Abstract
The extracellular Staphylococcus hyicus lipase was expressed under the control of different promoters in Lactococcus lactis and Bacillus subtilis. Its expression at high and moderate levels is toxic for the former and the latter hosts, respectively. In L. lactis, the lipase was expressed at a high level, up to 30% of the total cellular proteins, under the control of the inducible promoter PnisA. About 80% of the lipase remained associated with the cells. Close to half of this amount remained associated with the inner side of the cytoplasmic membrane as unprocessed pre-pro-lipase. The other half was trapped by the cell wall and partially degraded at the N-terminal end. This result suggests that extracellular proteases degrade the lipase. Surprisingly, the kinetics and the pattern of lipase degradation were different in the two L. lactis subspecies, L. lactis subsp. cremoris and L. lactis subsp. lactis. The extracellular proteolytic systems that degrade lipase are thus different in these closely related subspecies. The incorrect export of the lipase is not due to an inappropriate leader peptide but may be due to an inefficiency of several steps of lipase secretion. We propose that (i) the S. hyicus lipase may require a special accessory system to be correctly exported or (ii) the kinetics of lipase synthesis may be a critical factor for proper folding.
Lactic acid bacteria are widely used in the production and preservation of foodstuffs. Since these bacteria are generally regarded as safe, their use as delivery vehicles for foreign proteins in the field of medicine can be envisaged. With recent advances in the field of molecular biology, efficient expression vectors have been developed and allow the expression of heterologous proteins in Lactococcus lactis (53, 57, 64, 68). The aim of this work was to express a bacterial lipase in L. lactis in order to use lactococci as a lipase delivery vehicle. This goal could alleviate lipase deficiency in the digestive tract during digestion (steatorrhea) or improve flavor development in some cheese-making processes (23, 35, 56).
Among the best-characterized bacterial lipases are those of several Pseudomonas and Staphylococcus species (24). The genes encoding these lipases have been tested for heterologous expression in a variety of potential industrial production hosts. It appears that the Pseudomonas lipase cannot be overexpressed in heterologous hosts to commercially acceptable levels because it requires a helper protein to fold properly (15, 25). Moreover, the high GC content of Pseudomonas genes may require resynthesis of a gene to fit with the codon bias of the new host. On the other hand, the lipase of Staphylococcus hyicus already has been expressed successfully in Staphylococcus carnosus (66), Escherichia coli (16), and Lactobacillus curvatus (67). No chaperone or specialized helper proteins were reported to be required for the proper folding and secretion of this lipase (24). The GC content of the lip gene, 37%, is similar to that of the lactococcal genes and is comparable to that of low-GC-content gram-positive bacterial genes (16). Moreover, general codon usage is not very different in staphylococci and lactococci. This lipase was therefore chosen for overexpression in lactic acid bacteria.
The S. hyicus lip gene encodes a pre-pro-protein composed of a signal peptide of 38 amino acids (aa), a pro-peptide of 207 aa, and a mature lipase of 396 aa (66). The pro-protein is secreted by the general secretion machinery. The pro-lipase (86 kDa) is further cleaved into the mature lipase (46 kDa) in the culture medium by the metalloprotease ShpII (3). The mature lipase is threefold more active than the pro-lipase. In heterologous hosts, such as S. carnosus and E. coli, which lack ShpII, the 86-kDa form is produced (16, 66). The biochemical properties of this lipase are well characterized (66). It has a broad substrate specificity and hydrolyzes first and second ester bonds. It also has high phospholipase activities (A1 and A2), a unique feature for bacterial lipases. These properties are compatible with its use as an enzyme in some food-making processes or as an additive to alleviate lipase deficiency during digestion. However, the amounts of the enzyme needed for these purposes vary greatly. Only a small amount of lipase activity is required in cheese making, while a large amount is needed in treating steatorrhea (35, 56).
In this paper, we describe the cloning and expression of the lip gene in L. lactis at different levels. Strains producing stable lipase production at a moderate level can be used in cheese-making processes. Moreover, we achieved the overproduction of the S. hyicus lipase in an amount that could allow the use of L. lactis as a lipase delivery vector for digestive enzymes in the digestive tract. However, the ability of L. lactis to correctly secrete this lipase is limited, and the limiting steps were investigated.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. L. lactis strains were grown in M17 (59) with 0.5% glucose at 30°C. E. coli, Bacillus subtilis, and S. carnosus were grown in Luria-Bertani medium at 37°C. Chloramphenicol was used at a concentration of 10 μg/ml, and erythromycin was used at a concentration of 10 μg/ml for L. lactis and 1 mg/ml for E. coli. Nisin powder (ICN; 2.5% nisin content) was used at a concentration of 0.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| MTT19 | leuB6 trpC2 R− | Bacillus Genetic Stock Center |
| JIM7381 | MTT19 carrying pJIM2423 | This work |
| E. coli TG1 | supE thiD (lac-proAB) hsdD5 F+ traD36 proAB lacIZΔM15 | 10 |
| L. lactis | ||
| IL1403 | L. lactis subsp. lactis, plasmid free | 9 |
| NZ9000 | L. lactis subsp. cremoris, derivative of MG1363 pepN::nisR nisK | 12 |
| JIM7049 | IL1403 his::nisR nisK | S. Calero |
| JIM5496 | IL1403 carrying pJIM2429 | This work |
| JIM5497 | IL1403 carrying pJIM2430 | This work |
| JIM5498 | IL1403 carrying pJIM2431 | This work |
| JIM5928 | IL1403 carrying pJIM2098 | This work |
| JIM7022 | NZ9000 carrying pJIM2093 | This work |
| JIM7048 | JIM7049 carrying pJIM2093 | This work |
| Plasmids | ||
| pNEB193 | Ampr | New England Biolabs |
| pLipPS1 | Cmr, lip gene under the control of an S. carnosus promoter | 38 |
| pJIM2421 | Ampr, PstI fragment from pLipPS1 in pNEB193 | This work |
| pJIM2279 | Eryr, high- to low-copy-number vector for gram-positive bacteria | 47 |
| pBV502 | Eryr Cmr, contains the P5–cat-86 cassette | 4 |
| pGKV223 | Eryr Cmr, contains the P23–cat-86 cassette | 65 |
| pGKV232 | Eryr, Cmr, contains the P32–cat-86 cassette | 65 |
| pGKV244 | Eryr, Cmr, contains the P44–cat-86 cassette | 65 |
| pGKV259 | Eryr, Cmr, contains the P59–cat-86 cassette | 65 |
| pNuc11 | Eryr, contains the nuc gene with the leader sequence for Usp45 under the control of P59 | 36 |
| pNZ8008 | pNZ273 derivative carrying the gusA gene under the control of PnisA | 12 |
| pJIM2422 | Ampr Eryr, carrying the lip gene and obtained by fusion between pJIM2421 and pJIM2279 | This work |
| pJIM2423 | Eryr, carrying the lip gene under the control of Pres | This work |
| pJIM2424 | Eryr, pJIM2423 carrying the P5–cat-86 cassette upstream of the lip gene | This work |
| pJIM2425 | Eryr, pJIM2423 carrying the P23–cat-86 cassette upstream of the lip gene | This work |
| pJIM2426 | Eryr, pJIM2423 carrying the P32–cat-86 cassette upstream of the lip gene | This work |
| pJIM2427 | Eryr, pJIM2423 carrying the P44–cat-86 cassette upstream of the lip gene | This work |
| pJIM2428 | Eryr, pJIM2423 carrying the P59–cat-86 cassette upstream of the lip gene | This work |
| pJIM2429 | Eryr, pJIM2423 carrying the lip gene under the control of P23 | This work |
| pJIM2430 | Eryr, pJIM2423 carrying the lip gene under the control of P44 | This work |
| pJIM2431 | Eryr, pJIM2423 carrying the lip gene under the control of P59 | This work |
| pJIM2093 | Cmr, pNZ8008 carrying the lip gene under the control of PnisA | This work |
| pJIM2098 | pNuc11 containing the pro-lipase sequence instead of the mature nuc sequence | This work |
Isolation of plasmid DNA and enzyme analysis.
Plasmid DNA was isolated by the method of Holmes and Quigley (20) for E. coli and by the method of Anderson and McKay (2) for L. lactis, B. subtilis, and S. carnosus. S. carnosus was lysed after the addition of lysostaphin (4.5 U/ml). E. coli was transformed by the heat shock method (48). L. lactis was transformed by a procedure involving electroporation of cells grown in the presence of glycine to weaken the cell wall (21). B. subtilis was transformed by the standard method of Spizizen (55). Restriction and modification enzymes were purchased from Boehringer and used according to the instructions of the supplier.
Expression of the lip gene. (i) Construction of a promoter-probe vector with the lip gene.
A 3.5-kb PstI fragment of S. carnosus plasmid pLipPS1 (38) containing the lip gene, which encodes the pre-pro-lipase, was cloned into plasmid pNEB193 (New England Biolabs) to yield pJIM2421. This plasmid was digested with EcoRV and ligated into the general gram-positive cloning vector pJIM2279 (47) cleaved by PmeI, resulting in plasmid pJIM2422. A 3.7-kb fragment containing pNEB193 and a staphylococcal DNA fragment was then removed by SmaI-NruI digestion to give pJIM2423 (Fig. 1). This plasmid should be able to replicate in most gram-positive hosts and carries the lip coding sequence under the transcriptional control of the weak constitutive plasmid promoter Pres.
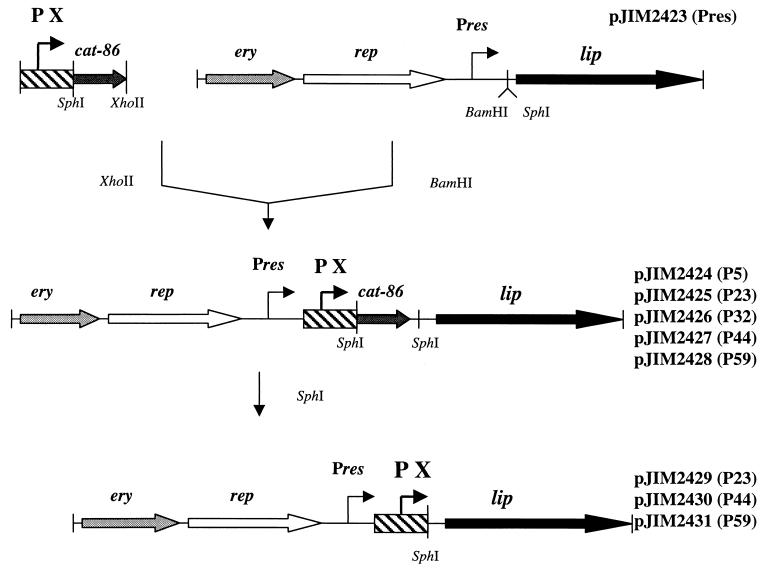
FIG. 1.
Strategy used to place the lip gene under the control of a strong constitutive promoter. Chloramphenicol resistance is used to select the first cloning step in L. lactis. PX, P5, P23, P32, P44, or P59.
(ii) Insertion of lactococcal promoters upstream of the lip gene.
The strong and constitutive promoters P5 (4) and P23, P32, P44, and P59 (65) were cloned upstream of the lip gene as outlined in Fig. 1. XhoII fragments containing the promoter–cat-86 cassette of pBV502 (4) and pGKV223, pGKV232, pGKV244, and pGKV259 (65) were inserted into the BamHI site of pJIM2423 to yield, respectively, pJIM2424, pJIM2425, pJIM2426, pJIM2427, and pJIM2428. Then, the cat-86 gene was deleted by SphI restriction, followed by ligation. This procedure put the lip gene directly under the transcriptional control of these promoters.
(iii) Expression of the lip gene under the control of the inducible promoter PnisA.
The XhoI-digested plasmid pNZ8008 (12) containing the inducible promoter PnisA was introduced into the BamHI site of pJIM2423 to yield pJIM2092. The initial vector carrying PnisA was then deleted by XhoI digestion, followed by ligation. The resulting plasmid, pJIM2093, was transformed in L. lactis strains containing nisRK, the genes necessary to induce PnisA. One strain is L. lactis subsp. cremoris NZ9000, a derivative of NZ3900 (12), and the other is L. lactis subsp. lactis JIM7049, a derivative of IL1403 (S. Calero, personal communication).
Substitution of the lipase signal peptide with the Usp45 signal peptide.
The oligonucleotides used to amplify the lip gene without the region encoding the signal peptide were 5′-GGCGTGGCAGATGCATATGATTCG-3′ and 5′-GTTTAAACCTGCGGCCGCAATTTTGA-3′; the 2.55-kb PCR fragment obtained was cloned by use of NsiI and NotI (italic letters), instead of the nuc gene on pNuc11 (36). In the resulting plasmid, pJIM2098, the pro-lipase is fused precisely with the leader peptide of Usp45 and is under the control of P59. The fusion was confirmed by sequence analysis. The product of the fused gene should be identical to that of the natural gene after secretion.
Preparation of cellular and supernatant fractions.
The cells were recovered from the medium by centrifugation for 10 min at 6,000 × g and treated as previously described (37). The supernatants were passed through filters (0.25-μm pore size) to remove eventual cellular debris. The extracellular proteins were precipitated by the addition of solid ammonium sulfate to the supernatant to 70% (wt/vol) saturation (66). After agitation for 2 h at 4°C, the precipitate was collected by centrifugation for 30 min at 10,000 × g and 4°C. The pellets were resuspended in 20 mM Tris-HCl buffer (pH 8) and dialyzed for 14 h at 4°C.
Enzymatic assays.
The lipase activity of lactococcal colonies was determined qualitatively by use of an agar plate assay medium containing 1% Tributyrin (Sigma) as a substrate and with nisin at 0.5 μg/ml for JIM7048 and JIM7022. Lipolysis is recognized as a zone of hydrolysis around the colonies. Lipase activity was determined quantitatively by monitoring the hydrolysis of tributyrin spectrophotometrically at 450 nm. The tributyrin was emulsified at 1% by sonication in a buffer consisting of 100 mM Tris-HCl (pH 8) and 25 mM CaCl2. The results obtained were converted to units per milliliter (1 U equals 1 μmol of liberated fatty acids/min). The lipase of Rhizopus arrhizus (Sigma; 50,000 U/ml) was used as a reference. Lactate dehydrogenase (LDH) activities were measured by monitoring the reduction of NAD in the presence of pyruvate spectrophotometrically at 340 nm (19).
Analysis of lipase and its cleavage products by Western blotting.
To monitor the production and the degradation products of lipase in L. lactis and B. subtilis, antilipase antibodies directed against the C-terminal (from aa 222 to the end) and N-terminal (from aa 39 to aa 222) parts of the pro-lipase were produced. The C- and N-terminal parts of the protein were fused to the E. coli maltose binding protein in order to purify it. The C-terminal part was cloned from a 1.45-kb SalI fragment of pJIM2423 and fused in frame with the maltose binding protein in the SalI site of pMal-p2 (New England Biolabs). The N-terminal part corresponding to the beginning of the gene without the signal peptide was cloned from a 0.8-kb PCR fragment ending with BamHI and SalI. The oligonucleotides used were 5′-CGGCTTATGGATCCGCGTCGTCGGTT-3′ (BamHI site in italic letters) and 5′-GGAACGTCGACTTGTTTCGGT-3′ (SalI site in italic letters). The fusion proteins MBP-CtermLIP and MBP-NtermLIP were overproduced after induction by isopropyl-β-d-thiogalactopyranoside (IPTG) and purified on affinity columns according to the instructions of the supplier. The purified fusion proteins were inoculated into rabbits to produce polyclonal antisera. Anti-CtermLIP and anti-NtermLIP sera do not reveal proteins from the wild-type L. lactis strains in Western blotting and are specific for the S. hyicus lipase.
Anti-PepC antibodies used as a control for cell lysis were kindly provided by M.-P. Chapot-Chartier (Laboratoire de Recherches sur les Protéines, Institut National de la Recherche Agronomique, Jouy en Josas, France).
Proteins were electrophoresed in sodium dodecyl sulfate-polyacrylamide gels (34) and then either stained with Coomassie blue or immunoblotted with polyclonal rabbit antibodies (1/1,000), followed by ECL kit detection (Amersham).
Localization of the heterologous lipase.
The culture was grown at 30°C to an optical density at 600 nm (OD600) of 0.5 and centrifuged for 10 min at 6,000 × g and 4°C to separate the supernatant from the cells. Cells were washed and resuspended in a buffer containing 0.5 M sucrose, 0.04 M magnesium acetate, 1 mM ammonium acetate, 0.04 M CaCl2, and 2 mg of lyzozyme (pH 7) per ml, incubated for 1 h at 37°C (50), and then divided into three aliquots. According to previously described protocols (1, 46), the first aliquot was kept for 30 min at 37°C as a control. The second aliquot was treated with trypsin (Sigma) (1 mg/ml) for 30 min at 37°C. In this sample, trypsin will digest only the proteins bound to the membrane and presented outside the cell. The third aliquot was treated with 1% Triton X-100 for 5 min and then with trypsin (1 mg/ml) for a further 30 min at 37°C. The Triton X-100 treatment will dissolve the membranes, and all proteins should be digested by trypsin. To terminate the trypsin digestion in the second and third aliquots, 4 mg of trypsin inhibitor (Sigma) per ml was added at the end of the incubation. The first two aliquots were then centrifuged for 10 min at 6,000 × g to collect the protoplasts and separate them from the cell wall extract (supernatant of the lyzozyme-treated cells). Finally, the protoplasts were suspended in a buffer provoking their osmotic lysis. The lysed samples were centrifuged for 20 min at 13,000 × g. The cytoplasmic fraction was in the supernatant, and the membrane-bound fraction was in the pellet.
Computer analysis.
The search for the codon bias of highly expressed genes in staphylococci and lactococci was made with the help of the Nakamura database (41) and with 139,261 codons for L. lactis and 172,616 codons for Staphylococcus aureus. The bias for the highly expressed glycolytic genes previously reported in L. lactis (8) was confirmed in this analysis. S. aureus was selected as a representative of the genus Staphylococcus because the number of sequenced genes in S. hyicus is not sufficient for statistical analysis. The bias for highly expressed genes in B. subtilis was kindly communicated by Y. Moszer.
RESULTS
Constitutive expression of the lipase at a high level is toxic to L. lactis cells.
Five strong promoters from L. lactis were cloned upstream of the lip gene, P5 (4) and P23, P32, P44, and P59 (65) (Fig. 1). P5 and P59 drive the expression of the rRNA operon, P23 drives the expression of a potential integral membrane protein, P32 drives the expression of the fructose 1-6 bisphosphate aldolase, and P44 drives the expression of a cell division protein, FtsA. The procedure is represented in Fig. 1. In the first step, an L. lactis promoter–cat-86 cassette was cloned upstream of the lip gene on a lactococcal replicative plasmid. The second step, removing the cat-86 gene and its terminator and placing the lip gene directly under the control of the promoter, was carried out successfully for P23 (pJIM2429), P44 (pJIM2430), and P59 (pJIM2431), resulting in strains JIM5496, JIM5497, and JIM5498, respectively (Table 2). In these strains, lipase production significantly affects the growth of the cells, since the generation time is 80 min, instead of the 45 min for wild-type strain IL1403 (data not shown). Deletion of the cat-86 gene failed for the other two promoters, P5 and P32.
TABLE 2.
Qualitative and quantitative lipolytic activities of different constructions carrying the S. hyicus lip gene in L. lactis IL1403a
| Plasmid | Cassette | Halos around coloniesb | Maximal lipolytic activity (U/ml)
|
|
|---|---|---|---|---|
| Intracellular | Extracellular | |||
| pJIM2422 | No promoter | − | <0.3 | <0.3 |
| pJIM2423 | Pres | + | <0.3 | <0.3 |
| pJIM2424 | P5–cat-86 | + | <0.3 | <0.3 |
| pJIM2425 | P23–cat-86 | − | <0.3 | <0.3 |
| pJIM2426 | P32–cat-86 | + | <0.3 | <0.3 |
| pJIM2427 | P44–cat-86 | − | <0.3 | <0.3 |
| pJIM2428 | P59–cat-86 | − | <0.3 | <0.3 |
| pJIM2429 | P23 | ++ | 9.5 | 0.77 |
| pJIM2430 | P44 | ++ | 8.3 | 0.52 |
| pJIM2431 | P59 | ++ | 3.9 | 0.3 |
| pJIM2093 | PnisA | +++ | 110 | 7 |
The limit of detection is 0.3 U/ml. The maximal quantitative lipolytic activities were determined during exponential growth for the constitutive promoters and 3 h after induction by nisin for the inducible one.
−, no halo; +, small; ++, medium; +++, large.
The fact that repeated attempts to delete the cat-86 gene from the two constructions containing P5 and P32 failed suggests that the lip gene under the control of these promoters is toxic for L. lactis cells. Indeed, the clones containing pJIM2424 or pJIM2426 gave halos, revealing lipase production despite the presence of the cat-86 gene and its terminator between P5 or P32 and the lip gene, while no halo was detected with the other cassettes (Table 2). This result shows that these two promoters are stronger than the others in this plasmid context and confirms that lipase overexpression is toxic for the cells.
Overexpression of the lipase under the control of an inducible promoter.
Since the expression of the lip gene under the control of constitutive promoters stronger than P23 was toxic for the cells (Table 2), we used an inducible promoter, PnisA (12) to increase the production of lipase. This promoter is tightly regulated, closed in the absence of nisin and opened in a dose-dependent manner in the presence of nisin in strains containing the nisRK regulators. Two such strains were used in this work, NZ9000, a derivative of L. lactis subsp. cremoris NZ3900 (12), and JIM7049, a derivative of L. lactis subsp. lactis IL1403 (S. Calero, personal communication). The lipase was induced by nisin as described in Materials and Methods at an OD600 of 0.3 in strains JIM7022 and JIM7048 (respectively, NZ9000 and JIM7049 carrying pJIM2093). This condition was determined previously as optimal for lipase overproduction (data not shown). Cell growth was severely affected after nisin induction in the presence of pJIM2093 (Fig. 2A), while it was not without nisin (generation time, 45 min); nisin had no significant effect on the parental strains lacking pJIM2093 (data not shown).
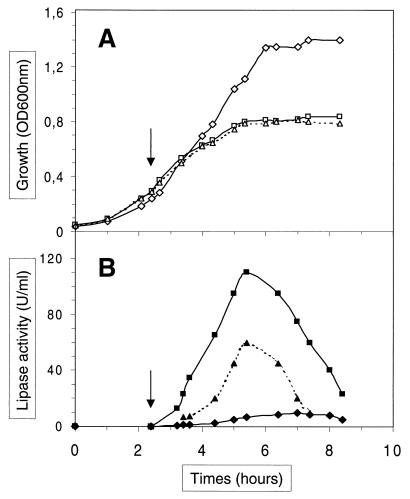
FIG. 2.
Growth (A) and lipolytic activities (B) determined for cell extracts of strains JIM5496 (diamonds), JIM7022 (squares), and JIM7048 (triangles). Strains JIM7022 and JIM7048 were induced by nisin at an OD600 of 0.3 (arrows).
The lipolytic activities were determined in units per milliliter of cell extracts or supernatants (Table 2). The maximal lipolytic activity was reached 3 h after induction and was about 10-fold higher for JIM7022 and 5-fold higher for JIM7048 than the activity obtained with the constitutive promoter P23 in JIM5496 (Table 2 and Fig. 2). The activities in the cellular fractions were approximately 5- to 10-fold higher than the extracellular ones. This result suggests that secretion is inefficient or that a large amount of lipase remains bound to the cells. Cell-associated lipase production was also monitored by staining proteins with Coomassie blue. Scanning of the gel and measurement of the band density (of the pre-pro-lipase at 91 kDa, the pro-lipase at 86 kDa, and the shorter products of the lipase ranging between 52 and 83 kDa; see also below) indicated that the lipase represents about 30% of the protein in the cell (Fig. 3). Quantification by successive dilution of total cell extracts of JIM7022 on a Western blot showed that the lipase was synthesized in a quantity 60-fold higher in this strain than in JIM5496 (P23) (data not shown).
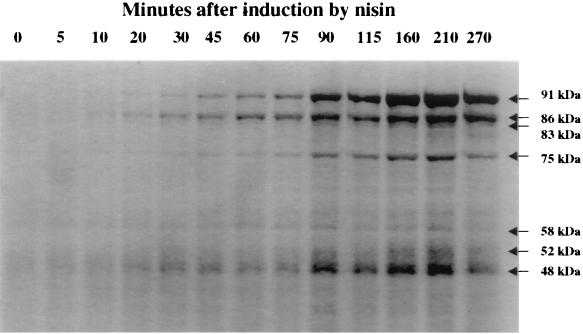
FIG. 3.
Coomassie blue staining of JIM7022 total cell extracts. The samples (0.1 U, corresponding to the quantity of cells in 100 μl of L. lactis culture at an OD600 of 1) were loaded at different times after nisin induction (in minutes) as shown in Fig. 2.
The lipase is degraded after membrane translocation.
To study the secretion of lipase in L. lactis, we monitored its production on Western blots using anti-CtermLIP sera because the amount of lipase produced under the control of our constitutive promoters (P23, P44 or P59) was too low to be detected in protein gels by Coomassie blue staining. The lipase was present in the total cell extracts in a quantity about 5-fold higher than that in the supernatants (Fig. 4A). A 91-kDa band, present only in the total cell extracts, could correspond to the pre-pro-lipase before it is translocated through the membrane. An 86-kDa band detected both in the supernatants and in the cells had the size expected for the pro-lipase after cleavage by the signal peptidase. Four additional truncated forms, at 81 kDa, 65 kDa, 54 kDa, and 52 kDa, larger than the mature lipase (46 kDa), were detected in significant amounts in the total cell extracts and in the supernatants.
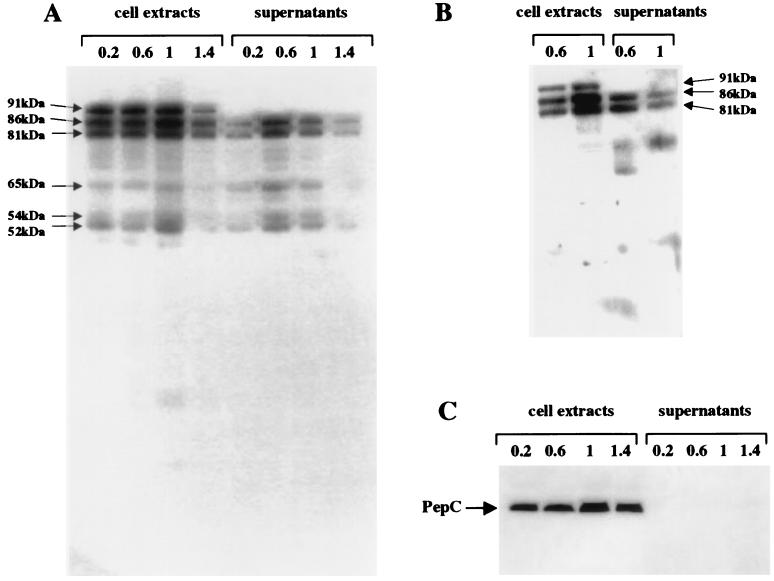
FIG. 4.
Western blots of total cell extract (0.1 U) and supernatant (0.5 U) proteins from lactococcal strain JIM5496 with anti-CtermLIP (A), anti-NtermLIP (B), and anti-PepC sera (C). The samples were taken at different stages of growth (OD600s of 0.2, 0.6, 1, and 1.4). See the legend to Fig. 3 for an explanation of U.
The truncated lipase forms could result from the premature termination of synthesis or from degradation by protease(s). To discriminate between these hypotheses, we used anti-NtermLIP sera. In the first case, the truncated forms detected with anti-CtermLIP antibodies should be detected with anti-NtermLIP antibodies. Moreover, new, smaller bands corresponding to a premature stop upstream of the C-terminal part of the lipase could also be detected with anti-NtermLIP antibodies. In the case of degradation by protease(s), bands corresponding to degradation products truncated in the N-terminal part should disappear, while new bands corresponding to degradation products containing the first part of the lipase should appear. Western blots with specific anti-NtermLIP antibodies detected the potential 86-kDa pro-lipase and the 81-kDa form in the cell extracts and the supernatants, while the truncated 65-, 54-, and 52-kDa forms were not revealed (Fig. 4B). Thus, these do not contain the N-terminal part of the lipase and are most likely degraded forms of the lipase.
The degraded lipase forms were detected both associated with the cells and in the supernatants. They could be liberated either after cell lysis or after secretion. To test the possibility of lysis, we searched for the presence of the intracellular enzymes LDH (by an enzymatic assay) and PepC (with anti-PepC sera) in the supernatants. LDH activity (data not shown) and PepC activity (Fig. 4C) were detected in the cell extracts only. Lipase degradation thus probably takes place after translocation.
The extracellular proteolytic systems of the two L. lactis subspecies are different.
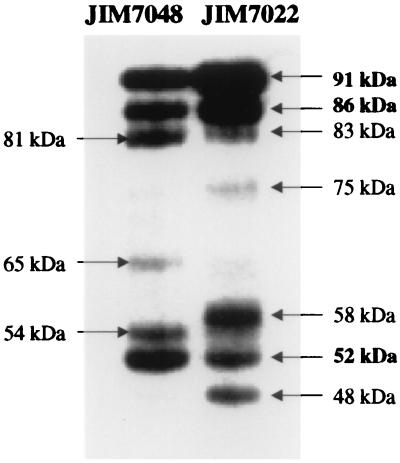
We compared lipase expression in the cell extracts of two strains, L. lactis subsp. cremoris JIM7022 (PnisA) and L. lactis subsp. lactis JIM7048 (PnisA), using Western blots with anti-CtermLIP sera (Fig. 5). The pattern of the degradation products seemed to be subspecies dependent. The bands observed with JIM7048 (PnisA) and JIM5496 (P23), both of which are L. lactis subsp. lactis but which produce different amounts of lipase, were the same: the pre-pro-lipase at 91 kDa, the pro-lipase at 86 kDa, and four degraded forms at 81, 65, 54, and 52 kDa. In L. lactis subsp. cremoris JIM7022 (PnisA), the degradation products of 81, 65, 54, and 52 kDa were not present, and four other bands appeared instead, at 83, 75, 58, and 48 kDa. We conclude that the proteases involved in lipase degradation in the two lactococcal strains have different cleavage specificities.
FIG. 5.
Comparison of the degradation products of the S. hyicus lipase produced in JIM7048 (L. lactis subsp. lactis) and JIM7022 (L. lactis subsp. cremoris), as revealed by immunostaining with anti-CtermLIP sera.
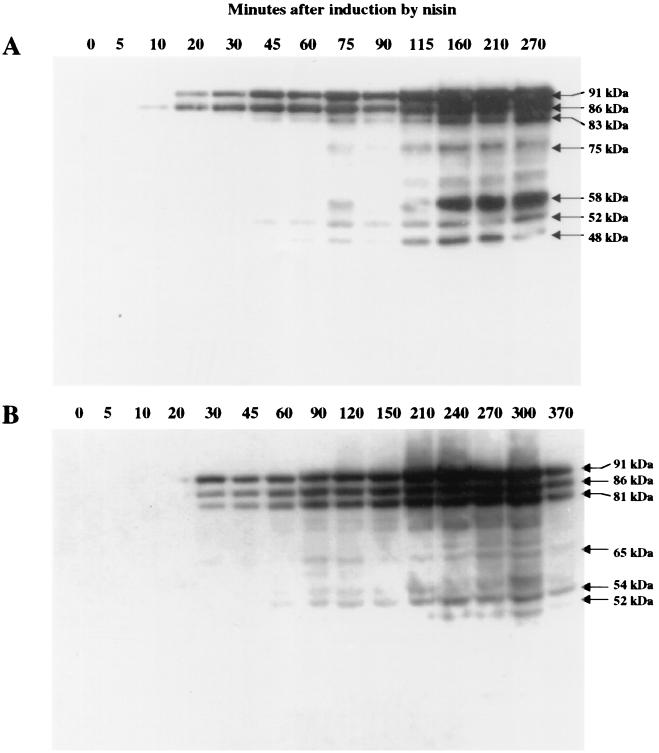
The kinetics of degradation of the lipase are different in the two strains, as deduced from the experiments shown in Fig. 6. In L. lactis subsp. cremoris JIM7022 (PnisA) (Fig. 6A), the bands corresponding to the lipase were visualized earlier after induction by nisin, leading to the production of a single band corresponding to the pro-lipase after 10 min. A second band corresponding to the pre-pro-lipase appeared after 20 min, and the intensity of these two bands increased progressively, to reach a maximum at about 150 min. The first degradation products were detected at very small amounts after 45 to 60 min, and the intensity of the corresponding bands increased progressively. In L. lactis subsp. lactis JIM7048 (PnisA) (Fig. 6B), the first bands appeared after 30 min. Since the early kinetics of induction by nisin are similar for the two strains with the luciferase gene as a reporter gene (data not shown), this result suggests that lipase is efficiently degraded at the early stage of production. Moreover, pre-pro-lipase, pro-lipase, and a degradation product are visualized simultaneously, confirming that degradation of the lipase occurs immediately upon its synthesis. Further degradation occurs later, as for the other strain.
FIG. 6.
Differences in the kinetics of lipase production and degradation of cell extracts (0.1 U) from JIM7022 (A) and JIM7048 (B), as revealed by immunostaining with anti-CtermLIP sera. Total cell extracts were loaded at different times after nisin induction (in minutes) as shown in Fig. 2. See the legend to Fig. 3 for an explanation of U.
Localization of the lipase and of its degradation products.
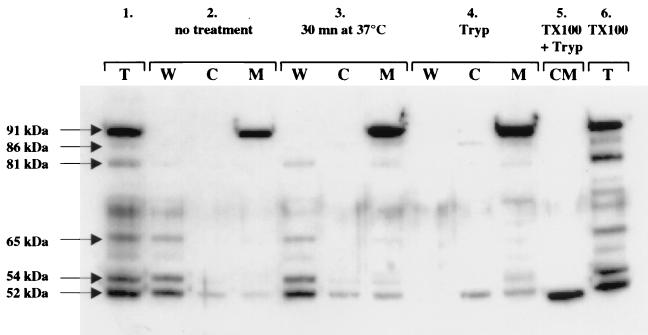
The S. hyicus lipase and its degradation products were localized by separating the cell components into three fractions corresponding to the cytoplasm, the membrane, and the cell wall. Interestingly, the different bands observed previously were not present in the same cell fractions (Fig. 7). The 81-, 65-, 54-, and 52-kDa bands and the pro-lipase were found in the cell wall extract (Fig. 7, lane 2W), while only the 52-kDa product, a possible contaminant from the cell wall due to incomplete wall digestion, was present in small amounts in the cytoplasm and in the membrane (lanes 2C and 2M and lanes 3C and 3M). These results confirm that the lipase is degraded by protease(s) after export. Finally, the pre-pro-lipase was found exclusively in the fraction containing the membrane (Fig. 7, lane 2M).
FIG. 7.
Localization of the S. hyicus lipase in strain JIM5496 (P23) by Western blotting with anti-CtermLIP sera. Panel 1, total cell extract; panel 2, sample without treatment; panel 3, sample incubated for 30 min at 37°C; panel 4, cell wall and protoplast fractions incubated with trypsin for 30 min at 37°C; panel 5, protoplasts incubated with Triton X-100 and trypsin for 30 min at 37°C; panel 6, total cell extract incubated with Triton X-100 for 30 min at 37°C. Abbreviations: T, total cell extract; W, cell wall fraction; C, cytoplasm fraction; M, membrane fraction; CM, protoplasts; Tryp, trypsin; TX100, Triton X-100.
To determine the orientation of the lipase in the membrane, we performed trypsin digestion of protoplasts with or without Triton X-100 treatment. Trypsin will digest only the proteins displayed on the outside membrane surface of nontreated protoplasts. Triton X-100 treatment dissolves the membranes, and all the proteins should be digested by trypsin. The pre-pro-lipase was not digested by trypsin when it was added to intact protoplasts (Fig. 7, compare lanes 3M and 4M). Only one band smaller than 52 kDa could be detected, and it was resistant to trypsin degradation (Fig. 7, lane 5). This result shows that most of the lipase precursor is anchored to the membrane inside the cell.
The low efficiency of lipase translocation is probably not leader peptide dependent.
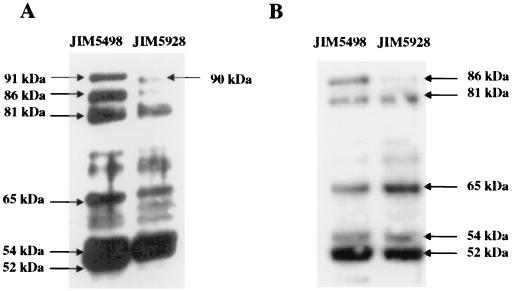
To investigate the reason for the poor export of the lipase, we replaced its leader peptide with a lactococcal one. The lip leader has all the features known to be required for protein export in gram-positive bacteria. However, we assumed that the S. hyicus leader may not be well recognized or may have some other properties which render it not fully functional in L. lactis. The lipase was thus fused to the leader peptide of Usp45, the most abundant protein secreted by L. lactis. The fusion removed precisely the lipase leader and replaced it with the L. lactis one. A shorter pre-pro-lipase should thus be obtained, due to the length difference between the signal peptides of Usp45 (27 aa) and the lipase (38 aa). No significant improvement in lipase export was observed (Fig. 8). Moreover, the same degradation pattern was obtained with the native and the Usp45 leader peptides. A similar experiment with the synthetic pro-peptide LEISSTCDA (37) gave similar results (data not shown; P. Langella, personal communication). This result strongly suggests that the poor export of lipase is not due to its leader peptide.
FIG. 8.
Effect of the Usp45 leader peptide on the secretion of lipase. Shown is a Western blot of total cell extract (0.1 U) (A) and supernatant (0.5 U) (B) proteins from lactococcal strains JIM5498 (native leader) and JIM5928 (Usp45 leader). The samples were taken at an OD600 of 0.8. See the legend to Fig. 3 for an explanation of U.
Limitation of S. hyicus lipase secretion in B. subtilis.
Lactococci do not naturally secrete many proteins and thus might lack a factor for efficient secretion of some heterologous proteins. We thus wanted to test if the low efficiency of lipase translocation that we encountered was specific for lactococci. Bacilli have a high capacity for secretion of proteins and are used industrially for the production of extracellular enzymes. Moreover, some Bacillus species are able to naturally produce extracellular lipases sharing high homology with the staphylococcal lipases (29, 49). In order to study the expression of the S. hyicus lipase in B. subtilis, the model gram-positive bacterium for the secretion of proteins, pJIM2423 (Pres), pJIM2429 (P23), pJIM2430 (P44), and pJIM2431 (P59) were used to transform MTT19 cells. Despite the facts that MTT19 cells were highly competent and that the plasmids were purified by the same method, transformed clones were obtained only for pJIM2423 (Pres). This plasmid expresses the lipase from a low-copy-number constitutive promoter, Pres. The production and localization of lipase were monitored by Western blotting with anti-Cterm-LIP sera (data not shown). As in L. lactis, a form corresponding to the pre-pro-lipase was present in the cells. Moreover, significant amounts of smaller forms of the lipase were linked to the cells (data not shown). Finally, small amounts of fragments corresponding to degradation products of the lipase could be detected in the supernatants. These results show that B. subtilis does not express the lipase properly, although it has a high capacity to secrete heterologous proteins.
DISCUSSION
The S. hyicus lip gene was expressed under the control of strong constitutive promoters in L. lactis. The strongest promoters, such as P32, a promoter driving the expression of a glycolytic gene, the fructose 1-6 bisphosphate aldolase gene, were toxic for the cells. This toxicity was even more pronounced in B. subtilis, although bacilli are naturally able to secrete large amounts of proteins, including lipases that share high homology with that of S. hyicus (29, 49). To overcome the problem of toxicity and increase the production of lipase in L. lactis, the lip gene was expressed from an inducible promoter controlled by the addition of nisin in the medium. This strategy led to a 10-fold increase in lipolytic activity and allowed production of the lipase to about 30% of total cell protein. While the lipase activity was increased 10-fold, the bulk of lipase was increased 60-fold compared to the level obtained with constitutive promoters. The sixfold difference between the two measures suggests that an important fraction of the produced lipase is inactive. In order to understand the bottleneck for lipase production in L. lactis, the different forms of the enzyme were characterized.
The use of immunodetection techniques allowed us to show that in addition to the pre-pro-lipase and the pro-lipase, large amounts of smaller lipase forms were produced in L. lactis and B. subtilis. In S. carnosus overexpressing the lipase from pLipPS1, only the pro-lipase can be detected in the supernatant (data not shown); in S. hyicus, only the mature lipase is detected in the supernatant (data not shown). Most of the smaller forms were present in the cell wall extracts and in the supernatants. They include the C-terminal part of the lipase, suggesting that proteolytic cleavages occur at its N-terminal part by cell wall-associated proteases. This degradation occurs during or shortly after membrane translocation, especially in L. lactis subsp. lactis. The proteolysis of heterologous proteins is a factor that is often reported to limit the yield of a product. A good example is the degradation of E. coli OmpA upon secretion in B. subtilis and S. carnosus (40). The difference in the degradation patterns of the lipases of the two L. lactis subspecies (and also B. subtilis) suggests that the proteases involved have different specificities. However, the two L. lactis strains used in this work are devoid of the extracellular proteinase, PrtP, that was shown to be involved in AcmA autolysin degradation (6). In the recently sequenced B. subtilis and L. lactis subsp. lactis genomes, genes encoding proteins homologous to E. coli DegP, involved in the degradation of unfolded or misfolded proteins trapped in the cell wall (43, 58), were detected (A. Bolotin, personal communication).
Different factors might induce the degradation of the lipase by specialized proteases, such as DegP. The fact that most of the secreted lipase remains associated with the cell wall suggests that the lipase has some difficulties crossing the cell wall. It is thus trapped in a location favoring its degradation by the membrane and cell wall-bound proteases. We do not know why the lipase is not able to pass efficiently through the cell wall, since L. lactis and B. subtilis are able to secrete proteins larger than lipase, such as a plasmid-encoded protease (18). To our knowledge, the cell wall of S. hyicus is not thinner than or has no special properties compared to those of L. lactis and B. subtilis. The poor ability of the lipase to cross the cell wall could be due to its lack of proper folding during secretion.
In addition to its inability to cross the cell wall, only 20% of the produced lipase is secreted outside the cytoplasmic membrane. This finding indicates that another step in secretion is also ineffective. Indeed, most of the lipase is still bound to the inner side of the membrane. This finding is probably not due to an inefficient signal peptide. First, the lipase gene exhibits a consensus signal sequence for gram-positive bacteria, such as L. lactis and B. subtilis. This sequence is composed of an initial hydrophilic segment (aa 1 to 16), followed by a hydrophobic sequence (aa 17 to 34), and ends with Ala-Glu-Ala, a classical sequence of the cleavage site recognized by the general signal peptidase (16). Second, replacement with the lactococcal signal peptide of Usp45, the most abundant protein secreted by L. lactis (62), did not change the pattern of secretion of the lipase and did not improve lipase export. The factor affecting secretion should thus be encoded downstream of the peptide leader in the lipase gene or be an additional factor that interacts with the lipase.
It was reported that the region adjacent to the leader peptide has an effect on secretion (37). The synthetic pro-peptide LEISSTCDA, known to enhance the secretion of the S. aureus nuc and Bacillus stearothermophilus α-amylase genes, was introduced between the Usp45 leader peptide and the lipase pro-peptide (P. Langella, personal communication). However, it did not lead to better secretion of the S. hyicus lipase, showing that the leader peptide and the adjacent lipase sequence were not responsible for the lipase secretion-limiting step (data not shown). The presence of a large pro-peptide may improve secretion and prevent protein degradation, for example, by guiding the folding of the secreted protein (5, 11, 22). Interestingly, the pro-peptide is not present in Bacillus lipases (49), suggesting that it might be specific for staphylococci. However, it improves the secretion of the E. coli OmpA protein in B. subtilis (40), suggesting a more subtle role during protein secretion.
From the structure of the lip leader peptide and current knowledge, it is expected that the lipase is secreted through the general pathway already described for L. lactis (37, 44, 45, 52). The factors known to be involved in the export and in the folding of the protein during secretion (33, 39) are present in the L. lactis genome (13, 30, 63; A. Bolotin, personal communication), although L. lactis secretes small amounts of proteins. A single secreted protein, Usp45, of unknown function, can be systematically identified in sodium dodecyl sulfate-polyacrylamide gels, while only traces of other proteins are detectable in L. lactis (62). These results suggest that L. lactis does not have a large potential for secreting proteins; thus, some components of the secretion machinery could be present in limiting amounts. An example of such a limiting factor has been documented for B. subtilis, the model organism for the secretion of protein in gram-positive bacteria. In this organism, the overproduction of PrsA allows an increase in the secretion of a single-chain antibody fragment (69). PrsA, a membrane-associated lipoprotein, plays the role of an extracellular chaperone, allowing the correct folding of secretory proteins after their translocation across the cytoplasmic membrane (31, 32). A homologue of this protein is present in L. lactis (45), and it could be of interest to test if it might be a limiting factor in the secretion of the lipase. However, the patterns of lipase production are similar when the lipase is expressed at different levels, in particular at a low level close to the limit of detection in Western blotting. Moreover, L. lactis is able to secrete large amounts of certain heterologous proteins, such as staphylococcal nuclease (36). These results do not support the hypothesis of a general factor limiting secretion.
Another possibility is that the lipase requires a specialized factor for its proper secretion. This is the case for Pseudomonas and Vibrio cholerae, where the secretion of the lipase encoded by lipA requires the expression of the linked lipB gene (15, 25, 42). Indeed, the S. hyicus lipase is well secreted in S. hyicus and S. carnosus but not in E. coli (16), L. lactis, and B. subtilis. Secretion efficiency in L. curvatus, a lactic acid bacterium, is not known, because the lipolytic activity was determined only by qualitative analysis (67). It is thus possible that staphylococci possess a specific foldase, not yet characterized but required for efficient lipase secretion.
Last, the rate of protein synthesis and the occurrence of pauses may have a determinant effect on the folding of the product (17, 26, 28, 61). Rate and pauses may be affected by codon bias (26, 54, 60) and codon context (7, 14). Highly expressed genes have a strong bias, allowing optimal synthesis (27, 51). The codon biases for highly expressed genes in L. lactis, B. subtilis, and S. aureus differ significantly (unpublished data; see Materials and Methods). In particular, UUA is the preferred leucine codon in S. aureus (80 to 90%) but is almost absent in L. lactis (less than 1%), while UUG and CUU are rare in S. aureus (1 and 5%, respectively) but frequent in L. lactis (38 and 53%, respectively). Not withstanding the absence of a cluster of rare codons in the lipase gene, it is possible that pauses or slowing down in the translation of the lip message takes place in different regions in L. lactis, B. subtilis, and S. hyicus. This process could lead to incorrect folding of the nascent peptide in the heterologous hosts and thus to an inactive and inefficiently exported lipase protein.
ACKNOWLEDGMENTS
We thank F. Götz for providing plasmid pLipPS1, O. P. Kuipers for providing plasmid pNZ8008 and strain NZ9000, and Y. Le Loir and P. Langella for providing plasmid pNuc11. We thank J. Anba, S. Bonneau, S. Calero, P. Langella, and I. Poquet for advice during this work and A. Bolotin for communication of the L. lactis genome sequence.
REFERENCES
- 1.Anba J, Pages J M, Lazdunski C. Mode of transfer of the phosphate-binding protein through the cytoplasmic membrane in Escherichia coli. FEMS Microbiol Lett. 1986;34:215–219. [Google Scholar]
- 2.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayora S, Lindgren P E, Götz F. Biochemical properties of a novel metalloprotease from Staphylococcus hyicus subsp. hyicus involved in extracellular lipase processing. J Bacteriol. 1994;176:3218–3223. doi: 10.1128/jb.176.11.3218-3223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bojovic B, Djordjevic G, Banina A, Topisirovic L. Mutational analysis of cat-86 gene expression controlled by lactococcal promoters in Lactococcus lactis subsp. lactis and Escherichia coli. J Bacteriol. 1994;176:6754–6758. doi: 10.1128/jb.176.21.6754-6758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun P, Tommassen J, Filloux A. Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol Microbiol. 1996;19:297–306. doi: 10.1046/j.1365-2958.1996.381908.x. [DOI] [PubMed] [Google Scholar]
- 6.Buist G, Venema G, Kok J. Autolysis of Lactococcus lactis is influenced by proteolysis. J Bacteriol. 1998;180:5947–5953. doi: 10.1128/jb.180.22.5947-5953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulmer M. The effect of context on synonymous codon usage in genes with low codon usage bias. Nucleic Acids Res. 1990;18:2869–2873. doi: 10.1093/nar/18.10.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancilla M R, Hillier A J, Davidson B E. Lactococcus lactis glyceraldehyde-3-phosphate dehydrogenase gene, gap: further evidence for strongly biased codon usage in glycolytic pathway genes. Microbiology. 1995;141:1027–1036. doi: 10.1099/13500872-141-4-1027. [DOI] [PubMed] [Google Scholar]
- 9.Chopin A, Chopin M C, Moillo-Batt A, Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984;11:260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- 10.Condon S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol Lett. 1987;46:269–280. [Google Scholar]
- 11.Demleitner G, Götz F. Evidence for importance of the Staphylococcus hyicus lipase pro-peptide in lipase secretion, stability and activity. FEMS Microbiol Lett. 1994;121:189–197. doi: 10.1111/j.1574-6968.1994.tb07098.x. [DOI] [PubMed] [Google Scholar]
- 12.De Ruyter P G G A, Kuipers O P, De Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton T, Shearman C, Gasson M J. Cloning and sequence analysis of the dnaK gene region of Lactococcus lactis. J Gen Microbiol. 1993;139:3253–3264. doi: 10.1099/00221287-139-12-3253. [DOI] [PubMed] [Google Scholar]
- 14.Folley L S, Yarus M. Codon contexts from weakly expressed genes reduce expression in vivo. J Mol Biol. 1989;209:359–378. doi: 10.1016/0022-2836(89)90003-x. [DOI] [PubMed] [Google Scholar]
- 15.Frenken L G, de Groot A, Tommassen J, Verrips C T. Role of the lipB gene product in the folding of the secreted lipase of Pseudomonas glumae. Mol Microbiol. 1993;9:591–599. doi: 10.1111/j.1365-2958.1993.tb01719.x. [DOI] [PubMed] [Google Scholar]
- 16.Götz F, Popp F, Korn E, Schleifer K H. Complete nucleotide sequence of the lipase gene from Staphylococcus hyicus cloned in Staphylococcus carnosus. Nucleic Acids Res. 1985;13:5895–5906. doi: 10.1093/nar/13.16.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guisez Y, Robbens J, Remaut E, Fiers W. Folding of the MS2 coat protein in Escherichia coli is modulated by translational pauses resulting from mRNA secondary structure and codon usage: a hypothesis. J Theor Biol. 1993;162:243–252. doi: 10.1006/jtbi.1993.1085. [DOI] [PubMed] [Google Scholar]
- 18.Haandrikman A J, Kok J, Laan H, Soemitro S, Ledeboer A M, Konings W N, Venema G. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J Bacteriol. 1989;171:2789–2794. doi: 10.1128/jb.171.5.2789-2794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillier A J, Jago G R. l-Lactate dehydrogenase, FDP-activated, from Streptococcus cremoris. Methods Enzymol. 1982;89:362–367. doi: 10.1016/s0076-6879(82)89065-4. [DOI] [PubMed] [Google Scholar]
- 20.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 21.Holo H, Nes I F. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inouye M. Intramolecular chaperone: the role of the pro-peptide in protein folding. Enzyme. 1991;45:314–321. doi: 10.1159/000468904. [DOI] [PubMed] [Google Scholar]
- 23.Jaeger K E, Reetz M T. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/s0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- 24.Jaeger K E, Ransac S, Dijkstra B W, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen S, Skov K W, Diderichsen B. Cloning, sequence, and expression of a lipase gene from Pseudomonas cepacia: lipase production in heterologous hosts requires two Pseudomonas genes. J Bacteriol. 1991;173:559–567. doi: 10.1128/jb.173.2.559-567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane J F. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 27.Karlin S, Mrazek J, Campbell A M. Codon usage in different gene classes of the Escherichia coli genome. Mol Microbiol. 1998;29:1341–1355. doi: 10.1046/j.1365-2958.1998.01008.x. [DOI] [PubMed] [Google Scholar]
- 28.Kepes F. The “+70 pause”: hypothesis of a translational control of membrane protein assembly. J Mol Biol. 1996;262:77–86. doi: 10.1006/jmbi.1996.0500. [DOI] [PubMed] [Google Scholar]
- 29.Kim H K, Park S Y, Lee J K, Oh T K. Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci Biotechnol Biochem. 1998;62:66–71. doi: 10.1271/bbb.62.66. [DOI] [PubMed] [Google Scholar]
- 30.Kim S G, Batt C A. Cloning and sequencing of the Lactococcus lactis subsp. lactis groESL operon. Gene. 1993;127:121–126. doi: 10.1016/0378-1119(93)90626-e. [DOI] [PubMed] [Google Scholar]
- 31.Kontinen V P, Saris P, Sarvas M. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol Microbiol. 1991;5:1273–1283. doi: 10.1111/j.1365-2958.1991.tb01901.x. [DOI] [PubMed] [Google Scholar]
- 32.Kontinen V P, Sarvas M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol. 1993;8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 33.Kumamoto C A. Molecular chaperones and protein translocation across the Escherichia coli inner membrane. Mol Microbiol. 1991;5:19–22. doi: 10.1111/j.1365-2958.1991.tb01821.x. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Layer P, Keller J. Pancreatic enzymes: secretion and luminal nutrient digestion in health and disease. J Clin Gastroenterol. 1999;28:3–10. doi: 10.1097/00004836-199901000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Le Loir Y. La nucléase de Staphylococcus aureus, protéine modèle pour l'étude de la sécrétion chez Lactococcus lactis. Ph.D. thesis. Orsay, France: University of Paris XI; 1996. [Google Scholar]
- 37.Le Loir Y, Gruss A, Ehrlich S D, Langella P. A nine-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J Bacteriol. 1998;180:1895–1903. doi: 10.1128/jb.180.7.1895-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebl W, Götz F. Studies on lipase directed export of Escherichia coli beta-lactamase in Staphylococcus carnosus. Mol Gen Genet. 1986;204:166–173. doi: 10.1007/BF00330205. [DOI] [PubMed] [Google Scholar]
- 39.Lund P A. The roles of molecular chaperones in vivo. Essays Biochem. 1995;29:113–123. [PubMed] [Google Scholar]
- 40.Meens J, Herbort M, Klein M, Freudl R. Use of the pre-pro part of Staphylococcus hyicus lipase as a carrier for secretion of Escherichia coli outer membrane protein A (OmpA) prevents proteolytic degradation of OmpA by cell-associated protease(s) in two different gram-positive bacteria. Appl Environ Microbiol. 1997;63:2814–2820. doi: 10.1128/aem.63.7.2814-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1998;26:334. doi: 10.1093/nar/26.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogierman M A, Fallarino A, Riess T, Williams S G, Attridge S R, Manning P A. Characterization of the Vibrio cholerae El Tor lipase operon lipAB and a protease gene downstream of the hly region. J Bacteriol. 1997;179:7072–7080. doi: 10.1128/jb.179.22.7072-7080.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Martinez G, Kok J, Venema G, Vandijl J M, Smith H, Bron S. Protein export elements from Lactococcus lactis. Mol Gen Genet. 1992;234:401–411. doi: 10.1007/BF00538699. [DOI] [PubMed] [Google Scholar]
- 45.Poquet I, Ehrlich S D, Gruss A. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J Bacteriol. 1998;180:1904–1912. doi: 10.1128/jb.180.7.1904-1912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puohiniemi R, Simonen M, Muttilainen S, Himanen J P, Sarvas M. Secretion of the Escherichia coli outer membrane proteins OmpA and OmpF in Bacillus subtilis is blocked at an early intracellular step. Mol Microbiol. 1992;6:981–990. doi: 10.1111/j.1365-2958.1992.tb02164.x. [DOI] [PubMed] [Google Scholar]
- 47.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Schmidt-Dannert C, Rua M L, Schmid R D. Two novel lipases from thermophile Bacillus thermocatenulatus: screening, purification, cloning, overexpression and properties. Methods Enzymol. 1997;284:194–220. doi: 10.1016/s0076-6879(97)84013-x. [DOI] [PubMed] [Google Scholar]
- 50.Shahbal S, Hemme D, Renault P. Characterization of a cell envelope-associated proteinase activity from Streptococcus thermophilus H-strains. Appl Environ Microbiol. 1993;59:177–182. doi: 10.1128/aem.59.1.177-182.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharp P M, Li W H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sibakov M, Koivula T, Vonwright A, Palva I. Secretion of TEM beta-lactamase with signal sequences isolated from the chromosome of Lactococcus lactis subsp lactis. Appl Environ Microbiol. 1991;57:341–348. doi: 10.1128/aem.57.2.341-348.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simons G, Rutten G, Hornes M, Nijhuis M, van Asseldonk M. Production of prochymosin in lactococci. Adv Exp Med Biol. 1991;306:115–119. doi: 10.1007/978-1-4684-6012-4_14. [DOI] [PubMed] [Google Scholar]
- 54.Sorensen M A, Kurland C G, Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 55.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleotide. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stead R. Microbial lipases: their characteristics, role in food spoilage and industrial uses. J Dairy Res. 1986;53:481–505. doi: 10.1017/s0022029900025103. [DOI] [PubMed] [Google Scholar]
- 57.Steidler L, Wells J M, Raeymaekers A, Vandekerckhove J, Fiers W, Remaut E. Secretion of biologically active murine interleukin-2 by Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1995;61:1627–1629. doi: 10.1128/aem.61.4.1627-1629.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strauch K L, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci USA. 1988;85:1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terzaghi B, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thanaraj T A, Argos P. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 1996;5:1973–1983. doi: 10.1002/pro.5560051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thanaraj T A, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Asseldonk M, De Vos W M, Simons G. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous alpha-amylase. Mol Gen Genet. 1993;240:428–434. doi: 10.1007/BF00280397. [DOI] [PubMed] [Google Scholar]
- 63.van Asseldonk M, Simons A, Visser H, De Vos W M, Simons G. Cloning, nucleotide sequence, and regulatory analysis of the Lactococcus lactis dnaJ gene. J Bacteriol. 1993;175:1637–1644. doi: 10.1128/jb.175.6.1637-1644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van de Guchte M, Kodde J, van der Vossen J M B M, Kok J, Venema G. Heterologous gene expression in Lactococcus lactis subsp. lactis: synthesis, secretion, and processing of the Bacillus subtilis neutral protease. Appl Environ Microbiol. 1990;56:2606–2611. doi: 10.1128/aem.56.9.2606-2611.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Vossen J M, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987;53:2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Oort M G, Deveer A M, Dijkman R, Tjeenk M L, Verheij H M, de Haas G H, Wenzig E, Gotz F. Purification and substrate specificity of Staphylococcus hyicus lipase. Biochemistry. 1989;28:9278–9285. doi: 10.1021/bi00450a007. [DOI] [PubMed] [Google Scholar]
- 67.Vogel R F, Gaier W, Hammes W P. Expression of the lipase gene from Staphylococcus hyicus in Lactobacillus curvatus Lc2-c. FEMS Microbiol Lett. 1990;57:289–292. doi: 10.1016/0378-1097(90)90082-2. [DOI] [PubMed] [Google Scholar]
- 68.Wells J M, Wilson P W, Norton P M, Gasson M J, Le Page R W F. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol. 1993;8:1155–1162. doi: 10.1111/j.1365-2958.1993.tb01660.x. [DOI] [PubMed] [Google Scholar]
- 69.Wu S C, Ye R, Wu X C, Ng S C, Wong S L. Enhanced secretory production of a single-chain antibody fragment from Bacillus subtilis by coproduction of molecular chaperones. J Bacteriol. 1998;180:2830–2835. doi: 10.1128/jb.180.11.2830-2835.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]