Abstract
Osteosarcoma (OS) differentially expressed genes (DEGs) have been predicted using the data portal of the Therapeutically Applicable Research to Generate Effective Treatments (TARGET). In this study, we sought to identify cell types that specially express key DEGs (MUC1, COL13A1, JAG2, and KAZALD1) in each of the nine identified cell populations derived from tissues of OS tumors with single-cell RNA-sequencing data. Gene expression levels were pairwise compared between cell clusters and a p value < 0.05 was considered differentially expressed. It was revealed that MUC1 is expressed at high levels in osteoblastic OS cells followed by carcinoma-associated fibroblasts (CAFs) and plasmocytes, respectively. COL13A1 is highly expressed in osteoblastic OS cells, CAFs, and endothelial cells (ECs), respectively. The KAZALD1 gene is expressed in CAFs and osteoblastic OS cells at high levels, but at very low levels in plasmocytes, osteoclasts, NK/T, myeloid cells 1, myeloid cells 2, ECs, and B cells. JAG2 is expressed at significantly high levels in ECs and osteoblastic OS cells, and at relatively lower levels in all other cell types. Interestingly, LSAMP, as an established gene in the development of OS shows high expression in osteoblastic OS cells and CAFs but low in other cells such as osteoclasts. Our findings here highlight the heterogeneity of OS cells and cell-type-dependent DEGs which have potential as therapeutic targets in OS.
Keywords: Osteosarcoma, TARGET, differentially expressed genes, single-cell sequencing, sarcoma, heterogeneity
Impact Statement
Osteosarcoma (OS) is a primary malignant bone tumor predominately affecting children and adolescence. Standard combination therapy involves surgery and chemotherapy; however, this treatment regimen faces risks of local relapse and drug resistance in the clinic. This study using single-cell genomics data characterizes the regulation of OS tumor cells by four differentially expressed genes (DEGs) whose precise expression is yet to be explored in-depth. These pivotal findings are clinically relevant and support the development of precision and personalized therapy in OS and strategies to overcome the drug-resistant nature of OS.
Introduction
Osteosarcoma (OS) is a highly malignant solid bone tumor that is characterized by malignant mesenchymal cells that produce pathological osteoid and/or bony matrix.1–3 Also known as osteogenic sarcoma, OS tumors are distinguished by rapid growth, invasion of nearby tissues, and a high tendency to metastasize to the lungs and distant bones. 4 Typically, OS occurs near the metaphysis of long bones in regions of rapid growth, such as the arms, legs, knees, and shoulders.5,6 OS is a relatively rare condition, representing only 0.2% of overall tumor burdens, but it is the most commonly diagnosed pediatric bone malignancy.2,7
Globally, OS affects 3.4 per million people per year and exhibits a high incidence rate among children, adolescents, and young adults.5,8,9 OS also has a second smaller peak incidence among elderly individuals.7,10 Since the introduction of neoadjuvant chemotherapy in the 1970s, the 5-year survival rate for children and adolescents with non-metastatic OS, although poor, has improved from a dismal 20% to 70%. 8 Nevertheless, survival outcomes have not shown considerable improvement in the last two decades, and therefore, new, less toxic therapies are needed. 11
Advances have shown that OS tumors are complex ecosystems composed of distinct populations of cells, such as endothelial cells (ECs), osteoblastic OS cells, and macrophages, which have different genotypes, phenotypes, and cell behavior. 12 However, the current knowledge of the etiology and molecular mechanisms of OS progression remains unclear and contentious.13,14 Recently, single-cell RNA sequencing (scRNA seq) has become a valuable tool in cancer research that has provided exciting insights into complex biological systems by identifying cell lineages, novel cell subpopulations, regulatory networks between genes, and cell-specific biological characteristics. 15 In particular, scRNA seq studies have revealed high intra-tumor heterogeneity of OS and varied expression of several genes across many OS cell types, which could not have been determined with conventional bulk sequencing data sets.12,16
The Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative is an open-source public database for the treatment of childhood cancers. 17 The objective of the TARGET project is to utilize genomic data for the development of effective and safe treatments. We have recently identified four differentially expressed genes (DEGs) in OS using the data portal of TARGET. 17 Given that OS cell types are expected to differ in gene expression profiles, the aim of this study is to detect and compare DEGs in tumor cell types from OS patients by bioinformatic analysis of data sets of single-cell gene expression profiles from the TARGET-OS project. This is the first study to characterize the expression of these DEGs in precise single OS cells. We hope that such efforts might provide critical insight into the underlying pathogenic mechanisms of OS and further improve clinical outcomes and diagnosis for patients with OS.
Materials and methods
ScRNA seq data acquisition and correlation to public data sets
The messenger RNA (mRNA) expression data used to identify four DEGs were downloaded from the TARGET database (https://ocg.cancer.gov/programs/target) as described previously. 17 The dissociation and single-cell RNA sequencing of OS tumors has been previously published by Liu. 12 Therefore, ethics approval or patient consent was exempted. The following scRNA seq data set GSE162454 was chosen for the analysis (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE162454). In total, 29,278 cells isolated from six primary tumor samples obtained from six OS patients were included in this data set. 12 We then screened four highly variable genes in single cells for downstream analysis.
Cell quality control
Data quality control was performed following the same quality control parameters of Seurat package (version 3.2.1) as the author’s previous paper. 12 To guarantee the quality of the data set cells with a gene number expressed between 300 and 4500 and mitochondrial gene percentage, less than 10% were filtered out. After filtering, there were a total of 29,278 cells available to identify DEGs. Finally, harmony package (version1.0; https://github.com/immunogenomics/Harmony) was used to consolidate the data.
Cell clustering and differential expression analysis in OS cells
We performed the Uniform Manifold Approximation and Projection (UMAP) cell clustering and visualization using the Dimplot function with parameter “dim = 1:30, resolution = 0.10.” We further adjusted the color using the “ggsci” package (version 2.9). The definition of cell types is consistent with our previous studies. 12 Next function FeaturePlot was implemented to show gene expression distribution.
Data processing
Count values of single-cell data were extracted and converted into Transcripts Per Kilobase Million (TPM) values by the following formula
Functional enrichment analysis
The Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis was performed in the DEGs between cell clusters using Metascape (http://metascape.Org). 18 GO and KEGG enrichment was used by researchers to explore the biological characteristics/function of DEGs. The statistical threshold for significance in GO and KEGG pathways was a false discovery rate (FDR) value–adjusted p value of <0.05.
Statistical analyses
All computational analysis and data visualization were performed using R version 3.6.3 (http://www.rproject.org). A p value lower than 0.05 was considered statistically significant. All statistical analysis results are summarized and presented in Supplementary Table 1.
Results
Characteristics of the OS tumor microenvironment
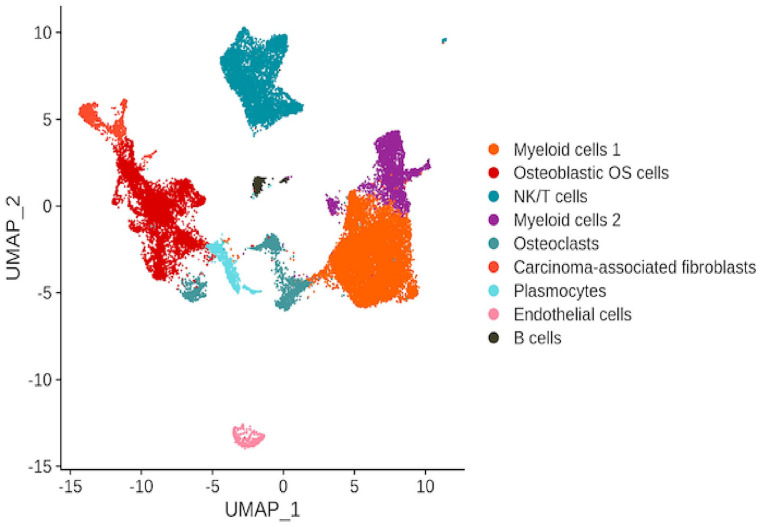
In cancer, high intra-tumoral heterogeneity and variation among cancer cells is tightly linked to invasive disease progression and therapeutic resistance. 19 Thus, to better characterize the cellular heterogeneity of OS, an analysis of the expression of DEGs in single cancer cells was performed. From 29,278 available cells, 9 main malignant cell clusters were identified by unique marker genes, including osteoblastic OS cells, NK/T cells, osteoclasts, carcinoma-associated fibroblasts (CAFs), plasmocytes, ECs, and B cells (Figure 1), and two subgroups of myeloid cells – myeloid cells 1 and myeloid cells 2 (Figure 1).
Figure 1.
Uniform Manifold Approximation and Projection (UMAP) depiction of scRNA seq data showing the nine main cell types in OS.
Identification of DEGs between OS cell clusters
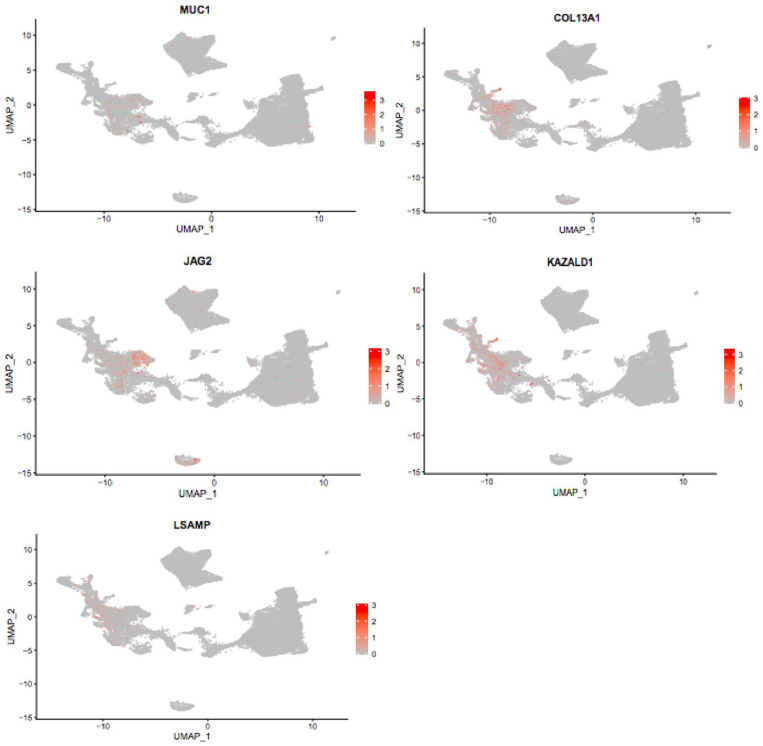
Previous bioinformatic analyses of RNA seq data indicate that the four genes, mucin 1-cell surface associated (MUC1), collagen type XIII alpha 1 chain (COL13A1), jagged canonical notch ligand 2 (JAG2), and kazal type serine peptidase inhibitor domain 1 (KAZALD1) are differentially expressed in OS. 17 However, these four OS DEGs have not been studied extensively at single-cell resolution and thus require further assessment. To more closely analyze the differences in expression patterns between the individual OS cell clusters, we selected the OS DEGs MUC1, COL13A1, JAG2, and KAZALD1 for further genetic analysis. The average expression per gene in each of the nine cell types was determined and compared pairwise. The expression patterns of each OS cell type were subsequently characterized by either higher or lower average expression of these selected genes. Our study demonstrated that significantly different gene expression patterns could be identified for these four OS DEG genes and the reference gene, LSAMP, for the nine cell clusters in the scRNA seq data, indicating cellular heterogeneity and transcriptional complexity in OS (Figure 2). The distinct gene expression patterns are characteristics of the tumors and not the patients. Our study revealed that particular genes did not differ in gene expression across many of the OS cell types and that many cells presented low expression of some genes. Interestingly, all four genes were highly expressed in the osteoblastic OS cell type. The COL13A1 gene, followed in order by JAG2, KAZALD1, and MUC1, respectively, showed the most significant differential expression changes among the OS cell clusters.
Figure 2.
Uniform Manifold Approximation and Projection (UMAP) plot showing the relative mRNA expression of MUC1, COL13A1, JAG2, and KAZALD1 in the nine main OS cell types.
DEG MUC1 is highly abundant in osteoblastic OS cells, CAFs, and plasmocytes
MUC1 expression was detected in all nine main cell types (Figure 3). Based on the cut-off criteria, the MUC1 gene was found to be expressed at higher levels in three of the OS cell clusters; these are plasmocytes, osteoblastic OS cells, and CAFs (Figure 3). In addition, according to our data, MUC1 was found to be expressed at relatively lower levels in four of the OS cell clusters; these are B cells, ECs, NK/T, myeloid 1, myeloid 2, and OCs (Figure 3). Notably, osteoblastic OS cells showed the highest average expression levels of MUC1 and the highest standard deviation among all other cell types (Figure 3). The second highest average MUC1 expression and standard deviation were found in the CAFs cluster (Figure 3). Relatively lower levels of MUC1 expression and a lower standard deviation were observed in plasmocytes compared with the clusters, CAFs and osteoblastic OS cells (Figure 3). A total of 36 pairwise comparisons between the nine major cell types were performed, and eight of these exhibited significant differences in MUC1 expression (Supplemental Table 1). We observed a significant difference in MUC1 expression between the following pairs of clusters: plasmocytes and osteoblastic OS cells (p < 0.0001), B cells and osteoblastic OS cells (p < 0.001), ECs and osteoblastic OS cells (p < 0.001), myeloid cells 1 and osteoblastic OS cells (p < 0.001), myeloid cells 2 and osteoblastic OS cells (p < 0.001), NK/T and osteoblastic OS cells (p < 0.001), osteoblastic OS cells and OCs (p < 0.001), and osteoblastic OS cells and plasmocytes (p < 0.001) (Supplemental Table 1).
Figure 3.
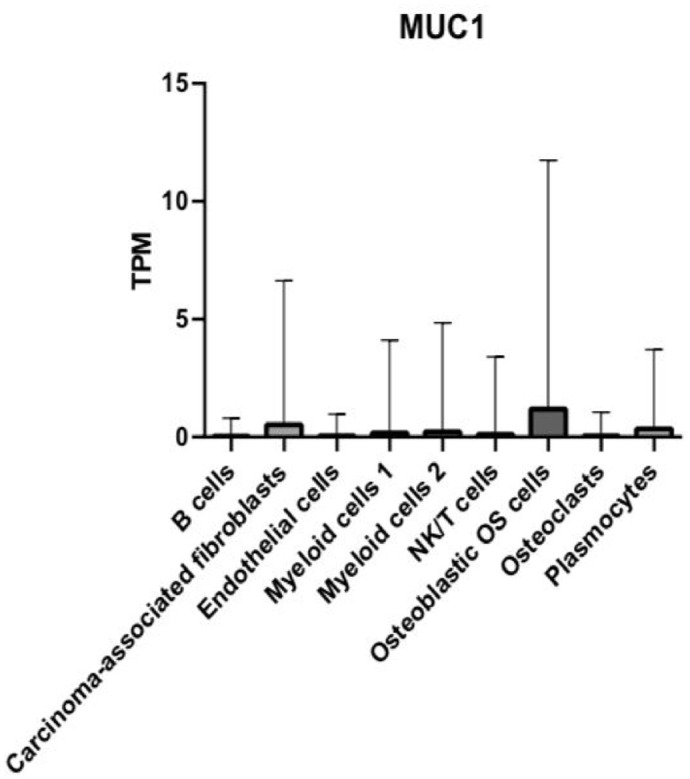
Bar graph depicting the relative mRNA expression of the MUC1 gene in TPM in the nine main OS cell types.
We found that MUC1 showed no significant difference in expression patterns between OCs and NK/T (p > 0.9999), OCs and ECs (p > 0.9999), OCs and B cells (p = 0.3029), NK/T and B cells (p = 0.1587), NK/T and ECs (p > 0.9999), plasmocytes and OCs (p = 0.3860), and ECs and B cells (p = 0.6674) (Supplemental Table 1). We also observed no significant difference in expression of MUC1 between plasmocytes and myeloid 2 (p = 0.7417), plasmocytes and myeloid 1 (p = 0.3051), ECs and plasmocytes (p = 0.7174), B cells and plasmocytes (p > 0.9999), B cells and myeloid 1 (p > 0.9999), B cells and myeloid 2 (p = 0.9999), ECs and myeloid 1 (p > 0.9999), ECs and myeloid 2 (0.9995), myeloid 1 and myeloid 2 (p = 0.9862), myeloid 1 and NK/T (p > 0.9999), myeloid 1 and OCs (p = 0.9958), myeloid 2 and OCs (p = 0.9958), and NK/T and plasmocytes (p = 0.2383) (Supplemental Table 1).
DEG COL13A1 is strongly enriched in osteoblastic OS cells, CAFs, and ECs
Next, we examined the average gene expression of COL13A1 in single OS cells. According to our analysis, COL13A1 displayed various levels of expression in all the nine cell types. Figure 4 highlights the relative abundance of COL13A1 expression in all cell types. COL13A1 expression is significantly higher in the three cell clusters CAFs, ECs, and osteoblastic OS cells compared with all other cell types (Figure 4). We also observed that, compared with all other cell types, COL13A1 was expressed at relatively low levels in the following six cell clusters, B cells, myeloid cells 1, myeloid cells 2, NK/T, OCs, and plasmocytes (Figure 4). Among the OS cell clusters, the most highly upregulated average expression of COL13A1 was found in the osteoblastic OS cell cluster (Figure 4). However, the highest standard deviation of COL13A1 expression was observed in the CAFs cluster (Figure 4). We performed a total of 36 pairwise comparisons of nine major cell types and 20 of these exhibited significant differential expression of COL13A1 (Supplemental Table 1). We observed a significant difference in the expression of COL13A1 between the following cell groups, B cells and ECs (p < 0.001), B cells and osteoblastic OS cells (p < 0.0001), B cells and CAFs (p < 0.0001), CAFs and myeloid cells 1 (p < 0.0001), CAFs and myeloid cells 2 (p < 0.0001), CAFs and NK/T cells (p < 0.0001), CAFs and osteoblastic OS cells (p < 0.0001), CAFs and OCs (p < 0.0001), and CAFs and plasmocytes (p < 0.0001) (Supplemental Table 1). We further observed significant difference in expression of COL13A1 between cell clusters ECs and myeloid cells 1 (p < 0.0001), ECs and myeloid cells 2 (p < 0.0001), ECs and NK/T (p < 0.0001), ECs and osteoblastic OS cells (p < 0.0001), ECs and OCs (p < 0.0001), ECs and plasmocytes (p < 0.0001), myeloid cells 1 and osteoblastic OS cells (p < 0.0001), myeloid cells 2 and osteoblastic OS cells (p < 0.0001), NK/T and osteoblastic OS cells (p < 0.0001), osteoblastic OS cells and OCs (p < 0.0001), and osteoblastic OS cells and plasmocytes (p < 0.0001) (Supplemental Table 1).
Figure 4.
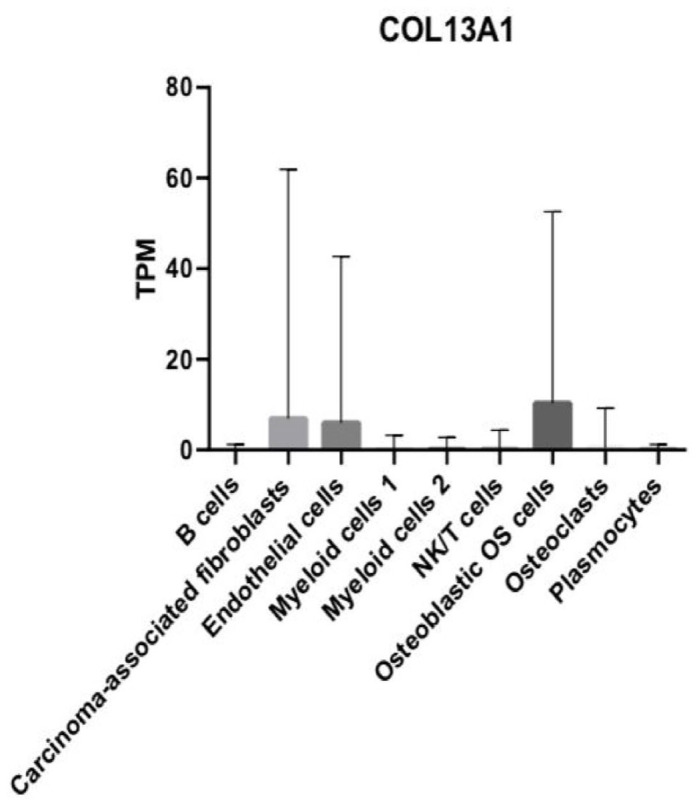
Bar graph depicting the relative mRNA expression of the COL13A1 gene in TPM in the nine main OS cell types.
DEG KAZALD1 is highly expressed in osteoblastic OS cells and CAFs
We quantified the average relative abundance of KAZALD1 in each of the tumor cell types and then compared the expression patterns of this gene between them. Our comparative analysis demonstrated that KAZALD1 is expressed in all nine cell types examined (Figure 5). We also found that KAZALD1 is expressed at higher levels in osteoblastic OS cells and CAFs than all other cell types (Figure 5). In addition, our data show that CAFs exhibited a higher standard deviation of average KAZALD1 expression than all other cell types (Figure 5). Notably, the expression of KAZALD1 was much weaker in the cell clusters of B cells, ECs, myeloid cells 1, myeloid cells 2, NK/T, OCs, and plasmocytes compared with all other cell types (Figure 5).
Figure 6.
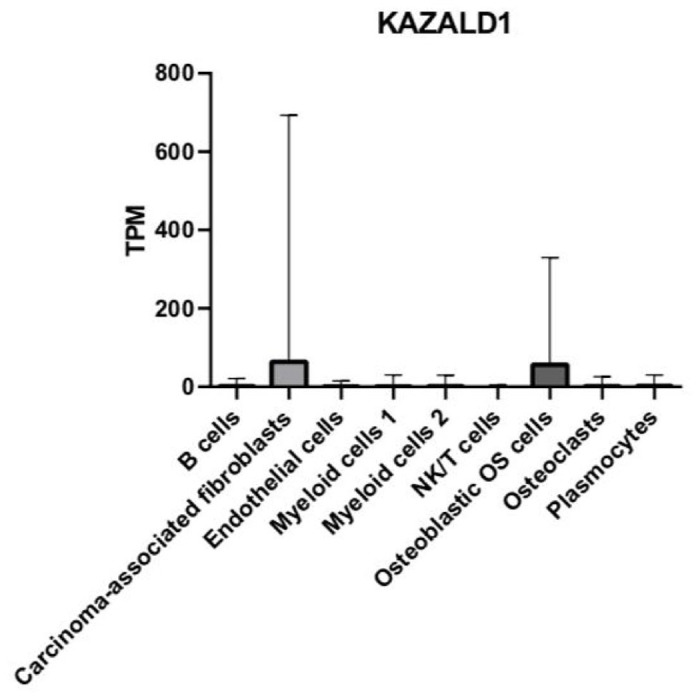
Bar graph depicting the relative mRNA expression of the JAG2 gene in TPM in the nine main OS cell types.
There was a total of 36 pairwise comparisons of nine major cell types, and 14 of these exhibited significant differential expression of KAZALD1 (Supplemental Table 1). We found significant differential expression of KAZALD1 between B cells and CAFs (p < 0.0001), B cells and osteoblastic OS cells (p < 0.0001), CAFs and ECs (p < 0.0001), CAFs and myeloid cells 1 (p < 0.0001), CAFs and myeloid cells 2 (p < 0.0001), CAFs and NK/T (p < 0.0001), CAFs and OCs (p < 0.0001), and CAFs and plasmocytes (p < 0.0001) (Supplemental Table 1). We also found significant differential expression of KAZALD1 between ECs and osteoblastic OS cells (p < 0.0001), myeloid cells 1 and osteoblastic OS cells (p < 0.0001), myeloid cells 2 and osteoblastic OS cells (p < 0.0001), NK/T and osteoblastic OS cells (p < 0.0001), osteoblastic OS cells and OCs (p < 0.0001), and osteoblastic OS cells and plasmocytes (p < 0.0001) (Supplemental Table 1).
We did not identify significant differential expression of KAZALD1 between B cells and ECs (p > 0.9999), B cells and myeloid cells 1 (p > 0.9999), B cells and myeloid cells 2 (p > 0.9999), B cells and NK/T cells (p > 0.9999), B cells and OCs (p > 0.9999), B cells and plasmocytes (p > 0.9999), CAFs and osteoblastic OS cells (p > 0.9999), ECs and myeloid cells 1 (p > 0.9999), ECs and myeloid cells 2 (p > 0.9999), and ECs and NK/T (p > 0.9999) (Supplemental Table 1). In addition, comparative analyses showed no significant differential expression between ECs and OCs (p > 0.9999), ECs and plasmocytes (p > 0.9999), myeloid cells 1 and myeloid cells 2 (p > 0.9999), myeloid cells 1 and NK/T (p > 0.9999), myeloid cells 1 and OCs (p > 0.9999), myeloid cells 2 and plasmocytes (p > 0.9999), NK/T and OCs (p = 0.9998), NK/T and plasmocytes (p = 0.9993), and OCs and plasmocytes (p > 0.9999) (Supplemental Table 1).
DEG JAG2 expression is increased in ECs and osteoblastic OS cells
Next, we determined the relative abundance of JAG2 and compared its expression between the different cell groups. JAG2 was expressed in all nine main cell types examined (Figure 6). Interestingly, JAG2 was highly enriched in both the EC and osteoblastic OS cell clusters and showed weaker expression in all other cell types (Figure 6). Notably, the highest elevation of JAG2 expression was in the EC cluster, followed by osteoblastic OS cells (Figure 6). There was a total of 36 pairwise comparisons of nine major cell types, and 15 of these exhibited significant differential expression of JAG2 (Supplemental Table 1).
Figure 5.
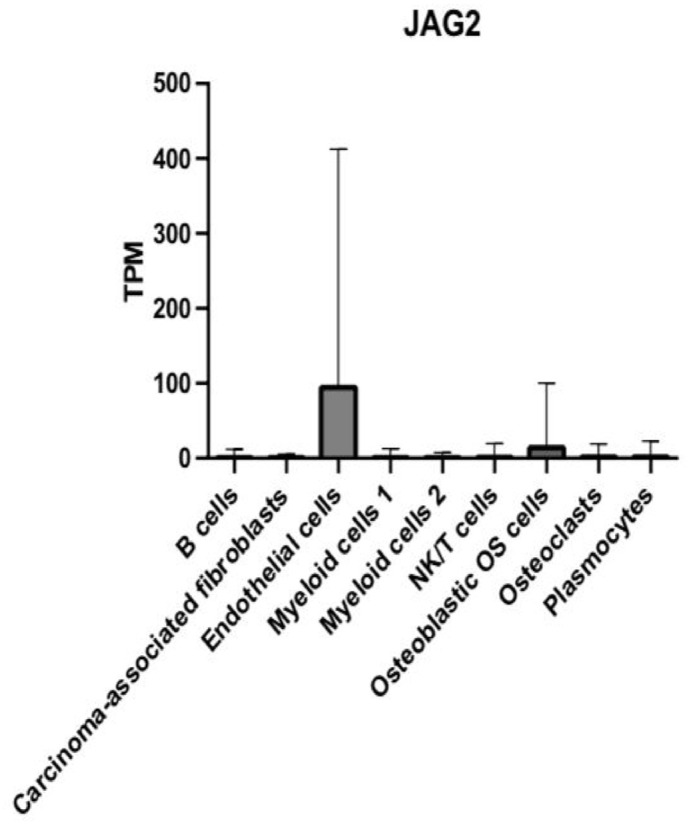
Bar graph depicting the relative mRNA expression of the KAZALD1 gene in TPM in the nine main OS cell types.
We observed significant differences in JAG2 expression between the following cell clusters: B cells and ECs (p < 0.0001), B cells and osteoblastic OS cells (p < 0.0001), CAFs and ECs (p < 0.0001), CAFs and osteoblastic OS cells (p < 0.0001), ECs and myeloid cells 1 (p < 0.0001), ECs and myeloid cells 2 (p < 0.0001), ECs and NK/T (p < 0.0001), ECs and osteoblastic OS cells (p < 0.0001), ECs and OCs (p < 0.0001), and ECs and plasmocytes (p < 0.0001) (Supplemental Table 1). We then found significant differences in JAG2 expression between the following cell clusters: myeloid cells 1 and osteoblastic OS cells (p < 0.0001), myeloid cells 2 and osteoblastic OS cells (p < 0.0001), NK/T cells and osteoblastic OS cells (p < 0.0001), osteoblastic OS cells and OCs (p < 0.0001), and osteoblastic OS cells and plasmocytes (p < 0.0001) (Supplemental Table 1). No significant differences were found in the expression of JAG2 between B cells and CAFs (p > 0.9999), B cells and myeloid cells 1 (p > 0.9999), B cells and myeloid cells 2 (p > 0.9999), B cells and NK/T (p = 0.9998), B cells and OCs (p = 0.9990), B cells and plasmocytes (p > 0.9999), and CAFs and myeloid cells 1 (p = 0.9997) (Supplemental Table 1). We also observed no significant differences between the clusters of CAFs and myeloid cells 2 (p > 0.9999), CAFs and NK/T (p > 0.9999), CAFs and OCs (p > 0.9999), CAFs and plasmocytes (p > 0.9999), myeloid cells 1 and myeloid cells 2 (p > 0.9999), myeloid cells 1 and NK/T (p = 0.9850), and myeloid cells 1 and OCs (p = 0.9806) (Supplemental Table 1). There were also no significant differences found between the cell clusters of myeloid cells 1 and plasmocytes (p > 0.9999), myeloid cells 2 and NK/T cells (p > 0.9999), myeloid cells 2 and OCs (p = 0.9990), myeloid cells 2 and plasmocytes (p > 0.9999), NK/T cells and OCs (p > 0.9999), NK/T cells and plasmocytes (p > 0.9999), and OCs and plasmocytes (p = 0.9996) (Supplemental Table 1).
LSAMP exhibits high expression in osteoblastic OS cells and CAFs
Next, we analyzed a well-established OS gene, limbic system–associated membrane protein (LSAMP), as the reference gene. Subsequently, we checked the transcription levels of LSAMP and pairwise compared gene expression patterns of LSAMP between the different OS tumor cell types. The mean expression values of LSAMP for all cell types are presented in Figure 7. Our results show that LSAMP is expressed at high levels in the osteoblastic OS cells and followed by CAFs cell cluster (Figure 7). In addition, LSAMP was expressed at relatively low levels in cell clusters, B cells, CAFs, ECs, myeloid cells 1, myeloid cells 2, NK/T, and plasmocytes (Figure 7). The expression patterns of LSAMP are consistent with its putative role in OS.20,21
Figure 7.
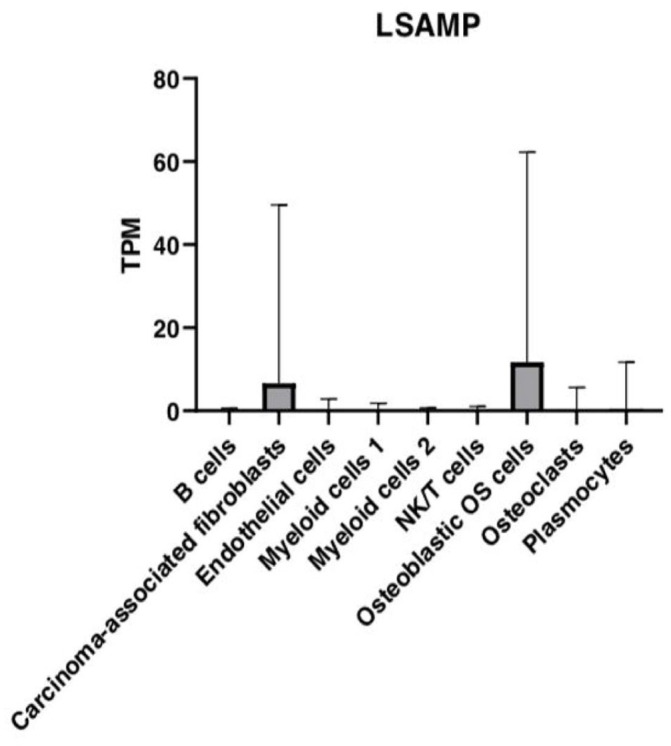
Bar graph depicting the relative mRNA expression of the LSAMP gene in TPM in the nine main OS cell types.
There were a total of 36 pairwise comparisons of nine major cell types and 15 of these exhibited significant differential expression of LSAMP (Supplemental Table 1). We observed significant differences in LSAMP expression between the cell clusters, B cells and CAFs (p < 0.0001), B cells and osteoblastic OS cells (p < 0.0001), CAFs and ECs (p < 0.0001), CAFs and myeloid cells 1 (p < 0.0001), CAFs and myeloid cells 2 (p < 0.0001), CAFs and NK/T (p < 0.0001), CAFs and osteoblastic OS cells (p < 0.0001), CAFs and OCs (p < 0.0001), and CAFs and plasmocytes (p < 0.0001) (Supplemental Table 1). In addition, we found significant differences in the expression of LSAMP between cell clusters, ECs and osteoblastic OS cells (p < 0.0001), myeloid cells 1 and osteoblastic OS cells (p < 0.0001), myeloid cells 2 and osteoblastic OS cells (p < 0.0001), NK/T and osteoblastic OS cells (p < 0.0001), osteoblastic OS cells and OCs, and osteoblastic OS cells and plasmocytes (p < 0.0001) (Supplemental Table 1).
LSAMP was shown to have no significant differences in expression between the cell clusters, B cells and ECs (p > 0.9999), B cells and myeloid cells 1 (p > 0.9999), B cells and myeloid cells 2 (p > 0.9999), B cells and NK/T cells (p > 0.9999), B cells and OCs (p > 0.9999), B cells and plasmocytes (p > 0.9999), ECs and myeloid cells 1 (p > 0.9999), and ECs and myeloid cells 2 (p > 0.9999) (Supplemental Table 1). We further observed no significant differences in LSAMP expression between cell clusters, ECs and NK/T (p > 0.9999), ECs and OCs (p > 0.9999), ECs and plasmocytes (p > 0.9999), myeloid cells 1 and myeloid cells 2 (p > 0.9999), myeloid cells 1 and NK/T cells (p > 0.9999), myeloid cells 1 and osteoclasts (p > 0.9999), and myeloid cells 1 and plasmocytes (p > 0.9999) (Supplemental Table 1). No significant differences in expression patterns of LSAMP were observed between the cell clusters, myeloid cells 2 and NK/T cells (p > 0.9999), myeloid cells 2 and OCs (p > 0.9999), myeloid cells 2 and plasmocytes (p > 0.9999), NK/T cells and OCs (p > 0.9999), NK/T and plasmocytes (p > 0.9999), and OCs and plasmocytes (p > 0.9999) (Supplemental Table 1).
GO and KEGG pathway enrichment analyses of the four survival OS DEGs
Finally, we performed KEGG analysis to seek out the enriched pathways and terms in the four DEGs. Functional enrichment analysis with Metascape showed that DEG, MUC1 were significantly enriched in interleukin-11 pathway, termination of O-glycan biosynthesis, O-linked glycosylation of mucins, post-translational protein modification, and protein metabolism (Supplemental Figure 1a). COL13A1 was significantly enriched in syndecan 1 pathway, collagen biosynthesis and modifying enzymes, integrins in angiogenesis, and extracellular matrix organization (Supplemental Figure 2a). The results of our analysis further showed that JAG2 was significantly enriched in signaling by NOTCH2, initiation of the second proteolytic cleavage of Notch receptor by receptor–ligand binding, activated NOTCH1 signaling in the nucleus (Supplemental Figure 3a). In addition, JAG2 was significantly enriched in p73 transcription factor network signaling by NOTCH and Notch signaling pathway. We also downloaded the network map of the four DEGs to show the relationship between terms (Supplemental Figures 1b, 2b, 3b, and 4a).
Discussion
Human OS is a highly malignant tumor with a poor prognosis. 22 scRNA seq is a method used in cancer research to identify tumor cells and analyze differences in gene expression at the single-cell resolution. 23 The variation in the transcriptional profile of specific OS cell types is considered reflective of the biological variation in OS tumors. Currently, there are few studies on the transcriptome of OS and no comprehensive study of gene expression in OS tumor cells. 12 Here, we analyzed and characterized the transcriptomic profiles of single OS cells using scRNA seq data of 29,278 cells from six patients downloaded from the TARGET-OS project. The expression changes of selected DEGs in different OS cells are likely to contribute to OS progression and metastasis and thus is worthy of further investigation. Based on p values, significant differences were observed in the expression of MUC1, COL13A1, JAG2, and KAZALD1 between several OS cell types following pairwise comparisons (Supplemental Table 1). The relevant DEGs and single OS cells are discussed briefly below with reference to published work on cancer and in particular OS.
MUC1 is a transmembrane mucin that is aberrantly overexpressed in many human cancer tissues, including breast, ovarian, and pancreatic cancer. 24 Research results have shown that MUC1 is most highly expressed in pancreatic, followed by lung, stomach, and endometrial cancer tissues, respectively (Supplemental Figure 5). The MUC1 protein is thought to have an important role in cancer invasion, metastasis, angiogenesis, and apoptosis. 24 MUC1 appears to contribute to metastasis progression through its O-glycosylated serine/threonine repeat region and intracellular domain (MUC1-CD). 25 In OS, a previous study has shown that expression of MUC1 correlated with worse overall survival and could be used as a prognostic biomarker and therapeutic target for OS. 17 Our study highlights that MUC1 appears to be highly expressed in osteoblastic OS cells. Therefore, MUC1 may contribute to tumor progression in osteoblastic OS cells; however, the functional significance of their elevated expression remains unknown.
In breast cancer, the COL13A1 gene is thought to be strongly associated with poor survival and metastasis. 26 In addition, reports have shown that COL13A1 is involved with high risk of disease progression and poor outcomes in human bladder cancer. 27 The highest levels of COL13A1 expression is thought to be found in thyroid followed by ovarian, pancreatic, and head and neck cancer tissues, respectively (Supplemental Figure 6). In OS, the functional role of COL13A1 is unclear, although in our study we have found that COL13A1 is highly expressed in CAFs, ECs, and osteoblastic OS cells of patient tumor samples. Previous studies also suggest that there was a positive correlation between expression of COL13A1 and poor survival in OS. 17 Further extensive functional studies will help to elucidate its function.
JAG2 encodes a zinc finger protein (ZNF) that is a marker for resistance to the alpha M1 oncolytic virus. 28 Interestingly, ZNFs may have a possible role in cancer development and progression in several cancer types.29,30 Published research has shown that JAG2 is upregulated in colorectal cancer cells.31,32 Modulated expression of JAG2 appears to disrupt migration and invasion of colorectal cancer cells. 31 In addition, over-expression of JAG2 has been linked with the progression of tumors in pancreatic, bladder, and lung cancers. 33 JAG2 was also associated with poor clinical outcomes in oral squamous cell carcinoma. 32 Highest expression of JAG2 is reported in head and neck cancer, followed by cervical, lung, and endometrial cancer, respectively (Supplemental Figure 7). According to our data, in OS JAG2 could be expressed at high levels in ECs and at lower levels in osteoblastic OS cells. In line with this, abnormal angiogenesis might contribute to OS development via the cross regulation with osteoblasts and osteoclasts.34,35 However, there are currently no published studies in OS highlighting the role of JAG2. Therefore, the function and mechanism of JAG2 in OS needs further exploration.
The KAZALD1 gene could promote malignant transformation in glioma by invasion and high proliferation of tumor cells. 36 In addition, low expression of the KAZALD1 gene in these tumors is associated with a better prognosis for glioma patients. 36 Interestingly, studies have shown that the highest expression levels of KAZALD1 are found in endometrial cancer followed by prostate, breast, and colorectal cancers, respectively (Supplemental Figure 8). While the role of KAZALD1 in OS is currently unclear, in this study, we have shown that in OS tumors CAFs and osteoblastic OS cells exhibit relatively high expression levels of KAZALD1 (Figure 5). Therefore, the KAZALD1 gene could be regarded as an attractive therapeutic target for OS and further research is essential to reveal its functional significance.
LSAMP also known as, LAMP, is a 64–68 kDa neuronal surface glycoprotein and member of the immunoglobin superfamily.37,38 Cell culture studies show that LSAMP might be involved in the regulation of neuronal growth and axon targeting in the brain. 39 Studies in cancer biology have also highlighted its possible role as a tumor suppressor gene in clear-cell renal cell carcinoma and epithelial ovarian cancer. 40 In prostate cancer, deletions of the LSAMP locus are associated with rapid disease progression. 41 LSAMP is most highly expressed in glioma, melanoma and prostate cancer tissues, respectively (Supplemental figure 9). Specifically in OS, research has successfully identified LSAMP as a novel candidate tumor suppressor gene using array comparative genomic hybridization. 20 LSAMP reportedly suppresses OS tumors by decreasing the rate of OS cell proliferation. 20 The low expression of LSAMP is strongly associated with poor outcomes in OS patients. Interestingly, the expression patterns of MUC1 gene and KAZALD1 gene are similar to that of LSAMP gene, suggestive of their roles in these cell types in OS.
Each of the distinct cell populations is considered to have a crucial role in the complexity of the tumor microenvironment of OS and subsequently tumor progression and metastasis. Recent published studies have shown that OCs have a role in OS-mediated osteolysis and pathogenesis of OS.12,42,43 Tumor-associated macrophages are involved in inflammatory responses and tissue homeostasis. 44 It has also been reported in the literature that ECs in OS tumors are associated with tumor angiogenesis, while CAFs have a role in metastasis and growth of solid tumors. 45 In addition, osteoblastic OS cells are suggested as the potential cell of origin and could regulate OC differentiation.12,46 Further research is necessary to investigate whether these genes may contribute to these cellular functions.
This study has limitations. First, we had a small sample size of six patients and minimal OS subtypes which may not fully represent the heterogeneity of OS and could potentially lead to bias in our results. Follow-up studies with additional tumor samples will help to improve the quality and validity of our findings. Second, our study did not screen for genes differentially expressed at different time points. Third, our analysis only examined DEGs between tumor samples from different patients and not within the same patient. Thus, it does not take into consideration inter-tumor heterogeneity. Finally, our results were generated by bioinformatic analysis, which is not sufficient, further molecular studies such as gene knockout are essential to validate and confirm our results. Despite of these limitations, our study by providing a comparative analysis of DEG expression in specific OS cell types will help to determine the molecular mechanisms underlying OS metastasis.
Conclusions
Taken together, our data provide in-depth insights into the genetic composition of populations of tumor cell types in OS. Our results suggest that various crucial OS DEGs exhibit different transcriptional patterns in different cell populations, thus possibly contributing to each cell type’s distinct functional role. This indicates that certain groups of tumor cells, such as osteoblastic OS cells, may have greater clinical significance. However, these genes have not been well characterized in cancer, specifically in OS. A more detailed understanding of the role of each cell population should help to uncover the precise biological functions of these OS DEGs. Several cellular subtypes have been recently identified, each of which have been found to vary in gene expression and functional roles. Future functional studies might also use protein–protein interaction (PPI) network analysis to determine whether these four OS DEGs are interrelated or are in independent pathways.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221080131 for Single-cell RNA-seq identification of four differentially expressed survival-related genes by a TARGET: Osteosarcoma database analysis by Mesalie Feleke, Wenyu Feng, Emel Rothzerg, Dezhi Song, Qingjun Wei, Sulev Kõks, David Wood, Yun Liu and Jiake Xu in Experimental Biology and Medicine
Footnotes
Authors’ Contributions: MF and WF contributed to the preparation of paper and mains figures and data analyses. MF contributed the first draft of the article. ER, WQ, and DS assisted data analysis. SK and DW discussed and revised the paper. YL and JX supervised the studies and data collections and revised paper. Mesalie Feleke and Wenyu Feng are equal contributors.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Data Availability: The data that support the findings of this study are available from the corresponding author upon reasonable request.
ORCID iDs: Emel Rothzerg  https://orcid.org/0000-0003-1957-930X
https://orcid.org/0000-0003-1957-930X
Sulev Kõks  https://orcid.org/0000-0001-6087-6643
https://orcid.org/0000-0001-6087-6643
Supplemental Material: Supplemental material for this article is available online.
References
- 1. de Azevedo JWV, de Medeiros Fernandes TAA, Fernandes JV, Jr, de Azevedo JCV, Lanza DCF, Bezerra CM, Andrade VS, de Araujo JMG, Fernandes JV. Biology and pathogenesis of human osteosarcoma. Oncol Lett 2020;19:1099–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Picci P. Osteosarcoma (osteogenic sarcoma). Orphanet J Rare Dis 2007;2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rothzerg E, Ho XD, Xu J, Wood D, Märtson A, Kõks S. Upregulation of 15 antisense long non-coding RNAs in osteosarcoma. Genes 2021;12:1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xin S, Wei G. Prognostic factors in osteosarcoma: a study level meta-analysis and systematic review of current practice. J Bone Oncol 2020;21:100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, Fiedorowicz M, Grieb P, Rutkowski P. Molecular biology of osteosarcoma. Cancers 2020;12:2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prater S, McKeon B. Osteosarcoma. Treasure Island, FL: StatPearls, 2019 [Google Scholar]
- 7. Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer 2009;115:1531–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernandes I, Melo-Alvim C, Lopes-Brás R, Esperança-Martins M, Costa L. Osteosarcoma pathogenesis leads the way to new target treatments. Int J Mol Sci 2021;22:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Misaghi A, Goldin A, Awad M, Kulidjian AA. Osteosarcoma: a comprehensive review. SICOT-J 2018;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma 2011;2011:548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin CH, Ji T, Chen C-F, Hoang BH. Wnt signaling in osteosarcoma. Curr Adv Osteosarcoma 2014;804:33–45 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y, Feng W, Dai Y, Bao M, Yuan Z, He M, Qin Z, Liao S, He J, Huang Q. Single-cell transcriptomics reveals the complexity of the tumor microenvironment of treatment-naive osteosarcoma. Front Oncol 2021; 11:709210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ta HT, Dass CR, Choong PF, Dunstan DE. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev 2009;28:247–63 [DOI] [PubMed] [Google Scholar]
- 14. Fuchs B, Pritchard DJ. Etiology of osteosarcoma. Clin Orthop Relat Res 2002;397:40–52 [DOI] [PubMed] [Google Scholar]
- 15. Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 2018;50:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gawel DR, Serra-Musach J, Lilja S, Aagesen J, Arenas A, Asking B, Bengnér M, Björkander J, Biggs S, Ernerudh J. A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Medicine 2019;11:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rothzerg E, Xu J, Wood D, Kõks S. 12 Survival-related differentially expressed genes based on the TARGET-osteosarcoma database. Exp Biol Med (Maywood) 2021;246:2072–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi A, Tanaseichuk O. 732 Benner C; Chanda SK, Metascape provides a biologist-oriented resource for the 733 analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun Yu Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol Sin 2015;36:1219–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kresse SH, Ohnstad HO, Paulsen EB, Bjerkehagen B, Szuhai K, Serra M, Schaefer KL, Myklebost O, Meza-Zepeda LA. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer 2009;48:679–93 [DOI] [PubMed] [Google Scholar]
- 21. Yen C-C, Chen W-M, Chen T-H, Chen Chen PC, Chiou HJ, Hung GY, Wu HT, Wei CJ, Shiau CY, Wu YC, Chao TC, Tzeng CH, Chen PM, Lin CH, Chen YJ, Fletcher JA. Identification of chromosomal aberrations associated with disease progression and a novel 3q13. 31 deletion involving LSAMP gene in osteosarcoma. Int J Oncol 2009;35:775–88 [DOI] [PubMed] [Google Scholar]
- 22. Mohseny AB, Machado I, Cai Y, Schaefer K-L, Serra M, Hogendoorn PC, Llombart-Bosch A, Cleton-Jansen A-M. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab Invest 2011;91:1195–205 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y, Wang D, Peng M, Tang L, Ouyang J, Xiong F, Guo C, Tang Y, Zhou Y, Liao Q. Single-cell RNA sequencing in cancer research. J Exp Clin Cancer Res 2021;40:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen W, Zhang Z, Zhang S, Zhu P, Ko JK-S, Yung KK-L. MUC1: structure, function, and clinic application in epithelial cancers. Int J Mol Sci 2021;22:6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horm TM, Schroeder JA. MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adh Migr 2013;7:187–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang H, Fredericks T, Xiong G, Qi Y, Rychahou PG, Li J-D, Pihlajaniemi T, Xu W, Xu R. Membrane associated collagen XIII promotes cancer metastasis and enhances anoikis resistance. Breast Cancer Res 2018;20:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyake M, Morizawa Y, Hori S, Tatsumi Y, Onishi S, Owari T, Iida K, Onishi K, Gotoh D, Nakai Y. Diagnostic and prognostic role of urinary collagens in primary human bladder cancer. Cancer Sci 2017;108:2221–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai J, Yan G. The identification and development of a novel oncolytic virus: alphavirus M1. Hum Gene Ther 2021;32:138–49 [DOI] [PubMed] [Google Scholar]
- 29. Jen J, Wang Y-C. Zinc finger proteins in cancer progression. J Biomed Sci 2016;23:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cassandri M, Smirnov A, Novelli F, Pitolli C, Agostini M, Malewicz M, Melino G, Raschellà G. Zinc-finger proteins in health and disease. Cell Death Discov 2017;3:17071–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He W, Tang J, Li W, Li Y, Mei Y, He L, Zhong K, Xu R. Mutual regulation of JAG2 and PRAF2 promotes migration and invasion of colorectal cancer cells uncoupled from epithelial–mesenchymal transition. Cancer Cell Int 2019;19:160–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hatano K, Saigo C, Kito Y, Shibata T, Takeuchi T. Overexpression of JAG2 is related to poor outcomes in oral squamous cell carcinoma. Clin Exp Dent Res 2020;6:174–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaish V, Kim J, Shim M. Jagged-2 (JAG2) enhances tumorigenicity and chemoresistance of colorectal cancer cells. Oncotarget 2017;8:53262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu S, Bennett S, Kuek V, Xiang C, Xu H, Rosen V, Xu J. Endothelial cells produce angiocrine factors to regulate bone and cartilage via versatile mechanisms. Theranostics 2020;10:5957–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y, Huang N, Liao S, Rothzerg E, Yao F, Li Y, Wood D, Xu J. Current research progress in targeted anti-angiogenesis therapy for osteosarcoma. Cell Prolif 2021;54:e13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H, Feng Y, Bao Z, Jiang C, Yan W, Wang Y, Zhang C, Liu Y, Zhang Q, Zhang W. Epigenetic silencing of KAZALD1 confers a better prognosis and is associated with malignant transformation/progression in glioma. Oncol Rep 2013;30:2089–96 [DOI] [PubMed] [Google Scholar]
- 37. Pimenta AF, Zhukareva V, Barbe MF, Reinoso BS, Grimley C, Henzel W, Fischer I, Levitt P. The limbic system-associated membrane protein is an Ig superfamily member that mediates selective neuronal growth and axon targeting. Neuron 1995;15:287–97 [DOI] [PubMed] [Google Scholar]
- 38. Pimenta AF, Fischer I, Levitt P. cDNA cloning and structural analysis of the human limbic-system-associated membrane protein (LAMP). Gene 1996;170:189–95 [DOI] [PubMed] [Google Scholar]
- 39. Innos J, Koido K, Philips MA, Vasar E. Limbic system associated membrane protein as a potential target for neuropsychiatric disorders. Front Pharmacol 2013;4:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petrovics G, Li H, Stümpel T, Tan SH, Young D, Katta S, Li Q, Ying K, Klocke B, Ravindranath L, Kohaar I, Chen Y, Ribli D, Grote K, Zou H, Cheng J, Dalgard CL, Zhang S, Csabai I, Kagan J, Takeda D, Loda M, Srivastava S, Scherf M, Seifert M, Gaiser T, McLeod DG, Szallasi Z, Ebner R, Werner T, Sesterhenn IA, Freedman M, Dobi A, Srivastava S. A novel genomic alteration of LSAMP associates with aggressive prostate cancer in African American men. EBioMedicine 2015;2:1957–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tan S-H, Babcock K, Kohaar I, Mohamed AA, Young D, Srivastava S, Dobi A. Biological functions of LSAMP, a gene frequently deleted in African American prostate cancers. AACR 2017;77:4464 [Google Scholar]
- 42. Ohba T, Cole HA, Cates JM, Slosky DA, Haro H, Ando T, Schwartz HS, Schoenecker JG. Bisphosphonates inhibit osteosarcoma-mediated osteolysis via attenuation of tumor expression of MCP-1 and RANKL. J Bone Miner Res 2014;29:1431–45 [DOI] [PubMed] [Google Scholar]
- 43. Kelleher FCO, O’Sullivan H. Monocytes, macrophages, and osteoclasts in osteosarcoma. J Adolesc Young Adult Oncol 2017;6:396–405 [DOI] [PubMed] [Google Scholar]
- 44. Cersosimo F, Lonardi S, Bernardini G, Telfer B, Mandelli GE, Santucci A, Vermi W, Giurisato E. Tumor-associated macrophages in osteosarcoma: from mechanisms to therapy. Int J Mol Sci 2020;21:5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang J-W, Wu X-F, Gu X-J, Jiang X-H. Exosomal miR-1228 from cancer-associated fibroblasts promotes cell migration and invasion of osteosarcoma by directly targeting SCAI. Oncol Res 2019;27:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mutsaers AJ, Walkley CR. Cells of origin in osteosarcoma: mesenchymal stem cells or osteoblast committed cells? Bone 2014;62:56–63 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221080131 for Single-cell RNA-seq identification of four differentially expressed survival-related genes by a TARGET: Osteosarcoma database analysis by Mesalie Feleke, Wenyu Feng, Emel Rothzerg, Dezhi Song, Qingjun Wei, Sulev Kõks, David Wood, Yun Liu and Jiake Xu in Experimental Biology and Medicine