Abstract
Background
Spatially-resolved transcriptomics has now enabled the quantification of high-throughput and transcriptome-wide gene expression in intact tissue while also retaining the spatial coordinates. Incorporating the precise spatial mapping of gene activity advances our understanding of intact tissue-specific biological processes. In order to interpret these novel spatial data types, interactive visualization tools are necessary.
Results
We describe spatialLIBD, an R/Bioconductor package to interactively explore spatially-resolved transcriptomics data generated with the 10x Genomics Visium platform. The package contains functions to interactively access, visualize, and inspect the observed spatial gene expression data and data-driven clusters identified with supervised or unsupervised analyses, either on the user’s computer or through a web application.
Conclusions
spatialLIBD is available at https://bioconductor.org/packages/spatialLIBD. It is fully compatible with SpatialExperiment and the Bioconductor ecosystem. Its functionality facilitates analyzing and interactively exploring spatially-resolved data from the Visium platform.
Supplementary Information
The online version contains supplementary material available at (10.1186/s12864-022-08601-w).
Keywords: Spatially-resolved transcriptomics, Interactive visualization, 10x Genomics Visium
Background
The rise of spatially-resolved transcriptomics technologies [1, 2] is providing new opportunities to answer questions about the structure and function of complex intact tissues [3–5]. However, this exciting opportunity requires the development of new computational methods and interactive software. These tools have to accommodate the analysis of transcriptomic data with spatial coordinates and high resolution images into previously developed methods for single cell RNA-sequencing (scRNA-seq) analysis, which lacked these components.
Currently there are limited options for interactive exploration of spatially-resolved transcriptomics data. The 10x Genomics Loupe Browser [6] utilizes a file created by the Space Ranger data pre-processing pipeline [6] and performs marker gene analysis, as well as unsupervised graph-based and k-means clustering. Loupe visualizes both the spatially-resolved transcriptomics data along with the high resolution imaging data to export gene expression maps. An alternative to Loupe is the Giotto pipeline [7], which contains both a data analysis and data visualization modules. The data analysis module has many functions ranging from pre-processing to more advanced analysis such as characterizing cell-cell interactions. The visualization module contains an interactive workspace for exploring multiple layers of information. However, Loupe and Giotto currently do not support visualizing more than one tissue section at a time, which is useful for comparing replicates and annotating observed spots on the Visium array across samples. Seurat [8] provides guidelines for processing and visualizing spatially-resolved transcriptomics data at https://satijalab.org/seurat/articles/spatial_vignette.html. Seurat [8] does not however provide interactive visualizations or the ability to create an interactive website. Furthermore, while unsupervised clustering methods are widely developed [9, 10], manual annotation of spots using known marker genes is important, as well as cross-sample dimension reduction techniques such as UMAP and t-SNE.
Implementation
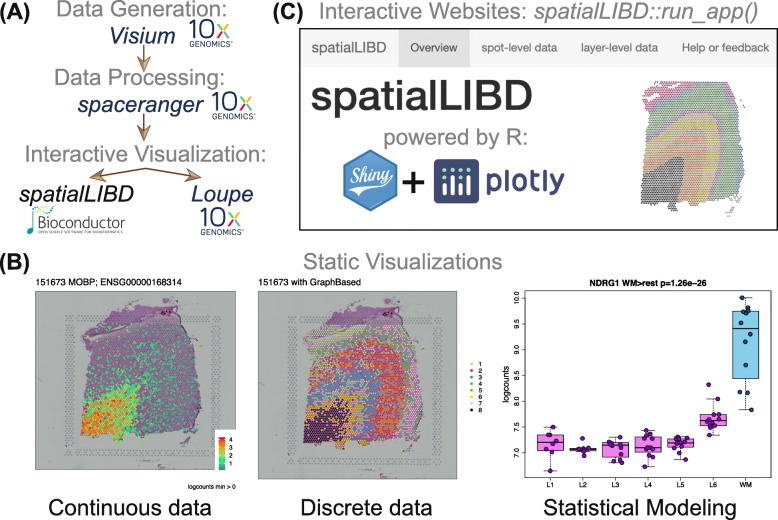
spatialLIBD is an R/Bioconductor [11] package that provides functions to import output from Space Ranger [6] including clustering (k-means, graph-based) and dimension reduction results (PCA, UMAP, t-SNE), interactively visualize and explore spatially-resolved transcriptomics data, and provides functionality to manually annotate spots. It was developed initially for visualizing multiple Visium tissue sections from the human brain [4], yet it is flexible enough to be used with other Visium data from different tissue types (Supplementary File 1, Fig. 1A).
Fig. 1.
Overview of spatialLIBD: A Using spatially-resolved transcriptomics data generated with the Visium platform and processed with spaceranger, both from 10x Genomics, spatialLIBD leverages the R/Bioconductor ecosystem and presents an alternative for interactive visualization to Loupe Browser. BspatialLIBD supports static data visualizations of both continuous and categorical measurements along with the histology images, as well as visualizing results from downstream analyses. CspatialLIBD supports interactive websites for exploring the data powered by shiny and plotly
The primary functions in spatialLIBD allow users (i) to visualize the spot-level spatial gene expression data or any other continuous variable, and data-driven clusters or any categorical variable, with the histology image in the background (Fig. 1B), and (ii) to inspect the data interactively, either on the user’s computer or through web hosting services such as shinyapps.io [12] (Fig. 1C). In addition, spatialLIBD enables users to visualize multiple samples at the same time, to export all static visualizations as PDF files or all interactive visualizations as PNG files, as well as all result tables as CSV files. Additional file 1 shows how to read in data processed with Space Ranger [6] into R, make plots or create an interactive website with spatialLIBD.
Additional file 2 illustrates how spatialLIBD can be used to visualize clustering results derived from multiple samples as well as other continuous covariates, using a human brain dataset with twelve samples [4]. In particular, clusters derived from all samples facilitate their interpretation since cluster numbers (and thus their meaning) will be the same across all samples. The companion interactive website for this dataset is available at http://spatial.libd.org/spatialLIBD. Additional file 1 shows the steps necessary to make your own interactive website, such as https://libd.shinyapps.io/spatialLIBD_Human_Lymph_Node_10x/.
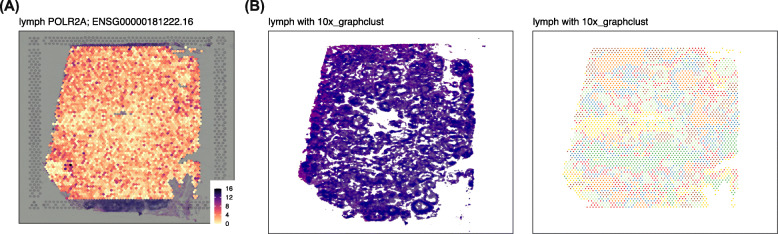
spatialLIBD has evolved over time and now provides features that further facilitate the visual inspection of the results. These include nine color scales, including colorblind-friendly scales as well as the option to reverse the color scale gradient order (Fig. 2A). The transparency of the spots can be controlled as well. Multiple images are supported and can be edited on the web application to increase brightness or perform other image manipulations that can reveal hidden patterns to the eye (Fig. 2B). Side by side plots of the images and the clusters or gene expression data can also be generated.
Fig. 2.
Enhanced visualization spatialLIBD features: Using the example spatialLIBD web application from https://libd.shinyapps.io/spatialLIBD_Human_Lymph_Node_10x/ we illustrate some recent enhancements. The data is from the human lymph node example dataset by 10x Genomics. A Expression of the POLR2A gene with a point size of 2.25 and using the reversed magma colorblind-friendly continuous scale: low values are in white. There are nine color scales options and each can be reversed. The transparency of the spots can be controlled as well. B A side-by-side view of the histology and the graph-based clusters produced by Space Ranger. The original lowres image from Space Ranger has been edited with spatialLIBD: enhance, normalize, equalize, transparent color pink, and transparent fuzz 40
spatialLIBD is designed to work with SpatialExperiment R/Bioconductor objects [13]. It uses shiny [14] and plotly [15] for the interactive website, ggplot2 [16] and cowplot [17] for the static images. Interactive image editing is powered by magick [18]. Colorblind-friendly colors are provided by viridis [19].
Discussion
spatialLIBD benefits from the open-source Bioconductor ecosystem [11] and relies on the infrastructure provided by SpatialExperiment [13]. It provides user-friendly functionality for visualizing continuous and categorical measurements along with the tissue images from spatially-resolved transcriptomics data from projects involving one or more samples measured on the 10x Genomics Visium platform. Since spatialLIBD is compatible with SpatialExperiment, spatialLIBD could be used for other spatially-resolved transcriptomic platforms as they become better supported by SpatialExperiment.
spatialLIBD provides a simple interactive website, which can be used for sharing data on the web, as well as manually annotating spots. spatialLIBD has limitations (Additional file 1) that are inherent to the methods used to implement it, such as (i) the memory per user required by a server for hosting the web application, (ii) response speeds for the interactive views due to the number of spots, (iii) the resolution of the images displayed limiting the usefulness to magnify specific spots, and (iv) customization of the web application by the end user. Despite these limitations, open-source software solutions like spatialLIBD enable faster data exploration and insights in spatially-resolved transcriptomics research projects [4] and can serve as a testing ground for ideas.
While some visualization software alternatives do not provide visualizations across multiple samples, static visualizations of multiple samples can be combined using R packages such as cowplot or patchwork or interactive versions using plotly, which is how spatialLIBD was implemented. As spatially-resolved transcriptomics becomes more widely available [20], new methods are being implemented in other languages, such as Python. One such example is squidpy [21], which is currently limited to one Visium sample as noted at https://github.com/theislab/squidpy/issues/417. squidpy does provide exciting and powerful features, and bridges the R and Python worlds.
Conclusion
spatialLIBD is fully integrated with the R/Bioconductor open source ecosystem and is available at https://bioconductor.org/packages/spatialLIBD. It provides user-friendly functionality for importing results from the 10x Genomics Space Ranger pipeline for processing spatially-resolved transcriptomics data from Visium. spatialLIBD provides different options for visualizing and interactively exploring the data from multiple Visium capture areas, and can create websites that can be self-hosted such as http://spatial.libd.org/spatialLIBD/ or hosted at shinyapps.io such as the example website https://libd.shinyapps.io/spatialLIBD_Human_Lymph_Node_10x/.
Availability and requirements
Project name:spatialLIBD: an R/Bioconductor package to visualize spatially-resolved transcriptomics data
Project home page: https://bioconductor.org/packages/spatialLIBD
Operating system(s): Platform independent
Programming language: R
Other requirements: Bioconductor version 3.13 or newer to access the latest features
License: Artistic-2.0
Any restrictions to use by non-academics: none
Supplementary Information
Additional file 1 Using spatialLIBD with 10x Genomics public datasets. Vignette document showing a detailed tutorial on how to use public 10x Genomics Visium data and visualize it using spatialLIBD.
Additional file 2 Visualizing multiple samples with spatialLIBD. R code and resulting PDF files for visualizing across all 12 human brain samples from Maynard et al [4] (a) clusters derived from a shared-nearest neighbor (k = 50) that was computed using the genomic data from all samples, (b) the percent of mitochondrial expression which has a spatial distribution that matches the cortical layers, and (c) the expression of MOBP which is higher in the white matter than other layers [4].
Acknowledgements
Jesús Vélez Santiago and Ricardo O Ramírez-Flores from Julio Saez-Rodriguez’s group at Heidelberg University tested the applicability of spatialLIBD to new datasets and improved the interactive website code.
Abbreviations
- scRNA-seq
single cell RNA sequencing
Authors’ contributions
BP and AS updated the software to make it usable with any Visium dataset, adapted it to use SpatialExperiment, and wrote the documentation for using publicly available data. LMW, SCH, AEJ, KM, and KRM provided input on the design of the software. LCT implemented the initial version of the software and maintains it. All authors contributed to writing the manuscript, and then read and approved the final manuscript.
Authors’ information
Andrew E Jaffe is currently an employee and shareholder of Neumora Therapeutics, which is unrelated to the contents of this manuscript.
Funding
This work was supported by the Lieber Institute for Brain Development, National Institutes of Health grant U01 MH122849 (LMW, SCH, AEJ, KM, KRM, LCT), and CZF2019-002443 (LMW, SCH) from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation. All funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
10x Genomics provides public data examples for their Visium platform, such as the lymph node example used here https://support.10xgenomics.com/spatial-gene-expression/datasets/1.1.0/V1_Human_Lymph_Node.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Brenda Pardo, Email: bpardo@lcgej.unam.mx.
Abby Spangler, Email: aspangle@gmail.com.
Lukas M. Weber, Email: lukas.weber@jhu.edu
Stephanie C. Page, Email: stephanie.page@libd.org
Stephanie C. Hicks, Email: shicks19@jhu.edu
Andrew E. Jaffe, Email: andrewejaffe@gmail.com
Keri Martinowich, Email: Keri.Martinowich@libd.org.
Kristen R. Maynard, Email: kristen.maynard@libd.org
Leonardo Collado-Torres, Email: lcolladotor@gmail.com.
References
- 1.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 2.Xia C, Fan J, Emanuel G, Hao J, Zhuang X. Spatial transcriptome profiling by merfish reveals subcellular rna compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci. 2019;116(39):19490–9. doi: 10.1073/pnas.1912459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maynard KR, Jaffe AE, Martinowich K. Spatial transcriptomics: putting genome-wide expression on the map. Neuropsychopharmacology. 2020;45(1):232–3. doi: 10.1038/s41386-019-0484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, Catallini JL, Tran MN, Besich Z, Tippani M, Chew J, Yin Y, Kleinman JE, Hyde TM, Rao N, Hicks SC, Martinowich K, Jaffe AE. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat Neurosci. 2021;24(3):425–36. doi: 10.1038/s41593-020-00787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuppe C, Ramirez Flores RO, Li Z, Hannani M, Tanevski J, Halder M, Cheng M, Ziegler S, Zhang X, Preisker F, Kaesler N, Xu Y, Hoogenboezem RM, Bindels EMJ, Schneider RK, Milting H, Costa IG, Saez-Rodriguez J, Kramann R. Spatial multi-omic map of human myocardial infarction. bioRxiv. 2020. 10.1101/2020.12.08.411686.
- 6.10x Genomics. Space Ranger and Loupe Browser. 2020. https://support.10xgenomics.com/spatial-gene-expression/software. Accessed 10 Mar 2021.
- 7.Dries R, Zhu Q, Dong R, Eng C-HL, Li H, Liu K, Fu Y, Zhao T, Sarkar A, Bao F, George RE, Pierson N, Cai L, Yuan G-C. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 2021;22(1):78. doi: 10.1186/s13059-021-02286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao E, Stone MR, Ren X, Guenthoer J, Smythe KS, Pulliam T, Williams SR, Uytingco CR, Taylor SEB, Nghiem P, Bielas JH, Gottardo R. Spatial transcriptomics at subspot resolution with BayesSpace. Nat Biotechnol. 2021;1–10. 10.1038/s41587-021-00935-2. [DOI] [PMC free article] [PubMed]
- 10.Biancalani T, Scalia G, Buffoni L, Avasthi R, Lu Z, Sanger A, Tokcan N, Vanderburg CR, Segerstolpe Ä, Zhang M, Avraham-Davidi I, Vickovic S, Nitzan M, Ma S, Subramanian A, Lipinski M, Buenrostro J, Brown NB, Fanelli D, Zhuang X, Macosko EZ, Regev A. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat Methods. 2021;18(11):1352–62. doi: 10.1038/s41592-021-01264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oles AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.RStudio. shinyapps.io. 2021. https://www.shinyapps.io/. Accessed 10 Dec 2021.
- 13.Righelli D, Weber LM, Crowell HL, Pardo B, Collado-Torres L, Ghazanfar S, Lun ATL, Hicks SC, Risso D. SpatialExperiment: infrastructure for spatially resolved transcriptomics data in R using Bioconductor. Bioinformatics. 2022;btac299. 10.1093/bioinformatics/btac299. [DOI] [PMC free article] [PubMed]
- 14.Chang W, Cheng J, Allaire J, Sievert C, Schloerke B, Xie Y, Allen J, McPherson J, Dipert A, Borges B. Shiny: Web Application Framework for R. 2021. R package version 1.6.0. https://CRAN.R-project.org/package=shiny. Accessed 20 Dec 2021.
- 15.Sievert C. Interactive Web-Based Data Visualization with R, Plotly, and Shiny. New York: Chapman and Hall/CRC; 2020. [Google Scholar]
- 16.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2016. [Google Scholar]
- 17.Wilke CO. Cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. 2020. R package version 1.1.1. https://CRAN.R-project.org/package=cowplot.
- 18.Ooms J. Magick: Advanced Graphics and Image-Processing in R. 2021. R package version 2.7.3. https://CRAN.R-project.org/package=magick.
- 19.Garnier, Simon, Ross, Noam, Rudis, Robert, Camargo, Pedro A, Sciaini, Marco, Scherer, Cédric. viridis - Colorblind-Friendly Color Maps for R. 2021. 10.5281/zenodo.4679424.
- 20.Moses L, Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022;1–13. 10.1038/s41592-022-01409-2. [DOI] [PubMed]
- 21.Palla G, Spitzer H, Klein M, Fischer D, Schaar AC, Kuemmerle LB, Rybakov S, Ibarra IL, Holmberg O, Virshup I, Lotfollahi M, Richter S, Theis FJ. Squidpy: a scalable framework for spatial omics analysis. Nat Methods. 2022;19(2):171–8. doi: 10.1038/s41592-021-01358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Using spatialLIBD with 10x Genomics public datasets. Vignette document showing a detailed tutorial on how to use public 10x Genomics Visium data and visualize it using spatialLIBD.
Additional file 2 Visualizing multiple samples with spatialLIBD. R code and resulting PDF files for visualizing across all 12 human brain samples from Maynard et al [4] (a) clusters derived from a shared-nearest neighbor (k = 50) that was computed using the genomic data from all samples, (b) the percent of mitochondrial expression which has a spatial distribution that matches the cortical layers, and (c) the expression of MOBP which is higher in the white matter than other layers [4].
Data Availability Statement
10x Genomics provides public data examples for their Visium platform, such as the lymph node example used here https://support.10xgenomics.com/spatial-gene-expression/datasets/1.1.0/V1_Human_Lymph_Node.