Abstract
Most genome-wide association study (GWAS)–identified breast cancer–associated causal variants remain uncharacterized. To provide a framework of understanding GWAS-identified variants to function, we performed a comprehensive study of noncoding regulatory variants at the NTN4 locus (12q22) and NTN4 gene in breast cancer etiology. We find that rs11836367 is the more likely causal variant, disrupting enhancer activity in both enhancer reporter assays and endogenous genome editing experiments. The protective T allele of rs11837367 increases the binding of GATA3 to the distal enhancer and up-regulates NTN4 expression. In addition, we demonstrate that loss of NTN4 gene in mice leads to tumor earlier onset, progression, and metastasis. We discover that NTN4, as a tumor suppressor, can attenuate the Wnt signaling pathway by directly binding to Wnt ligands. Our findings bridge the gaps among breast cancer–associated single-nucleotide polymorphisms, transcriptional regulation of NTN4, and breast cancer biology, which provides previously unidentified insights into breast cancer prediction and prevention.
Noncoding variant rs11836367 modulated NTN4 expression, which inhibits breast cancer through attenuating Wnt signaling.
INTRODUCTION
Breast cancer is a polygenic and complex disease (1). Genome-wide association studies (GWASs) have identified more than 200 breast cancer–associated single-nucleotide polymorphisms (SNPs) (2–6), which account for about 18% of the familial breast cancer risk (3). Most GWAS-identified SNPs are located in noncoding regions, especially in enhancers (7, 8), making it challenging to pinpoint the causal SNPs and their target genes. Previous studies predict the target genes of some breast cancer–associated SNPs according to expression quantitative trait loci (eQTL) (9), transcriptome-wide association studies (10), or comprehensive statistical approaches (11), while only a few such predictions have been functionally validated (12, 13).
GWAS meta-analyses suggested that rs17356907 (12q22) is robustly associated with breast cancer risk in East Asia and European ancestries (2–5, 14, 15) and with mammographic density, a strongly heritable trait and breast cancer–risk factor (16, 17). In addition, genetic fine mapping results suggested that rs17356907 and rs61938093 (odds ratio = 1.094, r2 = 1) are the most likely causal variant SNPs for breast cancer (11). However, this study only analyzed SNPs with high conditional P values (i.e., those within two orders of magnitude of the index variant), which could miss functional SNPs just below the significance threshold. rs17356907 and rs61938093 are located in a candidate enhancer region that physically interacts with the NTN4 gene. Silencing the enhancer overlapping with rs17356907 and rs61938093 leads to the down-regulation of NTN4 expression (18). However, only rs61938093 but not rs17356907 can change enhancer activity in reporter assays (18), suggesting that not all predicted risk variants play a functional role in NTN4 expression, and that identifying additional variants that contribute to NTN4 gene regulation will be necessary to improve the predictive power of the polygenic risk score for breast cancer (19, 20).
NTN4 encodes a secreted protein of the evolutionally conserved Netrin family (21), plays important roles in multiple tissues’ morphogenesis, such as lung and thalamocortical axon branching, and takes part in angiogenesis (22–24). NTN4 was down-regulated in breast cancer tissues compared with normal breast tissues. Low NTN4 expression is correlated with breast cancer proliferation and metastasis (18, 25, 26), and high NTN4 expression is suggested as a biomarker for predicting better survival of patients with breast cancer (25, 27). To date, the roles of NTN4 in breast cancer development remain to be demonstrated, and the molecular mechanism of NTN4 in tumor initiation and metastasis has not been explored.
Here, we describe further functional analyses of individual variants at this locus and transgenic mouse work to understand the mechanism by which NTN4 may affect breast cancer risk. Through a comprehensive characterization of all variants and haplotypes at the NTN4 locus, we identify another SNP rs11836367, in strong linkage disequilibrium (LD) with rs17356907, with a substantial effect on modulating NTN4 expression through its protective allele, which has a high binding capability with GATA3. We demonstrate that knockout Ntn4 promotes breast cancer onset, progression, and metastasis in vivo and reveal that NTN4 functions as a tumor suppressor gene by blocking Wnt/β-catenin signaling through interacting with Wnt ligands. Thus, we provide functional proof for an independent SNP rs11836367 that contributes to breast cancer onset and metastasis by attenuating Wnt/β-catenin signaling through regulating NTN4 expression.
RESULTS
rs11836367 plays a critical role in enhancer activity, NTN4 expression, and breast cancer risk
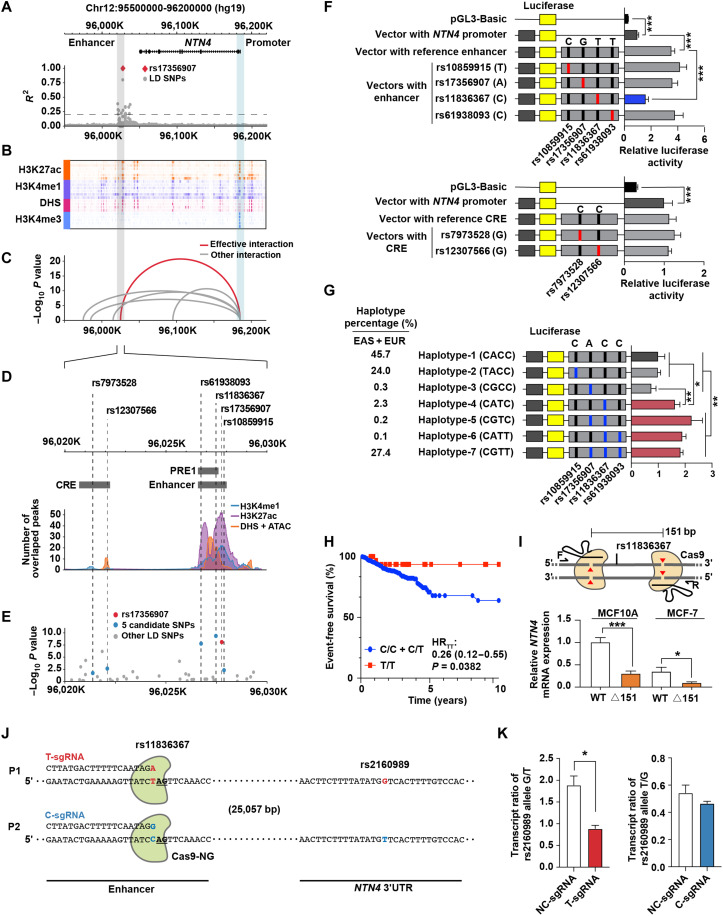
A previous study indicated NTN4 as the target gene of a breast cancer–related GWAS SNP rs17356907 (18), located in the downstream flanking region of NTN4 (approximately 157 kb downstream of NTN4; Fig. 1A). This distal region was highly enriched with enhancer marks [H3K27ac, H3K4me1, and deoxyribonuclease I (DNase I) hypersensitive sites (DHSs)] (Fig. 1B) and showed the most significant interactive features with the NTN4 promoter compared to other interaction regions with the NTN4 promoter (Fig. 1C). To filter the causal SNPs at this locus, we first used all its 28 LD-SNPs (R2 > 0.2; table S1) to overlap with 481 breast tissue–relevant epigenomic datasets [from Human Epigenomics Roadmap Project (Roadmap), The Encyclopedia of DNA Elements (ENCODE), and Cistrome database], including H3K4me1 and H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) peaks, DHSs, and assay for transposase accessible chromatin using sequencing (ATAC-seq) peaks (fig. S1A and table S2). We identified rs17356907 and its five LD-SNPs (rs11836367, rs10859915, rs61938093, rs7973528, and rs12307566) that overlapped with annotated peaks (Fig. 1D and table S1). Meanwhile, mammary gland eQTL data indicated that rs17356907 and its two LD-SNPs (rs11836367 and rs61938093) were more significantly associated with NTN4 expression (Fig. 1E), suggesting that these could be the potential functional SNPs at this locus.
Fig. 1. rs11836367 is associated with breast cancer risk and distally regulates NTN4 expression.
(A) The LocusZoom plot of LD illustrates rs17356907 and its LD-SNPs in the European population from the 1000 Genomes Project data. (B) The epigenetic heatmap displays H3K27ac, H3K4me1, and H3K4me3 ChIP-seq data and DHSs surrounding the NTN4 gene in breast-related cell lines from the Cistrome database. (C) The arc diagram shows Hi-C interactions in HMEC between the NTN4 promoter and distal elements. (D) The line plot displays the extent of NTN4-associated SNPs overlapped with ChIP-seq peaks (H3K4me1 and H3K27ac), DHS, and ATAC-seq peaks derived from the ENCODE, Roadmap, and Cistrome databases. CRE, cis-regulatory elements. (E) The LocusZoom plot of the eQTL displays NTN4-associated SNPs in normal breast tissue based on GTEX v8 data. (F and G) Luciferase reporter assays were performed in MCF10A cells using vectors containing different alleles of SNPs or different haplotypes in CREs (F). The percentages of each haplotype in East Asians (EAS) and European (EUR) populations are indicated on the left (G). (H) Kaplan-Meier plot showing the event-free survival of patients with breast cancer grouped by rs11836367 genotype. HR, hazard ratio. (I) Schematic illustrating the CRISPR-Cas9–mediated deletion of the 151-bp regulatory region (containing rs11836367, top). Quantitative reverse transcription PCR (qRT-PCR) was used to measure NTN4 mRNA expression between wild-type (WT) clones and deletion clones (△151, bottom). (J and K) Allele-specific T and C sgRNAs coupled with Cas9-NG preferentially introduced the T and C alleles, respectively, of rs11836367 (J). T and C allele-specific mutation of rs11836367 altered the allele-specific NTN4 transcript ratio determined by the reporter SNP rs2160989 G/T at the 3′ untranslated region (3′UTR) of NTN4 (K). Data are represented as means ± SEM of three independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
A prior study performed fine mapping of this region and concluded that rs61938093 is a functional SNP that can affect enhancer activity (18). We noted that another SNP, rs11836367, is closer to the center of the enhancer marks (Fig. 1D), which might be another potential causal SNP at this locus. Although rs11836367 was not considered as the credible causal variant (CCV) in prior analyses (11), it is in strong LD with the lead SNP, rs17356907 [r2 ≥ 0.8 in European and East Asians in which these associations were identified] and was also strongly associated with breast cancer risk based on Breast Cancer Association Consortium (BCAC) data (table S3). Since not all causal SNPs can be identified based on the statistical analyses alone (28), we decided to perform a comprehensive analysis of rs17356907 and the other five potential causal LD-SNPs (rs11836367, rs10859915, rs61938093, rs7973528, and rs12307566), which are located at two different regions with enhancer marks, using reporter assays (Fig. 1D and table S3). Enhancer sequence with the major/risk C allele at rs11836367 (C>T) exhibited significantly lower activity than that with the protective T allele in MCF10A (Fig. 1F) and MCF-7 cells (fig. S1B). Other SNPs in enhancer or another cis-regulatory element (CRE) did not affect luciferase activity (Fig. 1F and fig. S1B). Similarly, perturbation of enhancer with CRISPR can lead to significant changes of NTN4 gene expression but not another CRE (fig. S1, C to E). Consistent with the reporter assay results, eQTL data of the mammary gland demonstrated that the protective T allele of rs11836367 was most strongly associated with high NTN4 expression (Fig. 1E and table S3). To reconcile the discrepancies between the results of rs61938093 with no effect on altering enhancer activity in our study and the previous work showing rs61938093 as a functional SNP in reporter assays (18), we reproduced the reporter assay results for rs61938093 in their construct along with rs11836367 (fig. S1F). In addition to each SNP affecting luciferase activity in the MCF10A cell line, we showed a more profound joint effect of both SNPs in tested sequences on luciferase activity in both MCF10A and MCF-7 cell lines (fig. S1F). It is worth noting that the previous study tested the incomplete enhancer sequencing [putative regulatory elements 1 (PRE1)] compared to the ones used in our study (enhancer) (Fig. 1D), and the luciferase activity detected using the former construct is about twofold lower than the results of using our construct (fig. S1F), potentially explaining the inconsistency between the results of tests of rs61938093 function in both studies.
To further analyze the combinatorial effect of SNPs at this locus, we first examined enhancer activity with the common haplotypes of four SNPs (rs10859915, rs17356907, rs11836367, and rs61938093) in the enhancer. Compared to the most common haplotype CACC (45.7%), four other haplotypes [CGTT (27.4%), CATC (2.3%), CGTC (0.2%), and CATT (0.1%)] bearing the T allele of rs11836367 had higher luciferase activity (Fig. 1G and fig. S1G). Meanwhile, the other three SNPs (rs17356907, rs10859915, and rs61938093) had no or minimal additive effect on the enhancer activity (Fig. 1G and fig. S1G), emphasizing the regulatory roles of rs11836367 at this locus. To link these results to patients’ genotypes, we then genotyped these four SNPs in age-matched breast cancer cases and controls in Chinese Han women (table S4). Overall, three SNPs (rs11836367, rs61938093, and rs17356907) were significantly associated with breast cancer risk (table S5) in our study. By reconstructing haplotypes based on our genotyping data, the haplotypes CGTT (rs10859915 + rs17356907 + rs11836367 + rs61938093) and CATC, bearing the T allele at rs11836367, were significantly associated with decreased breast cancer risk compared with the most common haplotype CACC (table S6). Besides, two diplotypes (CACC-CGTT and CGTT-CGTT) were also associated with lower breast cancer risk, compared to the most common diplotype CACC-CACC (table S6). In addition, patients with the homozygous protective T allele at rs11836367 (genotype T/T) showed significantly longer event-free survival than patients with the nonprotective C allele (genotype C/C + C/T; Fig. 1H). These results collected from patient genotyping data confirm that rs11836367 is a critical SNP in breast cancer risk.
To further prove that the rs11836367 region is involved in regulating NTN4 expression, we deleted a 151–base pair (bp) fragment containing rs11836367 in both MCF10A and MCF-7 cell lines through CRISPR-Cas9 with flanking single guide RNAs (sgRNAs) (Fig. 1I, top). Three clones with bi-allelic deletion were generated in each cell line (fig. S1H). The deletion clones showed lower NTN4 expression than the wild-type cells (Fig. 1I, bottom). We noticed that rs11836367 was the first nucleotide adjacent to protospacer adjacent motif of Cas9-NG (Fig. 1J), and it was heterozygous in MCF10A. Therefore, we designed allele-specific sgRNAs (AS-sgRNAs) to target rs11836367 risk/major allele and protective/minor allele (Fig. 1J). Compared to MCF10A cells transduced with control sgRNA, those cells transduced with Cas9-NG coupled with T-sgRNA (protective allele) or C-sgRNA (risk allele) were demonstrated to preferentially mutate the T allele or C allele of rs11836367 (fig. S1I). Introducing its allele-specific mutation to either allele of rs11836367 decreased the transcripts of its target allele of NTN4 gene, which was determined by the ratio of the corresponding transcripts of NTN4 3′ untranslated region reporter SNP rs2160989 alleles (Fig. 1K). Compared with C-sgRNA, T-sgRNA caused a more significant change in its target allele transcripts (Fig. 1K). These results determine the effect of different alleles of rs11836367 on NTN4 expression at its endogenous genomic locus with its protective T allele having a stronger effect on NTN4 transcription than the C allele.
GATA3 preferentially binds to the protective allele rs11836367-T
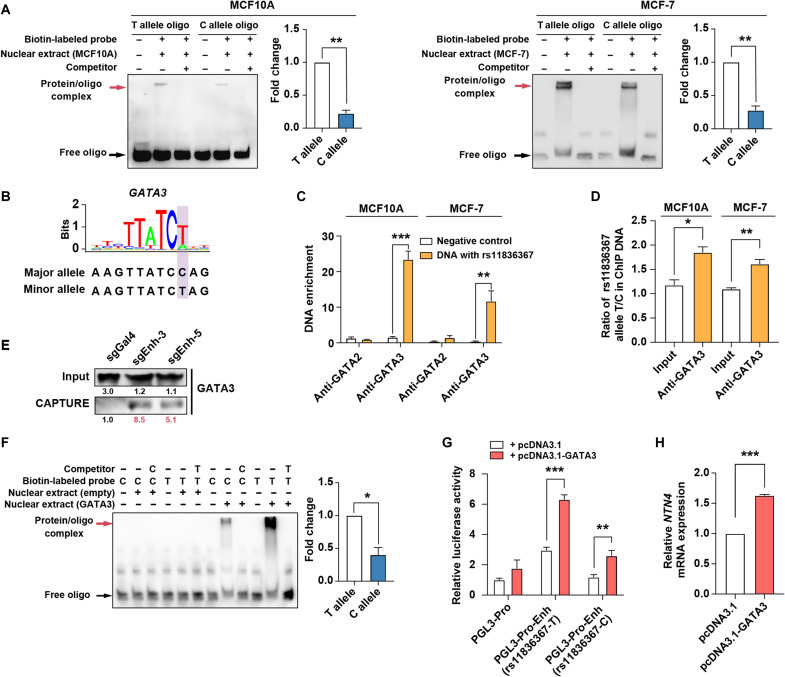
Differential transcription factor (TF) binding between SNP risk allele and protective allele is considered one of the mechanisms for functional SNPs affecting enhancer activities (29–31). We analyzed the effect of rs11836367 risk allele and protective allele on TF binding by electrophoretic mobility shift assay (EMSA). The result showed that nuclear extract had stronger binding capabilities with the rs11836367-T allele than with the rs11836367-C allele (Fig. 2A). We then scanned rs11836367 locus for potentially affected TF binding sites using Predicting Regulatory Functional Effect by Approximate P-value Estimation (PEREFECTPS-APE) (32) and found that GATA2 and GATA3 have a stronger binding capability with the rs11836367-T allele than with the rs11836367-C allele (Fig. 2B and fig. S2A). ChIP–quantitative polymerase chain reaction (qPCR) in MCF-7 and MCF10A demonstrated that GATA3 but not GATA2 bound to rs11836367 locus (Fig. 2C) with a stronger binding capability with the rs11836367-T allele than with the rs11836367-C allele in vivo (Fig. 2D), consistent with the results from both in situ CAPTURE assay (Fig. 2E and fig. S2B) and EMSA with ectopically expressed GATA3 and the oligos carrying T or C alleles at rs11836367 (Fig. 2F and fig. S2C).
Fig. 2. rs11836367 alleles specifically influence GATA3 binding affinity.
(A) The binding affinities between oligos containing the C allele or T allele of rs11836367 and nuclear extracts (from MCF10A and MCF-7 cells) were analyzed by EMSA, and shifted bands were quantified. (B) The risk C allele of rs11836367 interferes with a GATA3 binding motif. (C) ChIP-qPCR for GATA2 and GATA3 at the DNA fragment containing rs11836367 locus. (D) Allele-specific ChIP-qPCR for GATA3 at the DNA fragment containing rs11836367 locus. (E) Western blot showing enrichment of GATA3 at the rs11836367-containing enhancer in MCF10A by in situ CAPTURE assay. sgEnh-3 and sgEnh-5 target the enhancer (containing rs11836367), and sgGal4 is a negative control. (F) The binding affinities between oligos containing the C allele or T allele of rs11836367 and GATA3 were analyzed by EMSA (left), and shifted bands were quantified (right). Nuclear extracts were obtained from 293T cells transfected with either an empty vector or GATA3-overexpressing construct. (G) Luciferase reporter assay performed after transient cotransfection of MCF10A cells with the indicated constructs. The DNA fragment containing either the T or C allele of rs11836367 was cloned into the NTN4 promoter (PGL3-Pro)–driven luciferase reporter construct (PGL3-Pro-Enh-rs11836367-T vector or PGL3-Pro-Enh-rs11836367-C vector), which was cotransfected with either an empty vector (pcDNA3.1) or a GATA3-overexpressing vector (pcDNA3.1-GATA3). (H) qRT-PCR showing the transfection of MCF10A cells with pcDNA3.1-GATA3 increased NTN4 mRNA expression. Data are represented as means ± SEM of three independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
To further determine whether the allele-specific effect of rs11836367 on enhancer activity is through influencing GATA3 binding, we transfected cells with GATA3-overexpressing construct (pcDNA3.1-GATA3) and luciferase reporter constructs carrying the NTN4 promoter and enhancer, which contained either the T or C allele of rs11836367. We found that luciferase activity was significantly increased by cotransfection of pcDNA3.1-GATA3 with both rs11836367-T and rs11836367-C constructs (Fig. 2G). Notably, the T allele construct had a stronger capability in promoting luciferase activity than the C allele construct did (Fig. 2G). Furthermore, overexpression of GATA3 resulted in a significant up-regulation of NTN4 expression in MCF10A cells (Fig. 2H), consistent with elevated NTN4 expression in T-47D cell lines with GATA3 overexpression (GSE99479) (fig. S2D). Conversely, GATA3 knockdown decreased NTN4 expression in MCF-7 by CRISPR interference (CRISPRi) (fig. S2E). Besides, NTN4 expression was positively related to GATA3 expression in both normal breast tissues and breast tumor tissues (fig. S2F) in The Cancer Genome Atlas (TCGA) datasets. These results confirm that GATA3 preferentially bound to the T allele of rs11836367, which therefore increased NTN4 expression.
Ntn4 knockout enhances the onset, progression, and metastasis of mammary tumors
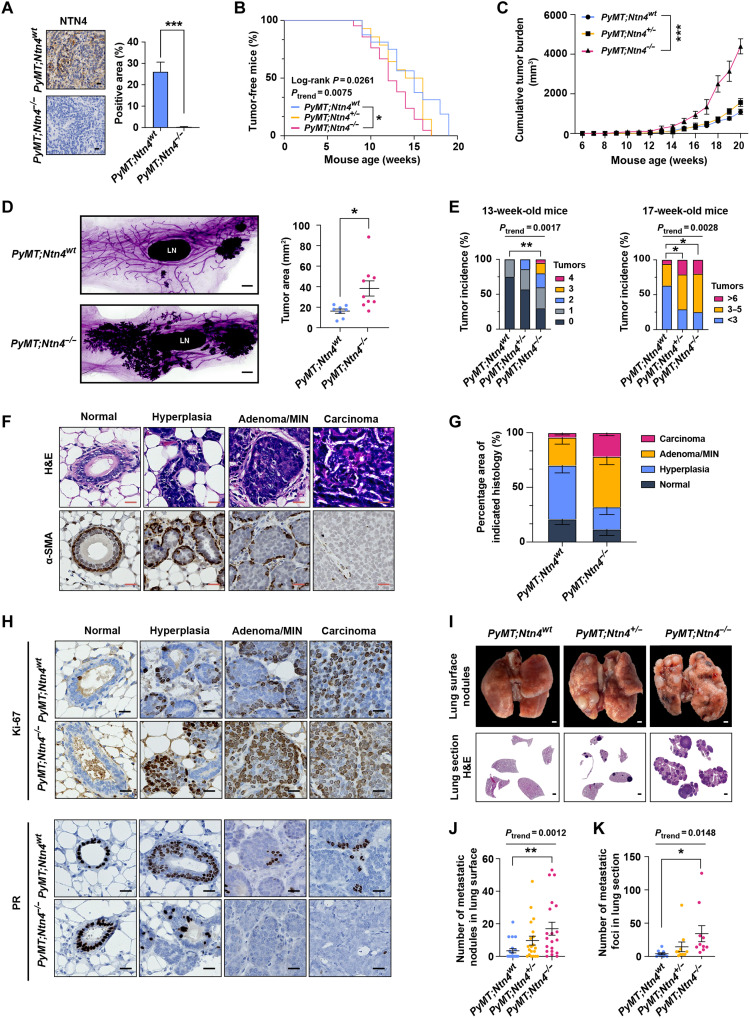
Previous studies indicate that NTN4 can inhibit breast cancer proliferation and metastasis based on the tumor cell lines or the xenograft model (18, 25, 26). To analyze the role of NTN4 in breast cancer onset and progression in vivo, we used an Ntn4 knockout mouse model, where the second exon of Ntn4 was knocked out by CRISPR-Cas9. The loss of Ntn4 mRNA and NTN4 protein expression in the mammary gland was confirmed in Ntn4−/− mice (fig. S3, A and B). The Ntn4−/− mice were viable and fertile and exhibited no gross phenotypic abnormalities compared with Ntn4 wild-type (Ntn4wt) littermates. In wild-type mice, NTN4 protein is detected in myoepithelial cells of the normal mammary gland (fig. S3B), and Ntn4 mRNA is expressed in flow-sorted mammary luminal and myoepithelial cells (fig. S3C). We crossed the Ntn4 knockout mice with MMTV-PyMT (PyMT) mice in the C57BL/6 background, which recapitulates the human breast cancer onset, progression, and metastasis (33) and has breast cancer phenotypes that are less aggressive than those in the FVB strain (34). The NTN4 protein expression was completely lost in the mammary tumors of PyMT;Ntn4−/− mice (Fig. 3A).
Fig. 3. Loss of Ntn4 in PyMT mice accelerates tumor onset, progression, and metastasis.
(A) Immunohistochemical (IHC) staining images (left) and quantification (right) of NTN4 in the late-stage tumors of PyMT;Ntn4wt and PyMT;Ntn4−/− mice. n = 3 for both groups. Scale bars, 20 μm. (B) Palpable tumor onset in PyMT;Ntn4wt (n = 16), PyMT;Ntn4+/− (n = 14), and PyMT;Ntn4−/− (n = 18) mice. (C) Tumor growth in PyMT;Ntn4wt, PyMT;Ntn4+/−, and PyMT;Ntn4−/− mice. n = 6 per group. (D) Whole-mount–stained fourth mammary gland tumors images (left) and quantification of tumor area (right) in 13-week-old PyMT;Ntn4wt (n = 8) and PyMT;Ntn4−/− (n = 9) mice. Scale bars, 1 mm. LN, inguinal lymph node. (E) Tumor multiplicity in PyMT;Ntn4wt (n = 16), PyMT;Ntn4+/− (n = 14), and PyMT;Ntn4−/− (n = 18) mice. (F) Hematoxylin and eosin (H&E) and IHC staining images comparing the normal mammary gland to mammary glands with various stages of PyMT tumors. Scale bars, 20 μm. (G) Tumor stages of fourth mammary glands from 13-week-old PyMT;Ntn4wt (n = 6) and PyMT;Ntn4−/− mice (n = 7). (H) IHC staining images for Ki-67 and PR in different stages of PyMT tumors from 13-week-old mice. Scale bars, 20 μm. (I) Representative images of lung surface metastatic nodules (top) and H&E staining of lung sections (bottom) from PyMT;Ntn4wt, PyMT;Ntn4+/−, and PyMT;Ntn4−/− mice. Scale bars, 1 mm. (J and K) The number of metastatic nodules (J) in PyMT;Ntn4wt (n = 16), PyMT;Ntn4+/− (n = 14), and PyMT;Ntn4−/− (n = 18) mice and metastatic foci (K) (n = 10) was quantified at the endpoint (20 to 25 weeks of age). Data are represented as means ± SEM. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
First, we assessed the effects of Ntn4 deficiency on mammary tumor initiation and progression. Compared to PyMT;Ntn4wt mice, PyMT;Ntn4−/− mice showed earlier tumor onset (Fig. 3B) and larger tumor burden (Fig. 3C). In 13-week-old mice, the whole-mount analysis revealed that most mammary gland regions were converted into lesions in PyMT;Ntn4−/− mice, while the lesions were mainly seen near nipples in PyMT;Ntn4wt mice (Fig. 3D). Besides, both heterozygous and homozygous Ntn4 knockout mice generated more mammary tumors at the age of 13 and 17 weeks (Fig. 3E). To assess the severity of tumors formed in WT and Ntn4 deletion mice, we staged these tumors as hyperplasia, adenoma/mammary intraepithelial neoplasia (MIN), and invasive carcinoma based on pathological features and molecular markers (35), such as α–smooth muscle actin (α-SMA) for detecting myoepithelial cells (Fig. 3F). The lesions of PyMT;Ntn4wt mice were mainly at the hyperplasia stage (Fig. 3G), while the lesions of PyMT;Ntn4−/− mice had progressed into the adenoma/MIN and carcinoma stage (Fig. 3G) at the age of 13 weeks with PyMT;Ntn4−/− tumors demonstrating higher Ki-67 labeling index (proliferative marker) and lower progesterone receptor (PR) protein expression in each stage (Fig. 3H and fig. S3D). Expression of estrogen receptor (ER) and HER2 had no significant difference between PyMT;Ntn4wt and PyMT;Ntn4−/− tumors (fig. S3D). Same as the findings from tumors of 13-week-old mice, tumors from late-stage mice also demonstrated higher Ki-67 labeling index and lower PR protein expression in PyMT;Ntn4−/− tumors than in PyMT;Ntn4wt tumors (fig. S3E).
Second, we assessed the effect of Ntn4 deficiency on mammary tumor metastasis. Mice with homozygous Ntn4 knockout had a significantly increased number and larger size of metastatic nodules in the lungs (Fig. 3, I to K, and fig. S3F) as well as a high Ki-67 labeling index compared with PyMT;Ntn4wt mice (fig. S3G). To determine the influence of Ntn4 deficiency on tumor intravasation, we examined circulating tumor cells (CTCs) in peripheral blood using quantitative reverse transcription PCR (qRT-PCR) analysis of molecular markers [Pymt; cytokeratin 8 (Krt8)]. The PyMT;Ntn4−/− mice showed more CTCs (fig. S3H) and a worse overall survival rate than PyMT;Ntn4wt mice (fig. S3I).
Third, we investigated the effect of Ntn4 depletion on cancer stem cell property. Ntn4-deficient tumors have a significant increase in CD24+CD90+ cancer stem cell population (fig. S3J). PyMT;Ntn4−/− primary tumor cells had an approximately fourfold increase in tumor-initiating ability (fig. S3K) and formed larger tumors after transplantation compared with wild-type primary tumor cells (fig. S3, L and M). Overall, our results present in vivo evidence that Ntn4 is a breast risk gene that affects tumor initiation, metastasis, and survival in gene knockout mouse models, which is consistent with the genetic and epidemiologic findings in human studies.
NTN4 inhibits cellular behaviors of human breast cancer
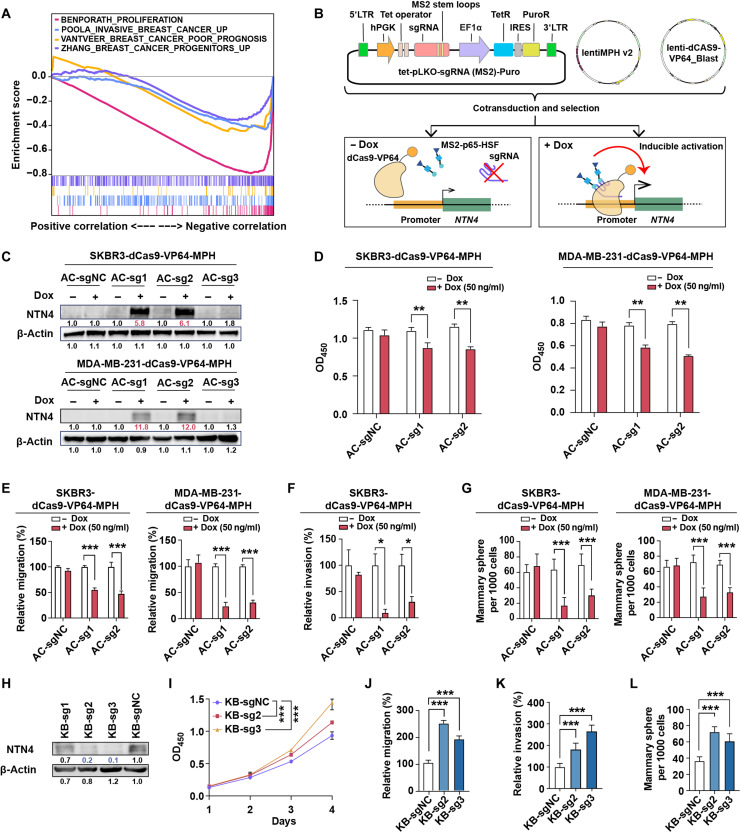
We further investigate the role of NTN4 in human breast cancer development and progression in clinical settings. NTN4 expression was down-regulated in human breast tumor tissues compared to normal breast tissues and showed similar down-regulation in multiple cancer types (fig. S4A). Compared with luminal breast cancers, NTN4 expression was lower in HER2-enriched breast cancer tissues and was even lower in triple-negative breast cancers (fig. S4B). Furthermore, NTN4 expression in human breast tumors was negatively correlated with a number of breast cancer–related gene sets, including proliferation, metastasis, stem cell property, and poor prognosis (Fig. 4A). Meanwhile, lower expression of NTN4 was consistently associated with worse relapse-free survival, overall survival, distant metastasis-free survival, and postprogression survival of patients with breast cancer (fig. S4C) using Kaplan-Meier survival analysis.
Fig. 4. NTN4 inhibits proliferation, migration, invasion, and cancer stem cell features in breast cancer cells.
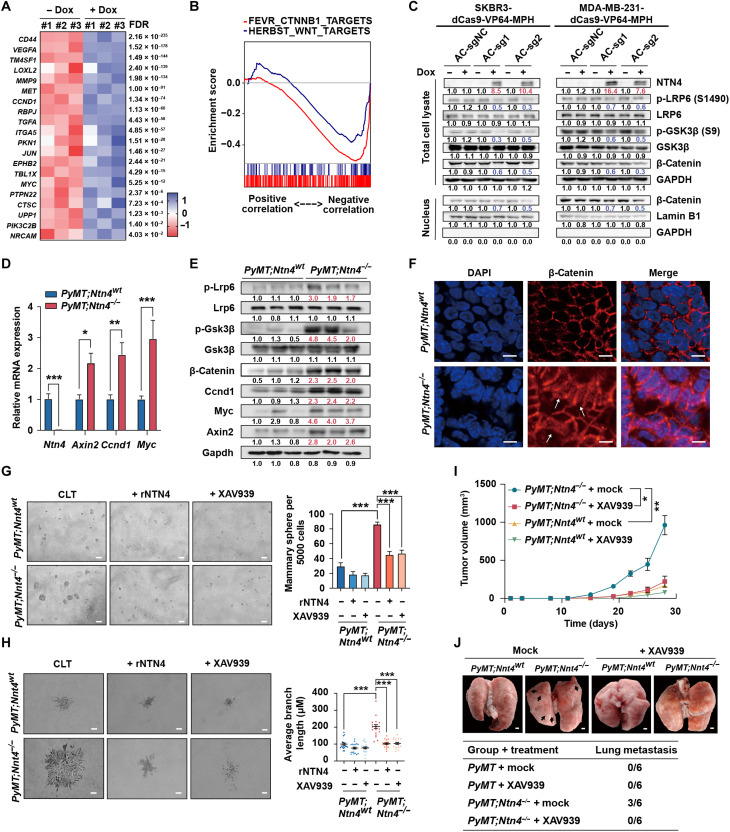
(A) Gene set enrichment analysis (GSEA) analysis of genes preranked by Pearson correlation to NTN4 expression for four breast cancer–related signatures, which were BENPORATH_PROLIFERATIION [red curve: normalized enrichment score (NES) = −3.87, false discovery rate (FDR) q value < 0.001], POOLA_INVASIVE_BREAST_CANCER_UP (blue curve: NES = −2.34, FDR q value < 0.001), VANTVEER_BREAST_CANCER_POOR_PROGNOSIS (yellow curve: NES = −1.81, FDR q value = 0.001), and ZHANG_BREAST_CANCER_PROGENITORS_UP (purple curve: NES = −2.01, FDR q value = 0.001) in RNA sequencing (RNA-seq) datasets from TCGA. (B) Schematic of dCas9-SAM–mediated doxycycline (Dox)–inducible endogenous overexpression of NTN4. TetR, Tet repressor; IRES, internal ribosomal entry site; 5’LTR, 5’ long terminal repeat; PuroR, puromycin resistent gene. (C) Validation of inducible overexpression of the NTN4 protein by Western blot analysis in SKBR3-dCas9-VP64-MPH and MDA-MB-231-dCas9-VP64-MPH cells following the addition of Dox (50 ng/ml). AC-sg1, AC-sg2, and AC-sg3 were designed to target the NTN4 promoter, and notargeting AC-sgNC was a negative control. (D) Cell proliferation was analyzed by Cell Counting Kit-8 (CCK-8) after 4-day culture with or without Dox treatment. OD450, optical density at 450 nm. (E) Cellular migration was analyzed using the Transwell assay with or without Dox treatment. (F) Cellular invasion was investigated using the three-dimensional spheroid invasion assay in Matrigel following a 6-day culture with or without Dox treatment. (G) Cancer stem cell features were analyzed using sphere formation assays after a 7-day culture with or without Dox treatment. (H) Validation of CRISPRi knockdown of NTN4 protein by Western blot analysis in SUM159-dCas9-KRAB cells. KB-sg1, KB-sg2 and KB-sg3 were designed to target the NTN4 promoter, and nontargeting KB-sgNC was used as a negative control. (I to L) Cellular proliferation (I), migration (J), invasion (K), and cancer stem cell features (L) were analyzed in NTN4 knockdown SUM159-dCas9-KRAB cell lines as described above. Data are represented as means ± SEM of three to six independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. Related representative images for (E) to (G) and (J) to (L) were shown in fig. S4.
The observations in clinical data and the mouse model prompted us to further assess the functional role of NTN4 in different human breast cancer cells. NTN4 expression was lower in breast cancer cell lines compared to that in the nontransformed mammary epithelial cell line MCF10A (fig. S4, D and E). In addition, NTN4 expression is lower in the human breast cancer stem cell CD24−CD44+ than in other tumor cells (fig. S4F). Doxycycline (Dox)–induced overexpression of endogenous NTN4 (Fig. 4, B and C) significantly suppresses proliferation, migration, invasion, and sphere formation (Fig. 4, D to G, and fig. S4, G to J) in SKBR3 and MDA-MB-231 cells. Conversely, knocking down NTN4 expression by CRISPRi in SUM159 with high NTN4 expression significantly increased tumor cell proliferation, migration, invasion, and sphere formation (Fig. 4, H to L, and fig. S4, K to M). These data illustrated that NTN4 inhibited human breast cancer cellular proliferation, migration, invasion, and stem cell–like properties, consistent with previous studies (18, 25, 26), which further support that NTN4 acts as a risk gene in human breast cancer.
NTN4 antagonizes Wnt/β-catenin signaling pathway
To date, the mechanism of NTN4 expression that affects breast cancer onset and metastasis remains elusive. We identified that several pathways were disturbed after NTN4 overexpression, including the Wnt signaling pathway (fig. S5A). Besides, multiple Wnt pathway target genes were down-regulated after NTN4 overexpression (Fig. 5A and fig. S5, B to D). In addition, the TCGA dataset showed that NTN4 expression in human breast tumors was negatively correlated with the expression of target genes in the Wnt/β-catenin signaling pathway (Fig. 5B). NTN4 overexpression had a notable suppressive impact on the phosphorylation of LRP6 and GSK3β as well as β-catenin expression and nuclear translocation in SKBR3 and MDA-MB-231 cells (Fig. 5C), while decreasing NTN4 expression had opposite effects in SUM159 (fig. S5E). Together, these results indicated that NTN4 could inhibit Wnt signaling in human breast cancer cells.
Fig. 5. NTN4 inhibits breast cancer initiation and metastasis by antagonizing Wnt/β-catenin signaling.
(A) The heatmap displays the RNA-seq result of differential expressions of Wnt/β-catenin target genes. (B) GSEA analysis of genes preranked by Pearson correlation to NTN4 expression for two Wnt target gene signatures, which were HERBEST_WNT_TARGETS (blue curve: NES = −1.87, FDR q value < 0.001) and FEVR_CTNNB1_TARGETS (red curve: NES = −2.84, FDR q value < 0.001) in breast cancer expression datasets from TCGA. (C) Western blots analyzing p-LRP6, LRP6, p-GSK3β, GSK3β, and β-catenin in total cell lysates and nuclear fractions from SKBR3-dCas9-VP64-MPH and MDA-MB-231-dCas9-VP64-MPH cells. GAPDH, glyceraldehyde phosphate dehydrogenase. (D) qRT-PCR analysis showing the expression of Ntn4 and the Wnt/β-catenin target genes Axin2, Ccnd1, and Myc in PyMT;Ntn4wt and PyMT;Ntn4−/− tumors. n = 3 per group. (E) Western blots analyzing p-LRP6, LRP6, p-GSK3β, GSK3β, β-catenin, CCND1, MYC, AXIN2, and GAPDH in total cell lysates from PyMT;Ntn4wt and PyMT;Ntn4−/− tumors. (F) Immunofluorescent staining images for β-catenin (red) and nuclei (blue) in PyMT;Ntn4wt and PyMT;Ntn4−/− tumors. Arrows, nucleus-localized β-catenin. Scale bars, 25 μm. DAPI, 4′,6-diamidino-2-phenylindole. (G) Sphere formation assays were performed in mammary tumor cells derived from PyMT;Ntn4wt and PyMT;Ntn4−/− tumor tissues in the presence of either recombinant NTN4 protein (rNTN4; 200 ng/ml), 10 μM XAV939, or vehicle control. n = 6. Scale bars, 50 μm. (H) Three-dimensional spheroid invasion assays were performed using organoids from PyMT;Ntn4wt and PyMT;Ntn4−/− mice in the presence of either rNTN4 (200 ng/ml), 10 μM XAV939, or vehicle control (n > 15 per group). Scale bars, 50 μm. (I) The volume measurement for xenografts of PyMT;Ntn4wt and PyMT;Ntn4−/− tumor cells treated with XAV939 (25 mg/kg) or vehicle. n = 6 per group. (J) Representative images (top) and quantification (bottom) of metastatic nodules on the lung surface in different mouse groups. Scale bar, 1 mm. Data are represented as means ± SEM. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Similar to human breast cancer cell lines, the loss of Ntn4 in the PyMT model significantly increased mRNA and protein expression of Wnt target genes, such as Myc, Ccnd1, and Axin2 (Fig. 5, D and E, and fig. S5F). In addition, knocking out Ntn4 increased phosphorylation of LRP6 and GSK3β and the expression of β-catenin (Fig. 5E). Moreover, we observed a substantial nuclear accumulation of β-catenin in PyMT;Ntn4−/− tumor cells compared to that in PyMT;Ntn4wt tumor cells (Fig. 5F). Both Wnt/β-catenin inhibitor XAV939 and recombinant NTN4 protein abrogated PyMT;Ntn4−/− tumor sphere formation and tumor organoid invasion in vitro (Fig. 5, G and H), and XAV939 can also affect tumor cells’ proliferation after transplantation (Fig. 5I) and lung metastasis in vivo (Fig. 5J). Overall, these data indicate that Wnt/β-catenin is a downstream target of NTN4 in mediating breast cancer cell proliferation and metastasis.
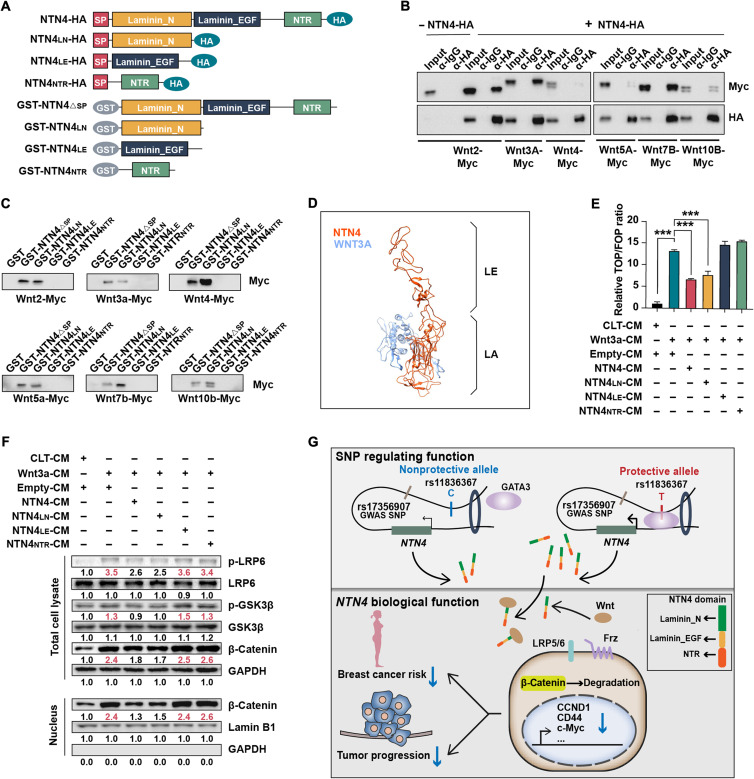
To delineate how NTN4 regulates the Wnt/β-catenin pathway, we examined the NTN4 domains and noticed that they contained a netrin-related (NTR) domain. As proteins from the secreted frizzled-related protein (SFRP) family and netrin family protein NTN1 could antagonize Wnt/β-catenin signaling by binding to Wnt ligands through their NTR domains (36), we first analyzed the interaction of NTN4 with different Wnt ligands (Fig. 6A). NTN4 bound strongly to WNT2, WNT3A, WNT7B, and WNT10B but less strongly to WNT4 and WNT5A (Fig. 6B). Instead of the NTR domain, NTN4 directly bound to Wnt ligands through Laminin_N domain (NTN4LN) (Fig. 6C). We then performed the structural simulation to explain the interaction between Wnt ligands and NTN4. We docked NTN4 with WNT3A proteins, which are both from SWISS-MODEL. The prediction suggested that the N terminus of NTN4 interacted with thumb and index finger domains of WNT3A, which is important for Wnt ligands to bind with Frizzled receptors (Fig. 6D).
Fig. 6. NTN4 protein modulates Wnt signaling through binding to Wnt ligands.
(A) Schematic diagram of the NTN4 protein’s full length (NTN4), Laminin_N domain (NTN4LN), Laminin_EGF domain (NTN4LE), and NTR domain (NTN4NTR) of the NTN4 protein with (SP) or without (ΔSP) signal peptide, used in coimmunoprecipitation, glutathione S-transferase (GST) pull-down assay, and conditional medium (CM) assays. (B) 293T cells were transiently cotransfected with the indicated plasmid. Coimmunoprecipitation was conducted using anti–HA-tag antibody–conjugated beads followed by immunoblotting with anti–HA-tag and anti–Myc-tag antibodies. The immunoglobulin G (IgG) antibody was used as a negative control. (C) GST pull-down assays were performed to determine the interaction of GST-tagged full-length or truncated NTN4 protein with indicated Myc-tagged Wnt ligands. The interaction was detected by immunoblotting with an anti–Myc-tag antibody. (D) Visualization of the NTN4-WNT3A docking model predicted by ClusPro. The protein structure of NTN4 and WNT3A was obtained from SWISS-MODEL. NTN4 (orange red) and WNT3A (blue) structures are shown as a ribbon diagram. (E) TOP/FOPFlash luciferase reporter assays were performed in MDA-MB-231 cell lines after treatment with different mixtures of CM. Data are represented as means ± SEM. ***P ≤ 0.001. n = 3. (F) Western blotting analyzing p-LRP6, LRP6, p-GSK3β, GSK3β, and β-catenin in total cell lysates and nuclear fractions prepared from the MDA-MB-231 cell line treated with different mixed CMs. (G) Graphical representation of NTN4 transcriptional regulation by the rs11836367 locus and its interaction with Wnt ligands. The protective T allele at the rs11836367 locus increases GATA3 binding to a distal enhancer of the NTN4 gene elevating NTN4 expression. NTN4 protein interacts with Wnt ligands and antagonizes Wnt/β-catenin signaling, decreasing breast cancer risk and tumor progression.
To investigate whether NTN4 could inhibit Wnt/β-catenin’s activity through the NTN4LN domain, we constructed several expression vectors containing full-length NTN4 and different domains of NTN4 (Fig. 6A) and then detected secreted corresponding proteins in the cell culture medium (fig. S6A). As WNT3A can activate Wnt/β-catenin’s activity, we incubated MDA-MB-231 cells with WNT3A conditional medium (CM) plus medium containing full-length or different domains of NTN4. TOP/FOPFlash luciferase reporter assays demonstrated that WNT3A can significantly increase luciferase activity, and full-length NTN4 and NTN4LN can attenuate WNT3A-induced luciferase activity, while the epidermal growth factor (EGF)–like domain (NTN4EGF) and the NTR domain (NTN4NTR) had no such effects (Fig. 6E). Furthermore, secreted full-length NTN4 and NTN4LN attenuated WNT3A-induced phosphorylation of LRP6 and GSK3β and β-catenin expression and nuclear translocation (Fig. 6F). Similar to full-length NTN4, NTN4LN attenuates proliferation, migration, and sphere formation of MDA-MB-231 cells (fig. S6, B to D). Together, these results suggest that NTN4 can suppress the Wnt/β-catenin pathway by binding to Wnt ligand through the NTN4LN domain.
DISCUSSION
GWASs have identified more than 200 SNPs associated with breast cancer risk (2–6), while the functional mechanisms of most SNPs are still not elucidated. Here, we systematically explored the 12q22 susceptibility locus for breast cancer and interrogated the function of NTN4 and its downstream effects using a combination of human genetic association data, enhancer assays in cell lines, and mouse models. As in prior analyses (18), we identified NTN4 as the most likely candidate gene at this locus. However, by carefully evaluating all the SNPs in functional assays, we also proposed rs11836367 as a causal variant. We also found that Ntn4 was a susceptibility gene in the mouse PyMT model of breast cancer. Furthermore, we proposed that NTN4 worked via direct effects on the Wnt/β-catenin pathway, ultimately influencing breast cancer onset and progression.
We found strong evidence that rs11836367 affects enhancer function more than any other SNP in the region and affects GATA3 binding. Although women with the haplotype containing the T allele of rs11836367 showed a lower breast cancer risk, rs61938093 was more significantly associated with breast cancer risk than rs11836367 in the largest genetic association studies done by BCAC (6). rs61938093 was also predicted as a CCV by comprehensive statistical approaches (11) with its functional role in enhancer activity demonstrated by luciferase assay with an incomplete enhancer sequence (18). In our study, we tested all variants with the full enhancer sequences and did not detect significant changes in enhancer activities with rs61938093 alone.
The disagreement between the statistical level of the association supporting either rs61938093 or rs17356907 and the functional studies supporting rs11836367 may be difficult to resolve. One unifying explanation may be that enhancer activity is altered by more than one variant in this locus. In general, most of the analyses generating a list of CCVs assume that only one causal variant underlies the locus. If the effect at this locus is due to multiple variants than the assumptions underlying the generation of CCVs, rs11836367 was not included in the previous CCV list (11). Alternatively, it is possible that the cell line analyses we performed do not model the transcriptional activity in vivo at the time of breast cancer development and that rs11836367 plays an important role in the cell lines but less so in vivo.
Our in vitro and in vivo studies revealed that rs11836367 can influence GATA3 binding and affect NTN4 expression in an allele-specific manner. GATA3 acts as a crucial transcriptional factor in breast development and inhibits breast cancer proliferation and metastasis (37, 38) and might be influenced by breast cancer–associated SNPs (12, 39). In addition, loss of GATA3 is one of the most common somatic mutations found in breast cancer (40). Here, we proposed that increased GATA3 binding with rs11836367 protective T allele might be a possible mechanism for GATA3 in inhibiting breast cancer risk.
NTN4 has important roles in tissue development (22, 24, 41) and tumor progression (18, 23, 26, 42, 43). However, the exact function of NTN4 appears to be dependent on the tissue types and cancer types. For example, NTN4 promotes axon branching and stimulates proliferation and invasion of gastric cancer cells (24, 44). In contrast, NTN4 inhibits lung branching and salivary gland formation and suppresses proliferation and invasion of colorectal cancer and breast cancer cells (18, 22, 23, 25, 26, 41). Prior studies have demonstrated that NTN4 can increase tumor formation in mouse xenograft models, but they did not demonstrate that it underlies breast cancer initiation. Our transgenic model work demonstrated that loss of Ntn4 promotes breast cancer initiation. We also found that NTN4 inhibited breast cancer progression and metastasis. Correlative analyses of expression in humans demonstrated that NTN4 expression was down-regulated in breast cancer tissues and positively correlated with patients’ prognosis (25, 27).
Mechanistically, NTN4 has been indicated to affect tumor proliferation via multiple signaling pathways. NTN4 activates Jak/Stat, PI3K/Akt, and ERK/MAPK pathways through the receptor Neogenin in gastric cancer, while NTN4 mediates antiangiogenic activity through the receptor Neogenin in colorectal cancer (23, 44). NTN4 could mediate protumor and antitumor effects through receptor integrins β4 and UNC5B, respectively, in glioblastoma (43). In breast cancer, NTN4 suppresses tumor cell migration and invasion via inhibiting epithelial-to-mesenchymal transition (EMT)–related gene expression (25) and inhibits breast cancer metastases via decreasing basement membrane stiffness (26). Here, we demonstrated a previously unknown mechanism for NTN4 in breast cancer, namely, NTN4 suppressed the Wnt/β-catenin signaling via direct binding with Wnt ligands. The Wnt/β-catenin pathway plays a major role in breast cancer onset and progression (45, 46). NTN1, a member of the Netrin family, inhibits Wnt signaling by binding to Wnt ligands through its NTR domain (36). We found that NTN4 protein bound to Wnt ligand through its laminin_N domain, a domain also shared by the laminin family. Another laminin_N domain–containing protein, Laminin-511, has also been reported to inhibit the Wnt pathway (47), but whether its inhibition is through the laminin_N domain needs to be investigated. Moreover, structural analysis has shown that NTN4 protein could not interact with known receptors for the netrin family (48). Here, we propose that NTN4 protein binding with Wnt ligands is a novel molecular mechanism that acts as a tumor suppressor in breast cancer.
In summary, this study elucidated a mechanism of a previously unidentified causal SNP affecting breast cancer risk, in which the protective T allele of rs11836367 increased GATA3 binding and NTN4 expression through a distal enhancer-promoter interaction mechanism. Up-regulated NTN4 blocked the Wnt/β-catenin pathway by interacting with Wnt ligands, thus ultimately inhibiting breast cancer onset and progression (Fig. 6G). We bridged the gap between GWAS-identified SNP rs17356907 and breast cancer risk and further characterized molecular biomarkers for breast cancer risk prediction and breast cancer prevention.
MATERIALS AND METHODS
Ethics statement
This study was approved by the Peking University Institutional Review Board (IRB; reference no. IRB00001052-11029). Written informed consents were obtained from all control samples. Breast cancer samples were collected for research purposes in the tissue/blood biobank. Written consents were obtained from patients with breast cancer who can read and write. For patients with breast cancer who cannot read and write, verbal consent was obtained and written consent was signed by their next of kin. IRB approved the written consent procedure. The data and samples were used anonymously. Peking University IRB approved our application to waive informed re-consent for the already collected breast cancer samples in the tissue/blood biobank.
Study population
The case-control study included 873 female patients with breast cancer and 962 cancer-free controls. All 873 patients were pathologically diagnosed with primary infiltrating ductal carcinoma of the breast at the Beijing Cancer Hospital in China during the period 1995–2007. Their epidemiological information was obtained from their clinical records, including age at diagnosis, height, weight, age at menarche and/or menopause, menopause status, age at first full-term pregnancy, and family history of cancer in first-degree relatives (table S4). Event-free survival time was defined as the time from surgery to the breast cancer events such as breast carcinoma recurrence, metastasis, and death caused by breast cancer. Cases were censored if the patients were still alive, voluntarily withdrew, or died of a cause other than breast cancer before 31 August 2010 (49). The 1336 controls were selected from cancer-free women participating in a community-based screening program for noninfectious diseases conducted in Beijing, China. The selection criteria included no history of cancer, Chinese Han ethnic background, and age-matched to cases (49). All eligible controls completed an epidemiological questionnaire.
Mouse models
The Ntn4 knockout mice with a C57BL/6 genetic background (C57BL/6J-Ntn4em1Smoc) were generated by deleting the genomic fragment containing exon 2 of the Ntn4 using CRISPR-Cas9 RNA-guided nucleases at Shanghai Biomodel Organism. We first bred B6.FVB-Tg (MMTV-PyVT) 634Mul/LellJ (PyMT, C57BL/6 background) mice from the Jackson Laboratory with Ntn4−/− mice and then bred the PyMT;Ntn4+/− males with Ntn4+/− females to obtain a cohort of female littermates with PyMT;Ntn4wt, PyMT;Ntn4+/−, and PyMT;Ntn4−/− genotypes for the mammary tumor studies. Mice were genotyped by PCR using genomic DNA extracted from mouse tails. The primers used for PCR genotyping Ntn4-deficient mice are listed in table S7. The PyMT transgene was identified using primer sequences from the Jackson Laboratory. Additional animal experiments were described in the Supplementary Materials. All mice used in this study were females. Mice were housed at the Laboratory Animal Center of the Peking University Health Science Center with a standard 12-hour light/dark schedule and fed standard chow and water. All mouse studies were approved by the Peking University Biomedical Ethics Committee.
Cell lines
MCF10A, MDA-MB-231, BT-549, T-47D, SKBR3, 293T, and L cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). MCF-7 and L Wnt-3A cells were purchased from the Cell Culture Collection of Chinese Academy of Medical Sciences (Shanghai, China). The SUM159 cell line was purchased from Asterand (Detroit, MI, USA). MCF10A cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), insulin (10 μg/ml), EGF (20 ng/ml), hydrocortisone (500 ng/ml), and cholera toxin (100 ng/ml). L cells, L Wnt-3A cells, MDA-MB-231, MCF-7, BT-549, SKBR3, and 293T cell lines were maintained in DMEM, supplemented with 10% FBS. For L Wnt-3A cells, G-418 (0.4 mg/ml) was added to the culture medium. SUM159 and T-47D cells were grown in RPMI 1640 with 10% FBS. Various derivative cell lines generated in the present study are described in the Supplementary Materials. Cell lines were authenticated using the short tandem repeat method. All cell lines were determined to be free of mycoplasma contamination by PCR assay (Yeasen, Shanghai, China).
Reagents, plasmids, and antibodies
The reagents, plasmids, and antibodies are listed in table S8.
Functional annotation of LD-SNPs
For GWAS SNP rs17356907, LD was calculated between rs17356907 and all other SNPs (in the 500-kilobase window) in East Asia and European ancestries based on 1000 Genomes Project’s data using the snpStats package. LD-SNPs for each signal were defined as r2 > 0.2.
LD-SNPs were overlapped with epigenetic enhancer marks. Mapped read files of H3K27ac ChIP-seq, H3K4me3 ChIP-seq, H3K4me1 ChIP-seq, and DHS of breast tissue or breast cell lines from the Human Epigenomics Roadmap Project (Roadmap) or ENCODE (table S2) were downloaded, and peaks were calculated using MACS 1.4.2 using parameters -p 1e-9, –keep-dup = auto, and -g hs with no control library. H3K27ac, H3K4me3, H3K4me1, DHS, and ATAC-seq peak files of breast cell lines were also downloaded from the Cistrome database (table S2) with no redundancy using data from Roadmap or ENCODE. All peak files were established on UCSC hg19 or converted to hg19 using the UCSC liftOver tool. For each SNP, the total number of overlapping peaks for each mark was calculated. Gene expression–related SNPs overlapping more than one enhancer mark (H3K27ac, H3K4me1, DHS, and ATAC-seq peak) were defined as modified SNPs.
LD-SNPs were also overlapped with mammary gland eQTL data in significant SNP-gene pairs dataset from GTEx v8 (50). For significant gene expression–related SNPs, chromatin interaction mapping analysis was performed to further identify SNPs directly interacting with potentially regulated genes’ promoters. Significant interactions from Hi-C data of Human Mammary Epithelial Cells (HMEC) (GSE63525) were calculated using specialized Hi-C programs in HOMER (51) using default parameters on the hg19 genome.
TF binding site prediction
The nucleotide sequence for the region (chr12:96,027,442-96,027,492, hg19) containing rs11836367 was extracted. The TF binding capability of rs11836367 was predicted using PrEdicting Regulatory Functional Effect by Approximate P-value Estimation software coupled with TFBS motif collection of HOMER. P value < 0.001 and fold change > 10 were used as parameters. To select candidate TFs, each TF was filtered by (i) showing high expression levels in normal breast tissues or breast cancer tumors based on RNA sequencing (RNA-seq) data TCGA and (ii) having been reported as related to breast cancer.
Chromatin immunoprecipitation assay
The ChIP experiments were performed as previously described with modification (52). A total of 1 × 107cells were fixed with 1% formaldehyde at room temperature for 10 min and quenched with glycine. Cell pellets were resuspended with radioimmunoprecipitation assay (RIPA) 0 buffer [10 mM tris-HCl, 1 mM EDTA, 0.1% NaDOC, 0.1% SDS, and 1% Triton X-100 (pH 8.0) with 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor cocktail] and rotated for 15 min at 4°C. After centrifugation, nuclei were suspended in 500 μl of 0.5% SDS lysis buffer [0.5% SDS, 1 mM EDTA, and 50 mM tris-HCl (pH 8.0) with 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail] and fragmented by sonication to an average size between 200 and 500 bp. Forty-four microliters of 10% Triton X-100 was added to the sonicated nuclei lysate and centrifuged at 16,100g for 10 min at 4°C. The supernatant was transferred to a new Eppendorf tube, followed by the addition of NaCl to a final concentration of 150 mM. Twenty microliters of the residual supernatant was saved as the input control. The remaining supernatant was divided into aliquots and incubated with 5 μg of the antibody or control immunoglobulin G (IgG) overnight at 4°C. After incubation, protein A/G beads (Bimake, Shanghai, China) were used to capture antibodies, and beads were washed with 1 ml of low salt buffer [50 mM Hepes, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, and 0.1% NaDOC (pH 7.5)], high salt buffer [50 mM Hepes, 1 mM EDTA, 500 mM NaCl, 1% Triton X-100, and 0.1% NaDOC (pH 7.5)], LiCl buffer [250 mM LiCl, 0.5% NP-40, 0.5% NaDOC, 1 mM EDTA, and 10 mM tris-HCl (pH 8.1)], and TE buffer [10 mM tris-HCl and 1 mM EDTA (pH 8.0)]. The chromatin was eluted in elution buffer (1% SDS and 0.1 M NaHCO3), followed by reversing cross-linking at 65°C overnight. Eluted DNA was treated with ribonuclease (RNase) at 37°C, followed by incubation with proteinase K at 65°C, and purification using QIAquick spin columns (QIAGEN, Hilden, Germany). The primer sequences used in ChIP-qPCR are listed in table S7. The antibodies used for ChIP are as follows: GATA2 (Abcam, ab109241), GATA3 (Abcam, ab199428), and control IgG (AC005, ABclonal). The results are presented as fold enrichment (normalized to IgG).
Luciferase reporter assay
The pGL3-promoter construct was generated by inserting a 1059-bp NTN4 promoter sequence (chr12:96184432-96185491, hg19) and amplified from MCF10A into the Xho I and Mlu I sites of pGL3-Basic (Promega, Madison, WI, USA). Then, PCR products of the enhancer fragment (1335 bp, chr12:96,026,578-96,027,912, hg19), CRE fragment (1439 bp, chr12:96,020,720-96,022,158, hg19), and PRE1 fragment (1010 bp, chr12:96,026,561-96,027,570) from MCF10A were inserted into the Bam HI and Sal I sites of the pGL3-promoter construct separately. The different alleles of individual SNPs were introduced into the enhancer or CRE by overlap extension PCR using HiFi HotStart DNA Polymerase (KAPA Biosystems, Wilmington, MA, USA). All plasmids were confirmed for correct variant incorporation by Sanger sequencing.
Breast cells were plated 1 day before transfection in 96-well plates. For normal luciferase assay, each well was transfected with 0.3 μl of ViaFect transfection reagent (Promega, Madison, WI, USA), 100 ng of experimental firefly luciferase reporter plasmid, and 2.5 ng of thymidine kinase promoter-Renilla luciferase (pRL-TK) plasmid (Promega, Madison, WI, USA). For the luciferase assay coupled with overexpression of GATA3, pcDNA3.1-GATA3 was first constructed by amplifying the GATA3 sequence from MCF10A cDNA and then cloned into a linearized pcDNA3.1 vector using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Then, each well was transfected with 0.3 μl of ViaFect transfection reagent (Promega, Madison, WI, USA), 50 ng of firefly luciferase reporter plasmid, 50 ng of pcDNA3.1-GATA3, and 2.5 ng of pRL-TK plasmid. The total amount of transfected DNA was balanced with the pcDNA3.1 empty vector. Cells were harvested 24 hours after transfection and assayed with the Dual-Glo luciferase assay system (Promega, Madison, WI, USA) on the Luminoskan (Thermo Fisher Scientific, Waltham, MA, USA) or Flexstation 3 (Molecular Devices, San Jose, CA, USA).
DNA isolation and genotyping assay
Genomic DNA for controls was isolated from peripheral blood leukocytes, whereas cases’ genomic DNA was extracted from formalin-fixed paraffin-embedded normal fallopian tube tissue. Genotyping was conducted with the QuantStudio 6 (Thermo Fisher Scientific, Waltham, MA, USA) using the TaqMan Assay in compliance with the manufacturer’s instructions. Primers and probes (FAM and VIC labeled) were supplied by ABI Incorporation, and the PCR reaction used ChamQ Geno-SNP Probe Master Mix (Vazyme, Nanjing, China) according to the manufacturer’s instructions.
Electrophoretic mobility shift assay
Nuclear extracts were prepared according to previously published standard protocols (53). Five micrograms of nuclear extract proteins was incubated with 0.2 μM double-strand biotin-labeled probe for 25 min at room temperature in 16 μl of binding reaction buffer [10 mM tris, 50 mM KCl, and 1 mM DTT (pH 7.5)] with 50 ng of poly(dI:dC). For the competition experiments, the 20 μM unlabeled probe was also added to the reaction buffer. The samples were separated on 10% tris-borate EDTA polyacrylamide gels with 20% polyethylene glycol. Duplex-bound complexes were transferred onto the Hybond N+ nylon membrane (Cytiva, Marlborough, MA, USA) and cross-linked under 254-nm ultraviolet light. The membrane was processed with the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA, USA) and detected with the Immobilon Western chemiluminescent horseradish peroxidase substrate. The details of the oligo sequences used in the EMSA assay are listed in table S7.
Lentivirus production and transduction
Lentiviral plasmid (7.46 μg), together with 3.72 μg of pMD2.G (Addgene plasmid no. 12259) and 5.74 μg of psPAX2 (Addgene plasmid no. 12260), was cotransfected into 293T cells in a 100-mm dish using polyethyleneimine as the transfection reagent. The virus-containing culture medium was collected after 48 hours, spun for 10 min at 3000 rpm, and filtered through a 0.45-μm filter. Lentiviral transfection was carried out in 12-well plates by mixing 500 μl of the virus supernatant with 500 μl of medium containing 8 μg of polybrene at a multiplicity of infection < 1.
Generation of CRISPR activation cell lines
Individual KB-sgRNAs were constructed by annealing each pair of oligos and ligating into a Bsm BI–linearized lentiviral plasmid pLV-hU6-sgRNA-hUbC-dCas9-KRAB-T2a-Puro (Addgene plasmid no. 71236) (54). Detailed information for each sgRNA sequence is listed in table S7. Lentivirus production and transduction are described above. The successfully transfected cells were selected and maintained using puromycin at a concentration of 1 μg/ml for SUM159 and MCF-7 and regularly passaged for further expression study or functional analysis.
Generation of CRISPR activation cell lines
MCF10A, SKBR3, and MDA-MB-231 cells stably expressing dCas9-VP64 and MS-P65-HSG1 were generated by transducing a lentiviral plasmid containing lenti-dCAS-VP64-Blast (Addgene plasmid no. 61425) and lenti-EF1a-MS2-p65-HSF1-2A-Hygro-WPRE (Addgene plasmid no. 89308), followed by selection, and maintained using blasticidin (10 μg/ml) and hygromycin (200 μg/ml).
Individual AC-sgRNAs used in the enhancer activation assay were constructed by annealing each pair of oligos and ligated into the Bsm BI–linearized lentiviral plasmid, lenti-sgRNA (MS2)-puro (Addgene plasmid no. 73795) (54). Detailed information for each sgRNA sequence is listed in table S7. The production and transduction of lentivirus are described above. The successfully transfected cells were selected and maintained with puromycin at a concentration of 0.5 μg/ml for MCF10A-dCas9-VP64-MPH until ready for the expression study.
For the Dox-induced NTN4 overexpression assay, a Dox-inducible sgRNA (MS2) vector [tet-pLKO-sgRNA (MS2)-puro] coexpressing the Tet repressor was first modified from the tet-pLKO-puro vector (Addgene plasmid no. 21915) by replacing the sequences encoding short hairpin RNA with sgRNA(MS2) and the human phosphoglycerate kinase eukaryotic (hPGK) promoter with the elongation factor 1-α ( (EF1α) promoter using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Individual AC-sgRNAs used in Dox-induced NTN4 overexpression assay were constructed by annealing each pair of oligos and ligating into the Bsm BI–linearized lentiviral plasmid, tet-pLKO-sgRNA (MS2)-puro. Detailed information for each sgRNA sequence is listed in table S7. The production and transduction of lentivirus are described above. The successfully transfected cells were selected and maintained with puromycin at a concentration of 0.5 μg/ml for SKBR3-dCas9-VP64-MPH and 1 μg/ml for MDA-MB-231-dCas9-VP64-MPH and regularly passaged for further analysis.
CRISPR-Cas9–mediated genome editing assay
To knock out a 151-bp genomic DNA sequence containing rs11836367, we applied a dual-guide RNA strategy using two Cas9-guide RNA constructs with different selection markers. We first constructed a PX459-Hygro vector by replacing the DNA sequence encoding puromycin N-acetyltransferase of PX459-puro (Addgene plasmid no. 62988) with hygromycin B phosphotransferase using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Two sgRNAs used in the knockout assay were constructed by annealing each pair of oligos and ligated into Bbs I–linearized lentiviral plasmid PX459-puro or PX459-Hygro, respectively. Details of the sgRNA sequences used are listed in table S7. MCF10A and MCF-7 cells were transiently cotransfected with two sgRNAs, which were then cloned into PX459-puro or PX459-Hygro, respectively, followed by selection at a concentration of 1 μg/ml puromycin and 200 μg/ml hygromycin for 2 days. Individual clones were selected after 14 days. Correctly targeted clones were identified by genomic PCR with primers outside the targeting region and further confirmed with Sanger sequencing. To avoid off-target effects of the Cas9 nuclease, three wild-type and three biallelic 151-bp deletion clones were selected in each cell line for further study.
To study the allele-specific effect of rs11836367 on NTN4 expression, allele-specific editing was performed as previously described with modification (30). Briefly, the AS-sgRNA targeting each allele of rs953413 was directly constructed by annealing each pair of oligos and ligated into a Bsm BI–linearized lentiviral plasmid, pCC_02-hU6-Bsm BI-sgRNA(E+F)-barcode-EFS-Cas9NG-NLS-2A-Puro-WPRE (Addgene plasmid no. 139087). Details of the sgRNA sequences used are listed in table S7. The production and transduction of lentivirus are described above. The successfully transfected cells were selected and maintained with puromycin and regularly passaged until ready for study.
Immunoblotting
Cells or mouse tumor tissues were lysed in RIPA lysis buffer (Applygen, Beijing, China) containing protease inhibitors and phosphatase inhibitors (Targetmol, Shanghai, China). Nuclear proteins were extracted using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China) according to the manufacturer’s instruction. Primary antibodies used are listed in table S8.
Tumor onset, progression, and metastasis studies in GEM models
The time period before developing a tumor in mice was determined by palpation. Tumor growth was monitored by measuring the volume of each individual mammary tumor for each PyMT;Ntn4wt, PyMT;Ntn4+/−, and PyMT;Ntn4−/− mouse once a week for up to 20 weeks with a caliper. Tumor volumes were calculated using the formula: V = (L/2) × (W)2, where V represents the volume, L represents the length, and W represents the width of the tumor. Mice were anesthetized with isoflurane when they met the institutional euthanasia criteria for tumor size (2 cm in diameter) or overall health. Whole tumors and lung tissues were collected, weighed, and processed for histopathological analysis. Lung metastases were analyzed via examination of freshly dissected lungs and hematoxylin and eosin (H&E)–stained lung sections. The detection of circulating cancer cells was performed as described previously with modification (55, 56). Briefly, 500 μl of blood sample was collected from each tumor-bearing mouse at the end stage of the experiment, via cardiac puncture under anesthesia with isoflurane. The red blood cells were depleted by 0.64% NH4Cl treatment, and the remaining cells were lysed by TRIzol. mRNA was extracted according to the manufacturer’s instructions. The HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) was used for cDNA preparation. qPCR was performed using the SYBR qPCR Mix (TOYOBO, Osaka, Japan), and Pymt and Krt8 transcript levels were normalized to the Hprt transcript level. The primer sequences used are listed in table S7. For early tumor progression analysis of 13-week-old mice, tumors were evaluated by staging them into normal, hyperplasia, adenoma, and carcinoma based on H&E and immunohistochemical (IHC) staining of α-SMA as previously reported (35). Morphometric analysis of early PyMT tumor growth was performed using whole-mount staining. Briefly, inguinal mammary glands were fixed in Carnoy’s fixative buffer overnight at 4°C. After rehydration, mammary glands were then stained at 0.2% carmine alum overnight and dehydrated. Mammary glands were then cleared in xylene and mounted in Permount for further analysis. Investigators were blinded to the group allocation when assessing the outcome.
RNA-seq analysis
The SKBR3-dCas9-VP64-MPH-AC-sg1 cell line was established as described in the Supplementary Materials. Total RNA from SKBR3-dCas9-VP64-MPH-AC-sg1 cells treated with or without Dox (50 ng/ml; three biological repeats for both groups) was extracted using TRIzol according to the manufacturer’s instructions. The quality of RNA samples was examined by Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). The samples were subjected to RNA-seq analysis on the BGISEQ-500 system by Beijing Genomics Institute (China). The clean-tag reads were aligned to the reference genome and reference genes using HISAT2 and Bowtie2. The matched reads were calculated and then normalized using RSEM software, and differential gene expression was analyzed using DESeq2. Significantly differential expression genes were characterized according to signaling pathways using PANTHER version 17.0. Expression data are accessible through Gene Expression Omnibus (GEO; GSE197635).
Gene set enrichment analysis
A gene set enrichment analysis (GSEA) software tool (version 4.0.1) was used to analyze the rank gene set, as described previously (57). For the RNA-seq data of 113 normal breast samples and 1109 breast cancer samples from TCGA, the genes were ranked by the Pearson correlations of NTN4 expression and expression of 20,250 genes within this dataset. The curated sets of BENPORATH_PROLIFERATIION, POOLA_INVASIVE_BREAST_CANCER_UP, VANTVEER_BREAST_CANCER_POOR_PROGNOSIS, and ZHANG_BREAST_CANCER_PROGENITORS_UP were obtained from the Molecular Signatures Database. The curated sets of WNT targets derived from Nusse and colleagues (web.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes), Fevr and colleagues (58), and Herbst and colleagues (59) listed in table S9 were also used in GSEA analysis.
Co-immunoprecipitation assay
pcDNA3.1-NTN4-HA was constructed by amplifying the NTN4 sequence from cDNA of MCF10A and cloning into a PCR-linearized pcDNA3.1 with C-terminal HA-tag. NTN4LN (amino acids 34 to 260), NTN4LE (amino acids 261 to 498), and NTN4NTR (amino acids 499 to 628) were obtained by PCR on full-length NTN4 and cloned into PCR-linearized pcDNA3.1 with the NTN4 signal peptide (amino acids 1 to 33) sequence at the N terminus and HA-tag at the C terminus using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. WNT3A, WNT4, WNT5A, WNT7B, and WNT10B were amplified from the cDNA of L Wnt-3A cells. MCF10A and Wnt2 were amplified from pCMV-SPORT6-Mnt2-Wnt2 (a gift from Ziyunbio Co. Ltd., Hangzhou, China). The amplified sequences of Wnt ligands were cloned into PCR-linearized pcDNA3.1 with Myc-tag at the C terminus using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions.
Co-immunoprecipitation of NTN4 protein with different Wnt ligands was conducted as previously described with modification (60). Briefly, 293T cells were plated into 100-mm plates on the day before transfection. Six micrograms of the construct expressing HA-tagged NTN4 and 12 μg of the construct expressing different Myc-tagged Wnt ligands were cotransfected into 293T cells in a 100-mm dish using polyethyleneimine as the transfection reagent. On the following days, cells were lysed in 1 ml of lysis buffer [150 mM NaCl, 50 mM tris, 1% NP-40, and 1× protease inhibitor cocktail (pH 7.5)]. After precleaning with protein A/G beads (Bimake, Shanghai, China), a small sample of lysate was saved as input. The remaining lysate was divided into aliquots and incubated with anti–Myc-tag antibody (Proteintech, 16286-1-AP) or negative control IgG (ABclonal, AC005) overnight at 4°C. After incubation, the lysate was further incubated with protein A/G beads (Bimake, Shanghai, China) for another 2 hours at 4°C. After three washes with washing buffer [150 mM NaCl, 50 mM tris, 0.5% NP-40, and 1× protease inhibitor cocktail (pH 7.5)] and one wash with phosphate-buffered saline (PBS), the beads were resuspended in 50 μl of 2× loading buffer and boiled. Fifteen microliters of each sample was loaded onto 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blot using the anti–HA-tag antibody (1:1000; Proteintech, 51064) for detection of NTN4 and anti–Myc-tag antibody (1:1000; Proteintech, 16286-1-AP) for detection of Wnt ligands.
GST pull-down assay
The glutathione S-transferase (GST) pull-down assay was performed as previously reported (60, 61). NTN4△SP, NTN4LN, NTN4LE, and NTN4NTR sequences were cloned into the pGEX-6P-1 plasmid using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. GST and GST fusion proteins were expressed and purified from Escherichia coli BL21 stain using Glutathione Sepharose 4B (GE Healthcare, Chicago, IL, USA). Wnt ligand plasmids were transfected into 293T cells using polyethyleneimine as the transfection reagent. After 24 hours, cells were lysed using 1× lysis buffer [150 mM NaCl, 50 mM tris, 1% NP-40, and protease inhibitors (pH 7.5)]. The lysates were then incubated with GST or GST-fused protein at 4°C overnight. The beads were washed three times with PBS and further boiled in 50 μl of 2× loading buffer for 5 min. Prepared samples were analyzed by Western blotting.
Primary organoids and cell models
Mammary carcinoma cell organoids were obtained as described previously with modification (62). Briefly, mammary tumors from age-matched virgin PyMT;Ntn4wt or PyMT;Ntn4−/− mice were minced and digested in DMEM/F12 containing 0.2% trypsin, collagenase type IV (300 U/ml), 5% FBS, and insulin (10 μg/ml) for 30 min at 37°C with rotation. Mammary tumor organoid suspensions were applied to a 100-μm cell strainer and washed in serum-free DMEM/F12. Organoids were separated from single cells with differential centrifugation before in vitro culture. For isolation of primary tumor single cells, tumor tissues from PyMT;Ntn4wt or PyMT;Ntn4−/− were dissected from age-matched virgin mice, minced with a razor blade, and digested in DMEM/F12 culture medium containing hydrocortisone (500 ng/ml), EGF (10 ng/ml), and 5% FBS supplemented with collagenase type IV (300 U/ml) and hyaluronidase (100 U/ml) for 2 hours at 37°C with rotation. After lysis of the red blood cells with 0.64% NH4Cl, a single suspension was obtained by sequential dissociation of the fragment in 0.25% trypsin for 2 min and dispase (5 mg/ml) with DNase I (0.1 mg/ml) for 2 min with gentle pipetting followed by filtration through a 40-μm cell strainer.
Bioinformatics analysis of clinical data
Data for TCGA-breast invasive carcinoma were accessed through UCSC Xena and used to analyze gene expression in normal breast and breast cancer tissues. NTN4 expression in different tumor tissues and the correlation between NTN4 expression and breast cancer molecular subtypes were analyzed by UALCAN. Kaplan-Meier Plotter (KMPlot) was used to analyze the prognostic value of genes in breast cancer. Gene Expression Profiling Interactive Analysis (GEPIA) was used to analyze the correlation between the expression of NTN4 and the expression of GATA3 in normal breast and breast cancer tissues.
RNA extraction, cDNA synthesis, and qPCR
For gene expression analysis, total RNA from human cell lines or mouse tissues was isolated using TRIzol reagent according to the manufacturer’s instruction. cDNA synthesis from total RNA was performed using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). Real-time PCR and data collection were performed with SYBR qPCR Mix (TOYOBO, Osaka, Japan) on the 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) or the CFX96 Real-Time PCR Detection system (Bio-Rad, Hercules, CA, USA). The relative mRNA expression level was calculated using the 2−ΔΔCT method. Primer sequences are listed in table S7.
Allele-specific qPCR (AS-qPCR) was performed as previously described (30). Primers for AS-qPCR were designed on the basis of the mismatch amplification mutation assay (63) and listed in table S7. AS-qPCR of rs11836367 was used to validate the allelic imbalance binding of GATA3 in the ChIP experiment and allelic imbalance editing using AS-sgRNA coupled with Cas9-NG. AS-qPCR of rs2160989 was used to evaluate the allelic imbalance in NTN4 expression. The AS-qPCR reactions were performed using SYBR qPCR Mix (TOYOBO, Osaka, Japan) on the CFX96 Real-Time PCR Detection system (Bio-Rad, Hercules, CA, USA).
Genomic sequencing–based phase construction
The procedure of phase construction between rs11836367 and the reporter SNP rs2160989 was performed as described previously with modification (29). Genomic DNA from wild-type MCF10A was amplified using HiFi HotStart DNA Polymerase (KAPA Biosystems, Wilmington, MA, USA). PCR products were ligated into the PCR-linearized pUC19 vector using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The phase between rs11836367 and rs2160989 was manually determined on the basis of the genotype of a linked heterozygous SNP in the overlapping region between fragments. Details about the PCR primer sequences used are listed in table S7.
In situ CAPTURE assay
The in situ CAPTURE assay was performed as previously described with modification (52). Briefly, the lentivirus vector Lenti-pEF1a-BirA-V5-Hygro was first modified from the lentiMPH v2 construct (Addgene plasmid no. 89308) by replacing the sequences encoding MS2-p65-HSF1 with sequences encoding BirA-V5 from the pEF1a-BirA-V5-neo construct (Addgene plasmid no. 100548), while Lenti-pEF1a-FB-dCas9-Blast was modified from the Lenti-dCAS-VP64-Blast plasmid (Addgene plasmid no. 61425) by replacing sequences encoding dCas9-VP64 with sequences encoding FB-dCas9 from the pEF1a-FB-dCas9-puro construct (Addgene plasmid no. 100547) using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Sequence-specific sgRNA was incorporated into the lentivirus vector pSLQ1651-sgRNA(F+E)-sgGal4 (Addgene plasmid no. 100549). The detailed information for each sgRNA sequence is listed in table S7. MCF10A cells stably expressing FB-dCas9, BirA-V5, and sgRNA were generated by transducing lentiviral plasmids containing Lenti-pEF1a-FB-dCas9-Blast, Lenti-pEF1a-BirA-V5-Hygro, and pSLQ1651-sgRNA(F+E)-sgRNA, followed by selection and maintaining the following concentrations: blasticidin (10 μg/ml), hygromycin (200 μg/ml), and puromycin (0.5 μg/ml). Sequence-specific or nontargeting control sgRNA was incorporated into the lentivirus vector pSLQ1651-sgRNA(F+E)-sgGal4 (Addgene plasmid no. 100549). The specificity and efficiency of CAPTURE were verified by ChIP-qPCR. For Western blot analysis of captured protein, 2.5 × 108 cells were fixed with 2% formaldehyde at room temperature for 10 min and quenched with glycine. Cell pellets were resuspended with cell lysis buffer [25 mM tris-HCl, 85 mM KCl, 0.1% Triron X-100 (pH 7.4), 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail] and rotated for 30 min at 4°C. After centrifugation, nuclei were suspended with cell lysis buffer containing RNase A (1 ng/μl) and 1× protease inhibitor cocktail followed by incubation at 37°C for 30 min with rotation. After centrifugation, nuclei were then lysed in 400 μl of nuclear lysis buffer [50 mM tris-HCl, 1 mM EDTA, 0.5% SDS (pH 8.1), 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail] mixed with 1.2 ml of 8 M urea buffer (10 mM tris-HCl, 1 mM EDTA, and 8 M urea). After centrifugation, the pelleted chromatin was then washed twice with cell lysis buffer and suspended in immunoprecipitation (IP) binding buffer with NaCl (20 mM tris-HCl, 1 mM EDTA, 0.1% NP-40, and 10% glycerol, with 1× protease inhibitor cocktail) and sonicated to an average size of 500 bp. Fragmented chromatin was centrifuged and NaCl was added to a final concentration of 150 mM. The balanced streptavidin agarose slurry (Thermo Fisher Scientific, Waltham, MA, USA) was added to the soluble chromatin and incubated at 4°C overnight. The beads were then washed five times with 1 ml of IP binding buffer [20 mM tris-HCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol, 150 mM NaCl, and 1× protease inhibitor cocktail (pH 7.5)]. After centrifugation, 50 μl of loading buffer was added to the spin beads followed by incubation at 100°C for 10 min. The protein was separated by SDS-PAGE and analyzed by Western blotting.
Flow cytometry analysis
Flow cytometry analysis of tumor stem cells in the PyMT model was performed as described previously (64). Briefly, single tumor cells isolated from PyMT and PyMT;Ntn4−/− tumors were incubated with biotin-conjugated lineage antibodies including anti-CD31 (1:200; Thermo Fisher Scientific, 13-0311-81), anti-CD45 (1:400; Thermo Fisher Scientific, 13-0451-82), and anti-TER119 (1:100; Thermo Fisher Scientific, 13-5921-82) antibodies for 15 min at 4°C and then incubated with anti-biotin microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) for 15 min at 4°C. Lin− (CD31−CD45−TER119−) tumor single cells were selected using an LS column adapter (Miltenyi Biotec, Bergisch Gladbach, Germany). Single-cell suspensions of tumor cells were suspended in staining buffer (2% bovine serum albumin in PBS) and stained with phycoerythrin (PE)–conjugated anti-CD24 (1:500; Thermo Fisher Scientific, 12-0242-82) and PE-Cyanine7–conjugated anti-CD90 (1:200; Thermo Fisher Scientific, 25-0902-82). 7-Aminoactinomycin D (Thermo Fisher Scientific) was immediately added before fluorescence-activated cell sorting analysis. Cells were analyzed using the CytoFLEX S Flow Cytometer (Beckton Dickinson, Franklin Lakes, NJ, USA). Data were analyzed using the software FlowJo 10.
Cell proliferation analysis
Cell proliferation was analyzed using the Cell Counting Kit-8 (CCK-8) (Yeasen, Shanghai, China). Briefly, breast cancer cells were plated at a density of 2 × 103 cells per well in the 96-well plate and maintained with or without treatment. After 24, 48, 72, and 96 hours, CCK-8 solution was added to each well according to the manufacturer’s instructions, and cells were maintained in the incubator for another 1 hour at 37°C. After incubation, absorbance readings were obtained at 450 nm.
Transwell migration assay
Transwell migration assays were performed using a 24-well Transwell chamber (Corning, NY, USA). Cancer cells were plated in the insert at a density of 5 × 104 cells per well, maintained in culture media containing 0.5% FBS, and allowed to migrate for 24 hours at 37°C with or without treatment. Migrated cells were fixed with 4% formaldehyde, stained with crystal violet, and counted under the microscope.
Three-dimensional invasion assay
For studying breast cancer cell invasion into the ECM using Matrigel, assays were performed as previously described with modification (65). Briefly, spheroids of breast cancer cells were generated for 48 hours starting with 3 × 103 cells in 0.25% Matrigel on ultra-low attachment plates. Invasion was initiated with the addition of Matrigel at a 1:1 volume ratio with spheroid medium, and spheroids were allowed to invade for 6 days with or without treatment. Bright-field images were taken with an Olympus IX71 microscope (Olympus, Shinjuku City, Tokyo, Japan) and analyzed with the software ImageJ Analyze Spheroid Cell Invasion In 3D matrix. For studying mammary tumor organoid invasion, isolated mammary organoids from age-matched PyMT;Ntn4wt and PyMT;Ntn4−/− were embedded in type I collagen (2.2 mg/ml) and were cultured in invasion media [DMEM/F12 containing Insulin-Transferrin-Selenium (ITS), insulin (10 μg/ml), FGF-2 (50 ng/μl), and 1% penicillin/streptomycin] with or without treatment. Tumor organoid branch length was measured after 2.5 days from bright-field micrographs by ImageJ software coupled with photoshop.
Tumor sphere formation assay
The tumor sphere assay for human breast cancer cells was performed as previously described (66). Breast cancer cells were plated as a single-cell suspension in 96-well ultra-low attachment plates in serum-free sphere medium for human breast cell lines [DMEM or RPMI 1640 medium supplemented with EGF (20 ng/ml), insulin (10 μg/ml), hydrocortisone (0.5 μg/ml), and B-27]. After 7 days, spheres larger than 50 μm were counted in each group. For the primary PyMT tumor cell sphere formation assay, primary tumor single cells isolated from age-matched PyMT;Ntn4wt and PyMT;Ntn4−/− were plated at a density of 5000 cells per well in 96-well ultra-low attachment plates. Cells were grown in serum-free sphere media for primary tumor cells [DMEM/F12 medium supplemented with B-27, EGF (20 ng/ml), FGF-2 (20 ng/ml), heparin (4 μg/ml), and 1% penicillin/streptomycin] with or without the treatment. After 7 days, spheres larger than 50 μm were counted in each group.
Limiting dilution assay
To assess breast cancer stemness resulting from the deletion of Ntn4, the in vivo limiting dilution assay was performed as previously described with modification (62). Briefly, 8-week-old female wild-type C57BL/6 mice were anesthetized with isoflurane and randomly assigned to the experimental groups. Fifty, 500, and 5000 single primary tumor cells isolated from PyMT or PyMT;Ntn4−/− mice were mixed in 20 μl of growth factor–reduced Matrigel and injected into the fourth mammary fat pad (n = 8 mice each group) of wild-type C57BL/6 mice under anesthesia. The wounds were closed with metal clips, and the animals were placed under a heat lamp until full recovery. Mice were anesthetized with isoflurane for analysis at 4 or 6 weeks after injection. Tumor initiation, cell frequency, and statistical calculations were analyzed using the Extreme Limiting Dilution Analysis Program (67).
Tumor transplantation assay
To test the ability of NTN4-Wnt/β-catenin signaling in tumor growth and metastasis, 1 × 104 primary tumor cells from age-matched PyMT;Ntn4wt and PyMT;Ntn4−/− mice were mixed in 20 μl of growth factor–reduced Matrigel (Corning) and injected into the fourth mammary fat pad of female wild-type C57BL/6 mice (8 weeks old and randomly assigned to the experimental groups) under anesthesia with isoflurane. Wnt/β-catenin XAV939 was administered intraperitoneally in corn oil (the control was corn oil alone) into mice at 25 mg/kg body weight. Treatment was initiated on the day after tumor inoculation (day 1) and administered every 2 days for 4 weeks. Tumor volume was measured twice a week. Mice were euthanized after 4 weeks. Tumors and lungs from each group were collected for subsequent analysis. Investigators were blinded to the group allocation when assessing the outcome.
Preparation of CM
WNT3A CM and control medium were harvested from L Wnt-3A cells and control L cells according to the ATCC manufacturer’s instructions. NTN4− and its different domain-generated CMs were harvested as previously described (36). Briefly, 293T cells were transiently transfected with empty pcDNA3.1 or pcNDA3.1-NTN4-HA, pcNDA3.1-NTN4LN-HA, pcNDA3.1-NTN4LE-HA, and pcNDA3.1-NTN4NTR-HA. CMs and control CMs from these transfected cells were harvested after 2 days of transfection and clarified via centrifugation. The secretion of NTN4 and its different domains was confirmed by Western blotting of ultra-filtrated CMs. As previously described (68), NTN4− or its different domain-derived CMs were mixed with WNT3A CM and incubated at 37°C for 1 hour. After incubation, the mixture was ready for further use.
TOP/FOPFlash reporter assay
Breast cancer cells were seeded in a 96-well plate and transfected with 100 ng of TOPFlash reporter vector (Addgene plasmid no. 12456) or FOPFlash reporter vector (Addgene plasmid no. 12457) coupled with 2.5 ng of pRL-TK (Promega, Madison, WI, USA). All transfections were performed with the ViaFect transfection reagent (Promega, Madison, WI, USA) according to the manufacturer’s protocol. After 6 hours, cell medium was changed with or without treatment. For the CM assay, cells were incubated with control medium or mixed with the CM as described above. After 24 hours, luciferase activity was measured using Dual-Glo luciferase assay system (Promega, Madison, WI, USA) on the Luminoskan (Thermo Fisher Scientific, Waltham, MA, USA). Values were normalized to untreated controls.
Histology
Tumors and lungs from mice were fixed for 24 h in 10% neutral buffered formalin and stored in 70% ethanol before embedding in paraffin. Sections were stained with H&E for pathological analysis. For IHC staining, the slides were deparaffinized and pretreated with a pressure cooker. The sections were developed with the Vectastain ABC Kit (Vector Laboratories, Burlingame, CA, USA) and the DAB Substrate Kit (ZSGB-BIO, Beijing, China). Hematoxylin was used as a counterstain. For immunofluorescent staining, deparaffinized sections were pretreated with a pressure cooker, incubated with primary antibodies followed by an Alexa Fluor 55–conjugated secondary antibody, and then counterstained with DAPI (4′,6-diamidino-2-phenylindole) in mounting media. All fluorescence images were acquired with a confocal microscope (LSM 700, Zeiss, Oberkochen, Germany). The following antibodies were used: antibodies against AXIN2 (1:200; ABclonal, A2513), cyclin D1 (1:200; ABclonal, A19038), Ki-67 (1:400; Abcam, ab16667), Ntn4 (1:100; R&D, AF1132), active β-catenin (1:200; Millipore, 05-665), PR (1:200; Genetech, GT216029), ER (1:800; Sino Biological, 106132-T08), HER2 (1:500; Cell Signaling Technology, 4290T), CK14 (1:400; ABclonal, A19039), and α-SMA (1:400; Abcam, ab7817). For Ki-67, CCND1, ER, and PR, the percentage of positively stained cells was evaluated. Assessment of staining for HER2 was performed as follows: 0 = no staining; 1 = rare/minimal positive staining; 2 = occasional positive staining; 3 = moderate positive staining; 4 = widespread positive staining. For AXIN2, the percentage of positive areas with AXIN2 staining was analyzed using the IHC profiler in ImageJ.
Quantification and statistical analysis
Data are expressed as the means ± SEM. Statistical analysis and graphs were generated using SPSS Statistics software (SPSS), R software, and GraphPad Prism 8 software. Sample size (n) was indicated in the figure legends, which are based on our previous works and those commonly used in the field. Data were compared using the two-sided chi-square test, Student’s t tests, and one- or two-way analysis of variance (ANOVA). Dunnett or Bonferroni correction was used in instances where multiple comparisons were performed. Hardy-Weinberg equilibrium was evaluated for each SNP using a one–degree of freedom goodness-of-fit test among the controls using a cutoff threshold of 0.05. The Cochran-Armitage trend test was performed to estimate the association between breast cancer risk and allele dose in each SNP (P trend). Furthermore, odds ratios with 95% confidence intervals were calculated to evaluate the effects of genotypes or haplotypes on breast cancer risk using chi-square or multivariate unconditional logistical regression models, adjusted for age at menarche, age of first birth, and family history of cancer in first-degree relatives. In the analysis of repeated measures of tumor volume, two-way repeated-measures ANOVA with Bonferroni correction was used. Survival curves of mice were calculated according to the Kaplan-Meier method, and statistical tests were performed using the log-rank test. All information regarding the statistical tests performed and “n” and P values used in each experiment can be found in the figures and figure legends. A P < 0.05 was considered as the indication of a statistically significant difference, with *P < 0.05, **P < 0.01, and ***P < 0.001 unless otherwise indicated in the figures.
Acknowledgments
We are grateful to X.Z. Lang, H.-Y. Chen, and C. Beaman for assistance with graphics and insightful scientific discussions. We thank the personnel of the Histology, Flow Cytometry, Cell Culture, Genomics and Animal Facility Shared Resources at Peking University Health Science Center for technical assistance.
Funding: This work was supported by grants to X.-X.T. and W.-G.F. from the National Natural Science Foundation of China (nos. 81872382, 81672790, and 81621063). E.Z. was supported by the National Cancer Institute (K24CA169004).
Author contributions: H.Y. conceptualized the idea, designed and performed the experiments, analyzed the data, and wrote the paper. X.T. and Y.-H.G. performed the experiments, analyzed the data, and wrote the paper. J.L.N. and Y.-F.H. performed the experiments and analyzed the data. Y.-T.Z., Y.H., Y.-Q.Y., X.-Y.Y., and X.-F.L. performed the experiments. W.-G.F., H.Z., E.Z., and Y.X. oversaw the project, designed the experiments, and revised the paper. X.-X.T. and Y.S. conceptualized the idea, oversaw the project, designed the experiments, analyzed the data, and revised the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA-seq data discussed in this publication have been deposited in NCBI’s GEO and are accessible through GEO series accession number GSE197635 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197635). The Ntn4−/− mice can be provided by X.-X.T., Peking University Health Science Center, pending scientific review and a completed material transfer agreement. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S9
REFERENCES AND NOTES
- 1.Walsh T., King M. C., Ten genes for inherited breast cancer. Cancer Cell 11, 103–105 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R. L., Schmidt M. K., Chang-Claude J., Bojesen S. E., Bolla M. K., Wang Q., Dicks E., Lee A., Turnbull C., Rahman N.; Breast and Ovarian Cancer Susceptibility Collaboration, Fletcher O., Peto J., Gibson L., Dos Santos Silva I., Nevanlinna H., Muranen T. A., Aittomaki K., Blomqvist C., Czene K., Irwanto A., Liu J., Waisfisz Q., Meijers-Heijboer H., Adank M., Hereditary B.; Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON), van der Luijt R. B., Hein R., Dahmen N., Beckman L., Meindl A., Schmutzler R. K., Muller-Myhsok B., Lichtner P., Hopper J. L., Southey M. C., Makalic E., Schmidt D. F., Uitterlinden A. G., Hofman A., Hunter D. J., Chanock S. J., Vincent D., Bacot F., Tessier D. C., Canisius S., Wessels L. F., Haiman C. A., Shah M., Luben R., Brown J., Luccarini C., Schoof N., Humphreys K., Li J., Nordestgaard B. G., Nielsen S. F., Flyger H., Couch F. J., Wang X., Vachon C., Stevens K. N., Lambrechts D., Moisse M., Paridaens R., Christiaens M. R., Rudolph A., Nickels S., Flesch-Janys D., Johnson N., Aitken Z., Aaltonen K., Heikkinen T., Broeks A., Veer L. J., van der Schoot C. E., Guenel P., Truong T., Laurent-Puig P., Menegaux F., Marme F., Schneeweiss A., Sohn C., Burwinkel B., Zamora M. P., Perez J. I., Pita G., Alonso M. R., Cox A., Brock I. W., Cross S. S., Reed M. W., Sawyer E. J., Tomlinson I., Kerin M. J., Miller N., Henderson B. E., Schumacher F., Le Marchand L., Andrulis I. L., Knight J. A., Glendon G., Mulligan A. M.; kConFab Investigators; Australian Ovarian Cancer Study Group, Lindblom A., Margolin S., Hooning M. J., Hollestelle A., van den Ouweland A. M., Jager A., Bui Q. M., Stone J., Dite G. S., Apicella C., Tsimiklis H., Giles G. G., Severi G., Baglietto L., Fasching P. A., Haeberle L., Ekici A. B., Beckmann M. W., Brenner H., Muller H., Arndt V., Stegmaier C., Swerdlow A., Ashworth A., Orr N., Jones M., Figueroa J., Lissowska J., Brinton L., Goldberg M. S., Labreche F., Dumont M., Winqvist R., Pylkas K., Jukkola-Vuorinen A., Grip M., Brauch H., Hamann U., Bruning T., Network G., Radice P., Peterlongo P., Manoukian S., Bonanni B., Devilee P., Tollenaar R. A., Seynaeve C., van Asperen C. J., Jakubowska A., Lubinski J., Jaworska K., Durda K., Mannermaa A., Kataja V., Kosma V. M., Hartikainen J. M., Bogdanova N. V., Antonenkova N. N., Dork T., Kristensen V. N., Anton-Culver H., Slager S., Toland A. E., Edge S., Fostira F., Kang D., Yoo K. Y., Noh D. Y., Matsuo K., Ito H., Iwata H., Sueta A., Wu A. H., Tseng C. C., Van Den Berg D., Stram D. O., Shu X. O., Lu W., Gao Y. T., Cai H., Teo S. H., Yip C. H., Phuah S. Y., Cornes B. K., Hartman M., Miao H., Lim W. Y., Sng J. H., Muir K., Lophatananon A., Stewart-Brown S., Siriwanarangsan P., Shen C. Y., Hsiung C. N., Wu P. E., Ding S. L., Sangrajrang S., Gaborieau V., Brennan P., McKay J., Blot W. J., Signorello L. B., Cai Q., Zheng W., Deming-Halverson S., Shrubsole M., Long J., Simard J., Garcia-Closas M., Pharoah P. D., Chenevix-Trench G., Dunning A. M., Benitez J., Easton D. F., Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361, 361e1-2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michailidou K., Lindstrom S., Dennis J., Beesley J., Hui S., Kar S., Lemacon A., Soucy P., Glubb D., Rostamianfar A., Bolla M. K., Wang Q., Tyrer J., Dicks E., Lee A., Wang Z., Allen J., Keeman R., Eilber U., French J. D., Chen X. Q., Fachal L., McCue K., McCart Reed A. E., Ghoussaini M., Carroll J. S., Jiang X., Finucane H., Adams M., Adank M. A., Ahsan H., Aittomaki K., Anton-Culver H., Antonenkova N. N., Arndt V., Aronson K. J., Arun B., Auer P. L., Bacot F., Barrdahl M., Baynes C., Beckmann M. W., Behrens S., Benitez J., Bermisheva M., Bernstein L., Blomqvist C., Bogdanova N. V., Bojesen S. E., Bonanni B., Borresen-Dale A. L., Brand J. S., Brauch H., Brennan P., Brenner H., Brinton L., Broberg P., Brock I. W., Broeks A., Brooks-Wilson A., Brucker S. Y., Bruning T., Burwinkel B., Butterbach K., Cai Q., Cai H., Caldes T., Canzian F., Carracedo A., Carter B. D., Castelao J. E., Chan T. L., Cheng T. Y. D., Chia K. S., Choi J. Y., Christiansen H., Clarke C. L.; NBCS Collaborators, Collee M., Conroy D. M., Cordina-Duverger E., Cornelissen S., Cox D. G., Cox A., Cross S. S., Cunningham J. M., Czene K., Daly M. B., Devilee P., Doheny K. F., Dork T., Dos-Santos-Silva I., Dumont M., Durcan L., Dwek M., Eccles D. M., Ekici A. B., Eliassen A. H., Ellberg C., Elvira M., Engel C., Eriksson M., Fasching P. A., Figueroa J., Flesch-Janys D., Fletcher O., Flyger H., Fritschi L., Gaborieau V., Gabrielson M., Gago-Dominguez M., Gao Y. T., Gapstur S. M., Garcia-Saenz J. A., Gaudet M. M., Georgoulias V., Giles G. G., Glendon G., Goldberg M. S., Goldgar D. E., Gonzalez-Neira A., Alnaes G. I. G., Grip M., Gronwald J., Grundy A., Guenel P., Haeberle L., Hahnen E., Haiman C. A., Hakansson N., Hamann U., Hamel N., Hankinson S., Harrington P., Hart S. N., Hartikainen J. M., Hartman M., Hein A., Heyworth J., Hicks B., Hillemanns P., Ho D. N., Hollestelle A., Hooning M. J., Hoover R. N., Hopper J. L., Hou M. F., Hsiung C. N., Huang G., Humphreys K., Ishiguro J., Ito H., Iwasaki M., Iwata H., Jakubowska A., Janni W., John E. M., Johnson N., Jones K., Jones M., Jukkola-Vuorinen A., Kaaks R., Kabisch M., Kaczmarek K., Kang D., Kasuga Y., Kerin M. J., Khan S., Khusnutdinova E., Kiiski J. I., Kim S. W., Knight J. A., Kosma V. M., Kristensen V. N., Kruger U., Kwong A., Lambrechts D., Le Marchand L., Lee E., Lee M. H., Lee J. W., Lee C. N., Lejbkowicz F., Li J., Lilyquist J., Lindblom A., Lissowska J., Lo W. Y., Loibl S., Long J., Lophatananon A., Lubinski J., Luccarini C., Lux M. P., Ma E. S. K., MacInnis R. J., Maishman T., Makalic E., Malone K. E., Kostovska I. M., Mannermaa A., Manoukian S., Manson J. E., Margolin S., Mariapun S., Martinez M. E., Matsuo K., Mavroudis D., McKay J., McLean C., Meijers-Heijboer H., Meindl A., Menendez P., Menon U., Meyer J., Miao H., Miller N., Taib N. A. M., Muir K., Mulligan A. M., Mulot C., Neuhausen S. L., Nevanlinna H., Neven P., Nielsen S. F., Noh D. Y., Nordestgaard B. G., Norman A., Olopade O. I., Olson J. E., Olsson H., Olswold C., Orr N., Pankratz V. S., Park S. K., Park-Simon T. W., Lloyd R., Perez J. I. A., Peterlongo P., Peto J., Phillips K. A., Pinchev M., Plaseska-Karanfilska D., Prentice R., Presneau N., Prokofyeva D., Pugh E., Pylkas K., Rack B., Radice P., Rahman N., Rennert G., Rennert H. S., Rhenius V., Romero A., Romm J., Ruddy K. J., Rudiger T., Rudolph A., Ruebner M., Rutgers E. J. T., Saloustros E., Sandler D. P., Sangrajrang S., Sawyer E. J., Schmidt D. F., Schmutzler R. K., Schneeweiss A., Schoemaker M. J., Schumacher F., Schurmann P., Scott R. J., Scott C., Seal S., Seynaeve C., Shah M., Sharma P., Shen C. Y., Sheng G., Sherman M. E., Shrubsole M. J., Shu X. O., Smeets A., Sohn C., Southey M. C., Spinelli J. J., Stegmaier C., Stewart-Brown S., Stone J., Stram D. O., Surowy H., Swerdlow A., Tamimi R., Taylor J. A., Tengstrom M., Teo S. H., Terry M. B., Tessier D. C., Thanasitthichai S., Thone K., Tollenaar R., Tomlinson I., Tong L., Torres D., Truong T., Tseng C. C., Tsugane S., Ulmer H. U., Ursin G., Untch M., Vachon C., van Asperen C. J., Van Den Berg D., van den Ouweland A. M. W., van der Kolk L., van der Luijt R. B., Vincent D., Vollenweider J., Waisfisz Q., Wang-Gohrke S., Weinberg C. R., Wendt C., Whittemore A. S., Wildiers H., Willett W., Winqvist R., Wolk A., Wu A. H., Xia L., Yamaji T., Yang X. R., Yip C. H., Yoo K. Y., Yu J. C., Zheng W., Zheng Y., Zhu B., Ziogas A., Ziv E., Investigators A., ConFab A. I., Lakhani S. R., Antoniou A. C., Droit A., Andrulis I. L., Amos C. I., Couch F. J., Pharoah P. D. P., Chang-Claude J., Hall P., Hunter D. J., Milne R. L., Garcia-Closas M., Schmidt M. K., Chanock S. J., Dunning A. M., Edwards S. L., Bader G. D., Chenevix-Trench G., Simard J., Kraft P., Easton D. F., Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne R. L., Kuchenbaecker K. B., Michailidou K., Beesley J., Kar S., Lindstrom S., Hui S., Lemacon A., Soucy P., Dennis J., Jiang X., Rostamianfar A., Finucane H., Bolla M. K., McGuffog L., Wang Q., Aalfs C. M.; ABCTB Investigators, Adams M., Adlard J., Agata S., Ahmed S., Ahsan H., Aittomaki K., Al-Ejeh F., Allen J., Ambrosone C. B., Amos C. I., Andrulis I. L., Anton-Culver H., Antonenkova N. N., Arndt V., Arnold N., Aronson K. J., Auber B., Auer P. L., Ausems M., Azzollini J., Bacot F., Balmana J., Barile M., Barjhoux L., Barkardottir R. B., Barrdahl M., Barnes D., Barrowdale D., Baynes C., Beckmann M. W., Benitez J., Bermisheva M., Bernstein L., Bignon Y. J., Blazer K. R., Blok M. J., Blomqvist C., Blot W., Bobolis K., Boeckx B., Bogdanova N. V., Bojesen A., Bojesen S. E., Bonanni B., Borresen-Dale A. L., Bozsik A., Bradbury A. R., Brand J. S., Brauch H., Brenner H., Bressac-de Paillerets B., Brewer C., Brinton L., Broberg P., Brooks-Wilson A., Brunet J., Bruning T., Burwinkel B., Buys S. S., Byun J., Cai Q., Caldes T., Caligo M. A., Campbell I., Canzian F., Caron O., Carracedo A., Carter B. D., Castelao J. E., Castera L., Caux-Moncoutier V., Chan S. B., Chang-Claude J., Chanock S. J., Chen X., Cheng T. D., Chiquette J., Christiansen H., Claes K. B. M., Clarke C. L., Conner T., Conroy D. M., Cook J., Cordina-Duverger E., Cornelissen S., Coupier I., Cox A., Cox D. G., Cross S. S., Cuk K., Cunningham J. M., Czene K., Daly M. B., Damiola F., Darabi H., Davidson R., De Leeneer K., Devilee P., Dicks E., Diez O., Ding Y. C., Ditsch N., Doheny K. F., Domchek S. M., Dorfling C. M., Dork T., Dos-Santos-Silva I., Dubois S., Dugue P. A., Dumont M., Dunning A. M., Durcan L., Dwek M., Dworniczak B., Eccles D., Eeles R., Ehrencrona H., Eilber U., Ejlertsen B., Ekici A. B., Eliassen A. H.; EMBRACE, Engel C., Eriksson M., Fachal L., Faivre L., Fasching P. A., Faust U., Figueroa J., Flesch-Janys D., Fletcher O., Flyger H., Foulkes W. D., Friedman E., Fritschi L., Frost D., Gabrielson M., Gaddam P., Gammon M. D., Ganz P. A., Gapstur S. M., Garber J., Garcia-Barberan V., Garcia-Saenz J. A., Gaudet M. M., Gauthier-Villars M., Gehrig A.; GEMO Study Collaborators, Georgoulias V., Gerdes A. M., Giles G. G., Glendon G., Godwin A. K., Goldberg M. S., Goldgar D. E., Gonzalez-Neira A., Goodfellow P., Greene M. H., Alnaes G. I. G., Grip M., Gronwald J., Grundy A., Gschwantler-Kaulich D., Guenel P., Guo Q., Haeberle L., Hahnen E., Haiman C. A., Hakansson N., Hallberg E., Hamann U., Hamel N., Hankinson S., Hansen T. V. O., Harrington P., Hart S. N., Hartikainen J. M., Healey C. S.; HEBON, Hein A., Helbig S., Henderson A., Heyworth J., Hicks B., Hillemanns P., Hodgson S., Hogervorst F. B., Hollestelle A., Hooning M. J., Hoover B., Hopper J. L., Hu C., Huang G., Hulick P. J., Humphreys K., Hunter D. J., Imyanitov E. N., Isaacs C., Iwasaki M., Izatt L., Jakubowska A., James P., Janavicius R., Janni W., Jensen U. B., John E. M., Johnson N., Jones K., Jones M., Jukkola-Vuorinen A., Kaaks R., Kabisch M., Kaczmarek K., Kang D., Kast K.; kConFab/AOCS Investigators, Keeman R., Kerin M. J., Kets C. M., Keupers M., Khan S., Khusnutdinova E., Kiiski J. I., Kim S. W., Knight J. A., Konstantopoulou I., Kosma V. M., Kristensen V. N., Kruse T. A., Kwong A., Laenkholm A. V., Laitman Y., Lalloo F., Lambrechts D., Landsman K., Lasset C., Lazaro C., Le Marchand L., Lecarpentier J., Lee A., Lee E., Lee J. W., Lee M. H., Lejbkowicz F., Lesueur F., Li J., Lilyquist J., Lincoln A., Lindblom A., Lissowska J., Lo W. Y., Loibl S., Long J., Loud J. T., Lubinski J., Luccarini C., Lush M., MacInnis R. J., Maishman T., Makalic E., Kostovska I. M., Malone K. E., Manoukian S., Manson J. E., Margolin S., Martens J. W. M., Martinez M. E., Matsuo K., Mavroudis D., Mazoyer S., McLean C., Meijers-Heijboer H., Menendez P., Meyer J., Miao H., Miller A., Miller N., Mitchell G., Montagna M., Muir K., Mulligan A. M., Mulot C., Nadesan S., Nathanson K. L., Collaborators N., Neuhausen S. L., Nevanlinna H., Nevelsteen I., Niederacher D., Nielsen S. F., Nordestgaard B. G., Norman A., Nussbaum R. L., Olah E., Olopade O. I., Olson J. E., Olswold C., Ong K. R., Oosterwijk J. C., Orr N., Osorio A., Pankratz V. S., Papi L., Park-Simon T. W., Paulsson-Karlsson Y., Lloyd R., Pedersen I. S., Peissel B., Peixoto A., Perez J. I. A., Peterlongo P., Peto J., Pfeiler G., Phelan C. M., Pinchev M., Plaseska-Karanfilska D., Poppe B., Porteous M. E., Prentice R., Presneau N., Prokofieva D., Pugh E., Pujana M. A., Pylkas K., Rack B., Radice P., Rahman N., Rantala J., Rappaport-Fuerhauser C., Rennert G., Rennert H. S., Rhenius V., Rhiem K., Richardson A., Rodriguez G. C., Romero A., Romm J., Rookus M. A., Rudolph A., Ruediger T., Saloustros E., Sanders J., Sandler D. P., Sangrajrang S., Sawyer E. J., Schmidt D. F., Schoemaker M. J., Schumacher F., Schurmann P., Schwentner L., Scott C., Scott R. J., Seal S., Senter L., Seynaeve C., Shah M., Sharma P., Shen C. Y., Sheng X., Shimelis H., Shrubsole M. J., Shu X. O., Side L. E., Singer C. F., Sohn C., Southey M. C., Spinelli J. J., Spurdle A. B., Stegmaier C., Stoppa-Lyonnet D., Sukiennicki G., Surowy H., Sutter C., Swerdlow A., Szabo C. I., Tamimi R. M., Tan Y. Y., Taylor J. A., Tejada M. I., Tengstrom M., Teo S. H., Terry M. B., Tessier D. C., Teule A., Thone K., Thull D. L., Tibiletti M. G., Tihomirova L., Tischkowitz M., Toland A. E., Tollenaar R., Tomlinson I., Tong L., Torres D., Tranchant M., Truong T., Tucker K., Tung N., Tyrer J., Ulmer H. U., Vachon C., van Asperen C. J., Van Den Berg D., van den Ouweland A. M. W., van Rensburg E. J., Varesco L., Varon-Mateeva R., Vega A., Viel A., Vijai J., Vincent D., Vollenweider J., Walker L., Wang Z., Wang-Gohrke S., Wappenschmidt B., Weinberg C. R., Weitzel J. N., Wendt C., Wesseling J., Whittemore A. S., Wijnen J. T., Willett W., Winqvist R., Wolk A., Wu A. H., Xia L., Yang X. R., Yannoukakos D., Zaffaroni D., Zheng W., Zhu B., Ziogas A., Ziv E., Zorn K. K., Gago-Dominguez M., Mannermaa A., Olsson H., Teixeira M. R., Stone J., Offit K., Ottini L., Park S. K., Thomassen M., Hall P., Meindl A., Schmutzler R. K., Droit A., Bader G. D., Pharoah P. D. P., Couch F. J., Easton D. F., Kraft P., Chenevix-Trench G., Garcia-Closas M., Schmidt M. K., Antoniou A. C., Simard J., Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 49, 1767–1778 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M. J., Maranian M. J., Bolla M. K., Wang Q., Shah M., Perkins B. J., Czene K., Eriksson M., Darabi H., Brand J. S., Bojesen S. E., Nordestgaard B. G., Flyger H., Nielsen S. F., Rahman N., Turnbull C.; BOCS, Fletcher O., Peto J., Gibson L., dos-Santos-Silva I., Chang-Claude J., Flesch-Janys D., Rudolph A., Eilber U., Behrens S., Nevanlinna H., Muranen T. A., Aittomaki K., Blomqvist C., Khan S., Aaltonen K., Ahsan H., Kibriya M. G., Whittemore A. S., John E. M., Malone K. E., Gammon M. D., Santella R. M., Ursin G., Makalic E., Schmidt D. F., Casey G., Hunter D. J., Gapstur S. M., Gaudet M. M., Diver W. R., Haiman C. A., Schumacher F., Henderson B. E., Le Marchand L., Berg C. D., Chanock S. J., Figueroa J., Hoover R. N., Lambrechts D., Neven P., Wildiers H., van Limbergen E., Schmidt M. K., Broeks A., Verhoef S., Cornelissen S., Couch F. J., Olson J. E., Hallberg E., Vachon C., Waisfisz Q., Meijers-Heijboer H., Adank M. A., van der Luijt R. B., Li J., Liu J., Humphreys K., Kang D., Choi J. Y., Park S. K., Yoo K. Y., Matsuo K., Ito H., Iwata H., Tajima K., Guenel P., Truong T., Mulot C., Sanchez M., Burwinkel B., Marme F., Surowy H., Sohn C., Wu A. H., Tseng C. C., Van Den Berg D., Stram D. O., Gonzalez-Neira A., Benitez J., Zamora M. P., Perez J. I., Shu X. O., Lu W., Gao Y. T., Cai H., Cox A., Cross S. S., Reed M. W., Andrulis I. L., Knight J. A., Glendon G., Mulligan A. M., Sawyer E. J., Tomlinson I., Kerin M. J., Miller N.; kConFab Investigators; AOCS Group, Lindblom A., Margolin S., Teo S. H., Yip C. H., Taib N. A., Tan G. H., Hooning M. J., Hollestelle A., Martens J. W., Collee J. M., Blot W., Signorello L. B., Cai Q., Hopper J. L., Southey M. C., Tsimiklis H., Apicella C., Shen C. Y., Hsiung C. N., Wu P. E., Hou M. F., Kristensen V. N., Nord S., Alnaes G. I.; NBCS, Giles G. G., Milne R. L., McLean C., Canzian F., Trichopoulos D., Peeters P., Lund E., Sund M., Khaw K. T., Gunter M. J., Palli D., Mortensen L. M., Dossus L., Huerta J. M., Meindl A., Schmutzler R. K., Sutter C., Yang R., Muir K., Lophatananon A., Stewart-Brown S., Siriwanarangsan P., Hartman M., Miao H., Chia K. S., Chan C. W., Fasching P. A., Hein A., Beckmann M. W., Haeberle L., Brenner H., Dieffenbach A. K., Arndt V., Stegmaier C., Ashworth A., Orr N., Schoemaker M. J., Swerdlow A. J., Brinton L., Garcia-Closas M., Zheng W., Halverson S. L., Shrubsole M., Long J., Goldberg M. S., Labreche F., Dumont M., Winqvist R., Pylkas K., Jukkola-Vuorinen A., Grip M., Brauch H., Hamann U., Bruning T., Network G., Radice P., Peterlongo P., Manoukian S., Bernard L., Bogdanova N. V., Dork T., Mannermaa A., Kataja V., Kosma V. M., Hartikainen J. M., Devilee P., Tollenaar R. A., Seynaeve C., Van Asperen C. J., Jakubowska A., Lubinski J., Jaworska K., Huzarski T., Sangrajrang S., Gaborieau V., Brennan P., McKay J., Slager S., Toland A. E., Ambrosone C. B., Yannoukakos D., Kabisch M., Torres D., Neuhausen S. L., Anton-Culver H., Luccarini C., Baynes C., Ahmed S., Healey C. S., Tessier D. C., Vincent D., Bacot F., Pita G., Alonso M. R., Alvarez N., Herrero D., Simard J., Pharoah P. P., Kraft P., Dunning A. M., Chenevix-Trench G., Hall P., Easton D. F., Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 47, 373–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Ahearn T. U., Lecarpentier J., Barnes D., Beesley J., Qi G., Jiang X., O’Mara T. A., Zhao N., Bolla M. K., Dunning A. M., Dennis J., Wang Q., Ful Z. A., Aittomaki K., Andrulis I. L., Anton-Culver H., Arndt V., Aronson K. J., Arun B. K., Auer P. L., Azzollini J., Barrowdale D., Becher H., Beckmann M. W., Behrens S., Benitez J., Bermisheva M., Bialkowska K., Blanco A., Blomqvist C., Bogdanova N. V., Bojesen S. E., Bonanni B., Bondavalli D., Borg A., Brauch H., Brenner H., Briceno I., Broeks A., Brucker S. Y., Bruning T., Burwinkel B., Buys S. S., Byers H., Caldes T., Caligo M. A., Calvello M., Campa D., Castelao J. E., Chang-Claude J., Chanock S. J., Christiaens M., Christiansen H., Chung W. K., Claes K. B. M., Clarke C. L., Cornelissen S., Couch F. J., Cox A., Cross S. S., Czene K., Daly M. B., Devilee P., Diez O., Domchek S. M., Dork T., Dwek M., Eccles D. M., Ekici A. B., Evans D. G., Fasching P. A., Figueroa J., Foretova L., Fostira F., Friedman E., Frost D., Gago-Dominguez M., Gapstur S. M., Garber J., Garcia-Saenz J. A., Gaudet M. M., Gayther S. A., Giles G. G., Godwin A. K., Goldberg M. S., Goldgar D. E., Gonzalez-Neira A., Greene M. H., Gronwald J., Guenel P., Haberle L., Hahnen E., Haiman C. A., Hake C. R., Hall P., Hamann U., Harkness E. F., Heemskerk-Gerritsen B. A. M., Hillemanns P., Hogervorst F. B. L., Holleczek B., Hollestelle A., Hooning M. J., Hoover R. N., Hopper J. L., Howell A., Huebner H., Hulick P. J., Imyanitov E. N.; kConFab Investigators; ABCTB Investigators, Isaacs C., Izatt L., Jager A., Jakimovska M., Jakubowska A., James P., Janavicius R., Janni W., John E. M., Jones M. E., Jung A., Kaaks R., Kapoor P. M., Karlan B. Y., Keeman R., Khan S., Khusnutdinova E., Kitahara C. M., Ko Y. D., Konstantopoulou I., Koppert L. B., Koutros S., Kristensen V. N., Laenkholm A. V., Lambrechts D., Larsson S. C., Laurent-Puig P., Lazaro C., Lazarova E., Lejbkowicz F., Leslie G., Lesueur F., Lindblom A., Lissowska J., Lo W. Y., Loud J. T., Lubinski J., Lukomska A., MacInnis R. J., Mannermaa A., Manoochehri M., Manoukian S., Margolin S., Martinez M. E., Matricardi L., McGuffog L., McLean C., Mebirouk N., Meindl A., Menon U., Miller A., Mingazheva E., Montagna M., Mulligan A. M., Mulot C., Muranen T. A., Nathanson K. L., Neuhausen S. L., Nevanlinna H., Neven P., Newman W. G., Nielsen F. C., Nikitina-Zake L., Nodora J., Offit K., Olah E., Olopade O. I., Olsson H., Orr N., Papi L., Papp J., Park-Simon T. W., Parsons M. T., Peissel B., Peixoto A., Peshkin B., Peterlongo P., Peto J., Phillips K. A., Piedmonte M., Plaseska-Karanfilska D., Prajzendanc K., Prentice R., Prokofyeva D., Rack B., Radice P., Ramus S. J., Rantala J., Rashid M. U., Rennert G., Rennert H. S., Risch H. A., Romero A., Rookus M. A., Rubner M., Rudiger T., Saloustros E., Sampson S., Sandler D. P., Sawyer E. J., Scheuner M. T., Schmutzler R. K., Schneeweiss A., Schoemaker M. J., Schottker B., Schurmann P., Senter L., Sharma P., Sherman M. E., Shu X. O., Singer C. F., Smichkoska S., Soucy P., Southey M. C., Spinelli J. J., Stone J., Stoppa-Lyonnet D.; EMBRACE Study; GEMO Study Collaborators, Swerdlow A. J., Szabo C. I., Tamimi R. M., Tapper W. J., Taylor J. A., Teixeira M. R., Terry M., Thomassen M., Thull D. L., Tischkowitz M., Toland A. E., Tollenaar R., Tomlinson I., Torres D., Troester M. A., Truong T., Tung N., Untch M., Vachon C. M., van den Ouweland A. M. W., van der Kolk L. E., van Veen E. M., vanRensburg E. J., Vega A., Wappenschmidt B., Weinberg C. R., Weitzel J. N., Wildiers H., Winqvist R., Wolk A., Yang X. R., Yannoukakos D., Zheng W., Zorn K. K., Milne R. L., Kraft P., Simard J., Pharoah P. D. P., Michailidou K., Antoniou A. C., Schmidt M. K., Chenevix-Trench G., Easton D. F., Chatterjee N., Garcia-Closas M., Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat. Genet. 52, 572–581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., Wang H., Reynolds A. P., Sandstrom R., Qu H., Brody J., Shafer A., Neri F., Lee K., Kutyavin T., Stehling-Sun S., Johnson A. K., Canfield T. K., Giste E., Diegel M., Bates D., Hansen R. S., Neph S., Sabo P. J., Heimfeld S., Raubitschek A., Ziegler S., Cotsapas C., Sotoodehnia N., Glass I., Sunyaev S. R., Kaul R., Stamatoyannopoulos J. A., Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson R., Sandelin A., Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Guo X., Lin W., Bao J., Cai Q., Pan X., Bai M., Yuan Y., Shi J., Sun Y., Han M. R., Wang J., Liu Q., Wen W., Li B., Long J., Chen J., Zheng W., A comprehensive cis-eQTL analysis revealed target genes in breast cancer susceptibility loci identified in genome-wide association studies. Am. J. Hum. Genet. 102, 890–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L., Shi W., Long J., Guo X., Michailidou K., Beesley J., Bolla M. K., Shu X. O., Lu Y., Cai Q., Al-Ejeh F., Rozali E., Wang Q., Dennis J., Li B., Zeng C., Feng H., Gusev A., Barfield R. T., Andrulis I. L., Anton-Culver H., Arndt V., Aronson K. J., Auer P. L., Barrdahl M., Baynes C., Beckmann M. W., Benitez J., Bermisheva M., Blomqvist C., Bogdanova N. V., Bojesen S. E., Brauch H., Brenner H., Brinton L., Broberg P., Brucker S. Y., Burwinkel B., Caldes T., Canzian F., Carter B. D., Castelao J. E., Chang-Claude J., Chen X., Cheng T. D., Christiansen H., Clarke C. L.; NBCS Collaborators, Collee M., Cornelissen S., Couch F. J., Cox D., Cox A., Cross S. S., Cunningham J. M., Czene K., Daly M. B., Devilee P., Doheny K. F., Dork T., Dos-Santos-Silva I., Dumont M., Dwek M., Eccles D. M., Eilber U., Eliassen A. H., Engel C., Eriksson M., Fachal L., Fasching P. A., Figueroa J., Flesch-Janys D., Fletcher O., Flyger H., Fritschi L., Gabrielson M., Gago-Dominguez M., Gapstur S. M., Garcia-Closas M., Gaudet M. M., Ghoussaini M., Giles G. G., Goldberg M. S., Goldgar D. E., Gonzalez-Neira A., Guenel P., Hahnen E., Haiman C. A., Hakansson N., Hall P., Hallberg E., Hamann U., Harrington P., Hein A., Hicks B., Hillemanns P., Hollestelle A., Hoover R. N., Hopper J. L., Huang G., Humphreys K., Hunter D. J., Jakubowska A., Janni W., John E. M., Johnson N., Jones K., Jones M. E., Jung A., Kaaks R., Kerin M. J., Khusnutdinova E., Kosma V. M., Kristensen V. N., Lambrechts D., Le Marchand L., Li J., Lindstrom S., Lissowska J., Lo W. Y., Loibl S., Lubinski J., Luccarini C., Lux M. P., MacInnis R. J., Maishman T., Kostovska I. M., Mannermaa A., Manson J. E., Margolin S., Mavroudis D., Meijers-Heijboer H., Meindl A., Menon U., Meyer J., Mulligan A. M., Neuhausen S. L., Nevanlinna H., Neven P., Nielsen S. F., Nordestgaard B. G., Olopade O. I., Olson J. E., Olsson H., Peterlongo P., Peto J., Plaseska-Karanfilska D., Prentice R., Presneau N., Pylkas K., Rack B., Radice P., Rahman N., Rennert G., Rennert H. S., Rhenius V., Romero A., Romm J., Rudolph A., Saloustros E., Sandler D. P., Sawyer E. J., Schmidt M. K., Schmutzler R. K., Schneeweiss A., Scott R. J., Scott C. G., Seal S., Shah M., Shrubsole M. J., Smeets A., Southey M. C., Spinelli J. J., Stone J., Surowy H., Swerdlow A. J., Tamimi R. M., Tapper W., Taylor J. A., Terry M. B., Tessier D. C., Thomas A., Thone K., Tollenaar R., Torres D., Truong T., Untch M., Vachon C., Van Den Berg D., Vincent D., Waisfisz Q., Weinberg C. R., Wendt C., Whittemore A. S., Wildiers H., Willett W. C., Winqvist R., Wolk A., Xia L., Yang X. R., Ziogas A., Ziv E.; kConFab/AOCS Investigators, Dunning A. M., Pharoah P. D. P., Simard J., Milne R. L., Edwards S. L., Kraft P., Easton D. F., Chenevix-Trench G., Zheng W., A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat. Genet. 50, 968–978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fachal L., Aschard H., Beesley J., Barnes D. R., Allen J., Kar S., Pooley K. A., Dennis J., Michailidou K., Turman C., Soucy P., Lemacon A., Lush M., Tyrer J. P., Ghoussaini M., Marjaneh M. M., Jiang X., Agata S., Aittomaki K., Alonso M. R., Andrulis I. L., Anton-Culver H., Antonenkova N. N., Arason A., Arndt V., Aronson K. J., Arun B. K., Auber B., Auer P. L., Azzollini J., Balmana J., Barkardottir R. B., Barrowdale D., Beeghly-Fadiel A., Benitez J., Bermisheva M., Bialkowska K., Blanco A. M., Blomqvist C., Blot W., Bogdanova N. V., Bojesen S. E., Bolla M. K., Bonanni B., Borg A., Bosse K., Brauch H., Brenner H., Briceno I., Brock I. W., Brooks-Wilson A., Bruning T., Burwinkel B., Buys S. S., Cai Q., Caldes T., Caligo M. A., Camp N. J., Campbell I., Canzian F., Carroll J. S., Carter B. D., Castelao J. E., Chiquette J., Christiansen H., Chung W. K., Claes K. B. M., Clarke C. L.; GEMO Study Collaborators; EMBRACE Collaborators, Collee J. M., Cornelissen S., Couch F. J., Cox A., Cross S. S., Cybulski C., Czene K., Daly M. B., de la Hoya M., Devilee P., Diez O., Ding Y. C., Dite G. S., Domchek S. M., Dork T., Dos-Santos-Silva I., Droit A., Dubois S., Dumont M., Duran M., Durcan L., Dwek M., Eccles D. M., Engel C., Eriksson M., Evans D. G., Fasching P. A., Fletcher O., Floris G., Flyger H., Foretova L., Foulkes W. D., Friedman E., Fritschi L., Frost D., Gabrielson M., Gago-Dominguez M., Gambino G., Ganz P. A., Gapstur S. M., Garber J., Garcia-Saenz J. A., Gaudet M. M., Georgoulias V., Giles G. G., Glendon G., Godwin A. K., Goldberg M. S., Goldgar D. E., Gonzalez-Neira A., Tibiletti M. G., Greene M. H., Grip M., Gronwald J., Grundy A., Guenel P., Hahnen E., Haiman C. A., Hakansson N., Hall P., Hamann U., Harrington P. A., Hartikainen J. M., Hartman M., He W., Healey C. S., Heemskerk-Gerritsen B. A. M., Heyworth J., Hillemanns P., Hogervorst F. B. L., Hollestelle A., Hooning M. J., Hopper J. L., Howell A., Huang G., Hulick P. J., Imyanitov E. N.; KConFab Investigators; HEBON Investigators; ABCTB Investigators, Isaacs C., Iwasaki M., Jager A., Jakimovska M., Jakubowska A., James P. A., Janavicius R., Jankowitz R. C., John E. M., Johnson N., Jones M. E., Jukkola-Vuorinen A., Jung A., Kaaks R., Kang D., Kapoor P. M., Karlan B. Y., Keeman R., Kerin M. J., Khusnutdinova E., Kiiski J. I., Kirk J., Kitahara C. M., Ko Y. D., Konstantopoulou I., Kosma V. M., Koutros S., Kubelka-Sabit K., Kwong A., Kyriacou K., Laitman Y., Lambrechts D., Lee E., Leslie G., Lester J., Lesueur F., Lindblom A., Lo W. Y., Long J., Lophatananon A., Loud J. T., Lubinski J., MacInnis R. J., Maishman T., Makalic E., Mannermaa A., Manoochehri M., Manoukian S., Margolin S., Martinez M. E., Matsuo K., Maurer T., Mavroudis D., Mayes R., McGuffog L., McLean C., Mebirouk N., Meindl A., Miller A., Miller N., Montagna M., Moreno F., Muir K., Mulligan A. M., Munoz-Garzon V. M., Muranen T. A., Narod S. A., Nassir R., Nathanson K. L., Neuhausen S. L., Nevanlinna H., Neven P., Nielsen F. C., Nikitina-Zake L., Norman A., Offit K., Olah E., Olopade O. I., Olsson H., Orr N., Osorio A., Pankratz V. S., Papp J., Park S. K., Park-Simon T. W., Parsons M. T., Paul J., Pedersen I. S., Peissel B., Peshkin B., Peterlongo P., Peto J., Plaseska-Karanfilska D., Prajzendanc K., Prentice R., Presneau N., Prokofyeva D., Pujana M. A., Pylkas K., Radice P., Ramus S. J., Rantala J., Rau-Murthy R., Rennert G., Risch H. A., Robson M., Romero A., Rossing M., Saloustros E., Sanchez-Herrero E., Sandler D. P., Santamarina M., Saunders C., Sawyer E. J., Scheuner M. T., Schmidt D. F., Schmutzler R. K., Schneeweiss A., Schoemaker M. J., Schottker B., Schurmann P., Scott C., Scott R. J., Senter L., Seynaeve C. M., Shah M., Sharma P., Shen C. Y., Shu X. O., Singer C. F., Slavin T. P., Smichkoska S., Southey M. C., Spinelli J. J., Spurdle A. B., Stone J., Stoppa-Lyonnet D., Sutter C., Swerdlow A. J., Tamimi R. M., Tan Y. Y., Tapper W. J., Taylor J. A., Teixeira M. R., Tengstrom M., Teo S. H., Terry M. B., Teule A., Thomassen M., Thull D. L., Tischkowitz M., Toland A. E., Tollenaar R., Tomlinson I., Torres D., Torres-Mejia G., Troester M. A., Truong T., Tung N., Tzardi M., Ulmer H. U., Vachon C. M., van Asperen C. J., van der Kolk L. E., van Rensburg E. J., Vega A., Viel A., Vijai J., Vogel M. J., Wang Q., Wappenschmidt B., Weinberg C. R., Weitzel J. N., Wendt C., Wildiers H., Winqvist R., Wolk A., Wu A. H., Yannoukakos D., Zhang Y., Zheng W., Hunter D., Pharoah P. D. P., Chang-Claude J., Garcia-Closas M., Schmidt M. K., Milne R. L., Kristensen V. N., French J. D., Edwards S. L., Antoniou A. C., Chenevix-Trench G., Simard J., Easton D. F., Kraft P., Dunning A. M., Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat. Genet. 52, 56–73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunning A. M., Michailidou K., Kuchenbaecker K. B., Thompson D., French J. D., Beesley J., Healey C. S., Kar S., Pooley K. A., Lopez-Knowles E., Dicks E., Barrowdale D., Sinnott-Armstrong N. A., Sallari R. C., Hillman K. M., Kaufmann S., Sivakumaran H., Marjaneh M. M., Lee J. S., Hills M., Jarosz M., Drury S., Canisius S., Bolla M. K., Dennis J., Wang Q., Hopper J. L., Southey M. C., Broeks A., Schmidt M. K., Lophatananon A., Muir K., Beckmann M. W., Fasching P. A., Dos-Santos-Silva I., Peto J., Sawyer E. J., Tomlinson I., Burwinkel B., Marme F., Guenel P., Truong T., Bojesen S. E., Flyger H., Gonzalez-Neira A., Perez J. I., Anton-Culver H., Eunjung L., Arndt V., Brenner H., Meindl A., Schmutzler R. K., Brauch H., Hamann U., Aittomaki K., Blomqvist C., Ito H., Matsuo K., Bogdanova N., Dork T., Lindblom A., Margolin S., Kosma V. M., Mannermaa A., Tseng C. C., Wu A. H., Lambrechts D., Wildiers H., Chang-Claude J., Rudolph A., Peterlongo P., Radice, Olson J. E., Giles G. G., Milne R. L., Haiman C. A., Henderson B. E., Goldberg M. S., Teo S. H., Yip C. H., Nord S., Borresen-Dale A. L., Kristensen V., Long J., Zheng W., Pylkas K., Winqvist R., Andrulis I. L., Knight J. A., Devilee P., Seynaeve C., Figueroa J., Sherman M. E., Czene K., Darabi H., Hollestelle A., van den Ouweland A. M., Humphreys K., Gao Y. T., Shu X. O., Cox A., Cross S. S., Blot W., Cai Q., Ghoussaini M., Perkins B. J., Shah M., Choi J. Y., Kang D., Lee S. C., Hartman M., Kabisch M., Torres D., Jakubowska A., Lubinski J., Brennan P., Sangrajrang S., Ambrosone C. B., Toland A. E., Shen C. Y., Wu P. E., Orr N., Swerdlow A., McGuffog L., Healey S., Lee A., Kapuscinski M., John E. M., Terry M. B., Daly M. B., Goldgar D. E., Buys S. S., Janavicius R., Tihomirova L., Tung N., Dorfling C. M., van Rensburg E. J., Neuhausen S. L., Ejlertsen B., Hansen T. V., Osorio A., Benitez J., Rando R., Weitzel J. N., Bonanni B., Peissel B., Manoukian S., Papi L., Ottini L., Konstantopoulou I., Apostolou P., Garber J., Rashid M. U., Frost D.; EMBRACE, Izatt L., Ellis S., Godwin A. K., Arnold N., Niederacher D., Rhiem K., Bogdanova-Markov N., Sagne C., Stoppa-Lyonnet D., Damiola F., Collaborators G. S., Sinilnikova O. M., Mazoyer S., Isaacs C., Claes K. B., De Leeneer K., De la Hoya M., Caldes T., Nevanlinna H., Khan S., Mensenkamp A. R.; HEBON, Hooning M. J., Rookus M. A., Kwong A., Olah E., Diez O., Brunet J., Pujana M. A., Gronwald J., Huzarski T., Barkardottir R. B., Laframboise R., Soucy P., Montagna M., Agata S., Teixeira M. R.; kConFab Investigators, Park S. K., Lindor N., Couch F. J., Tischkowitz M., Foretova L., Vijai J., Offit K., Singer C. F., Rappaport C., Phelan C. M., Greene M. H., Mai P. L., Rennert G., Imyanitov E. N., Hulick P. J., Phillips K. A., Piedmonte M., Mulligan A. M., Glendon G., Bojesen A., Thomassen M., Caligo M. A., Yoon S. Y., Friedman E., Laitman Y., Borg A., von Wachenfeldt A., Ehrencrona H., Rantala J., Olopade O. I., Ganz P. A., Nussbaum R. L., Gayther S. A., Nathanson K. L., Domchek S. M., Arun B. K., Mitchell G., Karlan B. Y., Lester J., Maskarinec G., Woolcott C., Scott C., Stone J., Apicella C., Tamimi R., Luben R., Khaw K. T., Helland A., Haakensen V., Dowsett M., Pharoah P. D., Simard J., Hall P., Garcia-Closas M., Vachon C., Chenevix-Trench G., Antoniou A. C., Easton D. F., Edwards S. L., Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 48, 374–386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrenson K., Kar S., McCue K., Kuchenbaeker K., Michailidou K., Tyrer J., Beesley J., Ramus S. J., Li Q., Delgado M. K., Lee J. M., Aittomäki K., Andrulis I. L., Anton-Culver H., Arndt V., Arun B. K., Arver B., Bandera E. V., Barile M., Barkardottir R. B., Barrowdale D., Beckmann M. W., Benitez J., Berchuck A., Bisogna M., Bjorge L., Blomqvist C., Blot W., Bogdanova N., Bojesen A., Bojesen S. E., Bolla M. K., Bonanni B., Børresen-Dale A.-L., Brauch H., Brennan P., Brenner H., Bruinsma F., Brunet J., Buhari S. A., Burwinkel B., Butzow R., Buys S. S., Cai Q., Caldes T., Campbell I., Canniotto R., Chang-Claude J., Chiquette J., Choi J.-Y., Claes K. B. M., Collonge-Rame M.-A., Damette A., Barouk-Simonet E., Bonnet F., Bubien V., Sevenet N., Longy M., Berthet P., Vaur D., Castera L., Ferrer S. F., Bignon Y.-J., Uhrhammer N., Coron F., Faivre L., Baurand A., Jacquot C., Bertolone G., Lizard S., Leroux D., Dreyfus H., Rebischung C., Peysselon M., Peyrat J.-P., Fournier J., Révillion F., Adenis C., Vénat-Bouvet L., Léone M., Boutry-Kryza N., Calender A., Giraud S., Verny-Pierre C., Lasset C., Bonadona V., Barjhoux L., Sobol H., Bourdon V., Noguchi T., Remenieras A., Coupier I., Pujol P., Sokolowska J., Bronner M., Delnatte C., Bézieau S., Mari V., Gauthier-Villars M., Buecher B., Rouleau E., Golmard L., Moncoutier V., Belotti M., de Pauw A., Elan C., Fourme E., Birot A.-M., Saule C., Laurent M., Houdayer C., Lesueur F., Mebirouk N., Coulet F., Colas C., Soubrier F., Warcoin M., Prieur F., Lebrun M., Kientz C., Muller D., Fricker J.-P., Toulas C., Guimbaud R., Gladieff L., Feillel V., Mortemousque I., Bressac-de-Paillerets B., Caron O., Guillaud-Bataille M., Cook L. S., Cox A., Cramer D. W., Cross S. S., Cybulski C., Czene K., Daly M. B., Damiola F., Dansonka-Mieszkowska A., Darabi H., Dennis J., Devilee P., Diez O., Doherty J. A., Domchek S. M., Dorfling C. M., Dörk T., Dumont M., Ehrencrona H., Ejlertsen B., Ellis S., Gregory H., Miedzybrodzka Z., Morrison P. J., Donaldson A., Rogers M. T., Kennedy M. J., Porteous M. E., Brady A., Barwell J., Foo C., Lalloo F., Side L. E., Eason J., Henderson A., Walker L., Cook J., Snape K., Murray A., McCann E., Engel C., Lee E., Evans D. G., Fasching P. A., Feliubadalo L., Figueroa J., Flesch-Janys D., Fletcher O., Flyger H., Foretova L., Fostira F., Foulkes W. D., Fridley B. L., Friedman E., Frost D., Gambino G., Ganz P. A., Garber J., García-Closas M., Gentry-Maharaj A., Ghoussaini M., Giles G. G., Glasspool R., Godwin A. K., Goldberg M. S., Goldgar D. E., González-Neira A., Goode E. L., Goodman M. T., Greene M. H., Gronwald J., Guénel P., Haiman C. A., Hall P., Hallberg E., Hamann U., Hansen T. V. O., Harrington P. A., Hartman M., Hassan N., Healey S., Rookus M. A., van Leeuwen F. E., van der Kolk L. E., Schmidt M. K., Russell N. S., de Lange J. L., Wijnands R., Collée J. M., Hooning M. J., Seynaeve C., van Deurzen C. H. M., Obdeijn I. M., van Asperen C. J., Tollenaar R. A. E. M., van Cronenburg T. C. T. E. F., Kets C. M., Ausems M. G. E. M., van der Pol C. C., van Os T. A. M., Waisfisz Q., Meijers-Heijboer H. E. J., Gómez-Garcia E. B., Oosterwijk J. C., Mourits M. J., de Bock G. H., Vasen H. F., Siesling S., Verloop J., Overbeek L. I. H., Heitz F., Herzog J., Høgdall E., Høgdall C. K., Hogervorst F. B. L., Hollestelle A., Hopper J. L., Hulick P. J., Huzarski T., Imyanitov E. N., Fox S., Kirk J., Lindeman G., Price M., Bowtell D., DeFazio A., Webb P., Isaacs C., Ito H., Jakubowska A., Janavicius R., Jensen A., John E. M., Johnson N., Kabisch M., Kang D., Kapuscinski M., Karlan B. Y., Khan S., Kiemeney L. A., Kjaer S. K., Knight J. A., Konstantopoulou I., Kosma V.-M., Kristensen V., Kupryjanczyk J., Kwong A., de la Hoya M., Laitman Y., Lambrechts D., Le N., De Leeneer K., Lester J., Levine D. A., Li J., Lindblom A., Long J., Lophatananon A., Loud J. T., Lu K., Lubinski J., Mannermaa A., Manoukian S., Le Marchand L., Margolin S., Marme F., Massuger L. F. A. G., Matsuo K., Mazoyer S., Guffog L. M., Lean C. M., Neish I. M., Meindl A., Menon U., Mensenkamp A. R., Milne R. L., Montagna M., Moysich K. B., Muir K., Mulligan A. M., Nathanson K. L., Ness R. B., Neuhausen S. L., Nevanlinna H., Nord S., Nussbaum R. L., Odunsi K., Offit K., Olah E., Olopade O. I., Olson J. E., Olswold C., O’Malley D., Orlow I., Orr N., Osorio A., Park S. K., Pearce C. L., Pejovic T., Peterlongo P., Pfeiler G., Phelan C. M., Poole E. M., Pylkäs K., Radice P., Rantala J., Rashid M. U., Rennert G., Rhenius V., Rhiem K., Risch H. A., Rodriguez G., Rossing M. A., Rudolph A., Salvesen H. B., Sangrajrang S., Sawyer E. J., Schildkraut J. M., Schmidt M. K., Schmutzler R. K., Sellers T. A., Seynaeve C., Shah M., Shen C.-Y., Shu X.-O., Sieh W., Singer C. F., Sinilnikova O. M., Slager S., Song H., Soucy P., Southey M. C., Stenmark-Askmalm M., Stoppa-Lyonnet D., Sutter C., Swerdlow A., Tchatchou S., Teixeira M. R., Teo S. H., Terry K. L., Terry M. B., Thomassen M., Tibiletti M. G., Tihomirova L., Tognazzo S., Toland A. E., Tomlinson I., Torres D., Truong T., Tseng C.-C., Tung N., Tworoger S. S., Vachon C., van den Ouweland A. M. W., van Doorn H. C., van Rensburg E. J., Van’t Veer L. J., Vanderstichele A., Vergote I., Vijai J., Wang Q., Wang-Gohrke S., Weitzel J. N., Wentzensen N., Whittemore A. S., Wildiers H., Winqvist R., Wu A. H., Yannoukakos D., Yoon S.-Y., Yu J.-C., Zheng W., Zheng Y., Khanna K. K., Simard J., Monteiro A. N., French J. D., Couch F. J., Freedman M. L., Easton D. F., Dunning A. M., Pharoah P. D., Edwards S. L., Chenevix-Trench G., Antoniou A. C., Gayther S. A., Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast–ovarian cancer susceptibility locus. Nat. Commun. 7, 12675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng W., Zhang B., Cai Q., Sung H., Michailidou K., Shi J., Choi J. Y., Long J., Dennis J., Humphreys M. K., Wang Q., Lu W., Gao Y. T., Li C., Cai H., Park S. K., Yoo K. Y., Noh D. Y., Han W., Dunning A. M., Benitez J., Vincent D., Bacot F., Tessier D., Kim S. W., Lee M. H., Lee J. W., Lee J. Y., Xiang Y. B., Zheng Y., Wang W., Ji B. T., Matsuo K., Ito H., Iwata H., Tanaka H., Wu A. H., Tseng C. C., Van Den Berg D., Stram D. O., Teo S. H., Yip C. H., Kang I. N., Wong T. Y., Shen C. Y., Yu J. C., Huang C. S., Hou M. F., Hartman M., Miao H., Lee S. C., Putti T. C., Muir K., Lophatananon A., Stewart-Brown S., Siriwanarangsan P., Sangrajrang S., Shen H., Chen K., Wu P. E., Ren Z., Haiman C. A., Sueta A., Kim M. K., Khoo U. S., Iwasaki M., Pharoah P. D., Wen W., Hall P., Shu X. O., Easton D. F., Kang D., Common genetic determinants of breast-cancer risk in East Asian women: A collaborative study of 23 637 breast cancer cases and 25 579 controls. Hum. Mol. Genet. 22, 2539–2550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira M. A., Gamazon E. R., Al-Ejeh F., Aittomaki K., Andrulis I. L., Anton-Culver H., Arason A., Arndt V., Aronson K. J., Arun B. K., Asseryanis E., Azzollini J., Balmana J., Barnes D. R., Barrowdale D., Beckmann M. W., Behrens S., Benitez J., Bermisheva M., Bialkowska K., Blomqvist C., Bogdanova N. V., Bojesen S. E., Bolla M. K., Borg A., Brauch H., Brenner H., Broeks A., Burwinkel B., Caldes T., Caligo M. A., Campa D., Campbell I., Canzian F., Carter J., Carter B. D., Castelao J. E., Chang-Claude J., Chanock S. J., Christiansen H., Chung W. K., Claes K. B. M., Clarke C. L.; EMBRACE Collaborators; GC-HBOC Study Collaborators; GEMO Study Collaborators, Couch F. J., Cox A., Cross S. S., Czene K., Daly M. B., de la Hoya M., Dennis J., Devilee P., Diez O., Dork T., Dunning A. M., Dwek M., Eccles D. M., Ejlertsen B., Ellberg C., Engel C., Eriksson M., Fasching P. A., Fletcher O., Flyger H., Friedman E., Frost D., Gabrielson M., Gago-Dominguez M., Ganz P. A., Gapstur S. M., Garber J., Garcia-Closas M., Garcia-Saenz J. A., Gaudet M. M., Giles G. G., Glendon G., Godwin A. K., Goldberg M. S., Goldgar D. E., Gonzalez-Neira A., Greene M. H., Gronwald J., Guenel P., Haiman C. A., Hall P., Hamann U., He W., Heyworth J., Hogervorst F. B. L., Hollestelle A., Hoover R. N., Hopper J. L., Hulick P. J., Humphreys K., Imyanitov E. N., Investigators A., Investigators H., Investigators B., Isaacs C., Jakimovska M., Jakubowska A., James P. A., Janavicius R., Jankowitz R. C., John E. M., Johnson N., Joseph V., Karlan B. Y., Khusnutdinova E., Kiiski J. I., Ko Y. D., Jones M. E., Konstantopoulou I., Kristensen V. N., Laitman Y., Lambrechts D., Lazaro C., Leslie G., Lester J., Lesueur F., Lindstrom S., Long J., Loud J. T., Lubinski J., Makalic E., Mannermaa A., Manoochehri M., Margolin S., Maurer T., Mavroudis D., McGuffog L., Meindl A., Menon U., Michailidou K., Miller A., Montagna M., Moreno F., Moserle L., Mulligan A. M., Nathanson K. L., Neuhausen S. L., Nevanlinna H., Nevelsteen I., Nielsen F. C., Nikitina-Zake L., Nussbaum R. L., Offit K., Olah E., Olopade O. I., Olsson H., Osorio A., Papp J., Park-Simon T. W., Parsons M. T., Pedersen I. S., Peixoto A., Peterlongo P., Pharoah P. D. P., Plaseska-Karanfilska D., Poppe B., Presneau N., Radice P., Rantala J., Rennert G., Risch H. A., Saloustros E., Sanden K., Sawyer E. J., Schmidt M. K., Schmutzler R. K., Sharma P., Shu X. O., Simard J., Singer C. F., Soucy P., Southey M. C., Spinelli J. J., Spurdle A. B., Stone J., Swerdlow A. J., Tapper W. J., Taylor J. A., Teixeira M. R., Terry M. B., Teule A., Thomassen M., Thone K., Thull D. L., Tischkowitz M., Toland A. E., Torres D., Truong T., Tung N., Vachon C. M., van Asperen C. J., van den Ouweland A. M. W., van Rensburg E. J., Vega A., Viel A., Wang Q., Wappenschmidt B., Weitzel J. N., Wendt C., Winqvist R., Yang X. R., Yannoukakos D., Ziogas A., Kraft P., Antoniou A. C., Zheng W., Easton D. F., Milne R. L., Beesley J., Chenevix-Trench G., Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer. Nat. Commun. 10, 1741 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brand J. S., Humphreys K., Thompson D. J., Li J., Eriksson M., Hall P., Czene K., Volumetric mammographic density: Heritability and association with breast cancer susceptibility loci. J. Natl. Cancer Inst. 106, dju334 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Stone J., Thompson D. J., dos Santos Silva I., Scott C., Tamimi R. M., Lindstrom S., Kraft P., Hazra A., Li J., Eriksson L., Czene K., Hall P., Jensen M., Cunningham J., Olson J. E., Purrington K., Couch F. J., Brown J., Leyland J., Warren R. M. L., Luben R. N., Khaw K.-T., Smith P., Wareham N. J., Jud S. M., Heusinger K., Beckmann M. W., Douglas J. A., Shah K. P., Chan H.-P., Helvie M. A., Le Marchand L., Kolonel L. N., Woolcott C., Maskarinec G., Haiman C., Giles G. G., Baglietto L., Krishnan K., Southey M. C., Apicella C., Andrulis I. L., Knight J. A., Ursin G., Alnaes G. I. G., Kristensen V. N., Borresen-Dale A.-L., Gram I. T., Bolla M. K., Wang Q., Michailidou K., Dennis J., Simard J., Pharoah P., Dunning A. M., Easton D. F., Fasching P. A., Pankratz V. S., Hopper J. L., Vachon C. M., Novel associations between common breast cancer susceptibility variants and risk-predicting mammographic density measures. Cancer Res. 75, 2457–2467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beesley J., Sivakumaran H., Marjaneh M. M., Shi W., Hillman K. M., Kaufmann S., Hussein N., Kar S., Lima L. G., Ham S., Möller A., Chenevix-Trench G., Edwards S. L., French J. D., eQTL colocalization analyses identify NTN4 as a candidate breast cancer risk gene. Am. J. Hum. Genet. 107, 778–787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y., Lu Q., Powles R., Yao X., Yang C., Fang F., Xu X., Zhao H., Leveraging functional annotations in genetic risk prediction for human complex diseases. PLoS Comput. Biol. 13, e1005589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goumidi L., Thibord F., Wiggins K. L., Li-Gao R., Brown M. R., van Hylckama Vlieg A., Souto J.-C., Soria J.-M., Ibrahim-Kosta M., Saut N., Daian D., Olaso R., Amouyel P., Debette S., Boland A., Bailly P., Morrison A. C., Mook-Kanamori D. O., Deleuze J.-F., Johnson A., de Vries P. S., Sabater-Lleal M., Chiaroni J., Smith N. L., Rosendaal F. R., Chasman D. I., Trégouët D.-A., Morange P.-E., Association between ABO haplotypes and the risk of venous thrombosis: Impact on disease risk estimation. Blood 137, 2394–2402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajasekharan S., Kennedy T. E., The netrin protein family. Genome Biol. 10, 239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Stein E., Oliver T., Li Y., Brunken W. J., Koch M., Tessier-Lavigne M., Hogan B. L., Novel role for Netrins in regulating epithelial behavior during lung branching morphogenesis. Curr. Biol. 14, 897–905 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eveno C., Broqueres-You D., Feron J. G., Rampanou A., Tijeras-Raballand A., Ropert S., Leconte L., Levy B. I., Pocard M., Netrin-4 delays colorectal cancer carcinomatosis by inhibiting tumor angiogenesis. Am. J. Pathol. 178, 1861–1869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayano Y., Sasaki K., Ohmura N., Takemoto M., Maeda Y., Yamashita T., Hata Y., Kitada K., Yamamoto N., Netrin-4 regulates thalamocortical axon branching in an activity-dependent fashion. Proc. Natl. Acad. Sci. U.S.A. 111, 15226–15231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Yan Q., Wang Y., Dong X., NTN4 is associated with breast cancer metastasis via regulation of EMT-related biomarkers. Oncol. Rep. 37, 449–457 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Reuten R., Zendehroud S., Nicolau M., Fleischhauer L., Laitala A., Kiderlen S., Nikodemus D., Wullkopf L., Nielsen S. R., McNeilly S., Prein C., Rafaeva M., Schoof E. M., Furtwängler B., Porse B. T., Kim H., Won K. J., Sudhop S., Zornhagen K. W., Suhr F., Maniati E., Pearce O. M. T., Koch M., Oddershede L. B., Van Agtmael T., Madsen C. D., Mayorca-Guiliani A. E., Bloch W., Netz R. R., Clausen-Schaumann H., Erler J. T., Basement membrane stiffness determines metastases formation. Nat. Mater. 20, 892–903 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Esseghir S., Kennedy A., Seedhar P., Nerurkar A., Poulsom R., Reis-Filho J. S., Isacke C. M., Identification of NTN4, TRA1, and STC2 as prognostic markers in breast cancer in a screen for signal sequence encoding proteins. Clin. Cancer Res. 13, 3164–3173 (2007). [DOI] [PubMed] [Google Scholar]
- 28.van de Bunt M., Cortes A., Consortium I., Brown M. A., Morris A. P., McCarthy M. I., Evaluating the performance of fine-mapping strategies at common variant GWAS loci. PLOS Genet. 11, e1005535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soldner F., Stelzer Y., Shivalila C. S., Abraham B. J., Latourelle J. C., Barrasa M. I., Goldmann J., Myers R. H., Young R. A., Jaenisch R., Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan G., Cavalli M., Carlsson B., Skrtic S., Kumar C., Wadelius C., rs953413 regulates polyunsaturated fatty acid metabolism by modulating ELOVL2 expression. iScience 23, 100808 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua J. T., Ahmed M., Guo H., Zhang Y., Chen S., Soares F., Lu J., Zhou S., Wang M., Li H., Larson N. B., McDonnell S. K., Patel P. S., Liang Y., Yao C. Q., van der Kwast T., Lupien M., Feng F. Y., Zoubeidi A., Tsao M.-S., Thibodeau S. N., Boutros P. C., He H. H., Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell 174, 564–575.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 32.I. E. Vorontsov, I. V. Kulakovskiy, G. Khimulya, D. D. Nikolaeva, V. J. Makeev, in International Conference on Bioinformatics Models, Methods and Algorithms (SCITEPRESS, 2015), vol. 2, pp. 102–108. [Google Scholar]
- 33.Lin E. Y., Jones J. G., Li P., Zhu L., Whitney K. D., Muller W. J., Pollard J. W., Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davie S. A., Maglione J. E., Manner C. K., Young D., Cardiff R. D., MacLeod C. L., Ellies L. G., Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res. 16, 193–201 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duivenvoorden H. M., Spurling A., O’Toole S. A., Parker B. S., Discriminating the earliest stages of mammary carcinoma using myoepithelial and proliferative markers. PLOS ONE 13, e0201370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Rios J., Esteve P., Ruiz J. M., Bovolenta P., The Netrin-related domain of Sfrp1 interacts with Wnt ligands and antagonizes their activity in the anterior neural plate. Neural Dev. 3, 19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dydensborg A. B., Rose A. A., Wilson B. J., Grote D., Paquet M., Giguere V., Siegel P. M., Bouchard M., GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 28, 2634–2642 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Asselin-Labat M.-L., Sutherland K. D., Barker H., Thomas R., Shackleton M., Forrest N. C., Hartley L., Robb L., Grosveld F. G., van der Wees J., Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Manjunath M., Zhang S., Chasman D., Roy S., Song J. S., Integrative genomic analysis predicts causative Cis-regulatory mechanisms of the breast cancer-associated genetic variant rs4415084. Cancer Res. 78, 1579–1591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The Cancer Genome Atlas Network , Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneiders F. I., Maertens B., Bose K., Li Y., Brunken W. J., Paulsson M., Smyth N., Koch M., Binding of netrin-4 to laminin short arms regulates basement membrane assembly. J. Biol. Chem. 282, 23750–23758 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Nacht M., Martin T. B. S., Byrne A., Klinger K. W., Teicher B. A., Madden S. L., Jiang Y., Netrin-4 regulates angiogenic responses and tumor cell growth. Exp. Cell Res. 315, 784–794 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Hu Y., Ylivinkka I., Chen P., Li L., Hautaniemi S., Nyman T. A., Keski-Oja J., Hyytiäinen M., Netrin-4 promotes glioblastoma cell proliferation through integrin β4 signaling. Neoplasia 14, 219–IN23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv B., Song C., Wu L., Zhang Q., Hou D., Chen P., Yu S., Wang Z., Chu Y., Zhang J., Yang D., Liu J., Netrin-4 as a biomarker promotes cell proliferation and invasion in gastric cancer. Oncotarget 6, 9794–9806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wellenstein M. D., Coffelt S. B., Duits D. E. M., van Miltenburg M. H., Slagter M., de Rink I., Henneman L., Kas S. M., Prekovic S., Hau C. S., Vrijland K., Drenth A. P., de Korte-Grimmerink R., Schut E., van der Heijden I., Zwart W., Wessels L. F. A., Schumacher T. N., Jonkers J., de Visser K. E., Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature 572, 538–542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yue Z., Yuan Z., Zeng L., Wang Y., Lai L., Li J., Sun P., Xue X., Qi J., Yang Z., Zheng Y., Fang Y., Li D., Siwko S., Li Y., Luo J., Liu M., LGR4 modulates breast cancer initiation, metastasis, and cancer stem cells. FASEB J. 32, 2422–2437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritie L., Spenle C., Lacroute J., Bolcato-Bellemin A. L., Lefebvre O., Bole-Feysot C., Jost B., Klein A., Arnold C., Kedinger M., Bagnard D., Orend G., Simon-Assmann P., Abnormal Wnt and PI3Kinase signaling in the malformed intestine of lama5 deficient mice. PLOS ONE 7, e37710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reuten R., Patel T. R., McDougall M., Rama N., Nikodemus D., Gibert B., Delcros J. G., Prein C., Meier M., Metzger S., Zhou Z., Kaltenberg J., McKee K. K., Bald T., Tuting T., Zigrino P., Djonov V., Bloch W., Clausen-Schaumann H., Poschl E., Yurchenco P. D., Ehrbar M., Mehlen P., Stetefeld J., Koch M., Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes. Nat. Commun. 7, 13515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J.-Y., Wang H., Xie Y.-T., Li Y., Zheng L.-Y., Ruan Y., Song A.-P., Tian X.-X., Fang W.-G., Association of germline variation in CCNE1 and CDK2 with breast cancer risk, progression and survival among Chinese Han women. PLOS ONE 7, e49296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.GTEx Consortium , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinz S., Texari L., Hayes M. G. B., Urbanowski M., Chang M. W., Givarkes N., Rialdi A., White K. M., Albrecht R. A., Pache L., Marazzi I., García-Sastre A., Shaw M. L., Benner C., Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X., Zhang Y., Chen Y., Li M., Zhou F., Li K., Cao H., Ni M., Liu Y., Gu Z., Dickerson K. E., Xie S., Hon G. C., Xuan Z., Zhang M. Q., Shao Z., Xu J., In situ capture of chromatin interactions by biotinylated dCas9. Cell 170, 1028–1043.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fillebeen C., Wilkinson N., Pantopoulos K., Electrophoretic mobility shift assay (EMSA) for the study of RNA-protein interactions: The IRE/IRP example. J. Vis. Exp. , 52230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thakore P. I., D’Ippolito A. M., Song L., Safi A., Shivakumar N. K., Kabadi A. M., Reddy T. E., Crawford G. E., Gersbach C. A., Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuchnio A., Moens S., Bruning U., Kuchnio K., Cruys B., Thienpont B., Broux M., Ungureanu A. A., Leite de Oliveira R., Bruyere F., Cuervo H., Manderveld A., Carton A., Hernandez-Fernaud J. R., Zanivan S., Bartic C., Foidart J. M., Noel A., Vinckier S., Lambrechts D., Dewerchin M., Mazzone M., Carmeliet P., The cancer cell oxygen sensor PHD2 promotes metastasis via activation of cancer-associated fibroblasts. Cell Rep. 12, 992–1005 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Feinberg T. Y., Zheng H., Liu R., Wicha M. S., Yu S. M., Weiss S. J., Divergent matrix-remodeling strategies distinguish developmental from neoplastic mammary epithelial cell invasion programs. Dev. Cell 47, 145–160.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blaj C., Bringmann A., Schmidt E. M., Urbischek M., Lamprecht S., Fröhlich T., Arnold G. J., Krebs S., Blum H., Hermeking H., Jung A., Kirchner T., Horst D., ADNP is a therapeutically inducible repressor of WNT signaling in colorectal cancer. Clin. Cancer Res. , 2769–2780 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Fevr T., Robine S., Louvard D., Huelsken J., Wnt/β-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herbst A., Jurinovic V., Krebs S., Thieme S. E., Blum H., Göke B., Kolligs F. T., Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC genomics 15, 1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernascone I., Gonzalez T., Barea M. D., Carabana C., Hachimi M., Bosch-Fortea M., Santamaria S., Martin R., Tarnick J., Garcia-Sanz J. A., Martin-Belmonte F., Sfrp3 modulates stromal-epithelial crosstalk during mammary gland development by regulating Wnt levels. Nat. Commun. 10, 2481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheng C., Yao C., Wang Z., Chen H., Zhao Y., Xu D., Huang H., Huang W., Chen S., Cyclophilin J limits inflammation through the blockage of ubiquitin chain sensing. Nat. Commun. 9, 4381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lv C., Li F., Li X., Tian Y., Zhang Y., Sheng X., Song Y., Meng Q., Yuan S., Luan L., Andl T., Feng X., Jiao B., Xu M., Plikus M. V., Dai X., Lengner C., Cui W., Ren F., Shuai J., Millar S. E., Yu Z., MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat. Commun. 8, 1036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cha R. S., Zarbl H., Keohavong P., Thilly W. G., Mismatch amplification mutation assay (MAMA): Application to the c-H-ras gene. PCR Methods Appl. 2, 14–20 (1992). [DOI] [PubMed] [Google Scholar]
- 64.Yu J.-m., Sun W., Wang Z.-h., Liang X., Hua F., Li K., Lv X.-x., Zhang X.-w., Liu Y.-y., Yu J.-j., Liu S.-s., Shang S., Wang F., Yang Z.-n., Zhao C.-x., Hou X.-y., Li P.-p., Huang B., Cui B., Hu Z.-W., TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat. Commun. 10, 5720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sperlich J., Teusch N., Pseudopterosin inhibits proliferation and 3D invasion in triple-negative breast cancer by agonizing glucocorticoid receptor alpha. Molecules 23, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi J., Wang Y., Zeng L., Wu Y., Deng J., Zhang Q., Lin Y., Li J., Kang T., Tao M., Rusinova E., Zhang G., Wang C., Zhu H., Yao J., Zeng Y. X., Evers B. M., Zhou M. M., Zhou B. P., Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25, 210–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Y., Smyth G. K., ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Wolf V., Endo Y., Rubin J. S., Purification and Wnt-inhibitory activities of secreted frizzled-related proteins. Methods Mol. Biol. 468, 31 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S9