Summary
A compact protein with a size of < 1,000 amino acids, the CRISPR-associated protein CasX is a fundamentally distinct RNA-guided nuclease compared to Cas9 and Cas12a. Although it can induce RNA-guided genome editing in mammalian cells, the activity of CasX is less robust than that of the widely used S. pyogenes Cas9. Here, we show that structural features of two CasX homologues and their guide RNAs affect the R-loop complex assembly and DNA cleavage activity. Cryo-EM-based structural engineering of either the CasX protein or the guide RNA produced two new CasX genome editors (DpbCasX-R3-v2 and PlmCasX-R1-v2) with significantly improved DNA manipulation efficacy. These results advance both the mechanistic understanding of CasX and its application as a genome editing tool.
Keywords: CRISPR, CasX, Cas12e, Genome editing, DNA cleavage, Cryo-EM, Structural engineering
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins comprise adaptive immune systems used by prokaryotes and some giant phage to fight against invading nucleic acids (Koonin et al., 2017; Mojica and Rodriguez-Valera, 2016). The entire immune response is typically comprised of three steps: integration of fragments from invading nucleic acids, synthesis of a ribonucleoprotein (RNP) interference complex, and nucleic acid interference (Hille et al., 2018; Le Rhun et al., 2019). During the last step of nucleic acid interference, a Cas protein is guided by its CRISPR RNA (crRNA), which is synthesized from the CRISPR array, to cleave a complementary DNA or RNA target. The programmability of CRISPR systems thus holds tremendous potential as transformative tools for genome editing (Doudna, 2020; Doudna and Charpentier, 2014; Hille et al., 2018; Wright et al., 2016). After years of effort, only a few types of CRISPR-Cas nucleases have been widely used for efficient genome editing, such as Cas9 and Cas12a (Jiang and Doudna, 2017; Makarova et al., 2019; Zetsche et al., 2015). While efficient for genome editing, the large size of Cas9 and Cas12a (1,000-1,500 amino acids (aa)) precludes their ability to be delivered via adeno-associated virus (AAV), which is useful for therapeutic delivery but has a limited transgene size of just 4.7 kilobase pairs (kbp).
A subtype of compact CRISPR nucleases, CasX (type V Cas12e, <1000 aa) has two homologous systems, CasX from Deltaproteobacteria (hereafter DpbCasX) and CasX from Planctomycetes (hereafter PlmCasX), that share 56% sequence similarity and expand the CRISPR-Cas genome editing family by offering a class of smaller programmable nucleases as additional therapeutic options (Burstein et al., 2017; Cao et al., 2021; Roberson, 2019). Compared to Cas9 or Cas12a, CasX is small enough to be delivered via a single AAV, with additional room for multiplexed single guide RNAs (sgRNA) or protein domain fusions (Liu et al., 2019; Yang and Patel, 2019). Previous biochemical analysis showed that DpbCasX cleaves double-stranded DNA (dsDNA) with a protospacer adjacent motif (PAM) of 5’-TTCN (Burstein et al., 2017; Liu et al., 2019). Structural analysis further showed that DpbCasX cuts the non-target strand (NTS) DNA and target strand (TS) DNA sequentially, using a single nuclease active site with the help of a large sgRNA scaffold (hereafter sgRNAv1) (Liu et al., 2019). Though DpbCasX is highly effective for bacterial interference, the genome editing activity in mammalian cells is modest relative to the widely used S. pyogenes Cas9. PlmCasX, although not well explored in vitro due to difficulty in protein expression and purification, showed equivalent or sometimes greater genome editing activity in mammalian cells compared to DpbCasX (Liu et al., 2019). Therefore, we aimed to determine the biochemical and structural mechanism of DNA cleavage by PlmCasX and further improve the genome editing capacity of CasX nucleases by structure-based engineering.
In this study, we expressed and purified PlmCasX protein with similar quality as DpbCasX via an improved workflow. While PlmCasX showed minimal dsDNA cleavage in vitro, consistent with our previous observation, PlmCasX efficiently disrupted GFP expression in a HEK293 fluorescent reporter cell assay at a similar or even higher rate compared to DpbCasX (Liu et al., 2019). Cryo-EM studies of the dPlmCasX-sgRNAv1-dsDNA ternary complex identified three distinct conformational states, including one that displays high flexibility of the Helical-II domain. The existence of this dynamic state suggests that the Helical-II domain assists with assembly of the ternary (R-loop) complex and ensures effective dsDNA cleavage via direct interaction with the sgRNA scaffold stem. Structural comparison of DpbCasX and PlmCasX suggests that three nucleotide-binding loops within CasX may play beneficial roles for PAM-proximal region recognition, sgRNA interaction and DNA substrate loading, which may contribute to the different biochemical and mammalian cell DNA cleavage efficacies between the two systems. Chimeric versions of CasX containing those beneficial loops showed improved DNA cleavage activity in vitro. Further, by rational sgRNA design based on new structural information, we improved the genome editing activities of both DpbCasX and PlmCasX using a sgRNA we have termed sgRNAv2. With synergetic improvement to both the protein and sgRNA, the new CasX nucleases (DpbCasX-R3-v2 and PlmCasX-R1-v2) showed ~10-fold and ~20-fold improvement in biochemical dsDNA cleavage kinetics, and ~53% and ~78% median editing efficacy (~2 to 3-fold improvement) for ten different GFP-targeting sgRNAs within human cells, respectively. In summary, these results yield fundamental knowledge and a practical improvement of CasX nucleases. Given the compact protein size of less than 1000 amino acids and the unique domain architecture relative to other Cas nucleases, CasX nucleases offer substantial advantages that expand the genome editing toolbox (Burstein et al., 2017; Liu et al., 2019; Makarova et al., 2019; Zhang et al., 2020).
Results
PlmCasX shows minimal biochemical activity but functions robustly in mammalian cells
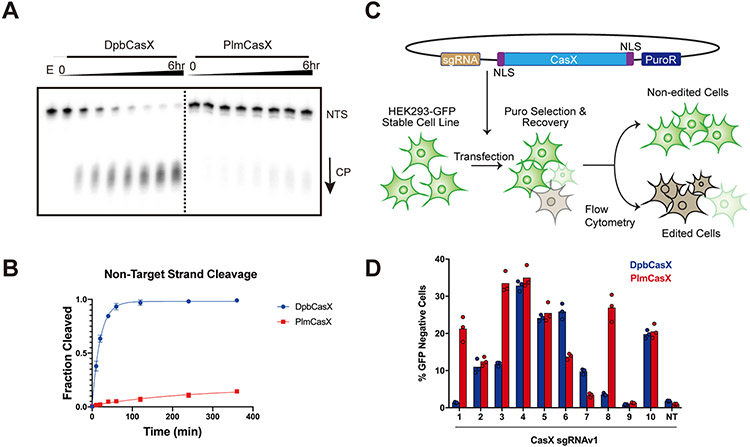
We used an improved protocol (see Method Details) to purify wildtype (wt)PlmCasX with similar purity and yield as wtDpbCasX (Figures S1A and S1B). PlmCasX eluted 0.3 mL earlier via size exclusion chromatography (Figure S1A), which suggests apo-PlmCasX (112.66 kDa) is less compact than apo-DpbCasX (112.93 kDa) and may lead to the increased difficulty observed during expression and purification. In vitro, PlmCasX cleaved just 10% of both the NTS and TS DNA (Figures 1A and 1B; Figures S1C and S1D) compared to DpbCasX with the previously reported sgRNA scaffold – sgRNAv1 (Liu et al., 2019). However, DpbCasX and PlmCasX showed similar linearization activity on pUC19 (Figure S1E), which may be due to the supercoiling-induced denaturation bubbles within plasmids (Adamcik et al., 2012). In HEK293 cells stably expressing GFP, plasmid transfection of PlmCasX showed adequate, and in some cases even higher, genome editing activity compared to DpbCasX using different GFP-targeting sgRNAv1s (Figures 1C and 1D; Figure S1F), which suggests PlmCasX is more proficient for genome editing by plasmid transfection. The vastly different in vitro and cell-based behavior further motivated us to understand the molecular difference between DpbCasX and PlmCasX. We therefore explored the structural details of PlmCasX and used this information to improve its biochemical and genome editing capacity through molecular engineering.
Figure 1. Comparison of DNA cleavage efficacy between DpbCasX and PlmCasX.
(A) In vitro dsDNA cleavage activity comparison between DpbCasX and PlmCasX revealed by denaturing PAGE. NTS denotes the non-target strand which was 32P labeled on the 5’ end. CP indicates the cleavage product. The fractions were collected at 0 min, 10 mins, 20 mins, 40 mins, 1 hr, 2 hrs, 4 hrs and 6 hrs, respectively. E indicates an empty well with labeled DNA but no CasX enzyme. (B) The plot of DNA cleavage kinetics analyzed based on the NTS band density from fractions compared to the input NTS band density at the reaction time of 0 min (n = 6, mean ± SD). One-phase association in Prism 7 was used to model the kinetics here and in following experiments. The single turnover rate constant k values (fraction cleaved per minute) for DpbCasX and PlmCasX were 0.05031 and 0.004137 (fraction/minute), respectively. (C) The workflow for human cell genome editing experiments, which were based on the disruption of constitutive GFP expression in HEK293 cells. (D) Human cell genome editing by DpbCasX and PlmCasX with sgRNAv1, measured 10 days after plasmid transfection. The GFP disruption efficacies for 10 GFP-targeting guides both for DpbCasX and PlmCasX are shown (n = 3, the mean of three technical replicates is shown). NT indicates the non-targeting sgRNAv1.
The mobility of the Helical-II domain impairs DNA cutting by PlmCasX
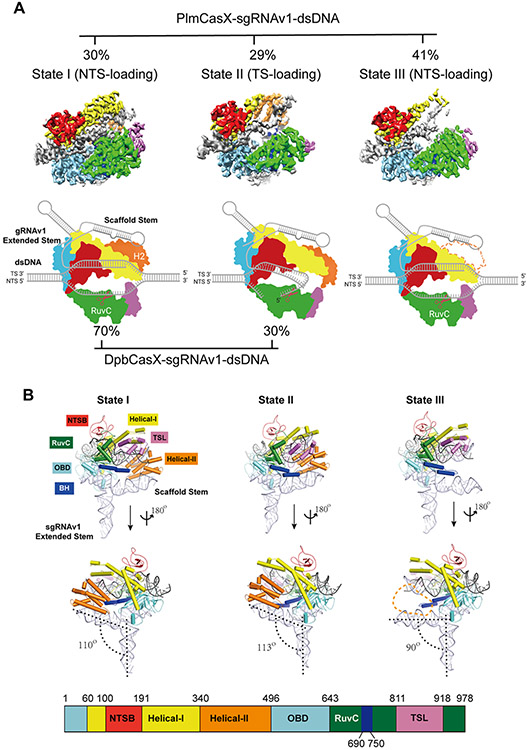
We reconstituted a ternary complex containing deactivated PlmCasX (D659A, E756A, D922A; dPlmCasX), sgRNAv1 (122 nucleotides (nt)) and a complementary DNA substrate (40 base pairs (bp)), but found that the majority of ternary complex disassembled during cryo-EM grid preparation (Figure S2A). Crosslinking the complex using BS3 significantly improved the holo-complex stability for single particle cryo-EM analysis (Figure S2B). 3D classification and refinement identified three conformational populations of the cross-linked complex that were resolved at resolutions of 2.9 Å, 3.4 Å and 3.2 Å (State I, State II and State III, respectively) (Figure 2A; Figures S2B-S2F). The cryo-EM density maps for States I and II both accounted for the entire complex, with all six CasX protein domains (Figure 2B). They correspond to a NTS DNA cleavage state and a TS DNA cleavage state, respectively (Figure 2A; Figure S3A). Comparison of these two conformations revealed a large structural rearrangement of the Helical-II (H2) domain, which may help to bend the sgRNA-DNA duplex and push the TS DNA into the RuvC catalytic domain (Movie S1). This structural rearrangement and stepwise DNA loading mechanism is highly similar to the mechanism we previously described for the dDpbCasX ternary complex (Liu et al., 2019).
Figure 2. Overall structures of the dPlmCasX-sgRNAv1-dsDNA complex.
(A) The different structural states of the dPlmCasX ternary complex with the sgRNAv1 scaffold revealed by single particle cryo-EM. The top views of refined EM maps for States I, II and III are shown in the top panel. The three maps are shown at contour thresholds of 6 to 9 times sigma. The cartoon model for each map is presented in the bottom panel for better elucidation of substrate DNA loading and cleavage. Referring to the published DpbCasX maps (Liu et al., 2019), the NTSB domain is colored in red, Helical-I in yellow, Helical-II in orange, OBD in aquamarine, RuvC in green, TSL in pink and the bridge helix (BH) in blue. The sgRNAv1 is in light gray and the dsDNA is in dark gray. The invisible Helical-II (H2) domain in State III is represented with a dashed line. The particle proportions for all functional states within the PlmCasX complex (determined in this study) and DpbCasX complex (Liu et al., 2019) are presented with percentages. (B) The atomic models of the dPlmCasX-sgRNAv1-dsDNA complex in three states shown in a front and back view. The domain architecture of the PlmCasX amino acid sequence is shown in the bottom panel. The protein domains in the atomic models share the same color codes as in A. The angle between the sgRNAv1 scaffold stem and extended stem (defined by RNA helix rotation axis, black dashed line) was calculated in PyMol. The Helical-II domain region is outlined with an orange dashed line in State III.
In State III of the dPlmCasX ternary complex, the NTS DNA appears loaded into the RuvC domain as in State I, but the density for the H2 domain is missing, most likely due to high flexibility (Figure 2A; Figure S2E; Figure S3A). By losing the interaction with the H2 domain, the sgRNA scaffold stem in State III is fully exposed and bent about 20° and 23° downward relative to States I and II, respectively (Figure 2B). Notably, State III accounted for 41% of the entire population of dPlmCasX ternary complexes (Figure 2A; Figure S2B). For many type V CRISPR nucleases, a stable H2 domain (also termed the REC2 domain) in the ternary complex is structurally important to maintain the active DNA R-loop conformation and assist with DNA cleavage (Liu et al., 2019; Yamano et al., 2016; Yang et al., 2016). Thus, we hypothesized that the presence of State III, with its highly mobile H2 domain, could explain the reduced DNA editing capability of PlmCasX in vitro (Figures 1A and 1B). To test this hypothesis, we truncated the H2 domain in DpbCasX (DpbCasX ΔH2), which resulted in decreased DNA cleavage activity down to a level similar to wtPlmCasX (Figures S3B-S3E). On the other hand, truncation of the H2 domain in PlmCasX (PlmCasX ΔH2) had little to no effect on DNA cleavage as compared to wtPlmCasX (Figures S3B-S3E). These results suggest that the high mobility of the H2 domain in wtPlmCasX largely decreases its in vitro cleavage capability to a minimal level similar to H2 truncation constructs. We then tested whether PlmCasX ΔH2 and DpbCasX ΔH2 are still capable of genome editing in human cells. Truncation of the H2 domain in both CasX enzymes led to insignificant GFP disruption in HEK293 cells, which demonstrated the necessity of the H2 domain for effective genome editing in cells (Figures S3F and S3G).
Nucleotide-binding loops in CasX contribute to R-loop assembly and DNA cutting
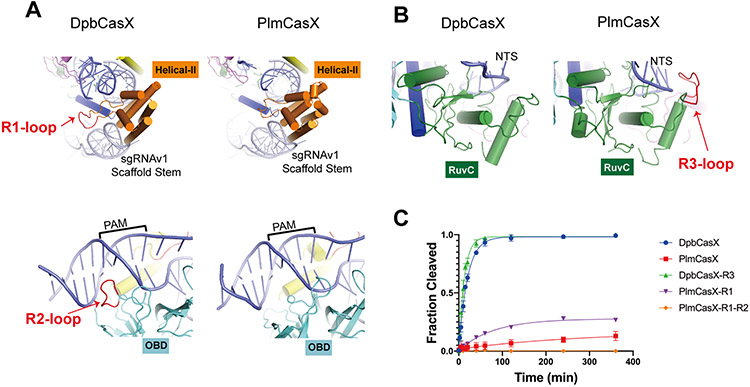
To further understand the structural details that led to unstable assembly and a mobile H2 domain within the PlmCasX ternary complex, we conducted a comprehensive analysis of the sequence and structural differences between PlmCasX and DpbCasX in State I. PlmCasX and DpbCasX share 56% sequence identity overall, with a structural similarity Z score of 33.8 as calculated by the Dali Server (Holm and Laakso, 2016). We identified the protein domains (OBD, Helical-I, Helical-II, RuvC, TSL and BH) of PlmCasX that correspond to those in the DpbCasX structure and redefined the protein sequence corresponding to the BH domain based on the better resolved structural details in PlmCasX (Figure 2B). Within the context of the same protein architecture, we found three nucleotide-binding loops that exist exclusively in either PlmCasX or DpbCasX and could have relevance to R-loop complex assembly and DNA cleavage (Figures 3A and 3B). We found that the region 1 loop (R1, K390~L396) in the DpbCasX H2 domain, which together with the H2 domain helices forms a deep pocket for tight binding of the sgRNA scaffold stem, likely contributes to the stable assembly of the R-loop complex (Figure 3A; Figures S4A and S4B). R1 is shortened in wtPlmCasX, giving rise to a shallower binding pocket that likely leads to weaker H2 domain-sgRNA binding and eventually the assembly of a less stable R-loop complex (Figure 3A; Figure S4A and S4B). A chimeric PlmCasX with the DpbCasX R1 loop (PlmCasX-R1) showed about 3-fold higher DNA cleavage kinetics in vitro (Figure 3C; Figure S4C; Figures S5A-C), but similar DNA editing activity in HEK293 cells compared to that of wtPlmCasX (Figure S5D). The region 2 loop (R2, G520~I526) is only present in the DpbCasX OBD domain and structurally interacts with the PAM proximal region (Figure 3A; Figures S4A and S4B), which may be important for initial steps of dsDNA substrate loading. However, adding R2 to PlmCasX-R1 (PlmCasX-R1-R2) completely disrupted DNA cleavage in vitro and editing in mammalian cells (Figure 3C; Figure S4C; Figure S5A-D). This result suggests that for R2, interactions with both DNA and the surrounding protein elements are likely important for the proper ternary complex assembly (Figure 3A). The region 3 loop (R3, Q945~G951) is exclusively present in PlmCasX, and similar to R1, forms a deep active pocket together with the remaining part of the RuvC domain that likely helps to faithfully accommodate and degrade ssDNA substrates (Figure 3B; Figures S4A and S4B). In contrast, the DpbCasX RuvC lacks R3 and contains a shallow active pocket that may have a lower affinity interaction with a ssDNA substrate (Figure 3B; Figure S4B). A chimeric DpbCasX with the PlmCasX R3 (DpbCasX-R3) had about 1.6-fold higher DNA cleavage kinetics in our biochemical cleavage assays (Figure 3C; Figure S4C; Figures S5A-C), and a 1.6-fold increase in median genome editing efficacy of HEK293 cells across three sgRNAv1s, compared to wtDpbCasX (Figure S5E).
Figure 3. Structural comparison between DpbCasX and PlmCasX.
(A) Region 1 (R1) and region 2 (R2) loops are located within DpbCasX but are absent from PlmCasX. The protein domains are colored as seen in Figure 2; the sgRNAv1 is colored in light gray and the dsDNA (with PAM region labeled) in blue. (B) The region 3 (R3) loop is located within PlmCasX but is absent from DpbCasX. (C) Biochemical dsDNA cleavage activity comparison between CasX chimeras with sgRNAv1 (n = 3, mean ± SD), based on cleavage of the NTS DNA. The rate constant k values for DpbCasX, PlmCasX, DpbCasX-R3 and PlmCasX-R1 were 0.05042, 0.003569, 0.07993 and 0.012503 (fraction/minute), respectively.
A new sgRNA scaffold promotes CasX R-loop assembly and DNA cleavage
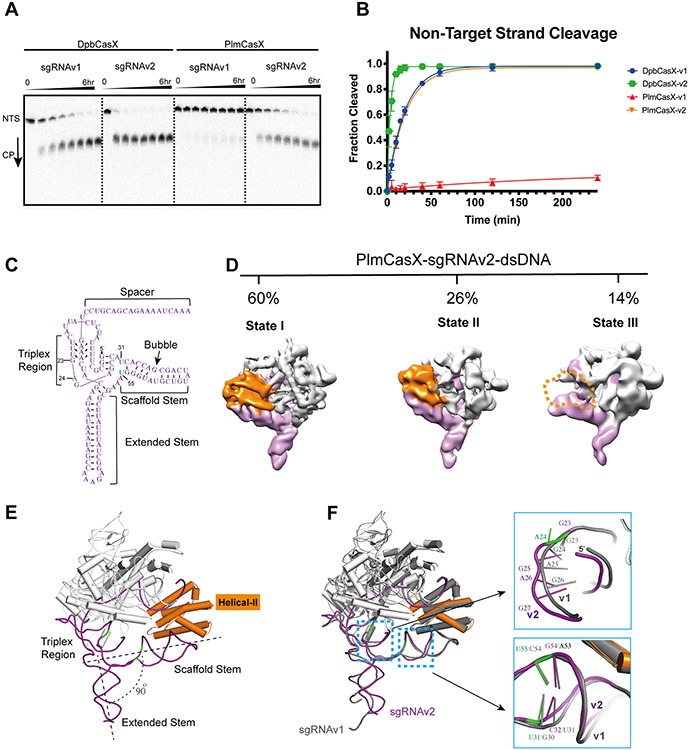
Our cryo-EM structures indicate that the weak interaction between the H2 domain and sgRNA scaffold stem likely interferes with R-loop complex assembly and thus decreases the DNA cleavage activity of PlmCasX. In addition to engineering the CasX protein, we were curious as to whether we could redesign the sgRNA sequence to stabilize the scaffold stem for better interaction with the H2 domain and further improve DNA cleavage activity. Based on secondary structure prediction and available atomic structures, adding an additional U at the 5’ end of sgRNAv1 could form a new base pairing interaction with A29 and thus limit the mobility of the scaffold stem without changing the structure (hereafter sgRNAv1-2) (Figures S6A-C). However, DpbCasX showed lower DNA cleavage activity with sgRNAv1-2 (Figures S6D and S6E). Instead, CasX may require a certain level of flexibility within the sgRNA to adopt the necessary conformational changes during the multi-step assembly of the ternary complex (Liu et al., 2019). By structural inspection, disruption of the G30-C54 base pairing and adding nucleotides after G23 to increase the single stranded linker may increase the flexibility of sgRNA scaffold stem while preserving its predicted secondary structure (Figures S6B and S6F). RNA profiling showed that the native PlmCasX tracrRNA sequence also contains additional nucleotides compared to sgRNAv1, which was designed based on the native DpbCasX tracrRNA sequence (Figure S6A). Referring to this structural interpretation and the PlmCasX tracrRNA sequence, we revised the sgRNA design by adding an additional nucleotide A after G23 and swapping the G30-C54 pair to U31-U55 (Figures S6C and S6F). The new sgRNA (hereafter sgRNAv2) enhanced both DpbCasX and PlmCasX dsDNA cleavage kinetics by 5.6 and 11-fold, respectively (Figures 4A and 4B). Again, adding a U or more nucleotides to the 5’ end of sgRNAv2 decreased the dsDNA cleavage activity of PlmCasX (sgRNAv2-2 and sgRNAv2-3) (Figures S6D-F). Both DpbCasX and PlmCasX also showed increased plasmid linearization activity using sgRNAv2 compared to sgRNAv1 (Figure S1E; Figure S6G).
Figure 4. In vitro biochemical cleavage behavior of CasX using sgRNAv2.
(A) In vitro dsDNA cleavage activity comparison between DpbCasX and PlmCasX using sgRNAv1 and sgRNAv2 revealed by denaturing PAGE. The fractions were collected at 0 min, 10 mins, 20 mins, 40 mins, 1 hr, 2 hrs, 4 hrs and 6 hrs, respectively. (B) Cleavage fraction analysis based on the NTS band density compared to the input NTS band density at the reaction time of 0 min (n = 5, mean ± SD). CasX-v1 denotes the CasX complex using sgRNAv1, while CasX-v2 denotes the CasX complex using sgRNAv2. The rate constant k values for DpbCasX-v1, PlmCasX-v1, DpbCasX-v2 and PlmCasX-v2 were 0.05065, 0.004433, 0.2817 and 0.04858 (fraction/minute), respectively. (C) The secondary architecture of sgRNAv2 revealed by cryo-EM. The key nucleic acid variants in sgRNAv2 compared to sgRNAv1 are marked in green. The nucleotide numbers for G23, A24, U31 and U55 are labeled. (D) The different structural states of the dPlmCasX ternary complex with the sgRNAv2 scaffold revealed by single particle cryo-EM. The back views of refined EM maps for State I, State II and State III are shown in the top panel. The three maps were low-pass filtered at 6 Å and shown at contour thresholds of 6 to 9 times sigma for clear presentation and comparison. The Helical-II domain is colored in orange and the sgRNAv2 in purple. Other parts of the complex are colored in light gray. The invisible Helical-II domain in State III is represented with a dashed outline. The particle proportions for all functional states within the dPlmCasX-sgRNAv2-dsDNA complex are presented with percentages. (E) Atomic model of dPlmCasX-sgRNAv2-dsDNA in State I. The CasX protein is colored in light gray and the sgRNAv2 is shown in purple. The Helical-II domain is emphasized by highlighting in orange. (F) Structural comparison between dPlmCasX-sgRNAv1-dsDNA (all in gray) and dPlmCasX-sgRNAv2-dsDNA (CasX in light gray and sgRNAv2 in purple) complexes in State I. The two structures were aligned in PyMol referring to the PlmCasX protein and dsDNA. The dsDNA models are hidden for better presentation. The zoomed in features for the sgRNA triplex region (top) and scaffold stem (bottom) are shown in the right panels, with the number of key nucleotides labeled.
To further investigate whether and how sgRNAv2 helped with the overall stability of the R-loop complex (Figure 4C), we performed single particle cryo-EM analysis on the dPlmCasX-sgRNAv2-dsDNA (40 bp) complex. Indeed, the new complex appeared more stable without the need for crosslinking during cryo-EM sample preparation (Figure S7A). 3D classification showed that only 14% of the dPlmCasX-sgRNAv2-dsDNA complexes were present in State III, a sharp decrease from 41% for the dPlmCasX-sgRNAv1-dsDNA complexes, presumably due to the higher affinity interaction of the H2 domain with sgRNAv2 (Figure 4D; Figure S7B). Further 3D variability analysis for particles from State I of dPlmCasX-sgRNAv2-dsDNA indicated that the extended stem of sgRNAv2 adopts a continuum of states (Movie S2), that may contribute to the limited resolution of the EM map (Figures S7C and S7D). Structural comparison of dPlmCasX-sgRNAv1-dsDNA and dPlmCasX-sgRNAv2-dsDNA showed that the addition of an A after G23 increased the curvature in the single strand RNA linker, and swapping the G30-C54 pair to U31-U55 generated a minor distortion at the end of the sgRNAv2 scaffold stem (Figures 4E and 4F; Figures S7E and S7F; Movie S3). Meanwhile, the angle between the sgRNA extended stem and scaffold stem decreased from 110° to 90° (Figure 2B; Figure 4E). Notably, the structures of the PlmCasX proteins appear indistinguishable between the two complexes (Figure 4F). Overall, sgRNAv2 increases the stability of the R-loop complex, which could explain the observed increase in DNA cleavage activity when complexed with both DpbCasX and PlmCasX.
Improved versions of CasX for mammalian genome editing
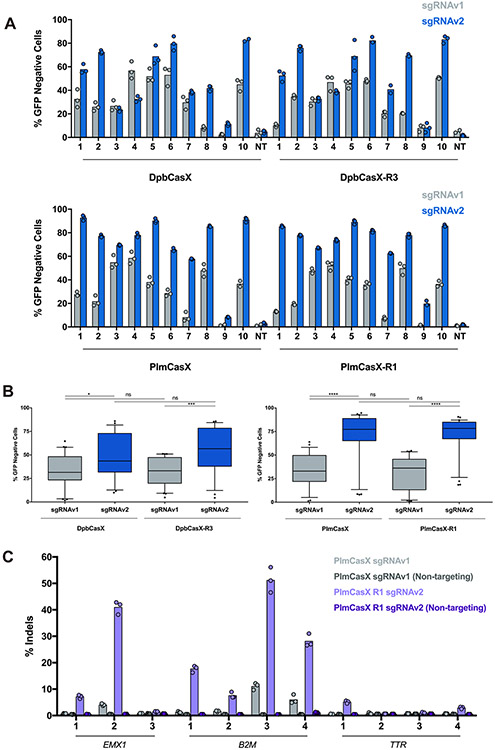
Using structure-based engineering of both the CasX protein and sgRNA, we were able to improve DNA cleavage by CasX in vitro (Figure 3C; Figure 4B). We further tested the newly designed sgRNA for mammalian cell genome editing and observed a considerable improvement in DNA editing efficacy for both DpbCasX and PlmCasX using ten different sgRNAs targeting HEK293 cells stably expressing GFP (Figure 5A). The median editing efficacy for DpbCasX and PlmCasX with sgRNAv2 (DpbCasX-v2 and PlmCasX-v2) was 43.50% and 77.25%, respectively, a significant improvement from 31.45% for DpbCasX and 32.95% for PlmCasX when using sgRNAv1 (DpbCasX-v1 and PlmCasX-v1) (Figure 5B).
Figure 5. Improved genome editing by engineered DpbCasX and PlmCasX.
(A) Human cell genome editing by DpbCasX, DpbCasX-R3, PlmCasX and PlmCasX-R1 using sgRNAv1 or sgRNAv2 revealed by disruption of genetically encoded GFP. The GFP disruption efficacies for all ten GFP guides are shown (n = 3 (except PlmCasX sgRNAv1 spacers 9, 10, NT; DpbCasX-R3 sgRNAv1 NT; DpbCasX sgRNAv2 spacer 10 and DpbCasX-R3 sgRNAv2 spacer 7; n = 2), mean). NT indicates the non-targeting sgRNA. (B) Genome editing efficacies for all ten GFP-targeting sgRNAs as a box and whisker plot: the box represents the 25th, 50th, and 75th percentile, the whiskers represent the 10th and 90th percentile, and outliers are plotted individually. Significances were determined via one-way ANOVA followed by Tukey’s multiple comparisons test. ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. (C) Editing of the human genes EMX1, B2M, and TTR by PlmCasX-v1 or PlmCasX-R1-v2 with multiple spacer sequences in HEK293T cells. The sequences of all spacers are listed in Table S1.
Next, we were curious as to whether combining both protein chimeras and the new sgRNAv2 could make a yet more effective CasX genome editing tool. Indeed, a combination of DpbCasX-R3 and sgRNAv2 (DpbCasX-R3-v2) outperformed all other combinations of CasX and sgRNA constructs in in vitro dsDNA cleavage activity (Figures S8A and S8B) and works robustly for genome editing (median editing efficacy of 56.60%) (Figure 5B). A combination of PlmCasX-R1 and sgRNAv2 (PlmCasX-R1-v2) showed improved dsDNA cleavage kinetics in vitro (~20-fold increase compared to PlmCasX with sgRNAv1 (PlmCasX-v1)) (Figures S8A and S8B) and showed the highest median editing efficacy (78.20%) and smallest interquartile range (18.33%) across multiple spacers compared to all other combinations of CasX and sgRNAs in HEK293 cells (Figure 5B). Unlike type II CRISPR nucleases like Cas9, sequence specific cis-cleavage by type V Cas12 nucleases activates non-specific ssDNA trans-cleavage(Chen et al., 2018; Li et al., 2019; Pausch et al., 2020). Our previous data indicated that DpbCasX with sgRNAv1 (DpbCasX-v1) shows minimal trans-activity compared to LbCas12a (Liu et al., 2019). Cleavage assays investigating indiscriminate ssDNA trans-cleavage revealed that the trans-activities of the new CasX enzymes and sgRNAs remain minimal, similar to the original DpbCasX-v1 (Figures S8C).
We further explored the capacity of PlmCasX-R1-v2, which showed the highest editing efficacy in our fluorescent reporter assay, for endogenous genome editing by targeting the EMX1 gene and clinically relevant B2M and TTR genes via plasmid transfection. Next generation sequencing revealed that PlmCasX-R1-v2 generated insertions and deletions (indels) at the targeted gene, and notably, showed as high as 10-fold higher activity than PlmCasX-v1 (Figure 5C). Interestingly, at the two endogenous targets with the highest levels of indels, PlmCasX-R1-v2 generated larger indels than seen with other class II CRISPR nucleases, such as Cas9, Cas12a, or Cas12f, with the most prevalent indel being a 15 or 19 bp deletion (Figures S8D and S8E) (Kim et al., 2021; Ran et al., 2015).
Discussion
In this study, we explored the biochemical and structural mechanism of DNA cleavage by PlmCasX and revealed the structural differences between DpbCasX and PlmCasX that correlate with their genome editing behaviors. By designing chimeric versions of CasX and new sgRNAs, we created two significantly improved versions of CasX as a DNA editing tool (DpbCasX-R3-v2 and PlmCasX-R1-v2) that offer small, yet efficient RNA-guided nucleases. PlmCasX-R1-v2 worked robustly in human cells, showing up to 90.5% editing in our fluorescent reporter assay and 56.1% editing at an endogenous human gene. In addition, CasX may offer substantial advantages compared to other CRISPR nucleases. First, the compact size of CasX would allow for delivery via a single AAV. The safety, efficacy and cell-specific tropism of AAVs have made them the leader for in vivo gene delivery, culminating in around 150 clinical trials and two FDA approved therapies within the United States alone (Kuzmin et al., 2021; Samulski and Muzyczka, 2014; Wang et al., 2020). However, a major limitation for AAV delivery is the minimal DNA packaging size, which prevents the ability of encoding S. pyogenes Cas9 within a single vector, let alone Cas9 fused to other functional domains. The compact size and structural flexibility of CasX could also be beneficial for functional domain insertions, creating tools such as epigenetic editors and base editors that still fit within this packaging capacity (Cao et al., 2021; Kleinstiver et al., 2019; Li et al., 2018). Previous studies have additionally shown the presence of pre-existing humoral and cellular immunity against the commonly used Cas9 nucleases from S. pyogenes and S. aureus in patients, presumably because these enzymes originate from common human commensal or pathogenic bacteria (Charlesworth et al., 2019; Crudele and Chamberlain, 2018).While the extent to which this pre-existing immunity may be a challenge for in vivo genome editing has yet to be fully elucidated, CRISPR nucleases from non-human associated sources such as CasX from Deltaproteobacteria or Planctomycetes could circumvent this potential issue. Moreover, though the off-target specificity has yet to be validated using these new CasX genome editing tools, recently published work showed that DpbCasX has a lower mismatch tolerance compared to Cas9 and Cas12a, suggesting that DpbCasX, and likely PlmCasX, has high fidelity and low off-target editing, an important property within the burgeoning clinical genome editing field (Zhang et al., 2020).
Recently, our group and others have described additional hypercompact CRISPR-Cas nucleases, including CasΦ-2 (Cas12j; 757 aa), AsCas12f1 (422 aa) and Cas14a1 (Un1Cas12f1; 537 aa). While CasΦ-2 is smaller than PlmCasX (984 aa), the CasΦ-2 nuclease showed only up to 33% editing of GFP in our fluorescent reporter assay compared to PlmCasX-R3-v2, which reached as high as 90.5% editing (Pausch et al., 2020). Extensive engineering of both the protein and sgRNA has dramatically increased the editing seen by Cas12f nucleases, which represent some of the smallest CRISPR effectors to date (Kim et al., 2021; Wu et al., 2021; Xu et al., 2021). However, the editing efficacy at endogenous genes by Cas12f in human cells varied greatly, ranging from a mean of ~5% to ~26%, which is comparable to the mean of ~15% seen with PlmCasX-R1-v2. Regarding delivery of the protein and sgRNA as an RNP, recent work has demonstrated that the Cas12f nuclease functions as an asymmetric homodimer to cleave dsDNA, which makes the effective RNP complex similar in size to CasX (Cas12f dimer: 800-1000 aa; CasX monomer: 984 aa) (Takeda et al., 2021; Xiao et al., 2021). Similar sequence-wide, high-throughput screening approaches to further engineer the CasX protein and sgRNA have yet to be explored. Based on the success of this strategy with other CRISPR nucleases, we anticipate this could be a promising approach to further minimize the size and improve the editing efficacy of CasX.
Limitations of the study
In this study, structural analysis revealed three nucleotide binding loops which contribute to DNA cleavage by CasX. While chimeric designs with the R1 or R3 loop insertion increased the DNA cleavage activity of CasX, the R2 loop insertion eliminated activity. Our current design of the R2 insertion is therefore non-optimal, and most likely disrupted the interaction between PlmCasX and the PAM DNA instead of stabilizing the interaction. In future work, the R2 region could be a potential spot for further improvement of PlmCasX by rational design or directed evolution screening. Additionally, our data suggests sgRNAv2 stabilizes the R-loop (ternary) complex and increases DNA cleavage activity by CasX; however, the mechanism by which sgRNAv2 affected the RNP (binary) complex assembly and thereby DNA unwinding and loading is unknown. We hypothesized the increase in flexibility of sgRNAv2 compared to sgRNAv1 was responsible for the significant improvement in activity, though more detailed studies are required to explore the structural states of the sgRNAs alone. Finally, while PlmCasX-R1-v2 proved to be a significantly improved genome editor at a fluorescent reporter gene and at endogenous genes, all experiments performed in this study were done in transformed cell lines. Future work is needed to test these improved versions of CasX within more difficult environments such as primary cells or animal models, along with delivery by methods such as AAV.
STAR★Methods
Resource availability
Lead contact
Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to the lead contact Jun-Jie Gogo Liu (junjiegogoliu@tsinghua.edu.cn).
Materials availability
Plasmids generated in this study have been deposited to Addgene or are available upon request.
Data and code availability
The electron density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-32389, EMD-32390, EMD-32391, and EMD-32392 and are publicly available as of the date of publication. The atomic coordinates and structural data have been deposited to the Protein Data Bank (PDB) under the accession numbers 7WAY, 7WAZ, 7WB0 and 7WB1 and are publicly available as of the date of publication. All the accession numbers are also listed in the key resources table. The raw cryo-EM micrographs and movies used in this study are available from the lead contact upon request.
This study did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli Rosetta2 | J. A. Doudna Lab and J.J. G. Liu Lab | N/A |
| Escherichia coli Mach1 T1 | Thermo Fisher | Cat#C862003 |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin | Sigma-Aldrich | Cat#A9518 |
| Phosphatase inhibitor cocktail | Roche | Cat#4906837001 |
| Phenylmethylsulfonyl fluoride (PMSF) | Roche | Cat#10837091001 |
| Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) | Sigma-Aldrich | Cat#C4706 |
| Tobacco Etch Virus (TEV) protease | J. A. Doudna Lab and J.J. G. Liu Lab | N/A |
| Q5 High-Fidelity DNA Polymerase | NEB | Cat#M0491 |
| T7 polymerase | J. A. Doudna Lab and J.J. G. Liu Lab | N/A |
| RNase inhibitor | Promega | Cat#N2615 |
| RNase-Free DNase I | Promega | Cat#M6101 |
| ATP, [g-32P]- 3000Ci/mmol | Perkin Elmer | Cat#BLU002A001MC |
| T4 PNK | NEB | Cat#M0236S |
| Proteinase K | Sangon Biotech | Cat#A600451-0050 |
| BbsI-HF | NEB | Cat#R3539L |
| AgeI | NEB | Cat#R3552L |
| BamHI | NEB | Cat#R0136L |
| KpnI | NEB | Cat#R3142L |
| PciI | NEB | Cat#R0655L |
| Dulbecco’s Modified Eagle’s Medium, high glucose | Gibco | Cat#11995073 |
| Opti-MEM I Reduced Serum Medium | Gibco | Cat#31985070 |
| Fetal Bovine Serum | VWR | Cat#89510-186 |
| Trypsin-EDTA (0.25%), phenol red | Thermo-Fisher | Cat#25200056 |
| Penicillin-Streptomycin | Gibco | Cat#10378016 |
| Puromycin Dihydrochloride | Gibco | Cat#A1113803 |
| BS3 cross-linker | Sigma-Aldrich | Cat#S5799 |
| Graphene-oxide | Sigma-Aldrich | Cat#777676 |
| Critical commercial assays | ||
| MycoAlert Mycoplasma Detection Kit | Lonza | Cat#LT07-318 |
| QuickExtract DNA Extraction Solution | Lucigen | Cat#QE09050 |
| QIAquick PCR Purification Kit | Qiagen | Cat#28104 |
| In-Fusion Snap Assembly Master Mix | Takara | Cat#638948 |
| Cloning Enhancer | Takara | Cat#639615 |
| Lipofectamine 3000 Transfection Reagent | Life Technologies | Cat#L3000001 |
| NucleoSpin Gel and PCR Cleanup Kit | Takara | Cat#740986.20 |
| Deposited data | ||
| Uncropped gels (Mendeley data) | This paper | DOI:10.17632/w6gfw3g5dt.1 |
| Coordinates of the dPlmCasX-sgRNAv1-dsDNA complex (State I) | This paper | PDB: 7WAY |
| Cryo-EM density map of the dPlmCasX-sgRNAv1-dsDNA complex (State I) | This paper | EMDB: EMD-32389 |
| Coordinates of the dPlmCasX-sgRNAv1-dsDNA complex (State II) | This paper | PDB: 7WAZ |
| Cryo-EM density map of the dPlmCasX-sgRNAv1-dsDNA complex (State II) | This paper | EMDB: EMD-32390 |
| Coordinates of the dPlmCasX-sgRNAv1-dsDNA complex (State III) | This paper | PDB: 7WB0 |
| Cryo-EM density map of the dPlmCasX-sgRNAv1-dsDNA complex (State III) | This paper | EMDB: EMD-32391 |
| Coordinates of the dPlmCasX-sgRNAv2-dsDNA complex (State I) | This paper | PDB: 7WB1 |
| Cryo-EM density map of the dPlmCasX-sgRNAv2-dsDNA complex (State I) | This paper | EMDB: EMD-32392 |
| Experimental models: Cell lines | ||
| HEK293T cells | UC Berkeley Cell Culture Facility | N/A |
| GFP HEK293 cells | Laboratory of Juan Hurtado | N/A |
| Oligonucleotides | ||
| ssDNA oligos (see Table S2 for sequences) | IDT | N/A |
| ssRNA oligos (see Table S2 for sequences) | IDT | N/A |
| Recombinant DNA | ||
| His-MBP-TEV-DpbCasX, expression vector | This paper | pJJGL001 Addgene plasmid #180605 |
| His-MBP-TEV-PlmCasX, expression vector | This paper | pJJGL002 Addgene plasmid #180606 |
| His-MBP-TEV-DpbCasX-H2 truncation, expression vector | This paper | N/A |
| His-MBP-TEV-PlmCasX-H2 truncation, expression vector | This paper | N/A |
| His-MBP-TEV-DpbCasX+Plm R3 insertion, expression vector | This paper | pJJGL003 Addgene plasmid #180607 |
| His-MBP-TEV-PlmCasX+Dpb R1 insertion, expression vector | This paper | pJJGL004 Addgene plasmid #180608 |
| His-MBP-TEV-PlmCasX+Dpb R1+2 insertion, expression vector | This paper | N/A |
| U6-sgRNAv1-CAG-DpbCasX-PuroR | Liu et al., 2019 | pBLO62.4 Addgene plasmid #123123 |
| U6-sgRNAv1-CAG-PlmCasX-PuroR | Liu et al., 2019 | pBLO62.5 Addgene plasmid #123124 |
| U6-sgRNAv1-CAG-DpbCasX(ΔH2)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-DpbCasX(R3)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-PlmCasX(ΔH2)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-PlmCasX(R1)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-PlmCasX(R1+2)-PuroR | This paper | N/A |
| U6-sgRNAv2-CAG-DpbCasX-PuroR | This paper | pCAT079 Addgene plasmid #180509 |
| U6-sgRNAv2-CAG-DpbCasX(R3)-PuroR | This paper | pCAT105 Addgene plasmid #180510 |
| U6-sgRNAv2-CAG-PlmCasX-PuroR | This paper | pCAT077 Addgene plasmid #180511 |
| U6-sgRNAv2-CAG-PlmCasX(R1)-PuroR | This paper | pCAT100 Addgene plasmid #180512 |
| U6-sgRNAv1-CAG-PlmCasX-mNeonGreen-PuroR | This paper | pCAT526 Addgene plasmid #180513 |
| U6-sgRNAv2-CAG-PlmCasX(R1)-mNeonGreen-PuroR | This paper | pCAT527 Addgene plasmid #180514 |
| Software and algorithms | ||
| Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ImageQuant TL | GE Healthcare | N/A |
| cryoSparc | Punjani et al., 2017 | https://cryosparc.com |
| Relion | Kimanius et al., 2016 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| PyMol | Schrodinger LLC, 2010 | https://pymol.org/2/ |
| UCSF-Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| PHENIX | Liebschner et al., 2019 | https://phenix-online.org/documentation/reference/refinement.html |
| Coot | Casañal et al., 2020 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| FlowJo | BD | https://www.flowjo.com |
| CRISPResso2 | Clement et al. 2019 | https://www.crispresso.pinellolab.partners.org |
| Other | ||
| Magnetic solid phase reversible immobilization (SPRI) beads | UC Berkeley Sequencing Core | N/A |
| 30 kDa MWCO concentrator | Amicon Ultra, Merck | Cat#UFC9030 |
| 3 kDa MWCO concentrator | Amicon Ultra, Merck | Cat#UFC8003 |
| Ni-NTA agarose beads | QIAGEN | Cat#30210 |
| Quantifoil 1.2/1.3 grids | EMS | Cat#Q310CR-14 |
| C-flat 2/2 grids | EMS | Cat#CF-224C-100 |
| HiTrap Heparin HP Columns | GE Healthcare | Cat#17040701 |
| Superdex 200 10/300 column | GE Healthcare | Cat#28990944 |
| QIAprep Spin Miniprep kit | Qiagen | Cat#27106 |
Experimental model and subject details
Culture of human cell lines
GFP HEK293 and HEK293T cells (UC Berkeley Cell Culture Facility) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (VWR) and 1% penicillin-streptomycin (Gibco). Cells were maintained at 37°C and 5% CO2 at sub-confluent conditions. The MycoAlert Mycoplasma Detection Kit (Lonza) was used to routinely test cells for mycoplasma.
Method Details
CasX protein expression and purification
The wildtype and engineered CasX proteins were expressed using Rosetta E.coli cells. The E. Coli cells were transformed by mixing competent cells (100 μL) with CasX encoding plasmids (100 ng) and incubating for 30 minutes on ice. The tube containing the plasmid and cells was incubated at 42°C for 35 seconds to induce the transformation. After 5 minutes of resting on ice, Luria broth (LB) (950 μl) was added to the solution and incubated at 37°C for 1 hour to recover. The cells were transferred to a flask containing LB and 50 mg/mL ampicillin (1:1000) and incubated at 37°C overnight. 2.7% of the grown culture was added to the main culture containing Terrific broth and 50 mg/mL ampicillin (1:1000). The main culture was incubated at 37°C until it reached an OD of 0.5-0.6. The culture was cooled on ice and protein expression was induced by addition of IPTG to a final concentration of 1 mM and incubated at 16°C overnight. Cells were harvested by centrifugation (4000 rpm, 4°C) and resuspended in lysis buffer (600 mM sodium chloride, 20 mM HEPES, pH 7.5, 10% glycerol, 50 mM imidazole, 1 mM TCEP). PMSF (0.5 mM) and 4 tablets of protease inhibitor cocktail (Roche) were added per 100 mL of mixture. The cell suspension was lysed by sonication and pelleted by ultra-centrifugation at 35,000 g for 30 minutes. The soluble lysate was mixed with equilibrated Ni-NTA agarose beads at 4°C for 2 hours. Using a gravity-flow column, the Ni-NTA agarose beads were washed using lysis buffer. To elute the construct, the Ni-NTA beads were incubated overnight at 4°C in lysis buffer and TEV protease (final concentration of 1 mg protease/ 20 mg purified protein). Using the gravity-flow column, the protein of interest was eluted using lysis buffer with 300 mM imidazole. The flow-through was collected and concentrated using a 30 kDa MWCO concentrator (Amicon Ultra, Merck). The solution containing the protein was mixed with lower salt buffer (200 mM sodium chloride, 20 mM HEPES, pH 7.5, 10% glycerol, 1mM TCEP) and applied to a heparin column on an Akta FPLC (GE). The protein was eluted using a potassium chloride gradient up to 1 M. The combined fractions were concentrated using a 30 kDa MWCO concentrator (Amicon Ultra, Merck) and applied to a Superdex 200 10/300 column (GE healthcare/Cytiva) using SEC buffer (400 mM potassium chloride, 20 mM HEPES, pH 7.5, 10% glycerol, 1 mM TCEP). The protein was concentrated, flash frozen in liquid nitrogen and stored at −80°C to use in assays. All the engineered and wildtype CasX proteins were expressed and purified using the same method. Compared to the original purification protocol for DpbCasX, we increased the sodium chloride concentration from 500 mM to 600 mM, and added 50 mM imidazole in the lysis buffer, which helped to decrease the non-specific protein and nucleic acid contamination. Since apo PlmCasX is less stable than DpbCasX, it should be kept in buffer with ≥400mM sodium chloride or potassium chloride during the entire purification process, and the purification should be ideally finished within 24 hours.
sgRNA preparation
All the sgRNAs were produced using in vitro transcription. First, to make the DNA template, primers were ordered from Integrated DNA Technologies (IDT) and PCR amplified using Q5 polymerase (New England Biolabs). The DNA template (50 μg) along with 10x IVT buffer (300 mM Tris-HCl (pH 8.1), 250 mM MgCl2, 0.1% Triton X-100, 20 mM Spermidine, add 100 mM DTT before use), 5x NTPs (25 mM NTP mixture, pH 7.5), T7 polymerase, RNase inhibitor (Promega) and DEPC-treated water were incubated on a 37°C heat block for 3-4 hours. The solutions were then treated with RNase-Free DNase I (Promega) by addition of 10x Reaction Buffer (400 mM Tris-HCl, pH 8.0 at 25°C, 100 mM MgSO4, 10 mM CaCl2) and RNase-free DNase I and incubated on a 37°C heat block for 30 minutes. The sample was spun down at 4°C and the soluble fraction was moved to a new tube. After adding 2x formamide (95% formamide, 0.02% SDS, 0.02% bromophenol blue,0.01% xylene cyanol FF, 1 mM EDTA), samples were gel purified using a 15% urea-PAGE gel. The band containing the sgRNA was cut out and incubated in water and 1/30 NaOAc at 4°C overnight. Samples were then filtered using a 0.22 μm Corning filter into 50 mL tubes. sgRNA samples were concentrated using a 3 kDa MWCO concentrator (Amicon Ultra, Merck). 100% ethanol was added to sgRNA samples to precipitate the sgRNA. Precipitated sgRNA was pelleted via centrifugation and washed using 70% ethanol. sgRNA samples were resuspended in DEPC-treated water and stored at −80°C to be used for cleavage assays.
In vitro cleavage assays
For dsDNA cleavage assays, DNA substrates were 5’ labeled using T4 PNK (NEB) by adding γ-32P-ATP. CasX proteins were diluted to 2 μM using 1x reaction buffer (400 mM potassium chloride, 5% glycerol, 20 mM Tris-HCl, pH 7.8, 5 mM magnesium chloride, 1 mM DTT). sgRNAs were diluted to 3 μM with 1x reaction buffer. The sgRNA and protein samples were then mixed and incubated at room temperature for 1 hour to reconstitute the RNP complex. The final concentration of the CasX-sgRNA was 300 nM and the concentration of radiolabeled probe was 2 nM. Reactions were initiated by mixing CasX-sgRNA and radiolabeled DNA on a 37°C heat block. Sample aliquots were taken at the following time points: 0, 2, 5, 10, 15, 20, 30, 60, 120, 240, and 360 minutes. The aliquots were mixed with 2x formamide loading buffer (95% formamide, 10 mM EDTA, 0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol FF) and quencher (50 μg/mL heparin, 25 mM EDTA) and were incubated in 95°C heat blocks for 5 minutes to stop the cleavage reaction. Samples were run on 12% urea-PAGE gels before being dried and visualized using a phosphoimager (Amersham Typhoon, GE Healthcare).
For plasmid cleavage assays, the target DNA sequence was cloned into the pUC19 plasmid. For each 100 μL cleavage reaction, 400 nM CasX-sgRNA RNP and 20 nM pUC19 plasmid DNA were incubated in 1x reaction buffer (500 mM sodium chloride, 5% glycerol, 20 mM Tris-HCl, pH 7.8, 10 mM magnesium chloride, 1 mM DTT) at 37°C. Sample aliquots were taken at the following time points: 0, 10, 20, 30, 60, 120, 240, and 360 minutes. The aliquots were mixed with 6x DNA loading buffer (30mM EDTA, 36% (v/v) glycerol, 0.05% (w/v) bromophenol blue, 0.05% (w/v) xylene Cyanol FF) and then digest with 100 μg/mL Proteinase K (from Tritirachium album, Sangon Biotech) for 1h at 37°C to quench the reaction. A 1% agarose gel was used to analyze cleavage products.
For the trans-cleavage activity assay, a random 50 nucleotide oligonucleotide substrate was labeled using T4 PNK (NEB) by adding γ-32P-ATP. Each reaction included 300 nM CasX protein, 360 nM sgRNA, 450 nM activator, and 2 nM substrate. The trans-cleavage assay was performed and analyzed similarly to the dsDNA cleavage assay, described above.
Plasmid Construction
For human genome editing experiments, DpbCasX plasmid pBLO62.4 (Addgene plasmid #123123) and PlmCasX plasmid pBLO62.5 (Addgene plasmid #123124) were utilized or modified, which were codon-optimized for expression in human cells and contain a SV40 nuclear localization sequence on both termini (Liu et al., 2019). Short oligonucleotides (IDT) containing the sgRNA spacer sequence were annealed and phosphorylated prior to Golden Gate assembly (BbsI restriction sites) for insertion just downstream of the CasX guide RNA scaffold within the plasmids. CasX protein mutants were constructed by PCR amplification of the CasX sequence in two pieces, with primers containing the deletion or insertion sequences. pBLO62.4 and pBLO62.5 were digested with AgeI and BamHI (NEB) and gel electrophoresis was utilized to separate the digested components. The plasmid backbone was excised from the gel and purified with the QIAquick PCR Purification Kit (Qiagen) or the NucleoSpin Gel and PCR Cleanup Kit (Takara) according to the manufacturer’s protocol. In-Fusion cloning (Takara) with the Cloning Enhancer was used to insert the PCR amplified mutant CasX sequences within the digested backbone according to the manufacturer's protocol. Plasmids encoding mutant CasX sgRNA scaffolds were constructed similarly to CasX mutant protein plasmids. Plasmids encoding engineered or wildtype CasX proteins were digested using KpnI and PciI (NEB). Gel electrophoresis was used to isolate the digested plasmid backbone. Digested backbone was excised from the gel and purified with the PCR QIAquick PCR Purification Kit (Qiagen) or the NucleoSpin Gel and PCR Cleanup Kit (Takara) according to the manufacturer’s protocol. Mutant sgRNA scaffolds were ordered as gBlocks from IDT and cloned into the digested backbone using In-Fusion cloning (Takara). Cloned plasmids were sequence verified by capillary Sanger sequencing (UC Berkeley DNA Sequencing Facility). For the endogenous genome editing experiments, an mNeonGreen fluorescent protein was genetically encoded between the CasX gene and puromycin resistance gene, each separated by self-cleaving 2A peptide sequences. Plasmids were cleaved with BamHI and In-Fusion cloning was utilized as described above to insert a gBlock (IDT) encoding mNeonGreen. Plasmids were propagated in Mach1 T1 competent cells (Thermo Fisher) and purified using a QIAprep Spin Miniprep kit (Qiagen) according to the manufacturer’s protocol.
Genome editing in fluorescent reporter human cells
GFP HEK293 reporter cells were seeded into 96-well plates and transfected 12-18 hours later at 60-70% confluency according to the manufacturer’s protocol with lipofectamine 3000 (Life Technologies) and 200 ng of plasmid DNA encoding the wildtype or engineered CasX plasmids. 24 hours post-transfection, GFP HEK293 reporter cells that were successfully transfected were selected for by adding 1.5 μg/mL puromycin to the cell culture media for 48 hours. Cell culture media was replaced with media containing fresh 1.5 μg/mL puromycin for an additional 24 hours before replacing with cell culture media without puromycin. Cells were passaged regularly to maintain sub-confluent conditions and then analyzed in 96- well round bottom plates on an Attune NxT Flow Cytometer with an autosampler. Cells were analyzed on the flow cytometer after 5, 7, and 10 days to track the disruption of the GFP gene in cells. The sequences of all spacers used in this study are listed in Table S1.
Endogenous genome editing
HEK293T cells (UC Berkeley Cell Culture Facility) were cultured in DMEM (Gibco) supplemented with 10% FBS (VWR) and 1% penicillin-streptomycin (Gibco). The MycoAlert Mycoplasma Detection Kit (Lonza) was used to routinely test cells for mycoplasma. HEK293T cells were plated in 96-well plates and allowed to grow overnight to ~60-70% confluency before transfecting with 200 ng of plasmid and lipofectamine 3000 according to the manufacturer’s protocol. 24 hours post-transfection, HEK293T cells that were successfully transfected were selected for by adding 1.5 μg/mL puromycin to the cell culture media for 48 hours. Cell culture media was replaced with media containing fresh 1.5 μg/mL puromycin for an additional 24 hours before replacing with cell culture media without puromycin. Media was removed from the cells and 50 μL of QuickExtract (Lucigen) was added to each well and incubated at room temperature for 10-15 minutes. Cell extracts were then thermocycled at 65°C for 20 minutes followed by 95°C for 20 minutes. Amplicons containing the targeted site were amplified via PCR with Q5 polymerase (NEB) and primers containing Illumina adaptor sequences. Amplicons were cleaned with magnetic solid phase reversible immobilization (SPRI) beads (UC Berkeley Sequencing Core) and were further library prepped and loaded onto an Illumina MiSeq by the Center for Translational Genomics (Innovative Genomics Institute, UC Berkeley). Over 20,000 reads per sample were routinely achieved. 300 bp paired-end reads were analyzed using CRISPResso2 (crispresso.pinellolab.partners.org), using a quantification window centered at −3 bp, a quantification window size of 8 bp (to account for the large, staggered cleavage pattern of CasX), and a plot window size of 30 bp (to visualize large indels). Cells treated with PlmCasX-v1 or PlmCasX-R1-v2 with a non-targeting sgRNA were evaluated at every spacer sequence within every amplicon as a control. Percentage of indels plotted was based on the percentage of modified reads from the CRISPResso2 output. For the indel size distribution plots, sequencing reads of a particular deletion length (regardless of insertions or substitutions) were grouped and plotted. The remaining reads were grouped and plotted based on insertion length (regardless of substitutions). For clarity, unmodified reads (indel length of 0 bp) were plotted as 0% of the total reads. The sequences of all spacers used in this study are listed in Table S1.
Cryo-EM sample preparation and data collection
The PlmCasX-sgRNA complex was assembled by incubating protein with a 1.25-fold excess of sgRNA for 30 min at room temperature. The ternary complexes were assembled by incubating dPlmCasX-sgRNA with a 1.5-fold excess of annealed dsDNA target for 30 min at room temperature. After the complexes were assembled, they were purified by size-exclusion chromatography using a Superdex200 10/300 column. PlmCasX complexes at 10 μM concentration in a buffer containing 20 mM HEPES, pH 7.5, 300 mM potassium chloride, 1 mM DTT, and 0.25% glycerol were aliquoted and stored in LN2 for further usage.
For EM sample preparation of dPlmCasX-sgRNAv1-dsDNA, the complex (final concentration 1 μM) was mixed with BS3 cross-linker (final concentration 1 mM) and incubated on ice for 1 hour. 3.7 μL droplets of the sample were placed onto Quantifoil grids (1.2/1.3 μm) with freshly coated graphene-oxide film (https://www.biorxiv.org/content/10.1101/2021.03.08.434344v1). After a 1-minute incubation, the grids were blotted for 3 seconds with a blot force of 4 and immediately plunged into liquid ethane using a FEI Vitrobot MarkIV maintained at 8°C and 100% humidity. Data was acquired using a Thermo Fisher Titan Krios transmission electron microscope operated at 300 keV with an energy filter (GIF quantum 1967), and images were taken at a nominal magnification of ×135,000 (0.9 Å pixel size) with defocus ranging from −0.7 to −2.1 μm. Micrographs were recorded using SerialEM on a Gatan K3 Summit direct electron detector operated in super-resolution mode (Mastronarde, 2003). We collected a 5s exposure fractionated into 50, 100 ms frames with a dose of 10 e- Å−2s−1. In total, 8,675 movies were collected for this sample.
For EM sample preparation of dPlmCasX-sgRNAv2-dsDNA, complex (non-crosslinked) at a concentration of 5 μM was used. Immediately after glow-discharging the grid for 14 seconds using a Solaris plasma cleaner, 3.6 μL droplets of the sample were placed onto C-flat grids (2/2 μm). The grids were blotted for 4 seconds with a blot force of 8 and rapidly plunged into liquid ethane using a FEI Vitrobot MarkIV maintained at 8°C and 100% humidity. Data was acquired by following the same protocol as described above but using 3 exposures per hole. In total, 4,171 movies were collected for this sample.
Single particle cryo-EM analysis
46 frames (the first 2 and last 2 frames were skipped) of each image stack in super-resolution mode were aligned, decimated, summed and dose-weighted using Motioncor2 (Zheng et al., 2017). They were then imported into cryoSparc (Punjani et al., 2017) for patched CTF estimation and particle picking using 2D class-averages of DpbCasX from our previous study(Liu et al., 2019) as templates. 3,652,583 raw particles were picked from dPlmCasX-sgRNAv1-dsDNA dataset, and 1,764,600 particles were picked from dPlmCasX-sgRNAv2-dsDNA dataset. Particle extraction, ab-initio reconstruction, and 3D classification were performed without 2D classification. Good models from 3D classification were further refined using homogenous refinement. In cases when the post-processing in cryoSparc over-sharpened the map, half-maps generated by cryoSparc were imported into Relion (Kimanius et al., 2016) for post-processing. The workflows and more details are summarized in Figure S2 and Figure S3.
Atomic model building and refinement
For dPlmCasX-sgRNAv1-dsDNA, an initial model of PlmCasX was first constructed using homology modeling in the Swiss-model server with the DpbCasX structure (PDB:6NY2) as reference. The sgRNAv1-DNA part was adopted from the DpbCasX structure (PDB:6NY2) with manual revision in Coot. The two parts were fitted into the density map of State I of dPlmCasX-sgRNAv1-dsDNA (2.9 Å resolution) and then manually modified in Coot to better fit the density. The entire model was subjected to PHENIX real space refinement (global minimization and ADP refinement) with secondary structure, Ramachandran, rotamer, and nucleic-acid restraints (Liebschner et al., 2019). The final model was validated using Molprobity (Chen et al., 2010). The atomic model of dPlmCasX-sgRNAv1-dsDNA State II was obtained by running flexible fitting of the State I atomic model against the State II cryo-EM map (3.4 Å resolution) with secondary structure restraints using MDFF (Trabuco et al., 2009). The output model was manually rebuilt in Coot (Casañal et al., 2020) and PHENIX real space refinement was used to improve backbone geometry. The State III atomic model was directly adopted from State I by deleting the Helical-II domain, followed by PHENIX real space refinement against the State III cryo-EM map (3.2 Å resolution).
For model building of dPlmCasX-sgRNAv2-dsDNA in State I, the dPlmCasX-sgRNAv1-dsDNA model in State I was used as the starting model. Then, the sgRNA sequence was modified and the structures were manually rebuilt in Coot. PHENIX real space refinements against dPlmCasX-sgRNAv2-dsDNA EM maps were used to improve the models. The final model was validated using Molprobity.
Quantification and statistical analysis
All statistical analysis was performed using GraphPad Prism 7. The number of independent technical replicates (n) for each experiment are listed in the respective figure legends. For cleavage kinetics plots, error bars represent the standard deviation between replicates and the data were fitted using one-phase association to yield the single turnover rate constant k values (fraction cleaved per minute). For cellular editing bar plots, individual technical replicates (n) were plotted with the bar representing the mean. For box and whisker plots the box represents the 25th, 50th, and 75th percentile, the whiskers represent the 10th and 90th percentile, and outliers are plotted individually. Significances were determined via one-way ANOVA followed by Tukey’s multiple comparisons test. ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
Supplementary Material
Structural rearrangement between State I and State II, related to Figure 2. The atomic coordinates of dPlmCasX-sgRNAv1-dsDNA complexes at State I and State II were used for this analysis. The structural alignment and simulation were performed in UCSF-Chimera. The domains are colored and labeled identical to Figure 2.
Structural dynamics within sgRNAv2, related to Figure 4. 3D variability analysis of the dPlmCasX-sgRNAv2-dsDNA complex in State I was performed in cryoSparc and presented in UCSF-Chimera. The Helical-II domain, sgRNAv2 scaffold stem, sgRNAv2 extended stem and dsDNA are labeled.
Simulation of the structural change from sgRNAv1 to gRNAv2, related to Figure 4. The sgRNAv1 and sgRNAv2 within the R-loop complexes of State I were used for this analysis. The sgRNA is colored in blue with the U31 and U55 nucleotides colored in red. The structural alignment and simulation were performed in UCSF-Chimera.
Table 1.
Cryo-EM data collection, refinement, and validation statistics
| Complex and State | dPlmCasX- sgRNAv1- dsDNA State I |
dPlmCasX- sgRNAv1- dsDNA State II |
dPlmCasX- sgRNAv1- dsDNA State III |
dPlmCasX- sgRNAv2- dsDNA State I |
dPlmCasX- sgRNAv2- dsDNA State II |
dPlmCasX- sgRNAv2- dsDNA State III |
|---|---|---|---|---|---|---|
| EMDB code | EMD-32389 | EMD-32390 | EMD-32391 | EMD-32392 | N/A | N/A |
| PDB code | 7WAY | 7WAZ | 7WB0 | 7WB1 | N/A | N/A |
| Data collection and processing | ||||||
| Magnification | 135,000 | 135,000 | 135,000 | 135,000 | 135,000 | 135,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | ~50 | ~50 | ~50 | ~50 | ~50 | ~50 |
| Defocus range (μm) | 0.5~2.0 | 0.5~2.0 | 0.5~2.0 | 0.5~2.0 | 0.5~2.0 | 0.5~2.0 |
| Pixel size (Å) | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 |
| Symmetry imposed | C1 | C1 | C1 | C1 | C1 | C1 |
| Final particle images (no.) | 520,115 | 502,778 | 710,824 | 616,493 | 267,147 | 143,849 |
| Map resolution (Å) | 2.9 | 3.4 | 3.2 | 3.7 | N/A | N/A |
| FSC threshold | at 0.143 | at 0.143 | at 0.143 | at 0.143 | ||
| Map resolution range (Å) | 2.5~6 | 3~7 | 3~7 | 3~7 | ||
| Refinement | ||||||
| Model resolution (Å) | 2.9 | 3.4 | 3.2 | 3.7 | ||
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | ||
| Model resolution range (Å) | 2.5~6 | 3~7 | 3~7 | 3~7 | ||
| Map sharpening B factor (Å2) | −70 | −120 | −100 | −137 | ||
| Model composition | 11453 | 11034 | 10096 | 11524 | ||
| Non-hydrogen atoms | ||||||
| Protein residues | 960 | 952 | 797 | 960 | ||
| Nucleotides | 175 | 157 | 175 | 178 | ||
| B factors-Mean (Å2) | ||||||
| Protein | 65.72 | 116.41 | 83.97 | 123.99 | ||
| Nucleotide | 106.17 | 161.06 | 157.90 | 157.80 | ||
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.004 | 0.002 | 0.003 | 0.003 | ||
| Bond angles (°) | 0.542 | 0.589 | 0.579 | 0.590 | ||
| Validation | ||||||
| MolProbity score | 1.45 | 2.43 | 1.65 | 1.90 | ||
| Clashscore | 5.49 | 11.37 | 7 | 11.07 | ||
| Poor rotamers (%) | 0.00 | 5.46 | 0 | 0 | ||
| Ramachandran plot | ||||||
| Favored (%) | 97.17 | 95.69 | 96.57 | 95.18 | ||
| Allowed (%) | 2.83 | 4.10 | 3.3 | 4.72 | ||
| Disallowed (%) | 0.00 | 0.21 | 0.1 | 0.1 |
Acknowledgements
EM data were collected at the Tsinghua Cryo-EM facility and the Cal-Cryo facility at UC Berkeley. The data were analyzed using the Bio-Computation platform at the Tsinghua University Branch of the Chinese National Center for Protein Sciences (Beijing). We thank D. B. Toso, J.L. Lei and X.M. Li for expert electron microscopy assistance. We thank T. Yang, Y.K. Wang, A. Chintangal and P. Tobias for computational support. We thank J. Hurtado for providing the GFP HEK293 cell line. We thank N. Krishnappa and the Center for Translational Genomics (Innovative Genomics Institute, UC Berkeley) for assistance with Illumina sequencing. We thank Y. Xue and X.Y. Fang for help analyzing the sgRNA structure. This project was supported by the Chunfeng Fund (project no. 2021Z99CFY020) and start-up funds from Tsinghua University, Beijing (J.J.G.L.); NSF grant no. 1244557 (J.A.D.); NIH grant no. P01GM051487 (J.A.D. and E.N.). C.A.T. is supported by Campus Executive Grants 2101705 and 1655264 through Sandia National Laboratories and an NIH NRSA F31 Pre-doctoral Fellowship (NHLBI, 1F31HL156468-01). J.A.D. and E.N. are Howard Hughes Medical Institute Investigators.
Footnotes
Declaration of interests
J.A.D., E.N., J.J.G.L, C.A.T., M.S.D and E.O. have filed a related patent on CasX mutations and new guide RNAs described herein with the United States Patent and Trademark Office. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics and Mammoth Biosciences, and a Director of Johnson & Johnson. J.A.D is a scientific advisor to Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego and Inari.
References
- Adamcik J, Jeon J-H, Karczewski KJ, Metzler R, and Dietler G (2012). Quantifying supercoiling-induced denaturation bubbles in DNA. Soft Matter 8, 8651–8658. [Google Scholar]
- Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, and Banfield JF (2017). New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Yao L, Li A, Zhang Q, Zhang Z, Wang X, Gani Y, Liu Y, and Zhang Q (2021). A CRISPR/dCasX-mediated transcriptional programming system for inhibiting the progression of bladder cancer cells by repressing c-MYC or activating TP53. Clinical and translational medicine 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casañal A, Lohkamp B, and Emsley P (2020). Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Science 29, 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, and Bode NM (2019). Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nature medicine 25, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, and Doudna JA (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crudele JM, and Chamberlain JS (2018). Cas9 immunity creates challenges for CRISPR gene editing therapies. Nature communications 9, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA (2020). The promise and challenge of therapeutic genome editing. Nature 578, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, and Charpentier E (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346. [DOI] [PubMed] [Google Scholar]
- Hille F, Richter H, Wong SP, Bratovič M, Ressel S, and Charpentier E (2018). The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259. [DOI] [PubMed] [Google Scholar]
- Holm L, and Laakso LM (2016). Dali server update. Nucleic acids research 44, W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, and Doudna JA (2017). CRISPR–Cas9 structures and mechanisms. Annual review of biophysics 46, 505–529. [DOI] [PubMed] [Google Scholar]
- Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, Kim D, Koo T, Ko J-H, and Kim Y-S (2021). Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nature Biotechnology, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, and Scarfò I (2019). Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nature biotechnology 37, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, and Zhang F (2017). Diversity, classification and evolution of CRISPR-Cas systems. Current opinion in microbiology 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin DA, Shutova MV, Johnston NR, Smith OP, Fedorin VV, Kukushkin YS, van der Loo JC, and Johnstone EC (2021). The clinical landscape for AAV gene therapies. Nature reviews Drug Discovery. [DOI] [PubMed] [Google Scholar]
- Le Rhun A, Escalera-Maurer A, Bratovič M, and Charpentier E (2019). CRISPR-Cas in Streptococcus pyogenes. RNA biology 16, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li S, Wu N, Wu J, Wang G, Zhao G, and Wang J (2019). HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS synthetic biology 8, 2228–2237. [DOI] [PubMed] [Google Scholar]
- Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, Lu Z, Zhang Y, Wu J, and Huang X (2018). Base editing with a Cpf1–cytidine deaminase fusion. Nature biotechnology 36, 324–327. [DOI] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung L-W, Jain S, and McCoy AJ (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallographica Section D: Structural Biology 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KL, Chuck J, Tan D, Knott GJ, and Harrington LB (2019). CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJ, Charpentier E, Cheng D, Haft DH, and Horvath P (2019). Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nature Reviews Microbiology, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2003). SerialEM: a program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microscopy and Microanalysis 9, 1182–1183. [Google Scholar]
- Mojica FJ, and Rodriguez-Valera F (2016). The discovery of CRISPR in archaea and bacteria. The FEBS journal 283, 3162–3169. [DOI] [PubMed] [Google Scholar]
- Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, and Doudna JA (2020). CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature methods 14, 290. [DOI] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, and Makarova KS (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED (2019). A catalog of CasX genome editing sites in common model organisms. BMC genomics 20, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski RJ, and Muzyczka N (2014). AAV-mediated gene therapy for research and therapeutic purposes. Annual review of virology 1, 427–451. [DOI] [PubMed] [Google Scholar]
- Takeda SN, Nakagawa R, Okazaki S, Hirano H, Kobayashi K, Kusakizako T, Nishizawa T, Yamashita K, Nishimasu H, and Nureki O (2021). Structure of the miniature type VF CRISPR-Cas effector enzyme. Molecular Cell 81, 558–570. e553. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, and Schulten K (2009). Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang F, and Gao G (2020). CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell 181, 136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, and Doudna JA (2016). Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44. [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhang Y, Yu H, Pan D, Wang Y, Wang Y, Li F, Liu C, Nan H, and Chen W (2021). Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nature Chemical Biology, 1–7. [DOI] [PubMed] [Google Scholar]
- Xiao R, Li Z, Wang S, Han R, and Chang L (2021). Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR–Cas12f nuclease. Nucleic acids research 49, 4120–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chemparathy A, Zeng L, Kempton HR, Shang S, Nakamura M, and Qi LS (2021). Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Molecular Cell. [DOI] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, and Koonin EV (2016). Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gao P, Rajashankar KR, and Patel DJ (2016). PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 167, 1814–1828. e1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, and Patel DJ (2019). CasX: a new and small CRISPR gene-editing protein. Cell research 29, 345–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, and Regev A (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Rube HT, Vakulskas CA, Behlke MA, Bussemaker HJ, and Pufall MA (2020). Systematic in vitro profiling of off-target affinity, cleavage and efficiency for CRISPR enzymes. Nucleic acids research 48, 5037–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural rearrangement between State I and State II, related to Figure 2. The atomic coordinates of dPlmCasX-sgRNAv1-dsDNA complexes at State I and State II were used for this analysis. The structural alignment and simulation were performed in UCSF-Chimera. The domains are colored and labeled identical to Figure 2.
Structural dynamics within sgRNAv2, related to Figure 4. 3D variability analysis of the dPlmCasX-sgRNAv2-dsDNA complex in State I was performed in cryoSparc and presented in UCSF-Chimera. The Helical-II domain, sgRNAv2 scaffold stem, sgRNAv2 extended stem and dsDNA are labeled.
Simulation of the structural change from sgRNAv1 to gRNAv2, related to Figure 4. The sgRNAv1 and sgRNAv2 within the R-loop complexes of State I were used for this analysis. The sgRNA is colored in blue with the U31 and U55 nucleotides colored in red. The structural alignment and simulation were performed in UCSF-Chimera.
Data Availability Statement
The electron density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-32389, EMD-32390, EMD-32391, and EMD-32392 and are publicly available as of the date of publication. The atomic coordinates and structural data have been deposited to the Protein Data Bank (PDB) under the accession numbers 7WAY, 7WAZ, 7WB0 and 7WB1 and are publicly available as of the date of publication. All the accession numbers are also listed in the key resources table. The raw cryo-EM micrographs and movies used in this study are available from the lead contact upon request.
This study did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli Rosetta2 | J. A. Doudna Lab and J.J. G. Liu Lab | N/A |
| Escherichia coli Mach1 T1 | Thermo Fisher | Cat#C862003 |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin | Sigma-Aldrich | Cat#A9518 |
| Phosphatase inhibitor cocktail | Roche | Cat#4906837001 |
| Phenylmethylsulfonyl fluoride (PMSF) | Roche | Cat#10837091001 |
| Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) | Sigma-Aldrich | Cat#C4706 |
| Tobacco Etch Virus (TEV) protease | J. A. Doudna Lab and J.J. G. Liu Lab | N/A |
| Q5 High-Fidelity DNA Polymerase | NEB | Cat#M0491 |
| T7 polymerase | J. A. Doudna Lab and J.J. G. Liu Lab | N/A |
| RNase inhibitor | Promega | Cat#N2615 |
| RNase-Free DNase I | Promega | Cat#M6101 |
| ATP, [g-32P]- 3000Ci/mmol | Perkin Elmer | Cat#BLU002A001MC |
| T4 PNK | NEB | Cat#M0236S |
| Proteinase K | Sangon Biotech | Cat#A600451-0050 |
| BbsI-HF | NEB | Cat#R3539L |
| AgeI | NEB | Cat#R3552L |
| BamHI | NEB | Cat#R0136L |
| KpnI | NEB | Cat#R3142L |
| PciI | NEB | Cat#R0655L |
| Dulbecco’s Modified Eagle’s Medium, high glucose | Gibco | Cat#11995073 |
| Opti-MEM I Reduced Serum Medium | Gibco | Cat#31985070 |
| Fetal Bovine Serum | VWR | Cat#89510-186 |
| Trypsin-EDTA (0.25%), phenol red | Thermo-Fisher | Cat#25200056 |
| Penicillin-Streptomycin | Gibco | Cat#10378016 |
| Puromycin Dihydrochloride | Gibco | Cat#A1113803 |
| BS3 cross-linker | Sigma-Aldrich | Cat#S5799 |
| Graphene-oxide | Sigma-Aldrich | Cat#777676 |
| Critical commercial assays | ||
| MycoAlert Mycoplasma Detection Kit | Lonza | Cat#LT07-318 |
| QuickExtract DNA Extraction Solution | Lucigen | Cat#QE09050 |
| QIAquick PCR Purification Kit | Qiagen | Cat#28104 |
| In-Fusion Snap Assembly Master Mix | Takara | Cat#638948 |
| Cloning Enhancer | Takara | Cat#639615 |
| Lipofectamine 3000 Transfection Reagent | Life Technologies | Cat#L3000001 |
| NucleoSpin Gel and PCR Cleanup Kit | Takara | Cat#740986.20 |
| Deposited data | ||
| Uncropped gels (Mendeley data) | This paper | DOI:10.17632/w6gfw3g5dt.1 |
| Coordinates of the dPlmCasX-sgRNAv1-dsDNA complex (State I) | This paper | PDB: 7WAY |
| Cryo-EM density map of the dPlmCasX-sgRNAv1-dsDNA complex (State I) | This paper | EMDB: EMD-32389 |
| Coordinates of the dPlmCasX-sgRNAv1-dsDNA complex (State II) | This paper | PDB: 7WAZ |
| Cryo-EM density map of the dPlmCasX-sgRNAv1-dsDNA complex (State II) | This paper | EMDB: EMD-32390 |
| Coordinates of the dPlmCasX-sgRNAv1-dsDNA complex (State III) | This paper | PDB: 7WB0 |
| Cryo-EM density map of the dPlmCasX-sgRNAv1-dsDNA complex (State III) | This paper | EMDB: EMD-32391 |
| Coordinates of the dPlmCasX-sgRNAv2-dsDNA complex (State I) | This paper | PDB: 7WB1 |
| Cryo-EM density map of the dPlmCasX-sgRNAv2-dsDNA complex (State I) | This paper | EMDB: EMD-32392 |
| Experimental models: Cell lines | ||
| HEK293T cells | UC Berkeley Cell Culture Facility | N/A |
| GFP HEK293 cells | Laboratory of Juan Hurtado | N/A |
| Oligonucleotides | ||
| ssDNA oligos (see Table S2 for sequences) | IDT | N/A |
| ssRNA oligos (see Table S2 for sequences) | IDT | N/A |
| Recombinant DNA | ||
| His-MBP-TEV-DpbCasX, expression vector | This paper | pJJGL001 Addgene plasmid #180605 |
| His-MBP-TEV-PlmCasX, expression vector | This paper | pJJGL002 Addgene plasmid #180606 |
| His-MBP-TEV-DpbCasX-H2 truncation, expression vector | This paper | N/A |
| His-MBP-TEV-PlmCasX-H2 truncation, expression vector | This paper | N/A |
| His-MBP-TEV-DpbCasX+Plm R3 insertion, expression vector | This paper | pJJGL003 Addgene plasmid #180607 |
| His-MBP-TEV-PlmCasX+Dpb R1 insertion, expression vector | This paper | pJJGL004 Addgene plasmid #180608 |
| His-MBP-TEV-PlmCasX+Dpb R1+2 insertion, expression vector | This paper | N/A |
| U6-sgRNAv1-CAG-DpbCasX-PuroR | Liu et al., 2019 | pBLO62.4 Addgene plasmid #123123 |
| U6-sgRNAv1-CAG-PlmCasX-PuroR | Liu et al., 2019 | pBLO62.5 Addgene plasmid #123124 |
| U6-sgRNAv1-CAG-DpbCasX(ΔH2)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-DpbCasX(R3)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-PlmCasX(ΔH2)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-PlmCasX(R1)-PuroR | This paper | N/A |
| U6-sgRNAv1-CAG-PlmCasX(R1+2)-PuroR | This paper | N/A |
| U6-sgRNAv2-CAG-DpbCasX-PuroR | This paper | pCAT079 Addgene plasmid #180509 |
| U6-sgRNAv2-CAG-DpbCasX(R3)-PuroR | This paper | pCAT105 Addgene plasmid #180510 |
| U6-sgRNAv2-CAG-PlmCasX-PuroR | This paper | pCAT077 Addgene plasmid #180511 |
| U6-sgRNAv2-CAG-PlmCasX(R1)-PuroR | This paper | pCAT100 Addgene plasmid #180512 |
| U6-sgRNAv1-CAG-PlmCasX-mNeonGreen-PuroR | This paper | pCAT526 Addgene plasmid #180513 |
| U6-sgRNAv2-CAG-PlmCasX(R1)-mNeonGreen-PuroR | This paper | pCAT527 Addgene plasmid #180514 |
| Software and algorithms | ||
| Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ImageQuant TL | GE Healthcare | N/A |
| cryoSparc | Punjani et al., 2017 | https://cryosparc.com |
| Relion | Kimanius et al., 2016 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| PyMol | Schrodinger LLC, 2010 | https://pymol.org/2/ |
| UCSF-Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| PHENIX | Liebschner et al., 2019 | https://phenix-online.org/documentation/reference/refinement.html |
| Coot | Casañal et al., 2020 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| FlowJo | BD | https://www.flowjo.com |
| CRISPResso2 | Clement et al. 2019 | https://www.crispresso.pinellolab.partners.org |
| Other | ||
| Magnetic solid phase reversible immobilization (SPRI) beads | UC Berkeley Sequencing Core | N/A |
| 30 kDa MWCO concentrator | Amicon Ultra, Merck | Cat#UFC9030 |
| 3 kDa MWCO concentrator | Amicon Ultra, Merck | Cat#UFC8003 |
| Ni-NTA agarose beads | QIAGEN | Cat#30210 |
| Quantifoil 1.2/1.3 grids | EMS | Cat#Q310CR-14 |
| C-flat 2/2 grids | EMS | Cat#CF-224C-100 |
| HiTrap Heparin HP Columns | GE Healthcare | Cat#17040701 |
| Superdex 200 10/300 column | GE Healthcare | Cat#28990944 |
| QIAprep Spin Miniprep kit | Qiagen | Cat#27106 |