLE) datasets (35,46). We observed that the methylation of AHR promoter is negatively correlated with AHR mRNA expression in this set of cell lines and that in U2OS cells AHR promoter is highly methylated and AHR mRNA expression is low (Figure 5D) (data are not available for the 143B cell line in this database). The methylation of AHR promoter in U2OS cells was further confirmed by interrogating single CpG methylation analyses from the CCLE (Figure 5E). We thus treated U2OS, U2OS-GFP and U2OS-GFP-HK2 cells with DNA demethylating agent 5-aza-2′-deoxycytidine for 48 h and monitored AHR expression by RT-qPCR. We detected AHR expression in 5-aza-2′-deoxycytidine-treated U2OS and U2OS-GFP cells at 48 h while it was hardly detectable in vehicle-treated cells. In contrast, expression of AHR was detectable in U2OS-GFP-HK2 cells in treated and untreated conditions, at roughly similar levels (Figure 5F). These results indicate that AHR expression is regulated by DNA methylation in U2OS and U2OS-GFP cells. Consistent with this conclusion, RT-qPCR analyses showed that in 143B, 143B-GFP and 143B-GFP-HK2 cells, which already express AHR in untreated conditions, 5-aza-2′-deoxycytidine treatment for 48 h moderately increases AHR expression (Figure 5G).
To further confirm the role of DNA methylation in the regulation of AHR expression we assessed the methylation status of the AHR promoter by methylated-DNA immunoprecipitation (MeDIP) in U2OS, 143B and their derivative cell lines (Figure 5H, I). As controls, we used GAPDH promoter as an unmethylated region and TSH2B promoter as a heavily methylated region as indicated by the manufacturer (Diagenode). The methylation status of control regions was in line with what was expected (Figure 5H, I). For the AHR promoter region we observed high levels of CpG methylation in U2OS and U2OS-GFP cells and low methylation levels in U2OS-GFP-HK2 cells as well as in 143B, 143B-GFP and 143B-GFP-HK2 cells (Figure 5H, I). These data indicate that over-expression of HK2 is associated with the demethylation of the AHR promoter in U2OS cells which may explain the enhanced expression of AHR.
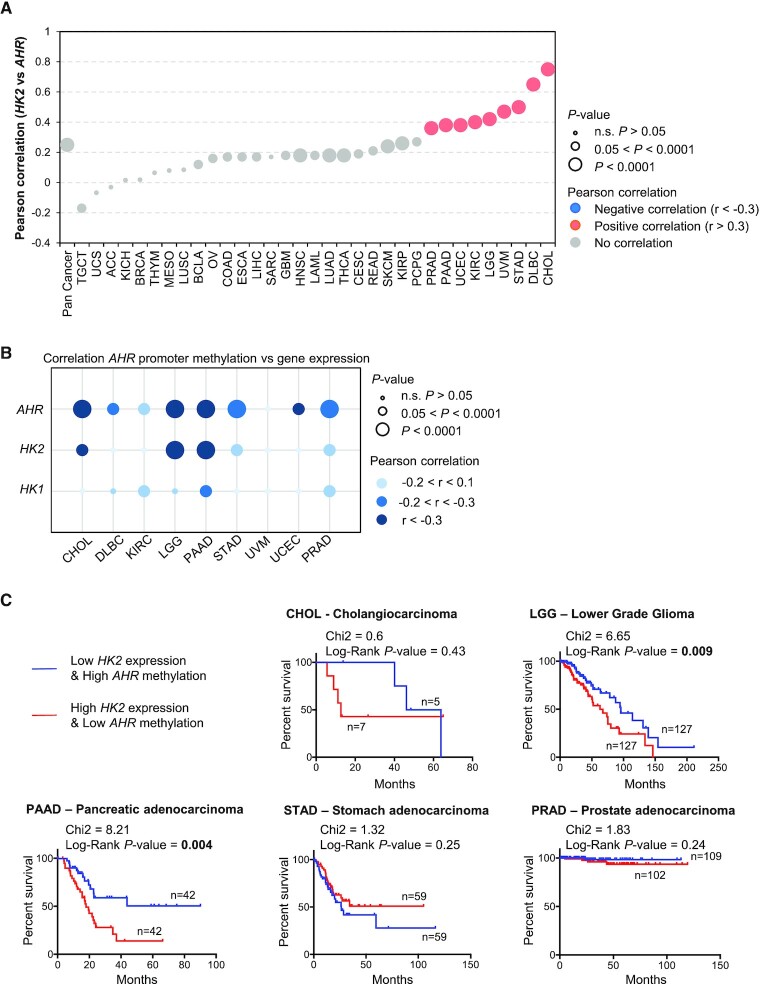
HK2 expression negatively correlates with the methylation of AHR promoter in several cancers
We next explored the relevance of the HK2/AHR axis in cancer by investigating the correlation between the expression level of HK2 and the expression level of AHR using the data of 33 types of cancer available through The Cancer Genome Atlas (TCGA) (47,48,56). We found that HK2 and AHR mRNA expression levels are positively correlated in cancer, notably in nine cancer types (Figure 6A). In addition to TCGA datasets, we also analysed two different osteosarcoma (OS) cohorts (NCI/TARGET/OS and GSE21257) publicly available and commonly used by the community (49). We found no significant correlation between AHR and HK2 mRNA expression in OS tumours in the two cohorts (Supplementary Figure S6).
Figure 6.
Relationship between the expression of HK2 and the methylation of AHR promoter in cancer. (A) Relationship between the expression levels of HK2 and AHR retrieved in 33 different types of cancer from the TCGA. Significant correlation, Pearson r coefficient >0.3 or <–0.3 are indicated in colours (blue, negative correlation; salmon, positive correlation) (B) Relationship between the expression levels of AHR, HK2 and HK1 and the methylation level of AHR promoter in cancers. (C) Prognostic value of the AHR/HK2 axis in five different types of cancer exhibiting a negative correlation between HK2 expression and AHR promoter methylation.
Focusing on the nine cancer types exhibiting a clear correlation between HK2 and AHR expression, we then investigated a possible correlation between HK2 expression and the methylation of AHR promoter. We observed a significant negative correlation between HK2 levels and AHR promoter methylation in cholangio-carcinoma (CHOL), lower grade glioma (LGG), pancreatic cancer (PAAD), stomach cancer (STAD) and prostate cancer (PRAD) (Figure 6B). Furthermore, we did not find such correlation between the expression level of HK1 and the methylation of AHR, indicating some level of specificity (Figure 6B). Finally, in lower grade glioma (LGG) and pancreatic cancer (PAAD), tumours exhibiting high levels of HK2 and low levels of AHR promoter methylation are correlated with shorter overall survival (Figure 6C).
These data indicate that HK2 expression negatively correlates with the methylation of AHR promoter in specific cancer types, which may have clinical implication.
DISCUSSION
It is well recognized that AHR signalling activation has variable effects on the transcription of its targets. Nonetheless, the details of the proteins and pathways involved in the regulation of AHR signalling activity are still far from complete and it might depend on features proper to each cellular context such as possible genetic mutations, exposure to exogenous cues and stage of cell differentiation (7). In here, we provide evidences that HK2 is a transcriptional target of AHR, as suggested by previous large-scale gene expression studies (22,57–59), and studies indicating that AHR might be an important regulator of glycolytic genes expression and glycolysis end-points (3,7,51,60–62). In MCF-7 cells, we identified a cluster of AHR binding sites in the vicinity of exon 3 of HK2 gene; and binding of AHR at the promoter of HK2 in two other cell lines GM17212 and HepG2. Using a ChIP-qPCR analysis we demonstrated similar AHR binding sites in U2OS cells treated with TCDD including binding sites nearby exon 3 and at the promoter of HK2. The binding at the promoter is detected 1 h after TCDD addition, but not at a later time point (24 h), indicating a transient binding of AHR at HK2 promoter. In contrast, the binding nearby exon 3, and at the CYP1A1 promoter, is observed up to 24 h and might reflect cis-regulatory elements mediating long-term and persistent effects of TCDD. It would thus be interesting to further dissect the role of these different cis-regulatory elements in the HK2 locus to better define their functions in the regulation of HK2 expression upon AHR agonists exposure. We indeed showed that in response to AHR agonists exposure, HK2 expression is enhanced, although the fold-increase is quite variable depending on the duration of the exposure, the concentration and the nature of the ligand. All these findings highlight that the regulation of HK2 expression by AHR is context-specific and might rely on the binding of additional cofactors of AHR, either at the promoter or nearby exon 3 (1,50,51). Using ENCODE ChIP-sequencing datasets we identified binding sites for cMYC and FOXO factors which may cooperate with AHR in the regulation of HK2 expression (50,51). The context-specific regulation of HK2 by AHR may alternatively be explained by our findings that HK2 regulates AHR transcriptional activity and thus that AHR or some of its targets might further modulate the kinetic and induction of HK2 expression upon AHR activation. For instance, transglutaminase 2 (TGM2), which is less abundant in U2OS-GFP-HK2 cells compared to controls, is also a known regulator of HK2 expression (63,64). Similarly, retinoblastoma protein 1 (RB1), also affected by over-expression of HK2, is a known partner of AHR in the regulation of gene expression (65). Thus, different layers of regulation, both intrinsic to cell type (i.e. mutations) or exogenous (i.e. environmental cues) may have different outcomes on the HK2/AHR axis and HK2 expression.
By investigating the molecular consequences of HK2 over-expression we observed that numerous proteins contributing to AHR pathway activity, or regulated by this pathway, are mis-regulated upon HK2 over-expression including cell cycle regulators, metabolic enzymes, signalling proteins and AHR targets. Whether additional AHR targets are also regulated by HK2 remains possible as the proteomic approach is not exhaustive. For instance, it has been shown that TCDD regulates expression of RANKL, CXCR4, CXCL2, COX2 and PGE2 in osteosarcoma MG63 cancer cells (66). None of these proteins as well as most of the CYP enzymes and AHR itself, were efficiently detected, quantified and thus analysed in our study. This is most likely due to technical limitations as peptides for some of these proteins were actually detected in a limited number of samples. It is thus unclear whether such classes of AHR targets are also mis-regulated in U2OS-GFP-HK2 compared to control cells. Following up on this proteomic data we unveiled that U2OS-GFP-HK2 cells present heightened expression levels of AHR protein and of its target genes, as well as enhanced response to BaP and FICZ, compared to U2OS-GFP control cells. Intriguingly, in 143B-GFP-HK2 cells we also observed heightened levels of AHR target genes expression under basal condition but did not observe a better response to AHR agonists. There might thus be a direct link between HK2 gene expression and AHR transcriptional activity. In that scenario, expression levels of HK2 might mitigate, or in contrary amplify, the consequences of AHR agonists on cell fate. Intriguingly, it has been shown that glucose can modulate AHR activity in aortic endothelial cells and that pyruvate kinase muscle isoform M2 (PKM2) regulates AHR transcriptional activity (67,68). It is thus likely that HK2 might alter AHR activity at multiple levels, as suggested by our data, to regulate cell biology and fate. Importantly, we observed that 2-DG treatment, which inhibits hexokinase activity and glycolysis, did not block the transcriptional response to AHR agonists, suggesting that HK2 role in glycolysis may have minor effect on the regulation of AHR signalling.
In addition, we documented a function of HK2 on the regulation of the activity of the AHR promoter. It has previously been shown that AHR promoter is regulated by DNA methylation in specific cell types (19). We obtained compelling evidences that over-expression of HK2 leads to AHR promoter demethylation in U2OS cells, and that it correlates with AHR expression level. This observation probably explains the strong response of U2OS-GFP-HK2 cells to AHR agonists tested, such as BaP and FICZ, compared to control cells. While HK2 over-expression triggers DNA demethylation, indicating a link between HK2 and AHR expression we did not clearly pinpoint the mechanism, although we can speculate based on previous studies that DNA demethylation might facilitate the binding of specificity protein 1 (SP1) that activates AHR expression (19,69). Intriguingly, numerous glycolytic enzymes can actually shuttle into the nucleus including glyceraldehyde 3-phosphate dehydrogenase (GAPDH), PKM2 and phosphoglycerate kinase 1 (PGK1) and regulate nuclear processes as diverse as gene expression, DNA replication and chromatin organisation (68,70–72). It is tempting to speculate that HK2 might also shuttle into the nucleus, and as PKM2, regulate the transcriptional activity of AHR or the activity of its promoter. Consistent with such scenario, upon pro-inflammatory stress and glucose deprivation, HK2 translocates in the nucleus of glioblastoma cells and regulates the expression of xanthine oxidoreductase gene in combination with transcription factor NRF2 (73). This remains the sole example of a potential role of human HK2 in the nucleus, needless to say in a quite specific context. Intriguingly, in yeast, low glucose environment also induces the nuclear localisation of the human HK2 homolog, Hxk2, where it regulates gene expression (74). Thus, further studies will be needed to clarify whether HK2 might regulate gene expression in high glucose environment as in our cellular models and how it may control DNA methylation levels at the AHR promoter.
This new axis HK2/AHR might also be important in the context of cancer. Indeed, using TCGA datasets, in 9 of the 33 cancer types that we analysed, we found that HK2 expression shows a positive correlation with AHR expression and in a subset of these 9 cancer types a negative correlation with AHR promoter methylation levels. Furthermore, tumours with high HK2 expression and low AHR methylation exhibit a poor prognosis in lower grade glioma (LGG) and pancreatic cancer (PAAD). Importantly, we did not see similar correlation with HK1 which shares high degree of similarity at the protein level and at the biochemical level with HK2 (26). We also did not document a recurrent binding of AHR at the promoter of HK1 indicating again a preferential relationship between HK2 and AHR. HK1 is expressed in most tissues while HK2 expression is restricted to muscle and adipocytes and it is over-expressed in most cancers. Accordingly, a large body of literature has demonstrated that HK2 is regulated by numerous micro-RNAs and transcription factors including oncogenes (26). Here, we revealed and characterized an additional regulator of HK2 expression, AHR, which can either act as an oncogene or tumour suppressor in different cancer types (13–15). Intriguingly, in osteosarcoma (OS) tumours, we did not observe a correlation between HK2 and AHR expression in two independent cohorts of patients. A possible explanation to this observation might be that OS tumours are quite heterogeneous at the genomic and cellular level and that additional criteria need to be taken into consideration to unveil a function of the AHR/HK2 axis in these tumours.
Our study is thus adding HK2 to the list of AHR targets that also regulate its function. It is well known that CYP enzymes are induced upon AHR agonists exposure and that they contribute to the detoxification or activation of most of these AHR agonists, which directly regulates AHR activation over time (1,10,14). Similarly, AHR repressor (AHRR) and TCDD inducible poly(ADP-ribose) polymerase (TIPARP) are induced by AHR upon activation and contribute to reduce AHR transcriptional activity by protein/protein interaction and/or protein degradation (1,23–25). Strikingly, as opposed to these later examples, HK2 is a direct target of AHR that positively contributes to AHR activity and/or expression depending on cell context under basal condition and in response to AHR agonists. Our study further highlights the need to identify and characterize additional AHR signalling factors to fully comprehend the molecular basis of AHR context-dependent cellular effects.
DATA AVAILABILITY
AHR ChIP-sequencing data were previously published and are publicly available on NCBI GEO servers with accession numbers GSE90550, GSE127649 and GSE116638. All software used for data analysis and visualization are freely usable and their references are indicated in the materials and methods. The databases used to collect and generate gene-chemical relationships and gene expression, chromatin and DNA methylation information in human cell lines and cancer patients (e.g. ENCODE, ChIP-Atlas, CTD, CellMinerDB, TCGA, TARGET Osteosarcoma) are publicly available and the publication embargoes on the different datasets included in our analyses have expired. The proteomics data produced in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with accession number PXD028992. All values and statistical tests reported in the figures and tables of the manuscript are accessible in Supplementary Table S5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the facilities at Institut Cochin (Paris, France) for trainings and help with data acquisition, processing and analysis, notably Marjorie Leduc, François Guillonneau and Morgane Le Gall for proteomic data production and analysis. We acknowledge the animal facility of CDTA (Center for Animal Distribution, Typing and Archiving, CNRS, Orléans, France) and the animal core facility of BioMed Tech facilities (Campus Saint Germain des Prés, INSERM US36, CNRS UMS2009, Université Paris Cité, Paris, France). We thank members of the ‘Epigenetic, DNA replication and Cancer’ (Institut Cochin, Paris, France) and ‘Signalling in environmental and drug toxicity’ (Université Paris Cité) laboratories for helpful comments and criticisms during the course of this work. We are grateful to Olivia Fromigué (Institut Gustave Roussy, Villejuif, France) and Frédérique Verdier (Institut Cochin, Paris, France) for sharing reagents. The Galaxy server that was used for some calculations is in part funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research (BMBF grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, 031L0106 de.STAIR (de.NBI)). Parts of the results were also based upon freely re-usable data generated by TCGA Research Network, ENCODE consortium and other projects funded by the National Institute of Health and National Cancer Institute (USA).
Notes
Present address: Solène Huard, Institut Curie – PSL Research University, Translational Research Department, Breast Cancer Biology Group, 75005 Paris, France.
Present address: Ludmila Juricek, Asfalia Biologics, 18 rue Charcot, 75013 Paris, France.
Contributor Information
Manon Watzky, Université Paris Cité, Institut Cochin, INSERM, U1016, CNRS, UMR8104, F-75014 Paris, France.
Solène Huard, Université Paris Cité, Institut Cochin, INSERM, U1016, CNRS, UMR8104, F-75014 Paris, France.
Ludmila Juricek, METATOX, T3S, Toxicologie Environnementale, Cibles thérapeutiques, Signalisation cellulaire et Biomarqueurs, INSERM UMR-S1124, F-75006 Paris, France.
Julien Dairou, Université Paris Cité, UFR des Sciences Fondamentales et Biomédicales, Paris, France; Laboratoire de Chimie et Biochimie Pharmacologiques et Toxicologiques, CNRS, UMR 8601, Université Paris Cité, F-75006 Paris, France.
Caroline Chauvet, METATOX, T3S, Toxicologie Environnementale, Cibles thérapeutiques, Signalisation cellulaire et Biomarqueurs, INSERM UMR-S1124, F-75006 Paris, France; Université Paris Cité, UFR des Sciences Fondamentales et Biomédicales, Paris, France.
Xavier Coumoul, METATOX, T3S, Toxicologie Environnementale, Cibles thérapeutiques, Signalisation cellulaire et Biomarqueurs, INSERM UMR-S1124, F-75006 Paris, France; Université Paris Cité, UFR des Sciences Fondamentales et Biomédicales, Paris, France.
Anne Letessier, Université Paris Cité, Institut Cochin, INSERM, U1016, CNRS, UMR8104, F-75014 Paris, France.
Benoit Miotto, Université Paris Cité, Institut Cochin, INSERM, U1016, CNRS, UMR8104, F-75014 Paris, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Laboratory of B.M. is partner of Labex ‘Who am I?’ [ANR-11-LABX-0071, ANR-11-IDEX-005-02] and is supported by Fondation pour la Recherche Medicale [AJE20151234749]; INSERM; CNRS; University Paris Cité; INCa-Plan Cancer [ASC15018KSA]; Groupement des Entreprises Ile-de-France contre le cancer - GEFLUC [RAK19154KKA]; Ligue Régionale Contre le Cancer Ile-de-France [RAB20191KKA]. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gargaro M., Scalisi G., Manni G., Mondanelli G., Grohmann U., Fallarino F.. The landscape of AhR regulators and coregulators to fine-tune AhR functions. Int. J. Mol. Sci. 2021; 22:E757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kolonko M., Greb-Markiewicz B.. bHLH-PAS proteins: their structure and intrinsic disorder. Int. J. Mol. Sci. 2019; 20:E3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dere E., Lo R., Celius T., Matthews J., Zacharewski T.R.. Integration of genome-wide computation DRE search, AhR chip-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 2011; 12:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sartor M.A., Schnekenburger M., Marlowe J.L., Reichard J.F., Wang Y., Fan Y., Ma C., Karyala S., Halbleib D., Liu X.et al.. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ. Health Perspect. 2009; 117:1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang S.Y., Ahmed S., Satheesh S.V., Matthews J.. Genome-wide mapping and analysis of aryl hydrocarbon receptor (AHR)- and aryl hydrocarbon receptor repressor (AHRR)-binding sites in human breast cancer cells. Arch. Toxicol. 2018; 92:225–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neavin D.R., Lee J.-H., Liu D., Ye Z., Li H., Wang L., Ordog T., Weinshilboum R.M.. Single nucleotide polymorphisms at a distance from aryl hydrocarbon receptor (AHR) binding sites influence AHR ligand-dependent gene expression. Drug Metab. Dispos. Biol. Fate Chem. 2019; 47:983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prokopec S.D., Houlahan K.E., Sun R.X., Watson J.D., Yao C.Q., Lee J., P’ng C., Pang R., Wu A.H., Chong L.C.et al.. Compendium of TCDD-mediated transcriptomic response datasets in mammalian model systems. BMC Genomics. 2017; 18:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kou Z., Dai W.. Aryl hydrocarbon receptor: its roles in physiology. Biochem. Pharmacol. 2021; 185:114428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xue P., Fu J., Zhou Y.. The aryl hydrocarbon receptor and tumor immunity. Front. Immunol. 2018; 9:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larigot L., Juricek L., Dairou J., Coumoul X.. AhR signaling pathways and regulatory functions. Biochim. Open. 2018; 7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murray I.A., Patterson A.D., Perdew G.H.. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer. 2014; 14:801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murray I.A., Perdew G.H.. How ah receptor ligand specificity became important in understanding its physiological function. Int. J. Mol. Sci. 2020; 21:E9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leclerc D., Staats Pires A.C., Guillemin G.J., Gilot D.. Detrimental activation of AhR pathway in cancer: an overview of therapeutic strategies. Curr. Opin. Immunol. 2021; 70:15–26. [DOI] [PubMed] [Google Scholar]
- 14. Paris A., Tardif N., Galibert M.-D., Corre S.. AhR and cancer: from gene profiling to targeted therapy. Int. J. Mol. Sci. 2021; 22:E752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Safe S., Lee S.-O., Jin U.-H.. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as a drug target. Toxicol. Sci. Off. J. Soc. Toxicol. 2013; 135:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Torti M.F., Giovannoni F., Quintana F.J., García C.C.. The aryl hydrocarbon receptor as a modulator of Anti-viral immunity. Front. Immunol. 2021; 12:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frericks M., Meissner M., Esser C.. Microarray analysis of the AHR system: tissue-specific flexibility in signal and target genes. Toxicol. Appl. Pharmacol. 2007; 220:320–332. [DOI] [PubMed] [Google Scholar]
- 18. Harper P.A., Riddick D.S., Okey A.B.. Regulating the regulator: factors that control levels and activity of the aryl hydrocarbon receptor. Biochem. Pharmacol. 2006; 72:267–279. [DOI] [PubMed] [Google Scholar]
- 19. Mulero-Navarro S., Carvajal-Gonzalez J.M., Herranz M., Ballestar E., Fraga M.F., Ropero S., Esteller M., Fernandez-Salguero P.M.. The dioxin receptor is silenced by promoter hypermethylation in human acute lymphoblastic leukemia through inhibition of Sp1 binding. Carcinogenesis. 2006; 27:1099–1104. [DOI] [PubMed] [Google Scholar]
- 20. Englert N.A., Turesky R.J., Han W., Bessette E.E., Spivack S.D., Caggana M., Spink D.C., Spink B.C.. Genetic and epigenetic regulation of AHR gene expression in MCF-7 breast cancer cells: role of the proximal promoter GC-rich region. Biochem. Pharmacol. 2012; 84:722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khanal T., Choi K., Leung Y.-K., Wang J., Kim D., Janakiram V., Cho S.-G., Puga A., Ho S.-M., Kim K.. Loss of NR2E3 represses AHR by LSD1 reprogramming, is associated with poor prognosis in liver cancer. Sci. Rep. 2017; 7:10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nault R., Fader K.A., Ammendolia D.A., Dornbos P., Potter D., Sharratt B., Kumagai K., Harkema J.R., Lunt S.Y., Matthews J.et al.. Dose-Dependent metabolic reprogramming and differential gene expression in TCDD-Elicited hepatic fibrosis. Toxicol. Sci. Off. J. Soc. Toxicol. 2016; 154:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evans B.R., Karchner S.I., Allan L.L., Pollenz R.S., Tanguay R.L., Jenny M.J., Sherr D.H., Hahn M.E.. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol. Pharmacol. 2008; 73:387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacPherson L., Ahmed S., Tamblyn L., Krutmann J., Förster I., Weighardt H., Matthews J.. Aryl hydrocarbon receptor repressor and TiPARP (ARTD14) use similar, but also distinct mechanisms to repress aryl hydrocarbon receptor signaling. Int. J. Mol. Sci. 2014; 15:7939–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ahmed S., Bott D., Gomez A., Tamblyn L., Rasheed A., Cho T., MacPherson L., Sugamori K.S., Yang Y., Grant D.M.et al.. Loss of the Mono-ADP-ribosyltransferase, tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J. Biol. Chem. 2015; 290:16824–16840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ciscato F., Ferrone L., Masgras I., Laquatra C., Rasola A.. Hexokinase 2 in cancer: a prima donna playing multiple characters. Int. J. Mol. Sci. 2021; 22:4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tanner L.B., Goglia A.G., Wei M.H., Sehgal T., Parsons L.R., Park J.O., White E., Toettcher J.E., Rabinowitz J.D.. Four key steps control glycolytic flux in mammalian cells. Cell Syst. 2018; 7:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patra K.C., Wang Q., Bhaskar P.T., Miller L., Wang Z., Wheaton W., Chandel N., Laakso M., Muller W.J., Allen E.L.et al.. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013; 24:213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jalili V., Afgan E., Gu Q., Clements D., Blankenberg D., Goecks J., Taylor J., Nekrutenko A.. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020; 48:W395–W402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langmead B., Salzberg S.L.. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Project Consortium ENCODE, Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A.et al.. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020; 583:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mi H., Ebert D., Muruganujan A., Mills C., Albou L.-P., Mushayamaha T., Thomas P.D.. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021; 49:D394–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Navarro Gonzalez J., Zweig A.S., Speir M.L., Schmelter D., Rosenbloom K.R., Raney B.J., Powell C.C., Nassar L.R., Maulding N.D., Lee C.M.et al.. The UCSC genome browser database: 2021 update. Nucleic Acids Res. 2021; 49:D1046–D1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C.. ChIP-Atlas: a data-mining suite powered by full integration of public chip-seq data. EMBO Rep. 2018; 19:e46255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., Barretina J., Gelfand E.T., Bielski C.M., Li H.et al.. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchal C., de Dieuleveult M., Saint-Ruf C., Guinot N., Ferry L., Olalla Saad S.T., Lazarini M., Defossez P.-A., Miotto B.. Depletion of ZBTB38 potentiates the effects of DNA demethylating agents in cancer cells via CDKN1C mRNA up-regulation. Oncogenesis. 2018; 7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miotto B., Ji Z., Struhl K.. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E4810–E4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., Wiegers J., Wiegers T.C., Mattingly C.J.. Comparative toxicogenomics database (CTD): update 2021. Nucleic Acids Res. 2021; 49:D1138–D1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wiśniewski J.R., Zougman A., Nagaraj N., Mann M.. Universal sample preparation method for proteome analysis. Nat. Methods. 2009; 6:359–362. [DOI] [PubMed] [Google Scholar]
- 40. Ishihama Y., Rappsilber J., Mann M.. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J. Proteome Res. 2006; 5:988–994. [DOI] [PubMed] [Google Scholar]
- 41. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M.. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics MCP. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 43. Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C.et al.. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, cytoscape and enrichmentmap. Nat. Protoc. 2019; 14:482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sellick C.A., Hansen R., Stephens G.M., Goodacre R., Dickson A.J.. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat. Protoc. 2011; 6:1241–1249. [DOI] [PubMed] [Google Scholar]
- 45. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E.et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rajapakse V.N., Luna A., Yamade M., Loman L., Varma S., Sunshine M., Iorio F., Sousa F.G., Elloumi F., Aladjem M.I.et al.. CellMinerCDB for integrative cross-database genomics and pharmacogenomics analyses of cancer cell lines. Iscience. 2018; 10:247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V.et al.. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018; 173:400–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saghafinia S., Mina M., Riggi N., Hanahan D., Ciriello G.. Pan-Cancer landscape of aberrant DNA methylation across human tumors. Cell Rep. 2018; 25:1066–1080. [DOI] [PubMed] [Google Scholar]
- 49. Buddingh E.P., Kuijjer M.L., Duim R.A.J., Bürger H., Agelopoulos K., Myklebost O., Serra M., Mertens F., Hogendoorn P.C.W., Lankester A.C.et al.. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011; 17:2110–2119. [DOI] [PubMed] [Google Scholar]
- 50. Klotz L.-O., Steinbrenner H.. Cellular adaptation to xenobiotics: interplay between xenosensors, reactive oxygen species and FOXO transcription factors. Redox Biol. 2017; 13:646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lafita-Navarro M.C., Perez-Castro L., Zacharias L.G., Barnes S., DeBerardinis R.J., Conacci-Sorrell M.. The transcription factors aryl hydrocarbon receptor and MYC cooperate in the regulation of cellular metabolism. J. Biol. Chem. 2020; 295:12398–12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao B., Degroot D.E., Hayashi A., He G., Denison M.S.. CH223191 is a ligand-selective antagonist of the ah (dioxin) receptor. Toxicol. Sci. Off. J. Soc. Toxicol. 2010; 117:393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhuo B., Li Y., Li Z., Qin H., Sun Q., Zhang F., Shen Y., Shi Y., Wang R.. PI3K/Akt signaling mediated hexokinase-2 expression inhibits cell apoptosis and promotes tumor growth in pediatric osteosarcoma. Biochem. Biophys. Res. Commun. 2015; 464:401–406. [DOI] [PubMed] [Google Scholar]
- 54. Song J., Wu X., Liu F., Li M., Sun Y., Wang Y., Wang C., Zhu K., Jia X., Wang B.et al.. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem. Biophys. Res. Commun. 2017; 490:217–224. [DOI] [PubMed] [Google Scholar]
- 55. Pajak B., Siwiak E., Sołtyka M., Priebe A., Zieliński R., Fokt I., Ziemniak M., Jaśkiewicz A., Borowski R., Domoradzki T.et al.. 2-Deoxy-d-Glucose and Its analogs: from diagnostic to therapeutic agents. Int. J. Mol. Sci. 2019; 21:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hoadley K.A., Yau C., Hinoue T., Wolf D.M., Lazar A.J., Drill E., Shen R., Taylor A.M., Cherniack A.D., Thorsson V.et al.. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018; 173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ambolet-Camoit A., Ottolenghi C., Leblanc A., Kim M.J., Letourneur F., Jacques S., Cagnard N., Guguen-Guillouzo C., Barouki R., Aggerbeck M.. Two persistent organic pollutants which act through different xenosensors (alpha-endosulfan and 2,3,7,8 tetrachlorodibenzo-p-dioxin) interact in a mixture and downregulate multiple genes involved in human hepatocyte lipid and glucose metabolism. Biochimie. 2015; 116:79–91. [DOI] [PubMed] [Google Scholar]
- 58. Huc L., Rissel M., Solhaug A., Tekpli X., Gorria M., Torriglia A., Holme J.A., Dimanche-Boitrel M.-T., Lagadic-Gossmann D.. Multiple apoptotic pathways induced by p53-dependent acidification in benzo[a]pyrene-exposed hepatic F258 cells. J. Cell. Physiol. 2006; 208:527–537. [DOI] [PubMed] [Google Scholar]
- 59. Halappanavar S., Wu D., Williams A., Kuo B., Godschalk R.W., Van Schooten F.J., Yauk C.L.. Pulmonary gene and microRNA expression changes in mice exposed to benzo(a)pyrene by oral gavage. Toxicology. 2011; 285:133–141. [DOI] [PubMed] [Google Scholar]
- 60. Diani-Moore S., Marques Pedro T., Rifkind A.B.. Organ-specific effects on glycolysis by the dioxin-activated aryl hydrocarbon receptor. PloS One. 2020; 15:e0243842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sutter C.H., Olesen K.M., Bhuju J., Guo Z., Sutter T.R.. AHR regulates metabolic reprogramming to promote SIRT1-dependent keratinocyte differentiation. J. Invest. Dermatol. 2019; 139:818–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mascanfroni I.D., Takenaka M.C., Yeste A., Patel B., Wu Y., Kenison J.E., Siddiqui S., Basso A.S., Otterbein L.E., Pardoll D.M.et al.. Metabolic control of type 1 regulatory t cell differentiation by AHR and HIF1-α. Nat. Med. 2015; 21:638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kumar S., Donti T.R., Agnihotri N., Mehta K.. Transglutaminase 2 reprogramming of glucose metabolism in mammary epithelial cells via activation of inflammatory signaling pathways. Int. J. Cancer. 2014; 134:2798–2807. [DOI] [PubMed] [Google Scholar]
- 64. Ko K.-W., Choi B., Park S., Arai Y., Choi W.C., Lee J.-M., Bae H., Han I.-B., Lee S.-H.. Down-regulation of transglutaminase 2 stimulates redifferentiation of dedifferentiated chondrocytes through enhancing glucose metabolism. Int. J. Mol. Sci. 2017; 18:E2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Puga A., Barnes S.J., Dalton T.P., Chang C., Knudsen E.S., Maier M.A.. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J. Biol. Chem. 2000; 275:2943–2950. [DOI] [PubMed] [Google Scholar]
- 66. Yang S.-C., Wu C.-H., Tu Y.-K., Huang S.-Y., Chou P.-C.. Exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin increases the activation of aryl hydrocarbon receptor and is associated with the aggressiveness of osteosarcoma MG-63 osteoblast-like cells. Oncol. Lett. 2018; 16:3849–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dabir P., Marinic T.E., Krukovets I., Stenina O.I.. Aryl hydrocarbon receptor is activated by glucose and regulates the thrombospondin-1 gene promoter in endothelial cells. Circ. Res. 2008; 102:1558–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Matsuda S., Adachi J., Ihara M., Tanuma N., Shima H., Kakizuka A., Ikura M., Ikura T., Matsuda T.. Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Res. 2016; 44:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang X., Li K., Liu L., Shi Q., Song P., Jian Z., Guo S., Wang G., Li C., Gao T.. AHR promoter variant modulates its transcription and downstream effectors by allele-specific AHR-SP1 interaction functioning as a genetic marker for vitiligo. Sci. Rep. 2015; 5:13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tristan C., Shahani N., Sedlak T.W., Sawa A.. The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 2011; 23:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li X., Qian X., Jiang H., Xia Y., Zheng Y., Li J., Huang B.-J., Fang J., Qian C.-N., Jiang T.et al.. Nuclear PGK1 alleviates ADP-dependent inhibition of CDC7 to promote DNA replication. Mol. Cell. 2018; 72:650–660. [DOI] [PubMed] [Google Scholar]
- 72. Gao X., Wang H., Yang J.J., Liu X., Liu Z.-R.. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell. 2012; 45:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sheikh T., Gupta P., Gowda P., Patrick S., Sen E.. Hexokinase 2 and nuclear factor erythroid 2-related factor 2 transcriptionally coactivate xanthine oxidoreductase expression in stressed glioma cells. J. Biol. Chem. 2018; 293:4767–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vega M., Riera A., Fernández-Cid A., Herrero P., Moreno F.. Hexokinase 2 is an intracellular glucose sensor of yeast cells that maintains the structure and activity of mig1 protein repressor complex. J. Biol. Chem. 2016; 291:7267–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
AHR ChIP-sequencing data were previously published and are publicly available on NCBI GEO servers with accession numbers GSE90550, GSE127649 and GSE116638. All software used for data analysis and visualization are freely usable and their references are indicated in the materials and methods. The databases used to collect and generate gene-chemical relationships and gene expression, chromatin and DNA methylation information in human cell lines and cancer patients (e.g. ENCODE, ChIP-Atlas, CTD, CellMinerDB, TCGA, TARGET Osteosarcoma) are publicly available and the publication embargoes on the different datasets included in our analyses have expired. The proteomics data produced in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with accession number PXD028992. All values and statistical tests reported in the figures and tables of the manuscript are accessible in Supplementary Table S5.