Abstract
Acid resistance (AR) is important to survival of Escherichia coli O157:H7 in acidic foods and may play a role during passage through the bovine host. In this study, we examined the role in AR of the rpoS-encoded global stress response regulator ςS and its effect on shedding of E. coli O157:H7 in mice and calves. When assayed for each of the three AR systems identified in E. coli, an rpoS mutant (rpoS::pRR10) of E. coli O157:H7 lacked the glucose-repressed system and possessed reduced levels of both the arginine- and glutamate-dependent AR systems. After administration of the rpoS mutant and the wild-type strain (ATCC 43895) to ICR mice at doses ranging from 101 to 104 CFU, we found the wild-type strain in feces of mice given lower doses (102 versus 103 CFU) and at a greater frequency (80% versus 13%) than the mutant strain. The reduction in passage of the rpoS mutant was due to decreased AR, as administration of the mutant in 0.05 M phosphate buffer facilitated passage and increased the frequency of recovery in feces from 27 to 67% at a dose of 104 CFU. Enumeration of E. coli O157:H7 in feces from calves inoculated with an equal mixture of the wild-type strain and the rpoS mutant demonstrated shedding of the mutant to be 10- to 100-fold lower than wild-type numbers. This difference in shedding between the wild-type strain and the rpoS mutant was statistically significant (P ≤ 0.05). Thus, ςS appears to play a role in E. coli O157:H7 passage in mice and shedding from calves, possibly by inducing expression of the glucose-repressed RpoS-dependent AR determinant and thus increasing resistance to gastrointestinal stress. These findings may provide clues for future efforts aimed at reducing or eliminating this pathogen from cattle herds.
Escherichia coli O157:H7 is a member of the enterohemorrhagic group of pathogenic E. coli (EHEC). This agent has emerged as a food-borne and waterborne pathogen of humans that causes hemorrhagic colitis, hemolytic-uremic syndrome, and thrombotic thrombocytopenic purpura. Outbreaks involving undercooked ground beef (1) and a variety of other foods including salami (7) and apple cider (3) have been documented.
Most human disease outbreaks caused by this organism have been linked to bovine fecal contamination of food or water. E. coli O157:H7 inhabits the intestinal tract of cattle, but it is unclear whether it actually colonizes the bovine intestinal tract (31). This organism seldom causes disease in cattle (10). Prevalence in individual beef and dairy cattle in the United States is low, from 0.3 to 2.2% (14, 20, 21). E. coli O157:H7 exists on about 7% of farms (14). Typically, shedding of serotype O157:H7 strains from cattle is sporadic and of limited duration, lasting for approximately 1 month (2, 31).
A number of animal models have been used to assess the pathogenesis and shedding of E. coli O157:H7 strains, including greyhounds (15), mice (33), neonatal calves (11), gnotobiotic pigs (16), and sheep (24). A modification of the streptomycin-treated mouse model described by Myhal and coworkers (29, 30) has been used to compare the colonization of E. coli O157:H7 strain 933 with that of strain 933cu, which was cured of pO157 (33). Results from these animal models have been useful in elucidating the mechanisms of virulence and pathology.
Most E. coli O157:H7 strains contain the eae locus, which encodes the adhesin protein intimin (12). The eae gene also is found in the enteropathogenic group of E. coli (23). The recent finding that intimin is required for E. coli O157:H7 attachment in the bovine intestinal tract in neonatal calves has led one group of investigators to suggest that an anti-intimin vaccine might be effective for reducing the level of this pathogen in herds (12). In addition to intimin, E. coli O157:H7 also produces Shiga-like toxins, which may play a role in the lesions seen in the gut and kidneys of infected humans. E. coli O157:H7 is thought to have a low infectious dose (≤200 organisms) in humans (19). As with other enteric pathogens, the infectious dose for E. coli O157:H7 is thought to correlate with acid resistance (AR) (18). Because E. coli O157:H7 survives in acidic foods and has a low infectious dose, we hypothesize that it can resist gastric acidity in cattle and that AR is another virulence mechanism of this pathogen.
Two recent studies have investigated the effect of diet on AR of E. coli shed from cattle. Diez-Gonzalez and coworkers (13) examined the effect of high-grain diets on shedding of acid-resistant E. coli from cattle. They found that the proportion of acid-resistant E. coli increased in cattle fed a high-grain diet, but that a switch to a diet of hay decreased the number of acid-resistant bacteria. It is unclear if the results observed for E. coli in these experiments are relevant to serotype O157 strains of E. coli. Hovde et al. (22) examined the AR and the duration of shedding of E. coli O157:H7 from experimentally inoculated cattle fed hay or grain. They found no difference in the proportion of acid-resistant E. coli O157:H7 between these two groups but observed that animals fed hay shed E. coli O157:H7 longer than animals fed grain. Further studies are needed to clarify the role played by AR on E. coli O157:H7 shedding from cattle.
EHEC strains contain three distinct AR systems (26). The oxidative or glucose-repressed system (AR system 1) is active when the cells are growing aerobically or anaerobically in the absence of glucose. In contrast, the glutamate-dependent (AR system 2) and arginine-dependent (AR system 3) systems are active during fermentation. All three systems are active during stationary-phase growth, which suggests the involvement of the alternate, stationary-phase sigma factor ςS, which is encoded by rpoS. ςS is now known to play a protective role when E. coli O157:H7 is exposed to a variety of environmental stresses, including acid (8).
In the present study, the role of resistance to acid stress in E. coli O157:H7 gastrointestinal passage in mice and shedding in calves was examined. We tested the hypothesis that rpoS regulates one or more of the AR systems in E. coli O157:H7 and that a mutation in rpoS would affect the ability of the organism to survive in mice and calves. The results indicate that rpoS regulates especially the glucose-repressed AR system and is important to passage and shedding of E. coli O157:H7 in mice and calves, respectively, possibly by inducing resistance to gastrointestinal acid stress.
(Portions of this work have been presented elsewhere [J. Minter, S. C. Richardson, F. J. DeGraves, J. C. Wright, T. A. Penfound, J. W. Foster, and S. B. Price, Abstr. 98th Gen. Meet. Am. Soc. for Microbiol. 1998, abstr. B-169, p. 81, 1998].)
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this study are listed in Table 1. E. coli O157:H7 strain ATCC 43895, originally isolated from raw hamburger meat implicated in a hemorrhagic colitis outbreak (strain 933 [34]), was used throughout the study. An rpoS mutant of this strain (FRIK 816-3 [8]) was constructed by insertional inactivation with pRR10, which carries the gene encoding β-lactamase. A gadA mutant of ATCC 43895 was constructed in a similar manner as described by Castanie-Cornet et al. (6). The location of the gadA insertion was confirmed using PCR and primers that amplified the correct fragment size only if the insertion had occurred in gadA. Spontaneous mutants of the rpoS and gadA strains resistant to nalidixic acid and rifampin were generated to aid in recovery from calf fecal specimens. Ampicillin (50 μg/ml) was added to cultures of these mutants to maintain pRR10.
TABLE 1.
E. coli strains used in this study
| Strain | Serotype or genotypea | Reference |
|---|---|---|
| ATCC 43895 | O157:H7 | 34 |
| EK274 | ATCC 43895 Nar Rfr | This work |
| FRIK 816-3 | ATCC 43895 rpoS::pRR10 (Ap) | 8 |
| EK275 | FRIK 816-3 Nar Rfr | This work |
| EF501 | 43895 gadA::pRR10 (Ap) Nar Rfr | This work |
| EK272 (B2F1) | O91:H21 | 27 |
Abbreviations for antibiotics: Na, nalidixic acid; Rf, rifampin; Ap, ampicillin.
AR assays.
Cells were grown overnight in one of several media: LBG (Luria-Bertani [LB] plus 0.4% glucose), BHIG (brain heart infusion [BHI] plus 0.4% glucose), LB or BHI buffered with either 100 mM morpholinepropanesulfonic acid [pH 8] or 100 mM morpholineethanesulfonic acid [pH 5.5] [28]), and minimal E glucose (EG [32]). Cultures were grown in 3 ml of the appropriate medium in 13-mm test tubes with shaking (240 rpm) at 37°C to stationary phase (22 h). The glucose-repressed system was tested using cells grown overnight in pH 5.5 buffered LB or BHI followed by 1:1,000 dilution into prewarmed (37°C) pH 2.5 EG. The glutamate and arginine systems were tested using stationary-phase cells grown in LBG followed by 1:1,000 dilution into prewarmed pH 2.5 EG supplemented with 1.5 mM glutamate or 0.6 mM arginine, respectively. Viable-cell counts were determined at 0, 2, and 4 h post-acid challenge by diluting cells in LB, plating cells onto LB agar, and incubating plates for 20 h at 37°C. Values given are representative of the results of triplicate experiments reproducible to within 50%.
Mouse inoculation studies.
A modified version of a previously described procedure (33) was used for oral administration of E. coli O157:H7 strains 43895 (RpoS+) and FRIK 816-3 (RpoS−). Bacteria were grown in Trypticase soy broth (BBL Microbiology Systems, Cockeysville, Md.) at 37°C with shaking (150 rpm) to stationary phase. The cells were harvested by centrifugation (10,000 × g, 10 min) and diluted to the appropriate concentration in a 10% sucrose solution or 0.05 M phosphate buffer (7.2 g of Na2HPO4, 1.2 g of KH2PO4 [pH 7.4]) containing 10% (wt/vol) sucrose. ICR mice (ca. 20 g; Harlan Sprague Dawley Inc., Madison, Wis.) were housed individually in cages with grated floors so that feces could be collected from sterile liners on trays below the cages. The mice were deprived of feed for 24 h preinoculation and then provided a sterile plate containing the sucrose cell suspension (0.5 ml) with numbers of E. coli O157:H7 cells ranging from 0 cells (control) to 104 CFU. Some mice received strain FRIK 816-3 in 10% sucrose containing 0.05 M phosphate buffer to ensure that susceptibility to acid and not another factor was the cause for reduction in gastrointestinal passage in mice. The mice were observed to make sure that they consumed the entire inoculum. To allow for clearance of the inoculum through the stomach and to avoid any protection to gastric acidity, the mice were not provided feed for 4 h after administration of the inoculum.
For each trial, five mice were administered the specified strain and dosage of E. coli O157:H7. Three trials were conducted. Fecal samples were collected on the 3 consecutive days after inoculation and tested for the presence or absence of E. coli O157:H7 cells. A 1:10 suspension was made (0.5 g of feces/4.5 ml of peptone [0.1%]) and mixed thoroughly using a vortex mixer. A 0.5-ml portion of this suspension was then spread plated onto five plates (0.1 ml/plate) of MacConkey sorbitol agar (MSA; Difco, Detroit, Mich.) supplemented with potassium tellurite (2.5 μg/ml; Sigma Chemical Co., St. Louis, Mo.) and cefixime (0.05 μg/ml; Lederle Laboratories, Pearl River, N.Y.) (MSA+ [35]). The plates were incubated at 42°C for 24 h and examined for sorbitol-negative E. coli O157:H7 colonies. Suspect colonies were confirmed as E. coli O157 by agglutination (RIM E. coli O157:H7; Remel, Inc., Lenexa, Kans.). The minimum detection level of this procedure was approximately 20 CFU/g. Mice with a positive fecal sample on any of the 3 days postinoculation were scored positive, meaning that the O157:H7 strain had survived passage through the gastrointestinal tract at the dose used.
Calf inoculation studies.
Weaned 6- to 8-week-old calves were acclimated outdoors for 1 week and then moved to an indoor, climate-controlled BL-2 containment facility, where they were acclimated for an additional week prior to inoculation. Pairs of calves were housed together on pine bedding and fed grain and hay in the morning and evening, with water provided ad libitum. Outdoor and indoor pens were cleaned twice daily. Each calf was cultured for E. coli O157:H7 three times before inoculation to ensure that any calves shedding wild strains of the organism were excluded from the study. At the completion of each experiment, the calves were euthanized with sodium pentobarbital and incinerated. Housing and care of the calves followed the guidelines of the American Association for Laboratory Animal Care.
Bacterial strains for inoculation into calves were grown in 12.5 ml of BHI broth (pH 5.5) to stationary phase. Cell pellets were harvested by centrifugation, washed, and suspended in 0.85% NaCl. Calves were inoculated by gastric lavage with a 50-ml inoculum of 0.85% NaCl containing 1010 CFU of an equal mixture of EK274 (RpoS+) and EK275 (RpoS−), followed by 500 ml of 0.85% NaCl. In a control experiment designed to confirm that pRR10 had no effect on shedding, a mixture of equal amounts of EK274 and EF501 (gadA) was inoculated into four calves.
Following inoculation, calf fecal samples were cultured daily for 16 days for the presence of E. coli O157:H7. Fifty-gram specimens were collected each morning and immediately transported back to the laboratory for culture. Quantitative culture of the specimens was performed by adding 1 g of feces to 9 ml of phosphate buffer followed by serial 10-fold dilution. A 0.1-ml volume of each dilution was plated in duplicate onto MSA plates containing nalidixic acid (35 μg/ml), which selected for EK274 and EK275 (or EF501), and MSA plates containing ampicillin (50 μg/ml), which selected for EK275 or EF501. Following 16 to 20 h of incubation at 37°C, sorbitol-negative colonies were counted manually. To determine the quantity of EK274 present in specimens, the number of ampicillin-resistant colonies was subtracted from the total number of nalidixic acid-resistant colonies.
To detect the organism in feces containing <103 CFU/g, fecal swabs were enriched in BHI containing novobiocin (20 μg/ml), potassium tellurite (2.5 μg/ml), and rifampin (25 μg/ml). Following overnight incubation, the BHI cultures were streaked onto Rainbow Agar O157 (Biolog, Inc., Hercules, Calif.) containing ampicillin or nalidixic acid. This medium is an E. coli O157-selective and differential medium on which colonies of this pathogen turn grey-black. Using this enrichment technique, the presence in fecal samples of as few as three organisms per fecal swab could be detected. Serologic confirmation of E. coli O157:H7 suspect colonies in both the direct plating and enrichment culture protocols was made using a commercial latex agglutination kit (Remel).
Statistical analysis.
Data from the mouse experiment were analyzed by analysis of variance using the SAS statistical analysis system (SAS Institute, Inc., Cary, N.C.). Fecal shedding data from the calf study were entered into a spreadsheet program (Microsoft Excel 97 for Windows, version 4.0) and analyzed using SAS. Daily values (CFU per gram of feces) for each strain were converted to log10 for analysis. Total CFU per gram shed over days 1 to 7 was calculated using area under the curve following the trapezoidal rule (17). EK275 (rpoS) shedding as a percentage of EK274 shedding was calculated by dividing area under the curve results from EK275 shedding by area under the curve EK274 shedding and multiplying the result by 100. EF501 (gadA) shedding as a percentage of EK274 shedding was calculated in a similar manner. Differences in percent shedding between EK275 and EF501 were analyzed using Student's t test (36).
RESULTS
Effect of rpoS on acid resistance of O157.
The E. coli O157:H7 parent (EK274) and rpoS mutant (EK275) were tested for the AR systems previously identified (25). The data presented in Table 2 show that insertional inactivation of rpoS eliminated the glucose-repressed system (AR system 1) and reduced the arginine- and glutamate-dependent systems to various levels when cells were adapted in LB (pH 5.5) and LBG, respectively. Previous results obtained with an E. coli K12 rpoS mutant were similar to the results reported here with the serotype O157:H7 rpoS mutant, EK275 (26). Although an rpoS mutation caused decreased glutamate- and arginine-dependent AR, these systems do not have an absolute requirement for RpoS. This was evident when adaptation was made in BHIG rather than LBG. After growth in BHIG, EK275 exhibited near-normal levels of glutamate- and arginine-dependent AR (Table 2). Yet, when the rpoS mutant was adapted in BHI (pH 5.5), it remained defective in glucose-repressed AR. Clearly, rpoS inactivation selectively prevents the induction of AR system 1, showing that this glucose-repressed system is also RpoS dependent. The effect of adaptation in BHI on AR was also observed in another EHEC strain (O91:H21) originally thought to lack acid resistance (27). Strain O91:H21 exhibited no AR when adaptations were performed in LB (Table 3). However, when adaptations were performed using BHI medium, this strain exhibited significant levels of all three AR systems (Table 3).
TABLE 2.
Effect of rpoS on each of the three AR systems in E. coli O157:H7
| Adaptation mediuma | Challenge medium (pH 2.5) | % Survivalb
|
||
|---|---|---|---|---|
| EK274 (rpoS+) | EK275 (rpoS) | EF501 (gadA) | ||
| LB | ||||
| pH 8 | EG | <0.005 | <0.006 | <0.004 |
| pH 5.5 | EG | 13.6 | <0.005 | 13 |
| LBG | EG | <0.004 | <0.007 | <0.005 |
| EG + Glu | 35 | 0.4 | 75 | |
| EG + Arg | <0.004 | <0.007 | ND | |
| BHI | ||||
| pH 8 | EG | <0.004 | <0.004 | <0.003 |
| pH 5.5 | EG | 4.2 | 0.09 | 5.8 |
| BHIG | EG | <0.004 | <0.006 | <0.004 |
| EG + Glu | 72 | 23 | 60 | |
| EG + Arg | 2 | 1.3 | 10 | |
Adaptation involved overnight growth in the medium indicated.
Measured by determining the number of surviving cells after 4 h of acid treatment. Experiments were conducted in triplicate, and results varied within 50% of the stated value. ND, not determined.
TABLE 3.
AR of E. coli O91:H21 strain
| Adaptation mediuma | Challenge medium (pH 2.5) | % Survivalb |
|---|---|---|
| LB, pH 5.5 | EG | <0.005 |
| BHI, pH 5.5 | EG | 9 |
| LBG | EG | <0.006 |
| EG + Glu | <0.06 | |
| EG + Arg | <0.01 | |
| BHIG | EG | <0.004 |
| EG + Glu | 29 | |
| EG + Arg | 6 |
Adaptation involved overnight growth in the medium indicated.
Measured by determining the number of surviving cells after 4 h of acid treatment. Experiments were conducted in triplicate, and results varied within 50% of the stated value.
Passage of E. coli O157:H7 RpoS+ and RpoS− strains in mice.
Oral administration of E. coli O157:H7 strains ATCC 43895 (RpoS+ wild-type strain) and FRIK 816-3 (RpoS−) did not result in visible signs of disease in ICR mice. Feed intake remained constant (average, 8.3 ± 0.5 g/day), and mice produced a daily average of 2.2 g of feces, of which 0.5 g was tested for the presence of the E. coli O157:H7 strains. The plating of fecal samples from mice (control and preinoculation) on MSA+ resulted in low numbers of background bacteria that were similar to colonies of serotype O157:H7 strains (i.e., sorbitol negative).
Of the 405 fecal samples cultured from inoculated mice, 38 samples (9%) tested positive for E. coli O157:H7 at day 1 after dosage, and two of these mice were still fecal positive at day 2. No fecal samples tested positive at day 3 after dosage. The results from the mouse inoculation studies are shown in Table 4. Control mice which received the sucrose or sucrose-phosphate buffer solution tested negative for E. coli O157:H7. Strain 43895 was recovered from 5 of 15 mice (33%; total from three trials) which received 100 CFU, while 12 of 15 mice (80%) administered 1,000 CFU tested positive. None of the mice given 10 CFU tested positive. In comparison, the rpoS mutant strain FRIK 816-3 was not detected in mice receiving 100 CFU and was found in only 2 of 15 (13%) and 4 of 15 (27%) mice administered 1,000 CFU and 10,000 CFU, respectively. Results from inoculation of 100 CFU and 1,000 CFU of strain 816-3 were significantly different (P < 0.05 and P < 0.001, respectively) from the results obtained with the parent strain 43895 administered at the same dose (Table 4). These results indicate that there was a reduction in the ability of strain 816-3 to survive passage through the gastrointestinal tract of mice in comparison to strain 43895.
TABLE 4.
Recovery of E. coli O157:H7 strains ATCC 43895 and FRIK 816-3 from feces following oral administration to ICR micea
| E. coli dose (CFU) | No. of mice with ≥1 fecal sample positive for E. coli O157:H7/5 mice examined in trial (no. of mice with ≥1 fecal sample positive for E. coli O157:H7/15 mice examined in 3 trials)
|
|||||
|---|---|---|---|---|---|---|
| Trial with 10% sucrose
|
Trial with 0.05 M PB–10% sucrose
|
|||||
| I | II | III | I | II | III | |
| Control | 0 | 0 | 0 (0) | 0 | 0 | 0 (0) |
| Strain 43895 | ||||||
| 101 | 0 | 0 | 0 (0) | ND | ND | ND ND |
| 102 | 0 | 3 | 2 (5) | ND | ND | ND ND |
| 103 | 5 | 4 | 3 (12) | ND | ND | ND ND |
| Strain 816-3 | ||||||
| 102 | 0 | 0 | 0 (0)b | 0 | 0 | 0 0 |
| 103 | 1 | 0 | 1 (2)c | 2 | 2 | 1 (5) |
| 104 | 1 | 2 | 1 (4)d | 3 | 4 | 3 (10) |
Cell pellets (strain ATCC 43895 or FRIK 816-3) were suspended and diluted to the appropriate concentration in 10% sucrose or 0.05 M phosphate buffer (PB) in 10% sucrose. Each mouse was administered 0.5 ml of the sucrose-cell suspension or sucrose-phosphate-cell suspension. Control mice were fed 0.5 ml of 10% sucrose or 0.05 M phosphate buffer containing 10% sucrose with no E. coli O157:H7. Results are from fecal samples collected 24 h after administration. ND, not determined.
Significantly different (P < 0.05) from results obtained with the parent strain (ATCC 43895) inoculated at a dose of 102 CFU.
Significantly different (P < 0.001) from results obtained with the parent strain (ATCC 43895) inoculated at a dose of 103 CFU.
Significantly different (P < 0.05) from results obtained with strain 816-3 inoculated at a dose of 104 CFU in phosphate buffer.
To determine if acid susceptibility rather than another factor was responsible for the reduced passage noted with strain 816-3, cells were suspended in 10% sucrose containing phosphate buffer. Suspension of strain 816-3 in phosphate buffer significantly increased (P < 0.05) the number of mice shedding at a dose of 10,000 CFU (Table 4). At a dose of 1,000 CFU, suspension of 816-3 in phosphate buffer increased the number of mice shedding the pathogen from 2 of 15 to 5 of 15, but this increase was not statistically significant. Although there was a significant increase in passage when strain 816-3 was administered in phosphate buffer with sucrose in comparison to sucrose alone, the wild-type strain 43895 was detected at similar frequencies following administration of 10-fold-fewer cells (e.g., 100 and 1,000 CFU). Suspension and administration of strain 816-3 in bicarbonate buffer as described previously (5) was detrimental to its survival (data not shown).
Shedding of E. coli O157:H7 RpoS+ and RpoS− strains in calves.
A quantitative approach was taken to examine the effect of rpoS on E. coli O157:H7 shedding patterns from experimentally inoculated calves. In these experiments, equal amounts of the wild-type (EK274; RpoS+) and RpoS− (EK275) strains were inoculated into three calves. The animals remained healthy throughout the experiment. As has been observed previously with inoculation of E. coli O157:H7 into calves, there was considerable variation of shedding among the calves (10). The reason for this variation is unknown.
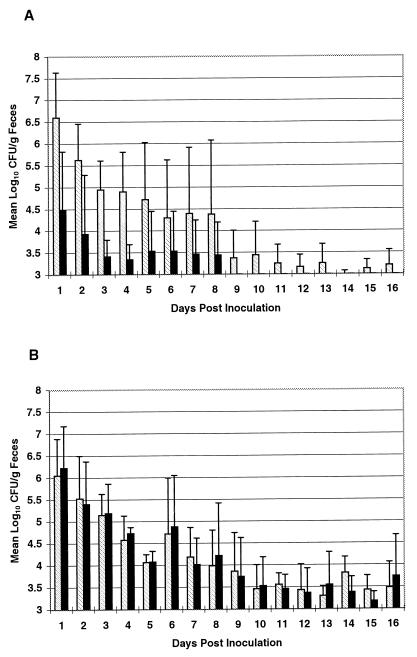
In each calf, the wild-type (RpoS+) strain was shed in large amounts within 24 h from all three calves, with a minimum of 2.7 × 105 to a maximum of 2.6 × 107 CFU/g of feces (Fig. 1). Shedding of >103 CFU of this strain per g of feces continued for a variable amount of time (from 6 to 16 days) in each calf. Generally, the number of organisms shed decreased over time (Fig. 1).
FIG. 1.
Shedding levels over time of E. coli O157:H7 wild-type (□) and RpoS− (■) (A) or wild-type (□) and GadA− (■) (B) strains. Calves were inoculated with 1010 to 1011 total CFU containing equal numbers of wild-type and mutant strains on day 0. Fecal samples were cultured daily beginning 1 day postinoculation. Colony counts are displayed as the means for three (A) or four (B) calves. Bars indicate standard deviation of shedding among calves. Specimens containing <103 CFU/g of feces were below the level needed for accurate enumeration (<10 CFU/10−1 dilution plate).
The RpoS− mutant strain was shed in significantly decreased amounts from all three calves compared to wild-type shedding (P ≤ 0.05). Although shedding of the RpoS− strain dropped below 103 CFU/g 8 days earlier than did the RpoS+ strain (Fig. 1), the mutant could be detected in feces from experimentally infected calves for 15 to 39 days, comparable to the length of parent strain shedding, which ranged from 17 to 43 days (data not shown).
To confirm that shedding differences observed were due to the inactivation of rpoS and not the plasmid (pRR10) insert, a mutant containing pRR10 inserted into the gene for glutamate decarboxylase (gadA) was constructed. The gadA mutant (EF501) was fully acid resistant (Table 2). The difference in shedding in four calves between the parent and the gadA mutant was not significantly different (P > 0.05; Fig. 1), indicating that pRR10 itself had no effect on shedding. This finding also illustrates the consistency of results when one is comparing two strains with equal ability to survive passage through the bovine gastrointestinal tract.
DISCUSSION
In the present study, the role of rpoS in AR and the subsequent impact on shedding of E. coli O157:H7 from mice and calves were examined. Our working hypothesis is that AR in E. coli O157:H7 is induced during passage and growth of the organism in the intestinal tract of cattle (26). This hypothesis has been supported by recent data from Hovde et al. (22), who showed that acid-resistant E. coli O157:H7 was shed from cattle. We have shown previously that once induced, all three AR systems (rpoS dependent, glutamate dependent, and arginine dependent) will persist for at least a month at 4°C (26). Combined, the published data indicate that AR can be induced in the intestinal tract, and growth of acid-resistant E. coli O157:H7 on a contaminated beef carcass is unnecessary for persistence of the phenotype.
The in vitro AR studies reported here extend the findings of a previous study by Cheville et al. (8), which described the construction and characterization of acid tolerance in an rpoS mutant of E. coli O157:H7 (FRIK 816-3). A marked difference in the glucose-repressed AR system between the wild-type and RpoS mutant was observed, confirming a role for rpoS in induction of this system (Table 2). Interestingly, an rpoS effect on the arginine- and glutamate-dependent AR systems in LBG was not found when the wild-type and RpoS mutant strains were acid challenged in BHIG. This finding was not limited to E. coli O157:H7 EHEC, as another EHEC strain, O91:H2, also showed differences in AR in LBG and BHIG.
ICR mice were used in studies to evaluate the influence of the rpoS regulon and acid tolerance on gastrointestinal passage in mice. Mice were used as one of the animals for testing our hypothesis due to the ease of handling and the increased size of groups which this species affords. However, the streptomycin-treated mouse model used by others (29, 33) for colonization and pathogenicity studies was not used because this study concerned passage through the gastric barrier and not colonization. In fact, colonization and subsequent replication would have made results more difficult to interpret. Positive fecal samples from mice were detected at day 1 after inoculation, and only two mice remained fecal positive at day 2. No mice tested positive 3 days after inoculation, which indicates that there was no colonization of mice by either E. coli O157:H7 strain (ATCC 43895 or FRIK 816-3).
Results from inoculation of mice with levels of wild-type strain ATCC 43895 or FRIK 816-3 ranging from 101 to 104 CFU demonstrated that a functional rpoS system promoted survival during gastrointestinal passage in mice. Inoculation of lower numbers of strain 43895 than of FRIK 816-3 resulted in positive fecal samples. Also, the frequency of positive fecal samples was greater in mice inoculated with 43895 when the same number of wild-type and mutant strain was administered (Table 4). For example, 12 of 15 mice (80%) had a positive fecal sample when administered 103 CFU of the wild-type strain, whereas, 2 of 15 (13%) mice given 103 CFU of the rpoS mutant strain FRIK 816-3 tested positive. To ensure that the reduced recovery of FRIK 816-3 was due to acid sensitivity and not another factor detrimental to passage, the mutant strain was suspended in 0.05 M phosphate buffer with 10% sucrose and used to dose mice. Suspension of FRIK 816-3 in phosphate buffer at doses of 103 and 104 CFU increased the number of positive fecal samples from 2 of 15 (13%) to 5 of 15 (33%) and from 4 of 15 (27%) to 10 of 15 (67%), respectively (Table 4). Early studies defining the infectious dose of Vibrio cholerae also demonstrated the importance of the gastric barrier, as suspension of V. cholerae in bicarbonate buffer reduced the infectious dose in humans from 108 to 104 organisms (5). Our study of E. coli O157:H7 used phosphate buffer instead of bicarbonate buffer because the latter reduced viable numbers of FRIK 816-3 (data not shown). The data from this study indicate that protection from acid afforded by rpoS-regulated systems promotes gastric passage.
A calf model of E. coli O157:H7 shedding was used to examine the effect of the rpoS system on the survival and shedding from the gastrointestinal tract of cattle. As in other studies using a calf model (4, 9, 10), an inoculum of 1010 CFU of the organism resulted in appreciable shedding during the first week postinoculation. When the RpoS+ and RpoS− strains were simultaneously administered to calves, we were able to compare numbers of each strain shed concurrently.
The RpoS− mutant (EK275) was reproducibly shed in lower numbers from the calves than was its RpoS+ parent (EK274) (Fig. 1), and this difference was significant (P < 0.05). This finding indicates that rpoS plays a role in E. coli O157:H7 shedding in calves, possibly by inducing resistance to gastrointestinal stress, including acid stress offset by the glucose-repressed RpoS-dependent AR system.
The observation that strains of E. coli O157:H7 were shed in detectable numbers for several weeks reflects the fact that these calves were inoculated with high numbers of E. coli O157:H7 (ca. 1010 CFU) in order to achieve extended periods of shedding at appropriate numbers for detection. In the natural setting, cattle most likely ingest fewer organisms. Indeed, field observations indicate that exposure of cattle does not result in shedding of high numbers of E. coli O157:H7 strains for extended periods of time (31). What was unexpected about these results was the finding that both the parent and rpoS strains were shed for essentially the same length of time (data not shown). This observation may indicate that E. coli O157:H7 survivors that reach and colonize the lower intestinal tract, where the pH is neutral to alkaline, no longer require AR for survival.
The long-term goal of this study is to further define the role of AR systems in gastrointestinal passage and to develop intervention strategies that may inactivate one or more of these systems and possibly reduce or eliminate this pathogen from the bovine intestinal tract. Such an approach might be complementary to other strategies such as vaccination of herds with intimin, as suggested by Dean-Nystrom et al. (12), or manipulation of feed content (13, 22) as part of a complete program to exclude E. coli O157:H7 from cattle by reducing its ability to compete or survive in this complex digestive system. In the present work, a global regulator of acid and other stress resistance was shown to influence E. coli O157:H7 shedding from mice and calves. We are presently continuing experiments aimed at dissecting the role of each AR determinant in E. coli O157:H7.
ACKNOWLEDGMENTS
This work was supported by grants 97-02329 to J.W.F. and 94-37201-1025 to C.W.K. from the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture. C.W.K. was also supported by the College of Agriculture and Life Sciences, University of Wisconsin—Madison. S.B.P., J.C.W., and F.J.D. were supported by the Alabama Agricultural Experiment Station and the College of Veterinary Medicine, Auburn University.
We thank David Baumler, Mark Freeman, Tony Fuller, and Joey Minter for providing excellent technical assistance.
REFERENCES
- 1.Bell B P, Goldoft M, Griffin P M, Davis M A, Gordon D C, Tarr P I, Bartleson C A, Lewis J H, Barrett T J, Wells J G, Baron R B, Kobayashi J. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA. 1994;272:1349–1353. [PubMed] [Google Scholar]
- 2.Besser T E, Hancock D D, Pritchett L C, McRae E M, Rice D H, Tarr P I. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J Infect Dis. 1997;175:726–729. doi: 10.1093/infdis/175.3.726. [DOI] [PubMed] [Google Scholar]
- 3.Besser T E, Lett S M, Weber J T, Doyle M P, Barrett T J, Wells J G, Griffin P M. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. JAMA. 1993;269:2217–2220. [PubMed] [Google Scholar]
- 4.Brown C A, Harmon B G, Zhao T, Doyle M P. Experimental Escherichia coli O157:H7 carriage in calves. Appl Environ Microbiol. 1997;63:27–32. doi: 10.1128/aem.63.1.27-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cash R A, Music S I, Libonati J P, Snyder M J, Wenzel R P, Hornick R B. Response of man to infection with Vibrio cholerae. I. Clinical, serologic, and bacteriologic responses to a known inoculum. J Infect Dis. 1974;129:45–52. doi: 10.1093/infdis/129.1.45. [DOI] [PubMed] [Google Scholar]
- 6.Castanie-Cornet M-P, Penfound T A, Smith D, Elliott J F, Foster J W. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California 1994. Morbid Mortal Weekly Rep. 1995;44:157–160. [PubMed] [Google Scholar]
- 8.Cheville A M, Arnold K W, Buchrieser C, Cheng C-M, Kaspar C W. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1996;62:1822–1824. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cray W C, Casey T A, Bosworth B T, Rasmussen M A. Effect of dietary stress on fecal shedding of Escherichia coli O157:H7 in calves. Appl Environ Microbiol. 1998;64:1975–1979. doi: 10.1128/aem.64.5.1975-1979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cray W C, Moon H W. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean-Nystrom E A, Bosworth B T, Cray W C, Jr, Moon H W. Pathogenicity of Escherichia coli O157 in the intestines of neonatal calves. Infect Immun. 1997;65:1842–1848. doi: 10.1128/iai.65.5.1842-1848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean-Nystrom E A, Bosworth B T, Moon H W, O'Brien A D. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun. 1998;66:4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diez-Gonzalez F, Callaway T R, Kizoulis M G, Russell J B. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science. 1998;281:1666–1668. doi: 10.1126/science.281.5383.1666. [DOI] [PubMed] [Google Scholar]
- 14.Faith N G, Shere J A, Brosch R, Arnold K W, Ansay S E, Lee M S, Luchansky J B, Kaspar C W. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl Environ Microbiol. 1996;62:1519–1525. doi: 10.1128/aem.62.5.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenwick B W, Cowan L A. Canine model of hemolytic-uremic syndrome. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: ASM Press; 1998. pp. 268–277. [Google Scholar]
- 16.Francis D H, Collins J E, Duimstra J R. Infection of gnotobiotic pigs with an Escherichia coli O157:H7 strain associated with an outbreak of hemorrhagic colitis. Infect Immun. 1986;51:953–956. doi: 10.1128/iai.51.3.953-956.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrielsson J, Weiner D. Pharmacokinetic and pharmacodynamic data analysis. Stockholm, Sweden: Swedish Pharmaceutical Press; 1994. p. 94. [Google Scholar]
- 18.Gorden J, Small P L. Acid resistance in enteric bacteria. Infect Immun. 1993;61:364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–761. [Google Scholar]
- 20.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 21.Hancock D D, Besser T E, Kinsel M L, Tarr P I, Rice D H, Paros M G. The prevalence of Escherichia coli O157:H7 in dairy and beef cattle in Washington State. Epidemiol Infect. 1994;113:199–207. doi: 10.1017/s0950268800051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hovde C J, Austin P R, Cloud K A, Williams C J, Hunt C W. Effect of cattle diet on Escherichia coli O157:H7 acid resistance. Appl Environ Microbiol. 1999;65:3233–3235. doi: 10.1128/aem.65.7.3233-3235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudva I T, Hatfield P G, Hovde C J. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl Environ Microbiol. 1995;61:1363–1370. doi: 10.1128/aem.61.4.1363-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J, Lee I S, Frey J, Slonczewski J L, Foster J W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Smith P, Chapin K C, Baik H S, Bennett G N, Foster J W. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindgren S W, Melton A R, O'Brien A D. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect Immun. 1993;61:3832–3842. doi: 10.1128/iai.61.9.3832-3842.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 29.Myhal M L, Cohen P S, Laux D C. Altered colonizing ability for mouse large intestine of a surface mutant of a human faecal isolate of Escherichia coli. J Gen Microbiol. 1983;129:1549–1558. doi: 10.1099/00221287-129-5-1549. [DOI] [PubMed] [Google Scholar]
- 30.Myhal M L, Laux D C, Cohen P S. Relative colonizing abilities of human fecal and K12 strains of Escherichia coli in the large intestine of streptomycin-treated mice. Eur J Clin Microbiol. 1982;1:186–192. doi: 10.1007/BF02019621. [DOI] [PubMed] [Google Scholar]
- 31.Shere J A, Bartlett K J, Kaspar C W. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl Environ Microbiol. 1998;64:1390–1399. doi: 10.1128/aem.64.4.1390-1399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 33.Wadolkowski E A, Burris J A, O'Brien A D. Mouse model for the colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1990;58:2438–2445. doi: 10.1128/iai.58.8.2438-2445.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells J G, Davis B R, Wachsmuth I K, Riley L W, Remis R S, Sokolow R, Morris G K. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zadik P M, Chapman P A, Siddons C A. Use of tellurite for the selection of verocytotoxigenic Escherichia coli O157:H7. J Med Microbiol. 1993;39:155–158. doi: 10.1099/00222615-39-2-155. [DOI] [PubMed] [Google Scholar]
- 36.Zar J H. Biostatistical analysis. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1984. pp. 185–205. [Google Scholar]