Abstract
The high mortality and financial burden associated with prostate cancer can be partly attributed to a lack of sensitive screening methods for detection and staging of the disease. Guided by in silico docking studies using the crystal structure of PSMA, we designed and synthesized a series of PSMA-targeted 99mTc–chelate complexes for imaging PSMA-expressing human prostate cancer cells (LNCaP cell line). Of the six targeted radioimaging agents synthesized, three were found to bind LNCaP cells with low nanomolar affinity. Moreover, the same three PSMA-targeted imaging agents were shown to localize primarily to LNCaP tumor xenografts in nu/nu mice, with an average of 9.8 ± 2.4% injected dose/g tissue accumulating in the tumor and only 0.11% injected dose/g tissue retained in the muscle at 4 h postinjection. Collectively, these high affinity, PSMA-specific radioimaging agents demonstrate significant potential for use in localizing prostate cancer masses, monitoring response to therapy, detecting prostate cancer recurrence following surgery, and selecting patients for subsequent PSMA-targeted chemotherapy.
Keywords: Prostate-specific membrane antigen, PSMA-targeted radioimaging, 99mTc-radioimaging agents, diagnosis of prostate cancer
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related deaths in American men, amounting to over 30,000 fatalities/year.1 The immense cost of PCa in lives and medical expenses can be attributed, at least in part, to inadequacies in early detection of the disease.2–7 In fact, current methods for diagnosis of PCa often reveal malignant tissue only after it has metastasized,8 emphasizing the need for new diagnostic techniques that can detect both primary and metastatic disease in its earliest stages.
To circumvent problems associated with existing diagnostic techniques, we have looked for a cancer biomarker that is solely expressed on PCa cells. Immunohistochemical analysis of benign and malignant prostate tissues has demonstrated that prostate-specific membrane antigen (PSMA, a plasma membrane-associated protein) is overexpressed on almost all PCa cells. Moreover, the expression level of PSMA has been shown to increase as the stage and grade of the tumor progresses,9–12 especially in hormone refractory PCa13,14 and in prostate tumors that have metastasized to the lymph nodes, bone, rectum, and lung tissues.15–17 PSMA is also expressed in extraprostatic tissues such as the proximal tubules of the kidney, some cells of the intestinal brush-border membrane and the brain.18–20 However, the expression levels of PSMA in these tissues are reported to be several hundred fold lower than in PCa cells.11 Finally, PSMA is detected in the tumor-associated neovasculature of most other solid tumors, yet largely absent from the vasculature of healthy tissues.21 Taken together, PSMA constitutes a good marker for use in PCa-targeted imaging.
Motivated by the above desirable properties, much effort has been devoted to the development of PSMA-targeted imaging agents for PCa.11,22 ProstaScint, the only Food and Drug Administration (FDA) approved imaging agent for PCa, is an 111In-labeled PSMA-targeted monoclonal antibody that recognizes the intracellular domain of the PSMA. Unfortunately, its value in PCa detection has been questioned due to its high false positive rate and poor tumor-to-normal tissue contrast (e.g., tumor-to-muscle ratio = 3:1).22–25 While a second generation antibody to the extracellular domain of PSMA is currently undergoing clinical trials,11,26,27 such antibody-based imaging agents may generally suffer from common drawbacks such as slow clearance from normal tissues, poor tumor penetration, high cost of production, and cumbersome labeling chemistries.28,29
Recently, a variety of low molecular weight inhibitors of PSMA have been radiolabeled and tested as imaging agents in murine models of PCa.30–33 While these radioimaging agents have not yet been approved for clinical use, they have demonstrated considerable advantages over antibody-based imaging agents in both tumor specificity and pharmacokinetics. In this paper, we use the molecular dynamics program, GLIDE, to design a ligand to optimally fit the active site and entry tunnel of PSMA, and we attach this ligand to a 99mTc-chelating moiety for use in imaging PCa. We demonstrate that this newly designed PCa imaging agent displays better tumor to background ratio and more sustained retention in PCa masses than previously described PCa imaging agents.
Experimental Section
Chemicals.
All amino acids and triphosgene were purchased from Chem-Impex Int (Chicago, IL). N-Hydroxy-benzotriazole (HOBt) and O-benzotriazole-N,N,N′,N′-tet-ramethyl-uronium-hexafluoro-phosphate(HBTU)wereobtained from Peptide Int. (Louisville, KY). Sodium pertechnetate (99mTc) was purchased from Cardinal Health (Indianapolis, IN). Palladium–carbon and stannous chloride dihydrate were obtained from Sigma-Aldrich (St. Louis, MO). HC Matrigel was purchased from BD Bioscience (San Jose, CA). All other chemicals were obtained from major suppliers.
General Methods.
Moisture and oxygen sensitive reactions were carried out under argon atmosphere. Solid phase peptide synthesis (SPPS) was performed using a standard peptide synthesis apparatus. Dichloromethane (DCM) was distilled under nitrogen from calcium hydride. Flash chromatography purifications were conducted using silica gel, and TLC was performed on silica gel TLC plates and visualized with UV light and iodine stain. All peptides and peptide conjugates were purified using a preparative reverse phase (RP)-HPLC (Waters, xTerra C18 10 μm; 19 × 250 mm) and analyzed by analytical RP-HPLC (Waters, X-Bridge C18 5 μm; 3.0 × 50 mm). 1H spectra were acquired using a Bruker 500 MHz NMR spectrometer equipped with a TXI cryoprobe. Samples were run in 5 mm NMR tubes using CDCl3 or DMSO-d6/D2O. Presaturation was used to reduce the intensity of the residual H2O peak. All 1H signals are recorded in ppm with reference to residual CHCl3 (7.27 ppm) or DMSO (2.50 ppm), and data are reported as s = singlet, d = doublet, t = triplet, q = quartet, and m = multiplet or unresolved, b = broad, with coupling constants in Hz. Electron impact (EI) high resolution mass spectrometry (HRMS) measurements were obtained in the peak matching mode using a Finnigan MAT XL95 (Bremen, Germany) mass spectrometer. Electrospray ionization (ESI) high resolution mass measurements were performed utilizing the appropriate polypropylene glycol standards. Radioactivity was counted on a Packard γ-counter (Packard Instrument Company, Meriden, CT). The tumor imaging was performed using a Kodak Image Station (In-Vivo FX, Eastman Kodak Company, New Haven, CT).
Cell Culture and Animals.
LNCaP cells were obtained from American Type Culture Collection (Rockville, MD) and grown as a monolayer using 1640 RPMI medium containing 10% heat-inactivated fetal bovine serum (HIFBS), sodium pyruvate (100 mM) and 1% penicillin streptomycin in a 5% carbon dioxide:95% air-humidified atmosphere at 37 °C.
Athymic male nu/nu mice were purchased from NCI Charles River Laboratories (Wilmington, MA), maintained on normal rodent chow, and housed in a sterile environment on a standard 12 h light–dark cycle for the duration of the study.
Docking Studies.
NAALDase inhibitors were assembled in silico, and energy minimizations were conducted using Molecular Operating Environment (MOE) program (Chemical Computational Group, Montreal, Canada) with Optimized Potential for Liquid Simulation (OPLS) force field. Energy minimization was performed until the system reached a root-mean-square (rms) gradient less than or equal to 0.01 (multiple rotamers were used for each radioimaging agent). The minimized structure for each compound was used in the docking simulation. The coordinates of PSMA (GCPII) protein were obtained from the crystal structure of the protein complex with GPI-18431 inhibitor34 taken from the protein data bank (PDB code: 2C6C). The protein coordinates were prepared by adding hydrogen atoms and assigning partial charges from the OPLS-AA (All Atom)35 force field using the Grid-based Ligand Docking with Energetics (GLIDE, FirstDiscovery v7.5, Schrödinger, Inc.)36,37 program. A coarse energy minimization with a small number of steps was performed to relax side chains and the newly added hydrogen atoms for the protein complex with the GPI-18431 inhibitor. The inhibitor was used to determine the location of a docking grid box, and it was removed prior to grid generation. A docking grid enclosing the entire area of the binding site was generated and docking simulations were carried out based on the docking grid. The reproducibility of the docking protocol was evaluated by redocking the GPI-18431 into the catalytic site using the GLIDE XP (extra-precision) docking procedure to ensure that the crystal structure was obtained.
The extra-precision (XP) docking procedure in GLIDE was used to perform the docking simulation of the small molecules into the binding site of the protein. Multiple conformations were generated for each molecule by exhaustive enumeration of the energy minima for rotatable torsion angles, and prescreened for docking by eliminating high-energy conformers.35–37 Due to limitations of the GLIDE program on the number of allowed flexible docking simulations, the conformers of radioimaging compounds were generated by MOE, then docked into the protein active site, and evaluated by GLIDE. The radioimaging moiety (99mTc-DaP-Asp-Cys) was held rigidly during the docking simulation. The binding modes were further confirmed by MOE docking simulations. cLogP was calculated using a fragment-based method implemented in MOE.
Synthesis of Core Ligand: 2-[3-(3-Benzyloxycarbonyl-1-tert-butoxycarbonyl-propyl)-ureido]pentanedioic Acid Di-tert-butyl Ester (9).
Synthesis of 9 was performed according to a reported procedure with minor modification.38 To a solution of l-glutamate di-tert-butyl ester hydrochloride (1.0 g, 3.39 mmol) and triphosgene (329.8 mg, 1.12 mmol) in DCM (25.0 mL) at −78 °C, triethylamine (TEA, 1.0 mL, 8.19 mmol) was added. After stirring for 2 h at −78 °C under argon, a solution of l-Glu(OBn)-OtBu (1.2 g, 3.72 mmol) and TEA (600 μL, 4.91 mmol) in DCM (5.0 mL) was added. The reaction mixture was allowed to come to room temperature (rt) over a period of 1 h and stirred at rt overnight. The reaction was quenched with 1 M HCl, and the organic layer was washed with brine and dried over Na2SO4. The crude product was purified using flash chromatography (hexane:EtOAc = 1:1) to yield 9 (1.76 g, 90.2%) as a colorless oil and crystallized using hexane:DCM. Rf = 0.67 (hexane:EtOAc = 1:1). 1H NMR (CDCl3): δ 1.43 (s, 9H, CH3-tBu); 1.44 (s, 9H, CH3-tBu); 1.46 (s, 9H, CH3-tBu); 1.85 (m, 1H, Glu-H); 1.87 (m, 1H, Glu-H); 2.06 (m, 1H, Glu-H); 2.07 (m, 1H, Glu-H); 2.30 (m, 2H, Glu-H); 2.44 (m, 2H, Glu-H); 4.34 [s (broad), 1H, αH]; 4.38 [s (broad), 1H, α-H]; 5.10 (s, 2H, CH2-Ar); 5.22 [s (broad), 2H, Urea-H); 7.34 (m, 5H, Ar-H). EI-HRMS (m/z): (M + H)+ calcd for C30H47N2O9, 579.3282; found, 579.3289.
Deprotection of Benzyl Group on Core Ligand: 2-[3-(1,3-Bis-tert-butoxycarbonyl-propyl)-ureido]pentanedioic Acid 1-tert-Butyl Ester (10).
To a solution of 9 (250 mg, 432 mmol) in DCM, 10% Pd/C was added. The reaction mixture was hydrogenated at 1 atm for 24 h at rt. Pd/C was filtered through a Celite pad and washed with DCM. The crude product was purified using flash chromatography (hexane: EtOAc = 40:60) to yield 10 (169 mg, 80.2%) as a colorless oil, and crystallized using hexane:DCM. Rf = 0.58 (hexane: EtOAc = 40:60). 1H NMR (CDCl3): δ 1.46 (m, 27H, CH3-tBu); 1.91 (m, 2H,Glu-H); 2.07 (m, 1H, Glu-H); 2.18 (m, 1H, Glu-H); 2.33 (m, 2H, Glu-H); 2.46 (m, 2H, Glu-H); 4.31 (s (broad), 1H, αH); 4.35 (s (broad), 1H, α-H); 5.05 (t, 2H, Urea-H); EI-HRMS (m/z): (M + H)+ calcd for C23H41N2O9, 489.2812; found, 489.2808.
General Procedure for Solid Phase Peptide Synthesis of Peptide 11, That Is, the Peptide for Radiotracer 5.
Fmoc-Cys(4-methoxytrityl)-Wang resin (100 mg, 0.43 mM) was swollen with DCM (3 mL) followed by (dimethyl forma-mide) (DMF, 3 mL). A solution of 20% piperidine in DMF (3 × 3 mL) was added to the resin, and argon was bubbled for 5 min. The resin was washed with DMF (3 × 3 mL) and isopropyl alcohol (i-PrOH, 3 × 3 mL). Formation of free amine was assessed by the Kaiser test. After swelling the resin in DMF, a solution of Fmoc-Asp(OtBu)-OH (2.5 equiv), HBTU (2.5 equiv), HOBt (2.5 equiv), and DIPEA (4.0 equiv) in DMF was added. Argon was bubbled for 2 h, and resin was washed with DMF (3 × 3 mL) and i-PrOH (3 × 3 mL). The coupling efficiency was assessed by the Kaiser Test. The above sequence was repeated for 5 more coupling steps. Final compound was cleaved from the resin using a trifluoroacetic acid (TFA):H2O:triisopropylsilane:ethanedithiol cocktail (92.5:2.5:2.5:2.5) and concentrated under vacuum. The concentrated product was precipitated in diethyl ether and dried under vacuum. The crude product was purified using preparative RP-HPLC [λ = 220 nm; solvent gradient: 1% B to 50% B in 25 min, 80% B wash 30 min run; A = 0.1% TFA, pH = 2; B = acetonitrile (ACN)]. ACN was removed under vacuum, and pure fractions were freeze-dried to yield 11 as a white solid. Analytical RP-HPLC: tR = 7.8 min [A = 0.1 TFA, B = acetonitrile (ACN), solvent gradient: 5% B to 80% B in 10 min, 80% B wash 15 min run]. 1H NMR: δ 0.93 (m, 2H); 1.08 (m, 5H); 1.27 (m, 5H); 1.69 (m, 2H); 1.90 (m, 2H); 1.94 (m, 2H); 2.10 (m, 2H); 2.24 (q, 2H); 2.62 (m, 2H); 2.78 (m, 4H); 2.88 (m, 1H); 2.96 (t, J = 6.8 Hz, 2H); 3.01 (m, 1H); 3.31 (dd, J = 13.9, 5.9 Hz, 1H); 3.62 (dd, J = 14.0, 5.9 Hz, 1H); 3.80 (q, J = 6.1 Hz, 1H); 4.07 (m, 1H); 4.37 (m, 1H); 4.42 (m, 2H); 4.66 (m, 1H); 7.18 (m, 10H). ESI-HRMS (m/z): (M + H)+ calcd for C47H66N9O17S, 1060.4297; found, 1060.4290. UV/vis: λmax = 257 nm.
Characterization of peptide for radiotracer 3:
Characterization of peptide for radiotracer 3: white solid. Analytical RP-HPLC: tR = 6.3 min (1% B to 50% B in 10 min, 80% B wash 15 min run). 1H NMR: δ 1.70 (m, 2H); 1.92 (m, 2H); 2.17 (m, 2H); 2.23 (m, 2H); 2.57 (dd, J = 9.0, 7.9 Hz, 1H); 2.77 (m, 4H); 3.45 (dd, J = 9.7, 4.6 Hz, 1H); 3.54 (dd, J = 8.0, 6.3 Hz, 1H); 3.83 (t, 1H, J = 5.5 Hz, αH); 4.06 (m, 1H, αH); 4.38 (t, J = 5.9 Hz, 1H, α-H); 4.63 (t, J = 6.6 Hz, 1H, α-H). ESI-HRMS (m/z): (M + H)+ calcd for C21H33N6O14S, 625.1776; found, 625.1784. UV/vis: λmax = 220 nm.
Characterization of peptide for radiotracer 4:
Characterization of peptide for radiotracer 4: white solid. Analytical HPLC: tR = 8.2 min (1% B to 50% B in 10 min, 80% B wash 15 min run; A = 10 mM NH4OAc, pH = 5; B = ACN). 1H NMR: δ 1.62 (m, 1H); 1.70 (m, 2H); 1.80 (m, 1H); 1.90 (m, 2H); 2.09 (m, 2H); 2.17 (m, 2H); 2.24 (m, 2H); 2.60 (dd, J = 8.7, 8.5 Hz, 1H); 2.75 (m, 4H); 2.81 (m, 1H); 2.97 (dd, J = 9.2, 4.4 Hz, 1H); 3.33 (dd, J = 8.1, 5.9 Hz, 1H); 3.60 (dd, J = 8.0, 6.0 Hz, 1H); 3.81 (t, J = 6.0 Hz, 1H, αH); 4.07 (m, 2H, αH); 4.33 (m, 1H, α-H); 4.39 (t, J = 5.9 Hz, 1H, α-H); 4.65 (m, 1H, α-H); 7.20 (m, 5H, Ar-H). ESI-HRMS (m/z): (M + H)+ calcd for C35H49N8O18S, 901.2886; found, 901.2892. UV/vis: λmax = 220 nm.
Characterization of peptide for radiotracer 6:
Characterization of peptide for radiotracer 6: white solid. Analytical HPLC: tR = 7.4 min (1% B to 80% B in 10 min, 80% B wash 15 min run; A = 10 mM NH4OAc, pH = 5; B = ACN). 1H NMR: δ 0.94 (m, 2H); 1.08 (m, 5H); 1.27 (m, 5H); 1.66 (m, 2H); 1.70 (m, 2H); 1.79 (m, 1H); 1.90 (m, 2H); 2.09 (m, 2H); 2.74 (m, 2H); 2.84 (m, 1H); 2.95 (t, J = 6.7 Hz, 3H); 3.07 (bd, 2H); 3.23 (m, 1H); 3.43 (m, 1H); 3.52 (m, 1H); 3.78 (m, 1H, αH); 3.81 (m, 1H, αH); 3.88 (m, 1H, αH); 4.11 (m, 1H, αH); 4.39 (m, 2H, α-H); 7.14 (m, 1H, Ar-H); 7.21 (m, 4H, Ar-H). ESI-HRMS (m/z): (M + H)+ calcd for C43H64N9O19S, 1042.4039; found, 1042.4049. UV/vis: λmax = 257 nm.
Characterization of peptide for radiotracer 7:
Characterization of peptide for radiotracer 7: white solid. Analytical HPLC: tR = 8.2 min (1% B to 50% B in 10 min, 80% B wash 15 min run; A = 0.1% TFA, B = ACN). 1H NMR: δ 1.55 (m, 1H); 1.73 (m, 4H); 1.89 (m, 1H); 1.97 (m, 1H); 2.06 (m, 2H); 2.17 (t, J = 7.15 Hz, 2H); 2.24 (q, J = 7.3 Hz, 2H); 2.75 (m, 8H); 3.00 (m, 3H); 3.32 (dd, J = 7.7, 5.8 Hz, 1H); 3.63 (m, 4H); 3.80 (q, J = 5.5 Hz, 2H, αH); 3.94 (q, J = 5.3 Hz, 1H, αH); 4.06 (m, 1H, αH); 4.38 (m, 4H, α-H); 4.46 (dd, J = 5.2, 4.7 Hz, 1H, α-H); 4.65 (t, J = 6.6 Hz, 1H, α-H); 4.68 (t, J = 4.5 Hz, 1H, α-H); 7.18 (m, 15H, Ar-H). ESI-HRMS (m/z): (M + H)+ calcd for C55H70N11O21S, 1252.4463; found, 1252.4414. UV/vis: λmax = 257 nm.
Characterization of peptide for radiotracer 8:
Characterization of peptide for radiotracer 8: colorless solid. Analytical HPLC: tR = 8 min (1% B to 50% B in 10 min, 80% B wash 15 min run; A = 0.1% TFA, B = ACN). 1H NMR: δ 1.66 (m, 2H); 2.07 (m, 4H); 2.31 (t, J = 6.2 Hz, 1H); 2.43 (m, 1H); 2.77 (m, 2H); 2.98 (dd, J = 9.3, 4.3 Hz, 1H); 3.14 (t, J = 5.7 Hz, 2H); 3.24 (d, J = 6.1 Hz, 1H); 3.40 (m, 4H, PEG-H); 3.46 (s, 24H, PEG-H); 3.78 (t, J = 5.5 Hz, 1H); 3.81 (t, J = 5.5 Hz, 1H); 4.03 (m, 1H, α-H); 4.40 (m, 2H, α-H); 7.16 (m, 1H, Ar-H); 7.22 (m, 4H, Ar-H). ESI-HRMS (m/z): (M + H)+ calcd for C45H71N8O22S, 1107.4406; found, 1107.4377. UV/vis: λmax = 257 nm.
General Procedure for Preparation of Nonradioactive Formulation and Procedure for Labeling with 99mTc.
A solution of stannous chloride dihydrate (0.8 mg, 0.003 mmol) in 0.02 M HCl (0.8 mL) was added to a solution of sodium α-d-glucoheptonate dihydrate (800 mg, 2.815 mmol) in argon purged water (5.0 mL). Requisite peptide (0.001 mmol) was then added to the reaction mixture while purging with argon. After adjusting the pH of the solution to 6.8 ± 0.2 using 0.1 N NaOH, argon purged water was added to achieve a total volume 10.0 mL. The solution mixture was dispensed into 5 mL vials (1.0 mL/ vial) under argon atmosphere and lyophilized for 36 h. The vials were sealed under argon atmosphere to yield the nonradioactive formulation kits, which were stored at −20 °C until use.
A solution of sodium pertechnetate 99mTc (1.0 mL, 15 mCi) was added to a vial, heated in a boiling water bath for 18 min, and then cooled to room temperature before use. Radiochemical purity was analyzed using RP-HPLC.
General Procedure for LNCaP Cell Binding Studies.
LNCaP cells (150,000 cells/well) were seeded into 24-well Falcon plates and allowed to form confluent monolayers over 48 h. Spent medium in each well was replaced with fresh medium (0.5 mL) containing increasing concentrations of the PSMA-targeted test article. After incubating for 1 h at 37 °C, cells were rinsed with culture medium (2 × 1.0 mL) and tris buffer (1 × 1.0 mL) to remove any unbound radioactivity. Cells were then resuspended in tris buffer (0.5 mL), and cell bound radioactivity was counted using a γ-counter. The dissociation constant (KD) was calculated using a plot of cell bound radioactivity versus the concentration of the radiotracer using GraphPad Prism 4 and assuming a noncooperative single site binding equilibrium.
Generation of Subcutaneous LNCaP Tumors in Athymic nu/nu Mice and Their Use in Imaging and Biodistribution Studies.
Five-week-old male nu/nu mice were inoculated subcutaneously on the shoulder with LNCaP cells (5.0 × 106 cells/mouse in 50% HC Matrigel). Growth of the tumors was measured in two perpendicular directions every 2 days using a caliper, and the volumes of the tumors were calculated as 0.5 × L × W2 (L = measurement of longest axis, and W = measurement of axis perpendicular to L in millimeters). Body weights were monitored on the same schedule as tumor volume measurements. Once tumors reached between 400 and 500 mm3 in volume, animals were treated (ip) with the desired PSMA-targeted test article (67 nmol, 150 μCi) in saline (100 μL). After 4 h, animals were sacrificed by CO2 asphyxiation and images were acquired with a Kodak Imaging Station coupled to a CCD camera using Kodak molecular imaging software (version 4.0). Radioimages: illumination source = radio isotope, acquisition time = 3 min, f-stop = 4, focal plane = 5, FOV = 160, binning = 4. White light images: illumination source = white light transillumination, acquisition time = 0.05 s, f-stop = 16, focal plane = 5, FOV = 160 with no binning.
Following imaging, animals were dissected and selected tissues were collected into preweighed γ-counter tubes. Radioactivity of weighed tissues and test articles were counted in a γ-counter. CPM values were decay corrected and results were calculated as % injected dose per gram of wet tissue (% ID/g).
Results
Selection of a PSMA-Targeting Ligand Using a Docking Simulation.
Although the function of PSMA on PCa cells remains unknown, it has been established to play a pivotal role in the central nervous system, where it hydrolyzes N-acetyl-aspartyl glutamate (NAAG) to glutamate plus N-acetyl-aspartate and is consequently known as NAALADase or glutamate carboxypeptidase II.11 Because high levels of the resulting glutamate can damage neuronal cells and cause neurological disorders, efforts have been made to develop low molecular weight inhibitors of NAALADase.30,39 The fact that PSMA and NAALADase40 are in fact the same protein motivated us to investigate whether these high affinity NAALADase inhibitors could be adapted for use in targeting imaging and therapeutic agents to PSMA-positive PCa cells.
In order to select the optimal targeting ligand for PSMA, in silico docking studies were conducted using a high resolution crystal structure of the ectodomain of NAALADase in complex with one of its high affinity inhibitors (GPI-18431).34 To validate the docking method, the crystal complex was first regenerated using the GLIDE program36,37 and an OPLS-AA force field,35 and the above inhibitor (GPI-18431) was redocked into the catalytic site using a GLIDE XP docking procedure. Superimposition of the redocked GPI-18431 with the complex’s crystal structure demonstrated that the docking program could accurately reproduce the known orientation of GPI-18431 in the enzyme’s active site (rms deviation <0.5 Å, see Supplementary Figure 2a in the Supporting Information). A series of other NAALADase inhibitors were then docked using the same protocol to investigate the conformational fits of functionally related chemical groups and to determine which chemical moieties might be essential for high affinity binding. Analysis of the docking results indicated that phosphorus and urea based inhibitors would bind similarly in the catalytic site and generate high affinity interactions (see Supplementary Figure 2b–f in the Supporting Information). From examination of over 100 inhibitors, 2-[3-(1,3-dicarboxypropyl)-ureido]pentanedioic acid (DUPA) attracted the most interest, because it was found to share the greatest number of docking interactions with GPI-18431 (Figure 1a,b) and because it was known to bind NAALADase with high affinity.30 Thus, the oxygen atom of the urea moiety in DUPA was found to directly coordinate with the zinc atom(s) in the active site. Moreover, the α and α′-carboxylic acids of DUPA were seen to interact with Arg210 and Arg534 of NAALADase, and the γ-carboxylic acid of DUPA was found to form a third salt bridge with Lys699 of the enzyme. Because additional hydrophobic and hydrogen bonding interactions were calculated to reinforce this latter interaction, DUPA was predicted to bind PSMA avidly without need for involvement of its γ′-carboxylic acid in the binding interaction. Maintenance of the γ′-carboxylic acid free from contact with NAALADase was important, since it permitted use of this fourth carboxylic acid in conjugation reactions with imaging and therapeutic agents.
Figure 1.
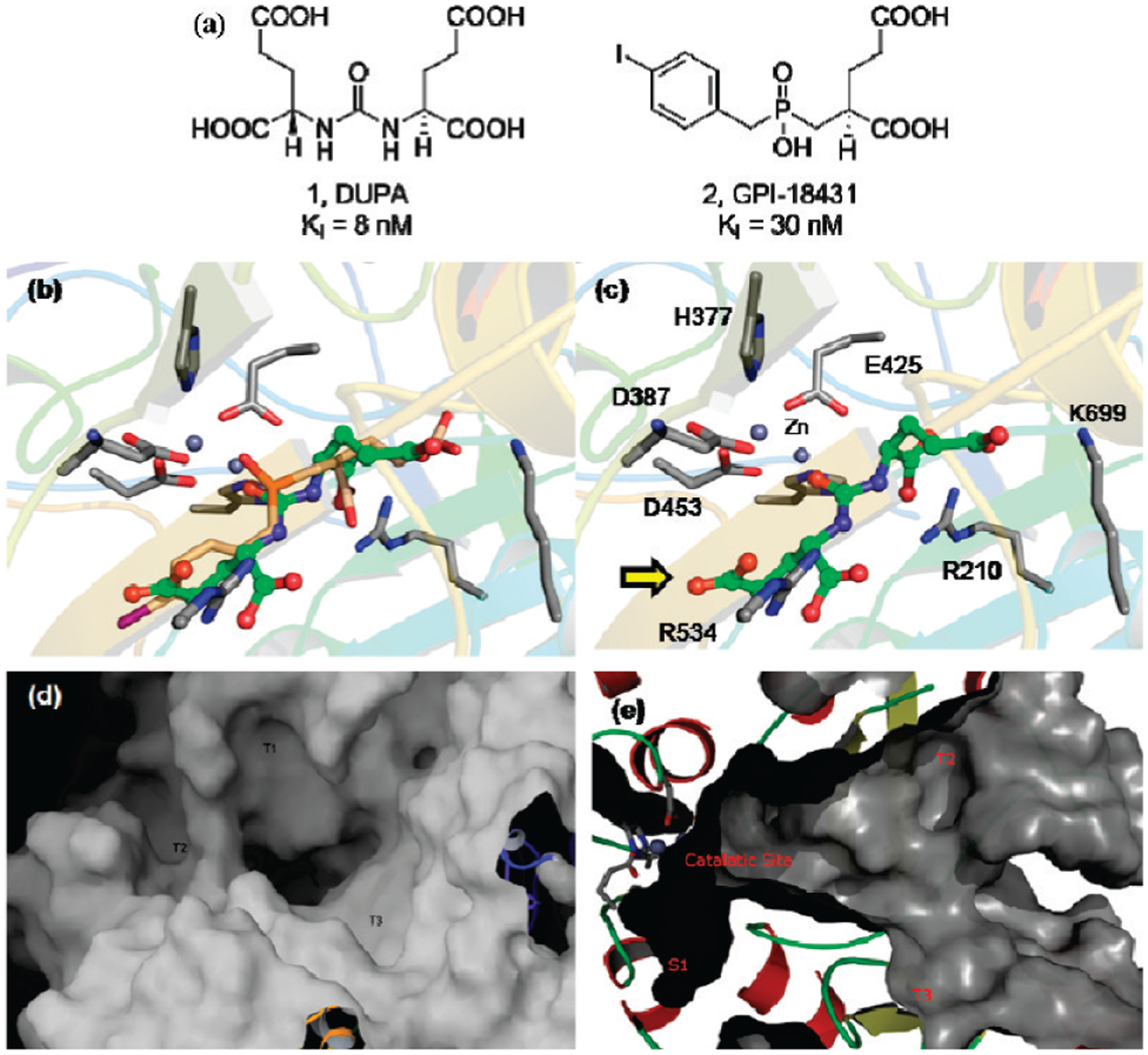
(a) Structures of DUPA and GPI-18431. (b–d) Structures were docked into the active site of NAALADase (PSMA) using the computer program GLIDE as described in the Experimental Section. Superimposition of (b) docked DUPA (green) with GPI-18431 (brown) and (c) docking interactions of DUPA (green) with residues in the active site of NAALADase. The protein is depicted in ribbon-cartoon mode, and side chains (indicated in standard one letter code) are depicted in stick mode. Yellow arrow indicates γ′-carboxylic acid of DUPA. The surface representation of the gradually narrowing 20 Å deep tunnel that extends from the protein surface to the active site [(d) top view and (e) side view]. T1, T2, and T3 label the cavities along the tunnel in panel d.
Design of a 99mTc-Based Radioimaging Agent.
As a result of its ease of preparation, low cost, convenient half-life (t1/2 = 6.01 h), negligible toxicity, and desirable emission energy (141 keV, 89% abundance),41 99mTc is the major radionuclide used in nuclear medicine today. Because the coordination chemistry of the Dap-Asp-Cys moiety allows chelation of 99mTc with high affinity and stability,42 a strategy for linking DUPA to DaP-Asp-Cys without compromising DUPA’s affinity for NAALADase was investigated.
Examination of the crystal structure of NAALADase (PSMA) revealed that access to the DUPA binding site could only be reached through a gradually narrowing 20 Å deep tunnel34 lined with two hydrophobic pockets and an Arg cluster (Figure 1c,d). Therefore, to design a DUPA conjugate with no steric overlap but optimal complementarity to the contours and chemistry of the tunnel, a series of conjugates were assembled in silico containing DUPA as the targeting ligand linked to DaP-Asp-Cys via spacers of varying lengths and chemistries (Figure 2). Optimal binding was found to occur with a spacer containing two Phe residues at appropriate positions between DUPA and the chelating moiety. Several closely related DUPA conjugates were therefore synthesized for evaluation of these predictions.
Figure 2.
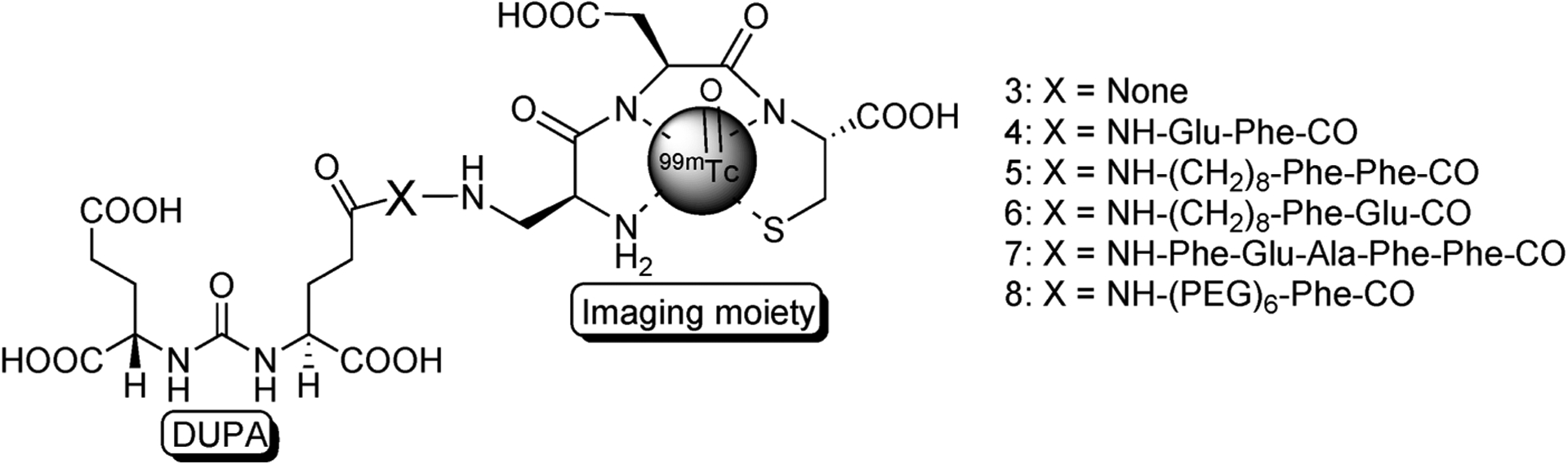
Structures of PSMA-targeted radiotracers. X = peptide spacer, in which amino acids are indicated using their three letter abbreviations.
Synthesis of Radioimaging Agents.
The synthesis of tert-butyl protected DUPA is shown in Scheme 1. Commercially available tert-butyl-protected glutamic acid was reacted with triphosgene to produce the isocyanate intermediate followed by addition of γ-benzoylated glutamic acid to furnish compound 9 in high yield. Deprotection of the benzyl group of 9 with activated palladium–carbon yielded 10. Assembly of DUPA-spacer-Dap-Asp-Cys was then accomplished using standard fluorenylmethoxycarbonyl (Fmoc) solid phase peptide synthesis (SPPS) starting from Fmoc-Cys(Trityl)-Wang resin according to the synthesis shown in Scheme 2. The final product was purified using preparative RP-HPLC and characterized using analytical RP-HPLC, 1H NMR, and HRMS.
Scheme 1.
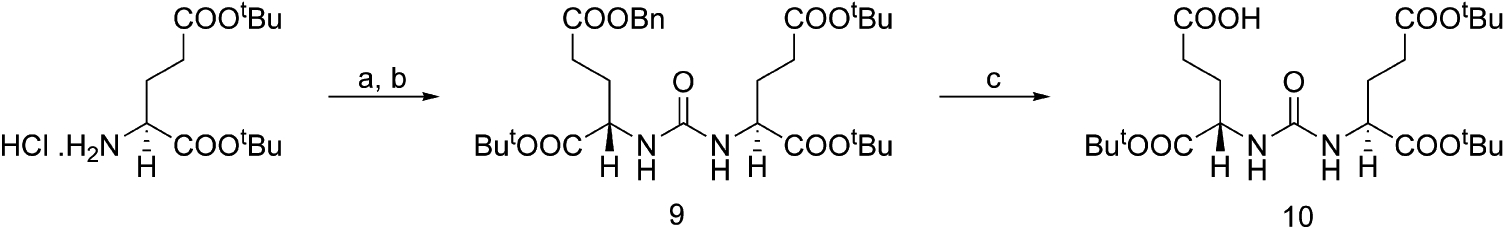
a Reagents and conditions: (a) triphosgene, TEA/ DCM, −78 °C; (b) H-l-Glu(OBn)-OtBu • HCl; (c) H2; Pd–C/DCM.
Scheme 2.
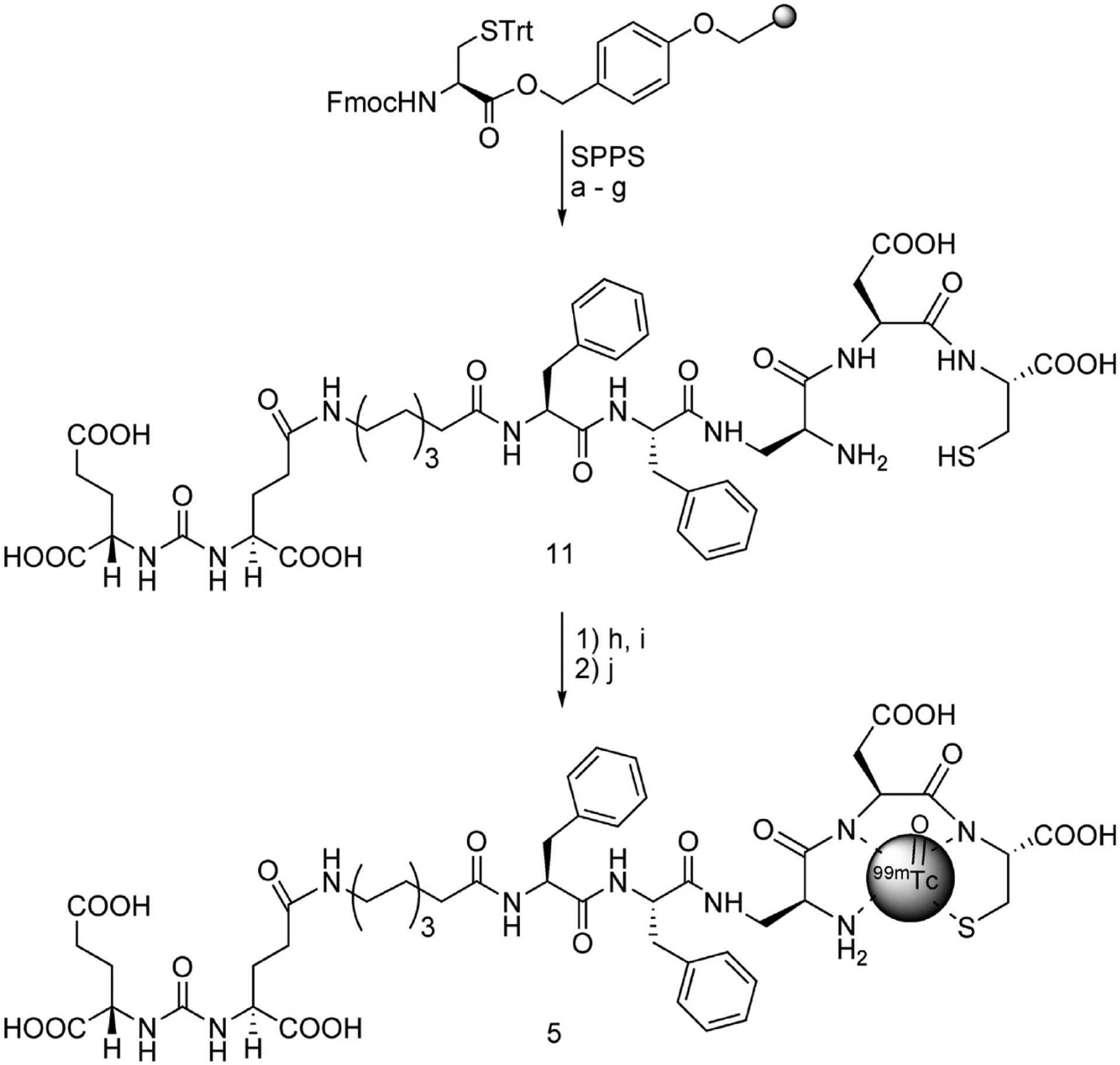
a Reagents and conditions: (a) (i) 20% piperidine/DMF, rt, 10 min; (ii) Fmoc-Asp(OtBu)-OH, HBTU, HOBt, DIPEA, 2 h; (b) (i) 20% piperidine/DMF, rt, 10 min; (ii) Fmoc-diaminopropionic (DAP) acid, HBTU, HOBt, DIPEA, 2 h; (c) (i) 20% piperidine/DMF, rt, 10 min; (ii) Fmoc-Phe-OH, HBTU, HOBt, DIPEA, 2 h; (d) (i) 20% piperidine/DMF, rt, 10 min; (ii) Fmoc-Phe-OH, HBTU, HOBt, DIPEA, 2 h; (e) (i) 20% piperidine/DMF, rt, 10 min; (ii) Fmoc-8-aminooctanoic (EAO) acid, HBTU, HOBt, DIPEA, 2 h; (f) (i) 20% piperidine/DMF, rt, 10 min; (ii) 9, HBTU, HOBt, DIPEA, 2 h; (g) TFA/H2O/TIPS/EDT (92.5:2.5:2.5:2.5), 1 h; (h) SnCl2, sodium glucoheptonate/H2O; (i) aq NaHCO3/pH = 6.8–7.2; (j) sodium pertechnetate/saline, 100 °C, 18 min.
Because of the short half-life of 99mTc, a formulation kit was required that would enable rapid and efficient labeling of the DUPA conjugate directly prior to injection. This kit was prepared using a lyophilized mixture of DUPA-spacer-Dap-Asp-Cys, stannous chloride to reduce 99mTc-pertechnetate, and sodium α-d-glucoheptonate to stabilize the Sn(II) and 99mTc(IV) intermediates. After addition of Na99mTcO4−, complexation of the radiotracer was achieved by heating the solution for 18 min to boiling and then cooling to room temperature. The desired product was obtained in high yield (>98%) and high specific radioactivity (purity >98%).
Analysis of Binding Affinity to PSMA-Positive Human PCa (LNCaP Cell Line) Cells.
The affinities of the various radioimaging agents for PSMA were evaluated by measuring association of each test article with PSMA-expressing LNCaP cells. Representative binding curves for radioimaging agents 3–8 are shown in Figure 3a, and their dissociation constants (KD) derived from Scatchard analysis are provided in Table 1. Radioimaging agents 4–6 were found to bind LNCaP cells with an affinity comparable to unmodified DUPA, and for all three conjugates, binding could be quantitatively inhibited by competition with 100-fold excess PMPA (potent inhibitor of NAALADase39) (Figure 3b). Moreover, incubation of radioimaging agent 5 with LNCaP cells at both 4 °C (where endocytosis cannot occur) and 37 °C (where endocytosis is permitted) demonstrated internalization of the DUPA conjugate at physiological temperature (Figure 3c). As anticipated, radioimaging agent 3, containing no spacer, exhibited weak affinity for PSMA, probably due to steric overlap between the bulky 99mTc- Dap-Asp-Cys moiety and residues of the tunnel. Imaging agent 8 containing a PEG spacer also showed weak affinity for LNCaP cells, probably at least in part due to the absence of phenylalanine residues that would otherwise have occupied the hydrophobic pockets lining the tunnel. Similarly, the somewhat lower affinities of radiotracers 4 and 6 (Figure 4e) may have arisen from the presence of only one rather than two phenyl rings in the spacer decreasing their hydrophobicity. Importantly, the most avid interaction was found with radioimaging agent 5, which not only showed the highest degree of superimposition with DUPA and GPI-18341 in the active site (Figure 4a) but also allowed the bulky 99mTc-Dap-Asp-Cys moiety to reside at the end of the tunnel where it would encounter no steric interference (Figure 4b,c,f).
Figure 3.
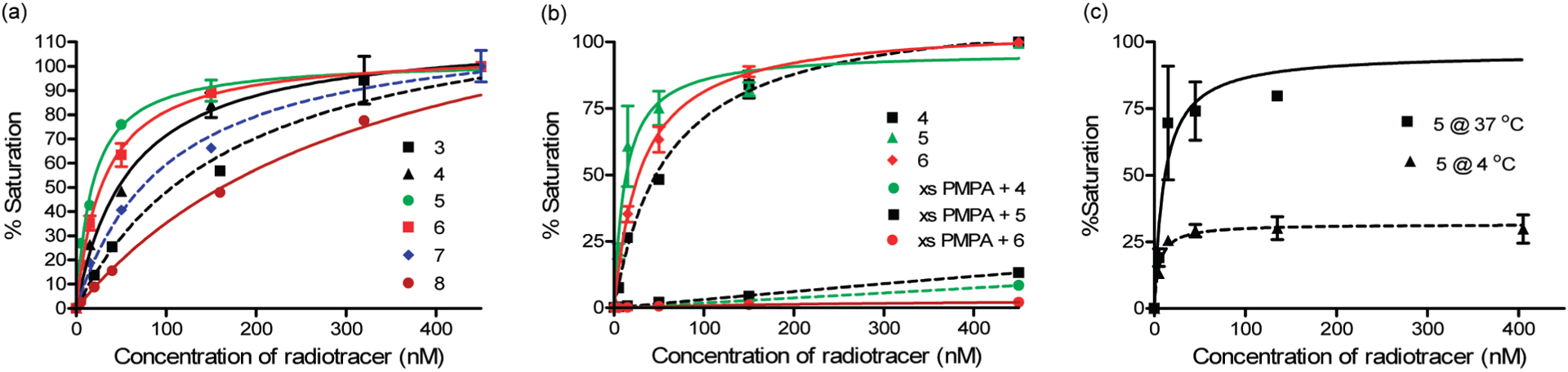
(a) Binding of PSMA-targeted radiotracers containing different spacers to PSMA-positive human prostate cancer cells (LNCaP) in culture. (b) Competitive inhibition of the binding of the 3 best DUPA conjugates (compounds 4, 5, and 6) with excess PMPA is shown in the middle panel. This nearly quantitative competition demonstrates that DUPA conjugate binding is highly specific. (c) Binding of compound 5 to cultured LNCaP cells for 1 h at 4 and 37 °C was compared as a means of assessing the rate of internalization of the DUPA-targeted conjugate. Error bars represent SD (n = 3).
Table 1.
Binding Affinities of Radioimaging Agents to PSMA-Positive LNCaP in Culture and Calculated cLogP Values for PSMA-Targeted Radioimaging Agents
| compound | peptide spacer (X) | cLogPa | KD (nM) |
|---|---|---|---|
| 3 | none | −3.94 | 176 |
| 4 | NH-Glu-Phe-CO | −3.45 | 60 |
| 5 | NH-(CH2)8-Phe-Phe-CO | −0.27 | 13.8 ± 3.9b |
| 6 | NH-(CH2)8-Phe-Glu-CO | −2.11 | 31 |
| 7 | NH-Phe-Glu-Ala-Phe-Phe-CO | −4.64 | 102 |
| 8 | NH-PEG6-Phe-CO | −2.16 | 338 |
cLogP = calculated log (partition coefficient).
SD (n = 3).
Figure 4.
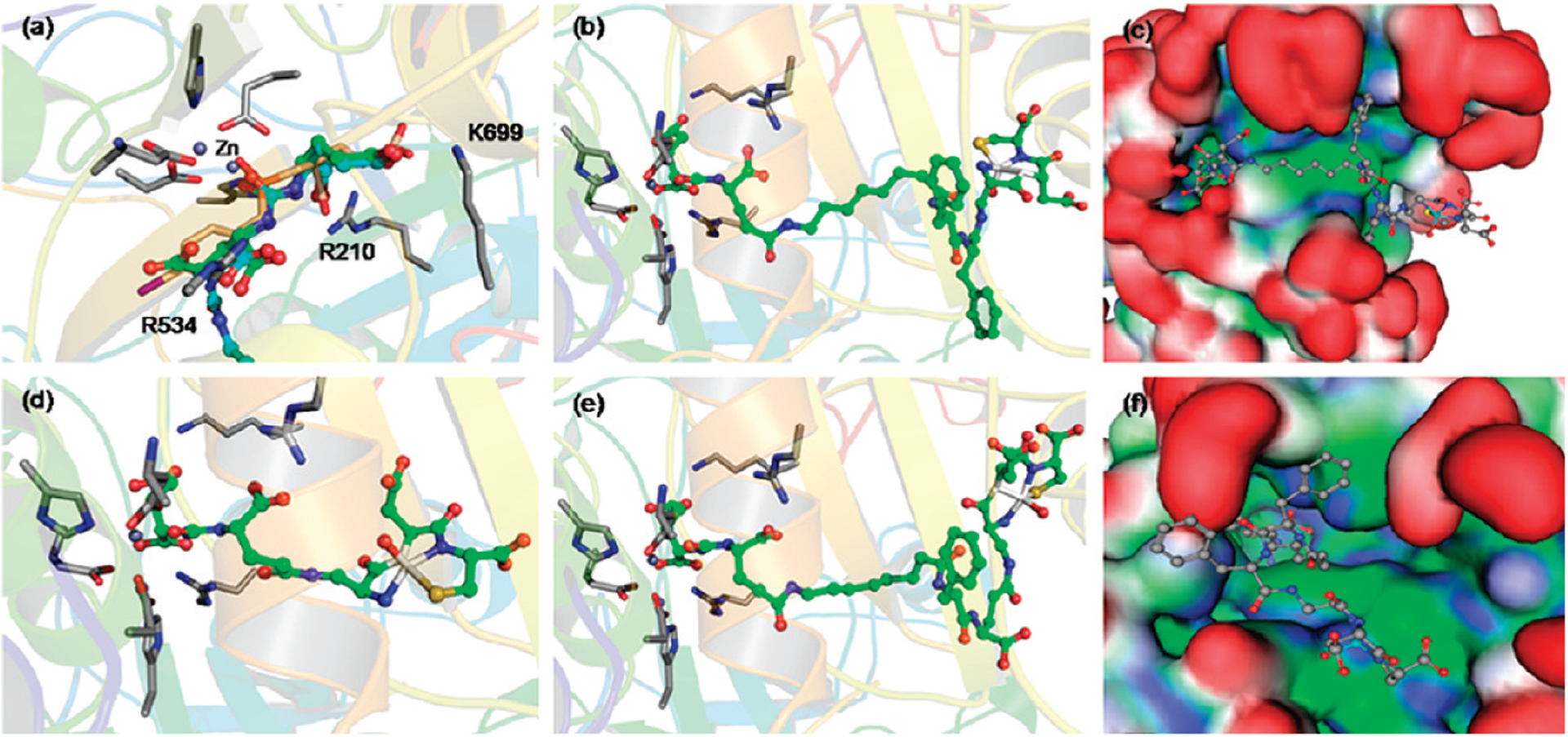
(a) Molecular dynamics prediction of the superimposition of radioimaging agent 5 (cyan) with DUPA (green) and GPI-18314 (brown) in the active site of the protein. The protein is depicted in ribbon-cartoon mode, and side chains are depicted in stick mode (indicated in standard one letter code). Binding modes of compounds 5 [(b), (c) side and (f) top view (hydrophobic sites = green, hydrophilic sites = blue, surfaces exposed to solvent = red)], (d) compound 3 and (e) compound 6.
Whole Body Imaging and Biodistribution of DUPA Conjugates in Tumor-Bearing Mice.
In order to evaluate the specificities of the various DUPA-targeted radioimaging agents in vivo, each agent was injected intraperitoneally into athymic nu/nu mice bearing LNCaP tumors and the distribution of retained radioactivity was examined 4 h later by radioimaging and γ-counting. As seen in Figure 5, radiotracers 4 (b), 5 (c, d), 6 (e), and 7 (f) accumulated predominantly in the PSMA-positive LNCaP tumors, with no substantial radioactivity in other organs except the kidneys. In contrast, tumor-specific accumulation of radiotracers 3 (Figure 5a) and 8 was not observed. More importantly, tumor uptake of compound 5 was shown to be PSMA-mediated, since no uptake was seen in two PSMA-negative human tumor xenografts43 and since coinjection of excess PMPA eliminated LNCaP tumor uptake (panel d).
Figure 5.
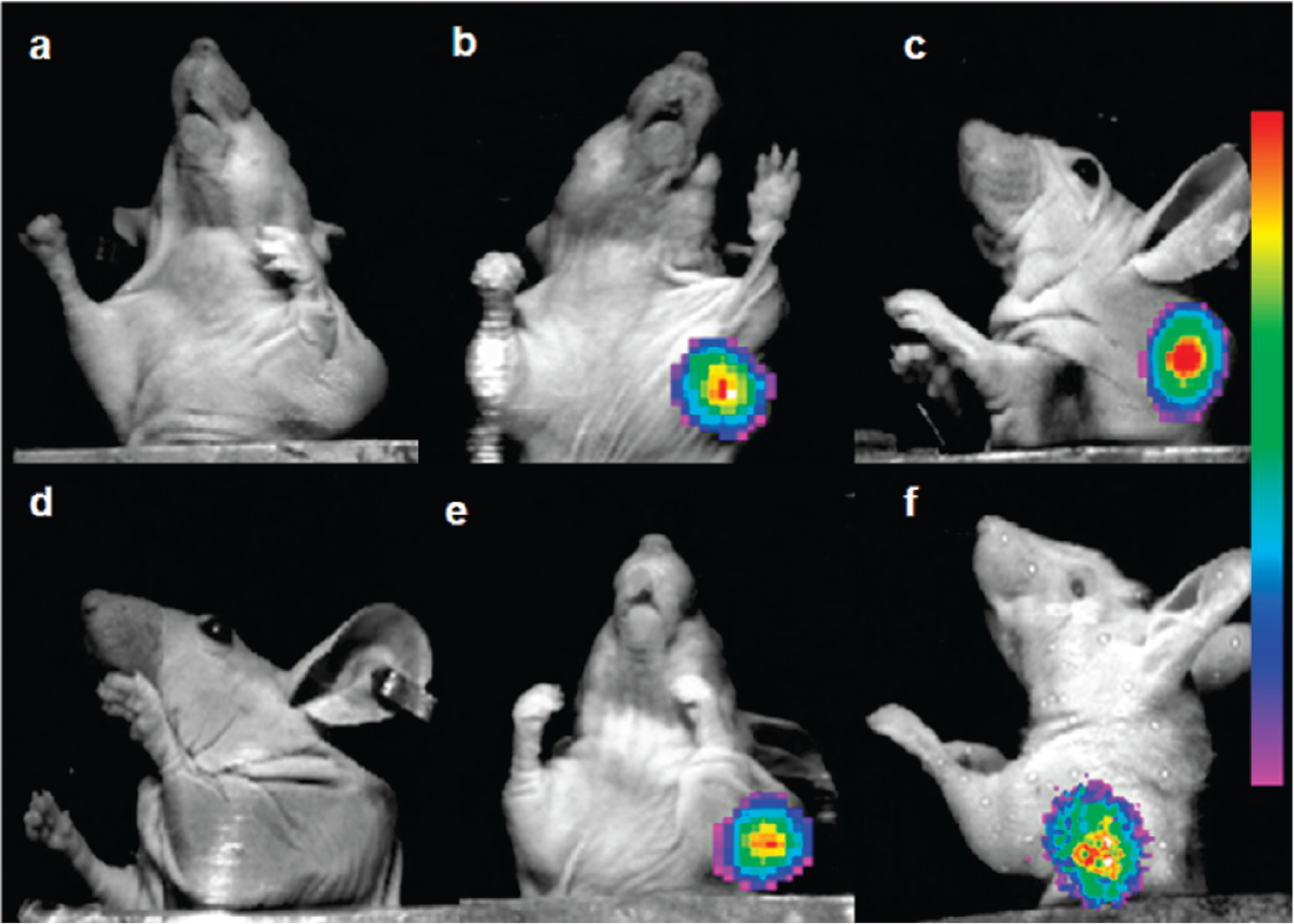
Overlay of whole-body radioimages (rainbow colors) on white light photographs of mice bearing LNCaP tumor xenografts that were treated with radioimaging agents (a) 3, (b) 4, (c) 5, (d) 5 (competition with 100-fold excess PMPA), (e) 6, and (f) 7. Kidneys were shielded with a lead plate.
Analysis of tissue biodistribution was performed on the same animals by sacrificing each mouse, removing its organs and tissues, and quantitating their radioactivity in a γ-counter. As expected, radiotracers 3 and 8 did not accumulate in the PSMA-positive tumors (Figure 6). However, remarkable tumor uptake (average % ID/g tissue = 9.8 ± 2.4) was observed for radiotracers 4, 5, and 6 with little accumulation in other tissues except the kidneys (Table 2). Significant uptake in mouse kidneys was not unanticipated, since murine kidneys have been known for some time to express high levels of PSMA.44 In addition, as we communicated else-where kidney uptake was completely inhibited by excess of PMPA indicating accumulation of radioactivity also PSMA mediated.43 In contrast, as mentioned in the Introduction, human kidneys express low levels of PSMA,11 suggesting that interference from kidney radioactivity may not compromise PCa imaging in humans. Importantly, the imaging agents were found to rapidly clear from all other tissues, resulting in tumor-to-background ratios appropriate for whole body imaging (Table 2: tumor-to-muscle >70).
Figure 6.
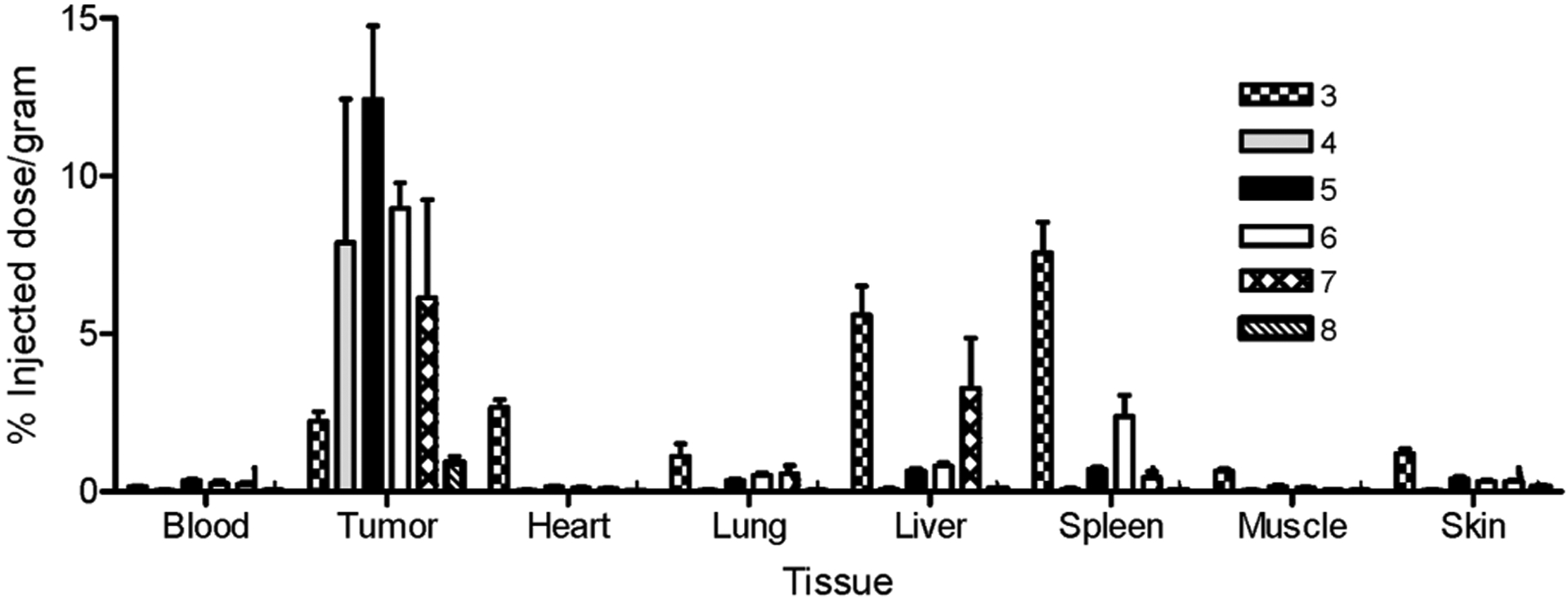
Biodistribution of radioimaging agents in nu/nu mice bearing LNCaP tumor xenografts. Error bars represent SD (n = 4).
Table 2.
Accumulation of Radioimaging Agents 4, 5, and 6 in LNCaP in Tumor Xenografts and in Kidneys and Their Tumor-to-Normal Tissue Ratios
| % ID/ga | |||
|---|---|---|---|
| 4 | 5 | 6 | |
| tumor | 7.9 | 12.4 | 9 |
| kidney | 3.4 | 37.8 | 42.9 |
| Tumor:Tissueb | |||
| blood | 91.7 | 36.4 | 51.4 |
| heart | 112.2 | 80.9 | 93.3 |
| lung | 101.8 | 35.2 | 18.9 |
| liver | 62.9 | 20.2 | 11.6 |
| spleen | 66.7 | 18.4 | 4.5 |
| muscle | 115.1 | 78.9 | 74.2 |
| skin | 92.3 | 30.7 | 28.7 |
% ID/g = percent injected dose per gram of wet tissue.
Tumor:tissue = [% ID/g of tumor]/[% ID/g of tissue].
Discussion
The objective of this study was to develop a PSMA-targeted radioimaging agent for the sensitive detection and localization of malignant disease in suspected PCa patients. Three DUPA conjugates, with 7 to 16 atom spacers, were observed to target PSMA with nanomolar affinity and high specificity, displaying strong tumor uptake and little or no accumulation in other tissues except kidneys. Because DUPA conjugates with longer or shorter spacers displayed much weaker affinity and lower specificity, we suggest that interactions along the 20 Å tunnel leading to the DUPA docking site can be optimized to achieve maximal imaging agent affinity and specificity. Indeed, linking DUPA to an 8-aminooctanoic acid moiety (to avoid steric overlap within the narrow regions at the base of the tunnel) followed by two phenylalanine residues (for maximal interaction with hydrophobic pockets near the mouth of the tunnel) created an imaging with excellent potential for eventual clinical use.
As mentioned in the Introduction, other researchers have also undertaken to develop low molecular weight PSMA-targeted imaging agents.30–33 Indeed, during preparation of this manuscript, a similar PSMA-specific ligand linked to a different chelating moiety via an unrelated spacer was described in the literature.32 Although good tumor uptake (7.9% ID/g tumor) was observed in xenografts of their PSMA+ PC3 PIP cell line (a PSMA-transfected prostate cancer cell line) at 30 min postinjection, tumor accumulation at longer times declined precipitously, with 3.9% ID/g tumor reported at 1 h, 2.3% ID/g tumor seen at 2 h, and 0.8% ID/g tumor at 5 h postinjection. In contrast, the DUPA conjugate (5) reported here displayed 12.4% ID/g tumor at 4 h postinjection. This significantly higher tumor uptake and more prolonged tumor retention are important for clinical imaging applications for two reasons. First, higher tumor uptake affords stronger tumor signals, resulting in more sensitive detection of small tumor masses. Second, longer tumor retention allows more time for clearance of unbound imaging agent from PSMA-negative tissues, permitting greater contrast between malignant and nonmalignant masses. Not surprisingly, the tumor-to-nontumor ratios seen with the DUPA conjugates reported here exceed the same ratios for any previously described PSMA-targeted imaging agent. However, it should also be noted that the PC3 tumors used in the work of Banerjee and colleagues32 differ from the LNCaP tumors used in our study in their sites of origin (PC3 cells are derived from a bone marrow aspirate whereas LNCaP cells are derived from a cervical lymph node metastasis), tumor formation, and invasiveness.45–47
While prolonged tumor retention can improve the quality of a radioimaging agent significantly, it is even more important for most therapeutic applications. Thus, the efficacy of radiotherapy correlates strongly with radiation dosimetry,48,49 and therefore sustained radiotherapeutic agent retention in a tumor may be critical for tumor cell killing. Similarly, the duration of exposure of a cancer cell to a chemotherapeutic drug will impact strongly the probability of tumor cell death, suggesting that a targeting construct that assures protracted retention in the cancer cell will generally yield a better therapeutic outcome.
Clinical candidates for tumor imaging not only must yield good tumor to normal tissue contrast but also should exhibit other properties that might render their use more compatible with medical needs. Thus, ideal tumor imaging agents should display rapid tumor accumulation and clearance from nonmalignant tissues, facile radiolabeling chemistry, high radiochemical purity, good stability during storage and preparation, and a reasonable procedure for its synthesis and purification. Importantly, compound 5 appears to exhibit essentially all of these attributes, suggesting it deserves consideration as a possible candidate for further development. However, whether compound 5, or some other highly specific PSMA-targeted radioimaging agent, eventually achieves clinical application, the prospect for use of a low molecular weight PSMA-targeted radionuclide to image primary PCa masses, locate metastatic disease, monitor response to therapy, detect disease recurrence following surgery, and select patients for subsequent PSMA-targeted chemotherapy seems very bright.
Supplementary Material
Acknowledgment.
We thank Wilfredo Ayala-Lopez for assisting with the Kodak Imaging Station studies, and P. Beck for assisting with LC–MS. This work was supported by a grant from Endocyte, Inc.
Footnotes
Supporting Information Available: Docking studies of phosphorus and urea based inhibitors (selected). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Jemal A; Siegel R; Ward E Cancer statistics 2008. Ca–Cancer J. Clin 2008, 58, 71–96. [DOI] [PubMed] [Google Scholar]
- (2).Zeller JL Grading of prostate cancer. JAMA, J. Am. Med. Assoc 2007, 298, 1596. [DOI] [PubMed] [Google Scholar]
- (3).Chodak MD; Keller P; Schoenberg HW Assessment of screening for prostate cancer using the digital rectal examination. J. Urol 1989, 141, 1136–1138. [DOI] [PubMed] [Google Scholar]
- (4).Essink-Bot M; de Koning HJ; Nijs HGJ; Kirkels WJ; van der Mass PJ; Schroder FH Short term effects of population-based screening for prostate cancer on health-related quality of life. J. Natl. Cancer Inst 1998, 90, 925–931. [DOI] [PubMed] [Google Scholar]
- (5).Linn MM; Ball RA; Maradiegue A Prostate-specific antigen screening: Friend or foe? Urol. Nurs 2007, 27, 481–489. [PubMed] [Google Scholar]
- (6).D’Amico AV; Roehrborn CG Effect of 1 mg.day finasteride on concentration of serum prostate specific antigen in men with androgenic alopecia: a randomized controlled trial. Lancet Oncol. 2007, 8, 21–25. [DOI] [PubMed] [Google Scholar]
- (7).Holves AM; Heesakkers RAM; Adang EM The diagnostic accuracy of CT and MRI in the pelvis of lymph nodes in patients with prostate cancer: a meta-analysis. Clin. Radiol 2008, 63, 387–95. [DOI] [PubMed] [Google Scholar]
- (8).Nie S; Xing Y; Kim GJ; Simons JW Nanotechnlogy applications in cancer. Annu. ReV. Biomed. Eng 2007, 9, 257–288. [DOI] [PubMed] [Google Scholar]
- (9).Chang SS; Reuter VE; Heston WD; Bander NH; Grauer LS; Gaudin PB Five different anti-prostate-specific membrane antigen (PSMA) antibodies confirm PSMA expression in tumor-associated neovasculature. Cancer Res. 1999, 59, 3192–3198. [PubMed] [Google Scholar]
- (10).Kawakami M; Nakayama J Enhanced expression of prostate-specific membrane antigen gene in prostate cancer as revealed by in situ hybridization. Cancer Res. 1997, 57, 2321–2324. [PubMed] [Google Scholar]
- (11).Ghosh A; Heston WD Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. J. Cell Biochem 2004, 91, 528–539. [DOI] [PubMed] [Google Scholar]
- (12).Ross JS; Sheehan CE; Fisher HA Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin. Cancer Res 2003, 9, 6357–6362. [PubMed] [Google Scholar]
- (13).Wright GL; Grob BM; Haley C Upregulation of prostate-specific membrane antigen after androgen-deprivation therapy. Urology 1996, 48, 326–334. [DOI] [PubMed] [Google Scholar]
- (14).Sweat SD; Pacelli A; Murphy GP; Bostwick DG Prostate-specific membrane antigen expression is greatest in prostate adenocarcinoma and lymph node metastases. Urology 1998, 52, 637–640. [DOI] [PubMed] [Google Scholar]
- (15).Chang SS; Reuter VE; Heston WD; Gaudin PB Comparison of anti-prostate-specific membrane antigen antibodies and other immunomarkers in metastatic prostate carcinoma. Urology 2001, 57, 1179–1183. [DOI] [PubMed] [Google Scholar]
- (16).Murphy GP; Barren RJ; Erickson SJ Evaluation and comparison of two new prostate carcinoma markers. Free-prostate specific antigen and prostate specific membrane antigen. Cancer 1996, 78, 809–818. [DOI] [PubMed] [Google Scholar]
- (17).Lopes AD; Davis WL; Rosenstraus MJ; Uveges AJ; Gilman SC Immunohistochemical and pharmacokinetic characterization of the site-specific immunoconjugate CYT-356 derived from antiprostate monoclonal antibody 7E11-C5. Cancer Res. 1990, 50, 6423–6429. [PubMed] [Google Scholar]
- (18).Troyer JK; Beckett ML; Wright GL Jr. Detection and characterization of the prostate-specific membrane antigen (PSMA) in tissue extracts and body fluids. Int. J. Cancer 1995, 62, 552–558. [DOI] [PubMed] [Google Scholar]
- (19).Carter RE; Feldman AR; Coyle JT Prostate-specific membrane antigen is a hydrolase with substrate and pharmacologic characteristics of a neuropeptidase. Proc. Natl. Acad. Sci. U.S.A 1996, 93, 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Silver DA; Pellicer I; Fair WR; Heston WD; Cordon-Cardo C Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res 1997, 3, 81–85. [PubMed] [Google Scholar]
- (21).Chang SS; O’Keefe DS; Bacich DJ; Reuter VE; Heston WD; Gaudin PB Prostate-specific membrane antigen is produced in tumor-associated neovasculature. Clin. Cancer Res 1999, 5, 2674–2681. [PubMed] [Google Scholar]
- (22).Liu H; Moy P; Kim S; Xia Y; Rajasekaran A; Navarro V; Knudsen B; Bander NH Monoclonal antibodies to the extracellular domain of prostate-specific membrane antigen also react with tumor vascular endothelium. Cancer Res. 1997, 57, 3629–3634. [PubMed] [Google Scholar]
- (23).Sodee DB; Ellis RJ; Samuels MA; Spirnak JP; Poole WF; Riester C; Martanovic DM; Stonecipher R; Bellon EM Prostate cancer and prostate bed SPECT imaging with ProstaScint: Semiquantitative correlation with prostatic biopsy results. Prostate 1998, 37, 140–148. [DOI] [PubMed] [Google Scholar]
- (24).Ponsky LE; Cherullo EE; Starkey R; Nelson D; Neumann D; Zipper CP Evaluation of preoperative ProstaScint scans in the prediction of nodal disease. Prostate Cancer Prostatic Dis. 2002, 5, 132–135. [DOI] [PubMed] [Google Scholar]
- (25).Tasch J; Gong M; Sadelain M; Heston WD A unique folate hydrolase, prostate-specific membrane antigen (PSMA): a target for immunotherapy. Crit. Rev. Immunol 2001, 21, 249–261. [PubMed] [Google Scholar]
- (26).Milowsky MI; Nanus DM; Kostakoglu L; Vallabhajosula S; Goldsmith SJ; Bander NH Phase I trial of Yittrium-90-labeled anti-prostate-specific membrane antigen monoclonal antibody J591 for androgen-independent prostate cancer. J. Clin. Oncol 2004, 22, 2522–2531. [DOI] [PubMed] [Google Scholar]
- (27).Milowsky MI; Nanus DM; Kostakoglu L; Sheehan CE; Vallabhajosula S; Goldsmith SJ; Ross J; Bander NH Vascular targeted therapy with anti-prostate-specific membrane antigen monoclonal antibody J591 in advanced solid tumors. J. Clin. Oncol 2007, 25, 540–547. [DOI] [PubMed] [Google Scholar]
- (28).Seccamani E; Tattanelli M; Mariani M; Spranzi E; Scassellati GA; Siccardi AG A simple qualitative determination of human antibodies to murine immunoglobulins (HAMA) in serum samples. Nucl. Med. Biol 1989, 16, 167–170. [DOI] [PubMed] [Google Scholar]
- (29).Colcher D; Bird R; Rosell M In vivo tumor targeting of a recombinant single-chain antigen-binding protein. J. Natl. Cancer Inst 1990, 82, 1191–1197. [DOI] [PubMed] [Google Scholar]
- (30).Zhou J; Neale JH; Pomper M,G; Kozikowski AP NAAG peptidase inhibitors and their potential for diagnosis and therapy. Nat. ReV 2005, 4, 1015–1026. [DOI] [PubMed] [Google Scholar]
- (31).Mease RC; Dusich CL; Foss CA N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-4-[18F]fluorobenzyl-Lcysteine, [18F]D-CFBC: a new imaging probe for prostate cancer. Clin. Cancer Res 2008, 14, 3036–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Banerjee SR; Foss AC; Castanares M Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA). J. Med. Chem 2008, 51, 4504–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chen Y; Foss CA; Byun Y Radiohalogenated prostate-specific membrane antigen (PSMA)-based ureas as imaging agents for prostate cancer. J. Med. Chem 2008, 51, 7933–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Mesters JR; Barinka C; Li W Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer. EMBO J. 2006, 25, 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Jorgensen WL; Maxwell DS; Tirado-Rives J Development and testing of the OPLS all-atom force field on conformational energetic and properties of organic liquids. J. Am. Chem. Soc 1996, 108, 11225–11236. [Google Scholar]
- (36).Friesner RA; Banks JL; Murphy RB; Halgern TA; Klicic JJ; Mainz DT; Repasky MP; Knoll EH; Shelly M; Perry JK; Shaw DE; Francis P; Shenkin PS Glide: a new approach for rapid, accurate docking and scoring. Part 1: method and assessment of docking accuracy. J. Med. Chem 2004, 47, 1739–1749. [DOI] [PubMed] [Google Scholar]
- (37).Halgren TA; Murphy RB; Friesner RA; Beard HS; Frye LL; Pollard WT; Banks JL Glide: a new approach for rapid, accurate docking and scoring. Part 2: enrichment factors in database screening. J. Med. Chem 2004, 47, 1750–1759. [DOI] [PubMed] [Google Scholar]
- (38).Kozikowski AP; Nan F; Conti P; Zhang J; Ramadan JT J. Med. Chem 2001, 44, 298–301. [DOI] [PubMed] [Google Scholar]
- (39).Jackson PF; Slusher BS Design of NAALADase: A novel neuroprotective strategy. Curr. Med. Chem 2001, 8, 949–957. [DOI] [PubMed] [Google Scholar]
- (40).O’Keefe DS; Su SL; Bacich DJ Mapping, genomic organization and promoter analysis of the human prostate-specific membrane antigen gene. Biochim. Biophys. Acta 1998, 1443, 113–127. [DOI] [PubMed] [Google Scholar]
- (41).Liu S; Edwards DS 99mTc-labeled small peptides as diagnostic radiopharmaceuticals. Chem. ReV 1999, 99, 2235–2268. [DOI] [PubMed] [Google Scholar]
- (42).Leamon CP; Parker MA; Vlahoc IR Synthesis and biological evaluation of EC20: a new folate-derived 99mTc-based radiopharmacetical. Bioconjugate Chem. 2002, 13, 1200–1210. [DOI] [PubMed] [Google Scholar]
- (43).Kularatne SA; Wang K; Santhapuram H-KR; Low PS Prostate-specific membrane antigen targeted imaging and therapy of prostate cancer using a PSMA inhibitor as a homing ligand. Mol. Pharmaceutics DOI 10.1021/mp900069d. [DOI] [PubMed] [Google Scholar]
- (44).Slusher BS; Tsai G; Yoo G; Coyle JT Immunocytochemical localization of the N-acetyl-aspartyl-glutamate (NAAG) hydrolyzing enzyme N-acetylated α-linked acidic dipeptidase (NAALA-Dase). J. Comp. Neurol 1992, 315, 217–229. [DOI] [PubMed] [Google Scholar]
- (45).Zheng QH; Gardner TA; Raikwar S; Kao C; Stone KL; Martinez TD; Mock BH; Fei X; Wang JQ; Hutchins GD [11C]Choline as a PET biomarker for assessment of prostate cancer tumor models. Bioorg. Med. Chem 2004, 12, 2887–2893. [DOI] [PubMed] [Google Scholar]
- (46).Yang M; Loda M; Sytkowski AJ Identification of genes expressed differentially by LNCaP or PC-3 prostate cancer cell lines. Cancer Res. 1998, 58, 3732–3735. [PubMed] [Google Scholar]
- (47).Chaiswing L; Bourdeau-Heller JM; Zhong W; Oberley TD Characterization of redox state of two human prostate carcinoma cell lines with different degrees of aggressiveness. Free Radical Biol. Med 2007, 43, 202–215. [DOI] [PubMed] [Google Scholar]
- (48).Liu S; Edwards DS Bifunctional chelators for therapeutic lanthanide radiopharmaceuticals. Bioconjugate Chem. 2001, 12, 7–34. [DOI] [PubMed] [Google Scholar]
- (49).Smith CJ; Gail H; Sieckman GL; Higginbotham C; Volkerf WA; Hoffman TJ Bioconjugate Chem. 2003, 14, 93–102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


