Abstract
RNAs are closely associated with human diseases; however, immune-related genes (IRGs) and their potential regulatory networks in relation to spinal cord injury (SCI) are still poorly understood. Here, we investigated the key IRGs as well as the competing endogenous RNA (ceRNA) mechanisms that are associated with SCI pathogenesis based on microarray datasets and the use of a rat SCI model. Specifically, four independent SCI microarray datasets from Gene Expression Omnibus (GEO) database were analyzed and, thereafter, differentially expressed IRGs were annotated via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Furthermore, based on the GEO datasets, differentially expressed RNAs (DERNAs), including DEcircRNAs, DEmiRNAs, and DEmRNAs were identified and interactions between them were also predicted using online databases, and to construct a circular RNA (circRNA) mediated ceRNA network, candidate RNAs were also identified. Furthermore, the support vector machine (SVM) and least absolute shrinkage and selection operator (LASSO) methods were used for the identification of critical DERNAs, while differential gene expression was validated using the GSE20907 dataset. Our results were as follows. In the SCI microarray datasets, 32, 58, and 74 DEIRGs, DEcircRNAs, and DEmiRNAs were identified, respectively. In addition, GO and KEGG analyses showed that the DEIRGs were primarily enriched in neutrophil-mediated immunity and nuclear factor-kappa B (NF-κB) and hypoxia-inducible factor-1 (HIF-1) signaling pathways, and based on LASSO and SVM screening, PLXNB2 was identified as a DEIRG, while hsa_circ_0026646 was identified as the key circRNA, showing a higher SCI expression. Furthermore, our results proved that PLXNB2 and hsa_circ_0026646 were upregulated in SCI, whereas miR-331-3p was downregulated, and, interestingly, similar expression profiles were confirmed using the rat SCI model. Furthermore, fluorescent reporter assay indicated that both hsa_circ_0026646 and PLXNB2 have miR-331-3p target sites, and the ceRNA hypothesis suggested the dysregulation of hsa_circ_0026646, miR-331-3p, and PLXNB2 in SCI. Thus, our results suggested that in SCI pathogenesis, hsa_circ_0026646 correlates with PLXNB2 by targeting miR-331-3p.
Keywords: Spinal cord injury, immune-related genes, competing endogenous RNA, microRNA, circular RNA, GEO database
Impact statement
Spinal cord injury (SCI), with a great societal burden, is a disease that seriously threatens human health, and so far, it has no effective treatments. Thus, further exploring its pathological mechanism is of great significance. In this study, we explored the competing endogenous (ceRNA) mechanisms associated with SCI. First, we constructed a circular RNA/micro RNA/messenger RNA (circRNA/miRNA/mRNA) regulatory network via bioinformatics analysis of microarray data and explored the potential mechanism of SCI pathogenesis. Thereafter, we used two machine learning methods (support vector machine [SVM] and least absolute shrinkage and selection operator [LASSO]) to screen key RNAs. Our results proved that hsa_circ_0026646 acts a role of ceRNA in upregulating PLXNB2 expression by adsorbing miR-331-3p. Furthermore, the results corresponding to the rat SCI model as well as those based on the dual-Luciferase reporter system were consistent with the bioinformatics-based prediction results. Our findings provide new evidence for the SCI regulatory mechanisms as well as the potential therapeutic targets.
Introduction
The clinical management of spinal cord injury (SCI) is quite challenging owing to the absence of effective treatments. In 2016, a total of 0.9 million SCI cases were recorded worldwide. The age-standardized incidence rate of SCI was 13 per million population. 1 In addition, at the moment, approximately, 3 million people suffer from SCI globally, and approximately, 180,000 new cases are reported every year. 2 It has also been estimated that in China, within the 2014–2018 period, SCI-associated mortality increased by 148.3%. 1 So far, the main SCI management methods include operation therapies, drug therapies, cell therapies, and genetic therapies.3,4 However, these different treatment strategies exhibit limited efficacy.
The prognosis of SCI is related to cellular and molecular mechanisms. The mechanical strike initially injures spinal cord tissue, including neurons and non-neuronal cells. Thereafter, the resulting secondary inflammation further damages the microenvironment of spinal cord. During the acute stage of SCI (2–48 h after injury), a series of events take place, including calcium influx, inflammatory factors accumulation, ion imbalance, and lipid peroxidation. 5 Furthermore, during this phase, the released inflammatory factors recruit inflammatory cells into the spinal cord, which worsen the injury. 6 From this perspective, inflammation is of great importance in SCI pathogenesis. Thus, inflammation response is a double-edged sword for SCI progression. One side, it removes necrotic tissues, and the other side, it causes secondary injury. 7 Given that immune-related genes (IRGs) regulate inflammatory responses, it is necessary to investigate their regulatory mechanisms as they relate to the pathogenesis of SCI.
In addition, the molecular events in SCI are poorly understood. In most studies, the focus has primarily been on single molecules, while their interactions have largely been neglected. In recent years, bioinformatics has emerged as an effective method that can be used to construct a co-expression network via gene expression signature, and compared with large panel gene sequencing and functional identification methods, bioinformatics methods are more cost-effective, and offer the possibility for the rapid identification of candidate therapeutic targets or biomarkers for a given disease.
circRNA, a special kind of non-coding RNA (ncRNA) with a closed circle structure, is unaffected by RNA exonuclease. Compared to miRNA, the expression level of circRNA is stable and not easy to degrade. 8 Studies have shown that it extensively participates in the disease processes of the nervous system and can stably exist in cerebrospinal fluid, passing through the blood–brain barrier (BBB). 9 Furthermore, in the nervous system, circRNAs can directly affect nervous system development and regulate the proliferation, differentiation, and apoptosis of neurons. 10 Furthermore, they have a great many of miRNA binding sites that can exert regulatory functions by competitively binding with miRNAs and regulate the target genes expression by promoting or inhibiting miRNAs. 11
In this study, we selected gene expression microarray chips related to SCI from different databases. Thereafter, we identified differently expressed RNAs via bioinformatics analysis and determined core RNAs using machine learning methods. Furthermore, by analyzing the target binding prediction and gene expression profiles, we hypothesized that the selected RNAs do interact. Furthermore, we also confirmed this hypothesis using a rat SCI model as well as dual-luciferase analysis; we also constructed a circRNA-mediated ceRNA network to explore the underlying regulation mechanism of IRGs in SCI. The results may provide more precise treatment targets for SCI.
Materials and methods
Study design and data acquisition
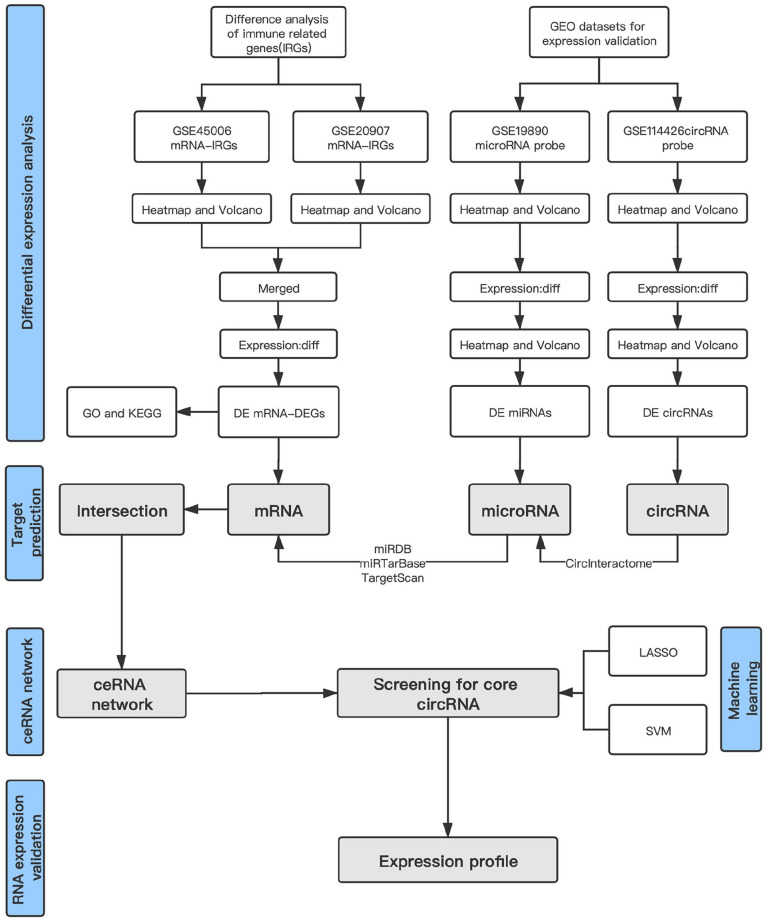
Microarray data were searched from the Gene Expression Omnibus (GEO) database of National Center for Biotechnology Information (NCBI). We downloaded four SCI microarray datasets (circRNA microarray set, GSE114426; miRNA microarray set, GSE19890; and mRNA microarray sets, GSE45006 and GSE20907) that corresponded to the Rattus norvegicus SCI model on Day 3 (Table 1). Furthermore, batch normalization was performed on the GSE45006 and GSE20907 datasets using the R package “sva” to obtain standardized data, and for mRNA analysis, GSE45006 was used as the training set, while the GSE20907 microarray was used as the test set. Data were normalized for further validation (Figure 1 shows the flowchart for our bioinformatics analysis).
Table 1.
Details and sample size of the studied GEO datasets.
Figure 1.
Flowchart of bioinformatic analysis. (A color version of this figure is available in the online journal.)
Screening differentially expressed RNAs
Differentially expressed RNAs (DERNAs), including DEcircRNAs, DEmiRNAs, and DEmRNAs, were used to construct the circRNA/miRNA/mRNA network, and to obtain differentially expressed immune-related genes (DEIRGs), the DEmRNAs and the immune gene set were merged. Based on the R package “limma,” the microarrays had cut-off value of fold change (FC) ⩾ 1.5, p < 0.05 was considered statistically significant. Normalization and log2 transformation were also performed.
Prediction of target binding sites
To predict the target DEmiRNAs of the DEcircRNAs, the circRNA interactome was used. Similarly, to predict the target DEmRNAs of the DEmiRNAs, we used TargetScan, miRTarBase and miRDB. Among the identified mRNAs, only those that showed consistency in at least these two databases were selected.
Enrichment analysis of DEmRNAs
The Gene Ontology (GO) analysis, such as biological process (BP), cellular component (CC) and molecular function (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEmRNAs were performed via DAVID database. 12
Feature selection based on machine learning methods
To screen vital RNAs, we used fivefold cross-validation with the R package “caret” to perform the SVM analysis. In all SVM progress, we set the random seed to 39. Moreover, we selected genes in the IRG set, which were obtained from the enrichment analysis. Finally, Venn’s method was used to identify key DEIRG(s) based on the SVM results, gene expression signatures, and immune gene set analyses results. Furthermore, to identify the critical ceRNAs that are regulated by the key IRG(s), other DERNAs were screened via SVM and expression-level analyses. 13 In addition, we used the LASSO method to identify gene signatures and obtain the respective coefficient values, and to fit the logistic LASSO regression, the glmnet package (version 2.0–16) was used.
Establishment of SCI model
The female Sprague–Dawley (SD) rats (age, 10–12 weeks; weight, 200–250 g; Xi’an Jiaotong University Health Science Center, Xi’an, China) were used. Rats were divided into the SCI and Sham groups (n = 4 in each group) randomly, and thereafter, they were anesthetized using 3% isoflurane, which was maintained at 1.5–2% during surgery. Rats in the SCI group underwent modified Allen’s weight-dropping injury, while those in the Sham group, they only underwent vertebral laminectomy. On day 1 after surgery, the BBB scores were used to ensure that the SCI model was made successfully, and at 3 days after injury and laminectomy in the SCI and control rats, respectively, the spinal cords of the rats in both groups were removed, and 10 mm long segments containing the injury site were quickly obtained for quantitative real-time polymerase chain reaction (qRT-PCR) analyses. 14 All the procedures involving the animals were approved by Xi’an Jiaotong University Ethics Committee and were performed in compliance with the Guide for the Care and Use of Laboratory Animals (8th Ed., 2011, The National Academies, USA).
Total RNA extraction and quantitative real-time RT-PCR
TRIzol (Invitrogen, Carlsbad, CA, USA) was used to isolate total RNA from the spinal cord segments. Furthermore, the First-Strand cDNA Synthesis Kits (Sangon Biotech, Shanghai, China) were used for the reverse transcription of mRNA, circRNA, and miRNA. The primers were as follows: hsa_circ_0026646 (forward, 5′-GATCTGCGTGGTCATGTTGG-3′; reverse, 5′-CAAAGATGACCGGAGAGCTG-3′), PLXNB2 (forward, 5′-CTTCAGCCTGATCCAGAGGTTTG-3′; reverse, 5′-GTGGAACACGTAGTCTGTACCC-3; Sangon Biotech). Furthermore, the primers of miR-331-3p were provided by GeneCopoeia (Guangzhou, China), and circPrimer2.0 was used to verify the primer specificity of hsa-circ_0026646. Furthermore, qRT-PCR was performed using the 2 × SG Fast qPCR Master Mix (Sangon, China), and the expression levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as an internal control for mRNAs and miRNAs, respectively. Gene expression analyses were performed using the 2−ΔΔCt method.
Luciferase activity assays
We used dual-luciferase reporter system to validate hsa_circ_0026646 with wild and mutant binding sites for miR-331-3p and the promoter region of PLXNB2. In addition, Renilla and firefly were transfected with miRNA transfection reagent (Invitrogen, Carlsbad, CA, USA) for 48 h, after which the luciferase intensity was measured. 15
Statistical analyses
R software 3.6.2 (The R Foundation for Statistical Computing, Vienna, Austria) and SPSS software 22.0 (SPSS Inc., Chicago, IL, USA) were used for statistical analyses. GraphPad Prism software 9.0 (GraphPad Prism Inc., San Diego, CA, USA)) and R software were used for the analysis of graphics. Student’s t-tests were used to assess statistical significance, the chi-square test was used for enrichment analyses, p < 0.05 was considered statistically significant.
Results
Identification of DEIRGs, DEmiRNAs, and DEcircRNAs
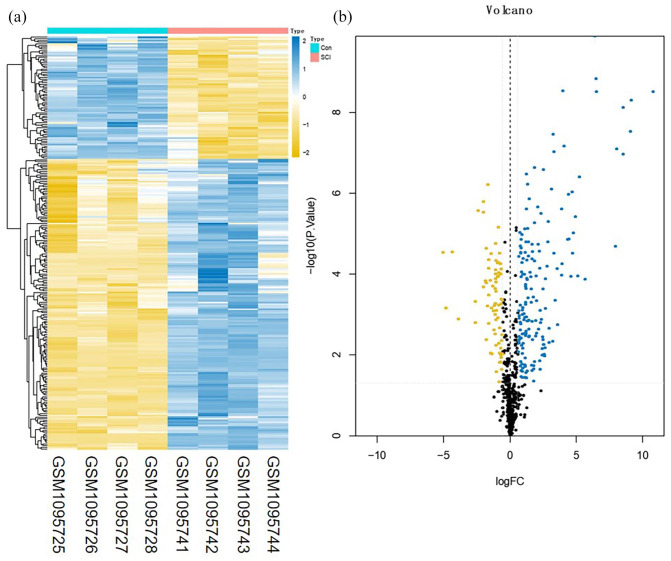
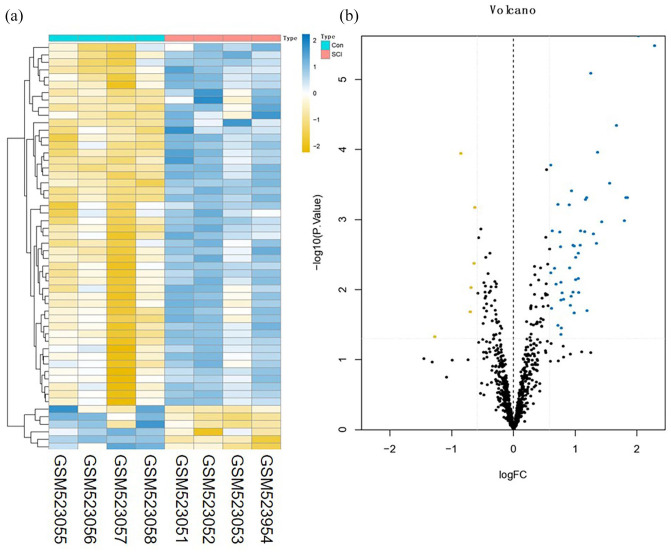
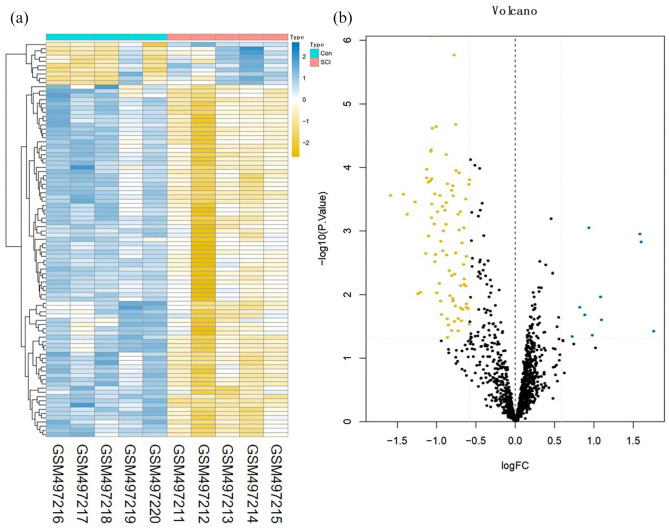
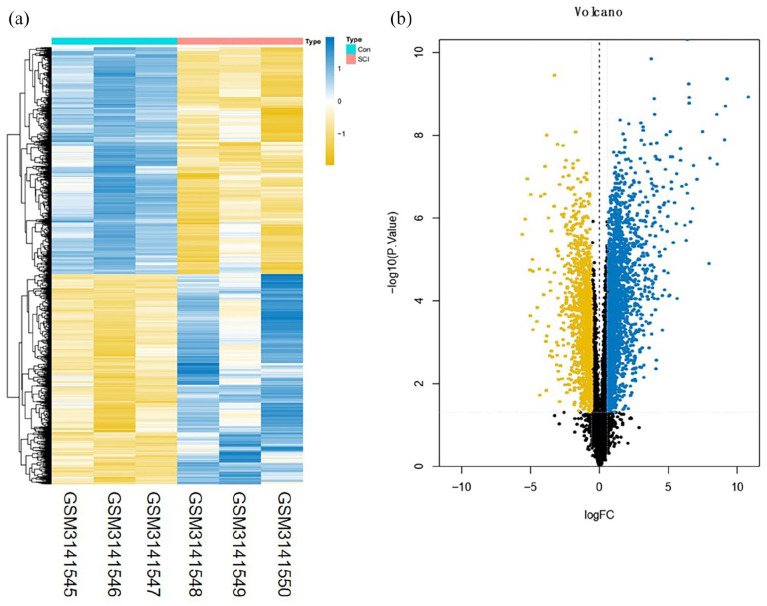
A total of 3273 DEmRNAs, including 246 DEIRGs were directly identified in the GSE45006 dataset, while a total of 357 DEmRNAs, including 54 DEIRGs were directly identified in the GSE20907 dataset. Furthermore, the determination of the intersection between the GSE45006 and GSE20907 datasets led to the identification of 32 uniformly DEIRGs (Figures 2 to 4 and Supplementary Table1). Furthermore, 74 DEmiRNAs were identified in the GSE19890 dataset, while 58 DEcircRNAs were identified on GSE114426. Notably, all the identified RNAs were human and rats co-expressed RNAs.
Figure 2.
Difference analysis of the GSE45006 dataset. (a) mRNA heat map of the GSE45006 dataset and (b) volcano plot of the GSE45006 dataset. The blue and yellow dots represent statistically significant high and low RNA expression levels based on a comparison of the SCI and control groups, respectively. The black dots represent differences without statistical significance. (A color version of this figure is available in the online journal.)
Figure 4.
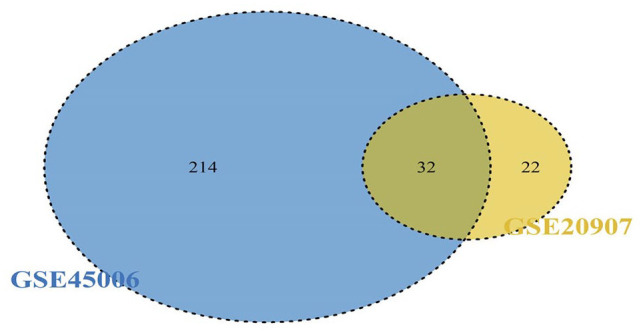
Intersection between the GSE45006 and GSE20907 datasets. (A color version of this figure is available in the online journal.)
Figure 3.
Difference analysis of the GSE20907 dataset. (a) mRNA heat map of the GSE20907 dataset and (b) volcano plot of the GSE20907 dataset. The blue and yellow dots represent statistically significant high and low RNA expression levels based on a comparison of the SCI and control groups, respectively. The black dots represent differences without statistical significance. (A color version of this figure is available in the online journal.)
Functional enrichment analyses
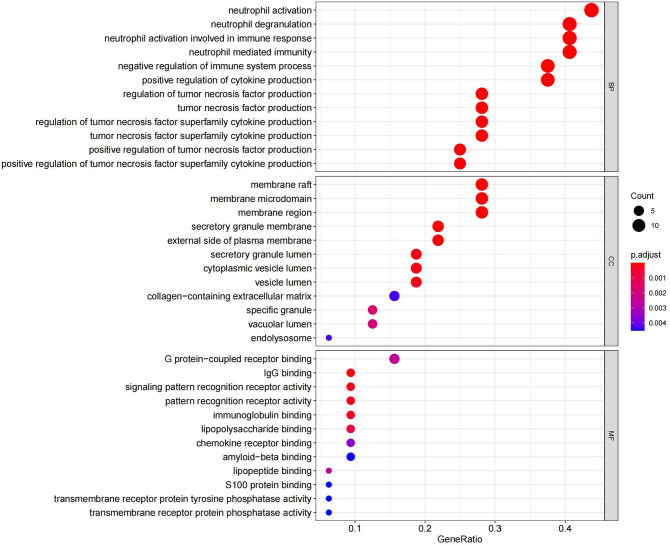
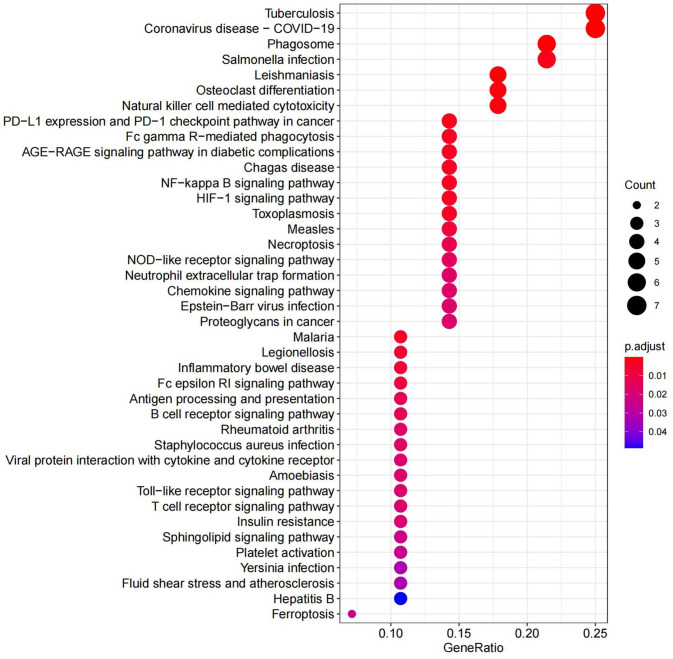
GO analysis listed the top 36 terms of DEmRNAs, and in terms of BP, the DEmRNAs were found to be primarily enriched in neutrophil-related immune response and regulation of cytokine production. Furthermore, in terms of CC, the DEmRNAs were found to be primarily enriched in membrane rafts, membrane microdomains, and membrane regions. Furthermore, the DEmRNAs were significantly enriched in the secretory granule membrane and the external side of plasma membrane. Some enrichments related to MF, such as immunoglobulin G (IgG) binding and pattern recognition receptor activity, were also observed (Figure 5 and Supplementary Table 2). In addition, KEGG pathway analysis indicated that DEmRNAs were mainly enriched in NF-κB and HIF-1 pathways (Figure 6 and Supplementary Table 3).
Figure 5.
Top enriched GO terms corresponding to DEmRNAs. (A color version of this figure is available in the online journal.)
GO: gene ontology; BP: biological process; CC: cellular component; MF: molecular function.
Figure 6.
Top enriched KEGG pathways corresponding to DEmRNAs (KEGG, Kyoto Encyclopedia of Genes and Genomes). (A color version of this figure is available in the online journal.)
Target prediction for DEcircRNAs, DEmiRNA, and DEIRGs
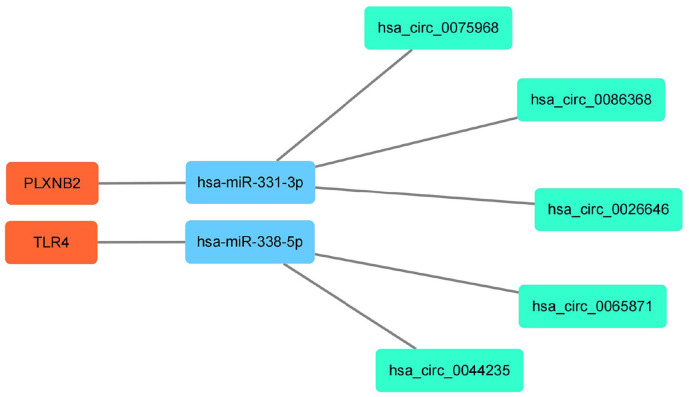
Of the 58 DEcircRNAs, five could bind with target miRNAs. However, when the miRDB, miRTarBase, and TargetScan databases were used, 4060 target mRNAs were predicted by 13 miRNAs in all 74 DEmiRNAs. Furthermore, among the 32 DEmRNAs, two which bind to two DEmiRNAs, were identified as the overlapping genes. Finally, we observed that hsa-miR-331-3p and hsa-miR-338-5p were DEmiRNAs predicted by the DEcircRNAs. Thus, focusing on miR-331-3p and miR-338-5p, five upstream circRNAs that could bind to it, including hsa_circ_0075968, hsa_circ_0086368, hsa_circ_0026646, hsa_circ_0065871, and hsa_circ_0044235. Furthermore, two downstream mRNAs, PLXNB2, and TLR4, related to miR-331-3p and miR-338-5p, were predicted, and to explore the potential regulation signatures of SCI, we constructed a circRNA/miRNA/mRNA network using the overlapping DEcircRNAs and DEIRGs. This constructed ceRNA network displayed five miRNA/circRNA links and two miRNA/mRNA links (Figures 7 to 9).
Figure 7.
Difference analysis of the GSE19890 dataset. (a) miRNA heat map of the GSE19890 dataset and (b) volcano plot of the GSE19890 dataset. The blue and yellow dots represent statistically significant high and low RNA expression levels based on a comparison of the SCI and control groups, respectively. The black dots represent differences without statistical significance. (A color version of this figure is available in the online journal.)
Figure 9.
A circRNA/miRNA/mRNA network in SCI. Green, blue, and orange represent circRNAs, miRNAs, and mRNAs, respectively. (A color version of this figure is available in the online journal.)
Figure 8.
Difference analysis of the GSE114426 dataset. (a) circRNA heat map of the GSE114426 dataset and (b) volcano plot of the GSE114426 dataset. The blue and yellow dots represent statistically significant high and low RNA expression levels based on a comparison of the SCI and control groups, respectively. The black dots represent differences without statistical significance. (A color version of this figure is available in the online journal.)
Selection of core DERNAs based on SVM and LASSO methods
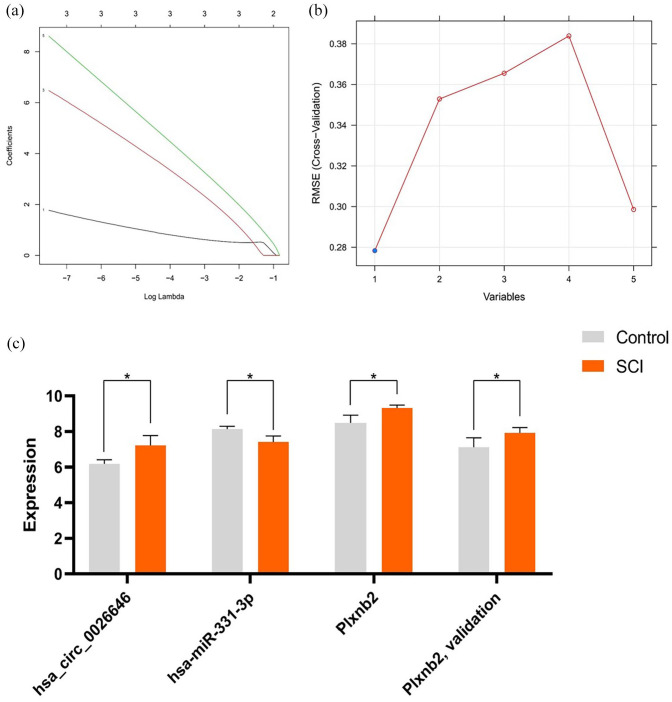
In the GSE45006 and GSE20907 datasets, we identified 32 overlapping DEIRGs, and the immune gene set was found to consist of 1793 genes. Furthermore, based on training using the SVM and LASSO methods, hsa_circ_0026646 was identified as the core DEcircRNA, while PLXNB2 was identified as the core DEIRG (Figure 10(a) and (b)). The further verification of the prediction results in the database showed that hsa_circ_0026646 and PLXNB2 were upregulated, whereas miRNA-331-3p was downregulated (Figure 10(c) and Supplementary Table 4).
Figure 10.
Screening for key IRGs and associated ceRNAs and the application SVM and LASSO to screen core DEcircRNA. (a) Application of LASSO to screen core circRNAs and (b) application of SVM to screen core circRNAs and (c) expression profiles of hsa_circ_0026646, miR-331-3p, and PLXNB2 in selected GEO datasets. *p < 0.05. (A color version of this figure is available in the online journal.)
Construction and confirmation of circRNA/miRNA/mRNA interactions
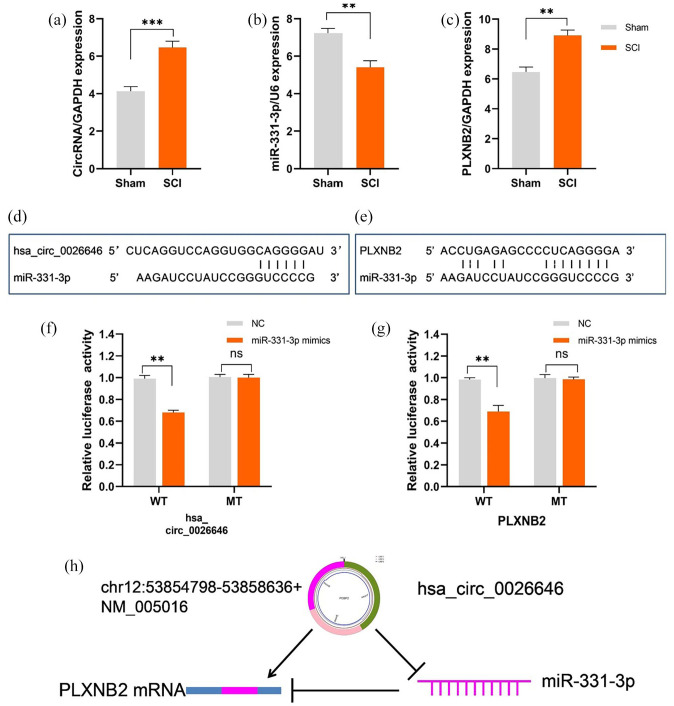
The relative expression levels of RNAs were measured via qRT-PCR. On day 3 after SCI, the miRNA expression level corresponding to the rats in the SCI group was markedly lower than that corresponding to those in the Sham group. However, their hsa_circ_0026646 and PLXNB2 expression levels were markedly higher than those corresponding to rats of Sham group (Figure 11(a) to (c)). In addition, consistent with our prediction, fluorescent reporter assay showed that both hsa_circ_0026646 and PLXNB2 had miR-331-3p target sites (Figure 11(d) to (g)). The schematic diagram displaying the hsa_circ_0026646/miR-331-3p/PLXNB2 regulatory axis is shown in Figure 11(h).
Figure 11.
circRNA/miRNA/mRNA network analysis. Relative expression levels of: (a) hsa_circ_0026646, (b) miR-331-3p, and (c) PLXNB2 in spinal cord tissues determined via qRT-PCR on day 3 post-SCI. (d) The miR-331-3p target site in the 3′UTR of hsa_circ_002664, (e) the miR-331-3p target site in the 3′UTR of PLXNB2, (f) relative luciferase intensity of wild-type (WT) and mutant-type (MT) hsa_circ_0026646 bearing luciferase vectors co-transfected with miR-331-3p expression vectors, (g) relative luciferase intensity of WT and MT PLXNB2 UTR-bearing luciferase vectors co-transfected with miR-331-3p expression vectors, (h) schematic diagram of the hsa_circ_0026646/miR-331-3p/PLXNB2 regulatory axis. **p < 0.01; ***p < 0.001. (A color version of this figure is available in the online journal.)
Discussion
Based on ongoing research on the pathological mechanism of SCI, it has been reported that ncRNAs, including miRNAs, circRNAs, and lncRNAs, play important roles, such as inflammation regulation, glial cell repair, axon regeneration, and angiogenesis, in acute SCI.16–18 It has also been reported that IRGs participate in a variety of human disease processes, and immune responses represent one of the key processes in SCI, especially in secondary injury after acute trauma. However, the key genes that are associated with this immune response and their ceRNA mechanism are still unclear.
In our study, we constructed a ceRNA regulation network to explore the key DEIRGs as well as their potential regulatory mechanisms in SCI pathogenesis. First, using SCI microarray datasets, we identified the DEIRGs between the SCI and control groups, and GO and KEGG analyses showed that the functions of these DEIRGs were primarily affected by neutrophil-related immune responses as well as the NF-κB and HIF-1 pathways. In addition, we identified 58 DEcircRNAs and 74 DEmiRNAs from the GEO microarray datasets, and after online database prediction, two DEmiRNAs and five DEcircRNAs were obtained. Furthermore, miR-331-3p was identified as the only DEmiRNA that was correctly predicted for circRNA/miRNA and miRNA/mRNA binding.
In this study, 32 overlapping DEIRGs and 1793 genes were also defined using the immune gene set. Specifically, hsa_circ_0026646 was identified as the core circRNA based on the results of SVM and LASSO analysis. Furthermore, PLXNB2 was identified as the key IRG, and predication based on the online database showed that hsa_circ_0026646 directly targets miR-331-3p, while miR-331-3p also directly targets PLXNB2, indicating that hsa_circ_0026646, miR-331-3p, and PLXNB2 possibly form an axis that regulates immune response in SCI. In addition, based on microarray data, gene expression validation further confirmed the credibility of this axis. Our results based on the SCI rat model also indicated that hsa_circ_0026646, miR-331-3p, and PLXNB2 formed an axis in SCI, indicating that miR-331-3p expression was downregulated in SCI, while PLXNB2 expression was upregulated. Moreover, the expression of hsa_circ_0026646, acting as a “molecular sponge” adsorbing miR-331-3p, was significantly up regulated during SCI.
Plexin-B2 (PLXNB2), which was first identified in malignant brain tumors, 19 participates in glioma malignancy and neuronal development.20–22 It is located on chromosome 22 and consists of 37 coding exons. Furthermore, PLXNB2 is a transmembrane protein, which consists of 1838 amino acids. It has a PDZ motif domain in the cell, as well as the rho GTPase-binding domain (RBD) and the GTPase activation protein (GAP) domain that interacts with the PDZ domain, which can regulate cell biological functions via intracellular signal transduction. It has also been reported that PLXNB2 is upregulated during development, while its expression in adult tissues is limited, primarily in neural stem cells. 23 In addition, it is primarily expressed in endothelial and epithelial cells as well as in immune cells, including monocytes, dendritic cells, and macrophages. Studies have demonstrated that it is constitutively expressed in mouse keratinocytes and has higher expression levels during wound repair. 23 After CD100 binds to its receptors, PLXNB1 and PLXNB2, it can activate the Rho GTPase nucleotide exchange receptor protein, RhoGEF, promote the activities of RhoA, RhoC, and Rac1, and regulate cell biological functions via signal transduction. 24
Previous studies have proved that Rho GTPase activates NF-κB signaling and that PLXNB2 regulates RhoA and Rac1 signaling to promote tumor cell invasion and migration. 21 It has also been demonstrated that blocking Rac1 GTPase can decrease the stability of HIF-1α and inhibit the expression of HIF-1α target molecules, PKM2 and GLUT1, in neuronal cells, thereby inhibiting neuronal glycolysis and metabolism. 25 In other words, a high PLXNB2 expression level can promote HIF-1α expression in neurons, thereby exerting neuroprotection effects. In addition, PLXNB2 also regulates the neurogenesis and neuroprotective activities of angiogenin. 26 Coincidentally, another study by our research team showed that PLXNB2 is upregulated in injury-activated microglia and macrophages (IAMs), it can protect tissues from damage and promote axon regeneration after SCI. 27
The abovementioned findings are consistent with our results in this study. Specifically, here, we identified PLXNB2 as a differential gene, whose expression was significantly upregulated after SCI. GO analysis also showed that it participates in the NF-κB and HIF-1α signaling pathways. However, few studies have reported the role of PLXNB2 in SCI so far; thus, the specific mechanism still needs to be confirmed via follow-up studies.
Reportedly, miRNAs participate in the pathological process of SCI, including inflammation regulation, glial cell repair, nerve repair, and angiogenesis, during SCI.28–31 Presently, research on miR-331-3p is mainly focused on its effect on tumors.32–34 Although one study has shown that miR-331-3p could attenuate neuropathic pain following SCI by targeting RAP1A, 31 studies on its specific role in the neuron repair process are scarce. Some studies have been conducted to investigate its function in the nervous system; however, the results are inconsistent. For example, Liu and Lei 35 reported that in patients with Alzheimer’s disease (AD) and in Abeta (1–40)-treated SH-SY5Y cells, miR-331-3p expression is significantly downregulated and shows correlation with the mini-mental state examination (MMSE) scores and proinflammatory cytokine levels in AD patients, indicating that possibly, it plays a neuroprotective role. However, a study conducted by Chen et al. led to an opposite conclusion. Basically, Chen et al. 36 observed that the overexpression of miR-331-3p in the SH-SY5Y cell line impairs autophagic activity and promotes AD progression. Therefore, the role of miR-331-3p in the nervous system still needs to be further investigated.
In addition, the roles and functions of circRNAs, such as circRim2 and circPlxnd1, in the central nervous system have been sufficiently explored; 37 however, studies with a focus on their effects on SCI are limited. In this study, based on bioinformatics analysis, we first identified hsa_circ_0026646, which was significantly upregulated in SCI, as a DEcircRNA, and via targeting site prediction, we observed that both PLXNB2 and hsa_circ_0026646 have miRNA-331-3p binding sites. Our analysis also indicated that PLXNB2 was upregulated in SCI, while miRNA-331-3p was downregulated. These same expression profiles were observed in the rat SCI models via qRT-PCR, and fluorescent reporter assay also confirmed that both hsa_circ_0026646 and PLXNB2 have miR-331-3p target sites. Therefore, we speculated that hsa_circ_0026646 exerts a neuroprotective effect by binding to miRNA-331-3p.
Based on bioinformatics analysis as well as experimental analysis based on the rat SCI model, this study indicated that DERNAs are closely correlated with post-SCI pathophysiology processes. Our findings provided new evidence of the existence of a ceRNA regulation mechanism in SCI. circRNAs are characterized by variety, a stable structure, sequence conservation, and cell- or tissue-specific expression, and compared with miRNAs, they are more likely to be an intervention strategy for SCI management. In future, we will explore the neuroprotective and axon regeneration effects of has_circ_0026646, and it is expected that the results obtained would further confirm its potential therapeutic effect.
Conclusions
In summary, we established a hsa_circ_0026646/miR-331-3p/PLXNB2 network. This provides new insights into the regulatory mechanism associated with SCI as well as potential therapeutic targets for its management.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221082929 for Identification of a circRNA-mediated comprehensive ceRNA network in spinal cord injury pathogenesis by Chao Zu, Jingyuan Li, Xijing He, Le Ji and Xia Li in Experimental Biology and Medicine
Footnotes
Authors’ Contributions: CZ and JYL collected the microarray data and CZ wrote the manuscript. CZ, JYL, and LJ analyzed the data, XL provided technical support, and XJH designed the study.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Natural Science Foundation of Shaanxi Province (grant number 2021JQ-746), the Social Development Project of Shaanxi Province (grant numbers 2021SF-285 and 2020SF-092), the Fundamental Research Funds for the Central Universities (grant number xzy012019111), the China Postdoctoral Science Foundation (grant number 2019M653736), the National Key Research and Development Program of China (grant number 2018YFE0114200), the National Natural Science Foundation of China (grant number 81771349), and the Key Research and Development Project of Shaanxi Province (grant number 2017ZDCXL-SF-01-05).
ORCID iD: Xijing He  https://orcid.org/0000-0002-0465-5376
https://orcid.org/0000-0002-0465-5376
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Zheng XY, Yi Q, Xu XJ, Meng RL, Ma SL, Tang SL, Xu HF, Xu YS, Xu YJ, Yang Y. Trends and external causes of traumatic brain injury and spinal cord injury mortality in south China, 2014-2018: an ecological study. BMC Public Health 2021;21:2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee BB, Cripps RA, Fitzharris M, Wing PC. The global map for traumatic spinal cord injury epidemiology: update 2011, global incidence rate. Spinal Cord 2014;52:110–6 [DOI] [PubMed] [Google Scholar]
- 3. Samantaray S, Das A, Matzelle DC, Yu SP, Wei L, Varma A, Ray SK, Banik NL. Administration of low dose estrogen attenuates gliosis and protects neurons in acute spinal cord injury in rats. J Neurochem 2016;136:1064–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Przekora A, Juszkiewicz L. The effect of autologous adipose tissue-derived mesenchymal stem cells’ therapy in the treatment of chronic posttraumatic spinal cord injury in a domestic ferret patient. Cell Transplant 2020;29:963689720928982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alizadeh A, Dyck SM, Karimi-Abdolrezaee S. Traumatic spinal cord injury: an overview of pathophysiology, models and acute injury mechanisms. Front Neurol 2019;10:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahuja CS, Wilson JR, Nori S, Kotter MRN, Druschel C, Curt A, Fehlings MG. Traumatic spinal cord injury. Nat Rev Dis Primers 2017;3:17018. [DOI] [PubMed] [Google Scholar]
- 7. Beck KD, Nguyen HX, Galvan MD, Salazar DL, Woodruff TM, Anderson AJ. Quantitative analysis of cellular inflammation after traumatic spinal cord injury: evidence for a multiphasic inflammatory response in the acute to chronic environment. Brain 2010;133:433–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhuo CJ, Hou WH, Jiang DG, Tian HJ, Wang LN, Jia F, Zhou CH, Zhu JJ. Circular RNAs in early brain development and their influence and clinical significance in neuropsychiatric disorders. Neural Regen Res 2020;15:817–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014;9:1966–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng P, Zhang B, Huang J, Xing C, Liu W, Sun C, Guo W, Yao S, Ruan W, Ning G, Kong X, Feng S. Identification of a circRNA-miRNA-mRNA network to explore the effects of circRNAs on pathogenesis and treatment of spinal cord injury. Life Sci 2020;257:118039. [DOI] [PubMed] [Google Scholar]
- 13. Cao S, Li J, Yang K, Li H. Major ceRNA regulation and key metabolic signature analysis of intervertebral disc degeneration. BMC Musculoskelet Disord 2021;22:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fu X, Shen Y, Wang W, Li X. MiR-30a-5p ameliorates spinal cord injury-induced inflammatory responses and oxidative stress by targeting Neurod 1 through MAPK/ERK signalling. Clin Exp Pharmacol Physiol 2018;45:68–74 [DOI] [PubMed] [Google Scholar]
- 15. Wang W, He D, Chen J, Zhang Z, Wang S, Jiang Y, Wei J. Circular RNA Plek promotes fibrogenic activation by regulating the miR-135b-5p/TGF-βR1 axis after spinal cord injury. Aging (Albany NY) 2021;13: 13211–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi Z, Zhou H, Lu L, Li X, Fu Z, Liu J, Kang Y, Wei Z, Pan B, Liu L, Kong X, Feng S. The roles of microRNAs in spinal cord injury. Int J Neurosci 2017;127:1104–15 [DOI] [PubMed] [Google Scholar]
- 17. Ning B, Gao L, Liu RH, Liu Y, Zhang NS, Chen ZY. microRNAs in spinal cord injury: potential roles and therapeutic implications. Int J Biol Sci 2014;10:997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang WZ, Li J, Liu L, Zhang ZD, Li MX, Li Q, Ma HX, Yang H, Hou XL. Role of circular RNA expression in the pathological progression after spinal cord injury. Neural Regen Res 2021;16:2048–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shinoura N, Shamraj OI, Hugenholz H, Zhu JG, McBlack P, Warnick R, Tew JJ, Wani MA, Menon AG. Identification and partial sequence of a cDNA that is differentially expressed in human brain-tumors. Cancer Lett 1995;89:215–21 [DOI] [PubMed] [Google Scholar]
- 20. Daviaud N, Chen K, Huang Y, Friedel RH, Zou H. Impaired cortical neurogenesis in plexin-B1 and-B2 double deletion mutant. Dev Neurobiol 2016;76:882–99 [DOI] [PubMed] [Google Scholar]
- 21. Le AP, Huang Y, Pingle SC, Kesari S, Wang H, Yong RL, Zou H, Friedel RH. Plexin-B2 promotes invasive growth of malignant glioma. Oncotarget 2015;6:7293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deng S, Hirschberg A, Worzfeld T, Penachioni JY, Korostylev A, Swiercz JM, Vodrazka P, Mauti O, Stoeckli ET, Tamagnone L, Offermanns S, Kuner R. Plexin-B2, but not Plexin-B1, critically modulates neuronal migration and patterning of the developing nervous system in vivo. J Neurosci 2007;27:6333–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saha B, Ypsilanti AR, Boutin C, Cremer H, Chedotal A. Plexin-B2 regulates the proliferation and migration of neuroblasts in the postnatal and adult subventricular zone. J Neurosci 2012;32:16892–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu W, Goncalves KA, Li S, Kishikawa H, Sun G, Yang H, Vanli N, Wu Y, Jiang Y, Hu MG, Friedel RH, Hu G-f. Plexin-B2 mediates physiologic and pathologic functions of angiogenin. Cell 2017;171:849–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malik MFA, Ye L, Jiang WG. The plexin-B family and its role in cancer progression. Histol Histopathol 2014;29:151–65 [DOI] [PubMed] [Google Scholar]
- 26. Guntert T, Gassmann M, Ogunshola OO. Temporal Rac1-HIF-1 crosstalk modulates hypoxic survival of aged neurons. Brain Res 2016;1642:298–307 [DOI] [PubMed] [Google Scholar]
- 27. Zhou X, Wahane S, Friedl MS, Kluge M, Friedel CC, Avrampou K, Zachariou V, Guo L, Zhang B, He X, Friedel RH, Zou H. Microglia and macrophages promote corralling, wound compaction and recovery after spinal cord injury via Plexin-B2. Nat Neurosci 2020;23:337–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howe JR, 6th, Li ES, Streeter SE, Rahme GJ, Chipumuro E, Russo GB, Litzky JF, Hills LB, Rodgers KR, Skelton PD, Luikart BW. MiR-338-3p regulates neuronal maturation and suppresses glioblastoma proliferation. PLoS ONE 2017;12:e0177661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li F, Zhou MW. MicroRNAs in contusion spinal cord injury: pathophysiology and clinical utility. Acta Neurol Belg 2019;119:21–7 [DOI] [PubMed] [Google Scholar]
- 30. Tang Y, Ling ZM, Fu R, Li YQ, Cheng X, Song FH, Luo HX, Zhou LH. Time-specific microRNA changes during spinal motoneuron degeneration in adult rats following unilateral brachial plexus root avulsion: ipsilateral vs. contralateral changes. Bmc Neurosci 2014;15:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Guo H, Xie A, Liao O, Ju F. microRNA-331-3p attenuates neuropathic pain following spinal cord injury via targeting RAP1A. J Biol Regul Homeost Agents 2020;34:25–37 [DOI] [PubMed] [Google Scholar]
- 32. Chang RM, Yang H, Fang F, Xu JF, Yang LY. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology 2014;60:1251–63 [DOI] [PubMed] [Google Scholar]
- 33. Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem 2009;284:24696–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fujii T, Shimada K, Asano A, Tatsumi Y, Yamaguchi N, Yamazaki M, Konishi N. MicroRNA-331-3p suppresses cervical cancer cell proliferation and E6/E7 expression by targeting NRP2. Int J Mol Sci 2016;17:1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Q, Lei C. Neuroprotective effects of miR-331-3p through improved cell viability and inflammatory marker expression: correlation of serum miR-331-3p levels with diagnosis and severity of Alzheimer’s disease. Exp Gerontol 2021;144:111187. [DOI] [PubMed] [Google Scholar]
- 36. Chen ML, Hong CG, Yue T, Li HM, Duan R, Hu WB, Cao J, Wang ZX, Chen CY, Hu XK, Wu B, Liu HM, Tan YJ, Liu JH, Luo ZW, Zhang Y, Rao SS, Luo MJ, Yin H, Wang YY, Xia K, Tang SY, Xie H, Liu ZZ. Inhibition of miR-331-3p and miR-9-5p ameliorates Alzheimer’s disease by enhancing autophagy. Theranostics 2021;11:2395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015;58:870–85 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221082929 for Identification of a circRNA-mediated comprehensive ceRNA network in spinal cord injury pathogenesis by Chao Zu, Jingyuan Li, Xijing He, Le Ji and Xia Li in Experimental Biology and Medicine