Abstract
Background and Aim:
Staphylococcus pseudintermedius is a zoonotic bacterium commonly found in animals, especially dogs. These bacteria can survive on environmental surfaces for several months. The infection of S. pseudintermedius from the environment is possible, but properly cleaning surface objects can prevent it. This study aimed to investigate the prevalence of methicillin-resistant S. pseudintermedius (MRSP) in the environment of a recently constructed veterinary hospital in Southern Thailand, where we hypothesized that the prevalence of MRSP might be very low.
Materials and Methods:
At three different time points, 150 samples were collected from different environmental surfaces and wastewater across the veterinary hospital. The collection was done after the hospital’s cleaning. Bacteria were purified in the culture before being identified as species by biochemical tests and polymerase chain reaction (PCR). Next, the antimicrobial-resistant profile was performed using an automated system (Vitek 2). Finally, the antimicrobial resistance genes were identified using PCR.
Results:
Fifteen colonies of S. pseudintermedius were isolated from the surfaces of eight floors, four tables, two chairs, and one rebreathing tube. Fourteen colonies (93.3%) were multidrug-resistant (MDR) and carried the blaZ gene (93.3%). The majority of colonies were resistant to benzylpenicillin (93.3%), cefovecin (93.3%), ceftiofur (93.3%), kanamycin (93.3%), and neomycin (93.3%). Notably, only four colonies (26.7%) were methicillin-susceptible S. pseudintermedius, whereas 11 colonies (73.3%) were MRSP and carried both the mecA and blaZ genes. Five MRSP (45.5%) were resistant to at least 14 antimicrobial drugs, represented as extensively drug-resistant (XDR) bacteria. Ten of eleven MRSP (90.9%) were Staphylococcal chromosomal mec type V, while another displayed untypeable. Despite the routine and extensive cleaning with detergent and disinfectant, MRSP isolates were still detectable.
Conclusion:
Many isolates of MRSP were found in this veterinary hospital. Almost all of them were MDR, and nearly half were XDR, posing a threat to animals and humans. In addition, the current hospital cleaning procedure proved ineffective. Future research should be conducted to determine the bacterial biofilm properties and bacterial sensitivity to certain detergents and disinfectants.
Keywords: antimicrobial resistance, cleaning, methicillin-resistant Staphylococcus pseudintermedius, veterinary hospital
Introduction
Staphylococcus pseudintermedius is a common pathogen that can be found in the environment, particularly on the surfaces of veterinary hospitals [1,2] and houses [3]. This organism is an opportunistic bacterium that is frequently identified in many companion animals such as dogs, cats, avians, and rabbits [4,5]. The colonization of S. pseudintermedius on canine skin can be persistent, sporadic, or transient depending on the dos [3,6]. These bacteria can cause numerous diseases in dogs, such as pyoderma, otitis externa, reproductive tract infection, respiratory tract infection, and urinary tract infection [4,7]. Methicillin-resistant S. pseudintermedius (MRSP) can be chronic and severe, and bacteria are typically detected on the infected lesion after a week of antimicrobial treatment [6,8]. Moreover, MRSP is regularly transferred between dogs and the environment throughout the veterinary hospital, which includes a rebreathing circuit, clippers, and floors [2,3,9]. Other staphylococci, including S. aureus, can also be present on the surface of veterinary hospitals and are capable of transferring the resistome to S. pseudintermedius [10]. Although S. pseudintermedius is zoonotic, human infections are rare and mainly limited to bite wounds; a few cases of septicemia resulting in brain abscess, endocarditis, sinusitis, otitis, arthritis, and pneumonia are reported [11-14].
Globally, the incidence of MRSP has been increasingly reported [15]. This pathogen carries the mecA gene and the Staphylococcal chromosomal mec (SCCmec), a large mobile genetic element shares among staphylococci [1,16] and contains extensively drug-resistant (XDR) genes [17]. Many MRSP also carried the blaZ gene, which produces penicillinase capable of degrading beta-lactam antibiotic drugs [18]. Most MRSPs are multidrug-resistant (MDR), causing treatment difficult and threatening a dog’s life, but MRSP infection is uncommon in cats [4,7,19]. The transmission of resistant genes among staphylococci is one of the crucial factors in the emergence of antimicrobial XDR and MDR bacteria that are potentially detrimental to human health [20].
Although reports of MRSP in veterinary hospitals in Thailand are rare and limited to older hospitals in central Thailand, those investigations suggest that MRSP is frequently discovered on the hospital’s environmental surface [1,2,18]. Therefore, we hypothesized that the high prevalence of MRSP might be associated with the age of veterinary hospitals. This study aimed to determine the prevalence of MRSP on the environmental surface of a recently constructed veterinary hospital in southern Thailand, where we believed that the prevalence of MRSP might be very low. This study will provide the knowledge to understand the colonization site and antimicrobial profile of S. pseudintermedius, leading to sanitation, hygiene, and treatment plans.
Materials and Methods
Ethical approval
This study was approved by the Walailak University-Institutional Biosafety Committee (IBC) (WU-IBC-63-027).
Study period and location
The samples were collected from December 2020 to April 2021. A study site was a veterinary hospital located in Nakhon Si Thammarat Province, Southern Thailand.
Sample collection
This hospital started operations in October 2018 and was open daily from 8.30 AM to 4.30 PM, with an average of approximately ten animals treated per day. The building was two stories tall, with the first-floor housing medical rooms for post-operative care, vaccinations, and an intensive care unit. The second level was dedicated to the surgical unit, including a preparation room and two operating rooms. Daily routine cleaning was done after the hospital closed at 4.30 PM using detergent by the cleaning company.
The samples were collected by swabbing 50 different environmental surfaces across the hospital. Sample collections were done 3 times at different time points (1.5 months for each collection). The first sample collection was done after the routine cleaning; all surfaces, including floors, doors, tables, chairs, and equipment, were cleaned with the company’s detergent for 30 min in each room. For the second sample collection, all surfaces were cleaned with detergent and disinfectant (Dermodacyn Disinfecting Solution, CA, USA) twice in every room. Finally, the third sample collection occurred after the cleaning with detergent and disinfectant twice and later 30 min of fumigation (Dermodacyn Disinfecting Solution) in each room on the second floor, while the first floor was cleaned with detergent and disinfectant.
Each collection included 50 samples from frequent hand-touch locations and wastewater (before recycling). On the first floor, samples were taken from 17 floor surfaces, ten table surfaces, two chair surfaces, two doorknobs, one refrigerator handle, and two wastewater sites. On the second floor, 16 samples were collected from the preparation room and surgery room, eight samples from the floor, four samples from tables, and four samples from equipment.
Before sample collection, sterile cotton swabs were moistened with sterile tryptic soy broth (TSB) (Oxoid, Hampshire, UK). On the floor, surface swab sampling was performed at 1 cm2 per site, whereas on tables and equipment, surface swab sampling was performed by rolling a cotton bud around the surface [21]. Samples were kept on ice and then transported within a sealed box to a microbiology laboratory for identification within 2 h.
Bacterial identification
Samples in TSB were incubated at 37°C for 24 h. Next, one suspension loop was placed on mannitol salt agar (Oxoid) supplemented with 0.5 μg/mL of oxacillin (Oxoid). Three staphylococcus-like colonies were picked and placed on blood agar for bacterial identification. Only colonies expressing hemolysin were chosen for biochemical identification using the VITEK 2 Compact, automated ID/AST instrument (Biomeriex, Marcy l’Etoile, France) and finally confirmed with multiplex polymerase chain reaction (PCR) as previously described by Sasaki et al. [22].
PCR
For DNA extraction, the selected colony was put into 1 mL of TSB (Oxoid) and incubated at 37°C for 24 h. Bacterial DNA was extracted using Presto™ Mini gDNA Bacteria Kit (Geneaid, New Taipei City, Taiwan). DNA templates were used for species identification, the mecA gene detection, the blaZ gene detection, and the SCCmec detection as shown in Table-1 [22-25].
Table 1.
The primer of Staphylococcal chromosomal mec typing.
| Primer | Sequence (5’–3’) | Size of PCR product (bp) | References |
|---|---|---|---|
| au-F3 | TCGCTTGCTATGATTGTGG | 359 | [22] |
| au-R | GCCAATGTTCTACCATAGC | ||
| in-F | CATGTCATATTATTGCGAATGA | 430 | |
| in-R3 | AGGACCATCACCATTGACATATTGAAACC | ||
| sch-F | AATGGCTACAATGATAATCACTAA | 526 | |
| sch-R | CATATCTGTCTTTCGGCGCG | ||
| hy-F1 | CATTATATGATTTGAACGTG | 793 | |
| hy-R1 | GAATCAATATCGTAAAGTTGC | ||
| pse-F2 | TRGGCAGTAGGATTCGTTAA | 926 | |
| pse-R5 | CTTTTGTGCTYCMTTTTGG | ||
| mecA-F | AAAATCGATGGTAAAGGTTGGC | 532 | [23] |
| mecA-R | AGTTCTGCAGTACCGGATTTGC | ||
| blaZ-F | ACTTCAACACCTGCTGCTTTC | 173 | [24] |
| blaZ-R | TGACCACTTTTATCAGCAACC | ||
| M-PCR 1 (for amplification of ccr gene complex with mecA) | [25] | ||
| mA1 | GCTATCCACCCTCAAACAGG | 286 | |
| mA2 | ACGTTGTAACCACCCCAAGA | ||
| α1 | AACCTATATCATCAATCAGTACGT | 695 | |
| α2 | TAAAGGCATCAATGCACAAACACT | 937 | |
| α3 | AGCTCAAAAGCAAGCAATAGAAT | 1791 | |
| βc | ATTGCCTTGATAATAGCCITCT | 1287 | |
| α4.2 | GTATCAATGCACCAGAACTT | ||
| β4.2 | TTGCGACTCTCTTGGCGTTT | ||
| γR | CGTCTATTACAAGATGTTAAGGATAAT | 518 | |
| γF | CCTTTATAGACTGGATTATTCAAAATAT | ||
| M-PCR 2 (for amplification of mec gene complex class) | |||
| mI6 | CATAACTTCCCATTCTGCAGATG | 1963 | |
| IS7 | ATGCTTAATGATAGCATCCGAATG | 2827 | |
| IS2 | TGAGGTTATTCAGATATTTCGATGT | ||
| mA7 | ATATACCAAACCCGACAACTACA | 804 | |
M-PCR=Multiplex polymerase chain reaction
PCR products were validated with electrophoresis on 1.5% agarose gel in 1x Tris-acid-EDTA buffer (Vivantis, Selangor Darul Ehsan, Malaysia) at 135 V/cm for 20 min. The DNA bands were visualized under UV light with the G-BOX F3 Gel imaging machine (Syngene, Cambridge, UK).
Antimicrobial-resistant profile
All mecA-positive isolates were examined for antimicrobial resistance (AMR) profiles using the AST-GP80 card with e the VITEK 2 Compact system based on minimal inhibitory concentration (MIC). Sixteen antimicrobial drugs were tested, including benzylpenicillin, cefovecin, ceftiofur, gentamicin, kanamycin, neomycin, enrofloxacin, marbofloxacin, pradofloxacin, erythromycin, clindamycin, doxycycline, tetracycline, nitrofuran, chloramphenicol, and trimethoprim/sulfamethoxazole. Most of the antimicrobial drugs in this study were routinely used in this hospital. The MIC was determined using VET: Clinical and Laboratory Standards Institute-based plus natural resistance data provided by the VITEK 2 Compact machine.
Statistical analysis
A descriptive analysis of the defining bacteria distribution variable in this hospital was performed using Jamovi software version 2.0 [26]. The population of pathogens and XDR bacteria were described by percentile. MDR, extensively XDR, and pan drug-resistance (PDR) bacteria were categorized according to Magiorakos et al. [27]. The SCCmec types were interpreted as I to V types or untypable [25].
Results
Isolation
In total, bacteria were observed on 144 of 150 swabs (96%). Bacteria were detected in 31 of 50 samples (62.0%) from the first sample collection, 18 samples (36.0%) from the second sample collection, and 21 samples from the third sample collection (42.0%).
Specifically, 110 staphylococci-like colonies (73.3%) from 150 environmental samples were found on mannitol salt agar. Then 52 of 110 staphylococci-like colonies (47.3%) were confirmed as staphylococci by PCR. However, staphylococci were not detected in various areas, including dive floors, one stethoscope, one surgery equipment, one doorknob, and two wastewaters throughout three sample collections.
Fifteen colonies from 150 environmental samples (10.0%) and 52 staphylococci isolates (28.8%) were verified by PCR as S. pseudintermedius. The prevalence of S. pseudintermedius in the veterinary hospital environment is shown in Figure-1. Most of the S. pseudintermedius isolates were found on the first floors (12/15, 80.0%), and the majority of isolates were obtained from the floor surface (8/15, 53.3%). The remaining were found on the surfaces of chairs, tables, and a rebreathing circuit.
Figure-1.
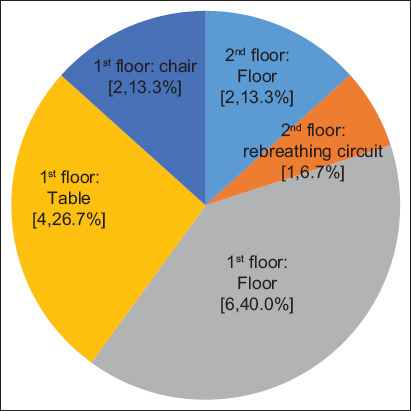
The prevalence of Staphylococcus pseudintermedius collected from the different environments in the veterinary hospitals. The numbers in the brackets represent the number of bacterial isolates and their percentage.
MRSP
The data for MRSP and methicillin-susceptible S. pseudintermedius (MSSP) are shown in Table-2. Most colonies were MRSP (11/15, 73.3%), while the minority were MSSP (4/15, 26.7%). Most MRSP was detected on the floors (5/11, 45.5%), then tables (4/11, 36.4%), and waiting chairs (2/11, 18.2%), respectively. All MRSP colonies (positive for the mecA gene) were also positive for the blaZ gene. In addition, most MRSP isolates carried the SCCmec type V (10/11, 90.9%), while another displayed an untypable (NT) type.
Table 2.
The number of Staphylococcus pseudintermedius colonies discovered during each collection period.
| Sample site | 1st collection (Detergent) | 2nd collection (Disinfectant) | 3rd collection (Disinfectant) | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| MSSP | MRSP | MSSP | MRSP | MSSP | MRSP | |
| Second floor | 2 | 1 | ||||
| First floor | 1 | 1 | 10 | |||
*Wiping and fumigation with disinfectant, MSSP=Methicillin-susceptible Staphylococcus pseudintermedius, MRSP=Methicillin-resistant Staphylococcus pseudintermedius
Antimicrobial-resistant profile
The AMR profile is shown in Table-2. The highest resistant drugs were benzylpenicillin, cefovecin, ceftiofur, kanamycin, and neomycin (14/15, 93.33%), then erythromycin, tetracycline, and gentamycin (13/15, 86.7%). Conversely, the antimicrobial drugs with the highest sensitive drugs were nitrofurantoin (15/15, 100%), then chloramphenicol (13/15, 86.67%), and clindamycin (12/15, 80.0%), respectively.
MDR
Fourteen colonies (93.3%) were MDR bacteria, including 11 MRSP and 3 MSSP colonies (Table-3). Five extensively XDR bacteria (33.3%) were found, including 4 MRSP colonies resistant to 15 antimicrobial drugs and one MSSP colony resistant to 14 drugs. All 11 MRSP colonies were MDR, with 36.4% (4/11) being XDR, whereas 75.0% (3/4) of MSSP colonies were MDR, with 25.0% (1/4) being XDR.
Table 3.
Collection places, MRSP identification, blaZ detection, and antimicrobial-resistant profiles of Staphylococcus pseudintermedius in this veterinary hospital.
| Places | MRSP (mecA) | BlaZ positive | Benzylpenicillin | Cefovecin | Ceftiofur | Gentamicin | Kanamycin | Neomycin | Enrofloxacin | Marbofloxacin |
|---|---|---|---|---|---|---|---|---|---|---|
| Floor in surgery room | MSSP | POS | R | R | R | R | R | R | S | S |
| Rebreathing circuit | MSSP | POS | R | R | R | R | R | R | S | R |
| Floor in preparation room | MRSP | POS | R | R | R | R | R | R | R | R |
| Floor in medication room 2 | MSSP | POS | R | R | R | R | R | R | R | R |
| Floor in medication room 2 | MRSP | POS | R | R | R | R | R | R | R | R |
| Examination table in medication room 2 | MRSP | POS | R | R | R | R | R | R | R | R |
| Medication table in medication room 2 | MRSP | POS | R | R | R | R | R | R | R | R |
| Equipment table in medication room 2 | MRSP | POS | R | R | R | R | R | R | R | R |
| Floor in medication room 3 | MRSP | POS | R | R | R | S | R | S | R | R |
| Floor in medication room 3 | MSSP | NEG | S | S | S | S | S | R | S | S |
| Medication table in medication room 3 | MRSP | POS | R | R | R | R | R | R | R | R |
| Floor in admission room 1 | MRSP | POS | R | R | R | R | R | R | R | R |
| Floor in admission room 1 | MRSP | POS | R | R | R | R | R | R | R | R |
| Waiting chair 1 | MRSP | POS | R | R | R | R | R | R | R | R |
| Waiting chair 2 | MRSP | POS | R | R | R | R | R | R | R | R |
| Percent of resistant results (%) | 93.3 | 93.3 | 93.3 | 86.7 | 93.3 | 93.3 | 93.3 | 93.3 | ||
|
| ||||||||||
| Places | Pradofloxacin | Inducible clindamycin resistance | Erythromycin | Clindamycin | Doxycycline | Tetracycline | Nitrofurantoin | Chloramphenicol | trimethoprim sulfamethoxazole | |
|
| ||||||||||
| Floor in surgery room | S | NEG | S | S | S | R | S | S | S | |
| Rebreathing circuit | S | NEG | S | S | S | R | S | S | S | |
| Floor in preparation room | R | NEG | R | R | S | S | S | S | R | |
| Floor in medication room 2 | R | POS | R | S | R | R | S | S | R | |
| Floor in medication room 2 | R | POS | R | S | R | R | S | S | R | |
| Examination table in medication room 2 | R | POS | R | S | R | R | S | S | R | |
| Medication table in medication room 2 | R | POS | S | S | R | R | S | S | R | |
| Equipment table in medication room 2 | R | POS | R | S | R | R | S | S | R | |
| Floor in medication room 3 | R | NEG | R | S | R | R | S | R | R | |
| Floor in medication room 3 | S | NEG | S | S | S | S | S | S | S | |
| Medication table in medication room 3 | R | NEG | R | R | R | R | S | S | R | |
| Floor in admission room 1 | R | POS | R | S | R | R | S | S | R | |
| Floor in admission room 1 | R | NEG | R | S | R | R | S | R | R | |
| Waiting chair 1 | R | POS | R | S | R | R | S | S | R | |
| Waiting chair 2 | R | NEG | R | R | R | R | S | S | R | |
| Percent of resistant results (%) | 93.3 | 86.7 | 0.2 | 73.3 | 86.7 | 0 | 13.3 | 0.8 | ||
MSSP = Methicillin-susceptible Staphylococcus pseudintermedius, MRSP = Methicillin-resistant Staphylococcus pseudintermedius
Relation of cleaning and S. pseudintermedius
At the first collection time (cleaning with detergent), 2 MSSP colonies were discovered on the second floor, and they were positive for the blaZ gene. Then, for the second sample collection (after cleaning with detergent and disinfectant), only 1 MSSP colony without the blaZ gene was found on the first floor. For the final collection, 1 MRSP colony was displayed on the second floor (after cleaning with detergent, disinfectant, and fumigation); unexpectedly, many colonies (10 MRSP and 1 MSSP) were found on the first floor (after cleaning with detergent and disinfectant) (Table-2).
Discussion
This study demonstrated that most areas (73%) of the recently constructed veterinary hospital could harbor S. pseudintermedius. The majority of colonies were MRSP and MDR, although the cleaning was done using a variety of methods. Almost all MRSP isolates carried the mecA and blaZ genes, as well as SCCmec type V.
The environment plays an important role in maintaining and transmitting staphylococci between animals and humans [28,29]. In the present study, the majority of MRSP isolates were discovered on frequent contact areas such as floors and veterinary tables, which is consistent with the previous studies [30,31]. Seriously, two colonies of MSSP were obtained from the surgical environment in the present study, although this environment is expected to be sterile to prevent surgical site infections. Staphylococci contamination of the surgical environment is occasionally found in veterinary hospitals and may cause surgical site infections [2,32]. Fortunately, S. pseudintermedius infection was not observed in any of the dogs operated in this hospital (unpublished data).
It has been noted that Staphylococci can remain on these environmental surfaces for at least 4 months before causing infection and the development of methicillin-resistant staphylococci (MRS) [33-36]. During hospitalization, dogs may come into contact with MRS, and the transmission between dogs, humans, and the environment can result in hospital- and community-acquired infections [37,38]. Moreover, the long-term circulation or persistence of S. pseudintermedius in pets and the environment can increase the probability of resistant genes being transmitted to other staphylococci, including S. aureus, an important pathogen in humans [10,37,38].
In this study, most colonies of S. pseudintermedius were MDR bacteria. They were resistant to several drug groups, such as beta-lactams, aminoglycosides, and fluoroquinolones. These groups of antimicrobial drugs are also commonly used in this veterinary hospital (e.g., amoxicillin/clavulanic acid, cefalexin, and enrofloxacin) and other hospitals [39-41]. The beta-lactam drugs, including penicillin, cefovecin, and ceftiofur, are the most commonly used antimicrobial drugs worldwide and in Thailand [1,8,42], and bacteria can be resistant to these drugs via several genes, such as the mecA and blaZ genes [43]. The co-existence of the mecA and blaZ genes was frequently observed in this study and other studies [15,43]; both genes can enhance bacterial AMR. However, no evidence of infection caused by S. pseudintermedius has been found in this hospital to date (personal communication). In the event of a future S. pseudintermedius infection, beta-lactamase inhibitors (to inhibit bacterial beta-lactamase) and nitrofurantoin (the most sensitive drug in this study) are recommended for therapy.
Surprisingly, many MRSP isolates were found on the first floor in this study, although the floor was cleaned with detergent and disinfectant. The disinfectant solution employed in this investigation contained 0.002% sodium hypochlorite and 0.013% hypochlorous acid, both of which have been shown in several papers to be effective against staphylococci when used as a liquid solution for in vitro studies [44-46]. Conversely, several reports argue that sodium hypochlorite does not effectively destroy staphylococci in the hospital environment [47-50], which might result from its instability after 24 h [48]. Another study shows that hypochlorous acid is inferior to chlorine dioxide as a disinfectant for hospital environments [50]. The conflicting results for hypochlorite as a disinfectant may be due to the varied quantities employed in each experiment, as many bacteria are eradicated at high concentrations (>0.5%) of sodium hypochlorite, including MRSA in human hospitals and MRSP in a veterinary hospital [49,51,52]. In this study, the disinfectant concentration might be lower than the recommendation [52], and hypochlorite and hypochlorous acid are not recommended for fumigation for eradicating bacteria. For fumigation, the theoretical options for sporicidal fumigants are formaldehyde, ethylene oxide, methyl bromide, hydrogen peroxide vapor, and chlorine dioxide [53]. Formaldehyde, in particular has been used to reduce bacteria in the surgery room [54]. Taking this together, using the high concentration of hypochlorite and hypochlorous acid as a liquid disinfectant might be recommended for cleaning this veterinary hospital [52].
Limited research has been conducted on the antibacterial efficacy of detergent or disinfectant products against S. pseudintermedius [52,55,56]. MRSP contamination in the environment can be reduced using household cleaning detergent and bleach, as well as antimicrobial treatment in infected dogs [55]. Furthermore, combining a high dose of chlorhexidine digluconate with cathelicidin (antimicrobial peptide) and incubating for 30-60 min can neutralize S. pseudintermedius [56]. A recent study in Thailand revealed that most MRSP obtained from dogs admitted to the veterinary hospital were strong biofilm producers [18]. It is possible that MRSP in this study could have the ability to form biofilms and develop resistance to the disinfectant. Further studies, such as biofilm formation assay, testing the effectiveness of detergents and disinfectants, must be done in the future to identify the cause of the existence of MRSP in the hospital and verify the most effective chemicals to eradicate it. Furthermore, the United States Environmental Protection Agency (https://www.epa.gov) has certified a number of commercial products as effective agents against MRSA; still, these products must be validated for their ability to eradicate MRSP.
Interestingly, the population of MRSP at a newly built veterinary hospital in southern Thailand was substantially lower than in prior findings in central Thailand, where the institution was almost 20 years old [1]. An older facility may be more susceptible to bacteria and antimicrobial drug accumulation than a new hospital. In addition, many factors may be involved in emerging antimicrobial-resistant bacteria, such as the number of patients, staff, location, and cleaning management.
Conclusion
The present study demonstrated the presence of MDR MRSP in a newly constructed veterinary hospital, with the floors and veterinary tables being the most contaminated regions. MRSP appears to be resistant to the detergent and disinfectant used in this hospital. The limitation of this study is the connection between the pathogen, animal patient, owner, and hospital staff. In addition, the mechanism by which S. pseudintermedius tolerates detergents and disinfectants should be studied in the future, as well as the efficacy of various detergents and disinfectants against is pathogen.
Authors’ Contributions
PF: Conception and designed the study, laboratory works, data analysis, and drafted the manuscript. NS and WB: Sample collection. KB: Laboratory works. TW: Conception, supervised the study, and revised manuscript. All authors have read and approved the final manuscript.
Acknowledgments
This study was fully supported by Walailak University personal research grant, Thailand (WU-IRG-64-001), and partially supported by the Thailand Science Research and Innovation Fund (WU-FF64104).
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Chanchaithong P, Perreten V, Schwendener S, Tribuddharat C, Chongthaleong A, Niyomtham W, Prapasarakul N. Strain typing and antimicrobial susceptibility of methicillin-resistant coagulase-positive staphylococcal species in dogs and people associated with dogs in Thailand. J. Appl. Microbiol. 2014;117(2):572–586. doi: 10.1111/jam.12545. [DOI] [PubMed] [Google Scholar]
- 2.Fungwithaya P, Brikshavana P, Chanchaithong P, Prapasarakul N. Distribution of methicillin-resistant coagulase-positive staphylococci (MRCoPS) in a surgical unit and cystotomy operation sites in a veterinary teaching hospital. J. Vet. Med. Sci. 2017;79(2):359–365. doi: 10.1292/jvms.16-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laarhoven L.M, de Heus, P, van Luijn J, Duim B, Wagenaar J.A, van Duijkeren E. Longitudinal study on methicillin-resistant Staphylococcus pseudintermedius in households. PLoS One. 2011;6(11):e27788. doi: 10.1371/journal.pone.0027788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch S.A, Helbig K.J. The complex diseases of Staphylococcus pseudintermedius in canines:Where to next? Vet. Sci. 2021;8(1):11. doi: 10.3390/vetsci8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin J.E, Ball K.R, Chirino-Trejo M. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from various animals. Can. Vet. J. 2011;52(2):153–157. [PMC free article] [PubMed] [Google Scholar]
- 6.Fungwithaya P, Chanchaithong P, Phumthanakorn N, Muaungkong P, Bumpenpol P, Kaewparuehaschai M, Tribuddharat C, Prapasarakul N. Association between cephalexin administration and emergence of methicillin-resistant coagulase-positive staphylococci (MRCoPS) in dogs. Thai J. Vet. Med. 2016;46(1):59–65. [Google Scholar]
- 7.Ruscher C, Lubke-Becker A, Wleklinski C.G, Soba A, Wieler L.H, Walther B. Prevalence of methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet. Microbiol. 2009;136(1-2):197–201. doi: 10.1016/j.vetmic.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Fungwithaya P, Chanchaithong P, Phumthanakorn N, Prapasarakul N. Nasal carriage of methicillin-resistant Staphylococcus pseudintermedius in dogs treated with cephalexin monohydrate. Can. Vet. J. 2017;58(1):73–77. [PMC free article] [PubMed] [Google Scholar]
- 9.Windahl U, Gren J, Holst B.S, Borjesson S. Colonisation with methicillin-resistant Staphylococcus pseudintermedius in multi-dog households:A longitudinal study using whole-genome sequencing. Vet. Microbiol. 2016;189:8–14. doi: 10.1016/j.vetmic.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz-Ripa L, Simon C, Ceballos S, Ortega C, Zarazaga M, Torres C, Gomez-Sanz E. S. pseudintermedius and S. aureus lineages with transmission ability circulate as causative agents of infections in pets for years. BMC Vet. Res. 2021;17(1):42. doi: 10.1186/s12917-020-02726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somayaji R, Priyantha M.A.R, Rubin J.E, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin:Report of 24 cases. Diagn. Microbiol. Infect. Dis. 2016;85(4):471–476. doi: 10.1016/j.diagmicrobio.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Weese J.S, van Duijkeren E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet. Microbiol. 2010;140(3-4):418–429. doi: 10.1016/j.vetmic.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 13.Stegmann R, Burnens A, Maranta C.A, Perreten V. Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. J. Antimicrob. Chemother. 2010;65(9):2047–2048. doi: 10.1093/jac/dkq241. [DOI] [PubMed] [Google Scholar]
- 14.Talan D.A, Goldstein E.J, Staatz D, Overturf G.D. Staphylococcus intermedius:Clinical presentation of a new human-dog bite pathogen. Ann. Emerg. Med. 1989;18(4):410–413. doi: 10.1016/s0196-0644(89)80582-7. [DOI] [PubMed] [Google Scholar]
- 15.Silva V, Oliveira A, Manageiro V, Canica M, Contente D, Capita R, Alonso-Calleja C, Carvalho I, Capelo J.L, Igrejas G, Poeta P. Clonal diversity and antimicrobial resistance of methicillin-resistant Staphylococcus pseudintermedius isolated from canine pyoderma. Microorganisms. 2021;25(3):482. doi: 10.3390/microorganisms9030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Chen D, Peters B.M, Li L, Li B, Xu Z, Shirliff M.E. Staphylococcal chromosomal cassettes mec (SCCmec):A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2016;101:56–67. doi: 10.1016/j.micpath.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 17.Hanssen A.M, Sollid J.U.E. SCCmec in staphylococci:Genes on the move. FEMS Microbiol. Immunol. 2006;46(1):8–20. doi: 10.1111/j.1574-695X.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- 18.Jantorn P, Heemmamad H, Soimala T, Indoung S, Saising J, Chokpaisarn J, Wanna W, Tipmanee V, Saeloh D. Antibiotic resistance profile and biofilm production of Staphylococcus pseudintermedius isolated from dogs in Thailand. Pharmaceuticals. 2021;14(6):592. doi: 10.3390/ph14060592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegener A, Broens E.M, Zomer A, Spaninks M, Wagenaar J.A, Duim B. Comparative genomics of phenotypic antimicrobial resistance in methicillin-resistant Staphylococcus pseudintermedius of canine origin. Vet. Microbiol. 2018;225:125–131. doi: 10.1016/j.vetmic.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Frosini S.M, Bond R, McCarthy A.J, Feudi C, Schwarz S, Lindsay J.A, Loeffler A. Genes on the move:In vitro transduction of antimicrobial resistance genes between human and canine staphylococcal pathogens. Microorganisms. 2020;18(12):2031. doi: 10.3390/microorganisms8122031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dancer S.J. How do we assess hospital cleaning?A proposal for microbiological standards for surface hygiene in hospitals. J. Hosp. Infect. 2004;56(1):10–15. doi: 10.1016/j.jhin.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, Kawakami T, Fukata T, Hiramatsu K. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J. Clin. Microbiol. 2010;48(3):765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocchetti T.T, Martins K.B, Martins P.Y.F, de Oliveira R.A, Mondelli A.L, Fortaleza C.M.C, da Cunha M.D.R. Detection of the mecA gene and identification of Staphylococcus directly from blood culture bottles by multiplex polymerase chain reaction. Braz. J. Infect. Dis. 2018;22(2):99–105. doi: 10.1016/j.bjid.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duran N, Ozer B, Duran G.G, Onlen Y, Demir C. Antibiotic resistance genes and susceptibility patterns in staphylococci. Indian J. Med. Res. 2012;135(3):389–396. [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo Y, Ito T, Ma X.X, Watanabe S, Kreiswirth B.N, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment:Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007;51(1):264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Jamovi Project. Jamovi. 2021. [Retrieved on 20-10-2021]. Available from: https://www.jamovi.org .
- 27.Magiorakos A.P, Srinivasan A, Carey R.B, Carmeli Y, Falagas M.E, Giske C.G, Harbarth S, Hindler J.F, Kahlmeter G, Olsson-Liljequist B, Paterson D.L, Rice L.B, Stelling J, Struelens M.J, Vatopoulos A, Weber J.T, Monnet D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria:An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 28.Hoet A.E, Johnson A, Nava-Hoet R.C, Bateman S, Hillier A, Dyce J, Gebreyes W.A, Wittum T.E. Environmental methicillin-resistant Staphylococcus aureus in a veterinary teaching hospital during a non-outbreak period. Vector Borne Zoonotic Dis. 2011;11(6):609–615. doi: 10.1089/vbz.2010.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rojas I, Barquero-Calvo E, van Balen J.C, Rojas N, Munoz-Vargas L, Hoet A.E. High prevalence of multidrug-resistant community-acquired methicillin-resistant Staphylococcus aureus at the largest veterinary teaching hospital in Costa Rica. Vector Borne Zoonotic Dis. 2017;17(9):645–653. doi: 10.1089/vbz.2017.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishihara K, Shimokubo N, Sakagami A, Ueno H, Muramatsu Y, Kadosawa T, Yanagisawa C, Hanaki H, Nakajima C, Suzuki Y, Tamura Y. Occurrence and molecular characteristics of methicillin-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus pseudintermedius in an academic veterinary hospital. Appl. Environ. Microbiol. 2010;76(15):5165–5174. doi: 10.1128/AEM.02780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins A.V, Sellon D.C, Gay J.M, Lofgren E.T, Moore D.A, Jones L.P, Davis M.A. Prevalence of methicillin-resistant Staphylococcus pseudintermedius on hand-contact and animal-contact surfaces in companion animal community hospitals. Can. Vet. J. 2020;61(6):613–620. [PMC free article] [PubMed] [Google Scholar]
- 32.Bierowiec K, Miszczak M, Korzeniowska-Kowal A, Wzorek A, Plokarz D, Gamian A. Epidemiology of Staphylococcus pseudintermedius in cats in Poland. Sci. Rep. 2021;11(1):18898. doi: 10.1038/s41598-021-97976-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces?A systematic review. BMC Infect. Dis. 2006;6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergström K, Aspán A, Landén A, Johnston C, Grönlund U. The first nosocomial outbreak of methicillin-resistant Staphylococcus aureus in horses in Sweden. Acta Vet. Scand. 2012;54(1):11. doi: 10.1186/1751-0147-54-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta M, Bisesi M, Lee J. Comparison of survivability of Staphylococcus aureus and spores of Aspergillus niger on commonly used floor materials. Am. J. Infect. Control. 2017;45(7):717–722. doi: 10.1016/j.ajic.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Chaoui L, Mhand R, Mellouki F, Rhallabi N. Contamination of the surfaces of a health care environment by multidrug-resistant (MDR) bacteria. Int. J. Microbiol. 2019;2019:3236526. doi: 10.1155/2019/3236526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergstrom A, Gustafsson C, Leander M, Fredriksson M, Gronlund U, Trowald-Wigh G. Occurrence of methicillin-resistant staphylococci in surgically treated dogs and the environment in a Swedish animal hospital. J. Small Anim. Pract. 2012;53(7):404–410. doi: 10.1111/j.1748-5827.2012.01238.x. [DOI] [PubMed] [Google Scholar]
- 38.Shoen H.R.C, Rose S.J, Ramsey S.A, de Morais H, Bermudez L.E. Analysis of Staphylococcus infections in a veterinary teaching hospital from 2012 to 2015. Comp. Immunol. Microbiol. Infect. Dis. 2019;66:101332. doi: 10.1016/j.cimid.2019.101332. [DOI] [PubMed] [Google Scholar]
- 39.Wayne A, McCarthy R, Lindenmayer J. Therapeutic antibiotic use patterns in dogs:Observations from a veterinary teaching hospital. J. Small Anim. Pract. 2011;52(6):310–318. doi: 10.1111/j.1748-5827.2011.01072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hur B.A, Hardefeldt L.Y, Verspoor K.M, Baldwin T, Gilkerson J.R. Describing the antimicrobial usage patterns of companion animal veterinary practices;free text analysis of more than 4.4 million consultation records. PLoS One. 2020;15(3):e0230049. doi: 10.1371/journal.pone.0230049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein E.Y, Van Boeckel T.P, Martinez E.M, Pant S, Gandra S, Levin S.A, Goossens H, Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U. S. A. 2018;115(15):E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thai Working Group on Health Policy and Systems Research on Antimicrobial Resistance (HPSR-AMR) Consumption of antimicrobial agents in Thailand in 2017, First Report. Vol. 1. International Health Policy Program, Ministry of Public Health, Nonthaburi. 2017. [Retrieved on 20-10-2021]. Available from: -http://www.ihppthaaigov.net/wpdm-package/thai-working-group-on-health-policy-and-systems-research-on-antimicrob .
- 43.Bagcigil A.F, Taponen S, Koort J, Bengtsson B, Myllyniemi A.L, Pyorala S. Genetic basis of penicillin resistance of S. aureus isolated in bovine mastitis. Acta Vet. Scand. 2012;54(1):69. doi: 10.1186/1751-0147-54-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stroman D.W, Mintun K, Epstein A.B, Brimer C.M, Patel C.R, Branch J.D, Najafi-Tagol K. Reduction in bacterial load using hypochlorous acid hygiene solution on ocular skin. Clin. Ophthalmol. 2017;11:707–714. doi: 10.2147/OPTH.S132851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eriksson S, van der Plas M.J.A, Morgelin M, Sonesson A. Antibacterial and antibiofilm effects of sodium hypochlorite against Staphylococcus aureus isolates derived from patients with atopic dermatitis. Br. J. Dermatol. 2017;177(2):513–521. doi: 10.1111/bjd.15410. [DOI] [PubMed] [Google Scholar]
- 46.Block M.S, Rowan B.G. Hypochlorous acid:A review. J. Oral Maxillofac. Surg. 2020;78(9):1461–1466. doi: 10.1016/j.joms.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almatroudi A, Tahir S, Hu H, Chowdhury D, Gosbell I.B, Jensen S.O, Whiteley G.S, Deva A.K, Glasbey T, Vickery K. Staphylococcus aureus dry-surface biofilms are more resistant to heat treatment than traditional hydrated biofilms. J. Hosp. Infect. 2018;98(2):161–167. doi: 10.1016/j.jhin.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Rios-Castillo A.G, Umana F.F, Rodriguez-Jerez J.J. Long-term antibacterial efficacy of disinfectants based on benzalkonium chloride and sodium hypochlorite tested on surfaces against resistant gram-positive bacteria. Food Control. 2018;93:219–225. [Google Scholar]
- 49.Kawamura M, Fujimura S, Tokuda K, Aoyagi T, Endo S, Kanamori H, Watanabe A, Kaku M. Mutant selection window of disinfectants for Staphylococcus aureus and Pseudomonas aeruginosa. J. Glob. Antimicrob. Resist. 2019;17:316–320. doi: 10.1016/j.jgar.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Lu M.C, Chen P.L, Huang D.J, Liang C.K, Hsu C.S, Liu W.T. Disinfection efficiency of hospital infectious disease wards with chlorine dioxide and hypochlorous acid. Aerobiologia. 2021;37:29–38. doi: 10.1007/s10453-020-09670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahamat A, Brooker K, Dauresa J.P, Gould I.M. Impact of hypochlorite disinfection on meticillin-resistant Staphylococcus aureus rate. J. Hosp. Infect. 2011;78(3):243–245. doi: 10.1016/j.jhin.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Pariser M, Gard S, Gram D, Schmeitzel L. An in vitro study to determine the minimal bactericidal concentration of sodium hypochlorite (bleach) required to inhibit meticillin-resistant Staphylococcus pseudintermedius strains isolated from canine skin. Vet Dermatol. 2013;24(6):632–634. doi: 10.1111/vde.12079. [DOI] [PubMed] [Google Scholar]
- 53.World Health Organization. Anthrax in Humans and Animals. 4th ed. Geneva: Disinfection, Decontamination, Fumigation, Incineration. World Health Organization; 2008. [PubMed] [Google Scholar]
- 54.Bali R, Sharma P, Nagrath S, Gupta P. Microbial isolations from maxillofacial operation theatre and its correlation to fumigation in a teaching hospital in India. J. Maxillofac. Oral Surg. 2014;13(2):128–132. doi: 10.1007/s12663-012-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frosini S.M, Bond R, King R, Feudi C, Schwarz S, Loeffler A. Effect of topical antimicrobial therapy and household cleaning on methicillin-resistant Staphylococcus pseudintermedius carriage in dogs. Vet. Rec. 2021;27:e937. doi: 10.1002/vetr.937. [DOI] [PubMed] [Google Scholar]
- 56.Santoro D, Kher L, Chala V, Navarro C. Evaluation of the effects of chlorhexidine digluconate with and without cBD103 or cCath against multidrug-resistant clinical isolates of Staphylococcus pseudintermedius. Vet. Dermatol. 2022;33(1):17–e6. doi: 10.1111/vde.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]


